
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 11 November 2021
Sec. Plant Physiology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.742504
This article is part of the Research TopicEnvironmental and molecular control of bud dormancy and bud break in woody perennials: An integrative approachView all 23 articles
 Kunjing Wu1†
Kunjing Wu1† Xiaojing Duan1,2†
Xiaojing Duan1,2† Zhonglong Zhu1,3,4
Zhonglong Zhu1,3,4 Ziyang Sang5
Ziyang Sang5 Yutong Zhang1,3,4
Yutong Zhang1,3,4 Haiying Li3,4
Haiying Li3,4 Zhongkui Jia4,6*
Zhongkui Jia4,6* Luyi Ma1,3,4*
Luyi Ma1,3,4*Magnolia wufengensis (Magnoliaceae) is a deciduous landscape species, known for its ornamental value with uniquely shaped and coloured tepals. The species has been introduced to many cities in south China, but low temperatures limit the expansion of this species in cold regions. Bud dormancy is critical for plants to survive in cold environments during the winter. In this study, we performed transcriptomic analysis of leaf buds using RNA sequencing and compared their gene expression during endodormancy, endodormancy release, and ecodormancy. A total of 187,406 unigenes were generated with an average length of 621.82 bp (N50 = 895 bp). In the transcriptomic analysis, differentially expressed genes involved in metabolism and signal transduction of hormones especially abscisic acid (ABA) were substantially annotated during dormancy transition. Our results showed that ABA at a concentration of 100 μM promoted dormancy maintenance in buds of M. wufengensis. Furthermore, the expression of genes related to ABA biosynthesis, catabolism, and signalling pathway was analysed by qPCR. We found that the expression of MwCYP707A-1-2 was consistent with ABA content and the dormancy transition phase, indicating that MwCYP707A-1-2 played a role in endodormancy release. In addition, the upregulation of MwCBF1 during dormancy release highlighted the enhancement of cold resistance. This study provides new insights into the cold tolerance of M. wufengensis in the winter from bud dormancy based on RNA-sequencing and offers fundamental data for further research on breeding improvement of M. wufengensis.
Owing to the instability in global climate, many perennial plants have suffered from abnormal weather conditions, including extreme temperatures in winter. Bud dormancy is the temporary suspension of visible growth in plant buds and represents a protective strategy for perennial plants to survive unfavourable climatic changes during winter (Rohde and Bhalerao, 2007). Bud dormancy is traditionally categorised into three phases: paradormancy (PD), inhabited by substances generated from another part of the plant; endodormancy (ED), controlled by internal factors; and ecodormancy (ECD), regulated by the external environment (Lang et al., 1987; Considine and Considine, 2016). Plants cannot resume growth in a favourable environment until ED release (Rohde and Bhalerao, 2007). To break ED, plants need to fulfil chilling requirements (CRs) after accumulating sufficient chilling hours (Arora et al., 2003), as insufficient cold accumulation may delay dormancy release, influence flower morphology, and even impair growth and production (Atkinson et al., 2013). Therefore, it is necessary to evaluate bud dormancy status and assess CRs in perennial trees. Three models are mainly used to calculate CRs in woody perennials: 0–7.2°C model (Weinberger, 1950), Utah model (Richardson, 1974), and dynamic model (Fishman et al., 1987a, b).
Temperature and photoperiod are important environmental signals controlling the seasonal dormancy cycle in perennials (Anderson et al., 2010; Maurya and Bhalerao, 2017). Short photoperiods induce bud formation, bud dormancy induction and apical meristem cessation of shoots (Weiser, 1970; Singh et al., 2017). Moreover, dormancy release requires sufficient chilling accumulation in winter as low temperature mostly regulate dormancy release and bud break (Heide, 2008; Yamane et al., 2011). According to the two different dormancy-related environmental factors, plants can be classified into three types: temperature-sensitive, photoperiod-sensitive, and temperature- and photoperiod-sensitive (Bai et al., 2016).
Phytohormones are a crucial factor influencing bud dormancy in perennials, and endogenous hormones and their balance regulate the induction of and release from dormancy (Sonnewald and Sonnewald, 2014; Liu and Sherif, 2019). Some conventional hormones such as gibberellin (GA), abscisic acid (ABA), and auxin (IAA) participate in the dormancy cycle (Horvath et al., 2003; Vimont et al., 2021). In general, an increase in the ABA content accompanied by a decrease in the GA3 and IAA content is observed during the dormant induction phase, whereas the opposite trend is observed during dormancy release in plants (Liu and Sherif, 2019). High levels of IAA and GA3 accelerate dormancy release (Rinne et al., 2011; Zhuang et al., 2013) whereas ABA maintains dormancy (Li et al., 2018; Tylewicz et al., 2018). In addition, exogenous ABA application results in a delay in bud break in, for example, Pyrus pyrifolia (pear) (Li et al., 2018), Vitis vinifera (grape) (Zheng et al., 2015), and Betula pendula (birch) (Rinne et al., 1994).
The role of ABA in dormancy has been widely studied at physiological and molecular levels and evidence has indicated that ABA biosynthesis, catabolism, and signalling pathway are involved in the regulation of bud dormancy (Zheng et al., 2018). A rate-limiting enzyme involved in ABA biosynthesis, 9-cis-epoxycarotenoid dioxygenase (NCED), has been indicated to control dormancy at the transcriptional level (Zheng et al., 2015; Li et al., 2018). During catabolism, ABA is degraded by ABA 8’-hydroxylase, which is encoded by cytochrome P450 CYP707A, and the relationship between CYP707A and ABA content has been widely investigated (Cutler and Krochko, 1999; Saito et al., 2004). The ABA signalling pathway consists of two groups of ABA regulators: Protein Phosphatase 2c (PP2Cs) and SNF1-Related Protein Kinase 2 (SnRK2s). Besides, the ABA receptors were identified as Pyrabactin Resistance (PYRs), Pyrabactin Resistance-Like (PYLs), and Regulatory Component of ABA Receptor (RCARs) (Hubbard et al., 2010). ABA binds to PYR/PYL/RCARs and forms PP2C complexes, which inhibit the activity of PP2Cs. PP2Cs can suppress SNF1-related protein kinase 2 (SnRK2s) function via dephosphorylation, which negatively affects ABA signalling, allowing SnRK2s to activate the downstream ABRE-binding factor (AREB/ABF) transcription factors (TFs) (Umezawa et al., 2009; Soon et al., 2011). Several studies have shown that genes related to ABA signalling are involved in dormancy regulation (Yang et al., 2020). In Hybrid Aspen, short days induce high levels of ABA which suppresses PICKLE (PKL) to induce the expression of SVP-like (SVL), which is an orthologue of short vegetative phase (SVP) and then SVL induces callose synthase 1 (CALS1) expression to promote the establishment of dormancy (Tylewicz et al., 2018; Singh et al., 2019).
Bud dormancy is an important overwintering process, and many studies have shown that bud dormancy is associated with winter cold resistance at the molecular level. C-repeat binding factor (CBF) belongs to the APETALA2/-ETHYLENE RESPONSE FACTOR (AP2/ERF) gene family, regulates many genes related to cold response and tolerance and can be induced by inducer of CBF expression (ICE) (Chinnusamy et al., 2007; Guo et al., 2018). Dormancy-associated MADS-box (DAM)/SVP/SVL genes are known to control bud dormancy in many species (Singh et al., 2018; Yang et al., 2018; Gao et al., 2021). Therefore, the relationship between DAM and CBF links between bud dormancy and cold resistance. PmCBFs are known to bind to the promoter of PmDAM6 and activate the expression of PmCBFs in P. mume (Zhao et al., 2018a, b). In P. pyrifolia, the expression of PpDAMs is directly induced by the accumulation of CBF by binding to CRT/DRE motifs (Niu et al., 2015; Saito et al., 2015). Li et al. (2019) reported that low temperature induces PpCBF1-PpDAM2 regulon to function during ED transition (Li et al., 2019). Thus, bud dormancy may be associated with cold tolerance during the winter.
Magnoliaceae plants have high ornamental value and are widely cultivated globally. Magnolia denudata, as a common species in north China, has been widely cultivated for its prominent cold tolerance (Yang et al., 2015). Magnolia wufengensis (Supplementary Figure 1), a new species of Magnoliaceae, was discovered growing in Wufeng County, Hubei Province, People’s Republic of China (Ma et al., 2006). As a deciduous landscape species with uniquely shaped and colours of the tepals, M. wufengensis has been introduced to many cities in south China for its rich biological characteristics and will have a place in global horticultural plants (Shi et al., 2021). However, this is difficult in north China where temperatures can be extremely low, because M. wufengensis is more sensitive to the cold and with a deeper dormancy level than other Magnoliaceae species such as M. denudata (Yang et al., 2015; Deng et al., 2019; Duan et al., 2019). Bud dormancy is an important biological process that helps plants survive cold temperature in winter. RNA sequencing (RNA-seq) has been recently used to study bud dormancy in many species such as pear (P. pyrifolia) (Bai et al., 2013), tea (Camellia sinensis) (Hao et al., 2017), sweet cherry (Prunus avium L.) (Vimont et al., 2019), and wintersweet (Chimonanthus praecox) (Li et al., 2020). In this study, using RNA-seq, we aimed to explore: (i) the cycle period between ED and ECD and the effects of different meteorological factors on dormancy release of M. wufengensis, (ii) which key genes and pathways were involved in regulation of different dormancy phases, (iii) the role that hormones, especially ABA, play in endodormancy maintenance, and (iv) the relationship between cold tolerance and different phases of dormancy. This study will provide a foundation for improving cold resistance and thus allowing normal growth in winter and expanding the northern boundary of M. wufengensis cultivation.
Eight-year-old M. wufengensis and M. denudata were cultivated in Jiufeng National Forest Park (Beijing, China; 40°3′25″N, 116°6′39″E). The trees were not clipped or chemically treated before sampling. In 2019–2020, one-year shoots with one apical bud were collected from M. wufengensis on 2 November (19N1), 23 November (19N2), 6 December (19D1), 14 December (19D2), 21 December (19D3), 30 December (19D4), 12 January (20J1), 18 January (20J2), and 19 February (20F1). In 2020–2021, M. wufengensis and M. denudata shoots and buds were collected on 5 October (20O1), 20 October (20O2), 5 November (20N1), 20 November (20N2), 5 December (20D1), 20 December (20D2), 5 January (21J1), and 20 January (21J2). 12 trees were divided into three replicates, and all samples were collected from the same 4 trees at each stage. All leaf bud samples of M. wufengensis were stored in liquid nitrogen immediately after collection and then at –80°C until RNA extraction.
The bud dormancy status of leaf buds was evaluated as previously described (Liu et al., 2012) with some modifications. In 2019–2020 and 2020–2021, 1-year-old shoots with one apical bud about 10 cm long (n = 7) were sampled and inserted in wet flower mud in a box full of water and allowed to grow in the climate chamber at 25 ± 1.0°C during the day and 22 ± 1.0°C during the night, with a photoperiod of 14 h light/10 h dark and 60% relative humidity. Twenty-one leaf buds grown in three boxes of flower mud were divided into three biological replicates. The water was changed and the base of the shoots was cut every 3–4 days. Dormancy status was determined by the bud break percentage (BBP) after 32 days. We defined the beginning of bud break when the as green leaf tips were enclosing visible leaves. If the buds were at more than 50% bud break after 32 days, then the buds were considered to be released from ED (Yooyongwech et al., 2009).
Maximum (Tmax), minimum (Tmin), and average temperatures (Tavg) were recorded every 15 min using the weather station (WeatherHawk, Campbell Scientific, UT, United States) located in Jiufeng National Forest Park.
Chilling units (CUs) of M. wufengensis and M. denudata leaf buds were calculated based on 0–7.2°C (Weinberger, 1950) and Utah models (Richardson, 1974).
The extraction, purification, and determination of endogenous abscisic acid and gibberellin (GA1, GA3 and GA4) were performed using an enzyme-linked immunosorbent assay (ELISA) according to manufacturer’s instructions. The fresh samples (1 g bud) were homogenised in liquid nitrogen and extracted in pre-cold 80% (v/v) methanol with butylated hydroxytoluene (BHT) (1 mmol/L) and kept at 4°C overnight. The samples were centrifuged for 15 min at 5,000 rpm (4°C). Afterward, the extracts were passed through a C18 Sep-Pak Cartridge (Waters, Milford, MA, United States) and dried with N2. Then the residues were dissolved in 0.01 mol L–1 PBS (pH 7.4) to determine the levels of ABA and GAs content. Calculations of plant hormones by ELISA followed the protocol described in Zhao et al. (2006). The ELISA kits used for the assay were purchased from Saipei Biotechnology Co., Ltd. (Wuhan, China). Each experiment contained three biological and technical replicates.
RNA for RNA-seq was isolated separately from the 19N1, 19D1, and 19D2 samples using Plant RNA Purification Reagent for plant tissue according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, United States) and genomic DNA was removed using DNase I (Takara Bio, Shiga, Japan). Then, RNA quality was determined using the 2100 Bioanalyzer Instrument (Agilent Technologies, Santa Clara, CA, United States) and quantified using NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, United States). Only high-quality RNA samples (optical density (OD)260/280 = ∼1.8–2.2, OD260/230 ≥ 2.0, RNA integrity number ≥ 6.5, 28S:18S ≥ 1.0, quantity > 1 μg) were used to construct the sequencing library.
RNA purification, reverse transcription, library construction, and sequencing were performed at Shanghai Majorbio Bio-pharm Biotechnology Co., Ltd. (Shanghai, China) according to the manufacturer’s instructions (Illumina, San Diego, CA, United States). The RNA-seq transcriptome libraries of M. wufengensis were prepared using TruSeq RNA Sample Prep Kit (Illumina). Poly(A) mRNA was purified from total RNA using oligo-dT-attached magnetic beads (Invitrogen) and then fragmented using the fragmentation buffer. Using these short fragments as templates, double-stranded complementary DNA (cDNA) was synthesised using SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen) with random hexamer primers (Illumina). Subsequently, the synthesised cDNA was subjected to end-repair, phosphorylation, and “A” base addition according to Illumina’s library construction protocol. Libraries were selected for size using cDNA target fragments of 200–300 bp on 2% Low Range Ultra Agarose (Bio-Rad) followed by PCR amplification using Phusion DNA polymerase (New England Biolabs, Boston, MA, United States) for 15 PCR cycles. After quantification using TBS380, two RNA-seq libraries were sequenced in a single lane on NovaSeq 6000 Sequencing System (Illumina) for 2 × 150 bp paired-end reads. Each experiment included three biological replicates.
The raw paired-end reads were trimmed and quality controlled using SeqPrep1 and Sickle2 with default parameters. Subsequently, clean data from M. wufengensis were used to perform de novo assembly with Trinity3 (Grabherr et al., 2011). All the assembled transcripts were searched against the National Center for Biotechnology Information protein non-redundant (NR), Clusters of Orthologous Genes (COG), and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases using BLASTX to identify the proteins that had the highest sequence similarity with the given transcripts to retrieve their function annotations and typical cut-off E-values were set as less than 1.0 × 10––5. Blast2GO4 (Conesa et al., 2005) programme was used to obtain gene ontology (GO) annotations of uniquely assembled transcripts for describing their biological processes, molecular functions, and cellular components. Metabolic pathway analysis was performed using KEGG5 (Ogata et al., 1999).
To identify differentially expressed genes (DEGs) between two different samples, the expression level of each transcript was calculated according to the transcripts per million reads method. RSEM6 (Li and Dewey, 2011) was used to quantify gene abundance. DEG analysis was performed using DESeq2 (Love et al., 2014) and EdgeR (Robinson et al., 2009) with DEGs |log2FC| > 1 and Q value ≤ 0.05 (DESeq2 or EdgeR) considered to be significant. In addition, functional enrichment analysis using GO and KEGG were performed to identify which DEGs were significantly enriched in GO terms and metabolic pathways with a Bonferroni-corrected p value ≤ 0.05 compared to the whole-transcriptome background. GO functional enrichment and KEGG pathway analyses were carried out using Goatools7 and KOBAS8 (Xie et al., 2011).
Venn diagrams were drawn and trend analysis was performed using Venny 2.19 and Short Time-series Expression Miner software (STEM) (Ernst and Bar-Joseph, 2006), respectively.
For ABA treatment, nine shoots of M. wufengensis were collected from the 12 trees from November 2020 to January 2021 and sprayed with 100, 200, or 300 μM ABA (Aidlab, Beijing, China) and 0.2% ethanol (mock treatment) at approximately 13:00 for three consecutive days. Each treatment was executed with three biological and technical replicates. Buds were collected at 7, 14, 21, and 28 days after ABA treatment and stored immediately in liquid nitrogen and then at –80°C. Other shoots that grew in the chamber environment mentioned above for 32 days were used to measure BBP.
To ensure DEGs identified by RNA-seq involve in cold tolerance of M. wufengensis, a cold acclimation experiment was conducted to valid their functions. Three apical buds were selected randomly and used for experiments during the whole cold acclimation treatment in September 2017 (autumn). The experiment included three temperatures for analysis. A room temperature of 22°C in a low-temperature incubator (3M, United States) served as the control. The samples in other groups of M. wufengensis buds were treated for seven days sequentially in low-temperatures incubators at the following two different experimental temperatures: low temperature of 12 and 4°C. Two days were left for cooling slowly in temperatures incubator between two groups (Supplementary Figure 2).
RNA of M. wufengensis buds for quantitative PCR (qPCR) was extracted using the HiPure HP Plant RNA Mini Kit (Magen, Shanghai, China) according to the manufacturer’s instructions and genomic DNA was removed using DNase I. cDNA used for qPCR was reverse transcribed from 2 μg of purified RNA in a 20 μL reaction volume based on the manufacturer’s instructions (G592, Applied Biological Materials, Richmond, BC, Canada). The qPCR primers were designed using Beacon Designer 7 (PREMIER Biosoft International, Palo Alto, CA, United States) and passed the specificity test. qPCR was carried out on a StepOnePlus Real-Time PCR System (Applied Biosystems, United States) with a final reaction volume of 10 μL containing 5 μL TB Green Premix Ex Taq (Tli RNaseH Plus; Takara Bio) (2X), 0.2 μL each of ROX Reference Dye (50X) (Takara Bio), upstream primer, and downstream primer, 1.0 μL cDNA, and 3.4 μL double-distilled water. MwACTIN was used for a reference gene for analysis. The primer sequences used in qPCR are listed in Supplementary Table 1. Each sample included three biological and technical replicates.
The study was conducted with a completely randomised design. The data were analysed using one-way analysis of variance followed by least significant difference test and p value <0.05 was considered significant. Graphs were constructed using SigmaPlot version 10 (Systat Software, San Jose, CA, United States) and R Project (R Foundation for Statistical Computing, Vienna, Austria). All data were analysed using SPSS Statistics version 20 (IBM, Armonk, NY, United States).
To study the relationships between bud dormancy and cold tolerance, it is imperative to define the status of bud dormancy. As is shown in Figure 1A, no bud breaking was observed in M. wufengensis and M. denudata on 20O2; however, BBP increased with progress in chilling accumulation mainly in November and December. In M. wufengensis, the apical leaf buds were determined in the ED phase before 20N2 and ED release between 20N2 and 20D1, when the number of CUs reached 62–214 CUs and 480–548 CUs based on the 0–7.2°C and Utah models, respectively. In addition, the ED release occurred between 5 December and 20 December in M. denudata, which was later than that in M. wufengensis with CUs reaching 214–294 CUs and 548–589.5 CUs based on the 0–7.2°C and Utah models, respectively (Figure 1B).
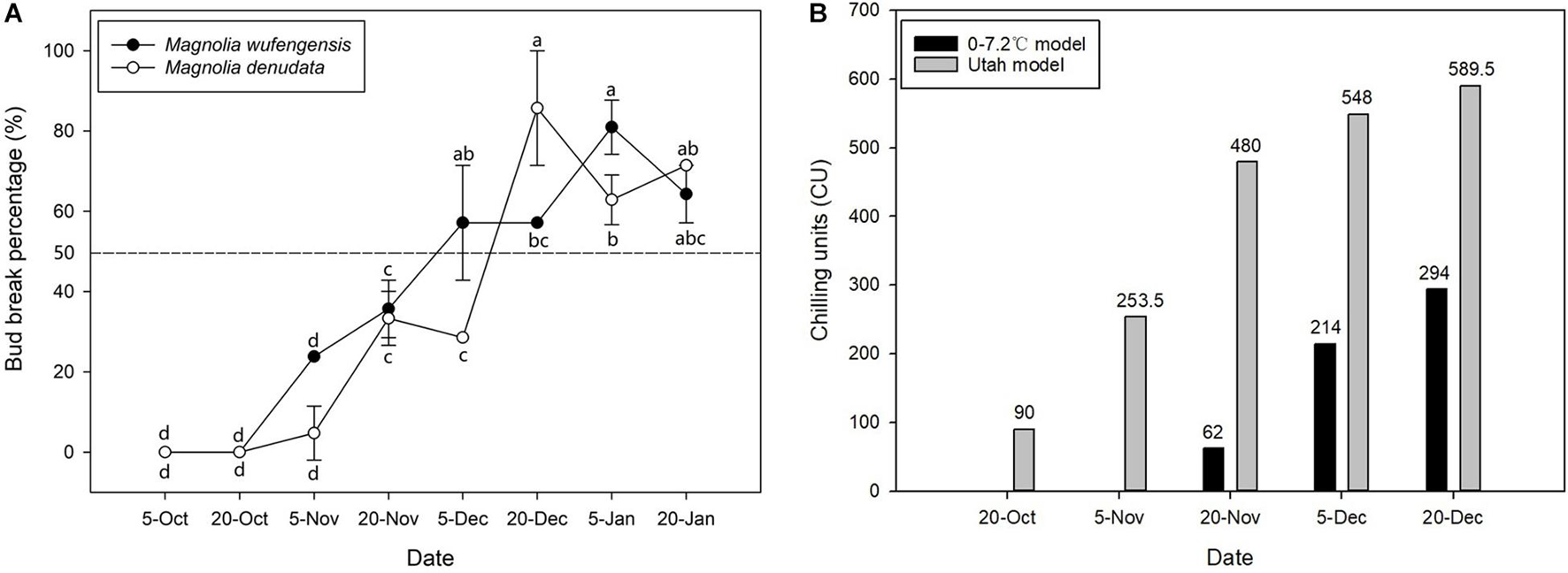
Figure 1. Bud break percentage and chilling units of Magnolia wufengensis and Magnolia denudata during 2020–2021 dormancy phase under natural conditions. (A) Bud break percentage of Magnolia wufengensis and Magnolia denudata was assessed after 32 days in 2020–2021. (B) Chilling units were calculated based on 0–7.2°C and Utah models beginning on 14 November and 29 September 2020, respectively. Each experiment was performed with three biological replicates. Different letters above bars indicate a significant difference among bud break percentage according to ANOVA and LSD test (p < 0.05).
GA3 content was low before 21J1 and peaked at 21J2, and a small peak appeared at 20N2 (before ED release) (Figure 2A) and a similar result was observed regarding the content of GA1 (Figure 2B). In addition, the content of GA4 decreased during ED release phase and a peak at 20D2 was observed (Figure 2B). Moreover, before ED release, the concentration of ABA kept increasing until at 20O2 and then decreased rapidly to the lowest level at 20N2 with dormancy release, with a slight increase at 20D1 and considerable reduction immediately at 20D2. After ED release, ABA levels sharply increased after 20D2 and increased further at 21J2 (Figure 2C). In addition, the ratio of content of ABA/GA3 increased before ED release and experienced a sharp decrease after ED release (Figure 2D).
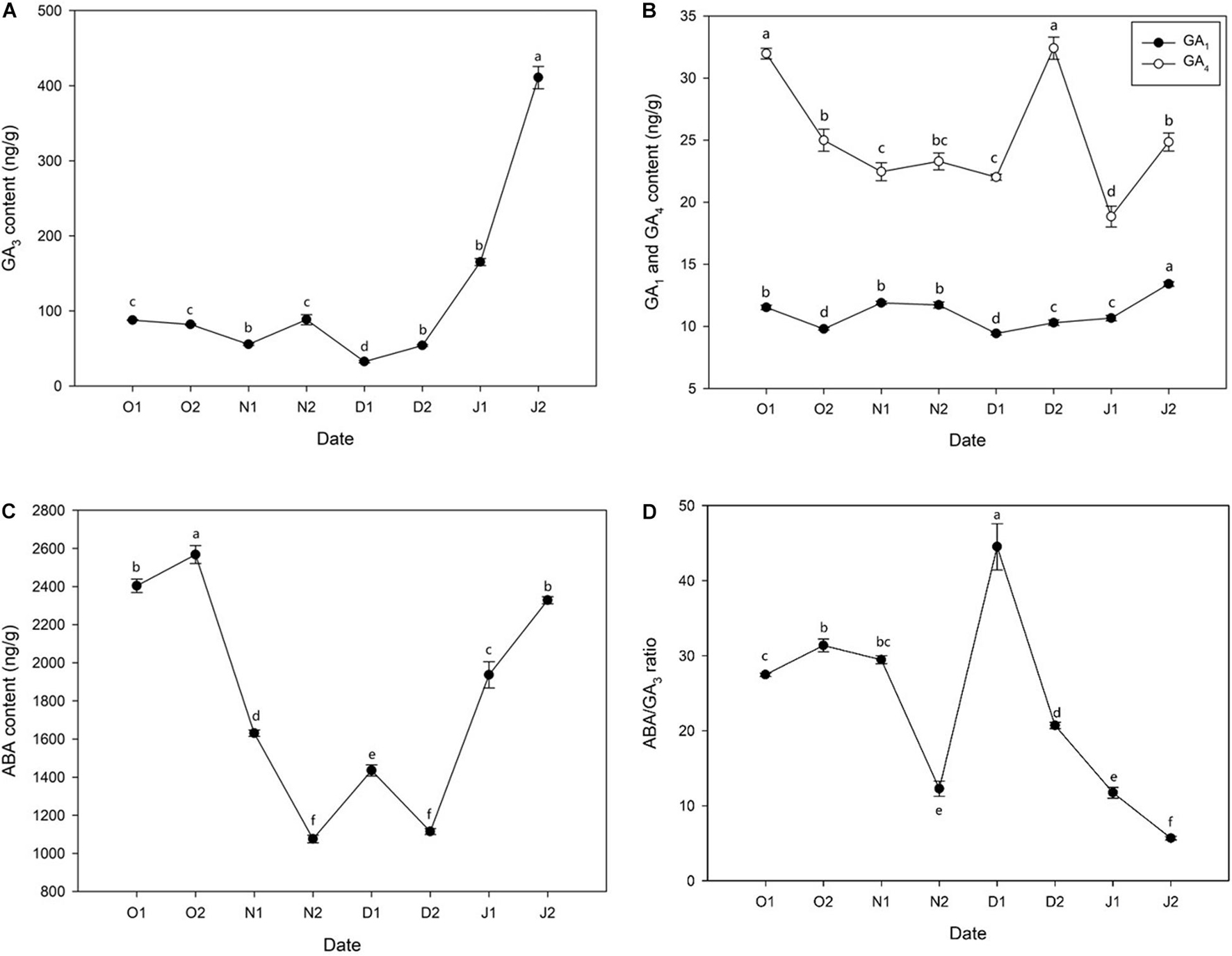
Figure 2. Concentration of phytohormones [(A) gibberellin 3 (GA3), (B) gibberellin 1 (GA1) and gibberellin 4 (GA4), (C) abscisic acid (ABA), (D) ABA/GA3 ratio] in Magnolia wufengensis during the season transition in 2020–2021. Each experiment was performed with three biological replicates. Different letters above bars indicate a significant difference among hormones according to ANOVA and LSD test (p < 0.05).
Three libraries 19N1 (ED, control, BBP = 0%), 19D1 (ED release phase, 0% < BBP < 50%), and 19D2 (ECD, BBP > 50%) (Supplementary Figure 3) were constructed from cDNA obtained from more than three apical buds and sequenced on the NovaSeq 6000 platform. Approximately 59.75 GB of clean reads was obtained after quality control, and Q30 percentage and guanine and cytosine content (GC) percentage were more than 92.14 and 46.92% in the nine samples, respectively (Table 1). The de novo assembly using Trinity yielded 187,406 unigenes ranging from 201 bp to 14,669 bp with an average length of 621.82 bp, and N50 of 895 bp (Table 2). In general, the number of unigenes decreased with the increase in gene length, and the largest proportion of unigenes was between 200 bp and 500 bp (123,562, 66%), followed by 501 bp to 1,000 bp (35,770, 19%), and 1,001 bp to 1,500 bp (11,895, 6%) (Supplementary Figure 4A).
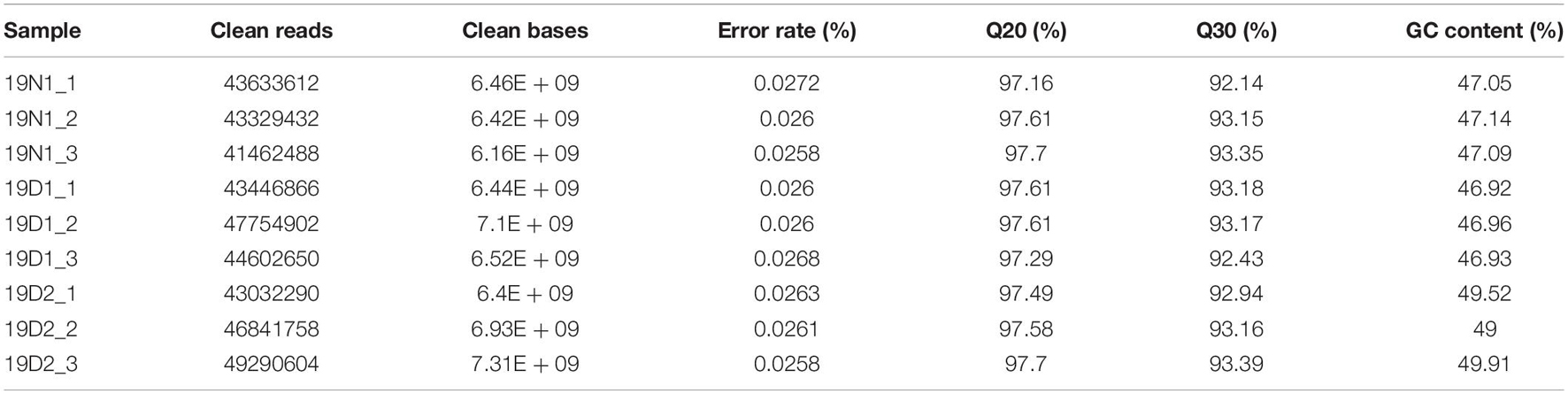
Table 1. Sequencing the Magnolia wufengensis transcriptome from nine apical leaf samples from plants from endodormancy (ED; 19N1_1, 19N1_2, 19N1_3), before ED release (19D1_1, 19D1_2, 19D1_3), and ecodormancy (ECD; 19D2_1, 19D2_2, 19D2_3).
The assembled unigenes were compared against NR, Swiss-Prot, Pfam, COG, GO, and KEGG databases using BLASTX (E-value < 1e–5). Simultaneously, we found that the number of unigenes successfully annotated to the NR database was the highest (78,142; 41.7%), followed by COG (71,437; 38.1%), KEGG (61,840; 33.0%), Pfam (55,774; 29.8%), Swiss-Prot (55,692; 29.7%), and GO (43,500; 23.2%) (Supplementary Figure 4B). With respect to species, the unigene sequences were most similar to genes from Quercus suber (29,500), Cinnamomum micranthum (11,576), Carpinus fangiana (5,871), Nelumbo nucifera (3,592), and V. vinifera (3,052) using BLASTX matches (Supplementary Figure 4C).
KEGG analysis revealed 61,840 unigenes to be significantly mapped to 139 KEGG pathways and classified into six categories: Metabolism, Genetic Information Processing, Environmental Information Processing, Cellular Processes, Organismal Systems, and Human Diseases. The highest unigenes representation pathways in “Metabolism” were carbohydrate metabolism (4,090) and amino acid metabolism (2,911), those in “Genetic Information Processing” were translation (4,895) and folding, sorting, and degradation (3,342). Signal transduction (823), transport and catabolism (2,138), environmental adaptation (852), and endocrine and metabolic disease (116) were most associated with “Environmental Information Processing,” “Cellular Processes,” “Organismal Systems,” and “Human Diseases,” respectively (Supplementary Figure 4D).
Based on GO analysis, 43,500 unigenes were classified into three main categories: biological process, cellular component, and molecular function. Biological process was mainly comprised of proteins involved in cellular process (29,572), metabolic process (27,167), and biological regulation (8,340). The most represented cellular components were cell part (29,063), membrane part (22,285), and organelle (16,681). In addition, we found a high number of unigenes involved in binding (36,757), catalytic activity (35,199), and transporter activity (5,244) in molecular function (Supplementary Figure 4E).
Unigenes with p value < 0.05 or |log2FC| ≥ 1 were defined as DEGs. Among the DEGs, 8,565 and 30,321 were upregulated and 7,675 and 13,672 genes were downregulated at 19D1 and 19D2, respectively. Moreover, 28,849 upregulated and 9,715 downregulated unigenes were identified between 19D2 and 19D1. The number of DEGs of 19D2 versus 19N1 were the highest, followed by 19D2 versus 19D1 and 19D1 versus 19N1 (Figure 3A). To further explore DEGs related to dormancy release under natural conditions, a Venn diagram was drawn between 19D1 versus 19N1, 19D2 versus 19N1, and 19D2 versus 19D1, and 4,286 DEGs were found to intersect all three groups (Figure 3B). To distinguish the changing patterns in gene expression, gene expression profile clustering was performed. From this, 4,286 genes were assigned to 16 different profiles by STEM and six profiles that were significantly enriched from 19N1 to 19D2 were identified (Figure 3C).
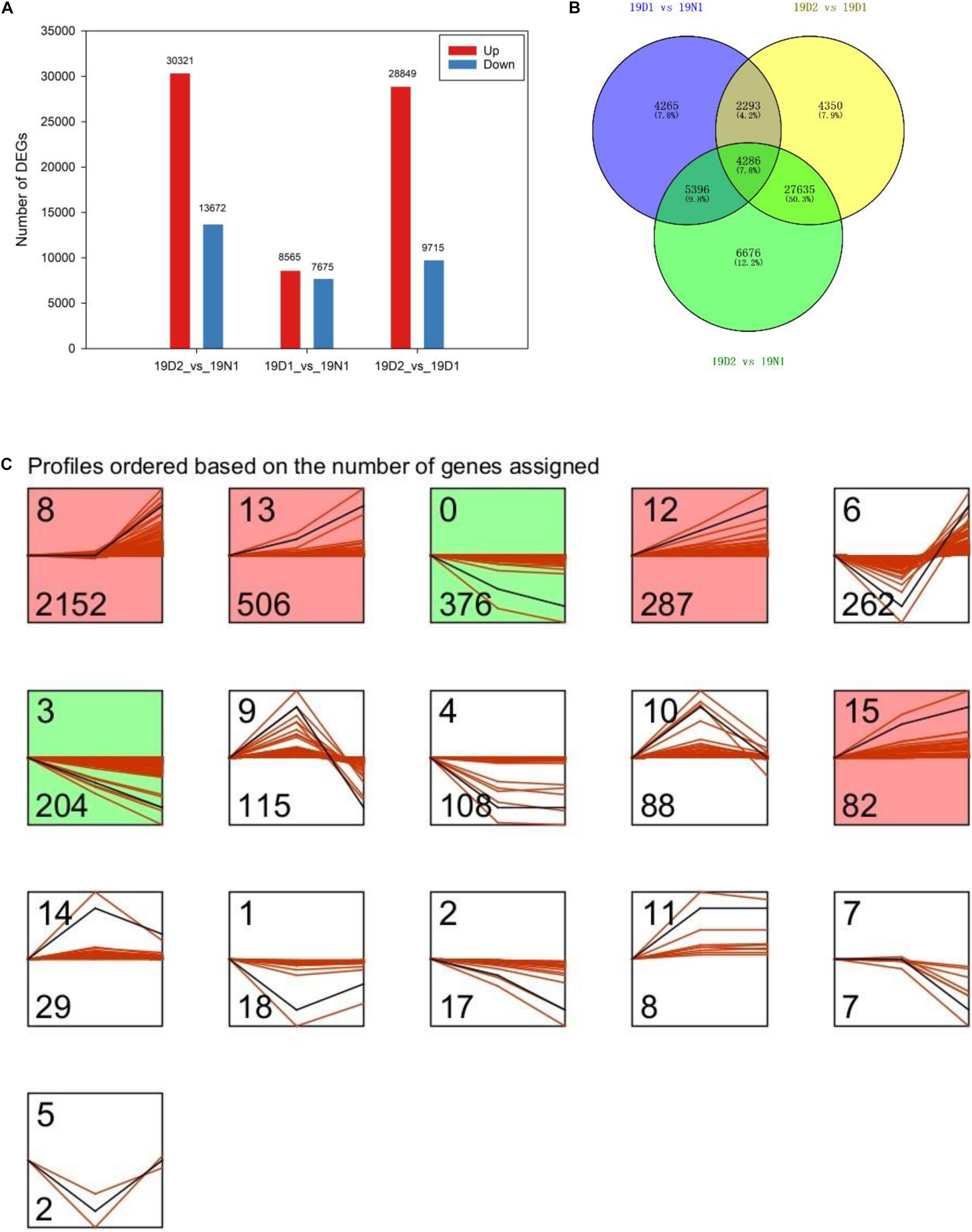
Figure 3. Expression patterns of differentially expressed genes (DEGs) during different dormancy phases in Magnolia wufengensis. (A) Changes in gene expression profile during different dormancy phases. (B) Venn diagram of DEGs between 19D1 versus 19N1, 19D2 versus 19N1, and 19D2 versus 19D1. (C) Trend analysis of 4,286 intersecting DEGs in panel (B).
Compared to 19N1, GO analysis of DEGs at 19D1 demonstrated that genes related to membrane structure and transcription were overexpressed. Biological process, cellular component, and molecular function, “single-organism transport,” “cellular component,” and “oxidoreductase activity” were the most enriched GO categories. At 19D2, in biological process, the major subcategories were “metabolic process” and “single-organism process.” In cellular component, “cellular component,” “cell part,” and “intracellular part” were the most representative subcategories, and “oxidoreductase activity,” “transporter activity,” and “RNA binding” were the top three subcategories compared to 19N1.
Compared to 19N1, KEGG pathway enrichment analysis for DEGs indicated that four pathways – “ribosome (map03010),” “oxidative phosphorylation (map00190),” “plant-pathogen interaction (map04626)” and “plant hormone signal transduction (map04075)” were significantly enriched at 19D1. At 19D2 versus 19N1, “spliceosome (map03040)” followed by “oxidative phosphorylation (map00190),” and “plant hormone signal transduction (map04075)” were significantly enriched. Compared with D1, “ribosome (map03010),” “oxidative phosphorylation (map00190),” and “spliceosome (map03040),” were significantly enriched at 19D2 (Supplementary Table 2).
Based on KEGG annotation, DEGs related to phytohormones play an important role in dormancy transition. A total of 51 DEGs related to plant hormone signal transduction (Supplementary Table 3) divided into three main gene clusters as shown in Figure 4. Cluster A (13 genes) was highly expressed at the 19N1 (ED) stage, and showed low expression levels during dormancy release, which indicated that the genes in this cluster may be involved in breaking ED. Among the DEGs in cluster A, two genes involved in ABA signalling, PP2C and ABF, and five auxin-related genes, including three IAA, and one each of GH3 and AUX, were found, which indicates that ABA and auxin signalling were activated during dormancy release. Cluster B contained 14 genes that showed low expression level at N1, and sharply increased at 19D1before decreasing to a relatively low level at 19D2. Among them, DEGs associated with auxin (IAA, SAUR, ARF, and T1R1), ethylene (EBF and ETR), and brassinosteroid (TCH4) regulation were most abundant in this expression profile cluster. Cluster C, with the largest number of DEGs (24 genes), exhibited a low expression level at 19N1, which gradually increased at 19D1 and part of 19D2. Among them, DEGs that responded to jasmonic acid (COI1, JAZ, and JAR1), cytokinin (ARR-A), auxin (SAUR, ARF, IAA and AUX1), and brassinosteroid (TCH4 and BZR1) exhibited a similar expression pattern to that in cluster C.
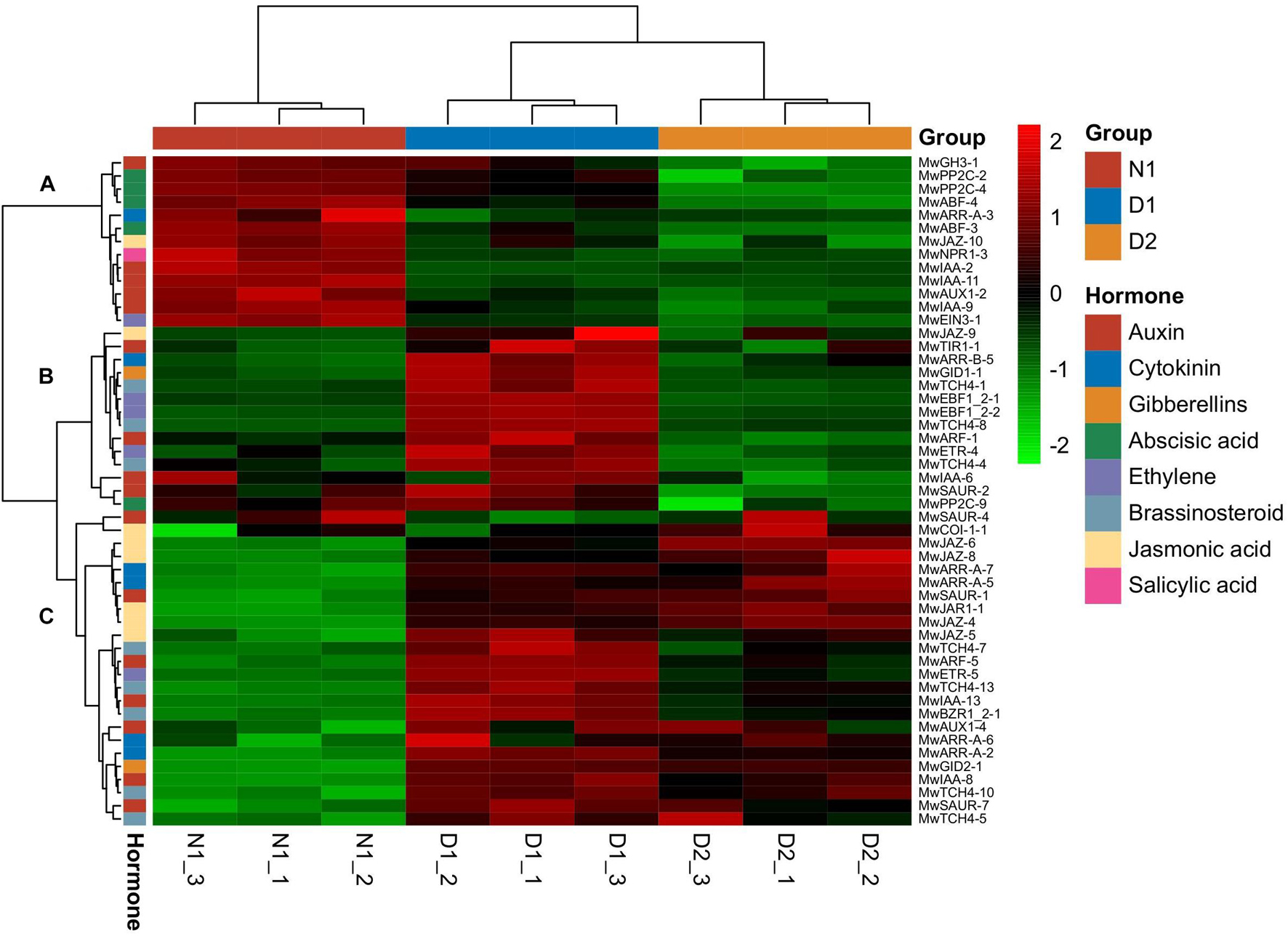
Figure 4. Heatmap of differentially expressed genes (DEGs) involved in plant hormone signal transduction (map04075) during different dormancy phases at 2019–2020 in Magnolia wufengensis.
A total of 1,001 TF genes (552 upregulated and 449 downregulated) that were active during dormancy transition were identified. These TFs were mainly concentrated in MYB_superfamily, C2H2, C2C2, bHLH, bZIP, AP2/ERF, NAC, and WRKY. Among all the evaluated genes, the number of genes of the MYB_superfamily was the highest with 131 genes (59 upregulated and 72 downregulated), followed by C2H2 with 103 genes (90 upregulated and 13 downregulated), C2C2 with 93 genes (56 upregulated and 37 downregulated), bHLH with 86 genes (45 upregulated and 41 downregulated), bZIP with 80 genes (64 upregulated and 16 downregulated), AP2/ERF with 66 genes (35 upregulated and 31 downregulated), NAC with 55 genes (25 upregulated and 30 downregulated), and WRKY with 48 genes (26 upregulated and 22 downregulated) (Supplementary Table 4).
Ten DEGs were randomly selected to demonstrate the reliability of RNA-seq using qPCR. The trends of genes during different dormancy phases using qPCR were consistent with the RNA-seq results, indicating favourable reliability of RNA-seq (Supplementary Figure 5).
Based on the transcriptional analysis, we analysed and inferred that hormone metabolism, signal transduction, and especially ABA may play important roles in dormancy transition. Therefore, to further figure out the function of ABA with respect to ED release, three different exogenous concentrations of ABA (100, 200, and 300 μM) were applied to apical buds on 20N1, 20N2, 20D1, 20D2, 21J1, and 21J2 in M. wufengensis, and their BBP with exogenous ABA and mock treatments (0.2% ethanol) was compared after 32 days. Based on the evaluated dormancy status, 20N1 and 20N2 were in ED and ready to release from ED, and 20D1, 20D2, 21J1, and 21J2 were in ECD. BBP on 20N1 and 20D1 significantly decreased after treatment with 100 μM ABA (Figure 5), whereas ABA at concentration had almost no effect on the germination rate in ECD (20D2, 21J1 and 21J2). This implies that during ED or the ED release phase, ABA at a concentration of 100 μM played a positive role in ED maintenance and was therefore selected for further investigation.
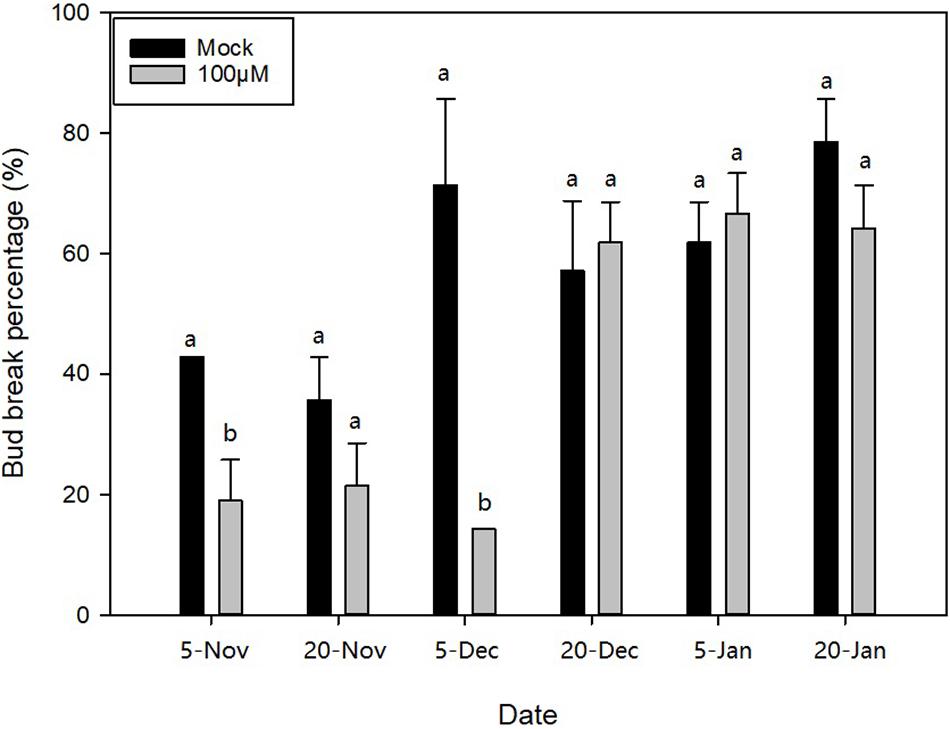
Figure 5. Bud break percentage after abscisic acid (ABA) treatment during endodormancy (ED) to ecodormancy (ECD) during 2020–2021 in Magnolia wufengensis. Each experiment was performed with three biological replicates. Different letters above bars indicate a significant difference among bud break percentage under different concentration of ABA according to ANOVA and LSD test (p < 0.05).
To further study the molecular mechanism of action of ABA on ED release, one NCED, three CYP707A, two PYL, five PP2C, one SNRK2, and three ABI DEGs related to ABA synthesis, metabolism, and signalling were identified using RNA-seq. Among these genes, the expression of MwNCED-3 was downregulated and showed a low expression before dormancy release, and then increased during dormancy release (Figure 6A). The expression of MwCYP707A-1-2 declined toward ED release and steadily increased after ED release, which is consistent with the content of ABA, whereas almost no expression of MwCYP707A-1-1 and MwCYP707A-2 was observed during the entire dormancy release phase (Figure 6B). Moreover, the expression patterns of genes related to ABA signalling were also determined. The expression levels of MwPYL-1/3 genes decreased before dormancy release and increased rapidly during dormancy release, and then decreased steadily at ECD. The same expression pattern was observed in MwPP2C-6. On the contrary, MwSNRK2-10 slowly increased before dormancy release, peaked on 20N2, decreased rapidly during dormancy release, and increased thereafter at ECD. In addition, MwPP2C-24 and MwABI-5 increased before dormancy release and dropped with ECD development (Figure 6C).
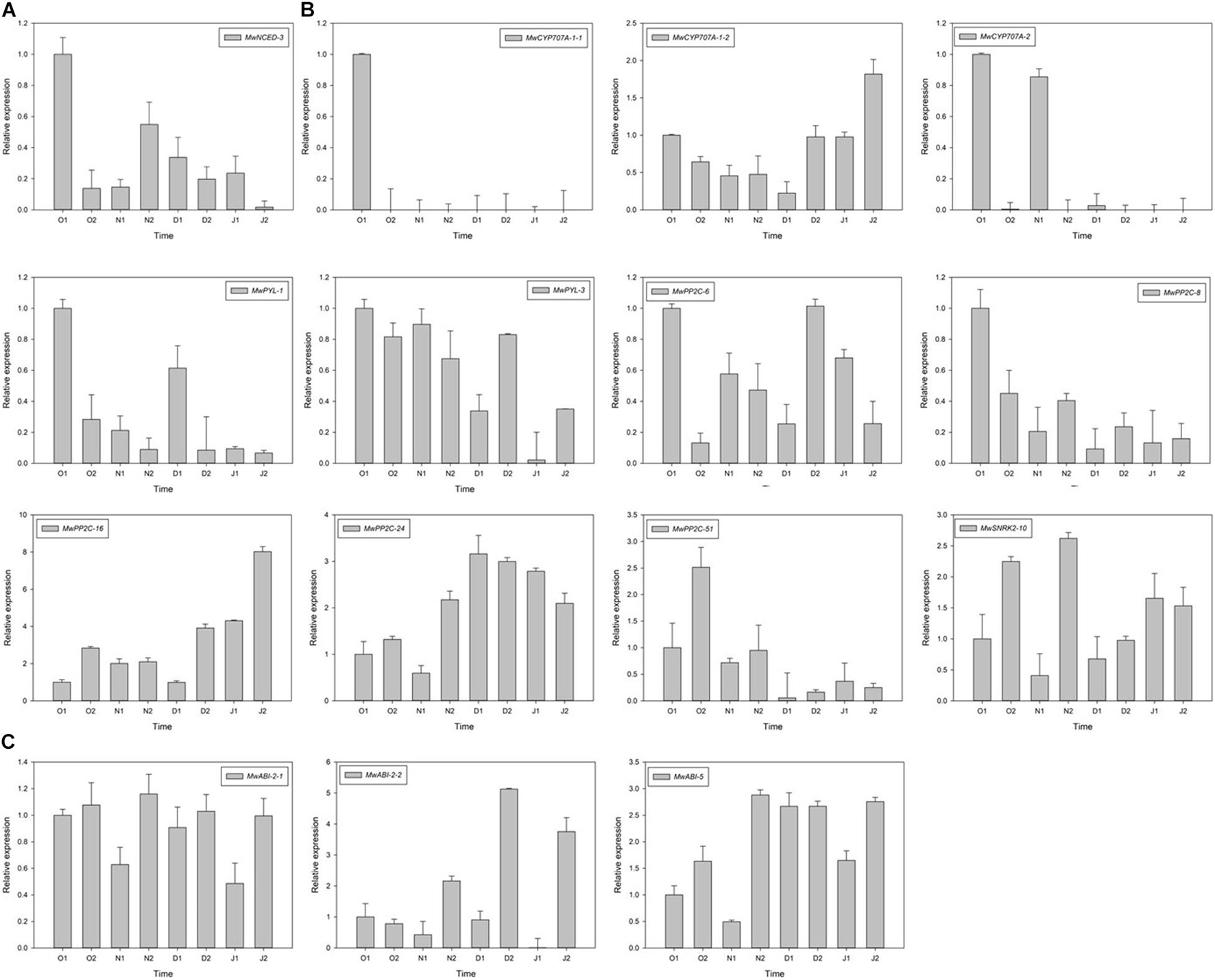
Figure 6. Relative gene expression levels of differentially expressed genes (DEGs) involved in abscisic acid (ABA) biosynthesis (A), metabolism (B), and signal transduction (C) under natural condition during 2020–2021 in Magnolia wufengensis. Each experiment was performed with three biological replicates. Each bar represents the mean ± SEM of three biological replicates.
To understand the relationship between dormancy transition and cold tolerance in winter, we identified one CBF1 and two ICE1 genes in our transcriptome data and measured their expression patterns during the natural ED process in M. wufengensis. To confirm their functions on cold resistance, we conducted an experiment under cold acclimation and found that MwCBF-1 and MwICE-1-1 were induced under cold stress, indicating that the two genes were associated with cold tolerance (Supplementary Figure 6). Among these genes, the expression of MwCBF-1 was upregulated to 20N2 and showed a high expression before dormancy release, and then decreased but still kept a high expression after ED release. The expression of MwICE-1-1 decreased to N1 and then increased slowly at ECD. The expression level of MwICE-1-2 showed a high expression at 20D2 but maintained a relatively low expression at other phases (Figure 7).
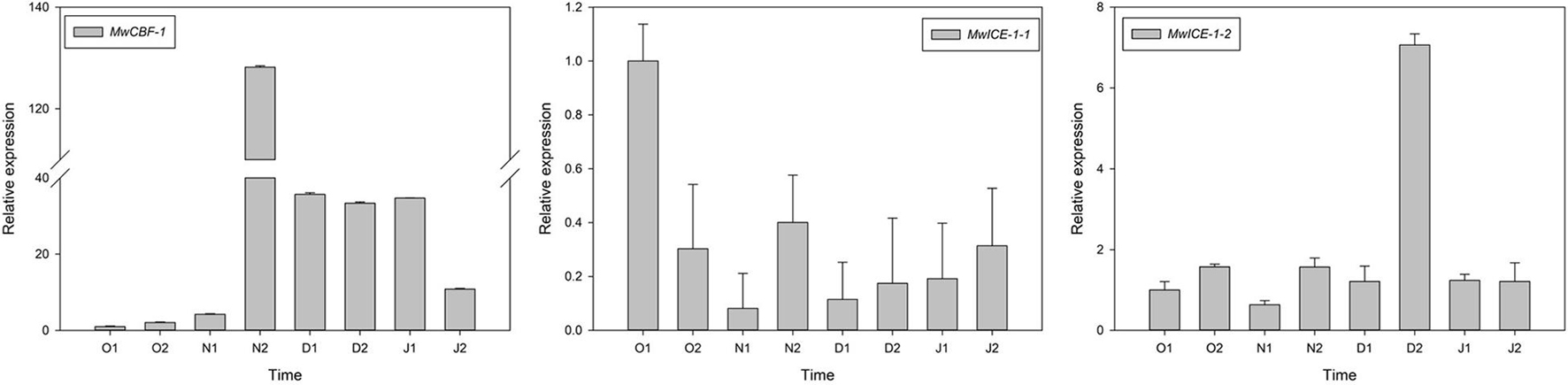
Figure 7. Relative gene expression levels of differentially expressed genes (DEGs) involved in cold resistance under natural condition during 2020–2021 in Magnolia wufengensis. Each experiment was performed with three biological replicates. Each bar represents the mean ± SEM of three biological replicates.
The significantly decreased BBP indicated that ABA promoted ED maintenance. To further determine the function of ABA in the maintenance of ED, the responses of buds collected at 20D1 were compared between mock and ABA treatment group. Genes related to dormancy transition under natural conditions were focussed on (Figure 8). The expression of MwCBF-1 induced by ABA was considerably upregulated compared to that in the mock treatment and a similar increase was observed in MwPYL-1 and MwABI-5. Fluctuations were observed in the expression of MwNCED-3 and MwPYL-3 during the whole treatment time. In addition, a slight decreasing trend can be seen in the expression of the two PP2C genes in the previous 21 days, while there was a significant increase in MwPP2C-6 on the 28th day after treatment which was the opposite to the trend observed for MwSnRK-2-10. The expression of MwCYP707A-1-2 was similar to that of MwPP2C-6, showing a high expression at the 7th and 28th days and maintaining a relatively low expression at the 14th and 21st days after ABA treatment.
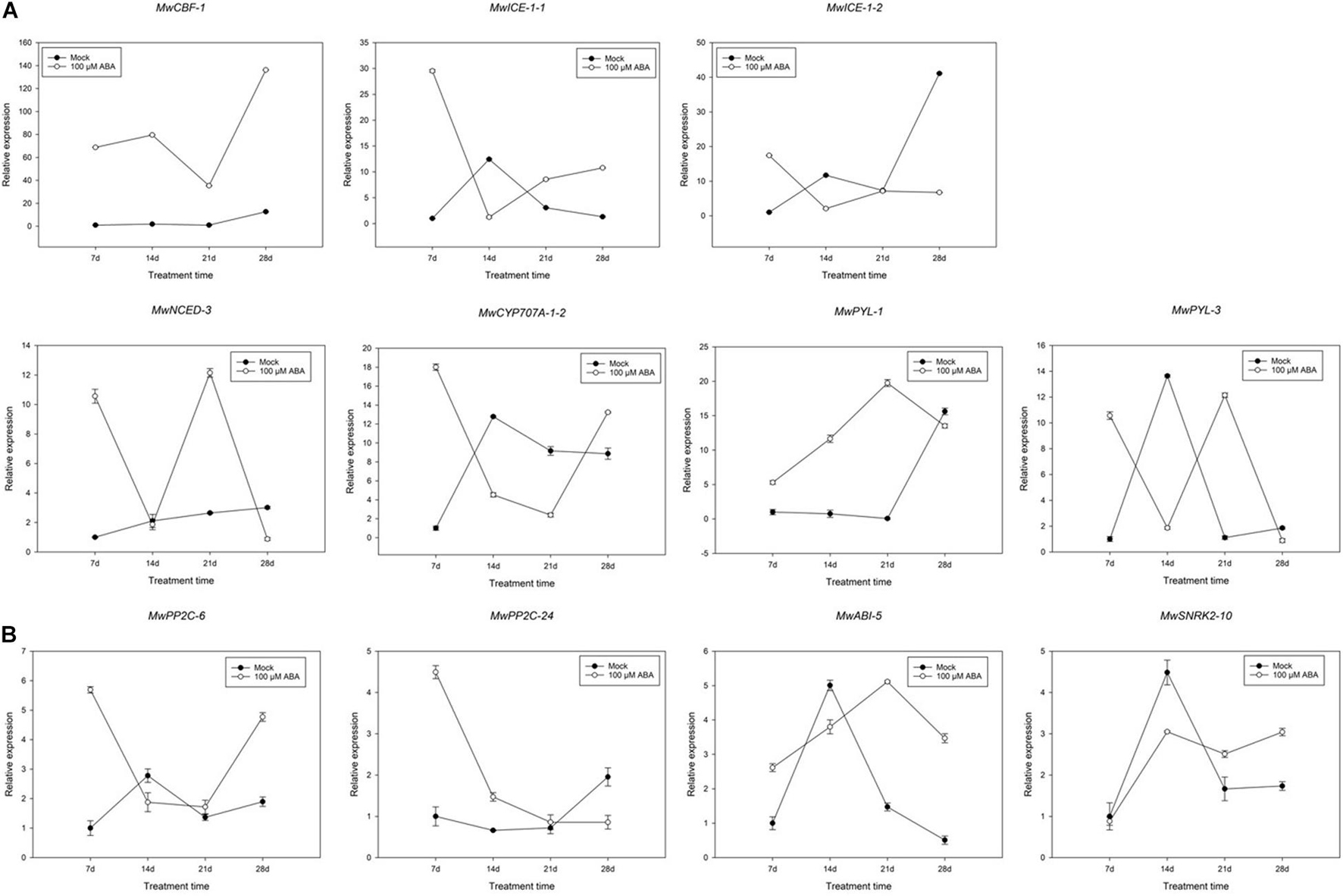
Figure 8. Relative gene expression levels of specific differentially expressed genes (DEGs) involved in abscisic acid (ABA) (A) and cold resistance (B) under ABA treatment during 2020–2021 in Magnolia wufengensis. Each experiment was performed with three biological replicates. Each bar represents the mean ± SEM of three biological replicates.
In addition to analysing the expression pattern of ABA and cold-related genes, several genes such as D-type cyclin (CYCD), PKL and CALS1 involving in bud dormancy have also been identified. Among the genes, one CYCD3, one PKL and two CALS1 genes were differently expressed during dormancy transition. To further study whether a similar model induced by ABA exists in M. wufengensis, we measured the expression of the dormancy-related genes under ABA treatment (Figure 9). The expression of MwCYCD-3 increased in the previous 21 days and was considerably promoted by ABA and a similar trend exists in MwCALS-1-1 in the first 21 days. In addition, the expression of MwPKL was depressed by ABA throughout the treatment time except the 21st day. During dormancy release, the expression MwCYCD3 and MwCALS-1-1/2 were promoted by ABA, but MwPKL expression was depressed by ABA.
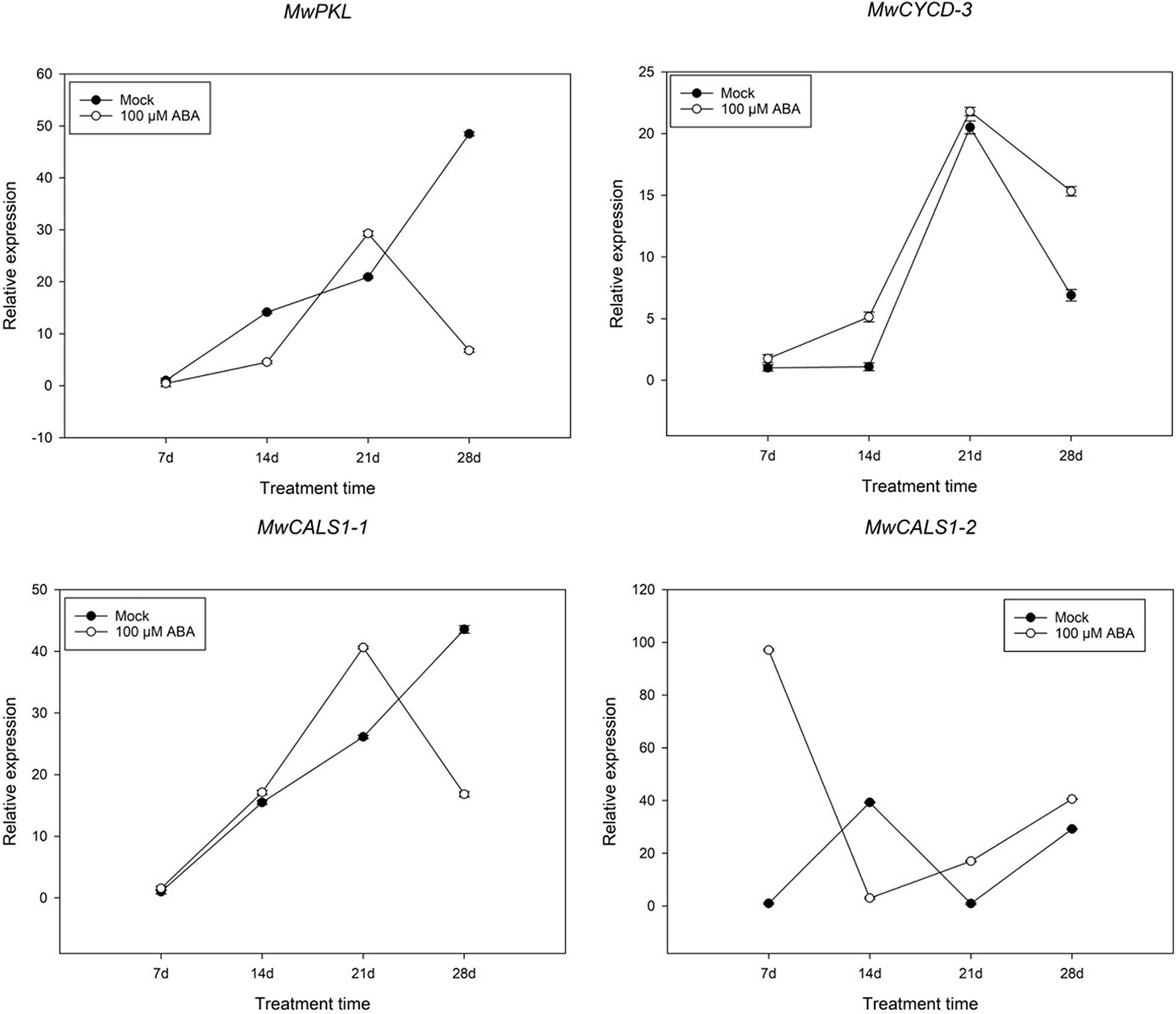
Figure 9. Relative gene expression levels of specific differentially expressed genes (DEGs) involved in bud dormancy under ABA treatment during 2020–2021 in Magnolia wufengensis. Each experiment was performed with three biological replicates. Each bar represents the mean ± SEM of three biological replicates.
The 0–7.2°C and Utah models are widely used to assess CRs in perennial trees (Yang et al., 2021). As shown in Figure 1B, temperature steadily dropped to 7.2°C relatively late, so no CUs were accumulated before 20N1 under the 0–7.2°C model. The Utah model is often used for cold regions such as north China, as it takes into consideration the accumulation effect of different temperatures (Erez et al., 1990). Our results were thus consistent with those of previous studies. Based on the Utah model, M. wufengensis underwent lesser CR to break dormancy than M. denudata. In addition, we found that the lower the temperature, the higher the BBP. Based on our results, the buds accumulated enough CUs to break ED before 20D1.
Bud dormancy is associated with phytohormones in many species (Cooke et al., 2012). GA and ABA regulate dormancy induction and release (Yue et al., 2018; Yang et al., 2019). In general, ABA levels increase with the establishment of dormancy (PD to ED) and decrease during dormancy release (ED to ECD) while GA3 exhibits opposite trends during dormancy transition. For example, in wintersweet, the content of ABA increases with the chilling accumulation from PD to ED and then decreases after dormancy release, and that of GA3 decreases with the length of dormancy (Li et al., 2020). Similar results have been observed in many other perennial species such as pear (P. pyrifolia) (Tuan et al., 2017; Ito et al., 2021), peach (Prunus persica) (Wang et al., 2016), grape (V. vinifera) (Zheng et al., 2015), and leafy spurge (Euphorbia esula) (Chao et al., 2017). In addition, besides changes of GAs content, we also found that the content of GA3 was higher than the other two GAs, indicating an important role GA3 plays in dormancy transition of M. wufengensis.
Many DEGs were significantly annotated to hormones during dormancy transition based on KEGG annotation. Therefore, the effect of hormones on dormancy was focussed on. In M. wufengensis, the ABA content decreased during dormancy release but increased rapidly from 20D2 and 21J2, and GA3 content steadily increased after dormancy release, similar those observed in other species, which suggests a relationship between content of ABA and GA3 and the depth of ED. Based on our records, the average temperature during this stage was –4.4°C. Therefore, we hypothesised that a protective strategy to cope with low temperatures may exist in M. wufengensis. In addition, a homeostatic network of various hormones is thought to be at the centre of dormancy transition (Shu et al., 2013; Zhang et al., 2018). In C. praecox, the ratio of ABA/GA3 increases with dormancy breaking (induced by chilling) and decreases during dormancy release in P. mume (Wen et al., 2016; Li et al., 2020). Thus, these results are consistent with those of other studies.
Studies of exogenous hormone application have shown that exogenous ABA application can effectively promote dormancy establishment and maintain ED (Zheng et al., 2015; Li and Dami, 2016). Based on previous studies, inhibitory effect of exogenous ABA does not always work all the time and depends on status of buds. In pear (Pyrus fauriei), ABA inhibitory effect on bud break could be affected by chilling accumulation (Tamura et al., 2002) and in grape (V. vinifera), ABA application to buds which are released from dormancy did not reduce BBP (Zheng et al., 2015). Similar results were found in the present study as 100 μM ABA delayed dormancy release before ECD, whereas 200 and 300 μM ABA could not effectively inhibit BBP. On the one hand, inhibition was not dependent on concentration, which is not consistent with the results observed in grape whose BBP is more efficiently inhibited by high concentration of ABA (Zheng et al., 2015). On the other hand, high concentration of ABA could not suppress bud break in M. wufengensis during dormancy release, which is different from results in pear whose BBP with 100, 200, and 300 μM ABA was similar (Li et al., 2018). Compared to 100 μM whose concentration is relatively low, we infer that 200 or 300 μM ABA may damage and then stimulus the defence system of buds in M. wufengensis, so higher concentration of ABA treatment has not depressed bud break percentage. These results suggest that a non-ABA-regulated controlling mechanism of dormancy may exist in M. wufengensis, and the mechanism needs further study. However, as our experiment was conducted in a climate chamber after buds were cut from trees, whether similar results can be observed in field experiments remains to be investigated.
In addition to physiological data about hormones, we further analysed the data at the molecular level. The content of ABA in plants is not dominated by a single factor but by a balance of biosynthesis and metabolism, and function through various signalling pathways. Therefore, we further analysed the changes in ABA content during dormancy by evaluating ABA biosynthesis, metabolism, and signalling. NCED and CYP707A are two of the main genes involved in ABA biosynthesis and metabolism, respectively. Overexpression of NCED promotes seed dormancy and delays germination in tobacco (Nicotiana plumbaginifolia) (Qin and Zeevaart, 2002). Li et al. (2018) found that PpNCED-2 and PpNCED-3 are highly expressed during ED and decrease rapidly during dormancy release. In this transcriptome data, we identified one NCED DEG and named the gene MwNCED-3. The expression pattern was similar to PpNCED-3 and consistent with the decrease in ABA content during ED released.
Furthermore, CYP707A is known to be highly expressed during dormancy release in peach and pear (Wang et al., 2016; Tuan et al., 2017). Recent studies have shown that CYP707A is involved in dormancy release regulation. In potato, ABA content changes are correlated with NCED and CYP707A gene families and associated with dormancy release in tubers (Destefano-Beltrán et al., 2006). In the present study, the expression of MwCYP707A-1-2 was downregulated during dormancy release and immediately upregulated during ECD (Figure 6B). This result is not consistent with those of the abovementioned research in peach or pear. Based on these results, we infer that MwCYP707A-1-2 may be a key gene involved in ED release, and the role of MwCYP707A-1-2 in bud dormancy of M. wufengensis needs to be further studied.
ABA signal transduction is also involved in dormancy regulation and bud dormancy. In pear, Bai et al. (2013) found that PpPP2Cs were upregulated while PpSNRK2s were downregulated after dormancy release. Similar results were observed by Li et al. (2018) who found the expression of PpPYLs, PpSNRK2s, and PpABIs to be upregulated from PD to ED, but the expression of PpPP2Cs was low during ED and increased with the decrease of ABA content during dormancy release. The expression of VvPP2Cs higher at PD than at ED (Tuan et al., 2017). From our results, genes associated with ABA signalling showed different expression patterns during dormancy transition. MwPP2C-6/8/51, MwPYL-1/3, and MwSNRK2-10 showed a relatively high expression in ED and ED release, while the expression of MwPP2C-16/24 and MwABI-5/2-2 in ECD was higher than that in ED. However, the consistency between the trends of their expression and dormancy transition was not significant and their functions need to be further studied.
In addition, ABA could not only influence the expression of ABA-related genes, but also involve in regulation of many dormancy-related genes. In the present study, these dormancy-related genes such as CYCD, PKL and CALS1 were differentially affected by ABA: MwCYCD-3 and MwCALS-1-1/2 were promoted by ABA and MwPKL was depressed by ABA which indicated that a similar ABA-centred model of H. Aspen exists in dormancy release M. wufengensis. In addition, functions of the dormancy-related genes need to be further study.
Bud dormancy, an important biological process for plants to survive the winter, is strongly related to cold hardiness enhancement in winter. Li et al. (2003) found that cold tolerance is enhanced before dormancy development in silver birch (B. pendula). In the winter, perennial plants tend to enhance cold tolerance through natural cold acclimation to survive under long-term cold conditions (Uemura et al., 1995; Lee and Thomashow, 2012). The CBF-dependent signalling pathway is an important cold signalling pathway in plants. CBFs/DREBs bind to cis-elements of cold resistance genes and activate their expression, thus improving cold resistance in plants (Park et al., 2015; Ding et al., 2020). ICE1, a positive regulator of cold response, can activate the expression of CBF (Chinnusamy, 2003; Lee et al., 2005). DAM/SVP can regulate the bud dormancy cycle in perennial trees (Falavigna et al., 2019). Recently, the relationship between CBF and DAM co-regulating dormancy has been widely studied in many species (Yang et al., 2021). In P. mume, PmDAM6 and PmCBFs mainly respond to chilling temperature (below 20°C) and freezing cold (0°C), respectively. Additionally, eight PmCBFs were upregulated under the stimulus of a cold signal, which then induced the expression of all six DAM genes during dormancy development (Zhang et al., 2018; Zhao et al., 2018a). In the Populus hybrid, PtCBF1 and PtDAM1 induction was found to be related to ED development (Boldizsár et al., 2021). In pear, Niu et al. (2015) found that PpCBF can induce the expression of PpDAM and PpDAM and subsequently inhibit PpFT, which then stimulates growth cessation and promote dormancy maintenance. In the present study, we identified one DREB1B/CBF1 gene (TRINITY_DN8378_c0_g1) and two ICE1 genes (TRINITY_DN20323_c1_g1 and TRINITY_DN20323_c0_g2). MwCBF-1 was highly expressed during overwintering, so we suspected that MwCBF-1 was a positive regulator for cold resistance in M. wufengensis. Before dormancy release, MwCBF-1 achieved a peak at 20N2, while the temperature was still above 0°C (Supplementary Figure 7). In the coldest two months, which included D1, D2, J1, and J2, MwCBF-1 decreased but maintained a high expression to cope with cold stress when dormancy was released (Figure 8). Furthermore, the expression of MwCBF-1 can be efficiently induced by exogenous ABA application. We suspected that M. wufengensis can efficiently enhance cold resistance during the dormancy release phase to survive the winter, so it is important to maintain ED and extend the time of the dormancy release phase. Above all, a hypothesis of molecular model for ABA and its biosynthesis, metabolism and signalling pathway; cold tolerance and acclimation; and dormancy during overwintering was proposed (Figure 10). In addition, the role that ECD plays in cold tolerance enhancement during overwintering needs to be further study.
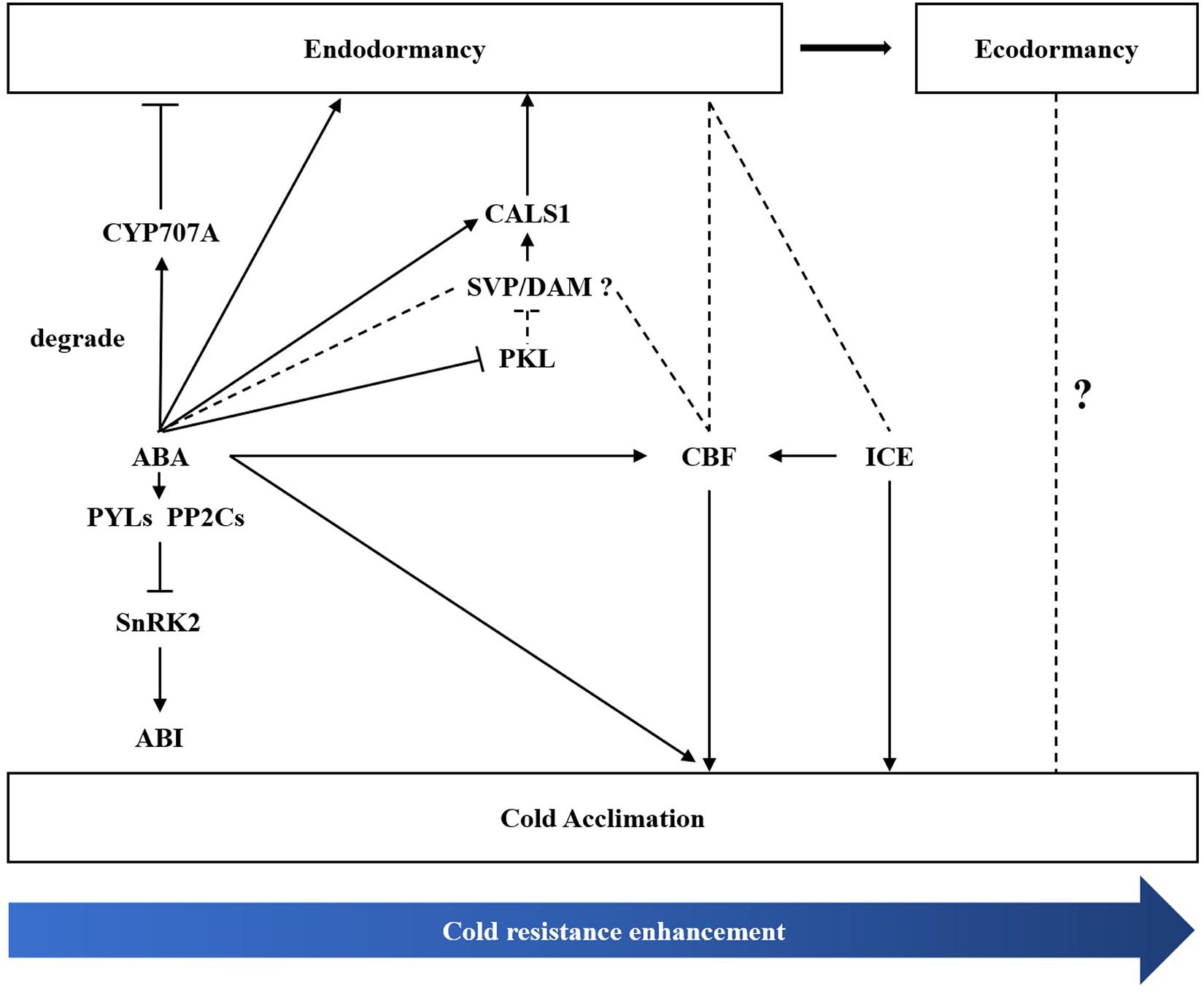
Figure 10. A hypothesis of molecular regulation model between bud dormancy and cold acclimation during winter in Magnolia wufengensis.
Overall, this study provides fundamental insights into the bud dormancy cycle and CR in two Magnoliaceae plants and we hypothesised that M. wufengensis and M. denudata are both sensitive to low temperature and short day based on meteorological data. The content of ABA and GA3, and the ABA/GA3 ratio significantly changed during dormancy release and ECD. A M. wufengensis dataset containing 187,406 unigenes was constructed to observe the dynamic changes in gene expression under different dormancy phases using RNA-seq. Comparison with ED led to the identification of 16,240 and 43,993 DEGs during ED release (19D1) and ECD (19D2), respectively. Among the DEGs, many key genes and metabolic pathways, especially those of plant hormones, were identified using KEGG and GO analyses. Based on heatmap analysis of plant hormone transduction, we found that auxin- and ABA-related genes showed high expression in ED. Thus, auxin and ABA may regulate dormancy transition in M. wufengensis. Application of 100 μM of exogenous ABA before dormancy release could effectively maintain dormancy. Seventeen DEGs involved in ABA biosynthesis, metabolism, and signal transduction were identified based on RNA-seq data. We conducted qPCR on the 17 ABA-related DEGs and found that MwCYP707A-1-2 may be involved in dormancy regulation. Besides, MwCBF-1 was highly expressed during dormancy release, suggesting a relationship between cold tolerance and bud dormancy. Thus, our findings shed light on the mechanism underlying dormancy release and further our understanding of overwintering from bud dormancy in M. wufengensis.
The original contributions presented in the study are publicly available. This data can be found here: National Center for Biotechnology Information (NCBI) BioProject database under accession number PRJNA695868 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA695868).
KW performed most of the experiment, analysed the data, and drafted the manuscript. KW and XD designed the experiment, did the bioinformatics analysis, and contributed to the writing of this article. ZZ and ZS provided the experimental materials and participated in data analysis. YZ involved in conducting experiments. HL analysed the data. ZJ and LM initiated and supervised the study. All authors contributed to the article and approved the submitted version.
This research was supported by lateral research of Beijing Forestry University (662004284).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are grateful to Forestry Bureau of Wufeng County and Boling Technology for providing materials.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.742504/full#supplementary-material
Anderson, J. V., Horvath, D. P., Chao, W. S., and Foley, M. E. (2010). “Bud dormancy in perennial plants: a mechanism for survival,” in Dormancy and Resistance in Harsh Environments. Topics in Current Genetics, eds E. Lubzens, J. Cerda, and M. Clark (Berlin, Heidelberg: Springer), 69–90.
Arora, R., Rowland, L. J., and Tanino, K. (2003). Induction and release of bud dormancy in woody perennials: a science comes of age. Hortscience 38, 911–921. doi: 10.21273/HORTSCI.38.5.911
Atkinson, C., Brennan, R. M., and Jones, H. (2013). Declining chilling and its impact on temperate perennial crops. Environ. Exper. Bot. 91, 48–62. doi: 10.1016/j.envexpbot.2013.02.004
Bai, S., Niu, Q., and Teng, Y. (2016). Advances in the research of the regulatory mechanism of endodormancyin pear (in chinese). J. Fruit Sci. 33, 1–9.
Bai, S., Saito, T., Sakamoto, D., Ito, A., Fujii, H., and Moriguchi, T. (2013). Transcriptome analysis of japanese pear (pyrus pyrifolia nakai) flower buds transitioning through endodormancy. Plant Cell Physiol. 54, 1132–1151. doi: 10.1093/pcp/pct067
Boldizsár, Á, Soltész, A., Tanino, K., Kalapos, B., Marozsán-Tóth, Z., Monostori, I., et al. (2021). Elucidation of molecular and hormonal background of early growth cessation and endodormancy induction in two contrasting populus hybrid cultivars. BMC Plant Biol. 21:111. doi: 10.1186/s12870-021-02828-7
Chao, W. S., Doğramacı, M., Horvath, D. P., Anderson, J. V., and Foley, M. E. (2017). Comparison of phytohormone levels and transcript profiles during seasonal dormancy transitions in underground adventitious buds of leafy spurge. Plant Mol. Biol. 94, 281–302. doi: 10.1007/s11103-017-0607-7
Chinnusamy, V. (2003). Ice1: a regulator of cold-induced transcriptome and freezing tolerance in arabidopsis. Genes Dev. 17, 1043–1054. doi: 10.1101/gad.1077503
Chinnusamy, V., Zhu, J., and Zhu, J. (2007). Cold stress regulation of gene expression in plants. Trends Plant Sci. 12, 444–451. doi: 10.1016/j.tplants.2007.07.002
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., and Robles, M. (2005). Blast2go: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Considine, M. J., and Considine, J. A. (2016). On the language and physiology of dormancy and quiescence in plants. J. Exper. Bot. 67, 3189–3203. doi: 10.1093/jxb/erw138
Cooke, J. E. K., Eriksson, M. E., and Junttila, O. (2012). The dynamic nature of bud dormancy in trees: environmental control and molecular mechanisms. Plant Cell Environ. 35, 1707–1728.
Cutler, A. J., and Krochko, J. E. (1999). Formation and breakdown of aba. Trends Plant Sci. 4, 472–478. doi: 10.1016/S1360-1385(99)01497-1
Deng, S., Ma, J., Zhang, L., Chen, F., Sang, Z., Jia, Z., et al. (2019). De novo transcriptome sequencing and gene expression profiling of magnolia wufengensis in response to cold stress. BMC Plant Biol. 19:321. doi: 10.1186/s12870-019-1933-5
Destefano-Beltrán, L., Knauber, D., Huckle, L., and Suttle, J. C. (2006). Effects of postharvest storage and dormancy status on aba content, metabolism, and expression of genes involved in aba biosynthesis and metabolism in potato tuber tissues. Plant Mol. Biol. 61, 687–697. doi: 10.1007/s11103-006-0042-7
Ding, Y., Shi, Y., and Yang, S. (2020). Molecular regulation of plant responses to environmental temperatures. Mol. Plant 13, 544–564. doi: 10.1016/j.molp.2020.02.004
Duan, X., Cai, C., Yang, Y., Chen, F., Sang, Z., and Ma, L. (2019). Fall ethephon application enhances the freezing tolerance of magnolia wufengensis during overwintering. Forests 10:868. doi: 10.3390/f10100868
Erez, A., Fishman, S., Linsley-Noakes, G. C., and Allan, P. (1990). The dynamic model for rest completion in peach buds. Leuven, Belgium: International Society for Horticultural Science (ISHS).
Ernst, J., and Bar-Joseph, Z. (2006). Stem: a tool for the analysis of short time series gene expression data. BMC Bioinformatics 7:191. doi: 10.1186/1471-2105-7-191
Falavigna, V., Costes, E., and Andrés, F. (2019). I want to (bud) break free: the potential role of dam and svp-like genes in regulating dormancy cycle in temperate fruit trees. Front. Plant Sci. 9:1990. doi: 10.3389/fpls.2018.01990
Fishman, S., Erez, A., and Couvillon, G. A. (1987a). The temperature dependence of dormancy breaking in plants: mathematical analysis of a two-step model involving a cooperative transition. J. Theor. Biol. 124, 473–483. doi: 10.1016/S0022-5193(87)80221-7
Fishman, S., Erez, A., and Couvillon, G. A. (1987b). The temperature dependence of dormancy breaking in plants: computer simulation of processes studied under controlled temperatures. J. Theor. Biol. 126, 309–321. doi: 10.1016/S0022-5193(87)80237-0
Gao, Y., Yang, Q., Yan, X., Wu, X., Yang, F., Li, J., et al. (2021). High-quality genome assembly of ‘cuiguan’ pear (pyrus pyrifolia) as a reference genome for identifying regulatory genes and epigenetic modifications responsible for bud dormancy. Horticulture Research 8:197. doi: 10.1038/s41438-021-00632-w
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from rna-seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Guo, X., Liu, D., and Chong, K. (2018). Cold signaling in plants: insights into mechanisms and regulation. J. Integ. Plant Biol. 60, 745–756. doi: 10.1111/jipb.12706
Hao, X., Yang, Y., Yue, C., Wang, L., Horvath, D. P., and Wang, X. (2017). Comprehensive transcriptome analyses reveal differential gene expression profiles of camellia sinensis axillary buds at para-, endo-, ecodormancy, and bud flush stages. Front. Plant Sci. 8:553. doi: 10.3389/fpls.2017.00553
Heide, O. M. (2008). Interaction of photoperiod and temperature in the control of growth and dormancy of prunus species. Sci. Horticult. 115, 309–314. doi: 10.1016/j.scienta.2007.10.005
Horvath, D. P., Anderson, J. V., Chao, W. S., and Foley, M. E. (2003). Knowing when to grow: signals regulating bud dormancy. Trends Plant Sci. 8, 534–540. doi: 10.1016/j.tplants.2003.09.013
Hubbard, K. E., Nishimura, N., Hitomi, K., Getzoff, E. D., and Schroeder, J. I. (2010). Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev. 24, 1695–1708. doi: 10.1101/gad.1953910
Ito, A., Tuan, P. A., Saito, T., Bai, S., Kita, M., and Moriguchi, T. (2021). Changes in phytohormone content and associated gene expression throughout the stages of pear (pyrus pyrifolia nakai) dormancy. Tree Physiol. 41, 529–543. doi: 10.1093/treephys/tpz101
Lang, G. A., Early, J. D., Darnell, R., and Martin, G. C. (1987). Endo-, para-, and eco-dormancy: physiological terminology and classification for dormancy research. Hortscience 22, 371–377.
Lee, B., Henderson, D. A., and Zhu, J. (2005). The arabidopsis cold-responsive transcriptome and its regulation by ice1. Plant Cell 17, 3155–3175. doi: 10.1105/tpc.105.035568
Lee, C. M., and Thomashow, M. F. (2012). Photoperiodic regulation of the c-repeat binding factor (cbf) cold acclimation pathway and freezing tolerance in arabidopsis thaliana. Proc. Natl. Acad. Sci. 109, 15054–15059. doi: 10.1073/pnas.1211295109
Li, B., and Dewey, C. N. (2011). Rsem: accurate transcript quantification from rna-seq data with or without a reference genome. BMC Bioinform. 12:323. doi: 10.1186/1471-2105-12-323
Li, C., Junttila, O., Ernstsen, A., Heino, P., and Palva, E. T. (2003). Photoperiodic control of growth, cold acclimation and dormancy development in silver birch (betula pendula) ecotypes. Physiol. Plantarum 117, 206–212. doi: 10.1034/j.1399-3054.2003.00002.x
Li, J., Xu, Y., Niu, Q., He, L., Teng, Y., and Bai, S. (2018). Abscisic acid (aba) promotes the induction and maintenance of pear (pyrus pyrifolia white pear group) flower bud endodormancy. Int. J. Mol. Sci. 19:310. doi: 10.3390/ijms19010310
Li, J., Yan, X., Yang, Q., Ma, Y., Yang, B., Tian, J., et al. (2019). Ppcbfs selectively regulate ppdams and contribute to the pear bud endodormancy process. Plant Mol. Biol. 99, 575–586. doi: 10.1007/s11103-019-00837-7
Li, S., and Dami, I. E. (2016). Responses of vitis vinifera ‘pinot gris’ grapevines to exogenous abscisic acid (aba): i. Yield, fruit quality, dormancy, and freezing tolerance. J. Plant Growth Regul. 35, 245–255. doi: 10.1007/s00344-015-9529-2
Li, Z., Liu, N., Zhang, W., Wu, C., Jiang, Y., Ma, J., et al. (2020). Integrated transcriptome and proteome analysis provides insight into chilling-induced dormancy breaking in chimonanthus praecox. Horticult. Res. 7:198. doi: 10.1038/s41438-020-00421-x
Liu, G., Li, W., Zheng, P., Xu, T., Chen, L., Liu, D., et al. (2012). Transcriptomic analysis of ‘suli’ pear (pyrus pyrifolia white pear group) buds during the dormancy by rna-seq. BMC Genom. 13:700. doi: 10.1186/1471-2164-13-700
Liu, J., and Sherif, S. M. (2019). Hormonal orchestration of bud dormancy cycle in deciduous woody perennials. Front. Plant Sci. 10:1136. doi: 10.3389/fpls.2019.01136
Love, M., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for rna-seq data with deseq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Ma, L. Y., Wang, L. R., He, S. C., Liu, X., and Wang, X. Q. (2006). A new species of magnolia (magnoliaceae) from hubei, china. Bull. Bot. Res. 26, 4–7.
Maurya, J. P., and Bhalerao, R. P. (2017). Photoperiod- and temperature-mediated control of growth cessation and dormancy in trees: a molecular perspective. Ann. Bot. 120, 351–360. doi: 10.1093/aob/mcx061
Niu, Q., Li, J., Cai, D., Qian, M., Jia, H., Bai, S., et al. (2015). Dormancy-associated mads-box genes and micrornas jointly control dormancy transition in pear (pyrus pyrifolia white pear group) flower bud. J. Exper. Bot. 67, 239–257. doi: 10.1093/jxb/erv454
Ogata, H., Goto, S., Sato, K., Fujibuchi, W., Bono, H., and Kanehisa, M. (1999). Kegg: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 27, 29–34. doi: 10.1093/nar/27.1.29
Park, S., Lee, C., Doherty, C. J., Gilmour, S. J., Kim, Y., and Thomashow, M. F. (2015). Regulation of the arabidopsis cbf regulon by a complex low-temperature regulatory network. Plant J. 82, 193–207. doi: 10.1111/tpj.12796
Qin, X., and Zeevaart, J. A. D. (2002). Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiol. 128:544. doi: 10.1104/pp.010663
Richardson, E. A. (1974). A model for estimating the completion of rest for ‘redhaven’ and ‘elberta’ peach trees. Hortscience 9, 331–332.
Rinne, P., Saarelainen, A., and Junttila, O. (1994). Growth cessation and bud dormancy in relation to aba level in seedlings and coppice shoots of betula pubescens as affected by a short photoperiod, water stress and chilling. Physiol. Plantarum 90, 451–458. doi: 10.1111/j.1399-3054.1994.tb08801.x
Rinne, P. X., Ivi, L. H., Welling, A., Vahala, J., Ripel, L., Ruonala, R., et al. (2011). Chilling of dormant buds hyperinduces flowering locus t and recruits ga-inducible 1,3-beta-glucanases to reopen signal conduits and release dormancy in populus. Plant Cell 23, 130–146.
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2009). Edger: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rohde, A., and Bhalerao, R. P. (2007). Plant dormancy in the perennial context. Trends Plant Sci. 12, 217–223.
Saito, S., Hirai, N., Matsumoto, C., Ohigashi, H., Ohta, D., Sakata, K., et al. (2004). Arabidopsis cyp707as encode (+)-abscisic acid 8’-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol. 134, 1439–1449. doi: 10.1104/pp.103.037614
Saito, T., Bai, S., Imai, T., Ito, A., Nakajima, I., and Moriguchi, T. (2015). Histone modification and signalling cascade of the dormancy-associated mads-box gene, ppmads13-1, in japanese pear (pyrus pyrifolia) during endodormancy. Plant Cell Environ. 38, 1157–1166. doi: 10.1111/pce.12469
Shi, X., Yin, Q., Sang, Z., Zhu, Z., Jia, Z., and Ma, L. (2021). Prediction of potentially suitable areas for the introduction of magnolia wufengensis under climate change. Ecol. Indicat. 127:107762. doi: 10.1016/j.ecolind.2021.107762
Shu, K., Huawei, Z., Wang, S., Chen, M., Wu, Y., Tang, S., et al. (2013). Abi4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in arabidopsis. Plos Genet. 9:e1003577. doi: 10.1371/journal.pgen.1003577
Singh, R. K., Maurya, J. P., Azeez, A., Miskolczi, P., Tylewicz, S., Stojkoviè, K., et al. (2018). A genetic network mediating the control of bud break in hybrid aspen. Nat. Commun. 9:4173. doi: 10.1038/s41467-018-06696-y
Singh, R. K., Miskolczi, P., Maurya, J. P., and Bhalerao, R. P. (2019). A tree ortholog of short vegetative phase floral repressor mediates photoperiodic control of bud dormancy. Curr. Biol. 29, 128–133. doi: 10.1016/j.cub.2018.11.006
Singh, R. K., Svystun, T., AlDahmash, B., Jönsson, A. M., and Bhalerao, R. P. (2017). Photoperiod- and temperature-mediated control of phenology in trees – a molecular perspective. New Phytol. 213, 511–524. doi: 10.1111/nph.14346
Sonnewald, S., and Sonnewald, U. (2014). Regulation of potato tuber sprouting. Planta 239, 27–38. doi: 10.1007/s00425-013-1968-z
Soon, F., Ng, L., Zhou, X., West, G., Kovach, A., Tan, M., et al. (2011). Molecular mimicry regulates aba signaling by snrk2 kinases and pp2c phosphatases. Science 335, 85–88. doi: 10.1126/science.1215106
Tamura, F., Tanabe, K., and Itai, A. (2002). Regulation of endodormancy in japanese pear. Acta Horticult. 587, 325–336. doi: 10.17660/ActaHortic.2002.587.44
Tuan, P. A., Bai, S., Saito, T., Ito, A., and Moriguchi, T. (2017). Dormancy-associated mads-box (dam) and the abscisic acid pathway regulate pear endodormancy through a feedback mechanism. Plant Cell Physiol. 58, 1378–1390. doi: 10.1093/pcp/pcx074
Tylewicz, S., Petterle, A., Marttila, S., Miskolczi, P., Azeez, A., Singh, R. K., et al. (2018). Photoperiodic control of seasonal growth is mediated by aba acting on cell-cell communication. Science 360, 212–215. doi: 10.1126/science.aan8576
Uemura, M., Joseph, R. A., and Steponkus, P. L. (1995). Cold acclimation of arabidopsis thaliana (effect on plasma membrane lipid composition and freeze-induced lesions). Plant Physiol. 109, 15–30. doi: 10.1104/pp.109.1.15
Umezawa, T., Sugiyama, N., Mizoguchi, M., Hayashi, S., Myouga, F., Yamaguchi-Shinozaki, K., et al. (2009). Type 2c protein phosphatases directly regulate abscisic acid-activated protein kinases in arabidopsis. Proc. Natl. Acad. Sci. 106:17588. doi: 10.1073/pnas.0907095106
Vimont, N., Fouché, M., Campoy, J. A., Tong, M., Arkoun, M., Yvin, J., et al. (2019). From bud formation to flowering: transcriptomic state defines the cherry developmental phases of sweet cherry bud dormancy. BMC Genom. 20:97.4. doi: 10.1186/s12864-019-6348-z
Vimont, N., Schwarzenberg, A., Domijan, M., Donkpegan, A. S. L., Beauvieux, R., le Dantec, L., et al. (2021). Fine tuning of hormonal signaling is linked to dormancy status in sweet cherry flower buds. Tree Physiol. 41, 544–561. doi: 10.1093/treephys/tpaa122
Wang, D., Gao, Z., Du, P., Xiao, W., Tan, Q., Chen, X., et al. (2016). Expression of aba metabolism-related genes suggests similarities and differences between seed dormancy and bud dormancy of peach (prunus persica). Fron. Plant Sci. 6:1248. doi: 10.3389/fpls.2015.01248
Weinberger, J. H. (1950). Chilling requirements of peach varieties. Proc. Amer. Soc. Hort. Sci. 56, 122–128.
Weiser, C. J. (1970). Cold resistance and injury in woody plants. Science 169, 1269–1278. doi: 10.1126/science.169.3952.1269
Wen, L. H., Zhong, W. J., Huo, X. M., Zhuang, W. B., Ni, Z. J., and Gao, Z. H. (2016). Expression analysis of aba- and ga-related genes during four stages of bud dormancy in japanese apricot (prunus mume sieb. Et zucc). J. Horticult. Sci. Biotechnol. 91, 362–369. doi: 10.1080/14620316.2016.1160546
Xie, C., Mao, X., Huang, J., Ding, Y., Wu, J., Dong, S., et al. (2011). Kobas 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–W322. doi: 10.1093/nar/gkr483
Yamane, H., Ooka, T., Jotatsu, H., Hosaka, Y., Sasaki, R., and Tao, R. (2011). Expressional regulation of ppdam5 and ppdam6, peach (prunus persica) dormancy-associated mads-box genes, by low temperature and dormancy-breaking reagent treatment. J. Exper. Bot. 62, 3481–3488. doi: 10.1093/jxb/err028
Yang, Q., Gao, Y., Wu, X., Moriguchi, T., Bai, S., and Teng, Y. (2021). Comparative analysis of natural cold acclimation and deacclimation of two magnolia species with different winter hardiness. Horticult. Res. 8:575. doi: 10.1038/s41438-021-00575-2
Yang, Q., Niu, Q., Li, J., Zheng, X., Ma, Y., Bai, S., et al. (2018). Pphb22, a member of hd-zip proteins, activates ppdam1 to regulate bud dormancy transition in ‘suli’ pear (pyrus pyrifolia white pear group). Plant Physiol. Biochem. 127, 355–365. doi: 10.1016/j.plaphy.2018.04.002
Yang, Q., Niu, Q., Tang, Y., Ma, Y., Yan, X., Li, J., et al. (2019). Ppygast1 is potentially involved in bud dormancy release by integrating the ga biosynthesis and aba signaling in ‘suli’ pear (pyrus pyrifolia white pear group). Environ. Exper. Bot. 162, 302–312. doi: 10.1016/j.envexpbot.2019.03.008
Yang, Q., Yang, B., Li, J., Wang, Y., Tao, R., Yang, F., et al. (2020). Aba-responsive abre-binding factor3 activates dam3 expression to promote bud dormancy in asian pear. Plant Cell Environ. 43, 1360–1375. doi: 10.1111/pce.13744
Yang, Y., Jia, Z., Chen, F., Sang, Z., and Ma, L. (2015). Comparative analysis of natural cold acclimation and deacclimation of two magnolia species with different winter hardiness. Acta Physiol. Plantarum 37:129. doi: 10.1007/s11738-015-1883-y
Yooyongwech, S., Sugaya, S., Sekozawa, Y., and Gemma, H. (2009). Differential adaptation of high- and low-chill dormant peaches in winter through aquaporin gene expression and soluble sugar content. Plant Cell Rep. 28:1709. doi: 10.1007/s00299-009-0770-7
Yue, C., Cao, H., Hao, X., Zeng, J., Qian, W., Guo, Y., et al. (2018). Differential expression of gibberellin- and abscisic acid-related genes implies their roles in the bud activity-dormancy transition of tea plants. Plant Cell Rep. 37, 425–441. doi: 10.1007/s00299-017-2238-5
Zhang, Z., Zhuo, X., Zhao, K., Zheng, T., Han, Y., Yuan, C., et al. (2018). Transcriptome profiles reveal the crucial roles of hormone and sugar in the bud dormancy of prunus mume. Sci. Rep. 8:5090. doi: 10.1038/s41598-018-23108-9
Zhao, J., Li, G., Yi, G., Wang, B., Deng, A., Nan, T., et al. (2006). Comparison between conventional indirect competitive enzyme-linked immunosorbent assay (icelisa) and simplified icelisa for small molecules. Anal. Chim. Acta 571, 79–85. doi: 10.1016/j.aca.2006.04.060
Zhao, K., Zhou, Y., Ahmad, S., Yong, X., Xie, X., Han, Y., et al. (2018a). Pmcbfs synthetically affect pmdam6 by alternative promoter binding and protein complexes towards the dormancy of bud for prunus mume. Sci. Rep. 8:4527. doi: 10.1038/s41598-018-22537-w
Zhao, K., Zhou, Y., Li, Y., Zhuo, X., Ahmad, S., Han, Y., et al. (2018b). Crosstalk of pmcbfs and pmdams based on the changes of phytohormones under seasonal cold stress in the stem of prunus mume. Int. J. Mol. Sci. 19:15. doi: 10.3390/ijms19020015
Zheng, C., Acheampong, A., Shi, Z., Mugzech, A., Halaly Basha, T., Shaya, F., et al. (2018). Abscisic acid catabolism enhances dormancy release of grapevine buds. Plant Cell Environ. 41:13371. doi: 10.1111/pce.13371
Zheng, C., Halaly, T., Acheampong, A. K., Takebayashi, Y., Jikumaru, Y., Kamiya, Y., et al. (2015). Abscisic acid (aba) regulates grape bud dormancy, and dormancy release stimuli may act through modification of aba metabolism. J. Exper. Bot. 66, 1527–1542. doi: 10.1093/jxb/eru519
Keywords: bud dormancy, RNA-seq, Magnolia wufengensis, ABA, dormancy release, cold tolerance
Citation: Wu K, Duan X, Zhu Z, Sang Z, Zhang Y, Li H, Jia Z and Ma L (2021) Transcriptomic Analysis Reveals the Positive Role of Abscisic Acid in Endodormancy Maintenance of Leaf Buds of Magnolia wufengensis. Front. Plant Sci. 12:742504. doi: 10.3389/fpls.2021.742504
Received: 16 July 2021; Accepted: 15 October 2021;
Published: 11 November 2021.
Edited by:
Songling Bai, Zhejiang University, ChinaReviewed by:
Xinyuan Hao, Tea Research Institute, Chinese Academy of Agricultural Sciences (CAAS), ChinaCopyright © 2021 Wu, Duan, Zhu, Sang, Zhang, Li, Jia and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongkui Jia, amlhemtAYmpmdS5lZHUuY24=; Luyi Ma, bWFsdXlpQGJqZnUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.