- 1College of Bioscience and Biotechnology, Yangzhou University, Yangzhou, China
- 2Joint International Research Laboratory of Agriculture and Agri-Product Safety, The Ministry of Education of China, Yangzhou University, Yangzhou, China
- 3Key Laboratory of Plant Functional Genomics of the Ministry of Education, College of Agriculture, Yangzhou University, Yangzhou, China
Rice Growth-Regulating Factors (GRFs) were originally identified to be gibberellin (GA)-induced, but the nature of GA induction has remained unknown because most reports thereafter focused on revealing their roles in growth-promoting activities. GRFs have the WRC (Trp, Arg, Cys) domain to target DNA and contain the QLQ (Gln, Leu, Gln) domain to interact with GRF-Interacting Factor (GIF), which recruits ATP-dependent DNA translocase Switch/Sucrose Non-fermenting (SWI/SNF) for chromatin remodeling. Both GRFs and GIFs exhibit transcriptional activities but GIFs lack a DNA-binding domain. So, GRFs act like a navigator in the GRF-GIF-SWI/SNF complex, determining when and where the complex should work on. The levels of most rice GRFs can be sensitively regulated by miR396, which responds to many developmental and environmental factors. Recent clues from several studies highlight the original question of how GRFs participate in GA signaling. DELLA (contain DELLA motif) protein plays dual roles in controlling the level of GRFs by regulating the level of miR396 and interacting with GRFs. Here we address the question of why this complex plays an essential role in controlling plant growth focusing on the action of GA signaling pivot, DELLA.
Introduction
Two decades ago, the first member of the Growth-Regulating Factor (GRF) family, OsGRF1, was identified as a gibberellin (GA)-induced gene in the internodes of rice (van der Knaap and Kende, 1995; van der Knaap et al., 2000). Subsequently, 12 GRF members were found in rice and most of them were proved to be GA-induced (Choi et al., 2004). The GRF family has been identified experimentally or computationally in many plant species and is deemed as a class of deeply conserved transcription factors in land plants (Omidbakhshfard et al., 2015; Kim, 2019). So far, growing evidence has shown that GRFs promote the growth of various plant organs, such as leaf expansion, stem and root elongation, seed and flower formation, etc. Very few were reported to inhibit growth (Omidbakhshfard et al., 2015; Kim, 2019). Since the last comprehensive review articles mainly focused on collecting and summarizing the individual roles of this family, here we present a new landscape of how this family and partners work together to take part in GA signaling. Our updated outline could provide new insights into a better understanding of the functions of the miR396-GRF-GIF-SWI/SNF module.
GRFS: Navigator in the Positive Complex of GRF- GRF-Interacting Factor (GIF)- Switch/Sucrose Non-Fermenting (SWI/SNF)
The roles of GRFs as transcription factors may be attributed to their WRC (Trp, Arg, Cys) domains in the N-terminus which have DNA-binding abilities as well as their transcriptional activities in C-terminus (van der Knaap et al., 2000; Kim et al., 2003; Choi et al., 2004). Another characteristic domain in GRFs is the QLQ (Gln, Leu, Gln) motif which is used for interacting with GIFs (Kim and Kende, 2004; Horiguchi et al., 2005). GRFs prompt growth via working with GIFs, which recruit the ATP-dependent DNA translocase SWI/SNF for chromatin remodeling (Vercruyssen et al., 2014). GIF genes are seemed to be more deeply conserved than GRF genes. GIFs exist in most eukaryotic organisms, such as embryophytes, “green algae,” and metazoans, implying probably more molecular roles (Kim and Kende, 2004; de Bruijn and Geurts van Kessel, 2006). Furthermore, unlike GRFs which contain 8–20 members in “higher” plant genomes, GIFs usually comprise lower copies of about 2–5 members in plant species (Omidbakhshfard et al., 2015). Besides interacting with GRFs, GIF may also work with other factors in promoting plant growth (Debernardi et al., 2014; Vercruyssen et al., 2014; Nelissen et al., 2015; Zhang et al., 2018). GIF has been also proved to have transcriptional activities, but it lacks a DNA-binding domain, suggesting its role in transcription may be a co-regulator (Vercruyssen et al., 2014; Zhang et al., 2018).
The functions of some identified targets of GRFs seemed to be diverse, for instance, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN2A (DREB2A) of AtGRF7 (Kim et al., 2012) and OBP3-RESPONSIVE GENE3 (ORG3) of AtGRF9 (Omidbakhshfard et al., 2018) in Arabidopsis. However, numerous clues indicated that GRFs execute their functions mainly through prompting cell proliferation in a broad range of organs (Ercoli et al., 2016). The time course of gene expression and analysis of cell division activity in the intercalary meristem suggest that OsGRF1 may mediate an early event in the cell cycle (van der Knaap et al., 2000). Nowadays, with a deepening understanding, it has been known that the positive roles of GRFs in regulating growth also require the partners of GIFs and SWI/SNF. To facilitate gene expressions that are responsible for the transition from stem cells to rapidly dividing daughter cells, the transcriptional machinery depends on chromatin remodeling complexes, such as SWI/SNF, to loosen the association between DNA and histone octamers (Figure 1A) (Clapier and Cairns, 2009). In this regard, the GRF-GIF-SWI/SNF module acts as a growth-promoting complex, in which GRF serves as a navigator determining, through its WRC domain, which places in the genome is supposed to be targeted. In rice and Arabidopsis, besides some common tissues such as shoot apical meristem (SAM) where almost all members of this family prefer to accumulate (Kim et al., 2003; Choi et al., 2004), the expression patterns of different GRFs overall tend to display a space- or time-specific manner (van der Knaap et al., 2000; Choi et al., 2004; Lu et al., 2020). The accumulation of a given GRF member in a specific tissue usually defines its growth-promoting roles in that organ.
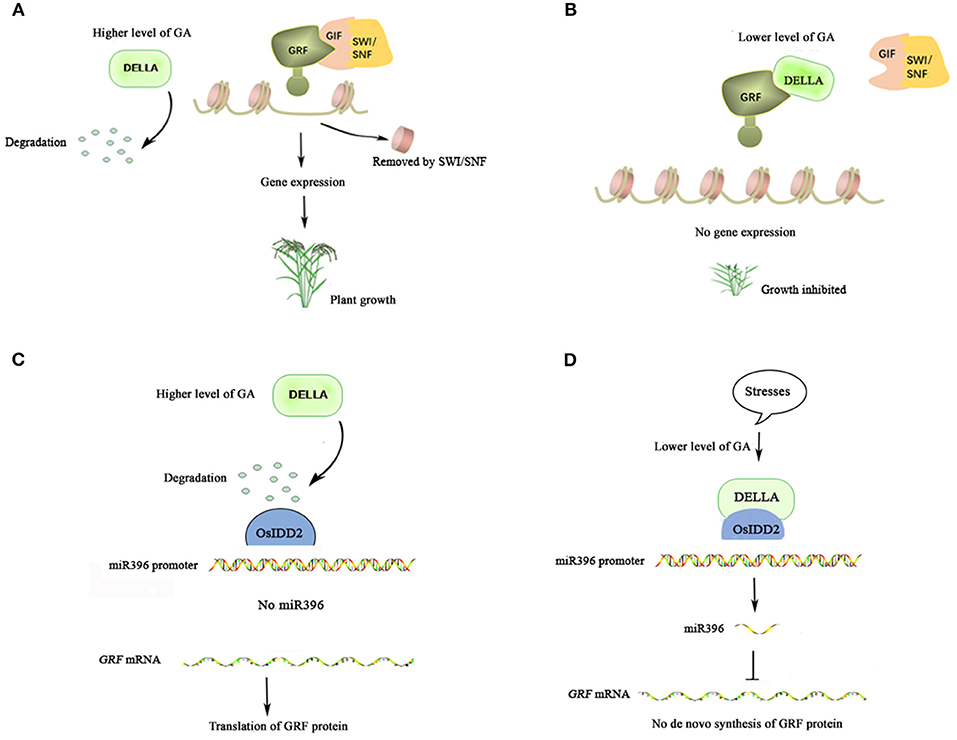
Figure 1. Rice miR396-growth regulating factors (GRF)-GRF Interacting factor (GIF)- Switch/Sucrose Non-fermenting SWI/SNF module in gibrellin (GA) signaling. (A) GRF-GIF-SWI/SNF module promotes plant growth. DELLA is degraded in the presence of a higher level of GA at meristematic organs or under suitable conditions, then GRF interacts with GIF which recruits SWI/SNF to form a positive complex for promoting plant growth. (B) DELLA antagonizes GRF under adverse conditions. A higher level of DELLA mediated by a lower level of GA under adverse conditions interacts with GRF. The separation between GRF and GIF mediated by DELLA leads to inhibition of plant growth. (C) GRFs are induced by GA. At meristematic organs or under suitable conditions along with a higher level of GA, degraded DELLA leads to a lower miR396 level which in turn results in a higher level of GRFs mRNA for translating protein. (D) miR396 suppresses GRFs mRNA under stress. A higher level of DELLA mediated by lower GA under stresses interacts with OsIDD2 to promote the expression of miR396 which in turn knocks down GRFs mRNA.
Della: Antagonizing GRFs Under Adverse Conditions
Even though GRFs are deemed as positive factors in promoting growth, surprisingly, they have been recently found to be able to interact with a negative regulator of GA signaling, DELLA (Li et al., 2018; Lantzouni et al., 2020). DELLA proteins act as growth repressors by inhibiting GA signaling in response to many conditions. GA, as one kind of classic plant hormone, regulates key processes of plant development and determines many traits of agronomic performances (Davière and Achard, 2013). Similar to GIFs, DELLA also lacks a DNA-binding domain but acts as transcriptional co-regulators (Ito et al., 2018). A number of studies demonstrated that DELLA proteins can interact with a list of DELLA-interacting proteins (DIPs) via transactivation or sequestration (Van De Velde et al., 2017). “Transactivation” means that DELLA can work with some transcription factors to promote the expression of the related genes, usually of growth repressors, while “sequestration” means that DELLA can prevent transcript factors from promoting the expression of the related genes, usually of growth activators (Van De Velde et al., 2017). Interestingly, several recent studies showed that DELLA can control GRFs via these two means (Li et al., 2018; Lantzouni et al., 2020; Lu et al., 2020).
OsGRF4 was discovered to improve agricultural performances by increasing grain size and yield (Che et al., 2015; Duan et al., 2015; Li et al., 2016), which could be ascribed to its ability to prompt inorganic nitrogen uptake and carbon fixation (Li et al., 2018). This contrasts with the property of DELLA, which inhibits them. Further evidence demonstrated that DELLA can interact with OsGRF4 and competitively inhibits the OsGRF4-OsGIF1 interaction. Interestingly, OsGRF4 abundance is self-promoted and DELLA inhibits that promotion even though the mechanism is unclear (Li et al., 2018). Furthermore, extensive interactions between OsSLR1 and OsGRFs were observed, although their physiologic meanings were not specified (Li et al., 2018). It is well-known that higher and stable levels of DELLAs exist in plants in the absence of GA (Ito et al., 2018). So, under lower GA concentrations, the interactions between abundant DELLAs, and GRFs would limit the growth-promoting roles of the GRF-GIF complex (Figure 1B). This view has been further supported by a study that shows that the extensive interactions of DELLAs and ATGRFs exist in response to cold stress (Lantzouni et al., 2020). In the meantime, the expression levels of some GA biosynthesis genes and endogenous levels of the bioactive GAs are decreased under low temperatures (Sakata et al., 2014). In fact, besides low temperature, various stresses, including salt and osmotic stress, can also reduce GA levels, which in turn increase the level of DELLA proteins (Colebrook et al., 2014). Given that, the interactions between DELLAs and GRFs facilitated by reduced levels of GAs under various stresses would result in growth inhibition and thus, probably be a strategy for plants to survive in adverse conditions.
Della-Controlled miR396: A Sensitive Regulatory Switch of GRF Complex
The interactions of SLR1 and OsGRFs at the protein level still cannot fully address the original question of why the expression of OsGRFs mRNA occurs in a low level under a low level of GA. So it is possible there might be another regulatory way that controls the expression of GRFs under the influence of GA. miR396 is an evolutionarily conserved miRNA that negatively regulates the abundance of GRFs mRNA (Jones-Rhoades and Bartel, 2004; Rodriguez et al., 2010, 2016). While miR396 also regulates other targets in groups of related species, only the miR396–GRF node is conserved in seed plants (Chorostecki et al., 2012). As a class of regulatory molecules, miR396 is up-regulated by various stress conditions, including drought, salinity, cold, and ultra-violet (UV)-B irradiation (Casadevall et al., 2013; Omidbakhshfard et al., 2015; Beltramino et al., 2018). It is possible that the increase in miR396 levels and the concomitant decrease in the level of GRFs mRNA are a part of the plant's strategy to reduce growth activities under various stresses, which are usually associated with decreased GA and thus increased levels of DELLAs.
Based on these clues, another study found that the negative response of miR396 to GA was a real cause of the positive response of OsGRFs to GA (Lu et al., 2020). There are 8 miR396 members in rice and 11 of 12 OsGRFs are miR396' targets. Under higher concentrations of GA in which SLR1 abundance is lower, the expression of OsmiR396 is also reduced, which in turn leads to higher levels of OsGRFs mRNA (Figure 1C). The positive correlation between the expression of miR396 and SLR1 was further confirmed by detecting miR396 levels in the two key mutants, gid1 (gibberellin insensitive dwarf1) and eui (elongated uppermost internode). GID1, a receptor of biologically active GAs, mediates the degradation of SLR1 under higher levels of active GAs. So, the level of SLR1 always keeps stable and can avoid being degraded in the gid1 mutant (Ueguchi-Tanaka et al., 2005, 2008). EUI, a GA deactivating enzyme, helps to maintain higher levels of inactive GAs. So the level of SLR1 is reduced in the eui mutant due to higher levels of biologically active GAs (Zhu et al., 2006). OsmiR396' expression is higher in the gid1 mutant and is lower in the eui mutant. The time course of gene expression also indicated that miR396 acts downstream of SLR1 and upstream of GA-induced cell-cycle genes (Lu et al., 2020).
Using the promoter of OsmiR396a as bait, rice transcription factor INDETERMINATE DOMAIN2 (OsIDD2), was found to directly bind the promoter of OsmiR396a and can interact with SLR1 (Lu et al., 2020). As a family of transcription factors conserved among all land plants, indeterminate domains (IDDs) are defined by encoding four zinc-finger motifs utilized for binding DNA (Colasanti et al., 1998, 2006). IDDs have been reported to control diverse processes at various stages of plant development (Coelho et al., 2018). Overexpressing OsIDD2 also led to a dwarf phenotype which is similar to transgenic lines overexpressing miR396 with a concomitant decrease in the level of OsGRFs mRNA (Huang et al., 2018; Lu et al., 2020). The interaction of OsIDD2 and SLR1 is indispensable for the expression of miR396 (Figure 1D) (Lu et al., 2020). A number of AtIDDs have been also shown to be capable of interacting with DELLA proteins via transactivation to regulate downstream genes in Arabidopsis (Feurtado et al., 2011; Fukazawa et al., 2014; Yoshida et al., 2014). So, SLR1 promoting miR396' expression with a subsequent decrease in the level of OsGRFs mRNA may provide more means for DELLA to decline GRFs' functions under stress. Because stress-induced DELLA, as the GA signaling hub, would be subjected to a large consumption due to extensive interactions between it and many other proteins. But DELLA may save its consumption by eliminating GRFs mRNA by promoting the expression of miR396 and thus curb the translation of GRFs which usually contain multiple members in plants.
Conclusions and Future Perspective
Growing evidence has shown that GRFs play important roles in regulating the growth of a wide range of plant organs, usually through promoting the meristematic potential of primordial cells (Rodriguez et al., 2010, 2016). The positive regulatory roles of GRFs also depend on the partners of GIFs and SWI/SNF. However, plant growth and development are processes being subjected to exquisite regulation which relies on the balance of both negative and positive conditions. In the GRF-GIF-SWI/SNF module, GRFs are adjustable at both mRNA and protein levels (Figure 1). Hormones, specifically associated with the internal or external conditions, are usually at the upmost part of the signaling cascades in controlling genes' expression. DELLAs, which play pivotal roles in GA signaling and whose abundance is negatively regulated by GA, control the function of GRFs by means of both transactivation and sequestration. Through transactivation, DELLA can work with other factors (for example OsIDD2 in rice) to stimulate the expression of miR396 to finally decrease the abundance of GRFs mRNA (Figure 1D). For the pre-existing GRF proteins, DELLA can prevent them from interacting with GIF-SWI/SNF by sequestration (through a competitive manner).
It may still need more biochemistry works in the future to reveal full details of the interaction between DELLAs and GRFs, including what are key domains needed for the interaction and how DELLA and GIFs competitively interact with GRFs. Even though every component of this module is deeply conserved in “higher” plants, whether these are some nuances in the mechanism of this module working in other plant species still needs to be elucidated.
Author Contributions
QL and YL conceived the review topics. YL wrote the manuscript. QL scrutinized the whole text. JZ made the figures. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Natural Science Foundation of China (No. 31271623) and the open fund of Key Laboratory of Plant Functional Genomics of the Ministry of Education (No. ML202004).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Dr. Xiaofeng Cui for their critical comments and helpful discussion on this manuscript. We also apologize to any authors and work that was not cited due to restrictions in the size of the manuscript.
References
Beltramino, M., Ercoli, M. F., Debernardi, J. M., Goldy, C., Rojas, A., Nota, F., et al. (2018). Robust increase of leaf size by Arabidopsis thaliana GRF3-like transcription factors under different growth conditions. Sci. Rep. 8:13447. doi: 10.1038/s41598-018-29859-9
Casadevall, R., Rodriguez, R. E., Debernardi, J. M., Palatnik, J. F., and Casati, P. (2013). Repression of growth regulating factors by the microRNA396 inhibits cell proliferation by UV-B radiation in Arabidopsis leaves. Plant Cell. 25, 3570–3583. doi: 10.1105/tpc.113.117473
Che, R., Tong, H., Shi, B., Liu, Y., Fang, S., Liu, D., et al. (2015). Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants 2:15195. doi: 10.1038/nplants.2015.195
Choi, D., Kim, J. H., and Kende, H. (2004). Whole genome analysis of the OsGRF gene family encoding plant-specific putative transcription activators in rice (Oryza sativa L.). Plant Cell Physiol. 45, 897–904. doi: 10.1093/pcp/pch098
Chorostecki, U., Crosa, V. A., Lodeyro, A. F., Bologna, N. G., Martin, A. P., Carrillo, N., et al. (2012). Identification of new microRNA-regulated genes by conserved targeting in plant species. Nucleic Acids Res. 40, 8893–8904. doi: 10.1093/nar/gks625
Clapier, C. R., and Cairns, B. R. (2009). The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304. doi: 10.1146/annurev.biochem.77.062706.153223
Coelho, C. P., Huang, P., Lee, D. Y., and Brutnell, T. P. (2018). Making roots, shoots, and seeds: IDD gene family diversification in plants. Trends Plant Sci. 23, 66–78. doi: 10.1016/j.tplants.2017.09.008
Colasanti, J., Tremblay, R., Wong, A. Y. M., Coneva, V., Kozaki, A., and Mable, B. K. (2006). The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants. BMC Genomics 7:158. doi: 10.1186/1471-2164-7-158
Colasanti, J., Yuan, Z., and Sundaresan, V. (1998). The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 93, 593–603. doi: 10.1016/S0092-8674(00)81188-5
Colebrook, E. H., Thomas, S. G., Phillips, A. L., and Hedden, P. (2014). The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 217, 67–75. doi: 10.1242/jeb.089938
Davière, J. M., and Achard, P. (2013). Gibberellin signaling in plants. Development 140, 1147–1151. doi: 10.1242/dev.087650
de Bruijn, D. R., and Geurts van Kessel, A. (2006). Common origin of the human synovial sarcoma associated SS18 and SS18L1 gene loci. Cytogenet. Genome Res. 112, 222–226. doi: 10.1159/000089874
Debernardi, J. M., Mecchia, M. A., Vercruyssen, L., Smaczniak, C., Kaufmann, K., Inze, D., et al. (2014). Post-transcriptional control of GRF transcription factors by microRNA miR396 and GIF co-activator affects leaf size and longevity. Plant J. 79, 413–426. doi: 10.1111/tpj.12567
Duan, P., Ni, S., Wang, J., Zhang, B., Xu, R., Wang, Y., et al. (2015). Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat. Plants 2:15203. doi: 10.1038/nplants.2015.203
Ercoli, M. F., Rojas, A. M., Debernardi, J. M., Palatnik, J. F., and Rodriguez, R. E. (2016). Control of cell proliferation and elongation by miR396. Plant Signal. Behav. 11:e1184809. doi: 10.1080/15592324.2016.1184809
Feurtado, J. A., Huang, D., Wicki-Stordeur, L., Hemstock, L. E., Potentier, M. S., Tsang, E. W., et al. (2011). The Arabidopsis C2H2 zinc finger INDETERMINATE DOMAIN1/ENHYDROUS promotes the transition to germination by regulating light and hormonal signaling during seed maturation. Plant Cell 23, 1772–1794. doi: 10.1105/tpc.111.085134
Fukazawa, J., Teramura, H., Murakoshi, S., Nasuno, K., Nishida, N., Ito, T., et al. (2014). DELLAs function as coactivators of GAI-ASSOCIATED FACTOR1 in regulation of gibberellin homeostasis and signaling in Arabidopsis. Plant Cell 26, 2920–2938. doi: 10.1105/tpc.114.125690
Horiguchi, G., Kim, G. T., and Tsukaya, H. (2005). The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 43, 68–78. doi: 10.1111/j.1365-313X.2005.02429.x
Huang, P., Yoshida, H., Yano, K., Kinoshita, S., Kawai, K., Koketsu, E., et al. (2018). OsIDD2, a zinc finger and INDETERMINATE DOMAIN protein, regulates secondary cell wall formation. J. Integr. Plant Biol. 60, 130–143. doi: 10.1111/jipb.12557
Ito, T., Okada, K., Fukazawa, J., and Takahashi, Y. (2018). DELLA-dependent and -independent gibberellin signaling. Plant Signal. Behav. 13:e1445933. doi: 10.1080/15592324.2018.1445933
Jones-Rhoades, M. W., and Bartel, D. P. (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 14, 787–799. doi: 10.1016/j.molcel.2004.05.027
Kim, J. H (2019). Biological roles and an evolutionary sketch of the GRF-GIF transcriptional complex in plants. BMB Rep. 52, 227–238. doi: 10.5483/BMBRep.2019.52.4.051
Kim, J. H., Choi, D., and Kende, H. (2003). The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 36, 94–104. doi: 10.1046/j.1365-313X.2003.01862.x
Kim, J. H., and Kende, H. (2004). A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 13374–13379. doi: 10.1073/pnas.0405450101
Kim, J. S., Mizoi, J., Kidokoro, S., Maruyama, K., Nakajima, J., Nakashima, K., et al. (2012). Arabidopsis growth-regulating factor7 functions as a transcriptional repressor of abscisic acid- and osmotic stress-responsive genes, including DREB2A. Plant Cell 24, 3393–3405. doi: 10.1105/tpc.112.100933
Lantzouni, O., Alkofer, A., Falter-Braun, P., and Schwechheimer, C. (2020). Growth-regulating factors interact with dellas and regulate growth in cold stress. Plant Cell 32, 1018–1034. doi: 10.1105/tpc.19.00784
Li, S., Gao, F., Xie, K., Zeng, X., Cao, Y., Zeng, J., et al. (2016). The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice. Plant Biotechnol. J. 14, 2134–2146. doi: 10.1111/pbi.12569
Li, S., Tian, Y., Wu, K., Ye, Y., Yu, J., Zhang, J., et al. (2018). Modulating plant growth-metabolism coordination for sustainable agriculture. Nature 560, 595–600. doi: 10.1038/s41586-018-0415-5
Lu, Y., Feng, Z., Meng, Y., Bian, L., Xie, H., Mysore, K. S., et al. (2020). Slender Rice1 and Oryza sativa indeterminate Domain2 regulating OsmiR396 Are Involved in Stem Elongation. Plant Physiol. 182, 2213–2227. doi: 10.1104/pp.19.01008
Nelissen, H., Eeckhout, D., Demuynck, K., Persiau, G., Walton, A., van Bel, M., et al. (2015). Dynamic changes in Angustifolia3 complex composition reveal a growth regulatory mechanism in the maize leaf. Plant Cell 27, 1605–1619. doi: 10.1105/tpc.15.00269
Omidbakhshfard, M. A., Fujikura, U., Olas, J. J., Xue, G. P., Balazadeh, S., and Mueller-Roeber, B. (2018). Growth-regulating factor 9 negatively regulates arabidopsis leaf growth by controlling ORG3 and restricting cell proliferation in leaf primordia. PLoS Genet. 14:e1007484. doi: 10.1371/journal.pgen.1007484
Omidbakhshfard, M. A., Proost, S., Fujikura, U., and Mueller-Roeber, B. (2015). Growth-Regulating Factors (GRFs): a small transcription factor family with important functions in plant biology. Mol. Plant. 8, 998–1010. doi: 10.1016/j.molp.2015.01.013
Rodriguez, R. E., Mecchia, M. A., Debernardi, J. M., Schommer, C., Weigel, D., and Palatnik, J. F. (2010). Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137, 103–112. doi: 10.1242/dev.043067
Rodriguez, R. E., Schommer, C., and Palatnik, J. F. (2016). Control of cell proliferation by microRNAs in plants. Curr. Opin. Plant Biol. 34, 68–76. doi: 10.1016/j.pbi.2016.10.003
Sakata, T., Oda, S., Tsunaga, Y., Shomura, H., Kawagishi-Kobayashi, M., Aya, K., et al. (2014). Reduction of gibberellin by low temperature disrupts pollen development in rice. Plant Physiol. 164, 2011–2019. doi: 10.1104/pp.113.234401
Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E., Kobayashi, M., et al. (2005). Gibberellin insensitive DWARF1 encodes a soluble receptor for gibberellin. Nature 437, 693–698. doi: 10.1038/nature04028
Ueguchi-Tanaka, M., Hirano, K., Hasegawa, Y., Kitano, H., and Matsuoka, M. (2008). Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant. Plant Cell 20, 2437–2446. doi: 10.1105/tpc.108.061648
Van De Velde, K., Ruelens, P., Geuten, K., Rohde, A., and Van Der Straeten, D. (2017). Exploiting DELLA signaling in cereals. Trends Plant Sci. 22, 880–893. doi: 10.1016/j.tplants.2017.07.010
van der Knaap, E., and Kende, H. (1995). Identification of a gibberellin-induced gene in deepwater rice using differential display of mRNA. Plant Mol. Biol. 28, 589–592. doi: 10.1007/BF00020405
van der Knaap, E., Kim, J. H., and Kende, H. (2000). A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 122, 695–704. doi: 10.1104/pp.122.3.695
Vercruyssen, L., Verkest, A., Gonzalez, N., Heyndrickx, K. S., Eeckhout, D., Han, S. K., et al. (2014). Angustifolia3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell 26, 210–229. doi: 10.1105/tpc.113.115907
Yoshida, H., Hirano, K., Sato, T., Mitsuda, N., Nomoto, M., Maeo, K., et al. (2014). DELLA protein functions as a transcriptional activator through the DNA binding of the indeterminate domain family proteins. Proc. Natl. Acad. Sci. U.S.A. 111, 7861–7866. doi: 10.1073/pnas.1321669111
Zhang, D., Sun, W., Singh, R., Zheng, Y., Cao, Z., Li, M., et al. (2018). GRF-interacting factor1 regulates shoot architecture and meristem determinacy in maize. Plant Cell 30, 360–374. doi: 10.1105/tpc.17.00791
Keywords: GRF, GIF, SWI/SNF, DELLA, GA signaling
Citation: Lu Y, Zeng J and Liu Q (2022) The Rice miR396-GRF-GIF-SWI/SNF Module: A Player in GA Signaling. Front. Plant Sci. 12:786641. doi: 10.3389/fpls.2021.786641
Received: 30 September 2021; Accepted: 13 December 2021;
Published: 11 January 2022.
Edited by:
Hirokazu Tsukaya, The University of Tokyo, JapanReviewed by:
Weibing Yang, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (CAS), ChinaJun Fang, Northeast Institute of Geography and Agroecology, Chinese Academy of Sciences (CAS), China
Jeong Hoe Kim, Kyungpook National University, South Korea
Copyright © 2022 Lu, Zeng and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuzhu Lu, bHV5dXpodUB5enUuZWR1LmNu; Qiaoquan Liu, cXFsaXVAeXp1LmVkdS5jbg==
 Yuzhu Lu
Yuzhu Lu Jia Zeng1
Jia Zeng1 Qiaoquan Liu
Qiaoquan Liu