
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 05 January 2022
Sec. Plant Breeding
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.778008
This article is part of the Research TopicMolecular Insights in Plant Reproductive Isolation BarriersView all 6 articles
 Sun Ha Kim1†
Sun Ha Kim1† Shi-Dong Ji1†‡
Shi-Dong Ji1†‡ Hyun-Sook Lee1‡
Hyun-Sook Lee1‡ Yun-A Jeon1
Yun-A Jeon1 Kyu-Chan Shim1
Kyu-Chan Shim1 Cheryl Adeva1
Cheryl Adeva1 Ngoc Ha Luong1
Ngoc Ha Luong1 Pingrong Yuan1‡
Pingrong Yuan1‡ Hyun-Jung Kim2
Hyun-Jung Kim2 Thomas H. Tai3,4
Thomas H. Tai3,4 Sang-Nag Ahn1*
Sang-Nag Ahn1*A novel hybrid weakness gene, DTE9, associated with a dark tip embryo (DTE) trait, was observed in CR6078, an introgression line derived from a cross between the Oryza sativa spp. japonica “Hwayeong” (HY) and the wild relative Oryza rufipogon. CR6078 seeds exhibit protruding embryos and flowers have altered inner floral organs. DTE9 was also associated with several hybrid weakness symptoms including decreased grain weight. Map-based cloning and transgenic approaches revealed that DTE9 is an allele of OsMADS8, a MADS-domain transcription factor. Genetic analysis indicated that two recessive complementary genes were responsible for the expression of the DTE trait. No sequence differences were observed between the two parental lines in the OsMADS8 coding region; however, numerous single nucleotide polymorphisms were detected in the promoter and intronic regions. We generated overexpression (OX) and RNA interference (RNAi) transgenic lines of OsMADS8 in HY and CR6078, respectively. The OsMADS8-OX lines showed the dark tip embryo phenotype, whereas OsMADS8-RNAi recovered the normal embryo phenotype. Changes in gene expression, including of ABCDE floral homeotic genes, were observed in the OsMADS8-OX and OsMADS8-RNAi lines. Overexpression of OsMADS8 led to decreased expression of OsEMF2b and ABA signaling-related genes including OsVP1/ABI3. HY seeds showed higher ABA content than CR6078 seeds, consistent with OsMADS8/DTE9 regulating the expression of genes related ABA catabolism in CR6078. Our results suggest that OsMADS8 is critical for floral organ determination and seed germination and that these effects are the result of regulation of the expression of OsEMF2b and its role in ABA signaling and catabolism.
Hybrid weakness (HW) or hybrid breakdown is a postembryonic barrier in the F2 or later generations of interspecific or intraspecific crosses (Stebbins, 1950). In rice, F1 hybrids between indica and japonica display various reproductive barriers including hybrid sterility (Oka, 1974; Ikehashi and Araki, 1986) and hybrid weakness (Oka, 1957; Sato and Morishima, 1987). Hybrid weakness, a form of hybrid incompatibility during the postembryonic stage of plant development, is frequently observed in many other plant species including Arabidopsis thaliana (Bomblies et al., 2007). Hybrid weakness manifests in rice through characteristic dwarfing, chlorotic phenotype, stunted growth, and poor seed setting (Ichitani et al., 2011; Chen et al., 2013). Nearly all studies have reported that hybrid weakness is controlled by the complementary interaction of unlinked loci (Fukuoka et al., 1998, 2005; Kubo and Yoshimura, 2002; Matsubara et al., 2007; Yamamoto et al., 2007). Recently, three causal genes for hybrid weakness at two loci, Hwi1 and Hwi2, have been cloned in rice (Chen et al., 2014). Hwi1, a locus with two leucine-rich repeat receptor-like kinase (LRR-RLK) genes, induces hybrid weakness while Hwi2 encodes a secreted putative subtilisin-like protease. When combined, Hwi1 and Hwi2 activate autoimmune responses in the basal nodes of hybrids, interrupting root formation and impairing shoot growth. Although hybrid weakness is associated with abnormal growth and development, there have been no reports indicating a role of plant hormones in regulating this disorder in rice.
In angiosperms, the flowers are typically comprised of sepals, petals, stamens, and pistils arranged in concentric whorls. The concept of whorls underlies the classical ABC model of floral organ development and, subsequently, the ABCDE model (Coen and Meyerowitz, 1991; Gutierrez-Cortines and Davies, 2000; Jeon et al., 2000; Honma and Goto, 2001; Lee et al., 2003; Yoshida and Nagato, 2011; Murai, 2013; Callens et al., 2018; Ali et al., 2019; Supplementary Figure S1). According to the ABC model, class A, B, and C genes together specify the identity of sepals, petals, stamens, and carpels (Angenent and Colombo, 1996; Pelaz et al., 2000). Since the early 2000s, many studies have reported that two additional classes of floral homeotic genes, D and E (SEPALLATA, SEP), are also necessary for specifying floral organ identity (Pelaz et al., 2000; Murai, 2013; Song et al., 2018). While the class D genes are crucial for ovule development, class E genes are required for specifying all floral organs. Other classes of genes involved in flower development include FRIZZY PANICLE (FZP), SUPERNUMERARY BRACT (SNB), MULTI-FLORET SPIKELET1 (MFS1), and APETALA2/ethylene-responsive factor (AP2/ERF) gene families (Komatsu et al., 2003; Lee et al., 2007a; Bai et al., 2016; Pasriga et al., 2019).
Most of the genes in the ABCDE model encode the MIKCC-type MADS-domain transcription factors (Riechmann and Meyerowitz, 1997; Egea-Cortines et al., 1999; Becker et al., 2000; Honma and Goto, 2001; Theissen, 2001). In rice, there are five different SEP-like genes (Arora et al., 2007): OsMADS1 (Jeon et al., 2000; Chen et al., 2006), OsMADS5 and OsMADS34 (also called OsMADS19) are likely members of the SEP1/2/4 clade of genes, and OsMADS7 and OsMADS8 are SEP3-like genes (Kang and An, 1997; Zahn et al., 2005). The functions of the monocot SEP-like genes are unknown, except for OsMADS1 (Agrawal et al., 2005; Prasad et al., 2005).
Several genes that are not classified as a family of MADS genes are related to determining floral organ identity. DROOPING LEAF (DL), a member of the YABBY gene family, controls the lemma development and carpel specification in rice (Nagasawa et al., 2003; Ishikawa et al., 2009; Li et al., 2011). OPEN BEAK (OPB) and DEGENERATED HULL1 (DH1) are required for the normal lemma and palea development (Horigome et al., 2009; Xiao et al., 2009). In Arabidopsis, the polycomb repressive complex 2 (PRC2) is known to regulate flower development through the inhibition of expression of the C function flower homologous gene, AGAMOUS (AG; Goodrich et al., 1997). The loss of either of the PRC2 components or EMBRYONIC FLOWER 2 (EMF2) results in a mild floral phenotype in Arabidopsis (Deng et al., 2017). Although the function of plant PRC2 is well-characterized in Arabidopsis, its specific function in other plant species remains unknown. Conrad et al. (2014) confirmed through transcriptome analysis that the E-function genes OsMADS1, OsMADS6, and OsMADS34 were differentially expressed in wild-type and emf2b mutants.
It is well known that abscisic acid (ABA) promotes seed dormancy and suppresses seed germination and seedling growth. Chen et al. (2017) reported that OsEMF2b regulates seed dormancy and seedling growth by activating or inhibiting the expression of ABA signaling response genes. Downregulation of OsEMF2b expression leads to vivipary and a decrease in the expression level of OsVP1, an ABI3 ortholog in rice. ABI3, a component of the ABA signal transduction pathway, plays a major role in seed development and dormancy inception. Disruption of the OsVP1 transcription reduces the seed dormancy or viability (Chen et al., 2017).
In this study, a novel interspecific hybrid weakness gene, dark tip embryo (DTE9), was discovered in CR6078, an introgression line derived from a cross between the Oryza sativa spp. japonica “Hwayeong” (HY) and the wild relative Oryza rufipogon. In addition to presenting the typical symptoms of hybrid weakness in rice, such as dwarfing, plants exhibiting the DTE trait showed a significant association with several deleterious traits including the ripening ratio (i.e., fertility). We characterized the phenotypic and physiological properties of DTE as a newly discovered hybrid weakness. The genetic locus of DTE was analyzed, and the gene responsible for this phenotype was fine mapped within a 27.7 kb region on chromosome nine containing two candidate genes. Gene expression and transgenic analysis indicated that OsMADS8, a MADS-domain transcription factor, controls the DTE phenotype by affecting the expression of class A, B, C, D, and E floral homeotic genes as well as viviparous genes related to ABA biosynthesis or signaling.
Previously, an introgression line population was developed from a cross between “Hwayeong” (O. sativa ssp. japonica), a Korean elite rice cultivar, and W1944 (O. rufipogon Griff.), a wild relative of cultivated rice from China (Lee et al., 2005). A DTE phenotype was observed in a line, CR6078 (BC3F3), which contained eight W1944 chromosome segments in the Hwayeong (HY) genetic background (Supplementary Figures S2, S3). CR6078 was backcrossed to HY, and the BC1 plants were self-pollinated to produce 204 BC4F2:3 lines. Preliminary mapping with eight simple sequence repeat (SSR) markers, one per W1944 segment, indicated that the gene for the DTE trait was located on chromosome 9 (designated DTE9) between RM553 and RM24637. Six BC4F3 plants that were heterozygous for the target region at RM553 and RM24637 were self-pollinated to produce a BC4F4 population for fine mapping DTE9. In total, 4,500 BC4F4 plants were genotyped with RM553 and RM24637, and 50 recombinants between the two markers were detected. These 50 BC4F4:5 recombinant lines were used to narrow down the location of the gene for DTE9 by substitution mapping.
A total of 204 BC4F3, 4,500 BC4F4 plants, 50 BC4F4 recombinant lines, and the parents (HY and CR6078) were grown at the experimental field of Chungnam National University, Daejeon during the summers of 2008, 2009, and 2010, respectively. The 204 BC4F3 and 4,500 BC4F4 plants were planted at 15 cm intervals within and 30 cm intervals between the rows. The 50 BC4F4 lines were represented by a single row of 30-day-old seedlings planted at an interval of 15 cm within and 30 cm between the rows. The BC4F3 population was evaluated for ripening ratio (RR), 1,000-grain weight (TGW), and grain weight per plant (GW). RR, a measure of fertility, was calculated as a percentage of the number of filled spikelets divided by the number of spikelets per panicle. The number of spikelets per panicle was measured by averaging the values of three major panicles per plant. The grains were dried naturally after harvesting, and the partially filled or unfilled seeds were removed with water (i.e., density separation). The filled seeds were re-dried in an oven at 30°C for 24 h. The TGW was evaluated by measuring the weight of 100 randomly selected filled grains averaged over two replications. The grain yield per plant (GW) was evaluated by averaging the grain yield (g) at 13% moisture content of five plants.
The germination rate, measured as a percentage, was compared among the seeds of HY, W1944, CR6078, and OsMADS8 transgenic plants. Germination tests were conducted as described by Nguyen et al. (2012) and Shim et al. (2019) with some modifications. Seeds were collected at 45 days after flowering. After breaking the seed dormancy at 50°C for 5 days, two sets of 30 seeds per line were germinated at 30°C under dark condition. Germination tests were replicated three times. Three panicles of HY and CR6078 were sampled at 1 or 2 days before flowering to determine pollen viability. Pollen collection and staining were carried out as described by Jiang et al. (2018). The anthers were crushed into powder and used to observe fertility with a light microscope. Pollen grains that were round and stained black were considered to be viable.
DNA extraction from the leaf tissues, PCR amplification of SSR markers, size separation using polyacrylamide gel electrophoresis, and marker detection by silver staining were performed as described by Shim et al. (2019). Silver staining kits were purchased from Bioneer Co., Daejeon, Korea.1 The linkage and orientation of the SSR markers were based on SSR maps (McCouch et al., 2002). Using the publicly available sequences for Nipponbare2 and 93-113 reference genomes, insertion and deletion markers were developed using the PRIMER 3.0 program (Supplementary Table S1). The quantitative trait loci (QTL) were fine mapped by comparing the phenotypic means of the genotypic classes of recombinants within the target region using the analysis of variance (ANOVA) feature in the Data Desk 4.0 and based on the interval analysis (p ≤ 0.01 and/or LOD ≥ 3.0) for the markers within the target region using QGENE (Nelson, 1997).
RNA was isolated from the seeds of the three lines (HY, W1944, and CR6078) and 7-day-old seedlings of transgenic plants using a NucleoSpin RNA kit (Macherey Nagel, Duren, Germany) according to the manufacturer’s instructions. Following reverse transcription into the first-strand cDNA with SmartGene Mixed cDNA synthesis kit (SJ Bioscience, Daejeon, Korea), real-time PCR was performed using a CFX Connect Real-Time System (Bio-Rad, Hercules, CA, United States). The real-time PCR protocol was as follows: 15 min at 95°C denaturation and enzyme activation, followed by 40 cycles at 95°C for 20 s, 60–55°C for 40 s (depending on the primer annealing temperature), and 72°C for 30 s. The rice tumor protein homolog (LOC_Os11g43900, OsTMP) was used for normalization, and the relative expression levels were compared using Student’s t-test and Tukey’s test. The primers used in this study are listed in Supplementary Table S1.
To clone the full-length cDNA of OsMADS8 gene from W1944, a pair of gene-specific primer (Forward: 5'-ATGGGGAGAGGGAGGGTGG-3' and Reverse: 5'-GGGGTAG CCATGTCGGCATG-3'), was synthesized based on the sequence of Nipponbare OsMADS8 gene from the RAP-DB.4 The PCR product was cloned into the pGEM-T Easy vector (Promega, Madison, WI, United States) and was sequenced to confirm fidelity. To construct the plant expression vectors, we used the Gateway cloning system (Invitrogen, Carlsbad, CA, United States). The PCR products were used for the second PCR reaction with primers containing attB sites (5'-GTACAAAAAAGCAGGCTATGGGGAGAGGGAGGGTGG-3') and (5'-ACCACTTTGTACAAGAAAGC TGGGTCGGGGGTAGCCATGTCGGCATG-3'). The linear fragments flanked by the attB sequences were subjected to site-specific recombination with the entry vector, pDONR207 (Invitrogen), which contains the ccdB gene flanked by attP sites; the reactions catalyzed using BP Clonase yielded entry clones that were used to transform competent DH5a (Escherichia coli) cells. Using LR Clonase enzyme mix (Invitrogen), the pDONR207- OsMADS8 clones were subjected to site-specific recombination with the plant expression vector pGWB5, which contains the cauliflower mosaic virus 35S promoter and the C-terminal GFP epitope. The resulting expression constructs (OsMADS8-OX) were transformed into the Agrobacterium tumefaciens strain GV3101 using the freeze-thaw method (Hofgen and Willmitzer, 1988).
To construct the OsMADS8 RNAi vectors, the OsMADS8-specific primers were designed from the sequence of OsMADS8, containing the following EcoRI and XhoI restriction sites: 5' -GGATCCCAGGCAGGTGACGTTCGCG-3' and 5'-GAGCTCCTTTTATGCCAAGTGTCCC-3'. The PCR product was digested with EcoRI and XhoI and ligated into the pENTR11 vector (Invitrogen). The pENTR11-OsMADS8 clones were then subjected to site-specific recombination with a plant RNAi expression vector, pH7GWIWG2(I), together with the cauliflower mosaic virus 35S promoter (Kim et al., 2012) using LR Clonase enzyme mix (Invitrogen). The resulting expression constructs (OsMADS8-RNAi) were transformed into the A. tumefaciens strain GV3101 using the freeze-thaw method.
The overexpression (OX) and RNA interference (RNAi) constructs were constructed and transformed into HY (hereafter OsMADS8-OX) and CR6078 (hereafter OsMADS8-RNAi), respectively, using the piercing and vacuum infiltration method (Lin et al., 2009).
To obtain embryo sections, seeds of the three lines (HY, W1944, and CR6078) and transgenic plants were incubated in distilled water for 4 h at 4°C. Embryos were manually thin-sectioned in a petri dish using a razor blade (DORCO, Seoul, Korea). The sections (9–12 μm thick) were stained with Perls’ Prussian blue (2% hydrochloric acid mixed with 2% potassium ferrocyanide) for 10 min followed by gently washing with distilled water for 2 min. Imaging was performed with an Olympus BX40 microscope and a Carl Zeiss AxioCam MRc 5 digital camera operated by the Zeiss AxioVision AC system.
The sequence of DTE9 was compared among 137 Asian cultivated rice accessions (O. sativa L.) from the KRICE_CORE (Kim et al., 2016) and sequence variant data were provided by Kongju National University. This mini core collection is comprised of 81 japonica, 43 indica, seven aus, four admixture, and two aromatic accessions (Supplementary Table S2). To compare the OsMADS8 sequence between W1944 and W1943, a putative ancestor of japonica rice (Huang et al., 2012), the 9-kb upstream and downstream of OsMADS8 genomic sequence of W1943 was downloaded from the EnsemblPlants database.5
The ABA content was measured using a Phytodetek® ABA Kit for ABA (Agdia, Elkhart, IN, United States) according to the manufacturer’s instructions. For this ELISA-based assay, seeds were ground in liquid nitrogen, and ABA was extracted in 80% acetone for 24 h at −20°C under dark condition. The supernatant was collected after centrifugation at 13,475 g for 10 min and diluted 10-fold with 1× TBS (Trizma Base 3.03 g/L, NaCl 5.84 g/L, magnesium chloride hexahydrate 0.20 g/L, NaN3 0.20 g/L) for use with the Phytodetek® kit. The absorbance was detected at 405 nm using a Multiskan SkyHigh microplate spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States). Each assay was performed in duplicate.
The data were analyzed using a one-way ANOVA. Tukey’s test was used to compare the mean values. All the statistical analyses were performed using the Statistical Package for the Social Sciences (SPSS 12; SPSS Inc., Chicago, IL, United States), and statistical significance was set at either p < 0.05, p < 0.01, or p < 0.001.
Dark tip embryos were observed in the seeds of a BC3F3 introgression line CR6078, which originated from a cross between HY and the O. rufipogon accession W1944 (Figure 1A). CR6078 showed a significantly higher occurrence of DTE (approximately 42%) compared to the parental lines HY (approximately 1%) and W1944 (approximately 0%; Figure 1B). CR6078 seeds were intermediate in shape and size compared to HY and W1944 and had protruding embryos.
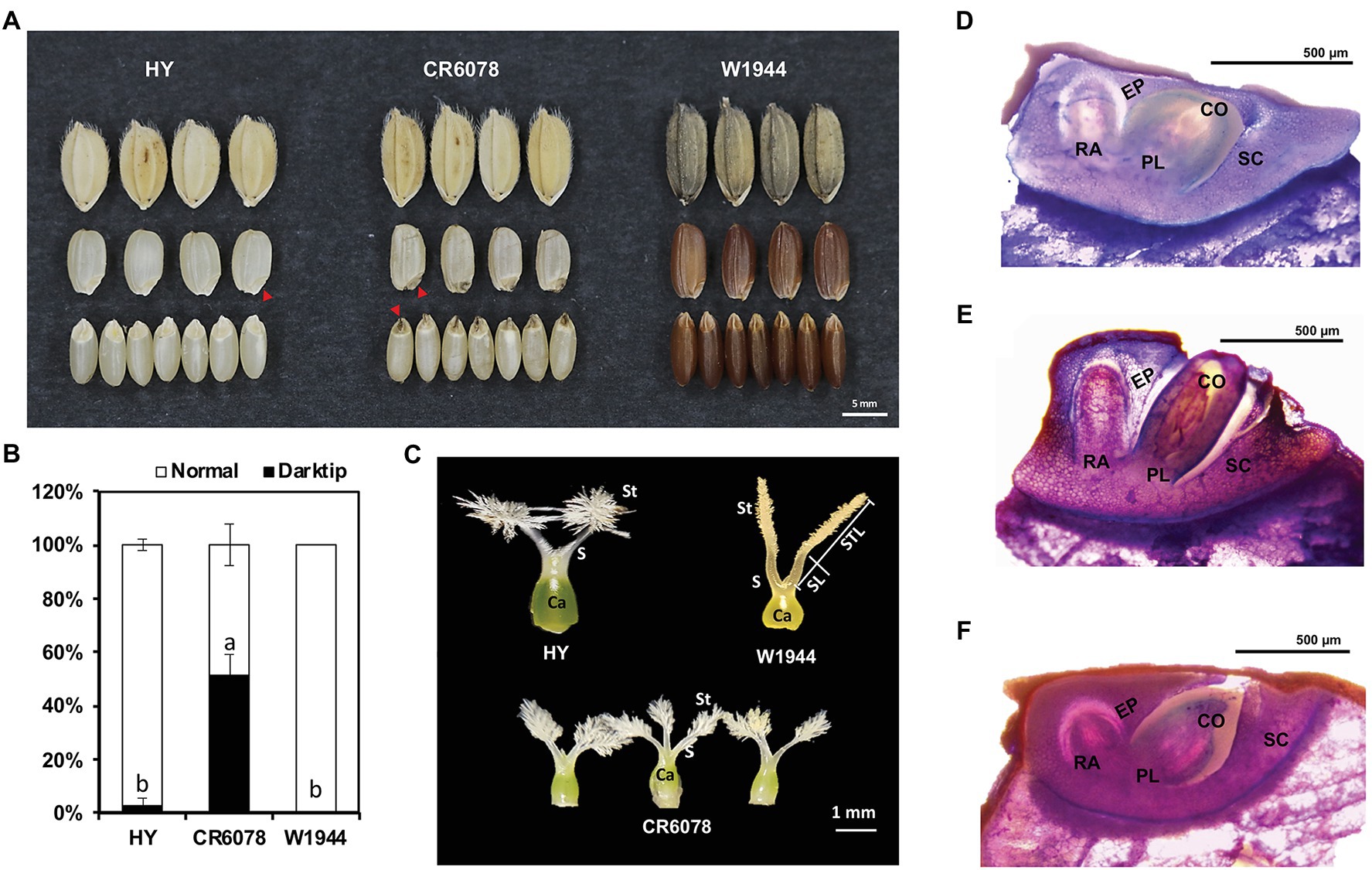
Figure 1. Phenotypic differences among HY, W1944, and CR6078. (A) Seed morphology. Red arrowhead indicates the position of dark tip embryo (DTE) in CR6078. (B) Comparison of the dark tip embryo rate (%). (C) Characterization of the pistil phenotype in HY, W1944, and CR6078. The normal pistil with one carpel (Ca) and two stigmas (St) in HY and W1944. The aberrant pistils of CR6078. One carpel with three or four stigmas. (D–F) Section analysis. The seed longitudinal section was stained with 0.05% toluidine blue to compare the embryo shape of HY (D), CR6078 (E), and W1944 (F). Ca, carpel; St, stigma; S, style; CO, coleoptile; PL, plumule; RA, radicle; SC, scutellum; EP, epidermis; STL, stigma length; SL, style length.
Differences in the number and morphology of the floral organs among the three accessions were also observed. CR6078 had one carpel with three or four stigmas, whereas HY and W1944 had normal pistils with one carpel and two stigmas (Figure 1C). We investigated the stigma characteristics of these three accessions. W1944 had longer stigmas than HY and CR6078, which is consistent with the finding that eight accessions of O. rufipogon and four of O. nivara had CC alleles at OsSYL2 and OsSYL3, contributing to an increase in the stigma length in these wild Oryza species (Dang et al., 2020). Histological analysis of thin sections from these CR6078 embryos showed a cleavage between the coleoptile and epidermis without any tissue covering the coleoptile (Figure 1E). Cleavages were not observed and the coleoptiles were covered by the surrounding tissues in both HY and W1944 (Figures 1D,F). Another cleavage was observed between the coleoptile and scutellum in CR6078, and the scutellum on the radicle side was swollen in CR6078 (Figure 1E). These cleavages suggest that CR6078 seeds initiate a precocious germination process with coleoptile protrusion, but germination is arrested perhaps because of environmental conditions, such as water deficit in the panicle.
For genetic analysis of the DTE trait, CR6078 was crossed with HY, and the F1 plants (BC4F1) were self-pollinated to produce BC4F2 seeds. All the BC4F2 seeds had the HY phenotype, suggesting that DTE was present in the maternal tissue. In the BC4F3 population (204 plants), a segregation of 3 (normal): 1 (DTE) was observed, suggesting that DTE is controlled by a single recessive gene in this population from the HY/CR6078 cross (Table 1). However, the results did not explain the finding that DTE was only observed in CR6078 but not in the two parents.
To better understand the genetics of DTE, a second cross was made between an indica rice cultivar Milyang23 and CR6078. A total of 370 F2 plants were grown, and F3 seeds were obtained from each F2 plant. All the F2 seeds were Milyang23 type, and the F3 population segregated 347 normal and 23 DTE plants, suggesting that DTE was controlled by two recessive complementary genes (Table 1). When more than 300 F3 plants from the cross between HY and Milyang23 were screened, no F3 line with the DTE phenotype higher than 1% was detected. The results suggested that two or more non-allelic genes regulate the DTE trait, which is not detected in the parents, and that the W1944 allele is required to display DTE.
The pollen viability of HY and CR6078 was also evaluated. The pollen grains of HY and CR6078 were uniform in size and shape and approximately 90% were viable, indicating that the low fertility was not associated with pollen viability of CR6078 (Supplementary Figure S4).
Fifteen SSR markers on the chromosomes 1, 2, 5, 7, 8, 9, and 11 detected W1944 segments in CR6078 (Supplementary Figure S2). A BC4F3 population of 204 individuals derived from a cross between HY and CR6078 was evaluated for DTE and some agronomic traits, such as TGW (g), RR (%), and GW (g) per plant. The frequency distribution of the DTE trait in the BC4F3 population followed a Mendelian ratio of 3 normal:1 DTE (χ2 = 0.53, 0.1 < p < 0.5), indicating that this trait is controlled by a single recessive gene. The gene was mapped to the interval between RM553 and RM24637 on chromosome 9 and designated DTE9 (Figure 2A). The frequency distribution of TGW, RR, and GW per plant, also followed a 3:1 segregation ratio (Table 2). The single-point analysis showed that the QTL for all four traits were linked to DTE9. These results suggest that DTE9OR (O. rufipogon W1944 allele) has a pleiotropic effect (Table 2).
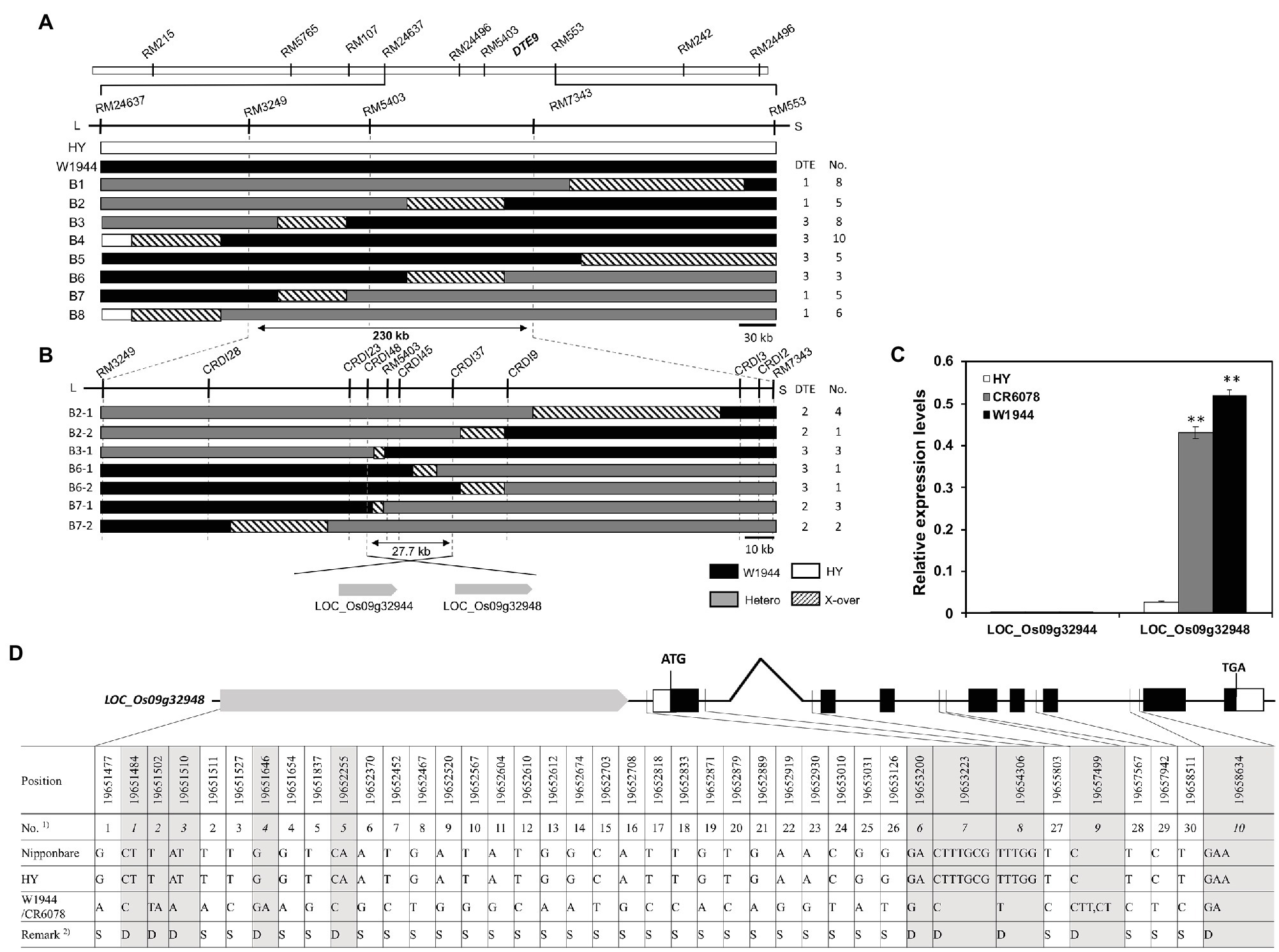
Figure 2. Graphical genotypes and substitution mapping of DTE9. (A) Substitution mapping of DTE9OR using F5 lines. Comparison of the genotypes of lines delimiting DTE9OR between the markers RM3249 and RM7343. Physical distance is about 230-kb based on the Nipponbare sequence. (B) High-density mapping of DTE9. Comparison of the genotypic recombinants delimiting DTE9 between the markers CRDI37 and CRDI48. White, black, and gray bars indicate homozygous and heterozygous alleles for HY and W1944 and hatched areas indicate regions where the crossing-over occurred on chromosome 9. Two genes were predicted in the target 27.7 kb region. (C) Expression levels of the DTE9 candidate genes in the three cultivars. Total RNA was isolated from 7-day-old seedlings of the three lines. Relative expression level of each gene was quantified using real-time PCR and normalized using OsTMP1 as an internal control. The relative transcript levels of DTE9-involved candidate genes (LOC_Os09g32944, LOC_Os09g32948). The significant differences between the HY and CR6078 plants and the HY and W1944 at **p < 0.01. (D) Comparison of promoter and genomic sequence of OsMASD8 in HY and W1944. A total of 30 single nucleotide polymorphisms (SNPs) and 10 InDels was identified between HY and W1944. 1)SNP and InDel are separately numbered in bold and italics, respectively. 2)S: SNP, D: InDel.
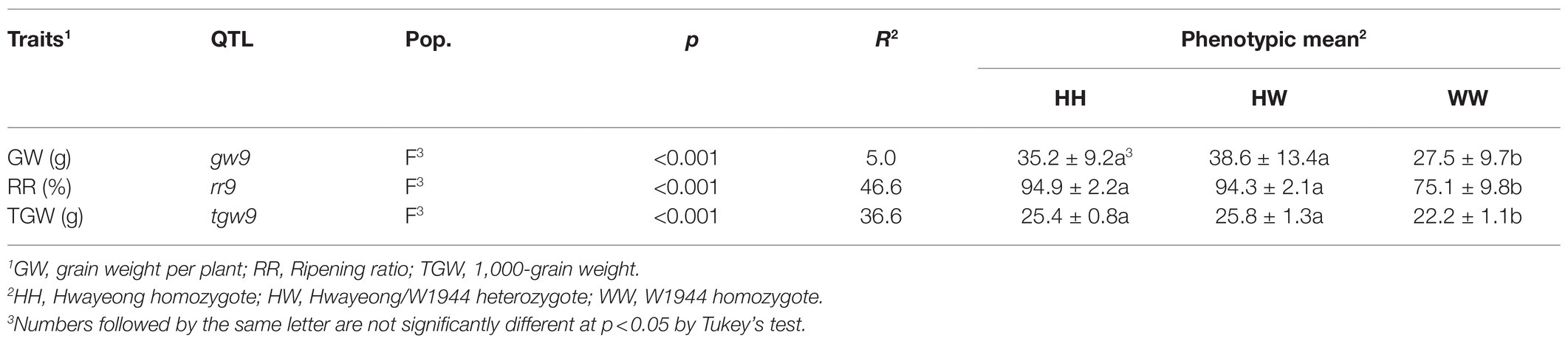
Table 2. Quantitative trait loci (QTL) for the three traits based on a single-point analysis in the F3 population.
For fine mapping of DTE9, six heterozygous plants at RM553 and RM24637 were selected from the BC4F3 population to produce a BC4F4 population, which was screened with the SSR markers RM553 and RM24637 to identify recombinants. Substitution mapping using 50 BC4F4 recombinants indicated that DTE9OR was located between the RM7343 and RM3249. This 230-kb interval has 38 genes based on the Nipponbare reference genome (Figure 2A). Sixty insertion/deletion (InDel) markers were designed by comparing the Nipponbare sequences of 38 genes with that of 93-11 sequences. Eight markers showed polymorphisms between HY and W1944 (Supplementary Table S1). Genotyping of 15 recombinants with eight InDel markers delimited DTE9OR to a 27.7 kb interval flanked by markers CRDI48 and CRDI37. This interval contains two predicted genes, LOC_Os09g32944 and LOC_Os09g32948 (Figure 2B). LOC_Os09g32944 encodes the SQUAMOSA promoter-binding-like protein 18 (OsSPL18), which controls ligule development, grain weight, and grain number in rice (Lee et al., 2007b; Yuan et al., 2019). LOC_Os09g32948 encodes OsMADS8/24, a member of the MADS-box transcription factor gene family in rice, which is involved in floral organ specification, plant growth, and development (Arora et al., 2007).
The expression of the two candidate genes was compared among HY, W1944, and CR6078 (Figure 2C). No differences in OsSPL18 expression were observed among the three lines. In contrast, OsMADS8 was expressed at higher levels in both CR6078 and W1944 compared to HY suggesting that OsMADS8 is more likely the candidate gene for DTE9OR (Figure 2C). Thirty single nucleotide polymorphisms (SNPs; 26 and four in the promoter and intron regions, respectively) and 10 InDels (seven and three in the promoter and intron regions, respectively) were detected in the OsMADS8 sequences between HY and W1944 (Supplementary Table S2). The lack of sequence differences in the eight exon regions suggests that the variation in the OsMADS8 promoter region underlies the differential expression of this gene in HY and W1944/CR6078.
Forty OsMADS8 sequence variants between HY and W1944 were detected in the promoter and intron regions. Two hotspots were observed in the promoter region between the nucleotides 19,651,477 and 19,651,511 (SNP1-2 and InDel1-3) and between nucleotides 19,653,010 and 19,654,306 (SNP24-26 and InDel6-8; Figure 2D). Considering that the promoter is responsible for regulating the expression of OsMADS8, the difference between the two variable promoter regions might be associated with the variation of DTE trait in CR6078.
When the comparison was expanded to 137 accessions from the KRICE_CORE collection, a total of 81 SNPs and 35 InDels were detected in the promoter, 5' UTR, and intron regions of OsMADS8 (Supplementary Tables S3–S6). Eighteen out of 40 SNPs in the promoter and intergenic regions were indica or japonica-specific (Supplementary Table S5). Among these subspecies-specific SNPs, W1944 had different SNPs from those of W1943, a putative ancestor of japonica rice (Huang et al., 2012). For SNP5, W1944 with an A-allele was different from all 137 accessions with a G-allele. In addition, 14 and 21 InDels were observed in the promoter and intron regions, respectively (Supplementary Table S4). Ten InDels were observed in the 137 accessions and half of the 10 InDels were species-specific. Six InDels were W1944-specific compared to W1943 (Supplementary Table S6).
To confirm the relationship between OsMADS8 and the abnormal embryo phenotype (DTE), the W1944 full-length OsMADS8 open reading frame was expressed under the control of the cauliflower mosaic virus 35S promoter. Overexpression (OX) and knockdown (RNAi) constructs were developed and transformed into HY (hereafter OsMADS8-OX) and CR6078 (hereafter OsMADS8-RNAi), respectively. Six independent OsMADS8-OX lines and 46 independent OsMADS8-RNAi lines were generated (Figures 3A,B). The T2 plants of OsMADS8-OX lines #77 and #88 showed significantly higher expression of OsMADS8, whereas T2 plants of OsMADS8-RNAi lines #38-1 and #40-4 showed significantly reduced transcript levels of OsMADS8 (Figures 3C,D). Significant differences between HY and OsMADS8-OX and CR6078 and OsMADS8-RNAi were observed for grain weight and ripening ratio (i.e., fertility), but not for the other traits (Supplementary Table S7). When the seeds of HY and OsMADS8-OX were compared, OX lines showed a higher rate of occurrence of DTE (about 22–33%) than HY (~3%). As expected, the two RNAi lines had lower DTE rates (~0.2%) than CR6078 (~42%; Figure 3D). Taken together, these results indicate that OsMADS8 is associated with the DTE phenotype (Figures 3A–D). The histological analysis showed that the coleoptile, plumule, and radicle of OsMADS8-OX embryos were protruded, showing clear cleavage from the scutellum tissue (Figures 3E–G). In contrast, the embryos of the HY and the OsMADS8-RNAi lines had coleoptiles embedded in the endosperm and appeared normal (Figures 3H–J). These results indicated that DTE9 is an allele of OsMADS8.
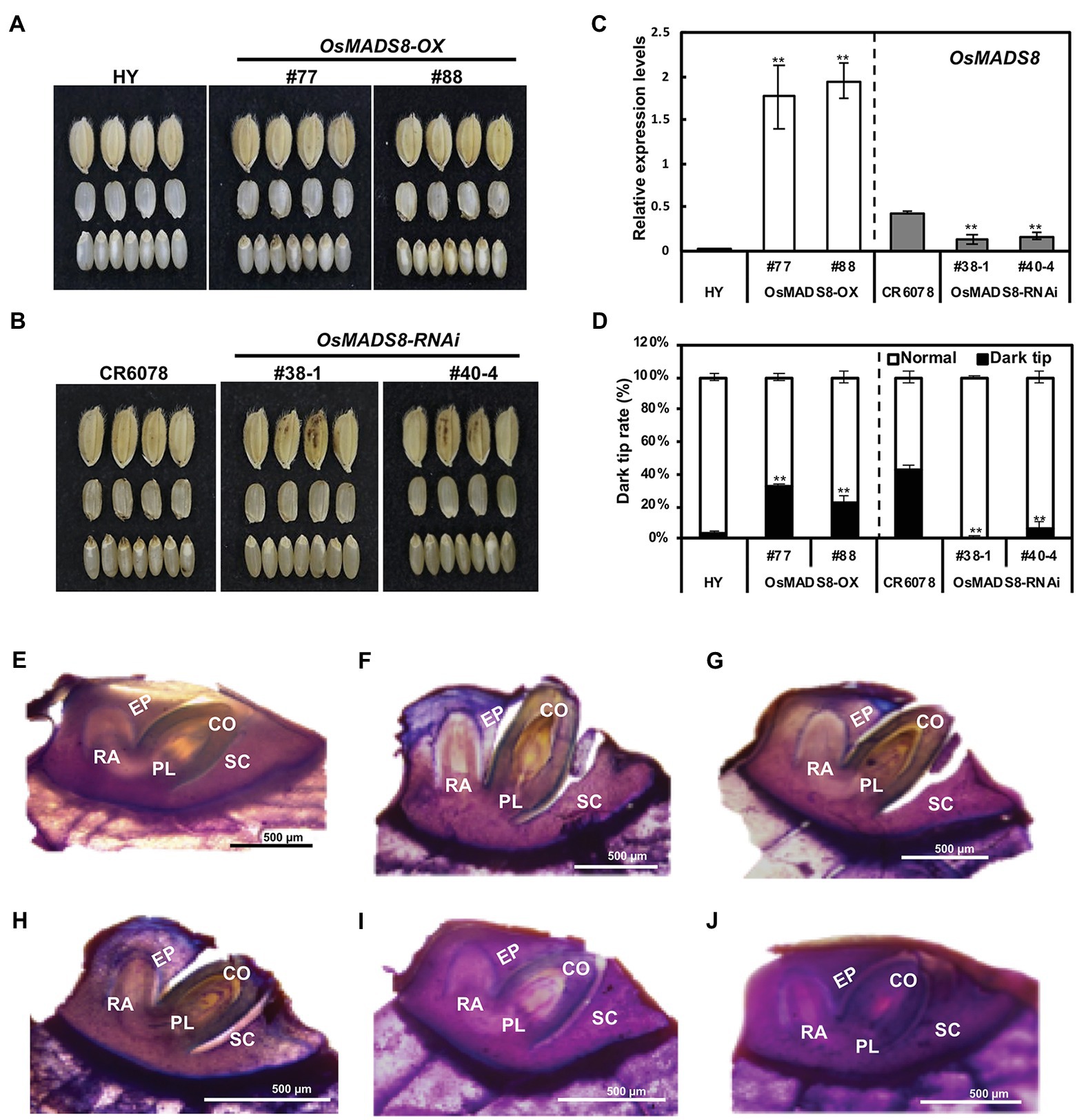
Figure 3. Characterization of the OsMADS8 transgenic lines. (A) Comparison of the grain between HY and OsMADS8-OX (#77, #88) lines. (B) Comparison of the grain between CR6078 and OsMADS8-RNAi (#38–1, #40–4) lines. (C) The expression level of the OsMADS8 gene in the transgenic plants. (D) Dark tip embryo rate (%). Histological analysis of the embryo shape of HY (E) and two OX lines (F,G) and CR6078 (H) and OsMADS8-RNAi lines (I,J). SC, scutellum; CO, coleoptile; PL, plumule; EP, epiblast; and RA, radicle. ** indicates significant difference based on the student’s t-test at p < 0.01.
CR6078 has more styles and stigmas than HY and W1944, indicating that the floral organ identity genes were differentially regulated in this line (Figure 1C). Therefore, the expression of genes possibly interacting with OsMADS8, an E-class floral identity group gene, was analyzed. Among five other E-class genes, the expression of OsMADS5, OsMADS7, and OsMADS34 was the highest in W1944, followed by CR6078 and HY (Figure 4A; Supplementary Figure S1). The higher expression of these three genes in CR6078 compared to HY suggests a background effect from W1944 in CR6078. Interestingly, CR6078 showed higher expression of OsMADS1 and OsMADS17 than HY and W1944, indicating that OsMADS8 might regulate OsMADS1 and OsMADS17 expression. This finding is consistent with the DTE trait in CR6078 resulting from the combined effect of the two complementary genes, the W1944 allele of OsMADS8 and an unknown gene from HY (Figure 4A).
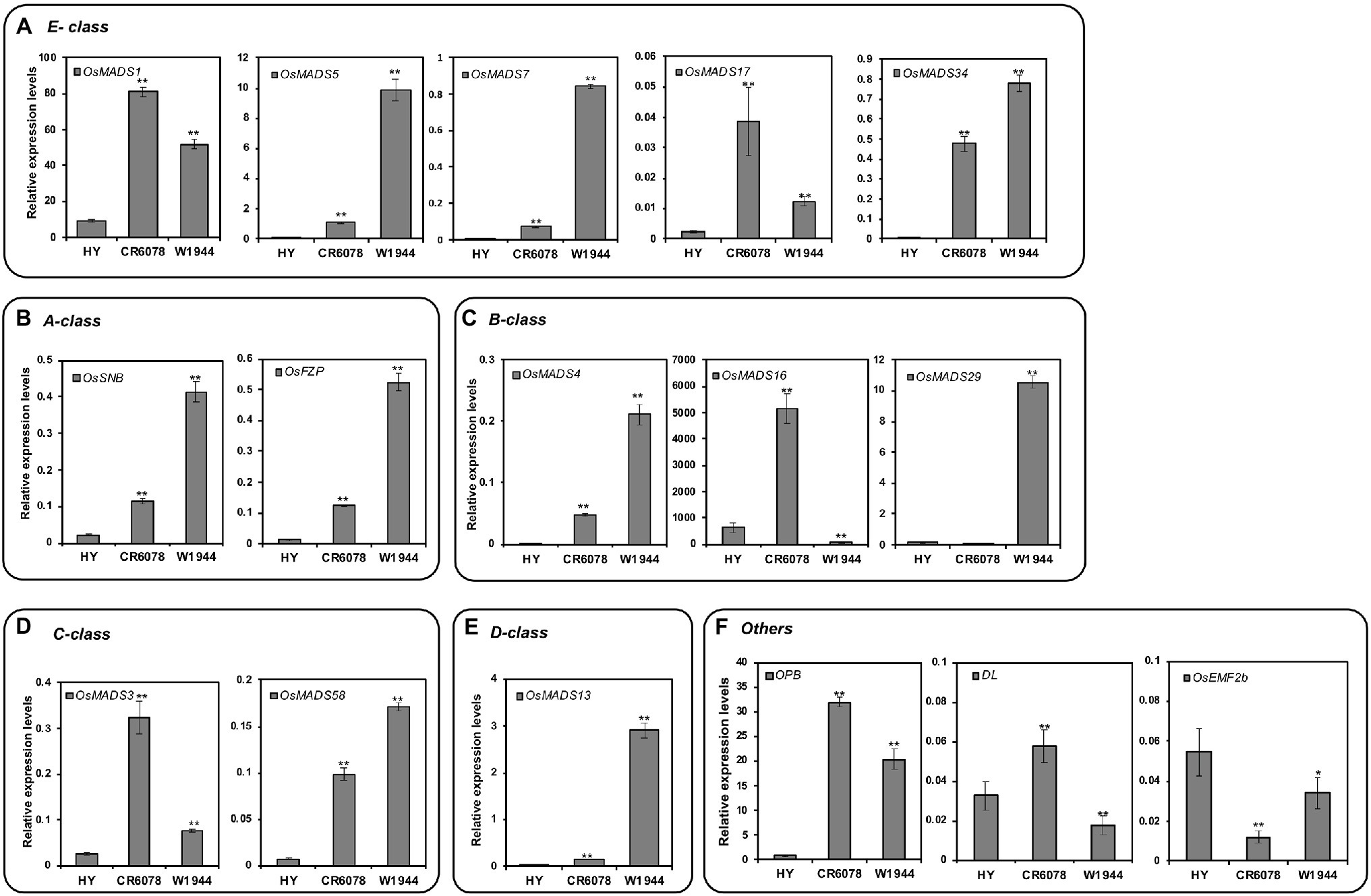
Figure 4. Expression patterns of floral organ determination genes in HY, CR6078, and W1944 plants. (A) E-class, (B) A-class, (C) B-class. (D) C-class, (E) D-class genes, and (F) other genes. Total RNA was isolated from 7-day-old seedlings. Relative expression level of each gene was quantified using real-time PCR and normalized using OsTMP1 as an internal control. The error bars represent mean ± SD (n = 3). Significant differences between the HY and each OX line, and the CR6078 plants and each RNAi line at *p < 0.05 and **p < 0.01.
To examine potential interactions with other floral homeotic genes, we analyzed the expression of the ABCD group genes. OsSNB, OsMADS4, and OsFZP were expressed at higher levels in W1944 than in CR6078 and HY (Figures 4B,C). It is noteworthy that OsMADS16 was expressed at higher levels in CR6078 than in HY and W1944, whereas OsMADS29 was expressed at a lower level in CR6078 than in HY and W1944 (Figure 4C). The expression of the C-class gene OsMADS3 was remarkably higher in CR6078 than in HY and W1944 (Figure 4D).
The floral organ identity genes that exhibited higher or lower expression in CR6078 compared to the parental lines HY and W1944 were then investigated in the transgenic plants (Figure 5). The expression of these four genes, OsMADS1, 3, 16, and 17, was higher in OsMADS8-OX than in HY. The OsMADS8-RNAi lines also showed lower expression of OsMADS1 and OsMADS16 than in CR6078. Unexpectedly, the OsMADS8-RNAi lines showed higher expression of OsMADS3 and OsMADS17 than CR6078 (Figures 5B,D). These results indicate that OsMADS8 directly or indirectly regulates the expression of these genes, which may underlie the formation of the abnormal floral structure including pistil in CR6078.
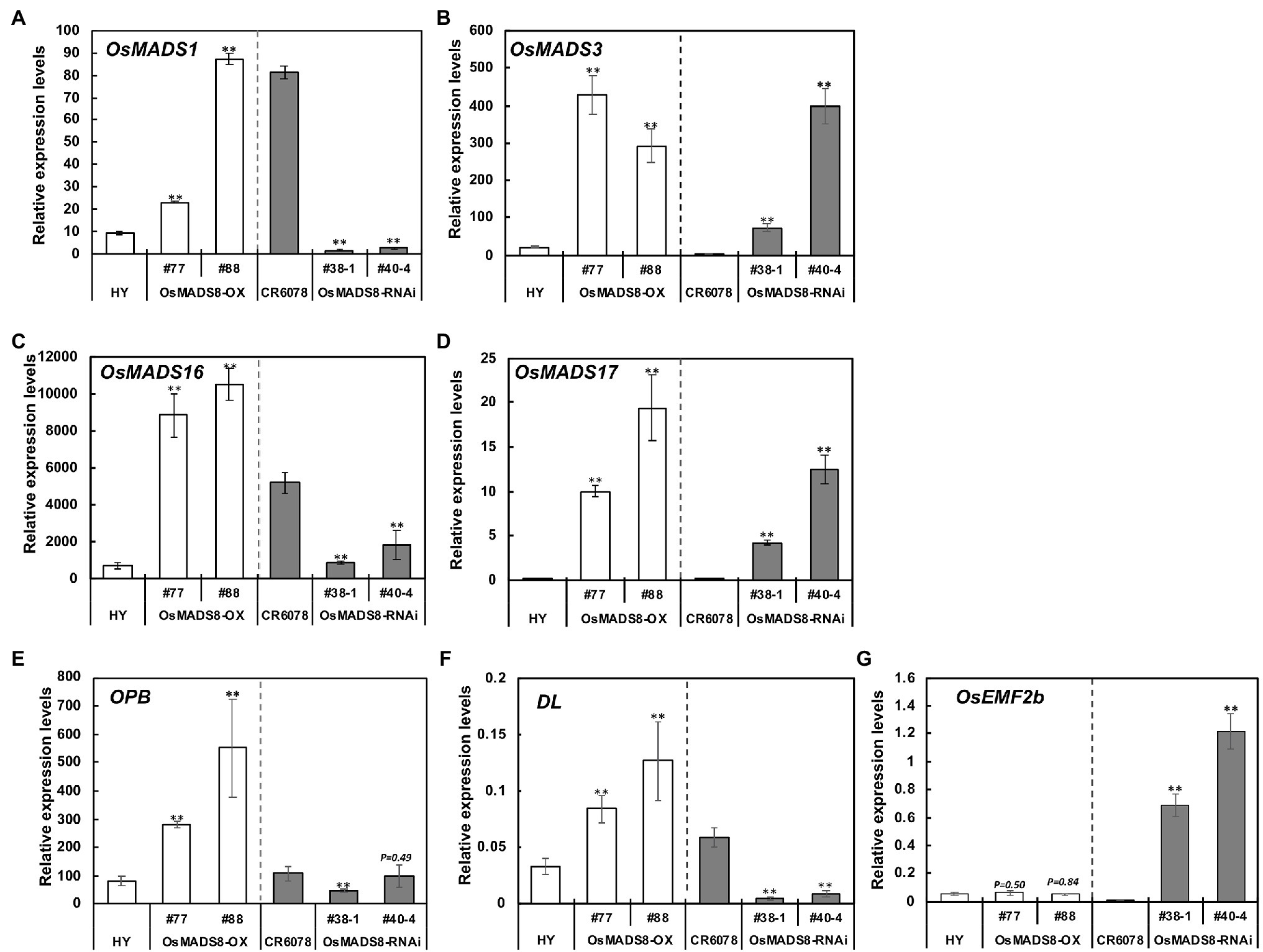
Figure 5. Expression of seven floral development genes in the OsMADS8 transgenic lines. (A) OsMADS1, (B) OsMADS3, (C) OsMADS16, (D) OsMADS17, (E) OPB, (F) DL, and (G) OsEMF2b. Total RNA was isolated from the 7-day-old seedlings of the OsMADS8 transgenic lines. Relative expression level of each gene was quantified using real-time PCR and normalized using OsTMP1 as an internal control. The error bars represent mean ± SD (n = 3). The significant differences between the HY and each OX line and the CR6078 plants and each RNAi line were evident at **p < 0.01.
Expression of the other ABCDE functional floral identity group genes was also examined in the transgenic lines. Among the A- and B-class genes, OsSNB and OsFZP exhibited higher expression in the OsMADS8-OX lines than in HY. The expression of OsSNB and OsFZP was at higher level in the OsMADS8-RNAi lines compared to CR6078, however, unexpectedly OsMADS4 and OsMADS29 showed lower expression levels in CR6078 than in the OsMADS8-RNAi lines (Supplementary Figures S5A–D). Among the C-class genes, OsMADS58 was expressed at higher levels both in the OX and RNAi transgenic lines, whereas the expression of OsMADS13 was remarkably higher in the OsMADS8-OX lines than in HY (Supplementary Figures S5E,F). Among the E-class genes, OsMADS5 and OsMADS34 expression was remarkably higher in the OsMADS8-OX lines than in HY (Supplementary Figures S5G,H).
No differences in the number of stigmas and styles were observed between HY and OsMADS8-OX lines (Supplementary Figure S6). In addition, the OsMADS8-RNAi lines did not show any difference in the number of floral organs compared to CR6078 plants (Supplementary Table S8; Supplementary Figure S7). These results suggest that the OsMADS8 expression in the OsMADS8-OX and OsMADS8-RNAi lines was not sufficient to induce phenotypic changes in the floral organs.
Previous studies have shown that other genes are also involved in determining the carpel and style or stigma identity (Cui et al., 2010; Li et al., 2011; Conrad et al., 2014; Deng et al., 2017; Zhang et al., 2018). Interestingly, the transcription levels of Open Beak (OPB) and Drooping Leaf (DL) in CR6078 were higher than those in HY and W1944 (Figure 4F). In contrast, OsEMF2b was expressed at lower levels in CR6078 than in HY and W1944. In the OsMADS8-OX plants, the transcript levels of OPB and DL were higher than those in HY, whereas the expression of OsEMF2b was not significantly different (Figures 5E–G). As expected, the transcript levels of OPB and DL in the OsMADS8-RNAi plants were lower than those in CR6078, whereas the OsEMF2b expression was higher (Figures 5E–G). Thus, OsMADS8 in CR6078 altered the expression of OPB, DL, and OsEMF2b in addition to the homeotic genes of the ABCDE model of flower development.
To explain the DTE trait exhibited by CR6078, OsEMF2b was investigated based on the previous finding that this gene directly regulates the expression of OsVP1/ABI3 and the ABA signaling genes regulating seed germination (Chen et al., 2017). CR6078 displayed reduced expression levels of OsVP1/ABI3, ABI4, and ABI5 compared to that of HY and W1944 (Figures 6A–C). Among the three genes regulating ABA catabolism, the OsABA8’ox1 expression levels were higher in CR6078 than in HY and W1944, while no clear pattern was observed in the expression of OsABA8’ox2 and OsABA8’ox3 (Figures 6D–F), suggesting that DTE might be associated with altered expression of OsEMF2b and OsABA8’ox1, which regulates seed dormancy and germination.
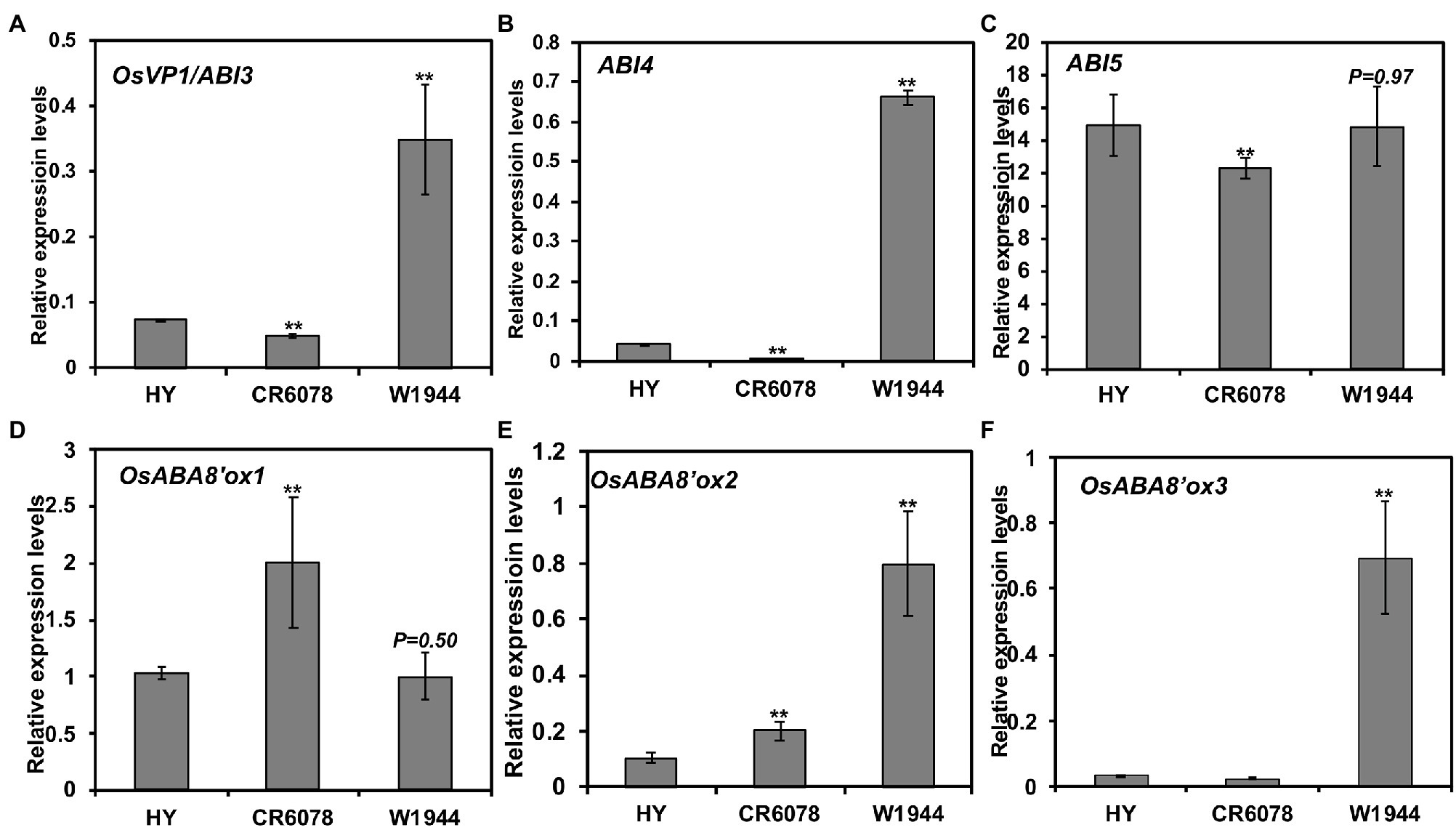
Figure 6. Expression of abscisic acid (ABA) signal transduction and ABA catabolism-related genes in three accessions (A–F). Seeds from the three lines were used to isolate the total RNA. Relative expression level of each gene was quantified using real-time PCR and normalized using OsTMP1 as an internal control. The error bars represent mean ± SD (n = 3). The significant differences between the HY and each OX line and the CR6078 plants and each RNAi line were evident at **p < 0.01.
To determine whether OsEMF2b is associated with changes in embryonic traits, the expression of genes regulating the ABA signaling pathway and catabolism in OsMADS8-OX and RNAi plants was examined. The expression of ABI3 and ABI4 was significantly reduced in OsMADS8-OX compared to that of HY (Figure 6). However, the expression of ABI5 was significantly reduced in HY compared to that in OsMADS8-OX (Supplementary Figures S8A–C). All three genes (OsABA8’ox1, OsABA8’ox2, and OsABA8’ox3) related to ABA catabolism showed higher expression in the OsMADS8-OX plants than in HY (Supplementary Figures S8D–F). The expression of ABI3, ABI4, and ABI5 was significantly increased in OsMADS8-RNAi compared to that of CR6078 (Supplementary Figures S8A–C). By contrast, the expression of OsABA8’ox1 was lower in the OsMADS8-RNAi plants than in CR6078, whereas OsABA8’ox3 was expressed at a higher level in the OsMADS8-RNAi than in CR6078 (Figures 6D–F). Taken together, these results suggest that OsMADS8 negatively regulates the expression of OsEMF2b, a positive regulator of seed dormancy, by overexpressing OsABA8’ox1.
To determine the effect of OsMADS8 on germination, the germination ratio of HY, CR6078, W1944, and the transgenic lines were compared. CR6078 and the OsMADS8-OX lines started germination at 24 h after sowing with a germination ratio of 82% while the germination ratios of HY and OsMADS8-RNAi lines were less than 3% (Figures 7A,B). The ABA content in the seeds of HY, CR6078, and transgenic lines was analyzed to determine whether the decreased germination ratio was related to the ABA content. The ABA level of OsMADS8-OX plants (385–377 pmoles ml−1) was significantly lower than that of HY (919 pmoles ml−1; Figure 7C). The two OsMADS8-RNAi lines had significantly higher ABA contents than CR6078 (Figure 7C). These results support the idea that in the CR6078 line, OsMADS8 regulates the expression of genes involved in ABA catabolism (Figure 7).
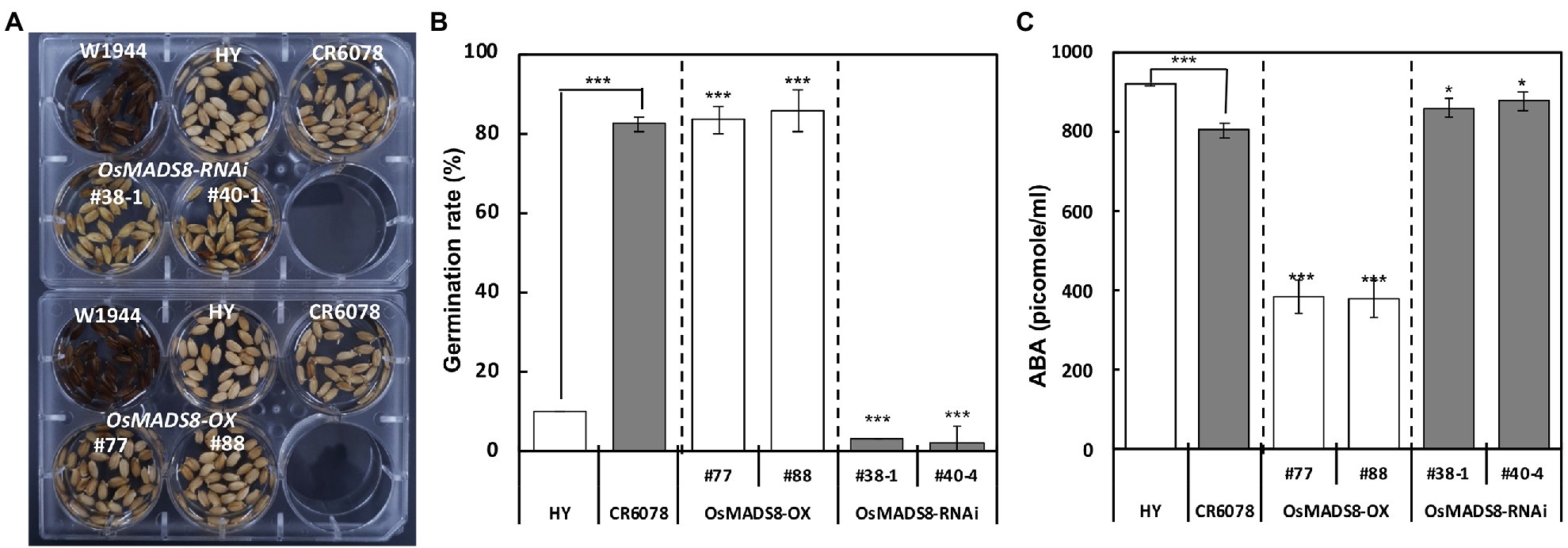
Figure 7. Comparison of germination ratio at 30°C and ABA contents in the transgenic and control plants. (A) The CR6078 and OsMADS8-OX lines showed a significantly higher germination ratio than the HY and OsMADS8-RNAi at 24 h after sowing. (B) The significant differences between the HY and CR6078, the HY and each OX line, and the CR6078 plants and each RNAi line were evident at ***p < 0.001. The germination rate (based on the length of radicles >1 mm) was recorded, and values are represented as mean ± SD (n = 3). (C) Analysis of the ABA content in the OsMADS8 transgenic lines and controls. The error bars represent mean ± SD (n = 3). Data represent the average of three replicates. The comparison was made between the HY and CR6078, and the HY and each OX line, and the CR6078 and each RNAi line. Significant differences were evident at *p < 0.05 and ***p < 0.001.
Hybrid weakness, the poor development of hybrids compared to their parents, hinders gene exchange between different species at the postzygotic stage. Hybrid weakness has been reported in rice at both the interspecific or intraspecific crosses, and most studies have reported that this phenomenon is mainly controlled by dominant or recessive complementary interactions of unlinked loci (Fukuoka et al., 1998, 2005; Kubo and Yoshimura, 2002; Matsubara et al., 2007; Yamamoto et al., 2007; Jiang et al., 2008). Characterization of the genetic basis of hybrid weakness contributes to our knowledge of the underlying mechanisms of reproductive isolation and has practical significance with regard to the maximal utilization of invaluable genes from the interspecific or intraspecific crosses in plant breeding (Chen et al., 2014). This study characterized a newly discovered gene for hybrid weakness in rice, DTE9. The DTE trait was observed in CR6078, an introgression line derived from an interspecific cross between O. sativa spp. japonica “Hwayeong” and the wild relative O. rufipogon.
To understand the genetic basis of DTE, a population derived from a cross between HY and CR6078 was developed. In the BC4F3 population, a segregation of 3 (normal): 1 (DTE) was observed, suggesting that DTE is controlled by a single recessive gene in this cross. However, a single recessive gene mode of inheritance for DTE is not compatible with the finding that the two parental lines do not display DTE. The F3 population from a second cross between an indica rice cultivar Milyang23 and CR6078 displayed a segregation ratio of 15:1 (normal:DTE). These results support the involvement of two complementary recessive genes, which is consistent with the previous findings that two complementary recessive genes regulate hybrid weakness in rice (Sato, 1997; Fukuoka et al., 1998; Kubo and Yoshimura, 2002; Ichitani et al., 2007; Matsubara et al., 2007; Kuboyama et al., 2009). Based on our genetic analysis of the progeny from two crosses, we hypothesize that two or more genes (QTLs), OsMADS8 and other unknown genetic factors control DTE in rice (Table 2). Another interesting feature of DTE was that all F2 seeds from the F1 plants of the two crosses showed normal Milyang23 and HY embryo types with segregation observed in the F3 population. This suggests that the DTE trait is of maternal tissue origin.
Dark tip embryo was significantly associated with several deleterious traits including reduced GW (Table 2). Considering that there are no reports or annotations for traits of agronomic importance in or near this region, DTE9 is likely to be associated with the deleterious phenotypes. Based on the DTE phenotype, it is possible that the CR6078 seeds, which have significantly lower ABA content than that of HY, began preharvest germination with coleoptile protrusion. However, germination was arrested perhaps due to environmental conditions, such as water deficit in the panicle. During the germination process, CR6078 consumed carbohydrates in grains, leading to a decrease in the GW. Given the association of DTE9 with several deleterious traits, the interaction between OsMADS8 and the other gene would likely not be selected for during breeding and domestication.
CR6078 displayed an abnormal floral organ phenotype with more styles and stigmas than those of HY and W1944. Gene expression analyses showed that the alteration of OsMADS8 expression in CR6078 and transgenic plants not only changed the expression of the various ABCDE model genes, but also that of genes, such as OPB and OsEMF2b (Figure 4). It is noteworthy that among the ABCDE model genes, the expression of OsMADS1, 3, 16, and 17 was higher in CR6078 than in HY and W1944. Additionally, OPB and DL displayed similar expression patterns, whereas CR6078 showed lower expression of OsEMF2b than HY and W1944. A similar expression pattern was also observed for the genes associated with the ABA signaling pathway and catabolism in CR6078, supporting the idea that the two complementary genes, OsMADS8, and another unknown gene in HY, control the DTE in this population (Figure 6). However, the OsMADS8-RNAi lines showed higher expression of OsMADS3 and OsMADS17 than CR6078 (Figure 5). The expression patterns of several genes were not the same in the transgenic plants as in CR6078. For example, OsMADS3 and OsMADS17 showed higher expression in the OsMADS8-RNAi lines than in CR6078, and ABI5 and OsABA8’ox3 displayed higher expression in the OsMADS8-OX lines than in HY. This is probably due to interactions between transgenes and genes associated with the floral organ phenotype in the background of HY and CR6078. Moreover, CR6078 harbors eight O. rufipogon segments in the HY background.
OsMADS8-OX plants failed to show phenotypic changes compared to HY, possibly owing to the low expression of OsEMF2b in the OsMADS8-OX lines. In a previous study, the loss of function of EMF2b resulted in abnormal changes in the internal floral organ structure. When the phenotype of the emf2b mutant was compared with that of CR6078, the emf2b mutant showed more carpels and stigmas and had lower expression of genes including OsMADS1, OPB, and DL than the wild type (Conrad et al., 2014; Deng et al., 2017). In contrast, CR6078 showed higher expression of OsMADS1, OPB, and DL and lower expression of OsEMF2b than HY, leading to phenotypic changes in CR6078. These results raise the possibility that OsMADS8 directly or indirectly regulates the expression of the B-, E-, and C-class genes as well as other genes, such as OPB and OsEMF2b, and this regulation is responsible for the changes in the inner floral organ phenotype observed in CR6078. It is rational to infer that OsMADS8 alone induces a weak phenotypic change (Cui et al., 2010).
OsEMF2b regulates seed dormancy and seedling growth by activating and inhibiting the expression of the ABA signaling response genes (Chen et al., 2017). ABA promotes seed dormancy and suppresses seed germination and seedling growth. Downregulation of OsEMF2b leads to vivipary and decreased expression levels of OsVP1, an ortholog of ABI3, which is a component of the ABA signaling pathway in Arabidopsis. CR6078 displayed reduced expression levels of OsEMF2b, OsVP1/ABI3, ABI4, and ABI5 compared to that of HY and W1944 (Figures 4F, 6). Additionally, the expression of genes related to ABA catabolism was differentially regulated in CR6078 (Figures 6D–F). Expression of these genes was examined in the OsMADS8-OX and RNAi transgenic plants (Supplementary Figure S8), revealing that OsMADS8 negatively regulates the expression of OsEMF2b, which is related to the activation and repression of expression of the ABA signal responsive genes. Downregulation of OsEMF2b expression might be associated with vivipary and decreased expression levels of OsVP1, which is related to the seed ABA signaling pathway and seed dormancy. Taken together, our results indicate that OsMADS8 plays an important role in regulating the expression of OsEMF2b associated with floral organ determination and seed germination via the ABA signaling pathway and ABA catabolism.
Several studies have reported the role of plant hormones in hybrid weakness (Reiber and Neuman, 1999; Yamada et al., 2001; Yamada and Marubashi, 2003; Hannah et al., 2007; Kobori et al., 2007). Our finding that HY had significantly higher seed ABA content than CR6078 is consistent with the role of OsMADS8 in controlling DTE in CR6078 by directly influencing the expression of genes regulating ABA catabolism. Thus, the hybrid weakness exhibited by CR6078 underscores the importance of plant hormones in this phenomenon (Yamada et al., 2001; Kobori et al., 2007).
In this study, a novel interspecific hybrid weakness gene controlling DTE phenotype was discovered in an introgression line derived from a cross between the japonica variety “Hwayeong” and the common wild rice W1944 (O. rufipogon). We characterized the phenotypic and physiological properties of this newly discovered expression of interspecific hybrid weakness and isolated the causal gene, DTE9, responsible for the DTE and several other traits of hybrid weakness. Based on the finding that two recessive complementary genes are responsible for full expression of the DTE trait, further studies to identify this second gene are in progress. These results contribute to a better understanding of the molecular mechanism causing hybrid weakness and will benefit rice breeding programs by assisting in strategies to maximize exploitation of invaluable genes from the interspecific or intraspecific crosses.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
SK, S-DJ, and S-NA designed the experiments and wrote the manuscript. TT edited the manuscript and provided advice on the experiments. H-SL, K-CS, CA, and NL conducted the agronomic traits investigation and qRT-PCR analysis. S-DJ, PY, and H-JK performed fine mapping of DTE9. Y-AJ and SK analyzed the sequencing data. All authors contributed to the article and approved the submitted version.
This work was carried out with the support of “Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ015757)” Rural Development Administration, Republic of Korea.
H-JK was employed by the company LG Chem., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.778008/full#supplementary-material
Agrawal, G. K., Abe, K., Yamazaki, M., Miyao, A., and Hirochika, H. (2005). Conservation of the E-function for floral organ identity in rice revealed by the analysis of tissue culture-induced loss-of-function mutants of the OsMADS1 gene. Plant Mol. Biol. 59, 125–135. doi: 10.1007/s11103-005-2161-y
Ali, Z., Raza, Q., Atif, R. M., Aslam, U., Ajmal, M., and Chung, G. (2019). Genetic and molecular control of floral organ identity in cereals. Int. J. Mol. Sci. 20:2743. doi: 10.3390/ijms20112743
Angenent, G. C., and Colombo, L. (1996). Molecular control of ovule development. Trends Plant Sci. 1, 228–232. doi: 10.1016/S1360-1385(96)86900-0
Arora, R., Agarwal, P., Ray, S., Singh, A. K., Singh, V. P., Tyagi, A. K., et al. (2007). MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8:242. doi: 10.1186/1471-2164-8-242
Bai, X., Huang, Y., Mao, D., Wen, M., Zhang, L., and Xing, Y. (2016). Regulatory role of FZP in the determination of panicle branching and spikelet formation in rice. Sci. Rep. 6:19022. doi: 10.1038/srep19022
Becker, A., Winter, K. U., Meyer, B., Saedler, H., and Theissen, G. (2000). MADS box gene diversity in seed plants 300 million years ago. Mol. Biol. Evol. 17, 1425–1434. doi: 10.1093/oxfordjournals.molbev.a026243
Bomblies, K., Lempe, J., Epple, P., Warthmann, N., Lanz, C., Dangl, J. L., et al. (2007). Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 5:e0050236. doi: 10.1371/journal.pbio.0050236
Callens, C., Tucker, M. R., Zhang, D., and Wilson, Z. A. (2018). Dissecting the role of MADS-box genes in monocot floral development and diversity. J. Exp. Bot. 69, 2435–2459. doi: 10.1093/jxb/ery086
Chen, C., Chen, H., Lin, Y. S., Shen, J. B., Shan, J. X., and Qi, P. (2014). A two-locus interaction causes interspecific hybrid weakness in rice. Nat. Commun. 5:3357. doi: 10.1038/ncomms4357
Chen, C., Chen, H., Shan, J. X., Zhu, M. Z., Shi, M., and Gao, J. P. (2013). Genetic and physiological analysis of a novel type of interspecific hybrid weakness in rice. Mol. Plant 6, 716–728. doi: 10.1093/mp/sss146
Chen, Z. X., Wu, J. G., Ding, W. N., Chen, H. M., Wu, P., and Shi, C. H. (2006). Morphogenesis and molecular basis on naked seed rice, a novel homeotic mutation of OsMADS1 regulating transcript level of AP3 homologue in rice. Planta 223, 882–890. doi: 10.1007/s00425-005-0141-8
Chen, M., Xie, S., Ouyang, Y., and Yao, J. (2017). Rice PcG gene OsEMF2b controls seed dormancy and seedling growth by regulating the expression of OsVP1. Plant Sci. 260, 80–89. doi: 10.1016/j.plantsci.2017.04.005
Coen, E. S., and Meyerowitz, E. M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37. doi: 10.1038/353031a0
Conrad, L. J., Khanday, I., Johnson, C., Guiderdoni, E., An, G., Vijayraghavan, U., et al. (2014). The polycomb group gene EMF2B is essential for maintenance of floral meristem determinacy in rice. Plant J. 80, 883–894. doi: 10.1111/tpj.12688
Cui, R., Han, J., Zhao, S., Su, K., Wu, F., Du, X., et al. (2010). Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa). Plant J. 61, 767–781. doi: 10.1111/j.1365-313X.2009.04101.x
Dang, X., Yang, Y., Zhang, Y., Chen, X., Fan, Z., Liu, Q., et al. (2020). OsSYL2 AA, an allele identified by gene-based association, increases style length in rice (Oryza sativa L.). Plant J. 104, 1491–1503. doi: 10.1111/tpj.15013
Deng, L., Zhang, S., Wang, G., Fan, S., Li, M., Chen, W., et al. (2017). Down-regulation of OsEMF2b caused semi-sterility due to anther and pollen development defects in rice. Front. Plant Sci. 8:1998. doi: 10.3389/fpls.2017.01998
Egea-Cortines, M., Saedler, H., and Sommer, H. (1999). Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18, 5370–5379. doi: 10.1093/emboj/18.19.5370
Fukuoka, S., Namai, H., and Okuno, K. (1998). RFLP mapping of the genes controlling hybrid breakdown in rice (Oryza sativa L.). Theor. Appl. Genet. 97, 446–449. doi: 10.1007/s001220050915
Fukuoka, S., Newingham, M. C. V., Ihitiaq, M., Nagamine, T., Kawase, M., and Okuno, K. (2005). Identification and mapping of two new loci for hybrid breakdown in cultivated rice. Rice Genet. Newsl. 22, 29–31.
Goodrich, J., Puangsomlee, P., Martin, M., Long, D., Meyerowitz, E. M., and Coupland, G. (1997). A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386, 44–51. doi: 10.1038/386044a0
Gutierrez-Cortines, M. E., and Davies, B. (2000). Beyond the ABCs: ternary complex formation in the control of floral organ identity. Trends Plant Sci. 5, 471–476. doi: 10.1016/S1360-1385(00)01761-1
Hannah, M. A., Krämer, K. M., Geffroy, V., Kopka, J., Blair, M. W., Erban, A., et al. (2007). Hybrid weakness controlled by the dosagedependent lethal (DL) gene system in common bean (Phaseolus vulgaris) is caused by a shoot-derived inhibitory signal leading to salicylic acid-associated root death. New Phytol. 176, 537–549. doi: 10.1111/j.1469-8137.2007.02215.x
Hofgen, R., and Willmitzer, L. (1988). Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 16:9877. doi: 10.1093/nar/16.20.9877
Honma, T., and Goto, K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409, 525–529. doi: 10.1038/35054083
Horigome, A., Nagasawa, N., Ikeda, K., Ito, M., Itoh, J. I., and Nagato, Y. (2009). Rice OPEN BEAK is a negative regulator of class 1 Knox genes and a positive regulator of class B floral homeotic gene. Plant J. 58, 724–736. doi: 10.1111/j.1365-313X.2009.03823.x
Huang, X., Kurata, N., Wei, X., Wang, Z., Wang, A., Zhao, Q., et al. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501. doi: 10.1038/nature11532
Ichitani, K., Namigoshi, K., Sato, M., Taura, S., Aoki, M., Matsumoto, Y., et al. (2007). Fine mapping and allelic dosage effect of Hwc1, a complementary hybrid weakness gene in rice. Theor. Appl. Genet. 114, 1407–1415. doi: 10.1007/s00122-007-0526-0
Ichitani, K., Taura, S., Tezuka, T., Okiyama, Y., and Kuboyama, T. (2011). Chromosomal location of HWA1 and HWA2, complementary hybrid weakness genes in rice. Rice 4, 29–38. doi: 10.1007/s12284-011-9062-2
Ikehashi, H., and Araki, H. (1986). “Genetics of F1 sterility in remote crosses of rice,” in Rice Genetics. ed. J. Banta Stephen (Manila: International Rice Research Institute-IRRI).
Ishikawa, M., Ohmori, Y., Tanaka, W., Hirabayashi, C., Murai, K., Ogihara, Y., et al. (2009). The spatial expression patterns of DROOPING LEAF orthologs suggest a conserved function in grasses. Genes Genet. Syst. 84, 137–146. doi: 10.1266/ggs.84.137
Jeon, J. S., Jang, S., Lee, S., Nam, J., Kim, C., Lee, S. H., et al. (2000). Leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12, 871–884. doi: 10.1105/tpc.12.6.871
Jiang, D., Wang, Y., and He, Y. (2008). Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS One 3:e3404. doi: 10.1371/journal.pone.0003404
Jiang, P., Wang, S., Ikram, A., Jiang, H., Cheng, B., and Ding, Y. (2018). SDG721 and SDG705 are required for rice growth. Acta Bot. Sin. 60, 530–535. doi: 10.1111/jipb.12644
Kang, H. G., and An, G. (1997). Isolation and characterization of a rice MADS box gene belonging to the AGL2 gene family. Mol. Cells 7, 45–51.
Kim, S. H., Ahn, Y. O., Ahn, M. J., Lee, H. S., and Kwak, S. S. (2012). Down-regulation of β-carotene hydroxylase increases β-carotene and total carotenoids enhancing salt stress tolerance in transgenic cultured cells of sweet potato. Phytochemistry 74, 69–78. doi: 10.1016/j.phytochem.2011.11.003
Kim, T. S., He, Q., and Kim, K. W. (2016). Genome-wide resequencing of KRICE_CORE reveals their potential for future breeding, as well as functional and evolutionary studies in the post-genomic era. BMC Genomics 17:408. doi: 10.1186/s12864-016-2734-y
Kobori, S., Masuda, Y., Horii, M., and Marubashi, W. (2007). High levels of the cytokinin BAP suppress programmed cell death in hybrid tobacco cells (Nicotiana suaveolens x N. tabacum) expressing hybrid lethality. Plant Biotechnol. 24, 375–381. doi: 10.5511/plantbiotechnology.24.375
Komatsu, M., Chujo, A., Nagato, Y., Shimamoto, K., and Kyozuka, J. (2003). FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Dev. Dent. 130, 3841–3850. doi: 10.1242/dev.00564
Kubo, T., and Yoshimura, A. (2002). Genetic basis of hybrid breakdown in a japonica/indica cross of rice, Oryza sativa L. Theor. Appl. Genet. 105, 906–911. doi: 10.1007/s00122-002-1059-1
Kuboyama, T., Saito, T., Matsumoto, T., Wu, J., Kanamori, H., Taura, S., et al. (2009). Fine mapping of HWC2, a complementary hybrid weakness gene, and haplotype analysis around the locus in rice. Rice 2, 93–103. doi: 10.1007/s12284-009-9026-y
Lee, S., Jeon, J. S., An, K., Moon, Y. H., Lee, S., Chung, Y. Y., et al. (2003). Alteration of floral organ identity in rice through ectopic expression of OsMADS16. Planta 217, 904–911. doi: 10.1007/s00425-003-1066-8
Lee, D. Y., Lee, J., Moon, S., Park, S. Y., and An, G. (2007a). The rice heterochronic gene SUPERNUMERARY BRACT regulates the transition from spikelet meristem to floral meristem. Plant J. 49, 64–78. doi: 10.1111/j.1365-313X.2006.02941.x
Lee, S., Oh, C. S., Suh, J. P., McCouch, S. R., and Ahn, S. N. (2005). Identification of QTLs for domestication-related and agronomic traits in an Oryza sartiva x O. rufipogon BC1F7 population. Plant Breed. 124, 209–219. doi: 10.1111/j.1439-0523.2005.01092.x
Lee, J., Park, J., Kim, S. L., Yim, J., and An, G. (2007b). Mutations in the rice liguleless gene result in a complete loss of the auricle, ligule, and laminar joint. Plant Mol. Biol. 65, 487–499. doi: 10.1007/s11103-007-9196-1
Li, H., Liang, W., Yin, C., Zhu, L., and Zhang, D. (2011). Genetic interaction of OsMADS3, DROOPING LEAF, and OsMADS13 in specifying rice floral organ identities and meristem determinacy. Plant Physiol. 156, 263–274. doi: 10.1104/pp.111.172080
Lin, J., Zhou, B., Yang, Y., Mei, J., Zhao, X., Guo, X., et al. (2009). Piercing and vacuum infiltration of the mature embryo: a simplified method for Agrobacterium-mediated transformation of Indica rice. Plant Cell Rep. 28, 1065–1074. doi: 10.1007/s00299-009-0706-2
Matsubara, K., Ando, T., Mizubayashi, T., Tto, S., and Yano, M. (2007). Identiication and linkage mapping of complementary recessive genes causing hybrid breakdown in an intraspeciWc rice cross. Theor. Appl. Genet. 115, 179–186. doi: 10.1007/s00122-007-0553-x
McCouch, S. R., Teytelman, L., Xu, Y., Lobos, K. B., Clare, K., Walton, M., et al. (2002). Development and mapping of 2240 new SSR markers for Rice (Oryza sativa L.). DNA Res. 9, 199–207. doi: 10.1093/dnares/9.6.199
Murai, K. (2013). Homeotic genes and the ABCDE model for floral organ formation in wheat. Plan. Theory 2, 379–395. doi: 10.3390/plants2030379
Nagasawa, N., Miyoshi, M., Sano, Y., Satoh, H., Hirano, H., Sakai, H., et al. (2003). SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Dev. Dent. 130, 705–718. doi: 10.1242/dev.00294
Nelson, J. C. (1997). QGENE: software for marker-based genomic analysis and breeding. Mol Breeding 3, 239–245. doi: 10.1023/A:1009604312050
Nguyen, H. N., Park, I. K., Yeo, S. M., Yun, Y. T., and Ahn, S. N. (2012). Mapping quantitative trait loci controlling low-temperature germinability in rice. Korean J. Agric. Sci. 39, 477–482. doi: 10.7744/cnujas.2012.39.4.477
Oka, H. I. (1957). Phylogenetic differentiation of cultivated rice, XV. Complementary lethal genes in rice. Jpn. J. Genet. 32, 83–87. doi: 10.1266/jjg.32.83
Oka, H. I. (1974). Analysis of genes controlling F1 sterility in rice by the use of isogenic lines. Genetics 77, 521–534. doi: 10.1093/genetics/77.3.521
Pasriga, R., Yoon, J., Cho, L. H., and An, G. (2019). Overexpression of RICE FLOWERING LOCUS T1 (RFT1) induces extremely early flowering in rice. Mol. Cells 42, 406–417. doi: 10.14348/molcells.2019.0009
Pelaz, S., Ditta, G. S., Baumann, E., Wisman, E., and Yanofsky, M. F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203. doi: 10.1038/35012103
Prasad, K., Parameswaran, S., and Vijayraghavan, U. (2005). OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and Palea and is an early-acting regulator of inner floral organs. Plant J. 43, 915–928. doi: 10.1111/j.1365-313X.2005.02504.x
Reiber, J. M., and Neuman, D. S. (1999). Hybrid weakness in Phaseolus vulgaris L. I. Disruption of development and hormonal allocation. J. Plant Growth Regul. 18, 101–106. doi: 10.1007/PL00007056
Riechmann, J. L., and Meyerowitz, E. M. (1997). MADS domain proteins in plant development. Biol. Chem. 378, 1079–1101.
Sato, Y. I. (1997). “Mode of inheritance of inviability,” in Science of the Rice Plant, 3, Genetics (Tokyo: Food and Agriculture Policy Research Center).
Sato, Y. I., and Morishima, H. (1987). Studies on the distribution of complementary genes causing F1 weakness in common rice and its wild relatives. 2. Distribution of two complementary genes, Hwc-1 and Hwc-2 gene in native cultivars and its wild relatives of tropical Asia. Euphytica. eds. T. Matsuo and K. Hoshikawa 36, 425–431. doi: 10.1007/BF00041485
Shim, K. C., Kim, S. H., Le, A. Q., Lee, H. S., Adeva, C., Jeon, Y. A., et al. (2019). Fine mapping of a low-temperature germinability QTL qLTG1 using introgression lines derived from Oryza rufipogon. Plant Breed. Biotechnol. 7, 141–150. doi: 10.9787/PBB.2019.7.2.141
Song, S., Wang, G., Hu, Y., Liu, H., Bai, X., Qin, R., et al. (2018). OsMFT1 increases spikelets per panicle and delays heading date in rice by suppressing Ehd1, FZP and SEPALLATA-like genes. J. Exp. Bot. 69, 4283–4293. doi: 10.1093/jxb/ery232
Stebbins, G. L. (1950). “Isolation and the origin of species,” in Variation and Evolution in Plants. eds. L. C. Dunn, H. T. Clarke, S. R. Detwiler, T. Dobzhansky, and F. Schrader (New York: Columbia University Press).
Theissen, G. (2001). Development of floral organ identity: stories from the MADS house. Curr. Opin. Plant Biol. 4, 75–85. doi: 10.1016/S1369-5266(00)00139-4
Xiao, H., Tang, J., Li, Y., Wang, W., Li, X., Jin, L., et al. (2009). STAMENLESS 1, encoding a single C2H2 zinc finger protein, regulates floral organ identity in rice. Plant J. 59, 789–801. doi: 10.1111/j.1365-313X.2009.03913.x
Yamada, T., and Marubashi, W. (2003). Overproduced ethylene causes programmed cell death leading to temperature-sensitive lethality in hybrid seedlings from the cross Nicotiana suaveolens x N. tabacum. Planta 217, 690–698. doi: 10.1007/s00425-003-1035-2
Yamada, T., Marubashi, W., Nakamura, T., and Niwa, M. (2001). Possible involvement of auxin-induced ethylene in an apoptotic cell death during temperature-sensitive lethality expressed by hybrid between Nicotiana glutinosa and N. repanda. Plant Cell Physiol. 42, 923–930. doi: 10.1093/pcp/pce114
Yamamoto, E., Takashi, T., Morinaka, Y., Lin, S., Kitano, H., Matsuoka, M., et al. (2007). Interaction of two recessive genes, hbd2 and hbd3, induces hybrid breakdown in rice. Theor. Appl. Genet. 115, 187–194. doi: 10.1007/s00122-007-0554-9
Yoshida, H., and Nagato, Y. (2011). Flower development in rice. J. Exp. Bot. 62, 4719–4730. doi: 10.1093/jxb/err272
Yuan, H., Qin, P., Hu, L., Zhan, S., Wang, S., Gao, P., et al. (2019). OsSPL18 controls grain weight and grain number in rice. J. Genet. Genomics 46, 41–51. doi: 10.1016/j.jgg.2019.01.003
Zahn, L. M., Kong, H., Leebens-Mack, J. H., Kim, S., Soltis, P. S., Landherr, L. L., et al. (2005). The evolution of the SEPALLATA subfamily of MADS-box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics 169, 2209–2223. doi: 10.1534/genetics.104.037770
Keywords: hybrid weakness, dark tip embryo, rice, floral organ determination, seed germination, abscisic acid
Citation: Kim SH, Ji S-D, Lee H-S, Jeon Y-A, Shim K-C, Adeva C, Luong NH, Yuan P, Kim H-J, Tai TH and Ahn S-N (2022) A Novel Embryo Phenotype Associated With Interspecific Hybrid Weakness in Rice Is Controlled by the MADS-Domain Transcription Factor OsMADS8. Front. Plant Sci. 12:778008. doi: 10.3389/fpls.2021.778008
Received: 16 September 2021; Accepted: 03 December 2021;
Published: 05 January 2022.
Edited by:
Sean Mayes, University of Nottingham, United KingdomReviewed by:
Yunhe Jiang, China Agricultural University, ChinaCopyright © 2022 Kim, Ji, Lee, Jeon, Shim, Adeva, Luong, Yuan, Kim, Tai and Ahn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sang-Nag Ahn, YWhuc25AY251LmFjLmty
†These authors have contributed equally to this work
‡Present address: Shi-Dong Ji, School of Life Science and Agricultural Engineering, Nanyang Normal University, Nanyang City, China
Hyun-Sook Lee, Crop Breeding Division, National Institute of Crop Science, Wanju-Gun, South Korea
Pingrong Yuan, Institute of Food Crop Research, Yunnan Academy of Agricultural Sciences, Kunming, China
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.