- 1Centre for Agriculture and the Bioeconomy, Queensland University of Technology, Brisbane, QLD, Australia
- 2School of Agriculture and Food Sciences, University of Queensland, Brisbane, QLD, Australia
- 3Department of Plant Pathology, University of Wisconsin-Madison, Madison, WI, United States
Autophagy is a genetically regulated, eukaryotic cellular degradation system that sequestrates cytoplasmic materials in specialised vesicles, termed autophagosomes, for delivery and breakdown in the lysosome or vacuole. In plants, autophagy plays essential roles in development (e.g., senescence) and responses to abiotic (e.g., nutrient starvation, drought and oxidative stress) and biotic stresses (e.g., hypersensitive response). Initially, autophagy was considered a non-selective bulk degradation mechanism that provides energy and building blocks for homeostatic balance during stress. Recent studies, however, reveal that autophagy may be more subtle and selectively target ubiquitylated protein aggregates, protein complexes and even organelles for degradation to regulate vital cellular processes even during favourable conditions. The selective nature of autophagy lends itself to potential manipulation and exploitation as part of designer protein turnover machinery for the development of stress-tolerant and disease-resistant crops, crops with increased yield potential and agricultural efficiency and reduced post-harvest losses. Here, we discuss our current understanding of autophagy and speculate its potential manipulation for improved agricultural performance.
Introduction
Plants must intricately balance their energy needs for growth and development with survival and stress responses. Autophagy contributes to this balance by trafficking and degrading/recycling unwanted cytoplasmic materials in the vacuole (plants) or lysosome (mammals; Masclaux-Daubresse et al., 2017; Bozhkov, 2018; Yoshimoto and Ohsumi, 2018). With roles in cancer, ageing, diabetes and numerous neurodegenerative diseases, the identification of autophagy components and regulatory machinery is one of the most research-intensive fields in mammalian biology (Hayat, 2017). Likewise, autophagy is crucial for the proper regulation of plant metabolism and nutrient remobilisation in response to biotic and abiotic stress and a housekeeping capacity. With climate change models suggesting more frequent droughts and unpredictable weather patterns in the future, the importance of autophagy in plant stress responses is gaining significant momentum (Liu et al., 2009; Tang and Bassham, 2018; Gou et al., 2019). The dual role of non-selective and selective autophagy allows bulk or targeted degradation of protein complexes and organelles (Kellner et al., 2017; Wang et al., 2018). The manipulation of plant autophagy pathways for the development of stress-tolerant and disease-resistant crops with increased yields and agricultural efficiency, and minimal post-harvest losses, is now feasible and may play a significant role in sustaining agriculture in changing climates. Here, we discuss the roles of autophagy in plant stress biology and potential ways to manipulate the process for improved crop performance.
Autophagy in Crop Stress Responses
The role of autophagy in response to plant stress is well-established (Kabbage et al., 2017; Avin-Wittenberg et al., 2018; Tang and Bassham, 2018; Avin-Wittenberg, 2019; Thanthrige et al., 2020). Stress directly damages proteins and membranes; it also causes the accumulation of unfolded proteins and endoplasmic reticulum (ER) stress that trigger programmed cell death (PCD) pathways. To avoid unwanted cell death, cells initiate corrective measures that remove misfolded or damaged proteins, including the unfolded protein response (UPR) and ER-associated degradation (ERAD) in a coordinated manner with the ubiquitin proteasome system (UPS) and autophagy (Li et al., 2020; Qu et al., 2021). Ubiquitin-mediated protein degradation occurs through proteasome or autophagy pathways (Su et al., 2020; Singh et al., 2021). While the UPS targets the degradation of ubiquitylated, short-lived, individual misfolded or regulatory polypeptides, autophagy eliminates individual misfolded proteins and bulk structures, such as large protein complexes, insoluble protein aggregates and organelles (Singh et al., 2021). Due to its specificity, the degradation of protein aggregates by the UPS is inefficient during prolonged stress (Toyooka et al., 2006). Therefore, autophagy represents a robust mechanism for the complete and efficient large-scale degradation of aggregated proteins during severe or prolonged stress conditions (Toyooka et al., 2006; Xiong et al., 2007). In the next few sections, we discuss how autophagy plays a role in plant stress responses.
Abiotic and biotic stresses impede electron transport chains and cause the accumulation of reactive oxygen species (ROS) that directly damage proteins, causing their partial denaturation and aggregation. Plant autophagy pathways are highly attuned to the oxidative state of the cell as part of housekeeping mechanisms to modulate ROS damage (Tang and Bassham, 2018). During stress and the accumulation of ROS, the oxidation of the autophagy gene, ATG4, triggers corrective autophagy pathways that mitigate cellular ROS levels by removing ROS generators, such as dysfunctional mitochondria or chloroplasts and removing oxidised proteins from the cell (Xiong et al., 2007; Tang and Bassham, 2018). In addition to ROS, many stresses, such as drought, cause the cessation of photosynthesis resulting in caloric and other nutritional deficiencies that also trigger autophagy pathways (Williams et al., 2015).
Drought and salinity induce the expression of autophagy genes (e.g., ATG18a), and Arabidopsis knockdown plants with defective autophagy pathways are sensitive to abiotic stresses (Liu et al., 2009). Autophagy’s efficient degradation of select proteins and large aggregates or organelles facilitates tolerance against severe stress states, such as desiccation (Williams et al., 2015; Oliver et al., 2020). Recent studies demonstrate that the native Australian resurrection grass, Tripogon loliiformis, uses trehalose metabolism to promote and maintain autophagy pathways that prevent senescence and PCD (Williams et al., 2015; Asami et al., 2019; Okemo et al., 2021). A similar role for autophagy in desiccation tolerance occurs for the resurrection plant, Boea hygrometrica (Zhu et al., 2015). Of particular note, the shoots and roots of T. loliiformis use different desiccation response strategies. Tripogon loliiformis shuts down photosynthesis during the early stages of drying and uses autophagy to transport resources from the shoots to the roots. The photosynthetic shutdown also slows down transpiration and water loss. While the remobilisation of resources helps T. loliiformis roots maintain energy homeostasis, mitigating the need to implement harsher survival measures, including autophagy (Asami et al., 2019). Whether a similar process occurs in Poikilochlorophyllous resurrection plants that remobilise their nitrogen sources via the degradation of chlorophyll remains to be seen confirmed (Costa et al., 2017; Asami et al., 2018).
Autophagy is a double-edged sword with survival and cell death roles in plant-pathogen interactions (Lenz et al., 2011; Thanthrige et al., 2020). Autophagy modulates defence responses regulated by salicylic acid (SA) and jasmonic acid (JA), thereby influencing plant basal resistance to both biotrophic and necrotrophic pathogens (Liao and Bassham, 2020). During necrotrophic fungal infection, autophagy positively regulates plant defences, serving an anti-death role to limit the disease lesion by suppressing ROS-mediated accumulation of oxidised compounds and subsequent lesion development and disease containment (Lenz et al., 2011). Autophagy-deficient mutants are susceptible to disease (Lai et al., 2011; Lenz et al., 2011). Conversely, the pro-death attributes of autophagy support host defence against viruses. In host plant-virus interactions, host-regulated autophagy kills surrounding uninfected cells, limiting disease spread (Hofius et al., 2009). The benefits of autophagy death for biotrophs in general are unclear (Leary et al., 2019).
Autophagy plays a role in plant responses to pathogens by positively regulating SA accumulation (Sun et al., 2018a,b). Overexpression of ATG18a in apple enhances SA levels and improve resistance to the fungal pathogen Diplocarpon mali (Sun et al., 2018a,b). The transcription factor WRKY33 interacts with ATG18a to regulate autophagy. The induction of ATG18 and autophagy was reduced in wrky33 mutants. Furthermore, autophagy mutants contained dysfunctional JA-mediated signalling pathways and were more susceptible to Botrytis cinerea (Lai et al., 2011). Similarly, autophagy defective banana are more susceptible to Fusarium wilt; the exogenous application of SA and JA can rescue the sensitive phenotype (Wei et al., 2017). The hypersensitive response (HR) is a localised form of PCD that occurs during plant-microbe interactions (Liu et al., 2005). Autophagy regulates the HR, and autophagy mutants display runaway cell death (Liu et al., 2005). Thus, autophagy plays an essential ‘pro-survival’ function in plants that effectively control the pathogen spread without resulting in ‘unwanted’ cell death in innocent uninfected bystander cells (Liu et al., 2005). Conversely, a pro-death function of autophagy during hypersensitive cell death has also been demonstrated (Hofius et al., 2009). In Arabidopsis, atg knockout mutants (atg7 and atg9) display delayed HR PCD induced by Pseudomonas syringae pv. tomato DC3000 harbouring the avirulence genes AvrRps4 (Hofius et al., 2009). Subsequent studies showed that Bax inhibitor-1 (BI-1) interacts with ATG6 to positively regulate autophagy (Xu et al., 2017). Silencing of BI-1 reduces the autophagic response to TMV, whereas overexpression of BI-1 increased autophagic activity and enhanced defence to viral infection (Xu et al., 2017).
Autophagy in Symbiotic ‘Friend Versus FOE’ Relationships
Autophagy plays a role in symbiotic relationships, including rhizobia-legume interactions. As an established inducer of autophagy, trehalose accumulates during symbiotic plant interactions (Brechenmacher et al., 2010; Williams et al., 2015; Li et al., 2016b). Silencing of trehalase, the enzyme that breaks down trehalose in common bean (Phaseolus vulgaris L.), increases trehalose content and supports augmented bacteroid number, nodule biomass and nitrogenase activity (Barraza et al., 2013). Transgenic TRE1-RNAi nodules accumulate more ATG3 transcripts, further suggesting a role for autophagy in nodule formation (Barraza et al., 2013). Studies on transgenic soybean expressing the CED-9 from Caenorhabditis elegans display reduced nodulation and organogenesis, possibly by interfering with autophagy and vesicle trafficking (Robert et al., 2014). Similarly, studies in mammalian cells demonstrated an association between CED-9/Bcl-2 and Beclin-1/ATG6 to show a link between the anti-apoptotic activities of CED-9 and autophagy (Takacs-Vellai et al., 2005). However, further studies must be conducted to consolidate these links and what implications they have for plant autophagy.
Autophagy in Nutrient Remobilisation and Plant Development
Autophagy and the UPS play pivotal roles in nutrient recycling and remobilisation and are distinguished by their capacity to function over distance (Wang et al., 2017; Avin-Wittenberg et al., 2018; Tang and Bassham, 2018). The UPS degrades individual proteins/protein aggregates and is not a suitable target for large-scale nutrient remobilisation. In optimal conditions, with low levels of aggregated proteins, the UPS mediates the majority of targeted protein degradation. By contrast, autophagy can function over the entire cell and degrade complete organelles. Plants often require rapid recycling, large-scale remobilisation and reabsorption of nutrients. Therefore, upon nutrient starvation and environmental stress, autophagy functions as a brute force yet efficient system to remobilise nutrients at scale (Doelling et al., 2002; Thompson et al., 2005).
Manipulation of Autophagy for Improved Stress Tolerance
Plants are continuously exposed to multiple stressors in their environment and modulate their growth and development accordingly; autophagy plays comprehensive roles in many of these responses. Furthering our knowledge about autophagy roles in stress responses provides possible routes for new strategies and candidates for crop improvement, promoting stress resistance and yield.
Autophagy can be regulated genetically (i.e., genetically modified or edited crops) or pharmacologically (Avin-Wittenberg et al., 2018). Before use as a biotechnological strategy for improving crops, however, reliable systems for the precise manipulation of autophagy must be available. Next, we discuss the potential manipulation of autophagy pathways under different stress conditions and speculate on potential targets of plant autophagy machinery for crop improvement (Figure 1).
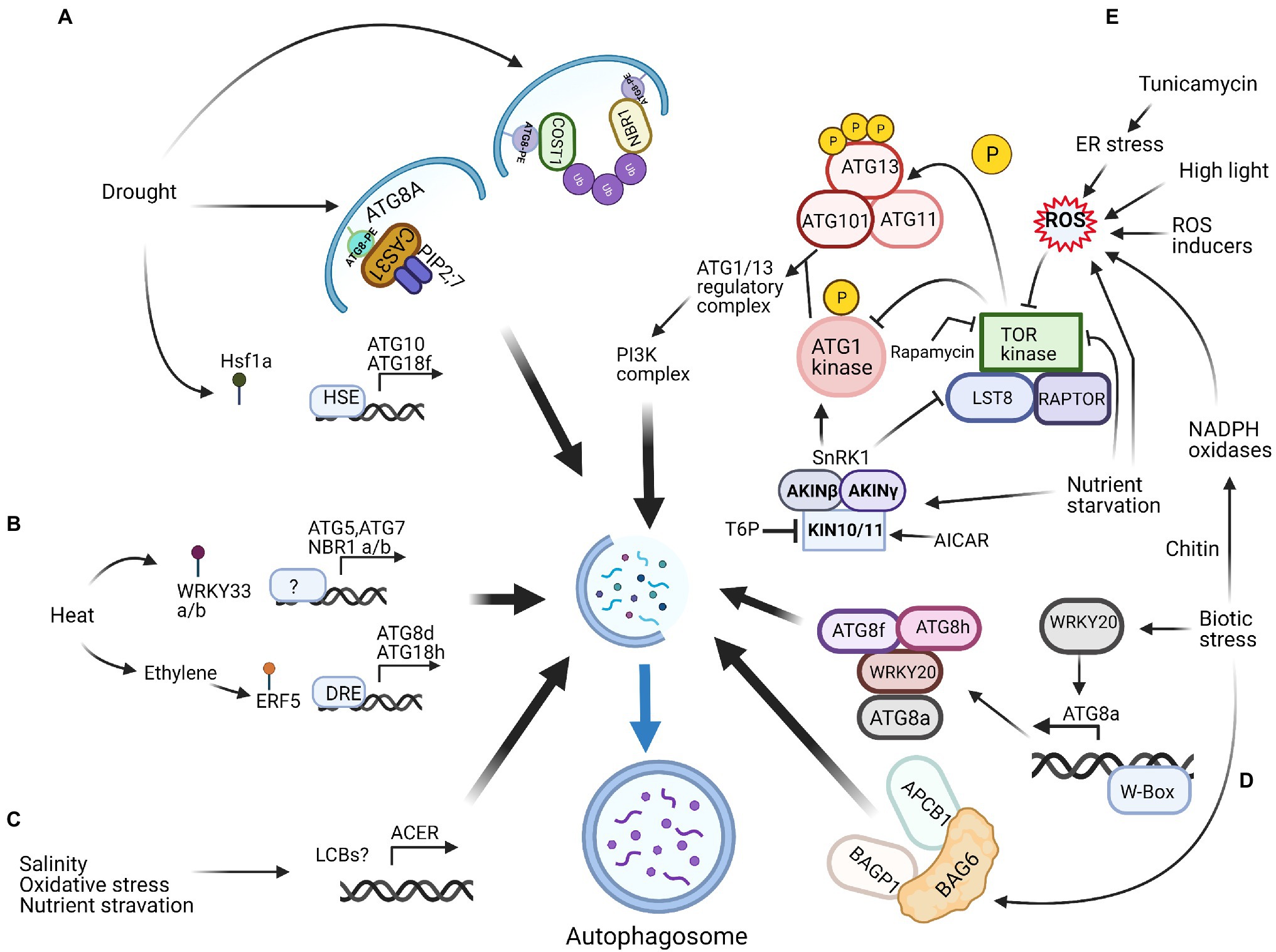
Figure 1. Potential targets for manipulation of autophagy in plants. Identified regulators and potential ATG gene targets during abiotic stresses such as drought stress (A), heat stress (B), salinity, oxidative and nutrient starvation stresses (C), and biotic stresses (D) and stresses caused by reactive oxygen species (E). Refer text for the detailed information. NBR1, Neighbour of BRCA1; COST1, Constitutively Stressed 1; HSE, Heat shock element; ATG, Autophagy related; PE, Phosphatidylethanolamine; PIPs, Plasma membrane intrinsic proteins; Ub, Ubiquitin; HsfA1a, Heat-shock transcription factor A1a; ERF5, Ethylene response factor 5; DRE, Drought-responsive elements; ACER, Alkaline ceramidase; LCBs, Long‐chain bases; PI3K, Phosphatidylinositol-3-phosphate kinase; SnRK1, Sucrose nonfermenting1-related protein kinase1; TOR, Target of rapamycin; T6P, Trehalose-6-phosphate; AICAR, 5-Aminoimidazole-4-carboxamide ribonucleoside monophosphate; BAGP1, BAG-Associated GRAM Protein; APCB1, Aspartyl protease cleaving BAG; ROS, Reactive oxygen species. This figure was created with Biorender.
Overexpression of Autophagy Genes
There is significant potential to manipulate the regulation of autophagy pathways for improved crop tolerance to abiotic stresses. For example, drought activates autophagy pathways in crops and transgenic crops, such as apple, wheat, tomato, foxtail millet and barley, overexpressing ATG genes display significantly improved drought tolerance (Kuzuoglu-Ozturk et al., 2012; Pei et al., 2014; Li et al., 2015b, 2016a; Wang et al., 2015, 2017; Zeng et al., 2017; Mamun et al., 2018; Sun et al., 2018c). Notably, the expression of ATG genes in crops resulted in no detrimental effects in non-stressed conditions. In addition to ATGs, genes that modulate autophagy pathways are suitable targets for crop improvement. However, as opposed to the expression of ATG genes, a compromise between yield and tolerance must often be satisfied when expressing autophagy regulators to minimise detrimental effects and gain agronomical benefits.
As an inducer or a suppressor, the dual role of autophagy in biotic stress responses lends itself to the generation of pathogen resistance (Liu et al., 2005). Furthermore, and similar to abiotic stress responses, other than ATG6, the expression of autophagy genes does not have detrimental effects on plant growth and yield; thus, ATGs are a good target for developing resistant crops. Plants overexpressing autophagy genes (e.g., ATG5 and ATG7) display improved stress tolerance and resistance against pathogens (Minina et al., 2018). A set of glyceraldehyde-3-phosphate dehydrogenases (GAPDHs) interact with ATG3 to negatively regulated disease resistance in Arabidopsis, Nicotiana benthamiana and cassava. Contrastingly, silencing of cytosolic glycolytic GAPDHs (GAPCs) enhanced disease resistance (Han et al., 2015; Henry et al., 2015; Zeng et al., 2018).
Another potential gene target for crop improvement is alkaline ceramidase (ACER), an essential enzyme in the sphingolipid metabolic pathway. Silencing of ACER inhibits autophagy. Conversely, its overexpression promotes autophagy under nutrient starvation, salinity and oxidative stresses (Zheng et al., 2018). Plant hormone signalling pathways are also targets for the regulation of autophagy. For example, the overexpression of ERF5 (an ethylene response factor) improves drought tolerance in tomato (Pan et al., 2012). ERF5 directly binds to the promoters of ATG8d and ATG18h and activates gene expression to promote autophagy, which is essential for ethylene-mediated drought resistance (Zhu et al., 2018; Figure 1).
Transcription Factors as Targets
Similar to drought, the modulation of autophagy regulators can alleviate heat stress in plants. The WRKYs are a large family of transcription factors that modulate many plant physiological processes, such as growth, development and responses to abiotic and biotic stresses (Chen et al., 2017a). Heat tolerance in plants requires WRKY33 (Li et al., 2011; Zhou et al., 2014). Silencing of WRKY33 genes compromised heat tolerance and reduced heat-induced ATG gene expression and autophagosome formation (Zhou et al., 2014). Like COST1, however, the silencing of transcription factors, such as the WRKY family, can also have detrimental effects on agronomic performance.
Recent studies show that the transcription factor, HY5 (elongated hypocotyl 5), regulates plant autophagy in response to light-to-dark conversion and nitrogen starvation in plants. Under nitrogen-sufficient or light conditions, HY5 interacts with and recruits HDA9 (histone deacetylase 9) to ATG5 and ATG8e to repress the gene expression by deacetylation of the Lys9 and Lys27 of histone 3 and inhibit autophagy. Conversely, upon dark conditions or nitrogen starvation, HY5 undergoes 26S proteasome-mediated degradation leading to dissociation of HDA9 from their target genes, thereby resulting in enhanced acetylation levels and upregulated expression of the ATGs and autophagy (Yang et al., 2020). Furthermore, overexpression of the transcription factor, TGA9 (TGACG (TGA) motif-binding protein 9) transcriptionally upregulates expression of autophagy genes to activate autophagy under stress conditions. Thus, TGA9 acts as a positive regulator of autophagy and is a possible target for improving plant stress tolerance (Wang et al., 2020).
Recently, more transcription factors have been identified in plants which can be potentially used to manipulate the autophagy pathway for stress tolerance. Brassinosteroids (BRs) regulate plant growth, development and stress responses by activating the core transcription factor BRI1-EMS-SUPPRESSOR1 (BES1), whose degradation occurs through the proteasome and autophagy pathways (Nolan et al., 2017; Wang et al., 2021). BES1 is targeted for autophagy-mediated degradation by direct interaction with ubiquitin-binding receptor protein DOMINANT SUPPRESSOR OF KAR 2 (DSK2) and is targeted to the autophagy pathway during stress via the interaction of DSK2 with ATG8 (Nolan et al., 2017). Recently, it has been found that ubiquitin ligase BES1-ASSOCIATED F-BOX 1 (BAF1) interacts with BES1 and mediates its ubiquitination and degradation via selective autophagy (Wang et al., 2021).
For instance, the plant-unique COST1 (Constitutively Stressed 1) protein negatively regulates plant drought tolerance by directly interacting with the autophagy receptor protein ATG8. Even though the loss of COST1 improves drought tolerance by activating autophagy, it also detrimentally affects plant growth and development (Bao et al., 2020). Therefore, COST1, via its role in autophagy regulation, is a vital mediator that helps control the balance between growth, development and stress responses in plants (Bao, 2020; Bao et al., 2020). The high conservation of COST proteins throughout the plant kingdom indicates its potential as a gene target to enhance drought tolerance in crops (Bao et al., 2020; Bao and Bassham, 2020). Dehydrin, CAS31 (cold acclimation-specific 31) in Medicago truncatula, is a positive regulator of drought responses and plays a crucial role in autophagic degradation (Li et al., 2020). Recent studies show that aquaporin, PIP2;7, functions as a negative regulator of drought response (Li et al., 2020). CAS31 participates in drought-induced autophagic degradation as a cargo receptor and facilitates the autophagic degradation of PIP2; 7 and reduced root hydraulic conductivity, thus reducing water loss and improving drought tolerance (Li et al., 2020). Therefore, it is also a potential target for manipulation to improve crop tolerance.
Manipulation of Autophagy to Improve Nutrient Remobilisation
Autophagy affects plant metabolism and is involved in efficient nutrient remobilisation from leaves to developing seeds (Marshall and Vierstra, 2018). Therefore, the manipulation of autophagy for improved nutrient recycling presents an intriguing strategy to increase the yield of seed-bearing food and biofuel crops, especially under unfavourable growth conditions or in situations where soil fertilisation is cost-prohibitive or ecologically unsound. The suppression of senescence can improve stress tolerance and yield. Sorghum stay-green hybrid cultivars have significant yield advantages under postanthesis drought compared to senescent hybrid lines (Borrell et al., 2000a,b). In addition to increasing yield potential, nutrient recycling may lower the requirement of chemical inputs, providing both environmental and economic benefits. The manipulation of the metabolome via the targeted degradation of specific constituents can also increase agronomic productivity in crop species (Marshall and Vierstra, 2018). Studies show that transcription factors, such as Hsf1A, an essential component of plant drought responses, activate ATG expression upon stress conditions and are potential gene targets for crop improvement (Wang et al., 2015).
Plants containing mutated ATG8-ATG12 conjugation pathways display accelerated leaf senescence and hypersensitivity to carbon and nitrogen limitation (Doelling et al., 2002; Yoshimoto et al., 2004; Hofius et al., 2009; Chung et al., 2010). Conversely, the overexpression of ATG (autophagy stimulation) enhances plant growth, fitness, seed set and stress tolerance (Xia et al., 2012; Wang et al., 2016; Minina et al., 2018; Sun et al., 2018c). Another potential strategy for the suppression of senescence and improved stress tolerance is the overexpression of KIN 10, the catalytic subunit of SnRK1. Expression of KIN10 promotes autophagy, delays senescence and increase tolerance to nutrient starvation conditions (Chen et al., 2017b; Soto-Burgos and Bassham, 2017; Huang et al., 2019).
Autophagy is a strong determinant of seed quality and a critical component of the micronutrient seed filling (Pottier et al., 2019). Arabidopsis thaliana autophagy mutants are inefficient at translocating iron from vegetative organs to the seeds (Pottier et al., 2019). Plants also use senescent vegetative organs to sequester manganese and zinc into seeds. The lower amounts of zinc and manganese in the seeds of autophagy mutants suggest that their translocation depends on autophagy (Pottier et al., 2019). Future work should investigate the potential manipulation of autophagy in crops at the seed filling stage to increase the pool of nutrients available for subsequent translocation to seeds.
Recently, researchers demonstrated an essential role for the autophagy-related kinase, PI3K, in symbiotic interactions between common bean plants and rhizobia bacteria or arbuscular mycorrhizal fungi (Estrada-Navarrete et al., 2016). Transgenic PI3K-RNAi plants display decreased root hair growth and curling (Estrada-Navarrete et al., 2016). Additionally, infection thread growth, nodule number and symbiosome formation were severely affected (Estrada-Navarrete et al., 2016). However, more work is required to conclude that autophagy is directly involved in symbiotic interactions because the PI3K complex is involved in many other biological processes (Tang and Bassham, 2018). Manipulation of autophagy pathways could potentially enhance symbiotic interactions with legumes and subsequently reduce fertiliser inputs (Blair et al., 2016).
Manipulation of Autophagy to Modulate Plant Development
Autophagy is highly involved in plant development, pollen maturation, lipid metabolism and nutrient supply in anthers. It is crucial in angiosperm sexual reproductive development and is potentially involved in the degradation of intracellular components, such as plastids and lipid bodies and the regulation of lipid metabolism during pollen maturation (Hanamata et al., 2014; Kurusu et al., 2014; Kurusu and Kuchitsu, 2017).
During Arabidopsis seed maturation, transcript levels of almost all autophagy-related genes increase in the silique (Di Berardino et al., 2018). Similarly, maize developing endosperms contain high amounts of lipidated ATG8 (ATG8-PE; Chung et al., 2009). Furthermore, transcriptome analyses show increased ATG transcript accumulation in the endosperm, but not the embryo, suggesting that autophagy participates in the endosperm’s maturation and death during seed development (Li et al., 2015a). The manipulation of autophagy pathways may improve seed vigour and storage.
Photoperiod regulates autophagy, and thus, autophagy may be an intriguing target for the development of determinant crops or crops with shortened or prolonged lifecycles. Day length and autophagy determine the number of fertile florets, and developmentally generated sugar starvation triggers floret autophagy in wheat (Ghiglione et al., 2008). Long days intensify this process due to the increased carbohydrate consumption caused by accelerated plant development (Ghiglione et al., 2008). It is feasible to manipulation flowering time by either activating or suppressing autophagy pathways. The activation of autophagy delays senescence and flowering, while plants with dysfunctional autophagy pathways display accelerated senescence.
Collectively, these findings display substantial potential for manipulating autophagy as a target for the development of more resilient crops that can withstand future inclement environmental conditions.
Pharmacological Elicitation of Plant Autophagy Pathways
Compared with animals, the pharmacological manipulation of autophagy with stimulants, such as rapamycin, fumonisin B1, tunicamycin, polyamines and suppressors wortmannin concanamycin A in agricultural applications, remains untested (Avin-Wittenberg et al., 2018). It is infeasible to spray agricultural fields with these chemicals; however, the use of nanoparticle carriers to deliver such autophagy regulators may be possible, and researchers have trialled such approaches in human cells (Chen et al., 2018; Fu et al., 2018) by inducing responses that prepare the plants for future more severe stresses (Signorelli et al., 2019). There is a potential to use nanoparticles and encapsulated autophagy modulators to deliver mild stress inducers, such as H2O2 and NaCl, or autophagy elicitors, for priming autophagy to prevent susceptibility of crop plants to abiotic stresses. Using chemical elicitors to manipulate autophagy pathways has some disadvantages, such as off-target effects, drug instability and cell permeability (Avin-Wittenberg et al., 2018). However, this approach has some benefits too.
Before using chemical autophagy modulators for agricultural benefits, it is important to investigate their specificity. Recently, Dauphinee et al designed a novel four phase pipeline to identify autophagy-modulating chemicals (Dauphinee et al., 2019). In addition to identifying novel chemical regulators, the pipeline provides in-depth mechanistic understanding of the modulator’s activity and information for optimisation of specificity and potency relevant for agricultural applications (Dauphinee et al., 2019).
The use of chemical modulators of autophagy does not require extensive plant breeding. It also circumvents many of the regulatory and public perception hurdles observed with genetically modified (GM) crops (Avin-Wittenberg et al., 2018).
Is selective autophagy as a promising strategy to improve crop fitness under stress?
In contrast to bulk degradation, autophagy can be targeted to identify and engulfment particular organelles, protein complexes, protein aggregates or pathogens into autophagosomes (Izumi et al., 2017; Wang et al., 2017; Marshall and Vierstra, 2018). This form of autophagy is termed selective autophagy and is defined as autophagy that requires additional autophagic receptors (e.g., NBR1) that help deliver specific cellular cargos (damaged, degraded, misfolded proteins and organelles) to the autophagosome for degradation in the vacuole (Svenning et al., 2014; Michaeli et al., 2016). The combination of bulk degradation with selectivity enables autophagy to mediate precise and context-dependent responses intricately. For example, a healthy cellular mitochondrial population is key to maintaining energy levels. However, misfunctioning mitochondria are a significant ROS source that directly damages cellular components and potentially trigger cell death. As mentioned, the accumulation of ROS triggers autophagy. Suppose this is not sufficient to protect the cell. In that case, selective autophagy pathways are activated that degrade dysfunctional mitochondria (the ROS source), termed ‘Mitophagy’, to mitigate stress and facilitate homeostasis (Kanki et al., 2015; Broda et al., 2018; Ma et al., 2021; Nakamura et al., 2021a,b; Ren et al., 2021). In contrast, bulk turnover of cytoplasmic components occurs in cells during development to remove cell debris and to replenish needed pools of amino acids, sugars, fatty acids and nucleotides during nutrient deprivation; it typically involves the random uptake of cytoplasm into the phagophore (Marshall and Vierstra, 2018; Signorelli et al., 2019). Combining bulk and selective autophagy may provide the versatility required for plants to respond to various stresses of different severities and duration.
Identification of selective autophagy in plants provided insight into how plants target unwanted or damaged organelles for degradation as a housekeeping mechanism and promote nutrient recycling required for plant growth and development. It also revealed new tools for innate immunity by which cargo receptors help eliminate invading pathogens. Selectivity in autophagy is conferred by cargo receptor proteins, which are able to simultaneously interact with the cargo and ATG8 family proteins on the autophagosomal membrane (Zaffagnini and Martens, 2016). In addition to interaction via the ATG8-interacting motif (AIM), recent research has identified an alternative site that promotes interaction between the cargo receptor and ATG8 via a new binding site that exploits ubiquitin-interacting motif (UIM)-like sequences, which interacts with the UIM-docking site (UDS; Marshall et al., 2019). This discovery expands on the complexity of the autophagy system and the pathways involved in selective cargo recruitment. The UDS targets non-functional CDC48/p97 complexes to the ATG8 decorated autophagosomes. CDC48/p97 turnover is vital in eukaryotes because it is essential for endoplasmic reticulum-associated degradation (ERAD) and other protein quality control pathways related to several human diseases (Ye et al., 2001; Tang and Xia, 2016). Therefore, the UDS interface is critical for maintaining eukaryotic proteostasis (Marshall et al., 2019). Several autophagy receptors are present in plants and are extensively reviewed in the literature (Marshall and Vierstra, 2018; Avin-Wittenberg, 2019). The knowledge of selective autophagy mechanisms can help guide the manipulation of autophagy to develop robust, high yielding crops.
Conclusion and Future Directions
Over the past decade, autophagy research has expanded from model plants to crop species. The roles of autophagy in stress response, plant microbial interactions and development are established in multiple plant species. Recently, selective autophagy for the targeted degradation of organelles or protein complexes has gained significant attention. The manipulation of autophagy pathways shows excellent promise for the improvement of crop productivity under challenging environmental conditions. To harness the full potential of selective autophagy in agriculture, however, it is essential to identify and characterise the adapters/receptor proteins or other new compounds that help mediate the process in plants and the signals that act in specific cargo recruitment. We still need clarification on species-specific differences in selective autophagy pathways in crop plants and their responses to stress conditions. This information could elude to essential factors of stress perception and adaptation in plants. The manipulation of autophagy can improve environmental stress tolerance, nutrient remobilisation (i.e., growth and yield), pathogen resistance in the field and mitigate post-harvest losses. Identifying the regulatory mechanics of plant autophagy presents unique and exciting opportunities for plant biologists and agricultural scientists to understand how plant cells perceive stress and improve yield potential and stress tolerance in future predicted inclement environments.
Author Contributions
NT and BW contributed to conception, design, and drafting the manuscript. SB, BF, MK, SM, and BW contributed to the critical revision of the article. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Queensland University of Technology (QUT) scholarship, and an Advance QLD Research Fellowship package AQRF14816-17RD2, AQRF04016-17RD2, and AQRF14716-17RD2.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Asami, P., Mundree, S., and Williams, B. (2018). Saving for a rainy day: control of energy needs in resurrection plants. Plant Sci. 271, 62–66. doi: 10.1016/j.plantsci.2018.03.009
Asami, P., Rupasinghe, T., Moghaddam, L., Njaci, I., Roessner, U., Mundree, S., et al. (2019). Roots of the resurrection plant tripogon loliiformis survive desiccation without the activation of autophagy pathways by maintaining energy reserves. Front. Plant Sci. 10:459. doi: 10.3389/fpls.2019.00459
Avin-Wittenberg, T. (2019). Autophagy and its role in plant abiotic stress management. Plant Cell Environ. 42, 1045–1053. doi: 10.1111/pce.13404
Avin-Wittenberg, T., Baluška, F., Bozhkov, P. V., Elander, P. H., Fernie, A. R., Galili, G., et al. (2018). Autophagy-related approaches for improving nutrient use efficiency and crop yield protection. J. Exp. Bot. 69, 1335–1353. doi: 10.1093/jxb/ery069
Bao, Y. (2020). Links between drought stress and autophagy in plants. Plant Signal. Behav. 15:1779487. doi: 10.1080/15592324.2020.1779487
Bao, Y., and Bassham, D. C. (2020). COST1 balances plant growth and stress tolerance via attenuation of autophagy. Autophagy 16, 1157–1158. doi: 10.1080/15548627.2020.1752981
Bao, Y., Song, W.-M., Wang, P., Yu, X., Li, B., Jiang, C., et al. (2020). COST1 regulates autophagy to control plant drought tolerance. Proc. Natl. Acad. Sci. 117, 7482–7493. doi: 10.1073/pnas.1918539117
Barraza, A., Estrada-Navarrete, G., Rodriguez-Alegria, M. E., Lopez-Munguia, A., Merino, E., Quinto, C., et al. (2013). Down-regulation of PvTRE1 enhances nodule biomass and bacteroid number in the common bean. New Phytol. 197, 194–206. doi: 10.1111/nph.12002
Blair, M. W., Wu, X., Bhandari, D., Zhang, X., and Hao, J. (2016). “Role of legumes for and as horticultural crops in sustainable agriculture,” in Organic Farming for Sustainable Agriculture: Sustainable Development and Biodiversity. Vol 9. ed. D. Nandwani (Cham: Springer).
Borrell, A. K., Hammer, G. L., and Douglas, A. C. (2000a). Does maintaining green leaf area in sorghum improve yield under drought? I. Leaf growth and senescence. Crop. Sci. 40, 1026–1037. doi: 10.2135/cropsci2000.4041026x
Borrell, A. K., Hammer, G. L., and Henzell, R. G. (2000b). Does maintaining green leaf area in sorghum improve yield under drought? II. Dry matter production and yield. Crop. Sci. 40, 1037–1048. doi: 10.2135/cropsci2000.4041037x
Brechenmacher, L., Lei, Z., Libault, M., Findley, S., Sugawara, M., Sadowsky, M. J., et al. (2010). Soybean metabolites regulated in root hairs in response to the symbiotic bacterium Bradyrhizobium japonicum. Plant Physiol. 153, 1808–1822. doi: 10.1104/pp.110.157800
Broda, M., Millar, A. H., and Van Aken, O. (2018). Mitophagy: a mechanism for plant growth and survival. Trends Plant Sci. 23, 434–450. doi: 10.1016/j.tplants.2018.02.010
Chen, F., Hu, Y., Vannozzi, A., Wu, K., Cai, H., Qin, Y., et al. (2017a). The WRKY transcription factor family in model plants and crops. Crit. Rev. Plant Sci. 36, 311–335. doi: 10.1080/07352689.2018.1441103
Chen, L., Su, Z.-Z., Huang, L., Xia, F.-N., Qi, H., Xie, L.-J., et al. (2017b). The AMP-activated protein kinase KIN10 is involved in the regulation of autophagy in Arabidopsis. Front. Plant Sci. 8:1201. doi: 10.3389/fpls.2017.01201
Chen, X., Tong, R., Shi, Z., Yang, B., Liu, H., Ding, S., et al. (2018). MOF nanoparticles with encapsulated autophagy inhibitor in controlled drug delivery system for antitumor. ACS Appl. Mater. Interfaces 10, 2328–2337. doi: 10.1021/acsami.7b16522
Chung, T., Phillips, A. R., and Vierstra, R. D. (2010). ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J. 62, 483–493. doi: 10.1111/j.1365-313X.2010.04166.x
Chung, T., Suttangkakul, A., and Vierstra, R. D. (2009). The ATG autophagic conjugation system in maise: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol. 149, 220–234. doi: 10.1104/pp.108.126714
Costa, M.-C. D., Artur, M. A., Maia, J., Jonkheer, E., Derks, M. F., Nijveen, H., et al. (2017). A footprint of desiccation tolerance in the genome of Xerophyta viscosa. Nat. Plants 3, 1–10. doi: 10.1038/nplants.2017.38
Dauphinee, A. N., Cardoso, C., Dalman, K., Ohlsson, J. A., Fick, S. B., Robert, S., et al. (2019). Chemical screening pipeline for identification of specific plant autophagy modulators. Plant Physiol. 181, 855–866. doi: 10.1104/pp.19.00647
Di Berardino, J., Marmagne, A., Berger, A., Yoshimoto, K., Cueff, G., Chardon, F., et al. (2018). Autophagy controls resource allocation and protein storage accumulation in Arabidopsis seeds. J. Exp. Bot. 69, 1403–1414. doi: 10.1093/jxb/ery012
Doelling, J. H., Walker, J. M., Friedman, E. M., Thompson, A. R., and Vierstra, R. D. (2002). The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277, 33105–33114. doi: 10.1074/jbc.M204630200
Estrada-Navarrete, G., Cruz-Mireles, N., Lascano, R., Alvarado-Affantranger, X., Hernández-Barrera, A., Barraza, A., et al. (2016). An autophagy-related kinase is essential for the symbiotic relationship between Phaseolus vulgaris and both rhizobia and arbuscular mycorrhizal fungi. Plant Cell 28, 2326–2341. doi: 10.1105/tpc.15.01012
Fu, H., Li, X., and Tan, J. (2018). NIPAAm-MMA nanoparticle-encapsulated visnagin ameliorates myocardial ischemia/reperfusion injury through the promotion of autophagy and the inhibition of apoptosis. Oncol. Lett. 15, 4827–4836. doi: 10.3892/ol.2018.7922
Ghiglione, H. O., Gonzalez, F. G., Serrago, R., Maldonado, S. B., Chilcott, C., Curá, J. A., et al. (2008). Autophagy regulated by day length determines the number of fertile florets in wheat. Plant J. 55, 1010–1024. doi: 10.1111/j.1365-313X.2008.03570.x
Gou, W., Li, X., Guo, S., Liu, Y., Li, F., and Xie, Q. (2019). Autophagy in plant: a new orchestrator in the regulation of the phytohormones homeostasis. Int. J. Mol. Sci. 20:2900. doi: 10.3390/ijms20122900
Han, S., Wang, Y., Zheng, X., Jia, Q., Zhao, J., Bai, F., et al. (2015). Cytoplastic glyceraldehyde-3-phosphate dehydrogenases interact with ATG3 to negatively regulate autophagy and immunity in Nicotiana benthamiana. Plant Cell 27, 1316–1331. doi: 10.1105/tpc.114.134692
Hanamata, S., Kurusu, T., and Kuchitsu, K. (2014). Roles of autophagy in male reproductive development in plants. Front. Plant Sci. 5:457. doi: 10.3389/fpls.2014.00457
Hayat, M. (2017). Autophagy: Cancer, Other Pathologies, Inflammation, Immunity, Infection, and Aging. San Diego: Academic Press, 1–71.
Henry, E., Fung, N., Liu, J., Drakakaki, G., and Coaker, G. (2015). Beyond glycolysis: GAPDHs are multi-functional enzymes involved in regulation of ROS, autophagy, and plant immune responses. PLoS Genet. 11:e1005199. doi: 10.1371/journal.pgen.1005199
Hofius, D., Schultz-Larsen, T., Joensen, J., Tsitsigiannis, D. I., Petersen, N. H., Mattsson, O., et al. (2009). Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137, 773–783. doi: 10.1016/j.cell.2009.02.036
Huang, X., Zheng, C., Liu, F., Yang, C., Zheng, P., Lu, X., et al. (2019). Genetic analyses of the Arabidopsis ATG1 kinase complex reveal both kinase-dependent and independent autophagic routes during fixed-carbon starvation. Plant Cell 31, 2973–2995. doi: 10.1105/tpc.19.00066
Izumi, M., Ishida, H., Nakamura, S., and Hidema, J. (2017). Entire photodamaged chloroplasts are transported to the central vacuole by autophagy. Plant Cell 29, 377–394. doi: 10.1105/tpc.16.00637
Kabbage, M., Kessens, R., Bartholomay, L. C., and Williams, B. (2017). The life and death of a plant cell. Annu. Rev. Plant Biol. 68, 375–404. doi: 10.1146/annurev-arplant-043015-111655
Kanki, T., Furukawa, K., and Yamashita, S.-I. (2015). Mitophagy in yeast: molecular mechanisms and physiological role. Biochim. Biophys. Acta 1853, 2756–2765. doi: 10.1016/j.bbamcr.2015.01.005
Kellner, R., De La Concepcion, J. C., Maqbool, A., Kamoun, S., and Dagdas, Y. F. (2017). ATG8 expansion: A driver of selective autophagy diversification? Trends Plant Sci. 22, 204–214. doi: 10.1016/j.tplants.2016.11.015
Kurusu, T., Koyano, T., Hanamata, S., Kubo, T., Noguchi, Y., Yagi, C., et al. (2014). OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development. Autophagy 10, 878–888. doi: 10.4161/auto.28279
Kurusu, T., and Kuchitsu, K. (2017). Autophagy, programmed cell death and reactive oxygen species in sexual reproduction in plants. J. Plant Res. 130, 491–499. doi: 10.1007/s10265-017-0934-4
Kuzuoglu-Ozturk, D., Yalcinkaya, O. C., Akpinar, B. A., Mitou, G., Korkmaz, G., Gozuacik, D., et al. (2012). Autophagy-related gene, TdAtg8, in wild emmer wheat plays a role in drought and osmotic stress response. Planta 236, 1081–1092. doi: 10.1007/s00425-012-1657-3
Lai, Z., Wang, F., Zheng, Z., Fan, B., and Chen, Z. (2011). A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 66, 953–968. doi: 10.1111/j.1365-313X.2011.04553.x
Leary, A. Y., Savage, Z., Tumtas, Y., and Bozkurt, T. O. (2019). Contrasting and emerging roles of autophagy in plant immunity. Curr. Opin. Plant Biol. 52, 46–53. doi: 10.1016/j.pbi.2019.07.002
Lenz, H. D., Haller, E., Melzer, E., Kober, K., Wurster, K., Stahl, M., et al. (2011). Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J. 66, 818–830. doi: 10.1111/j.1365-313X.2011.04546.x
Li, F., Chung, T., Pennington, J. G., Federico, M. L., Kaeppler, H. F., Kaeppler, S. M., et al. (2015a). Autophagic recycling plays a central role in maise nitrogen remobilisation. Plant Cell 27, 1389–1408. doi: 10.1105/tpc.15.00158
Li, S., Fu, Q., Chen, L., Huang, W., and Yu, D. (2011). Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 233, 1237–1252. doi: 10.1007/s00425-011-1375-2
Li, W., Chen, M., Wang, E., Hu, L., Hawkesford, M. J., Zhong, L., et al. (2016a). Genome-wide analysis of autophagy-associated genes in foxtail millet (Setaria italica L.) and characterisation of the function of SiATG8a in conferring tolerance to nitrogen starvation in rice. BMC Genomics 17:797. doi: 10.1186/s12864-016-3113-4
Li, W.-W., Chen, M., Zhong, L., Liu, J.-M., Xu, Z.-S., Li, L.-C., et al. (2015b). Overexpression of the autophagy-related gene SiATG8a from foxtail millet (Setaria italica L.) confers tolerance to both nitrogen starvation and drought stress in Arabidopsis. Biochem. Biophys. Res. Commun. 468, 800–806. doi: 10.1016/j.bbrc.2015.11.035
Li, X., Liu, Q., Feng, H., Deng, J., Zhang, R., Wen, J., et al. (2020). Dehydrin MtCAS31 promotes autophagic degradation under drought stress. Autophagy 16, 862–877. doi: 10.1080/15548627.2019.1643656
Li, Y., Kabbage, M., Liu, W., and Dickman, M. B. (2016b). Aspartyl protease-mediated cleavage of BAG6 is necessary for autophagy and fungal resistance in plants. Plant Cell 28, 233–247. doi: 10.1105/tpc.15.00626
Liao, C.-Y., and Bassham, D. C. (2020). Combating stress: The interplay between hormone signaling and autophagy in plants. J. Exp. Bot. 71, 1723–1733. doi: 10.1093/jxb/erz515
Liu, Y., Schiff, M., Czymmek, K., Tallóczy, Z., Levine, B., and Dinesh-Kumar, S. (2005). Autophagy regulates programmed cell death during the plant innate immune response. Cell 121, 567–577. doi: 10.1016/j.cell.2005.03.007
Liu, Y., Xiong, Y., and Bassham, D. C. (2009). Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 5, 954–963. doi: 10.4161/auto.5.7.9290
Ma, J., Liang, Z., Zhao, J., Wang, P., Ma, W., Mai, K. K., et al. (2021). Friendly mediates membrane depolarization-induced mitophagy in Arabidopsis. Curr. Biol. 31, 1931.e4–1944.e4. doi: 10.1016/j.cub.2021.02.034
Mamun, M., Tang, C., Sun, Y., Islam, M., Liu, P., Wang, X., et al. (2018). Wheat gene TaATG8j contributes to stripe rust resistance. Int. J. Mol. Sci. 19:1666. doi: 10.3390/ijms19061666
Marshall, R. S., Hua, Z., Mali, S., Mcloughlin, F., and Vierstra, R. D. (2019). ATG8-binding UIM proteins define a new class of autophagy adaptors and receptors. Cell 177, 766.e24–781.e24. doi: 10.1016/j.cell.2019.02.009
Marshall, R. S., and Vierstra, R. D. (2018). Autophagy: the master of bulk and selective recycling. Annu. Rev. Plant Biol. 69, 173–208. doi: 10.1146/annurev-arplant-042817-040606
Masclaux-Daubresse, C., Chen, Q., and Havé, M. (2017). Regulation of nutrient recycling via autophagy. Curr. Opin. Plant Biol. 39, 8–17. doi: 10.1016/j.pbi.2017.05.001
Michaeli, S., Galili, G., Genschik, P., Fernie, A. R., and Avin-Wittenberg, T. (2016). Autophagy in plants–what's new on the menu? Trends Plant Sci. 21, 134–144. doi: 10.1016/j.tplants.2015.10.008
Minina, E. A., Moschou, P. N., Vetukuri, R. R., Sanchez-Vera, V., Cardoso, C., LIU, Q., et al. (2018). Transcriptional stimulation of rate-limiting components of the autophagic pathway improves plant fitness. J. Exp. Bot. 69, 1415–1432. doi: 10.1093/jxb/ery010
Nakamura, S., Hagihara, S., and Izumi, M. (2021a). Mitophagy in plants. Biochim. Biophys. Acta Gen. Subj. 1865:129916. doi: 10.1016/j.bbagen.2021.129916
Nakamura, S., Hagihara, S., Otomo, K., Ishida, H., Hidema, J., Nemoto, T., et al. (2021b). Autophagy contributes to the quality control of leaf mitochondria. Plant Cell Physiol. 62, 229–247. doi: 10.1093/pcp/pcaa162
Nolan, T. M., Brennan, B., Yang, M., Chen, J., Zhang, M., Li, Z., et al. (2017). Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev. Cell 41, 33.e7–46.e7. doi: 10.1016/j.devcel.2017.03.013
Okemo, P., Long, H., Cheng, Y., Mundree, S., and Williams, B. (2021). Stachyose triggers apoptotic like cell death in drought sensitive but not resilient plants. Sci. Rep. 11:7099. doi: 10.1038/s41598-021-86559-7
Oliver, M. J., Farrant, J. M., Hilhorst, H. W., Mundree, S., Williams, B., and Bewley, J. D. (2020). Desiccation tolerance: avoiding cellular damage during drying and rehydration. Annu. Rev. Plant Biol. 71, 435–460. doi: 10.1146/annurev-arplant-071219-105542
Pan, Y., Seymour, G. B., Lu, C., Hu, Z., Chen, X., and Chen, G. (2012). An ethylene response factor (ERF5) promoting adaptation to drought and salt tolerance in tomato. Plant Cell Rep. 31, 349–360. doi: 10.1007/s00299-011-1170-3
Pei, D., Zhang, W., Sun, H., Wei, X., Yue, J., and Wang, H. (2014). Identification of autophagy-related genes ATG4 and ATG8 from wheat (Triticum aestivum L.) and profiling of their expression patterns responding to biotic and abiotic stresses. Plant Cell Rep. 33, 1697–1710. doi: 10.1007/s00299-014-1648-x
Pottier, M., Dumont, J., Masclaux-Daubresse, C., and Thomine, S. (2019). Autophagy is essential for optimal translocation of iron to seeds in Arabidopsis. J. Exp. Bot. 70, 859–869. doi: 10.1093/jxb/ery388
Qu, J., Zou, T., and Lin, Z. (2021). The roles of the ubiquitin–proteasome system in the endoplasmic reticulum stress pathway. Int. J. Mol. Sci. 22:1526. doi: 10.3390/ijms22041526
Ren, K., Feng, L., Sun, S., and Zhuang, X. (2021). Plant mitophagy in comparison to mammals: what is still missing? Int. J. Mol. Sci. 22:1236. doi: 10.3390/ijms22031236
Robert, G., Muñoz, N., Melchiorre, M., Sanchez, F., and Lascano, R. (2014). Expression of animal anti-apoptotic gene Ced-9 enhances tolerance during Glycine max L.–Bradyrhizobium japonicum interaction under saline stress but reduces nodule formation. PLoS One 9:e101747. doi: 10.1371/journal.pone.0101747
Signorelli, S., Tarkowski, Ł. P., Van Den Ende, W., and Bassham, D. C. (2019). Linking autophagy to abiotic and biotic stress responses. Trends Plant Sci. 24, 413–430. doi: 10.1016/j.tplants.2019.02.001
Singh, A., Phogat, J., Yadav, A., and Dabur, R. (2021). The dependency of autophagy and ubiquitin proteasome system during skeletal muscle atrophy. Biophys. Rev. 13, 203–219. doi: 10.1007/s12551-021-00789-7
Soto-Burgos, J., and Bassham, D. C. (2017). SnRK1 activates autophagy via the TOR signaling pathway in Arabidopsis thaliana. PLoS One 12:e0182591. doi: 10.1371/journal.pone.0182591
Su, T., Yang, M., Wang, P., Zhao, Y., and Ma, C. (2020). Interplay between the ubiquitin proteasome system and ubiquitin-mediated autophagy in plants. Cell 9:2219. doi: 10.3390/cells9102219
Sun, X., Huo, L., Jia, X., Che, R., Gong, X., Wang, P., et al. (2018a). Overexpression of MdATG18a in apple improves resistance to Diplocarpon mali infection by enhancing antioxidant activity and salicylic acid levels. Hortic. Res. 5, 1–10. doi: 10.1038/s41438-018-0059-5
Sun, X., Jia, X., Huo, L., Che, R., Gong, X., Wang, P., et al. (2018c). MdATG18a overexpression improves tolerance to nitrogen deficiency and regulates anthocyanin accumulation through increased autophagy in transgenic apple. Plant Cell Environ. 41, 469–480. doi: 10.1111/pce.13110
Sun, X., Wang, P., Jia, X., Huo, L., Che, R., and Ma, F. (2018b). Improvement of drought tolerance by overexpressing MdATG18a is mediated by modified antioxidant system and activated autophagy in transgenic apple. Plant Biotechnol. J. 16, 545–557. doi: 10.1111/pbi.12794
Svenning, S., Lamark, T., Krause, K., and Johansen, T. (2014). Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 7, 993–1010. doi: 10.4161/auto.7.9.16389
Takacs-Vellai, K., Vellai, T., Puoti, A., Passannante, M., Wicky, C., Streit, A., et al. (2005). Inactivation of the autophagy gene bec-1 triggers apoptotic cell death in C. elegans. Curr. Biol. 15, 1513–1517. doi: 10.1016/j.cub.2005.07.035
Tang, J., and Bassham, D. C. (2018). Autophagy in crop plants: what's new beyond Arabidopsis? Open Biol. 8:180162. doi: 10.1098/rsob.180162
Tang, W. K., and Xia, D. (2016). Mutations in the human AAA+ chaperone p97 and related diseases. Front. Mol. Biosci. 3:79. doi: 10.3389/fmolb.2016.00079
Thanthrige, N., Jain, S., Bhowmik, S. D., Ferguson, B. J., Kabbage, M., Mundree, S., et al. (2020). Centrality of BAGs in plant PCD, stress responses, and host defense. Trends Plant Sci. 25, 1131–1140. doi: 10.1016/j.tplants.2020.04.012
Thompson, A. R., Doelling, J. H., Suttangkakul, A., and Vierstra, R. D. (2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138, 2097–2110. doi: 10.1104/pp.105.060673
Toyooka, K., Moriyasu, Y., Goto, Y., Takeuchi, M., Fukuda, H., and Matsuoka, K. (2006). Protein aggregates are transported to vacuoles by macroautophagic mechanism in nutrient-starved plant cells. Autophagy 2, 96–106. doi: 10.4161/auto.2.2.2366
Wang, Y., Cai, S., Yin, L., Shi, K., Xia, X., Zhou, Y., et al. (2015). Tomato HsfA1a plays a critical role in plant drought tolerance by activating ATG genes and inducing autophagy. Autophagy 11, 2033–2047. doi: 10.1080/15548627.2015.1098798
Wang, P., Mugume, Y., and Bassham, D. C. (2018). New advances in autophagy in plants: regulation, selectivity and function. Semin. Cell Dev. Biol. 80, 113–122. doi: 10.1016/j.semcdb.2017.07.018
Wang, P., Nolan, T. M., Clark, N. M., Jiang, H., Montes-Serey, C., Guo, H., et al. (2021). The F-box E3 ubiquitin ligase BAF1 mediates the degradation of the brassinosteroid-activated transcription factor BES1 through selective autophagy in Arabidopsis. The Plant Cell koab210. doi: 10.1093/plcell/koab210
Wang, P., Nolan, T. M., Yin, Y., and Bassham, D. C. (2020). Identification of transcription factors that regulate ATG8 expression and autophagy in Arabidopsis. Autophagy 16, 123–139. doi: 10.1080/15548627.2019.1598753
Wang, P., Sun, X., Jia, X., and Ma, F. (2017). Apple autophagy-related protein MdATG3s afford tolerance to multiple abiotic stresses. Plant Sci. 256, 53–64. doi: 10.1016/j.plantsci.2016.12.003
Wang, P., Sun, X., Jia, X., Wang, N., Gong, X., and Ma, F. (2016). Characterisation of an autophagy-related gene MdATG8i from apple. Front. Plant Sci. 7:720. doi: 10.3389/fpls.2016.00720
Wei, Y., Liu, W., Hu, W., Liu, G., Wu, C., Liu, W., et al. (2017). Genome-wide analysis of autophagy-related genes in banana highlights MaATG8s in cell death and autophagy in immune response to Fusarium wilt. Plant Cell Rep. 36, 1237–1250. doi: 10.1007/s00299-017-2149-5
Williams, B., Njaci, I., Moghaddam, L., Long, H., Dickman, M. B., Zhang, X., et al. (2015). Trehalose accumulation triggers autophagy during plant desiccation. PLoS Genet. 11:e1005705. doi: 10.1371/journal.pgen.1005705
Xia, T., Xiao, D., Liu, D., Chai, W., Gong, Q., and Wang, N. N. (2012). Heterologous expression of ATG8c from soybean confers tolerance to nitrogen deficiency and increases yield in Arabidopsis. PLoS One 7:e37217. doi: 10.1371/journal.pone.0037217
Xiong, Y., Contento, A. L., Nguyen, P. Q., and Bassham, D. C. (2007). Degradation of oxidised proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 143, 291–299. doi: 10.1104/pp.106.092106
Xu, G., Wang, S., Han, S., Xie, K., Wang, Y., Li, J., et al. (2017). Plant Bax Inhibitor-1 interacts with ATG6 to regulate autophagy and programmed cell death. Autophagy 13, 1161–1175. doi: 10.1080/15548627.2017.1320633
Yang, C., Shen, W., Yang, L., Sun, Y., Li, X., Lai, M., et al. (2020). HY5-HDA9 module transcriptionally regulates plant autophagy in response to light-to-dark conversion and nitrogen starvation. Mol. Plant 13, 515–531. doi: 10.1016/j.molp.2020.02.011
Ye, Y., Meyer, H. H., and Rapoport, T. A. (2001). The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414, 652–656. doi: 10.1038/414652a
Yoshimoto, K., Hanaoka, H., Sato, S., Kato, T., Tabata, S., Noda, T., et al. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16, 2967–2983. doi: 10.1105/tpc.104.025395
Yoshimoto, K., and Ohsumi, Y. (2018). Unveiling the molecular mechanisms of plant autophagy—from autophagosomes to vacuoles in plants. Plant Cell Physiol. 59, 1337–1344. doi: 10.1093/pcp/pcy112
Zaffagnini, G., and Martens, S. (2016). Mechanisms of selective autophagy. J. Mol. Biol. 428, 1714–1724. doi: 10.1016/j.jmb.2016.02.004
Zeng, H., Xie, Y., Liu, G., Lin, D., He, C., and Shi, H. (2018). Molecular identification of GAPDHs in cassava highlights the antagonism of MeGAPCs and MeATG8s in plant disease resistance against cassava bacterial blight. Plant Mol. Biol. 97, 201–214. doi: 10.1007/s11103-018-0733-x
Zeng, X., Zeng, Z., Liu, C., Yuan, W., Hou, N., Bian, H., et al. (2017). A barley homolog of yeast ATG6 is involved in multiple abiotic stress responses and stress resistance regulation. Plant Physiol. Biochem. 115, 97–106. doi: 10.1016/j.plaphy.2017.03.013
Zheng, P., Wu, J. X., Sahu, S. K., Zeng, H. Y., Huang, L. Q., Liu, Z., et al. (2018). Loss of alkaline ceramidase inhibits autophagy in Arabidopsis and plays an important role during environmental stress response. Plant Cell Environ. 41, 837–849. doi: 10.1111/pce.13148
Zhou, J., Wang, J., Yu, J.-Q., and Chen, Z. (2014). Role and regulation of autophagy in heat stress responses of tomato plants. Front. Plant Sci. 5:174. doi: 10.3389/fpls.2014.00174
Zhu, Y., Wang, B., Phillips, J., Zhang, Z.-N., Du, H., Xu, T., et al. (2015). Global transcriptome analysis reveals acclimation-primed processes involved in the acquisition of desiccation tolerance in Boea hygrometrica. Plant Cell Physiol. 56, 1429–1441. doi: 10.1093/pcp/pcv059
Keywords: autophagy, pathogen resistance, crop improvement, abiotic stress, senescence, programmed cell death, stress response
Citation: Thanthrige N, Bhowmik SD, Ferguson BJ, Kabbage M, Mundree SG and Williams B (2021) Potential Biotechnological Applications of Autophagy for Agriculture. Front. Plant Sci. 12:760407. doi: 10.3389/fpls.2021.760407
Edited by:
Tamar Avin-Wittenberg, Hebrew University of Jerusalem, IsraelReviewed by:
Taijoon Chung, Pusan National University, South KoreaZhibing Lai, Huazhong Agricultural University, China
Ping Wang, Iowa State University, United States
Copyright © 2021 Thanthrige, Bhowmik, Ferguson, Kabbage, Mundree and Williams. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brett Williams, Yi53aWxsaWFtc0BxdXQuZWR1LmF1
 Nipuni Thanthrige
Nipuni Thanthrige Sudipta Das Bhowmik
Sudipta Das Bhowmik Brett J. Ferguson
Brett J. Ferguson Mehdi Kabbage
Mehdi Kabbage Sagadevan G. Mundree
Sagadevan G. Mundree Brett Williams
Brett Williams