- Zhejiang Provincial Key Laboratory for Genetic Improvement and Quality Control of Medicinal Plants, Hangzhou Normal University, Hangzhou, China
TCP transcription factors play important roles in diverse aspects of plant development as transcriptional activators or repressors. However, the functional mechanisms of TCPs are not well understood, especially in cotton fibers. Here, we identified a total of 37 non-redundant TCP proteins from the diploid cotton (Gossypium raimondii), which showed great diversity in the expression profile. GrTCP11, an ortholog of AtTCP11, was preferentially expressed in cotton anthers and during fiber initiation and secondary cell wall synthesis stages. Overexpression of GrTCP11 in Arabidopsis thaliana reduced root hair length and delayed flowering. It was found that GrTCP11 negatively regulated genes involved in jasmonic acid (JA) biosynthesis and response, such as AtLOX4, AtAOS, AtAOC1, AtAOC3, AtJAZ1, AtJAZ2, AtMYC2, and AtERF1, which resulted in a decrease in JA concentration in the overexpressed transgenic lines. As with the JA-deficient mutant dde2-2, the transgenic line 4-1 was insensitive to 50 μM methyl jasmonate, compared with the wild-type plants. The results suggest that GrTCP11 may be an important transcription factor for cotton fiber development, by negatively regulating JA biosynthesis and response.
Introduction
TCP transcription factors are plant-specific developmental regulators, named according to the first four characterized members of the family, namely maize (Zea mays) TEOSINTE BRANCHED1, snapdragon (Antirrhinum majus) CYCLOIDEA, and rice (Oryza sativa) PROLIFERATING CELL NUCLEAR ANTIGEN FACTOR1 and 2. The main feature of TCP proteins is a conserved non-canonical basic-Helix-Loop-Helix structure near the N-terminus, known as the TCP domain, which is consisting of approximately 60 amino-acid residues (Cubas et al., 1999). The TCP transcription family can be categorized into two subfamilies, namely class I and class II, based on sequence similarities of the TCP domain, which act as activators or repressors by the formation of homodimers or heterodimers (Kosugi and Ohashi, 2002). They have been shown to be involved in a wide range of biological processes throughout the entire life span of plants. One of the founding members of the TCP family, AmCYC, controls floral asymmetry (Luo et al., 1996). AtTCP14 and AtTCP15 redundantly modulate internode elongation and leaf shape in Arabidopsis (Kieffer et al., 2011). GhTCP12, GhTCP13, and GhTCP18 are functionally redundant in controlling branching in cotton and Arabidopsis (Diao et al., 2019). AtTCP2, AtTCP3, AtTCP11, and AtTCP15 were found to participate in the regulation of circadian clock by interacting with different components (Giraud et al., 2010). The expression of a repressor form of AtTCP11 resulted in smaller and curly leaves, shorter petioles, pedicels, and siliques, as well as a higher proportion of abnormal seeds and pollens. The presence of a threonine residue at position 15 of the TCP domain is responsible for the distinct DNA-binding properties of AtTCP11 (Viola et al., 2011). Knockdown of AsPCF5, AsPCF6, AsPCF8, and AsTCP14 by overexpression of miR319 increases tolerance to dehydration and salinity stress in creeping bentgrass (Zhou et al., 2013). However, the molecular mechanisms by which TCPs are involved in plant development are largely unknown.
Recent studies have revealed that many TCP proteins seem to play central roles in biological processes by regulating phytohormone signals. PsBRC1 plays a key role in integrating strigolactone and cytokinin signals in pea to control shoot branching (Braun et al., 2012). CIN limits excess cell proliferation and maintains the flatness of the leaf surface by directly regulating the expression of cytokinin- and auxin-related genes in Antirrhinum majus (Das Gupta et al., 2014). AtTCP14 regulates seed germination by affecting response to abscisic acid and gibberellin signals (Tatematsu et al., 2008). AtTCP15 modulates gynoecium development by influencing the balance between auxin and cytokinin levels (Lucero et al., 2015). AtTCP1 mediates plant growth and development by directly inducing the expression of DWF4 and thus promoting brassinosteroid biosynthesis (Gao et al., 2015). Contrary to class II TCP transcription members AtTCP2, AtTCP4 and AtTCP10, the class I TCP transcription factor AtTCP20 induce the expression of AtLOX2 by binding to its promoter, and therefore decrease the level of jasmonic acid (JA) to inhibit leaf senescence (Schommer et al., 2008; Danisman et al., 2012). Identifying more TCPs will be beneficial in unraveling the precise regulatory mechanisms by which they control plant growth and development.
As one of the most economically important crops, cotton provides the main natural fibers for the textile industry. Cotton fibers are highly elongated ovule epidermal cells that undergo four continuous but overlapping stages: initiation, elongation, secondary cell wall (SCW) biosynthesis and dehydration/maturation (Basra and Malik, 1984; Haigler et al., 2012). It has been shown that the regulation of cotton fiber development depends on multiple plant hormone signaling processes. Auxins, gibberellins and brassinosteroids are required for fiber initiation and elongation both in vivo and in in vitro ovule culture (Beasley and Ting, 1973; Luo et al., 2007; Xiao et al., 2010; Zhang et al., 2011). Ethylene plays a major role in promoting cotton fiber elongation (Shi et al., 2006). Previous reports also showed that JA suppressed cotton fiber initiation and elongation with sustained high concentrations, while promoted fiber elongation with an appropriate concentration (Hao et al., 2012; Tan et al., 2012). However, the regulatory mechanism upstream of phytohormone signaling in cotton fibers is still not well understood. Our previous research had revealed that GbTCP, homologous to AtTCP15, played positive roles in cotton fiber and Arabidopsis root hair elongation by stimulating JA biosynthesis and response (Hao et al., 2012). GhTCP14, which is the ortholog of AtTCP14 in Arabidopsis, promoted the differentiation and elongation of trichome and root hair cells in Arabidopsis by directly binding to the promoters of auxin-related genes (Wang et al., 2013). These results imply that members of the cotton TCP transcription factor family may be involved in the regulation of fiber development by affecting the synthesis of or response to phytohormones.
Because of their possible roles in fiber development, members of the TCP transcription factor family have been identified and analyzed in different cotton species through an extensive genome-wide survey. A total of 38 and 36 non-redundant TCP genes were identified in diploid Gossypium raimondii and G. arboreum, respectively (Ma et al., 2014, 2016). A total of 74 and 75 TCP transcription factor members were identified in two allotetraploid cottons, namely G. hirsutum and G. barbadense, respectively. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) results indicated that many GhTCPs and GbTCPs were preferentially or specifically expressed in fibers at various developmental stages of cotton. GhTCP14a and GhTCP22 can interact with several transcription factors, which are involved in fiber development (Li et al., 2017; Zheng et al., 2018). GhTCP4, a miR319-targeted TCP gene, inhibited fiber cell elongation and promoted cell-wall thickening in cotton (Cao et al., 2020). Heterologous overexpression of the class II TCP genes GbTCP4 and GbTCP5 increased root hair length, root hair and trichome density, and the lignin content in transgenic Arabidopsis (Wang et al., 2019, 2020). The genome-wide analysis of the TCP transcription factor gene family will lay a solid foundation for future studies into the functional characterization of TCP proteins in cotton fiber development.
In the present study, we performed the genome-wide identification and expression analysis of the TCP family genes in G. raimondii slightly different from the previous report (Ma et al., 2014). A class I TCP transcription factor (designated GrTCP11) was cloned based on the phylogenetic and expression analysis, which was preferentially expressed in the stages of cotton fiber initiation and SCW synthesis. An ectopic expression strategy was used to investigate the effect of GrTCP11 on Arabidopsis root hair development, as Arabidopsis has been employed successfully as a model system for functional characterization of several cotton fiber-specific genes (Wang et al., 2004; Hao et al., 2012). Overexpression of GrTCP11 in Arabidopsis down-regulated expression levels of JA-related genes, which led to lower JA concentration and shorter root hair cells. In addition, we found that GrTCP11 also affects the bolting and flowering time of Arabidopsis. Our findings provide important insights into the role and mechanism of GrTCP11 in cotton fiber and root hair development.
Materials and Methods
Plant Materials and Growth Conditions
Cotton plants (G. raimondii and G. hirsutum cv. TM-1) were cultivated in the field under standard agronomic conditions at Hangzhou Normal University, in Hangzhou, China. Roots, stems and leaves were collected from 15-day-old seedlings. Petals, anthers and stigmas were harvested from flowers at 0 days post-anthesis (DPA). Fibers were detached gently from the ovules at 10 to 25 DPA. The collected materials were immediately frozen in liquid nitrogen and stored at –80°C prior to use.
The Arabidopsis thaliana wild-type (WT) was a Columbia-0 accession and the JA-deficient mutant dde2-2 was derived from Columbia-0. Seeds were first vernalized at 4°C for at least 2 days and then germinated in an illuminated growth chamber (22°C, 16-/8-h light/dark cycle regime) in soil or on agar plates (after surface sterilization of the seeds) containing half-strength Murashige and Skoog (MS) salts.
Identification of TCP Genes From Gossypium raimondii
The reference genome of diploid cotton G. raimondii was downloaded from the Joint Genome Institute (JGI) Phytozome.1 The TCP protein sequences of Arabidopsis were downloaded from NCBI2 and used as reference sequences for building a Hidden Markov Model (HMM) to identify the G. raimondii TCP proteins using HMMER 3.1.3 All the putative TCPs were further subjected to SMART4 and InterPro5 analyses to identify their conserved domains as previously described (Long et al., 2020). The Compute pI/Mw tool6 was used to calculate the molecular weight (Mw) and theoretical isoelectric point (pI). Multiple sequence alignments were performed using Clustal X (version 1.83) (Thompson et al., 1997). The phylogenetic tree was generated from the aligned sequences, using the neighbor-joining method in MEGA6, with 1000 bootstrap replicates (Tamura et al., 2011).
Heat-Map Analysis
Transcriptome data of G. hirsutum cv. TM-1 were downloaded from SRA databases (Accession code: PRJNA248163) (Zhang et al., 2015). Various tissues (root, stem, leaf, petal, pistil, stamen, ovule, and fiber) and abiotic stress (cold, hot, drought and salt stress for 1, 3, 6, and 12 h) transcriptome datasets were employed. The heat map was generated according to the method described previously (Gao et al., 2018). The reads per kilobase of transcript per million mapped reads values representing the expression levels of TCPs were collected from the “TM-1” cultivar transcriptome data downloaded from NCBI. The gene expression data were analyzed using the Genesis 1.8.1 program to generate heat maps. The cluster analysis, which was developed using the K-means method on the expression profiles of all 37 TCP genes, was also performed using the Genesis program.
RNA Extraction and Quantitative Reverse Transcription Polymerase Chain Reaction Analysis
Total RNA was extracted from 0.1 g fresh weight samples with the OmniPlant RNA Kit (DNase I) CW2598 (CWBIO, Beijing, China) according to the manufacturer’s instructions. HiFiScript gDNA Removal cDNA Synthesis Kit CW2582 (CWBIO, Beijing, China) was used to achieve first-strand cDNA synthesis from approximately 1 μg of total RNA. qRT-PCR was performed using the iQ SYBR Green Supermix (Bio-Rad, Hercules, CA, United States) and run on the ABI Prism 7000 system (Applied Biosystems, Foster City, CA, United States). GhUB7 and AtACT2 were used as the reference housekeeping genes in cotton and Arabidopsis, respectively. The relative expression level of the target genes was normalized against the reference housekeeping genes (Long et al., 2019). The error bars represent the standard deviation of three biological replicates. The primers used in the qRT-PCR are listed in Supplementary Table 1.
Vector Construction and Plant Transformation
The open reading frame of GrTCP11 was amplified from G. raimondii fiber cDNA with primers GW-GrTCP11F and GW-GrTCP11R (Supplementary Table 2), and inserted into pB7WG2D,1 with the CaMV 35S promoter to generate the overexpression vector via the Gateway BP and LR reactions. The construct was electroporated into the GV3101 strain of Agrobacterium tumefaciens and used for Arabidopsis transformation (Clough and Bent, 1998).
Observation and Measurements of Root Hair Length
For root hair length measurements, seedlings from surface-sterilized seeds were grown on plates held upright on half-strength MS medium, as previously reported (Hao et al., 2012). Root hairs on 7-day-old seedlings were photographed using an inverted microscope (Nikon Eclipse Ti, Tokyo, Japan), and measured with ImageJ software7. At least ten roots were measured for each independent experiment. Data were analyzed by Student’s t-test.
Extraction and Quantitative Analysis of Jasmonic Acid
One-month-old seedlings (500 mg fresh weight) were extracted with 80% cold methanol (v/v) by shaking overnight at 4°C. Each sample was extracted twice to ensure adequate extraction. The organic phase was evaporated to dryness with N2 and the residue dissolved in 0.4 mL methanol. The samples were stored at –20°C before being assayed. JA was quantified using an Applied Biosystems 4000Q-TRAR high-performance liquid chromatography–tandem mass spectrometry (HPLC–MS/MS) system with JA (Sigma, St. Louis, MO, United States) as the external standard. Three biological replicates were performed.
Root Growth Inhibition Assays
Surface-sterilized Arabidopsis seeds of the WT, 4-1 and dde2-2 were grown on plates held upright on half-strength MS medium plates with or without 50 μM methyl jasmonate (MeJA) at 22°C under a 16-h light/8-h dark photoperiod. The effect of MeJA on root growth inhibition was scored after 12 days of growth. Root lengths were measured using a ruler. At least 20 roots were measured for each independent experiment. Data were analyzed by Student’s t-test.
Results
Identification and Expression Profiles of TCP Genes in Gossypium raimondii
The TCP transcription factor family is a group of plant-specific transcription factors that have versatile functions in diverse aspects of plant development, including cotton fiber initiation and elongation (Hao et al., 2012; Wang et al., 2013). Based on an extensive genome-wide survey, we isolated a total of 37 putative non-redundant TCP genes in diploid G. raimondii (Table 1 and Supplementary Table 3), different from previous report of 38 non-redundant TCP genes identified from diploid G. raimondii (Ma et al., 2014). The deduced protein sequences alignment showed that the counterpart of GrTCP16 was reduced in our identification. Compared with Cotton_D_gene_10033515, the counterpart of GrTCP16 had the identical amino acid sequences, except for a truncation of 136 amino acids at the C-terminus. So, the counterpart of GrTCP16 was discarded as a redundant sequence of Cotton_D_gene_10033515 in our identification. The identities of other TCP proteins exhibit a good correspondence, excluding slight differences between the six proteins (GrTCP1, GrTCP6, GrTCP18a, GrTCP20c, GrTCP22, and GrTCP23) and their counterparts.
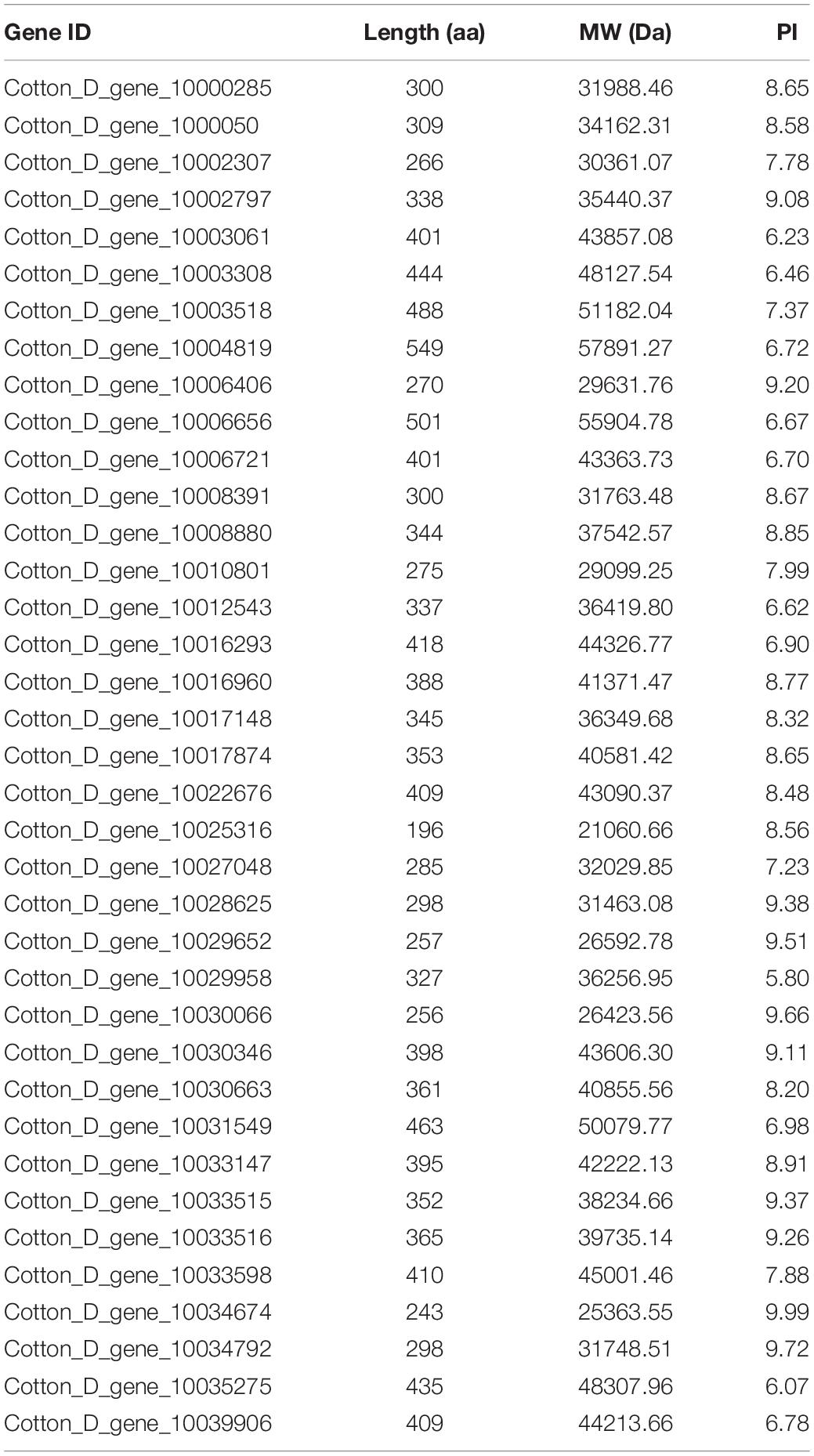
The expression levels of the 37 putative TCP genes in different tissues and in response to abiotic stress were analyzed using previously published transcriptome data to understand the possible roles of TCPs (Zhang et al., 2015). The reads per kilobase of transcript per million mapped reads values are listed in Supplementary Tables 4, 5. These values were used to create a heat-map of TCP genes’ expression. As indicated in Figure 1, substantial diversity is manifested in the tissue expression profile of TCP members. A few genes (with Gene ID of Cotton_D_gene_10006656, Cotton_D_gene_10017874, Cotton_D_gene_10030346, and Cotton_D_gene_10030663) were expressed at very low level across all 20 tissues, which suggested that they might be primarily expressed under particular conditions. More than half of the remaining TCP genes were predominantly expressed in different stages of cotton fiber development, which suggested that these genes might play important roles in fiber development. For example, the genes with Gene ID of Cotton_D_gene_10006406, Cotton_D_gene_10033147 or Cotton_D_gene_10008391 were preferentially expressed during the stages of fiber initiation, elongation or SCW synthesis, respectively. Two genes (with Gene ID of Cotton_D_gene_10000500 and Cotton_D_gene_10027048) had high expression in floral organs but low expression in all other tissues, which indicated they might specifically regulate the reproductive development of cotton. Additionally, the gene with Gene ID of Cotton_D_gene_10031549 was constitutively expressed in every tissue tested at very high level, which implied its possible roles at multiple development stages (Figure 1). However, most TCP members are not very sensitive to abiotic stresses, including cold, hot, drought and salt. For example, four genes (with Gene ID of Cotton_D_gene_10025316, Cotton_D_gene_10030346, Cotton_D_gene_10017874, and Cotton_D_gene_10030663) were not detected in any treatment, which suggested that these genes might not be involved in the stress responses (Figure 2).
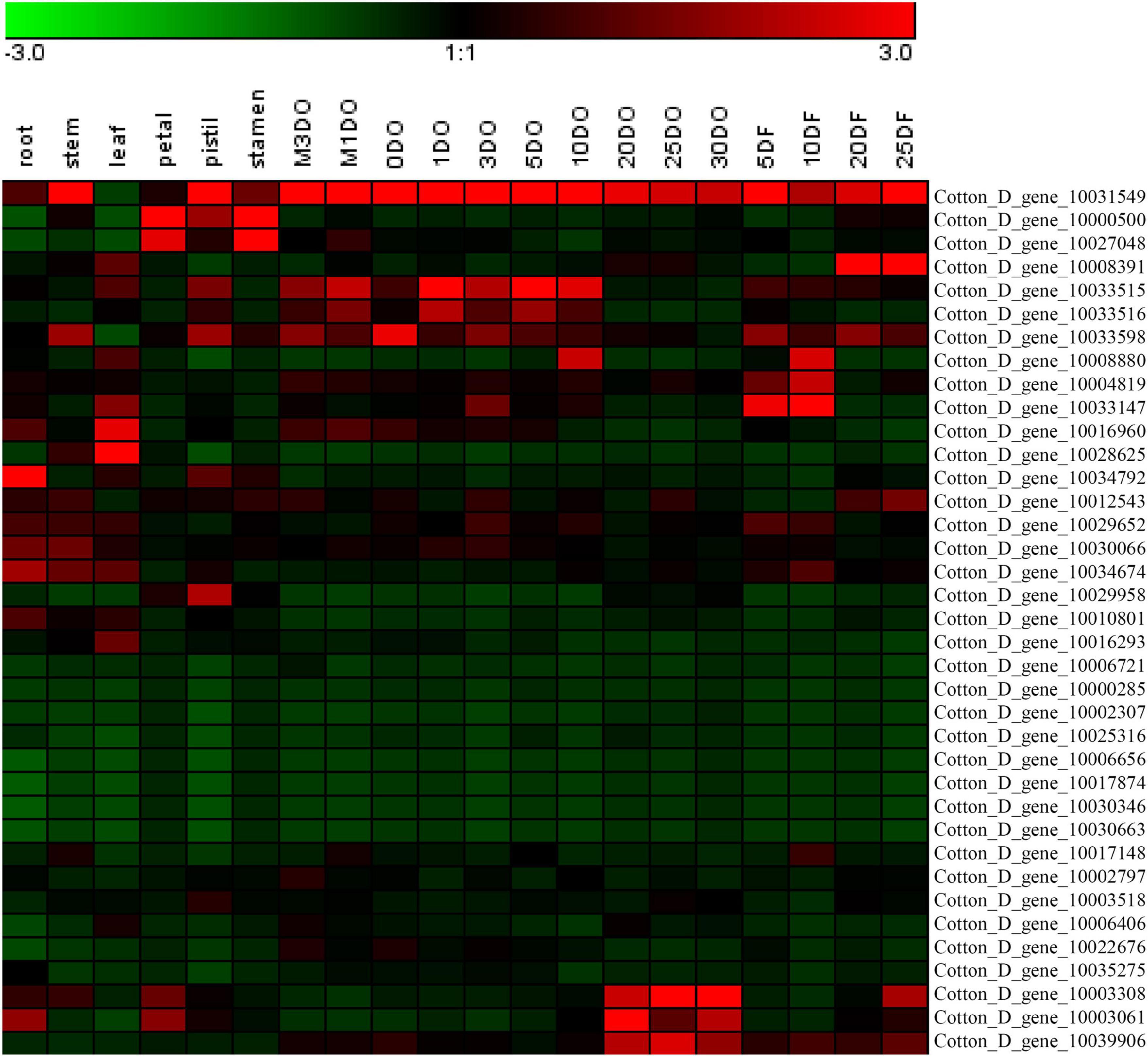
Figure 1. Heat map representation for expression patterns of Gossypium raimondii TCP genes in 20 representative tissues. The heat map, generated with Genesis, shows the hierarchical clustering of GrTCPs in root, stem, leaf, petal, pistil, stamen, ovule and fiber tissues. M3DO, M1DO, 0DO, 1DO, and 3DO: ovules attached with fibers at -3, -1, 0, 1, and 3 days post-anthesis; 5DO, 10DO, 20DO, 25DO, and 30DO: ovules without fibers at 5, 10, 20, 25, and 30 days post-anthesis; 5DF, 10DF, 20DF, and 25DF: fibers at 5, 10, 20, and 25 days post-anthesis. The reads per kilobase of transcript per million mapped reads values were log10 transformed and indicated the expression level of GrTCPs genes, while the gradient color (red/black/green) reflects the expression levels (high to low).
Figure 2. Heat map representation for expression patterns of Gossypium raimondii TCP genes in cotton leaves under abiotic stress. The heat map, generated with Genesis, shows the hierarchical clustering of GrTCPs in response to cold (4°C), hot (37°C), drought (20% PEG) and salt (200 mM NaCl) stress, respectively. 1HCOLD, 3HCOLD, 6HCOLD, and 12HCOLD: cold treatment for 1, 3, 6 and 12 h; 1HHOT, 3HHOT, 6HHOT, and 12HHOT: hot treatment for 1, 3, 6, and 12 h; 1HPEG, 3HPEG, 6HPEG, and 12HPEG: drought treatment for 1, 3, 6, and 12 h; 1HSALT, 3HSALT, 6HSALT, and 12HSALT: drought treatment for 1, 3, 6, and 12 h. The reads per kilobase of transcript per million mapped reads values were log10 transformed and indicated the expression level of GrTCPs genes, while the gradient color (red/black/green) reflects the expression levels (high to low).
GrTCP11 Was Preferentially Expressed in Anthers and During Stages of Fiber Initiation and Secondary Cell Wall Synthesis
Due to the expression profiles analysis, GrTCP11 (Gene ID: Cotton_D_gene_10006406) was cloned for further study as one of the fiber-specific expressed genes. Phylogenetic analysis was used to investigate the evolutionary relationship between the TCP proteins from G. raimondii and Arabidopsis. GrTCP11 was found to belong to class I TCP proteins as a unique homolog of AtTCP11 (Figure 3A). The amino acid sequence alignment showed that the identity of GrTCP11 with AtTCP11 full-length amino acids was 38%, and their TCP domains were 86% identical. Like most class I TCP proteins, the position 15 of the TCP domain in GrTCP11 is arginine, which is different from the threonine in AtTCP11 (Figure 3B). The coding sequence of GrTCP11 is 813 bp in length and encodes a putative polypeptide of 270 amino-acid residues with a calculated molecular weight of 29.6 kDa and an isoelectric point of 9.20. Based on the alignment between the coding sequence and the genomic DNA sequence, GrTCP11 was found to consist of two exons and a 366-bp intron. qRT-PCR analysis showed that the transcript level of GrTCP11 was the highest in anthers, while its transcript level was lower in roots, stems, leaves, stigmas and petals. qRT-PCR also showed that GrTCP11 was expressed in cotton fibers at different developmental stages. It had the highest transcript level at 0 DPA and 15 DPA, and could not be detected at other stages of fiber development (Figure 3C).
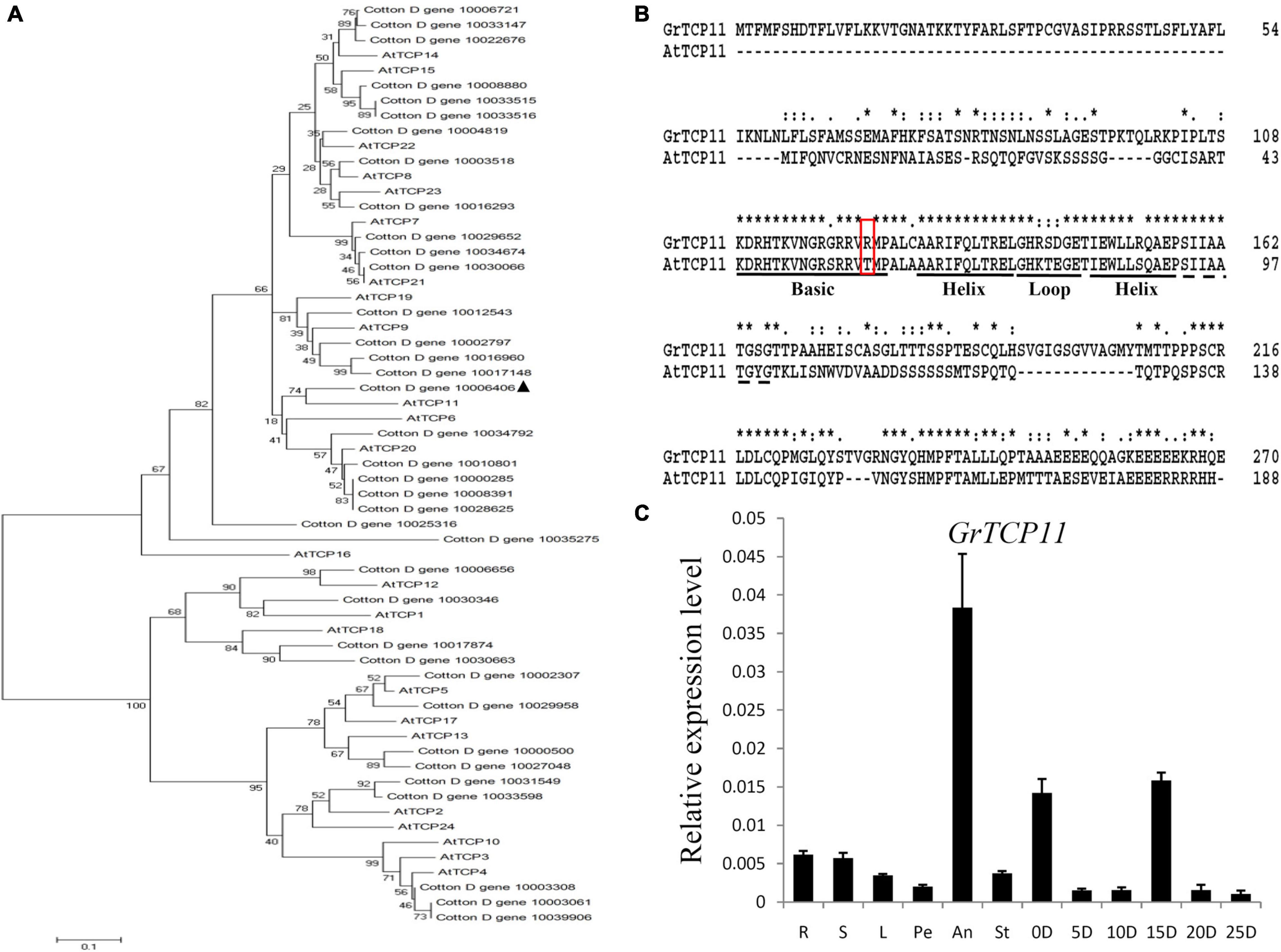
Figure 3. Phylogenetic analysis, sequence alignment and expression analysis of GrTCP11. (A) Phylogenetic relationships of TCP transcription factors from Gossypium raimondii and Arabidopsis. The unrooted phylogenetic tree was constructed using MEGA 6.0 with the neighbor-joining method and the bootstrap test involved 1000 iterations. GrTCP11 is marked with a solid triangle. (B) Alignment of the amino acid sequences of GrTCP11 and AtTCP15. The horizontal lines indicate the conserved teosinte branched1/cycloidea/proliferating cell factor1 (TCP) domain. The red box indicates the amino acid residue at position 15 of the TCP protein domain. (C) Relative expression level of GrTCP11 in various tissues, including root (R), stem (S), leaf (L), petal (Pe), anther (An), stigma (St) and fibers (0D and 5D: ovules with fibers at 0 and 5 days post-anthesis; 10D, 15D, 20D, and 25 D: fibers at 10, 15, 20, and 25 days post-anthesis) from Gossypium hirsutum cv.TM-1; expression of GrTCP11 was calculated relative to GhUB7 expression. Error bar on each mean value represents the standard deviation of three biological replicates.
Overexpression of GrTCP11 in Arabidopsis Inhibited Root Hair Elongation and Delayed Flowering Timing
To verify the function of GrTCP11, we generated ectopic GrTCP11-overexpressed transgenic Arabidopsis plants, to bypass the difficulty and delay associated with conventional cotton transformation. Several independent transformants were isolated and the expression levels were measured using qRT-PCR. Five independent T3 transgenic homozygous lines (3-1, 4-1, 5-4, 8-3, and 10-4) with different expression levels were further analyzed (Figure 4A). Overexpression of GrTCP11 in Arabidopsis inhibited root hair elongation. The mean length of the root hairs was significantly shorter in transgenic overexpressing lines 4-1 (171.2 μm), 8-3 (332.6 μm), 10-4 (448.3 μm), 5-4 (507.2 μm) and 3-1 (568.1 μm) than the root hairs of WT plants (650.3 μm) (Figures 4B,C). Overexpression of GrTCP11 in Arabidopsis also influenced the timing of flowering. The flowering time of the WT plants was earlier than in the transgenic lines (Figure 5). But there was no difference in their maturity time (Supplementary Figure 1).
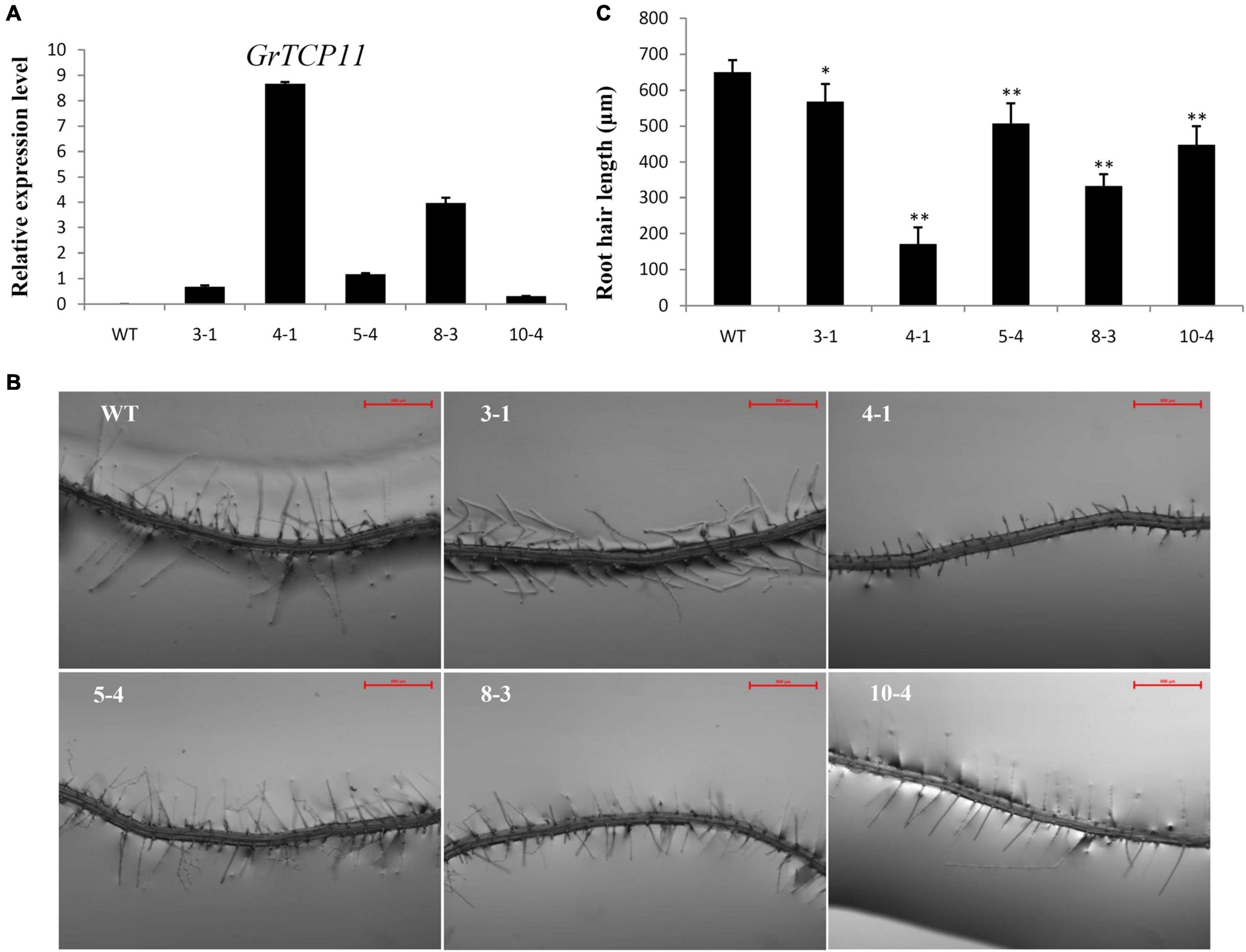
Figure 4. Expression level and morphological alterations of the root hairs in Arabidopsis transgenic plants overexpressing GrTCP11. (A) Relative expression level of GrTCP11 in transgenic lines (3-1, 4-1, 5-4, 8-3, and 10-4) and wild-type plants; expression of GrTCP11 was calculated relative to reference AtACT2 expression; error bars represent the standard deviation of three biological replicates. (B) Root hairs in the mature area of taproots of 7-day-old transgenic lines and wild-type plants; scale bars = 500 μm. (C) Length of the root hairs of 7-day-old seedlings within the mature zone; data are mean of 100 root hairs; *P < 0.05; **P < 0.01 relative to the wild-type, using Student’s t test.

Figure 5. The flowering time in 1-month-old wild-type and GrTCP11-overexpressed transgenic lines (3-1, 4-1, 5-4, 8-3, and 10-4).
Up-Regulation of GrTCP11 Affected Transcription of Genes Involved in Jasmonic Acid Biosynthesis and Response
Previous studies indicated that TCP transcription factors could regulate the development of cotton fibers and Arabidopsis root hairs through mediating phytohormone biosynthesis and signal transduction (Hao et al., 2012; Wang et al., 2013). Therefore, the expression levels of genes involved in phytohormone biosynthesis and response were determined in transgenic overexpressing lines and WT plants by qRT-PCR. As shown in Figure 6, many genes associated with JA biosynthesis (AtLOX4 and AtAOC3) and response (AtMYC2, AtJAZ1, AtJAZ2, and AtERF1) were significantly down-regulated in GrTCP11-overexpressed transgenic lines, compared with the WT plants. However, there was no significant difference in the expression levels of genes associated with other phytohormones, such as AtACO2, AtETR1, AtCTR1, AtTAA1, AtPIN1, AtIAA3, AtITP1, AtAHK3, and AtARR1 (Supplementary Figure 2).
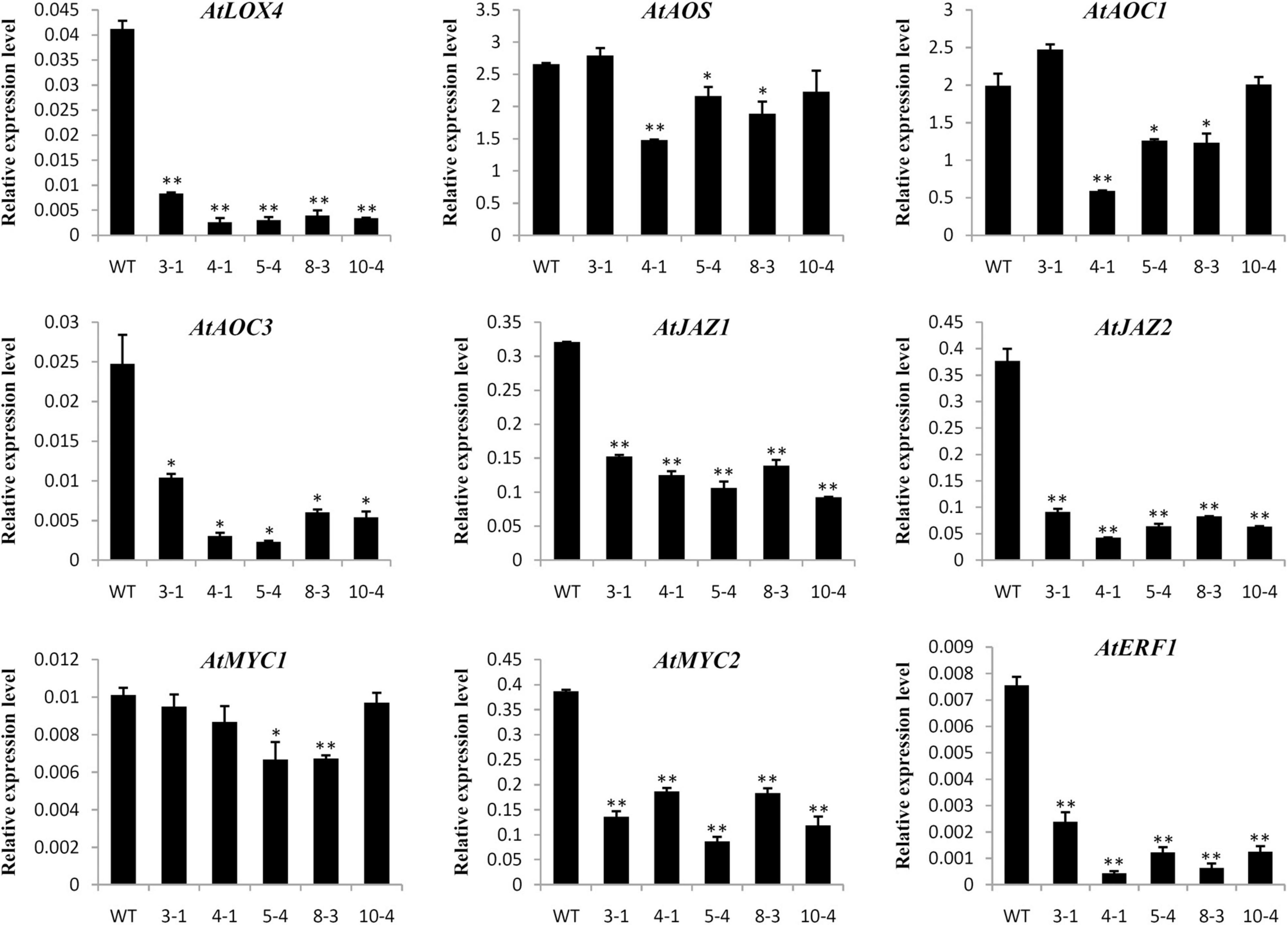
Figure 6. Relative expression level of the genes related to JA biosynthesis and response in wild-type and GrTCP11-overexpressed transgenic lines (3-1, 4-1, 5-4, 8-3, and 10-4). The accession numbers of the genes are as follows: AtLOX4, AT1G72520; AtAOS, AT5G42650; AtAOC1, AT3G25760; AtAOC3, AT3G25780; AtJAZ1, AT1G19180; AtJAZ2, AT1G74950; AtMYC1, AT4G00480; AtMYC2, AT1G32640; AtERF1, AT3G23240. Gene expression values are relative to reference AtACT2 expression; error bars represent the standard deviation of three biological replicates. Asterisks indicate a significant difference (*P < 0.05; **P < 0.01) relative to the corresponding wild-type, using Student’s t test.
Induced Expression of GrTCP11 Reduced Jasmonic Acid Concentrations in Arabidopsis
Due to changes in the expression of JA biosynthesis and signaling-related genes, it was hypothesized that GrTCP11 might affect root hair development by regulating JA concentrations. To test this hypothesis, we extracted and determined the concentration of JA in transgenic lines overexpressing GrTCP11, dde2-2 and WT plants, using HPLC–MS/MS. The results showed that the concentrations of JA were significantly lower in the overexpressed lines, compared with the WT controls. In particular, the JA concentration in line 4-1, which had the lowest expression level of JA-related genes, was similar to that in the mutant dde2-2 (Figure 7).
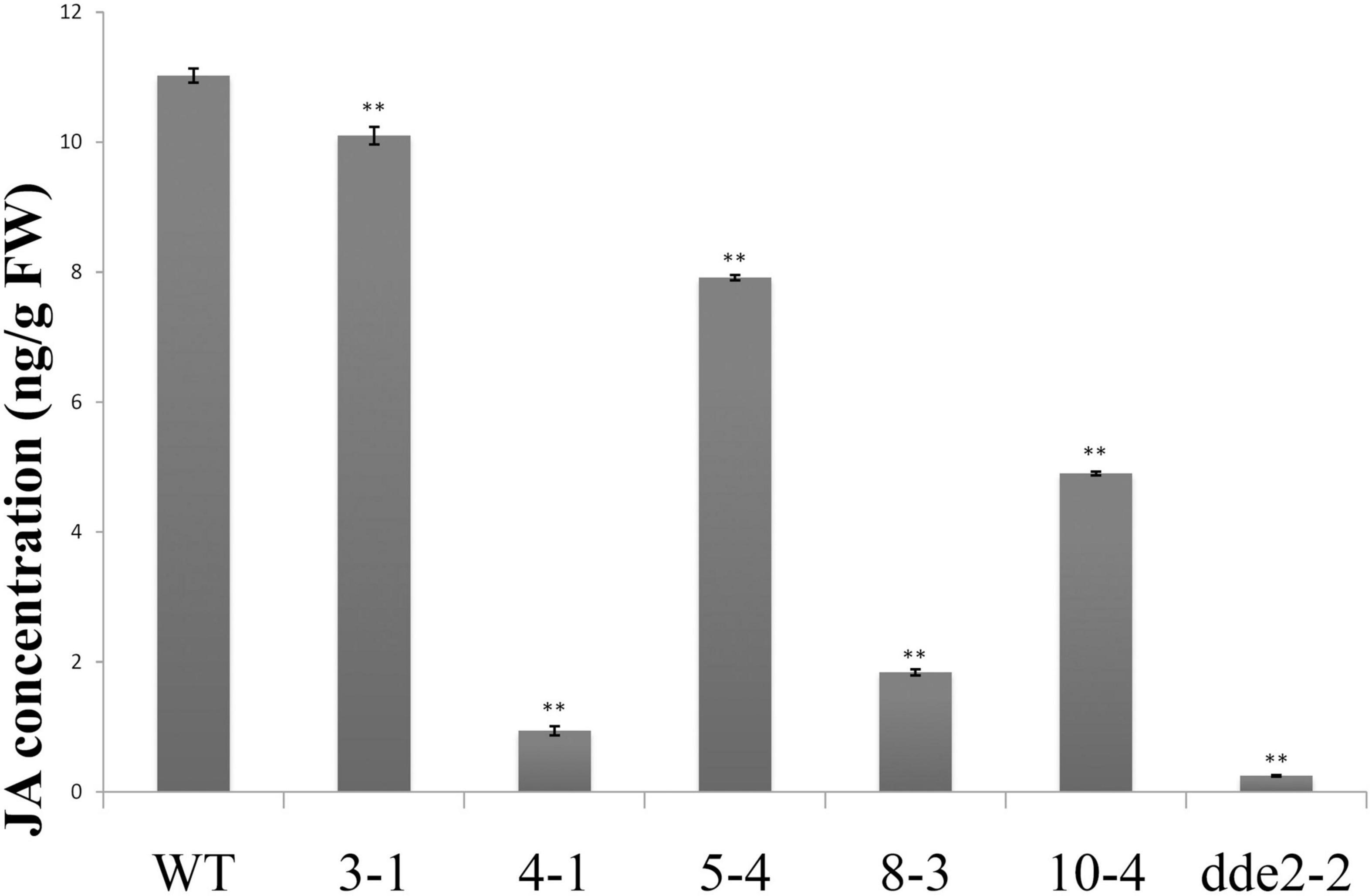
Figure 7. JA concentration in wild-type, GrTCP11-overexpressed lines (3-1, 4-1, 5-4, 8-3, and 10-4) and the JA-insensitive mutant dde2-2. Three independent experiments were performed; error bars represent the standard deviation. Significant differences between the wild-type and other lines were calculated by Student’s t-test analysis (**P < 0.01).
Induced Expression of GrTCP11 Decreased the Sensitivity of Root Growth to Methyl Jasmonate
To further test the effect of GrTCP11 on JA biosynthesis in Arabidopsis, 12-day-old seedlings of WT, 35S:GrTCP11 transgenic line 4-1 and dde2-2 were treated with 50 μM MeJA. As shown in Figures 8A,B, before MeJA treatment, 4-1 and dde2-2 were similar to WT plants in terms of root length. In the presence of 50 μM MeJA, the root growth of 4-1 was as insensitive to MeJA as was dde2-2. However, WT plants were hypersensitive to MeJA, with significantly shorter roots.
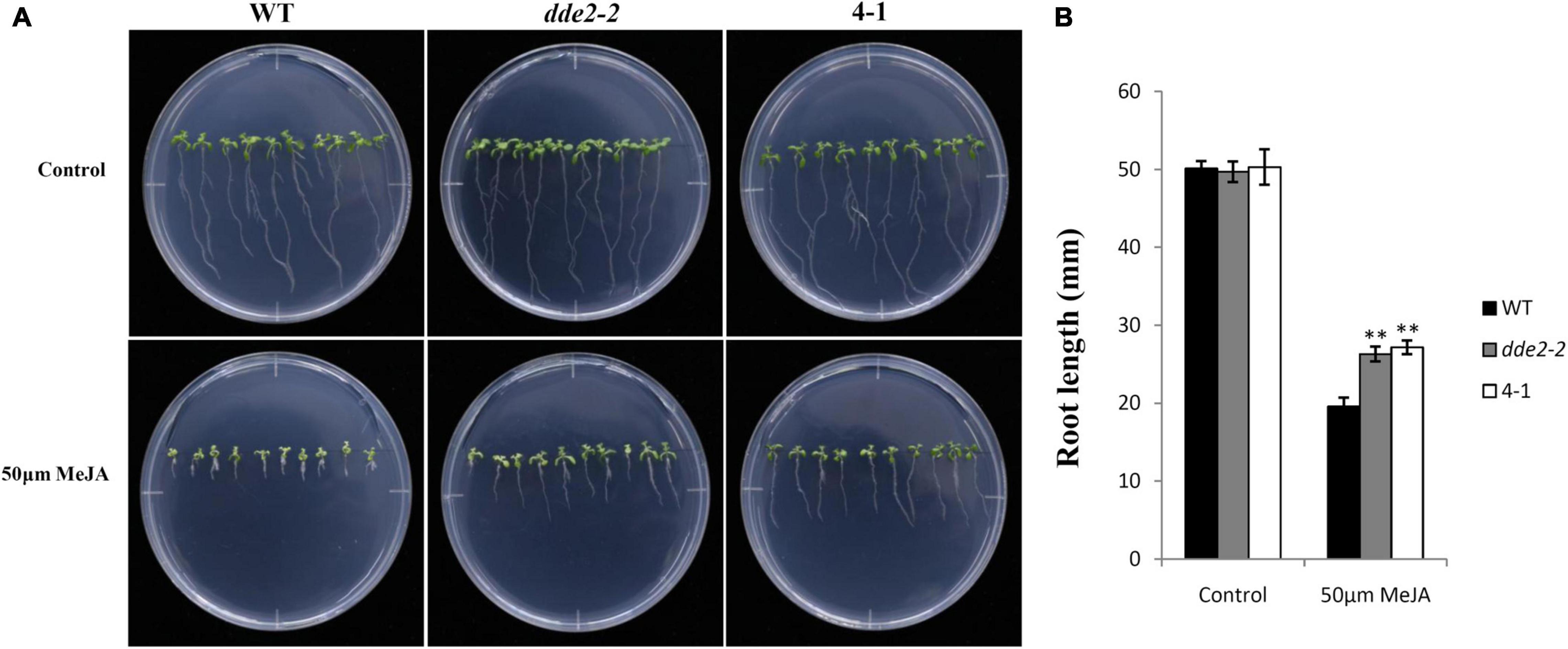
Figure 8. Sensitivity analysis of wild-type, transgenic line 4-1 and JA-insensitive mutant dde2-2 in response to methyl jasmonate (MeJA). (A) 12-day-old seedlings were grown on agar plates supplemented with (50 μM MeJA) or without (Control) MeJA. (B) Measurement of the root length in wild-type, 4-1 and dde2-2 grown for 12 days on agar plates held upright and containing or not containing 50 μM MeJA. Data are mean of at least 20 roots; error bars represent the standard deviation of 20 biological replicates; **P < 0.01 relative to the wild-type, using Student’s t test.
Discussion
GrTCP11 Might Regulate Cotton Fiber Development by Directly Down-Regulating Jasmonic Acid Biosynthesis and Response
Several TCPs have been shown to function in the control of trichome and root hair development (Hao et al., 2012; Vadde et al., 2018; Wang et al., 2019, 2020). In the present study, a cotton class I TCP transcription factor GrTCP11 was characterized, which was homologous to AtTCP11. Heat-map and qRT-PCR analysis showed that GrTCP11 was preferentially expressed during the stages of fiber initiation and SCW synthesis, rather than in the fiber elongation stage, which indicated that GrTCP11 had a spatio-temporal regulatory effect on cotton fiber development. Ectopic overexpression of GrTCP11 in Arabidopsis led to shorter root hair length. Previous studies have shown that there may be similar regulatory mechanism operating between the elongation of cotton fibers and Arabidopsis root hairs (Hao et al., 2012). We speculated that GrTCP11 might also inhibit cotton fiber elongation.
Plant hormones play vital roles in fiber and root hair development, as previously described. The expression levels of genes associated with JA biosynthesis and response (AtLOX4, AtAOC3, AtJAZ1, AtJAZ2, and AtMYC2) were significantly down-regulated in the transgenic lines compared with the WT controls, in association with a decrease in JA concentration. Changes in JA concentration were further validated with respect to JA sensitivity and flowering time. It has been reported that MeJA inhibits root growth in Arabidopsis, whereas the sensitivity to MeJA can be alleviated in some JA-deficient mutants (Staswick et al., 1992; Lorenzo et al., 2004). Compared to the wild type, the transgenic line 4-1 exhibited reduced sensitivity of root growth to 50 μM MeJA, similar to that of the JA-deficient mutant dde2-2. Jasmonic acid has been implicated in regulating flowering time (Pak et al., 2009). The flowering times of transgenic lines 4-1 and 8-3 were later than the other lines, which might be associated with the decreased JA concentration in these transgenic lines. AtERF1, a common downstream responsive element of the jasmonate and ethylene pathways, was also significantly suppressed by GrTCP11 (Lorenzo et al., 2003). However, no significant difference was observed in the expression levels between the transgenic lines and WT plants of other hormone-related genes, involved in the biosynthesis and response of ethylene, auxin or cytokinin. The above results revealed that GrTCP11 might inhibit fiber and root hair elongation by directly suppressing JA biosynthesis and response. JA is a crucial factor which suppresses cotton fiber initiation, with an appropriate concentration promoting fiber elongation as described above. Therefore, the expression pattern of GrTCP11 is associated with fiber initiation and elongation. Overall, spatio-temporal specific GrTCP11 may be an important regulator for cotton fiber development through negative regulation of JA biosynthesis and response.
The Possible Molecular Mechanism of TCP Transcription Factors in Developing Cotton Fibers
Cotton is one of the most important economic crops in the world, as its main product, cotton fibers, are the main raw material for the natural textile industry. Previous studies had revealed that transcription factors were widely involved in the regulation of cotton fiber development (Samuel Yang et al., 2006; Hande et al., 2017). Most of the plant-specific R2R3-MYBs were shown to regulate various developmental stages in cotton fiber, such as GhMYB109, GhMYB25 and GhMYB25-like (Pu et al., 2008; Machado et al., 2009; Walford et al., 2011). A R3-MYB transcription factor GhCPC delayed fiber initiation and inhibited early elongation by a potential CPC-MYC1-TTG1/4 complex. GhMYC1 could bind to the E-box cis-elements and the promoter of GhHOX3, which suggested that GhHOX3 may be downstream gene of the regulatory complex (Liu et al., 2015). GbTCP and GhTCP14 were shown to play vital roles in fiber initiation and/or elongation through regulating JA- and auxin-related genes, respectively, opening up research into the role of TCP transcription factors on cotton fiber development (Hao et al., 2012; Wang et al., 2013). Genome-wide analysis of the TCP family in a number of cotton varieties showed that they might be involved in different physiological processes of cotton fiber development. It was found that GhTCP22 and GhTCP14a could interact with several transcription factors related to cotton fiber growth and development, such as GhSLR1, GhGL3, GhARF6, GhTTG1, GhMYB23, and GhMYB25. In addition, GhTCP14a could also interact with GhEIN3, GhBZR1, and GhMYB25-Like proteins (Li et al., 2017). GhTCP4 interacted with GhHOX3 to coordinate fiber cell elongation and SCW biosynthesis, two events that are key to cotton fiber traits (Cao et al., 2020). Overexpression of GbTCP4 increased root hair length, root hair and trichome density, and the lignin content by binding directly to the AtCPC and AtCAD5 promoters in Arabidopsis (Wang et al., 2019). GbTCP5 regulated root hair development and SCW formation by binding to the promoters of the AtGL3, AtEGL3, AtCPC, AtMYB46, AtLBD30, AtCesA4, AtVND7, AtCCOMT1, and AtCAD5 genes to upregulate their expression in Arabidopsis (Wang et al., 2020). Here, we found that GrTCP11 possibly regulate fiber development by inhibiting JA biosynthesis and response in fiber initiation and SCW synthesis. The results indicated that TCP transcription factors possibly regulated fiber development by interacting with other fiber-related transcription factors, or binding to promoters to activate or inhibit their expressions. Protein complexes or targeted genes may regulate fiber development by affecting the homeostasis of phytohormones.
TCP Proteins Show Functional Diversity in Different Species
A growing number of TCP transcription factors have been characterized and confirmed to be widely involved in the regulation of plant growth, architecture and development. TCP transcription factors have some evolutionarily conserved roles in a range of plant species, such as regulation of branching, floral symmetry and leaf development (Danisman, 2016). For example, TEOSINTE BRANCHED1 in maize and its orthologs in rice, Arabidopsis, pea and poplar have a conserved function of suppressing branching (Doebley et al., 1997; Aguilar-Martinez et al., 2007; Braun et al., 2012; Muhr et al., 2016). On the other hand, there are suggestions that TCP proteins may have some new evolutionary roles in different species. Two closely related Arabidopsis TCP transcription factors, TCP14 and TCP15, were shown to be redundant in promoting cell proliferation in internodes and trichomes, and repressing cell proliferation in leaves and flower tissues (Kieffer et al., 2011). They were also necessary for seed germination by gibberellin-dependent activation (Resentini et al., 2015). AtTCP15 could modulate gynoecium development by participating in a feedback loop that helps to adjust the balance between auxin levels and cytokinin responses (Lucero et al., 2015). However, their homologous proteins in cotton had new biological functions. GbTCP, a protein orthologous to AtTCP15 in Sea-island cotton, promoted cotton fiber and Arabidopsis root hair elongation, as well as plant branching by directly activating the biosynthesis of and response to JA and then affecting downstream complex signal regulatory networks (Hao et al., 2012). Ectopic expression of GhTCP14 from upland cotton, which is homologous to AtTCP14, promoted the differentiation and elongation of trichomes and root hair cells through alteration of auxin homeostasis (Wang et al., 2013). AtTCP11 influences the growth of leaves, stems and petioles as well as pollen development in Arabidopsis (Viola et al., 2011). In the current work, a homolog of AtTCP11, named GrTCP11, was characterized in G. raimondii. High transcript abundance in anthers implied that GrTCP11 may have the same conserved function in pollen development as did AtTCP11. Our results showed that GrTCP11 may affect fiber and root hair elongation by directly inhibiting JA biosynthesis and response. The above research indicated that TCPs share some common functions but have also partially diverged, evolving distinct roles in different species through different regulatory mechanisms.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
JH and MX designed the experiments. PL, YH, ZC, and JC performed the experiments. JN, YY, and ZJ performed the data analyzes. JH wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant number 31601343, 2016–2019), Hangzhou Normal University Xinmiao Talent Program (Grant number 2021R426077), Scientific Research Fund of Zhejiang Province Education Department (Grant number Y201533081), State Key Laboratory of Cotton Biology Open Fund (Grant number CB2016A11), Natural Science Foundation of Zhejiang Province (Grant numbers LY19C150005 and LY19C020003), Major Increase or Decrease Program in the Central Finance Level (Grant number 2060302), and Zhejiang Provincial Key Research and Development Project Grants (Grant numbers 2017C02011 and 2018C02030).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.769675/full#supplementary-material
Supplementary Figure 1 | The maturity in 2-month-old wild-type (WT) and GrTCP11-overexpressed transgenic lines (3-1, 4-1, 5-4, 8-3 and 10-4).
Supplementary Figure 2 | Relative expression level of the genes related to phytohormones biosynthesis and response in wild-type (WT) and GrTCP11-overexpressed transgenic lines (3-1, 4-1 and 8-3). Gene expression values are relative to reference AtACT2 expression; error bars represent the standard deviation of three biological replicates.
Footnotes
- ^ https://phytozome.jgi.doe.gov
- ^ http://www.ncbi.nlm.nih.gov/
- ^ http://hmmer.janelia.org/
- ^ http://smart.embl-heidelberg.de/
- ^ http://www.ebi.ac.uk/interpro/
- ^ http://web.expasy.org/compute_pi/
- ^ http://rsbweb.nih.gov/ij/
References
Aguilar-Martinez, J. A., Poza-Carrion, C., and Cubas, P. (2007). Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19, 458–472. doi: 10.1105/tpc.106.048934
Basra, A. S., and Malik, C. P. (1984). Development of the cotton fiber. Int. Rev. Cytol. 89, 65–113. doi: 10.1016/S0074-7696(08)61300-5
Beasley, C. A., and Ting, I. P. (1973). The effects of plant growth substances on in vitro fiber development from fertilized cotton ovules. Am. J. Bot. 60, 130–139. doi: 10.1002/j.1537-2197.1973.tb10209.x
Braun, N., De Saint Germain, A., Pillot, J.-P., Boutet-Mercey, S., Dalmais, M., Antoniadi, I., et al. (2012). The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiol. 158, 225–238. doi: 10.1104/pp.111.182725
Cao, J. F., Zhao, B., Huang, C. C., Chen, Z. W., Zhao, T., Liu, H. R., et al. (2020). The miR319-targeted GhTCP4 promotes the transition from cell elongation to wall thickening in cotton fiber. Mol. Plant 13, 1063–1077. doi: 10.1016/j.molp.2020.05.006
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cubas, P., Lauter, N., Doebley, J., and Coen, E. (1999). The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18, 215–222. doi: 10.1046/j.1365-313x.1999.00444.x
Danisman, S. (2016). TCP transcription factors at the interface between environmental challenges and the plant’s growth responses. Front. Plant Sci. 7:1930. doi: 10.3389/fpls.2016.01930
Danisman, S., Van Der Wal, F., Dhondt, S., Waites, R., De Folter, S., Bimbo, A., et al. (2012). Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 159, 1511–1523. doi: 10.1104/pp.112.200303
Das Gupta, M., Aggarwal, P., and Nath, U. (2014). CINCINNATA in Antirrhinum majus directly modulates genes involved in cytokinin and auxin signaling. New Phytol. 204, 901–912. doi: 10.1111/nph.12963
Diao, Y., Zhan, J., Zhao, Y., Liu, L., Liu, P., Wei, X., et al. (2019). GhTIE1 regulates branching through modulating the transcriptional activity of TCPs in cotton and Arabidopsis. Front. Plant Sci. 10:1348. doi: 10.3389/fpls.2019.01348
Doebley, J., Stec, A., and Hubbard, L. (1997). The evolution of apical dominance in maize. Nature 386, 485–488. doi: 10.1038/386485a0
Gao, W., Xu, F. C., Guo, D. D., Zhao, J. R., Liu, J., Guo, Y. W., et al. (2018). Calcium-dependent protein kinases in cotton: insights into early plant responses to salt stress. BMC Plant Biol. 18:15. doi: 10.1186/s12870-018-1230-8
Gao, Y., Zhang, D., and Li, J. (2015). TCP1 modulates DWF4 expression via directly interacting with the GGNCCC motifs in the promoter region of DWF4 in Arabidopsis thaliana. J. Genet. Genomics 42, 383–392. doi: 10.1016/j.jgg.2015.04.009
Giraud, E., Ng, S., Carrie, C., Duncan, O., Low, J., Lee, C. P., et al. (2010). TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. Plant Cell 22, 3921–3934. doi: 10.1105/tpc.110.074518
Haigler, C. H., Betancur, L., Stiff, M. R., and Tuttle, J. R. (2012). Cotton fiber: a powerful single-cell model for cell wall and cellulose research. Front. Plant Sci. 3:104. doi: 10.3389/fpls.2012.00104
Hande, A. S., Katageri, I. S., Jadhav, M. P., Adiger, S., Gamanagatti, S., Padmalatha, K. V., et al. (2017). Transcript profiling of genes expressed during fibre development in diploid cotton (Gossypium arboreum L.). BMC Genomics 18:675. doi: 10.1186/s12864-017-4066-y
Hao, J., Tu, L., Hu, H., Tan, J., Deng, F., Tang, W., et al. (2012). GbTCP, a cotton TCP transcription factor, confers fibre elongation and root hair development by a complex regulating system. J. Exp. Bot. 63, 6267–6281. doi: 10.1093/jxb/ers278
Kieffer, M., Master, V., Waites, R., and Davies, B. (2011). TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. 68, 147–158. doi: 10.1111/j.1365-313X.2011.04674.x
Kosugi, S., and Ohashi, Y. (2002). DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30, 337–348. doi: 10.1046/j.1365-313x
Li, W., Li, D. D., Han, L. H., Tao, M., Hu, Q. Q., Wu, W. Y., et al. (2017). Genome-wide identification and characterization of TCP transcription factor genes in upland cotton (Gossypium hirsutum). Sci. Rep. 7:10118. doi: 10.1038/s41598-017-10609-2
Liu, B., Zhu, Y., and Zhang, T. (2015). The R3-MYB gene GhCPC negatively regulates cotton fiber elongation. PLoS One 10:e0116272. doi: 10.1371/journal.pone.0116272
Long, L., Yang, W. W., Liao, P., Guo, Y. W., Kumar, A., and Gao, W. (2019). Transcriptome analysis reveals differentially expressed ERF transcription factors associated with salt response in cotton. Plant Sci. 281, 72–81. doi: 10.1016/j.plantsci.2019.01.012
Long, L., Zhao, J. R., Guo, D. D., Ma, X. N., Xu, F. C., Yang, W. W., et al. (2020). Identification of NHXs in Gossypium species and the positive role of GhNHX1 in salt tolerance. BMC Plant Biol. 20:147. doi: 10.1186/s12870-020-02345-z
Lorenzo, O., Chico, J. M., Sanchez-Serrano, J. J., and Solano, R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16, 1938–1950. doi: 10.1105/tpc.022319
Lorenzo, O., Piqueras, R., Sanchez-Serrano, J. J., and Solano, R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15, 165–178. doi: 10.1105/tpc.007468
Lucero, L. E., Uberti-Manassero, N. G., Arce, A. L., Colombatti, F., Alemano, S. G., and Gonzalez, D. H. (2015). TCP15 modulates cytokinin and auxin responses during gynoecium development in Arabidopsis. Plant J. 84, 267–282. doi: 10.1111/tpj.12992
Luo, D., Carpenter, R., Vincent, C., Copsey, L., and Coen, E. (1996). Origin of floral asymmetry in Antirrhinum. Nature 383, 794–799. doi: 10.1038/383794a0
Luo, M., Xiao, Y., Li, X., Lu, X., Deng, W., Li, D., et al. (2007). GhDET2, a steroid 5alpha-reductase, plays an important role in cotton fiber cell initiation and elongation. Plant J. 51, 419–430. doi: 10.1111/j.1365-313X.2007.03144.x
Ma, J., Liu, F., Wang, Q. L., Wang, K. B., Jones, D. C., and Zhang, B. H. (2016). Comprehensive analysis of TCP transcription factors and their expression during cotton (Gossypium arboreum) fiber early development. Sci. Rep. 6:21535. doi: 10.1038/srep21535
Ma, J., Wang, Q., Sun, R., Xie, F., Jones, D. C., and Zhang, B. (2014). Genome-wide identification and expression analysis of TCP transcription factors in Gossypium raimondii. Sci. Rep. 4:6645. doi: 10.1038/srep06645
Machado, A., Wu, Y., Yang, Y., Llewellyn, D. J., and Dennis, E. S. (2009). The MYB transcription factor GhMYB25 regulates early fibre and trichome development. Plant J. 59, 52–62. doi: 10.1111/j.1365-313X.2009.03847.x
Muhr, M., Prufer, N., Paulat, M., and Teichmann, T. (2016). Knockdown of strigolactone biosynthesis genes in Populus affects BRANCHED1 expression and shoot architecture. New Phytol. 212, 613–626. doi: 10.1111/nph.14076
Pak, H., Guo, Y., Chen, M., Chen, K., Li, Y., Hua, S., et al. (2009). The effect of exogenous methyl jasmonate on the flowering time, floral organ morphology, and transcript levels of a group of genes implicated in the development of oilseed rape flowers (Brassica napus L.). Planta 231, 79–91. doi: 10.1007/s00425-009-1029-9
Pu, L., Li, Q., Fan, X., Yang, W., and Xue, Y. (2008). The R2R3 MYB transcription factor GhMYB109 is required for cotton fiber development. Genetics 180, 811–820. doi: 10.1534/genetics.108.093070
Resentini, F., Felipo-Benavent, A., Colombo, L., Blazquez, M. A., Alabadi, D., and Masiero, S. (2015). TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana. Mol. Plant 8, 482–485. doi: 10.1016/j.molp.2014.11.018
Samuel Yang, S., Cheung, F., Lee, J. J., Ha, M., Wei, N. E., Sze, S. H., et al. (2006). Accumulation of genome-specific transcripts, transcription factors and phytohormonal regulators during early stages of fiber cell development in allotetraploid cotton. Plant J. 47, 761–775. doi: 10.1111/j.1365-313X.2006.02829.x
Schommer, C., Palatnik, J. F., Aggarwal, P., Chetelat, A., Cubas, P., Farmer, E. E., et al. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6:e230. doi: 10.1371/journal.pbio.0060230
Shi, Y. H., Zhu, S. W., Mao, X. Z., Feng, J. X., Qin, Y. M., Zhang, L., et al. (2006). Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 18, 651–664. doi: 10.1105/tpc.105.040303
Staswick, P. E., Su, W., and Howell, S. H. (1992). Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. U.S.A. 89, 6837–6840. doi: 10.1073/pnas.89.15.6837
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tan, J., Tu, L., Deng, F., Wu, R., and Zhang, X. (2012). Exogenous jasmonic acid inhibits cotton fiber elongation. J. Plant Growth Regul. 31, 599–605. doi: 10.1007/s00344-012-9260-1
Tatematsu, K., Nakabayashi, K., Kamiya, Y., and Nambara, E. (2008). Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J. 53, 42–52. doi: 10.1111/j.1365-313X.2007.03308.x
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Vadde, B. V. L., Challa, K. R., and Nath, U. (2018). The TCP4 transcription factor regulates trichome cell differentiation by directly activating GLABROUS INFLORESCENCE STEMS in Arabidopsis thaliana. Plant J. 93, 259–269. doi: 10.1111/tpj.13772
Viola, I. L., Uberti Manassero, N. G., Ripoll, R., and Gonzalez, D. H. (2011). The Arabidopsis class I TCP transcription factor AtTCP11 is a developmental regulator with distinct DNA binding properties due to the presence of threonine at position 15 of the TCP domain. Biochem. J. 435, 143–155. doi: 10.1042/BJ20101019
Walford, S. A., Wu, Y., Llewellyn, D. J., and Dennis, E. S. (2011). GhMYB25-like: a key factor in early cotton fibre development. Plant J. 65, 785–797. doi: 10.1111/j.1365-313X.2010.04464.x
Wang, M. Y., Zhao, P. M., Cheng, H. Q., Han, L. B., Wu, X. M., Gao, P., et al. (2013). The cotton transcription factor TCP14 functions in auxin-mediated epidermal cell differentiation and elongation. Plant Physiol. 162, 1669–1680. doi: 10.1104/pp.113.215673
Wang, S., Wang, J. W., Yu, N., Li, C. H., Luo, B., Gou, J. Y., et al. (2004). Control of plant trichome development by a cotton fiber MYB gene. Plant Cell 16, 2323–2334. doi: 10.1105/tpc.104.024844
Wang, Y., Yu, Y., Chen, Q., Bai, G., Gao, W., Qu, Y., et al. (2019). Heterologous expression of GbTCP4, a class II TCP transcription factor, regulates trichome formation and root hair development in Arabidopsis. Genes 10:726. doi: 10.3390/genes10090726
Wang, Y., Yu, Y., Wang, J., Chen, Q., and Ni, Z. (2020). Heterologous overexpression of the GbTCP5 gene increased root hair length, root hair and stem trichome density, and lignin content in transgenic Arabidopsis. Genes 758:144954. doi: 10.1016/j.gene.2020.144954
Xiao, Y. H., Li, D. M., Yin, M. H., Li, X. B., Zhang, M., Wang, Y. J., et al. (2010). Gibberellin 20-oxidase promotes initiation and elongation of cotton fibers by regulating gibberellin synthesis. J. Plant Physiol. 167, 829–837. doi: 10.1016/j.jplph.2010.01.003
Zhang, M., Zheng, X., Song, S., Zeng, Q., Hou, L., Li, D., et al. (2011). Spatiotemporal manipulation of auxin biosynthesis in cotton ovule epidermal cells enhances fiber yield and quality. Nat. Biotechnol. 29, 453–458. doi: 10.1038/nbt.1843
Zhang, T., Hu, Y., Jiang, W., Fang, L., Guan, X., Chen, J., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotech. 33, 531–537. doi: 10.1038/nbt.3207
Zheng, K., Ni, Z., Qu, Y., Cai, Y., Yang, Z., Sun, G., et al. (2018). Genome-wide identification and expression analyses of TCP transcription factor genes in Gossypium barbadense. Sci. Rep. 8: 14526. doi: 10.1038/s41598-018-32626-5
Keywords: TCP transcription factor, Gossypium raimondii, fiber, Arabidopsis thaliana, root hair, jasmonic acid
Citation: Hao J, Lou P, Han Y, Chen Z, Chen J, Ni J, Yang Y, Jiang Z and Xu M (2021) GrTCP11, a Cotton TCP Transcription Factor, Inhibits Root Hair Elongation by Down-Regulating Jasmonic Acid Pathway in Arabidopsis thaliana. Front. Plant Sci. 12:769675. doi: 10.3389/fpls.2021.769675
Received: 02 September 2021; Accepted: 29 October 2021;
Published: 22 November 2021.
Edited by:
Jian Li Yang, Zhejiang University, ChinaReviewed by:
Jiameng Xu, Shandong University, ChinaSoledad Francisca Undurraga, Pontificia Universidad Católica de Chile, Chile
Copyright © 2021 Hao, Lou, Han, Chen, Chen, Ni, Yang, Jiang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maojun Xu, eHVtYW9qdW5oekAxNjMuY29t
 Juan Hao
Juan Hao Panpan Lou
Panpan Lou Yidie Han
Yidie Han Zhehao Chen
Zhehao Chen Jun Ni
Jun Ni Yanjun Yang
Yanjun Yang Maojun Xu
Maojun Xu