
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 25 November 2021
Sec. Plant Biotechnology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.752246
This article is part of the Research Topic Ca2+ Signalling in Plant Biotic Interactions View all 6 articles
Plant growth, development, and ultimately crop productivity are largely impacted by the interaction of plants with different abiotic and biotic factors throughout their life cycle. Perception of different abiotic stresses, such as salt, cold, drought, heat, and heavy metals, and interaction with beneficial and harmful biotic agents by plants lead to transient, sustained, or oscillatory changes of [calcium ion, Ca2+]cyt within the cell. Significant progress has been made in the decoding of Ca2+ signatures into downstream responses to modulate differential developmental and physiological responses in the whole plant. Ca2+ sensor proteins, mainly calmodulins (CaMs), calmodulin-like proteins (CMLs), and others, such as Ca2+-dependent protein kinases (CDPKs), calcineurin B-like proteins (CBLs), and calmodulin-binding transcription activators (CAMTAs) have played critical roles in coupling the specific stress stimulus with an appropriate response. This review summarizes the current understanding of the Ca2+ influx and efflux system in plant cells and various Ca2+ binding protein-mediated signal transduction pathways that are delicately orchestrated to mitigate abiotic and biotic stresses. The probable interactions of different components of Ca2+ sensor relays and Ca2+ sensor responders in response to various external stimuli have been described diagrammatically focusing on established pathways and latest developments. Present comprehensive insight into key components of the Ca2+ signaling toolkit in plants can provide an innovative framework for biotechnological manipulations toward crop improvability in near future.
Calcium ion (Ca2+) acts as a critically important relay hub for many biological messages. Spatio-temporal changes in Ca2+ concentration inside cellular compartments and the whole cell itself have an important function in the signal transduction networks of all eukaryotes. The presence of Ca2+ pumps in the most primitive bacteria suggests that from the very beginning of life, sensing and transporting of Ca2+ have emerged as an absolute homeostatic necessity for cells to combat cytotoxicity (Verkhratsky and Parpura, 2014). Due to its sessile nature, plants have developed sophisticated adaptation mechanisms for counteracting environmental cues, such as abiotic and biotic factors (Kissoudis et al., 2014). In normal conditions, plant cells maintain a relatively low concentration of free cytosolic calcium ([Ca2+]cyt), of around 100 nM compared to extracellular surroundings and intracellular storage compartments where Ca2+ concentration ranges in the mM scale (Trewavas and Malhó, 1998). [Ca2+]cyt level is strictly regulated at the cellular level and tiny fluctuations in [Ca2+]cyt can be decoded as useful external inklings for activation of downstream signaling components that include various proteins.
Upon sensing aggravation, the ubiquitous change that happens within the plant system is the shift in free [Ca2+]cyt which is raised either from the external medium or from the intracellular/sub-cellular compartments having higher Ca2+ concentration compared to the [Ca2+]cyt (Srivastava et al., 2013). Based on electrophysiological properties, there are three distinct Ca2+ transporting channels, categorized as hyperpolarization-activated calcium channels (HACCs), depolarization-activated calcium channels (DACCs), and voltage-independent calcium channels (VICCs) (Swarbreck et al., 2013). Non-selective cation channels (NSCCs) comprised of depolarization-activated NSCCs (DA-NSCCs), voltage-independent NSCCs (VI-NSCCs), and hyperpolarization-activated NSCCs (HA-NSCCs) are the ubiquitous group of ion channels that do not discriminate between essential and toxic cations of either monovalent or divalent nature and are found in the plasma membrane, tonoplast, and another endomembrane of plant tissues (Demidchik and Maathuis, 2007). [Ca2+]cyt elevation is mainly a result of the Ca2+ influx through membrane-localized Ca2+ accessible ion channels, such as cyclic nucleotide-gated channels (CNGCs), ionotropic glutamate receptors (GLRs), two-pore channel 1 (TPC1), annexins, mechanosensitive-like channels (MSLs), mid1-complementing activity channels (MCA), and hyperosmolarity-induced [Ca2+]cyt increase channels (OSCAs) (Kudla et al., 2018). Along with the plasma membrane, vacuole, the largest intracellular Ca2+ reservoir, is imperative for determining the concentration of [Ca2+]cyt in plants. Several channels have been detected in the vacuolar tonoplast to be involved in Ca2+ influx in the cytosol, such as voltage-dependent slow vacuolar/TPC1 (SV/TPC1) and CNGCs. Moreover, the channels for inositol-1,4,5-trisphosphate (IP3) and cyclic-ADP ribose (cADPR) are expected to be involved in determining the concentration of [Ca2+]cyt in plants, but the molecular elucidation is not yet clear (Schönknecht, 2013). On the other hand, efflux in a cell is mostly controlled by two types of active Ca2+ transport systems, namely, P-type Ca2+-ATPase (Huda et al., 2013) and cation exchanger (CAX) family Ca2+:H+ exchangers (Schönknecht, 2013; Pittman and Hirschi, 2016).
Certain external stimuli mediate specific transient boosts in [Ca2+]cyt along with stimulus-dependent amplitude and shape (e.g., sinusoidal and square-wave) in a temporal manner (Qudeimat and Frank, 2009). These transient boosts in [Ca2+]cyt, known as Ca2+ signatures, depend on Ca2+ channels available in cell membranes or in the sub-cellular compartments within the cell along with the network of Ca2+ transporting proteins (Qudeimat and Frank, 2009). These Ca2+ signatures are eventually sensed, decoded, and transmitted to the downstream signaling components. Plant Ca2+ sensor proteins are classified into two groups, namely, sensor relays and sensor responders. Plant proteins, namely, calmodulins (CaMs), calmodulin-like proteins (CMLs), and calcineurin B-like proteins (CBLs) are the components of sensor relays that do not possess any enzymatic functions but they possess one or more E and F helices (EF) hands for binding with Ca2+ ions and subsequent modulation in their protein conformation (Meena et al., 2019). Contrarily, sensor responders, such as calcium-dependent protein kinases (CDPKs or CPKs) and CBL-interacting protein kinases (CIPKs), possess enzymatic functions for interacting with specific downstream proteins through phosphorylation and dephosphorylation (Hashimoto and Kudla, 2011).
Plant genomes maintain a diverse array of Ca2+ signaling components in different plant species. Arabidopsis genome contains at least 7 CaM, 50 CML, 10 CBL, and 34 CDPK genes (Zeng et al., 2017). Ca2+ ion and CaM can interact and regulate the activity of many transcription factors (TFs) in the plant and among the reported TFs, calmodulin-binding transcription activators (CAMTAs) are the largest well-characterized CaM-binding TF in all multicellular organisms, more or less conserved from plants to human (Finkler et al., 2007). Ca2+/CaMs binding regulates transcriptional activity of CAMTAs on several downstream genes involved in abiotic stress tolerance and disease resistance (Du et al., 2009).
In this review, we have attempted to highlight the role of Ca2+ ion and associated signaling armor viz., CaM, CMLs, CDPKs, CBLs, and CAMTAs, toward harmonizing plant responses to mitigate various abiotic and biotic stresses.
Taking lessons of the escalating environmental vagaries, attempts have been made to underpin the impact of different Ca2+ signatures and their downstream components toward elevating abiotic stress tolerance in plants.
Salt stress creates an unfavorable environment for plant growth through osmotic and ionic stresses. Higher concentrations of soluble ions in the rhizosphere reduce the soil water potential hindering water uptake by plant roots concurrently creating osmotic stress or dehydration stress. Eventually, the gradual accumulation of salt ions inside the plant tissue causes ionic stress to the plant. The salt overlay sensitive (SOS) pathway is a well-established Ca2+-dependent signaling cascade for Na+ ion homeostasis. Na+ ion probably enters into the cell through anatomical leaks or embroiling hyperpolarization-activated NSCC to eventuate a rapid spike in the calcium-gated channel (Ji et al., 2013). The spike in the Ca2+-gated channel activates calcium-binding protein (CBP) SOS3 through myristoylation, a post-translational modification of SOS3, to mediate subsequent binding and activation of a serine/threonine kinase protein, SOS2 (Mahajan et al., 2008). Finally, the SOS2-SOS3 complex activates SOS1 available in the cell membrane to remove excess Na+ ions from the cell (Mahajan et al., 2008). In Arabidopsis, calcineurin B-like 4 (CBL4) acts as a SOS3 protein and CBL-interacting protein kinase 24 (CIPK24), a serine/threonine protein kinase, performs as SOS2 (Yang et al., 2019). Further, the study identified a SOS3-like calcium-binding protein 8 (SCaBP8), later named calcineurin B-like 10 (CBL10), that can bind with the SOS2 protein and intercedes phosphorylation for subsequent downstream signaling (Quan et al., 2007; Lin et al., 2009). CBL4 is mostly functional in root tissue, while CBL10 is mostly linked with the shoot and critically associated with the reproductive tissue development of different plants and the signaling of both CBL4 and CBL10 under salt stress work independently (Yang et al., 2019). CBL10 directly acts on translocon of the outer membrane of chloroplasts 34 (TOC34) protein and reduces the GTPase activity of TOC34 (Cho et al., 2016). For TOC34 protein translocation in the plastid, GTP binding and hydrolysis are highly essential while transportation of several proteins inside the chloroplast is mediated by Ca2+/CaM signaling (Chigri et al., 2005). Since CBL10 and TOC34 are expressed in different types of plastids in plants (Cho et al., 2016), it can be speculated that salt stress signaling modulates the functioning of various proteins in the cytoplasm and other organelles. Moreover, another finding reveals that the Ca2+ dynamics in the cytosol and nucleus are differentially regulated to control overall stress signaling in Arabidopsis (Huang et al., 2017). CBL10 also interacts with CIPK24 in addition to various other CIPKs and activates several transporters in vacuoles like H+/Ca2+ antiporter (also known as CAX1), H+-ATPase transporter, and Na+/H+ transporter (NHX1) in Arabidopsis (Yang et al., 2019) for maintaining Na+ and K+ ion homeostasis in the plant cell. Besides, Annexin1 has been found to regulate Ca2+ influx in the root epidermal tissue of Arabidopsis upon sensing salt stress (Laohavisit et al., 2013).
Another crosstalk has been detected in Arabidopsis and other plants to combat salt stress through phosphatidic acid (PA)-mediated signaling cascade (Yu et al., 2010; McLoughlin et al., 2012). Some membrane phospholipids, namely, phosphatidylcholine and phosphatidylethanolamine, are hydrolyzed by phospholipase D (PLD) to produce PA and the residual head group. Moreover, the hydrolysis of phosphatidylinositol lipid is carried out by phospholipase C to produce diacylglycerol (DAG) that is eventually phosphorylated by DAG kinase to produce PA (Yao and Xue, 2018). Various stresses animate transient increase in PA which eventually bind with mitogen-activated protein kinase 6 (MPK6) to trigger phosphorylation of SOS1 protein, facilitating Na+ extrusion from the cell and other TFs, having a direct effect on salt-stress alleviation (Yu et al., 2010). Additionally, PA binds with two sucrose non-fermenting-1 (SNF1)-related protein kinase 2 (SnRK2.4 and SnRK2.10) proteins for shaping root architecture under salt stress (McLoughlin et al., 2012; Julkowska et al., 2015). A comprehensive study unravels that the binding of mitogen-activated protein kinase kinase 7 (MKK7) and MKK9 with PA is crucial for their translocation to the membrane. These interactions trigger MPK6 to activate SOS1 in the membrane (Shen et al., 2019).
The nuclear Ca2+ signature is mediated in plants by CaM and CDPK which are basically EF-hand motif-containing Ca2+ sensors (Kim et al., 2009). In response to salt stress in common ice plants (Mesembryanthemum crystallinum), a CDPK (McCDPK1) has been isolated containing a nuclear localization signal (NLS) for translocation of McCDPK1 into the nucleus to mitigate salt stress (Patharkar and Cushman, 2000). Several Ca2+ binding TFs viz., AtNIG1 (Arabidopsis thaliana NaCl-inducible gene 1), have been detected for decoding of Ca2+ signaling in both cytosol and the nucleus (Kim and Kim, 2006), which is an integral part of the promoter region of various salinity stress-responsive genes (Chinnusamy et al., 2003). CaM isoforms have also been discovered in soybeans that modulated the salt-induced Ca2+ signaling by activating an MYB-type TF, which is an upstream regulator of several salt and dehydration responsive genes (Yoo et al., 2005). The overall Ca2+ signaling under salt stress is depicted in Figure 1.
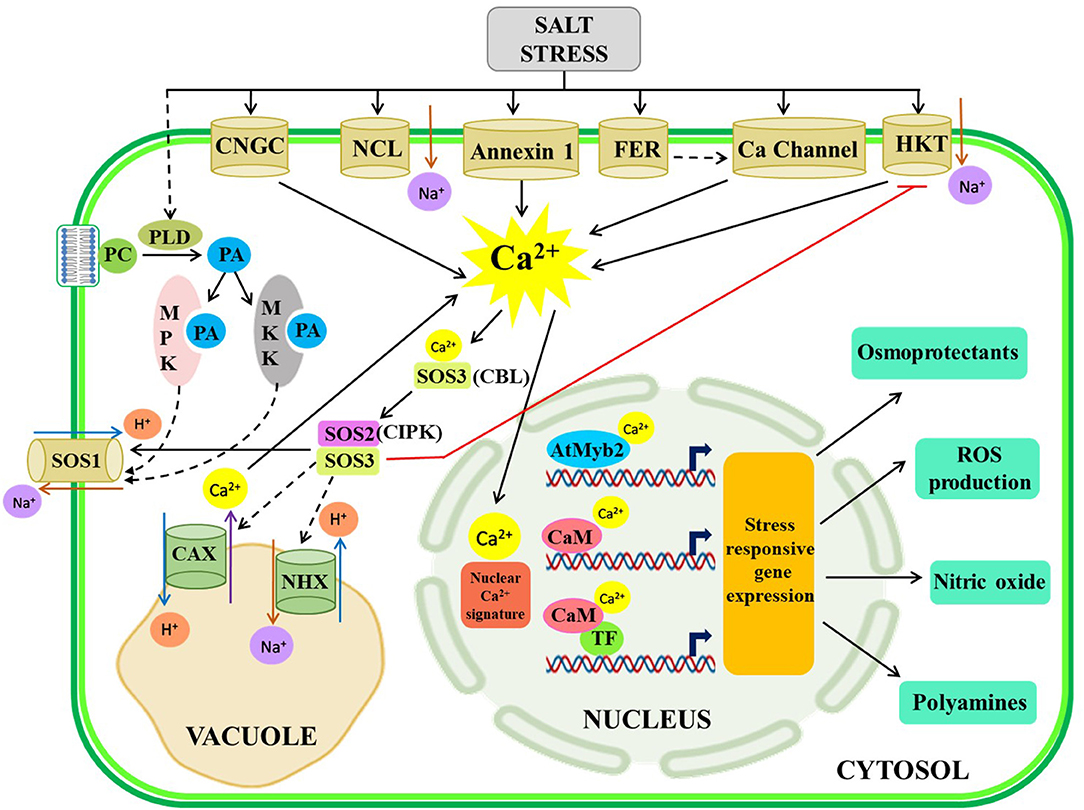
Figure 1. Illustrative representation of crosstalk among Ca2+ signaling components under the salt-stressed condition. During the salt-stressed condition, Na+ is transported into the cytoplasm through different plasma membrane-localized channels leading to a spike in [Ca2+]cyt, which activates SOS pathway and PA-mediated signaling, necessary for extrusion of Na+ from the cytosol to the vacuole and extracellular space and subsequent stress-responsive gene expression for production of osmoprotectants, ROS, nitric oxide, and polyamines. The solid arrow represents direct activation; the dotted arrow represents indirect activation via some intermediates. NCL-Na+/Ca2+exchanger like protein, HKT, high-affinity K+ transport, FER-FERONIA, PCphosphatidylcholine; SOS, salt overlay sensitive; ROS, reactive oxygen species.
Drought is one of the most challenging abiotic stresses that impart severe detrimental effects on growth and crop production. Plants encounter drought stress if the plant-available water is limited in the soil or when plants undergo higher transpiration losses compared to the root water uptake (Salehi-Lisar and Bakhshayeshan-Agdam, 2016). Roots are the first organs to sense drought stress and several sensory proteins, such as histidine kinases, receptor-like kinases, and G-protein-coupled receptors receive the external signal and amplify it through secondary messengers, such as Ca2+, reactive oxygen species (ROS), and IP3 (Jain et al., 2018). These secondary messengers will subsequently activate complex regulatory network controlling root growth under drought stress by regulating hormonal pathways and the expression of several TFs, such as APETALA 2/ethylene-responsive element binding factor (AP2/ERF), myeloblastosis viral oncogene homolog TF (MYB), basic leucine zipper (bZIP), and NAM/ATAF1/CUC2 (NAC) TF (Valliyodan and Nguyen, 2006; Jung et al., 2017). Similar to SOS signaling during salt stress, some CBL proteins (SOS3 homolog) interact with CIPK (SOS2 homolog) to confer abscisic acid (ABA) and drought stress signaling. An earlier study in Arabidopsis explains that a CBP SCaBP5 (CBL1) interacts with a protein kinase, PKS3 (CIPK15), to regulate ABA-dependent seed germination, stomatal opening, and subsequent gene expression (Guo et al., 2002). Additionally, PKS3 physically binds with ABI1 (ABA-insensitive 1) and ABI2, which are protein phosphatase 2C (PP2C) protein and are supposed to negatively regulate certain features of the ABA cascade, such as stomatal closure (Guo et al., 2002). In Arabidopsis, ABA-dependent stomatal closure is positively regulated by SnRK2 protein kinase (SRK2E/OST1/SnRK2.6) containing two C-terminal domains where domain I work in an ABA-independent manner and domain II is activated in presence of ABA (Yoshida et al., 2006). It has been found that SRK2E/OST1 physically interacts with ABI1 through domain II and is involved in ABA and osmotic stress signaling to control stomatal closure in Arabidopsis. Further studies identify that CBL9 interacts with CIPK3 and forms a module for ABA signaling and in addition to that abscisic acid repressor 1 (ABR1) acts as a downstream component of CIPK3 (Pandey et al., 2008; Sanyal et al., 2017). Another study in Arabidopsis identifies CML20 as a negative regulator of ABA-induced stomatal opening while the mutant Arabidopsis lines (cml20) show relatively less water loss from leaves and more drought tolerance (Wu et al., 2017). Moreover, drought and ABA stress upregulate transcript expressions of several stress-responsive genes, such as MYB2, RAB18, ERD10, COR47, and RD29A in the cml20 lines. On contrary, another CBP (CBP60g) acts as a positive regulator of drought stress in Arabidopsis (Wan et al., 2012). Classical model on ABA-mediated drought stress signaling depicts that upon binding with ABA, pyrabactin resistance 1/PYR1-Like/Regulatory components of ABA receptors (PYR/PYL/RCAR) inactivate PP2C which eventually activates SnRK2s, other ion channels, and TFs to turn on ABA-responsive genes (Hubbard et al., 2010). The mechanistic action of different environmental cues controlling the ABA signaling and other CBPs is nicely reviewed by Kumar et al. (2019). Hypo-methylation of CaM might be changing their binding partners and subsequent downstream signaling to depict salt and dehydration tolerance (Banerjee et al., 2013). Several CDPKs are also found to be involved in drought stress signaling (Xu et al., 2010; Ho et al., 2013; Pandey et al., 2013). Functional characterization of OsCDPK1 in rice (Oryza sativa) reveals negative regulation of gibberellic acid biosynthesis while it activates a 14-3-3 protein (GF14c) and improves drought tolerance in transgenic seedlings during constitutive expression (Ho et al., 2013). Another study in Arabidopsis states that CAMTA1 mutant (camta1) plants are highly susceptible to drought stress, and CAMTA1 further regulates the expression of several stress-responsive genes and TFs, such as RD26, ERD7, RAB18, LTPs, COR78, CBF1, heat shock proteins (HSPs), and AP2-EREBP (Pandey et al., 2013). An interesting study in maize (Zea mays) establishes that ZmCCaMK (calcium/CaMs-dependent protein kinase) phosphorylates a NAC TF (ZmNAC84) and induces antioxidant defense by activating downstream genes to enhance drought tolerance (Zhu et al., 2016). The overall Ca2+ signaling under drought stress is portrayed in Figure 2.
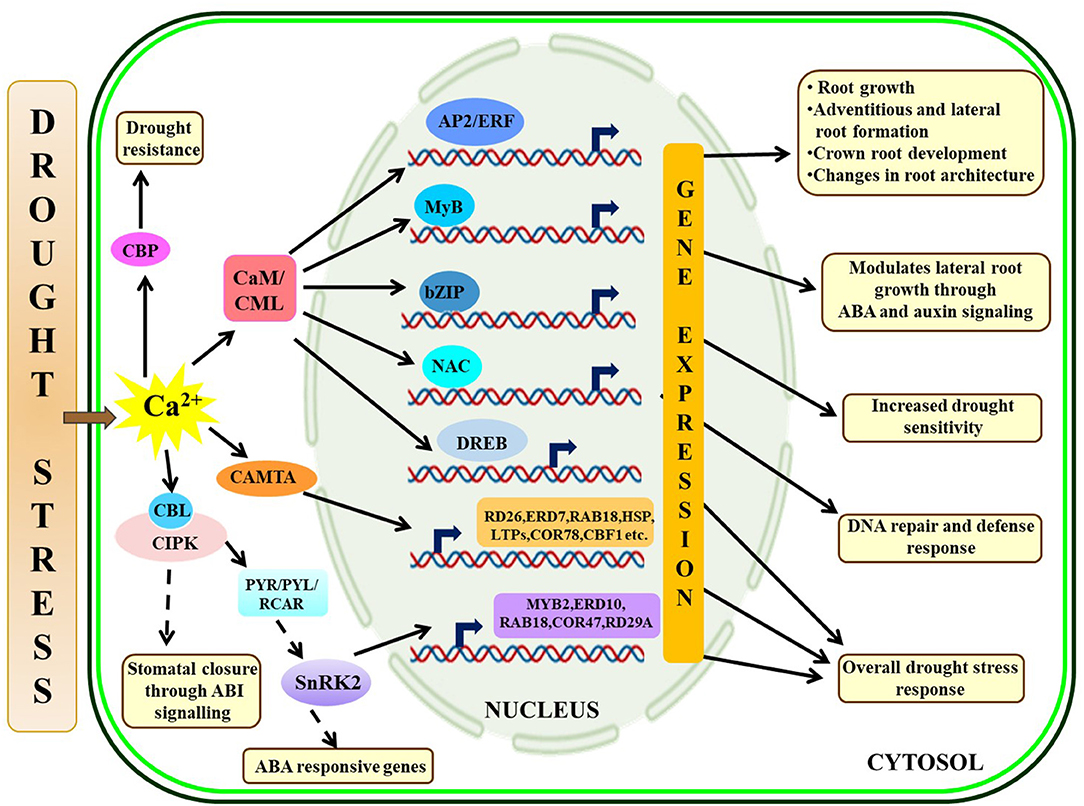
Figure 2. Illustrative representation of crosstalk among Ca2+ signaling components under drought-stressed condition, Drought stress-mediated [Ca2+]cyt fluctuations activate Ca2+-interacting partners and provide response against the stress through varying mechanisms like CaM- and CAMTA-mediated gene expressions through several TFs conferring drought tolerance, CBL-CIPK interaction controls water loss through stomatal closure, PYR/PYL/RCAR regulates ABA pathway critical in stress-management. The solid arrow represents direct activation; the dotted arrow represents indirect activation via some intermediates. CaM, calmodulins; CAMTA, calmodulin-binding transcription activators; CIPK, CBL-interacting protein kinases.
Calcium ion and CaM are important components in mediating the heat shock (HS) response in plants. Similar to other stresses, the increase in [Ca2+]cyt is the first step in the HS signal transduction pathway. Transgenic tobacco (Nicotiana tabacum) plants expressing aequorin, a luminescent protein, exhibit an increase in [Ca2+]cyt upon heat treatment. Additionally, exogenous application of Ca2+ results in improved thermo-tolerance, whereas the effect is diminished in the presence of Ca2+ chelators and inhibitors (Gong et al., 1998). Plants cells when exposed to high temperatures mobilize HSPs or molecular chaperones to aid in proper protein folding for mitigating the damage caused by heat stress. Laser confocal and fluorescence microscopic studies in wheat demonstrated that intracellular Ca2+ levels were increased within 1 min upon exposure to heat stress at 37°C. Furthermore, HS at 37°C upregulates CaM 1-2 expression after 10 min, while two HSP genes, hsp26 and hsp70, are induced later on. The expression of these HSP genes is upregulated after exogenous Ca2+ application, and their expressions are downregulated in the presence of CaM antagonists, Ca2+ chelators, and Ca2+ channel blockers (Liu et al., 2003). Similarly, maize seedlings pre-treated with CaCl2 showed enhanced thermotolerance coupled with a remarkable increase in CaM accumulation upon heat stress exposures (Zeng et al., 2015). The purified heat-inducible homolog of Hsp70 (DgHsp70) from orchard grass (Dactylis glomerata) possesses ATPase, holdase, and ATP-dependent foldase activity along with a CaM-binding domain (CaMBD). Moreover, binding to AtCaM2 in the presence of Ca2+ affects the ATPase and foldase activities of DgHsp70 protein (Cha et al., 2012). AtCaM3 plays a pivotal role in heat stress signaling by regulating the activity of CaM-binding protein kinase (AtCBK3) which eventually controls HS TFs (AtHSFA1a) and HS protein genes through phosphorylation or dephosphorylation modulating heat stress response in Arabidopsis (Zhang et al., 2009). Furthermore, AtCaM3 acts as a downstream factor in nitric oxide (NO) signaling resulting in the activation of heat shock factors (HSFs), accumulation of HSPs, and development of thermotolerance (Xuan et al., 2010). In a similar manner, Ca2+/CaM signaling is involved in HS response and an increase in the expression of Ca2+/HS-related proteins in rice (Wu et al., 2012). In Arabidopsis, AtPP7, a Ser/Thr phosphatase, is found to be induced under heat stress and known to interact with CaM in a Ca2+-dependent manner (Zeng et al., 2015). Moreover, AtPP7 is also found to interact with AtHSF1, an HS TF, thereby, suggesting its possible involvement in the regulation of the HSP genes (Liu et al., 2007). Vitis amurensis (V. amurensis) is a wild species of grapevine having high resistance to cold and disease. Overexpression of VaCPK29 improves heat tolerance in Arabidopsis (Dubrovina et al., 2017). Another finding in V. amurensis depicts the differential regulations of various VaCaMs and VaCMLs in response to an array of abiotic stresses, such as salt, cold, heat, and osmotic stress (Dubrovina et al., 2019). Altogether, 79 CML genes have been identified in the Chinese cabbage (Brassica rapa ssp. pekinensis), and expressions of many BrCMLs, particularly BrCML21–1, are upregulated under heat stress (Nie et al., 2017). However, comprehensive research is warranted to elucidate the specific mechanisms by which these genes are regulated in different plants.
Heat stress causes a rise in intracellular levels of [Ca2+]cyt which then combines with CaM and activates L-glutamate decarboxylase (GAD) for subsequent decarboxylation of glutamate (Glu) into γ-amino butyrate (GABA). However, calcium and CaM inhibitors block the GABA accumulation in Arabidopsis seedlings (Locy et al., 2000). CaM binding to GAD plays an important role in the normal development of plants while GABA is found to impart partial protection against heat stress by improving the leaf turgor, increasing the concentration of protective osmolytes, and reduction in the oxidative damage (Nayyar et al., 2014). Furthermore, transcriptomic analyses of rice reveal that OsMSR2 (multi-stress-responsive gene 2), a novel CML is strongly upregulated under various abiotic stresses, such as cold, heat, and drought stress (Xu et al., 2011).
Plant responses to cold stress involve complex crosstalk through activation of several signaling pathways that could improve the chilling and freezing tolerance of plants. Cold temperature is perceived by various cold sensors localized in the plasma membrane which eventually led to a transient increase in the cytosolic or nuclear Ca2+ levels followed by activation of the downstream signaling cascade (Yuan et al., 2018). The rate of cooling also plays an important role in temperature sensing, a faster rate of cooling results in increased sensitivity along with increased Ca2+ response, whereas prolonged or repeated exposure to low temperatures attenuates the response (Knight and Knight, 2012). An earlier study revealed that CAX1 might have played a vital role in the cold stress response of Arabidopsis (Dodd et al., 2010). Additionally, two calcium-permeable mechano-sensitive channels have been detected as a regulator of cold-induced increment of cytosolic Ca2+ levels (Mori et al., 2018). In Arabidopsis, Yang et al. (2010) reported CRLK1 as a novel plasma membrane-localized calcium/CaM-regulated receptor-like kinase 1 containing two CaM-binding sites, which is stimulated in response to cold and oxidative stress and induces various cold-responsive (COR) genes, such as CBF1, RD29A, COR15a, and KIN1. Further research studies affirmed that an increase in cellular Ca2+ level caused CRLK1 to phosphorylate a specific mitogen-activated protein kinase kinase kinase (MEKK1), followed by the activation of protein kinase kinase 2 (MKK2) and MAPK signaling cascade leading to the activation of cold response genes (Furuya et al., 2013, 2014). Several Ca2+/CaM-binding proteins, such as PsCCaMK in pea (Pisum sativum), CaM-binding membrane transporter-like proteins (MCamb1 and MCamb2) in Physcomitrella patens, OsCPK24 in rice and TCH, and a CaM-related protein, in Arabidopsis, show upregulation upon cold stress (Takezawa and Minami, 2004; Zeng et al., 2015; Liu et al., 2018). In common bean (Phaseolus vulgaris), the Ca2+-CaM-dependent NAD kinase (NADK) activity has increased after 60 min and 120 min of cold-shock (Ruiz et al., 2002). Further study speculates that CPKs induces a signaling cascade involving ROS, NO, and MPKs for ABA-dependent cold adaptation (Liu et al., 2018; Lv et al., 2018). The mechanistic action of CPKs under cold stress has been studied in a wide range of crops. The ZmCPK1 in maize acts as a negative regulator of the cold stress response (Weckwerth et al., 2015); while PeCPK10 in Populus euphratica (Chen et al., 2013), OsCPK7, OsCPK13, and OsCPK17 in rice (Saijo et al., 2000; Komatsu et al., 2007; Almadanim et al., 2017), and AtCPK1 in Arabidopsis act as positive regulator conferring cold tolerance (Almadanim et al., 2017). Furthermore, CaM3, an isoform of CaM, mediate reduced expression of the COR genes, such as KIN1/2 and LT178, which affirmed that CaM3 exerted a negative effect on the COR expression probably by the activation of the Ca2+-ATPase (Townley and Knight, 2002). Finally, Ca2+-binding TF like CAMTA3 could bind to the promoter region of the CBF2/DREB1c gene and positively regulates their expression to confer freezing tolerance in plants (Doherty et al., 2009). The well-documented basic helix-loop-helix (bHLH) TF inducer of CBF expression 1 (ICE1) is the modulator of CBF genes during cold stress (Chinnusamy et al., 2003; Lindemose et al., 2013). When MYB15 and ICE1 interacted, they bound to the DREB1A/CBF3 promoter to regulate cold stress tolerance. Another plausible role of ICE1 is to act as a co-receptor for the cooling induction of Bon1-associated protein 1 (BAP1), which is responsive to cold stress (Zhu et al., 2011). The overall Ca2+ signaling under heat and cold stress is depicted in Figure 3.
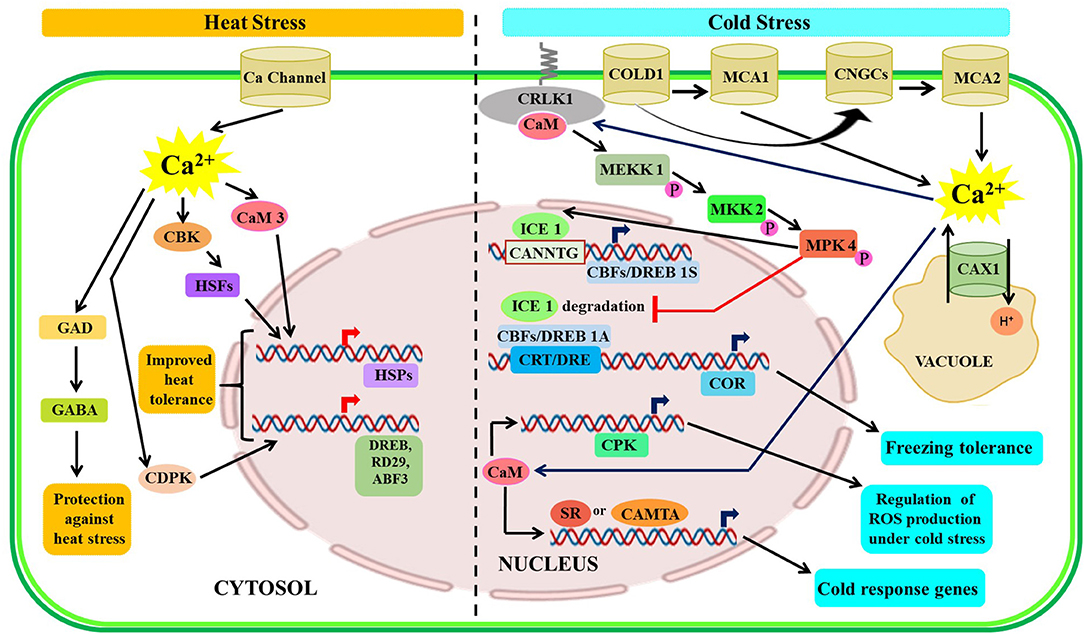
Figure 3. Illustrative representation of crosstalk among Ca2+ signaling components under temperature stressed condition. Upon sensing heat stress, plants respond through increased expression of several HSPs and other proteins like DREB, etc., along with activation of the GAD-GABA pathway. During cold stress, freezing tolerance is achieved through upregulated expression COR, ICE1, etc. CRLK, calcium/calmodulin-regulated receptor-like kinase 1; ICE1, inducer of CBF expression 1; CRT/DRE-C, repeat/dehydration-responsive element; SR, serine/arginine-rich; HSPs, heat shock protein; GAD, L-glutamate decarboxylase; GABA, γ-amino butyrate.
Ever-increasing anthropogenic activities in recent years, such as industrialization and urbanization along with non-judicious use of inorganic fertilizer and pesticide containing heavy metals in agricultural fields, have led to severe heavy metal/metalloid pollution in soil and environment, which is malicious to the local ecosystem. The most common heavy metals/metalloid are lead (Pb), chromium (Cr), iron (Fe), arsenite (AsIII), arsenate (AsV), zinc (Zn), cadmium (Cd), copper (Cu), mercury (Hg), aluminum (Al), and nickel (Ni) (Wuana and Okieimen, 2011).
Lead ions (Pb2+) are capable of binding to the Ca-binding sites of CaM, eventually gaining entry into plant cells through Ca2+ permeable channels (Gorkhali et al., 2016). Arabidopsis CNGCs and tobacco NtCBP4 are involved in the transport of Ca2+ ions during signal transduction and overexpressing NtCBP4 confers Ni+ tolerance, but Pb2+ hypersensitivity (Moon et al., 2019). Interestingly, the expression of a truncated NtCBP4 gene, which lacked a major part of the C-terminal half containing the cyclic nucleotide-binding domain and CaMBD in transgenic tobacco plants resulted in increased Pb2+ tolerance associated with reduced Pb2+ accumulation, while overexpression of full-length NtCBP4 led to Pb hypersensitivity along with overaccumulation of Pb. In rice roots, Pb2+ treatment stimulates ROS production, Ca2+ accumulation, and activation of MAPKs in addition to cell death and stunted root growth. Conversely, pre-treatment of the roots with an antioxidant, CDPK-inhibitor (W7), and an nicotinamide adenine dinucleotide phosphate (NADPH) oxidase inhibitor effectively reduce the Pb2+-induced cell death and MAPK activity depicting the interplay between CDPK and ROS signaling under Pb2+ stress (Huang and Huang, 2008).
Cadmium is extremely phytotoxic even in low concentrations resulting in reduced photosynthetic rate, impeded stomatal conductance, and adverse effects on several biochemical and physiological activities in plants (Guo et al., 2019). Cd-stress depicted a detrimental effect in root growth of white clover (Trifolium repens) and Arabidopsis seedlings (Brunetti et al., 2011; Stravinskiene and Račaite, 2014). Arabidopsis root hair growth is also inhibited by Cd2+ as it affects the cytosolic Ca2+ gradient required for root growth (Fan et al., 2011). Cd2+ can also displace Ca2+ from CaM proteins, thus affecting calcium-dependent signaling (Baliardini et al., 2015). An increase in [Ca2+]cyt levels occurs after Cd2+ exposure, which in turn activates the NADPH oxidase enzyme and transient H2O2 accumulation followed by superoxide () accumulation in the mitochondria causing oxidative damage (Garnier et al., 2006). Baliardini et al. (2015) identified the probable role of the CAX1 gene in Cd2+ tolerance. Several reports revealed that the application of Ca2+ played a crucial role in alleviating the damages caused by Cd2+ uptake in plants (Zorrig, 2012; Moon et al., 2019).
In a similar manner, pre-treatment with Ca2+ alone or in combination with other compounds can ameliorate the toxicity of Al, Cr, and Ni in different plants (Mozafari, 2013; Fang et al., 2014; Hossain et al., 2014). Ca2+ oscillation in rice roots was monitored using a Ca2+ indicator, and ROS-specific dyes revealed a significant increase in Ca2+ after exposure to arsenate [As(V)]. Further, transcriptome analysis of rice roots upon As(V) exposure reveals overexpression of various calcium-related proteins and kinases, such as CaMs, CBLs, CDPKs, and CIPKs, coupled with several TFs, MAPKs, NADPH oxidases along with genes involved in abiotic stresses (Huang et al., 2012). Additionally, transcript analyses depict upregulation of different Ca2+/CaM-binding partners, such as CDPKs in rice upon Cr6+ stress, CaM in basidiomycetous yeast species, Cryptococcus humicola, consequent to Al stress, and another CaM in zucchini (Cucurbita pepo) upon Ni stress (Huang et al., 2014; Zhang et al., 2016; Valivand et al., 2019). Transcriptomic analysis of Tibetan Plateau annual wild barley (Hordeum spontaneum) affirms that multiple CIPKs are playing a pivotal role to mitigate heavy metal toxicity (Hg, Cd, Cr, Pb, and Cu) and other climatic vagaries such as salt, drought, ABA, cold, and heat stress (Pan et al., 2018). Lanthanum (LaIII), a rare earth element, is reported to trigger an increase in Ca2+ levels in the cell, besides interacting with CaM, consequently affecting the expression and conformation of CaM (Wang et al., 2016; Zhang et al., 2016). At low concentrations, LaIII could bind with the two Ca-binding sites of the extracellular CaM by electrostatic attraction to improve CaM function and vice versa (Wang et al., 2016).
The ability of plants to discriminate between harmful biotic agents (pathogens and pests) and beneficial symbiont subsequently exerting differential response either by limiting invasion or by promoting association is the key to survival.
Most of our current understanding of symbiosis signaling is based on the work done mainly on the association of rhizobia and mycorrhiza with Lotus japonicus and Medicago truncatula. Besides rhizobia and mycorrhiza, beneficial interactions also occur with endophytic fungi and plant-growth-promoting bacteria which colonize at the root of the host and result in better plant performance (Khare et al., 2018). Symbiotic interactions start through sequential cytoplasmic and nuclear Ca2+ elevations in the plant cell, and the responses depend on the amplitude, duration, frequency, and location of Ca2+ signature and selective activation of specific calcium channels in cellular membranes (Whalley and Knight, 2013; Yuan et al., 2017).
Plant root-secreted flavonoids and strigolactones are sensed by Rhizobium sp. and mycorrhizal fungi, respectively. Upon recognizing these compounds, symbionts produce modified lipo-chitooligosaccharides, such as nodulation (Nod) factors in the case of rhizobial symbiosis, and mycorrhizal (Myc) factors in the case of arbuscular mycorrhizal fungi symbiosis (Oldroyd, 2013). Upon recognition and binding with the Nod factor, LysM receptor-like kinases form heterocomplex on the cell membrane and activate the cytosolic kinase domain of LysM, triggering cytosolic Ca2+-oscillation within seconds to minutes. Nuclear Ca2+ oscillation is generated through some yet unidentified secondary messenger activated by LysM receptor-like kinases (Oldroyd, 2013). Successful formation of both root nodule and arbuscular mycorrhizal symbiosis requires activation of a common symbiotic pathway composed of eight components, SYMRK (in L. japonicus)/DMI2 (in M. truncatula), POLLUX/DMI1, CASTOR, NENA, NUP85, NUP133, CCaMK, and CYCLOPS/IPD3 (Singh and Parniske, 2012; Oldroyd, 2013). Nuclear membrane-localized Ca2+-regulated cation channels, such as CASTOR and POLLUX in L. japonicus (Charpentier et al., 2008) and DMI1 in M. truncatula (Ané et al., 2004), have been identified as crucial for symbiotic Ca2+ oscillations in the root cell nucleus (Singh et al., 2014). Besides these, three components of the nuclear pore complex, i.e., NUP133, NUP85, and NENA, are also essential for symbiotic signaling (Binder and Parniske, 2014). Nucleus-localized CCaMK, calcium- and CaM-dependent serine/threonine protein kinase, directly interacts with CYCLOPS and phosphorylates it, which is critical for decoding the calcium oscillations (Singh et al., 2014). The mechanism by which CCaMK-CYCLOPS activates specific downstream signaling for nodulation or mycorrhiza formation yet remains unresolved.
Transcription factors, such as nodulation signaling pathway 1 (NSP1) and NSP2, are critical for rhizobium-specific gene expression as it interacts with the promoters of rhizobium-induced genes, such as nodule inception (NIN) and ERN1 (Cerri et al., 2012). Both the CCaMK–CYCLOPS complex and the NSP1–NSP2 complex are vital for driving specific genes expression associated with successful nodule formation. In the case of mycorrhizal signaling, expression of mycorrhiza-specific RAM2 (required for arbuscular mycorrhization 2) gene is controlled by the same signaling complexes with a substitution of NSP1 by RAM1 (Gobbato et al., 2013).
Plant's first line of defense is activated by interaction either between plasma membrane bound pattern-recognition receptors (PRRs) with several elicitors, such as pathogen-associated molecular patterns (PAMPs), microbe-associated molecular patterns (MAMPs), endogenous damage-associated molecular patterns (DAMPs), or compounds produced from cell damage due to pathogenic activity (Dodds and Rathjen, 2010). PRR initiates an overall defense response through activating PAMP-triggered immunity (PTI), followed by rapid activation of MAPKs, oxidative burst, and Ca2+ influx at the plasma membrane (Ranf et al., 2011) and finally augmenting biosynthesis of plant defense hormones, namely, salicylic acid (SA), jasmonic acid (JA), and their derivatives (Vlot et al., 2009). Microbe-induced transient increase of Ca2+ flux is the most important upstream signaling event leading to SA biosynthesis (Du et al., 2009). CaM-binding CBP60 family TFs, CBP60g, and systemic-acquired resistance deficient 1 (SARD1) act as a positive regulator of isochorismate synthase 1 (ICS1) expression, a key enzyme in the SA biosynthesis necessary for inducing systemic-acquired resistance (SAR) (Zhang et al., 2010). The importance of CaM binding of CBP60g and other related proteins (CBP60a) in defense signaling has been well-studied in Arabidopsis (Wang et al., 2009; Truman et al., 2013). The crucial role of CAMTA as an early integration node of both PTI and ETI signaling pathways came from a transcriptomics study by comparative analysis of different CAMTA conditional mutant transgenic lines of Arabidopsis (Jacob et al., 2018). CAMTA3 negatively regulates expression of enhanced disease susceptibility 1 (EDS1) and brassinosteroid insensitive 1-associated kinase 1 (BAK1) interfering with SA and JA signaling, respectively (Du et al., 2009; Rahman et al., 2016). Plants with CAMTA3 mutation exhibit greater resistance to pathogenic bacteria (Pseudomonas syringae, Botrytis cinereal, and Sclerotinia sclerotiorum) but become more susceptible to insect attack (Trichoplusia ni) (Galon et al., 2008; Rahman et al., 2016). Similarly, AtCAMTA3 negatively regulates plant immunity by conferring non-host resistance to Xanthomonas oryzae (Rahman et al., 2016).
The first evidence linking CMLs with plant defense came from overexpression of soybean (Glycine max) CML1 and CML2 (previously, SCaM-4 and SCaM-5) in tobacco, Arabidopsis, and soybean causing increased resistivity against a wide range of virulent and avirulent pathogens (Heo Do et al., 1999; Park et al., 2004; Rao et al., 2014). Validation from autologous and heterologous systems highlights the role of CaM/CMLs in plant immune responses through modulating the hypersensitive response (HR) (Chiasson et al., 2005; Ma et al., 2008). Arabidopsis CML8 and CML9 are also acting as positive regulators of defense mechanisms against different strains of P. syringae through activation of SA-dependent defense responses (Zhu et al., 2017). Additionally, CML9 interacts with a plant-specific TF, pseudo-response regulator 2 (PRR2) in planta which is a player in SA-dependent immunity response in the plant (Cheval et al., 2017). Furthermore, CML37 and CML42 play contrasting roles in insect herbivory-induced resistance in positive and negative manners, respectively (Vadassery et al., 2012; Scholz et al., 2014).
Advances in functional genomics in recent times shed light on the importance of CDPKs in plant immune responses. As a family, CDPKs exert both positive and negative regulations on the immune response. Several members of Arabidopsis CDPKs, i.e., CPK4, 5, 6, and 11 are transiently activated within a few minutes after recognition of common 22-amino-acid long bacterial flagellin epitope flg22 (Boudsocq et al., 2010). These CPKs along with MAPKs positively regulate innate immune signaling perceived by PAMP receptor FLS2 (Boudsocq et al., 2010). In vitro phosphorylation studies revealed that CPKs can also phosphorylate various WRKY TF-controlling downstream immunity-related crucial gene expressions (Boudsocq et al., 2010; Gao et al., 2013). Arabidopsis mutant screening identified two CDPKs (AtCPK3 and AtCPK13) as important transcriptional regulators of plant defensing gene PDF1.2. HSF HsfB2a promotes transcriptional activation of PDF1.2, after being phosphorylated by CPK3-or CPK13 in response to wounding by herbivores, such as Spodoptera littoralis (Kanchiswamy et al., 2010). Pathogen-induced Ca2+ elevations in cytosol activate potato CDPKs (StCPK4 and StCPK5) which in turn stimulates respiratory burst oxidase homolog protein B (RBOHB) (Kobayashi et al., 2007). RBOH gene family encodes integral membrane protein NADPH oxidases, which are a key component of innate immunity due to its central role in ROS production and subsequent oxidative burst and CPK5-mediated in vivo phosphorylation of RBOH homolog D in Arabidopsis (Dubiella et al., 2013). Moreover, CDPKs control cell death in tobacco after recognizing Cladosporium fulvum through interaction between pathogen race-dependent elicitor Avr4/Avr9 and disease-resistant gene Cf9 (Romeis et al., 2001). In the japonica rice cultivar Nipponbare, overexpression of CDPK enhanced blast resistance by preventing Magnaporthe oryzae penetration (Bundó and Coca, 2016). In addition to enhancing plant resistance, CDPKs also perform a negative role in plant defense (Freymark et al., 2007; Asano et al., 2012; Monaghan et al., 2014). However, the function of CDPKs remains unresolved due to duplicity and functional redundancy in higher plants which further warrants comprehensive research to uncover the role of this protein toward plant defense mechanisms. The intricate Ca2+ signaling pathways toward regulating plant-biotic factors interaction are illustrated in Figure 4.
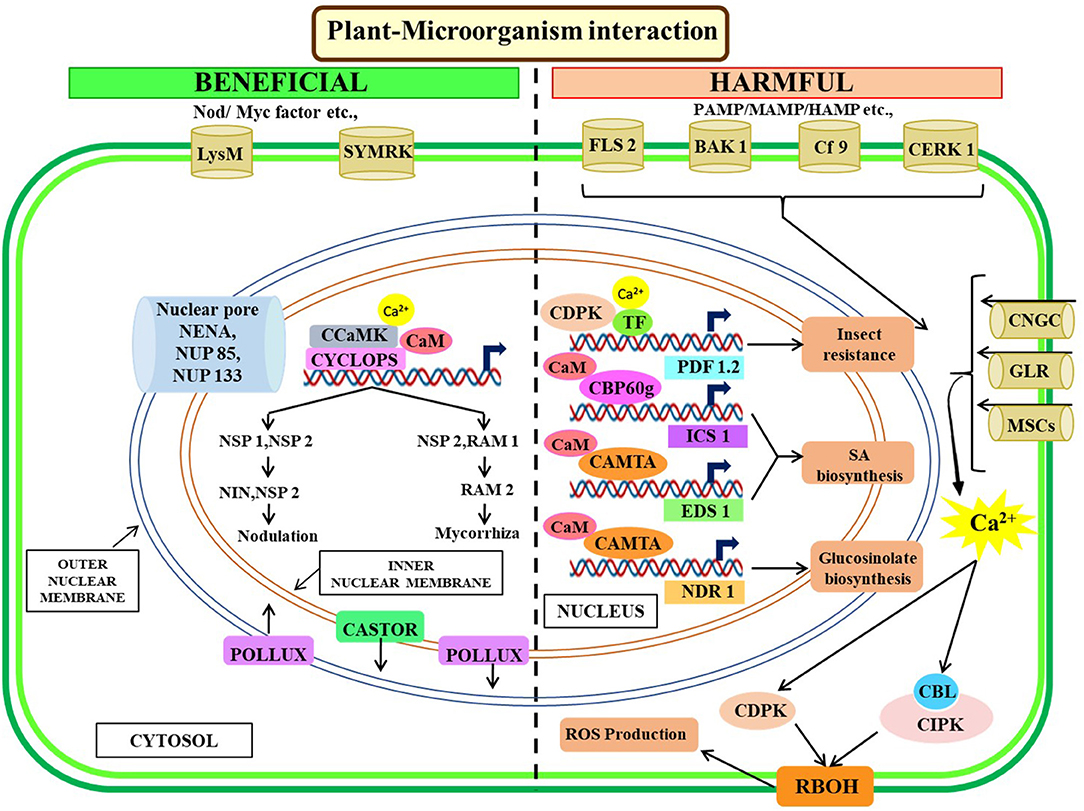
Figure 4. Illustrative representation of crosstalk among Ca2+ signaling components for plant's biotic interactions. Symbiosis signaling starts with the perception of Nod/Myc factors and subsequent Ca2+ signaling mediated activation of either nodulation through the expression of NIN, NSP1, NSP2, or mycorrhiza through RAM1, RAM2 expression. Nuclear membrane localized calcium channels (POLLUX, CASTOR) and nuclear pore complex are essential for plant-microorganism symbiosis. PAMP/MAMP/HAMP activates resistance pathways agents, biotic agents, through SA, glucosinolate biosynthesis, and ROS production during pathogenesis. The membrane-localized transporters trigger Ca2+ in the cytoplasm upon interaction with harmful biotic agents which eventually triggers the CBL-CIPK-CAMTA-mediated signaling cascade for downstream gene expression. NDR, non-race-specific disease resistance1; MSC, mechanosensitive ion channels; SYMRK, symbiosis receptor-like kinase; FLS2, flagellin sensing 2; CERK1, chitin elicitor receptor kinase 1; CAMTA, calmodulin-binding transcription activators; CIPK, CBL-interacting protein kinases; CBLs, calcineurin B-like proteins.
Plants with complex systems and sessile nature have to face multiple stresses when the growing environment is suboptimal and ultimately hinders the expression of complex traits, such as yield and other developmental activities. On the verge of climatic aberrations, plants are becoming more vulnerable toward biotic and abiotic impulses which further elicit changes in the physiological and molecular levels within the plant system to cope up with the vagaries without compensating yield. As plant responses toward multiple stresses are mostly additive in nature, the understanding of the crosstalk between multiple signaling components is the need of the hour to ameliorate the issue in a pragmatic way. The involvements of Ca2+/CaM-related multiple pathways, sensory components, regulatory proteins, and TFs have a plausible role in plant defense mechanisms against biotic and abiotic cues to maintain plant homoeostasis.
For plant growth and development, nitrate () is one of the vital forms of inorganic nitrogen uptake by crop plants. The plethora of sensors has been documented in Arabidopsis. Among these, NRT1.1 is well-studied and presumed to act upstream of several signaling pathways (Sun and Zheng, 2015). Typically, NRT1.1 is the vital component of nitrate peptide transporter family (NPF) related to membrane transporters (Léran et al., 2014) and governs a range of responses related to primary response coupled with germination, flower initiation, development of root and shoot growth, and tolerance or sensitivity to various -dependent abiotic cues viz., temperature extremities, moisture stress, salinity, , and Cd toxicities (Mao et al., 2014; O'Brien et al., 2016; Álvarez-Aragón and Rodríguez-Navarro, 2017; Jian et al., 2018; Teng et al., 2019). Therefore, NRT1.1 has been recognized as a “transceptor” that contemplates the dual role of transporter and receptor related to transport and sensing function (Ho et al., 2009; Gojon et al., 2011). Interestingly, NRT1.1 has a dual affinity of and acts as both low-affinity and high-affinity transport systems (Wang et al., 2012). During this biphasic control of signaling and transport, NRT1.1 forms a dimer by inducing/through crosstalk with CBL9-activated kinase, CIPK23 (Sun and Zheng, 2015; Wen and Kaiser, 2018). Studies have revealed that NRT1.1 ameliorated drought and salinity stress in both ABA-dependent and independent manners through modulating the activity of CIPK23. Furthermore, it has been found that CIPK23 interacts and phosphorylates the NRT1.1 along with ammonium transporter AMT1 toward triggering their activity to maneuver the ammonium toxicity (Ho et al., 2009; Straub et al., 2017; Tian et al., 2021). In addition to the crosstalk between the NRT1.1 sensor and Ca2+/CaM signaling components, Calmodulin-lysine N-methyltransferase (CaM KMT) has been identified as an important modulator of several signal transductions (Banerjee et al., 2013). CaM KMT trimethylates CaM at a specific lysyl residue and the methylation status of CaM plays an important role in the determination of CaM-binding partners in presence of Ca2+ (Magnani et al., 2010, 2012). The earlier report depicted that undermethylation of CaM in CaM KMT knockout mutant plants made it relatively more tolerant to salt, drought, and thermal stresses compared to CaM KMT overexpressed and control Arabidopsis plants through possible changes in CaM-binding partners (Banerjee et al., 2013). Similarly, another major regulatory TF, CAMTA, is involved in the regulation of multiple stress-related genes upon exposure to drought, temperature extremities, and biotic factors (Doherty et al., 2009; Du et al., 2009; Pandey et al., 2013). The overall Ca2+/CaM-mediated signaling upon exposure to multiple stimuli is depicted in Figure 5.
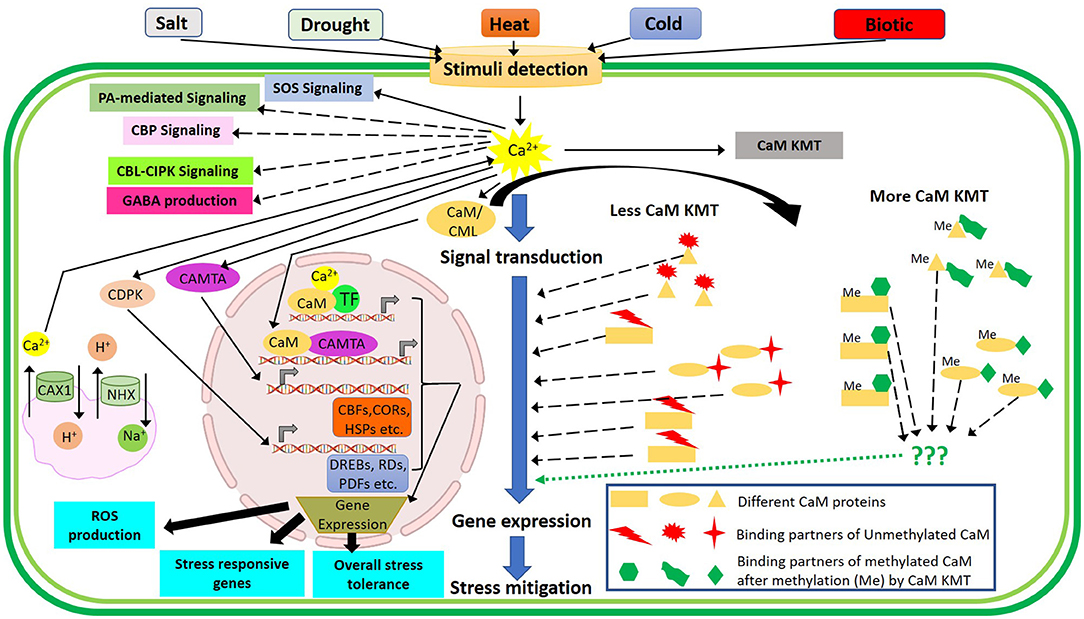
Figure 5. Illustrative representation of crosstalk among Ca2+ signaling components associated with multiple stimuli perceived by the plants. Spike in [Ca2+]cyt triggers several pathways like SOS signaling, PA-mediated signaling, CBP signaling, CBL-CIPK signaling, GABA production, CAX1, and NHX regulation and activates CDPK, CAMTA, CaM, and CML for subsequent expression of stress-responsive genes in the nucleus depending on the types and magnitudes of stimuli. Calmodulin-lysine N-methyltransferase (CaM KMT) methylates CaM proteins in the cytosol while the methylated CaM (Me) and unmethylated CaM depicts differential interactions with CaM-binding partners (depicted in green and red shapes, respectively) and subsequently regulate the downstream signaling components to mitigate various stresses. CaM, calmodulin; CIPK, CBL-interacting protein kinases; CBLs, calcineurin B-like proteins; CDPKs, Ca2+-dependent protein kinases; CML, calmodulin-like proteins.
Unprecedented progress in modern molecular biology coupled with novel “OMICS” tools has enabled researchers to unravel the physiological and molecular mechanisms to combat stress through direct and unbiased monitoring of the complex interplay among stress signaling and response by plants. Modulating the gene activity through various biotechnological approaches, such as overexpression strategies, RNA interference-mediated gene silencing, or genome-editing technologies, has hastened crop improvement since the last three decades. Biotechnological interventions on various Ca2+/CaM-binding proteins viz., CaM, CML, CBP, CDPK, TFs, and others had resulted in the generation of stress-tolerant lines to mitigate the adverse effects of various abiotic and biotic stresses (Table 1). Although the specificity of an individual Ca2+/CaM-binding partner toward specific stress is obscure, out of the several Ca2+/CaM-binding proteins, CDPK/CIPK regulations were reported with several abiotic or biotic factors possibly due to their action in the upstream part of the signaling cascade (Pan et al., 2018). CAMTAs, being a TF also regulate several downstream genes associated with signaling cascade to regulate several stressors (Doherty et al., 2009; Rahman et al., 2016). Furthermore, another master regulator of Ca2+/CaM signaling is CaM KMT, and expressional regulation of CaM KMT in Arabidopsis depicts the involvement of CaM KMT in salt, drought, and cold stress signaling (Banerjee et al., 2013). Hence, the identification of master switch is of utmost priority to the scientific community to cope up with the inevitable abiotic and biotic stressors.
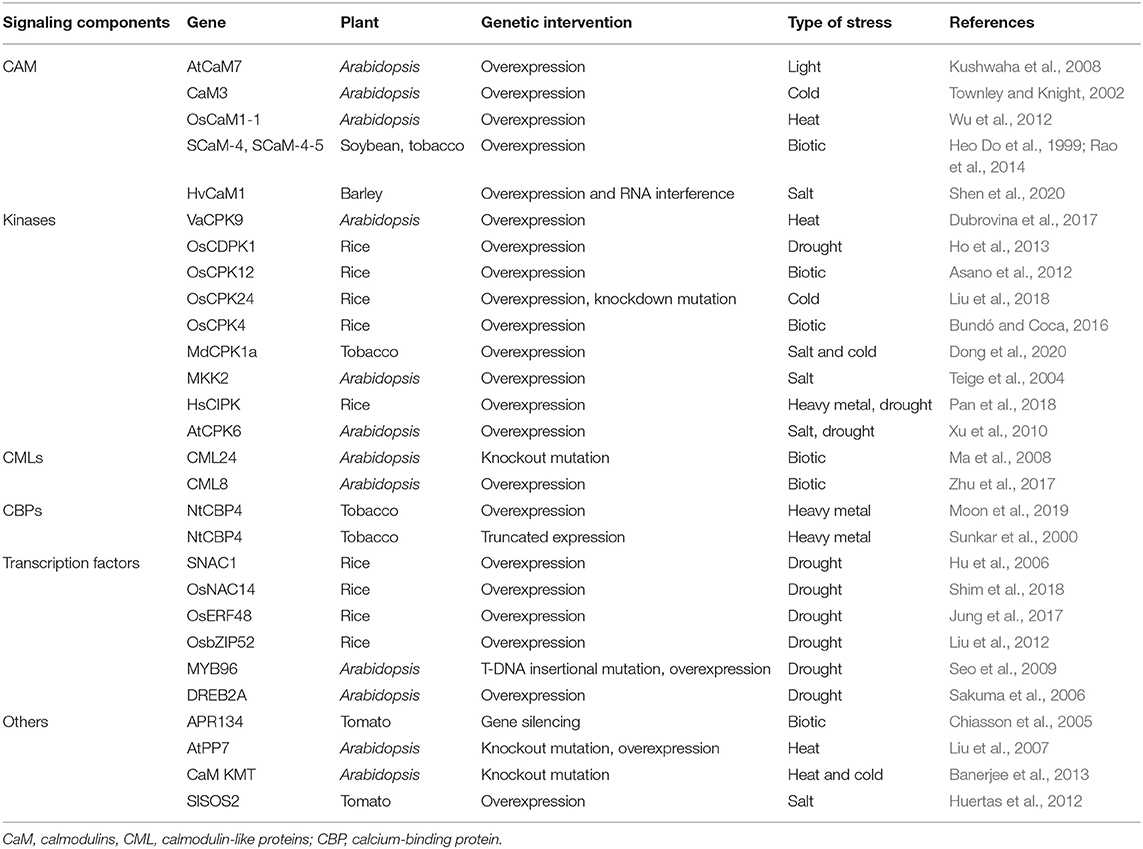
Table 1. Functional genomics studies on Ca2+/CaM-signaling components for conferring stress tolerance in plants.
The agriculture sector is facing an unnerving challenge due to rapid climate change. In crop breeding programs, researchers are more focused on developing improved stress resilient varieties for better sustenance of mankind. Notable progress has been made in recent decades in the field of both basic and applied research studies highlighting the role of Ca2+ signaling as the core mechanisms and processes underlying abiotic and biotic stress adaptation in crop plants and decoding Ca2+ signature by employing various Ca2+-binding proteins viz., CaM, CML, CDPK, and others (Du et al., 2009; Hashimoto and Kudla, 2011; Banerjee et al., 2013; Meena et al., 2019) through intended genetic modifications. Functional genomics and mutation study in Arabidopsis deciphered the biological significance of several Ca2+/CaM signaling components associated with abiotic and biotic signal transduction processes but the picture is unclear in most of the food crops. As there is a huge redundancy for CaMs, CMLs, CAMTAs, and CDPKs in the plant genome, neither siRNA-mediated nor artificial microRNA (amiRNA)-mediated gene-silencing strategies would be sufficient to knockdown the effect of respective isoforms. In spite of overexpressing a single Ca2+/CaM-player to mitigate stress, gene stacking or gene pyramiding of multiple positive alleles would be more relevant to cope up with the situation. Furthermore, simultaneous editing (either gene knockdown or overexpression or both of several pathway components associated with particular stress) of multiple genes could alleviate certain stresses which would eventually improve crop fitness. Although stress is a complex phenomenon, and multiple stresses are encountered by the plant at the same time, Ca2+/CaM signaling pathways are ubiquitous in every stress signaling. Hence, overexpression and genome editing might be the most suitable approach to regulate the master regulator (CaM KMT or CAMTAs) for stress mitigation in the climate-resilient condition to fetch better yield in agricultural crops.
NP, AD, and JB conceptualized the review. NP and SH prepared the draft manuscript. AD, MM, and JB reviewed and edited the manuscript. HG made the figure after revision and corrected the manuscript. All authors contributed to the article and approved the submitted version.
This publication was supported by SERB, DST, Govt. of India (File no. ECR/2018/000328) and Indian Institute of Technology Kharagpur as well as SRIC, IIT Kharagpur.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Almadanim, M. C., Alexandre, B. M., Rosa, M. T. G., Sapeta, H., Leitão, A. E., Ramalho, J. C., et al. (2017). Rice calcium-dependent protein kinase OsCPK17 targets plasma membrane intrinsic protein and sucrose-phosphate synthase and is required for a proper cold stress response. Plant Cell Environ. 40, 1197–1213. doi: 10.1111/pce.12916
Álvarez-Aragón, R., and Rodríguez-Navarro, A. (2017). Nitrate-dependent shoot sodium accumulation and osmotic functions of sodium in Arabidopsis under saline conditions. Plant J. 91, 208–219. doi: 10.1111/tpj.13556
Ané, J. M., Kiss, G. B., Riely, B. K., Penmetsa, R. V., Oldroyd, G. E., Ayax, C., et al. (2004). Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303, 1364–1367. doi: 10.1126/science.1092986
Asano, T., Hayashi, N., Kobayashi, M., Aoki, N., Miyao, A., Mitsuhara, I., et al. (2012). A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance. Plant J. 69, 26–36. doi: 10.1111/j.1365-313X.2011.04766.x
Baliardini, C., Meyer, C. L., Salis, P., Saumitou-Laprade, P., and Verbruggen, N. (2015). CATION EXCHANGER1 cosegregates with cadmium tolerance in the metal hyperaccumulator Arabidopsis halleri and plays a role in limiting oxidative stress in Arabidopsis Spp. Plant Physiol. 169, 549–559. doi: 10.1104/pp.15.01037
Banerjee, J., Magnani, R., Nair, M., Dirk, L. M., DeBolt, S., Maiti, I. B., et al. (2013). Calmodulin-mediated signal transduction pathways in Arabidopsis are fine-tuned by methylation. Plant Cell 25, 4493–4511. doi: 10.1105/tpc.113.119115
Binder, A., and Parniske, M. (2014). Analysis of the Lotus japonicus nuclear pore NUP107-160 subcomplex reveals pronounced structural plasticity and functional redundancy. Front. Plant Sci. 4:552. doi: 10.3389/fpls.2013.00552
Boudsocq, M., Willmann, M. R., McCormack, M., Lee, H., Shan, L., He, P., et al. (2010). Differential innate immune signalling via Ca 2+ sensor protein kinases. Nature 464, 418–422. doi: 10.1038/nature08794
Brunetti, P., Zanella, L., Proia, A., De Paolis, A., Falasca, G., Altamura, M. M., et al. (2011). Cadmium tolerance and phytochelatin content of Arabidopsis seedlings over-expressing the phytochelatin synthase gene AtPCS1. J. Exp. Bot. 62, 5509–5519. doi: 10.1093/jxb/err228
Bundó, M., and Coca, M. (2016). Enhancing blast disease resistance by overexpression of the calcium-dependent protein kinase OsCPK4 in rice. Plant Biotechnol. J. 14, 1357–1367. doi: 10.1111/pbi.12500
Cerri, M. R., Frances, L., Laloum, T., Auriac, M. C., Niebel, A., Oldroyd, G. E., et al. (2012). Medicago truncatula ERN transcription factors: regulatory interplay with NSP1/NSP2 GRAS factors and expression dynamics throughout rhizobial infection. Plant Physiol. 160, 2155–2172. doi: 10.1104/pp.112.203190
Cha, J. Y., Su'udi, M., Kim, W. Y., Kim, D. R., Kwak, Y. S., and Son, D. (2012). Functional characterization of orchardgrass cytosolic Hsp70 (DgHsp70) and the negative regulation by Ca2+/AtCaM2 binding. Plant Physiol. Biochem. 58, 29–36. doi: 10.1016/j.plaphy.2012.06.006
Charpentier, M., Bredemeier, R., Wanner, G., Takeda, N., Schleiff, E., and Parniske, M. (2008). Lotus japonicus CASTOR and POLLUX are ion channels essential for perinuclear calcium spiking in legume root endosymbiosis. Plant Cell 20, 3467–3479. doi: 10.1105/tpc.108.063255
Chen, J., Xue, B., Xia, X., and Yin, W. (2013). A novel calcium-dependent protein kinase gene from Populus euphratica, confers both drought and cold stress tolerance. Biochem. Biophys. Res. Commun. 441, 630–636. doi: 10.1016/j.bbrc.2013.10.103
Cheval, C., Perez, M., Leba, L. J., Ranty, B., Perochon, A., Reichelt, M., et al. (2017). PRR2, a pseudo-response regulator, promotes salicylic acid and camalexin accumulation during plant immunity. Sci Rep. 7:6979. doi: 10.1038/s41598-017-07535-8
Chiasson, D., Ekengren, S. K., Martin, G. B., Dobney, S. L., and Snedden, W. A. (2005). Calmodulin-like proteins from Arabidopsis and tomato are involved in host defense against Pseudomonas syringae pv. tomato. Plant Mol. Biol. 58, 887–897. doi: 10.1007/s11103-005-8395-x
Chigri, F., Soll, J., and Vothknecht, U. C. (2005). Calcium regulation of chloroplast protein import. Plant J. 42, 821–831. doi: 10.1111/j.1365-313X.2005.02414.x
Chinnusamy, V., Ohta, M., Kanrar, S., Lee, B. H., Hong, X., Agarwal, M., et al. (2003). ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17, 1043–1054. doi: 10.1101/gad.1077503
Cho, J. H., Lee, J. H., Park, Y. K., Choi, M. N., and Kim, K.-N. (2016). Calcineurin B-like protein CBL10 directly interacts with TOC34 (translocon of the outer membrane of the chloroplasts) and decreases its GTPase activity in Arabidopsis. Front. Plant Sci. 7:1911. doi: 10.3389/fpls.2016.01911
Demidchik, V., and Maathuis, F. J. M. (2007). Physiological roles of nonselective cation channels in plants: from salt stress to signalling and development. New Phytol. 175, 387–404. doi: 10.1111/j.1469-8137.2007.02128.x
Dodd, A. N., Kudla, J., and Sanders, D. (2010). The language of calcium signaling. Annu. Rev. Plant Biol. 61, 593–620. doi: 10.1146/annurev-arplant-070109-104628
Dodds, P. N., and Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Doherty, C. J., Van Buskirk, H. A., Myers, S. J., and Thomashow, M. F. (2009). Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21, 972–984. doi: 10.1105/tpc.108.063958
Dong, H., Wu, C., Luo, C., Wei, M., Qu, S., and Wang, S. (2020). Overexpression of MdCPK1a gene, a calcium dependent protein kinase in apple, increase tobacco cold tolerance via scavenging ROS accumulation. PLoS ONE 15:e0242139. doi: 10.1371/journal.pone.0242139
Du, L., Ali, G. S., Simons, K. A., Hou, J., Yang, T., Reddy, A. S. N., et al. (2009). Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457, 1154–1158. doi: 10.1038/nature07612
Dubiella, U., Seybold, H., Durian, G., Komander, E., Lassig, R., Witte, C. P., et al. (2013). Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. U.S.A. 110, 8744–8749. doi: 10.1073/pnas.1221294110
Dubrovina, A. S., Aleynova, O. A., Ogneva, Z. V., Suprun, A. R., Ananev, A. A., and Kiselev, K. V. (2019). The effect of abiotic stress conditions on expression of calmodulin (CaM) and calmodulin-like (CML) genes in wild-growing grapevine Vitis amurensis. Plants 8, 1–18. doi: 10.3390/plants8120602
Dubrovina, A. S., Kiselev, K. V., Khristenko, V. S., and Aleynova, O. A. (2017). The calcium-dependent protein kinase gene VaCPK29 is involved in grapevine responses to heat and osmotic stresses. Plant Growth Regul. 82, 79–89. doi: 10.1007/s10725-016-0240-5
Fan, J. L., Wei, X. Z., Wan, L. C., Zhang, L. Y., Zhao, X. Q., Liu, W. Z., et al. (2011). Disarrangement of actin filaments and Ca2+ gradient by CdCl2 alters cell wall construction in Arabidopsis thaliana root hairs by inhibiting vesicular trafficking. J. Plant Physiol. 168, 1157–1167. doi: 10.1016/j.jplph.2011.01.031
Fang, H., Jing, T., Liu, Z., Zhang, L., Jin, Z., and Pei, Y. (2014). Hydrogen sulfide interacts with calcium signaling to enhance the chromium tolerance in Setaria italica. Cell Calcium 56, 472–481. doi: 10.1016/j.ceca.2014.10.004
Finkler, A., Ashery-Padan, R., and Fromm, H. (2007). CAMTAs: calmodulin-binding transcription activators from plants to human. FEBS Lett. 581, 3893–3898. doi: 10.1016/j.febslet.2007.07.051
Freymark, G., Diehl, T., Miklis, M., Romeis, T., and Panstruga, R. (2007). Antagonistic control of powdery mildew host cell entry by barley calcium-dependent protein kinases (CDPKs). Mol. Plant-Microbe Interact. 20, 1213–1221. doi: 10.1094/MPMI-20-10-1213
Furuya, T., Matsuoka, D., and Nanmori, T. (2013). Phosphorylation of Arabidopsis thaliana MEKK1 via Ca2+ signaling as a part of the cold stress response. J. Plant Res. 126, 833–840. doi: 10.1007/s10265-013-0576-0
Furuya, T., Matsuoka, D., and Nanmori, T. (2014). Membrane rigidification functions upstream of the MEKK1-MKK2-MPK4 cascade during cold acclimation in Arabidopsis thaliana. FEBS Lett. 588, 2025–2030. doi: 10.1016/j.febslet.2014.04.032
Galon, Y., Nave, R., Boyce, J. M., Nachmias, D., Knight, M. R., and Fromm, H. (2008). Calmodulin-binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS lett. 582, 943–948. doi: 10.1016/j.febslet.2008.02.037
Gao, X., Chen, X., Lin, W., Chen, S., Lu, D., Niu, Y., et al. (2013). Bifurcation of Arabidopsis NLR immune signaling via Ca2+-dependent protein kinases. PLoS Pathog. 9, e1003127. doi: 10.1371/journal.ppat.1003127
Garnier, L., Simon-plas, F., Thuleau, P., Agnel, J. P., Blein, J. P., Ranjeva, R., et al. (2006). Cadmium affects tobacco cells by a series of three waves of reactive oxygen species that contribute to cytotoxicity. Plant Cell Environ. 29, 1956–1969. doi: 10.1111/j.1365-3040.2006.01571.x
Gobbato, E., Wang, E., Higgins, G., Bano, S. A., Henry, C., Schultze, M., et al. (2013). RAM1 and RAM2 function and expression during arbuscular mycorrhizal symbiosis and Aphanomyces euteiches colonization. Plant Signal Behav. 8:e26049. doi: 10.4161/psb.26049
Gojon, A., Krouk, G., Perrine-Walker, F., and Laugier, E. (2011). Nitrate transceptor (s) in plants. J. Exp. Bot. 62, 2299–2308. doi: 10.1093/jxb/erq419
Gong, M., van der Luit, A. H., Knight, M. R., and Trewavas, A. J. (1998). Heat-shock-induced changes in intracellular Ca2+ level in tobacco seedlings in relation to thermotolerance. Plant Physiol. 116, 429–437. doi: 10.1104/pp.116.1.429
Gorkhali, R., Huang, K., Kirberger, M., and Yang, J. J. (2016). Defining potential roles of Pb2+ in neurotoxicity from a calciomics approach. Metallomics 8, 563–578. doi: 10.1039/C6MT00038J
Guo, J., Qin, S., Rengel, Z., Gao, W., Nie, Z., Liu, H., et al. (2019). Cadmium stress increases antioxidant enzyme activities and decreases endogenous hormone concentrations more in Cd-tolerant than Cd-sensitive wheat varieties. Ecotoxicol. Environ. Saf. 172, 380–387. doi: 10.1016/j.ecoenv.2019.01.069
Guo, Y., Xiong, L., Song, C. P., Gong, D., Halfter, U., and Zhu, J.-K. (2002). A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev. Cell. 3, 233–244. doi: 10.1016/S1534-5807(02)00229-0
Hashimoto, K., and Kudla, J. (2011). Calcium decoding mechanisms in plants. Biochimie 93, 2054–2059. doi: 10.1016/j.biochi.2011.05.019
Heo Do, W., Lee, S. H., Kim, M. C., Kim, J. C., Chung, W. S., Chun, H. J., et al. (1999). Involvement of specific calmodulin isoforms in salicylic acid-independent activation of plant disease resistance responses. Proc. Natl. Accad. Sci. U.S.A. 96, 766–771. doi: 10.1073/pnas.96.2.766
Ho, C. H., Lin, S. H., Hu, H. C., and Tsay, Y. F. (2009). CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194. doi: 10.1016/j.cell.2009.07.004
Ho, S. L., Huang, L. F., Lu, C. A., He, S. L., Wang, C. C., Yu, S. P., et al. (2013). Sugar starvation- and GA-inducible calcium-dependent protein kinase 1 feedback regulates GA biosynthesis and activates a 14-3-3 protein to confer drought tolerance in rice seedlings. Plant Mol. Biol. 81, 347–361. doi: 10.1007/s11103-012-0006-z
Hossain, M. A., Ashrafuzzaman, M., Hossain, A. K. M., Ismail, M., and Koyama, H. (2014). Role of accumulated calcium in alleviating aluminum injury in wheat plants. Sci. World J. 2014:457187. doi: 10.1155/2014/457187
Hu, H., Dai, M., Yao, J., Xiao, B., Li, X., Zhang, Q., et al. (2006). Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. U.S.A. 103, 12987–12992. doi: 10.1073/pnas.0604882103
Huang, F., Luo, J., Ning, T., Cao, W., Jin, X., Zhao, H., et al. (2017). Cytosolic and nucleosolic calcium signaling in response to osmotic and salt stresses are independent of each other in roots of Arabidopsis seedlings. Front. Plant Sci. 8:1648. doi: 10.3389/fpls.2017.01648
Huang, T. L., and Huang, H. J. (2008). ROS and CDPK-like kinase-mediated activation of MAP kinase in rice roots exposed to lead. Chemosphere 71, 1377–1385. doi: 10.1016/j.chemosphere.2007.11.031
Huang, T. L., Huang, L. Y., Fu, S. F., Trinh, N. N., and Huang, H. J. (2014). Genomic profiling of rice roots with short-and long-term chromium stress. Plant Mol. Biol. 86, 157–170. doi: 10.1007/s11103-014-0219-4
Huang, T. L., Nguyen, Q. T. T., Fu, S. F., Lin, C. Y., Chen, Y. C., and Huang, H. J. (2012). Transcriptomic changes and signalling pathways induced by arsenic stress in rice roots. Plant Mol. Biol. 80, 587–608. doi: 10.1007/s11103-012-9969-z
Hubbard, K. E., Nishimura, N., Hitomi, K., Getzoff, E. D., and Schroeder, J. I. (2010). Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev. 24, 1695–1708. doi: 10.1101/gad.1953910
Huda, K. M. K., Banu, M. S. A., Tuteja, R., and Tuteja, N. (2013). Global calcium transducer P-type Ca2+-ATPases open new avenues for agriculture by regulating stress signalling. J. Exp. Bot. 64, 3099–3109. doi: 10.1093/jxb/ert182
Huertas, R., Olías, R., Eljakaoui, Z., Gálvez, F. J., Li, J., De Morales, P. A., et al. (2012). Overexpression of SlSOS2 (SlCIPK24) confers salt tolerance to transgenic tomato. Plant Cell Environ. 35, 1467–1482. doi: 10.1111/j.1365-3040.2012.02504.x
Jacob, F., Kracher, B., Mine, A., Seyfferth, C., Blanvillain-Baufumé, S., Parker, J. E., et al. (2018). A dominant-interfering camta3 mutation compromises primary transcriptional outputs mediated by both cell surface and intracellular immune receptors in Arabidopsis thaliana. New Phytol. 217, 1667–1680. doi: 10.1111/nph.14943
Jain, M., Nagar, P., Goel, P., Singh, A. K., Kumari, S., and Mustafiz, A. (2018). “Second messengers: central regulators in plant abiotic stress response,” in Abiotic Stress-Mediated Sensing and Signaling in Plants: An Omics Perspective, eds S. Zargar and M. Zargar (Singapore: Springer), 47–94. doi: 10.1007/978-981-10-7479-0_2
Ji, H., Pardo, J. M., Batelli, G., Van Oosten, M. J., Bressan, R. A., and Li, X. (2013). The salt overly sensitive (SOS) pathway: established and emerging roles. Mol. Plant 6, 275–286. doi: 10.1093/mp/sst017
Jian, S., Liao, Q., Song, H., Liu, Q., Lepo, J. E., Guan, C., et al. (2018). NRT1. 1-related NH4+ toxicity is associated with a disturbed balance between NH4+ uptake and assimilation. Plant Physiol. 178, 1473–1488. doi: 10.1104/pp.18.00410
Julkowska, M. M., McLoughlin, F., Galvan-Ampudia, C. S., Rankenberg, J. M., Kawa, D., Klimecka, M., et al. (2015). Identification and functional characterization of the Arabidopsis Snf1-related protein kinase SnRK2.4 phosphatidic acid-binding domain. Plant Cell Environ. 38, 614–624. doi: 10.1111/pce.12421
Jung, H., Chung, P. J., Park, S. H., Redillas, M. C. F. R., Kim, Y. S., Suh, J.-W., et al. (2017). Overexpression of OsERF48 causes regulation of OsCML16, a calmodulin-like protein gene that enhances root growth and drought tolerance. Plant Biotechnol. J. 15, 1295–1308. doi: 10.1111/pbi.12716
Kanchiswamy, C. N., Takahashi, H., Quadro, S., Maffei, M. E., Bossi, S., Bertea, C., et al. (2010). Regulation of Arabidopsis defense responses against Spodoptera littoralis by CPK-mediated calcium signaling. BMC Plant Biol. 10:97. doi: 10.1186/1471-2229-10-97
Khare, E., Mishra, J., and Arora, N. K. (2018). Multifaceted interactions between endophytes and plant: developments and prospects. Front. Microbiol. 9:2732. doi: 10.3389/fmicb.2018.02732
Kim, J., and Kim, H. Y. (2006). Functional analysis of a calcium-binding transcription factor involved in plant salt stress signaling. FEBS Lett. 580, 5251–5256. doi: 10.1016/j.febslet.2006.08.050
Kim, M. C., Chung, W. S., Yun, D. J., and Cho, M. J. (2009). Calcium and calmodulin-mediated regulation of gene expression in plants. Mol. plant 2, 13–21. doi: 10.1093/mp/ssn091
Kissoudis, C., van de Wiel, C., Visser, R. G., and van der Linden, G. (2014). Enhancing crop resilience to combined abiotic and biotic stress through the dissection of physiological and molecular crosstalk. Front. Plant Sci. 5:207. doi: 10.3389/fpls.2014.00207
Knight, M. R., and Knight, H. (2012). Low-temperature perception leading to gene expression and cold tolerance in higher plants. New Phytol. 195, 737–751. doi: 10.1111/j.1469-8137.2012.04239.x
Kobayashi, M., Ohura, I., Kawakita, K., Yokota, N., Fujiwara, M., Shimamoto, K., et al. (2007). Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell 19, 1065–1080. doi: 10.1105/tpc.106.048884
Komatsu, S., Yang, G., Khan, M., Onodera, H., Toki, S., and Yamaguchi, M. (2007). Over-expression of calcium-dependent protein kinase 13 and calreticulin interacting protein 1 confers cold tolerance on rice plants. Mol. Genet. Genomics 277, 713–723. doi: 10.1007/s00438-007-0220-6
Kudla, J., Becker, D., Grill, E., Hedrich, R., Hippler, M., Kummer, U., et al. (2018). Advances and current challenges in calcium signaling. New Phytol. 218, 414–431. doi: 10.1111/nph.14966
Kumar, M., Kesawat, M. S., Ali, A., Lee, S. C., Gill, S. S., and Kim, A. H. U. (2019). Integration of abscisic acid signaling with other signaling pathways in plant stress responses and development. Plants. 8:592. doi: 10.3390/plants8120592
Kushwaha, R., Singh, A., and Chattopadhyay, S. (2008). Calmodulin7 plays an important role as transcriptional regulator in Arabidopsis seedling development. Plant Cell 20, 1747–1759. doi: 10.1105/tpc.107.057612
Laohavisit, A., Richards, S. L., Shabala, L., Chen, C., Colaço, R. D. D. R., Swarbreck, S. M., et al. (2013). Salinity-induced calcium signaling and root adaptation in Arabidopsis require the calcium regulatory protein annexin1. Plant Physiol. 163, 253–262. doi: 10.1104/pp.113.217810
Léran, S., Varala, K., Boyer, J. C., Chiurazzi, M., Crawford, N., Daniel-Vedele, F., et al. (2014). A unified nomenclature of nitrate transporter 1/peptide transporter family members in plants. Trends Plant Sci. 19, 5–9. doi: 10.1016/j.tplants.2013.08.008
Lin, H., Yang, Y., Quan, R., Mendoza, I., Wu, Y., Du, W., et al. (2009). Phosphorylation of SOS3-like calcium binding protein8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. Plant Cell 21, 1607–1619. doi: 10.1105/tpc.109.066217
Lindemose, S., O'Shea, C., Jensen, M. K., and Skriver, K. (2013). Structure, function and networks of transcription factors involved in abiotic stress responses. Int. J. Mol. Sci. 14, 5842–5878. doi: 10.3390/ijms14035842
Liu, C., Wu, Y., and Wang, X. (2012). bZIP transcription factor OsbZIP52/RISBZ5: a potential negative regulator of cold and drought stress response in rice. Planta 235, 1157–1169. doi: 10.1007/s00425-011-1564-z
Liu, H. T., Li, B., Shang, Z. L., Li, X. Z., Mu, R. L., Sun, D. Y., et al. (2003). Calmodulin is involved in heat shock signal transduction in wheat. Plant Physiol. 132, 1186–1195. doi: 10.1104/pp.102.018564
Liu, H. T., Li, G. L., Chang, H., Sun, D. Y., Zhou, R. G., and Li, B. (2007). Calmodulin-binding protein phosphatase PP7 is involved in thermotolerance in Arabidopsis. Plant Cell Environ. 30, 156–164. doi: 10.1111/j.1365-3040.2006.01613.x
Liu, Y., Xu, C., Zhu, Y., Zhang, L., Chen, T., Zhou, F., et al. (2018). The calcium-dependent kinase OsCPK24 functions in cold stress responses in rice. J. Integr. Plant Biol. 60, 173–188. doi: 10.1111/jipb.12614
Locy, R. D., Wu, S.-J., Bisnette, J., Barger, T. W., McNabb, D., Zik, M., et al. (2000). “The regulation of GABA accumulation by heat stress in Arabidopsis,” in Plant Tolerance to Abiotic Stresses in Agriculture: Role of Genetic Engineering, eds J. H. Cherry, R. D. Locy, and A. Rychter (Dordrechteds: Springer), 39–52. doi: 10.1007/978-94-011-4323-3_3
Lv, X., Li, H., Chen, X., Xiang, X., Guo, Z., Yu, J., et al. (2018). The role of calcium-dependent protein kinase in hydrogen peroxide, nitric oxide and ABA-dependent cold acclimation. J. Exp. Bot. 69, 4127–4139. doi: 10.1093/jxb/ery212
Ma, W., Smigel, A., Tsai, Y. C., Braam, J., and Berkowitz, G. A. (2008). Innate immunity signaling: cytosolic Ca2+ elevation is linked to downstream nitric oxide generation through the action of calmodulin or a calmodulin-like protein. Plant Physiol. 148, 818–828. doi: 10.1104/pp.108.125104
Magnani, R., Chaffin, B., Dick, E., Bricken, M. L., Houtz, R. L., and Bradley, L. H. (2012). Utilization of a calmodulin lysine methyltransferase co-expression system for the generation of a combinatorial library of post-translationally modified proteins. Protein Expr. Purif. 86, 83–88. doi: 10.1016/j.pep.2012.09.012
Magnani, R., Dirk, L., Trievel, R., and Houtz, R. (2010). Calmodulin methyltransferase is an evolutionarily conserved enzyme that trimethylates Lys-115 in calmodulin. Nat. Commun. 1:43. doi: 10.1038/ncomms1044
Mahajan, S., Pandey, G. K., and Tuteja, N. (2008). Calcium-and salt-stress signaling in plants: shedding light on SOS pathway. Arch. Biochem. Biophys. 471, 146–158. doi: 10.1016/j.abb.2008.01.010
Mao, Q. Q., Guan, M. Y., Lu, K. X., Du, S. T., Fan, S. K., Ye, Y. Q., et al. (2014). Inhibition of nitrate transporter 1.1-controlled nitrate uptake reduces cadmium uptake in Arabidopsis. Plant Physiol. 166, 934–944. doi: 10.1104/pp.114.243766
McLoughlin, F., Galvan-Ampudia, C. S., Julkowska, M. M., Caarls, L., Does, D., Laurière, C., et al. (2012). The Snf1-related protein kinases SnRK2.4 and SnRK2.10 are involved in maintenance of root system architecture during salt stress. Plant J. 72, 436–449. doi: 10.1111/j.1365-313X.2012.05089.x
Meena, M. K., Sardar, A., and Chattopadhyay, D. (2019). Decoding and relay of calcium signals by CBL-CIPK module in plants. Proc. Indian Natl. Sci. Acad. 85, 143–156. doi: 10.16943/ptinsa/2018/49503
Monaghan, J., Matschi, S., Shorinola, O., Rovenich, H., Matei, A., Segonzac, C., et al. (2014). The calcium-dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turnover. Cell Host Microbe 16, 605–615. doi: 10.1016/j.chom.2014.10.007
Moon, J. Y., Belloeil, C., Ianna, M. L., and Shin, R. (2019). Arabidopsis CNGC family members contribute to heavy metal ion uptake in plants. Int. J. Mol. Sci. 20:413. doi: 10.3390/ijms20020413
Mori, K., Renhu, N., Naito, M., Nakamura, A., Shiba, H., Yamamoto, T., et al. (2018). Ca2+-permeable mechanosensitive channels MCA1 and MCA2 mediate cold-induced cytosolic Ca2+ increase and cold tolerance in Arabidopsis. Sci. Rep. 8:550. doi: 10.1038/s41598-017-17483-y
Mozafari, H. (2013). Calcium and L-histidine interaction on growth improvement of three tomato cultivars under nickel stress. Acta Biol. Szeged. 57, 131–144. doi: 10.1080/01904167.2015.1061549
Nayyar, H., Kaur, R., Kaur, S., and Singh, R. (2014). γ-Aminobutyric acid (GABA) imparts partial protection from heat stress injury to rice seedlings by improving leaf turgor and upregulating osmoprotectants and antioxidants. J. Plant Growth Regul. 33, 408–419. doi: 10.1007/s00344-013-9389-6
Nie, S., Zhang, M., and Zhang, L. (2017). Genome-wide identification and expression analysis of calmodulin-like (CML) genes in Chinese cabbage (Brassica rapa L. ssp. pekinensis). BMC Genomics 18, 1–12. doi: 10.1186/s12864-017-4240-2
O'Brien, J. A., Vega, A., Bouguyon, E., Krouk, G., Gojon, A., Coruzzi, G., et al. (2016). Nitrate transport, sensing, and responses in plants. Mol. Plant 9, 837–856. doi: 10.1016/j.molp.2016.05.004
Oldroyd, G. E. D. (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11, 252–263. doi: 10.1038/nrmicro2990
Pan, W., Shen, J., Zheng, Z., Yan, X., Shou, J., Wang, W., et al. (2018). Overexpression of the tibetan plateau annual wild barley (Hordeum spontaneum) HsCIPKs enhances rice tolerance to heavy metal toxicities and other abiotic stresses. Rice 11:51. doi: 10.1186/s12284-018-0242-1
Pandey, G. K., Grant, J. J., Cheong, Y. H., Kim, B. G., Li le, G., and Luan, S. (2008). Calcineurin-B-like protein CBL9 interacts with target kinase CIPK3 in the regulation of ABA response in seed germination. Mol. Plant 1, 238–248. doi: 10.1093/mp/ssn003
Pandey, N., Ranjan, A., Pant, P., Tripathi, R. K., Ateek, F., Pandey, H. P., et al. (2013). CAMTA 1 regulates drought responses in Arabidopsis thaliana. BMC Genomics. 14:216. doi: 10.1186/1471-2164-14-216
Park, C. Y., Heo, W. D., Yoo, J. H., Lee, J. H., Kim, M. C., et al. (2004). Pathogenesis-related gene expression by specific calmodulin isoforms is dependent on NIM1, a key regulator of systemic acquired resistance. Mol. Cells 18, 207–213.
Patharkar, O. R., and Cushman, J. C. (2000). A stress-induced calcium-dependent protein kinase from Mesembryanthemum crystallinum phosphorylates a two-component pseudo-response regulator. Plant J. 24, 679–691. doi: 10.1046/j.1365-313x.2000.00912.x
Pittman, J. K., and Hirschi, K. D. (2016). CAX-ing a wide net: Cation/H(+) transporters in metal remediation and abiotic stress signalling. Plant Biol. 18, 741–749. doi: 10.1111/plb.12460
Quan, R., Lin, H., Mendoza, I., Zhang, Y., Cao, W., Yang, Y., et al. (2007). SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 19, 1415–1431. doi: 10.1105/tpc.106.042291
Qudeimat, E., and Frank, W. (2009). Ca2+ signatures the role of Ca 2+-ATPases. Plant Signal. Behav. 4, 350–352. doi: 10.4161/psb.4.4.8218
Rahman, H., Xu, Y. P., Zhang, X. R., and Cai, X. Z. (2016). Brassica napus genome possesses extraordinary high number of CAMTA genes and CAMTA3 contributes to PAMP triggered immunity and resistance to Sclerotinia sclerotiorum. Front. Plant Sci. 7:581. doi: 10.3389/fpls.2016.00581
Ranf, S., Eschen-Lippold, L., Pecher, P., Lee, J., and Scheel, D. (2011). Interplay between calcium signalling and early signalling elements during defence responses to microbe-or damage-associated molecular patterns. Plant J. 68, 100–113 doi: 10.1111/j.1365-313X.2011.04671.x
Rao, S. S., El-Habbak, M. H., Havens, W. M., Singh, A., Zheng, D., Vaughn, L., et al. (2014). Overexpression of GmCaM4 in soybean enhances resistance to pathogens and tolerance to salt stress. Mol. Plant Pathol. 15, 145–160. doi: 10.1111/mpp.12075
Romeis, T., Ludwig, A. A., Martin, R., and Jones, J. D. (2001). Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J. 20, 5556–5567. doi: 10.1093/emboj/20.20.5556
Ruiz, J. M., Sánchez, E., García, P. C., López-Lefebre, L. R., Rivero, R. M., and Romero, L. (2002). Proline metabolism and NAD kinase activity in greenbean plants subjected to cold-shock. Phytochemistry 59, 473–478. doi: 10.1016/S0031-9422(01)00481-2
Saijo, Y., Hata, S., Kyozuka, J., Shimamoto, K., and Izui, K. (2000). Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. 23, 319–327. doi: 10.1046/j.1365-313x.2000.00787.x
Sakuma, Y., Maruyama, K., Osakabe, Y., Qin, F., Seki, M., Shinozaki, K., et al. (2006). Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18, 1292–1309. doi: 10.1105/tpc.105.035881
Salehi-Lisar, S. Y., and Bakhshayeshan-Agdam, H. (2016). “Drought stress in plants: causes, consequences, and tolerance,” in Drought Stress Tolerance in Plants, Vol. 1, eds M. Hossain, S. Wani, S. Bhattacharjee, D. Burritt, L. S. S. Tran (Cham: Physiology and Biochemistry Springer), 1–16. doi: 10.1007/978-3-319-28899-4_1
Sanyal, S. K., Kanwar, P., Yadav, A. K., Sharma, C., Kumar, A., and Pandey, G. K. (2017). Arabidopsis CBL interacting protein kinase 3 interacts with ABR1, an APETALA2 domain transcription factor, to regulate ABA responses. Plant Sci. 254, 48–59. doi: 10.1016/j.plantsci.2016.11.004
Scholz, S. S., Vadassery, J., Heyer, M., Reichelt, M., Bender, K. W., Snedden, W. A., et al. (2014). Mutation of the Arabidopsis calmodulin-like protein CML37 deregulates the jasmonate pathway and enhances susceptibility to herbivory. Mol. Plant 7, 1712–1726. doi: 10.1093/mp/ssu102
Schönknecht, G. (2013). Calcium signals from the vacuole. Plants 2, 589–614. doi: 10.3390/plants2040589
Seo, P. J., Xiang, F., Qiao, M., Park, J. Y., Lee, Y. N., Kim, S. G., et al. (2009). The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 151, 275–289. doi: 10.1104/pp.109.144220
Shen, L., Zhuang, B., Wu, Q., Zhang, H., Nie, J., Jing, W., et al. (2019). Phosphatidic acid promotes the activation and plasma membrane localization of MKK7 and MKK9 in response to salt stress. Plant Sci. 287:110190. doi: 10.1016/j.plantsci.2019.110190
Shen, Q., Fu, L., Su, T., Ye, L., Huang, L., Kuang, L., et al. (2020). Calmodulin HvCaM1 negatively regulates salt tolerance via modulation of HvHKT1s and HvCAMTA4. Plant Physiol. 183, 1650–1662. doi: 10.1104/pp.20.00196
Shim, J. S., Oh, N., Chung, P. J., Kim, Y. S., Choi, Y. D., and Kim, J. K. (2018). Overexpression of OsNAC14 improves drought tolerance in rice. Front. Plant Sci. 9:310. doi: 10.3389/fpls.2018.00310
Singh, S., Katzer, K., Lambert, J., Cerri, M., and Parniske, M. (2014). CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 15, 139–152. doi: 10.1016/j.chom.2014.01.011
Singh, S., and Parniske, M. (2012). Activation of calcium- and calmodulin-dependent protein kinase (CCaMK), the central regulator of plant root endosymbiosis. Curr. Opin. Plant Biol. 15, 444–453. doi: 10.1016/j.pbi.2012.04.002
Srivastava, A. K., Rai, A. N., Patade, V. Y., and Suprasanna, P. (2013). “Calcium signaling and its significance in alleviating salt stress in plants,” in Salt Stress in Plants, eds P. Ahmad, M. M. Azooz, M. N. V. Prasad (New York, NY: Springer), 197–218. doi: 10.1007/978-1-4614-6108-1_9
Straub, T., Ludewig, U., and Neuhaeuser, B. (2017). The kinase CIPK23 inhibits ammonium transport in Arabidopsis thaliana. Plant Cell 29, 409–422. doi: 10.1105/tpc.16.00806
Stravinskiene, V., and Račaite, M. (2014). Impact of cadmium and zinc on the growth of white clover (Trifolium repens L.) shoots and roots. Pol. J. Environ. Stud. 23, 1355–1359.
Sun, J., and Zheng, N. (2015). Molecular mechanism underlying the plant NRT1. 1 dual-affinity nitrate transporter. Front. Physiol. 6:386. doi: 10.3389/fphys.2015.00386
Sunkar, R., Kaplan, B., Bouché, N., Arazi, T., Dolev, D., Talke, I. N., et al. (2000). Expression of a truncated tobacco NtCBP4 channel in transgenic plants and disruption of the homologous Arabidopsis CNGC1 gene confer Pb2+ tolerance. Plant J. 24, 533–542. doi: 10.1046/j.1365-313x.2000.00901.x
Swarbreck, S. M., Colaço, R., and Davies, J. M. (2013). Plant calcium-permeable channel. Plant Physiol. 163, 514–522. doi: 10.1104/pp.113.220855
Takezawa, D., and Minami, A. (2004). Calmodulin-binding proteins in bryophytes: Identification of abscisic acid-, cold-, and osmotic stress-induced genes encoding novel membrane-bound transporter-like proteins. Biochem. Biophys. Res. Commun. 317, 428–436. doi: 10.1016/j.bbrc.2004.03.052
Teige, M., Scheikl, E., Eulgem, T., Dóczi, R., Ichimura, K., Shinozaki, K., et al. (2004). The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol Cell. 15, 141–152. doi: 10.1016/j.molcel.2004.06.023
Teng, Y., Liang, Y., Wang, M., Mai, H., and Ke, L. (2019). Nitrate Transporter 1.1 is involved in regulating flowering time via transcriptional regulation of FLOWERING LOCUS C in Arabidopsis thaliana. Plant Sci. 284, 30–36. doi: 10.1016/j.plantsci.2019.04.002
Tian, W. H., Ye, J. Y., Cui, M. Q., Chang, J. B., Liu, Y., Li, G. X., et al. (2021). A transcription factor STOP1-centered pathway coordinates ammonium and phosphate acquisition in Arabidopsis. Mol. Plant 14, 1554–1568. doi: 10.1016/j.molp.2021.06.024
Townley, H. E., and Knight, M. R. (2002). Calmodulin as a potential negative regulator of Arabidopsis COR gene expression. Plant Physiol. 128, 1169–1172. doi: 10.1104/pp.010814
Trewavas, A. J., and Malhó, R. (1998). Ca2+ signalling in plant cells: the big network! Curr. Opin. Plant Biol. 1, 428–433. doi: 10.1016/S1369-5266(98)80268-9
Truman, W., Sreekanta, S., Lu, Y., Bethke, G., Tsuda, K., Katagiri, F., et al. (2013). The CALMODULIN-BINDING PROTEIN60 family includes both negative and positive regulators of plant immunity. Plant Physiol. 163, 1741–1751. doi: 10.1104/pp.113.227108
Vadassery, J., Reichelt, M., Hause, B., Gershenzon, J., Boland, W., and Mithöfer, A. (2012). CML42-mediated calcium signaling coordinates responses to Spodoptera herbivory and abiotic stresses in Arabidopsis. Plant Physiol. 159, 1159–1175. doi: 10.1104/pp.112.198150
Valivand, M., Amooaghaie, R., and Ahadi, A. (2019). Interplay between hydrogen sulfide and calcium/calmodulin enhances systemic acquired acclimation and antioxidative defense against nickel toxicity in zucchini. Environ Exp Bot. 158, 40–50. doi: 10.1016/j.envexpbot.2018.11.006
Valliyodan, B., and Nguyen, H. T. (2006). Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr. Opin. Plant Biol. 9, 189–195. doi: 10.1016/j.pbi.2006.01.019
Verkhratsky, A., and Parpura, V. (2014). Calcium signalling and calcium channels: evolution and general principles. Eur. J. Pharmacol. 739, 1–3. doi: 10.1016/j.ejphar.2013.11.013
Vlot, A., Dempsey, D. A., and Klessig, D. F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. doi: 10.1146/annurev.phyto.050908.135202
Wan, D., Li, R., Zou, B., Zhang, X., Cong, J., Wang, R., et al. (2012). Calmodulin-binding protein CBP60g is a positive regulator of both disease resistance and drought tolerance in Arabidopsis. Plant Cell Rep. 31, 1269–1281. doi: 10.1007/s00299-012-1247-7
Wang, L., Cheng, M., Chu, Y., Li, X., Chen, D. D., Huang, X., et al. (2016). Responses of plant calmodulin to endocytosis induced by rare earth elements. Chemosphere 154, 408–415. doi: 10.1016/j.chemosphere.2016.03.106
Wang, L., Tsuda, K., Sato, M., Cohen, J. D., Katagiri, F., and Glazebrook, J. (2009). Arabidopsis CaM binding protein CBP60g contributes to MAMP-induced SA accumulation and is involved in disease resistance against Pseudomonas syringae. PLoS Pathog. 5:e1000301. doi: 10.1371/journal.ppat.1000301
Wang, Y. Y., Hsu, P. K., and Tsay, Y. F. (2012). Uptake, allocation and signaling of nitrate. Trends Plant Sci. 17, 458–467. doi: 10.1016/j.tplants.2012.04.006
Weckwerth, P., Ehlert, B., and Romeis, T. (2015). ZmCPK1, a calcium-independent kinase member of the Zea mays CDPK gene family, functions as a negative regulator in cold stress signalling. Plant Cell Environ. 38, 544–558. doi: 10.1111/pce.12414
Wen, Z., and Kaiser, B. N. (2018). Unraveling the functional role of NPF6 transporters. Front. Plant Sci. 9:973. doi: 10.3389/fpls.2018.00973
Whalley, H. J., and Knight, M. R. (2013). Calcium signatures are decoded by plants to give specific gene responses. New Phytol. 197, 690–693. doi: 10.1111/nph.12087
Wu, H. C., Luo, D. L., Vignols, F., and Jinn, T. L. (2012). Heat shock-induced biphasic Ca2+ signature and OsCaM1-1 nuclear localization mediate downstream signalling in acquisition of thermotolerance in rice (Oryza sativa L.). Plant Cell Environ. 35, 1543–1557. doi: 10.1111/j.1365-3040.2012.02508.x
Wu, X., Qiao, Z., Liu, H., Acharya, B. R., Li, C., and Zhang, W. (2017). CML20, an Arabidopsis calmodulin-like protein, negatively regulates guard cell ABA signaling and drought stress tolerance. Front. Plant Sci. 8:824. doi: 10.3389/fpls.2017.00824
Wuana, R. A., and Okieimen, F. E. (2011). Heavy metals in contaminated soils: a review of sources, chemistry, risks and best available strategies for remediation. ISRN Ecol. 2011, 1–20. doi: 10.5402/2011/402647
Xu, G. Y., Rocha, P. S., Wang, M. L., Xu, M. L., Cui, Y. C., Li, L. Y., et al. (2011). A novel rice calmodulin-like gene, OsMSR2, enhances drought and salt tolerance and increases ABA sensitivity in Arabidopsis. Planta 234, 47–59. doi: 10.1007/s00425-011-1386-z
Xu, J., Tian, Y. S., Peng, R. H., Xiong, A. S., Zhu, B., Jin, X. F., et al. (2010). AtCPK6, a functionally redundant and positive regulator involved in salt/drought stress tolerance in Arabidopsis. Planta 231, 1251–1260. doi: 10.1007/s00425-010-1122-0
Xuan, Y., Zhou, S., Wang, L., Cheng, Y., and Zhao, L. (2010). Nitric oxide functions as a signal and acts upstream of AtCaM3 in thermotolerance in Arabidopsis seedlings. Plant Physiol. 153, 1895–1906. doi: 10.1104/pp.110.160424
Yang, T., Chaudhuri, S., Yang, L., Du, L., and Poovaiah, B. W. (2010). A calcium/calmodulin-regulated member of the receptor-like kinase family confers cold tolerance in plants. J. Biol. Chem. 285, 7119–7126. doi: 10.1074/jbc.M109.035659
Yang, Y., Zhang, C., Tang, R. J., Xu, H. X., Lan, W. Z., Zhao, F., et al. (2019). Calcineurin B-like proteins CBL4 and CBL10 mediate two independent salt tolerance pathways in Arabidopsis. Int. J. Mol. Sci. 20:2421. doi: 10.3390/ijms20102421
Yao, H. Y., and Xue, H. W. (2018). Phosphatidic acid plays key roles regulating plant development and stress responses. J. Integr. Plant Biol. 60, 851–863. doi: 10.1111/jipb.12655
Yoo, J. H., Park, C. Y., Kim, J. C., Do Heo, W., Cheong, M. S., Park, H. C., et al. (2005). Direct interaction of a divergent CaM isoform and the transcription factor, MYB2, enhances salt tolerance in Arabidopsis. J. Biol. Chem. 280, 3697–3706. doi: 10.1074/jbc.M408237200
Yoshida, R., Umezawa, T., Mizoguchi, T., Takahashi, S., Takahashi, F., and Shinozaki, K. (2006). The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J. Biol. Chem. 281, 5310–5318. doi: 10.1074/jbc.M509820200
Yu, L., Nie, J., Cao, C., Jin, Y., Yan, M., Wang, F., et al. (2010). Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 188, 762–773. doi: 10.1111/j.1469-8137.2010.03422.x
Yuan, P., Jauregui, E., Du, L., Tanaka, K., and Poovaiah, B. W. (2017). Calcium signatures and signaling events orchestrate plant–microbe interactions. Curr. Opin. Plant Biol. 38, 173–183. doi: 10.1016/j.pbi.2017.06.003
Yuan, P., Yang, T., and Poovaiah, B. W. (2018). Calcium signaling-mediated plant response to cold stress. Int. J. Mol Sci. 19, 1–11. doi: 10.3390/ijms19123896
Zeng, H., Xu, L., Singh, A., Wang, H., Du, L., and Poovaiah, B. W. (2015). Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front. Plant Sci. 6, 600. doi: 10.3389/fpls.2015.00600
Zeng, H., Zhang, Y., Zhang, X., Pi, E., and Zhu, Y. (2017). Analysis of EF-hand proteins in soybean genome suggests their potential roles in environmental and nutritional stress signaling. Front. Plant Sci. 8:877. doi: 10.3389/fpls.2017.00877
Zhang, W., Zhou, R. G., Gao, Y. J., Zheng, S. Z., Xu, P., Zhang, S. Q., et al. (2009). Molecular and genetic evidence for the key role of AtCaM3 in heat-shock signal transduction in Arabidopsis. Plant Physiol. 149, 1773–1784. doi: 10.1104/pp.108.133744
Zhang, X., Wang, L., Zhou, A., Zhou, Q., and Huang, X. (2016). Alterations in cytosol free calcium in horseradish roots simultaneously exposed to lanthanum (III) and acid rain. Ecotoxicol. Environ. Saf. 126, 62–70. doi: 10.1016/j.ecoenv.2015.12.014
Zhang, Y., Xu, S., Ding, P., Wang, D., Cheng, Y. T., He, J., et al. (2010). Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc. Natl. Acad. Sci. U.S.A. 107, 18220–18225. doi: 10.1073/pnas.1005225107
Zhu, X., Perez, M., Aldon, D., and Galaud, J. P. (2017). Respective contribution of CML8 and CML9, two Arabidopsis calmodulin-like proteins, to plant stress responses. Plant Signal. Behav. 12:e1322246. doi: 10.1080/15592324.2017.1322246
Zhu, Y., Yan, J., Liu, W., Liu, L., Sheng, Y., Sun, Y., et al. (2016). Phosphorylation of a NAC transcription factor by a calcium/calmodulin-dependent protein kinase regulates abscisic acid-induced antioxidant defense in maize. Plant Physiol. 171, 1651–1664. doi: 10.1104/pp.16.00168
Zhu, Y., Yang, H., Mang, H. G., and Hua, J. (2011). Induction of BAP1 by a moderate decrease in temperature is mediated by ICE1 in Arabidopsis. Plant Physiol. 155, 580–588. doi: 10.1104/pp.110.169466
Keywords: calcium, calmodulin, genetic intervention, signaling, stress
Citation: Patra N, Hariharan S, Gain H, Maiti MK, Das A and Banerjee J (2021) TypiCal but DeliCate Ca++re: Dissecting the Essence of Calcium Signaling Network as a Robust Response Coordinator of Versatile Abiotic and Biotic Stimuli in Plants. Front. Plant Sci. 12:752246. doi: 10.3389/fpls.2021.752246
Received: 02 August 2021; Accepted: 27 September 2021;
Published: 25 November 2021.
Edited by:
Wusirika Ramakrishna, Central University of Punjab, IndiaReviewed by:
Abdul Azeez, Washington State University, United StatesCopyright © 2021 Patra, Hariharan, Gain, Maiti, Das and Banerjee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joydeep Banerjee, amJhbmVyamVlYmlvdGVjaEBnbWFpbC5jb20=; am95ZGVlcEBhZ2ZlLmlpdGtncC5hYy5pbg==; Arpita Das, YXJwaXRhY29oQGdtYWlsLmNvbQ==
†ORCID: Neelesh Patra orcid.org/0000-0003-1867-4807
Mrinal K. Maiti orcid.org/0000-0001-7803-4356
Joydeep Banerjee orcid.org/0000-0003-3856-88762
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.