
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 27 January 2022
Sec. Plant Abiotic Stress
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.806195
This article is part of the Research Topic Identification and Functional Dissection of Stress-responsive Genes in Cotton View all 26 articles
A subfamily of transcription factors known as HD-ZIP III plays distinct roles in the secondary cell wall biosynthesis, which could be attributed to the quality of cotton fiber and adaptation to drought stress. In this study, 18 HD-ZIP III genes were identified as genome wide from the upland cotton (Gossypium hirsutum). These genes are distributed on 14 different chromosomes, and all of them have undergone segmental duplications. Numerous cis-elements were identified in the promoter regions, which are related to phytohormone responses and abiotic stresses. Expression profiling of these genes by quantitative real-time (qRT)-PCR illustrated their differential spatial expression, with preferential expression in cotton fiber. Among these genes, GhHB8-5D was predicted to encode a protein that is targeted to the cell nucleus and having self-activation ability. In addition, the ectopic expression of GhHB8-5D or its synonymous mutant GhHB8-5Dm in Arabidopsis resulted in stunted plant growth, curly leaves, and twisted inflorescence stems. Microscopy examination revealed that the morphology of vascular bundles and deposition of secondary wall had substantially altered in stems, which is concomitant with the significant alteration in the transcription levels of secondary wall-related genes in these transgenic Arabidopsis. Further, ectopic expression of GhHB8-5D or GhHB8-5Dm in Arabidopsis also led to significant increase in green seedling rate and reduction in root length relative to wild type when the plants were grown under mimicked drought stress conditions. Taken together, our results may shed new light on the functional roles of GhHB8-5D that is attributable for secondary cell wall thickening in response to drought stress. Such a finding may facilitate a novel strategy for improving plant adaptations to environmental changes via regulating the biosynthesis of secondary cell wall.
As the most important crop producing natural fibers, upland cotton (Gossypium hirsutum) is broadly cultivated in the temperate regions in the world and the quality of fiber is of paramount economic significance (Paterson et al., 2012). Fiber development undergoes four distinctive but overlapping stages: initiation, elongation (primary cell wall biosynthesis), secondary cell wall (SCW) thickening (cellulose biosynthesis), and maturation (Haigler et al., 2012). The formation of SCW in fiber cells has been recognized as one of the most crucial steps that directly affect cotton fiber quality (Li et al., 2015).
In mature cotton fiber, SCW is mainly composed of cellulose that is a polysaccharide synthesized by cellulose synthase (CESA) complex (McFarlane et al., 2014; Hernandez-Gomez et al., 2015). Transcriptional regulation network controlling SCW biosynthesis has been extensively elucidated in the model plant Arabidopsis (Arabidopsis thaliana), in which numerous transcription factors (TFs) such as NAC acting as master switch coordinate the expression of CESAs by coupling with MYB and others (Taylor-Teeples et al., 2015; Zhong and Ye, 2015). However, in cotton, only a limited number of TFs that regulate SCW synthesis have been identified, while their functionality and intricate relationship remain poorly understood (Zhang et al., 2018a; Huang et al., 2019, 2021; Cao et al., 2020). This is despite the recent premise of a model of a four-layered transcriptional regulatory network consisting of GhTCP4, GhMYB7, GhFSN1, and GhMYB46_D13 regulating fiber SCW GhCesA genes (Huang et al., 2021). Such a network model may still fall short in sophistication, as many other TFs that function in concert in SCW formation, such as HD-ZIP III TFs, remain unexplored (Robischon et al., 2011; Du et al., 2015).
HD-ZIP III subfamily contains five members in the A. thaliana genome, including REVOLUTA/INTERFASCICULAR FIBERLESS1, ATHB8, PHAVOLUTA/ATHB9, PHABULOSA/ATHB14, and CORONA/ATHB15, all of which harbor a leucine zipper motif (LZ) downstream of the homeodomain (HD) (Schena and Davis, 1992). The HD is responsible for the specific binding to target DNA, whereas LZ acts as a dimerization motif (Ariel et al., 2007). The conserved amino acids then form a START (steroidogenic acute regulatory protein-related lipid transfer) domain and an adjacent conserved region known as SAD (START-adjacent domain). HD-ZIP III genes are posttranscriptionally regulated by miR165/166 that targets the START domain of the subfamily (McConnell et al., 2001; Kidner and Martienssen, 2004; Kim et al., 2005). Additionally, all members of this subfamily have a conserved domain known as MEKHLA in their C-termini, which shares significant similarity with the PAS domain and is involved in light, oxygen, and redox potential sensing (Mukherjee and Burglin, 2006).
Previous studies showed that all the members in the HD-ZIP III subfamily were required for xylem cells differentiation and secondary wall biosynthesis. For example, REV is a positive regulator of secondary wall deposition in interfascicular fibers, and its defective mutant rev lacked normal interfascicular fibers in stems (Zhong et al., 1997; Zhong and Ye, 1999). REV is also negatively regulated by KNAT7 and BLH6, the expression of which promoted SCW deposition in the knat7/blh6 double knockout mutant (Liu et al., 2014). ATHB8, a gene positively regulated by auxin (Baima et al., 1995), was considered as an early marker of the procambial and cambial cells during vascular development. Ectopic expression of ATHB8 in Arabidopsis increased the production of xylem tissues (Baima et al., 2001). Similarly, a Populus trichocarpa HD-ZIP III gene, PtrHB7, was preferentially expressed in the cambial zone; on the other hand, PtrHB7-suppressed plants displayed significant changes in vascular tissues with a reduction in xylem but increase in the phloem (Zhu et al., 2013). In the phylogenetic analysis of A. thaliana genes, ATHB9, ATHB14, and REV comprised a clade and exhibited similar expression patterns in the vasculature, with ATHB9 and ATHB14 as a sister pair (McConnell et al., 2001; Emery et al., 2003). Mutations in the ATHB9 and ATHB14 genes enhanced the vascular defects of the rev mutant (Prigge et al., 2005). The coordinated expression of REV, ATHB9, and ATHB14 are necessary for xylem cell specification and secondary wall biosynthesis (Carlsbecker et al., 2010). The overexpression of OsHB4, a member of the rice HD-ZIP III subfamily, resulted in leaf rolling and altered stem xylem in rice, and the polysaccharide synthesis-related genes could be regulated by miR166-OsHB4 as revealed by transcriptomic analysis (Zhang et al., 2018b). Therefore, OsHB4 may contribute to cell wall formation and vascular development in rice. In Arabidopsis, overexpression of a miRNA-resistant ATHB15 resulted in moderate dwarfing, upcurling leaves, and a drastic reduction in xylem and lignified interfascicular tissues (Kim et al., 2005). Transgenic Populus expressing a synthetic miRNA targeting ATHB15 led to abnormal lignification in cells of the pith, while the overexpression of a miRNA-resistant ATHB15 caused delayed lignification of xylem and phloem fibers during secondary growth (Du et al., 2011), hence ATHB15 was believed to be involved in secondary wall transcriptional pathway by regulating wall-related TFs and synthetic genes (Du et al., 2015).
The plant root xylem is a specialized tissue that distributes water to the shoot. In Arabidopsis, water deficiency enhanced the levels of miR165, which in turn negatively affected HD-ZIP III expression and impacted on xylem development and hydraulic conductivity of root (Ramachandran et al., 2018). On the other hand, the overexpression of a miR166-resistant form of OsHB4 contributed to cell wall formation, vascular development, and drought resistance in rice (Zhang et al., 2018b), which was well in line with the overexpression of OsHOX32 in rice that displayed narrow rolled leaves, reduced stomatal conductance, and transpiration rate, leading to the improvement in water use efficiency (Li et al., 2016). In cotton, deep sequencing of salt- and drought-treated small RNA libraries, led to the identification of ghr-miR166a-j that was downregulated in both drought and salinity treatments, and its target gene ghr-HD-ZIP IIIs was significantly upregulated as a result (Xie et al., 2015).
In this study, we performed a comprehensive genome-wide analysis of HD-ZIP III genes in cotton and presented characteristics of this subfamily. In addition, we further investigated the function of GhHB8-5D in Arabidopsis, overexpression of which altered the morphology of vasculature, secondary wall deposition, and drought tolerance. This study may not only provide new insight on the functions of GhHB8-5D but also facilitate additional useful tools for genetic improvements of cotton fiber quality and drought resistance.
Five protein sequences of Arabidopsis HD-ZIP III members were obtained from TAIR1 as queries to search cotton (Gossypium hirsutum L. acc. TM-1) genome database2. The conserved HD (PF00046), START domain (PF01852), and MEKHLA domain (PF08670) of HD-ZIP III subfamily were found by using Pfam3, HMMER4, and the Batch Web CD-Search5. The molecular features and subcellular localization of the cotton HD-ZIP III proteins were predicted using the ProtParam tool6 and Plant-mPLoc7, respectively.
The genomic and coding sequences of HD-ZIP III genes were compared by Gene Structure Display Server8 to investigate the distribution of exon/intron. A total number of 18 HD-ZIP III protein sequences were used to predict the conserved motifs by using the MEME online program9, which were further validated by the Batch Web CD-Search, Pfam, and Batch SMART10.
Chromosomal location information of HD-ZIP III genes was extracted from the cotton genome database. All the members of the HD-ZIP III subfamily were mapped to their respective locus of chromosomes and visualized using TBtools (Chen et al., 2020). For collinearity analysis, the protein sequences of 18 HD-ZIP III genes were served as queries for BLASTP to search the cotton genome database. Duplicated sequences of HD-ZIP III genes were identified as previously described. The alignment covers > 80% of the longer gene and the aligned region has an identity > 80% at the nucleotide level (Wang et al., 2020; Zhang et al., 2020). MCScanX software was employed to estimate the collinear pairs and gene duplication type and the result was visualized by using Circos software (Krzywinski et al., 2009; Wang et al., 2012).
A fragment of 2 kb upstream region of the transcriptional start site of each HD-ZIP III gene was retrieved as the promoter sequence and analyzed using the PlantCARE11 for cis-element prediction.
The putative protein sequences of HD-ZIP III derived from A. thaliana, Gossypium arboretum, G. hirsutum, G. raimondii, Oryza Sativa, Populus trichocarpa, and Zinnia elegans genes were aligned using ClustalX and a phylogenetic tree was constructed by using MEGA7 and the maximum likelihood method with 1000 bootstrap replications (Larkin et al., 2007; Kumar et al., 2016).
Gossypium hirsutum cv. ZM24 plants were grown to maturation in a controlled growth chamber under 30°C with a 16 h photoperiod. The seeds of A. thaliana ecotype Col-0 were surface-sterilized and sown on half strength Murashige and Skoog medium supplemented with 2% sucrose. After a vernalization period at 4°C for 48 h, the plates containing Arabidopsis seeds were transferred to a plant growth incubator for a further 7 days before being transplanted to soil in a greenhouse at 22°C with 16 h photoperiod.
The entire coding region of GhHB8-5D was PCR amplified and subcloned into pBI121 under the transcriptional control of the CaMV 35S promoter. Mutations at the miR166a target site were introduced by fusion PCR amplification to construct the GhHB8-5Dm overexpression vector. Two overlapping primers (Supplementary Table 5) that mismatched the miRNA binding sites without changing the protein coding sequence were used to generate mutations in the GhHB8-5D cDNA sequence. The mutated cDNA was then inserted into pBI121 behind the CaMV 35S promoter. The vectors were transferred into Agrobacterium tumefaciens and introduced in A. thaliana Col-0 by floral dip method (Zhang et al., 2006). Transgenic Arabidopsis seedlings were selected by germinating seeds on kanamycin-supplemented Murashige and Skoog (MS) agar plates. Two independent lines with a single transgene were bred to homozygosity and used for further analysis.
The coding sequence of GhHB8-5D was cloned to a pBI-eGFP vector that was then introduced into A. tumefaciens for transient expression in the leaf tissues of Nicotiana benthamiana as previously described (Sparkes et al., 2006). Green fluorescent protein (GFP) in agro-infiltrated N. benthamiana leaf cells was detected under an SP5 Meta confocal laser microscope (Leica Microsystems, Mannheim, Germany). To investigate the transcriptional activity of GhHB8-5D, both the full-length and truncated coding sequences of GhHB8-5D were inserted into the pGBKT7 vector, which were then introduced into baker’s yeast Saccharomyces cerevisiae strain AH109 using the high-efficiency lithium acetate transformation procedure. Yeast transformants were streaked on a selective medium lacking tryptophan to assay transcriptional activity. the β-galactosidase activity was also assayed by colony-lift filter assay using 5-bromo-4-chloro-3-indolyl β-D-galactopyranoside (X-gal) as substrate.
Freehand slicing and paraffin-embedded section of Arabidopsis stems and secondary wall staining were performed as previously described (Huang et al., 2019).
Cotton RNA was extracted from different cotton tissues, and Arabidopsis RNA was isolated from 6-week-old stems using RNAprep Pure Plant Plus Kit (TIANGEN, Beijing, China) according to the manufacturer’s instructions. Expression profiles of cotton and Arabidopsis genes were analyzed by qRT-PCR using GhUBI1 (EU604080) and AtActin2 (AT3G18780.1) as reference genes according to a previously described method (Xu et al., 2013). All primers were listed in Supplementary Table 5.
Thirty surface-sterilized seeds from wild type or each transgenic Arabidopsis line were placed for germination on the MS medium supplemented with or without different concentrations of mannitol and placed at 4°C for 2 days, prior to being moved to a growth room at 22°C and 16 h photoperiod. The number of seedlings with green cotyledons was counted after 2 days. Each experiment was repeated three times.
For assaying primary root length, the seeds of wild type or transgenic Arabidopsis lines were germinated and maintained on MS medium supplemented with different concentrations of mannitol for 7 days. The length of primary roots of seedlings was measured and compared. All experiments were repeated at least three times.
In order to identify HD-ZIP III genes in cotton, five Arabidopsis HD-ZIP III protein sequences were used as queries to blast search the cotton genome database. The putative HD-ZIP III protein sequences were analyzed by Pfam, HMMER, and the Batch Web CD-Search to find conserved domains. As a result, 18 genes were identified as the members of HD-ZIP III subfamily in cotton. Every HD-ZIP III protein encompasses three conserved domains, including the HOMEOBOX domain, START domain, and MEKHLA domain (Supplementary Table 1). The 18 cotton HD-ZIP III genes were designated according to their chromosomal positions. The protein length and molecular weights are about 840 amino acids and 92 KDa, respectively, in all these HD-ZIP III proteins except GhHB8-5A, these two features of which are 776 amino acids and 85.675 KDa, respectively. The isoelectric point (pI) of HD-ZIP IIIs varies from 5.74 to 6.19. Bioinformatics analysis predicted that all cotton HD-ZIP III proteins may be located in the nucleus (Supplementary Table 2).
To get a further understanding of the cotton HD-ZIP III subfamily, gene structure and conserved motifs of each HD-ZIP III member were investigated. Most of the HD-ZIP III genes possess 17 introns, while GhHB8-5A, GhHB8-7A, and GhHB8-7D have 15, 16, and 16 introns, respectively (Figure 1A). Such a distribution pattern of exon/intron is consistent with that of HD-ZIP III genes in rice (Agalou et al., 2008). As shown in Figure 1B, there are ten different motifs in each HD-ZIP III based on the sequence conservation as predicted by using MEME. Among them, motifs 1 and 4 are the core sequences of the HD and LZ domains, respectively. Motif 2, 3, 5, and 9 compose the START domain. The SAD domain is constitutive of motif 7 and 8. Motif 6 and 10 belong to the MEKHLA domain. The fact that all cotton HD-ZIP IIIs share the same batch of motifs is suggestive of functional versatility and potential redundancy in this subfamily.
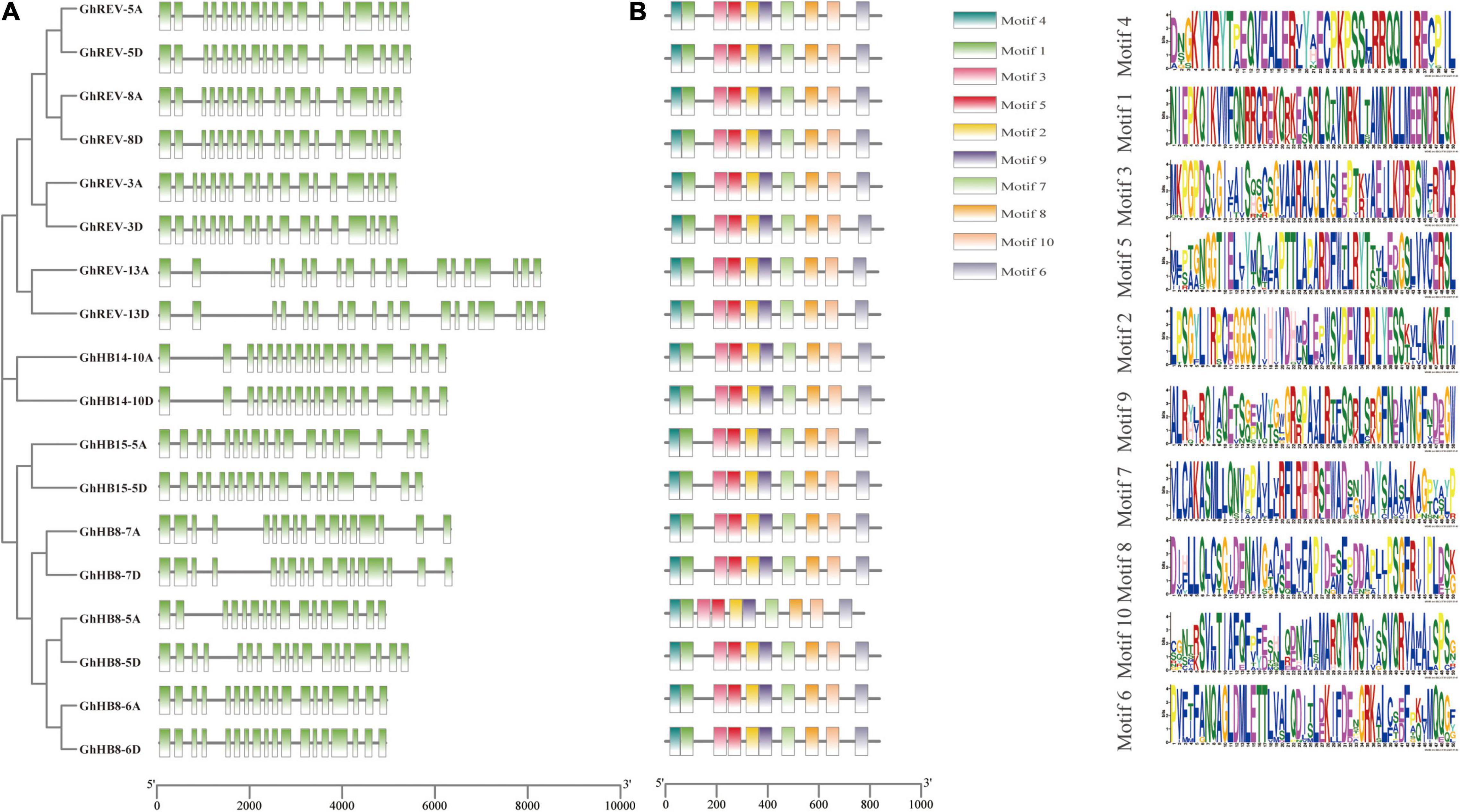
Figure 1. Gene structure and conserved motifs of HD-ZIP III subfamily in cotton (Gossypium hirsutum). (A) The exon/intron distribution of HD-ZIP III genes. Green boxes and black lines represent exons and introns, respectively. (B) Conserved motifs of each HD-ZIP III protein. Each conserved motif is indicated by different color boxes. The length of each gene and protein can be estimated using the scale at the bottom.
The promoter regions of cotton HD-ZIP III genes were analyzed to study their biological function in depth. As shown in Figure 2, in the 2 kb sequences upstream of the start codon, many cis-elements were identified, including ABRE, TGA-element, GARE-motif, AuxRE, AuxRR-core, TGA-box, TCA-element, P-box, TATC-box, CGTCA-motif, and TGACG-motif, which were reported to respond to the induction of various phytohormones, while LTR (cis-acting element involved in low-temperature responsiveness), TC-rich repeats (cis-acting element involved in defense and stress responsiveness), and MBS (MYB binding site involved in drought inducibility) were involved in abiotic stresses responsiveness. From the distribution of these cis-elements in each HD-ZIP III promoter, it is conceivable that almost all cotton HD-ZIP IIIs could be involved in phytohormone responsiveness, such as auxin, gibberellin, and abscisic acid, except GhHB15-5A. More than half of this subfamily member could be responsive to drought and low-temperature stresses.
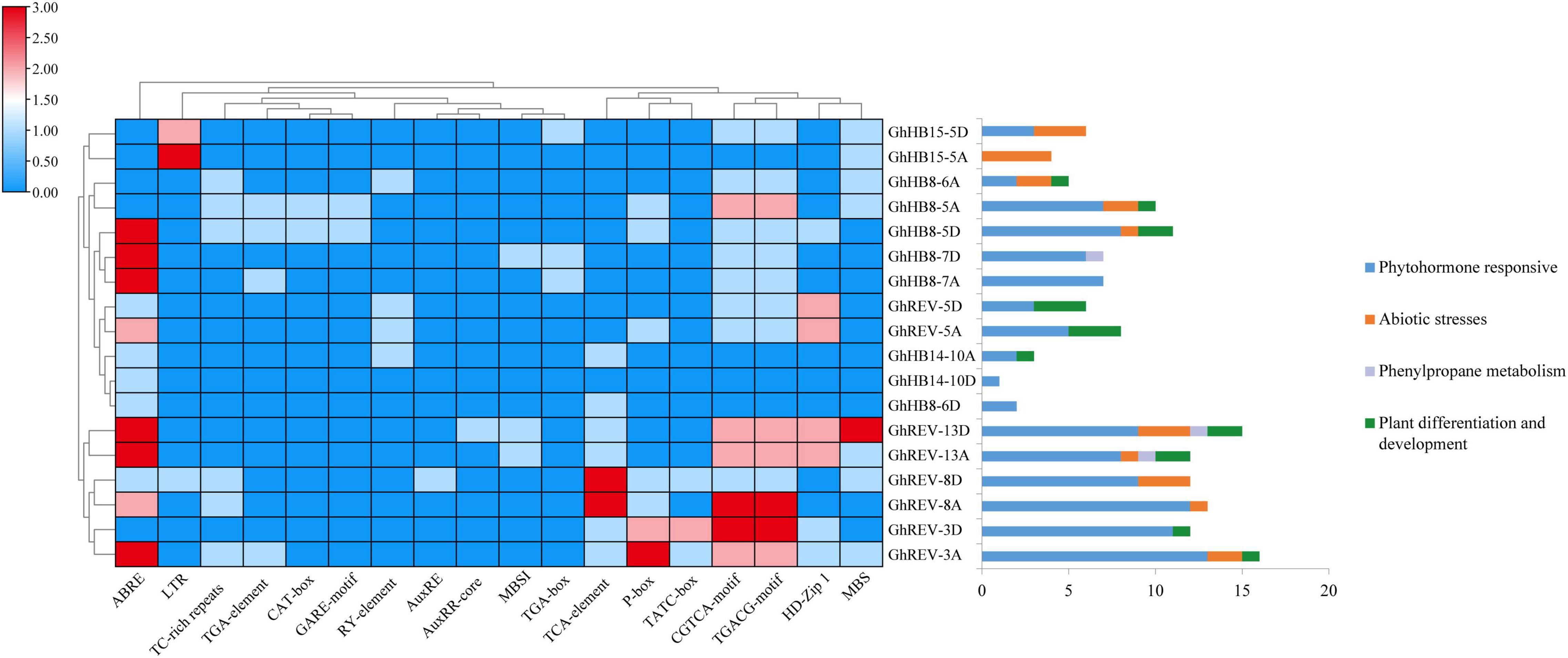
Figure 2. Statistics of cis-elements in the promoter region of cotton (Gossypium hirsutum) HD-ZIP III genes. The left part shows statistics on the number of specific cis-elements contained in each HD-ZIP III gene promoter region. The right part exhibits statistics on the function of cis-elements possessed in each HD-ZIP III gene promoter region.
The physical location of cotton HD-ZIP IIIs on chromosomes was determined using the chromosomal location information extracted from the cotton genome database (Supplementary Figure 1), and it was found that the 18 HD-ZIP IIIs were unevenly distributed on different chromosomes but symmetrically in A and D-subgenomes. Chromosome 5 contains most, i.e., six HD-ZIP III genes, while the remaining 12 genes are evenly distributed on six other chromosomes. A total of 23 pairs of collinearity genes were detected, all of which were located on different chromosomes (Figure 3), suggesting that gene segmental duplication may be the primary route of HD-ZIP III subfamily expansion and evolution.
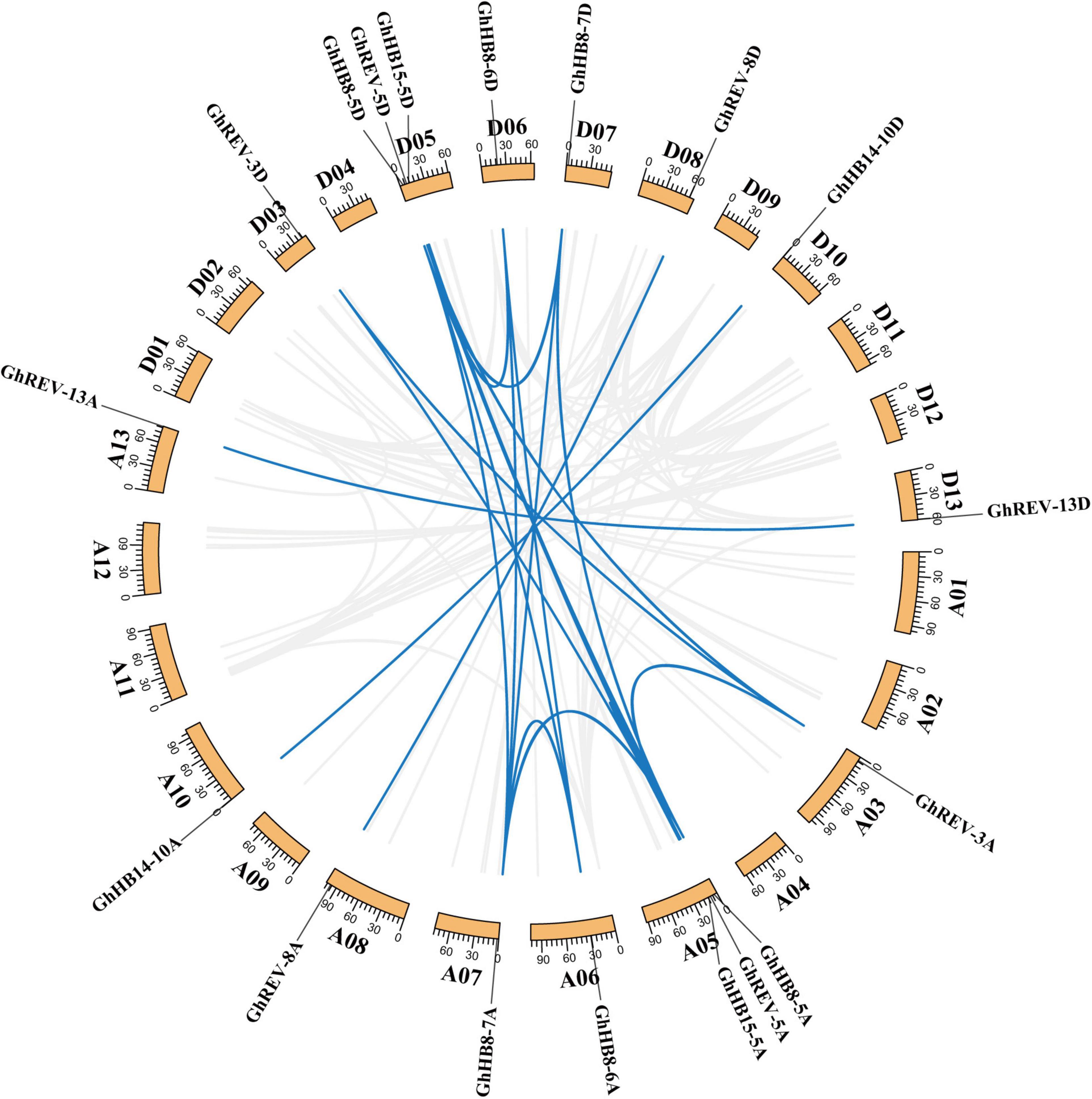
Figure 3. Chromosomal distribution and gene duplication of HD-ZIP III genes in cotton (Gossypium hirsutum). The value on each chromosome represents chromosome length. Blue lines indicate duplicated HD-ZIP III gene pairs.
To understand the evolutionary relationship of cotton HD-ZIP III genes and their orthologs in different plant species, we construct a phylogenetic tree from the alignment of 18 G. hirsutum, 9 G. arboretum, 9 G. raimondii, 5 A. thaliana, 5 O. sativa, 8 P. trichocarpa, 4 Z. elegans HD-ZIP III protein sequences (Supplementary Table 3). As shown in Figure 4, the HD-ZIP III subfamily can be divided into four groups: REV, HB8, HB14, and HB15. The members of G. hirsutum, G. arboretum, G. raimondii, A. thaliana, and P. trichocarpa HD-ZIP IIIs were dispersed in each group, whereas the O. sativa homolog was not present in the HB8 group and the Z. elegans homolog was not included in the group HB14, suggestive of lineage-specific gene loss in these two species. Except for HB14, single REV, AtHB8, and CNA genes were found in the subclades of REV, HB8, and HB15 in Arabidopsis, whereas multiple genes were found in tetraploid G. hirsutum, which may indicate that the duplication of HD-ZIP III genes had occurred in G. hirsutum after the divergence of G. hirsutum and Arabidopsis.
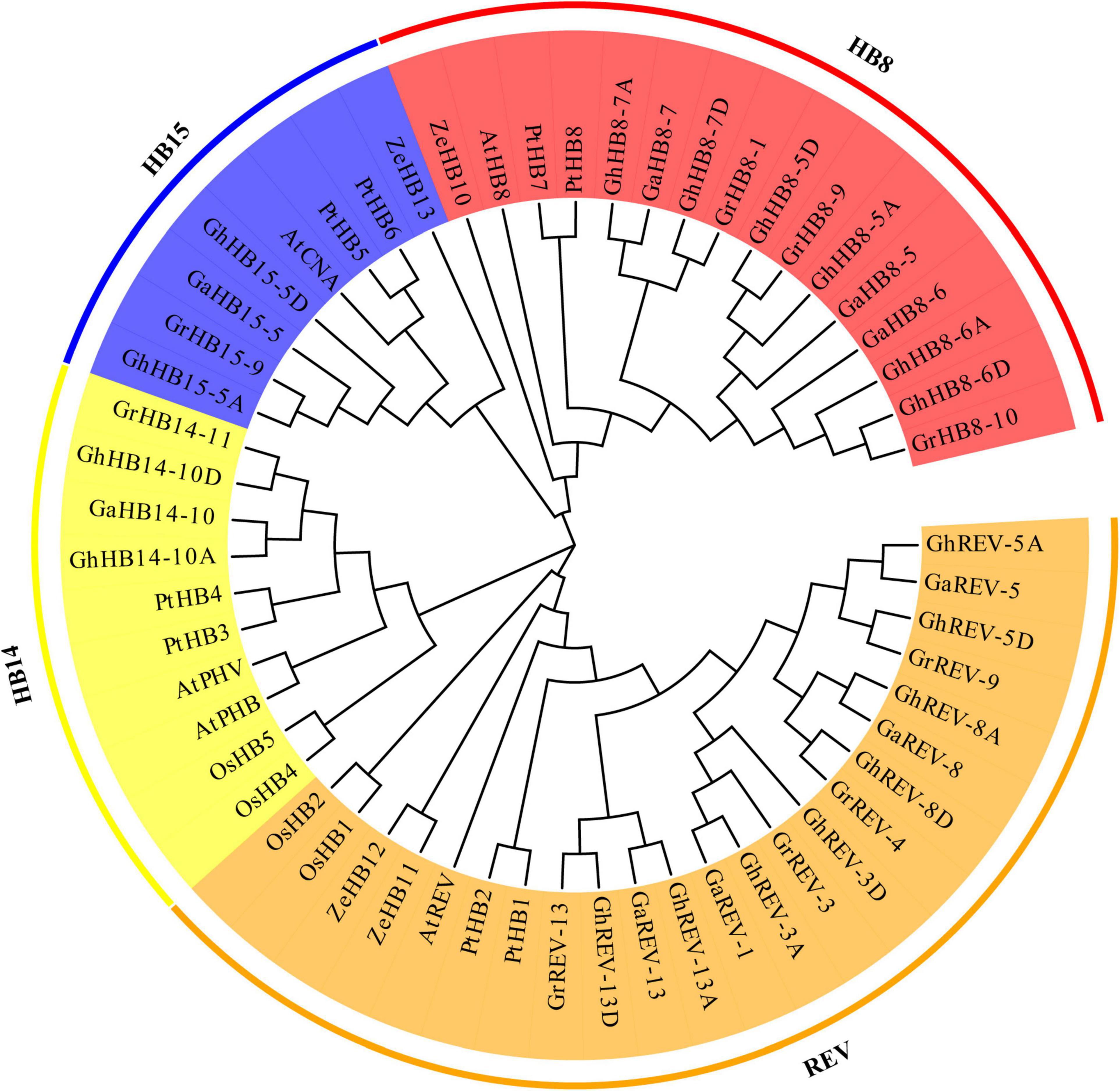
Figure 4. Phylogenetic tree of Arabidopsis, Gossypium arboretum, Gossypium hirsutum, Gossypium raimondii, Oryza Sativa, Populus trichocarpa, and Zinnia elegans HD-ZIP III proteins. All the protein sequences were aligned by ClustalX and the phylogenetic tree was constructed by MEGA7 using the maximum likelihood method with 1000 bootstrap replications. The four groups are represented with different colors.
To get insight into the expression profiles of the cotton HD-ZIP III subfamily, we performed qRT-PCR using the RNAs derived from various cotton tissues. Because the sequences of two homologous genes from the A and D subgenome are very similar, we designed a pair of consensus primers for the two genes (Supplementary Table 5). It was shown that GhREV-3, 5, 8, and 13 were highly expressed in both vegetative and reproductive organs, whereas GhHB8-6, 7 and GhHB14-10 were mainly expressed in root, stem, and 20 days post anthesis (DPA) ovule. GhHB8-5 and GhHB15-5 were expressed preferentially in 25 DPA fibers (Figure 5). The diversified expression patterns of HD-ZIP III genes in cotton may indicate their functional diversity and versatility relevant to cotton growth and development.
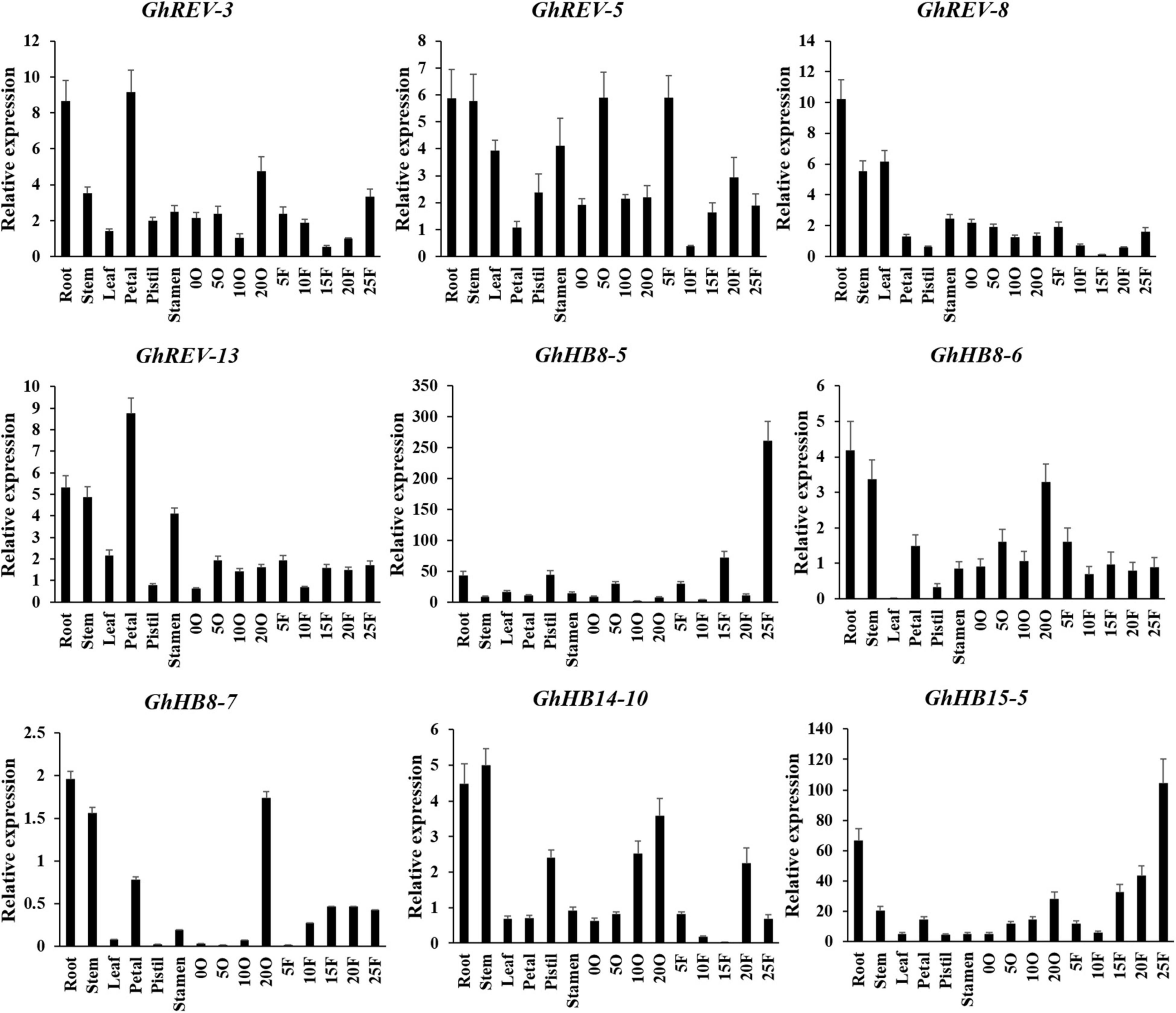
Figure 5. Expression profiles of cotton HD-ZIP III genes in different cotton tissues. GhUBI1 was used as the internal control for normalization. Values represent the mean ± SD of three biological replicates. 0O, ovule in anthesis; 5O, ovules in 5 days post anthesis (DPA); 10O, ovules in 10 DPA; 20O, ovules in 20 DPA; 5F, fibers in 5 DPA; 10F, fibers in 10 DPA; 15F, fibers in 15 DPA; 20F, fibers in 20 DPA; 25F, fibers in 25 DPA.
In order to further clarify the expression of GhHB8-5A and 5D, the two homoeologous genes of GhHB8-5, first, we compared the cis-elements in the promoter regions of GhHB8-5A and 5D, and their variations were documented in Supplementary Table 4. Then, we designed primers based on the most variable regions in order to differentiate the homoeologous genes (Supplementary Table 5) for qRT-PCR. As shown in Supplementary Figure 2, the expression of GhHB8-5A was abundant in root, flower, 20 and 25 DPA fiber. GhHB8-5D was predominantly expressed in fiber, and its expression level increased along with the progress of fiber development. Hence, GhHB8-5D might be a key player in fiber development and was selected for further study. In addition, we found that GhHB8-5A and 5D displayed similar expression patterns during fiber development, which suggested that there may be functional redundancy between them.
To further determine whether GhHB8-5D is a transcriptional activator, we transiently expressed GhHB8-5D-eGFP fusion protein in N. benthamiana leaf cells, which demonstrated that GFP signals were only detectable in the nucleus (Figures 6A–C). Additionally, various versions of GhHB8-5D, including full-length (F), both SAD- and MEKHLA-domain-truncated (ΔSM) and MEKHLA domain-truncated (ΔM), were individually inserted into the pGBKT7 vector and expressed in yeast strain AH109. Empty pGBKT7 vector was used as a negative control. The transformed yeast cells all grew well on the selective SD/-Trp medium, but only positive control and transformed yeast cells harboring full-length GhHB8-5D turned blue in color in the presence of X-gal, indicating that GhHB8-5D has transcriptional activation activity and the activation domain is located in the MEKHLA domain (Figures 6D,E).
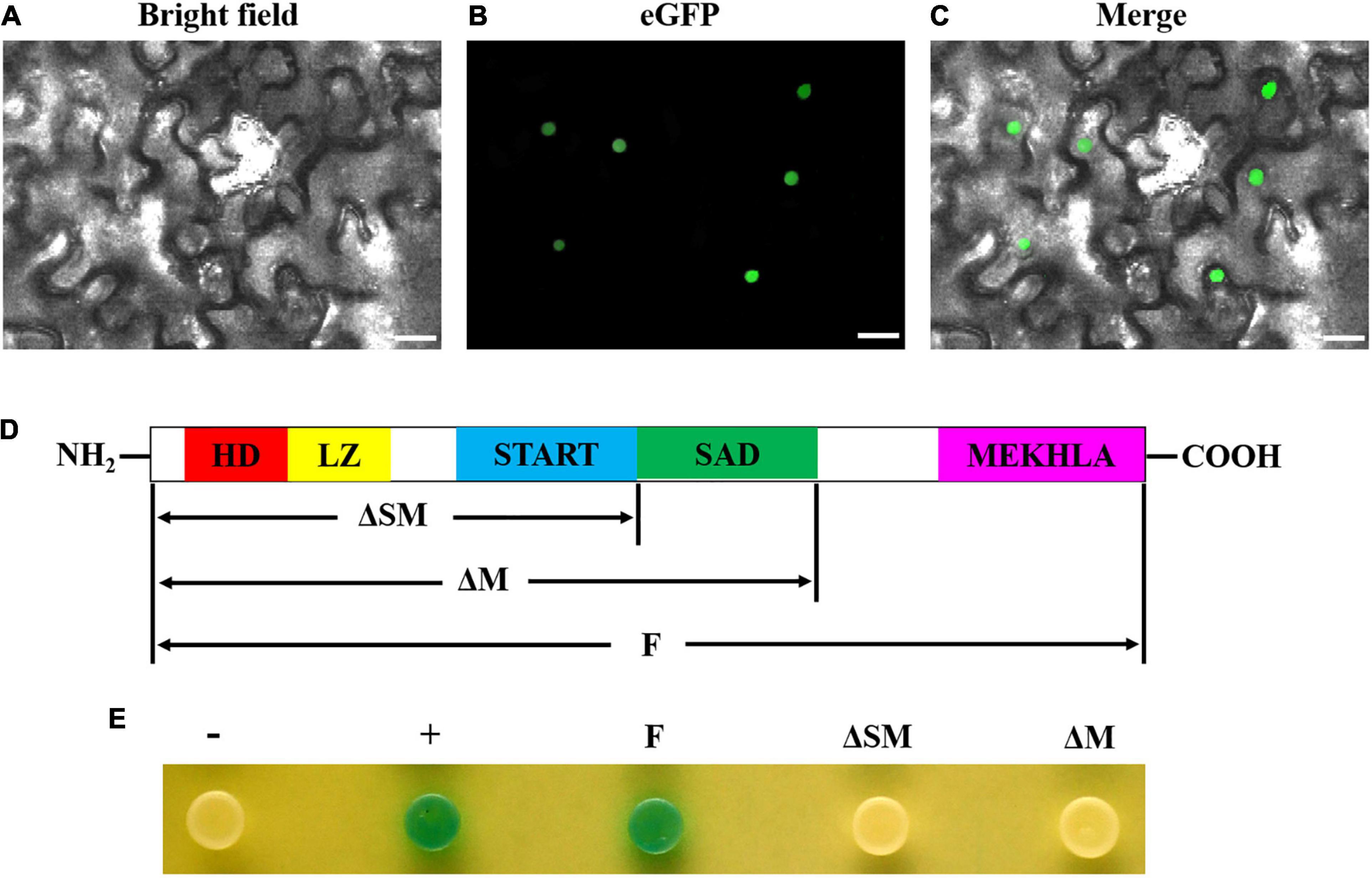
Figure 6. GhHB8-5D is a typical transcriptional activator. (A–C) The subcellular location of GhHB8-5D in epidermal cells of tobacco leaves. Bars = 50 μm. (D) Schematic diagram shows domain constructs of GhHB8-5D. (E) Assay of GhHB8-5D transcriptional activation activity in yeast. –, negative control; +, positive control; F, full-length of GhHB8-5D; ΔSM, a truncated version of GhHB8-5D lacking SAD and MEKHLA domains; ΔM, a truncated version of GhHB8-5D lacking MEKHLA domain.
To further study the physiological function of GhHB8-5D and avoid the interference of microRNA to its function, we first aligned the START domain sequences of GhHB8-5D and AtHB8, which revealed that their microRNA binding sites were identical (Supplementary Figure 3). Then, vectors overexpressing GhHB8-5D or GhHB8-5Dm (synonymous mutation of the miR binding site, Supplementary Figure 4) were constructed and used to transform Arabidopsis. Transgenic A. thaliana plants were verified by using semiquantitative RT-PCR and two independent homozygous lines overexpressing GhHB8-5D or GhHB8-5Dm were selected for further analyses (Supplementary Figure 5). Compared with the wild type, 3-week-old transgenic seedlings were obviously smaller with curly leaves (Figures 7A–E). Six-week-old transgenic plants were significantly shorter than wild type (Figures 7F–J) and their stems were twisted (Figures 7K–O). Subsequently, cross-sections of the 6-week-old stems from wild type and transgenic plants were stained by using toluidine blue, or phloroglucinol or Pontamine Fast Scarlet 4B (S4B). As shown in Figure 8, Arabidopsis plants overexpressing GhHB8-5D displayed ectopic deposition of the secondary wall in some parenchymatous cells (Figures 8B,C) compared with the wild type, in which secondary wall deposition existed only in the xylem and interfascicular fibers (Figure 8A). Staining with phloroglucinol or S4B or lignin autofluorescence revealed that GhHB8-5D overexpression caused ectopic deposition of secondary wall components, including lignin and cellulose (Figures 8G,H,L,M,Q,R). It is worthy of noting that some irregular xylem vessels appeared in the GhHB8-5D overexpression lines (Figures 8B,C,H,L,M,R). In wild type, xylems and interfascicular fibers were arranged in a ring shape. But such a regular organization was disrupted in the GhHB8-5Dm transgenic lines due to the aberrant placement of amphivasal vascular bundles. Moreover, there was no deposition of the secondary wall at the position of interfascicular fibers (Figures 8D,E,I,J,N,O,S,T). Interestingly, a few ectopic depositions of the secondary wall appeared in the epidermis and phloems of the transgenic line #9 expressing GhHB8-5Dm (Figures 8E,J,O,T).

Figure 7. The phenotype of wild type and GhHB8-5D and GhHB8-5Dm transgenic lines. (A–E) Three-week-old seedlings of wild type and transgenic lines. (F–J) Six-week-old plants of wild type and transgenic lines. (K–O) Magnified images of the inflorescence stem from 6-week-old wild type and transgenic lines. WT represent wild type plants. L6 and L7 indicate two independent lines of GhHB8-5D overexpression plants. L3 and L9 denote two independent lines of GhHB8-5Dm overexpression plants in which the miR166 binding site was executed synonymous mutation. Bars = 1 cm.
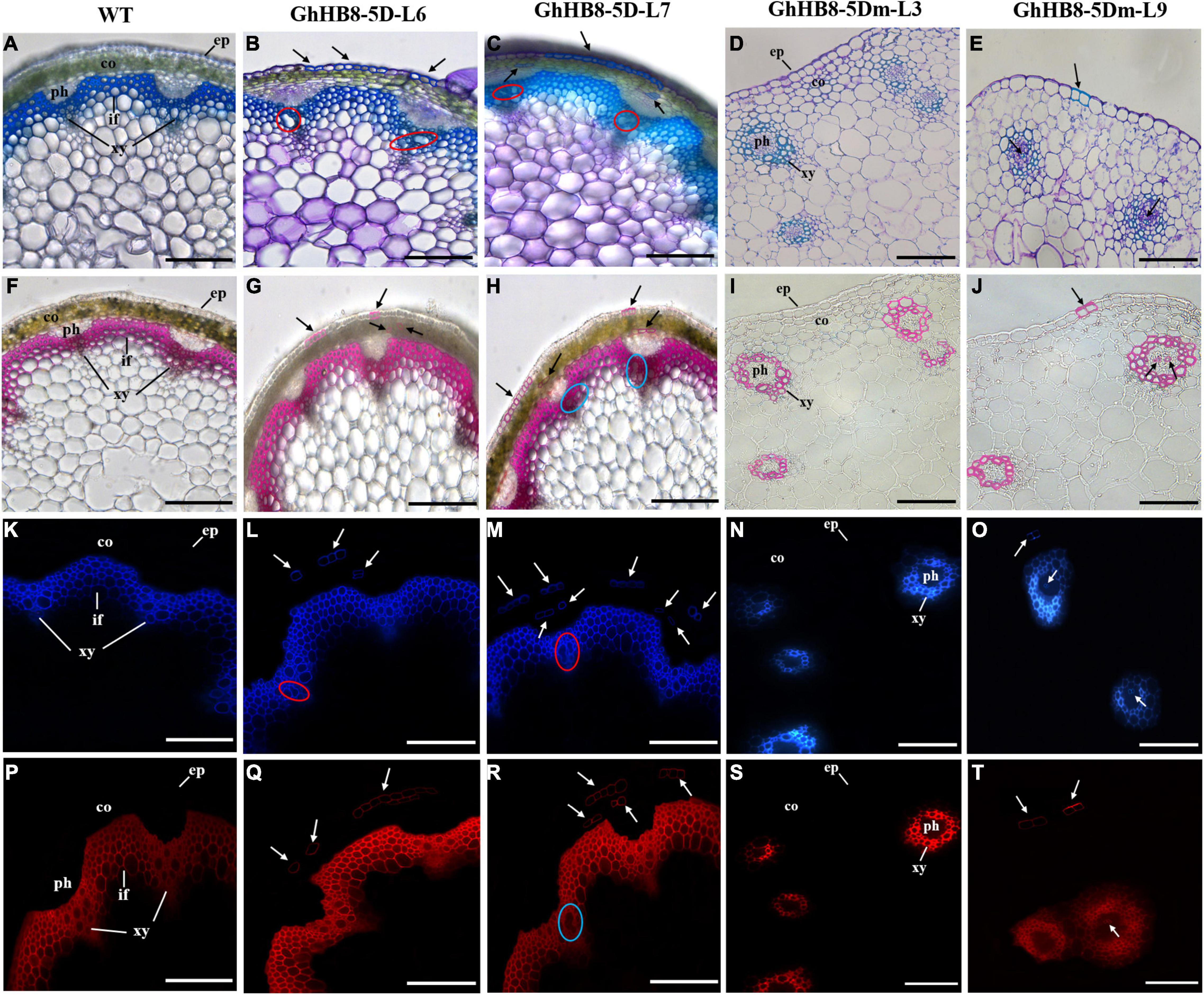
Figure 8. Cross-sections of the 6-week-old stem from wild type and transgenic lines. (A–E) Toluidine blue staining of 6-week-old stems from wild type and transgenic lines. The blue regions indicate the deposition of the secondary wall; (F–J) phloroglucin staining of 6-week-old stems from wild type and transgenic lines. The red regions indicate the deposition of lignin. (K–O) Lignin autofluorescence of 6-week-old stems from wild type and transgenic lines. Regions with blue fluorescence indicate the deposition of lignin. (P–T) Pontamine Fast Scarlet 4B (S4B) staining of 6-week-old stems from wild type and transgenic lines. Regions with red fluorescence indicate the deposition of cellulose. Black arrows show ectopic deposition of the secondary wall. Red and blue circles show irregular xylem vessels. ep, epidermis; co, cortex; ph, phloem; xy, xylem; if, interfascicular fiber. Bars = 100 μm.
To better understand the mechanism underlying the ectopic secondary wall deposition as a result of overexpressing GhHB8-5D, we investigated whether GhHB8-5D could induce the expression of secondary wall-related genes in transgenic Arabidopsis. Consistent with the ectopic deposition of secondary walls, GhHB8-5D overexpression led to upregulating the transcription level of most secondary wall biosynthetic genes significantly, except AtNST1 and AtCESA7 (Figure 9). However, overexpression of GhHB8-5Dm could repress the expression of nine secondary wall-related genes, including three master switches for the biosynthesis of the secondary wall (AtNST1, AtVND7, and AtMYB46), three cellulose synthase genes (AtCESA4, AtCESA7, and AtCESA8), and three lignin biosynthetic genes (At4CL1, AtCCoAOMT1, and AtCCR1). These results indicated that GhHB8-5D may affect the deposition of the secondary wall by regulating the expression of secondary wall-related genes.
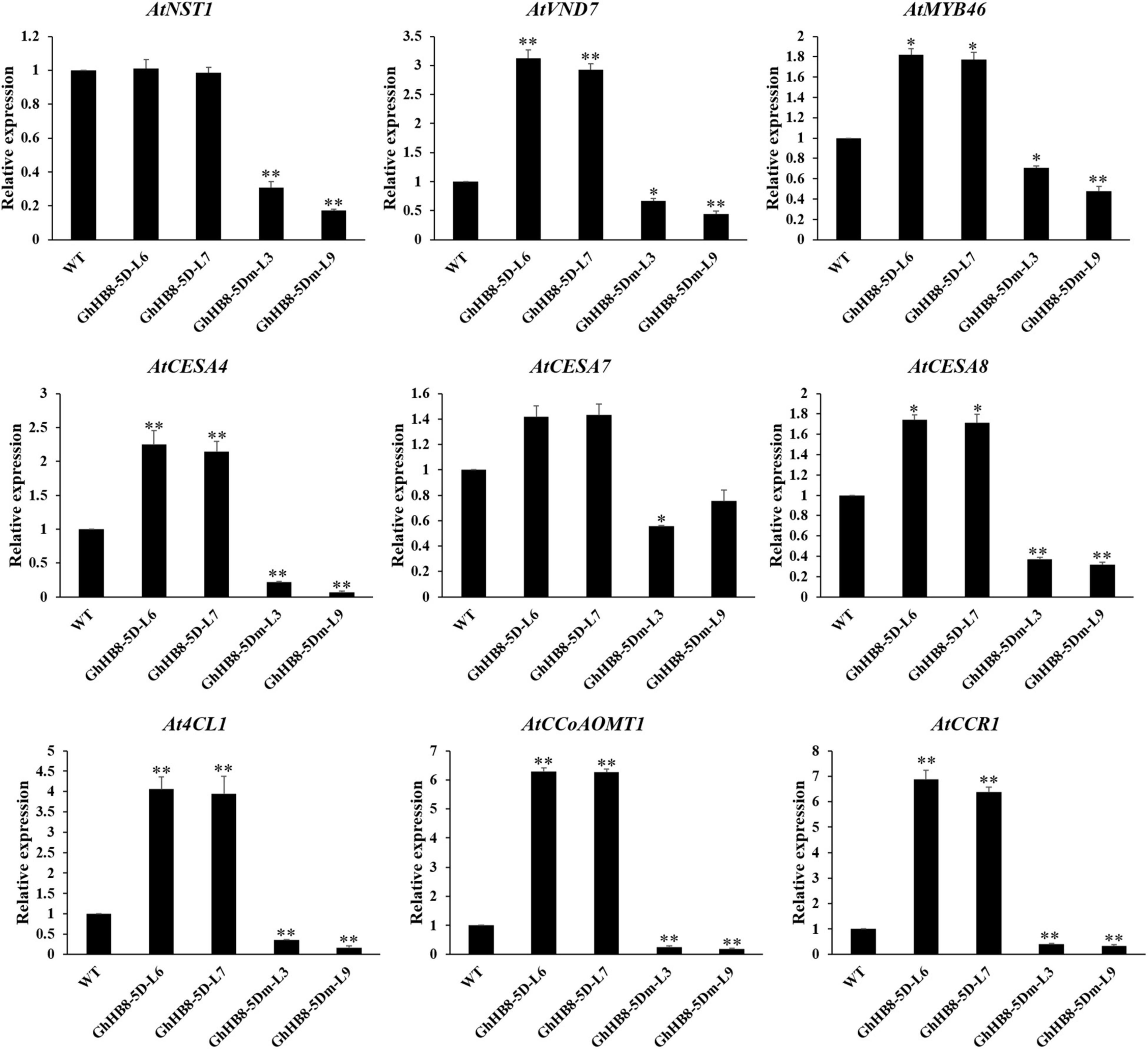
Figure 9. Expression of secondary wall-related genes in 6-week-old stems of wild type and transgenic lines. The AtActin2 was used as an internal control for normalization. Values represent the mean ± SD of three biological replicates. Student’s t-tests demonstrated that there were significant differences (*p < 0.05, **P < 0.01) between the transgenic lines and the wild type.
As the Arabidopsis HD-ZIP III genes were also involved in the morphogenesis of the vascular system, would the overexpressing GhHB8-5D influence the xylem morphology by interfering with the expressions of HD-ZIP III genes in Arabidopsis? In order to answer such a question, we examined the expression levels of Arabidopsis HD-ZIP III genes in wild type and transgenic lines. As shown in Supplementary Figure 6, GhHB8-5D overexpression led to downregulating the transcription level of AtHB8 and AtPHV but upregulating the transcription level of AtREV. Overexpression of GhHB8-5Dm could induce the expression of AtPHB and AtPHV but represses the expression of AtHB8, AtCNA, and AtREV. These results implied that GhHB8-5D may influence the xylem morphology by interfering with the expression of Arabidopsis HD-ZIP III genes.
It has been reported that the members of the HD-ZIP III subfamily are associated with water utilization efficiency and abiotic stresses (Xie et al., 2015; Ramachandran et al., 2018; Zhang et al., 2018b). In this study, we have identified a drought-induced cis-element in the promoter region of GhHB8-5D (Figure 2). To investigate whether GhHB8-5D is related to drought tolerance, we observed seed germination and counted the rate of green seedlings on MS medium supplemented with different concentrations of mannitol to mimic the drought stress. GhHB8-5D and GhHB8-5Dm overexpression seeds showed normal germination as did wild type seeds under normal conditions. The green seedling rate of both wild type and transgenic lines reached almost 100% after 4 days (Figures 10A,G). When sowing on MS medium containing 50 mM mannitol, green seedling rate of GhHB8-5D overexpression lines was slightly higher compared with that of wild type, but the rate of GhHB8-5Dm overexpression lines was significantly higher than that of wild type (Figures 10B,H). In the presence of 100 mM mannitol, the green seedling rate was 71.1%, but it was significantly raised in the range of 84.4–87.8% in the transgenic lines (Figures 10C,I). Similarly, four transgenic lines showed an obviously higher green seedling rate on MS medium with 150 mM and 200 mM mannitol, compared with that of wild type in the same conditions (Figures 10D,E,J,K).

Figure 10. Assay of the green seedling rate of wile type and transgenic lines overexpressing GhHB8-5D and GhHB8-5Dm under mannitol treatment. (A–E) Postgermination green seedling analysis of the wild type and transgenic lines overexpressing GhHB8-5D and GhHB8-5Dm grown on MS medium supplemented with 0, 50, 100, 150, and 200 mM mannitol for 4 days. (F) Schematic diagram of wild type and transgenic seedlings distribution on the MS medium. (G–K) Statistical analysis of the green seedling rate of wild type and transgenic seedlings grown on MS medium containing 0, 50, 100, 150, and 200 mM mannitol for 4 days. Mean values and SD are shown from three biological replicates. Independent t-tests demonstrate that there are significant (p < 0.05) or very significant (p < 0.01) differences in green seedling rate between wild type and transgenic lines. *means significant differences (p < 0.05); **means very significant differences (p < 0.01).
In addition, the seeds of wild type and transgenic lines were sowed on MS medium supplemented with 0, 50, 100, 150, and 200 mM mannitol for 7 days prior to the measurement of the primary root lengths. As shown in Figures 11A,F, there was no statistically meaningful difference in the primary root length between wild type and transgenic lines without mannitol treatment. In contrast, in the presence of 50 mM mannitol, the primary root length of transgenic lines overexpressing GhHB8-5Dm was significantly shorter than that of wild type (Figures 11B,G). With the increase of mannitol concentration (100, 150, and 200 mM), root growth of both wild type and transgenic lines was suppressed dramatically, and the primary root length of the wild type was remarkably longer than that of transgenic lines (Figures 11C–E,H–J). These results indicated that GhHB8-5D may be induced by drought and involved in drought resistance.
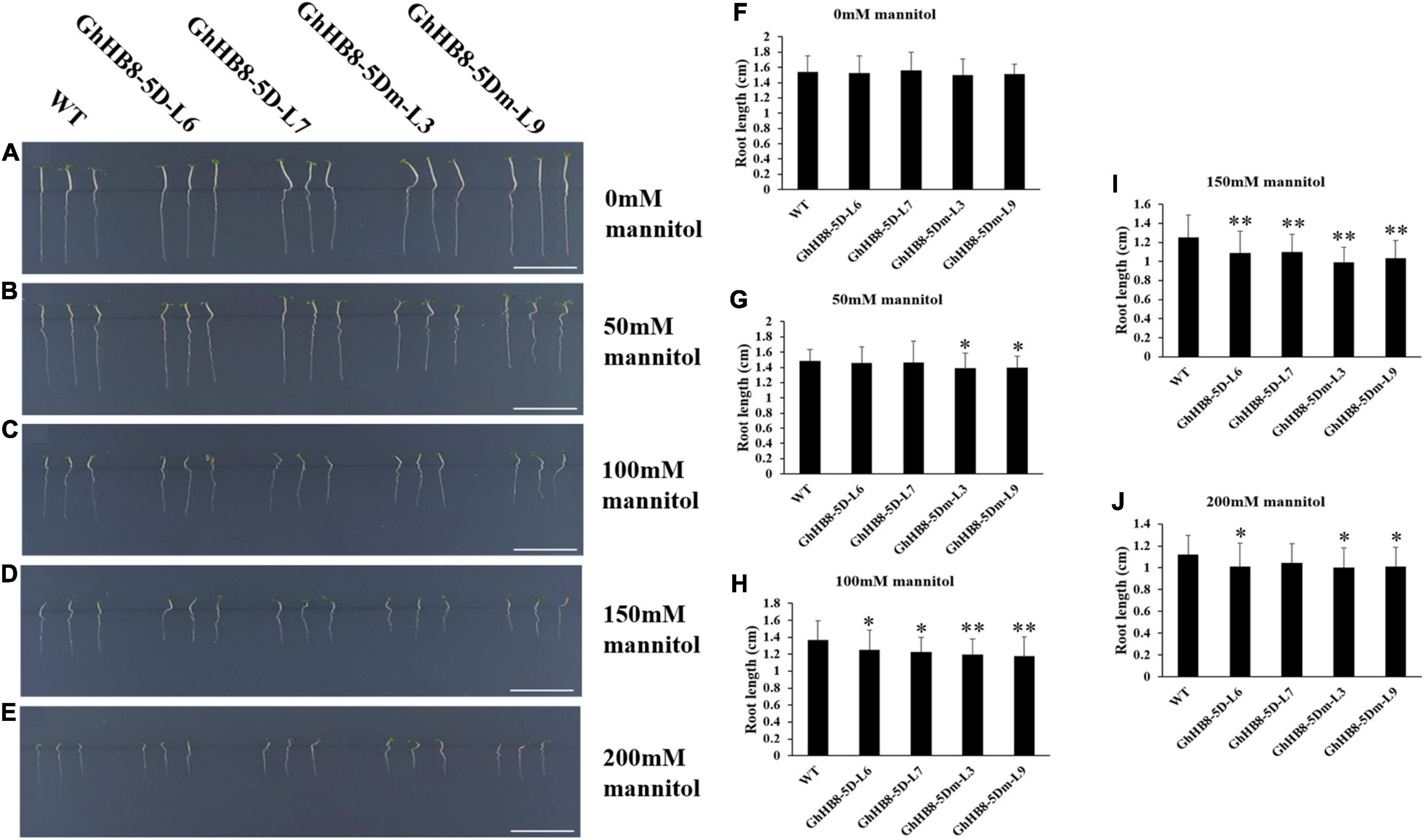
Figure 11. Analysis of root length of wild type and transgenic lines overexpressing GhHB8-5D and GhHB8-5Dm under mannitol treatment. (A–E) The phenotype of wild type and transgenic seedlings under different concentrations of mannitol treatment for 7 days. Bars = 1 cm. (F–J) Statistical analysis of root length of wild type and transgenic seedlings under different concentrations of mannitol treatment for 7 days. Mean values and SD are shown from three biological replicates. Independent t-tests demonstrate that there are significant (p < 0.05) or very significant (p < 0.01) differences in root length between wild type and transgenic lines. *means significant differences (p < 0.05); **means very significant differences (p < 0.01).
HD-ZIP III belongs to a plant-specific and highly conserved protein subfamily, the members of which play vital roles in plant differentiation and development. Although HD-ZIP III has been studied in several plants (Chai et al., 2018; Li et al., 2019; Sharif et al., 2020), this study represents the first comprehensive investigation on this subfamily of genes in cotton. In this study, we identified 18 HD-ZIP III genes in the tetraploid upland cotton genome, which could be divided into four distinct groups. HD-ZIP III and HD-ZIP IV subfamilies share a similar domain arrangement but HD-ZIP IV lacks the MEKHLA domain, suggesting their common origin and relatively recent divergence from a common lineage (Schrick et al., 2004). However, HD-ZIP I and HD-ZIP II only have the HD and LZ domains with spacing different from those of HD-ZIP III and HD-ZIP IV, indicating that their juxtaposition may have evolved independently (Sessa et al., 1998).
The result of the cis-element analysis showed that the members of cotton HD-ZIP III may be involved in phytohormone response, abiotic stresses tolerance, phenylpropane metabolism, and plant differentiation and development (Figure 2). It has been reported that phytohormones can regulate the expression of NAC and MYB TFs, which enables them to play a role in secondary wall formation. The application of cytokinin was shown to inhibit the expression of VND6 and VND7, whereas auxin inhibited VND6 alone. However, when both hormones were applied together, they promoted the expression of both VNDs. Similarly, complex effects were obtained with the combination of auxin, cytokinin, and brassinolide. The application of brassinolide alone upregulated VND6, while brassinolide in combination with auxin had no effect on the expression of VNDs. The combination of cytokinin and brassinolide caused the transcriptional repression of VND6 and VND7, whereas a mixture of auxin, cytokinin, and brassinolide promoted the expression of these VNDs (Kubo et al., 2005). The auxin treatment was also shown to downregulate MYB26 expression and suppress precocious lignification, as demonstrated by the afb1/myb26 double mutant, which failed to show lignification, manifesting that auxin may act as a negative regulator of lignification via the downregulation of MYB26 (Cecchetti et al., 2013). In addition, abiotic stresses have been found to be responsible for increasing secondary wall substances, especially lignin (Lee et al., 2007), which not only provide terrestrial plants with rigidity against compressive forces but also form a mechanical barrier against drought stress (Moura et al., 2010). The aromatic properties of lignin make the secondary wall impermeable to water, which reduces transpiration and assists with maintaining normal turgor pressures under drought stress (Yao et al., 2021). Therefore, it is conceivable that lignification represents an initial form of protection against drought stress. Several studies have shown that the process of lignification is important for drought tolerance. Overexpression of IbLEA14 in sweet potato (Park et al., 2011), OsTF1L in rice (Bang et al., 2019), VlbZIP30 in grapevine (Tu et al., 2020), and PoCCoAOMT in tobacco (Zhao et al., 2021) all enhanced lignin biosynthesis and drought resistance. In maize, drought-tolerant inbred lines showed higher lignification than drought-sensitive lines, suggesting that lignification is an important adaptation to drought stress (Hu et al., 2009).
Gene duplication is a primary source of “raw material” for evolutionary innovations because redundant paralogs have fewer selective constrains and are ready to evolve new functions than non-redundant genes (Prigge and Clark, 2006). Collinearity analysis of the HD-ZIP III subfamily revealed that all HD-ZIP III members have undergone segmental duplication (Figure 3). This may be the reason for functional redundancy and diversity among HD-ZIP III proteins. Phylogenetic analysis using HD-ZIP III homologs from diverse plant species identified three divergent clades that occurred during plant evolution and only one monocot HD-ZIP III gene (OsHB3) appeared in the HB8/HB15 clade (Figure 4). The three clades of HD-ZIP III members were consistent with the analysis of monocot- and eudicot-derived genes indicating that the genome of the ancestral angiosperm plant contained REV, HB14, and HB8/HB15 paralogs. The relationship of monocot HD-ZIP III to the eudicot HB8/HB15 clade suggested that the HB8/HB15 gene duplication event may have taken place after the monocot-eudicot split (Prigge and Clark, 2006). Additionally, most homologs of cotton HD-ZIP IIIs were related to xylem differentiation and secondary wall formation in their respective species (Ohashi-Ito et al., 2002; Ko et al., 2006; Zhu et al., 2013; Du et al., 2015; Zhang et al., 2018b; Chen et al., 2021). Considering the conservation of this subfamily in different plants, we inferred that the genes of the same subfamily in cotton (Gossypium hirsutum) may have similar functions.
GhHB8-5 showed D-subgenome biased expression, possibly due to the promoter difference, as we compared the promoter sequences of GhHB8-5A and 5D and identified variations in their cis-elements (Supplementary Table 4). In addition, the GhmiR166 binding site can be found in the START domain of both GhHB8-5A and 5D. The different cleavage efficiency of GhmiR166 to GhHB8-5A and 5D may also lead to their differential expression.
Specific to the influence on the vasculature and cell wall, the HD-ZIP III subfamily can not only change the vascular patterning and organization but also affect the xylem differentiation, which leads to abnormal secondary wall deposition. Overexpression of AtHB8 in Arabidopsis caused rolled-up leaves, a strong reduction of inflorescence stem elongation, and shorter plant height. The anatomical structure of transgenic plant stem showed an increase in the production of phloem fiber sclereids and the lignified tissues (Baima et al., 2001), which were similar to the phenotype of ectopic expression of GhHB8-5D in Arabidopsis, as illustrated in this study (Figures 7, 8). Furthermore, several genes that are known to play significant roles during xylem differentiation and secondary wall formation were upregulated as a result of PtrHB7 (homolog of AtHB8) overexpression in Populus and Arabidopsis (Zhu et al., 2013). Similarly, ectopic expression of GhHB8-5D in Arabidopsis also increased the transcription levels of secondary wall-related TFs, cellulose synthase genes, and lignin biosynthetic genes (Figure 9). Taken together, our results indicated that GhHB8-5D is a positive regulator of xylem differentiation and secondary wall biosynthesis.
However, with the emergence of new reports, the idea that AtHB8 is a negative regulator of xylem differentiation and secondary wall formation was supported by empirical observation. In the process of vascular bundles formation, REV played a role in promoting xylem differentiation and secondary wall deposition. While xylem vessels were present in rev mutants, xylary fibers were reduced in weaker alleles and absent in strong alleles. This loss of fibers resulted in large reductions of the secondary wall (Zhong et al., 1997; Zhong and Ye, 1999). Genetic analysis has also supported the above conclusion. REV can bind to the promoter of VND7, a master regulator for xylem and secondary wall formation and promote its activity (Endo et al., 2015). Studies on the genetic redundancy between REV and the other HD-ZIP III TFs showed that phb and phv are strong enhancers of the rev phenotype in the xylem. In contrast, the defects of rev mutant were restored in anth8 cna rev triple mutants. The overexpression of AtHB8 and CNA driven by REV promoter in the rev mutant background was not able to rescue the phenotypic defects of rev. Taken together, these observations suggested that PHB and PHV genes performed overlapping functions with REV, but AtHB8 and CNA played roles antagonistic to REV in xylem differentiation (Prigge et al., 2005). In addition, the phenotypes of gain-of-function phb-1d and phv-1d showed amphivasal vascular bundles with xylem surrounding phloem (McConnell et al., 2001). In transgenic Arabidopsis lines overexpressing GhHB8-5Dm, the expression levels of AtPHB and AtPHV were significantly higher than those in wild type (Supplementary Figure 6). This may explain why amphivasal vascular bundles appeared in these transgenic plants (Figure 8). Under such phenotypic changes, there must be a sophisticated regulatory network. ACL5 (ACAULIS 5) is a gene encoding a thermospermine synthase, which has been shown to regulate xylem differentiation and secondary wall deposition in a negative manner (Muñiz et al., 2008). AtHB8 acts together with auxin as a direct positive regulator of ACL5, which slows xylem differentiation and secondary wall formation, in part by negative regulation of REV (Baima et al., 2014). This may explain the altered expression patterns of those genes involved in secondary wall-related genes, as a result of overexpressing GhHB8-5D and GhHB8-5Dm in Arabidopsis (Figure 9).
Recent reports have revealed that the HD-ZIP III subfamily participated in the regulation of response to drought stress. Rice plants overexpressing OsHB4, a member of the HD-ZIP III subfamily and a major target of miR166, resembled the phenotypes of the miR166 knockdown plants showing enhanced drought resistance (Zhang et al., 2018b). The suppression of miR166 was found to be coincidental with the upregulation of HD-ZIP III as a result of drought treatment (Xie et al., 2015). Further, water-limiting conditions caused major changes to the root xylem morphology (Ramachandran et al., 2018). Similar changes have also been observed in poplar and soybean, where drought stress resulted in an increase in vessel number and vessel wall thickness, thereby conferring resistance to water deficiency (Awad et al., 2010; Prince et al., 2017). Using the combination of mannitol treatment and statistical analysis, we showed that transgenic Arabidopsis overexpressing GhHB8-5D and GhHB8-5Dm had a higher green seedling rate but shorter root length compared with wild type (Figures 10, 11). GhHB8-5D can affect xylem morphology and secondary wall biosynthesis and could, therefore, be responsible for the improvement in drought resistance and alteration in root length in transgenic plants.
In this study, we comprehensively analyzed the conservative domains, cis-elements contained in the promoter regions, location and duplication, evolutionary relationship, and expression of HD-ZIP III subfamily in cotton. Furthermore, we established GhHB8-5D as a transcriptional activator and its C-terminus as the transactivation domain. GhHB8-5D could be involved in drought resistance by influencing xylem morphology and secondary wall biosynthesis. Our findings may provide a novel strategy for improving plant adaptations to environmental changes via regulating plant cell wall synthesis.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
WX and FL designed the experiments. JZ, YG, and MF performed the experiments. JZ, YC, SL, and LL analyzed the data. YW prepared the plant materials. JZ, WX, and FL wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (Grant Nos. 32170579 and 31671735) and China Postdoctoral Science Foundation (Grant No. 2019M660890).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.806195/full#supplementary-material
Agalou, A., Purwantomo, S., Overnäs, E., Johannesson, H., Zhu, X., Estiati, A., et al. (2008). A genome-wide survey of HD-Zip genes in rice and analysis of drought-responsive family members. Plant Mol. Biol. 66, 87–103. doi: 10.1007/s11103-007-9255-7
Ariel, F. D., Manavella, P. A., Dezar, C. A., and Chan, R. L. (2007). The true story of the HD-ZIP family. Trends Plant. Sci. 12, 419–426. doi: 10.1016/j.tplants.2007.08.003
Awad, H., Barigah, T., Badel, E., Cochard, H., and Herbette, S. (2010). Poplar vulnerability to xylem cavitation acclimates to drier soil conditions. Physiol. Plantarum. 139, 280–288. doi: 10.1111/j.1399-3054.2010.01367.x
Baima, S., Forte, V., Possenti, M., Peñalosa, A., Leoni, G., Salvi, S., et al. (2014). Negative feedback regulation of auxin signaling by ATHB8/ACL5-BUD2 transcription module. Mol. Plant 7, 1006–1025. doi: 10.1093/mp/ssu051
Baima, S., Nobili, F., Sessa, G., Lucchetti, S., Ruberti, I., and Morelli, G. (1995). The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121, 4171–4182. doi: 10.1111/j.1365-2303.1995.tb00491.x
Baima, S., Possenti, M., Matteucci, A., Wisman, E., Altamura, M. M., Ruberti, I., et al. (2001). The Arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol. 126, 643–655. doi: 10.1104/pp.126.2.643
Bang, S. W., Lee, D. K., Jung, H., Chung, P. J., Kim, Y. S., Choi, Y. D., et al. (2019). Overexpression of OsTF1L, a rice HD-Zip transcription factor, promotes lignin biosynthesis and stomatal closure that improves drought tolerance. Plant Biotechnol. J. 17, 118–131. doi: 10.1111/pbi.12951
Cao, J. F., Zhao, B., Huang, C. C., Chen, Z. W., Zhao, T., Liu, H. R., et al. (2020). The miR319-targeted GhTCP4 promotes the transition from cell elongation to wall thickening in cotton fiber. Mol. Plant. 13, 1063–1077. doi: 10.1016/j.molp.2020.05.006
Carlsbecker, A., Lee, J. Y., Roberts, C. J., Dettmer, J., Lehesranta, S., Zhou, J., et al. (2010). Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465, 316–321. doi: 10.1038/nature08977
Cecchetti, V., Altamura, M. M., Brunetti, P., Petrocelli, V., Falasca, G., Ljung, K., et al. (2013). Auxin controls Arabidopsis anther dehiscence by regulating endothecium lignification and jasmonic acid biosynthesis. Plant J. 74, 411–422. doi: 10.1111/tpj.12130
Chai, W., Si, W., Ji, W., Qin, Q., Zhao, M., and Jiang, H. (2018). Genome-wide investigation and expression profiling of HD-Zip transcription factors in Foxtail millet (Setaria italica L.). Biomed. Res. Int. 2018:8457614. doi: 10.1155/2018/8457614
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, H., Fang, R., Deng, R., and Li, J. (2021). The OsmiRNA166b-OsHox32 pair regulates mechanical strength of rice plants by modulating cell wall biosynthesis. Plant Biotechnol. J. 19, 1468–1480. doi: 10.1111/pbi.13565
Du, J., Miura, E., Robischon, M., Martinez, C., and Groover, A. (2011). The Populus Class III HD ZIP transcription factor POPCORONA affects cell differentiation during secondary growth of woody stems. PLoS One 6:e17458. doi: 10.1371/journal.pone.0017458
Du, Q., Avci, U., Li, S., Gallego-Giraldo, L., Pattathil, S., Qi, L., et al. (2015). Activation of miR165b represses AtHB15 expression and induces pith secondary wall development in Arabidopsis. Plant J. 83, 388–400. doi: 10.1111/tpj.12897
Emery, J. F., Floyd, S. K., Alvarez, J., Eshed, Y., Hawker, N. P., Izhaki, A., et al. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13, 1768–1774. doi: 10.1016/j.cub.2003.09.035
Endo, H., Yamaguchi, M., Tamura, T., Nakano, Y., Nishikubo, N., Yoneda, A., et al. (2015). Multiple classes of transcription factors regulate the expression of VASCULAR-RELATED NAC-DOMAIN7, a master switch of xylem vessel differentiation. Plant Cell Physiol. 56, 242–254. doi: 10.1093/pcp/pcu134
Haigler, C. H., Betancur, L., Stiff, M. R., and Tuttle, J. R. (2012). Cotton fiber: a powerful single-cell model for cell wall and cellulose research. Front. Plant Sci. 3:104. doi: 10.3389/fpls.2012.00104
Hernandez-Gomez, M. C., Runavot, J. L., Guo, X., Bourot, S., Benians, T. A., Willats, W. G., et al. (2015). Heteromannan and heteroxylan cell wall polysaccharides display different dynamics during the elongation and secondary cell wall deposition phases of cotton fiber cell development. Plant Cell Physiol. 56, 1786–1797. doi: 10.1093/pcp/pcv101
Hu, Y., Li, W. C., Xu, Y. Q., Li, G. J., Liao, Y., and Fu, F. L. (2009). Differential expression of candidate genes for lignin biosynthesis under drought stress in maize leaves. J. Appl. Genet. 50, 213–223. doi: 10.1007/BF03195675
Huang, J., Chen, F., Guo, Y., Gan, X., Yang, M., Zeng, W., et al. (2021). GhMYB7 promotes secondary wall cellulose deposition in cotton fibres by regulating GhCesA gene expression through three distinct cis-elements. New Phytol. 232, 1718–1737. doi: 10.1111/nph.17612
Huang, J., Guo, Y., Sun, Q., Zeng, W., Li, J., Li, X., et al. (2019). Genome-wide identification of R2R3-MYB transcription factors regulating secondary cell wall thickening in cotton fiber development. Plant Cell Physiol. 60, 687–701. doi: 10.1093/pcp/pcy238
Kidner, C. A., and Martienssen, R. A. (2004). Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428, 81–84. doi: 10.1038/nature02366
Kim, J., Jung, J. H., Reyes, J. L., Kim, Y. S., Kim, S. Y., Chung, K. S., et al. (2005). microRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J. 42, 84–94. doi: 10.1111/j.1365-313X.2005.02354.x
Ko, J. H., Prassinos, C., and Han, K. H. (2006). Developmental and seasonal expression of PtaHB1, a Populus gene encoding a class III HD-Zip protein, is closely associated with secondary growth and inversely correlated with the level of microRNA (miR166). New Phytol. 169, 469–478. doi: 10.1111/j.1469-8137.2005.01623.x
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kubo, M., Udagawa, M., Nishikubo, N., Horiguchi, G., Yamaguchi, M., Ito, J., et al. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19, 1855–1860. doi: 10.1101/gad.1331305
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lee, B. R., Kim, K. Y., Jung, W. J., Avice, J. C., Ourry, A., and Kim, T. H. (2007). Peroxidases and lignification in relation to the intensity of water-deficit stress in white clover (Trifolium repens L.). J. Exp. Bot. 58, 1271–1279. doi: 10.1093/jxb/erl280
Li, F., Fan, G., Lu, C., Xiao, G., Zou, C., Kohel, R. J., et al. (2015). Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 33, 524–530. doi: 10.1038/nbt.3208
Li, L., Zheng, T., Zhuo, X., Li, S., Qiu, L., Wang, J., et al. (2019). Genome-wide identification, characterization and expression analysis of the HD-Zip gene family in the stem development of the woody plant Prunus mume. PeerJ 7:e7499. doi: 10.7717/peerj.7499
Li, Y. Y., Shen, A., Xiong, W., Sun, Q. L., Luo, Q., Song, T., et al. (2016). Overexpression of OsHox32 results in pleiotropic effects on plant type architecture and leaf development in rice. Rice 9:46. doi: 10.1186/s12284-016-0118-1
Liu, Y., You, S., Taylor-Teeples, M., Li, W. L., Schuetz, M., Brady, S. M., et al. (2014). Bel1-like homeodomain6 and knotted arabidopsis thaliana7 interact and regulate secondary cell wall formation via repression of Revoluta. Plant Cell 26, 4843–4861. doi: 10.1105/tpc.114.128322
McConnell, J. R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M. K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713. doi: 10.1038/35079635
McFarlane, H. E., Döring, A., and Persson, S. (2014). The cell biology of cellulose synthesis. Annu. Rev. Plant Biol. 65, 69–94. doi: 10.1146/annurev-arplant-050213-040240
Moura, J. C., Bonine, C. A., de Oliveira Fernandes Viana, J., Dornelas, M. C., and Mazzafera, P. (2010). Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 52, 360–376. doi: 10.1111/j.1744-7909.2010.00892.x
Mukherjee, K., and Burglin, T. R. (2006). MEKHLA, a novel domain with similarity to PAS domains, is fused to plant homeodomain-leucine zipper III proteins. Plant Physiol. 140, 1142–1150. doi: 10.1104/pp.105.073833
Muñiz, L., Minguet, E. G., Singh, S. K., Pesquet, E., Vera-Sirera, F., Moreau-Courtois, C. L., et al. (2008). ACAULIS5 controls Arabidopsis xylem specification through the prevention of premature cell death. Development 135, 2573–2582. doi: 10.1242/dev.019349
Ohashi-Ito, K., Demura, T., and Fukuda, H. (2002). Promotion of transcript accumulation of novel Zinnia immature xylem-specific HD-Zip III homeobox genes by brassinosteroids. Plant Cell Physiol. 43, 1146–1153. doi: 10.1093/pcp/pcf135
Park, S. C., Kim, Y. H., Jeong, J. C., Kim, C. Y., Lee, H. S., Bang, J. W., et al. (2011). Sweetpotato late embryogenesis abundant 14 (IbLEA14) gene influences lignification and increases osmotic- and salt stress-tolerance of transgenic calli. Planta 233, 621–634. doi: 10.1007/s00425-010-1326-3
Paterson, A. H., Wendel, J. F., Gundlach, H., Guo, H., Jenkins, J., Jin, D., et al. (2012). Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427. doi: 10.1038/nature11798
Prigge, M. J., and Clark, S. E. (2006). Evolution of the class III HD-Zip gene family in land plants. Evol. Dev. 8, 350–361. doi: 10.1111/j.1525-142X.2006.00107.x
Prigge, M. J., Otsuga, D., Alonso, J. M., Ecker, J. R., Drews, G. N., and Clark, S. E. (2005). Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell. 17, 61–76. doi: 10.1105/tpc.104.026161
Prince, S. J., Murphy, M., Mutava, R. N., Durnell, L. A., and Nguyen, H. T. (2017). Root xylem plasticity to improve water use and yield in water-stressed soybean. J. Exp. Bot. 68, 2027–2036. doi: 10.1093/jxb/erw472
Ramachandran, P., Wang, G., Augstein, F., Vries, J. D., and Carlsbecker, A. (2018). Continuous root xylem formation and vascular acclimation to water deficit involves endodermal ABA signalling via miR165. Development 145:dev159202. doi: 10.1242/dev.159202
Robischon, M., Du, J., Miura, E., and Groover, A. (2011). The Populus Class III HD ZIP, popREVOLUTA, influences cambium initiation and patterning of woody stems. Plant Physiol. 155, 1214–1225. doi: 10.1104/pp.110.167007
Schena, M., and Davis, R. W. (1992). HD-Zip proteins: members of an Arabidopsis homeodomain protein superfamily. Proc. Natl. Acad. Sci. USA 89, 3894–3898. doi: 10.1073/pnas.89.9.3894
Schrick, K., Nguyen, D., Karlowski, W. M., and Mayer, K. F. (2004). START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol. 5:R41. doi: 10.1186/gb-2004-5-6-r41
Sessa, G., Steindler, C., Morelli, G., and Ruberti, I. (1998). The Arabidopsis Athb-8, -9 and -14 genes are members of a small gene family coding for highly related HD-ZIP proteins. Plant Mol. Biol. 38, 609–622. doi: 10.1023/a:1006016319613
Sharif, R., Xie, C., Wang, J., Cao, Z., Zhang, H., Chen, P., et al. (2020). Genome wide identification, characterization and expression analysis of HD-ZIP gene family in Cucumis sativus L. under biotic and various abiotic stresses. Int. J. Biol. Macromol. 158, 502–520. doi: 10.1016/j.ijbiomac.2020.04.124
Sparkes, I. A., Runions, J., Kearns, A., and Hawes, C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025. doi: 10.1038/nprot.2006.286
Taylor-Teeples, M., Lin, L., De Lucas, M., Turco, G., Toal, T. W., Gaudinier, A., et al. (2015). An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517, 571–575. doi: 10.1038/nature14099
Tu, M., Wang, X., Yin, W., Wang, Y., Li, Y., Zhang, G., et al. (2020). Grapevine VlbZIP30 improves drought resistance by directly activating VvNAC17 and promoting lignin biosynthesis through the regulation of three peroxidase genes. Hortic. Res. 7:150. doi: 10.1038/s41438-020-00372-3
Wang, T. T., Yu, T. F., Fu, J. D., Su, H. G., Chen, J., Zhou, Y. B., et al. (2020). Genome-wide analysis of the GRAS gene family and functional identification of GmGRAS37 in drought and salt tolerance. Front Plant Sci. 11:604690. doi: 10.3389/fpls.2020.604690
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Xie, F., Wang, Q., Sun, R., and Zhang, B. (2015). Deep sequencing reveals important roles of microRNAs in response to drought and salinity stress in cotton. J. Exp. Bot. 66, 789–804. doi: 10.1093/jxb/eru437
Xu, W. L., Zhang, D. J., Wu, Y. F., Qin, L. X., Huang, G. Q., Li, J., et al. (2013). Cotton PRP5 gene encoding a proline-rich protein is involved in fiber development. Plant Mol. Biol. 82, 353–365. doi: 10.1007/s11103-013-0066-8
Yao, T., Feng, K., Xie, M., Barros, J., Tschaplinski, T. J., and Tuskan, G. A. (2021). Phylogenetic occurrence of the phenylpropanoid pathway and lignin biosynthesis in plants. Front. Plant Sci. 12:704697. doi: 10.3389/fpls.2021.704697
Zhang, J., Huang, G. Q., Zou, D., Yan, J. Q., Li, Y., Hu, S., et al. (2018a). The cotton (Gossypium hirsutum) NAC transcription factor (FSN1) as a positive regulator participates in controlling secondary cell wall biosynthesis and modification of fibers. New Phytol. 217, 625–640. doi: 10.1111/nph.14864
Zhang, J., Zhang, H., Srivastava, A. K., Pan, Y., Bai, J., Fang, J., et al. (2018b). Knockdown of rice microRNA166 confers drought resistance by causing leaf rolling and altering stem xylem development. Plant Physiol. 176, 2082–2094. doi: 10.1104/pp.17.01432
Zhang, J. B., Wang, X. P., Wang, Y. C., Chen, Y. H., Luo, J. W., Li, D. D., et al. (2020). Genome-wide identification and functional characterization of cotton (Gossypium hirsutum) MAPKKK gene family in response to drought stress. BMC Plant Biol. 20:217. doi: 10.1186/s12870-020-02431-2
Zhang, X., Henriques, R., Lin, S. S., Niu, Q. W., and Chua, N. H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. doi: 10.1038/nprot.2006.97
Zhao, D., Luan, Y., Shi, W., Zhang, X., Meng, J., and Tao, J. (2021). A Paeonia ostii caffeoyl-CoA O-methyltransferase confers drought stress tolerance by promoting lignin synthesis and ROS scavenging. Plant Sci. 303:110765. doi: 10.1016/j.plantsci.2020.110765
Zhong, R., Taylor, J. J., and Ye, Z. H. (1997). Disruption of interfascicular fiber differentiation in an Arabidopsis mutant. Plant Cell. 9, 2159–2170. doi: 10.1105/tpc.9.12.2159
Zhong, R., and Ye, Z. H. (1999). IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell 11, 2139–2152. doi: 10.1105/tpc.11.11.2139
Zhong, R., and Ye, Z. H. (2015). Secondary cell walls: biosynthesis, pattern deposition and transcriptional regulation. Plant Cell Physiol. 56, 195–214. doi: 10.1093/pcp/pcu140
Keywords: cotton (Gossypium hirsutum), HD-ZIP III, ectopic expression, secondary wall, drought resistance
Citation: Zhang J, Gao Y, Feng M, Cui Y, Li S, Liu L, Wang Y, Xu W and Li F (2022) Genome-Wide Identification of the HD-ZIP III Subfamily in Upland Cotton Reveals the Involvement of GhHB8-5D in the Biosynthesis of Secondary Wall in Fiber and Drought Resistance. Front. Plant Sci. 12:806195. doi: 10.3389/fpls.2021.806195
Received: 31 October 2021; Accepted: 20 December 2021;
Published: 27 January 2022.
Edited by:
Bhumi Nath Tripathi, Indira Gandhi National Tribal University, IndiaReviewed by:
Kai Guo, Southwest University, ChinaCopyright © 2022 Zhang, Gao, Feng, Cui, Li, Liu, Wang, Xu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenliang Xu, d2VubGlhbmd4dUBtYWlsLmNjbnUuZWR1LmNu; Fuguang Li, YXlsaWZ1Z0BjYWFzLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.