- Dipartimento di Bioscienze, Università degli Studi di Milano, Milano, Italy
Anthocyanins represent the major red, purple, and blue pigments in many flowers, fruits, vegetables, and cereals. They are also recognized as important health-promoting components in the human diet with protective effects against many chronic diseases, including cardiovascular diseases, obesity, and cancer. Anthocyanin biosynthesis has been studied extensively, and both biosynthetic and key regulatory genes have been isolated in many plant species. Here, we will provide an overview of recent progress in understanding the anthocyanin biosynthetic pathway in plants, focusing on the transcription factors controlling activation or repression of anthocyanin accumulation in cereals and fruits of different plant species, with special emphasis on the differences in molecular mechanisms between monocot and dicot plants. Recently, new insight into the transcriptional regulation of the anthocyanin biosynthesis, including positive and negative feedback control as well as epigenetic and post-translational regulation of MYB-bHLH-WD40 complexes, has been gained. We will consider how knowledge of regulatory mechanisms has helped to produce anthocyanin-enriched foods through conventional breeding and metabolic engineering. Additionally, we will briefly discuss the biological activities of anthocyanins as components of the human diet and recent findings demonstrating the important health benefits of anthocyanin-rich foods against chronic diseases.
Introduction
Anthocyanin synthesis is one of the most studied biosynthetic pathways in plants. Besides providing the major red, purple, violet, and blue pigmentation in flowers and fruits for attracting pollinators and seed dispersers, anthocyanins act as antioxidants in plants and are involved in both abiotic and biotic stresses, such as UV radiation, cold temperatures, drought, and in defense against pathogens and herbivores (Sarma and Sharma, 1999; Lorenc-Kukuła et al., 2005; Gould and Lister, 2006). Anthocyanins are health-protecting components of our daily diet found to activate endogenous antioxidant defenses and suppress inflammatory mediators (Speciale et al., 2013; Lee et al., 2014; Ullah et al., 2019; Kozłowska and Dzierżanowski, 2021). A number of studies suggest that they have protective effects against cardiovascular diseases, obesity, cancer, and neurodegenerative diseases (Tsuda, 2012; Li et al., 2017b; Salehi et al., 2020). For this reason, there has been growing interest in the identification of regulatory genes controlling anthocyanin biosynthesis in major crops as targets for both metabolic engineering and breeding programs.
Anthocyanins are the final products of a specific branch of flavonoid biosynthesis, also producing flavonols, phlobaphenes, and proanthocyanidins (Figure 1). Flavonols, proanthocyanidins, and anthocyanins are water-soluble and almost ubiquitous, whereas phlobaphenes are alcohol-soluble and water-insoluble phenolics mostly produced in maize, wheat, and sorghum (Casas et al., 2014; Ibraheem et al., 2015; Lachman et al., 2017). The first biosynthetic genes of the anthocyanin pathway (i.e., CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; F3′H, flavanone 3′-hydroxylase; F3′5′H, flavanone 3′5′-hydroxylase), which have been termed early biosynthetic genes (EBGs), are involved in the synthesis of dihydroflavonols (i.e., dihydrokaempferol, dihydroquercetin, and dihydromyricetin). Dihydroflavonols are then converted into anthocyanidins (i.e., pelargonidin, cyanidin, and delphinidin) by the late biosynthetic genes (LBGs), encoding dihydroflavonol reductase (DFR), flavonol synthase (FLS), and anthocyanidin synthase/leucoanthocyanidin dioxygenase (ANS/LDOX), and to the anthocyanidin derivatives peonidin, malvidin, and petunidin by methyltransferases (MT; Figure 1). Anthocyanidins are glycosylated by UDP-glucose flavonoid 3-O-glucosyltransferase (UFGT), acylated by anthocyanin acyltransferase (AAT), and then transferred to the vacuole by glutathione S-transferase (GST). The stability of different anthocyanins is highly influenced by glycosylation and acylation, both aromatic and aliphatic, and influence their color and degradation in plant tissues (Glover and Martin, 2012). Anthocyanin 3-monosaccharides (e.g., 3-glucoside) are the most common, anthocyanin 3-disaccharides (e.g., 3-rutinoside) are generally more stable than 3-monosaccharides, whereas anthocyanin 3,5 disaccharides are also common, but less stable (Zhang et al., 2014a). The sugar residues of anthocyanins are often acylated with aromatic (e.g., p-coumaric, caffeic, and ferulic) or aliphatic acids (e.g., malonic and acetic acid). Anthocyanin acylation increases the stability, allowing intra- and inter-molecular co-pigmentation, and causes a shift toward the blue color (Zhang et al., 2014a).
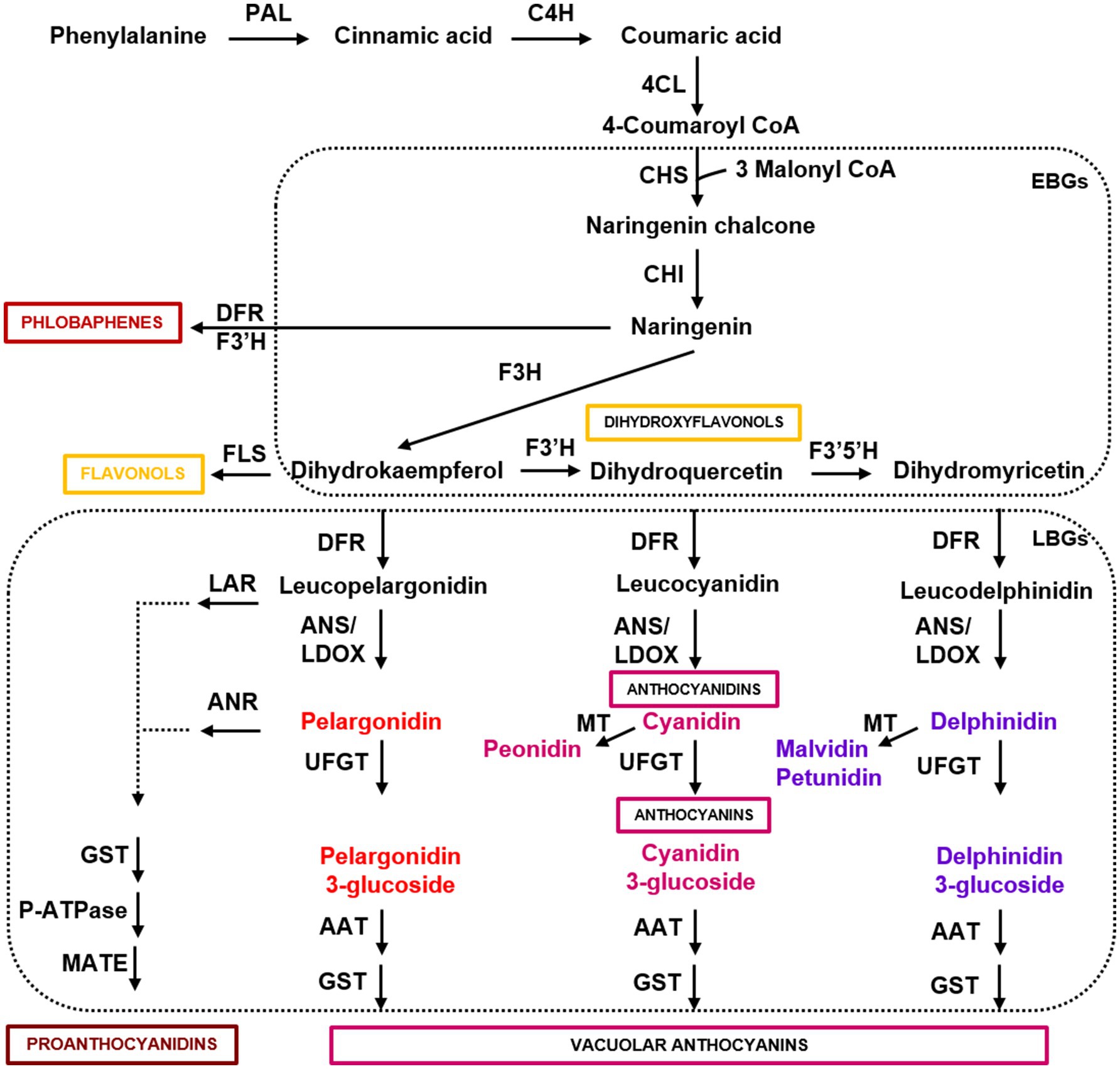
Figure 1. Simplified scheme of the flavonoid pathway, comprising the general phenylpropanoid pathway, the anthocyanin branch, and other subgroups of flavonoid end-products. Enzyme names are abbreviated as follows: PAL, phenylalanine ammonia lyase; C4H, cinnamic acid 4-hydroxylase; 4CL, 4 coumarate CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; F3′H, flavanone 3′-hydroxylase; F3′5′H, flavanone 3′5′-hydroxylase; DFR, dihydroflavonol reductase; FLS, flavonol synthase; ANS/LDOX, anthocyanidin synthase/leucoanthocyanidin dioxygenase; UFGT, UDP-flavonoid glucosyl transferase; ANR, anthocyanidin reductase; LAR, anthocyanidin reductase; MT, methyltransferase; AAT, anthocyanin acyltransferase; GST, glutathione S-transferase; P-ATPase, P-type ATPase proton pump; MATE, MATE-type transporter. Boxes indicate the early and late biosynthetic genes (EBGs and LBGs) of the flavonoid pathway in dicots.
Anthocyanin biosynthesis has been studied extensively, and key regulatory genes have been identified in many plant species (Petroni and Tonelli, 2011; Patra et al., 2013c; Chaves-Silva et al., 2018; Liu et al., 2018). Here, we focus on recent progress in understanding the positive and negative regulatory mechanisms of anthocyanin biosynthesis. Activators and repressors of anthocyanins in maize and Arabidopsis will be firstly described as examples of the regulatory systems in monocots and dicots, respectively, with particular emphasis on the important post-transcriptional mechanisms of regulation involving miRNAs identified in Arabidopsis and thereafter in other dicots. Finally, recent findings demonstrating the important health benefits of anthocyanin-rich food against chronic diseases will be discussed.
Activators of Anthocyanin Biosynthesis in Maize and Arabidopsis
In most species, anthocyanin biosynthesis is activated by the interaction of R2R3-MYB regulatory proteins in combination with bHLH and WD40-type regulatory proteins to form a ternary MBW complex (Figure 2; Allan et al., 2008; Petroni and Tonelli, 2011; Chaves-Silva et al., 2018).
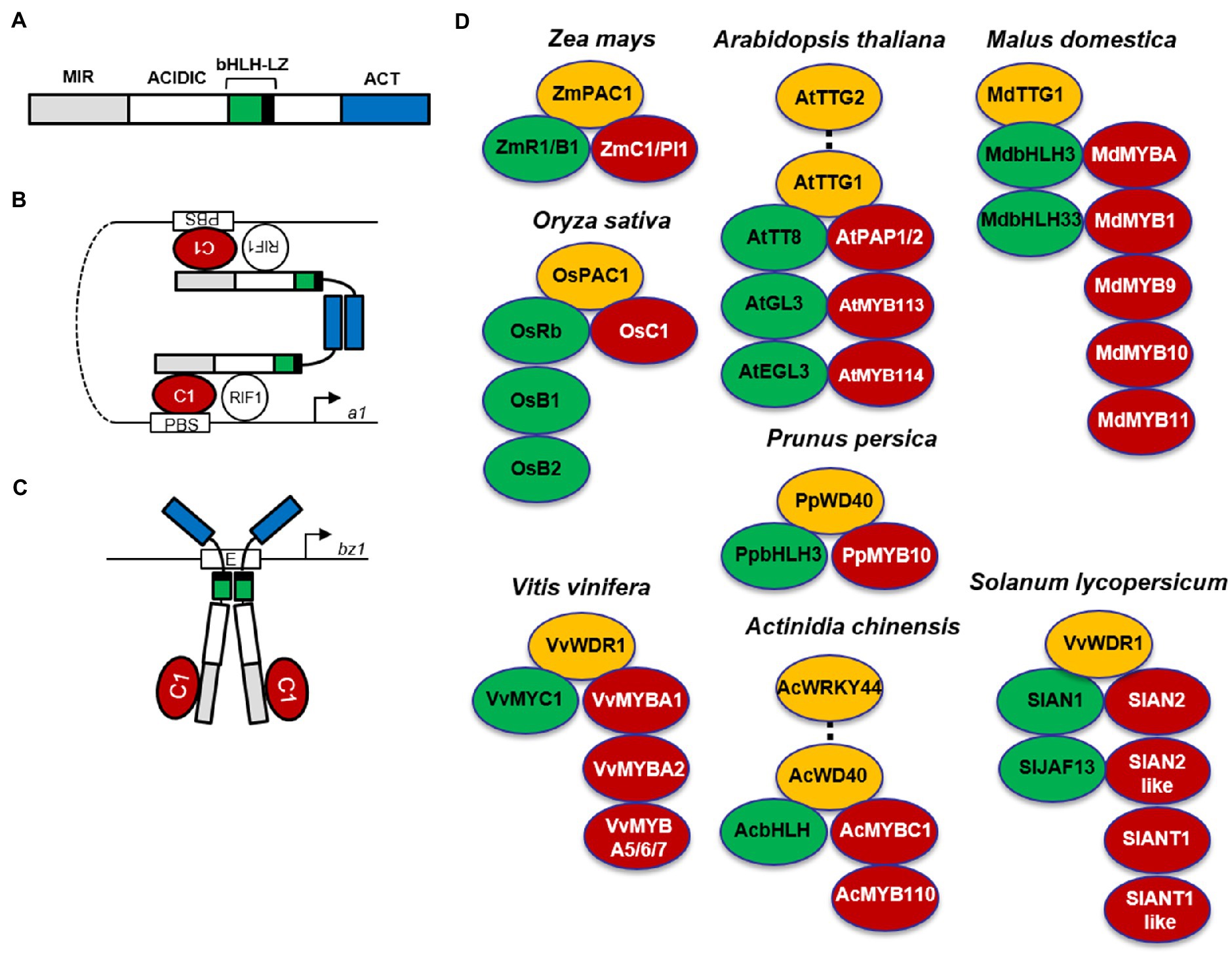
Figure 2. MBW complexes in different plant species. Alternative R1/C1 complexes activating anthocyanin biosynthesis genes in maize. (A) Schematic representation of the R1 functional domains. MIR, MYB interacting region; ACIDIC, acidic region; bHLH, basic helix-loop-helix domain; LZ, leucine zipper; ACT, ACT-like domain. (B) In the first R1/C1 complex, R1 homodimerizes via the ACT-like domain, the bHLH region of R1 interacts with R-interacting factor 1 (RIF1), and the N-terminal acidic region interacts with C1, which binds the P1-binding sites (PBS) found in promoters of some anthocyanin biosynthesis genes, such as a1. (C) In the second R1/C1 complex, R1 homodimerizes via the bHLH-LZ domain, which binds E-box elements, such as those present in the bz1 promoter, and interacts with the C1 protein, providing a strong transcriptional activation domain. (Schemes modified from Kong et al., 2012). (D) MYB-bHLH-WD40 (MBW) complexes involved in anthocyanin biosynthesis in different plant species. MYBs are indicated in red, bHLHs in green, and WD40s in yellow. The Arabidopsis MBWW complex (PAP1-TT8-TTG1-TTG2) controlling a subset of proanthocyanidin LBGs is indicated by a dotted line connecting TTG2 and TTG1. The MBWW complex (AcMYBC1-AcbHLH-AcWD40-AcWRKY44) controlling F3′H and F3′5′H in kiwifruit (Actinidia chinensis) is indicated by a dotted line connecting AcWD40-AcWRKY44.
In the monocot maize, the regulatory genes of anthocyanin biosynthesis are divided in two families (C1/Pl1 and R1/B1), encoding MYB and bHLH transcription factors, respectively. The R1/B1 genes (R1, Red1; B1, Booster1; Sn1, Scutellar node1; Lc1, Leaf color1; Hopi1), whose expression is tissue-specific, determine the distribution of pigment production in different tissues, whereas Colorless1 (C1) in the seed or Purple plant1 (Pl1) in plant tissues regulates anthocyanin accumulation during development and in response to light. In maize seed, the dominant R1 C1 genes activate anthocyanin pigmentation of the aleurone layer in blue corn, whereas the dominant B1 Pl1 genes induce anthocyanin synthesis in the pericarp of purple corn (Petroni et al., 2014). MYB and bHLH transcription factors form a heterodimer activating all anthocyanin biosynthetic genes in a coordinate manner (Hernandez et al., 2004). By contrast, the phlobaphene branch only requires the maize MYB PERICARP COLOR1 (P1) to activate a subset of biosynthetic genes (i.e., CHS, CHI, DFR, and FLS; Ferreyra et al., 2010). In addition, the PALE ALEURONE COLOR1 (PAC1) WD40 protein cooperates with B1 or R1 proteins for full activation of anthocyanin biosynthetic genes in the seed, probably via a ternary MBW complex similar to that identified in other species (Figure 2D; Carey et al., 2004).
Despite being activated coordinately, anthocyanin biosynthetic genes lack conserved cis-regulatory elements. Therefore, their coordinate activation is proposed to be achieved by two different R1/C1 complexes, each able to bind specific cis-elements and thus to activate a subset of anthocyanin biosynthetic genes (Tuerck and Fromm, 1994). These alternative R1/C1 complexes depend on two different dimerization domains found in bHLH proteins (Figure 2A). In the proposed model, when R1 homodimerizes via the ACT-like domain (Figure 2B), the bHLH region of R1 interacts with R-interacting factor 1 (RIF1), an EMSY-like maize nuclear factor, necessary to the R1/C1 complex for transcriptional activation of anthocyanin biosynthetic genes through increased histone acetylation of the promoter region (Hernandez et al., 2007), whereas the N-terminal acidic region interacts with the MYB C1, necessary to bind the P1-binding sites (PBS) in the a1 promoter (Sainz et al., 1997). When R1 homodimerizes via the bHLH-LZ domain (Figure 2C), it directly binds E-box elements in the bronze1 (bz1) promoter, and interacts with the C1 protein, necessary to provide a strong transcriptional activation domain (Kong et al., 2012).
In the dicot Arabidopsis, EBGs leading to the production of flavonols (Figure 1) are activated by functionally redundant R2R3-MYB regulatory genes (MYB11/12/111), whereas the activation of LBGs, leading to the production of proanthocyanidins in seeds and anthocyanins in vegetative tissues (Figure 1), requires the MYB-bHLH-WD40 complex (Figure 2D; Baudry et al., 2004; Stracke et al., 2007; Gonzalez et al., 2008). The MBW complex activating anthocyanin synthesis in vegetative tissues includes the WD40 factor TRANSPARENT TESTA GLABRA1 (TTG1) and different bHLH and MYB transcription factors (Figure 2D). The bHLH TRANSPARENT TESTA8 (TT8), GLABRA3 (GL3), and ENHANCER OF GLABRA3 (EGL3) genes have partially redundant roles in the regulation of anthocyanins in seedlings (Zhang et al., 2003; Gonzalez et al., 2008). Among R2R3-MYB factors, MYB75/PRODUCTION OF ANTHOCYANIN PIGMENT1 (PAP1), MYB90/PRODUCTION OF ANTHOCYANIN PIGMENT2 (PAP2), MYB113, and MYB114 can form multiple MBW complexes with EGL3, GL3, and TT8 (Gonzalez et al., 2008). On the other hand, seed-specific activation of proanthocyanidin synthesis requires the activity of an MBW complex comprising the R2R3-MYB protein MYB123/TRANSPARENT TESTA2 (TT2), TT8, and TTG1 (Ramsay and Glover, 2005). A newly identified complex requires the additional interaction between TTG1 and TTG2 WRKY transcription factors, thus forming an MBWW complex necessary to activate a subset of LBGs (i.e., TRANSPARENT TESTA12, TT12 and Autoinhibited H(+)-ATPase Isoform 10, AHA10) encoding a MATE type transporter and a P-type ATPase proton pump, functioning as vacuolar proanthocyanidin transporters (Figure 2D; Gonzalez et al., 2016; Lloyd et al., 2017). When overexpressed, PAP1 and PAP2 also weakly activate EBGs (Tohge et al., 2005), but in pap1 knock-out plants or when all four MYBs (PAP1, PAP2, MYB113, and MYB114) were silenced through RNAi, only LBGs were found to be significantly reduced (Gonzalez et al., 2008).
Interestingly, a two-step activation of anthocyanin biosynthesis has been proposed in Arabidopsis (Albert et al., 2014). TT8 is regulated at the transcriptional level and can regulate its own expression through a positive feedback mechanism. In etiolated seedlings, an initial PAP1/EGL3/TTG1 complex may activate TT8, which can then form MBW complexes that autoactivates TT8 through a positive feedback mechanism and confers a strong induction of LBGs and anthocyanin accumulation (Baudry et al., 2006). In seedlings, TT8 expression is therefore controlled by three different MBW complexes, including PAP1, TTG1, and one of three bHLHs (TT8/EGL3/GL3), whereas three other MBW complexes consisting of TTG1, TT2/MYB5, and TT8/EGL3 control proanthocyanidin accumulation in seeds (Xu et al., 2015). Overall, it appears that despite the variable function of transcription factors in the MBW complex, the complex is conserved in monocots and dicots, but the mechanisms of regulation of the regulatory genes have diverged. In the monocot maize, each regulatory gene is independently regulated, whereas in dicots a regulatory loop exists, in which one regulatory gene controls the expression of another in a tissue- and developmental-specific manner (Carey et al., 2004; Xu et al., 2015). Consistent with this divergence, based on phylogenetic analyses, the R2R3-MYBs from Arabidopsis (i.e., MYB75/PAP1, MYB90/PAP2, MYB113, and MYB114) and many other angiosperms belong to subgroup 6, whereas the R2R3-MYBs C1 and Pl1 from maize are more similar to R2R3-MYBs from subgroup 5 (i.e., TT2), which usually activate proanthocyanidins (Dubos et al., 2010).
Considerable research on the upstream activators of MBW complexes that mediate the response to environmental signals has been reported in many reviews (Jaakola, 2013; Zoratti et al., 2014; Xu et al., 2015; Chaves-Silva et al., 2018; Liu et al., 2018), but only ELONGATED HYPOCOTYL 5 (HY5) has been demonstrated as a direct activator of PAP1 in response to light through direct binding to the G-box and ACE element in the PAP1 promoter (Shin et al., 2013). Therefore, HY5 appears to regulate anthocyanin biosynthesis in two ways: via direct binding to the promoter regions of the biosynthetic genes, and by positive regulation of PAP1 transcription (Shin et al., 2013). Other candidates in the light-regulated activation of PAP1 are LIGHT-REGULATED ZINC FINGER PROTEIN 1 (LZF1), which is under control of HY5 (Chang et al., 2008), the NAC domain transcription factor, ANAC078, that also activates TT2, TT8, GL3, and EGL3 in addition to PAP1 (Morishita et al., 2009), and MYB112 which activates the expression of PAP1 upon perception of high light and salt stress, but acts negatively toward MYB12 and MYB111, which both control flavonol biosynthesis (Lotkowska et al., 2015).
Interestingly, the MBW complexes are then stabilized by the transcription factor TCP3, which interacts with R2R3-MYBs and reinforces their transcriptional activation capacity when bound to TT8, thus promoting the expression of LBGs (Figure 3A). TCP3 also interacts with R2R3-MYBs controlling EBGs (MYB11/MYB12/MYB111), thereby enhancing both flavonol and anthocyanin production (Li and Zachgo, 2013). In yeast 2-hybrid, TCP3 was also found to form a heterodimer with the negative regulator R3-MYB protein MYB-LIKE2 (MYBL2; see below), suggesting that TCP3 may stabilize the MBW complex by sequestering MYBL2 and preventing its binding to the bHLH proteins (Figure 3A; Li and Zachgo, 2013). Therefore, TCP3 can be considered an anthocyanin activator with a positive activation mode, based on the interaction with the MBW complex to enhance its transcriptional activity, and a passive activation mode, based on the sequestration of the MYBL2 repressor, known to fine-tune anthocyanin accumulation (Figure 3A).
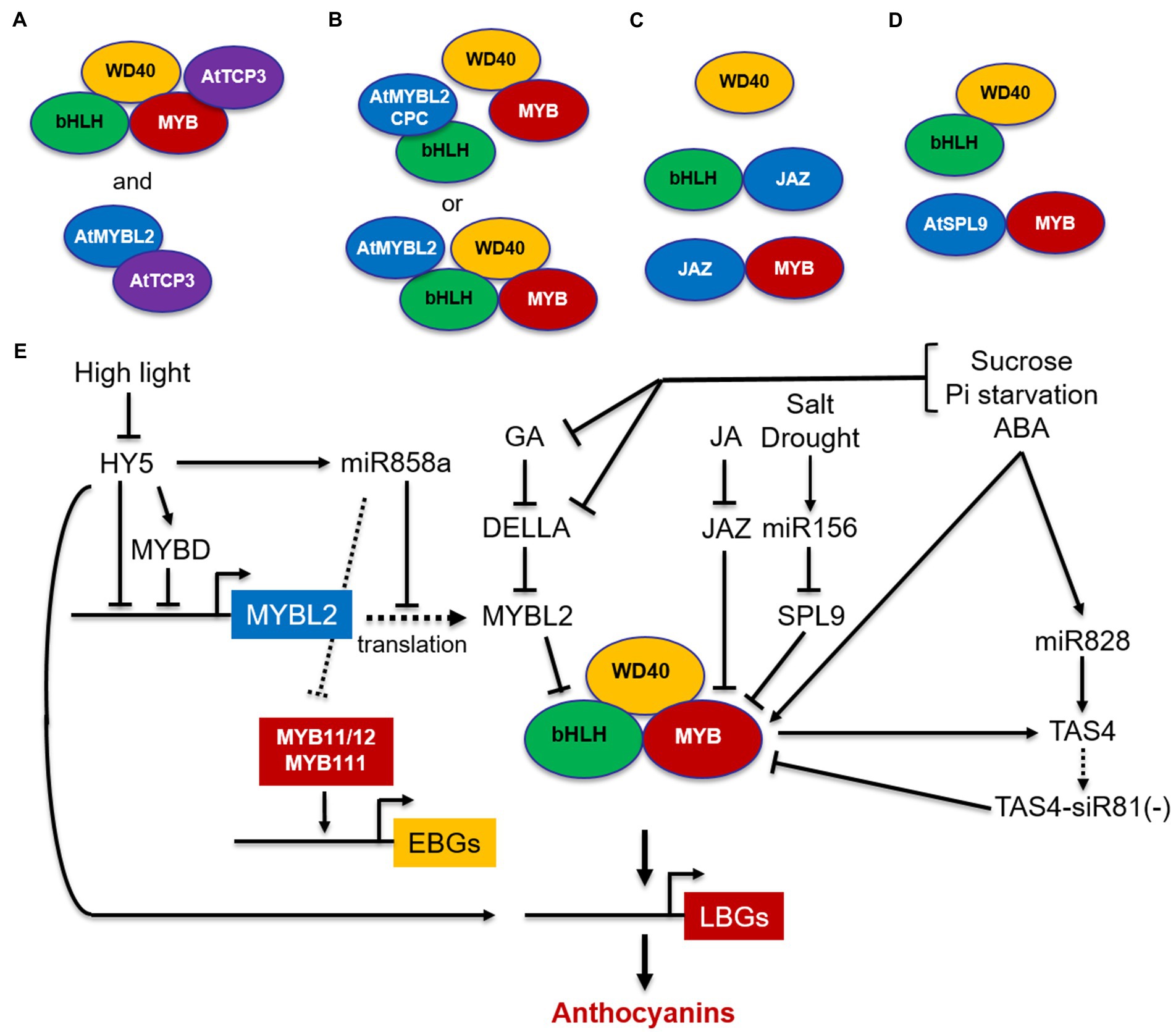
Figure 3. Schematic model of positive and negative regulation of MBW complexes in Arabidopsis. The MYB-bHLH-WD40 (MBW) complex activates the expression of LBGs. (A) TCP3 stabilizes the MBW complex and reinforces its transcriptional capacity by associating with MYBs and by interacting with the MYBL2 repressor, thus preventing its binding to the bHLH proteins; (B) MYBL2 disrupts the MBW complex by interacting with bHLHs or participates in the MBW complex and down-regulates LBGs through its repression domain; (C) JAZ proteins disrupt the MBW complex by interacting with bHLHs or R2R3-MYBs; (D) SPL9 disrupts the MBW complex by interacting with MYBs; (E) simplified model for negative regulation of MYB11/12/111genes and the MBW complex through MYBL2, JAZ, SPL9, and miRNAs. MYBs are indicated in red, bHLHs in green, and WD40s in yellow. TCP3 activator is indicated in violet, whereas MYBL2 and SPL9 repressors are indicated in light blue. Arrows, positive regulation; blunt ends, negative regulation; dashed arrow, translation.
Repression of Anthocyanin Biosynthesis in Maize and Arabidopsis
Significant progress in understanding the negative regulators controlling flavonoid biosynthesis has been made (Li, 2014; Xu et al., 2015; Chen et al., 2019; LaFountain and Yuan, 2021). Most anthocyanin repressors act by disrupting the MBW complexes, but there is increasing evidence of direct repression of biosynthetic genes, post-transcriptional regulation through miRNAs (see also Section “Post-transcriptional Regulation of Anthocyanin Biosynthesis in Arabidopsis and Other Dicots”) and post-translational modifications that fine-tune the activity of MBW complexes.
In maize, the INTENSIFIER1 (IN1) gene encodes a bHLH protein, which is most closely related to the clade bHLH-2 within subgroup IIIf of the bHLH family (Feller et al., 2011). The bHLH-1 proteins regulate anthocyanin biosynthesis, whereas in many plants the bHLH-2 proteins, besides being essential for anthocyanin biosynthesis (Albert et al., 2021), are also involved in regulating proanthocyanidin biosynthesis and vacuolar acidification (Spelt et al., 2002; Butelli et al., 2019; Strazzer et al., 2019). In maize, the IN1 gene inhibits the anthocyanin pathway, possibly by interfering with R1 binding in the MBW complex, since IN1 does not appear to control the transcript level of the B1 and C1 genes (Carey et al., 2004; Feller et al., 2011).
In dicots, the first negative R2R3-MYB regulators identified, AmMYB308 and AmMYB330 from Antirrhinum majus, reduced anthocyanin biosynthesis when overexpressed in tobacco flowers (Tamagnone et al., 1998), whereas in Arabidopsis AtMYB4 was the first R2R3-MYB shown to act as a repressor of phenylpropanoid metabolism (Jin et al., 2000). Both contain an ETHYLENE RESPONSE FACTOR (ERF)-associated amphiphilic repression (EAR) motif in the C-terminal domain and belong to subgroup 4, where many other plant R2R3-MYB repressors are included (Dubos et al., 2010; Chen et al., 2019).
In addition to R2R3-MYB repressors, in Arabidopsis two types of R3-MYB repressors, MYBL2 and CAPRICE (CPC), have been identified. MYBL2 is an R3-MYB repressor derived from subgroup 4 R2R3-MYBs after a partial loss of the R2 domain and characterized by an R3 domain including the bHLH interacting motif and a C-terminal domain containing an EAR motif and a TLLLFR repression motif (Dubos et al., 2008; Matsui et al., 2008). Conversely, CPC consists of a short protein, containing the highly conserved bHLH-interacting motif, but no repressor domains, and belongs to a distinct clade that emerged before the divergence between monocots and dicots (Albert et al., 2014). Specifically, CPC is known to regulate root hair differentiation, trichome initiation, and stomatal formation (Wada et al., 1997), but it has also been shown to be involved in the regulation of anthocyanin biosynthesis in Arabidopsis (Zhu et al., 2009). These two types of R3-MYB repressors are characterized by two different mechanisms of action: CPC only exerts passive repression by titrating the bHLH partners from the MBW complex (Zhu et al., 2009), whereas MYBL2 exerts both a passive repression by preventing the formation of an MBW complex through interaction with the bHLH proteins and an active repression by participating in the MBW complex and actively repressing transcription of LBGs via an EAR repression domain (Figure 3B; Dubos et al., 2008; Matsui et al., 2008). In addition, MYBL2 represses the expression level of MYB and bHLH regulatory genes in seedlings (e.g., TT8, PAP1, and PAP2; Dubos et al., 2008; Matsui et al., 2008) and in turn is activated by TT8, thus reducing the positive feedback loop of TT8 (Matsui et al., 2008).
MYBL2 also appears to mediate the response to environmental stresses and hormones. It is down-regulated by high light and up-regulated by low light, resulting in increased or reduced anthocyanin synthesis, respectively, through its regulation of TT8, PAP1/PAP2 (Dubos et al., 2008; Rowan et al., 2009). MYBL2 response to light depends on HY5 and occurs via three different mechanisms (Figure 3E). First, MYB-like Domain transcription factor (MYBD), which is induced in response to light and is a target gene of HY5, is a direct transcriptional repressor of MYBL2, able to reduce the acetylation of lysine 9 of histone 3 (H3K9) in MYBL2 promoter (Nguyen et al., 2015). Second, HY5 is also able to directly bind the MYBL2 promoter and to repress its expression through an epigenetic mechanism, consisting of increased levels of H3K9 acetylation and H3K4 trimethylation at HY5 binding sites (Wang et al., 2016). Third, HY5 directly activates miR858a, which then inhibits MYBL2 by translational repression (Wang et al., 2016). Overall, MYBL2 is negatively regulated by transcriptional repression by HY5 as well as by miR858a via translational repression (see also Section “Post-transcriptional Regulation of Anthocyanin Biosynthesis in Arabidopsis and Other Dicots”) and affects the expression of biosynthetic genes and anthocyanin accumulation through the interaction with MBW complexes. An additional level of epigenetic regulation involves the enrichment of the histone 2 variant H2A.Z in the promoter of anthocyanin biosynthetic genes in normal conditions as well as in high light and drought, in order to reduce an excessive anthocyanin accumulation. H2A.Z inhibits the expression of biosynthetic genes by reducing the accessibility and the H3K4 trimethylation around the transcription start site of genes (Cai et al., 2019).
Besides MYBL2, other transcription factors acting as repressors of anthocyanin biosynthesis in response to hormonal stimuli have been identified, including the JASMONATE ZIM-DOMAIN (JAZ) proteins (Qi et al., 2011) and the SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 9 (SPL9; Gou et al., 2011). Despite their interactions with different proteins of the MBW complex, they share similar repressor mechanisms through ubiquitination and degradation by the 26S proteasome. JAZ proteins, which are the master regulators of jasmonic acid (JA) signaling, negatively regulate anthocyanin accumulation by interacting with bHLHs and R2R3-MYBs and disrupting the MBW complex (Figure 3C). Upon JA signaling, JAZ proteins are degraded through ubiquitination and degradation by the 26S proteasome and the reconstituted MBW complex activate anthocyanin biosynthesis (Qi et al., 2011). MYBL2 and JAZ proteins were also found to mediate gibberellic acid (GA)-inhibited anthocyanin biosynthesis in Arabidopsis (Xie et al., 2012, 2016). In the absence of GA, DELLA proteins, which are known repressors of GA signaling, are degraded (Jiang et al., 2007), but in the presence of GA they directly sequester MYBL2 and JAZ repressors, leading to the release of bHLH/MYB subunits and subsequently to the formation of an active MBW complex, which then activates the anthocyanin pathway. Possibly, a similar mechanism involves DELLA, MYBL2, and JAZs in response to abiotic stress-induced anthocyanin biosynthesis (Xie et al., 2012). Phosphate starvation and sucrose were also shown to reduce the concentration of GA or to specifically inhibit the GA-dependent degradation of DELLA proteins respectively, thus inducing anthocyanin accumulation in Arabidopsis (Hsieh et al., 2009). Similar to JAZ proteins, SPL9 acts as negative regulator of anthocyanin biosynthesis by interacting with R2R3-MYBs (PAP1 and MYB113) and disrupting the formation of the MBW complex with TT8 (Figure 3D; Gou et al., 2011; Cui et al., 2014).
Curiously, brassinosteroids (BR) negatively regulate the JA-induced anthocyanin accumulation by reducing the expression of PAP1, PAP2, and GL3 (Peng et al., 2011). Whether this suppression is mediated by JAZ proteins or by MYBL2, which has been found to participate in the down-regulation of BR-repressed genes, is presently unknown (Ye et al., 2012). Furthermore, ethylene inhibits anthocyanin accumulation induced by sucrose and light by suppressing the expression of transcription factors that positively regulate anthocyanin biosynthesis, including GL3, TT8, and PAP1, while activating the negative regulator MYBL2 (Jeong et al., 2010).
A NAC-type transcription factor ANAC032 has been proposed as an indirect negative regulator of the expression of LBGs (i.e., DFR, ANS/LDOX) and TT8 regulatory gene under stress conditions (i.e., sucrose treatment, high light, and oxidative stress) through activation of the negative regulators of anthocyanin biosynthesis, MYBL2 and SPL9 (Mahmood et al., 2016). Furthermore, three members of the LATERAL ORGAN BOUNDARY DOMAIN (LBD) protein family (LBD37, 38, and 39) are strongly induced by nitrogen and function as transcriptional repressor of PAP1 and PAP2, thereby suppressing anthocyanin biosynthesis in response to nitrogen (Zhou et al., 2012). Very recently, it has been highlighted the signaling pathway of strigolactones, which are carotenoid-derived hormones known to increase anthocyanin biosynthesis (Wang et al., 2020). More specifically, strigolactones trigger ubiquitination and degradation of SMAX-like 6 from Arabidopsis (AtSMXL6), a repressor interacting with PAP1, which is then released and allowed to activate anthocyanin biosynthesis (Wang et al., 2020).
Overall, the negative regulators mediating hormonal and environmental response (i.e., MYBL2, JAZ, and SPL9) act through a passive repression which disrupts the MBW complex by interacting with bHLHs (i.e., MYBL2, Figure 3B), with MYBs (i.e., SPL9, Figure 3D) or both (i.e., JAZ, Figure 3C). MYBL2 represents an exception, since it also actively represses transcription as part of the MBW complex (Figure 3B). Anthocyanin activation in response to hormones and abiotic stresses then occurs through a common mechanism, which involves the release of one or more of these negative regulators from the interaction with MYBs and/or bHLHs (Figures 3B–D), thus restoring a functional MBW complex that activates LBGs (Figure 3E).
Finally, some post-translational modifications of MBW proteins have been identified that negatively modulate the activity of MBW complexes. PAP1/PAP2, TT8/GL3/EGL3, and TTG1 are all short-lived and degraded by ubiquitin/26S proteasome (UPS)-dependent proteolysis (Maier et al., 2013; Patra et al., 2013a,b). For GL3/EGL3, the UPS targeting for degradation is part of a negative regulatory feedback loop likely functioning to reduce their hyperactivation (Patra et al., 2013b). For PAP1/PAP2, such degradation occurs in the dark and is mediated by the CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1)/SPA E3 ligase, indicating that the light-regulated activation of PAP1/PAP2 is in part due to protein stabilization (Maier et al., 2013). A recent study has revealed that MAP-Kinase 4 (MAPK4) phosphorylation of PAP1 also increases its stability and is essential for light-induced anthocyanin accumulation (Li et al., 2016).
Activators and Repressors of Anthocyanin Biosynthesis in Fruits, Vegetables, and Other Cereals
Transcription factors orthologous to the MBW complex of maize and Arabidopsis have been isolated in cereals and many fruits and vegetables, including grape, apple, kiwi, pear, peach and strawberry, with grape, apple, and tomato being the best studied in terms of mechanism of regulation of anthocyanin synthesis (Petroni and Tonelli, 2011; Jaakola, 2013; Chaves-Silva et al., 2018; Liu et al., 2018; Chen et al., 2019). Despite the fact that the MBW is highly conserved in fruits (Figure 2D), some R2R3-MYBs, such as VvMYBF1 in grapevine and SlMYB12 in tomato, are known that control the flavonol synthesis without a bHLH partner and are activators of EBGs (Czemmel et al., 2009; Ballester et al., 2010).
In rice, the Purple leaf (Pl) complex locus, including the bHLH OsB1 and OsB2 genes, was proposed to control the anthocyanin biosynthesis in rice leaves (Sakamoto et al., 2001). However, recent studies determined that the predominant MBW complex activating anthocyanin biosynthesis in purple-leaf rice includes the R2R3-MYB OsC1, the bHLH OsRb, and the WD40 OsPAC1, which contributes to the full activation of anthocyanin biosynthesis, similar to the maize PAC1 gene (Figure 2D). Consistent with this, OsRb was found to be much higher expressed in leaves compared to OsB1 and OsB2, whose expression level was extremely low (Zheng et al., 2019). Instead, OsB2 and OsC1 (also named Kala4 and Kala3, respectively) have a predominant role in anthocyanin pigmentation of rice pericarp in black-grained rice (Maeda et al., 2014; Oikawa et al., 2015; Sun et al., 2018; Zheng et al., 2019) and are found to be negatively regulated by ethylene in the absence of light (Kumar et al., 2019), whereas the bHLH gene, OsRc, controls proanthocyanidin synthesis in pericarp of red rice varieties (Sweeney et al., 2006). Another R2R3-MYB gene, OsP1, closely related to the maize P1 and the Arabidopsis MYB11, MYB12, and MYB112, was recently identified in rice. Similar to maize, OsP1 specifically activates a subset of anthocyanin biosynthetic genes (i.e., CHS, CHI, and F3H), with partial functional overlap with the rice MBW (Zheng et al., 2019). In other cereals, such as wheat, only the Pp (Purple pericarp) gene controlling anthocyanin pigmentation in seeds has been identified so far (McIntosh et al., 2003).
In grapevine, a complex locus with two MYB genes (VvMYBA1 and VvMYBA2), homologs of Arabidopsis PAP1, PAP2, MYB113, and MYB114 is specifically expressed in ripening berries, where it promotes the synthesis of tri-hydroxylated anthocyanins (i.e., delphinidin, malvidin, and petunidin), whereas the closely related VvMYBA5, VvMYBA6, and VvMYBA7 are predominantly expressed in vegetative tissues and almost exclusively promote the synthesis of di-hydroxylated anthocyanins (i.e., cyanidin and peonidin; Azuma et al., 2008; Matus et al., 2017). Consistently, these regulators share the activation of late biosynthetic and modification/transport-related genes, but only VvMYBA1 and VvMYBA2 activate F3′5′H leading to tri-hydroxylated anthocyanins. In addition, VvMYBA1 in fruits and VvMYBA6-A7 in both plantlets and fruits were found to be responsive to UV-B in an HY5-dependent manner, similar to PAP1 in Arabidopsis (Matus et al., 2017). The existence of an MBW complex is suggested by the identification of VvWDR1 as an activator of anthocyanin synthesis when overexpressed in Arabidopsis (Matus et al., 2010) and by the interaction of VvMYC1 with all grape MYBs inducing anthocyanins (i.e., VvMYBA1/A2/A6/A7), proanthocyanidins (i.e., VvMYBPA1/PA2; Matus et al., 2017), or both (i.e., VvMYB5a/5b; Figure 2D; Hichri et al., 2010). Similar to TT8 in Arabidopsis, VvMYC1 is involved in a positive feedback regulation of its own expression (Hichri et al., 2010). A retrotransposon insertion in the promoter of VvMYBA1 (Kobayashi et al., 2004) and point mutations in VvMYBA2 coding regions (Walker et al., 2007) are present in some white cultivars.
In apple, allelic homologs of R2R3-MYBs are responsible for the light-dependent anthocyanin pigmentation of red-skin apple cultivars (i.e., MdMYB1 and MdMYBA; Takos et al., 2006; Ban et al., 2007) or determine red pigmentation in apple fruit and leaves (i.e., MdMYB10; Espley et al., 2007). MdMYB1 protein is degraded in the dark by the UPS, suggesting that the light-mediated anthocyanin accumulation in apple skin is due, at least in part, to the stabilization of these factors in light, similar to the Arabidopsis PAP1/PAP2 (Li et al., 2012c). On the other hand, the MdMYB10 expression results from an autoregulatory loop mediated by five tandem repeats of an MdMYB10 binding motif in its promoter (Espley et al., 2009). Other R2R3-MYBs activating anthocyanins are MdMYB3 (Vimolmangkang et al., 2013), MdMYB9, and MdMYB11, also controlling proanthocyanidins (An et al., 2015), and MdMYB110a, a paralog of MdMYB10 resulting from a whole-genome duplication event (Chagné et al., 2013). In apple, the MBW complex includes two bHLHs, MdbHLH3 and MdbHLH33, interacting with MdMYB1, MdMYB9, MdMYB10, and MdMYB11 (Espley et al., 2007; Xie et al., 2012; An et al., 2015) and an apple WD40 protein, MdTTG1, able to interact with bHLH proteins, but not MYBs (Brueggemann et al., 2010; An et al., 2012).
Interestingly, the apple R2R3-MYBs appear to regulate anthocyanin biosynthesis in response to low temperature and plant hormones through transcriptional and post-translational mechanisms similar to those highlighted in Arabidopsis. Upon exposure to low temperature, MdbHLH3 is transcriptionally activated and, once translated, the MdbHLH3 protein is phosphorylated to further enhance its transcription activation of MdMYB1, and form with it the MBW complex able to activate the genes of the anthocyanin biosynthetic pathway (Xie et al., 2012). On the other hand, MdbHLH3 protein was found to be sequestered by JAZ proteins, which are then degraded following JA exposure, allowing MdbHLH3 to act as direct activator of MdMYB9 and MdMYB11 and to form with them an MBW complex for activation of anthocyanin biosynthesis (An et al., 2015). Similarly, MdMYB10 is sequestered by the auxin-responsive factor MdARF13, which destabilizes the MBW complex and indirectly inhibits the anthocyanin biosynthesis, but it is also able to directly repress anthocyanin biosynthetic genes. Specifically, under low auxin concentration, the MdIAA121 repressor interacts with MdARF13 and prevents its binding to promoters of anthocyanin biosynthetic genes or to MdMYB10, whereas under high auxin concentration MdIAA121 is degraded by the 26S proteasome, thus allowing MdARF13 to repress anthocyanin biosynthesis (Wang et al., 2018). Finally, MdMYB1 was found to interact with the promoter of ETHYLENE RESPONSE FACTOR3 (ERF3), a key regulator of ethylene biosynthesis, thus providing a positive feedback on ethylene biosynthesis and as a consequence an increase in anthocyanin accumulation (An et al., 2018).
Homologs of apple MdMYB10 have been isolated in many other Rosaceae, including PpMYB10, FaMYB10, PyMYB10, and PavMYB10.1 from peach, strawberry, pear, and cherry, respectively (Feng et al., 2010; Lin-Wang et al., 2010; Jin et al., 2016). In most of these fruits, an MBW complex has been identified. In peach (Prunus persica), PpMYB10, PpbHLH3, and PpWD40 are involved in the regulation of anthocyanin biosynthesis by forming a MBW complex (Figure 2D; Liu et al., 2015). In red-fleshed peaches, PpMYB10 is activated by the NAC transcription factors BLOOD (BL) and PpNAC1, which are under control of the PpSPL1 repressor (Zhou et al., 2015). In addition, low temperature-dependent DNA demethylation has been recently observed in the promoter of anthocyanin biosynthetic genes, except for PpGST (Zhu et al., 2020). Instead, the expression and activity of PpGST, directly activated by PpMYB10 and PpbHLH3, were found to be a key step for anthocyanin regulation in a red-fleshed peach cultivar, resulting in a reduction of both anthocyanin accumulation and expression of biosynthetic and regulatory genes upon PpGST silencing (Zhao et al., 2020). In strawberry, FaMYB9/FaMYB11, FabHLH3, and FaTTG1 from the octoploid Fragaria ananassa form an MBW complex specifically activating proanthocyanidins (Schaart et al., 2013), whereas in a transient assay FvMYB10 and FvbHLH33 from the diploid Fragaria vesca were found to activate the anthocyanin-specific DFR and UFGT promoters (Lin-Wang et al., 2014). Recently, a RAV (related to ABI3/viviparous 1) transcription factor, FaRAV1, has been demonstrated to stimulate anthocyanin accumulation both by up-regulating FaMYB10 and by directly binding to promoters of anthocyanin biosynthetic genes (Zhang et al., 2020a). In pear, an MBW complex has not been identified, but PyMYB114 and PybHLH3 were found to interact with PyERF3 in red pear fruit. In addition, PyMYB10 and PyMYB114 showed an additive effect in activating anthocyanins when co-expressed in tobacco and strawberry (Yao et al., 2017).
In kiwifruit, AcMYB10 and AcMYB110 from Actinidia chinensis activate the CHS, DFR, flavonoid-3-galattosyltransferase (F3GT) and LDOX genes (Li et al., 2017a; Herath et al., 2020), but the regulation of the branch points F3′H and F3′5′H, which determine the production of cyanidin- or delphinidin-based anthocyanins, respectively, has been attributed to MYBC1 and WRKY44, which are also activators of the biosynthetic genes leading to proanthocyanidins (i.e., FLS, LAR, and ANR; Peng et al., 2020). In the proposed model, the MBW complex including MYB110 activates CHS, DFR, and F3GT, but when MYBC1 interacts with bHLH, WD40, and WRKY44, an MBWW complex (Figure 2D) activates the F3′H and F3′5′H genes, allowing both the anthocyanin and proanthocyanidin synthesis (Peng et al., 2020). Recently, AcMYB123 and AcbHLH42 have been also identified as activators of ANS, F3GT and anthocyanin biosynthesis in the inner pericarp of A. chinensis (Wang et al., 2019).
In tomato, two tightly linked R2R3-MYB genes, SlANT1 and SlAN2, were considered as the main positive regulators controlling anthocyanin levels in the skin of fruits (Mes et al., 2008; Sapir et al., 2008). Both activated anthocyanin biosynthesis when overexpressed in tomato fruits, leaves, and flowers (Mathews et al., 2003; Schreiber et al., 2012; Meng et al., 2015), but only SlAN2 was shown to act as a positive regulator of anthocyanin synthesis in vegetative tissues under high light or low temperature conditions through the control of the expression of the bHLHs SlAN1 and SlJAF13 (Kiferle et al., 2015). Expression of the WDR gene SlAN11 seems to be constitutive and does not require SlAN2, whereas the expression of SlANT1 is not detectable in wild-type tomato (Kiferle et al., 2015). This is consistent with studies in other species showing that MYB proteins regulate the transcription of their bHLH partners and subsequently form with the bHLH protein and the ubiquitously expressed WDR one an MBW complex that activates the anthocyanin genes. Recent studies have highlighted that the dominant ANTHOCYANIN FRUIT (Aft) gene, which was introgressed in cultivated tomatoes from the wild species Solanum chilense, co-segregates not only with SlAN2 and SlANT1 (Mes et al., 2008; Sapir et al., 2008), but also with two other R2R3-MYB genes, named SlAN2like and SlANT1like (Sun et al., 2019; Colanero et al., 2020; Yan et al., 2020). These studies proved that (i) only SlAN2like is necessary for anthocyanin synthesis (Sun et al., 2019); (ii) in cultivated wild-type tomatoes splicing mutations in SlAN2like lead to aberrant mRNAs producing shorter proteins lacking the most R2R3-MYB domain and the C-terminal domain, which are unable to interact with bHLH proteins in the MBW complex (Colanero et al., 2020), and (iii) the recently characterized CPC-like R3-MYB repressor ATROVIOLACEA (ATV) competes with SlAN2like in binding the bHLH SlAN1, thus contributing to a negative feedback loop that fine-tunes excessive anthocyanin synthesis (Colanero et al., 2018; Sun et al., 2019).
In other Solanum species, such as eggplant, pepper, and potato, no differences were observed in the expression level of WD40 genes (i.e., SmWD40, CaTTG1, and StAN11, respectively) in pigmented compared to non-pigmented tissues, whereas the expression of MYB and bHLH genes was correlated to anthocyanin accumulation (Stommel and Dumm, 2015; Liu et al., 2016; Tang et al., 2020). Despite different R2R3-MYB putative activators have been reported in eggplant (Solanum melongena), only SmMYB1 and SmMYB75 have been demonstrated to activate anthocyanin biosynthesis in the fruit and to interact with SmTT8 in activating SmCHS (Zhang et al., 2016; Shi et al., 2021). In potato (Solanum tuberosum), the R2R3-MYB transcription factor StAN1 and the bHLHs StbHLH1 or StJAF13 are necessary for anthocyanin biosynthesis in tuber skin and flesh (Payyavula et al., 2013; Liu et al., 2016). In addition to StAN1, StMYBA1 and StMYB113 were also found to activate anthocyanin biosynthesis in tobacco by transient assays (Liu et al., 2016). Very recently, a genome-wide association study has identified two closely related R2R3-MYB genes, StMYB88 and StMYB89, representing potential regulators of anthocyanin biosynthesis in tuber flesh (Li et al., 2021b). In pepper, the upregulation of CaMYBA and CabHLH has been correlated with the expression of anthocyanin biosynthetic genes in anthocyanin-pigmented fruits (Ohno et al., 2020). Recently, the R2R3-MYB CaANT1, CaANT2, and CaAN1 genes have been proposed to interact with CaTTG1 in an MMBW complex able to regulate the expression of LBGs in purple pepper (Tang et al., 2020).
In carrot (Daucus carota), the R2R3-MYB transcription factor DcMYB7 has been demonstrated the main positive regulator of anthocyanin biosynthesis in purple roots by activating DcbHLH3 and the biosynthetic genes (Iorizzo et al., 2019; Xu et al., 2019). Despite the overexpression of DcMYB6 resulted in anthocyanin accumulation in Arabidopsis (Xu et al., 2017a), its overexpression in carrot did not determine anthocyanin accumulation in roots and high transcript levels were found in both pigmented and non-pigmented roots (Xu et al., 2019). Recently, DcMYB113 was found to activate DcbHLH3 and anthocyanin biosynthetic genes, including those responsible for anthocyanin glycosylation and acylation (Xu et al., 2020). Other MYB, bHLH, and WD40 genes have been associated with anthocyanin accumulation in carrot roots, but their functional validation is presently lacking (Kodama et al., 2018).
Similar to Arabidopsis, many subgroup 4 R2R3-MYB repressors have been identified in fruit species (Chen et al., 2019; LaFountain and Yuan, 2021). Based on phylogenetic analyses, these R2R3-MYBs are divided into AtMYB4-like repressors, which control the phenylpropanoid metabolism by directly inhibiting biosynthetic genes, and FaMYB1-like repressors, which regulate anthocyanin biosynthesis by disrupting the MBW complex (Chen et al., 2019; LaFountain and Yuan, 2021). FaMYB1 from strawberry is the first identified R2R3-MYB containing an EAR motif in the C-terminal domain in fruit species. Since then, other FaMYB1-like proteins acting upon MBW complexes have been identified. The FaMYB1 gene is expressed at high levels only at ripe fruit stages, consistent with its role of anthocyanin repressor in the latter stages of strawberry fruit maturation (Aharoni et al., 2001; Lin-Wang et al., 2010). PhMYB27, and PhMYBx from petunia were found to act upon MBW complexes to repress anthocyanin synthesis (Koes et al., 2005; Albert et al., 2014), and similar mechanisms have been shown for FaMYB1, PpMYB18, SmMYB86, VvMYBC2, and VvMYB114 (Paolocci et al., 2011; Cavallini et al., 2015; Tirumalai et al., 2019; Zhou et al., 2019; Li et al., 2021a). A newly identified anthocyanin R2R3-MYB repressor, StMYB44, containing an EAR-repression domain, represses the anthocyanin biosynthesis in tuber flesh in response to high temperature without interacting with a bHLH factor. In fact, StMYB44 down-regulates the expression of StAN1, StbHLH1, and anthocyanin biosynthetic genes, thus resulting in a redirection of metabolic flux into chlorogenic acid or lignin biosynthesis (Liu et al., 2019). Interestingly, the R2R3-MYB27 repressor from kiwifruit is transcriptionally inhibited by high sugar concentrations (i.e., trehalose 6-phosphate), whereas it is consistently up-regulated upon carbon starvation, and has been proposed to mediate the reduced anthocyanin accumulation in red-fleshed fruits in response to sugar depletion (Nardozza et al., 2020).
A VvMYB4-like transcription factor from grape, highly expressed in the skin of berries, resulted in down-regulation of LBGs when overexpressed in tobacco flowers (Pérez-Díaz et al., 2016). As an exception, the AtMYB4-like MdMYB16 acts as homodimer to directly inhibit the anthocyanin biosynthetic genes via its EAR repressor domain (Xu et al., 2017b), but may also be able to weaken the interaction of MdMYB10-MdbHLH3/MdbHLH33 complex acting on anthocyanin genes (Lin-Wang et al., 2010).
Post-Transcriptional Regulation of Anthocyanin Biosynthesis in Arabidopsis and Other Dicots
In recent years, several studies have highlighted the important role of miRNAs involved in fine-tuning flavonoid biosynthesis in Arabidopsis. Based on initial studies and recent validations, MYB11, MYB12, and MYB111 have been confirmed as targeted by miR858 in Arabidopsis (Figure 3E), thereby regulating flavonol production (Sharma et al., 2016; Liu et al., 2021). On the other hand, the expression of TCP3, and therefore its activity as anthocyanin enhancer through the interaction with the MBW complex and the MYBL2 repressor (Figure 3A), is feedback inhibited by miR319 (Nag et al., 2009).
In Arabidopsis, SPL9, which negatively regulates anthocyanin biosynthesis through interfering with the formation of the MBW complex, is also under the negative control of miR156 and determines the acropetal accumulation of anthocyanins in Arabidopsis stem. Increased miR156 activity at the junction between rosette and stem promotes high levels of anthocyanins, whereas reduced miR156 activity in the upper part of stem results in SPL9 repression of anthocyanin synthesis and redirection of metabolic flux to high levels of flavonols (Gou et al., 2011). In addition, when miR156 is induced by salt and drought stress, SPL9 is repressed, resulting in the activation of the anthocyanin pathway through PAP1 activity (Figure 3E), whereas in non-stressed conditions, miR156 is suppressed and SPL9 repression of anthocyanins is restored (Cui et al., 2014). In grape, miR156 expression was found to be modulated by multiple hormonal signals (i.e., induced by JA and abscisic acid, ABA; repressed by GA and auxin) and to modulate anthocyanin synthesis through SPL9 (Su et al., 2021).
Several studies showed that miR828 is also involved in the negative regulation of anthocyanin biosynthesis in response to different signals (Hsieh et al., 2009; Luo et al., 2012; Yang et al., 2013). Sucrose, phosphate starvation, and ABA have been shown to directly induce Trans-acting siRNA gene 4 (TAS4) and miR828 and indirectly through activation of PAP1, PAP2, and MYB113 expression. TAS4 cleavage is then triggered by miR828 into TAS4-siR81(−), which in turn down-regulates PAP1, PAP2, MYB113, and MYB82 in a regulatory feedback loop that fine-tunes anthocyanin biosynthesis (Figure 3E). Interestingly, miR828 also directly targets MYB113, suggesting a close relationship between these MYBs, miR828, and TAS4 (Hsieh et al., 2009; Luo et al., 2012; Yang et al., 2013). Similarly, miR828 negatively controls anthocyanin biosynthesis by repressing the expression of BrPAP1, BrPAP2, and BrMYB82 through BrTAS4 in Brassica rapa (Zhou et al., 2020). In apple, Md-TAS4-siR81(−) is activated by MdMYB1 during the late fruit maturation stage and in response to high temperature and reduces anthocyanin biosynthesis by targeting MdbHLH3 (Zhang et al., 2020b).
In contrast, in Arabidopsis miR408 and miR858a positively regulate anthocyanin biosynthesis in seedlings. HY5 and SPL7 have been found to co-regulate several genes in response to light and copper, respectively, including anthocyanin biosynthetic genes through coordinate activation of miR408, which promotes anthocyanin accumulation through an undefined mechanism (Zhang et al., 2014b). As previously reported, miR858a has also been demonstrated to enhance anthocyanin biosynthesis in seedlings by inhibiting the expression of MYBL2 through translational repression (Wang et al., 2016). miR858 together with miR828 inhibits the anthocyanin repressor VvMYB114 in grape resulting in the activation of anthocyanin biosynthesis (Tirumalai et al., 2019). However, in kiwifruit and tomato, miR858 negatively regulates the anthocyanin pathway by repressing AaMYBC1 (Li et al., 2019) or by modulating SlMYB48-like and SlMYB7-like putative activators, respectively (Jia et al., 2015).
Although the mature sequence of miR858 is very similar, if not identical, in different species, these findings indicate that miR858 is associated with negative regulation in tomato and kiwifruit and positive regulation in Arabidopsis. Similarly, in grape miR828 and TAS4-siR81(−) activate anthocyanin biosynthesis by inhibiting a MYB repressor, whereas in Arabidopsis, B. rapa and apple they down-regulate anthocyanin accumulation by inhibiting MYB repressors or bHLH proteins. Overall, this suggests that the miR858/miR828-mediated mechanisms underlying anthocyanin biosynthesis may be distinctive in different species and depend on the downstream targets affected.
Interestingly, two long noncoding natural antisense RNAs (lncNATs), named asDcMYB6 and asDcMYB7, have been shown to be transcribed in opposite direction to DcMYB6 and DcMYB7, respectively, and to be highly expressed in purple carrots compared to orange ones, like their corresponding genes. These lncNATs may represent a novel player in post-transcriptional regulation of anthocyanin biosynthesis in carrot (Chialva et al., 2021).
Enhancing Anthocyanin Content in Our Plant Food
The health benefits of anthocyanins make them important targets for improving existing commercial varieties for anthocyanin-rich functional foods, which help consumers to achieve a greater content of anthocyanins in their diets. Enrichment of bioactives could be particularly useful to assess the nutritional properties of different bioactives by comparing near-isogenic plant-based foods that vary only in the type and quantity of the bioactives under analysis (Martin et al., 2011).
Most efforts have been focused on increasing anthocyanins by introducing or inducing the expression of MBW transcription factors to activate the endogenous anthocyanin biosynthetic genes either by conventional breeding or by metabolic engineering.
The introgression of dominant mutations (i.e., Aft and Aubergine, Abg) through interspecific crosses with wild species transferred the ability to produce anthocyanins in the peel of cultivated tomatoes (Jones et al., 2003; Mes et al., 2008). Furthermore, the recessive gene atroviolacea (atv) and the constitutive photoresponsive high-pigment-1 (hp-1) allele in combination with Aft or Abg have been shown to stimulate a higher production of anthocyanins in the peel (Povero et al., 2011; Catola et al., 2017). Similarly, taking advantage of geographic accessions of the MYB and bHLH regulatory gene families (C1/Pl1 and B1/R1, respectively) from Andean corn cultivars, maize hybrids carrying different levels of anthocyanins in kernels have been obtained (Petroni et al., 2014).
In order to increase the anthocyanin content in tomato fruits, anthocyanin regulatory genes from different plant species have been expressed in tomato most successfully when the bHLH Delila and MYB Rosea1 genes from snapdragon were overexpressed under the control of a tomato fruit-specific promoter, to give purple tomato fruits containing large amounts of anthocyanins (i.e., delphinidin) and flavonols (i.e., myricetin) both in the peel and flesh (Butelli et al., 2008). High expression of AtMYB12, controlling flavonol synthesis in Arabidopsis, together with Delila and Rosea1 further enhanced anthocyanin production in tomato by activating pathways of primary metabolism (glycolysis, the TCA cycle, the oxidative pentose phosphate pathway and the shikimate pathway) toward the production of substrates, ATP and reducing power for the phenylpropanoid pathway (Zhang et al., 2015). A similar strategy was used to obtain tomato fruits with high levels of resveratrol and genistein, reaching levels comparable to those present in soy-based products, like tofu (Zhang et al., 2015). Based on a similar concept, arogenate dehydratases (ADT) genes, controlling the level of phenylalanine, have been proposed as new targets for metabolic engineering to modulate anthocyanin content in plants (Chen et al., 2016). In general, metabolic engineering that combines multi-level transcriptional regulation and pathway rerouting offers an excellent strategy for biofortification of foods, for the production of plant-derived phytochemicals and ingredients, and for establishing materials for comparative nutrition studies. Such comparisons should lead to much clearer understanding of the health benefits of foods rich in specific polyphenolic phytonutrients in the diet, and shed light on their mechanisms of action (Zhang et al., 2015; Fu et al., 2018).
Novel regulatory targets for enhancing anthocyanin biosynthesis in plant food could include the COP1-mediated degradation of MYB transcription factors involved in anthocyanin production, such as MdMYB1 in apple (Li et al., 2012c), which could provide a very efficient and specific approach to increasing anthocyanin levels while maintaining otherwise normal light signaling. Alternatively, the control of expression of MYB transcription factors by small RNAs, like miR828/TAS4 and miR858 or other negative regulators, such PpSPL1 in peach (Zhou et al., 2015) and R2R3-MYB repressors found to reduce anthocyanin biosynthesis in fruits and vegetables, might be exploited to increase anthocyanin synthesis in our foods, perhaps using genome editing techniques. Beyond this, an understanding of the mechanisms that determine anthocyanin stability, such as anthocyanin decoration (i.e., glycosylation and acylation) or pH in vacuoles, as well as the fading processes operational in some flowers, has been suggested as additional tools for both biotechnological approaches and marker-assisted breeding (Zhang et al., 2015; Passeri et al., 2016).
Assessing Health Benefits of Anthocyanin-Rich Functional Foods
There are several reviews of epidemiological and preclinical intervention studies reporting the beneficial effects of anthocyanins on health (He and Giusti, 2010; Tsuda, 2012; Pojer et al., 2013; Wallace, 2013; Wallace and Giusti, 2015; Li et al., 2017b; Mattioli et al., 2020; Salehi et al., 2020; Kozłowska and Dzierżanowski, 2021). Despite this, in developed countries, there is an increasing trend in eating energy-dense foods, rich in sugar and saturated fatty acids. The consumption of fruits and vegetables has declined substantially over the past 30years, with a consequent increase in obesity, especially childhood obesity, defined as a body mass index (BMI) ≥30 (Martin et al., 2011). Increased consumption of anthocyanin-rich fruits and vegetables could have positive effects on health. This is even more important considering that purified anthocyanins consumed as dietary supplements do not have the same beneficial effects as anthocyanins in a natural food matrix (Prior et al., 2008; Titta et al., 2010; Martin et al., 2011). In this context, the development of near-isogenic genotypes of common foods, devoid or rich in anthocyanins is of importance for four reasons. First, the production of near-isogenic plant foods allows to reduce some of the complexity of food in the diet–health relationship and provide model foods that can be used for both animal feeding studies and human intervention trials for assessing the role of plant bioactives in the diet. Specifically, being near-isogenic, the anthocyanin-free genotypes represent a matched control for assessing the health-protective effects of anthocyanins, allowing the identification of their specific mechanisms of action compared to those of other polyphenols and phytonutrients present in the food matrix. Second, different anthocyanin-rich foods can be used to assess whether the consumption of comparable amounts of anthocyanins in different food matrices gives the same health benefits against different specific diseases. Third, anthocyanin-rich foods can be used in in animal models or directly in human intervention studies, if there are no safety concerns, to validate their health benefits. Fourth, once assessed, they can add back health-promoting anthocyanins to the diet. Anthocyanin-rich and anthocyanin-free comparator foods have been developed successfully either by conventional breeding (i.e., anthocyanin-rich corn and orange) or by metabolic engineering (i.e., anthocyanin-rich tomato and apple) and used to test the health benefits of anthocyanins in animal models and in some pilot intervention studies. Below we provide a summary of their beneficial effects (also reported in Table 1).
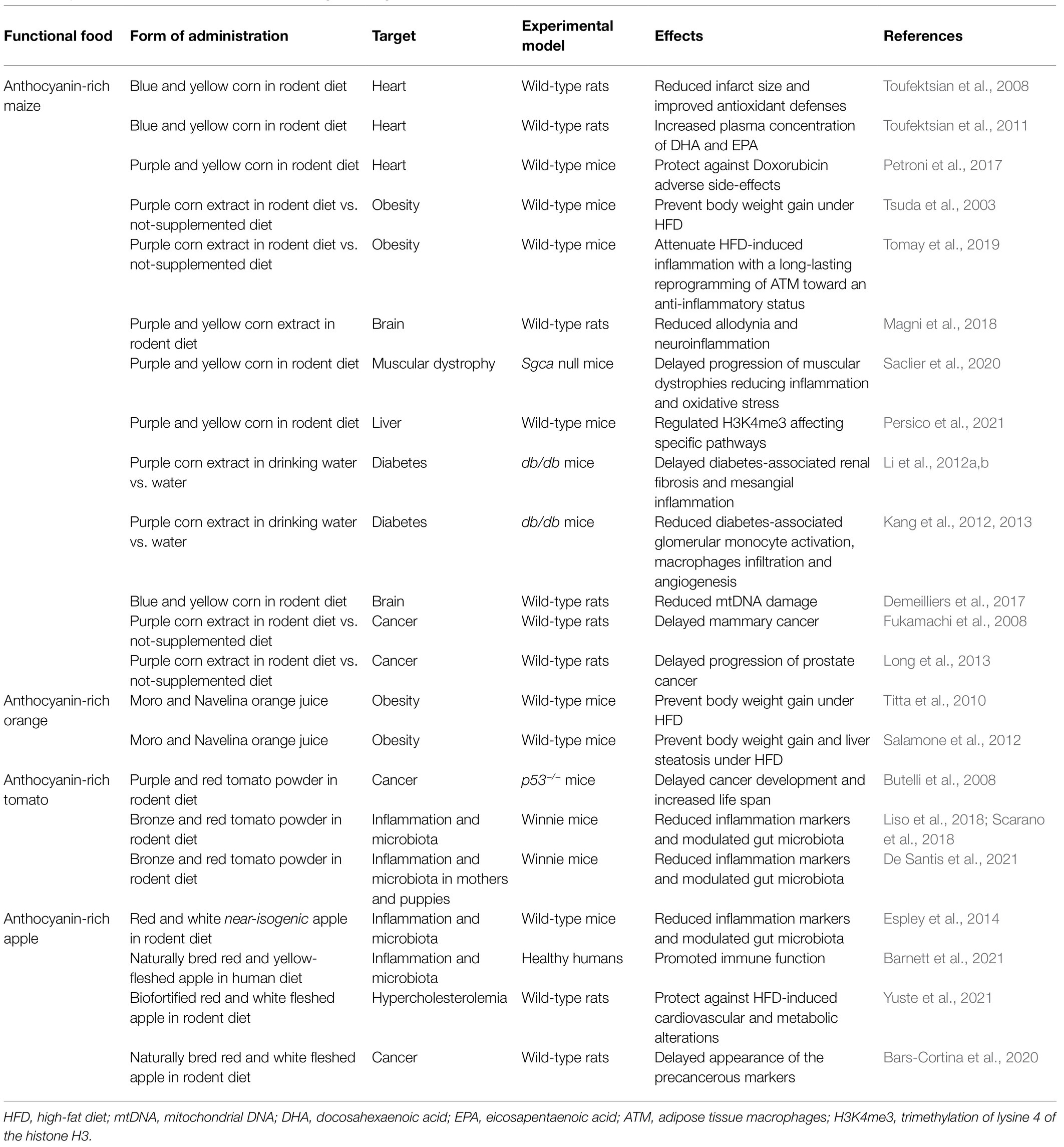
Anthocyanin-Rich Maize
Anthocyanin-rich maize (i.e., blue and purple corn) originates from South America, where it is largely used also as colorant for food and beverages. It mainly contains cyanidin 3-glucoside and, to a small extent, pelargonidin 3-glucoside and peonidin 3-glucoside (Pedreschi and Cisneros-Zevallos, 2007; Petroni et al., 2017). The beneficial effects of purple corn have been recognized for a very long time. Aztecs used to prepare a beverage rich in purple corn extract, called Tlaolli, used to treat a number of illnesses.
There are indications that a daily intake of anthocyanins in quantities comparable to those consumed in a Mediterranean diet is protective against cardiac injuries and pathologies. The cardioprotective effects of an anthocyanin-rich diet were tested using an ex vivo model of isolated perfused rat heart (Toufektsian et al., 2008). The infarct size in rats fed with anthocyanin-rich R1 C1 blue corn diet for 8weeks was reduced compared with those of rats fed with the near-isogenic r1 c1 yellow corn diet, meaning that anthocyanins can induce a state of myocardial resistance. Moreover, the anthocyanin-rich diet was able to increase the total and reduced glutathione in preischemic heart, suggesting that the protection against ischemia-reperfusion injury might be related, at least in part, to an improvement in endogenous antioxidant defenses. Most importantly, dietary anthocyanins from blue corn were shown to modulate the metabolism of (n-3) polyunsaturated fatty acids (PUFA) and increase plasma concentrations of eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA), two fatty acids known to have a very important role of protection against heart disease complications (Toufektsian et al., 2011). Hence, the comparison of these isogenic corn lines showed that anthocyanins might exert their beneficial effects in two ways: directly, e.g., increasing the endogenous antioxidant defenses, and/or indirectly, as signaling molecules modulating other metabolic pathways.
Dietary anthocyanins from purple corn have been shown to be protective against the cardiotoxic side effects of chemotherapeutic drugs, like Doxorubicin (DOXO). A recent study (Petroni et al., 2017) demonstrated that mice fed with a purple corn-rich diet (Red diet, RD) were more resistant to DOXO-induced cardiac alterations (i.e., disorganized myofibrils, mitochondrial fragmentation/degradation, and defects in sarcolemma junctions) than mice fed with an isogenic yellow corn diet (YD). Moreover, the mid-term DOXO-induced mortality was significantly attenuated in mice on the RD compared to mice fed with YD. In addition, a purple corn extract did not interfere with the chemotherapeutic activity of DOXO in tumor cell lines. The mechanisms by which purple corn protects against DOXO side effects are still under investigation.
Dietary anthocyanins from blue corn have also been shown to be protective from brain mitochondrial DNA (mtDNA) damage induced by ethanol. Oxidative stress due to ethanol metabolism is known to cause damage to mtDNA. Rats, divided into 4 experimental groups, were fed for 8weeks with anthocyanin-rich or anthocyanin-free diets while receiving 12% ethanol or water as beverages. Mice consuming the anthocyanin-free diet and ethanol showed increased reactive oxygen species (ROS) and mtDNA damage in their brains, whereas consumption of ethanol with the anthocyanin-rich diet did not show the accumulation of damaged mtDNA. Again, this may be due to the induction of antioxidant defense responses promoted by the blue corn diets (Demeilliers et al., 2017).
Other studies have demonstrated that anthocyanins from purple corn have preventive effects on the development of obesity and hyperglycemia induced by the consumption of a high-fat diet (HFD). When fed for 12weeks with a HFD, addition of purple corn extracts to the diet prevented weight gain and hypertrophy of adipocytes, which is an increase in cell size of adipocytes generally associated with increased cellular stress in the adipose tissue and with systemic diabetes (Tsuda et al., 2003). Moreover, in cell cultures of human adipocytes, cyanidin 3-glucoside (C3G) positively regulated obesity and type 2 diabetes markers increasing adiponectin and down-regulating PAI-1 (Plasminogen Activator Inhibitor-1) and IL-6 (Interleukin-6; Tsuda et al., 2006). Adiponectin is the most important adipocytokine, and its expression is inversely correlated to the amount of fat tissue in the body: In conditions of obesity and type 2 diabetes, it is downregulated, while it is up-regulated during starvation (Kadowaki et al., 2006; Lee and Shao, 2006). Elevated levels of PAI-1 and IL-6 are characteristics of obesity and type-2 diabetes (Jung and Choi, 2014). Another study on the adipose-tissue macrophages (ATM) confirmed the protective effect of purple corn against obesity-related inflammation. Macrophages of mice fed with HFD and purple corn extract (HFD+RED) for 12weeks showed an M2 anti-inflammatory phenotype associated with increasing production of anti-inflammatory markers and tissue repair (i.e., Arginase I, ArgI; Found in inflammatory zone 1, Fizz1; Transforming growth factor β, TGFβ), while mice receiving HFD and water had M1 macrophages that produce pro-inflammatory cytokines and encourage inflammation and tissue destruction (i.e., IL-6; Interleukin-1β, IL-1β; Tumor necrosis factor α, TNF-α; Cyclooxygenase-2, COX-2). Moreover, adipose tissue M2 macrophages obtained from HFD+RED mice and treated with lipopolysaccharide (LPS) ex vivo, maintained the anti-inflammatory phenotype, indicating a long-lasting effect of anthocyanins (Tomay et al., 2019).
The ability of anthocyanins to exert antioxidant and anti-inflammatory activity makes them eligible for the study of different pathologies characterized by both oxidative stress and inflammation. A study conducted on Sgca null dystrophic mice, with a severe degenerative myopathy similar to Duchenne, has demonstrated that anthocyanins from purple corn counteracted the progression of muscular dystrophy (early and late-stage) acting on both oxidative and inflammatory status without affecting regeneration. Purple diet, but not the near-isogenic yellow diet, ameliorated tissue morphology, fibrosis, and muscle performance, promoted a metabolic shift to an oxidative fiber metabolism, and increased the mitochondrial amount counteracting the progression of the disease. Finally, mice fed with purple corn diet presented less macrophage infiltration compared with the yellow diet counterpart (Saclier et al., 2020).
Anthocyanins from purple corn reduced the orofacial pain induced by the inflammation of the trigeminal nerve, by preventing the macrophage infiltration in the trigeminal ganglion and the activation of microglia (i.e., macrophages resident in the nervous central system) both in vivo and in vitro. In a rat model of trigeminal sensitization drinking near-isogenic yellow or purple corn extract, anthocyanins and acetyl salicylic acid (ASA) equally reduced allodynia and macrophage infiltration, but only purple corn extract inhibited microglial activation in vivo and reverted LPS-induced inflammation in vitro resulting in lower production of pro-inflammatory mediators (IL-6, TNF-α, IL-1β; Monocyte Chemoattractant Protein-1, MCP-1; inducible nitric oxide synthase, iNOS) and in an increase in the anti-inflammatory ones (Interleukin-10, IL-10; Interleukin 13, IL-13; Arg-1; Fizz1, YM-1; Magni et al., 2018).
The nutriepigenetic effect of anthocyanins was also recently investigated. Anthocyanins, as many other phytonutrients, can alter the phenotype through epigenetic modifications. One of these modification includes histone tail modifications that can alter structure of the chromatin and modify gene expression and function. Many histone modifications have been identified, and the most studied one is the trimethylation of lysine 4 of histone H3 (H3K4me3), which is associated with transcribed genes in mice and humans. Persico et al. (2021) analyzed the effect of five different diets (standard, caloric restriction, high fat, purple corn, and yellow corn) on H3K4me3 in mice liver, highlighting that anthocyanins from purple corn regulated H3K4me3 affecting different pathways like the integrin-like kinase signaling, involved in inflammation, and the metabolism of the pyruvate and amino acids (Persico et al., 2021).
Purple corn extract can also ameliorate diabetes-associated diseases, retarding diabetic nephropathy, ameliorating hyperglycemia, counteracting renal filtration dysfunctions in db/db mice and inhibiting high-glucose-induced fibrosis and inflammation in vitro (Li et al., 2012a,b). Moreover, purple corn extract reduced the clinical manifestations of diabetic nephropathy, since it lowered diabetes-associated glomerular mesangial expansion (i.e., the accumulation of extracellular matrix proteins in the mesangial interstitial space) and macrophage infiltration into diabetic glomeruli (Kang et al., 2012) and counteracted glomerular angiogenesis (Kang et al., 2013), thus representing a potential complementary therapy for diabetes-associated glomerulosclerosis, inflammation, and angiogenesis.
There is some evidence that purple corn has anticancer activity as well. Model rats treated with a heterocyclic amine carcinogen showed that purple corn extract had an anti-tumor activity exerted through the modulation of cell proliferation and apoptosis in the mammary neoplastic lesions, due to the reduction of RAS protein level, which is commonly higher in tumors since it promotes cell growth through the Phosphatidyl Inositol 3-Kinase/Akt (PI3K/Akt) and Extracellular signal-regulated kinase 1/2 (Erk1/2) pathways. The decrease in RAS and phosphorylated Akt correlated with the increase of the cleaved caspase 3 which induced apoptosis (Fukamachi et al., 2008). More recently, purple corn extract was reported to retard the progression of the tumor, reducing the percentage of adenocarcinoma in a dose-dependent manner in Transgenic Rats for Adenocarcinoma of Prostate (TRAP) model rats. The most effective anthocyanins appeared to be both cyanidin 3-glucoside and pelargonidin 3-glucoside (Long et al., 2013).
Anthocyanin-Rich Orange
Tarocco, Moro, and Sanguinello are the three major blood varieties that differ from the other varieties of the sweet orange group (Valencia Late, Washington navel, and Navelina) for the presence of anthocyanins, mainly represented by C3G. Sweet orange is an interspecific hybrid that has no sexual segregation, so that all varieties of this group can be considered near-isogenic (Butelli et al., 2017). Tarocco is from Italy, and it is medium-sized seedless and very flavorful. It is also called “half-blood” because the flesh is less red-pigmented than the other two varieties. The Moro oranges produce the highest levels of anthocyanins, and they are called “deep blood orange” and they originate from Italy. Sanguinello comes from Spain, but is also cultivated in Sicily; it is called “full-blood” orange and has similar characteristics to Moro (Grosso et al., 2013). All three blood varieties of sweet orange have a common ancestor, since they all carry the same Copia-like retrotransposon in the subgroup 6 R2R3-MYB Ruby gene, responsible for the cold-dependent fruit-specific activation of anthocyanin biosynthesis, and arose through selection of bud mutations (Butelli et al., 2012). Moro and the common sweet orange Navelina contain comparable concentrations of vitamin C, flavanones, and hydroxycinnamic acids, thus allowing their use in comparative nutritional studies with the aim of testing their obesity-preventing activities (Titta et al., 2010).
Dietary supplementation of Moro juice significantly reduced body weight gain and fat accumulation in mice. Mice fed for 12weeks with a standard diet (SD, 3.3kcal/g, mainly composed of carbohydrates) together with Moro juice, gained less weight than mice drinking water or Navelina juice. Moro juice was also effective in almost abolishing weight gain induced by a HFD (5.24kcal/g, with 60% fat supplement), reducing the abdominal and inguinal fat mass by approximately 50%, while showing a marked reduction in adipocyte cell size and lipid accumulation. Mice fed a HFD with drinking water or Navelina juice showed no such reduction (Titta et al., 2010). In addition, glucose, fatty acid, and triglyceride blood levels were not altered in mice drinking Moro juice on a HFD. Analysis of the transcriptomes of adipocytes of the mice on the different diets revealed that the Moro juice can counteract the effects of the HFD on adipocytes by altering gene expression. Indeed, the gene expression profiles of mice on the HFD regimen drinking Moro juice, but not of those drinking Navelina juice, were similar to those of mice fed with the SD, preventing the change in expression of 21% of the up-regulated and the 55% of the down-regulated genes in response to the HFD (Titta et al., 2010). In agreement with this study, mice fed with HFD and Moro juice showed a reduction in body weight gain compared to mice in the same condition but drinking water. Moreover, Moro juice could counteract liver steatogenesis in HFD fed mice (Salamone et al., 2012).
Analyses performed using Moro anthocyanin-rich extracts confirmed the beneficial effects in reducing fat accumulation in mice, but the Moro extract was less effective than crude Moro juice. The administration of purified C3G did not show any effect on weight gain (Titta et al., 2010). This suggested that other components of the Moro juice, in addition to anthocyanins, may contribute to the anti-obesity effects. For these reasons functional foods are extremely important and the consumption of fresh fruit has to be preferred to fruit extracts or fruit-derived supplementations.
A study in healthy human volunteers consuming either blood orange juice or blond juice, showed that consumption of either type of orange juice decreased the pro-coagulant activity of whole blood, an indicator of cardiovascular risk. This suggested a role of antioxidants independent of the anthocyanin content of blood orange juice (Napoleone et al., 2013). Another study on healthy humans showed potential protective effects of both blood and blond orange juice on the low-grade pro-inflammatory status induced by the consumption of a standardized fatty meal (Cerletti et al., 2015). Consumption of a fatty meal can induce an acute inflammatory status, defined by an increase in platelet and leukocyte counts and in myeloperoxidase (MPO) degranulation of granulocytes. Granulocytes release MPO, a peroxidase enzyme, into the extracellular space in the inflammatory locus, increasing inflammation. Frequent fatty meal consumption may lead to chronic low-grade inflammation and to a series of events that may develop into atherothrombosis (Cerletti et al., 2016). Consumption of both blood and blond orange juice prevented neutrophil MPO degranulation, used as a marker of cell activation induced by the fatty meal, but did not modify other leukocyte cellular markers. High anthocyanin, blood orange juice was effective in reducing total cholesterol in plasma, unaccompanied by high-density lipoprotein (HDL) changes. Both juices reduced blood glucose levels (Cerletti et al., 2015). Other preclinical studies have confirmed that consumption of blood oranges can have anti-inflammatory effects and limit body weight gain, enhance insulin sensitivity and decrease serum triglycerides and total cholesterol in mice (Grosso et al., 2013).
Purple Tomato
Tomato is among the most important vegetables consumed world-wide. It is rich in vitamins, flavonoids, and other health-promoting compounds, but usually it does not contain anthocyanins, except in some tomato wild species, such as S. chilense (Jones et al., 2003). The red color, in fact, is due to the presence of carotenoids, including lycopene and the orange-colored β-carotene. Since tomato is the second most consumed among vegetables in the human diet, the importance of this fruit as a vehicle for nutrients and bioactive compounds for improving human health, is clear. Biotechnological and conventional breeding approaches have been used to engineer anthocyanin production in tomatoes (Gonzali et al., 2009).
Tomatoes genetically engineered to produce high levels of delphinidin and petunidin were produced through the expression of Delila and Rosea1 regulatory genes from A. majus, specifically in fruit (Butelli et al., 2008). To investigate whether the anthocyanin levels reached were enough to promote health, diets supplemented with 10% red or purple tomato powder were fed to cancer-prone Trp53−/− mice. Mice lacking p53 are prone to develop soft tissue carcinoma at an early age. Mice fed with purple tomato powder supplemented diets showed a significant extension of life span compared to mice fed diets supplemented with red wild-type tomato powder or SD without supplementation. This demonstrated that dietary consumption of high levels of anthocyanins can extend the life span of Trp53−/− cancer-prone mice by as much as 30% (Butelli et al., 2008).
Recent studies indicate an association between purple tomato and a reduction in the severity of symptoms of inflammatory bowel disease (IBD), a chronic inflammation of the gut including Crohn’s disease and ulcerative colitis (Liso et al., 2018; Scarano et al., 2018; De Santis et al., 2021). Tomato lines with different combinations of polyphenols have been tested in a mouse model of IBD, demonstrating that tomato enriched in flavonols, anthocyanins, and stilbenoids (named Bronze) were able to reduce/delay the symptoms as well as the dysbiotic intestinal microbiota associated with dextran sodium sulfate (DSS)-induced colitis, and showed significantly diminished pro-inflammatory mediators IL-6 and TNF-α levels. Interestingly, the combination of different polyphenols was more effective than single flavonoid classes (Liso et al., 2018; Scarano et al., 2018), and determined a reduction of mother’s dysbiosis and prevented/reduced IBD development in puppies, when supplied during pregnancy and lactation (De Santis et al., 2021). Overall, these studies indicate that tomato extracts enriched in multiple classes of flavonoids, including anthocyanins, display not only a direct anti-inflammatory role, but also a change on the gut microbiota that prevents a chronic inflammation status of the gut.
Anthocyanin-Rich Apple
Apple and apple-related products are some of the most important dietary sources of polyphenols. Moreover, recent discoveries suggest that apple consumption reduces the risk of a number of chronic diseases (Boyer and Liu, 2004; Hyun and Jang, 2016). Anthocyanins accumulate preferentially in the peel, where they are responsible for the color of apples. In addition, some consumers are used to peel apples and other fruits before eating them, thus limiting the consumption of anthocyanins from these dietary sources. There are a number of wild red-fleshed apples, and intense breeding has created red-fleshed apple varieties, because of the increasing interest in developing commercial red-fleshed apple varieties. Extensive crossbreeding programs involving good flavored, white-fleshed apples, have managed to improve the poor taste of the wild red-fleshed apple, producing a number of good tasting red-fleshed apples (Bars-Cortina et al., 2017). Recently, the discovery that the red flesh color in apple is under the control of the MdMYB10 gene allowed the development of an alternative approach to produce a red-fleshed apple variety which could be compared to an isogenic, white-fleshed variety, by direct integration of the dominant MdMYB10 allele into the “Royal Gala” line (Espley et al., 2009, 2013). Sensory and volatile profile analysis of these apples revealed no differences in flavor and aroma between MdMYB10-modified apple and the near-isogenic Royal Gala apple (Espley et al., 2013). However, consumption of red-fleshed MdMYB10-modified apple affected inflammatory pathway and gut microbiota in mice. After 7days of diet supplemented with MdMYB10-modified apple, expression of a group of cytokine genes linked to inflammation (Interleukin-2 receptor B, Il2rb; CC motif chemokine receptor 2 and 10, Ccr2 and Ccr10; C-X-C motif chemokine ligand 10, Cxcl10) was decreased by twofold compared to mice fed with a diet supplemented with non-transformed Royal Gala apples. After 21days, mice fed with MdMYB10-modified apple showed a tenfold decrease in prostaglandin E2 (PGE2) and a non-significant decrease in leukotriene B4 (LTB4) plasma levels, compared with mice fed with Royal Gala apple. PGE2 and LTB4 are both inflammatory mediators derived from the arachidonic acid metabolism: the first, synthesized by cyclooxygenase 1 and 2 (COX-1, COX-2), is involved in the cardiovascular event associated with inflammation, while the second, a lipoxygenase product, is involved in the chemotaxis of leukocytes. Moreover, the gut bacterial flora changed in relation with the diet, and mice fed with MdMYB10-modified apple showed a significant decrease in Lactobacillus spp., whereas mice fed with Royal Gala apple experienced an increase in Bifidobacterium spp. (Espley et al., 2014). A similar study was conducted on 25 healthy adults in a randomized cross-over controlled trial using naturally bred red-fleshed apples or white-fleshed control apples. The analyses of fecal microbiota and of gene expression in peripheral blood mononuclear cells (PBMC, which consist of peripheral blood cells having a round nucleus, like lymphocytes and monocytes) have shown minimal differences between the two groups, but genes regulated by red-fleshed apples were immunoglobulin-related, suggesting a potential role in modulating the immune function (Barnett et al., 2021). Finally, both red- and white-fleshed apples seemed to have beneficial effects in hypercholesterolemic rats (Yuste et al., 2021) or in model rats with adenocarcinoma (Bars-Cortina et al., 2020). The benefits of apple and anthocyanin consumption are well established (Knekt et al., 2002; Boyer and Liu, 2004; Cassidy et al., 2015, 2016; Hyun and Jang, 2016), and apples with high anthocyanin contents could offer effective functional foods, to reduce the incidence of chronic diseases when part of a normal diet.
Concluding Remarks
A considerable body of research has been devoted to identify the MYB-bHLH-WD40 transcription factors involved in the MBW complex activating the anthocyanin biosynthesis in many crop species, with the final aim of improving the anthocyanin content of plant-derived foods by means of conventional breeding or by metabolic engineering. Additional regulators of the anthocyanin pathway have been identified recently, acting as repressors disrupting the MBW complex or activators stabilizing the MBW complex. In addition, new levels of regulation have been described, in which the activity of repressors and activators is controlled by post-translational regulation. Anthocyanin production is also under epigenetic and post-transcriptional regulation (histone acetylation and miRNAs, respectively).
Despite research on positive and negative regulators has been extensive in the past decade, there are some gaps that could be filled. In some species, MBW complexes activating anthocyanins and related negative regulators still need to be identified. Are the negative regulatory systems identified in Arabidopsis, apple, and peach (e.g., MYBL2, SPL, JAZ, LBD, ANAC032, and AtSMX6) conserved in other species? Is TCP3 and its role as enhancer of MBW function and passive repressor of MYBL2 also conserved? Can these systems be exploited to enhance anthocyanin content in edible organs? How are R2R3-/R3-MYB repressors regulated in edible organs of fruits and vegetables and how can we modulate their expression in order to enhance anthocyanins, while avoiding their excessive undesirable accumulation? Further studies are required to understand the role of DNA methylation and histone modifications in anthocyanin repression both in model and crop species and eventually verified as a new possible approach to enhance organ-specific anthocyanin biosynthesis. Can miRNAs (e.g., miR828, and miR858) be silenced in edible organs to enhance anthocyanin accumulation? Are the lncRNAs newly identified in carrot conserved in other species? Can they be employed to enhance anthocyanin accumulation?
Advancing our knowledge of anthocyanin biosynthesis will allow the development of new biotechnological tools for the generation of value-added plants with increased anthocyanin content, which help consumers to achieve the desired amount of anthocyanins in their daily diet. Enrichment of foods in anthocyanins will also be particularly useful for the comparison of the nutritional properties of these bioactives from different food sources. Furthermore, the precise identification of biosynthetic genes encoding decorating enzymes in a specific plant food and a better understanding of their regulation will help in designing plant foods enriched in specific types of glycosylated/acylated anthocyanins. This may contribute to define which is the bioavailability and contribution to health-promoting properties of the single anthocyanin species present in a plant food, of their combination and eventually of the specific decorating groups in a food context.
Author Contributions
All authors contributed equally to the manuscript, and read and approved the final manuscript. KP and CT contributed to conceptualization. FC, AM, MT, KP, and CT contributed to writing, reviewing, and editing.
Funding
Financial support was provided by Fondazione Umberto Veronesi (Grant 2014 to KP; Grant Fellowships to FC and AM), EU FP7 project ATHENA (FP7-KBBE-2009-3/245121) to CT and the project MIND FoodS HUB (Milano Innovation District Food System Hub): Innovative concept for the eco-intensification of agricultural production and for the promotion of dietary patterns for human health and longevity through the creation in MIND of a digital Food System Hub, cofunded by POR FESR 2014-2020_BANDO Call HUB Ricerca e Innovazione, Regione Lombardia to KP and CT.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors wish to thank Cathie Martin for critical reading of the manuscript.
References
Aharoni, A., De Vos, C. H., Wein, M., Sun, Z., Greco, R., Kroon, A., et al. (2001). The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 28, 319–332. doi: 10.1046/j.1365-313X.2001.01154.x
Albert, N. W., Butelli, E., Moss, S. M. A., Piazza, P., Waite, C. N., Schwinn, K. E., et al. (2021). Discrete BHLH transcription factors play functionally overlapping roles in pigmentation patterning in flowers of Antirrhinum majus. New Phytol. 231, 849–863. doi: 10.1111/nph.17142
Albert, N. W., Davies, K. M., and Schwinn, K. E. (2014). Gene regulation networks generate diverse pigmentation patterns in plants. Plant Signal. Behav. 9:e29526. doi: 10.4161/psb.29526
Allan, A. C., Hellens, R. P., and Laing, W. A. (2008). MYB transcription factors that colour our fruit. Trends Plant Sci. 13, 99–102. doi: 10.1016/j.tplants.2007.11.012
An, X. H., Tian, Y., Chen, K. Q., Liu, X. J., Liu, D. D., Xie, X. B., et al. (2015). MdMYB9 and MdMYB11 are involved in the regulation of the Ja-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol. 56, 650–662. doi: 10.1093/pcp/pcu205
An, X. H., Tian, Y., Chen, K. Q., Wang, X. F., and Yu Jin Hao, Y. J. (2012). The apple WD40 protein MdTTG1 interacts with BHLH but not MYB proteins to regulate anthocyanin accumulation. J. Plant Physiol. 169, 710–717. doi: 10.1016/j.jplph.2012.01.015
An, J. P., Wang, X. F., Li, Y. Y., Song, L. Q., Zhao, L. L., You, C. X., et al. (2018). EIN3-LIKE1, MYB1, and ETHYLENE RESPONSE FACTOR3 act in a regulatory loop that synergistically modulates ethylene biosynthesis and anthocyanin accumulation. Plant Physiol. 178, 808–823. doi: 10.1104/pp.18.00068
Azuma, A., Kobayashi, S., Mitani, N., Shiraishi, M., Yamada, M., Ueno, T., et al. (2008). Genomic and genetic analysis of Myb-related genes that regulate anthocyanin biosynthesis in grape berry skin. Theor. Appl. Genet. 117, 1009–1019. doi: 10.1007/s00122-008-0840-1
Ballester, A.-R., Molthoff, J., De Vos, R., Te, B., Hekkert, L., Orzaez, D., et al. (2010). Biochemical and molecular analysis of pink tomatoes: deregulated expression of the gene encoding transcription factor SlMYB12 leads to pink tomato fruit color. Plant Physiol. 152, 71–84. doi: 10.1104/pp.109.147322
Ban, Y., Honda, C., Hatsuyama, Y., Igarashi, M., Bessho, H., and Moriguchi, T. (2007). Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol. 48, 958–970. doi: 10.1093/pcp/pcm066
Barnett, M. P. G., Young, W., Armstrong, K., Brewster, D., Cooney, J. M., Ellett, S., et al. (2021). A polyphenol enriched variety of apple alters circulating immune cell gene expression and faecal microbiota composition in healthy adults: a randomized controlled trial. Nutrients 13:1092. doi: 10.3390/nu13041092
Bars-Cortina, D., Macià, A., Iglesias, I., Romero, M. P., and Motilva, M. J. (2017). Phytochemical profiles of new red-fleshed apple varieties compared with traditional and new white-fleshed varieties. J. Agric. Food Chem. 65, 1684–1696. doi: 10.1021/acs.jafc.6b02931
Bars-Cortina, D., Martínez-Bardají, A., Macià, A., Motilva, M. J., and Piñol-Felis, C. (2020). Consumption evaluation of one apple flesh a day in the initial phases prior to adenoma/adenocarcinoma in an azoxymethane rat colon carcinogenesis model. J. Nutr. Biochem. 83:108418. doi: 10.1016/j.jnutbio.2020.108418
Baudry, A., Caboche, M., and Lepiniec, L. (2006). TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and BHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 46, 768–779. doi: 10.1111/j.1365-313X.2006.02733.x
Baudry, A., Heim, M. A., Dubreucq, B., Caboche, M., Weisshaar, B., and Lepiniec, L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39, 366–380. doi: 10.1111/j.1365-313X.2004.02138.x
Boyer, J., and Liu, R. H. (2004). Apple phytochemicals and their health benefits. Nutr. J. 3:5. doi: 10.1186/1475-2891-3-5
Brueggemann, J., Weisshaar, B., and Sagasser, M. (2010). A WD40-repeat gene from malus × domestica is a functional homologue of Arabidopsis thaliana TRANSPARENT TESTA GLABRA1. Plant Cell Rep. 29, 285–294. doi: 10.1007/s00299-010-0821-0
Butelli, E., Garcia-Lor, A., Licciardello, C., Casas, G. L., Hill, L., Recupero, G. R., et al. (2017). Changes in anthocyanin production during domestication of citrus. Plant Physiol. 173, 2225–2242. doi: 10.1104/pp.16.01701
Butelli, E., Licciardello, C., Ramadugu, C., Durand-Hulak, M., Celant, A., Recupero, G. R., et al. (2019). Noemi controls production of flavonoid pigments and fruit acidity and illustrates the domestication routes of modern citrus varieties. Curr. Biol. 29, 158.e2–164.e2. doi: 10.1016/j.cub.2018.11.040
Butelli, E., Licciardello, C., Zhang, Y., Liu, J., Mackay, S., Bailey, P., et al. (2012). Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 24, 1242–1255. doi: 10.1105/tpc.111.095232
Butelli, E., Titta, L., Giorgio, M., Mock, H. P., Matros, A., Peterek, S., et al. (2008). Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 26, 1301–1308. doi: 10.1038/nbt.1506
Cai, H., Zhang, M., Chai, M., He, Q., Huang, X., Zhao, L., et al. (2019). Epigenetic regulation of anthocyanin biosynthesis by an antagonistic interaction between H2A.Z and H3K4me3. New Phytol. 221, 295–308. doi: 10.1111/nph.15306
Carey, C. C., Strahle, J. T., Selinger, D. A., and Chandler, V. L. (2004). Mutations in the pale aleurone color1 regulatory gene of the Zea mays anthocyanin pathway have distinct phenotypes relative to the functionally similar transparent testa glabra1 gene in Arabidopsis thaliana. Plant Cell 16, 450–464. doi: 10.1105/tpc.018796
Casas, M. I., Duarte, S., Doseff, A. I., and Grotewold, E. (2014). Flavone-rich maize: an opportunity to improve the nutritional value of an important commodity crop. Front. Plant Sci. 5:440. doi: 10.3389/fpls.2014.00440
Cassidy, A., Bertoia, M., Chiuve, S., Flint, A., Forman, J., and Rimm, E. B. (2016). Habitual intake of anthocyanins and flavanones and risk of cardiovascular disease in men. Am. J. Clin. Nutr. 104, 587–594. doi: 10.3945/ajcn.116.133132
Cassidy, A., Rogers, G., Peterson, J. J., Dwyer, J. T., Lin, H., and Jacques, P. F. (2015). Higher dietary anthocyanin and flavonol intakes are associated with anti-inflammatory effects in a population of US adults. Am. J. Clin. Nutr. 102, 172–181. doi: 10.3945/ajcn.115.108555
Catola, S., Castagna, A., Santin, M., Calvenzani, V., Petroni, K., Mazzucato, A., et al. (2017). The dominant allele aft induces a shift from flavonol to anthocyanin production in response to UV-B radiation in tomato fruit. Planta 246, 263–275. doi: 10.1007/s00425-017-2710-z
Cavallini, E., Matus, J. T., Finezzo, L., Zenoni, S., Loyola, R., Guzzo, F., et al. (2015). The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine. Plant Physiol. 167, 1448–1470. doi: 10.1104/pp.114.256172
Cerletti, C., De Curtis, A., Bracone, F., Digesù, C., Morganti, A. G., Iacoviello, L., et al. (2016). Dietary anthocyanins and health: data from FLORA and ATHENA EU projects. Br. J. Clin. Pharmacol. 83, 103–106. doi: 10.1111/bcp.12943
Cerletti, C., Gianfagna, F., Tamburrelli, C., De Curtis, A., D’Imperio, M., Coletta, W., et al. (2015). Orange juice intake during a fatty meal consumption reduces the postprandial low-grade inflammatory response in healthy subjects. Thromb. Res. 135, 255–259. doi: 10.1016/j.thromres.2014.11.038
Chagné, D., Lin-Wang, K., Espley, R. V., Volz, R. K., How, N. M., Rouse, S., et al. (2013). An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol. 161, 225–239. doi: 10.1104/pp.112.206771
Chang, C.-S., Li, Y.-H., Chen, L.-T., Chen, W.-C., Hsieh, W.-P., Shin, J., et al. (2008). LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. Plant J. 54, 205–219. doi: 10.1111/j.1365-313X.2008.03401.x
Chaves-Silva, S., dos Santos, A. L., Chalfun-Júnior, A., Zhao, J., Peres, L. E. P., and Benedito, V. A. (2018). Understanding the genetic regulation of anthocyanin biosynthesis in plants – tools for breeding purple varieties of fruits and vegetables. Phytochemistry 153, 11–27. doi: 10.1016/j.phytochem.2018.05.013
Chen, L., Hu, B., Qin, Y., Hu, G., and Zhao, J. (2019). Advance of the negative regulation of anthocyanin biosynthesis by MYB transcription factors. Plant Physiol. Biochem. 136, 178–187. doi: 10.1016/j.plaphy.2019.01.024
Chen, Q., Man, C., Li, D., Tan, H., Xie, Y., and Huang, J. (2016). Arogenate dehydratase isoforms differentially regulate anthocyanin biosynthesis in Arabidopsis thaliana. Mol. Plant 9, 1609–1619. doi: 10.1016/j.molp.2016.09.010
Chialva, C., Blein, T., Crespi, M., and Lijavetzky, D. (2021). Insights into long non-coding RNA regulation of anthocyanin carrot root pigmentation. Sci. Rep. 11:4093. doi: 10.1038/s41598-021-83514-4
Colanero, S., Perata, P., and Gonzali, S. (2018). The atroviolacea gene encodes an R3-MYB protein repressing anthocyanin synthesis in tomato plants. Front. Plant Sci. 9:830. doi: 10.3389/fpls.2018.00830
Colanero, S., Tagliani, A., Perata, P., and Gonzali, S. (2020). Alternative splicing in the anthocyanin fruit gene encoding an R2R3 MYB transcription factor affects anthocyanin biosynthesis in tomato fruits. Plant Commun. 1:100006. doi: 10.1016/j.xplc.2019.100006
Cui, L. G., Shan, J. X., Shi, M., Gao, J. P., and Lin, H. X. (2014). The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 80, 1108–1117. doi: 10.1111/tpj.12712
Czemmel, S., Stracke, R., Weisshaar, B., Cordon, N., Harris, N. N., Walker, A. R., et al. (2009). The grapevine R2R3-MYB transcription factor VvMYBF1 regulates flavonol synthesis in developing grape berries. Plant Physiol. 151, 1513–1530. doi: 10.1104/pp.109.142059
De Santis, S., Scarano, A., Liso, M., Calabrese, F. M., Verna, G., Cavalcanti, E., et al. (2021). Polyphenol enriched diet administration during pregnancy and lactation prevents dysbiosis in ulcerative colitis predisposed littermates. Front. Cell. Infect. Microbiol. 11:622327. doi: 10.3389/fcimb.2021.622327
Demeilliers, C., Toufektsian, M.-C., Salen, P., Laporte, F., Petroni, K., and de Lorgeril, M. (2017). Ethanol drinking, brain mitochondrial DNA, polyunsaturated fatty acids and effects of dietary anthocyanins. Clin. Nutr. Exp. 12, 11–19. doi: 10.1016/j.yclnex.2017.01.003
Dubos, C., Le Gourrierec, J., Baudry, A., Huep, G., Lanet, E., Debeaujon, I., et al. (2008). MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 55, 940–953. doi: 10.1111/j.1365-313X.2008.03564.x
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Espley, R. V., Bovy, A., Bava, C., Jaeger, S. R., Tomes, S., Norling, C., et al. (2013). Analysis of genetically modified red-fleshed apples reveals effects on growth and consumer attributes. Plant Biotechnol. J. 11, 408–419. doi: 10.1111/pbi.12017
Espley, R. V., Brendolise, C., Chagné, D., Kutty-Amma, S., Green, S., Volz, R., et al. (2009). Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 21, 168–183. doi: 10.1105/tpc.108.059329
Espley, R. V., Butts, C. A., Laing, W. A., Martell, S., Smith, H., McGhie, T. K., et al. (2014). Dietary flavonoids from modified apple reduce inflammation markers and modulate gut microbiota in mice. J. Nutr. 144, 146–154. doi: 10.3945/jn.113.182659
Espley, R. V., Hellens, R. P., Putterill, J., Stevenson, D. E., Kutty-Amma, S., and Allan, A. C. (2007). Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 49, 414–427. doi: 10.1111/j.1365-313X.2006.02964.x
Feller, A., MacHemer, K., Braun, E. L., and Grotewold, E. (2011). Evolutionary and comparative analysis of MYB and BHLH plant transcription factors. Plant J. 66, 94–116. doi: 10.1111/j.1365-313X.2010.04459.x
Feng, S., Wang, Y., Yang, S., Xu, Y., and Chen, X. (2010). Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 232, 245–255. doi: 10.1007/s00425-010-1170-5
Ferreyra, M. L. F., Rius, S., Emiliani, J., Pourcel, L., Feller, A., Morohashi, K., et al. (2010). Cloning and characterization of a UV-B-inducible maize flavonol synthase. Plant J. 62, 77–91. doi: 10.1111/j.1365-313X.2010.04133.x
Fu, R., Martin, C., and Zhang, Y. (2018). Next-generation plant metabolic engineering, inspired by an ancient Chinese irrigation system. Mol. Plant 11, 47–57. doi: 10.1016/j.molp.2017.09.002
Fukamachi, K., Imada, T., Ohshima, Y., Xu, J., and Tsuda, H. (2008). Purple corn color suppresses ras protein level and inhibits 7,12-dimethylbenz[a]anthracene-induced mammary carcinogenesis in the rat. Cancer Sci. 99, 1841–1846. doi: 10.1111/j.1349-7006.2008.00895.x
Glover, B. J., and Martin, C. (2012). Anthocyanins. Curr. Biol. 22, 147–150. doi: 10.1016/j.cub.2012.01.021
Gonzalez, A., Brown, M., Hatlestad, G., Akhavan, N., Smith, T., Hembd, A., et al. (2016). TTG2 controls the developmental regulation of seed coat tannins in Arabidopsis by regulating vacuolar transport steps in the proanthocyanidin pathway. Dev. Biol. 419, 54–63. doi: 10.1016/j.ydbio.2016.03.031
Gonzalez, A., Zhao, M., Leavitt, J. M., and Lloyd, A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/BHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827. doi: 10.1111/j.1365-313X.2007.03373.x
Gonzali, S., Mazzucato, A., and Perata, P. (2009). Purple as a tomato: towards high anthocyanin tomatoes. Trends Plant Sci. 14, 237–241. doi: 10.1016/j.tplants.2009.02.001
Gou, J. Y., Felippes, F. F., Liu, C. J., Weigel, D., and Wang, J. W. (2011). Negative regulation of anthocyanin biosynthesis in Arabidopsis by a MiR156-targeted SPL transcription factor. Plant Cell 23, 1512–1522. doi: 10.1105/tpc.111.084525
Gould, K. S., and Lister, C. E. (2006). “Flavonoid function in plants,” in Flavonoids: Chemistry, Biochemistry and Applications. eds. Ø. M. Andersen and K. R. Markham (Boca Raton, Florida: CRC Press), 397–441.
Grosso, G., Galvano, F., Mistretta, A., Marventano, S., Nolfo, F., Calabrese, G., et al. (2013). Red orange: experimental models and epidemiological evidence of its benefits on human health. Oxidative Med. Cell. Longev. 2013:157240. doi: 10.1155/2013/157240
He, J., and Giusti, M. M. (2010). Anthocyanins: natural colorants with health-promoting properties. Annu. Rev. Food Sci. Technol. 1, 163–187. doi: 10.1146/annurev.food.080708.100754
Herath, D., Wang, T., Peng, Y., Allan, A. C., Putterill, J., and Varkonyi-Gasic, E. (2020). An improved method for transformation of Actinidia arguta utilized to demonstrate a central role for MYB110 in regulating anthocyanin accumulation in kiwiberry. Plant Cell Tissue Organ Cult. 143, 291–301. doi: 10.1007/s11240-020-01915-1
Hernandez, J. M., Feller, A., Morohashi, K., Frame, K., and Grotewold, E. (2007). The basic helix-loop-helix domain of maize r links transcriptional regulation and histone modifications by recruitment of an EMSY-related factor. Proc. Natl. Acad. Sci. U. S. A. 104, 17222–17227. doi: 10.1073/pnas.0705629104
Hernandez, J. M., Heine, G. F., Irani, N. G., Feller, A., Kim, M. G., Matulnik, T., et al. (2004). Different mechanisms participate in the R-dependent activity of the R2R3 MYB transcription factor C1. J. Biol. Chem. 279, 48205–48213. doi: 10.1074/jbc.M407845200
Hichri, I., Heppel, S. C., Pillet, J., Léon, C., Czemmel, S., Delrot, S., et al. (2010). The basic helix-loop-helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine. Mol. Plant 3, 509–523. doi: 10.1093/mp/ssp118
Hsieh, L. C., Lin, S. I., Shih, A. C. C., Chen, J. W., Lin, W. Y., Tseng, C. Y., et al. (2009). Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 151, 2120–2132. doi: 10.1104/pp.109.147280
Hyun, T. K., and Jang, K. I. (2016). Apple as a source of dietary phytonutrients: an update on the potential health benefits of apple. EXCLI J. 15, 565–569. doi: 10.17179/excli2016-483
Ibraheem, F., Gaffoor, I., Tan, Q., Shyu, C. R., and Chopra, S. (2015). A sorghum MYB transcription factor induces 3-deoxyanthocyanidins and enhances resistance against leaf blights in maize. Molecules 20, 2388–2404. doi: 10.3390/molecules20022388
Iorizzo, M., Cavagnaro, P. F., Bostan, H., Zhao, Y., Zhang, J., and Simon, P. W. (2019). A cluster of MYB transcription factors regulates anthocyanin biosynthesis in carrot (Daucus carota L.) root and petiole. Front. Plant Sci. 9:1927. doi: 10.3389/fpls.2018.01927
Jaakola, L. (2013). New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 18, 477–483. doi: 10.1016/j.tplants.2013.06.003
Jeong, S. W., Das, P. K., Jeoung, S. C., Song, J. Y., Lee, H. K., Kim, Y. K., et al. (2010). Ethylene suppression of sugar-induced anthocyanin pigmentation in Arabidopsis. Plant Physiol. 154, 1514–1531. doi: 10.1104/pp.110.161869
Jia, X., Shen, J., Liu, H., Li, F., Ding, N., Gao, C., et al. (2015). Small tandem target mimic-mediated blockage of microRNA858 induces anthocyanin accumulation in tomato. Planta 242, 283–293. doi: 10.1007/s00425-015-2305-5
Jiang, C., Gao, X., Liao, L., Harberd, N. P., and Fu, X. (2007). Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol. 145, 1460–1470. doi: 10.1104/pp.107.103788
Jin, H., Cominelli, E., Bailey, P., Parr, A., Mehrtens, F., Jones, J., et al. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19, 6150–6161. doi: 10.1093/emboj/19.22.6150
Jin, W., Wang, H., Li, M., Wang, J., Yang, Y., Zhang, X., et al. (2016). The R2R3 MYB transcription factor PavMYB10.1 involves in anthocyanin biosynthesis and determines fruit skin colour in sweet cherry (Prunus avium L.). Plant Biotechnol. J. 14, 2120–2133. doi: 10.1111/pbi.12568
Jones, C. M., Mes, P., and Myers, J. R. (2003). Characterization and inheritance of the anthocyanin fruit (Aft) tomato. J. Hered. 94, 449–456. doi: 10.1093/jhered/esg093
Jung, U. J., and Choi, M.-S. (2014). Obesity and its metabolic complications: the role of adipokines and the relationship between obesity, inflammation, insulin resistance, dyslipidemia and nonalcoholic fatty liver disease. Int. J. Mol. Sci. 15, 6184–6223. doi: 10.3390/ijms15046184
Kadowaki, T., Yamauchi, T., Kubota, N., Hara, K., Ueki, K., and Tobe, K. (2006). Adiponectin and adiponectin receptors in insulin resistance, diabetes, and the metabolic syndrome. J. Clin. Investig. 116, 1784–1792. doi: 10.1172/JCI29126
Kang, M. K., Li, J., Kim, J. L., Gong, J. H., Kwak, S. N., Park, J. H. Y., et al. (2012). Purple corn anthocyanins inhibit diabetes-associated glomerular monocyte activation and macrophage infiltration. Am. J. Physiol. Ren. Physiol. 303, 1060–1069. doi: 10.1152/ajprenal.00106.2012
Kang, K. M., Lim, S. S., Lee, J.-Y., Yeo, K. M., and Kang, Y. K. (2013). Anthocyanin-rich purple corn extract inhibit diabetes-associated glomerular angiogenesis. PLoS One 8:79823. doi: 10.1371/journal.pone.0079823
Kiferle, C., Fantini, E., Bassolino, L., Povero, G., Spelt, C., Buti, S., et al. (2015). Tomato R2R3-MYB proteins SlANT1 and SlAN2: same protein activity, different roles. PLoS One 10:e0136365. doi: 10.1371/journal.pone.0136365
Knekt, P., Kumpulainen, J., Järvinen, R., Rissanen, H., Heliövaara, M., Reunanen, A., et al. (2002). Flavonoid intake and risk of chronic diseases. Am. J. Clin. Nutr. 76, 560–568. doi: 10.1093/ajcn/76.3.560
Kobayashi, S., Goto-Yamamoto, N., and Hirochika, H. (2004). Retrotransposon-induced mutations in grape skin color. Science 304:982. doi: 10.1126/science.1095011
Kodama, M., Brinch-Pedersen, H., Sharma, S., Holme, I. B., Joernsgaard, B., Dzhanfezova, T., et al. (2018). Identification of transcription factor genes involved in anthocyanin biosynthesis in carrot (Daucus carota L.) using RNA-Seq. BMC Genomics 19:811. doi: 10.1186/s12864-018-5135-6
Koes, R., Verweij, W., and Quattrocchio, F. (2005). Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 10, 236–242. doi: 10.1016/j.tplants.2005.03.002
Kong, Q., Pattanaik, S., Feller, A., Werkman, J. R., Chai, C., Wang, Y., et al. (2012). Regulatory switch enforced by basic helix-loop-helix and ACT-domain mediated dimerizations of the maize transcription factor R. Proc. Natl. Acad. Sci. U. S. A. 109, 2091–2097. doi: 10.1073/pnas.1205513109
Kozłowska, A., and Dzierżanowski, T. (2021). Targeting inflammation by anthocyanins as the novel therapeutic potential for chronic diseases: an update. Molecules 26:4380. doi: 10.3390/molecules26144380
Kumar, D., Sarmah, B. K. S., and Das, P. S. (2019). Ethylene mediates repression of anthocyanin accumulation in black rice pericarps in the absence of light. J. Plant Physiol. 236, 34–38. doi: 10.1016/j.jplph.2019.02.011
Lachman, J., Martinek, P., Kotíková, Z., Orsák, M., and Šulc, M. (2017). Genetics and chemistry of pigments in wheat grain – a review. J. Cereal Sci. 74, 145–154. doi: 10.1016/j.jcs.2017.02.007
LaFountain, A. M., and Yuan, Y. W. (2021). Repressors of anthocyanin biosynthesis. New Phytol. 231, 933–949. doi: 10.1111/nph.17397
Lee, S. G., Kim, B., Yang, Y., Pham, T. X., Park, Y. K., Manatou, J., et al. (2014). Berry anthocyanins suppress the expression and secretion of proinflammatory mediators in macrophages by inhibiting nuclear translocation of NF-ΚB independent of NRF2-mediated mechanism. J. Nutr. Biochem. 25, 404–411. doi: 10.1016/j.jnutbio.2013.12.001
Lee, B., and Shao, J. (2006). Adiponectin and energy homeostasis. Rev. Endocr. Metab. Disord. 15, 149–156. doi: 10.1007/s11154-013-9283-3
Li, S. (2014). Transcriptional control of flavonoid biosynthesis. Plant Signal. Behav. 9:e27522. doi: 10.4161/psb.27522
Li, Y., Cui, W., Wang, R., Lin, M., Zhong, Y., Sun, L., et al. (2019). MicroRNA858-mediated regulation of anthocyanin biosynthesis in kiwifruit (Actinidia arguta) based on small RNA sequencing. PLoS One 14:e0217480. doi: 10.1371/journal.pone.0217480
Li, W., Ding, Z., Ruan, M., Yu, X., Peng, M., and Liu, Y. (2017a). Kiwifruit R2R3-MYB transcription factors and contribution of the novel AcMYB75 to red kiwifruit anthocyanin biosynthesis. Sci. Rep. 7:16861. doi: 10.1038/s41598-017-16905-1
Li, L., He, Y., Ge, H., Liu, Y., and Chen, H. (2021a). Functional characterization of SmMYB86, a negative regulator of anthocyanin biosynthesis in eggplant (Solanum melongena L.). Plant Sci. 302:110696. doi: 10.1016/j.plantsci.2020.110696
Li, J., Kang, M. K., Kim, J. K., Kim, J. L., Kang, S. W., Lim, S. S., et al. (2012a). Purple corn anthocyanins retard diabetes-associated glomerulosclerosis in mesangial cells and Db/Db mice. Eur. J. Nutr. 51, 961–973. doi: 10.1007/s00394-011-0274-4
Li, Y., Liang, J., Zeng, X., Guo, H., Luo, Y., Kear, P., et al. (2021b). Genome-wide analysis of MYB gene family in potato provides insights into tissue-specific regulation of anthocyanin biosynthesis. Hortic. Plant J. 7, 129–141. doi: 10.1016/j.hpj.2020.12.001
Li, J., Lim, S. S., Lee, J. Y., Kim, J. K., Kang, S. W., Kim, J. L., et al. (2012b). Purple corn anthocyanins dampened high-glucose-induced mesangial fibrosis and inflammation: possible renoprotective role in diabetic nephropathy. J. Nutr. Biochem. 23, 320–331. doi: 10.1016/j.jnutbio.2010.12.008
Li, Y. Y., Mao, K., Zhao, C., Zhao, X. Y., Zhang, H. L., Shu, H. R., et al. (2012c). MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol. 160, 1011–1022. doi: 10.1104/pp.112.199703
Li, S., Wang, W., Gao, J., Yin, K., Wang, R., Wang, C., et al. (2016). MYB75 phosphorylation by MPK4 is required for light-induced anthocyanin accumulation in Arabidopsis. Plant Cell 28, 2866–2883. doi: 10.1105/tpc.16.00130
Li, D., Wang, P., Luo, Y., Zhao, M., and Chen, F. (2017b). Health benefits of anthocyanins and molecular mechanisms: update from recent decade. Crit. Rev. Food Sci. Nutr. 57, 1729–1741. doi: 10.1080/10408398.2015.1030064
Li, S., and Zachgo, S. (2013). TCP3 interacts with R2R3-MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana. Plant J. 76, 901–913. doi: 10.1111/tpj.12348
Lin-Wang, K., Bolitho, K., Grafton, K., Kortstee, A., Karunairetnam, S., Mcghie, T. K., et al. (2010). An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 10:50. doi: 10.1186/1471-2229-10-50
Lin-Wang, K., McGhie, T. K., Wang, M., Liu, Y., Warren, B., Storey, R., et al. (2014). Engineering the anthocyanin regulatory complex of strawberry (Fragaria vesca). Front. Plant Sci. 5:651. doi: 10.3389/fpls.2014.00651
Liso, M., De Santis, S., Scarano, A., Verna, G., Dicarlo, M., Galleggiante, V., et al. (2018). A bronze-tomato enriched diet affects the intestinal microbiome under homeostatic and inflammatory conditions. Nutrients 10:1862. doi: 10.3390/nu10121862
Liu, Y., Lin-Wang, K., Espley, R. V., Wang, L., Li, Y., Liu, Z., et al. (2019). StMYB44 negatively regulates anthocyanin biosynthesis at high temperatures in tuber flesh of potato. J. Exp. Bot. 70, 3809–3824. doi: 10.1093/jxb/erz194
Liu, Y., Lin-Wang, K., Espley, R. V., Wang, L., Yang, H., Yu, B., et al. (2016). Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J. Exp. Bot. 67, 2159–2176. doi: 10.1093/jxb/erw014
Liu, J., Liu, X., Zhang, S., Liang, S., Luan, W., and Ma, X. (2021). TarDB: an online database for plant miRNA targets and miRNA-triggered phased siRNAs. BMC Genomics 22:348. doi: 10.1186/s12864-021-07680-5
Liu, T., Song, S., Yuan, Y., Wu, D., Chen, M., Sun, Q., et al. (2015). Improved peach peel color development by fruit bagging. Enhanced expression of anthocyanin biosynthetic and regulatory genes using white non-woven polypropylene as replacement for yellow paper. Sci. Hortic. 184, 142–148. doi: 10.1016/j.scienta.2015.01.003
Liu, Y., Tikunov, Y., Schouten, R. E., Marcelis, L. F. M., Visser, R. G. F., and Bovy, A. (2018). Anthocyanin biosynthesis and degradation mechanisms in solanaceous vegetables: a review. Front. Chem. 6:52. doi: 10.3389/fchem.2018.00052
Lloyd, A., Brockman, A., Aguirre, L., Campbell, A., Bean, A., Cantero, A., et al. (2017). Advances in the MYB-BHLH-WD repeat (MBW) pigment regulatory model: addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 58, 1431–1441. doi: 10.1093/pcp/pcx075
Long, N., Suzuki, S., Sato, S., Naiki-Ito, A., Sakatani, K., Shirai, T., et al. (2013). Purple corn color inhibition of prostate carcinogenesis by targeting cell growth pathways. Cancer Sci. 104, 298–303. doi: 10.1111/cas.12078
Lorenc-Kukuła, K., Jafra, S., Oszmiański, J., and Szopa, J. (2005). Ectopic expression of anthocyanin 5-o-glucosyltransferase in potato tuber causes increased resistance to bacteria. J. Agric. Food Chem. 53, 272–281. doi: 10.1021/jf048449p
Lotkowska, M. E., Tohge, T., Fernie, A. R., Xue, G. P., Balazadeh, S., and Mueller-Roeber, B. (2015). The Arabidopsis transcription factor MYB112 promotes anthocyanin formation during salinity and under high light stress. Plant Physiol. 169, 1862–1880. doi: 10.1104/pp.15.00605
Luo, Q. J., Mittal, A., Jia, F., and Rock, C. D. (2012). An autoregulatory feedback loop involving PAP1 and TAS4 in response to sugars in Arabidopsis. Plant Mol. Biol. 80, 117–129. doi: 10.1007/s11103-011-9778-9
Maeda, H., Yamaguchi, T., Omoteno, M., Takarada, T., Fujita, K., Murata, K., et al. (2014). Genetic dissection of black grain rice by the development of a near isogenic line. Breed. Sci. 64, 134–141. doi: 10.1270/jsbbs.64.134
Magni, G., Marinelli, A., Riccio, D., Lecca, D., Tonelli, C., Abbracchio, M. P., et al. (2018). Purple corn extract as anti-allodynic treatment for trigeminal pain: role of microglia. Front. Cell. Neurosci. 12:378. doi: 10.3389/fncel.2018.00378
Mahmood, K., Xu, Z., El-Kereamy, A., Casaretto, J. A., and Rothstein, S. J. (2016). The Arabidopsis transcription factor ANAC032 represses anthocyanin biosynthesis in response to high sucrose and oxidative and abiotic stresses. Front. Plant Sci. 7:1548. doi: 10.3389/fpls.2016.01548
Maier, A., Schrader, A., Kokkelink, L., Falke, C., Welter, B., Iniesto, E., et al. (2013). Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J. 74, 638–651. doi: 10.1111/tpj.12153
Martin, C., Butelli, E., Petroni, K., and Tonelli, C. (2011). How can research on plants contribute to promoting human health? Plant Cell 23, 1685–1699. doi: 10.1105/tpc.111.083279
Mathews, H., Clendennen, S. K., Caldwell, C. G., Liu, L., Connors, K., Matheis, N., et al. (2003). Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 15, 1689–1703. doi: 10.1105/tpc.012963
Matsui, K., Umemura, Y., and Ohme-Takagi, M. (2008). AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 55, 954–967. doi: 10.1111/j.1365-313X.2008.03565.x
Mattioli, R., Francioso, A., Mosca, L., and Silva, P. (2020). Anthocyanins: a comprehensive review of their chemical properties and health effects on cardiovascular and neurodegenerative diseases. Molecules 25:3809. doi: 10.3390/molecules25173809
Matus, J., Cavallini, E., Loyola, R., Finezzo, L., Santo, S. D., Vialet, S., et al. (2017). A group of grapevine MYBA transcription factors located in chromosome 14 control anthocyanin synthesis in vegetative organs with different specificities compared with the berry color locus. Plant J. 91, 220–236. doi: 10.1111/tpj.13558
Matus, J. T., Poupin, M. J., Cañón, P., Bordeu, E., Alcalde, J. A., and Arce-Johnson, P. (2010). Isolation of WDR and BHLH genes related to flavonoid synthesis in grapevine (Vitis vinifera L.). Plant Mol. Biol. 72, 607–620. doi: 10.1007/s11103-010-9597-4
McIntosh, R. A., Yanmazaki, Y., Devos, K., Dubcovsky, J., Rogers, J., and Apples, R. (2003). Catalogue of gene symbols for wheat. Available at: https://shigen.nig.ac.jp/wheat/komugi/genes/symbolClassList.jsp
Meng, X., Wang, J.-R., Wang, G.-D., Liang, X.-Q., Li, X.-D., and Meng, Q.-W. (2015). An R2R3-MYB gene, LeAN2, positively regulated the thermo-tolerance in transgenic tomato. J. Plant Physiol. 175, 1–8. doi: 10.1016/j.jplph.2014.09.018
Mes, P. J., Boches, P., Myers, J. R., and Durst, R. (2008). Characterization of tomatoes expressing anthocyanin in the fruit. J. Am. Soc. Hortic. Sci. 133, 262–269. doi: 10.21273/JASHS.133.2.262
Morishita, T., Kojima, Y., Maruta, T., Nishizawa-Yokoi, A., Yabuta, Y., and Shigeoka, S. (2009). Arabidopsis NAC transcription factor, ANAC078, regulates flavonoid biosynthesis under high-light. Plant Cell Physiol. 50, 2210–2222. doi: 10.1093/pcp/pcp159
Nag, A., King, S., and Jack, T. (2009). MiR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 106, 22534–22539. doi: 10.1073/pnas.0908718106
Napoleone, E., Cutrone, A., Zurlo, F., Di Castelnuovo, A., D’Imperio, M., Giordano, L., et al. (2013). Both red and blond orange juice intake decreases the procoagulant activity of whole blood in healthy volunteers. Thromb. Res. 132, 288–292. doi: 10.1016/j.thromres.2013.06.022
Nardozza, S., Boldingh, H. L., Kashuba, M. P., Feil, R., Jones, D., Thrimawithana, A. H., et al. (2020). Carbon starvation reduces carbohydrate and anthocyanin accumulation in red-fleshed fruit via trehalose 6-phosphate and MYB27. Plant Cell Environ. 43, 819–835. doi: 10.1111/pce.13699
Nguyen, N. H., Jeong, C. Y., Kang, G. H., Yoo, S. D., Hong, S. W., and Lee, H. (2015). MYBD employed by HY5 increases anthocyanin accumulation via repression of MYBL2 in Arabidopsis. Plant J. 84, 1192–1205. doi: 10.1111/tpj.13077
Ohno, S., Ueno, M., and Doi, M. (2020). Differences in the CaMYBA genome between anthocyanin-pigmented cultivars and non-pigmented cultivars in pepper (Capsicum annuum). Hortic. J. 89, 30–36. doi: 10.2503/hortj.UTD-097
Oikawa, T., Maeda, H., Oguchi, T., Yamaguchi, T., Tanabe, N., Ebana, K., et al. (2015). The birth of a black rice gene and its local spread by introgression. Plant Cell 27, 2401–2414. doi: 10.1105/tpc.15.00310
Paolocci, F., Robbins, M. P., Passeri, V., Hauck, B., Morris, P., Rubini, A., et al. (2011). The strawberry transcription factor FaMYB1 inhibits the biosynthesis of proanthocyanidins in lotus corniculatus leaves. J. Exp. Bot. 62, 1189–1200. doi: 10.1093/jxb/erq344
Passeri, V., Koes, R., and Quattrocchio, F. M. (2016). New challenges for the design of high value plant products: stabilization of anthocyanins in plant vacuoles. Front. Plant Sci. 7:153. doi: 10.3389/fpls.2016.00153
Patra, B., Pattanaik, S., and Yuang, L. (2013a). Ubiquitin protein ligase 3 mediates the proteasomal degradation of GLABROUS 3 and ENHANCER of GLABROUS 3, regulators of trichome development and flavonoid biosynthesis in Arabidopsis. Plant J. 74, 435–447. doi: 10.1111/tpj.12132
Patra, B., Pattanaik, S., and Yuan, L. (2013b). Proteolytic degradation of the flavonoid regulators, TRANSPARENT TESTA8 and TRANSPARENT TESTA GLABRA1, in Arabidopsis is mediated by the ubiquitin/26sproteasome system. Plant Signal. Behav. 8, 6–9. doi: 10.4161/psb.25901
Patra, B., Schluttenhofer, C., Wu, Y., Pattanaik, S., and Yuan, L. (2013c). Transcriptional regulation of secondary metabolite biosynthesis in plants. Biochim. Biophys. Acta 1829, 1236–1247. doi: 10.1016/j.bbagrm.2013.09.006
Payyavula, R. S., Singh, R. K., and Navarre, D. A. (2013). Transcription factors, sucrose, and sucrose metabolic genes interact to regulate potato phenylpropanoid metabolism. J. Exp. Bot. 64, 5115–5131. doi: 10.1093/jxb/ert303
Pedreschi, R., and Cisneros-Zevallos, L. (2007). Phenolic profiles of andean purple corn (Zea mays L.). Food Chem. 100, 956–963. doi: 10.1016/j.foodchem.2005.11.004
Peng, Z., Han, C., Yuan, L., Zhang, K., Huang, H., and Ren, C. (2011). Brassinosteroid enhances jasmonate-induced anthocyanin accumulation in Arabidopsis seedlings. J. Integr. Plant Biol. 53, 632–640. doi: 10.1111/j.1744-7909.2011.01042.x
Peng, Y., Thrimawithana, A. H., Cooney, J. M., Jensen, D. J., Espley, R. V., and Allan, A. C. (2020). The proanthocyanin-related transcription factors MYBC1 and WRKY44 regulate branch points in the kiwifruit anthocyanin pathway. Sci. Rep. 10:14161. doi: 10.1038/s41598-020-70977-0
Pérez-Díaz, J. R., Pérez-Díaz, J., Madrid-Espinoza, J., González-Villanueva, E., Moreno, Y., and Ruiz-Lara, S. (2016). New member of the R2R3-MYB transcription factors family in grapevine suppresses the anthocyanin accumulation in the flowers of transgenic tobacco. Plant Mol. Biol. 90, 63–76. doi: 10.1007/s11103-015-0394-y
Persico, G., Casciaro, F., Marinelli, A., Tonelli, C., Petroni, K., and Giorgio, M. (2021). Comparative analysis of histone H3k4me3 distribution in mouse liver in different diets reveals the epigenetic efficacy of cyanidin-3-o-glucoside dietary intake. Int. J. Mol. Sci. 22:6503. doi: 10.3390/ijms22126503
Petroni, K., Pilu, R., and Tonelli, C. (2014). Anthocyanins in corn: a wealth of genes for human health. Planta 240, 901–911. doi: 10.1007/s00425-014-2131-1
Petroni, K., and Tonelli, C. (2011). Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 181, 219–229. doi: 10.1016/j.plantsci.2011.05.009
Petroni, K., Trinei, M., Fornari, M., Calvenzani, V., Marinelli, A., Micheli, L. A., et al. (2017). Dietary cyanidin 3-glucoside from purple corn ameliorates doxorubicin-induced cardiotoxicity in mice. Nutr. Metab. Cardiovasc. Dis. 27, 462–469. doi: 10.1016/j.numecd.2017.02.002
Pojer, E., Mattivi, F., Johnson, D., and Stockley, C. S. (2013). The case for anthocyanin consumption to promote human health: a review. Compr. Rev. Food Sci. Food Saf. 12, 483–508. doi: 10.1111/1541-4337.12024
Povero, G., Gonzali, S., Bassolino, L., Mazzucato, A., and Perata, P. (2011). Transcriptional analysis in high-anthocyanin tomatoes reveals synergistic effect of Aft and Atv genes. J. Plant Physiol. 168, 270–279. doi: 10.1016/j.jplph.2010.07.022
Prior, R. L., Wu, X., Gu, L., Hager, T. J., Hager, A., and Howard, L. R. (2008). Whole berries versus berry anthocyanins: interactions with dietary fat levels in the C57BL/6J mouse model of obesity. J. Agric. Food Chem. 56, 647–653. doi: 10.1021/jf071993o
Qi, T., Song, S., Ren, Q., Wu, D., Huang, H., Chen, Y., et al. (2011). The jasmonate-ZIM-domain proteins interact with the WD-repeat/BHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23, 1795–1814. doi: 10.1105/tpc.111.083261
Ramsay, N. A., and Glover, B. J. (2005). MYB-BHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10, 63–70. doi: 10.1016/j.tplants.2004.12.011
Rowan, D. D., Cao, M., Lin-Wang, K., Cooney, J. M., Jensen, D. J., Austin, P. T., et al. (2009). Environmental regulation of leaf colour in red 35S:PAP1 Arabidopsis thaliana. New Phytol. 182, 102–115. doi: 10.1111/j.1469-8137.2008.02737.x
Saclier, M., Bonfanti, C., Antonini, S., Angelini, G., Mura, G., Zanaglio, F., et al. (2020). Nutritional intervention with cyanidin hinders the progression of muscular dystrophy. Cell Death Dis. 11:127. doi: 10.1038/s41419-020-2332-4
Sainz, M. B., Grotewold, E., and Chandleragl, V. L. (1997). Evidence for direct activation of an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related Myb domain proteins. Plant Cell 9, 611–625. doi: 10.1105/tpc.9.4.611
Sakamoto, W., Ohmori, T., Kageyama, K., Miyazaki, C., Saito, A., Murata, M., et al. (2001). The Purple leaf (Pl) locus of rice: the Pl(w) allele has a complex organization and includes two genes encoding basic helix-loop-helix proteins involved in anthocyanin biosynthesis. Plant Cell Physiol. 42, 982–991. doi: 10.1093/pcp/pce128
Salamone, F., Volti, G. L., Titta, L., Puzzo, L., Barbagallo, I., La Delia, F., et al. (2012). Moro orange juice prevents fatty liver in mice. World J. Gastroenterol. 18, 3862–3868. doi: 10.3748/wjg.v18.i29.3862
Salehi, B., Sharifi-Rad, J., Cappellini, F., Reiner, Z., Zorzan, D., Imran, M., et al. (2020). The therapeutic potential of anthocyanins: current approaches based on their molecular mechanism of action. Front. Pharmacol. 11:1300. doi: 10.3389/fphar.2020.01300
Sapir, M., Oren-Shamir, M., Ovadia, R., Reuveni, M., Evenor, D., Tadmor, Y., et al. (2008). Molecular aspects of anthocyanin fruit tomato in relation to high pigment-1. J. Hered. 99, 292–303. doi: 10.1093/jhered/esm128
Sarma, A. D., and Sharma, R. (1999). Anthocyanin-DNA copigmentation complex: mutual protection against oxidative damage. Phytochemistry 52, 1313–1318. doi: 10.1016/S0031-9422(99)00427-6
Scarano, A., Butelli, E., De Santis, S., Cavalcanti, E., Hill, L., De Angelis, M., et al. (2018). Combined dietary anthocyanins, flavonols, and stilbenoids alleviate inflammatory bowel disease symptoms in mice. Front. Nutr. 4:75. doi: 10.3389/fnut.2017.00075
Schaart, J. G., Dubos, C., De La Fuente, I. R., Van Houwelingen, A. M., De Vos, R. C. H., Jonker, H. H., et al. (2013). Identification and characterization of MYB-BHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (fragaria × ananassa) fruits. New Phytol. 197, 454–467. doi: 10.1111/nph.12017
Schreiber, G., Reuveni, M., Evenor, D., Oren-Shamir, M., Ovadia, R., Sapir-Mir, M., et al. (2012). ANTHOCYANIN1 from Solanum chilense is more efficient in accumulating anthocyanin metabolites than its Solanum lycopersicum counterpart in association with the ANTHOCYANIN FRUIT phenotype of tomato. Theor. Appl. Genet. 124, 295–307. doi: 10.1007/s00122-011-1705-6
Sharma, D., Tiwari, M., Pandey, A., Bhatia, C., Sharma, A., and Trivedi, P. K. (2016). MicroRNA858 is a potential regulator of phenylpropanoid pathway and plant development. Plant Physiol. 171, 944–959. doi: 10.1104/pp.15.01831
Shi, S., Liu, Y., He, Y., Li, L., Li, D., and Chen, H. (2021). R2R3-MYB transcription factor SmMYB75 promotes anthocyanin biosynthesis in eggplant (Solanum melongena L.). Sci. Hortic. 282:110020. doi: 10.1016/j.scienta.2021.110020
Shin, D., Choi, M., Kim, K., Bang, G., Cho, M., Choi, S.-B., et al. (2013). HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 587, 1543–1547. doi: 10.1016/j.febslet.2013.03.037
Speciale, A., Anwar, S., Canali, R., Chirafisi, J., Saija, A., Virgili, F., et al. (2013). Cyanidin-3-O-glucoside counters the response to TNF-alpha of endothelial cells by activating Nrf2 pathway. Mol. Nutr. Food Res. 57, 1979–1987. doi: 10.1002/mnfr.201300102
Spelt, C., Quattrocchio, F., Mol, J., and Koes, R. (2002). ANTHOCYANIN1 of petunia controls pigment synthesis, vacuolar PH, and seed coat development by genetically distinct mechanisms. Plant Cell 14, 2121–2135. doi: 10.1105/tpc.003772
Stommel, J. R., and Dumm, J. M. (2015). Coordinated regulation of biosynthetic and regulatory genes coincides with anthocyanin accumulation in developing eggplant fruit. J. Am. Soc. Hortic. Sci. 140, 129–135. doi: 10.21273/JASHS.140.2.129
Stracke, R., Ishihara, H., Huep, G., Barsch, A., Mehrtens, F., Niehaus, K., et al. (2007). Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50, 660–677. doi: 10.1111/j.1365-313X.2007.03078.x
Strazzer, P., Spelt, C. E., Li, S., Bliek, M., Federici, C. T., Roose, M. L., et al. (2019). Hyperacidification of citrus fruits by a vacuolar proton-pumping P-ATPase complex. Nat. Commun. 10:744. doi: 10.1038/s41467-019-08516-3
Su, Z., Wang, X., Xuan, X., Sheng, Z., Jia, H., Emal, N., et al. (2021). Characterization and action mechanism analysis of VvmiR156b/c/d-VvSPL9 module responding to multiple-hormone signals in the modulation of grape berry color formation. Foods 10:986. doi: 10.3390/foods10040896
Sun, C., Deng, L., Du, M., Zhao, J., Chen, Q., Huang, T., et al. (2019). A transcriptional network promotes anthocyanin biosynthesis in tomato flesh. Mol. Plant 13, 42–58. doi: 10.1016/j.molp.2019.10.010
Sun, X., Zhang, Z., Chen, C., Wu, W., Ren, N., Jiang, C., et al. (2018). The C-S-A gene system regulates hull pigmentation and reveals evolution of anthocyanin biosynthesis pathway in rice. J. Exp. Bot. 69, 1485–1498. doi: 10.1093/jxb/ery001
Sweeney, M. T., Thomson, M. J., Pfeil, B. E., and McCouch, S. (2006). Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 18, 283–294. doi: 10.1105/tpc.105.038430
Takos, A. M., Jaffé, F. W., Jacob, S. R., Bogs, J., Robinson, S. P., and Walker, A. R. (2006). Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 142, 1216–1232. doi: 10.1104/pp.106.088104
Tamagnone, L., Merida, A., Parr, A., Mackay, S., Culianez-Macia, F. A., Roberts, K., et al. (1998). The AmMYB308 and AmMYB330 transcription factors from antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10, 135–154. doi: 10.1105/tpc.10.2.135
Tang, B., Li, L., Hu, Z., Chen, Y., Tan, T., Jia, Y., et al. (2020). Anthocyanin accumulation and transcriptional regulation of anthocyanin biosynthesis in purple pepper. J. Agric. Food Chem. 68, 12152–12163. doi: 10.1021/acs.jafc.0c02460
Tirumalai, V., Swetha, C., Nair, A., Pandit, A., and Shivaprasad, P. V. (2019). MiR828 and MiR858 regulate VvMYB114 to promote anthocyanin and flavonol accumulation in grapes. J. Exp. Bot. 70, 4775–4791. doi: 10.1093/jxb/erz264
Titta, L., Trinei, M., Stendardo, M., Berniakovich, I., Petroni, K., Tonelli, C., et al. (2010). Blood orange juice inhibits fat accumulation in mice. Int. J. Obes. 34, 578–588. doi: 10.1038/ijo.2009.266
Tohge, T., Nishiyama, Y., Hirai, M. Y., Yano, M., Nakajima, J. I., Awazuhara, M., et al. (2005). Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 42, 218–235. doi: 10.1111/j.1365-313X.2005.02371.x
Tomay, F., Marinelli, A., Leoni, V., Caccia, C., Matros, A., Mock, H.-P., et al. (2019). Purple corn extract induces long-lasting reprogramming and M2 phenotypic switch of adipose tissue macrophages in obese mice. J. Transl. Med. 17:237. doi: 10.1186/s12967-019-1972-6
Toufektsian, M.-C., De Lorgeril, M., Nagy, N., Salen, P., Donati, M. B., Giordano, L., et al. (2008). Chronic dietary intake of plant-derived anthocyanins protects the rat heart against ischemia-reperfusion injury. J. Nutr. 138, 747–752. doi: 10.1093/jn/138.4.747
Toufektsian, M. C., Salen, P., Laporte, F., Tonelli, C., and De Lorgeril, M. (2011). Dietary flavonoids increase plasma very long-chain (n-3) fatty acids in rats. J. Nutr. 141, 37–41. doi: 10.3945/jn.110.127225
Tsuda, T. (2012). Dietary anthocyanin-rich plants: biochemical basis and recent progress in health benefits studies. Mol. Nutr. Food Res. 56, 159–170. doi: 10.1002/mnfr.201100526
Tsuda, T., Horio, F., Uchida, K., Aoki, H., and Osawa, T. (2003). Dietary cyanidin 3-O-β-D-glucoside-rich purple corn color prevents obesity and ameliorates hyperglycemia in mice. J. Nutr. 133, 2125–2130. doi: 10.1093/jn/133.7.2125
Tsuda, T., Ueno, Y., Yoshikawa, T., Kojo, H., and Osawa, T. (2006). Microarray profiling of gene expression in human adipocytes in response to anthocyanins. Biochem. Pharmacol. 71, 1184–1197. doi: 10.1016/j.bcp.2005.12.042
Tuerck, J. A., and Fromm, M. E. (1994). Elements of the maize A1 promoter required for transactivation by the anthocyanin BICI or phlobaphene P regulatory genes. Plant Cell 6, 1655–1663. doi: 10.1105/tpc.6.11.1655
Ullah, R., Khan, M., Shah, S. A., Saeed, K., and Kim, M. O. (2019). Natural antioxidant anthocyanins—a hidden therapeutic candidate in metabolic disorders with major focus in neurodegeneration. Nutrients 11:1195. doi: 10.3390/nu11061195
Vimolmangkang, S., Han, Y., Wei, G., and Korban, S. S. (2013). An apple MYB transcription factor, MdMYB3, is involved in regulation of anthocyanin biosynthesis and flower development. BMC Plant Biol. 13:176. doi: 10.1186/1471-2229-13-176
Wada, T., Tachibana, T., Shimura, Y., and Okada, K. (1997). Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277, 1113–1116. doi: 10.1126/science.277.5329.1113
Walker, A. R., Lee, E., Bogs, J., McDavid, D. A. J., Thomas, M. R., and Robinson, S. P. (2007). White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 49, 772–785. doi: 10.1111/j.1365-313X.2006.02997.x
Wallace, T. C. (2013). “Anthocyanins in cardiovascular disease prevention,” in Anthocyanins in Health and Disease. eds. T. C. Wallace and M. Giusti (Boca Raton: CRC Press), 165–197.
Wang, L., Tang, W., Hu, Y., Zhang, Y., Sun, J., Guo, X., et al. (2019). A MYB/BHLH complex regulates tissue-specific anthocyanin biosynthesis in the inner pericarp of red-centered kiwifruit Actinidia chinensis cv. Hongyang. Plant J. 99, 359–378. doi: 10.1111/tpj.14330
Wang, Y., Wang, Y., Song, Z., and Zhang, H. (2016). Repression of MYBL2 by both microRNA858a and HY5 leads to the activation of anthocyanin biosynthetic pathway in Arabidopsis. Mol. Plant 9, 1395–1405. doi: 10.1016/j.molp.2016.07.003
Wang, Y. C., Wang, N., Xu, H. F., Jiang, S. H., Fang, H. C., Su, M. Y., et al. (2018). Auxin regulates anthocyanin biosynthesis through the Aux/IAA–ARF signaling pathway in apple. Hortic. Res. 5:59. doi: 10.1038/s41438-018-0068-4
Wang, L., Wang, B., Yu, H., Guo, H., Lin, T., Kou, L., et al. (2020). Transcriptional regulation of strigolactone signalling in Arabidopsis. Nature 583, 277–281. doi: 10.1038/s41586-020-2382-x
Xie, X. B., Li, S., Zhang, R. F., Zhao, J., Chen, Y. C., Zhao, Q., et al. (2012). The BHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 35, 1884–1897. doi: 10.1111/j.1365-3040.2012.02523.x
Xie, Y., Tan, H., Ma, Z., and Huang, J. (2016). DELLA proteins promote anthocyanin biosynthesis via sequestering MYBL2 and JAZ suppressors of the MYB/BHLH/WD40 complex in Arabidopsis thaliana. Mol. Plant 9, 711–721. doi: 10.1016/j.molp.2016.01.014
Xu, W., Dubos, C., and Lepiniec, L. (2015). Transcriptional control of flavonoid biosynthesis by MYB-BHLH-WDR complexes. Trends Plant Sci. 20, 176–185. doi: 10.1016/j.tplants.2014.12.001
Xu, Z. S., Feng, K., Que, F., Wang, F., and Xiong, A. S. (2017a). A MYB transcription factor, DcMYB6, is involved in regulating anthocyanin biosynthesis in purple carrot taproots. Sci. Rep. 7:45324. doi: 10.1038/srep45324
Xu, H., Wang, N., Liu, J., Qu, C., Wang, Y., Jiang, S., et al. (2017b). The molecular mechanism underlying anthocyanin metabolism in apple using the MdMYB16 and MdbHLH33 genes. Plant Mol. Biol. 94, 149–165. doi: 10.1007/s11103-017-0601-0
Xu, Z. S., Yang, Q. Q., Feng, K., and Xiong, A. S. (2019). Changing carrot color: insertions in DcMYB7 alter the regulation of anthocyanin biosynthesis and modification. Plant Physiol. 181, 195–207. doi: 10.1104/pp.19.00523
Xu, Z. S., Yang, Q. Q., Feng, K., Yu, X., and Xiong, A. S. (2020). DcMYB113, a root-specific R2R3-MYB, conditions anthocyanin biosynthesis and modification in carrot. Plant Biotechnol. J. 18, 1585–1597. doi: 10.1111/pbi.13325
Yan, S., Chen, N., Huang, Z., Li, D., Zhi, J., Yu, B., et al. (2020). Anthocyanin fruit encodes an R2R3-MYB transcription factor, SlAN2-like, activating the transcription of SlMYBATV to fine-tune anthocyanin content in tomato fruit. New Phytol. 225, 2048–2063. doi: 10.1111/nph.16272
Yang, F., Cai, J., Yang, Y., and Liu, Z. (2013). Overexpression of microRNA828 reduces anthocyanin accumulation in Arabidopsis. Plant Cell Tissue Organ Cult. 115, 159–167. doi: 10.1007/s11240-013-0349-4
Yao, G., Ming, M., Allan, A. C., Gu, C., Li, L., Wu, X., et al. (2017). Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J. 92, 437–451. doi: 10.1111/tpj.13666
Ye, H., Li, L., Guo, H., and Yin, Y. (2012). MYBL2 is a substrate of GSK3-like kinase BIN2 and acts as a corepressor of BES1 in brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 109, 20142–20147. doi: 10.1073/pnas.1205232109
Yuste, S., Ludwig, I. A., Romero, M.-P., Piñol-Felis, C., Catalán, U., Pedret, A., et al. (2021). Metabolic fate and cardiometabolic effects of phenolic compounds from red-fleshed apple in hypercholesterolemic rats: a comparative study with common white-fleshed apple. The AppleCOR study. Mol. Nutr. Food Res. 65:e2001225. doi: 10.1002/mnfr.202001225
Zhang, Y., Butelli, E., Alseekh, S., Tohge, T., Rallapalli, G., Luo, J., et al. (2015). Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat. Commun. 6:8635. doi: 10.1038/ncomms9635
Zhang, Y., Butelli, E., and Martin, C. (2014a). Engineering anthocyanin biosynthesis in plants. Curr. Opin. Plant Biol. 19, 81–90. doi: 10.1016/j.pbi.2014.05.011
Zhang, Y., Chu, G., Hu, Z., Gao, Q., Cui, B., Tian, S., et al. (2016). Genetically engineered anthocyanin pathway for high health-promoting pigment production in eggplant. Mol. Breed. 36, 1–14. doi: 10.1007/s11032-016-0454-2
Zhang, F., Gonzalez, A., Zhao, M., Payne, C. T., and Lloyd, A. (2003). A network of redundant BHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130, 4859–4869. doi: 10.1242/dev.00681
Zhang, Z., Shi, Y., Ma, Y., Yang, X., Yin, X., Zhang, Y., et al. (2020a). The strawberry transcription factor FaRAV1 positively regulates anthocyanin accumulation by activation of FaMYB10 and anthocyanin pathway genes. Plant Biotechnol. J. 18, 2267–2279. doi: 10.1111/pbi.13382
Zhang, B., Yang, H. J., Yang, Y. Z., Zhu, Z. Z., Li, Y. N., Qu, D., et al. (2020b). Mdm-MiR828 participates in the feedback loop to regulate anthocyanin accumulation in apple peel. Front. Plant Sci. 11:608109. doi: 10.3389/fpls.2020.608109
Zhang, H., Zhao, X., Li, J., Cai, H., Deng, X. W., and Li, L. (2014b). Microrna408 is critical for the HY5-SPl7 gene network that mediates the coordinated response to light and copper. Plant Cell 26, 4933–4953. doi: 10.1105/tpc.114.127340
Zhao, Y., Dong, W., Zhu, Y., Allan, A. C., Lin-Wang, K., and Xu, C. (2020). PpGST1, an anthocyanin-related glutathione S-transferase gene, is essential for fruit coloration in peach. Plant Biotechnol. J. 18, 1284–1295. doi: 10.1111/pbi.13291
Zheng, J., Wu, H., Zhu, H., Huang, C., Liu, C., Chang, Y., et al. (2019). Determining factors, regulation system, and domestication of anthocyanin biosynthesis in rice leaves. New Phytol. 223, 705–721. doi: 10.1111/nph.15807
Zhou, B., Leng, J., Ma, Y., Fan, P., Li, Y., Yan, H., et al. (2020). BrmiR828 targets BrPAP1, BrMYB82, and BrTAS4 involved in the light induced anthocyanin biosynthetic pathway in Brassica rapa. Int. J. Mol. Sci. 21:4326. doi: 10.3390/ijms21124326
Zhou, H., Lin-Wang, K., Wang, F., Espley, R. V., Ren, F., Zhao, J., et al. (2019). Activator-type R2R3-MYB genes induce a repressor-type R2R3-MYB gene to balance anthocyanin and proanthocyanidin accumulation. New Phytol. 221, 1919–1934. doi: 10.1111/nph.15486
Zhou, H., Lin-Wang, K., Wang, H., Gu, C., Dare, A. P., Espley, R. V., et al. (2015). Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 82, 105–121. doi: 10.1111/tpj.12792
Zhou, L.-L., Shi, M.-S., and Xie, D.-Y. (2012). Regulation of anthocyanin biosynthesis by nitrogen in TTG1-GL3/TT8-PAP1-programmed red cells of Arabidopsis thaliana. Planta 236, 825–837. doi: 10.1007/s00425-012-1674-2
Zhu, H. F., Fitzsimmons, K., Khandelwal, A., and Kranz, R. G. (2009). CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis. Mol. Plant 2, 790–802. doi: 10.1093/mp/ssp030
Zhu, Y. C., Zhang, B., Allan, A. C., Lin-Wang, K., Zhao, Y., Wang, K., et al. (2020). DNA demethylation is involved in the regulation of temperature-dependent anthocyanin accumulation in peach. Plant J. 102, 965–976. doi: 10.1111/tpj.14680
Keywords: MYB, bHLH, WD40, transcription factors, anthocyanins, chronic diseases
Citation: Cappellini F, Marinelli A, Toccaceli M, Tonelli C and Petroni K (2021) Anthocyanins: From Mechanisms of Regulation in Plants to Health Benefits in Foods. Front. Plant Sci. 12:748049. doi: 10.3389/fpls.2021.748049
Edited by:
Igor Cesarino, University of São Paulo, BrazilReviewed by:
Carolyn Lister, The New Zealand Institute for Plant and Food Research Ltd., New ZealandChris Rock, Texas Tech University, United States
Copyright © 2021 Cappellini, Marinelli, Toccaceli, Tonelli and Petroni. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katia Petroni, a2F0aWEucGV0cm9uaUB1bmltaS5pdA==
†ORCID: Francesca Cappellini, orcid.org/0000-0002-9472-424X
Alessandra Marinelli, orcid.org/0000-0002-6501-5364
Chiara Tonelli, orcid.org/0000-0002-9505-3118
Katia Petroni, orcid.org/0000-0003-3318-0442
 Francesca Cappellini
Francesca Cappellini Alessandra Marinelli
Alessandra Marinelli Marta Toccaceli
Marta Toccaceli Chiara Tonelli
Chiara Tonelli Katia Petroni
Katia Petroni