- MOE Key Laboratory for Cellular Dynamics, School of Life Sciences, Division of Molecular and Cell Biophysics, Division of Life Sciences and Medicine, Hefei National Science Center for Physical Sciences at the Microscale, University of Science and Technology of China, Hefei, China
Phosphatidylinositol (PtdIns) is one type of phospholipid comprising an inositol head group and two fatty acid chains covalently linked to the diacylglycerol group. In addition to their roles as compositions of cell membranes, phosphorylated PtdIns derivatives, termed phosphoinositides, execute a wide range of regulatory functions. PtdIns can be phosphorylated by various lipid kinases at 3-, 4- and/or 5- hydroxyls of the inositol ring, and the phosphorylated forms, including PtdIns3P, PtdIns4P, PtdIns5P, PtdIns(3,5)P2, PtdIns(4,5)P2, can be reversibly dephosphorylated by distinct lipid phosphatases. Amongst many other types, the SUPPRESSOR OF ACTIN (SAC) family of phosphoinositide phosphatases recently emerged as important regulators in multiple growth and developmental processes in plants. Here, we review recent advances on the biological functions, cellular activities, and molecular mechanisms of SAC domain-containing phosphoinositide phosphatases in plants. With a focus on those studies in the model plant Arabidopsis thaliana together with progresses in other plants, we highlight the important roles of subcellular localizations and substrate preferences of various SAC isoforms in their functions.
Introduction
The biological membrane doesn’t only separate the cell from the outer environment, but also defines specific territory for subcellular compartments. It is mainly composed of sterols, sphingolipids and structural glycerophospholipids together with regulatory phospholipids including phosphatidylinositol and its phosphorylated derivatives phosphatidylinositol phosphates (PtdInsPs, also referred to as phosphoinositides) (Ejsing et al., 2009; Andreyev et al., 2010; Colin and Jaillais, 2020). Despite representing a small fraction of total phospholipids, these negatively charged phosphoinositides play a vital role in various cellular activities, including membrane trafficking and cellular dynamics (Gerth et al., 2017; Noack and Jaillais, 2020). Different phosphoinositides are enriched in different membranes and execute specific cellular functions (Noack and Jaillais, 2020). By application of fluorescent probes, PtdIns3P was found to distribute at multivesicular bodies/late endosomes/prevacuolar compartment (MVB/LE/PVC) and the vacuolar membrane (Vermeer et al., 2006; Simon et al., 2014); PtdIns(3,5)P2 localizes predominantly in MVB/LE (Hirano et al., 2017) and PtdIns(4,5)P2 mainly at the plasma membrane (PM; Van Leeuwen et al., 2007). These subcellular localizations may vary according to distinct cell types. In Arabidopsis root hairs, PtdIns(3,5)P2 localizes to the PM along the shank region and is involved in the hardening of the shank, whereas PtdIns(4,5)P2 was observed to accumulate at the apex and it is required for tip growth (Hirano et al., 2018). In root epidermal cells, PtdIns4P exhibited a gradient distribution from the highest concentration at the PM, intermediate concentration in post-Golgi/endosomal compartments to the lowest level in Golgi (Simon et al., 2014). Although both PtdIns(4,5)P2 and PtdIns4P accumulate at the PM, they contribute differently to PM surface charges, which give rise to different electrostatic filed along the membrane (Simon et al., 2016; Dubois and Jaillais, 2021). It is exactly these special biophysiochemical properties that determine membrane specificity and thus contribute to the organellar identity (Noack and Jaillais, 2017, 2020; Platre et al., 2018). In addition, these PtdInsPs can also recruit different lipid-binding proteins, ensuring their subcelluar localizations and functions at local membranes.
In eukaryotes, phosphoinositides can be interconverted through corresponding modifying enzymes, phosphoinositide kinases and phosphatases respectively. In plants, phosphoinositide phosphatases can be categorized into three major families, the PHOSPHATASE AND TENSIN homologs (PTEN) family, 5-Phosphatases (5-PTases) and Suppressor of Actin (SAC) domain containing phosphatases (hereafter SAC phosphatases) (Gerth et al., 2017). Here, we summarize and discuss the recent advances on this SAC family, especially the subcellular localization, substrate specificity, as well as their interacting proteins and regulators.
Overview of Plant SAC Family
The SAC phosphatase Sac1p was first identified in a genetic screen for the suppressor of actin defective mutant in yeast. The loss of Sac1p led to increased sensitivity to cold treatment, disorganized intracellular actin as well as decreased secretion rate in yeast (Novick et al., 1989), whereas absence of SAC1 resulted in embryonic and preimplantation lethality in Drosophila and mice (Wei et al., 2003; Liu et al., 2008). In yeast and mammals, SAC domain proteins can be further divided into two subfamilies. Members of the first subfamily contain the SAC domain and the type II phosphatidylinositol phosphatase 5-phosphatase domain, including yeast Inp51p, Inp52p, and Inp53p, and mammalian synaptojanin1 and synaptojanin2. The other subfamily includes yeast Sac1p and Fig4p, and mammalian counterparts Sac1, Sac2/INPP5f, and Sac3 (Hughes et al., 2000a; Manford et al., 2010). Based on sequence homology, nine SAC phosphatases (SAC1∼SAC9) have been found in both Arabidopsis thaliana and rice (Oryza sativa L.) genome, respectively, forming three subgroups (Table 1 and Figure 1A; Zhong and Ye, 2003; Novakova et al., 2014): (I) AtSAC1 ∼ AtSAC5 form a clade with medium protein size, exhibiting high similarity to yeast Fig4p (Zhong et al., 2005; Novakova et al., 2014). (II) The Sac1p closest homologs AtSAC6/AtSAC1b, AtSAC7/ROOT HAIR DEFFECTIVE4 (RHD4)/NON-CYTOLEDON PHENOTYPE2 (NCP2)/AtSAC1c, and AtSAC8/AtSAC1a all contain the two transmembrane domains (TMD) at their C-termini (Despres et al., 2003; Zhong and Ye, 2003; Thole et al., 2008). Notably, ectopic expression of AtSAC6 to AtSAC8 rescued the cold-sensitive phenotype associated with ablation of yeast Sac1p (Despres et al., 2003). The recently characterized rice GRAIN NUMBER AND PLANT HEIGHT (OsGH1) was also classified into this subclade according to its protein size and domain organization (Guo et al., 2020; Figure 1B). (III) The third clade only contains one member, AtSAC9, which possesses a larger protein size and a unique protein sequence, a WW protein-protein interaction domain following the N-terminal SAC catalytic domain plus around 1,100 amino acid residues at the C-terminus with unknown function (Zhong and Ye, 2003; Williams et al., 2005; Figure 1B).
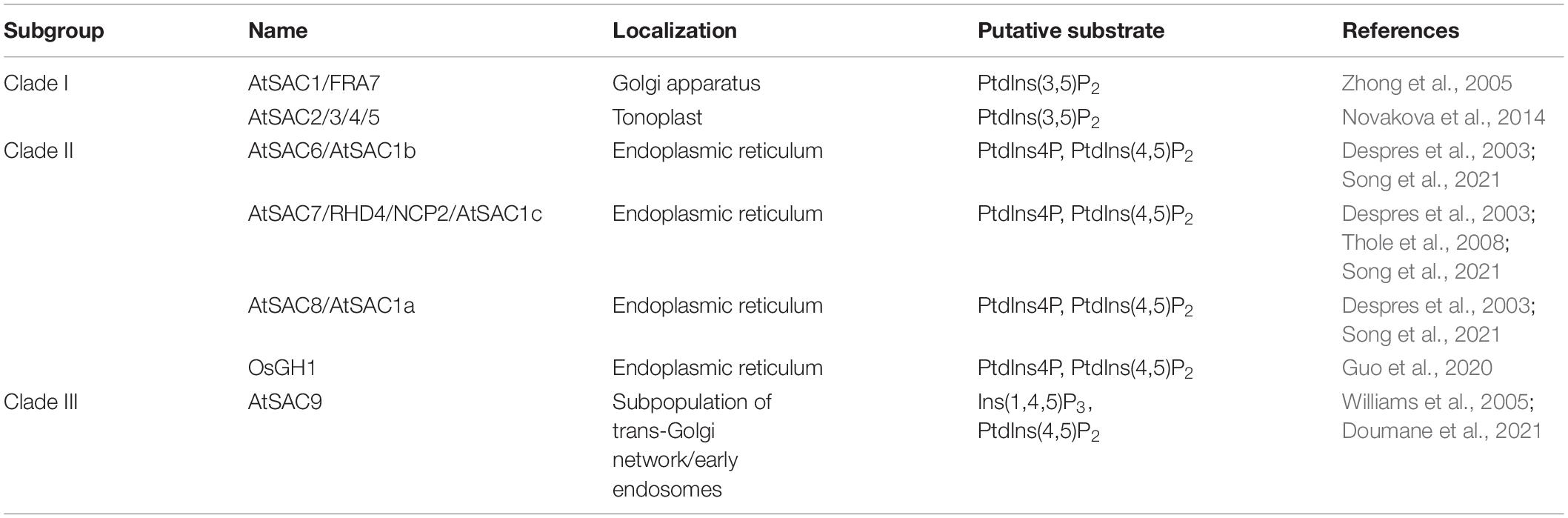
Table 1. Subcellular localization and substrate specificity of currently characterized plant SAC family members.
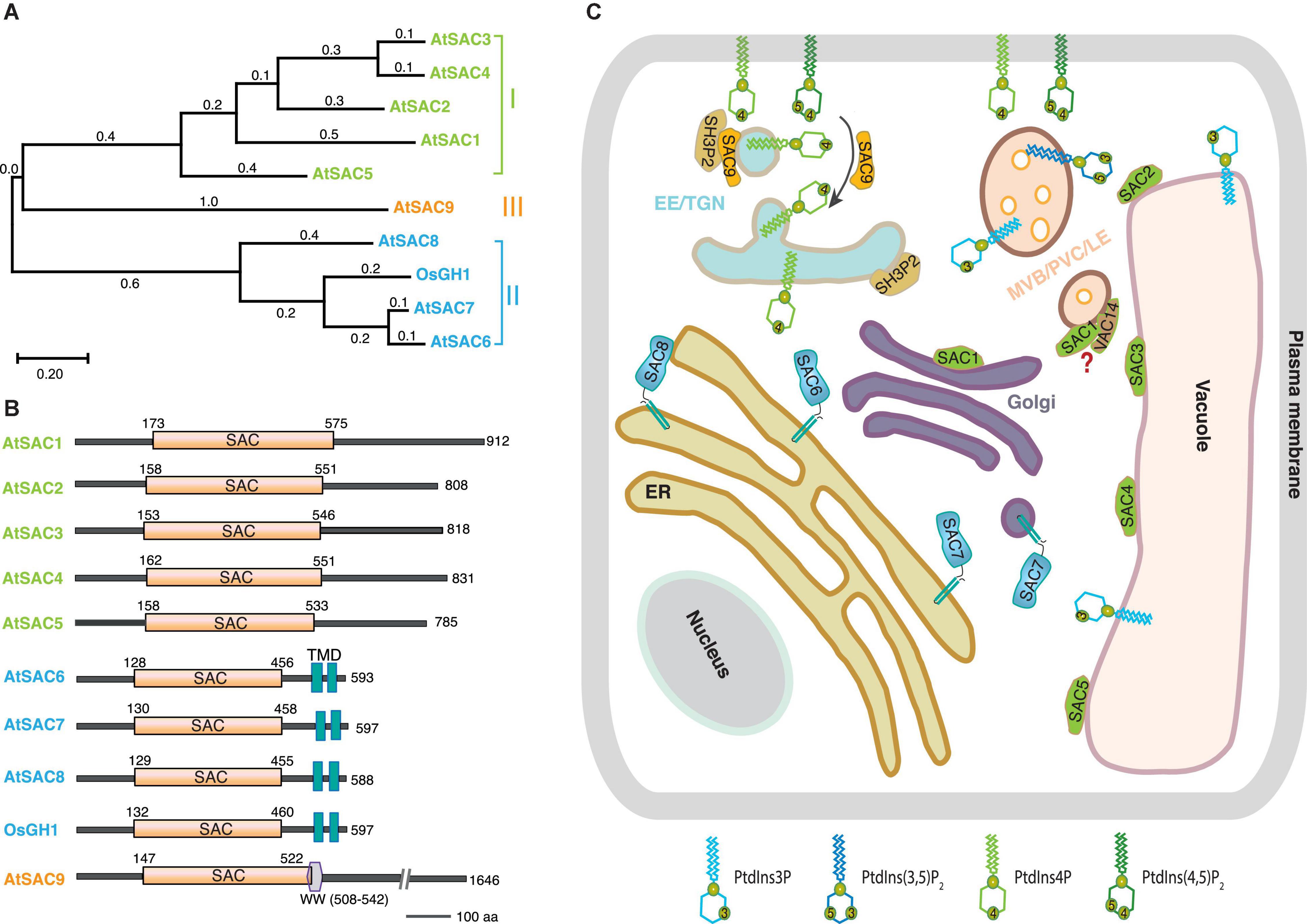
Figure 1. The current overview of plant SACs.(A) Phylogenetic analysis and (B) schematic domain structures of the experimentally characterized plant SACs. The phylogenetic tree was conducted by MEGA X10.2.4 with a maximum likelihood method and a Poisson correction model. The aligned protein sequences were based on 10 plant SAC members, AtSAC1 (At1g22620.1), AtSAC2 (At3g14205.1), AtSAC3 (At3g43220.1), AtSAC4 (At5g20840.1), AtSAC5 (At1g17340.1), AtSAC6 (At5g66020.1), AtSAC7 (At3g51460.1), AtSAC8 (At3G51830.1), AtSAC9 (At3g59770.3), and OsGH1 (Os02g0554300). Domain structures were determined by using the PROSITE program (https://prosite.expasy.org/). Abbreviations, SAC, suppressor of actin domain; TMD, transmembrane domain; WW, a domain with two conserved tryptophan residues. The numbers denote the amino acid residue positions. (C) Schematic representation of sub-cellular distribution of SACs and their putative substrates in the plant cell. AtSAC1 (green) localizes at the Golgi, whereas all the other members (AtSAC2∼AtSAC5) from subgroup I locate in the tonoplast. AtSAC6∼AtSAC8 (blue) from subgroup II are all integrated on ER through two transmembrane motifs. Additionally, AtSAC7 also localizes in post-Golgi secretory compartments. The subclade III member AtSAC9 (orange) localizes in a subpopulation of TGN/EEs close to cell cortex and thus also regulates the distribution pattern of PtdIns(4,5)P2. Meanwhile, AtSAC9 interacts and colocalizes with the endocytic component SH3P2. AtSAC1 colocalizes with VAC14 in MVB/LE/PVC in pollen, but it is still unclear if they can form a complex or not in plants. Abbreviations: TGN/EE, trans-Golgi network/early endosome; ER, endoplasmic reticulum; MVB/LE/PVC, multivesicular body/late endosome/pre-vacuolar compartments. The black arrow indicates the conversion of PtdIns(4,5)P2 to PtdIns4P during endocytosis regulated by AtSAC9.
Substrate Specificity
The SAC domain is highly conserved and profiled with PtdInsP phosphatase activity among eukaryotes (Del Bel and Brill, 2018). In vitro biochemical studies revealed that mouse Sac1 and yeast Sac1p are capable of dephosphorylating PtdIns3P, PtdIns4P and PtdIns(3,5)P2. Whereas the levels of PtdIns3P only slightly increased (around 1.5-fold) in yeast sac1 mutant, 8- to 10-fold elevated levels of PtdIns4P were detected in this mutant (Guo et al., 1999; Hughes et al., 2000b; Nemoto et al., 2000). These data indicated that PtdIns4P, rather than PtdIns3P, might be a preferential substrate for Sac1p, and that enzyme specificities do not always correlate between in vitro and in vivo assays. This could be due to the differences in the subcellular distributions of phospholipids and SAC1 enzymes, or potential additional components involved in the catalysis in vivo.
In Arabidopsis, AtSAC1 exhibited phosphatase activity toward PtdIns(3,5)P2 as observed by an in vitro activity assay (Zhong et al., 2005). Overexpression of AtSAC2∼AtSAC5 resulted in the reduction of PtdIns(3,5)P2, whereas the level of PtdIns(3,5)P2 was unchanged in the sac3 sac4 sac5 triple mutant, perhaps due to a compensatory response. Nevertheless, together with the abundance of PtdIns3P in the tonoplast, it is implied that AtSAC2∼AtSAC5 are responsible for the conversion of PtdIns(3,5)P2 to PtdIns3P (Zhong et al., 2005; Novakova et al., 2014). Overall, AtSAC1∼AtSAC5 share high similarity to the homologous yeast Fig4p on the phosphatase activity toward PtdIns(3,5)P2 (Rudge et al., 2004; Duex et al., 2006).
In line with the in vitro data that AtSAC7 displayed a preference for PtdIns4P, around a 50% increase of PtdIns4P was detected in the root of the sac7 mutant compared to wild type (Thole et al., 2008). By use of an ion chromatography system, both PtdIns4P and PtdIns(4,5)P2 contents were found to dramatically increase in sac6/7, sac6/8 and sac7/8 double mutants, while a slight increase was detected in single mutants (Song et al., 2021), suggesting that PtdIns4P and PtdIns(4,5)P2 might be the substrates for AtSAC6/7/8. In rice, the close related homolog OsGH1 specifically dephosphorylated PtdIns4P and PtdIns(4,5)P2 from both in vitro phosphatase activity assay and binding studies. Moreover, the OsGH1 knockout plant showed a significant increase in endogenous PtdIns4P and PtdIns(4,5)P2 levels, whereas OsGH1-overexpressing plants exhibited reduced PtdIns4P and PtdIns(4,5)P2 levels (Guo et al., 2020), differently from the yeast homolog Sac1p and mammalian SAC1, indicating the functional diversity among the eukaryotes. These findings suggested that the plant SAC family clade II displayed phosphatase activity toward PtdIns4P and PtdIns(4,5)P2. In addition, AtSAC9 was also reported to act as a PtdIns(4,5)P2 phosphatase based on the observation of approximately 4-fold higher PtdIns(4,5)P2 levels in root extracts of loss-of-function sac9 mutant (Williams et al., 2005). Nevertheless, the elevated level of Ins(1,4,5)P3 was also detected in sac9 (Williams et al., 2005), indicating the catalytic diversity of AtSAC9. The overall substrate preference is summarized here based on the previous research (Table 1), but more direct biochemical evidence is still required to validate the substrate specificity for each SAC enzyme.
The Expression Pattern and Subcellular Localization of SACs
In Arabidopsis, transcripts of all the nine AtSACs were detected in most tissues (Despres et al., 2003; Zhong and Ye, 2003; Song et al., 2021), implying pleotropic functions in various life activities. Among the members of subgroup I, AtSAC1 was reported to colocalize with the Golgi marker AVP2-ECFP when co-expressed in carrot protoplasts. The truncation of its C terminus by the fra7 mutation resulted in its mislocalization at the cytoplasm, but the phosphatase catalytic activity was unaffected. These results suggest that the sub-cellular localization of AtSAC1 might be essential for its biological functions (Zhong et al., 2005). Interestingly, in pollen and pollen tubes, over-expression of AtSAC1-GFP under the LAT52 promoter was found to reside at the PVC (Zhang et al., 2018), implying the subcellular distribution pattern may vary in distinct cell types. Furthermore, AtSAC2∼AtSAC5 were all observed to localize in the tonoplast, and this thus ensures the abundance of PtdIns3P in the tonoplast for appropriate maintenance of vacuolar morphology (Novakova et al., 2014). Similarly, the yeast homolog Fig4p was also identified to localize in the limiting membrane of yeast vacuoles, controlling the level of PtdIns(3,5)P2 upon hyperosmotic shock (Rudge et al., 2004), suggesting a conserved role of SACs from this subgroup in vacuole development among eukaryotes.
In yeast, Sac1p is an integral protein residing at endoplasmic reticulum (ER) and Golgi apparatus membrane, and regulates the PtdIns4P pool that is important for vacuolar morphology and Golgi trafficking (Whitters et al., 1993; Foti et al., 2001). The endoplasmic reticulum localization was also identified for mammalian and Drosophila Sac1 (Nemoto et al., 2000; Liu et al., 2008; Forrest et al., 2013). Consistently, the rice homolog OsGH1 was reported to co-localize with the ER marker in rice protoplasts. Disruption of OsGH1 destroyed homeostasis of membrane PtdIns4P and PtdIns(4,5)P2, thus affecting organelle morphology and cell development (Guo et al., 2020). In Arabidopsis, AtSAC6/7/8 targeted to the ER compartment when expressed in tobacco BY2 cells together with a C-terminally fused GFP (Despres et al., 2003), and they are required for both the maintenance of PtdIns(4,5)P2 polarity and the restriction of PtdIns4P at PM in root hairs (Thole et al., 2008; Song et al., 2021). Intriguingly, the N-terminal fusion version EYFP-AtSAC7 which is functional, as demonstrated by rescuing the phenotype of the sac7 loss-of-function mutant, localized at the post-Golgi secretory compartments and regulated the accumulation of PtdIns4P on membrane compartments at the tips of growing root hairs (Thole et al., 2008). Future experimental validations are thus needed to address whether all these three SAC isoforms share the same subcellular localization. A recent study has shown that AtSAC9 localizes to a subpopulation of trans-Golgi network/early endosomes in close vicinity to the PM in meristematic epidermal cells of Arabidopsis roots, restricting the distribution of its substrate PtdIns(4,5)P2 at the PM for proper regulation of endocytosis (Doumane et al., 2021; Table 1 and Figure 1C).
Functions and Mechanisms of SACs and Phosphoinositides
The anionic lipids, PtdIns4P, PtdIns(3,5)P2, and PtdIns(4,5)P2, regulate many cellular processes, including endocytosis, vacuolar trafficking and actin dynamics, though in low abundance (Gerth et al., 2017; Hirano and Sato, 2019; Noack and Jaillais, 2020). Deficiency in these phosphoinositides or dysfunction of their related metabolic enzymes SAC phosphatases usually causes defects in endocytic trafficking and results in abnormal vacuole, Golgi and endosome morphology (Rudge et al., 2004; Liu et al., 2008; Whitley et al., 2009; He et al., 2017). In Arabidopsis, the AtSAC1-truncated mutant fra7 (fragile fiber7) displayed a wide range of defects in plant growth and architecture, including a decrease of cell wall thickness and cell length in fiber cells and vessel elements, and aberrant actin organization (Zhong et al., 2005). However, how loss of AtSAC1 and accumulation of PtdIns(3,5)P2 are related with these various phenotypes is still elusive. Interestingly, seedlings overexpressing AtSAC2 ∼AtSAC5 or the sac2 sac3 sac4 sac5 quadruple mutant and sac3 sac4 sac5 triple mutant all exhibited arrested growth on medium without sucrose. It is proposed that fragmented vacuoles and abnormal protein trafficking toward the vacuole in the sac mutants might explain for the decreased viability in seedlings (Novakova et al., 2014). In yeast, the vacuole-localized Fig4p physically associates with an adaptor-like protein Vac14p in FAB1/PIKfyve protein complex and thus regulates the biosynthesis and turnover of PtdIns(3,5)P2, controlling the vacuolar size (Rudge et al., 2004; Duex et al., 2006; Jin et al., 2008). Moreover, Vac14 can also control Fig4p localization in yeast (Rudge et al., 2004), indicating that the subcellular distribution of the phosphatase is directed by its interacting partner. AtSAC1 was also found to colocalize with the yeast Vac14p homolog VAC14 at PVC in wortmannin (an inhibitor of PI3K)-treated pollen tubes in Arabidopsis (Zhang et al., 2018).
Consistent with the functional redundancy among the SAC family, the single mutant of sac6, sac7, and sac8 didn’t show any obvious defects in plant development except the short-bulged root hairs in sac7/rhd4-1 (Schiefelbein and Somerville, 1990; Thole et al., 2008). However, in the double and triple mutants, significantly delayed embryonic development or even lethal embryos were observed, correlated with a decreased auxin distribution (Song et al., 2021). Furthermore, the retrograde trafficking of auxin efflux carrier PIN-FORMED1 (PIN1) and PIN2 proteins was also affected in the loss of AtSAC7/8, suggesting a role of AtSAC6-AtSAC8 in auxin-mediated development (Song et al., 2021). In rice, over-accumulation of PtdIns4P and PtdIns(4,5)P2 caused by dysfunction of OsGH1 disrupted the activity of actin-related protein 2/3 (Arp2/3) complex for actin nucleation, thus resulting in the reduced plant height, panicle and grain size. In addition, the morphology of Golgi apparatus and chloroplast was also perturbed by elevated PtdInsP4 and PtdIns(4,5)P2 levels (Guo et al., 2020), highlighting the conserved role of PtdInsP homeostasis in organellar organization in rice. In Drosophila, Vesicle-associated membrane protein (VAMP)-Associated Protein (DVAP) physically interacts with SAC1 in controlling phosphoinositide metabolism (Forrest et al., 2013), providing the evidence for the potential working mechanism of SACs in vesicle trafficking.
In Arabidopsis, the endocytic component Src Homology 3 Domain Protein 2 (SH3P2) was identified as a AtSAC9-interacting partner through yeast two-hybrid screening. Absence of AtSAC9 resulted in mis-localization of PtdIns(4,5)P2, and also triggered the altered SH3P2 localization and reduced clathrin-mediated endocytosis rate (Doumane et al., 2021). Moreover, sac9 knock-out mutant exhibited cell wall defects and overall stressed phenotype, including dwarfism, closed stomata, anthocyanin accumulation, and increased transcription of stress response genes (Williams et al., 2005; Vollmer et al., 2011). These results suggest that the AtSAC9 phosphatase is required for modulating phosphoinositide distribution during stress response and cell wall deposition.
Conclusion
In plants, three types of phosphoinositides, PtdIns4P, PtdIns(3,5)P2, and PtdIns(4,5)P2, have been identified so far as substrates of SAC phosphatases within distinct endomembrane compartments. Malfunction of SACs severely disrupts PtdInsP homeostasis reflected on both contents and distribution patterns, thus affecting endomembrane trafficking and organellar organization, and results in a wide range of developmental defects. However, it is still less understood how SACs execute their catalytic functions and associate with membrane in the case of those isoforms lacking TMDs. Future studies might be required to determine the regulatory molecules and interacting partners to better understand cellular pathways in which SACs are involved.
Author Contributions
YM and ST conceptualized and wrote the manuscript. YM made the figures. Both authors contributed to the article and approved the submitted version.
Funding
This work was supported by Youth Innovation Fund of University of Science and Technology of China (No. WK9100000021).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We acknowledge University of Science and Technology of China for support. We are sorry for those whose work is not included due to limited space.
References
Andreyev, A. Y., Fahy, E., Guan, Z., Kelly, S., Li, X., McDonald, J. G., et al. (2010). Subcellular organelle lipidomics in TLR-4-activated macrophages1[S]. J. Lipid Res. 51, 2785–2797. doi: 10.1194/jlr.M008748
Colin, L. A., and Jaillais, Y. (2020). Phospholipids across scales: lipid patterns and plant development. Curr. Opin. Plant Biol. 53, 1–9. doi: 10.1016/j.pbi.2019.08.007
Del Bel, L. M., and Brill, J. A. (2018). Sac1, a lipid phosphatase at the interface of vesicular and nonvesicular transport. Traffic 19, 301–318. doi: 10.1111/tra.12554
Despres, B., Bouissonnié, F., Wu, H.-J., Gomord, V., Guilleminot, J., Grellet, F., et al. (2003). Three SAC1-like genes show overlapping patterns of expression in Arabidopsis but are remarkably silent during embryo development. Plant J. 34, 293–306. doi: 10.1046/j.1365-313X.2003.01720.x
Doumane, M., Lebecq, A., Fangain, A., Bayle, V., Rozier, F., Marquès-Bueno, M. D. M., et al. (2021). The Arabidopsis SAC9 Enzyme defines a cortical population of early endosomes and restricts PI(4,5)P<sub>2</sub> to the Plasma Membrane. bioRxiv [Preprint]. doi: 10.1101/2021.09.10.459735
Dubois, G. A., and Jaillais, Y. (2021). Anionic phospholipid gradients: an uncharacterized frontier of the plant endomembrane network. Plant Physiol. 185, 577–592. doi: 10.1093/plphys/kiaa056
Duex, J. E., Nau, J. J., Kauffman, E. J., and Weisman, L. S. (2006). Phosphoinositide 5-phosphatase Fig 4p is required for both acute rise and subsequent fall in stress-induced phosphatidylinositol 3,5-bisphosphate levels. Eukaryot. Cell 5, 723–731. doi: 10.1128/ec.5.4.723-731.2006
Ejsing, C. S., Sampaio, J. L., Surendranath, V., Duchoslav, E., Ekroos, K., Klemm, R. W., et al. (2009). Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 106, 2136–2141. doi: 10.1073/pnas.0811700106
Forrest, S., Chai, A., Sanhueza, M., Marescotti, M., Parry, K., Georgiev, A., et al. (2013). Increased levels of phosphoinositides cause neurodegeneration in a Drosophila model of amyotrophic lateral sclerosis. Hum. Mol. Genet. 22, 2689–2704. doi: 10.1093/hmg/ddt118
Foti, M., Audhya, A., and Emr, S. D. (2001). Sac1 lipid phosphatase and Stt4 phosphatidylinositol 4-kinase regulate a pool of phosphatidylinositol 4-phosphate that functions in the control of the actin cytoskeleton and vacuole morphology. Mol. Biol. Cell 12, 2396–2411. doi: 10.1091/mbc.12.8.2396
Gerth, K., Lin, F., Menzel, W., Krishnamoorthy, P., Stenzel, I., Heilmann, M., et al. (2017). Guilt by Association: a Phenotype-Based View of the Plant Phosphoinositide Network. Annu. Rev. Plant Biol. 68, 349–374. doi: 10.1146/annurev-arplant-042916-041022
Guo, S., Stolz, L. E., Lemrow, S. M., and York, J. D. (1999). SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J. Biol. Chem. 274, 12990–12995. doi: 10.1074/jbc.274.19.12990
Guo, T., Chen, H. C., Lu, Z. Q., Diao, M., Chen, K., Dong, N. Q., et al. (2020). A SAC Phosphoinositide Phosphatase Controls Rice Development via Hydrolyzing PI4P and PI(4,5)P2. Plant Physiol. 182, 1346–1358. doi: 10.1104/pp.19.01131
He, K., Marsland Iii, R., Upadhyayula, S., Song, E., Dang, S., Capraro, B. R., et al. (2017). Dynamics of phosphoinositide conversion in clathrin-mediated endocytic traffic. Nature 552, 410–414. doi: 10.1038/nature25146
Hirano, T., Konno, H., Takeda, S., Dolan, L., Kato, M., Aoyama, T., et al. (2018). PtdIns(3,5)P2 mediates root hair shank hardening in Arabidopsis. Nat. Plants 4, 888–897. doi: 10.1038/s41477-018-0277-8
Hirano, T., Munnik, T., and Sato, M. H. (2017). Inhibition of phosphatidylinositol 3,5-bisphosphate production has pleiotropic effects on various membrane trafficking routes in Arabidopsis. Plant Cell Physiol. 58, 120–129. doi: 10.1093/pcp/pcw164
Hirano, T., and Sato, M. H. (2019). Diverse Physiological Functions of FAB1 and Phosphatidylinositol 3,5-Bisphosphate in Plants. Front. Plant Sci. 10:274. doi: 10.3389/fpls.2019.00274
Hughes, W. E., Cooke, F. T., and Parker, P. J. (2000a). Sac phosphatase domain proteins. Biochem. J. 350, 337–352. doi: 10.1042/bj3500337
Hughes, W. E., Woscholski, R., Cooke, F. T., Patrick, R. S., Dove, S. K., McDonald, N. Q., et al. (2000b). SAC1 encodes a regulated lipid phosphoinositide phosphatase, defects in which can be suppressed by the homologous Inp52p and Inp53p phosphatases. J. Biol. Chem. 275, 801–808. doi: 10.1074/jbc.275.2.801
Jin, N., Chow, C. Y., Liu, L., Zolov, S. N., Bronson, R., Davisson, M., et al. (2008). VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P2 in yeast and mouse. EMBO J. 27, 3221–3234. doi: 10.1038/emboj.2008.248
Liu, Y., Boukhelifa, M., Tribble, E., Morin-Kensicki, E., Uetrecht, A., Bear, J. E., et al. (2008). The Sac1 Phosphoinositide Phosphatase Regulates Golgi Membrane Morphology and Mitotic Spindle Organization in Mammals. Mol. Biol. Cell 19, 3080–3096. doi: 10.1091/mbc.e07-12-1290
Manford, A., Xia, T., Saxena, A. K., Stefan, C., Hu, F., Emr, S. D., et al. (2010). Crystal structure of the yeast Sac1: implications for its phosphoinositide phosphatase function. EMBO J. 29, 1489–1498. doi: 10.1038/emboj.2010.57
Nemoto, Y., Kearns, B. G., Wenk, M. R., Chen, H., Mori, K., Alb, J. G., et al. (2000). Functional Characterization of a Mammalian Sac1 and Mutants Exhibiting Substrate-specific Defects in Phosphoinositide Phosphatase Activity*. J. Biol. Chem. 275, 34293–34305. doi: 10.1074/jbc.M003923200
Noack, L. C., and Jaillais, Y. (2017). Precision targeting by phosphoinositides: how PIs direct endomembrane trafficking in plants. Curr. Opin. Plant Biol. 40, 22–33. doi: 10.1016/j.pbi.2017.06.017
Noack, L. C., and Jaillais, Y. (2020). Functions of Anionic Lipids in Plants. Annu. Rev. Plant Biol. 71, 71–102. doi: 10.1146/annurev-arplant-081519-035910
Novakova, P., Hirsch, S., Feraru, E., Tejos, R., van Wijk, R., Viaene, T., et al. (2014). SAC phosphoinositide phosphatases at the tonoplast mediate vacuolar function in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 111, 2818–2823. doi: 10.1073/pnas.1324264111
Novick, P., Osmond, B. C., and Botstein, D. (1989). Suppressors of yeast actin mutations. Genetics 121, 659–674. doi: 10.1093/genetics/121.4.659
Platre, M. P., Noack, L. C., Doumane, M., Bayle, V., Simon, M. L. A., Maneta-Peyret, L., et al. (2018). A Combinatorial Lipid Code Shapes the Electrostatic Landscape of Plant Endomembranes. Dev. Cell 45, 465–480.e11. doi: 10.1016/j.devcel.2018.04.011
Rudge, S. A., Anderson, D. M., and Emr, S. D. (2004). Vacuole size control: regulation of PtdIns(3,5)P2 levels by the vacuole-associated Vac14-Fig4 complex, a PtdIns(3,5)P2-specific phosphatase. Mol. Biol. Cell 15, 24–36. doi: 10.1091/mbc.e03-05-0297
Schiefelbein, J. W., and Somerville, C. (1990). Genetic Control of Root Hair Development in Arabidopsis thaliana. Plant Cell 2, 235–243. doi: 10.1105/tpc.2.3.235
Simon, M. L., Platre, M. P., Marques-Bueno, M. M., Armengot, L., Stanislas, T., Bayle, V., et al. (2016). A PtdIns(4)P-driven electrostatic field controls cell membrane identity and signalling in plants. Nat. Plants 2:16089. doi: 10.1038/nplants.2016.89
Simon, M. L. A., Platre, M. P., Assil, S., van Wijk, R., Chen, W. Y., Chory, J., et al. (2014). A multi-colour/multi-affinity marker set to visualize phosphoinositide dynamics in Arabidopsis. Plant J. 77, 322–337. doi: 10.1111/tpj.12358
Song, L., Wang, Y., Guo, Z., Lam, S. M., Shui, G., and Cheng, Y. (2021). NCP2/RHD4/SAC7, SAC6 and SAC8 phosphoinositide phosphatases are required for PtdIns4P and PtdIns(4,5)P2 homeostasis and Arabidopsis development. New Phytol. 231, 713–725. doi: 10.1111/nph.17402
Thole, J. M., Vermeer, J. E., Zhang, Y., Gadella, T. W. Jr., and Nielsen, E. (2008). Root hair defective4 encodes a phosphatidylinositol-4-phosphate phosphatase required for proper root hair development in Arabidopsis thaliana. Plant Cell 20, 381–395. doi: 10.1105/tpc.107.054304
Van Leeuwen, W., Vermeer, J. E. M., Gadella, T. W. J. Jr., and Munnik, T. (2007). Visualization of phosphatidylinositol 4,5-bisphosphate in the plasma membrane of suspension-cultured tobacco BY-2 cells and whole Arabidopsis seedlings. Plant J. 52, 1014–1026. doi: 10.1111/j.1365-313X.2007.03292.x
Vermeer, J. E. M., van Leeuwen, W., Tobeña-Santamaria, R., Laxalt, A. M., Jones, D. R., Divecha, N., et al. (2006). Visualization of PtdIns3P dynamics in living plant cells. Plant J. 47, 687–700. doi: 10.1111/j.1365-313X.2006.02830.x
Vollmer, A. H., Youssef, N. N., and DeWald, D. B. (2011). Unique cell wall abnormalities in the putative phosphoinositide phosphatase mutant AtSAC9. Planta 234, 993–1005. doi: 10.1007/s00425-011-1454-4
Wei, H.-C., Sanny, J., Shu, H., Baillie, D. L., Brill, J. A., Price, J. V., et al. (2003). The Sac1 Lipid Phosphatase Regulates Cell Shape Change and the JNK Cascade during Dorsal Closure in Drosophila. Curr. Biol. 13, 1882–1887. doi: 10.1016/j.cub.2003.09.056
Whitley, P., Hinz, S., and Doughty, J. (2009). Arabidopsis FAB1/PIKfyve Proteins Are Essential for Development of Viable Pollen. Plant Physiol. 151, 1812–1822. doi: 10.1104/pp.109.146159
Whitters, E. A., Cleves, A. E., McGee, T. P., Skinner, H. B., and Bankaitis, V. A. (1993). SAC1p is an integral membrane protein that influences the cellular requirement for phospholipid transfer protein function and inositol in yeast. J. Cell Biol. 122, 79–94. doi: 10.1083/jcb.122.1.79
Williams, M. E., Torabinejad, J., Cohick, E., Parker, K., Drake, E. J., Thompson, J. E., et al. (2005). Mutations in the Arabidopsis phosphoinositide phosphatase gene SAC9 lead to overaccumulation of PtdIns(4,5)P2 and constitutive expression of the stress-response pathway. Plant Physiol. 138, 686–700. doi: 10.1104/pp.105.061317
Zhang, W.-T., Li, E., Guo, Y.-K., Yu, S.-X., Wan, Z.-Y., Ma, T., et al. (2018). Arabidopsis VAC14 Is Critical for Pollen Development through Mediating Vacuolar Organization. Plant Physiol. 177, 1529–1538. doi: 10.1104/pp.18.00495
Zhong, R., Burk, D. H., Nairn, C. J., Wood-Jones, A., Morrison, W. H. III, and Ye, Z. H. (2005). Mutation of SAC1, an Arabidopsis SAC domain phosphoinositide phosphatase, causes alterations in cell morphogenesis, cell wall synthesis, and actin organization. Plant Cell 17, 1449–1466. doi: 10.1105/tpc.105.031377
Keywords: phosphoinositides, SAC phosphatases, subcellular localization, trafficking, Arabidopsis
Citation: Mao Y and Tan S (2021) Functions and Mechanisms of SAC Phosphoinositide Phosphatases in Plants. Front. Plant Sci. 12:803635. doi: 10.3389/fpls.2021.803635
Received: 28 October 2021; Accepted: 30 November 2021;
Published: 16 December 2021.
Edited by:
Ruixi Li, Southern University of Science and Technology, ChinaReviewed by:
Martin Potocký, Institute of Experimental Botany, Academy of Sciences of the Czech Republic, CzechiaTakashi Aoyama, Kyoto University, Japan
Copyright © 2021 Mao and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanbo Mao, eWJtYW9AdXN0Yy5lZHUuY24=; Shutang Tan, c3R0YW5AdXN0Yy5lZHUuY24=
 Yanbo Mao*
Yanbo Mao* Shutang Tan
Shutang Tan