
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 29 November 2021
Sec. Plant Abiotic Stress
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.759245
This article is part of the Research Topic Identification and Functional Dissection of Stress-responsive Genes in Cotton View all 26 articles
In environmental conditions, crop plants are extremely affected by multiple abiotic stresses including salinity, drought, heat, and cold, as well as several biotic stresses such as pests and pathogens. However, salinity, drought, and wilt diseases (e.g., Fusarium and Verticillium) are considered the most destructive environmental stresses to cotton plants. These cause severe growth interruption and yield loss of cotton. Since cotton crops are central contributors to total worldwide fiber production, and also important for oilseed crops, it is essential to improve stress tolerant cultivars to secure future sustainable crop production under adverse environments. Plants have evolved complex mechanisms to respond and acclimate to adverse stress conditions at both physiological and molecular levels. Recent progresses in molecular genetics have delivered new insights into the regulatory network system of plant genes, which generally includes defense of cell membranes and proteins, signaling cascades and transcriptional control, and ion uptake and transport and their relevant biochemical pathways and signal factors. In this review, we mainly summarize recent progress concerning several resistance-related genes of cotton plants in response to abiotic (salt and drought) and biotic (Fusarium and Verticillium wilt) stresses and classify them according to their molecular functions to better understand the genetic network. Moreover, this review proposes that studies of stress related genes will advance the security of cotton yield and production under a changing climate and that these genes should be incorporated in the development of cotton tolerant to salt, drought, and fungal wilt diseases (Verticillium and Fusarium).
Cotton (Gossypium spp.) is one of the most important cellulosic fiber crops, contributing about 35% of total fiber worldwide and also important for oilseed crops. Cotton crops grow well in in the tropics and subtropics in over 80 countries, and cotton is considered the leading crop in about 30 of these countries (Abdelraheem et al., 2019). China is the top cotton fiber producing country. Approximately, 2/3 of cotton fiber derives from China, India, the United States, Pakistan, and Brazil (Khan et al., 2020). According to prediction models for the next 50–100 years, surface temperatures will increase by 3–5°C, radically disturbing agricultural systems worldwide (Solomon et al., 2007). Rising temperatures will increase the frequency of drought, flood, and soil salinization areas, and decrease cultivable land for agriculture. Drought alone is currently reported to affect about 45% of agricultural land worldwide; likewise, about 19.5% of the cultivable agricultural lands are under salinity stress (Dos Reis et al., 2012). Ullah et al. (2017) reported that cotton fiber production affected by drought and heat stress may lead to yield loss of about 34%. Drought and salinity combined may reduce >50% of arable land on average in the next 20 years (Abdelraheem et al., 2019). In addition, crop plants are subjected to various pests and pathogens, such as fungi, viruses, bacteria, nematodes, and herbivorous insects. Fusarium and Verticillium wilt diseases caused by soil-borne fungal pathogens of cotton plants consistently cause extreme yield losses in cotton producing countries including China (Cun et al., 2002; Davis et al., 2006). These two diseases were likely introduced into China in the 1930s, and expanded throughout the main cotton planting areas by the 1970s (Li X. et al., 2017). In the early 1980s, these diseases caused serious problems for cotton production, resulting in >150 thousand tons of lint cotton per year in China (Chen et al., 1985). At present, these two wilt diseases are considered the main impediments for producing quality cotton with sustainable yields in China (Pei et al., 2020).
Cotton plants are exposed to combinations of stress factors in all growth environments. Each stress factor stimulates a complex cellular and molecular network in the crop plants to avoid injury and provide defense, while preserving growth and production (Herms and Mattson, 1992) (Figure 1). To better understand plant responses, Mittler and colleagues proposed a “stress matrix” to identify the complex interactions among multiple abiotic and biotic stresses (Mittler, 2006; Suzuki et al., 2014; Pandey et al., 2017). Stress tolerance genes are activated by a variety of factors such as salinity, drought, heat, cold, and active oxygen balance, and they include membrane permeability, hormone signal transduction, and osmotic regulation (Noctor et al., 2014; Tian et al., 2015). Crop plants trigger a definite and distinctive stress mechanism when exposed to combined stresses (Rizhsky et al., 2004). Multiple stress factors result in overproduction of reactive oxygen species (ROS), such as H2O2, causing widespread cell damage and suppressing photosynthesis. Generally, crop plants use a complex antioxidative defense system to repair or prevent damage through exciting multiple stress-related genes (Ciarmiello et al., 2011) (Figure 1). Genes associated with the antioxidative defense system are divided into three major groups: (1) genes that participate directly in the defense of cell membranes and proteins, such as late embryogenesis abundant (LEA) proteins, heat shock proteins (HSPs) or chaperones, antifreeze proteins, osmoprotectants, free-radical scavengers, and detoxification enzymes (Wang and Jiao, 2000; Vinocur and Altman, 2005); (2) genes intricate in signaling cascades and transcriptional control, e.g., mitogen-activated protein kinase (MAPK), phospholipases, calcium-dependent protein kinase (CDPK), SOS kinase, phospholipases, and transcription factors (TFs) (Frank et al., 2000; Wang et al., 2003; Ludwig et al., 2004; Vinocur and Altman, 2005); and (3) genes that participate in ion uptake and transport (Vinocur and Altman, 2005).
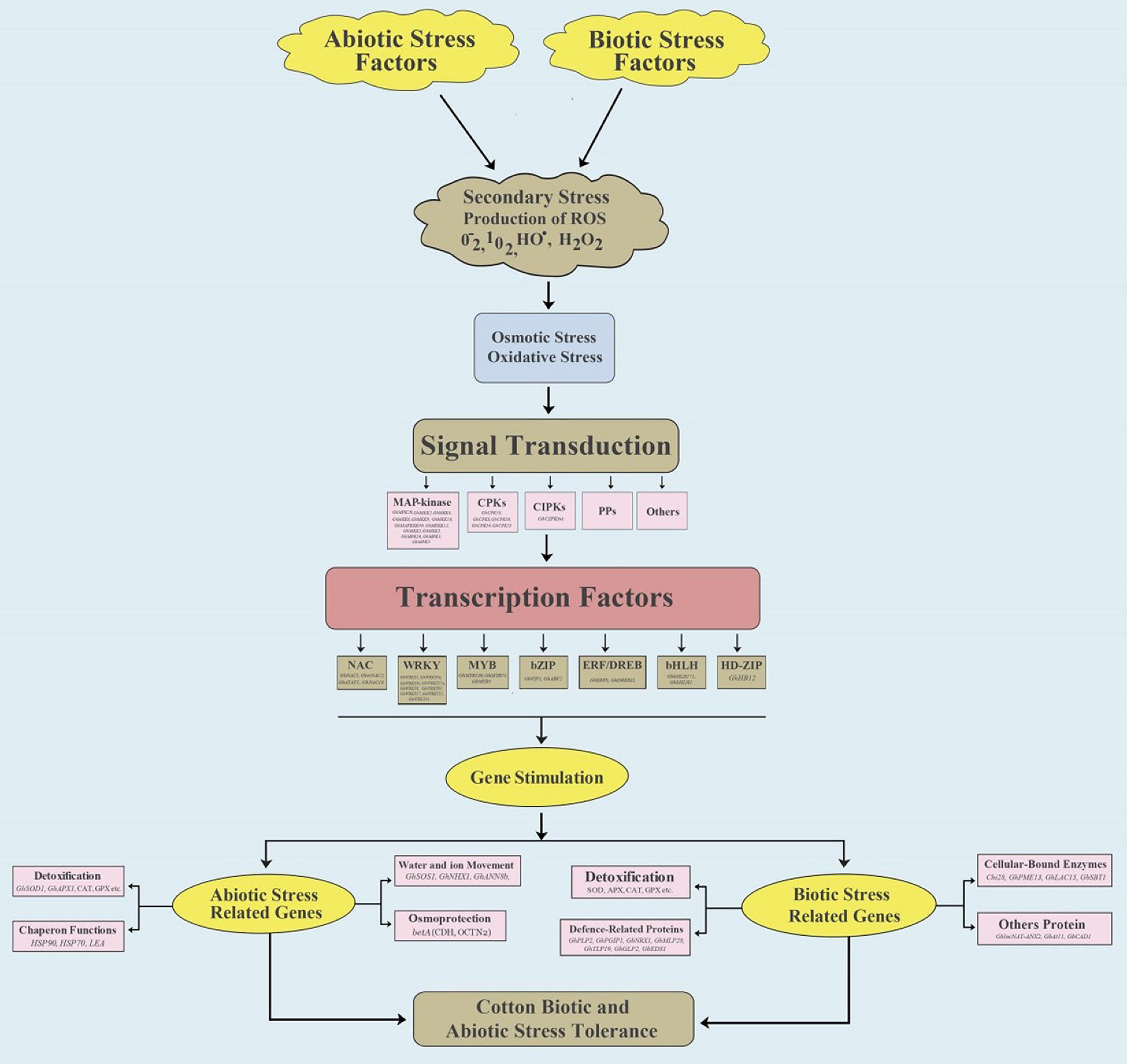
Figure 1. Overall representation of cotton gene regulatory networks in response to abiotic (Salt and Drought) and biotic (Verticillium and Fusarium) stresses. In the MAP-kinase signaling, abiotic stress genes include GhMAPKKK49, GhMEKK12, GhMKK1, GhMKK3, GhMPK16, GhMPK3, and GbMPK3, while biotic stress genes include GhMPK20, GhMKK10, GhMKK9, GhMKK6, GhMKK4, and GhMKK2. In the CPKs family, GhCPK8, GhCPK38, GhCPK54, and GhCPK55 are involved in abiotic stress, and GhCPK33 is involved in biotic stress. In the CIPKs family, GhCIPK6a is involved in abiotic stress. In the transcriptional factors, for abiotic and biotic stress, NAC includes GhirNAC2, GhATAF1, GhNAC18, and GbNAC1, MYB includes GhMYB73, GbMYB5, GhMYB108, and GhMYB108, WRKY includes GhWRKY46, GhWRKY41, GhWRKY27a, GhWRKY6, GhWRKY91, GhWRKY17, GhWRKY25, GhWRKY33, and GbWRKY1, bZIB includes GhABF2 and GbVIP1, ERF/DREB includes GhERF2, GhDREB1L, and GbERFb, and bHLH includes GhbHLH1 and GbbHLH171. HD-ZIP includes GhHB12 for biotic stress.
It is necessary to identify and characterize stress-inducible genes to understand molecular functions as well as generate stress tolerant crops through gene manipulation. In cotton, several genes and gene families have been identified, related to salt stress (Yao et al., 2011; Dongdong et al., 2016; He et al., 2016; Liang et al., 2016; Gao et al., 2018), drought stress (Luo et al., 2013; Dongdong et al., 2016; Liang et al., 2016; Li F. et al., 2017; Wang W. et al., 2019), and wilt diseases of Fusarium (Wang et al., 2009, 2010; Liu N. et al., 2017; Pei et al., 2020; Wang C. et al., 2020) and Verticillium (Li et al., 2014; Xu et al., 2016; Tang et al., 2019; Feng et al., 2021), but their molecular mechanisms are still unknown. Our understanding of advanced molecular mechanisms and signaling pathways of genes in response to biotic and abiotic stresses is still limited, but recent gene discoveries provide a foundation for future research. In this review, we mainly summarize recent studies of cotton genes differentiated according to their molecular functions in response to salt, drought, and Fusarium and Verticillium wilt diseases. The objective of this review is to update knowledge about regulation of cotton genes in response to salt, drought, and wilt diseases and describe recent advances in these stress-response mechanisms.
Mitogen-activated protein kinase cascades are evolutionarily preserved signal transduction pathways involved in transducing extra-cellular cues to the nucleus for proper cellular regulation through phosphorylation of downstream signaling marks into eukaryotic cells. MAPK cascades are divided into three kinases: mitogen activated protein (MAP) kinase kinase kinases (MAPKKKs), MAP kinase kinases (MKKs), and MAPKs. MAPKs are located in the cytoplasm and nucleus and are involved in various cellular processes like growth, development, and multiple stress stimulus (Danquah et al., 2015; Wang et al., 2015). MAPK cascades play multiple roles as both positive and negative regulators in environmental stress (Lin et al., 2021).
In upland cotton (Gossypium hirsutum), 52 GhMAPKs, 23 GhMAPKKs, 166 GhMAPKKKs, and in G. raimondii 28 putative MAPK cascade genes, were identified (Zhang X. et al., 2014; Yin et al., 2021), while very few genes were characterized over the salt and drought stresses.
Dongdong et al. (2016) suggested that cotton GhMAPKKK49 responds to multiple external stresses and may be involved in jasmonic acid (JA), ethyl- ene (ET), salicylic acid (SA), abscisic acid (ABA) and H2O2-mediated signaling pathways. Gene-silenced GhRAF4 and GhMEKK12 cotton plants exhibited decreased drought tolerance by the rapid accumulation of malondialdehyde (MDA), superoxide dismutase (SOD), and peroxidase (POD) (Zhang et al., 2020). Conversely, GhRaf19, a Raf-like MAPKKK gene, controls cotton plant tolerance to drought and salt by reducing cellular ROS (Jia et al., 2016). MAP kinase cascade is reported to phosphorylate and stimulate a key WRKY TF, exposing a regulation module, including GhMAP3K15-GhMKK4-GhMPK6-GhWRKY59-GhDREB2, that has a role in modulating cotton drought resistance (Li F. et al., 2017). GhMKK1 was highly induced by treatment of salt, drought, and H2O2, whereas Overexpression of GhMKK1 in tobacco improved resistance to salinity and drought, which was determined by the regulation of ROS scavenging capability (Lu et al., 2013). Silencing GhMKK3 in cotton resulted in susceptibility to drought stress. In contrast, overexpression of GhMKK3 in N. benthamiana initiated drought resistance by contributing to regulation of ABA-induced stomatal closure and root hair growth (Wang C. et al., 2016).
In addition, GhMKK3 and GhPIP1 act together with GhMPK7 to regulate drought and ABA-activated MAPK elements (Danquah et al., 2015). Therefore, GhMKK5 had a negative role in response to salt and drought stress in transgenic tobacco (Zhang L. et al., 2012). GhMPK16 might be involved in several signal transduction pathways, participating in both biotic and abiotic stress signaling pathways. GhMPK16 exhibited significant resistance to fungi and bacteria in transgenic Arabidopsis, but resulted in sensitivity to drought tolerance and rapid H2O2 accumulation (Shi et al., 2011). In overexpressing transgenic Arabidopsis, GhMPK17 was found to contribute in the plant response to high salinity and osmotic stresses and ABA-mediated signaling pathways (Zhang J. et al., 2014). GbMPK3 may also be a positive regulator of drought tolerance through regulating ROS (Long et al., 2014). Recently, GhMPK3 was identified and characterized from upland cotton. Silencing GhMPK3 increased drought tolerance in cotton plants, whereas overexpression improved plant resistance to drought, cold, and salt stress (Sadau et al., 2021). These findings help in better understanding of the regulatory network of MAPKs under salt and drought stresses, and offer another strategy for improving stress tolerance in cotton crop production.
Transcription factors are crucial in gene expression of plants, including stress-response, hormones, cell division, and organ development. Of the more than 80 TF families, only NAC, MYB, apetala2/ethylene responsive factor (AP2/ERF), basic leucine zipper (bZIP), basic helix-loop-helix (bHLH), WRKY families have been studied to understand their roles in response to salt and drought stresses. Transcription factors either negatively or positively control gene expression, which determines plant survival under environmental stress (Ijaz et al., 2020). Therefore, in order to comprehend the mechanism of stress tolerance, it is critical to investigate the transcription factors involved in regulating gene expression. Few reports about TF genes in cotton have been published but results of available studies have shown that FTs plays an important role in responding to cotton salinity and drought (Evans et al., 2016; Baillo et al., 2019; Debbarma et al., 2019; Li Z. et al., 2019; Zhao et al., 2019).
In total, 495 NAC genes were identified in three cotton species and the evolution and diversity of these genes was explored in cotton plants (Fan et al., 2018). The expression patterns, co-expression network, and transactivation of GhNAC were studied in response to salt and drought stresses (Sun et al., 2018). Overexpression of SNAC1 has a significant role in drought and salt tolerance in cotton through improving root development and decreasing transpiration rates (Liu G. et al., 2014). Both gain and loss of function studies revealed that ABA inducible GhirNAC2 has a positive role in cotton drought tolerance via regulating stomata closure and GhNCED3a/3c expression (Shang et al., 2020). Moreover, GhATAF1, a stress-responsive NAC TF, functions directly in the response to salinity stress with the activation of SA-mediated signaling but suppression of JA-mediated signaling (He et al., 2016). Evans et al. (2016) reported that GhNAC18 was induced in leaf senescence by treatment with various phytohormones including methyl jasmonate (MeJA), salicylic acid (SA), and ethylene (ET) but was down-regulated by abscisic acid (ABA). In addition, GhNAC18 was upregulated by drought but suppressed by high salinity stress.
Myeloblastosis (MYB) TFs are extensive and considered the most functionally diverse gene family of all TFs in plants. MYB TFs work as dynamic regulators, modulating the response of abiotic stress in crop plants. A total of 524 MYB TF encoding genes in G. hirsutum and 205 putative R2R3-MYB genes in G. raimondii have been identified (Salih et al., 2016). However, much less is known about MYB proteins in cotton in response to drought and salt stresses.
GrMYB020, GrMYB074, GrMYB163, GrMYB170, and GrMYB201 exhibited significant increasing patterns under stresses of drought and/or salt, while GrMYB121, GrMYB169, GrMYB 176, GrMYB188, and GrMYB190 were induced in response to salt and drought treatment (Dongdong et al., 2016). GhMYB73 clearly improved tolerance to salt and ABA stress in transgenic Arabidopsis (Zhao et al., 2019). GbMYB5 decreased plant water loss capability by regulating the expression of dehydration-responsive genes in the ABA-mediated signaling pathway, sustained the maintenance of plant cells and proteins by activating biosynthesis of osmolytes and LEA proteins, and efficiently detoxified ROS (Chen T. et al., 2015). Moreover, various physico-chemical characteristics of the GhMYB108-like gene have been described, suggesting that GhMYB108-like is a crucial regulatory gene under drought and salinity stresses (Ullah et al., 2020).
WRKY proteins are about 60 amino acids long and contain one or two highly conserved WRKYGQK motifs as well as a typical zinc-finger structure. They can identify and bind to W-box cis-regulatory elements. WRKY is large TF family of transcriptional regulators, whose members are involved in diverse processes in plants responding to both biotic and abiotic stress. WRKY family genes function in ROS regulation and in mitigating the adverse effects of oxidative stress in cotton and can have positive or negative roles in response to salt and drought stresses. A total of 102, 112, and 109 WRKY genes were, respectively identified in G. hirsutum, G. raimondii, and G. arboreum and their functions were also characterized (Dou et al., 2014; Ding et al., 2015). Overexpression of GhWRKY46 in Arabidopsis improved tolerance to salt and drought by enhancing survival rates, chlorophyll content, and biomass content (Li Y. et al., 2021). Moreover, the constitutive expression of GhWRKY41 in transgenic tobacco plants advances salt and drought tolerance by regulating stomatal closure in ABA-mediated pathways (Chu et al., 2015). Overexpression of GhWRKY27a reduces the drought tolerance of transgenic tobacco plants by enhancing stomatal opening and attenuating expression of ABA (Yan et al., 2015).
Recently, Li Z. et al. (2019) reported that GhWRKY6 overexpression in Arabidopsis enhanced salt and drought sensitivity by regulating stomatal aperture, enriching ROS, reducing proline content, and increasing electrolyte and MDA contents in ABA signaling pathways. A transient dual-luciferase reporter method confirmed that GhWRKY91 stimulated the expression of GhWRKY17 and negatively regulated natural and stress-induced leaf senescence with ABA signals and ROS production (Gu et al., 2019). Overexpression of GhWRKY25 in tobacco decreased the plant’s resistance to drought stress, but improves resistance to salt stress, suggesting that this gene has both positive and negative functions in response to abiotic stresses (Liu et al., 2016). Under drought stress, GhWRKY33 overexpressing transgenic Arabidopsis plants were emaciated much more quickly than wild-type plants because of earlier water loss. Additionally, GhWRKY33 transgenic plants exhibited more tolerance in ABA-mediated media (Wang N.N. et al., 2019).
Basic leucine zipper TF family genes have significant roles in diverse biological processes in response to biotic and abiotic stresses (Alves et al., 2013; Sornaraj et al., 2016). Some bZIP genes have been characterized in crops other than cotton in response to salt and drought stresses. A total of 228 bZIP genes in G. hirsutum, 91 in G. arboreum, and 86 in G. raimondii have been identified (Khanale et al., 2020). GHbZIPs involved in endoplasmic reticulum (ER) stress were supposed to stimulate abiotic stress signals through interaction with other GHbZIPs (Wang et al., 2020b). GhABF2, a bZIP TF significantly enhanced salt and drought stress tolerance in transgenic Arabidopsis and cotton plants, while suppression of GhABF2 modulated transgenic cotton sensitive to drought and salinity stress. Moreover, GhABF2 expression was induced by ABA treatment but was inhibited by high salinity (Liang et al., 2016). ABP9 is another maize bZIP encoding gene that improves salt and drought tolerance by altering physiological and biochemical processes as well as stress-related gene expression, and it may induce the ABA signaling in transgenic cotton (Wang C. et al., 2017).
Ethylene-responsive factor/dehydration-responsive element-binding (ERF/DREB) proteins are in the AP2/ERF (APETALA 2/ethylene-responsive element binding factor) TF family and make up a large TF subfamily, which was first documented in Arabidopsis (Jofuku et al., 1994; Sakuma et al., 2002). The ERF/DREB subfamily contains stress-responsive factors, and several of these genes are participated in both biotic and abiotic stress responses (Zhang et al., 2013). ERF/DREB TFs are essential in ABA-independent signaling pathways, which modulate stress-induced genes (Baillo et al., 2019), to form an inter-connected stress controlling network. Many AP2/ERF genes respond to plant stress hormones such as ABA and to help regulate ABA and ET dependent and independent stress responsive genes (Xie et al., 2019). However, a total of 504 AP2/EREBPs in G. hirsutum and 269 AP2/EREBP genes in G. raimondii (D5) were identified through a genome wide association study (GWAS) (Liu and Zhang, 2017), and their functions in response to abiotic stress in cotton were proposed.
The novel cotton gene GhDREB1L might have a significant role in response to drought and high salinity through binding to the DRE cis-element (Huang et al., 2007). RNA blot evaluation confirmed that the GhDREB gene was induced by high salt, drought, and cold stresses in cotton seedlings. Similarly, GhDREB in transgenic wheat confers promoted tolerance to high salt, drought, and freezing stresses (Gao et al., 2009). Overexpression of StDREB2 in cotton might improve drought tolerance by up-regulating GhERF2, GhDREB1B, GhDREB1A, and antioxidant genes (Baillo et al., 2019). GhDREB40D and GhDREB7A from G. hirsutum have a positive role in responding to drought stress in G. herbaceum. Moreover, GhERF38 acts as a novel regulator and is involved in response to salt and drought stress and ABA signaling by regulating stomatal aperture of guard cells during plant development (Ma et al., 2017).
The basic helix-loop-helix (bHLH) is a functionally diverse group of TFs found in both plants and animals. bHLH TFs have been demonstrated to contribute in regulating several abiotic stresses in plants (Khan et al., 2018), though very little is known about cotton bHLH proteins. Expression of GhbHLH1 in leaves was rapidly induced by ABA and drought (PEG) treatments, suggesting that bHLH may function as a regulator of ABA signaling and drought stress in cotton (Meng et al., 2009). However, these findings suggest that transcriptional regulation of stress-responsive genes is an important step in determining the mechanisms underlying salt and drought stress responses, and that these transcription factors may be key targets for the development of cotton crops with enhanced salt and drought stress tolerance.
The antioxidant system is central to maintaining cell activity in plants by detoxifying ROS under stress conditions. Plants evolved antioxidant defense systems that can not only detoxify ROS but also adjust ROS levels required for proper cell signaling (Gupta et al., 2019). Here we discuss genes involved in enzymatic and non-enzymatic systems for ROS scavenging that regulate the state of detoxification and homeostasis in plant cells in response to salt and drought conditions. These genes annex ascorbate peroxidase (APX), catalase (CAT), glutathione reductase (GR), glutathione peroxidase (GPX), glutathione S-transferase (GST), monodehydroascorbate reductase (MDHAR), dehydroascorbate reductase (DHAR), myo-inositol monooxygenease (MIOX), peroxiredoxin (PRX), proline synthesis, and superoxide dismutase (SOD). The information regarding their functions and regulatory mechanisms in cotton are limited.
Ascorbate peroxidase is required for the first step of the AsA-GSH cycle, which scavenges ROS and protects the plant cell from stress damage (Prashanth et al., 2008). A recent study investigated the role of APX in protecting cellular oxidative homeostasis of stomatal guard cells and in regulating cotton photosynthesis (Guo et al., 2020). In allotetraploid cotton, a total of 26 APX genes were found (Tao et al., 2018). GhAPX8/9/10 is a new APX gene that is not found in rice and Arabidopsis, but its molecular function is still unknown. APX-silenced cotton fibers displayed more sensitivity to oxidative stress than wild-type plants, and the overexpression of GhAPX1 enhanced tolerance of fibers to oxidative stress in cotton (Guo K. et al., 2016). Therefore, simultaneously overexpressing GhSOD1 and GhAPX1 showed no effect to methyl viologen and salt stress (Luo et al., 2013).
Catalases gene family members are considered to be great ROS-scavenging proteins associated with various physiological functions in plant growth, development, and stress responses (Wang W. et al., 2019). As noted for other genes, there is very little known about CAT genes in cotton. A total of seven CAT genes have been identified in the genomes of G. hirsutum and G. barbadense (Wang W. et al., 2019).
Superoxide dismutases mainly convert highly reactive superoxide radicals into hydrogen peroxide and molecular oxygen and are associated with a group of proteins that play an important role in the stress response of plants (Wang et al., 2016a). SODs are designated as the frontline defence within the plant systems against ROS and are categorized by the metal ions that are bound to their active sites such as iron (FeSOD), copper/zinc (Cu/ZnSOD), and manganese (MnSOD). A total of 18 SOD genes have been identified from G. hirsutum, G. raimondii, and G. arboreum (Wang et al., 2016a; Wang W. et al., 2017). In cotton, a previous study revealed that overexpression of SODs improved tolerance to salt stress and oxidizing stress induced by methyl viologen, showing that SODs increased cotton resistance to abiotic stress (Luo et al., 2013).
Peroxidases are involved in various plant physiological systems, including cell elongation, cross-linking of cell wall components, auxin metabolism, lignin and suberin formation, phytoalexin synthesis, defense against biotic or abiotic stress, and metabolism of reactive nitrogen species and ROS from germination to senescence (Marjamaa et al., 2006; Almagro et al., 2009). Duan et al. (2019) identified 198 non-redundant GhPOD genes. A recent expression study on cotton POD genes proposed that they are crucial under high salt stress (Li S. et al., 2020).
The plant glutathione peroxidase (GPX) family includes multiple isoenzymes and has a significant role in ROS hemostasis by catalyzing the decline of H2O2 and other organic hydro-peroxides to defend plant cells under environmental stress responses. A total of 13 putative GPXs from the genome of G. hirsutum (GhGPXs) were identified and the expression patterns of GhGPX transcripts were observed under short-term exposure to salt, osmotic, and ABA-induced stresses to understand their role in these stresses. Additionally, in terms of their role under abiotic stresses, gpx3Δ (H2O2-sensitive mutant) of Saccharomyces cerevisiae was complemented with the GhGPXs, revealing their participation in the oxidative stress response (Chen M. et al., 2017). There are no reports available that provide insights on expression profiling or functional validation of GPXs in cotton under salt and drought stresses.
Glutathione S-transferases are ancient and ubiquitous proteins that are part of a large gene family and have great versatility in organisms. Based on gene association and amino acid sequence the plant GSTs can be divided into four classes, including Phi (F), Tau (U), Lambda (L), and dehydroascorbate reductase (DHAR) (Dong et al., 2016). GST together with Glutathione (GSH) can decrease Peroxidase POX activity through scavenging in the cell. When plants are subjected to abiotic stress, this enzyme is highly induced (Nadarajah, 2020). Over 100 GST genes from maize, soybean, and Arabidopsis have been identified (Pandey P. et al., 2015), with multiple functions such as apoptosis, cellular metabolism, hormone homeostasis, cellular detoxification, and responses to various other biotic and abiotic stresses (Wang L. et al., 2016), but in cotton their roles are limited. Transcriptome analysis of 40 selected GST genes showed tissue-specific expression patterns and salt stress either induced or suppressed their expression levels. These findings provide insight into the function and evolution of the GST gene family in cotton in response to salt stress (Dong et al., 2016). A GST gene (Gst-cr1) from cotton was introduced into tobacco plants, and overexpressing Gst-cr1 exhibited enhanced resistance to oxidative stress induced by methyl viologen (Yu et al., 2003).
Monodehydroascorbate reductase is a key enzyme in ascorbate-glutathione recycling that regulates ascorbic acid (AsA)-mediated reduction/oxidation (redox) regulation and thus plays critical roles in plant cell growth, development, and physiological and molecular responses to environmental stress (Zhou et al., 2021). The MDHAR defensive system protects plant cells against oxidative stress damages (Gill and Tuteja, 2010). In cotton, information regarding MDHAR function and regulatory mechanisms in response to abiotic stress is limited. The identification and universal bioinformatic analysis of 36 MDHAR family genes in G. hirsutum, G. arboreum, G. raimondii, and G. barbadense were conducted. GhMDHAR expression pattern analysis in different cotton tissues, as well as under abiotic stress and phytohormone treatment, revealed a diverse of expression features (Zhou et al., 2021). These findings provide a comprehensive understanding of cotton plant antioxidant gene families and lay the foundations for decoding the molecular mechanisms of these genes in response to salt and drought stress.
Calcium (Ca2+) is a universal secondary messenger in cell signal transduction pathways that functions directly in physiological and molecular processes. Ephemeral alterations of the cytoplasmic Ca2+ level in response to multiple stresses are recognized and interpreted by various Ca2+ sensors or Ca2+ binding proteins, which pass the signals into downstream response processes such as stimulation of gene expression patterns and phosphorylation cascades (Tuteja and Mahajan, 2007). The regulation of gene expression by calcium is critical for plant defense against abiotic stress. Transient changes in cytoplasmic Ca2+ levels have been demonstrated in response to salinity, drought, cold, wounding, and pathogens (Hetherington and Brownlee, 2004). Plant Ca2+ binding proteins can be divided into four classes: calcium-dependent protein kinases (CDPK), calcineurin B-like proteins (CBL), calmodulins (CaM), and calmodulin-like proteins (CaML) (Cheng et al., 2002; Kolukisaoglu et al., 2004).
Among them, CDPKs are the best categorized and are of specific interest to cotton plants, which comprise a large multi-gene family and their roles in response to various stresses have been described. CDPKs from different plants have been identified, and their regulatory mechanisms in plant development or stress responses have been investigated (Nadarajah, 2020), but again, little is known about responses to salt and drought stresses in cotton plants. A total of 98 predicted CDPK genes from G. hirsutum and 41 from G. raimondii were identified (Liu W. et al., 2014; Gao et al., 2018). An earlier study proposed that CDPK gene expression in response to various abiotic stresses would be useful for identifying GhCDPKs, which may have important roles in cotton adaptation to abiotic stresses (Li et al., 2015). A total of 19 CPKs were identified for their rapid transcriptional responses to salt stress, the majority of which were also induced by ethephon, indicating that the salinity and ethylene responses overlapped. Moreover, silencing of four CPK genes (GhCPK8, GhCPK38, GhCPK54, and GhCPK55) severely decreased tolerance to salt stress (Gao et al., 2018), which suggested that the sensing and regulatory network of CPKs in cotton are involved in the response to salt stress. CPK11 from Arabidopsis has recently been shown to phosphorylate drought-induced protein 19 (GhDi19–1 and GhDi19–2) in cotton plants (Qin et al., 2016).
Calcineurin B-like protein-interacting protein kinase (CIPK) is modulated by calcineurin B-like protein (CBL) and is an important component of Ca2+ signal transduction with a significant role in plant abiotic stress. CIPKs are well documented to act as vital elements in plant salt and drought stress signaling pathways (Pandey G.K. et al., 2015; Luo et al., 2017). Through genome wide analysis, in G. hirsutum and G. barbadense, a total of 80 and 78 CIPK genes were identified, respectively (Cui et al., 2020). The analysis of CIPK transcriptome sequence data under abiotic stresses (drought, salt, and low temperature) in different tissues at the trefoil stage revealed that these stresses induced CIPK expression in cotton (Wang J. et al., 2016). GhCIPK6a overexpression lines demonstrated higher salt tolerance, which was achieved through involvement in ROS scavenging and MAPK pathways (Su et al., 2020). Thus, Ca2+ transporters and binding proteins have the potential to be used in the cotton regulatory network and breeding to improve stress-tolerance.
Plant Na+/H+ antiporters (NHXs) are membrane transporter proteins that regulate cellular Na+/K+ and pH homeostasis levels. In plant development and salt stress response, considerable evidence has underlined the crucial roles of the NHX family (Bassil and Blumwald, 2014). Physiological and molecular evidence showed that co-overexpression of AVP1 and AtNHX1 in transgenic cotton further improves drought and salt tolerance (Shen et al., 2015). GhSOS1 is a plasma membrane Na + /H + antiporter gene that improves salt tolerance in transgenic Arabidopsis by increasing the expression of stress-related marker genes, while silencing GhSOS1 reduced cotton tolerance to salt (Chen X. et al., 2017). In a recent study, a vacuolar localized protein, GhNHX1, was induced by salt stress in cotton and loss of function of GhNHX1 showed enhanced sensitivity in cotton seedlings to high salt concentrations. This finding suggests that GhNHX1 positively regulates salt stress to cotton (Long et al., 2020). PYR/PYL/RCAR proteins are putative ABA receptors with vital roles in both plant biotic and abiotic stresses (Santiago et al., 2009).
Annexins are a multi-gene family that is highly conserved in plants, animals, and fungi (Davies, 2014) whose members mediate calcium transport and Ca2+ conductance in plant cells (Demidchik et al., 2018). Little is known about phosphatases that dephosphorylate annexins, especially during salinity stress-induced Ca2+ influx. A cotton phosphatase GhDsPTP3a and an annexin protein GhANN8b interact and conversely regulate Ca2+ and Na+ fluxes under salinity stress in cotton (Mu et al., 2019). Most of the controlling mechanisms of auxin are regulated by auxin-responsive genes, which are mainly involved in homeostasis based on catalytic activities. These genes are divided into three categories: Aux/IAA, SAUR, and GH3 (Goda et al., 2004). However, few studies on these genes in relation to environmental stresses have been conducted (Yuan et al., 2013). The functional characterization of Gh A08G1120 (GH3.5) plants using virus-induced gene silencing (VIGS) revealed that silenced plants were more sensitive to drought and salt stresses than wild types (Kirungu et al., 2019). In upland cotton, a total of 27 predicted PYL proteins were identified. Overexpression of GhPYL10, GhPYL12, and GhPYL26 in Arabidopsis increase sensitivity to ABA but confer tolerance to drought stress in transgenic plants (Chen Y. et al., 2017).
Detoxification efflux carriers (DTX)/multidrug and toxic compound extrusion (MATE) transporters are important in the translocation of ABA, a phytohormone with functions in plants under multiple abiotic stresses (Tiwari et al., 2014). Gh_D06G0281 (DTX/MATE) overexpressing Arabidopsis improved tolerance to salt, drought, and cold stress with a high level of antioxidant enzyme production and significantly lower levels of oxidant (Lu et al., 2019). Additionally, recent genome wide studies of PIN-FORMED (PIN), respiratory burst oxidase homolog (Ghrboh), Valine-glutamine (VQ) gene family, heat shock protein 20 (Hsp20), at-hook motif containing nuclear localized (AHL), leaf senescence, protein phosphatases (PP2C), 9-cisepoxycarotenoiddioxygenase (NCED), and Myo-inositol oxygenase (MIOX) have laid a foundation to predict the regulatory network of cotton genes in response to various abiotic stresses (Ma et al., 2016; He et al., 2017; Elasad et al., 2018; Shazadee et al., 2019; Chen et al., 2020; Li Z. et al., 2020; Wang W. et al., 2020; Zhao et al., 2020; Li Q. et al., 2021). To gain a better understanding, we summarized the regulatory networks of cotton genes in response to salt and drought in this review (Table 1).
Plants can develop resistance to Verticillium and Fusarium through a variety of mechanisms, including cell wall modifications, extracellular enzymes, pattern recognition receptors, TFs, and signal transduction pathways related to SA/JA/ET (Song et al., 2020). Several studies have been conducted over the last decade on the physiological and molecular mechanisms of plant resistance to V. dahliae and F. oxysporum wilt. Many resistance-related genes are summarized in this review to provide a theoretical foundation for a better understanding of the molecular genetic mechanisms underlying plant resistance to Verticillium and Fusarium wilt disease (Table 2). Moreover, this review is intended to be a resource for future research on the development of genetic resistance mechanisms to combat fungal wilt diseases.
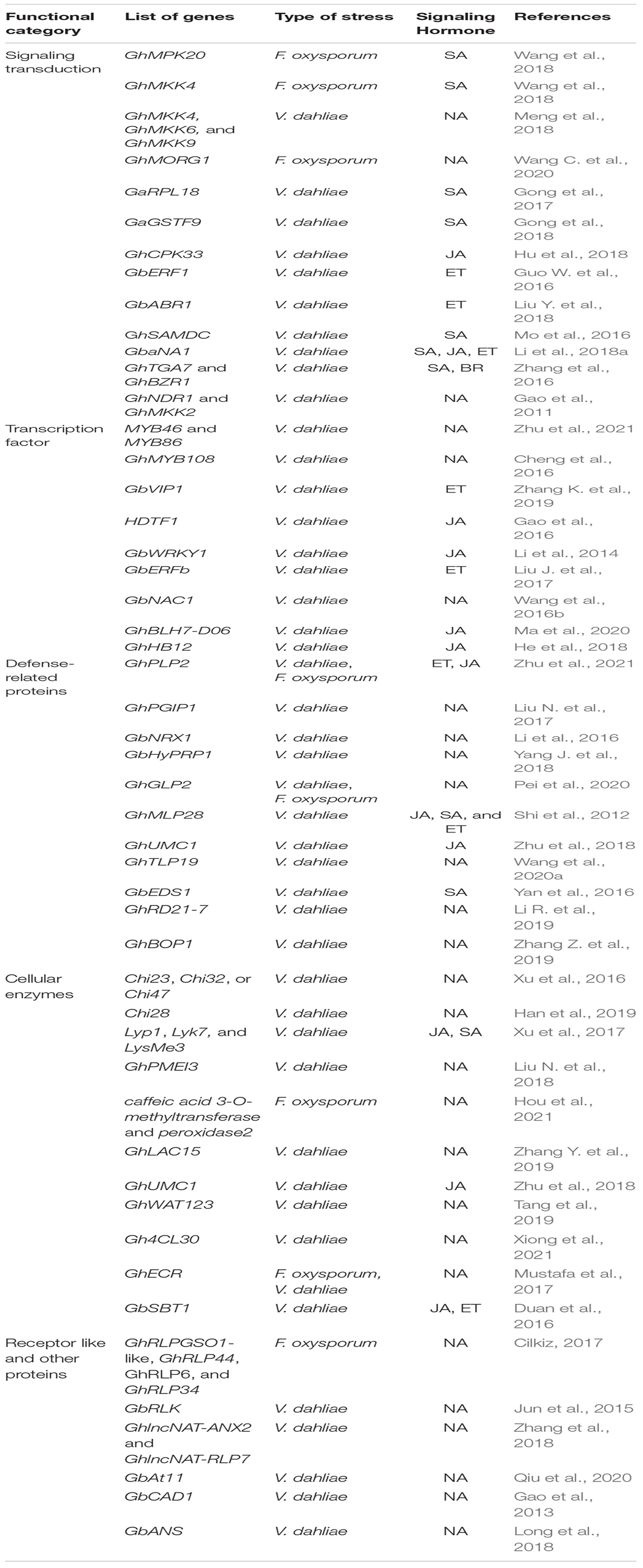
Table 2. Summary of cotton genes regulatory networks in response to Verticillium and Fusarium wilt disease.
A series of complex signal transduction processes and phytohormones function directly to control plant immunity systems. Amongst the intricate signaling networks, MAPK cascades are the primary modules responsible for classifying and amplifying external signals into intracellular components (Meng and Zhang, 2013). They are significant in both biotic and abiotic stress (Shi et al., 2011). Different signal transduction pathways operate independently while also exhibiting significant crosstalk (Gull et al., 2019). It complicates their comprehension of biotic stimuli. Multiple genes that are affected by biotic stresses suggest that there may not be a single stress tolerance marker.
Expression of GhMPK20 is significantly induced by F. oxysporum. GhMPK20 silencing in cotton increased tolerance to F. oxysporum, whereas ectopic GhMPK20 overexpression in tobacco decreased F. oxysporum resistance by interfering with the SA-mediated defense pathway. Moreover, GhMKK4 and GhWRKY40 silencing improved F. oxysporum resistance in cotton, and GhMKK4-GhMPK20 function was revealed to be required for F. oxysporum-induced GhWRKY40 expression (Wang et al., 2018). More importantly, using gene silencing techniques, GhNDR1 and GhMKK2 have been shown to be essential for Verticillium resistance in cotton (Gao et al., 2011). MKK members in MAPK signaling cascades also play dual functions in delicately modulating cotton plant resistance to fungal wilt; GhMKK4, GhMKK6, and GhMKK9 positively regulate cotton Verticillium resistance, while GhMKK10 negatively regulates it (Meng et al., 2018). Recently, a cotton MAPK scaffold protein (GhMORG1) was shown to interact with GhMKK6 and GhMPK4, and the overexpression of GhMORG1 in cotton protoplasts significantly increased the activity of the GhMKK6-GhMPK4 cascade that positively regulates the resistance of cotton to F. oxysporum (Wang C. et al., 2020).
The SA-mediated glutathione S-transferase GaGSTF9 was a positive regulator to Verticillium wilt based on VIGS and overexpression in Arabidopsis (Gong et al., 2018). Expression of the ribosomal protein, GaRPL18, is induced by SA treatment, suggesting an association of GaRPL18 in the SA signal transduction pathway. Importantly, due to a considerable decrease in the amount of immune-related molecules, wilt-resistant cotton species in which GaRPL18 was silenced became more susceptible to V. dahliae than control plants. In contrast, overexpressing GaRPL18 resulted in more resistance to V. dahliae infections (Gong et al., 2017). Expression of cyclin-dependent kinase E (GhCDKE) in cotton was induced by V. dahliae infection and MeJA treatment, and silencing of GhCDKE led to enhanced susceptibility to V. dahliae in cotton, while overexpression of GhCDKE improved resistance to this pathogen in Arabidopsis (Li et al., 2018). A calcium-dependent protein kinase, GhCPK33, derived from upland cotton functions as a negative regulator of V. dahliae resistance, which is induced by JA biosynthesis. Knockdown of GhCPK33 enhanced resistance to V. dahliae (Hu et al., 2018).
Knock-down of SA-related Spermine (Spm) proteins, Spm synthase (GhSPMS), and S-adenosylmethionine decarboxylase (GhSAMDC), damages plant resistance to V. dahliae infection in cotton. In contrast, enhanced resistance to transgenic Arabidopsis suggests that GhSAMDC contributes in plant resistance to V. dahliae via SA and leucine-related signaling pathways and mediates Spm biosynthesis (Mo et al., 2016). GhPAO expression in Arabidopsis improves resistance to V. dahliae and affects the accumulation of high levels of H2O2, SA, and camalexin (a phytoalexin), implying that GhPAO contributes to plant resistance to V. dahliae by activating Spm and camalexin signaling pathways (Mo et al., 2015). Walls are thin (WAT) promotes the resistance of crops to a wide range of pathogens by regulating SA metabolism and signaling transduction through affecting the polar transport of auxin (Denancé et al., 2013). GhWATs knockdown increased SA content accumulation, triggered SA pathway-related gene expression, and increased lignin accumulation in xylem sections, all of which accelerated plant resistance to Verticillium wilt (Tang et al., 2019).
Ethylene-responsive factors (ERFs) are generally required for pathogen defense responses. However, only a few ERF genes have been characterized in cotton in response to fungal wilt. GbABR1 is a member of the AP2 family and an ERF subfamily B4 member from G. barbadense. Silencing GbABR1 in cotton plants resulted in a higher disease index, showing that this gene positively contributes to Verticillium wilt resistance (Liu Y. et al., 2018). GbERF1-like, ET response-related factor contributes to plant resistance against V. dahliae by positively regulating lignin synthesis (Guo W. et al., 2016). Nucleotide-binding site leucine-rich repeat (NBS-LRR) proteins play an important role in plant defense against fungal pathogens. A NBS-LRR gene GbaNA1 derived from Gossypium barbadense can be induced by V. dahliae and by the phytohormones SA, ET, and JA participating in island cotton resistance to V. dahliae (Li et al., 2018b). Prominently, overexpression of GbaNA1 increases ROS content in Arabidopsis and the expression of genes associated with the ET signaling pathway (Li et al., 2018a). Additionally, eight differentially expressed candidate genes in SA (GhPUB17, GhTGA7, and GhPR1), JA (GhJAZ10 and GhbHLH18), ET (GhEBF1), cytokinine (GhE13L13), and BR (GhBZR1) signal pathways were investigated using VIGS techniques in the transcriptome with V. dahliae infection and non-infection. Knock-down of up-regulated genes GhJAZ10, GhPUB17, GhbHLH18, and GhEBF1 significantly enhanced susceptibility of resistant varieties to V. dahliae, while silencing down-regulated genes GhTGA7 and GhBZR1 significantly improved resistance of susceptible varieties to V. dahliae (Zhang et al., 2016). This revealed that genes from different hormone signaling pathways have important roles in response to fungal wilt infection.
Plant TF responses to biotic stress are extremely complex, with several TF families clearly linked to single or multiple stresses, as well as complex cross-talk between different signal transduction pathways. Proteins of the MYB family function as TFs involved in defense against pathogen infection. Two TFs, MYB46 and MYB86, are probably involved in the accumulation and synthesis of lignin suggesting that they can be used to detect Fusarium wilt resistant cotton (Zhu et al., 2021). Knockdown of GhMYB108 expression conferred enhanced susceptibility of cotton plants to V. dahliae, whereas overexpression of GhMYB108 in Arabidopsis led to improved tolerance (Cheng et al., 2016). Silencing of a home domain transcription factor gene (HDTF1) derived from cotton significantly improved cotton plant resistance to V. dahliae via activation of the JA-mediated signaling pathway (Gao et al., 2016). A stress-responsive HD-ZIP ∣ TF GhHB12 in cotton was induced by JA and V. dahliae infection, and cotton plant susceptibility to the fungal pathogens Botrytis cinerea and V. dahliae was increased by overexpression of GhHB12, which was coupled with suppression of the JA-response genes GhJAZ2 and GhPR3 (He et al., 2018). bHLH is another TF that functions against plant pathogens. As such, GbbHLH171 cooperates with and is phosphorylated by a defense-related receptor-like kinase (GbSOBIR1) in G. barbadense, and had a positive role on cotton resistance to V. dahliae (Zhou et al., 2019). GbWRKY1 is a key regulator that mediates the plant defense-to-development transition by activating JAZ1 expression during V. dahliae infection, and it has been shown to be a negative regulator of the JA-mediated defense pathway, participating in plant resistance against V. dahliae and B. cinerea (Li et al., 2014).
GbVIP1 (VirE2 interaction protein 1), which encodes a bZIP TF protein, was cloned in G. barbadense. Inoculation with V. dahliae and exogenous ET treatment both increased GbVIP1 expression. VIGS showed that silencing of GbVIP1 decreased cotton resistance to Verticillium wilt, while ectopic expression of GbVIP1 in tobacco improved resistance to Verticillium wilt by up-regulating PR1, PR1-like, and HSP70 genes (Zhang K. et al., 2019). The BEL1-Like TF GhBLH7-D06, which is commonly expressed in vascular tissues, functions in formation of secondary cells and also responds to V. dahliae infection, is induced by phytohormone JA treatment. The loss of function expression of GhBLH7-D06 could increase the resistance of cotton plants against Verticillium wilt. This resistance may be primarily due to the notable overexpression of genes involved in lignin biosynthesis and the JA signaling pathway, which also suggests that GhBLH7-D06 negatively controls cotton resistance to Verticillium wilt (Ma et al., 2020). Additionally, knock-down of GbNAC1 TFs showed that cotton was susceptible to Verticillium wilt, and GbNAC1-overexpressed in transgenic Arabidopsis plants enhanced resistance to V. dahliae compared to wild type (Wang et al., 2016b). GbERFb, a AP2/ERF type TF, can also improve cotton disease resistance (Liu J. et al., 2017). The findings from previous reports suggest that TF genes could be used to improve biotic stress tolerance/resistance in important cotton crops; though, more research is needed to understand the mechanisms of these TFs.
Plants’ resistance to fungal pathogens is greatly influenced by defense-related proteins. Plants have evolved intricate sensory mechanisms to detect biotic invasion and overcome the negative effects on growth, yield, and survival (Iqbal et al., 2021). Therefore, plants have evolved a plethora of responses to defend themselves against a wide range of pests and pathogens. Patatin-like proteins (PLPs) are defensive proteins with non-specific lipid acyl hydrolyze activity, which can hydrolyze membrane lipids into fatty acids and lysophospholipids. The importance of PLPs in plant growth and abiotic stress has been extensively studied (Jenks and Wood, 2009; Kim et al., 2014; Cheng et al., 2019; Gao et al., 2021), but the molecular function of PLPs in the plant defense system against fungal wilt is still poorly known.
GhPLP2, a cotton PLP protein located in the cell wall and plasma membrane was highly induced by treatment with V. dahliae, F. oxysporum, and signaling molecules ET and JA in cotton plants. Silence of GhPLP2 cotton plants showed reduced resistance to V. dahliae infection, whereas overexpression of GhPLP2 in Arabidopsis enhanced resistance to V. dahliae, with mild symptoms and lower disease index and fungal biomass. Moreover, GhPLP2-transgenic plants had higher accumulation of JA and JA synthesis precursor linoleic acid and α-linolenic acid than control plants, showing that PLPs have a positive role against fungal pathogenicity and have a significant role in the pathogenicity of V. dahliae (Zhu et al., 2021). Overexpression of CkPGIP1 from Cynanchum komarovii and GhPGIP1 from G. hirsutum can improve cotton resistance to V. dahliae by increasing the expression of pathogenesis-related proteins (PRs) and increasing disease susceptibility, as well as phytoalexin-deficient and isochorismate synthase genes that upregulate xylem lignification (Liu N. et al., 2017). GbNRX1 is an apoplastic thioredoxin protein found in Verticillium wilt-resistant island cotton, which is connected to an increase in abundance in response to infection with V. dahliae. The higher accumulation of ROS in apoplastic and reduced V. dahliae resistance in GbNRX1-silenced plants show that GbNRX1 can improve immune response against this fungus (Li et al., 2016).
BLADE-ON-PETIOLE1 (BOP1) and BOP2 are two BTB-ankyrin proteins that are specifically expressed in lateral-organ boundaries (LOBs). Silencing and overexpression studies show that GhBOP1 is a positive regulator of plant resistance to V. dahliae. Moreover, GhBOP1 works in tandem with GhBP1 to modulate lignin biosynthesis, conferring enhanced resistance to V. dahliae in cotton plants (Zhang Z. et al., 2019). Cotton GbHyPRP1 encodes a protein with both proline-rich repetitive and pollen ole e I domains. Cotton resistance to V. dahliae was improved in HyPRP1-silent plants through cell wall thickening and ROS accumulation. Overexpression of HyPRP1 in transgenic Arabidopsis plants significantly enhanced resistance to V. dahliae (Yang J. et al., 2018).
A defense-related major latex protein (GhMLP28) derived from upland cotton was induced by V. dahliae infection, JA, SA, and ET treatment. Knock-down of GhMLP28 increases cotton plant susceptibility to V. dahliae infection, while GhMLP28 ectopic overexpression improves disease resistance in tobacco. GhDIR1 encodes a putative dirigent protein, and its overexpression increases lignin content in transgenic cotton plants, resulting in increased tolerance to V. dahliae infection (Shi et al., 2012). Both gain and loss of function analyses revealed that GhUMC1, a cotton umecyanin-like gene, is involved in V. dahliae resistance via regulation of the JA signaling pathway and lignin metabolism (Zhu et al., 2018). The defense regulator enhanced disease susceptibility 1 (EDS1), encoding a lipase-like protein induced by SA, has a crucial role against pathogens. GbEDS1, overexpression in Arabidopsis increased SA and H2O2 production, leading to increased disease resistance to V. dahliae. GbEDS1-silencing in G. barbadense significantly reduced SA and H2O2 accumulation, resulting in increased susceptibility (Yan et al., 2016).
Moreover, papain-like cysteine proteases (PLCPs), a large plant family, are thought to play a role in plant defense against pathogens. Transcriptome analysis revealed that GhRD21-7 genes in cotton were significantly up-regulated in response to V. dahliae. More importantly, over-expression of GhRD21-7 improved resistance, while RNAi lines were more susceptible to V. dahliae in cotton (Li R. et al., 2019). Recently, germin-like proteins (GLPs), a diverse and ubiquitous family of plant glycoproteins were identified as part of the cupin super family; they have notable roles in plant defense against various abiotic and biotic stresses. Silencing of GhGLP2 in upland cotton enhanced susceptibility to V. dahliae and F. oxysporum with resulting severe wilt on leaves, enhanced vascular browning, and inhibited callose deposition. Overexpression in Arabidopsis exhibited significant resistance to V. dahliae and F. oxysporum, with decreased mycelial growth, increased callose deposition and cell wall lignification at infection sites on leaves (Pei et al., 2020). Thaumatin-like proteins (TLPs), another type of defense related protein (PR-5) in a large multigene family, have important roles in biotic and abiotic stress. When GhTLP19 was silenced in cotton the plants were more sensitive to V. dahliae, with increased MDA content and decreased CAT content, and as well as increased disease index (DI) and hyphae accumulation (Wang et al., 2020a). These studies describe cotton defense-related proteins, as well as their putative mechanisms of action, pathogen targets, and biotechnological implications.
Extracellular enzymes in plants are the first line of protection against fungal pathogens. The available research supports the fact that extracellular enzyme is not only a part of the adaptations that help cotton plants cope with pathogen infections, but it is also a key metabolic enzyme that enhances cotton plants’ growth and development. Many studies have shown that chitinase (Chi) has a housekeeping role in plasticizing the cell wall as a key hydrolytic enzyme, which destroys the fungal cell wall (Xu et al., 2016). Moreover, Chi expression can be instigated in response to biotic and abiotic stress in plants (Cheng et al., 2017). A total of 47, 49, 92, and 116 Chis from four sequenced cotton species, diploid G. raimondii and G. arboreum and tetraploid G. hirsutum and G. barbadense were, respectively identified. Phylogenetic classification classified these Chis into six groups. Cotton resistance to V. dahliae was significantly reduced when Chi23, Chi32, or Chi47 genes were knockdown, indicating that these genes function as positive regulators of V. dahliae (Xu et al., 2016). Chi28 includes the class IV chitinase subfamily. Silencing of CRR1 or Chi28 led to cotton plants more susceptible to V. dahliae infection, while overexpression of CRR1 increased V. dahliae resistance (Han et al., 2019). In plants, lysin motif (LysM)-containing proteins play a key role in chitin recognition as well as the control of defense responses against fungal pathogen attack. The genes Lyp1, Lyk7, and LysMe3 were found in the plasma membrane, and knockdown of their expression in cotton significantly reduced SA, JA, and ROS generation, decreased defense gene activation, and negotiated resistance to V. dahliae (Xu et al., 2017).
Pectins are the major elements of the primary plant cell wall, and play an important role in pathogen defense mechanisms. Pectin methylesterases (PMEs) have protective roles in the plant cell wall through catalyze dimethyl esterification of the homogalacturonan domains of pectin. Silencing GhPMEI3 in upland cotton results in increased susceptibility to V. dahliae infection, while ectopic expression of GhPMEI3 increased pectin methyl esterification and limited fungal disease by modulating root elongation. In addition, GhPMEI3 and GhPME may be involved in protein-protein interactions and are important for plant evolution to resist fungal diseases (Liu N. et al., 2018). Furthermore, erasure of two pectin lyase genes (VdPL3.1 and VdPL3.3) reduced wilt virulence to cotton. This study shows that the V. dahliae exoproteome plays an important role in the development of wilting and necrosis symptoms, primarily through pathogenic mechanisms of plant cell wall degradation as part of host plant infection (Chen J.Y. et al., 2016). Cotton resistance to fungal wild diseases is largely determined by lignin synthesis. The resistant cotton cultivars accumulate a significant amount of lignin and lignin-like phenolic polymers. Increasing evidence shows that lignin content is positively correlated with resistance to fungal wilt (Xu et al., 2011). Two coding genes in cotton, caffeic acid 3-O-methyltransferase and peroxidase2, are probably involved in the accumulation and synthesis of lignin in response to Fusarium wilt (Hou et al., 2021). Hence, quantification of lignin can be used as a selection tool to identify Fusarium resistant cotton.
A laccase gene, GhLAC15, has been found to be highly inducible by pathogens. Additionally, Transgenic expression enhances Verticillium wilt resistance by increasing defense-induced lignification, arabinose, and xylose accumulation in the cotton cell wall (Zhang Y. et al., 2019). GhUMC1, a blue copper-binding protein, is involved in cotton resistance to V. dahliae via lignin synthesis and in cell wall remodeling through the JA signaling pathway (Zhu et al., 2018). Three concurrently silenced GhWATs (GhWAT123-silenced), repressed plant growth and increased plant resistance to V. dahliae by increasing lignin deposition in the xylem (Tang et al., 2019). Knockdown of cotton lignin biosynthetic gene Gh4CL30 led to decreased content of flavonoids, lignin, and S monomer but an increased content of G monomer, G/S lignin monomer, caffeic acid, and ferulic acid, providing new insights into cotton resistance to V. dahliae (Xiong et al., 2021).
The cotton enoyl-CoA reductase (GhECR) gene functions directly in very-long-chain fatty acid formation. VIGS analysis exposed that GhECR-silenced plants are more sensitive to V. dahliae and F. oxysporum infection, showing that the GhECR gene is linked to cotton resistance to various V. dahliae and F. oxysporum strains (Mustafa et al., 2017). GbSBT1, a subtilase like protein derived from G. barbadense located on the cell membrane, is highly induced by V. dahliae, JA, and ET treatment. Moreover, silencing the GbSBT1 gene decreases the tolerance to V. dahliae infection. Notably, in Arabidopsis, overexpression of GbSBT1 stimulates the expression of defense-related genes and enhances resistance to F. oxysporum and V. dahliae (Duan et al., 2016). These findings revealed that the induced defensive enzymes, which are produced in response to an attack, provide remarkable protection against pathogens via defense mechanisms.
Plant receptor-like proteins are involved in diverse of biological processes, including development, innate immunity, cell differentiation and patterning, nodulation, and self-incompatibility (Yang et al., 2012). Receptor-like proteins are on the front lines of the plant-pathogen battle because they are present at the plasma membrane and detect signature molecules from either the invading pathogen or damaged plant tissue. Cell-surface-associated PRRs are essential in fungal pathogen recognition. PRRs are receptor-like kinases (RLKs) and receptor-like proteins (RLPs) found in plants. Using the gene silencing approach, it was suggested that GhRLPGSO1-like, GhRLP44, GhRLP6, and GhRLP34 might be needed for defense against F. oxysporum in cotton plants (Cilkiz, 2017). An RLK gene (GbRLK) from the disease-resistant cotton G. barbadense, is stimulated with the infection of V. dahliae. In addition, transgenic cotton and Arabidopsis plants of GbRLK confer resistance to V. dahliae infection (Jun et al., 2015). NBS-LRR (nucleotide-binding site leucine-rich repeats) proteins play an important role in plant pathogen defense. A genome-wide association study revealed that TIR-NBS-LRR domains containing CG02 are the most likely candidate associated to cotton resistance against V. dahliae. According to Real-time quantitative PCR and VIGS analysis, CG02 was specific to up-regulation in the resistant genotype, and silenced plants were more susceptible to V. dahliae (Li T. et al., 2017).
Furthermore, the Ve R-gene locus contributes to Verticillium resistance by encoding RLPs with extracellular leucine-rich repeats (Nazar et al., 2018). Several studies explored how Ve1 is involved in the cotton resistance to Verticillium wilt infection (Zhang B. et al., 2012; Chen T. et al., 2016; Song et al., 2017, 2020; Yang Y. et al., 2018). GhlncNAT-ANX2 and GhlncNAT-RLP7 are conserved long non-coding RNAs, and their silencing in cotton promotes resistance to V. dahliae, which may be connected to the upregulated expression of lipoxygenase 1 and lipoxygenase 2 (Zhang et al., 2018). GbAt11 (AXMN Toxin Induced Protein-11) was found to be highly resistant to Verticillium wilt in G. barbadense. Moreover, FLS2, BAK1, and other disease resistance genes can be up-regulated by GbAt11 overexpression (Qiu et al., 2020). GhPUB17, a U-box E3 ubiquitin ligase that interacts with and is inhibited by the antifungal protein GhCyP3, negatively regulates cotton resistance to the Verticillium wilt pathogen (Qin et al., 2019). Moreover, anthocyanin and production of gossypol is sufficient to influence V. dahliae infection. As such, cotton GbANS is involved in anthocyanin biosynthesis, and silencing GbANS significantly decreases anthocyanin production as well as cotton plant tolerance to V. dahliae (Long et al., 2018). Silencing of GbCAD1, which encodes a key enzyme contributed in gossypol biosynthesis, compromises cotton plant resistance to V. dahliae (Gao et al., 2013).
Due to increasing incidences of both biotic and abiotic stresses, sustainability of crop production is a serious challenge under field conditions. The cotton mechanisms that exhibit tolerance to various biotic and abiotic stresses appear to be interrelated and may have overlapping genetic elements. Both abiotic and biotic stresses negatively affect molecular, biochemical, and physiological processes, ultimately resulting in suppressed growth and development in cotton, such as reduced photosynthetic rate, plant height, leaf and root size, biomass, yield, and yield components, and poorer fiber quality. Currently, many phytohormone-based growth regulators are commercially used in agriculture to improve the resistance of plants to abiotic and biotic stresses. This indicates that identification and characterization of genetic components such as defense of cell membranes and proteins, signaling cascades and transcriptional control, and ion uptake and transport and their relevant biochemical pathways and multiple signal factors, are necessary to provide important clues to understand basic molecular mechanism/network of plant response and the development of plants with better resistance to adverse conditions. Nevertheless, due to the complexity of stress conditions and the difficulty of phenotyping, the genetic basis of this tolerance is not fully understood, because it is affected by multiple gene regulatory systems with environmental influences. However, drought alone impacts 45% of the world’s agricultural land; additionally, 19.5% of irrigated agricultural lands are classified as saline (Abdelraheem et al., 2019). A combination of two or more abiotic stresses, such as drought and salinity, results in greater yield loss than either stress alone. Drought and salinization are expected to cause up to 50% of arable land loss globally. Moreover, Verticillium and fusarium wilt are caused by soil-borne pathogenic fungi, and are major constraints to cotton production (Pei et al., 2020). Therefore, one of the most practical solutions is the development of abiotic (drought and/or salt) and biotic (Verticillium and Fusarium) stress tolerant cultivars. In recent decades, numerous genes responsive to drought, salt, and Verticillium and Fusarium wilt diseases in cotton have been identified, some of which were further studied using transgenic approaches, but none of the genes have been utilized in commercial cotton breeding programs. In this review, we summarized cotton genes related to salt, drought, and wilt disease resistance on the basis of their molecular functions. The review provides researchers with good theoretical knowledge and identifies gene networks that can help in discovering other resistance-related genes in order to better understand the molecular genetic mechanisms of cotton resistance to these stresses.
MB, FL, and ZY conceptualized the review. MB wrote the original draft. ZY and FL investigated, revised, and edited the draft manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by funding from the National Natural Science Foundation of China (grants 31801416 to ZY and 31621005 to FL) and the Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-IVFCAAS to FL and ZY).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors would like to acknowledge “Abdelraheem et al. (2019)” for their review article that helped us to build the foundation of our manuscript.
Abdelraheem, A., Esmaeili, N., O’Connell, M., and Zhang, J. (2019). Progress and perspective on drought and salt stress tolerance in cotton. Industr. Crops Product. 130, 118–129. doi: 10.1016/j.indcrop.2018.12.070
Almagro, L., Gómez Ros, L., Belchi-Navarro, S., Bru, R., Ros Barceló, A., and Pedreno, M. (2009). Class III peroxidases in plant defence reactions. J. Exp. Bot. 60, 377–390. doi: 10.1093/jxb/ern277
Alves, M. S., Dadalto, S. P., Gonçalves, A. B., De Souza, G. B., Barros, V. A., and Fietto, L. G. (2013). Plant bZIP transcription factors responsive to pathogens: a review. Int. J. Mol. Sci. 14, 7815–7828. doi: 10.3390/ijms14047815
Baillo, E. H., Kimotho, R. N., Zhang, Z., and Xu, P. (2019). Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes 10:771. doi: 10.3390/genes10100771
Bassil, E., and Blumwald, E. (2014). The ins and outs of intracellular ion homeostasis: NHX-type cation/H+ transporters. Curr. Opin. Plant Biol. 22, 1–6. doi: 10.1016/j.pbi.2014.08.002
Chen, J.-Y., Xiao, H.-L., Gui, Y.-J., Zhang, D.-D., Li, L., Bao, Y.-M., et al. (2016). Characterization of the Verticillium dahliae exoproteome involves in pathogenicity from cotton-containing medium. Front. Microbiol. 7:1709. doi: 10.3389/fmicb.2016.01709
Chen, M., Li, K., Li, H., Song, C.-P., and Miao, Y. (2017). The glutathione peroxidase gene family in Gossypium hirsutum: genome-wide identification, classification, gene expression and functional analysis. Sci. Rep. 7, 1–15. doi: 10.1038/srep44743
Chen, P., Cheng, S., Ma, L., Wang, H., Zhang, M., Mao, G., et al. (2020). A comprehensive analysis of cotton VQ gene superfamily reveals their potential and extensive roles in regulating cotton abiotic stress. BMC Genom. 21:1–16. doi: 10.1186/s12864-020-07171-z
Chen, Q., Ji, X., and Sun, W. (1985). Identification of races of cotton wilt Fusarium in China. Agric. Sci. China 6, 1–6.
Chen, T., Kan, J., Yang, Y., Ling, X., Chang, Y., and Zhang, B. (2016). A Ve homologous gene from Gossypium barbadense, Gbvdr3, enhances the defense response against Verticillium dahliae. Plant Physiol. Biochem. (Mosc.) 98, 101–111. doi: 10.1016/j.plaphy.2015.11.015
Chen, T., Li, W., Hu, X., Guo, J., Liu, A., and Zhang, B. (2015). A cotton MYB transcription factor, GbMYB5, is positively involved in plant adaptive response to drought stress. Plant Cell Physiol. 56, 917–929. doi: 10.1093/pcp/pcv019
Chen, X., Lu, X., Shu, N., Wang, D., Wang, S., Wang, J., et al. (2017). GhSOS1, a plasma membrane Na+/H+ antiporter gene from upland cotton, enhances salt tolerance in transgenic Arabidopsis thaliana. PLoS One 12:e0181450. doi: 10.1371/journal.pone.0181450
Chen, X., Wang, J., Zhu, M., Jia, H., Liu, D., Hao, L., et al. (2015). A cotton Raf-like MAP3K gene, GhMAP3K40, mediates reduced tolerance to biotic and abiotic stress in Nicotiana benthamiana by negatively regulating growth and development. Plant Sci. 240, 10–24. doi: 10.1016/j.plantsci.2015.08.012
Chen, Y., Feng, L., Wei, N., Liu, Z.-H., Hu, S., and Li, X.-B. (2017). Overexpression of cotton PYL genes in Arabidopsis enhances the transgenic plant tolerance to drought stress. Plant Physiol. Biochem. (Mosc.) 115, 229–238. doi: 10.1016/j.plaphy.2017.03.023
Cheng, H.-Q., Han, L.-B., Yang, C.-L., Wu, X.-M., Zhong, N.-Q., Wu, J.-H., et al. (2016). The cotton MYB108 forms a positive feedback regulation loop with CML11 and participates in the defense response against Verticillium dahliae infection. J. Exp. Bot. 67, 1935–1950. doi: 10.1093/jxb/erw016
Cheng, J., Song, N., and Wu, J. (2019). A patatin-like protein synergistically regulated by jasmonate and ethylene signaling pathways plays a negative role in Nicotiana attenuata resistance to Alternaria alternata. Plant Divers. 41, 7–12. doi: 10.1016/j.pld.2018.12.001
Cheng, S.-H., Willmann, M. R., Chen, H.-C., and Sheen, J. (2002). Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 129, 469–485. doi: 10.1104/pp.005645
Cheng, X.-X., Zhao, L.-H., Klosterman, S. J., Feng, H.-J., Feng, Z.-L., Wei, F., et al. (2017). The endochitinase VDECH from Verticillium dahliae inhibits spore germination and activates plant defense responses. Plant Sci. 259, 12–23. doi: 10.1016/j.plantsci.2017.03.002
Chu, X., Wang, C., Chen, X., Lu, W., Li, H., Wang, X., et al. (2015). The cotton WRKY gene GhWRKY41 positively regulates salt and drought stress tolerance in transgenic Nicotiana benthamiana. PLoS One 10:e0143022. doi: 10.1371/journal.pone.0143022
Ciarmiello, L. F., Woodrow, P., Fuggi, A., Pontecorvo, G., Carillo, P. J., and Petronia, C. (2011). “Plant genes for abiotic stress,” in Abiotic Stress in Plants–Mechanisms Adaptations, eds A. Shanker and B. Venkateswarlu (London: IntechOpen), 283–308.
Cilkiz, M. (2017). Genome-Wide Bioinformatic and Functional Analysis of Receptor-Like Proteins in Cotton. Master’s thesis. College Station, TX: Texas A & M University.
Cui, Y., Su, Y., Wang, J., Jia, B., Wu, M., Pei, W., et al. (2020). Genome-wide characterization and analysis of CIPK gene family in two cultivated allopolyploid cotton species: sequence variation, association with seed oil content, and the role of GhCIPK6. Int. J. Mol. Sci. 21:863. doi: 10.3390/ijms21030863
Cun, M., Guiliang, J., and Chuanlin, Z. (2002). The advances in cotton breeding resistance to Fusarium and Verticillium wilts in China during past fifty years. Zhongguo Nongye Kexue 35, 508–513.
Danquah, A., de Zélicourt, A., Boudsocq, M., Neubauer, J., Frei dit Frey, N., Leonhardt, N., et al. (2015). Identification and characterization of an ABA-activated MAP kinase cascade in Arabidopsis thaliana. Plant J. 82, 232–244. doi: 10.1111/tpj.12808
Davies, J. M. (2014). Annexin-mediated calcium signalling in plants. Plants 3, 128–140. doi: 10.3390/plants3010128
Davis, R., Colyer, P., Rothrock, C., and Kochman, J. (2006). Fusarium wilt of cotton: population diversity and implications for management. Plant Dis. 90, 692–703. doi: 10.1094/PD-90-0692
Debbarma, J., Sarki, Y. N., Saikia, B., Boruah, H. P. D., Singha, D. L., and Chikkaputtaiah, C. (2019). Ethylene response factor (ERF) family proteins in abiotic stresses and CRISPR–Cas9 genome editing of ERFs for multiple abiotic stress tolerance in crop plants: a review. Mol. Biotechnol. 61, 153–172. doi: 10.1007/s12033-018-0144-x
Demidchik, V., Shabala, S., Isayenkov, S., Cuin, T. A., and Pottosin, I. (2018). Calcium transport across plant membranes: mechanisms and functions. New Phytol. 220, 49–69. doi: 10.1111/nph.15266
Denancé, N., Ranocha, P., Oria, N., Barlet, X., Rivière, M. P., Yadeta, K. A., et al. (2013). Arabidopsis wat1 (walls are thin1)-mediated resistance to the bacterial vascular pathogen, Ralstonia solanacearum, is accompanied by cross-regulation of salicylic acid and tryptophan metabolism. Plant J. 73, 225–239. doi: 10.1111/tpj.12027
Ding, M., Chen, J., Jiang, Y., Lin, L., Cao, Y., Wang, M., et al. (2015). Genome-wide investigation and transcriptome analysis of the WRKY gene family in Gossypium. Mol. Genet. Genom. 290, 151–171. doi: 10.1007/s00438-014-0904-7
Dong, Y., Li, C., Zhang, Y., He, Q., Daud, M. K., Chen, J., et al. (2016). Glutathione S-transferase gene family in Gossypium raimondii and G. arboreum: comparative genomic study and their expression under salt stress. Front. Plant Sci. 7:139. doi: 10.3389/fpls.2016.00139
Dongdong, L., Ming, Z., Lili, H., Xiaobo, C., Yang, G., Xingqi, G., et al. (2016). GhMAPKKK49, a novel cotton (Gossypium hirsutum L.) MAPKKK gene, is involved in diverse stress responses. Acta Physiol. Plant. 38:13. doi: 10.1007/s11738-015-2029-y
Dos Reis, S. P., Lima, A. M., and De Souza, C. R. B. (2012). Recent molecular advances on downstream plant responses to abiotic stress. Int. J. Mol. Sci. 13, 8628–8647. doi: 10.3390/ijms13078628
Dou, L., Zhang, X., Pang, C., Song, M., Wei, H., Fan, S., et al. (2014). Genome-wide analysis of the WRKY gene family in cotton. Mol. Genet. Genom. 289, 1103–1121. doi: 10.1007/s00438-014-0872-y
Duan, P., Wang, G., Chao, M., Zhang, Z., and Zhang, B. (2019). Genome-wide identification and analysis of class iii peroxidases in allotetraploid cotton (Gossypium hirsutum L.) and their responses to pk deficiency. Genes 10:473. doi: 10.3390/genes10060473
Duan, X., Zhang, Z., Wang, J., and Zuo, K. (2016). Characterization of a novel cotton subtilase gene GbSBT1 in response to extracellular stimulations and its role in Verticillium resistance. PLoS One 11:e0153988. doi: 10.1371/journal.pone.0153988
Elasad, M., Ondati, E., Wei, H., Wang, H., Su, J., Fan, S., et al. (2018). Functional analysis of nine cotton genes related to leaf senescence in Gossypium hirsutum L. Physiol. Mol. Biol. Plants 24, 729–739. doi: 10.1007/s12298-018-0561-0
Evans, O., Dou, L., Guo, Y., Pang, C., Wei, H., Song, M., et al. (2016). GhNAC18, a novel cotton (Gossypium hirsutum L.) NAC gene, is involved in leaf senescence and diverse stress responses. Afr. J. Biotechnol. 15, 1233–1245. doi: 10.5897/AJB2016.15224
Fan, K., Li, F., Chen, J., Li, Z., Lin, W., Cai, S., et al. (2018). Asymmetric evolution and expansion of the NAC transcription factor in polyploidized cotton. Front. Plant Sci. 9:47. doi: 10.3389/fpls.2018.00047
Feng, H., Li, C., Zhou, J., Yuan, Y., Feng, Z., Shi, Y., et al. (2021). A cotton WAKL protein interacted with a DnaJ protein and was involved in defense against Verticillium dahliae. Int. J. Biol. Macromol. 167, 633–643. doi: 10.1016/j.ijbiomac.2020.11.191
Frank, W., Munnik, T., Kerkmann, K., Salamini, F., and Bartels, D. (2000). Water deficit triggers phospholipase D activity in the resurrection plant Craterostigma plantagineum. Plant Cell 12, 111–123. doi: 10.1105/tpc.12.1.111
Gao, G., Zhang, X., Zhao, K., Zhao, K., Cao, D., Ma, Q., et al. (2021). Genome wide identification and expression analysis of patatin-like protein family members in peanut (Arachis hypogaea L.). Reprod. Breed. 1, 48–54. doi: 10.1016/j.repbre.2021.03.002
Gao, S.-Q., Chen, M., Xia, L.-Q., Xiu, H.-J., Xu, Z.-S., Li, L.-C., et al. (2009). A cotton (Gossypium hirsutum) DRE-binding transcription factor gene, GhDREB, confers enhanced tolerance to drought, high salt, and freezing stresses in transgenic wheat. Plant Cell Rep. 28, 301–311. doi: 10.1007/s00299-008-0623-9
Gao, W., Long, L., Xu, L., Lindsey, K., Zhang, X., and Zhu, L. (2016). Suppression of the homeobox gene HDTF1 enhances resistance to Verticillium dahliae and Botrytis cinerea in cotton. J. Integrat. Plant Biol. 58, 503–513. doi: 10.1111/jipb.12432
Gao, W., Long, L., Zhu, L.-F., Xu, L., Gao, W.-H., Sun, L.-Q., et al. (2013). Proteomic and virus-induced gene silencing (VIGS) analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae. Mol. Cell. Proteom. 12, 3690–3703. doi: 10.1074/mcp.M113.031013
Gao, W., Xu, F.-C., Guo, D.-D., Zhao, J.-R., Liu, J., Guo, Y.-W., et al. (2018). Calcium-dependent protein kinases in cotton: insights into early plant responses to salt stress. BMC Plant Biol. 18, 1–15. doi: 10.1186/s12870-018-1230-8
Gao, X., Wheeler, T., Li, Z., Kenerley, C. M., He, P., and Shan, L. (2011). Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J. 66, 293–305. doi: 10.1111/j.1365-313X.2011.04491.x
Gill, S. S., and Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. (Mosc.) 48, 909–930. doi: 10.1016/j.plaphy.2010.08.016
Goda, H., Sawa, S., Asami, T., Fujioka, S., Shimada, Y., and Yoshida, S. (2004). Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 134, 1555–1573. doi: 10.1104/pp.103.034736
Gong, Q., Yang, Z., Chen, E., Sun, G., He, S., Butt, H. I., et al. (2018). A phi-class glutathione S-transferase gene for Verticillium wilt resistance in Gossypium arboreum identified in a genome-wide association study. Plant Cell Physiol. 59, 275–289. doi: 10.1093/pcp/pcx180
Gong, Q., Yang, Z., Wang, X., Butt, H. I., Chen, E., He, S., et al. (2017). Salicylic acid-related cotton (Gossypium arboreum) ribosomal protein GaRPL18 contributes to resistance to Verticillium dahliae. BMC Plant Biol. 17, 1–15. doi: 10.1186/s12870-017-1007-5
Gu, L., Ma, Q., Zhang, C., Wang, C., Wei, H., Wang, H., et al. (2019). The cotton GhWRKY91 transcription factor mediates leaf senescence and responses to drought stress in transgenic Arabidopsis thaliana. Front. Plant Sci. 10:1352. doi: 10.3389/fpls.2019.01352
Gull, A., Lone, A. A., and Wani, N. U. I. (2019). “Biotic and abiotic stresses in plants,” in Abiotic Biotic Stress in Plants, ed. A. B. de Oliveira (London: IntechOpen), 1–19. doi: 10.5772/intechopen.85832
Guo, K., Du, X., Tu, L., Tang, W., Wang, P., Wang, M., et al. (2016). Fibre elongation requires normal redox homeostasis modulated by cytosolic ascorbate peroxidase in cotton (Gossypium hirsutum). J. Exp. Bot. 67, 3289–3301. doi: 10.1093/jxb/erw146
Guo, K., Li, Z., Tian, H., Du, X., Liu, Z., Huang, H., et al. (2020). Cytosolic ascorbate peroxidases plays a critical role in photosynthesis by modulating reactive oxygen species level in stomatal guard cell. Front. Plant Sci. 11:446. doi: 10.3389/fpls.2020.00446
Guo, W., Jin, L., Miao, Y., He, X., Hu, Q., Guo, K., et al. (2016). An ethylene response-related factor, GbERF1-like, from Gossypium barbadense improves resistance to Verticillium dahliae via activating lignin synthesis. Plant Mol. Biol. 91, 305–318. doi: 10.1007/s11103-016-0467-6
Gupta, S., Dong, Y., Dijkwel, P. P., Mueller-Roeber, B., and Gechev, T. S. (2019). Genome-wide analysis of ROS antioxidant genes in resurrection species suggest an involvement of distinct ROS detoxification systems during desiccation. Int. J. Mol. Sci. 20:3101. doi: 10.3390/ijms20123101
Han, L.-B., Li, Y.-B., Wang, F.-X., Wang, W.-Y., Liu, J., Wu, J.-H., et al. (2019). The cotton apoplastic protein CRR1 stabilizes chitinase 28 to facilitate defense against the fungal pathogen Verticillium dahliae. Plant Cell 31, 520–536. doi: 10.1105/tpc.18.00390
He, P., Zhao, P., Wang, L., Zhang, Y., Wang, X., Xiao, H., et al. (2017). The PIN gene family in cotton (Gossypium hirsutum): genome-wide identification and gene expression analyses during root development and abiotic stress responses. BMC Genom. 18, 1–10. doi: 10.1186/s12864-017-3901-5
He, X., Wang, T., Zhu, W., Wang, Y., and Zhu, L. (2018). GhHB12, a HD-ZIP I transcription factor, negatively regulates the cotton resistance to Verticillium dahliae. Int. J. Mol. Sci. 19:3997. doi: 10.3390/ijms19123997
He, X., Zhu, L., Xu, L., Guo, W., and Zhang, X. (2016). GhATAF1, a NAC transcription factor, confers abiotic and biotic stress responses by regulating phytohormonal signaling networks. Plant Cell Rep. 35, 2167–2179. doi: 10.1007/s00299-016-2027-6
Herms, D. A., and Mattson, W. J. (1992). The dilemma of plants: to grow or defend. Quar. Rev. Biol. 67, 283–335. doi: 10.1086/417659
Hetherington, A. M., and Brownlee, C. (2004). The generation of Ca2+ signals in plants. Annu. Rev. Plant Biol. 55, 401–427. doi: 10.1146/annurev.arplant.55.031903.141624
Hou, J., Yang, X., Li, W., Xie, D., Tang, Z., Lv, S., et al. (2021). Lignin synthesis related genes with potential significance in the response of upland cotton to fusarium wilt identified by transcriptome profiling. Tropical Plant Biol. 14, 106–119. doi: 10.1007/s12042-020-09278-9
Hu, Q., Zhu, L., Zhang, X., Guan, Q., Xiao, S., Min, L., et al. (2018). GhCPK33 negatively regulates defense against Verticillium dahliae by phosphorylating GhOPR3. Plant Physiol. 178, 876–889. doi: 10.1104/pp.18.00737
Huang, B., Jin, L., and Liu, J. (2007). Molecular cloning and functional characterization of a DREB1/CBF-like gene (GhDREB1L) from cotton. Sci. China Ser. C Life Sci. 50, 7–14. doi: 10.1007/s11427-007-0010-8
Ijaz, M., Shahzadi, R., Masoud, M. S., Iqbal, M., and Umirbekovna, I. A. (2020). “Transcription factors and plant abiotic stress responses,” in Plant Ecophysiology and Adaptation Under Climate Change: Mechanisms and Perspectives I, ed. M. Hasanuzzaman (Singapore: Springer), 663–687. doi: 10.1007/978-981-15-2156-0_23
Iqbal, Z., Iqbal, M. S., Hashem, A., Abd_Allah, E. F., and Ansari, M. I. (2021). Plant defense responses to biotic stress and its interplay with fluctuating dark/light conditions. Front. Plant Sci. 12:297. doi: 10.3389/fpls.2021.631810
Jenks, M. A., and Wood, A. J. (2009). Genes for Plant Abiotic Stress. Hoboken, NJ: John Wiley & Sons. doi: 10.1002/9780813809380
Jia, H., Hao, L., Guo, X., Liu, S., Yan, Y., and Guo, X. (2016). A Raf-like MAPKKK gene, GhRaf19, negatively regulates tolerance to drought and salt and positively regulates resistance to cold stress by modulating reactive oxygen species in cotton. Plant Sci. 252, 267–281. doi: 10.1016/j.plantsci.2016.07.014
Jofuku, K. D., Den Boer, B., Van Montagu, M., and Okamuro, J. K. (1994). Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6, 1211–1225. doi: 10.1105/tpc.6.9.1211
Jun, Z., Zhang, Z., Gao, Y., Zhou, L., Fang, L., Chen, X., et al. (2015). Overexpression of GbRLK, a putative receptor-like kinase gene, improved cotton tolerance to Verticillium wilt. Sci. Rep. 5, 1–12. doi: 10.1038/srep15048
Khan, M. A., Wahid, A., Ahmad, M., Tahir, M. T., Ahmed, M., Ahmad, S., et al. (2020). “World cotton production and consumption: an overview,” in Cotton Production Uses, eds S. Ahmad and M. Hasanuzzaman (Singapore: Springer), 1–7. doi: 10.1007/978-981-15-1472-2_1
Khan, S.-A., Li, M.-Z., Wang, S.-M., and Yin, H.-J. (2018). Revisiting the role of plant transcription factors in the battle against abiotic stress. Int. J. Mol. Sci. 19:1634. doi: 10.3390/ijms19061634
Khanale, V., Bhattacharya, A., Satpute, R., and Char, B. (2020). Version 4.3-12/08/20 Cotton bZIP transcription factors: characterization of the bZIP family from Gossypium hirsutum, Gossypium arboreum and Gossypium raimondii. Res. Square [Preprint]. doi: 10.21203/rs.3.rs-70685/v1
Kim, D. S., Jeun, Y., and Hwang, B. K. (2014). The pepper patatin-like phospholipase CaPLP1 functions in plant cell death and defense signaling. Plant Mol. Biol. 84, 329–344. doi: 10.1007/s11103-013-0137-x
Kirungu, J. N., Magwanga, R. O., Lu, P., Cai, X., Zhou, Z., Wang, X., et al. (2019). Functional characterization of Gh_A08G1120 (GH3. 5) gene reveal their significant role in enhancing drought and salt stress tolerance in cotton. BMC Genet. 20, 1–17. doi: 10.1186/s12863-019-0756-6
Kolukisaoglu, Ü, Weinl, S., Blazevic, D., Batistic, O., and Kudla, J. (2004). Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 134, 43–58. doi: 10.1104/pp.103.033068
Li, C., He, X., Luo, X., Xu, L., Liu, L., Min, L., et al. (2014). Cotton WRKY1 mediates the plant defense-to-development transition during infection of cotton by Verticillium dahliae by activating JASMONATE ZIM-DOMAIN1 expression. Plant Physiol. 166, 2179–2194. doi: 10.1104/pp.114.246694
Li, F., Li, M., Wang, P., Cox, K. L. Jr., Duan, L., Dever, J. K., et al. (2017). Regulation of cotton (Gossypium hirsutum) drought responses by mitogen-activated protein (MAP) kinase cascade-mediated phosphorylation of Gh WRKY 59. New Phytol. 215, 1462–1475. doi: 10.1111/nph.14680
Li, L.-b., Yu, D.-w., Zhao, F.-l., Pang, C.-y., Song, M.-z., Wei, H.-l., et al. (2015). Genome-wide analysis of the calcium-dependent protein kinase gene family in Gossypium raimondii. J. Integrat. Agricult. 14, 29–41. doi: 10.1016/S2095-3119(14)60780-2
Li, N.-Y., Ma, X. F., Short, D. P., Li, T. G., Zhou, L., Gui, Y. J., et al. (2018b). The island cotton NBS-LRR gene GbaNA1 confers resistance to the non-race 1 Verticillium dahliae isolate Vd991. Mol. Plant Pathol. 19, 1466–1479. doi: 10.1111/mpp.12630
Li, N.-Y., Zhou, L., Zhang, D.-D., Klosterman, S. J., Li, T.-G., Gui, Y.-J., et al. (2018a). Heterologous expression of the cotton NBS-LRR gene GbaNA1 enhances Verticillium wilt resistance in Arabidopsis. Front. Plant Sci. 9:119. doi: 10.3389/fpls.2018.00119
Li, Q., Yu, X., Chen, L., Zhao, G., Li, S., Zhou, H., et al. (2021). Genome-wide identification and expression analysis of the NCED family in cotton (Gossypium hirsutum L.). PLoS One 16:e0246021. doi: 10.1371/journal.pone.0246021
Li, R., Liu, C., Zhao, R., Wang, L., Chen, L., Yu, W., et al. (2019). CRISPR/Cas9-Mediated SlNPR1 mutagenesis reduces tomato plant drought tolerance. BMC Plant Biol. 19:38. doi: 10.1186/s12870-018-1627-4
Li, S., Chen, H., Hou, Z., Li, Y., Yang, C., Wang, D., et al. (2020). Screening of abiotic stress-responsive cotton genes using a cotton full-length cDNA overexpressing Arabidopsis library. J. Integr. Plant Biol. 62, 998–1016. doi: 10.1111/jipb.12861
Li, T., Ma, X., Li, N., Zhou, L., Liu, Z., Han, H., et al. (2017). Genome-wide association study discovered candidate genes of Verticillium wilt resistance in upland cotton (Gossypium hirsutum L.). Plant Biotechnol. J. 15, 1520–1532. doi: 10.1111/pbi.12734
Li, X., Pei, Y., Sun, Y., Liu, N., Wang, P., Liu, D., et al. (2018). A cotton Cyclin-dependent kinase E confers resistance to Verticillium dahliae mediated by Jasmonate-responsive pathway. Front. Plant Sci. 9:642. doi: 10.3389/fpls.2018.00642
Li, X., Zhang, Y. n., Ding, C., Xu, W., and Wang, X. (2017). Temporal patterns of cotton Fusarium and Verticillium wilt in Jiangsu coastal areas of China. Sci. Rep. 7, 1–8. doi: 10.1038/s41598-017-12985-1
Li, Y., Chen, H., Li, S., Yang, C., Ding, Q., Song, C.-P., et al. (2021). GhWRKY46 from upland cotton positively regulates the drought and salt stress responses in plant. Environ. Exp. Bot. 186:104438. doi: 10.1016/j.envexpbot.2021.104438
Li, Y.-B., Han, L.-B., Wang, H.-Y., Zhang, J., Sun, S.-T., Feng, D.-Q., et al. (2016). The thioredoxin GbNRX1 plays a crucial role in homeostasis of apoplastic reactive oxygen species in response to Verticillium dahliae infection in cotton. Plant Physiol. 170, 2392–2406. doi: 10.1104/pp.15.01930
Li, Z., Li, L., Zhou, K., Zhang, Y., Han, X., Din, Y., et al. (2019). GhWRKY6 acts as a negative regulator in both transgenic Arabidopsis and cotton during drought and salt stress. Front. Genet. 10:392. doi: 10.3389/fgene.2019.00392
Li, Z., Liu, Z., Wei, Y., Liu, Y., Li, P., Xing, L., et al. (2020). Genome-wide identification and characterization of the MIOX gene family in cotton. Res. Square [Preprint]. doi: 10.21203/rs.3.rs-130287/v1
Liang, C., Meng, Z., Meng, Z., Malik, W., Yan, R., Lwin, K. M., et al. (2016). GhABF2, a bZIP transcription factor, confers drought and salinity tolerance in cotton (Gossypium hirsutum L.). Sci. Rep. 6, 1–14. doi: 10.1038/srep35040
Lin, L., Wu, J., Jiang, M., and Wang, Y. (2021). Plant mitogen-activated protein kinase cascades in environmental stresses. Int. J. Mol. Sci. 22:1543. doi: 10.3390/ijms22041543
Liu, C., and Zhang, T. (2017). Expansion and stress responses of the AP2/EREBP superfamily in cotton. BMC Genom. 18, 1–16. doi: 10.1186/s12864-017-3517-9
Liu, G., Li, X., Jin, S., Liu, X., Zhu, L., Nie, Y., et al. (2014). Overexpression of rice NAC gene SNAC1 improves drought and salt tolerance by enhancing root development and reducing transpiration rate in transgenic cotton. PLoS One 9:e86895. doi: 10.1371/journal.pone.0086895
Liu, J., Wang, Y., Zhao, G., Zhao, J., Du, H., He, X., et al. (2017). A novel Gossypium barbadense ERF transcription factor, GbERFb, regulation host response and resistance to Verticillium dahliae in tobacco. Physiol. Mol. Biol. Plants 23, 125–134. doi: 10.1007/s12298-016-0402-y
Liu, N., Sun, Y., Pei, Y., Zhang, X., Wang, P., Li, X., et al. (2018). A pectin methylesterase inhibitor enhances resistance to Verticillium wilt. Plant Physiol. 176, 2202–2220. doi: 10.1104/pp.17.01399
Liu, N., Zhang, X., Sun, Y., Wang, P., Li, X., Pei, Y., et al. (2017). Molecular evidence for the involvement of a polygalacturonase-inhibiting protein, GhPGIP1, in enhanced resistance to Verticillium and Fusarium wilts in cotton. Sci. Rep. 7, 1–18. doi: 10.1038/srep39840
Liu, W., Li, W., He, Q., Daud, M. K., Chen, J., and Zhu, S. (2014). Genome-wide survey and expression analysis of calcium-dependent protein kinase in Gossypium raimondii. PLoS One 9:e98189. doi: 10.1371/journal.pone.0098189
Liu, X., Song, Y., Xing, F., Wang, N., Wen, F., and Zhu, C. (2016). GhWRKY25, a group I WRKY gene from cotton, confers differential tolerance to abiotic and biotic stresses in transgenic Nicotiana benthamiana. Protoplasma 253, 1265–1281. doi: 10.1007/s00709-015-0885-3
Liu, Y., Liu, X., Long, L., Wang, W., Sun, Q., Li, B., et al. (2018). GbABR1 is associated with Verticillium wilt resistance in cotton. Biologia 73, 449–457. doi: 10.2478/s11756-018-0058-x
Long, L., Gao, W., Xu, L., Liu, M., Luo, X., He, X., et al. (2014). GbMPK3, a mitogen-activated protein kinase from cotton, enhances drought and oxidative stress tolerance in tobacco. Plant Cell Tissue Organ Cult. 116, 153–162. doi: 10.1007/s11240-013-0392-1
Long, L., Zhao, J.-R., Guo, D.-D., Ma, X.-N., Xu, F.-C., Yang, W.-W., et al. (2020). Identification of NHXs in Gossypium species and the positive role of GhNHX1 in salt tolerance. BMC Plant Biol. 20, 1–13. doi: 10.1186/s12870-020-02345-z
Long, L., Zhao, J.-R., Xu, F.-C., Yang, W.-W., Liao, P., Gao, Y., et al. (2018). Silencing of GbANS reduces cotton resistance to Verticillium dahliae through decreased ROS scavenging during the pathogen invasion process. Plant Cell Tissue Organ Cult. 135, 213–221. doi: 10.1007/s11240-018-1457-y
Lu, P., Magwanga, R. O., Kirungu, J. N., Hu, Y., Dong, Q., Cai, X., et al. (2019). Overexpression of cotton a DTX/MATE gene enhances drought, salt, and cold stress tolerance in transgenic Arabidopsis. Front. Plant Sci. 10:299. doi: 10.3389/fpls.2019.00299
Lu, W., Chu, X., Li, Y., Wang, C., and Guo, X. (2013). Cotton GhMKK1 induces the tolerance of salt and drought stress, and mediates defence responses to pathogen infection in transgenic Nicotiana benthamiana. PLoS One 8:e68503. doi: 10.1371/journal.pone.0068503
Ludwig, A. A., Romeis, T., and Jones, J. D. (2004). CDPK-mediated signalling pathways: specificity and cross-talk. J. Exp. Bot. 55, 181–188. doi: 10.1093/jxb/erh008
Luo, Q., Wei, Q., Wang, R., Zhang, Y., Zhang, F., He, Y., et al. (2017). BdCIPK31, a calcineurin B-like protein-interacting protein kinase, regulates plant response to drought and salt stress. Front. Plant Sci. 8:1184. doi: 10.3389/fpls.2017.01184
Luo, X., Wu, J., Li, Y., Nan, Z., Guo, X., Wang, Y., et al. (2013). Synergistic effects of GhSOD1 and GhCAT1 overexpression in cotton chloroplasts on enhancing tolerance to methyl viologen and salt stresses. PLoS One 8:e54002. doi: 10.1371/journal.pone.0054002
Ma, L., Hu, L., Fan, J., Amombo, E., Khaldun, A., Zheng, Y., et al. (2017). Cotton GhERF38 gene is involved in plant response to salt/drought and ABA. Ecotoxicology 26, 841–854. doi: 10.1007/s10646-017-1815-2
Ma, Q., Wang, N., Ma, L., Lu, J., Wang, H., Wang, C., et al. (2020). The cotton BEL1-Like transcription factor GhBLH7-D06 negatively regulates the defense response against Verticillium dahliae. Int. J. Mol. Sci. 21:7126. doi: 10.3390/ijms21197126
Ma, W., Zhao, T., Li, J., Liu, B., Fang, L., Hu, Y., et al. (2016). Identification and characterization of the GhHsp20 gene family in Gossypium hirsutum. Sci. Rep. 6, 1–13. doi: 10.1038/srep32517
Marjamaa, K., Hildén, K., Kukkola, E., Lehtonen, M., Holkeri, H., Haapaniemi, P., et al. (2006). Cloning, characterization and localization of three novel class III peroxidases in lignifying xylem of Norway spruce (Picea abies). Plant Mol. Biol. 61, 719–732. doi: 10.1007/s11103-006-0043-6
Meng, C.-M., Zhang, T.-Z., and Guo, W.-Z. (2009). Molecular cloning and characterization of a novel Gossypium hirsutum L. bHLH gene in response to ABA and drought stresses. Plant Mol. Biol. Rep. 27, 381–387. doi: 10.1007/s11105-009-0112-5
Meng, J., Gao, H., Zhai, W., Shi, J., Zhang, M., Zhang, W., et al. (2018). Subtle regulation of cotton resistance to Verticillium wilt mediated by MAPKK family members. Plant Sci. 272, 235–242. doi: 10.1016/j.plantsci.2018.05.003
Meng, X., and Zhang, S. (2013). MAPK cascades in plant disease resistance signaling. Ann. Rev. Phytopathol. 51, 245–266. doi: 10.1146/annurev-phyto-082712-102314
Mittler, R. (2006). Abiotic stress, the field environment and stress combination. Trends Plant Sci. 11, 15–19. doi: 10.1016/j.tplants.2005.11.002
Mo, H., Wang, X., Zhang, Y., Zhang, G., Zhang, J., and Ma, Z. (2015). Cotton polyamine oxidase is required for spermine and camalexin signalling in the defence response to Verticillium dahliae. Plant J. 83, 962–975. doi: 10.1111/tpj.12941
Mo, H.-J., Sun, Y.-X., Zhu, X.-L., Wang, X.-F., Zhang, Y., Yang, J., et al. (2016). Cotton S-adenosylmethionine decarboxylase-mediated spermine biosynthesis is required for salicylic acid-and leucine-correlated signaling in the defense response to Verticillium dahliae. Planta 243, 1023–1039. doi: 10.1007/s00425-015-2463-5
Mu, C., Zhou, L., Shan, L., Li, F., and Li, Z. (2019). Phosphatase GhDs PTP 3a interacts with annexin protein Gh ANN 8b to reversely regulate salt tolerance in cotton (Gossypium spp.). New Phytol. 223, 1856–1872. doi: 10.1111/nph.15850
Mustafa, R., Hamza, M., Kamal, H., Mansoor, S., Scheffler, J., and Amin, I. (2017). tobacco rattle virus-based silencing of enoyl-CoA reductase gene and its role in resistance against cotton wilt disease. Mol. Biotechnol. 59, 241–250. doi: 10.1007/s12033-017-0014-y
Na, Y.-j., Choi, H.-k., Park, M. Y., Choi, S.-w., Xuan Vo, K. T., Jeon, J.-S., et al. (2019). OsMAPKKK63 is involved in salt stress response and seed dormancy control. Plant Signal. Behav. Genet. 14:e1578633. doi: 10.1080/15592324.2019.1578633
Nadarajah, K. K. (2020). ROS homeostasis in abiotic stress tolerance in plants. Int. J. Mol. Sci. 21:5208. doi: 10.3390/ijms21155208
Nazar, R. N., Xu, X., Kurosky, A., and Robb, J. (2018). Antagonistic function of the Ve R-genes in tomato. Plant Mol. Biol. 98, 67–79. doi: 10.1007/s11103-018-0764-3
Noctor, G., Mhamdi, A., and Foyer, C. H. (2014). The roles of reactive oxygen metabolism in drought: not so cut and dried. Plant Physiol. 164, 1636–1648. doi: 10.1104/pp.113.233478
Pandey, G. K., Kanwar, P., Singh, A., Steinhorst, L., Pandey, A., Yadav, A. K., et al. (2015). Calcineurin B-like protein-interacting protein kinase CIPK21 regulates osmotic and salt stress responses in Arabidopsis. Plant Physiol. 169, 780–792. doi: 10.1104/pp.15.00623
Pandey, P., Irulappan, V., Bagavathiannan, M. V., and Senthil-Kumar, M. (2017). Impact of combined abiotic and biotic stresses on plant growth and avenues for crop improvement by exploiting physio-morphological traits. Front. Plant Sci. 8:537. doi: 10.3389/fpls.2017.00537
Pandey, P., Singh, J., Achary, V., and Reddy, M. K. (2015). Redox homeostasis via gene families of ascorbate-glutathione pathway. Front. Environ. Sci. 3:25. doi: 10.3389/fenvs.2015.00025
Pei, Y., Zhu, Y., Jia, Y., Ge, X., Li, X., Li, F., et al. (2020). Molecular evidence for the involvement of cotton GhGLP2, in enhanced resistance to Verticillium and Fusarium wilts and oxidative stress. Sci. Rep. 10, 1–15. doi: 10.1038/s41598-020-68943-x
Prashanth, S., Sadhasivam, V., and Parida, A. (2008). Over expression of cytosolic copper/zinc superoxide dismutase from a mangrove plant Avicennia marina in indica rice var Pusa Basmati-1 confers abiotic stress tolerance. Transg. Res. 17, 281–291. doi: 10.1007/s11248-007-9099-6
Qin, L.-X., Nie, X.-Y., Hu, R., Li, G., Xu, W.-L., and Li, X.-B. (2016). Phosphorylation of serine residue modulates cotton Di19-1 and Di19-2 activities for responding to high salinity stress and abscisic acid signaling. Sci. Rep. 6, 1–15. doi: 10.1038/srep20371
Qin, T., Liu, S., Zhang, Z., Sun, L., He, X., Lindsey, K., et al. (2019). GhCyP3 improves the resistance of cotton to Verticillium dahliae by inhibiting the E3 ubiquitin ligase activity of GhPUB17. Plant Mol. Biol. 99, 379–393. doi: 10.1007/s11103-019-00824-y
Qiu, T., Wang, Y., Jiang, J., Zhao, J., Wang, Y., and Qi, J. (2020). GbAt11 gene cloned from Gossypium barbadense mediates resistance to Verticillium wilt in Gossypium hirsutum. J. Cott. Res. 3, 1–10. doi: 10.1186/s42397-020-00047-3
Rizhsky, L., Liang, H., Shuman, J., Shulaev, V., Davletova, S., and Mittler, R. (2004). When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol. 134, 1683–1696. doi: 10.1104/pp.103.033431
Sadau, S. B., Ahmad, A., Tajo, S. M., Ibrahim, S., Kazeem, B. B., Wei, H., et al. (2021). Overexpression of GhMPK3 from cotton enhances cold, drought, and salt stress in Arabidopsis. Agronomy 11:1049. doi: 10.3390/agronomy11061049
Sakuma, Y., Liu, Q., Dubouzet, J. G., Abe, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration-and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009. doi: 10.1006/bbrc.2001.6299
Salih, H., Gong, W., He, S., Sun, G., Sun, J., and Du, X. (2016). Genome-wide characterization and expression analysis of MYB transcription factors in Gossypium hirsutum. BMC Genet. 17, 1–12. doi: 10.1186/s12863-016-0436-8
Santiago, J., Rodrigues, A., Saez, A., Rubio, S., Antoni, R., Dupeux, F., et al. (2009). Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 60, 575–588. doi: 10.1111/j.1365-313X.2009.03981.x
Shang, X., Yu, Y., Zhu, L., Liu, H., Chai, Q., and Guo, W. (2020). A cotton NAC transcription factor GhirNAC2 plays positive roles in drought tolerance via regulating ABA biosynthesis. Plant Sci. 296:110498. doi: 10.1016/j.plantsci.2020.110498
Shazadee, H., Khan, N., Wang, J., Wang, C., Zeng, J., Huang, Z., et al. (2019). Identification and expression profiling of protein phosphatases (pp2c) gene family in Gossypium hirsutum L. Int. J. Mol. Sci. 20:1395. doi: 10.3390/ijms20061395
Shen, G., Wei, J., Qiu, X., Hu, R., Kuppu, S., Auld, D., et al. (2015). Co-overexpression of AVP1 and AtNHX1 in cotton further improves drought and salt tolerance in transgenic cotton plants. Plant Mol. Biol. Rep. 33, 167–177. doi: 10.1007/s11105-014-0739-8
Shi, H., Liu, Z., Zhu, L., Zhang, C., Chen, Y., Zhou, Y., et al. (2012). Overexpression of cotton (Gossypium hirsutum) dirigent1 gene enhances lignification that blocks the spread of Verticillium dahliae. Acta Biochim. Biophys. Sin. 44, 555–564. doi: 10.1093/abbs/gms035
Shi, J., An, H.-L., Zhang, L., Gao, Z., and Guo, X.-Q. (2010). GhMPK7, a novel multiple stress-responsive cotton group C MAPK gene, has a role in broad spectrum disease resistance and plant development. Plant Mol. Biol. 74, 1–17. doi: 10.1007/s11103-010-9661-0
Shi, J., Zhang, L., An, H., Wu, C., and Guo, X. (2011). GhMPK16, a novel stress-responsive group D MAPK gene from cotton, is involved in disease resistance and drought sensitivity. BMC Mol. Biol. 12, 1–15. doi: 10.1186/1471-2199-12-22
Solomon, S., Manning, M., Marquis, M., and Qin, D. (2007). Climate Change 2007-the Physical Science Basis: Working Group I Contribution to the Fourth Assessment Report of the IPCC. Cambridge: Cambridge University Press.
Song, R., Li, J., Xie, C., Jian, W., and Yang, X. (2020). An overview of the molecular genetics of plant resistance to the Verticillium wilt pathogen Verticillium dahliae. Int. J. Mol. Sci. 21:1120. doi: 10.3390/ijms21031120
Song, Y., Zhang, Z., Seidl, M. F., Majer, A., Jakse, J., Javornik, B., et al. (2017). Broad taxonomic characterization of Verticillium wilt resistance genes reveals an ancient origin of the tomato Ve1 immune receptor. Mol. Plant Pathol. 18, 195–209. doi: 10.1111/mpp.12390
Sornaraj, P., Luang, S., Lopato, S., and Hrmova, M. (2016). Basic leucine zipper (bZIP) transcription factors involved in abiotic stresses: a molecular model of a wheat bZIP factor and implications of its structure in function. Biochim. Biophys. Acta Gen. Sub. 1860, 46–56. doi: 10.1016/j.bbagen.2015.10.014
Su, Y., Guo, A., Huang, Y., Wang, Y., and Hua, J. (2020). GhCIPK6a increases salt tolerance in transgenic upland cotton by involving in ROS scavenging and MAPK signaling pathways. BMC Plant Biol. 20, 1–19. doi: 10.1186/s12870-020-02548-4
Sun, H., Hu, M., Li, J., Chen, L., Li, M., Zhang, S., et al. (2018). Comprehensive analysis of NAC transcription factors uncovers their roles during fiber development and stress response in cotton. BMC Plant Biol. 18, 1–15. doi: 10.1186/s12870-018-1367-5
Suzuki, N., Rivero, R. M., Shulaev, V., Blumwald, E., and Mittler, R. (2014). Abiotic and biotic stress combinations. New Phytol. 203, 32–43. doi: 10.1111/nph.12797
Tang, Y., Zhang, Z., Lei, Y., Hu, G., Liu, J., Hao, M., et al. (2019). Cotton WATs modulate SA biosynthesis and local lignin deposition participating in plant resistance against Verticillium dahliae. Front. Plant Sci. 10:526. doi: 10.3389/fpls.2019.00526
Tao, C., Jin, X., Zhu, L., Xie, Q., Wang, X., and Li, H. (2018). Genome-wide investigation and expression profiling of APX gene family in Gossypium hirsutum provide new insights in redox homeostasis maintenance during different fiber development stages. Mol. Genet. Genom. 293, 685–697. doi: 10.1007/s00438-017-1413-2
Tian, M., Lou, L., Liu, L., Yu, F., Zhao, Q., Zhang, H., et al. (2015). The RING finger E3 ligase STRF1 is involved in membrane trafficking and modulates salt-stress response in Arabidopsis thaliana. Plant J. 82, 81–92. doi: 10.1111/tpj.12797
Tiwari, M., Sharma, D., Singh, M., Tripathi, R. D., and Trivedi, P. K. (2014). Expression of OsMATE1 and OsMATE2 alters development, stress responses and pathogen susceptibility in Arabidopsis. Sci. Rep. 4, 1–12. doi: 10.1038/srep03964
Tuteja, N., and Mahajan, S. (2007). Calcium signaling network in plants: an overview. Plant Signal. Behav. 2, 79–85. doi: 10.4161/psb.2.2.4176
Ullah, A., Qamar, M. T. U., Nisar, M., Hazrat, A., Rahim, G., Khan, A. H., et al. (2020). Characterization of a novel cotton MYB gene, GhMYB108-like responsive to abiotic stresses. Mol. Biol. Rep. 47, 1573–1581. doi: 10.1007/s11033-020-05244-6
Ullah, A., Sun, H., Yang, X., and Zhang, X. (2017). Drought coping strategies in cotton: increased crop per drop. Plant Biotechnol. J. 15, 271–284. doi: 10.1111/pbi.12688
Vinocur, B., and Altman, A. (2005). Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr. Opin. Biotechnol. 16, 123–132. doi: 10.1016/j.copbio.2005.02.001
Wang, C., Guo, H., He, X., Zhang, S., Wang, J., Wang, L., et al. (2020). Scaffold protein GhMORG1 enhances the resistance of cotton to Fusarium oxysporum by facilitating the MKK6-MPK4 cascade. Plant Biotechnol. J. 18, 1421–1433. doi: 10.1111/pbi.13307
Wang, C., He, X., Li, Y., Wang, L., Guo, X., and Guo, X. (2018). The cotton MAPK kinase GhMPK20 negatively regulates resistance to Fusarium oxysporum by mediating the MKK4–MPK20–WRKY40 cascade. Mol. Plant Pathol. 19, 1624–1638. doi: 10.1111/mpp.12635
Wang, C., Lu, G., Hao, Y., Guo, H., Guo, Y., Zhao, J., et al. (2017). ABP9, a maize bZIP transcription factor, enhances tolerance to salt and drought in transgenic cotton. Planta 246, 453–469. doi: 10.1007/s00425-017-2704-x
Wang, C., Lu, W., He, X., Wang, F., Zhou, Y., Guo, X., et al. (2016). The cotton mitogen-activated protein kinase kinase 3 functions in drought tolerance by regulating stomatal responses and root growth. Plant Cell Physiol. 57, 1629–1642. doi: 10.1093/pcp/pcw090
Wang, G., Liang, Y.-h., Zhang, J.-y., and Cheng, Z.-M. (2020). Cloning, molecular and functional characterization by overexpression in Arabidopsis of MAPKK genes from grapevine (Vitis vinifera). BMC Plant Biol. 20:194. doi: 10.1186/s12870-020-02378-4
Wang, J., Lu, X., Yin, Z., Mu, M., Zhao, X., Wang, D., et al. (2016). Genome-wide identification and expression analysis of CIPK genes in diploid cottons. Genet Mol. Res. 15:4238. doi: 10.4238/gmr15048852
Wang, J., Pan, C., Wang, Y., Ye, L., Wu, J., Chen, L., et al. (2015). Genome-wide identification of MAPK, MAPKK, and MAPKKK gene families and transcriptional profiling analysis during development and stress response in cucumber. BMC Genom. 16, 1–22. doi: 10.1186/s12864-015-1621-2
Wang, L., Ding, L., Yu, Z., Zhang, T., Ma, S., and Liu, J. (2016). Intracellular ROS scavenging and antioxidant enzyme regulating capacities of corn gluten meal-derived antioxidant peptides in HepG2 cells. Food Res. Int. 90, 33–41. doi: 10.1016/j.foodres.2016.10.023
Wang, N.-N., Xu, S.-W., Sun, Y.-L., Liu, D., Zhou, L., Li, Y., et al. (2019). The cotton WRKY transcription factor (GhWRKY33) reduces transgenic Arabidopsis resistance to drought stress. Sci. Rep. 9, 1–13. doi: 10.1038/s41598-018-37035-2
Wang, P., Su, L., Qin, L., Hu, B., Guo, W., and Zhang, T. (2009). Identification and molecular mapping of a Fusarium wilt resistant gene in upland cotton. Theor. Appl. Genet. 119, 733–739. doi: 10.1007/s00122-009-1084-4
Wang, P.-Z., Shi, L.-F., Li, S., and Hu, B.-M. (2010). Quantitative trait loci for resistance against Fusarium wilt based on three cotton F2 populations. Agricult. Sci. China 9, 1799–1806. doi: 10.1016/S1671-2927(09)60278-9
Wang, S. Y., and Jiao, H. J. (2000). Scavenging capacity of berry crops on superoxide radicals, hydrogen peroxide, hydroxyl radicals, and singlet oxygen. J. Agricult. Food Chem. 48, 5677–5684. doi: 10.1021/jf000766i
Wang, W., Chen, D., Liu, D., Cheng, Y., Zhang, X., Song, L., et al. (2020). Comprehensive analysis of the Gossypium hirsutum L. respiratory burst oxidase homolog (Ghrboh) gene family. BMC Genom. 21, 1–19. doi: 10.1186/s12864-020-6503-6
Wang, W., Cheng, Y., Chen, D., Liu, D., Hu, M., Dong, J., et al. (2019). The catalase gene family in cotton: genome-wide characterization and bioinformatics analysis. Cells 8:86. doi: 10.3390/cells8020086
Wang, W., Vinocur, B., and Altman, A. (2003). Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218, 1–14. doi: 10.1007/s00425-003-1105-5
Wang, W., Xia, M., Chen, J., Deng, F., Yuan, R., Zhang, X., et al. (2016a). Genome-wide analysis of superoxide dismutase gene family in Gossypium raimondii and G. arboreum. Plant Gene 6, 18–29. doi: 10.1016/j.plgene.2016.02.002
Wang, W., Yuan, Y., Yang, C., Geng, S., Sun, Q., Long, L., et al. (2016b). Characterization, expression, and functional analysis of a novel NAC gene associated with resistance to Verticillium wilt and abiotic stress in cotton. G3 Genes Genom Genet. 6, 3951–3961. doi: 10.1534/g3.116.034512
Wang, W., Zhang, X., Deng, F., Yuan, R., and Shen, F. (2017). Genome-wide characterization and expression analyses of superoxide dismutase (SOD) genes in Gossypium hirsutum. BMC Genom. 18, 1–25. doi: 10.1186/s12864-017-3768-5
Wang, X., Cui, Y., Qiao, K., Fu, Z. L., Fan, S., and Ma, Q. (2020a). Comprehensive genome-wide analysis of thaumatin-like gene family in four cotton species and functional identification of GhTLP19 involved in regulating tolerance to Verticillium dahlia and drought. Front. Plant Sci. 11:1597. doi: 10.3389/fpls.2020.575015
Wang, X., Lu, X., Malik, W. A., Chen, X., Wang, J., Wang, D., et al. (2020b). Differentially expressed bZIP transcription factors confer multi-tolerances in Gossypium hirsutum L. Int. J. Biol. Macromol. 146, 569–578. doi: 10.1016/j.ijbiomac.2020.01.013
Xie, Z., Nolan, T. M., Jiang, H., and Yin, Y. (2019). AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front. Plant Sci. 10:228. doi: 10.3389/fpls.2019.00228
Xiong, X.-P., Sun, S.-C., Zhu, Q.-H., Zhang, X.-Y., Li, Y.-J., Liu, F., et al. (2021). The cotton lignin biosynthetic gene Gh4CL30 regulates lignification and phenolic content and contributes to Verticillium wilt resistance. Mol. Plant Microbe Interact. 34, 240–254. doi: 10.1094/MPMI-03-20-0071-R
Xu, J., Wang, G., Wang, J., Li, Y., Tian, L., Wang, X., et al. (2017). The lysin motif-containing proteins, Lyp1, Lyk7 and LysMe3, play important roles in chitin perception and defense against Verticillium dahliae in cotton. BMC Plant Biol. 17, 1–18. doi: 10.1186/s12870-017-1096-1
Xu, J., Xu, X., Tian, L., Wang, G., Zhang, X., Wang, X., et al. (2016). Discovery and identification of candidate genes from the chitinase gene family for Verticillium dahliae resistance in cotton. Sci. Rep. 6, 1–12. doi: 10.1038/srep29022
Xu, L., Zhu, L., Tu, L., Liu, L., Yuan, D., Jin, L., et al. (2011). Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J. Exp. Bot. 62, 5607–5621. doi: 10.1093/jxb/err245
Yan, Y., Jia, H., Wang, F., Wang, C., Liu, S., and Guo, X. (2015). Overexpression of GhWRKY27a reduces tolerance to drought stress and resistance to Rhizoctonia solani infection in transgenic Nicotiana benthamiana. Front. Physiol. 6:265. doi: 10.3389/fphys.2015.00265
Yan, Z., Xingfen, W., Wei, R., Jun, Y., and Zhiying, M. (2016). Island cotton enhanced disease susceptibility 1 gene encoding a lipase-like protein plays a crucial role in response to Verticillium dahliae by regulating the SA level and H2O2 accumulation. Front. Plant Sci. 7:1830. doi: 10.3389/fpls.2016.01830
Yang, J., Zhang, Y., Wang, X., Wang, W., Li, Z., Wu, J., et al. (2018). HyPRP1 performs a role in negatively regulating cotton resistance to V. dahliae via the thickening of cell walls and ROS accumulation. BMC Plant Biol. 18:1–18. doi: 10.1186/s12870-018-1565-1
Yang, X., Deng, F., and Ramonell, K. M. (2012). Receptor-like kinases and receptor-like proteins: keys to pathogen recognition and defense signaling in plant innate immunity. Front. Biol. 7:155–166. doi: 10.1007/s11515-011-1185-8
Yang, Y., Chen, T., Ling, X., and Ma, Z. (2018). Gbvdr6, a gene encoding a receptor-like protein of cotton (Gossypium barbadense), confers resistance to Verticillium wilt in Arabidopsis and upland cotton. Front. Plant Sci. 8:2272. doi: 10.3389/fpls.2017.02272
Yao, D., Zhang, X., Zhao, X., Liu, C., Wang, C., Zhang, Z., et al. (2011). Transcriptome analysis reveals salt-stress-regulated biological processes and key pathways in roots of cotton (Gossypium hirsutum L.). Genomics 98, 47–55. doi: 10.1016/j.ygeno.2011.04.007
Yin, Z., Zhu, W., Zhang, X., Chen, X., Wang, W., Lin, H., et al. (2021). Molecular characterization, expression and interaction of MAPK, MAPKK and MAPKKK genes in upland cotton. Genomics 113, 1071–1086. doi: 10.1016/j.ygeno.2020.11.004
Yu, T., Li, Y. S., Chen, X. F., Hu, J., Chang, X., and Zhu, Y. G. (2003). Transgenic tobacco plants overexpressing cotton glutathione S-transferase (GST) show enhanced resistance to methyl viologen. J. Plant Physiol. 160, 1305–1311. doi: 10.1078/0176-1617-01205
Yuan, H., Zhao, K., Lei, H., Shen, X., Liu, Y., Liao, X., et al. (2013). Genome-wide analysis of the GH3 family in apple (Malus× domestica). BMC Genom. 14, 1–14. doi: 10.1186/1471-2164-14-297
Zhang, B., Yang, Y., Chen, T., Yu, W., Liu, T., Li, H., et al. (2012). Island cotton Gbve1 gene encoding a receptor-like protein confers resistance to both defoliating and non-defoliating isolates of Verticillium dahliae. PLoS One 7:e51091. doi: 10.1371/journal.pone.0051091
Zhang, H., Huang, Y., Zhang, H., and Huang, Y. (2013). “Genome-wide survey and characterization of greenbug induced nac transcription factors in sorghum [Sorghum bicolor (L.) Moench],” in Proceedings of the XXI Annual International Plant & Animal Genome Conference, January 12–16, 2013, San Diego, CA, 0191.
Zhang, H., Zhang, W., Jian, G., Qi, F., and Si, N. (2016). The genes involved in the protective effects of phytohormones in response to Verticillium dahliae infection in Gossypium hirsutum. J. Plant Biol. 59, 194–202. doi: 10.1007/s12374-016-0568-4
Zhang, J., Zou, D., Li, Y., Sun, X., Wang, N.-N., Gong, S.-Y., et al. (2014). GhMPK17, a cotton mitogen-activated protein kinase, is involved in plant response to high salinity and osmotic stresses and ABA signaling. PLoS One 9:e95642. doi: 10.1371/journal.pone.0095642
Zhang, J.-B., Wang, X.-P., Wang, Y.-C., Chen, Y.-H., Luo, J.-W., Li, D.-D., et al. (2020). Genome-wide identification and functional characterization of cotton (Gossypium hirsutum) MAPKKK gene family in response to drought stress. BMC Plant Biol. 20, 1–14. doi: 10.1186/s12870-020-02431-2
Zhang, K., Pei, Z., Hongmei, W., Yunlei, Z., Wei, C., Haiyan, G., et al. (2019). Isolation and characterization of the GbVIP1 gene and response to Verticillium wilt in cotton and tobacco. J. Cotton Res. 2, 1–11. doi: 10.1186/s42397-019-0019-0
Zhang, L., Li, Y., Lu, W., Meng, F., Wu, C.-a., and Guo, X. (2012). Cotton GhMKK5 affects disease resistance, induces HR-like cell death, and reduces the tolerance to salt and drought stress in transgenic Nicotiana benthamiana. J. Exp. Bot. 63, 3935–3951. doi: 10.1093/jxb/ers086
Zhang, L., Wang, M., Li, N., Wang, H., Qiu, P., Pei, L., et al. (2018). Long noncoding RNA s involve in resistance to Verticillium dahliae, a fungal disease in cotton. Plant Biotechnol. J. 16, 1172–1185. doi: 10.1111/pbi.12861
Zhang, X., Wang, L., Xu, X., Cai, C., and Guo, W. (2014). Genome-wide identification of mitogen-activated protein kinase gene family in Gossypium raimondii and the function of their corresponding orthologs in tetraploid cultivated cotton. BMC Plant Biol. 14, 1–17. doi: 10.1186/s12870-014-0345-9
Zhang, Y., Wu, L., Wang, X., Chen, B., Zhao, J., Cui, J., et al. (2019). The cotton laccase gene GhLAC15 enhances Verticillium wilt resistance via an increase in defence-induced lignification and lignin components in the cell walls of plants. Mol. Plant Pathol. 20, 309–322. doi: 10.1111/mpp.12755
Zhang, Z., Wang, P., Luo, X., Yang, C., Tang, Y., Wang, Z., et al. (2019). Cotton plant defence against a fungal pathogen is enhanced by expanding BLADE-ON-PETIOLE1 expression beyond lateral-organ boundaries. Commun. Biol. 2, 1–15. doi: 10.1038/s42003-019-0468-5
Zhao, L., Lü, Y., Chen, W., Yao, J., Li, Y., Li, Q., et al. (2020). Genome-wide identification and analyses of the AHL gene family in cotton (Gossypium). BMC Genom. 21:69. doi: 10.1186/s12864-019-6406-6
Zhao, Y., Yang, Z., Ding, Y., Liu, L., Han, X., Zhan, J., et al. (2019). Over-expression of an R2R3 MYB Gene, GhMYB73, increases tolerance to salt stress in transgenic Arabidopsis. Plant Sci. 286, 28–36. doi: 10.1016/j.plantsci.2019.05.021
Zhou, F., Zheng, B., Wang, F., Cao, A., Xie, S., Chen, X., et al. (2021). Genome-wide analysis of MDHAR gene family in four cotton species provides insights into fiber development via regulating AsA redox homeostasis. Plants 10:227. doi: 10.3390/plants10020227
Zhou, Y., Sun, L., Wassan, G. M., He, X., Shaban, M., Zhang, L., et al. (2019). Gb SOBIR 1 confers Verticillium wilt resistance by phosphorylating the transcriptional factor Gbb HLH 171 in Gossypium barbadense. Plant Biotechnol. J. 17, 152–163. doi: 10.1111/pbi.12954
Zhu, W., Gao, E., Shaban, M., Wang, Y., Wang, H., Nie, X., et al. (2018). GhUMC1, a blue copper-binding protein, regulates lignin synthesis and cotton immune response. Biochem. Biophys. Res. Commun. 504, 75–81. doi: 10.1016/j.bbrc.2018.08.128
Keywords: cotton, genes, network, drought, salt, Verticillium, Fusarium wilt
Citation: Billah M, Li F and Yang Z (2021) Regulatory Network of Cotton Genes in Response to Salt, Drought and Wilt Diseases (Verticillium and Fusarium): Progress and Perspective. Front. Plant Sci. 12:759245. doi: 10.3389/fpls.2021.759245
Received: 16 August 2021; Accepted: 13 October 2021;
Published: 29 November 2021.
Edited by:
Xiaojuan Li, Beijing Forestry University, ChinaReviewed by:
Khayalethu Ntushelo, University of South Africa, South AfricaCopyright © 2021 Billah, Li and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fuguang Li, YXlsaWZ1Z0AxNjMuY29t; Zhaoen Yang, eWFuZ3poYW9lbjA5MjVAMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.