- 1State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources, Guangdong Key Laboratory for Innovative Development and Utilization of Forest Plant Germplasm, College of Forestry and Landscape Architecture, South China Agricultural University, Guangzhou, China
- 2Guangdong Key Laboratory for Innovative Development and Utilization of Forest Plant Germplasm, College of Forestry and Landscape Architectures, South China Agricultural University, Guangzhou, China
- 3Guangdong Laboratory of Lingnan Modern Agriculture, Guangzhou, China
Nitrogen (N) is one of the abundant and essential elements for plant growth and development, and N deficiency (ND) affects plants at both physiological and transcriptomic levels. Neolamarckia cadamba is a fast-growing woody plant from the Rubiaceae family. However, the physiological and molecular impacts of ND on this species have not been well investigated. Here, we studied how N. cadamba responds to ND under hydroponic conditions. In a physiological aspect, ND led to a reduction in biomass, chlorophyll content, and photosynthetic capacity. ND also impaired the assimilation of N as the activities of glutamine synthetase (GS) and nitrate reductase (NR) were decreased in the root. Interestingly, the lignin content of stem increased progressively during the ND stress. The main transcription factors, the transcription factors that are important to N regulation has been found to be upregulated, including Nodule inception-like protein 7 (NLP7), TGACG motif-binding factor 1 (TGA1), basic helix-loop-helix protein 45 (BHLH45), NAM, ATAF1,2, CUC2 (NAC) transcription factor 43 (NAC43), and basic leucine zipper pattern 44 (bZIP44). The expression of N transporters, such as nitrate transporter 2.4 (NRT2.4), ammonium transporter 3 (AMT3), and amino acid transporter protein 3 (AAP3), was also upregulated. In addition, phosphorus- and calcium-related genes such as phosphate starvation response 2 (PHR2) and cyclic nucleotide-gated ion channel 15 (CNGC15) were expressed more abundantly in response to ND stress. Our results reveal the physiological and molecular mechanisms by which woody plants respond to ND.
Introduction
Nitrogen (N) is the most important inorganic nutrient in plants. It is a major component of proteins, nucleic acids, cofactors, and secondary metabolites. N influences all aspects of plants, for example, metabolism, resource allocation, growth, and development (Stitt and Krapp, 1999). High N availability is required to ensure high productivity of crops, including woody crops such as fast-growing Poplars (Mamashita et al., 2015). However, in natural ecosystems and agroecosystems, the availability of N is often limited, leading to abiotic plant stress—nitrogen deficiency (ND) (Martin et al., 2002). Large amounts of N fertilizer are being used in agricultural soils to meet the increasing demand for food, raw materials, and biofuels from a growing global population (Robertson and Vitousek, 2009). About 75% of N fertilizer is not taken up by plants and is lost to the environment, leading to an eutrophication of water bodies and an enrichment of atmospheric N oxide gases (Gutierrez, 2012). To maintain high yields and reduce N application rates, it is necessary to better understand the molecular mechanisms that regulate the morphological and physiological adaptations of crops to N effectiveness (Rennenberg et al., 2010; Miller et al., 2012).
In nature, N in the soil exists mainly in two forms: organic and inorganic N. Urea, amino acids, and amides are examples of organic N. Plants, on the other hand, primarily absorb N in inorganic forms such as nitrate nitrogen (NO3–) and ammonium nitrogen (NH4+). N uptake by plants is mainly dependent on nitrate transporters (NRTs) and ammonium transporters (AMTs). Plant NRT proteins were discovered and functionally described more than two decades ago. They are encoded by at least four gene families, NRT1 (NPF), NRT2, CLC, and SLAC1/SLAH (Chopin et al., 2007; Geiger et al., 2009; Feng et al., 2011; Maierhofer et al., 2014; Qin et al., 2019). N assimilation is the process by which plants absorb inorganic N from the environment and then undergo a series of reactions within the plant to synthesize amino acids. The amount of NO3–– absorbed by plant cells is reduced to nitrite by cytoplasmic nitrate reductase (NR) (Masclaux-Daubresse et al., 2010) and delivered into the chloroplast, where it is converted to NH4+ by nitrite reductase (NiR) before being further utilized. Ammonium, either absorbed directly from the soil or converted from nitrate, is assimilated via a glutamine synthetase (GS)/glutamate synthase (GOGAT) cycle (Lam et al., 1996).
Nitrogen deficiency affects crop growth and development, with symptoms, including leaf loss, plant dwarfing, thickening of root cell walls, and reduced yield (Coruzzi and Bush, 2001; Papadopoulos and Pougioula, 2010; Kusano et al., 2011). Reduced chlorophyll content, decreased anthocyanin accumulation in leaves, and restricted root growth, particularly of lateral roots, were typical signs in young seedlings (Scheible et al., 2004). Recent studies in maize have shown that the gene regulatory network controlling the root development also has a role in ND response (He et al., 2016). In N-deficient plants, there was an increase in reactive oxygen species (ROS), which caused damages to the cell membranes. To cope up with ROS, plants actively produced ROS scavengers such as the superoxide dismutase (SOD) and the peroxidase (POD) (Yang et al., 2019). In addition, ascorbic acid synthesis was increased in cucumber seedlings and fruits under ND conditions, supporting the role of ROS in ND response (Zhao et al., 2015). N stress results in reduced amino acid synthesis, protein biosynthesis (Liu et al., 2019), and reduced activities of NR, GS, and GOGAT in the leaves and roots of potato (Zhang et al., 2020). The response of leaf photosynthesis to ND is related to chlorophyll. Because N is a raw material for chlorophyll and its biosynthetic enzymes, ND resulted in reduced chlorophyll production and, as a result, a decrease in leaf chlorophyll content (Mu et al., 2016). In plant leaves, photosystems use approximately half of N, while the light harvesting system, dominated by chlorophyll a and b, consumes the other third (Martin et al., 2007). The quantity of N in the light harvesting system of maize leaves was reduced under ND conditions. The photosynthetic rate was reduced in N-deficit plants, and the light energy absorbed by the light trapping antennae cannot be fully utilized for CO2 fixation, so the remaining excitation energy was lost through thermal dissipation, photorespiration, and cyclic electron transfer (Frank et al., 2001; Jiang et al., 2006). A transcriptome study of cucumber leaves subjected to ND was carried out to better understand the putative regulatory mechanisms of chlorophyll reduction and cell wall remodeling. ND suppressed the transcription of chlorophyll a/b-binding proteins (CAB) genes, hence increasing chlorophyll breakdown, whereas the BR signaling system regulated cucumber cell wall reformation (Zhao et al., 2015). In addition, previous findings have shown that hormone signaling is involved in the regulation of N metabolism (Ristova et al., 2016). In conclusion, there are a wide variety of changes in the different aspects of plants under ND stress.
The main transcription factors involved in the regulation of nitrate transport were Nodule inception-like protein 7 (NLP7), NLP6, TGACG motif-binding factor 1 (TGA1), TGA4, Arabidopsis nitrate regulated 1 (ANR1), basic leucine zipper 1 (bZIP1), LOB domain-containing protein 37 (LBD37), LBD38, PCF/TCP-domain family protein 20 (TCP20), NAC domain-containing protein 4 (NAC4), squamosa promoter binding protein-like 9 (SPL9), etc. (O’Brien et al., 2016). APETALA2/ethylene responsive factor (AP2/ERF), WRKY, basic helix-loop-helix protein (BHLH), NAC, and MYB transcription factors, which were associated with abiotic stress in plants and were also frequently expressed at elevated levels under ND conditions (Lea et al., 2007). A high proportion of ND tolerance-related transcription factors were found to be differently expressed in the maize and spinach as revealed by transcriptome analysis, particularly from APETALA2/ethylene responsive element binding proteins (AP2/EREBP), BHLH, and WRKY families. These transcription factors may play an important role in plant N stress (Chen et al., 2015; Joshi et al., 2020). Phosphorus and calcium response genes were also found to be involved in the nitrate response. NIN-LIKE PROTEIN 7 (NLP7) was a major regulator of the nitrate-dependent transcriptional response (Marchive et al., 2013; Alvarez et al., 2020) and nitrate assimilation process (Castaings et al., 2009; Konishi and Yanagisawa, 2013; Marchive et al., 2013). In Arabidopsis, CBL-interacting protein kinase 23 (CIPK23) and CALCINEURIN B-LIKE PROTEIN (CBL1/9) regulate NRT1.1/NPF6.3 phosphorylation to transport nitrate, which in turn promotes calcium influx into the cell. Calcium-dependent protein kinases (CPK10/30/32) promote NLP7 phosphorylation (Wang et al., 2021). At the low levels of nitrate, SPX (SPX domain-containing proteins) inhibits the expression of N- and phosphorus-responsive genes by binding to NLP3/phosphate starvation response 2 (PHR2) to block the entry of NLP3 and PHR2 into the nucleus. In rice, NRT1.1B, the E3 ligase NBIP1 and SPX4 form a complex at high levels of NO3– conditions that leads to the ubiquitination and degradation of SPX4, thereby allowing NLP3 to enter the nucleus (Hu et al., 2019). The gene expression of Arabidopsis NLP7 and rice NLP3 induces nitrate response in the nucleus (Fichtner et al., 2021).
Neolamarckia cadamba is a tall evergreen tree that grows extremely fast (Liu et al., 2021), widely distributed in tropical and subtropical regions (such as South China, Indonesia, India, and Malaysia), intolerant of low temperatures and susceptible to insect damage. At the World Forestry Congress in 1972, it was recognized as a “miracle tree” for fast growth. Moreover, the tree provides an ideal raw material for plywood, fiberboard, pulp, and paper (Zhao et al., 2014, 2017; Ouyang et al., 2016). Neolamarckia cadamba has received more attention in recent years as a source of biomass for bioenergy production (Zhao et al., 2017).
However, as a fast-growing species, N. cadamba has a high demand for water and nutrients (Chu et al., 2017), and adequate nutrition is critical for its normal growth. So, in this study, the response of N. cadamba to ND stress was investigated at the physiological and molecular level. Specifically, differences in the transcriptome profiles of the roots and leaves of N. cadamba seedlings upon ND stress of 6 and 12 days were studied using RNA-sequencing (RNA-seq) to investigate the mechanism of fertilizer utilization. To our knowledge, this study is the first comprehensive report on the transcriptomic response of N. cadamba to ND stress.
Materials and Methods
Plant Materials and Nitrogen Deficiency Treatment
The experiments were conducted in the growth room at 25°C with 16 h light and 8 h dark. Some excellent clones of N. cadamba were obtained through a large-scale selection. Then, we picked up the best one for tissue culture. All seedlings used here were reproduced by tissue culture techniques for the propagation of those clones. The seedlings were cultured in a rooting medium for 15 days and then grown on outdoors for 7 days. The 4–5 cm tall seedlings were removed from the flasks, rinsed with tap water, and then fixed in a 6-L hydroponic box (length: 42 cm, width: 29 cm, and height: 7.5 cm) with a 1 cm3 volume sponge and foam board and placed in a growth room. The plants were incubated for 7 days in 1/4 Hoagland nutrient solution after 7 days of incubation in tap water as described by Dai et al. (2020) and Liu et al. (2021). The uniformed plants were finally selected and transferred into hydroponic boxes (length: 39 cm, width: 12 cm, and height: 10 cm) with a capacity of 2.5 L. Each box was fixed with a volume of 1 cm3 sponge and foam board to fix 5 seedlings, 5 seedlings for one replicate, and three replicates each for the control and treatment groups at the 6th and 12th day, so 60 seedlings in total once time were aerated by an oxygen pump (10 h/day) and changed every 3 days. The nutrient solution was changed every 3 days. The nutrient solution was composed of 4 mM Ca (NO3)2⋅4H2O, 6 mM KNO3, 2 mM MgSO4⋅7H2O, 1 mM NH4H2PO4, 80 μM NaFe-EDTA, 46.3 μM H3BO3,95 μM MnSO4⋅4H2O,0.8 μM ZnSO4⋅7H2O2, 0.3 μM CuSO4, and 0.02 μM (NH4)6Mo7O24⋅4H2O. For ND treatment, Ca (NO3)2⋅4H2O and KNO3 were replaced with CaCl2 and KCl for the bulk elements, NH4H2PO4 was replaced with KH2PO4, and (NH4)6Mo7O24⋅4H2O was removed, pH = 5.8. The concentration of N in the nutrient solution for the ND treatment was 0 mM, and the total nitrogen (CK) nutrient solution was 15 mM. Leaves and roots were taken at the 6th and 12th day for physiological index measurements and transcriptome sequencing. All experiments were repeated at least three times.
Measurement of Plant Physiological Parameters
Samples were randomly taken from plants after 6 and 12 days of the normal and ND treatments, plant height was measured with a straightedge, data were recorded and photographs were taken to observe the phenotype; root architecture was scanned with a root scanner (Epson Expression 12000XL Scanner, Nagano, Japan) and root structure (root length, root volume, and root surface area) was analyzed with the Win RHIZO root analysis software.
The seedlings to be tested were rinsed three times with deionized water, blotted dry with paper towels, weighed as a whole, and then placed in kraft bags to be baked in an oven to a constant weight and weighed as a dry weight. Each indicator was measured in three replicates.
The enzyme activities of leaves and roots were determined using SOD, POD, and ascorbate oxidase (APX) content kits (Suzhou Comin Biotechnology Co., Suzhou, China) (Yao et al., 2017).
The first, second, and third stem nodes (counting from top to bottom) of 6- and 12-day seedlings were embedded in 3% (mass fraction) agarose, sliced to a thickness of 40 μm using a vibrating microtome (Leica VT1000S, Nussloch, Germany) and placed on slides, stained with 10% KOH for 4 min, rinsed three to five times with pure water, stained with 1M HCl for 4 min, and rinsed three to five times with pure water. The sections were then stained with 0.02% toluidine blue for 10 s and rinsed three times with pure water. The films were observed and photographed under an automatic digital scanning imaging system (Beijing Wanbang Junyi M8, Beijing, China).
Chlorophyll Content and Fluorescence Determination
The chlorophyll fluorescence of leaves was measured in vivo using an IMAGING-PAM chlorophyll fluorescence imaging system (WALZ, Effeltrich, Germany) on three representative plants, avoiding the leaf veins as much as possible. The measurements were repeated three times, and the fluorescence parameters were analyzed.
The chlorophyll content of the leaves was determined by a 80% acetone extraction. About 0.1 g of the second pair of old leaves of the plant was taken and weighed in a 10-ml centrifuge tube after removing the veins, 5 ml of 80% acetone was added, and the leaves were soaked for 1 week under dark conditions until they turned white, and the chlorophyll content was determined by spectrophotometry (Wellburn, 1994). Three replicates of each index were determined.
Indicators of N Metabolism Measurement
After dry weight measurement, 0.1 g of the tissue was weighed to determine the CK and protein content using Kjeldahl N determination (Hong et al., 2013). Fresh samples were obtained by random sampling of normal and N-deficient plants at the 6th and 12th day, then 0.1 g of the tissue was ground in liquid N and weighed to determine the activity of enzymes related to N metabolism using NR and GS kits (Suzhou Comin Biotechnology Co., Suzhou, China).
RNA Extraction, Sequencing, and Differential Expression Analysis
Plants treated with ND and CK for 6 and 12 days were randomly sampled in three replicates per treatment to obtain leaf and root tissues, which were ground to a powder using liquid N. Then, the total RNA was extracted using the RNAprep Pure Assay kit (TIANGEN, Beijing, China). RNA quality was examined by electrophoresis using a 1% agarose gel and a NanoPhotometer® spectrophotometer (IMPLEN, Westlake Village, CA, United States) and the Bioanalyzer 2100 system’s RNA Nano 6000 assay kit (Agilent Technologies, Santa Clara, CA, United States) was used to assess, respectively, the purity and integrity of the RNA. Transcriptome analysis was commissioned to Novogene Company (Beijing, China). The RNA-seq analysis was performed using the edgeR package in R software v. 3.18. Differential expression analysis for both treatments was performed by the edgeR package in R software v. 3.18. with the Benjamini and Hochberg method to adjust for significant values, using a differential expression threshold of the value of p < 0.05. TBtools was used to depict a heat map based on the differentially expressed gene (DEG) results (Chen et al., 2020). In addition, RNA-seq data were submitted to https://ngdc.cncb.ac.cn/gsa/browse/CRA004153 with the submission number: CRA004153.
Functional Enrichment Analysis
Gene Ontology (GO) functional enrichment analysis of the DEG set was performed using the ClusterProfile software. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was performed on the differential expressed gene sets using ClusterProfile package in R software, a comprehensive database that integrates genomic, chemical, and phylogenetic information. The threshold of significant enrichment was p < 0.05.
Quantitative Real-Time-PCR Analysis
TaKaRa PrimeScriptTM RT reagent kit with gDNA Eraser (Perfect Real Time, Takara, Japan) was used to synthesize complementary DNA (cDNA) for real-time quantitative PCR (qRT-PCR) on a LightCycler 480 instrument (Beiqing et al., 2016). The information of primers for the genes for quantitative PCR (qPCR) are listed in Supplementary Table 3. SAMDC was chosen as the housekeeping gene (Dai et al., 2020). The expression data were calculated by the 2–ΔΔCt method (Pfaffl, 2001).
Results
Morphology and Growth Parameters of Neolamarckia cadamba Upon Nitrogen Deficiency
To evaluate the effects of ND stress on the plant growth, N. cadamba seedlings were investigated. At the 6th day, the root length under ND stress was the same as in the CK but with fewer lateral roots, while at the 12th day, the root length was shorter than in the control and leaves started to fade from fresh green to yellow (Figure 1A). The biomass of N. cadamba seedlings was also examined (Figures 1B–G). In the ND treatment group, fresh weight, dry weight, plant height, and root system (length, surface area, and volume) were not significantly different from the control at the 6th day. All these physiological indicators were lower than those in the control (CK) at the 12th day, with fresh weight reduced by 11.3%, dry weight reduced by 27%, plant height reduced by 6.5%, and root length, root surface area, and root volume reduced by 21.9, 25.9, and 29.6%, respectively (Figures 1B–G). The antioxidant enzyme activity in N. cadamba also responded differently to ND stress (Figures 2A–F). SOD and POD activities were affected differently depending on tissues and the time duration under ND stress. To be specific, POD was most significantly affected in roots at the 6th day and leaves at the 12th day as its content increased by 21.1 and 79.8%, respectively, while APX, on the other hand, decreased significantly in roots, by 75.2 and 59.2% at the 6th and 12th day, respectively (Figures 2A–F).
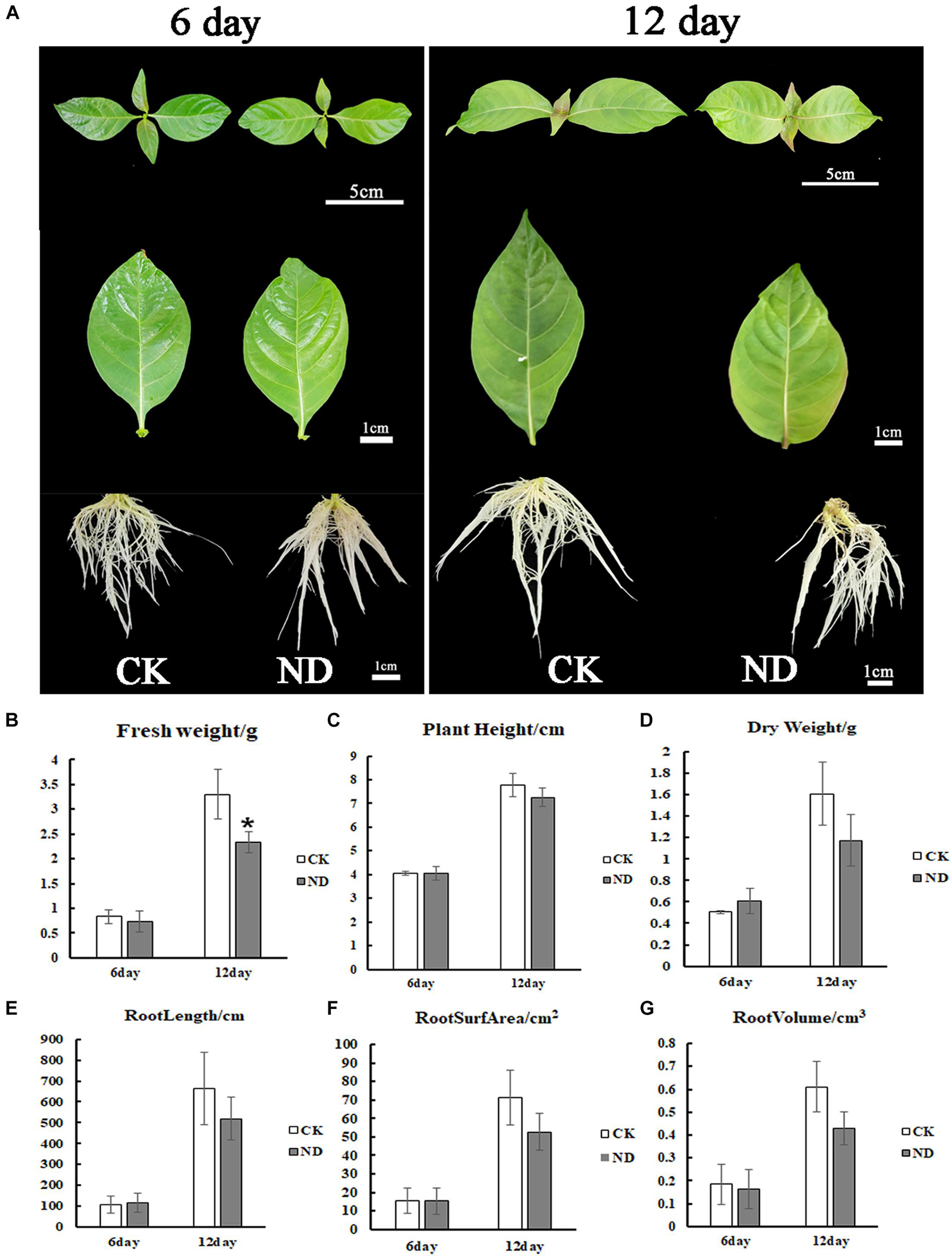
Figure 1. The growth of Neolamarckia cadamba under treatments of normal nutrition [total nitrogen (CK)] and nitrogen deficiency (ND). (A) The phenotypes of N. cadamba under total nitrogen (CK) and ND treatments in a growth room were shown at the 6th and 12th day, respectively. Fresh weight (B), plant height (C), dry weight (D), root length (E), root surface area (F), root volume (G) under CK and ND conditions were determined at the 6th and 12th day, respectively. Compared with CK, although all the parameters were reduced under ND treatment at the 12th day, only fresh weight showed a significant decrease. Data are means ± SD (n = 3). Differences between the mean values of CK and ND were compared using the t-test (* denote significant differences at p < 0.05).
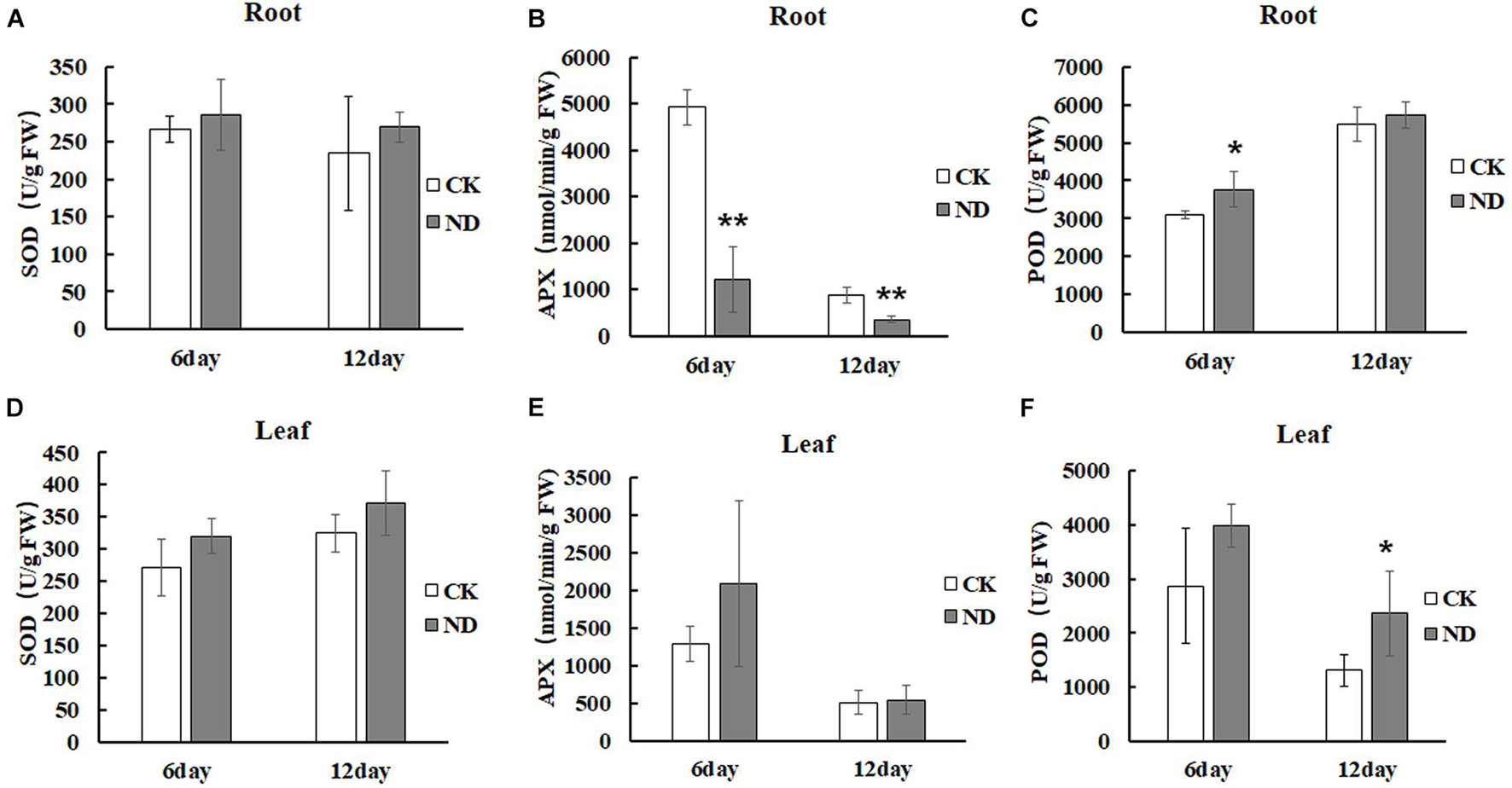
Figure 2. Enzyme activity in roots and leaves, superoxide dismutase (SOD) (A,D), ascorbate oxidase (APX) (B,E), peroxidase (POD) (C,F) was determined at indicated times. Enzymatic activities were measured in roots and leaves under the ND and CK treatment. Compared with CK, there was a significant difference in the physiological parameters of ND at the last stages. Data are means ± SD (n = 3). Differences between the mean values of CK and ND were compared using t-test (* and ** denote significant differences at p < 0.05 and p < 0.01).
Transcriptome Changes in Response to Nitrogen Deficiency
Total RNA was extracted from the roots and leaves of N. cadamba at the 6th and 12th day, and eight data sets were obtained, namely N6L, C6L, N6R, C6R, N12L, C12L, N12R, and C12R for transcriptome sequencing. The low quality and short reads were removed. Clean reads mapped over 75% of the N. cadamba reference genome (Supplementary Table 1). Principal component analysis (PCA) clustered transcripts into four groups according to tissues and treatments, indicating that there were good correlations and differences among the samples (Figures 3A,B).
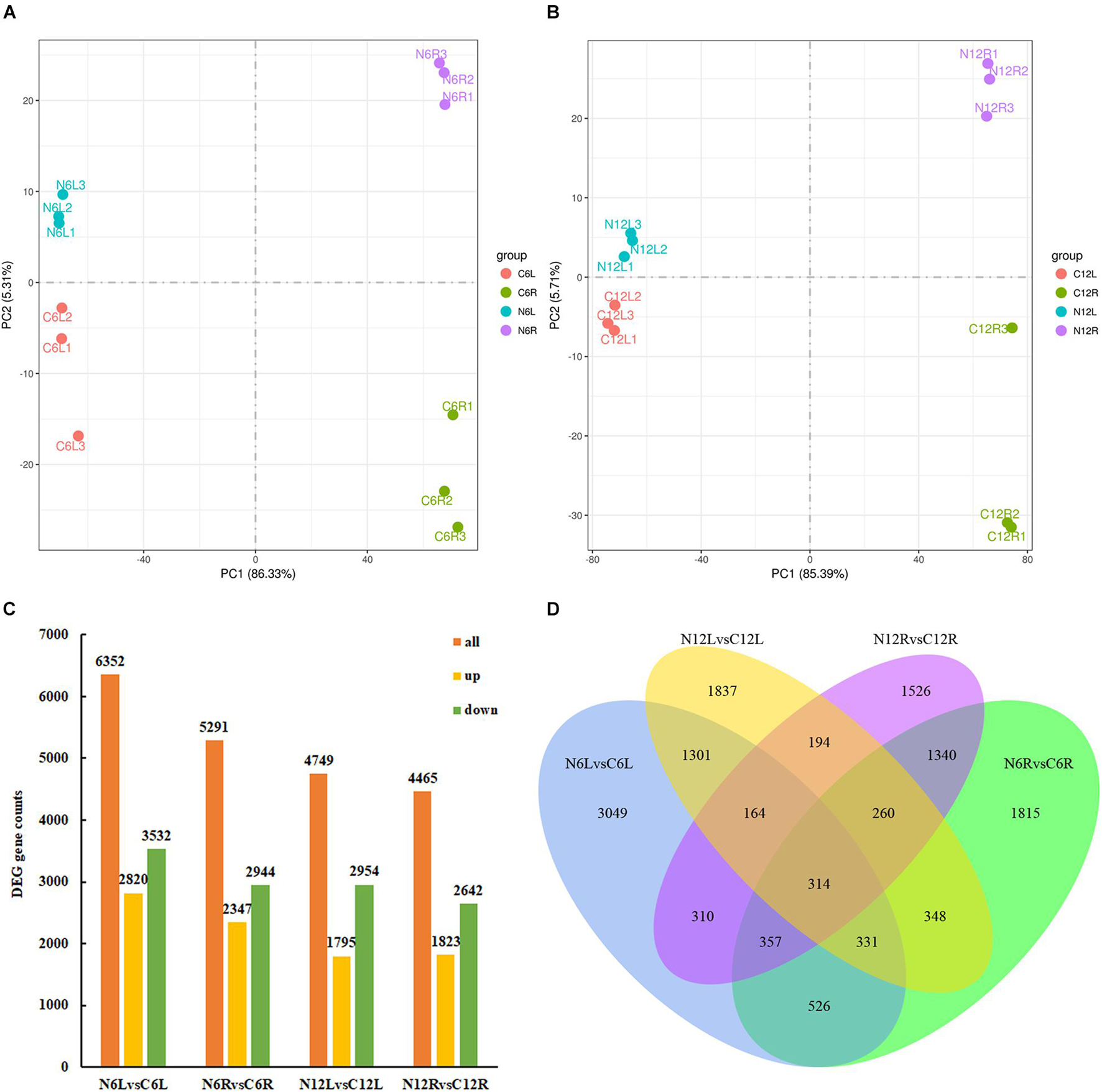
Figure 3. Differentially expressed genes (DEGs) in N. cadamba of control and ND conditions. The transcriptome of N. cadamba were sequenced and analyzed from the different samples of roots and leaves under ND and CK. Then, principal component analysis (PCA) and DEGs were analyzed. (A) PCA of DEGs at the 6th day. (B) PCA of DEGs at the 12th day. (C) The orange bar represents all DEGs, downregulated DEGs are in green, and upregulated DEGs are in yellow, FDR < 0.05. (D) This Venn diagram converges the DEGs of CK vs. ND at different time points (6 and 12 days) and different tissues (root and leaf). The overlapping area represents the DEGs with a common regulation mode between different treatments.
The comparative transcriptome analysis of N. cadamba under ND stress provided a basis for further elucidation of gene function and metabolic pathways. The number of DEGs (p ≤ 0.05) between groups differed significantly, i.e., 6,352 DEGs for N6L vs. C6L, 5,291 for N6R vs. C6R, 4,749 for N12L vs. C12L, and 4,465 for N12R vs. C12R (Figure 3C). The significant DEGs at different times and tissues under ND stress were used in the functional enrichment analysis, i.e., GO and KEGG databases. The corrected p ≤ 0.05 was defined as prominent enriched GO terms (Supplementary Figure 1) response to oxidative stress (GO:0006979) and response to stress (GO:0006950) were significant GO terms associated with biological processes in roots, while in leaves GO terms associated with photosynthesis (GO:0015979), peptide biosynthetic process (GO:0043043), and amide biosynthetic process (GO:0043604) were significantly enriched. For cellular components, photosynthetic membrane (GO:0034357), photosystem (GO:0009521), and cell wall (GO:0005618) were enriched in roots while in leaves GO terms associated with ribosome (GO:0005840), thylakoid (GO:0009579), photosynthetic membrane (GO:0034357), and photosystem (GO:0009521) were significantly enriched. For molecular functions, heme binding (GO:0020037), peroxide activity (GO:0004601), and tetrapyrrole binding (GO:0046906) were significant GO terms in roots while structural molecule activity (GO:0005198), iron–sulfur cluster binding (GO:0051536), and metal cluster binding (GO:0051540) were significantly enriched in leaves.
After the KEGG enrichment analysis for CK and ND at the 6th day was completed, significant KEGG pathways enriched in roots were identified, including mainly phenylpropanoid biosynthesis, carbon fixation in photosynthetic organisms, N metabolism, MAPK signaling pathway-plant, and plant hormone signal transduction. While for leaf samples carbon fixation in photosynthetic organisms was the most significantly enriched pathway. Interestingly, in the 12 day samples, porphyrin and chlorophyll metabolism, carbon fixation in photosynthetic organisms, N metabolism, photosynthesis, and photosynthesis-antenna proteins were significantly enriched pathways in leaves while in roots phenylpropanoid biosynthesis, N metabolism, and MAPK signaling pathway-plant were significantly enriched.
There were 314 differential expressed genes that were shared by all ways of comparison (Figure 3D). We were mainly interested in DEGs associated with “carbon metabolism” and “N metabolism” pathways by KEGG, and GO terms of these DEGs were in oxidoreductase activity (GO:0016616) and hexose metabolic process (GO:0019318).
To confirm the transcriptome results, 15 genes were randomly selected for qRT-PCR analysis. These genes are responsible for nitrate transporting, hormone response, photosynthesis, chlorophyll synthesis, and amino acid transporter proteins (AAPs). qRT-PCR results demonstrated that all gene expression trends were consistent with our RNA-seq data, indicating that our transcriptome analysis was reliable and ready for further analysis (Figure 4).
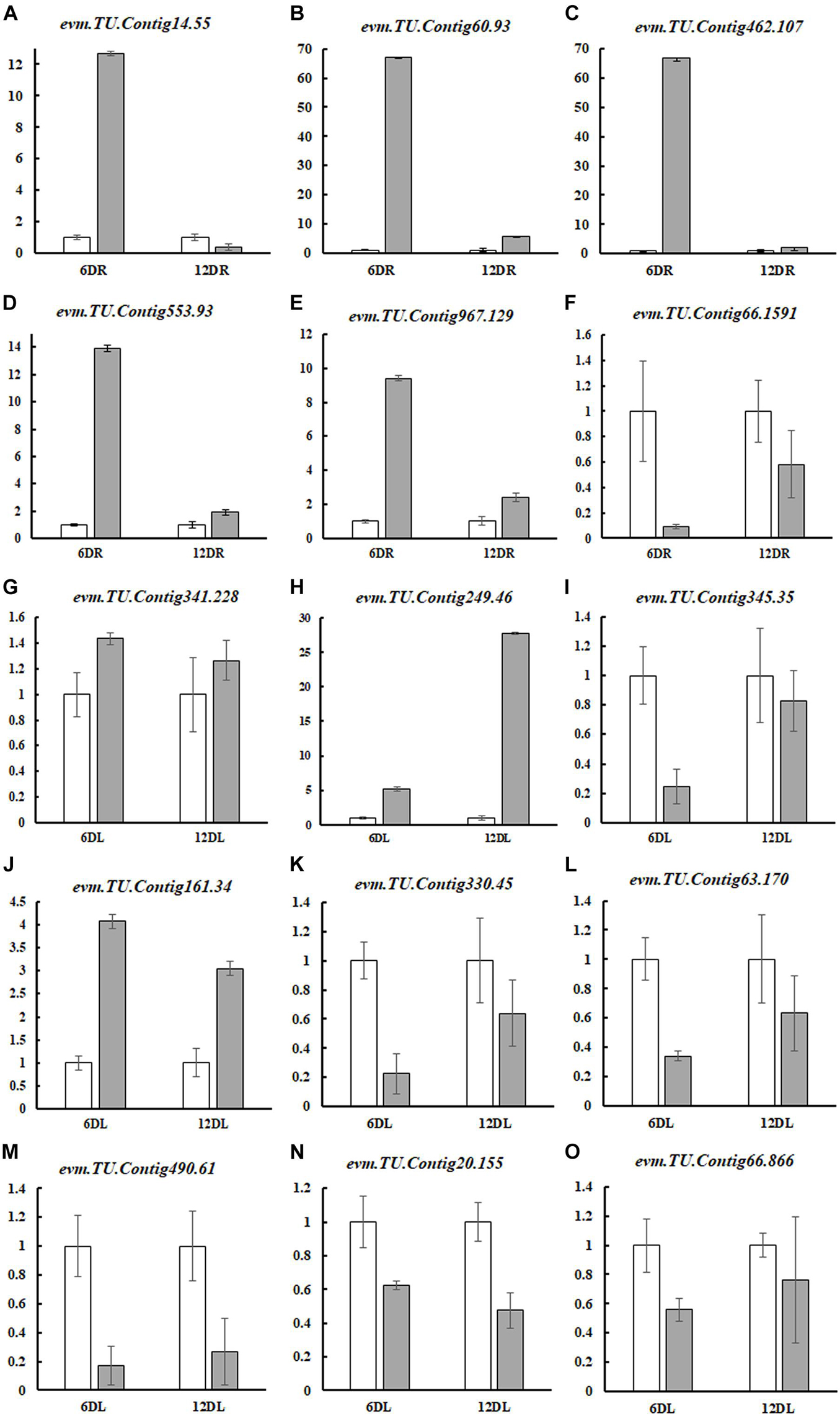
Figure 4. Quantitative real-time PCR validation of 15 DEGs. Normal nutrition control (CK), ND, root (R), and leaf (L) were labeled, and the pairs of CK and ND were compared at different time points (6DR: CKR6d vs. NDR6d; 12DR: CKR12d vs. NDR12d; 6DL: CKL6d vs. NDL6d; and 12DL: CKL12d vs. NDL12d). The experiments were repeated four times. The expression level of each gene in the CK was “1”. These genes included nitrate transporter (NRT) proteins (evm.TU.Contig14.55, evm.TU.Contig60.93, evm.TU.Contig462.107, evm.TU.Contig66.1591), hormone response (evm.TU.Contig553.93, evm.TU.Contig967.129), transcription factors (evm.TU.Contig341.228, evm.TU.Contig249.46), photosynthesis (evm.TU.Contig345.35, evm.TU.Contig20.155, evm.TU.Contig66.866), chlorophyll synthesis (evm.TU.Contig330.45, evm.TU.Contig63.170, evm.TU.Contig490.61), and amino acid transporter proteins (AAPs) (evm.TU.Contig161.34). Gene-specific primers used for real-time PCR are listed in Supplementary Table 3.
Chlorophyll Content and Photosynthetic Parameters Decreased Under Nitrogen Deficiency Stress in Neolamarckia cadamba
As yellowing leaf was found in the treatment at the 12th day, the efficiency of leaf photosynthesis was measured by imaging-PAM (Figure 5A) and some important parameters related to photosynthesis, such as Y(I), ETR(I), Y(II), qN, qP, and qL, were also measured to evaluate the effects of ND on photosynthesis (Supplementary Table 2). It was obvious that Y(I) and ETR(I) were significantly reduced at both the 6th and 12th day under ND. At the 6th day, although the leaves did not turn yellow significantly as much as the 12th day, Y(II), qN, qP, and qL all decreased significantly (Supplementary Table 2). At the 12th day, besides Y(II), qN, qP, and qL, chlorophyll fluorescence imaging was also decreased. However, Y(NPQ) increased at the 6th and 12th day (Figure 5A). In addition, the contents of chlorophyll a, chlorophyll b, total chlorophyll, and carotenoids were measured at the 1st, 3rd, 6th, 9th, and 12th day, and the results showed that these contents under ND began to decrease at the 6th day, and the decrease reached a larger level at the 12th day after the yellowing of the leaves appeared (Figure 5B).
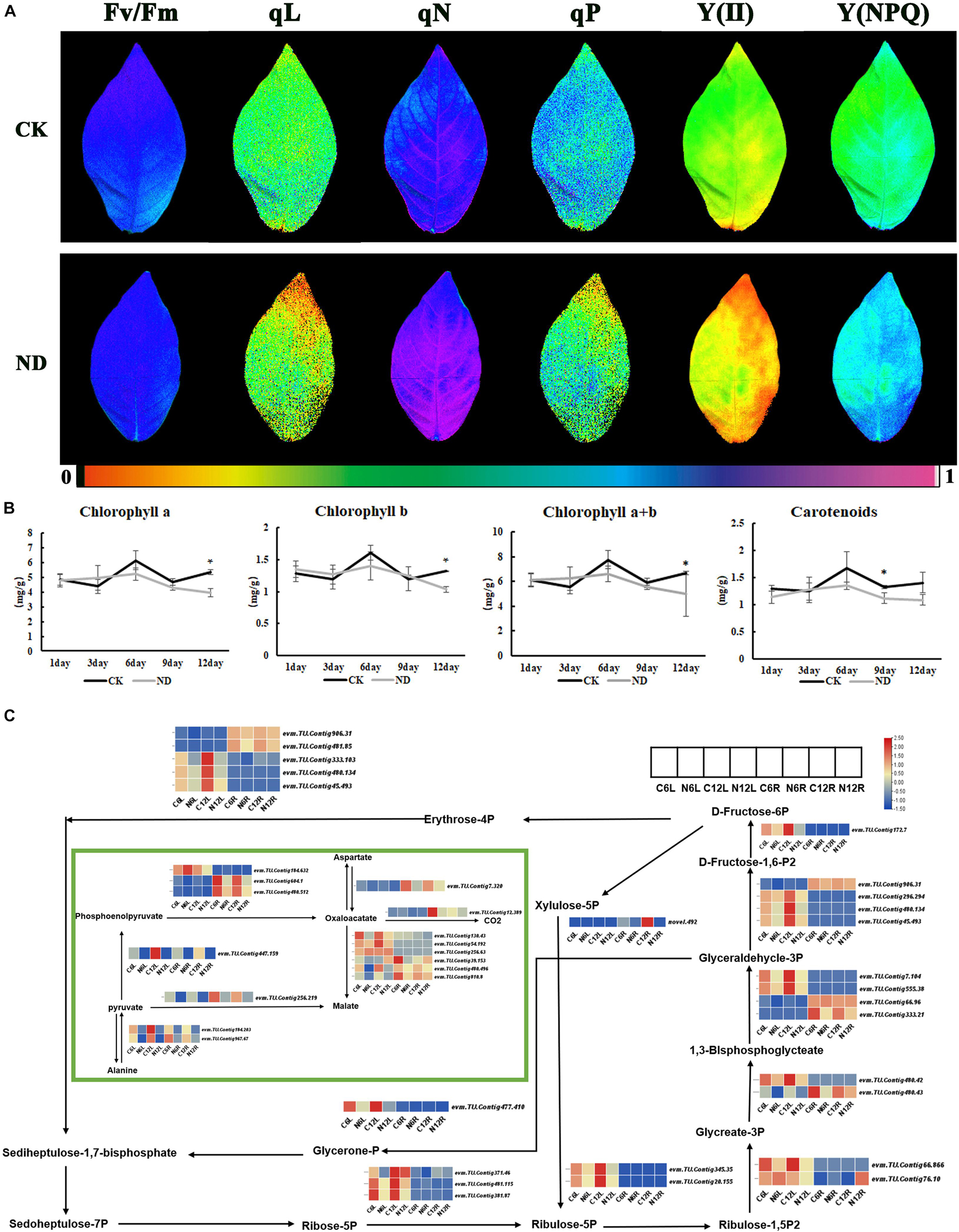
Figure 5. Response of ND to photosynthesis in N. cadamba at the physiological and molecular level. (A) The chlorophyll fluorescence imaging is shown in Fv/Fm, qL, qN, qP, Y(II), and Y(NPQ), which were examined at the 12th day. (B) The content of Chlorophyll a, Chlorophyll b, Chlorophyll a + b, and Carotenoids at 1, 3, 6, 9, and 12 days was measured. Data are means ± SD (n = 3). Differences between the mean values of CK and ND were compared using student’s t-test (* denote significant differences at p < 0.05). (C) RNA-sequencing (RNA-seq) data showed the DEGs of carbon fixation in photosynthesis pathway (KO00710). Heatmap of columns and rows represent samples and genes, respectively. Chlorophyll fluorescence parameters were listed in Supplementary Table 2.
In the transcriptome DEGs data, the carbon fixation in photosynthetic organisms under ND stress was enriched with a high expression in leaves such as evm.TU.Contig 333.103, evm.TU.Contig 480.134, evm.TU.Contig 45.493, and evm.TU.Contig 477.410. At the same time, there were a few genes that were preferentially expressed in roots, such as evm.TU.Contig 604.1, evm.TU.Contig 480.512, and evm.TU.Contig 256.219. As expected, all these gene expressions were downregulated in ND comparing with CK (Figure 5C), suggesting that ND does affect photosynthesis.
Nitrate Reductase, Nitrite Reductase, Glutamine Synthetase, Nitrogen Content, and Nitrogen Metabolism Response to Nitrogen Deficiency Stress
To determine the responses of plants to ND, some important parameters participating in N metabolism, such as GS, NR, NiR, N content, and protein content, were measured (Figure 6A). NR and NiR catalyzed the reduction of nitrate to nitrite and nitrite to ammonia, respectively. So, NR is the first rate-limiting enzyme in the assimilation of nitrate into plants. Under ND stress, the activity of the NR kept falling in roots till 12 days while in leaves it significantly increased at the 6 day, implying that NR activities might have been compensated in leaves (Figure 6A). Further, evm.TU.Contig 201.33, which encodes NiR and was significantly downregulated under ND conditions at the 6th and 12th day (Figure 6B). GS and GOGAT can convert glutamate and ammonia to glutamine. In our measurement, the activity of the GS was significantly reduced at the 6th and 12th day in roots while in leaves it increased initially at the 6th day and then decreased at the 12th day (Figure 6B). The transcriptome analysis showed that the expressions of evm.TU.Contig 45.180 and evm.TU.Contig 298.77, both encoding GS, were downregulated and consistent with the reduced GS activity phenotype. Meanwhile, evm.TU.Contig 471.255, evm.TU.Contig 96.644, and evm.TU.Contig 96.645, encoding GOGAT, were significantly downregulated in roots at both the 6th and 12th day, and evm.TU.Contig 4.19 was significantly downregulated in leaves at the 12th day (Figure 6B).
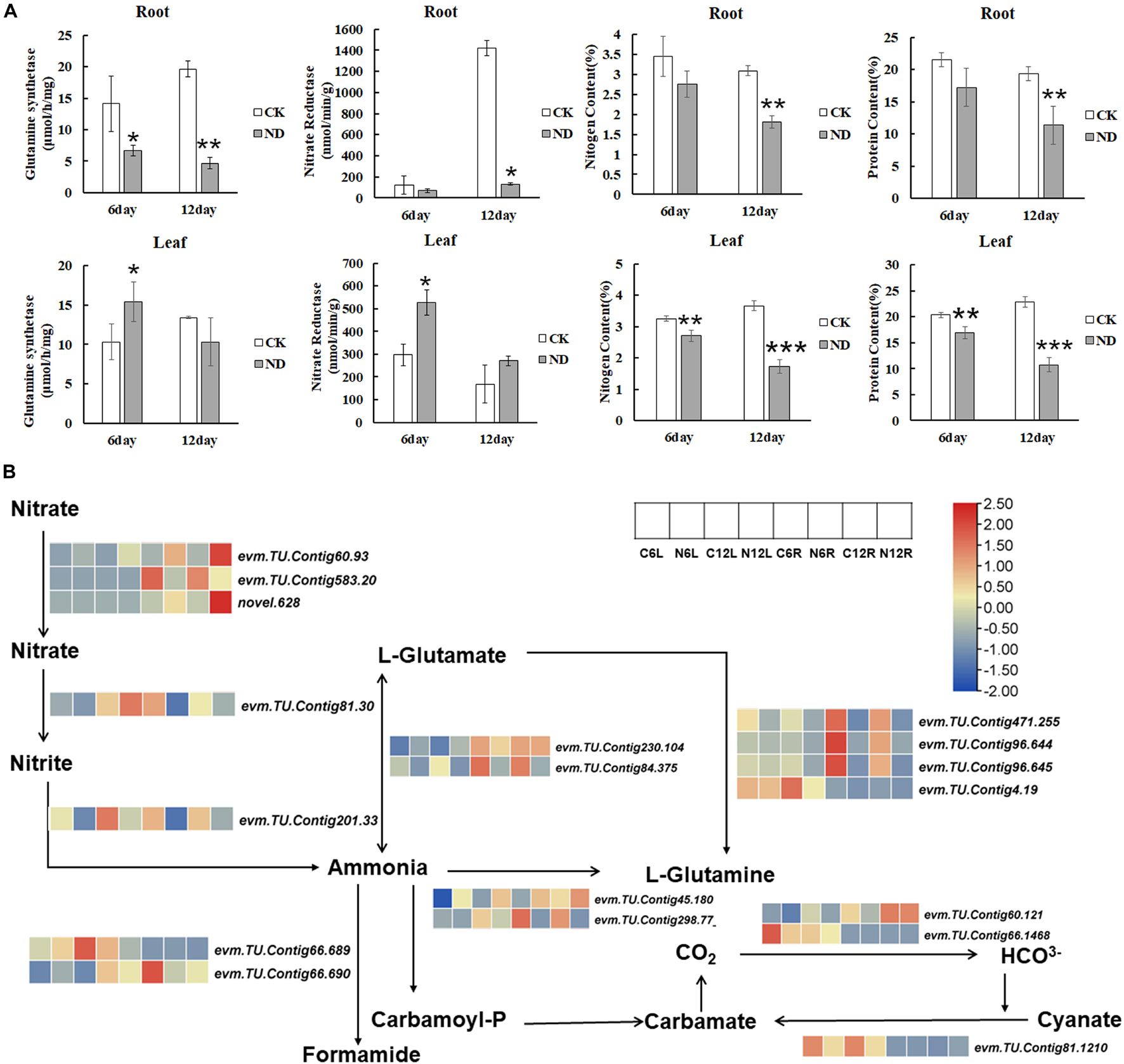
Figure 6. Nitrogen (N) metabolism in response to ND in N. cadamba. (A) The changes of the N. cadamba under CK and ND treatments in glutamine synthetase (GS), nitrate reductase (NR), N content, and protein content. Data are means ± SD (n = 3). Differences between the mean values of CK and ND were compared using student’s t-test (*, **, *** denote significant differences at p < 0.05, p < 0.01, and p < 0.001). (B) Transcriptome data showing the expression of genes related to N metabolism in N. cadamba after ND. A heatmap of columns and rows represent samples and genes, respectively.
As ND stress inevitably affects both the N and protein content of plants, these two components are measured. Compared to CK, the samples under ND showed a reduction of 16.61% in leaves and 19.94% in roots in the N content at the 6th day. At the 12th day, a reduction of 41.28% in roots and a reduction of 53% in leaves were observed. Because protein content was converted from N content, the changes of these two indicators were consistent.
Relative Transporters and Transcription Factors Were Mainly Upregulated
Transporters are important factors related to N absorption. The DEGs identified showed that the NRTs, including NRT1 (NPF), NRT2, CLC and SLAC1/SLAH, and AMT were upregulated in both roots and leaves under ND stress (Figure 7A). For example, evm.TU.Contig 60.93, evm.TU.Contig 583.20, and novel.628, all encoding NRT2 genes, were upregulated under ND conditions (Figure 6B). At the same time, we also identified a number of amino acid transporters, such as AAP7, AAP6, AAP3, ANTL1, and ANTL2, which were mostly upregulated (Supplementary Figure 3A). GABA transporters, such as GAT1 and GAT2, were also upregulated in response to ND stress in N. cadamba. In addition, auxin transporter-like protein (LAX), transmembrane AAP, vacuolar amino acid transporter 1, lysine histidine transporter (LHT), and vesicular inhibitory amino acid transporter (VIAAT) were upregulated.
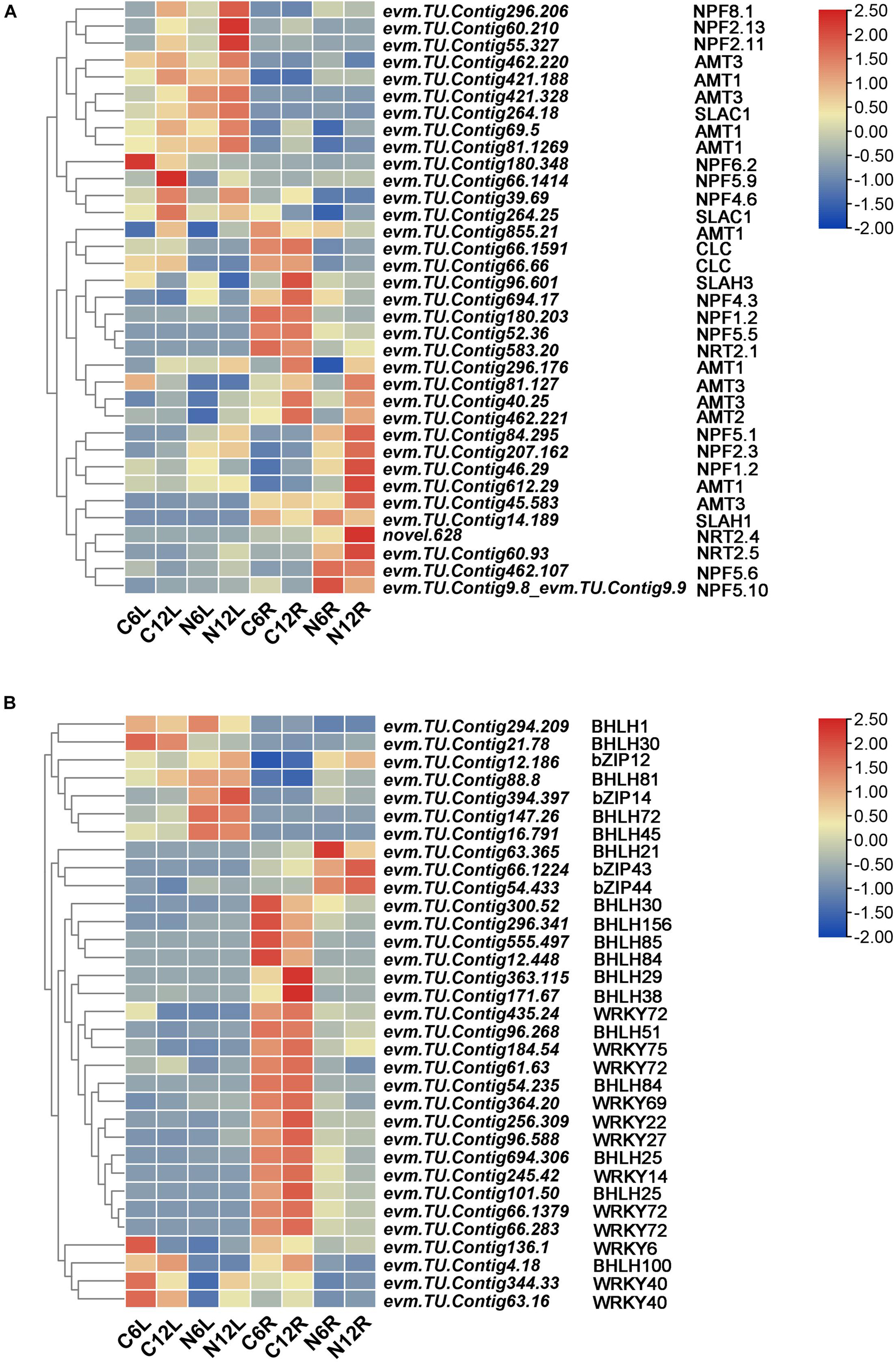
Figure 7. Nitrogen deficiency stress induces the expression of related transporter proteins and transcription factors in the leaf and root. (A) A heatmap of ND stress-induced expression of related transcription factors. (B) A Heatmap of NRTs and ammonium transporters (AMT) expression induced by ND stress. The threshold of FDR < 0.05, | log2 (FoldChange)| > 1 was used to screen the changes of DEGs between CK and ND in treatments. A heatmap of columns and rows represent samples and genes, respectively. Sample and DEGs names were displayed below the heatmap. The color bar is the scale for the expression levels of each gene.
To investigate changes in transcription factors upon ND stress, DEGs of transcription factors (BHLH, bZIP, MYB, and WRKY, etc.) at different tissues and time points under ND stress were identified. BHLHs, bZIPs, and WRKYs with 2-fold differences under ND were selected (Figure 7B). BHLH1, BHLH30, bZIP12, bZIP14, bZIP43, bZIP44, BHLH81, BHLH72, BHLH45, and BHLH21 were upregulated, while the members of WRKY family were mostly downregulated. Additionally, transcription factors, including NLP7, NLP6, TCP20, TGA1, ERF/RAP2, and ERF72, were also responded to ND stress (Supplementary Figure 3B).
Nitrogen Deficiency Stress Stimulates Hormone Gene Expression
Several plant hormones, such as salicylic acid (SA) and abscisic acid (ABA), have been reported to be associated with the metabolism and accumulation of N (Vega et al., 2019). In this study, DEGs (Supplementary Table 4) associated with hormones were identified, including nine auxin-related genes and five of which belong to the PIN family. evm.TU.Contig 600.155 (PIN2) and evm.TU.Contig 16.409 (PIN2) were downregulated in the root. Novel.433 (PIN5) and novel.551 (PIN5) were both upregulated in roots. In addition, genes related to gibberellin, cytokinin, ABA, and SA were also differentially expressed (Supplementary Table 4). This suggests that these hormones were engaged in the response of N. cadamba plants to ND.
The Phenylpropanoid Biosynthetic Pathway Changes in Nitrogen Deficiency Stress
Stem sections at the 6th and 12th day revealed (Supplementary Figure 2A) an increase in lignification and cell wall thickening from the top to bottom of stem. In our transcriptomic data, KEGG pathway revealed an enrichment in “Phenylpropanoid biosynthesis” in roots. Further analysis of the genes in this pathway exhibited that evm.TU.Contig 14.317 [4-coumarate-CoA ligase (4CL)], evm.TU.Contig 296.60 [cinnamate 4-hydroxylase (C4H)], evm.TU.Contig 14.271 [cinnamyl alcohol dehydrogenase (CAD)], novel.2274 [cinnamoyl-CoA reductase (CCR)], evm.TU.Contig 149.105 [caffeic acid O-methyltransferase (COMT)], evm.TU.Contig 45.482 [ferulate 5-hydroxylase (F5H)], evm.TU.Contig 766.86 [phenylalanine ammonia-lyase (PAL)], and POD protein genes were upregulated (Supplementary Figure 2B).
The Response of Phosphorus- and Calcium-Related Genes to Nitrogen Deficiency in Neolamarckia cadamba
Nitrogen and phosphorus are the two mineral nutrients that are highly in demand by plants. N/P supply ratios affect N/P uptake by activating the phosphorus signaling pathway (Hu et al., 2019). The SPX domain-containing protein (SPX) gene, which regulates the cellular response to phosphate starvation, was significantly downregulated in leaves, but in roots, more SPX genes were upregulated more than downregulated (Supplementary Table 5). The majority of the phosphate transporter protein-related genes were downregulated.
Furthermore, Ca2+, a ubiquitous second messenger in plants, also plays a role in the nitrate response (Riveras et al., 2015). There were another expression trends in calcium-related genes. cyclic nucleotide-gated ion channel (CNGC) a cyclic nucleotide-gated channel with calcium channel activity, was downregulated in roots, just like the CPK and CBL-interacting protein kinase, while the CBL-interacting serine-/threonine-protein kinase was upregulated in leaves and roots.
Discussion
Nitrogen is one of the most abundant elements and is required for the biosynthesis of amino acids, proteins, and enzymes in plant cells (Ganie et al., 2017). Neolamarckia cadamba is a fast-growing species, and its rapid growth must be dependent on an adequate supply of N. Therefore, ND certainly affects the rapid growth of N. cadamba, but previous studies have not elucidated the underlying mechanisms. Therefore, we combined the relative physiological parameters and transcriptome analysis to gain insights into the growth of N. cadamba on ND stress. The present study showed that the growth of N. cadamba seedlings was limited under ND. A reduction in fresh weight, dry weight, plant height, and root system (surface area, length, and volume) was recorded at the 12th day of ND treatment compared to CK (Figure 1), which was consistent with the Eucalyptus plants grown under low N conditions (Nguyen et al., 2003), indicating that N plays a critical important role in woody plant growth.
Photosynthetic capacity is an important indicator of plant growth (Blankenship, 2002; Zhang et al., 2014). In our study, although the maximal quantum yield of PSII photochemistry (Fv/Fm) did not change significantly between the ND and CK conditions, the photochemical quenching (qP) of the leaves of N. cadamba seedlings under ND stress was significantly lower than those of CK, and the actual photochemical efficiency of leaf PSII [Y(II)] and qL decreased vastly in ND, implying that the photosynthetic efficiency of N. cadamba leaves decreased in ND. Transcriptome analysis showed that the KEGG enrichment pathway for “porphyrin and chlorophyll metabolism” was downregulated in leaves after 6 days of ND stress, which is consistent with the downregulation of chlorophyll synthesis and carbon fixation in the chloroplast after 6 days of ND stress (Figure 5C). Although photosynthetic pathway under ND decreased at the 6th day, but the phenotype of yellowing leaf was not observed till the 12th day, suggesting a lag between the transcriptional changes and the phenotypic transition (Figure 1A). Meanwhile, the photosynthetic genes, such as evm.TU.Contig 45.493 [fructose-bisphosphate aldolase 2 (FBA2)], evm.TU.Contig 345.35 (PRK), evm.TU.Contig 172.7 (FBPase), and evm.TU.Contig 604.1 (PEPC), were downregulated in leaves under ND (Figure 5C), indicating that downregulation of photosynthesis-related genes under ND causes physiological yellowing of leaves. Furthermore, the reduction of dry weight in ND treatment could be related to photosynthesis inhibition, since less photosynthetic products were accumulated.
Transporters play critical roles in the first steps of N assimilation in plants. According to the transcriptome analysis, both NRT and AMT were upregulated to maintain N balance against external ND stress, which may indicate that the plant requires a higher expression of the N transporters to promote nitrate and ammonium accumulation. The activities of critical enzymes of N metabolism, such as NR, NiR, GS, and GOGAT, indirectly reflect the rate of the N assimilation and accumulation in plants (Zhang and Chu, 2020). Evm.TU.Contig 81.30 (NR2), which can convert the nitrate to nitrite, was downregulated in roots compared to the CK (Figure 6B), consistent with the NR activity reduction in roots. The expression of N metabolism-related genes was downregulated in plants, implying that the scale of N assimilation was reduced, ultimately leading to a decrease in N and protein content in plants (Figure 6A). Under N-limiting conditions, Poplar induces transcripts of many N-transporter genes in their roots, allowing them to adapt to soil ND by reducing N assimilation (Luo et al., 2015; Meng et al., 2018). This also explained why the root biomass did not change significantly at the 6th day but started to decline at the 12th day. Furthermore, amino acid transporters, including AAP3, GAT1, and LAX5, were upregulated in response to ND stress in N. cadamba (Supplementary Figure 3A). Amino acid transporters’ response to ND has been previously identified in tea plants. The gene expression of these transporters increased under ND conditions suggesting that they occur in various organs (Li et al., 2020). The effects of ND on enzyme activity and amino acid transporters described above will ultimately reduce the N and protein content of N. cadamba (Figure 6A). Likewise, the reduction in N content is consistent with Eucalypts and Poplar at low N (Nguyen et al., 2003; Song et al., 2019).
Hormonal signaling has also been linked to N metabolism (Ristova et al., 2016). Here, the KEGG pathway “plant hormone signal transduction” was upregulated at the 6th and 12th day in the leaves of N. cadamba after ND stress, indicating that the hormone compensated to ND stress. We identified genes with | log2foldchange| > 1 (Supplementary Table 4) and found five hormones that responded to ND stress. The expression of SA-related gene (SABP2) was downregulated. SA was reported to reduce the nitrate accumulation caused by N starvation (Yendrek et al., 2010; Conesa et al., 2020). ABA and auxin are thought to be signaling molecules that bind to NO3– and regulate root growth and development (Signora et al., 2001). ABAH and PYL, which are associated with ABA, were upregulated. ARF and PIN5 both related to auxin were upregulated. In addition, G2OX2 and CKX3 were downregulated, suggesting that the gibberellin and cytokinin signaling pathways were also affected by ND stress (Supplementary Table 4).
Thinner fibrous walls were observed in the secondary xylem of hybrid poplars (Populus trichocarpa × deltoides H11-11) under high N conditions relative to plants with sufficient N (Plavcova et al., 2013). Our results showed an increase of lignification and thickening of the cell walls in the stem (Supplementary Figure 2). Root transcriptome data also revealed that most genes in the phenylpropanoid pathway were upregulated. Enzymes involved in phenylpropanoid metabolism, such as 4-coumaric acid-CoA ligase (4CL), CCR, COMT, and CAD, were increased under N-limited conditions (Scheible et al., 2004). ROS can stimulate lignification and induce SOD and POD activities. In our experiments, ND treatment increased both SOD and POD activities in roots and leaves (Figure 2), which is consistent with the fact that ND stress promotes a significant increase in SOD and POD activities in cucumber leaves (Yang et al., 2019). Therefore, ND stress might stimulate lignification in plants but an exact mechanism is still not clear.
A previous study reported the main transcription factors that participate in the regulation of nitrate transport were Nodule inception-like protein 7 (NLP7), NLP6, TGA1, TGA4, ANR1, and bZIP1, etc. (O’Brien et al., 2016). Under N starvation, the expression of BHLH, a transcription factor, increases and mediates anthocyanin accumulation (Lea et al., 2007). In our study, the expression of transcription factors, such as BHLH, bZIP, WRKY, and NLP7, were induced (Figure 7B), while WRKY40, WRKY72, WRKY22, WRKY27, and WRKY14 were downregulated. Meanwhile, NLP7, NLP6, and TGA1 were upregulated under ND condition in N. cadamba (Supplementary Figure 3B).
We discovered phosphorus- and calcium-related genes in our data (Supplementary Table 5), the SPX genes that were upregulated are more than those downregulated in roots at the 12th day, indicating that NLP3 cannot responsive to nitrate gene expression. Furthermore, PHR2 was found to be upregulated, implying that phosphate-related genes work to alleviate the nutritional suppression caused by ND. Furthermore, CNGC and CPK were downregulated in roots. Under ND, CNGC15 and NRT1.1, which form a complex on the cytoplasmic membrane, lacked a calcium channel activity and were unable to encode a calcium channel for the nitrate nutrient response (Wang et al., 2021). It was proposed that ND caused dwarfism in N. cadamba and hampered its rapid growth, most likely due to the combined effect of Ca- and P-related genes (Supplementary Table 5).
Based on our results, we hypothesize the following pattern of the response to ND stress in N. cadamba (Figure 8). To compensate for ND stress, roots take up NH4+ via AMT and NO3– via NRT from the soil, whose expression was upregulated. However, due to limited substrate availability, i.e., NH4+ and NO3–, the expression of the internal genes encoding the N assimilating enzymes (NR and GS) was decreased, resulting in a decrease in amino acid and protein biosynthesis. Meanwhile, under low N conditions, SPX4 forms a complex with NLP3/PHR2, preventing NLP3 and PHR2 from entering the nucleus and thereby inhibiting the expression of N and phosphorus-responsive genes in N. cadamba. The expression of CNGC15 was upregulated to promote Ca2+ uptake. Therefore, these results suggested that N assimilation was linked with calcium and phosphorus absorption in N. cadamba. To deal with N stress, N. cadamba upregulated the expression of genes related to the phenylpropanoid biosynthetic pathway (including PAL, C4H, 4CL, and CCR) in the stem and eventually increased its lignification. Decreased expression of glutamate and chlorophyll biosynthesis genes results in consecutive downregulation of the genes involved in the Calvin cycle, such as FBA, phosphoglycerate kinase (PGK), and ribulose bisphosphate carboxylase small chain (RBCS1A), resulting in the inhibition of leaf photosynthesis and thus in a decrease of N. cadamba fresh weights. Our research provides a foundation to further study the physiological consequences of N. cadamba growth under nutrient deficiency.
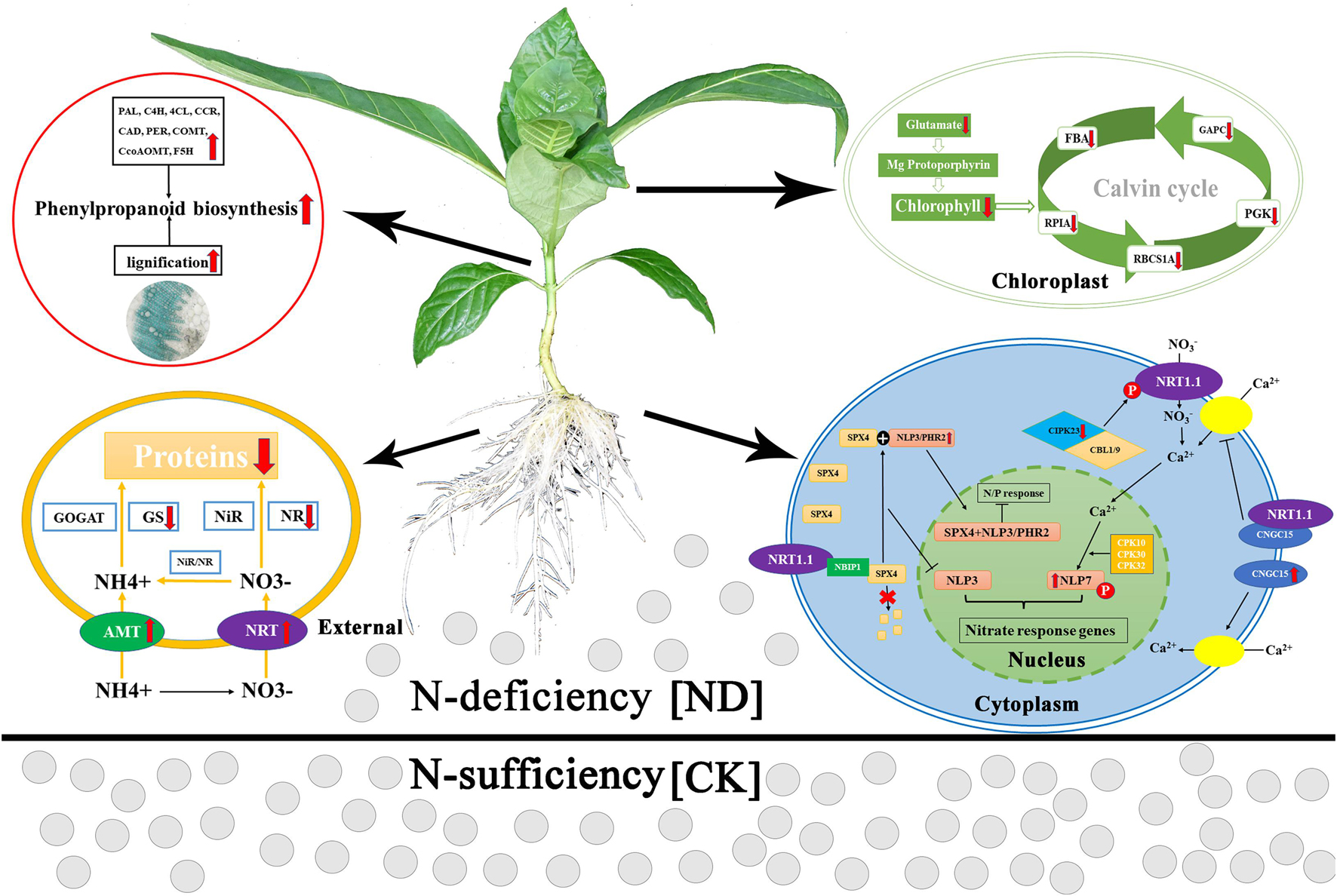
Figure 8. A hypothetical model of the effects of ND on the physiological and molecular mechanisms of N. cadamba. Phenotypes and transcriptome changes in roots, stems, and leaves treated with ND are indicated by the red arrows. Red arrows indicate an increase or a decrease in physiological indicators or gene expression. PAL, phenylalanine ammonia-lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate-CoA ligase; CCR, cinnamoyl-CoA reductase; CAD, cinnamyl alcohol dehydrogenase; COMT, caffeic acid O-methyltransferase; CCoAOMT, caffeoyl-CoA O-methyltransferase; F5H, ferulate 5-hydroxylase; NR, nitrate reductase; NiR, nitrite reductase; GS, glutamine synthetase; GOGAT, glutamate synthase; AMT, ammonium transporter; NRT, nitrate transporter; FBA, fructose-bisphosphate aldolase; GAPC, glyceraldehyde-3-phosphate dehydrogenase; PGK, phosphoglycerate kinase; RBCS1A, ribulose bisphosphate carboxylase small chain; RPIA, ribose 5-phosphate isomerase A; SPX4, SPX domain-containing proteins 4; NLP7/3, nodule inception-like protein 7/3; PHR2, phosphate starvation response 2; CBL1/9, calcineurin B-like protein; CIPK23, CBL-interacting protein kinase 23; CNGC15, cyclic nucleotide-gated ion channel 15.
Conclusion
The present study reveals that ND inhibited the growth of N. cadamba. N and protein content in roots and leaves were both reduced. Chlorophyll and carotenoid contents were reduced as were chlorophyll fluorescence parameters. Genes encoding transcription factors, transport proteins, and oxidative stress-related proteins were significantly upregulated, suggesting that the nutrient transporting and the hormonal regulation may play an important role for N. cadamba in response to N starvation.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: RNA-seq data were submitted to (http://bigd.big.ac.cn/gsa/with) submission number: CRA004153.
Author Contributions
A-MW and HL conceived the project. LLi, YZ, LLu, NY, YL, and MQ conducted the experiments and analyzed the data under the supervision of A-MW and HL. LLu, YZ, and MQ wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Key Project of Guangzhou Science and Technology Plan (201904020014), the National Natural Science Foundation of China (Grant Nos. 31870653 and 31670670).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank YL, Baojia Dai, and YZ from South China Agricultural University for excellent technical assistance and discussions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.747121/full#supplementary-material
References
Alvarez, J. M., Schinke, A. L., Brooks, M. D., Pasquino, A., Leonelli, L., Varala, K., et al. (2020). Transient genome-wide interactions of the master transcription factor nlp7 initiate a rapid nitrogen-response cascade. Nat. Commun. 11:1157. doi: 10.1038/s41467-020-14979-6
Beiqing, K., Xianhai, Z., Chun, Z., Wei, Z., Junli, R., Ebert, B., et al. (2016). Role of udp-glucuronic acid decarboxylase in xylan biosynthesis in arabidopsis. Mol. Plant 9, 1119–1131.
Blankenship, R. E. (2002). Molecular Mechanisms of Photosynthesis. Hoboken, NJ: Blackwell Science Ltd.
Castaings, L., Camargo, A., Pocholle, D., Gaudon, V., Texier, Y., Boutet-Mercey, S., et al. (2009). The nodule inception-like protein 7 modulates nitrate sensing and metabolism in arabidopsis. Plant J. 57, 426–435. doi: 10.1111/j.1365-313X.2008.03695.x
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). Tbtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, Q., Liu, Z., Wang, B., Wang, X., Lai, J., Tian, F., et al. (2015). Transcriptome sequencing reveals the roles of transcription factors in modulating genotype by nitrogen interaction in maize. Plant Cell Rep. 34, 1761–1771. doi: 10.1007/s00299-015-1822-9
Chopin, F., Orsel, M., Dorbe, M. F., Chardon, F., Truong, H. N., Miller, A. J., et al. (2007). The arabidopsis atnrt2.7 nitrate transporter controls nitrate content in seeds. Plant Cell 19, 1590–1602. doi: 10.1105/tpc.107.050542
Chu, S. S., Tong, X., Wang, W. R., Mo, Q. F., Chen, X. Y., Zeng, S. C., et al. (2017). Effects of sewage sludge compost on the growth and element uptake of neolamarckia cadamba seedlings. Ying Yong Sheng Tai Xue Bao 28, 1550–1556. doi: 10.13287/j.1001-9332.201705.036
Conesa, C. M., Saez, A., Navarro-Neila, S., de Lorenzo, L., Hunt, A. G., Sepúlveda, E. B., et al. (2020). Alternative polyadenylation and salicylic acid modulate root responses to low nitrogen availability. Plants 9:251. doi: 10.3390/plants9020251
Coruzzi, G., and Bush, D. R. (2001). Nitrogen and carbon nutrient and metabolite signaling in plants. Plant Physiol. 125, 61–64. doi: 10.1104/pp.125.1.61
Dai, B., Chen, C., Liu, Y., Liu, L., Qaseem, M. F., Wang, J., et al. (2020). Physiological, biochemical, and transcriptomic responses of neolamarckia cadamba to aluminum stress. Int. J. Mol. Sci. 21:9624. doi: 10.3390/ijms21249624
Feng, H., Yan, M., Fan, X., Li, B., Shen, Q., Miller, A. J., et al. (2011). Spatial expression and regulation of rice high-affinity nitrate transporters by nitrogen and carbon status. J. Exp. Bot. 62, 2319–2332. doi: 10.1093/jxb/erq403
Fichtner, F., Dissanayake, I. M., Lacombe, B., and Barbier, F. (2021). Sugar and nitrate sensing: a multi-billion-year story. Trends Plant Sci. 26, 352–374. doi: 10.1016/j.tplants.2020.11.006
Frank, H. A., Das, S. K., Bautista, J. A., Bruce, D., Vasil’Ev, S., Crimi, M., et al. (2001). Photochemical behavior of xanthophylls in the recombinant photosystem ii antenna complex, cp26. Biochemistry 40, 1220–1225. doi: 10.1021/bi001160q
Ganie, A. H., Ahmad, A., Yousuf, P. Y., Pandey, R., Ahmad, S., Aref, I. M., et al. (2017). Nitrogen-regulated changes in total amino acid profile of maize genotypes having contrasting response to nitrogen deficit. Protoplasma 254, 2143–2153.
Geiger, D., Scherzer, S., Mumm, P., Stange, A., Marten, I., Bauer, H., et al. (2009). Activity of guard cell anion channel slac1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc. Natl. Acad. Sci. U.S.A. 106, 21425–21430. doi: 10.1073/pnas.0912021106
Gutierrez, R. A. (2012). Systems biology for enhanced plant nitrogen nutrition. Science 336, 1673–1675. doi: 10.1126/science.1217620
He, X., Ma, H., Zhao, X., Nie, S., Li, Y., Zhang, Z., et al. (2016). Comparative rna-seq analysis reveals that regulatory network of maize root development controls the expression of genes in response to n stress. PLoS One 11:e151697. doi: 10.1371/journal.pone.0151697
Hong, L., Jing, L., Yalong, H., Shaojun, L., Zongsuo, L., Changhui, P., et al. (2013). Changes in carbon, nutrients and stoichiometric relations under different soil depths, plant tissues and ages in black locust plantations. Acta Physiol. Plant. 35, 2951–2964. doi: 10.1007/s11738-013-1326-6
Hu, B., Jiang, Z., Wang, W., Qiu, Y., Zhang, Z., Liu, Y., et al. (2019). Nitrate-nrt1.1b-spx4 cascade integrates nitrogen and phosphorus signalling networks in plants. Nat. Plants 5, 401–413. doi: 10.1038/s41477-019-0384-1
Jiang, C., Gao, H., Zou, Q., Jiang, G., and Li, L. (2006). Leaf orientation, photorespiration and xanthophyll cycle protect young soybean leaves against high irradiance in field. Environ. Exp. Bot. 55, 87–96. doi: 10.1016/j.envexpbot.2004.10.003
Joshi, V., Joshi, M., and Penalosa, A. (2020). Comparative analysis of tissue-specific transcriptomic responses to nitrogen stress in spinach (spinacia oleracea). PLoS One 15:e232011. doi: 10.1371/journal.pone.0232011
Konishi, M., and Yanagisawa, S. (2013). Arabidopsis nin-like transcription factors have a central role in nitrate signalling. Nat. Commun. 4:1617. doi: 10.1038/ncomms2621
Kusano, M., Fukushima, A., Redestig, H., and Saito, K. (2011). Metabolomic approaches toward understanding nitrogen metabolism in plants. J. Exp. Bot. 62, 1439–1453. doi: 10.1093/jxb/erq417
Lam, H. M., Coschigano, K. T., Oliveira, I. C., Melo-Oliveira, R., and Coruzzi, G. M. (1996). The molecular-genetics of nitrogen assimilation into amino acids in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 569–593. doi: 10.1146/annurev.arplant.47.1.569
Lea, U. S., Slimestad, R., Smedvig, P., and Lillo, C. (2007). Nitrogen deficiency enhances expression of specific myb and bhlh transcription factors and accumulation of end products in the flavonoid pathway. Planta 225, 1245–1253. doi: 10.1007/s00425-006-0414-x
Li, F., Li, H., Dong, C., Yang, T., Zhang, S., Bao, S., et al. (2020). Theanine transporters are involved in nitrogen deficiency response in tea plant (camellia sinensis l.). Plant Signal. Behav. 15:1728109. doi: 10.1080/15592324.2020.1728109
Liu, X., Wen, J., Chen, W., Du, H., and Amsler, C. (2019). Physiological effects of nitrogen deficiency and recovery on the macroalga gracilariopsis lemaneiformis (rhodophyta). J. Phycol. 55, 830–839. doi: 10.1111/jpy.12862
Liu, Y., Yin, Q., Dai, B., Wang, K., Lu, L., Qaseem, M. F., et al. (2021). The key physiology and molecular responses to potassium deficiency in neolamarckia cadamba. Ind. Crops Prod. 162:113260. doi: 10.1016/j.indcrop.2021.113260
Luo, J., Zhou, J., Li, H., Shi, W., Polle, A., Lu, M., et al. (2015). Global poplar root and leaf transcriptomes reveal links between growth and stress responses under nitrogen starvation and excess. Tree Physiol. 35, 1283–1302.
Maierhofer, T., Lind, C., Huttl, S., Scherzer, S., Papenfuss, M., Simon, J., et al. (2014). A single-pore residue renders the arabidopsis root anion channel slah2 highly nitrate selective. Plant Cell 26, 2554–2567. doi: 10.1105/tpc.114.125849
Mamashita, T., Larocque, G. R., DesRochers, A., Beaulieu, J., Thomas, B. R., Mosseler, A., et al. (2015). Short-term growth and morphological responses to nitrogen availability and plant density in hybrid poplars and willows. Biomass Bioenergy 81, 88–97. doi: 10.1016/j.biombioe.2015.06.003
Marchive, C., Roudier, F., Castaings, L., Brehaut, V., Blondet, E., Colot, V., et al. (2013). Nuclear retention of the transcription factor nlp7 orchestrates the early response to nitrate in plants. Nat. Commun. 4:1713. doi: 10.1038/ncomms2650
Martin, R. E., Asner, G. P., and Sack, L. (2007). Genetic variation in leaf pigment, optical and photosynthetic function among diverse phenotypes of metrosideros polymorpha grown in a common garden. Oecologia 151, 387–400. doi: 10.1007/s00442-006-0604-z
Martin, T., Oswald, O., and Graham, I. A. (2002). Arabidopsis seedling growth, storage lipid mobilization, and photosynthetic gene expression are regulated by carbon:nitrogen availability. Plant Physiol. 128, 472–481. doi: 10.1104/pp.010475
Masclaux-Daubresse, C., Daniel-Vedele, F., Dechorgnat, J., Chardon, F., Gaufichon, L., Suzuki, A., et al. (2010). Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann. Bot. 105, 1141–1157. doi: 10.1093/aob/mcq028
Meng, S., Wang, S., Quan, J., Su, W., Lian, C., Wang, D., et al. (2018). Distinct carbon and nitrogen metabolism of two contrasting poplar species in response to different n supply levels. Int. J. Mol. Sci. 19:2302.
Miller, A. J., Xu, G., and Fan, X. (2012). Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 63, 153–182. doi: 10.1146/annurev-arplant-042811-105532
Mu, X., Chen, Q., Chen, F., Yuan, L., and Mi, G. (2016). Within-leaf nitrogen allocation in adaptation to low nitrogen supply in maize during grain-filling stage. Front. Plant Sci. 7:699. doi: 10.3389/fpls.2016.00699
Nguyen, T. N., Kazuo, N., Pravat, K. M., Julian, T., and Kounosuke, F. (2003). Effect of nitrogen deficiency on biomass production, photosynthesis, carbon partitioning, and nitrogen nutrition status of melaleuca and eucalyptus species. Soil Sci. Plant Nutr. 49, 99–109.
O’Brien, J. A., Vega, A., Bouguyon, E., Krouk, G., Gojon, A., Coruzzi, G., et al. (2016). Nitrate transport, sensing, and responses in plants. Mol. Plant 9, 837–856. doi: 10.1016/j.molp.2016.05.004
Ouyang, K., Li, J., Zhao, X., Que, Q., Li, P., Huang, H., et al. (2016). Transcriptomic analysis of multipurpose timber yielding tree neolamarckia cadamba during xylogenesis using rna-seq. PLoS One 11:e159407. doi: 10.1371/journal.pone.0159407
Papadopoulos, A. N., and Pougioula, G. (2010). Mechanical behaviour of pine wood chemically modified with a homologous series of linear chain carboxylic acid anhydrides. Bioresour. Technol. 101, 6147–6150. doi: 10.1016/j.biortech.2010.02.079
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time rt-pcr. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Plavcova, L., Hacke, U. G., Almeida-Rodriguez, A. M., Li, E., and Douglas, C. J. (2013). Gene expression patterns underlying changes in xylem structure and function in response to increased nitrogen availability in hybrid poplar. Plant Cell Environ. 36, 186–199. doi: 10.1111/j.1365-3040.2012.02566.x
Qin, L., Walk, T. C., Han, P., Chen, L., Zhang, S., Li, Y., et al. (2019). Adaption of roots to nitrogen deficiency revealed by 3d quantification and proteomic analysis. Plant Physiol. 179, 329–347. doi: 10.1104/pp.18.00716
Rennenberg, H., Wildhagen, H., and Ehlting, B. (2010). Nitrogen nutrition of poplar trees. Plant Biol. 12, 275–291. doi: 10.1111/j.1438-8677.2009.00309.x
Ristova, D., Carre, C., Pervent, M., Medici, A., Kim, G. J., Scalia, D., et al. (2016). Combinatorial interaction network of transcriptomic and phenotypic responses to nitrogen and hormones in the arabidopsis thaliana root. Sci. Signal. 9:rs13. doi: 10.1126/scisignal.aaf2768
Riveras, E., Alvarez, J. M., Vidal, E. A., Oses, C., Vega, A., Gutiérrez, R. A., et al. (2015). The calcium ion is a second messenger in the nitrate signaling pathway of arabidopsis. Plant Physiol. 169, 1397–1404. doi: 10.1104/pp.15.00961
Robertson, G. P., and Vitousek, P. M. (2009). Nitrogen in agriculture:balancing the cost of an essential resource. Annu. Rev. Environ. Resourc. 34, 97–125. doi: 10.1146/annurev.environ.032108.105046
Scheible, W. R., Morcuende, R., Czechowski, T., Fritz, C., Osuna, D., Palacios-Rojas, N., et al. (2004). Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of arabidopsis in response to nitrogen. Plant Physiol. 136, 2483–2499. doi: 10.1104/pp.104.047019
Signora, L., De Smet, I., Foyer, C. H., and Zhang, H. (2001). Aba plays a central role in mediating the regulatory effects of nitrate on root branching in arabidopsis. Plant J. 28, 655–662. doi: 10.1046/j.1365-313x.2001.01185.x
Song, H., Cai, Z., Liao, J., Tang, D., and Zhang, S. (2019). Sexually differential gene expressions in poplar roots in response to nitrogen deficiency. Tree Physiol. 39, 1614–1629.
Stitt, M., and Krapp, A. (1999). The interaction between elevated carbon dioxide and nitrogen nutrition: the physiological and molecular background. Plant Cell Environ. 22, 583–621. doi: 10.1046/j.1365-3040.1999.00386.x
Vega, A., O’Brien, J. A., and Gutierrez, R. A. (2019). Nitrate and hormonal signaling crosstalk for plant growth and development. Curr. Opin. Plant Biol. 52, 155–163. doi: 10.1016/j.pbi.2019.10.001
Wang, X., Feng, C., Tian, L., Hou, C., Tian, W., Hu, B., et al. (2021). A transceptor-channel complex couples nitrate sensing to calcium signaling in arabidopsis. Mol. Plant 14, 774–786. doi: 10.1016/j.molp.2021.02.005
Wellburn, A. R. (1994). The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. Urban Fischer 144, 307–313. doi: 10.1016/S0176-1617(11)81192-2
Yang, Z., Wang, Z., Yang, C., Yang, Z., Li, H., Wu, Y., et al. (2019). Physiological responses and small rnas changes in maize under nitrogen deficiency and resupply. Genes Genomics 41, 1183–1194. doi: 10.1007/s13258-019-00848-0
Yao, Y., He, R. J., Xie, Q. L., Zhao, X. H., Deng, X. M., He, J. B., et al. (2017). Ethylene response factor 74 (erf74) plays an essential role in controlling a respiratory burst oxidase homolog d (rbohd)-dependent mechanism in response to different stresses in arabidopsis. New Phytol. 213, 1667–1681.
Yendrek, C. R., Lee, Y. C., Morris, V., Liang, Y., Pislariu, C. I., Burkart, G., et al. (2010). A putative transporter is essential for integrating nutrient and hormone signaling with lateral root growth and nodule development in medicago truncatula. Plant J. 62, 100–112. doi: 10.1111/j.1365-313X.2010.04134.x
Zhang, H., Liu, H., Niedzwiedzki, D. M., Prado, M., Jiang, J., Gross, M. L., et al. (2014). Molecular mechanism of photoactivation and structural location of the cyanobacterial orange carotenoid protein. Biochemistry 53, 13–19. doi: 10.1021/bi401539w
Zhang, J., Wang, Y., Zhao, Y., Zhang, Y., Zhang, J., Ma, H., et al. (2020). Transcriptome analysis reveals nitrogen deficiency induced alterations in leaf and root of three cultivars of potato (solanum tuberosum l.). PLoS One 15:e240662. doi: 10.1371/journal.pone.0240662
Zhang, Z., and Chu, C. (2020). Nitrogen-use divergence between indica and japonica rice: variation at nitrate assimilation. Mol. Plant 13, 6–7. doi: 10.1016/j.molp.2019.11.011
Zhao, W., Yang, X., Yu, H., Jiang, W., Sun, N., Liu, X., et al. (2015). Rna-seq-based transcriptome profiling of early nitrogen deficiency response in cucumber seedlings provides new insight into the putative nitrogen regulatory network. Plant Cell Physiol. 56, 455–467. doi: 10.1093/pcp/pcu172
Zhao, X., Ouyang, K., Gan, S., Zeng, W., Song, L., Zhao, S., et al. (2014). Biochemical and molecular changes associated with heteroxylan biosynthesis in neolamarckia cadamba (rubiaceae) during xylogenesis. Front. Plant Sci. 5:602. doi: 10.3389/fpls.2014.00602
Keywords: nitrogen deficiency, Neolamarckia cadamba, transcriptome analysis, physiology, stress response
Citation: Lu L, Zhang Y, Li L, Yi N, Liu Y, Qaseem MF, Li H and Wu A-M (2021) Physiological and Transcriptomic Responses to Nitrogen Deficiency in Neolamarckia cadamba. Front. Plant Sci. 12:747121. doi: 10.3389/fpls.2021.747121
Received: 25 July 2021; Accepted: 12 October 2021;
Published: 23 November 2021.
Edited by:
Marouane Baslam, Niigata University, JapanReviewed by:
Takuji Ohyama, Tokyo University of Agriculture, JapanMohammad-Reza Hajirezaei, Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), Germany
Copyright © 2021 Lu, Zhang, Li, Yi, Liu, Qaseem, Li and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huiling Li, bGlobEBzY2F1LmVkdS5jbg==; Ai-Min Wu, d3VhaW1pbkBzY2F1LmVkdS5jbg==
 Lu Lu1,2
Lu Lu1,2 Mirza Faisal Qaseem
Mirza Faisal Qaseem Huiling Li
Huiling Li Ai-Min Wu
Ai-Min Wu