
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 30 September 2021
Sec. Plant Biotechnology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.754325
This article is part of the Research Topic Phenylpropanoid Systems Biology and Biotechnology View all 34 articles
Blueberries (Vaccinium corymbosum) contain large amounts of flavonoids, which play important roles in the plant’s ability to resist stress and can also have beneficial effects on human health when the fruits are eaten. However, the molecular mechanisms that regulate flavonoid synthesis in blueberries are still unclear. In this study, we combined two different transcriptome sequencing platforms, single-molecule real-time (SMRT) and Illumina sequencing, to elucidate the flavonoid synthetic pathways in blueberries. We analyzed transcript quantity, length, and the number of annotated genes. We mined genes associated with flavonoid synthesis (such as anthocyanins, flavonols, and proanthocyanidins) and employed fluorescence quantitative PCR to analyze the expression of these genes and their correlation with flavonoid synthesis. We discovered one R2R3 MYB transcription factor from the sequencing library, VcMYB1, that can positively regulate anthocyanin synthesis in blueberries. VcMYB1 is mainly expressed in colored (mature) fruits. Experiments showed that overexpression and transient expression of VcMYB1 promoted anthocyanin synthesis in Arabidopsis, tobacco (Nicotiana benthamiana) plants and green blueberry fruits. Yeast one-hybrid (Y1H) assay, electrophoretic mobility shift assay, and transient expression experiments showed that VcMYB1 binds to the MYB binding site on the promoter of the structural gene for anthocyanin synthesis, VcMYB1 to positively regulate the transcription of VcDFR, thereby promoting anthocyanin synthesis. We also performed an in-depth investigation of transcriptional regulation of anthocyanin synthesis. This study provides background information and data for studying the synthetic pathways of flavonoids and other secondary metabolites in blueberries.
Blueberries (Vaccinium corymbosum, Ericaceae) are one of the most popular fruits globally. In North America, blueberries are grown along the Atlantic coast of the United States and in southeastern Canada. In China, blueberries are cultivated in more than 27 provinces, including Shandong, Liaoning, and Guizhou. By the end of 2020, China’s blueberry cultivation area had reached 66,400 hm2, and total production was 347,200 tons (Li et al., 2021). Thus, blueberries are becoming one of the most important fruit crops in China.
Blueberries contain large amounts of anthocyanins, proanthocyanidins, and flavonols, which are the three most common flavonoids. These flavonoids play important roles in fruit coloring, they span from functions in regulating plant development, pigmentation, to an array of roles in defense and signaling between plants and microorganisms, and flavonoid-rich mixed berries maintain and improve cognitive function over a 6 h period in young healthy adults (Ulrike, 2018; Adrian et al., 2019). Anthocyanin is the most common flavonoid and is responsible for the blue and red colors of the fruit. Therefore, elucidating the molecular mechanisms of anthocyanin synthesis would be valuable as a way to develop tools for modifying fruit quality and specifically enriching the formation of fruit color.
Usually MBW protein complexes, consisting of R2R3 MYB, basic helix-loop-helix (bHLH), and WD-repeat proteins regulate the synthesis of plant flavonoids and this has been verified in many plants. The R2R3 MYB transcription factor plays an important role in this protein complex. The plant R2R3 MYB gene family is extremely huge and their functions in other plants were also extensively studied. In apples, MdMYB1 and MdMYB10 can regulate the synthesis of anthocyanins (An et al., 2017a, An et al., 2020b). PyMYB114 and PybHLH3 can regulate anthocyanin synthesis in peaches (Yao et al., 2017). In bilberry (Vaccinium myrtillus L.), Wu C. et al. (2021) describe the complex MYBA locus and identify the key regulating MYB genes that determine anthocyanin production. In American cranberry (Vaccinium macrocarpon Ait.), the subgroup-6 (SG-6 or sg6) R2R3 MYB transcription factors likely act as anthocyanin biosynthesis regulators (Diaz et al., 2021). In blueberries, VcMYBPA can regulate proanthocyanidin synthesis (Zifkin et al., 2012). However, there are currently no studies on the R2R3 MYB gene that regulates anthocyanin synthesis in blueberries.
Most studies on flavonoid synthesis have been in model plants such as apples and peaches, whereas fewer studies have used blueberries (Zhou et al., 2015; An et al., 2017a; An et al., 2020b). One reason that little is known about the genetic mechanisms regulating flavonoid production in blueberries is that these plants are polyploid and have high heterozygosity; thus, genome information and analyses of gene function in blueberry has lagged behind other species (Costich et al., 1993; Rowland et al., 2012). In recent years, with the development of high-throughput sequencing technologies, transcriptome sequencing has become an important method for studying the regulation of gene expression. Transcriptome sequencing can be used to rapidly obtain almost all transcript information of a specific tissue or organ under certain conditions. The use of RNA sequencing (RNA-seq) combined with bioinformatics has provided new research ideas and routes for transcriptomics research. This combined technique is particularly suited for species for which genomic data are not yet available and has been used widely to perform studies on the formation of quality fruits and the mechanisms of stress resistance in many species, such as peaches (Choi et al., 2021), grapes (Xu et al., 2021), sweet oranges (Yao et al., 2020), bananas (Kaushal et al., 2021), and rice (Bakade et al., 2021).
Some studies have employed transcriptome sequencing (Illumina sequencing) technologies and expressed sequence tag (EST) library methods to analyze the metabolic mechanisms (Liang et al., 2020; Wu P. et al., 2021). Jaakola et al. (2002) and Zifkin et al. (2012) analyzed the correlation between expression of genes related to flavonoid synthesis at different developmental stages and flavonoid content in highbush blueberries (V. corymbosum “Rubel”) and wild blueberries (Vaccinium myrtillus), respectively. Li Y. Y. et al. (2012) and Sun et al. (2015) both employed de novo transcriptome sequencing to analyze the differential expression of genes related to flavonoid synthesis in red and white fruits of cranberries (V. macrocarpon “Bergman”) and in mature blueberry skins and pulp (V. corymbosum “Northland”), respectively. Li et al. (2016) also employed de novo transcriptome sequencing to analyze the expression of structural genes related to anthocyanin biosynthesis during different color stages of blueberries (V. corymbosum “Sierra”). Still, there is less genomic, transcriptomic, and proteomic molecular data on blueberries compared with other species. In addition, Although Illumina sequencing technology has high accuracy, due to the limitation of reading length, the assembled transcripts are incomplete and the accuracy is low (Xu et al., 2015; Li et al., 2017). As an alternative, PacBio single-molecule real-time sequencing (SMRT) technology can be used to construct libraries with different inserts and has an advantage of ultra-long read lengths (5–8 kb). SMRT technology has been widely applied to multiple species (Minoche et al., 2015; Du et al., 2021). SMRT has also been applied in combination with Illumina sequencing in many studies (Xu et al., 2015; Li et al., 2017; Qian et al., 2020). Here, we aimed to combine SMRT and Illumina sequencing technologies to study transcriptome data in different developmental stages of blueberry (V. corymbosum “Duke”) to identify the genes that regulate flavonoid synthesis. “Duke” was selected because it is one of the main cultivated blueberry varieties in Shandong and Liaoning provinces in China. In addition, this cultivar has relatively higher flavonoid content and antioxidant activity (Prior et al., 1998).
This work will provide a platform for future research on improving fruit coloring and understanding the regulatory mechanisms for flavonoid synthesis in blueberries.
The experiments were carried out at the College of Horticulture Sciences, Shandong Agricultural University (Tai’an, Shandong, China) and Ministry of Agriculture Key Laboratory for utilization of horticultural crop germplasm resources, Fruit Tree Research Institute, Chinese Academy of Agricultural Sciences (Xingcheng, Liaoning, China) from April 2018 to January 2021. The experimental material were 9-year-old blueberry (V. corymbosum “Duke”) seedlings that were collected from the small berry garden in the Fruit Tree Research Institute, Chinese Academy of Agricultural Sciences. Fruits were collected 40 days (green fruits), 65 days (pink fruits), or 80 days (blue fruits) after flowering. To ensure consistency across the materials, fruits were collected from the tips of inflorescences (first to mature), snap-frozen in liquid nitrogen, and stored at –80°C until analysis.
Plant total RNA extraction kits (TaKaRa, Beijing, China, No. 9769S) were used for extraction of total RNA from fruits of each of the three stages. Agarose gel electrophoresis (Agilent 2100 bioanalyzer, Agilent),1 and spectrophotometry (NanoDrop spectrophotometer, Thermo Fisher Scientific)2 were used to examine the integrity, purity, and quality of total RNA. Oligo (dT) magnetic beads were used for mRNA enrichment and the SMART PCR cDNA Synthesis Kit was used to synthesize full-length cDNA from mRNA. BluePippin (Sage Science)3 was used to screen full-length cDNA fragments and three types of insert libraries were constructed (1–2, 2–3, and 3–6 kb). PCR amplification was carried out on full-length, screened cDNA. End-terminal repair was carried out on full-length cDNA and the cDNAs were ligated to SMRT bell adapters before digestion using exonucleases. Secondary screening of the libraries was carried out using BluePippin to obtain the sequencing libraries. After library construction, Qubit 2.0 (Thermo Fisher Scientific) was used for quantitative analysis. The Agilent 2100 bioanalyzer (Agilent 2100 bioanalyzer) was used for examination of the insert fragments in the libraries. After the libraries passed the quality check, the PacBio RS II platform was used for full-length transcriptome sequencing. Two SMRT cells were used for sequencing the 1–2 and 3–6 kb fragment libraries, and three SMRT cells were used for sequencing the 2–3 kb fragment libraries. The LoRDEC software was used to correct the sequencing errors in the consensus transcripts using Illumina reads as the reference (Salmela and Rivals, 2014).
The methods for total RNA extraction from fruits and examination of total RNA integrity, purity, and quality were carried out as described above. After DNAse I digestion of total RNA from fruit samples, oligo (dT) magnetic beads were used for mRNA enrichment. After mRNAs were made into short fragments, first-strand cDNA and second-strand cDNA were synthesized, purified and recovered, and the sticky ends were repaired. An “A” adapter was added to the 3′ ends of the cDNA before selection of fragment sizes. Finally, PCR amplification was used to construct the libraries. Qubit 2.0 (Thermo Fisher Scientific; see text footnote 2) and Agilent 2100 bioanalyzers were used for examination of library concentration and the fragment sizes of the inserts in the constructed libraries. We used Quantitative PCR to verify quantitation of the effective concentrations of the libraries. After the libraries passed the quality check, the Illumina HiSeq 2500 sequencer was used for sequencing and the sequencing read length was 125 bp paired-ends. RNA samples from green, pink, and blue fruits were sequenced in duplicates.
The IsoSeqTM protocol (Pacific Biosciences, United States) was used to process the sequenced reads using circular consensus sequencing RNA BIOLOGY 5. Effective subreads were obtained using the P_Fetch and P_Filter function (parameters: miniLength = 50, read Score = 0.75, artifact = –1,000) in the SMRT Analysis software suite.4 CCS was obtained from the P_CCS module using the parameter Min Complete Passes = 2 and Min Predicted Accuracy = 0. After examining for poly (A) signal and 5′ and 3′ adaptors, only the CCS with all three signals was considered as a FLNC read (Du et al., 2021). Unmerged subreads were also examined for the three signals, and those with three signals were incorporated into the final FLNC read set.
Blueberry transcriptome data from Illumina and SMRT sequencing were used for alignment with COG, GO, and NR databases to obtain VcMYB genes. Phylogenetic analysis of all the excavated VcMYB protein sequences and Arabidopsis MYB protein sequences showed that only one VcMYB sequence has extremely high homology with the Arabidopsis sg6 R2R3 MYB protein, and it was named VcMYB1.
Three RNA samples were isolated from fruits of each of the three fruit development stages of blueberries. RNA samples were isolated from leaves of Arabidopsis. Single-stranded cDNA was obtained using a reverse transcription kit (TaKaRa, Shiga, Japan). All qRT-PCR analyses were performed with three biological repeats and three techincal repeats. Arabidopsis polyubiquitin 10 (AtUBQ10) was used as housekeeping gene. Supplementary Table 4 shows the primers used.
The extraction methods for anthocyanins, flavonols, and proanthocyanidins were referenced from Prior et al. (2001); Lätti et al. (2010), and Pertuzatti et al. (2016), respectively. High performance liquid chromatography (HPLC) separation, identification and quantitation of anthocyanins, proanthocyanidins, and flavonols were performed on an Agilent 1100 Series system (Agilent, Germany), equipped with DAD (G1315B) and a LC/MSD Trap VL (G2445C VL) electrospray ionization mass spectrometry (ESIMSn) system that was coupled to an Agilent ChemStation (version B.01.03) data-processing station. The mass spectra data were processed with the Agilent LC/MS Trap software (Version 5.3).
ESI-MSn was used to identify the anthocyanins, proanthocyanidins, and flavonols profile and the following parameters were employed: positive ionization mode; dry gas, N2, 11 mL/min; drying temperature, 350°C; nebulizer, 65 psi; capillary, –2500 V; capillary exit offset, 70 V; skimmer1, 20 V; skimmer 2, 6 V; compound stability, 100%; scan range, 50–1,200 m/z. For the purposes of quantitation, For quantification, DAD chromatograms were extracted at 520 nm for anthocyanins, proanthocyanidins at 280 nm, and flavonols at 360 nm.
The Protein BLAST program5 was used to obtain homologs of Arabidopsis, Vitis vinifera and Malus domestica MYB. The amino-acid secondary structure of VcMYB was predicted using the Simple Modular Architecture Research Tool (SMART) software program.6
The ORF (Open Reading Frame) was cloned and inserted into the pRI101-AN vector, and used to transform Agrobacterium tumefaciens GV3101. Arabidopsis plants were transformed with Agrobacterium by the floral dipping method. T1 VcMYB1 transgenic plants were selected on Murashige and Skoog (MS) media containing 50 mg L–1 kanamycin. Kanamycin-resistant T1 seedlings were transferred to soil and grown at 22°C in a growth chamber with a 16 h day length. T2 seedlings were selected on MS media containing 50 mg L–1 kanamycin.
The Agrobacterium GV3101 strains containing the pRI101-VcMYB1 were infiltrated into the abaxial leaf surface of tobacco according to Espley et al. (2007). Each infiltration was performed using three leaves from the same plants. Photographs were taken 4–7 days after infiltration. To control for leaf-to-leaf variability, at least two plants were used for infiltration, and each leaf included negative controls (Agrobacterium with pRI101 empty vector).
Fruit injection assays were carried out according to Li X. Y. et al. (2012), Li Y. Y. et al. (2012), and An et al. (2017b). The VcMYB1-pGreenII62-SK vector was generated by cloning the ORF of VcMYB1 into the pGreenII62-SK vector. The mixed vectors and the A. tumefaciens solutions were injected into the fruit peels.
Chromosome walking techniques were used to isolate the promoter sequence for VcDFR and the method was referenced from the Genome Walking Kit (TaKaRa).7 Specific primers (SP1, SP2, and SP3) were designed according to the open reading frame of VcDFR. Specific primers and four degenerate primers in the reagent kit were used for three rounds of thermal asymmetric PCR. Finally, a 834 bp fragment was obtained through three rounds of PCR.
Y1H assays were performed with the yeast strain Y1HGold (Clontech).8 The full-length cDNA of VcMYB1 was cloned and inserted into the pGADT7 vector. The promoter fragments of VcDFR were cloned into the pAbAi vector to generated the construct pAbAi-VcDFRpro, and then transformed into Y1HGold using a PEG/LiAc method. Yeast cells were cultured on the medium lacking Ura (SD/-Ura). Yeast colony PCR for confirming that positive plasmid has integrated into the Y1HGold genome, and then transformed pGADT7-VcMYB1 into Y1HGold-pAbAi-VcDFRpro. Yeast cells were cultured on media lacking Leu (SD/-Leu) with Aureobasidin A to identify possible interactions.
The EMSA was performed using the LightShift Chemiluminescent EMSA Kit (Thermo Fisher Scientific, Waltham, MA, United States). Briefly, biotin-labeled probes were incubated in a 1 × binding buffer containing 5 mM MgCl2, 50 mM KCl and 2.5% glycerol with or without VcMYB1-His fusion proteins at room temperature for 20 min. An unlabeled probe was added to the reactions for unlabeled probe competition.
Transient expression assay in tobacco leaves were performed according to Shang et al. (2010). The VcDFR promoter was amplified and cloned into pGreenII0800-LUC vector, which generated the reporter constructs VcDFRpro:Luc. The effector (35Spro:VcMYB1) was constructed by cloning the ORF of VcMYB1 into the pGreenII62-SK vector. Transformed leaves were sprayed with 100 mM luciferin, after which they were placed in darkness for 5 min before examining for luminescence. A charge-coupled device-imaging apparatus (NightOWL LB983 in conjunction with Indigo software) was used to collect the LUC images.
The Agrobacterium GV3101 strains containing the pRI101-VcMYB1 generated effector constructs. Reporter constructs were generated using the promoter sequences of VcDFR cloned upstream of the β-glucuronidase (GUS) reporter gene in the pCAMBIA1301 vector, and used to transform Agrobacterium GV3101. For the transient expression assay, Agrobacterium GV3101 strains containing the pRI101-VcMYB1 were co-infiltrated into the abaxial leaf surface of tobacco. Each infection was performed in three biological replicates. After grow in a growth chamber for 3–4 days, the infected leaves were used to analyze GUS activity. Proteins were extracted from infected leaves and fluorescence was measured using a fluorometer (VersaFluor Fluorometer, Bio-Rad)9 as performed according to Jefferson et al. (1987). The values of GUS activity were calculated from three biological replicates.
The Illumina and SMRT sequencing platforms were used to analyze transcriptome data of blueberries. First, the Hiseq 2500 platform was used for sequencing of three samples from blueberry fruits of each of three developmental stage, green (diameter 8–10 mm), pink (12–14 mm), and blue (12–14 mm) (Figure 1A). Samples were sequenced in duplicates. After removal of sequencing adapters and primer sequences the high-quality reads (clean data) totaled 112,625,479 (Table 1) and the total number of bases was 28,376,811,360. After Trinity assembly, a total of 104,068 Unigenes were obtained (Supplementary Table 1).
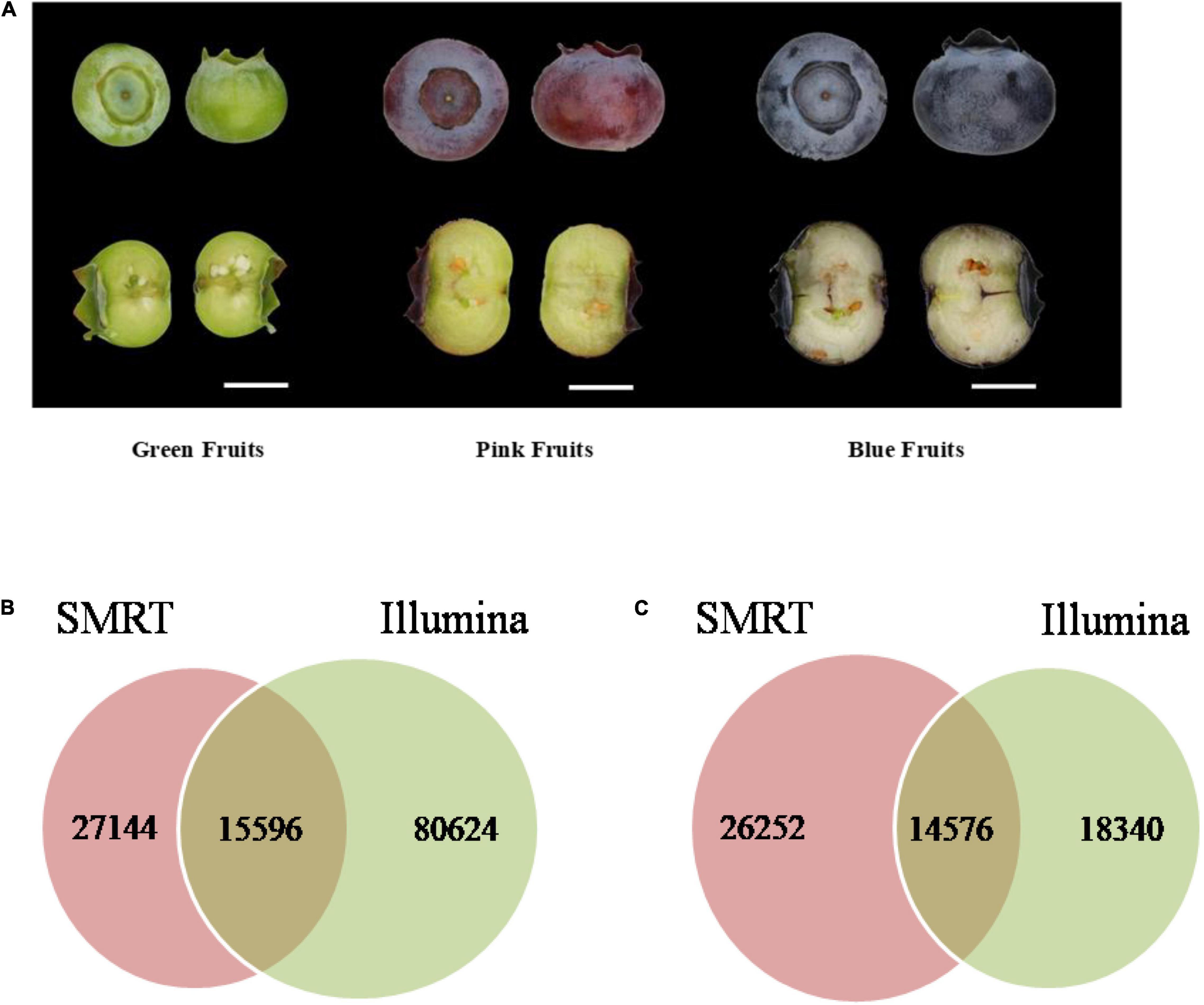
Figure 1. Blueberry (Vaccinium corymbosum “Duke”) fruits of different developmental stages were used in molecular and chemical analyses and Statistical analysis of sequencing data. (A) Green (40 days since flowering), pink (65 days), and blue (80 days) fruits were used in the analyses. Scale bar = 7 mm. Venn diagrams of (B) the number of common and unique sequences and (C) the number of common and unique annotated genes obtained in Illumina and SMRT sequencing.
Next, SMRT sequencing was carried out on the same samples. Equal amounts of six types of mRNAs from the three fruit stages were mixed to construct a sample pool. Three insert libraries were constructed and seven SMRT cell (two cells for 1–2 kb fragments, three cells for 2–3 kb fragments, and two cells for 3–6 kb fragments) sequencing microarrays were used. After removal of adapters and low-quality reads, 572,495 subreads (1.9 billion) were obtained. These subreads were screened to obtain 458,964 reads of insert (RoIs), of which there were 209,715 full-length non-chimeric reads (FLNCs) with 5′ and 3′ primer sequences and poly (A) sequences (Table 1). The average length of RoIs and FLNC reads were 2,938 and 2,785 bp, respectively. The iterative isoform-clustering (ICE) algorithm and Quiver software were used for sequence calibration and sequences with accuracy greater than 0.99 were retained to obtain 117,784 consensus reads. Then, CD-HIT EST software was used to remove redundant sequences from the above high-quality transcripts. We obtained 42,740 non-redundant sequences with an average length of 2,452 bases (Supplementary Table 1).
Although Illumina sequencing has extremely high coverage, the transcripts assembled by Trinity cannot represent full-length cDNA. In this study, the Illumina sequencing results of blueberry messenger RNA (mRNA) samples showed that approximately 37.81% of Unigenes were shorter than 300 bases. The number of Unigenes that were greater than 1, 2, and 3 kb were 18,849, 7,042, and 2,476, respectively, with an average length of 677 bases. In the SMRT sequencing data, the percentage of RoIs that were smaller than 300 bases was only 5.6%. The number of FLNCs that were greater than 1, 2, and 3 kb were 42,646, 24,312, and 9,896, respectively. The average length of FLNC reads in the three insert libraries (1–2, 2–3, and 3–6 kb) were 1,757, 2,823, and 3,775 bases. Overall, sequences obtained through SMRT sequencing were longer and the transcripts were more intact.
Blast alignment analysis of the corresponding relationship between 104,068 Unigene sequences obtained from Illumina sequencing and 42,740 non-redundant sequences from SMRT sequencing showed that the sequences do not show one-to-one correspondence as a single SMRT transcript corresponds to a single or multiple Illumina transcripts. There were 15,596 transcripts present in both Illumina and SMRT sequencing; 27,144 transcripts were only found in SMRT sequencing, and 80,624 transcripts were only found in Illumina sequencing (Figure 1B). The Unigene sequences obtained from Illumina sequencing and the non-redundant sequences obtained from SMRT sequencing were used for data alignment in Clusters of Orthologous Groups (COG), Gene Ontology (GO), and non-redundant (NR) databases. The number of annotated genes obtained through Illumina sequencing was lower than that obtained through SMRT sequencing. There were 32,916 annotated genes that were obtained through Illumina sequencing, which accounted for 31.6% of Unigenes and there were 40,828 annotated genes that were obtained through SMRT sequencing, which accounted for 95.5% of non-redundant sequences. Of the obtained annotated genes, 14,576 genes were common to both Illumina and SMRT sequencing; 18,340 genes were only found in Illumina sequencing, and 26,252, genes were only found in SMRT sequencing (Figure 1C).
We used three methods [coding-non-coding index (CNCI), coding potential calculator (CPC), and Pfamscan (PFAM)] to predict long non-coding RNAs (lncRNAs) longer than 200 bases. We obtained 6,251, 3,322, and 7,610 potential lncRNA sequences, respectively, using these three methods. Among these sequences, 2,171 potential lncRNA sequences simultaneously appeared in all three methods and these sequences were regarded as candidate lncRNAs in the target dataset (Supplementary Figure 1).
Plants employ the phenylalanine metabolic pathway to synthesize flavonoids such as anthocyanins, proanthocyanidins, and flavonols. Here, blueberry transcriptome data from Illumina and SMRT sequencing were used for alignment with COG, GO, and NR databases to obtain genes that regulate flavonoid synthesis.
To study the correlation between expression patterns of flavonoid synthesis genes and accumulated flavonoids in fruits, this study employed HPLC-DAD-ESI-MS to quantitate anthocyanin, flavonol, and proanthocyanidin content in fruits from three developmental stages. Total anthocyanin content showed an increasing trend by development stage, while the total flavonol and proanthocyanidin content decreased (Figure 2A).
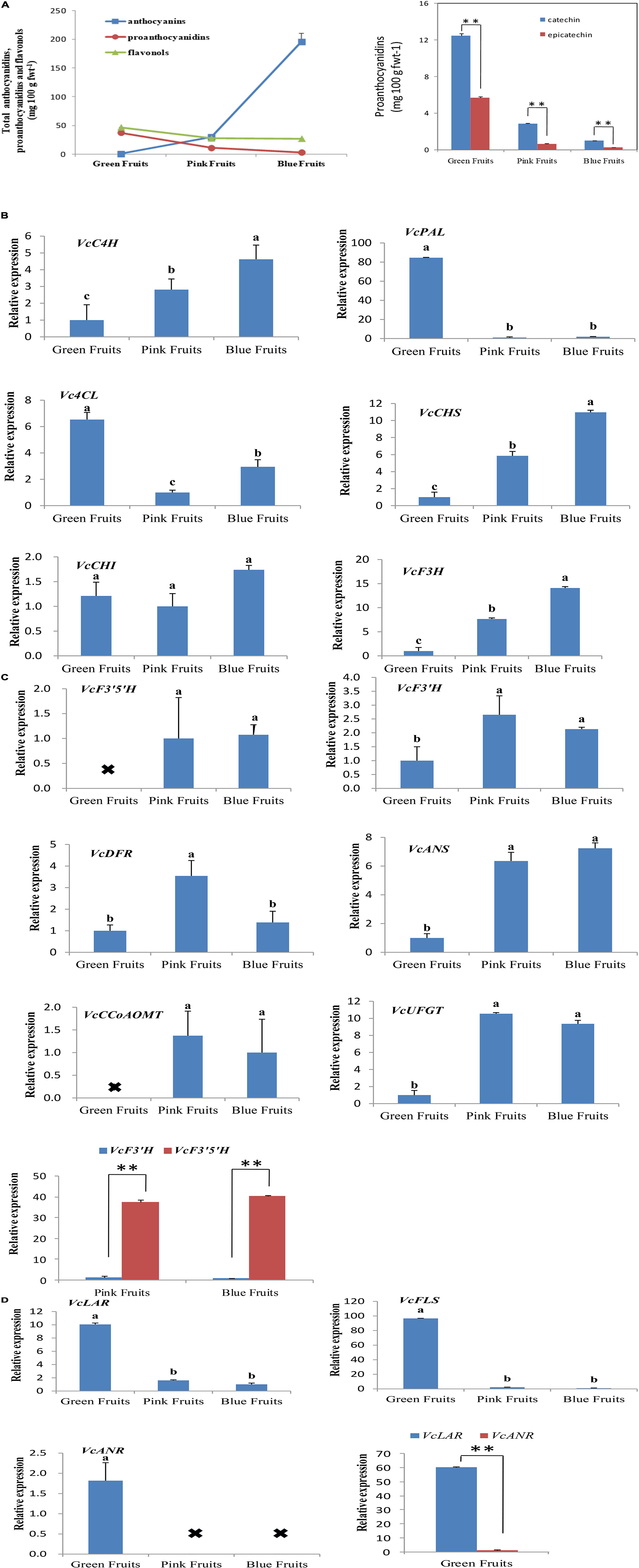
Figure 2. Analysis of the expression of genes associated with flavonoid synthesis and accumulated flavonoids. (A) Liquid chromatography was used for quantitation of flavonoids content. Liquid chromatography-mass spectrometry was used for qualitative testing of flavonoids. Proanthocyanidin content at different developmental stages in blueberries. fwt, fresh weight. Asterisks denote t-test significance: ∗∗P < 0.01. The mean ± SE of biological triplicates were taken for every value. (B–D) All values were calculated based on the geometric mean of the housekeeping genes, VcGADPH and VcSAND, at each stage. Asterisks denote t-test significance: ∗∗P < 0.01. Different English letters represents that the one-way ANOVA difference was significant (P < 0.05). The mean ± SE of biological triplicates were taken for every value.
The anthocyanin synthesis pathway is an important branch of the flavonoid synthesis pathway, which is relatively conserved in higher plants, and its process involves multiple complex enzymatic reactions (Supplementary Figure 2). Most studies on flavonoid synthesis have been in model plants such as apples and peaches, whereas fewer studies have used blueberries (Zhou et al., 2015; An et al., 2017a, An et al., 2020b). To validate the bioinformatics analysis results of annotated genes in the sequencing libraries and study the expression of genes in the phenylalanine metabolic pathways in blueberries, we used homology alignment and phylogenetic analysis to analyze structural genes of 15 main enzymes involved in anthocyanin synthesis pathway: PAL, C4H, 4CL, CHS, CHI (chalcone isomerase), F3H (flavanone 3′-hydroxylase), F3′H (flavonoid 3′ -hydroxylase), F3′5′H (flavonoid 3′, 5′-hydroxylase), DFR, ANS/LDOX (leucoanthocyanidin dioxygenase), UFGT (UDP-glucose: flavonoid 3-O-glucosyltransferase), CCoAOMT (caffeoyl-CoA O-methyltransferase), FLS (flavonol synthase), ANR (anthocyanidin reductase) and LAR (leucoanthocyanidin reductase). These genes have the largest FPKM (Fragments Per Kilobase of transcript per Million mapped reads).
Fluorescence quantitative PCR was used to quantitate the expression status of the structural genes and regulatory genes in flavonoid synthesis during the three developmental stages of blueberries. In this study, we referenced the results from the literature (Zhou et al., 2015) and selected VcGADPH and VcSAND as internal reference genes. The geometric means of the Ct values of these two genes were used as calibration values to analyze the relative expression of other genes. Supplementary Table 2 shows the annotated functions and FPKM (fragments per kb per million) values obtained from transcriptome sequencing of these genes. Supplementary Table 3 shows the primer sequences for qPCR.
Fluorescence qPCR results showed that the genes for flavonoid synthesis exhibited varied trends in expression during the three developmental stages of fruits. We first analyzed the expression of VcC4H, VcPAL, Vc4CL, VcCHS, VcCHI, and VcF3H (Figure 2B). These genes are located upstream of the phenylalanine metabolic pathway and the enzymes encoded by them can catalyze the synthesis of anthocyanins, proanthocyanidins, and flavonols. The results of qPCR showed that with the exception of VcPAL and Vc4CL, the expression of the remaining four genes was highest in the most mature fruits (blue fruits). VcPAL had the highest expression in the least mature (green) fruits and extremely low expression in pink and blue fruits. The expression of Vc4CL was the highest in green fruits, and lowest in pink fruits (Figure 2B).
Next, we analyzed six genes that encode for anthocyanin biosynthesis (VcF3′H, VcF3′5′H, VcDFR, VcANS, VcUFGT, and VcCCoAOMT). In the middle of the anthocyanin synthetic pathway, VcF3′5′H and VcF3′H are branch points as VcF3′5′H can regulate the biosynthesis of delphinidin, petunidin, and malvidin in blueberries whereas VcF3′H can regulate the synthesis of cyanidin and peonidin. In all three fruit developmental stages, VcF3′5′H and VcANS had persistently elevated levels whereas VcF3′H, VcDFR, VcUFGT, and VcCCoAOMT varied; levels were low in immature (green) fruit and high in mature (blue) fruit, then even higher in intermediate (pink) fruits (Figure 2C). Interestingly, in pink and blue fruits, the expression of VcF3′5′H was 31- and 40-fold that of VcF3′H (Figure 2C). This was also verified in quantitation of anthocyanin contents. In blue fruits, the total content of delphinidin, petunidin, and malvidin was 8.1 times that of the total content of cyanidin and peonidin (Table 2). These results suggest that anthocyanin biosynthesis mainly occurs during the fruit maturation phase in “Duke” blueberries, particularly the synthesis of delphinidin, petunidin, and malvidin, and also express the VcF3′5′H gene was upregulated in colored fruits, which changed the ratio of different anthocyanins.
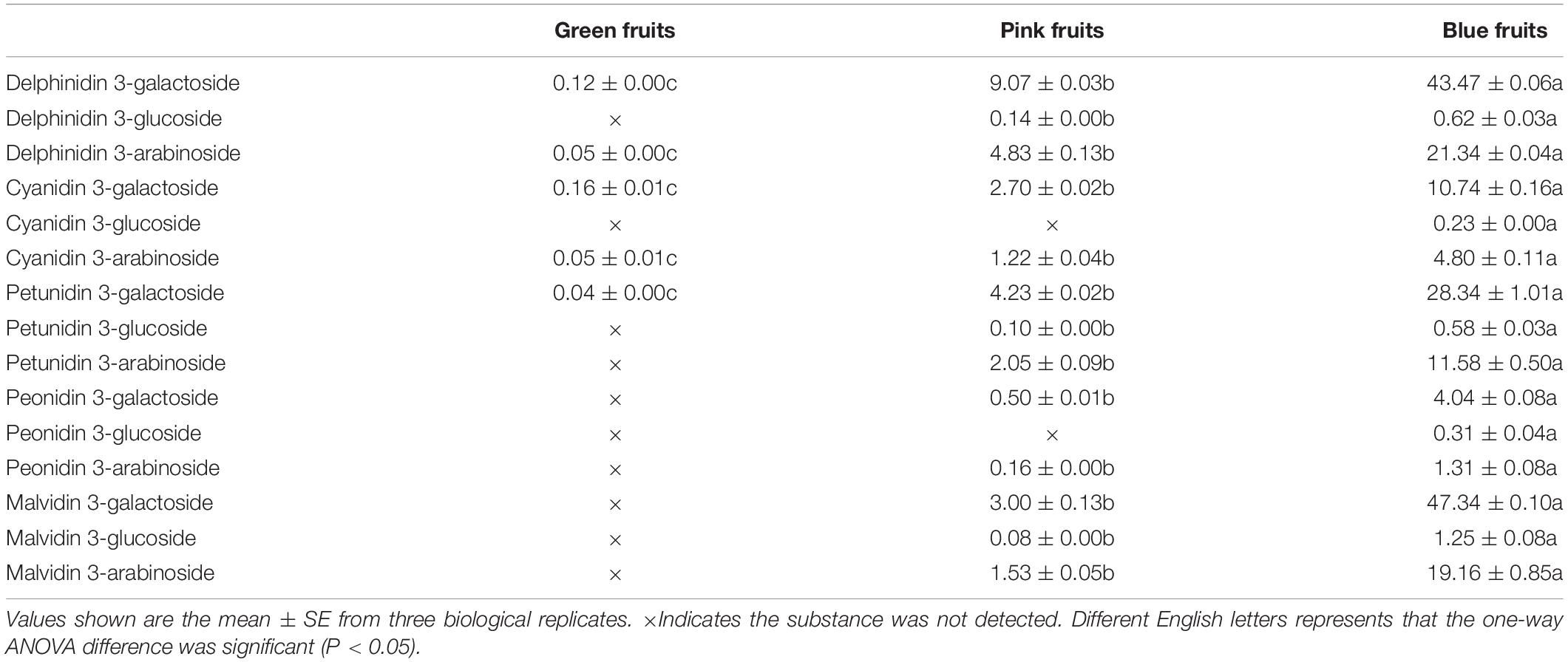
Table 2. Types and contents of anthocyanins in fruits at different developmental stages in blueberries.
Then, we analyzed the VcFLS gene that regulates flavonol synthesis. VcFLS had the highest expression in green fruits and extremely low expression in pink and blue fruits (Figure 2D). This result was consistent with quantitation of flavonol content, which showed that flavonol contents in green fruits were approximately 1.69 times greater than in pink or blue fruits (Figure 2A). Subsequently, we analyzed two genes that regulate proanthocyanidin biosynthesis, VcANR and VcLAR, which were mainly expressed in green fruits and the expression of VcLAR was approximately 60 times that of VcANR (Figure 2D). The quantitation results of proanthocyanidins in fruits showed that catechin content was 2.19 times that of epicatechin content in green fruits (Figure 2A). The results suggest that the differential expression of VcLAR and VcANR may be responsible for differences in catechin and epicatechin contents. These results suggest that the biosynthetic characteristics of flavonols and proanthocyanidins are relatively similar as they are mainly carried out in immature fruits and there is almost little or no biosynthesis in mature fruits. Of these enzymes, VcLAR may play a dominant role in proanthocyanidin synthesis in green fruits compared with VcANR.
In the blueberry transcriptome data, we obtained a total of 45 blueberry MYB gene sequences named VcMYB1-45, respectively. A heat map analysis of these gene sequences revealed that 14 MYB genes were up-regulated in the stages of green fruit to pink fruit or from green fruit to blue fruit, and 31 MYB genes were down-regulated at the above stage (Figure 3A). The amino acid sequences of R2R3 MYB transcription factors in other plants that regulate anthocyanin synthesis, such as grapes (VvMYBA1), apples (MdMYB10), and Arabidopsis (PAP1) were used as probes for alignment and screening of the MYB amino acid sequence of up-regulated gene expression in blueberries. It was found that only VcMYB1 contained the sg6-motif among the genes up-regulated in blueberries (Figure 3B). Studies have shown that the sg6 motif can promote the synthesis of plant anthocyanins (Stracke et al., 2001; Muñoz et al., 2021). For further verification, Phylogenetic analysis of all the excavated VcMYB protein sequences and homologous sequences of other 138 species of plants showed that only the VcMYB1 gene has extremely high homology with the Arabidopsis SG-6 R2R3 MYB protein (Figure 3C).

Figure 3. Sequence alignment and phylogenetic analysis of VcMYB. (A) Heat map depicting the various MYBs, qRT-PCR analysis was also carried out for the genes in red. (B) Protein alignment of Vc MYB and its homologs in Arabidopsis, Vitis vinifera and Malus domestica, The sg6-motif in boxed. (C) Phylogenetic analysis of VcMYB and 138 other plants MYB protein sequences obtained from the NCBI database.VcMYB1 is denoted by the black spot. MYB of Arabidopsis (AtMYB0: At3g27920; AtMYB1: At3g09230; AtMYB10: At3g12820; AtMYB100: At2g25230; AtMYB101: At2g32460; AtMYB102: At4g21440; AtMYB103: At1g63910; AtMYB104: At2g26950; AtMYB105: At1g69560; AtMYB106: At3g01140; AtMYB107: At3g02940; AtMYB108: At3g06490; AtMYB109: At3g55730; AtMYB11: At3g62610;AtMYB110: At3g29020 AtMYB111: At5g49330; AtMYB112: At1g48000; AtMYB113: At1g66370; AtMYB114: At1g66380; AtMYB115: At5g40360; AtMYB116: At1g25340; AtMYB117: At1g26780; AtMYB118: At3g27780; AtMYB119: At5g58850; AtMYB12: At2g47460; AtMYB120: At5g55020; AtMYB121: At3g30210; AtMYB122: At1g74080; AtMYB123: At5g35550; AtMYB124: At1g14350; AtMYB125: At3g60460; AtMYB13: At1g06180; AtMYB14: At2g31180; AtMYB15: At3g23250; AtMYB16: At5g15310; AtMYB17:At3g61250; AtMYB18: At4g25560; AtMYB19: At5g52260; AtMYB2: At2g47190; AtMYB20: At1g66230; AtMYB21: At3g27810; AtMYB22: At5g40430; AtMYB23: At5g40330; AtMYB24: At5g40350; AtMYB25: At2g39880; AtMYB26: At3g13890; AtMYB27: At3g53200; AtMYB28: At5g61420; AtMYB29: At5g07690; AtMYB3: At1g22640; AtMYB30: At3g28910; AtMYB31: At1g74650; AtMYB32: At4g34990; AtMYB33: At5g06100; AtMYB34: At5g60890; AtMYB35: At3g28470; AtMYB36: At5g57620; AtMYB37: At5g23000; AtMYB38: At2g36890; AtMYB39: At4g17780; AtMYB4: At4g38620; AtMYB40: At5g14340; AtMYB41: At4g28110; AtMYB42: At4g12350; AtMYB43: At5g16600; AtMYB44: At5g67300; AtMYB45: At3g48920; AtMYB46: At5g12870; AtMYB47: At1g18710; AtMYB48: At3g46130; AtMYB49: At5g54230; AtMYB5: At3g13540; AtMYB50: At1g57560; AtMYB51: At1g18570; AtMYB52: At1g17950; AtMYB53: At5g65230; AtMYB54: At1g73410; AtMYB55: At4g01680; AtMYB56: At5g17800; AtMYB57: At3g01530; AtMYB58: At1g16490; AtMYB59: At5g59780; AtMYB6: At4g09460; AtMYB60: At1g08810; AtMYB61: At1g09540; AtMYB62: At1g68320; AtMYB63: At1g79180; AtMYB64: At5g11050; AtMYB65: At3g11440; AtMYB66: At5g14750; AtMYB67: At3g12720; AtMYB68: At5g65790; AtMYB69: At4g33450; AtMYB7: At2g16720; AtMYB70: At2g23290; AtMYB71: At3g24310; AtMYB72: At1g56160; AtMYB73: At4g37260; AtMYB74: At4g05100; AtMYB75: At1g56650; AtMYB76: At5g07700; AtMYB77: At3g50060; AtMYB78: At5g49620; AtMYB79: At4g13480; AtMYB8: At1g35515; AtMYB80: At5g56110; AtMYB81: At2g26960; AtMYB82: At5g52600; AtMYB83: At3g08500; AtMYB84: At3g49690; AtMYB85: At4g22680; AtMYB86: At5g26660; AtMYB87: At4g37780; AtMYB88: At2g02820; AtMYB89: At5g39700; AtMYB9: At5g16770; AtMYB90: At1g66390; AtMYB91: At2g37630; AtMYB92: At5g10280; AtMYB93: At1g34670; AtMYB94: At3g47600; AtMYB95: At1g74430;AtMYB96: At5g62470; AtMYB97: At4g26930; AtMYB98: At4g18770; AtMYB99: At5g62320; MdMYB10: M. domestica, ABB84753; LjTT2a: Lotus japonicas, BAG12893; PtMYB134: P. tremuloides, FJ573151; VvMYBPA2: Vitis vinifera, EU919682; BnTT2-1: Brassica napus, DQ778643; MdMYB1: Malus domestica, DQ886414.
To further verify the reliability of the blueberry transcriptome data. Fluorescence qPCR results using 9 blueberry VcMYB transcription factors with larger FPKM showed that the expression trends of VcMYB1 were similar to the expression trends of the six genes that regulate anthocyanin synthesis, such as VcDFR and VcF3′5′ H, which are mainly expressed in colored fruits. VcMYB23, VcMYB26, and VcMYB28 are almost only expressed in green fruits. VcMYB25, VcMYB27, and VcMYB29 exhibited a continuously declining trend. VcMYB24 levels varied; in green fruit VcMYB24 was high; in mature fruits it was lower. VcMYB22 expression varied; it was highest in immature (green) fruits, lower in pink fruits, and higher in the mature (blue) fruits (Supplementary Figure 3). The results suggest that the upregulated expression of VcMYB1 in pink and blue fruits may promote the synthesis of anthocyanins.
Given that the expression patterns of VcMYB1 and anthocyanin-synthesis genes are similar, i.e., relatively high expression in colored fruits and VcMYB1 has relatively high homology and similarity to MYB transcription factors in other plants that regulate anthocyanin synthesis (Figures 3B,C), we hypothesized that VcMYB1 may positively regulate anthocyanin synthesis. To investigate whether VcMYB1 can promote anthocyanin synthesis, we used a heterologous expression system and validated the function of VcMYB1 in Arabidopsis (ecotype Columbia) under the control of CaMV-35S. Results showed that under the same conditions, red pigmentation was present in Arabidopsis seeds (8–10 days after flowering), cotyledons, and hypocotyls overexpressed 35Spro:VcMYB1 whereas red pigmentation was absent in wild-type (WT) Arabidopsis in these three tissues (Figures 4A,C). Anthocyanin contents in transgenic plants and control plants were 311.61 mg/kg fresh weight and 2.82 mg/kg fresh weight, respectively. Thus, anthocyanin contents in plants overexpressing VcMYB1 was approximately 110 times the expression levels in control plants (Figure 4B). To further understand the molecular mechanisms by which VcMYB1 regulates anthocyanin synthesis in Arabidopsis, we extracted RNA from 35Spro:VcMYB1 Arabidopsis and WT Arabidopsis. Fluorescence qPCR was used to measure the expression of structural genes for anthocyanin synthesis in Arabidopsis such as AtPAL, AtCHS and AtDFR, and VcMYB1. Except for At4CL, expression of all other genes in the transgenic plants was significantly higher than in WT plants (Figure 4D). This suggests that VcMYB1 affects anthocyanin synthesis in Arabidopsis by regulating structural genes that are related to anthocyanin synthesis.

Figure 4. Functional analysis of VcMYB1 in a heterologous system. (A) Arabidopsis thaliana (ecotype Columbia) plants overexpressing (i) 35S:VcMYB1 and (ii) wild-type (WT) A. thaliana. (B) Anthocyanin content of A. thaliana plants overexpressing 35S:VcMYB1 and wild-type A. thaliana. Asterisks denote t-test significance: ∗∗P < 0.01. The mean ± SE of biological triplicates were taken for every value. (C) Seeds of A. thaliana plants overexpressing (i) 35S:VcMYB1 and (ii) WT A. thaliana. (D) Relative expression of VcMYB1, AtPAL, AtCHS, At4CL, AtCHI, AtANS, AtF3′H, and AtDFR in A. thaliana plants overexpressing 35S:VcMYB1 and wild-type A. thaliana. All values were calculated based on the housekeeping gene AtUBQ. Asterisks denote t-test significance: ∗∗P < 0.01. n.s means no significant differences. The mean ± SE of biological triplicates were taken for every value. (A,C) Are one of the four T2 transgenic stains and all strains exhibit the phenotype shown by (A,C). (E) Tobacco (Nicotiana benthamiana) leaves on day 7 after injection. (i) 35S:VcMYB1, (ii) Wild-type. (F) Anthocyanin content in tobacco leaves on Day 7 after injection. The mean ± SE of biological triplicates were taken for every value. Asterisks denote t-test significance: ∗∗P < 0.01.
To confirm the function of VcMYB1, we conducted transient expression experiments and validated the function of VcMYB1 in tobacco (Nicotiana benthamiana). 35Spro:VcMYB1 was transformed into Agrobacterium before injecting into tobacco leaves. Agrobacterium that were transformed with empty vectors were used as controls. Results showed that 4 days after injection, tobacco leaves overexpressing VcMYB1 were colored whereas no color changes were observed in the control (Figure 4E). Anthocyanin content measurement results showed that the total anthocyanin content in tobacco leaves overexpressing VcMYB1 was 6.8-fold higher than in the control leaves (Figure 4F). The above data shows that VcMYB1 can positively regulate anthocyanin synthesis.
Moreover, vector-mediated overexpression was conducted using 45 days blueberries. VcMYB1-pGreenII62-SK vectors (VcMYB1) were generated, and pGreenII62-SK were used as a control. Compared with the empty vector control (pGreenII62-SK), overexpression of VcMYB1 promoted anthocyanin accumulation in the blueberry shin around the injection sites (Figures 5A,B), The total content of delphinidin, petunidin, and malvidin was higher than the total content of cyanidin and peonidin, consistent with Supplementary Table 2. The expression levels of VcMYB1, VcDFR3, VcANS1 and VcUFGT1 were elevated in the VcMYB1-pGreenII62-SK vectors injection areas compared with the controls (Figure 5C). Therefore, these results demonstrated that VcMYB1 played a positive role in anthocyanin synthesis and fruit coloration.
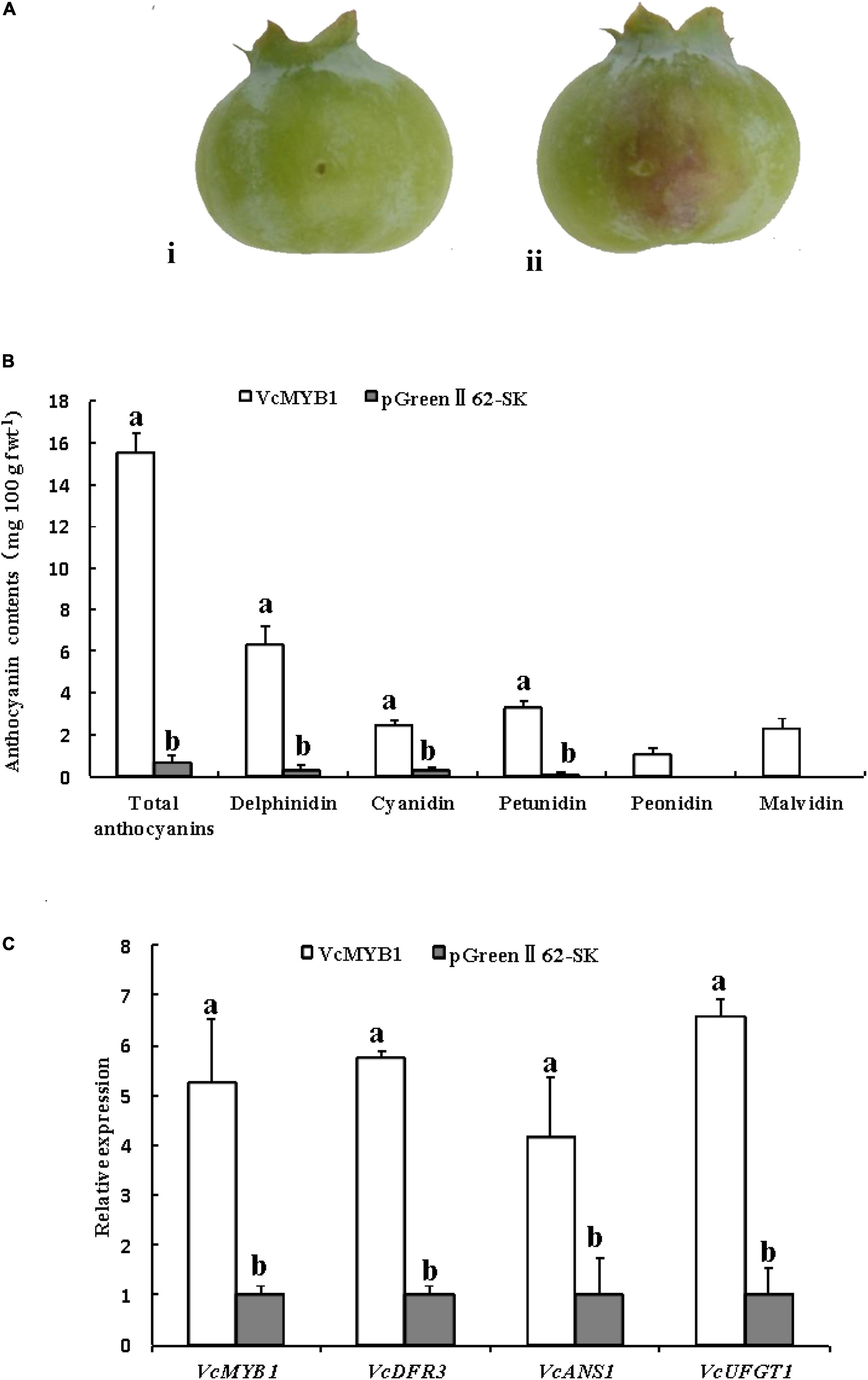
Figure 5. Functional analysis of VcMYB1 in blueberry. (A) blueberry overexpressing (i) pGreenII62-SK fruit and (ii) VcMYB1-pGreenII62-SK (VcMYB1) fruit. (B) Anthocyanin content of fruits overexpressing VcMYB1-pGreenII62-SK (VcMYB1) and pGreenII62-SK fruit. The mean ± SE of biological triplicates were taken for every value. Different English letters represents that the t-test difference was significant. P < 0.05. (C) Relative expression of VcMYB1, VcDFR3, VcANS1, and VcUFGT1 in fruits overexpressing VcMYB1-pGreenII62-SK (VcMYB1) and pGreenII62-SK fruit. The mean ± SE of biological triplicates were taken for every value. Different English letters represents that the t-test difference was significant P < 0.05.
Because VcMYB1 can promote anthocyanin synthesis, we predicted that VcMYB1 may directly regulate the expression of structural genes in anthocyanin synthesis to regulate anthocyanin synthesis. Previous studies have reported that MYB function by binding to the promoters of their target genes to modulate their expressions (Takos et al., 2006; Ban et al., 2007; Espley et al., 2007; An et al., 2020a). We employed chromosome walking techniques to clone the promoter sequence of VcDFR, VcF3′5′ H and VcUFGT, which was found to have a length of 834, 1,828, and 2,362 bp respectively. Unfortunately, the promoter sequence of VcANS is not available. This promoter was inserted into a pAbAi vector before transformation into a Y1HGoldstrain. The open reading frame of VcMYB1 was inserted into a pGADT7 vector. The Y1H assay was used to analyze whether VcMYB1 can interact with these promoters. Results showed that VcMYB1 can bind the MYB binding site (MBS) motifs in the VcDFR promoter (Figure 6A). For further research, we used the promoter prediction website10 to predict and analyze the VcDFR, VcF3′5′H and VcUFGT promoter sequences. It was found that only the VcDFR promoter sequence contained the MBS motif. In the anthocyanin synthesis pathway, the action site of VcDRF is adjacent to the action sites of the other three structural genes. Therefore, it is speculated that the expression of VcDRF will promote their expression. We also used a prokaryotic expression system to induce and purify the VcMYB1 protein and used electrophoretic mobility shift assay (EMSA) to validate the results of the Y1H assay. Results showed that VcMYB1 can bind to the biotinylated MBS motif on the VcDFR promoter (Figure 6B).
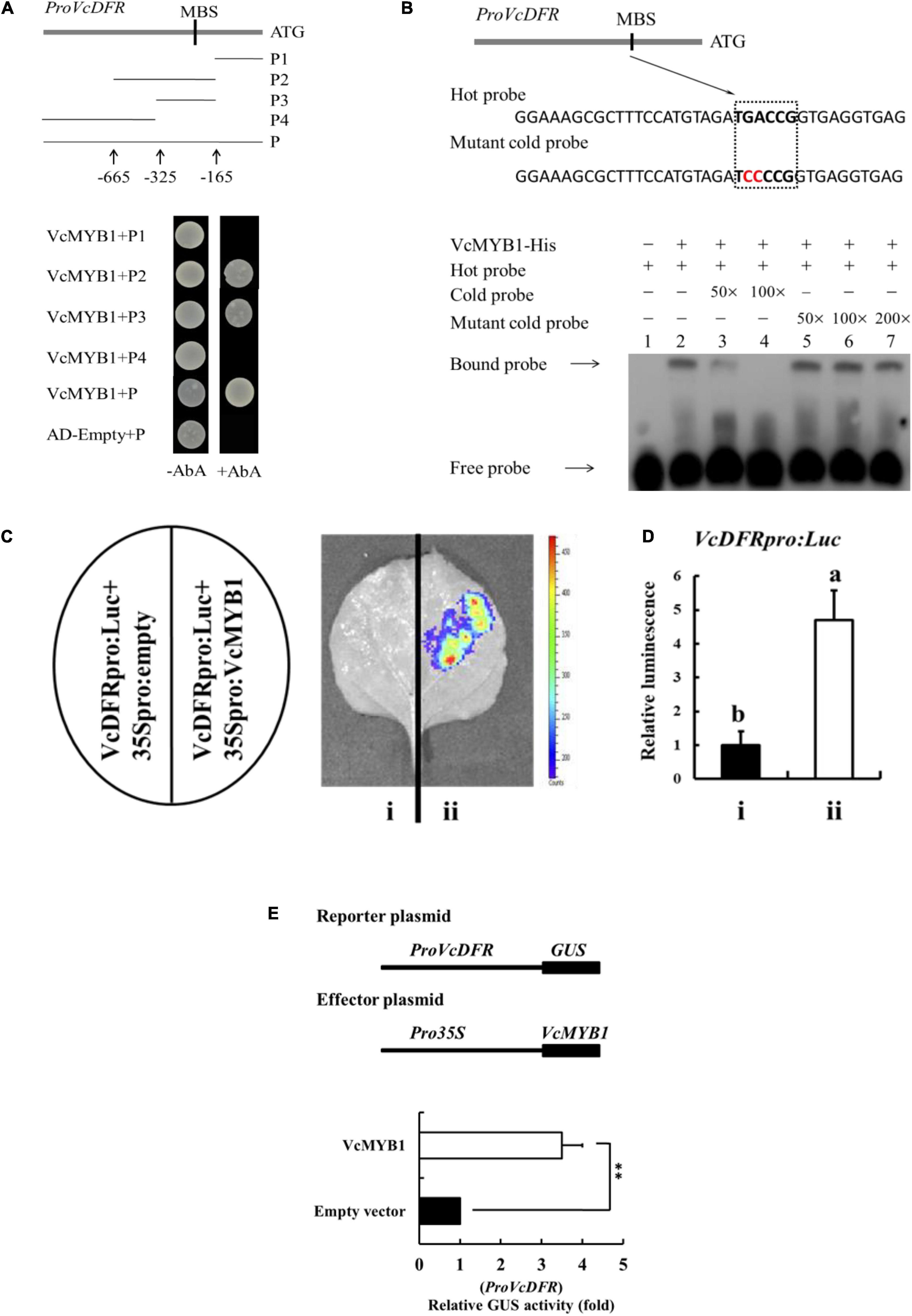
Figure 6. VcMYB1 can directly bind to and activate the VcDFR promoter. (A) Y1H analysis showed that VcMYB1 can bind to VcDFR promoter fragments that contain the MBS motif. The VcDFR promoter was divided into four segments (P1–P4). AbA (Aureobasidin A) is a growth inhibitor of yeast cells and was used as a screening marker. The screening concentration for AbA was 100 ng/mL. The empty vector and the full-length VcDFR promoter were used as a negative control. (B) EMSA analysis showed that VcMYB1 can bind to the MBS motif on the VcDFR promoter. The hot probe contains a VcDFR promoter with a biotinylated MBS motif and the unbiotinylated cold probe was used as a competitive probe (concentration of 50- and 100-fold to the hot probe). The mutant cold probe is an unbiotinylated hot probe in which three nucleotides were mutated. (C) Firefly luciferase complementation assay analysis showed that VcMYB1 can activate VcDFR promoter activity in tobacco (N. benthamiana) leaves. (i) VcDFRpro:Luc + 35Spro:empty, (ii) VcDFRpro:Luc + 35Spro:VcMYB1. (D) Quantitative analysis of luminescence intensity in C. The value for a VcDFRpro:Luc was set to 1. The mean ± SE of biological triplicates were taken for every value. Different English letters represents that the t-test difference was significant. P < 0.01 (E) GUS activity analysis showed that VcMYB1 can activate VcDFR promoter activity in tobacco (N. benthamiana) leaves. VcMYB1 effector vector and a reporter vector containing the VcDFR promoter were co-injected into wild-type tobacco leaves to analysis their effects on GUS activity. Asterisks denote t-test significance: ∗∗P < 0.01. The mean ± SE of biological triplicates were taken for every value.
To investigate whether VcMYB1 can activate the VcDFR promoter, we carried out firefly luciferase (Luc) complementation experiments for validation. We inserted the VcDFR promoter upstream of the Luc gene to generate VcDFRpro:Luc. VcMYB1 was inserted downstream of the 35S promoter to generate 35Spro:VcMYB1. Both vectors were transformed into Agrobacterium before co-injection into tobacco leaves. Results showed that luminescence signals could be detected in tobacco plants that were co-injected with VcDFRpro:Luc and 35Spro:VcMYB1 whereas no luminescence signals were detected in the control (VcDFRpro:Luc + 35Spro:empty) (Figures 6C,D). Following that, we validated the regulation of the VcDFR promoter by VcMYB1 in tobacco leaves by analyzing the activity of the β-glucuronidase (GUS) reporter gene. The results showed that co-transforming VcDFRpro:Luc and 35Spro:VcMYB1 into tobacco leaves elevated VcDFR promoter activity (Figure 6E). Therefore, we believe that VcMYB1 directly binds to the promoter of VcDFR to activate the expression of this gene, thereby promoting anthocyanin synthesis.
Blueberries contain large amounts of diverse flavonoids, which reduce reactive oxygen species generation and they enhance the ability of plants to resist biological and non-biological stressors (Cascaes Márcia et al., 2021; Mu et al., 2021). But there were very few studies on the molecular mechanisms of their synthesis or their metabolism. This study is the first to employ SMRT combined with Illumina sequencing technology to carry out transcriptome sequencing of three fruit developmental stages in blueberries to improve the abundance of transcripts as much as possible in order to understand the mechanisms for flavonoid synthesis in fruits.
This study is the first to employ SMRT combined with Illumina sequencing technology to carry out transcriptome sequencing of three fruit developmental stages in blueberries. Results showed that compared with the assembled data obtained from Illumina sequencing, better data quality and longer transcripts were obtained from SMRT sequencing. This was also found to be the case in studies by Xu et al. (2015) and Li et al. (2017), i.e., The amount of data acquired by Illumina sequencing was approximately 2.4 times that acquired by SMRT sequencing. This was not consistent with the results by Li et al. (2017) who found a greater number of annotated genes by Illumina sequencing compared with SMRT sequencing. This inconsistency may be due to species specificity. Therefore, combining these two types of sequencing techniques can be used to obtain abundant and more intact transcriptome information of blueberries during fruit development. Our study obtained a large number of structural genes that participate in flavonoid synthesis through combination of Illumina and SMRT sequencing methods. The number of genes involved in flavonoid synthesis that were obtained by Sun et al. (2015); Li Y. Y. et al. (2012), and Li et al. (2016) using Illumina sequencing technologies were lower than that obtained in this study (Supplementary Table 4). The results showed that compared with single assembly using the established Illumina sequencing method, SMRT sequencing greatly improved the assembly results of Illumina fragmentation. Compared with using only one sequencing technology (Illumina or SMRT), the integrated use of these two sequencing technologies can significantly enrich transcriptome information and increase the integrity of transcriptome data. The combination of these two sequencing technologies can thus enable more effective and greater acquisition of target transcriptome information. Specifically, we obtained full-length sequences of genes that are involved in flavonoid synthesis, showing this technique can aid the discovery and analysis of functional genes.
The results of the study showed that anthocyanin contents continuously increased, across the three fruit development stages (Figure 2A). In addition, this study also screened out a MYB transcription factor that regulates anthocyanin synthesis, VcMYB1. From this, we can see that “Duke” blueberries mainly synthesize anthocyanins in colored fruits (pink and blue fruits). The anthocyanin synthesis genes (VcF3′H, VcF3′5′H, VcDFR, VcANS, VcUFGT and VcCCoAOMT) were all mainly expressed in colored fruits (Figure 2C). This result was consistent with the results of other studies, i.e., the upregulated expression of these six genes can promote plant coloring and increase anthocyanin content (Bogs et al., 2006; Yao et al., 2017; Ma et al., 2021). The differential expression of F3′5′H and F3′H genes can affect the type of anthocyanins synthesized in plants and different colors are presented (Holton et al., 1993a; Han et al., 2010). Our study found that VcF3′5′H and VcF3′H are mainly expressed in colored fruits in “Duke” blueberries, but expression is vastly different (Figure 2C). This explains why the total content of delphinidin, petunidin, and malvidin in pink and blue fruits was significantly higher than the total content of cyanidin and peonidin (Table 2). This result is supported by previous studies of flavonoid content in blueberries (Lätti et al., 2010; Zifkin et al., 2012).
The WBM (WD40, BHLH, and MYB) complex regulates flavonoid synthesis through interactions between its components as well as binding to the promoters of structural genes for anthocyanin, flavonol, and proanthocyanidin synthesis, such as DFR and UFGT (Grotewold et al., 2000; Mehrtens et al., 2005; Stracke et al., 2007; Xie et al., 2012; Zifkin et al., 2012). Among the R2R3 MYB transcription factors, AtMYBs (AtMYB75, AtMYB90, AtMYB113, and AtMYB144), MdMYB1 and MdMYB10 can promote the expression of genes encoding key anthocyanin biosynthetic enzymes, such as DFR and ANS (Dooner et al., 1991; Takos et al., 2006; Ban et al., 2007; Espley et al., 2007; Antonio et al., 2008; Li et al., 2014; An et al., 2018; An et al., 2020a). Our study screened out one R2R3 MYB transcription factor, VcMYB1 from the transcriptome library. VcMYB1 and these MYB transcription factors have a high degree of homology (Figure 3A). Fluorescence quantitative PCR analysis results showed that the expression pattern of VcMYB1 is similar to the expression patterns of multiple genes that regulate anthocyanin synthesis, which are mainly highly expressed in colored fruits. Functional validation experiments also proved that VcMYB1 can promote anthocyanin synthesis in Arabidopsis, tobacco plants and green blueberry fruits (Figures 4, 5). In addition, Y1H assay, EMSA, luciferase complementation, and GUS activity experiments showed that VcMYB1 can bind to the promoter of VcDFR, which activates the promoter. This is similar to the results from many studies (Ban et al., 2007; James et al., 2017). VcMYB1 can promote the synthesis of anthocyanins by binding to the 5′-TGACCG-3′ sequence of the MBS motif of the DFR promoter. In Arabidopsis, the general MYB/bHLH protein complex binds to the DFR promoter sequence to promote anthocyanin synthesis (Ilona et al., 2004). In rice, MYB can bind to the promoter sequence of DFR, the sequences are CC(T/A)ACC (CCAACC) or AC(C/A)C(T/A)A(C/A)C (ACCTACC) (Kim et al., 2017). In apples, MdMYB1 can also bind to the MBS motif of the DFR promoter, and its binding sequence is 5′-GCCAGG-3′ which is different from VcMYB1 (An et al., 2018, 2020a). We speculate that this is related to the specificity of the species. Blueberries are non-climacteric plants, apples are climacteric plants, and rice and Arabidopsis do not have high-quality fleshy fruits. Therefore, we believe that in “Duke” blueberries, VcMYB1 can bind and activate the promoters of structural genes involved in anthocyanin synthesis, thereby positively regulating the biosynthesis of anthocyanins.
This study also carried out studies on genes involved in flavonol biosynthesis in blueberries. We found that VcFLS can regulate the biosynthesis of quercetin, myricetin and other flavonols. In this study, VcFLS is mainly expressed in green fruits and the expression of VcFLS in colored fruits is extremely low (Figure 2D). This causes flavonols to be mainly synthesized in green fruits (Figure 2A). This result was similar to the results by Holton et al. (1993b); Stracke et al. (2009), and Zifkin et al. (2012). VcLAR and VcANR can regulate proanthocyanidin synthesis (Tanner et al., 2003; Pang et al., 2013). In this study, the VcLAR and VcANR genes that regulate proanthocyanidin synthesis were only expressed in green fruits, with the expression of VcLAR being 60 times that of VcANR (Figure 2D). In addition, the contents of catechin were significantly higher than epicatechin. This shows that the high expression of VcLAR in green fruits resulted in accumulated of higher contents of proanthocyanidins. This result was similar to the results by Wang et al. (2017).
In summary, our study shows that flavonoid biosynthetic pathways are regulated in developing blueberry fruits, and that this regulation occurs at the level of gene expression. Our combinatorial approach to transcriptome analysis enabled us to generate full-length transcripts as well as providing evidence for flavonoid biosynthesis. Our work provides a new method for investigating the full-length transcriptome and secondary metabolism in other plants.
The original contributions presented in the study are publicly available. These data can be found here: National Center for Biotechnology Information (NCBI) BioProject database under BioProject ID: PRJNA757138.
YS conceived and designed the experiments. QT and YS performed the research and wrote the manuscript. QT, YS, F-MC, H-DL, and H-JZ analyzed the data. All authors contributed to the article and approved the submitted version.
This work was supported by the Natural Science Foundation of Liaoning Province (2019-MS-348) and the Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2016-RIP).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Xiu-hong An (Hebei Agricultural University) and Jian-ping AN (Shandong Agricultural University) for experimental helping.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.754325/full#supplementary-material
Supplementary Figure 1 | Venn diagram of long non-coding RNA (lncRNA) sequences predicted by three different methods.
Supplementary Figure 2 | Schematic diagram of flavonoid synthesis in blueberries, including anthocyanin, proanthocyanidin, and flavonol synthesis.
Supplementary Figure 3 | Transcript abundance of MYB genes at different developmental stages in blueberries.
Supplementary Table 1 | Statistics of sequences length from Illumina and SMRT sequencing data.
Supplementary Table 2 | FPKM values and annotation of flavonoid-synthesis genes in blueberries.
Supplementary Table 3 | List of primers used in this study.
Supplementary Table 4 | Comparison of flavonoid synthesis-related genes in blueberries.
Adrian, R. W., Nancy, C., Laurie, T. B., Daniel, J. L., and Claire, M. W. (2019). Flavonoid-Rich Mixed Berries Maintain and Improve Cognitive Function Over a 6h Period in Young Healthy Adults. Nutrients 11, 2685–2698. doi: 10.3390/nu11112685
An, J. P., Qu, F. J., Yao, J. F., Wang, X. N., You, C. X., Wang, X. F., et al. (2017a). The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Horticul. Ress. 4, 17023–17031. doi: 10.1038/hortres.2017.23
An, J. P., Yao, J. F., Wang, X. N., You, C. X., Wang, X. F., and Hao, Y. J. (2017b). MdHY5 positively regulates cold tolerance via CBF-dependent and CBF-independent pathways in apple. J. Plant Physiol. 218, 275–281. doi: 10.1016/j.jplph.2017.09.001
An, J. P., Wang, X. F., Espley, R. V., Lin, W. K., Bi, S. Q., and You, C. X. (2020a). An Apple B-Box Protein MdBBX37 Modulates Anthocyanin Biosynthesis and Hypocotyl Elongation Synergistically with MdMYBs and MdHY5. Plant Cell Physiol. 61, 130–143. doi: 10.1093/pcp/pcz185
An, J. P., Yao, J. F., Xu, R. R., You, C. X., Wang, X. F., and Hao, Y. J. (2018). Apple bZIP transcription factor MdbZIP44 regulates abscisic acid-promoted anthocyanin accumulation. Plant Cell Env. 41, 2678–2692. doi: 10.1111/pce.13393
An, J. P., Zhang, X. W., Bi, S. Q., You, C. X., Wang, X. F., and Hao, Y. J. (2020b). The ERF transcription factor MdERF38 promotes drought stress-induced anthocyanin biosynthesis in apple. Plant J. 101, 573–589. doi: 10.1111/tpj.14555
Antonio, G., Zhao, M. Z., John, M. L., and Alan, M. L. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827. doi: 10.1111/j.1365-313X.2007.03373.x
Bakade, R., Ingole, K. D., Deshpande, S., Pal, G., Patil, S. S., Bhattacharjee, S., et al. (2021). Comparative Transcriptome Analysis of Rice Resistant and Susceptible Genotypes to Xanthomonas oryzae pv. oryzae Identifies Novel Genes to Control Bacterial Leaf Blight. Mole. Biotechnol. 63, 719–731. doi: 10.1007/s12033-021-00338-3
Ban, Y., Honda, C., Hatsuyama, Y., Igarashi, M., Bessho, H., and Moriguchi, T. (2007). Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol. 48, 958–970. doi: 10.1093/pcp/pcm066
Bogs, J., Ebadi, A., McDavid, D., and Robinson, S. P. (2006). Identification of the flavonoid hydroxylases from grapevine and their regulation during fruit development. Plant Physiol. 140, 279–291. doi: 10.1104/pp.105.073262
Cascaes Márcia, M., Guilhon Giselle, M. S. P., Zoghbi Maria das Graças, Andrade Eloisa, Helena, A., Santos Lourivaldo, S., et al. (2021). Flavonoids, antioxidant potential and antimicrobial activity of Myrcia rufipila mcvaugh leaves (myrtaceae). Nat. Prod. Res. 35, 1717–1721. doi: 10.1080/14786419.2019.1629912
Choi, H. R., Jeong, M. J., Baek, M. W., Choi, J. H., Lee, H. C., Jeong, C. S., et al. (2021). Transcriptome Analysis of Pre-Storage 1-MCP and High CO2-Treated ‘Madoka’ Peach Fruit Explains the Reduction in Chilling Injury and Improvement of Storage Period by Delaying Ripening. Internat. J. Mole. Sci. 22, 4437–4453. doi: 10.3390/IJMS22094437
Costich, D. E., Ortiz, R., Meagher, T. R., Bruederle, L. P., and Vorsa, N. (1993). Determination of ploidy level and nuclear DNA content in blueberry by flow cytometry. Theor. Apple Genet. 86, 1001–1006. doi: 10.1007/BF00211053
Diaz, G. L., Garcia, O. L. F., González, R. M., Delaye, L., Iorizzo, M., and Zalapa, J. (2021). Chromosome-Level Genome Assembly of the American Cranberry (Vaccinium macrocarpon Ait.) and Its Wild Relative Vaccinium microcarpum. Front. Plant Sci. 12:633310. doi: 10.3389/FPLS.2021.633310
Dooner, H. K., Robbins, T. P., and Jorgensen, R. A. (1991). Genetic and Developmental Control of Anthocyanin Biosynthesis. Ann. Rev. Genet. 25, 173–199. doi: 10.1146/ANNUREV.GE.25.120191.001133
Du, Z. M., Sun, L., Lin, Y. L., Yang, F. Y., and Cai, Y. M. (2021). The use of PacBio SMRT technology to explore the microbial network and fermentation characteristics of woody silage prepared with exogenous carbohydrate additives. J. Appl. Microb. 2021:15124. doi: 10.1111/jam.15124
Espley, R. V., Hellens, R. P., Putterill, J., Stevenson, D. E., Kutty-Amma, S., and Allan, A. C. (2007). Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 49, 414–427. doi: 10.1111/j.1365-313X.2006.02964.x
Grotewold, E., Saine, M. B., Tagliani, L., Hernandez, J. M., Bowen, B., and Chandler, V. L. (2000). Identification of the residues in the Myb domain of maize C1 that specify the interaction with the bHLH cofactor R. Proc. Natl. Acad. Sci. U S A. 97, 13579–13584. doi: 10.1073/pnas.250379897
Han, Y. P., Vimolmangkang, S., Soria-Guerra, R. E., Rosales-Mendoza, S., Zheng, D. M., Lygin, A. V., et al. (2010). Ectopic expression of apple F3′H genes contributes to anthocyanin accumulation in the Arabidopsistt7 mutant grown under nitrogen stress. Plant Physiol. 153, 806–820. doi: 10.1104/PP.109.152801
Holton, T. A., Brugliera, F., Lester, D. R., Tanaka, Y., Hyland, C. D., Mentlng, J. G. T., et al. (1993a). Cloning and expression of cytochrome P450 genes controlling flower colour. Nature 366, 276–279. doi: 10.1038/366276a0
Holton, T. A., Brugliera, F., and Yanaka, Y. (1993b). Cloning and expression of flavonol synthase from Petunia hybrida. Plant J. 4, 1003–1010. doi: 10.1046/j.1365-313X.1993.04061003
Ilona, M. Z., Marc, A. H., Bernd, W., and Joachim, F. U. (2004). Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 40, 22–34. doi: 10.1111/j.1365-313X.2004.02183.x
Jaakola, L., Määttä, K., Pirttilä, A. M., Törrönen, R., Kärenlampi, S., and Hohtola, A. (2002). Expression of genes involved in anthocyanin biosynthesis in relation to anthocyanin, proanthocyanidin, and flavonol levels during bilberry fruit development. Plant Physiol. 130, 729–739. doi: 10.1104/PP.006957
James, A. M., Ma, D. W., Mellway, R., Gesell, A., Yoshida, K., Walker, V., et al. (2017). Poplar MYB115 and MYB134 transcription factors regulate proanthocyanidin synthesis and structure. Plant Physiol. 174, 154–171. doi: 10.1104/pp.16.01962
Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987) GUS fusions: b-glucuronidase as a sensitive and versatile gene fusion marker in higher palnts. The EMBO journal. 6, 3901–3907.
Kaushal, M., Mahuku, G., and Swennen, R. (2021). Comparative Transcriptome and Expression Profiling of Resistant and Susceptible Banana Cultivars during Infection by Fusarium oxysporum. Internat. J. Mole. Sci. 22, 3002–3030. doi: 10.3390/IJMS22063002
Kim, J., Lee, H. J., Jung, Y. J., Kang, K. K., Tyagi, W., Kovach, M., et al. (2017). Functional properties of an alternative, tissue-specific promoter for rice NADPH-dependent dihydroflavonol reductase. PLoS One 12:e0183722. doi: 10.1371/journal.pone.0183722
Lätti, A. K., Jaakola, L., Riihinen, K. R., and Kainulainen, K. (2010). Anthocyanin and flavonol variation in Bog Bilberries (Vaccinium uliginosum L.) in Finland. J. Agric. Food Chem. 58, 427–433. doi: 10.1021/jf903033m
Li, L. L., Zhang, H. H., Liu, Z. S., Cui, X. Y., Zhang, T., Li, Y. F., et al. (2016). Comparative transcriptome sequencing and de novo analysis of Vaccinium corymbosum during fruit and color development. BMC Plant Biol. 16:223. doi: 10.1186/s12870-016-0866-5
Li, T., Jia, K. P., Lian, H. L., Yang, X., Li, L., and Yang, H. Q. (2014). Jasmonic acid enhancement of anthocyanin accumulation is dependent on phytochrome A signaling pathway under far-red light in Arabidopsis. Biochem. Biophys. Res. Comm. 454, 78–83. doi: 10.1016/j.bbrc.2014.10.059
Li, X. Y., Sun, H. Y., Pei, J. B., Dong, Y. Y., Wang, F. W., Chen, H., et al. (2012). De novo sequencing and comparative analysis of the blueberry transcriptome to discover putative genes related to antioxidants. Gene. 511, 54–61. doi: 10.1016/j.gene.2012.09.021
Li, Y. D., Pei, J. B., Chen, L., and Sun, H. Y. (2021). China Blueberry Industry Report 2020. J. Jilin Agricult. Univ. 43, 1–8. doi: 10.13327/j.jjlau.2021.1071
Li, Y. P., Dai, C., Hu, C. G., Liu, Z. C., and Kang, C. Y. (2017). Global identification of alternative splicing via comparative analysis of SMRT- and Illumina-based RNA-seq in strawberry. Plant J. 90, 164–176. doi: 10.1111/tpj.13462
Li, Y. Y., Mao, K., Zhao, C., Zhao, X. Y., Zhang, H. L., Shu, H. R., et al. (2012). MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol. 160, 1011–1022. doi: 10.1104/pp.112.199703
Liang, J. P., Li, W. Q., Jia, X. Y., Zhang, Y., and Zhao, J. P. (2020). Transcriptome sequencing and characterization of Astragalus membranaceus var. mongholicus root reveals key genes involved in flavonoids biosynthesis. Genes Genom. 42, 901–914. doi: 10.1007/s13258-020-00953-5
Ma, D. W., Tang, H., Reichelt, M., Piirtola, E. M., Salminen, J. P., Gershenzon, J., et al. (2021). Poplar MYB117 promotes anthocyanin synthesis and enhances flavonoid B-ring hydroxylation by upregulating the flavonoid 3’,5’-hydroxylase gene. J. Exp. Bot. 72, 3864–3880. doi: 10.1093/JXB/ERAB116
Mehrtens, F., Kranz, H., Bednarek, P., and Weisshaar, B. (2005). The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 138, 1083–1096. doi: 10.1104/pp.104.058032
Minoche, A. E., Dohm, J. C., Schneider, J., Holtgräwe, D., Viehöver, P., Montfort, M., et al. (2015). Exploiting single-molecule transcript sequencing for eukaryotic gene prediction. Genome Biol. 16:184. doi: 10.1186/s13059-015-0729-7
Mu, P. Y., Hu, Y. M., Ma, X., Shi, J. R., Zhong, Z. D., and Huang, L. Y. (2021). Total flavonoids of Rhizoma Drynariae combined with calcium attenuate osteoporosis by reducing reactive oxygen species generation. Exp. Ther. Med. 21, 618–627. doi: 10.3892/ETM.2021.10050
Muñoz, G. S., Suárez, B. H., Alzate, J. F., González, F., and Pabón, M. N. (2021). Evolution of the Subgroup 6 R2R3-MYB Genes and Their Contribution to Floral Color in the Perianth-Bearing Piperales. Front. Plant Sci. 12:633227. doi: 10.3389/FPLS.2021.633227
Pang, Y. Z., Abeysinghe, I. S. B., He, J., He, X. Z., Huhman, D., Mewan, K. M., et al. (2013). Functional characterization of proanthocyanidin pathway enzymes from tea and their application for metabolic engineering. Plant Physiol. 161, 1103–1116. doi: 10.1104/pp.112.212050
Pertuzatti, P. B., Barcia, M. T., Rebello, L. P. G., Gómez-Alonso, S., Duarte, R. M. T., Duarte, M. C. T., et al. (2016). Antimicrobial activity and differentiation of anthocyanin profiles of rabbiteye and highbush blueberry using HPLC-DAD-ESI-MS and multivariate analysis. J. Funct. Foods 26, 506–516. doi: 10.1016/j.jff.2016.07.026
Prior, R. L., Cao, G. H., Martin, A., Sofic, E., McEwen, J., O’Brien, C., et al. (1998). Antioxidant capacity as influenced by total phenolic and anthocyanin content, maturity, and variety of Vaccinium species. J. Agric. Food Chem. 43, 2686–2693. doi: 10.1021/jf980145d
Prior, R. L., Lazarus, S. A., Cao, G. H., Muccitelli, H., and Hammerstone, J. F. (2001). Identification of procyanidins and anthocyanins in blueberries and cranberries (Vaccinium Spp.) using high-performance liquid chromatography/mass spectrometry. J. Agricult. Food Chem. 49, 1270–1276. doi: 10.1021/jf001211q
Qian, Y. G., Cao, L. W., Zhang, Q., Maurice, A., Chen, K., and Chen, L. (2020). SMRT and Illumina RNA sequencing reveal novel insights into the heat stress response and crosstalk with leaf senescence in tall fescue. BMC Plant Biolog. 20:366–384. doi: 10.1186/s12870-020-02572-4
Rowland, L. J., Alkharouf, N., Darwish, O., Ogden, E. L., Polashock, J. J., Bassio, N. V., et al. (2012). Generation and analysis of blueberry transcriptome sequceces from leaves, developing fruit, and flower buds from cold acclimation through deacclimation. BMC Plant Biol. 12:46. doi: 10.1186/1471-2229-12-46
Salmela, L., and Rivals, E. (2014). LoRDEC: accurate and efficient long read error correction. Bioinformatics 30, 3506–3514. doi: 10.1093/bioinformatics/btu538
Shang, Y., Yan, L., Liu, Z. Q., Cao, Z., Mei, C., Xin, Q., et al. (2010). The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22, 1909–1935. doi: 10.1105/tpc.110.073874
Stracke, R., De Vos, R. C. H., Bartelniewoehner, L., Ishihara, H., Sagasser, M., Martens, S., et al. (2009). Metabolomic and genetic analyses of flavonols synthesis in Arabidopsis thaliana support the in vivo involvement of leucoanthocyanidin dioxygenase. Planta 229, 427–445. doi: 10.1007/s00425-008-0841-y
Stracke, R., Ishihara, H., Huep, G., Barsch, A., Mehrtens, F., Niehaus, K., et al. (2007). Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50, 660–677. doi: 10.1111/j.1365-313X.2007.03078.x
Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. doi: 10.1016/S1369-5266(00)00199-0
Sun, H. Y., Liu, Y. S., Gai, Y. Z., Geng, J. M., Chen, L., Liu, H. D., et al. (2015). De novo sequencing and analysis of the cranberry fruit transcriptome to identify putative genes involved in falconoid biosynthesis, transport and regulation. BMC Genomics 16:652. doi: 10.1186/s12864-015-1842-4
Takos, A. M., Jaffeì, F. W., Jacob, S. R., Bogs, J., Robinson, S. P., and Walker, A. R. (2006). Light-Induced Expression of a MYB Gene Regulates Anthocyanin Biosynthesis in Red Apples. Plant Physiol. 142, 1216–1232. doi: 10.1104/PP.106.088104
Tanner, G. J., Francki, K. T., Abrahams, S., Watson, J. M., Larkin, P. J., and Ashton, A. R. (2003). Proanthocyanidin biosynthesis in plants. Purification of legume leucoanthocyanidin reductase and molecular cloning of its cDNA. J. Biolog. Chem. 278, 31647–31656. doi: 10.1074/jbc.M302783200
Ulrike, M. (2018). Flavonoid Functions in Plants and Their Interactions with Other Organisms. Plants 7, 30–32. doi: 10.3390/plants7020030
Wang, N., Xu, H. F., Jiang, S. H., Zhang, Z. Y., Lu, N. L., Qiu, H. R., et al. (2017). MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple. Plant J. 90, 276–292. doi: 10.1111/tpj.13487
Wu, C., Deng, C., Hilario, E., Albert, N. W., Lafferty, D., Grierson, E. R., et al. (2021). A chromosome-scale assembly of the bilberry genome identifies a complex locus controlling berry anthocyanin composition. Mole Ecol. Resour. 00, 1–16. doi: 10.1111/1755-0998.13467
Wu, P., Liu, A. L., and Li, L. J. (2021). Metabolomics and transcriptome analysis of the biosynthesis mechanism of flavonoids in the seeds of Euryale ferox Salisb at different developmental stages. Mole. Genet. Genom. 296, 953–970. doi: 10.1007/s00438-021-01790-1
Xie, X. B., Li, S., Zhang, R. F., Zhao, J., Chen, Y. C., Zhao, Q., et al. (2012). The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Env. 35, 1884–1897. doi: 10.1111/j.1365-3040.2012.02523
Xu, M. Q., Zhang, X. Y., Li, D. D., Gu, X. Y., Godana, E. A., Dhanasekaran, S., et al. (2021). Transcriptome analysis of postharvest grapes in response to Talaromyces rugulosus O1 infection. Postharvest Biol. Tech. 178, 111542–111554. doi: 10.1016/J.POSTHARVBIO.2021
Xu, Z. C., Peters, R. J., Weirather, J., Luo, H. M., Liao, B. S., Zhang, X., et al. (2015). Full-length transcriptome sequences and splice variants obtained by a combination of sequencing platforms applied to different root tissues of Salvia miltiorrhiza and tanshinone biosynthesis. Plant J. 82, 951–961. doi: 10.1111/tpj.12865
Yao, G. F., Ming, M. L., Allan, A. C., Gu, C., Li, L. T., Wu, X., et al. (2017). Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J. 92, 437–457. doi: 10.1111/tpj.13666
Yao, S. X., Wang, Z. M., Cao, Q., Xie, J., Wang, X. R., Zhang, R., et al. (2020). Molecular basis of postharvest granulation in orange fruit revealed by metabolite, transcriptome and methylome profiling. Postharvest Biol. Tech. 166, 111205–111219. doi: 10.1016/j.postharvbio.2020.111205
Zhou, H., Wang, K. L., Wang, H. L., Gu, C., Dare, A. P., Espley, R. V., et al. (2015). Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 82, 105–121. doi: 10.1111/tpj.12792
Zifkin, M., Jin, A., Ozga, J. A., Zaharia, L. I., Schernthaner, J. P., Gesell, A., et al. (2012). Gene expression and metabolite profiling of developing highbush blueberry fruit indicates transcriptional regulation of flavonoid metabolism and activation of abscisic acid metabolism. Plant Physiol. 158, 200–224. doi: 10.1104/pp.111.180950
Keywords: transcriptome, fruit development, SMRT sequencing, RNA-seq, flavonoid biosynthesis, R2R3 MYB, Vaccinium corymbosum
Citation: Tang Q, Chi F-M, Liu H-D, Zhang H-J and Song Y (2021) Single-Molecule Real-Time and Illumina Sequencing to Analyze Transcriptional Regulation of Flavonoid Synthesis in Blueberry. Front. Plant Sci. 12:754325. doi: 10.3389/fpls.2021.754325
Received: 06 August 2021; Accepted: 08 September 2021;
Published: 30 September 2021.
Edited by:
Meng Xie, Brookhaven National Laboratory, United StatesReviewed by:
Yali Sun, Oak Ridge National Laboratory, United StatesCopyright © 2021 Tang, Chi, Liu, Zhang and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Song, c29uZ3lhbmdAY2Fhcy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.