Corrigendum: Characterization of Fusarium Spp. Inciting Vascular Wilt of Tomato and Its Management by a Chaetomium-Based Biocontrol Consortium
- 1ICAR-Indian Agricultural Research Institute, New Delhi, India
- 2ICAR-National Institute of Plant Biotechnology, New Delhi, India
- 3ICAR-Central Coastal Agricultural Research Institute, Goa, India
Though the vascular wilt of tomato caused by the species of Fusarium is globally reported to be a complex disease in certain countries, for example, India, our studies indicated that the disease is caused by either Fusarium oxysporum f. spp. lycopersici (Fol) or Fusarium solani (FS) with the Fol being widely prevalent. In assessing the genetic diversity of 14 Fol strains representing the four Indian states by the unweighted pair group method with arithmetic averaging using Inter Simple Sequence Repeat (ISSR) amplicons, the strains distinguished themselves into two major clusters showing no correlation with their geographic origin. In pot experiments under polyhouse conditions, the seed dressing and soil application of a talc-based formulation of a biocontrol treatment, TEPF-Sungal-1 (Pseudomonas putida) + S17TH (Trichoderma harzianum) + CG-A (Chaetomium globosum), which inhibited Fol, was equally effective like the cell suspensions and was even better than the fungicidal mixture (copper oxychloride-0.25% + carbendazim-0.1%) in promoting the crop growth (52.3%) and reducing vascular wilt incidence (75%) over the control treatment, despite the challenge of inoculation with a highly pathogenic TOFU-IHBT strain. This was associated with significant expressions of the defense genes, indicating the induction of host resistance by a biocontrol consortium. In field experiments on two locations, the bioconsortium was highly effective in recording maximum mean fruit yields (54.5 and 60%) and a minimum mean vascular wilt incidence (37.5%) in comparison to the untreated control. Thus, Chaetomium-based bioconsortium demonstrated consistency in its performance across the two experiments in 2 years under the two field conditions.
Introduction
Successful cultivation of tomato (Lycopersicon esculentum Mill) is limited by the vascular wilt disease caused by Fusarium oxysporum f. spp. lycopersici (Fol) W. C. Synder and H. N. Hans (Sidharthan et al., 2018, 2019). The disease inflicts a substantial yield loss ranging from 25 to 55% in many tomato growing areas of the country (Nirmaladevi et al., 2016) besides deteriorating the quality of fruits. Management practices have little impact on vascular wilt incidence mainly due to highly resistant chlamydospores produced by the pathogen under unfavorable conditions (Shanmugam and Kanoujia, 2011) and limited studies on the genetic variability of the fungal pathogen (Akbar et al., 2018). The soil application of fungicides besides failing to control the disease (Nirmaladevi et al., 2016) under field conditions, the appearance of fungicide-resistant pathogen strains, toxicity, and negative environmental effects of fungicides are of great concern (Shanmugam and Kanoujia, 2011). Although resistant tomato cultivars bearing major gene (I/I-1, I-2, and I-3) resistance to Fusarium wilt have been developed elsewhere (Huang and Lindhout, 1997) in India, no resistant varieties have been reported. Hence, the biological management of the disease could be a potential and attractive alternate management strategy that needs to be exploited.
In the biomanagement of Fusarium wilt of tomato, the potential of some plant-growth promoting rhizobacteria microbes such as the species of Bacillus, Pseudomonas, and Trichoderma has been reported under glasshouse conditions (Shanmugam et al., 2011a,b, 2015; Shanmugam and Kanoujia, 2011). Of late, the exploration of new biocontrol agents with an ample potential for disease control in plants leads to the identification of several species of Chaetomium as useful bioagents (Aggarwal et al., 2014). The filamentous fungi belonging to the class Ascomycetes and the family Chaetomiaceae usually survive as saprophytes in organic compost or soil or as endophytes in host plants (Aggarwal et al., 2008). Though some species are exploited in biotechnological industries for the case of being good laccase and cellulose producers (Abdel-Azeem et al., 2016), the species Chaetomium globosum has largely been identified as a potential biocontrol agent against damping off and several seed and soil borne diseases. Like other biocontrol agents, the biocontrol mechanisms in C. globosum are attributed to a competition for nutrients and substrates, mycoparasitism, antibiosis, or their combinations (Zhang and Yang, 2007). Besides exerting a direct antifungal activity, the biocontrol fungus is also known to induce systemic resistance in crop plants (Biswas et al., 2000; Aggarwal et al., 2016). The induction of defense enzymes has mostly been implicated in such plant-mediated resistance (Soytong et al., 2001). Despite the exploitation of C. globosum in biocontrol, its potential for the biomanagement of Fusarium wilt of tomato has not been reported.
Though the biological management of plant diseases using a single biocontrol agent is effective under controlled conditions, the performance of the bioagents is inconsistent in the field due to complex soil environments (Shanmugam and Kanoujia, 2011). The use of a microbial consortium is a potential alternative approach for the uncertainty experienced in biocontrol under field conditions. The strains constituting the consortia need to be carefully selected in such a way that they express different plant-growth promoting and biocontrol traits and that they complement each other to express the traits together (Shanmugam et al., 2013). Microbial consortia that perform better than individual strains in improving crop productivity by promoting plant growth directly or indirectly through their effects on the pathogen have been well documented in several crops (Shanmugam et al., 2011a,b, 2013; Shanmugam and Kanoujia, 2011). Several of these microbial consortia mainly constituted the species of Bacillus, Pseudomonas, Serratia, Penicillium, and Trichoderma (Shanmugam et al., 2013; Yang et al., 2014). The use of C. globosum for the biomanagement of plant diseases has mostly been resorted to using a single strain (Aggarwal, 2015) and not in a consortium. Microbial consortia consisting of antagonistic bacteria and fungi have an added advantage of protecting the plants under different conditions or at different times by occupying complementary or different niches (Larkin and Fravel, 1998).
In the present study, we, therefore, aimed to characterize the Fol inciting the vascular wilt of tomato to understand its genetic variability and to develop and test carrier-based formulations of microbial consortia comprising antagonistic Bacillus subtilis (S2BC-1), Pseudomonas putida (TEPF-Sungal-1), Trichoderma harzianum (S17TH), and C. globosum (CG-A) in varying combinations under glasshouse conditions. The induction of resistance in the crop plant by the consortium was assessed based on the expression of candidate defense genes. The potential consortium identified from the studies was then validated under field conditions for Fusarium wilt control.
Materials and Methods
Pathogen Isolates
Field surveys were carried out in the tomato fields of IARI Farm and National Capital Region (NCR) along the Yamuna plain (six locations) during 2017–2018, and Fusarium-infected plant samples were collected from each of the locations. The isolation of the fungus was made on a potato dextrose agar (PDA) medium from the infected tissues by the direct plating method as described earlier (Shanmugam et al., 2013). These isolates together with putative Fusarium isolates obtained from vascular wilt-infected tomato samples and available in the culture collections of the laboratory were used for these studies. The isolates were purified by a single spore isolation method, and their morphological characters (Nelson et al., 1983) were assessed to establish their identities. For pathogenicity assay, a conidial suspension was prepared as described earlier (Shanmugam et al., 2013) by pouring 20 ml of sterile tap water onto 7-day-old culture of each of the isolates. The aerial mycelium was then gently scraped using a sterile glass slide, and the fungal suspension was filtered using a sterile double-layer muslin cloth. The suspension was quantified with a hemocytometer, and the spore density was adjusted to 1 × 104 conidia ml–1 with sterile tap water. The inoculation of the spore suspension was performed on 4-week-old tomato (cv. Pusa Rohini) plants maintained in a polyhouse by dipping the wounded roots in the conidial suspension for 10 min. Tomato plants dipped in sterile tap water served as control. The treated seedlings were then planted in 10-cm plastic pots containing 1.5 kg sterile soil. The wilt incidence was determined as percent disease index (PDI) using the scale described earlier (Shanmugam et al., 2011a) as 0 = no symptoms; 1 ≤ 25% of the leaves with symptoms; 2 = 26–50% of the leaves with symptoms; 3 = 51–75% of the leaves with symptoms; and 4 = 76–100% of the leaves with symptoms. The PDI was calculated as follows: Disease index = [Σ(rating × number of plants rated)/Total number of plants × highest rating] × 100. The pathogenic fungi were then reisolated from the diseased tissues exhibiting vascular browning by plating them on PDA after surface sterilization (1% sodium hypochlorite) and Koch’s postulates were confirmed. The fungus inciting a maximum wilt incidence was rated as a highly pathogenic isolate. The identity of the pathogenic isolates was reconfirmed by PCR amplification and partial sequencing of the Internal Transcribed Spacer (ITS) region and transcription elongation factor (tef) gene from the genomic DNA as described earlier (Divakara et al., 2014; Sidharthan et al., 2018). To isolate the genomic DNA, two individual colonies of each of the fungal isolates grown on PDA plates were inoculated separately in 100 ml of potato dextrose broth (PDB). The inoculated flasks were incubated on a rotary shaker at 28 ± 2°C and 180 rpm for 5 days. The mycelium was filtered out on a sterile Whatman No. 1 filter paper, washed three times with sterile distilled water to remove traces of the medium, and air-dried. The dried mycelial mat was ground with a mortar and pestle in liquid nitrogen, and the DNA was isolated from the ground mycelium by a commercial kit (Sigma, Burlington, MA, United States) following the manufacturer’s protocol. For the molecular characterization of the pathogen isolates, a region of the nuclear rRNA gene containing ITS regions 1 and 4 and the 5.8S rRNA gene was amplified from the genomic DNA by PCR using the primers, ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) to get an amplicon size of 500–550 bp. PCR amplification of the tef gene (600 bp) from the genomic DNA was done with the primers, EF-F [5′-ATGGGTAAGGA(A/G)GACAAGAC-3′] and TEF-R [5′-GGA(G/A)GTACCAGT(G/C)ATCATGTT-3′]. The amplification of either of the genes was carried out in a 50 μl reaction volume consisting of 10X buffer, 5.0 μl; 2 mM dNTPs, 2.0 μl; 3 U/μl Taq DNA polymerase, 1.0 μl; 100 ng/μl of each primer, 2 μl; 50–100 ng template DNA, 3 μl; and H2O 32.0 μl in a Biorad thermal cycler using the PCR conditions of 94°C for 4 min (initial denaturation), and 35 cycles of 94°C for 1 min (denaturation), 55°C for 1 min (annealing), and 72°C for 2 min (extension). The PCR product was purified, was cloned into a pGEM-T easy vector (Promega, Madison, WI, United States) following the manufacturer’s protocol, and was sequenced. The deduced sequences were evaluated for a homology with the NCBI database, and the sequences were deposited in GenBank. The pathogenic isolates were stored as 15% glycerol stocks at −80°C in the laboratory culture collections.
Genetic Diversity of Fol Strains
The deduced sequences of the ITS region were aligned using the LALIGN program and were grouped based on the levels of similarities in the consensus sequences. PCR amplifications of the Inter Simple Sequence Repeat (ISSR) region of genomic DNA were done with the primers, ISSR9 5′-GAGAGAGAGAGAGAGAGAC-3′, and ISSR10 5′-GAGAGAGAGAGAGAGAGAT-3′ (Nirmaladevi et al., 2016) and UBC855 (5′-ACACACACACACACACYT-3′ and UBC 856 (5′-ACACACACACACACACYA-3′) (Lin et al., 2012) in a Applied BiosystemsTM A24811 thermocycler using the conditions of 94°C for 5 min, 94°C for 30 s, 50°C for 1 min, and 72°C for 2 min for 35 cycles, with a final extension of 72°C for 10 min. The reaction mixture (25 μl) consisted of 14.5 μl of MilliQ water, 2.5 μl of 10X PCR buffer, 0.5 μl of 10 mM dNTPs, 2 μl of primer, 0.3 μl of 3 U of Taq DNA polymerase, and 100 ng genomic DNA as the template. The reaction mixture without the genomic DNA was served as the control. The PCR mixture was electrophoresed in 1% agarose gel, and the bands appearing consistently in the three PCR amplifications were evaluated.
Biocontrol Strains and Evaluation for Antagonism
Four biocontrol strains, S2BC-1 (B. subtilis, GenBank No. AM268039), TEPF-Sungal-1 (P. putida, GenBank No. MZ363827), S17TH (T. harzianum, GenBank No. GU048855), and CG-A (C. globosum GenBank No. AY429049) available in the culture collections of the laboratory were used for these studies. The antifungal activity of the biocontrol strains against the fungal pathogen was established by a dual-culture assay on PDA (Shanmugam et al., 2011a,b). The biocontrol strains were either inoculated after 48 h of pathogen inoculation (S2BC-1 and TEPF-Sungal-1) or co-inoculated (S17TH and CG-A) with the pathogen on the same day in triplicate. PDA plates inoculated with the pathogen alone served as control. Upon incubating at 28°C ± 2°C and when the control plate displayed a full growth, the zone of inhibition between the colony margins was measured. A mutual inhibition among the biocontrol strains was also assessed by the dual-culture assay in triplicate.
Preparation of Carrier-Based Formulations of Antagonistic Strains
The biocontrol strains and strain mixtures were mass multiplied in talc-based formulations to contain 1 cfu g–1 × 108 cfu g–1 and 3 cfu g–1 × 106 cfu g–1 of antagonistic bacteria and fungi, respectively (Shanmugam et al., 2011b). Briefly, the bacterial inoculum was prepared by suspending the cells (3 cfu ml–1 × 109 cfu ml–1) grown in 150 ml of nutrient (S2BC-1) or King’s B (TEPF-Sungal-1) broth at 28°C ± 2°C on a rotary shaker for 48 h and collected by centrifugation at 6,000 g for 15 min at 4°C in 0.2 M sodium phosphate buffer (pH 7.0). The fungal inoculum was prepared by suspending the spores (3 cfu ml–1 × 109 cfu ml–1) collected from 150 ml of potato dextrose broth after separating the mycelium by passing the inoculum through a muslin cloth. The broth was earlier inoculated with a mycelial disk (5 mm) of S17TH or CG-A and incubated at 28°C ± 2°C on a rotary shaker for 10 days. To prepare the inoculum for strain mixtures, the designated strains were cultured on their respective media as described, and equal volumes (v/v) of the strains were used. The carrier-based formulation for each of the treatments was then prepared by aseptically mixing 400 ml of the inoculum with autoclaved (121°C for 20 min for 2 consecutive days) talc powder (1 kg) containing 15 g CaCO3 and 10 g carboxymethyl cellulose (CMC). The moisture content in the product was reduced to less than 20% by shade drying before storage at 28 ± 2°C. The population of the rhizobacteria and antagonistic fungi in the bio-formulation was 1 cfu g–1 × 108cfu g–1 and 3 cfu g–1 × 106cfu g–1.
Pot Culture Studies on Fusarium Wilt Management
The experiment was carried out on tomato cv. Pusa Rohini as described (Shanmugam et al., 2011a, 2015) for a period of 120 days from sowing in a polyhouse using a completely randomized block design with three replicates containing five plants per treatment. For seed treatment, the seeds were surface sterilized with sodium hypochlorite (1%) for 5 min and then soaked in the cell or spore suspensions (3 cfu ml–1 × 109 cfu ml–1) of the antagonistic bacteria and fungi, and sterile distilled water containing talc-based formulations (20 g–1) were applied as seed treatment. After overnight (12 h) incubation at room temperature (28°C ± 2°C) in dark, the suspension was drained off and the seeds were dried under shade for 30 min. Ten seeds were sown in individual 20 cm depth and 20 cm diameter plastic pots containing 6.5 kg steam sterilized soil (121°C, 30 min for 2 consecutive days). Thirty-day-old seedlings were thinned to six per pot and 15 days after thinning, the plants were infected by incorporating 50 ml of conidial suspension (1 × 104 conidia ml–1) of a highly pathogenic TOFU-IHBT strain in soil. The cell or spore suspension or talc-based formulation (8 g) containing antagonistic cells was applied two times to the soil at a 45- and 60-day old plant. Seed treatment and soil application with carbendazim (0.1%; 1 g/L) + copper oxychloride (0.25%; 2.5 g/L) were maintained as a fungicide control. Seeds treated with pathogen alone and untreated seeds served as control. The experimental setup was maintained at 20–30°C and 90–95% relative humidity. The severity of wilt incidence and PDI were determined as described earlier. The root and shoot lengths were recorded at the time of harvest. The cultivar developed systemic infection from 4 to 6 weeks of the inoculation.
Tissue Collection, Enzyme Extraction, and Assays
For the potential biocontrol treatment and the uninoculated control, three plants were uprooted without injury after 6 days of pathogen inoculation and the root samples were processed for gene expression studies. The samples were initially washed under running tap water and then rinsed in distilled water. The samples were homogenized with liquid nitrogen in a prechilled mortar and pestle, and the homogenized root tissues were stored at −80°C. Total RNA was isolated from 100 mg of the frozen samples using Tri-reagent (Invitrogen, Waltham, MA, United States) following the manufacturer’s protocol. cDNA synthesis was carried out with 1 μg of RNA by adding 1 μl of oligo DT, 0.5 μl dNTPs, 2 μl 10X RT buffer, 1 μl Firescript reverse transcriptase, 0.5 μl RNAse inhibitor, and reverse transcription was carried out at 37°C for 30 min and 85°C for 5 min. The amounts of cDNA in the samples were balanced by employing actin (ToActin F 5′-AGGCAGGATTTGCTGGTGATGATGCT-3′ and ToActin R 5′-ATACGCATC CTTCTGTCCCATTCCGA-3′) as a house-keeping gene. Quantitative reverse transcription PCR (RT-PCR) was carried out in a 30 μl reaction volume consisting of cDNA (2 μl), SYBR Green (15 μl), and 100 nM final concentration of defense gene primers (1 μl) (Table 1). The assays were conducted in a (Roche, Basel, Switzerland) Light cycler 480 with an initial denaturation for 180 s at 95°C, 40 cycles of 95°C for 15 s, 60°C for 20 s, and 72°C for 20 s. Each of the samples along with no template control was used in triplicate, and the experiment was done in duplicate.
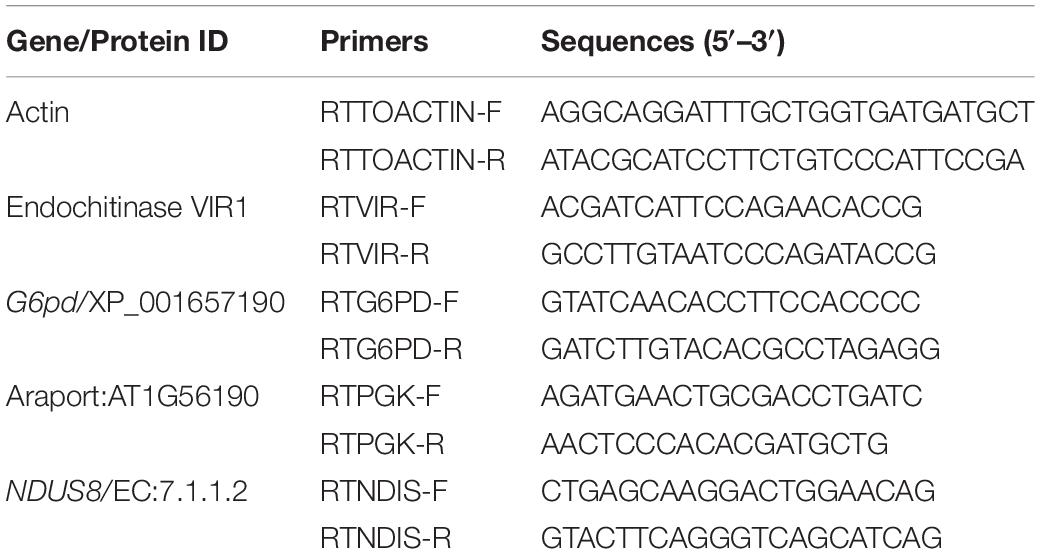
Table 1. Primers used in qRT-PCR assay to assess the expressions of defense genes in tomato induced by the biocontrol treatments on challenge inoculation with the Fusarium oxysporum f. sp. lycopersici strain, TOFU-IHBT.
Biocontrol Under Field Conditions
The carrier-based formulations of the best performing individual strains and strain mixtures identified from polyhouse studies were further assessed for their performances by the two field experiments at the host institute, an endemic location for the vascular wilt disease. Both the field experiments on tomato cv. Pusa Rohini were carried out from mid-October to mid-March during 2018 and 2019. The soil texture in the field was clay loam with a pH of 5.8 and an EC of 0.095 mmhos cm–1. The organic matter content was 4.07%, and the amount of N:P:K was 222.66:39.73:562.01 kg ha–1). The tomato seeds were soaked overnight in a water solution made with the talc-based formulations (20 g L–1) of the biocontrol consortia. The seeds were separated from the biocontrol suspension by draining off the latter, dried overnight under aseptic conditions, and then planted in 20 cm diameter plastic pots after drying overnight. One-month-old seedlings of 16 numbers were transplanted in plots of 6′ (1.8 m) × 4′ (1.2 m) size at 40 cm × 40 cm spacing accommodating four rows of four seedlings each. The plots were replicated three times and arranged in a randomized block design. The formulation was also applied to the soil (200 g bed–1) three times at 45, 60, and 75 days of planting by mixing with an equal quantity of farmyard manure. Copper oxychloride-0.25% + carbendazim-0.1% were applied as seedling treatment and soil application, and the combination was used as a fungicide control. Seedlings treated with formulation slurry followed by soil application and untreated seedlings served as control. Agronomic practices, such as irrigation, fertilization, and other cultural practices like weeding and hoeing were followed as a recommendation to the farmers. Ten plants were chosen at random from each plot and assessed for natural incidence (PDI) of yellows 65 days after sowing. Fruits were collected regularly and weighed, and 180-day cumulative yields were calculated as fruit number and weight per treatment. The yield parameters on shoot length and root length were recorded at the time of harvest. The Fusarium population in the rhizospheres of inoculated plants was determined at 0- (control), 30-, 60-, and 90-day post-inoculation. Isolation was made by a serial dilution plate technique on Fo-G1 (Nishimura, 2007), and the fungal number was determined by the plate counting method. The isolates were purified and checked for pathogenicity as described earlier.
Data Analyses
The NTSYS.PC (Numerical Taxonomy System Applied Biostatistics, Setauket, NY, United States) computer program was employed for the analysis of the fingerprints. The data (presence or absence of band) were introduced in the form of a binary matrix, and a pairwise similarity matrix was constructed using the Jaccard coefficient. The dendrogram was generated by the Unweighted Pair-Group Method with Arithmetical Averages (UPGMA) method using the NTSYS program. For data analysis, the presence or absence of a given band was scored as 1 and 0, respectively. The pot culture experiments were repeated two times with similar results. Hence, one representative trial is reported. Statistical analyses for ANOVA were conducted using the IRRISTAT version 92-1 program (IRRI, 1992) developed by the biometrics unit at the International Rice Research Institute, the Philippines. Differences between the mean values of the treatments were determined using an LSD test at a 0.05 probability level.
Results
Identification of the Fusarium Isolates
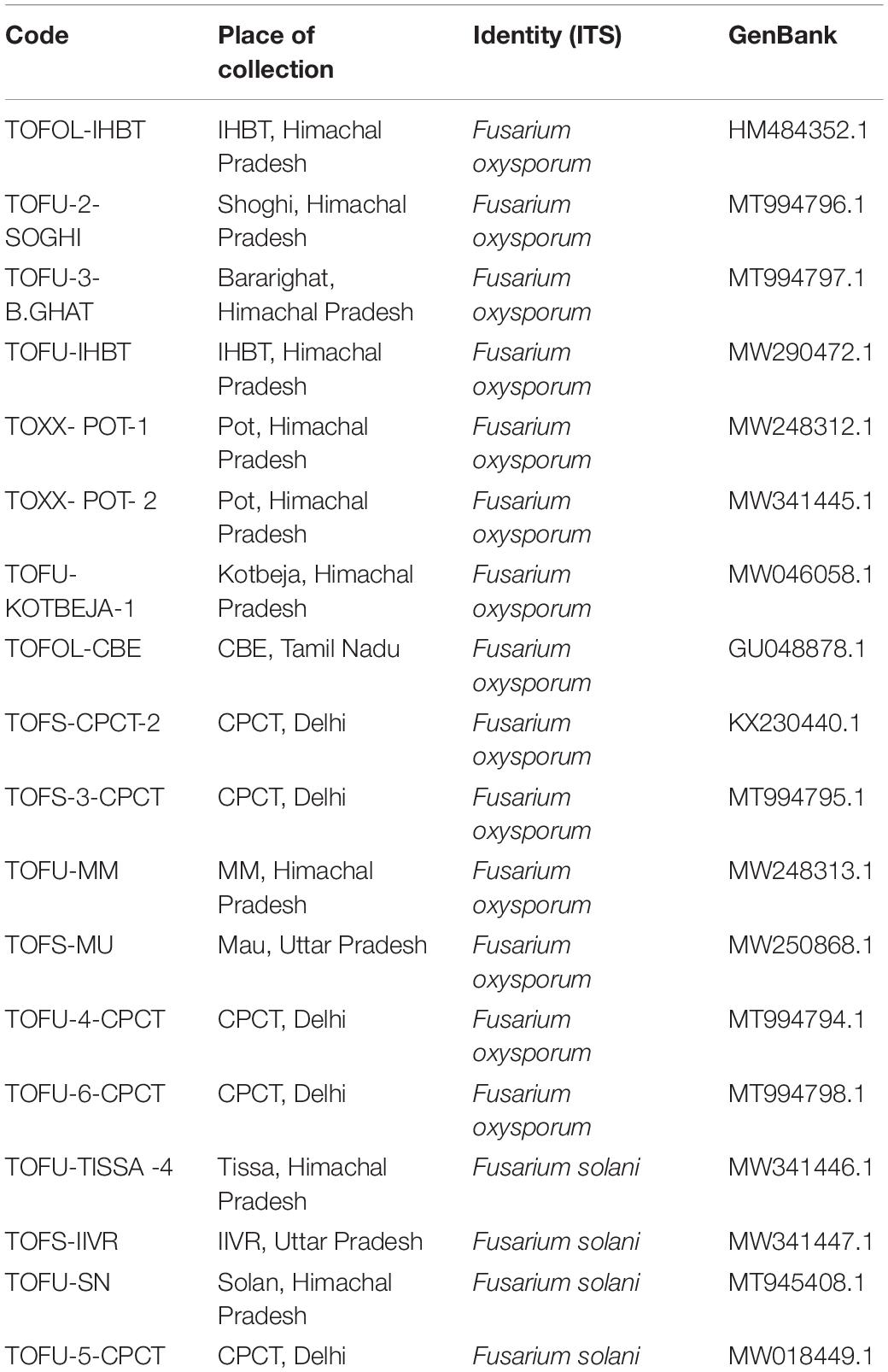
Fusarium wilt of tomato is universally present to the extent of 17–32% in the tomato growing regions surveyed in the states (Uttar Pradesh and Tamil Nadu) of India. Based on the morphological characterization of the fungal isolates on potato carrot agar, only the association of F. oxysporum. (Nelson et al., 1983) was observed in the infected stem vascular tissues of Tamil Nadu. On the contrary, one sample of Uttar Pradesh yielded Fusarium solani (FS) in addition to Fol. Among the laboratory collections, except for the two samples of Himachal Pradesh and one sample of Delhi, Fol was predominant (Table 2). On PDA, Fol displayed white aerial mycelium tinged with loosely floccose, delicate, purple pigmentation with occasional bluish violet sclerotial bodies (Figure 1). Conidiophores are branched and unbranched monophialides and are short with abundant microconidia. The microconidia are largely single-celled, kidney or oval-shaped, and are produced in false heads, which distinguish the fungus from the closely related Fusarium species. Macroconidia are abundant, thin-walled and delicate, slightly curved with a foot-shaped basal cell, and an attenuated apical cell, 4–6 septate. Chlamydospores appear singly or in pairs. The growth of FS on PDA was rapid with white and dense aerial mycelium with abundant macroconidia and microconidia. Microconidia were lemon to pear-shaped with 0–1 septum. Macroconidia were sickle-shaped, and their basal cells were distinctly notched or foot-shaped. Like Fol, the chlamydospores also appear singly or in chains. In the artificial inoculation assays to prove pathogenicity, the plants inoculated with either of the fungal species on incubating at 85% humidity and 28°C developed vascular wilt symptoms in 7–14 days, and the symptoms were similar to those of the diseased tomato plants. The diseased plant tissues on reisolation yielded the fungal species displaying typical conidial and colony morphologies characteristic to the respective inoculated fungus. The asymptomatic and uninoculated control plants did not yield any fungus. Though all the 14 Fol and four Fs isolates were pathogenic, one isolate, TOFU-IHBT, characterized as Fol and could inflict vascular wilt symptoms (100 per cent PDI) within 3 weeks is identified as a highly pathogenic one and used for pot culture studies on biocontrol. In sequencing the ITS region of the 18 isolates to confirm their identity, the Fol and Fs isolates displayed 99–100% sequence identities to those of the strains, WZ-176 of F. oxysporum (GenBank Accession No. MN856310.1) and CP1 of FS (GenBank Accession No. MH729019.1) from the databases. The identity of the isolates was further confirmed by sequencing the tef gene. Both the isolates of Fol and Fs displayed 99–100% sequence identities to those of the strains, A117-W-YY of F. oxysporum (Accession MT313924.1) and NRRL 22639 of FS (Accession No. MK818416) from the databases. Thus, the identity of all the 18 isolates was established based on colony characteristics by proving Koch’s postulates and by the analyses of the ITS and tef sequences.
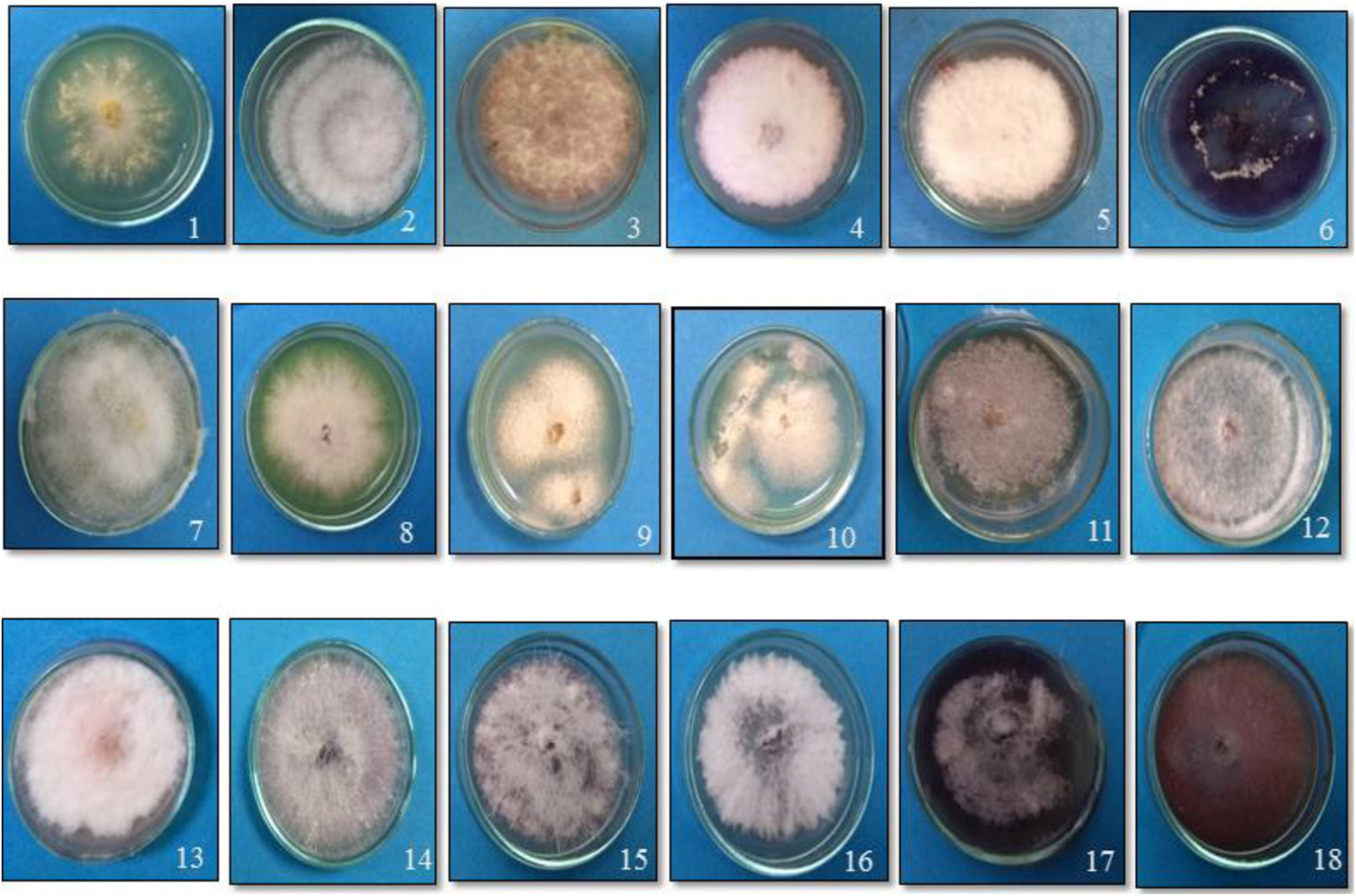
Figure 1. Growth of the putative Fusarium isolates of tomato on potato dextrose agar medium. (1) TOFOL-IHBT; (2) TOFU-2-SOGHI; (3) TOFU-3-B.GHAT; (4) TOFU-IHBT; (5) TOXX- POT-1; (6) TOXX- POT- 2; (7) TOFU-KOTBEJA-1; (8) TOFOL-CBE; (9) TOFS-CPCT-2; (10) TOFS-3-CPCT; (11) TOFU-MM; (12) TOFS-MU; (13) TOFU-4-CPCT; (14) TOFU- 6-CPCT; (15)TOFU-TISSA-4; (16) TOFS-IIVR; (17) TOFU-SN; (18) TOFU-5.CPCT.
Genetic Diversity of Fusarium oxysporum Strains
In the phylogenetic analyses of the ITS sequences, the strains of Fol and Fs form separate clusters and were grouped along with the reference strains of F. oxysporum (Accession No. KT211527.1) and FS (Accession No. KX650831.1), respectively, as a single cluster (Figure 2). The extent of genetic diversity among the strains was further assessed by studying the level of polymorphisms exhibited by the ISSR region. In PCR amplification of genomic DNA of the pathogenic strains, four primers, UBC 855, UBC856, ISSR9, and ISSR10, gave specific, reproducible, and scorable bands with a high percentage of polymorphisms (40–60%). The number of polymorphic bands varied from 15 to 20 with 200–3,500 bp size range. No monomorphic bands were observed in all the primers. Among the primers, only the pattern generated by UBC855 and ISSR10 differentiated the strains of F. oxysporum (Figure 3) generating 60% polymorphisms. The Jaccard’s similarity coefficients among the strains varied from 0.36 to 0.79%. The maximum similarity coefficient was observed for the Fol strains and the strains, and TOFS-CPCT-2 and TOFU-4-CPCT showed 100% similarity. In the UPGMA analysis, the genetic relatedness among the Fol strains revealed by the dendrogram (Figure 4) as two major clusters. Cluster I consist of three Fol strains, TOFOL-IHBT, TOXX-POT2, and TOFU-6-CPCT. Cluster II was further divided into two sub-clusters. Sub-cluster IIa consists of seven strains, which are further grouped into two separate clusters comprising four and three strains. The four strains, TOFU-2-SOGHI, TOFU-3B. GHAT, TOFU-IHBT, and TOXX-POT1, exhibited similarity indices of 60–72%. In the latter, though the strains exhibited 45% similarity among themselves, the strains TOFS-CPCT2 and TOFU-4-CPCT exhibited 100% similarity. Four strains consisting of TOFU-KOTBEJA, TOFS-3-CPCT, TOFU-MM, and TOFS-MU formed the second sub-cluster IIb with similarity indices of 50–70%. Interestingly, the clustering did not correlate with the geographic origins of the strains, and the two largest clusters contained strains from all the regions.
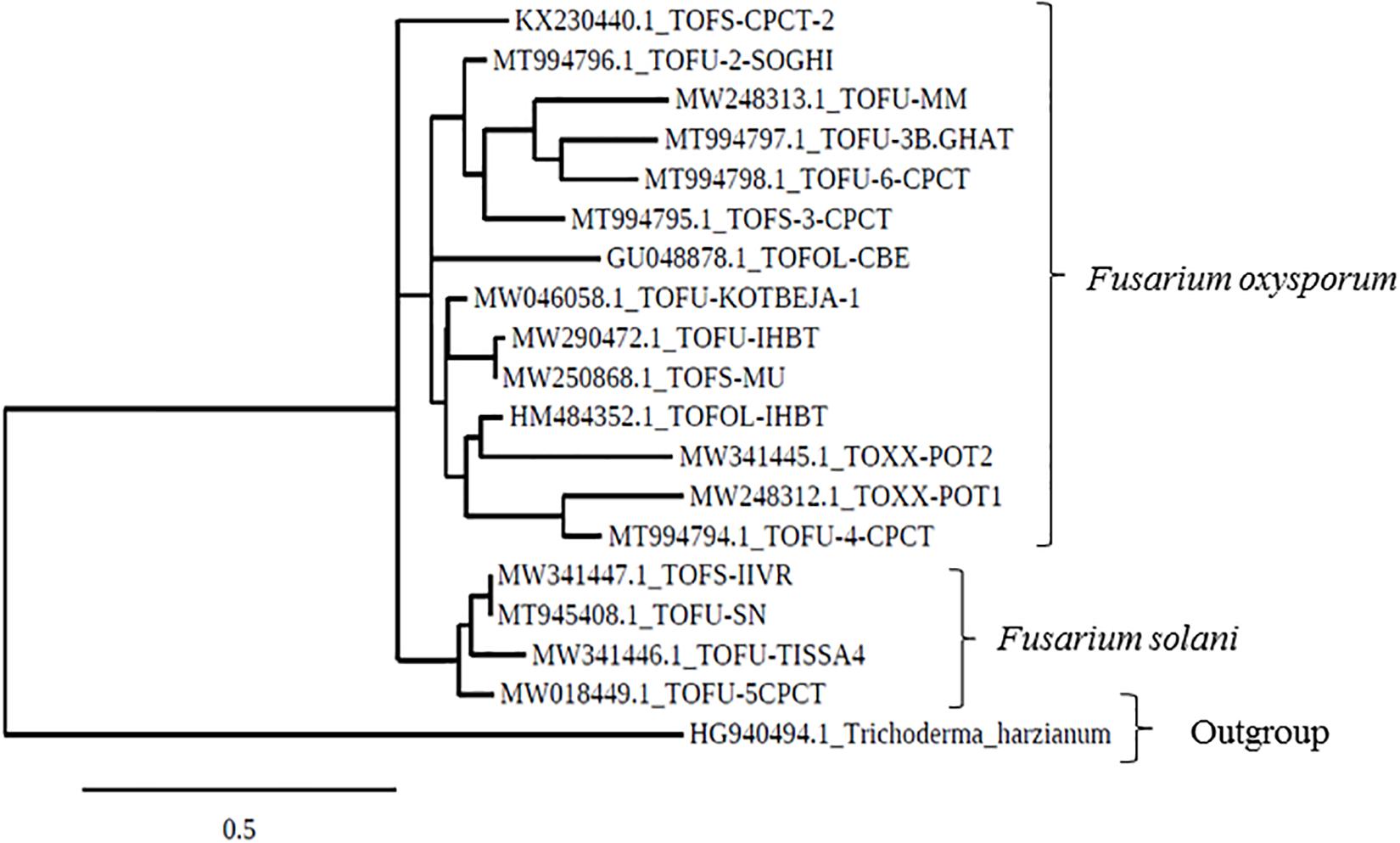
Figure 2. Phylogenetic tree based on ITS gene sequences, drawn using the Maximum likelihood method and showing the relation between Fusarium strains.
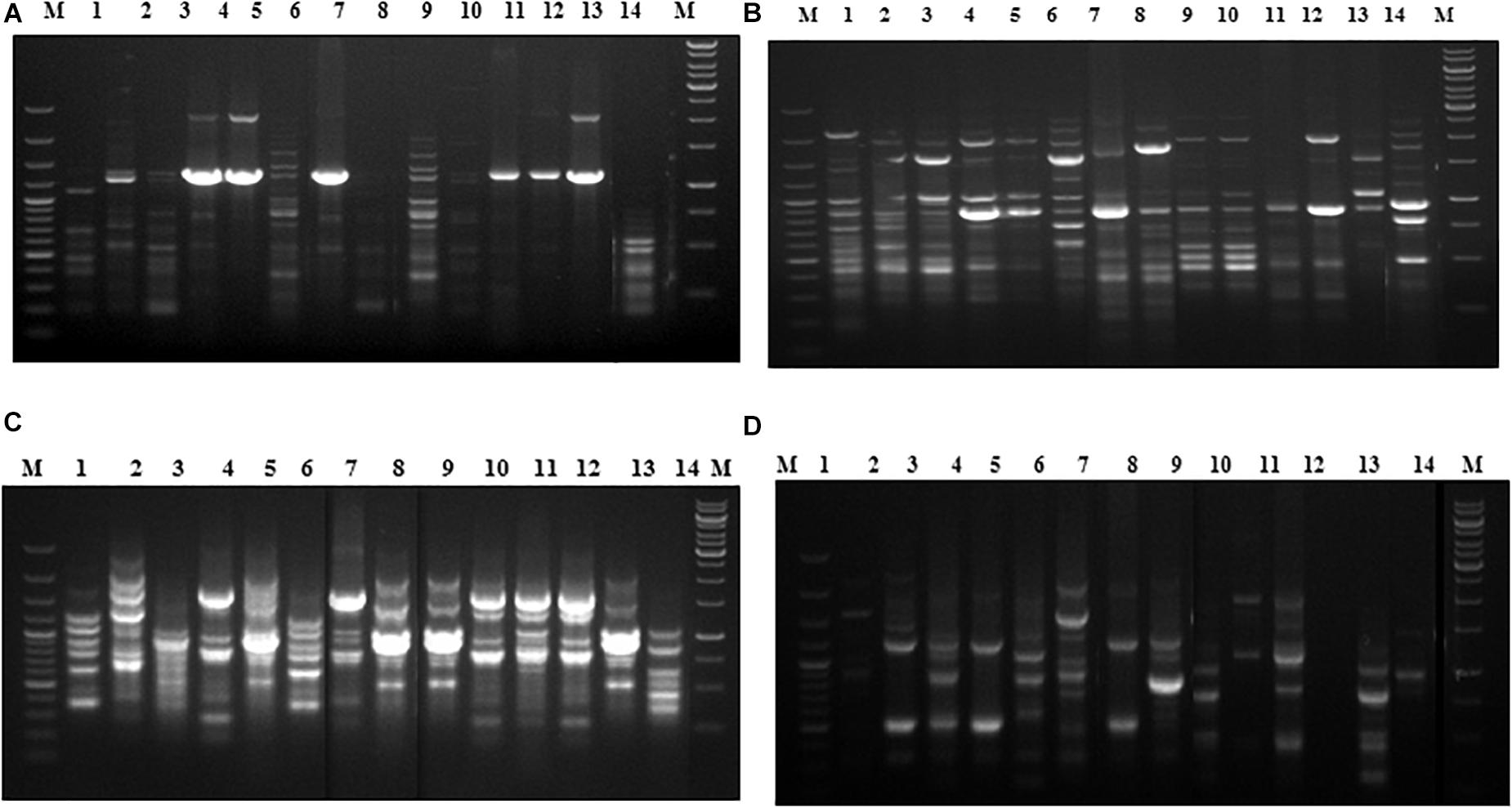
Figure 3. Fingerprinting of Fusarium oxysporum strains with ISSR specific primers, (A) UBC855, (B) UBC 856, (C) ISSR 9, (D) ISSR 10. Lanes (1) 100 bp plus ladder (Fermentas); (2) TOFOL-IHBT; (3) TOFU-2-SOGHI; (4) TOFU-3 B.GHAT; (5) TOFU-IHBT; (6) TOXX- POTI; (7) TOXX-POT2; (8) TOFU-KOTBEJA; (9) TOFOL-CBE; (10) TOFS-CPCT2; (11) TOFS-3-CPCT; (12) TOFU-XM; (13) TOFS-MU; (14) TOFU-4CPCT; (15) TOFU-6-CPCT; (16) 1 kb ladder (Fennentas).
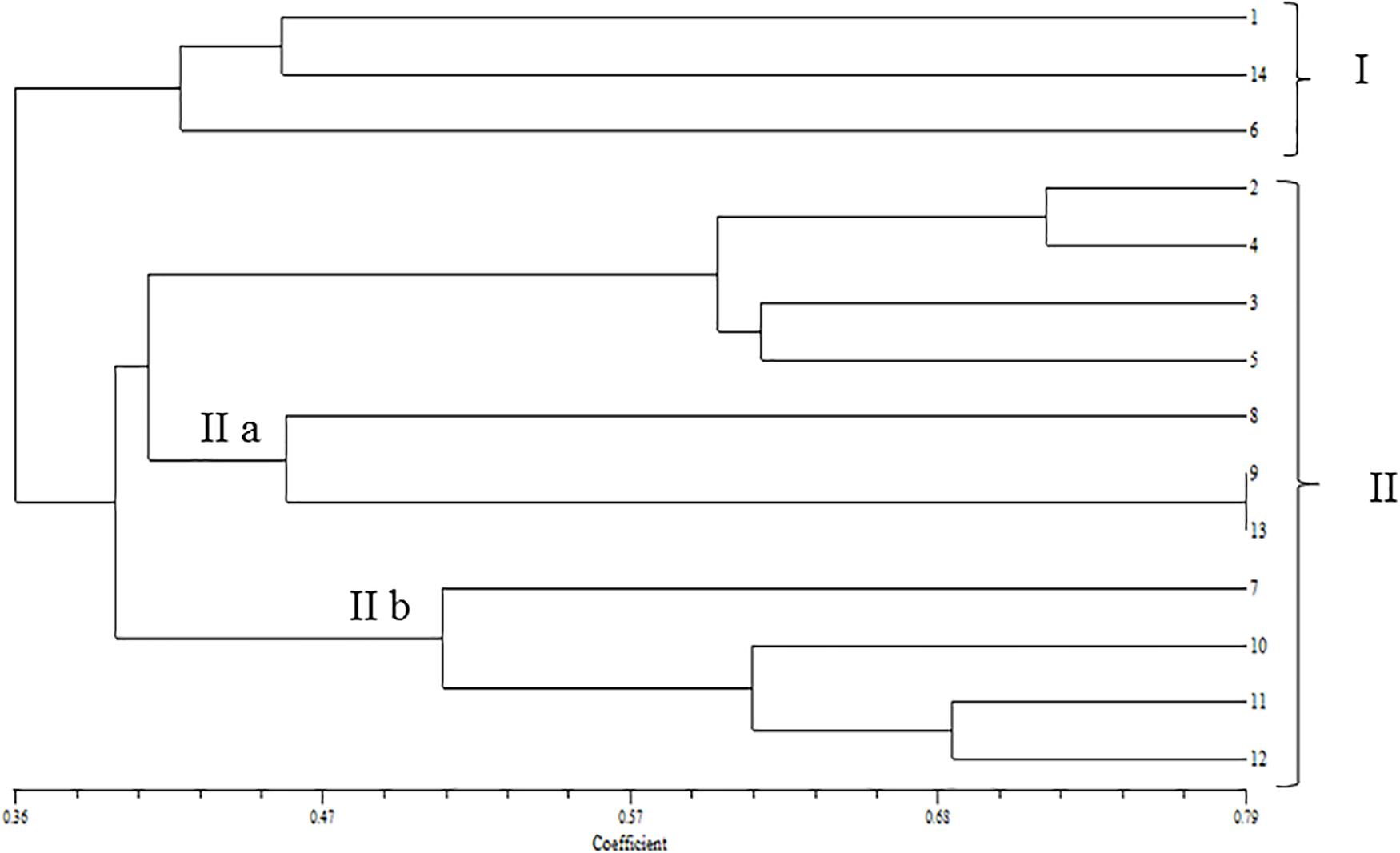
Figure 4. Dendrogram of UPGMA cluster analysis constructed by using DICE similarity coefficients calculated for the Fusarium oxysporum strains based on UBC 855; UBC 856; ISSR 9 and ISSR 10. (1) TOFOL-IHBT; (2) TOFU-2- SOGHI; (3) TOFU-3 B.GHAT; (4) TOFU-IHBT; (5) TOXX- POTI; (6) TOXX-POT2; (7) TOFU-KOTBEJA; (8) TOFOL-CBE; (9) TOFS-CPCT2; (10) TOFS-3-CPCT; (11) TOFU-MM; (12) TOFS-MU; (13) TOFU-4CPCT; (14) TOFU-6-CPCT.
Evaluation of the Microbial Strains for Antagonism Against the Pathogen
A dual-culture assay for antifungal activity indicated that the biocontrol strains significantly inhibit the mycelial growth of the pathogen with 37.7–42.6% efficiency over pathogenic control with varying levels of antagonism. Among S2BC-1, TEPF-Sungal-1 and CG-A strains exhibiting the zone of inhibition, S2BC-1 followed by CG-A exhibited a maximum mycelial growth inhibition, displaying 9- and 11.9-mm inhibition zones on co-inoculation. In the TEPF-Sungal-1 treatment, the pathogen recorded a mycelial growth of 55.6 mm with a 34.4% reduction over control. The Trichoderma strain S17TH exhibiting mycoparasitic activity was also observed to be highly antagonistic for displaying class I level of antagonism (Figure 5).
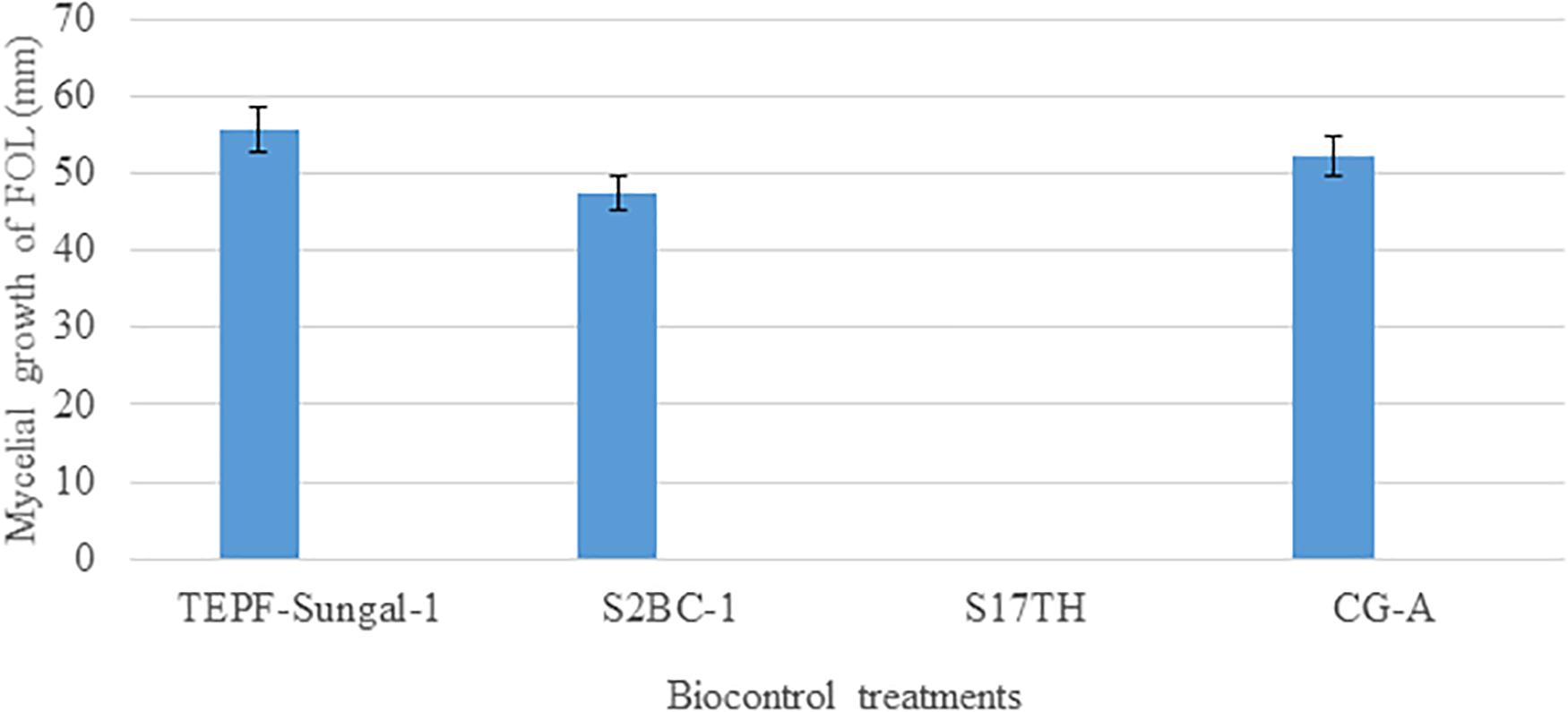
Figure 5. Antagonism of the candidate biocontrol strains against the Fusarium oxysporum strain, TOFU- IHBT in dual culture assay. No mycelial growth of the pathogen was observed in the dual culture assay with the Trichoderma harzianum strain S17TH. Error bars indicate Standard Deviation (SD).
Testing of Compatibility Between the Antagonistic Strains
In the confrontation assays by dual inoculation, none of the antagonistic strains inhibited the growth (data not shown) of the other indicating positive interactions among them. Both the bacterial strains did not show mycelial growth inhibition of S17TH and CG-A in a dual-culture assay. Likewise, S17TH and CG-A did not exhibit mutual antagonism between them. In the co-inoculation assay for the bacterial strains, the strains did not display inhibition zones indicating a lack of antagonism among them. Hence, the strains were selected to develop microbial consortia.
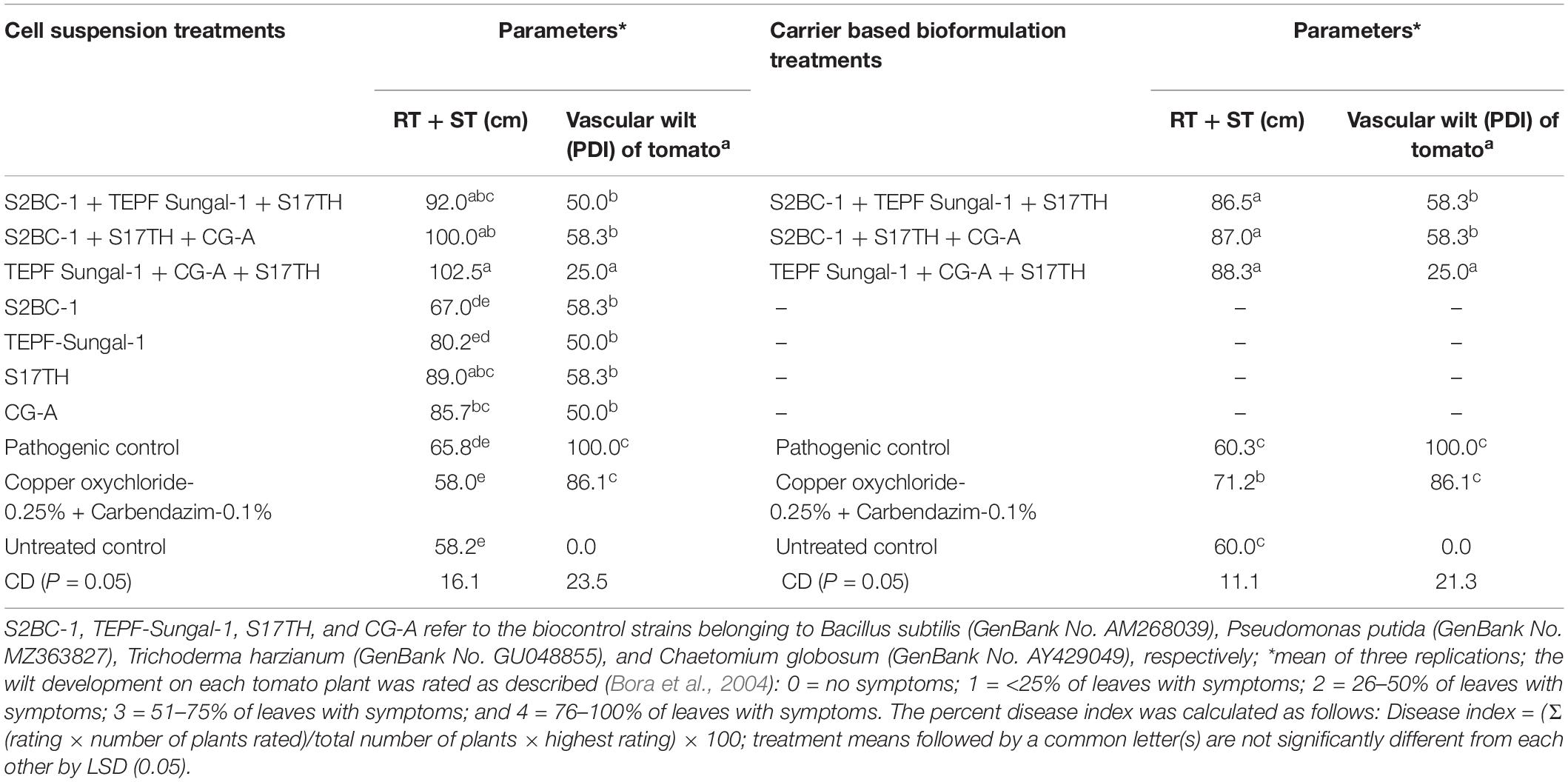
Polyhouse Evaluation of Selected Antagonists
In the pot experiments, the seed and soil applications of the antagonistic microbes as cell suspensions and carrier-based formulations on the challenge of inoculation with Fol significantly reduced the wilt incidence on the tomato to 41.7–75 and 50–75%, respectively, besides promoting the crop growth to 21.8–55.7 and 49.1–52.3%, respectively, over the pathogenic control treatment. The efficacies of the antagonistic microbes in disease reduction and growth promotion were also significant in recording 13–22.7% efficacies, respectively, in relation to the fungicide treatment (Table 3). Among the antagonistic microbial treatments, in comparison with the individual strains (S2BC-1, TEPF-Sungal-1, S17TH, and CG-A), the application of strain mixtures (S2BC-1 + TEPF-Sungal-1 + S17TH, S2BC-1 + S17TH + CG-A, and TEPF-Sungal-1 + S17TH + CG-A) as cell suspensions and carrier-based formulations significantly reduced the wilt incidence to 25–58.3% and 50–75%, respectively, and increased the plant growth to 39.7–55.7% and 49.1–52.3%, respectively, in comparison to the pathogen control (Table 3). Among the strain mixture treatments, the mixture of TEPF-Sungal-1 + S17TH + CG-A as cell suspensions and carrier-based formulations recorded a minimum wilt incidence (25% and 25%, respectively) with increased plant growth (55.7 and 52.3%, respectively) over the pathogen control (Figure 6 and Table 3). In comparison to the microbial treatments, the disease incidence for the fungicide treatment was comparable (86.1%) to the pathogen control. This treatment recorded a relatively higher plant growth with a 22.7% increase over the pathogen control.

Figure 6. Polyhouse evaluation of the biocontrol agents to manage Fusarium wilt of tomato on challenge inoculated with the highly virulent Fusarium oxysporum strain, TOFU-IHBT (A) overview (B) shoot length (C) root length. (1) pot experiments with cell suspensions (i) S2BC-1 + TEPF-Sungal-1 + S17TH; (ii) S2BC-1 + S17TH + CG-A; (iii) TEPF-Sungal-1 + CG-A + S17TH; (iv) S2BC-1; (v) TEPF-Sungal-1; (vi) S17TH; (vii) CG-A; (viii) pathogenic (TOFU-IHBT) control; (ix) fungicide (copper oxychloride-0.25%-rcarbendazim-0.1%) control; (x) untreated control. (2) pot experiments with carrier based bioformulations (i) S2BC-1 + TEPF-Sungal-1 + S17TH; (ii) S2BC-1 + S17TH + CG-A; (iii) TEPF-Sungal-l + CG- A + S17TH; (iv) pathogenic (TOFU-IHBT) control; (v) fungicide (copper oxychloride-0.25%-rcarbendazim-0.1%) control; (vi) untreated control.
Role of Host Defense Genes in Biocontrol of Fusarium Wilt
In the quantitative RT-PCR assays to assess the expression of four host genes in response to the potential biocontrol treatment, the seed and soil applications of the strain mixture TEPF-Sungal-1 + CG-A + S17TH during the challenge of inoculation with Fol induced four genes more than the pathogenic control treatment. The biocontrol treatment stimulated endochitinase (vir1), glucosamine-6-phosphate (Gln-6P) deaminase (g6pd), phosphoglycerate kinase (PGK), and NADH dehydrogenase [ubiquinone] iron-sulfur protein 8 (NDUS8) genes by 2. 4-, 1. 5-, 2. 7-, and 2.5-fold, respectively, more than those of the pathogen control (Figure 7). Among the genes, the phosphoglycerate kinase followed by the endochitinase showed a maximum upregulation of 2.7–2.4-fold, respectively, in comparison to the control. The induction of the defense genes indicated that the reduction in vascular wilt control by the biocontrol strains can be partly plant-mediated (Figure 7).
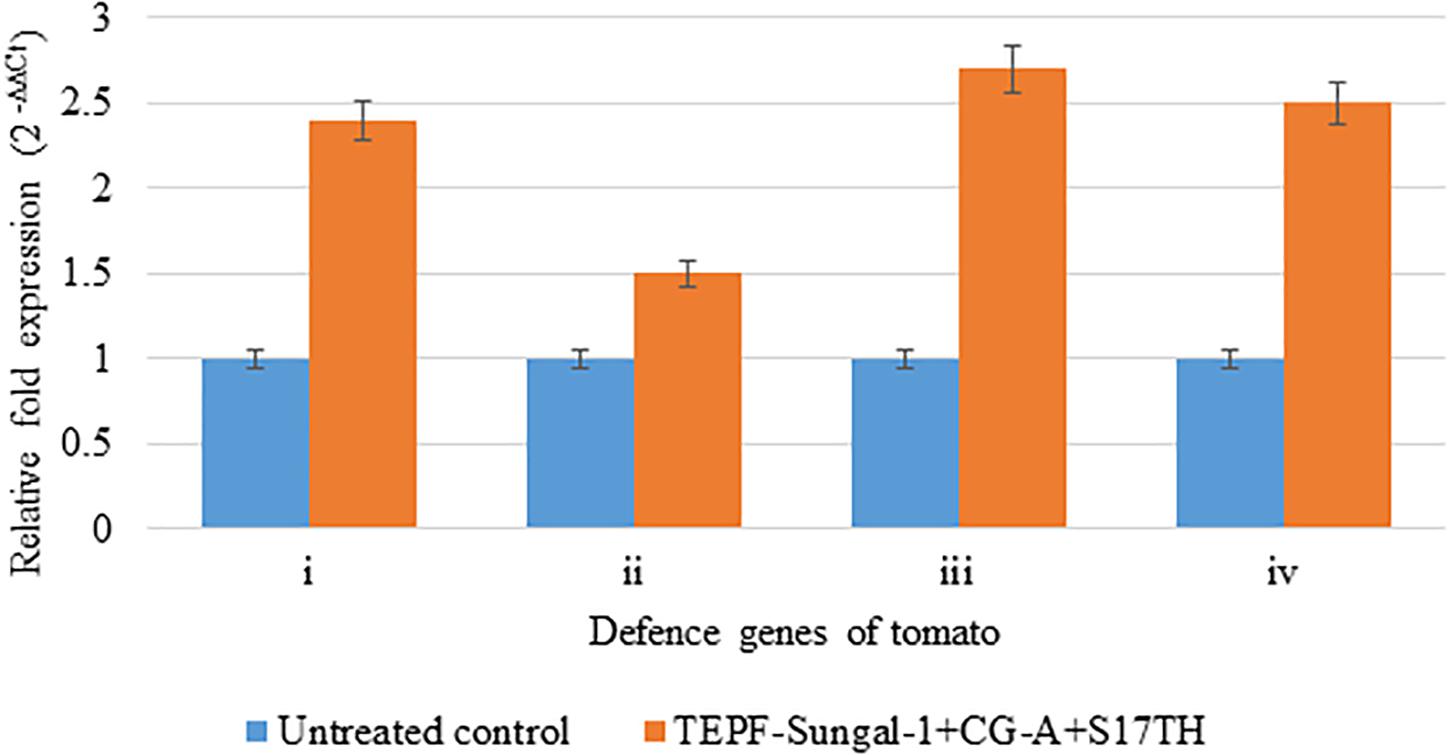
Figure 7. Relative expression of defense genes in the tomato plants cv. Pusa Rohini induced by the potential biocontrol treatment. TEPF-Sungal-1 (Pseudomonas putida) + CG-A (Chaetomium globosum) + S17TH (Trichoderma harzianum) challenge inoculated with the Fusarium oxysporum f. sp. lycopersici strain, TOFU-IHBT over the untreated control, (i) vzrJ [endochitinase]; (ii) g6pd [glucosamine-6-phosphate (Gln-6P) deaminase]; (iii) PGK [phosphoglycerate kinase]; (iv) XDUS8 [NADH dehydrogenase [ubiquinone] iron-sulfur protein 8]. The amount of cDNA in the sample were normalized employing actin as an internal control. Error bars indicate Standard Deviation (SD).
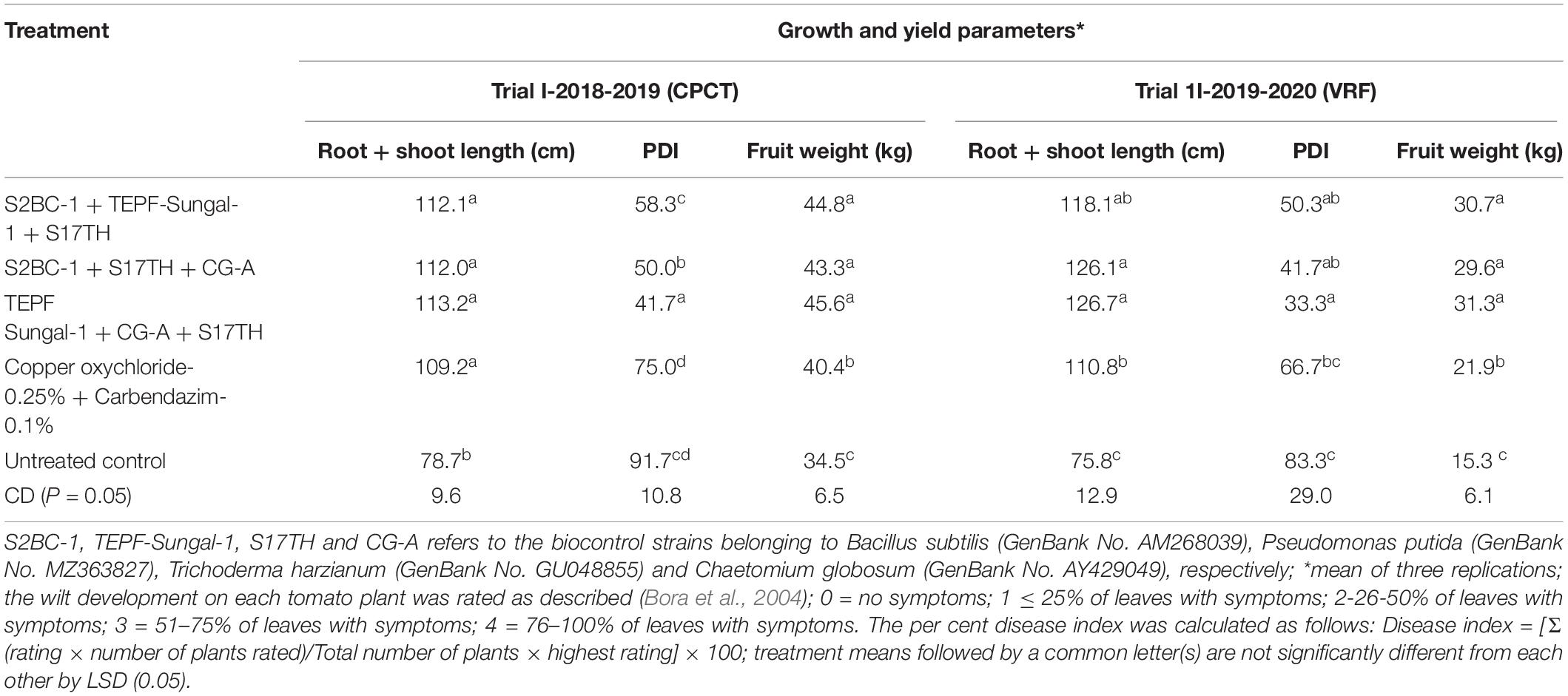
Field Evaluation of Biocontrol Formulations for Fusarium Wilt Management
The soil characteristics were analyzed to possess clay loam texture with an organic matter content of 4.07% and N:P:K of 250:50:250 kg/ha. The pH and EC of the soil were analyzed to be 7.5 and 0.3 mmhos/cm–1. Both the field trials in 2018–2019 and 2020 indicated that the wilt incidence is low (41.7 and 33.3%) in the microbial treated plots relative to the untreated control. Besides the microbial treated plots of 2018–2019 and 2020 also recorded higher fruit yields of over 32.4 and 105%, respectively, over the untreated control. Among the microbial consortia treatments, the strain mixture, TEPF-Sungal-1 + CG-A + S17TH, performed better than the untreated control. The strain mixture produced lesser wilt incidence with a 54.5 and 60% mean reduction over the untreated control, and the performances were even better than those of the fungicide treatment (18.2%). Besides the treatment also increased the fruit yield by 30.25% over the control. Interestingly, the untreated control recorded a lower fruit yield (34.5 and 15.3 kg) and was followed by the fungicide control (40.4 and 21.9 kg). No pathogenic Fusarium population was recorded in the rhizosphere soils treated with the strain mixture, whereas the Fusarium population was denser in the rhizospheres of the untreated control plots (Table 4 and Figures 8, 9) and was pathogenic.
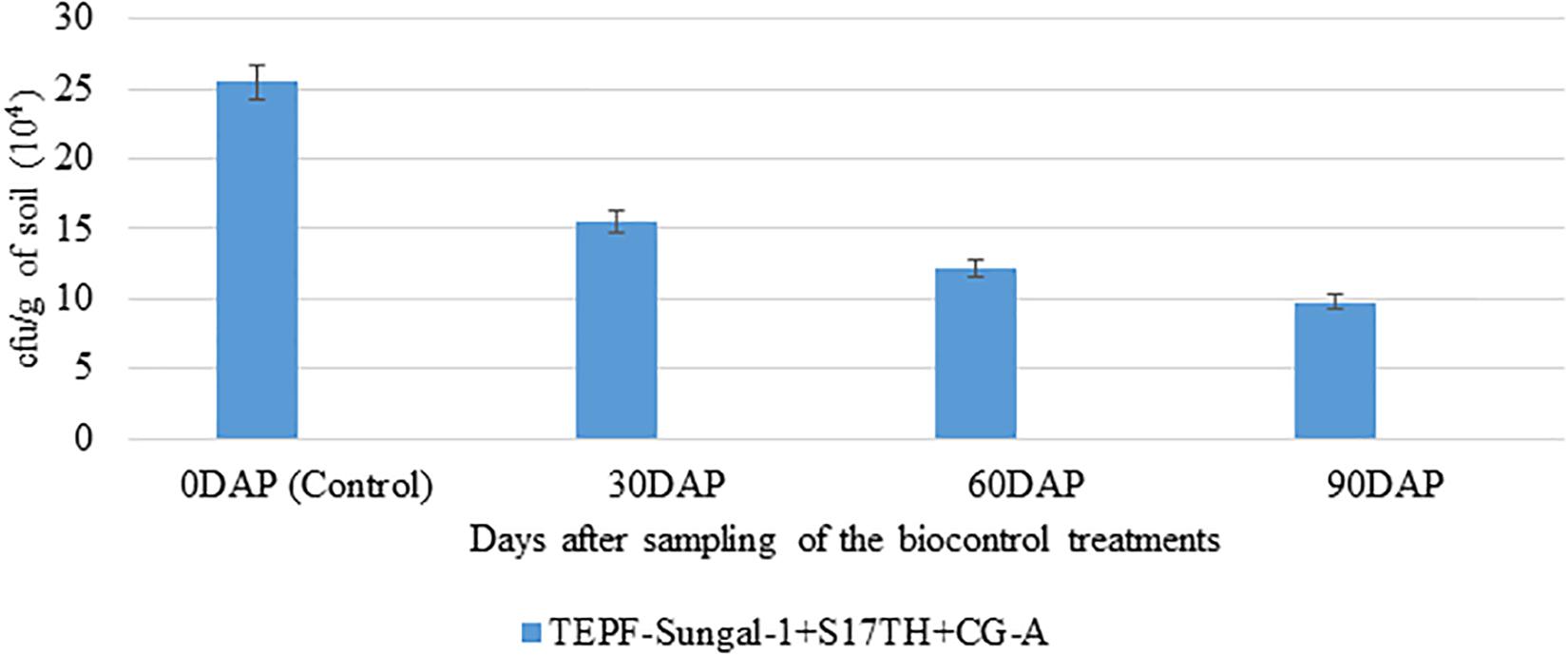
Figure 8. Enumeration of Fusarium population in the rhizosphere soils of tomato treated with TEPF-Sungal-1 (Pseudomonas putida), S17TH (Trichoderma harzianum) and CG-A (Chaetomium globosum) under field conditions [VRF, I ARI]; *mean of three replications; the fungal colonies appeared after 3 days of plating on Fusarium selective media, Fo-Gl. Error bars indicate Standard Deviation (SD).

Figure 9. Field evaluation [CPCT, I ARI] of the biocontrol agents to control Fusarium wilt of tomato (cv. Pusa Rohini) during harvest for (A) root growth, (B) fruit yield, (C) vascular wilt (i) S2BC-1 + TEPF-Sungal-1 + S17TH; (ii) S2BC-1 + S17TH + CG-A; (iii) TEPF-Sungal-1 + CG-A + S17TH; (iv) fungicide control (copper oxychloride- 0.25% + carbendazim-0.1%); (v) untreated control.
Discussion
Globally, fungal vascular wilt of tomato is known to be caused by the three different species of Fusarium, Fol, Fs, and F. oxysporum f. spp. radicis lycopersici (Kouki et al., 2012). However, in India, Fusarium wilt of tomato is caused by either Fol or FS (Chandra et al., 1983; Shanmugam et al., 2011a). We earlier reported the association of Fol with vascular wilt in the selected locations of the states (Himachal Pradesh and Delhi) of India (Sidharthan et al., 2019). In this study, we further studied the etiology of the disease in more tomato growing regions of the country at selected locations of north and south India. The results indicated that the disease is predominated by Fol, and the association of FS has been limited to select locations in the newly surveyed regions. Though mixed infections due to Fol and Fs have been reported globally (Abdesselem et al., 2016) in India, no mixed infections have been reported (Sidharthan et al., 2019). In assessing the genetic diversity of 18 isolates of the two species representing the four Indian states using ITS sequences, the isolates of either of the species did form a distinct group. Recently, based on the molecular phylogenetic analysis of Fusarium species associated with tomato, Imazaki and Kadota (2015) revealed Fs as a species complex distinct from that of Fol. In further analyses of the Fol population by the unweighted pair group method with arithmetic averaging using amplicons of ISSR primers, which do not require prior gene sequence information, the strains distinguished themselves into two major clusters and the level of clustering among the isolates of F. oxysporum clearly revealed a greater genetic variability within the population, indicating evolutionary adaptation, which may help in the survival of the species with a change in the environment (McDonald and Linde, 2002; Chandra et al., 2008; Sharma et al., 2014). A high genetic diversity of Fusarium spp. infected tomato had earlier been reported (Edel-Hermann et al., 2011). Besides genetic polymorphism, no correlation with the geographical delimitation has been observed in the dendrogram developed for the ISSR profiles. No correlation between the genetic diversity and geographic origins of pathogen isolates had been observed for Fusarium species infecting several crops (Weller, 1988; Najafiniya and Sharma, 2011). Such a lack of correlation indicates that local ecological niches might be influencing more than the geographical isolation.
Plants living in close association with microbes and rhizosphere soil are identified to be a major microbial reservoir for being influenced by the secretions of roots (Kennedy and de Luna, 2005). Plant health can be improved by engineering the soil microbiota through the host, introducing microbes into the soil or plants and through the manipulation of native microbes by the modification of agricultural practices. The introduction of microbes into the soil for improving crop productivity under field conditions has mostly been accomplished by using single biocontrol agents (Larkin and Fravel, 1998). However, a majority of the strains fail to perform satisfactorily in the field with limited reproducibility upon testing at different locations. A possible reason for the low success and limited reproducibility at different locations could be that the introduced microbial inoculant competes less efficiently with the local microbial community, whose members are extensively diverse and have been well adapted to the environment (Compant et al., 2019). The use of a microbial consortium is suggested as a potential alternative approach for the uncertainty experienced in biocontrol under field conditions. We earlier demonstrated the usefulness of microbial consortia consisting of varying combinations of plant growth-promoting microbes (PGPMs), Bacillus, Pseudomonas, and Trichoderma in controlling serious soilborne diseases of gladiolus (Shanmugam et al., 2011b) and ginger (Shanmugam et al., 2013) under field conditions. In this study, we tested the microbial consortia consisting of these PGPMs in combination with C. globosum, CG-A, a potential fungal biocontrol strain but relatively less explored for disease control under field conditions. The biocontrol potential of CG-A has earlier been demonstrated for the control of diseases in crop plants under polyhouse (Sibounnavong et al., 2011) and field conditions (Cullen et al., 1984). The strains constituting the consortia were carefully selected in such a way that they express different biocontrol traits besides being compatible among themselves. Though the rhizobacterial strains, S2BC-1 and TEPF-Sungal-1, and the T. harzianum strain, S17TH were reported to produce lytic enzymes (Shanmugam et al., 2011a,b, 2013; Shanmugam and Kanoujia, 2011), the CG-A strain produced antibiotics, chaetoglobosin, and chaetomin (Aggarwal et al., 2004, 2007). The strains constituting the consortia were also ensured to exhibit growth over a wide range of temperatures ranging from 7°C ± 2°C to 45°C ± 2°C under controlled conditions. In the in vitro dual-culture assays, the time of inoculation of the bacterial and fungal biocontrol strains was selected in such a way to favor the pathogen and less conducive to the biocontrol strains. Accordingly, as standardized earlier (Shanmugam and Kanoujia, 2011; Shanmugam et al., 2013) and the bacterial biocontrol strains are fast growing than the fungal biocontrol strains, they were inoculated in the plates after 48 h of pathogen inoculation. On the contrary, the fungal biocontrol strains were co-inoculated with the pathogen since they exhibited a relatively slower growth than the bacterial biocontrol strains but a similar growth to that of the fungal pathogen. The growth rate of each of the strains were standardized earlier in monocultures. To increase the efficiency of the microbial consortia in biocontrol, the microbial strains and their combinations were effectively formulated using a talc-based formulation that could sustain the microbial populations over a period of time during storage (Shanmugam et al., 2013). Accordingly, in the polyhouse studies, the talc-based formulations of the biocontrol treatments, which inhibits Fol, were equally effective like the cell suspensions and were even better than the fungicidal mixture in promoting the crop growth and reducing vascular wilt incidence over the control treatment to significant levels despite the challenge of inoculation with the pathogen. Interestingly, in both the experiments, the application of the mixture of S2BC-1 (B. subtilis) + TEPF-Sungal-1 (P. putida) as a seed dressing and also a soil application recorded a maximum plant growth promotion and minimum disease incidence in comparison to the uninoculated control. The efficacy of the best performing bioconsortium was further tested in the field at two locations, IARI-CPCT, and IARI-VRF colonized by the two highly pathogenic Fol isolates, TOFU-IHBT and TOFU-CPCT6, for 2 consecutive years. In both the field experiments, the bioconsortium was effective in reducing the vascular wilt incidence to 54.5 and 60% over the untreated control, respectively. Besides the treatment also resulted in improved (32.4 and 45.8%) fruit production higher than the control. Enhanced plant growth and disease suppression by the carrier-based formulations of CG-A have been reported for other diseases (Selvakumar et al., 2001). However, the use of carrier-based formulations of Chaetomium-based bioconsortium for disease management in crop plants appears to be the first report. Though many biocontrol agents are potent enough to control plant diseases, only commercial formulations of a few exhibit consistently stable and high efficiency in managing the plant diseases in the field. Chaetomium-based bioconsortium demonstrates consistency in its performance across the four experiments in 2 years under the two field conditions.
Understanding the mode of action of biocontrol agents is one of the strategies to improve the success of the biocontrol of plant diseases (Shanmugam et al., 2011b). Multiple antifungal traits known to be produced by the strains constituting the consortium might have accounted for its direct antifungal activity. To understand the role of plant-mediated resistance induced by the biocontrol consortium in vascular wilt control, the induction of candidate defense genes (vir1, g6pd, PGK, and NDUS8) by the biocontrol treatment was studied. The endochitinase is known to degrade the cell wall components of fungal pathogens (Shanmugam, 2005; Gruber et al., 2011), whereas the g6pd, PGK, and NDUS8 genes are involved in various defense reaction pathways of plants (Petriacq et al., 2012, 2013; Rubio et al., 2012). In the polyhouse studies, significant expressions of the four defense genes coupled with the lower vascular wilt incidence and higher plant growth were observed in the bioconsortium treatment over the uninoculated control. Hence, it is speculated that the biocontrol treatment also exerts its antifungal effect through the host plant, and these candidate genes might be involved in the disease suppression. Earlier, the induction of plant defense genes, chitinase, and β-1,3-glucanase leading to decreased disease incidences has been reported (Shanmugam et al., 2011a,b; Shanmugam and Kanoujia, 2011).
It is concluded that the present study has revealed the genetic variability of the Fusarium isolates of tomato in the selected tomato growing areas of India, which have been exploited to design and deploy biocontrol agents producing multifunctional traits and occupying different host niches. The potential talc-based formulation of the Chaetomium-based bioconsortium can be a component of an integrated disease management system upon commercialization to achieve the goal of practical applications of biocontrol, which are under progress.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
GP: experimentation, data analysis, and manuscript drafting. ZH: assistance in Fusarium characterization. AKS: assistance in biocontrol experiments. AUS: assistance in qPCR experiments. RA: assistance in biocontrol experiments and manuscript editing. RR: assistance in data analysis. VS: conceptualization, resource, data analysis, and manuscript editing and revision. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank the Director, ICAR-Indian Agricultural Research Institute for his support and encouragement during this investigation. The authors would also like to thank the trainees, Priya Muduli and Manisha Mishra, for assistance in assessing the pathogen population in the soil samples. This publication is supported by IARI Publication No. PME/Plant-Path/2000021.
References
Abdel-Azeem, A. M., Gherbawy, Y. A., and Sabry, A. M. (2016). Enzyme profiles and genotyping of Chaetomium globosum isolates from various substrates. Plant Biosyst. 150, 420–428. doi: 10.1080/11263504.2014.984791
Abdesselem, S. M., Nisserine, H. K., Mebrouk, K., Jamal, E. H., Jos Eacute, S., Eduardo, G., et al. (2016). Characterization of Fusarium oxysporum isolates from tomato plants in Algeria. Afr. J. Microbiol. Res. 10, 1156–1163. doi: 10.5897/ajmr2016.8161
Aggarwal, R. (2015). Chaetomium globosum: a potential biocontrol agent and its mechanism of action. Indian Phytopathol. 68, 8–24.
Aggarwal, R., Gupta, S., Sharma, S., and Shukla, R. (2014). Development of conventional and real time PCR assay for rapid detection and quantification of a biocontrol agent. Plant Pathol. J 46, 477–485.
Aggarwal, R., Sharma, S., Gupta, S., Singh, K. B. M., and Shanmugam, V. (2016). Role of defence enzymes in biocontrol of spot blotch and leaf rust of wheat (Triticum sp. L.) by Chaetomium globosum. J. Pure Appl. Microbiol. 10, 2071–2078.
Aggarwal, R., Sharma, V., Kharbikan, L. L., and Renu (2008). Molecular characterization of Chaetomium spp. using URP-PCR. Genet. Mol. Bio. 31, 943–946. doi: 10.1590/s1415-47572008005000014
Aggarwal, R., Tiwari, A. K., Dureja, P., and Srivastava, K. D. (2007). Quantitative analysis of secondary metabolites produced by Chaetomium globosum Krunze ex Fr. J. Biol Control 21, 163–168.
Aggarwal, R., Tiwari, A. K., Srivastava, K. D., and Singh, D. V. (2004). Role of antibiosis in the biological control of spot blotch (Cochliobolus sativus) of wheat by Chaetomium globosum. Mycopathologia 157, 369–377. doi: 10.1023/b:myco.0000030446.86370.14
Akbar, A., Hussain, S., Ullah, K., Fahim, M., and Ali, G. S. (2018). Detection, virulence and genetic diversity of Fusarium species infecting tomato in Northern Pakistan. PLoS One 13:e0203613. doi: 10.1371/journal.pone.0203613
Biswas, S. K., Srivastava, K. D., Aggarwal, R., Prem, D., and Singh, D. V. (2000). Antagonism of Chaetomium globosum to Drechslera sorokiniana the spot blotch pathogen of wheat. Indian Phytopathol. 53, 436–440.
Bora, T., Ozaktan, H., Gore, E., and Aslan, E. (2004). Biological control of Fusarium oxysporum f. sp. melonis by wettable powder formulations of the two strains of Pseudomonas putida. J. Phytopathol. 152, 471–475.
Chandra, S., Raizada, M., and Gaur, A. K. S. (1983). Pathological variability in Fusarium oxysporum and Fusarium solani. Indian Phytopathol. 36, 36–40.
Chandra, N. S., Udaya Shankar, A. C., Niranjana, S. R., and Prakash, H. S. (2008). Molecular detection and characterisation of Fusarium verticillioides in maize (Zea mays. L) grown in southern India. Ann. Microbiol. 58, 359–367.
Compant, S., Samad, A., Faist, H., and Sessitsch, A. (2019). A review on the plant microbiome: ecology, functions, and emerging trends in microbial application. J. Adv. Res. 19, 29–37. doi: 10.1016/j.jare.2019.03.004
Cullen, D., Barbee, F. M., and Andrews, J. H. (1984). Chaetomium globosum antagonizes the apple scab pathogen, Venturia inaequalis, under field conditions. Can. J. Bot. 62, 1814–1818. doi: 10.1139/b84-245
Divakara, S. T., Santosh, P., Aiyaz, M., Venkata Ramana, M., Hariprasad, P., and Nayaka, S. C. (2014). Molecular identification and characterization of Fusarium spp. associated with sorghum seeds. J. Sci. Food Agric. 94, 1132–1139. doi: 10.1002/jsfa.6380
Edel-Hermann, V., Gautheron, N., and Steinberg, C. (2011). Genetic diversity of Fusarium oxysporum and related species pathogenic on tomato in Algeria and other Mediterranean countries. Plant Pathol. 61, 787–800. doi: 10.1111/j.1365-3059.2011.02551.x
Gruber, S., Kubicek, C. P., and Seidl-Seiboth, V. (2011). Differential regulation of orthologous chitinase genes in mycoparasitic Trichoderma species. Appl. Environ. Microbiol. 77, 7217–7226. doi: 10.1128/AEM.06027-11
Huang, C. C., and Lindhout, P. (1997). Screening for resistance in wild Lycopersicon species to Fusarium oxysporum f.sp. lycopersici race 1 and race 2. Euphytica 93, 145–153. doi: 10.1023/A:1002943805229
Imazaki, I., and Kadota, I. (2015). Molecular phylogeny and diversity of Fusarium endophytes isolated from tomato stems. FEMS Microbiol. Ecol. 91:fiv098. doi: 10.1093/femsec/fiv098
IRRI (1992). IRRISTAT User’s Manual. Version 92–1. Metro Manila: International Rice Research Institute.
Kennedy, A. C., and de Luna, L. Z. (2005). “Rhizosphere,” in Encyclopedia of soils in the environment, ed. D. Hillel (Oxford: Elsevier), 399–406. doi: 10.1016/B0-12-348530-4/00163-6
Kouki, S., Saidi, N., Ben Rajeb, A., Brahmi, M., Bellila, A., Fumio, M., et al. (2012). Control of Fusarium wilt of tomato caused by Fusarium oxysporum f.sp. radicis-lycopersici using mixture of vegetable and Posidonia oceanica compost. Appl. Environ. Soil Sci. 2012:239639. doi: 10.1155/2012/239639
Larkin, R. P., and Fravel, D. R. (1998). Efficacy of various fungal and bacterial biocontrol organisms for control of Fusarium wilt of tomato. Plant Dis. 82, 1022–1028. doi: 10.1094/PDIS.1998.82.9.1022
Lin, Y., Fuping, L., Shanshan, L., Xiao, W., Ruixuan, Z., Ziqin, L., et al. (2012). ISSR analysis of Fusarium oxysporum Schl. in Hebei province. Proc. Environ. Sci. 12, 1237–1242. doi: 10.1016/j.proenv.2012.01.414
McDonald, B. A., and Linde, C. (2002). Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 40, 349–379. doi: 10.1146/annurev.phyto.40.120501.101443
Najafiniya, M., and Sharma, P. (2011). Specific PCR-based marker for detection of pathogenic groups of Fusarium oxysporum f.sp. cucumerinum in India. J. Genet. Eng. Biotechnol. 9, 29–34. doi: 10.1016/j.jgeb.2011.05.009
Nelson, P. E., Toussoun, T. A., and Marasas, W. F. (1983). Fusarium Species: An Illustrated Manual for Identification. University Park, TX: Pennsylvania State University Press.
Nirmaladevi, D., Venkataramana, M., Srivastava, R. K., Uppalapati, S. R., Gupta, V. K., and Yli-Mattila, T. (2016). Molecular phylogeny, pathogenicity and toxigenicity of Fusarium oxysporum f.sp. lycopersici. Sci. Rep. 6:21367. doi: 10.1038/srep21367
Nishimura, N. (2007). Selective media for Fusarium oxysporum. J. Gen. Plant Pathol. 73, 342–348. doi: 10.1007/s10327-007-0031-y
Petriacq, P., De Bont, L., Hager, J., Didierlaurent, L., Mauve, C., Guerard, F., et al. (2012). Inducible NAD overproduction in Arabidopsis alters metabolic pools and gene expression correlated with increased salicylate content and resistance to Pst-AvrRpm1. Plant J. 70, 650–665. doi: 10.1111/j.1365-313X.2012.04920.x
Petriacq, P., De Bont, L., Tcherkez, G., and Gakiere, B. (2013). NAD: not just a pawn on the board of plant-pathogen interactions. Plant Signal. Behav. 8:e22477. doi: 10.4161/psb.22477
Rubio, M. B., Dominguez, S., Monte, E., and Hermosa, R. (2012). Comparative study of Trichoderma gene expression in interactions with tomato plants using high-density oligonucleotide microarrays. Microbiology 158, 119–128. doi: 10.1099/mic.0.052118-0
Selvakumar, R., Srivastava, K. D., Aggarwal, R., and Singh, D. V. (2001). Biocontrol of spot blotch of wheat using Chaetomium globosum. Ann. Plant Prot. Sci. 9, 286–291.
Shanmugam, V. (2005). “Chitinases in defence against phytopathogenic fungi,” in Crop Protection-Management Strategies, ed. D. Prasad (New Delhi: Daya Publishing House), 403.
Shanmugam, V., Atri, K., Gupta, S., Kanoujia, N., and Naruka, D. S. (2011a). Selection and differentiation of Bacillus spp. antagonistic to Fusarium oxysporum f.sp. lycopersici and Alternaria solani infecting tomato. Folia Microbiol. 56, 170–177. doi: 10.1007/s12223-011-0031-3
Shanmugam, V., Chugh, P., and Sharma, P. (2015). Cold-tolerant Trichoderma species for the management of Fusarium wilt of tomato plants. Ann. Microbiol. 65, 543–551. doi: 10.1007/s13213-014-0890-3
Shanmugam, V., Gupta, S., and Dohroo, N. P. (2013). Selection of a compatible biocontrol strain mixture based on co-cultivation to control of rhizome rot of ginger. Crop Prot. 43, 119–127. doi: 10.1016/j.cropro.2012.08.012
Shanmugam, V., Kanouji, N., Singh, M., Sing, S., and Prasad, R. (2011b). Biocontrol of vascular wilt and corm rot of gladiolus caused by Fusarium oxysporum f.sp. gladioli using plant growth promoting rhizobacterial mixture. Crop Prot. 30, 807–813. doi: 10.1016/j.cropro.2011.02.033
Shanmugam, V., and Kanoujia, N. (2011). Biological management of vascular wilt of tomato caused by Fusarium oxysporum f.sp. lycopersici by plant growth-promoting mixture. Biol. Control 57, 85–93. doi: 10.1016/j.biocontrol.2011.02.001
Sharma, M., Nagavardhini, A., and Thudi, M. (2014). Development of DArT markers and assessment of diversity in Fusarium oxysporum f.sp. ciceris, wilt pathogen of chickpea (Cicer arietinum L.). BMC Genomics 15:454. doi: 10.1186/1471-2164-15-454
Sibounnavong, P., Charoenporn, C., Kanokmedhakul, S., and Soytong, K. (2011). Antifungal metabolites from antagonistic fungi used to control tomato wilt fungus Fusarium oxysporum f.sp. lycopersici. Afr. J. Biotechnol. 10, 19714–19722. doi: 10.5897/AJB11.3343
Sidharthan, K. V., Aggarwal, R., and Shanmugam, V. (2018). Selection and characterization of the virulent Fusarium oxysporum f.sp. lycopersici isolate inciting vascular wilt of tomato. Int. J. Curr. Microbiol. Appl. Sci. 7, 1749–1756. doi: 10.20546/ijcmas.2018.702.212
Sidharthan, V. K., Aggarwal, R., and Shanmugam, V. (2019). “Fusarium wilt of crop plants: disease development and management,” in Wilt Diseases of Crops and their Management, eds A. Bhattacharyya, B. N. Chakraborty, R. N. Pandey, D. Singh, and S. C. Dubey (New Delhi: Today and Tomorrow Printers and Publisher), 519–533.
Soytong, K., Kanokmedhakul, S., Kukongviriyapa, V., and Isobel, M. (2001). Application of Chaetomium species (Ketomium§) as a new broad spectrum biological fungicide for plant disease control. Fungal Diver. 7, 1–15.
Weller, D. M. (1988). Biological control of soilborne plant pathogens in the rhizosphere with bacteria. Annu. Rev. Phytopathol. 26, 379–407. doi: 10.1146/annurev.py.26.090188.002115
Yang, W., Zheng, L., Liu, H. X., Wang, K. B., Wang, Y. P., and Luo, Y. M. (2014). Evaluation of the effectiveness of a consortium of three plant-growth promoting rhizobacteria for biocontrol of cotton Verticillium wilt. Biocontrol. Sci. Technol. 24, 489–502. doi: 10.1080/09583157.2013.873389
Keywords: Fusarium wilt, tomato, Chaetomium, Trichoderma, PGPR, consortium, biocontrol
Citation: Pothiraj G, Hussain Z, Singh AK, Solanke AU, Aggarwal R, Ramesh R and Shanmugam V (2021) Characterization of Fusarium Spp. Inciting Vascular Wilt of Tomato and Its Management by a Chaetomium-Based Biocontrol Consortium. Front. Plant Sci. 12:748013. doi: 10.3389/fpls.2021.748013
Received: 29 July 2021; Accepted: 18 October 2021;
Published: 19 November 2021.
Edited by:
Sabrina Sarrocco, University of Pisa, ItalyReviewed by:
Sotiris Tjamos, Agricultural University of Athens, GreeceDzarifah Zulperi, Universiti Putra Malaysia, Malaysia
Copyright © 2021 Pothiraj, Hussain, Singh, Solanke, Aggarwal, Ramesh and Shanmugam. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Veerubommu Shanmugam, c2hhbnBhdGhvQHlhaG9vLmNvbQ==
 Govindan Pothiraj1
Govindan Pothiraj1 Zakir Hussain
Zakir Hussain Awani Kumar Singh
Awani Kumar Singh Amolkumar U. Solanke
Amolkumar U. Solanke Rashmi Aggarwal
Rashmi Aggarwal Raman Ramesh
Raman Ramesh Veerubommu Shanmugam
Veerubommu Shanmugam