
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 01 October 2021
Sec. Plant Biotechnology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.757152
This article is part of the Research TopicTargeted Genome Editing for Crop ImprovementView all 20 articles
 Chaolei Liu1†
Chaolei Liu1† Yiwei Cao2†
Yiwei Cao2† Yufeng Hua1†
Yufeng Hua1† Guijie Du2
Guijie Du2 Qing Liu1
Qing Liu1 Xin Wei1
Xin Wei1 Tingting Sun1
Tingting Sun1 Jianrong Lin1
Jianrong Lin1 Mingguo Wu1
Mingguo Wu1 Zhukuan Cheng2
Zhukuan Cheng2 Kejian Wang1,3*
Kejian Wang1,3*Manipulation of the distribution and frequency of meiotic recombination events to increase genetic diversity and disrupting genetic interference are long-standing goals in crop breeding. However, attenuation of genetic interference is usually accompanied by a reduction in recombination frequency and subsequent loss of plant fertility. In the present study, we generated null mutants of the ZEP1 gene, which encodes the central component of the meiotic synaptonemal complex (SC), in a hybrid rice using CRISPR/Cas9. The null mutants exhibited absolute male sterility but maintained nearly unaffected female fertility. By pollinating the zep1 null mutants with pollen from indica rice variety 93-11, we successfully conducted genetic analysis and found that genetic recombination frequency was greatly increased and genetic interference was completely eliminated in the absence of ZEP1. The findings provided direct evidence to support the controversial hypothesis that SC is involved in mediating interference. Additionally, the remained female fertility of the null mutants makes it possible to break linkage drag. Our study provides a potential approach to increase genetic diversity and fully eliminate genetic interference in rice breeding.
Plant breeding aims to develop superior varieties to suit the needs of farmers and consumers (Moose and Mumm, 2008). However, innovations in breeding materials strongly depend on creating novel allele combinations that bring together advantageous alleles and remove linked, disadvantageous alleles. This is traditionally limited by the number of crossovers (COs) during meiosis (Taagen et al., 2020). To generate sufficient genetic diversity, breeders and geneticists are exploring approaches to increase the CO frequency, alter CO distribution, or induce COs between non-homologous chromosomal regions (Mieulet et al., 2018; Blary and Jenczewski, 2019).
The synaptonemal complex (SC) is a meiosis-specific structure involved in CO formation and chromosome segregation (Gao and Colaiácovo, 2018). SC assembly starts from early prophase I and forms a tripartite structure at pachytene, which consists of axial or lateral elements, transverse filaments, and central element (Moses, 1969; de Boer and Heyting, 2006; Fraune et al., 2016). SC dynamics are tightly regulated by its components for the proper SC assembly (Gao and Colaiácovo, 2018). Among them, TF proteins locate at the central of SC and act important roles in bridging the parallel homologous axes (Dubois et al., 2019). Usually, the N-terminal domain of the TF protein is positioned in the middle of the SC, while its C terminus is located next to the lateral elements (Anderson et al., 2005; Schild-Prüfert et al., 2011). Such special organization pattern is critical for the function of the SC. The first reported TF protein gene is ZIP1, which was identified in budding yeast (Sym et al., 1993; Storlazzi et al., 1996). zip1 mutants exhibit defects in SC formation and genetic recombination; however, its homologous genes in other organisms appear to play opposite roles on CO frequency. Sycp1 encodes a TF protein in mouse, and Sycp1−/− mice shows 90% of COs disappeared (de Vries et al., 2005). Mutation of c(3)G, the TF protein gene in Drosophila, causes the loss of all COs (Page and Hawley, 2001). Unexpectedly, the syp-1 mutants in the roundworm Caenorhabditis elegans exhibit a severe reduction in COs (MacQueen et al., 2002), while knockdown of SYP-1 by RNA interference (RNAi) increases the CO frequency (Libuda et al., 2013). The ZIP1 orthologs have also been identified in plant species including Arabidopsis thaliana, rice (Oryza sativa), maize (Zea mays), wheat (Triticum aestivum), and barley (Hordeum vulgare; Higgins et al., 2005; Wang et al., 2010; Golubovskaya et al., 2011; Khoo et al., 2012; Barakate et al., 2014). Suppression of ZYP1 in Arabidopsis thaliana or in barley (H. vulgare) led to reduced CO frequency (Higgins et al., 2005; Barakate et al., 2014). In contrast with Arabidopsis and barley, the Tos17 insertion mutant lines of ZEP1 in rice had more COs than the wild type (WT; Wang et al., 2010; Wang K. et al., 2015).
Genetic interference, a phenomenon where the occurrence of one crossover (CO) inhibits the formation of other COs nearby on the same chromosome pair, imparts a remarkable level of regulation of the number and distribution of COs per chromosome in eukaryotes (Sun et al., 2017; Otto and Payseur, 2019). Therefore, reduction or disruption of genetic interference can be considered to increase genetic diversity in crop breeding (Taagen et al., 2020). Presently, a number of studies have revealed that the strength of genetic interference increased as the extent of synapsis increased (Libuda et al., 2013; Zhang et al., 2014; Wang K. et al., 2015). However, in some organisms, evidence indicates that genetic interference is exerted prior to the SC assembly (Bishop and Zickler, 2004; Fung et al., 2004). Partly because of the lack of null TF mutants, the exact role of SC in CO interference is still enigmatic. Most recently, the null mutants of zyp1 have been generated in Arabidopsis thaliana, and cytological and genetic analysis reveals that ZYP1 is required for CO interference (Capilla-Pérez et al., 2021; France et al., 2021). However, researches of null mutants of TF protein genes in cereal crops are scarce to date.
In the present study, we selected the inter-subspecific hybrid rice variety Chunyou84 (CY84), an elite inter-subspecific hybrid rice from a cross between the maternal Chunjiang 16 A (16 A), a japonica male-sterile line, and the paternal C84, an indica-japonica intermediate-type line, as the acceptor and used the CRISPR/Cas9 technology to generate zep1 null mutants. By genetic analysis, we revealed that genetic interference is completely eliminated in the absence of ZEP1, indicating an important role of SC in mediating interference in rice. In addition, the way of generating zep1 null mutants with increased genetic recombination frequency and eliminated genetic interference is a promising approach to increase genetic diversity in rice breeding.
The CRISPR-Cas9 vector for complete knockout of ZEP1 was constructed by the isocaudomer ligation method, as described in Wang C. et al. (2015). The annealed ZEP1 g++/ZEP1 g-- oligonucleotides (Supplementary Table 1) were ligated into the SK-sgRNA vector that digested with AarI. Then, the sgRNA of ZEP1 (digested with KpnI and BglII) was assembled into the pC1300-Actin:Cas9 binary vector (digested with KpnI and BamHI) to obtain the vector pC1300-Actin:Cas9-sgRNAZEP1 for generation of zep1 null mutants.
The binary vector pC1300-Actin:Cas9-sgRNAZEP1 was introduced into Agrobacterium tumefaciens (Strain EHA105). The Agrobacterium was then transformed into calluses derived from CY84 to generate transgenic lines. Fifteen independent transgenic plants of pC1300-Actin:Cas9-sgRNAZEP1 were obtained and grown in summer in the transgenic paddy fields of the China National Rice Research Institute in Hangzhou, China.
Genomic DNA of transgenic plants was extracted from approximately 50mg fresh leaf tissue via the cetyltrimethylammonium bromide (CTAB) method. PCR was performed with KOD FX DNA polymerase (Toyobo, Japan) to amplify the fragments surrounding ZEP1 target site. Primers ZEP1 F and ZEP1 R are listed in Supplementary Table 1. The mutants were detected according to the published Hi-TOM procedure (Liu et al., 2019).
Panicles at meiosis stage were harvested and fixed in Carnoy’s solution (ethanol:glacial acetic acid, 3:1) for more than 24h at room temperature. Microsporocytes were squashed on a slide with a needle and covered with a coverslip. Then, slides were frozen in liquid nitrogen, and the coverslips were removed rapidly with a blade. Chromosomes were stained with 4′,6-diamidino-2-phenylindole (DAPI) in an antifade solution (Vector Laboratories, Burlingame, CA, United States) as described in (Wang et al., 2009). Fluorescence microscopy was conducted using an Olympus BX61 fluorescence microscope fitted with a micro charge-coupled device camera.
Fresh young rice panicles were fixed in 4% (w/v) paraformaldehyde for 45min at room temperature. Anthers at proper meiosis stage were squashed with a needle in PBS solution and covered with a coverslip. After soaking in liquid nitrogen and removing the coverslip, slides were then incubated in a humid chamber at 37°C for 4h with anti-REC8 (Mouse) and anti-ZEP1 (rabbit) polyclonal antibodies (diluted 1:500 in TNB buffer: 0.1M Tris–HCl, pH 7.5, 0.15M NaCl, and 0.5% blocking reagent). After three rounds of washing in PBS, Texas red-conjugated goat anti-mouse antibody and fluorescein isothiocyanate-conjugated sheep anti-rabbit antibody (1:1,000) were added to the slides. The chromosomes were counterstained with DAPI in an antifade solution (Vector Laboratories, Burlingame, CA, United States). Fluorescence microscopy was performed as described above.
Single nucleotide polymorphisms (SNPs) markers were developed on the basis of the whole-genome sequences of C84 and chunjiang16A (Wang et al., 2019). The primers are listed in Supplementary Table 1. The Hi-TOM method was performed for the genotyping as previously described in (Liu et al., 2019).
The analysis of interference strength was described in Wang K. et al. (2015). For each pair of adjacent intervals, the coefficient of coincidence analysis compared the observed frequency of CO occurring in both intervals with the frequency expected if the CO occurred independently in the two intervals, with the interference strength calculated as (1-observed/expected). The expected number of chromosomes with CO occurring in both intervals (XY and YZ) was calculated as: (recombination frequency between X and Y)×(recombination frequency between Y and Z)×(number of chromosomes examined). See the details in Supplementary Tables 2 and 3.
The WT (CY84) and zep1-KO plants were crossed with the indica variety 93-11 to generate the segregation population. The hybrid seeds were soaked in deionized water at 37°C in the dark for 2days and then transferred to a net floating on deionized water for further 5days. The seedlings were cultured in a half-strength Kimura B nutrient solution (pH 5.4; Liu et al., 2020). The genotypes of each plant in the segregation population were identified by Hi-TOM. The genotypes of each female gamete of CY84 and zep1-KO plants were deduced and listed in Supplementary Tables 4–7. Then, they were used for the frequency and distribution of genetic recombination analysis. For each pair of adjacent markers (X and Y), the recombination frequency was calculated as: (number of chromosomes with one CO between X and Y/number of chromosomes examined).
ZEP1 has a full length of 9.478-kbp, distributed in 21 exons and 20 introns and encodes a transverse filament protein containing 869 amino acids (Figure 1A). ZEP1 comprises coiled-coil proteins with globular domains at its N termini from amino acids 1 to 63 and C termini from amino acids 714 to 869. The C-terminal globular domain of 156 residues putatively binds DNA, while the N-terminal globular domain of 63 residues is basic (Wang et al., 2010). The Tos17 insertion mutant lines of ZEP1 used in previous studies are partial loss-of-function mutants (Wang et al., 2010; Wang K. et al., 2015), which still contain the N-terminal residues. To generate zep1 knockout mutants, we employed the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) system to edit ZEP1 and chose the 19th to the 38th nucleotides in the first exon near the initiation codon as our target site (Figure 1A). We successfully constructed the CRISPR-Cas9 vector pC1300-Actin:Cas9-sgRNAZEP1 (Figure 1B) and then introduced it into the elite inter-subspecific hybrid rice variety Chunyou84 (CY84) via Agrobacterium. A total of fifteen independent transgenic plants were obtained in the T0 generation with six homozygous mutations (#1, #4, #7, #9, #11, and #12) and five biallelic mutations (#2, #3, #6, #13, and #14; Figure 1C).
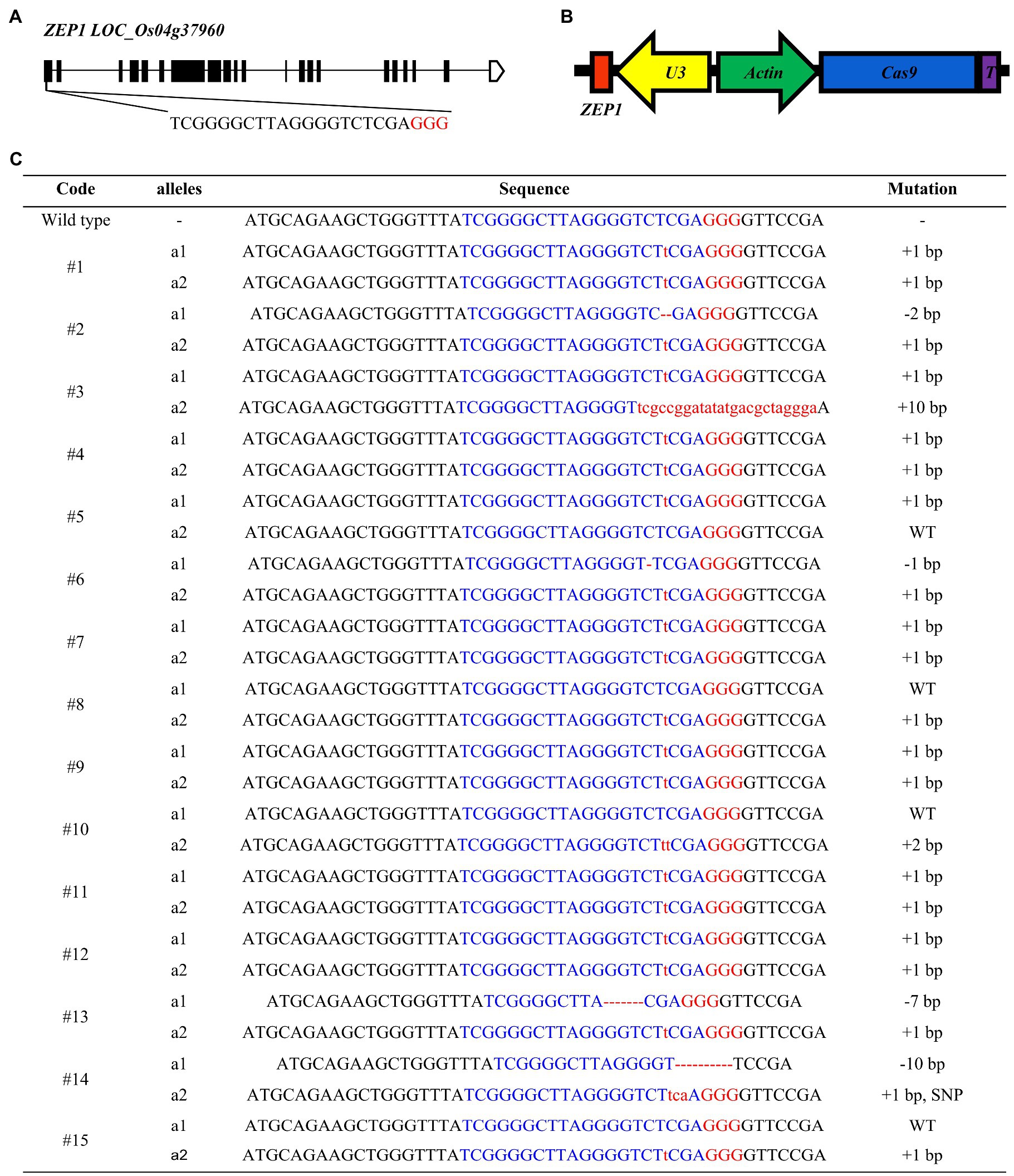
Figure 1. Genome editing of ZEP1 in hybrid rice. (A) ZEP1 gene structure and gRNA target site. (B) The structure of CRISPR-Cas9 vector targeting ZEP1. (C) Mutations in ZEP1 editing lines in the Chunyou84 (CY84) background. The protospacer adjacent motif sequence is highlighted in red. Red lowercase letters and dashed lines indicate inserted or deleted nucleotides, respectively.
To clearly uncover the editing mutations of ZEP1 on genetic recombination frequency and genetic interference, we selected the 1-bp insertion homozygous mutants for further researches. As shown in Figure 2A, the 1-bp insertion caused premature termination of ZEP1 transcription and ultimately encoded a severely truncated protein containing 48 residues, of which only 12 residues were consistent with that of WT. Because of the basic of ZEP1 N-terminal globular domain, we speculated the 1-bp insertion mutants are ZEP1 knockout lines, which differ from the previous partial loss-of-function zep1 mutants (Wang et al., 2010; Wang K. et al., 2015). To confirm the absolute absence of ZEP1, we conducted immunostaining by using anti-REC8 and anti-ZEP1 antibodies. REC8 is a component of the cohesion complex and is required for sister chromatid cohesion, axial element formation, and homolog pairing and is used as a marker to monitor early meiotic events during prophase I here (Zhang et al., 2006). In the WT plants, ZEP1 signals were robust at pachytene, while in the mutants, no signals except the background were detected in any observed meiocytes (n=500; Figure 2B). These results implied that ZEP1 was completely knocked out in the mutants. Hereafter, we renamed these mutants as zep1-KO.
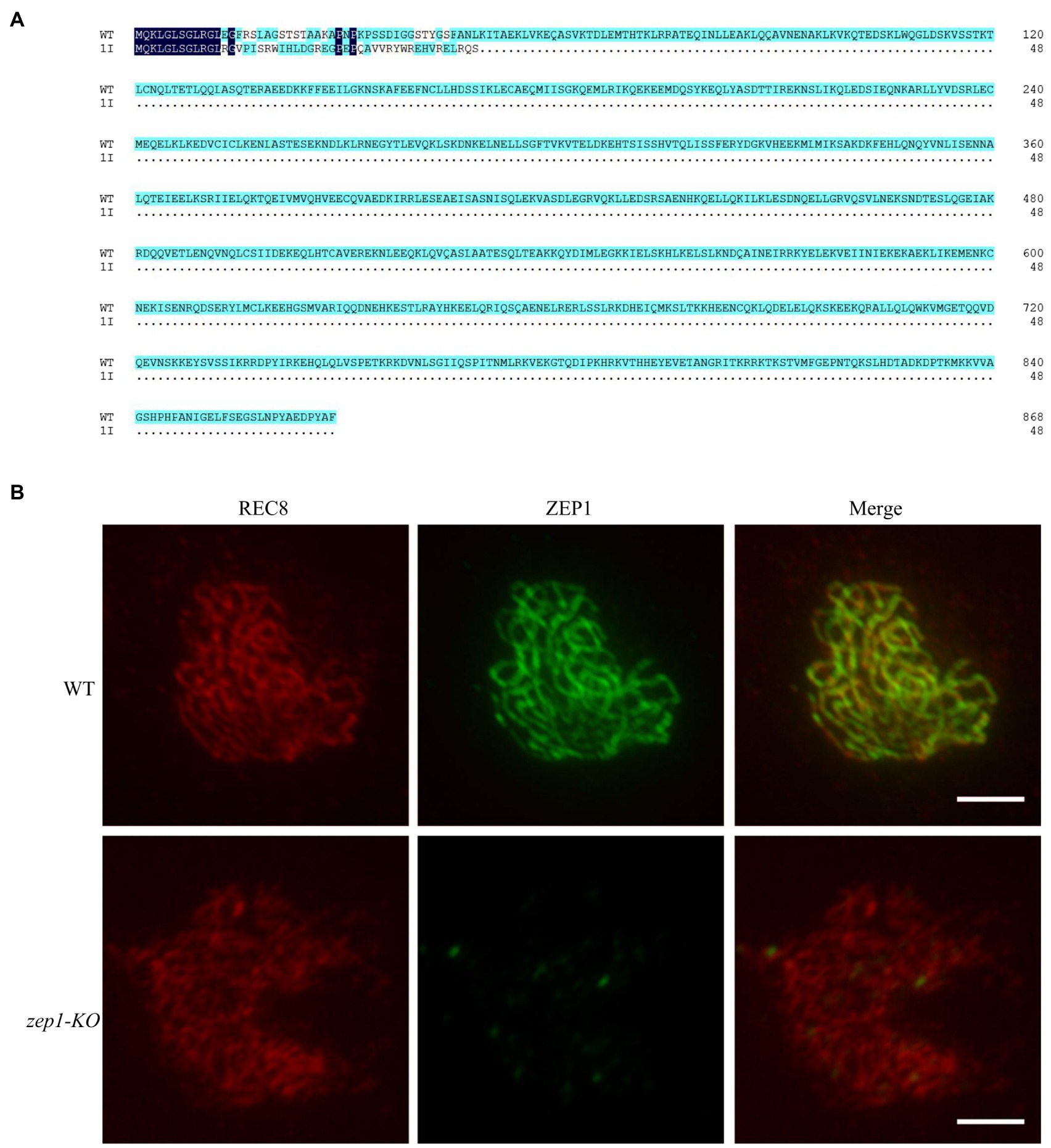
Figure 2. Generation of zep1 null mutants. (A) Amino acid sequence alignment of ZEP1 protein of WT and mutants. The ZEP1 gene encodes a transverse filament protein containing 869 amino acids, while 1bp insertion caused the premature transcription termination of ZEP1. (B) Immunostaining of REC8 (red) and ZEP1 (green) at pachytene stage in the WT and zep1-KO plants. Bar=5μm. REC8 was used to indicate the meiotic chromosomes.
Fertility is an important agronomic trait for genetic recombination analysis and plant breeding (Wang K. et al., 2015). There were no discernible differences in plant morphology between the WT and the zep1-KO lines (Figure 3A), but the anthers of zep1-KO plants were light white while those of the WT were bright yellow (Figure 3B). We further confirmed the suspected male sterility by 1% I2-KI solution staining and observed no staining in the pollens from the zep1-KO plants (Figure 3C). Therefore, unlike the previously described partial fertility of the zep1 mutants (Wang et al., 2010; Wang K. et al., 2015), complete knockout of ZEP1 caused absolute male sterility. In contrast to the anthers, the zep1-KO pistils seemed unaffected, similar to those of WT plants (Figure 3B). To determine whether the female gametes were fertile, we pollinated the zep1-KO and the control WT plants with pollen from WT plants. Both the WT and the zep1-KO plants could set seeds (Figure 3D), with a seed-setting rate of 38.1±5.8% and 34.4±8.5% for the WT and zep1-KO, respectively. These results suggested that the female meiocytes of zep1-KO mutants were not severely affected.

Figure 3. Effects of ZEP1 knockout mutation on sterility. (A) Comparison of WT and zep1-KO plants at maturity. Bar=20cm. (B) Comparison of anthers and pistils of WT and zep1-KO plants at heading date. Bar=1mm. (C) Iodine-iodide kalium staining of pollen from the WT and zep1-KO plants. Bar=5μm (pollen). (D) Cross the WT and zep1-KO plants (as maternal plant) with pollen from WT plants. Bar=0.5cm.
The complete male sterility in the zep1-KO plants might result from defects in meiosis. To test this hypothesis, we investigated the chromosome behaviors of male meiocytes by DAPI staining. In WT plants, all 12 bivalents in the pollen mother cells (PMCs) lined up on the equatorial plate at metaphase I (Figure 4A). However, univalents, bivalents, and multivalents were observed in the zep1-KO PMCs, numbering 4.40±1.96, 8.37±1.30, and 1.53±0.97 (n=30), respectively (Figure 4). Therefore, the presence of univalents and multivalents likely caused the male sterility of the zep1-KO lines.
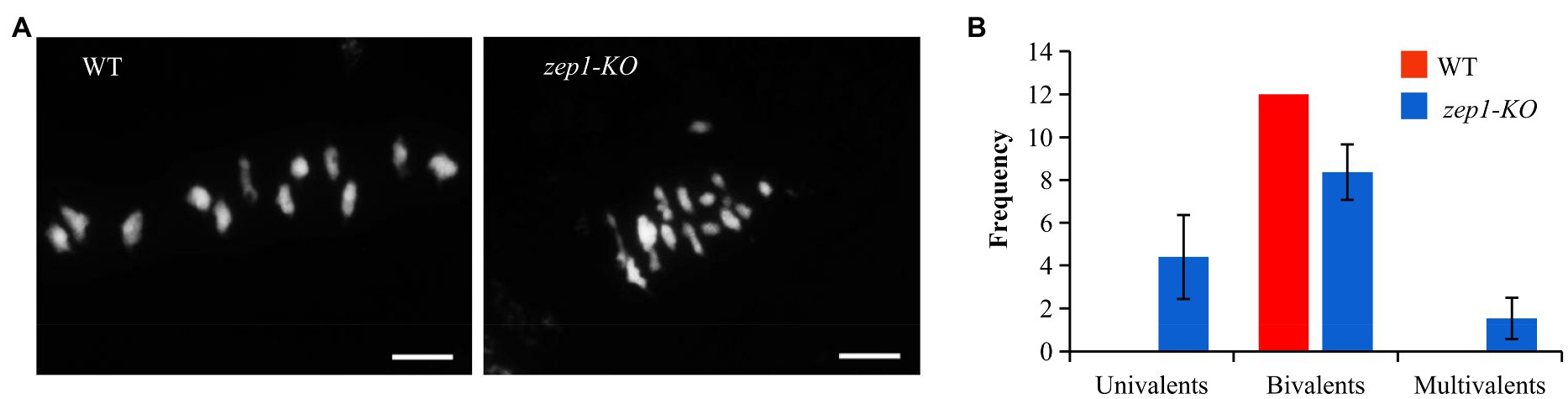
Figure 4. Chromosome behaviors of male meiocytes in WT and zep1-KO plants. (A) DAPI staining of the meiotic chromosomes of pollen mother cells (PMCs) from the WT and zep1-KO plants. Bar=5μm. (B) Frequency distribution of univalents, bivalents, and multivalents in the pollen mother cells (PMCs) from WT and zep1-KO plants. (n=30).
To explore whether the female recombination frequency was affected by disruption of ZEP1, we firstly crossed the WT (the CY84 hybrid) and zep1-KO lines with the indica rice variety 93-11 and then examined the genetic recombination events of WT×93-11 and zep1-KO × 93-11 F1 individuals via genotyping. To this end, we developed 10 nearly uniformly distributed single nucleotide polymorphism (SNP) markers along each of chromosomes 1 and 8 (Figure 5A) that were polymorphic between the parental lines of the CY84 hybrid. Then, we genotyped 364 individual F1 progeny of WT×93-11 and 525 individual F1 progeny of zep1-KO × 93-11 using those markers. After this, we tracked the recombination events that had occurred in the WT or zep1-KO parental plants (CY84 hybrid background) based on a previous method (Wang K. et al., 2015), using our SNP markers to deduce the genotype of each chromatid following meiosis and thus infer the number of COs that had occurred to produce each chromatid. In the WT plants, chromosome 1 chromatids with zero, single, double, triple, and quadruple COs occurred at frequencies of 20, 34, 34, 11, and 1%, respectively. By contrast, the zep1-KO mutants exhibited a decreased frequency of zero (8%), single (25%), and double (28%) COs but a higher frequency of triple (20%), quadruple (14%), quintuple (3%), sextuple (1%), and septuple (1%) COs (Figure 5B). Similar results were detected on chromosome 8, with 19% zero, 34% single 25% double, 16% triple, 4% quadruple, and 2% quintuple COs occurred in zep1-KO mutants, while the frequencies of zero, single, double, and triple COs occurred in WT plants were 23, 49, 25, and 2%, respectively (Figure 5C).
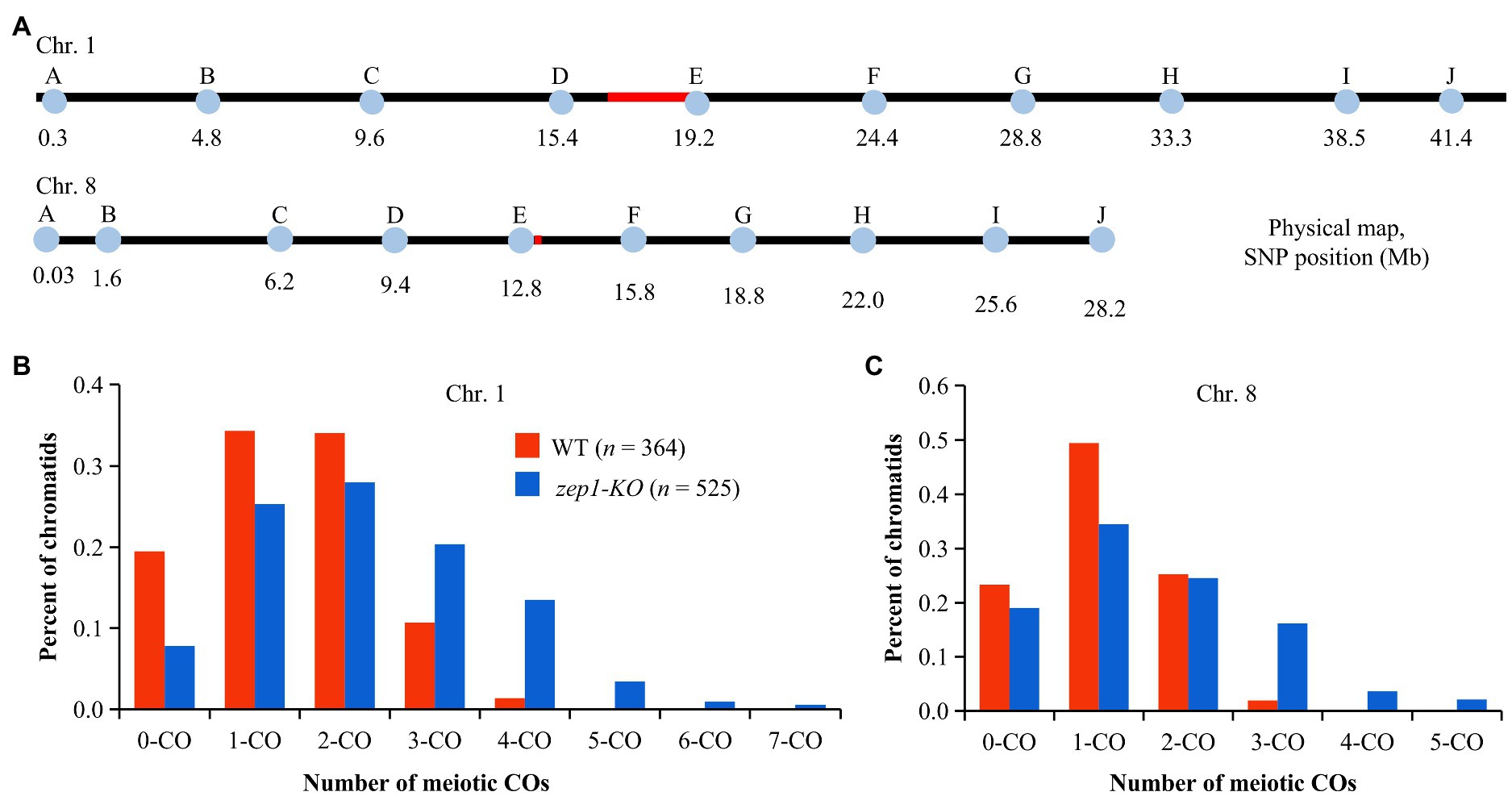
Figure 5. Analysis of chromatids in WT plants and zep1 null mutants. (A) Physical positions of the SNP markers along chromosomes 1 and 8. (B) Percentage of total chromatids (n) for different types of COs on chromosome 1. (C) Percentage of total chromatids (n) for different types of COs on chromosome 8.
We further calculated the recombination frequency of zep1-KO and WT plants. The average number of COs per chromatid on chromosomes 1 and 8 in WT plants was 1.40 and 1.06, respectively, while that of zep1-KO mutants was significantly higher, with 2.33 and 1.57 COs per chromatid on chromosomes 1 and 8, respectively (Figure 6). The recombination frequency was elevated in all identified intervals in the zep1-KO lines compared to that of the WT, varying from 1.01- to 3.57-fold (Figure 6). In particular, the recombination frequency of the DE interval on chromosome 1 and the EF interval on chromosome 8, both of which span the centromere, was elevated by 1.83- and 3.29-fold, which was higher than the average recombination frequency of entire chromosomes: 1.63- and 1.58-fold for chromosomes 1 and 8, respectively (Figure 6).
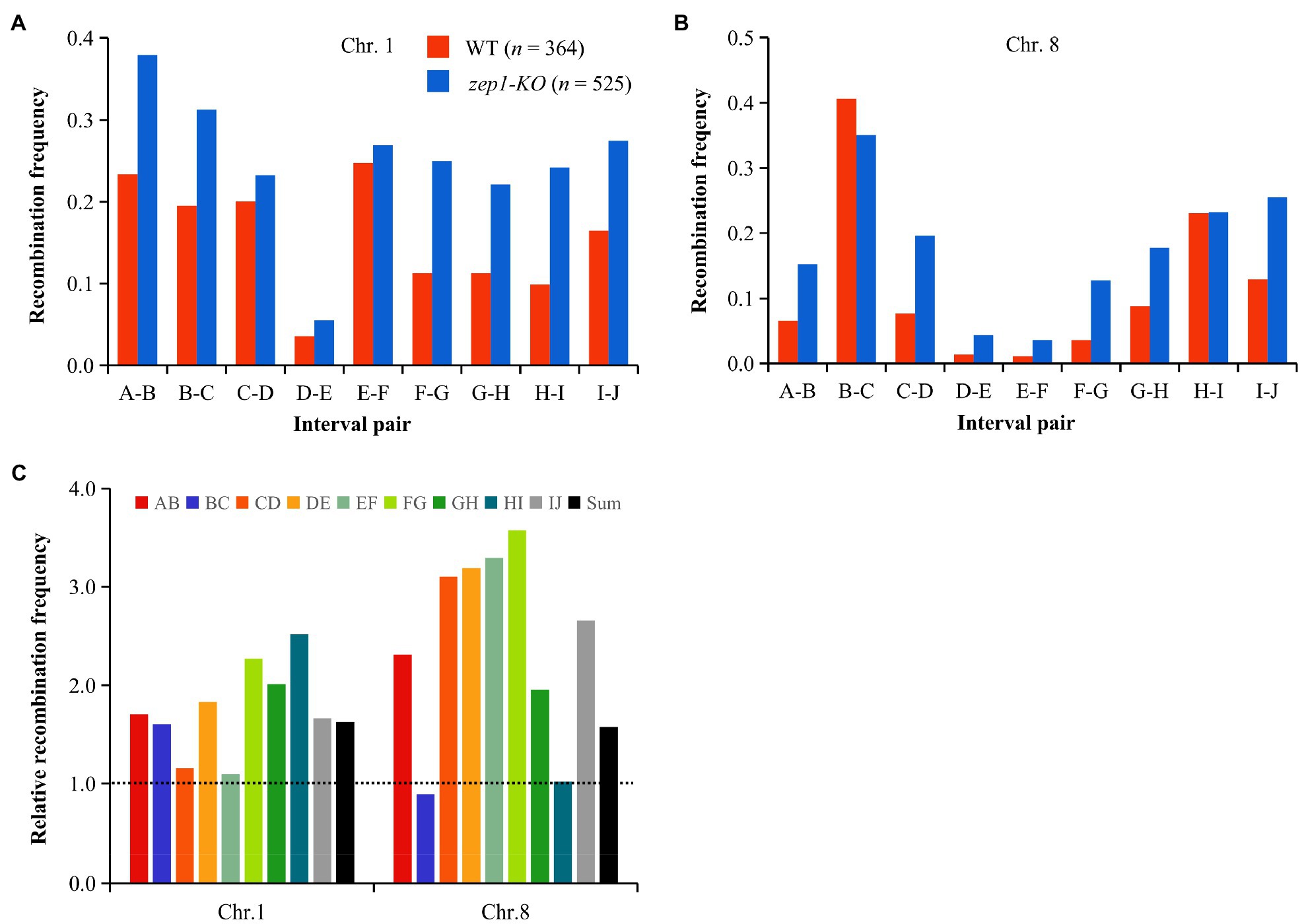
Figure 6. zep1 null mutation increases recombination frequencies. (A) Recombination frequencies measured between adjacent SNP markers on chromosomes 1. (B) Recombination frequencies measured between adjacent SNP markers on chromosomes 8. (C) Relative recombination frequency (zep1-KO/WT) measured between adjacent SNP markers on the chromosomes 1 and 8.
To assess whether genetic interference was affected by the null mutation of the ZEP1 gene, we conducted coefficient of coincidence analysis according to the method described for C. elegans (Libuda et al., 2013). In the analysis, the strength of interference (I) equal to 1 is indicative of complete interference, while an I equal to zero is indicative of an absence of interference (Fernandes et al., 2018). In WT plants, the strength of interference (I) in eight adjacent interval pairs of chromosome 1 ranged from 0.37 to 1.00, indicating the existence of strong genetic interference between neighboring COs. By contrast, the zep1-KO mutants showed reduced I for those interval pairs, with I values of −0.18 to 0.14 (Figure 7A). For chromosome 8, the I value of the four measured interval pairs in the WT ranged from 0.48 to 0.71, whereas that in zep1-KO lines ranged from −0.17 to 0.05 (Figure 7B). These results suggest that genetic interference is likely fully eliminated in zep1-KO plants.
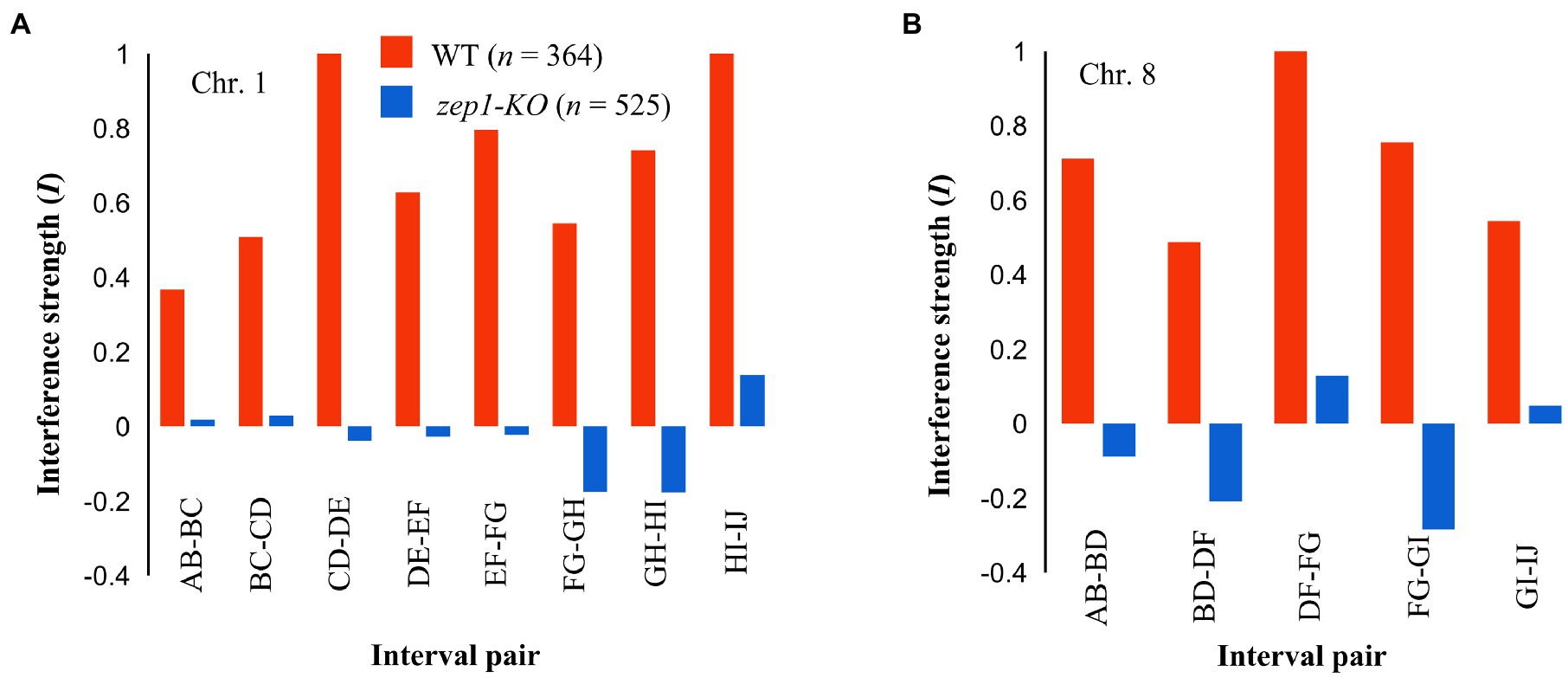
Figure 7. zep1 null mutation disrupts crossover (CO) interference. Interference strength (I) values for different adjacent interval pairs of chromosomes 1 (A) and 8 (B) in WT and zep1-KO plants.
ZEP1 is the central element protein of the SC that regulates genetic recombination frequency in rice (Wang et al., 2010). The partial loss of function of this gene can increase genetic recombination frequency and attenuate CO interference (Wang K. et al., 2015), but the effects of ZEP1 null mutations on CO formation and interference have not been reported comprehensively yet. The N-terminal domain of the ZEP1 is located in the middle of the SC, and this special organization pattern is critical for SC assembly (Anderson et al., 2005; Wang et al., 2010). In this study, we successfully generated null mutation of ZEP1 in hybrid rice by genome editing. Genetic analysis of the null mutants revealed that genetic recombination frequency was greatly increased, and CO interference was completely eliminated in the absence of ZEP1 in CY84 variety background. Compared with the partial loss of function of ZEP1 mutants (Wang et al., 2010; Wang K. et al., 2015), the new null allele in this study showed stronger phenotype than those of previous weak alleles, indicating the residuary N-terminal domain of the ZEP1 in previous weak alleles might still play some roles in meiosis, although the SC cannot be assembled in the weak mutant.
Interference is a phenomenon that reduces the likelihood of a CO occurring adjacent to another CO along regions of a chromosome (Taagen et al., 2020). However, the relationship between the SC and genetic interference has long been a controversy in organisms. In budding yeast, the data of synapsis initiation complex (SIC) are assembled prior to SC and their proper position in the absence of SC formation demonstrated an aspect of interference that is independent of synapsis (Fung et al., 2004). By contrast, in Caenorhabditis elegans, it was found that partial depletion of the central region proteins of SC attenuated genetic interference (Libuda et al., 2013). The partial loss of function of ZEP1 mutants in rice showed similar results like that in Caenorhabditis elegans (Wang K. et al., 2015). Nonetheless, the exact role of SC in CO interference is still enigmatic because of the lack of null TF mutants. Recently, the zyp1 null mutants in Arabidopsis thaliana were generated. Cytological and genetic analysis of the null mutants revealed that ZYP1 is required for CO interference (Capilla-Pérez et al., 2021; France et al., 2021). In this study, we found that genetic interference was eliminated in some detected genomic regions in our obtained rice ZEP1 null mutants. The phenotype of ZEP1 weak and null alleles in rice supports that the strength of genetic interference increased as the extent of synapsis increased.
Meiotic crossovers shuffle chromosomes to produce unique combinations of alleles that are transmitted to offspring (Mieulet et al., 2018). However, meiotic crossovers are tightly regulated by numerous of genes, typically one to three per chromosome (Fernandes et al., 2018). To combine favorable alleles into elite varieties, breeders and geneticists are exploring approaches to increase meiotic crossovers. Mutation of anti-crossover factors, such as FANCM, RECQ4, and FIGL1 in plants, induces a large increase in crossover frequency (Crismani et al., 2012; Girard et al., 2015; Séguéla-Arnaud et al., 2015; Mieulet et al., 2018). Reducing CO interference alters CO distribution, which also shows potential to enhance crop breeding efficiency (Taagen et al., 2020). However, it is very difficult to elevate genetic recombination frequency and eliminate genetic interference simultaneously in breeding process. Here, we used CRISPR/Cas9 to knock out ZEP1, encoding a component of the SC, in hybrid rice. The edited mutants concurrently exhibited disrupted CO interference and an elevated recombination frequency. Our study provides an effective approach to enrich genetic diversity and accelerate rice breeding. Of course, as the background play important roles in the formation of COs, more studies in different varieties are required to test its application potential in the future.
In summary, we used CRISPR/Cas9 to completely knock out the ZEP1 gene, encoding the central component of the SC, in hybrid rice. The mutants exhibited eliminated genetic interference and elevated genetic recombination frequency on chromosomes 1 and 8 in CY84 variety background. Our study of female gamete revealed the important role of the SC in mediating genetic interference and limiting COs. Very recently, null mutants of ZYP1 in Arabidopsis were generated and a virtual absence of CO interference had been detected (Capilla-Pérez et al., 2021; France et al., 2021). Therefore, all of these studies support the long-term debating role of SC in mediating CO interference. Moreover, it is potential to break linkage drag and enrich genetic diversity by using zep1 null mutants during rice breeding.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
KW managed the project. CL, YC, YH, GD, XW, TS, JL, and MW performed the experiments. YH, QL, and CL analyzed the data. CL and YH wrote the manuscript. KW and ZC revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (32025028 and U20A2030), Central Public-interest Scientific Institution Basal Research Fund (Y2020XK17), the Agricultural Science and Technology Innovation Program (CAAS-ZDRW202001), and China National Rice Research Institute Key Research and Development Project (CNRRI-2020-01).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.757152/full#supplementary-material
Anderson, L. K., Royer, S. M., Page, S. L., McKim, K. S., Lai, A., Lilly, M. A., et al. (2005). Juxtaposition of C(2)M and the transverse filament protein C(3)G within the central region of Drosophila synaptonemal complex. Proc. Natl. Acad. Sci. U. S. A. 102, 4482–4487. doi: 10.1073/pnas.0500172102
Barakate, A., Higgins, J. D., Vivera, S., Stephens, J., Perry, R. M., Ramsay, L., et al. (2014). The synaptonemal complex protein ZYP1 is required for imposition of meiotic crossovers in barley. Plant Cell 26, 729–740. doi: 10.1105/tpc.113.121269
Bishop, D. K., and Zickler, D. (2004). Early decision: meiotic crossover interference prior to stable strand exchange and synapsis. Cell 117, 9–15. doi: 10.1016/S0092-8674(04)00297-1
Blary, A., and Jenczewski, E. (2019). Manipulation of crossover frequency and distribution for plant breeding. Theor. Appl. Genet. 132, 575–592. doi: 10.1007/s00122-018-3240-1
Capilla-Pérez, L., Durand, S., Hurel, A., Lian, Q., Chambon, A., Taochy, C., et al. (2021). The synaptonemal complex imposes crossover interference and heterochiasmy in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 118:e2023613118. doi: 10.1073/pnas.2023613118
Crismani, W., Girard, C., Froger, N., Pradillo, M., Santos, J. L., Chelysheva, L., et al. (2012). FANCM limits meiotic crossovers. Science 336, 1588–1590. doi: 10.1126/science.1220381
de Vries, F. A., de Boer, E., van den Bosch, M., Baarends, W. M., Ooms, M., Yuan, L., et al. (2005). Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev. 19, 1376–1389. doi: 10.1101/gad.329705
de Boer, E., and Heyting, C. (2006). The diverse roles of transverse filaments of synaptonemal complexes in meiosis. Chromosoma 115, 220–234. doi: 10.1007/s00412-006-0057-5
Dubois, E., De Muyt, A., Soyer, J. L., Budin, K., Legras, M., Piolot, T., et al. (2019). Building bridges to move recombination complexes. Natl. Acad. Sci. U. S. A. 116, 12400–12409. doi: 10.1073/pnas.1901237116
Fernandes, J. B., Séguéla-Arnaud, M., Larchevêque, C., Lloyd, A. H., and Mercier, R. (2018). Unleashing meiotic crossovers in hybrid plants. Proc. Natl. Acad. Sci. U. S. A. 115, 2431–2436. doi: 10.1073/pnas.1713078114
France, M. G., Enderle, J., Röhrig, S., Puchta, H., Franklin, F., and Higgins, J. D. (2021). ZYP1 is required for obligate cross-over formation and cross-over interference in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 118:e2021671118. doi: 10.1073/pnas.2021671118
Fraune, J., Brochier-Armanet, C., Alsheimer, M., Volff, J. N., Schücker, K., and Benavente, R. (2016). Evolutionary history of the mammalian synaptonemal complex. Chromosoma 125, 355–360. doi: 10.1007/s00412-016-0583-8
Fung, J. C., Rockmill, B., Odell, M., and Roeder, G. S. (2004). Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell 116, 795–802. doi: 10.1016/S0092-8674(04)00249-1
Gao, J., and Colaiácovo, M. P. (2018). Zipping and unzipping: protein modifications regulating synaptonemal complex dynamics. Trends Genet. 34, 232–245. doi: 10.1016/j.tig.2017.12.001
Girard, C., Chelysheva, L., Choinard, S., Froger, N., Macaisne, N., Lemhemdi, A., et al. (2015). AAA-ATPase FIDGETIN-LIKE 1 and helicase FANCM antagonize meiotic crossovers by distinct mechanisms. PLoS Genet. 11:e1005369. doi: 10.1371/journal.pgen.1005369
Golubovskaya, I. N., Wang, C. J., Timofejeva, L., and Cande, W. Z. (2011). Maize meiotic mutants with improper or non-homologous synapsis due to problems in pairing or synaptonemal complex formation. J. Exp. Bot. 62, 1533–1544. doi: 10.1093/jxb/erq292
Higgins, J. D., Sanchez-Moran, E., Armstrong, S. J., Jones, G. H., and Franklin, F. C. (2005). The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 19, 2488–2500. doi: 10.1101/gad.354705
Khoo, K. H., Able, A. J., and Able, J. A. (2012). The isolation and characterisation of the wheat molecular ZIPper I homologue, TaZYP1. BMC. Res. Notes 5:106. doi: 10.1186/1756-0500-5-106
Libuda, D. E., Uzawa, S., Meyer, B. J., and Villeneuve, A. M. (2013). Meiotic chromosome structures constrain and respond to designation of crossover sites. Nature 502, 703–706. doi: 10.1038/nature12577
Liu, C. L., Gao, Z. Y., Shang, L. G., Yang, C. H., Ruan, B. P., Zeng, D. L., et al. (2020). Natural variation in the promoter of OsHMA3 contributes to differential grain cadmium accumulation between Indica and japonica rice. J. Integr. Plant Biol. 62, 314–329. doi: 10.1111/jipb.12794
Liu, Q., Wang, C., Jiao, X., Zhang, H., Song, L., Li, Y., et al. (2019). Hi-TOM: a platform for high-throughput tracking of mutations induced by CRISPR/Cas systems. Sci. China Life Sci. 62, 1–7. doi: 10.1007/s11427-018-9402-9
MacQueen, A. J., Colaiácovo, M. P., McDonald, K., and Villeneuve, A. M. (2002). Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 16, 2428–2442. doi: 10.1101/gad.1011602
Mieulet, D., Aubert, G., Bres, C., Klein, A., Droc, G., Vieille, E., et al. (2018). Unleashing meiotic crossovers in crops. Nat. Plants 4, 1010–1016. doi: 10.1038/s41477-018-0311-x
Moose, S. P., and Mumm, R. H. (2008). Molecular plant breeding as the foundation for 21st century crop improvement. Plant Physiol. 147, 969–977. doi: 10.1104/pp.108.118232
Otto, S. P., and Payseur, B. A. (2019). Crossover interference: shedding light on the evolution of recombination. Annu. Rev. Genet. 53, 19–44. doi: 10.1146/annurev-genet-040119-093957
Page, S. L., and Hawley, R. S. (2001). c(3)G encodes a Drosophila synaptonemal complex protein. Genes Dev. 15, 3130–3143. doi: 10.1101/gad.935001
Schild-Prüfert, K., Saito, T. T., Smolikov, S., Gu, Y., Hincapie, M., Hill, D. E., et al. (2011). Organization of the synaptonemal complex during meiosis in Caenorhabditis elegans. Genetics 189, 411–421. doi: 10.1534/genetics.111.132431
Séguéla-Arnaud, M., Crismani, W., Larchevêque, C., Mazel, J., Froger, N., Choinard, S., et al. (2015). Multiple mechanisms limit meiotic crossovers: TOP3α and two BLM homologs antagonize crossovers in parallel to FANCM. Proc. Natl. Acad. Sci. U. S. A. 112, 4713–4718. doi: 10.1073/pnas.1423107112
Storlazzi, A., Xu, L., Schwacha, A., and Kleckner, N. (1996). Synaptonemal complex (SC) component Zip1 plays a role in meiotic recombination independent of SC polymerization along the chromosomes. Proc. Natl. Acad. Sci. U. S. A. 93, 9043–9048. doi: 10.1073/pnas.93.17.9043
Sun, L., Wang, J., Sang, M., Jiang, L., Zhao, B., Cheng, T., et al. (2017). Landscaping crossover interference across a genome. Trends Plant Sci. 22, 894–907. doi: 10.1016/j.tplants.2017.06.008
Sym, M., Engebrecht, J. A., and Roeder, G. S. (1993). ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72, 365–378. doi: 10.1016/0092-8674(93)90114-6
Taagen, E., Bogdanove, A. J., and Sorrells, M. E. (2020). Counting on crossovers: controlled recombination for plant breeding. Trends Plant Sci. 25, 455–465. doi: 10.1016/j.tplants.2019.12.017
Wang, C., Liu, Q., Shen, Y., Hua, Y., Wang, J., Lin, J., et al. (2019). Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes. Nat. Biotechnol. 37, 283–286. doi: 10.1038/s41587-018-0003-0
Wang, C., Shen, L., Fu, Y., Yan, C., and Wang, K. (2015). A simple CRISPR/Cas9 system for multiplex genome editing in rice. J. Genet. Genomics 42, 703–706. doi: 10.1016/j.jgg.2015.09.011
Wang, K., Tang, D., Wang, M., Lu, J., Yu, H., Liu, J., et al. (2009). MER3 is required for normal meiotic crossover formation, but not for presynaptic alignment in rice. J Cell Sci. 122, 2055–2063. doi: 10.1242/jcs.049080
Wang, K., Wang, C., Liu, Q., Liu, W., and Fu, Y. (2015). Increasing the genetic recombination frequency by partial loss of function of the synaptonemal complex in rice. Mol. Plant 8, 1295–1298. doi: 10.1016/j.molp.2015.04.011
Wang, M., Wang, K., Tang, D., Wei, C., Li, M., Shen, Y., et al. (2010). The central element protein ZEP1 of the synaptonemal complex regulates the number of crossovers during meiosis in rice. Plant Cell 22, 417–430. doi: 10.1105/tpc.109.070789
Zhang, L., Espagne, E., de Muyt, A., Zickler, D., and Kleckner, N. E. (2014). Interference-mediated synaptonemal complex formation with embedded crossover designation. Proc. Natl. Acad. Sci. U. S. A. 111, E5059–E5068. doi: 10.1073/pnas.1416411111
Keywords: genetic diversity, genetic interference, genetic recombination, ZEP1 , synaptonemal complex, hybrid rice, genome editing
Citation: Liu C, Cao Y, Hua Y, Du G, Liu Q, Wei X, Sun T, Lin J, Wu M, Cheng Z and Wang K (2021) Concurrent Disruption of Genetic Interference and Increase of Genetic Recombination Frequency in Hybrid Rice Using CRISPR/Cas9. Front. Plant Sci. 12:757152. doi: 10.3389/fpls.2021.757152
Received: 11 August 2021; Accepted: 09 September 2021;
Published: 01 October 2021.
Edited by:
Tao Zhang, Yangzhou University, ChinaReviewed by:
Yingxiang Wang, Fudan University, ChinaCopyright © 2021 Liu, Cao, Hua, Du, Liu, Wei, Sun, Lin, Wu, Cheng and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kejian Wang, d2FuZ2tlamlhbkBjYWFzLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.