
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 07 January 2022
Sec. Plant Symbiotic Interactions
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.779302
 Xiaodan Li1,2†
Xiaodan Li1,2† Songfeng Wang1,2†
Songfeng Wang1,2† Yating Fan1,2
Yating Fan1,2 Zhe Zhou1,2
Zhe Zhou1,2 Sheng Xu1,2
Sheng Xu1,2 Penglei Zhou3
Penglei Zhou3 Jiayu Zhou1,2*
Jiayu Zhou1,2* Ren Wang1,2*
Ren Wang1,2*
Corydalis yanhusuo, a precious herb of the Papaveraceae family, is widely used in multiple traditional Chinese medicines for the treatment of many painful conditions, and its medicinal part is the dried tuber. Yet how to improve this plant’s medicinal yield as well as its economic efficiency remains a key problem in its cultivation. The planting of C. yanhusuo in rotation with peanut (Arachis hypogaea L.) aims to improve land utilization efficiency, but the total production of tubers is severely reduced relative to fields without rotation. However, an increased yield was observed in C. yanhusuo plants grown in previously flooded fields (HR field) compared to the ones grown in the fields that had been used to cultivate peanut (PL field) or in fields without rotation or flooding (N field). Based on these phenomena, in this study, we explored the potential factors responsible for the altered growth/yield of C. yanhusuo under different field conditions. Soil physicochemical properties and the diversity and community of rhizobacteriome of C. yanhusuo were both analyzed. By testing several soil physicochemical properties, we found that the cation exchange capacity (CEC), soil organic matter (SOM), total nitrogen (TN), and pH value differed significantly among these three types of fields. 16S rRNA amplicon sequencing revealed stark differences in the composition, diversity, and potential functions of the bacterial community in the rhizosphere of C. yanhusuo plants grown in field with the peanut rotation or flooding. Notably, the Acidobacteria were enriched in the HR field, while Actinobacteria were enriched in the PL field. More importantly, further analysis showed that changed soil physicochemical properties could be one reason for why the rhizospheric bacterial community has changed; hence, soil physicochemical properties might also be affecting plant performance indirectly by regulating the rhizospheric bacterial community. The RDA analysis distinguished CEC as the most important soil physicochemical property influencing the structure and composition of the C. yanhusuo rhizobacteriome. In summary, our results suggest peanut rotation- and flooding-induced soil physicochemical properties changes would further impact the rhizobacteriome of C. yanhusuo albeit differentially, culminating in opposite effects upon the plant growth and medicinal yield of C. yanhusuo.
Rhizoma Corydalis (RC), the dried tubers of the Corydalis yanhusuo plant (commonly known as “Yuanhu” in China), is a well-known Chinese herbal medicine that is widely prescribed for treating headaches, spastic and abdominal pains, in addition to relieving pain from injury (Tang and Nie, 2006). Modern medical research has revealed that the C. yanhusuo extract has significant inhibitory effects upon acetylcholinesterase (AChE) and cancer cell proliferation, as well as beneficial effects for the cardiovascular system (Ling et al., 2006; Xiao et al., 2011; Zhao Y. et al., 2014; Chen et al., 2016). To date, more than 100 compounds have been isolated and identified from RC, including fatty acids, organic acids, amino acids, sugars, steroids, anthraquinones, volatile compounds, and alkaloids (Hung and Wu, 2016; Abdallah et al., 2017; Hassan et al., 2017). In particular, over 80 types of alkaloids have been isolated and identified (Tian et al., 2020), these being major pharmacologically active compounds for the treatment of blood vessel diseases, tumors, and various pains (Li et al., 2014; Wu et al., 2016). Tetrahydropalmatine is a tetrahydroprotoberberine diisoquinoline alkaloid and the primary active constituent of RC; tetrahydropalmatine has been used as a drug with analgesic, hypnotic, sedative and other effects (Gong et al., 2016; Yu et al., 2016; Liu et al., 2021).
Driven by its large market demand and because of its short planting cycle, many plantations of C. yanhusuo have been quickly established for economic gain (Wang Y.M. et al., 2019). The suitable period for planting C. yanhusuo is from late September to mid-October, with its harvest in May (Xu, 2005). To make the most use of this land, implementing a crop rotation, generally done with peanut, in the other 4 to 5 months may be economically advantageous. However, to the best of our knowledge, the C. yanhusuo-peanut rotation is facing serious problems. After peanut planting, the growth of C. yanhusuo plants is severely impaired compared to their cultivation in fields without such a rotation, leading to their reduced medicinal yield. We also found that C. yanhusuo plants in flooded fields grew better than those in the monocrop fields without flooding.
We know that the rhizospheric microbiome plays important roles in benefiting plant health and productivity (Philippot et al., 2013; Trivedi et al., 2020), by promoting plants growth, enhancing their nutrient acquisition capacity, and also increasing their resistance to both biotic and abiotic stresses (Maarastawi et al., 2018). Nevertheless, much is still unknown about the rhizospheric microbiome of C. yanhusuo. Accordingly, their potential influence on the growth and ensuing medicinal yield of C. yanhusuo is also unclear. To fill these knowledge gaps, C. yanhusuo plants and rhizospheric soil from three different types of fields were collected and analyzed in this study. For the HR field, during the rainy season, its soil was flooded for 1 month. For the N field, no other plants were planted during the last growing season until C. yanhusuo seedlings were planted. In the PL field, C. yanhusuo seedlings were planted in the three individual fields that had been used to cultivate peanut in the preceding growing season. Before harvest, both the biomass of C. yanhusuo plants and the content of tetrahydropalmatine in their tubers were determined. Further, certain soil physicochemical properties, such as soil moisture, sodium content, and pH value, can strongly influence both the composition and diversity of rhizospheric microbial communities, Peiffer et al. (2013), Edwards et al. (2015), Lüneberg et al. (2018). Therefore, it would be both interesting and timely to explore the interactions among C. yanhusuo plants, their rhizospheric bacterial community (rhizobacteriome), and environmental factors.
Given the above documented phenomena in extant fields, this study aimed to assess the changes in soil physicochemical properties and rhizobacteriome in the three different types of C. yanhusuo cultivation fields [the N field (mono-crop), the PL field (rotation with peanut), and the HR field (flooded before planting)], to find out the potential soil and microbial factors that could affect the plant growth and yield of C. yanhusuo. Our findings provide insight into the complex interactions among medicinal plants, the soil environment, and the rhizospheric microbiome and are useful for guiding the cultivation of C. yanhusuo in southeastern China.
Individuals of C. yanhusuo were planted in September, 2020 and collected in April 2021 (i.e., 1 month before harvest). The three field types were located in the Xuyi District (118° 23′ E, 33° 14′ N), Huai’an, Jiangsu Province, China, and consisted of an “HR” field (mono-crop of C. yanhusuo that had been flooded in July, 2020), a “PL” field (rotation of peanut and C. yanhusuo) and an “N” field (mono-crop of C. yanhusuo). Each field type had three replicates, and each replicate was about 300 m2. Specifically, for the HR field, C. yanhusuo seedlings were planted in three individual fields near the Huai River, and flooded for 1 month in July 2020 for which the submerged water depth was 5–8 cm. For the PL field, C. yanhusuo seedlings were planted in three individual fields that were already used to cultivate peanut crops during the prior growing season. For the N field, which served as the control, its three individual fields were left fallow during last growing season (i.e., no crops planted). These three types of fields were adjacent to each other and all managed using the same cultivation practices. These included the application of fertilizer and suitable planting depth and distance as follows: before planting, compound fertilizer (1,050 kg ha–1) and urea (45 kg ha–1) were applied as base fertilizer in each field (three fields per treatment, nine fields in total); the planting depth was 5–6 cm, and planting distance and row spacing were ∼10 and ∼15 cm, respectively.
Twenty-five individual plants at five positions—the four corners and center—of each replicate field were collected and pooled as a biological replicate sample. Rhizospheric soil samples were collected as described before (Sun et al., 2018; Zuo et al., 2021). Briefly, the roots and tubers were shaken vigorously to remove any loose soil; remaining soils firmly attached to roots and tubers were designated rhizospheric soil, and this material collected with sterile brushes. All samples of rhizospheric soil were then sieved through 2 mm mesh to remove any plant debris. Each soil sample was divided into three portions, with one portion immediately stored at −80°C for DNA extraction followed by 16S rRNA amplicon sequencing; one portion was stored at 4°C to later isolate any culturable bacteria; and another portion was used for the analysis of soil physicochemical properties.
Four C. yanhusuo plants were collected from each field type and each plant served as a biological replicate. Their tubers were oven-dried at 50°C to a constant weight and then ground into powder. From each sample, exactly 0.3 g of powdered tuber was weighed and extracted in 20 mL of a methanol-ammonia solution (20: 1, v/v) for 1 h in an ultrasonic bath at room temperature. Each extraction was filtered through a Millipore filter with a 0.22-μm pore size and stored at 4°C until its high-performance liquid chromatography (HPLC) analysis.
The standard of tetrahydropalmatine (purity ≥ 98%; Shanghai Yuanye Bio-Technology Co., Ltd.) was accurately weighed, dissolved to generate a stock solution at 4.6 mg mL–1, and then diluted with methanol into a series of working solutions. For tetrahydropalmatine’s quantification, 20 μL of each sample was injected into an LC-20A HPLC system equipped with an SPD-M20A Photodiode Array Detector (Shimadzu Corporation, Tokyo, Japan). An InertSustain C18 reverse-phase column (5 μm; 200 mm × 4.6 mm I.D.) was used. The optimized parameters were set as follows: the mobile phase consisted of eluent A [0.1% phosphoric acid in water (v/v), adjusted to pH 6.0 with triethylamine] and eluent B (methanol). The flow rate was 0.8 mL min–1, and column temperature maintained at 30°C with detections made at 280 nm. The optimal elution program consisted of 0–30 min, using 75% B.
The collected rhizospheric soil was air-dried at room temperature; any visible roots or other debris were manually removed; the soil was then ground and sieved through 2 mm before its characteristics were analyzed. The analyses of different soil physicochemical properties were carried out as described in previous studies. Briefly, soil pH value was measured with a pH-meter using a soil suspension (soil: 0.01 M CaCl2 = 1:2.5, w/v) (Lu, 2000). Soil organic matter (SOM) was determined using the K2CrO7 oxidation titration method, as described by Lu (2000). Soil total nitrogen (TN) was measured with an element analyzer according to Kjedahl method (Lu, 2000). Soil was digested by H2SO4/HClO4 and total phosphorus (TP) then determined by the molybdenum blue method. Total potassium (TK) in soil was quantified using flame photometry after its aqua-regia digestion (Xiao et al., 2015). Soil cation exchange capacity (CEC) was determined by the exchange with ammonium acetate (NH4OAc, 1.0 M, pH 7.0) and titration with 0.05 M HCl (Dai et al., 1998). Soil texture—percentages of sand, silt, and clay components—was determined by a laser particle size analyzer (Master-Sizer 3000, Malvern Panalytical, Malvern, United Kingdom) according to the Stokes law, for which classification names were assigned following the “textural triangle” developed by the USDA (Zhao, 1981).
Soil samples were resuspended and serially diluted in sterile water and plated on nutrient agar (NA) plates. All plates were incubated at 30°C for 48–60 h, after which the number of colony-forming units (CFUs) was counted per plate. Culturable bacteria concentrations were calculated and expressed as CFU per gram soil (CFU g–1 soil). Next, every single visible bacterial colony was picked, purified on fresh NA medium, and stored in 15% glycerol at –80°C. From each, its total DNA was extracted using the TIANamp Bacteria DNA Kit (Tiangen Biotech Co., Ltd., Beijing, China) according to manufacturer’s protocol. The primer pair of 27F (5′-AGAGTTTGATCCTGGCTCA-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) was used to amplify the 16S rRNA gene. The sequence of each bacterial colony was BLASTed against the NCBI database and identified accordingly.
Rhizospheric bacterial DNA was extracted using the HiPure Soil DNA Kit (Guangzhou Magen Biotechnology Co., Ltd., Guangzhou, China) and following the manufacturer’s protocols. The 16S rDNA V3-V4 region was amplified by PCR, using the primers 341F (5′-CCTACGGGNGGCWGCAG-3′) and 806R (5′-GGACTACHVGGGTATCTAAT-3′). The ensuing PCR amplification product was recovered by using an E.Z.N.A. Gel Extraction Kit (Omega bio-tek Inc., GA, United States), for which Illumina Misequence PE 250 (Beijing Allwegene Biotechnology Co., Ltd., Beijing, China) was then used for its sequencing. Raw sequences were deposited in the SRA database under the accession number PRJNA762442. Based on their distribution, nearly all the tags were within the length range of 440–470 bp and rather high in quality. To avoid the bias from different depths of sequencing for the alpha and beta diversity measurements, all libraries were normalized to the same size (104,909 tags in PL-2), based on the library with the lowest number of sequences.
Raw reads were filtered using fastp (v 0.18.0) to remove any reads containing more than 10% unknown nucleotides and reads which contained less than 50% of bases with quality values (Q values) above 20 (Chen et al., 2018). Paired end clean reads were merged as raw tags by using the FLASH tool (v 1.2.11) with a minimum overlap of 10 bp and a mismatch error rate of 2% (Magoè and Salzberg, 2011). Noisy sequences of raw tags were filtered under specific filtering conditions (Bokulich et al., 2013). The clean tags were clustered into operational taxonomic units (OTUs) of ≥ 97% similarity using the UPARSE (v 9.2.64) pipeline (Edgar, 2013). All chimeric tags were removed using the UCHIME algorithm (Edgar et al., 2011). The representative OTU sequences were classified by a naïve Bayesian model using the RDP classifier (v 2.2) (Wang et al., 2007) based on SILVA database (v 132) (Pruesse et al., 2007), with the confidence threshold value of 0.8. Venn analysis was performed using the R project VennDiagram package (v 1.6.16) to identify unique and common OTUs in each group (Chen and Boutros, 2011). Circular layout representations of species abundance were graphed using Circos (v 0.69-3) (Krzywinski et al., 2009). Taxonomic composition of bacteria that are present in different samples was visualized using a stack bar plot at different taxonomic level using ggplot2 package (v 2.2.1) (Wickham, 2011). Alpha diversity indexes and species richness estimators in each group was evaluated in QIIME (v 1.9.1) (Caporaso et al., 2010). Phylogenetic diversity-whole tree (PD tree) was calculated in Picante (v 1.8.2) (Kembel et al., 2010). PCoA (principal coordinates analysis) of (Un) weighted unifrac was generated in the R project Vegan package (v 2.5.3) (Oksanen et al., 2010) and plotted in the R project ggplot2 package (v 2.2.1) (Wickham, 2011). Biomarker features in each group were screened by LEfSe software (v 1.0) (Segata et al., 2011). The KEGG pathway analysis of the OTUs was inferred using Tax4Fun (v 1.0) (Aßhauer et al., 2015). Analysis of function difference between groups was calculated by Welch’s t-test in the R project Vegan package (v 2.5.3) (Oksanen et al., 2010).
Redundancy analysis (RDA) was implemented to test whether and which soil physicochemical properties contributed significantly (p < 0.05) to variation in the bacterial community of the C. yanhusuo rhizosphere. RDA and Mantel test was executed in R project Vegan package (v 2.5.3) (Oksanen et al., 2010) to clarify the influence of environmental factors on community composition of C. yanhusuo rhizosphere. All these analyses of 16S rRNA amplicon sequencing data and according figures were generated using Omicsmart1, a dynamic real-time interactive online platform for data analysis (He et al., 2021).
Analyses were carried out using SPSS 13.0 (SPSS Inc., Chicago, IL, United States) and Microsoft Excel 2019 software programs. A threshold of p < 0.05 was used to define statistically significant results.
In this study, C. yanhusuo plants of the same age but under different field cultivation conditions (HR, PL, and N) were harvested. The C. yanhusuo plants in the N and HR fields were healthy and their tubers were big, plump, and solid, but those plants in the PL field were small and appeared feeble (Figure 1A). Moreover, compared with those of N or HR plants, the tubers of PL plants were significantly smaller which directly diminished their yield as the medicinal part (Figure 1B). Accordingly, the total yield of C. yanhusuo tubers from the HR field was at least 24,150 kg ha–1, while that from the N and PL fields, respectively, was 17,250 and 10,950 kg ha–1 (Figure 1B). These results indicated that flooding could promote the growth of C. yanhusuo and its ensuing medicinal yield, whereas its rotation with peanut could be disadvantageous. Further, there was no significant difference in the content of tetrahydropalmatine in C. yanhusuo plants among the three field types (Supplementary Table 1). Building on this result, we next explored the mechanisms underlying the disparities in plant performance under the different cultivation conditions.

Figure 1. The effect of different field types upon the Corydalis yanhusuo tuber yield. (A) Photographs of C. yanhusuo plants from the three different fields. (B) The total tuber yield per hectare of C. yanhusuo tuber from each field type. PL, plants from the PL field (field under peanut–C. yanhusuo rotation); N, plants from the N field (C. yanhusuo field without rotation and flooding); HR, plants from the HR field (C. yanhusuo field that had been flooded in July, 2020).
The TN in soil was significantly higher at PL than the other two fields (Table 1), and both N and PL fields harbored greater amount of SOM than HR did (Table 1). By contrast, the TP was similar among all soils, as was TK (Table 1).
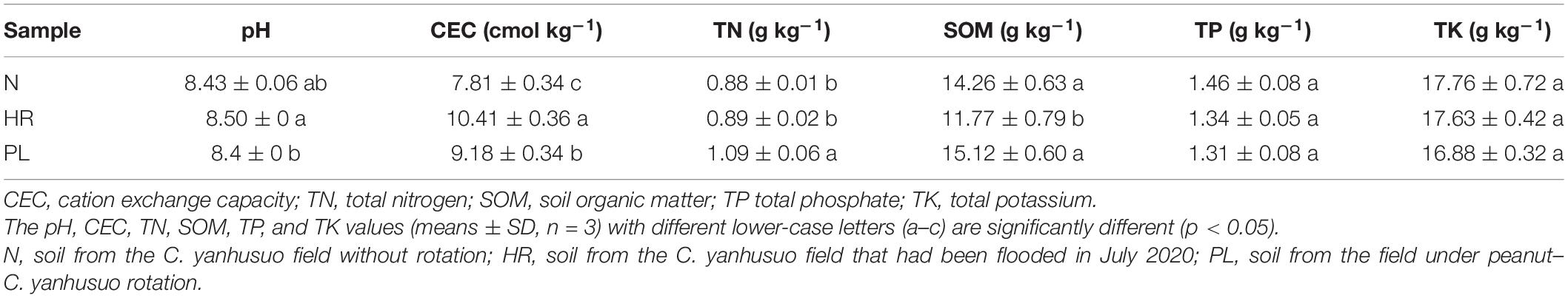
Table 1. Physicochemical properties of soils from three different cultivation fields of C. yanhusuo in the Xuyi District, eastern China.
Soil pH values were significantly different between the HR and PL fields (p < 0.05) (Table 1). Compared with the N field, the soil pH in the HR and PL fields were 0.83% higher and 0.36% lower, respectively. The strongest effect sizes were found for the CEC of different soils, which were ranked as follows: HR > PL > N (Table 1). Although changes to soil physicochemical properties across the fields were detected, the direct influences of soil characteristics cannot fully explain the changed plant growth and tuber yields of C. yanhusuo.
The concentration of culturable bacteria evidently varied among the different soils (Supplementary Figure 1). Compared with soil from the N (29.2–38.1 × 107 CFU g–1 soil) and HR (24–27.5 × 107CFU g–1 soil) fields, the bacterial CFU number in soil of the PL field (approximately 44.1–39.9 × 107 CFU g–1 soil) was significantly higher (p < 0.05). Hence, the inhibition of plant growth observed in the PL field might have arisen from the enrichment of some harmful bacteria in its rhizospheric soil. To gauge and understand which bacterial taxa were enriched and depleted in the different soils, each bacterial strain was identified based on 16S rDNA sequencing.
From all soil samples, a total of 194 strains were identified that belonged to 61 genera. Bacillus (24%), Arthrobacter (8%) and Microbacterium (6.7%) were the most abundant genera in the PL soil (Supplementary Figure 2). Although Bacillus (16.1%), Arthrobacter (17.7%), and Microbacterium (8.1%) were the most abundant bacterial genera in the N soil as well, their relative proportions differed considerably from those in PL soil. The top three genera present in HR soil were Bacillus (10.6%), Arthrobacter (10.6%), and Streptosporangium (8.5%). Therefore, these compositional changes in the rhizobacteriome might explain the altered performance of C. yanhusuo across the three field types.
The bacterial community in the rhizosphere of C. yanhusuo plants grown in each field type was further characterized by 16S rRNA amplicon sequencing, whose results corroborated the changed rhizospheric bacteriome community structure of plants growing in different soils across the fields. The number of OTUs varied from 5,497 to 6,428 per sample (mean = 5,890) (Supplementary Table 2). The number of bacterial OTUs differed significantly in abundance among the different rhizospheric soils. As the Venn diagram shows (Figure 2A), overall, 2,448 OTUs were detected in the rhizosphere of plants grown in all three fields’ soil. Notably, plants grown in PL soil harbored 1,899 unique OTUs, far more that in plants cultivated in the N (1,371 OTUs) and HR (1,248 OTUs) soils. Circos plots (Figure 2B) revealed that samples from N and PL soils were associated with RB41 (ca. 20%) and Sphingomonas (ca. 20%), while their association with Massilia (< 10%) and Nocardioides (<10%) appeared less pronounced. Rhizospheric soil samples from the HR field sustained a higher abundance of RB41 (ca. 40%) in comparison with the N and PL fields. Despite sharing a similar microbial community composition, evidently there were some key differences in the abundance of certain genera among the rhizospheric soil samples from different cultivation fields. Circos plots also showed that the PL field presented a higher association with Massilia (>15%) (Figure 2B). Additionally, Pseudomonas seemed to be more abundant in the N field (Figure 2B).
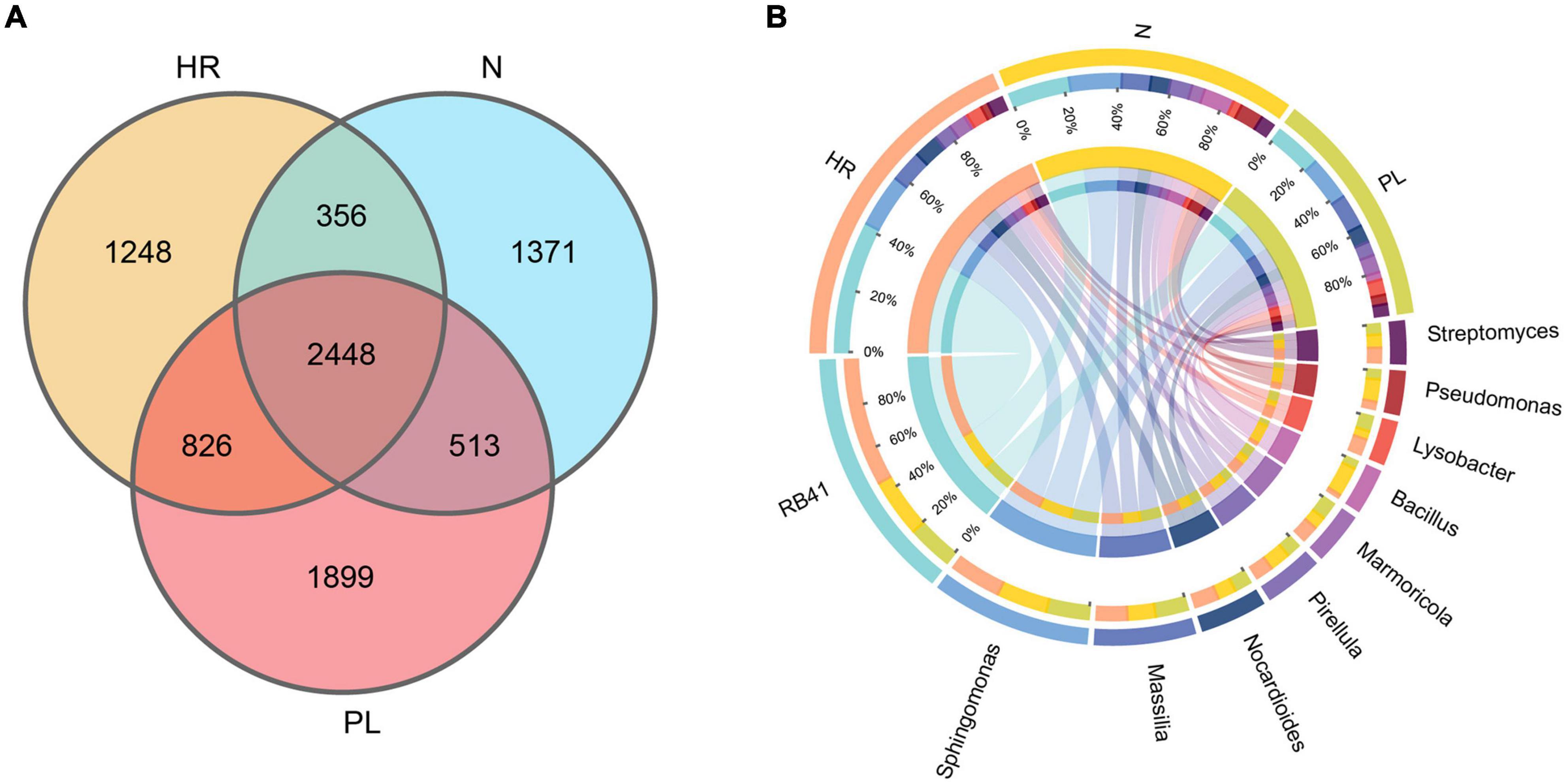
Figure 2. Venn diagram of rhizosphere-enriched bacterial OTUs in rhizospheric soils from the three field types. (A) Venn diagram representing the bacterial OTUs associated with the rhizosphere of C. yanhusuo. (B) CIRCOS diagram illustrating the taxonomic representation of differences between N, HR and PL fields. The width of each ribbon represents the abundance of each taxon. N, soil from the C. yanhusuo field without rotation; HR, soil from the C. yanhusuo field that had been flooded in July 2020; PL, soil from the field under peanut–C. yanhusuo rotation.
The most abundant bacteria at different taxonomic levels in three fields are presented in Figure 3. Acidobacteria, Proteobacteria, and Actinobacteria were the most abundant phyla in all the soils (Figure 3A), but the most abundant phylum differed among the field types. The relative abundance of Acidobacteria was highest in HR field (26.8%) followed by PL (17.3%) and N (15.5%) fields. The relative abundance of Proteobacteria (26.7%) was greatest in PL field, followed by N (26.5%) and HR (23.9%) fields; PL field also possessed the highest relative abundance of Actinobacteria (17.7%), whose abundance was lower in the two other fields: HR (13.7%) and N (13.2%). At the class level, the relative abundances of Alphaproteobacteria and Bacteroidia in HR field were low, while Subgroup_6 and Blastocatellia_Subgroup_4 were significantly more abundant in HR field (Figure 3B). At the order level (Figure 3C), the relative abundance of Pyrinomonadales in the N (4%) or PL (3.1%) field was low, while the relative abundance in HR (8.7%) was significantly higher. Betaproteobacteriales (belonging to Gammaproteobacteria) was the most abundant order. Gemmatimonadales was relatively lower in abundance in PL (2.8%) than that in either HR (3.5%) or N (4.9%). For both Rhizobiales and Micrococcales, the relative abundance of each was lower in HR field. Yet at the family level, Pyrinomonadaceae was most abundant in HR (8.7%), while in N and PL, its relative abundance was 4.0 and 3.1%, respectively (Figure 3D).
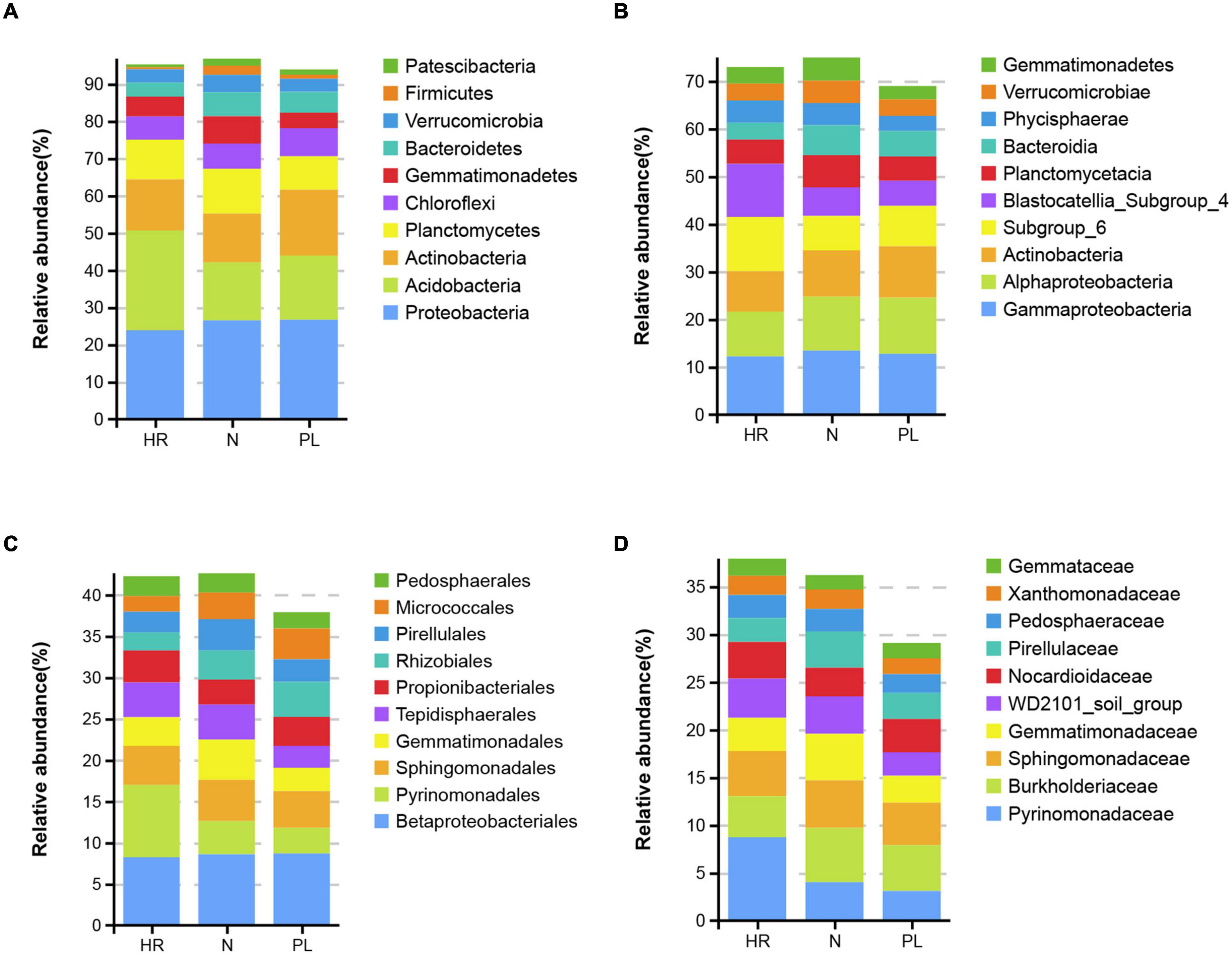
Figure 3. Top 10 relative abundances of bacterial communities classified at the phylum (A), class (B), order (C), and family (D) levels in different rhizospheric soils. N, soil from the C. yanhusuo field without rotation; HR, soil from the C. yanhusuo field that had been flooded in July 2020; PL, soil from the field under peanut–C. yanhusuo rotation.
It is noteworthy that the α diversity indexes (Shannon, and Simpson), diversity estimator (PD-tree), species richness estimators (ACE and Chao 1), the observed species (sobs) all showed a similar pattern in different fields (Figure 4). The rarefaction curve showing Good’s coverage appears in Supplementary Figure 3. The rhizospheric microbiome of the PL field had significantly higher α diversity in comparison to the other two fields (N and HR); correspondingly, species richness estimators (ACE and Chao 1) were lower in the N and HR microbiomes. The Shannon index of PL was 10.66 ± 0.03, being significantly higher than that of N (10.28 ± 0.07) and HR (10.31 ± 0.01) (Figure 4D). Moreover, Simpson index, PD-tree, and the species richness estimators including Chao 1, ACE, all showed similar trends (Figure 4). Taken together, these results indicated the composition and diversity of the bacterial community in the rhizosphere of C. yanhusuo varied non-randomly among the tested soil samples. Evidently, using a peanut rotation could increase the α diversity of rhizobacteriome. Still, how the presence of more diverse bacteria in the rhizosphere might be linked to a negative influence on plant performance needs further explanation.
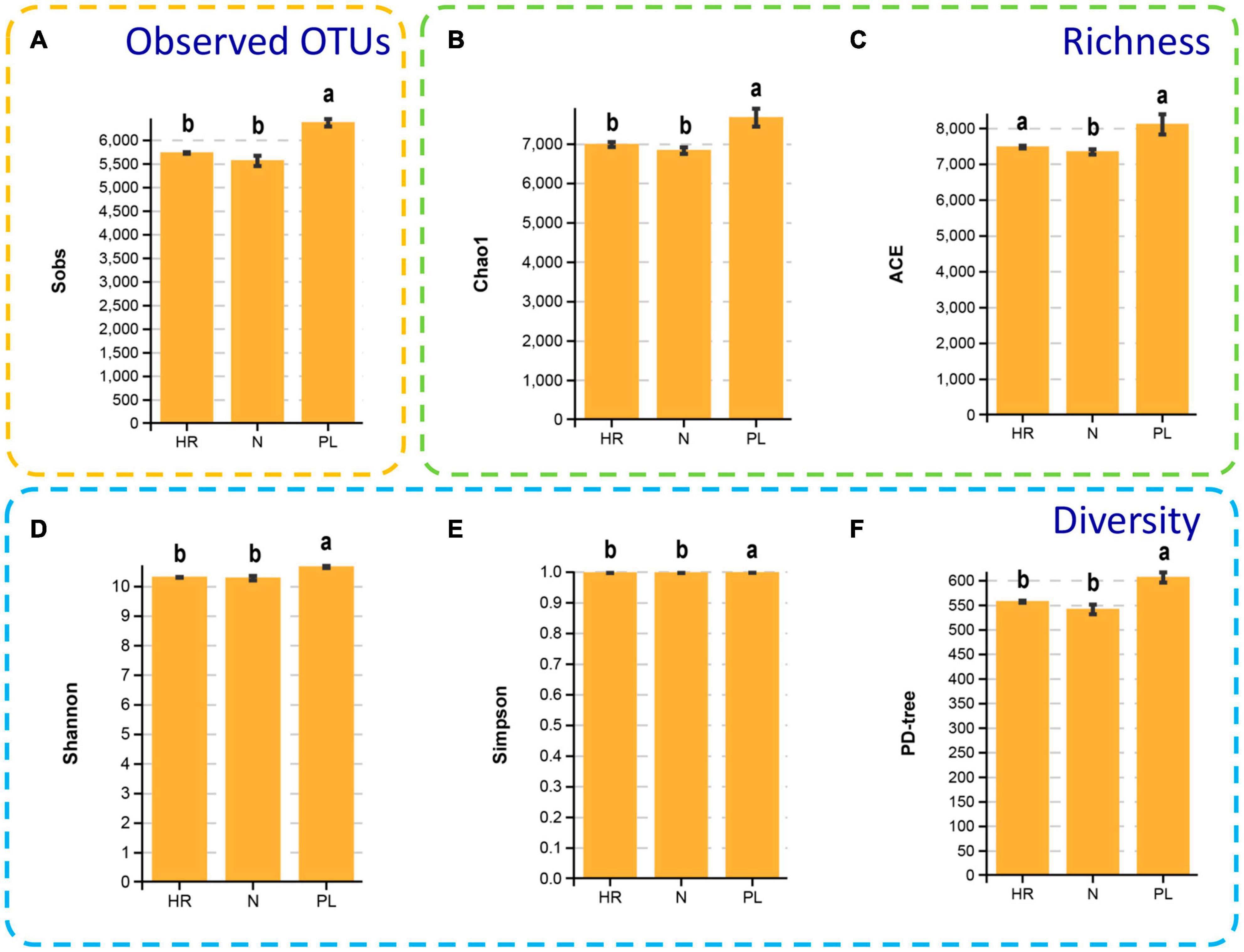
Figure 4. Alpha diversity of the microbial communities of the rhizospheric bacterial communities of the N, HR, and PL field samples. Sobs (A), Chao1 (B), ACE (C), Shannon (D), Simpson (E), and PD-tree (F). Different letters indicate significant differences (Tukey-HSD, p < 0.05). N, soil from the C. yanhusuo field without rotation; HR, soil from the C. yanhusuo field that had been flooded in July 2020; PL, soil from the field under peanut–C. yanhusuo rotation.
Samples from the different fields were clearly separated in the pooled principal coordinates analysis (PCoA) at the OTU level, whether based on unweighted UniFrac (UU) or weighted UniFrac (WU) distances (Figure 5). This suggested that flooding or a peanut rotation could significantly and differentially influence the rhizospheric bacterial diversity of C. yanhusuo. Using the UU distance, a distinct pattern was discernible for the rhizospheric bacterial communities. The two axes explained 22.15 and 19.56% of the total variation in rhizobacteriome (Figure 5A); however, when based on the WU distance, the first and second axes were now able to respectively explain 49.88 and 27.71% of that variation (Figure 5B).
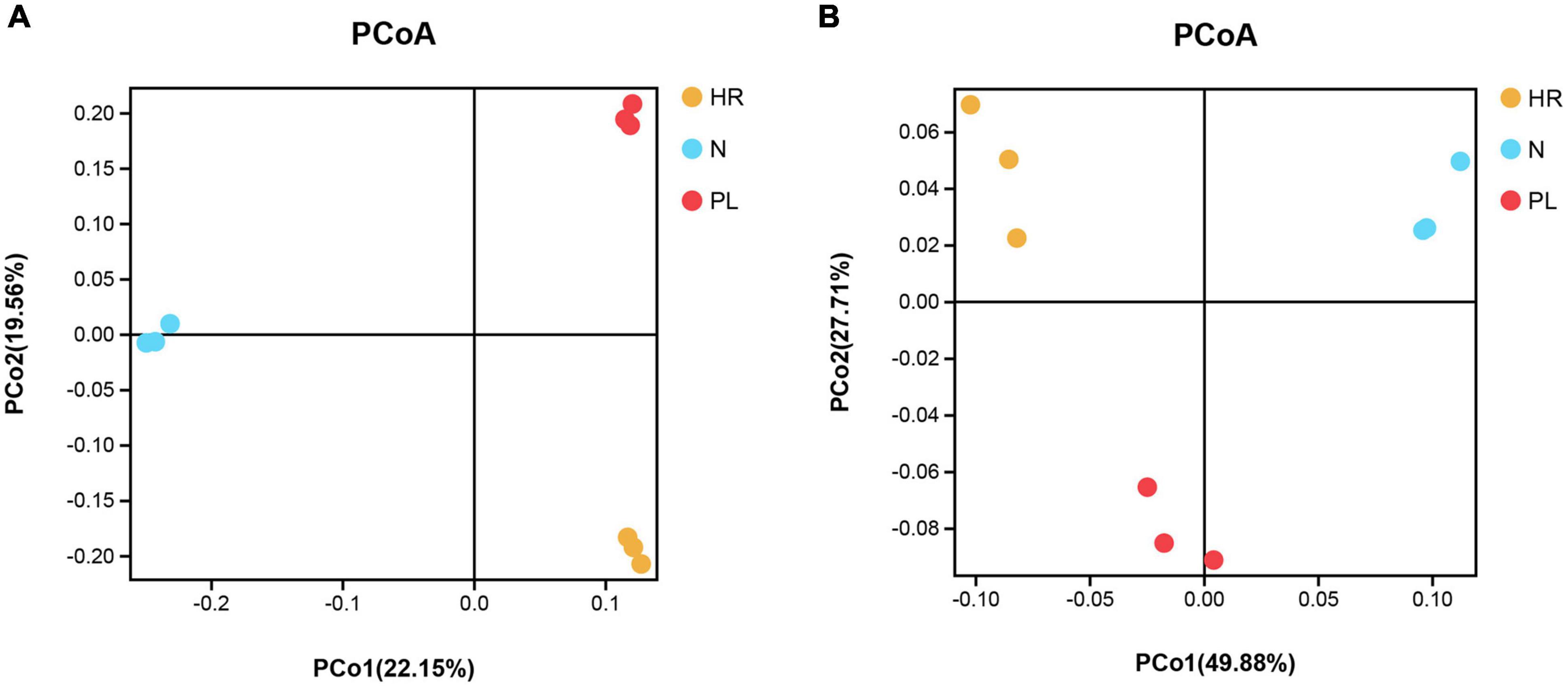
Figure 5. Principal coordinates analysis (PCoA) based on the unweighted UniFrac (UU) and weighted UniFrac (WU) distances among different rhizospheric soil bacterial communities of β-diversity: (A) UU distance; (B) WU distance. Symbols with unique colors denote different samples: yellow circle for HR, soil from the C. yanhusuo field that had been flooded in July 2020; blue circle for N, soil from the C. yanhusuo field without rotation; red circle for PL, soil from the field under peanut–C. yanhusuo rotation).
Redundancy analysis (RDA) let us evaluate whether the changed soil physicochemical properties were paramount driver of the varied rhizospheric bacterial community among the three different fields (Figure 6A). When applied to the main bacterial genera, the RDA showed that the first two axes accounted for 90.13% of the total variance in bacterial community composition, with the first axis capturing 74.52% of that variance. Among all the environmental factors examined using the Mantel test, CEC (r = 0.798, p = 0.002), SOM (r = 0.382, p = 0.036), and TP (r = 0.308, p = 0.047) were significantly correlated with microbial community structure at the genus level (Supplementary Figures 4B,D,F), whereas the other soil factors were negligibly correlated with it (p > 0.05) (Supplementary Figures 4A,C,E). These results indicated that CEC was the most critical soil factor potentially shaping the structure of rhizosphere microbial communities. At the genus level, CEC were associated with an excess of MND1, and significantly associated with less of Bacillus, Pedobacter, Pir4_lineage, Variovorax, and Allorhizobium–Neorhizobium–Pararhizobium–Rhizobium (Figure 6B); hence, these bacterial taxa could have potential influence on the growth of C. yanhusuo.
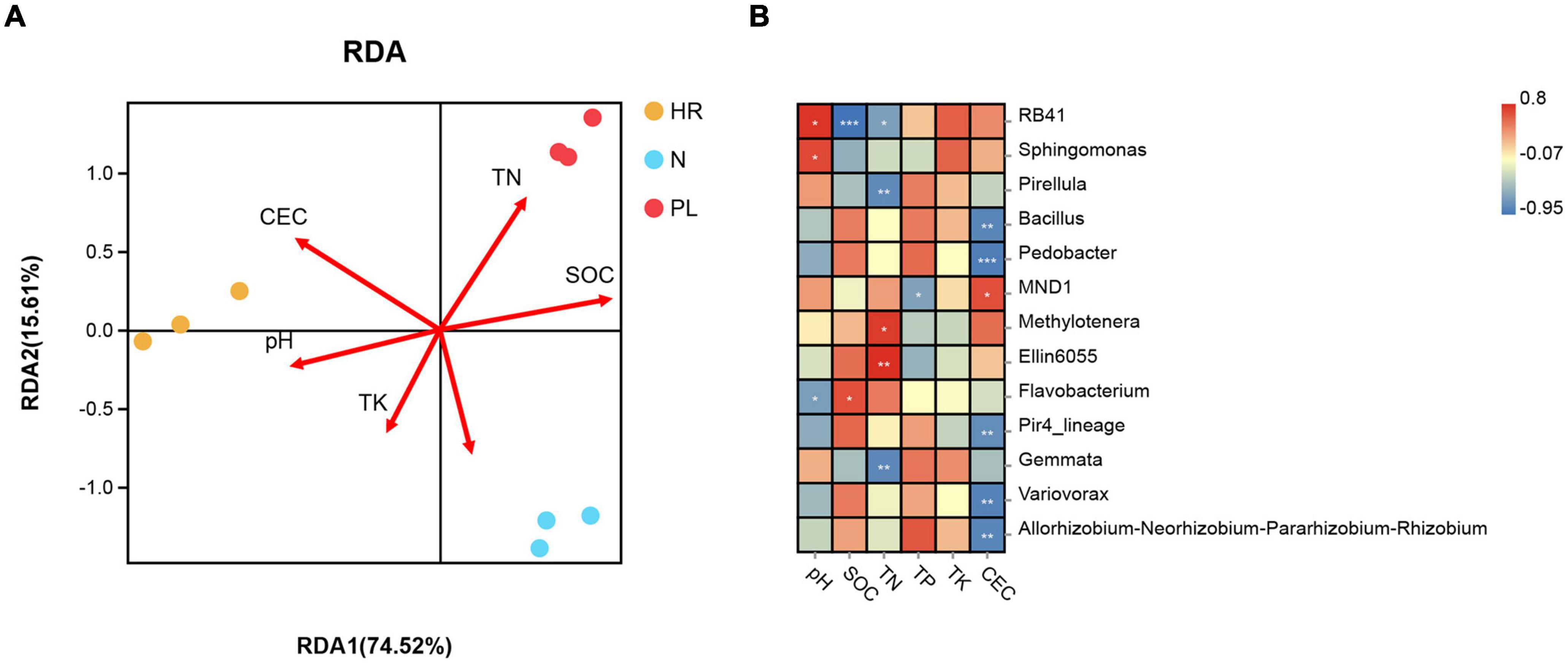
Figure 6. Relationships between microbial community and soil properties (A) Redundancy analysis (RDA) of soil properties and bacterial abundance in the C. yanhusuo rhizosphere. The angle and length of the arrows indicate the direction and strength of the relationships of soil factors vis-à-vis the microbial populations. CEC, cation exchange capacity, SOM, soil organic matter, TN, total nitrogen, TP, total phosphorus, TK, total K. The CEC (r2 = 0.798, p = 0.002), SOM (r2 = 0.382, p = 0.036) and TP (r2 = 0.308, p = 0.047) were significant factors that influenced the microbial community structure of the C. yanhusuo rhizosphere. (B) Genus influenced by different soil properties in the rhizosphere of C. yanhusuo (*p < 0.05, **p < 0.01, ***p < 0.001; Spearman correlations). N, soil from the C. yanhusuo field without rotation; HR, soil from the C. yanhusuo field that had been flooded in July 2020; PL, soil from the field under peanut–C. yanhusuo rotation.
To identify biomarkers in the rhizosphere of each field type, a linear discriminant analysis effect size (LEfSe) analysis was carried out (Figure 7). We found fewer biomarkers for the PL than the N and HR groups. The LEfSe analysis—using a linear discriminant analysis (LDA) score threshold of 4.0, at p < 0.05—showed that the Rhizobiales occurred only in PL soil (Figure 7), which must somehow be related to its peanut rotation. Logically then, the bacteria belonging to Rhizobiales could not have had a beneficial effect upon C. yanhusuo plants. Bacteroidetes, Bacteroidia, Sphingobacteriales, Gemmatimonadetes, and Gemmatimonadales were biomarkers in the N group, while Acidobacteria, Pyrinomonadaceae, RB41, Pyrinomonadales, and Subgroup_6 were biomarkers in the HR group (Figure 7).
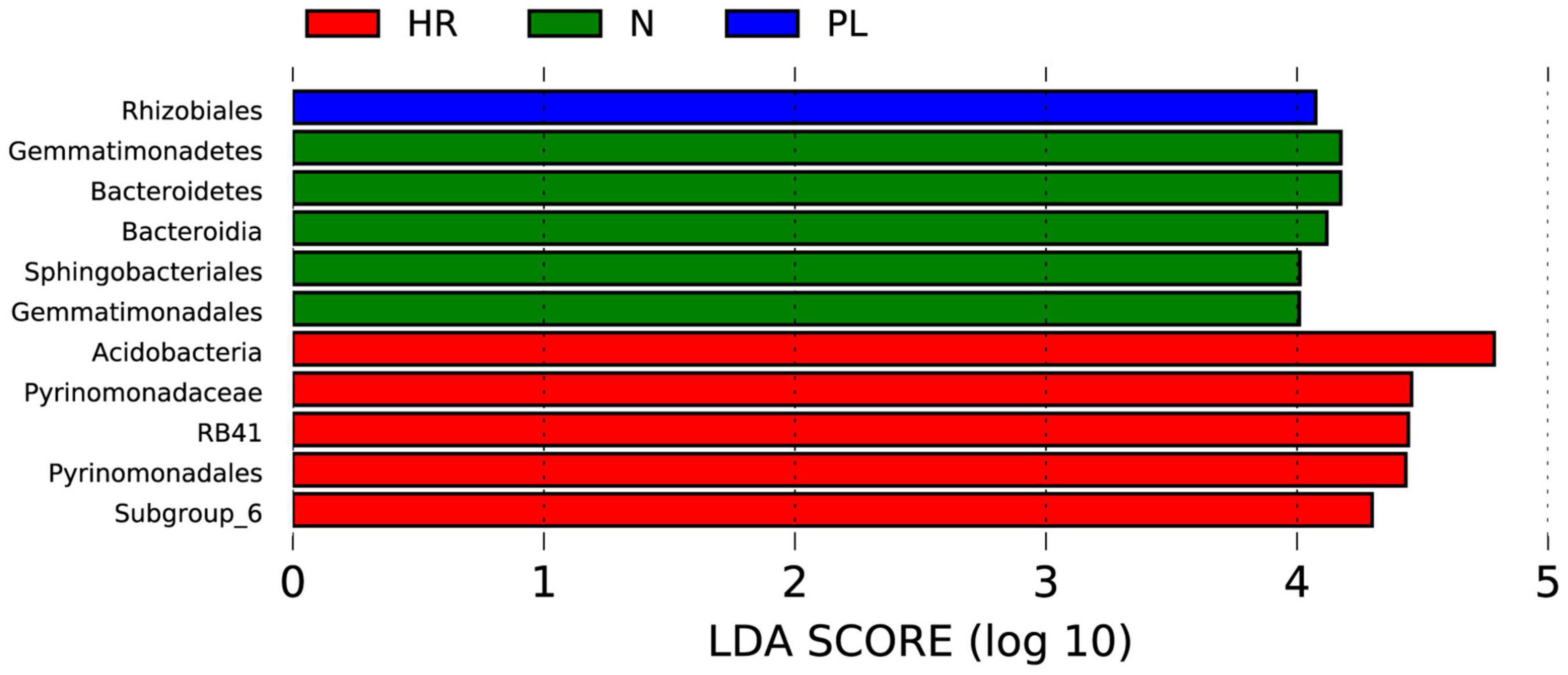
Figure 7. Linear discriminant analysis Effect Size (LEfSe) analysis of microbial abundance in the HR, N, and PL fields. The histogram of linear discriminant analysis (LDA) scores computed for differentially abundant microbes among different samples (these identified at a threshold value of 4.0). N, soil from the C. yanhusuo field without rotation; HR, soil from the C. yanhusuo field that had been flooded in July 2020; PL, soil from field under peanut–C. yanhusuo rotation.
To further assess how changed soil properties might have influenced the functional profiles of rhizospheric bacterial communities in the three different fields, a Tax4Fun analysis was carried out to predict bacterial functioning (Wang X.J. et al., 2020). As seen in Supplementary Figure 5A, more metabolic pathways of bacterial communities in the N and HR fields emerged at level 2, including amino acid metabolism, biosynthesis of other secondary metabolites, replication and repair, signal transduction, glycan biosynthesis and metabolism, cell motility, and translation pathways, among others, but the opposite pattern characterized the PL soil. These prediction results demonstrated that the potential bacterial functions shifted drastically among the N, HR and PL soil samples. The Tax4Fun-predicted significant abundances of carbohydrate metabolism in PL soil when compared with N soil or HR soil (p < 0.05, Welch’s t-test) (Supplementary Figures 5B,D). This suggested that carbohydrate metabolism might play a role in the adaptation of rhizospheric bacteria in the soil with C. yanhusuo-peanut rotation.
When C. yanhusuo plants were grown in the PL field, we found the yield of their tubers and thus medicinal parts were severely decreased when compared to those harvested from plants cultivated in the N field. By contrast, enhanced growth of the C. yanhusuo plants occurred in the HR field, a phenomenon common in southeast China. To explore why disparities in plant growth and tuber yield were observed under the different field conditions, firstly, local soil physicochemical properties were measured, some of which could directly influence plant growth while may indirectly influence by regulating the rhizospheric bacterial community of C. yanhusuo. Secondly, we investigated the bacterial community in the rhizosphere of C. yanhusuo plants cultivated in the different fields. This latter analysis showed significantly higher bacterial diversity in rhizosphere of C. yanhusuo plants from the PL field than the other two fields (Figure 4), with Rhizobiales exclusively enriched in the PL field (Figure 7). It is plausible that the enrichment of Rhizobiales might lead its members to occupy too much of an ecologic niche shared with other bacterial taxa in the rhizosphere, thus limit the growth of other plant-growth promoting microbes. Moreover, the changed soil physicochemical properties were found capable of affecting the composition and diversity of the rhizospheric bacterial community (Figure 6). Thus, this study showed that peanut rotation and flooding induced different alterations in soil physicochemical properties, some of which could further differentially affect the rhizospheric microbiome of C. yanhusuo. Given that the rhizospheric microbiome is known to play an important role in shaping plant phenotypes (Panke-Buisse et al., 2015), its modification likely underpinned the opposite responses observed in plant growth and medicinal yield of C. yanhusuo in the PL and HR fields.
Soil is one of the richest microbial ecosystems on Earth and it serves as a reservoir for myriad colonizers that form plant root-associated microbiomes (Zuo et al., 2021). A previous study estimated that there were more than 2,000 bacterial species in 0.5 g of soil (Wagg et al., 2014). Rhizospheric microbes have close relationships with the roots of plants, colonizing their surface or even their interior; these microbes could benefit plant adaptation and growth in various ways, mainly by secreting hormones, dissolving plant inaccessible nutrients, and resisting phytopathogens (Wang X. et al., 2020). Moreover, it was recently concluded that soil factors are key determinants influencing the rhizobacteriome in multiple stressed environments, including those prone to flooding (Thomas-Barry et al., 2021). Thus, it is reasonable to suppose the contrasting plant performances observed in the PL and HR fields were due to a modified rhizobacteriome as induced by the peanut rotation and flooding, respectively. In recent research, flooding stress was suggested as a factor that may increase bacterial activity and energy metabolism (Mavrodi et al., 2018; Liu et al., 2020), while rotation of crops can differentially impact rhizosphere microbial communities (Peng et al., 2016). Hence, here we speculated that the peanut rotation and flooding each induced specific changes in soil physicochemical properties and then, accordingly, in the rhizobacteriome, whose modification further influenced the growth of C. yanhusuo.
A variety of soil characteristics like clay content, organic carbon content, pH value, water holding capacity, and so forth have been reported to shape the soil microbial community (Zhao J. et al., 2014; Bai et al., 2017). Our results suggest the soils in all the three field types were sandy loam soils according to the soil texture triangle technique developed by the USDA (Supplementary Table 3). We also found that the low SOM content (11.77 ± 0.79 g kg–1) and high CEC (10.41 ± 0.36 cmol kg–1) of the HR soil might indirectly benefit the growth of C. yanhusuo vis-à-vis the other two soils (Table 1). The SOM is determined by the dynamic balance between microbial metabolism and plants’ carbon input (Janzen, 2015). Accordingly, we observed that HR soil had lower bacterial abundance, which might explain its low SOM. Arguably, the interactions between soil properties and rhizospheric bacterial community are bidirectional and complex. The PL soil had a high SOM, TN, and microbial abundance due to the peanut rotation. A significant positive correlation between CEC and bacterial species distribution was observed (Supplementary Figure 4), which implied that variables associated with CEC may be crucial determinants of bacterial community structure in the C. yanhusuo rhizosphere. Recent work has proven that CEC is relatively higher in soils featuring a higher buffering capacity against acidification (Shu et al., 2019; Thomas-Barry et al., 2021), and this is significantly correlated with rhizobacteriome community assembly and bacterial functioning (de Carvalho Mendes et al., 2012). Thus, our findings agree with those of a previous study which suggested CEC is a common global variable for explaining microbial community structure across different terrestrial biomes (Docherty et al., 2015). Moreover, SOM and CEC were each greatest in HR soil, indicating that flooding might have positive effects on both edaphic factors. Moreover, Bastida et al. (2017) reported soil water was a key factor that could impact the bacterial community, which is consistent with our study’s results.
The rhizosphere zone serves as a vital interface for plant-microbe interactions and many physiological and ecological processes occur there, which are essential for plants’ adaption, growth, and development (Mendes et al., 2014; Sohn et al., 2021). Microbes have faster adaptive responses to changing edaphoclimatic conditions and could thereby gain competitive evolutionary advantages because of their shorter life cycles (Waller et al., 2005). As such, the composition and diversity of bacterial community in the rhizosphere of C. yanhusuo was quickly modified in response to the peanut rotation. Specifically, the Sobs, Chao1 (species richness estimators) along with Simpson and Shannon indexes, all increased in PL soil. Another study also reported on changes in the soil bacterial community associated with plant growth and development in response to crop rotation (Panke-Buisse et al., 2015). We also found that the OTU number and Shannon diversity index was much higher for rhizospheric communities of C. yanhusuo than of other medicinal plants, or even peanut (Gao et al., 2019; Wang X.L. et al., 2019). The OTUs and Shannon diversity index of rhizospheric soil would be jointly affected by multiple factors (cultivation site, season, rainfall, soil type, etc.), hence, we speculate that some specific factors existing in the rhizosphere of C. yanhusuo fostered the higher OTU number and Shannon index observed in this study, a view that concurs with another study concerning a high OTU number and Shannon index in bacterial community in the rhizosphere of C. yanhusuo grown under different environmental conditions (Li et al., 2019).
Acidobacteria and Proteobacteria are two dominant phyla present in many soil environments (Clark et al., 2009; Prober et al., 2015). According to our results, the dominant bacterial phyla (i.e., relative abundance > 3%) in the rhizosphere of C. yanhusuo cultivated in the soils from all three field types were Proteobacteria, Acidobacteria, Actinobacteria, Bacteroidetes, Planctomycetes, Chloroflexi, Gemmatimonadetes, Bacteroidetes, and Verrucomicrobia (Figure 3A). It has been shown that bacteria could rapidly adapt to abiotic stresses (heavy metal pollution or flooding), possibly due to their faster metabolism and more extensive substrate utilization (Unger et al., 2009). In our study, three major groups at the phylum level (Proteobacteria, Acidobacteria, and Actinobacteria) were dominant in the bacterial community. Other work suggested the phylum Proteobacteria is prevalent in wetland soils (Ligi et al., 2014). Although the bacterial communities in the rhizosphere of C. yanhusuo grown in the three field types did differ in their abundance of rhizospheric bacterial taxa, we found they were nonetheless concentrated in Proteobacteria, Acidobacteria, and Actinobacteria (Figure 3A). The composition of rhizospheric bacteria communities was not constant among C. yanhusuo plants grown in the different fields. Moreover, the percentage of Acidobacteria decreased in N and PL soils compared to HR soil. Acidobacteria, which are widespread and thrive in nutrient-poor soil, are reportedly able to reduce levels of nitrates and nitrites (Schmidt, 2019). We used the indicator value (IndVal) coefficient analysis developed by Dufrêne and Legendre (1997) to identify the bacterial taxa that significantly contributed to differences among the three field types. According to the indicator value, both Pyrinomonadaceae and Rhodanobacteraceae were significantly abundant in HR soil (Supplementary Figure 6). In a recent study, Pyrinomonadaceae exerted a strong positive influence upon plant development (Obermeier et al., 2020). Thus, we speculate the enrichment of some beneficial bacterial taxa is what fostered better plant growth in the HR field. Furthermore, we are currently conducting studies that focus on screening specific bacteria isolated from the rhizosphere of C. yanhusuo plants cultivated in different fields. Compared with culture-independent (NGS) data, we found that 19.1% of bacterial genera were isolated. This isolation rate is admittedly not high, which could be due to limitations of the isolation method we used. In future research steps planned, we will improve our isolation methods to isolate more bacteria, including anaerobic/microaerophilic bacteria, which escaped detection and thus identification in this study. That work aims to further elucidate and explain the complex mechanistic interactions among the soil environment, rhizospheric microbiome, and plant growth.
Overall, this study reveals that the composition and diversity of the bacterial community dwelling in the rhizosphere of C. yanhusuo is significantly influenced by peanut rotation and flooding, which can further affect this plant’s performance and its eventual medicinal yield. Our results also illustrate how the influences of peanut rotation and flooding upon C. yanhusuo rhizospheric bacterial communities are related to changed soil properties. The soil micro-ecological environment and soil nutrient imbalance may lead to serious declines in the biomass of C. yanhusuo grown in fields with a peanut rotation. In summary, this study has helped to fill knowledge gaps in the rhizospheric bacterial community of C. yanhusuo and it provides a basis for the subsequent mining of microbial functions. Moreover, the findings advance our understanding of rotation effects in C. yanhusuo’s cultivation and could prove useful for developing promising strategies to improve its medicinal yield and thereby enhance its economic benefit to farmers.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA762442.
RW designed the study. XL and PZ performed the sample collection. YF performed the bacteria isolation and identification. SW performed the soil characters measurements. SX and ZZ performed the tetrahydropalmatine content determination. XL performed the statistical analyses and manuscript preparation. JZ revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the National Natural Science Foundation of China (Grant Nos. 32000068, 21806061, and 31800427); High-level Talents Project of Six Talent Peaks in Jiangsu Province (Grant No. NY-081); and the Natural Science Foundation of Jiangsu Province (Grant No. BK20180318).
PZ was employed by the company Jiangsu Jiangtong Agricultural Science and Technology Development.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like thank to Guangzhou Genedenovo Biotechnology Co., Ltd., for assisting in sequencing and bioinformatics analysis.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.779302/full#supplementary-material
Abdallah, I. A., Huang, P., Liu, J., Lee, D. Y., Liu-Chen, L. Y., and Hassan, H. E. (2017). Development and validation of a high performance liquid chromatography quantification method of levo-tetrahydropalmatine and its metabolites in plasma and brain tissues: application to a pharmacokinetic study. Biomed. Chromatogr. 31:e3850. doi: 10.1002/bmc.3850
Aßhauer, K. P., Wemheuer, B., Daniel, R., and Meinicke, P. (2015). Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 31, 2882–2884.
Bai, R., Wang, J. T., Deng, Y., He, J. Z., Feng, K., and Zhang, L. M. (2017). Microbial community and functional structure significantly varied among distinct types of paddy soils but responded differently along gradients of soil depth layers. Front. Microbiol. 8:945. doi: 10.3389/fmicb.2017.00945
Bastida, F., Torres, I. F., Romero-Trigueros, C., Baldrian, P., Vìtrovskı, T., Bayona, J. M., et al. (2017). Combined effects of reduced irrigation and water quality on the soil microbial community of a citrus orchard under semi-arid conditions. Soil. Biol. Biochem. 104, 226–237. doi: 10.1016/j.soilbio.2016.10.024
Bokulich, N. A., Subramanian, S., Faith, J. J., Gevers, D., Gordon, J. I., Knight, R., et al. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57–59. doi: 10.1038/nmeth.2276
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chen, H., and Boutros, P. C. (2011). VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 12:35. doi: 10.1186/1471-2105-12-35
Chen, J. X., Lu, X. G., Lu, C. H., Wang, C. Y., Xu, H. Z., Xu, X. L., et al. (2016). 13-Methyl-palmatrubine induces apoptosis and cell cycle arrest in A549 cells in vitro and in vivo. Oncol. Rep. 36, 2526–2534. doi: 10.3892/or.2016.5093
Chen, S. F., Zhou, Y. Q., Chen, Y. R., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Clark, J. S., Campbell, J. H., Grizzle, H., Acosta-Martìnez, V., and Zak, J. C. (2009). Soil microbial community response to drought and precipitation variability in the Chihuahuan Desert. Microb. Ecol. 57, 248–260. doi: 10.1007/s00248-008-9475-7
Dai, Z. H., Liu, Y. X., Wang, X. J., and Zhao, D. W. (1998). Changes in pH, CEC and exchangeable acidity of some forest soils in Southern China during the last 32-35 years. Water Air Soil Pollut. 108, 377–390. doi: 10.1023/A:1005015526022
de Carvalho Mendes, I., Fernandes, M. F., Chaer, G. M., and Junior, F. B. R. (2012). Biological functioning of Brazilian Cerrado soils under different vegetation types. Plant Soil 359, 183–195. doi: 10.1007/s11104-012-1195-6
Docherty, K. M., Borton, H. M., Espinosa, N., Gebhardt, M., Gil-Loaiza, J., Gutknecht, J. L., et al. (2015). Key edaphic properties largely explain temporal and geographic variation in soil microbial communities across four biomes. PLoS One 10:e0135352. doi: 10.1371/journal.pone.0135352
Dufrêne, M., and Legendre, P. (1997). Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol. Monogr. 67, 345–366.
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr38
Edwards, J., Johnson, C., Santos-Medellín, C., Lurie, E., Podishetty, N. K., Bhatnagar, S., et al. (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. U.S.A. 112, E911–E920. doi: 10.1073/pnas.1414592112
Gao, L., Liu, X. M., Du, Y. M., Zong, H., and Shen, G. M. (2019). Effects of tobacco–peanut relay intercropping on soil bacteria community structure. Ann. Microbiol. 69, 1531–1536. doi: 10.1007/s13213-019-01537-9
Gong, X. K., Yue, K., Ma, B. M., Xing, J., Gan, Y., Wang, D., et al. (2016). Levo-tetrahydropalmatine, a natural, mixed dopamine receptor antagonist, inhibits methamphetamine self-administration and methamphetamine-induced reinstatement. Pharmacol. Biochem. Behav. 144, 67–72. doi: 10.1016/j.pbb.2016.01.010
Hassan, H. E., Kelly, D., Honick, M., Shukla, S., Ibrahim, A., Gorelick, D. A., et al. (2017). Pharmacokinetics and safety assessment of letrahydropalmatine in cocaine users: a randomized, double-blind, placebo-controlled study. J. Clin. Pharmacol. 57, 151–160. doi: 10.1002/jcph.789
He, C., Wang, H., Yu, C., Peng, C., Shu, X., Liao, W. D., et al. (2021). Alterations of gut microbiota in patients with intestinal tuberculosis that different fom crohn’s Disease. Front. Bioeng. Biotechnol. 9:673691. doi: 10.3389/fbioe.2021.673691
Hung, H. Y., and Wu, T. S. (2016). Recent progress on the traditional Chinese medicines that regulate the blood. J. Food Drug Anal. 24, 221–238.
Janzen, H. H. (2015). Beyond carbon sequestration: soil as conduit of solar energy. Eur. J. Soil Sci. 66, 19–32. doi: 10.1111/ejss.12194
Kembel, S. W., Cowan, P. D., Helmus, M. R., Cornwell, W. K., Morlon, H., Ackerly, D. D., et al. (2010). Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464. doi: 10.1093/bioinformatics/btq166
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645.
Li, J., Qiu, D. M., Chen, S. H., Cao, S. P., and Xia, X. L. (2014). Suppression of human breast cancer cell metastasis by coptisine in vitro. Asian Pac. J. Cancer Prev. 15, 5747–5751.
Li, W. C., Sheng, H. Y., Yang, H. M., Liu, Y. Y., and Zhang, R. (2019). Responses of Corydalis yanhusuo rhizosphere soil bacterial community diversity and structure to gap size in a moso bamboo forest. Chinese J. Ecol. 38, 1716–1724.
Ligi, T., Oopkaup, K., Truu, M., Preem, J. K., Nõlvak, H., Mitsch, W. J., et al. (2014). Characterization of bacterial communities in soil and sediment of a created riverine wetland complex using high-throughput 16S rRNA amplicon sequencing. Ecol. Eng. 72, 56–66. doi: 10.1016/j.ecoleng.2013.09.007
Ling, H., Wu, L., and Li, L. (2006). Corydalis yanhusuo rhizoma extract reduces infarct size and improves heart function during myocardial ischemia/reperfusion by inhibiting apoptosis in rats. Phytother. Res. 20, 448–453. doi: 10.1002/ptr.1875
Liu, F. D., Mo, X., Kong, W. J., and Song, Y. (2020). Soil bacterial diversity, structure, and function of Suaeda salsa in rhizosphere and non-rhizosphere soils in various habitats in the Yellow River Delta, China. Sci. Total Environ. 740:140144. doi: 10.1016/j.scitotenv.2020.140144
Liu, L., Liu, M., Zhao, W., Zhao, Y. L., and Wang, Y. (2021). Levo-tetrahydropalmatine: a new potential medication for methamphetamine addiction and neurotoxicity. Exp. Neurol. 344:113809. doi: 10.1016/j.expneurol.2021.113809
Lu, R. K. (2000). Soil Agriculture Chemistry Analytical Methods. Beijing: Chinese Agriculture Science and Technology Press.
Lüneberg, K., Schneider, D., Siebe, C., and Daniel, R. (2018). Drylands soil bacterial community is affected by land use change and different irrigation practices in the Mezquital Valley, Mexico. Sci. Rep. 8:1413. doi: 10.1038/s41598-018-19743-x
Maarastawi, S. A., Frindte, K., Linnartz, M., and Knief, C. (2018). Crop rotation and straw application impact microbial communities in Italian and Philippine soils and the rhizosphere of Zea mays. Front. Microbiol. 9:1295. doi: 10.3389/fmicb.2018.01295
Magoè, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Mavrodi, D. V., Mavrodi, O. V., Elbourne, L. D. H., Tetu, S., Bonsall, R. F., Parejko, J., et al. (2018). Long-term irrigation affects the dynamics and activity of the wheat rhizosphere microbiome. Front. Plant Sci. 9:345. doi: 10.3389/fpls.2018.00345
Mendes, L. W., Kuramae, E. E., Navarrete, A. A., van Veen, J. A., and Tsai, S. M. (2014). Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J. 8, 1577–1587. doi: 10.1038/ismej.2014.17
Obermeier, M. M., Minarsch, E. M. L., Raj, A. C. D., Rineau, F., and Schrder, P. (2020). Changes of soil-rhizosphere microbiota after organic amendment application in a Hordeum vulgare L. short-term greenhouse experiment. Plant Soil 455, 489–506. doi: 10.1007/s11104-020-04637-7
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., O’Hara, R. B., Simpson, G. L., et al. (2010). Vegan: Community Ecology Package. R Package Version 1.17-4.
Panke-Buisse, K., Poole, A. C., Goodrich, J. K., Ley, R. E., and Kao-Kniffin, J. (2015). Selection on soil microbiomes reveals reproducible impacts on plant function. ISME J. 9, 980–989. doi: 10.1038/ismej.2014.196
Peiffer, J. A., Spor, A., Koren, O., Jin, Z., Tringe, S. G., Dangl, J. L., et al. (2013). Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. U.S.A. 110, 6548–6553.
Peng, C. J., Lai, S. S., Luo, X. S., Lu, J. W., Huang, Q. Y., and Chen, W. L. (2016). Effects of long term rice straw application on the microbial communities of rapeseed rhizosphere in a paddy-upland rotation system. Sci. Total. Environ. 556-557, 231–239.
Philippot, L., Raaijmakers, J. M., Lemanceau, P., and van der Putten, W. H. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. doi: 10.1038/nrmicro3109
Prober, S. M., Leff, J. W., Bates, S. T., Borer, E. T., Firn, J., Harpole, W. S., et al. (2015). Plant diversity predicts beta but not alpha diversity of soil microbes across grasslands worldwide. Ecol. Lett. 18, 85–95. doi: 10.1111/ele.12381
Pruesse, E., Quast, C., Knittel, K., Fuchs, B. M., Ludwig, W., Peplies, J., et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196. doi: 10.1093/nar/gkm864
Schmidt, T. M. (2019). “Acidobacteria,” in Encyclopedia of Microbiology, Fourth Edn, eds E. E. Kuramae and O. Y. de Assis Costa (Cambridge, MA: Academic Press), 1–8.
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:1. doi: 10.1186/gb-2011-12-6-r60
Shu, X., Zhang, K. R., Zhang, Q. F., and Wang, W. B. (2019). Ecophysiological responses of Jatropha curcas L. seedlings to simulated acid rain under different soil types. Ecotoxicol. Environ. Saf. 185:109705. doi: 10.1016/j.ecoenv.2019.109705
Sohn, S. I., Ahn, J. H., Pandian, S., Oh, Y. J., Shin, E. K., Kang, H. J., et al. (2021). Dynamics of bacterial community structure in the rhizosphere and root nodule of soybean: Impacts of growth stages and varieties. Int. J. Mol. Sci. 22:5577. doi: 10.3390/ijms22115577
Sun, C. L., Liu, G. B., and Xue, S. (2018). Interaction between plant competition and rhizospheric bacterial community influence secondary succession of abandoned farmland on the Loess Plateau of China. Front. Plant Sci. 9:898. doi: 10.3389/fpls.2018.00898
Tang, F. Y., and Nie, A. G. (2006). Overview of studies on Corydalis yanhusuo. J. Clin. Exp. Med. 5, 185–186.
Thomas-Barry, G., St Martin, C. C. G., Lynch, M. D. J., Ramsubhag, A., Rouse-Miller, J., and Charles, T. C. (2021). Driving factors influencing the rhizobacteriome community structure of plants adapted to multiple climatic stressors in edaphic savannas. Sci. Total. Environ. 769:145214. doi: 10.1016/j.scitotenv.2021.145214
Tian, B., Tian, M., and Huang, S. M. (2020). Advances in phytochemical and modern pharmacological research of Rhizoma Corydalis. Pharm. Biol. 58, 265–275.
Trivedi, P., Leach, J. E., Tringe, S. G., Sa, T., and Singh, B. K. (2020). Plant-microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol. 18, 607–621. doi: 10.1038/s41579-020-0412-1
Unger, I. M., Kennedy, A. C., and Muzika, R. M. (2009). Flooding effects on soil microbial communities. Appl. Soil Ecol. 42, 1–8.
Wagg, C., Bender, S. F., Widmer, F., and van der Heijden, M. G. A. (2014). Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proc. Natl. Acad. Sci. U.S.A. 111, 5266–5270.
Waller, F., Achatz, B., Baltruschat, H., Fodor, J., Becker, K., Fischer, M., et al. (2005). The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. Proc. Natl. Acad. Sci. U. S. A. 102, 13386–13391. doi: 10.1073/pnas.0504423102
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang, X., Whalley, W. R., Miller, A. J., White, P. J., Zhang, F. S., and Shen, J. B. (2020). Sustainable cropping requires adaptation to a heterogeneous rhizosphere. Trends Plant Sci. 25, 1194–1202. doi: 10.1016/j.tplants.2020.07.006
Wang, X. J., Zhang, Z. C., Yu, Z. Q., Shen, G. F., Cheng, H. F., and Tao, S. (2020). Composition and diversity of soil microbial communities in the alpine wetland and alpine forest ecosystems on the Tibetan Plateau. Sci. Total Environ. 747:141358. doi: 10.1016/j.scitotenv.2020.141358
Wang, X. L., Wang, Z. K., Jiang, P., He, Y. L., Mu, Y. D., Lv, X. H., et al. (2019). Bacterial diversity and community structure in the rhizosphere of four Ferula species. Sci. Rep. 8:5345. doi: 10.1038/s41598-018-22802-y
Wang, Y. M., Chen, X. X., Yu, B., Jin, Y. H., Zhang, C. C., Li, S. Q., et al. (2019). Effects of different cropping patterns on the yield and quality formation of Corydalis Rhizoma. Chin. J. Mod. Appl. Pharm. 36, 1042–1049. doi: 10.13748/j.cnki.issn1007-7693.2019.09.003
Wu, J., Xiao, Q., Zhang, N., Xue, C., Leung, A. W., Zhang, H., et al. (2016). Palmatine hydrochloride mediated photodynamic inactivation of breast cancer MCF-7 cells: effectiveness and mechanism of action. Photodiagn. Photodyn. Ther. 15, 133–138.
Xiao, H. T., Peng, J., Liang, Y., Yang, J., Bai, X., Hao, X. Y., et al. (2011). Acetylcholinesterase inhibitors from Corydalis yanhusuo. Nat. Prod. Res. 25, 1418–1422. doi: 10.1080/14786410802496911
Xiao, S. Z., Xiong, K. N., Lan, J. C., Zhang, H., and Yang, L. (2015). Impact of rocky desertification treatment on underground water chemistry and dissolved inorganic carbon isotope in karst areas. Huan Jing Ke Xue 36, 1590–1597. doi: 10.13227/j.hjkx.2015.05.011
Xu, C. M. (2005). Studies on Technique System of Yanhusuo Cultivating for High Quality and Yield. Master Thesis. Hangzhou: Zhejiang University.
Yu, J., Zhao, L., Liu, L., Yang, F., Zhu, X., and Cao, B. (2016). Tetrahydropalmatine protects rat pulmonary endothelial cells from irradiation-induced apoptosis by inhibiting oxidative stress and the calcium sensing receptor/phospholipase C-γ1 pathway. Free Radic. Res. 50, 611–626. doi: 10.3109/10715762.2016.1154549
Zhao, J., Zhang, R. F., Xue, C., Xun, W. B., Sun, L., Xu, Y. C., et al. (2014). Pyrosequencing reveals contrasting soil bacterial diversity and community structure of two main winter wheat cropping systems in China. Microb. Ecol. 67, 443–453. doi: 10.1007/s00248-013-0322-0
Zhao, Y., Gao, J. L., Ji, J. W., Gao, M., Yin, Q. S., Qiu, Q. L., et al. (2014). Cytotoxicity enhancement in MDA-MB-231 cells by the combination treatment of tetrahydropalmatine and berberine derived from Corydalis yanhusuo W. T. Wang. J Intercult. Ethnopharmacol. 3, 68–72. doi: 10.5455/jice.20140123040224
Zhao, Q. G. (1981). Physical and Chemical Analysis of Soil. Shanghai: Shanghai Science and Technology Press.
Keywords: Corydalis yanhusuo, flooding, medicinal yield, peanut rotation, rhizosphere, 16S rRNA amplicon sequencing
Citation: Li X, Wang S, Fan Y, Zhou Z, Xu S, Zhou P, Zhou J and Wang R (2022) Peanut Rotation and Flooding Induce Rhizobacteriome Variation With Opposing Influences on the Growth and Medicinal Yield of Corydalis yanhusuo. Front. Plant Sci. 12:779302. doi: 10.3389/fpls.2021.779302
Received: 21 September 2021; Accepted: 16 December 2021;
Published: 07 January 2022.
Edited by:
Katharina Pawlowski, Stockholm University, SwedenReviewed by:
Hugo Cesar Ramirez-Saad, Autonomous Metropolitan University, MexicoCopyright © 2022 Li, Wang, Fan, Zhou, Xu, Zhou, Zhou and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiayu Zhou, emhvdWppYXl1QGNuYmcubmV0; Ren Wang, cndhbmdAY25iZy5uZXQ=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.