
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Plant Sci. , 12 January 2022
Sec. Plant Biotechnology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.766506
This article is part of the Research Topic Plant Cell Wall Polysaccharides as Biofuels and Biomaterials View all 19 articles
Efficient separation of the plant cell wall polymers during lignocellulose processing has been historically challenging due to insolubility of the polymers and their propensity for recalcitrant reassembly. Methods, such as “lignin first” extraction techniques, have advanced efficient biomass use, but the molecular mechanisms for recalcitrance remain enigmatic. Here, we discuss how solid-state Nuclear Magnetic Resonance (NMR) approaches report on the 3D organization of cellulose, xylan, and lignin in the plant cell wall. Recent results illustrate that the organization of these polymers varies across biomass sources and sample preparation methods, with even minimal physical processing causing significant effects. These structural differences contribute to variable extraction efficiencies for bioproducts after downstream processing. We propose that solid-state NMR methods can be applied to follow biomass processing, providing an understanding of the polymer rearrangements that can lead to poor yields for the desired bioproducts. The utility of the technique is illustrated for mechanical processing using lab-scale vibratory ball milling of Sorghum bicolor.
Plant lignocellulosic biomass is a sustainable, renewable feedstock for producing bio-based fuels, chemicals, and materials (U.S. Department of Energy, 2016; Li and Takkellapati, 2018). There are two types of plant cell walls: the thin and expandable primary cell wall surrounding all cells and the thicker secondary cell wall deposited at the cessation of cell expansion in some cell types. Due to its thickness, the secondary cell wall forms the vast majority of lignocellulosic biomass (Marriott et al., 2016). It is predominantly composed of the polysaccharide cellulose, a class of polysaccharides called hemicelluloses, and the aromatic polymer, lignin (Marriot et al., 2016). These polymers can then be converted via chemical or biological routes into fuels and other valuable chemicals that are conventionally derived from fossil resources (van Putten et al., 2013; Li and Takkellapati, 2018; Baral et al., 2019; Genuino et al., 2019; Priharto et al., 2020).
A major bottleneck in the extraction of chemical precursors from lignocellulosic biomass is the high recalcitrance of the plant cell wall (Marriott et al., 2016; Gilna et al., 2017). Strategies to decrease this intrinsic recalcitrance include the development of plants with altered biomass composition (Pauly and Keegstra, 2010) and novel deconstruction methods, such as the use of new solvents (Kim et al., 2018) and enzyme cocktails (Lopes et al., 2018), which target key polymer linkages. Deconstruction methods can also introduce recalcitrance, for example, by the deposition of more condensed lignin following solubilization (Li et al., 2016). Efforts to reduce recalcitrance include pretreating biomass before deconstruction (Yao et al., 2018; Baig et al., 2019) and “lignin first” deconstruction (Li et al., 2016; Li and Takkellapati, 2018). However, an understanding of how mechanical processing can cause biomass recalcitrance is needed (Gilna et al., 2017; Li and Takkellapati, 2018; Baig et al., 2019).
Mechanical processing is commonly used to reduce biomass particle size to increase solvent accessibility and polymer solubilization (Zhao et al., 2016; Zhai et al., 2019). Lab-scale vibratory ball milling achieves this goal by rapidly vibrating a chamber containing lignocellulosic biomass with either steel or zirconium balls. An overview of this process is provided in Figure 1A. However, milling leading to recalcitrance is frequently reported during lignocellulosic biomass conversion efforts, impeding the efficiency of subsequent processing and separation steps (Tolbert et al., 2014; Li et al., 2016; Wang et al., 2021). Milling-induced recalcitrance could be due to the production of reactive lignin species (Zhao et al., 2016) promoting aberrant hemicellulose-lignin crosslinks (Terrett and Dupree, 2019) as the lignin recondenses (Li et al., 2016; Kang et al., 2019). Additionally, increased amorphous cellulose content (Ling et al., 2019) could induce reorganization of hemicellulose-cellulose contacts due to their multiple modes of interaction (Simmons et al., 2016; Gao et al., 2020). Importantly, past studies on the plant cell wall structure have used mechanical milling to prepare samples for analysis, so the outcomes may be influenced by non-native interactions and contacts between these polymers (Foston et al., 2012; Park and Cosgrove, 2012; Zhao et al., 2016; Li and Takkellapati, 2018; Ishak et al., 2020).
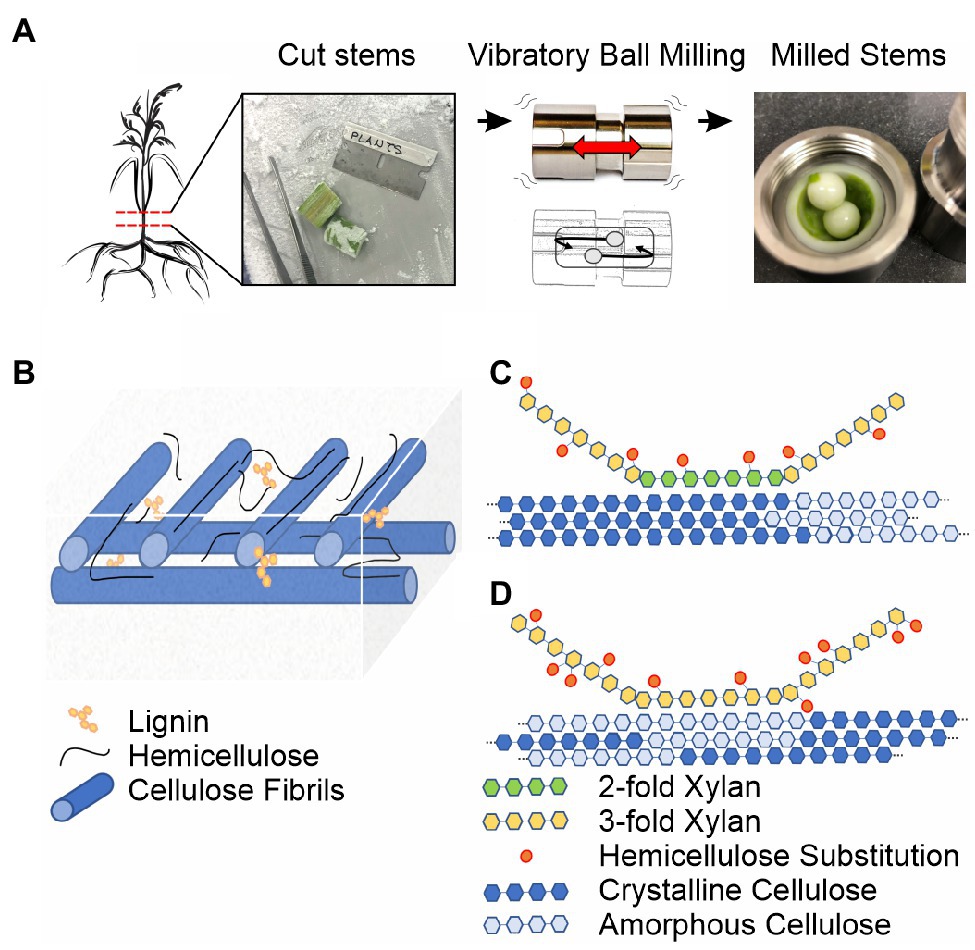
Figure 1. Ball milling approach and overview of the secondary plant cell wall. (A) 13C-enriched plant stems are harvested, frozen in liquid nitrogen, sectioned on a dry ice cooled surface, and subjected to vibrational ball milling using a zirconium chamber and balls. (B) The macroscopic organization of the secondary plant cell wall based on the studies reviewed in Marriot et al. (2016) and Zhang et al. (2021a). Cellulose fibrils are blue, lignin interspersed in the plant cell wall matrix and on carbohydrate surfaces is yellow, and unbound and cellulose-associated hemicellulose is black. (C) Solid-state NMR-based model for hemicellulose-cellulose interactions in eudicot Arabidopsis based on Simmons et al. (2016). (D) Solid-state NMR-based model for hemicellulose-cellulose interactions in monocot sorghum based on Gao et al. (2020). A major pattern of xylan substitutions is represented as orange ovals in (C,D). Even patterning of glucuronic acid dictates 2-fold xylan associates with crystalline cellulose in eudicots (Simmons et al., 2016; Grantham et al., 2017). The irregular L-arabinosyl xylan substitutions in monocots allow 3-fold xylan and amorphous cellulose interactions (Gao et al., 2020).
Recent advances in the incorporation of 13C isotopes into living plant tissues allow characterization of the native plant cell wall structure at unprecedented resolution using solid-state Nuclear Magnetic Resonance (NMR; Dick-Pérez et al., 2011; Gao and Mortimer, 2020). Current efforts reveal significant diversity in the organization of plant cell wall polymers across plant species and tissue types (Dick-Pérez et al., 2011; Dupree et al., 2015; Simmons et al., 2016; Kang et al., 2019; Terrett et al., 2019; Gao et al., 2020). After summarizing the current experimental understanding of the plant cell wall architecture with emphasis on solid-state NMR methods, we illustrate the potential of solid-state NMR measurements to assess molecular changes during mechanical preprocessing using Sorghum bicolor (sorghum) biomass as an example. The future use of solid-state NMR-based methods to monitor biomass structure during deconstruction will provide an understanding of the molecular mechanisms of recalcitrance, guiding the development of rationally designed deconstruction and crop engineering approaches to enable efficient production of chemical precursors from plants.
Complexity and insolubility are major barriers for in situ characterization of the native plant cell wall structure (Agarwal, 2019; Zhao et al., 2020). The chemical composition of the secondary plant cell wall has been primarily defined using liquid chromatography (Sluiter et al., 2008), mass spectrometry (MS) (Jung et al., 2012; Qi and Volmer, 2019; Zhang et al., 2021b), and solution-state NMR measurements on solvent-extracted polymers (Kim and Ralph, 2010; Foston et al., 2016; Ding et al., 2019) and solid-state NMR (Dick-Pérez et al., 2011; Zhao et al., 2020), vibrational spectroscopy (Agarwal, 2019), and X-ray diffraction (Nishiyama et al., 2002) measurements on native (intact) plant tissues. To help the reader appreciate the potential for polymer reorganization during processing, we present an overview of the current understanding of the secondary plant cell wall chemical composition and architecture.
Figures 1B–D provide an overview of the major polymers present in the secondary plant cell wall and the current understanding of their general macroscopic organization. Interspersed cellulose fibrils provide an immobilized framework for a plant cell wall matrix containing water, soluble proteins, lignin, and hemicellulose polymers (Marriot et al., 2016; Zhang et al., 2021a). The individual cellulose fibrils are built from linear β-(1,4)-glucan polymers composed of crystalline and amorphous arrangements, which differ primarily in their water content and hydrogen bonding patterns, the precise details of which are still under investigation (Song et al., 2020). Hemicellulose polymers both rigidly associate with cellulose fibrils and extend into the matrix environment where they exhibit significant molecular motion and can interact with lignin polymers (Scheller and Ulvskov, 2010; Li et al., 2016; Ding et al., 2019; Kang et al., 2019). Differences across monocot and eudicot plant species have been observed, such as variable substitution patterns on hemicellulose (Simmons et al., 2016; Grantham et al., 2017; Zhang et al., 2019; Gao et al., 2020) and variable hemicellulose-lignin contacts (Scheller and Ulvskov, 2010; Terrett and Dupree, 2019).
The details of lignin structure are particularly challenging to analyze due to its high heterogeneity and mobility, so partial extraction of the polymer is often necessary to assess it (Foston et al., 2012; Li and Takkellapati, 2018; Zhao et al., 2020; Zhang et al., 2021b). Lignin is a polyphenolic network formed from the oxidative cross-linking of the monolignols p-coumaryl alcohol (H), coniferyl alcohol (G), and sinapyl alcohol (S) (Li and Takkellapati, 2018). Lignin exists within the plant cell matrix, interfacing with both hemicellulose and cellulose (Foston et al., 2012, Figure 1B). Solution-state NMR in combination with MS, which relies on swelling ball-milled plant cell walls with deuterated solvents, provides detailed information on the types, functionalization, abundance of lignin linkages present, and linkage size (Kim and Ralph, 2010; Pu et al., 2011; Balakshin and Capanema, 2015; Foston et al., 2016; Bergs et al., 2020; Hyväkkö et al., 2020; Zhang et al., 2021b). However, due to the necessity for drying, mechanical treatment, dissolution, or solvent extraction in these techniques, they do not report on recalcitrance in the intact (native) secondary plant cell wall.
Like MRI, NMR is a non-invasive way to probe the chemical environment in tissues. Unlike MRI, NMR is inherently an atomic resolution technique, as the observed signals derive from nuclear spin magnetic moments located at precise locations in the molecules under study. In contrast to solution-state NMR, which requires solubilization of the sample, solid-state NMR methods allow for analysis of intact plant tissues. Here, we discuss how solid-state NMR and access to cost effective 13C labeling has contributed to our understanding of plant cell wall structure.
The requirement of NMR-active 13C isotopes was a major hurdle for characterizations of native plant cell wall structure. 13C has a low natural abundance (~1%) and the cost of early efforts at isotope incorporation restricted their use. Relatively low (~11%) 13C enrichment enabled early studies on hardwood (Lesage et al., 1999). The ability to detect the relative populations of rigid polymers (e.g., cellulose and a fraction of hemicellulose) was then applied to samples of pure cellulose and heterogeneous assemblies containing cellulose (VanderHart and Atalla, 1984; Newman and Hemmingson, 1995; Lesage et al., 1997; Wickholm et al., 1998; Bootten et al., 2004; Fayon et al., 2005).
While early X-ray diffraction studies demonstrated the crystalline nature of cellulose in plant cell walls (Hauser, 1929; Sisson, 1935; Ward, 1950; Gardner and Blackwell, 1974; Sarko and Muggli, 1974), solid-state NMR measurements provided more detail, such as the pattern of hydrogen bond interactions responsible for the macroscopic shape of in situ cellulose fibers (VanderHart and Atalla, 1984; Nishiyama et al., 2002). A set of 1D cross polarization (CP) measurements have been successfully applied to crystalline cellulose in birch and spruce biomass and offer the possibility of detecting exterior and interior cellulose components in macroscopic cellulose fibers (Wickholm et al., 1998; Fernandes et al., 2011). These straightforward 1D experiments were also useful for characterizing amorphous cellulose after ionic liquid processing of crystalline cellulose fibrils (Mori et al., 2012). The 2D Incredible Natural Abundance Double Quantum Transfer Experiment (INADEQUATE) method reports on directly bonded carbon atoms within polymers and has been useful for probing rigid structure, for example, resolving C2, C3, and C5 signals of cellulose in Populus euramericana hardwood samples (Lesage et al., 1999) and characterizing the structure of amorphous cellulose (Mori et al., 2012).
A major breakthrough occurred when a highly efficient method of 13C incorporation (13C glucose feeding to cultured plant cells) was coupled with multi-dimensional solid-state NMR to investigate the primary plant cell wall structure (Dick-Pérez et al., 2011). This series of studies provided both a compositional and architectural description of the primary plant cell wall (Dick-Pérez et al., 2011; Wang et al., 2012, 2013, 2014, 2015; White et al., 2014; Phyo et al., 2017, 2018; Zhao et al., 2020). Hemicellulose-cellulose interactions were found to be much less prevalent in the primary plant cell wall than suggested by earlier models based on solvent-extracted hemicellulose (Keegstra et al., 1973; Talmadge et al., 1973; Pauly et al., 1999; Dick-Pérez et al., 2011; White et al., 2014; Phyo et al., 2017). Furthermore, it was also revealed that the hemicellulose xyloglucan interacts mainly with the flat surfaces of crystalline cellulose fibers (Dick-Pérez et al., 2011; White et al., 2014), expanding on the idea of xyloglucan associating, cross-linking, and embedding into cellulose fibrils (Bootten et al., 2004). Semiquantitative distance measurements recorded with the Proton Driven Spin Diffusion (PDSD) experiments substantiated the organization of cellulose, xyloglucan, and pectin in primary cell walls of both monocot and eudicot cell species (Wang et al., 2012, 2014; White et al., 2014). These advances in 13C enrichment have shaped our understanding of the primary plant cell wall architecture (Wang and Hong, 2016).
The strategy of 13C glucose feeding is not suitable to the study of the secondary plant cell wall because the plants need to be grown to relative maturity, which is prohibitively expensive and complicated by respiration-dependent glucose synthesis. The development of inexpensive growth chambers, utilizing 13C enriched carbon dioxide as the sole carbon source, which support the growth of plants throughout their lifecycle, enabled the efficient incorporation of 13C isotopes (>90%) into plant tissues (Chen et al., 2011; Dupree et al., 2015; Gao and Mortimer, 2020). Use of these growth chambers revealed significant differences in the dominant hemicellulose-cellulose contacts in different plant species (Simmons et al., 2016; Terrett et al., 2019; Gao et al., 2020). For example, in eudicot Arabidopsis thaliana (Arabidopsis), 2-fold screw conformations of hemicellulose, dictated by even patterns of substitution on xylan, enable a close association with crystalline cellulose (Simmons et al., 2016; Grantham et al., 2017, Figure 1C). In contrast, in monocot sorghum, the high degree and irregularity of arabinose substitution patterns on xylan dictate a 3-fold screw conformation, enabling the association with amorphous cellulose (Gao et al., 2020, Figure 1D). Additionally, in softwoods, cellulose fibrils can be tethered by both xylan and mannan hemicellulose, increasing the strength of the plant cell wall (Terrett et al., 2019). For the sorghum case, limitations in biochemical techniques prevent the analysis of carbohydrate substitutions on xylan and the solid-state NMR measurements therefore describe the xylan-cellulose interaction that is otherwise unobtainable using other methods (Gao et al., 2020, Figure 1D).
Carbon dioxide 13C labeling and new applications of advanced solid-state NMR techniques have helped elucidate the structure of lignin in the secondary plant cell wall. Signal enhancement by dynamic nuclear polarization (DNP) demonstrated lignin directly bridges hemicellulose polymers that interact strongly with cellulose fibers in uniformly labeled switchgrass, highlighting the role of lignin in supporting the 3D organization of hemicellulose and cellulose (Kang et al., 2019). However, effective penetration of the DNP reagent into the plant cell wall for this signal enhancement required 15–20 min of milling (Kang et al., 2019), which could perturb native lignin structure. Direct polarization experiments utilizing PDSD (Addison et al., 2020) performed on 13C enriched poplar stems highlight a potential avenue to probe lignin contacts and spatial proximities through selective excitation and magnetization transfer from lignin to other polymers, which provide support for the putative organization of lignin in switchgrass (Kang et al., 2019) and Arabidopsis (Dupree et al., 2015). Further development of selective excitation and other solid-state NMR methods to probe biomass with minimal sample manipulation have the potential to provide a more complete picture of the secondary plant cell wall structure and how established sample preparation methods influence that structure.
Although a wide variety of solid-state NMR methods can be applied to highly 13C enriched plant tissues, two methods provide rapid and straightforward characterization of the polymer organization in them. First, the INADEQUATE approach provides an avenue for the characterization of the polymers present within a secondary cell wall sample at relatively high resolution and can distinguish at least three populations of amorphous and crystalline cellulose, in addition to three populations of xylan (Simmons et al., 2016; Gao et al., 2020). Second, 13C-13C recoupling methods, such as PDSD and Dipolar Assisted Rotational Resonance (DARR), report on the spatial proximity of cellulose, hemicellulose, and lignin (Simmons et al., 2016; Grantham et al., 2017; Kang et al., 2019; Terrett et al., 2019; Gao et al., 2020). These experiments provide a more complete picture of contacts between the polymers in the plant cell wall then has ever been obtainable before. For example, the differences hemicellulose-cellulose interactions (Busse-Wicher et al., 2016; Gao et al., 2020) that likely underlie the recalcitrance observed in deconstruction efforts can direct plant selection the biofuel and biomaterials industry.
Examining the reorganization of the secondary plant cell wall polymers due to mechanical conversion is important for the development of the plant cell wall model and effective utilization of biomass without recalcitrance (Marriott et al., 2016; Zhao et al., 2020; Zhang et al., 2021a). Solid-state NMR measurements on commercial cotton balls subjected to 15–120 min of vibrational ball milling readily show the conversion of crystalline to amorphous cellulose (Ling et al., 2019). The results are consistent with X-ray diffraction and vibrational spectroscopy measurements that show loss of crystallinity within the cellulose fibers of up to 60% during this same time period (Ling et al., 2019). However, relatively little work has been done on monitoring the native plant cell wall during the milling process. Conversion techniques commonly utilize mechanical milling for times typically as short as 2 min and can exceed 4 h (Kim and Ralph, 2010). As a result, these experiments report on cell wall structure after reorganization of the plant cell wall polymers occurs during mechanical preprocessing. For example, solid-state NMR measurements on maize biomass after mechanical and solvent processing methods support lignin association with the surface of hemicellulose coated cellulose fibers in the cell wall (Foston et al., 2012), a different result than those obtained from recent solid-state NMR measurements on less processed grass and other plant species biomass (Kang et al., 2019; Terrett et al., 2019; Gao et al., 2020). However, these experiments used different sample preparation methods and are from different species, so further investigation is required to make conclusive and precise statements about the influence of these processing methods on plant cell wall structure.
Currently, we are examining the structural rearrangements that occur in native secondary plant cell walls during mechanical preprocessing using 13C sorghum stems subjected to ball milling by solid-state NMR. Milling of stem tissue at 30 Hz for 2 min was selected to allow direct comparison to DMSO swelling studies employing the same milling time (Kim and Ralph, 2010). Figure 2A shows the percent difference in integrated signal intensities from 2D CP-INADEQUATE spectra of the ball milled sample and unprocessed control samples, normalized to minimize the difference in the integrals of the C1-C2 signals of cellulose. Samples were obtained from plants flash frozen in liquid nitrogen after harvest and sectioned on a dry ice cooled surface (Figure 1A). Control experiments show that a cycle of flash freezing does not alter the solid-state NMR spectra of our samples, at least beyond any influence of the initial flash freezing process. Figure 2B shows the polysaccharide region of the control sample spectrum. Signals in these spectra arise from highly immobilized polymers in the secondary plant cell wall (Lesage et al., 1997, 1999). Signal intensities from arabinose all significantly decrease by at least 30% after ball milling while 3-fold xylan C4 and C5 signal intensities increase. The signals from other carbons in xylan, arabinose substituted xylan, and the amorphous and crystalline cellulose intensities do not show a clear trend. Interpretation of these signals is less straightforward due to resonance overlap for these carbohydrates (Gao et al., 2020). The field emission scanning electron microscopy (FE-SEM) images of the control and milled samples in Figure 2C show the macroscopic structure of the plant biomass is lost even after 2 min of ball milling, yet at higher magnification, the individual fibers within the sample are largely similar, with a slightly rougher texture. Although more sophisticated analyses and a wider variety of NMR experiments are possible, this simple example highlights the ability of solid-state NMR to detect structural changes during lignocellulosic biomass conversion.
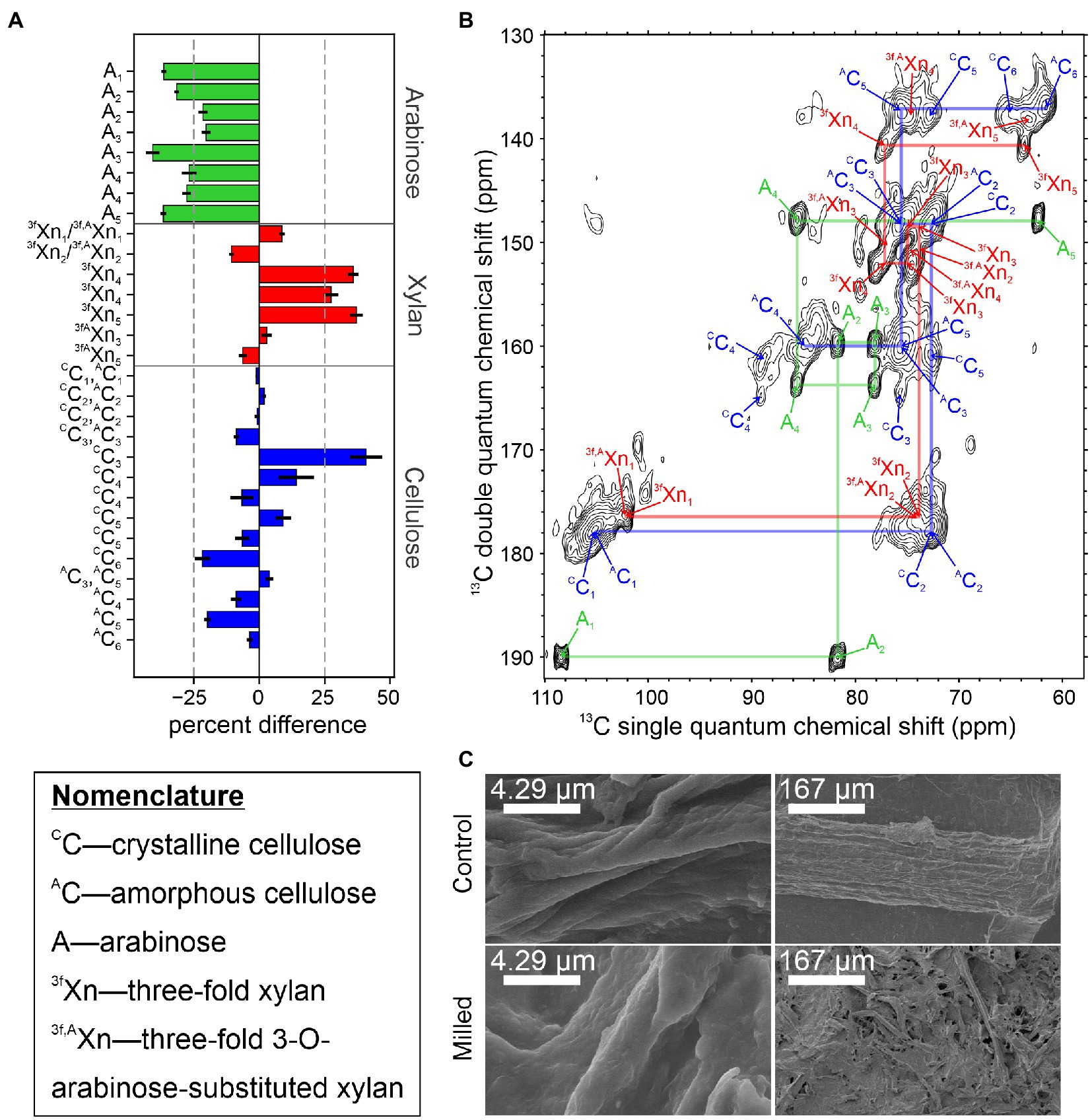
Figure 2. Changes in the rigid structure of sorghum stem tissue after 2 min of ball milling. (A) Percent difference in integrated signal intensities from the 2 min ball-milled sample, relative to the unprocessed control sample. Due to the nature of the CP-INADEQUATE experiment, two signals are observed for most sites. Severely overlapping signals were omitted from this analysis. Error bars derive from the root-mean-squared noise in the spectra. (B) The CP-INADEQUATE spectrum of the unprocessed control sample. Semi-transparent lines are drawn to show the connections between signals within amorphous cellulose (blue), 3-fold xylan (red), and arabinosyl units from xylan (green). The spectrum was recorded at a 1H frequency of 500 MHz with 10 kHz magic angle spinning and high power 1H decoupling. (C) Field emission SEM images of the 2 min ball-milled and control samples. For all experiments, sorghum Tx430 was grown to maturity in a custom 13CO2 chamber as previously described (Gao and Mortimer, 2020), and the material used here had at least 92% 13C incorporation. Ball milling was adapted from Kim and Ralph (2010), INADEQUATE experiments were performed according to Gao et al. (2020), and samples prepared for FE-SEM and imaged according to Zheng et al. (2020). NMR data were processed in NMRPipe (Delaglio et al., 1995) and plotted in Sparky (Lee et al., 2015).
Although solid-state NMR has the huge advantage of being able to characterize molecular structure of plant cell wall polymers within intact plant material, there are careful considerations to be aware of when planning for these experiments. The time required for 2D solid-state NMR can range from a few hours to almost a day, therefore requiring a few days of experimental time for an extensive set of spectra to be recorded. In our laboratory, we minimize the potential for sample degradation to influence the results of long 2D experiments by interweaving 1D control measurements into the data acquisition. Experiments can be stopped once significant changes in these 1D spectra are observed. Since multiple samples can be obtained from a single plant, dividing acquisition of different data sets between several identical samples is not a problem. Furthermore, the requirement of 13C isotope labeled tissue limits the range of plant types and processing methods that can be investigated. We see a bright outlook in this regard with DNP technology, where the future development of non-invasive polarization agents can permit high signal-to-noise data to be obtained from samples with natural abundance 13C isotope distributions (Wang et al., 2013; Fernando et al., 2020).
Efficient 13C labeling of living plants has opened the door for understanding how biopolymer organization within the plant cell wall confers biomass recalcitrance. Since solid-state NMR does not require solubilization of the plant cell wall, access to 13C labeled plant material allows for the native secondary plant cell wall architecture to be tracked during biomass conversion at high resolution. Here, we present a method for quantitative comparison using normalized integration of 2D spectra before and after milling for the refocused CP-INADEQUATE. Although initial results clearly indicate a >30% decrease in arabinose substitutions of xylan, other results remain inconclusive. Future experiments will explore, for example, structural changes in rigid cellulose and the mobile components of hemicellulose and lignin. Contrasts between tissue types containing more or less secondary plant cell wall (e.g., comparing stem and leaf tissue) will also aid in assessing lignin associated recalcitrance. Genetic engineering efforts based on a molecular understanding of recalcitrance will surely also prove invaluable (Carpita and McCann, 2020). In conclusion, recalcitrance remains to be defined within the 3D structure of the plant cell wall and solid-state NMR will be an invaluable tool to investigate native secondary plant cell wall structure and for monitoring structural changes due to genetic engineering and biomass conversion approaches.
The raw data supporting the conclusions of this article will be made available by the authors upon request, without undue reservation.
CRM and DTM recorded and interpreted NMR spectra. CRM recorded the FE-SEM images. YG and JCM produced the 13C labeled sorghum plants. CRM, YG, JCM, and DTM wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was made possible by generous start-up funding from the University of California, Davis. The NMR experiments made use of the University of California, Davis NMR Campus Core Facility and Keck Spectral Imaging Facility with funding from the National Science Foundation through NSF EAR0213546. Part of this work was conducted through the DOE Joint BioEnergy Institute (http://www.jbei.org) supported by the U. S. Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U. S. Department of Energy.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Ping Yu of the University of California, Davis for assistance with the 500 MHz NMR spectrometer.
MS, Mass Spectrometry; CP, Cross Polarization; INADEQUATE, Incredible Natural Abundance Double Quantum Transfer Experiment; NMR, Nuclear Magnetic Resonance; PDSD, Proton Driven Spin Diffusion; DARR, Dipolar Assisted Rotational Resonance; FE-SEM, Field Emission Scanning Electron Microscopy.
Addison, B., Stengel, D., Bharadwaj, V. S., Happs, R. M., Doeppke, C., Wang, T., et al. (2020). Selective One-Dimensional 13C–13C Spin-Diffusion Solid-State Nuclear Magnetic Resonance Methods to Probe Spatial Arrangements in Biopolymers Including Plant Cell Walls, Peptides, and Spider Silk. J. Phys. Chem. 124, 9870–9883. doi: 10.1021/acs.jpcb.0c07759
Agarwal, U. P. (2019). Analysis of cellulose and lignocellulose materials by Raman spectroscopy: a review of the current status. Molecules 24:1659. doi: 10.3390/molecules24091659
Baig, K. S., Wu, J., and Turcotte, G. (2019). Future prospects of delignification pretreatments for the lignocellulosic materials to produce second generation bioethanol. Int. J. Energy Res. 43, 1411–1427. doi: 10.1002/er.4292
Balakshin, M., and Capanema, E. (2015). On the quantification of lignin hydroxyl groups with 31P and 13C NMR spectroscopy. J. Wood Chem. Technol. 35, 220–237. doi: 10.1080/02773813.2014.928328
Baral, N. R., Sundstrom, E. R., Das, L., Gladden, J., Eudes, A., Mortimer, J. C., et al. (2019). Approaches for more efficient biological conversion of lignocellulosic feedstocks to biofuels and bioproducts. ACS Sustain. Chem. Eng. 7, 9062–9079. doi: 10.1021/acssuschemeng.9b01229
Bergs, M., Do, X. T., Rumpf, J., Kusch, P., Monakhova, Y., Konow, C., et al. (2020). Comparing chemical composition and lignin structure of Miscanthus × Giganteus and Miscanthus nagara harvested in autumn and spring and separated into stems and leaves. RSC Adv. 10, 10740–10751. doi: 10.1039/C9RA10576J
Bootten, T. J., Harris, P. J., Melton, L. D., and Newman, R. H. (2004). Solid-state 13C-NMR spectroscopy shows that the xyloglucans in the primary cell walls of mung bean (Vigna radiata L.) occur in different domains: a new model for xyloglucan–cellulose interactions in the cell wall. J. Exp. Bot. 55, 571–583. doi: 10.1093/jxb/erh065
Busse-Wicher, M., Li, A., Silveira, R. L., Pereira, C. S., Tryfona, T., Gomes, T. C. F., et al. (2016). Evolution of xylan substitution patterns in gymnosperms and angiosperms: implications for xylan interaction with cellulose. Plant Physiol. 171, 2418–2431. doi: 10.1104/pp.16.00539
Carpita, N. C., and McCann, M. C. (2020). Redesigning plant cell walls for the biomass-based bioeconomy. J. Biol. Chem. 295, 15144–15157. doi: 10.1074/jbc.REV120.014561
Chen, W., Yang, X., Harms, G. L., Gray, W. M., Hegeman, A. D., and Cohen, J. D. (2011). An automated growth enclosure for metabolic labeling of Arabidopsis thaliana with 13C-carbon dioxide – an in vivo labeling system for proteomics and metabolomics research. Proteome Sci. 9:9. doi: 10.1186/1477-5956-9-9
Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J., and Bax, A. (1995). NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293. doi: 10.1007/BF00197809
Dick-Pérez, M., Zhang, Y., Hayes, J., Salazar, A., Zabotina, O. A., and Hong, M. (2011). Structure and interactions of plant cell-wall polysaccharides by two- and three-dimensional magic-angle-spinning solid-state NMR. Biochemistry 50, 989–1000. doi: 10.1021/bi101795q
Ding, J., Yoo, C. G., Pu, Y., Meng, X., Bhagia, S., Yu, C., et al. (2019). Cellulolytic enzyme-aided extraction of hemicellulose from switchgrass and its characteristics. Green Chem. 21, 3902–3910. doi: 10.1039/C9GC00252A
Dupree, R., Simmons, T. J., Mortimer, J. C., Patel, D., Iuga, D., Brown, S. P., et al. (2015). Probing the molecular architecture of Arabidopsis thaliana secondary cell walls using two- and three-dimensional 13C solid state nuclear magnetic resonance spectroscopy. Biochemistry 54, 2335–2345. doi: 10.1021/bi501552k
Fayon, F., Massiot, D., Levitt, M. H., Titman, J. T., Gregory, D. H., Duma, L., et al. (2005). Through-space contributions to two-dimensional double-quantum J correlation NMR spectra of magic-angle-spinning solids. J. Chem. Phys. 122:194313. doi: 10.1063/1.1898219
Fernandes, A. N., Thomas, L. H., Altaner, C. M., Callow, P., Forsyth, V. T., Apperley, D. C., et al. (2011). Nanostructure of cellulose microfibrils in spruce wood. Proc. Natl. Acad. Sci. U. S. A. 108, E1195–E1203. doi: 10.1073/pnas.1108942108
Fernando, L. D., Zhao, W., Widanage, M. C. D., Metink-Vigier, F., and Wang, T. (2020). Solid-state NMR and DNP investigations of carbohydrates and cell-wall biomaterials. eMagRes 9, 251–258. doi: 10.1002/pnas.9780470034590.emrstm1624
Foston, M., Katahira, R., Gjersing, E., Davis, M. F., and Ragauskas, A. J. (2012). Solid-state selective 13C excitation and spin diffusion NMR to resolve spatial dimensions in plant cell walls. J. Agric. Food Chem. 60, 1419–1427. doi: 10.1021/jf204853b
Foston, M., Samuel, R., He, J., and Ragauskas, A. J. (2016). A review of whole cell wall NMR by the direct-dissolution of biomass. Green Chem. 18, 608–621. doi: 10.1039/C5GC02828K
Gao, Y., Lipton, A. S., Wittmer, Y., Murray, D. T., and Mortimer, J. C. (2020). A grass-specific cellulose–xylan interaction dominates in sorghum secondary cell walls. Nat. Commun. 11:6081. doi: 10.1038/s41467-020-19837-z
Gao, Y., and Mortimer, J. C. (2020). “Chapter 7 - Unlocking the architecture of native plant cell walls via solid-state nuclear magnetic resonance,” in Methods in Cell Biology. Vol. 160. eds. C. T. Anderson, E. S. Haswell, and R. Dixit (Cambridge, MA: Academic Press), 121–143.
Gardner, K. H., and Blackwell, J. (1974). The structure of native cellulose. Biopolymers 13, 1975–2001. doi: 10.1002/bip.1974.360131005
Genuino, H. C., Muizebelt, I., Heeres, A., Schenk, N. J., Winkelman, J. G. M., and Heeres, H. J. (2019). An improved catalytic pyrolysis concept for renewable aromatics from biomass involving a recycling strategy for co-produced polycyclic aromatic hydrocarbons. Green Chem. 21, 3802–3806. doi: 10.1039/C9GC01485C
Gilna, P., Lynd, L. R., Mohnen, D., Davis, M. F., and Davison, B. H. (2017). Progress in understanding and overcoming biomass recalcitrance: a BioEnergy Science Center (BESC) perspective. Biotechnol. Biofuels 10:285. doi: 10.1186/s13068-017-0971-1
Grantham, N. J., Wurman-Rodrich, J., Terrett, O. M., Lyczakowski, J. J., Stott, K., Iuga, D., et al. (2017). An even pattern of xylan substitution is critical for interaction with cellulose in plant cell walls. Nat. Plants 3, 859–865. doi: 10.1038/s41477-017-0030-8
Hauser, E. A. (1929). What the X-ray tells us of the structure of cellulose. Ind. Eng. Chem. 21, 124–125. doi: 10.1021/ie50230a006
Hyväkkö, U., Maltari, R., Kakko, T., Kontro, J., Mikkilä, J., Kilpeläinen, P., et al. (2020). On the effect of hot-water pretreatment in sulfur-free pulping of aspen and wheat straw. ACS Omega 5, 265–273. doi: 10.1021/acsomega.9b02619
Ishak, W. H., Rosli, N. A., and Ahmad, I. (2020). Influence of amorphous cellulose on mechanical, thermal, and hydrolytic degradation of poly(lactic acid) biocomposites. Sci. Rep. 10:11342. doi: 10.1038/s41598-020-68274-x
Jung, S., Foston, M., Kalluri, U. C., Tuskan, G. A., and Ragauskas, A. J. (2012). 3D chemical image using TOF-SIMS revealing the biopolymer component spatial and lateral distributions in biomass. Angew. Chem. Int. Ed. 51, 12005–12008. doi: 10.1002/anie.201205243
Kang, X., Kirui, A., Widanage, M. C. D., Mentink-Vigier, F., Cosgrove, D. J., and Wang, T. (2019). Lignin-polysaccharide interactions in plant secondary cell walls revealed by solid-state NMR. Nat. Commun. 10:347. doi: 10.1038/s41467-018-08252-0
Keegstra, K., Talmadge, K. W., Bauer, W. D., and Albersheim, P. (1973). The structure of plant cell walls. Plant Physiol. 51, 188–197. doi: 10.1104/pp.51.1.188
Kim, K. H., Dutta, T., Sun, J., Simmons, B., and Singh, S. (2018). Biomass pretreatment using deep eutectic solvents from lignin derived phenols. Green Chem. 20, 809–815. doi: 10.1039/C7GC03029K
Kim, H., and Ralph, J. (2010). Solution-state 2D NMR of ball-milled plant cell wall gels in DMSO-D6/pyridine-D5. Org. Biomol. Chem. 8, 576–591. doi: 10.1039/B916070A
Lee, W., Tonelli, M., and Markley, J. L. (2015). NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327. doi: 10.1093/bioinformatics/btu830
Lesage, A., Auger, C., Caldarelli, S., and Emsley, L. (1997). Determination of through-bond carbon−carbon connectivities in solid-state NMR using the INADEQUATE experiment. J. Am. Chem. Soc. 119, 7867–7868. doi: 10.1021/ja971089k
Lesage, A., Bardet, M., and Emsley, L. (1999). Through-bond carbon−carbon connectivities in disordered solids by NMR. J. Am. Chem. Soc. 121, 10987–10993. doi: 10.1021/ja992272b
Li, M., Pu, Y., and Ragauskas, A. J. (2016). Current understanding of the correlation of lignin structure with biomass recalcitrance. Front. Chem. 4:45. doi: 10.3389/fchem.2016.00045
Li, T., and Takkellapati, S. (2018). The current and emerging sources of technical lignins and their applications. Biofuels Bioprod. Biorefin. 12, 756–787. doi: 10.1002/bbb.1913
Ling, Z., Wang, T., Makarem, M., Cintrón, M. S., Cheng, H. N., Kang, X., et al. (2019). Effects of ball milling on the structure of cotton cellulose. Cellulose 26, 305–328. doi: 10.1007/s10570-018-02230-x
Lopes, A. M., Filho, E. X. F., and Moreira, L. R. S. (2018). An update on enzymatic cocktails for lignocellulose breakdown. J. Appl. Microbiol. 125, 632–645. doi: 10.1111/jam.13923
Marriott, P. E., Gómez, L. D., and McQueen-Mason, S. J. (2016). Unlocking the potential of lignocellulosic biomass through plant science. New Phytol. 209, 1366–1381. doi: 10.1111/nph.13684
Mori, T., Chikayama, E., Tsuboi, Y., Ishida, N., Shisa, N., Noritake, Y., et al. (2012). Exploring the conformational space of amorphous cellulose using NMR chemical shifts. Carbohydr. Polym. 90, 1197–1203. doi: 10.1016/j.carbpol.2012.06.027
Newman, R. H., and Hemmingson, J. A. (1995). Carbon-13 NMR distinction between categories of molecular order and disorder in cellulose. Cellulose 2, 95–110. doi: 10.1007/BF00816383
Nishiyama, Y., Langan, P., and Chanzy, H. (2002). Crystal structure and hydrogen-bonding system in cellulose Iβ from synchrotron X-ray and neutron fiber diffraction. J. Am. Chem. Soc. 124, 9074–9082. doi: 10.1021/ja0257319
Park, Y. B., and Cosgrove, D. J. (2012). A revised architecture of primary cell walls based on biomechanical changes induced by substrate-specific endoglucanases. Plant Physiol. 158, 1933–1943. doi: 10.1104/pp.111.192880
Pauly, M., Albersheim, P., Darvill, A., and York, W. S. (1999). Molecular domains of the cellulose/xyloglucan network in the cell walls of higher plants. Plant J. 20, 629–639. doi: 10.1046/j.1365-313X.1999.00630.x
Pauly, M., and Keegstra, K. (2010). Plant cell wall polymers as precursors for biofuels. Curr. Opin. Plant Biol. 13, 304–311. doi: 10.1016/j.pbi.2009.12.009
Phyo, P., Wang, T., Xiao, C., Anderson, C. T., and Hong, M. (2017). Effects of pectin molecular weight changes on the structure, dynamics, and polysaccharide interactions of primary cell walls of Arabidopsis thaliana: insights from solid-state NMR. Biomacromolecules 18, 2937–2950. doi: 10.1021/acs.biomac.7b00888
Phyo, P., Wang, T., Yang, Y., O’Neill, H., and Hong, M. (2018). Direct determination of hydroxymethyl conformations of plant cell wall cellulose using 1H polarization transfer solid-state NMR. Biomacromolecules 19, 1485–1497. doi: 10.1021/acs.biomac.8b00039
Priharto, N., Ronsse, F., Yildiz, G., Heeres, H. J., Deuss, P. J., and Prins, W. (2020). Fast pyrolysis with fractional condensation of lignin-rich digested stillage from second-generation bioethanol production. J. Anal. Appl. Pyrolysis 145:104756. doi: 10.1016/j.jaap.2019.104756
Pu, Y., Cao, S., and Ragauskas, A. J. (2011). Application of quantitative 31P NMR in biomass lignin and biofuel precursors characterization. Energy Environ. Sci. 4, 3154–3166. doi: 10.1039/c1ee01201k
Qi, Y., and Volmer, D. A. (2019). Chemical diversity of lignin degradation products revealed by matrix-optimized MALDI mass spectrometry. Anal. Bioanal. Chem. 411, 6031–6037. doi: 10.1007/s00216-019-01984-y
Sarko, A., and Muggli, R. (1974). Packing analysis of carbohydrates and polysaccharides. III. Valonia cellulose and cellulose II. Macromolecules 7, 486–494. doi: 10.1021/ma60040a016
Scheller, H. V., and Ulvskov, P. (2010). Hemicelluloses. Annu. Rev. Plant Biol. 61, 263–289. doi: 10.1146/annurev-arplant-042809-112315
Simmons, T. J., Mortimer, J. C., Bernardinelli, O. D., Pöppler, A., Brown, S. P., Eduardo, R., et al. (2016). Folding of xylan onto cellulose fibrils in plant cell walls revealed by solid-state NMR. Nat. Commun. 7:13902. doi: 10.1038/ncomms13902
Sisson, W. A. (1935). X-ray studies of crystallite orientation in cellulose fibers natural fibers. Ind. Eng. Chem. 27, 51–56. doi: 10.1021/ie50301a012
Sluiter, A., Hames, B., Ruiz, R., Scarlata, C., Sluiter, J., Templeton, D., et al. (2008). Determination of Structural Carbohydrates and Lignin in Biomass. Golden, CO: National Renewable Energy Laboratory.
Song, B., Zhao, S., Shen, W., Collings, C., and Ding, S. (2020). Direct measurement of plant cellulose microfibril and bundles in native cell walls. Front. Plant Sci. 11:479. doi: 10.3389/fpls.2020.00479
Talmadge, K. W., Keegstra, K., Bauer, W. D., and Albersheim, P. (1973). The structure of plant cell walls: I. The macromolecular components of the walls of suspension-cultured sycamore cells with a detailed analysis of the pectic polysaccharides. Plant Physiol. 51, 158–173. doi: 10.1104/pp.51.1.158
Terrett, O. M., and Dupree, P. (2019). Covalent interactions between lignin and hemicelluloses in plant secondary cell walls. Curr. Opin. Biotechnol. 56, 97–104. doi: 10.1016/j.copbio.2018.10.010
Terrett, O. M., Lyczakowski, J. J., Yu, L., Iuga, D., Franks, W. T., Brown, S. P., et al. (2019). Molecular architecture of softwood revealed by solid-state NMR. Nat. Commun. 10:4978. doi: 10.1038/s41467-019-12979-9
Tolbert, A., Akinosho, H., Khunsupat, R., Naskar, A. K., and Ragauskas, A. J. (2014). Characterization and analysis of the molecular weight of lignin for biorefining studies. Biofuels Bioprod. Biorefin. 8, 836–856. doi: 10.1002/bbb.1500
U.S. Department of Energy (2016). “2016 billion-ton report: advancing domestic resources for a thriving bioeconomy,” in Economic Availability of Feedstocks. Vol. 1. eds. M. H. Langholtz, B. J. Stokes, and L. M. Eaton (Oak Ridge, TN: Oak Ridge National Laboratory), 448.
van Putten, R., van der Waal, J. C., de Jong, E., Rasrendra, C. B., Heeres, H. J., and de Vries, J. G. (2013). Hydroxymethylfurfural, a versatile platform chemical made from renewable resources. Chem. Rev. 113, 1499–1597. doi: 10.1021/cr300182k
VanderHart, D. L., and Atalla, R. H. (1984). Studies of microstructure in native celluloses using solid-state carbon-13 NMR. Macromolecules 17, 1465–1472. doi: 10.1021/ma00138a009
Wang, T., and Hong, M. (2016). Solid-state NMR investigations of cellulose structure and interactions with matrix polysaccharides in plant primary cell walls. J. Exp. Bot. 67, 503–514. doi: 10.1093/jxb/erv416
Wang, T., Park, Y. B., Caporini, M. A., Rosay, M., Zhong, L., Cosgrove, D. J., et al. (2013). Sensitivity-enhanced solid-state NMR detection of expansin’s target in plant cell walls. Proc. Natl. Acad. Sci. U. S. A. 110, 16444–16449. doi: 10.1073/pnas.1316290110
Wang, T., Park, Y. B., Cosgrove, D. J., and Hong, M. (2015). Cellulose-pectin spatial contacts are inherent to never-dried arabidopsis primary cell walls: evidence from solid-state nuclear magnetic resonance. Plant Physiol. 168, 871–884. doi: 10.1104/pp.15.00665
Wang, T., Salazar, A., Zabotina, O. A., and Hong, M. (2014). Structure and dynamics of brachypodium primary cell wall polysaccharides from two-dimensional 13C solid-state nuclear magnetic resonance spectroscopy. Biochemistry 53, 2840–2854. doi: 10.1021/bi500231b
Wang, T., Zabotina, O. A., and Hong, M. (2012). Pectin–cellulose interactions in the Arabidopsis primary cell wall from two-dimensional magic-angle-spinning solid-state nuclear magnetic resonance. Biochemistry 51, 9846–9856. doi: 10.1021/bi3015532
Wang, Z., Zhu, X., and Deuss, P. J. (2021). The effect of ball milling on birch, pine, reed, walnut shell enzymatic hydrolysis recalcitrance and the structure of the isolated residual enzyme lignin. Ind. Crop. Prod. 167:113493. doi: 10.1016/j.indcrop.2021.113493
Ward, K. (1950). Crystallinity of cellulose and its significance for the fiber properties. Text. Res. J. 20, 363–372. doi: 10.1177/004051755002000601
White, P. B., Wang, T., Park, Y. B., Cosgrove, D. J., and Hong, M. (2014). Water–polysaccharide interactions in the primary cell wall of Arabidopsis thaliana from polarization transfer solid-state NMR. J. Am. Chem. Soc. 136, 10399–10409. doi: 10.1021/ja504108h
Wickholm, K., Larsson, P. T., and Iversen, T. (1998). Assignment of non-crystalline forms in cellulose I by CP/MAS 13C NMR spectroscopy. Carbohydr. Res. 312, 123–129. doi: 10.1016/S0008-6215(98)00236-5
Yao, L., Yang, H., Yoo, C. G., Pu, Y., Meng, X., Muchero, W., et al. (2018). Understanding the influences of different pretreatments on recalcitrance of populus natural variants. Bioresour. Technol. 265, 75–81. doi: 10.1016/j.biortech.2018.05.057
Zhai, Q., Li, F., Wang, F., Feng, J., Jiang, J., and Xu, J. (2019). Ultrafine grinding of poplar biomass: effect of particle morphology on the liquefaction of biomass for methyl glycosides and phenolics. Cellulose 26, 3685–3701. doi: 10.1007/s10570-019-02332-0
Zhang, L., Gao, C., Mentink-Vigier, F., Tang, L., Zhang, D., Wang, S., et al. (2019). Arabinosyl deacetylase modulates the arabinoxylan acetylation profile and secondary wall formation. Plant Cell 31, 1113–1126. doi: 10.1105/tpc.18.00894
Zhang, B., Gao, Y., Zhang, L., and Zhou, Y. (2021a). The plant cell wall: biosynthesis, construction, and functions. J. Integr. Plant Biol. 63, 251–272. doi: 10.1111/jipb.13055
Zhang, R., Qi, Y., Ma, C., Ge, J., Hu, Q., Fu-Jun Yue, F., et al. (2021b). Characterization of lignin compounds at the molecular level: mass spectrometry analysis and raw data processing. Molecules 26:178. doi: 10.3390/molecules26010178
Zhao, W., Fernando, L. D., Kirui, A., Deligey, F., and Wang, T. (2020). Solid-state NMR of plant and fungal cell walls: a critical review. Solid State Nucl. Magn. Reason. 107:101660. doi: 10.1016/j.ssnmr.2020.101660
Zhao, X., Zhang, Y., Hu, H., Huang, Z., Yang, M., Chen, D., et al. (2016). Effect of mechanical activation on structure changes and reactivity in further chemical modification of lignin. Int. J. Biol. Macromol. 91, 1081–1089. doi: 10.1016/j.ijbiomac.2016.06.074
Keywords: solid-state nuclear magnetic resonance, Sorghum bicolor, cellulose, hemicellulose, lignin, ball milling, recalcitrance (saccharification), preprocessing
Citation: Munson CR, Gao Y, Mortimer JC and Murray DT (2022) Solid-State Nuclear Magnetic Resonance as a Tool to Probe the Impact of Mechanical Preprocessing on the Structure and Arrangement of Plant Cell Wall Polymers. Front. Plant Sci. 12:766506. doi: 10.3389/fpls.2021.766506
Received: 29 August 2021; Accepted: 25 November 2021;
Published: 12 January 2022.
Edited by:
John Paul Moore, Stellenbosch University, South AfricaReviewed by:
Chang Geun Yoo, SUNY College of Environmental Science and Forestry, United StatesCopyright © 2022 Munson, Gao, Mortimer and Murray. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dylan T. Murray, ZHRtdXJyYXlAdWNkYXZpcy5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.