
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 26 October 2021
Sec. Plant Biotechnology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.747476
This article is part of the Research TopicTargeted Genome Editing for Crop ImprovementView all 20 articles
New breeding technologies have not only revolutionized biological science, but have also been employed to generate transgene-free products. Genome editing is a powerful technology that has been used to modify genomes of several important crops. This review describes the basic mechanisms, advantages and disadvantages of genome editing systems, such as ZFNs, TALENs, and CRISPR/Cas. Secondly, we summarize in detail all studies of the CRISPR/Cas system applied to potato and other tuber crops, such as sweet potato, cassava, yam, and carrot. Genes associated with self-incompatibility, abiotic-biotic resistance, nutrient–antinutrient content, and post-harvest factors targeted utilizing the CRISPR/Cas system are analyzed in this review. We hope that this review provides fundamental information that will be useful for future breeding of tuber crops to develop novel cultivars.
Cells have various inherent mechanisms for the repair of double-stranded DNA breaks (DSBs), which are caused by endogenous and exogenous genotoxic agents (Wang and Xu, 2017; Vats et al., 2019). Cell type, cell state, and nature of the DSBs influence multiple DNA repair mechanisms. The failure and misrepair of DSBs can result in mutations and occur genome instability, and even can lead to cell death (Mcvey and Sang, 2008). However, during some essential physiological processes, DSBs are used to generate genetic diversity, such as the immune repertoire generation and the recombination of alleles during meiosis (Le Guen et al., 2015). Moreover, the loss of genetic diversity would results in an evolutionary dead end. So the DNA repair mechanism needs to cause active maintenance of genome stability and also generate genetic diversity in populations. Therefore, the high accuracy of DNA repair processes is an important phenomena in all organisms (Guirouilh-Barbat et al., 2014).
The DSB repair uses three general type of strategies. Non-homologous end joining (NHEJ) is generally regarded as error-prone, and results in mutations such as smaller insertions, deletions, and substitutions at the DSB site. It is the dominant and most active pathway in eukaryotes for repairing DSBs in all phases of the cell cycle (Anzalone et al., 2020). Homologous recombination (HR) is error-prone often with point and fragment mutations, insertions, and deletions (Rodgers and McVey, 2016). It is believed that HR is the foundation of genome engineering. In HR, the identical sister chromatids are served as templates to repair DSBs in cells during the late S/G2 phases of the cell cycle (Wang and Xu, 2017; Afzal et al., 2020). The HR, in theory, can lead to knock-in, protein-domain swapping, new gene functions, or alteration in gene regulation. HR is commonly considered as an error-free process, whereas NHEJ is considered to be error-prone process. However, Bétermier et al. (2014) and Guirouilh-Barbat et al. (2014) concluded that HR and NHEJ have a double effect, which on the one hand, they are essential for genomic stability maintenance and diversity, but on the other hand can damage the maintenance of genomic integrity. Microhomology-mediated end joining (MMEJ) is an error-prone repair system that repairs DNA breaks through using of substantial microhomologous sequence (4–25 bp) and always lead to small insertions or deletions (Mcvey and Sang, 2008; Uranga et al., 2021). The MMEJ is an active repair system during the G1and early S phases of the cell cycle (Taleei and Nikjoo, 2013).
Genome editing (GE), a recent technological innovation in the life sciences, is an example of techniques that are used to explore biological phenomena (Vats et al., 2019). In this technique, DNA is modified through cell-repair mechanisms that introduce precise breaks in the genome at specific sites (Sun et al., 2016). Genome-edited plants are not similar to conventional transgenic plants, as they may not incorporate foreign DNA. This distinction makes genome editing a unique and powerful breeding tool that has promising applications in agriculture development, especially when genome-edited crops are not adjusted as genetically-modified (GM) plants. This technique requires the introduction of sequence-specific nucleases (SSNs) into plants through the following three major methods of transformation: Agrobacterium-mediated transformation (Zhang et al., 2020), biolistic-mediated transformation (Hamada et al., 2018; Rustgi and Luo, 2020), and polyethylene glycol (PEG)-mediated protoplast transfection (Sant’Ana et al., 2020). Potato and most tuber crops are members of the dicotyledonous family and the most popular method for genetic transformation of dicotyledonous plants is Agrobacterium-mediated gene transfer.
From both a scientific and a regulatory perspective, it is beneficial if the integration of DNA in the edited plant genome is avoided (Andersson et al., 2018). In general, several strategies have been developed for obtaining transgene-free CRISPR/Cas9-edited tuber crops. They are the selection of transgene-free genome-edited plants by Mendelian segregation in T1 generation of potato obtained by using Agrobacterium-mediated delivery method (Ye et al., 2018; Enciso-Rodriguez et al., 2019) CRISPR/Cas9 cytidine base editor through the Agrobacterium-mediated delivery method (Veillet et al., 2019b), transient expression of CRISPR/Cas9 construct by the PEG-mediated transfection method (Andersson et al., 2017; Chatukuta and Rey, 2020), and ribonucleoprotein (RNP)-based genome editing (Andersson et al., 2018; González et al., 2020). In the case of Mendelian segregation, one of the limiting factors is the need of hybridization and selection of transgene-free plants from T1 segregating generations which are time consuming and laborious. The alternative delivery methods, including biolistic, PEG-mediated protoplast transfection, and RNP- based methods have been considered to be easy ways to result in non-transgenic mutants. However, these approaches always show high sensitivity and are limited to some species due to bottlenecks in the regeneration process.
Genome editing provides agriculture productivity for the world population, which has reached 7.8 billion and is estimated to exceed 10 billion by 2055 (FAO, 2017; Li and Yan, 2020). Up until 2018, GE crops have been planted approximately 191.7 million acres throughout 26 countries and adopted by approximately 17 million farmers (Yu et al., 2019; Przezbórska-Skobiej and Siemiński, 2020). The legal status between genetically modified organisms (GMO) and genome-edited products is still in discussion. However, the United States and Japan have regulatory systems at the highest political levels, whereas European countries adopted a strict policy toward genome-edited products (Menz et al., 2020). So that scientists are giving evidence that genome editing is different from genetic modification because the DNA editing method is the same as the conventional breeding method or natural biological evolution (Turnbull et al., 2021).
Current advancement of SSNs, such as meganuclease I-SceI (1944), zinc-finger nucleases (ZFNs,1996–2003), transcription activator-like effector nucleases (TALENs, 2009–2010), and Cas nuclease (160 kDa protein, 2013-present) offer alternative approaches for trait improvement in crop plants (Butler et al., 2015; Schiml et al., 2015; Neela and Fanta, 2019; Turk, 2019). All SSNs have been widely used in the genome editing studies for trait improvement of various plant species such as Arabidopsis, tobacco, and maize (reviewed in Zhang et al., 2018).
Zinc-finger nucleases consist of two functional fused domains, including a non-specific DNA-cleavage domain and a DNA-binding domain composed of Cys2His2 zinc fingers, which target three base pairs and a non-specific catalytic domain of the FokI endonuclease (Bibikova et al., 2002; Nargesi et al., 2021). ZFNs were first used in model plant Arabidopsis thaliana (Lloyd et al., 2005). Since then, ZFNs has become increasingly popular for the genome modifications in various crop species, such as rice (Cantos et al., 2014) and maize (Gao et al., 2010). However, ZFNs have several limitations, including complexity of the contact between zinc fingers and DNA, an inherent drawback, and challenges in authenticating such proteins for a particular DNA locus of context.
Similar to ZFNs, TALENs are fusions of transcriptional activator-like effectors (TALE) repeats built from arrays of 33–35 amino acid modules and the FokI restriction enzyme from the plant pathogen Xanthomonas (Schiml et al., 2015; Ye et al., 2018; Afzal et al., 2020). The Fok1 nuclease is a common functional part between ZFNs and TALENs. However, designing TALENs is more affordable than ZFNs (Nargesi et al., 2021). The TALENs are simpler to construct and authenticate and thus provide a cost effective and faster method of genome editing. The challenges associated with synthesis, protein design, and corroboration remain as obstacles to more extensive use in genome-editing applications (Bétermier et al., 2014).
Clustered regularly interspaced short palindromic repeats (CRISPR/Cas) is an essential genome-editing technique based on the type II prokaryotic adaptive immune system from Streptococcus pyogenes Cas9 (SpCas9) (Schiml et al., 2015; Veillet et al., 2019a). Jennifer Doudna and Emmanuelle Charpentier first adapted gene-editing by using CRISPR/Cas9 system to function in the test tube in 2012, and were awarded the Nobel Prize for a significant contribution to Chemistry in 2020. In nature, bacteria and archaea use the RNA-guided endonucleases as part of an adaptive immune system to direct the degradation foreign nucleic acids (Anzalone et al., 2020). Although SpCas9 is predominantly used for plant functional genomics various other Cas enzymes from different bacteria have been successfully utilized as an alternative for genome editing system in plants, including the Staphylococcus aureus Cas9 (SaCas9) (Kaya et al., 2016), Streptococcus thermophilus (CRISPR1, CRISPR2, CRISPR3, and CRISPR4) (Sapranauskas et al., 2011), Francisella novicida Cas12a (Cpf1, FnCas12a) (Wu et al., 2020), Brevibacillus laterosporus (BlatCas9) (Gao et al., 2020), and Leptotrichia shahii (LshCas13a, Cas13a) (Veillet et al., 2019a).
The CRISPR/Cas9 genome-editing system is a two-component complex composing of single-guide RNA (gRNA) and Cas9 enzyme. The customizable gRNA, a small piece of the pre-designed RNA sequence of 20 bases, binds to Cas9 and specifies a target sequence (locus) within the genome. The Cas9 enzyme acts as a pair of molecular scissor that cut the two strands of DNA at a specific location in the genome defined by the gRNA sequence, then section of DNA can be modified by adding or removing (Hongbao et al., 2018; Veillet et al., 2020). Schematic representation of plant genome editing with CRISPR/Cas is shown in Figure 1. In August 2013, three studies were published in the same issue of Nature Biotechnology showing that Cas9 works in plant cells. One of the genes considered for editing was phytoene desaturase gene (PDS3) in the model plants A. thaliana and N. benthamiana and in the crop plant O. sativa by transient expression system in protoplasts with the plant codon-optimized SpCas9 (pcoCas9) (Li et al., 2013; Nekrasov et al., 2013; Shan et al., 2013). In these studies other genes including flagellin sensitive 2 gene (AtFLS2) in Arabidopsis (Li et al., 2013), three rice genes (OsBADH2, Os02g23823 and OsMPK2), and a wheat gene (TaMLO) (Shan et al., 2013) have been targeted. Higher mutation efficiency was demonstrated with the use of plant codon optimized Cas9.
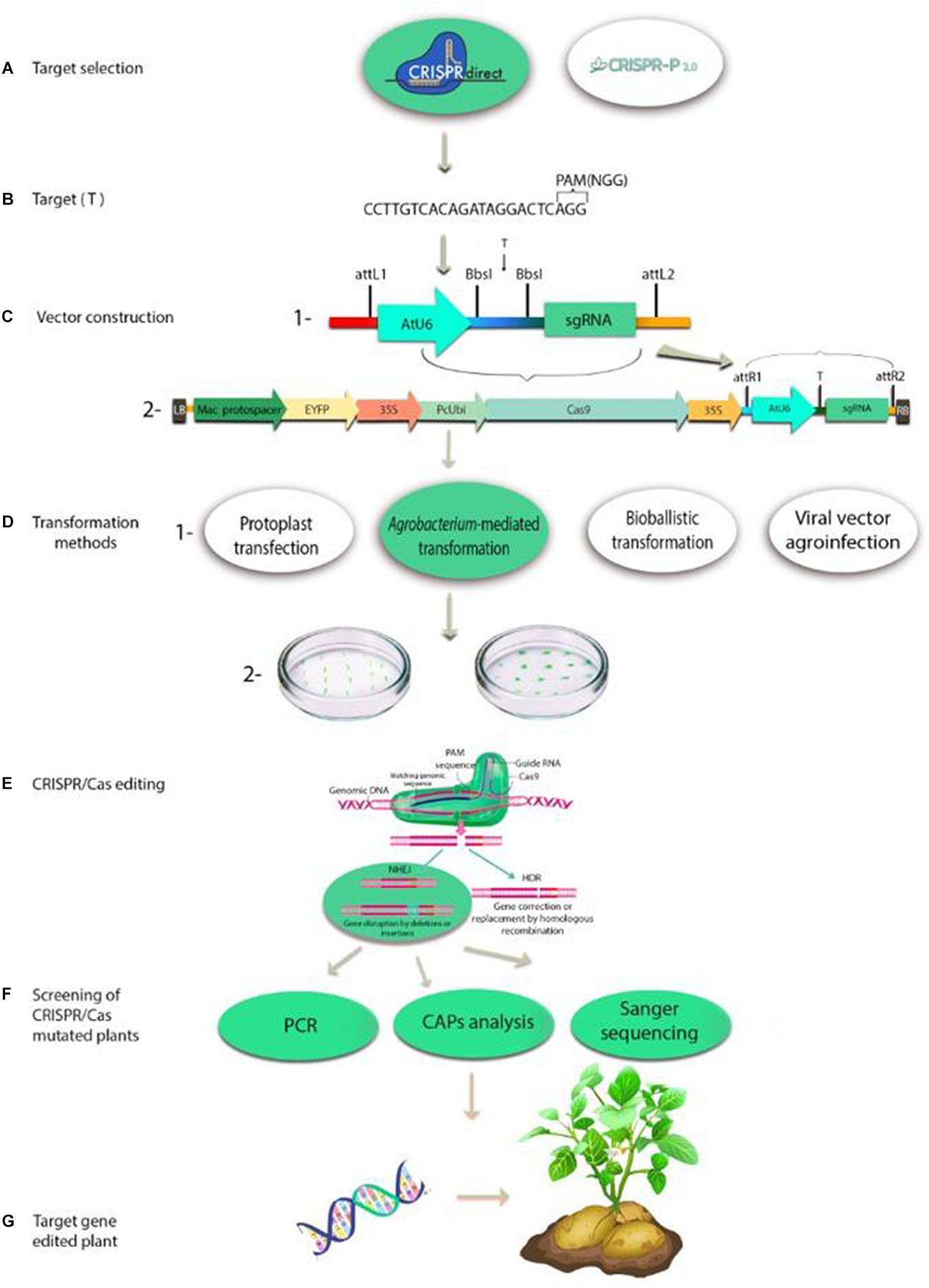
Figure 1. Schematic representation of plant genome editing with CRISPR/Cas: (A) Examples of software tools available for CRISPR sgRNA design; (B) The 20-bp target sequence with protospacer adjacent motif (PAM); (C) (1) Representation of cloning the 20-bp target sequence into the pair of BbsI sites in gRNA cloning vector; (2) Representation of cloning the gRNA expression construct into all-in-one binary vector by the recombination of attL and attR sites; (D) (1) Plant transformation methods are used for delivery of CRISPR/Cas9 system: protoplast transfection with polyethylene glycol (PEG) or via electroporation; Agrobacterium-mediated transformation; bioballistic transformation and viral vector agroinfection. (2) Examples of plant tissues used as the explants for transformation; (E) Diagrammatic representation of the CRISPR/Cas editing system: CRISPR/Cas induces double-stranded breaks in genomic DNA strands. The PAM sequence is primarily required for the Cas9 to cleave DNA. A stretch of 20 bases of sgRNA defines the binding specificity. Two methods repair the DSBs in DNA: non-homologous end-joining (NHEJ), which is not precise and permanently results in a gene knockout and homology-directed repair (HDR), which is activated in the presence of a template and results in knockin or gene replacement; (F) Screening of CRISPR/Cas mutated plants by polymerase chain reaction (PCR) analysis, cleaved amplified polymorphic sequences (CAPs) analysis and Sanger sequencing; (G) Target gene-edited plant in a potato example.
The CRISPR/Cas9 system has been mainly used to generate knockout mutants by single gene editing. The development and advantage of base editors has expanded the scope of genome editing by introducing base substitutions without requiring donor DNA or induction of a DSB (Nadakuduti et al., 2018; Choi et al., 2021). Thus far, two kinds of base editors have been developed. Cytosine base editors (CBEs) catalyze the conversion of C and G base pairs to T and A base pairs, and adenine base editors (ABEs), which catalyze A and T to G and C conversions (Anzalone et al., 2020). For CBE, the Cas9 nickase (nCas9) or catalytically dead Cas protein (dCas9) is fused with a cytidine deaminase that catalyzes the deamination of cytosine (C) in an arrow window of the non-targeted strand. Deamination converts the original C to U, which is recognized as T during DNA replication, ultimately resulting in a C and G to T and A single-base substitution (Bharat et al., 2020). Base editing in the CRISPR/Cas9 system for various plant species was reviewed by Li et al. (2021). For example, Veillet et al. (2019a, 2020) developed a S. aureus cytosine base editor in the granule-bound starch synthase StGBSSI (PGSC0003DMG400012111) and StDMR6-1 (PGSC0003DMG400000582) genes through a SanCas9 (D10A) that mediated transition (C to T) and transversion (C to G) mutations in potato.
Constant improvements in CRISPR/Cas systems have enabled more ambitious applications that aim to improve plant productivity or develop other desirable traits. One long-standing aim has been the induction of targeted chromosomal rearrangements (crossovers, inversions, or translocations) (Enciso-Rodriguez et al., 2019). Chromosome breaks in dicentric chromosomes whose centromeres are separated by >20 kb are repaired via pathways that rely mainly on sequence homology (HR, SSA) (Cook et al., 2021).
CRISPR/Cas is a flexible, simple, effective and affordable system, and it does not require any protein engineering steps in the CRISPR/Cas system construction (Amancio et al., 2017; Lino et al., 2018). As a consequence, these advantages make the CRISPR/Cas a more usable genome-editing system compare with ZFNs and TALENs. However, CRISPR/Cas still has some disadvantages and limitations, such as PAM restriction and the need for more diversity in CRISPR tools to mediate different simultaneous catalytic activities (Veillet et al., 2019a). The option of using the CRISPR/Cas tool and the general editing strategy depends on some factors such as cell type, cellular environment, a form of the agent, method of delivery each forces different constraints, and different propensities for unwanted genome modification events (Anzalone et al., 2020). All these methods including ZFNs, TALENs, and CRISPR/Cas have been applied for trait improvement in Arabidopsis, barley, Brachypodium, maize, tobacco, rice, soybean, tomato, wheat, and potato (reviewed in the references) (Malzahn et al., 2017; Nathalia and Patron, 2017; Afzal et al., 2020). The use of CRISPR/Cas9 in vivo and in vitro for crop improvement via nutritional enhancement and production of biotic and abiotic stress-tolerant plants have also been described (Afzal et al., 2020).
Starchy root and tuber crops (RTC), including potato, cassava, sweet potato, yam, and other minor healthiest root vegetables are essential to the agriculture and food security of many countries, and are a dietary supplement of 2.2 billion people in developing countries1. The nutritional composition of some RTCs have been reviewed (Chandrasekara and Josheph Kumar, 2016).
Cassava, potato, and sweet potato account for 90% of the world’s tuber crop production at an annual level of approximately 836 million tons. The primary producer of the RTCs is Asia, followed by Africa, Europe, and America. In 2017, 494.6 million tons of RTC were produced worldwide (Neela and Fanta, 2019). This review covers the technological advancement, limitations, and future prospects of the CRISPR/Cas system and summarizes the application of this system in potato and other RTCs such as sweet potato (Ipomoea batatas), cassava (Manihot esculenta Crantz), yam (Dioscorea spp.), and carrot (Daucus carota subsp. sativus).
Potato (Solanum tuberosum L.) is one of the most consuming crops globally after rice (Oryza sativa L.), wheat (Triticum spp.), and maize (Zea mays L.) (Amir et al., 2018; Gonzales et al., 2020; Reyniers et al., 2020; Das et al., 2021). Most potato cultivars are tetraploid (2n = 4x = 48), highly heterozygous, and extremely genetically diverse with a very high single nucleotide polymorphism (SNP) frequency (Johansen et al., 2019b). Potato is grown on approximately 18.6 million ha in 150 countries, with an estimated production of 400 million tons annually (Singh, 2008). Potato occupies a wide eco-geographical range, including the northern latitudes of North America, Europe, and Asia (the long-day photoperiod) after originated in the equatorial Andean region of South America (a short-day photoperiod) (Gonzales et al., 2020). Potato grows in a temperature range of 15–29oC and an altitude range of 1500–3200 m a.s.l (Minda et al., 2018). The annual rainfall range requirement of the potato crop is 400–1500 mm, while the pH requirement is 5–6.5. Potato can be grown below 5 up to 4.5 pH levels.
Potatoes have potential of extraordinarily high yield and are an excellent source of more carbohydrates, proteins, minerals, and vitamins than other potential food crops (Hameed et al., 2018). These aspects have received increasing attention from scientific and many research studies have been performed on yield increase, various stress adaptation, and post-harvest conservation and processing (Sevestre et al., 2020). Unique morphological, developmental, and compositional features have been characterized for nearly 70,000 species of potato. The nuclear and organelles genome of a homozygous doubled-monoploid potato breeding line (S. tuberosum group Phureja DM1-3 516 R44) and a heterozygous diploid line (S. tuberosum group Tuberosum RH89-039-16) have been sequenced by the Potato Genome Sequencing Consortium in 2011 and predicted 39 031 protein-coding genes. This provides a platform for genetic improvement, including molecular and gene editing research in potato2. Historically, conventional breeding techniques have been used to increase agronomic yield, processing, and improve storage quality. Unfortunately, the conventional breeding and genetic analysis of cultivated potato has some challenging related to tetrasomic inheritance, high heterozygosity, and self-incompatibility (Nadakuduti et al., 2018). In this study, we reviewed the recent developments of CRISPR/Cas system for the potato and discuss the future perspectives (Table 1).
Plant genetic resources are the most essential natural resources in the world. Many crops species are recognizable as hybrids that have been domesticated from the wild type polyploidy plants or generated by various conventional or modern biotechnological breeding program (Alix et al., 2017; Bevan et al., 2017). The conventional mutagenesis-based breeding processes are time-consuming and laborious, especially for polyploid crop breeding (Liu et al., 2021). Re-domestication of diploid potatoes applying modern agricultural technologies has made good progress in unlocking the genetic diversity of potatoes. However, most diploid potato species have gametophytic self-incompatible, which has been a limiting factor for the development of inbred lines in potatoes. The genetic basis of self-incompatible mechanism in flowering plants, including the Solanaceae family was reviewed in the some researches (Kao and McCubbin, 1996; Silva and Goring, 2001). These technologies include genetic engineering, molecular marker technology, cis-genesis, and more recently gene editing (Beumer et al., 2021).
Self-incompatibility is genetically determined by the highly polymorphic S-locus, which consists of strongly linked genes, termed as the pistil-expressed S-RNase (S-locus RNase) and the pollen-expressed F-box proteins (SLF or S locus F-box). The pollen tube growth is one of the main self-incompatible indicators, and a multi-allelic RNase in the pistil blocks incompatible pollen tube growth. To generate self-compatible diploid potato lines, the CRISPR/Cas9 system was used to target the first and second exons of the S-RNase gene and self-compatibility was confirmed in T1 lines (Ye et al., 2018; Enciso-Rodriguez et al., 2019). Ye et al. (2018) revealed that a self-incompatible diploid potato could be reinvented with a heterozygous mutation by knocking out the S-RNase alleles, and only a single mutation was transmitted to the T1 generation. Further research revealed that self-compatibility was achieved in the S-RNase knockout T0 lines, which contained bi-allelic and homozygous deletions/insertions in self-incompatible genotypes, and transmitted self-compatibility to T1 progeny with a potential transgenerational deletion (Enciso-Rodriguez et al., 2019). This study also confirmed that S-RNase is located within a pericentromeric region of 6.1 and 18.9 Mb of chromosome 1 in the physical map.
Biotic stress in plants is occurred by living organisms, commonly viruses, bacteria, fungi, nematodes, insects, arachnids, and weeds (Singla and Krattinger, 2016; Hashem et al., 2019). Potato crops are suffering several pests and diseases, such as late blight (caused by the oomycete pathogen Phytophthora infestans), Colorado potato beetle (Leptinotarsa decemlineata), bacterial wilt (caused by Ralstonia solanacearum), early blight (caused by the fungus Alternaria solani), and potato blacklegs (bacterial infection) (Kieu et al., 2021)3. Various pathogen infections affect potato yield with high economic losses of 30–64% every year (Torrance and Talianksy, 2020), even it is reached 88% (Kolychikhina et al., 2021). So the identification and characterization of disease resistance genes in the major crops genome have been a main focusing issue for the research community.
CRISPR/Cas has been used to enhance resistance to potato late blight, which remains the most significant disease of potato cultivation globally (Hameed et al., 2018). Late blight is a fungal disease that can destroy the leaves, stems, and tubers of potato plants (Tsedaley, 2014) and exhibits rapid multiplication and dissemination. Tiwari et al. (2021) published late blight resistance genes (R3a, RGA2, RGA3, R1B-16, Rpi-blb2, Rpi, and Rpi-vnt1) in potato. Susceptibility genes (S-genes) are important for increasing pathogen resistance (Tiwari et al., 2021). Potato pathogen susceptible genes StMLO1 (Mildew locus O), HDS gene homologue (DMG400008050), AtTTM2 gene homolog (DMG400025117), StDND1 (DMG400001441), StCHL1 (DMG400000711), and other two DMR6 homologues (DMG400000582 and DMG401026923) were edited with CRISPR/Cas9. The mutant lines encoding StMLO1, HDS, and AtTTM2 genes were as susceptible to late blight as wild type, whereas StDND1, StCHL1, and StDMR6-1 tetra-allelic deletion mutants exhibited enhanced resistance to late blight (Kieu et al., 2021).
Potato virus X (PVX, genus potexvirus), potato virus Y (PVY, genus potyvirus), potato virus A (PVA), potato virus M (PVM), potato virus S (PVS), and potato leafroll virus (PLRV, genus polerovirus) have been recorded in all potato growing zones and are probably the most diverse and economically harmful viruses infecting the world’s potato production (Halabi et al., 2021). The PVY have positive-sense (+), single-stranded RNA genome, with an average size of 9.7 kb in length. Upon translation, the RNA genome encodes a single large polyprotein of around 3061 amino acid residues. The single large polyprotein is processed by three virus-specific proteases domains into 11 mature proteins, namely P1, HC-Pro, P3, P3N-PIPO, 6K1, VPg, CI, 6K2, NIa (VPg plus Pro), NIb (viral replicase), and CP (capsid protein) (Quenouille et al., 2013; Cui and Wang, 2016). Zhang et al. successfully applied the CRISPR/Cas13a system by programming sgRNA (named pP3, pCI, pNIb, and pCP) specific for the RNA genome of three PVY strains (PVYO, PVYN, and PVYN:O). When transgenic lines with high transgene expression levels were assessed for PVY resistance with the PVYo virus, PVY mosaic symptoms were detected in leaves of infected WT plants at 25 days post-inoculation, whereas, no disease symptoms were detected in any of the transgenic plants. ELISA and qRT-PCR results indicated that transgenic plants accumulated much lower levels of the virus in systemic leaves than WT plants after PVY inoculation (Zhan et al., 2019).
One of the genes associated with plant stress is coilin. The mechanisms associated with the protective response to stress are unclear. Coilin is a major structural scaffold protein necessary for Cajal bodies, which are dynamic subnuclear compartments involved in the biogenesis of ribonucleoproteins. Makhotenko et al. (2019) applied the CRISPR/Cas9 system on the coilin gene in the potato cultivar Chicago. The edited lines (named F25 and D15) showed significantly higher resistance than control plants to biotic (ordinary strain PVY-O) and abiotic (salt and osmotic) stresses (Makhotenko et al., 2019).
Development of herbicide-resistant crops has resulted in remarkable changes to sustainable development of agronomy, one of which is the adoption of practical, effortless, and scientific crop-production systems with green technology (Vencill et al., 2012). The recent advantages of CRISPR/Cas9-mediated gene editing allows to target endogenous genes such as acetolactate synthase (ALS), 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS), cellulose synthase A catalytic subunit 3 (CESA3), and splicing factor 3B subunit 1 (SF3B1) (Han and Kim, 2019). ALS, also called acetohydroxyacid synthase, is found in plants and microorganisms and catalyzes the biosynthesis of various amino acids, including valine, leucine, and isoleucine (Ay et al., 1998). Resistance to ALS inhibitors is often attributed to an amino acid substitution in ALS (Svitashev et al., 2015). One of the first applications of the CRISPR/Cas system for ALS gene editing in potato was performed by Butler and colleagues (Butler et al., 2015, 2016). The StALS1 gene of the diploid and tetraploid potato was modified using CRISPR/Cas and TALEN with transformation of a binary T-DNA vector [conventional T-DNA and modified geminivirus (GVR) T-DNA]. Transformed events modified with GVRs introduced point mutations that supported a more herbicide-resistant phenotype and inheritable mutations, while events transformed with conventional T-DNAs had no detectable mutations and had same appearance as wild type. The ALS gene was also successfully and efficiently targeted in tomato and potato plants by CBE using Agrobacterium-mediated transformation. This resulted in chlorsulfuron-resistant mutant plants, as reported transgene-free edited plants were generated in the first offspring (Veillet et al., 2019b).
The extreme levels of abiotic stresses such as drought, flooding, salinity, mineral deficiency, low or high temperatures, high light, post-harvest problems, and accumulation of reducing sugars during cold storage have negative impact in crop yield (Le Gall et al., 2015). Abiotic stresses usually lead to superoxide overproduction in mitochondria (Dourmap et al., 2019). The mitochondrial respiratory chain in higher plants consists of the ATP-coupling cytochrome pathway (CP) and the cyanide (CN)-resistant respiratory pathway. The relationship of this chain to abiotic stress has been studied (Watanabe et al., 2010; Afzal et al., 2020). Plant mitochondria have an alternative oxidase (AOX), which is encoded by a small family of nuclear genes (Saha et al., 2016). AOX catalyzes the ATP-uncoupling cyanide (CN)-resistant pathway and can command the synthesis of important signaling molecules, such as hydrogen peroxide (H2O2), superoxide (O2–), and nitric oxide (NO). AOX is present in plants, fungi, protozoa, and some invertebrates. At the beginning of the 20th century, the AOX was first discovered from thermogenic plants during anthesis (Saha et al., 2016). Later it was found that it exists in the inner mitochondrial membrane of plants, fungi, protozoa, and some invertebrates (Araújo Castro et al., 2017). AOX activity can protect cells from reactive oxygen species (ROS) such as O2–, H2O2, and OH, particularly during different abiotic stress (Wang and Vanlerberghe, 2013). A CRISPR vector for the functional gene StAOX of the potato cyanide-resistant respiratory pathway was designed to examine the role of cyanide-resistant respiration in potato against high-light stress. The results indicated that high-light stress induced expression of AOX and accelerated cyanide-resistant respiration, and that loss of StAOX directly led to inhibition of cyanide-resistant respiration (Hua et al., 2020).
CRISPR/Cas9 technology has been widely applied for plant genomics research, with a focus on not only loss-of-function and gain-of-function analysis, but also on plant gene association analysis. In the potato genome, the association of transcription factor StCDF1 with the antisense transcript StFLORE to enhance drought tolerance was detected by mutation of the StFLORE promoter with CRISPR/Cas9 system. Although indicated that the overexpression of StFLORE or the downregulation of StCDF1 increased tolerance to drought through the regulation of stomata size and number, and StFLORE is regulated by StCDF1 (Gonzales et al., 2020).
Several studies have described improvements in the nutritional value of potato tubers, for example by enhancing essential amino acid compounds of seed protein (lysine, methionine, cysteine, and tyrosine contents), triacylglycerols, and vitamin E by overexpressing AmA1, PrLeg, WRINKLED1, DIACYLGLYCEROL ACYLTRANSFERASE 1, OLEOSIN, At-HPPD and At-HPT genes (Raina and Datta, 1992; Chakraborty et al., 2000, 2010; Crowell et al., 2008; Goo et al., 2013; Xu et al., 2020). Potato is also one of the major crops grown for starch production. Starch granules of potatoes have typical microscopic structures with different shapes and sizes, neutral taste, and good paste clarity (Singh et al., 2016). The starch content in potato, sweet potato, cassava, yam and maize endosperm consists of 65–90% of the absolute dry weight (Tigabu and Desta, 2018). Technology for starch isolation from natural sources is relatively simple and the range of industrial starch-based products is quite extensive and diverse. Starch extracted from potatoes has many utilization in food and technical-industrial applications, as potato starch has a variety of preferable molecular and intermolecular features (purity, molecular weight, phosphorylation, branching) (Khlestkin and Eltsov, 2021). Starch contains two major different carbohydrate, namely amylose (20–30%) and amylopectin (70–80%). Due to the low solubility of amylose, amylose solutions are generally unstable and easily to retrograde, gel, and turn opaque. In contrast, amylopectin is highly soluble in water, which characteristically makes amylopectin solutions very stable and clear (Visser et al., 1997). Changing the ratio of these two components significantly alters the properties of the starch. Therefore, starch has been the target of numerous genetic modification studies for a variety of purposes. Mutations in granule-bound starch synthase (GBSS) and two starch synthase (SS) genes (SSII and SSIII) resulted in starch with short-chain amylopectin and gelatinization (Khlestkin et al., 2017). In potato, the GBSS enzyme is encoded by a single locus (GBSSI) with four alleles in cultivated potato. Granule-bound starch synthase (GBSS) plays an essential role in amylose synthesis (Miao et al., 2014). In cultivated potatoes, the GBSS enzyme is encoded by a single locus (GBSSI) with four alleles. Waxy or amylose-free (amf) starch is a new type of starch made uniquely out of amylopectin molecules. Due to its superior viscosity and elasticity, amf starch has found applications in many areas of the food industry, such as stabilizers and thickeners. Waxy potato has been generated by silencing GBSSI gene function using traditional mutational breeding (Visser et al., 1991; Kuipers et al., 1994; Muth et al., 2010), RNAi (Andersson et al., 2003) and modern gene editing technology (Andersson et al., 2018; Veillet et al., 2019b). The StGBSS gene in the cultivar Kuras has been targeted for full knockout of GBSS enzyme activity using transient expression of CRISPR/Cas9 components in protoplasts, either as DNA plasmids or as ribonucleoprotein (RNP) complexes. As a result, the starch was confirmed as being of amylopectin quality in the line with all four alleles mutated (Andersson et al., 2018). In the study of Kusano, to the high-level expression of the CAS9 gene, two traditional enhancers were used for the efficient mutation of GBSSI. One of these enhancers was a translational enhancer dMac3 sequence (161 nucleotides), which consisting of a portion of the OsMac3 mRNA 5′-untranslated region (UTR). Another one was 5′-UTR from the alcohol dehydrogenase (ADH) gene. As the result, transformants containing dMac3 showed a higher frequency of mutations compare with transformants containing ADH enhancer and no enhancer sequence (Kusano et al., 2018).
Furthermore, multiple of unknown proteins associated with potato starch granules and multiple protease inhibitors were identified. Among these proteins, a still unknown isoform of starch synthase (SS6 gene coordinates on chromosome 7) demonstrated a great potential to be a key enzyme of the starch biosynthetic pathway in potato (Helle et al., 2018). Knockout mutagenesis of SS6 resulted in deletions and the function of SS6 was deciphered. The properties of the starch generated by this enzyme has great potential in industrial applications (Sevestre et al., 2020). CRISPR/Cas9 editing efficiency may be improved by using endogenous plant-specific U6 promoters, which contributes to increased sgRNA levels. Replacement of the regularly used Arabidopsis AtU6-1 promoter with endogenous potato StU6 promoters resulted in enhanced editing efficiency of the GBSS gene, with editing frequencies of 30–70% in protoplasts and 35% full allelic gene editing (Johansen et al., 2019b).
Another research priority for scientists and breeders is to improve potato tuber quality by removing anti-nutritional compounds, such as steroidal glycoalkaloids, acrylamide, and food toxins (Hameed et al., 2018). Potato accumulates the poisonous and bitter-tasting solanidane glycoalkaloids α-solanine and α-chaconine. These substances are harmful to many living things (Akiyama et al., 2021). It is known that St16DOX is the single gene responsible for steroid 16α hydroxylation in SGA biosynthesis and is an ideal research objective for generation CRISPR/Cas9-mediated mutagenesis to SGA-free potato plants. The knockout of St16DOX gene resulted in multiple mutations, including chromosomal fragment deletion at different sites, which resulted in abolition of St16DOX wild-type sequences in tetraploid potato (Nakayasu et al., 2018).
The post-harvest storage of potato tubers is also one of the key factors that determine the processing quality and final products. This long-term storage always accompanies with various storage diseases, such as soft rot, black dot, and Fusarium dry rot, which significantly affect tuber quality and makes it unsuitable for further processing (Barley Disease Control, 2009; Hameed et al., 2018). Potatoes are usually subjected to cold storage (4–8oC) to achieve a more sustainable supply to consumers throughout the year. However, cold temperature increases the reducing sugars accumulation in potato tubers. Upon high-temperature processing, these reducing sugars react with free amino acids to produce a brown and bitter-tasting products, and elevated levels of acrylamide, which is considered a potential carcinogen (Clasen et al., 2016). Enzymatic browning of potatoes is a major problem that arises during harvest and post-harvest procedures such as transport, storage, distribution, and blanching. In enzymatic browning, polyphenol oxidases (PPOs) catalyze the rapid polymerization of o-quinones from natural phenols. As a result, the chemical reaction leads to the formation of dark-colored precipitates in fruits and vegetables with negative effects on color, taste, flavor, and nutritional value (Yoruk and Marshall, 2010).
Four genes are mainly responsible for PPO activity in the potato tuber. The StPPO2 (PGSC0003DMG400018916) gene is the principal contributor to PPO total protein content (55% of the total enzyme), followed by StPPO1 (PGSC0003DMG400029575; 25–30%) and StPPO4 (PGSC0003DMG400018917) and StPPO3 (PGSC0003DMG400018914) (both < 15%). According to some reports, the down-regulation of multiple StPPO genes may harm the other functions of the enzyme in the plant. González et al. (2020) developed a CRISPR/Cas-based genome-editing approach to target StPPO2. Mutations induced in the four alleles of StPPO2 gene showed a significant reduction (up to 69%) in tuber PPO activity and enzymatic browning of (73%) in compared to the control (González et al., 2020). In this study, we also summarized CRISPR/Cas studies for other tuber crops and discuss the future perspectives (Table 2).
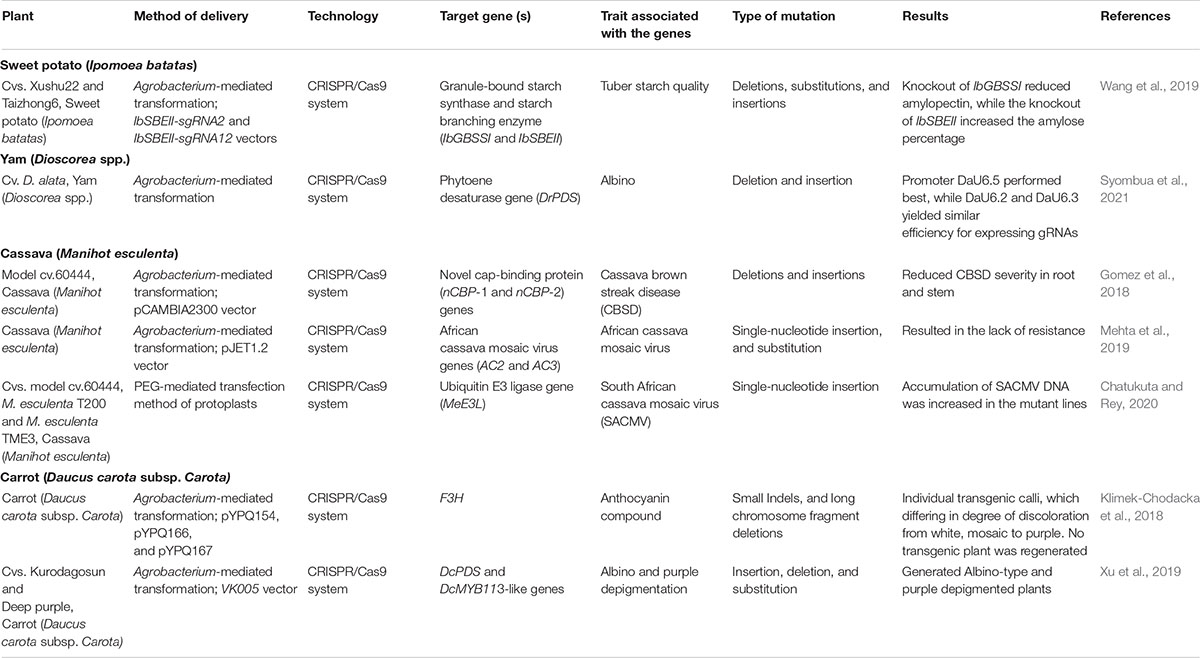
Table 2. Application of CRISPR/Cas system in sweet potato (Ipomoea batatas), cassava (Manihot esculenta Crantz), yam (Dioscorea spp.), and carrot (Daucus carota subsp. sativus).
Sweet potato (Ipomoea batatas) is one of the main starch-rich tuber crops and is a source of carbohydrates, β-carotene, vitamin A, C, B, and E complex, calcium, and iron. Cultivated forms of sweet potato have 2n = 6x = 90 chromosomes and are widely grown in 111 countries, particularly Southeast Asia is the major contributor, following by Oceania, and China. Sweet potato originated from either the Central or South American lowlands (Saranraj et al., 2019). Based on nutritional qualities and sensory acceptability, the color of sweet potato flesh varies from white, yellow, purple, and orange (Neela and Fanta, 2019). In contrast to other staple food crops, sweet potato possess special attributes, such as adaptability to wider topography, good productivity in short duration, and balanced nutritional composition (Trancoso-Reyes et al., 2016). To improve starch quality in sweet potato, the genes encoding starch biosynthesis (IbGBSSI- and IbSBEII) were subjected to CRISPR/Cas9-based mutagenesis by Wang et al. (2019). Most of mutations were nucleotide substitutions that cause amino acid changes and, less frequently, to stop codons. This study demonstrated the efficiency of CRISPR/Cas9 technology for improvement of starch qualities in sweet potato (Wang et al., 2019).
Yam (Dioscorea spp.) is a multi-species tuber crop that supplies food and income to millions of people around the world, particularly in Africa. The “yam belt” in West Africa includes Nigeria, Benin, Togo, Ghana, and Cote d’Ivoire and produces 92% of the 72.6 million tons of worldwide global yam production (NARIT; http://narit.or.th/images/category/dioscorea-spp-yam-4f349d). Yams are consumed raw, as a cooked soup, or used as a powder and flour in food preparations. Yam tubers are not only considered as a fundamental carbohydrate with relatively high protein and ascorbic acid content, but also have many bioactive components, such as mucin, dioscin, dioscorin, allantoin, choline, polyphenols, diosgenin, and vitamins such as carotenoids and tocopherols (Iwu et al., 1990). The phytoene desaturase gene (PDS) is essential for chlorophyll biosynthesis and is involved in converting phytoene into the carotenoid precursors phytofluene and carotene (Mann et al., 1994). PSD knockout mutations have an albino phenotype and thus are usually used as a visual marker to approve genome editing in various plants, including rice (Banakar et al., 2020), Arabidopsis, M. truncatula, N. benthamiana (Li et al., 2013; Wolabu et al., 2020), and potato (Butler et al., 2020). The CRISPR/Cas9 system was successfully used to target the PDS gene in yam. The efficiency of yam-derived promoters was identified by expression of gRNAs for PDS gene editing. Promoter DaU6.5 performed the best efficiency, while DaU6.2 and DaU6.3 had similar efficiency as revealed by fluorescence scores (Syombua et al., 2021).
Cassava (Manihot esculenta) is one of the most major staple crops for around 800 million people in tropical and sub-tropical regions of the world (Alicai et al., 2016). Africa presents over 50% of the world cassava production of 233.8 million metric tons. Cassava brown streak disease (CBSD) is the most devastating cassava disease in the Eastern, Central, and Southern Africa. Nowadays, West Africa including Nigeria, the largest cassava producer in the world, is also suffering from this disease (Alicai et al., 2019).
Cassava brown streak disease is caused by two species of positive-sense RNA viruses belonging to the family Potyviridae and genus Ipomovirus, specifically CBSV and Ugandan cassava brown streak virus (UCBSV) (Winter et al., 2010). The CBSV genome contains a polyprotein of 2902 amino acids that is proteolytically cleaved into 10 mature proteins (Mbanzibwa et al., 2009). Diseases caused by the family Potyviridae require the interaction of viral genome-linked protein (VPg) and host eukaryotic translation initiation factor 4E (eIF4E) isoforms. The eIF4E protein family assumes a fundamental par in the initiation of cap-dependent mRNA translation. It is known that cassava encodes the five eIF4E proteins such as eIF4E, eIF(iso)4E-1, eIF(iso)4E-2, novel cap-binding protein-1 (nCBP-1), and nCBP-2. Protein–protein interaction experiments consistently demonstrate that VPg proteins associate with cassava nCBPs. CRISPR/Cas9-mediated genome editing was applied to generate ncbp-1, ncbp-2, and ncbp-1/ncbp-2 mutants in cassava cultivar 60444. Compared to wild-type, the ncbp-1/ncbp-2 double mutants showed more resistance to CBSD severity in root and stem (Gomez et al., 2018). Among the viruses that infect cassava, cassava mosaic disease (CMD) is the most dangerous and widespread in the cassava-growing field. At least seven cassava mosaic geminivirus (CMG) species, including South African cassava mosaic virus (SACMV) limit crop production in Africa (Patil and Fauquet, 2010). The complete nucleotide sequences of infectious clones of SACMV were determined and this enabled the selection of putative host interacting genes for different investigations (Berrie et al., 2001). CRISPR/Cas9 technology was used in cassava with the aim of engineering resistance to SACMV. The sgRNAs of AC2 gene that is coding for the multifunctional TrAP protein and AC3 gene that is coding for the REn protein were targeted, but a clear disease-resistance phenotype was not observed in mutant lines (Mehta et al., 2019).
Plant gene association analysis as described above in potato plants has also been applied in cassava. For example, the association of SACMV with the ubiquitin E3 ligase gene (MeE3L) was studied by CRISPR/Cas9-mediated system of cassava gene MeE3L in SACMV-infected cassava protoplasts (Chatukuta and Rey, 2020).
Carrot (Daucus carota subsp. carota L.) is an economically important toot vegetable crop. Carrots are grouped into the carotene or western (D. carota ssp. sativus var. sativus) and the anthocyanin or eastern groups (D. carota ssp. sativus var. atrorubens Alef.). Purple carrot cultivars accumulate rich cyanidin-based anthocyanins, while orange, yellow, and red carrot cultivars accumulate rich carotenoids in the taproots (Xu et al., 2017). Carotenoids and anthocyanins both provide various health benefits to humans. Anthocyanins are an enormous group of more than 500 pigments abundant in the plant kingdom that accumulate in the cell vacuole of certain some organs. Many of the genes that encode enzymes involved in anthocyanin biosynthesis have been well characterized. For the first time, CRISPR/Cas9-mediated loss of function of the gene F3H, which is critical for anthocyanin biosynthesis, was reported by Klimek-Chodacka et al. (2018). They studied three codon-optimization variants of SpCas9 genes and demonstrated that AteCas9 has high efficiency in carrot cells for producing mutations, followed by zCas9, and Cas9p. The knockout of F3H gene affected the discoloration of calli, however, no transgenic plant was regenerated.
Albino and purple depigmentation mutants were regenerated when DcPDS and DcMYB113-like genes in orange and purple carrot were targeted by CRISPR/Cas9, respectively (Xu et al., 2019). The function of PDS genes is described above, while the DcMYB113-like changes critical functions in anthocyanin biosynthesis, and knockout mutations were expected to cause depigmentation phenotype. In addition, four different promoters (AtU3b, AtU3d, AtU6-1, and AtU6-29), which individually drive four sgRNA expression cassettes, were tested for mutation efficiency rate. The highest efficiency of mutagenesis was noticed in the loci targeted by AtU6-29-driven sgRNAs in both DcPDS and DcMYB113-like knockout T0 plants (Xu et al., 2019).
CRISPR/Cas-mediated genome editing technology can selectively modify any part of a genome to target genes controlling stress tolerance and nutritional quality of tuber crops. The simplicity and versatility of this technology make it a powerful tool for precise crop improvement via generation of knockout mutations in the form of insertions, deletions, and substitutions. The development of base editors, which generate base substitutions without requiring donor DNA or DSB induction, has expanded the possibilities of genome editing. The cultivated potato and most tuber crops have a complex genetic structure due to autotetraploidy and heterozygosity. Genetic improvement of these crops presents numerous challenges using traditional breeding techniques. To this end, the genome sequences of potato and other tuber crops provides a platform for genetic improvement via gene-editing technology. As mentioned above the CRISPR/Cas9 system with increased mutagenesis frequency has been established by using the improved CRISPR/Cas9 vector containing a translational enhancer sequence dMac3 in potato (Kusano et al., 2018) and through the use of codon optimized U6 promoter according species (Johansen et al., 2019a; Syombua et al., 2021).
Although significant progress in the last 5 years has made it possible to increase the efficiency and target specificity of CRISPR technology by using marker gene such as PDS, more work remains to be done to improve this technology for agriculturally important traits of commercial cultivars. In potato and other tuber crops, it is possible use the fluorescence marker-assisted selection for transgene-free genome-edited plants by the co-expression of fluorescence marker genes, as has been done for rice, tomato, and Arabidopsis (Aliaga-Franco et al., 2019). All the more already investigated the potentials of green fluorescent protein as a marker to select transgenic hybrid potato tubers (Palumbo and Veilleux, 2007) and red fluorescent protein to identify the localization of key enzymes of carotenoid biosynthesis after stable transformation in potato (tubers) (Nick, 2013). However, the fluorescence marker facilitates them of the transgene-free plants, but it does not improve the ratio of transgene-free plants. The self-eliminating and Transgene Killer CRISPR system proposed by He et al. (2018) and He and Zhao (2020) may accelerate identification of transgene-free and CRISPR-edited plants. For that they have modified the CRISPR/Cas9 construct with the bacterial BARNASE gene under the control of the rice REG2 promoter to express the suicide transgene during early embryo development to kill all of the CRISPR/Cas9-containing pollen and embryos produced by T0 rice plants that can be used for tuber crops in perspective.
One of the main limiting factors is the low embryogenic competency of local crops to tissue culture. It would be relevant to use the CRISPR/Cas technology to control endogenous plant hormones controlling and triggering plant morphogenesis, including somatic embryogenesis and organogenesis. Another major limiting factor is the size of Cas9, which prevents the use of plant viral vectors to deliver CRISPR/Cas components into a plant genome. Further research in this direction would widespread use the CRISPR/Cas technology to improve agro-important traits of local crop cultivars.
The studies reported in this review indicate that genome improvement in existing potato and other tuber crops is possible with this technology. The use of CRISPR/Cas for gene knockouts and base-editing systems for expression regulation of any gene of interest will facilitate development of non-transgenic crops. Recent developments in CRISPR-based genome editing technology for analyzed crops allow not only targeting of single genes in the genome, but also modification of multiple genes at once. One of the problems was observed that is utilization of genome-editing technology for model cultivars, due to the low regeneration efficiency of local cultivars in tissue culture. By considering the use of CRISPR/Cas technology as reported in this review, scientists may be able to design efficient vectors and transformation systems for employing CRISPR/Cas technology in tuber crops.
TD and SM wrote the manuscript together. Both authors contributed to the article and approved the submitted version.
This work was supported by grants from the Ministry of Education and Science of the Republic of Kazakhstan (Grant Number: AP09259964).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Afzal, S., Sirohi, P., and Singh, N. K. (2020). A review of CRISPR associated genome engineering: application, advances and future prospects of genome targeting tool for crop improvement. Biotechnol. Lett. 42, 1611–1632. doi: 10.1007/s10529-020-02950-w
Akiyama, R., Watanabe, B., Nakayasu, M., Lee, H. J., Kato, J., Umemoto, N., et al. (2021). The biosynthetic pathway of potato solanidanes diverged from that of spirosolanes due to evolution of a dioxygenase. Nat. Commun. 12:1300. doi: 10.1038/s41467-021-21546-0
Aliaga-Franco, N., Zhang, C., Presa, S., Srivastava, A. K., Granell, A., Alabadí, D., et al. (2019). Identification of transgene-free CRISPR-edited plants of rice, tomato, and Arabidopsis by monitoring DsRED fluorescence in dry seeds. Front. Plant Sci. 10:1150. doi: 10.3389/fpls.2019.01150
Alicai, T., Ndunguru, J., Sseruwagi, P., Tairo, F., Okao-Okuja, G., Nanvubya, R., et al. (2016). has a rapidly evolving genome: implications for virus speciation, variability, diagnosis and host resistance. Sci. Rep. 6:36164. doi: 10.1038/srep36164
Alicai, T., Szyniszewska, A. M., Omongo, C. A., Abidrabo, P., and Gilligan, C. A. (2019). Expansion of the cassava brown streak pandemic in Uganda revealed by annual field survey data for 2004 to 2017. Sci. Data 6:327. doi: 10.1038/s41597-019-0334-9
Alix, K., Gérard, P. R., Schwarzacher, T., and Heslop-Harrison, J. S. (2017). Polyploidy and interspecific hybridization: partners for adaptation, speciation and evolution in plants. Ann. Bot. 120, 183–194. doi: 10.1093/aob/mcx079
Amancio, S., Nakajima, I., Ban, Y., Azuma, A., Onoue, N., Moriguchi, T., et al. (2017). CRISPR/Cas9-mediated targeted mutagenesis in grape. PLoS One 12:e0177966. doi: 10.1371/journal.pone.0177966
Amir, H., Shan-E-Ali, Z. S., Sara, S., and Shahid, M. (2018). Applications of new breeding technologies for potato improvement. Front. Plant Sci. 9:925. doi: 10.3389/fpls.2018.00925
Andersson, M., Trifonova, A., Andersson, A. B., Johansson, M., Bülow, L., and Hofvander, P. (2003). A novel selection system for potato transformation using a mutated AHAS gene. Plant Cell Rep. 22, 261–267. doi: 10.1007/s00299-003-0684-8
Andersson, M., Turesson, H., Nicolia, A., Fält, A. S., Samuelsson, M., and Hofvander, P. (2017). Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep. 36, 117–128. doi: 10.1007/s00299-016-2062-3
Andersson, M., Turesson, H., Olsson, N., Fält, A.-S., Ohlsson, P., Gonzalez, M. N., et al. (2018). Genome editing in potato via CRISPR-Cas9 ribonucleoprotein delivery. Physiol. Plant. 164, 378–384.
Anzalone, A. V., Koblan, L. W., and Liu, D. R. (2020). Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 38, 824–844.
Araújo Castro, J., Gomes Ferreira, M. D., Santana Silva, R. J., Andrade, B. S., and Micheli, F. (2017). Alternative oxidase (AOX) constitutes a small family of proteins in Citrus clementina and Citrus sinensis L. Osb. PLoS One 12:e0176878. doi: 10.1371/journal.pone.0176878
Ay, D., Barak, Z., and Schloss, J. V. (1998). Biosynthesis of 2-aceto-2-hydroxy acids: acetolactate synthases and acetohydroxyacid synthases. Biochim. Biophys. Acta BBA Protein Struct. Mol. Enzymol. 1385, 401–419. doi: 10.1016/s0167-4838(98)00083-1
Banakar, R., Schubert, M., Collingwood, M., Vakulskas, C., and Wang, K. (2020). Comparison of CRISPR-Cas9/Cas12a ribonucleoprotein complexes for genome editing efficiency in the rice phytoene desaturase (OsPDS) gene. Rice 13:4. doi: 10.1186/s12284-019-0365-z
Barley Disease Control (2009). Technical Note TN6XX. Ludwigshafen: SAS, BASF, the chemical company. Available online at: https://www.sruc.ac.uk/media/r2mfdbup/tn644-haulm-destruction.pdf
Berrie, L. C., Rybicki, E. P., and Rey, M. (2001). Complete nucleotide sequence and host range of South African cassava mosaic virus: further evidence for recombination amongst begomoviruses. J. Gen. Virol. 82, 53–58. doi: 10.1099/0022-1317-82-1-53
Bétermier, M., Bertrand, P., and Lopez, B. S. (2014). Is non-homologous end-joining really an inherently error-prone process? PLoS Genet. 10:e1004086. doi: 10.1371/journal.pgen.1004086
Beumer, K., Stemerding, D., and Swart, J. A. A. (2021). Innovation and the commons: lessons from the governance of genetic resources in potato breeding. Agric. Hum. Values 38, 525–539.
Bevan, M. W., Uauy, C., Wulff, B. B. H., Zhou, J., Krasileva, K., and Clark, M. D. (2017). Genomic innovation for crop improvement. Nature 543, 346–354.
Bharat, S. S., Li, S., Li, J., Yan, L., and Xia, L. (2020). Base editing in plants: current status and challenges. Crop J. 8, 384–395.
Bibikova, M., Golic, M., Golic, K. G., and Carroll, D. (2002). Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 161, 1169–1175. doi: 10.1093/genetics/161.3.1169
Butler, N. M., Atkins, P. A., Voytas, D. F., Douches, D. S., and Hodaka, F. (2015). Generation and inheritance of targeted mutations in potato (Solanum tuberosum L.) using the CRISPR/Cas system. PLoS One 10:e0144591. doi: 10.1371/journal.pone.0144591
Butler, N. M., Baltes, N. J., Voytas, D. F., and Douches, D. S. (2016). Geminivirus-mediated genome editing in potato (Solanum tuberosum L.) using sequence-specific nucleases. Front. Plant Sci. 7:1045. doi: 10.3389/fpls.2016.01045
Butler, N. M., Jansky, S. H., and Jiang, J. (2020). First-generation genome editing in potato using hairy root transformation. Plant Biotechnol. J. 18, 2201–2209. doi: 10.1111/pbi.13376
Cantos, C., Francisco, P., Trijatmiko, K. R., Slamet-Loedin, I., and Chadha-Mohanty, P. K. (2014). Identification of “safe harbor” loci in indica rice genome by harnessing the property of zinc-finger nucleases to induce DNA damage and repair. Front. Plant Sci. 5:302. doi: 10.3389/fpls.2014.00302
Chakraborty, S., Chakraborty, N., Agrawal, L., Ghosh, S., Narula, K., Shekhar, S., et al. (2010). Next-generation protein-rich potato expressing the seed protein gene AmA1 is a result of proteome rebalancing in transgenic tuber. Proc. Natl. Acad. Sci. U.S.A. 107, 17533–17538. doi: 10.1073/pnas.1006265107
Chakraborty, S., Chakraborty, N., and Datta, A. (2000). Increased nutritive value of transgenic potato by expressing a nonallergenic seed albumin gene from Amaranthus hypochondriacus. Proc. Natl. Acad. Sci. U.S.A. 97, 3724–3729. doi: 10.1073/pnas.050012697
Chandrasekara, A., and Josheph Kumar, T. (2016). Roots and tuber crops as functional foods: a review on phytochemical constituents and their potential health benefits. Int. J. Food Sci. 2016:3631647. doi: 10.1155/2016/3631647
Chatukuta, P., and Rey, M. E. C. (2020). A cassava protoplast system for screening genes associated with the response to South African cassava mosaic virus. Virol. J. 17:184. doi: 10.1186/s12985-020-01453-4
Choi, M., Yun, J. Y., Kim, J. H., Kim, J. S., and Kim, S. T. (2021). The efficacy of CRISPR-mediated cytosine base editing with the RPS5a promoter in Arabidopsis thaliana. Sci. Rep. 11:8087. doi: 10.1038/s41598-021-87669-y
Clasen, B. M., Stoddard, T. J., Luo, S., Demorest, Z. L., Li, J., Cedrone, F., et al. (2016). Improving cold storage and processing traits in potato through targeted gene knockout. Plant Biotechnol. J. 14, 169–176. doi: 10.1111/pbi.12370
Cook, D., Long, S., Stanton, J., Cusick, P., and Bloom, K. (2021). Behavior of dicentric chromosomes in budding yeast. PLoS Genet. 17:e1009442. doi: 10.1371/journal.pgen.1009442
Crowell, E., McGrath, J., and Douches, D. (2008). Accumulation of vitamin E in potato (Solanum tuberosum) tubers. Transgenic Res. 17, 205–217.
Cui, H., and Wang, A. (2016). Plum pox virus 6K1 protein is required for viral replication and targets the viral replication complex at the early stage of infection. J. Virol. 90, 5119–5131. doi: 10.1128/JVI.00024-16
Das, S., Mitra, B., Luthra, S. K., Saha, A., and Hossain, A. (2021). Study on morphological, physiological characteristics and yields of twenty-one potato (Solanum tuberosum L.) cultivars grown in eastern sub-himalayan plains of India. Agronomy 11:335.
Dourmap, C., Roque, S., Morin, A., Caubrière, D., Kerdiles, M., Béguin, K., et al. (2019). Stress signalling dynamics of the mitochondrial electron transport chain and oxidative phosphorylation system in higher plants. Ann. Bot. 125, 721–736. doi: 10.1093/aob/mcz184
Enciso-Rodriguez, F., Manrique-Carpintero, N. C., Nadakuduti, S. S., Buell, C. R., Zarka, D., and Douches, D. (2019). Overcoming self-incompatibility in diploid potato using CRISPR-Cas9. Front. Plant Sci. 10:376. doi: 10.3389/fpls.2019.00376
FAO (2017). The Future of Food and Agriculture - Trends and Challenges. Rome: Food and Agriculture Organization of the United Nations. Available online at: https://www.fao.org/3/i6583e/I6583E.pdf
Gao, H., Smith, J., Yang, M., Jones, S., Djukanovic, V., Nicholson, M. G., et al. (2010). Heritable targeted mutagenesis in maize using a designed endonuclease. Plant J. 61, 176–187.
Gao, N., Zhang, C., Hu, Z., Li, M., and Liu, H. (2020). Characterization of Brevibacillus laterosporus Cas9 (BlatCas9) for mammalian genome editing. Front. Cell Dev. Biol. 8:583164. doi: 10.3389/fcell.2020.583164
Gomez, M. A., Lin, Z. D., Moll, T., Chauhan, R. D., and Bart, R. S. (2018). Simultaneous CRISPR/Cas9-mediated editing of cassava eIF4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnol. J. 17, 421–434. doi: 10.1111/pbi.12987
Gonzales, L. R., Li, S., Bergonzi, S., Oortwijn, M., and Bachem, C. (2020). Potato Cycling Dof Factor1 and its lncRNA counterpart StFLORE, link tuber development and drought response. Plant J. 105, 855–869. doi: 10.1111/tpj.15093
González, M., Massa, G. A., Andersson, M., Turesson, H., and Feingold, S. E. (2020). Reduced enzymatic browning in potato tubers by specific editing of a polyphenol oxidase gene via ribonucleoprotein complexes delivery of the CRISPR/Cas9 system. Front. Plant Sci. 10:1649. doi: 10.3389/fpls.2019.01649
Goo, Y.-M., Kim, T.-W., Lee, M.-K., and Lee, S.-W. (2013). Accumulation of PrLeg, a perilla legumin protein in potato tuber results in enhanced level of sulphur-containing amino acids. C. R. Biol. 336, 433–439. doi: 10.1016/j.crvi.2013.09.002
Guirouilh-Barbat, J., Lambert, S., Bertrand, P., and Lopez, B. S. (2014). Is homologous recombination really an error-free process? Front. Genet. 5:175. doi: 10.3389/fgene.2014.00175
Halabi, M. H., Oladokun, J. O., Baldodiya, G. M., Borah, B. K., and Nath, P. D. (2021). Identification, prevalence and genetic diversity study of potato viruses in Northeastern states of India. Ann. Appl. Biol. 179, 185–194.
Hamada, H., Liu, Y., Nagira, Y., Miki, R., Taoka, N., and Imai, R. (2018). Biolistic-delivery-based transient CRISPR/Cas9 expression enables in planta genome editing in wheat. Sci. Rep. 8:14422. doi: 10.1038/s41598-018-32714-6
Hameed, A., Zaidi, S. S.-E.-A., Shakir, S., and Mansoor, S. (2018). Applications of new breeding technologies for potato improvement. Front. Plant Sci. 9:925.
Han, Y. J., and Kim, J. I. (2019). Application of CRISPR/Cas9-mediated gene editing for the development of herbicide-resistant plants. Plant Biotechnol. Rep. 13, 447–457. doi: 10.3390/genes12060912
Hashem, A., Tabassum, B., and Abd_Allah, E. F. (2019). Bacillus subtilis: a plant-growth promoting rhizobacterium that also impacts biotic stress. Saudi J. Biol. Sci. 26, 1291–1297. doi: 10.1016/j.sjbs.2019.05.004
He, Y., and Zhao, Y. (2020). Technological breakthroughs in generating transgene-free and genetically stable CRISPR-edited plants. aBIOTECH 1, 88–96.
He, Y., Zhu, M., Wang, L., Wu, J., Wang, Q., Wang, R., et al. (2018). Programmed self-elimination of the CRISPR/Cas9 construct greatly accelerates the isolation of edited and transgene-free rice plants. Mol. Plant 11, 1210–1213. doi: 10.1016/j.molp.2018.05.005
Helle, S., Bray, F., Verbeke, J., Devassine, S., and Szydlowski, N. (2018). Proteome analysis of potato starch reveals the presence of new starch metabolic proteins as well as multiple protease inhibitors. Front. Plant Sci. 9:746. doi: 10.3389/fpls.2018.00746
Hua, D., Ma, M., Ge, G., Suleman, M., and Li, H. (2020). The role of cyanide-resistant respiration in Solanum tuberosum L. against high light stress. Plant Biology 22, 425–432. doi: 10.1111/plb.13098
Iwu, M., Okunji, C., Ohiaeri, G., Akah, P., Corley, D., and Tempesta, M. (1990). Hypoglycaemic activity of dioscoretine from tubers of Dioscorea dumetorum in normal and alloxan diabetic rabbits. Planta Med. 56, 264–267. doi: 10.1055/s-2006-960952
Johansen, I. E., Liu, Y., Jrgensen, B., Bennett, E. P., and Petersen, B. L. (2019b). High efficacy full allelic CRISPR/Cas9 gene editing in tetraploid potato. Sci. Rep. 9:17715. doi: 10.1038/s41598-019-54126-w
Johansen, I. E., Liu, Y., Jørgensen, B., Bennett, E. P., Andreasson, E., Nielsen, K. L., et al. (2019a). High efficacy full allelic CRISPR/Cas9 gene editing in tetraploid potato. Sci. Rep. 9:17715.
Kao, T. H., and McCubbin, A. G. (1996). How flowering plants discriminate between self and non-self pollen to prevent inbreeding. Proc. Natl. Acad. Sci. U.S.A. 93, 12059–12065.
Kaya, H., Mikami, M., Endo, A., Endo, M., and Toki, S. (2016). Highly specific targeted mutagenesis in plants using Staphylococcus aureus Cas9. Sci. Rep. 6:26871. doi: 10.1038/srep26871
Khlestkin, V. K., Peltek, S. E., and Kolchanov, N. A. (2017). Target genes for development of potato (Solanum tuberosum l.)Cultivars with desired starch properties (review). Sel’skokhozyaistvennaya Biol. 52, 25–36.
Khlestkin, V., and Eltsov, I. (2021). Different reactivity of raw starch from diverse potato genotypes. Molecules 26:226. doi: 10.3390/molecules26010226
Kieu, N. P., Lenman, M., Wang, E. S., Petersen, L., and Andreasson, E. (2021). Mutations introduced in susceptibility genes through CRISPR/Cas9 genome editing confer increased late blight resistance in potatoes. Sci. Rep. 11:4487. doi: 10.1038/s41598-021-83972-w
Klimek-Chodacka, M., Oleszkiewicz, T., Lowder, L. G., Qi, Y., and Baranski, R. (2018). Efficient CRISPR/Cas9-based genome editing in carrot cells. Plant Cell Rep. 37, 575–586. doi: 10.1007/s00299-018-2252-2
Kolychikhina, M. S., Beloshapkina, O. O., and Phiri, C. (2021). Change in potato productivity under the impact of viral diseases. IOP Conf. Ser. 663:012035. doi: 10.1016/j.jviromet.2016.05.009
Kuipers, A., Soppe, W., Jacobsen, E., and Visser, R. (1994). Field evaluation of transgenic potato plants expressing an antisense granule-bound starch synthase gene: increase of the antisense effect during tuber growth. Plant Mol. Biol. 26, 1759–1773. doi: 10.1007/BF00019490
Kusano, H., Ohnuma, M., Mutsuro-Aoki, H., Asahi, T., Ichinosawa, D., Onodera, H., et al. (2018). Establishment of a modified CRISPR/Cas9 system with increased mutagenesis frequency using the translational enhancer dMac3 and multiple guide RNAs in potato. Sci. Rep. 8:13753. doi: 10.1038/s41598-018-32049-2
Le Gall, H., Philippe, F., Domon, J. M., Gillet, F., Pelloux, J., and Rayon, C. (2015). Cell wall metabolism in response to abiotic stress. Plants (Basel) 4, 112–166.
Le Guen, T., Ragu, S., Guirouilh-Barbat, J., and Lopez, B. S. (2015). Role of the double-strand break repair pathway in the maintenance of genomic stability. Mol. Cell Oncol. 2:e968020.
Li, J. F., Norville, J. E., Aach, J., Mccormack, M., Zhang, D., Bush, J., et al. (2013). Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31, 688–691. doi: 10.1038/nbt.2654
Li, Q., and Yan, J. (2020). Sustainable agriculture in the era of omics: knowledge-driven crop breeding. Genome Biol. 21, 1–5. doi: 10.1186/s13059-020-02073-5
Li, Y., Li, W., and Li, J. (2021). The CRISPR/Cas9 revolution continues: from base editing to prime editing in plant science. J. Genet. Genomics 48, 661–670. doi: 10.1016/j.jgg.2021.05.001
Lino, C. A., Harper, J. C., Carney, J. P., and Timlin, J. A. (2018). Delivering CRISPR: a review of the challenges and approaches. Drug Deliv. 25, 1234–1257.
Liu, Q., Yang, F., Zhang, J., Liu, H., Rahman, S., Islam, S., et al. (2021). Application of CRISPR/Cas9 in crop quality improvement. Int. J. Mol. Sci. 22:4206.
Lloyd, A., Plaisier, C. L., Carroll, D., and Drews, G. N. (2005). Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 102, 2232–2237.
Makhotenko, A. V., Khromov, A. V., Snigir, E. A., Makarova, S. S., and Taliansky, M. E. (2019). Functional analysis of coilin in virus resistance and stress tolerance of potato Solanum tuberosum USING CRISPR/Cas9 editing. Dokl. Akad. Nauk 484, 772–776. doi: 10.1134/S1607672919010241
Malzahn, A., Lowder, L., and Qi, Y. (2017). Plant genome editing with TALEN and CRISPR. Cell Biosci. 7:21.
Mann, V., Pecker, I., and Hirschberg, J. (1994). Cloning and characterization of the gene for phytoene desaturase (Pds) from tomato (Lycopersicon esculentum). Plant Mol. Biol. 24, 429–434. doi: 10.1007/BF00024111
Mbanzibwa, D. R., Tian, Y., Mukasa, S. B., and Valkonen, J. (2009). Cassava brown streak virus (Potyviridae) encodes a putative Maf/HAM1 pyrophosphatase implicated in reduction of mutations and a P1 proteinase that suppresses RNA silencing but contains No HC-Pro. J. Virol. 83, 6934–6940. doi: 10.1128/JVI.00537-09
Mcvey, M., and Sang, E. L. (2008). MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet. 24, 529–538. doi: 10.1016/j.tig.2008.08.007
Mehta, D., Stürchler, A., Anjanappa, R. B., Zaidi, S., Hirsch-Hoffmann, M., Gruissem, W., et al. (2019). Linking CRISPR-Cas9 interference in cassava to the evolution of editing-resistant geminiviruses. Genome Biol. 20:80. doi: 10.1186/s13059-019-1678-3
Menz, J., Modrzejewski, D., Hartung, F., Wilhelm, R., and Sprink, T. (2020). Genome edited crops touch the market: a view on the global development and regulatory environment. Front. Plant Sci. 11:586027. doi: 10.3389/fpls.2020.586027
Miao, H., Sun, P., Liu, W., Xu, B., and Jin, Z. (2014). Identification of genes encoding granule-bound starch synthase involved in amylose metabolism in banana fruit. PLoS One 9:e88077. doi: 10.1371/journal.pone.0088077
Minda, T. T., Van, D., Struik, P. C., Marie, C., Jiménez, P., Khan, M. S., et al. (2018). The combined effect of elevation and meteorology on potato crop dynamics. Agric. For. Meteorol. 262, 166–177.
Muth, J., Hartje, S., Twyman, R. M., Hofferbert, H. R., Tacke, E., and Prüfer, D. (2010). Precision breeding for novel starch variants in potato. Plant Biotechnol. J. 6, 576–584. doi: 10.1111/j.1467-7652.2008.00340.x
Nadakuduti, S. S., Buell, C. R., Voytas, D. F., Starker, C. G., and Douches, D. S. (2018). Genome editing for crop improvement – applications in clonally propagated polyploids with a focus on potato (Solanum tuberosum L.). Front. Plant Sci. 9:2161–2163. doi: 10.3389/fpls.2018.01607
Nakayasu, M., Akiyama, R., Lee, H. J., Osakabe, K., Osakabe, Y., Watanabe, B., et al. (2018). Generation of α-solanine-free hairy roots of potato by CRISPR/Cas9 mediated genome editing of the St16DOX gene. Plant Physiol. Biochem. 131, 70–77. doi: 10.1016/j.plaphy.2018.04.026
Nargesi, S., Kaboli, S., Thekkiniath, J., Heidari, S., Keramati, F., Seyedmousavi, S., et al. (2021). Recent advances in genome editing tools in medical mycology research. J. Fungi 7:257. doi: 10.3390/jof7040257
Nathalia, V., and Patron, N. J. (2017). CRISPR-based tools for plant genome engineering. Emerg. Topics Life Sci. 1, 135–149.
Neela, S., and Fanta, S. W. (2019). Review on nutritional composition of orange-fleshed sweet potato and its role in management of vitamin A deficiency. Food Sci. Nutr. 7, 1920–1945. doi: 10.1002/fsn3.1063
Nekrasov, V., Staskawicz, B., Weigel, D., Jones, J., and Kamoun, S. (2013). Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31, 691–693. doi: 10.1038/nbt.2655
Palumbo, R. E., and Veilleux, R. E. (2007). Variation of GFP expression in diploid and tetraploid transgenic potato. Am. J. Potato Res. 84, 393–401.
Patil, B. L., and Fauquet, C. M. (2010). Cassava mosaic geminiviruses: actual knowledge and perspectives. Mol. Plant Pathol. 10, 685–701. doi: 10.1111/j.1364-3703.2009.00559.x
Przezbórska-Skobiej, L., and Siemiński, P. (2020). Gmos and global food security. Ann. Polish Assoc. Agric. Agribus. Econ. XXII, 173–182.
Quenouille, J., Vassilakos, N., and Moury, B. (2013). Potato virus Y: a major crop pathogen that has provided major insights into the evolution of viral pathogenicity. Mol. Plant Pathol. 14, 439–452. doi: 10.1111/mpp.12024
Raina, A., and Datta, A. (1992). Molecular cloning of a gene encoding a seed-specific protein with nutritionally balanced amino acid composition from Amaranthus. Proc. Natl. Acad. Sci. U.S.A. 89, 11774–11778.
Reyniers, S., Ooms, N., Gomand, S. V., and Delcour, J. A. (2020). What makes starch from potato (Solanum tuberosum L.) tubers unique: a review. Compr. Rev. Food Sci. Food Saf. 19, 2588–2612. doi: 10.1111/1541-4337.12596
Rodgers, K., and McVey, M. (2016). Error-prone repair of DNA double-strand breaks. J. Cell Physiol. 231, 15–24.
Rustgi, S., and Luo, H. (eds). (2020). Biolistic DNA Delivery in Plants and Protocols: Methods and Protocols. Berlin/Heidelberg: Springer Science+Business Media. doi: 10.1007/978-1-0716-0356-7
Saha, B., Borovskii, G., and Panda, S. K. (2016). Alternative oxidase and plant stress tolerance. Plant Signal. Behav. 11:e1256530.
Sant’Ana, R., Caprestano, C. A., Nodari, R. O., and Agapito-Tenfen, S. Z. (2020). PEG-delivered CRISPR-Cas9 ribonucleoproteins system for gene-editing screening of maize protoplasts. Genes 11:1029. doi: 10.3390/genes11091029
Sapranauskas, R., Gasiunas, G., Fremaux, C., Barrangou, R., Horvath, P., and Siksnys, V. (2011). The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 39, 9275–9282. doi: 10.1093/nar/gkr606
Saranraj, P., Behera, S. S., and Ray, R. C. (2019). “Traditional foods from tropical root and tuber crops,” in Innovations in Traditional Foods, ed. C. M. Galanakis (London: Woodhead Publishing), 159–191.
Schiml, S., Fauser, F., and Puchta, H. (2015). The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J. 80, 1139–1150. doi: 10.1111/tpj.12704
Sevestre, F., Facon, M., Wattebled, F., and Szydlowski, N. (2020). Facilitating gene editing in potato: a single-nucleotide polymorphism (SNP) map of the Solanum tuberosum L. cv. desiree genome. Sci. Rep. 10:2045. doi: 10.1038/s41598-020-58985-6
Shan, Q., Wang, Y., Li, J., Zhang, Y., Chen, K., Liang, Z., et al. (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31, 686–688.
Silva, N. F., and Goring, D. R. (2001). Mechanisms of self-incompatibility in flowering plants. Cell Mol. Life Sci. 58, 1988–2007.
Singh, H. P. (2008). Policies and strategies conducive to potato development in Asia and the Pacific region. Rap Publ. 164, 554–560.
Singh, J., Colussi, R., McCarthy, O. J., and Kaur, L. (2016). “Chapter 8 – potato starch and its modification,” in Advances in Potato Chemistry and Technology (Second Edition), 195-247, eds J. Singh and L. Kaur (San Diego, CA: Academic Press).
Singla, J., and Krattinger, S. G. (2016). “Biotic stress resistance genes in wheat,” in Encyclopedia of Food Grains, eds C. W. Wrigley, J. Faubion, H. Corke, and K. Seetharaman (Oxford: Elsevier), 388–392.
Sun, Y., Li, J., and Xia, L. (2016). Precise genome modification via sequence-specific nucleases-mediated gene targeting for crop improvement. Front. Plant Sci. 7:1928. doi: 10.3389/fpls.2016.01928
Svitashev, S., Young, J., Schwartz, C., Gao, H., Falco, S. C., and Cigan, A. M. (2015). Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using CAS9 and guide RNA. Plant Physiol. 169, 931–945. doi: 10.1104/pp.15.00793
Syombua, E. D., Zhang, Z., Tripathi, J. N., Ntui, V. O., Kang, M., George, O. O., et al. (2021). A CRISPR/Cas9-based genome-editing system for yam (Dioscorea spp.). Plant Biotechnol. J. 19, 645–647. doi: 10.1111/pbi.13515
Taleei, R., and Nikjoo, H. (2013). Biochemical DSB-repair model for mammalian cells in G1 and early S phases of the cell cycle. Mutat. Res. 756, 206–212. doi: 10.1016/j.mrgentox.2013.06.004
Tigabu, Y. T., and Desta, T. A. (2018). Starch Production, Consumption, Challenges and Investment Potentials in Ethiopia: The Case of Potato Starch. Berlin: Research Gate.
Tiwari, J. K., Rawat, S., Luthra, S. K., Zinta, R., Sahu, S., Varshney, S., et al. (2021). Genome sequence analysis provides insights on genomic variation and late blight resistance genes in potato somatic hybrid (parents and progeny). Mol. Biol. Rep. 48, 623–635.
Torrance, L., and Talianksy, M. E. (2020). Potato virus Y emergence and evolution from the andes of South America to become a major destructive pathogen of potato and other solanaceous crops worldwide. Viruses 12:1430. doi: 10.3390/v12121430
Trancoso-Reyes, N., Ochoa-Martinez, L. A., Bello-Perez, L. A., Morales-Castro, J., Estevez-Santiago, R., and Olmedilla-Alonso, B. (2016). Effect of pre-treatment on physicochemical and structural properties, and the bioaccessibility of β-carotene in sweet potato flour. Food Chem. 200, 199–205. doi: 10.1016/j.foodchem.2016.01.047
Tsedaley, B. (2014). Late blight of potato (Phytophthora infestans) biology, economic importance and its management approaches. J. Biol. Agric. Healthcare 4, 215–225.
Turk, H. (2019). Chitosan-induced enhanced expression and activation of alternative oxidase confer tolerance to salt stress in maize seedlings. Plant Physiol. Biochem. 141, 415–422. doi: 10.1016/j.plaphy.2019.06.025
Turnbull, J. W., Johnston, E. L., and Clark, G. F. (2021). Evaluating the social and ecological effectiveness of partially protected marine areas. Conserv. Biol. 35, 921–932. doi: 10.1111/cobi.13677
Uranga, M., Aragonés, V., Selma, S., Vázquez-Vilar, M., Orzáez, D., and Daròs, J.-A. (2021). Efficient Cas9 multiplex editing using unspaced sgRNA arrays engineering in a Potato virus X vector. Plant J. 106, 555–565. doi: 10.1111/tpj.15164
Vats, S., Kumawat, S., Kumar, V., Patil, G. B., and Deshmukh, R. (2019). Genome editing in plants: exploration of technological advancements and challenges. Cells 8:1386.
Veillet, F., Kermarrec, M. P., Chauvin, L., Chauvin, J. E., and Nogué, F. (2020). CRISPR-induced indels and base editing using the Staphylococcus aureus Cas9 in potato. PLoS One 15:e0235942. doi: 10.1371/journal.pone.0235942
Veillet, F., Perrot, L., Chauvin, L., Kermarrec, M.-P., Guyon-Debast, A., Chauvin, J.-E., et al. (2019b). Transgene-free genome editing in tomato and potato plants using agrobacterium-mediated delivery of a CRISPR/Cas9 cytidine base editor. Int. J. Mol. Sci. 20:402. doi: 10.3390/ijms20020402
Veillet, F., Chauvin, L., Kermarrec, M. P., Sevestre, F., and Chauvin, J. E. (2019a). The Solanum tuberosum GBSSI gene: a target for assessing gene and base editing in tetraploid potato. Plant Cell Rep. 38, 1065–1080. doi: 10.1007/s00299-019-02426-w
Vencill, W. K., Nichols, R. L., Webster, T. M., Soteres, J. K., Mallory-Smith, C., Burgos, N. R., et al. (2012). Herbicide resistance: toward an understanding of resistance development and the impact of herbicide-resistant crops. Weed Sci. 60, 2–30.
Visser, R. G. F., Suurs, L. C. J. M., Bruinenberg, P. M., Bleeker, I., and Jacobsen, E. (1997). Comparison between amylose-free and amylose containing potato starches. Starch Stärke 49, 438–443.
Visser, R., Somhorst, I., Kuipers, G. J., Ruys, N. J., Feenstra, W. J., and Jacobsen, E. (1991). Inhibition of the expression of the gene for granule-bound starch synthase in potato by antisense constructs. Mol. Gen. Genet. 225, 289–296.
Wang, H., and Xu, X. (2017). Microhomology-mediated end joining: new players join the team. Cell Biosci. 7:6. doi: 10.1186/s13578-017-0136-8
Wang, H., Wu, Y., Zhang, Y., Yang, J., Fan, W., Zhang, H., et al. (2019). CRISPR/Cas9-based mutagenesis of starch biosynthetic genes in sweet potato (Ipomoea batatas) for the improvement of starch quality. Int. J. Mol. Sci. 20:4702. doi: 10.3390/ijms20194702
Wang, J., and Vanlerberghe, G. C. (2013). A lack of mitochondrial alternative oxidase compromises capacity to recover from severe drought stress. Physiol. Plant 149, 461–473. doi: 10.1111/ppl.12059
Watanabe, C. K., Hachiya, T., Terashima, I., and Noguchi, K. (2010). The lack of alternative oxidase at low temperature leads to a disruption of the balance in carbon and nitrogen metabolism, and to an up-regulation of antioxidant defence systems in Arabidopsis thaliana leaves. Plant Cell Environ. 31, 1190–1202. doi: 10.1111/j.1365-3040.2008.01834.x
Winter, S., Koerbler, M., Stein, B., Pietruszka, A., Paape, M., and Butgereitt, A. (2010). Analysis of cassava brown streak viruses reveals the presence of distinct virus species causing cassava brown streak disease in East Africa. J. Gen. Virol. 91, 1365–1372. doi: 10.1099/vir.0.014688-0
Wolabu, T. W., Park, J. J., Chen, M., Cong, L., and Wang, Z. (2020). Improving the genome editing efficiency of CRISPR/Cas9 in Arabidopsis and Medicago truncatula. Planta 252:15. doi: 10.1007/s00425-020-03415-0
Wu, Y., Yuan, Q., Zhu, Y., Gao, X., and Yin, Z. (2020). Improving FnCas12a genome editing by exonuclease fusion. CRISPR J. 3, 503–511. doi: 10.1089/crispr.2020.0073
Xu, X.-Y., Akbar, S., Shrestha, P., Venugoban, L., Devilla, R., Hussain, D., et al. (2020). A synergistic genetic engineering strategy induced triacylglycerol accumulation in potato (Solanum tuberosum) leaf. Front. Plant Sci. 11:215. doi: 10.3389/fpls.2020.00215
Xu, Z. S., Feng, K., and Xiong, A. S. (2019). CRISPR/Cas9-mediated multiply targeted mutagenesis in orange and purple carrot plants. Mol. Biotechnol. 61, 191–199. doi: 10.1007/s12033-018-00150-6
Xu, Z. S., Feng, K., Que, F., Wang, F., and Xiong, A. S. (2017). A MYB transcription factor, DcMYB6, is involved in regulating anthocyanin biosynthesis in purple carrot taproots. Sci. Rep. 7:45324.
Ye, M., Peng, Z., Tang, D., Yang, Z., Li, D., Xu, Y., et al. (2018). Generation of self-compatible diploid potato by knockout of. Nat Plants 4, 651–654.
Yoruk, R., and Marshall, M. R. (2010). Physicochemical properties and function of plant polyphenol oxidase: a review. J. Food Biochem. 27, 361–422.
Yu, C., Deng, H., and Hu, R. (2019). Attitude GAPS WITH RESPECT to GM non-food crops and GM food crops and confidence in the government’s management of biotechnology: evidence from beijing consumers, chinese farmers, journalists, and government officials. Sustainability 12:324.
Zhan, X., Zhang, F., Zhong, Z., Chen, R., Wang, Y., Chang, L., et al. (2019). Generation of virus-resistant potato plants by RNA genome targeting. Plant Biotechnol. J. 17, 1814–1822. doi: 10.1111/pbi.13102
Zhang, Y., Massel, K., Godwin, I. D., and Gao, C. (2018). Applications and potential of genome editing in crop improvement. Genome Biol. 19:210.
Keywords: double-stranded DNA breaks, genome editing, sequence-specific nucleases, CRISPR/Cas system, tuber crops
Citation: Tussipkan D and Manabayeva SA (2021) Employing CRISPR/Cas Technology for the Improvement of Potato and Other Tuber Crops. Front. Plant Sci. 12:747476. doi: 10.3389/fpls.2021.747476
Received: 26 July 2021; Accepted: 04 October 2021;
Published: 26 October 2021.
Edited by:
Kejian Wang, China National Rice Research Institute, Chinese Academy of Agricultural Sciences (CAAS), ChinaReviewed by:
Qinlong Zhu, South China Agricultural University, ChinaCopyright © 2021 Tussipkan and Manabayeva. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuga A. Manabayeva, bWFuYWJheWV2YUBiaW9jZW50ZXIua3o=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.