
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 04 January 2022
Sec. Plant Breeding
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.794373
This article is part of the Research Topic Mechanisms of Abiotic Stress Responses and Tolerance in Plants: Physiological, Biochemical and Molecular Interventions, volume II View all 37 articles
 Ali Raza1†
Ali Raza1† Javaria Tabassum2
Javaria Tabassum2 Zainab Zahid3
Zainab Zahid3 Sidra Charagh2†
Sidra Charagh2† Shanza Bashir3
Shanza Bashir3 Rutwik Barmukh4
Rutwik Barmukh4 Rao Sohail Ahmad Khan5
Rao Sohail Ahmad Khan5 Fernando Barbosa Jr.6
Fernando Barbosa Jr.6 Chong Zhang1
Chong Zhang1 Hua Chen1
Hua Chen1 Weijian Zhuang1*
Weijian Zhuang1* Rajeev K. Varshney1,4,7*
Rajeev K. Varshney1,4,7*
Food safety has emerged as a high-urgency matter for sustainable agricultural production. Toxic metal contamination of soil and water significantly affects agricultural productivity, which is further aggravated by extreme anthropogenic activities and modern agricultural practices, leaving food safety and human health at risk. In addition to reducing crop production, increased metals/metalloids toxicity also disturbs plants’ demand and supply equilibrium. Counterbalancing toxic metals/metalloids toxicity demands a better understanding of the complex mechanisms at physiological, biochemical, molecular, cellular, and plant level that may result in increased crop productivity. Consequently, plants have established different internal defense mechanisms to cope with the adverse effects of toxic metals/metalloids. Nevertheless, these internal defense mechanisms are not adequate to overwhelm the metals/metalloids toxicity. Plants produce several secondary messengers to trigger cell signaling, activating the numerous transcriptional responses correlated with plant defense. Therefore, the recent advances in omics approaches such as genomics, transcriptomics, proteomics, metabolomics, ionomics, miRNAomics, and phenomics have enabled the characterization of molecular regulators associated with toxic metal tolerance, which can be deployed for developing toxic metal tolerant plants. This review highlights various response strategies adopted by plants to tolerate toxic metals/metalloids toxicity, including physiological, biochemical, and molecular responses. A seven-(omics)-based design is summarized with scientific clues to reveal the stress-responsive genes, proteins, metabolites, miRNAs, trace elements, stress-inducible phenotypes, and metabolic pathways that could potentially help plants to cope up with metals/metalloids toxicity in the face of fluctuating environmental conditions. Finally, some bottlenecks and future directions have also been highlighted, which could enable sustainable agricultural production.
Over the last few decades, intensive anthropogenic activities and modern farming practices have led to the contamination of ecosystems by toxic metals/metalloids (Rai et al., 2019), an alarming global concern. Toxic metals/metalloids are ubiquitous in the earth’s crust and possess multiple benefits but can be harmful to the ecosystem when present in excess amounts (Gashi et al., 2020). Abiotic stresses (e.g., toxic metals/metalloids contamination, drought, salinity, etc.) are amongst some important factors affecting the growth and productivity of crop plants, resulting in up to 70% yield losses (Rai et al., 2019; Raza et al., 2020, 2021a; Roorkiwal et al., 2020; Varshney et al., 2021c). Climate changes give rise to several environmental stresses, including toxic metals/metalloids. Consequently, climate change significantly impacts the toxic metals/metalloids pollutions based on bioavailability, fate, and toxicity (Wijngaard et al., 2017; Wu et al., 2017; Oyewo et al., 2020). Among various environmental stresses, drought stress may cause an upsurge in eutrophication and toxic metals/metalloids meditations. Whereas the flooding stress may cause more toxic metals/metalloids meditations owing to desorption or re-suspension signifying that climate change determined impacts on toxic metals/metalloids transport is a composite and dynamic environmental problem, demanding a systematic understanding of toxic metals/metalloids accessibility, transport, and uptake pathways (Wijngaard et al., 2017; Wu et al., 2017; Oyewo et al., 2020).
Plants require mineral nutrients for proper growth and development. For instance, they need macro-nutrients and micro-nutrients to carry out primary functions like metabolism, photosynthesis, synthesis of enzymes, DNA and pigments, chlorophyll (Chl) functioning, nitrogen (N) fixation, etc. (Malar et al., 2016; Patel et al., 2020; Salim and Raza, 2020). Among these nutrients, some metals/metalloids such as copper (Cu), zinc (Zn), nickel (Ni), iron (Fe), selenium (Se), etc. are required in trace amounts (Karthika et al., 2018; Hasanuzzaman et al., 2020a), and can be toxic when taken up in excess by the plants (Pandey et al., 2019; Hasanuzzaman et al., 2020a). Besides, other non-essential metals/metalloid elements, like arsenic (As), cadmium (Cd), lead (Pb), mercury (Hg), are toxic to plant even at low amounts (Asati et al., 2016; Mawia et al., 2020; Raza et al., 2021a). The toxicity of non-essential toxic metals/metalloids in plants can lead to severe damages, including reduced or inhibited growth, low biomass and reduced production, chlorosis, water and nutrient imbalance, denaturation of essential enzymes and proteins, production of reactive oxygen species (ROS), disturbed electron transport chain, lipid peroxidation, and at times ultimately plant death (Rucińska-Sobkowiak, 2016; Venkatachalam et al., 2017; Jalmi et al., 2018; Tiwari and Lata, 2018; Gashi et al., 2020; Salehi et al., 2021). As a mechanism of toxicity, some non-essential toxic metals/metalloids tend to replace the essential ones in enzymes and pigments, thus disrupting their functioning (Sarwar et al., 2017; Erickson et al., 2019). Thus, plants need these metals/metalloids at a low amount to perform their metabolic activities.
Various biotechnological techniques are being implied to get an insight and in-depth understanding of the mechanisms and pathways involved in plant responses and tolerance toward toxic metals/metalloids toxicity. “Omics” approaches such as genomics (genes and complete DNA), transcriptomics (coding RNA and its types), proteomics (proteins), metabolomics (profiling of metabolites), ionomics (micro and macro ions profiling), miRNAomics (non-coding RNA), and phenomics (plant phenotype) are being widely implied for this purpose (Jamla et al., 2021a; Khan et al., 2021; Raza et al., 2021a). Molecular regulators (genes, RNA, metabolites, and proteins) and their related activities like replication, translation, post-translation, transcription, etc., play a pivotal role in the performance and maintenance of critical plant functions. Since they also determine plant responses to toxic metals/metalloids stress, the understanding of regulatory principles at the genetic level is necessary. Genomics and transcriptomics approaches hold the potential to provide insights into such complex and intricate processes during plant development (Pirzadah et al., 2019). Proteomics offers insights into the stress-inducible proteins and their involvement in mitigation against toxic metals/metalloids toxicity (Wen et al., 2019). It also provides cues into protein profiles from the cellular to organ level and offers insights into protein behavior under stress conditions (Wen et al., 2019). Metabolomics enables an understanding of the differentially regulated metabolites and complex metabolic activities occurring within the plant under diverse conditions (Razzaq et al., 2019; Raza, 2020). Furthermore, ionomics provides insights into the nutrient and trace element composition of the plant, as well as activities and mechanisms involved in uptake, storage, assimilation, and the plants’ responses to toxic metals (Stich et al., 2020). MicroRNAs (miRNAs), a group of single-stranded, non-coding micro RNAs, have been shown to regulate gene expression at a post-transcriptional level. Plants respond to stress (including toxic metals/metalloids) by triggering the miRNAs that work by cleaving or neutralizing the transcribed mRNA according to the plant needs under stressed conditions (Dubey et al., 2021; Jamla et al., 2021b).
In this review, we start by discussing the physiological, biochemical, and molecular responses of the plants toward toxic metals/metalloids toxicity. We then describe recent innovations in “omics” approaches such as genomics, transcriptomics, proteomics, metabolomics, ionomics, miRNAomics, and phenomics that could empower the development of toxic metals/metalloids tolerant plants. Further, we summarize some important online databases/tools for the integration of omics data. Additionally, an overview of some plant-based remediation approaches has also been discussed. We aim to offer a comprehensive overview of intricate plant responses to toxic metal stress and highlight the potential of omics approaches for developing toxic metals/metalloids tolerant plants in future.
Since plants constitute the primary level of the food chain, harmful metals/metalloids may enter the food chain through plants, thereby posing a threat of exposure to animals and especially humans, and may cause several physiological, morphological, and metabolic disorders and abnormalities (Rai et al., 2019). Increased metals/metalloids toxicity has resulted in reduced crop and food production by disturbing the demand and supply equilibrium, demanding an urgent need to develop strategies for combating the situation and enhancing plant tolerance toward toxic metals/metalloids (Rai et al., 2019; Gashi et al., 2020).
Plants being sessile organisms, have no escape when it comes to unfavorable environmental conditions. Several physiological, biochemical, and molecular responses are generated in the plant due to toxic metals/metalloids toxicity, as these toxicants interfere with several natural processes (Figure 1; Raza et al., 2021a). The interference of toxic metals/metalloids with plant processes takes place mainly through: (i) competition at the root absorption surface level with the essential metal ions for uptake; (ii) displacement of ions from specific binding sites and essential biomolecules; (iii) displacement of a critical functional group from proteins (e.g., sulfhydryl group, -SH), denaturing them and making them inactive; and (iv) the formation of ROS which interacts with and deteriorates biomolecules and disrupts the metabolic activities (Ghori et al., 2019; Mishra S. et al., 2019).
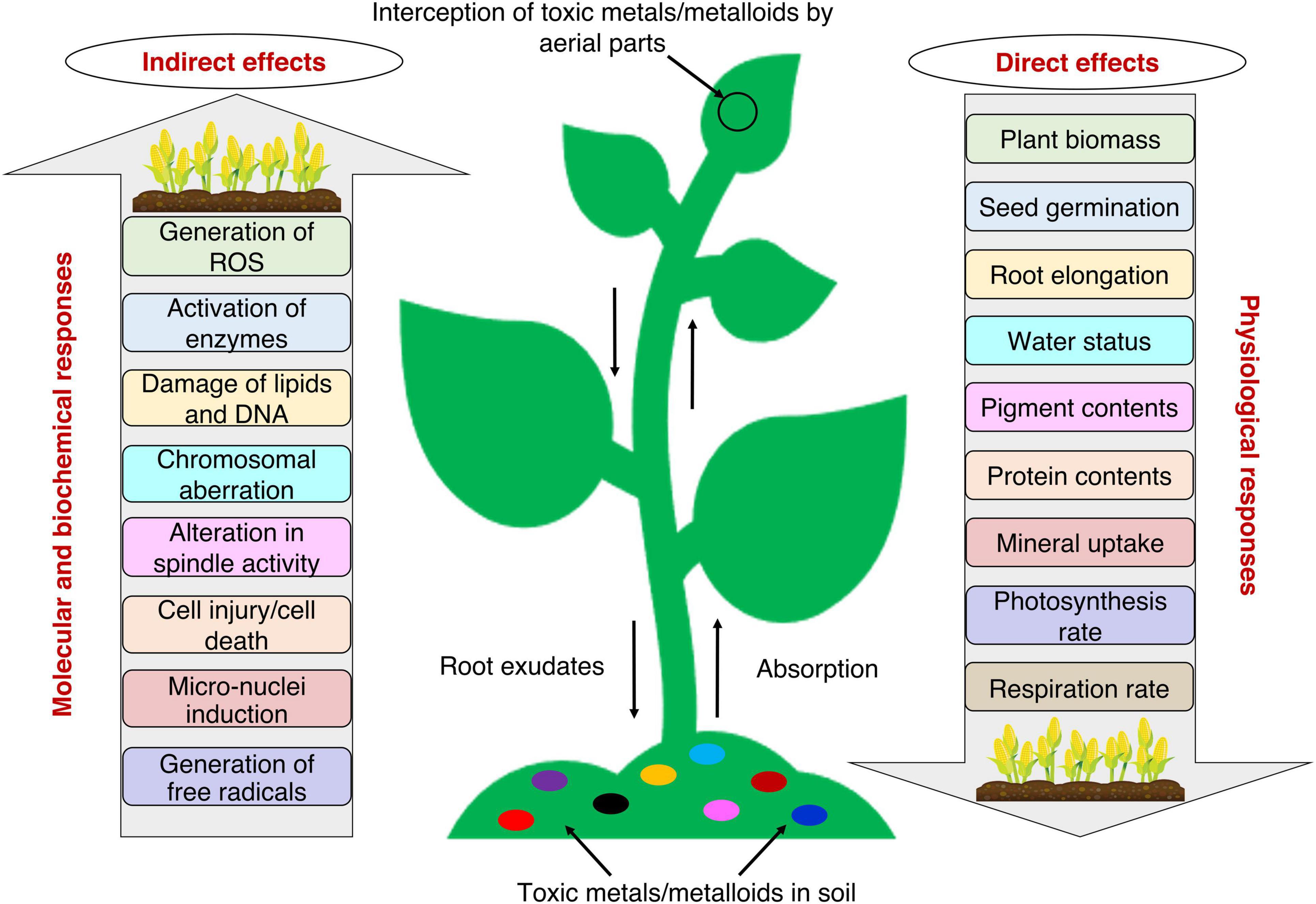
Figure 1. Plant responses to toxic metals/metalloids toxicity with possible direct and indirect effects on crop productivity. Plants interact with toxic metals/metalloids via above-ground and/or below-ground parts. The toxic effects of several toxic metals/metalloids decrease the physiological responses and increase the molecular and biochemical responses.
The early responses of the plant to toxic metals/metalloids stress are easier to study, whereas signal transduction at later stages of exposure is difficult to detect (Jalmi et al., 2018). Detection of stress triggers plants’ defense mechanisms through various pathways (Raza et al., 2021a). For instance, root endings detect the presence of metals and send signals from the roots throughout the plant cells and into different cell organelles (Dalvi and Bhalerao, 2013). Plants first attempt to prevent the entry of metal ions into the body by immobilizing ions with the help of their mycorrhizal associations and converting them to complexes of organic acids, amino acids, etc., secreted from the root endings (Dalvi and Bhalerao, 2013; Jalmi et al., 2018). Toxic metals/metalloids that enter the plant body may get accumulated within vacuoles and/or get bound to the cell wall and proteins, thereby causing a further modification of physiological, biochemical, and molecular activities when transported to different plant parts (Figure 1; Jalmi et al., 2018).
Toxic metals/metalloids toxicity induces and alters many physiological responses in plants. One of the most observed plant responses is the reduction or inhibition of plant growth and development (Rucińska-Sobkowiak, 2016; Ghori et al., 2019; Raza et al., 2020, 2021a). For instance, Zn, Cd, Cu, Hg, As, Ni, chromium (Cr), and many other prominent metal toxicants have been reported to retard plant growth, seed germination and cause other morphological modifications (Ghori et al., 2019). Moreover, Pb has been reported to reduce growth in alfalfa (Hattab et al., 2016) and Acalypha indica (Venkatachalam et al., 2017). Increasing concentrations of Hg from 5 to 80 μg mL–1 decreased roots, shoots, and leaves content in Jatropha curcas (Marrugo-Negrete et al., 2016). Nickel has been reported to reduce seed germination in sunflower due to lower α-amylase and protease activities (Ashraf et al., 2011). Furthermore, adverse effects on leaf area due to Cd contamination were observed in tomato (Rehman et al., 2011), and production of chlorosis by Cd was reported in Phaseolus vulgaris (Bahmani et al., 2012). An account of the effect on pollen germination has been found in Pisum sativum by Cd and Cu toxicity (Sabrine et al., 2010).
Since roots represent the first plant parts to encounter toxic metals/metalloids stress in the soil, a decrease in root growth, root hair surface, and enhanced root dieback has been observed (Rucińska-Sobkowiak, 2016). Root cells tend to harden their cell walls and inhibit growth by avoiding the entrance of Cu ions in Festuca arundinacea and Lolium perenne (Zhao et al., 2010). Root cells further stimulated the entered metals/metalloids ions into the vacuole to prevent their interference with the organelle functions. For instance, Eleftheriou et al. (2015) reported Cr sequestration in the cell wall and vacuole in Arabidopsis thaliana. The plasma membranes in cells work in a highly controlled fashion, preventing unwanted materials from entering the organelles and mediating the harmful effects of various stresses. Despite this, Cd has been reported to disrupt the structure and functionality of the plasma membrane (Janicka-Russak et al., 2012). Such effects can further produce many undesirable outcomes, such as disruption of water and nutrient supply.
Although few metals/metalloids such as Zn, Fe, Cu, cobalt (Co), molybdenum (Mo), and manganese (Mn) are considered as essential elements for photosynthesis (Schmidt et al., 2020), their increased concentration has been shown to alter photosynthetic activities in plants (Chandra and Kang, 2016). Some metals/metalloids (e.g., Fe) act as necessary co-factors for various enzymes involved in the photosynthetic process (Balk and Schaedler, 2014); however, they tend to retard normal activities when present at higher concentrations (Asati et al., 2016). For instance, Pb and Cd were found to inhibit photosynthetic pigments in Davidia involucrate (Yang Y. et al., 2020). Toxic metals/metalloids toxicity also affects the overall photosynthetic apparatus and the rate of photosynthesis, causing a reduction in Chl content and mesophyll thickness (Pereira et al., 2016; Schmidt et al., 2020). In wheat, Cr has been reported to reduce the electron transport chain rate and negatively affect light-harvesting complex of photosystem II (PSII) by reducing the number of active reaction sites (Mathur et al., 2016).
Toxic metals/metalloids toxicity tends to affect the nutrient uptake, water uptake, and their translocation and assimilation in plants. Several alterations in plant functions, including dehydration due to inhibition of water translocation (Hussain et al., 2013), reduced transpiration, and altered stomatal activity, have also been observed (Wani et al., 2018). Decreased transpiration was observed for poplar hybrid in response to Cd, Cu, Cr, and Zn exposure (Chandra and Kang, 2016). In contrast, reduced stomatal activity was observed in Pistacia vera due to Zn toxicity (Tavallali, 2017). Moreover, xylem and phloem functionality were affected due to toxic metals/metalloids contamination, e.g., reduction in root xylem area was observed in Salix caprea in response to Zn and Cd exposure (Vaculík et al., 2012). Toxic ions have been found to compete with essential ions for translocation throughout the plant (Rucińska-Sobkowiak, 2016). For example, Cd was identified to compete with essential ions such as Fe, Mg, and calcium (Ca), among others (Raza et al., 2020).
Toxic metals/metalloids usually trigger biochemical responses by displacing/replacing essential ions or blocking functional groups. They compete with crucial nutrients at bindings sites of enzymes, rendering them denatured and inhibiting their activity (Ghori et al., 2019). For instance, the biochemical response of Cr on mung bean seeds showed a significant decline in protein, Chl, and starch content under 50% metal contaminated soil (Rath et al., 2019). Similarly, Se-stressed rice plants were found to promote Chl and protein degradation in response to metal accumulation. Alteration in these biochemical processes caused significant physiological toxicity symptoms in plants (Gouveia et al., 2020). Different ornamental plant species grown under urban Pb-contaminated soil showed a decline in their net photosynthetic rate and root activity due to stress responses. In contrast, the levels of soluble sugars, leaf proline content, and membrane stability index, altogether maintain osmotic adjustment, form metal-proline complexes and identify the extent of membrane damage, respectively, were found to increase significantly (Song et al., 2020b). The biochemical responses of kenaf (Hibiscus cannabinus L.) revealed Cu-induced damage in root and shoot tissues as observed through excess production of malondialdehyde (MDA), hydrogen peroxide (H2O2), and electrolyte leakage. Its negative impacts on the plant were mediated through antioxidative enzyme activities, which increase with enhancing metal concentration (Saleem et al., 2020).
The assessment of cowpea subjected to Fe toxicity showed an increase in the MDA content (Ifie et al., 2020). Another study on halophytic phytoremediator of Cu, Sesuvium portulacastrum, also reported similar findings where plants were affected by the toxic effects of Cu at higher concentrations. The MDA production was mediated through an active antioxidant mechanism in plants to counteract the damage caused by ROS production (Lokhande et al., 2020). The antioxidants work by searching, neutralizing, and removing the reactive species (Hasanuzzaman et al., 2020b). An assessment of biochemical responses in alfalfa plants showed high peroxidase (POD) and glutathione S-transferase (GST) activity upon its exposure to Ni. The data on antioxidant enzymes was also in corroboration with the expression transcripts of the Prx1C gene, showing elevated biochemical activity in plants as a function of plant defense (Helaoui et al., 2020).
Furthermore, carbon dioxide (CO2) fixation and assimilation activities are also greatly affected by metal toxicity. For instance, inhibition of CO2 assimilation was reported in wheat by excessive concentrations of Cd and Zn (Paunov et al., 2018). Rubisco activity was observed to be reduced due to toxic metals/metalloids stress, as they react with the thiol group of the enzyme and render denaturing it (Son et al., 2014). Nitrogen metabolism is a crucial mechanism in plant growth and development that suffers from metal stress. For example, Cr lowered nitrate and ammonia assimilation enzymes in Cyamopsis tetragonoloba by enhancing their destruction through protease increase (Sangwan et al., 2014).
As a defense mechanism to toxic metals/metalloids stimuli, root growth is reduced and inhibited by terminating the mitotic activity (Campos et al., 2018). Pavlova (2017) detected a decrease in meristem mitosis in Plantago lanceolate due to Ni toxicity, inhibiting root elongation. Similarly, Cr has also been described to inhibit mitotic cell division by delaying and extending the cell cycle in rice (Sundaramoorthy et al., 2010). ROS production in response to toxic metals/metalloids stress can cause damage to cellular membranes, nucleic acids, lipids, and proteins (Hasanuzzaman et al., 2020b). For example, Cu and Zn caused severe damage to cellular structures in the Populus species (Benyó et al., 2016). Plants also produce various metal binding structures, chelating agents or ligands as a defense or detoxification mechanism, such as organic acids, amino acids, phytochelatins (PCs), and metallothionein (MT), among others (Raza et al., 2020, 2021a).
Although plants have different mechanisms to prevent metal ions from reaching the nucleus, the entry of metals/metalloids causes cross-linking of DNA and proteins, DNA mutation (deletion, addition, modification), alter the base structure of the DNA, or cause DNA strands to break (Emamverdian et al., 2015). They further disrupt lipids, damage Chl, disturb cell homeostasis, and also interfere with the electron transport chain and energy production and assimilation through ATP molecules, leading to programmed cell death (Parmar et al., 2013).
Exposure to Cd was found to display an enhanced expression of Chit5 (one of the chitinase encoding genes) in roots by 7. 3-, 3. 9-, and 3.7-fold, compared with its expression in shoots in Amaranth. cruentus, A. hypochondriacus × A. hybridus, and A. hypochondriacus × A. hybridus, respectively (Lancíková et al., 2020). Alfalfa plant experienced a 25- and 29-fold increase in detoxifying enzyme superoxide dismutase (SOD) transcripts in response to bulk Cu and nano Cu, respectively (Cota-Ruiz et al., 2020). Research has established melatonin expression to help alleviate Cd stress in radish plants (Xu L. et al., 2020). Gao et al. (2020) demonstrated PtrMTP (Populus trichocarpa metal tolerance proteins) gene to play a significant role in inducing tolerance toward metal contamination in roots, stems, and leaves of Populus trichocarpa through homeostasis and detoxification mechanisms.
MicroRNAs have been reported to play a significant role in plant response and tolerance to toxic metals/metalloids stress by regulating chelation, antioxidant response, auxin signaling, and cytokinin signaling, among others (Ding et al., 2020). For instance, Casarrubia et al. (2020) reported microRNA to play a significant role in combination with the mycorrhizal association for regulating the response of Vaccinium myrtillus against Cd contamination. MicroRNA expression in terms of biosynthesis/biogenesis of various secondary metabolites has been found to play a major role in enhancing Al and Cd tolerance in transgenic tobacco (Cedillo-Jimenez et al., 2020). Notably, the expression of different microRNAs, including miR156, miR162, miR164, miR166, miR172, miR398, and miR408, were found to regulate the expression of their respective target genes in Carthamus tinctorius as a result of Cd exposure (Kouhi et al., 2020).
Plant responses to toxic metals/metalloids toxicity rely on the regulation of molecular factors. Therefore, an integrated omics approach has been extensively used to comprehend the plant’s biological interactions and molecular mechanisms against toxic metals/metalloids toxicity. Regardless of the incredible advancements in genomics, it is essential to evaluate other omics tools for wide-ranging knowledge at the molecular level (Figure 2). Scientific investigations and existing information derived by omics tools target signaling pathways, key molecular regulators, and integrated mechanisms to enhance tolerance toward toxic metals/metalloids toxicity for crop improvement.
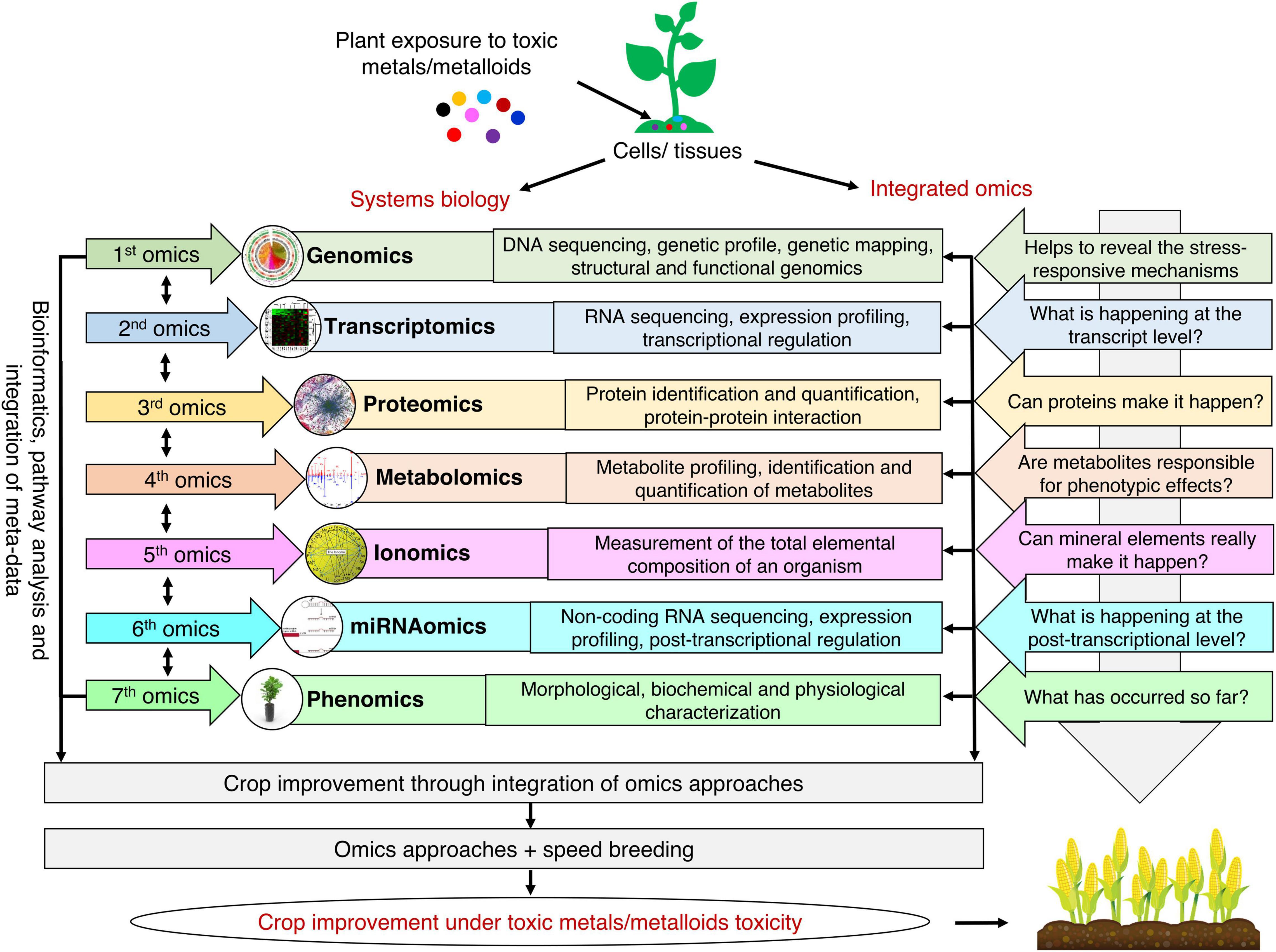
Figure 2. Integrated omics approach for developing toxic metals/metalloids tolerant plants. The use of multi-omics approach can help to reveal stress-responsive mechanisms at the genomic level, understand what is happening at the transcript and proteome level, provide clues about the interaction of metabolites with the phenotype, understand the role of different mineral elements, and unravel phenotypic changes in plants in response to toxic metals/metalloids toxicity. Integrating state-of-the-art omics approaches with speed breeding will help to meet the challenge of feeding a burgeoning human population.
Genomics includes characterization, data sequencing, structural organization, genetic alignments, interactions, and functions of a whole organism (plant) (Gilliham et al., 2017). Genomics is interrelated to the terms like transcriptomics, metabolomics, transgenomics, epigenomics, and phenomics. These approaches, associated with high throughput (HTP) technologies, have made significant advancements in plant genomics research and have enabled the improvement of multiple important crop plants (Varshney et al., 2020, 2021a). Genomics can help to identify genes, enzymes, or other molecular factors involved in stimulating toxic metals/metalloids stress. Genomics-based findings and online genomic data sets provide a way forward to open new windows for multi-omics technologies and genome editing tools.
Various trait mapping methods, such as quantitative trait locus (QTL) mapping, QTL-sequencing (QTL-seq), and RNA-sequencing (RNA-seq), have been established to switch the fine-mapping course as it can distinguish candidate genes within major QTLs quickly. For example, the genetic architecture and regulation of Cd tolerance in barley plants identified two QTLs, one minor-effect and other major-effect QTL, with a phenotypic variance of 47.24 and 38.59%, respectively, associated with Cd tolerance. This analysis also identified 16 candidate genes linked with Cd tolerance (Derakhshani et al., 2020). Marker-trait association study was conducted for As related traits using 704 SNPs. About 9 QTLs were identified with a phenotypic variance ranged from 8.6 to 12.6%, out of which six QTLs (qAsS2, qAsS5.1, qAsS5.2, qAsS6, qAsS9.1, and qAsS9.2) for As content in shoots were mapped on chromosome 2, 5, 6, and 9, two QTLs in roots (qAsR8.1 and qAsR8.2) on chromosome 8 and one QTL (qRChlo1) for relative Chl content was mapped on chromosome 1. Using these QTL intervals, 25 associated candidate genes were identified, showing transcription regulation for As toxicity-related traits (Murugaiyan et al., 2019). A total of four QTLs (qAsTSL8, qAsTSL12, qAsTRL8, and qAsTRSB8) for As phytotoxicity tolerance were found on chromosomes 8 and 12 using a composite interval mapping approach. A significantly effective QTL for As phytotoxicity tolerance in root contributing 24.9% phenotypic discrepancies was found on chromosome 8 with apparent pleiotropic effect on root-shoot biomass and shoot length (Syed et al., 2016).
QTL mapping in bread wheat for Al stress response revealed 79 QTLs, out of which 22 were putative, showing a range of phenotypic variance from 4.38 to 12.24%. Identified stable QTLs related to days to heading and grain yield were co-located with those of Al concentration with zero × additive environment interaction (Farokhzadeh et al., 2020b). Two bi-parental populations were used for QTL identification against Al toxicity in rice accessions. Fourty eight regions with a phenotypic variation of 57% were identified for Al tolerance, out of which four were co-localized with previously reported QTLs and three new regions (ART1, STAR2, Nrat1) on chromosome 1, 9, and 12 found inducing Al sensitive rice mutants. The study revealed that mapping with Indica/Japonica background identified QTLs where indica parent enhanced Al tolerance in Japonica background (Famoso et al., 2011). In another QTL map-based study of rice against Al resistance, a QTL named Alt12.1, was identified for Al resistance and ultimately considered ART1 as a primary candidate gene for this QTL region. The presence of the ART1 allele in different parental backgrounds has reportedly affected the expression of many genes in rice against Al tolerance (Arbelaez et al., 2017). A recent study in tomato identified 103 QTLs, out of which six QTLs were for fruit and leaf Fe contents while eight QTLs were for yield Fe content (FeUEc). Also, two relevant candidate genes encoding for specific proteins of tomato xylem sap were identified under Fe deficiency, thus affecting fruit yield and quality traits (Asins et al., 2020). Eight QTLs were detected for Al resistance in common bean (Phaseolus vulgaris) with a phenotypic variation of 7.6–14.7%. QTLs found were related to traits such as root length (RL), root dry weight (RDW), and shoot dry weight (SDW). This study identified the resistant Al activated transporter candidate gene (Phvul.007G025900) underlying the target QTL (Njobvu et al., 2020). QTL mapping for toxic metals/metalloids stress tolerance in rice identified one QTL related to Cu and Hg, three QTLs for As, and two QTLs for Fe and Zn contents. Candidate genes underlying the target QTLs for Zn (LOC_Os01g14440, LOC_Os01g18584, LOC_Os01g20160) and Fe (LOC_Os04g34600) were predicted to improve Zn and Fe stress tolerance in rice (Zhu et al., 2020). A total of 40 QTLs for Al stress tolerance were mapped on the A genome in wheat using composite interval mapping (CIM) and mixed composite interval mapping (MCIM) algorithms, which showed significant QTL × environment interactions (Farokhzadeh et al., 2020c). Another study in wheat revealed 58 QTLs associated with Al stress tolerance affecting spikelet-related traits (Farokhzadeh et al., 2020a). A further list of QTLs mapped for toxic metals/metalloids tolerance in different plant species is provided in Table 1.
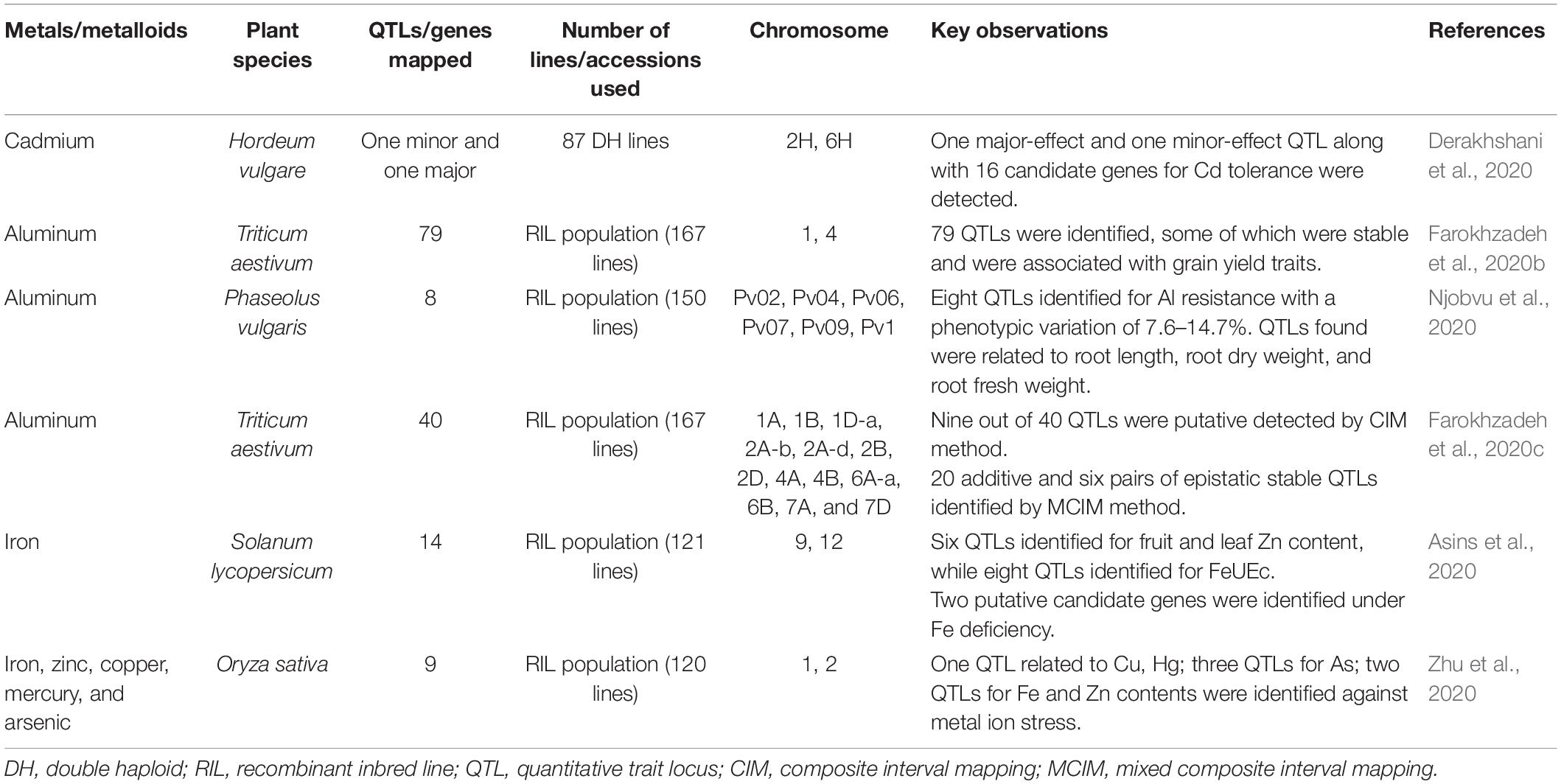
Table 1. Summary of QTL/gene mapping for toxic metals/metalloids tolerance in different plant species.
Genome-wide association study (GWAS) is a method utilized in genetics to relate definite genetic variations with certain traits in different individuals. It overcomes many constraints of traditional trait mapping (QTL) by offering advanced resolution, often to the gene level, and using trials from formerly examined populations in which frequently occurring genetic variations can be coupled with a phenotypic difference. For example, a GWA study for low Cd accumulation in rice identified OsABCB24 gene underlying a novel QTL (qCd1-3) (Pan et al., 2020). A recent GWA study in wheat identified five loci (qSCd−1A, qSCd−1D, qZn−2B1, qZn−2B2, and qFe−6D) associated with Cd stress tolerance (Safdar et al., 2020a). Analysis of a large collection of rapeseed accessions identified four QTLs and underlying candidate genes, including GSTUs, BCATs, UBP13, TBR, and HIPP01, responsible for Pb tolerance (Zhang et al., 2020a). GWAS investigation on rice grains for Fe and Zn traces have identified novel marker-trait associations with a phenotypic variation of 2.1–53%, which could be helpful to identify candidate genes for improving Fe and Zn tolerance (Bollinedi et al., 2020). Rice crop has also been studied for Fe toxicity using a GWAS approach, and three linkage disequilibrium (LD) blocks were found to mainly contribute to Fe omission. These LD blocks were detected on chromosomes 1, 2, 3, 4, and 7 for vegetative and generative stages, facilitating tolerance against Fe toxicity (Utami et al., 2020). Integration of approaches like genome-wide association, genome-wide epistasis (GWE), and gene expression proved to be an effective strategy for identifying novel QTLs related to Fe tolerance (69 genomic regions) across 19 chromosomes (Assefa et al., 2020). GWA study in barley for potassium (K) tolerance identified three primary QTLs responsible for K translocation and also identified some candidate genes for improving potassium-use efficiency (Ye et al., 2020).
Furthermore, a GWA study in bread wheat identified 534 significant MTAs for K-related traits, which included 11 stable loci and 16 M-QTLs. This study also identified potential candidate genes involved in critical pathways associated with stress tolerance, nutrient uptake, and sugar metabolism, which possessed the potential to develop K stress tolerant wheat cultivars (Safdar et al., 2020b). Zhao Z. et al. (2018) performed a GWA study and found six SNPs associated with four non-redundant QTLs significantly related to As accumulation. The loci, localized from 25.71 to 25.77 Mb on chromosome 1, co-localized with already reported QTLs (CAsA1/CAsS1), and just one candidate gene GRMZM2G130987 was discovered. The identified gene encodes a protein with P-P-bond hydrolysis-driven protein transmembrane transporter activity and contributes to As ion transport. Other SNPs located on chromosome 2 were found within the BAsA2/XAsA2 QTL. Five candidate genes are present in this QTL, and the GRMZM2G125495 candidate gene encodes a protein with extracellular glutamate-gated ion channel activity. This study provides a solid reason to further study the gene’s function in As accumulation (Zhao Z. et al., 2018). Liu X. et al. (2019) performed a GWA study against toxic metals in rice grain and found 22, 17, and 21 QTLs for grain associated with As, Cd, and Pb toxicity, respectively. The authors examined the candidate gene in qGAS1, a QTL for grain arsenic, with the best P-value found for the whole population. Toxin extrusion and transport protein of the multidrug might be the candidate gene for this QTL (Liu X. et al., 2019). Examples of GWA studies conducted in different crop plants for identifying toxic metals/metalloids tolerance loci have been described in Table 2.
Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) system is a valuable editing tool with high efficiency, specificity, and possessing a wide range of applications (Zafar et al., 2020). Targeted knock-in/out, deletion, insertion, and substitution mutations generated by CRISPR/Cas9 system have explored regulatory functions of genes and their impact on other biochemical processes and have helped to improve many crops by increasing their scavenging capacity under toxic metals/metalloids toxicity (Table 3).
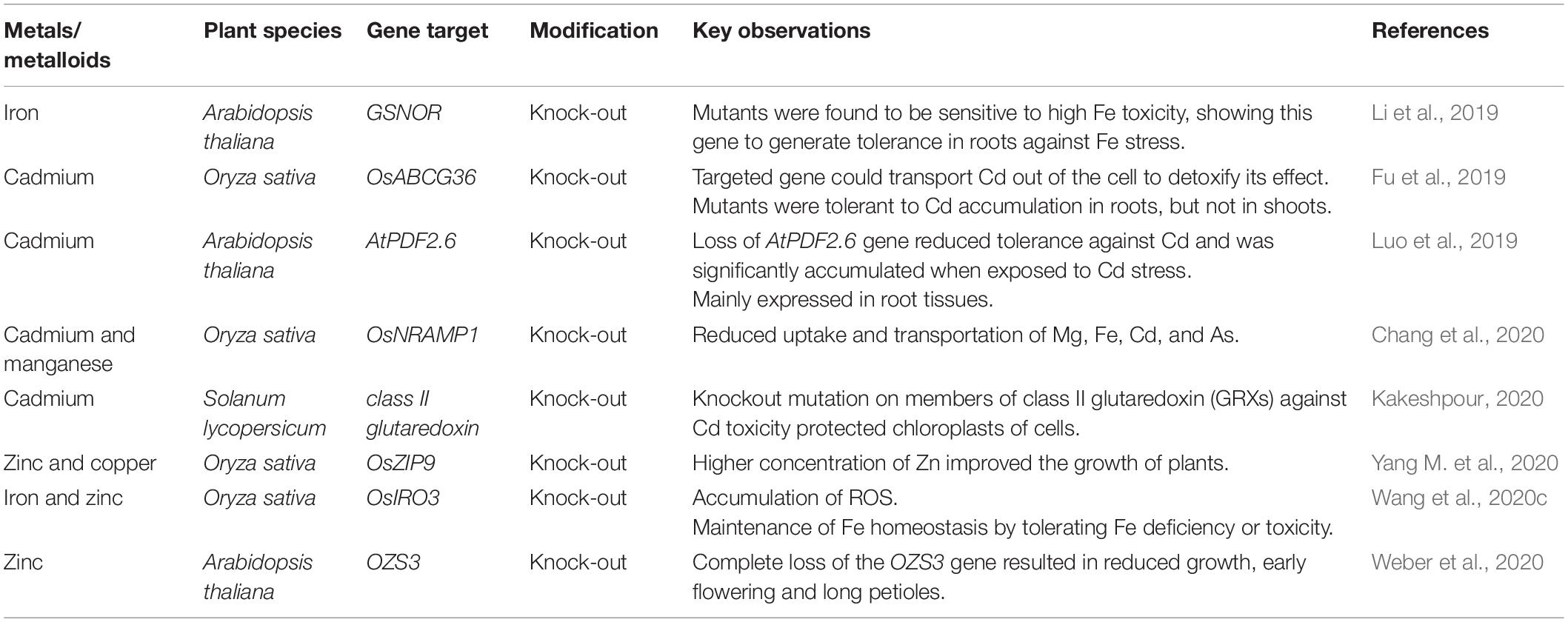
Table 3. Summary of genome editing studies for toxic metals/metalloids tolerance in different plant species.
Knock-out of OsNRAMP1 gene responsible for the uptake of metals like Cd, Fe, As, and Mn, significantly reduced the uptake of Cd and Mg and their storage in rice shoots and grains (Chang et al., 2020). Furthermore, OsIRO3 gene knock-out using the CRISPR/Cas9 approach highlighted the function of this gene in the regulation of Fe homeostasis. OsIRO3 gene mutants accumulated ROS, and their growth and development were affected under Fe deficiency (Wang et al., 2020c). Single, double, and triple mutants developed against Cd toxicity in tomato by CRISPR/Cas9 system were found to protect chloroplasts by mutating members of class II glutaredoxin (GRXs) from Cd toxicity (Kakeshpour, 2020). Knock-out mutant lines generated by CRISPR/Cas9 system in rice targeting OsZIP9 gene against Zn toxicity showed a substantial reduction in growth, which revealed Zn as an essential element for growth (Yang M. et al., 2020). In Arabidopsis, OZS3 gene knock-out reduced the growth of roots and elusive development of plants, further damaged embryos, and caused early flowering (Weber et al., 2020). Cd accumulation was studied in rice by knocking out a segment of the OsABCG3 gene via CRISPR/Cas9 technology. Knock-out mutants accumulated Cd in roots cells and routed Cd contents out of the cell sap to detoxify its effect, thus generating tolerance against Cd (Fu et al., 2019). In Arabidopsis, the variant GSNOR has been observed to generate tolerance and promote root growth against Fe toxicity. The mutant lines produced using the CRISPR/Cas9 vector were also found to be highly sensitive toward high Fe content, which affected the whole seedlings and produced leaves that were smaller than the wild type (Li et al., 2019). Loss of AtPDF2.6 gene has been reported to reduce tolerance against Cd along with its significant accumulation in root cells when exposed to Cd stress (Luo et al., 2019). Genome editing using the CRISPR/Cas system has a promising future for sustainable agricultural production to feed the world’s growing population. In the near future, more genome editing works need to be performed under the toxicity of different metals/metalloids like As, Pb, Hg, Ni, Cr, etc., in different crop plants to explore the stress resistance mechanisms.
The term transcriptomics refers to a set of techniques that are used to study the RNA transcripts in an organism. Several transcriptomic techniques are widely used to measure the abundance of the transcripts of interest (Lowe et al., 2017; Mehmood et al., 2021; Raza et al., 2021c). Studies have shown that stress-induced changes in gene expression may lead to the synthesis of novel proteins, stress mediating metabolic compounds, or encode transcription factors (TFs), which help to regulate the stress-responsive genes (Table 4).
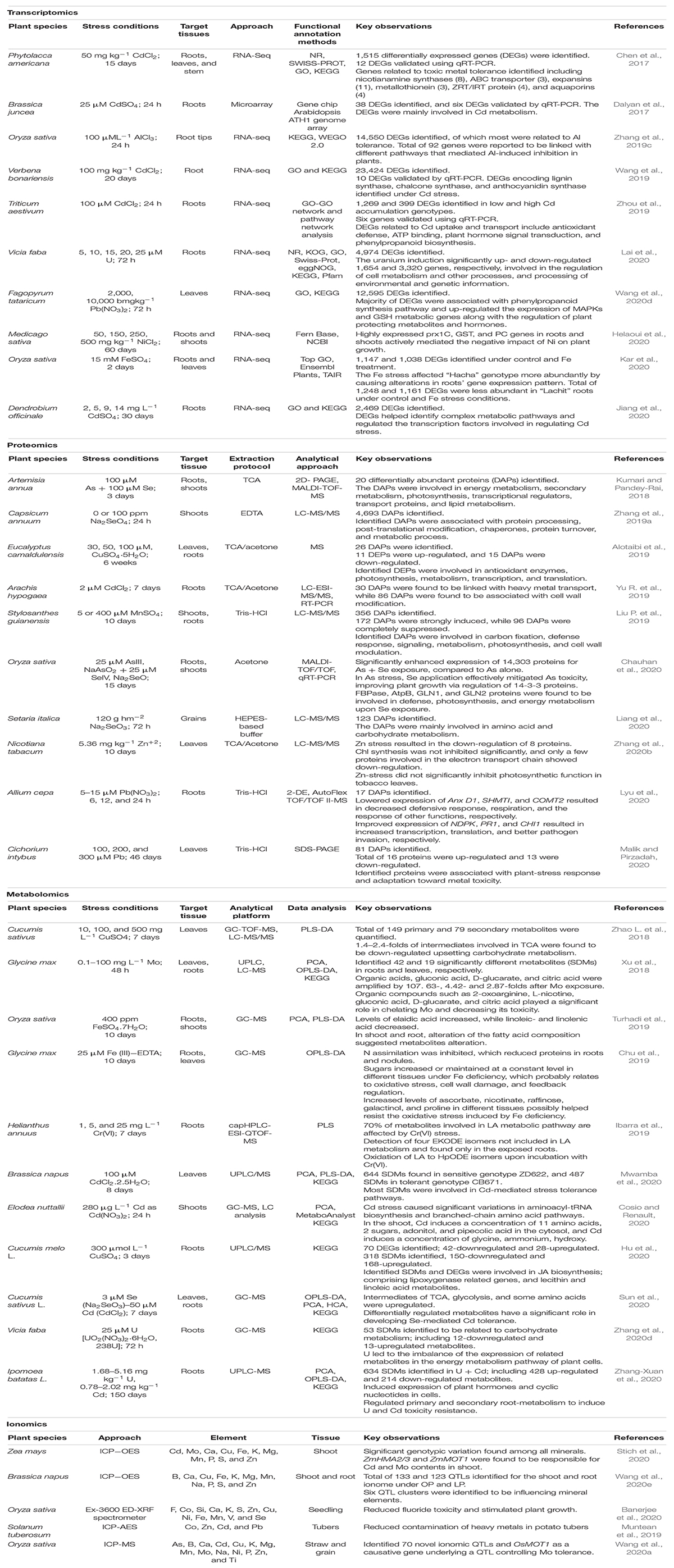
Table 4. Summary of key transcriptomics, proteomics, metabolomics, and ionomics studies under toxic metals/metalloids toxicity in different plant species.
Transcriptomic techniques represent an effective method to enhance toxic metals/metalloids tolerance in plants by better understanding signaling mechanisms and gene ontology (GO). The transcriptomic profile screening of two contrasting varieties of rice (Fe susceptible “Hatcha” and Fe tolerant “Lachit”) revealed 22 out of 35 metal homeostasis genes in the tolerant variety. Furthermore, leaf transcriptome showed a more pronounced response in Hatcha, leading to a high degree of differential gene regulation (Kar et al., 2020). High and low grain Zn and Fe containing wheat genotypes revealed enrichment of GO terms such as Zn and Fe binding, Chl synthesis, ATP-synthase coupled transport, and oxidoreductase activity (Mishra V. K. et al., 2019). The molecular insights into transcriptome expression profiling of chickpea under toxic metals/metalloids [As(III), Cr(III), and Cd(II)] showed induction of crucial metabolic pathways under stressed conditions. Moreover, nine genes that played a major role in regulating these metabolic pathways were found to be differentially expressed in response to stress conditions (Yadav et al., 2019). Recently, Di et al. (2021) studied the stress-responsive genes of upland rice exposed to As(III) and As(V). Under metals toxicity, many genes were down-regulated compared to those that were up-regulated. Arsenic treatment resulted in unique transcriptome profiles changes, and a novel set of typical response genes were found. The antioxidant enzyme activities were consistent with the antioxidant enzyme-related genes expression, and several transports and defense enzyme-related pathways were identified (Di et al., 2021). Shukla T. et al. (2018) studied the response of Arabidopsis accessions under As(V) toxicity. Differential transcriptome modulation was found in sensitive “Slavi-1” and tolerant “Col-0” accessions. The results showed that As-induced genes are linked with stress response and detoxification pathways (Shukla T. et al., 2018).
According to Komarkova et al. (2020), Cd resulted in alteration of poplar’s gene transcripts, which not only regulated the plant defense mechanism through differential gene expression but also led to the production of phytohormones. Additionally, long-term exposure to stress was found to reduce the Cd toxicity in plants (Komarkova et al., 2020). Another transcriptomic study carried out on Chinese flowering cabbage (Brassica parachinensis) showed that Cd toxicity was mediated through some important tolerance-inducing genes, including HMA3, HMA4, and Nramp1 (Wang et al., 2017). Furthermore, the genetic insights in two contrasting wheat genotypes (low Cd accumulating L17 and high Cd accumulating H17) revealed 1,269 genes to be differentially expressed. These genes showed heme-binding as the most active GO network, followed by metal binding. In contrast, phenylpropanoid biosynthesis and glutathione metabolism were found to be the major pathways active under Cd stress (Zhou et al., 2019). The analysis of the transcriptional data set of Verbena bonariensis under Cd stress revealed ROS scavenging system, photosynthesis, transpiration mechanism, chelating reaction, and production of secondary metabolites associated with DEGs as analyzed through GO and KEGG pathways (Wang et al., 2019).
The transcriptional analysis of the creeping bentgrass identified four transcription factors (bZIP, WRKY, MYB, and ERF) linked with Cd stress (Yuan et al., 2018). In a Cd-hyperaccumulator, Siegesbeckia orientalis, the comparative transcriptome analysis of roots with and without Cd treatment revealed a high number of DEGs, indicating the involvement of multiple biological pathways to cope up with Cd stress at the molecular level (Xu X. et al., 2020). Among such functional pathways, the ubiquitin-proteasome system (UPS) in plants has been reported to act through sequential actions of a cascade of enzymes. A comparative transcriptional study on a Cd hyperaccumulator Viola baoshanensis and its non-tolerant counterpart Viola inconspicua showed overexpression of genes involved in the UPS pathway under Cd exposure in V. baoshanensis, thereby supporting the hypothesis that high transcript levels of the genes involved in the UPS pathway can enhance tolerance to Cd toxicity (Shu et al., 2019).
Another study on the transcriptomic response of alfalfa plants, while assessing its phytoremediation potential for Ni, showed high expression of peroxiredoxin-1C, glutathione-S-transferase (GST), and phytochelatins (PCs). These genes were linked to an antioxidative response, prevention of cell damage, and Ni detoxification through its binding with PCs forming Ni-PC complexes, respectively (Helaoui et al., 2020). Kök et al. (2020) evaluated B accumulator Puccinellia distans for Se tolerance. Here, RNA-seq data showed induction of Se assimilation and stress response genes under stress conditions, which in turn altered the expression of gene transcripts involved in developmental (2.2%), transcriptional and translational (7.3%) processes, biotic (2.2%), and abiotic (17.7%) stress responses in P. distans. It further restricted the movement of Se by trapping it in the cell wall through the up-regulation of lignin production-related transcripts (Kök et al., 2020). Transcriptomic analysis of the Co stress response in Salix babylonica showed 2,002 DEGs, out of which 1,165 were identified in root and 837 in shoots. In addition, 107 transcription factors were identified from the DEGs, and most of them were reported to belong to the NAC and ERC families (Wang et al., 2020f). Another technique, translating ribosome affinity purification (TRAP) has been used to study transcriptional program in Arabidopsis against Fe stress. Using this technique, the excess Fe was sensed in opt3 leaves. This study highlighted the first tissue, vasculature, in comparison with roots and leaves to respond against Fe deprivation and supply, thus maintaining Fe homeostasis through xylem and phloem. Moreover, a total of 1,143 DEGs were identified with more than 2-fold change, including 539 DEGs in roots and 604 DEGs in leaves, thus an up-regulation of iron deficiency responsive genes (Khan et al., 2018).
Laf vasculature from Arabidopsis leaves have also been used to execute single-cell RNA analysis and identified 19 clusters of all cell types, including mesophyll, guard cells, vascular cells, hydathodes, and some metabolic pathways to recognize their roles. The results identified potential roles of these clusters in sugar transport, amino acid transport, hormone biosynthesis, and defense-related responses (Kim et al., 2021). Single-cell transcriptome analysis has not been studied so far. Yet, there is a dire need to implement single-cell transcriptome analysis against the toxic metals/metalloids toxicity and may provide key resources and molecular insights to develop new strategies regulating the flux of ions, signals, and metabolites. Taken together, the analysis of transcriptomics data obtained through various studies suggests that it not only brings useful insights to functional genomics but also has a great potential to support molecular breeding, genetic engineering, phytoremediation, and metal complexation pathways. Meanwhile, there is a need to make smart use of comprehensive tools to integrate transcriptomics with other omics data and get useful insights into unidentified linkages. Summary of some experiments performed using transcriptomic approaches under toxic metals/metalloids toxicity is discussed in Table 4.
Proteomics comprehensively covers the encoded proteins in living organisms at a particular instance and plays a vital role in understanding all cellular routes at the molecular level (Mehmood et al., 2021). Recently, proteomics has appeared as a vital tool to convey data on the survival of plants and adaptation toward toxic metals/metalloids toxicity in several plants (Table 4). From a methodological viewpoint, proteomics technology has progressed quite fast from the first generation (i.e., two-dimensional electrophoresis-mass spectrometry (2DE-MS); to second-generation (i.e., isobaric/isotopic tagging); to third-generation (i.e., shotgun and gel/label-free approaches); and lastly to fourth generation (i.e., mass western, targeted, SRM/MRM approaches) (Jorrin-Novo et al., 2019). Innovative proteomic systems provide a comprehensive understanding of metal-responsive proteins for plant stress tolerance (Table 4).
For instance, plant vascular systems like xylem sap transport minerals, water, and toxic metals (Cd) from roots to shoots. Proteomic variations in xylem sap significantly detoxify Cd in plants (Luo and Zhang, 2019). Brassica seedlings were treated with Cd (0 and 10 μM) for 3 days, and the collected xylem sap was subjected to lyophilization. Notably, 672 proteins from the xylem sap of Cd-treated rapeseed plants were identified through proteomic analysis (LC-MS/MS, shotgun). Most affected metabolic pathways between these proteins were found to be linked with stress/oxidoreductases, protein/lipid metabolism, and cell wall modification. Protein-like defensins, BnPDFL, found in xylem sap behaved as a Cd-chelating agent, confirming its positive role in regulating Cd tolerance (Luo and Zhang, 2019). In another study, the iTRAQ approach illustrated dynamic changes in root proteome in maize seedlings (Wen et al., 2019). Plant’s roots were exposed to 200 mg L–1 CdCl2, and both root and shoot growth were found to be severely inhibited in the first 72 h. The different numbers of differentially abundant proteins (DAPs; 733, 307, 499, and 576) were isolated after 12, 24, 48, and 72 h, respectively. These DAPs displayed numerous functions like energy and carbohydrate metabolism, ribosomal synthesis, cellular metabolism, ROS homeostasis, and cell wall organization. Amongst these GSTs, GRMZM2G308687 showed extra abundance after 12, 48, and 48 h Cd-treatment. GST protein was found to be mainly tangled in PCs generation for Cd tolerance (Wen et al., 2019). Label-free proteomic analysis of Cd treated Iris lactea revealed 163 and 196 DAPs expressed in shoots and roots, respectively. Bioinformatic studies revealed that these DAPs, which are responsive to Cd, were majorly involved in redox reactions, signal transduction, ion transport, and other biochemical mechanisms. They were not only involved in lignin and amino acid biosynthesis pathways but also assisted GSH and glycerolipid metabolism. From I. lactea, a mannose-specific lectin (Cd-induced) was found to increase Cd-sensitivity and enhance Cd-accumulation in yeast (Liu et al., 2020).
Chauhan et al. (2020) performed a transcriptomic and proteomic analysis in rice to explore the molecular cross-talk involving Se-mediated tolerance of As-toxicity. Se supplementation restored the structural deformities caused by As, which comprised the cell wall and membrane disintegration. As-transporter gene expression viz., ABCG5, NIP1;1, NIP2;1, TIP2;2, NRAMP1, NRAMP5, and sulfate transporters SULTR3;1, SULTR3;6 were found to be high in As + Se treated plants when compared to As alone. This not only resulted in low As concentration but also reduced the toxicity. During As + Se exposure, the GST, GRX, and PRX up-regulation also confirmed that elevation of As resulted in oxidative stress (Chauhan et al., 2020). Proteomic analysis (LC-MS/MS) of Se-treated pepper seedlings revealed up-regulation of 172 proteins, while 28 proteins were found to be down-regulated. Identified DAPs were mainly associated with metabolic processes, protein turnover, protein processing, post-translational modifications, and chaperones. Furthermore, various heat shock proteins (HSPs) were also identified as DAPs, which helped to cope with metal toxicity (Zhang et al., 2019a). Zeng et al. (2019) performed quantitative proteomics (iTRAQ) to study differential protein expression in Se-enriched and non-Se-enriched rice seedlings. Overall, 3235 proteins were detected, of which 3,161 proteins were quantified from 401 DAPs. Interestingly, 77 targeted significant DAPs were screened further and classified into 10 sets comprising of actin, synthetases, hydrolases, tubulin, ligases, lyases, isomerases, heat shock proteins, oxidoreductases, and transferases. These findings indicated that active oxygen metabolism, anti-stress, anti-oxidation, amino acid, and carbohydrate metabolism of Se-enriched rice seedlings was higher as compared to non-Se-enriched plants (Zeng et al., 2019).
In another study, proteomic analysis of Cu-treated young seedlings of Eucalyptus camaldulensis revealed 26 targets taking part in protein expression. Elevated Cu levels up-regulated the expression of 11 proteins and down-regulated expression of 15 proteins. Identified proteins were associated with antioxidant enzymes, photosynthesis, metabolism, transcription, and translation (Alotaibi et al., 2019). Furthermore, Ceballos-Laita et al. (2018) used two different proteomic approaches, i.e., shotgun and 2-DE, to study the effects of Mn toxicity on tomato root proteome. The shotgun approach identified 367 reliable proteins, while 2-DE yielded 340 consistent spots. A total of 54 proteins were detected using a 2-DE approach, which was found to be altered in relative abundance, while shotgun found variations in 118 proteins. Only 7% of DAPs were found to be common in both methods. The most affected metabolic pathways were signaling, protein metabolism, and oxidoreductases. Further findings suggested that Mn-toxicity mediated protein-turnover impaired the roots for energy production, leading to changes in oxidative phosphorylation, glycolysis, TCA, and pyruvate metabolism. Root proteome indicated a slowdown of metabolic activities, including call wall integrity, protein turnover, and energy production (Ceballos-Laita et al., 2018). Nevertheless, Mn interaction with Cd and attenuation of the toxic effects on plants are also found to play a major role in photosynthesis. Oliveira et al. (2020) studied the mechanisms of Mn-response in Cd toxicity mitigation in young plants of cacao by analyzing the alterations in DAPs and exclusive proteins (EP). Few significant proteins were produced in the presence of Cd while repressed in the presence of Cd + Mn, and vice versa. These findings suggested that Mn mitigated the adverse effects of Cd on cacao plants (Oliveira et al., 2020).
Two Brassica napus cultivars, ZS758 and ZD622, were treated with high As concentrations, and their response to As toxicity has been studied through iTRAQ-based proteomics analysis. The chlorophyll fluorescence attributes revealed that As pressure significantly lowered the photochemical efficiency of photosystem I and photosystem II and closed stomata detected under scanning electron microscopy. Metabolic pathways, followed by ribosome and secondary metabolites biosynthesis, were the dominant functional annotation among the differentially expressed proteins. Many genes involved in stress defense and primary metabolism were As-responsive DAPs (Farooq et al., 2021). Alka et al. (2021) performed histological and proteome analysis and found that Microbacterium foliorum lowered the As toxicity in Melastoma malabathricum. 2D gel electrophoresis and transmission electron microscopy were used to conduct the histological and proteome analysis. When As-treated cells were compared to untreated cells, considerable changes were discovered. Compared to control, root cells ultra-structure showed intact cell wall, cytoplasm, and vacuole under As + bacteria. To further understand the As + bacteria, proteome profiling of root cells was analyzed. It has been found that proteins involved in photosynthesis, defense, signaling, and protein biogenesis were higher in As + bacteria than As alone (Alka et al., 2021).
Metabolomics is considered as an emerging field that broadly detects and quantifies all exogenous and endogenous molecules of low molecular weight (<1 kDa), including metabolites present in living organisms (Razzaq et al., 2019; Raza, 2020). Multiple analytical techniques have been developed to understand plant metabolic responses like inductively joined MS, liquid, and gas chromatography-MS (LC-MS, GC-MS), and nuclear magnetic resonance spectroscopy (NMR) (Razzaq et al., 2019; Raza, 2020). Table 4 shows some examples of metabolomics studies under toxic metals/metalloids toxicity in different plants.
Xie et al. (2019) performed LC-MS/MS and HPLC analysis for metabolites and thiol compounds in Amaranthus hypochondriacus under Cd stress. A. hypochondriacus leaves accumulated Cd levels 40 times more than the control plant and also enhanced the PCs contents. Out of 12,084 metabolites identified, 41 were found to be significantly different metabolites (SDMs) among two groups and known to take part in seven different metabolic pathways. Among them, 12 SDMs related to PCs were associated with three different pathways, namely Arg and Pro metabolism, Val, Leu and Ile biosynthesis, and Ala, Asp, and Glu metabolism (Xie et al., 2019). An HPLC coupled with MS-based metabolome profiles of rice was analyzed in response to Cd and Cu toxicity. A total of 112 metabolites were identified, of which 97 metabolites were subsequently confirmed under Cd toxicity. Importantly, the secondary metabolism, amino acid metabolism-like purine, carbon, and glycerolipid metabolism pathways were found to be greatly affected. Furthermore, reduction in plant growth, photosynthetic capacity, and induction of defense systems to protect cell damage have also been observed (Navarro-Reig et al., 2017). In yet another study, GC-TOF-MS and LC-MS/MS-based metabolome profiling were performed to study the effect of Cu stress on cucumber plants (Zhao L. et al., 2018). Metabolomics helped in the identification of 149 primary and 79 secondary metabolites. Down-regulation of TCA intermediates (up to 1.4–2.4-fold) was observed, indicating disturbed carbohydrate metabolism. Excess Cu affected the aldarate and ascorbate metabolism and shikimate phenylpropanoid biosynthesis (Zhao L. et al., 2018). In soil, excess Se may appear destructive to plants. Zhang et al. (2019b) carried out an untargeted metabolome analysis of Se-treated and control celery seedlings and identified 24 sulfur and seleno-compound metabolic unigenes to be differentially expressed. Moreover, 1,774 metabolites and 237 SDMs were found through UHPLC-MS/MS. Results revealed that identified metabolites could be associated with significant biological pathways regulating Se tolerance (Zhang et al., 2019b). Furthermore, an untargeted metabolomics method was used to study the effects of 5 days application of 100 μmol L–1 selenate on broccoli sprouts metabolome. Multivariate statistical analysis displayed that tyrosine, D-erythronolactone, serine, and melezitose were up-regulated, while citric acid, D-glyceric acid, and succinic acid were down-regulated after selenate application. Selenate application also affected the metabolism of GSH, β-alanine, and plant-metabolite biosynthesis associated with glucosinolate precursors (Tian et al., 2018).
The metabolomic responses of tea plants toward Zn stress showed that Zn excess or deficiency differentially affected the metabolic pathways in tea leaves (Zhang et al., 2017). Zn-deficiency influenced carbohydrates metabolism, whereas Zn-excess affected the metabolism of flavonoids. Furthermore, it was observed that both Zn-excess and Zn-deficiency led to lowered nicotinamide concentrations, which accelerated NAD+ breakdown and resulted in low energy metabolism (Zhang et al., 2017). Furthermore, UHPLC/Q-TOF based metabolic investigation was conducted to study the effects of 100 mM NaCl and 100 mM ZnSO4 on lettuce root metabolic profiling (Rouphael et al., 2016). Most of the compounds identified in ZnSO4/NaCl environments were lipids, carbohydrates, glucosinolate, phenolics, or hormones. Results showed that osmotic stress and redox-imbalance have an essential role in defining lettuce root metabolic response; while polyamines and polyamine conjugates were found to be elicited as a precise reaction to ZnSO4 (Rouphael et al., 2016). Qualea grandiflora plants were grown in Murashige and Skoog (MS) medium with or without Al supplementation for 120 days. Metabolite profiling of Qualea grandiflora plants was performed through GC-MS under Al stress. Q. grandiflora plants with starved Al showed shorter roots and shoots, chlorotic leaves, and low biomass. Al was found to be critical for lignin synthesis, cell wall, processing of genetic information, and organic acid metabolism. Furthermore, Al was predicted to help plants uptake phosphorous (Cury et al., 2020).
Cosio and Renault (2020) studied physiology and shoot metabolomic profiles of Elodea nuttallii exposed to methyl-Hg (30 ng L–1), inorganic Hg (70 ng L–1), and Cd (280 μg L–1) for 24 h. KEGG pathway analysis revealed that MeHg exposure resulted in numerous biochemical changes like aminoacyl-tRNA biosynthesis, serine, glycine, threonine, proline, arginine, and nitrogen metabolism. In contrast, Cd-stress resulted in important alterations in aminoacyl-tRNA biosynthesis and branched-chain amino acid pathways. Data supported the argument that MeHg impacts N homeostasis, Cd caused an osmotic stress-like pattern, and inorganic Hg had less impact on both features (Cosio and Renault, 2020). In a different study, Pb-treated vetiver plants grown in a hydroponic setup were used for LC/MS/MS-based metabolome analysis of shoot, and root tissues. Multivariate metabolite analysis displayed tremendous induction of vital metabolic pathways like amino acid metabolism, sugar metabolism, enhanced osmoprotectants production like polyols and betaine, and metal chelating organic acids (Pidatala et al., 2016). For Pb remediation, vetiver grass is considered a superior choice due to its Pb-hyperaccumulation ability. Pidatala et al. (2018) performed comparative metabolic profiling of maize and vetiver under Pb-stress conditions. Vetiver plants displayed a massive increase of metabolites under Pb-stress, including coenzymes, amino acids, and organic acids; whereas, maize displayed a modest increase in the same metabolites with no significant effect on other metabolites (Pidatala et al., 2018). Furthermore, Zhang et al. (2019d) performed metabolic profiling of Malus halliana leaves under Fe-deficiency through GC/MS and identified a total of 18, 39, and 17 metabolites in three pairs L12h vs. L0h, L3d vs. L0h, and L3d vs. L12h, respectively. The findings showed that trehalose and sucrose are the most abundant metabolites in glucose metabolism and have a role in balancing photosynthetic activity in M. halliana leaves with the utilization of photo-assimilate. Overall, identified metabolites were found to play a major role in the positive regulation of Fe-deficiency response (Zhang et al., 2019d).
Das et al. (2021) investigated the differential expression of two Andrographis paniculata genotypes under As stress. The As distribution was found to be higher in roots than in other tissues with TF <1. APMS (wild collection) was more tolerant and accumulated less As than APwC (mass selection line). HPLC was used to quantify the metabolites in sample extract, and metabolites detection was performed at 223 nm by photodiode array detector (PDA). Arsenic enhanced flavonoids production like 5,7,2′,3′-tetramethoxyflavone in A. paniculata. The main secondary metabolites, oxidative enzymes, and nutrient uptake showed a significant difference in the detoxification process under As stress. The improved Ap2 expression proposed its involvement in metabolic flux channeling toward the ent-LRDs biosynthesis under As stress (Das et al., 2021). NMR-based metabolomics approach provides a quick snapshot of metabolites without prior knowledge of organisms. Thus, Arora et al. (2018) validated the applicability of NMR-based metabolomics using freshwater microalga Scenedesmus sp. Using NMR spectroscopy, the authors identified and quantified around 45 metabolites, comprising organic acids, sugars, amino acids, nucleotides, phosphagens, and osmolytes. These findings revealed that microalga tolerated the As toxicity by accumulating these diverse metabolites (Arora et al., 2018).
Plants require minerals in appropriate amounts to perform their functions properly. Ionome or ionomics are the mineral elements needed in trace amounts for plant growth, development, and removal of toxic metals/metalloids (Stich et al., 2020). Nitrogen is an essential plant nutrient and forms an integral part of nucleic acids, proteins, vitamins, and hormones. It reduces toxic metals/metalloids toxicity by synthesizing Chl, N-containing antioxidants, and metabolites (Lin T. et al., 2011). Punshon et al. (2013) used the ionomic approach of multi-element imaging synchrotron X-ray fluorescence microscopy (SXRF) with A. thaliana for studying the phenotype, gene identification, and screening. Su et al. (2017) performed Fourier transform infrared spectrometry (FTIR) analysis to examine the subcellular distribution, accumulation, alteration in metabolic activity, and physiology of S. polyrhiza under Cd stress (5, 10 μM for 4 days). In another study, ICP-MS ionomic approach was applied to sunflower growth under Cd stress (50, 350, and 750 mg as CdCl2). Increased doses of Cd caused necrosis and chlorosis and led to disproportion in Cu/Zn homeostasis (Júnior et al., 2014). Some recent examples of ionomics studies under toxic metals/metalloids toxicity are provided in Table 4.
Zhu et al. (2010) reported 16 mM N fertilizer (NH4)2SO4 to reduce the Cd toxicity in Sedum. In S. nigrum, the fertilizers, (NH4)2SO4 and CH4N2O application increased plant biomass by up to 2.0 and 2.1-folds, respectively, as compared to control under Cd stress (2 mg kg–1) (Yang et al., 2019). Mg, which is an essential component of Chl, reduced Cd toxicity by enhancing the activity of antioxidant enzymes (Chou et al., 2011). In Japanese mustard spinach, Cd stress and accumulation caused by a dose of 0.25 μM was reduced by up to 40% after the application of 10 mM Mg (Kashem and Kawai, 2007). Calcium not only regulates the metabolic activity of plants but also reduces the toxicity caused by toxic metals/metalloids. For instance, 30 mM Ca reduced Cd quantity from 46.7 to 17.4 μg in A. thaliana (Suzuki, 2005). Feng et al. (2017) performed inductively coupled plasma-MS (ICP-MS) determination of Cd/As to demonstrate a distinct transport of elements to 14 parts of 21 different brown rice genotypes. Interestingly, sixteen other elements were also determined in roots, and associated with the highest Cd accumulation fraction and increased glycolytic activity. Cu, Zn, Mg, and Co distribution was found to be associated with Cd concentration in roots and nodes. Ionomic profile indicated that all elements are correlated with different plant parts, and distribution occurs through them, while node1 had the highest accumulation rate of Cd (Feng et al., 2017).
The ionome of maize shoot has been studied under the Fe regime using recombinant inbred line population, and significant genotypic variance among 12 mineral elements was identified using inductively coupled plasma optical emission spectrometry (ICP−OES) (Stich et al., 2020). Besides, ZmHMA2/3 and ZmMOT1 were also proposed to cause genetic variation of Cd and Mo contents in maize shoots. Another ionome study targeting shoots and roots of Brassica species identified a total of 133 and 123 QTLs under optimal and low P (OP/LP) conditions (Wang et al., 2020e). These QTL clusters are predicted to reveal the uptake and transport mechanisms of mineral elements. Ionome study in rice seedlings, when mediated by nano-Si-priming (SiNP), generated fluoride tolerance by reducing fluoride uptake and bioaccumulation. This improved photosynthetic activity and uptake of mineral nutrients by lowering oxidative damage, thus ensuring a safe rice cultivation method (Banerjee et al., 2020). During 3 consecutive years, a decreasing trend of low contamination of toxic metals/metalloids was observed in an ionome study of potato tubers (Muntean et al., 2019). A study on rice grain and straw identified 70 novel ionomic QTLs for 15 nutrient elements. The study identified a molybdate transporter gene OsMOT1;1 underlying qMo8 QTL controlling Mo concentration in both grain and straw tissues (Wang et al., 2020a). Ionomic variations of rice had also been compared to Arabidopsis against sodium, molybdenum, and nitrogen using genome-wide association studies from 529 accessions. This comprehensive study with 6.4 million SNPs identified 72 loci for ionomic variation and further identified candidate genes, sodium transporter gene Os-HKT1, Os-MOLYBDATE TRANSPORTER, and thus grain number, plant height, and heading date thus featured the molecular mechanism controlling rice ionome (Yang et al., 2018). Mišúthová et al. (2021) studied the growth and developments of maize plants in response to silicon (Si) and As metalloids. The experimental findings revealed increased As (75 and 150 μM) concentration reduced the biomass and root length, and 2.5 μM Si supplementation to the medium did not affect root growth. Further, Si application did not affect the uptake of micro-and macro-elements into the roots (Ni, Ca, Zn, P, Cu, Mo, and K). While, Si significantly reduced the presence of hydrogen peroxide and superoxide in roots suffering from As toxicity and increased root antioxidant enzymes (Mišúthová et al., 2021). In another study, Xiao et al. (2021) investigated the transcriptomic variations and ionomic profiles to understand the molecular mechanism of alleviating the As(III) stress. Under As(III) stressed and control conditions, a total of 29 elements were found through inductively coupled plasma mass spectrometry (ICP-MS) in the rice roots and shoots. Strong connections were observed between transcriptome and ionome in the rice shoots. Overall, 3,812 DEGs were discovered in the As(III)-applied shoots than control shoots, and these DEGs were mainly associated with transmembrane transport and ion binding. The rice plants preferred to translocate more nutrients to cope with arsenic toxicity (Xiao et al., 2021).
miRNAs are tiny non-coding RNA species and key post-transcriptional regulators. They have also emerged as an important technique like other omics approaches to develop toxic metals/metalloids tolerant plants (Dubey et al., 2021; Jamla et al., 2021b). Some successful studies have been completed to identify metals/metalloids responsive miRNAs. For instance, miRNA against Cd toxicity have been reported in many crop plants, including wheat (Qiu et al., 2016), rice (Huang et al., 2009), maize (Gao et al., 2019), barley (Yu J. et al., 2019) and rapeseed (Jian et al., 2018). These successful studies depicted miRNAs regulating toxic metal uptake and transport, inducing oxidative stress and antioxidative defense, thus maintaining homeostasis. The exploration of miRNAs in plants, along with other omics approaches and computational tools, may act as potential targets for improving plants to confer toxic metals/metalloids tolerance (Dubey et al., 2021). Some recent reports of miRNAs against toxic metals/metalloids stress and their impact on plants are mentioned below.
The role of miR156j from miR156 family was observed in rice for physiological growth stages against As toxicity. Expression analysis revealed the functional role of Osa-miR156j at various developmental stages and plant tissues under As(V) exposure. Further, stress-responsive cis-acting regulatory elements confirmed its involvement during stress conditions. The results altogether showed the obvious role of miR156j in metabolic activities under As toxicity and can be used to develop As stress-tolerant varieties (Pandey et al., 2020). The genome-wide identification analysis in Brassica juncea identified some novel miRNAs, including 11 miRNAs and 56 transcripts against Cd stress response, using transcriptomic and sRNA sequencing technology. This study found bra-miR172b-3p regulating ATCCS playing a significant role against Cd stress and highlighted the regulatory relationship between miRNAs and transcripts, thus providing insights into molecular mechanisms in response to Cd stress in plants (Liu et al., 2021). A library of more than 2,000 artificial microRNAs (amiRNAs) has been developed in Arabidopsis to overcome the limitation for dissecting As responsive mechanisms. The research identified six transformant lines in response to As(V) and Cd exposure. However, further characterization of miRNA line closely related to homologous CBF and ERF transcriptional factor genes revealed ERF34 and ERF35 as important resistant TFs against Cd stress. The CBF1, CBF2, and CBF3 TFs were mediating negative regulation under As exposure and can be further used in the future to identify Cd and As induced transcriptional control networks (Xie et al., 2021).
Another miRNA named miR1511 for the MIR1511 gene responded against Al toxicity in common bean and then targeted against ALS3 (Aluminum Sensitive Protein 3) gene. Both the genes opposed each other for sensitive genotypes as decreased expression of miR1511 showed increased ALS3 transcript level under Al toxicity and vice versa, thus revealing the prominent role of miR1511 for inducing resistance against Al stress in plants (Ángel et al., 2021). Overexpression of miR156 had significantly reduced the endogenous ROS in Arabidopsis and enhanced tolerance against Cd stress by reducing the effect of Cd-related transporters (Zhang et al., 2020c). In Miscanthus sinensis, high-throughput miRNA sequencing for Cr toxicity identified a total of 104 conserved miRNAs and 158 non-conserved miRNAs, out of which 45 differentially expressed miRNAs were linked with roots while 13 differentially expressed miRNAs were linked with leaves under Cr stress. GO, and KEGG analysis identified miR167a, novel-miR15, and novel-miR22 as potential candidates that were involved in Cr transportation and chelation. Moreover, miR156a, miR164, miR396d, and novel miR155 were involved in physiological and biochemical metabolisms and detoxifying the toxic effect of Cr in plants (Nie et al., 2021). Overall, these studies showed that miRNA significantly regulates genes expression and plays a key role in mitigating the adverse effect of toxic metals/metalloids. Thus, miRNAomics should be more exploited in identifying tiny key players (miRNAs) in regulating the stress-responsive mechanisms against a variety of toxic metals in different crop plants.
A phenome is a set of all biological, physical, and biochemical processes expressed by an organism in the form of phenotypes (qualitative and quantitative features) in a specific living condition. Plant phenomics depicts the phenotypic and genotypic expression within a particular living condition (Pratap et al., 2019). The phenotype of a plant may have a diverse manifestation within the same genotype due to different environmental conditions, i.e., abiotic stresses. The plant phenotype can be identified by using accomplishments made via genomic innovations and bioinformatics with the application of analytical techniques/phenomics technologies determining the relationship between plant genotype and environmental stress (Walter et al., 2015). The significance of phenomics is highly appreciable in the post-genomic era and is noticeable after using approaches like genomic selection, marker-assisted selection, GWAS, and QTL mapping, which are highly dependent on HTP for crop improvement. HTP technologies collect large data samples with automated digital analysis, interpret and process the data accurately with adequate statistical processing (Awada et al., 2018). The use of recent approaches for genetic dissection, knowledge of plant structure, and investigation of all functional processes enable the identification of plant phenotypic expression (Pratap et al., 2019). Some reported and widely used tools for phenotyping plants are provided in Supplementary Table 1. The integration of phenomics with other omics approaches forms the basis of crop breeding to identify plants with the best acceptable phenotype. It is the most promising approach for achieving sustainability in agriculture by understanding the plant’s response to multiple stresses, including toxic metals/metalloids toxicity.
Recently, root system architecture (RSA) traits have been studied in soybean using the phenome approach to identify genetic diversity since it is difficult to analyze RSA traits manually for a large number of accessions (Falk et al., 2020). Genotype and phenotype-based clusters of accessions showed similarity, in which genotype-based clusters depicted co-relation by geographical origins. This analysis provides opportunities for the breeding of root traits using beneficial functional genetic diversity that can be used in the future for crop improvements (Falk et al., 2020). In A. thaliana, a dataset of stress-responsive signals has been developed under a wide range of biotic and abiotic stresses, including Al and Fe toxicity, temperature, light, osmotic, and oxidative stresses. This research highlighted several shared and unshared biological processes, molecular functions, metabolic pathways, and phenomic characteristics that may help develop improved varieties using genome editing techniques (Naika et al., 2013). Since minimal applications of phenomics are reported against toxic metals/metalloids toxicity in plants, there is a margin to fulfill this gap by utilizing the phenomics approach to develop tolerant varieties by breeding crop plants against metals/metalloids toxicity.
The diversity and large volumes of omics data present a need for various tools required for data analysis. Several omics platforms have been developed to meet the needs and to integrate better the generated multi-omics data (Hernández-de-Diego et al., 2018). These tools are useful to decipher plant responses and behavior through results from the varying levels of molecular information. A few selected databases and tools/software for effective integration of the multi-omics data are discussed in this section. Some other examples, along with their functionality, have been provided in Supplementary Table 2.
One of the web databases, the Plant Omics Data Centre (PODC), allows gene mining by acquiring publicly available transcriptomics data obtained through RNA-sequencing of different model and crop plants. It also comprehensively integrates the gene expression networks and provides some essential features on inter species diversity by comparing gene expression networks, manual addition of information on published genes, and functional gene annotation through text mining (Shenton et al., 2019). Another web database, CATchUP, deals with the Spatio-temporal gene expression, i.e., genes expressed under specific conditions, by identification of the maximum expression difference of the transcripts. Owing to its unique data mining properties under particular treatment/conditions, it is mainly useful to find tissue-specific genes expressed at a certain developmental stage (Shenton et al., 2019). The National Genomics Data Centre (NGDC) processes a wide range of genomics-related data available through a family of database resources. This suite of databases has allowed data integration and curation, which can be accessed in either of three categories of resources, namely meta and sequenced data, standardized information, and processed data. Some prominent databases maintained by NGDC include manually curated Genome-Wide Variant-Trait Association (GWAS) Atlas, Genome Warehouse (GWH), Genome Variation Map (GVM), Plant Editosome Database (PED), and The Methylation Bank/MethBank (National Genomics Data Center Members and Partners, 2020).
Studies have shown that different tools and techniques involving joint visualization of the multi-omics data provide molecular context and a better understanding of the molecular interconnections (Hernández-de-Diego et al., 2018). One of the metabolic pathway-based data visualization tools, KaPPa-View, allows the integration of transcript and metabolite data into pathway maps (Hernández-de-Diego et al., 2018) and hence helps to find a correlation with the biological knowledge. Similarly, a protein classification and annotation framework, MapMan4, supports multiple omics platforms, including transcriptomics, proteomics, and metabolomics (Schwacke et al., 2019). Another web-based tool, PaintOmics3, lays a framework for interactive exploration of multi-omics data via the KEGG pathway diagram and has enabled researchers to understand different levels of regulation in a biological system (Hernández-de-Diego et al., 2018). MetaBridge is a database that uses data from KEGG and MetaCyc metabolic pathway databases. It is useful for data integration and gene mapping for specific metabolites (Blimkie et al., 2020). The Miodin (MultI-Omics Data INtegration) R package allows users to integrate the omics data in a streamlined workflow-based syntax either based on vertical (across experiments on the same samples) or horizontal (across studies on the same variables) integration of samples. It supports the multi-omics modalities and enables workflows to import efficiently, process, and analyze the multi-omics data (Ulfenborg, 2019).
Besides the fact that these tools and databases have proved useful for researchers, the fast-evolving multi-omics platforms have resulted in the lack of uniformity among the available tools. This necessitates committed efforts for quality assessment and validation from scientific communities across the world.
Rapid industrialization has resulted in a large number of toxic metals/metalloids pollutants into the soil, water, and air, which then enter crops directly or indirectly through various pathways, posing significant health and wildlife risks (Rai et al., 2019; Raza et al., 2020, 2021a). Nevertheless, different plant-based remediation techniques have been developed to solve these issues, and some of the major techniques have been briefly discussed in the subsequent sections.
Bioremediation, particularly biochar amended phytoremediation, is one of the most promising remediation strategies that is widely used to remediate toxic metals/metalloids in the soil (Shukla A. et al., 2018; Wang et al., 2021). Biochar has many significant properties that make it a good remediation material for contaminated soils, including a large internal surface area, negative charge, and resistance to degradation. It is also the most important bioremediation strategy since it not only serves to remediate soil pollution and improve soil quality but is also a cost-effective product that decreases the global environmental challenge of solid waste management (Shukla A. et al., 2018; Wang et al., 2021). A recent study shows that the addition of 10% of biochar generated from rice straw to Cu-contaminated soil reduced Cu availability by 96% (Rehman et al., 2019). Furthermore, biochar made from rice husk pyrolysis was found to be a useful agricultural by-product and an excellent immobilizer of metals contaminants such as Zn, Pb, and Cd (Dhiman et al., 2021). Sulfur-modified rice husk biochar advanced its sulfur content and was utilized as a “green bioremediation approach” to stabilize Hg-polluted soil (O’Connor et al., 2018). Metal pollutants are taken in and concentrated by hyperaccumulators and organic soil remedies, resulting in their precipitation within the root’s innards (Huang et al., 2018). Cassia alata, a promising photo stabilizer with biochar aid to improve soil pH, nitrogen content, and total carbon, reduces Pb and Cu metal contamination in mine tailings and also leads to increased plant shoot and root biomass (Huang et al., 2018).
Phytostabilization is the process in which plant species with high metal tolerance are used to immobilize toxic metals/metalloids below ground and reduce their bioavailability, thus preventing metals from migrating into the environment and reducing the probability of metals/metalloids entering the food chain (Raza et al., 2020; Yan et al., 2020). In this approach, toxic metals/metalloids are physically and chemically immobilized in the rhizosphere through sorption on roots of plants, fixation with soil amendments, complexation, precipitation, or reduction in metals valence (Mthembu et al., 2020). Phytostabilization requires the identification of appropriate plant species with high resistance to toxic metals/metalloids (Yan et al., 2020). Because of their better tolerance to toxic metals/metalloids, territorial expansion, and evapotranspiration, some plant species, particularly crop grasses and willows (Salix spp.), are effective for phytostabilization (Sylvain et al., 2016). Plants such as Festuca spp. and Agrostis spp. are commonly used to phytostabilize Pb, Zn, and Cu contaminants in soil (Galende et al., 2014). Lands with increased metals/metalloids contamination or physical disturbances lack natural flora; hence, phytostabilization techniques can be used to rebuild a vegetative cover. Wind erosion of surface soils and leaching to groundwater is reduced when metal-tolerant plants are planted in such areas (Mthembu et al., 2020).
Phytoextraction is the process of absorbing, transporting, and accumulating toxic metals/metalloids in the biomass of harvestable plant parts using specialized and highly adapted hyperaccumulators (Diarra et al., 2021). The electrochemical characteristics of the metals being extracted, as well as their absorption, transport, and accumulation in the plants, all influence the efficiency of the phytoextraction process (Saini and Dhania, 2020). The most important factor determining the viability and success of phytoextraction is plant selection criteria; therefore, hyperaccumulators with exceptional tolerance and bioaccumulation of a broad range of toxic metals/metalloids are preferred (Diarra et al., 2021). Over 500 plant species from 45 different families, including Lamiaceae, Caryophyllaceae, Poaceae, Asteraceae, Fabaceae, and Brassicaceae, have been identified as hyperaccumulators so far. Brassicaceae is the most common family among those identified (Gul et al., 2021). In a recent study, the flowering plant Lantana camara L. exhibited all of the desired criteria of a hyperaccumulator plant, including a translocation factor (TF) >1 and Cd content in aerial portions of >100 mg kg–1. As a result, it was proposed as a Cd hyperaccumulator (Liu S. et al., 2019). Artemisia vulgaris was recently identified as a promising plant for metal phytoremediation, with TF > 11 for As, Cd, Cu, Cr, Ni, and Pb, while Silene vulgaris had a TF greater than 1 for Cd (Antoniadis et al., 2020).
Climate change and agricultural production are highly correlated, as climate change is the leading cause of several environmental stresses, including toxic metals/metalloids toxicity in the soil and environment. Increasing metals/metalloids toxicity poses a severe threat to agricultural production by hampering plant growth and yield. The toxic effects of these toxic metals/metalloids also significantly affect the plant’s physiological, biochemical, and molecular mechanisms, which are vital for healthy plant growth and improved yield. Several plant-based remediation approaches have also been successfully used to remediate the toxicity of the toxic metals/metalloids in the soil and environment. Under metals/metalloids toxicity, plants adjust themselves by modifying genes, proteins, and metabolites expression levels to cope with unfavorable conditions. Thus, a comprehensive understanding of the biological processes is required to cope with toxic metals/metalloids toxicity in the emerging technological era. During the last few years, significant progress has been made in utilizing state-of-the-art “omics” approaches to develop climate-smart plants. Interestingly, genome-wide omics analysis holds the potential to identify stress-related genes, metabolites, proteins, minor and major elements, miRNAs, stress regulators, and metabolic pathways precisely associated with plant phenotype under stressful environments.
Nevertheless, some persisting bottlenecks in exploiting omics approaches demand urgent attention (Figure 3; Raza et al., 2021b,d). Polygenic inheritance and genetic diversity are significant bottlenecks among the existing ones. Therefore, releasing these bottlenecks using molecular tools will help us to feat the new strategy to develop stress resistance plants; and consecutively will guarantee global food safety. For instance, recent developments in high-throughput technologies enable the analysis of global genetic diversity. Genomic selection and haplotype-based breeding are expected to lessen the genetic diversity of a breeding plan in the long term. Consequently, sustaining genetic diversity in breeding programs will be essential for maintaining genetic gains from breeding revolutions (Varshney et al., 2021a,b, c). Therefore, integrating ideal selection contributions with these methodologies may help preserve genetic diversity while enhancing genetic gains. Furthermore, effective crop breeding programs with an improved genetic base will speed up fast-forward breeding, proposed by Varshney et al. (2021b). Briefly, genomics-assisted breeding, including haplotype-GWAS, haplotype-assisted breeding, and haplotype-assisted genomic selection, will effectively utilize superior haplotypes to accelerate future breeding advancement (Bhat et al., 2021). Moreover, modern machine learning tools and artificial intelligence methodologies systematically incorporate the overflow of data rolling through multi-omics approaches (van Dijk et al., 2020; Jung et al., 2021). Hence, system-level knowledge will help clarify functional diversity and regulatory networks underlying intricate phenotypes of agricultural value.
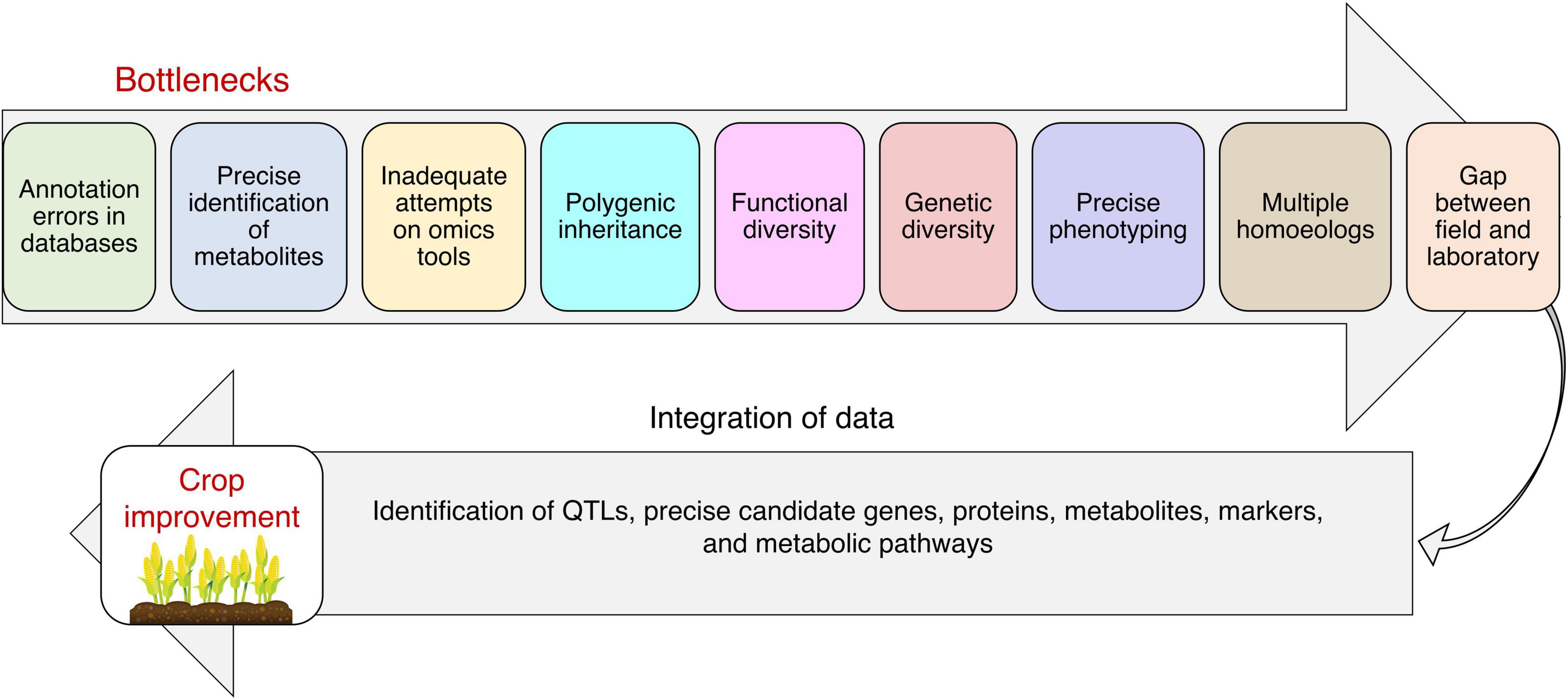
Figure 3. Persisting bottlenecks in the exploitation of omics approaches (Raza et al., 2021b). Although an outstanding revolution has been accomplished in the biotechnological era, many queries and bottlenecks presently limit the utilization of omics approaches for stress tolerance research in several plant species. The amendment of these bottlenecks using molecular tools will help us to exploit the innovative manifesto in developing toxic metals/metalloids tolerant plants.
Genome editing by CRISPR/Cas systems and genetic engineering of stress-associated genes could be essential prospects for improving stress tolerance. Likewise, the engineering of metabolic pathways could also provide new avenues for the development of climate-resilient plants. Recently, speed breeding has emerged as the most powerful and time-saving tool to shorten the breeding cycle and enhance genetic gains in plants. Therefore, omics approaches could be galvanized by utilizing speed breeding to facilitate plant breeders to keep pace with increasing environmental fluctuations and burgeoning human population.
AR conceived the idea and prepared the figures. AR, JT, ZZ, SC, SB, RB, CZ, and HC contributed in writing. AR, JT, ZZ, and SC designed the tables. AR, RB, FBJ, RSAK, RKV, and WZ reviewed and edited the manuscript. All authors have read and approved the final version of the manuscript.
This work was supported by grants from the National Science Foundation (NSF) of China (U1705233 to WZ) and the Fujian Agriculture and Forestry University, Fuzhou, China. Further, RKV is thankful to Bill & Melinda Gates Foundation, USA, and Australia-India Strategic Research Fund from the Department of Biotechnology, Government of India, for supporting this work in part.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
AR would like to thank the Fujian Agriculture and Forestry University (FAFU) and the Fujian Provincial Government for providing Post-doc Research Fellowship. We are grateful to many scientists and colleagues for scientific discussions, which enabled the development of this up-to-date comprehensive review. We apologize to colleagues whose relevant work could not be cited due to space limitations.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.794373/full#supplementary-material
Alka, S., Shahir, S., Ibrahim, N., Rahmad, N., Haliba, N., and Abd Manan, F. (2021). Histological and proteome analyses of Microbacterium foliorum-mediated decrease in arsenic toxicity in Melastoma malabathricum. 3 Biotech 11, 1–17. doi: 10.1007/s13205-021-02864-y
Alotaibi, M. O., Mohammed, A. E., Almutairi, T. A., and Elobeid, M. M. (2019). Morpho-physiological and proteomic analyses of Eucalyptus camaldulensis as a bioremediator in copper-polluted soil in Saudi Arabia. Plants 8:43. doi: 10.3390/plants8020043
Ángel, M. R. J., Ariani, A., Leija, A., Elizondo, A., Fuentes, S. I., Ramirez, M., et al. (2021). Phaseolus vulgaris MIR1511 genotypic variations differentially regulate plant tolerance to aluminum toxicity. Plant J. 105, 1521–1533. doi: 10.1111/tpj.15129
Antoniadis, V., Shaheen, S. M., Stark, H. J., Wennrich, R., Levizou, E., Merbach, I., et al. (2020). Phytoremediation potential of twelve wild plant species for toxic elements in a contaminated soil. Environ. Int. 146:106233. doi: 10.1016/j.envint.2020.106233
Arbelaez, J. D., Maron, L. G., Jobe, T. O., Piñeros, M. A., Famoso, A. N., Rebelo, A. R., et al. (2017). ALUMINUM RESISTANCE TRANSCRIPTION FACTOR 1 (ART1) contributes to natural variation in aluminum resistance in diverse genetic backgrounds of rice (O. sativa). Plant Direct. 1:4. doi: 10.1002/pld3.14
Arora, N., Dubey, D., Sharma, M., Patel, A., Guleria, A., Pruthi, P. A., et al. (2018). NMR-based metabolomic approach to elucidate the differential cellular responses during mitigation of arsenic (III, V) in a green microalga. ACS Omega 3, 11847–11856. doi: 10.1021/acsomega.8b01692
Asaari, M. S. M., Mertens, S., Dhondt, S., Inzé, D., Wuyts, N., and Scheunders, P. (2019). Analysis of hyperspectral images for detection of drought stress and recovery in maize plants in a high-throughput phenotyping platform. Comput. Electron. Agric. 162, 749–758.
Asati, A., Pichhode, M., and Nikhil, K. (2016). Effect of heavy metals on plants: an overview. Int. J. Appl. Innov. Eng. Manag. 5, 56–66.
Ashraf, M. Y., Sadiq, R., Hussain, M., Ashraf, M., and Ahmad, M. S. A. (2011). Toxic effect of nickel (Ni) on growth and metabolism in germinating seeds of sunflower (Helianthus annuus L.). Biol. Trace Element Res. 143, 1695–1703. doi: 10.1007/s12011-011-8955-7
Asins, M., Raga, M., Torrent, D., Roca, D., and Carbonell, E. (2020). QTL and candidate gene analyses of rootstock-mediated tomato fruit yield and quality traits under low iron stress. Euphytica 216, 1–19. doi: 10.9734/cjast/2019/v38i630452
Assefa, T., Zhang, J., Chowda-Reddy, R., Lauter, A. N. M., Singh, A., O’Rourke, J. A., et al. (2020). Deconstructing the genetic architecture of iron deficiency chlorosis in soybean using genome-wide approaches. BMC Plant Biol. 20:42. doi: 10.1186/s12870-020-2237-5
Awada, L., Phillips, P. W., and Smyth, S. J. (2018). The adoption of automated phenotyping by plant breeders. Euphytica 214:148.
Bahmani, R., Bihamta, M., Habibi, D., Forozesh, P., and Ahmadvand, S. (2012). Effect of cadmium chloride on growth parameters of different bean genotypes (Phaseolus vulgaris L.). ARPN J. Agric. Biol. Sci. 7, 35–40.
Balk, J., and Schaedler, T. A. (2014). Iron cofactor assembly in plants. Ann. Rev. Plant Biol. 65, 125–153. doi: 10.1146/annurev-arplant-050213-035759
Banerjee, A., Singh, A., Sudarshan, M., and Roychoudhury, A. (2020). Silicon nanoparticle-pulsing mitigates fluoride stress in rice by fine-tuning the ionomic and metabolomic balance and refining agronomic traits. Chemosphere 262:127826. doi: 10.1016/j.chemosphere.2020.127826
Benyó, D., Horváth, E., Németh, E., Leviczky, T., Takács, K., Lehotai, N., et al. (2016). Physiological and molecular responses to heavy metal stresses suggest different detoxification mechanism of Populus deltoides and P. x canadensis. J. Plant Physiol. 201, 62–70. doi: 10.1016/j.jplph.2016.05.025
Bhat, J. A., Yu, D., Bohra, A., Ganie, S. A., and Varsheny, R. K. (2021). Features and applications of haplotypes in crop breeding. Commun. Biol. 4:1266. doi: 10.1038/s42003-021-02782-y
Blimkie, T., Lee, A. H. Y., and Hancock, R. E. (2020). MetaBridge: an integrative multi-omics tool for metabolite-enzyme mapping. Curr. Protoc. Bioinform. 70:e98. doi: 10.1002/cpbi.98
Bollinedi, H., Yadav, A. K., Vinod, K., Krishnan, S. G., Bhowmick, P. K., Nagarajan, M., et al. (2020). Genome-wide association study reveals novel marker-trait associations (MTAs) governing the localization of Fe and Zn in the rice grain. Front. Genet. 11:213. doi: 10.3389/fgene.2020.00213
Borjigin, C., Schilling, R. K., Bose, J., Hrmova, M., Qiu, J., Wege, S., et al. (2020). A single nucleotide substitution in TaHKT1; 5-D controls shoot Na+ accumulation in bread wheat. bioRxiv [Preprint]. doi: 10.1101/2020.01.21.909887
Campos, N. V., Arcanjo-Silva, S., Freitas-Silva, L., de Araújo, T. O., Souza-Fernandes, D. P., and Azevedo, A. A. (2018). Arsenic hyperaccumulation in Pityrogramma calomelanos L.(Link): adaptive traits to deal with high metalloid concentrations. Environ. Sci. Pollut. Res. 25, 10720–10729. doi: 10.1007/s11356-017-1085-9
Canzler, S., Schor, J., Busch, W., Schubert, K., Rolle-Kampczyk, U. E., Seitz, H., et al. (2020). Prospects and challenges of multi-omics data integration in toxicology. Arch. Toxicol. 94, 371–388. doi: 10.1007/s00204-020-02656-y
Carroll, A. A., Clarke, J., Fahlgren, N., Gehan, M. A., Lawrence-Dill, C. J., and Lorence, A. (2019). NAPPN: who we are, where we are going, and why you should join us! Plant Phenome J. 2, 1–4.
Casarrubia, S., Martino, E., Daghino, S., Kohler, A., Morin, E., Khouja, H.-R., et al. (2020). Modulation of plant and fungal gene expression upon cd exposure and symbiosis in ericoid mycorrhizal Vaccinium myrtillus. Front. Microbiol. 11:341. doi: 10.3389/fmicb.2020.00341
Caspi, R., Billington, R., Ferrer, L., Foerster, H., Fulcher, C. A., Keseler, I. M., et al. (2016). The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 44, D471–D480.
Ceballos-Laita, L., Gutierrez-Carbonell, E., Imai, H., Abadía, A., Uemura, M., Abadía, J., et al. (2018). Effects of manganese toxicity on the protein profile of tomato (Solanum lycopersicum) roots as revealed by two complementary proteomic approaches, two-dimensional electrophoresis and shotgun analysis. J. Proteom. 185, 51–63. doi: 10.1016/j.jprot.2018.06.016
Cedillo-Jimenez, C. A., García-Ortega, L. F., Martínez-Reséndiz, M., Arné-Robles, D., Cruz-Hernandez, A., and Guevara-Gonzalez, R. G. (2020). Response of transgenic tobacco overexpressing the CchGLP gene to cadmium and aluminium: phenotypic and microRNAs expression changes. Physiol. Mol. Biol. Plant 26, 3–13. doi: 10.1007/s12298-019-00716-x
Chandra, R., and Kang, H. (2016). Mixed heavy metal stress on photosynthesis, transpiration rate, and chlorophyll content in poplar hybrids. Forest Sci. Technol. 12, 55–61. doi: 10.1080/21580103.2015.1044024
Chang, J. D., Huang, S., Yamaji, N., Zhang, W., Ma, J. F., and Zhao, F. J. (2020). OsNRAMP1 transporter contributes to cadmium and manganese uptake in rice. Plant Cell Environ. 43, 2476–2491. doi: 10.1111/pce.13843
Chauhan, R., Awasthi, S., Indoliya, Y., Chauhan, A. S., Mishra, S., Agrawal, L., et al. (2020). Transcriptome and proteome analyses reveal selenium mediated amelioration of arsenic toxicity in rice (Oryza sativa L.). J. Hazard. Mater. 390:122122. doi: 10.1016/j.jhazmat.2020.122122
Chen, L., Wan, H., Qian, J., Guo, J., Sun, C., Wen, J., et al. (2018). Genome-wide association study of cadmium accumulation at the seedling stage in rapeseed (Brassica napus L.). Front. Plant Sci. 9:375. doi: 10.3389/fpls.2018.00375
Chen, Y., Zhi, J., Zhang, H., Li, J., Zhao, Q., and Xu, J. (2017). Transcriptome analysis of Phytolacca americana L. in response to cadmium stress. PLoS One 12:e0184681. doi: 10.1371/journal.pone.0184681
Chong, J., Soufan, O., Li, C., Caraus, I., Li, S., Bourque, G., et al. (2018). MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 46, W486–W494. doi: 10.1093/nar/gky310
Chou, T.-S., Chao, Y.-Y., Huang, W.-D., Hong, C.-Y., and Kao, C. H. (2011). Effect of magnesium deficiency on antioxidant status and cadmium toxicity in rice seedlings. J. Plant Physiol. 168, 1021–1030. doi: 10.1016/j.jplph.2010.12.004
Chu, Q., Sha, Z., Maruyama, H., Yang, L., Pan, G., Xue, L., et al. (2019). Metabolic reprogramming in nodules, roots, and leaves of symbiotic soybean in response to iron deficiency. Plant Cell Environ. 42, 3027–3043. doi: 10.1111/pce.13608
Colmer, J., O’Neill, C. M., Wells, R., Bostrom, A., Reynolds, D., Websdale, D., et al. (2020). SeedGerm: a cost-effective phenotyping platform for automated seed imaging and machine-learning based phenotypic analysis of crop seed germination. New Phytol. 228, 778–793. doi: 10.1111/nph.16736
Cosio, C., and Renault, D. (2020). Effects of cadmium, inorganic mercury and methyl-mercury on the physiology and metabolomic profiles of shoots of the macrophyte Elodea nuttallii. Environ. Pollut. 257:113557. doi: 10.1016/j.envpol.2019.113557
Cota-Ruiz, K., Ye, Y., Valdes, C., Deng, C., Wang, Y., Hernández-Viezcas, J. A., et al. (2020). Copper nanowires as nanofertilizers for alfalfa plants: understanding nano-bio systems interactions from microbial genomics, plant molecular responses and spectroscopic studies. Sci. Total Environ. 742:140572. doi: 10.1016/j.scitotenv.2020.140572
Cury, N. F., Silva, R. C., Andre, M. S., Fontes, W., Ricart, C. A., Castro, M. S., et al. (2020). Root proteome and metabolome reveal a high nutritional dependency of aluminium in Qualea grandiflora Mart. (Vochysiaceae). Plant Soil 446, 125–143.
Dalvi, A. A., and Bhalerao, S. A. (2013). Response of plants towards heavy metal toxicity: an overview of avoidance, tolerance and uptake mechanism. Ann. Plant Sci. 2, 362–368.
Dalyan, E., Yüzbaşıoğlu, E., Keskin, B. C., Yıldızhan, Y., Memon, A., Ünal, M., et al. (2017). The identification of genes associated with Pb and Cd response mechanism in Brassica juncea L. by using Arabidopsis expression array. Environ. Exp. Bot. 139, 105–115. doi: 10.1016/j.envexpbot.2017.05.001
Das, P., Khare, P., Singh, R. P., Yadav, V., Tripathi, P., Kumar, A., et al. (2021). Arsenic-induced differential expression of oxidative stress and secondary metabolite content in two genotypes of Andrographis paniculata. J. Hazard. Mater. 406:124302. doi: 10.1016/j.jhazmat.2020.124302
de Alvarenga Mudadu, M., and Zerlotini, A. (2020). Machado: open source genomics data integration framework. bioRxiv [Preprint]. doi: 10.1101/2020.05.08.084731
Derakhshani, B., Jafary, H., Maleki Zanjani, B., Hasanpur, K., Mishina, K., Tanaka, T., et al. (2020). Combined QTL mapping and RNA-Seq profiling reveals candidate genes associated with cadmium tolerance in barley. PLoS One 15:e0230820. doi: 10.1371/journal.pone.0230820
Dhiman, S., Ibrahim, M., Devi, K., Sharma, N., Kapoor, N., Kaur, R., et al. (2021). “Biochar assisted remediation of toxic metals and metalloids.” in Handbook of Assisted and Amendment: Enhanced Sustainable Remediation Technology, ed. M. N. V. Prasad (Hoboken, NJ: Wiley-Blackwell), 131–162. doi: 10.1002/9781119670391.ch7
Di, X., Zheng, F., Norton, G. J., Beesley, L., Zhang, Z., Lin, H., et al. (2021). Physiological responses and transcriptome analyses of upland rice following exposure to arsenite and arsenate. Environ. Exp. Bot. 183:104366. doi: 10.1016/j.envexpbot.2020.104366
Diarra, I., Kotra, K. K., and Prasad, S. (2021). Assessment of biodegradable chelating agents in the phytoextraction of heavy metals from multi–metal contaminated soil. Chemosphere 273:128483. doi: 10.1016/j.chemosphere.2020.128483
Ding, Y., Ding, L., Xia, Y., Wang, F., and Zhu, C. (2020). Emerging roles of microRNAs in plant heavy metal tolerance and homeostasis. J. Agric. Food Chem. 68, 1958–1965. doi: 10.1021/acs.jafc.9b07468
Dubey, S., Shri, M., and Chakrabarty, D. (2021). MicroRNA mediated regulation of gene expression in response to heavy metals in plants. J. Plant Biochem. Biotechnol. [Epub ahead of print]. doi: 10.1007/s13562-021-00718-5
Eleftheriou, E. P., Adamakis, I.-D. S., Panteris, E., and Fatsiou, M. (2015). Chromium-induced ultrastructural changes and oxidative stress in roots of Arabidopsis thaliana. Int. J. Mol. Sci. 16, 15852–15871. doi: 10.3390/ijms160715852
Emamverdian, A., Ding, Y., Mokhberdoran, F., and Xie, Y. (2015). Heavy metal stress and some mechanisms of plant defense response. Sci. World J. 2015, 756120. doi: 10.1155/2015/756120
Erickson, T. B., Brooks, J., Nilles, E. J., Pham, P. N., and Vinck, P. (2019). Environmental health effects attributed to toxic and infectious agents following hurricanes, cyclones, flash floods and major hydrometeorological events. J. Toxicol. Environ. Health, Part B 22, 157–171. doi: 10.1080/10937404.2019.1654422
Falk, K. G., Jubery, T. Z., O’Rourke, J. A., Singh, A., Sarkar, S., Ganapathysubramanian, B., et al. (2020). Soybean root system architecture trait study through genotypic, phenotypic, and shape-based clusters. Plant Phenomics 2020:1925495. doi: 10.34133/2020/1925495
Famoso, A. N., Zhao, K., Clark, R. T., Tung, C. W., Wright, M. H., Bustamante, C., et al. (2011). Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome-wide association analysis and QTL mapping. PLoS Genet. 7:e1002221. doi: 10.1371/journal.pgen.1002221
Farokhzadeh, S., Fakheri, B. A., Nezhad, N. M., Tahmasebi, S., Mirsoleimani, A., and Heidari, B. (2020b). Mapping QTLs associated with grain yield and yield-related traits under aluminum stress in bread wheat. Crop Past. Sci. 71, 429–444.
Farokhzadeh, S., Fakheri, B. A., Nezhad, N. M., Tahmasebi, S., Mirsoleimani, A., and McIntyre, C. L. (2020c). Genetic control of some plant growth characteristics of bread wheat (Triticum aestivum L.) under aluminum stress. Genes Genomics 42, 245–261. doi: 10.1007/s13258-019-00895-7
Farokhzadeh, S., Fakheri, B. A., Nezhad, N. M., Tahmasebi, S., and Mirsoleimani, A. (2020a). Genetic dissection of spike-related traits in wheat (Triticum aestivum L.) under aluminum stress. Genet. Resour. Crop Evol. 67, 1221–1243. doi: 10.1007/s10722-020-00907-6
Farooq, M. A., Hong, Z., Islam, F., Noor, Y., Hannan, F., Zhang, Y., et al. (2021). Comprehensive proteomic analysis of arsenic induced toxicity reveals the mechanism of multilevel coordination of efficient defense and energy metabolism in two Brassica napus cultivars. Ecotoxicol. Environ. Saf. 208:111744. doi: 10.1016/j.ecoenv.2020.111744
Feng, X., Han, L., Chao, D., Liu, Y., Zhang, Y., Wang, R., et al. (2017). Ionomic and transcriptomic analysis provides new insight into the distribution and transport of cadmium and arsenic in rice. J. Hazard. Mater. 331, 246–256. doi: 10.1016/j.jhazmat.2017.02.041
Fu, S., Lu, Y., Zhang, X., Yang, G., Chao, D., Wang, Z., et al. (2019). The ABC transporter ABCG36 is required for cadmium tolerance in rice. J. Exp. Bot. 70, 5909–5918. doi: 10.1093/jxb/erz335
Galende, M. A., Becerril, J. M., Barrutia, O., Artetxe, U., Garbisu, C., and Hernández, A. (2014). Field assessment of the effectiveness of organic amendments for aided phytostabilization of a Pb-Zn contaminated mine soil. J. Geochem. Explor. 145, 181–189. doi: 10.1016/j.scitotenv.2016.04.080
Gao, J., Luo, M., Peng, H., Chen, F., and Li, W. (2019). Characterization of cadmium-responsive MicroRNAs and their target genes in maize (Zea mays) roots. BMC Mol. Biol. 20:14. doi: 10.1186/s12867-019-0131-1
Gao, Y., Yang, F., Liu, J., Xie, W., Zhang, L., Chen, Z., et al. (2020). Genome-wide identification of metal tolerance protein genes in Populus trichocarpa and their roles in response to various heavy metal stresses. Int. J. Mol. Sci. 21:1680. doi: 10.3390/ijms21051680
Gashi, B., Osmani, M., Aliu, S., Zogaj, M., and Kastrati, F. (2020). Risk assessment of heavy metal toxicity by sensitive biomarker δ-aminolevulinic acid dehydratase (ALA-D) for onion plants cultivated in polluted areas in Kosovo. J. Environ. Sci. Health B 55, 462–469. doi: 10.1080/03601234.2020.1721229
Ghori, N.-H., Ghori, T., Hayat, M., Imadi, S., Gul, A., Altay, V., et al. (2019). Heavy metal stress and responses in plants. Int. J. Environ. Sci. Technol. 16, 1807–1828.
Ghosh, S., Datta, A., Tan, K., and Choi, H. (2019). SLIDE–a web-based tool for interactive visualization of large-scale–omics data. Bioinformatics 35, 346–348. doi: 10.1093/bioinformatics/bty534
Gilliham, M., Able, J. A., and Roy, S. J. (2017). Translating knowledge about abiotic stress tolerance to breeding programmes. Plant J. 90, 898–917. doi: 10.1111/tpj.13456
Gouveia, G. C. C., Galindo, F. S., Lanza, M. G. D. B., da Rocha Silva, A. C., de Brito Mateus, M. P., da Silva, M. S., et al. (2020). Selenium toxicity stress-induced phenotypical, biochemical and physiological responses in rice plants: characterization of symptoms and plant metabolic adjustment. Ecotoxicol. Environ. Saf. 202:110916. doi: 10.1016/j.ecoenv.2020.110916
Gui, S., Yang, L., Li, J., Luo, J., Xu, X., Yuan, J., et al. (2020). ZEAMAP, a comprehensive database adapted to the maize multi-omics era. iScience 23:101241. doi: 10.1016/j.isci.2020.101241
Gul, I., Manzoor, M., Hashim, N., Shah, G. M., Waani, S. P. T., Shahid, M., et al. (2021). Challenges in microbially and chelate-assisted phytoextraction of cadmium and lead–A review. Environ. Pollut. 287:117667. doi: 10.1016/j.envpol.2021.117667
Hasanuzzaman, M., Bhuyan, M. B., Raza, A., Hawrylak-Nowak, B., Matraszek-Gawron, R., Al Mahmud, J., et al. (2020a). Selenium in plants: boon or bane? Environ. Exp. Bot. 178:104170. doi: 10.1016/j.envexpbot.2020.104170
Hasanuzzaman, M., Bhuyan, M., Zulfiqar, F., Raza, A., Mohsin, S. M., Mahmud, J. A., et al. (2020b). Reactive oxygen species and antioxidant defense in plants under abiotic stress: revisiting the crucial role of a universal defense regulator. Antioxidants 9:681. doi: 10.3390/antiox9080681
Hattab, S., Hattab, S., Flores-Casseres, M. L., Boussetta, H., Doumas, P., Hernandez, L. E., et al. (2016). Characterisation of lead-induced stress molecular biomarkers in Medicago sativa plants. Environ. Exp. Bot. 123, 1–12.
Helaoui, S., Boughattas, I., Hattab, S., Mkhinini, M., Alphonse, V., Livet, A., et al. (2020). Physiological, biochemical and transcriptomic responses of Medicago sativa to nickel exposure. Chemosphere 249:126121. doi: 10.1016/j.chemosphere.2020.126121
Hernández-de-Diego, R., Tarazona, S., Martínez-Mira, C., Balzano-Nogueira, L., Furió-Tarí, P., and Pappas, G. J. Jr. et al., (2018). PaintOmics 3: a web resource for the pathway analysis and visualization of multi-omics data. Nucleic Acids Res. 46, W503–W509. doi: 10.1093/nar/gky466
Hu, Z., Fu, Q., Zheng, J., Zhang, A., and Wang, H. (2020). Transcriptomic and metabolomic analyses reveal that melatonin promotes melon root development under copper stress by inhibiting jasmonic acid biosynthesis. Hortic. Res. 7, 1–15. doi: 10.1038/s41438-020-0293-5
Huang, L., Li, Y., Zhao, M., Chao, Y., Qiu, R., Yang, Y., et al. (2018). Potential of Cassia alata L. coupled with biochar for heavy metal stabilization in multi-metal mine tailings. Int. J. Environ. Res. Public Health 15:494. doi: 10.3390/ijerph15030494
Huang, S. Q., Peng, J., Qiu, C. X., and Yang, Z. M. (2009). Heavy metal-regulated new microRNAs from rice. J. Inor. Biochem. 103, 282–287. doi: 10.1016/j.jinorgbio.2008.10.019
Hussain, M. B., Ali, S., Azam, A., Hina, S., Farooq, M. A., Ali, B., et al. (2013). Morphological, physiological and biochemical responses of plants to nickel stress: a review. Afr. J. Agric. Res. 8, 1596–1602.
Ibarra, A. A. G., Wrobel, K., Barrientos, E. Y., Escobosa, A. R. C., Corona, J. F. G., Donis, I. E., et al. (2019). Impact of Cr (VI) on the oxidation of polyunsaturated fatty acids in Helianthus annuus roots studied by metabolomic tools. Chemosphere 220, 442–451. doi: 10.1016/j.chemosphere.2018.12.145
Ifie, J., Ifie-Etumah, S., and Ikhajiagbe, B. (2020). Physiological and biochemical responses of selected cowpea (Vigna unguiculata (L.) Walp.) accessions to iron toxicity. Acta Agric. Slovenica 115, 25–38.
Jalmi, S. K., Bhagat, P. K., Verma, D., Noryang, S., Tayyeba, S., Singh, K., et al. (2018). Traversing the links between heavy metal stress and plant signaling. Front. Plant Sci. 9:12. doi: 10.3389/fpls.2018.00012
Jamla, M., Khare, T., Joshi, S., Patil, S., Penna, S., and Kumar, V. (2021a). Omics approaches for understanding heavy metal responses and tolerance in plants. Curr. Plant Biol. 27:100213. doi: 10.1016/j.cpb.2021.100213
Jamla, M., Patil, S., Joshi, S., Khare, T., and Kumar, V. (2021b). MicroRNAs and their exploration for developing heavy metal-tolerant plants. J. Plant Growth Regul. [Epub ahead of print]. doi: 10.1007/s00344-021-10476-2
Jang, Y., Yu, N., Seo, J., Kim, S., and Lee, S. (2016). MONGKIE: an integrated tool for network analysis and visualization for multi-omics data. Biol. Direct 11, 1–9. doi: 10.1186/s13062-016-0112-y
Janicka-Russak, M., Kabała, K., and Burzyński, M. (2012). Different effect of cadmium and copper on H+-ATPase activity in plasma membrane vesicles from Cucumis sativus roots. J. Exp. Bot. 63, 4133–4142. doi: 10.1093/jxb/ers097
Jian, H., Yang, B., Zhang, A., Ma, J., Ding, Y., Chen, Z., et al. (2018). Genome-wide identification of microRNAs in response to cadmium stress in oilseed rape (Brassica napus L.) using high-throughput sequencing. Int. J. Mol. Sci. 19:1431. doi: 10.3390/ijms19051431
Jiang, W., Wu, Z., Wang, T., Mantri, N., Huang, H., Li, H., et al. (2020). Physiological and transcriptomic analyses of cadmium stress response in Dendrobium officinale seedling. Plant Physiol. Biochem. 148, 152–165. doi: 10.1016/j.plaphy.2020.01.010
Jorrin-Novo, J. V., Komatsu, S., Sanchez-Lucas, R., and de Francisco, L. E. R. (2019). Gel electrophoresis-based plant proteomics: past, present, and future. Happy 10th anniversary Journal of Proteomics! J. Proteom. 198, 1–10. doi: 10.1016/j.jprot.2018.08.016
Jung, J., Maeda, M., Chang, A., Bhandari, M., Ashapure, A., and Landivar-Bowles, J. (2021). The potential of remote sensing and artificial intelligence as tools to improve the resilience of agriculture production systems. Curr. Opin. Biotechnol. 70, 15–22. doi: 10.1016/j.copbio.2020.09.003
Júnior, C. A. L., Mazzafera, P., and Arruda, M. A. Z. (2014). A comparative ionomic approach focusing on cadmium effects in sunflowers (Helianthus annuus L.). Environ. Exp. Bot. 107, 180–186. doi: 10.1016/j.envexpbot.2014.06.002
Kakeshpour, T. (2020). Tomato Class II Glutaredoxin Mutants Generated Via Multiplex CRISPR/Cas9 Genome Editing Technology are Susceptible to Multiple Abiotic Stresses. Doctoral dissertation. Manhattan, KS: Kansas State University.
Kanehisa, M., Sato, Y., Furumichi, M., Morishima, K., and Tanabe, M. (2019). New approach for understanding genome variations in KEGG. Nucleic Acids Res. 47, D590–D595. doi: 10.1093/nar/gky962
Kar, S., Mai, H.-J., Khalouf, H., Abdallah, H. B., Flachbart, S., Fink-Straube, C., et al. (2020). Transcriptomic expression patterns of two contrasting lowland rice varieties reveal high iron stress tolerance. bioRxiv [Preprint]. doi: 10.1101/2020.05.01.070516
Karthika, K., Rashmi, I., and Parvathi, M. (2018). “Biological functions, uptake and transport of essential nutrients in relation to plant growth,” in Plant Nutrients and Abiotic Stress Tolerance, eds M. Hasanuzzaman, M. Fujita, H. Oku, K. Nahar, and B. Hawrylak-Nowak (Berlin: Springer), 1–49. doi: 10.1007/978-981-10-9044-8_1
Kashem, M. A., and Kawai, S. (2007). Alleviation of cadmium phytotoxicity by magnesium in Japanese mustard spinach. Soil Sci. Plant Nutr. 53, 246–251. doi: 10.1111/j.1747-0765.2007.00129.x
Khan, M. A., Castro-Guerrero, N. A., McInturf, S. A., Nguyen, N. T., Dame, A. N., Wang, J., et al. (2018). Changes in iron availability in Arabidopsis are rapidly sensed in the leaf vasculature and impaired sensing leads to opposite transcriptional programs in leaves and roots. Plant Cell Environ. 41, 2263–2276. doi: 10.1111/pce.13192
Khan, M. I. R., Chopra, P., Chhillar, H., Ahanger, M. A., Hussain, S. J., and Maheshwari, C. (2021). Regulatory hubs and strategies for improving heavy metal tolerance in plants: chemical messengers, omics and genetic engineering. Plant Physiol. Biochem. 164, 260–278. doi: 10.1016/j.plaphy.2021.05.006
Kim, J. Y., Symeonidi, E., Pang, T. Y., Denyer, T., Weidauer, D., Bezrutczyk, M., et al. (2021). Distinct identities of leaf phloem cells revealed by single cell transcriptomics. Plant Cell 33, 511–530. doi: 10.1093/plcell/koaa060
Kök, A. B., Mungan, M. D., Doğanlar, S., and Frary, A. (2020). Transcriptomic analysis of selenium accumulation in Puccinellia distans (Jacq.) Parl., a boron hyperaccumulator. Chemosphere 245:125665. doi: 10.1016/j.chemosphere.2019.125665
Komarkova, M., Chromy, J., Pokorna, E., Soudek, P., and Machova, P. (2020). Physiological and transcriptomic response of grey Poplar (Populus× Canescens Aiton Sm.) to cadmium stress. Plants 9:1485. doi: 10.3390/plants9111485
Kouhi, F., Sorkheh, K., and Ercisli, S. (2020). MicroRNA expression patterns unveil differential expression of conserved miRNAs and target genes against abiotic stress in safflower. PLoS One 15:e0228850. doi: 10.1371/journal.pone.0228850
Kumari, A., and Pandey-Rai, S. (2018). Enhanced arsenic tolerance and secondary metabolism by modulation of gene expression and proteome profile in Artemisia annua L. after application of exogenous salicylic acid. Plant Physiol. Biochem. 132, 590–602. doi: 10.1016/j.plaphy.2018.10.010
Lai, J.-L., Liu, Z.-W., and Luo, X.-G. (2020). A metabolomic, transcriptomic profiling, and mineral nutrient metabolism study of the phytotoxicity mechanism of uranium. J. Hazard. Mater. 386:121437. doi: 10.1016/j.jhazmat.2019.121437
Lancíková, V., Tomka, M., Žiarovská, J., Gažo, J., and Hricová, A. (2020). Morphological responses and gene expression of grain Amaranth (Amaranthus spp.) growing under Cd. Plants 9:572. doi: 10.3390/plants9050572
Li, B., Sun, L., Huang, J., Göschl, C., Shi, W., Chory, J., et al. (2019). GSNOR provides plant tolerance to iron toxicity via preventing iron-dependent nitrosative and oxidative cytotoxicity. Nat. Commun. 10, 1–13. doi: 10.1038/s41467-019-11892-5
Liang, K., Liang, S., and Zhu, H. (2020). Comparative proteomics analysis of the effect of selenium treatment on the quality of foxtail millet. Lebensm. Wiss. Technol. 131:109691.
Lin, K., Kools, H., de Groot, P. J., Gavai, A. K., Basnet, R. K., Cheng, F., et al. (2011). MADMAX–Management and analysis database for multiple~ omics experiments. J. Integr. Bioinfor. 8, 59–74.
Lin, T., Zhu, X., Zhang, F., and Wan, X. (2011). The detoxification effect of nitrogen on cadmium stress in Populus yunnanensis. Bot. Res. J. 4, 13–19.
Liu, H., Wang, F., Xiao, Y., Tian, Z., Wen, W., Zhang, X., et al. (2016). MODEM: multi-omics data envelopment and mining in maize. Database 2016:baw117. doi: 10.1093/database/baw117
Liu, L., Yin, H., Liu, Y., Shen, L., Yang, X., Zhang, D., et al. (2021). Analysis of cadmium-stress-induced microRNAs and their targets reveals bra-miR172b-3p as a potential Cd2+-specific resistance factor in Brassica juncea. Processes 9:1099. doi: 10.3390/pr9071099
Liu, P., Huang, R., Hu, X., Jia, Y., Li, J., Luo, J., et al. (2019). Physiological responses and proteomic changes reveal insights into Stylosanthes response to manganese toxicity. BMC Plant Biol. 19:212. doi: 10.1186/s12870-019-1822-y
Liu, Q., Zhang, Y., Wang, Y., Wang, W., Gu, C., Huang, S., et al. (2020). Quantitative proteomic analysis reveals complex regulatory and metabolic response of Iris lactea Pall. var. chinensis to cadmium toxicity. J. Hazard. Mater. 400:123165. doi: 10.1016/j.jhazmat.2020.123165
Liu, S., Ali, S., Yang, R., Tao, J., and Ren, B. (2019). A newly discovered Cd-hyperaccumulator Lantana camara L. J. Hazard Mater. 371, 233–242. doi: 10.1016/j.jhazmat.2019.03.016
Liu, X., Chen, S., Chen, M., Zheng, G., Peng, Y., Shi, X., et al. (2019). Association study reveals genetic loci responsible for arsenic, cadmium and lead accumulation in rice grain in contaminated farmlands. Front. Plant Sci. 10:61. doi: 10.3389/fpls.2019.00061
Lokhande, V. H., Patade, V. Y., Srivastava, S., Suprasanna, P., Shrivastava, M., and Awasthi, G. (2020). Copper accumulation and biochemical responses of Sesuvium portulacastrum (L.). Mater. Today Proc. 31, 679–684. doi: 10.1016/j.matpr.2020.07.117
Lowe, R., Shirley, N., Bleackley, M., Dolan, S., and Shafee, T. (2017). Transcriptomics technologies. PLoS Comput. Biol. 13:e1005457. doi: 10.1371/journal.pcbi.1005457
Luo, J.-S., Gu, T., Yang, Y., and Zhang, Z. (2019). A non-secreted plant defensin AtPDF2.6 conferred cadmium tolerance via its chelation in Arabidopsis. Plant Mol. Biol. 100, 561–569. doi: 10.1007/s11103-019-00878-y
Luo, J.-S., and Zhang, Z. (2019). Proteomic changes in the xylem sap of Brassica napus under cadmium stress and functional validation. BMC Plant Biol. 19:280. doi: 10.1186/s12870-019-1895-7
Luo, W., Pant, G., Bhavnasi, Y. K., Blanchard, S. G. Jr., and Brouwer, C. (2017). Pathview Web: user friendly pathway visualization and data integration. Nucleic Acids Res. 45, W501–W508. doi: 10.1093/nar/gkx372
Lyu, G., Li, D., Li, S., Ning, C., and Qin, R. (2020). Genotoxic effects and proteomic analysis on Allium cepa var. agrogarum L. root cells under Pb stress. Ecotoxicology 29, 959–972. doi: 10.1007/s10646-020-02236-x
Malar, S., Vikram, S. S., Favas, P. J., and Perumal, V. (2016). Lead heavy metal toxicity induced changes on growth and antioxidative enzymes level in water hyacinths [Eichhornia crassipes (Mart.)]. Bot. Stud. 55, 1–11. doi: 10.1186/s40529-014-0054-6
Malik, B., and Pirzadah, T. B. (2020). Lead induced changes in biomarkers and proteome map of Chicory (Cichorium intybus L.). Curr. Bot. 11, 111–116.
Marrugo-Negrete, J., Durango-Hernández, J., Pinedo-Hernández, J., Enamorado-Montes, G., and Díez, S. (2016). Mercury uptake and effects on growth in Jatropha curcas. J. Environ. Sci. 48, 120–125. doi: 10.1016/j.jes.2015.10.036
Mathur, S., Kalaji, H., and Jajoo, A. (2016). Investigation of deleterious effects of chromium phytotoxicity and photosynthesis in wheat plant. Photosynthetica 54, 185–192.
Mawia, A. M., Hui, S., Zhou, L., Li, H., Tabassum, J., Lai, C., et al. (2020). Inorganic arsenic toxicity and alleviation strategies in rice. J. Hazard. Mater. 408:124751. doi: 10.1016/j.jhazmat.2020.124751
Mehmood, S. S., Lu, G., Luo, D., Hussain, M. A., Raza, A., Zafar, Z., et al. (2021). Integrated analysis of transcriptomics and proteomics provides insights into the molecular regulation of cold response in Brassica napus. Environ. Exp. Bot. 187:104480.
Mishra, S., Bharagava, R. N., More, N., Yadav, A., Zainith, S., Mani, S., et al. (2019). “Heavy metal contamination: an alarming threat to environment and human health,” in Environmental Biotechnology: For Sustainable Future, eds R. C. Sobti, N. K. Arora, and R. Kothari (Cham: Springer), 103–125.
Mishra, V. K., Gupta, S., Chand, R., Yadav, P. S., Singh, S. K., Joshi, A. K., et al. (2019). Comparative transcriptomic profiling of High-and Low-grain Zinc and Iron containing Indian wheat genotypes. Curr. Plant Biol. 18:100105. doi: 10.1016/j.cpb.2019.100105
Mišúthová, A., Slováková, L’, Kollárová, K., and Vaculík, M. (2021). Effect of silicon on root growth, ionomics and antioxidant performance of maize roots exposed to As toxicity. Plant Physiol. Biochem. 168, 155–166. doi: 10.1016/j.plaphy.2021.10.012
Montague, E., Stanberry, L., Higdon, R., Janko, I., Lee, E., Anderson, N., et al. (2014). MOPED 2.5—an integrated multi-omics resource: multi-omics profiling expression database now includes transcriptomics data. Omics J. Integr. Biol. 18, 335–343. doi: 10.1089/omi.2014.0061
Mthembu, M. S., Odinga, C. A., Bux, F., and Swalaha, F. M. (2020). “Constructed wetlands: an eco-sustainable phytotechnology for degradation and detoxification of industrial wastewaters,” in Bioremediation of Industrial Waste for Environmental Safety, eds G. Saxena and R. N. Bharagava (Singapore: Springer), 291–312.
Muntean, E., Muntean, N., Duda, M., and Michalski, R. (2019). Heavy metals’ uptake from soil in potato tubers: an ionomic approach. ProEnvironment 12, 159–162.
Murugaiyan, V., Ali, J., Mahender, A., Aslam, U. M., Jewel, Z. A., Pang, Y., et al. (2019). Mapping of genomic regions associated with arsenic toxicity stress in a backcross breeding populations of rice (Oryza sativa L.). Rice 12:61. doi: 10.1186/s12284-019-0321-y
Mwamba, T., Islam, F., Ali, B., Lwalaba, J., Gill, R., Zhang, F., et al. (2020). Comparative metabolomic responses of low-and high-cadmium accumulating genotypes reveal the cadmium adaptive mechanism in Brassica napus. Chemosphere 250:126308. doi: 10.1016/j.chemosphere.2020.126308
Nagel, K. A., Lenz, H., Kastenholz, B., Gilmer, F., Averesch, A., Putz, A., et al. (2020). The platform GrowScreen-Agar enables identification of phenotypic diversity in root and shoot growth traits of agar grown plants. Plant Methods 16, 1–17. doi: 10.1186/s13007-020-00631-3
Naika, M., Shameer, K., and Sowdhamini, R. (2013). Comparative analyses of stress-responsive genes in Arabidopsis thaliana: insight from genomic data mining, functional enrichment, pathway analysis and phenomics. Mol. BioSyst. 9, 1888–1908. doi: 10.1039/c3mb70072k
National Genomics Data Center Members and Partners (2020). Database resources of the National Genomics Data Center in 2020. Nucleic Acids Res. 48, D24–D33. doi: 10.1093/nar/gkz913
Navarro-Reig, M., Jaumot, J., Piña, B., Moyano, E., Galceran, M. T., and Tauler, R. (2017). Metabolomic analysis of the effects of cadmium and copper treatment in Oryza sativa L. using untargeted liquid chromatography coupled to high resolution mass spectrometry and all-ion fragmentation. Metallomics 9, 660–675. doi: 10.1039/c6mt00279j
Nie, G., Liao, Z., Zhong, M., Zhou, J., Cai, J., Liu, A., et al. (2021). MicroRNA-mediated responses to chromium stress provides insight toward to tolerance characteristics of Miscanthus sinensis. Front. Plant Sci. 12:666117. doi: 10.3389/fpls.2021.666117
Njobvu, J., Hamabwe, S. M., Munyinda, K., Kelly, J. D., and Kamfwa, K. (2020). Quantitative trait loci mapping of resistance to aluminum toxicity in common bean. Crop Sci. 60, 1294–1302. doi: 10.1007/s00122-009-1051-0
O’Connor, D., Peng, T., Li, G., Wang, S., Duan, L., Mulder, J., et al. (2018). Sulfur-modified rice husk biochar: a green method for the remediation of mercury contaminated soil. Sci. Total Environ. 621, 819–826. doi: 10.1016/j.scitotenv.2017.11.213
Oliveira, B. R. M., de Almeida, A.-A. F., Pirovani, C. P., Barroso, J. P., Neto, C. H. C., Santos, N. A., et al. (2020). Mitigation of Cd toxicity by Mn in young plants of cacao, evaluated by the proteomic profiles of leaves and roots. Ecotoxicology 29, 340–358. doi: 10.1007/s10646-020-02178-4
Oyewo, O. A., Adeniyi, A., Bopape, M. F., and Onyango, M. S. (2020). “Heavy metal mobility in surface water and soil, climate change, and soil interactions,” in Climate Change and Soil Interactions, eds M. N. V. Prasad and M. Pietrzykowski (Amsterdam: Elsevier), 51–88. doi: 10.1016/b978-0-12-818032-7.00004-7
Pan, X., Li, Y., Liu, W., Liu, S., Min, J., Xiong, H., et al. (2020). QTL mapping and candidate gene analysis of cadmium accumulation in polished rice by genome-wide association study. Sci. Rep. 10:11791. doi: 10.1038/s41598-020-68742-4
Pandey, A. K., Gautam, A., and Dubey, R. S. (2019). Transport and detoxification of metalloids in plants in relation to plant-metalloid tolerance. Plant Gene 17:100171. doi: 10.1007/s11356-020-10293-2
Pandey, A. K., Gedda, M. R., and Verma, A. K. (2020). Effect of arsenic stress on expression pattern of a rice specific miR156j at various developmental stages and their allied co-expression target networks. Front. Plant Sci. 11:752. doi: 10.3389/fpls.2020.00752
Papoutsoglou, E. A., Faria, D., Arend, D., Arnaud, E., Athanasiadis, I. N., Chaves, I., et al. (2020). Enabling reusability of plant phenomic datasets with MIAPPE 1.1. New Phytol. 227, 260–273. doi: 10.1111/nph.16544
Parmar, P., Dave, B., Sudhir, A., Panchal, K., and Subramanian, R. (2013). Physiological, biochemical and molecular response of plants against heavy metals stress. Int. J. Curr. Res. 5, 80–89.
Patel, M., Rangani, J., Kumari, A., and Parida, A. K. (2020). Mineral nutrient homeostasis, photosynthetic performance, and modulations of antioxidative defense components in two contrasting genotypes of Arachis hypogaea L. (peanut) for mitigation of nitrogen and/or phosphorus starvation. J. Biotechnol. 323, 136–158.
Paunov, M., Koleva, L., Vassilev, A., Vangronsveld, J., and Goltsev, V. (2018). Effects of different metals on photosynthesis: cadmium and zinc affect chlorophyll fluorescence in durum wheat. Int. J. Mol. Sci. 19:787. doi: 10.3390/ijms19030787
Pavlova, D. (2017). Nickel effect on root-meristem cell division in Plantago lanceolata (Plantaginaceae) seedlings. Aust. J. Bot. 65, 446–452. doi: 10.1071/bt17054
Pereira, M. P., de Almeida Rodrigues, L. C., Corrêa, F. F., de Castro, E. M., Ribeiro, V. E., and Pereira, F. J. (2016). Cadmium tolerance in Schinus molle trees is modulated by enhanced leaf anatomy and photosynthesis. Trees 30, 807–814. doi: 10.1007/s00468-015-1322-0
Pidatala, V. R., Li, K., Sarkar, D., Ramakrishna, W., and Datta, R. (2016). Identification of biochemical pathways associated with lead tolerance and detoxification in Chrysopogon zizanioides L. Nash (Vetiver) by metabolic profiling. Environ. Sci. Technol. 50, 2530–2537. doi: 10.1021/acs.est.5b04725
Pidatala, V. R., Li, K., Sarkar, D., Wusirika, R., and Datta, R. (2018). Comparative metabolic profiling of vetiver (Chrysopogon zizanioides) and maize (Zea mays) under lead stress. Chemosphere 193, 903–911. doi: 10.1016/j.chemosphere.2017.11.087
Pinu, F. R., Beale, D. J., Paten, A. M., Kouremenos, K., Swarup, S., Schirra, H. J., et al. (2019). Systems biology and multi-omics integration: viewpoints from the metabolomics research community. Metabolites 9:76. doi: 10.3390/metabo9040076
Pirzadah, T. B., Malik, B., and Hakeem, K. R. (2019). “Integration of “Omic” approaches to unravel the heavy metal tolerance in plants,” in Essentials of Bioinformatics, ed. J. Xiong (Berlin: Springer), 79–92. doi: 10.1007/978-3-030-19318-8_4
Pratap, A., Gupta, S., Nair, R. M., Gupta, S., Schafleitner, R., Basu, P., et al. (2019). Using plant phenomics to exploit the gains of genomics. Agronomy 9:126. doi: 10.3389/fpls.2021.626565
Punshon, T., Ricachenevsky, F. K., Hindt, M. N., Socha, A. L., and Zuber, H. (2013). Methodological approaches for using synchrotron X-ray fluorescence (SXRF) imaging as a tool in ionomics: examples from Arabidopsis thaliana. Metallomics 5, 1133–1145. doi: 10.1039/c3mt00120b
Qiu, Z., Hai, B., Guo, J., Li, Y., and Zhang, L. (2016). Characterization of wheat miRNAs and their target genes responsive to cadmium stress. Plant Physiol. Biochem. 101, 60–67. doi: 10.1016/j.plaphy.2016.01.020
Rai, P. K., Lee, S. S., Zhang, M., Tsang, Y. F., and Kim, K.-H. (2019). Heavy metals in food crops: health risks, fate, mechanisms, and management. Environ. Int. 125, 365–385. doi: 10.1016/j.envint.2019.01.067
Rath, B. P., Hota, S., Subhadarshini, S., Dash, D., and Das, P. K. (2019). Consequence of chromium-tainted soil on physical and biochemical responses of Vigna radiata L. J. Appl. Biol. Biotechnol. 7, 35–41.
Raza, A. (2020). Metabolomics: a systems biology approach for enhancing heat stress tolerance in plants. Plant Cell Rep. [Epub ahead of print]. doi: 10.1007/s00299-020-02635-8
Raza, A., Habib, M., Charagh, S., and Kakavand, S. N. (2021a). “Genetic engineering of plants to tolerate toxic metals and metalloids,” in Handbook of Bioremediation, eds M. Hasanuzzaman and M. N. V. Prasad (Cambridge, MA: Academic Press), 411–436. doi: 10.1016/b978-0-12-819382-2.00026-0
Raza, A., Habib, M., Kakavand, S. N., Zahid, Z., Zahra, N., Sharif, R., et al. (2020). Phytoremediation of cadmium: physiological, biochemical, and molecular mechanisms. Biology 9:177. doi: 10.3390/biology9070177
Raza, A., Razzaq, A., Mehmood, S. S., Hussain, M. A., Wei, S., He, H., et al. (2021b). Omics: the way forward to enhance abiotic stress tolerance in Brassica napus L. GM Crops Food 12, 251–281. doi: 10.1080/21645698.2020.1859898
Raza, A., Su, W., Hussain, M. A., Mehmood, S. S., Zhang, X., Cheng, Y., et al. (2021c). Integrated analysis of metabolome and transcriptome reveals insights for cold tolerance in rapeseed (Brassica napus L.). Front. Plant Sci. 12:721681. doi: 10.3389/fpls.2021.721681
Raza, A., Tabassum, J., Kudapa, H., and Varshney, R. K. (2021d). Can omics deliver temperature resilient ready-to-grow crops? Crit. Rev. Biotechnol. 7, 1–24. doi: 10.1080/07388551.2021.1898332
Razzaq, A., Sadia, B., Raza, A., Khalid Hameed, M., and Saleem, F. (2019). Metabolomics: a way forward for crop improvement. Metabolites 9:303. doi: 10.3390/metabo9120303
Rehman, F., Khan, F., Varshney, D., Naushin, F., and Rastogi, J. (2011). Effect of cadmium on the growth of tomato. Biol. Med. 3, 187–190.
Rehman, M., Liu, L., Bashir, S., Saleem, M. H., Chen, C., Peng, D., et al. (2019). Influence of rice straw biochar on growth, antioxidant capacity and copper uptake in ramie (Boehmeria nivea L.) grown as forage in aged copper-contaminated soil. Plant Physiol. Biochem. 138, 121–129. doi: 10.1016/j.plaphy.2019.02.021
Rohart, F., Gautier, B., Singh, A., and Lê Cao, K.-A. (2017). mixOmics: an R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 13:e1005752. doi: 10.1371/journal.pcbi.1005752
Roorkiwal, M., Bharadwaj, C., Barmukh, R., Dixit, G. P., Thudi, M., Gaur, P. M., et al. (2020). Integrating genomics for chickpea improvement: achievements and opportunities. Theor. Appl. Genet. 133, 1703–1720. doi: 10.1007/s00122-020-03584-2
Rouphael, Y., Colla, G., Bernardo, L., Kane, D., Trevisan, M., and Lucini, L. (2016). Zinc excess triggered polyamines accumulation in lettuce root metabolome, as compared to osmotic stress under high salinity. Front. Plant Sci. 7:842. doi: 10.3389/fpls.2016.00842
Rucińska-Sobkowiak, R. (2016). Water relations in plants subjected to heavy metal stresses. Acta Physiol. Plant 38:257.
Sabrine, H., Afif, H., Mohamed, B., Hamadi, B., and Maria, H. (2010). Effects of cadmium and copper on pollen germination and fruit set in pea (Pisum sativum L.). Sci. Hortic. 125, 551–555. doi: 10.1016/j.scienta.2010.05.031
Safdar, L. B., Almas, F., Sarfraz, S., Ejaz, M., Ali, Z., Mahmood, Z., et al. (2020a). Genomewide association study identifies five new cadmium uptake loci in wheat. Plant Genome 13:e20030. doi: 10.1002/tpg2.20030
Safdar, L. B., Andleeb, T., Latif, S., Umer, M. J., Tang, M., Li, X., et al. (2020b). Genome-wide association study and QTL meta-analysis identified novel genomic loci controlling potassium use efficiency and agronomic traits in bread wheat. Front. Plant Sci. 11:70. doi: 10.3389/fpls.2020.00070
Saini, S., and Dhania, G. (2020). “Cadmium as an environmental pollutant: ecotoxicological effects, health hazards, and bioremediation approaches for its detoxification from contaminated sites,” in Bioremediation of Industrial Waste for Environmental Safety, eds R. N. Bharagava and G. Saxena (Singapore: Springer), 357–387.
Sakurai, N., Ara, T., Ogata, Y., Sano, R., Ohno, T., Sugiyama, K., et al. (2011). KaPPA-View4: a metabolic pathway database for representation and analysis of correlation networks of gene co-expression and metabolite co-accumulation and omics data. Nucleic Acids Res. 39, D677–D684. doi: 10.1093/nar/gkq989
Saleem, M. H., Fahad, S., Rehman, M., Saud, S., Jamal, Y., Khan, S., et al. (2020). Morpho-physiological traits, biochemical response and phytoextraction potential of short-term copper stress on kenaf (Hibiscus cannabinus L.) seedlings. PeerJ 8:e8321. doi: 10.7717/peerj.8321
Salehi, H., Chehregani Rad, A., Raza, A., and Chen, J. T. (2021). Foliar application of CeO2 nanoparticles alters generative components fitness and seed productivity in bean crop (Phaseolus vulgaris L.). Nanomaterials 11:862. doi: 10.3390/nano11040862
Salim, N., and Raza, A. (2020). Nutrient use efficiency (NUE) for sustainable wheat production: a review. J. Plant Nutr. 43, 297–315. doi: 10.1080/01904167.2019.1676907
Sangwan, P., Kumar, V., and Joshi, U. (2014). Effect of chromium (VI) toxicity on enzymes of nitrogen metabolism in clusterbean (Cyamopsis tetragonoloba L.). Enzyme Res. 2014:784036. doi: 10.1155/2014/784036
Sarwar, N., Imran, M., Shaheen, M. R., Ishaque, W., Kamran, M. A., Matloob, A., et al. (2017). Phytoremediation strategies for soils contaminated with heavy metals: modifications and future perspectives. Chemosphere 171, 710–721. doi: 10.1016/j.chemosphere.2016.12.116
Schmidt, S. B., Eisenhut, M., and Schneider, A. (2020). Chloroplast transition metal regulation for efficient photosynthesis. Trends Plant Sci. 25, 817–828. doi: 10.1016/j.tplants.2020.03.003
Schwacke, R., Ponce-Soto, G. Y., Krause, K., Bolger, A. M., Arsova, B., Hallab, A., et al. (2019). MapMan4: a refined protein classification and annotation framework applicable to multi-omics data analysis. Mol. Plant. 12, 879–892. doi: 10.1016/j.molp.2019.01.003
Shenton, M., Kudo, T., Kobayashi, M., Nakamura, Y., Ohyanagi, H., and Yano, K. (2019). “Plant omics data center and CATchUP: web databases for effective gene mining utilizing public RNA-Seq-based transcriptome data,” in The Model Legume Medicago Truncatula, ed. F. J. de Bruijn (Hoboken, NJ: Wiley), 1137–1146.
Shu, H., Zhang, J., Liu, F., Bian, C., Liang, J., Liang, J., et al. (2019). Comparative transcriptomic studies on a cadmium hyperaccumulator Viola baoshanensis and its non-tolerant counterpart V. inconspicua. Int. J. Mol. Sci. 20:1906. doi: 10.3390/ijms20081906
Shukla, A., Srivastava, S., and D’Souza, S. F. (2018). An integrative approach toward biosensing and bioremediation of metals and metalloids. Inter. J. Environ. Sci. Technol. 15, 2701–2712.
Shukla, T., Khare, R., Kumar, S., Lakhwani, D., Sharma, D., Asif, M. H., et al. (2018). Differential transcriptome modulation leads to variation in arsenic stress response in Arabidopsis thaliana accessions. J. Hazard Mat. 351, 1–10. doi: 10.1016/j.jhazmat.2018.02.031
Son, J. A., Narayanankutty, D. P., and Roh, K. S. (2014). Influence of exogenous application of glutathione on rubisco and rubisco activase in heavy metal-stressed tobacco plant grown in vitro. Saudi J. Biol. Sci. 21, 89–97. doi: 10.1016/j.sjbs.2013.06.002
Song, X., Nie, F., Chen, W., Ma, X., Gong, K., Yang, Q., et al. (2020a). Coriander genomics database: a genomic, transcriptomic, and metabolic database for coriander. Hortic. Res. 7, 1–10.
Song, X., Zhang, C., Chen, W., Zhu, Y., and Wang, Y. (2020b). Growth responses and physiological and biochemical changes in five ornamental plants grown in urban lead-contaminated soils. Plant Environ. Int. 1, 29–47. doi: 10.1002/pei3.10013
Stich, B., Benke, A., Schmidt, M., Urbany, C., Shi, R., and von Wirén, N. (2020). The maize shoot ionome: its interaction partners, predictive power, and genetic determinants. Plant Cell Environ. 43, 2095–2111. doi: 10.1111/pce.13823
Su, C., Jiang, Y., Li, F., Yang, Y., Lu, Q., Zhang, T., et al. (2017). Investigation of subcellular distribution, physiological, and biochemical changes in Spirodela polyrhiza as a function of cadmium exposure. Environ. Exp. Bot. 142, 24–33.
Sun, H., Wang, X., Li, H., Bi, J., Yu, J., Liu, X., et al. (2020). Selenium modulates cadmium-induced ultrastructural and metabolic changes in cucumber seedlings. RSC Adv. 10, 17892–17905. doi: 10.1039/d0ra02866e
Sundaramoorthy, P., Chidambaram, A., Ganesh, K. S., Unnikannan, P., and Baskaran, L. (2010). Chromium stress in paddy:(i) nutrient status of paddy under chromium stress;(ii) phytoremediation of chromium by aquatic and terrestrial weeds. Comptes Rendus Biol. 333, 597–607. doi: 10.1016/j.crvi.2010.03.002
Suzuki, N. (2005). Alleviation by calcium of cadmium-induced root growth inhibition in Arabidopsis seedlings. Plant Biotechnol. 22, 19–25.
Syed, M. A., Iftekharuddaula, K., Mian, M. K., Rasul, M. G., Rahmam, G. M., Panaullah, G. M., et al. (2016). Main effect QTLs associated with arsenic phyto-toxicity tolerance at seedling stage in rice (Oryza sativa L.). Euphytica 209, 805–814. doi: 10.1007/s10681-016-1683-5
Sylvain, B., Mikael, M. H., Florie, M., Emmanuel, J., Marilyne, S., Sylvain, B., et al. (2016). Phytostabilization of As, Sb and Pb by two willow species (S. viminalis and S. purpurea) on former mine technosols. Catena 136, 44–52. doi: 10.1016/j.catena.2015.07.008
Tavallali, V. (2017). Interactive effects of zinc and boron on growth, photosynthesis, and water relations in pistachio. J. Plant Nutr. 40, 1588–1603. doi: 10.1080/01904167.2016.1270308
Tian, M., Xu, X., Liu, F., Fan, X., and Pan, S. (2018). Untargeted metabolomics reveals predominant alterations in primary metabolites of broccoli sprouts in response to pre-harvest selenium treatment. Food Res. Int. 111, 205–211. doi: 10.1016/j.foodres.2018.04.020
Tiwari, S., and Lata, C. (2018). Heavy metal stress, signaling, and tolerance due to plant-associated microbes: an overview. Front. Plant Sci. 9:452. doi: 10.3389/fpls.2018.00452
Turhadi, T., Hamim, H., Ghulamahdi, M., and Miftahudin, M. (2019). Iron toxicity-induced physiological and metabolite profile variations among tolerant and sensitive rice varieties. Plant Signal. Behav. 14:1682829. doi: 10.1080/15592324.2019.1682829
Ulfenborg, B. (2019). Vertical and horizontal integration of multi-omics data with miodin. BMC Bioinform. 20:649. doi: 10.1186/s12859-019-3224-4
Utami, D. W., Rosdianti, I., Chrisnawati, L., Subardi, S., Nurani, S., and Suwarno, S. (2020). Identification of iron tolerant candidate loci in rice determined through genome-wide association study. Indonesian J. Agric. Sci. 21, 17–29. doi: 10.1186/s12864-017-4221-5
Vaculík, M., Konlechner, C., Langer, I., Adlassnig, W., Puschenreiter, M., Lux, A., et al. (2012). Root anatomy and element distribution vary between two Salix caprea isolates with different Cd accumulation capacities. Environ. Pollut. 163, 117–126. doi: 10.1016/j.envpol.2011.12.031
van Dijk, A. D. J., Kootstra, G., Kruijer, W., and de Ridder, D. (2020). Machine learning in plant science and plant breeding. Iscience 24:101890. doi: 10.1016/j.isci.2020.101890
Varshney, R. K., Bohra, A., Yu, J., Graner, A., Zhang, Q., and Sorrells, M. E. (2021c). Designing future crops: genomics-assisted breeding comes of age. Trends Plant Sci. 26, 631–649. doi: 10.1016/j.tplants.2021.03.010
Varshney, R. K., Bohra, A., Roorkiwal, M., Barmukh, R., Cowling, W. A., Chitikineni, A., et al. (2021b). Fast-forward breeding for a food-secure world. Trends Genet. 37, 1124–1136. doi: 10.1016/j.tig.2021.08.002
Varshney, R. K., Barmukh, R., Roorkiwal, M., Qi, Y., Kholova, J., Tuberosa, R., et al. (2021a). Breeding custom-designed crops for improved drought adaptation. Adv. Genet. 2:e202100017.
Varshney, R. K., Sinha, P., Singh, V. K., Kumar, A., Zhang, Q., and Bennetzen, J. L. (2020). 5Gs for crop improvement. Curr. Opin. Plant Biol. 56, 190–196. doi: 10.1016/j.pbi.2019.12.004
Venkatachalam, P., Jayalakshmi, N., Geetha, N., Sahi, S. V., Sharma, N. C., Rene, E. R., et al. (2017). Accumulation efficiency, genotoxicity and antioxidant defense mechanisms in medicinal plant Acalypha indica L. under lead stress. Chemosphere 171, 544–553. doi: 10.1016/j.chemosphere.2016.12.092
Walter, A., Liebisch, F., and Hund, A. (2015). Plant phenotyping: from bean weighing to image analysis. Plant Methods 11, 1–11. doi: 10.1186/s13007-015-0056-8
Wang, C., Tang, Z., Zhuang, J.-Y., Tang, Z., Huang, X.-Y., and Zhao, F.-J. (2020a). Genetic mapping of ionomic quantitative trait loci in rice grain and straw reveals OsMOT1; 1 as the putative causal gene for a molybdenum QTL qMo8. Mol. Genet. Genomics 295, 391–407. doi: 10.1007/s00438-019-01632-1
Wang, D., Fan, W., Guo, X., Wu, K., Zhou, S., Chen, Z., et al. (2020b). MaGenDB: a functional genomics hub for Malvaceae plants. Nucleic Acids Res. 48, D1076–D1084. doi: 10.1093/nar/gkz953
Wang, F., Itai, R. N., Nozoye, T., Kobayashi, T., Nishizawa, N. K., and Nakanishi, H. (2020c). The bHLH protein OsIRO3 is critical for plant survival and iron (Fe) homeostasis in rice (Oryza sativa L.) under Fe-deficient conditions. Soil Sci. Plant Nutr. 66, 579–592.
Wang, J., Li, H., Zou, D., Zhao, J., Fan, L., and Wu, T. (2017). Transcriptome profile analysis of cadmium tolerance in Chinese flowering cabbage. Hortic. Environ. Biotechnol. 58, 56–65. doi: 10.1007/s13580-017-0075-7
Wang, L., Rinklebe, J., Tack, F. M., and Hou, D. (2021). A review of green remediation strategies for heavy metal contaminated soil. Soil Use Manag. 37, 936–963.
Wang, L., Zheng, B., Yuan, Y., Xu, Q., and Chen, P. (2020d). Transcriptome profiling of Fagopyrum tataricum leaves in response to lead stress. BMC Plant Biol. 20:54. doi: 10.1186/s12870-020-2265-1
Wang, M.-Q., Bai, Z.-Y., Xiao, Y.-F., Li, Y., Liu, Q.-L., Zhang, L., et al. (2019). Transcriptomic analysis of Verbena bonariensis roots in response to cadmium stress. BMC Genomics 20:877. doi: 10.1186/s12864-019-6152-9
Wang, W., Ding, G., White, P. J., Wang, M., Zou, J., Xu, F., et al. (2020e). Genetic dissection of the shoot and root ionomes of Brassica napus grown with contrasting phosphate supplies. Ann. Bot. 126, 119–140. doi: 10.1093/aob/mcaa055
Wang, X., Zhang, J., He, S., Gao, Y., Ma, X., Gao, Y., et al. (2018). HMOD: an omics database for herbal medicine plants. Mol. Plant 11, 757–759. doi: 10.1016/j.molp.2018.03.002
Wang, Y.-M., Yang, Q., Xu, H., Liu, Y.-J., and Yang, H.-L. (2020f). Physiological and transcriptomic analysis provide novel insight into cobalt stress responses in willow. Sci. Rep. 10, 1–12. doi: 10.1038/s41598-020-59177-y
Wani, W., Masoodi, K. Z., Zaid, A., Wani, S. H., Shah, F., Meena, V. S., et al. (2018). Engineering plants for heavy metal stress tolerance. Rend. Lincei Sci. Fis. Nat. 29, 709–723.
Weber, M., Beyene, B., Nagler, N., Herfert, J., Schempp, S., Klecker, M., et al. (2020). A mutation in the essential and widely conserved DAMAGED DNA BINDING1-Cullin4 ASSOCIATED FACTOR gene OZS3 causes hypersensitivity to zinc excess, cold and UV stress in Arabidopsis thaliana. Plant J. 103, 995–1009. doi: 10.1111/tpj.14779
Wen, R., Ya, L., Zhou, M.-Y., Zi, S., Wang, T.-Y., Zhao, J.-R., et al. (2019). Dynamic changes of root proteome reveal diverse responsive proteins in maize subjected to cadmium stress. J. Integr. Agric. 18, 2193–2204.
Wijngaard, R. R., van der Perk, M., van der Grift, B., de Nijs, T. C., and Bierkens, M. F. (2017). The impact of climate change on metal transport in a lowland catchment. Water Air Soil Pollut. 228:107. doi: 10.1007/s11270-017-3261-4
Wu, Q., Qi, J., and Xia, X. (2017). Long-term variations in sediment heavy metals of a reservoir with changing trophic states: implications for the impact of climate change. Sci. Total Environ. 609, 242–250. doi: 10.1016/j.scitotenv.2017.04.041
Xiao, W., Liu, P., Wang, K., Yang, Z., and Wang, L. (2021). Relationship between ionomics and transcriptomics of rice plant in response to arsenite stress. Environ. Exp. Bot. 189:104565.
Xie, M., Chen, W., Lai, X., Dai, H., Sun, H., Zhou, X., et al. (2019). Metabolic responses and their correlations with phytochelatins in Amaranthus hypochondriacus under cadmium stress. Environ. Pollut. 252, 1791–1800. doi: 10.1016/j.envpol.2019.06.103
Xie, Q., Yu, Q., Jobe, T. O., Pham, A., Ge, C., Guo, Q., et al. (2021). An amiRNA screen uncovers redundant CBF and ERF34/35 transcription factors that differentially regulate arsenite and cadmium responses. Plant Cell Environ. 44, 1692–1706. doi: 10.1111/pce.14023
Xu, L., Zhang, F., Tang, M., Wang, Y., Dong, J., Ying, J., et al. (2020). Melatonin confers cadmium tolerance by modulating critical heavy metal chelators and transporters in radish plants. J. Pineal Res. 69:e12659. doi: 10.1111/jpi.12659
Xu, S., Hu, C., Hussain, S., Tan, Q., Wu, S., and Sun, X. (2018). Metabolomics analysis reveals potential mechanisms of tolerance to excess molybdenum in soybean seedlings. Ecotoxicol. Environ. Saf. 164, 589–596. doi: 10.1016/j.ecoenv.2018.08.062
Xu, X., Zhang, S., Cheng, Z., Li, T., Jia, Y., Wang, G., et al. (2020). Transcriptome analysis revealed cadmium accumulation mechanisms in hyperaccumulator Siegesbeckia orientalis L. Environ. Sci. Pollut. Res. 27, 18853–18865. doi: 10.1007/s11356-020-08387-y
Yadav, B. S., Singh, S., Srivastava, S., Singh, N. K., and Mani, A. (2019). Whole transcriptome expression profiling and biological network analysis of chickpea during heavy metal stress. J. Plant Biochem. Biotechnol. 28, 345–352. doi: 10.1007/s13562-019-00486-3
Yan, A., Wang, Y., Tan, S. N., Mohd Yusof, M. L., Ghosh, S., and Chen, Z. (2020). Phytoremediation: a promising approach for revegetation of heavy metal-polluted land. Front. Plant Sci. 11:359. doi: 10.3389/fpls.2020.00359
Yang, M., Li, Y., Liu, Z., Tian, J., Liang, L., Qiu, Y., et al. (2020). A high activity zinc transporter OsZIP9 mediates zinc uptake in rice. Plant J. 103, 1695–1709. doi: 10.1111/tpj.14855
Yang, M., Lu, K., Zhao, F. J., Xie, W., Ramakrishna, P., Wang, G., et al. (2018). Genome-wide association studies reveal the genetic basis of ionomic variation in rice. Plant Cell 30, 2720–2740. doi: 10.1105/tpc.18.00375
Yang, W., Dai, H., Dou, X., Zhang, Q., and Wei, S. (2019). Effect and mechanism of commonly used four nitrogen fertilizers and three organic fertilizers on Solanum nigrum L. hyperaccumulating Cd. Environ. Sci. Pollut. Res. 26, 12940–12947. doi: 10.1007/s11356-019-04848-1
Yang, Y., Zhang, L., Huang, X., Zhou, Y., Quan, Q., Li, Y., et al. (2020). Response of photosynthesis to different concentrations of heavy metals in Davidia involucrata. PLoS One 15:e0228563. doi: 10.1371/journal.pone.0228563
Ye, Z., Zeng, J., Ye, L., Long, L., and Zhang, G. (2020). Genome-wide association analysis of potassium uptake and translocation rates under low K stress in Tibetan wild barley. Euphytica 216:35.
Yu, R., Jiang, Q., Xv, C., Li, L., Bu, S., and Shi, G. (2019). Comparative proteomics analysis of peanut roots reveals differential mechanisms of cadmium detoxification and translocation between two cultivars differing in cadmium accumulation. BMC Plant Biol. 19:137. doi: 10.1186/s12870-019-1739-5
Yu, J., Wu, L., Fu, L., Shen, Q., Kuang, L., Wu, D., et al. (2019). Genotypic difference of cadmium tolerance and the associated microRNAs in wild and cultivated barley. Plant Growth Regul. 87, 389–401. doi: 10.1007/s10725-019-00479-1
Yuan, J., Bai, Y., Chao, Y., Sun, X., He, C., Liang, X., et al. (2018). Genome-wide analysis reveals four key transcription factors associated with cadmium stress in creeping bentgrass (Agrostis stolonifera L.). PeerJ 6:e5191. doi: 10.7717/peerj.5191
Zafar, S. A., Zaidi, S., Gaba, Y., Singla-Pareek, S. L., Dhankher, O. P., Li, X., et al. (2020). Engineering abiotic stress tolerance via CRISPR/Cas-mediated genome editing. J. Exp. Bot. 71, 470–479. doi: 10.1093/jxb/erz476
Zeng, R., Farooq, M. U., Wang, L., Su, Y., Zheng, T., Ye, X., et al. (2019). Study on differential protein expression in natural selenium-enriched and non-selenium-enriched rice based on iTRAQ quantitative proteomics. Biomolecules 9:130. doi: 10.3390/biom9040130
Zhang, C., Xu, B., Geng, W., Shen, Y., Xuan, D., Lai, Q., et al. (2019a). Comparative proteomic analysis of pepper (Capsicum annuum L.) seedlings under selenium stress. PeerJ 7:e8020. doi: 10.7717/peerj.8020
Zhang, C., Xu, B., Zhao, C.-R., Sun, J., Lai, Q., and Yu, C. (2019b). Comparative de novo transcriptomics and untargeted metabolomic analyses elucidate complicated mechanisms regulating celery (Apium graveolens L.) responses to selenium stimuli. PLoS One 14:e0226752. doi: 10.1371/journal.pone.0226752
Zhang, P., Zhong, K., Zhong, Z., and Tong, H. (2019c). Mining candidate gene for rice aluminum tolerance through genome wide association study and transcriptomic analysis. BMC Plant Biol. 19:490. doi: 10.1186/s12870-019-2036-z
Zhang, X.-Y., Jia, X.-M., Zhang, R., Zhu, Z.-L., Liu, B., Gao, L.-Y., et al. (2019d). Metabolic analysis in Malus halliana leaves in response to iron deficiency. Sci. Hortic. 258:108792.
Zhang, F., Xiao, X., Xu, K., Cheng, X., Xie, T., Hu, J., et al. (2020a). Genome-wide association study (GWAS) reveals genetic loci of lead (Pb) tolerance during seedling establishment in rapeseed (Brassica napus L.). BMC Genomics 21:139. doi: 10.1186/s12864-020-6558-4
Zhang, H., Xu, Z., Guo, K., Huo, Y., He, G., Sun, H., et al. (2020b). Toxic effects of heavy metal Cd and Zn on chlorophyll, carotenoid metabolism and photosynthetic function in tobacco leaves revealed by physiological and proteomics analysis. Ecotoxicol. Environ. Saf. 202:110856. doi: 10.1016/j.ecoenv.2020.110856
Zhang, L., Ding, H., Jiang, H., Wang, H., Chen, K., Duan, J., et al. (2020c). Regulation of cadmium tolerance and accumulation by miR156 in Arabidopsis. Chemosphere 242:125168. doi: 10.1016/j.chemosphere.2019.125168
Zhang, Y., Lai, J.-L., Ji, X.-H., and Luo, X.-G. (2020d). Unraveling response mechanism of photosynthetic metabolism and respiratory metabolism to uranium-exposure in Vicia faba. J. Hazard. Mater. 398:122997. doi: 10.1016/j.jhazmat.2020.122997
Zhang, Y., Wang, Y., Ding, Z., Wang, H., Song, L., Jia, S., et al. (2017). Zinc stress affects ionome and metabolome in tea plants. Plant Physiol. Biochem. 111, 318–328. doi: 10.1016/j.plaphy.2016.12.014
Zhang-Xuan, D., Xiao-Hui, J., and Xue-Gang, L. (2020). Absorption and Interaction mechanisms of uranium & cadmium in purple sweet potato (Ipomoea batatas L.). J. Hazard. Mater. 400:123264. doi: 10.1016/j.jhazmat.2020.123264
Zhao, L., Pan, Y., Dong, Z., Zheng, Y., Liu, J., Geng, J., et al. (2020). Investigation and genome-wide association study of grain copper content in Chinese common wheat. J. Cereal Sci. 95:102991.
Zhao, S., Liu, Q., Qi, Y., and Duo, L. (2010). Responses of root growth and protective enzymes to copper stress in turfgrass. Acta Biol. Cracoviensia Series Bot. 52, 7–11.
Zhao, Z., Zhang, H., Fu, Z., Chen, H., Lin, Y., Yan, P., et al. (2018). Genetic-based dissection of arsenic accumulation in maize using a genome-wide association analysis method. Plant Biotechnol. J. 16, 1085–1093. doi: 10.1111/pbi.12853
Zhao, L., Huang, Y., Paglia, K., Vaniya, A., Wancewicz, B., and Keller, A. A. (2018). Metabolomics reveals the molecular mechanisms of copper induced cucumber leaf (Cucumis sativus) senescence. Environ. Sci. Technol. 52, 7092–7100. doi: 10.1021/acs.est.8b00742
Zhou, M., Zheng, S., Liu, R., Lu, J., Lu, L., Zhang, C., et al. (2019). Comparative analysis of root transcriptome profiles between low-and high-cadmium-accumulating genotypes of wheat in response to cadmium stress. Funct. Integr. Genomics 19, 281–294. doi: 10.1007/s10142-018-0646-4
Zhu, E., Liu, D., Li, J., Li, T., Yang, X., He, Z., et al. (2010). Effect of nitrogen fertilizer on growth and cadmium accumulation in Sedum alfredii Hance. J. Plant Nutr. 34, 115–126.
Keywords: abiotic stress, CRISPR/Cas system, genomics, metabolomics, proteomics, speed breeding, miRNAomics
Citation: Raza A, Tabassum J, Zahid Z, Charagh S, Bashir S, Barmukh R, Khan RSA, Barbosa F Jr, Zhang C, Chen H, Zhuang W and Varshney RK (2022) Advances in “Omics” Approaches for Improving Toxic Metals/Metalloids Tolerance in Plants. Front. Plant Sci. 12:794373. doi: 10.3389/fpls.2021.794373
Received: 13 October 2021; Accepted: 22 November 2021;
Published: 04 January 2022.
Edited by:
Shabir Hussain Wani, Sher-e-Kashmir University of Agricultural Sciences and Technology, IndiaReviewed by:
Suprasanna Penna, Bhabha Atomic Research Centre (BARC), IndiaCopyright © 2022 Raza, Tabassum, Zahid, Charagh, Bashir, Barmukh, Khan, Barbosa, Zhang, Chen, Zhuang and Varshney. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weijian Zhuang, d2VpamlhbnpAZmFmdS5lZHUuY24=; Rajeev K. Varshney, Ui5LLlZhcnNobmV5QGNnaWFyLm9yZw==; cmFqZWV2LnZhcnNobmV5QG11cmRvY2guZWR1LmF1
†ORCID: Ali Raza, orcid.org/0000-0002-5120-2791; Sidra Charagh, orcid.org/0000-0002-8077-7324
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.