
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 22 November 2021
Sec. Plant Biotechnology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.773656
This article is part of the Research Topic Targeted Genome Editing for Crop Improvement View all 20 articles
 Wan-Chin Yeap1*
Wan-Chin Yeap1* Norkhairunnisa Che Mohd Khan 1
Norkhairunnisa Che Mohd Khan 1 Norfadzilah Jamalludin1
Norfadzilah Jamalludin1 Muhammad Rashdan Muad2
Muhammad Rashdan Muad2 David Ross Appleton1
David Ross Appleton1 Harikrishna Kulaveerasingam2
Harikrishna Kulaveerasingam2The clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 (Cas9) system has emerged as a powerful tool for the precise editing of plant genomes for crop improvement. Rapid in vitro methods for the determination of guide RNA (gRNA) cleavage efficiency and an efficient DNA delivery system is essential for gene editing. However, we lack an efficient gene-editing system for palm species. In this study, we described the development of a transient oil palm protoplast assay to rapidly evaluate the cleavage efficiency of CRISPR/Cas9 mutagenesis and the generation of stable transformed oil palms using biolistic particle bombardment in immature embryos. Using the phytoene desaturase (EgPDS) gene, we found cleavage frequency of up to 25.49% in electro-transfected protoplast, which enables the production of transgenic oil palm shoots exhibiting chimeric albino phenotypes as a result of DNA insertions, deletions (InDels), and nucleotide substitutions, with a mutation efficiency of 62.5–83.33%. We further validated the mutagenesis efficiency and specificity of the CRISPR/Cas9 system in oil palm by targeting the brassinosteroid-insensitive 1 (EgBRI1) gene, which resulted in nucleotide substitutions in EgBRI1 with premature necrosis phenotype in oil palm transgenic shoots and stunted phenotype resulting from DNA InDels. Taken together, our results showed that effective and efficient editing of genes using the CRISPR/Cas9 system can be achieved in oil palm by optimizing the selection of efficient gRNA and DNA delivery methods. This newly designed strategy will enable new routes for the genetic improvement in oil palm and related species.
Genome editing technology is emerging as a powerful tool to introduce accurately targeted mutations for plant gene function studies and provide new avenues for crop improvement due to its simplicity, flexibility, consistency, accuracy, and high efficiency (Lozano-Juste and Cutler, 2014; Voytas and Gao, 2014; Bortesi and Fischer, 2015). It has been made simpler with clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 (Cas9) system due to its versatility, effectiveness, and efficiency (Feng et al., 2013; Ma et al., 2015; Jiang and Doudna, 2017). The CRISPR/Cas9 system has been successfully established and widely applied in various economically important crops, including wheat, rice, maize, brassica, sweet potato, banana, and grapevine, to enhance crop productivity, grain quality, nutritional value, disease tolerance, resilience to climate change, and herbicide resistance (Shi et al., 2017; Macovei et al., 2018; Okuzaki et al., 2018; Zhang et al., 2018; Wang et al., 2019; Kaur et al., 2020; Li et al., 2020).
Clustered regularly interspaced short palindromic repeat/Cas9 mediates precise genome modification by gene knock in, knock out, base editing, transcriptional activation or repression, epigenetic modification, and RNA editing (Feng et al., 2013; Konermann et al., 2015; Zong et al., 2017; Mao et al., 2018; Papikian et al., 2019). The CRISPR/Cas9 endonuclease protein is directed to a specific chromosomal DNA site by a short sequence-specific single guide RNA (gRNA) to induce DNA double-stranded breaks (DSBs) and enable precise editing of the target DNA sequence with mutation events, including insertions, deletions (InDels), and nucleotide substitutions induced by the cell repair mechanisms, non-homologous end-joining (NHEJ), or homology-directed repair (HDR) (Jiang and Doudna, 2017). In plants, the efficiency of CRISPR/Cas9-mediated mutagenesis is affected by the editing components (Cas9, promoter, gRNA design, and specificity), the transformation or delivery method into plant cells, the ability of plants to regenerate, and the sensitivity of the mutation detection method (Ma et al., 2015; Liu et al., 2016; Thyme et al., 2016; Wang et al., 2018; Montecillo et al., 2020). The CRISPR/Cas9 system can be delivered into plant cells using several methods, including plant transformation of CRISPR/Cas9 and gRNA expression cassettes, virus-mediated gRNA delivery, DNA-free CRISPR–Cas9 ribonucleoprotein (RNP) delivery to protoplasts, and de novo meristem induction bypassing tissue culture process (Xu et al., 2014; Ali et al., 2015; Woo et al., 2015; Wang et al., 2018; Montecillo et al., 2020). The efficiency of CRISPR/Cas9 is variable in different plant species and requires fine-tuning of the conditions to achieve successful and highly efficient genome editing.
Global food demand is expected to increase by 60% in 2050, driven by the rapid expansion of the human population reaching approximately 9–11 billion and global dietary shifts intensifying wellness and value addition (Fróna et al., 2019). Oil palm is well recognized as the most efficient oil crop that contributes to 35% of global vegetable oil; thus, it can offer a critical solution for global food insecurity to feed the growing global population (Mielke, 2017). Despite the yield and oil quality improvement achieved through conventional breeding and marker-assisted breeding in the past decades (Soh et al., 2017), the annual growth in palm oil production is likely to slow down in the coming years due to a deceleration in land expansion, yield stagnation, climate change, declining labor force, increase in production costs, and pest and disease issues. Thus, the application of genome editing technology can provide opportunities to accelerate and enhance breeding programs with high precision in oil palm to achieve food security and sustainable agriculture.
Limitations in an efficient genetic transformation system, a plant regeneration process, and an in vitro testing system hindered the establishment of the CRISPR/Cas9 system in oil palm. Here, we report an establishment of the CRISPR/Cas9 system that enables efficient genome editing in oil palm. We investigated the efficacy and efficiency of a codon-optimized CRISPR/Cas9 by targeting oil palm phytoene desaturase (EgPDS) gene as a phenotypic marker to facilitate rapid screening of mutant lines as mutation of the PDS gene disrupts the carotenoid pathway and results in albino and dwarf phenotypes (Odipio et al., 2017). We first established a rapid in vitro detection method for testing gRNA efficiency in oil palm using protoplast electro-transfection. Subsequently, we demonstrated the delivery of CRISPR/Cas9 and gRNA cassettes into oil palm immature embryos using biolistic particle bombardment-mediated transformation and validated the mutagenesis effects of CRISPR/Cas9, both genotype and phenotype in regenerated oil palms. We further validated the mutagenesis efficiency and specificity of the CRISPR/Cas9 system in oil palm by targeting oil palm brassinosteroid-insensitive 1 (EgBRI1). We showed that mutations in the kinase domain of BRI1 are associated with palms displaying stunted phenotype and necrosis at the leaf apex.
Oil palm dura (Deli dura) immature embryos extracted from the fresh oil palm fruits at 12 weeks after pollination (WAP) were supplied by Oil Palm Breeding, Sime Darby Plantation Research Sdn. Bhd., Banting, Malaysia, and used as the starting material for this study. Before particle bombardment with plasmids at 200 replicates/constructs, the immature embryos were cultured on Murashige and Skoog (1962) media with sucrose and vitamins. Unopened oil palm spear leaf was used for protoplast isolation.
The Streptococcus pyogenes Cas9 nuclease gene (Jinek et al., 2012) was codon optimized (Supplementary Data Sheet 1) by replacing the tandem rare codons in native gene to codon usage bias in oil palm using the OptimumGene™ codon optimization analysis (GenScript USA Inc., Nanjing, China) to enhance the efficiencies in Cas9 gene expression and translation in oil palm. The codon-optimized Cas9 was synthesized together with the Gateway attP1 site and nuclear localization signal (NLS) fused to the 5′ of SpCas9 and Gateway attP2 site fused to the 3′ of the gene (GenScript USA Inc., Nanjing, China). The synthesized gene construct was shuttled into the pXHb7SNFI-UBIL vector (Vlaams Instituut voor Biotechnologie, Ghent, Belgium) fused to the Ubiquitin promoter and β-glucuronidase (GUS) reporter using Gateway® BP Clonase™ II enzyme mix (Thermo Fisher Scientific, Waltham, MA, United States). The resulting construct is known as pUbil-Cas9 (Supplementary Figure 1). The potential target sequences of gRNAs for oil palms EgPDS (accession no. XP_010937221) and EgBRI1 (accession no. XP_010927763) were analyzed and designed using Benchling1 and CCTop2 (Stemmer et al., 2015). These gRNAs were selected based on their position in the gene, the predicted efficacy score, and the potential for off-target mutation. These gRNAs (Supplementary Table 1) designed to target different exon regions of the EgPDS gene (Figure 1A) were cloned into a modified pCAMBIA1201 vector, fused to rice OsU3 promoter and gRNA scaffold cassette (Supplementary Figure 1).
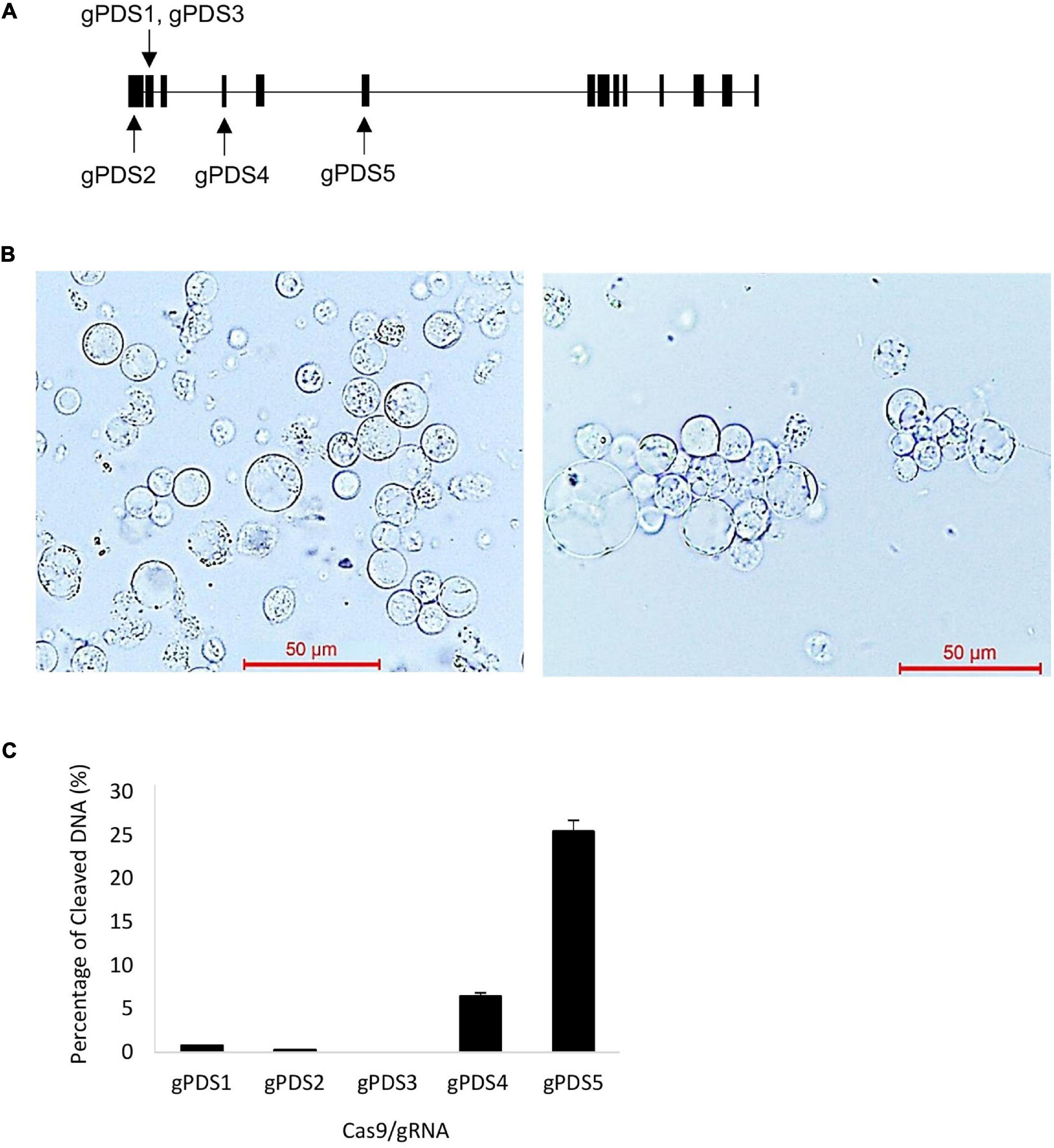
Figure 1. Efficiency testing of clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 (Cas9) and guide RNA (gRNA) activity using electro-transfection in oil palm protoplasts. (A) Schematic map of gRNA target site in phytoene desaturase (EgPDS) gene. (B) Oil palm protoplast before (left) and protoplast agglutination after (right) electro-transfection with Cas9/gRNA. (C) Cleavage efficiency of Cas9/gRNA in oil palm protoplast following electro-transfection. Data are average values ± SE from four independent experiments.
The oil palm mesophyll protoplasts were isolated from the unopened spear leaf as described previously (Masani et al., 2013). These protoplasts were co-transformed with 2 μg plasmids of pUbil-Cas9 and individual gRNA constructed for EgPDS and EgBRI1 genes using electroporation. A single vector of pUbil-Cas9 or gRNA was transformed as controls. The protoplast and plasmid mixture in HEPES-buffered saline solution was electroporated using Gene Pulser Xcell electroporation system (Bio-Rad Laboratories, Hercules, CA, United States) with a square wave pulse program: 550 V/cm (220 V setting/0.4 cm width), 10 ms for two times, and a 20 ms interval for poring pulse. The electroporated protoplasts were incubated in HEPES-buffered saline solution under dark conditions at 25°C with gentle swirling for 72 h. The protoplasts were washed using a washing solution, and the genomic DNA of transformed protoplasts were extracted using QuickExtract™ DNA Extraction Solution (Lucigen, Middleton, WI, United States) according to the instructions of the manufacturer. The electro-transfection efficiency was determined using absolute quantitative real-time PCR. The DNA copies of the GUS reporter gene transformed into protoplast were compared against the total DNA copies of the endogenous gene, Cyclophilin 2 in protoplast, both DNA copies were determined from plasmid dilution standard curve. According to the instructions of the manufacturer, the target site region, flanking the gRNAs of EgPDS and EgBRI1 genes, was amplified, respectively, from the transformed and wild-type protoplasts DNA using region-specific primers (Supplementary Table 2) and iProof™ High-Fidelity DNA Polymerase (Bio-Rad Laboratories, Hercules, CA, United States). The mutations were detected by comparing amplified fragments from the transformed and wild-type protoplasts (50:50 ratio) using the AccuCleave™ T7CE Kit and detection using CRISPR Discovery Gel Kit and Fragment Analyzer™ Automated CE System (Agilent Technologies, Palo Alto, CA, United States). The mutation frequency of the protoplast population was determined based on the percentage of cleavage efficiency calculated using the PROsize Data Analysis Software (Agilent Technologies, Palo Alto, CA, United States).
The pUbil-Cas9 and two efficient gRNA constructs for EgPDS and EgBRI1 (2 μg each) were coated on the surface of 3 mg gold particles in 50% glycerol and mixed with 1M CaCl2 and 16 mM spermidine. The mixture was washed with absolute ethanol three times and was resuspended in 60 μl absolute ethanol. For each bombardment, 10 μl of DNA-microcarrier mixture was placed on the microcarrier and bombarded two times into oil palm immature embryos using the PDS-1000/Hepta Biolistic Particle Delivery System (Bio-Rad Laboratories, Hercules, CA, United States) with 1,100 psi rupture disks, vacuum at 27 mmHg pressure, and 9 cm distance from the tissue. Individual pUbil-Cas9, gRNA, and the combinations of both constructs were bombarded or co-bombarded into 200 replicates of immature embryos. The pUbil-Cas9 and individual gRNAs were used as controls. The transformed immature embryos were maintained in MS agar with sucrose and vitamins (Sigma-Aldrich, St. Louis, MO, United States) supplemented with 50 μg/ml of hygromycin B. Regenerated plants were transferred into MS agar without antibiotic or herbicide and cultured under 16-h light/8-h dark photoperiod for 6–8 months.
Transgenic plants were identified by their resistance to hygromycin at the early phase of tissue culture. Three transformed embryos were randomly selected from the controls and confirmed using β-glucuronidase staining after 2 months on selection media. Putative transgenic plants were confirmed using the Phire Plant Direct PCR Kit (Thermo Fisher Scientific, Waltham, MA, United States) and specific primers targeting Cas9 and OsU3 promoter sequences (Supplementary Table 3). Five transgenic oil palm mutants with altered phenotypes were randomly selected for sequence analysis. Their genomic DNA was extracted using QuickExtract™ DNA Extraction Solution (Lucigen, Middleton, WI, United States). The target site of EgPDS and EgBRI1 was amplified using region-specific primers (Supplementary Table 2) and iProof™ High-Fidelity DNA Polymerase (Bio-Rad Laboratories, Hercules, CA, United States). Amplicons were purified and analyzed using Sanger sequencing (Applied Biosystems, Foster City, CA, United States) for the detection of specific InDels and substitution. Mutation efficiency for each gRNA was evaluated based on the number of T0 transgenic plants with mutation compared to the number of genotyped T0 transgenic plants. The mutation event and frequency were analyzed and detected using TIDE3 (Brinkman et al., 2014). These amplified fragments were cloned into pJET1.2 (Thermo Fisher Scientific, Waltham, MA, United States), and 20 colonies were randomly selected for single-colony plasmid sequencing for chimeric mutation assessment. The sequences were analyzed by aligning to wild-type sequences as reference.
To establish a highly efficient CRISPR/Cas9 system in oil palm, we targeted EgPDS as genetic lesions will result in visible phenotypes. Five gRNAs were designed to target mutagenesis at different exon regions of the EgPDS gene (Figure 1A). First, we examined the functionality of codon-optimized SpCas9 and the effectiveness of the five gRNAs to identify efficient gRNA for further phenotypic validation in the transgenic oil palm, using a fast electroporation-mediated protoplast transient expression system. The protoplast electro-transfection efficiencies ranged from 17 to 26%. Agglutination among protoplasts was observed after electroporation with target constructs without damages (Figure 1B). In EgPDS mutagenesis, mutations were detected in protoplasts transformed with Cas9/gPDS4 and Cas9/gPDS5 with cleavage frequencies of 6.49 ± 0.7 and 25.49 ± 4.11%, respectively (Figure 1C). In contrast, we found minimal mutagenesis activity for gPDS1, gPDS2, and gPDS3 gRNAs, and no mutation activity was detected in the control protoplast pool. These results suggest that the codon-optimized Cas9 and gRNAs are an efficient system for mutagenesis in oil palm and that their efficiency can be easily determined by protoplast electro-transfection.
To examine the efficiency of CRISPR/Cas9 in oil palm, we generated stable transgenic plants by the transformation of pUbil-Cas9 and highly efficient gRNAs for EgPDS (gPDS4 and gPDS5) using the biolistic particle bombardment method. Putatively transformed embryos were selected from non-transformants based on their survival on a hygromycin selection medium. A total of 14 putative transformants were generated using pUbil-Cas9 and gPDS4 transformation, whereas 13 putative transformants were generated using pUil-Cas9 and gPDS5 transformation, with a transformation efficiency of 7 and 6.5%, respectively (Table 1). The transformation of a single vector cassette for Cas9 or individual gRNA into immature embryos resulted in a relatively higher transformation efficiency of up to 19.5%. We randomly selected the putative transformed immature embryos for GUS histochemical assay (Figure 2A). The hygromycin-resistant embryos were later transferred to negative selection media to induce shoot regeneration. Notably, we found that the regeneration efficiencies for EgPDS-edited plants ranged from 44 to 77%.
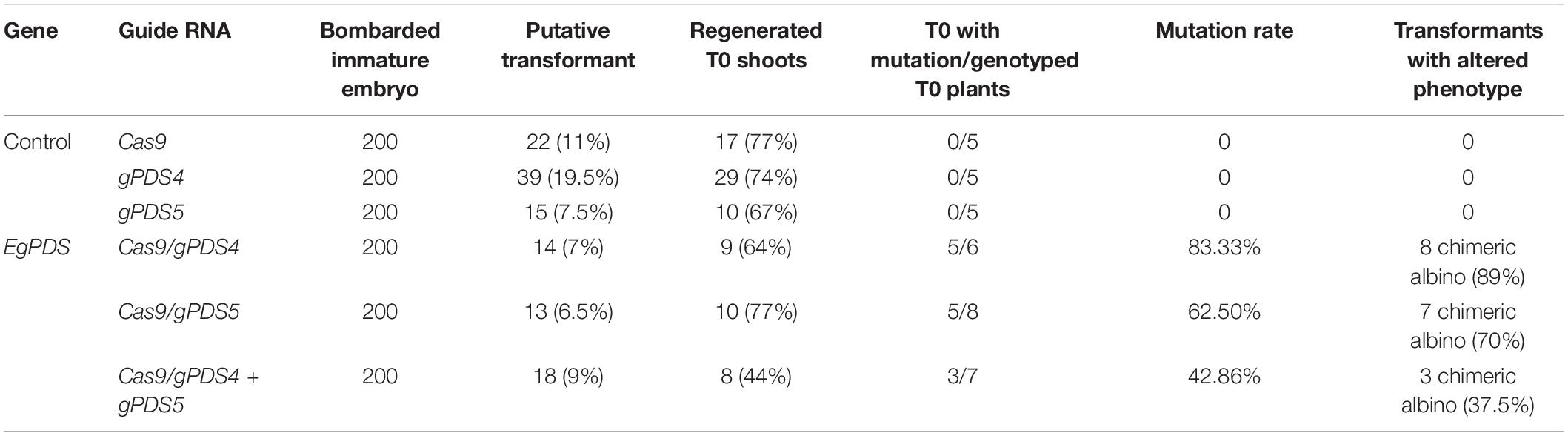
Table 1. The efficiency of clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 (Cas9) and guide RNA (gRNA) transformation targeting phytoene desaturase (EgPDS) gene in oil palm and the phenotypes of the transgenic plants.
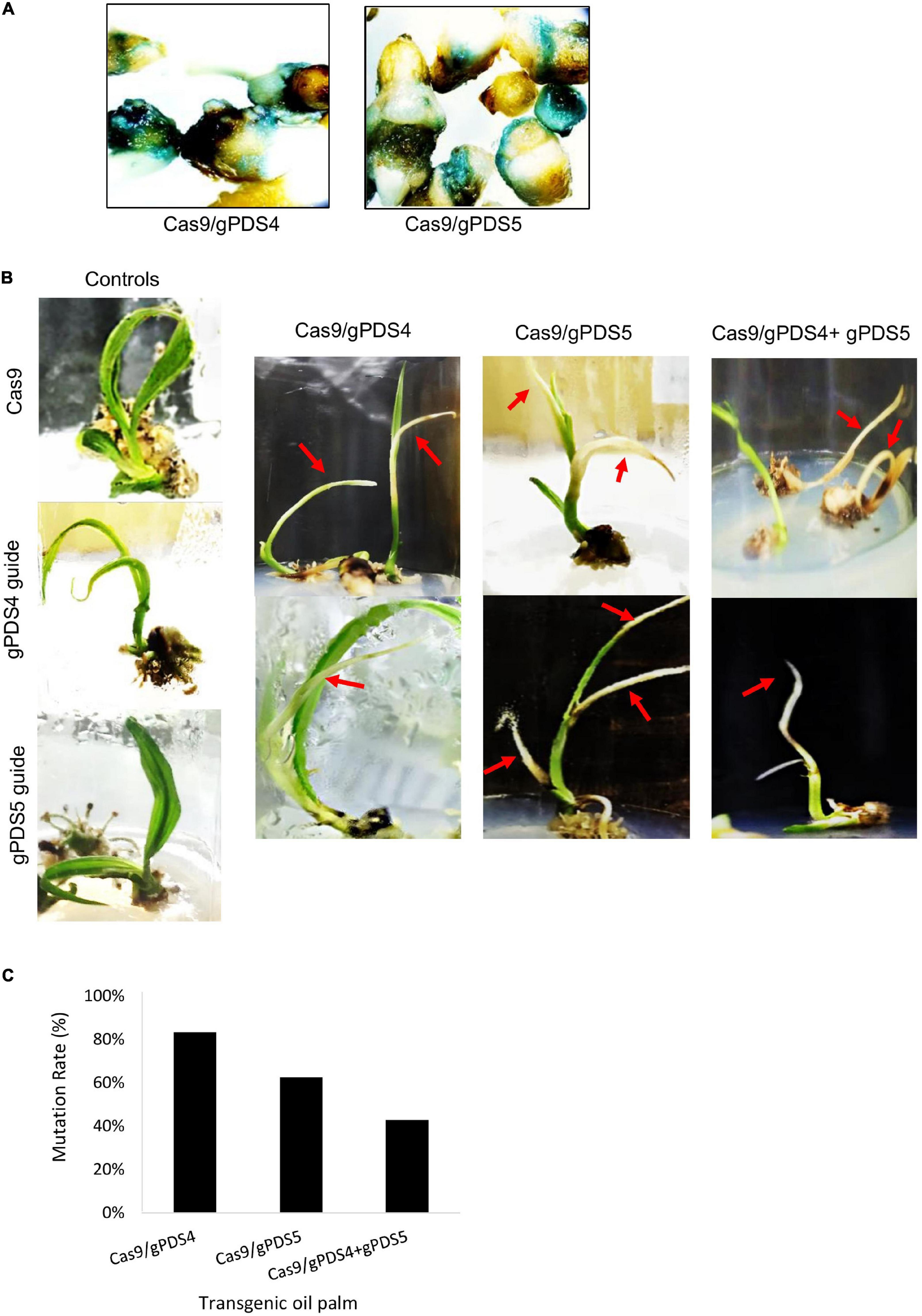
Figure 2. Mutagenesis mediated by CRISPR/Cas9 and gRNA of EgPDS in in vitro transgenic oil palm. (A) Confirmation of genetic transformation by β-glucuronidase (GUS) assay in immature embryos transformed with Cas9/gPDS4 and Cas9/gPDS5. (B) Phenotypes of the Cas9/gPDS4 or gPDS5-mediated mutagenesis and control transgenic in vitro oil palms. Arrow indicating shoots with albino phenotypes. (C) Mutation frequencies of CRISPR/Cas9 mutagenesis in in vitro transgenic oil palms.
Alteration of PDS gene function is predicted to result in albino phenotypes due to defects in carotenoid biosynthesis (Mann et al., 1994). We found that the shoots of regenerated oil palms transformed with pUbil-Cas9/gPDS4 and pUbil-Cas9/gPDS5 displayed albino sectors indicating mosaicism (Figure 2B). These transgenic plantlets were scored based on their albino phenotype to provide a preliminary mutation efficiency for each gRNA module. Mutagenesis mediated by pUbil-Cas9/gPDS4 resulted in 83.3% mutation with chimeric albinism phenotype (Figure 2C) observed in 8 out of 9 regenerated oil palm shoots (Table 1). A total of 7 out of 10 regenerated oil palm shoots exhibited a similar chimeric albinism phenotype with Cas9/gPDS5 mutagenesis. Control plantlets transformed with a single construct of pUbil-Cas9/gRNA did not display signs of albinism. To investigate the effect of multiple mutageneses on plant regeneration and phenotype, we generated mutants transformed with pUbil-Cas9 and both gRNA (gPDS4 and gPDS5) constructs. Notably, only eight hygromycin-resistant immature embryos with gRNA double mutations regenerated into shoots. The EgPDS double mutants showed delayed shoot regeneration compared with the single gRNA mutants and non-edited control equivalents, nevertheless, the albino phenotype was noticeable in the shoots of the three mutants (Table 1 and Figure 2B). All regenerated shoots with albino phenotypes were tested positive for the presence of both Cas9 and gRNA using PCR analysis. To confirm the gene-editing events, we performed PCR amplification and Sanger sequencing analysis using shoots exhibiting a clear EgPDS-edited albino phenotype (Figures 3A–C). These sequence traces were first analyzed using the TIDE algorithm (Brinkman et al., 2014) to identify the occurrence of InDels from the DSBs (Supplementary Figure 2). The sequence data indicated multiple mutations including InDels and single base substitutions in the EgPDS gene of the genotyped regenerated shoots, and the mutated sequences were validated using gene cloning. Mutations by Cas9/gPDS4 induced independent events with 71% occurrence of deletions (Figure 3D) ranging from −1 to −24 bp and 29% of nucleotide substitutions upstream of the protospacer adjacent motif sequence (PAM) region of the target site. The largest deletion (−24 bp) was detected at the boundary of intron 3, and exon 4 of the EgPDS gene may cause disruption in the splice-regulatory sequences and modulate exon skipping or alter splicing of pre-mRNA. Insertion (+1 bp), deletion (−1, −2, and −9 bp), and substitution (1–3 bp) were detected in Cas9/gPDS5 mutants at an occurrence frequency of 29, 43, and 29%, respectively. The nucleotide indels led to disruption in amino acid sequence and exon skipping in one of the mutants, whereas amino acid substitutions resulted from the nucleotide substitutions. In double mutation shoots, deletion (−2 and −16 bp) (75% occurrence) and insertion (+8 bp) (25% occurrence) were detected at the gPDS4 and gPDS5 target regions. These mutations led to amino acid frameshifts and the introduction of multiple stop codons near target regions. Collectively, our data suggest that the optimization of vectors for efficient Cas9 and gRNA activity combined with a rapid transformation system is a critical factor for an efficient gene-editing system in oil palm.
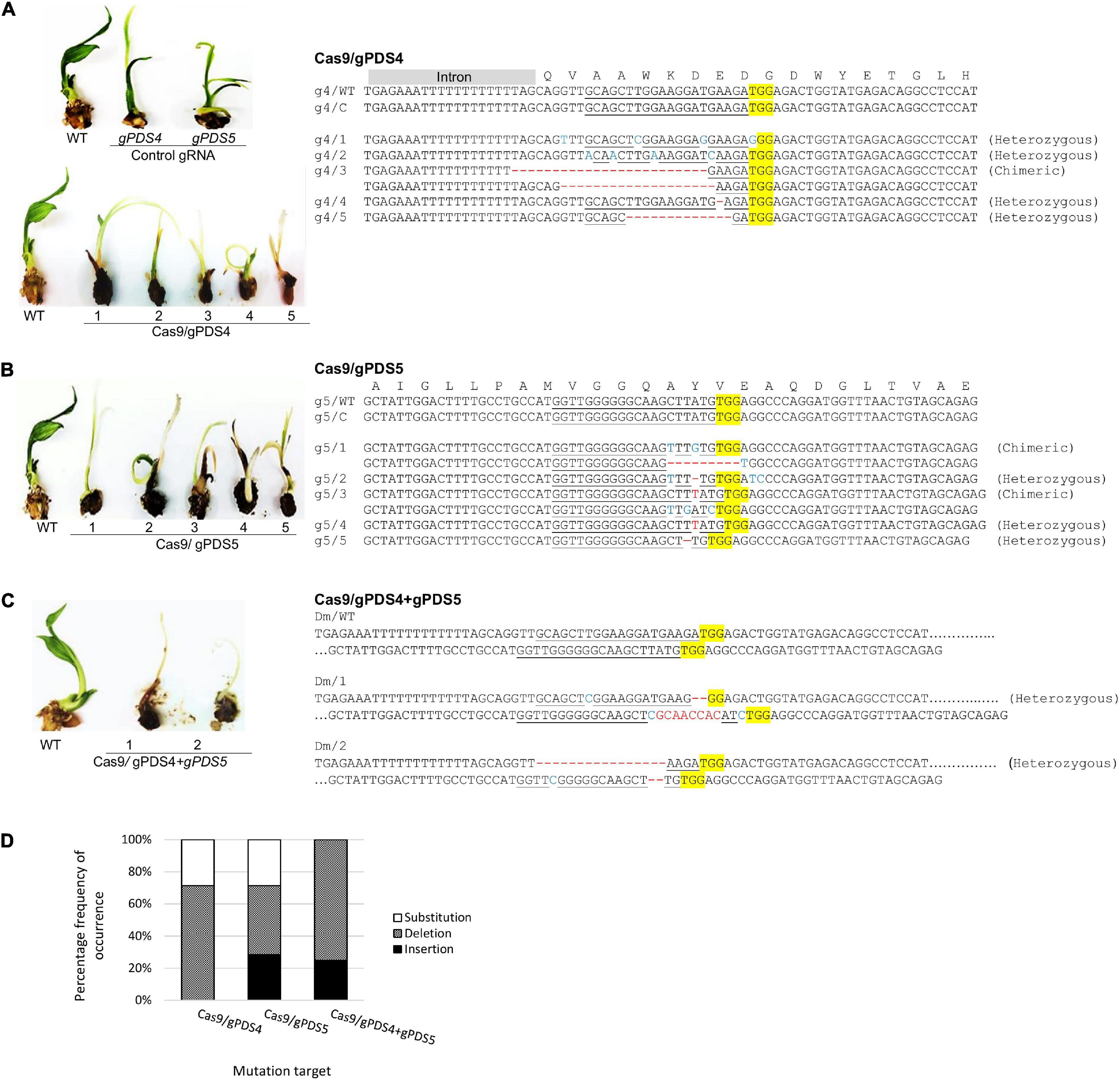
Figure 3. Mutation events of targeted CRISPR/Cas9 mutagenesis of EgPDS gene in in vitro transgenic oil palm. Mutation events detected in the transgenic oil palms exhibiting the chimeric albino phenotypes using sequencing-based detection at the target sites of (A) gPDS4, (B) gPDS5, and (C) both gPDS4 and gPDS5 in the EgPDS gene and (D) the percentage frequency of occurrence for each type of mutation event. Protospacer adjacent motif sequence (PAM) is highlighted in yellow, InDels sequences of the target site are shown in red, and nucleotide substitution are marked in blue. g4, gPDS4; WT, wild-type; C, control; g5, gPDS5; Dm, double mutant; 1–5, plantlet number.
BRI1 gene encodes a brassinosteroid receptor and defective in BRI1 results in dwarf phenotype in Arabidopsis, rice, and maize (Clouse et al., 1996; Noguchi et al., 1999; Yamamuro et al., 2000; Kir et al., 2015). Hence, we selected dwarfism as another visual phenotypic marker for the validation of the genome editing system in oil palm. To manipulate the brassinosteroid responses, we selected EgBRI1 (accession no. XP_010927763) for targeted mutagenesis. The EgBRI1 genomic DNA contains one intronless open reading frame of 3390 bp, and gRNAs were designed based on PAM sites (Figure 4A). The activity of the CRISPR/Cas9 system and the efficiencies of gRNAs targeting the EgBRI1 gene were first verified using in vitro electro-transfection system in protoplast. Mutagenesis of EgBRI1 gene mediated by Cas9 with gBRI1-1, gBRI1-2, and gBRI1-5 showed cleavage frequencies of 9.8 ± 1.21, 18.32 ± 1.4, and 5.1 ± 0.53%, respectively, in protoplast with electro-transfection efficiencies ranging from 12 to 17% (Figure 4B). However, cleavage was not detectable with the remaining gRNAs tested in the protoplast system. The two most efficient gRNAs, gBRI1-1 and gBRI1-2, were selected for transformation in the oil palm. The transformation with pUbil-Cas9 and gBRI1-1 resulted in 24 putative transformants (with a 12% transformation efficiency), while the transformation with pUbil-Cas9 and gBRI1-2 resulted in 23 putative transformants (with an 11.5% transformation efficiency) (Table 2). Overall, the mutagenesis efficiency for EgBRI1 ranged between 58.82 and 100%. Targeted mutagenesis of the EgBRI1 resulted in two distinct phenotypes found in most of the regenerated plants (75–87% regeneration efficiency), reduced plant elongation (stunted), and apex necrosis (Figures 4C,D) with mutation efficiency of 58.82 and 66.67% using pUbil-Cas9/gBRI1-1 and pUbil-Cas9/gBRI1-2, respectively. Sequence analysis of the targeted region detected a wide range of mutations in regenerated transgenic oil palm shoots. Those that arose from pUbil-Cas9/gBRI1-1 and pUbil-Cas9/gBRI1-2 had multiple nucleotide insertions (+1, +8 bp), nucleotide deletions (−1 to −25 bp), and nucleotide substitutions at the sequence targeted at an occurrence frequency of 18–20, 30–36, and 45–50%, respectively (Figures 5A,B). These nonsense mutations may cause the frameshift and aberrant amino acid sequences with premature stop codon introduction and lead to protein truncation. Likewise, the impaired growth phenotypes in mutants generated using paired-gRNAs resulted primarily in deletions (−1, −2, −18, and −39 bp) and insertions (+4 bp) (Figure 5C). Nucleotide substitution mutations are predicted to affect the activity of the leucine-rich repeat receptor-like protein kinase domain of BRI1 (Figure 5D) and are responsible for the apex necrosis phenotype observed in these plants.
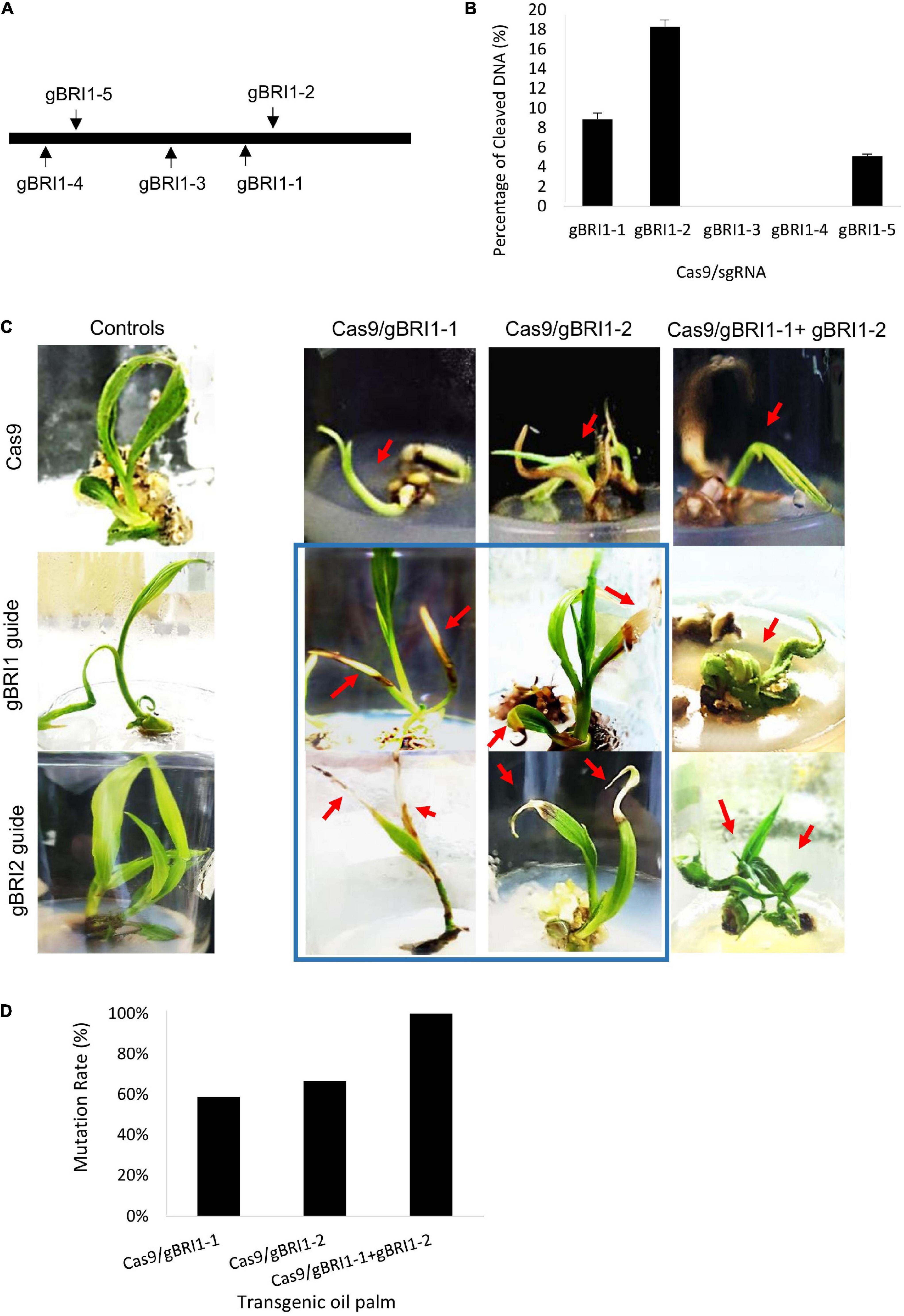
Figure 4. Clustered regularly interspaced short palindromic repeat/Cas9-targeted mutagenesis of brassinosteroid-insensitive 1(EgBRI1) gene in in vitro oil palm plantlets. (A) Schema of gRNA targeting intronless EgBRI1 gene in oil palm. (B) Cleavage efficiency of Cas9/gRNA targeting EgBRI1 gene in oil palm protoplast. Data are average values ± SE from four independent experiments. (C) CRISPR/Cas9-induced EgBRI1 mutations at different gRNA target sites showing stunted growth and leaf apex necrosis phenotypes (boxed in blue) in transgenic oil palm shootlets and controls transformed with either Cas9 or gRNA alone. (D) Mutation frequencies of CRISPR/Cas9 mutagenesis based on the occurrence of stunted or necrotic leaf apex phenotypes in transgenic oil palm.
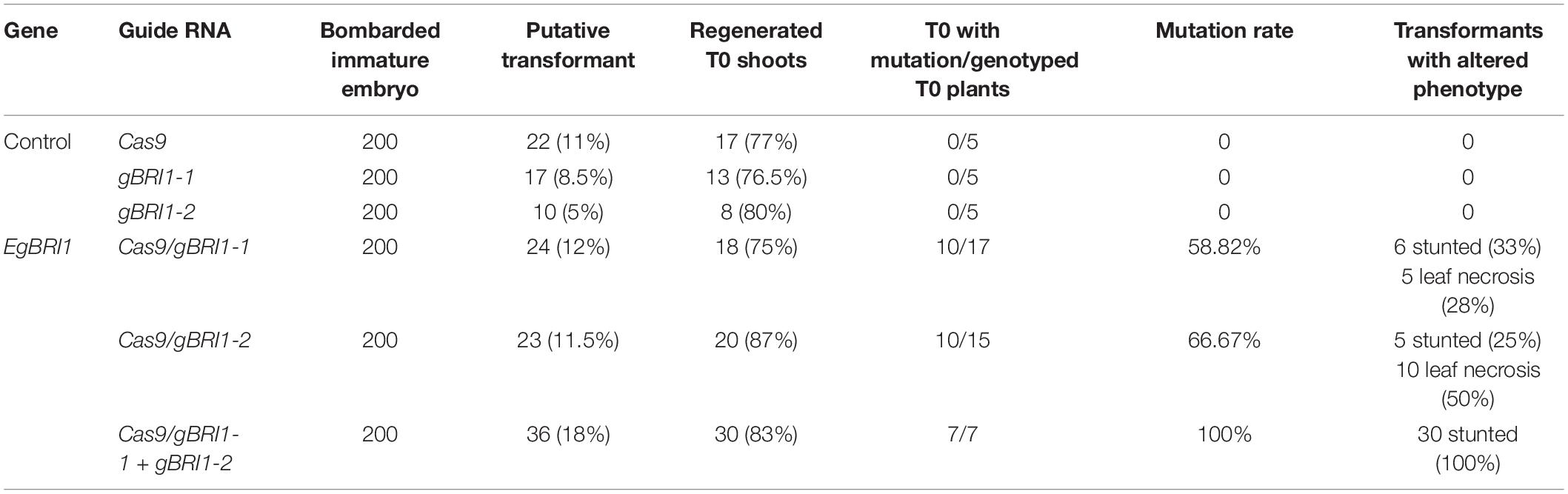
Table 2. The efficiency of CRISPR/Cas9 and gRNA transformation targeting brassinosteroid-insensitive 1 (EgBRI1) gene in oil palm and the phenotypes of the transgenic plants.

Figure 5. Clustered regularly interspaced short palindromic repeat/Cas9-mediated mutation events and phenotypes of EgBRI1 mutant transgenic oil palm. (A) The overall percentage frequency of occurrence for each type of mutation event detected at gBRI1-1 and gBRI1-2 targeted sites. The mutation events detected in the transgenic oil palms exhibiting (B) stunted growth phenotype induced by CRISPR/Cas9 and gBRI1-1, gBRI1-2, or (C) dual gRNAs mutation induction in the EgBRI1 gene, and (D) necrotic shoot apex phenotype. PAM is highlighted in yellow, InDels sequences of the target site are shown in red, and nucleotide substitution is marked in blue. g1, gBRI1-1; g2, gBRI1-2; WT, wild-type; C, control; Dm, double mutant; 1–5, plantlet number.
Genome editing provides opportunities for the improvement of the trait in crops, yet its implementation requires an efficient genetic transformation and regeneration system. In oil palm, limitations in the DNA delivery method, inefficient clonal propagation, and regeneration processes constrain rapid progress in genetic engineering. In this study, we developed an efficient system for CRISPR/Cas9 genome editing in oil palm by targeting EgPDS and EgBRI1 genes. Mutation rates mediated by CRISPR/Cas9 varied depending on the specificity and effectiveness of gRNAs to target sequences, delivery methods, and sensitivity of the detection method. We first established an in vitro protoplast electro-transfection method to identify mutagenic and efficient gRNAs. Electroporation of Cas9 and gRNA constructs using our optimized conditions resulted in cleavage frequencies of up to 25.49% in viable oil palm protoplasts, suggesting that the codon-optimized Cas9 was effective in inducing cleavage. Several gRNAs failed to elicit cleavage despite high GC-content has been associated with higher Cas9 editing efficiency (Liu et al., 2016). This may be contributed by the sequence composition or secondary structure of several target gRNAs that potentially cause failure to recognize target sequence or formation of functional Cas9-gRNA complex in protoplast (Thyme et al., 2016). In cabbage, neon electro-transfection at 1,000 V was 1.4 times more efficient than PEG-mediated transfection in delivering CRISPR-Cas9 RNP into protoplast and resulted in 3.4% InDels mutation frequency in the PDS1 gene (Lee et al., 2020). Despite a previously reported PEG-mediated transfection method for protoplast, the protoplast viability and reproducibility are significantly reduced due to toxicity effects of PEG (Masani et al., 2014). Our method for electro-transfection of genome editing tools in oil palm protoplasts provides an effective alternative strategy for gRNA cleavage activity and potential opportunity to develop DNA-free genome-edited palms. However, limitations with oil palm protoplast regeneration require further studies for improvement to enable deployment of the protoplast system for DNA-free gene editing.
High transformation efficiency gene editing has been reported in monocot plant species including 57% in maize, 80% in wheat, 64% in rice, and 14.3% in sorghum (Xu et al., 2014; Char et al., 2017, 2020; Macovei et al., 2018; Zhang et al., 2018). Previous studies in oil palm achieved 0.7–1.5% transformation efficiencies using Agrobacterium-mediated and microprojectile bombardment transformation methods in embryogenic callus and regeneration by indirect somatic embryogenesis (Parveez et al., 1998; Masli et al., 2012; Dayang Izawati et al., 2015). In this study, we obtained 5–19.5% transformation efficiencies using particle bombardment into zygotic embryos, and up to 87% of embryos regenerated into shoots through direct embryogenesis. Our results suggest that the optimization of transformation protocol, selection of explant, and regeneration methods are effective in improving oil palm transformation efficiency. Furthermore, a direct embryogenesis approach was employed to enable rapid regeneration with minimized risk of culture-induced genetic variation that commonly occurs during callusing phase (Weckx et al., 2019). Although chimeric albinism phenotype was observed in the transgenic EgPDS-edited palms using our method, nevertheless, homozygous mutants and transgene removal can be obtained by genetic segregation and backcrossing.
We achieved high editing efficiency (42.86–100%) at target sites of EgPDS and EgBRI1 genes in transgenic oil palms with our CRISPR/Cas9 system. In rice, high mutagenesis efficiency of 87–100% (di-allelic edits) was achieved using rice codon-optimized SpCas9 compared with the optimized SpCas9 from bacterial, human, and Chlamydomonas in T0 transgenic plants (Zhou et al., 2014). High editing efficiency in our system may be attributed to the oil palm codon-optimized Cas9 gene driven by ubiquitin, an active promoter in monocot (Schledzewski and Mendel, 1994), efficient gRNAs, and delivery method for Cas9/gRNA. Our transgenic plants showed insertions (18–40%), deletions (30–75%), and high levels of nucleotide substitutions (29–50%) in EgPDS and EgBRI1 genes. Although InDels are the most common mutation induced by error-prone NHEJ repair mechanism that involves direct rejoining of DSB ends regardless of sequence homology (Rodgers and McVey, 2016), high frequency of NHEJ-induced base substitutions are also prevalent in plants and substantial evidence have been reported in rice, cassava, citrus, soybean, and melon (Sun et al., 2015; Odipio et al., 2017; Macovei et al., 2018; Hooghvorst et al., 2019; Dutt et al., 2020). For instance, a high frequency of base substitutions has been reported in melon (91%) and rice (25–45%) (Macovei et al., 2018; Hooghvorst et al., 2019). In our study, InDels in the EgBRI1 gene resulted in reduced plant elongation phenotype, while leaf apex necrosis was generated from base substitution events in transgenic palms. Early studies in Arabidopsis and rice reported dwarf and shortened internode phenotypes in the bri1 loss-of-function mutant plants (Clouse et al., 1996; Noguchi et al., 1999; Yamamuro et al., 2000). Choe et al. (1999) further reported that mutation in Arabidopsis bri1 results in a retarded leaf senescence phenotype. Phenotypes of our EgBRI1 transgenic mutant palms are substantially affected by the types of NHEJ-induced mutations, and the specificity of the repair mechanism for DSBs is critical to driving accurate genomic changes without errors. Interestingly, we observed a low frequency of nucleotide substitutions using pUbil-Cas9/paired-gRNAs in both genes. Blunt end cleavage site generated using single gRNA is commonly repaired by error-prone NHEJ mechanism, but precise end-joining repairs have been reported in human cells through simultaneous DNA DSBs by Cas9/paired gRNAs, suggesting a direct ligation without end processing (Zheng et al., 2014). Moreover, previous studies in mammalian cells reported that the accuracy of the NHEJ repair mechanism is reduced when the distance between two collinear DSBs is beyond 1 kb (Boubakour-Azzouz and Ricchetti, 2008; Guirouilh-Barbat et al., 2016; Guo et al., 2018). Simultaneous DSBs by Cas9/paired-gRNAs at short distance may improve the specificity of the repair mechanism; however, the current understanding of this precise mechanism remains to be determined. Hence, the dynamic interaction between Cas9 cleavage and the endogenous DNA repair mechanism needs further study to enhance the efficiency and specificity of CRISPR/Cas9 system applications in plants.
Furthermore, we adapted the paired-gRNA using multi-cassette (monocistronic) transformation to increase the mutation frequency and chromosomal fragment deletion in the targeted gene. The assayed pUbil-Cas9/paired-gRNAs were mutagenic and active in inducing InDels in two target genes; however, no large chromosomal fragment deletion was detected. Chromosomal fragment deletion has been reported in kiwifruit with 755 and 271 bp deletions using polycistronic tRNA-gRNA (PTG)/Cas9 system, which is 10-fold higher than conventional CRISPR/Cas9 expression cassette (Wang et al., 2018). The PTG/Cas9 system enables a simultaneous expression of multiple gRNAs up to 31 times higher than monocistronic cassette and improves mutagenesis of multiple chromosomal fragment deletions in rice protoplast (Xie et al., 2015). Moreover, large chromosomal segments of more than 100 kb deletions between two genomic loci were achieved in rice protoplasts using the multiplex gRNA expression system (Zhou et al., 2014). Chromosomal fragment deletion is a potential strategy for chromosomal engineering to remove undesired loci with detrimental traits in crops and could be achieved by multiplex gRNA expression or PTG/Cas9 systems using gRNAs with similar efficiency at both target sites.
Our study demonstrated that the CRISPR/Cas9 genome editing system is effective and efficient in editing oil palm genes. In this study, the established in vitro electro-transfection assay provides a rapid assessment and evaluation of gRNA efficiency in oil palm protoplast to reduce the time and cost for transformation and regeneration in oil palm using inefficient gRNAs. An efficient transformation system in oil palm or methods to generate non-transgenic gene-edited mutants are highly desired to enhance the occurrence of targeted mutagenesis with the CRISPR/Cas9 system. This enabling platform for genome editing may accelerate the exploration of gene function for trait improvement in oil palm, in particular, the agronomic traits that address challenges in oil palm cultivation including basal stem rot or Ganoderma disease, abiotic stress tolerance to reduce losses due to climate change, and ease of harvesting traits such as dwarf, long stalk, and virescens. In addition, our study offers new insights to other related palm species including date palm and coconut that shared similar limitations in the DNA delivery method, low efficacy of genetic transformation, and inefficient tissue culture propagation and regeneration processes. The availability of full genome sequence opened a new opportunity for genetic improvement in date palm and coconut through the CRISPR/Cas9 genome editing approach. This approach will be helpful to develop date palm and coconut resistance against biotic and abiotic stresses.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
W-CY, DRA, and KH conceived the experiments. W-CY constructed the vectors, performed oil palm protoplast experiments, analyzed the data, and wrote the manuscript. NCMK, NJ, and MRM conducted the oil palm transformation and tissue culture and regenerated transgenic oil palm. All authors contributed to the article and approved the submitted version.
The authors declare that this study received funding from Sime Darby Plantation Berhad. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article, or the decision to submit it for publication.
All authors were employed by the company Sime Darby Plantation Berhad.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank Sime Darby Research, Banting, Selangor, Malaysia, for the oil palm samples; Anusha Nair and Nurliyana Ruzlan for technical assistance; and Jose Gutierrez-Marcos (University of Warwick) for providing advice and reviewing. The authors also thank the reviewers for their critical comments.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.773656/full#supplementary-material
Ali, Z., Abul-Faraj, A., Li, L., Ghosh, N., Piatek, M., Mahjoub, A., et al. (2015). Efficient virus-mediated genome editing in plants using the CRISPR/Cas9 system. Mol. Plant 8, 1288–1291. doi: 10.1016/j.molp.2015.02.011
Bortesi, L., and Fischer, R. (2015). The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 33, 41–52. doi: 10.1016/j.biotechadv.2014.12.006
Boubakour-Azzouz, I., and Ricchetti, M. (2008). Low joining efficiency and non-conservative repair of two distant double-strand breaks in mouse embryonic stem cells. DNA Repair 7, 149–161. doi: 10.1016/j.dnarep.2007.09.005
Brinkman, E. K., Chen, T., Amendola, M., and van Steensel, B. (2014). Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168–e168. doi: 10.1093/nar/gku936
Char, S. N., Neelakandan, A. K., Nahampun, H., Frame, B., Main, M., Spalding, M. H., et al. (2017). An Agrobacterium-delivered CRISPR/Cas9 system for high-frequency targeted mutagenesis in maize. Plant Biotechnol. J. 15, 257–268. doi: 10.1111/pbi.12611
Char, S. N., Wei, J., Mu, Q., Li, X., Zhang, Z. J., Yu, J., et al. (2020). An Agrobacterium-delivered CRISPR/Cas9 system for targeted mutagenesis in sorghum. Plant Biotechnol. J. 18:319. doi: 10.1111/pbi.13229
Choe, S., Dilkes, B. P., Gregory, B. D., Ross, A. S., Yuan, H., Noguchi, T., et al. (1999). The Arabidopsis dwarf1 mutant is defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiol. 119, 897–908. doi: 10.1104/pp.119.3.897
Clouse, S. D., Langford, M., and McMorris, T. C. (1996). A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 111, 671–678. doi: 10.1104/pp.111.3.671
Dayang Izawati, A. M., Abdul Masani, M. Y., Ismail, I., and Ghulam Kadir, A. P. (2015). Evaluation on the effectiveness of 2-deoxyglucose-6-phosphate phosphatase (DOGR1) gene as a selectable marker for oil palm (Elaeis guineensis Jacq.) embryogenic calli transformation mediated by Agrobacterium tumefaciens. Front. Plant Sci. 6:727. doi: 10.3389/fpls.2015.00727
Dutt, M., Mou, Z., Zhang, X., Tanwir, S. E., and Grosser, J. W. (2020). Efficient CRISPR/Cas9 genome editing with Citrus embryogenic cell cultures. BMC Biotechnol. 20, 1–7. doi: 10.1186/s12896-020-00652-9
Feng, Z., Zhang, B., Ding, W., Liu, X., Yang, D.-L., Wei, P., et al. (2013). Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 23, 1229–1232. doi: 10.1038/cr.2013.114
Fróna, D., Szenderák, J., and Harangi-Rákos, M. (2019). The challenge of feeding the world. Sustainability 11:5816. doi: 10.3390/su11205816
Guirouilh-Barbat, J., Gelot, C., Xie, A., Dardillac, E., Scully, R., and Lopez, B. S. (2016). 53BP1 protects against CtIP-dependent capture of ectopic chromosomal sequences at the junction of distant double-strand breaks. PLoS Genet. 12:e1006230. doi: 10.1371/journal.pgen.1006230
Guo, T., Feng, Y.-L., Xiao, J.-J., Liu, Q., Sun, X.-N., Xiang, J.-F., et al. (2018). Harnessing accurate non-homologous end joining for efficient precise deletion in CRISPR/Cas9-mediated genome editing. Genome Biol. 19, 1–20. doi: 10.1186/s13059-018-1518-x
Hooghvorst, I., López-Cristoffanini, C., and Nogués, S. (2019). Efficient knockout of phytoene desaturase gene using CRISPR/Cas9 in melon. Sci. Rep. 9, 1–7. doi: 10.1038/s41598-019-53710-4
Jiang, F., and Doudna, J. A. (2017). CRISPR–Cas9 structures and mechanisms. Annu. Rev. Biophys. 46, 505–529. doi: 10.1146/annurev-biophys-062215-010822
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Kaur, N., Alok, A., Kumar, P., Kaur, N., Awasthi, P., Chaturvedi, S., et al. (2020). CRISPR/Cas9 directed editing of lycopene epsilon-cyclase modulates metabolic flux for β-carotene biosynthesis in banana fruit. Metab. Eng. 59, 76–86. doi: 10.1016/j.ymben.2020.01.008
Kir, G., Ye, H., Nelissen, H., Neelakandan, A. K., Kusnandar, A. S., Luo, A., et al. (2015). RNA interference knockdown of BRASSINOSTEROID INSENSITIVE1 in maize reveals novel functions for brassinosteroid signaling in controlling plant architecture. Plant Physiol. 169, 826–839. doi: 10.1104/pp.15.00367
Konermann, S., Brigham, M. D., Trevino, A. E., Joung, J., Abudayyeh, O. O., Barcena, C., et al. (2015). Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588. doi: 10.1038/nature14136
Lee, M. H., Lee, J., Choi, S. A., Kim, Y.-S., Koo, O., Choi, S. H., et al. (2020). Efficient genome editing using CRISPR–Cas9 RNP delivery into cabbage protoplasts via electro-transfection. Plant Biotechnol. Rep. 14, 695–702. doi: 10.1007/s11816-020-00645-2
Li, M.-Y., Jiao, Y.-T., Wang, Y.-T., Zhang, N., Wang, B.-B., Liu, R.-Q., et al. (2020). CRISPR/Cas9-mediated VvPR4b editing decreases downy mildew resistance in grapevine (Vitis vinifera L.). Hortic. Res. 7, 1–11. doi: 10.1038/s41438-020-00371-4
Liu, X., Homma, A., Sayadi, J., Yang, S., Ohashi, J., and Takumi, T. (2016). Sequence features associated with the cleavage efficiency of CRISPR/Cas9 system. Sci. Rep. 6, 1–9. doi: 10.1038/srep19675
Lozano-Juste, J., and Cutler, S. R. (2014). Plant genome engineering in full bloom. Trends Plant Sci. 19, 284–287. doi: 10.1016/j.tplants.2014.02.014
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., et al. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284. doi: 10.1016/j.molp.2015.04.007
Macovei, A., Sevilla, N. R., Cantos, C., Jonson, G. B., Slamet-Loedin, I., Čermák, T., et al. (2018). Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to Rice tungro spherical virus. Plant Biotechnol. J. 16, 1918–1927. doi: 10.1111/pbi.12927
Mann, V., Pecker, I., and Hirschberg, J. (1994). Cloning and characterization of the gene for phytoene desaturase (Pds) from tomato (Lycopersicon esculentum). Plant Mol. Biol. 24, 429–434. doi: 10.1007/BF00024111
Mao, Y., Yang, X., Zhou, Y., Zhang, Z., Botella, J. R., and Zhu, J.-K. (2018). Manipulating plant RNA-silencing pathways to improve the gene editing efficiency of CRISPR/Cas9 systems. Genome Biol. 19, 1–15. doi: 10.1186/s13059-018-1529-7
Masani, M. Y. A., Noll, G., Parveez, G. K. A., Sambanthamurthi, R., and Prüfer, D. (2013). Regeneration of viable oil palm plants from protoplasts by optimizing media components, growth regulators and cultivation procedures. Plant Sci. 210, 118–127. doi: 10.1016/j.plantsci.2013.05.021
Masani, M. Y. A., Noll, G. A., Parveez, G. K. A., Sambanthamurthi, R., and Prüfer, D. (2014). Efficient transformation of oil palm protoplasts by PEG-mediated transfection and DNA microinjection. PLoS One 9:e96831. doi: 10.1371/journal.pone.0096831
Masli, D. I. A., Kadir, A. P. G., and Yunus, A. M. M. (2012). Transformation of oil palm using Agrobacterium tumefaciens. Methods Mol. Biol. 847, 177–188. doi: 10.1007/978-1-61779-558-9_15
Mielke, T. (2017). “World markets of vegetable oils: status and prospects” in Encyclopedia of Sustainability Science and Technology Series. ed. R. A. Meyers (New York: Springer). 1–38.
Montecillo, J. A. V., Chu, L. L., and Bae, H. (2020). CRISPR-Cas9 system for plant genome editing: current approaches and emerging developments. Agronomy 10:1033. doi: 10.3390/agronomy10071033
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio agsays with tohaoco tissue cultures. Physiol. Plant 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Noguchi, T., Fujioka, S., Choe, S., Takatsuto, S., Yoshida, S., Yuan, H., et al. (1999). Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 121, 743–752. doi: 10.1104/pp.121.3.743
Odipio, J., Alicai, T., Ingelbrecht, I., Nusinow, D. A., Bart, R., and Taylor, N. J. (2017). Efficient CRISPR/Cas9 genome editing of phytoene desaturase in cassava. Front. Plant Sci. 8:1780. doi: 10.3389/fpls.2017.01780
Okuzaki, A., Ogawa, T., Koizuka, C., Kaneko, K., Inaba, M., Imamura, J., et al. (2018). CRISPR/Cas9-mediated genome editing of the fatty acid desaturase 2 gene in Brassica napus. Plant Physiol. Biochem. 131, 63–69. doi: 10.1016/j.plaphy.2018.04.025
Papikian, A., Liu, W., Gallego-Bartolomé, J., and Jacobsen, S. E. (2019). Site-specific manipulation of Arabidopsis loci using CRISPR-Cas9 SunTag systems. Nat. Commun. 10, 1–11. doi: 10.1038/s41467-019-08736-7
Parveez, G. K. A., Chowdhury, M., and Saleh, N. M. (1998). Biological parameters affecting transient GUS gene expression in oil palm (Elaeis guineensis Jacq.) embryogenic calli via microprojectile bombardment. Industr. Crops Prod. 8, 17–27. doi: 10.1016/S0926-6690(97)00077-0
Rodgers, K., and McVey, M. (2016). Error-prone repair of DNA double-strand breaks. J. Cell. Physiol. 231, 15–24. doi: 10.1002/jcp.25053
Schledzewski, K., and Mendel, R. R. (1994). Quantitative transient gene expression: comparison of the promoters for maize polyubiquitin1, rice actin1, maize-derived Emu and CaMV 35S in cells of barley, maize and tobacco. Transgen. Res. 3, 249–255. doi: 10.1007/BF02336778
Shi, J., Gao, H., Wang, H., Lafitte, H. R., Archibald, R. L., Yang, M., et al. (2017). ARGOS 8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 15, 207–216. doi: 10.1111/pbi.12603
Soh, A. C., Mayes, S., and Roberts, J. A. (2017). Oil Palm Breeding: genetics and Genomics. Atlanta: CRC Press.
Stemmer, M., Thumberger, T., del Sol Keyer, M., Wittbrodt, J., and Mateo, J. L. (2015). CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One 10:e0124633. doi: 10.1371/journal.pone.0124633
Sun, X., Hu, Z., Chen, R., Jiang, Q., Song, G., Zhang, H., et al. (2015). Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci. Rep. 5, 1–10. doi: 10.1038/srep10342
Thyme, S. B., Akhmetova, L., Montague, T. G., Valen, E., and Schier, A. F. (2016). Internal guide RNA interactions interfere with Cas9-mediated cleavage. Nat. Commun. 7, 1–7. doi: 10.1038/ncomms11750
Voytas, D. F., and Gao, C. (2014). Precision genome engineering and agriculture: opportunities and regulatory challenges. PLoS Biol. 12:e1001877. doi: 10.1371/journal.pbio.1001877
Wang, H., Wu, Y., Zhang, Y., Yang, J., Fan, W., Zhang, H., et al. (2019). CRISPR/Cas9-based mutagenesis of starch biosynthetic genes in sweet potato (Ipomoea Batatas) for the improvement of starch quality. Int. J. Mol. Sci. 20:4702. doi: 10.3390/ijms20194702
Wang, Z., Wang, S., Li, D., Zhang, Q., Li, L., Zhong, C., et al. (2018). Optimized paired-sgRNA/Cas9 cloning and expression cassette triggers high-efficiency multiplex genome editing in kiwifruit. Plant Biotechnol. J. 16, 1424–1433. doi: 10.1111/pbi.12884
Weckx, S., Inzé, D., and Maene, L. (2019). Tissue culture of oil palm: finding the balance between mass propagation and somaclonal variation. Front. Plant Sci. 10:722. doi: 10.3389/fpls.2019.00722
Woo, J. W., Kim, J., Kwon, S. I., Corvalán, C., Cho, S. W., Kim, H., et al. (2015). DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 33, 1162–1164. doi: 10.1038/nbt.3389
Xie, K., Minkenberg, B., and Yang, Y. (2015). Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. 112, 3570–3575. doi: 10.1073/pnas.1420294112
Xu, R., Li, H., Qin, R., Wang, L., Li, L., Wei, P., et al. (2014). Gene targeting using the Agrobacterium tumefaciens-mediated CRISPR-Cas system in rice. Rice 7, 1–4. doi: 10.1186/s12284-014-0005-6
Yamamuro, C., Ihara, Y., Wu, X., Noguchi, T., Fujioka, S., Takatsuto, S., et al. (2000). Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12, 1591–1605. doi: 10.1105/tpc.12.9.1591
Zhang, S., Zhang, R., Song, G., Gao, J., Li, W., Han, X., et al. (2018). Targeted mutagenesis using the Agrobacterium tumefaciens-mediated CRISPR-Cas9 system in common wheat. BMC Plant Biol. 18:302. doi: 10.1186/s12870-018-1496-x
Zheng, Q., Cai, X., Tan, M. H., Schaffert, S., Arnold, C. P., Gong, X., et al. (2014). Precise gene deletion and replacement using the CRISPR/Cas9 system in human cells. Biotechniques 57, 115–124. doi: 10.2144/000114196
Zhou, H., Liu, B., Weeks, D. P., Spalding, M. H., and Yang, B. (2014). Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 42, 10903–10914. doi: 10.1093/nar/gku806
Keywords: genome editing, CRISPR/Cas9, oil palm, EgPDS, EgBRI1
Citation: Yeap W-C, Norkhairunnisa CMK, Norfadzilah J, Muhammad Rashdan M, Appleton DR and Harikrishna K (2021) An Efficient Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)/CRISPR-Associated Protein 9 Mutagenesis System for Oil Palm (Elaeis guineensis). Front. Plant Sci. 12:773656. doi: 10.3389/fpls.2021.773656
Received: 10 September 2021; Accepted: 18 October 2021;
Published: 22 November 2021.
Edited by:
Kejian Wang, China National Rice Research Institute, Chinese Academy of Agricultural Sciences (CAAS), ChinaReviewed by:
Peng-cheng Wei, Anhui Academy of Agricultural Sciences, ChinaCopyright © 2021 Yeap, Norkhairunnisa, Norfadzilah, Muhammad Rashdan, Appleton and Harikrishna. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wan-Chin Yeap, eWVhcC53YW4uY2hpbkBzaW1lZGFyYnlwbGFudGF0aW9uLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.