- Key Laboratory of Plant-Soil Interaction (MOE), Centre for Resources, Environment and Food Security, College of Resources and Environmental Sciences, China Agricultural University, Beijing, China
Zinc (Zn) deficiency, a globally predominant micronutrient disorder in crops and humans, reduces crop yields and adversely impacts human health. Despite numerous studies on the physiological mechanisms underlying Zn deficiency tolerance, its genetic basis of molecular mechanism is still poorly understood. Thus, the Zn efficiency of 20 maize inbred lines was evaluated, and a quantitative trait locus (QTL) analysis was performed in the recombination inbred line population derived from the most Zn-efficient (Ye478) and Zn-inefficient inbred line (Wu312) to identify the candidate genes associated with Zn deficiency tolerance. On this basis, we analyzed the expression of ZmZIP1-ZmZIP8. Thirteen QTLs for the traits associated with Zn deficiency tolerance were detected, explaining 7.6–63.5% of the phenotypic variation. The genes responsible for Zn uptake and transport across membranes (ZmZIP3, ZmHMA3, ZmHMA4) were identified, which probably form a sophisticated network to regulate the uptake, translocation, and redistribution of Zn. Additionally, we identified the genes involved in the indole-3-acetic acid (IAA) biosynthesis (ZmIGPS) and auxin-dependent gene regulation (ZmIAA). Notably, a high upregulation of ZmZIP3 was found in the Zn-deficient root of Ye478, but not in that of Wu312. Additionally, ZmZIP4, ZmZIP5, and ZmZIP7 were up-regulated in the Zn-deficient roots of Ye478 and Wu312. Our findings provide a new insight into the genetic basis of Zn deficiency tolerance.
Introduction
Maize (Zea mays L.) is the third most important cereal crop globally, after wheat and rice, and is a foundational model for genetics and genomics (Jiao et al., 2017). It accounts for about 64% of coarse grain and 27.1% of the total cereal production, providing at least 30% of food calories to more than 4.5 billion people in 94 developing countries (Shiferaw et al., 2011; Wu and Guclu, 2013). At present, China and other rapidly developing economies face the challenge of substantially increasing the yield of cereal grains (Chen et al., 2011). China must increase its agricultural output by 50% to meet its growing food demand in the future (Cui et al., 2014). China is the second-largest producer of maize after the United States (Xomphoutheb et al., 2020). Maize is an important crop to solve the cereal demand in China.
Zinc is an essential element in the range of physiological and biochemical mechanisms of plants and is needed for more than 300 plant enzymes, and functions as co-factors in the processes of photosynthesis, respiration, and other metabolic reactions. Within the broad category of mineral-related abiotic stresses, Zn deficiency is one of the most widespread limiting factors to crop production, affecting 50% of cereal crops soil (Nielsen, 2012). Zn deficiency in plants leads to leaf bronzing, stunted growth, shortened internodes and petioles, malformed leaves, delayed flowering and fruit maturity, spikelet sterility, and seedling mortality (Widodo et al., 2010; Gainza-Cortés et al., 2012; Mattiello et al., 2015), thus reducing the yield and quality of crops (Broadley et al., 2007; Graham et al., 2012). Zn is not only an important trace element for plants but also an indispensable nutrient for humans. As the second most abundant micronutrient, Zn serves as a structural component of at least 3,000 proteins in the body (Sasaki et al., 2018). Zn deficiency causes a variety of diseases (Kambe et al., 2015; King et al., 2015), which in the worst case may cause human death (Wessells and Brown, 2012). The WHO estimates that one-third of the world population suffers from Zn shortage, especially in developing countries (Swain et al., 2016). Therefore, understanding the mechanisms of Zn deficiency tolerance would be helpful to improve the tolerance of maize to Zn deficiency via conventional breeding using marker assistant selection or through transgenic technology.
The ability to tolerate Zn deficiency can also be termed Zn efficiency (ZE), which is evaluated by the ability of a plant to grow and yield well under Zn-deficient conditions (Impa et al., 2013a). Studies on the physiological mechanisms underlying ZE have been reported in several other crop species, including beans (Hacisalihoglu et al., 2001), barley (Genc et al., 2007), wheat (Rengel and Graham, 1996), and rice (Wissuwa et al., 2006). However, the mechanisms of ZE in maize are still unclear. Several studies implicated that some root processes could increase the bioavailability of soil Zn for root uptake to enhance ZE, mainly including the release of root exudates (Khoshgoftarmanesh et al., 2018) and morphological changes in roots (Mori et al., 2016). In addition to Zn uptake, ZE is probably related to root-to-shoot transport and the remobilization of Zn from old to young leaves (Impa et al., 2013b). Besides, the shoot-localized mechanism in ZE illustrated by Hacisalihoglu et al. (2003), contains the subcellular compartmentation of Zn in shoot cells and the biochemical utilization of Zn in the cells of shoots (Hacisalihoglu and Kochian, 2003; White and Broadley, 2011).
Among these physiological processes, increased Zn uptake from the soil, as a major physiological mechanism, requires Zn transporters which can transport Zn2+ across membranes (Kambe et al., 2004). These transporters get involved in the uptake of Zn from the soil, translocation inside plants, loading and unloading of xylem, sequestration in the vacuolar, and remobilization from the vacuole. To date, several metal transporters have been identified in plants, including zinc-regulated transporters, iron-regulated transporters-like protein (ZIP) family, heavy metal ATPases (HMA) family, natural resistance-associated macrophage protein (NRAMP) family, and cation diffusion facilitator (CDF) family. The regulation of the ZIP gene family is considered to be a major mechanism in tolerating Zn deficiency stress. Expression analysis showed that the transcripts of AtZIP1–AtZIP5, AtZIP9–AtZIP12 increased under Zn deficiency, suggesting that these genes may enhance Zn acquisition under Zn-deficient conditions in Arabidopsis (Krämer et al., 2007). AtIRT3 reverses growth defects in the Zn- and Fe-uptake-deficient yeast mutants, and the overexpression of AtIRT3 enhances the Zn accumulation in shoots (Lin et al., 2009). OsZIP1, OsZIP2, OsZIP4, OsZIP5, OsZIP6, OsZIP7, and OsZIP8 are induced in Zn-deficient roots, and function in the uptake, transport, or allocation of Zn in rice under Zn-deficient conditions (Chen et al., 2008; Lee and An, 2009; Kavitha et al., 2015; Sasaki et al., 2015). Six HvZIP genes (HvZIP3, HvZIP5, HvZIP7, HvZIP8, HvZIP10, and HvZIP13) in barley are highly induced in Zn-deficient roots, and their increased expression enhances the uptake and root-to-shoot translocation of Zn in response to Zn deficiency stress (Tiong et al., 2015).
Quantitative trait locus (QTL) analysis is a powerful method to identify the chromosomal regions encompassing genes controlling complex quantitative traits (Alonso-Blanco et al., 2009; Ueno et al., 2009). However, the genetic basis of ZE remains largely unknown, since most studies on QTL identification mainly focus on seed Zn concentration and content (Gu et al., 2015; Jin et al., 2015; Hindu et al., 2018) and only a few researchers have reported QTLs associated with performance under Zn deficiency. Wissuwa et al. (2006) reported the QTLs associated with the most severe and susceptible Zn deficiency symptoms of rice in the field, including leaf bronzing, plant mortality, and biomass reduction in Zn-deficient conditions. Similarly, Genc et al. (2009) have assessed the severity of Zn deficiency symptoms on a scale from 1 to 9 in wheat. The QTLs controlling the Zn score, shoot dry weight, and concentrations of Zn and Fe in shoots and grains were detected at two levels of Zn. By combining bi-parental QTL mapping and genome-wide association analysis, Lee et al. (2017) identified one putative candidate gene which was associated with grain yield and component traits, and confirm that the candidate gene Os06g44220 is strongly up-regulated in both Zn-deficient root and shoot.
Therefore, the aims of this study are to (1) select the most and least Zn-efficient maize inbred lines for further studies in the linkage analysis and candidate gene expressions; (2) reveal possible physiological mechanisms on ZE; (3) detect the QTLs for ZE in the recombinant inbred line (RIL) population derived from the most and least Zn-efficient maize inbred lines and identify the candidate genes in the QTL co-localization; (4) to analyze the expression patterns of the eight ZmZIP genes (ZmZIP1-ZmZIP8) in response to Zn deficiency.
Materials and Methods
Plant Material and Experiment Design
Experiment 1: Variations in the Zn Efficiency of Twenty Maize Inbred Lines
Twenty maize inbred lines (Ye478, CI7, Yu87-1, DE3, By815, Zheng58, KUI3, B77, SC55, SK, By804, Dan340, Chang7-2, X178, Mo17, Zong3, B73, HuangC, K22, and Wu312) which have generated linkage populations among them, were grown hydroponically in a mixed-crop system under Zn-deficient (-Zn) (0.3 μmol L–1 Zn-EDTA) and Zn-sufficient (CK) (10 μmol L–1 Zn-EDTA) conditions. Each treatment contained three replications. For each treatment, three replications for the twenty inbred lines were grown hydroponically in a 40 L tank (665 × 410 × 160 mm3, length × width × height).
Experiment 2: Quantitative Trait Locus Analysis of Zn Efficiency in Maize
The Zn-efficient (Ye478) and -inefficient inbred lines (Wu312) were selected from Experiment 1. A RIL population consisting of 218 lines were derived from Ye478 (female parent) and Wu312 (male parent), as described by Liu J. et al. (2011). The RIL population and their parents were hydroponically grown under -Zn (0.3 μmol L–1) and CK (10 μmol L–1) conditions. For each treatment, two independent experiments were conducted in randomized complete blocks with three replications each. Fifty-five RILs and two plants for each parent were grown in a 40 L tank. In total, 12 tanks were used for each experiment.
Experiment 3: Expression of ZIP Genes in Maize
The expression of eight ZmZIP genes (ZmZIP1–ZmZIP8) in the roots of the Zn-efficient (Ye478) and -inefficient inbred lines (Wu312) were analyzed in two treatments, Experiment 1 and 2. Ye478 and Wu312 were grown hydroponically in a mixed-crop system under -Zn (0.3 μmol L–1) and CK (10 μmol L–1) conditions. Each treatment contained three biological replications. Each biological replication contained two plants for each inbred line. Mixed cropping was performed on four plants in a 3.3-L tank. Three technical replications were performed for each biological replication.
Plant Culture in Hydroponics
The maize seeds were sterilized for 30 min in a 10% solution of hydrogen peroxide (H2O2), washed with distilled water, and soaked in saturated calcium sulfate (CaSO4) for 10 h, and then germinated on a moist filter paper in the dark at room temperature. Two days later, the germinated seeds were wrapped in a moist filter paper roll and grown. At the stage of two visible leaves, the seedlings were selected and transferred into a 40 L black tank (Experiment 1 and 2) or a 3.3 L tank (Experiment 3). The solution pH was set at 5.5–6. The adjusted Hoagland nutrient solution contained (mmol L–1): 0.5 NH4NO3, 0.5 CaCl2, 1.5 Ca(NO3)2, 0.75 K2SO4, 0.65 MgSO4, 0.1 KCl, 0.25 KH2PO4, 1.0 × 10–3 H3BO3, 0.35 Fe(II)-EDTA, 8.0 × 10–3 CuSO4, 1.2 × 10–2 MnSO4, 4.0 × 10–5 (NH4)Mo7O24, and 4.0 × 10–3 NiCl. The nutrient solution was renewed every 3 days and aerated by a pump. The maize seedlings were cultured with hydroponics in a growth chamber condition with controlled conditions, namely, 28°C during the 14 h light period from 8:00 to 22:00 and 22°C during the 10 h dark period, the average light intensity was 350 μmol m–2 s–1 that was measured at the canopy.
Data Collection
The hydroponic culture experiments were carried out to collect data in Experiments 1–3 at the seedling stage. Experiment 1 was terminated 21 days after transplanting, and all the samples were dried at 75°C till constant weight. The plant height and shoot and root dry weights were measured and the root to shoot (R/S) ratios and ZE were calculated. The concentrations of Zn, iron (Fe), manganese (Mn), and copper (Cu) in the shoots and roots were analyzed using an inductively coupled plasma-atomic emission spectroscopy (ICP-AES). The ZE, relative ratios of R/S efficiency, nutrient contents, uptake efficiency, and relative transport were estimated using the following equations from (1) to (5), respectively.
Experiment 2 was terminated 21 days after transplanting. All the samples were dried at 75°C over 72 h. The shoot and root dry weights were measured, and the R/S ratio and the ratio of -Zn/CK for each trait were also calculated. Based on the typical Zn deficiency symptoms in plants on the 15–21th day after transplanting, the Zn score for each plant has been visually recorded three times since the 15th day after transplanting. Three scales (0, 1, 2) were designed to assess the tolerance of the RILs in the Ye478 × Wu312 population to Zn deficiency under -Zn conditions (0.3 μmol L–1 Zn-EDTA) (Figure 1). The score-0 plants (two plants in the left in Figure 1) only developed four leaves and showed the severest Zn deficiency symptoms in the shoots, including stunted growth, shortened internodes, small malformed leaves, wrinkled leaf margins, and chlorosis and necrotic patches distributed on more than 50% of the areas on all leaves. The root growth of the score-0 plants was strongly suppressed, manifested in the great reductions in the root length and number of lateral roots and the large-area brown lesions on the roots. Compared with the score-0 plants, the score-1 plants showed a better growth without brown lesions on the roots, but still appeared to have shortened internodes, small malformed leaves, and chlorosis distributed on 10–30% of the areas on the middle and young leaves. Score 2 represented the green healthy plants under the Zn-deficient conditions, showing little Zn deficiency symptoms in both the shoots and roots when compared with the plants grown in the CK treatment.

Figure 1. Three scales for Zn score of maize in the RIL population under Zn deficiency. Based on the typical Zn-deficient symptoms in plants on the 15–21th day after transplanting, the Zn score for each plant has been visually recorded three times since the 15th day after transplanting. Three scales (0, 1, 2) are designed to assess the ability of RILs in the Ye478 × Wu312 population to tolerate Zn deficiency under Zn-deficient conditions (0.3 μmol L−1 Zn-EDTA). Score-0 plants (two plants on the left) only developed four leaves and showed the severest Zn deficiency symptoms in the shoots, including stunted growth, shortened internodes, small malformed leaves, wrinkled leaf margins and chlorosis and necrotic patches distributed on more than 50% of the areas on all leaves. The root growth of score-0 plants was strongly suppressed, manifested in the great reductions in the root length and number of lateral roots, large-area brown lesions on roots. Compared with the score-0 plants, the score-1 plants showed better growth without brown lesions on roots, but still appeared to have shortened internodes, small malformed leaves and chlorosis distributed on 10–30% of areas on the middle and young leaves. Score 2 represents green healthy plants under Zn-deficient conditions, showing little Zn-deficient symptoms in both shoots and roots when compared with the plants grown in the Zn-sufficient (CK) treatment. The representative shoots and roots are displayed. A ruler of 30 cm length is shown.
Experiment 3 was terminated 21 days after transplanting. The total RNA was isolated from the roots of Ye478 and Wu312 using TRIzol (Takara, Kusatsu, Shiga, Japan). We used 1.5 μg of the total RNA to synthesize the complementary DNA (cDNA). A quantitative real-time PCR (qRT-PCR) was performed using an SYBR Green Real-time RT-PCR (Applied Biosystems, Waltham, Massachusetts, United States) and an ABI7500 Fast Real-Time PCR System (Applied Biosystems). The primers used for the RT-PCR are shown in Supplementary Table 11.
Data Analysis
Statistical Analysis
In Experiments 1, 2, and 3, the means for each trait were compared using one-way ANOVA at a 0.05 level of probability followed by Turkey’s test using SPSS 20.0 (IBM, Armonk, New York, United States). The linear mixed effect function Imer from the lme4 package in R was fitted to each RIL to obtain the best linear unbiased prediction (BLUP) value for each trait: yi = μ + fi + ei + εi, where yi is the phenotypic value of individual i, μ is the grand mean for all environments, fi is the genetic effect, ei is the effect of different environments, and εi is the random error. The broad-sense heritability for each trait was calculated by H2 = σg2/(σg2 + σge2/e + σε2/re), where σg2 is the genetic variance, σge2 is the interaction of the genotype and treatment, σε2 is the residual error, while e and r are the number of replications and treatments, respectively. A principal component analysis (PCA) was conducted by CANOCO 4.5. In Experiment 3, the changes in expression were calculated via the ΔΔCt method.
Quantitative Trait Locus Mapping
In Experiment 2, a genetic linkage map with a total length of 2,084 cm consisting of 184 polymorphic markers, was constructed by Liu J. et al. (2011). The QTL identification was performed using composite interval mapping (CIM) in the Windows QTL Cartographer version 2.5. Model 6 was selected for detecting the QTLs and estimating their effects. The threshold logarithm of odds (LOD) values needed to declare the putative QTLs were estimated by permutation tests with a minimum of 1,000 replicates at a significant level of P < 0.05 (LOD = 3). The confidence interval for each QTL was determined using the 1-LOD interval method.
Annotation of Candidate Genes
According to the physical distance of the peak bins, the genes within the refined interval and their functional descriptions were identified using the maizeB73 reference genome assembly version 2 available on the MaizeGDB Genome database1.
Results
Variations in Zn Efficiency Among Twenty Maize Inbred Lines
Effects of Zinc Deficiency on Maize Inbred Lines
Under Zn-deficient conditions, most maize inbred lines showed chlorosis on young leaves and reduced leaf size. As shown in Figure 2, Wu312 appeared to have the severest symptoms of Zn deficiency, including stunted growth, shortened internodes and petioles, wrinkled leaf margins, and small malformed leaves. However, little difference was observed in Ye478 between the -Zn and CK treatments. Substantial variations in the ZE based on the shoot (Supplementary Figure 1A) and root (Supplementary Figure 1B) dry weights were observed within 20 inbred lines, ranging from 38.5 to 128% and 39.4–146%, respectively. The ZEs based on the shoot and root dry weights of Ye478 and CI7 were the highest, which were about three times as high as those of HuangC, K22, and Wu312.

Figure 2. The phenotypes of Ye478 and Wu312 under -Zn and CK conditions. In the -Zn (0.3 μmol L–1 Zn-EDTA) (A) and CK (10 μmol L–1 Zn-EDTA) (B) treatments, the maize inbred lines Ye478 (left) and Wu312 (right) were grown hydroponically for 21 days after transplanting. The representative shoots are displayed. Rulers of 30 cm length are shown in both (A,B).
In general, Zn-efficient genotypes are not only able to accumulate more Zn, but also produce more dry matter weights and grain yields. Our results indicated that Ye478 and CI7 not only showed little Zn deficiency symptoms but also produced higher plant heights (Supplementary Figure 2) and accumulated more shoot and root dry weights in comparison with K22 and Wu312 (Supplementary Table 1). The shoot dry weights of Ye478 and CI7 were more than five times higher than those of K22 and Wu312 under Zn deficiency. The root dry weights of Ye478 and CI7 were more than three times as high as those of K22 and Wu312 under -Zn conditions. No significant differences in the shoot dry weights of Ye478 and CI7 were observed between the -Zn and CK treatments. Zn deficiency markedly enhanced the root dry weights of Ye478 and CI7 by 46 and 32.8%, respectively. By contrast, Zn deficiency significantly decreased the shoot and root dry weights of K22 by 59.9 and 43.7%, and Wu312 by 61.5 and 48.6%, respectively. Additionally, Ye478 and CI7 maintained stable R/S ratios in response to Zn deficiency stress. Low Zn stress significantly increased the R/S ratio of K22 by 33.5%.
Zn deficiency decreased the Zn concentrations in the shoots and roots of 20 inbred lines by more than 50% (Supplementary Table 2). Most lines were Zn-deficient plants diagnosed by the shoot Zn concentration which ranged from 7.9 to 20.4 μg g–1. A Pearson correlation analysis indicated that the ZE based on the shoot and root dry weights of the 20 inbred lines were not correlated with the Zn concentrations in shoots (Figure 3A) and roots (Figure 3B). Therefore, the Zn concentrations in the shoots and roots were not used in Experiment 2. The Zn accumulation in the shoots and roots was markedly decreased by Zn deficiency. Nevertheless, regardless of the Zn supply, the shoot and root Zn contents of Ye478 and CI7 were remarkably higher than those of K22 and Wu312 (Supplementary Table 3). A greater than eightfold difference in the shoot Zn content occurred between Ye478 (49.9 μg plant–1) and Wu312 (6.2 μg plant–1). In addition, Zn deficiency resulted in a decrease in the Zn uptake efficiency and an increase in the relative Zn transport of maize inbred lines (Supplementary Table 4). Compared with Wu312, Ye478, and CI7 maintained higher levels of Zn uptake and transport under different conditions. In conclusion, Ye478 and Wu312 were screened out to be the most and least tolerant inbred lines to Zn deficiency, respectively, allowing for further molecular genetic studies in Experiments 2 and 3 (Figure 4A).
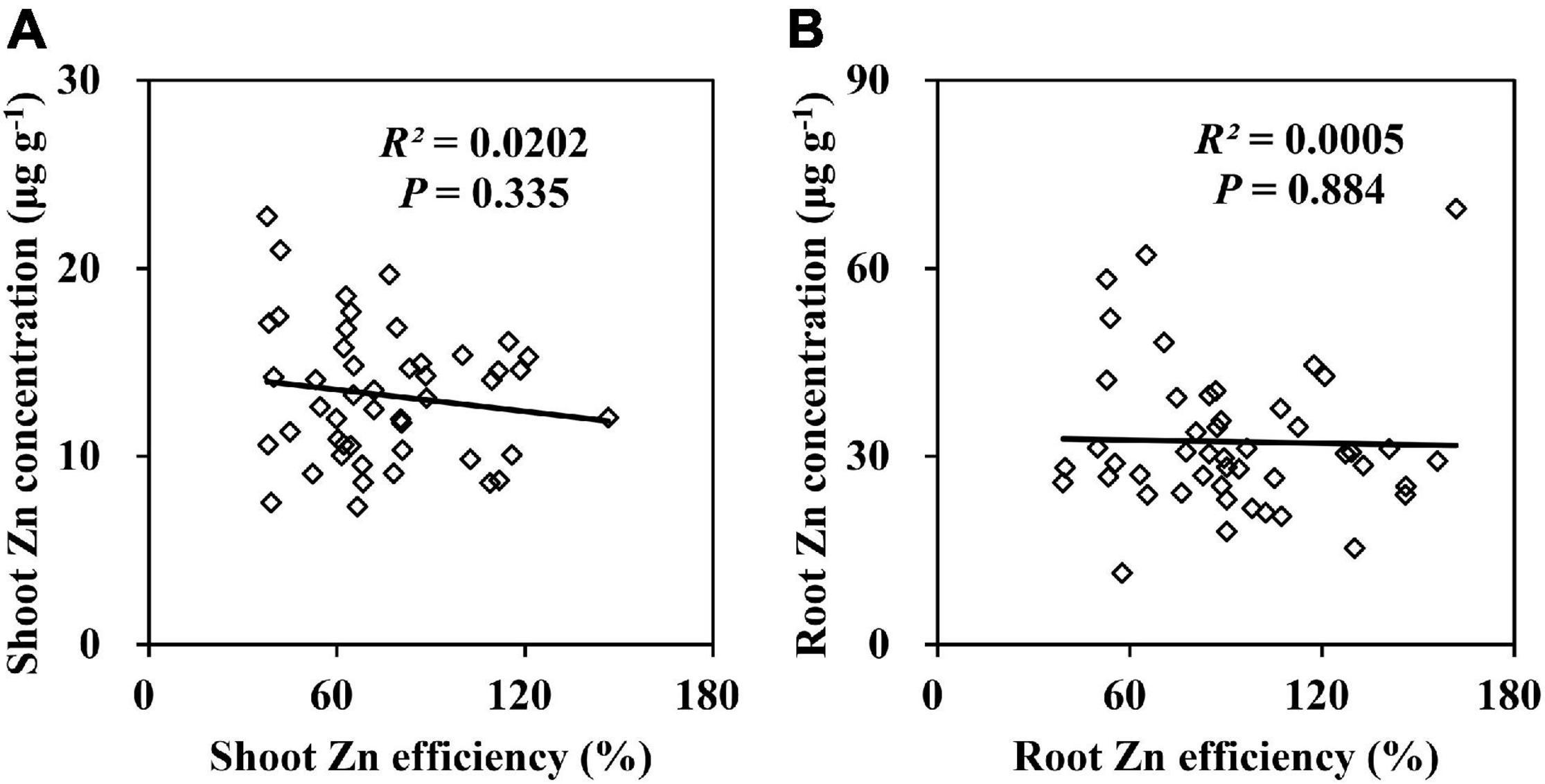
Figure 3. Relationships between Zn efficiencies (ZEs) based on the shoot (A) and root (B) dry weights and Zn concentrations in the shoot (A) and root (B) under Zn deficiency. P < 0.05 indicates significant differences.
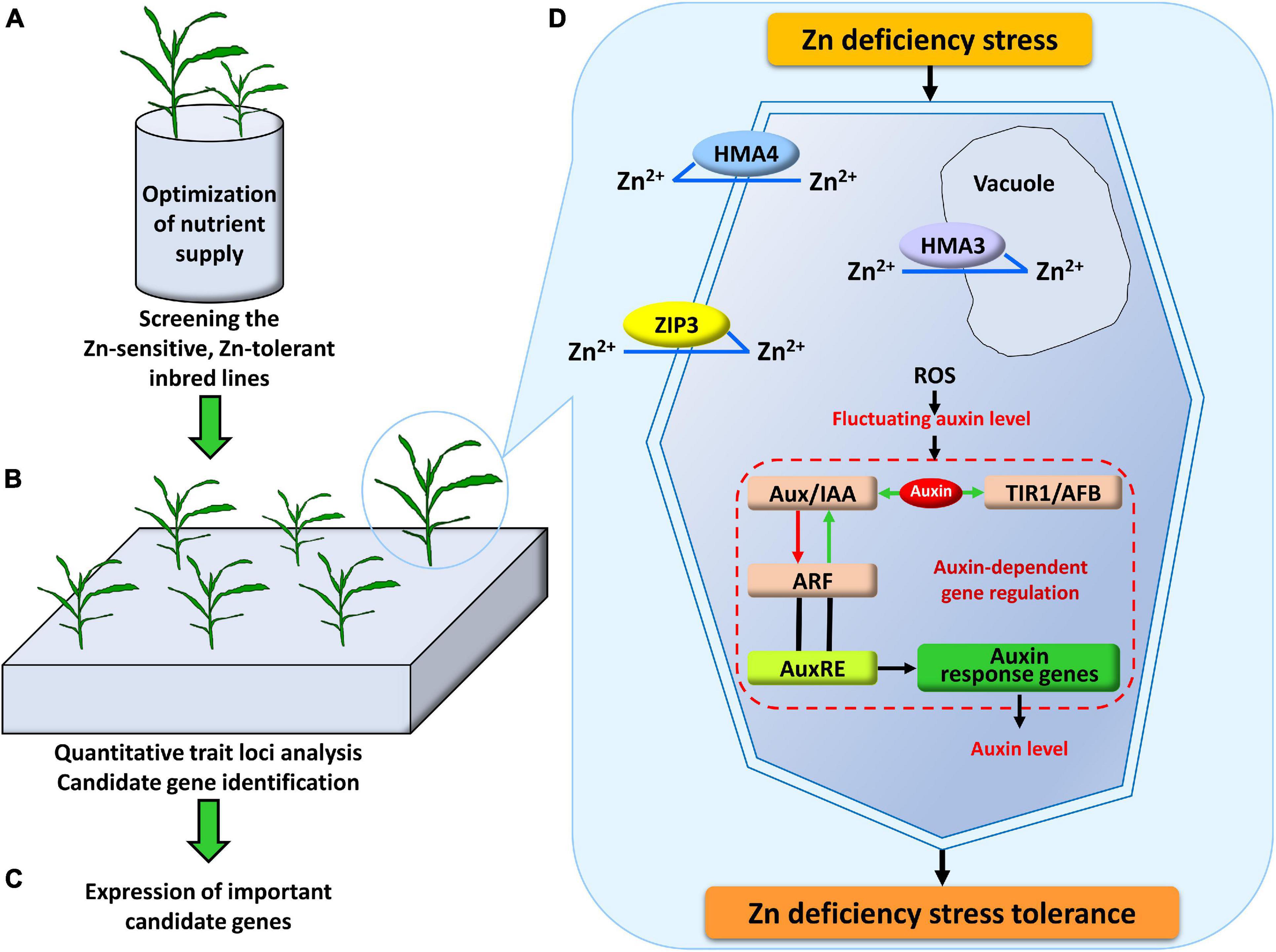
Figure 4. Systematic research design and schematic model for the mechanism of Zn deficiency tolerance in plants. (A) The nutrient solution supply was optimized by adjusting the NH4+/NO3– ratio and the supply level of other metal ions. On this basis, the most tolerant and the most sensitive parents under Zn deficiency stress were screened out. (B) Quantitative trait loci associated with Zn deficiency tolerance were detected in the linkage population derived from the parents screened out from (A), and candidate genes were identified within these loci. (C) Based on the results of physiological experiments and the candidate genes identified by the QTL analysis, the expression of eight ZIP genes (ZmZIP1-8) was analyzed under -Zn and CK conditions. (D) In the mechanisms of Zn deficiency stress tolerance, the important genes which may be involved in the uptake of Zn2+ from the soil to roots and preferential distribution to shoot (ZmZIP3), sequestering Zn2+ into vacuole (ZmHMA3), translocating Zn2+ from root to shoot (ZmHMA4), probably participate in the uptake, transport and redistribution of Zn. In addition, the gene that may repress transcription of ARF plays an essential role in the auxin-dependent gene regulation (ZmIAA17).
In terms of the effects on other mineral elements, Zn deficiency resulted in a 20.1–119.3% increase of Fe concentrations in the shoots (Supplementary Table 5) and a 7.4–60.5% increase of Mn concentrations in the roots (Supplementary Table 6) for most inbred lines. Additionally, much more Mn is retained in the shoots than in roots under Zn-deficient conditions, resulting in the enhancement of the shoot-to-root transport of Mn (Supplementary Table 8). Compared with the CK treatment, higher Cu concentrations were detected in both the shoots and roots in the -Zn treatment (Supplementary Tables 5, 6), which led to the enhancement of Cu uptake and transport (Supplementary Tables 7, 8).
Principal Components Analysis of Zn Efficiency
The PCA extracted two principal components that accounted collectively for 85.4% of the variance in ZE, relative R/S ratio efficiency, Zn uptake efficiency, and Zn transport efficiency under the Zn deficiency of 20 inbred lines (Figure 5). PC1 was negatively loaded with ZE while PC2 was positively loaded with relative R/S ratio efficiency and negatively loaded with Zn uptake and transport efficiency.
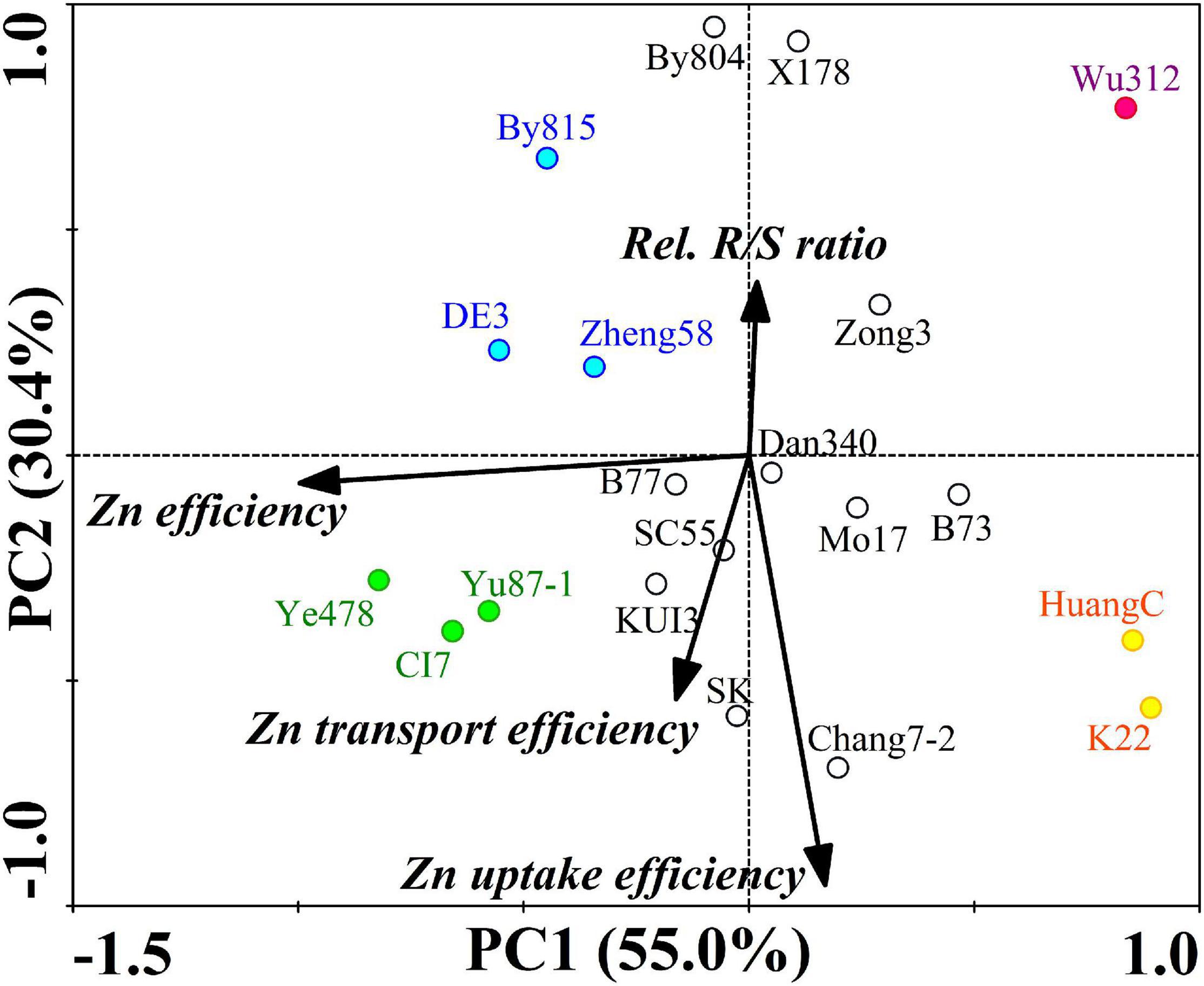
Figure 5. Principal component analysis of ZE based on the shoot dry weight, relative R/S ratio efficiency, Zn uptake efficiency, and Zn transport efficiency (relative Zn transport) under the Zn deficiency of 20 maize inbred lines. Two principal components (PC1 and PC2) were extracted and accounted collectively for 85.4% of the variance. Dots represent the position of the 20 inbred lines determined by their values on the given principal component. The dots in green, blue, orange, and red represent four typical types of maize inbred lines differing in Zn deficiency tolerance. Black lines indicate vectors quantifying the magnitude and direction of a trait’s contribution to the principal component.
Four typical types of inbred lines differing in ZE are shown in Figure 5. The most Zn-efficient inbred lines with the highest ZEs (109.3–128%), containing Ye478, CI7, and Yu87-1, were efficient in Zn uptake and transport, maintaining stable R/S ratios. Compared with Ye478, CI7, and Yu87-1, the CK inbred lines with lower ZEs (91.6–102.1%), including DE3, By815, and Zheng58, kept lower Zn uptake and transport efficiency as well as higher R/S ratio efficiency. The least Zn-efficient inbred line was Wu312 with the lowest ZE (38.5%) and the lowest Zn uptake and transport. Compared with Wu312, the -Zn inbred lines HuangC and K22, with higher ZEs (39.0–40.3%), had higher efficiency in Zn uptake and transport.
Quantitative Trait Locus Analysis of Zn Efficiency in Maize
Phenotypic Data Analysis
A RIL population consisting of 218 lines derived from Wu312 and Ye478 was used to identify the Zn efficiency-associated loci (ZEALs) in maize. Consistent with the results of Experiment 1, Wu312 showed the severest Zn-deficient symptoms like the score-0 plants shown in Figure 1, and Ye478 showed green healthy plants without symptoms of Zn deficiency similar with the score-2 plant. Large phenotypic variations were observed in Zn-deficient symptoms among different RILs (Figure 6).

Figure 6. Phenotypic variation in the recombinant inbred line (RIL) population. In the -Zn treatment (0.3 μmol L–1 Zn-EDTA), two plants for each parent and 55 RILs were grown in hydroponics for 21 days after transplanting in each tank. Large phenotypic variations of Zn-deficient symptoms were observed among RILs under Zn deficiency.
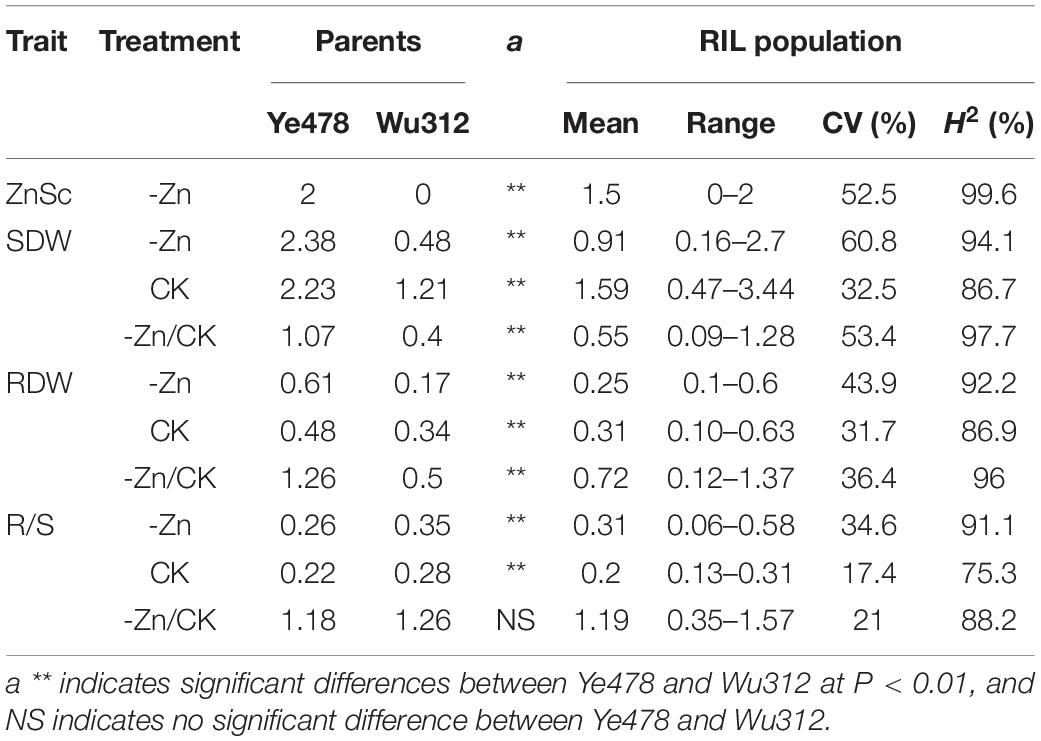
Phenotypic data were collected for each trait from the RILs in two experiments under different conditions (-Zn, CK, -Zn/CK). The phenotypic means and range of each trait for the parents and their RIL population are in Table 1. Our results indicated that Zn deficiency resulted in marked increases in the shoot and root dry weights of Ye478. On the contrary, Zn deficiency significantly decreased the shoot and root dry weights for Wu312 by 60.3 and 50%, respectively. Zn deficiency increased the R/S ratios for Ye478 and Wu312 by 18.2 and 25.0%, respectively. Each physiological trait displayed normal distribution based on the physiological traits among the RILs, and the coefficients of variation (CV) for each trait ranged from 17.4 to 60.8% (Table 1). In the -Zn and -Zn/CK treatments, the means for each trait of the population were between two parents. The broad-sense heritabilities (H2) of traits were higher than 75%, indicating that traits associated with Zn deficiency tolerance were highly controlled by heredity.
The correlations among different traits were examined and the coefficients of the correlation between each trait pair are shown in Supplementary Tables 9, 10. Spearman’s correlation analysis indicated that the Zn score was positively correlated with the shoot and root dry weights, and was negatively correlated with the R/S ratios (Supplementary Table 9). This was expected because the Zn score was visually scaled by the shoot and root growth in the plants under Zn-deficient conditions (Figure 1). In this study, ZE was mainly characterized by the ratios of the dry weights under -Zn conditions to the values under CK conditions. And these also implied that plants with higher zinc scores also have higher shoot and root dry weights, even higher Zn efficiencies, as well as lower R/S ratios. Additionally, Pearson’s correlation analysis showed that the shoot dry weight displayed a highly positive correlation with the root dry weight at the same Zn nutritional status (r > 0.772, P < 0.01) (Supplementary Table 10), indicating that plants with higher shoot biomass accumulation may also obtain better root growth. And a positive correlation between two Zn-efficiency indexes was found (r = 0.647, P < 0.01) (Supplementary Table 10), suggesting that plants with higher ZE based on shoot dry weight also have higher ZE based on root dry weight.
Quantitative Trait Locus Detection
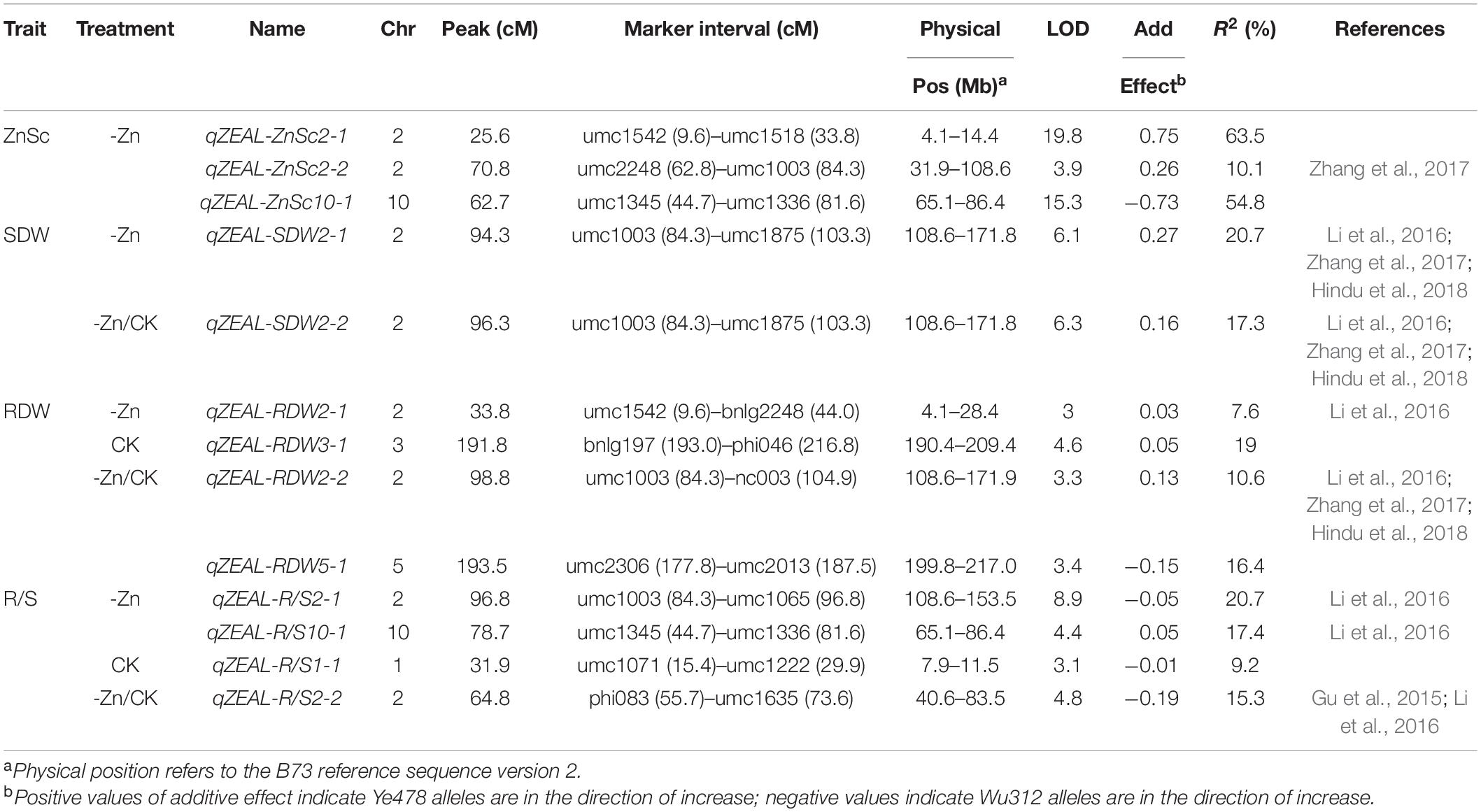
In this study, thirteen ZEALs were detected using the BLUP for each trait, namely, three for the Zn score, two for the shoot dry weight, and four each for the root dry weight and R/S ratio. These loci were distributed on chromosomes 1, 2, 3, 5, and 10, explaining 7.6–63.5% of the phenotypic variation. The information for each QTL is presented in Table 2, including the peak position, 1-LOD genetic interval and its corresponding physical interval, LOD value, and percentage of total phenotypic variance explained (R2), additive effects. The physical positions for each locus are displayed in Figure 7.
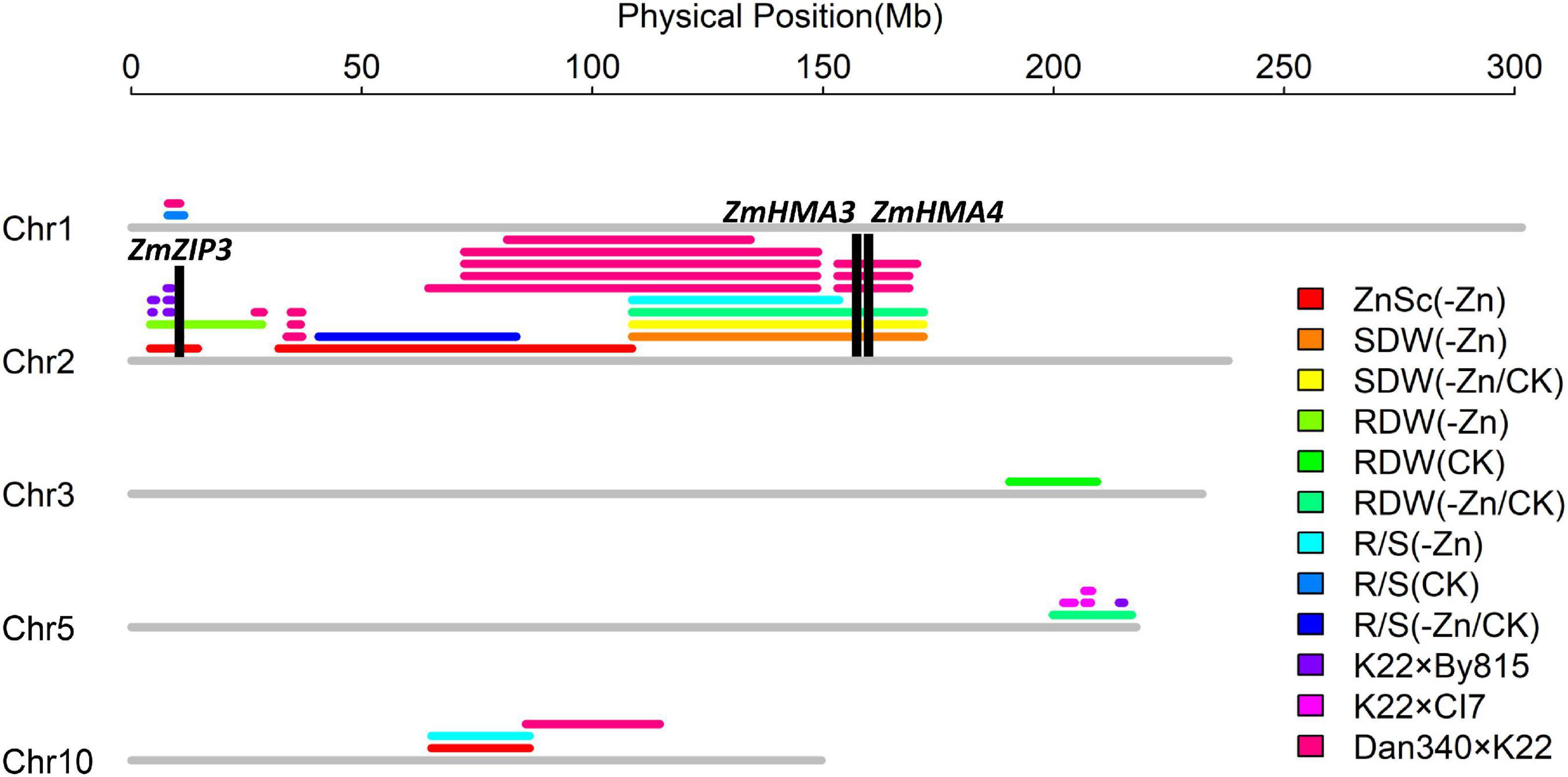
Figure 7. Thirteen loci detected on chromosomes 1, 2, 3, 5, 10 in the Ye478 × Wu312 RIL population and the co-localizations (unpublished data of our lab) identified in the K22 × By815, K22 × CI7, and Dan340 × K22 RIL populations. Important candidate genes (ZmZIP3, ZmHMA3, ZmHMA4) are depicted in black columns.
Quantitative Trait Locus for Zinc Score
Three QTLs controlling the Zn score were identified on chromosomes 2 and 10. qZEAL-ZnSc2-1 is a major QTL detected in the interval of umc1542–umc1518, explaining 63.5% of phenotypic variation. A minor-effect QTL qZEAL-ZnSc2-2 was identified between umc2248 and umc1003, explaining 10.1% of the phenotypic variation. The alleles from Ye478 at these two loci contributed to the increased Zn scores. Another major effect QTL, qZEAL-ZnSc10-1, was mapped in the interval of umc1345–umc1336 on chromosome 10, explaining 54.8% of the phenotypic variation. The additive effect at this locus was originated from Wu312.
Quantitative Trait Locus for the Shoot and Root Dry Weights
Six loci controlling the shoot and root dry weights were mapped on chromosomes 2, 3, and 5. Two moderate-effect QTLs, qZEAL-SDW2-1 and qZEAL-SDW2-2, were both located in the genomic region of umc1003–umc1875, explaining 20.7 and 17.3% of the phenotypic variation, respectively. Four QTLs controlling the root dry weights were identified on chromosomes 2, 3 and 5, explaining 7.6–19.0% of the phenotypic variations. Two minor-effect QTLs were identified on chromosome 2 and explained 7.6–10.6% of the phenotypic variation, including qZEAL-RDW2-1 which was mapped at 4.1–28.4 Mb, and qZEAL-RDW2-2 which was detected at 108.6–171.9 Mb. Two moderate-effect QTLs, qZEAL-RDW3-1 detected on chromosome 3 and qZEAL-RDW5-1 on chromosome 5, accounted for 19.0 and 16.4% of the phenotypic variation, respectively. Except for qZEAL-RDW5-1, the alleles from Ye478 at the five loci contributed to the enhanced shoot and root biomass accumulation under low Zn stress as well as ZE based on dry weights.
Quantitative Trait Locus for Root to Shoot Ratio
Four QTLs controlling the R/S ratio were identified on chromosomes 1, 2, and 10. Under Zn-deficient conditions, the moderate-effect QTL qZEAL-R/S2-1 was flanked by umc1003–umc1065 on chromosome 2 and explained 20.7% of the phenotypic variation. Another moderate-effect QTL qZEAL-R/S10-1 was mapped on chromosome 10 and explained 17.4% of the phenotypic variation. In the CK treatment, a minor-effect QTL qZEAL-R/S1-1 was detected on chromosome 1, accounting for 9.2% of the phenotypic variation. In the -Zn/CK treatment, a moderate-effect QTL qZEAL-R/S2-2 (R2 = 15.3%) was flanked by phi083–umc1635 on chromosome 2.
Quantitative Trait Locus Co-localization and Candidate Gene Identification
In the Ye478 × Wu312 RIL population, four QTL co-localizations were identified by ten QTLs for different traits. The LOD score profile for each QTL within the four QTL co-localizations is shown in Supplementary Figure 3. The major QTL qZEAL-ZnSc2-1 (R2 = 63.5%) was co-localized with the minor-effect QTL qZEAL-RDW2-1 on chromosome 2 (Supplementary Figure 3A). Additionally, qZEAL-ZnSc2-1 was co-localized with qZEAL-R/S2-2 on chromosome 2 (Supplementary Figure 3B). And on chromosome 10, a major-effect QTL qZEAL-ZnSc10-1 (R2 = 54.8%) was co-localized with a moderate-effect QTL qZEAL-R/S10-1 (Supplementary Figure 3D). The significant correlation (P < 0.01) between the Zn score and the shoot and root dry weight and the R/S ratio in the -Zn and -Zn/CK treatments, implied that these two major-effect QTL controlling Zn score may have pleiotropic effects on the biomass accumulation and their allocation in shoots and roots at low Zn nutritional status. In addition, two QTLs controlling the shoot dry weight (qZEAL-SDW2-1 qZEAL-SDW2-2) were co-localized with qZEAL-RDW2-2 and qZEAL-R/S2-1 which controlled the root dry weight and R/S ratio, respectively (Supplementary Figure 3C). There were strong positive correlations (P < 0.01) among the shoot and root dry weights and the R/S ratio in the -Zn and -Zn/CK treatments. These indicated that QTLs for the shoot dry weight may also affect the root growth and both of them may share the same genetic base in response to Zn deficiency stress.
Additionally, based on physical position, eleven genomic regions were identified in the QTL co-localization detected by the QTLs in this study and the QTLs in other three different RIL populations (K22 × By815, K22 × CI7, Dan340 × K22, unpublished data). As shown in Figure 7, on chromosome 2, the major QTL qZEAL-ZnSc2-1 and minor-effect QTL qZEAL-RDW2-1 were co-localized with three QTLs mapped in the K22 × By815 (KB) population which accounted for 9.2–12.0% of the phenotypic variation. Two intervals were identified at 4.4–4.9 and 7.7–8.8 Mb. Additional two intervals (26.8–28.7, 33.7–37.1 Mb) on chromosome 2 were narrowed by three loci which explained 9.3–13.1% of the phenotypic variation in the Dan340 × K22 (DK) population. At 108.6–134.3 Mb, the moderate-effect QTL qZEAL-SDW2-1 (R2 = 20.7%), together with qZEAL-SDW2-2, qZEAL-RDW2-2, and qZEAL-R/S2-1 were co-localized with four QTLs which accounted for 11.9–23.3% of the phenotypic variance in the DK and KB population. At 153.1–168.7 Mb, qZEAL-SDW2-1, qZEAL-RDW2-2, and qZEAL-SDW2-2 were co-localized with the locus which explained 21.2% of the phenotypic variation in the DK population. On chromosome 5, three genetic intervals (202.1–204.6, 206.6–208.1, 214.1–215.3 Mb) were located in the QTL co-localization of qZEAL-RDW5-1 with four loci detected in the KB and DK population (Figure 7). On chromosome 10, the moderate-effect QTL qZEAL-R/S10-1 (R2 = 17.4%) was co-localized with the major-effect QTL qZEAL-ZnSc10-1 (R2 = 54.8%) in this study.
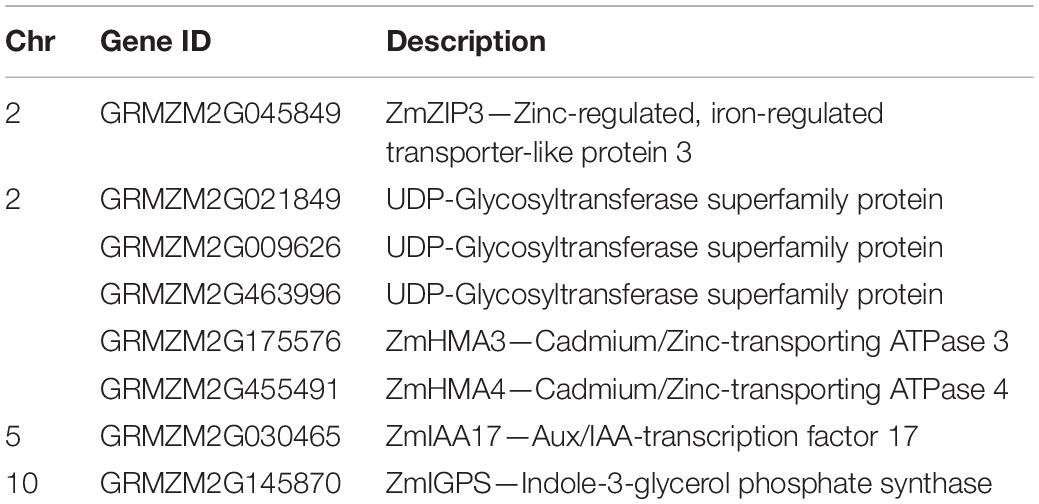
These genomic regions reduced by the co-localization identified on chromosomes 2, 5, and 10 were selected for candidate genes identification (Supplementary Table 12). In total, 601 candidate genes were identified within the eleven intervals reduced by the QTL co-localization: 317 genes on chromosome 2, 145 genes on chromosome 5, 139 genes on chromosome 10 (Supplementary Table 11). Among them, eight candidate genes were considered to be associated with Zn efficiency in maize, including ZmZIP3, ZmHMA3, and ZmHMA4 (Table 3).
Expression of ZIP Genes in Maize
The results of Experiment 1 have shown that the differential ZEs of the most tolerant (Ye478) and the most sensitive (Wu312) inbred lines were mainly due to their differences in Zn uptake and transport efficiency. In addition, ZmZIP3, a member of the ZIP gene family which encodes major Zn transporters in maize, was identified in the major QTL qZEAL-ZnSc2-1 (R2 = 63.5%) in the Ye478 × Wu312 RIL population. Therefore, based on Experiments 1 and 2 (Figures 4A,B), Experiment 3 was designed to analyze the expression of eight ZIP genes (ZmZIP1-ZmZIP8) under -Zn and CK conditions (Figure 4C).
The expression of ZmZIP 1-8 under -Zn and CK conditions was shown in Figure 8. ZmZIP3, ZmZIP4, ZmZIP5, ZmZIP7, and ZmZIP8 in the roots of Ye478 showed at least 39-, 6-, 22-, 3-, and 24-fold upregulation in response to Zn deficiency, respectively. Zn deficiency had no significant effects on the expressions of ZmZIP1, ZmZIP2, and ZmZIP6 in the roots of Ye478. The relative expression level of ZmZIP2, ZmZIP4, ZmZIP5, and ZmZIP7 in the Zn-deficient roots of Wu312 were increased by 1. 5-, 4. 5-, 9. 3-, and 1.9-fold, respectively. The other four genes (ZmZIP1, ZmZIP3, ZmZIP6, and ZmZIP8) were not upregulated in response to Zn deficiency. Notably, ZmZIP3 and ZmZIP8 were barely expressed in the roots of Ye478 under CK conditions but showed high gene expression under Zn deficiency. By contrast, there was no significant difference in the expression level of these two genes in the roots of Wu312 between -Zn and CK conditions. Additionally, the upregulations of ZmZIP4, ZmZIP5, and ZmZIP7 were higher in the Zn-deficient roots of the tolerant line (Ye478) than in those of the sensitive line (Wu312). These facts indicated that the five ZIP genes (ZmZIP3, ZmZIP4, ZmZIP5, ZmZIP7, and ZmZIP8) may be important for Zn deficiency tolerance variations in maize.
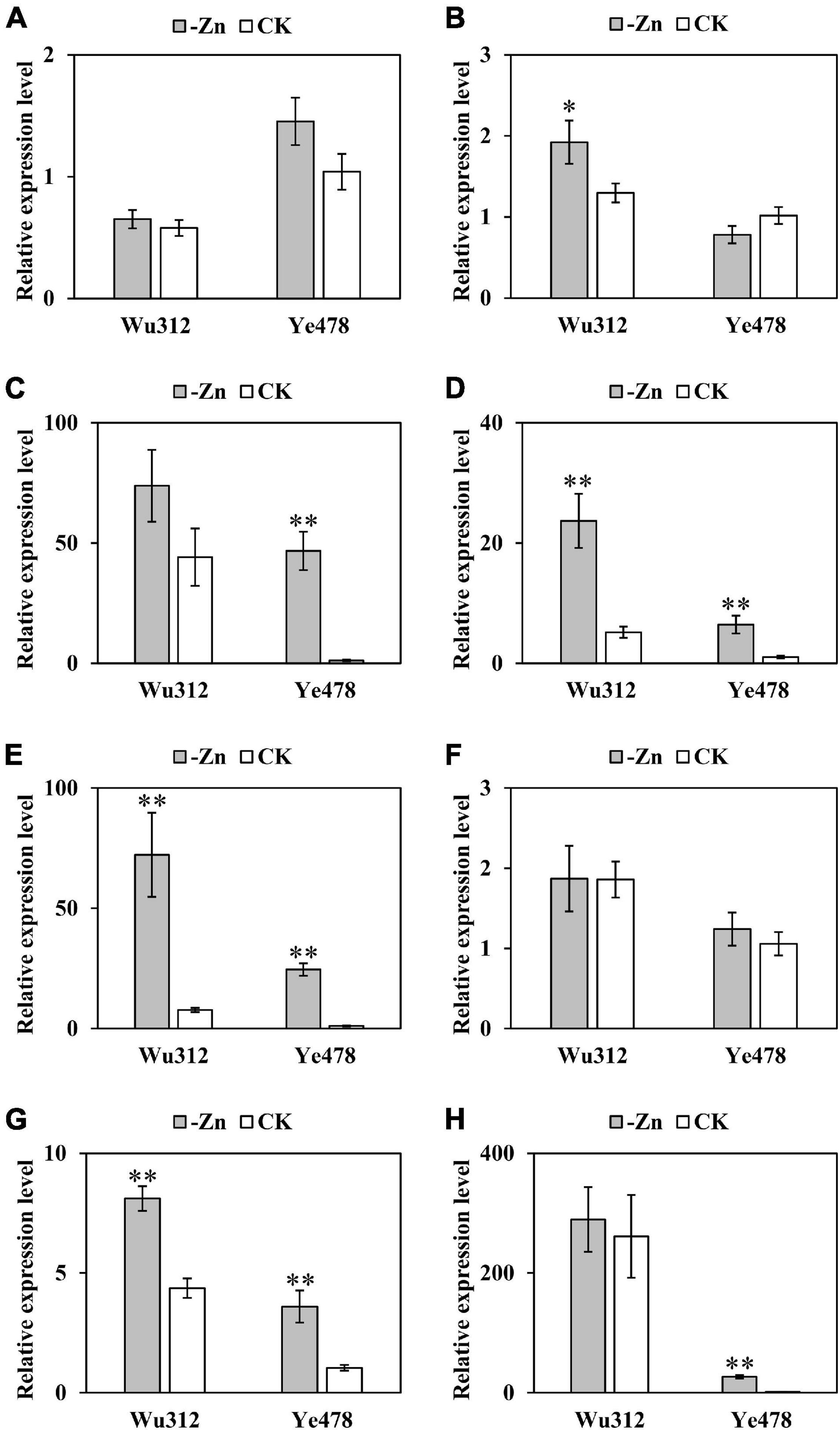
Figure 8. Expression of eight ZIP genes in maize. Relative expression levels of eight ZmZIPs (ZmZIP1- ZmZIP8) in the roots of Wu312 and Ye478 grown hydroponically in a mixed-crop system for 21 days after transplanting in the -Zn (0.3 μmol L–1 Zn-EDTA) and CK (10 μmol L–1 Zn-EDTA) treatment. (A–H) show the relative expression levels of ZmZIP1- ZmZIP8, respectively. ∗ and ∗∗ indicate the significant differences between the -Zn and CK treatments at P < 0.05 and P < 0.01, respectively.
Discussion
Physiological Mechanisms Underlying Zn Efficiency
Root to shoot ratio increase, an initial response to Zn deficiency, is also one of the most sensitive indicators to characterize ZE. This response indicates a compensation mechanism that plants could allocate the photosynthetic products produced by the shoot to root growth (Rengel and Graham, 1995), which may lead to greater morphological changes in the roots (Genc et al., 2007). Besides, the PCA analysis showed that Ye478 had a higher Zn uptake, transport efficiency, as well as lower R/S ratio efficiency, compared with Wu312 (Figure 5), indicating that the molecular mechanisms underlying Zn uptake and transport may be vital in explaining the differences in ZE between Ye478 and Wu312.
The Zn-efficient genotypes may not necessarily have higher Zn concentration in the tissue or grain (Graham et al., 1992; Cakmak et al., 1996). Our results further confirmed that the Zn concentrations in the shoots and roots were not correlated with ZE (Figure 3) and were not suitable to characterize ZE in maize at the seedling stage. The shoot Zn concentrations of the Zn-efficient inbred lines (Ye478, CI7) were not significantly higher than the Zn-inefficient inbred lines (Wu312, K22), mainly due to the dilute effect (Marschner, 1995). The Zn-efficient inbred lines, which do not maintain higher shoot Zn concentrations, can still produce more biomass than the Zn-inefficient inbred lines, possibly attributed to the advantages in the subcellular Zn compartmentation and homeostasis (Outten and O’Halloran, 2001; Hacisalihoglu et al., 2004) and the more efficient biochemical utilization of the cellular Zn (Cakmak, 2000; Hacisalihoglu et al., 2003). The Zn concentrations in the shoots and roots declined significantly under Zn-deficient conditions, but the Fe and Cu concentrations in the shoots and the Mn concentrations in the roots increased obviously. This may be due to the competitive interaction of the Zn deficiency-inducible transport proteins in the transport of Fe, Mn, Cu compared with Zn across the plasma membrane (Rengel and Graham, 1996; Tiong et al., 2015).
Comparison of Detected Quantitative Trait Locus With Previous Reports
There were few reports on the QTL mapping of Zn deficiency tolerance in maize and only a small number of researches related to Zn nutrition in maize, which were mostly concentrated on Zn and other mineral concentration or content in grain, have been reported (Šimić et al., 2012; Gu et al., 2015; Jin et al., 2015; Zhang et al., 2017; Hindu et al., 2018; Ju et al., 2018). On chromosome 2, five QTLs (qZEAL-ZnSc2-2, qZEAL-SDW2-1, qZEAL-SDW2-2, qZEAL-RDW2-2, qZEAL-R/S2-2) detected in our work were co-localized with the loci controlling kernel mineral concentration in maize (Table 2). Among these five QTLs, the overlapped genomic regions of qZEAL-SDW2-1, qZEAL-SDW2-2, and qZEAL-RDW2-2, were co-localized with the QTLs controlling Fe concentration in grain by linkage analysis, and the SNP associated with kernel Fe concentration by GWAS (Zhang et al., 2017; Hindu et al., 2018). Additionally, qZEAL-ZnSc2-2 and qZEAL-R/S2-2 were co-localized with the QTL for the grain Fe concentration and the QTL controlling the kernel Mn concentration, respectively (Gu et al., 2015; Zhang et al., 2017). Apart from that, on chromosomes 2 and 10, seven QTLs detected in the -Zn and -Zn/CK treatment of this study, were co-localized with the loci associated with the root dry weight and root morphological traits under high- and low-nitrogen levels (Li et al., 2016). These results implicate that these QTL regions may have pleiotropic effects on the mineral concentration of grains and root traits in maize. Overall, 62% of the QTLs have been identified to be co-localized by the loci detected in the previous studies (Table 2), suggesting that these QTLs detected by different traits may be highly reliable for candidate gene identification.
Expression Patterns of ZIPs in Maize
Despite ZIPs having been characterized in many plants, there are little researches concerning the expressions of ZIPs in maize. The expression patterns for ZmZIPs in our study showed that ZmZIP4, ZmZIP5 and ZmZIP7 were induced in Zn-deficient roots, which is consistent with Li et al. (2013) and Mager et al. (2018), indicating that ZmZIP4, ZmZIP5, ZmZIP7, and ZmZIP8 may play important roles in the uptake and transport of Zn. The ZIPs from maize and rice may share a common evolutionary ancestor, and they existed as orthologs, including ZmZIP3 and OsZIP3, ZmZIP4 and OsZIP4, ZmZIP5, ZmZIP7 and OsZIP7, and ZmZIP8 and OsZIP8 (Li et al., 2013). In rice, OsZIP4, OsZIP7, and OsZIP8 are strongly induced in Zn-deficient shoots and roots and complement the growth defect of the Zn-uptake yeast mutant, indicating that they are functional transporters of Zn (Ishimaru et al., 2005; Lee et al., 2010a; Kabir et al., 2017). OsZIP4, an ortholog of ZmZIP4, which is mainly expressed in the vascular bundles of Zn-deficient roots (Lee et al., 2010b) and meristem of Zn-deficient roots and shoots (Ishimaru et al., 2005), may be responsible for regulating Zn supply in developing young leaves and transporting Zn over a long distance from the old to young leaves (Lee et al., 2010b). OsZIP7 which is the ortholog for both ZmZIP5 and ZmZIP7 regulates the distribution of Zn within rice, including xylem loading into roots and inter-vascular transfer in nodes, to preferentially deliver Zn to developing tissues (Tan et al., 2019).
Candidate Genes Associated With Zinc Deficiency Tolerance
ZmZIP3, also known as GRMZM2G045849, was detected in the major QTL qZEAL-ZnSc2-1 which explained 63.5% of the phenotypic variation. It was reported that ZmZIP3 is induced in Zn-deficient roots and efficiently reverses the growth of yeast mutant defectives in Zn uptake caused by Zn deficiency (Li et al., 2013). Notably, distinct expression patterns of ZIP genes exist between Zn tolerant and sensitive genotypes under Zn deficiency (Khatun et al., 2018). In contrast to the mild upregulation of ZmZIP3 in Zn deficiency recorded by Mager et al. (2018), there was 39-fold upregulation of ZmZIP3 in the Zn-deficient roots of Ye478, whereas there was no significant difference of expression level in Wu312 between -Zn and CK conditions. This indicated that ZmZIP3 may be associated with the ZE difference between Zn-efficient and Zn-inefficient inbred lines.
OsZIP3, an ortholog of ZmZIP3, was detected in the vascular bundles and epidermal cells inside the stem in rice (Ramesh et al., 2003). More recently, OsZIP3 has been found to be located at the xylem, intervening the parenchyma cells and xylem transfer cell of the enlarged vascular bundle (EVB) in the nodes, and show a high expression in the nodes (Sasaki et al., 2015). The experiment with the Zn stable isotope and knockdown analysis confirmed that OsZIP3 is required for unloading Zn from the xylem of EVB, which initiates a preferential distribution to the developing tissues in the shoot (Sasaki et al., 2015). HvZIP3, which is 83% identical with OsZIP3, restores the growth of the yeast mutant defective in Zn2+ uptake grown under Zn2+-depleted conditions (Pedas et al., 2009). HvZIP3, an ortholog of ZmZIP3, is highly and consistently induced in Zn-deficient roots and plays a role in the uptake and root-to-shoot translocation of Zn under Zn deficiency, which is supported by the results of the tissue-specific expression in the roots (Tiong et al., 2015). Hence, the ZmZIP3 mapped in this research, which is the ortholog for OsZIP3 and HvZIP3, is suggested to play an essential role in Zn uptake and transport in maize.
In this research, ZmHAM3 and ZmHMA4 (GRMZM2G175576, GRMZM2G455491) encoding Zn transporters were identified in the co-localizations on chromosome 2. AtHMA2 which is the homologous gene of ZmHMA3 has N- and C-terminal domains that could bind Zn2+ with high affinity. AtHMA2 is expressed in the root vasculature and is responsible for the root-to-shoot translocation of Zn via xylem loading (Eren and Arguello, 2004; Wong et al., 2009). Hussain et al. (2004) have done research on hma2, hma4 double mutants, and found that hma2, hma4 double mutants have a severe shoot Zn-deficiency phenotype and accumulate Zn in roots. The results also showed that the levels of Zn, but not other essential elements, in the shoot tissues of a hma2, hma4 double mutant are decreased compared with the wild type.
OsHMA3, the homologous gene of ZmHMA3 in rice, is the first QTL for cadmium (Cd) accumulation in rice which has been cloned by forwarding genetics (Ueno et al., 2010; Liu et al., 2019). OsHMA3 is preferentially expressed in the roots and is involved in the sequestration of Cd into the vacuoles of the roots. Some rice cultivars possess weak or loss-of-function alleles of OsHMA3 and the difference in the messenger RNA (mRNA) expression level of OsHMA3, accumulating high levels of Cd in the shoots and grain (Miyadate et al., 2011; Yan et al., 2016; Cai et al., 2019). When OsHMA3 is overexpressed under the control of the ubiquitin or OsHMA2 promoter, not only Cd but also Zn in the roots and the root cell saps are increased (Shao et al., 2018). Several genes related to Zn transporters are upregulated in the OsHMA3-overexpressed lines (Sasaki et al., 2014). Zn sequestered by OsHMA3 in the roots provides an important source for the shoot under conditions of Zn deficiency (Yamaji et al., 2013; Sasaki et al., 2015). Up to now, many HMA genes have been identified and studied in Arabidopsis and rice. However, the analysis of the HMA gene family in maize is still scarce. Further characterization of these transporters is crucial to understand the mechanism underlying the uptake and transport of Zn in maize.
Under abiotic stress, plants scavenge reactive oxygen species (ROS) to restore redox metabolism and to keep cellular turgor and structures actively functioning (Yancey, 2005; Mittler, 2006). Zn deficiency causes the accumulation of ROS, resulting in the oxidative degradation of the indole-3-acetic acid (IAA) and repression in the shoot growth (Marschner, 1995). Furthermore, Zn is involved in the detoxification of superoxide radicals and the synthesis of phytohormones (Cakmak, 2000), directly participating in the synthesis of IAA (Alloway, 2008). Variations in hormones may be signals in response to nutrient stress, resulting in morphological and physiological changes in plants. It is reported that the IAA level of the shoot tips and young leaves in -Zn plants is about 50% lower than that in CK plants (Cakmak et al., 1989). The genes associated with the signaling and metabolism of IAA may be related to the mechanisms in tolerating Zn-deficiency stress.
To prevent auxin-responsive transcription in the path from the auxin signal perception to the altered gene expression, Aux/IAA proteins generally function as repressors (Korasick et al., 2014). This pathway requires three components: auxin-perceiving TIR1/AFB F-box proteins, Aux/IAA repressor proteins, and auxin response factors (ARF) transcription factors (Weijers and Wagner, 2016). When the auxin level is higher, ARFs are released from the inhibition of Aux/IAAs, which allows ARF proteins to reach their AuxRE targets to derepress/activate the early auxin response genes (such as GH3 and SAUR gene family). When the auxin concentration decreases below a threshold, the Aux/IAAs are associated with ARFs and those auxin-response genes are repressed (Roosjen et al., 2018; Figure 4D).
In maize, ZmAFB2 shared high amino acid sequence homology with Arabidopsis AtAFB2 and AtAFB3 and contained one F-Box region and four LRR regions (Yang et al., 2013). It has been reported that miR393 targets several auxin receptors for degradation such as TIR1, AFB2, and AFB3 (Navarro et al., 2006). The ZmAFB2 contained the miR393 cleaved site. Iglesias et al. (2011) demonstrated that salicylic acid (SA) might inhibit auxin signaling by the translational repression of TIR1/AFB proteins during stress. ZmAFB2 may be involved in the crosstalk between SA and auxin signal transduction (Yang et al., 2013). The auxin-responsive gene Zm00001d045203, which encodes an Aux/IAA inhibitor involved in the SCFTIR1/AFB-mediated auxin signaling pathway, is the homolog of Arabidopsis thaliana IAA29 (Zhang et al., 2019).
Among the 34 members of the maize Aux/IAA gene family, all but one (ZmIAA23) tested maize Aux/IAA genes were auxin-inducible, displaying two types of auxin induction within 3 h of treatment (Ludwig et al., 2013). Putative cis-acting regulatory DNA elements involved in auxin response, light signaling transduction, and abiotic stress adaption (such as dehydration-responsive element, low temperature-responsive element), were observed in the promoters of ZmIAA genes (Wang et al., 2010). The diversity of the cis-elements in the promoters of ZmIAA genes is related to the key role of Aux/IAA proteins in plant life. The maize lateral root primordia 1 (lrp1) encodes a transcriptional activator that is directly regulated by the Aux/IAA protein ROOTLESS WITH UNDETECTABLE MERISTEM 1 (RUM1 corresponding to ZmIAA10), which regulates the initiation of lateral and seminal root (von Behrens et al., 2011; Zhang et al., 2014; Wang et al., 2021). RUM1 acts as a transcriptional repressor interacting with ZmARF25 or ZmARF34, thereby regulating the transcription of auxin-responsive genes in pericycle cells of primary roots (Zhang et al., 2015).
An analysis of the conserved motifs indicated that 23 of 35 ZmARF proteins contain domains III and IV, which were also found in the C-terminus of Aux/IAAs (Liu Y. et al., 2011; Saidi and Hajibarat, 2020). These domains have been shown to mediate both homo- and hetero-dimerization between the members of the Aux/IAA and ARF families (Kim et al., 1997; Galli et al., 2018). Twelve maize ARFs with glutamine-rich middle regions could be activators in modulating the expression of auxin-responsive genes (Wang et al., 2012). The expression of ARF genes in maize is regulated by auxin and small RNAs. The dynamic expression patterns of ZmARF genes were observed in different stages of embryo development (Xing et al., 2011).
The GRMZM2G030465 on chromosome 5, also known as ZmIAA17 which is a member of the Aux/IAA gene family (Ludwig et al., 2013), was detected at the locus qZEAL-RDW5-1. AtIAA13, an orthologous of ZmIAA17 in Arabidopsis, is highly expressed in the cortex and lateral root primordia, suggesting that AtIAA13 functions in initiating the formation of the lateral roots (Yamauchi et al., 2019). OsIAA9, an ortholog of ZmIAA17 in rice, is also greatly induced by multiple hormones and abiotic stresses (Luo et al., 2015). The ectopic overexpression of OsIAA9 results in fewer crown and lateral roots and reduced the inhibition of root elongation by auxin, suggesting that OsIAA9 is a negative regulator of auxin-regulated root growth (Song, 2019). Additionally, OsIAA9 is confirmed to regulate gravitropic response by controlling granules accumulation and distribution in root tips (Luo et al., 2015).
GRMZM2G021849, GRMZM2G009626, and GRMZM2G463996 on chromosome 2, co-localized by four QTLs in the Ye478 × Wu312 and Dan340 × K22 population, were identified to be members of UDP-Glycosyltransferase superfamily, which are likely associated with IAA inactivation (Tanaka et al., 2014). Some UDP-glycosyltransferases (UGTs) catalyze the glucosylation of plant hormones, including auxin, ABA, cytokinins, and SA using UDP-glucose as a co-substrate (Lim and Bowles, 2004; Gachon et al., 2005). It was reported that UGT84B1, UGT74E2, and UGT74B1 catalyzed the conversion of IAA to IAA-Glc (Jackson et al., 2001; Grubb et al., 2004; Tognetti et al., 2010). Furthermore, GRMZM2G145870, also known as indole-3-glycerol phosphate synthase (ZmIGPS) in maize, was identified within qZEAL-ZnSc10-1, which was the second largest effect QTL. Indole-3-glycerol phosphate synthase is the only known enzyme to catalyze the formation of the indole ring, which is the key for the tryptophan-independent IAA synthesis pathway in plants (Normanly and Bartel, 1999; Ouyang et al., 2000). Taken together, the members of the UGT family, IAA9, and IGPS3 may be involve in the biosynthesis of IAA and likely affect plant growth and development. Nevertheless, specific functions for these candidate genes in maize are still lacking, and further systematic studies are required.
In summary, there are few systematic studies and explicit demonstrations of the molecular mechanism underlying the tolerance of Zn deficiency stress. In our study, members of the ZIP gene family and HMA gene family were detected by linkage analysis. ZIP3 is required for unloading Zn from the xylem of enlarged vascular bundles and for the preferential distribution to the developing tissues in the shoot (Sasaki et al., 2015). HMA3 is localized to the tonoplast of all root cells (Ueno et al., 2010) and sequester Zn2 + into the vacuole (Cai et al., 2019). HMA4 plays an essential role in the root-to-shoot translocation of Zn and as an efflux pump (Wong and Cobbett, 2009). Additionally, Zn deficiency causes ROS in plants, which leads to fluctuating auxin levels. In the auxin-dependent gene regulation pathway, Aux/IAA acts as a repressor of auxin transcription factors (Weijers and Wagner, 2016). Under low-auxin levels, Aux/IAA is bond to ARF functioning as a repressor to prevent transcription. Under high-auxin levels, Aux/IAA disassociates ARF and is bonded to TIR1/AFB with auxin, then ARF is released from the inhibition of Aux/IAA and derepresses/activates the regulation of downstream genes (Roosjen et al., 2018; Figure 4D). These genes mentioned above may play very important roles in the mechanism of Zn deficiency stress tolerance.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
JX and XW performed the experiments and analyzed the data. JX, XW, and HZ wrote the manuscript. FY designed the study and modified the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program of China (2016YFD0200405).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Xiaohong Yang (China Agricultural University) for providing 18 maize inbred lines. We also thank Fanjun Chen (China Agricultural University) for providing the inbred lines Wu312 and Ye478, and the Ye478 × Wu312 RIL population.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.739282/full#supplementary-material
Footnote
References
Alloway, B. J. (2008). Zinc in soil and crop nutrition. Belgium and Paris. France: International zinc association.
Alonso-Blanco, C., Aarts, M. G., Bentsink, L., Keurentjes, J. J., Reymond, M., Vreugdenhil, D., et al. (2009). What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell 21, 1877–1896. doi: 10.1105/tpc.109.068114
Broadley, M. R., White, P. J., Hammond, J. P., Zelko, I., and Lux, A. (2007). Zinc in plants. New Phytol. 173, 677–702. doi: 10.1111/j.1469-8137.2007.01996.x
Cai, H., Huang, S., Che, J., Yamaji, N., and Ma, J. F. (2019). The tonoplast-localized transporter OsHMA3 plays an important role in maintaining Zn homeostasis in rice. J. Exp. Bot. 70, 2717–2725. doi: 10.1093/jxb/erz091
Cakmak, I. (2000). Possible roles of zinc in protecting plant cells from damage by reactive oxygen species. New Phytol. 146, 185–205. doi: 10.1046/j.1469-8137.2000.00630.x
Cakmak, I., Marschner, H., and Bangerth, F. (1989). Effect of zinc nutritional status on growth, protein metabolism and levels of indole-3-acetic acid and other phytohormones in bean (Phaseolus vulgaris L.). J. Exp. Bot. 40, 405–412. doi: 10.1093/jxb/40.3.405
Cakmak, I., Yilmaz, A., Kalayci, M., Ekiz, H., Torun, B., Erenoğlu, B., et al. (1996). Zinc deficiency as a critical problem in wheat production in Central Anatolia. Plant Soil 180, 165–172. doi: 10.1007/BF00015299
Chen, W. R., Feng, Y., and Chao, Y. E. (2008). Genomic analysis and expression pattern of OsZIP1, OsZIP3, and OsZIP4 in two rice (Oryza sativa L.) genotypes with different zinc efficiency. Russ. J. Plant Physiol. 55, 400–409. doi: 10.1134/S1021443708030175
Chen, X. P., Cui, Z. L., Vitousek, P. M., Cassman, K. G., Matson, P. A., Bai, J. S., et al. (2011). Integrated soil-crop system management for food security. Proc. Natl. Acad. Sci. U. S. A. 108, 6399–6404. doi: 10.1073/pnas.1101419108
Cui, Z. L., Dou, Z. X., Chen, X. P., Ju, X. T., and Zhang, F. S. (2014). Managing agricultural nutrients for food security in China: past, present, and future. Agro. J. 106, 191–198. doi: 10.1002/jsfa.6098
Eren, E., and Arguello, J. M. (2004). Arabidopsis HMA2, a divalent heavy metal-transporting (PIB)-type ATPase, is involved in cytoplasmic Zn2+ homeostasis. Plant Physiol. 136, 3712–3723. doi: 10.1104/pp.104.046292
Gachon, C. M., Langlois-Meurinne, M., and Saindrenan, P. (2005). Plant secondary metabolism glycosyltransferases: the emerging functional analysis. Trends Plant Sci. 10, 542–549. doi: 10.1016/j.tplants.2005.09.007
Gainza-Cortés, F., Pérez-Dïaz, R., Pérez-Castro, R., Tapia, J., Casaretto, J. A., González, S., et al. (2012). Characterization of a putative grapevine Zn transporter, VvZIP3, suggests its involvement in early reproductive development in Vitis vinifera L. BMC Plant Biol. 12:111. doi: 10.1186/1471-2229-12-111
Galli, M., Khakhar, A., Lu, Z., Chen, Z., Sen, S., Joshi, T., et al. (2018). The DNA binding landscape of the maize AUXIN RESPONSE FACTOR family. Nat. Commun. 9:4526. doi: 10.1038/s41467-018-06977-6
Genc, Y., Huang, C. Y., and Langridge, P. (2007). A study of the role of root morphological traits in growth of barley in zinc-deficient soil. J. Exp. Bot. 58, 2775–2784. doi: 10.1093/jxb/erm142
Genc, Y., Verbyla, A. P., Torun, A. A., Cakmak, I., Willsmore, K., Wallwork, H., et al. (2009). Quantitative trait loci analysis of zinc efficiency and grain zinc concentration in wheat using whole genome average interval mapping. Plant Soil 314, 49–66. doi: 10.1007/s11104-008-9704-3
Graham, R. D., Ascher, J. S., and Hynes, S. C. (1992). Selecting zinc efficient cereal genotypes for soil of low zinc status. Plant Soil 146, 241–250. doi: 10.1016/j.jtemb.2009.05.002
Graham, R. D., Knez, M., and Welch, R. M. (2012). How much nutritional iron deficiency in humans globally is due to an underlying zinc deficiency? Adv. Agron. 115, 1–40. doi: 10.1016/B978-0-12-394276-0.00001-9
Grubb, C. D., Zipp, B. J., Ludwig-Müller, J., Masuno, M. N., Molinski, T. F., and Abel, S. (2004). Arabidopsis glucosyltransferase UGT74B1 functions in glucosinolate biosynthesis and auxin homeostasis. Plant J. 40, 893–908. doi: 10.1111/j.1365-313X.2004.02261.x
Gu, R., Chen, F., Liu, B., Wang, X., Liu, J., Li, P., et al. (2015). Comprehensive phenotypic analysis and quantitative trait locus identification for grain mineral concentration, content, and yield in maize (Zea mays L.). Theor. Appl. Genet. 128, 1777–1789. doi: 10.1007/s00122-015-2546-5
Hacisalihoglu, G., Hart, J. J., and Kochian, L. V. (2001). High- and low-affinity zinc transport systems and their Possible role in zinc efficiency in bread wheat. Plant Physiol. 125, 456–463. doi: 10.1104/pp.125.1.456
Hacisalihoglu, G., Hart, J. J., Vallejos, C. E., and Kochian, L. V. (2004). The role of shoot-localized processes in the mechanism of Zn efficiency in common bean. Planta 218, 704–711. doi: 10.1007/s00425-003-1155-8
Hacisalihoglu, G., Hart, J. J., Wang, Y. H., Cakmak, I., and Kochian, L. V. (2003). Zinc efficiency is correlated with enhanced expression and activity of zinc-requiring enzymes in wheat. Plant Physiol. 131, 595–602. doi: 10.1104/pp.011825
Hacisalihoglu, G., and Kochian, L. V. (2003). How do some plants tolerate low levels of soil zinc? Mechanisms of zinc efficiency in crop plants. New Phytol. 159, 341–350. doi: 10.1046/j.1469-8137.2003.00826.x
Hindu, V., Palacios-Rojas, N., Babu, R., Suwarno, W. B., Rashid, Z., Usha, R., et al. (2018). Identification and validation of genomic regions influencing kernel zinc and iron in maize. Theor. Appl. Genet. 131, 1443–1457. doi: 10.1007/s00122-018-3089-3
Hussain, D., Haydon, M. J., Wang, Y., Wong, E., Sherson, S. M., Young, J., et al. (2004). P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell 16, 1327–1339. doi: 10.1105/tpc.020487
Iglesias, M. J., Terrile, M. C., and Casalongué, C. A. (2011). Auxin and salicylic acid signalings counteract the regulation of adaptive responses to stress. Plant Signal Behav. 6, 452–454. doi: 10.4161/psb.6.3.14676
Impa, S. M., Gramlich, A., Tandy, S., Schulin, R., Frossard, E., and Johnson-Beebout, S. E. (2013a). Internal Zn allocation influences Zn deficiency tolerance and grain Zn loading in rice (Oryza sativa L.). Front. Plant Sci. 4:534. doi: 10.3389/fpls.2013.00534
Impa, S. M., Morete, M. J., Ismail, A. M., Schulin, R., and Johnson-Beebout, S. E. (2013b). Zn uptake, translocation and grain Zn loading in rice (Oryza sativa L.) genotypes selected for Zn deficiency tolerance and high grain Zn. J. Exp. Bot. 64, 2739–2751. doi: 10.1093/jxb/ert118
Ishimaru, Y., Suzuki, M., Kobayashi, T., Takahashi, M., Nakanishi, H., Mori, S., et al. (2005). OsZIP4, a novel zinc-regulated zinc transporter in rice. J. Exp. Bot. 56, 3207–3214. doi: 10.1093/jxb/eri317
Jackson, R. G., Lim, E. K., Li, Y., Kowalczyk, M., Sandberg, G., Hoggett, J., et al. (2001). Identification and biochemical characterization of an Arabidopsis indole-3-acetic acid glucosyltransferase. J. Biol. Chem. 276, 4350–4356. doi: 10.1074/jbc.M006185200
Jiao, Y., Peluso, P., Shi, J., Liang, T., Stitzer, M. C., Wang, B., et al. (2017). Improved maize reference genome with single-molecule technologies. Nature 546, 524–527. doi: 10.1038/nature22971
Jin, T., Chen, J., Zhu, L., Zhao, Y., Guo, J., and Huang, Y. (2015). Comparative mapping combined with homology-based cloning of the rice genome reveals candidate genes for grain zinc and iron concentration in maize. BMC. Genet. 16:17. doi: 10.1186/s12863-015-0176-1
Ju, C., Zhang, W., Liu, Y., Gao, Y., Wang, X., Yan, J., et al. (2018). Genetic analysis of seedling root traits reveals the association of root trait with other agronomic traits in maize. BMC Plant Biol. 18:171. doi: 10.1186/s12870-018-1383-5
Kabir, A. H., Hossain, M. M., Khatun, M. A., Sarkar, M. R., and Haider, S. A. (2017). Biochemical and molecular mechanisms associated with Zn deficiency tolerance and signaling in rice (Oryza sativa L.). J. Plant Interact. 12, 447–456. doi: 10.1080/17429145.2017.1392626
Kambe, T., Tsuji, T., Hashimoto, A., and Itsumura, N. (2015). The physiological, biochemical, and molecular roles of zinc transporters in zinc homeostasis and metabolism. Physiol. Rev. 95, 749–784. doi: 10.1152/physrev.00035.2014
Kambe, T., Yamaguchi-Iwai, Y., Sasaki, R., and Nagao, M. (2004). Overview of mammalian zinc transporters. Cell Mol. Life Sci. 61, 49–68. doi: 10.1007/s00018-003-3148-y
Kavitha, P. G., Kuruvilla, S., and Mathew, M. K. (2015). Functional characterization of a transition metal ion transporter, OsZIP6 from rice (Oryza sativa L.). Plant Physiol. Biochem. 97, 165–174. doi: 10.1016/j.plaphy.2015.10.005
Khatun, M. A., Hossain, M. M., Bari, M. A., Abdullahil, K. M., Parvez, M. S., Alam, M. F., et al. (2018). Zinc deficiency tolerance in maize is associated with the up-regulation of Zn transporter genes and antioxidant activities. Plant Biol. 20, 765–770. doi: 10.1111/plb.12837
Khoshgoftarmanesh, A. H., Afyuni, M., Norouzi, M., Ghiasi, S., and Schulin, R. (2018). Fractionation and bioavailability of zinc (Zn) in the rhizosphere of two wheat cultivars with different Zn deficiency tolerance. Geoderma 309, 1–6. doi: 10.1016/j.geoderma.2017.08.019
Kim, J., Harter, K., and Theologis, A. (1997). Protein–protein interactions among the Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 94, 11786–11791. doi: 10.1073/pnas.94.22.11786
King, J. C., Brown, K. H., Gibson, R. S., Krebs, N. F., Lowe, N. M., Siekmann, J. H., et al. (2015). Biomarkers of nutrition for development (BOND)-zinc review. J. Nutr. 146, 858S–885S. doi: 10.3945/jn.115.220079
Korasick, D. A., Westfall, C. S., Lee, S. G., Nanao, M. H., Dumas, R., Hagen, G., et al. (2014). Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc. Natl. Acad. Sci. U. S. A. 111, 5427–5432. doi: 10.1073/pnas.1400074111
Krämer, U., Talke, I. N., and Hanikenne, M. (2007). Transition metal transport. FEBS. Lett. 581, 2263–2272. doi: 10.1016/j.febslet.2007.04.010
Lee, J. S., Sajise, A. G. C., Gregorio, G. B., Kretzschmar, T., Ismail, A. M., and Wissuwa, M. (2017). Genetic dissection for zinc deficiency tolerance in rice using bi-parental mapping and association analysis. Theor. Appl. Genet. 130, 1903–1914. doi: 10.1007/s00122-017-2932-2
Lee, S., and An, G. (2009). Over-expression of OsIRT1 leads to increased iron and zinc accumulations in rice. Plant Cell Environ. 32, 408–416. doi: 10.1111/j.1365-3040.2009.01935.x
Lee, S., Jeong, H. J., Kim, S. A., Lee, J., Guerinot, M. L., and An, G. (2010a). OsZIP5 is a plasma membrane zinc transporter in rice. Plant Mol. Biol. 73, 507–517. doi: 10.1007/s11103-010-9637-0
Lee, S., Kim, S. A., Lee, J., Guerinot, M. L., and An, G. (2010b). Zinc deficiency-inducible OsZIP8 encodes a plasma membrane-localized zinc transporter in rice. Mol. Cells 29, 551–558. doi: 10.1007/s10059-010-0069-0
Li, P., Zhuang, Z., Cai, H., Cheng, S., Soomro, A. A., Liu, Z., et al. (2016). Use of genotype-environment interactions to elucidate the pattern of maize root plasticity to nitrogen deficiency. J. Integr. Plant Biol. 58, 242–253. doi: 10.1111/jipb.12384
Li, S., Zhou, X., Huang, Y., Zhu, L., Zhang, S., Zhao, Y., et al. (2013). Identification and characterization of the zinc-regulated transporters, iron-regulated transporter-like protein (ZIP) gene family in maize. BMC Plant Biol. 13:114. doi: 10.1186/1471-2229-13-114
Lim, E. K., and Bowles, D. J. (2004). A class of plant glycosyltransferases involved in cellular homeostasis. EMBO. J. 23, 2915–2922. doi: 10.1038/sj.emboj.7600295
Lin, Y. F., Liang, H. M., Yang, S. Y., Boch, A., Clemens, S., Chen, C. C., et al. (2009). Arabidopsis IRT3 is a zinc-regulated and plasma membrane localized zinc/iron transporter. New Phytol. 182, 392–404. doi: 10.1111/j.1469-8137.2009.02766.x
Liu, C. L., Gao, Z. Y., Shang, L. G., Yang, C. H., Ruan, B. P., Zeng, D. L., et al. (2019). Natural variation in the promoter of OsHMA3 contributes to differential grain cadmium accumulation between Indica and Japonica rice. J. Integr. Plant Biol. 62, 314–329. doi: 10.1111/jipb.12794
Liu, J., Cai, H., Chu, Q., Chen, X., Chen, F., Yuan, L., et al. (2011). Genetic analysis of vertical root pulling resistance (VRPR) in maize using two genetic populations. Mol. Breed. 28, 463–474. doi: 10.1007/s11032-010-9496-z
Liu, Y., Jiang, H., Chen, W., Qian, Y., Ma, Q., Cheng, B., et al. (2011). Genome-wide analysis of the auxin response factor (ARF) gene family in maize (Zea mays). Plant Growth Regul. 63, 225–234. doi: 10.1007/s10725-010-9519-0
Ludwig, Y., Zhang, Y., and Hochholdinger, F. (2013). The maize (Zea mays L.) AUXIN/INDOLE-3-ACETIC ACID gene family: phylogeny, synteny, and unique root-type and tissue-specific expression patterns during development. PLoS One 8:e78859. doi: 10.1371/journal.pone.0078859
Luo, S., Li, Q., Liu, S., Pinas, N. M., Tian, H., and Wang, S. (2015). Constitutive expression of OsIAA9 affects starch granules accumulation and root gravitropic response in Arabidopsis. Front. Plant Sci. 6:1156. doi: 10.3389/fpls.2015.01156
Mager, S., Schönberger, B., and Ludewig, U. (2018). The transcriptome of zinc deficient maize roots and its relationship to DNA methylation loss. BMC Plant Biol. 18:372. doi: 10.117086/s128-018-1603-z
Mattiello, E. M., Ruiz, H. A., Neves, J. C., Ventrella, M. C., and Araújo, W. L. (2015). Zinc deficiency affects physiological and anatomical characteristics in maize leaves. J. Plant Physiol. 183, 138–143. doi: 10.1016/j.jplph.2015.05.014
Mittler, R. (2006). Abiotic stress, the field environment and stress combination. Trends Plant Sci. 11, 15–19. doi: 10.1016/j.tplants.2005.11.002
Miyadate, H., Adachi, S., Hiraizumi, A., Tezuka, K., Nakazawa, N., Kawamoto, T., et al. (2011). OsHMA3, a P1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles. New Phytol. 189, 190–199. doi: 10.1111/j.1469-8137.2010.03459.x
Mori, A., Kirk, G. J., Lee, J. S., Morete, M. J., Nanda, A. K., Johnson-Beebout, S. E., et al. (2016). Rice genotype differences in tolerance of zinc-deficient soils: evidence for the importance of root-Induced changes in the rhizosphere. Front. Plant Sci. 6:1160. doi: 10.3389/fpls.2015.01160
Navarro, L., Dunoyer, P., Jay, F., Arnold, B., Dharmasiri, N., Estelle, M., et al. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439. doi: 10.1126/science.aae0382
Nielsen, F. H. (2012). History of zinc in agriculture. Adv. Nutr. 3, 783–789. doi: 10.3945/an.112.002881
Normanly, J., and Bartel, B. (1999). Redundancy as a way of life - IAA metabolism. Curr. Opin. Plant Biol. 2, 207–213. doi: 10.1016/s1369-5266(99)80037-5
Outten, C. E., and O’Halloran, T. V. (2001). Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292, 2488–2492. doi: 10.1126/science.1060331
Ouyang, J., Shao, X., and Li, J. (2000). Indole-3-glycerol phosphate, a branchpoint of indole-3-acetic acid biosynthesis from the tryptophan biosynthetic pathway in Arabidopsis thaliana. Plant J. 24, 327–333. doi: 10.1046/j.1365-313x.2000.00883.x
Pedas, P., Schjoerring, J. K., and Husted, S. (2009). Identification and characterization of zinc-starvation-induced ZIP transporters from barley roots. Plant Physiol. Biochem. 47, 377–383. doi: 10.1016/j.plaphy.2009.01.006
Ramesh, S. A., Shin, R., Eide, D. J., and Schachtman, D. P. (2003). Differential metal selectivity and gene expression of two zinc transporters from rice. Plant Physiol. 133, 126–134. doi: 10.1104/pp.103.026815
Rengel, Z., and Graham, R. D. (1995). Wheat genotypes differ in Zn efficiency when grown in chelate-buffered nutrient solution. Plant Soil 176, 307–316. doi: 10.1007/BF00011796
Rengel, Z., and Graham, R. D. (1996). Uptake of zinc from chelate-buffered nutrient solutions by wheat genotypes differing in zinc efficiency. J. Exp. Bot. 47, 217–226. doi: 10.1093/jxb/47.2.217
Roosjen, M., Paque, S., and Weijers, D. (2018). Auxin Response Factors: output control in auxin biology. J. Exp. Bot. 69, 179–188. doi: 10.1093/jxb/erx237
Saidi, A., and Hajibarat, Z. (2020). Computational study of environmental stress-related transcription factor binding sites in the promoter regions of maize auxin response factor (ARF) gene family. Not. Sci. Biol. 12, 646–657. doi: 10.15835/nsb12310823
Sasaki, A., Yamaji, N., and Ma, J. F. (2014). Overexpression of OsHMA3 enhances Cd tolerance and expression of Zn transporter genes in rice. J. Exp. Bot. 65, 6013–6021. doi: 10.1093/jxb/eru340
Sasaki, A., Yamaji, N., Mitani-Ueno, N., Kashino, M., and Ma, J. F. (2015). A node-localized transporter OsZIP3 is responsible for the preferential distribution of Zn to developing tissues in rice. Plant J. 84, 374–384. doi: 10.1111/tpj.13005
Sasaki, S., Tsukamoto, M., Saito, M., Hojyo, S., Fukada, T., Takami, M., et al. (2018). Disruption of the mouse Slc39a14 gene encoding zinc transporter ZIP14 is associated with decreased bone mass, likely caused by enhanced bone resorption. FEBS. Open. Bio. 8, 655–663. doi: 10.1002/2211-5463.12399
Shao, J. F., Xia, J., Yamaji, N., Shen, R. F., and Ma, J. F. (2018). Effective reduction of cadmium accumulation in rice grain by expressing OsHMA3 under the control of the OsHMA2 promoter. J. Exp. Bot. 69, 2743–2752. doi: 10.1093/jxb/ery107
Shiferaw, B., Prasanna, B., Hellin, J., and Bänziger, M. (2011). Crops that feed the world 6. Past successes and future challenges to the role played by maize in global food security. Food Secur. 3, 307–327. doi: 10.1007/s12571-011-0140-5
Šimić, D., Mladenović Drinić, S., Zdunić, Z., Jambrović, A., Ledencan, T., Brkić, J., et al. (2012). Quantitative trait loci for biofortification traits in maize grain. J. Hered. 103, 47–54. doi: 10.1093/jhered/esr122
Song, Y. L. (2019). The gene OsIAA9 encoding auxin/indole-3-acetic acid proteins is a negative regulator of auxin-regulated root growth in rice. Biol. Plantarum 63, 210–218. doi: 10.32615/bp.2019.024
Swain, P. S., Rao, S. B. N., Rajendran, D., Dominic, G., and Selvaraju, S. (2016). Nano zinc, an alternative to conventional zinc as animal feed supplement: A review. Anim. Nutr. 2, 134–141. doi: 10.1016/j.aninu.2016.06.003
Tan, L., Zhu, Y., Fan, T., Peng, C., Wang, J., Sun, L., et al. (2019). OsZIP7 functions in xylem loading in roots and inter-vascular transfer in nodes to deliver Zn/Cd to grain in rice. Biochem. Biophys. Res. Commun. 512, 112–118. doi: 10.1016/j.bbrc.2019.03.024
Tanaka, K., Hayashi, K., Natsume, M., Kamiya, Y., Sakakibara, H., Kawaide, H., et al. (2014). UGT74D1 catalyzes the glucosylation of 2-oxindole-3-acetic acid in the auxin metabolic pathway in Arabidopsis. Plant Cell Physiol. 55, 218–228. doi: 10.1093/pcp/pct173
Tiong, J., McDonald, G., Genc, Y., Shirley, N., Langridge, P., and Huang, C. Y. (2015). Increased expression of six ZIP family genes by zinc (Zn) deficiency is associated with enhanced uptake and root-to-shoot Translocation of Zn in barley (Hordeum vulgare). New Phytol. 207, 1097–1109. doi: 10.1111/nph.13413
Tognetti, V. B., Van Aken, O., Morreel, K., Vandenbroucke, K., van de Cotte, B., De Clercq, I., et al. (2010). Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell 22, 2660–2679. doi: 10.1105/tpc.109.071316
Ueno, D., Kono, I., Yokosho, K., Ando, T., Yano, M., and Ma, J. F. (2009). A major quantitative trait locus controlling cadmium translocation in rice (Oryza sativa). New Phytol. 182, 644–653. doi: 10.1111/j.1469-8137.2009.02784.x
Ueno, D., Yamaji, N., Kono, I., Huang, C. F., Ando, T., Yano, M., et al. (2010). Gene limiting cadmium accumulation in rice. Proc. Natl. Acad. Sci. U. S. A. 107, 16500–16505. doi: 10.1073/pnas.1005396107
von Behrens, I., Komatsu, M., Zhang, Y., Berendzen, K. W., Niu, X., Sakai, H., et al. (2011). Rootless with undetectable meristem 1 encodes a monocot-specific AUX/IAA protein that controls embryonic seminal and post-embryonic lateral root initiation in maize. Plant J. 66, 341–353. doi: 10.1111/j.1365-313X.2011.04495.x
Wang, H., Tang, X., Yang, X., Fan, Y., Xu, Y., Li, P., et al. (2021). Exploiting natural variation in crown root traits via genome-wide association studies in maize. BMC Plant Biol. 21:346. doi: 10.1186/s12870-021-03127-x
Wang, Y., Deng, D., Bian, Y., Lv, Y., and Xie, Q. (2010). Genome-wide analysis of primary auxin-responsive Aux/IAA gene family in maize (Zea mays. L.). Mol. Biol. Rep. 37, 3991–4001. doi: 10.1007/s11033-010-0058-6
Wang, Y., Deng, D., Shi, Y., Miao, N., Bian, Y., and Yin, Z. (2012). Diversification, phylogeny and evolution of auxin response factor (ARF) family: insights gained from analyzing maize ARF genes. Mol. Biol. Rep. 39, 2401–2415. doi: 10.1007/s11033-011-0991-z
Weijers, D., and Wagner, D. (2016). Transcriptional responses to the auxin hormone. Annu. Rev. Plant Biol. 67, 539–574. doi: 10.1146/annurev-arplant-043015-112122
Wessells, K. R., and Brown, K. H. (2012). Estimating the global prevalence of zinc deficiency: results based on zinc availability in national food supplies and the prevalence of stunting. PLoS ONE 7:e50568. doi: 10.1371/journal.pone.0050568
White, P. J., and Broadley, M. R. (2011). Physiological limits to zinc biofortification of edible crops. Front. Plant Sci. 2:80. doi: 10.3389/fpls.2011.00080
Widodo, B., Broadley, M. R., Rose, T., Frei, M., Pariasca-Tanaka, J., Yoshihashi, T., et al. (2010). Response to zinc deficiency of two rice lines with contrasting tolerance is determined by root growth maintenance and organic acid exudation rates, and not by zinc-transporter activity. New Phytol. 186, 400–414. doi: 10.1111/j.1469-8137.2009.03177.x
Wissuwa, M., Ismail, A. M., and Yanagihara, S. (2006). Effects of zinc deficiency on rice growth and genetic factors contributing to tolerance. Plant Physiol. 142, 731–741. doi: 10.1104/pp.106.085225
Wong, C. K. E., and Cobbett, C. S., (2009). HMA P-type ATPases are the major mechanism for root-to-shoot Cd translocation in Arabidopsis thaliana. New Phytol. 181, 71–78. doi: 10.1111/j.1469-8137.2008.02638.x
Wong, C. K. E., Jarvis, R. S., Sherson, S. M., and Cobbett, C. S. (2009). Functional analysis of the heavy metal binding domains of the Zn/Cd-transporting ATPase, HMA2, in Arabidopsis thaliana. New Phytol. 181, 79–88. doi: 10.1111/j.1469-8137.2008.02637.x
Wu, F., and Guclu, H. (2013). Global maize trade and food security: implications from a social network model. Risk Anal. 33, 2168–2178. doi: 10.1111/risa.12064
Xing, H., Pudake, R. N., Guo, G., Xing, G., Hu, Z., Zhang, Y., et al. (2011). Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genomics 12:178. doi: 10.1186/1471-2164-12-17
Xomphoutheb, T., Jiao, S., Guo, X., Mabagala, F. S., Sui, B., Wang, H., et al. (2020). The effect of tillage systems on phosphorus distribution and forms in rhizosphere and non-rhizosphere soil under maize (Zea mays L.) in Northeast China. Sci. Rep. 10:6574. doi: 10.1038/s41598-020-63567-7
Yamaji, N., Xia, J., Mitani-Ueno, N., Yokosho, K., and Ma, J. F. (2013). Preferential delivery of zinc to developing tissues in rice is mediated by P-type heavy metal ATPase OsHMA2. Plant Physiol. 162, 927–939. doi: 10.1104/pp.113.216564
Yamauchi, T., Tanaka, A., Inahashi, H., Nishizawa, N. K., Tsutsumi, N., Inukai, Y., et al. (2019). Fine control of aerenchyma and lateral root development through AUX/IAA- and ARF-dependent auxin signaling. Proc. Natl. Acad. Sci. U. S. A. 116, 20770–20775. doi: 10.1073/pnas.1907181116
Yan, J., Wang, P., Wang, P., Yang, M., Lian, X., Tang, Z., et al. (2016). A loss-of-function allele of OsHMA3 associated with high cadmium accumulation in shoots and grain of Japonica rice cultivars. Plant Cell Environ. 39, 1941–1954. doi: 10.1111/pce.12747
Yancey, P. H. (2005). Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J. Exp. Biol. 208, 2819–2830. doi: 10.1242/jeb.01730
Yang, C., Deng, W., Tang, N., Wang, X., Yan, F., Lin, D., et al. (2013). Overexpression of ZmAFB2, the maize homologue of AFB2 gene, enhances salt tolerance in transgenic tobacco. Plant Cell Tiss. Org. 112, 171–179. doi: 10.1007/s11240-012-0219-5
Zhang, H., Liu, J., Jin, T., Huang, Y., Chen, J., Zhu, L., et al. (2017). Identification of quantitative trait locus and prediction of candidate genes for grain mineral concentration in maize across multiple environments. Euphytica 213:90. doi: 10.1007/s10681-017-1875-7
Zhang, X., Zheng, L., Zhang, H., Huang, X., Liu, Y., Zhang, J., et al. (2019). Insights into the BR2/PGP1-mediated patterns for shoot and root growth in maize early seedling development by comparative transcriptome sequencing. J. Plant Biol. 62, 217–228. doi: 10.1007/s12374-018-0394-y
Zhang, Y., Paschold, A., Marcon, C., Liu, S., Tai, H., Nestler, J., et al. (2014). The Aux/IAA gene rum1 involved in seminal and lateral root formation controls vascular patterning in maize (Zea mays L.) primary roots. J. Exp. Bot. 65, 4919–4930. doi: 10.1093/jxb/eru249
Zhang, Y., von Behrens, I., Zimmermann, R., Ludwig, Y., Hey, S., and Hochholdinger, F. (2015). LATERAL ROOT PRIMORDIA 1 of maize acts as a transcriptional activator in auxin signalling downstream of the Aux/IAA gene rootless with undetectable meristem 1. J. Exp. Bot. 66, 3855–3863. doi: 10.1093/jxb/erv187
Keywords: maize (Zea mays L.), quantitative trait locus (QTL), zinc (Zn) deficiency tolerance, ZRT/IRT-like protein (ZIP), candidate genes
Citation: Xu J, Wang X, Zhu H and Yu F (2021) Identification and Analysis of Zinc Efficiency-Associated Loci in Maize. Front. Plant Sci. 12:739282. doi: 10.3389/fpls.2021.739282
Received: 10 July 2021; Accepted: 30 September 2021;
Published: 15 November 2021.
Edited by:
Durgesh Kumar Tripathi, Amity University, IndiaReviewed by:
Shutang Tan, University of Science and Technology of China, ChinaKailiang Bo, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (CAAS), China
Copyright © 2021 Xu, Wang, Zhu and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Futong Yu, ZnV0b25neXVAY2F1LmVkdS5jbg==
 Jianqin Xu
Jianqin Xu Xuejie Wang
Xuejie Wang Futong Yu
Futong Yu