
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 22 November 2021
Sec. Plant Bioinformatics
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.760379
This article is part of the Research TopicStructural Bioinformatics and Biophysical Approaches for Understanding the Plant Responses to Biotic and Abiotic StressView all 10 articles
The CONSTANS-LIKE (COL) genes are important signaling component in the photoperiod pathway and flowering regulation pathway. However, people still know little about their role in Brassica napus. To achieve a better understanding of the members of the BnaCOL gene family, reveal their evolutionary relationship and related functions involved in photoperiod regulation, we systematically analyzed the BnaCOL family members in B. napus genome. A total of 33 BnaCOL genes distributed unevenly on 16 chromosomes were identified in B. napus and could be classified into three subfamilies. The same subfamilies have relatively conservative gene structures, three-dimensional protein structures and promoter motifs such as light-responsive cis-elements. The collinearity analysis detected 37 pairs of repetitive genes in B. napus genome. A 67.7% of the BnaCOL genes were lost after B. napus genome polyploidization. In addition, the BnaCOL genes showed different tissue-specific expression patterns. A 81.8% of the BnaCOL genes were mainly expressed in leaves, indicating that they may play a conservative role in leaves. Subsequently, we tested the circadian expression profiles of nine homologous genes that regulate flowering in Arabidopsis. Most BnaCOL genes exhibit several types of circadian rhythms, indicating that these BnaCOL genes are involved in the photoperiod pathway. As such, our research has laid the foundation for understanding the exact role of the BnaCOL family in the growth and development of rapeseed, especially in flowering.
Flowering is an important link in the process of plant reproduction (Fitter and Fitter, 2002). In Arabidopsis thaliana, the photoperiod pathway, vernalization pathway, autonomous pathway and gibberellin pathway constitute a complex genetic network that regulates flowering time (Roux et al., 2006). In Arabidopsis, CONSTANS-like (CO/COL) and FLOWERING LOCUS T (FT) are two important network regulation centers in the photoperiod induction pathway. CO/COL activates the transcription of FT to move the FT protein from the leaf phloem to the shoot apex meristem, thereby promoting plant flowering (Shim et al., 2017). In rice, the Arabidopsis CO/COL homologous gene HEADING DATE 1 (Hd1) appears to be a bifunctional regulator. It induces FT homologous HEADING DATE 3a (Hd3a) gene expression to promote flowering under SD conditions while under LD conditions Hd1 functions as an inhibitor of Hd3a transcription and flowering (Hayama et al., 2003).
CO/COL is an important network center of the photoperiod flowering pathway, integrating together various environmental and internal signals (Shim et al., 2017). Structurally, CO/COL genes contain two conserved domains: a C-terminal CCT domain (also termed CO, CO-like, TOC1) and an N-terminal zinc finger B-box domain (Robson et al., 2001). The B-box domains are found in many kinds of animal proteins, including some transcription factors, ribonucleoprotein and proto-oncogene products (Reddy et al., 1992; Borden, 1998), and it acts as a protein-protein interaction domain in several transcription factors in animals (Borden, 1998). The CCT domain has the function of nuclear localization similar to the yeast HEME ACTIVATOR PROTEIN2 (HAP2) protein and participates in DNA binding (Wenkel et al., 2006). In Arabidopsis, the 17 AtCOL genes can be classified into three subgroups according to the difference in structural domains (Robson et al., 2001): (i) CO, COL1-COL5 form the first subgroup, and they all have two B-box domains and one CCT domain. (ii) The second subgroup consists of COL9-COL15 members. Compared with members in other groups, they have a zinc finger domain in addition to a B-box domain and a CCT domain. (iii) The third subgroup includes COL6-COL8 and COL16 with one B-box and one CCT domain (Robson et al., 2001; Griffiths et al., 2003). However, there are exceptions in structural domains, such as OsH and OsI in rice and HvCO9 in barley, which contain an intron and a CCT domain, but lack the B-box structure (Robson et al., 2001; Griffiths et al., 2003).
The COL gene family have been widely studied in angiosperms, such as monocotyledonous plants (rice, barley, maize, etc.) (Griffiths et al., 2003; Song et al., 2018) and dicotyledonous plants (Arabidopsis, soybean, cotton, tomato, etc.) (Robson et al., 2001; Wu et al., 2014; Cai et al., 2017; Yang et al., 2020). The COL genes function in all developmental stages of plants (Supplementary Table 1). In A. thaliana, AtCOL1-AtCOL2 have less effect on flowering time, but overexpression of AtCOL1 can shorten the cycle of circadian rhythm, and have a certain impact on the light input pathway (Ledger et al., 2001). Both AtCOL3 and AtCOL4 play an inhibitory role in flowering in both LD and SD (Datta et al., 2006; Steinbach, 2019). Besides, AtCOL3 also promotes red light signal transmission, lateral root growth, bud branching and anthocyanin accumulation (Datta et al., 2006), while mutation of AtCOL4 shows increased tolerance to ABA and salt stress (Min et al., 2015). Recently, AtCOL3 and AtCOL13 were found to be co-regulator of hypocotyl elongation under red light (Liu B. et al., 2021). Overexpression of AtCOL5 will bloom early under SD conditions (Hassidim et al., 2009). AtCOL7 not only affects the branching and shade response of A. thaliana (Wang et al., 2013), it is also a key factor linking light perception to auxin homeostasis (Zhang et al., 2014). And AtCOL8 and AtCOL9 transgenic plants flower late under LD conditions (Cheng and Wang, 2005; Takase et al., 2012). In rice, OsCOL3, OsCOL4, OsCOL9, OsCOL10, OsCOL13, OsCOL15, and OsCOL16 act as flowering inhibitor to delay flowering time (Kim et al., 2008; Lee et al., 2010; Liu H. et al., 2016; Sheng et al., 2016; Tan et al., 2016, 2017; Wu et al., 2017, 2018), except Ghd2, which regulates leaf senescence and drought resistance (Liu J. et al., 2016). Furthermore, CO/COL genes are also found to regulate flowering time in potato (González-Schain and Suárez-López, 2008), sugar beets (Chia et al., 2008; Dally et al., 2018), soybean (Wu et al., 2014), sorghum (Yang et al., 2014), and bamboo (Xiao et al., 2018).
Brassica napus is an important and worldwide cultivated oil crop with strong adaptability, wide use and high economic value. Due to the great different cultivation in latitude, longitude and climate, different ecological types of B. napus varieties are needed. Hence, it is also a good plant resource to research flowering pattern and photoperiod rhythm like spring ecotype, winter ecotype and semi-winter ecotype. And the CO/COL genes are important transcription element in photoperiod and flowering regulation pathway. At present, the functions of CO/COL gene family members have been comprehensively studied in the model plant Arabidopsis thaliana, but little is known about CO/COL genes preservation and functional differentiation in B. napus after polyploidy events. In this study, we have identified 33 BnaCOL gene family members and performed bioinformatics analysis on their physical and chemical properties, evolutionary relationships, chromosome location, gene structure, three-dimensional protein structures, cis-acting elements of the promoter, GO annotation enrichment analysis and gene duplication. We also studied the expression patterns of BnaCOL gene family in different tissues and their response to SD or LD light treatment. This study would provide important clues for the functional study of the COL gene family in the Cruciferae plants, and lay a foundation for further exploration of its functional and molecular mechanisms.
The genome sequences, protein sequences and gene annotation files of rapeseed were downloaded from the website (BnPIR)1 (Song et al., 2020). The Markov model of the two domains of CO-like CCT (PF06203) and zinc finger B-box (PF00643) was downloaded from Pfam database2. Using these two Markov models to preliminarily screen the protein sequences of rapeseed on the HMMER software, and the cut-off E-value were set to1e-4, respectively. Subsequently, all candidate proteins were submitted to three online websites, i.e., SMART3, NCBI CDD4 and PFAM (see text footnote 2) to screen out candidate COL proteins with both CCT and B-box conserved domains. The identified COL candidate genes were submitted to the ExPASy website5 for prediction analysis of protein molecular weight (MW) and isoelectric point (pI). And subcellular localization is predicted by WoLF PSORT6.
The chromosome location data of BnaCOLs comes from the BnPIR website (see text footnote 1). And then the MapChart software was used to analyze the distribution of the identified BnaCOLs on rapeseed chromosomes. The results were refined with Adobe Illustrator software.
According to the reported literature (Hu et al., 2018), the protein sequences of COL family members of Arabidopsis, B. oleracea, B. rapa, Capsella rubella, Oryza sativa, Raphanus sativus, and Zea mays were downloaded and Clustal W was used to analyze the COL protein sequences of these plants. In addition, the sequence alignment results were submitted to MEGA 7.0 software, and the neighbor joining method (NJ) was used to construct the evolutionary tree (Saitou and Nei, 1987; Kumar et al., 2016).
The exon and intron structures of COL genes in rapeseed were analyzed by Gene Structure Display Server 2.07 (Hu et al., 2015). The BnaCOL protein sequences were submitted to the MEME software to analyze the conserved domain of genes. Setting the maximum number of motifs to 10, the maximum number of motif amino acids to 20 and the minimum width to 6 and other settings to default. Finally, TBtools software was used to visualize the conserved motifs of BnaCOL proteins.
We submitted 33 protein sequences of BnaCOL to DNAMAN7.0 software for multiple sequence comparison. Subsequently, we used the online website Phyre28 to predict the three-dimensional structure of the protein.
The analysis of intra-species collinearity of BnaCOL genes in B. napus was performed with McScanX software and the relationship was plotted with Circos software. In addition, the collinearity analysis was plotted with Python version of McScanX software.
The 1,500 bp upstream sequences of BnaCOL genes were obtained from B. napus Whole Genome Information Resource Website (see text footnote 1). The online website Plant CARE9 was used to extract homeopathic a components, and then using the online website DSGS to visualize.
To shed light on the function of the BnaCOL genes, we used eggNOG database10 for the gene ontology (GO) annotation analysis. Subsequently, the GO annotation data was processed in TBtools.
At the online website BnTIR: Brassica napus transcriptome information resource11 (Liu D. et al., 2021), we downloaded RNA-seq data of different tissues including roots, cotyledons, leafs, sepals, petals, pollen, buds, siliques, and seeds. The data were submitted to the online tool12 to draw the expression heat map.
The seeds of Zhongshuang 11 were grown in a growth chamber with a temperature of 25°C/18°C, light for 16 h/darkness for 8 h and humidity of 80%. When the seedlings were at the five leaf stage, two different photoperiod treatments were applied: long daylight (LD, 16 h light/8 h dark) and short daylight (SD, 8 h light/16 h dark). We collected the third leaf of these seedlings at 0, 4, 8, 12, 16, 20, and 24 h after photoperiod treatment. Besides, we set up three biological replicates with samples collected. The collected leaves were immediately frozen in liquid nitrogen and then stored in −80°C refrigerator.
Total RNA was extracted from leaves treated with different photoperiodic treatments using polysaccharide polyphenol total RNA extraction kit (Tiangen Biochemical Technology Co., Ltd: DP201101X). The quantity and quality of RNA was determined by an ultramicroscopic spectrophotometer (Thermo Fisher, NanoDrop One). We used a reverse transcription kit to synthesize cDNA and diluted 100 times with ddH2O as templates for subsequent RT-qPCR experiments. Based on the coding sequences of BnaCOL genes, specific primers were designed using online website qPCR Primer Database13. All BnaCOL genes primers were listed in the Supplementary Table 1. SYBR® Premix Ex TaqTM (TaKaRa) was used for the real-time quantitative experiment. In this experiment, three biological replicates were collected and the samples without photoperiod treatment were used as controls. The gene relative expression analysis refers to the 2−ΔΔCt method.
We have identified 33 COL genes in B. napus and named them BnaCOL1-BnaCOL33 (Table 1). Subsequently, the physical and chemical properties of all members were analyzed and predicted. The lengths of the proteins encoded by BnaCOL genes varied from 289 to 414 amino acids, the MW ranged from 31.51 to 46.5 kDa, and the PI ranged from 5.08 to 8.10. The other information about all BnaCOL proteins were list in Table 1, including subcellular location prediction and coding sequence length.
Furthermore, the 33 BnaCOL genes were mapped on chromosomes (Figure 1). These 33 COL genes were distributed unevenly across 16 chromosomes in rapeseed genome and no gene distributed on chromosome A03, A08, and C08. There was only one gene located on chromosome A01, A04, A05, C01, C03, C04, and C06; and two genes on chromosome A06, A09, C05; three genes on chromosome A07, A10, C02, and C09; four genes on chromosome A02 and C07.
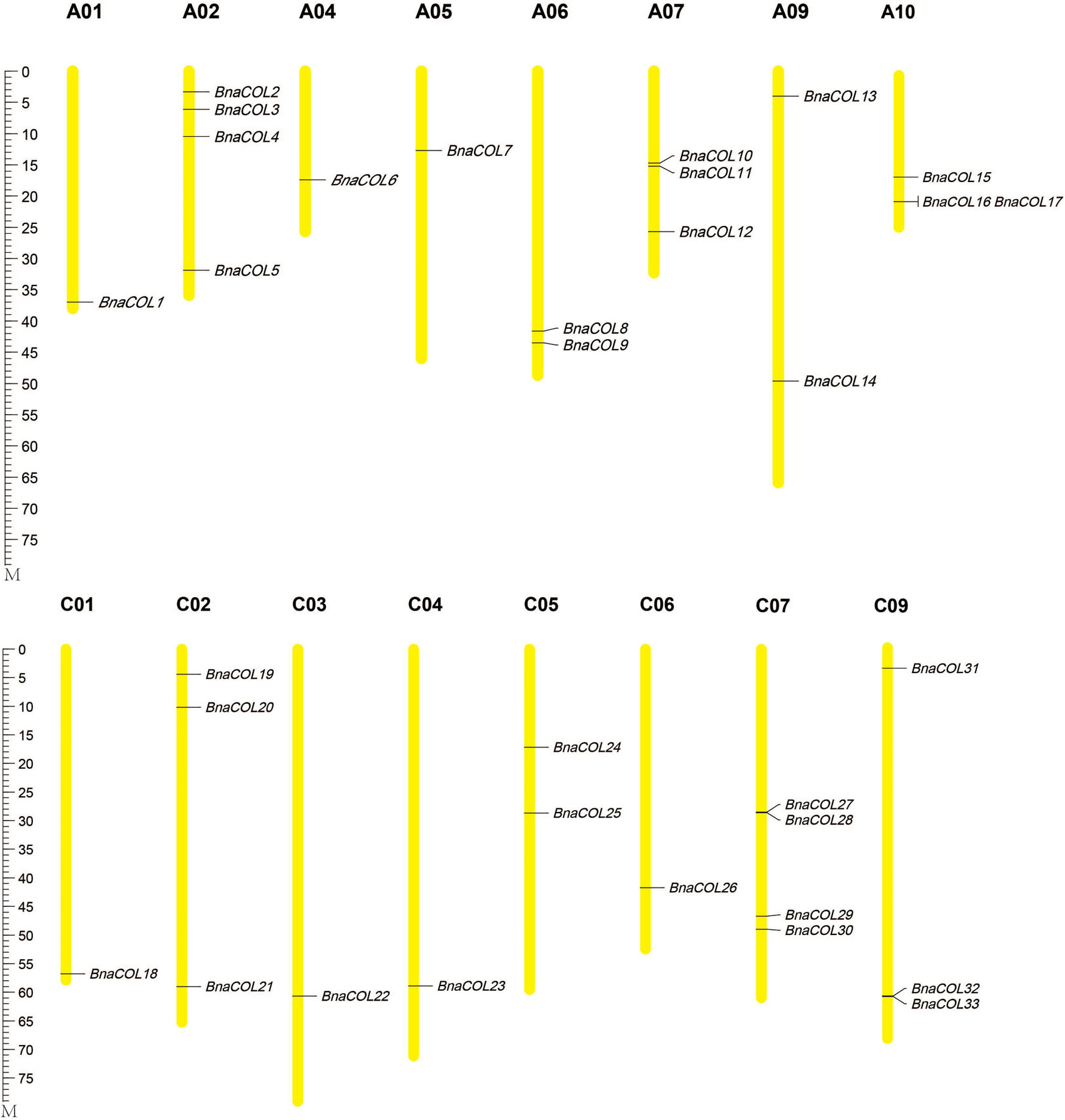
Figure 1. Chromosomal localization of BnaCOL genes in B. napus. The chromosomes with different sizes are represented by the yellow vertical bars with different lengths. The locations of genes are shown from top to bottom.
To gain a better understanding of the evolutionary relationship between the COL genes of different species, we constructed a phylogenetic tree using 137 COL proteins from seven species, including Arabidopsis, B. oleracea, B. rapa, B. nigra, rice, radish, maize (Protein sequences are shown in Supplementary Table 2). As shown in Figure 2A, these COL genes were classified into three groups. The first group consisting of seven species contained the most COL members, while the third group had the least numbers of COL genes. For the BnaCOL genes, there were 17, 5, and 11 members clustered in to the group 1, 2, and 3, respectively (marked with asterisks in Figure 2A). The BnaCOL members which were closely grouped may come from a common origin and have similar functions.
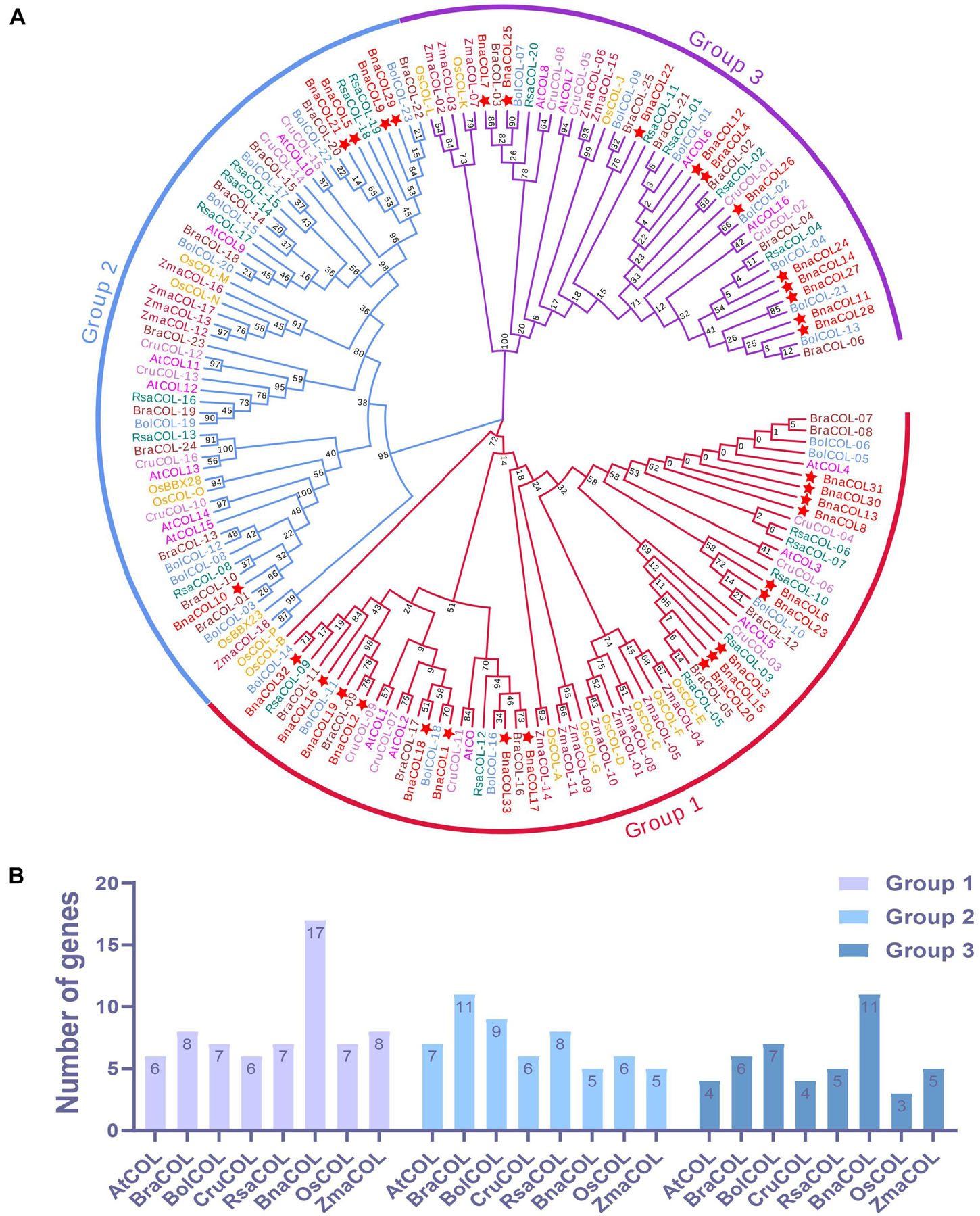
Figure 2. Phylogenetic analysis and number statistics of COL homologs in different species. (A) Phylogenetic tree using 137 COL proteins from 7 species, including rice, maize, Arabidopsis, B. napus, B. oleracea, B. rapa, and radish. The clades of group 1, 2, and 3 are marked in red, blue, and purple, respectively. Among them, BnaCOLs are represented by red five-pointed stars. The abbreviations represent the species as follows: Os, Oryza sativa; Zma, Zea mays; At, Arabidopsis thaliana; Bna, Brassica napus; Bol, Brassica oleracea; Bra, Brassica rapa; Cru, Capsella rubella, and Rsa, Raphanus sativus. (B) Statistics of the number of COL genes in each group. The ordinate is the number of genes and the abscissa is the COL genes of each species.
Subsequently, we counted the number of COL genes of each species in each group (Figure 2B). Based on the number of COL genes in A. thaliana, only about one copy of the COL genes were retained in each group of species. It should be pointed out that the COL genes of B. napus in Group 1 and Group 3 retain about 3 homologous copies.
To investigate the structural diversity of BnaCOL genes, we constructed a phylogenetic tree using 17 AtCOL protein sequences from A. thaliana and 33 BnaCOL protein sequences from B. napus. All of the COL genes were classified into three groups: 1, 2, and 3 (Figure 3A). And then their protein conserved domains and gene structure were further analyzed.
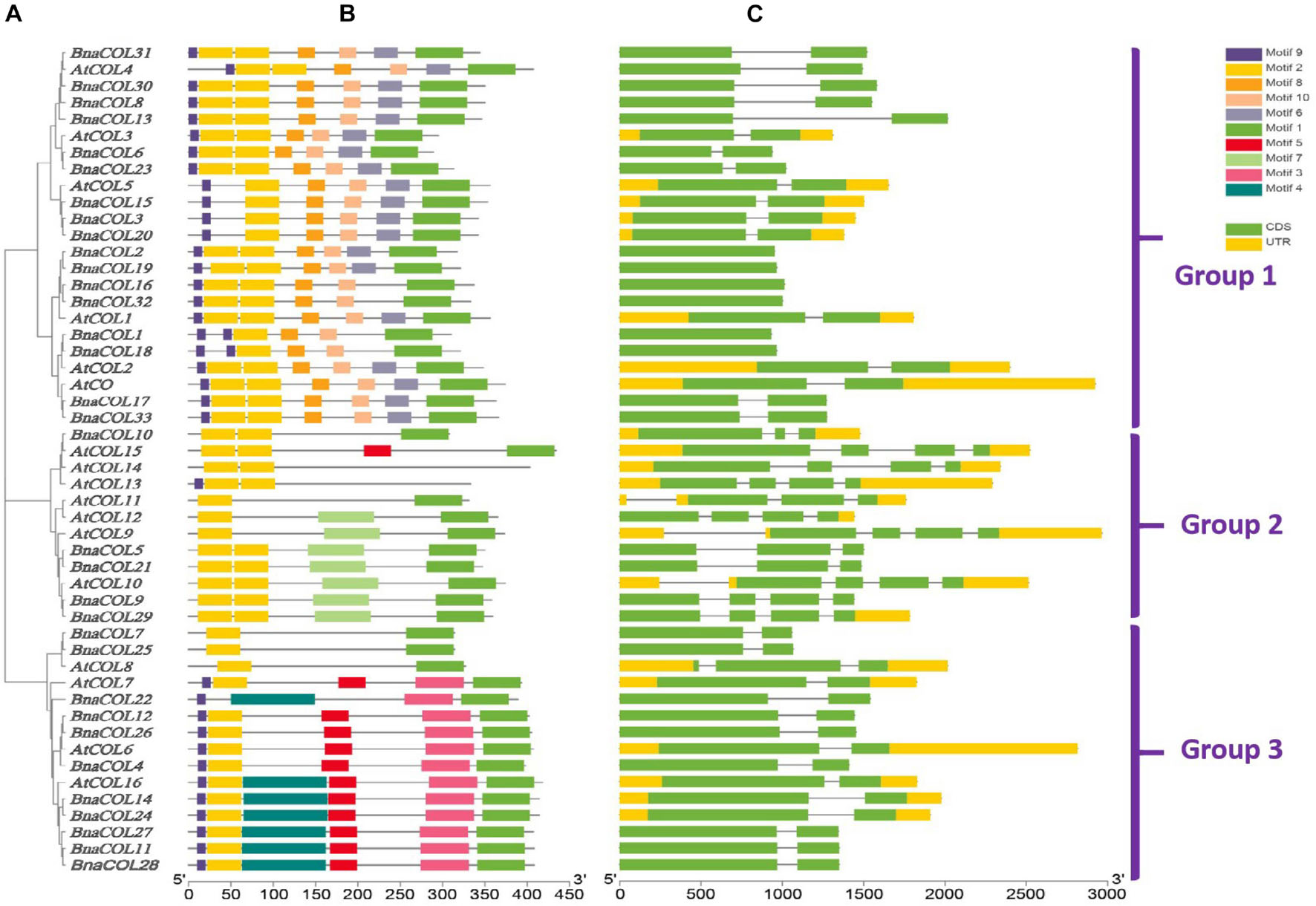
Figure 3. Comparative analysis for conserved domain and gene structure between AtCOL genes and BnaCOL genes. (A) The phylogenetic analysis of 17 AtCOL genes and 33 BnaCOL genes divided them into three groups: 1, 2, and 3. (B) The conserved domains of AtCOL and BnaCOL proteins. Schematic representation of the 10 conserved motifs in BnaCOL proteins, different colored boxes represent different motifs. (C) The gene structure of AtCOL and BnaCOL genes. The green box represents exons, the yellow box represents UTR and the black line represents introns.
The protein conserved domain analysis revealed a total of 10 different conservative motifs (Figure 3B). In general, all members contained motif 1 (CCT domain) and motif 2 (B-box domain), indicating that CCT domain and B-box domain are highly conserved in BnaCOL genes. Besides, similar conserved motifs were found in members of the same group. For example, all members of group 1 contained six motifs: motif 9, motif 2, motif 8, motif 10, motif 6, and motif 1 and the distribution and length of these motifs were consistent. Furthermore, most members of group 2 contained three motifs, among which motif 7 only existed in group 2, while the most BnaCOLs in group 3 contain six motifs, among which motif 5, motif 3 and motif 4 are unique to members of group 3. However, there were slight differences in the number and distribution of COL motifs in different groups. In group 2, four members, i.e., BnaCOL10, AtCOL13, AtCOL14, and AtCOL15 contained conserved motifs different from other members. In group 3, BnaCOL7, BnaCOL25, and AtCOL8 had three fewer motifs than other members.
The gene structure analysis showed that the BnaCOL genes in the same group usually had similar exons and introns (Figure 3C). Both group 1 and 3 contained two exons and one intron. But there was a little difference in the distribution and quantity of exons and introns in group 2.
To elucidate the structural characteristics of BnaCOL proteins, we carried out multiple sequence alignment (Supplementary Figure 1) and three-dimensional structure prediction analysis (Supplementary Figure 2). On the basis of these results we concluded that these proteins have highly conserved CCT and B-box 1 domains, but the sequence of B-box 2 is slightly different.
Then we further predicted the three-dimensional structure of the B-box domain, the results are in good consistent with previous research results (Li et al., 2020). We divided the BnaCOL proteins into three groups according to their genetic relationship. Most of the B-box structure is similar. It is worth noting that group C only contains the predicted B-box 1 domain but not the B-box 2 domain.
Genome wide replication analysis is of great importance for the origin, evolution and genome expansion of species. We hence analyzed the COL gene family replication events in B. napus to understand the causes of BnaCOL genes replication events. The results showed that 37 pairs of large fragment repeat genes were detected (Figure 4) and fragment repeats were found on 17 chromosomes except for A03 and C08. These results indicated that large fragment replication may be a major driving force for the amplification and evolution of COL genes in B. napus genome.
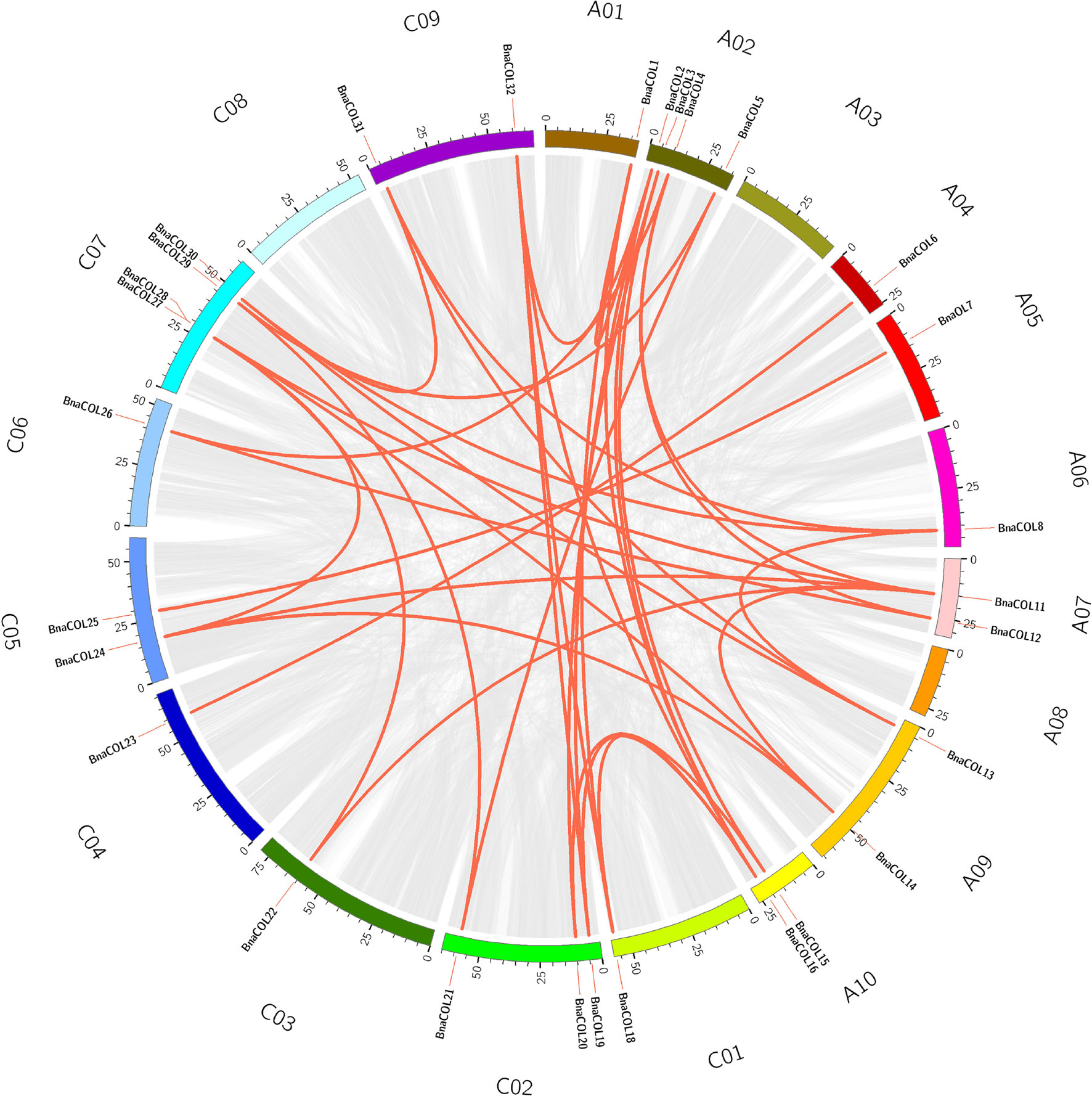
Figure 4. Collinearity analysis of the AC subgenome of COL genes in B. napus. Among them, the gray line represents the replication event of all genes in B. napus and the red line represents tandem repeat events within the BnaCOL genes.
In order to trace the evolutionary process of the COL gene family in Brassica, we analyzed the homologous relationship among Arabidopsis, B. napus (A and C subgenomes), B. rapa (A genome), and B. oleracea (C genome) (Figure 5). The collinearity analysis showed that there were a large number of orthologous COL genes in Arabidopsis, B. rapa, B. oleracea, and B. napus. There were 19 pairs of genes in Arabidopsis and B. rapa that showed collinearity, 13 B. rapa COL genes had homologous genes in Arabidopsis, among which 6 were multi-copy genes and 7 were single-copy genes. In addition, B. rapa lacked homologous genes of AtCOL2, AtCOL7, AtCOL11, and AtCOL14, which indicated that gene loss happened in B. rapa during evolution. Moreover, 17 pairs of genes in Arabidopsis and B. oleracea showed collinearity, and 12 B. oleracea COL genes had homologous genes in Arabidopsis while homologous genes of AtCOL5, AtCOL7, AtCOL11, AtCOL13, and AtCOL14 were not found. The A and C subgenomes of B. napus were mainly collinear with the corresponding diploid B. rapa and B. oleracea. The A genome of B. napus and B. rapa had 16 homologous gene pairs, while 14 homologous gene pairs were found between the C genome of B. napus and B. oleracea. For the evolution of COL gene family, although gene loss occurs, the vast majority of COL genes remain intact in B. napus.
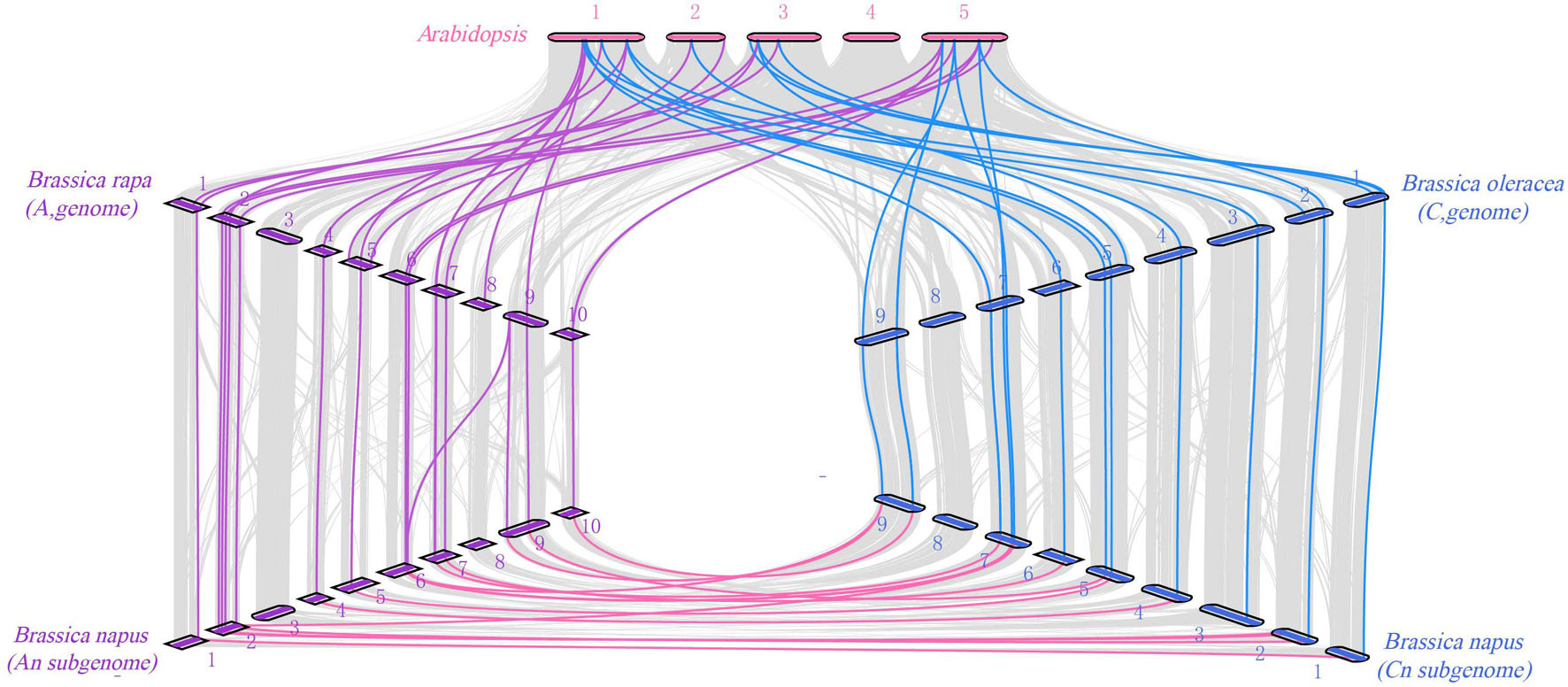
Figure 5. Syntenic relationship of COL genes in B. napus and three ancestral plant species. The figure shows the collinearity between Arabidopsis (A. thaliana), Brassica rape (B. rapa), Brassica oleracea (B. oleracea), and Brassica napus (B. napus).
The cis-acting element is the binding site of transcriptional regulators and regulates gene transcription. In order to investigate the potential function of the BnaCOL genes, we analyzed the cis-acting elements of the upstream sequence of the BnaCOL promoters at 1,500 bp and excluded the elements with unknown function and the general transcriptional regulatory elements (Table 2 and Figure 6). These cis-acting elements can be broadly classified into four categories, which involved in light response, hormonal response, growth regulation, and abiotic-stress response. Among them, the components involved in the light reaction include G-box, GATA-motif, Box4, TCT-motif, ATCT-motif, Circadian, AAAC-motif, AE-box, TCCC-motif, GT1-motif, 3-AF1 binding site and MRE. Hormone response elements include TCA-element, TGACG-motif and CGTCA-motif, ABRE. In addition, several stress response elements such as TC-rich repeats, LTR, MBS were observed. These results showed that most of the BnaCOL genes had photoresponsive elements indicating that BnaCOL genes may played a critical role in the regulation of photoreactivity.
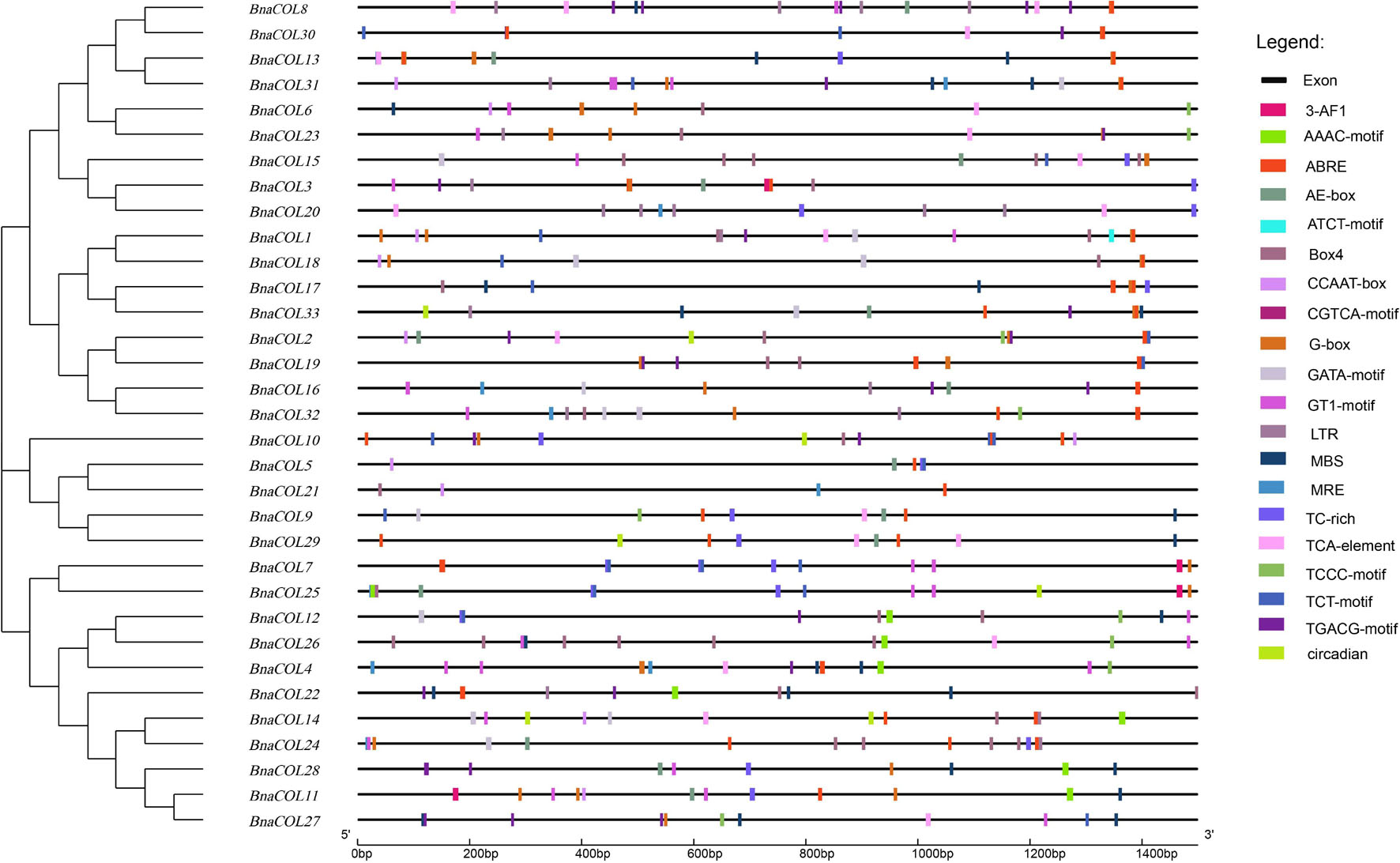
Figure 6. Cis-acting element analysis of COL gene family in B. napus. The boxes with different colors on the black line represent different cis elements.
Taking the above observations in account, we performed GO annotation and enrichment analysis of BnaCOL genes to gain a better understanding of their function. The analysis results mainly included three aspects: biological process (BP), molecular function (MF) and cellular component (CC) (Supplementary Table 4 and Supplementary Figure 3). In the biological process (BP), most genes were annotated in light signal response and transmission, photoperiod response, flowering regulation, circadian rhythm, etc. This is consistent with the observation from the cis-acting element. In the molecular function (MF), a total of 11 highly enriched items were detected, including the combination of DNA, protein and organic compounds and transcription regulator activity. Likewise, in the cellular component (CC), most gene annotations were located on the nucleus and organelles. This indicates a good consistency between the prediction of subcellular location and GO enrichment analysis.
To further study the expression patterns of the BnaCOL genes in different tissues of rapeseed, we used the online website (BnTIR: Brassica napus transcriptome information resource) to download the rapeseed genome-wide transcription data of different tissues. As show in Figure 7 and Supplementary Table 5, all BnaCOL genes showed different expression characteristics in various tissues, which indicated that BnaCOL genes were usually not tissue-specific genes. A 81.8% of BnaCOL genes showed high expression levels in leaves and sepals, while BnaCOL5, BnaCOL9, BnaCOL10, BnaCOL21, and BnaCOL29 were abundantly expressed in pollen and flower buds. Nevertheless, BnaCOL6, BnaCOL7, BnaCOL13, BnaCOL23, and BnaCOL31 were not only highly expressed in leaves and sepals, but also highly detected in siliques. BnaCOL6 and BnaCOL23 expressed the highest levels in siliques. In particular, the expression of BnaCOL22 was lower in other tissues, but highest in seeds. On the basis of these results we concluded that the BnaCOL gene family were critical for all stages of the development of rapeseed individuals and some members with similar expression characteristics may perform similar functions.
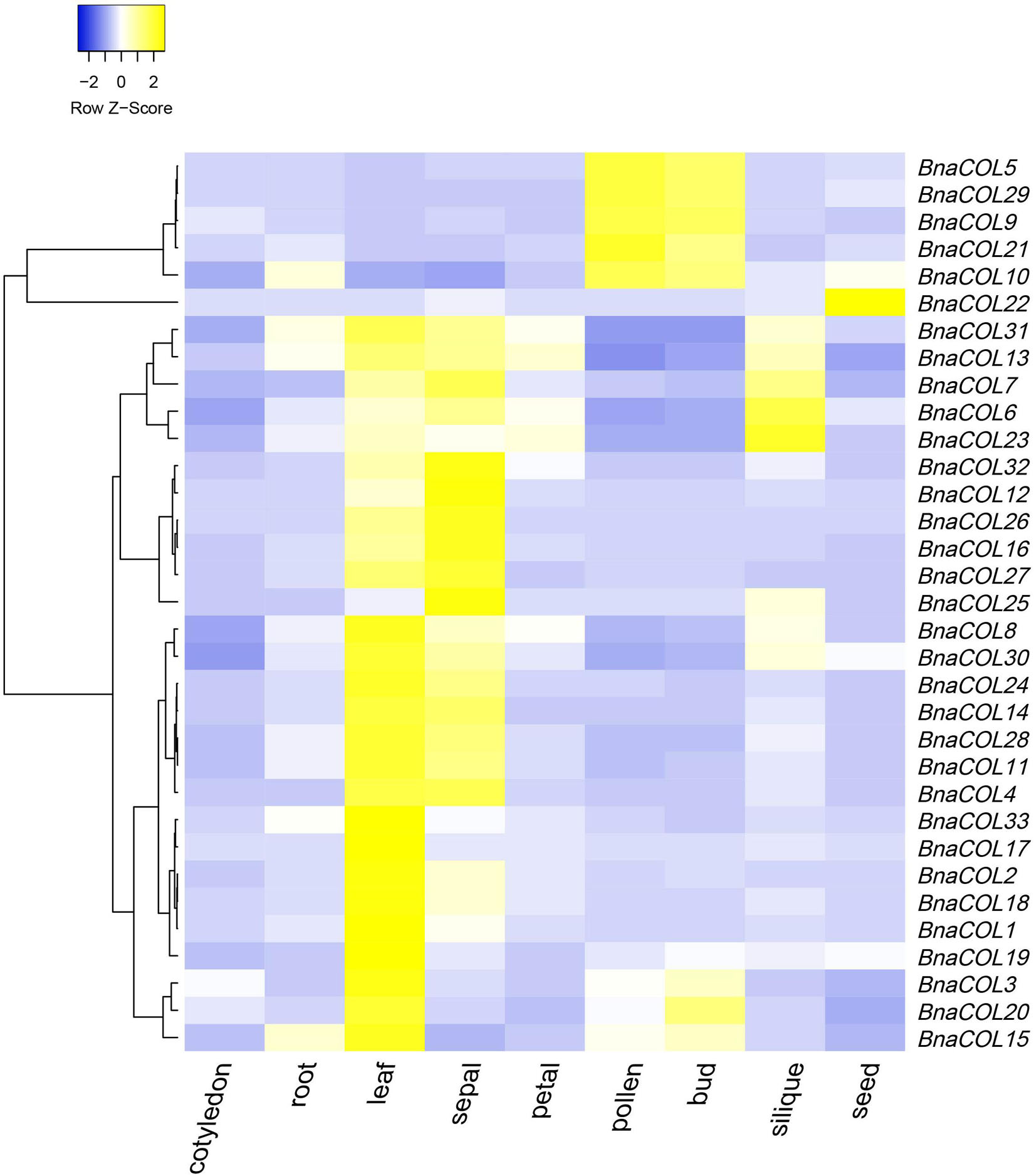
Figure 7. The heat map shows the expression profile of BnaCOL genes in different tissues. The transcriptome data origins from the online website (BnTIR: Brassica napus transcriptome information resource).
Previously, we analyzed the cis-acting elements in the upstream sequence of the BnaCOL promoters and found that most BnaCOL genes have light-responsive elements, indicating that they may involve in photoperiod regulation. To further identify the possible function, we selected nine homologous genes that regulate flowering in Arabidopsis (BnaCOL3, BnaCOL5, BnaCOL11, BnaCOL12, BnaCOL15, BnaCOL16, BnaCOL23, BnaCOL30, BnaCOL33) and tested the circadian expression profile of these nine genes within 24 h (Figures 8, 9).
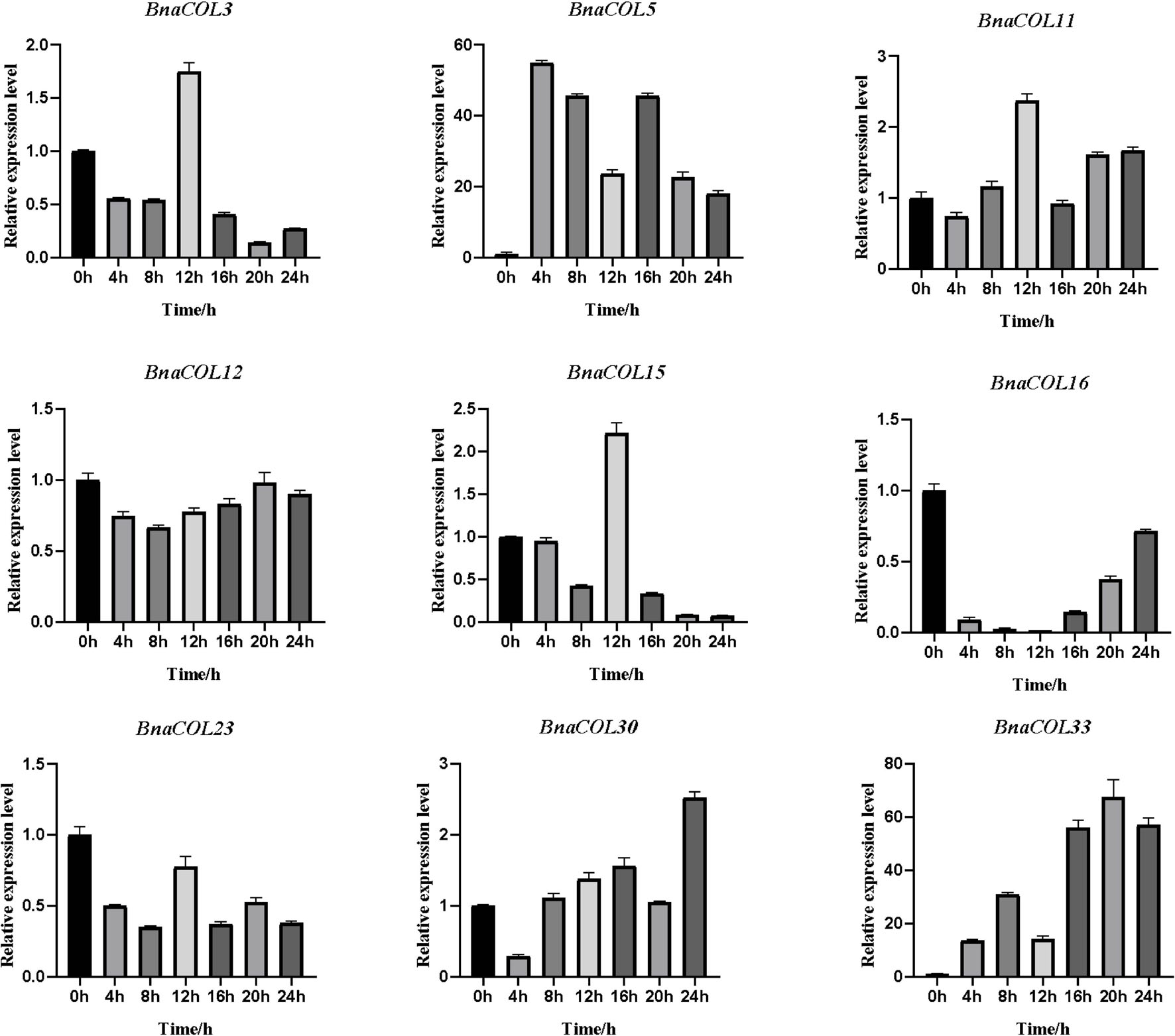
Figure 8. The expression pattern of BnaCOL genes under LD conditions. Under LD (16 h dark/8 h), the third true leaf was collected every 4 h. The error column represents the standard deviation of the three biological replicates.
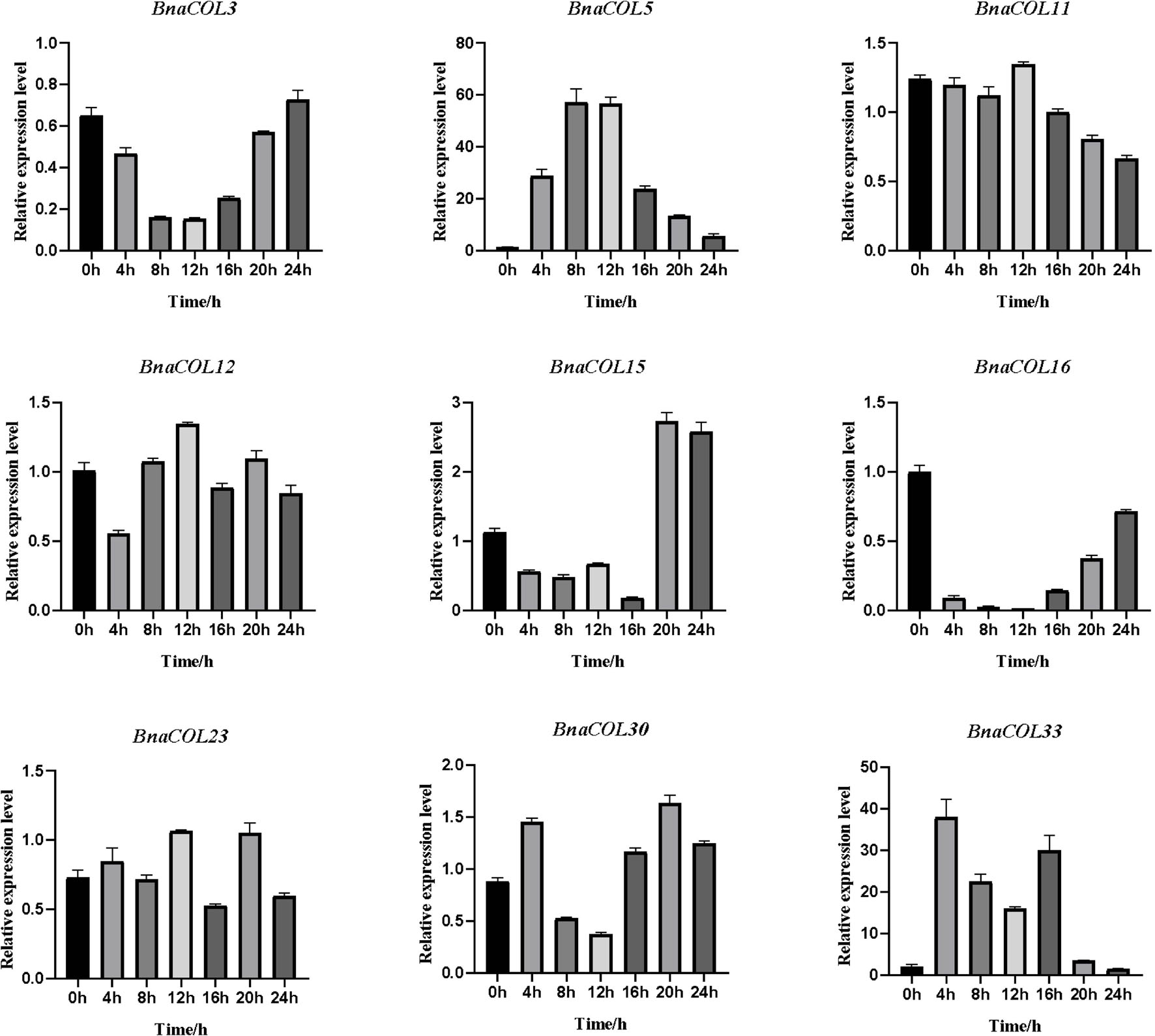
Figure 9. The expression pattern of BnaCOL genes under SD conditions. Under SD (8 h light/16 h dark), the third true leaf was collected every 4 h. The error column represents the standard deviation of the three biological replicates.
The circadian expression pattern of BnaCOL genes under LD illumination showed four types: (i) The expression of BnaCOL3, BnaCOL11, BnaCOL15, and BnaCOL23 increased slowly during illumination, peaked at 12 h and then rapidly decreased to a lower level. (ii) While BnaCOL30 and BnaCOL33 had similar expression patterns, their expression levels presented a stepwise increase pattern. Whether it is day or night, they reach their peak in 20 h or 24 h. (iii) On the contrary, BnaCOL5 was up-regulated before 4 h, then gradually declined. (iv) In particular, the expression level of BnaCOL16 showed light inhibition and dark induction. The expression reached its peak at 0 h and decreased rapidly to a lower level under light conditions, then slowly increased to dark. However, BnaCOL12 had no significant difference in expression level under LD illumination.
The circadian expression pattern of BnaCOL genes under SD illumination showed three types: (i) The expression level of BnaCOL3, BnaCOL16, and BnaCOL30 gradually decreased during the light period, decreased to a minimum at 8 h or 12 h and then slowly rised, showing U-shaped curve. (ii) However, the expression levels of BnaCOL5, BnaCOL11, BnaCOL12, and BnaCOL23 increased slowly from 0 h, reached a peak at 12 h, whereafter gradually dropped down. (iii) It was interesting to find that the expression level of BnaCOL15 was lower during the light period and reached the highest at 20 and 24 h under dark conditions, indicating that the dark treatment can activate the expression of BnaCOL15.
In general, most BnaCOL genes exhibited different diurnal expression patterns, indicating that these COL genes were involved in the photoperiod pathway. However, the expression patterns of BnaCOL12 and BnaCOL16 were similar under LD or SD conditions, indicating that these two genes may not be affected by photoperiod.
In this study, we conducted a biological analysis of the COL gene family of B. napus and related species, including their chromosomal location, phylogenetic analysis, gene structure, protein conserved domain analysis and three-dimensional structure prediction of protein. The results showed that 33 BnaCOL family members were unevenly distributed on 16 chromosomes of rapeseed. Based on phylogenetic analysis, these members were divided into three groups. And most of the genes in the same subgroup had similar gene structure, protein conserved domains and three-dimensional protein structure, which reflects the conservation of the BnaCOL gene family.
Most angiosperms (including monocots and dicots) have experienced whole-genome duplications (WGDs). In the dicotyledon A. thaliana, it has experienced three WGDS events: two ancient tetraploid events (β, α) and one ancient hexaploid (γ) event (Bowers et al., 2003; Tang et al., 2008). And previous studies have shown that the ancestral genomes of Brassica species (similar in structure to A. thaliana) have undergone genome-wide triploid replication events, indicating that they evolved from a common hexaploid ancestor (Lysak et al., 2005; Town et al., 2006). B. napus (A and C genomes) is the amphidiploid species formed by a cross between B. rapa (A genome) and B. oleracea (C genome). As such, the rape genome contains six times the ancestral genome (Parkin et al., 2002). During the evolution of Brassica, it remains question to be explored whether the retention and deletion of flowering and photoperiod genes like COL accompanying with the evolution of Brassica genome.
In the process of polyploidization, copy number variation is an important way of evolution. Taking the number of COL genes in Arabidopsis as a reference, the number of COL genes in B. oleracea, B. rapa, B. nigra, radish, rice and maize in each group only retained roughly one copy. We speculate that the COL genes have basically undergone recombination and fusion during the evolution process. It should be noted that the retention rate of BnaCOL genes in the three groups were quite different. In the first group, 47.2% of the BnaCOL genes were retained. In the second group, only 11.9% of the BnaCOL genes remained. The third group, the BnaCOL genes retention rate was 45.8%. On the basis of our findings, it can be concluded that the AtCOL genes of the first and third groups had about three orthologous genes in B. napus, while the AtCOL genes of the second group only retained one copy in B. napus, indicating that the COL gene family shrank during the diversification of various species. These missing COL genes may be redundant genes, gradually being replaced by other genes with similar functions.
To further explore the evolutionary process of the COL gene family in Brassica, we analyzed the replication events of the COL gene family in B. napus. The results revealed a total of 37 pairs of large-segment repetitive genes existed. We also analyzed the homology relationships among Arabidopsis, B. napus, B. rapa, and B. oleracea. The results of collinearity analysis showed that there were a large number of homologous COL genes in B. napus, Arabidopsis, B. rapa, and B. oleracea. However, B. rapa lacks homologous genes of AtCOL2, AtCOL7, AtCOL11, and AtCOL14, while B. oleracea lacks homologous genes of AtCOL5, AtCOL7, AtCOL11, AtCOL13, and AtCOL14, indicating that gene loss occurred in the process of evolution. This results are basically consistent with the previous phylogenetic tree analysis. As mentioned earlier, there should be six homologous copies of each Arabidopsis gene in B. napus genome. As 17 COL genes were identified in Arabidopsis (Robson et al., 2001). In theory, there should be 102 COL genes in B. napus after whole genome replication, but only 33 BnaCOL genes were identified in this study, indicating that about 67.7% of them were lost after whole genome replication. We hypothesized that during the evolution of B. napus, the COL genes may have undergone strict purification and selection, which play a key role in the maintenance of gene number.
We analyzed expressional pattern of 33 BnaCOL genes in the different tissues using public data. The results showed that they were expressed in different tissues and their expression patterns in different tissues were different. A 81.8% of BnaCOL genes were highly transcribed in leaves and sepals and main expression of five genes were found in pollen and buds (BnaCOL5, BnaCOL9, BnaCOL10, BnaCOL21, and BnaCOL29). BnaCOL6, BnaCOL7, and BnaCOL23 showed higher transcription levels in siliques while BnaCOL22 only highly expressed in seeds but lower in other tissues. These results indicate that the BnaCOL gene family may play a significant role in all stages of rapeseed growth and development. Since 81.8% of BnaCOL genes mainly expressed in leaves, they may have a potential key role in leaves to response to environmental factors like light condition. Previous studies had shown that CO initiated the transcriptional expression of FT, which in turn transferred FT proteins from leaf phloem to shoot tip meristem, thereby activate flowering (Shim et al., 2017). Hence, the high expression of BnaCOL genes help to initiate FT transcription, consequently promote flowering.
The study results of COL genes in Arabidopsis provide reference and clue for the BnaCOL genes function. Previous studies had identified that overexpression of AtCOL1 in Arabidopsis shorten the circadian cycle (Ledger et al., 2001). And AtCOL3, AtCOL4, AtCOL5, AtCOL8, and AtCOL9 were all involved in the regulation of flowering under different photoperiod conditions (Cheng and Wang, 2005; Datta et al., 2006; Hassidim et al., 2009; Takase et al., 2012; Steinbach, 2019). Hence, in order to further explore the role of BnaCOL genes in the photoperiod pathway, we selected nine Arabidopsis homologous genes that regulate flowering time (BnaCOL3, BnaCOL5, BnaCOL11, BnaCOL12, BnaCOL15, BnaCOL16, BnaCOL23, BnaCOL30, and BnaCOL33) and detected the diurnal expression profiles of these nine genes within 24 h under LD and SD illumination treatments. The RT-qPCR results showed that most members had different daily expression patterns between LD and SD conditions, revealing the functional difference and divergence of the BnaCOL family members in B. napus.
The genes of the same evolutionary branch between species are homologous or closely related and their biological functions are roughly the same. Notably, previous studies have shown that AtCOL1 and AtCOL4 transcription levels are regulated by circadian rhythms (Ledger et al., 2001; Steinbach, 2019). Our results showed that BnaCOL16, as a homologous gene of AtCOL1, was expressed at exactly the same level as AtCOL1 in LD and SD. Therefore, it is speculated that BnaCOL16 has similar functions to AtCOL1. Besides, AtCOL4 is not only a flowering inhibitor under LD and SD (Steinbach, 2019), but also participates in ABA and salt stress responses (Min et al., 2015). Our RT-qPCR results showed that BnaCOL30, as a homolog of AtCOL4, had a circadian rhythm expression profile consistent with that of AtCOL4. And the response to abiotic stimuli was also detected in the GO annotation of BnaCOL30 gene. This implied that BnaCOL30 may have the same function as AtCOL4. In the course of evolution, structurally similar genes may sometimes diverge functionally within or between species. Previous studies have shown that overexpression of AtCOL5 makes plants early flowering in SD light conditions (Hassidim et al., 2009). In our study, the expression levels of AtCOL5 and its homologous gene BnaCOL15 were completely opposite under SD light conditions. The BnaCOL15 gene may have functional differentiation and its specific function needs to be verified by subsequent experiments. In our results, the expression level of BnaCOL11 is regulated by the circadian rhythm. But the AtCOL7 gene, which is closely related to BnaCOL11, has only reported the function of regulating the branching and shading response of Arabidopsis thaliana (Wang et al., 2013), as well as linking light perception with auxin (Zhang et al., 2014). In addition, the BnaCOL genes exhibit several types of circadian rhythms, suggesting that functional differences in the BnaCOL family responsive to multiple aspects of plant development, including the regulation of flowering. Although the COL genes are highly conserved among species, it will also undergo functional differentiation in the course of evolution to adapt to environmental changes. The analysis of cis-acting elements in the upstream sequence of BnaCOL promoters revealed that BnaCOL genes contained elements of light response, hormone response, growth regulation and abiotic-stress response. However, only the role of the BnaCOL family in the photoperiod pathway was identified in this study, there are many additional unknown functions of the BnaCOL gene family required to be explored.
In summary, a total of 33 BnaCOL genes were identified in B. napus and these genes were distributed unevenly on 16 chromosomes. The phylogeny, gene structure, conserved motifs and three-dimensional structure of the COL proteins were analyzed. These genes were classified into three subfamilies and relatively conservative gene structures and motifs were found in the same subfamily. In addition, the BnaCOLs promoter region have light-responsive cis-elements, as well as a variety of cis-acting elements related to hormones and abiotic-stress response. Subsequently, GO annotation and enrichment analysis of BnaCOL genes lead us to conclude that most genes are annotated in light signal response and transmission, photoperiod response, flowering regulation, circadian rhythm, etc. The collinearity analysis found 37 pairs of large-segment repetitive genes in B. napus. Based on comparative genomics research, the COL genes of B. napus had undergone polyploidization and different degrees of loss and expansion. We also analyzed the expression patterns of the BnaCOL genes in different tissues of rapeseed, which indicated that the BnaCOL gene family were of great importance at various developmental stages in B. napus. Besides, we tested the diurnal expression profiles of 9 BnaCOL genes under LD and SD conditions. Most members showed different daily expression patterns between LD and SD conditions, revealing the functional differences of the BnaCOL family in B. napus. In general, this article comprehensively analyzed the conservation and divergence of BnaCOL family genes functions, which provided a biological basis for the further functional identification of COL genes in cruciferous plants.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
YC, WW, and JL designed the experiments. YC and RZ performed the experiments. YC, QH, and JL analyzed the data. YC wrote the manuscript. All authors reviewed and approved the manuscript.
This work was supported by the National Key Research and Development Program of China (2018YFE0108000), the Science and Technology Innovation Project of Chinese Academy of Agricultural Sciences (Group No. 118), and the Earmarked Fund for China Agriculture Research System (CARS-12, CAAS-ZDRW202105).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Our deepest gratitude goes to the reviewers for their careful work and thoughtful suggestions that have helped improve this manuscript substantially.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.760379/full#supplementary-material
Supplementary Figure 1 | Multiple sequence alignment of AtCOL and BnaCOL proteins.
Supplementary Figure 2 | Three-dimensional structure prediction of B-box domain of BnaCOL proteins. (A) Two B-box domains from group I. (B) A B-box domain and a second divergent B-box domain from group II. (C) One B-box domain from group III. The images are from dark blue to dark red, indicating from the N end to the C end.
Supplementary Figure 3 | GO annotation of BnaCOL genes in B. napus.
Borden, K. L. (1998). RING fingers and B-boxes: zinc-binding protein-protein interaction domains. Biochem. Cell Biol. 76, 351–358. doi: 10.1139/bcb-76-2-3-351
Bowers, J. E., Chapman, B. A., Rong, J., and Paterson, A. H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 433–438. doi: 10.1038/nature01521
Cai, D., Liu, H., Sang, N., and Huang, X. (2017). Identification and characterization of CONSTANS-like (COL) gene family in upland cotton (Gossypium hirsutum L.). PLoS One 12:e0179038. doi: 10.1371/journal.pone.0179038
Cheng, X. F., and Wang, Z. Y. (2005). Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. Plant J. 43, 758–768. doi: 10.1111/j.1365-313X.2005.02491.x
Chia, T. Y., Müller, A., Jung, C., and Mutasa-Göttgens, E. S. (2008). Sugar beet contains a large CONSTANS-LIKE gene family including a CO homologue that is independent of the early-bolting (B) gene locus. J. Exp. Bot. 59, 2735–2748. doi: 10.1093/jxb/ern129
Dally, N., Eckel, M., Batschauer, A., Höft, N., and Jung, C. (2018). Two CONSTANS-LIKE genes jointly control flowering time in beet. Sci. Rep. 8:16120. doi: 10.1038/s41598-018-34328-4
Datta, S., Hettiarachchi, G. H., Deng, X. W., and Holm, M. (2006). Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 18, 70–84. doi: 10.1105/tpc.105.038182
Fitter, A. H., and Fitter, R. S. (2002). Rapid changes in flowering time in British plants. Science 296, 1689–1691. doi: 10.1126/science.1071617
González-Schain, N. D., and Suárez-López, P. (2008). CONSTANS delays flowering and affects tuber yield in potato. Biol. Plant. 52:251. doi: 10.1111/j.1365-313X.2012.04909.x
Griffiths, S., Dunford, R. P., Coupland, G., and Laurie, D. A. (2003). The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol. 131, 1855–1867. doi: 10.1104/pp.102.016188
Hassidim, M., Harir, Y., Yakir, E., Kron, I., and Green, R. M. (2009). Over-expression of CONSTANS-LIKE 5 can induce flowering in short-day grown Arabidopsis. Planta 230, 481–491. doi: 10.1007/s00425-009-0958-7
Hayama, R., Yokoi, S., Tamaki, S., Yano, M., and Shimamoto, K. (2003). Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422, 719–722. doi: 10.1038/nature01549
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Hu, T., Wei, Q., Wang, W., Hu, H., Mao, W., Zhu, Q., et al. (2018). Genome-wide identification and characterization of CONSTANS-like gene family in radish (Raphanus sativus). PLoS One 13:e0204137. doi: 10.1371/journal.pone.0204137
Kim, S. K., Yun, C. H., Lee, J. H., Jang, Y. H., Park, H. Y., and Kim, J. K. (2008). OsCO3, a CONSTANS-LIKE gene, controls flowering by negatively regulating the expression of FT-like genes under SD conditions in rice. Planta 228, 355–365. doi: 10.1007/s00425-008-0742-0
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Ledger, S., Strayer, C., Ashton, F., Kay, S. A., and Putterill, J. (2001). Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. Plant J. 26, 15–22. doi: 10.1046/j.1365-313x.2001.01003.x
Lee, Y. S., Jeong, D. H., Lee, D. Y., Yi, J., Ryu, C. H., Kim, S. L., et al. (2010). OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB. Plant J. 63, 18–30. doi: 10.1111/j.1365-313X.2010.04226.x
Li, J., Gao, K., Yang, X., Khan, W. U., Guo, B., Guo, T., et al. (2020). Identification and characterization of the CONSTANS-like gene family and its expression profiling under light treatment in Populus. Int. J. Biol. Macromol. 161, 999–1010. doi: 10.1016/j.ijbiomac.2020.06.056
Liu, B., Long, H., Yan, J., Ye, L., Zhang, Q., Chen, H., et al. (2021). A HY5-COL3-COL13 regulatory chain for controlling hypocotyl elongation in Arabidopsis. Plant Cell Environ. 44, 130–142. doi: 10.1111/pce.13899
Liu, D., Yu, L., Wei, L., Yu, P., Wang, J., Zhao, H., et al. (2021). BnTIR: an online transcriptome platform for exploring RNA-seq libraries for oil crop Brassica napus. Plant Biotechnol. J. 19, 1895–1897. doi: 10.1111/pbi.13665
Liu, H., Gu, F., Dong, S., Liu, W., Wang, H., Chen, Z., et al. (2016). CONSTANS-like 9 (COL9) delays the flowering time in Oryza sativa by repressing the Ehd1 pathway. Biochem. Biophys. Res. Commun. 479, 173–178. doi: 10.1016/j.bbrc.2016.09.013
Liu, J., Shen, J., Xu, Y., Li, X., Xiao, J., and Xiong, L. (2016). Ghd2, a CONSTANS-like gene, confers drought sensitivity through regulation of senescence in rice. J. Exp. Bot. 67, 5785–5798. doi: 10.1093/jxb/erw344
Lysak, M. A., Koch, M. A., Pecinka, A., and Schubert, I. (2005). Chromosome triplication found across the tribe Brassiceae. Genome Res. 15, 516–525. doi: 10.1101/gr.3531105
Min, J. H., Chung, J. S., Lee, K. H., and Kim, C. S. (2015). The CONSTANS-like 4 transcription factor, AtCOL4, positively regulates abiotic stress tolerance through an abscisic acid-dependent manner in Arabidopsis. J. Integr. Plant Biol. 57, 313–324. doi: 10.1111/jipb.12246
Parkin, I. A., Lydiate, D. J., and Trick, M. (2002). Assessing the level of collinearity between Arabidopsis thaliana and Brassica napus for A. thaliana chromosome 5. Genome 45, 356–366. doi: 10.1139/g01-160
Reddy, B. A., Etkin, L. D., and Freemont, P. S. (1992). A novel zinc finger coiled-coil domain in a family of nuclear proteins. Trends Biochem. Sci. 17, 344–345. doi: 10.1016/0968-0004(92)90308-v
Robson, F., Costa, M. M., Hepworth, S. R., Vizir, I., Piñeiro, M., Reeves, P. H., et al. (2001). Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 28, 619–631. doi: 10.1046/j.1365-313x.2001.01163.x
Roux, F., Touzet, P., Cuguen, J., and Le Corre, V. (2006). How to be early flowering: an evolutionary perspective. Trends Plant Sci. 11, 375–381. doi: 10.1016/j.tplants.2006.06.006
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Sheng, P., Wu, F., Tan, J., Zhang, H., Ma, W., Chen, L., et al. (2016). A CONSTANS-like transcriptional activator, OsCOL13, functions as a negative regulator of flowering downstream of OsphyB and upstream of Ehd1 in rice. Plant Mol. Biol. 92, 209–222. doi: 10.1007/s11103-016-0506-3
Shim, J. S., Kubota, A., and Imaizumi, T. (2017). Circadian Clock and Photoperiodic Flowering in Arabidopsis: CONSTANS is a hub for signal Integration. Plant Physiol. 173, 5–15. doi: 10.1104/pp.16.01327
Song, J. M., Guan, Z., Hu, J., Guo, C., Yang, Z., Wang, S., et al. (2020). Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat. Plants 6, 34–45. doi: 10.1038/s41477-019-0577-7
Song, N., Xu, Z., Wang, J., Qin, Q., Jiang, H., Si, W., et al. (2018). Genome-wide analysis of maize CONSTANS-LIKE gene family and expression profiling under light/dark and abscisic acid treatment. Gene 673, 1–11. doi: 10.1016/j.gene.2018.06.032
Steinbach, Y. (2019). The Arabidopsis thaliana CONSTANS-LIKE 4 (COL4) - a modulator of flowering time. Front. Plant Sci. 10:651. doi: 10.3389/fpls.2019.00651
Takase, T., Kakikubo, Y., Nakasone, A., Nishiyama, Y., Yasuhara, M., Yoko, T. O., et al. (2012). Characterization and transgenic study of CONSTANS-LIKE8 (COL8) gene in Arabidopsis thaliana: expression of 35S:COL8 delays flowering under long-day conditions. Plant Tissue Cult. Lett. 28, 439–446. doi: 10.5511/plantbiotechnology.11.0823b
Tan, J., Jin, M., Wang, J., Wu, F., Sheng, P., Cheng, Z., et al. (2016). OsCOL10, a CONSTANS-Like Gene, functions as a flowering time repressor downstream of Ghd7 in rice. Plant Cell Physiol. 57, 798–812. doi: 10.1093/pcp/pcw025
Tan, J., Wu, F., and Wan, J. (2017). Flowering time regulation by the CONSTANS-Like gene OsCOL10. Plant Signal. Behav. 12:e1267893. doi: 10.1080/15592324.2016.1267893
Tang, H., Wang, X., Bowers, J. E., Ming, R., Alam, M., and Paterson, A. H. (2008). Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 18, 1944–1954. doi: 10.1101/gr.080978.108
Town, C. D., Cheung, F., Maiti, R., Crabtree, J., Haas, B. J., Wortman, J. R., et al. (2006). Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell 18, 1348–1359. doi: 10.1105/tpc.106.041665
Wang, H., Zhang, Z., Li, H., Zhao, X., Liu, X., Ortiz, M., et al. (2013). CONSTANS-LIKE 7 regulates branching and shade avoidance response in Arabidopsis. J. Exp. Bot. 64, 1017–1024. doi: 10.1093/jxb/ers376
Wenkel, S., Turck, F., Singer, K., Gissot, L., Le Gourrierec, J., Samach, A., et al. (2006). CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18, 2971–2984. doi: 10.1105/tpc.106.043299
Wu, F., Price, B. W., Haider, W., Seufferheld, G., Nelson, R., and Hanzawa, Y. (2014). Functional and evolutionary characterization of the CONSTANS gene family in short-day photoperiodic flowering in soybean. PLoS One 9:e85754. doi: 10.1371/journal.pone.0085754
Wu, W., Zhang, Y., Zhang, M., Zhan, X., Shen, X., Yu, P., et al. (2018). The rice CONSTANS-like protein OsCOL15 suppresses flowering by promoting Ghd7 and repressing RID1. Biochem. Biophys. Res. Commun. 495, 1349–1355. doi: 10.1016/j.bbrc.2017.11.095
Wu, W., Zheng, X. M., Chen, D., Zhang, Y., Ma, W., Zhang, H., et al. (2017). OsCOL16, encoding a CONSTANS-like protein, represses flowering by up-regulating Ghd7 expression in rice. Plant Sci. 260, 60–69. doi: 10.1016/j.plantsci.2017.04.004
Xiao, G., Li, B., Chen, H., Chen, W., Wang, Z., Mao, B., et al. (2018). Overexpression of PvCO1, a bamboo CONSTANS-LIKE gene, delays flowering by reducing expression of the FT gene in transgenic Arabidopsis. BMC Plant Biol. 18:232. doi: 10.1186/s12870-018-1469-0
Yang, S., Weers, B. D., Morishige, D. T., and Mullet, J. E. (2014). CONSTANS is a photoperiod regulated activator of flowering in sorghum. BMC Plant Biol. 14:148. doi: 10.1186/1471-2229-14-148
Yang, T., He, Y., Niu, S., Yan, S., and Zhang, Y. (2020). Identification and characterization of the CONSTANS (CO)/CONSTANS-like (COL) genes related to photoperiodic signaling and flowering in tomato. Plant Sci. 301:110653. doi: 10.1016/j.plantsci.2020.110653
Keywords: Brassica napus, CONSTANS-LIKE (COL) genes, genome-wide analysis, expression pattern, photoperiod
Citation: Chen Y, Zhou R, Hu Q, Wei W and Liu J (2021) Conservation and Divergence of the CONSTANS-Like (COL) Genes Related to Flowering and Circadian Rhythm in Brassica napus. Front. Plant Sci. 12:760379. doi: 10.3389/fpls.2021.760379
Received: 18 August 2021; Accepted: 25 October 2021;
Published: 22 November 2021.
Edited by:
Luis Fernando Saraiva Macedo Timmers, Universidade do Vale do Taquari, BrazilReviewed by:
Shoupu He, State Key Laboratory of Cotton Biology, Institute of Cotton Research, Chinese Academy of Agricultural Sciences (CAAS), ChinaCopyright © 2021 Chen, Zhou, Hu, Wei and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenliang Wei, d2h3ZW5saWFuZ0AxNjMuY29t; Jia Liu, bGl1amlhMDJAY2Fhcy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.