- 1Triticeae Research Institute, Sichuan Agricultural University, Chengdu, China
- 2Crop Research Institute, Sichuan Academy of Agricultural Sciences (SAAS), Chengdu, China
- 3Australia-China Joint Centre for Wheat Improvement, Western Australian State Agricultural Biotechnology Centre, College of Science, Health, Engineering and Education, Murdoch University, Perth, WA, Australia
- 4Institute of Germplasm Resources and Biotechnology, Jiangsu Academy of Agricultural Sciences (JAAS), Nanjing, China
- 5Key Laboratory of Wheat Biology and Genetic Improvement on Southwestern China, Ministry of Agriculture, Chengdu, China
- 6Institute of Biotechnology and Nuclear Technology Research, Sichuan Academy of Agricultural Sciences (SAAS), Chengdu, China
Durum wheat is one of the important food and cash crops. The main goals in current breeding programs are improving its low yield potential, kernel characteristics, and lack of resistance or tolerance to some biotic and abiotic stresses. In this study, a nascent synthesized hexaploid wheat Lanmai/AT23 is used as the female parent in crosses with its AB genome donor Lanmai. A tetraploid line YL-443 with supernumerary spikelets and high resistance to stripe rust was selected out from the pentaploid F7 progeny. Somatic analysis using multicolor fluorescence in situ hybridization (mc-FISH) revealed that this line is a disomic substitution line with the 4B chromosome pair of Lanmai replaced by the 4D chromosome pair of Aegilops tauschii AT23. Comparing with Lanmai, YL-443 shows an increase in the number of spikelets and florets per spike by 36.3 and 75.9%, respectively. The stripe rust resistance gene Yr28 carried on the 4D chromosome was fully expressed in the tetraploid background. The present 4D(4B) disomic substitution line YL-443 was distinguished from the previously reported 4D(4B) lines with the 4D chromosomes from Chinese Spring (CS). Our study demonstrated that YL-443 can be used as elite germplasm for durum wheat breeding targeting high yield potential and stripe rust resistance. The Yr28-specific PCR marker and the 4D chromosome-specific KASP markers together with its unique features of pubescent leaf sheath and auricles can be utilized for assisting selection in breeding.
Introduction
Tetraploid durum wheat (2n = 28, AABB, Triticum turgidum L. ssp. durum) appears to have been derived from wild emmer wheat (T. turgidum ssp. dicoccoides) about 10,000 years ago in southeast Turkey, most likely through artificial domestication and continuous evolution (Özkan et al., 2011; Tidiane Sall et al., 2019). Tetraploid durum subsequently hybridized with diploid Aegilops tauschii Coss. (2n = 14, DD), resulting in the allohexaploid wheat (Triticum aestivum L.) (Kihara, 1944; McFadden and Sears, 1946). The addition of the D genome of Ae. tauschii into durum wheat improves the plasticity of hexaploid wheat, conferring a higher adaption capacity than durum wheat, including broader adaptability to different photoperiod and vernalization conditions, improved abiotic tolerance, and increased biotic resistance (Dubcovsky and Dvorak, 2007; Li et al., 2014, 2018). The allopolyploidy also gives hexaploid wheat greater potential to produce a variety of food products, such as bread, biscuits, cakes, pasta, noodles, and even brewing wine (Shewry, 2009; Faltermaier et al., 2014). Compared with bread wheat, durum wheat only accounts for 5% of the cultivated area and is principally grown in the Mediterranean region, Canada, United States, Mexico, India, Australia, etc. (Tidiane Sall et al., 2019; Xynias et al., 2020). Despite its lower planting area compared with hexaploid wheat, the global durum wheat production has been forecast up to 42.7 million tons (MT) in 2020–2021 (Agriculture Agri-Food Canada, 2021). About 75% of the global durum wheat production is contributed by the Mediterranean region (Othmeni et al., 2019), where crops are mainly grown under environmental (high temperature, drought, and salinity) and biotic (pests and diseases) stresses (Tidiane Sall et al., 2019). In China, durum wheat cultivation was first started in the 1970s using varieties introduced from European countries such as Italy (Zhao et al., 2002). It was only grown in China sporadically in the beginning. Since then, the pasta flavor is getting more and more popular particularly among young people, resulting in huge demand for durum wheat. A total of 1.71 and 1.97 MT of durum wheat were imported in 2015 and 2019, respectively (Zhiyan, 2020). The high level of consumption resulted in expanded domestic cultivation. As a consequence, domestic cultivars with high yield and quality are required in the farming sector, making durum wheat breeding an important task in China.
Durum wheat improvement mainly focuses on aspects of kernel hardness, protein content, dough elasticity and extensibility, gluten strength, and yellow pigment concentration (Magallanes-López et al., 2017), which play a key role in processing and end-use quality. Hybrids between hexaploid and durum wheat are readily produced (Padmanaban et al., 2017), and hence, researchers have proposed using hexaploid wheat as a genetic resource to improve durum wheat (Ma et al., 2013; Padmanaban et al., 2017; Mastrangelo and Cattivelli, 2021). To date, a substantial number of hexaploid wheat genes have been successfully introgressed into durum wheat through pentaploids produced by hybridization. The Glu-D1 locus encoding high molecular weight (HMW) glutenin (Morris et al., 2019; Sissons et al., 2019) was transferred from common wheat mediated by the Chinese Spring (CS) mutant line ph1b (Sears, 1977) and durum wheat mutant line ph1c (Giorgi, 1978) into 1A chromosome of the recipient durum wheat for improving baking quality (Ceoloni et al., 1995, 1996; Vitellozzi et al., 1997; Gennaro et al., 2012). The endosperm softness control gene Puroindoline (Pin-a and Pin-b) on 5DS was transferred by ph1b-mediated translocation and generated soft kernel durum wheat (Bhave and Morris, 2008; Boehm et al., 2017). The durum wheat cultivar Langdon (LDN) and hexaploid wheat landrace CS have been used to develop a set of D-chromosome disomic substitution lines for transferring desirable genes (Joppa and Williams, 1988). Morris et al. (2011) developed a soft durum wheat by using the LDN 5D(5B) substitution line. The introgression of a 4D-chromosome fragment from the LDN 4D(4B) substitution line increased the Al3+ tolerance of durum wheat (Han et al., 2014, 2016). Calderón et al. (2012) successfully developed durum wheat-Hordeum chilense introgression lines by crossing a CS 4Hch(4B) substitution line with a LDN 4D(4B) substitution line, which may contain Septoria tritici blotch resistance and salt tolerance gene. In order to transfer abiotic stress and disease resistance genes from Amblyopyrum muticum into durum wheat, Othmeni et al. (2019) utilized bread wheat/Am. muticum hybrids to cross with durum wheat, resulting in D-genome and/or T-genome segment introgression lines.
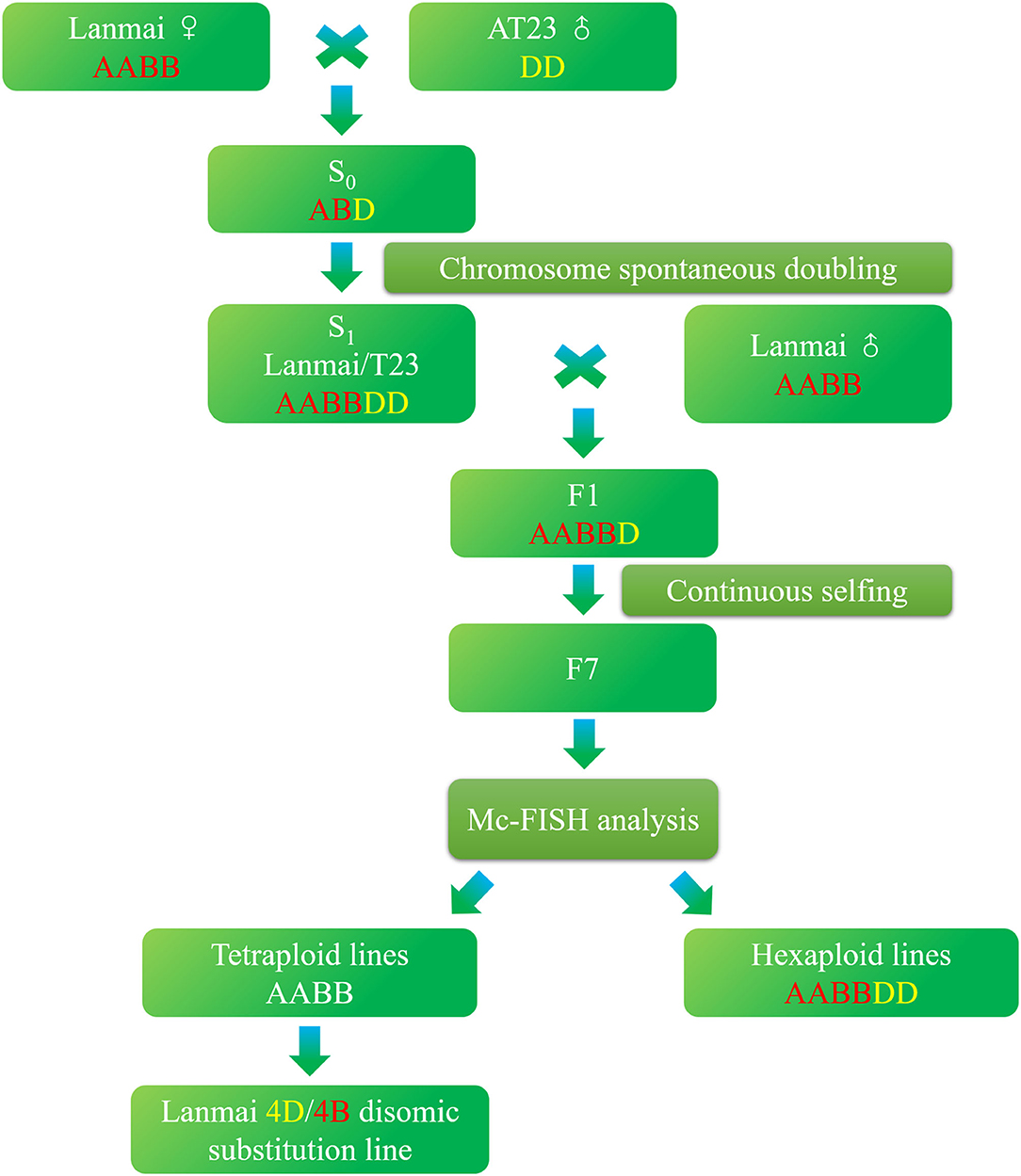
In this research, we first developed a nascent synthetic hexaploid wheat (SHW) Lanmai/AT23 line YL-724 using durum wheat Lanmai and Ae. tauschii accession AT23, which was subsequently crossed with Lanmai to produce a 4D(4B) disomic substitution line YL-443 after six consecutive generations of selfing (Figure 1). The developed line displayed supernumerary spikelets and high resistance to stripe rust. It can be used for durum wheat breeding to increase yield potential and disease resistance.
Materials and Methods
Production of Hybrids
The development of durum wheat/Ae. tauschii introgression lines began by crossing Lanmai (2n = 28, AABB) and Ae. tauschii accession AT23 (2n = 14, DD), followed by spontaneous chromosome doubling. Lanmai is a selected line from LDN (T. turgidum L. var. durum cv. Langdon) as 3–5 days early flowering. This variation may have been attributed to the spontaneous mutation of LDN or out-crossing of LDN with an unknown cultivar. The euploid SHW lines of Lanmai/AT23 with 42 chromosomes (AABBDD) were identified using multicolor fluorescence in situ hybridization (mc-FISH) assay. One of the euploid SHW lines, YL-724, was selected and used as female parent to cross with Lanmai. After crossing, six consecutive cycles of selfing were carried out until the F7 generation. Since Lanmai can promote the spontaneous doubling of chromosome of haplotype hybrids (ABD or ABDvNv) produced by crossing Lanmai with Ae. tauschii or Aegilops ventricosa (Hao et al., 2014; Zhang et al., 2020), the production of F1 hybrids did not require embryo rescue techniques or hormone treatment. During selfing, spikes of F2 hybrid plants were randomly selected and planted in rows. Individual plants of the F3 generation with desirable yield components, biotic resistance, and abiotic tolerance were selected for descent. All spikes in each generation were bagged. Finally, a total of 197 lines of the F7 generation were produced from the pentaploid-derived population, and the 4D(4B) disomic substitution line YL-443 was selected from the F7 generation using mc-FISH. Figure 1 shows the procedure followed to develop YL-443.
Lanmai and CS were obtained from the Sichuan Academy of Agricultural Sciences (SAAS), China. AT23 was provided by Dr. A. Mujeeb-Kazi, International Maize and Wheat Improvement Center (CIMMYT), Mexico, in 1995 and kept in SAAS. The accession number in CIMMYT is AE. SQUARROSA (309).
Mc-FISH Characterization
Multicolor fluorescence in situ hybridization was used to identify the chromosome constitutions of the introgression lines. Slides of root tips of Lanmai/AT23//Lanmai F7 generation and their parents Lanmai and AT23 were prepared as described by Zhang et al. (2018) and Kato et al. (2004). Two oligonucleotide probes, Oligo-pSc119.2 (hybridization with B genome chromosomes) and Oligo-pTa535 (hybridization with A and D genome chromosomes) (Tang et al., 2014), were labeled with 6-carboxyfluorescein (6-FAM) or 6-carboxytetramethylrhodamine (TAMRA) at the 5′-end (synthesized by Invitrogen Biotechnology Co. Ltd., Shanghai, China) and hybridized to the slides as reported by Fu et al. (2015). After hybridization, the slides were stained and imaged following the procedures by Zhang et al. (2020).
Yield Component Traits Survey
The yield component traits of Lanmai, YL-724, and YL-443 was examined at the Pidu experiment station of SAAS, Sichuan, China, in 2020 and 2021. A randomized complete block design was arranged with three replications. The seeds were sown as described by Wan et al. (2015). After maturity, the plant height (PH), spike length (SL), spikelet number per spike (SNS), floret number per spike (FNS), kernel number per spike (KNS), and seed setting rate (SR) (calculated through dividing KNS by FNS) were recorded with 10 replications of each material.
Mean ± SD was calculated using IBM SPSS Statistics (Version 26) (George and Mallery, 2019). Two-way ANOVA followed by Bonferroni's multiple comparisons test was performed using GraphPad Prism (Version 8.0.1) (Mavrevski et al., 2018) to compare the variances among different years and lines for all traits.
Morphological Trait Analysis
Hairy leaf sheath and auricles are the morphological trait studied in this study [Note: Most Ae. tauschii accessions possess this trait which could be transferred into common wheat through SHW-involved crossing (Yang et al., 1999), and its associated QTL has been mapped on 4DL (Dobrovolskaya et al., 2007; Wan et al., 2015). Therefore, hairy leaf sheath and auricles mark the authenticity of 4D(4B) substitution line]. During phenotyping, pubescence was examined on the leaf sheath and auricles of lines Lanmai, AT23, YL-443, and YL-724.
Adult Plant Resistance Assessment of Stripe Rust
Stripe rust was caused by Puccinia striiformis f. sp. tritici (Pst). Its resistance genes YrAS2388 and Yr28, which are believed to be the same gene, have been mapped on chromosome 4D in Ae. tauschii (Singh et al., 2000; Huang et al., 2011; Liu et al., 2013; Zhang R. et al., 2019). Combining with various stripe rust resistance phenotypes screened in the field, the PCR primer of YrAS2388 that was specified in Zhang C. et al. (2019) was used to identify the Pst resistance gene. The PCR and thermal cycles were performed as described by Zhang C. et al. (2019).
Adult plant resistance (APR) tests of stripe rust were conducted via a field trial at the Pidu experiment station of SAAS, Sichuan, China, in 2021. Each field plot of lines Lanmai, AT23, YL-443, and YL-724 and spreader rows were sown as reported by Rouse et al. (2011). The mixed urediniospores of Pst races, CYR32, CYR33, and CYR34 (Wan et al., 2007; Chen et al., 2009; Yang et al., 2019) that are currently prevalent in China, were inoculated onto the highly susceptible wheat Chuanyu12 (Chengdu Institute of Biology of Chinese Academy of Sciences, Sichuan, China), which was used as a spreader and control. Adult plant infection types (IT) were recorded when rust development reached 90% on Chuanyu12 using a 0–4 scale as described by Chen et al. (2009) and divided into five categories: immune (I, 0), near immune (NI, 0), high resistance (HR, 1), moderate resistance (MR, 2), moderate susceptibility (MS, 3), and susceptible (S, 4). The IT was scored and recorded once every 10 days, making a total of three times of recording for all field plots. The Pst races were provided by the Institute of Plant Protection, SAAS and the Institute of Plant Protection, Gansu Academy of Agricultural Sciences.
KASP Markers Screening of 4D and 4B Chromosomes
The Kompetitive Allele-Specific PCR (KASP) assay (Grewal et al., 2020) was used to screen the 4D and 4B chromosomes in this study. Each SNP-specific KASP primer set contains two allele-specific competing forward primers with unique tail sequences and one reverse primer. A total of 15 primer sets for chromosomes 4B and 16 primer sets for 4D were chosen based on the SNPs discovered on 4 hexaploid wheat and 23 wild relatives (Grewal et al., 2020) (Invitrogen Biotechnology Co. Ltd., Shanghai, China), to screen the chromosome substitution line YL-443. The parental lines Lanmai and AT23 and the landrace CS were used as controls. The KASP reaction system of 2 μl was modified from the “KASP genotyping chemistry User guide and manual” (https://biosearch-cdn.azureedge.net/assetsv6/KASP-genotyping-chemistry-User-guide.pdf), containing 1 μl of 2× KASP Master mix (standard ROX) (LGC Biosearch Technologies, Shanghai, China), 0.028 μl of primer mix (10 pmol/μl of each primer), and 1 μl of DNA (50 ng/μl). The KASP thermal cycling program was set based on the instructions in the user guide and manual.
All reactions were conducted using the GeneMatrix system (HC Scientific LLC., Chengdu, China). The collected data were analyzed by the companion Matrix Master Data Analysis & Management System.
Results
Chromosomal Characterization
The results of mc-FISH detection of the mitotic spread chromosomes of Lanmai, AT23, YL-443, and YL-724 via probes Oligo-pSc119.2 and Oligo-pTa535 are shown in Figure 2. The FISH karyotype of the SHW line YL-724 showed 42 chromosomes (Figures 2D,E), demonstrating an accurate distinction of the durum wheat and Ae. tauschii chromosomes using the above probes. The Oligo-pTa535 (red) highlights a pair of 4D chromosomes of Ae. tauschii with strong hybridization signals near the telomeric region of the chromosomal long arm and the pericentromeric region (Figures 2B,C,E). On the same chromosome, it also has an Oligo-pSc119.2 (green) signal at the end of the short arm (Figures 2B,C,E). Compared with the karyotype of CS (Tang et al., 2014), Lanmai (Figures 2A,E), AT23 (Figures 2B,E), and YL-724 (Figures 2D,E), line YL-443 (Figures 2C,E) clearly showed as a 4D(4B) disomic substitution line that contained a pair of 4D chromosomes plus the other 26 durum wheat chromosomes.
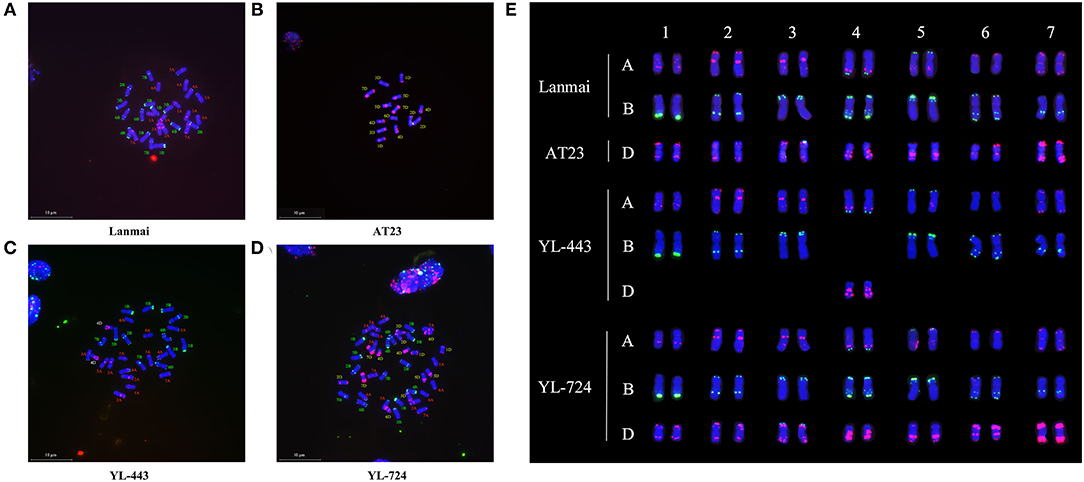
Figure 2. The mc-FISH karyotypes of Lanmai, AT23, YL-443, and YL-724 hybridized with probes Oligo-pSc119.2 (green) and Oligo-pTa535 (red). (A) Durum wheat Lanmai; (B) Ae. tauschii AT23; (C) Lanmai 4D(4B) disomic substitution line YL-443; (D) Lanmai/AT23 amphiploid line YL-724; (E) Comparison of chromosome constitution of all lines through cutting out chromosomes.
Yield Component Traits Survey
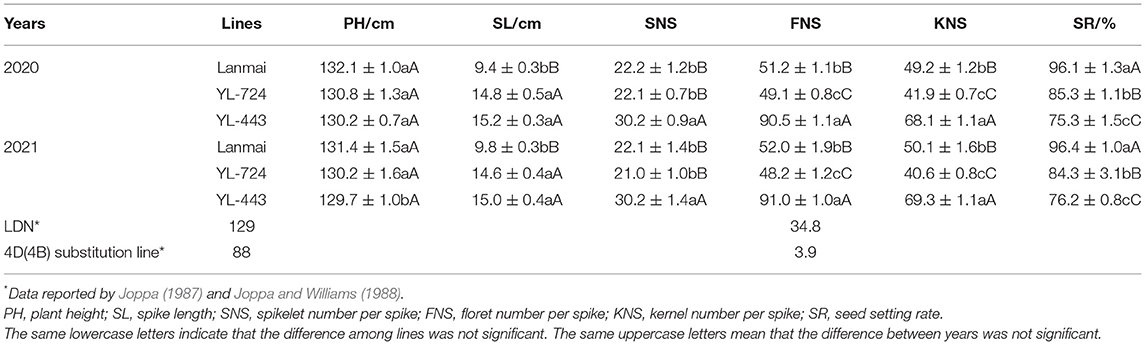
The yield component traits of Lanmai, YL-724, and YL-443 were phenotyped in 2020 and 2021 (Table 1). For all lines, there was no significant difference between years based upon the ANOVA results (Figure 3). All lines almost had the same plant height (Figure 3A). Even though the lines had similar spike length YL-443 displayed more spikelets per spike than that of YL-724 (Figures 3B, 4A,B). Meanwhile, YL-443 had a significantly higher spikelet number per spike (SNS) and floret number per spike (FNS) than the other lines (Table 1; Figures 3C,D, 4A,B). Compared with Lanmai, YL-443 had SNS and FNS higher than Lanmai by 36.3% and 75.9%, respectively. Although YL-443 had a lower seed setting rate than that of Lanmai and YL-724, it still had the highest kernel number per spike among all lines (Table 1; Figures 3E,F).
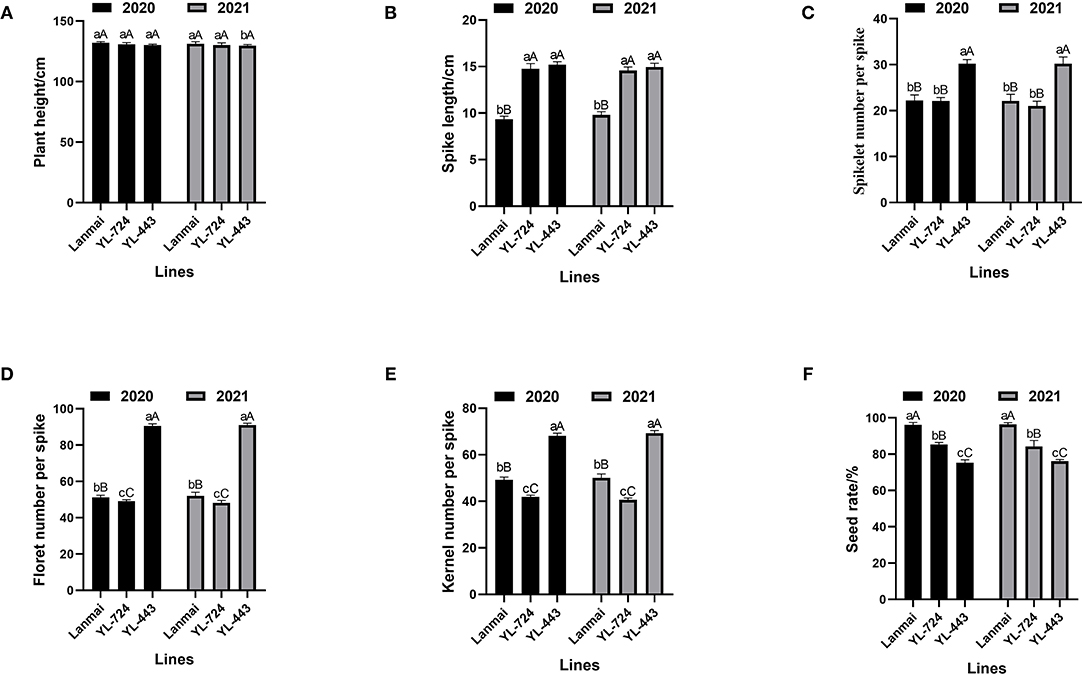
Figure 3. ANOVA of yield component trait of durum wheat Lanmai, Lanmai/AT23 amphiploid line YL-724, Ae. tauschii AT23, and Lanmai 4D(4B) disomic substitution line YL-443. (A) Plant height comparison. (B) Spike length comparison. (C) Spikelet number comparison. (D) Comparison of floret number per spike. (E) Comparison of kernel number per spike. (F) Seed setting rate comparison. The same lowercase letters indicate that the difference among lines was not significant. The same uppercase letters mean the difference between years was not significant.

Figure 4. The spike and spikelet comparison of durum wheat Lanmai, Lanmai/AT23 amphiploid line YL-724, Ae. tauschii AT23, and Lanmai 4D(4B) disomic substitution line YL-443. (A) Spike comparison. (B) Spikelet comparison.
Morphological Trait Analysis
Since our previous work has mapped the QTLs for hairy leaf sheath and auricles on chromosome 4DL (Wan et al., 2015), indicating that the 4D(4B) substitution line YL-443 should contain the hairy leaf sheath and auricles characteristics of AT23. The comparison of leaf sheath and auricles hairiness among lines Lanmai, AT23, YL-724, and YL-443 is shown in Figure 5. Lanmai had a glabrous leaf sheath and auricles, while AT23, YL-724, and YL-443 were all pubescent. This special characterization of YL-443 verified its authenticity as a 4D(4B) chromosome substitution. It is worth noting that the hairy leaf sheath and auricles characteristics can be used for assisting selection in crosses involving YL-443.

Figure 5. Comparison of leaf sheath and auricles hairiness among durum wheat Lanmai, Lanmai/AT23 amphiploid line YL-724, 4D(4B) disomic substitution line YL-443, and Ae. tauschii AT23. (A) Hairy auricles comparison. (B) Hairy leaf sheath comparison.
Stripe Rust Resistance Evaluation
The stripe rust resistance gene Yr28 has been mapped on chromosome 4D in Ae. tauschii (Singh et al., 2000; Huang et al., 2011; Liu et al., 2013; Zhang R. et al., 2019). As shown in Figure 6A, PCR using DNA from lines YL-443, YL-724, and AT23 produced a specific amplicon (727 bp) of Yr28, while PCR using DNA from Lanmai did not. This result indicated that the Yr28 gene has been transferred from Ae. tauschii AT23 to YL-724 and then to YL-443.
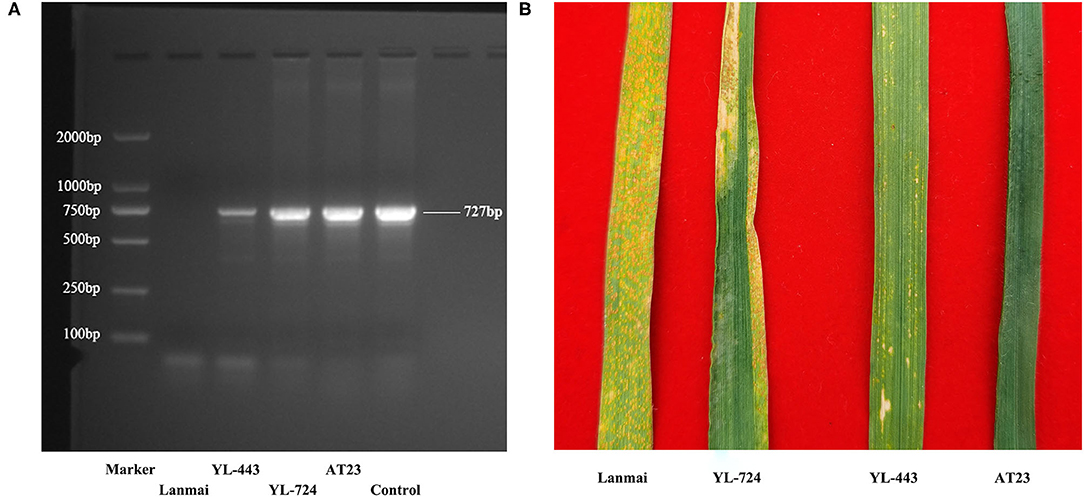
Figure 6. The Yr28 gene amplification results and different APR phenotypes of durum wheat Lanmai, Lanmai/AT23 amphiploid line YL-724, 4D(4B) disomic substitution line YL-443, and Ae. tauschii AT23 for stripe rust. (A) The Yr28 gene amplification results with positve products (727 bp) in all plants except Lanmai. (B) The APR test results of Lanmai (S), YL-724 (MR), YL-443 (HR), and AT23 (I) for stripe rust resistance. I, immune; NI, near immune; HR, high resistance; MR, moderate resistance; MS, moderate susceptibility; S, susceptible.
To identify the infection type of YL-443, inoculation of stripe rust was conducted in the field using the currently prevalent Pst races in China including CYR32, CYR33, and CYR34. Ae. tauschii AT23 showed an immune response to all Pst races, whereas durum wheat Lanmai was susceptible (Figure 6B). The amphiploid line YL-724 displayed an MR phenotype (Figure 6B), which is similar to another SHW line reported by Zhang R. et al. (2019). The developed substitution line YL-443 showed high resistance to all Pst races (Figure 6B). This indicates that the Yr28 gene can fully express in the 4D(4B) disomic substitution line in a tetraploid background but is partially suppressed in a hexaploid genetic background.
KASP Markers Identification
Results of the identification of the chromosomal substitution using the released specific KASP markers of chromosomes 4D (15 sets) and 4B (16 sets) developed by Grewal et al. (2020) were shown in Supplementary Data S1. For the 15 4B-specific markers, YL-443 and AT23 failed to give a positive result, while YL-724 showed the same results as plant Lanmai except WRC500 (with no result for Lanmai). Among the 16 4D-specific markers, seven showed no result for all plants, possibly due to the genetic variations compared with marker-based reference germplasm. For the remaining nine markers, Lanmai (without chromosome 4D) showed no positive results, while YL-443 and YL-724 both showed the same genotype as their parent AT23 and one marker WRC531 showing no result with CS. The results meant that the 4D(4B) substitution was verified by 23 specific KASP markers including 9 markers for 4D and 14 markers for 4B, suggesting that these markers can be used for 4D introgression line selection during durum wheat breeding.
Discussion
The D genome addition and inter/intra-specific introgressions cause a substantial difference between durum and bread wheat at the whole genome level (Dubcovsky and Dvorak, 2007; Cheng et al., 2019). Consequently, many end-use-related trait variations can be inserted, such as kernel hardness, grain protein content (gliadins and glutenins), and yellow pigment content (Mastrangelo and Cattivelli, 2021). On the other hand, the D-genome contains a high number of biotic resistance and abiotic tolerance genes, while the A or B genome usually lacks these genes (Padmanaban et al., 2017). Therefore, strategies to incorporate new alleles or traits from the D genome into the A or B genome have been followed for improving durum wheat. Previous researches have mainly focused on introgression by ph1b- and ph1c-mediated homologous recombination for developing translocations with chromosome fragments of the D genome (Sears, 1977; Ceoloni et al., 1995, 1996; Vitellozzi et al., 1997; Gennaro et al., 2012; Boehm et al., 2017; Morris et al., 2019; Sissons et al., 2019) or by crossing CS nullitetras with the durum wheat Langdon for developing D-genome substitutions (Joppa et al., 1983; Joppa and Williams, 1988). Most of the D-chromosomes introduced into durum wheat come from common wheat, such as CS (Joppa and Williams, 1988; Morris et al., 2011; Han et al., 2014, 2016). Chromosomes 4T-b and 4Hch from Am. muticum or H. chilense were also transferred into durum wheat, resulting in the development of 4T-b(4B) and 4Hch(4B) disomic substitution lines, respectively (Calderón et al., 2012; Othmeni et al., 2019).
Here, we reported a new procedure for developing introgression and/or substitution lines by using an allohexaploid wheat crossed with its tetraploid donor wheat. These introgressed segments or chromosomes under a single genetic background have an advantage for mapping genes and mining gene functions. By using this new procedure, a novel durum 4D(4B) disomic substitution line possessing a pair of 4D chromosomes from Ae. tauschii was developed.
According to the data reported by Joppa (1987) and Joppa and Williams (1988), durum wheat LDN had similar plant height and kernel number per spike, and its 4D(4B) substitution line with chromosome 4D from CS showed significantly lower plant height and kernel number per spike (Table 1). In contrast, due to the 4D chromosome compensation effect, the developed substitution line in this study has a super-large spike with a significantly (P < 0.001) increased spikelet number per spike and floret number per spike outperforming the Lanmai by 36.3 and 75.9%, respectively (Table 1; Figures 3C,D). These results indicated that the chromosome 4D of AT23 has a better compensatory effect than that of CS in a tetraploid background and possesses new genes for supernumerary spikelets. Although YL-443 had a lower seed setting rate than that of Lanmai and YL-724, its kernel number per spike was still the highest among all lines (Table 1; Figures 3E,F). These results demonstrate that the substitution line YL-443 can be further used for high-yield breeding by improving the seed setting rate.
To date, a range of genes or genetic loci that are responsible for supernumerary spikelets have been mapped on different chromosomes, such as 2A, 2B, 2D, 5B, 6A, 6B, 7A, and 7B (Li et al., 2012; Dobrovolskaya et al., 2015; Echeverry-Solarte et al., 2015; Kuzay et al., 2019; Ma et al., 2019; Rizwan et al., 2020; Du et al., 2021), but none has been reported on chromosome 4D. This study is the first report that chromosome 4D has strong effects on supernumerary spikelets. Further work is required to identify the gene underlying the supernumerary spikelets trait of the 4D chromosome of YL-443. In parallel, our previous research mapped a major hairy leaf sheath QTL on 4DL that were originated from Ae. tauschii and related with the enhanced grain yield trait in a SHW-derived wheat variety CM42 (Wan et al., 2015; Li et al., 2018). This indicates that the hairy leaf sheath genes in YL-443 may be related to its enhanced yield component traits, thus are potentially valuable in breeding aiming for yield improvement.
The resistance evaluation of stripe rust showed that the amphiploid line YL-724 had lower resistance than that of the developed substitution line YL-443, indicating a suppressed response to Pst races under a hexaploid genetic background, which was consistent with the results reported by Zhang C. et al. (2019). The stripe rust resistance gene Yr28 from the 4D chromosome of Ae. tauschii AT23 was transferred and completely expressed in the developed substitution line YL-443. Previous research also mapped the major salinity-tolerant gene Kna1 (Lindsay et al., 2004) and Al-tolerant gene TaALMT1 (Han et al., 2014, 2016) on 4D, which means that the 4D chromosome introduction may enhance the abiotic stress tolerance as well. Therefore, the 4D(4B) disomic substitution line YL-443 can potentially be an elite germplasm for enhancing the Pst resistance and Al tolerance of durum wheat. Moreover, YL-443 could also be used for cross-hybridization with rye for generating primary hexaploid triticale lines containing part of D genomes (Kwiatek and Nawracała, 2018). The Yr28-specific PCR marker, 4D chromosome-specific KASP markers, and special pubescent leaf sheath and auricles features can be utilized for assisting selection.
Considering the substantial and positive variations of the developed Lanmai-AT23 4D(4B) substitution line in this study, some tetraploid recombinant inbred lines (RIL) are currently under development by using YL-443 to cross with the CS and a synthesized hexaploid wheat Lanmai/AT18 (Ae. tauschii var. typica accession AT18). These RIL lines will help us understand the loss of 4D-chromosome functional genes after allohexaploid hybridization through fine-mapping the 4D-chromosome genes carried by Ae. tauschii AT23, such as genes that control supernumerary spikelets. On the other hand, although the whole chromosome substitution line reported in this study showed a range of enhanced phenotypes, it does not mean that whole chromosome substitution can be expanded to all 14 chromosomes for durum wheat trait enhancement. There are insufficient studies to support such a proposal. A thorough characterization is needed to judge the value of any substitution line in breeding.
Conclusion
A durum wheat-Ae. tauschii 4D(4B) disomic substitution line YL-443 carrying the 4D chromosome with high stripe rust resistance, large spike, and unique pubescent leaf sheath and auricles features was developed and verified by mc-FISH, Yr28 gene-specific PCR, and 4D- and 4B-specific KASP assays. The developed line can potentially be used for future durum wheat yield improvement. The Yr28-specific PCR marker, 4D chromosome-specific KASP markers, and pubescent leaf sheath and auricles phenotype can be used to assist selection in breeding.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
WY, WM, and YZ conceptualized the study. FY, QW, NY, SY, YW, and JZ contributed to the methodology. FY, QL, NY, and HL contributed to the formal analysis. NY, ZL, and WY contributed to the investigation. FY, QW, and SY contributed to the data curation. HW and WY contributed to the resources. XF, WY, and YZ contributed to the funding acquisition. JL and HW contributed to the project administration. FY contributed to the manuscript writing (original draft). WM, XF, and WY contributed to the manuscript writing (reviewing and editing). All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the China Scholarship Council, the Accurate Identification Project of Crop Germplasm from Sichuan Provincial Finance Department, the National Natural Science Foundation of China (Nos. 31870360, 31470305), the Second Tibetan Plateau Scientific Expedition and Research Program (STEP) (No. 2019QZKK0303), and the China Agriculture Research System of MOF and MARA (CARS-03). This work was supported by Sichuan Agricultural University in China, Sichuan Academy of Agricultural Sciences in China, and Murdoch University in Australia.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Junyan Feng in Sichuan Academy of Agricultural Sciences for his support and assistance on methodology.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.745290/full#supplementary-material
References
Agriculture Agri-Food Canada (2021). Canada: Outlook for Principal Field Crops. Available online at: https://agriculture.canada.ca/en/canadas-agriculture-sectors/crops/reports-and-statistics-data-canadian-principal-field-crops/canada-outlook-principal-field-crops-2021-07-20 (accessed 20 April 2021).
Bhave, M., and Morris, C. F. (2008). Molecular genetics of puroindolines and related genes: allelic diversity in wheat and other grasses. Plant Mol. Biol. 66, 205–219. doi: 10.1007/s11103-007-9263-7
Boehm, J. D., Zhang, M., Cai, X., and Morris, C. F. (2017). Molecular and cytogenetic characterization of the 5DS−5BS chromosome translocation conditioning soft kernel texture in durum wheat. Plant Genome 10:plantgenome2017.2004. doi: 10.3835/plantgenome2017.04.0031
Calderón, M. D. C., Ramírez, M. D. C., Martin, A., and Prieto, P. (2012). Development of Hordeum chilense 4Hch introgression lines in durum wheat: a tool for breeders and complex trait analysis. Plant Breed. 131, 733–738. doi: 10.1111/j.1439-0523.2012.02010.x
Ceoloni, C., Biagetti, M., Ciaffi, M., Forte, P., and Pasquini, M. (1996). Wheat chromosome engineering at the 4x level: the potential of different alien gene transfers into durum wheat. Euphytica 89, 87–97. doi: 10.1007/BF00015724
Ceoloni, C., Ciaffi, M., Lafiandra, D., and Giorgi, B. (1995). “Chromosome engineering as a means of transferring 1D storage protein genes from common to durum wheat,” in Proceeding on 8th International Wheat Genet. Symposium, 159–163.
Chen, W., Wu, L., Liu, T., Xu, S., Jin, S., Peng, Y., et al. (2009). Race dynamics, diversity, and virulence evolution in Puccinia striiformis f. sp. tritici, the causal agent of wheat stripe rust in China from 2003 to 2007. Plant Dis. 93, 1093–1101. doi: 10.1094/PDIS-93-11-1093
Cheng, H., Liu, J., Wen, J., Nie, X., Xu, L., Chen, N., et al. (2019). Frequent intra-and inter-species introgression shapes the landscape of genetic variation in bread wheat. Genome Biol. 20, 1–16. doi: 10.1186/s13059-019-1744-x
Dobrovolskaya, O., Pont, C., Sibout, R., Martinek, P., Badaeva, E., Murat, F., et al. (2015). FRIZZY PANICLE drives supernumerary spikelets in bread wheat. Plant Physiol. 167, 189–199. doi: 10.1104/pp.114.250043
Dobrovolskaya, O., Pshenichnikova, T., Arbuzova, V., Lohwasser, U., Röder, M., and Börner, A. (2007). Molecular mapping of genes determining hairy leaf character in common wheat with respect to other species of the Triticeae. Euphytica 155, 285–293. doi: 10.1007/s10681-006-9329-7
Du, D., Zhang, D., Yuan, J., Feng, M., Li, Z., Wang, Z., et al. (2021). FRIZZY PANICLE defines a regulatory hub for simultaneously controlling spikelet formation and awn elongation in bread wheat. New Phytol. 231, 814–833. doi: 10.1111/nph.17388
Dubcovsky, J., and Dvorak, J. (2007). Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316, 1862–1866. doi: 10.1126/science.1143986
Echeverry-Solarte, M., Kumar, A., Kianian, S., Mantovani, E. E., McClean, P. E., Deckard, E. L., et al. (2015). Genome-wide mapping of spike-related and agronomic traits in a common wheat population derived from a supernumerary spikelet parent and an elite parent. Plant Genome 8:plantgenome2014-12. doi: 10.3835/plantgenome2014.12.0089
Faltermaier, A., Waters, D., Becker, T., Arendt, E., and Gastl, M. (2014). Common wheat (Triticum aestivum L.) and its use as a brewing cereal—a review. J. Inst. Brew. 120, 1–15. doi: 10.1002/jib.107
Fu, S., Chen, L., Wang, Y., Li, M., Yang, Z., Qiu, L., et al. (2015). Oligonucleotide probes for ND-FISH analysis to identify rye and wheat chromosomes. Sci. Rep. 5, 1–7. doi: 10.1038/srep10552
Gennaro, A., Forte, P., Panichi, D., Lafiandra, D., Pagnotta, M. A., D'Egidio, M. G., et al. (2012). Stacking small segments of the 1D chromosome of bread wheat containing major gluten quality genes into durum wheat: transfer strategy and breeding prospects. Mol. Breed. 30, 149–167. doi: 10.1007/s11032-011-9606-6
George, D., and Mallery, P. (2019). IBM SPSS Statistics 26 Step by Step: A Simple Guide and Reference. Milton Park: Routledge. doi: 10.4324/9780429056765
Giorgi, B. (1978). A homoeologous pairing mutant isolated in Triticum durum cv. Cappelli. Mutat. Breed. Newsl 11, 4–5.
Grewal, S., Hubbart-Edwards, S., Yang, C., Devi, U., Baker, L., Heath, J., et al. (2020). Rapid identification of homozygosity and site of wild relative introgressions in wheat through chromosome-specific KASP genotyping assays. Plant Biotechnol. J. 18, 743–755. doi: 10.1111/pbi.13241
Han, C., Ryan, P. R., Yan, Z., and Delhaize, E. (2014). Introgression of a 4D chromosomal fragment into durum wheat confers aluminium tolerance. Ann. Bot. 114, 135–144. doi: 10.1093/aob/mcu070
Han, C., Zhang, P., Ryan, P. R., Rathjen, T. M., Yan, Z., and Delhaize, E. (2016). Introgression of genes from bread wheat enhances the aluminium tolerance of durum wheat. Theor. Appl. Genet. 129, 729–739. doi: 10.1007/s00122-015-2661-3
Hao, M., Luo, J., Zeng, D., Zhang, L., Ning, S., Yuan, Z., et al. (2014). QTug. sau-3B is a major quantitative trait locus for wheat hexaploidization. G3 Genes Genomes Genet. 4, 1943–1953. doi: 10.1534/g3.114.013078
Huang, L., Zhang, L.-Q., Liu, B.-L., Yan, Z.-H., Zhang, B., Zhang, H.-G., et al. (2011). Molecular tagging of a stripe rust resistance gene in Aegilops tauschii. Euphytica 179, 313–318. doi: 10.1007/s10681-010-0330-9
Joppa, L. (1987). “Aneuploid analysis in tetraploid wheat,” in Wheat and Wheat Improvement, Vol. 13, eds E. G. Heyne (New York, NY: Wiley), 255–267. doi: 10.2134/agronmonogr13.2ed.c11
Joppa, L., Khan, K., and Williams, N. (1983). Chromosomal location of genes for gliadin polypeptides in durum wheat Triticum turgidum L. Theor. Appl. Genet. 64, 289–293. doi: 10.1007/BF00274164
Joppa, L., and Williams, N. (1988). Langdon durum disomic substitution lines and aneuploid analysis in tetraploid wheat. Genome 30, 222–228. doi: 10.1139/g88-038
Kato, A., Lamb, J. C., and Birchler, J. A. (2004). Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Nat. Acad. Sci. 101, 13554–13559. doi: 10.1073/pnas.0403659101
Kihara, H. (1944). Discovery of the DD–analyzer, one of the analyzers of Triticum vulgare. Agric. Hortic. 19, 889–890.
Kuzay, S., Xu, Y., Zhang, J., Katz, A., Pearce, S., Su, Z., et al. (2019). Identification of a candidate gene for a QTL for spikelet number per spike on wheat chromosome arm 7AL by high-resolution genetic mapping. Theor. Appl. Genet. 132, 2689–2705. doi: 10.1007/s00122-019-03382-5
Kwiatek, M. T., and Nawracała, J. (2018). Chromosome manipulations for progress of triticale (× Triticosecale) breeding. Plant Breed. 137, 823–831. doi: 10.1111/pbr.12652
Li, A., Liu, D., Wu, J., Zhao, X., Hao, M., Geng, S., et al. (2014). mRNA and small RNA transcriptomes reveal insights into dynamic homoeolog regulation of allopolyploid heterosis in nascent hexaploid wheat. Plant Cell 26, 1878–1900. doi: 10.1105/tpc.114.124388
Li, A., Liu, D., Yang, W., Kishii, M., and Mao, L. (2018). Synthetic hexaploid wheat: yesterday, today, and tomorrow. Engineering 4, 552–558. doi: 10.1016/j.eng.2018.07.001
Li, J., Wang, Q., Wei, H., Hu, X., and Yang, W. (2012). SSR mapping for locus conferring on the triple-spikelet trait of the Tibetan triple-spikelet wheat (Triticum aestivum L. concv. tripletum). Triticeae Genomics Genet. 2, 1–6. doi: 10.5376/tgg.2011.02.0001
Lindsay, M. P., Lagudah, E. S., Hare, R. A., and Munns, R. (2004). A locus for sodium exclusion (Nax1), a trait for salt tolerance, mapped in durum wheat. Funct. Plant Biol. 31, 1105–1114. doi: 10.1071/FP04111
Liu, M., Zhang, C., Yuan, C., Zhang, L., Huang, L., Wu, J., et al. (2013). Stripe rust resistance in Aegilops tauschii germplasm. Crop Sci. 53, 2014–2020. doi: 10.2135/cropsci2013.01.0008
Ma, J., Ding, P., Liu, J., Li, T., Zou, Y., Habib, A., et al. (2019). Identification and validation of a major and stably expressed QTL for spikelet number per spike in bread wheat. Theor. Appl. Genet. 132, 3155–3167. doi: 10.1007/s00122-019-03415-z
Ma, J., Zhang, C., Yan, G., and Liu, C. (2013). Improving yield and quality traits of durum wheat by introgressing chromosome segments from hexaploid wheat. Genet. Mol. Res. 12, 6120–6129. doi: 10.4238/2013.December.2.9
Magallanes-López, A. M., Ammar, K., Morales-Dorantes, A., González-Santoyo, H., Crossa, J., and Guzmán, C. (2017). Grain quality traits of commercial durum wheat varieties and their relationships with drought stress and glutenins composition. J. Cereal Sci. 75, 1–9. doi: 10.1016/j.jcs.2017.03.005
Mastrangelo, A. M., and Cattivelli, L. (2021). What makes bread and durum wheat different? Trends Plant Sci. 26, 677–684. doi: 10.1016/j.tplants.2021.01.004
Mavrevski, R., Traykov, M., Trenchev, I., and Trencheva, M. (2018). Approaches to modeling of biological experimental data with GraphPad Prism software. WSEAS Trans. Syst. Control 13, 242–247. Available online at: https://www.wseas.org/multimedia/journals/control/2018/a625903-711.pdf
McFadden, E. S., and Sears, E. R. (1946). The origin of Triticum spelta and its free-threshing hexaploid relatives. J. Heredity 37, 81–89. doi: 10.1093/oxfordjournals.jhered.a105590
Morris, C. F., Kiszonas, A. M., Murray, J., Boehm, J. D., Ibba, M. I., Zhang, M., et al. (2019). Re-evolution of durum wheat by introducing the hardness and Glu-D1 loci. Front. Sustain. Food Syst. 3:103. doi: 10.3389/fsufs.2019.00103
Morris, C. F., Simeone, M. C., King, G., and Lafiandra, D. (2011). Transfer of soft kernel texture from Triticum aestivum to durum wheat, Triticum turgidum ssp. durum. Crop Sci. 51, 114–122. doi: 10.2135/cropsci2010.05.0306
Othmeni, M., Grewal, S., Hubbart-Edwards, S., Yang, C., Scholefield, D., Ashling, S., et al. (2019). The use of pentaploid crosses for the introgression of Amblyopyrum muticum and D-genome chromosome segments into durum wheat. Front. Plant Sci. 10:1110. doi: 10.3389/fpls.2019.01110
Özkan, H., Willcox, G., Graner, A., Salamini, F., and Kilian, B. (2011). Geographic distribution and domestication of wild emmer wheat (Triticum dicoccoides). Genet. Resour. Crop Evol. 58, 11–53. doi: 10.1007/s10722-010-9581-5
Padmanaban, S., Zhang, P., Hare, R. A., Sutherland, M. W., and Martin, A. (2017). Pentaploid wheat hybrids: applications, characterisation, and challenges. Front. Plant Sci. 8:358. doi: 10.3389/fpls.2017.00358
Rizwan, M., Mahboob, W., Faheem, M., Shimelis, H., Hameed, A., Sial, M., et al. (2020). Can we exploit supernumerary spikelet and spike branching traits to boost bread wheat (Triticum aestivum L.) yield? Appl. Ecol. Environ. Res. 18, 6243–6258. doi: 10.15666/aeer/1805_62436258
Rouse, M., Wanyera, R., Njau, P., and Jin, Y. (2011). Sources of resistance to stem rust race Ug99 in spring wheat germplasm. Plant Dis. 95, 762–766. doi: 10.1094/PDIS-12-10-0940
Sears, E. R. (1977). Genetics society of canada award of excellence lecture an induced mutant with homoeologous pairing in common wheat. Can. J. Genet. Cytol. 19, 585–593. doi: 10.1139/g77-063
Singh, R., Nelson, J., and Sorrells, M. (2000). Mapping Yr28 and other genes for resistance to stripe rust in wheat. Crop Sci. 40, 1148–1155. doi: 10.2135/cropsci2000.4041148x
Sissons, M., Pleming, D., Sestili, F., and Lafiandra, D. (2019). Effect of Glu-D1 gene introgression and amylose content on breadmaking potential of blends of durum and hexaploid wheat. Cereal Chem. 96, 193–206. doi: 10.1002/cche.10136
Tang, Z., Yang, Z., and Fu, S. (2014). Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119. 2, pTa-535, pTa71, CCS1, and pAWRC. 1 for FISH analysis. J. Appl. Genet. 55, 313–318. doi: 10.1007/s13353-014-0215-z
Tidiane Sall, A., Chiari, T., Legesse, W., Seid-Ahmed, K., Ortiz, R., Van Ginkel, M., et al. (2019). Durum wheat (Triticum durum Desf.): origin, cultivation and potential expansion in Sub-Saharan Africa. Agronomy 9:263. doi: 10.3390/agronomy9050263
Vitellozzi, F., Ciaffi, M., Dominici, L., and Ceoloni, C. (1997). Isolation of a chromosomally engineered durum wheat line carrying the common wheat Glu-D1d allele. Agronomie 17, 413–419. doi: 10.1051/agro:19970805
Wan, A. M., Chen, X. M., and He, Z. (2007). Wheat stripe rust in China. Aust. J. Agric. Res. 58, 605–619. doi: 10.1071/AR06142
Wan, H., Yang, Y., Li, J., Zhang, Z., and Yang, W. (2015). Mapping a major QTL for hairy leaf sheath introgressed from Aegilops tauschii and its association with enhanced grain yield in bread wheat. Euphytica 205, 275–285. doi: 10.1007/s10681-015-1457-5
Xynias, I. N., Mylonas, I., Korpetis, E. G., Ninou, E., Tsaballa, A., Avdikos, I. D., et al. (2020). Durum wheat breeding in the Mediterranean region: current status and future prospects. Agronomy 10:432. doi: 10.3390/agronomy10030432
Yang, M., Li, G., Wan, H., Li, L., Li, J., Yang, W., et al. (2019). Identification of QTLs for stripe rust resistance in a recombinant inbred line population. Int. J. Mol. Sci. 20:3410. doi: 10.3390/ijms20143410
Yang, W.-Y., Wu, B.-H., Hu, X.-R., Ye, Y., and Zhang, Y. (1999). Inheritance in hexaploid wheat of genes for hairy auricles and hairy leaf sheath derived from Aegilops tauschii Coss. Genet. Resour. Crop Evol. 46, 319–323. doi: 10.1023/A:1008796410782
Zhang, C., Huang, L., Zhang, H., Hao, Q., Lyu, B., Wang, M., et al. (2019). An ancestral NB-LRR with duplicated 3′ UTRs confers stripe rust resistance in wheat and barley. Nat. Commun. 10, 1–12. doi: 10.1038/s41467-019-11872-9
Zhang, J., Jiang, Y., Wang, Y., Guo, Y., Long, H., Deng, G., et al. (2018). Molecular markers and cytogenetics to characterize a wheat-Dasypyrum villosum 3V (3D) substitution line conferring resistance to stripe rust. PLoS ONE 13:e0202033. doi: 10.1371/journal.pone.0202033
Zhang, J., Yang, F., Jiang, Y., Guo, Y., Wang, Y., Zhu, X., et al. (2020). Preferential subgenome elimination and chromosomal structural changes occurring in newly formed tetraploid wheat—Aegilops ventricosa Amphiploid (AABBDvDvNvNv). Front. Genet. 11:330. doi: 10.3389/fgene.2020.00330
Zhang, R., Singh, R. P., Lillemo, M., He, X., Randhawa, M. S., Huerta-Espino, J., et al. (2019). Two main stripe rust resistance genes identified in synthetic-derived wheat line Soru# 1. Phytopathology 109, 120–126. doi: 10.1094/PHYTO-04-18-0141-R
Zhao, P., He, Z., Li, G., and Luo, H. (2002). General situation and developmental prospect on the breeding of hard wheat varieties in China. Shanxi J. Agric. Sci. 19−21.
Zhiyan (2020). Statistics of Import Amount and Value of Other Durum Wheat (10011900) in China from 2015 to 2019. Available online at: https://www.chyxx.com/shuju/202004/854208.html (accessed 21 April 2020).
Keywords: durum wheat, Aegilops tauschii, substitution, pentaploidy, supernumerary spikelets, stripe rust, interspecific hybridization
Citation: Yang F, Liu Q, Wang Q, Yang N, Li J, Wan H, Liu Z, Yang S, Wang Y, Zhang J, Liu H, Fan X, Ma W, Yang W and Zhou Y (2021) Characterization of the Durum Wheat-Aegilops tauschii 4D(4B) Disomic Substitution Line YL-443 With Superior Characteristics of High Yielding and Stripe Rust Resistance. Front. Plant Sci. 12:745290. doi: 10.3389/fpls.2021.745290
Received: 21 July 2021; Accepted: 01 September 2021;
Published: 30 September 2021.
Edited by:
Keiichi Okazaki, Niigata University, JapanReviewed by:
Hisashi Tsujimoto, Tottori University, JapanIan Dundas, University of Adelaide, Australia
Copyright © 2021 Yang, Liu, Wang, Yang, Li, Wan, Liu, Yang, Wang, Zhang, Liu, Fan, Ma, Yang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wujun Ma, Vy5NYUBtdXJkb2NoLmVkdS5hdQ==; Wuyun Yang, eWFuZ3d1eXVuQDEyNi5jb20=; Yonghong Zhou, emhvdXloQHNpY2F1LmVkdS5jbg==
†These authors have contributed equally to this work
 Fan Yang
Fan Yang Qier Liu3,4†
Qier Liu3,4† Qin Wang
Qin Wang Jun Li
Jun Li Honshen Wan
Honshen Wan Zehou Liu
Zehou Liu Ying Wang
Ying Wang Jie Zhang
Jie Zhang Hang Liu
Hang Liu Wujun Ma
Wujun Ma Wuyun Yang
Wuyun Yang Yonghong Zhou
Yonghong Zhou