- Key Laboratory of Xiamen Plant Genetics and State Key Laboratory of Cellular Stress Biology, School of Life Sciences, Xiamen University, Xiamen, China
Based on how plants respond to shade, we typically classify them into two groups: shade avoiding and shade tolerance plants. Under vegetative shade, the shade avoiding species induce a series of shade avoidance responses (SARs) to outgrow their competitors, while the shade tolerance species induce shade tolerance responses (STRs) to increase their survival rates under dense canopy. The molecular mechanism underlying the SARs has been extensively studied using the shade avoiding model plant Arabidopsis thaliana, while little is known about STRs. In Aarabidopsis, there is a PHYA-mediated negative feedback regulation that suppresses exaggerated SARs. Recent studies revealed that in shade tolerance Cardamine hirsuta plants, a hyperactive PHYA was responsible for suppressing shade-induced elongation growth. We propose that similar signaling components may be used by shade avoiding and shade tolerance plants, and different phenotypic outputs may result from differential regulation or altered dynamic properties of these signaling components. In this review, we summarized the role of PHYA and its downstream components in shade responses, which may provide insights into understanding how both types of plants respond to shade.
Introduction
Plants grown in complex and dynamic light environments are constantly in fierce competition with their surrounding neighbors for limited light sources. Once light reaches plant leaves, it is absorbed by chlorophylls and other pigments of the photosynthetic apparatus and used for photosynthesis. The upper leaves preferentially absorb blue (B, λ = 400–500 nm) and red (R, λ = 600–700 nm) light for photosynthesis while reflecting most of the far-red (FR) (λ = 700–800 nm) light (Vandenbussche et al., 2005; Franklin, 2008). As a result, the shading of upper leaves not only reduced the total intensity but also the R:FR ratio of light reaching lower leaves (Franklin, 2008). The reduction in R:FR serves as an indicator of local vicinity of vegetation (Vandenbussche et al., 2005; Franklin, 2008) and is perceived by phytochromes, a family of R\FR photoreceptors.
Depending on the strategy that is adopted by plant to cope with vegetative shade, we typically classify plants into two groups: shade avoiding or shade tolerance species. In response to shade, plants living in open habitats often induce a series of shade avoidance responses (SARs) to outgrow their competitors. On the other hand, species typically found in forest understories exhibit shade tolerance responses (STRs), which allows them being adapted to shade environment. The molecular mechanism that underlies the SARs have been extensively studied, while little is known about the STRs. Recent studies indicated that during the SARs, several negative regulators of the SARs, including PHYTOCHROME A (PHYA), were induced. PHYA functions to tune down the SARs under excessive or prolonged shade conditions, which may allow the shade avoiding species to become tolerant to shade (Song et al., 2020). Interestingly, PHYA was also reported to suppress hypocotyl elongation in Cardamine hirsuta, a shade tolerance plant (Molina-Contreras et al., 2019). It is thus possible that some signaling components are shared between the shade avoiding and shade tolerance species. In this review, we summarize results from recent studies on the role of PHYA in regulating low R:FR induced SARs, which may provide hints for understanding the potential mechanism of the STRs.
The Shade Avoidance Responses
The SARs encompass various phenotypic traits, including elongation of stems and petioles, upward leaf movement (increased hyponasty), enhanced apical dominance and reduced branching, which allows plants to reach out for more light (Morgan et al., 1980; Johnson, 1982; Franklin and Whitelam, 2005; Mullen et al., 2006; Franklin, 2008). These responses come at the cost of reduced development of leaf, root and storage organs, such as reduced leaf area, chlorophyll content and chlorophyll a:b ratio (Smith and Whitelam, 1997). Under prolonged shade, flowering is accelerated to allow seed set (Halliday et al., 1994; Casal, 2012). Furthermore, SARs are also accompanied by reduced resistance to various pathogens and symbiotic interactions with microorganisms (Moreno et al., 2009; Cerrudo et al., 2012; De Wit et al., 2013; Ballare, 2014; Konvalinková and Jansa, 2016).
The R/FR light signal is perceived by the phytochrome family of photoreceptors. Three major types of phytochromes, PHYA, PHYB, and PHYC, were identified in angiosperms (Sharrock and Quail, 1989). In Arabidopsis thaliana, there are five phytochromes (PHYA-PHYE) (Sharrock and Quail, 1989; Clack et al., 1994). Among them, PHYB is the major phytochrome repressing SARs under sun light. phyB mutants display constitutive shade avoidance phenotype (Somers et al., 1991; Devlin et al., 1992; Reed et al., 1993; Robson et al., 1993; Kerckhoffs et al., 2010). phyA mutant, on the other hand, displayed exaggerated hypocotyl extension in response to low R:FR signal, but was indistinguishable from the wild type under continuous white light (Johnson et al., 1994). PHYA is thus believed to mediate the negative feedback regulation of the SARs.
The Shade Tolerance Responses
In contrast to the well-defined SARs, the STRs vary a lot between species and are influenced by plant ontogeny and various biotic and abiotic factors (Valladares and Niinemets, 2008). The shade tolerance species usually have reduced elongation of stem and petiole in shade, as compared to the shade avoidance species (Smith, 1979; Niinemets, 1997; Niinemets and Valladares, 2004). However, shade tolerance is not just a lack of shade avoidance. Shade tolerance species induce STRs to optimize their survival under shade, such as lower growth rates, having thinner leaves, altered chlorophyll a:b ratio, reduced apical dominance (increased branching) (Beneragama and Goto, 2011; Casal, 2012; Gommers et al., 2013; Roig-Villanova and Martinez-Garcia, 2016). Several hypotheses have been proposed to explain the survival strategy of the shade tolerance species. The “carbon gain hypothesis” proposed that the shade tolerance may be achieved through optimizing light capture and lowering dark respiration rate, and therefore increasing maximum potential carbon gain (Givnish, 1988; Valladares and Niinemets, 2008). For example, shade tolerant species can optimize light capture and utilization by increasing PSII (photosystem II):PSI ratio (Melis and Harvey, 1981) and special leaf area [SLA, leaf area (m2) per leaf dry mass (kg)], lowering chlorophyll a:b ratio (Melis and Harvey, 1981; Evans and Poorter, 2001). Other studies suggested that it is the pattern of the relationship between leaf nitrogen and irradiance (Niinemets, 1997) or the relative growth rate (Reich et al., 1998) that correlates with the shade tolerance. An extension of the carbon gain hypothesis is the “trade-off hypothesis,” which predicts an inverse correlation between growth rates in high light and survival rates in low light (Valladares and Niinemets, 2008). The “Stress tolerance hypothesis,” on the other hand, proposed that survival in shade positively correlates with resistance to biotic and abiotic stress (Kitajima, 1994). Shade tolerance correlates positively with resistance to pathogens and diseases and increasing physical defense to the external environment (Augspurger, 1984). In deciduous trees, it was also found that the shade-tolerant species restrict carbon allocation toward defense and radial growth instead of increasing height and storage capacities (Giertych et al., 2015). In shade avoiding Arabidopsis, mutant that failed to induce elongation growth in shade still exhibited FR-induced attenuation in defense response, suggesting alteration in defense is not a simple trade-off response (Ballare, 2014; Moreno and Ballare, 2014). In summary, the STRs may be much more complex than the SARs, as different strategies/responses may be employed by plant species with variable shade tolerance ability, or by plants at different developmental stages, or by plants facing shade along with other stresses (Valladares and Niinemets, 2008). So far, there is limited number of studies on the molecular mechanisms of the STRs.
Phytochrome A-Mediated Inhibition of Shade Avoidance Responses
In nature, there are different degrees of shade. Light with reduced R:FR ratio can be generated under three situations: light reflection from nearby plants [neighbor detection, no direct vegetative shade, and little reduction in photosynthetically active radiations (PARs)]; direct, but unclosed shade (mild shade, modest reduction in R:FR and PARs); and dense canopy shade (strong shade, low R:FR, and PARs) (Casal, 2012; Martinez-Garcia et al., 2014). Most shade-tolerant plants are herbaceous species that live in the understory of the forest and usually receive strong canopy shade. For shade avoiding plants grown under dense canopy, the elongation growth of stems, a typical SAR, is also inhibited in a PHYA-dependent manner (Martinez-Garcia et al., 2014; Yang et al., 2018; Song et al., 2020). By measuring the hypocotyl growth rates under mild (R:FR ca. 0.7) and strong shade (R:FR ca. 0.1), Song et al. (2020) demonstrated that PHYA mainly functions after prolonged shade (24 h) and under very low R:FR ratio light. Through whole genome expression profiling, Devlin et al. (2003) also showed that there was an antagonistic effect of PHYA after prolonged shade treatment (24 h). Furthermore, PHYA is also involved in repressing FR-dependent senescence and leaf yellowing in shade (Brouwer et al., 2014; Lim et al., 2018). phyA mutant had reduced survival in deep vegetative shade (Yanovsky et al., 1995). Both the transcript and protein level of PHYA increased after low R:FR treatment, nuclear localization of PHYA was also enhanced by prolonged strong shade treatment (Devlin et al., 2003; Yang et al., 2018; Song et al., 2020). Thus, in shade avoiding plants, PHYA is activated by shade to inhibit some of the SARs and to prevent exaggerated responses to shade, especially under dense canopy.
Theoretically, it is plausible for shade-tolerant plants to develop some of the STRs through enhancing PHYA signaling pathway. Over-expressing PHYA in tomato, tobacco, rice, potato and creeping bentgrass, and zoysiagrass all inhibited stem elongation in shade (McCormac et al., 1992; Robson et al., 1996; Rousseaux et al., 1997; Kong et al., 2004; Garg et al., 2006; Ganesan et al., 2012). Recent studies showed that PHYA protein in a shade-tolerant plant C. hirsuta (a close relative of Arabidopsis) has stronger activity in inhibition of hypocotyl elongation than that of the Arabidopsis (Jose Molina-Contreras et al., 2019). It was reported that a single amino-acid change in PHYA caused a 100-fold shift in the threshold for FR light sensitivity (Maloof et al., 2001). Thus, natural variations in PHYA may allow plants to activate PHYA-mediated negative feedback regulation with different dynamics and to cope with a wide range of shade conditions. By comparing PHYA sequence in species with different degrees of shade tolerance may help us to understand if this a widely adopted strategy in regulating shade responses.
Constitutive Photomorphogenic 1 is a Key Regulator of the Phytochrome A Pathway
Under shade, PHYA was proposed to act through the FR-high irradiance response (Martinez-Garcia et al., 2014) and it may attenuate the SARs through antagonizing PHYB signaling (Johnson et al., 1994; Ciolfi et al., 2013). Results from the genome wide expression profiling suggested that PHYA acts partially through antagonizing the effect of PHYB, while there also exist shade responsive genes that are specifically targeted by PHYA alone (Devlin et al., 2003). Song et al. (2020) reported that shade-induced nuclear localization of COP1 (CONSTITUTIVE PHOTOMORPHOGENIC 1), a key negative regulator of photomorphogenesis, was suppressed in a PHYA-dependent manner after prolonger shade. Mutation in COP1 fully suppressed the long hypocotyl phenotype of phyA under strong shade, indicating it is a key regulator in PHYA-mediated elongation growth. COP1 interacts with the suppressor of phyA-105 (SPA) proteins and functions as an E3 ubiquitin ligase that promotes the ubiquitination and degradation of target proteins, including negative regulators of the SARs such as HY5 (ELONGATED HYPOCOTYL 5), HFR1 (LONG HYPOCOTYL IN FAR-RED) and PAR1,2 (PHYTOCHROME RAPIDLY REGULATED 1,2) and BBX21,22 (B-box-containing proteins 21,22) (Ang and Deng, 1994; Holm et al., 2002; Saijo et al., 2003; Jang et al., 2005; Zhu et al., 2008; Crocco et al., 2010; Zhou et al., 2014). Indeed, comparing HY5 protein level in the strong shade vs. that in the mild shade, a PHYA-dependent increase was observed. As COP1 acts downstream of both PHYA and PHYB, it may account for the antagonizing effect of PHYA.
Regulation of Phytochrome Interacting Factors by Phytochrome A
PHYTOCHROME INTERACTING FACTOR (PIF) transcriptional factors play a key role in regulating the expression of shade responsive genes and elongation growth. Mutation in PIF4,5,7 led to reduced SARs, including shade-induced hypocotyl elongation and response of shade marker genes. PIF4,5,7 accumulated rapidly after low R:FR treatment, which is proceeded by changes at protein phosphorylation level (Lorrain et al., 2008; Hornitschek et al., 2009, 2012; Li et al., 2012; Huang et al., 2018). After prolonged strong shade treatment, PIF4 protein level decreased in a PHYA-dependent manner (Song et al., 2020). Mutation in PIF4,5 partially suppressed the exaggerated hypocotyl growth of phyA mutant in strong shade, and the altered expression of PHYA downstream target genes (Song et al., 2020).
Little is known about how PHYA regulates PIF after prolonged strong shade treatment. Based on previous studies, we speculate that under strong shade, PHYA may regulate the stability, modification and activity of PIFs. First, COP1 is required for accumulation of PIF3 in darkness (Bauer et al., 2004). Ling et al. (2017) reported that COP1/SPA complex associates with and stabilizes PIF3 through repressing BIN2 (BRASSINOSTEROID-INSENSITIVE 2)-mediated phosphorylation of PIF3 and its subsequent degradation. Thus, Shade-activated PHYA may regulate the protein level of PIFs through COP1-dependent route. But not all PIFs are degraded under FR light. PIF8 is degraded in a COP1-dependent manner in dark. Under FR light, activation of PHYA led to stabilization of PIF8, which inhibits suppression of hypocotyl growth by PHYA in FR light (Oh et al., 2020). Whether PIF8 is involved in PHYA-mediated SARs remains to be investigated. Secondly, PHYA preferentially interacts with PIF1 and PIF3 in its Pfr form. In most cases, the interaction led to phosphorylation of PIFs and its subsequent degradation (Shen et al., 2005, 2007, 2008; Al-Sady et al., 2006). An Avena sativa phytochrome A (AsPHYA) was shown to directly phosphorylate PIF3, and a mutant version of AsPHYA with reduced kinase activity compromised FR-induced phosphorylation and degradation of PIF3 (Shin et al., 2016). Finally, shade induces the expression of several PIF-interacting proteins, such as HFR1, PAR1,2, which represses the transcriptional activity of PIFs (Roig-Villanova et al., 2007; Hornitschek et al., 2009; Galstyan et al., 2011; Hao et al., 2012). PHYA does not affect early induction of PAR1 and HFR1 by shade. After prolonged shade treatment, PHYA partially suppresses PAR1 expression, but does not affect HFR1 expression (Song et al., 2020). Thus, PHYA does not inhibit SARs through upregulating the transcript level of PAR1 and HFR1. FHY3 (FAR-RED ELONGATED HYPOCOTYL3) and FAR1 (FAR-RED-IMPAIRED RESPONSE1) are two homologous transcription factors that act downstream of PHYA in FR light signaling (Wang and Deng, 2002; Lin et al., 2007). fhy3far1 mutant displayed exaggerated hypocotyl elongation under shade, which was similar to phyA (Liu et al., 2019). They also directly interact with PIF3,5 and repress their transcriptional activity (Liu et al., 2020). It would be interesting to see if FHY3 and FAR1 mediate PHYA signaling in shade and how they are regulated. Recently, Fraser et al. (2021) showed that early evening expression of the central circadian clock components TOC1 (TIMING OF CAB EXPRESSION 1), PRR7 (PSEUDO RESPONSE REGULATOR 7), and ELF3,4 (EARLY FLOWERING 3,4) was elevated in photocycles of low R:FR and low PAR in a PHYA-dependent manner. And among them, TOC1, PRRs, and ELF3 also interact with PIFs and negatively regulate their transcriptional activities and/or DNA binding ability (Nieto et al., 2015; Soy et al., 2016; Martin et al., 2018; Jiang et al., 2019; Zhang et al., 2020). Together, these proteins suppress stem elongation under strong shade (Figure 1).
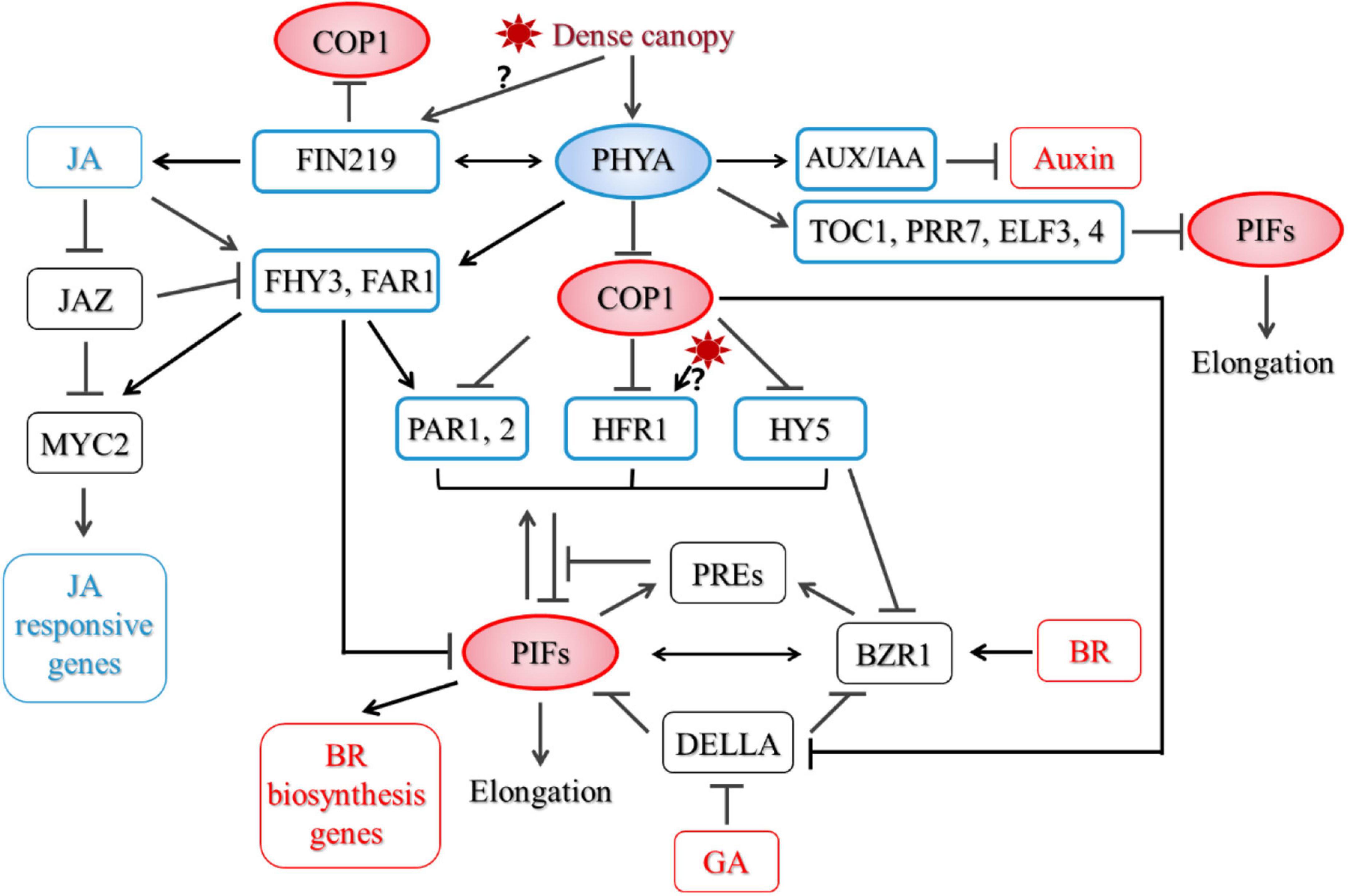
Figure 1. A proposed model for phytochrome A (PHYA)-mediated regulatory network in shade. Proteins outlined in blue are negative regulators of the shade avoidance responses (SARs), while those in red are positive regulators.
Thus, in addition to affecting PIFs through up-regulating PHYA level and/or activity, shade tolerance plants may also tune down the activity of PIFs through a PHYA-independent pathway, either through having PIFs with altered protein dynamics or activity, or by using negative regulators that can strongly reduce the activity or protein level of PIFs. Ciolfi et al. (2013) demonstrated that although in Arabidopsis, both PHYA and HFR1 are negative regulators of SARs, they act independently in regulating several shade-induced target genes. HFR1 was also shown to regulate shade-induced flowering independent of PHYA. HFR1 interacts with CO (CONSTANS) and PIF7, which inhibit their DNA binding activities to downstream target genes (Zhang et al., 2019). It was reported that HFR1 in the shade tolerant C. hirsuta was more stable than its counterpart in Arabidopsis due to its lower binding capacity to COP1. The enhanced HFR1 activity then caused a reduction in PIFs’ activity and attenuated PIF-mediated responses (Paulisic et al., 2021). Furthermore, it was reported that the shade avoiding Scots pine lacks FR high irradiance response, suggesting a different negative regulatory pathway in angiosperm, if there is any (Mohr, 1990).
Phytohormones and the Phytochrome A Signaling Pathways
In shade avoiding plants, changes in plant architecture, defense responses and metabolism are associated with both altered level and sensitivity of various phytohormones as previously reviewed (Yang and Li, 2017; Fernandez-Milmanda and Ballare, 2021; Liu et al., 2021). For example, in Arabidopsis, shade induced stem elongation requires growth promoting hormones: auxin, gibberellins (GAs) and brassinosteroids (BRs), while shade reduced expression of defense genes that are responsive to jasmonic acid (JA) and salicylic acid (SA). Here we summarize phytohormones that are influenced by PHYA under shade.
Biosynthesis of auxin increases in shade. Mutation in TAA1 (Trp aminotransferase in Arabidopsis), a key enzyme in Trp-dependent auxin biosynthesis, severely impaired hypocotyl elongation in shade (Tao et al., 2008). However, this mutation had limited effect on hypocotyl growth in phyA mutant, suggesting a TAA1-independent mechanism may be employed by PHYA. Both PHYA and PHYB can interact with a similar set of AUX/IAA (auxin/indole-3-acetic acid) proteins, and PHYA activated under deep shade can thus counteract the effect of PHYB through stabilizing these proteins and subsequently suppressing the auxin sensitivity and the SARs (Xu et al., 2018; Yang et al., 2018).
Both BR biosynthesis and signaling components are required for petiole and stem growth under low R/FR (Kim et al., 1998; Luccioni et al., 2002; Kozuka et al., 2010; Keller et al., 2011). Activation of the BR signal pathway lead to the dephosphorylation of BES1/BZR1 transcriptional factors, which then move into the nucleus and promote the expression of BR-responsive genes (Belkhadir and Jaillais, 2015). PHYA inhibits hypocotyl elongation partially through repressing key BR biosynthesis genes’ expression in hypocotyls, which is mediated by COP1, PIF4, and PIF5 (Song et al., 2020). In addition, BES1/BZR1 directly interacts with PIF4, they regulate downstream target genes and promote hypocotyl elongation interdependently in response to BR, darkness and heat (Oh et al., 2012). HY5 also directly interacts with the active form of BZR1 and attenuates its transcriptional activity (Li and He, 2016). HY5 represses BZR1 accumulation through enhancing BIN2 kinase activity (Li et al., 2020). After prolonged strong shade treatment, protein level of both PIF4 and the active form of BES1 were much higher in phyA mutant than those in the wild type, while HY5 protein level was repressed by PHYA (Song et al., 2020). Thus, PHYA may also repress BR signaling through regulating BES1/BZR1-interacting proteins (Figure 1).
Although little is known about how PHYA affects GA pathway, the dwarf phenotype of PHYA overexpressing-tobacco was related to a reduction in the active GA level (Jordan et al., 1995). DELLA proteins are transcriptional regulators that inhibit GA signaling. DELLAs are also targets of COP1, which promotes degradation of DELLAs through direct protein interactions and ubiquitination (Blanco-Tourinan et al., 2020). Furthermore, DELLAs interact with BZR1 and PIFs to repress their activities (Feng et al., 2008; Bai et al., 2012). BZR1 and PIF4 share common target genes, including PRE1 (PACLOBUTRAZOL RESISTANCE1), which is co-regulated by BR, GA, and auxin (Bai et al., 2012). By forming heterodimers with PAR1, PRE1 prevents PAR1 from interacting with PIF4 and represses its activity (Hao et al., 2012). These findings defined a regulating network including DELLAs, BZR1, PIFs, and downstream targets, which can integrate GA and BR signaling with light signal to fine-tune SARs. How this signaling network is influenced by PHYA and in the shade tolerance species remain to be illustrated.
Jasmonic acid is a lipid-derived plant hormone that is well known for its involvement in responses to wounding, necrotrophic pathogens and herbivores. Furthermore, JA participates in plant development regulation, including root growth inhibition, trichome initiation, male fertility, leaf senescence and photomorphogenesis. JA suppresses hypocotyl elongation and promotes cotyledon opening through inhibiting COP1 activity (Zheng et al., 2017). In Arabidopsis, both JA biosynthesis and sensitivity is reduced in shade due to inactivation of PHYB (De Wit et al., 2013; Leone et al., 2014; Fernandez-Milmanda et al., 2020). phyA seedlings grown in dark and FR contains higher level of OPDA [cis-(+)-12-oxophytodienoic acid], an intermediate of JA biosynthesis, than wild type (Robson et al., 2010). In addition, PHYA is required for inhibition of root growth by JA, and wound/JA-induced degradation of JAZ1 (JASMONATE-ZIM-DOMAIN PROTEIN 1), repressors of JA signaling (Robson et al., 2010), indicating that PHYA is also required for JA signaling. Arabidopsis FIN219 (FAR-RED INSENSITIVE219)/JAR1 (JASMONATE RESISTANT 1) is involved in PHYA-mediated FR light signaling (Hsieh et al., 2000). It encodes an enzyme catalyzing the final step of JA-Ile (an active form of JA) production. Jar1 mutant was hypersensitive to shade-induced hypocotyl elongation (Robson et al., 2010). In seedlings grown in FRc (continuous FR), FIN219/JAR1 directly interacts with COP1 and retains COP1 in the cytoplasm, which subsequently increases HY5 protein level (Wang et al., 2011). Under shade, FIN219/JAR1 protein level is reduced. It affects shade-induced accumulation of PHYA protein and shade-regulated expression of PIF5, PAR1 and the auxin responses gene IAA29 and SAUR68 (Swain et al., 2017). Interestingly, through analyzing phyAfin219 double mutants, it was proposed that FIN219 and PHYA synergistically regulating shade-induced hypocotyl elongation and gene expression, and FIN219-mediated repression of SARs is independent of PHYA-mediated high irradiance response (Swain et al., 2017). Thus, FIN219 may serve as a key node that can sense PHYA-independent shade signal and regulates downstream components of the PHYA signaling pathway.
Phytochrome A signaling components FHY3 and FAR1 were shown to repress hypocotyl growth through direct transcriptional activation of PAR1 and PAR2 and their transcriptional activities were attenuated by JAZ proteins through direct protein interactions (Liu et al., 2019). Interestingly, JA also promoted the accumulation of FHY3 protein and unleashed them from inhibition by JAZ proteins (Liu et al., 2019). FHY3 interacts with MYC2, a key transcription factor in the JA pathway, enhances its transcriptional activity on the expression of JA-responsive genes, and affects defense responses (Liu et al., 2019). FHY3 and FAR1 were thus proposed to be regulators that balance growth and defense. As shade rapidly induced accumulation of FHY3 protein (Liu et al., 2019), but repressed JA biosynthesis, a detailed characterization on the dynamics of FHY3 protein level in shade and how PHYA is involved is required to fully illustrate the role of FHY3. In addition to regulating hypocotyl growth, FHY3 and FAR1 are involved in an array of SAR-related biological processes, including senescence, starch metabolism, chloroplast development, circadian gating, shoot branching, flowering control etc. (McCormac et al., 1992; Allen et al., 2006; Stirnberg et al., 2012; Tang et al., 2012; Ma et al., 2016, 2017; Liu et al., 2019, 2020; Tian et al., 2020; Xie et al., 2020a,b). It would be interesting to examine if these processes are also inhibited under prolonged strong shade and how PHYA and FHY3, FAR1 are involved (Figure 1).
Through studying a shade-avoiding and a shade tolerance species of genus Geranium, Gommers et al. (2018) found that auxin and GA levels, but not BR, increased in elongating petioles through local perception of FR light in the shade-avoiding species, which did not occur in the shade-tolerance species (McCormac et al., 1992). In the shade-avoiding species, BR level decreased in petioles after long exposure (11.5 h) to FR-enriched light, it would be interesting to know if this reduction in BR and the lack of induction in auxin and GA level is PHYA-regulated. Transcriptional profiling data revealed up-regulation of ethylene and cytokinin pathway genes in shade tolerance Swarnaprabha rice (Panigrahy et al., 2019). In Arabidopsis, shade arrests leaf primordia development is associated with an auxin-dependent induction of AtCKX6, a gene encoding a cytokinin oxidase involved in cytokinin breakdown (Carabelli et al., 2007). Cytokinin level is reduced after shade treatment in soybean (Wu et al., 2017). The higher rate of panicle emergence of Swarnaprabha rice in shade may thus be related to its upregulated cytokinin pathway. Shade treatment increases ethylene level and promotes petiole elongation in Arabidopsis (Pierik et al., 2009). Similar to JA, ethylene also plays an important role in various defense and stress responses. How ethylene affects the STRs and survival of the shade tolerance species remains to be investigated.
Conclusion Remarks
Most of the crops are shade avoiding plants. When planted at high density, SARs are induced, which is often detrimental to yield (Smith, 1995; Duvick, 1997; Kebrom and Brutnell, 2007). Shade tolerance plants are naturally evolved to be adapted to grow under dense canopy. STRs are induced to achieve efficient light utilization and high survival rate under prolonged strong shade. Thus, suppressing SARs alone may not be sufficient to enhance the performance of crops planted at high density. Recent studies indicate that the shade avoiding plants processes a PHYA-mediated negative feedback regulation, which suppresses some of the SARs and may increase the survival of these plants under prolonged strong shade. Furthermore, some shade tolerance plants may also utilize the PHYA-mediated pathway to regulate their responses to shade. Understanding how the PHYA signaling pathway is regulated in both types of plants, what are the downstream components, and besides elongation growth, what and how other physiological processes are being regulated, can help us understand how plants survive under dense canopy. Modifying the shade responses of various crops using their existing components may be a shortcut to obtain high-yield shade tolerance crops.
Author Contributions
HX and YT designed the opinion. HX, PC, and YT jointly wrote the opinion. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation of China, 31770316 to YT and by the 111 Project B12001.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Allen, T., Koustenis, A., Theodorou, G., Somers, D. E., Kay, S. A., Whitelam, G. C., et al. (2006). Arabidopsis FHY3 specifically gates phytochrome signaling to the circadian clock. Plant Cell 18, 2506–2516. doi: 10.1105/tpc.105.037358
Al-Sady, B., Ni, W., Kircher, S., Schafer, E., and Quail, P. H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23, 439–446. doi: 10.1016/j.molcel.2006.06.011
Ang, L. H., and Deng, X. W. (1994). Regulatory hierarchy of photomorphogenic loci: allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell 6, 613–628. doi: 10.1105/tpc.6.5.613
Augspurger, C. K. (1984). Light requirements of neotropical tree seedlings: a comparative study of growth and survival. J. Ecol. 72, 777–795.
Bai, M. Y., Shang, J. X., Oh, E., Fan, M., Bai, Y., Zentella, R., et al. (2012). Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14, 810–817. doi: 10.1038/ncb2546
Ballare, C. L. (2014). Light regulation of plant defense. Annu. Rev. Plant Biol. 65, 335–363. doi: 10.1146/annurev-arplant-050213-040145
Bauer, D., Viczian, A., Kircher, S., Nobis, T., Nitschke, R., Kunkel, T., et al. (2004). Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16, 1433–1445. doi: 10.1105/tpc.021568
Belkhadir, Y., and Jaillais, Y. (2015). The molecular circuitry of brassinosteroid signaling. New Phytol. 206, 522–540. doi: 10.1111/nph.13269
Beneragama, C. K., and Goto, K. (2011). Chlorophyll a:b ratio increases under low-light in ‘shade-tolerant’ Euglena gracillis. Trop. Agric. Res. 22, 12–25. doi: 10.4038/tar.v22i1.2666
Blanco-Tourinan, N., Legris, M., Minguet, E. G., Costigliolo-Rojas, C., Nohales, M. A., Iniesto, E., et al. (2020). COP1 destabilizes DELLA proteins in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 117, 13792–13799. doi: 10.1073/pnas.1907969117
Brouwer, B., Gardestrom, P., and Keech, O. (2014). In response to partial plant shading, the lack of phytochrome a does not directly induce leaf senescence but alters the fine-tuning of chlorophyll biosynthesis. J. Exp. Bot. 65, 4037–4049. doi: 10.1093/jxb/eru060
Carabelli, M., Possenti, M., Sessa, G., Ciolfi, A., Sassi, M., Morelli, G., et al. (2007). Canopy shade causes a rapid and transient arrest in leaf development through auxin-induced cytokinin oxidase activity. Genes Dev. 21, 1863–1868. doi: 10.1101/gad.432607
Cerrudo, I., Keller, M. M., Cargnel, M. D., Demkura, P. V., De Wit, M., Patitucci, M. S., et al. (2012). Low red/far-red ratios reduce Arabidopsis resistance to Botrytis cinerea and jasmonate responses via a COI1-JAZ10-dependent, salicylic acid-independent mechanism. Plant Physiol. 158, 2042–2052. doi: 10.1104/pp.112.193359
Ciolfi, A., Sessa, G., Sassi, M., Possenti, M., Salvucci, S., Carabelli, M., et al. (2013). Dynamics of the shade-avoidance response in Arabidopsis. Plant Physiol. 163, 331–353. doi: 10.1104/pp.113.221549
Clack, T., Mathews, S., and Sharrock, R. A. (1994). The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mol. Biol. 25, 413–427. doi: 10.1007/BF00043870
Crocco, C. D., Holm, M., Yanovsky, M. J., and Botto, J. F. (2010). AtBBX21 and COP1 genetically interact in the regulation of shade avoidance. Plant J. 64, 551–562. doi: 10.1111/j.1365-313X.2010.04360.x
De Wit, M., Spoel, S. H., Sanchez-Perez, G. F., Gommers, C. M. M., Pieterse, C. M. J., Voesenek, L., et al. (2013). Perception of low red:far-red ratio compromises both salicylic acid- and jasmonic acid-dependent pathogen defences in Arabidopsis. Plant J. 75, 90–103. doi: 10.1111/tpj.12203
Devlin, P. F., Rood, S. B., Somers, D. E., Quail, P. H., and Whitelam, G. C. (1992). Photophysiology of the elongated internode (ein) mutant of Brassica rapa: ein mutant lacks a detectable phytochrome B-Like polypeptide. Plant Physiol. 100, 1442–1447. doi: 10.1104/pp.100.3.1442
Devlin, P. F., Yanovsky, M. J., and Kay, S. A. (2003). A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiol. 133, 1617–1629. doi: 10.1104/pp.103.034397
Evans, J. R., and Poorter, H. (2001). Photosynthetic acclimation of plants to growth irradiance: the relative importance of specific leaf area and nitrogen partitioning in maximizing carbon gain. Plant Cell Environ. 24, 755–767. doi: 10.1046/j.1365-3040.2001.00724.x
Feng, S., Martinez, C., Gusmaroli, G., Wang, Y., Zhou, J., Wang, F., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451, 475–479. doi: 10.1038/nature06448
Fernandez-Milmanda, G. L., and Ballare, C. L. (2021). Shade avoidance: expanding the color and hormone palette. Trends Plant Sci. 26, 509–523. doi: 10.1016/j.tplants.2020.12.006
Fernandez-Milmanda, G. L., Crocco, C. D., Reichelt, M., Mazza, C. A., Kollner, T. G., Zhang, T., et al. (2020). A light-dependent molecular link between competition cues and defence responses in plants. Nat. Plants 6, 223–230. doi: 10.1038/s41477-020-0604-8
Franklin, K. A., and Whitelam, G. C. (2005). Phytochromes and shade-avoidance responses in plants. Ann. Bot. 96, 169–175. doi: 10.1093/aob/mci165
Fraser, D. P., Panter, P. E., Sharma, A., Sharma, B., Dodd, A. N., and Franklin, K. A. (2021). Phytochrome a elevates plant circadian-clock components to suppress shade avoidance in deep-canopy shade. Proc. Natl. Acad. Sci. U.S.A. 118:e2108176118. doi: 10.1073/pnas.2108176118
Galstyan, A., Cifuentes-Esquivel, N., Bou-Torrent, J., and Martinez-Garcia, J. F. (2011). The shade avoidance syndrome in Arabidopsis: a fundamental role for atypical basic helix-loop-helix proteins as transcriptional cofactors. Plant J. 66, 258–267. doi: 10.1111/j.1365-313X.2011.04485.x
Ganesan, M., Han, Y.-J., Bae, T.-W., Hwang, O.-J., Chandrasekkhar, T., Shin, A.-Y., et al. (2012). Overexpression of phytochrome a and its hyperactive mutant improves shade tolerance and turf quality in creeping bentgrass and zoysiagrass. Planta 236, 1135–1150. doi: 10.1007/s00425-012-1662-6
Garg, A. K., Sawers, R. J., Wang, H., Kim, J. K., Walker, J. M., Brutnell, T. P., et al. (2006). Light-regulated overexpression of an Arabidopsis phytochrome a gene in rice alters plant architecture and increases grain yield. Planta 223, 627–636. doi: 10.1007/s00425-005-0101-3
Giertych, M. J., Karolewski, P., and Oleksyn, J. (2015). Carbon allocation in seedlings of deciduous tree species depends on their shade tolerance. Acta Physiol. Plant. 37:216.
Givnish, T. J. (1988). Adaptation to sun and shade: a whole-plant perspective. Funct. Plant Biol. 15, 63–92. doi: 10.1071/pp9880063
Gommers, C. M. M., Buti, S., Tarkowska, D., Pencik, A., Banda, J. P., Arricastres, V., et al. (2018). Organ-specific phytohormone synthesis in two Geranium species with antithetical responses to far-red light enrichment. Plant Direct 2:e00066. doi: 10.1002/pld3.66
Gommers, C. M., Visser, E. J., St Onge, K. R., Voesenek, L. A., and Pierik, R. (2013). Shade tolerance: when growing tall is not an option. Trends Plant Sci. 18, 65–71. doi: 10.1016/j.tplants.2012.09.008
Halliday, K. J., Koornneef, M., and Whitelam, G. C. (1994). Phytochrome B and at least one other phytochrome mediate the accelerated flowering response of Arabidopsis thaliana L. to Low Red/Far-Red Ratio. Plant Physiol. 104, 1311–1315. doi: 10.1104/pp.104.4.1311
Hao, Y., Oh, E., Choi, G., Liang, Z., and Wang, Z. Y. (2012). Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol. Plant 5, 688–697. doi: 10.1093/mp/sss011
Holm, M., Ma, L. G., Qu, L. J., and Deng, X. W. (2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16, 1247–1259. doi: 10.1101/gad.969702
Hornitschek, P., Kohnen, M. V., Lorrain, S., Rougemont, J., Ljung, K., Lopez-Vidriero, I., et al. (2012). Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71, 699–711. doi: 10.1111/j.1365-313X.2012.05033.x
Hornitschek, P., Lorrain, S., Zoete, V., Michielin, O., and Fankhauser, C. (2009). Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 28, 3893–3902. doi: 10.1038/emboj.2009.306
Hsieh, H. L., Okamoto, H., Wang, M., Ang, L. H., Matsui, M., Goodman, H., et al. (2000). FIN219, an auxin-regulated gene, defines a link between phytochrome a and the downstream regulator COP1 in light control of Arabidopsis development. Genes Dev. 14, 1958–1970.
Huang, X., Zhang, Q., Jiang, Y., Yang, C., Wang, Q., and Li, L. (2018). Shade-induced nuclear localization of PIF7 is regulated by phosphorylation and 14-3-3 proteins in Arabidopsis. Elife 7:e31636. doi: 10.7554/eLife.31636
Jang, I. C., Yang, J. Y., Seo, H. S., and Chua, N. H. (2005). HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome a signaling. Genes Dev. 19, 593–602. doi: 10.1101/gad.1247205
Jiang, Y., Yang, C., Huang, S., Xie, F., Xu, Y., Liu, C., et al. (2019). The ELF3-PIF7 interaction mediates the circadian gating of the shade response in Arabidopsis. iScience 22, 288–298. doi: 10.1016/j.isci.2019.11.029
Johnson, E., Bradley, M., Harberd, N. P., and Whitelam, G. C. (1994). Photoresponses of light-grown phya mutants of arabidopsis (phytochrome a is required for the perception of daylength extensions). Plant Physiol. 105, 141–149. doi: 10.1104/pp.105.1.141
Johnson, G. (1982). Photomorphogenesis in impatiens parviflora and other plant species under simulated natural canopy radiations. New Phytol. 90, 611–618. doi: 10.1111/j.1469-8137.1982.tb03270.x
Jordan, E. T., Hatfield, P. M., Hondred, D., Talon, M., Zeevaart, J. A., and Vierstra, R. D. (1995). Phytochrome a overexpression in transgenic tobacco. correlation of dwarf phenotype with high concentrations of phytochrome in vascular tissue and attenuated gibberellin levels. Plant Physiol. 107, 797–805. doi: 10.1104/pp.107.3.797
Jose Molina-Contreras, M., Paulisic, S., Then, C., Moreno-Romero, J., Pastor-Andreu, P., Morelli, L., et al. (2019). Photoreceptor activity contributes to contrasting responses to shade in cardamine and Arabidopsis seedlings. Plant Cell 31, 2649–2663. doi: 10.1105/tpc.19.00275
Kebrom, T. H., and Brutnell, T. P. (2007). The molecular analysis of the shade avoidance syndrome in the grasses has begun. J. Exp. Bot. 58, 3079–3089. doi: 10.1093/jxb/erm205
Keller, M. M., Jaillais, Y., Pedmale, U. V., Moreno, J. E., Chory, J., and Ballare, C. L. (2011). Cryptochrome 1 and phytochrome B control shade-avoidance responses in Arabidopsis via partially independent hormonal cascades. Plant J. 67, 195–207. doi: 10.1111/j.1365-313X.2011.04598.x
Kerckhoffs, L., Schreuder, M., Tuinen, A. V., Koornneef, M., and Kendrick, R. E. (2010). Phytochrome control of anthocyanin biosynthesis in tomato seedlings: analysis using photomorphogenic mutants. Photochem. Photobiol. 65, 374–381.
Kim, G. T., Tsukaya, H., and Uchimiya, H. (1998). The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev. 12, 2381–2391. doi: 10.1101/gad.12.15.2381
Kitajima, K. (1994). Relative importance of photosynthetic traits and allocation patterns as correlates of seedling shade tolerance of 13 tropical trees. Oecologia 98, 419–428. doi: 10.1007/BF00324232
Kong, S. G., Lee, D. S., and Kwak, S. N. (2004). Characterization of sunlight-grown transgenic rice plants expressing Arabidopsis phytochrome a. Mol. Breed. 14, 35–45.
Konvalinková, T., and Jansa, J. (2016). Lights off for Arbuscular mycorrhiza: on its symbiotic functioning under light deprivation. Front. Plant Sci. 7:782. doi: 10.3389/fpls.2016.00782
Kozuka, T., Kobayashi, J., Horiguchi, G., Demura, T., Sakakibara, H., Tsukaya, H., et al. (2010). Involvement of auxin and brassinosteroid in the regulation of petiole elongation under the shade. Plant Physiol. 153, 1608–1618. doi: 10.1104/pp.110.156802
Leone, M., Keller, M. M., Cerrudo, I., and Ballare, C. L. (2014). To grow or defend? low red : far-red ratios reduce jasmonate sensitivity in Arabidopsis seedlings by promoting DELLA degradation and increasing JAZ10 stability. New Phytol. 204, 355–367. doi: 10.1111/nph.12971
Li, J., Terzaghi, W., Gong, Y., Li, C., Ling, J. J., Fan, Y., et al. (2020). Modulation of BIN2 kinase activity by HY5 controls hypocotyl elongation in the light. Nat. Commun. 11:1592. doi: 10.1038/s41467-020-15394-7
Li, L., Ljung, K., Breton, G., Schmitz, R. J., Pruneda-Paz, J., Cowing-Zitron, C., et al. (2012). Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26, 785–790. doi: 10.1101/gad.187849.112
Li, Q. F., and He, J. X. (2016). BZR1 Interacts with HY5 to mediate brassinosteroid- and light-regulated cotyledon opening in Arabidopsis in darkness. Mol. Plant 9, 113–125. doi: 10.1016/j.molp.2015.08.014
Lim, J., Park, J. H., Jung, S., Hwang, D., Nam, H. G., and Hong, S. (2018). Antagonistic Roles of PhyA and PhyB in far-red light-dependent leaf senescence in Arabidopsis thaliana. Plant Cell Physiol. 59, 1753–1764. doi: 10.1093/pcp/pcy153
Lin, R., Ding, L., Casola, C., Ripoll, D. R., Feschotte, C., and Wang, H. (2007). Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318, 1302–1305. doi: 10.1126/science.1146281
Ling, J. J., Li, J., Zhu, D., and Deng, X. W. (2017). Noncanonical role of Arabidopsis COP1/SPA complex in repressing BIN2-mediated PIF3 phosphorylation and degradation in darkness. Proc. Natl. Acad. Sci. U.S.A. 114, 3539–3544. doi: 10.1073/pnas.1700850114
Liu, Y., Jafari, F., and Wang, H. (2021). Integration of light and hormone signaling pathways in the regulation of plant shade avoidance syndrome. aBiotech 2, 131–145. doi: 10.1007/s42994-021-00038-1
Liu, Y., Ma, M., Li, G., Yuan, L., Xie, Y., Wei, H., et al. (2020). Transcription Factors FHY3 and FAR1 regulate light-induced CIRCADIAN CLOCK ASSOCIATED1 gene expression in Arabidopsis. Plant Cell 32, 1464–1478. doi: 10.1105/tpc.19.00981
Liu, Y., Wei, H., Ma, M., Li, Q., Kong, D., Sun, J., et al. (2019). Arabidopsis FHY3 and FAR1 regulate the balance between growth and defense responses under shade conditions. Plant Cell 31, 2089–2106. doi: 10.1105/tpc.18.00991
Lorrain, S., Allen, T., Duek, P. D., Whitelam, G. C., and Fankhauser, C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53, 312–323. doi: 10.1111/j.1365-313X.2007.03341.x
Luccioni, L. G., Oliverio, K. A., Yanovsky, M. J., Boccalandro, H. N. E., and Casal, J. J. (2002). Brassinosteroid mutants uncover fine tuning of phytochrome signaling. Plant Physiol. 128, 173–181. doi: 10.1104/pp.128.1.173
Ma, L., Tian, T., Lin, R., Deng, X. W., Wang, H., and Li, G. (2016). Arabidopsis FHY3 and FAR1 regulate light-induced myo-inositol biosynthesis and oxidative stress responses by transcriptional activation of MIPS1. Mol. Plant 9, 541–557. doi: 10.1016/j.molp.2015.12.013
Ma, L., Xue, N., Fu, X., Zhang, H., and Li, G. (2017). Arabidopsis thaliana FAR-RED ELONGATED HYPOCOTYLS3 (FHY3) and FAR-RED-IMPAIRED RESPONSE1 (FAR1) modulate starch synthesis in response to light and sugar. New Phytol. 213, 1682–1696. doi: 10.1111/nph.14300
Maloof, J. N., Borevitz, J. O., Dabi, T., Lutes, J., Nehring, R. B., Redfern, J. L., et al. (2001). Natural variation in light sensitivity of Arabidopsis. Nat. Genet. 29, 441–446. doi: 10.1038/ng777
Martin, G., Rovira, A., Veciana, N., Soy, J., Toledo-Ortiz, G., Gommers, C. M. M., et al. (2018). Circadian waves of transcriptional repression shape PIF-regulated photoperiod-responsive growth in Arabidopsis. Curr. Biol. 28, 311–318 e315. doi: 10.1016/j.cub.2017.12.021
Martinez-Garcia, J. F., Gallemi, M., Molina-Contreras, M. J., Llorente, B., Bevilaqua, M. R., and Quail, P. H. (2014). The shade avoidance syndrome in Arabidopsis: the antagonistic role of phytochrome a and B differentiates vegetation proximity and canopy shade. PLoS One 9:e109275. doi: 10.1371/journal.pone.0109275
McCormac, A., Whitelam, G., and Smith, H. (1992). Light-grown plants of transgenic tobacco expressing an introduced oat phytochrome-a gene under the control of a constitutive viral promoter exhibit persistent growth-inhibition by far-red light. Planta 188, 173–181. doi: 10.1007/BF00216811
Melis, A., and Harvey, G. W. (1981). Regulation of photosystem stoichiometry, chlorophyll a and chlorophyll b content and relation to chloroplast ultrastructure. Biochim. Biophys. Acta Bioenerg. 637, 138–145. doi: 10.1016/0005-2728(81)90219-X
Mohr, E. F. H. (1990). Coaction of blue/ultraviolet-a light and light absorbed by phytochrome in controlling growth of pine (Pinussylvestris L.) seedlings. Planta 180, 212–216. doi: 10.1007/BF00193998
Molina-Contreras, M. J., Paulisic, S., Then, C., Moreno-Romero, J., Pastor-Andreu, P., Morelli, L., et al. (2019). Photoreceptor activity contributes to contrasting responses to shade in cardamine and arabidopsis seedlings. Plant Cell 31, 2649–2663. doi: 10.1105/tpc.19.00275
Moreno, J. E., and Ballare, C. L. (2014). Phytochrome regulation of plant immunity in vegetation canopies. J. Chem. Ecol. 40, 848–857. doi: 10.1007/s10886-014-0471-8
Moreno, J. E., Tao, Y., Chory, J., and Ballaré, C. L. (2009). Ecological modulation of plant defense via phytochrome control of jasmonate sensitivity. Proc. Natl. Acad. Sci. U.S.A. 106, 4935–4940. doi: 10.1073/pnas.0900701106
Morgan, D. C., O’brien, T., and Smith, H. (1980). Rapid photomodulation of stem extension in light-grownSinapis alba L: studies on kinetics, site of perception and photoreceptor. Planta 150, 95–101. doi: 10.1007/BF00582351
Mullen, J. L., Weinig, C., and Hangarter, R. P. (2006). Shade avoidance and the regulation of leaf inclination in Arabidopsis. Plant Cell Environ. 29, 1099–1106. doi: 10.1111/j.1365-3040.2005.01484.x
Nieto, C., Lopez-Salmeron, V., Daviere, J. M., and Prat, S. (2015). ELF3-PIF4 interaction regulates plant growth independently of the evening complex. Curr. Biol. 25, 187–193. doi: 10.1016/j.cub.2014.10.070
Niinemets, U. (1997). Role of foliar nitrogen in light harvesting and shade tolerance of four temperate deciduous woody species. Funct. Ecol. 11, 518–531. doi: 10.1046/j.1365-2435.1997.00109.x
Niinemets, U., and Valladares, F. (2004). Photosynthetic acclimation to simultaneous and interacting environmental stresses along natural light gradients: optimality and constraints. Plant Biol. 6, 254–268. doi: 10.1055/s-2004-817881
Oh, E., Zhu, J. Y., and Wang, Z. Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14, 802–809. doi: 10.1038/ncb2545
Oh, J., Park, E., Song, K., Bae, G., and Choi, G. (2020). PHYTOCHROME INTERACTING FACTOR8 inhibits phytochrome a-mediated far-red light responses in arabidopsis. Plant Cell 32, 186–205. doi: 10.1105/tpc.19.00515
Panigrahy, M., Ranga, A., Das, J., and Panigrahi, K. C. S. (2019). Shade tolerance in Swarnaprabha rice is associated with higher rate of panicle emergence and positively regulated by genes of ethylene and cytokinin pathway. Sci. Rep. 9:6817. doi: 10.1038/s41598-019-43096-8
Paulisic, S., Qin, W., Arora Veraszto, H., Then, C., Alary, B., Nogue, F., et al. (2021). Adjustment of the PIF7-HFR1 transcriptional module activity controls plant shade adaptation. EMBO J. 40:e104273. doi: 10.15252/embj.2019104273
Pierik, R., Djakovic-Petrovic, T., Keuskamp, D. H., De Wit, M., and Voesenek, L. A. (2009). Auxin and ethylene regulate elongation responses to neighbor proximity signals independent of gibberellin and della proteins in Arabidopsis. Plant Physiol. 149, 1701–1712. doi: 10.1104/pp.108.133496
Reed, J. W., Nagpal, P., Poole, D. S., Furuya, M., and Chory, J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5, 147–157. doi: 10.1105/tpc.5.2.147
Reich, P. B., Tjoelker, M. G., Walters, M. B., and Buschena, D. (1998). Close Association of RGR, leaf and root morphology, seed mass and shade tolerance in seedlings of nine boreal tree species grown in high and low light. Funct. Ecol. 12, 327–338. doi: 10.1046/j.1365-2435.1998.00208.x
Robson, F., Okamoto, H., Patrick, E., Harris, S. R., Wasternack, C., Brearley, C., et al. (2010). Jasmonate and phytochrome a signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell 22, 1143–1160. doi: 10.1105/tpc.109.067728
Robson, P. R. H., Mccormac, A. C., Irvine, A. S., and Smith, H. (1996). Genetic engineering of harvest index in tobacco through overexpression of a phytochrome gene. Nat. Biotechnol. 14, 995–998. doi: 10.1038/nbt0896-995
Robson, P., Whitelam, G. C., and Smith, H. (1993). Selected components of the shade-avoidance syndrome are displayed in a normal manner in mutants of Arabidopsis thaliana and Brassica rapa deficient in Phytochrome B. Plant Physiol. 102, 1179–1184. doi: 10.1104/pp.102.4.1179
Roig-Villanova, I., and Martinez-Garcia, J. F. (2016). Plant responses to vegetation proximity: a whole life avoiding shade. Front. Plant Sci. 7:236. doi: 10.3389/fpls.2016.00236
Roig-Villanova, I., Bou-Torrent, J., Galstyan, A., Carretero-Paulet, L., Portoles, S., Rodriguez-Concepcion, M., et al. (2007). Interaction of shade avoidance and auxin responses: a role for two novel atypical bHLH proteins. EMBO J. 26, 4756–4767. doi: 10.1038/sj.emboj.7601890
Rousseaux, M. C., Ballare, C. L., Jordan, E. T., and Vierstra, R. D. (1997). Directed overexpression of PHYA locally suppresses stem elongation and leaf senescence responses to far-red radiation. Plant Cell Environ. 20, 1551–1558.
Saijo, Y., Sullivan, J. A., Wang, H., Yang, J., Shen, Y., Rubio, V., et al. (2003). The COP1-SPA1 interaction defines a critical step in phytochrome a-mediated regulation of HY5 activity. Genes Dev. 17, 2642–2647. doi: 10.1101/gad.1122903
Sharrock, R. A., and Quail, P. H. (1989). Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev. 3, 1745–1757. doi: 10.1101/gad.3.11.1745
Shen, H., Moon, J., and Huq, E. (2005). PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J. 44, 1023–1035. doi: 10.1111/j.1365-313X.2005.02606.x
Shen, H., Zhu, L., Castillon, A., Majee, M., Downie, B., and Huq, E. (2008). Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20, 1586–1602. doi: 10.1105/tpc.108.060020
Shen, Y., Khanna, R., Carle, C. M., and Quail, P. H. (2007). Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 145, 1043–1051. doi: 10.1104/pp.107.105601
Shin, A. Y., Han, Y. J., Baek, A., Ahn, T., Kim, S. Y., Nguyen, T. S., et al. (2016). Evidence that phytochrome functions as a protein kinase in plant light signalling. Nat. Commun. 7:11545. doi: 10.1038/ncomms11545
Smith, D. (1979). A systematic relationship between phytochrome-controlled development and species habitat, for plants grown in simulated natural radiation. Planta 145, 253–258. doi: 10.1007/BF00454449
Smith, H. (1995). Physiological and ecological function within the phytochrome family. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 289–315.
Smith, H., and Whitelam, G. C. (1997). The shade avoidance syndrome: multiple responses mediated by multiple phytochromes. Plant Cell Environ. 20, 840–844. doi: 10.1046/j.1365-3040.1997.d01-104.x
Somers, D. E., Sharrock, R. A., Tepperman, J. M., and Quail, P. H. (1991). The hy3 long hypocotyl mutant of arabidopsis is deficient in phytochrome B. Plant Cell 3, 1263–1274. doi: 10.1105/tpc.3.12.1263
Song, B., Zhao, H., Dong, K., Wang, M., Wu, S., Li, S., et al. (2020). Phytochrome A inhibits shade avoidance responses under strong shade through repressing the brassinosteroid pathway in Arabidopsis. Plant J. 104, 1520–1534. doi: 10.1111/tpj.15018
Soy, J., Leivar, P., Gonzalez-Schain, N., Martin, G., Diaz, C., Sentandreu, M., et al. (2016). Molecular convergence of clock and photosensory pathways through PIF3-TOC1 interaction and co-occupancy of target promoters. Proc. Natl. Acad. Sci. U.S.A. 113, 4870–4875. doi: 10.1073/pnas.1603745113
Stirnberg, P., Zhao, S., Williamson, L., Ward, S., and Leyser, O. (2012). FHY3 promotes shoot branching and stress tolerance in Arabidopsis in an AXR1-dependent manner. Plant J. 71, 907–920. doi: 10.1111/j.1365-313X.2012.05038.x
Swain, S., Jiang, H. W., and Hsieh, H. L. (2017). FAR-RED INSENSITIVE 219/JAR1 contributes to shade avoidance responses of Arabidopsis seedlings by modulating key shade signaling components. Front. Plant Sci. 8:1901. doi: 10.3389/fpls.2017.01901
Tang, W., Wang, W., Chen, D., Ji, Q., Jing, Y., Wang, H., et al. (2012). Transposase-derived proteins FHY3/FAR1 interact with PHYTOCHROME-INTERACTING FACTOR1 to regulate chlorophyll biosynthesis by modulating HEMB1 during deetiolation in Arabidopsis. Plant Cell 24, 1984–2000. doi: 10.1105/tpc.112.097022
Tao, Y., Ferrer, J. L., Ljung, K., Pojer, F., Hong, F., Long, J. A., et al. (2008). Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133, 164–176. doi: 10.1016/j.cell.2008.01.049
Tian, T., Ma, L., Liu, Y., Xu, D., Chen, Q., and Li, G. (2020). Arabidopsis FAR-RED ELONGATED HYPOCOTYL3 integrates age and light signals to negatively regulate leaf senescence. Plant Cell 32, 1574–1588. doi: 10.1105/tpc.20.00021
Valladares, F., and Niinemets, Ü (2008). Shade tolerance, a key plant feature of complex nature and consequences. Annu. Rev. Ecol. Evol. Syst. 39, 237–257. doi: 10.1146/annurev.ecolsys.39.110707.173506
Vandenbussche, F., Pierik, R., Millenaar, F. F., Voesenek, L. A., and Van Der Straeten, D. (2005). Reaching out of the shade. Curr. Opin. Plant Biol. 8, 462–468. doi: 10.1016/j.pbi.2005.07.007
Wang, H., and Deng, X. W. (2002). Arabidopsis FHY3 defines a key phytochrome a signaling component directly interacting with its homologous partner FAR1. EMBO J. 21, 1339–1349. doi: 10.1093/emboj/21.6.1339
Wang, J. G., Chen, C. H., Chien, C. T., and Hsieh, H. L. (2011). FAR-RED INSENSITIVE219 modulates CONSTITUTIVE PHOTOMORPHOGENIC1 activity via physical interaction to regulate hypocotyl elongation in Arabidopsis. Plant Physiol. 156, 631–646.
Wu, Y., Gong, W., and Yang, W. (2017). Shade inhibits leaf size by controlling cell proliferation and enlargement in soybean. Sci. Rep. 7:9259.
Xie, Y., Liu, Y., Ma, M., Zhou, Q., Zhao, Y., Zhao, B., et al. (2020a). Arabidopsis FHY3 and FAR1 integrate light and strigolactone signaling to regulate branching. Nat. Commun. 11:1955.
Xie, Y., Zhou, Q., Zhao, Y., Li, Q., Liu, Y., Ma, M., et al. (2020b). FHY3 and FAR1 integrate light signals with the miR156-SPL module-mediated aging pathway to regulate arabidopsis flowering. Mol. Plant 13, 483–498.
Xu, F., He, S., Zhang, J., Mao, Z., Wang, W., Li, T., et al. (2018). Photoactivated CRY1 and phyB interact directly with AUX/IAA proteins to inhibit auxin signaling in Arabidopsis. Mol. Plant 11, 523–541.
Yang, C., Xie, F., Jiang, Y., Li, Z., Huang, X., and Li, L. (2018). Phytochrome A negatively regulates the shade avoidance response by increasing auxin/indole acidic acid protein stability. Dev. Cell 44, 29–41 e24.
Yanovsky, M., Casai, J., and Whitelam, G. (1995). Phytochrome A, phytochrome B and HY4 are involved in hypocotyl growth responses to natural radiation in Arabidopsis: weak de-etiolation of the phyA mutant under dense canopies. Plant Cell Environ. 18, 788–794.
Zhang, R., Yang, C., Jiang, Y., and Li, L. (2019). A PIF7-CONSTANS-centered molecular regulatory network underlying shade-accelerated flowering. Mol. Plant 12, 1587–1597.
Zhang, Y., Pfeiffer, A., Tepperman, J. M., Dalton-Roesler, J., Leivar, P., Gonzalez Grandio, E., et al. (2020). Central clock components modulate plant shade avoidance by directly repressing transcriptional activation activity of PIF proteins. Proc. Natl. Acad. Sci. U.S.A. 117, 3261–3269.
Zheng, Y., Cui, X., Su, L., Fang, S., Chu, J., Gong, Q., et al. (2017). Jasmonate inhibits COP1 activity to suppress hypocotyl elongation and promote cotyledon opening in etiolated Arabidopsis seedlings. Plant J. 90, 1144–1155.
Zhou, P., Song, M., Yang, Q., Su, L., Hou, P., Guo, L., et al. (2014). Both PHYTOCHROME RAPIDLY REGULATED1 (PAR1) and PAR2 promote seedling photomorphogenesis in multiple light signaling pathways. Plant Physiol. 164, 841–852.
Keywords: shade tolerance, shade avoidance, phytochrome A, phytohormones, Arabidopsis
Citation: Xu H, Chen P and Tao Y (2021) Understanding the Shade Tolerance Responses Through Hints From Phytochrome A-Mediated Negative Feedback Regulation in Shade Avoiding Plants. Front. Plant Sci. 12:813092. doi: 10.3389/fpls.2021.813092
Received: 11 November 2021; Accepted: 03 December 2021;
Published: 22 December 2021.
Edited by:
Lin Li, Fudan University, ChinaReviewed by:
Chuanwei Yang, Fudan University, ChinaGang Li, Shandong Agricultural University, China
Copyright © 2021 Xu, Chen and Tao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yi Tao, eWl0YW9AeG11LmVkdS5jbg==
†These authors have contributed equally to this work
 Huiying Xu
Huiying Xu Peirui Chen
Peirui Chen Yi Tao
Yi Tao