- Dahlem Centre of Plant Sciences (D), Institute of Biology/Applied Genetics, Freie Universität Berlin, Berlin, Germany
The photoperiod, which is the length of the light period in the diurnal cycle of 24 h, is an important environmental signal. Plants have evolved sensitive mechanisms to measure the length of the photoperiod. Photoperiod sensing enables plants to synchronize developmental processes, such as the onset of flowering, with a specific time of the year, and enables them to alleviate the impact of environmental stresses occurring at the same time every year. During the last years, the importance of the photoperiod for plant responses to abiotic and biotic stresses has received increasing attention. In this review, we summarize the current knowledge on the signaling pathways involved in the photoperiod-dependent regulation of responses to abiotic (freezing, drought, osmotic stress) and biotic stresses. A central role of GIGANTEA (GI), which is a key player in the regulation of photoperiod-dependent flowering, in stress responses is highlighted. Special attention is paid to the role of the photoperiod in regulating the redox state of plants. Furthermore, an update on photoperiod stress, which is caused by sudden alterations in the photoperiod, is given. Finally, we will review and discuss the possible use of photoperiod-induced stress as a sustainable resource to enhance plant resistance to biotic stress in horticulture.
Introduction
Eukaryotes, including plants, adapt numerous life processes to regular rhythms of light and darkness. Light and dark periods regularly alternate in a daily cycle of approximately 24 h due to the rotation of the Earth around its own axis. The duration of the light period during this 24 h day-night cycle determines the photoperiod, which varies with the season and latitude (Jackson, 2009). Plants synchronize their physiological decisions with the correct time of the year to maximize growth and to produce offspring (Casal et al., 2004). Thus, sensing of and responding to the photoperiod are important plant functions to adapt to their environment.
Among the most prominent plant responses influenced by the photoperiod are the regulation of flowering time (Carré, 2001; Song et al., 2015), tuberization (Sarkar, 2010), bud setting, and dormancy (Jackson, 2009; Singh et al., 2017). In annual plants, senescence is adjusted by the photoperiod (Serrano-Bueno et al., 2021) and in perennial plants like trees, the growth cessation is influenced by season-dependent photoperiods (Singh et al., 2017). In temperate climate zones but also in tropical regions, the photoperiod is the dominant environmental factor controlling the onset and end of the seasonal growing (Adole et al., 2019). Scent emission from flowers is also under the control of the photoperiod (Hendel-Rahmanim et al., 2007) to mention just a few examples of photoperiod-regulated developmental processes in plants.
Based on their flowering response to the photoperiod, plants can be classified into three groups: short-day, long-day, and day-neutral plants. This classification is based on the critical day length (CDL), which determines the ability of plants to respond to the photoperiod. Short-day-grown plants flower when the photoperiod is shorter than the CDL, while long-day plants flower only, when the photoperiod is longer than the CDL. Day-neutral plants do not respond to the photoperiod (Jackson, 2009). Besides the CDL, the plant developmental phase determines the ability to sense and subsequently respond to photoperiods. The flowering response of Arabidopsis plants is insensitive to photoperiods during their juvenile phase. Entering the adult phase makes Arabidopsis sensitive to photoperiods enabling responses to floral inducers (Matsoukas, 2014). Taken together, the synchronization of the photoperiod sensing and intrinsic developmental programs or developmental phases with the seasonal photoperiod is essential for the reproduction and survival of plants.
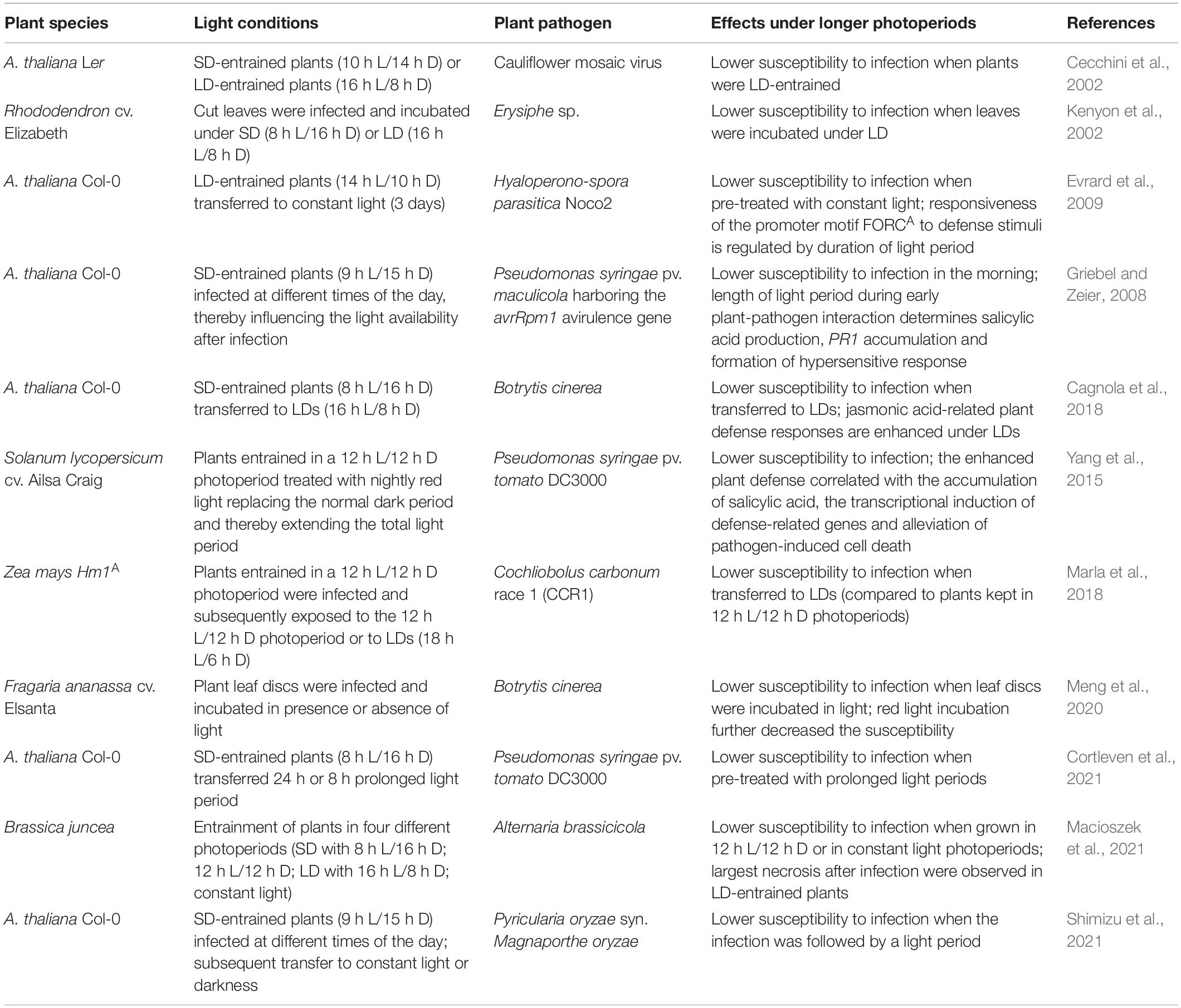
Photoperiod sensing not only enables plants to synchronize their developmental processes with a specific time of the year but also alleviates the impact of environmental stresses occurring at the same time every year. Recently, the interest in the effect of the photoperiod on the response to abiotic and biotic stresses has grown. For example, the role of shortening of days in cold acclimation to prepare for freezing winter temperatures has been uncovered (Ouellet and Charron, 2013). The photoperiod has also been shown to influence the plants’ resistance to drought stress (Iuchi et al., 2001; Han et al., 2013a) and salt stress (Kim et al., 2013; Park et al., 2016). In addition, increasing evidence suggests that the length of the light period is important for the outcome of plant-pathogen interactions (e.g., Griebel and Zeier, 2008). Thus, photoperiod sensing enables plants to improve their responses to diverse environmental stresses (Figure 1). However, sudden changes in the photoperiod can also result in stress. Although the molecular mechanisms underlying this new abiotic stress form are not yet completely resolved (Nitschke et al., 2016, 2017; Abuelsoud et al., 2020; Frank et al., 2020), experiments revealed that changes in the photoperiod elicit stress reactions in Arabidopsis plants, which resemble responses to pathogen attack (Cortleven et al., 2021). The establishment of systemic acquired resistance (SAR) in plants forms an important defense against future pathogen attacks (Conrath et al., 2006). As photoperiod stress provokes similar effects, this might open new horizons for the use of altered photoperiods as a sustainable tool to alleviate pathogen infections and thereby decrease yield losses in horticulture. In the following chapters, we will address the above-mentioned topics in more detail.
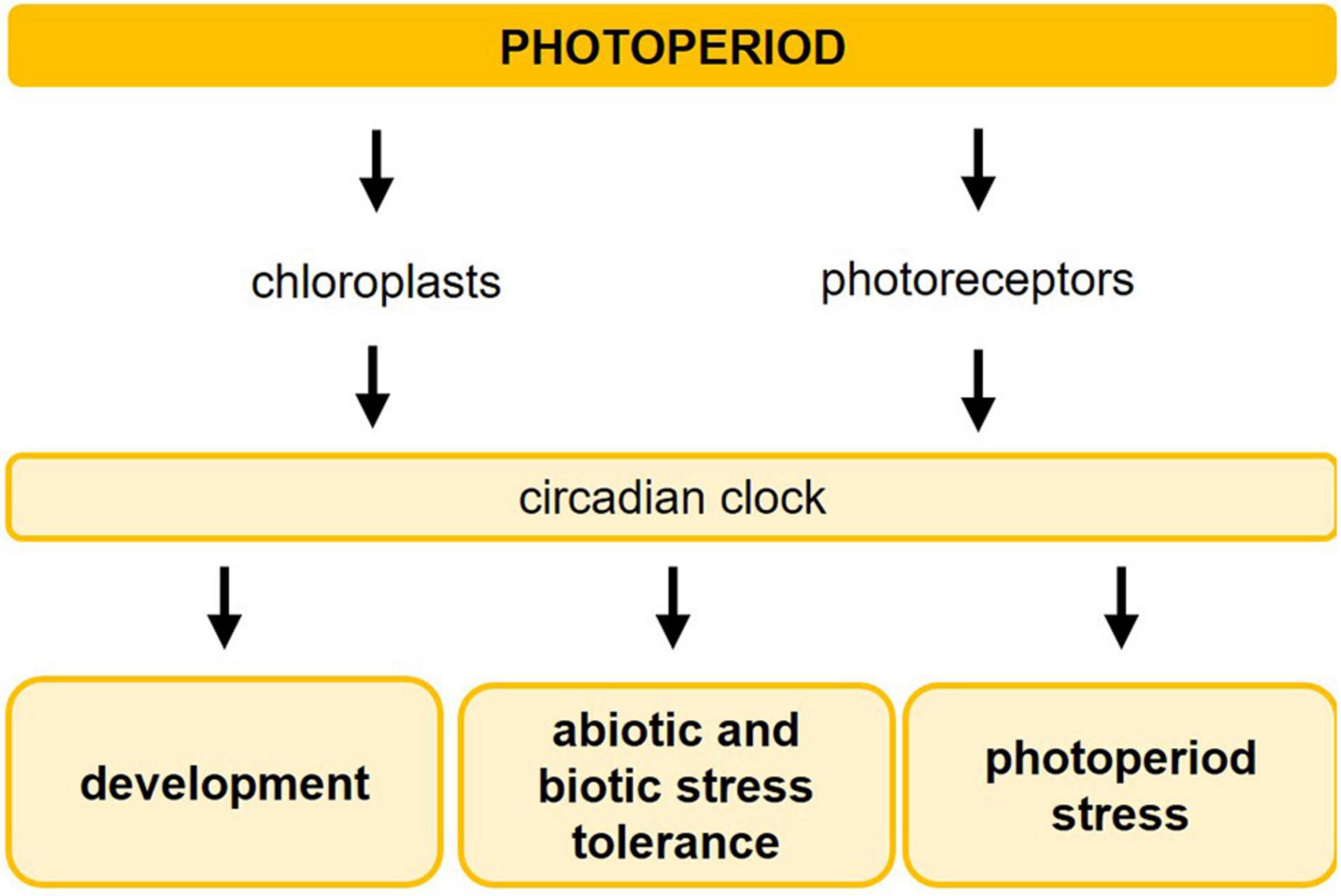
Figure 1. Photoperiod sensing influences development of plants, induces abiotic and biotic stress tolerance, and causes photoperiod stress. The length of the photoperiod is detected by a sensing mechanism consisting of chloroplasts and photoreceptors, which transfer the light information to the circadian clock. The interplay between the photoperiod and the circadian clock regulates developmental processes, such as flowering, tuberization, bud setting, dormancy, and senescence, and improves the plants’ tolerance to abiotic and biotic stresses. A sudden prolongation of the photoperiod results in photoperiod stress.
Molecular Mechanisms Involved in the Perception of Light and the Photoperiod
The perception of and response to photoperiods in plants require a sensing mechanism, which involves the detection of light (via photoreceptors or chloroplasts) and the measurement of time (via the circadian clock) (Jackson, 2009; Serrano-Bueno et al., 2021).
Light perception by photoreceptors and chloroplasts provides plants with comprehensive information concerning their surrounding light environment, such as quality (spectral composition, direction), quantity, intensity, and duration of incoming irradiation (Figure 1). In Arabidopsis thaliana, five photoreceptor families sense the light from different parts of the solar light spectrum: red/far-red light is detected by phytochromes (phyA to phyE). Blue light is perceived by cryptochromes (CRY1, CRY2, CRY3), phototropins (PHOT1, PHOT2), and F-box containing flavin-binding proteins ZEITLUPE (ZTL) and FLAVIN-BINDING KELCH REPEAT F-BOX1 (FKF1)/LOV KELCH PROTEIN2 (LKP2). UV light is sensed by the UVR8 photoreceptor (for review, see Sanchez et al., 2020; Roeber et al., 2021). All of the above-mentioned photoreceptor families are involved in the light entrainment of the circadian clock.
Besides the photoreceptors, chloroplasts operate as plant light sensors and respond to different photoperiods by altering their ultrastructure (Lepisto and Rintamaki, 2012). Chloroplasts of plants grown under long days exhibit smaller grana stacks and increased chlorophyll content. These features correspond to structural and photosynthetic characteristics typical of sun plants (Walters and Horton, 1995). Redox signals arising from chloroplasts determine the light intensity-dependent acclimation of plants (Pfannschmidt et al., 2009). Which signaling mechanisms are involved in the photoperiodic-dependent development of chloroplasts remains to be resolved. The redox state of the photosynthetic electron transport chain, ROS metabolism, and chloroplast-to-nucleus retrograde signaling are only few examples of possible pathways involved, all acting independent of the photoreceptors (for review, see Lepisto and Rintamaki, 2012; Feng et al., 2016). Besides the chloroplast ultrastructure, the photoperiod regulates the photosynthate partitioning to starch and the amount of carbohydrate (C) stored in chloroplasts (Zeeman et al., 2007). Under conditions where less C is available such as short photoperiods, a larger proportion of the fixed C is allocated into starch (Smith and Stitt, 2007). During the night the near-linear starch degradation is slowed down as compared to long-day-grown plants. This results in an almost but not completely exhausted starch content at dawn preventing C-starvation or C-excess at the end of the night period (Stitt and Zeeman, 2012; Moraes et al., 2019). This pattern of C-mobilization is robust across different photoperiods (Stitt and Zeeman, 2012; Moraes et al., 2019). Also here, the exact molecular mechanisms controlling the formation of starch under various photoperiods are not known, but possible feedback inhibition from the carbohydrate metabolism, redox regulation, transcriptional control of chloroplast enzymes, and circadian regulation might play a role.
The circadian clock enables plants to measure time by an endogenous time-keeping mechanism (Hsu and Harmer, 2014; Figure 2). The clock is set through daily entrainment, especially by light and temperature, in order to adjust the internal rhythm (McClung, 2006). In Arabidopsis thaliana, the circadian clock consists of multiple interlocked transcription-translation feedback loops (Hsu and Harmer, 2014). The MYB-domain transcription factor genes CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) are expressed in the morning (Schaffer et al., 1998; Wang and Tobin, 1998) and repress the expression of TIMING OF CAB EXPRESSION1 (TOC1) during the day (Alabadi et al., 2001). In turn, TOC1 represses the transcription of CCA1 and LHY (Gendron et al., 2012). Late at night, TOC1 transcription is down-regulated by an Evening Complex (EC), which is composed of three proteins: EARLY FLOWERING3 (ELF3), ELF4, and LUX ARRHYTHMO (LUX). This down-regulation enables transcription of LHY and CCA1 to resume the following dawn. PSEUDO-RESPONSE REGULATOR9 (PRR9), PRR7, and PRR5 are expressed in consecutive waves throughout the day and repress CCA1 and LHY expression (Nakamichi et al., 2012). Additional rhythmically expressed transcriptional activators, such as REVEILLE4 (RVE4), RVE6, and RVE8, the LIGHT-REGULATED WD1 (LWD1) and LWD2 proteins, and the transcription factors NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED GENE1 (LNK1) and LNK2 also contribute to the clock function (Rawat et al., 2011; Rugnone et al., 2013). The circadian clock contributes to the plants’ ability to respond to various environmental stresses but there is also a reciprocal influence of abiotic stresses on the clock function. More information about this can be found in the reviews of Sanchez et al. (2011), Kiełbowicz-Matuk and Czarnecka (2014), Grundy et al. (2015), Seo and Mas (2015), and Sharma et al. (2021). A novel webtool to investigate the transcriptional networks regulated by light and the circadian clock has been launched recently (de los Reyes et al., 2020). With ATTRACTOR (Arabidopsis Thaliana TRanscriptionAl Circadian neTwORk1), target genes of circadian regulators can be identified. This might contribute to a better understanding of the interaction between the circadian clock and plant responses to environmental stresses.
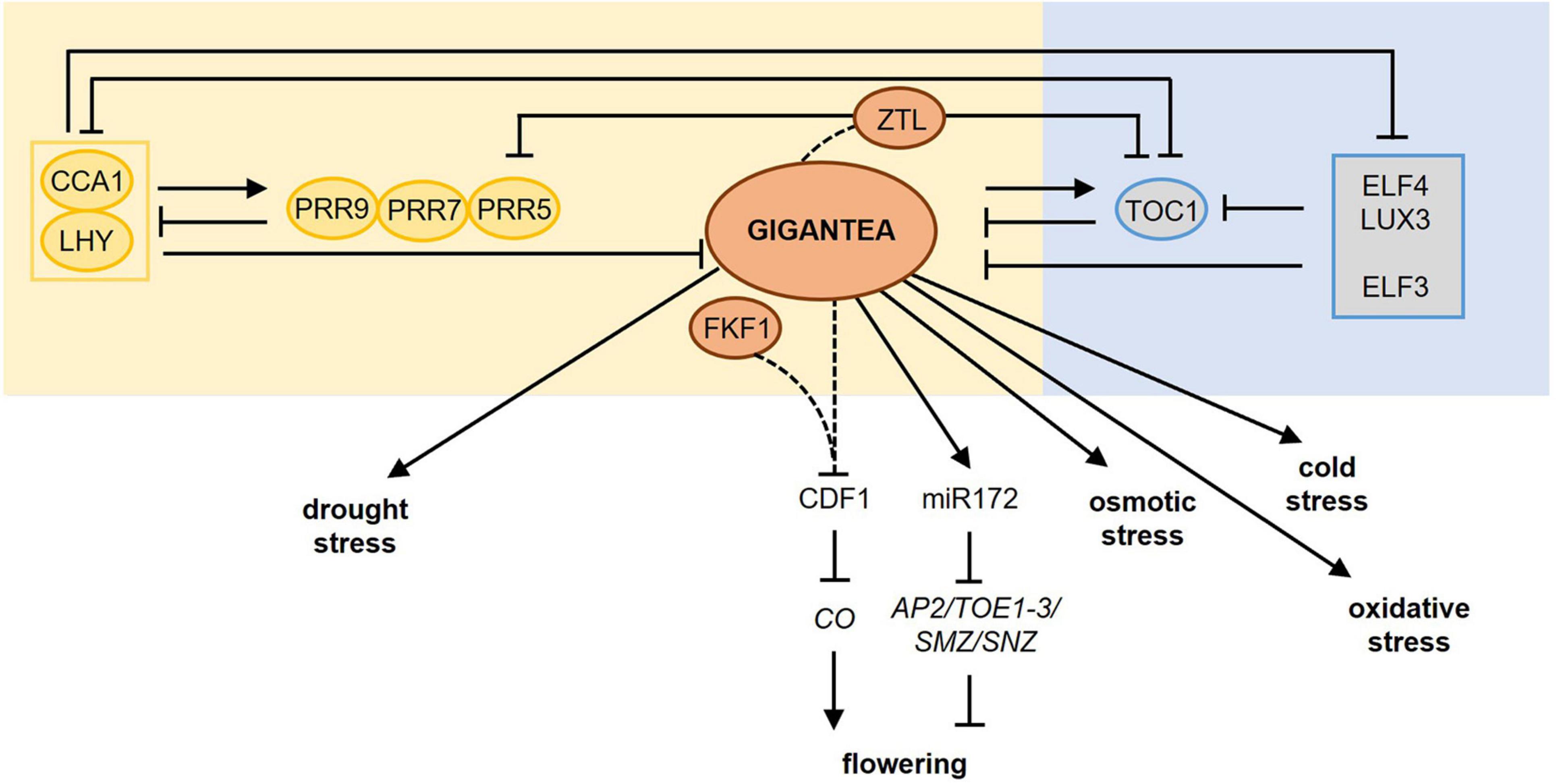
Figure 2. GIGANTEA plays a central role in photoperiod sensing and mediates the impact of photoperiod on stress responses. The circadian clock is an internal time-keeping mechanism involved in photoperiod sensing. The main regulatory components of the circadian clock are shown including their mutual transcription-translation feedback loops. GIGANTEA is expressed late in the afternoon and the protein improves the stability of ZEITLUPE (ZTL) upon blue light perception thereby targeting TOC1 and PRR5 for proteasomal degradation, reinforcing the entrainment of the clock. Upon blue light perception, GI interacts also with FKF1 causing the degradation of CYCLING DOF FACTOR1 (CDF1), which is a transcriptional repressor of CONSTANS (CO), encoding a central protein in photoperiod-dependent flowering. In addition, GI regulates the miR172-mediated post-transcriptional downregulation of several floral repressor genes. Besides its role in photoperiod-dependent flowering, GI has a central role in the photoperiod-dependent plant responses to drought, osmotic, cold, and oxidative stress. Dashed lines mark protein–protein interactions upon blue light perception. For more detailed information about the different pathways, please refer to section “Molecular Mechanisms Involved in the Perception of Light and the Photoperiod.” AP2, APETALA2; CCA1, CIRCADIAN CLOCK ASSOCIATED1; LHY, LATE ELONGATED HYPOCOTYL; PRR, PSEUDO-RESPONSE REGULATOR, TOC1, TIMING OF CAB EXPRESSION1; ELF3, EARLY FLOWERING3; ELF4, EARLY FLOWERING4; LUX, LUX ARRHYTHMO; FKF1, FLAVIN-BINDING KELCH REPEAT F-BOX1; TOE1, TARGET OF EAT1; TOE2, TARGET OF EAT2; SMZ, SCHLAFMÜTZE; SNZ, SCHNARCHZAPFEN.
One of the circadian clock-controlled genes that have a crucial role in the photoperiod sensing mechanism is GIGANTEA (GI) (Fowler et al., 1999). It encodes a large single-gene encoded protein with a chaperone activity (Cha et al., 2017). Upon blue light perception, the stability of the F-box protein ZEITLUPE (ZTL) improves due to interaction with GI. ZTL is an evening-phased E3 ubiquitin ligase targeting the clock components TOC1 and PRR5 for proteasomal degradation (Mas et al., 2003; Kiba et al., 2007). GI protein abundance peaks in the evening, thereby maintaining ZTL abundance high. Consequently, high amplitude oscillations of TOC1 and PRR5 are sustained (Kim et al., 2007). This reinforces the entrainment of the clock resulting in the correct setting of the phase of clock output genes such as CONSTANS (CO), encoding a central protein in photoperiod-dependent flowering (Suarez-Lopez et al., 2001; Shim et al., 2017). GI also interacts with FKF1 upon blue light perception causing the degradation of CYCLING DOF FACTOR1 (CDF1), which is a transcriptional repressor of CO (Figure 2; Sawa et al., 2007). The synchronization of the correct timing of protein stabilization during long days with the circadian-regulated expression of FKF1, GI, and CDF1 is essential for photoperiodic responses, such as flowering. Interesting to mention is that the CO-FT-GI-CDF hub is conserved among distantly related flowering plants (Serrano-Bueno et al., 2021).
GI also regulates the maturation of miR172 (Jung et al., 2007), which targets APETALA2 (AP2) and the AP2-like genes TARGET OF EAT1 (TOE1), TOE2, TOE3, SCHLAFMÜTZE (SMZ), and SCHNARCHZAPFEN (SNZ) (Figure 2). The miR172-mediated posttranscriptional downregulation of these floral repressors regulates flowering time and floral development in the shoot apical meristem (Mathieu et al., 2009) depending on the age of the plants (Aukerman and Sakai, 2003). In addition, GI controls the circadian clock-mediated photoperiod sensing together with EARLY FLOWERING3 (ELF3). In their absence, the circadian clock fails to properly respond to light signals, resulting in the breakdown of the photoperiod sensing mechanism (Anwer et al., 2020).
GI plays not only a central role in the photoperiod sensing mechanism but is also involved in mediating the impact of the photoperiod in response to diverse stresses (Figure 2), e.g., drought, oxidative, osmotic, and cold stress (Cao et al., 2005; Fornara et al., 2015), as will be outlined further below.
The Photoperiod Influences Responses to Abiotic and Biotic Stresses
Photoperiod and Freezing Tolerance
One of the best-known stress tolerances depending on the photoperiod is freezing tolerance (Figure 3A). The shortening of day length sensed by plants in autumn anticipates the effect of colder temperatures in winter and causes an increased freezing tolerance (Lee and Thomashow, 2012). For example, red-osier dogwood (Cornus sericea) responds to a shortening of the photoperiod by a decrease of the stem water content, which results in an increased freezing tolerance (Karlson et al., 2003). In hybrid aspen, the phyA-mediated apical bud formation under short days is the main switch turning metabolism from vegetative growth to dormancy and inducing freezing tolerance (Welling et al., 2002).
Figure 3. Molecular mechanisms involved in photoperiod-dependent responses to cold, drought, and osmotic stress. (A) During cold stress (indicated by the ice crystal), CBF gene expression is upregulated and induces the expression of COR genes resulting in cold acclimation. Under short-day (SD) conditions, CBF genes are strongly induced causing cold acclimation. Under warmer long-day (LD) conditions, PIF4 and PIF7, which are under the control of phyB, are higher expressed resulting in an inhibition of CBF gene expression. As days shorten, e.g., during autumn, this repression falls away resulting in cold acclimation. GI is also induced by colder temperatures and blocks the CBF genes, whereas CDF1 promotes the expression of CBF. GI also promotes freezing tolerance in a CBF-independent manner (dashed line). In addition, HOS1, another photoperiod-dependent flowering-inducing component inhibits CBF gene expression thereby blocking cold acclimation. Figure adapted from Roeber et al. (2021). (B) Drought stress results in biosynthesis of abscisic acid (ABA) leading to ABA-dependent gene regulation causing drought escape (left) and drought tolerance (right). The increased ABA levels promote earlier flowering (drought escape, left part) under LD but not under SD conditions. Under LD conditions, GI is activated and activates the expression of florigen genes (TSF and FT via CO) triggering the activation of SOC1 and inducing flowering. SOC1 in turn contributes to TSF upregulation boosting SOC1 activity. ABA also induces the expression and activity of ABF3 and ABF4. ABF3/4, together with their interacting partner, the NF-Y complex, binds to the SOC1 promoter and promotes its expression to accelerate flowering during drought escape. Under SD conditions, delay of flowering occurs during drought stress due to enhanced activity of repressors like FLC and SVP on SOC1 transcription. Under these SD conditions, GI is not activated (pale circle). Adapted from Riboni et al. (2013, 2016) and Hwang et al. (2019). In the drought tolerance signaling pathway (right part), GI forms a complex with EEL (ENHANCED EM LEVEL) thereby upregulating the diurnal expression of NCED3 (NINE-CIS-EPOXYCAROTENOID DIOXYGENASE3) encoding a rate-limiting enzyme in ABA synthesis. Furthermore, interaction between GI and miR172 results in a reduction of WRKY44 expression promoting sugar signaling and drought tolerance. Besides GI, also NUCLEAR FACTOR-Y (NF-Y) promotes drought tolerance. (C) In the absence of salt stress (-NaCl) GI represses SOS2 thus blocking the SOS pathway. Upon salt stress (+ NaCl), the proteasomal degradation of GI is promoted, releasing SOS2. Free SOS2 interacts with SOS3 to form an active SOS2–SOS3 protein kinase complex that translocates to the plasma membrane causing the phosphorylation and activation of SOS1 resulting in salt stress tolerance. Adapted from Kim et al. (2013). For more information concerning the different pathways, please refer to section “Photoperiod and Freezing Tolerance” for cold stress, section “Photoperiod and Drought Stress” for drought stress, and section “Photoperiod and Osmotic Stress” for osmotic stress. Yellow background marks pathways taking place in LD conditions, a gray background indicates pathways during SD conditions. Gray lines mark the direct influence of specific photoperiod sensing components on stress responses. LD, long day; SD, short day; phyB, phytochrome B; PIF, PHYTOCHROME INTERACTING FACTOR; CBF, C-repeat/dehydration-responsive element-binding factor; COR, COLD-REGULATED; GI, GIGANTEA; HOS1, HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1; CDF1, CYCLING DOF FACTOR1; ABA, abscisic acid; ABF, abscisic acid binding factor; CO, CONSTANS; TSF, TWIN SISTER OF FT; SOC1, SUPPRESSOR OF OVEREXPRESSOR OF CONSTANS; FLC, FLOWERING LOCUS C; SVP, SHORT VEGETATIVE PHASE; SOS, SALT OVERLAY SENSITIVE.
Increased freezing tolerance caused by shortening of the photoperiod also occurs in Arabidopsis thaliana. Geographical distant accessions of Arabidopsis exhibit differences in freezing tolerance, which can be related to the photoperiod conditions they are geographically exposed to (Alonso-Blanco et al., 2005). The C-repeat/dehydration-responsive element-binding factor (CBF/DREB) signaling cascade is the central molecular mechanism mediating these differences in response to day length. Cold temperatures stimulate the CBF genes resulting in the induction of COLD-REGULATED (COR) genes leading to freezing tolerance (Thomashow, 2010; Pareek et al., 2017). Under long days, the CBF regulon is repressed by phyB, PHYTOCHROME INTERACTING FACTOR4 (PIF4), and PIF7, which causes less freezing tolerance. Shortening of the days during autumn relieves this repression causing an increased expression of the CBF genes, thereby preparing plants for upcoming colder temperatures (Figure 3A; Lee and Thomashow, 2012).
Among the components involved in photoperiodic flowering, the GI-CDF module regulates also freezing tolerance in Arabidopsis (Fornara et al., 2015). GI expression is induced by cold (Fowler and Thomashow, 2002; Cao et al., 2005) and many cold-regulated genes in Arabidopsis are co-regulated by GI and CDFs (Figure 3A). In gi-100 mutants, mRNA of COR genes was present at higher levels than in wild type correlating with enhanced expression of CDF1, CDF2, CDF3 and CDF5 and increased freezing and oxidative stress tolerance. Consequently, this increase in COR gene expression was suppressed in gi-100 cdf1235 mutants (Fornara et al., 2015). In contrast, Cao et al. (2005) found that gi-3 mutants are hypersensitive to freezing. As no differences were found in the transcript levels of CBF genes upon cold stress, it was concluded that GI acts in a CBF-independent manner to promote freezing tolerance by altering the carbohydrate metabolism. The exact mechanisms are still unclear (Cao et al., 2005, 2006). Such divergences may be due to the use of gi mutant alleles in different ecotypes and/or different assay conditions (Fornara et al., 2015). However, gi loss-of-function mutants of Brassica rapa plants show increased freezing tolerance suggesting that the role of GI in resistance to freezing stress is conserved between species (Xie et al., 2015).
Another component involved in regulating both photoperiod flowering and freezing tolerance is HOS1 (HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1) (Figure 3A). HOS1 encodes a RING finger-containing E3 ubiquitin ligase controlling the abundance of CO thereby ensuring that the CO-dependent activation of FT occurs only when the light period reaches a certain length (Lazaro et al., 2012). HOS1 negatively regulates cold acclimation by mediating the ubiquitination and proteasomal degradation of ICE1 (INDUCER OF CBF EXPRESSION1) and thus negatively regulates the CBF regulon (Ishitani et al., 1998; Dong et al., 2006; Lee and Thomashow, 2012).
Photoperiod and Drought Stress
Drought has detrimental effects on plants limiting their performance and productivity. Upon the perception of drought signals, the endogenous abscisic acid (ABA) level increases resulting in closure of the stomata in order to decrease water loss via transpiration (Outlaw, 2003).
The drought escape is an adaptive strategy of plants to accelerate reproductive development (i.e., flowering) under drought stress (Figure 3B). This allows plants to finish their life cycle before severe stress results in lethality (McKay et al., 2003). Drought escape only occurs under inductive long-day conditions involving the photoperiodic response gene GI and the florigen genes FLOWERING LOCUS T (FT) and TWIN SISTER OF FT (TSF) (Riboni et al., 2013). Drought stress releases the transcriptional repression at the FT/TSF promotors in an ABA- and photoperiod (via GI)-dependent manner thereby promoting transcriptional upregulation of the florigen genes (Riboni et al., 2013). The ABA-dependent activation of FT, but not of TSF, requires CO (Riboni et al., 2016). Increased florigen levels trigger the activation of the floral integrator SUPPRESSOR OF OVEREXPRESSOR OF CONSTANS (SOC1) thereby initiating flowering. SOC1 activation contributes to TSF upregulation thus further increasing the florigen levels (Riboni et al., 2013). Under short-day conditions, ABA delays flowering under drought stress due to the repressive action of SHORT VEGETATIVE PHASE (SVP)/FLOWERING LOCUS C (FLC) on SOC1 (Riboni et al., 2016). Also the NUCLEAR FACTOR Y (NF-Y) subunit c, belonging to a family of transcription factors known to be involved in photoperiod-dependent flowering (Kumimoto et al., 2008, 2010), is implicated in drought escape. The ABA-response element (ABRE)-binding factors (ABFs) interact with NF-Y subunit c-3/4/9, thereby inducing SOC1 to promote flowering (Hwang et al., 2019). Besides Arabidopsis, also wheat and barley have a drought escape strategy (McMaster and Wilhelm, 2003; Gol et al., 2021), just like Avena barbata (Sherrard and Maherali, 2006) and Brassica rapus (Franks, 2011). Other species, such as rice, delay flowering upon drought stress to resume its life cycle when the stress is over (Galbiati et al., 2016). Also here, primary integrators of day length provide a molecular connection between stress and the photoperiodic flowering pathway. Taken together, drought escape is a photoperiod-depend developmental response as it is the direct consequence of the perception of the long-day photoperiod during drought stress.
Besides their role during drought escape, the photoperiod sensing components GI and NF-Y are known to additionally influence drought tolerance without any direct link to the perception of the photoperiod (Figure 3B). The synthesis and signaling of ABA are at least partially under photoperiodic control (Zeevaart, 1971). A recent study (Baek et al., 2020) revealed that GI forms a complex with the bZIP transcription factor ENHANCED EM LEVEL (EEL) involved in ABA signaling responses to regulate the expression of NINE-CIS-EPOXYCAROTENOID DIOXYGENASE3 (NCED3). NCED3 encodes a rate-limiting enzyme in ABA synthesis (Iuchi et al., 2001). The GI-EEL complex positively regulates the diurnal ABA synthesis by binding to the ABA-responsive element motif in the NCED3 gene promotor resulting in increased ABA synthesis and improved drought tolerance (Baek et al., 2020). The abundance of NCED3 transcript and ABA content decreased in gi-1 and eel mutants under dehydration, which correlates with their dehydration-sensitive phenotype (Baek et al., 2020). These results indicate that GI and EEL together enhance the plant tolerance to drought by regulating ABA homeostasis.
Another study described a role for GI during the drought stress response (Figure 3B; Han et al., 2013a). Upon drought stress, both level and function of mature miR172 are upregulated, with miRNA172e showing the strongest response to drought stress (Han et al., 2013a). Under long days and drought conditions, GI promotes the processing of pre-miRNA172 resulting in a suppression of WRKY44, which leads to drought tolerance. The exact underlying mechanism is not fully understood but might relate to sugar metabolism (Han et al., 2013a).
NF-Y transcription factors (Figure 3B) have been shown to improve drought tolerance in Arabidopsis (Li et al., 2013; Ni et al., 2013), maize (Nelson et al., 2007; Su et al., 2018), poplar (Han et al., 2013b), rice (Chen et al., 2015), and citrus (Pereira et al., 2018). In Arabidopsis, overexpression of NF-YA5 improved drought tolerance and micro-array analysis revealed that oxidative stress-responsive genes are strongly upregulated upon drought stress (Li et al., 2008). Transgenic Arabidopsis plants overexpressing the soybean NF-YA3 gene exhibited an increased expression of ABA biosynthesis, signaling, and stress-responsive genes (Ni et al., 2013). While this study suggested an ABA-dependent signaling resulting in improved drought resistance, overexpression of NF-YB1 in Arabidopsis also enhanced plant drought resistance independent of ABA signaling (Nelson et al., 2007). Besides their role in drought tolerance, overexpression of NF-Y transcription factors genes also improves freezing tolerance (Shi and Chan, 2014) and salt stress resistance (Li et al., 2008).
Photoperiod and Osmotic Stress
Osmotic stress leads to desiccation, due to the high osmotic potential of saline soils, and inhibits plant growth and development (Munns, 2002; Mahajan and Tuteja, 2005). To cope with salinity or osmotic stress, plants have developed adaptation strategies, such as decreasing the water loss by stomata closure, decreasing their growth, and activating antioxidant systems (Munns and Tester, 2008). The salt overly sensitive (SOS) pathway forms the first line of defense to salt stress in Arabidopsis plants (Ji et al., 2013). The SOS pathway, which depends on SOS1, SOS2, and SOS3, has been shown to regulate cellular signaling during salt stress to achieve ion homeostasis. SOS1 encodes a Na+/H+-antiporter located at the plant cell plasma membrane, which is responsible for the efflux of Na+ from the cytoplasm to the apoplast. SOS1 is activated by the calcium-regulated SOS2-SOS3 protein kinase complex (Shi et al., 2000; Qiu et al., 2002).
GI has been shown to be a major component of the salt stress adaptation pathway (Figure 3C; Kim et al., 2013). gi mutants are salt stress tolerant, while GI overexpression lines are extremely salt-sensitive. The underlying mechanism was revealed by Kim et al. (2013). Under non-stress conditions, GI prevents SOS2 from activating SOS1, thereby retaining the SOS system in a resting state. Upon salt stress, GI is degraded releasing SOS2 for interaction with SOS3, which causes in turn the activation of SOS1 to re-establish ion homeostasis (Kim et al., 2013; Park et al., 2016). No direct effect of the photoperiod on plant performance under salt stress is known. The involvement of the photoperiod-sensitive GI in the SOS pathway, however, suggests that the photoperiod might have a strong impact on salt stress tolerance.
Photoperiod and Biotic Stress Responses
Increasing evidence indicates that the length of the light period is also important for the response to diverse biotic stresses, including the responses to viruses, bacteria, and fungi (Table 1). The first observations showing that the photoperiod influences the response to pathogen infection were from Cecchini et al. (2002). They found that short-day-grown Arabidopsis (Ler) plants developed stronger disease symptoms than long-day-grown plants after infection with cauliflower mosaic virus (CaMV Cabb B-JI), although the virus replication was even higher under long-day conditions. Later on, Griebel and Zeier (2008) demonstrated that for Arabidopsis plants grown in different but constant light-dark cycles, the early disease resistance of Arabidopsis plants inoculated with Pseudomonas syringae pv. maculicola (Psm) ES4326 harboring the avrRpm1 avirulence gene is positively correlated with the length of the light period, underpinning the importance of the photoperiod. They also showed that the concentration of salicylic acid (SA) accumulated in Psm avrRpm1-infected Arabidopsis leaves, the early expression of the SA-regulated defense gene PATHOGENESIS-RELATED GENE1 (PR1), and the magnitude of the hypersensitive response-induced lesion formation are influenced by the duration of the light period. Long-day-entrained Arabidopsis plants exposed to constant light were less susceptible to infections with virulent Hyaloperonospora parasitica isolate Noco2 (Evrard et al., 2009) or P. syringae pv. tomato DC3000 (Cortleven et al., 2021). Transferring Arabidopsis from a short to a long photoperiod enhanced the resistance to the necrotrophic fungus Botrytis cinerea (Cagnola et al., 2018) and the hemibiotrophic fungus Pyricularia oryzae (syn. Magnaporthe oryzae) (Shimizu et al., 2021). In the latter case, the outcome of early plant–pathogen interactions was influenced by the length of the photoperiod following inoculation. The plant resistance to fungus penetration was enhanced, if a light period followed evening inoculations instead of the normal dark period (Shimizu et al., 2021).
The length of the light period influences plant responses to biotic stresses on the transcriptional level. Evrard et al. (2009) demonstrated that in Arabidopsis plants grown under 14 h day/10 h night cycles or under the same conditions but followed by 3 days of darkness, the transcriptional activity mediated by the hexameric promoter motif FORCA is suppressed by defense-related stimuli. In contrast, in constant light, the FORCA-mediated gene expression is enhanced resulting in increased defense. More generally, Baerenfaller et al. (2015) showed that the abundance of transcripts for biotic stress responses increased in Arabidopsis grown under long photoperiods compared to plants cultivated under short-day conditions. Similarly, Cagnola et al. (2018) revealed by transcriptome analysis of A. thaliana transferred from short- to long-day conditions that long photoperiods enhance the jasmonic acid (JA)-related plant defense responses.
An improved resistance to biotic stimuli under long photoperiods is also observed in other plant species than Arabidopsis. Kenyon et al. (2002) reported that long photoperiods enhanced the resistance of Rhododendron cv. Elizabeth cut leaves, as fewer hyphae of the fungus Erysiphe sp. were produced than under short photoperiods. In tomato plants (Solanum lycopersicum cv. Ailsa Craig), nightly red light treatment (replacing the normal dark period and thereby extending the duration of the total light period) enhanced the plant resistance against P. syringae pv. tomato DC3000 infection. The increased resistance was correlated with the accumulation of SA, increased abundance of defense-related transcripts and alleviated pathogen-induced cell death (Yang et al., 2015). The resistance of maize Hm1A seedlings (containing a partial loss-of-function mutation in the Hm1 gene, encoding HC-toxin reductase inactivating the HC-toxin produced by Cochliobolus carbonum, which causes leaf spot in maize) inoculated with C. carbonum race 1 was enhanced in plants grown after infection in extended light periods and might correlate to the energy status of the plant (Marla et al., 2018). Similarly, strawberry (Fragaria ananassa cv. Elsanta) plants inoculated with B. cinerea developed stronger disease symptoms, if plants were transferred to darkness after infection compared to plants kept under their normal light conditions (Meng et al., 2020). Macioszek et al. (2021) observed that growth under short-day conditions results in increased necrosis formation in Brassica juncea plants infected with the necrotrophic fungus Alternaria brassicicola.
Overall, these publications clearly highlight the importance of the duration of the photoperiod in plant responses to diverse biotic stresses. Generally, a longer photoperiod causes increased biotic stress resistance. Whether this is causally linked to an improved energy state of the plants under longer photoperiods has not been discussed in the above-mentioned publications, but would be an interesting direction for future research. The role of another parameter influenced by the length of the photoperiod, the plant redox state, will be discussed in the following chapter.
Photoperiod and the Plant Redox State
A possible reason for the impact of the photoperiod on responses to diverse stresses is its regulatory influence on the plant redox state (for review see Shim and Imaizumi, 2015). The plant redox state and regulation of redox reactions are connected with levels of reactive oxygen species (ROS). ROS have previously been considered only as toxic by-products of aerobic metabolism, but recent research highlights their importance as plant signaling molecules (for review see Mittler, 2017). In addition, ROS cause post-translational modifications of cysteine residues in redox-sensitive proteins and thus interfere with the redox regulatory network of the cell allowing fast responses of plants to environmental cues (Cejudo et al., 2021).
ROS include several oxygen-containing molecules, such as singlet oxygen (1O2), superoxide (O2–), hydroxyl radicals (OH–), and hydrogen peroxide (H2O2) (Shim and Imaizumi, 2015; Mittler, 2017). Of these, especially H2O2 levels are regulated by the photoperiod. In the algae Chlamydomonas reinhardtii, elevated H2O2 levels in the presence of light result from slower H2O2 degradation due to light-dependent inactivation of catalases (Shao et al., 2008). Similar observations were made in rye (Secale cereale) leaves (Hertwig et al., 1992).
Experiments with Arabidopsis catalase2 (cat2) mutants exposed to different day lengths were particularly informative about the influence of the photoperiod on oxidative stress responses. The photoperiod, in which the cat2 mutants are grown, is decisive for the oxidative stress response and regulates H2O2-induced gene expression as well as the severity of the cell death phenotype (Queval et al., 2007; Chaouch et al., 2010; Shim and Imaizumi, 2015). While short-day-grown cat2 mutants do not display any lesions, lesion formation is visible in long day-grown cat2 mutants pointing to elevated H2O2 levels. Further analysis revealed that increased peroxisomal H2O2 in cat2 triggers pathogen defense responses and enhances the plants’ resistance in a photoperiod-dependent manner. Also lesion simulating disease1 (lsd1) mutants formed lesions only in long photoperiods (Mateo et al., 2004; Shim and Imaizumi, 2015). LSD1 is a catalase-interacting protein, regulating catalase activity, as a consequence, catalase activity is decreased in lsd1 mutants. LSD1 and CATALASE genes interact genetically and their encoded proteins are part of a protein complex, which plays an important role in regulating programmed cell death (PCD) (Li et al., 2013).
Similar as observed for catalase, also other enzymes involved in ROS detoxification including ascorbate peroxidase and NAD-malate dehydrogenase showed in Arabidopsis a higher activity under long photoperiods (Becker et al., 2006). GI was shown to be a negative regulator of the expression of genes encoding enzymes detoxifying ROS. Indeed, the increased tolerance of gi-3 mutants to oxidative stress is caused by the constitutive activation of SUPEROXIDE DISMUTASE (SOD) and ASCORBATE PEROXIDASE (APX) genes (Cao et al., 2006).
Together these publications support a photoperiod-dependent regulation of the plant redox state. Photoperiod sensing is linked to redox regulation, allowing efficient light usage and redox balancing in short days and preventing oxidative damage in long days (Becker et al., 2006). Especially catalases but also SOD and APX seem to be important factors exerting photoperiod information on redox regulation (Shim and Imaizumi, 2015).
Changes in the Photoperiod Cause Stress
Sudden changes in the photoperiod, in particular its prolongation, cause photoperiod stress in short-day-adapted A. thaliana plants (Nitschke et al., 2016, 2017; Figure 4). The photoperiod stress response, which was originally observed after a prolongation of the light period by 24 h, is characterized by a typical course of events: During the night following an extended light period, the expression of stress marker genes, such as ZINC FINGER of ARABIDOPSIS THALIANA12 (ZAT12) and BON ASSOCIATED PROTEIN1 (BAP1), is induced, the concentration of the stress hormones JA and SA increase and oxidative stress occurs. The nightly increase in oxidative stress coincides with a strong decrease in the ascorbic acid (ASC) redox state and the formation of peroxides. The peroxide formation is associated with an increase of PEROXIDASE (PRX) gene expression as well as enhanced PRX and decreased catalase activities (Abuelsoud et al., 2020). During the next day, the photosystem II maximum quantum efficiency decreases, and eventually PCD ensues in the leaves (Nitschke et al., 2016, 2017).
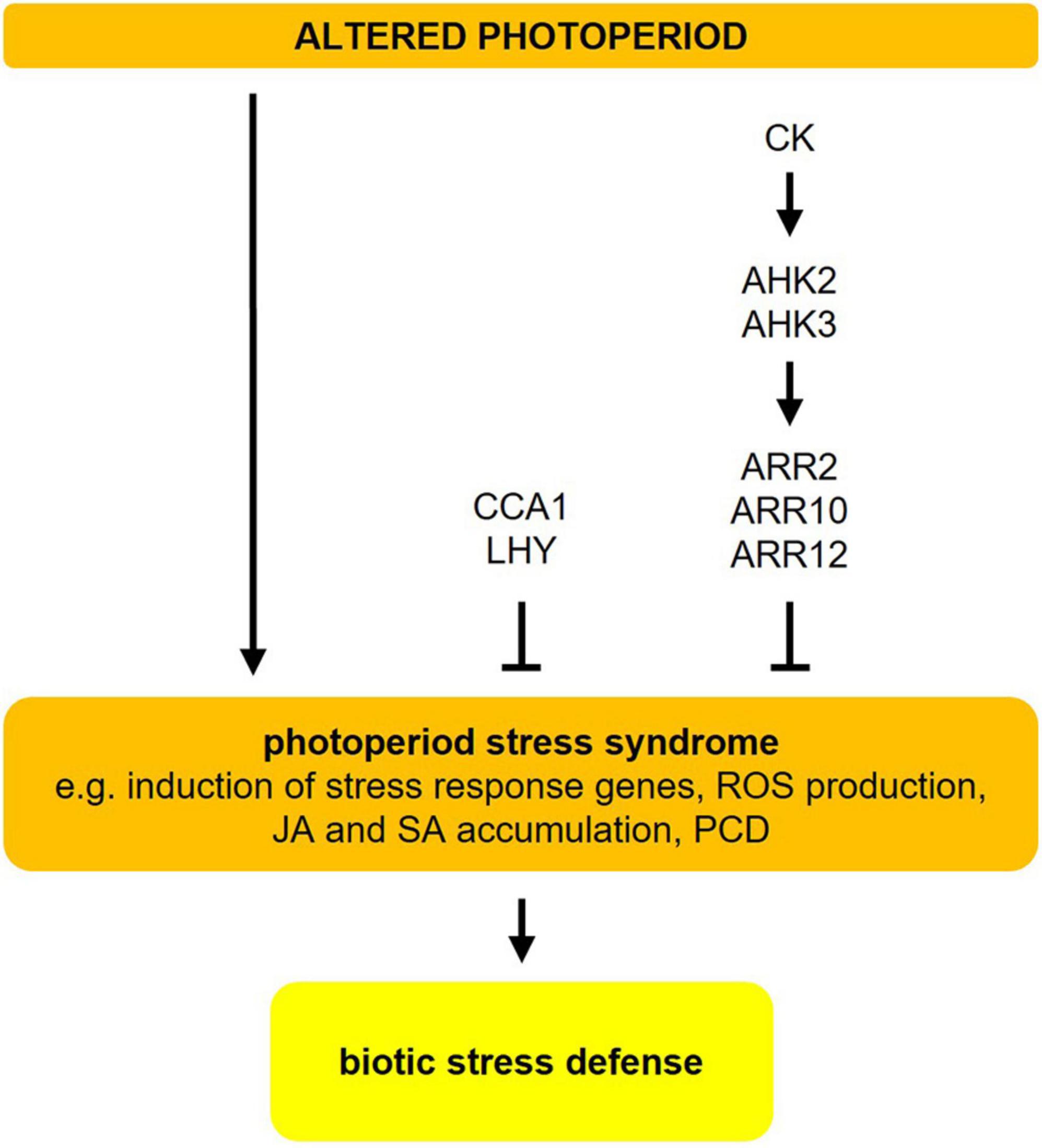
Figure 4. Photoperiod stress in Arabidopsis. Changes in the photoperiod, i.e., prolongation of the light period, result in a photoperiod stress syndrome, which is characterized by induction of stress response genes, ROS production, accumulation of jasmonic acid (JA) and salicylic acid (SA), and eventually programmed cell death (PCD). Both, cytokinin (CK) (mostly trans-zeatin) and CCA1/LHY are negative regulators of photoperiod stress. Photoperiod stress elicits a transcriptional response that resembles the response to ozone stress and pathogen infection. The resistance to an infection with P. syringae pv. tomato DC3000 is increased after a preceding photoperiod stress event. For more detailed information about the different pathways, please refer to sections “Changes in the Photoperiod Cause Stress” and “Photoperiod Stress Elicits a Similar Response as Pathogen Infection.” The figure has been adapted from Roeber et al. (2021).
Photoperiod stress was first noted in cytokinin (CK)-deficient Arabidopsis plants, which show a particularly strong stress response. Among the CKs, especially trans-zeatin has a protective function acting through the ARABIDOPSIS HISTIDINE KINASE3 (AHK3) receptor and the transcriptional regulators ARABIDOPSIS RESPONSE REGULATOR2 (ARR2), ARR10, and ARR12 (Figure 4; Frank et al., 2020). Certain clock mutants (e.g., cca1 lhy, elf3) also show a stronger molecular and phenotypical response to sudden prolongations of the photoperiod. Both photoperiod stress-sensitive clock mutants and CK-deficient plants have a lower expression or impaired function of CCA1 and LHY, two key regulators of the circadian clock. This indicates that a functional clock is required to cope with photoperiod stress (Figure 4; Nitschke et al., 2016, 2017).
In short-day-entrained Arabidopsis plants, a prolongation of the light period by only 4 h is sufficient to induce the production of ROS and the expression of stress marker genes during the following night. Longer prolongations of the light phase induce a gradually stronger stress response, which indicates that light duration has an impact on the strength of the photoperiod stress response (Abuelsoud et al., 2020). Shorter prolongations of the light period, which cause lower stress levels, are perceived as not harmful and may present a beneficial stress (eustress), while higher stress levels (by longer prolongations) induce a true stress (distress) (Krasensky-Wrzaczek and Kangasjarvi, 2018).
Photoperiod Stress Elicits a Similar Response as Pathogen Infection
RNA-seq analysis of 5-weeks-old short-day grown Arabidopsis plants exposed to a 24 h-prolongation of the light period revealed that photoperiod stress causes massive time-dependent transcriptomic changes during the night following the prolonged light period (Cortleven et al., 2021). Among the photoperiod stress-responsive genes are numerous genes related to ROS. The photoperiod stress transcript profile resembles that caused by ozone stress and pathogen attacks, which commonly elicit an apoplastic oxidative burst. Moreover, both SA and camalexin levels increased and transcript levels of genes involved in SA biosynthesis, such as ISOCHORISMATE SYNTHASE1 (ICS1), and genes involved in SAR, such as PATHOGENESIS RELATED1 (PR1) were induced by photoperiod stress (Cortleven et al., 2021).
Interestingly, photoperiod stress pre-treated wild-type plants showed less P. syringae pv. tomato DC3000 colony-forming units after infection in comparison to non-photoperiod stress-treated plants. This indicates that not only similar molecular pathways are activated in response to photoperiod stress and pathogen attack, but that photoperiod stress pre-treatment leads to improved pathogen immunity in Arabidopsis plants without an actual pathogen attack (Cortleven et al., 2021). Other reports support this conclusion. In tomato, it was shown that nightly red light treatment improved the resistance against P. syringae pv. tomato DC3000 (Yang et al., 2015). The improved resistance was associated with increased SA levels and the induction of defense-related genes, which is typical of a photoperiod stress response (Cortleven et al., 2021). The transfer of short-day-grown Arabidopsis plants to long-day conditions, causing essentially photoperiod stress by 8 h light prolongation, resulted in an improved resistance against B. cinerea (Cagnola et al., 2018). Prolonged light exposure due to transfer from short to long days resulted in lower nuclear abundance of CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1), thereby leading to stabilization of DELLA proteins and increased expression of the JA-signaling gene JASMONATE INSENSITIVE1 (MYC2) (Cagnola et al., 2018). Exposure of maize to a prolonged light period increased resistance against C. carbonum race 1 (Marla et al., 2018). These studies indicate that the response to photoperiod stress is conserved among different plant species and that it has similar effects on pathogen resistance. Future research needs to identify the specific mechanisms conferring the improved pathogen resistance in photoperiod stress-treated plants.
Conclusion and Future Research Perspectives
The photoperiod provides plants with information to synchronize their developmental program with the prevailing season. It is used to match the optimal conditions for offspring production and to alleviate the threats of seasonal stresses occurring at the same time every year. In this review, we have summarized the complex photoperiod sensing mechanisms (Figures 1, 2) and especially focussed on the role of the photoperiod in plant responses to cold, drought, osmotic and biotic stresses (Figure 3 and Table 1). While the molecular mechanisms of photoperiod-dependent regulation of cold, drought, and osmotic stress are at least partly elucidated, the impact of the photoperiod on biotic stress responses remains descriptive.
Recent studies (Nitschke et al., 2016, 2017) revealed that a sudden prolongation of the photoperiod causes a new form of abiotic stress, namely photoperiod stress, resulting in a nightly ROS accumulation in the apoplast and a stress response resembling pathogen infection (Abuelsoud et al., 2020; Cortleven et al., 2021). Photoperiod stress signals might have an adaptive value, for example by acting as a priming agent, which improves the plants’ performance to future stresses. The ecological relevance of photoperiod stress needs to be unveiled. It has been hypothesized that changes in intensity and ratios of wavelength during dawn and dusk that depend on weather conditions may modulate the output of the photoperiod sensing system (Abuelsoud et al., 2020), but the experimental proof is missing.
In view of the impact of the photoperiod on plant responses to pathogens, it may be envisaged that controlled changes of the photoperiod have a perspective for application in greenhouse farming. In these controlled environmental conditions the duration, intensity and wavelength composition of the illumination can be precisely regulated by light-emitting diodes (LEDs), which holds the potential to improve crop yield and quality (Jones, 2018; Lazzarin et al., 2021). LEDs can also be used for pest management. Several studies showed the influence of the light environment in greenhouses (including the length of the photoperiod and light quality) (for review, see Gallé et al., 2021; Lazzarin et al., 2021), on plant resilience against pathogens such as B. cinerea in strawberries (Meng et al., 2020) or tomatoes (Yang et al., 2015). Understanding how supplemental light through LEDs acts on plant growth and defense may lead to novel sustainable horticultural methods for pest management.
Author Contributions
VR and AC wrote the draft of the manuscript. VR, AC, and TS revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This project was funded by grants of the Deutsche Forschungsgemeinschaft to TS (Sfb 973 and Schm 814/29-1). Open access funding was provided by the Freie Universität Berlin.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Abuelsoud, W., Cortleven, A., and Schmülling, T. (2020). Photoperiod stress induces an oxidative burst-like response and is associated with increased apoplastic peroxidase and decreased catalase activities. J. Plant Physiol. 253:153252. doi: 10.1016/j.jplph.2020.153252
Adole, T., Dash, J., Rodriguez-Galiano, V., and Atkinson, P. M. (2019). Photoperiod controls vegetation phenology across Africa. Commun. Biol. 2:391. doi: 10.1038/s42003-019-0636-7
Alabadi, D., Oyama, T., Yanovsky, M. J., Harmon, F. G., Mas, P., and Kay, S. A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293, 880–883. doi: 10.1126/science.1061320
Alonso-Blanco, C., Gomez-Mena, C., Llorente, F., Koornneef, M., Salinas, J., and Martinez-Zapater, J. M. (2005). Genetic and molecular analyses of natural variation indicate CBF2 as a candidate gene for underlying a freezing tolerance quantitative trait locus in Arabidopsis. Plant Physiol. 139, 1304–1312. doi: 10.1104/pp.105.068510
Anwer, M. U., Davis, A., Davis, S. J., and Quint, M. (2020). Photoperiod sensing of the circadian clock is controlled by EARLY FLOWERING 3 and GIGANTEA. Plant J. 101, 1397–1410. doi: 10.1111/tpj.14604
Aukerman, M. J., and Sakai, H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15, 2730–2741. doi: 10.1105/tpc.016238
Baek, D., Kim, W. Y., Cha, J. Y., Park, H. J., Shin, G., Park, J., et al. (2020). The GIGANTEA-ENHANCED EM LEVEL complex enhances drought tolerance via regulation of abscisic acid synthesis. Plant Physiol. 184, 443–458. doi: 10.1104/pp.20.00779
Baerenfaller, K., Massonnet, C., Hennig, L., Russenberger, D., Sulpice, R., Walsh, S., et al. (2015). A long photoperiod relaxes energy management in Arabidopsis leaf six. Curr. Plant Biol. 2, 34–45. doi: 10.1016/j.cpb.2015.07.001
Becker, B., Holtgrefe, S., Jung, S., Wunrau, C., Kandlbinder, A., Baier, M., et al. (2006). Influence of the photoperiod on redox regulation and stress responses in Arabidopsis thaliana L. (Heynh.) plants under long- and short-day conditions. Planta 224, 380–393. doi: 10.1007/s00425-006-0222-3
Cagnola, J. I., Cerdan, P. D., Pacin, M., Andrade, A., Rodriguez, V., Zurbriggen, M. D., et al. (2018). Long-day photoperiod enhances jasmonic acid-related plant defense. Plant Physiol. 178, 163–173. doi: 10.1104/pp.18.00443
Cao, S., Jiang, S., and Zhang, R. (2006). The role of GIGANTEA gene in mediating the oxidative stress response in Arabidopsis. Plant Growth Regul. 48, 261–270. doi: 10.1007/s10725-006-0012-8
Cao, S., Ye, M., and Jiang, S. (2005). Involvement of GIGANTEA gene in the regulation of the cold stress response in Arabidopsis. Plant Cell Rep. 24, 683–690. doi: 10.1007/s00299-005-0061-x
Carré, I. A. (2001). Day-length perception and the photoperiodic regulation of flowering in Arabidopsis. J. Biol. Rhythms 16, 415–423. doi: 10.1177/074873001129002006
Casal, J. J., Fankhauser, C., Coupland, G., and Blazquez, M. A. (2004). Signalling for developmental plasticity. Trends Plant Sci. 9, 309–314. doi: 10.1016/j.tplants.2004.04.007
Cecchini, E., Geri, C., Love, A. J., Coupland, G., Covey, S. N., and Milner, J. J. (2002). Mutations that delay flowering in Arabidopsis de-couple symptom response from cauliflower mosaic virus accumulation during infection. Mol. Plant Pathol. 3, 81–90. doi: 10.1046/j.1464-6722.2001.00097.x
Cejudo, F. J., Sandalio, L. M., and Van Breusegem, F. (2021). Understanding plant responses to stress conditions: redox-based strategies. J. Exp. Bot. 72, 5785–5788. doi: 10.1093/jxb/erab324
Cha, J. Y., Kim, J., Kim, T. S., Zeng, Q., Wang, L., Lee, S. Y., et al. (2017). GIGANTEA is a co-chaperone which facilitates maturation of ZEITLUPE in the Arabidopsis circadian clock. Nat. Commun. 8:3. doi: 10.1038/s41467-016-0014-9
Chaouch, S., Queval, G., Vanderauwera, S., Mhamdi, A., Vandorpe, M., Langlois-Meurinne, M., et al. (2010). Peroxisomal hydrogen peroxide is coupled to biotic defense responses by ISOCHORISMATE SYNTHASE1 in a daylength-related manner. Plant Physiol. 153, 1692–1705. doi: 10.1104/pp.110.153957
Chen, M., Zhao, Y., Zhuo, C., Lu, S., and Guo, Z. (2015). Overexpression of a NF-YC transcription factor from bermudagrass confers tolerance to drought and salinity in transgenic rice. Plant Biotechnol. J. 13, 482–491. doi: 10.1111/pbi.12270
Conrath, U., Beckers, G. J., Flors, V., Garcia-Agustin, P., Jakab, G., Mauch, F., et al. (2006). Priming: getting ready for battle. Mol. Plant. Microbe Interact. 19, 1062–1071. doi: 10.1094/MPMI-19-1062
Cortleven, A., Roeber, V. M., Frank, M., Bertels, J., Lortzing, V., Beemster, G., et al. (2021). The transcriptomic landscape of the photoperiodic stress response in Arabidopsis thaliana resembles the response to pathogen infection. bioRxiv [Preprint]. doi: 10.1101/2021.04.13.439491
de los Reyes, P., Romero-Losada, A. B., and Romero-Campero, F. J. (2020). ATTRACTOR, Arabidopsis thaliana Transcriptional Circadian Network v1.0. Zenodo. doi: 10.5381/zenodo.3780022
Dong, C. H., Agarwal, M., Zhang, Y., Xie, Q., and Zhu, J. K. (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. U.S.A. 103, 8281–8286. doi: 10.1073/pnas.0602874103
Evrard, A., Ndatimana, T., and Eulgem, T. (2009). FORCA, a promoter element that responds to crosstalk between defense and light signaling. BMC Plant Biol. 9:2. doi: 10.1186/1471-2229-9-2
Feng, P., Guo, H., Chi, W., Chai, X., Sun, X., Xu, X., et al. (2016). Chloroplast retrograde signal regulates flowering. Proc. Natl. Acad. Sci. U.S.A. 113, 10708–10713. doi: 10.1073/pnas.1521599113
Fornara, F., de Montaigu, A., Sanchez-Villarreal, A., Takahashi, Y., Ver Loren van Themaat, E., Huettel, B., et al. (2015). The GI-CDF module of Arabidopsis affects freezing tolerance and growth as well as flowering. Plant J. 81, 695–706. doi: 10.1111/tpj.12759
Fowler, S., Lee, K., Onouchi, H., Samach, A., Richardson, K., Morris, B., et al. (1999). GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18, 4679–4688. doi: 10.1093/emboj/18.17.4679
Fowler, S., and Thomashow, M. F. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14, 1675–1690. doi: 10.1105/tpc.003483
Frank, M., Cortleven, A., Novak, O., and Schmülling, T. (2020). Root-derived trans-zeatin cytokinin protects Arabidopsis plants against photoperiod stress. Plant Cell Environ. 43, 2637–2649. doi: 10.1111/pce.13860
Franks, S. J. (2011). Plasticity and evolution in drought avoidance and escape in the annual plant Brassica rapa. New Phytol. 190, 249–257. doi: 10.1111/j.1469-8137.2010.03603.x
Galbiati, F., Chiozzotto, R., Locatelli, F., Spada, A., Genga, A., and Fornara, F. (2016). Hd3a, RFT1 and Ehd1 integrate photoperiodic and drought stress signals to delay the floral transition in rice. Plant Cell Environ. 39, 1982–1993. doi: 10.1111/pce.12760
Gallé, A., Czekus, Z., Toth, L., Galgoczy, L., and Poor, P. (2021). Pest and disease management by red light. Plant Cell Environ. 44, 3197–3210. doi: 10.1111/pce.14142
Gendron, J. M., Pruneda-Paz, J. L., Doherty, C. J., Gross, A. M., Kang, S. E., and Kay, S. A. (2012). Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. U.S.A. 109, 3167–3172. doi: 10.1073/pnas.1200355109
Gol, L., Haraldsson, E. B., and von Korff, M. (2021). Ppd-H1 integrates drought stress signals to control spike development and flowering time in barley. J. Exp. Bot. 72, 122–136. doi: 10.1093/jxb/eraa261
Griebel, T., and Zeier, J. (2008). Light regulation and daytime dependency of inducible plant defenses in Arabidopsis: phytochrome signaling controls systemic acquired resistance rather than local defense. Plant Physiol. 147, 790–801. doi: 10.1104/pp.108.119503
Grundy, J., Stoker, C., and Carré, I. A. (2015). Circadian regulation of abiotic stress tolerance in plants. Front. Plant Sci. 6:648. doi: 10.3389/fpls.2015.00648
Han, Y., Zhang, X., Wang, W., Wang, Y., and Ming, F. (2013a). The suppression of WRKY44 by GIGANTEA-miR172 pathway is involved in drought response of Arabidopsis thaliana. PLoS One 8:e73541. doi: 10.1371/journal.pone.0073541
Han, X., Tang, S., An, Y., Zheng, D. C., Xia, X. L., and Yin, W. L. (2013b). Overexpression of the poplar NF-YB7 transcription factor confers drought tolerance and improves water-use efficiency in Arabidopsis. J. Exp. Bot. 64, 4589–4601. doi: 10.1093/jxb/ert262
Hendel-Rahmanim, K., Masci, T., Vainstein, A., and Weiss, D. (2007). Diurnal regulation of scent emission in rose flowers. Planta 226, 1491–1499. doi: 10.1007/s00425-007-0582-3
Hertwig, B., Streb, P., and Feierabend, J. (1992). Light dependence of catalase synthesis and degradation in leaves and the influence of interfering stress conditions. Plant Physiol. 100, 1547–1553. doi: 10.1104/pp.100.3.1547
Hsu, P. Y., and Harmer, S. L. (2014). Global profiling of the circadian transcriptome using microarrays. Methods Mol. Biol. 1158, 45–56. doi: 10.1007/978-1-4939-0700-7_3
Hwang, K., Susila, H., Nasim, Z., Jung, J. Y., and Ahn, J. H. (2019). Arabidopsis ABF3 and ABF4 transcription factors act with the NF-YC complex to regulate SOC1 expression and mediate drought-accelerated flowering. Mol. Plant 2, 489–505. doi: 10.1016/j.molp.2019.01.002
Ishitani, M., Xiong, L., Lee, H., Stevenson, B., and Zhu, J. K. (1998). HOS1, a genetic locus involved in cold-responsive gene expression in Arabidopsis. Plant Cell 10, 1151–1161. doi: 10.1105/tpc.10.7.1151
Iuchi, S., Kobayashi, M., Taji, T., Naramoto, M., Seki, M., Kato, T., et al. (2001). Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 27, 325–333. doi: 10.1046/j.1365-313x.2001.01096.x
Jackson, S. D. (2009). Plant responses to photoperiod. New Phytol. 181, 517–531. doi: 10.1111/j.1469-8137.2008.02681.x
Ji, H., Pardo, J. M., Batelli, G., Van Oosten, M. J., Bressan, R. A., and Li, X. (2013). The Salt Overly Sensitive (SOS) pathway: established and emerging roles. Mol. Plant 6, 275–286. doi: 10.1093/mp/sst017
Jones, M. A. (2018). Using light to improve commercial value. Hortic. Res. 5:47. doi: 10.1038/s41438-018-0049-7
Jung, J. H., Seo, Y. H., Seo, P. J., Reyes, J. L., Yun, J., Chua, N. H., et al. (2007). The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19, 2736–2748. doi: 10.1105/tpc.107.054528
Karlson, D. T., Fujino, T., Kimura, S., Baba, K., Itoh, T., and Ashworth, E. N. (2003). Novel plasmodesmata association of dehydrin-like proteins in cold- acclimated Red-osier dogwood (Cornus sericea). Tree Physiol. 23, 759–767. doi: 10.1093/treephys/23.11.759
Kenyon, D. M., Dixon, G. R., and Helfer, S. (2002). Effects of relative humidity, light intensity and photoperiod on the colony development of Erysiphe sp. on Rhododendron. Plant Pathol. 51, 103–108. doi: 10.1046/j.0032-0862.2001.x
Kiba, T., Henriques, R., Sakakibara, H., and Chua, N. H. (2007). Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell 19, 2516–2530. doi: 10.1105/tpc.107.053033
Kiełbowicz-Matuk, A., and Czarnecka, J. (2014). “Interplays of plant circadian clock and abiotic stress response networks,” in Emerging Technologies and Management of Crop Stress Tolerance, eds P. Ahmad and S. Rasool (Cambridge, MA: Academic Press). doi: 10.1016/B978-0-12-800876-8.00020-5
Kim, W. Y., Ali, Z., Park, H. J., Park, S. J., Cha, J. Y., Perez-Hormaeche, J., et al. (2013). Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat. Commun. 4:1352. doi: 10.1038/ncomms2357
Kim, W. Y., Fujiwara, S., Suh, S. S., Kim, J., Kim, Y., Han, L., et al. (2007). ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449, 356–360. doi: 10.1038/nature06132
Krasensky-Wrzaczek, J., and Kangasjarvi, J. (2018). The role of reactive oxygen species in the integration of temperature and light signals. J. Exp. Bot. 69, 3347–3358. doi: 10.1093/jxb/ery074
Kumimoto, R. W., Adam, L., Hymus, G. J., Repetti, P. P., Reuber, T. L., Marion, C. M., et al. (2008). The nuclear factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta 228, 709–723. doi: 10.1007/s00425-008-0773-6
Kumimoto, R. W., Zhang, Y., Siefers, N., and Holt, B. F. III (2010). NF-YC3, NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. Plant J. 63, 379–391. doi: 10.1111/j.1365-313X.2010.04247.x
Lazaro, A., Valverde, F., Pineiro, M., and Jarillo, J. A. (2012). The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell 24, 982–999. doi: 10.1105/tpc.110.081885
Lazzarin, M., Meisenburg, M., Meijer, D., van Ieperen, W., Marcelis, L. F. M., Kappers, I. F., et al. (2021). LEDs make it resilient: effects on plant growth and defense. Trends Plant Sci. 26, 496–508. doi: 10.1016/j.tplants.2020.11.013
Lee, C. M., and Thomashow, M. F. (2012). Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 109, 15054–15059. doi: 10.1073/pnas.1211295109
Lepisto, A., and Rintamaki, E. (2012). Coordination of plastid and light signaling pathways upon development of Arabidopsis leaves under various photoperiods. Mol. Plant 5, 799–816. doi: 10.1093/mp/ssr106
Li, W. X., Oono, Y., Zhu, J., He, X. J., Wu, J. M., Iida, K., et al. (2008). The Arabidopsis NF-YA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20, 2238–2251. doi: 10.1105/tpc.108.059444
Li, Y., Chen, L., Mu, J., and Zuo, J. (2013). LESION SIMULATING DISEASE1 interacts with catalases to regulate hypersensitive cell death in Arabidopsis. Plant Physiol. 163, 1059–1070. doi: 10.1104/pp.113.225805
Macioszek, V. K., Sobczak, M., Skoczowski, A., Oliwa, J., Michlewska, S., Gapińska, M., et al. (2021). The effect of photoperiod on necrosis development, photosynthetic efficiency and ‘Green Islands’ formation in Brassica juncea infected with Alternaria brassicicola. Int. J. Mol. Sci. 22:8435. doi: 10.3390/ijms22168435
Mahajan, S., and Tuteja, N. (2005). Cold, salinity and drought stresses: an overview. Arch. Biochem. Biophys. 444, 139–158. doi: 10.1016/j.abb.2005.10.018
Marla, S. R., Chu, K., Chintamanani, S., Multani, D. S., Klempien, A., DeLeon, A., et al. (2018). Adult plant resistance in maize to northern leaf spot is a feature of partial loss-of-function alleles of Hm1. PLoS Pathog. 14:e1007356. doi: 10.1371/journal.ppat.1007356
Mas, P., Kim, W. Y., Somers, D. E., and Kay, S. A. (2003). Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426, 567–570. doi: 10.1038/nature02163
Mateo, A., Muhlenbock, P., Rusterucci, C., Chang, C. C., Miszalski, Z., Karpinska, B., et al. (2004). LESION SIMULATING DISEASE 1 is required for acclimation to conditions that promote excess excitation energy. Plant Physiol. 136, 2818–2830. doi: 10.1104/pp.104.043646
Mathieu, J., Yant, L. J., Murdter, F., Kuttner, F., and Schmid, M. (2009). Repression of flowering by the miR172 target SMZ. PLoS Biol. 7:e1000148. doi: 10.1371/journal.pbio.1000148
Matsoukas, I. G. (2014). Attainment of reproductive competence, phase transition, and quantification of juvenility in mutant genetic screens. Front. Plant Sci. 5:32. doi: 10.3389/fpls.2014.00032
McKay, J. K., Richards, J. H., and Mitchell-Olds, T. (2003). Genetics of drought adaptation in Arabidopsis thaliana: I. Pleiotropy contributes to genetic correlations among ecological traits. Mol. Ecol. 12, 1137–1151. doi: 10.1046/j.1365-294x.2003.01833.x
McMaster, G. S., and Wilhelm, W. W. (2003). Phenological responses of wheat and barley to water and temperature: improving simulation models. J. Agric. Sci. 141, 129–147. doi: 10.1017/S0021859603003460
Meng, L., Van Labeke, M. C., and Höfte, M. (2020). Timing of light quality affects susceptibility to Botrytis cinerea in strawberry leaves. J. Photochem. Photobiol. B 211:111988. doi: 10.1016/j.jphotobiol.2020.111988
Moraes, T. A., Mengin, V., Annunziata, M. G., Encke, B., Krohn, N., Hohne, M., et al. (2019). Response of the circadian clock and diel starch turnover to one day of low light or low CO2. Plant Physiol. 179, 1457–1478. doi: 10.1104/pp.18.01418
Munns, R. (2002). Comparative physiology of salt and water stress. Plant Cell Environ. 25, 239–250. doi: 10.1046/j.0016-8025.2001.00808.x
Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Nakamichi, N., Kiba, T., Kamioka, M., Suzuki, T., Yamashino, T., Higashiyama, T., et al. (2012). Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc. Natl. Acad. Sci. U.S.A. 109, 17123–17128. doi: 10.1073/pnas.1205156109
Nelson, D. E., Repetti, P. P., Adams, T. R., Creelman, R. A., Wu, J., Warner, D. C., et al. (2007). Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc. Natl. Acad. Sci. U.S.A. 104, 16450–16455. doi: 10.1073/pnas.0707193104
Ni, Z., Hu, Z., Jiang, Q., and Zhang, H. (2013). GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol. Biol. 82, 113–129. doi: 10.1007/s11103-013-0040-5
Nitschke, S., Cortleven, A., Iven, T., Feussner, I., Havaux, M., Riefler, M., et al. (2016). Circadian stress regimes affect the circadian clock and cause jasmonic acid-dependent cell death in cytokinin-deficient Arabidopsis plants. Plant Cell 28, 1616–1639. doi: 10.1105/tpc.16.00016
Nitschke, S., Cortleven, A., and Schmülling, T. (2017). Novel stress in plants by altering the photoperiod. Trends Plant Sci. 22, 913–916. doi: 10.1016/j.tplants.2017.09.005
Ouellet, F., and Charron, J.-B. (2013). “Cold acclimation and freezing tolerance in plants,” in eLS, Chichester: John Wiley & Sons, Ltd. doi: 10.1002/9780470015902.a0020093.pub2
Outlaw, J. W. H. (2003). Integration of cellular and physiological functions of guard cells. Crit. Rev. Plant Sci. 22, 503–529. doi: 10.1080/713608316
Pareek, A., Khurana, A., Sharma, A. K., and Kumar, R. (2017). An overview of signaling regulons during cold stress tolerance in plants. Curr. Genomics 18, 498–511. doi: 10.2174/1389202918666170228141345
Park, H. J., Kim, W. Y., and Yun, D. J. (2016). A new insight of salt stress signaling in plant. Mol. Cells 39, 447–459. doi: 10.14348/molcells.2016.0083
Pereira, S. L. S., Martins, C. P. S., Sousa, A. O., Camillo, L. R., Araujo, C. P., Alcantara, G. M., et al. (2018). Genome-wide characterization and expression analysis of citrus NUCLEAR FACTOR-Y (NF-Y) transcription factors identified a novel NF-YA gene involved in drought-stress response and tolerance. PLoS One 13:e0199187. doi: 10.1371/journal.pone.0199187
Pfannschmidt, T., Brautigam, K., Wagner, R., Dietzel, L., Schroter, Y., Steiner, S., et al. (2009). Potential regulation of gene expression in photosynthetic cells by redox and energy state: approaches towards better understanding. Ann. Bot. 103, 599–607. doi: 10.1093/aob/mcn081
Qiu, Q. S., Guo, Y., Dietrich, M. A., Schumaker, K. S., and Zhu, J. K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. U.S.A. 99, 8436–8441. doi: 10.1073/pnas.122224699
Queval, G., Issakidis-Bourguet, E., Hoeberichts, F. A., Vandorpe, M., Gakiere, B., Vanacker, H., et al. (2007). Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J. 52, 640–657. doi: 10.1111/j.1365-313X.2007.03263.x
Rawat, R., Takahashi, N., Hsu, P. Y., Jones, M. A., Schwartz, J., Salemi, M. R., et al. (2011). REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet. 7:e1001350. doi: 10.1371/journal.pgen.1001350
Riboni, M., Galbiati, M., Tonelli, C., and Conti, L. (2013). GIGANTEA enables drought escape response via abscisic acid-dependent activation of the florigens and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS. Plant Physiol. 162, 1706–1719. doi: 10.1104/pp.113.217729
Riboni, M., Robustelli, T. A., Galbiati, M., Tonelli, C., and Conti, L. (2016). ABA-dependent control of GIGANTEA signalling enables drought escape via up-regulation of FLOWERING LOCUS T in Arabidopsis thaliana. J. Exp. Bot. 67, 6309–6322. doi: 10.1093/jxb/erw384
Roeber, V. M., Bajaj, I., Rohde, M., Schmulling, T., and Cortleven, A. (2021). Light acts as a stressor and influences abiotic and biotic stress responses in plants. Plant Cell Environ. 44, 645–664. doi: 10.1111/pce.13948
Rugnone, M. L., Faigon Soverna, A., Sanchez, S. E., Schlaen, R. G., Hernando, C. E., Seymour, D. K., et al. (2013). LNK genes integrate light and clock signaling networks at the core of the Arabidopsis oscillator. Proc. Natl. Acad. Sci. U.S.A. 110, 12120–12125. doi: 10.1073/pnas.1302170110
Sanchez, A., Shin, J., and Davis, S. J. (2011). Abiotic stress and the plant circadian clock. Plant Signal. Behav. 6, 223–231. doi: 10.4161/psb.6.2.14893
Sanchez, S. E., Rugnone, M. L., and Kay, S. A. (2020). Light perception: a matter of time. Mol. Plant 13, 363–385. doi: 10.1016/j.molp.2020.02.006
Sarkar, D. (2010). Photoperiodic inhibition of potato tuberization: an update. Plant Growth Regul. 62, 117–125. doi: 10.1007/s10725-010-9502-9
Sawa, M., Nusinow, D. A., Kay, S. A., and Imaizumi, T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318, 261–265. doi: 10.1126/science.1146994
Schaffer, R., Ramsay, N., Samach, A., Corden, S., Putterill, J., Carré, I. A., et al. (1998). The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93, 1219–1229. doi: 10.1016/s0092-8674(00)81465-8
Seo, P. J., and Mas, P. (2015). STRESSing the role of the plant circadian clock. Trends Plant Sci. 20, 230–237. doi: 10.1016/j.tplants.2015.01.001
Serrano-Bueno, G., Sanchez de Medina Hernandez, V., and Valverde, F. (2021). Photoperiodic signaling and senescence, an ancient solution to a modern problem? Front. Plant Sci. 12:634393. doi: 10.3389/fpls.2021.634393
Shao, N., Beck, C. F., Lemaire, S. D., and Krieger-Liszkay, A. (2008). Photosynthetic electron flow affects H2O2 signaling by inactivation of catalase in Chlamydomonas reinhardtii. Planta 228, 1055–1066. doi: 10.1007/s00425-008-0807-0
Sharma, M., Irfan, M., Kumar, A., Kumar, P., and Datta, A. (2021). Recent insights into plant circadian clock response against abiotic stress. J. Plant Growth Regul. 221:112403. doi: 10.1007/s00344-021-10531-y
Sherrard, M. E., and Maherali, H. (2006). The adaptive significance of drought escape in Avena barbata, an annual grass. Evolution 60, 2478–2489. doi: 10.1554/06-150.1
Shi, H., and Chan, Z.-L. (2014). AtHAP5A modulates freezing stress resistance in Arabidopsis independent of the CBF pathway. Plant Signal. Behav. 9:e29109. doi: 10.4161/psb.29109
Shi, H., Ishitani, M., Kim, C., and Zhu, J. K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. U.S.A. 97, 6896–6901. doi: 10.1073/pnas.120170197
Shim, J. S., and Imaizumi, T. (2015). Circadian clock and photoperiodic response in Arabidopsis: from seasonal flowering to redox homeostasis. Biochemistry 54, 157–170. doi: 10.1021/bi500922q
Shim, J. S., Kubota, A., and Imaizumi, T. (2017). Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiol. 173, 5–15. doi: 10.1104/pp.16.01327
Shimizu, S., Yamauchi, Y., and Ishikawa, A. (2021). Photoperiod following inoculation of Arabidopsis with Pyricularia oryzae (syn. Magnaporthe oryzae) influences on the plant–pathogen interaction. Int. J. Mol. Sci. 22:5004. doi: 10.3390/ijms22095004
Singh, R. K., Svystun, T., AlDahmash, B., Jonsson, A. M., and Bhalerao, R. P. (2017). Photoperiod- and temperature-mediated control of phenology in trees - a molecular perspective. New Phytol. 213, 511–524. doi: 10.1111/nph.14346
Smith, A. M., and Stitt, M. (2007). Coordination of carbon supply and plant growth. Plant Cell Environ. 30, 1126–1249. doi: 10.1111/j.1365-3040.2007.01708.x
Song, Y. H., Shim, J. S., Kinmonth-Schultz, H. A., and Imaizumi, T. (2015). Photoperiodic flowering: time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 66, 441–464. doi: 10.1146/annurev-arplant-043014-115555
Stitt, M., and Zeeman, S. C. (2012). Starch turnover: pathways, regulation and role in growth. Curr. Opin. Plant Biol. 15, 282–292. doi: 10.1016/j.pbi.2012.03.016
Su, H., Cao, Y., Ku, L., Yao, W., Cao, Y., Ren, Z., et al. (2018). Dual functions of ZmNF-YA3 in photoperiod-dependent flowering and abiotic stress responses in maize. J. Exp. Bot. 69, 5177–5189. doi: 10.1093/jxb/ery299
Suarez-Lopez, P., Wheatley, K., Robson, F., Onouchi, H., Valverde, F., and Coupland, G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410, 1116–1120. doi: 10.1038/35074138
Thomashow, M. F. (2010). Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiol. 154, 571–577. doi: 10.1104/pp.110.161794
Walters, R. G., and Horton, P. (1995). Acclimation of Arabidopsis thaliana to the light environment: regulation of chloroplast composition. Planta 197, 475–481. doi: 10.1007/BF00196669
Wang, Z. Y., and Tobin, E. M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93, 1207–1217. doi: 10.1016/s0092-8674(00)81464-6
Welling, A., Moritz, T., Palva, E. T., and Junttila, O. (2002). Independent activation of cold acclimation by low temperature and short photoperiod in hybrid aspen. Plant Physiol. 129, 1633–1641. doi: 10.1104/pp.003814
Xie, Q., Lou, P., Hermand, V., Aman, R., Park, H. J., Yun, D. J., et al. (2015). Allelic polymorphism of GIGANTEA is responsible for naturally occurring variation in circadian period in Brassica rapa. Proc. Natl. Acad. Sci. U.S.A. 112, 3829–3834. doi: 10.1073/pnas.1421803112
Yang, Y. X., Wang, M. M., Yin, Y. L., Onac, E., Zhou, G. F., Peng, S., et al. (2015). RNA-seq analysis reveals the role of red light in resistance against Pseudomonas syringae pv. tomato DC3000 in tomato plants. BMC Genomics 16:120. doi: 10.1186/s12864-015-1228-7
Zeeman, S. C., Smith, S. M., and Smith, A. M. (2007). The diurnal metabolism of leaf starch. Biochem. J. 401, 13–28. doi: 10.1042/BJ20061393
Keywords: biotic stress, cold stress, drought stress, GIGANTEA, osmotic stress, photoperiod, photoperiod stress
Citation: Roeber VM, Schmülling T and Cortleven A (2022) The Photoperiod: Handling and Causing Stress in Plants. Front. Plant Sci. 12:781988. doi: 10.3389/fpls.2021.781988
Received: 23 September 2021; Accepted: 13 December 2021;
Published: 25 January 2022.
Edited by:
Chris Helliwell, Commonwealth Scientific and Industrial Research Organisation (CSIRO), AustraliaReviewed by:
Hyo-Jun Lee, Korea Research Institute of Bioscience and Biotechnology (KRIBB), South KoreaFrancisco J. Romero-Campero, Seville University, Spain
Copyright © 2022 Roeber, Schmülling and Cortleven. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anne Cortleven, YW5uZS5jb3J0bGV2ZW5AZnUtYmVybGluLmRl
 Venja M. Roeber
Venja M. Roeber Thomas Schmülling
Thomas Schmülling Anne Cortleven
Anne Cortleven