- 1Department of Plant Molecular Physiology, Polish Academy of Sciences, Institute of Bioorganic Chemistry, Poznań, Poland
- 2Dipartimento di Scienze Agro-Alimentari, Ambientali e Animali, University of Udine, Udine, Italy
- 3Institute of Plant and Microbial Biology, University of Zurich, Zurich, Switzerland
- 4Faculty of Science and Technology, Free University of Bozen Bolzano, Bolzano, Italy
- 5Department of Biochemistry and Biotechnology, Poznań University of Life Sciences, Poznań, Poland
- 6International Research Center for Environmental Membrane Biology, Foshan University, Foshan, China
Nitrogen (N) as well as Phosphorus (P) are key nutrients determining crop productivity. Legumes have developed strategies to overcome nutrient limitation by, for example, forming a symbiotic relationship with N-fixing rhizobia and the release of P-mobilizing exudates and are thus able to grow without supply of N or P fertilizers. The legume-rhizobial symbiosis starts with root release of isoflavonoids that act as signaling molecules perceived by compatible bacteria. Subsequently, bacteria release nod factors, which induce signaling cascades allowing the formation of functional N-fixing nodules. We report here the identification and functional characterization of a plasma membrane-localized MATE-type transporter (LaMATE2) involved in the release of genistein from white lupin roots. The LaMATE2 expression in the root is upregulated under N deficiency as well as low phosphate availability, two nutritional deficiencies that induce the release of this isoflavonoid. LaMATE2 silencing reduced genistein efflux and even more the formation of symbiotic nodules, supporting the crucial role of LaMATE2 in isoflavonoid release and nodulation. Furthermore, silencing of LaMATE2 limited the P-solubilization activity of lupin root exudates. Transport assays in yeast vesicles demonstrated that LaMATE2 acts as a proton-driven isoflavonoid transporter.
Introduction
One of the major challenges of sustainable agriculture comprises the production of high-quality plant material with preservation of soil components and reduced application of chemical fertilizers without penalizing yield. Nitrogen (N) and phosphorus (P) are limiting nutrients in most natural soils (Tilman, 1999). High input of N-fertilizers is required to sustain crop growth in conventional agriculture. However, this feature may contaminate soils and groundwater and markedly contribute to the release of greenhouse gases (Tilman et al., 2001). Much of the P in soils is not available to plants due to its tendency to interact with calcium and magnesium salts or iron and aluminum oxides. It is mainly present as sparingly soluble rock phosphate, or it is immobilized in slowly mineralizable P-containing organic compounds, such as phytates. Phosphorous is a non-renewable resource which is mined at an increasing rate to meet the demand for fertilizers (Vance et al., 2003).
Leguminous plants (Fabaceae) such as soybean and lupin, have evolved several strategies to survive in low nutrient soils. In the case of P, in many ecosystems and, in particular, in acidic soils, the plant’s response consists mainly in the association with mycorrhizal fungi or the formation of particular root structures, such as cluster roots (Purnell, 1960; Neumann and Martinoia, 2002; Lambers et al., 2015). Cluster roots release huge amounts of exudates into the rhizosphere which are mainly composed of carboxylates and flavonoids. Flavonoids are involved both in the mobilization of nutrients and in the modulation of soil microbial activities (Cesco et al., 2010). The production and release of flavonoids, are also essential for the establishment of the symbiotic interaction between legumes and N-fixing bacteria such as Ensifer, Bradyrhizobium or Mesorhizobium, leading to the fixation of atmospheric N. Moreover it has been hypothesized that flavonoids are involved in the initiation of the nodule through their action on the plant hormone auxin and could thus play a developmental role in addition to their action as nod gene regulators (Subramanian et al., 2006; Wasson et al., 2006; Li et al., 2016).
The mutualistic fungal and bacterial symbionts are striking examples of soil microorganisms that have successfully coevolved with their hosts since plants adapt to terrestrial ecosystems. They promote plant growth by facilitating the acquisition of scarce nutrients. The most commonly established symbiosis in plants is the mycorrhizal association, with 80–90% of all land-plant species able to enter this interaction. Around 100 Mio years ago, certain angiosperms evolved a bias toward the evolution of nodulation with the so-called N-fixing soil bacteria. Among those angiosperms are legumes (Fabales) and one non-legume genus, Parasponia (Cannabaceae, Rosales) which can establish mutualistic symbioses with Rhizobia, a polyphyletic group of proteobacteria and diverse group of plants belonging to the orders Fagales, Rosales, and Cucurbitales which can associate symbiotically with filamentous actinobacteria of the genus Frankia (Martin et al., 2017). By forming symbiotic associations, plants obtain mineral nutrients. In turn, they supply the symbiont with organic compounds, sugars and lipids in the case of mycorrhiza, mostly carboxylates to N-fixing bacteria (Udvardi and Poole, 2013; Jiang et al., 2017; Luginbuehl et al., 2017). The establishment of the symbiosis is a complex event and requires coordinated regulation of the corresponding genes and release of signaling molecules into the rhizosphere. For the legume-rhizobia symbiosis, it is expected that a flavonoid transporter must be present in the plasma membrane of root cells to release isoflavonoids into the rhizosphere (Sugiyama et al., 2007). Up to now, transporters for flavonoids have been mainly described at the vacuolar membrane (Zhao, 2015). Furthermore, an ABC (ATP-Binding Cassette) transporter from Medicago was shown to transport flavonoids. However, this transporter is localized in the vasculature (Biala et al., 2017). Using a biochemical approach Sugiyama et al. (2007) presented evidence that, in soybean, genistein transmembrane transport is mediated by an ABC-type transport system. But its contribution to genistein root release and the legume-rhizobia symbiosis establishment is still unclear.
Despite all the research performed on this symbiotic interaction, a transporter releasing flavonoids into the rhizosphere and initiating the first step of this symbiosis awaits its identification.
Materials and Methods
Plant Growth and Transformation
White lupin seeds (Lupinus albus L. cv. Amiga; Südwestdeutsche Saatzucht, Rastatt, Germany) were soaked for 24 h in aerated water and germinated on a plastic net placed at the surface of an aerated 0.5 mM CaSO4 solution in a growth chamber at 25°C in the dark. Thereafter, 7-day-old seedlings were transferred to a hydroponic system, containing a P-free nutrient solution (μM): 5000 Ca(NO3)2, 1,250 MgSO4, 1750 K2SO4, 250 KCl, 20 Fe(III)EDTA, 25 H3BO4, 1.25 MnSO4, 1.5 ZnSO4, 0.5 CuSO4, 0.025 (NH4)6Mo7O4. Phosphorus-deficient plants were grown on P-free nutrient solution, while 0.25 mM KH2PO4 were added to nutrient solution for P-sufficient condition. Plants were grown under controlled conditions for 4 weeks (day/night photoperiod, 16/8 h; radiation, 220 μE m−2 s−1; day/night temperature, 25/20°C; relative humidity, 70–80%).
In P-deficiency, lupin plants modify the root architecture developing particularly root structures, called cluster roots or proteoid roots. In order to differentiate the developmental stages of root clusters (Supplementary Figure S1), the root system was immerged in a pH-indicator solution (0.04% w/v bromocresol purple). Depending on their morphology and capability to acidify the solution, at the end of the growing period different regions of cluster root were sampled from P-deficient plants (juvenile, immature, mature, senescent), as described by Massonneau et al. (2001). Root apices and cluster root parts were sampled from a pool of 12–16 P-sufficient and P-deficient plants. The samples were immediately frozen in liquid nitrogen for the RNA extraction or were rinsed twice in 0.5 mM CaSO4 solution and the root exudates were collected for 1 hour in 0.5 mM CaSO4 10 mM 2-[N-Morpholino] ethanesulfonic acid (MES)-KOH pH 6.0 at a ratio 1:10 W/V. The samples were conserved at −80°C until processing. Six independent experiments were performed.
To investigate the plant response to N-deficiency, white lupin seeds were grown for 2 weeks on watered Whatman paper in Petri dishes and then transferred to the magenta boxes containing PFR N-free solution described by Strozycki et al. (2003). For N-sufficient conditions KNO3 (0.1 mM), NH4H2PO4 (5 mM), Ca(NO3)2 (2.46 mM) were added to the PFR medium. Plants were grown for further 2 weeks under controlled conditions (day/night photoperiod, 16/8 h; radiation, 220 μE m−2s−1; day/night temperature, 23/20°C; relative humidity, 70–80%). The samples were collected at 7 and 14 days after the transfer in N-deficient condition. Three independent experiments were performed; data shown are from the third experiment.
White lupin seedlings were transformed using Agrobacterium rhizogenes ARqua1 strain (Quandt et al., 1993) carrying binary vector pRedRoot::LaMATE2 RNAi or empty vector (EV) pRedRoot. After germination 5 mm root tips were removed from radicles. The sectioned surface was coated with A. rhizogenes and seedlings were placed on solid Fahraeus medium supplemented with kanamycin (15 mgL−1).
Nitrogen-deficient plants were grown and analyzed as described above. For P-deficiency, after 3 weeks post germination, plants were transferred to P-free nutrient solution (as described above). After 2 weeks, white lupin plants were moved in hydroponic solutions in P-free nutrient solution and grown for 3 additional weeks. LaMATE2 expression analyses were performed, and root release was collected as described previously on the fully developed cluster root (immature, mature). Six biological replicates were performed for each sample.
Nodulation and Effects of Genistein Exogenous Addition
Three-week-old lupin composite plants were transferred to the pots filled with sterile perlite (0.75l). Plants were nourished with PFR N-free solution. After 2 weeks of N-deficiency, plants were inoculated with B. japonicum (strain UPP 133 (Stępkowski et al., 2011)) and grown for further 2 weeks under controlled conditions (day/night photoperiod, 16/8h; radiation, 220 μE m−2s−1; day/night temperature, 23/20°C; relative humidity, 70–80%). For complementation experiments 1h before inoculation with B. japonicum plants were additionally supplemented with 1 μM genistein solution. Fourteen-day post-inoculation, plants were removed from pots, nodules were counted for each single root, and roots were collected for each plant. Afterwards all roots of single plant were grounded and divided into aliquots dedicated for gene expression and metabolomic analysis. Only plants revealing expression of marker gene encoding fluorescent protein DsRed were considered during analysis. Three independent transformations were used with sample sizes 6, 8 and 21.
Mobilization of P From Vivianite
The vivianite suspension (Fe3(PO4)2.8H2O) was prepared by mixing H3PO4 with FeSO4·as described by Eynard et al. (1992). A solution of 0.035 M H332PO4 and 0.05 M FeSO4·7H2O was brought to pH 6.0 with 0.05M KOH under stirring. A bluish suspension of vivianite was obtained, after precipitation via centrifugation (8,000 g, 10 min), the precipitate was washed (by 3 successive centrifugations and decantations) with deionized water. Mobilization of PO42− from Vivianite (Fe3(PO4)2.8H2O) by exudates after purification with the SPE methods, in order to remove carboxylates, from LaMATE2 RNAi or empty vector (EV) pRedRoot transformed P-deficient roots was performed as follows: 1ml of a suspension containing 32P-vivianite (70 μmol PO4, specific activity 60 KBq μmol−1 P) was transferred into a dialysis tube (ZelluTrans/Roth 6.0, ∅ 16 mm, exclusion limit of 4÷6kDa, ROTH) and mixed with 5 ml of mobilization solution [0.5 mM CaSO4, 20 mM Mes-KOH (pH 6.0), 50 μl methanol]. Thereafter, the dialysis tube was transferred into continuously mixed 34ml of mobilization solution. At the beginning of the experiment, exudates corresponding to the amount release for 1 h by 1 gram of juvenile cluster root tissues (Figure 1) were added to the external solution. After 15, 30, 45 and 60 min, samples from the solution outside the dialysis tube were collected and the amount of 32P was measured by liquid scintillation counting. The 32P mobilization was estimated from the difference between the 32P concentration measured in the presence of the mock and in the absence of exudates and was expressed as μmol P h−1.
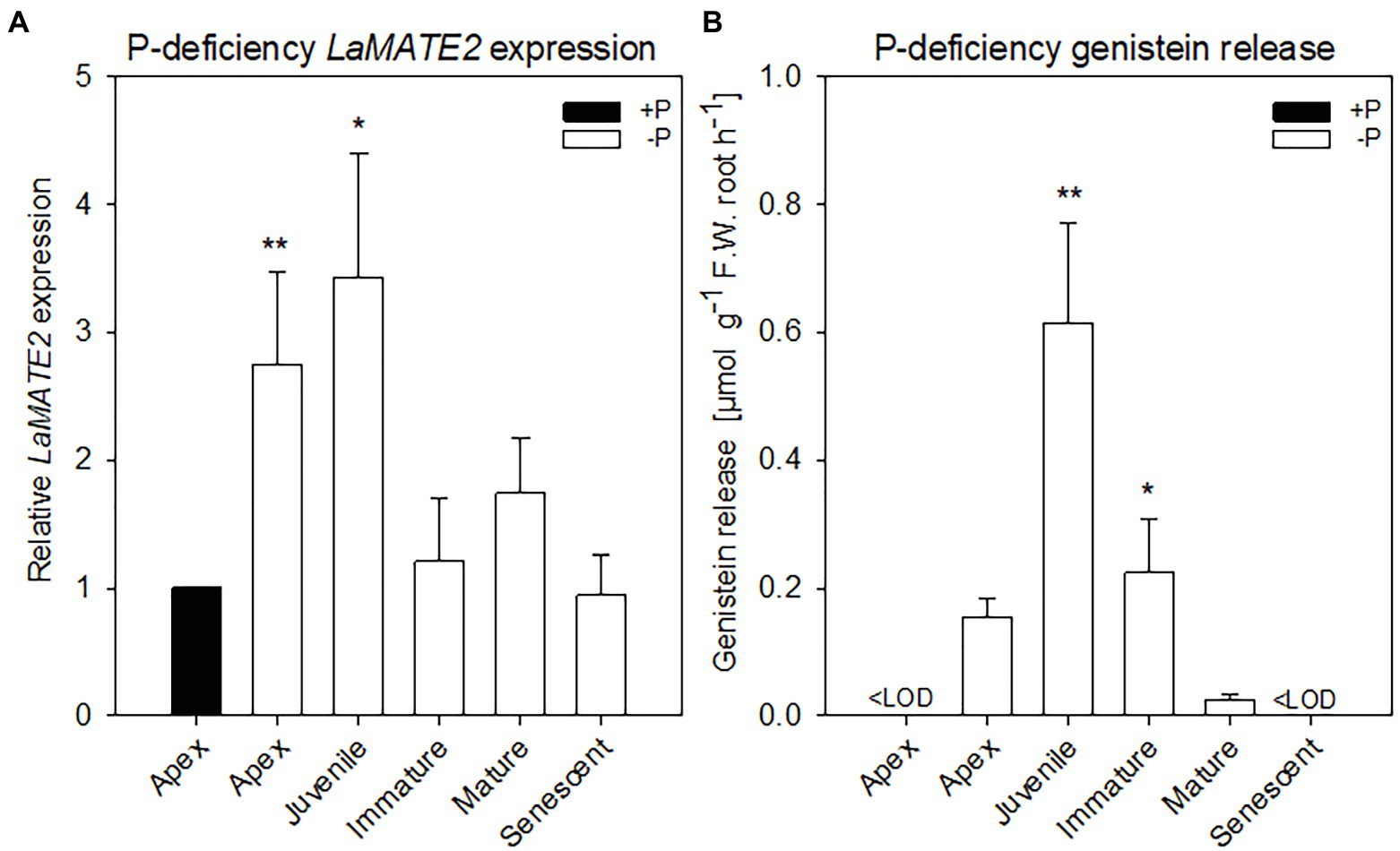
Figure 1. LaMATE2 expression level and genistein release in roots of P-deficient white lupin. LaMATE2 expression analyses in white lupin grown under control (+N + P, black bars) condition or P deficiency (+N-P, white bars) (A). Gene expression was evaluated in control apex (+N + P), P-deficient apex and cluster roots (separated depending on developing stages: Juvenile, Immature, Mature, Senescent cluster-root stages; example of cluster root shown in Supplementary Figure S1). Expression levels are shown relative to the LaMATE2 expression level of the root apex under control conditions (+N + P). Release of genistein from different root tissues of 4-week-old P-deficient plants, release from the control apex was below detectable value <LOD (B). Data are mean + SD (Asterisks refer to statistically significant differences among the mean value of the sample vs control or -P apex for genistein release, ANOVA Holm–Sidak, N=6, *p<0.05, **p<0.01; with 3 technical replicates for each real-time PCR).
Extraction and LC/MS Analysis
Frozen root tissue (500 mg) was grounded and extracted with 80% methanol. Root exudates were extracted from the medium by SPE (Solid Phase Extraction) method using octadecylosilane matrix and methanol according to Staszków et al. (2011). Luteolin was used as an internal standard. Samples were analyzed by liquid chromatography-electrospray ionization tandem mass spectrometry (LC/ESI/MS) using a Waters UPLC coupled with Bruker micrOTOF-Q mass spectrometer. The analysis was performed in a gradient mobile phase consisting of 0.5% formic acid (v/v) in water (A) and 0.5% formic acid (v/v) in acetonitrile (B). The m/z range of the recorded spectra was 50–1,000. Analyses were performed in the ion-positive mode.
RNA Extraction and cDNA Synthesis
RNA extractions were performed using the InviTrap Spin Plant RNA Mini Kit (Stratec Molecular, Berlin, Germany) following the manufacturer’s instructions and contaminant genomic DNA was removed using 10 U of DNase I (GE Healthcare, Munich, Germany). The quantity and the quality of RNA were checked using a spectrophotometer, followed by a migration in a 1% agarose gel. One microgram of total RNA for each sample was retro-transcribed using 1 pmol Oligo d(T)23 (Sigma Aldrich, Saint Louis, United States) and 10 UM-MulV RNase H (Finnzymes, Helsinki, Finland) following manufacturers’ instruction.
Isolation of the LaMATE2 Sequence
The partial sequence was isolated via a cDNA-AFLP approach starting from RNA extracted from juvenile cluster roots which were compared to mature and senescent cluster roots; for details, see Massonneau et al. (2001). The full Open Reading Frame (ORF) of LaMATE2 (LaMATE2ORF) was isolated from the cDNA of juvenile cluster root tissues from P-deficient plants using the 5'/3' RACE Kit (2nd generation, Roche Diagnostics S.p.a., Monza, Italy) following the manufacturer’s instructions. LaMATE2ORF sequence was cloned in pGEM-T easy vector (for primers, see Supplementary Table S1, Promega Italia Srl, Milan, Italy) and deposited in the NCBI database (KY464927).
Gene Expression Analysis
The reaction was performed by adding 0.1 μl of cDNA to RT complete reaction mix, Fluocycle™ sybr green (20-μl final volume, Euroclone, Pero, Italy). Specific primers were designed for the target, the two closest homologues (www.whitelupin.fr; Hufnagel et al., 2020) and the housekeeping gene using Primer3 software (Koressaar and Remm, 2007; Untergasser et al., 2012) and were synthesized by Sigma Aldrich (Supplementary Table S1). Gene expression analyses were performed using CFX96™ Real-Time System (C1000TM Thermal Cycler, BioRad) and CFX Manager™ Software (v 2.0, BioRad). The two closest homologues (Lalb_Chr06g0165721, Lalb_Chr02g0145611) have a low expression in the root tissues and their expression levels are shown in Supplementary Figure S5. Data were normalized in respect to the transcript level of the housekeeping gene (Ubiquitin gene, LaUBI) using the 2-ΔΔCT method (Livak and Schmittgen, 2001). The efficiency of amplification was calculated using R program (version 2.9.0)1 with the qPCR package (version 1.1–8), following the authors’ indications (Ritz and Spiess, 2008). Six biological replicates were performed for each sample with 3 technical replicates.
Genistein Transport by LaMATE2 in Saccharomyces Cerevisiae Vesicles
The construction of the yeast expression vector pNEV (Sauer and Stolz, 1994) containing LaMATE2ORF was performed by amplifying LaMATE2ORF sequence with primers having both NotI-restriction sites at the 5’end. The cloning of the PCR product (LaMATE2ORF) was performed into the NotI site of pNEV (pNEV::LaMATE2ORF), the orientation was verified by sequencing. The transformation of competent yeast cells (S. cerevisiae YPH499 strain) was performed following a standard procedure (Gietz and Woods, 2002) and transformants were selected on synthetic dextrose minimal medium lacking uracil (SD-Ura medium; Burke et al., 2000).
The yeast cells, strain YPH499, were transformed with the pNEV empty vector or pNEV::LaMATE2ORF and selected on liquid-SD Ura-medium. Thereafter, cells were incubated in YPD medium for 30 min, collected by centrifugation, and digested with lyticase (1,000 Ug−1 fresh weight cells; Sigma Aldrich), and subsequently microsomal vesicles were isolated as described by Klein et al. (2002). Transport assays were performed to study the genistein transport using the rapid filtration technique with nitrocellulose filters (0.45 μM pore size; Millipore, Millipore Co., Bedford, United States). The transport experiment was carried out in the presence of isolated vesicles, transport buffer (0.4 M glycerol, 0.1 M KCl, 1 mM DTT, 1 mM EDTA, 5 mM ATP, 10 mM MgCl2, 10 mM creatine phosphate, 0.1 mgml−1 creatine kinase, 20 mM Tris-MES pH 7.4) and 5 μM of labelled 3H-genistein (American Radiolabeled Chemicals, Saint Louis, United States; 1850 Bq filter−1). Only for kinetic experiments, the 3H-genistein concentration ranged from 5 μM up to 100 μM (5, 7, 10, 15, 33, 50 and 100 μM 3H-genistein) and the incubation time was 30 s. The concentration-dependency of 3H-genistein uptake were calculated by subtracting uptake rates recorded in the empty-vector vesicles. The concentration-dependency of 3H-genistein uptake were calculated between 7 and 100 μM by subtracting uptake rates recorded in the empty-vector vesicles using the Hanes–Woolf plot.
To test the dependence of genistein transport on a proton gradient, 25 mM NH4Cl was included in the transport buffer assay and the incubation time ranging between 15 and 120 s (15, 30, 60 and 120 s). Moreover, the capability of LaMATE2 to mediate the uptake of different flavonoids was tested by UPLC under the same experimental conditions reported above (5 μM of flavonoid: genistein, genistin, hydroxygenistein, biochanin A, daidzein, or kaempferol; the incubation time was 30 s). The mixture was loaded on a pre-wetted filter and removed by suction at the end of incubation time. The membranes were rapidly washed twice with 2ml of ice-cold transport buffer.
The radioactive measurements were determined with a beta-counter (Tri-Carb 1900CA, Packard, Downers Grove, United States). As standards, solutions with known amounts of 3H-genistein were used. Vesicle protein content was quantified with BioRad Protein Assay Dye Reagent (BioRad, Hercules, CA, United States), and the data are shown as pmol 3H-genistein μg−1 protein. Data are shown as net pmol 3H-genistein μg−1 protein after removing background, i.e., unspecific adsorption of genistein onto empty-vector yeast membrane. Three independent transformations were performed for each sample.
LaMATE2 Subcellular Localization in Arabidopsis Thaliana Protoplasts
For transient expression of LaMATE2ORF in Arabidopsis protoplasts, the plasmid harboring the sequence for the Green Fluorescent Protein (GFP) was fused at the C-terminus of LaMATE2ORF inside the pUC18-Sp-GFP6 vector (Komarova et al., 2008) using NheI and SphI restriction sites via PCR amplification. A plasmid harboring the sequence for mCherry-fluorescent protein was fused with AtPIP2a, a gene coding for an aquaporin used as a plasma membrane marker (Nelson et al., 2007). Arabidopsis protoplasts were co-transformed with both constructs, LaMATE2ORF-GFP and AtPIP2a-mCherry, using the polyethylene glycol method (Jin et al., 2001). Protoplasts were examined with a TCS SP5 confocal microscope (Leica Microsystems, Wetzlar, Germany), excited with an argon laser at 458 nm for GFP and 540–552 nm for mCherry (for GFP: excitation BP458, beamsplitter FT500, emission BP 492–511 nm; for mCherry: excitation BP 540–552, beamsplitter FT560, emission BP 575–640).
RNAi-Based Silencing of LaMATE2 Gene in White Lupin Roots
LaMATE2 silencing was adapted from Uhde-Stone et al. (2005) using binary transformation vectors, pRNAi and pRedRoot (Limpens et al., 2004). The target region (350 bp long, covering the 5' end of LaMATE2ORF) was first amplified with PCR, and subsequently cloned into pRNAi vector between the restriction sites NcoI–SwaI and BamHI–SpeI. The cloned sequence (LaMATE2 RNAi) was regulated by a double CaMV35s promoter and OCS-3' terminator. Using the KpnI–PacI restriction sites, the LaMATE2 RNAi cassette from the previously produced in pRNAi vector was transferred into the pRedRoot binary vector (pRedRoot::LaMATE2 RNAi).
Phylogenetic and Statistical Analyses
Phylogenetic analyses were conducted using MEGA software, version 6 (Tamura et al., 2013). The tree was constructed by aligning the protein sequences by Clustal-W and the evolutionary history was inferred using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown in Supplementary Figure S1 next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site.
The topology prediction of LaMATE2 was performed using the PROTTER program (Omasits et al., 2014), while the alignment of LaMATE2 protein sequence with orthologous MATE-protein sequences was generated by Clustal-WS using Jalview software version 2 (Waterhouse et al., 2009).
Statistical significance was determined by one-way analysis of variances (ANOVAs) using Holm–Sidak test, p<0.05 or 0.01. Statistical analyses were calculated using SigmaPlot (Systat Software Inc., San Jose, CA, United States).
Results and Discussion
Release of Flavonoids and Expression of LaMATE2
Former work from our laboratories showed that flavonoids are released mainly from the so-called juvenile, and immature (young states) cluster roots and that genistein and its derivative exudation was induced by phosphate (P) deficiency (Weisskopf et al., 2006). Furthermore, in 2001, we published research on genes differentially expressed amongst different stages of cluster roots in P-deficient white lupin (Massonneau et al., 2001). Within these genes, we identified two LaMATE-type transporters, LaMATE1 was predominantly expressed in mature (Uhde-Stone et al., 2005), citrate excreting cluster roots, while the second (LaMATE2) was predominantly present in juvenile cluster roots.
To identify a flavonoid exporter possibly playing an important role in the establishment of nodules in Fabaceae plants, we looked first whether a link between the expression of LaMATE2 and release of flavonoids exists. Under P-deficiency, the LaMATE2 was predominantly expressed in the root apex and in the juvenile stage of cluster roots (Figure 1A). Nitrogen (N) deficiency, on the other hand, induced LaMATE2 expression mainly in apex and nodule (Figure 2A). Analyses of the root exudates of white lupin revealed that genistein release was strongly induced by P deficiency, particularly in young cluster root tissues (Figure 1B), confirming previous results (Weisskopf et al., 2006). In order to see whether a similar behavior could be observed under N deficiency, plants were grown under N-deficient condition. In this case, cluster root formation was not induced. Hence, we compared the whole roots of plants grown in the presence or absence of N. Nevertheless, we could observe that under N deficiency, genistein release was only slightly higher after 1 and became significantly higher after 2 weeks (+123%; Figure 2B).
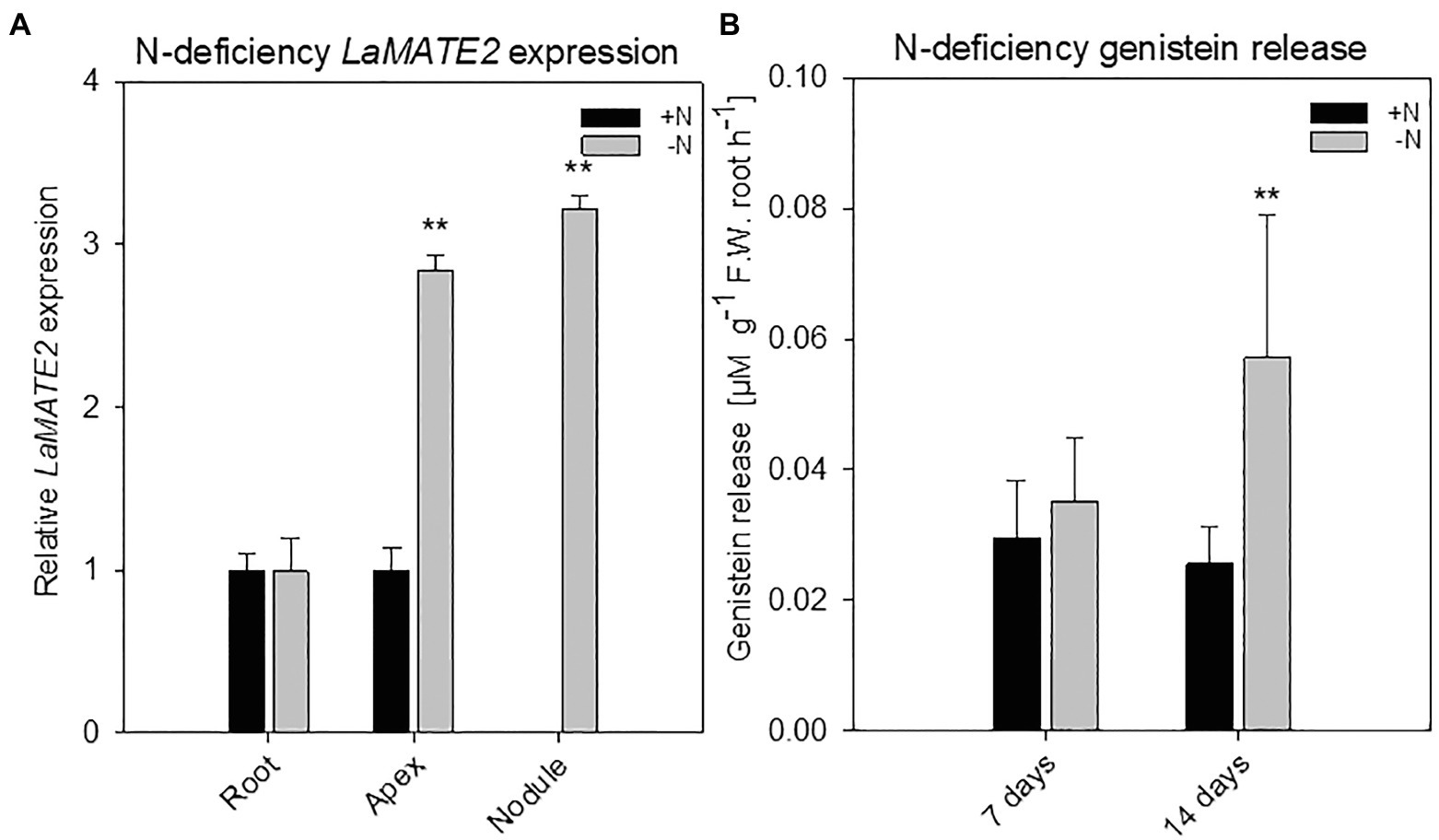
Figure 2. LaMATE2 expression analyses and genistein release in roots of N-deficient white lupin. LaMATE2 expression analyses in white lupin grown under control (+N + P, black bars) condition or after 2 weeks of N-deficiency (-N + P, dark grey bars) (A). Gene expression was evaluated in control and N-deficient (other) root, root apex and nodules. Expression level is shown relative to LaMATE2 expression in control (+N+P) root. Root release of genistein from white lupin plants grown under 1- or 2-week-old N-sufficient and -deficient condition (B). Genistein release in 7 and 14 days of N deficiency (-N+P) or sufficiency (+N + P). Data are mean + SD (Asterisks refers to statistically significant differences among the mean value of the sample vs control, ANOVA Holm–Sidak, N=6–8, ** p<0.01; with 3 technical replicates for each real-time PCR).
Characterization of LaMATE2
As already shown for other MATE transporters, LaMATE2 contains 12-transmembrane helical domains (Supplementary Figure S2) (He et al., 2010). Indeed, LaMATE2 exhibits a good homology to other plant MATE transporters (Figure 3; Supplementary Figure S2), including the functionally characterized vacuolar flavonoid transporters MtMATE1, MtMATE2 and AtTT12 (Marinova et al., 2007; Zhao and Dixon, 2010; Zhao et al., 2011). Up to date, only few MATE transporters have been characterized in roots and most of them mediate either the efflux of citrate, such as AtFRD3, HvAACT1, SbMATE1 (Durrett et al., 2007; Furukawa et al., 2007; Magalhaes et al., 2007) or act as vacuolar flavonoid transporters in Arabidopsis, Medicago or grapevine (for a review see Zhao and Dixon (2010) and Zhao (2015)). Interestingly all citrate transporters contain a large cytosolic loop. In white lupin roots and in agreement with our previous results (Massonneau et al., 2001), the expression of another MATE transporter, LaMATE1, a homolog of the citrate transporter AtFRD3, is highest at the mature stage where a burst of citrate exudation occurs (Uhde-Stone et al., 2005; Wang et al., 2014). Also, this transporter contains a large cytosolic loop.
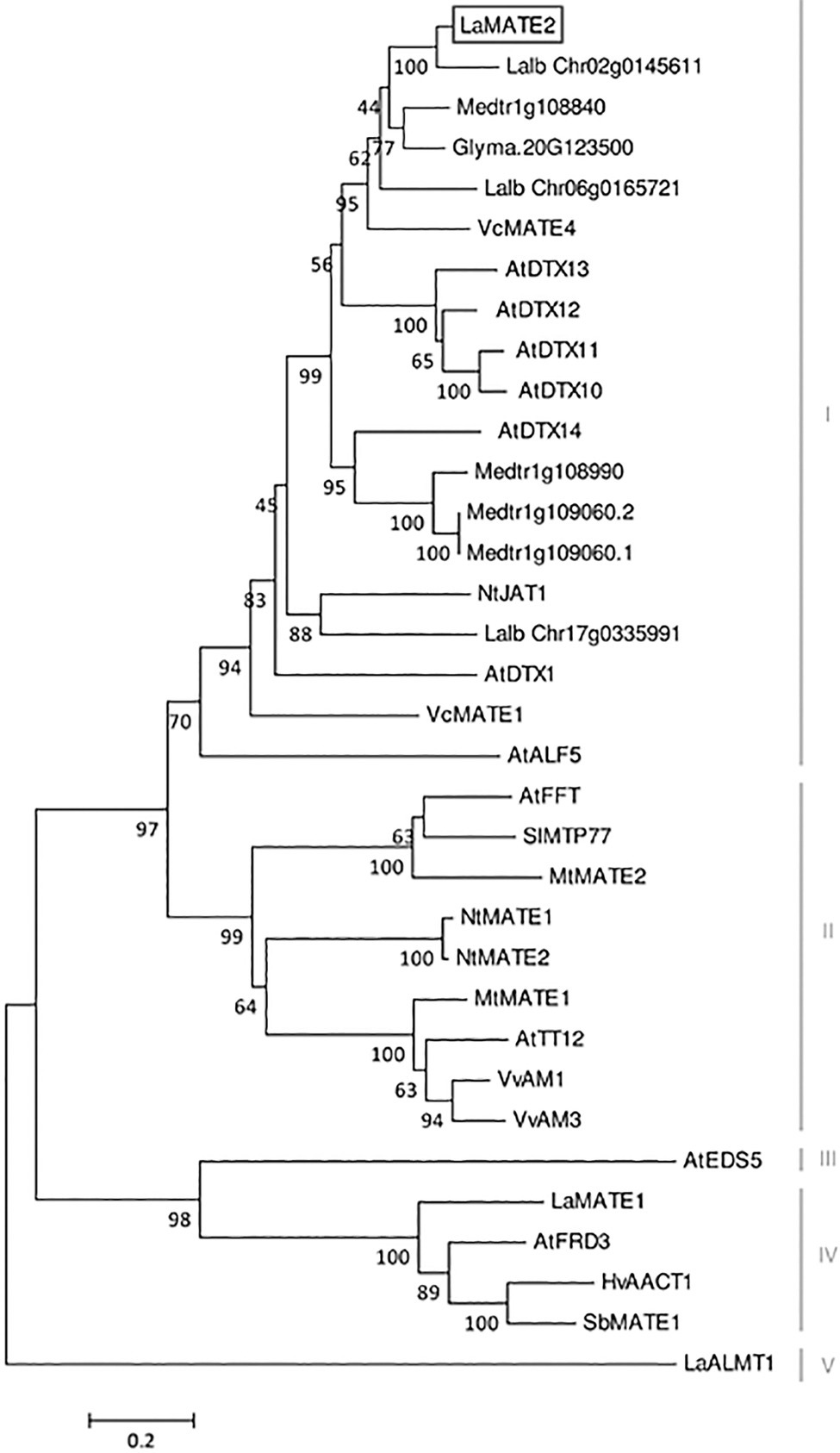
Figure 3. Phylogenetic tree of MATE transporters. A phylogenetic analysis was performed using the LaMATE2 amino acid sequences of Lupinus albus (LaMATE1, LaMATE2, Lalb_Chr17g0335991, Lalb_Chr06g0165721, Lalb_Chr02g0145611, LaALMT1); AtFRD3 AtEDS5, AtTT12, AtDTX1,10,11,12,13,14, AtALF5 and AtFFT of Arabidopsis thaliana; Glyma.10G267700 of Glycine max; HvAACT1 of Hordeum vulgare; MtMATE1, MtMATE2, Medtr1g108840, Medtr1g108990, Medtr1g109060.2, Medtr1g109060.1 of Medicago truncatula; NtJAT1, NtMATE1 and NtMATE2 of Nicotiana tabacum; SlMTP77 of Solanum lycopersicum; SbMATE1 of Sorghum bicolor; VvAM1 and VvAM3 of Vitis vinifera; VcMATE1 and VcMATE4 of Vaccinium corymbosum. The tree was constructed by aligning the protein sequences by Clustal-W and the evolutionary history was inferred using the Neighbour-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site.
At the amino acid level, LaMATE2 exhibits a high similarity to a cluster of MATE proteins, which includes a heterogeneous group of transporters (different substrates, subcellular localization and physiological role). Among the characterized members of this cluster, NtJAT1 and AtDTX1 are the closest homologs of LaMATE2, 55 and 50% of identities, respectively (Figure 3). Both are localized at the plasma membrane (PM) of root cells and function as efflux carriers for plant-derived alkaloids and other toxic compounds (Li et al., 2002; Morita et al., 2009).
Several ABC transporters and MATE proteins mediating the export of compounds into the soil have been described (Weston et al., 2012; Baetz and Martinoia, 2014). However, none of them was shown so far to be responsible for the root release of flavonoids and isoflavonoids. Our data showing a connection between flavonoid exudation and LaMATE2 expression together with the fact that LaMATE2 fused with GFP co-localizes with the PM marker AtPIP2a-mCherry (Figure 4) prompted us to investigate whether LaMATE2 acts as a flavonoid transporter.
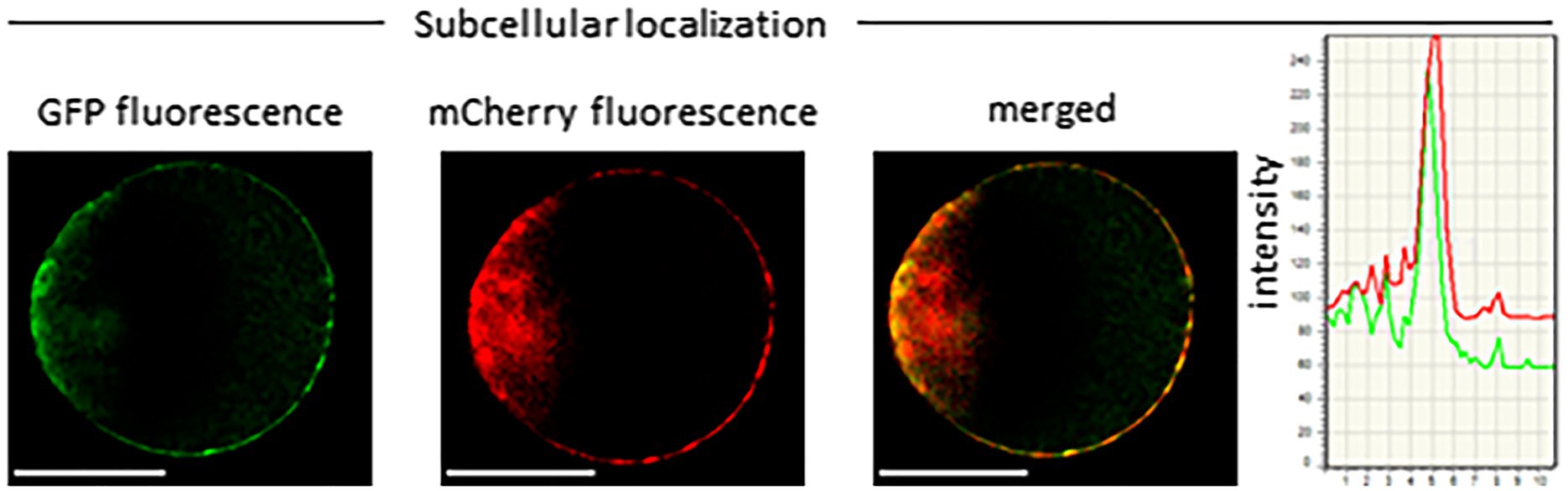
Figure 4. Subcellular localization of LaMATE2. Co-localization of LaMATE2 fused with green fluorescent protein (GFP) and mCherry-labeled plasma membrane marker AtPIP2A in an Arabidopsis mesophyll protoplast. White scale bars=20 μM. Fluorescence intensity over distance plot of LaMATE2-GFP (green) and AtPIP2a-mCherry (red).
LaMATE2 Silencing Reduces Genistein Release, P Mobilization and Nodule Number
Genistein is one of the major released isoflavonoids of Fabaceae inducing NOD genes in Rhizobium bacteria, attracting them and inducing nodule formation (Zhang and Smith, 1995; Lang et al., 2008; Liu and Murray, 2016). Therefore, to assess whether the LaMATE2 might affect genistein release and nodule formation, we have used RNA-dependent gene silencing (LaMATE2-RNAi) in roots of white lupin plants. The effectiveness of the silencing was confirmed by a significant reduction (approximatively −80 and 65% of LaMATE2 expression in LaMATE2-RNAi transformants grown in N and P deficiency, respectively (Figures 5A,G). Analysis of these roots revealed that the release of genistein was strongly reduced in these transformants (Figures 5B,H) and concomitantly isoflavonoids, the uppermost as glycosides, were accumulating in the cell content of silenced roots, which can be regarded as a direct consequence of the reduced genistein exudation (Figures 5D-F). To investigate whether the impaired genistein exudation influences nodulation, we used the LaMATE2-RNAi plants to compare the nodule number of silenced and control plants. Our results highlight the importance of LaMATE2-dependent isoflavonoid release in the early step of nodulation, since LaMATE2 silencing leads to a highly significant reduction (a. -80%) of nodules in LaMATE2-RNAi compared to empty-vector transformed roots (Figures 5C; 6B, Supplementary Figure S4). Interestingly exogenous application of genistein onto the LaMATE2-RNAi roots partially restored the nodulation efficiency (Figures 6A,B). Similarly, LaMATE2 expression, genistein release patterns and capability of root exudates to mobilize P were measured under P-deficient conditions in silenced plants (Figures 5G-I). Cluster roots of LaMATE2-RNAi transformants grown under P deficiency released 85% less genistein along with the lower level of LaMATE2 expression (a. −60%). The SPE-purified exudates from those roots also exhibited a limited (a. −66%) mobilization capability from a poorly soluble P source (vivianite). These results suggest that LaMATE2 is the major if not the sole genistein exporter from roots of white lupin plants and in P-deficient condition it contributes to the P solubilization process.
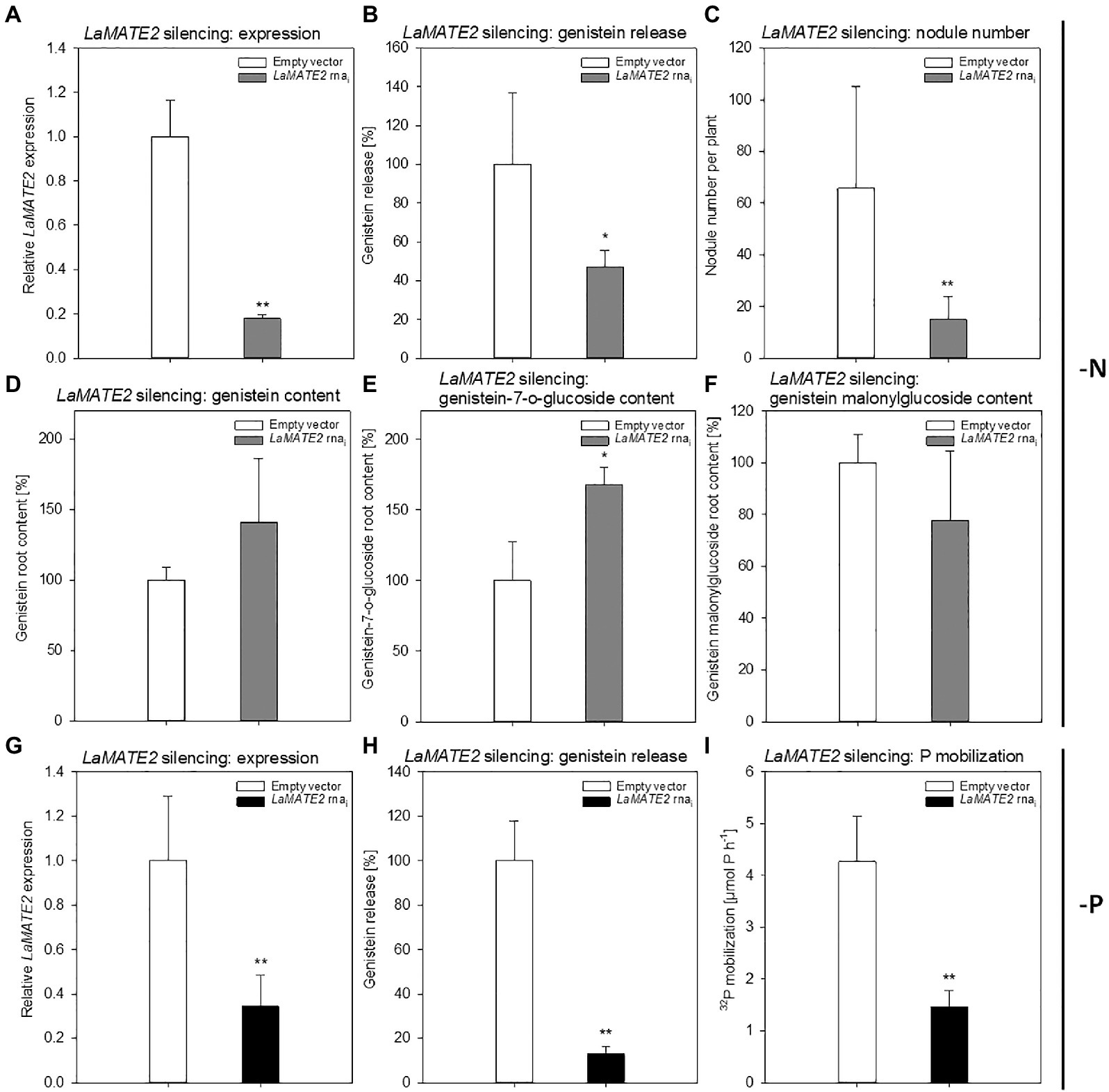
Figure 5. Effect of LaMATE2 silencing in N and P deficient roots. Alteration of LaMATE2 expression, flavonoid content and release, nodule number or P mobilization due to LaMATE2 silencing in N (A-F) or P (G-I) deficiency. LaMATE2 relative expression (a, g; 6 biological replicates), genistein release (B,H), number of nodules per plant (pictures shown in Supplementary Figure S4) (C), cell content: genistein (D), genistein 7-O-glucoside (E), genistein malonylglucoside (F), P mobilization from a poorly soluble P source (I). The analyses were performed on roots of N- or P-deficient lupin plants independently transformed with either pRedRoot::LaMATE2 RNAi (LaMATE2 RNAi) or empty-vector pRedRoot (Empty vector). All data are expressed relative to Empty-vector-transformed roots, with the exceptions of the number of nodule per plants and P mobilization (μmol P h−1). Data are mean + SD (Asterisks refers to statistically significant differences among the mean value of RNAi and Empty vector, ANOVA Holm–Sidak, N=6–21, *p<0.05; for **p<0.01; with 3 technical replicates for each real-time PCR).
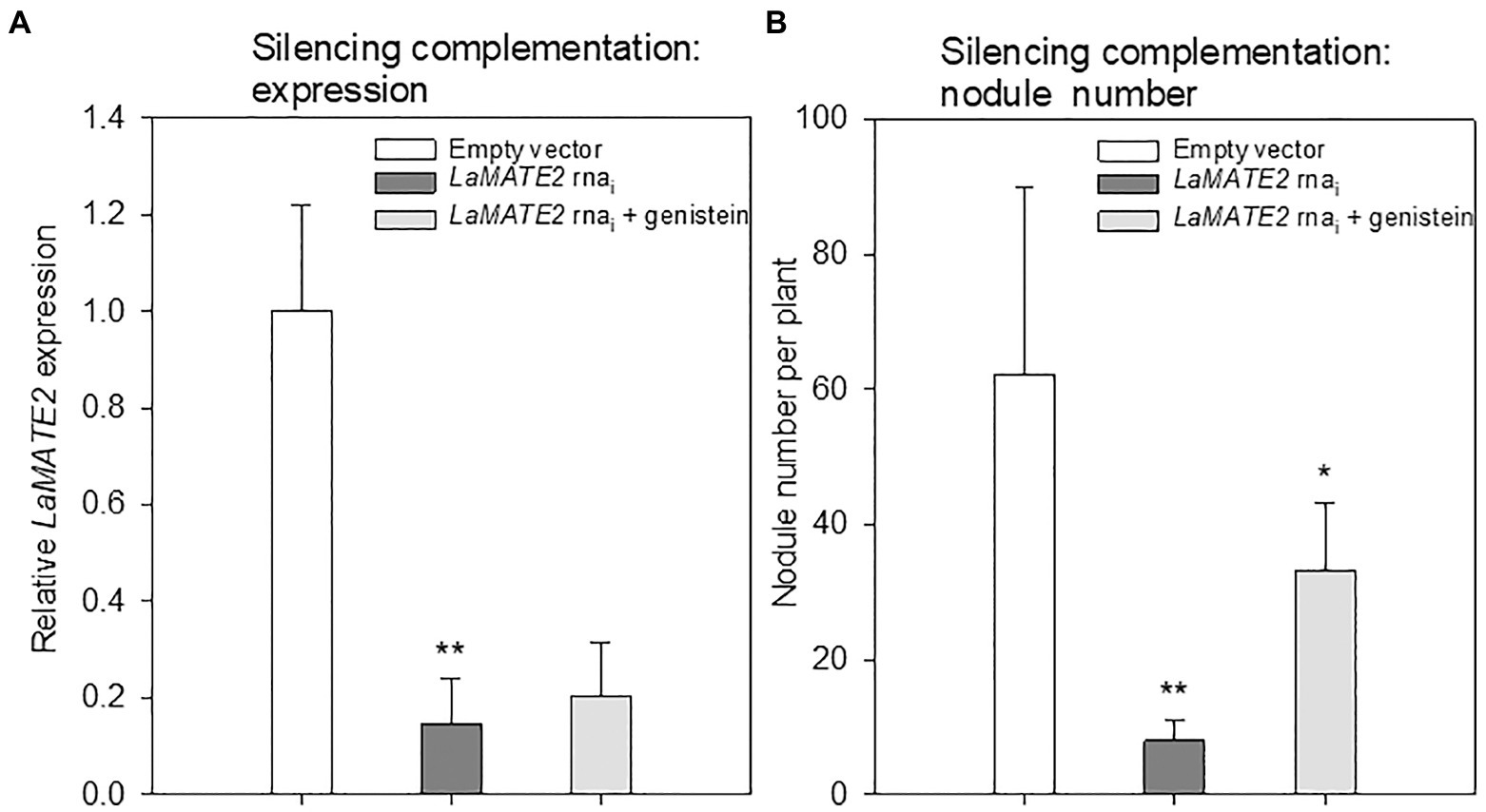
Figure 6. Effect of exogenous genistein on LaMATE2 expression and nodule numbers. LaMATE2 relative expression (A) and number of nodules per plant (B) in roots of N-deficient lupin plants independently transformed with either pRedRoot::LaMATE2 RNAi (LaMATE2 RNAi) or empty-vector pRedRoot (Empty vector) or RNAi roots treated with 1μM genistein (LaMATE2 RNAi + genistein). Expression data are shown relative to LaMATE2 expression level in empty vector-transformed roots. Data are mean + SD (Asterisks refers to statistically significant differences among the mean value of RNAi vs EV or RNAi+G vs RNAi values, ANOVA Holm–Sidak, N=6, *p<0.05; ** for p<0.01).
The results showing that genistein export is dependent on the presence of LaMATE2 indicates that within the root this transporter is at least partially localized in the cortex and/or in the epidermis. Attempts in our laboratory to get a more detailed picture on its localization, either using in-situ hybridization or a GUS-promoter construct unfortunately failed. However, it should be mentioned that interestingly, excretion of strigolactones into the soil was independent, whether the transporter was localized specifically in hypodermal passage cells as in Petunia or in the whole cortex as in Medicago (Kretzschmar et al., 2012; Banasiak et al., 2020).
Transport of Phenolics by LaMATE2
To demonstrate that the reduced genistein release of LaMATE2 silenced plants depends directly on the activity of LaMATE2, we expressed it heterologously in Saccharomyces cerevisiae and performed transport assays using isolated membrane vesicles (Figure 7). Indeed, using this system, we could observe that LaMATE2 mediates an efficient transport of genistein which was characterized by a saturable kinetic exhibiting an apparent Km of 16.2 μM (Figure 7B). This Km is in the same order of magnitude as determined for the vacuolar flavonoid transporters characterized so far (e.g., AtTT12 and MtMATE1: Km of 50 and 36 μM for epicatechin 3'-O-glucoside, respectively; MtMATE2: Km of 88 μM for cyanidin 3-O-glucoside) (Zhao and Dixon, 2010; Zhao et al., 2011). In plasmalemma vesicles isolated from soybean roots, genistein transport exhibited a Km of 160 μM and was strongly dependent on the presence of ATP (Sugiyama et al., 2007).
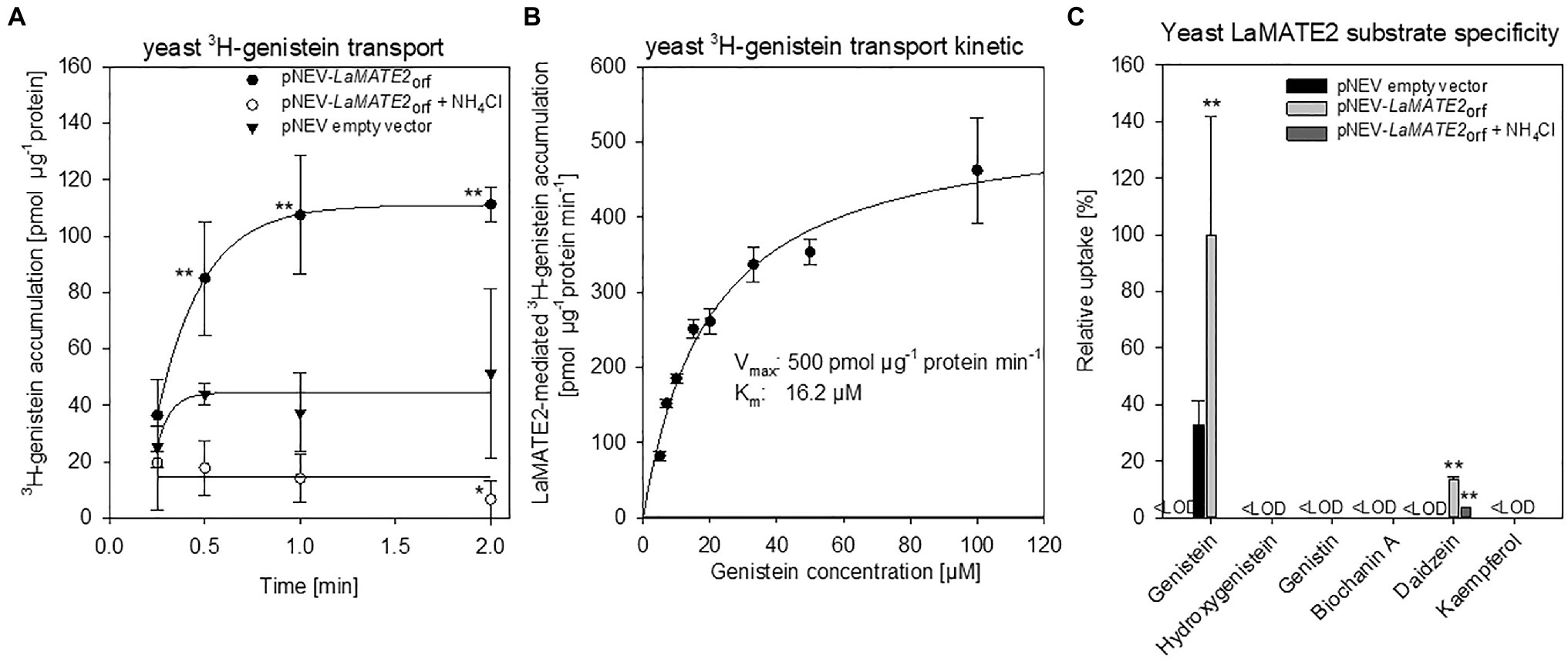
Figure 7. Transport of phenylpropanoid compounds mediated by LaMATE2. LaMATE2-mediated transport rate of phenylpropanoids in yeast microsomal membrane vesicles. (A) Time-dependent accumulation of 3H-genistein in yeast microsomal membrane vesicles. Membrane vesicles were isolated from yeast transformed with the empty vector (pNEV empty vector) or transformed with pNEV-LaMATE2ORF. The yeast vesicles were incubated up to 120 s in presence of 5 μM 3H-genistein: To determine the effect of the pH gradient, 25 mM NH4Cl was added in an assay solution (pNEV-LaMATE2ORF + NH4Cl), as uncoupler of the proton gradient. (B) Concentration-dependent transport of 3H-genistein by LaMATE2 in yeast vesicles. Vesicles were incubated for 30s in the assay solution containing 3H-genistein at different concentrations (from 5 to 100 μM). The kinetic parameters of 3H-genistein uptake were calculated by subtracting uptake rates recorded in the empty-vector vesicles using the Hanes–Woolf plot. (C) Substrate specificity of LaMATE2 transporter was evaluated in presence of genistein, hydroxygenistein, genistin (glycosylated genistein), biochanin A (methylgenistein), daidzein or kaempferol (5 μM, 30 s). All data are expressed relative to pNEV-LaMATE2ORF-transformed yeast. Data are mean ± SD of three independent experiments (Asterisks refers to statistically significant differences among the mean value in comparison to empty vector sample within each time point; capital letters refer to statistically significant differences among the mean value, ANOVA Holm–Sidak, N=6, **p<0.01, <LOD: below detectable value).
Up to date, certain MATEs have been shown to exhibit a broad range of substrate specificity (Takanashi et al., 2014), while LaMATE2 displays a strong specificity for the substrate genistein (Figure 7C). Besides genistein, LaMATE2 was able to transport only daidzein at a low rate, but not other tested flavonoids such as genistin (a glycosylated form of genistein), hydroxygenistein, biochanin A (a methylated form of genistein) or kaempferol. Previous observations in soybean showed that the presence of daidzein in the external media strongly limited genistein transport in plasma membrane vesicles of root cells, indicating a possible competition between those molecules (Sugiyama et al., 2007).
A feature of MATE proteins is to couple the substrate transport to an electrochemical gradient, working as H+ or Na+/substrate antiporters (He et al., 2010; Lu et al., 2013; Tanaka et al., 2013). Our results showed that ATP-dependent 3H-genistein accumulation in vesicles deriving from LaMATE2ORF-expressing yeast was strongly reduced in the presence of ammonium chloride, which dissipates the transmembrane proton gradient (Marinova et al., 2007; Zhao and Dixon, 2010) (Figure 7A), indicating that LaMATE2 acts as a substrate-proton co-transporter, similarly to AtTT12 (Marinova et al., 2007), AtDTX1 (Li et al., 2002) and NtJAT1 (Morita et al., 2009).
Conclusion
In this work, the long-sought-after isoflavonoid plasma membrane exporter required to attract symbiotic bacteria for N fixation has been identified. Different leguminous plants release different sets of isoflavonoids to induce nodulation (Liu and Murray, 2016). Released genistein has been indicated in faba bean and soybean as rhizobia attractant (Zhang and Smith, 1995; Li et al., 2016). Since Medicago truncatula and soybean both encode one MATE protein that exhibits high homology to LaMATE2, it is tempting to speculate that these homologues could also act as isoflavonoid exporters, possibly showing slightly different substrate preferences according to the isoflavonoid produced by the plant to initiate the symbiosis. However, it cannot be excluded that in addition to MATE-type transporters and ABC proteins, others transporters are also involved in isoflavonoid release. Since isoflavonoids can also release Pi from minerals (Cesco et al., 2010) and organic complexes, these transporters may play a dual role in N and P supply.
During the past years, we learned a lot about the signaling pathways and the effectors involved in establishing the legume-rhizobia symbiosis. Surprisingly molecular identity of membrane transporters responsible for the release of phenolic compounds initiating interactions was not known. With the identification of LaMATE2, we succeeded to identify the very initial step leading to this symbiosis. Since there is a huge interest to transfer this complex mechanism to other crop plants to grow them in the absence of artificial N-sources, we do believe that it is a valuable piece of the puzzle to consider in such an ambitious project.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
NT, EM, RP, SC, and MJ designed and oversaw the research. WB, LZ, SG, RF, SV, FV, TM, BB, and NT performed experiments and analyzed data. NT, EM, RP, and MJ wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
Research was supported by grants from Italian Ministry of University and Research-MIUR(FIRB-Programme Futuro in Ricerca, RBFR08L2ZT and RBFR127WJ9), from funds of the Zurich University; and statutory funds from the Polish Ministry of Science and Higher Education.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.758213/full#supplementary-material
Footnotes
References
Baetz, U., and Martinoia, E. (2014). Root exudates: The hidden part of plant defense. Trends Plant Sci. 19, 90–98. doi: 10.1016/j.tplants.2013.11.006
Banasiak, J., Borghi, L., Stec, N., Martinoia, E., and Jasiński, M. (2020). The full-size ABCG transporter of Medicago truncatula is involved in Strigolactone secretion, affecting Arbuscular mycorrhiza. Front. Plant Sci. 11:18. doi: 10.3389/fpls.2020.00018
Biala, W., Banasiak, J., Jarzyniak, K., Pawela, A., and Jasinski, M. (2017). Medicago truncatula ABCG10 is a transporter of 4-coumarate and liquiritigenin in the medicarpin biosynthetic pathway. J. Exp. Bot. 68, 3231–3241. doi: 10.1093/jxb/erx059
Burke, D., Dawson, D., and Stearns, T. (2000). Methods in yeast genetics: A cold Spring Harbor laboratory course manual, 2000 ed. Cold. Spring. Harbor. Press: Plainview. 17:205.
Cesco, S., Neumann, G., Tomasi, N., Pinton, R., and Weisskopf, L. (2010). Release of plant-borne flavonoids into the rhizosphere and their role in plant nutrition. Plant Soil 329, 1–25. doi: 10.1007/s11104-009-0266-9
Durrett, T. P., Gassmann, W., and Rogers, E. E. (2007). The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol. 144, 197–205. doi: 10.1104/pp.107.097162
Furukawa, J., Yamaji, N., Wang, H., Mitani, N., Murata, Y., Sato, K., et al. (2007). An Aluminum-activated citrate transporter in barley. Plant Cell Physiol. 48, 1081–1091. doi: 10.1093/pcp/pcm091
Gietz, R. D., and Woods, R. A. (2002). Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350, 87–96. doi: 10.1016/s0076-6879(02)50957-5
He, X., Szewczyk, P., Karyakin, A., Evin, M., Hong, W.-X., Zhang, Q., et al. (2010). Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature 467:991. doi: 10.1038/nature09408
Hufnagel, B., Marques, A., Soriano, A., Marquès, L., Divol, F., Doumas, P., et al. (2020). High-quality genome sequence of white lupin provides insight into soil exploration and seed quality. Nat. Commun. 11, 1–12. doi: 10.1038/s41467-019-14197-9
Jiang, Y., Wang, W., Xie, Q., Liu, N., Liu, L., Wang, D., et al. (2017). Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 356, 1172–1175. doi: 10.1126/science.aam9970
Jin, J. B., Kim, Y. A., Kim, S. J., Lee, S. H., Kim, D. H., Cheong, G. W., et al. (2001). A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13, 1511–1526. doi: 10.1105/TPC.000534
Klein, M., Mamnun, Y. M., Eggmann, T., Schüller, C., Wolfger, H., Martinoia, E., et al. (2002). The ATP-binding cassette (ABC) transporter Bpt1p mediates vacuolar sequestration of glutathione conjugates in yeast. FEBS Lett. 520, 63–67. doi: 10.1016/S0014-5793(02)02767-9
Komarova, N. Y., Thor, K., Gubler, A., Meier, S., Dietrich, D., Weichert, A., et al. (2008). AtPTR1 and AtPTR5 transport dipeptides in planta. Plant Physiol. 148, 856–869. doi: 10.1104/pp.108.123844
Koressaar, T., and Remm, M. (2007). Enhancements and modifications of primer design program Primer3. Bioinformatics 23, 1289–1291. doi: 10.1093/bioinformatics/btm091
Kretzschmar, T., Kohlen, W., Sasse, J., Borghi, L., Schlegel, M., Bachelier, J. B., et al. (2012). A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 483, 341–344. doi: 10.1038/nature10873
Lambers, H., Martinoia, E., and Renton, M. (2015). Plant adaptations to severely phosphorus-impoverished soils. Curr. Opin. Plant Biol. 25, 23–31. doi: 10.1016/j.pbi.2015.04.002
Lang, K., Lindemann, A., Hauser, F., and Göttfert, M. (2008). The genistein stimulon of Bradyrhizobium japonicum. Mol. Gen. Genomics. 279, 203–211. doi: 10.1007/s00438-007-0280-7
Li, L., He, Z., Pandey, G. K., Tsuchiya, T., and Luan, S. (2002). Functional cloning and characterization of a plant efflux carrier for multidrug and heavy metal detoxification. J. Biol. Chem. 277, 5360–5368. doi: 10.1074/jbc.M108777200
Li, B., Li, Y.-Y., Wu, H.-M., Zhang, F.-F., Li, C.-J., Li, X.-X., et al. (2016). Root exudates drive interspecific facilitation by enhancing nodulation and N2 fixation. Proc. Natl. Acad. Sci. 113, 6496–6501. doi: 10.1073/pnas.1523580113
Limpens, E., Ramos, J., Franken, C., Raz, V., Compaan, B., Franssen, H., et al. (2004). RNA interference in agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J. Exp. Bot. 55, 983–992. doi: 10.1093/jxb/erh122
Liu, C.-W., and Murray, J. (2016). The role of flavonoids in nodulation host-range specificity: An update. Plan. Theory 5:33. doi: 10.3390/plants5030033
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, M., Radchenko, M., Symersky, J., Nie, R., and Guo, Y. (2013). Structural insights into H+-coupled multidrug extrusion by a MATE transporter. Nat. Struct. Mol. Biol. 20, 1310–1317. doi: 10.1038/nsmb.2687
Luginbuehl, L. H., Menard, G. N., Kurup, S., Van Erp, H., Radhakrishnan, G. V., Breakspear, A., et al. (2017). Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 356, 1175–1178. doi: 10.1126/science.aan0081
Magalhaes, J. V., Liu, J., Guimaraes, C. T., Lana, U. G. P., Alves, V. M. C., Wang, Y. H., et al. (2007). A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat. Genet. 39, 1156–1161. doi: 10.1038/ng2074
Marinova, K., Kleinschmidt, K., Weissenbock, G., and Klein, M. (2007). Flavonoid biosynthesis in barley primary leaves requires the presence of the vacuole and controls the activity of Vacuolar flavonoid transport. Plant Physiol. 144, 432–444. doi: 10.1104/pp.106.094748
Martin, F. M., Uroz, S., and Barker, D. G. (2017). Ancestral alliances: Plant mutualistic symbioses with fungi and bacteria. Science 356:eaad4501. doi: 10.1126/science.aad4501
Massonneau, A., Langlade, N., Leon, S., Smutny, J., Vogt, E., Neumann, G., et al. (2001). Metabolic changes associated with cluster root development in white lupin (Lupinus albus L.): Relationship between organic acid excretion, sucrose metabolism and energy status. Planta 213, 534–542. doi: 10.1007/s004250100529
Morita, M., Shitan, N., Sawada, K., Van Montagu, M. C. E., Inzé, D., Rischer, H., et al. (2009). Vacuolar transport of nicotine is mediated by a multidrug and toxic compound extrusion (MATE) transporter in Nicotiana tabacum. Proc. Natl. Acad. Sci. 106, 2447–2452. doi: 10.1073/pnas.0812512106
Nelson, B. K., Cai, X., and Nebenführ, A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x
Neumann, G., and Martinoia, E. (2002). Cluster roots - an underground adaptation for survival in extreme environments. Trends Plant Sci. 7, 162–167. doi: 10.1016/s1360-1385(02)02241-0
Omasits, U., Ahrens, C. H., Müller, S., and Wollscheid, B. (2014). Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30, 884–886. doi: 10.1093/bioinformatics/btt607
Purnell, H. M. (1960). Studies of the family Proteaceae. Anatomy and morphology of the roots of some victorian species. Aust. J. Bot. 8, 38–50.
Quandt, H. J., Pühler, A., and Broer, I. (1993). Transgenic root nodules of Vicia hirsuta: A fast and efficient system for the study of gene expression in indeterminate-type nodules. MPMI-Mol. Plant. Microbe. Interact. 6, 699–706. doi: 10.1094/MPMI-6-699
Ritz, C., and Spiess, A. N. (2008). qpcR: an R package for sigmoidal model selection in quantitative real-time polymerase chain reaction analysis. Bioinformatics 24, 1549–1551. doi: 10.1093/bioinformatics/btn227
Sauer, N., and Stolz, J. (1994). SUC1 and SUC2: Two sucrose transporters from Arabidopsis thaliana; expression and characterization in baker’s yeast and identification of the histidine-tagged protein. Plant J. 6, 67–77. doi: 10.1046/j.1365-313X.1994.6010067.x
Staszków, A., Swarcewicz, B., Banasiak, J., Muth, D., Jasiński, M., and Stobiecki, M. (2011). LC/MS profiling of flavonoid glycoconjugates isolated from hairy roots, suspension root cell cultures and seedling roots of Medicago truncatula. Metabolomics 7, 604–613. doi: 10.1007/s11306-011-0287-2
Stępkowski, T., Żak, M., Moulin, L., Króliczak, J., Golińska, B., Narożna, D., et al. (2011). Bradyrhizobium canariense and Bradyrhizobium japonicum are the two dominant rhizobium species in root nodules of lupin and serradella plants growing in Europe. Syst. Appl. Microbiol. 34, 368–375. doi: 10.1016/j.syapm.2011.03.002
Strozycki, P. M., Skąpska, A., Szcześniak, K., Sobieszczuk, E., Briat, J.-F., and Legocki, A. B. (2003). Differential expression and evolutionary analysis of the three ferritin genes in the legume plant Lupinus luteus. Physiol. Plant. 118, 380–389. doi: 10.1034/j.1399-3054.2003.00081.x
Subramanian, S., Stacey, G., and Yu, O. (2006). Endogenous isoflavones are essential for the establishment of symbiosis between soybean and Bradyrhizobium japonicum. Plant J. 48, 261–273. doi: 10.1111/j.1365-313X.2006.02874.x
Sugiyama, A., Shitan, N., and Yazaki, K. (2007). Involvement of a soybean ATP-binding cassette-type transporter in the secretion of genistein, a signal flavonoid in legume-rhizobium Symbiosis. Plant Physiol. 144, 2000–2008. doi: 10.1104/pp.107.096727
Takanashi, K., Shitan, N., and Yazaki, K. (2014). The multidrug and toxic compound extrusion (MATE) family in plants. Plant Biotechnology 31, 417–430. doi: 10.5511/plantbiotechnology.14.0904a
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tanaka, Y., Hipolito, C. J., Maturana, A. D., Ito, K., Kuroda, T., Higuchi, T., et al. (2013). Structural basis for the drug extrusion mechanism by a MATE multidrug transporter. Nature 496, 247–251. doi: 10.1038/nature12014
Tilman, D. (1999). Global environmental impacts of agricultural expansion: The need for sustainable and efficient practices. Proc. Natl. Acad. Sci. 96, 5995–6000. doi: 10.1073/pnas.96.11.5995
Tilman, D., Fargione, J., Wolff, B., D’antonio, C., Dobson, A., Howarth, R., et al. (2001). Forecasting agriculturally driven global environmental change. Science 292, 281–284. doi: 10.1126/science.1057544
Udvardi, M., and Poole, P. S. (2013). Transport and metabolism in legume-rhizobia symbioses. Annu. Rev. Plant Biol. 64, 781–805. doi: 10.1146/annurev-arplant-050312-120235
Uhde-Stone, C., Liu, J., Zinn, K. E., Allan, D. L., and Vance, C. P. (2005). Transgenic proteoid roots of white lupin: A vehicle for characterizing and silencing root genes involved in adaptation to P stress. Plant J. 44, 840–853. doi: 10.1111/j.1365-313X.2005.02573.x
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3—new capabilities and interfaces. Nucleic Acids Res. 40:e115. doi: 10.1093/nar/gks596
Vance, C. P., Uhde-Stone, C., and Allan, D. L. (2003). Phosphorus acquisition and use: Critical adaptations by plants for securing a nonrenewable resource. New Phytol. 157, 423–447. doi: 10.1046/j.1469-8137.2003.00695.x
Wang, Z., Straub, D., Yang, H., Kania, A., Shen, J., Ludewig, U., et al. (2014). The regulatory network of cluster-root function and development in phosphate-deficient white lupin (Lupinus albus) identified by transcriptome sequencing. Physiol. Plant. 151, 323–338. doi: 10.1111/ppl.12187
Wasson, A. P., Pellerone, F. I., and Mathesius, U. (2006). Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. Plant Cell 18, 1617–1629. doi: 10.1105/tpc.105.038232
Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M., and Barton, G. J. (2009). Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. doi: 10.1093/bioinformatics/btp033
Weisskopf, L., Tomasi, N., Santelia, D., Martinoia, E., Langlade, N. B., Tabacchi, R., et al. (2006). Isoflavonoid exudation from white lupin roots is influenced by phosphate supply, root type and cluster-root stage. New Phytol. 171, 657–668. doi: 10.1111/j.1469-8137.2006.01776.x
Weston, L. A., Ryan, P. R., and Watt, M. (2012). Mechanisms for cellular transport and release of allelochemicals from plant roots into the rhizosphere. J. Exp. Bot. 63, 3445–3454. doi: 10.1093/jxb/ers054
Zhang, F., and Smith, D. L. (1995). Preincubation of Bradyrhizobium japonicum with Genistein accelerates nodule development of soybean at suboptimal root zone temperatures. Plant Physiol. 108, 961–968. doi: 10.1104/pp.108.3.961
Zhao, J. (2015). Flavonoid transport mechanisms: How to go, and with whom. Trends Plant Sci. 20, 576–585. doi: 10.1016/j.tplants.2015.06.007
Zhao, J., and Dixon, R. A. (2010). The ‘ins’ and ‘outs’ of flavonoid transport. Trends Plant Sci. 15, 72–80. doi: 10.1016/j.tplants.2009.11.006
Keywords:Bradyrhizobium, genistein, Lupinus albus, MATE transporter, nitrogen, phosphorus, plant-microbe interaction
Citation: Biała-Leonhard W, Zanin L, Gottardi S, de Brito Francisco R, Venuti S, Valentinuzzi F, Mimmo T, Cesco S, Bassin B, Martinoia E, Pinton R, Jasiński M and Tomasi N (2021) Identification of an Isoflavonoid Transporter Required for the Nodule Establishment of the Rhizobium-Fabaceae Symbiotic Interaction. Front. Plant Sci. 12:758213. doi: 10.3389/fpls.2021.758213
Edited by:
Gerald Alan Berkowitz, University of Connecticut, United StatesReviewed by:
Peter Ryan, Commonwealth Scientific and Industrial Research Organisation (CSIRO), AustraliaKojiro Takanashi, Shinshu University, Japan
Copyright © 2021 Biała-Leonhard, Zanin, Gottardi, de Brito Francisco, Venuti, Valentinuzzi, Mimmo, Cesco, Bassin, Martinoia, Pinton, Jasiński and Tomasi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicola Tomasi, bmljb2xhLnRvbWFzaUB1bml1ZC5pdA==; Michał Jasiński, amFzaW5za2lAaWJjaC5wb3puYW4ucGw=
†These authors have contributed equally to this work
 Wanda Biała-Leonhard
Wanda Biała-Leonhard Laura Zanin
Laura Zanin Stefano Gottardi2†
Stefano Gottardi2† Rita de Brito Francisco
Rita de Brito Francisco Silvia Venuti
Silvia Venuti Fabio Valentinuzzi
Fabio Valentinuzzi Tanja Mimmo
Tanja Mimmo Stefano Cesco
Stefano Cesco Enrico Martinoia
Enrico Martinoia Roberto Pinton
Roberto Pinton Michał Jasiński
Michał Jasiński Nicola Tomasi
Nicola Tomasi