- 1Triticeae Research Institute, Sichuan Agricultural University, Chengdu, China
- 2State Key Laboratory of Crop Gene Exploration and Utilization in Southwest China, Sichuan Agricultural University, Chengdu, China
- 3Qinghai Province Key Laboratory of Crop Molecular Breeding, Xining, China
- 4Key Laboratory of Adaptation and Evolution of Plateau Biota (AEPB), Northwest Institute of Plateau Biology, Chinese Academy of Sciences, Xining, China
Triticum boeoticum Boiss (AbAb, 2n = 2x = 14) is one of the sources of the blue grain trait controlled by blue aleurone layer 2 (Ba2). However, the underlying genes have not been cloned. In this study, a transcriptomic comparison between a blue-grained wheat-T. boeoticum substitution line and its wheat parent identified 41 unigenes related to anthocyanin biosynthesis and 29 unigenes related to transport. The bHLH transcription factor gene TbMYC4A showed a higher expression level in the blue-grained substitution line. TbMYC4A contained the three characteristic bHLH transcription factor domains (bHLH-MYC_N, HLH and ACT-like) and clustered with genes identified from other wheat lines with the blue grain trait derived from other Triticeae species. TbMYC4A overexpression confirmed that it was a functional bHLH transcription factor. The analysis of a TbMYC4A-specific marker showed that the gene was also present in T. boeoticum and T. monococcum with blue aleurone but absent in other Triticeae materials with white aleurone. These results indicate that TbMYC4A is a candidate gene of Ba2 controlling the blue aleurone trait. The isolation of TbMYC4A is helpful for further clarifying the genetic mechanism of the blue aleurone trait and is of great significance for breeding blue-grained wheat varieties.
Introduction
Colored-grain wheat is regarded as an ideal food for human health and has attracted increasing interest from both food manufacturers and researchers. Unlike purple-grained wheat, which stores anthocyanins in the seed coat, blue-grained wheat shows anthocyanin accumulation in the aleurone layer (Abdel-Aal and Hucl, 2003, Abdel-Aal et al., 2006). In addition, purple and blue wheat can be crossed to form black wheat (BW), which is rich in nutritional and bioactive compounds giving it great potential for the development of healthy functional foods (Dhua et al., 2021). Anthocyanins are an important component of the human diet and are widely distributed in vegetables and fruits. They plan an important role in human health as anti-inflammatory, antioxidant, anticancer and antidiabetic agents (Shipp and Abdel-Aal, 2010). Anthocyanidins require glycosylation, methylation, and acylation to form stable anthocyanins and over 400 anthocyanins have been described so far (Springob et al., 2003; Sasaki et al., 2013). There are six main anthocyanidins in plants: delphinidin, cyanidin, peonidin, pelargonidin, petunidin and malvidin (Kähkönen and Heinonen, 2003). Delphinidin is the major anthocyanidin in blue-grained wheat (Trojan et al., 2014).
Common wheat does not contain the blue grain trait, but the trait can be introduced by crossing it with some species of Triticeae to give it a blue aleurone layer (Burešová et al., 2015). Several genes conferring the blue aleurone trait have been transferred into wheat from homoeologous group 4 of Triticeae species (Burešová et al., 2015). Ba1 is derived from Thinopyrum ponticum, located on the long arm of chromosome 4Ag FL 0.71–0.8 (Zheng et al., 2006; Liu et al., 2018). ThMYC4E is a candidate Ba1 gene (Li et al., 2017). Ba2, originating from Triticum boeoticum and Triticum monococcum, was mapped to a location close to the centromere on the long arm of 4Ab or 4Am, respectively (Zeller et al., 1991; Zeven, 1991; Dubcovsky et al., 1996; Singh et al., 2007; Yu et al., 2017). BaThb is located on the chromosome 4J of Th. bessarabicum (Shen et al., 2013). In addition, the HvMYC2 gene was identified to be involved in anthocyanin synthesis in the barley aleurone layer (Strygina et al., 2017), and TsMYC2 has been correlated with the blue grain trait in triticale (Zong et al., 2019).
The anthocyanin biosynthesis pathway is generally an extension of the flavonoid pathway and is a component of secondary plant metabolism (Larter et al., 2019; Wheeler and Smith, 2019). In many plants, the anthocyanin biosynthesis pathway is a highly conserved network (Tanaka et al., 2008). Anthocyanin biosynthesis begins with the conversion of phenylalanine to coumaric CoA through a three-step enzymatic reaction. Then, the chalcone synthase (CHS)-mediated synthesis of naringin chalcone from malonyl-CoA and 4-coumaroyl-CoA occurs. Chalcone isomerase isomerizes (CHI) chalcone to naringenin. Naringin is hydroxylated to form dihydroflavanols, which is catalyzed by flavonoid-3-hydroxylase (F3H), and can be further hydroxylated by flavonoid 3′,5′-hydroxylase (F3′5′H) or 3′-hydroxylase (F3′H) to generate dihydromyricetin or dihydroquercetin, respectively. Colored anthocyanidins are formed by dihydroflavonol 4-reductase (DFR) and anthocyanin synthase (ANS/LDOX). Subsequently, various stable anthocyanins are formed by modification by acyltransferase (AT), methyltransferase (MT), and glucosyltransferase (GT) (Springob et al., 2003; Sasaki et al., 2013). After the synthesis of anthocyanins in the cytosol, they must enter the vacuole for their diverse colors to be observable (Winkel-Shirley, 2001). One of the most complete potential mechanisms of anthocyanin transport involves the cooperation of glutathione transferase (GST) and multidrug resistance-associated related proteins (MRPs) (Goodman et al., 2004; Hu et al., 2016). MYB, bHLH and WD40 transcription factors form the MBW complex and combine with promoters to activate the transcription of structural genes in the anthocyanin biosynthetic pathway, thereby driving the tissue-specific accumulation of pigments (Ravaglia et al., 2013; Lloyd et al., 2017). Studies have shown that the co-expression of MYB and bHLH transcription factors can induce the synthesis of anthocyanins, such as ZmC1 and ZmR in maize and TaMYB7D and TaMYC1 in purple wheat (Ahmed et al., 2003; Zong et al., 2017).
The key genes for anthocyanin biosynthesis have been isolated in distinct tissues of certain plants using high-throughput RNA sequencing (RNA-Seq) (Li et al., 2017; Strygina et al., 2017; Zong et al., 2019). In our previous study, the blue-grained wheat-T. boeoticum substitution line Z18-1244 was developed (Liu et al., 2020). However, the gene controlling the blue aleurone of T. boeoticum has not been identified. The objectives of the present study were (1) to compare the expression levels of transcripts in the aleurone of red- and blue-grained wheat and (2) to identify the candidate genes regulating the blue aleurone of the Z18-1244 substitution line.
Materials and Methods
Plant Materials
The wheat-T. boeoticum substitution line Z18-1244 and its parents (common wheat Crocus and T. boeoticum G52) were used for RNA-Seq analysis. A total of 24 genotypes were subjected to TbMYC4A gene detection, including 14 with white aleurone from Triticum turgidum, Aegilops tauschii, Triticum urartu, Triticum araraticum, and Triticum zhukovskyi and 10 with blue aleurone from T. boeoticum and its cultivated type, T. monococcum (Supplementary Table 1). Crocus and G52 were kindly provided by George Fedak at the Ottawa Research and Development Center in Canada. Lines with PI or CItr prefixes were kindly provided by USDA-ARS, United States, while AS lines were obtained from Sichuan Agricultural University. Triticum zhukovskyi TRI 7270-1 and TRI 7270-3 were kindly provided by Prof. Fangpu Han at the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences.
Genomic DNA, Total RNA, and cDNA Preparation
Genomic DNA was extracted from fresh leaves 22 days post-anthesis using a plant genomic DNA kit (Tiangen Biotech, Beijing Co. Ltd., Beijing, China). At 14, 22, 28, 35 days post-anthesis (dpa), the aleurone layer of the immature grains was carefully stripped and immediately placed in liquid nitrogen for total RNA extraction, with replicates of 10 grain aleurone layers for each material. Total RNA was extracted using the Tiangen RNAprep Pure Plant Kit (Tiangen Corporation, Beijing, China) according to the standard protocol. RNA concentration and purity were measured using a NanoDrop 2,000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, United States). cDNA was obtained from total RNA using the Thermo RevertAid First Strand cDNA Synthesis Kit (Thermo-Fisher Scientific, Shanghai, China).
Illumina Sequencing and Transcriptome Analysis
Total RNA at 22 days post-anthesis was purified and used to construct a Strand-Specific Normal Library for sequencing on the Illumina (HiSeq 2500/4000) platform (Biomarker Technologies Co., Ltd., China). The clustering of index-coded samples was performed on a cBot Cluster Generation System using the TruSeq PE Cluster Kit v4-cBot-HS (Illumina) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina platform, and paired-end reads were generated.
Raw data (raw reads) in FASTQ format were first processed with in-house Perl scripts. All downstream analyses were based on clean data with high quality. These clean reads were then mapped to the reference genome sequence. Only reads with a perfect match or one mismatch were further analyzed and annotated based on the reference genome. HISAT2 tools were used for mapping against the reference genome (ta_IWGS C_MIPSv2.2_HighConf_CDS_2014Jul18.fa) (Kim et al., 2015). The average mapping ratio of each sample reached 86.27%, which showed that the data were comparable between samples. After comparative analysis, String Tie comparison was performed to assemble and quantify the reads (Pertea et al., 2015). Differential expression analysis was performed using the DESeq2 R package (Love et al., 2014). Differentially expressed genes (DEGs) were identified according to a false discovery rate (FDR) threshold of < 0.05 and a | log2 (fold change)| ≥ 1. To annotate and classify genes, the assembled unigenes were subjected to searches in public databases, including Nr,1 SWISS-PROT,2 COG,3 KEGG,4 GO,5 KOG,6 Pfam,7 and eggNOG.8 Gene ontology (GO) and KEGG enrichment analyses were performed using ShinyGO v 0.61 (Ge et al., 2020). The functional categories of the genes were selected according to an enrichment FDR of p < 0.05. Genes related to anthocyanin synthesis were collected from KEGG and compared with unigenes sequenced from blue and white aleurone by the BlastX algorithm with an E-value of < 1e-5. The raw sequence reads were stored in the NCBI SRA database with the accession number PRJNA757248.
PCR Amplification
Primer 5 software (Premier Biosoft, Palo Alto, CA, United States) was used to design primers. PCR amplification was performed using Vereti96 Abi Life 2720 (Thermo Fisher Scientific) and i-Max Ultra-high-fidelity DNA polymerase (Hunan Innovagene Biotechnology Limited). The PCR reaction system and amplification program for all primers used in the manuscript are shown below: 2 μl DNA/cDNA (200 ng/μl), 1 μl F/R (10 mM), 1 μl i-Max,1 μl dNTPs (10 mM), 25 μl 2 × i-Max Buffer, add double distilled water to 50 μl and then denaturation at 95°C for 5 min, 35 cycles of 95°C for 30 s, 60°C for 30 s, an 72°C for 1 min and 40 s, followed by a final extension at 72°C for 5 min. The Tiangen TIAN gel Midi Purification Kit (Tiangen Co.) was used to purify the PCR products from 1% agarose gel. The PCR products were cloned into the pGEM-T Easy plasmid vector (Promega Corporation, Madison, WI, United States), and the recombinant plasmid was subsequently transformed into Escherichia coli DH5α cells, from which five positive clones were selected and sent to a commercial company (Tsingke Biotechnology Co., Ltd.) for sequencing. All primers used in this study are listed in Supplementary Table 2. The coding sequence of TbMYC4A was stored in National Center for Biotechnology Information9 under accession number MZ686958.
Bioinformatics Analysis
The conserved functional domains were predicted on https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi website. Phylogenetic trees of the amino acid sequences of bHLH transcription factors were constructed by using MEGA X via the adjacency matrix method (Kumar et al., 2018). The pairwise deletion parameters and the p-distance model were used. A bootstrap test of phylogeny was performed with 1,000 replicates.
Transient Expression
Transient expression vectors were constructed using TbMYC4AF2 and TbMYC4AR2 as adapter-specific primers. The expression vector pLGY-02:TbMYC4A contained the whole coding sequence of TbMYC4A with a maize ubiquitin promoter. pBRACT214:TaMYC1 and pBRACT214:TaMYB7D, provided by the Northwest Institute of Plateau Biology, Chinese Academy of Sciences, have been shown to act together to induce the production of large numbers of red cells in the white wheat coleoptile (Zong et al., 2017). Following the steps of the gene gun bombardment technique described by Ahmed et al. (2003), different vector combinations were bombarded into the white coleoptile of Opata wheat with a gene gun, after which they were incubated in light for 16 h after bombardment, and all treated coleoptiles were observed through a stereomicroscope (Leica Co., Oskar Barnack, Germany), photographed and counted.
Quantitative Real-Time PCR Analysis
GAPDH (Glyceraldehyde-3-phosphate dehydrogenase) was used as the reference to determine relative expression values (Scholtz and Visser, 2013), and quantitative real-time PCR (qPCR) was performed using the CFX96 PCR System. cDNA was synthesized from 1 mg of total RNA and diluted 50-fold in DEPC-treated water prior to qPCR analysis. The qPCR amplification was performed using TB Green® Premix Ex TaqTM II (TliRNaseH Plus), 10 μl of reaction mixture including 5 μl 2 × TB Green Premix Ex Taq II(Tli RNaseH Plus), 0.4 μl F/R (10 μM), 1 μl cDNA, and finally sterile double distilled water was added to the total volume. The qRT-PCR reaction procedure was at 95°C for 3 min, 39 cycles at 10 s and 58°C for 30 s. Melt curve was obtained after the last qRT-PCR cycle: 95°C for 5 s followed by a constant increase of 0.5°C in the temperature between 65 and 95°C. The changes in TbMYC4A expression in the seeds of Crocus and Z18-1244 at 14, 22, 28, and 35 dpa were examined. The relative quantitation method (2–ΔΔCT) was used to calculate the fold changes in the expression levels of the target genes (Schefe et al., 2006).
Results
Transcriptome Analyses of Blue (Z18-1244) and White Aleurone (Crocus)
After filtering out low-quality reads, 27,664,008 and 23,288,416 clean reads were found in Crocus and Z18-1244, with GC contents of 53.93 and 56.35% and Q30 percentages of 93.56 and 93.91%, respectively. The alignment efficiency was 85.44% in Crocus and 87.17% in Z18-1244 according to sequence alignment against the reference genome. The transcripts per kilobase of fragment per million fragments were used to estimate quantitative gene expression levels. There were 9820 DEGs between Z18-1244 and Crocus according to the parameters of an FDR < 0.05 and a | log2 (fold change)| ≥ 1 (Supplementary Figure 1).
GO assignments classify annotated DEGs in terms of biological processes, molecular functions, and cellular components. Since elevated anthocyanin contents are generally caused by gene upregulation, upregulated genes were subjected to enrichment analysis. The subcomponents with p-values < 0.05 in each of the categories are depicted in bar graphs (Supplementary Figure 2). In the molecular function category, the greatest enrichment was observed for the activity of various enzymes and transporters, such as glutathione transferases, oxidoreductases, ATPase, and quercetin glycosyltransferase. In the biological process category, the annotated DEGs were enriched in secondary metabolite biosynthetic processes, flavonoid biosynthetic processes, flavonoid glucuronidation and other processes related to plant metabolites. In contrast, only one component was highly significantly enriched in the cellular component category.
The top 20 items in the KEGG enrichment results were selected for mapping in order of their P-values (Supplementary Figure 3). KEGG is mainly enriched in pathways associated with various nutrients, such as biosynthesis of amino acids, starch and sucrose metabolism, fructose and mannose metabolism. To find the key genes that control the blue aleurone layer, 12 structural genes and two transcription factors from the anthocyanin biosynthesis pathway were identified in the KEGG database to perform BLAXT analysis of the DEGs. Anthocyanin modification-related transferases and anthocyanin transport proteins were combined with the above structural genes to build a roadmap of anthocyanin biosynthesis and transport (Figure 1). A total of 41 unigenes were associated with anthocyanin biosynthesis and 29 to transport. No genes homologous to C4H were found. Among these genes, LDOX, F3′5′H, UFGT, AT, GT, GST, and MRP expression was elevated. Since the 4Ab(4B) substitution line and Ba2 from T. boeoticum were mapped to the centromeric region of 4AL, three genes located on chromosome 4 were screened out, including one structural gene (4CL, newGene_9199) and two transcription factor genes (one MYB, newGene_21086, and one bHLH (TbMYC4A), newGene_34829) (Supplementary Table 3). The MYB transcription factor of Z18-1244 was 100% homologous to that of Chinese spring, while the bHLH transcription factor was only 89.93% homologous. The expression level of this bHLH transcription factor (TbMYC4A) was 0 in Crocus, and the log2FoldChange between Crocus and Z18-1244 reached 6.14. Therefore, TaMYC4A was inferred as a candidate key gene for determining the blue aleurone layer trait in the 4Ab(4B) substitution line Z18-1244 and T. boeoticum.
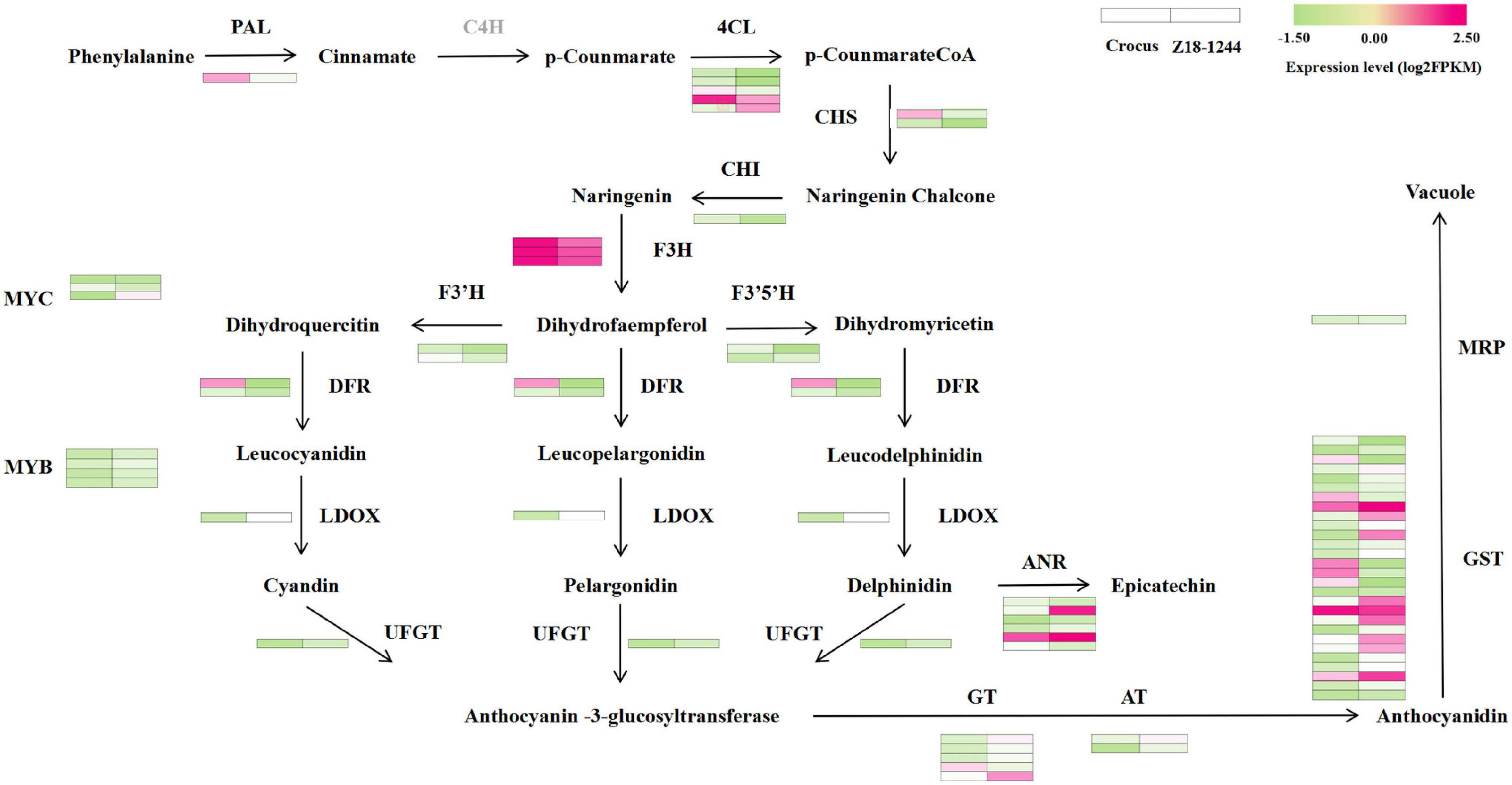
Figure 1. Expression profiles of DEGs [|log2(fold change)| ≥ 1, FDR < 0.05] related to the anthocyanin biosynthesis and transport processes in blue and white aleurone. The color scale from green (low) to purple (high) represents the log2FPKM values measured in white and blue aleurones.
Molecular Characteristics of TbMYC4A
To further understand the evolution of this gene, a phylogenetic tree was constructed using the full-length amino acid sequences of bHLH transcription factors associated with anthocyanin biosynthesis via the adjacency matrix method with a bootstrap test of phylogeny performed with 1,000 replicates (Figure 2). TbMYC4A, encoding a protein of 572 amino acids, belonged to a branch including the bHLH proteins regulating anthocyanin biosynthesis in Th. ponticum and H. vulgare. TbMYC4A did not cluster with sequences of T. urartu, Ae. tauschii, and T. aestivum. In the phylogenetic tree, TbMYC4A was most closely related to ThMYC4E from Th. ponticum and HvMYC2 from H. vulgare, which have been shown to be the key genes controlling the blue aleurone trait in the corresponding species. TbMYC4A also contained three complete functional domains, bHLH-MYC_N, HLH and ACT-like (Figure 3), which are essential for bHLH proteins to exert their transcriptional function.
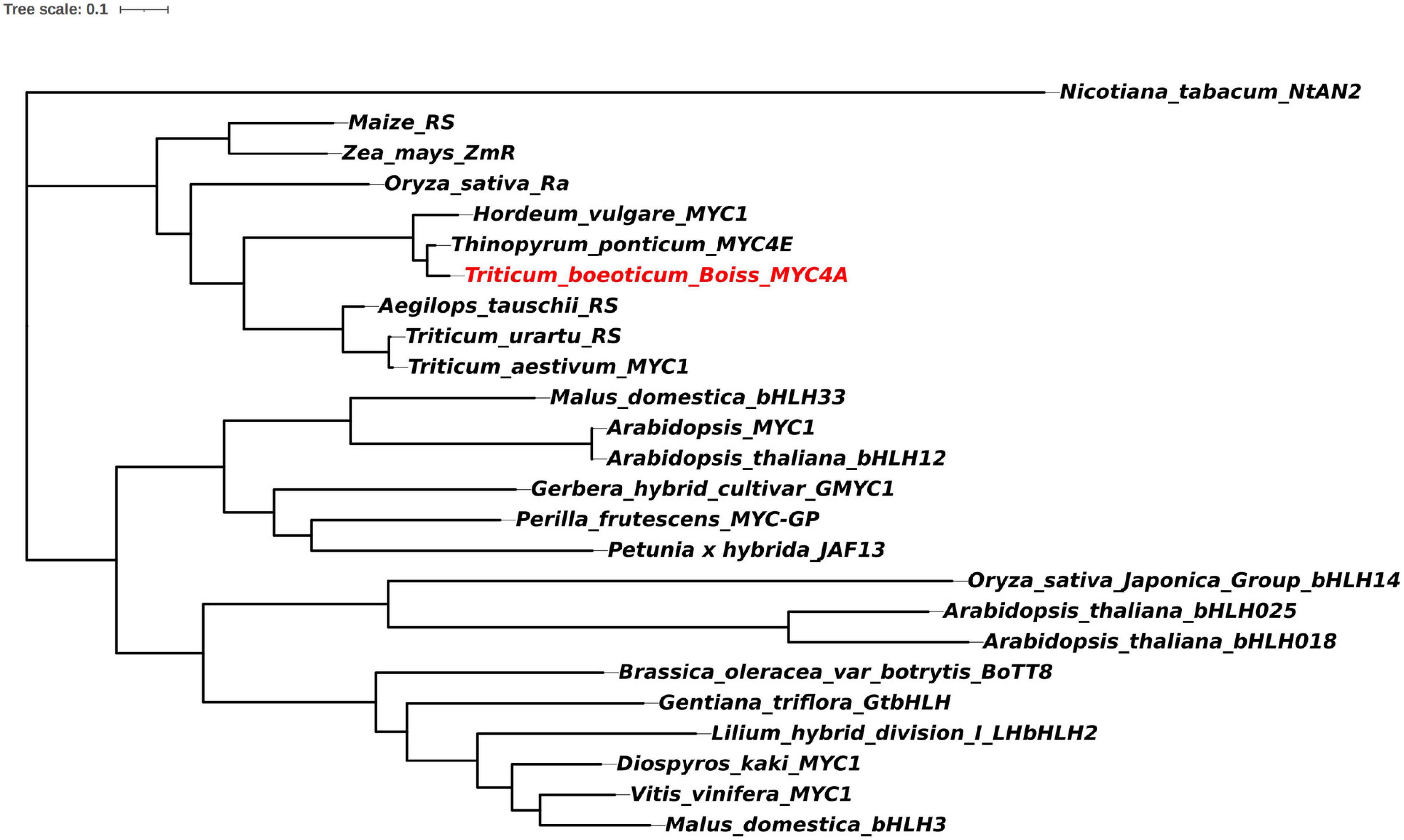
Figure 2. Phylogenetic tree of bHLH proteins regulating anthocyanin biosynthesis. The accession numbers of these proteins (or translated products) in the GenBank database are as follows: Nicotiana tabacum/NtAN2:ACO52472.1; Oryza/OsbHLH14:XP_015649359.1; Arabidopsis thaliana/bHLH025:Q9T072.2; Arabidopsis thaliana/bHLH018:Q1PF17.1; Brassica oleracea var.botrytis/BoTT8: ADP76654.1; Gentiana triflora/GTbHLH1:BAH03387.1; Lilium hybrid division I/LHbHLH2:BAE20058.1; Diospyros kaki/MYC1:AEC03343.1; Vitis vinifera/MYC1: ACC68685.1; Malus domestica/MdbHLH3:ADL36597.1; Malus domestica/MdbHLH33:AAC49219.1; Arabidopsis thaliana/MYC1:NP_191957; Arabidopsis thaliana/bHLH12:AAL55719.1; Gerbera hybrid cultivar/GMYC1: CAA07615.1; Perilla frutescens/MYC-GP:BAA75514.1; Petunia x hybrida/JAF13: AAC39455.1; Oryza/Ra:AAC49219; Maize/RS:NP_001106073; Zea mays/ZmR: NP_001105339.2; Hordeum_vulgare/MYC2:MF679157.1; Thinopyrum ponticum/MYC4E:KX914905.1; Aegilops tauschii/R-S:KD566857.1; Triticum urartu/RS: KD032825.1; and Triticum aestivum/MYC1:KY499900.1.

Figure 3. Comparison of the amino acid sequence of TbMYC4A with those of ThNYC4E, HvMYC2, and bHLH transcription factors known to regulate anthocyanin biosynthesis from Thinopyrum ponticum and Hordeum vulgare, with black lines representing bHLH-MYC_N, HLH and ACT-like domains. The accession numbers of these proteins (or translated product) in the GenBank database are as follows: Thinopyrum ponticum/MYC4E:KX914905.1 and Hordeum vulgare/MYC2:MF679157.1.
A bHLH transcription factor was shown to induce anthocyanin biosynthesis when coexpressed with the MYB transcription factor TaMYB7D (Zong et al., 2017). In this study, TaMYC1 and TbMYC4A induced anthocyanin biosynthesis in the presence of TaMYB7D in “Opata” coleoptile cells; however, TaMYC1, TbMYC4A or TaMYB7D could not induce anthocyanin biosynthesis alone (Figure 4). The average number of cells accumulating anthocyanins was 35 or 50 in the coleoptiles simultaneously transfected with the TaMYC1 and TaMYB7D or TbMYC4A and TaMYB7D expression vectors, respectively. Therefore, TbMYC4A should have a similar function to TaMYC1 in regulating anthocyanin biosynthesis.
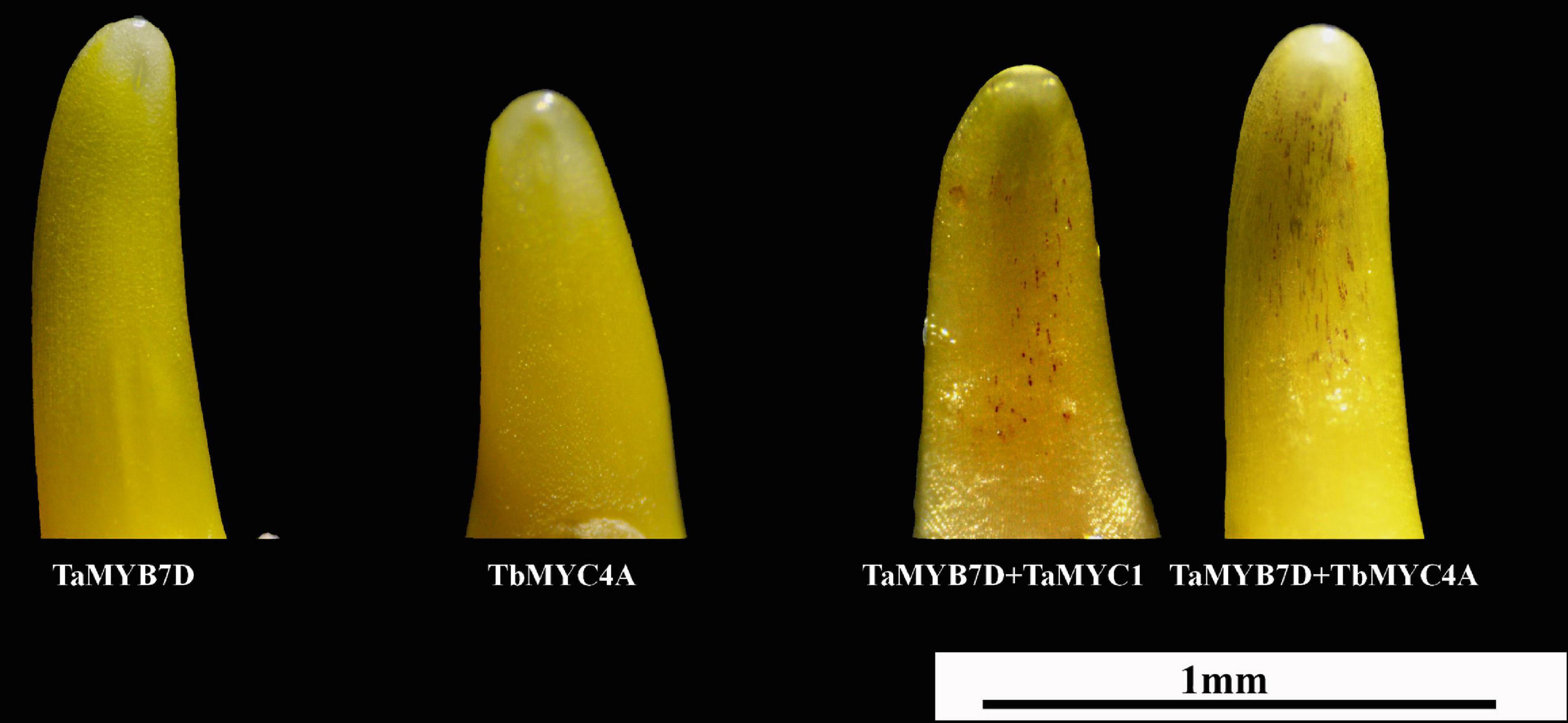
Figure 4. Wheat coleoptiles 16 h after bombardment with different plasmids. TaMYB7D, TbMYC4A, and TaMYC1 represent the constructed vectors pBRACT214:TaMYB7D, pLGY-02: TbMYC4A, and pBRACT214:TaMYC1, respectively.
Specific Marker Development for TbMYC4A and the Distribution of TbMYC4A in Natural Populations
To distinguish TbMYC4A from homologous genes in T. aestivum, the TbMYC4Aa1-F and TbMYC4Aa1-R primers were designed based on the coding sequences. The primers amplified a product of 310 bp from the genomic DNA of the blue-grained lines Z18-1244 and its male parent, T. boeoticum G52, but a 1543 bp product was amplified in white-aleurone Crocus (Figure 5A). Sequencing showed that the 310 bp amplification product of Z18-1244 and G52 was the same as the coding sequence of TbMYC4A, with an intron of 154 bp. It was found that Crocus had 1414 bp intron in the first and second exons, Z18-1244 had 154 bp in length and Ae. tauschii had 250 bp. A total of 24 genotypes, including 2 T. urartu, 4 Ae. tauschii, 4 turgidum, 2 T. araraticum, 2 T. zhukovskyi, 4 T. monococcum, and 6 T. boeoticum genotypes, were checked using the specific primers (Supplementary Table 1). Among the tested genotypes, the TbMYC4A sequence was also detected in other T. boeoticum accessions and its corresponding cultivated species, T. monococcum, which has the blue aleurone trait (Figure 5B). A product of slightly less length than TbMYC4A was detected in Ae. tauschii, which was sequenced to be 293 bp in length and contained a 250 bp intron. Since the homolog of TbMYC4A in wheat is localized on the D genome, no product were amplified in T. urartu (AuAu), and T. turgidum (AABB), but fragments of the same size as Crocus were amplified in T. araraticum (AtAtGG), and T. zhukovskyi (AAAtAtGG), which contains the G genome (Figure 5B).
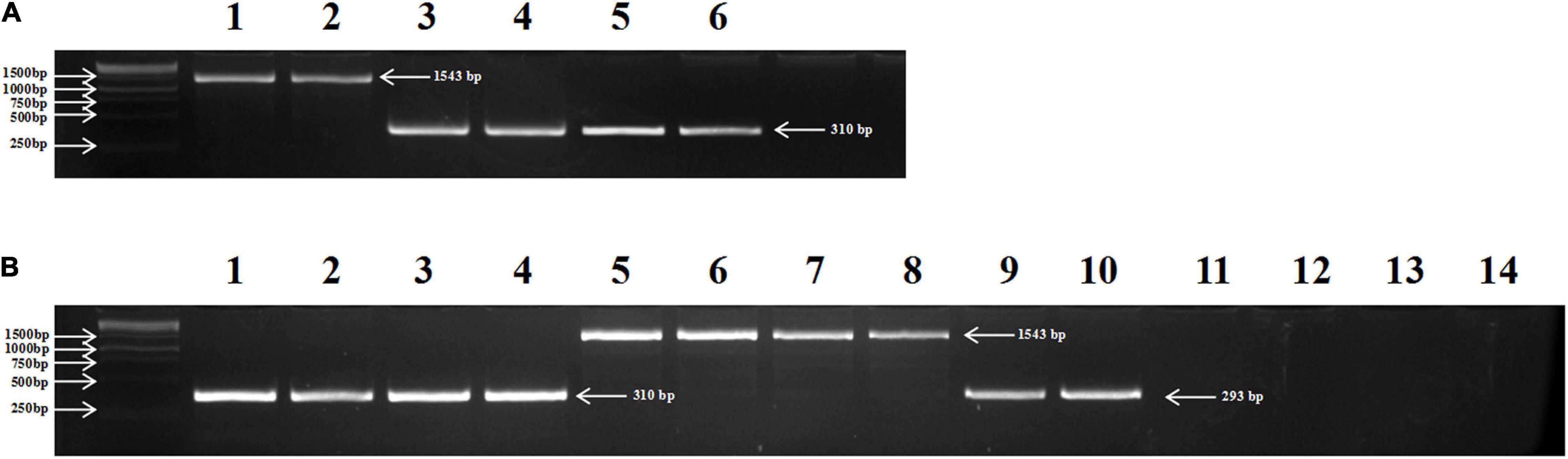
Figure 5. Distribution of TbMYC4A in Z18-1244, Crocus, G52, and 14 examples selected from 7 Triticeae species. (A) lanes 1 and 2: Crocus; lanes 3 and 4: T. boeoticum G52; lanes 5 and 6: Z18-1244; (B) T. monococcum accessions PI 10474T (1) and PI 168805 (2); T. boeoticum accessions PI 401416 (3) and PI 427506 (4); T. timopheevii accessions AS270 (5) and AS272 (6), T. zhukovskyi accessions TRI 7270-1 (7) and TRI 7270-3 (8); Ae. tauschii accessions AS60 (9) and AS62 (10); T. urartu accessions PI428224 (11) and PI428274 (12); and T. turgidum accessions PI525355 (13) and AS2239 (14).
Expression Profile Analysis of TbMYC4A
The expression of TbMYC4A in different developmental stages of seeds was observed by quantitative real-time PCR using immature seeds of Z18-1244 and Crocus at 14, 22, 28, and 35 dpa. The expression of TbMYC4A gradually increased at 14 dpa, 22 dpa, and 28 dpa and decreased considerably at 35 dpa in Z18-1244 (Figure 6). The expression level was 0 in all periods in Crocus, indicating that it does not express TbMYC4A, consistent with the transcriptomic results (Figure 6). The seeds of Crocus showed no color appearance in different developmental periods, while Z18-1244 showed a blue color visible to the naked eye at 22 dpa. Then the color deepened gradually, and the blue paste powder layer covered the whole seeds at 28 dpa. Then, the color did not change significantly, which was consistent with the qPCR results (Supplementary Figure 4).
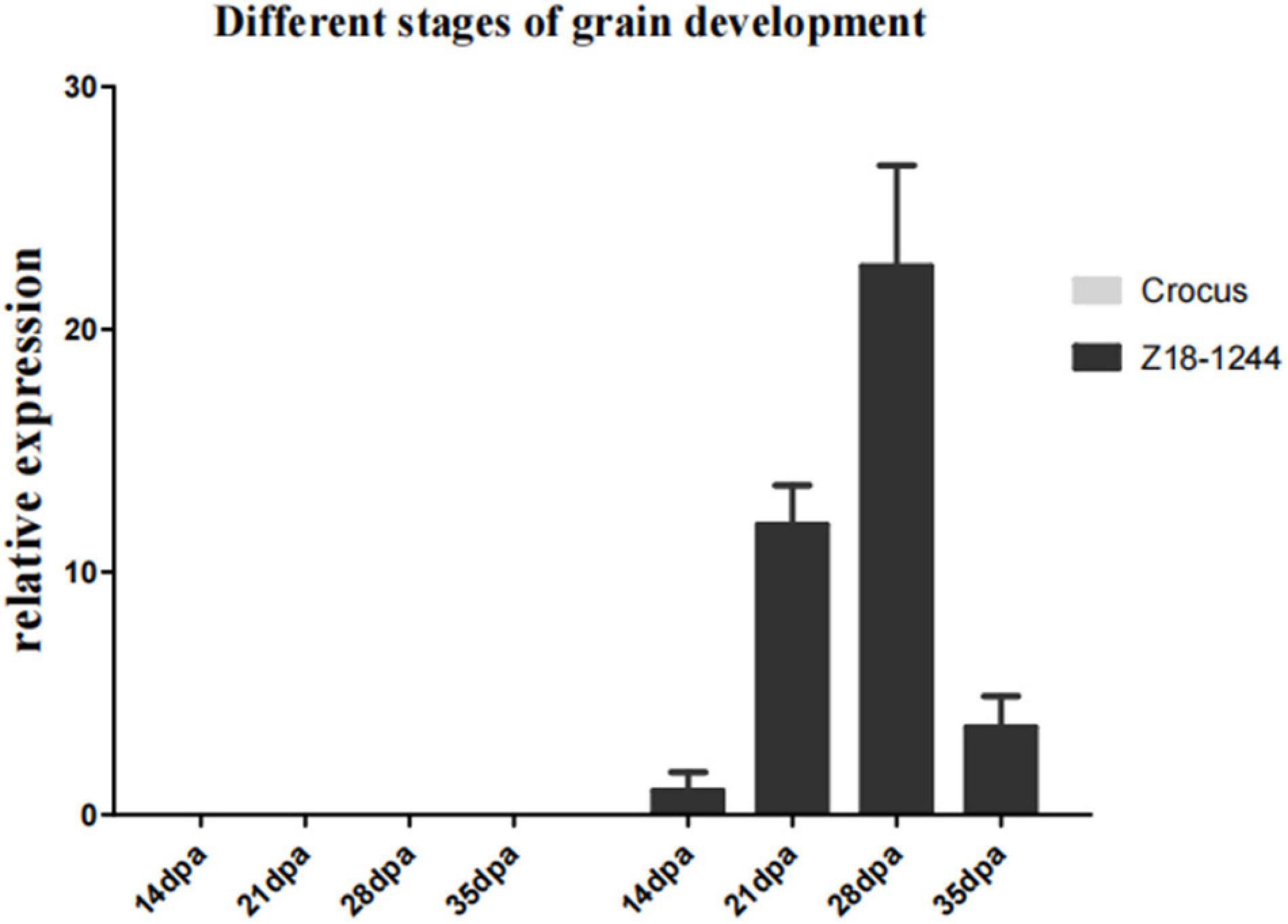
Figure 6. Relative expression of TbMYC4A in the seeds of Z18-1244 and Crocus at different developmental stages.
Discussion
Cyanidin-derived anthocyanins give some rice and maize varieties a red color, while no blue-grained trait has ever been found in rice or maize (Abdel-Aal et al., 2006; Sharma et al., 2011). However, the blue aleurone trait is present in several species of Triticeae, such as Th. ponticum, T. boeoticum, H. vulgare, and Secale species (Zeller et al., 1991; Li et al., 2017; Zong et al., 2019). Through transcriptome analysis, 77 genes related to anthocyanin biosynthesis were screened in an 4E additional line of common wheat-Th. ponticum, of which only eight genes were up-regulated in expression in blue paste-powder layer material, including five bHLH transcription factors and similarly. In addition, 78 genes were identified in blue-grained triticale, most of which were upregulated over the white aleurone layer and also included a MYB and a MYC transcription factor (TsMYC2, a candidate gene for control of wheat blue grain) (Li et al., 2017; Zong et al., 2019). In this study, RNA-Seq was used to identify the gene network controlling the blue aleurone trait of Z18-1244 wheat derived from T. boeoticum. In total, 41 unigenes were found to be associated with anthocyanin biosynthesis in Z18-1244. Similar to previous studies, the upregulated genes mainly included LDOX, F3′5′H, UFGT, AT, GT, most of which are late biosynthetic genes. LDOX, F3′5′H and UFGT are necessary for the synthesis of anthocyanins. Anthocyanins need to be transported to the vesicle for their bright color to be observed, and the most complete version of this process is probably carried out by a combination of GST located in the cytoplasm and MRP located in the vesicle membrane (Winkel-Shirley, 2001; Gu et al., 2019). Increases in anthocyanin contents are inextricably linked to these genes. To date, there have been few studies on the anthocyanin transport related genes. This study identified 29 anthocyanin transport related genes, most of which were up-regulated in blue aleurone layer materials relative to white aleurone layer materials.
bHLH transcription factor genes localized on chromosome 4 are considered the most likely candidates for various blue aleurone genes (Finch and Simpson, 1978; Zeven, 1991; Finch and Zong et al., 2019). This study identified the bHLH transcription factor TbMYC4A from the 4Ab(4B) substitution line. It had the characteristic bHLH-MYC_N, HLH and ACT-like domains. In the evolutionary tree composed of bHLH proteins regulating anthocyanin biosynthesis, TbMYC4A was most closely related to the blue grain candidate genes ThMYC4E in Th. ponticum and HvMYC2 in H. vulgare. The transient expression of TbMYC4A from T. boeoticum confirmed that it was a functional bHLH transcription factor with the ability to regulate anthocyanin biosynthesis. The TbMYC4A-specific marker was present in blue-grained common wheat Z18-1244, T. boeoticum and T. monococcum but absent in T. urartu, Ae. tauschii, T. turgidum, T. araraticum, and T. zhukovskyi. This indicates that the blue aleurone genes of T. boeoticum and its cultivated species, T. monococcum, are the same, consistent with previous studies (Zeller et al., 1991; Singh et al., 2007; Yu et al., 2017). qPCR experiments showed that the expression of TbMYC4A was rapidly elevated from 14 dpa to 28 dpa and decreased considerably at 35 dpa in Z18-1244, consistent with the anthocyanin accumulation pattern. TbMYC4A was not expressed in any stage of seed development in non-blue-grained Crocus wheat. Several studies have shown that amino acids, trace elements and other nutrients are increased in blue-grained wheat relative to common wheat, as is the anthocyanin content (Tian et al., 2018). In this study, GO and KEGG enrichment analyses of Z18-1244 and Crocus revealed that upregulated differentially expressed genes were enriched in various nutrient-related pathways, including flavonoid biosynthesis, amino acid biosynthesis, zinc and iron transport-related pathways, starch and sucrose metabolism, and nitrogen metabolism (Supplementary Figures 1, 2). It has been indicated that blue aleurone presents some advantages over white aleurone in terms of the accumulation of certain nutrients in addition to anthocyanins. This result implies that the anthocyanin pathway may affect other nutrient-related biosynthesis pathways.
Conclusion
In conclusion, a functional bHLH transcription factor, TbMYC4A, was isolated from the wheat-T. boeoticum substitution line Z18-1244, with the blue aleurone trait derived from T. boeoticum. All natural accessions of T. boeoticum and its cultivated species, T. monococcum, with the blue aleurone trait carried this gene. It was indicated that TbMYC4A was the candidate Ba2 gene that controlled the related trait. No blue-grained varieties containing the Ba2 gene have been released to date. These results will help to explore the molecular mechanism of blue aleurone formation in T. boeoticum and will help to breed wheat varieties with blue aleurone.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MZ686958.
Author Contributions
XL: formal analysis, investigation, data curation, writing—original draft, and visualization. MZ: formal analysis and investigation. XJ, HL, and ZJ: investigation. MH, BJ, LH, SN, ZY, XJC, and XC: supervision. DL: supervision, writing—review, and editing. BL: writing—review, and editing, supervision, and methodology. LZ: conceptualization, methodology, writing—review, editing, and project administration. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Key Research and Development Program of Sichuan Province, China (2021YFYZ0002) and the National Natural Science Foundation of China (31671682, 31661143007, and 31671689).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.762265/full#supplementary-material
Abbreviations
PAL, Phenylalanine lyase; 4CL, 4-coumarate–CoA ligase; CHS, Chalcone synthase; CHI, Chalcone isomerase; F3H, Flavanone 3-hydroxylase; F3′H, Flavonoid 3′-hydroxylase; F3′5′H, Flavonoid 3′5′-hydroxylase; DFR, Dihydroflavonol 4-reductase; LDOX, Leucoanthocyanidin dioxygenase; UFGT, Anthocyanidin3-O-glucosyltransferase; MYB, Myeloblastosis transcription factor; MYC, Basic helix-loop-helix transcription factor; ANR, Anthocyanidin reductase; GT, Glucosyltransferase; AT, Acyltransferase; GST, Glutathione transferase; MRP, Multidrug-resistance associated protein; dpa, Days post-anthesis.
Footnotes
- ^ ftp://ftp.ncbi.nih.gov/blast/db/
- ^ https://www.UniProt.org/
- ^ http://www.ncbi.nlm.nih.gov/COG/
- ^ https://www.genome.jp/kegg/
- ^ http://www.geneontology.org/
- ^ ftp://ftp.ncbi.nih.gov/pub/COG/KOG/
- ^ http://pfam.xfam.org/
- ^ http://eggnog5.embl.de/
- ^ http://www.ncbi.nlm.nih.gov/genbank/
References
Abdel-Aal, E. S. M., and Hucl, P. (2003). Composition and stability of anthocyanins in blue-grained wheat. J. Agric. Food Chem. 51, 2174–2180.
Abdel-Aal, E. S. M., Young, J. C., and Rabalski, I. (2006). Anthocyanin composition in black, blue, pink, purple, and red cereal grains. J. Agric. Food Chem. 54, 4696–4704. doi: 10.1021/jf0606609
Ahmed, N., Maekawa, M., Utsugi, S., Himi, E., Ablet, H., Rikiishi, K., et al. (2003). Transient expression of anthocyanin in developing wheat coleoptile by maize C1 and B-peru regulatory genes for anthocyanin synthesis. Breed. Sci. 53, 29–34. doi: 10.1270/jsbbs.53.29
Burešová, V., Kopeckı, D., Bartoš, J., Martinek, P., Watanabe, N., Vyhnánek, T., et al. (2015). Variation in genome composition of blue-aleurone wheat. Theor. Appl. Genet. 128, 273–282. doi: 10.1007/s00122-014-2427-3
Dhua, S., Kumar, K., Kumar, Y., Singh, L., and Sharanagat, V. S. (2021). Composition, characteristics and health promising prospects of black wheat: a review. Trends Food Sci. Technol. 112, 780–794. doi: 10.1016/j.tifs.2021.04.037
Dubcovsky, J., Luo, M. C., Zhong, G. Y., Bransteitter, R., Desai, A., Kilian, A., et al. (1996). Genetic map of diploid wheat, Triticum monococcum L., and its comparison with maps of Hordeum vulgare L. Genetics 143, 983–999.
Finch, R. A., and Simpson, E. (1978). New colours and complementary colour genes in barley. Z. Pflanzenzücht. 81, 40–53.
Ge, S. X., Jung, D., and Yao, R. (2020). ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36, 2628–2629. doi: 10.1093/bioinformatics/btz931
Goodman, C. D., Casati, P., and Walbot, V. (2004). A multidrug resistance–associated protein involved in anthocyanin transport in Zea mays. Plant Cell 16, 1812–1826. doi: 10.1105/tpc.022574
Gu, K. D., Wang, C. K., Hu, D. G., and Hao, Y. J. (2019). How do anthocyanins paint our horticultural products? Sci. Hortic. 249, 257–262. doi: 10.1016/j.scienta.2019.01.034
Hu, B., Zhao, J., Lai, B., Qin, Y., Wang, H., and Hu, G. (2016). LcGST4 is an anthocyanin-related glutathione S-transferase gene in Litchi chinensis Sonn. Plant Cell Rep. 35, 831–843. doi: 10.1007/s00299-015-1924-4
Kähkönen, M. P., and Heinonen, M. (2003). Antioxidant activity of anthocyanins and their aglycons. J. Agric. Food Chem. 51, 628–633. doi: 10.1021/jf025551i
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360.
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Larter, M., Dunbar-Wallis, A., Berardi, A. E., and Smith, S. D. (2019). Evolution of floral pigmentation and regulation of the anthocyanin pathway in Iochrominae. Inter. Comp. Biol. 59:E355.
Li, N., Li, S., Zhang, K., Chen, W., Zhang, B., Wang, D., et al. (2017). ThMYC4E, candidate Blue aleurone 1 gene controlling the associated trait in Triticum aestivum. PLoS One 12:e0181116. doi: 10.1371/journal.pone.0181116
Liu, L., Luo, Q., Li, H., Li, B., Li, Z., and Zheng, Q. (2018). Physical mapping of the blue-grained gene from Thinopyrum ponticum chromosome 4Ag and development of blue-grain-related molecular markers and a FISH probe based on SLAF-seq technology. Theor. Appl. Genet. 131, 2359–2370.
Liu, X., Feng, Z., Liang, D., Zhang, H., Hao, M., Liu, C., et al. (2020). Development, identification, and characterization of blue-grained wheat-Triticum boeoticum substitution lines. J. Appl. Genet. 61, 169–177.
Lloyd, A., Brockman, A., Aguirre, L., Campbell, A., Bean, A., Cantero, A., et al. (2017). Advances in the MYB–bHLH–WD repeat (MBW) pigment regulatory model: addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 58, 1431–1441. doi: 10.1093/pcp/pcx075
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21. doi: 10.1186/s13059-014-0550-8
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T. C., Mendell, J. T., and Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Ravaglia, D., Espley, R. V., Henry-Kirk, R. A., Andreotti, C., Ziosi, V., Hellens, R. P., et al. (2013). Transcriptional regulation of flavonoid biosynthesis in nectarine (Prunus persica) by a set of R2R3 MYB transcription factors. BMC Plant Biol. 13:68. doi: 10.1186/1471-2229-13-68
Sasaki, N., Matsuba, Y., Abe, Y., Okamura, M., Momose, M., Umemoto, N., et al. (2013). Recent advances in understanding the anthocyanin modification steps in carnation flowers. Sci. Hortic. 163, 37–45. doi: 10.1016/j.scienta.2013.07.029
Schefe, J. H., Lehmann, K. E., Buschmann, I. R., Unger, T., and Funke-Kaiser, H. (2006). Quantitative real-time RT-PCR data analysis: current concepts and the novel “gene expression’s CT difference” formula. Int. J. Mol. Med. 84, 901–910. doi: 10.1007/s00109-006-0097-6
Scholtz, J. J., and Visser, B. (2013). Reference gene selection for qPCR gene expression analysis of rust-infected wheat. Physiol. Mol. Plant Pathol. 81, 22–25.
Sharma, M., Cortes-Cruz, M., Ahern, K. R., McMullen, M., Brutnell, T. P., and Chopra, S. (2011). Identification of the Pr1 gene product completes the anthocyanin biosynthesis pathway of maize. Genetics 188, 69–79. doi: 10.1534/genetics.110.126136
Shen, Y., Shen, J., Zhuang, L., Wang, Y., Pu, J., Feng, Y., et al. (2013). Physical localization of a novel blue-grained gene derived from Thinopyrum bessarabicum. Mol. Breed. 31, 195–204. doi: 10.1007/s11032-012-9783-y
Shipp, J., and Abdel-Aal, E. S. M. (2010). Food applications and physiological effects of anthocyanins as functional food ingredients. Open Food Sci. J. 4, 7–22. doi: 10.2174/1874256401004010007
Singh, K., Ghai, M., Garg, M., Chhuneja, P., Kaur, P., Schnurbusch, T., et al. (2007). An integrated molecular linkage map of diploid wheat based on a Triticum boeoticum × T. monococcum RIL population. Theor. Appl. Genet. 115, 301–312. doi: 10.1007/s00122-007-0543-z
Springob, K., Nakajima, J. I., Yamazaki, M., and Saito, K. (2003). Recent advances in the biosynthesis and accumulation of anthocyanins. Nat. Prod. Rep. 20, 288–303. doi: 10.1039/b109542k
Strygina, K. V., Börner, A., and Khlestkina, E. K. (2017). Identification and characterization of regulatory network components for anthocyanin synthesis in barley aleurone. BMC Plant Biol. 17:184. doi: 10.1186/s12870-017-1122-3
Tanaka, Y., Sasaki, N., and Ohmiya, A. (2008). Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 54, 733–749. doi: 10.1111/j.1365-313X.2008.03447.x
Tian, S. Q., Chen, Z. C., and Wei, Y. C. (2018). Measurement of colour-grained wheat nutrient compounds and the application of combination technology in dough. J. Cereal Sci. 83, 63–67. doi: 10.1016/j.jcs.2018.07.018
Trojan, V., Musilová, M., Vyhnánek, T., Klejdus, B., Hanáček, P., and Havel, L. (2014). Chalcone synthase expression and pigments deposition in wheat with purple and blue colored caryopsis. J. Cereal Sci. 59, 48–55. doi: 10.1016/j.jcs.2013.10.008
Wheeler, L. C., and Smith, S. D. (2019). Computational modeling of anthocyanin pathway evolution: biases, hotspots, and trade-offs. Inter. Comp. Biol. 59, 585–598. doi: 10.1093/icb/icz049
Winkel-Shirley, B. (2001). Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126, 485–493. doi: 10.1104/pp.126.2.485
Yu, K., Liu, D., Wu, W., Yang, W., Sun, J., Li, X., et al. (2017). Development of an integrated linkage map of einkorn wheat and its application for QTL mapping and genome sequence anchoring. Theor. Appl. Genet. 130, 53–70. doi: 10.1007/s00122-016-2791-2
Zeller, F. J., Cermeno, M. C., and Miller, T. E. (1991). Cytological analysis on the distribution and origin of the alien chromosome pair conferring blue aleurone color in several European common wheat (Triticum aestivum L.) strains. Theor. Appl. Genet. 81, 551–558. doi: 10.1007/BF00219448
Zeven, A. C. (1991). Wheats with purple and blue grains: a review. Euphytica 56, 243–258. doi: 10.1007/BF00042371
Zheng, Q., Li, B., Mu, S. M., Zhou, H. P., and Li, Z. S. (2006). Physical mapping of the blue-grained gene (s) from Thinopyrum ponticum by GISH and FISH in a set of translocation lines with different seed colors in wheat. Genome 49, 1109–1114. doi: 10.1139/g06-073
Zong, Y., Li, G., Xi, X., Sun, X., Li, S., Cao, D., et al. (2019). A bHLH transcription factor TsMYC2 is associated with the blue grain character in triticale (Triticum× Secale). Plant Cell Rep. 38, 1291–1298. doi: 10.1007/s00299-019-02449-3
Keywords: Triticum boeoticum, anthocyanin biosynthesis, blue aleurone, transcriptome analysis, TbMYC4A
Citation: Liu X, Zhang M, Jiang X, Li H, Jia Z, Hao M, Jiang B, Huang L, Ning S, Yuan Z, Chen X, Chen X, Liu D, Liu B and Zhang L (2021) TbMYC4A Is a Candidate Gene Controlling the Blue Aleurone Trait in a Wheat-Triticum boeoticum Substitution Line. Front. Plant Sci. 12:762265. doi: 10.3389/fpls.2021.762265
Received: 21 August 2021; Accepted: 12 October 2021;
Published: 05 November 2021.
Edited by:
Petr Smıkal, Palackı University, Olomouc, CzechiaReviewed by:
Shuwei Liu, Shandong University (Qingdao), ChinaTomáš Vyhnánek, Mendel University in Brno, Czechia
Copyright © 2021 Liu, Zhang, Jiang, Li, Jia, Hao, Jiang, Huang, Ning, Yuan, Chen, Chen, Liu, Liu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Baolong Liu, YmxsaXVAbndpcGIuY2FzLmNu; Lianquan Zhang, emhhbmdsaWFucXVhbjE5NzdAMTI2LmNvbQ==; orcid.org/0000-0001-9005-8705
 Xin Liu1
Xin Liu1 Ming Hao
Ming Hao Bo Jiang
Bo Jiang Lin Huang
Lin Huang Zhongwei Yuan
Zhongwei Yuan Dengcai Liu
Dengcai Liu Baolong Liu
Baolong Liu Lianquan Zhang
Lianquan Zhang