
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 23 December 2021
Sec. Plant Cell Biology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.804356
This article is part of the Research Topic Phase Separation in Plants View all 6 articles
Tudor staphylococcal nucleases (TSNs) are evolutionarily conserved RNA binding proteins, which include redundant TSN1 and TSN2 in Arabidopsis. It has been showed TSNs are the components of stress granules (SGs) and regulate plant growth under salt stress. In this study, we find a binding protein of TSN1, RH31, which is a DEAD-box RNA helicase (RH). Subcellular localization studies show that RH31 is mainly located in the nucleus, but under salinity, it translocates to the cytoplasm where it accumulates in cytoplasmic granules. After cycloheximide (CHX) treatment which can block the formation of SGs by interfering with mRNP homeostasis, these cytoplasmic granules disappeared. More importantly, RH31 co-localizes with SGs marker protein RBP47. RH31 deletion results in salt-hypersensitive phenotype, while RH31 overexpression causes more resistant to salt stress. In summary, we demonstrate that RH31, the TSN1 binding protein, is a component of plant SGs and participates in regulation of salt-stress tolerance in Arabidopsis.
Abiotic stresses affect plant normal growth and lead to crop losses. Plants resist these environmental stresses by regulating mRNA translation and protein synthesis to alter the proteome rapidly in response to various stress signals. Stress granules (SGs) formation is thought to be required for the post-transcriptional regulation of stress-responsive mRNAs (Tsai et al., 2017; Omer et al., 2018).
SGs are condensates of proteins and RNAs assembled via liquid–liquid phase separation which involvement plays emerging roles in RNA-related cellular events under various stresses (Ivanov et al., 2019; Lin and Fang, 2021). SGs assembly is initiated by stress-induced eIF2α phosphorylation (Kedersha et al., 1999). When stress is encountered, SGs are formed to save energy by reprograming their translational machinery to allow selective expression of proteins. Some specific mRNAs translation initiation may occur in SGs (Thomas et al., 2011). SGs also function in the mammalian stress response by sequestering mRNAs and allowing for dynamic sorting of mRNAs for translation, storage, or degradation (Vanderweyde et al., 2013). Zebrafish cells lacking SGs will lose the ability to recover once the stress has ended (Zampedri et al., 2016). The components of SGs, AtTZF4, 5 and 6, are involved in light-, abscisic acid-, and gibberellic acid-mediated regulation of seed germination (Bogamuwa and Jang, 2013). SGs component VOZ2 functions as a transcriptional repressor of DREB2A to increase resistance to higher temperatures in Arabidopsis (Koguchi et al., 2017). CI and CII small heat shock proteins, which interact with many stress granule proteins, are both important for tolerance to severe heat stress in Arabidopsis (McLoughlin et al., 2016). Thus, SGs play critical roles under various stresses.
Tudor staphylococcal nucleases (TSNs) are evolutionarily conserved RNA binding proteins (Gao et al., 2015; Gutierrez-Beltran et al., 2016), which include redundant TSN1 and TSN2 in Arabidopsis thaliana. TSN is also a component of SGs (Yan et al., 2014). Previous studies revealed that TSN is essential for stress tolerance and stabilizes stress-responsive mRNAs expession (Dit Frey et al., 2010). Meanwhile, TSN functions in mRNA catabolism and links the formation of SGs and processing bodies (PBs)-cytoplasmic RNA granules which are believed to be sites of mRNA degradation (Arribere et al., 2011; Gutierrez-Beltran et al., 2015). In the latest study, TSN is established with an important role in stress signaling as a docking platform for stress granule proteins (Gutierrez-Beltran et al., 2021). Therefore, TSN is important for plant development and stress tolerance.
In the present study, we identified a binding protein of TSN1, RH31 (Aubourg et al., 1999), which belongs to the DEAD-box RNA helicase (RH) family. RHs function in a variety of RNA metabolism processes by catalyzing unwinding of RNA secondary structure in an ATP-dependent manner and are known as the best candidates for RNA chaperones (Richardson et al., 1998; Tanner and Linder, 2001; Carlotto et al., 2016). Increasing evidence suggests that RHs perform significant functions in innate immunity. As a transcription coactivator, nuclear DExD/H-box helicase 9 plays a critical role in the stimulation of NF-κB-mediated innate immunity against DNA virus infection in mice (Ng et al., 2018). DDX21 translocates from the nucleus to the cytoplasm and participates in the inhibition of Dengue Virus infection (Dong et al., 2016). The DEAD-box RNA helicase 51 aids cell cancer proliferation by regulating cell cycle progression via multiple pathways (Wang et al., 2015a). In addition, DEAD-box RHs also play central roles in plant growth, development and stress responses (Liu et al., 2002, 2008; Vashisht and Tuteja, 2006; Kant et al., 2007). In Sacchromyces cerevisiae, translation initiation factor 2 overexpression confers lithium tolerance in galactose medium (Montero-Lomeli et al., 2002). The DEAD-box RH8 positively regulates ABA signaling and increases drought tolerance via inhibiting PP2CA activity (Baek et al., 2018). A putative ATP-dependent DEAD-box RH, HVD1, is induced under salt stress, cold stress, and ABA treatment in sorghum (Nakamura et al., 2004).
In this study, we show that RH31 is binding protein of TSN1. We also prove that RH31 is localized to SGs under salt stress. Furthermore, rh31 mutants display salt-hypersensitive. More importantly, a pathogen-related (PR) gene, PROAtCAPE3 (Baek et al., 2010), and some salt tolerance-related genes including ABA-responsive element binding protein 1 (AREB1), Δ1-PYRROLINE-5-CARBOXYLATESYNTHASE 1 (P5CS1), and RD29B are downregulated in rh31 mutants. atcape3 mutants show the similar salt sensitivity to rh31. Thus, we conclude that TSN1 binding protein RH31 is a component of SGs and participates in regulation of salt-stress tolerance in Arabidopsis.
Arabidopsis ecotype Columbia (Col-0) was used as wild-type controls. Plant growth conditions were same with the conditions described in the previous study (Yan et al., 2014). Col-0 seed batches that were used for phenotype investigation in this work were collected at the same time and stored in the same conditions as the mutant and transgenic seed batches.
We used CRISPR/Cas9 technology to obtain the knockout mutant. For rh31, the lowest homology target (5'-CGAGGAAGAGCTCAGCAATTGGG-3') was selected. For atcape3, the lowest homology target (5'-TTGGGGTGGGACCCTTAAGATGG-3') was selected. Above targets were cloned into the pHEE2A-TRI vector (Wang et al., 2015b). Then, the construct was transformed into Col-0. The RH31 and PROAtCAPE3 fragments were amplified using the primers listed in Supplementary Table S1, and homozygotes were selected by restriction enzyme digestion and sequencing from the T1 generation. A specific pair of primers (zCas9-IDF3-2/rbcS_E9t-IDR) was used to identify non-transgenic lines of T2 generation. These non-transgenic lines were further examined on plates containing 25 mg L−1 hygromycin.
The full-length RH31 genomic sequence was amplified using the primers RH31-OE-LP and RH31-OE-RP (Supplementary Table S1) and then cloned into Super Promoter 1300 vector (pSuper 1300) to obtain overexpression lines (OEs).
The RH31-GFP construct described below was transformed into rh31 lines to generate RH31 complemented lines, and transgenic line (4–2) with similar mRNA levels as Col-0 was used for further study.
In subcellular localization assay, the full-length RH31 genomic sequence without the stop codon was amplified using the primers RH31-GFP-LP and RH31-GFP-RP (Supplementary Table S1), and then cloned into pSuper 1300-GFP vector. The resulting construct RH31-GFP was transformed into Col-0, and homozygotes were selected from the T3 generation, then the roots of 7-day-old seedlings were used to test subcellular localization of RH31. To test the effect of cycloheximide (CHX), 7-day-old seedlings were incubated in liquid medium (0.5 × Murashige and Skoog salts, 1% sucrose, and 0.5 g L−1 MES, pH 6.8) supplemented with 200 μg ml−1 CHX (Sigma-Aldrich) and shaken for 80 min as previously described (Goeres et al., 2007). To further confirm the localization of RH31, the RH31-GFP construct was used for transient expression assay, and GFP fusion proteins were observed with a Zeiss 710 Meta laser scanning confocal microscope (Zeiss, Oberkochen, Germany) as previously described (Kim and Somers, 2010). For CHX experiments, 100 μg ml−1 CHX was added to the respective protoplast suspension, and then gently mixed and incubated for 15 min. The cells were then either kept under control condition or subjected to 150 mM NaCl stress (Weber et al., 2008).
Three TSN1 fragments1 were generated as follows: tu1 I-1 (1–380 amino acids), including SN1 and SN2 domains and has a molecular weight of approximately 41.4 kD; tu1 I-2 (381–720 amino acids), including SN3 and SN4 domains and has a molecular weight of approximately 37.2 kD; tu1 II (721–970 amino acids), including C-terminal TSN (Tudor-SN) domain and has a molecular weight of 27.5 kD (Figure 1A). These three fragments were amplified using primers listed in Supplementary Table S1, then inserted into pGEX-4 T-1 vector and termed pGEX-tu1 I-1, pGEX-tu1 I-2 and pGEX-tu1 II, respectively. E. coli DE3 transformants were induced to express the fusion protein at 26°C for 8 h by the addition of IPTG to 1 mM. The expression of total GST fusion protein was detected using 10% SDS-PAGE. Protein concentration was determined using the Bio-Rad protein assay kit (Bio-Rad, Hercules, CA, United States), using bovine serum albumin as the standard. Buffer L (50 mM Tris–HCl, pH 8.0; 250 mM NaCl; 1 mM EDTA, pH8.0; and pH7.5) balanced Glutathione-Sepharose 4B were incubated with 100 μg GST fusion protein for 1 h at 4°C, and GST protein was used as negative control. Beads were centrifuged at 1000 × g for 2 min at 4°C. After removing the supernatant, beads were washed 3 × 5 min with Buffer L. Total protein of Arabidopsis thaliana seedlings was extracted from 0.5 g 2-week-old Col-0 lines using a Plant Protein Extraction Kit (CWBio, Beijing, China) according to manufacturer instructions. After sedimentation at 4°C for 1 h, the solution was centrifuged at 12000 × g for 30 min at 4°C, supernatants were collected, and again centrifuged. Total proteins of Arabidopsis thaliana seedlings were added into agarose, then the mixture was incubated overnight at 4°C; the mixture was centrifuged at 1000 g for 2 min at 4°C, after the supernatant was removed, the mixture was washed 4 × 5 min with Buffer L. Agarose was diluted with 100 μl Buffer L, and binding proteins were detected by 10% SDS-PAGE. The gel containing the binding proteins was cut off, and proteins were digested with trypsin and analyzed with LC–MS (Biological Mass Spectrometry Platform of China Agricultural University, Beijing, China). LC–MS data were searched using Matrix Server software against Arabidospsis TAIR10_pep_20101214 database.
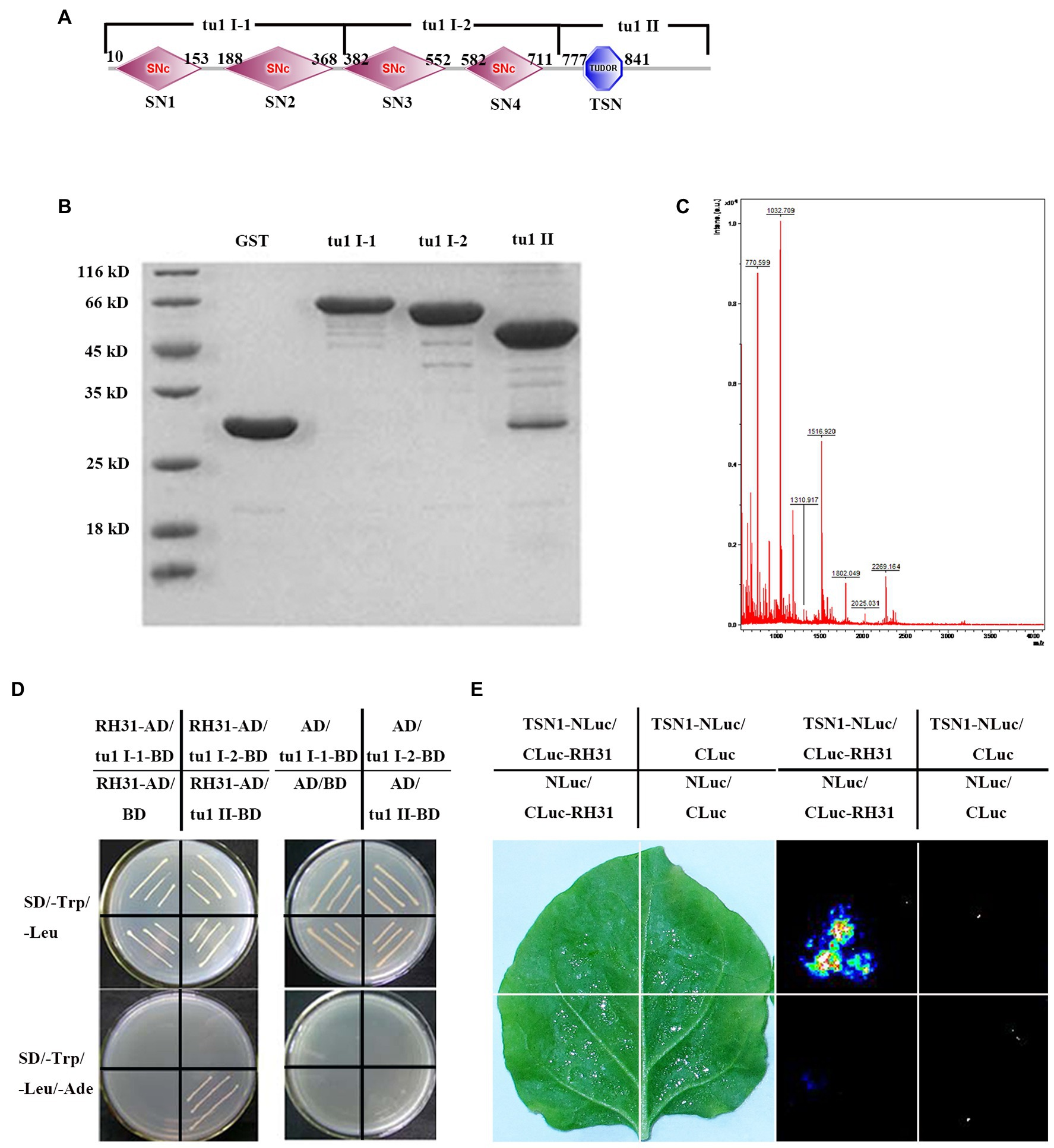
Figure 1. Identification of the RH31-TSN1 interaction. (A) The predicted protein domains of TSN1. The four SNc from left to right represent SN1, SN2, SN3, SN4, respectively. (B) GST Pull-down assay of truncated TSN1 protein. Three TSN1 fragments were generated as follows: tu1 I-1 (1–380 amino acids), including SN1 and SN2 domains; tu1 I-2 (381–720 amino acids), including SN3 and SN4 domains; tu1 II (721–970 amino acids), including C-terminal TSN (Tudor-SN) domain. GST fusion protein was used as bait to pull down the binding proteins of truncated TSN1 protein among total proteins of Arabidopsis seedlings. GST protein was used as a negative control. (C) Mass spectrometry analysis of tu1 II binding proteins. (D) Binding of tu1 II to RH31 by yeast-two-hybrid analysis. (E) Binding of TSN1 to RH31 by firefly luciferase complementation imaging.
The full-length RH31 genomic sequence was amplified using the primers RH31-hy-LP and RH31-hy-RP (Supplementary Table S1) and inserted into pGADT-7 vector. The construct was termed as RH31-AD. The tu1 I-1, tu1 I-2, and tu1 II fragments were digested with EcoRI and BamHI and then inserted into pGBKT-7 vector. The resulting constructs were termed as tu1 I-1-BD, tu1 I-2-BD, and tu1 II-BD, respectively. Empty pGADT-7 and pGBKT-7 vectors were used as negative controls. Constructs were co-transformed into yeast strain AH109. Yeast cells were cultured at 30°C on SD/−L-T plates. Positive clones were transformed on the selective medium SD/−L-T-A.
The full-length TSN1 and RH31 genomic sequences were amplified using the primers listed in Supplementary Table S1, then inserted into NLuc and CLuc vectors, respectively. Empty NLuc and CLuc vectors were used as negative controls. Constructs were co-transformed into A. tumefaciens strain GV3101. The Luciferase Complementation Imaging (LCI) assay was performed as previously described (Chen et al., 2008). Bacteria containing CLuc-RH31, TSN1-NLuc, and P19 (Shuhua Yang, State Key Laboratory of Plant Physiology and Biochemistry, China Agricultural University) were centrifuged together and the bacterial mixture was resuspended with activation buffer containing 10 mM MES, 10 mM MgCl2, and 150 mM acetosyringone to a final concentration of OD600 = 0.5, then incubated at 28°C for 3 h. Bacterial suspensions were infiltrated into young but fully expanded leaves of N. benthamiana plants using a needleless syringe. After infiltration, plants were placed at 23°C for 3 days. CCD imaging was used to measure LUC activity.
Total RNA was isolated from Arabidopsis as previously described (Oñatesánchez and Vicentecarbajosa, 2008). M-MLV reverse transcription system (TaKaRa, Beijing, China) was used to synthesize first-strand cDNA according to the manufacturer’s protocol. Quantitative RT-PCR was conducted using the SYBR Green I Master Mix (TaKaRa, Beijing, China) in a total reaction volume of 20 μl. The reaction was completed on the ABI7500 Fast Real-Time PCR system (Applied Biosystems, Foster City, CA, United States), and data were normalized with respect to At4g34270 (Czechowski et al., 2005). For semi-quantitative PCR, reactions were performed using Taq DNA polymerase (TaKaRa, Beijing, China) for 30 cycles in a total reaction volume of 25 μl. Actin2 was used as an internal control. The primers used for quantitative RT-PCR and semi-quantitative PCR are listed in Supplementary Table S1.
After 24 h of growth at 22°C on a 16-h light/8 h dark photoperiod on MS agar medium, synchronized growth seedlings (same root length) were transferred to MS medium for another 6 days or MS medium containing 150 mM NaCl or 250 mM mannitol for another 10 days. The root lengths of 7-day-old (normal) or 11-day-old (stress) seedlings were measured. We calculated relative root growth, which was determined as the ratio of root length under stress to root length under normal condition following a previously described method (Ezaki et al., 2007). Experiments were repeated three times with similar results, and at least 30 seedlings per genotype were measured. For fresh weight determination, the weight of at least 20 seedlings was measured. The relative fresh weight under stress conditions was calculated.
Seeds from the Col-0, rh31 and OEs were grown on 1/2 MS medium for 6 days and then transferred onto 1/2 MS medium supplemented with 200 mM NaCl for 5 days (Zhou et al., 2017). Experiments were repeated three times with similar results, and at least 30 seedlings for each genotype were examined per replicate. We calculated seedlings survival ratio according to visibly green leaves (Achard et al., 2006): seedlings with green leaves were counted as living, while seedlings with bleached white or albino leaves were counted as dead. Cell death in leaves was examined by trypan blue staining according to the method described by Bowling et al. (1997).
The RFP-RBP47 vector was generously provided by Dr. Markus Fauth (Weber et al., 2008). Arabidopsis mesophyll protoplasts were prepared from rosette leaves of 4-week-old Col-0 plants as previously described (Kim and Somers, 2010). Arabidopsis protoplasts were co-transformed with RH31-GFP and RFP-RBP47 and then incubated at 22°C for 20 h. Co-localization analysis was performed using a Zeiss 710 Meta laser scanning confocal microscope (Zeiss, Oberkochen, Germany) as previously described (Kim and Somers, 2010).
Yan et al. (2014) have previously proved that the RNA binding protein TSN1 is a component of stress granules (SGs) and involves in salt-stress adaptation (Yan et al., 2014). Here, TSN1-interacting proteins were identified. Three TSN1 fragments, tu1 I-1 which including SN1 and SN2 domains, tu1 I-2 which including SN3 and SN4 domains, tu1 II which including C-terminal TSN (Tudor-SN) domain were generated (Figure 1A). Several GST tagged TSN1 fragments (pGEX-tu1 I-1, pGEX-tu1 I-2 and pGEX-tu1 II) were extracted, and pull-down assays were performed to obtain the TSN1 binding proteins among total proteins of Arabidopsis seedlings (Figures 1A,B). We found that there are some proteins that could bind to pGEX-tu1 II, but no protein could bind to pGEX-tu1 I-1 or pGEX-tu1 I-2 (Figure 1B). A matching protein, which is a DEAD-box RNA helicase (At5g63630), RH31, was showed by mass spectrometry analysis (Figure 1C).
To further confirm the interaction between TSN1 and RH31, yeast two-hybrid assay was performed. When yeast cells were transformed with RH31-AD, tu1 I-1-BD, tu1 I-2-BD or tu1 II-BD, no colonies grew on SD/−L-T-A, while all transformants could grow on SD/−T plates. These results indicate that the transformants cannot self-activated and this yeast two-hybrid system can be used in our subsequent experiment. Then, RH31-AD was co-transformed with tu1 I-1-BD, tu1 I-2-BD, or tu1 II-BD. We found that only bait cells containing both the TSN1 C-terminal fragment tu1 II domain and RH31 could grow on the selective medium SD/−L-T-A, whereas other transformants could not grow (Figure 1D). These results suggest that RH31 can specially interact with C-terminal region of TSN1 in vitro.
The LCI assay further verified the interaction between RH31 and TSN1 in vivo. We found that co-infiltration of Agrobacteria containing CLuc-RH31 and TSN1-NLuc resulted in LUC complementation, while no LUC complementation was produced by expression of TSN1-NLuc construct and the empty 35STCLuc vector, CLuc-RH31 construct and the empty 35STNLuc vector, or empty 35STCLuc vector and empty 35STNLuc vector. These results demonstrate that RH31 can interact with TSN1 both in vitro and in vivo.
To investigate the subcellular localization of RH31, we constructed a RH31-GFP vector under control of the cauliflower mosaic virus (CaMV) 35S promoter (Weber et al., 2008; Koguchi et al., 2017) and expressed RH31 protein in Arabidopsis protoplasts and the RH31-GFP transgenic lines, respectively. The transgenic plants rescued the short root length and low fresh weight phenotype of rh31 mutant which indicated that RH31 protein was expressed (Supplementary Figure S4). RH31-GFP was transiently co-expressed with a nucleus-localized protein, red fluorescent fusion protein RFP-AHL22, in Arabidopsis protoplasts (Xiao et al., 2009). Under normal condition, RH31-GFP was mainly distributed in nucleus (Figure 2A, upper). However, after a short term treatment of salt, it rapidly shifted to the cytoplasm and redistributed to small granules (Figure 2A, below). Similarly, RH31-GFP mainly localized in cell nucleus in RH31-GFP transgenic lines under normal condition (Figure 2B, sample 1), but it shifted to the cytoplasm and formed some small granules under 150 mM NaCl treatment (Figure 2B, samples 2 and 3). Whereas GFP alone was present throughout the cell under all conditions (Figure 2B, samples 5 and 6).
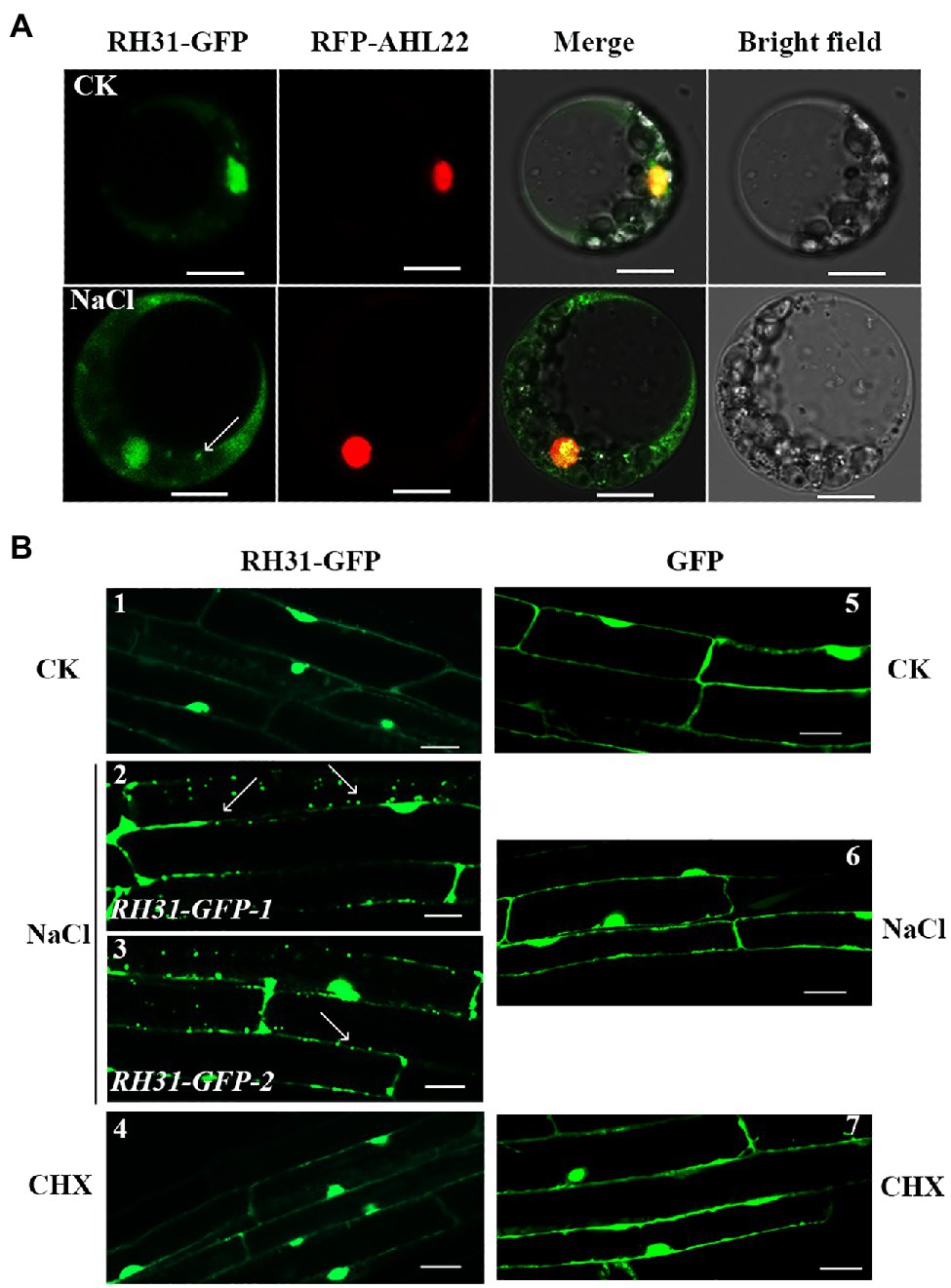
Figure 2. Subcellular localization of RH31-GFP. (A) Intracellular distribution of RH31-GFP and RFP-AHL22 in living Arabidopsis protoplasts. RH31-GFP mainly distributed in cell nucleus under normal condition (top row), but rapidly shifted to the cytoplasm and redistributed to small granules after 5 min treatment with 150 mM NaCl (bottom row). Scale bars = 10 μm. (B) Confocal micrographs of root elongation zones of 7-day-old seedlings. RH31-GFP mainly localized to cell nucleus under normal condition (1), but shifted to the cytoplasm and formed a number of small granules when seedling roots were treated with 150 mM NaCl (2 and 3). RH31-GFP seedling roots under salinity no longer showed small granules after treated with CHX (4). GFP transgenic lines under normal (5), salt stress (6), and CHX (7) conditions were used as controls. Scale bars = 20 μm. CK, Control; CHX, Cycloheximide. (2) and (3) represent two RH31-GFP lines. Arrows point to the granules.
Previous studies showed that TSN1 accumulates in SGs under stress (Yan et al., 2014). Since RH31 is a binding protein of TSN1, it is possible that the granules we observed were SGs in the RH31-GFP transgenic lines. SGs assembly depends on the release of untranslated mRNPs from polysomes. As an inhibitor of the translocation step during the elongation phase in protein synthesis, CHX traps mRNA in polysomes to block mRNA release and inhibits SG assembly in mammalian and plant cells (Kedersha et al., 1999; Weber et al., 2008; Grousl et al., 2009; Gutierrez-Beltran et al., 2015). As we expected, after CHX treatment, there is not the localized spots can be detected in the RH31-GFP transgenic lines under salinity (Figure 2B, sample 4).
To further confirm that these granules were indeed SGs in RH31-GFP transgenic lines, we co-expressed RH31-GFP and RFP-RBP47 under salt stress in Arabidopsis mesophyll protoplasts (Figure 3). RFP-RBP47 was primarily located in the nucleus (Figure 3A, lane 2), but was relocated to cytoplasmic granules with 150 mM NaCl treatment (Figures 3B,C, lane 2), which was consistent with previous study (Weber et al., 2008). Similarly, RH31-GFP was also primarily located in the nucleus under normal condition (Figure 3A, lane 1), but shifted into cytoplasmic granules in response to salt stress (Figures 3B,C, lane 1). Moreover, RH31-GFP fluorescence overlapped with RFP-RBP47 fluorescence. After CHX treatment, these cytoplasmic granules disappeared (Figure 3D). These results indicate that RH31 is a component of plant SGs and accumulates in SGs under salt stress.
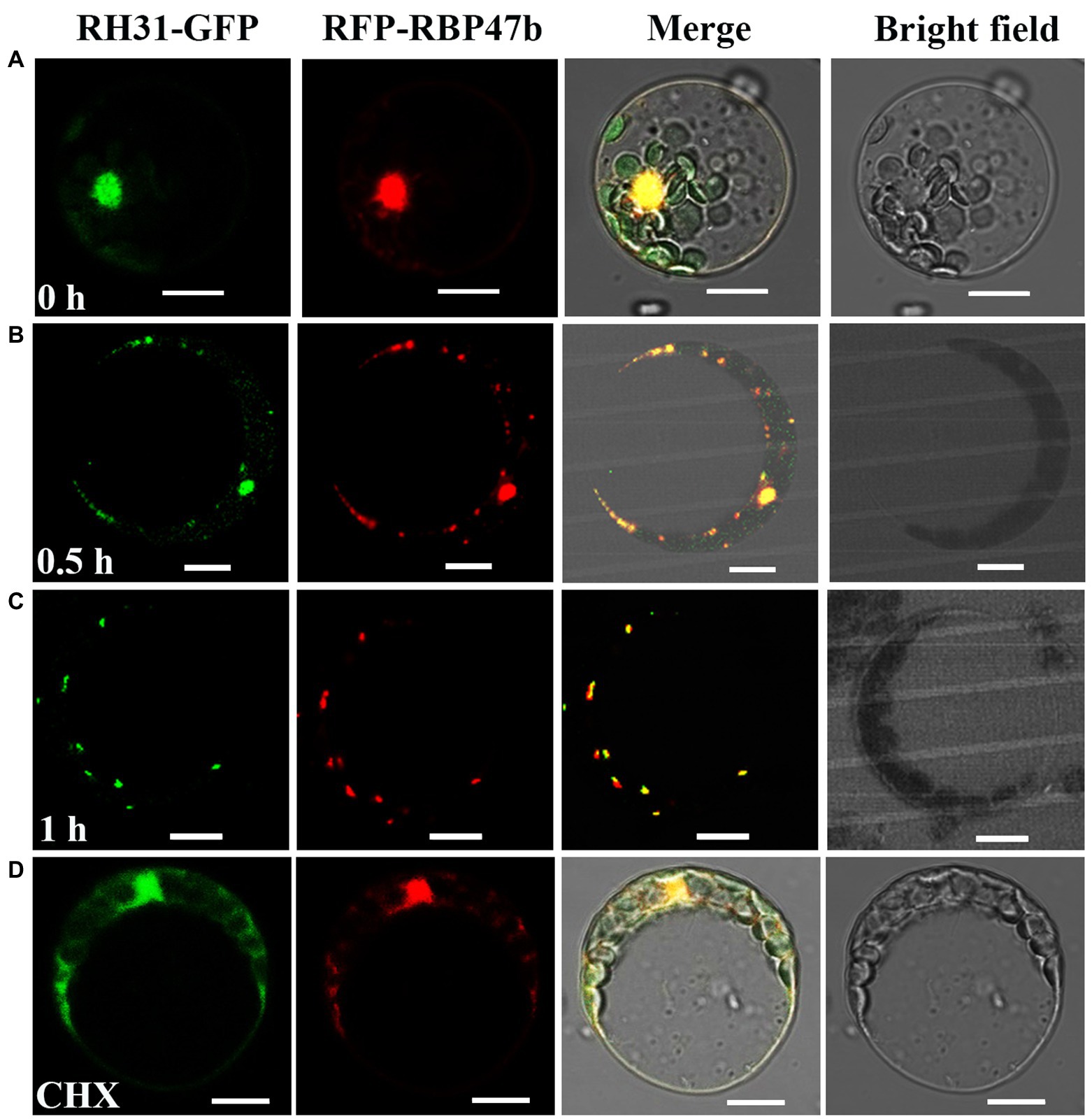
Figure 3. Co-localization analysis of RH31-GFP and RFP-RBP47 in SGs under salt stress in Arabidopsis mesophyll protoplasts. (A) RH31-GFP and RFP-RBP47 mainly located in nucleus under normal condition. Scale bars = 10 μm. (B,C) RH31-GFP and RFP-RBP47 relocated to cytoplasmic granules in response to 150 mM NaCl in different time point, and RH31-GFP fluorescence overlapped with RFP-RBP47 fluorescence. Scale bars = 10 μm. (D) After CHX treatment, there were no cytoplasmic granules when treated protoplasts with 150 mM NaCl. Scale bars = 10 μm. CHX, Cycloheximide.
To determine the role of RH31 under stress, rh31 mutant and RH31 OEs were generated. In terms of rh31 mutant, the lowest homology sgRNA-targeted RH31 was cloned into pHEE2A-TRI vector and then was transformed into Col-0. Mutants where loci were located upstream of the functional domain were selected (Figure 4A).2 Homozygous rh31 lines were identified among T2 progeny by sequencing. A single dTMP insertion was found within the 87th amino acid codon in rh31-14 mutant, while a single dAMP insertion was found within the 87th amino acid codon in rh31-41 mutant (Figures 4B,D); these two mutant loci were both upstream of the functional domain. A specific pair of primers (zCas9-IDF3-2/rbcS_E9t-IDR) was used to exclude any effect of the Cas9 gene (Figure 4C). We further confirmed hygromycin resistance in these lines (Figure 4E). The selected non-hygromycin resistant lines were used for further study.

Figure 4. Generation of rh31 mutants. (A) The predicted protein domains of RH31. (B) Schematic of the rh31 insertion mutation. Arrows point to the insertion site. (C) RT-PCR analysis of non-transgenic lines. Col-0 genomic DNA was served as a negative control. Plants 4, 7, 8 of rh31-14 and plant 12 of rh31-41 were non-transgenic, while other plants were Cas9 transgenic. (D) SANGER sequencing chromatography confirming insertion in rh31 mutants. Arrows point to the insertion site. (E) Screening non-Cas9 transgenic plants by hygromycin resistance. The details of target construction and mutant generation are described in the Materials and Methods section. Scale bars = 1 cm.
In terms of RH31 OEs, full length RH31 gene was introduced into Arabidopsis under control of CaMV 35S promoter. Four RH31-OEs were confirmed by both RT-PCR and qRT-PCR. OE1 and OE2 in which the transcript level of RH31 was increased by about 10-fold and 76-fold, respectively, were selected for further study (Supplementary Figure S1).
As the above studies showed that RH31 localized to SGs under salt stress, we hypothesized that RH31 might function in stress responses. To test this, synchronized growth of Col-0, rh31, and OEs were transferred to MS medium with or without NaCl or mannitol treatment. First of all, we designed serial concentrations of NaCl (0, 50, 100, 150, 200, 250 mM) for growth and survival experiments (data not shown), and the most obvious effects of 150 mM and 200 mM concentrations were determined for growth experiment and survival experiment, respectively. Under normal condition, rh31 seedlings had shorter primary root lengths and reduced fresh weight compared to Col-0, while OEs showed increased root growth and biomass accumulation (Figures 5A-C). Moreover, under salt stress (150 mM NaCl), the rh31 lines were even more severely impaired with distinct small, yellowish cotyledons, obviously shorter primary roots and less fresh weight after 10 days of NaCl treatment, while OEs were more resistant to NaCl (Figures 5D–F; Supplementary Figure S2). However, under 250 mM mannitol treatment, no significant difference was observed (Figures 5G–I). These show that rh31 mutant is relatively sensitive to salt, but not sensitive to osmotic stress.
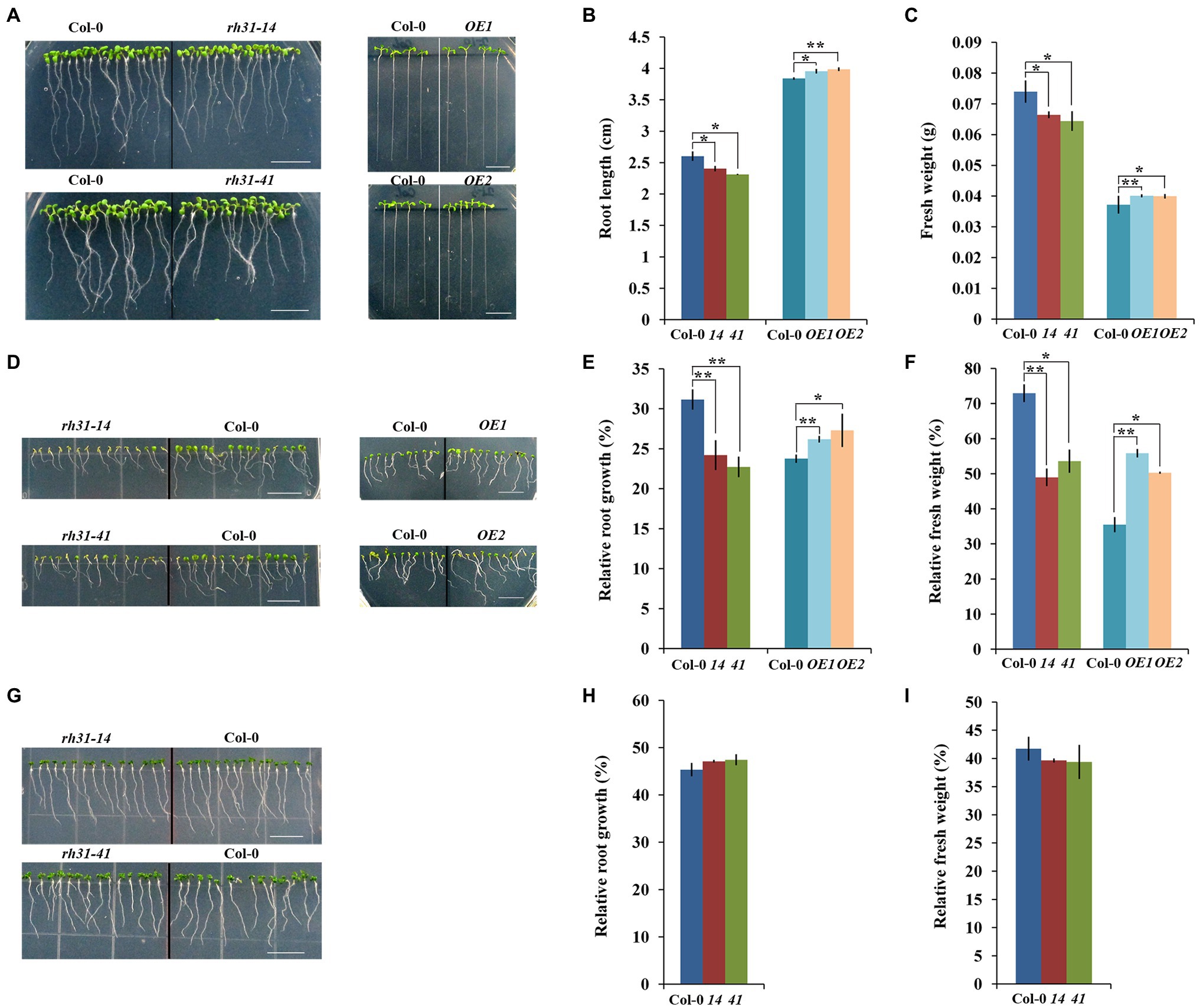
Figure 5. Growth phenotype characterization of rh31 and RH31 overexpression (OE1, OE2) lines under normal and stress conditions. (A) Phenotype of 7-day-old rh31 and 11-day-old OE seedlings under normal condition. After germinated for 24 h under normal condition, synchronized growth of Col-0, rh31, and OE seedlings were transferred to MS medium for another 6 days (rh31) or 10 days (OE). Experiments were repeated three times with similar results. Scale bars = 1 cm. (B) Root length of seedlings shown in (A). The primary roots of at least 30 seedlings were measured and reported as the mean length (n = 3 replicates). (C) Fresh weight of 11-day-old rh31 and OE seedlings under normal condition. 40 (rh31) and 20 (OE) seedlings were measured per replicate (n = 3 replicates). (D) Phenotype of rh31 and OE seedlings treated with 150 mM NaCl for 10 days. After germinated for 24 h under normal condition, synchronized growth of Col-0, rh31, and OE seedlings were transferred to MS medium with or without 150 mM NaCl for another 10 days. Experiments were repeated three times with similar results. Scale bars = 1 cm. (E) Relative root growth of seedlings shown in (D). The primary roots of at least 30 11-day-old seedlings were measured and relative growth was reported as the mean length (n = 3 replicates). (F) Relative fresh weight of seedlings shown in (D). 40 (rh31) and 20 (OE) seedlings were measured per replicate, and relative fresh weight was reported as the mean fresh weight (n = 3 replicates). (G) Phenotype of rh31 seedlings treated with 250 mM mannitol for 10 days. After germinated for 24 h under normal condition, synchronized growth of Col-0 and rh31 seedlings were transferred to MS medium with or without 250 mM mannitol for another 10 days. Experiments were repeated three times with similar results. Scale bars = 1 cm. (H) Relative root growth of rh31 seedlings shown in (G). The primary roots of at least 30 11-day-old seedlings were measured and relative growth was reported as the mean length (n = 3 replicates). (I) Relative fresh weight of seedlings shown in (G). 40 seedlings were measured per replicate, and relative fresh weight was reported as the mean fresh weight (n = 3 replicates). *p < 0.05 and **p < 0.01 (Student’s t-test) indicate significant differences between rh31, OE, and Col-0 plants. Error bars indicate the standard error for the average of three independent experiments.
When Col-0, rh31, and OE seedlings growing on 1/2 MS medium were transferred to 1/2 MS medium containing 200 mM NaCl for 5 days, the seedling survival rate of rh31 was significantly lower than that of Col-0 (Figure 6). The leaves of rh31 were chlorotic and significantly bleached as compared to those of Col-0, while the leaves of OEs displayed slight greenish than Col-0 (Figure 6A), and rh31 seedlings showed only 34.4% (rh31-14) and 32.1% (rh31-41) of plants surviving, compared with 62.5% of Col-0 seedlings surviving (Figure 6B). Using trypan blue staining assay, we found that a significant number of cells in rh31 mutants was positively stained after salt treatment, indicating more cells were dead, while for OEs, fewer positive cells were stained, indicating less cells were dead (Supplementary Figure S3).

Figure 6. Survival Phenotype characterization of rh31 and RH31 overexpression lines (OE1, OE2). (A) Survival phenotype of Col-0, rh31-14, rh31-41, and 35S: RH31 seedlings under salt treatment. Plants were grown on 1/2 MS medium for 6 days and then transferred onto 1/2 MS medium supplemented with 200 mM NaCl for 5 days. Experiments were repeated three times with similar results. Scale bars = 1 cm. (B) Survival rate of seedlings shown in (A). At least 30 seedlings were examined per replicate (n = 3 replicates). *p < 0.05 and **p < 0.01 (Student’s t-test) indicate significant differences between mutants or transgenic lines and Col-0 plants. Error bars indicate the standard error for the average of three independent experiments.
To confirm that the observed rh31 mutant phenotype indeed results from RH31 deletion, a complementation line of RH31 (4–2) was generated. As shown in Supplementary Figure S4, the phenotype of the 4–2 complementation line was similar to those of Col-0. The above results suggest that RH31 deletion results in hypersensitivity to salt, i.e., RH31 functions as a positive regulator of salt-stress tolerance in Arabidopsis.
Previous work indicated that TSN1 regulates growth under stress by modulating the transcriptional level of GA20ox3 in Arabidopsis (Yan et al., 2014). As its binding protein, RH31 might also be involved in this regulation pathway. We tested GA20ox3 mRNA level in rh31, but unexpectedly, no significant difference was observed between Col-0 and rh31 (Supplementary Figure S5), indicating that RH31 might function in other pathway than regulating GA20ox3 mRNA level.
To understand how RH31 regulates salt tolerance in Arabidopsis, the expression levels of various salt-inducible genes, based on transcriptome profiling analysis results (Peng et al., 2014), were tested in Col-0 and rh31 mutant seedlings. 7-day-old seedlings were treated with or without 150 mM NaCl for 24 h. We found that under normal condition, of these detected genes, PROAtCAPE3 (At4g33720), whose translation protein PROAtCAPE3 is classified as one of the cysteine-rich secretory proteins, antigen 5, and patho-genesis-related 1 proteins (CAP) superfamily (Chien et al., 2015), was decreased upon RH31 deletion. Quantitatively, in contrast with Col-0, the transcript levels of PROAtCAPE3 in rh31-14 and rh31-41 were reduced by 70 and 72%, respectively (Figure 7A). However, when seedlings were treated with 150 mM NaCl for 24 h, compared to Col-0, the PROAtCAPE3 transcript levels in rh31-14 and rh31-41 were reduced by 91 and 88%, respectively (Figure 7B), which was in consistent with the phenotype of rh31 grown under salinity (Figures 5D–F; Supplementary Figure S2). In addition, the transcript levels of PROAtCAPE3 were increased in RH31-OEs under both normal and salt-stress conditions (Figure 7C). These results indicate that PROAtCAPE3 mRNA levels can be significantly affected by RH31 deletion, an effect that is exacerbated under salinity. But the expression of other salt-inducible genes we tested such as At1g49570 and At5g19890, both encoding a peroxidase, and At1g60810, encoding subunit A of the trimeric enzyme ATP Citratelyase, and ATL31 (At5g27420) did not change significantly (Supplementary Figure S6). Nine potential PROAtCAPEs were identified as precursor candidates for AtCAPEs in Arabidopsis (Chien et al., 2015). We thus determined the mRNA levels of these nine PROAtCAPEs. Under normal condition, PROAtCAPE7 (At2g14580), PROAtCAPE8 (At5g26130), and PROAtCAPE9 (At2g14610) mRNA levels were not affected by RH31 deletion (Figure 7D). However, after treatment with 150 mM NaCl, the mRNA levels of both PROAtCAPE7 and PROAtCAPE8 were decreased by approximately twofold, and PROAtCAPE9 mRNA level was decreased by approximately 1.5-fold (Figure 7E). AREB1 (At1g45249; Fujita et al., 2005) in Col-0 was highly increased by salt treatment by approximately 6-fold as compared to normal condition, which was consistent with previous findings (Chien et al., 2015). However, rh31 mutation resulted in a slight decrease in AREB1 as compared to Col-0 under salinity (Figure 7E).
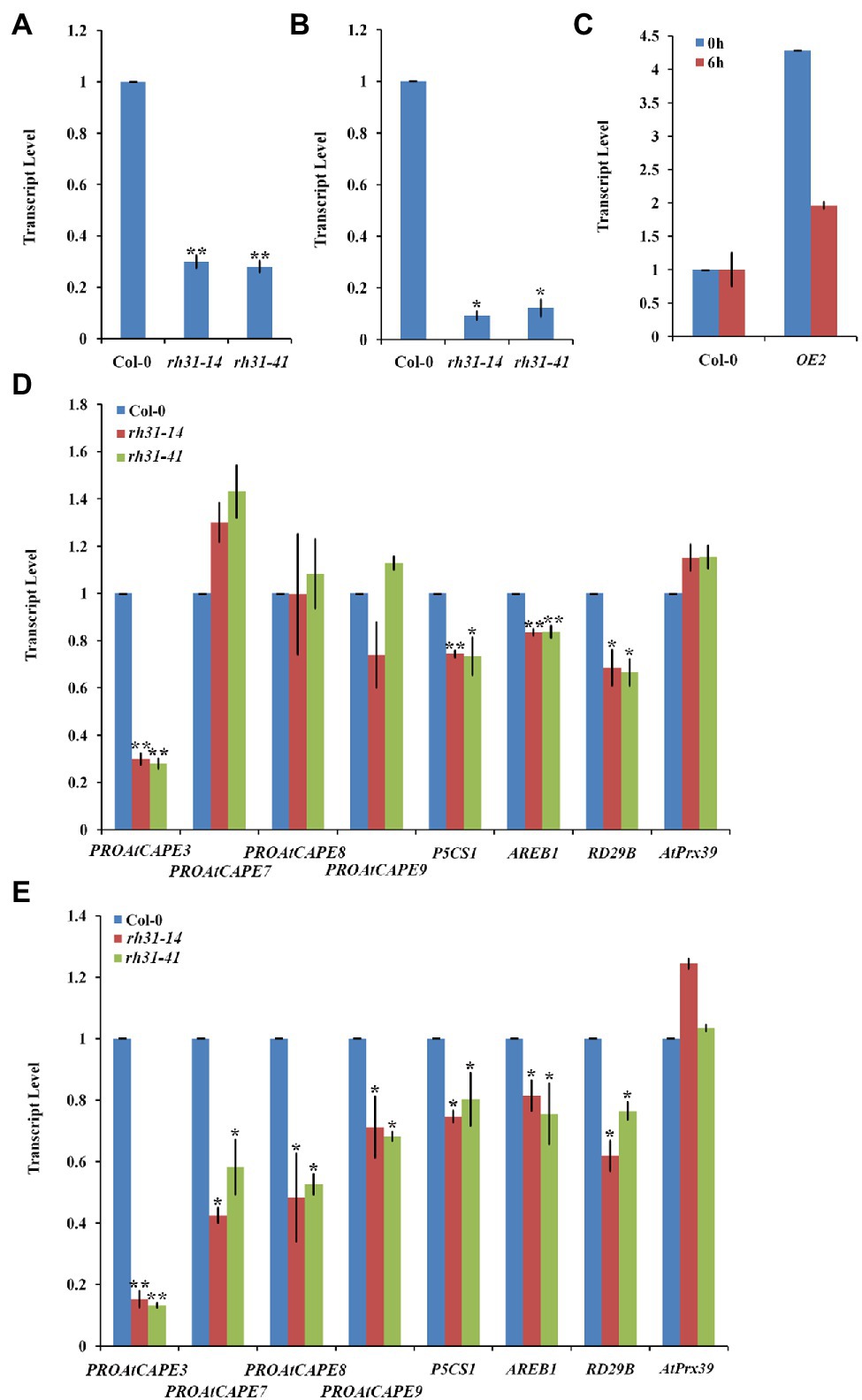
Figure 7. PROAtCAPE3 and salt-inducible gene expression analysis under normal and salt-stress conditions. (A,B) Quantitative RT-PCR of PROAtCAPE3 in rh31 and Col-0 lines under normal growth condition (A) or after treatment with 150 mM NaCl for 24 h (B). (C) Quantitative RT-PCR of PROAtCAPE3 in RH31 overexpression (OE2) and Col-0 lines under normal growth condition or after treatment with 150 mM NaCl for 6 h. (D,E) Quantitative RT-PCR of PROAtCAPE3 and salt-inducible genes in rh31 and Col-0 lines under normal growth condition (D) or after treatment with 150 mM NaCl for 3 h (E). Each value indicates relative quantity, with the genes expressed in Col-0 set at 1.0. *p < 0.05 and **p < 0.01 (Student’s t-test) indicate significant differences between mutants or transgenic lines and Col-0 plants. Error bars indicate the standard error for the average of three independent experiments.
As reported P5CS1 (At2g39800) and RD29B (At5g52300), involve in osmoprotectant biosynthesis, are high-salt-inducible downstream genes (Yoshiba et al., 1999; Uno et al., 2000). In our tests, P5CS1 and RD29B mRNA levels were also decreased by RH31 deletion (Figure 7E). The above results suggest that RH31 deletion affects the transcript levels of PROAtCAPE3 and some other salt-inducible genes.
The significant reduction of PROAtCAPE3 expression in rh31 prompted us to hypothesize that rh31 salt sensitivity might be mediated through PROAtCAPE3 downregulation. To test the role of PROAtCAPE3 in response to salt stress, we constructed PROAtCAPE3 knockout mutants atcape3-11 and atcape3-114 (Figures 8A,B) and compared the salt tolerance of Col-0 and atcape3 mutants under salinity (Figure 8C). In the presence of 200 mM NaCl, atcape3 seedlings showed only 49.6% and 46.5% of plants surviving after salt exposure, compared with 71.1% of Col-0 seedlings surviving (Figure 8D). These results indicate that PROAtCAPE3 acts positively in the salt tolerance response.
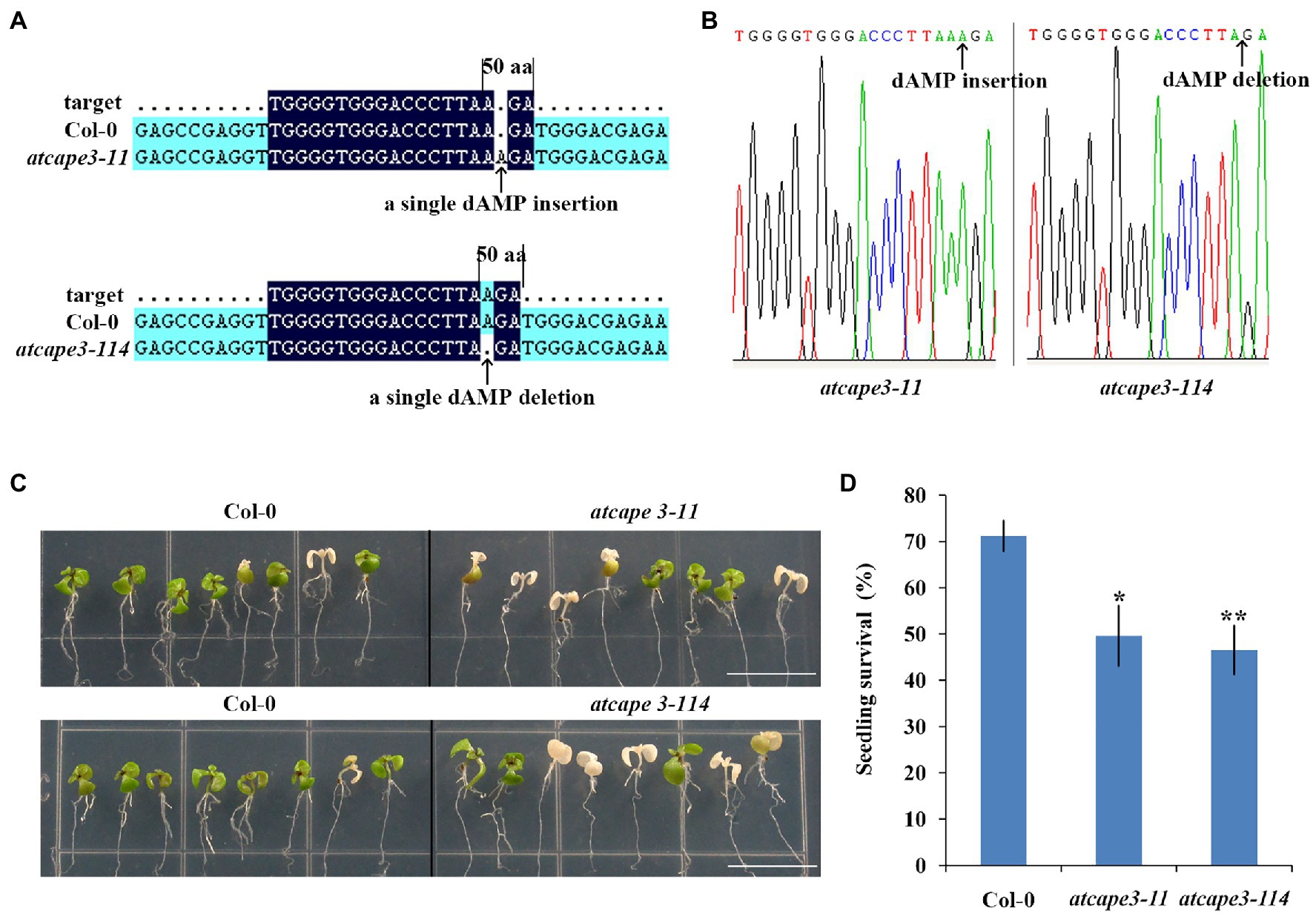
Figure 8. Generation of atcape3 mutants and seedling survival rate. (A) Schematic of the atcape3 mutant insertion (atcape3-11) site and deletion (atcape3-114) site. Arrows point to the insertion site and deletion site. (B) SANGER sequencing chromatography confirming insertion site and deletion site in atcape3 mutants. Arrows point to the insertion site and deletion site. (C) Survival phenotype of Col-0 and atcape3 seedlings under salt treatment. Plants were grown on 1/2 MS medium for 6 days and then transferred onto 1/2 MS medium supplemented with 200 mM NaCl for 5 days. Experiments were repeated three times with similar results. Scale bars = 1 cm. (D) Survival rate of seedlings shown in (C). At least 30 seedlings were examined per replicate (n = 3 replicates). *p < 0.05 and **p < 0.01 (Student’s t-test) indicate significant differences between mutants and Col-0 plants. Error bars indicate the standard error for the average of three independent experiments.
SGs are ubiquitous and assemble through protein-RNA, protein–protein, and RNA–RNA interactions. The composition of SGs differ according to stressor and cell type (Anderson and Kedersha, 2008), but some core components are conserved (Kedersha et al., 2002). SGs also contain RNA-binding proteins (Anderson and Kedersha, 2008) and RHs (Yu et al., 2011; Yasuda-Inoue et al., 2013), such as numerous ATP-dependent helicases and protein remodelers are identified in purified yeast and mammalian stress granule cores (Jain et al., 2016). Proteins involved in various aspects of mRNA metabolism and diverse cell signaling pathways are also the components of SGs (Kim et al., 2005).
As a major constituent of SGs, TSN functions in mRNA decapping during heat-stress tolerance (Gutierrez-Beltran et al., 2015). TSN is also involved in regulating transcription (Paukku et al., 2003; Yang et al., 2014), pre-mRNA splicing (Yang et al., 2007), mRNA stabilization (Paukku et al., 2008), RNA silencing (Caudy et al., 2003), and cleaving hyperedited double-stranded RNA (Scadden, 2005) in animals. Most relevant to the present study, TSN is established with an important role in stress signaling as a docking platform for stress granule proteins and is necessary for assembly and/or function of SGs (Gao et al., 2015; Gutierrez-Beltran et al., 2021).
Protein–protein interaction is important for RHs to accomplish their functions (Sugiura et al., 2007). In this study, we found a DEAD-box RH, RH31, which is a TSN1 binding protein (Figure 1). More importantly, RH31 is a component of plant SGs through fluorescence co-localization and CHX treatment experiments (Figure 3). Under stress condition, RH31 translocates from the nucleus to cytoplasm, where it localizes to SGs (Figures 2,3).
It has been reported that RHs function in a variety of RNA metabolism processes and are known as the best candidates for RNA chaperones (Richardson et al., 1998; Tanner and Linder, 2001; Carlotto et al., 2016). The DEAD-box RH AhRH47 maintains protein synthesis to enhance tolerance to salinity and mannitol-induced stresses (Mahesh and Udayakumar, 2018). DDX21 translocates from nucleus to cytoplasm, activating the innate immune response against Dengue Virus through stimulating IFN-β induction and consequently hinders Dengue Virus replication in the early phases of infection (Dong et al., 2016; Mohamed, 2021). As reported before, RNA binding protein CIP29 interacts with DEAD box RH DDX39 to enhance its helicase activity (Sugiura et al., 2007). Therefore, as a DEAD-box RH, RH31 may transfer from the nucleus to cytoplasm under salt stress, binding, and stabilizing adversity-related mRNAs, while the interacting partner-RNA binding protein TSN1 may enhance RH31 helicase activity and promote SGs formation.
The knockout mutant rh31 showed a salt-hypersensitive phenotype, while RH31-OEs conferred salt tolerance to Arabidopsis seedlings and the complemented transgenic line restored the wild-type phenotype (Figures 5,6; Supplementary Figures S2–S4). Thus, RH31 was identified as a positive regulator of salt tolerance in Arabidopsis. By qRT-PCR analysis, we determined genes that were changed significantly in RH31-OE, rh31, and Col-0 (Figure 7). Among the detected genes, the transcript level of PROAtCAPE3 was obviously reduced in rh31 (Figures 7A,B) while increased in RH31-OEs (Figure 7C).
As reported before, nine potential CAPs are identified in Arabidopsis, which are named PROAtCAPEs, and the peptide derived from PROAtCAPE is designated AtCAPE (Chen et al., 2014; Chien et al., 2015). As a member of CAP superfamily, Arabidopsis pathogenesis-related protein 1 (PR1, AtCAPE9) can resist Pseudomonas syringae pv. tomato strain DC3000 infection (Baek et al., 2010; Chen et al., 2014). Some PR genes are activated by abiotic stresses, suggesting that they play a role in cellular processes other than pathogen resistance (Seo et al., 2008). Additional evidence shows that OsPR4a overexpression in rice enhances tolerance to drought (Wang et al., 2011), and PR1 upregulation in Arabidopsis confers drought tolerance (Liu et al., 2013). Overexpression of tomato PROSYSTEMIN, the precursor of the anti-herbivore systemin peptide signal (Pearce et al., 1991), shows more tolerance to salt stress (Orsini et al., 2010). Thus, plant peptides function not only in regulating innate immunity, but also in abiotic stress tolerance responses. Studies by Peng et al. (2014) indicated that PROAtCAPE3 transcription is reduced in ein3eil1 mutants, but increased in EIN3ox, and this trend is exaggerated in ein3eil1 under salt stress. EIN3/EIL1 enhances salt tolerance by regulating myriad salt-inducible EIN3/EIL1-dependent genes. As an EIN3-induced gene, PROAtCAPE3 may play an important role in salt stress. Indeed, our work showed that atcape3 mutants displayed a reduced salt-tolerant phenotype (Figures 8C,D). Therefore, results of PROAtCAPE3 downregulation can be linked with salt hypersensitivity in rh31 mutants.
In addition to PROAtCAPE3, some other of the salt-inducible genes including AREB1, P5CS1, and RD29B were affected by RH31 deletion. The transcript level of AREB1, which controls the transcription of downstream ABA-dependent and salt-responsive genes and functions mainly at the vegetative stage (Nakashima and Yamaguchi-Shinozaki, 2013), is highly upregulated in Col-0 under salinity. However, RH31 deletion led to a slight decrease in AREB1 as compared to Col-0 (Figure 7E). As targets of AREB1, P5CS1, the gene for the enzyme involved in osmoprotectant biosynthesis, and the dehydration response gene, RD29B, were also reduced (Figure 7). These results indicate that RH31 is necessary to maintain the transcript levels of PROAtCAPE3 and some other salt-inducible genes.
SGs formation in mammals is involved in regulating mRNA stability and may be required for the optimal translation of stress-responsive mRNAs (Buchan and Parker, 2009; Vanderweyde et al., 2013). DEAD box RH DDX5 interacts with mettl3 to stabilize MSR1 mRNA in macrophages and plays a positive role in macrophage lipid uptake (Zhao et al., 2018). Human DEAD-box RH DDX6 acts as an oncogene in GC cells by associating with c-Myc mRNA and promoting c-Myc expression (Taniguchi et al., 2018). Thus, mRNA stabilization during cellular stress requires specific protein–mRNA interactions (Stohr et al., 2006). Here, we showed that RH31 was a component of plant SGs (Figure 3). In addition, we found that RH31 was involved in regulating PROAtCAPE3 mRNA levels under normal condition, an effect that was exacerbated under salinity (Figure 7). Furthermore, the atcape3 mutant showed the same salt hypersensitivity phenotype as rh31 mutant (Figures 6, 8C). Thus, one hypothesis is that, as a component of SGs, RH31 enhances salt tolerance by maintaining the optimal transcript levels of PROAtCAPE3 and some other salt tolerance genes under salt stress. However, at present, we cannot rule out the possibility that RH31 increases salt tolerance through the nuclear-localized portion. The potential mechanism by which RH31 regulates salt tolerance needs to be further investigated.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
YH conceived the research, supervised the experiment. YL designed and performed the experiments, analyzed the data, and prepared the figures. SL conducted the pull down and mass spectrometry analyses. HS, JM, and MJ provided technical assistance. YL and YH wrote the manuscript. All authors contributed to the article and approved the submitted version.
The study was supported by a grant from the National Natural Science Foundation of China (No. 31570254).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Prof. Qijun Chen (China Agricultural University, China) for providing pHEE401 vector and the assistance of CRISPR/Cas9 technology, and Prof. Shuhua Yang (China Agricultural University, China) for providing NLuc and CLuc vectors, and Dr. Zhen Li (China Agricultural University, China) for the assistance of LC–MS.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.804356/full#supplementary-material
Achard, P., Cheng, H., De Grauwe, L., Decat, J., Schoutteten, H., Moritz, T., et al. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311, 91–94. doi: 10.1126/science.1118642
Anderson, P., and Kedersha, N. (2008). Stress granules: the tao of RNA triage. Trends Biochem. Sci. 33, 141–150. doi: 10.1016/j.tibs.2007.12.003
Arribere, J. A., Doudna, J. A., and Gilbert, W. V. (2011). Reconsidering movement of eukaryotic mRNAs between polysomes and P bodies. Mol. Cell 44, 745–758. doi: 10.1016/j.molcel.2011.09.019
Aubourg, S., Kreis, M., and Lecharny, A. (1999). The DEAD box RNA helicase family in Arabidopsis thaliana. Nucleic Acids Res. 27, 628–636. doi: 10.1093/nar/27.2.628
Baek, W., Lim, C. W., and Lee, S. C. (2018). A DEAD-box RNA helicase, RH8, is critical for regulation of ABA signaling and the drought stress response via inhibition of PP2CA activity. Plant Cell Environ. 41, 1593–1604. doi: 10.1111/pce.13200
Baek, D., Pathange, P., Chung, J. S., Jiang, J. F., Gao, L. Q., Oikawa, A., et al. (2010). A stress-inducible sulphotransferase sulphonates salicylic acid and confers pathogen resistance in Arabidopsis. Plant Cell Environ. 33, 1383–1392. doi: 10.1111/j.1365-3040.2010.02156.x
Bogamuwa, S., and Jang, J. C. (2013). The Arabidopsis tandem CCCH zinc finger proteins AtTZF4, 5 and 6 are involved in light-, abscisic acid- and gibberellic acid- mediated regulation of seed germination. Plant Cell Environ. 36, 1507–1519. doi: 10.1111/pce.12084
Bowling, S. A., Clarke, J. D., Liu, Y. D., Klessig, D. F., and Dong, X. N. (1997). The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9, 1573–1584. doi: 10.2307/3870444
Buchan, J. R., and Parker, R. (2009). Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36, 932–941. doi: 10.1016/j.molcel.2009.11.020
Carlotto, N., Wirth, S., Furman, N., Solari, N. F., Ariel, F., Crespi, M., et al. (2016). The chloroplastic DEVH-box RNA helicase INCREASED SIZE EXCLUSION LIMIT 2 involved in plasmodesmata regulation is required for group II intron splicing. Plant Cell Environ. 39, 165–173. doi: 10.1111/pce.12603
Caudy, A. A., Ketting, R. F., Hammond, S. M., Denli, A. M., Bathoorn, A. M. P., Tops, B. B. J., et al. (2003). A micrococcal nuclease homologue in RNAi effector complexes. Nature 425, 411–414. doi: 10.1038/nature01956
Chen, Y. L., Lee, C. Y., Cheng, K. T., Chang, W. H., Huang, R. N., Nam, H. G., et al. (2014). Quantitative peptidomics study reveals that a wound-induced peptide from PR-1 regulates immune signaling in tomato. Plant Cell 26, 4135–4148. doi: 10.1105/tpc.114.131185
Chen, H. M., Zou, Y., Shang, Y. L., Lin, H. Q., Wang, Y. J., Cai, R., et al. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146, 368–376. doi: 10.2307/40065847
Chien, P. S., Nam, H. G., and Chen, Y. R. (2015). A salt-regulated peptide derived from the CAP superfamily protein negatively regulates salt-stress tolerance in Arabidopsis. J. Exp. Bot. 66, 5301–5313. doi: 10.1093/jxb/erv263
Czechowski, T., Stitt, M., Altmann, T., Udvardi, M. K., and Scheible, W. R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17. doi: 10.1104/pp.105.063743
Dit Frey, N. F., Muller, P., Jammes, F., Kizis, D., Leung, J., Perrot-Rechenmann, C., et al. (2010). RNA binding protein Tudor-SN is essential for stress tolerance and stabilizes levels of stress-responsive mRNAs encoding secreted proteins in Arabidopsis. Plant Cell 22, 1575–1591. doi: 10.1105/tpc.109.070680
Dong, Y. C., Ye, W., Yang, J., Han, P. J., Wang, Y., Ye, C. T., et al. (2016). DDX21 translocates from nucleus to cytoplasm and stimulates the innate immune response due to dengue virus infection. Biochem. Biophys. Res. Commun. 473, 648–653. doi: 10.1016/j.bbrc.2016.03.120
Ezaki, B., Kiyohara, H., Matsumoto, H., and Nakashima, S. (2007). Overexpression of an auxilin-like gene (F9E10.5) can suppress Al uptake in roots of Arabidopsis. J. Exp. Bot. 58, 497–506. doi: 10.1093/jxb/erl221
Fujita, Y., Fujita, M., Satoh, R., Maruyama, K., Parvez, M. M., Seki, M., et al. (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17, 3470–3488. doi: 10.1105/tpc.105.035659
Gao, X. J., Fu, X., Song, J., Zhang, Y., Cui, X. T., Su, C., et al. (2015). Poly(A)(+) mRNA-binding protein Tudor-SN regulates stress granules aggregation dynamics. FEBS J. 282, 874–890. doi: 10.1111/febs.13186
Goeres, D. C., Van Norman, J. M., Zhang, W. P., Fauver, N. A., Spencer, M. L., and Sieburth, L. E. (2007). Components of the Arabidopsis mRNA decapping complex are required for early seedling development. Plant Cell 19, 1549–1564. doi: 10.1105/tpc.106.047621
Grousl, T., Ivanov, P., Frydlova, I., Vasicova, P., Janda, F., Vojtova, J., et al. (2009). Robust heat shock induces eIF2α-phosphorylation-independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae. J. Cell Sci. 122, 2078–2088. doi: 10.1242/jcs.045104
Gutierrez-Beltran, E., Denisenko, T. V., Zhivotovsky, B., and Bozhkov, P. V. (2016). Tudor staphylococcal nuclease: biochemistry and functions. Cell Death Differ. 23, 1739–1748. doi: 10.1038/cdd.2016.93
Gutierrez-Beltran, E., Elander, P. H., Dalman, K., Dayhoff, G. W. II, Moschou, P. N., Uversky, V. N., et al. (2021). Tudor staphylococcal nuclease is a docking platform for stress granule components and is essential for SnRK1 activation in Arabidopsis. EMBO J. 40:e105043. doi: 10.15252/embj.2020105043
Gutierrez-Beltran, E., Moschou, P. N., Smertenko, A. P., and Bozhkov, P. V. (2015). Tudor staphylococcal nuclease links formation of stress granules and processing bodies with mRNA catabolism in Arabidopsis. Plant Cell 27, 926–943. doi: 10.1105/tpc.114.134494
Ivanov, P., Kedersha, N., and Anderson, P. (2019). Stress granules and processing bodies in translational control. Cold Spring Harb. Perspect. Biol. 11:a032813. doi: 10.1101/cshperspect.a032813
Jain, S., Wheeler, J. R., Walters, R. W., Agrawal, A., Barsic, A., and Parker, R. (2016). ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498. doi: 10.1016/j.cell.2015.12.038
Kant, P., Kant, S., Gordon, M., Shaked, R., and Barak, S. (2007). STRESS RESPONSE SUPPRESSOR1 and STRESS RESPONSE SUPPRESSOR2, two DEAD-box RNA helicases that attenuate Arabidopsis responses to multiple abiotic stresses. Plant Physiol. 145, 814–830. doi: 10.1104/pp.107.099895
Kedersha, N., Chen, S., Gilks, N., Li, W., Miller, I. J., Stahl, J., et al. (2002). Evidence that ternary complex (eIF2-GTP-tRNA (i) (met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell 13, 195–210. doi: 10.1091/mbc.01-05-0221
Kedersha, N. L., Gupta, M., Li, W., Miller, I., and Anderson, P. (1999). RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 147, 1431–1442. doi: 10.1083/jcb.147.7.1431
Kim, W. J., Back, S. H., Kim, V., Ryu, I., and Jang, S. K. (2005). Sequestration of TRAF2 into stress granules interrupts tumor necrosis factor signaling under stress conditions. Mol. Cell. Biol. 25, 2450–2462. doi: 10.1128/MCB.25.6.2450-2462.2005
Kim, J., and Somers, D. E. (2010). Rapid assessment of gene function in the circadian clock using artificial microRNA in Arabidopsis mesophyll protoplasts. Plant Physiol. 154, 611–621. doi: 10.1104/pp.110.162271
Koguchi, M., Yamasaki, K., Hirano, T., and Sato, M. H. (2017). Vascular plant one-zinc-finger protein 2 is localized both to the nucleus and stress granules under heat stress in Arabidopsis. Plant Sig. Behav. 12:e1295907. doi: 10.1080/15592324.2017.1295907
Lin, Y., and Fang, X. F. (2021). Phase separation in RNA biology. J. Genet. Genomics 48, 872–880. doi: 10.1016/j.jgg.2021.07.012
Liu, H. H., Liu, J., Fan, S. L., Song, M. Z., Han, X. L., Liu, F., et al. (2008). Molecular cloning and characterization of a salinity stress-induced gene encoding DEAD-box helicase from the halophyte Apocynum venetum. J. Exp. Bot. 59, 633–644. doi: 10.1111/j.1532-950x.2004.04023.x
Liu, H. Y., Nefsky, B. S., and Walworth, N. C. (2002). The Ded1 DEAD box helicase interacts with Chk1 and Cdc2. J. Biol. Chem. 277, 2637–2643. doi: 10.1074/jbc.M109016200
Liu, W. X., Zhang, F. C., Zhang, W. Z., Song, L. F., Wu, W. H., and Chen, Y. F. (2013). Arabidopsis Di19 functions as a transcription factor and modulates PR1, PR2, and PR5 expression in response to drought stress. Mol. Plant 6, 1487–1502. doi: 10.1093/mp/sst031
Mahesh, S., and Udayakumar, M. (2018). Peanut RNA helicase AhRH47 sustains protein synthesis under stress and improves stress adaptation in Arabidopsis. Plant Mol. Biol. Report. 36, 58–70. doi: 10.1007/s11105-017-1056-9
McLoughlin, F., Basha, E., Fowler, M. E., Kim, M., Bordowitz, J., Katiyar-Agarwal, S., et al. (2016). Class I and II small heat shock proteins together with HSP101 protect protein translation factors during heat stress. Plant Physiol. 172, 1221–1236. doi: 10.1104/pp.16.00536
Mohamed, A. M. A. (2021). DEAD-box RNA helicases: The driving forces behind RNA metabolism at the crossroad of viral replication and antiviral innate immunity. Virus Res. 296:198352. doi: 10.1016/j.virusres.2021.198352
Montero-Lomeli, M., Morais, B. L. B., Figueiredo, D. L., Neto, D. C. S., Martins, J. R. P., and Masuda, C. A. (2002). The initiation factor eIF4A is involved in the response to lithium stress in Saccharomyces cerevisiae. J. Biol. Chem. 277, 21542–21548. doi: 10.1074/jbc.M201977200
Nakamura, T., Muramoto, Y., Yokota, S., Ueda, A., and Takabe, T. (2004). Structural and transcriptional characterization of a salt-responsive gene encoding putative ATP-dependent RNA helicase in barley. Plant Sci. 167, 63–70. doi: 10.1016/j.plantsci.2004.03.001
Nakashima, K., and Yamaguchi-Shinozaki, K. (2013). ABA signaling in stress-response and seed development. Plant Cell Rep. 32, 959–970. doi: 10.1007/s00299-013-1418-1
Ng, Y. C., Chung, W. C., Kang, H. R., Cho, H. J., Park, E. B., Kang, S. J., et al. (2018). A DNA-sensing–independent role of a nuclear RNA helicase, DHX9, in stimulation of NF-κB–mediated innate immunity against DNA virus infection. Nucleic Acids Res. 46, 9011–9026. doi: 10.1093/nar/gky742
Omer, A., Patel, D., Lian, X. J., Sadek, J., Di Marco, S., Pause, A., et al. (2018). Stress granules counteract senescence by sequestration of PAI-1. EMBO Rep. 19:e44722. doi: 10.15252/embr.201744722
Oñatesánchez, L., and Vicentecarbajosa, J. (2008). DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC. Res. Notes 1, 93–93. doi: 10.1186/1756-0500-1-93
Orsini, F., Cascone, P., De Pascale, S., Barbieri, G., Corrado, G., Rao, R., et al. (2010). Systemin-dependent salinity tolerance in tomato: evidence of specific convergence of abiotic and biotic stress responses. Physiol. Plant. 138, 10–21. doi: 10.1111/j.1399-3054.2009.01292.x
Paukku, K., Kalkkinen, N., Silvennoinen, O., Kontula, K. K., and Lehtonen, J. Y. A. (2008). p100 increases AT1R expression through interaction with AT1R 3'-UTR. Nucleic Acids Res. 36, 4474–4487. doi: 10.1093/nar/gkn411
Paukku, K., Yang, J., and Silvennoinen, O. (2003). Tudor and nuclease-like domains containing protein p100 function as coactivators for signal transducer and activator of transcription 5. Mol. Endocrinol. 17, 1805–1814. doi: 10.1210/me.2002-0256
Pearce, G., Strydom, D., Johnson, S., and Ryan, C. A. (1991). A polypeptide from tomato leaves induces wound-inducible proteinase-inhibitor proteins. Science 253, 895–897. doi: 10.1126/science.253.5022.895
Peng, J. Y., Li, Z. H., Wen, X., Li, W. Y., Shi, H., Yang, L. S., et al. (2014). Salt-induced stabilization of EIN3/EIL1 confers salinity tolerance by deterring ROS accumulation in Arabidopsis. PLoS Genet. 10:e1004664. doi: 10.1371/journal.pgen.1004664
Richardson, A., Landry, S. J., and Georgopoulos, C. (1998). The ins and outs of a molecular chaperone machine. Trends Biochem. Sci. 23, 138–143. doi: 10.1016/S0968-0004(98)01193-1
Scadden, A. D. J. (2005). The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat. Struct. Mol. Biol. 12, 489–496. doi: 10.1038/nsmb936
Seo, P. J., Lee, A. K., Xiang, F. N., and Park, C. M. (2008). Molecular and functional profiling of Arabidopsis pathogenesis-related genes: insights into their roles in salt response of seed germination. Plant Cell Physiol. 49, 334–344. doi: 10.1093/pcp/pcn011
Stohr, N., Lederer, M., Reinke, C., Meyer, S., Hatzfeld, M., Singer, R. H., et al. (2006). ZBP1 regulates mRNA stability during cellular stress. J. Cell Biol. 175, 527–534. doi: 10.1083/jcb.200608071
Sugiura, T., Sakurai, K., and Nagano, Y. (2007). Intracellular characterization of DDX39, a novel growth-associated RNA helicase. Exp. Cell Res. 313, 782–790. doi: 10.1016/j.yexcr.2006.11.014
Taniguchi, K., Iwatsuki, A., Sugito, N., Shinohara, H., Kuranaga, Y., Oshikawa, Y., et al. (2018). Oncogene RNA helicase DDX6 promotes the process of c-Myc expression in gastric cancer cells. Mol. Carcinog. 57, 579–589. doi: 10.1002/mc.22781
Tanner, N. K., and Linder, P. (2001). DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell 8, 251–262. doi: 10.1016/S1097-2765(01)00329-X
Thomas, M. G., Loschi, M., Desbats, M. A., and Boccaccio, G. L. (2011). RNA granules: the good, the bad and the ugly. Cell. Sig. 23, 324–334. doi: 10.1016/j.cellsig.2010.08.011
Tsai, W. C., Reineke, L. C., Jain, A., Jung, S. Y., and Lloyd, R. E. (2017). Histone arginine demethylase JMJD6 is linked to stress granule assembly through demethylation of the stress granule-nucleating protein G3BP1. J. Biol. Chem. 292, 18886–18896. doi: 10.1074/jbc.M117.800706
Uno, Y., Furihata, T., Abe, H., Yoshida, R., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. U. S. A. 97, 11632–11637. doi: 10.1073/pnas.190309197
Vanderweyde, T., Youmans, K., Liu-Yesucevitz, L., and Wolozin, B. (2013). Role of stress granules and RNA-binding proteins in neurodegeneration: a mini-review. Gerontology 59, 524–533. doi: 10.1159/000354170
Vashisht, A. A., and Tuteja, N. (2006). Stress responsive DEAD-box helicases: a new pathway to engineer plant stress tolerance. J. Photochem. Photobiol. 84, 150–160. doi: 10.1016/j.jphotobiol.2006.02.010
Wang, X. J., Liu, H. L., Zhao, C. L., Li, W., Xu, H. B., and Chen, Y. Q. (2015a). The DEAD-box RNA helicase 51 controls non-small cell lung cancer proliferation by regulating cell cycle progression via multiple pathways. Sci. Rep. 6:26108. doi: 10.1038/srep26108
Wang, N. L., Xiao, B. Z., and Xiong, L. Z. (2011). Identification of a cluster of PR4-like genes involved in stress responses in rice. J. Plant Physiol. 168, 2212–2224. doi: 10.1016/j.jplph.2011.07.013
Wang, Z. P., Xing, H. L., Dong, L., Zhang, H. Y., Han, C. Y., Wang, X. C., et al. (2015b). Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 16, 1–12. doi: 10.1186/s13059-015-0715-0
Weber, C., Nover, L., and Fauth, M. (2008). Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J. 56, 517–530. doi: 10.1111/j.1365-313X.2008.03623.x
Xiao, C. W., Chen, F. L., Yu, X. H., Lin, C. T., and Fu, Y. F. (2009). Over-expression of an AT-hook gene, AHL22, delays flowering and inhibits the elongation of the hypocotyl in Arabidopsis thaliana. Plant Mol. Biol. 71, 39–50. doi: 10.1007/s11103-009-9507-9
Yan, C. X., Yan, Z. Y., Wang, Y. Z., Yan, X. Y., and Han, Y. Z. (2014). Tudor-SN, a component of stress granules, regulates growth under salt stress by modulating GA20ox3 mRNA levels in Arabidopsis. J. Exp. Bot. 65, 5933–5944. doi: 10.1093/jxb/eru334
Yang, J., Aittomaki, S., Pesu, M., Carter, K., Saarinen, J., Kalkkinen, N., et al. (2014). Identification of p100 as a coactivator for STAT6 that bridges STAT6 with RNA polymerase II. EMBO J. 21, 4950–4958. doi: 10.1093/emboj/cdf463
Yang, J., Valineva, T., Hong, J. X., Bu, T. X., Yao, Z., Jensen, O. N., et al. (2007). Transcriptional co-activator protein p100 with snRNP proteins and facilitates the assembly of the spliceosorne. Nucleic Acids Res. 35, 4485–4494. doi: 10.1093/nar/gkm470
Yasuda-Inoue, M., Kuroki, M., and Ariumi, Y. (2013). DDX3 RNA helicase is required for HIV-1 tat function. Biochem. Biophys. Res. Commun. 441, 607–611. doi: 10.1016/j.bbrc.2013.10.107
Yoshiba, Y., Nanjo, T., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1999). Stress-responsive and developmental regulation of Delta(1)-pyrroline-5-carboxylate synthetase 1 (P5CS1) gene expression in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 261, 766–772. doi: 10.1006/bbrc.1999.1112
Yu, S. F., Lujan, P., Jackson, D. L., Emerman, M., and Linial, M. L. (2011). The DEAD-box RNA helicase DDX6 is required for efficient encapsidation of a retroviral genome. PLoS Pathog. 7:e1002303. doi: 10.1371/journal.ppat.1002303
Zampedri, C., Tinoco-Cuellar, M., Carrillo-Rosas, S., Diaz-Tellez, A., Ramos-Balderas, J. L., Pelegri, F., et al. (2016). Zebrafish P54 RNA helicases are cytoplasmic granule residents that are required for development and stress resilience. Biol. Open 5, 1473–1484. doi: 10.1242/bio.015826
Zhao, W. T., Wang, Z., Sun, Z. W., He, Y. X., Jian, D. D., Hu, X. T., et al. (2018). RNA helicase DDX5 participates in oxLDL-induced macrophage scavenger receptor 1 expression by suppressing mRNA degradation. Exp. Cell Res. 366, 114–120. doi: 10.1016/j.yexcr.2018.03.003
Keywords: TSN1 binding protein, stress granules, DEAD-box RNA helicase, RH31, salt-stress tolerance
Citation: Liu Y, Liu S, Shi H, Ma J, Jing M and Han Y (2021) The TSN1 Binding Protein RH31 Is a Component of Stress Granules and Participates in Regulation of Salt-Stress Tolerance in Arabidopsis. Front. Plant Sci. 12:804356. doi: 10.3389/fpls.2021.804356
Received: 29 October 2021; Accepted: 02 December 2021;
Published: 23 December 2021.
Edited by:
Xiaofeng Fang, Tsinghua University, ChinaReviewed by:
Panagiotis N. Moschou, Swedish University of Agricultural Sciences, SwedenCopyright © 2021 Liu, Liu, Shi, Ma, Jing and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuzhen Han, aGFueXV6aGVuQGNhdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.