
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 23 November 2021
Sec. Plant Cell Biology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.761008
This article is part of the Research Topic Plant Epigenetics and Chromatin Dynamics - EPIPLANT 2021-2022 View all 9 articles
Phosphorylation of H2A at serine 95 (H2AS95ph) mediated by MLK4 promotes flowering and H2A.Z deposition. However, little is known about MLK1, MLK2, and MLK3 during the flowering time. Here, we systemically analyze the functions of MLK family in flowering time and development. Mutation in MLK3, but not MLK1 and MLK2, displayed late-flowering phenotype. Loss of MLK3 function enhanced the late-flowering phenotype of mlk4 mutant, but not reinforced the late-flowering phenotype of mlk1 mlk2 double mutants. MLK3 displayed the kinase activity to histone H2AS95ph in vitro. The global H2AS95ph levels were reduced in mlk3 mlk4, but not in mlk3 and mlk4 single mutant and mlk1 mlk2 double mutant, and the H2AS95ph levels in mlk1 mlk3 mlk4 and mlk2 mlk3 mlk4 were similar to those in mlk3 mlk4 double mutant. MLK3 interacted with CCA1, which binds to the promoter of GI. Correspondingly, the transcription levels and H2AS95ph levels of GI were reduced in mlk3 and mlk4 single mutant, and greatly decreased in mlk3 mlk4 double mutant, but not further attenuated in mlk1 mlk3 mlk4 and mlk2 mlk3 mlk4 triple mutant. Together, our results suggested that H2AS95ph deposition mediated by MLK3 and MLK4 is essential for flowering time in Arabidopsis.
The N-terminal tails of the core histones in nucleosomes are subjected to posttranslational modifications such as acetylation, methylation, phosphorylation, ubiquitination, glycosylation, ADP-ribosylation, carbonylation, and sumoylation. The phosphorylation generally occurs at the serine10, serine28, threonine3, and threonine11 at the N-terminus of H3, and these phosphorylation sites are primarily linked to chromosome condensation/segregation, transcription activation, and apoptosis (Wang et al., 2011; Pirrotta, 2015). Phosphorylation of histone H3 at threonine 3 (H3T3ph) by Haspin during mitosis is accompanied by chromatin condensation, and it provides a chromatin binding site for the chromosomal passenger complex at centromeres to regulate chromosome segregation (Kelly et al., 2010; Wang et al., 2011; Pirrotta, 2015). MSK1/2-mediated phosphorylation of H3 serine 10 and serine 28 is coupled with transcription activation through antagonizing polycomb silencing (Soloaga et al., 2003; Lau and Cheung, 2011). Protein-kinase-C-related kinase 1 (PRK1) phosphorylates histone H3 at threonine 11 (H3T11ph) upon ligand-dependent recruitment to androgen receptor target genes (Metzger et al., 2008). In contrast to phosphorylation of H3, H2A phosphorylation is mostly well known in DNA damage. In yeast, H2A phosphorylation at serine 129 is essential for the repair of DNA double-stranded breaks by recruiting the INO80 complex (Van Attikum et al., 2004). In addition to DNA damage, H2A phosphorylation at threonine 133 is associated with centromere function and maintenance during meiosis in maize (Dong and Han, 2012). Phosphorylation of H2A at serine 95 (H2AS95ph) is associated with transcription activation in Arabidopsis (Su et al., 2017).
The histone phosphorylation is linked with histone methylation, histone acetylation, and histone variants. Phosphorylation of H3T11 by PRK1 accelerates demethylation by the Jumonji C (JmjC)-domain-containing protein JMJD2C (Metzger et al., 2008). Phosphorylation of histone H3 at threonine 6 (H3T6ph) by protein kinase C beta I prevents LSD1 from demethylating H3K4 during AR-dependent gene activation (Metzger et al., 2010). In Chlamydomonas, phosphorylation of histone H3 at threonine 3 (H3T3ph) by MUT9p is responsible for repression of transgenes and transposons, and the loss of MUT9p function resulted in a reduction of H3T3ph levels with the induction of H3K4me2 and H3K4me3 (Casas-Mollano et al., 2008). Phosphorylation of H3 serine 10 is tightly linked to acetylation of H3K9 and H3K14 (Winter et al., 2008). In addition to histone methylation and acetylation, phosphorylation of histone H2A at serine 95 (H2AS95ph) promotes SWR1 complex recruitment and H2A.Z enrichment (Su et al., 2017).
Flowering at an appropriate time is crucial for plant life cycles. The flowering time is initially in response to endogenous controls and environmental cues (Boss et al., 2004; Amasino and Michaels, 2010). The two common environmental factors are day length (photoperiod) and temperature (Michaels et al., 2005; Amasino and Michaels, 2010). Plant perceives the seasonal changes in day length to regulate flowering time. Photoperiod is the primary environmental signal regulating flowering time in summer-annual Arabidopsis with flowering rapidly under long days (LD) and slowly under short days (SD). CONSTANS (CO) encodes a B-box zinc-finger protein that activates the expression of FT and acts as a suppressor of the overexpression of CO 1 (SOC1) (Putterill et al., 1995; Samach et al., 2000; Song et al., 2015). CO primarily accelerates flowering under LD; and loss of CO function resulted in late flowering under LD, but not SD (Putterill et al., 1995; Song et al., 2015). CO transcription levels were regulated by flavin-binding, Kelch repeat, F-box 1 (FKF1), and Gigantea (GI) (Sawa et al., 2007). FKF1 and GI form a complex in the late afternoon and promote the CO transcription; thus, the transcript levels of CO reach the peak at the end of a long day (Sawa et al., 2007; Song et al., 2015). Arabidopsis flowering time in photoperiod pathway also requires components involved in circadian rhythms. Circadian clock associated 1 (CCA1) and late elongated hypocotyl (LHY) encode MYB-like transcription factors (Wang and Tobin, 1998; Mizoguchi et al., 2002). CCA1 and LHY formed a central oscillator, and the loss of CCA1 and LHY function displayed dramatically earlier phases of expression of GI (Mizoguchi et al., 2002, 2005).
Casein kinase I, a serine/threonine protein kinase, is conserved from yeast to mammalian cells, and plants (Cruciat, 2014). Casein kinase I (CKI) members play a critical role in Wnt signaling, circadian rhythms, and multiple cellular processes (Lin et al., 2002; Cruciat, 2014; Su et al., 2017). In Arabidopsis, CKI is involved in root development, ethylene synthesis, flowering time, and circadian clock (Liu et al., 2003; Ogiso et al., 2010; Tan and Xue, 2014; Uehara et al., 2019). Chlamydomonas MUT9p is closely related to casein kinase I and possesses kinase activity to histone H2A and H3 (Casas-Mollano et al., 2008). Mapping of phosphorylation sites show that MUT9p prefers to phosphorylate histone H3 at threonine 3 (Casas-Mollano et al., 2008). Arabidopsis genome encodes four MUT9p-like kinase/photoregulatory protein kinases/Arabidopsis EL1-like kinases, herein referred to as the MLKs/AELs, namely, MLK1, MLK2, MLK3, and MLK4 (Casas-Mollano et al., 2008; Huang et al., 2016). MLK1 and MLK2 are involved in osmotic stress and hypocotyl elongation (Wang et al., 2015; Zheng and Ding, 2018; Zheng et al., 2018). MLK4 was firstly identified from the evening clock complex (Huang et al., 2016). The following study showed that MLK4 phosphorylated H2AS95ph in vitro and in vivo (Su et al., 2017). MLK4 interacts with CCA1, and loss of MLK4 functions resulted in late flowering and reduction of H2AS95ph and H2A.Z level at GI (Su et al., 2017). A recent study showed that MLK4 deposited H3T3ph at FLC/MAF (Wang et al., 2021). Phosphoproteomics showed that MLKs phosphorylated proteins in circadian clock, multiple metabolic, and signaling pathways in Arabidopsis (Wilson et al., 2021). MLKs/AELs are also involved in abscisic acid signaling pathway (Chen et al., 2018), suggesting that the functions of MLKs are complicated and need further study.
The amino acid of H2A is not conserved as that of H3 in different species (Kawashima et al., 2015). The sequence diversity of H2A determines the modification sites complexity from yeast to mammalian cells, and plants. Compared with the phosphorylation of H3, less is known about phosphorylation at H2A. Most importantly, the functions and phosphorylation sites of MLKs are elusive. Here, we comprehensively revealed the phosphorylation activity and flowering time of MLKs. Loss of MLK3 function exhibited late-flowering phenotype under LD and reinforced the late-flowering phenotype of mlk4. Mutations of MLK1 or MLK2 in mlk3 mlk4 double mutant do not enhance the late-flowering phonotype of mlk3 mlk4 plants. MLK3 phosphorylated histone H2AS95ph in vitro and in vivo. The global levels of H2AS95ph were reduced in mlk3 mlk4 double mutant, but were not further reduced in mlk1 mlk3 mlk4 or mlk2 mlk3 mlk4 triple mutants. In addition, MLK3 interacts with CCA1 and binds to the GI promoter. The transcription levels and H2AS95ph levels at GI were reduced in mlk3 mlk4 double mutants, and in mlk1 mlk3 mlk4 and mlk2 mlk3 mlk4 triple mutants. Together, our results showed that MLKs prefer to phosphorylate H2AS95ph in vitro and in vivo, and MLK3 and MLK4 are the major components in flowering time and histone 2A phosphorylation at serine 95.
Phylogenetic analysis showed MLK3 is closely related to MLK1, MLK2, and MLK4 (Supplementary Figure 1). To investigate the function of MLK3, we isolated the two T-DNA insertion mutants. Genotyping showed that the mlk3-1 and mlk3-2 alleles, each contain a T-DNA in the exon10 and exon12, respectively (Figures 1A,B). RT-PCR analyses revealed that mlk3-1 and mlk3-2 are null alleles (Figure 1C). The primary leaf number at bolting of mlk3-1 and mlk3-2 is more than that of the wild type (Supplementary Table 1). mlk3-1 and mlk3-2 exhibited late flowering-time phenotype under long day (LD) photoperiod, but not in short day (SD) photoperiod (Figures 1D,E and Supplementary Table 1), suggesting that MLK3 is involved in photoperiod pathway.
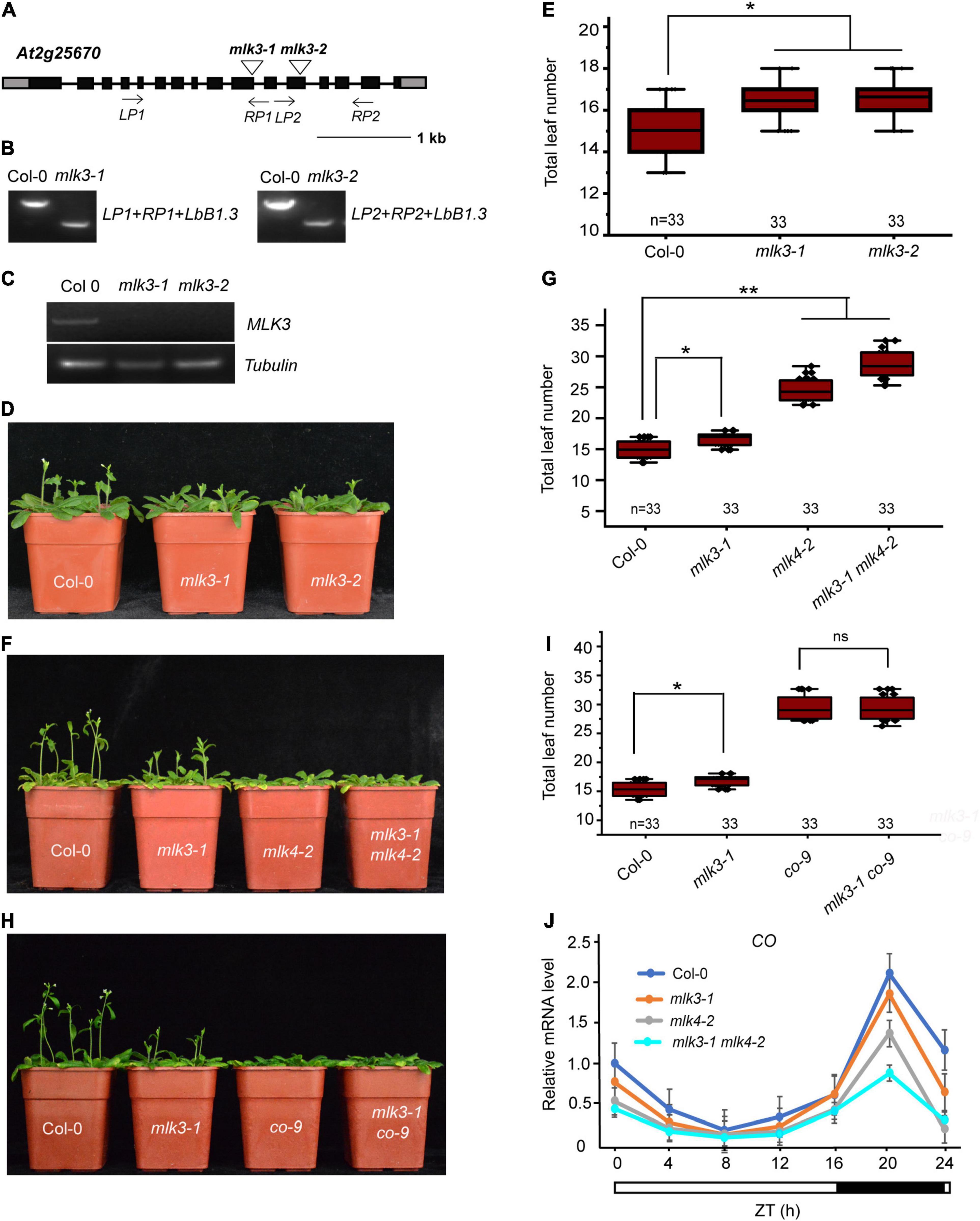
Figure 1. MLK3 is involved in photoperiod flowering time pathway. (A) Gene structure of MLK3, indicating exons (boxes), introns (lines), and T-DNA insertions (triangles). The primers used for genotype analysis are marked with arrows. (B) Genotypic analysis of the mlk3 mutants. The genotype was analyzed with the left genomic primer (LP), right genomic primer (RP), and the vector primer (LBb1.3). The positions of LP1, RP1, LP2, and RP2 are indicated in panel (A). (C) The full-length MLK3 transcript in the mlk3 mutants was examined by RT-PCR. (D) The representative phenotype of Col-0 and mlk3 mutants under a LD photoperiod. (E) The total leaf number of Col-0 and mlk3 mutants under a LD photoperiod. Flowering time was assessed by counting the number of rosette leaves and cauline leaves at bolting under LD. Values shown are mean ± SD of total leaves; 33 plants were scored for each line. Asterisks indicate P < 0.05 by t-test, ns indicates no significance. (F) The representative phenotype of Col-0, mlk3-1, mlk4-2, and mlk3-1 mlk4-2 plants under a LD photoperiod. (G) The total leaf number of Col-0, mlk3-1, mlk4-2, and mlk3-1 mlk4-2 mutants under a LD photoperiod. Values shown are mean ± SD of total leaves; 33 plants were scored for each line. Asterisks indicate P < 0.05 by t-test, ns indicates no significance. (H) The representative phenotype of Col-0, mlk3-1, co-9, and mlk3-1 co-9 plants under a LD photoperiod. (I) The total leaf number of Col-0, mlk3-1, co-9, and mlk3-1 co-9 mutants under a LD photoperiod. Values shown are mean ± SD of total leaves; 33 plants were scored for each line. Asterisks indicate P < 0.05 and ns indicates no significance by t-test. (J) The transcript levels of CO were examined in Col-0, mlk3-1, mlk4-2, and mlk3-1 mlk4-2 mutants. The white bar indicates the light periods, and the black bar indicates the dark period. ZT, Zeitgeber time. Experiments were repeated at least three times, and the representative experiments shown indicate the mean ± SE, n = 3 replicates.
To investigate the redundant functions of MLK3 and MLK4, we crossed mlk3-1 into mlk4-2 and generated the mlk3-1 mlk4-2 double mutant. The flowering time of mlk3-1 mlk4-2 double mutant is later than those of mlk3-1 and mlk4-2 in LD, but not in SD (Figures 1F,G, and Supplementary Table 1). mlk3-2 mlk4-3 double mutant further supported that the functions of MLK3 and MLK4 in flowering time (Supplementary Table 1). These results suggested that MLK3 and MLK4 have unequal redundancy in flowering time.
To confirm this, we crossed the mlk3-1 into co-9 and generated mlk3-1 co-9 double mutant. The flowering time of mlk3-1 co-9 is similar to that of co-9 (Figures 1H,I and Supplementary Table 1), suggesting that MLK3 is involved in CO-dependent flowering pathway. The transcripts of CO were examined, and the transcription levels of CO were reduced in mlk3 mutants, and further reduced in mlk3 mlk4 mutants (Figure 1J and Supplementary Figure 2A), suggesting that CO is downstream of MLK3 and MLK4. We then investigated transcripts of FT and found that the transcript levels of FT were reduced in mlk3, mlk4, and mlk3 mlk4 plants, which are positively correlated with transcript levels of CO (Supplementary Figure 2B). We then investigated the transcripts of MLK3 and found that MLK3 exhibits a circadian rhythm dependent expression pattern with a peak at the end of the 16-h light period, which is similar to the expression patterns of CO and FT (Supplementary Figures 2A–C). However, the transcript levels of FLC did not show an obvious difference in mlk3, mlk4 single mutants, and mlk3 mlk4 double mutants (Supplementary Figure 2D).
Mutation in MLK1 or MLK2 did not show any flowering-time phenotype, and mlk1 mlk2 double mutant displayed the late flowering under LD, but not SD (Zheng et al., 2018; Supplementary Figures 3A,B and Supplementary Table 1). We examined whether MLK1/2 is involved in the photoperiod pathway. We crossed mlk1-3 mlk2-3 into co-9 and found that the flowering time of mlk1-3 mlk2-3 co-9 triple mutant is similar to that of co-9 (Figures 2A,B, Supplementary Figure 3C, and Supplementary Table 1), suggesting that MLK/2 is involved in the photoperiod pathway. These results were confirmed with reduced CO transcription level in mlk1-3 mlk2-3 double mutant (Supplementary Figure 3D).
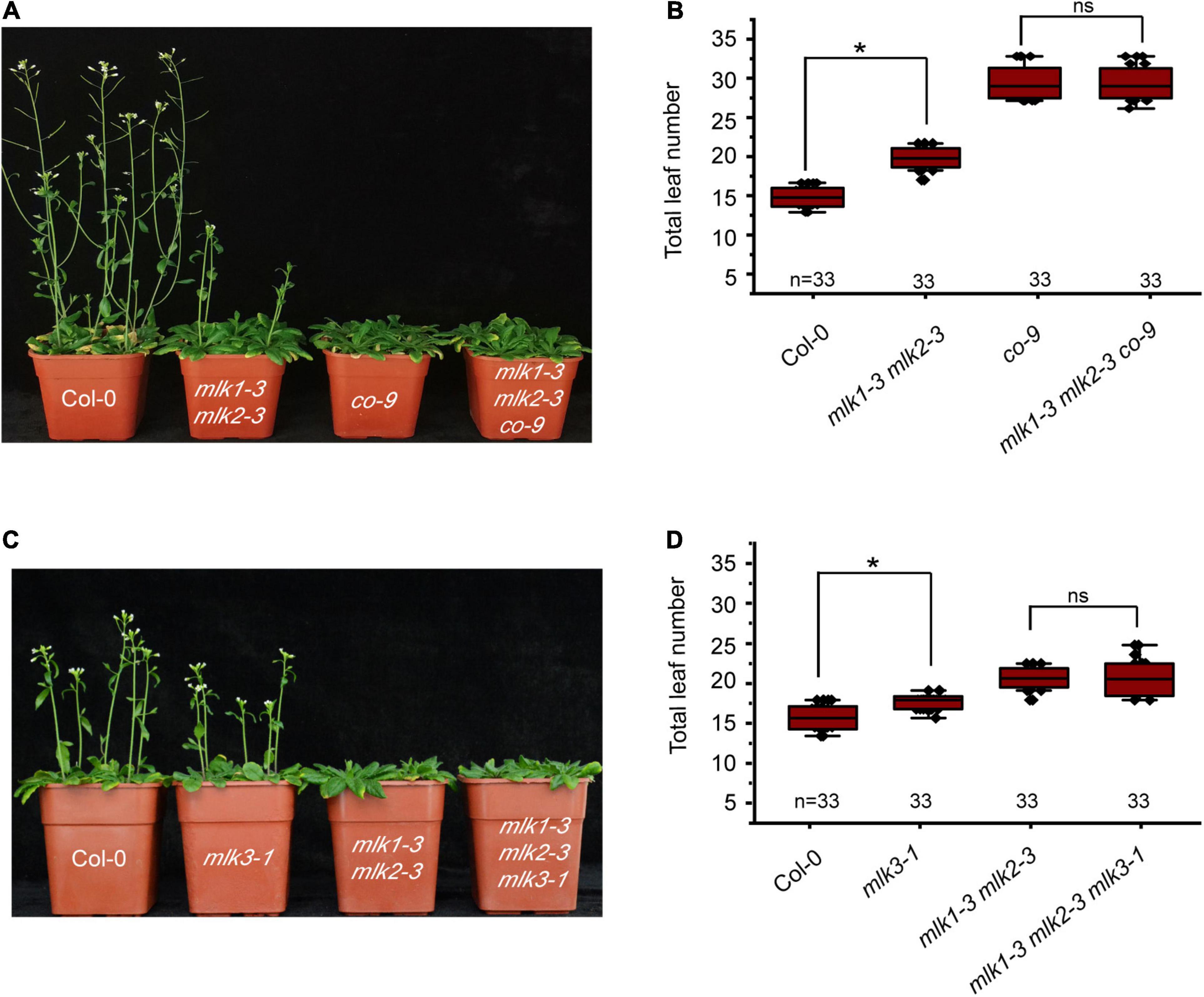
Figure 2. Loss of MLK1 and MLK2 function resulted in late-flowering phenotype. (A) The representative phenotype of Col-0, mlk1-3 mlk2-3, co-9, and mlk1-3 mlk2-3 co-9 plants under a LD photoperiod. (B) The total leaf number of Col-0, mlk1-3 mlk2-3, co-9, and mlk1-3 mlk2-3 co-9 mutants under a LD photoperiod. Values shown are mean ± SD of total leaves; 33 plants were scored for each line. Asterisks indicate P < 0.05 by t-test, ns indicates no significance. (C) The representative phenotype of Col-0, mlk1-3 mlk2-3, mlk3-1, and mlk1-3 mlk2-3 mlk3-1 plants under a LD photoperiod. (D) The total leaf number of Col-0, mlk1 mlk2, mlk3, and mlk1 mlk2 mlk3 mutants under a LD photoperiod. Values shown are mean ± SD of total leaves; 33 plants were scored for each line. Asterisks indicate P < 0.05 and ns indicates no significance by t-test.
To investigate the relationship between MLK3 with MLK1 and MLK2, we crossed mlk3-1 into mlk1-3 mlk2-3 double mutant and generated mlk1-3 mlk2-3 mlk3-1 triple mutant. The flowering time of mlk1-3 mlk2-3 mlk3-1 triple mutant is similar to that in mlk1-3 mlk2-3 double mutant in LD and SD (Figures 2C,D and Supplementary Table 1). The transcript levels support these results that the transcription levels of CO in mlk1 mlk2 double mutant are similar to those in mlk1 mlk2 mlk3 triple mutant (Supplementary Figure 3D).
We next individually examined the MLK1 and MLK2 in contribution to flowering time in mlk3 mlk4 double mutant. We introduced mlk1-3 and mlk2-3 into mlk3-1 mlk4-2, and generated mlk1-3 mlk3-1 mlk4-2 and mlk2-3 mlk3-1 mlk4-2 triple mutant, respectively. The flowering time of mlk1-3 mlk3-1 mlk4-2 and mlk2-3 mlk3-1 mlk4-2 is similar to that of mlk3-1 mlk4-2 under LD (Figures 3A–D and Supplementary Table 1). These observations suggested that mutations of MLK1 or MLK2 in mlk3 mlk4 double mutant failed to reinforce the late-flowering phenotype of mlk3 mlk4 under LD. Collecting the flowering-time of mlk3, mlk4, mlk3 mlk4, mlk1 mlk3 mlk4, and mlk2 mlk3 mlk4 mutants (Figures 1F,G, 2C,D, 3A–D), we concluded that MLK4 played critical roles in flowering time in MLK family.
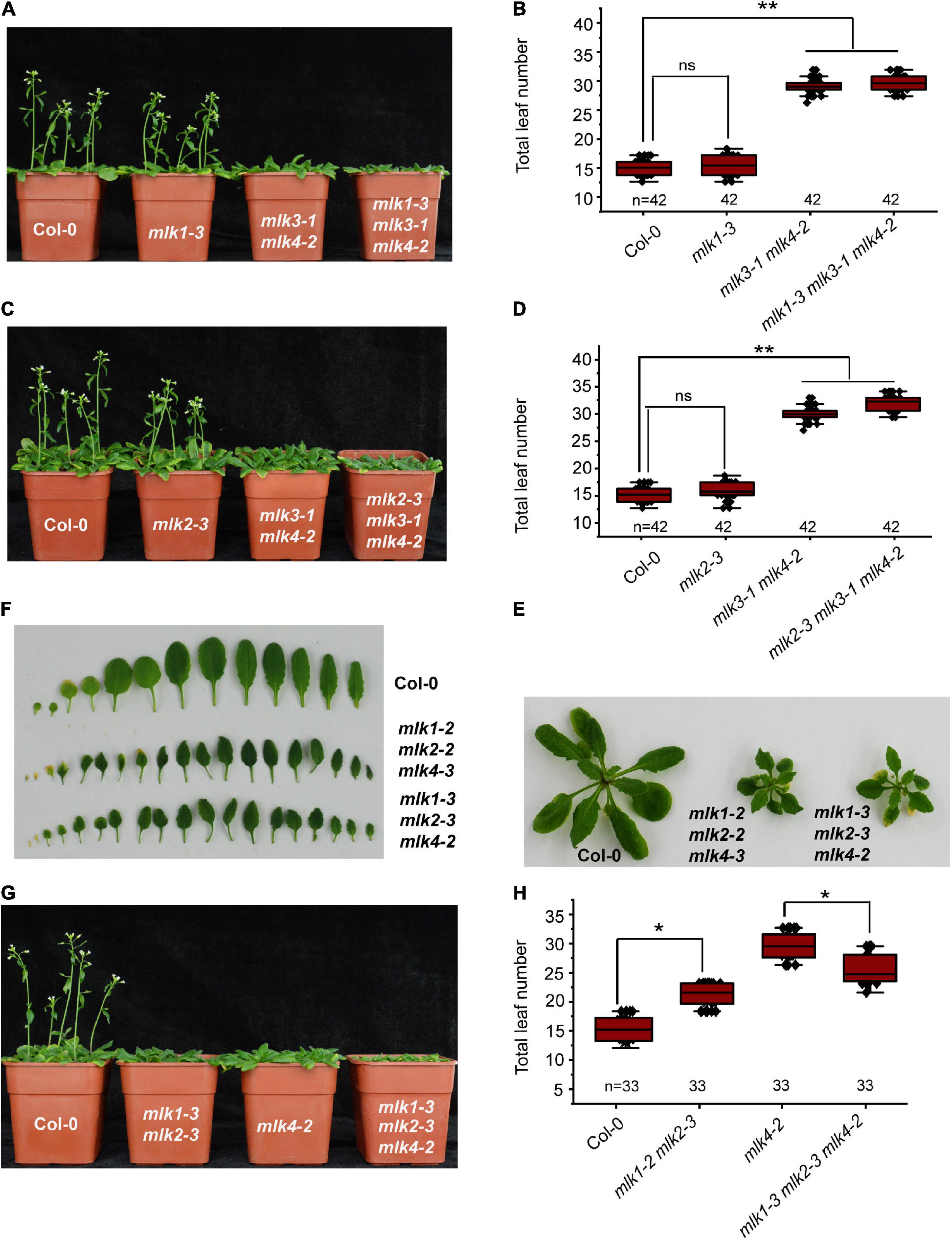
Figure 3. Loss of MLK1, MLK2, and MLK4 function resulted in growth defects. (A) The representative phenotype of Col-0, mlk1-3, mlk3-1 mlk4-2, and mlk1-3 mlk3-1 mlk4-2 plants under a LD photoperiod. (B) The total leaf number of Col-0, mlk1-3, mlk3-1 mlk4-2, and mlk1-3 mlk3-1 mlk4-2 mutants under a LD photoperiod. Values shown are mean ± SD of total leaves; 33 plants were scored for each line. Asterisks indicate P < 0.01 and ns indicates no significance by t-test. (C) The representative phenotype of Col-0, mlk2-3, mlk3-1 mlk4-2, and mlk2-3 mlk3-1 mlk4-2 plants under a LD photoperiod. (D) The total leaf number of Col-0, mlk2-3, mlk3-1 mlk4-2, and mlk2-3 mlk3-1 mlk4-2 mutants under a LD photoperiod. Values shown are mean ± SD of total leaves; 33 plants were scored for each line. Asterisks indicate P < 0.01 and ns indicates no significance by t-test. (E) The 3-week old mlk1-3 mlk2-3 mlk4-2 triple mutants exhibit the dwarf phenotype. (F) The representative leaves from 3-week old mlk1-3 mlk2-3 mlk4-2 plants are shown. (G) The representative phenotype of Col-0, mlk1-3 mlk2-3, mlk4-2, and mlk1-3 mlk2-3 mlk4-2 plants under a LD photoperiod. (H) The total leaf number of Col-0, mlk1-3 mlk2-3, mlk4-2, and mlk1-3 mlk2-3 mlk4-2 mutants under a LD photoperiod. Values shown are mean ± SD of total leaves; 33 plants were scored for each line. Asterisks indicate P < 0.05 by t-test.
The functions of MLK1/2 and MLK4 were then investigated. The mlk1-3 mlk2-3 displayed semidwarf phenotype, whereas mlk1-3 mlk2-3 mlk4-2 exhibited severe dwarf phenotype with short petiole, sharp, and curly leaf (Figures 3E,F and Supplementary Figure 3E); suggesting that MLK4 is involved in plant development with MLK1/2. The flowering-time of mlk1-3 mlk2-3 mlk4-2 is early than that of mlk4-2 under LD (Figures 3G,H). One possibility is due to strong growth defects in mlk1-3 mlk2-3 mlk4-2 (Figures 3E,F and Supplementary Figure 3E).
We investigated the MLK3 phosphorylation activity and found that MLK3 exhibited high activity to histone H2A (Figure 4A). MLK3 prefer to phosphorylate the serine of H2A and the six serines of H2A are located at amino acid positions 9, 17, 19, 20, 95, and 124. The H2A histones containing serine-to-alanine substitutions at 9, 17, 19, and 20 were strongly phosphorylated, but this phosphorylation activity was impaired when serine 95 and serine 124 were substituted with alanine (Figures 4B,C). Further study showed that serine 95 substituted by alanine primarily abolished the MLK3 activity (Figure 4C). Similar results were observed in MLK1/2 phosphorylation activity assays (Supplementary Figures 4A–F).
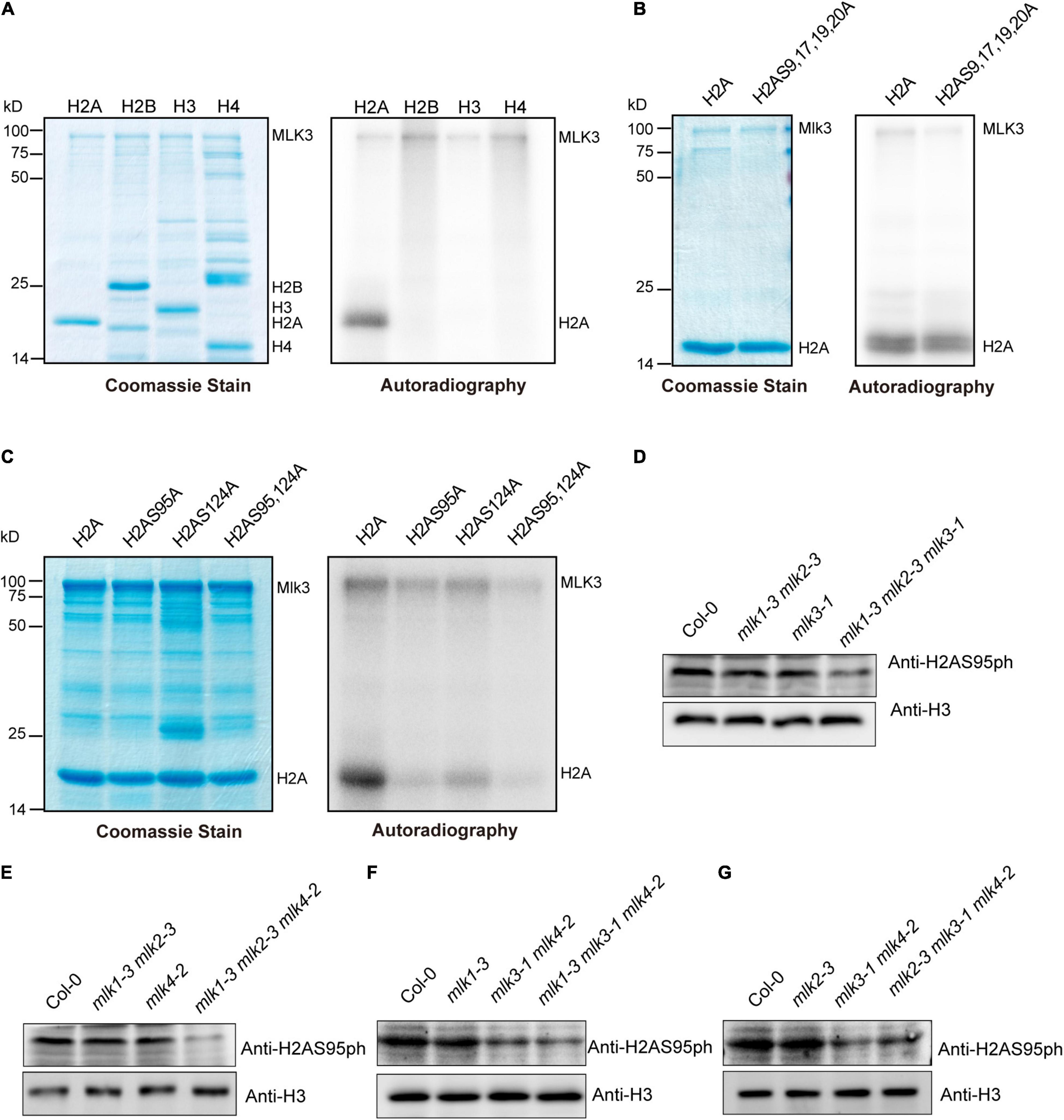
Figure 4. The global changes of H2AS95ph in mlks plants. (A) MLK3 phosphorylated H2A. The ability of MLK3 to phosphorylate H2A, H2B, H3, and H4 was assessed. The MLK3, H2A, H2B, H3, and H4 were marked. (B,C) The activity and specificity of the MLK3 kinase were assessed using different substrates. Wild-type H2A and H2A containing serine-to-alanine substitutions were used as various residues. (D–G) The global levels of H2AS95 phosphorylation were examined in mlk1, mlk2, mlk3, and mlk4 single mutants, mlk1 mlk2 and mlk3 mlk4 double mutants, and mlk1 mlk2 mlk3, mlk1 mlk2 mlk4, mlk1 mlk3 mlk4, and mlk2 mlk3 mlk4 triple mutants. H3 was used as internal control.
The global levels of H2AS95ph were examined and no obvious changes were observed in mlk1, mlk2, mlk3, and mlk4 single mutants and mlk1 mlk2 double mutants (Figures 4D,E). However, mutations in MLK3 or MLK4 in mlk1 mlk2 double mutant reduced global H2AS95ph levels (Figures 4D,E), suggesting that MLK3 and MLK4 are crucial in H2AS95ph deposition. These results confirmed that reduction of H2AS95ph levels in mlk3 mlk4 double mutants was observed (Figures 4F,G). In addition, H2AS95ph levels in mlk1 mlk3 mlk4 and mlk2 mlk3 mlk4 triple mutants are similar to those of mlk3 mlk4 double mutants (Figures 4F,G). These observations indicated that the downregulated H2AS95ph levels were correlated with late-flowering time phenotype in mlk3 mlk4, mlk1 mlk3 mlk4, and mlk2 mlk3 mlk4 plants. In addition to flowering time, MLK1 and MLK2 were also involved in plant development and osmotic stress resistance (Wang et al., 2015; Zheng et al., 2018), suggesting that MLK1 and MLK2 might function independently with MLK3 and MLK4.
Since MLK1, MLK2, and MLK4 also functioned as kinases to histone H3 at threonine 3 (Wang et al., 2015, 2021), we examined the H3T3ph levels. No obvious difference of H3T3ph levels were observed in wild type, mlk1 mlk2 mlk3, mlk1 mlk2 mlk4, mlk1 mlk3 mlk4, and mlk2 mlk3 mlk4 triple mutants (Supplementary Figure 5A). These results suggested that MLKs are predominately involved in H2AS95 phosphorylation, but not H3T3 phosphorylation.
Since MLK3 is involved in photoperiod pathway, we tested whether MLK3 interacts with any of the factors in the circadian rhythm pathway. A yeast two-hybrid result showed that MLK3 directly interacts with CCA1, but not with the controls (Figure 5A). These results were confirmed with pull-down and BiFC. Beads containing His fused with CCA1 (His-CCA1), but not His, bound to soluble GST fused with MLK3 (GST-MLK3) (Figure 5B). Reciprocally, beads containing GST-MLK3 were bound to soluble His-CCA1 (Figure 5C). These results were validated by BiFC, and reconstituted YFP fluorescence was observed in the nucleus with coexpressing MLK3 fused to the YFP N-terminus (MLK3-YFPN) and CCA1 fused to the YFP C-terminus (CCA1-YFPC) (Figure 5D). The co-immunoprecipitation (Co-IP) supported our finding that MLK3 directly binds to CCA1. HA fused with MLK3 (HA-MLK3) and FLAG fused with CCA1 (FLAG-CCA1) were coexpressed in Arabidopsis protoplasts and immunoprecipitated with anti-FLAG antibody. MLK3, but not control, could be coimmunoprecipitated with CCA1 (Figure 5E).
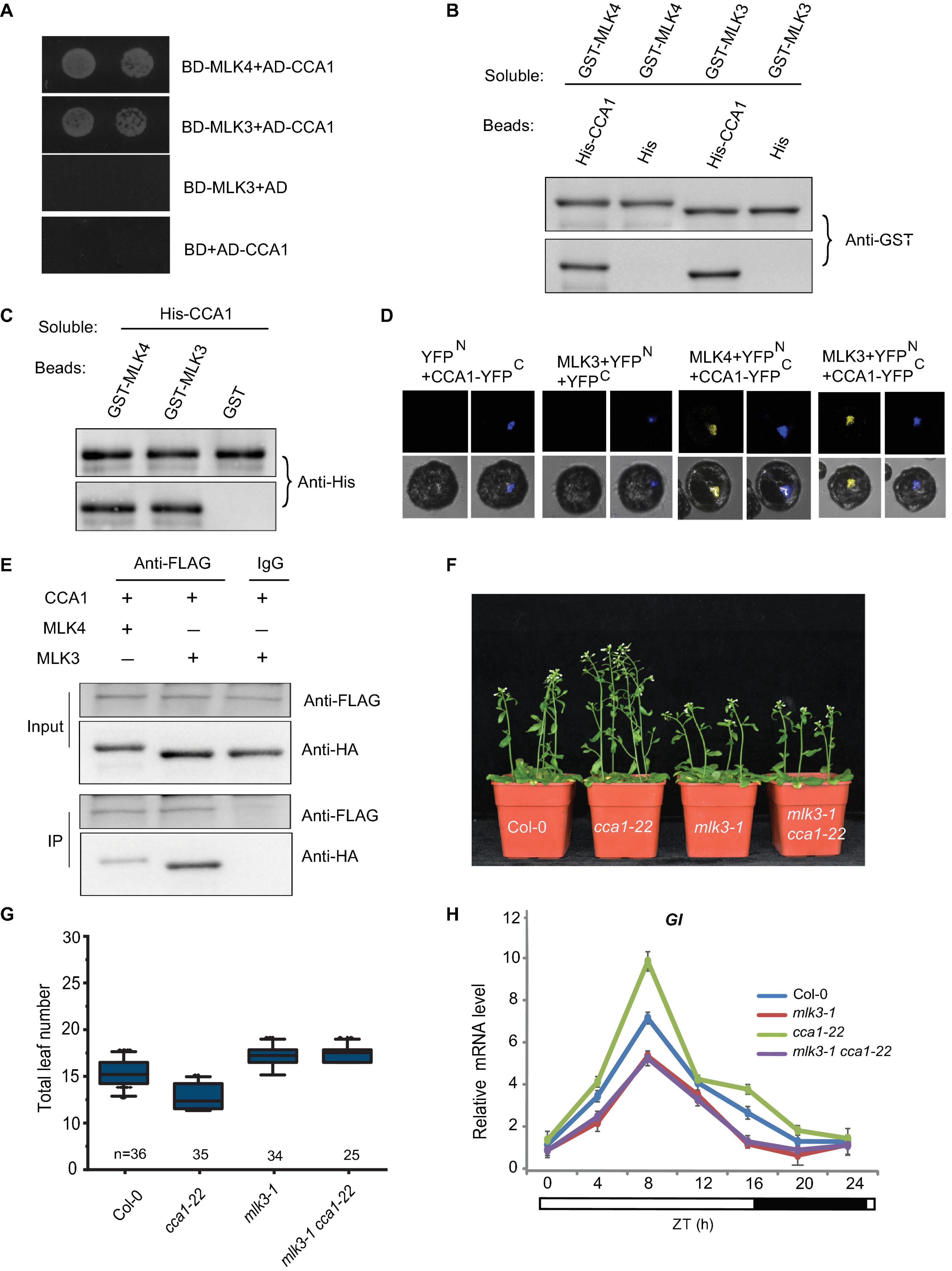
Figure 5. MLK3 interacts with CCA1 in vitro and in vivo. (A) A yeast two-hybrid assay revealed an interaction between MLK3 and CCA1. The growth of two dilutions (2 × 10–2 and 2 × 10–3) of the yeast culture on SD medium lacking Trp, Leu, His, and adenine is shown. (B) Beads containing a His-fused CCA1 were assayed for their ability to bind to soluble GST-fused MLK3. The input and bound proteins were detected with an antibody to GST (anti-GST). MLK4 was used as positive control, and His alone was used as negative control. (C) Beads containing a GST-fused MLK3 were assessed for their ability to bind to soluble His-fused CCA1 and detected with antibody to His (anti-His). (D) Either MLK3 fused to the N terminus of YFP or the N terminus of YFP alone was tested for their ability to bind to the C terminus of YFP fused to C terminus of YFP fused to CCA1. Yellow fluorescence and a bright-field image were recorded and the resulting images were merged. Twenty-five cells were examined for each transformation. Bar = 10 μm. (E) Co-immunoprecipitation of MLK3 and CCA1. FLAG-CCA1 and HA-MLK3 were cotransformed into Arabidopsis protoplasts, and the expressed proteins were immunoprecipitated using an anti-FLAG antibody, and detected with anti-Flag and anti-HA. In panels (A–E), experiments were repeated at least three times, and representative experiments are shown. (F) The representative phenotype of Col-0, mlk3-1, cca1-22, and mlk3-1cca1-22 plants under a LD photoperiod. (G) The total leaf number of Col-0, mlk3-1, cca1-22, and mlk3-1 cca1-22 plants under a LD photoperiod. Values shown are mean ± SD of total leaves; At least 25 plants were scored for each line. Asterisks indicate P < 0.05 and ns indicates no significance by t-test. (H) The transcript levels of GI were examined in Col-0, mlk3-1, cca1-22, and mlk3-1 cca1-22 mutants. The white bar indicates the light periods, and the black bar indicates the dark period. ZT, Zeitgeber time. Experiments were repeated at least three times, and the representative experiments shown indicate the mean ± SE, n = 3 replicates.
The mlk3 cca1 double mutant was then generated by crossing mlk3-1 into cca1-22. Mutations in MLK3 reversed the early-flowering time of cca1-22, and the flowering time of mlk3-1 cca1-22 is similar to mlk3-1 plants (Figures 5F,G). Since CCA1 could bind to the promoter of GI (Su et al., 2017), we then investigated the transcripts of GI and found transcription levels of GI in mlk3-1 cca1-22 are similar to those in mlk3-1 plants (Figure 5H).
To investigate the relationship between MLK3 and GI, we crossed gi-1 into mlk3-1 and generated the mlk3-1 gi-1 double mutant. The flowering time of mlk3-1 gi-1 is similar to gi-1 (Figures 6A,B and Supplementary Table 1). To confirm these, we generated mlk3-1 mlk4-2 gi-1 triple mutant and found that the flowering time of mlk3-1 mlk4-2 gi-1 is similar to that of gi-1 under LD (Figures 6C,D and Supplementary Table 1). These results suggested that MLK3 and MLK4 are redundantly involved in GI-dependent flowering time pathway.
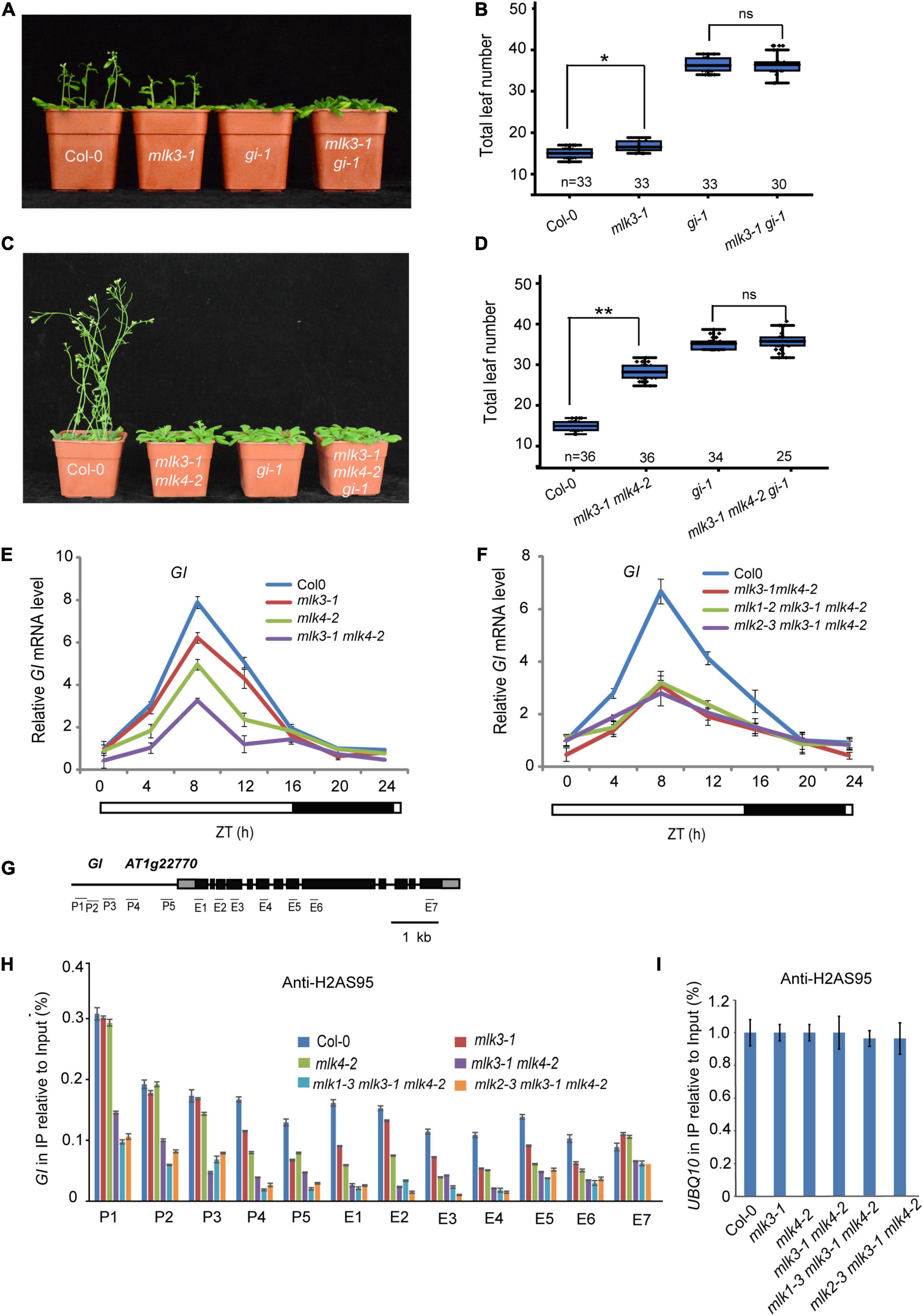
Figure 6. MLK3 and MLK4 are required for H2A phosphorylation at serine 95 at GI. (A) The representative phenotype of Col-0, mlk3-1, gi-1, and mlk3-1 gi-1 plants under a LD photoperiod is shown. (B) The total leaf number of Col-0, mlk3-1, gi-1, and mlk3-1 gi-1 plants under a LD photoperiod. Values shown are mean ± SD of total leaves; At least 30 plants were scored for each line. Asterisks indicate P < 0.05 and ns indicates no significance by t-test. (C) The representative phenotype of Col-0, mlk3-1mlk4-2, gi-1, and mlk3-1mlk4-2 gi-1 plants under a LD photoperiod. (D) The total leaf number of Col-0, mlk3-1mlk4-2, gi-1, and mlk3-1mlk4-2 gi-1 plants under a LD photoperiod. Values shown are mean ± SD of total leaves; At least 25 plants were scored for each line. Asterisks indicate P < 0.01 and ns indicates no significance by t-test. (E,F) The transcript levels of GI were examined in mlk3-1, mlk4-2 and mlk3-1 mlk4-2 mutants (E), and in mlk3-1 mlk4-2, mlk1-3 mlk3-1 mlk4-2, and mlk2-3 mlk3-1 mlk4-2 plants (F). The white bar indicates the light periods, and the black bar indicates the dark period. ZT, Zeitgeber time. Experiments were repeated at least three times, and the representative experiments shown indicate the mean ± SE, n = 3 replicates. (G) Gene structure of GI, indicating exons (boxes), introns (lines). The primers used for ChIP-PCR are marked with lines. (H,I) The amounts of H2AS95ph at different regions of GI were tested in the mlk3-1, mlk4-2, mlk3-1 mlk4-2, mlk1-3 mlk3-1 mlk4-2, and mlk2-3 mlk3-1 mlk4-2 plants (H). Ubiquitin 10 was used as internal control (I). Experiments were repeated at least three times, and the representative experiments shown indicate the mean ± SE, n = 3 replicates.
We then examined the transcription levels of GI. The decreased GI transcripts were further reduced in mlk3 mlk4 plants (Figure 6E and Supplementary Figure 6A). However, mutations of MLK1 or MLK2 in mlk3 mlk4 double mutants could not further reduce the attenuated GI transcripts in mlk3 mlk4 plants (Figure 6F). These results are consistent with the flowering time phenotype of mlk3 mlk4, mlk1 mlk3 mlk4, and mlk2 mlk3 mlk4 mutants.
We investigated the distribution of H2AS95ph by chromatin immunoprecipitation (ChIP), followed by the quantitative PCR measurement of DNA enrichment. The H2AS95ph was highly enriched at the promoter and the gene body at GI in wild type. The H2AS95ph enrichment was reduced in mlk3 and mlk4 single mutant, and further reduced in mlk3 mlk4 double mutant (Figures 6G–I and Supplementary Figure 6B). We then examined the H2AS95ph levels in triple mutants and found that the H2AS95ph levels in mlk134 and mlk234 triple mutants are similar to those in mlk3 mlk4 double mutant (Figures 6H,I), suggesting that MLK3 and MLK4 primarily deposited H2AS95ph at GI. These observations indicated that the transcription levels of GI are positively associated with the levels of H2AS95ph.
In this study, we revealed that MLKs redundantly participated in flowering time and H2AS95 phosphorylation. Loss of MLK3 function displayed late-flowering phenotype under LD, but not SD. Mutations of MLK3 enhanced the late-flowering phenotype of mlk4, but not mlk1 mlk2 double mutant. Loss of MLK1 or MLK2 functions in mlk3 mlk4 double mutant did not enhance the late-flowering phenotype of mlk3 mlk4. The transcript levels of GI were reduced in mlk3 mlk4 double mutant, but not further attenuated in mlk1 mlk3 mlk4 and mlk2 mlk3 mlk4 triple mutants. Accordingly, the H2AS95ph levels of GI is severely reduced in mlk3 mlk4 double mutant, but not further decreased in mlk1mlk3 mlk4 and mlk2 mlk3 mlk4 triple mutants. These results suggested that MLK3 and MLK4 are the major factors in flowering time and H2AS95 phosphorylation.
Among MLKs, MLK1, MLK2, and MLK4 are redundant in leaf sharp and height development. Loss of MLK4 function did not show any growth defects, whereas mlk1 mlk2 double mutant only exhibited semidwarf with short hypocotyls (Wang et al., 2015; Su et al., 2017; Zheng et al., 2018). The growth defects including short petiole, and sharp and curly leaf were observed in mlk1 mlk2 mlk4 triple mutant, but not other triple mutants such as mlk1 mlk2 mlk3, mlk1 mlk3 mlk4, and mlk2 mlk3 mlk4, suggesting that MLKs might evolve the divergent functions, and MLK4 might be a key factor in flowering time and development factor. The previous study showed that loss of MLK3 function resulted in early flowering (Kang et al., 2020). However, our observation showed that mlk3 plants displayed the late-flowering phenotype under LD, but not SD. In addition, loss of MLK3 function enhanced the late-flowering phenotype of mlk4 mutants, but not mlk1 mlk2 double mutant. MLK3 interacted with CCA1, and loss of function MLK3 reversed the early-flowering phenotype of cca1. The inductions of GI in cca1 were reversed in cca1 mlk3 mutants, suggesting that MLK3 is required for GI transcripts modulated by CCA1.
The in vitro and in vivo assays showed that MLK3 also contained kinase activity for the phosphorylation of H2AS95ph. H2AS95ph levels were not reduced in mlk3 and mlk4 single mutant, but reduced in mlk3 mlk4 double mutant. Further study showed that H2AS95ph levels in mlk1 mlk3 mlk4 and mlk2 mlk3 mlk4 triple mutant are similar to that in mlk3 mlk4 double mutant. The flowering time phenotype of mlk3 mlk4 double mutant is similar to that of mlk1 mlk3 mlk4 and mlk2 mlk3 mlk4 triple mutant. MUT9p could phosphorylate H2A and H3, and H3 at Threonine 3 is the best substrate in Chlamydomonas (Casas-Mollano et al., 2008). In our study, H3T3ph levels were not reduced in mlk1 mlk2 mlk3, mlk1 mlk2 mlk4, mlk1 mlk3 mlk4, and mlk2 mlk3 mlk4 triple mutant, suggesting that MLKs, unlike MUT9p, are primarily involved in H2AS95 phosphorylation.
Flowering at appropriate time determines whether plant successfully generate seeds in next generation; thus, the plant has evolutes the complicated system to regulate the flowering time. The histone modifications, including histone methylation, histone acetylation, and histone variants were observed at FLC, and these modifications play pivotal roles in autonomous and vernalization flowering time pathway in Arabidopsis (He and Amasino, 2005; Berry and Dean, 2015; Whittaker and Dean, 2017). The histone trimethylation of H3 lysine 27 was induced and histone trimethylation of H3 lysine 36 was reduced with prolonged cold treatment (Yang et al., 2014), suggesting that the histone methylations dynamically change in response to environmental change. Although the temperature and photoperiod are mostly common environment, little is known about how histone modification changes in response to photoperiod. Our study showed that defects in MLKs reduced H2AS95ph levels and transcription levels ofGI, which, in turn, decreased the transcription levels of CO and FT. CCA1, MLK4, and YAF9a form a protein complex and promote H2A.Z deposition at GI (Su et al., 2017). The correlation between H2AS95ph levels and transcription levels at GI was at least partially due to the attenuated H2A.Z deposition. Together, our study indicated that H2AS95ph levels of GI are closely linked with flowering time and transcription levels ofGI, and H2AS95 phosphorylation might be an essential activation marker in flowering time.
The Arabidopsis thaliana ecotype Col-0 was grown at 22°C under a LD photoperiod in a 16-h-light/8-h-dark cycle and light intensity of 140 μmol m–2 s–1 or a SD photoperiod with 8-h-light/16-h dark. The mutant strains obtained from the SALK collection were as follows: mlk1-2, Salkseq_132455; mlk1-3, SALK_039903; mlk2-2, SALK_149222; mlk2-3, SALK_064333; mlk3-1, SALK_017102; mlk3-2, SAIL_1151_E03; mlk4-2, SALK_201615c; mlk4-3, SAILseq_317_D02.1; cca1-22, SALKseq_120169; gi-1, CS3123; and co-9, CS870084.
The plasmids were constructed with the DNA primers and protocols described in Supplementary Data Set 1. All cloned DNAs were confirmed by DNA sequencing.
The yeast two-hybrid assay was performed according to the manufacturer’s protocol (Clontech; user manual 630489). Briefly, the Saccharomyces cerevisiae strain Y190 was transformed with the bait constructs pGBKT-MLK3 and pGADT7-CCA1. The yeast was scored for protein interaction based on their ability to grow on synthetic-defined medium lacking Trp, Leu, His, and adenine. The primers used to generate the constructs are shown in Supplementary Data Set 1.
The phosphorylation reaction assay was performed as described (Su et al., 2017). Briefly, 2–3 μg purified protein was incubated with 2.5 μCi γ–32 P ATP in a reaction buffer (50 mM Tris–HCl, pH 7.4, 10 mM MgCl2, 50 mM NaCl, 1 mM DTT, 2 mMEDTA, and 50 mM ATP) at 30°C for 1 h. The reaction products were separated by SDS-PAGE and autoradiographed with Storage Phosphor System.
For the pull-down assays, 3 μg bait and prey proteins were incubated overnight at 4°C. The beads were washed with a solution containing 20 mM Tris (pH 7.4), 150 mM NaCl, and 0.05% Tween 20 separated on a sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and analyzed by immunoblotting using an anti-GST antibody (GenScript, Nanjing, China; A00866-100, lot: 18A001413) or an anti-His antibody (Abmart, Shanghai, China; M30111M, lot: 293871). Co-IP was performed as described previously (Su et al., 2017). Briefly, 1 × 106 protoplasts were lysed with PEN-140 buffer (140 mM NaCl, 2.7 mM KCl, 25 mM Na2HPO4, 1.5 mM KH2PO4, 0.01 mM EDTA and 0.05% CA-630). The supernatant was precipitated with anti-FLAG (Sigma-Aldrich; H8592, lot: SLBV3799) antibodies, followed by washes with PEN-400 buffer (400 mM NaCl, 2.7 mM KCl, 25 mM Na2HPO4, 1.5 mM KH2PO4, 0.01 mM EDTA and 0.05% CA-630). The samples were analyzed by immunoblotting using anti-HA (Roche; 11867423001, lot: 13500600) and anti-FLAG antibodies.
Total RNA was isolated from 7-day seedlings grown under a LD photoperiod. RT-PCR analysis was performed with a CFX real-time PCR instrument (Bio-Rad) and SYBR Green mixture (Roche). The relative expression of the genes was quantified with the 2–ΔΔCT calculation using Ubiquitin 10 as the reference housekeeping gene for the expression analyses. The enrichment of DNA at specific genes was quantified with the 2–ΔΔCT calculation using Ubiquitin 10 as the reference housekeeping gene for ChIP assays. The gene-specific primers are shown in Supplementary Data Set 1.
Chromatin immunoprecipitation assay was performed as described previously (Su et al., 2017). Briefly, 7-day seedlings grown under a LD photoperiod were fixed with formaldehyde and quenched in glycine. The grounds were extracted with buffer I (0.4 M sucrose, 10 mM Tris (pH 8.0), 5 mM β-mercaptoethanol, 0.1 mM phenylmethylsulfonylfluoride (PMSF), and protease inhibitor cocktail), buffer II (0.25 M sucrose, 10 mM Tris (pH 8.0), 10 mM MgCl2, 1% Triton X-100, 5 mM β-mercaptoethanol, 0.1 mM PMSF and protease inhibitor cocktail), and buffer III (1.7 M sucrose, 10 mM Tris (pH 8.0), 10 mM MgCl2, 1% Triton X-100, 5 mM β-mercaptoethanol, 0.1 mM PMSF, and protease inhibitor cocktail). The specific antibodies anti-H2AS95ph were generated as described previously (Su et al., 2017), or control IgG serum, was added to the precleared supernatants for an overnight incubation at 4°C. The immunoprecipitated sample was extracted and analyzed by RT-PCR with the gene-specific primers shown in Supplementary Data Set 1.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: MLK1 (At5g18190), MLK2 (At3g03940), MLK3 (At2g25760), and MLK4 (At3g13670).
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
YD and TH conceived the study and designed the experiments. HeZ performed the MLK3 kinase activity experiments, western blot, and ChIP-PCR assays. HaZ performed the MLK1 and MLK2 kinase activity experiments, and generated the mlk1 mlk2 and mlk1 mlk2 co mutants. TH prepared the materials with the help of other authors and performed most of the experiments. YD wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (31871278 and U19A2021 to YD, 32000242 to YS, and 32000241 to HaZ), the Strategic Priority Research Program “Molecular Mechanisms of Plant Growth and Development” of CAS (grant no. XDB27030203), and the University Synergy Innovation Program of Anhui province (GXXT-2019-033).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.761008/full#supplementary-material
Amasino, R. M., and Michaels, S. D. (2010). The timing of flowering. Plant Physiol. 154, 516–520. doi: 10.1104/pp.110.161653
Berry, S., and Dean, C. (2015). Environmental perception and epigenetic memory: mechanistic insight through FLC. Plant J. 83, 133–148. doi: 10.1111/tpj.12869
Boss, P. K., Bastow, R. M., Mylne, J. S., and Dean, C. (2004). Multiple pathways in the decision to flower: enabling, promoting, and resetting. Plant Cell 16, S18–S31. doi: 10.1105/tpc.015958
Casas-Mollano, J. A., Jeong, B.-R., Xu, J., Moriyama, H., and Cerutti, H. (2008). The MUT9p kinase phosphorylates histone H3 threonine 3 and is necessary for heritable epigenetic silencing in Chlamydomonas. Proc. Natl. Acad. Sci. U.S.A. 105, 6486–6491. doi: 10.1073/pnas.0711310105
Chen, H.-H., Qu, L., Xu, Z.-H., Zhu, J.-K., and Xue, H.-W. (2018). EL1-like casein kinases suppress ABA signaling and responses by phosphorylating and destabilizing the ABA receptors PYR/PYLs in Arabidopsis. Mol. Plant 11, 706–719. doi: 10.1016/j.molp.2018.02.012
Cruciat, C.-M. (2014). Casein kinase 1 and Wnt/β-catenin signaling. Curr. Opin. Cell Biol. 31, 46–55. doi: 10.1016/j.ceb.2014.08.003
Dong, Q., and Han, F. (2012). Phosphorylation of histone H2A is associated with centromere function and maintenance in meiosis. Plant J. 71, 800–809. doi: 10.1111/j.1365-313x.2012.05029.x
He, Y., and Amasino, R. M. (2005). Role of chromatin modification in flowering-time control. Trends Plant Sci. 10, 30–35. doi: 10.1016/j.tplants.2004.11.003
Huang, H., Alvarez, S., Bindbeutel, R., Shen, Z., Naldrett, M. J., Evans, B. S., et al. (2016). Identification of evening complex associated proteins in Arabidopsis by affinity purification and mass spectrometry. Mol. Cell. Proteomics 15, 201–217. doi: 10.1074/mcp.M115.054064
Kang, J., Cui, H., Jia, S., Liu, W., Yu, R., Wu, Z., et al. (2020). Arabidopsis thaliana MLK3, a plant-specific casein kinase 1, negatively regulates flowering and phosphorylates histone H3 in vitro. Genes 11:345. doi: 10.3390/genes11030345
Kawashima, T., Lorković, Z. J., Nishihama, R., Ishizaki, K., Axelsson, E., Yelagandula, R., et al. (2015). Diversification of histone H2A variants during plant evolution. Trends Plant Sci. 20, 419–425. doi: 10.1016/j.tplants.2015.04.005
Kelly, A. E., Ghenoiu, C., Xue, J. Z., Zierhut, C., Kimura, H., and Funabiki, H. (2010). Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science 330, 235–239. doi: 10.1126/science.1189505
Lau, P. N. I., and Cheung, P. (2011). Histone code pathway involving H3 S28 phosphorylation and K27 acetylation activates transcription and antagonizes polycomb silencing. Proc. Natl. Acad. Sci. U.S.A. 108, 2801–2806. doi: 10.1073/pnas.1012798108
Lin, J.-M., Kilman, V. L., Keegan, K., Paddock, B., Emery-Le, M., Rosbash, M., et al. (2002). A role for casein kinase 2α in the Drosophila circadian clock. Nature 420, 816–820. doi: 10.1038/nature01235
Liu, W., Xu, Z. H., Luo, D., and Xue, H. W. (2003). Roles of OsCKI1, a rice casein kinase I, in root development and plant hormone sensitivity. Plant J. 36, 189–202. doi: 10.1046/j.1365-313X.2003.01866.x
Metzger, E., Imhof, A., Patel, D., Kahl, P., Hoffmeyer, K., Friedrichs, N., et al. (2010). Phosphorylation of histone H3T6 by PKCβ I controls demethylation at histone H3K4. Nature 464, 792–796. doi: 10.1038/nature08839
Metzger, E., Yin, N., Wissmann, M., Kunowska, N., Fischer, K., Friedrichs, N., et al. (2008). Phosphorylation of histone H3 at threonine 11 establishes a novel chromatin mark for transcriptional regulation. Nat. Cell Biol. 10, 53–60. doi: 10.1038/ncb1668
Michaels, S. D., Himelblau, E., Kim, S. Y., Schomburg, F. M., and Amasino, R. M. (2005). Integration of flowering signals in winter-annual Arabidopsis. Plant Physiol. 137, 149–156. doi: 10.1104/pp.104.052811
Mizoguchi, T., Wheatley, K., Hanzawa, Y., Wright, L., Mizoguchi, M., Song, H.-R., et al. (2002). LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2, 629–641. doi: 10.1016/S1534-5807(02)00170-3
Mizoguchi, T., Wright, L., Fujiwara, S., Cremer, F., Lee, K., Onouchi, H., et al. (2005). Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17, 2255–2270. doi: 10.1105/tpc.105.033464
Ogiso, E., Takahashi, Y., Sasaki, T., Yano, M., and Izawa, T. (2010). The role of casein kinase II in flowering time regulation has diversified during evolution. Plant Physiol. 152, 808–820. doi: 10.1104/pp.109.148908
Pirrotta, V. (2015). Histone marks direct chromosome segregation. Cell 163, 792–793. doi: 10.1016/j.cell.2015.10.043
Putterill, J., Robson, F., Lee, K., Simon, R., and Coupland, G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847–857. doi: 10.1016/0092-8674(95)90288-0
Samach, A., Onouchi, H., Gold, S. E., Ditta, G. S., Schwarz-Sommer, Z., Yanofsky, M. F., et al. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288, 1613–1616.
Sawa, M., Nusinow, D. A., Kay, S. A., and Imaizumi, T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318, 261–265. doi: 10.1126/science.1146994
Soloaga, A., Thomson, S., Wiggin, G. R., Rampersaud, N., Dyson, M. H., Hazzalin, C. A., et al. (2003). MSK2 and MSK1 mediate the mitogen-and stress-induced phosphorylation of histone H3 and HMG-14. EMBO J. 22, 2788–2797.
Song, Y. H., Shim, J. S., Kinmonth-Schultz, H. A., and Imaizumi, T. (2015). Photoperiodic flowering: time measurement mechanisms in leaves. Annu. Rev. plant Biol. 66, 441–464. doi: 10.1146/annurev-arplant-043014-115555
Su, Y., Wang, S., Zhang, F., Zheng, H., Liu, Y., Huang, T., et al. (2017). Phosphorylation of histone H2A at serine 95: a plant-specific mark involved in flowering time regulation and H2A. Z deposition. Plant Cell 29, 2197–2213. doi: 10.1105/tpc.17.00266
Tan, S.-T., and Xue, H.-W. (2014). Casein kinase 1 regulates ethylene synthesis by phosphorylating and promoting the turnover of ACS5. Cell Rep. 9, 1692–1702.
Uehara, T. N., Mizutani, Y., Kuwata, K., Hirota, T., Sato, A., Mizoi, J., et al. (2019). Casein kinase 1 family regulates PRR5 and TOC1 in the Arabidopsis circadian clock. Proc. Natl. Acad. Sci. U.S.A. 116, 11528–11536. doi: 10.1073/pnas.1903357116
Van Attikum, H., Fritsch, O., Hohn, B., and Gasser, S. M. (2004). Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119, 777–788.
Wang, F., Ulyanova, N. P., Van Der Waal, M. S., Patnaik, D., Lens, S. M., and Higgins, J. M. (2011). A positive feedback loop involving Haspin and Aurora B promotes CPC accumulation at centromeres in mitosis. Curr. Biol. 21, 1061–1069. doi: 10.1016/j.cub.2011.05.016
Wang, Z., Casas-Mollano, J. A., Xu, J., Riethoven, J.-J. M., Zhang, C., and Cerutti, H. (2015). Osmotic stress induces phosphorylation of histone H3 at threonine 3 in pericentromeric regions of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 112, 8487–8492. doi: 10.1073/pnas.1423325112
Wang, Z., Kang, J., Armando Casas-Mollano, J., Dou, Y., Jia, S., Yang, Q., et al. (2021). MLK4-mediated phosphorylation of histone H3T3 promotes flowering by transcriptional silencing of FLC/MAF in Arabidopsis thalian a. Plant J. 105, 1400–1412. doi: 10.1111/tpj.15122
Wang, Z.-Y., and Tobin, E. M. (1998). Constitutive expression of the circadian clock associated 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93, 1207–1217. doi: 10.1016/S0092-8674(00)81464-6
Whittaker, C., and Dean, C. (2017). The FLC locus: a platform for discoveries in epigenetics and adaptation. Annu. Rev. Cell Dev. Biol. 33, 555–575. doi: 10.1146/annurev-cellbio-100616-060546
Wilson, M. E., Tzeng, S.-C., Augustin, M. M., Meyer, M., Jiang, X., Choi, J. H., et al. (2021). Quantitative proteomics and phosphoproteomics support a role for Mut9-like kinases in multiple metabolic and signaling pathways in Arabidopsis. Mol. Cell. Proteomics 20:100063.
Winter, S., Simboeck, E., Fischle, W., Zupkovitz, G., Dohnal, I., Mechtler, K., et al. (2008). 14-3-3 proteins recognize a histone code at histone H3 and are required for transcriptional activation. EMBO J. 27, 88–99.
Yang, H., Howard, M., and Dean, C. (2014). Antagonistic roles for H3K36me3 and H3K27me3 in the cold-induced epigenetic switch at Arabidopsis FLC. Curr. Biol. 24, 1793–1797. doi: 10.1016/j.cub.2014.06.047
Zheng, H., and Ding, Y. (2018). MLK1 and MLK2 integrate gibberellins and circadian clock signaling to modulate plant growth. Plant Signal. Behav. 13:e1439654.
Keywords: histone phosphorylation, flowering time, GI, MLK family, phosphorylation of histone H2A at serine 95
Citation: Huang T, Zhang H, Zhou Y, Su Y, Zheng H and Ding Y (2021) Phosphorylation of Histone H2A at Serine 95 Is Essential for Flowering Time and Development in Arabidopsis. Front. Plant Sci. 12:761008. doi: 10.3389/fpls.2021.761008
Received: 19 August 2021; Accepted: 12 October 2021;
Published: 23 November 2021.
Edited by:
Cristel C. Carles, Université Grenoble Alpes, FranceReviewed by:
Soon-Ki Han, Daegu Gyeongbuk Institute of Science and Technology (DGIST), South KoreaCopyright © 2021 Huang, Zhang, Zhou, Su, Zheng and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Han Zheng, emhlbmdoYW4xMDA0QHFxLmNvbQ==; Yong Ding, ZGluZ3lvbmdAdXN0Yy5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.