
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 28 October 2021
Sec. Plant Breeding
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.734419
This article is part of the Research Topic Disease Resistance for Sustainable Agriculture in Brassica View all 18 articles
 Qian Li1
Qian Li1 Nadil Shah1
Nadil Shah1 Xueqing Zhou1
Xueqing Zhou1 Huiying Wang1
Huiying Wang1 Wenlin Yu1
Wenlin Yu1 Jiajie Luo2
Jiajie Luo2 Yajun Liu3
Yajun Liu3 Genze Li4
Genze Li4 Chao Liu1
Chao Liu1 Chunyu Zhang1
Chunyu Zhang1 Peng Chen1*
Peng Chen1*Clubroot disease, which is caused by the soil-borne pathogen Plasmodiophora brassicae War (P. brassicae), is one of the oldest and most destructive diseases of Brassica and cruciferous crops in the world. Plant microRNAs [micro ribonucleic acids (miRNAs)] play important regulatory roles in several developmental processes. Although the role of plant miRNAs in plant-microbe interaction has been extensively studied, there are only few reports on the specific functions of miRNAs in response to P. brassicae. This study investigated the roles of miRNAs and their targets during P. brassicae infection in a pair of Brassica napus near-isogenic lines (NILs), namely clubroot-resistant line 409R and clubroot-susceptible line 409S. Small RNA sequencing (sRNA-seq) and degradome-seq were performed on root samples of 409R and 409S with or without P. brassicae inoculation. sRNA-seq identified a total of 48 conserved and 72 novel miRNAs, among which 18 had a significant differential expression in the root of 409R, while only one miRNA was differentially expressed in the root of 409S after P. brassicae inoculation. The degradome-seq analysis identified 938 miRNA target transcripts, which are transcription factors, enzymes, and proteins involved in multiple biological processes and most significantly enriched in the plant hormone signal transduction pathway. Between 409R and 409S, we found eight different degradation pathways in response to P. brassicae infection, such as those related to fatty acids. By combining published transcriptome data, we identified a total of six antagonistic miRNA-target pairs in 409R that are responsive to P. brassicae infection and involved in pathways associated with root development, hypersensitive cell death, and chloroplast metabolic synthesis. Our results reveal that P. brassicae infection leads to great changes in miRNA pool and target transcripts. More interestingly, these changes are different between 409R and 409S. Clarification of the crosstalk between miRNAs and their targets may shed new light on the possible mechanisms underlying the pathogen resistance against P. brassicae.
Rapeseed (Brassica napus) is an important oilseed crop in the temperate climate zone of the world, providing edible oil and raw materials for the production of bioenergy (Zajac et al., 2016). China is the second-largest producer of rapeseed, which is the fourth leading cash crop after rice, wheat, and maize (Hu et al., 2017). Rapeseed is also widely cultivated in the European Union, Canada, and other parts of Asia (Zajac et al., 2016).
Clubroot is a disease caused by the soil-borne pathogen Plasmodiophora brassicae War (P. brassicae) and one of the oldest and most destructive diseases of Brassica and cruciferous crops in the world (Dixon, 2014; Hirani et al., 2016). It spreads in more than 60 countries and causes more than 20% yield loss in highly infested fields (Diederichsen et al., 2009; Bhattacharya et al., 2014; Chai et al., 2014; Rahman et al., 2014; Wallenhammar et al., 2014). The pathogen can cause the formation of galls or clubs on the roots of susceptible hosts, which prevents water and nutrient uptake from the soil and finally results in stunting, wilting, and immature death (Dixon, 2009; Hwang et al., 2011). Besides, this obligate biotrophic pathogen can survive on soil for more than 20 years, resulting in very difficult control of the disease with chemicals or other mechanical methods (Kageyama and Asano, 2009). Therefore, the development of P. brassicae-resistant varieties is considered the most economical and effective approach to control the clubroot disease. To date, a number of clubroot resistant (CR) loci have been identified, such as CRa, CRb, CRc, CRk (Matsumoto et al., 1998; Piao et al., 2004; Sakamoto et al., 2008), Crr1, Crr2, Crr3, Crr4 (Suwabe et al., 2003, 2006; Hirai et al., 2004), CRd (Pang et al., 2018), PbBa3.1, PbBa3.2, PbBa3.3, PbBa1.1, PbBa8.1 (Chen et al., 2013), Rcr1 (Chu et al., 2014), Rcr4, Rcr8, and Rcr9 (Yu et al., 2017). Among these loci, CRa and Crr1 are isolated from Chinese cabbage and encode toll-interleukin-1 receptor/nucleotide-binding site/leucine-rich-repeat (TNL/TIR-NBS-LRR) proteins (Hatakeyama et al., 2013, 2017). These resistance-related proteins (R proteins) are mostly intracellular receptors that interact with pathogen “effectors” to activate the effect or triggered immunity (ETI) of plants (Jones and Dangl, 2006). However, the specific mechanism for resistance to P. brassicae mediated by clubroot resistance (CR) genes remains unclear.
Micro ribonucleic acids (miRNAs) are a class of endogenous non-coding small RNAs usually with a length of 20–24 nucleotides (nt). miRNAs have been demonstrated to play important regulatory roles in several plant growth and development processes (Sunkar et al., 2012; Jin et al., 2013; Tang and Chu, 2017; Song et al., 2019). In plants, the miRNA-mediated regulation of gene expression occurs in three ways. First, miRNAs can directly target the messenger RNAs based on near-perfect sequence complementarity and lead to the cleavage of the targets (Song et al., 2004; Baumberger and Baulcombe, 2005; German et al., 2008; Carbonell et al., 2012; Fei et al., 2013). Second, miRNAs can also downregulate gene expression through translational repression that reduces protein level (Brodersen et al., 2008; Iwakawa and Tomari, 2013; Li et al., 2013; Reis et al., 2015). Third, besides the cleavage of target miRNAs and translational repression at the posttranscriptional level, miRNAs can also influence the level of transcripts through DNA methylation (Bao et al., 2004; Wu et al., 2010).
In the past few years, miRNAs have also been demonstrated to play crucial roles in mediating plant immune responses (Song et al., 2019; Kulshrestha et al., 2020). Generally, plants have two types of immune responses upon pathogen attack, which are known as the pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI) pathways (Jones and Dangl, 2006; Boller and Felix, 2009; Dangl, 2013; Peng et al., 2018). So far, at least 21 miRNA-target modules have been found to be involved in plant defense against pathogens through the regulation of PTI and ETI (Song et al., 2019). miR393 is the first miRNA identified in Arabidopsis for PTI induced by bacterial flagellin peptide, which negatively regulates auxin signaling by targeting the mRNAs of auxin receptors (Navarro et al., 2006). miR393 and miR166 are induced in the PTI of soybean roots upon infection by the fungus-like pathogen Phytophthora sojae (Wong et al., 2014). Hvu-miR398 is regulated by barley R genes Mla and Rom1 and acts as a repressor of HvSOD1 in response to the barley powdery mildew fungus (Xu et al., 2014). miRNA393* (derived from the lagging strand of pre-miR393), which is induced by avirulent P. syringae pv. Tomato DC3000 mediates the silencing of a Golgi-localized SNARE gene (MEMB12) and contributes to ETI in Arabidopsis (Zhang et al., 2011). However, R-protein-triggered ETI usually has a fitness cost for plant growth and, thus, is tightly controlled in the absence of pathogen attack and attenuated after defense (Tian et al., 2003; Deng et al., 2017; Greene and Dong, 2018; Wang et al., 2018; Liu et al., 2019; Cui et al., 2020). Recent studies have revealed that several miRNA families target the transcripts of R genes, which triggers the production of 21-nt phased siRNAs (phasiRNA), and prevent R-protein-triggered autoimmunity in the absence of pathogen infection (Zhai et al., 2011; Li et al., 2012; Shivaprasad et al., 2012; Liu et al., 2014; Gonzalez et al., 2015; Deng et al., 2018). Therefore, the miRNA-mediated regulation of R gene expression may be a conserved mechanism underlying pathogen-induced plant immunity (de Vries et al., 2015; Zhang et al., 2016).
Numerous studies have reported the functions of miRNAs in plant-microbe interactions. However, there have only been relatively few studies concerning the functions of miRNAs in response to P. brassicae infection in B. napus. Previous studies have reported miRNA expression profiles in B. napus or Brassica rapa under P. brassicae infection (Verma et al., 2014; Tang et al., 2015; Wei et al., 2016). A comparison of miRNA profiles between susceptible and resistant plants can help to dissect the mechanism underlying clubroot resistance. In our previous study, we introduced a dominant clubroot disease resistance locus (CRb) into the B. napus restorer line Bing 409 and obtained a pair of near-isogenic lines (NILs): a clubroot-resistant line (409R) and a clubroot-susceptible line (409S) (Li et al., 2021). In this study, sRNA-seq and degradome-seq were performed on the roots of 409R and 409S with or without P. brassicae infection to identify the critical miRNAs and corresponding target transcripts for clubroot resistance, aiming to establish a model for the miRNA-mediated regulatory network associated with the resistance of B. napus to P. brassicae.
In our previous study, we have introduced the CRb locus from CR Shinki (a Chinese cabbage material) to Bing409 (a Pol. CMS restorer line of B. napus) and obtained a pair of NILs with contrast phenotype of clubroot disease resistance through marker-assisted foreground selection and background selection (Li et al., 2021). The resulting lines, namely, the clubroot-resistant line carrying CRb locus (designated as 409R) and clubroot-susceptible line (designated as 409S), were sown and grown under a 16-h photoperiod at 25°C in an artificial growth chamber. The single sequence repeat genotyping of 409R revealed that 97% of the recurrent parent genome was recovered.
Rapeseed roots were inoculated with the P. brassicae strain collected from Zhijiang (Hubei, China, 30°43'00.00 N, “111°77'00.00” E). The P. brassicae strain we used in this study was collected and characterized as pathotypes 4 and Pb1 according to the Williams and sinitic clubroot differential classification systems, respectively (Williams, 1966; Pang et al., 2020). The homogenate of roots with galls or clubs was mixed with dried culture soil at a mass ratio of 1: 20 and sealed at 25°C for more than 48 h. Subsequently, 20 g of the prepared P. brassica-containing soil was put into the culture soil in each hole of the cavity tray, which was then filled with tap water (about 40 ml for each hole) and then sown with one to two seeds.
The inoculated roots (Int409R, Int409S) were collected 20 days post-inoculation (dpi), and un-inoculated roots (Mock409R, Mock409S) were collected as control samples. Tissues were immediately frozen in liquid nitrogen and stored at −80°C. The samples were prepared with three biological replicates.
Total RNA was isolated using the TRIzol (Invitrogen, Carlsbad, CA, United States) reagent according to the instructions of the manufacturer. To ensure the quality of RNA for library construction, an Agilent 2100 Bioanalyzer system was used for RNA quality control. Small RNA (sRNA) was separated from total RNA by NaCl-PEG8000 precipitation, as previously described (Lu et al., 2007). sRNAs in the size range of 18–30 nt were gel-purified and ligated to adapters. The sRNA library was generated by reverse transcription and sequenced using an Illumina HiSeq 2000 platform at Shanghai Personal Biotechnology Co., Ltd in China.
Degradome libraries were generated as previously described (Ma et al., 2010; Zhai et al., 2014). Samples from three biological replicates were pooled for degradome library construction. In brief, mRNA fragments with poly (A) sequences were annealed and captured with poly (T) magnetic beads; 5′ RNA adapters were ligated to RNAs containing 5′ monophosphates. The ligated products were then purified and reverse-transcribed to cDNA using biotinylated random primers. The cDNA was amplified by PCR to construct the degradome libraries. Sequencing was also performed using the Illumina HiSeq 2000 platform at LC-Bio Technologies Co., Ltd (Hangzhou, China).
For sRNA sequencing data, the raw reads were first filtered by removal of low-quality reads to obtain clean reads (sRNAs). The clean reads of each sample were screened within a certain range of length, from 18 to 30, and then mapped to the B. napus genome (https://www.ncbi.nlm.nih.gov/genome/?term=brassica+napus) using Bowtie to obtain read counts and genomic location information. The sRNAs were also aligned to the GenBank (ftp://ftp.ncbi.nlm.nih.gov/genbank/) and Rfam 11.0 (http://rfam.janelia.org/) databases for functional annotation. All sequences annotated as repeat, intron, exon, ribonucleic acid (rRNA), transfer RNA (tRNA), small cytoplasmic RNA (scRNA), small nuclear RNA (snRNA), or small nucleolar RNA (snoRNA) were removed in subsequent miRNA analyses. Unannotated sRNAs were then used to predict the secondary structure using miReap (http://sourceforge.net/projects/mireap/) combined with genome mapping information. sRNAs with classic miRNA secondary structure were then aligned to miRBase 21.0 (http://www.mirbase.org/ftp.shtml) to identify known miRNAs using miReap. In addition, sRNAs containing classic miRNA secondary structure but not included on miRBase were classified as “novel.” The clean reads for each miRNA were normalized using the following formula: normalized expression (TPM) = mapped read count/total reads * 1,000,000. Fold changes between the samples were calculated using log2 (TPM of sample 1/TPM of sample 2).
For degradome sequencing data, the raw reads were filtered to remove reads with adapters. Clean reads were aligned to the GenBank and Rfam 11.0 databases to obtain the annotation information for rRNA, tRNA, scRNA, snRNA, and snoRNA. sRNAs not associated with these annotated reads were further mapped to the B. napus genome (v2.0, https://www.ncbi.nlm.nih.gov/genome/?term=brassica+napus) to obtain the cDNA sense and antisense reads using Bowtie. The reads mapped to cDNA or mRNA sequences were then used to predict the sites of cleavage. Two software programs were used to predict the sites of cleavage of the targets: psRNATarget (http://plantgrn.noble.org/psRNATarget/) was used to predict miRNA targets; and CleaveLand3 (http://axtell-lab-psu.weebly.com/cleaveland.html) was used to summarize the information of cleaved sites, define categories (containing 0–4 categories), and plot T-plot figures.
The quantification of miRNAs was performed using miRcute Plus miRNA First-Strand cDNA Synthesis Kit (TIANGEN, China) according to the instructions of the manufacturer. Briefly, 2 μg of total RNA was mixed with an RT RNA reaction buffer and an RT Enzyme mix in a total volume of 20 μl. The reaction system was incubated at 42°C for 60 min and stopped at 95°C for 3 min. quantitative reverse transcription (qRT)-PCR was carried out using the miRcute Plus miRNA qPCR Detection Kit (TIANGEN Biotech Co Ltd, Beijing, China), with miRNA-specific forward primers (Supplementary Table 9) and a universal reverse primer. Briefly, 7.5 μl 2 × miRcute Plus miRNA Premix, 0.3 μl miRNA-specific forward primers, 0.3 μl universal reverse primer, and 6.9 μl 20 × diluted cDNA template were mixed in a total of 15 μl reaction volume. The Bio-Rad CFX96 Realtime System (Bio-Rad, Hercules, CA, United States) was used with the following PCR cycling parameters: 95°C for 15 min; 45 cycles of 94°C for 20 s followed by 60°C for 34 s. Reactions were performed in triplicates, and U6 rRNA was used as the internal reference. The relative expression of miRNAs was calculated according to a previous study (Livak and Schmittgen, 2001). The student's t-test was performed for the significance test.
To identify the miRNAs involved in the response of B. napus to P. brassicae infection in the resistant line 409R and the susceptible line 409S, 12 sRNA libraries generated from the P. brassicae inoculated roots at 20 dpi (Int409R and Int409S) and mock roots (Mock409R and Mock409S) were sequenced with the Illumina Solexa high-throughput sequencing technology. We obtained ca. 1G raw data per sample by SE50 mode (single end, read length 50 nt), with an average of 20.95 million reads (ranging from 17.5 to 40.5 million) for each library (Table 1). About 76.4% of the reads were mapped to the B. napus genome, resulting in an average of 16 million clean reads and million repeat reads (Table 1). The reads matched with rRNA, tRNA, snRNA, and snoRNA accounted for 34.25% of the sequences (Table 1). Besides the t/r/sn/snoRNAs, 2.03 million sRNA reads on average were identified from each library (Table 1). Reads corresponding to 18–30 nt sRNAs were selected for further analysis, with the majority of sRNAs exhibiting lengths of 21 and 24 nt (Figure 1A). 409R had relatively more 18-nt and 19-nt sRNAs than 409S; similarly, the inoculated samples (Int409R and Int409S) had more 21-nt sRNAs than the control samples (Mock409R or Mock409S), indicating that P. brassicae infection could induce some changes in the sRNA pool of B. napus (Figure 1A).
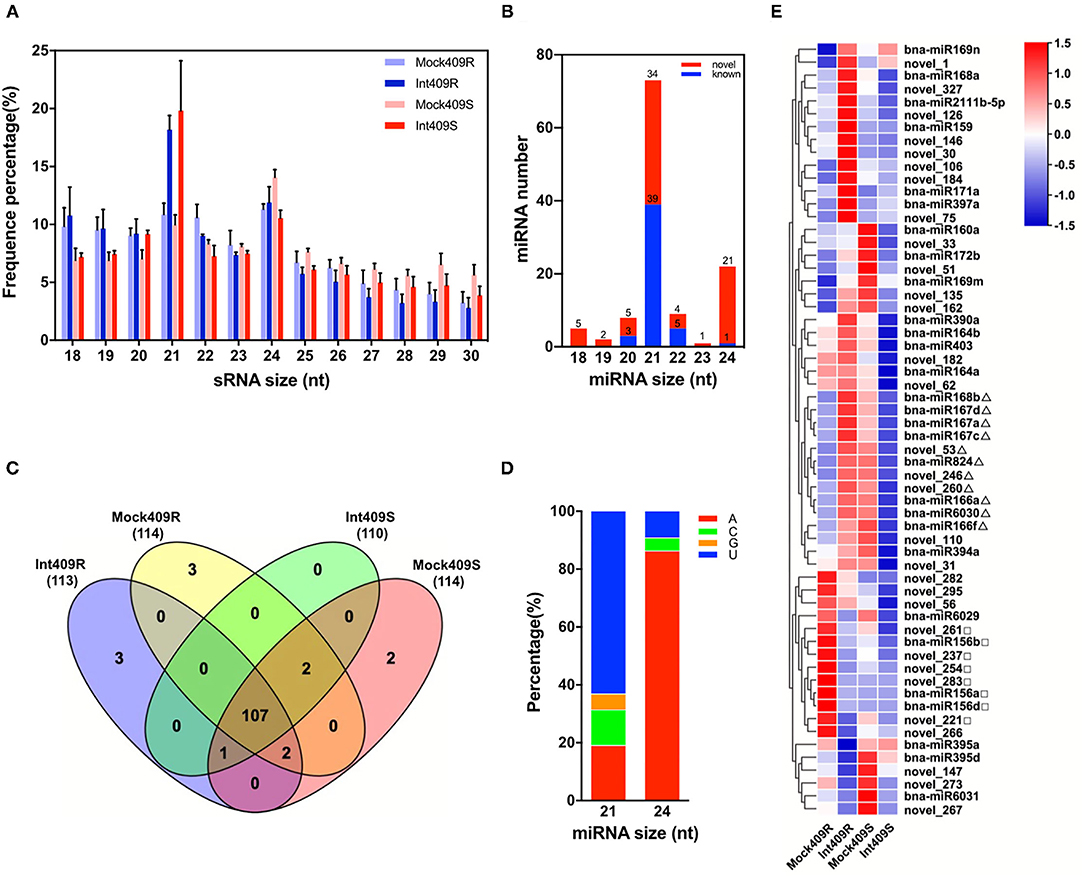
Figure 1. Identification and characterization of micro ribonucleic acids (miRNAs) in resistant and susceptible Brassica napus upon Plasmodiophora brassicae infection. Distribution of (A) Small RNA (sRNA) length in 12 sRNA libraries and (B) length distribution of miRNAs, (C) Venn diagram of the number of miRNAs in 409R and 409S with or without P. brassicae infection, (D) preference of the first nucleotide for 21 or 24-nt miRNAs, and (E) heat map of 60 highly expressed miRNAs. Δ and □ mark some miRNAs with different expression patterns between 409R and 409S in response to P. brassicae infection.
Based on a filtering pipeline designed to distinguish plant miRNAs (Zhai et al., 2011), a total of 120 miRNA precursors were identified, namely, 72 novel miRNAs and 48 known miRNAs (Figures 1B,C, Supplementary Table 1). The lengths of mature miRNAs ranged from 18 to 24 nt, and 21-nt miRNAs (39 known and 34 novel) and 24-nt miRNAs (1 known and 21 novel) were the two most abundant types (Figure 1B). An analysis of nucleotide preference revealed that “U” was preferred by the 21-nt miRNAs, while “A” was preferred by the majority of 24-nt miRNAs (Figure 1D). According to the miRbase database, the 44 known miRNAs belonged to 25 conserved miRNA families across diverse plant species (Supplementary Table 1a). Seventeen novel miRNAs were new members of 11 known miRNA families. For example, novel_147, novel_172, novel_202, novel_207, and novel_222 were new members of the MIR169_2 family (Supplementary Table 1b). A total of 55 novel miRNAs could not be associated with any known miRNA families, and were, thus, defined as “new miRNA candidates” (Supplementary Table 1c). Moreover, five miRNAs (bna-miR1140, bna-miR6032, bna-miR161, bna-miR860, and bna-miR824), which belonged to five miRNA families (MIR1140, MIR6032, MIR161, MIR860, and MIR824), were specifically present in Brassica (Table 2, Supplementary Table 2).
There were considerable differences in the expression levels of miRNAs detected in this study (Figure 1E), including highly expressed miRNAs (22.47%) with read numbers of more than 200 across all samples and lowly expressed miRNAs (26.59%) with read numbers below five, while the majority of miRNAs showed read numbers between 5 and 200 (Supplementary Figure 1). Based on the normalized expression of miRNAs, 60 highly expressed miRNAs were selected to construct the heat map for comparing the changes in the miRNAs upon P. brassicae infection (Figure 1E). The data revealed that the expression profiles of miRNAs differed greatly between 409R and 409S upon pathogen infection (Figure 1E). For example, bna-miR168b, bna-miR167, bna-miR166, bna-miR824, bna-miR6030, novel_53, novel_260, and novel_246 were upregulated in 409R upon infection, while an opposite trend was observed for 409S (Figure 1E). On the contrary, bna-miR156, novel_261, novel_237, novel_254, novel_221, and novel_283 were downregulated in 409R after infection, while their expression exhibited no change in 409S after P. brassicae infection (Figure 1E). Furthermore, 18 miRNAs were significantly differentially expressed in 409R upon P. brassicae infection, including nine upregulated (miR168b, miR169m, miR169n, novel98, novel246, novel_75, novel_180, novel_106, and novel_162) and nine downregulated (miR395d, miR6029, novel_221, novel_147, novel_237, novel_295, novel_254, novel_261, and novel_266) (Table 3). In 409S, only one miRNA (novel_1) was found to be upregulated upon P. brassicae infection (Table 3).
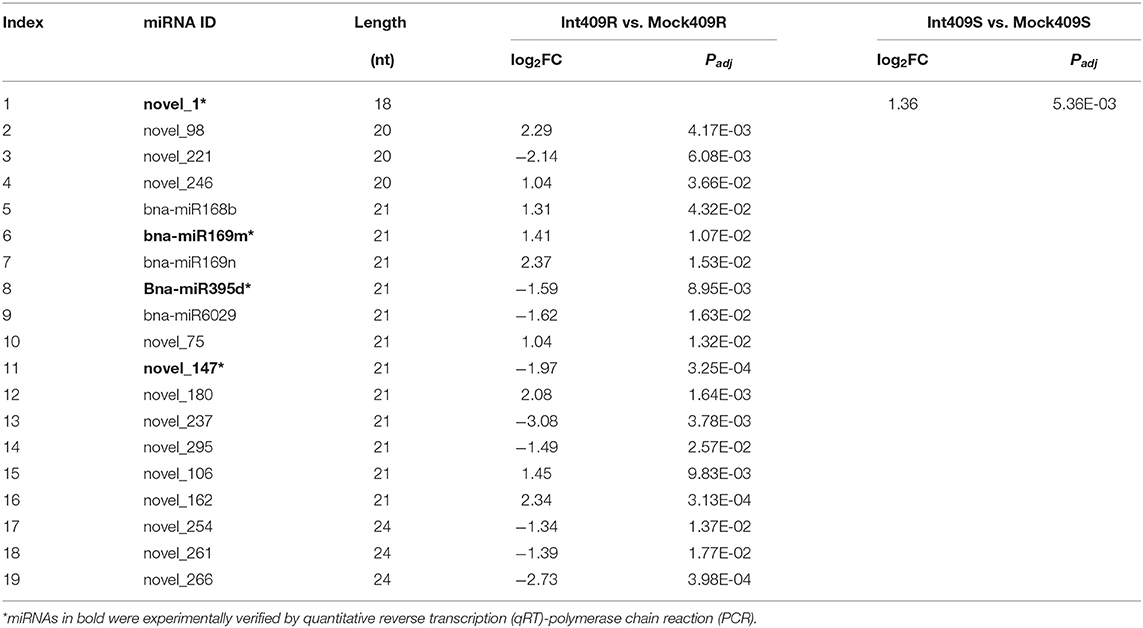
Table 3. Differentially expressed miRNAs in response to P. brassicae infection (p < 0.05, |log2FC| > 1).
These results suggested that P. brassicae infection could cause global changes in the miRNA pool of B. napus and that there are substantial differences between the resistant (409R) and susceptible (409S) lines. The differential expression of miRNAs may lead to subsequent changes in target transcripts, which may explain the phenotype differences in plant immune response to P. brassicae infection.
To validate these results, we examined the expression dynamics of miRNAs at different time points (15, 20, and 25 days) post P. brassicae infection by qRT-PCR. The expression levels of four miRNAs were analyzed, and the results are presented in Figure 2. Phase- and species-dependent changes were observed for specific miRNAs. For example, although bna-miR395d was downregulated in both 409R and 409S at 25 dpi, the tendency was different at 20 dpi between 409R and 409S (Figure 2A). In both 409R and 409S, the relative abundance of novel_147 decreased from 15 to 20 dpi and then increased from 20 to 25 dpi, but at each time point upon infection, novel_147 abundance decreased significantly in 409R, while the changes in 409S were not significant (Figure 2B), which was also consistent with the sRNA-seq data (Table 3). Overall, these results indicated dynamic changes in miRNA expression upon P. brassicae infection.
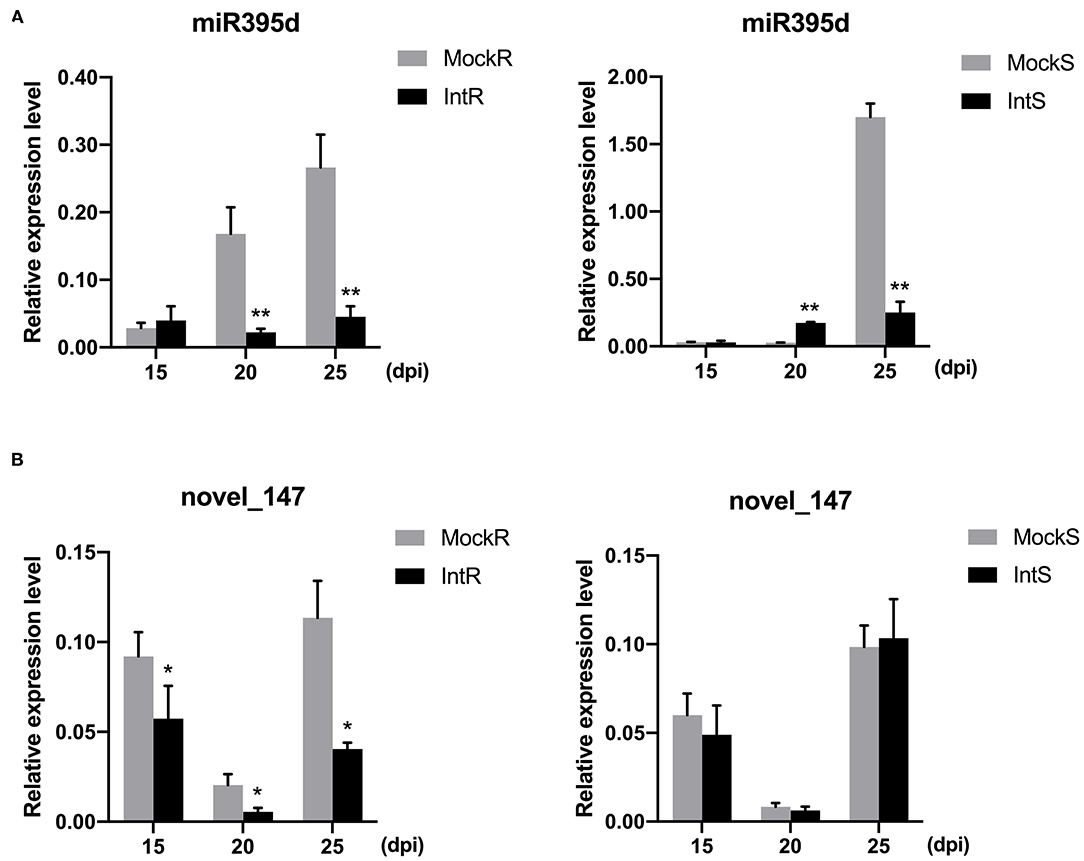
Figure 2. Validation of the relative expression level of (A) miRNA395d and (B) novel_147 in 409R and 409S in response to P. brassicae infection. Y-axis, relative expression level; X-axis, days post-inoculation (dpi). Data were obtained from at least three biological replicates. Bars represent SD (STDEV). *p ≤ 0.05, **p ≤ 0.01 by Student's t-test. MockR, 409R without inoculation; IntR, 409R with inoculation; MockS, 409S, without inoculation; IntS, 409S with inoculation.
Target identification is important for understanding the regulatory function of miRNAs. We constructed four degradome libraries using RNAs derived from Int409R, Int409S, Mock409R, and Mock409S roots to identify the target transcripts of critical miRNAs involved in the progression of clubroot disease. Sequencing of these libraries generated a total of 28 million raw reads and 9 million unique reads on average; 99.52% of the unique reads could be matched to the B. napus genome (Supplementary Table 3). As a result, a total of 1,513 miRNA target pairs involving 83 miRNAs (47 known and 36 novel) and 938 target transcripts were identified (Table 4, Supplementary Table 4). The number of target transcripts for a particular miRNA ranged from 1 to 81 (Supplementary Table 5). Interestingly, in some cases, target transcripts could be identified for a subset of miRNAs in 409R but not in 409S and vice versa (Supplementary Table 5). About 63.43% (595/938) of target transcripts showed a one-to-one association with miRNAs, while the rest had two to six matching miRNAs (Supplementary Figure 2). The miRNAs associated with the same transcript usually belonged to the same family (Supplementary Table 6).
According to gene function annotation, about 45% (421/938) of the target transcripts were related to transcription regulation (Figure 3A), including transcription factors such as auxin response factors (ARFs), growth-regulating factors (GRFs), ethylene-responsive transcription factors (AP2, TOE, and RAP), myeloblastosis (MYBs), basic helix-loop-helix, Teosinte Branched1-Cycloidea-Pcf, nuclear transcription factor Y subunit As (NFYAs), squamosa promoter-binding-like proteins (SPLs), scarecrow-like proteins (SCLs), and NAC domain-containing proteins (NACs) (Supplementary Table 6). In addition to transcription factors, a variety of enzymes (22.5%; 211/938) and proteins (12.5%; 117/938) involved in several biological processes were also detected by the degradome sequencing (Figure 3A, Supplementary Table 6). These enzymes or proteins participate in diverse cellular processes, namely, signal transduction, lipid transport and metabolism, inorganic ion transport and metabolism, RNA processing, and modification (Figure 3A, Supplementary Table 6).
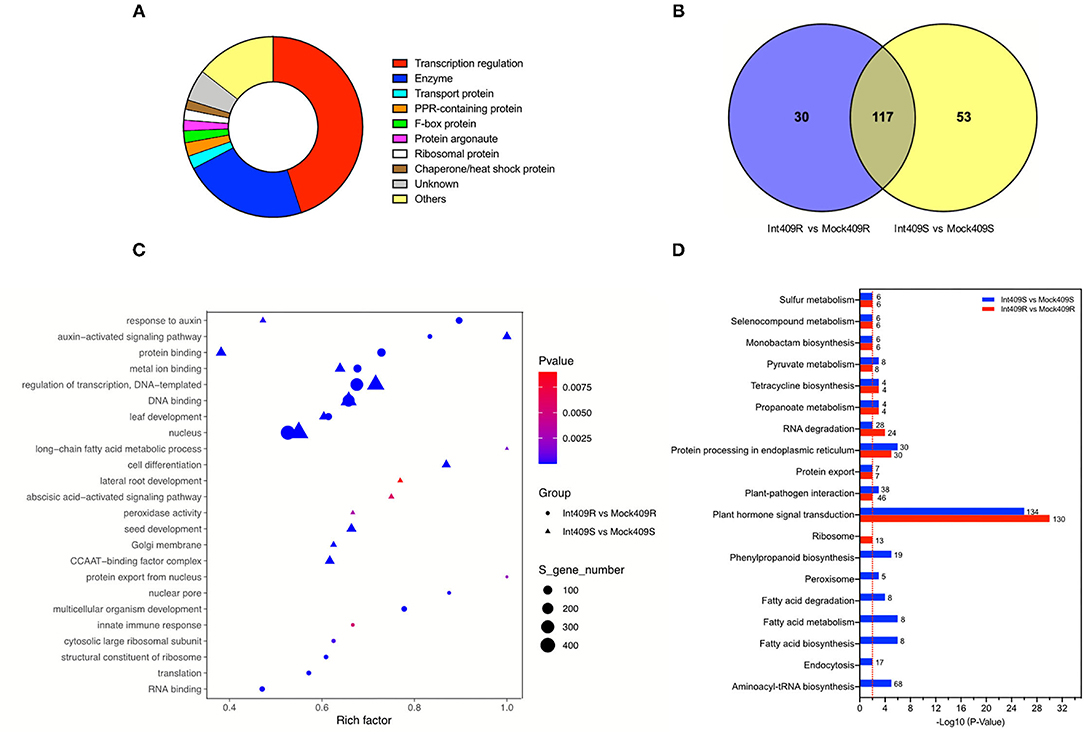
Figure 3. Function annotation and enrichment analysis of target transcripts. (A) Functional classification of all detected targets, (B) Venn diagram of the number of Gene Ontology (GO) terms, (C) GO enrichment, and (D) Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses in comparison between susceptible material (409S) and resistant material (409R) after P. brassicae inoculation. Numbers represent the number of target transcripts significantly enriched in the corresponding pathway. The red line means p = 0.01.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed to clarify the functions of the targets (Figures 3B–D). In total, 147 and 170 GO terms were significantly enriched in 409R and 409S, respectively, among which 30 and 53 GO terms were differentially enriched upon infection (Figure 3B). The GO enrichment analysis indicated that most of the enriched targets participate in diverse biological processes. The GO terms shared by 409R and 409S included DNA binding, metal ion binding, protein binding, and abscisic acid (ABA)- and auxin-activated signaling pathways. The GO terms specifically enriched in 409S included long-chain fatty acid metabolic process, cell differentiation, lateral root development, peroxidase activity, and ABA signaling pathways (Figure 3C), while different sets of GO terms were found for 409R, such as protein export from the nucleus, nuclear pore, and innate immune response (Figure 3C). These differences between 409R and 409S were also revealed by the KEGG enrichment analysis (Figure 3D). The most significantly enriched KEGG term was “plant hormone signal transduction,” which included 130 and 134 targets in 409R and 409S, respectively (Figure 3D). Seven pathways were uniquely identified for 409S after P. brassicae inoculation, namely, “phenylpropanoid biosynthesis, peroxisome, aminoacyl-tRNA biosynthesis, endocytosis, fatty acid biosynthesis, pyruvate and sulfur metabolism, and RNA degradation,” while “ribosome” was the only specific pathway enriched for 409R (Figure 3D, Table 5).
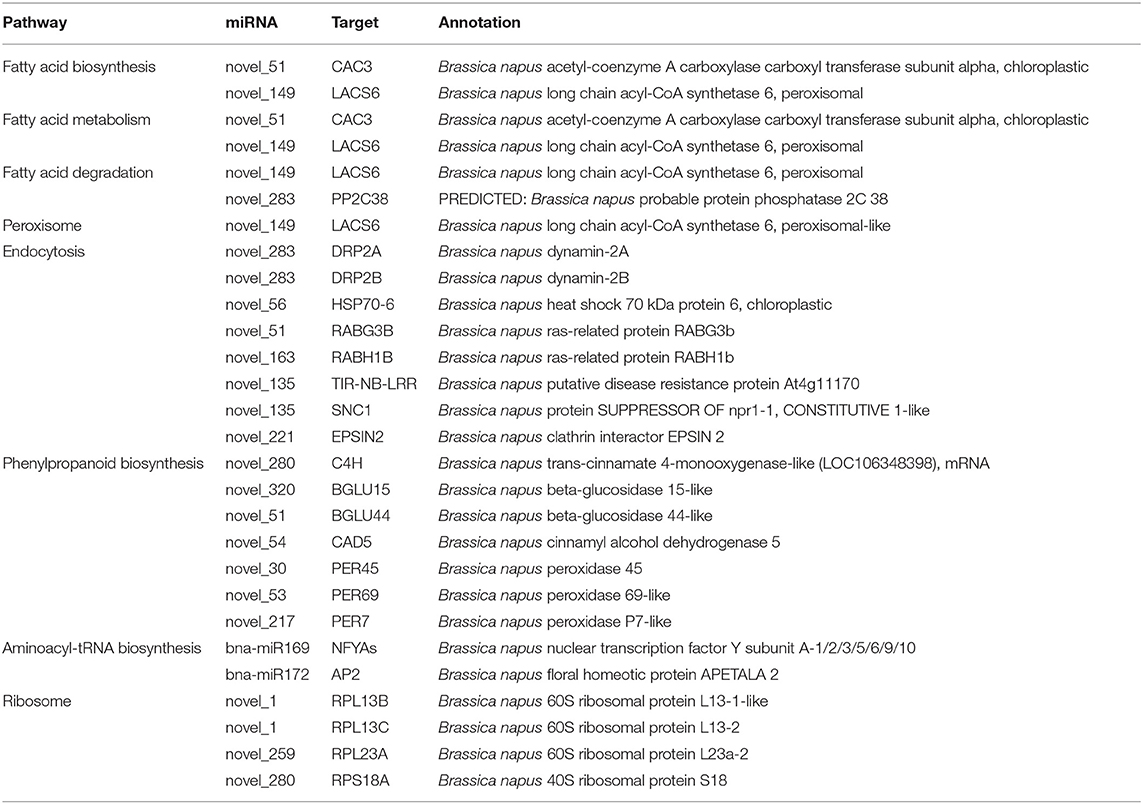
Table 5. Micro ribonucleic acids and targets involved in differential enrichment pathways between resistant and susceptible materials.
To gain deeper insights into the defense mechanisms of B. napus against P. brassicae infection, we selected the miRNA-target pairs with degradome category values below 2 and we constructed a miRNA-target network that included five subclusters based on functional category annotation (Figure 4, Supplementary Table 7).
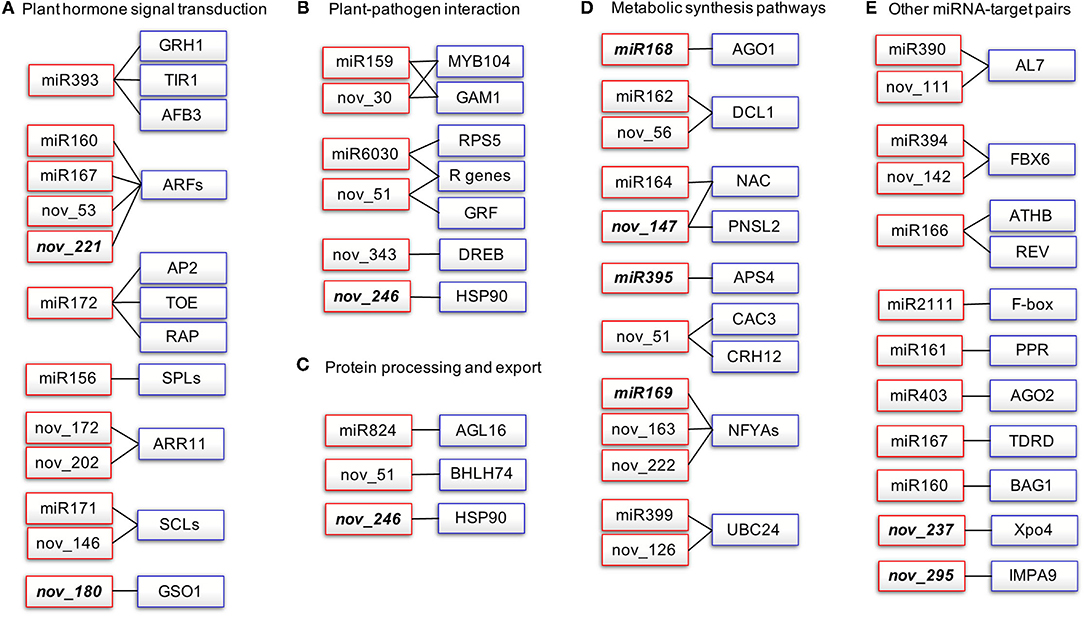
Figure 4. Network of degradome validated miRNA-target pairs associated with P. brassicae response in B. napus. Red rectangles represent the miRNAs and blue rectangles represent the target transcripts validated by degradome sequencing. Italics indicate miRNAs that are significantly differentially expressed after P. brassicae infection. AFB, auxin signaling F-box; AGO1, argonaute1; AP2, apetala2; APS, ATP sulfurylase; ARF, auxin response factor; MYB, myeloblastosis; NFYA, nuclear transcription factor Y subunit alpha; AGL, agamous-like MADS-box protein; AL7, PHD finger protein ALFIN-LIKE 7-like; BAG1. BAG family molecular chaperone regulator 1-like; CAC3, acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha; DCL1, endoribonuclease Dicer homolog 1; DREB, dehydration-responsive element-binding protein 2B-like; FBX6, F-box only protein 6; GRF, growth-regulating factor; HSP90, heat shock protein 90-2-like; PNSL2, photosynthetic NDH subunit of lumenal location 2, chloroplastic-like; PPR, pentatricopeptide repeat-containing protein; SCL, scarecrow-like protein; SPL, quamosa promoter-binding-like protein; TIR1, transport inhibitor response 1. (A) Plant hormone signal transduction, (B) Plant-Pathogen interaction, (C) Protein processing and export, (D) Metabolic synthesis pathways, (E) Other miRNA-target pairs.
A total of 16 kinds of miRNA-target pairs (cluster I) were associated with plant hormone signal transduction (ko04075, P-value: 1.48E-30; Figure 4A, Supplementary Table 7), namely, miR393-TIR1/AFB3/GRH1 and miR160/miR167/novel_53/novel_221-ARFs for auxin signaling, miR172-AP2/TOEs/RAP2-7 for ethylene response, miR156-SPLs for jasmonic acid (JA) signaling, and novel_172/novel_202-ARR11 for type-A response regulators in response to cytokinin (CK). miR171 and novel_146 both target SCLs required for quiescent center cell specification and maintenance in root meristem zone, and asymmetric cell division for radial pattern formation. Novel_180 targets GSO1 (protein brassinosteroid in sensitive 1-like), which is involved in the regulation of root development and root morphogenesis (Figure 4A, Supplementary Table 7). Cluster II comprised 10 pairs of miRNA-targets associated with plant-pathogen interaction (ko04626), namely, miR159 and novel_30 targeting both GAM1 and MYB104, miR6030, and novel_51 targeting both RPS5 and R genes, novel_246 targeting HSP90-2, and novel_343 targeting DREB2B (Figure 4B, Supplementary Table 7). Cluster III included three miRNA-target pairs for protein processing and export (ko04141, P-value: 9.07E-06; Figure 4C). Cluster IV consisted of miRNA-target pairs involved in metabolic synthesis pathways (Figure 4D): miR168 targets AGO1; miR162 and novel_56 target DCL1; both AGO1 and DCL1 are involved in RNA-mediated post-transcriptional gene silencing (PTGS); miR164 and novel_147 both target NACs involved in RNA degradation; novel_147 targets PNSL2 involved in photosynthesis; miR395 targets APS for sulfate-deficiency response, seleno-compound metabolism, sulfur metabolism, and monobactam biosynthesis; novel_51 targets CAC3 for fatty acid biosynthesis and metabolism, propanoate metabolism, pyruvate metabolism, and tetracycline biosynthesis; miR169, novel_163, and novel_222 target NFYAs involved in aminoacyl-tRNA biosynthesis; and miR399 and novel_126 target UBC24 associated with ubiquitin-mediated proteolysis (Figure 4D). miRNA-target pairs that did not fit into any of clusters I–IV are listed in cluster V (Figure 4E, Supplementary Table 7).
Combined with the transcriptome data, the expression profiles of both miRNAs and their targets responsive to P. brassicae infection were integrated to infer the regulatory role of miRNAs during P. brassicae infection. We obtained a total of 27 differentially expressed targets of eight miRNAs responsive to P. brassicae upon infection in 409R and 409S, with a cutoff value of p < 0.05 and |log2FC| of > 1 (Supplementary Table 8). To be more specific, there were six antagonistic miRNA-target pairs in 409R upon infection, such as miR395d-NM_001315829.1 (APS4), miR395d-XM_013888737.2 (uncharacterized), miR395d-XM_013820969.2 (uncharacterized), novel_147-XM_013818148.2 (NAC076) and novel_147-XM_013820748.2/XM_013857207.2 (PNSL2) (Table 6). Some targets cleaved by miRNAs were validated by degradome sequencing (Supplementary Figure 3).
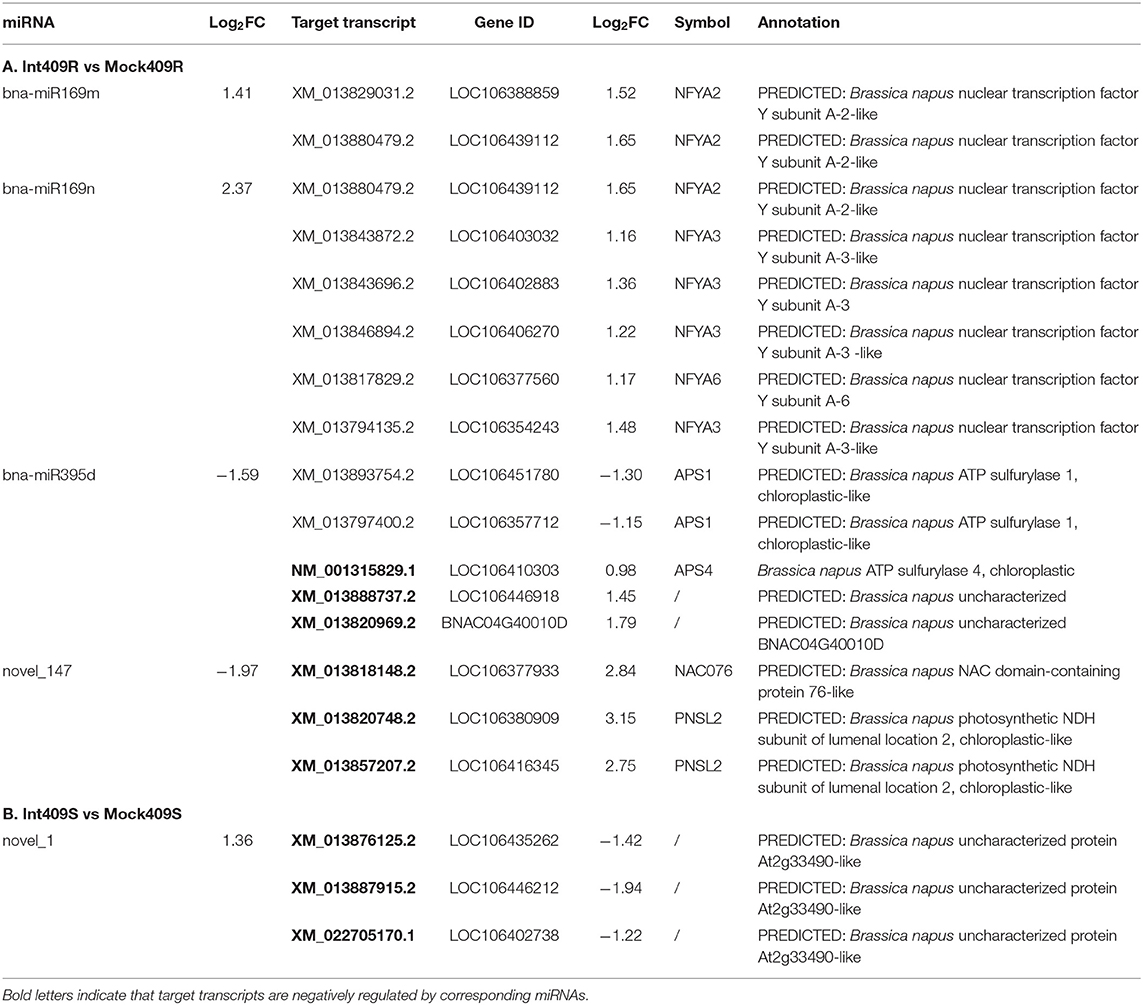
Table 6. Differentially expressed miRNA-target pairs in response to P. brassicae (p < 0.05, |log2FC| > 1).
Numerous studies have demonstrated that miRNAs are involved in plant-pathogen interactions. In this study, we systematically studied the regulatory roles of miRNAs and their targets in response to P. brassicae infection using the resistant line 409R and the susceptible line 409S of rapeseed, which possessed the same genetic background while exhibiting a contrasting phenotype of clubroot resistance. The aim of the study is to identify key candidate miRNAs and corresponding target transcripts involved in clubroot resistance of B. napus. The findings may build up a better understanding of the regulatory network underlying the immune response of B. napus to clubroot pathogen infection.
We noticed some differences in the abundance of particular miRNAs by sRNAseq compared with qRT-PCR (data not shown). Although these results were not presented, but might be encountered for similar studies using qRT-PCR to validate sRNAseq results. Difference RT strategies have been used for RNAseq and qRT of miRNAs, allowing discrimination between pri-, pre- and mature miRNAs (Verma et al., 2014; Wei et al., 2016). The cellular level of pri- or pre- precursors are very low and therefore difficult to quantify, in the current study we focus on the expression level of mature miRNAs and their regulation on target transcripts relevant for clubroot disease progression.
Although these results were not presented, they still might be anticipated for similar studies using both sRNAseq and qRT-PCR to quantify the abundance of miRNAs. These differences could be generated because of the different RT strategies used for RNAseq and qRT, and, indeed, might also reveal a difference in the cellular level of pri-, pre-, and mature miRNAs. As we know, miRNAs made from a primary transcript goes through processing to yield stem-loop structured pre-miRNA. Mature miRNA is generated by the Ago complex that results in 21- to 24-nt single-stranded sRNAs. A change in the mature miRNA level could be generated through pri- or pre- precursors; however, they are present in low abundance, easily degraded in defined cellular compartments and, therefore, difficult to quantify.
The sRNA-seq data revealed that 21-nt and 24-nt sRNAs accounted for the highest proportion of the total sRNAs. Interestingly, after P. brassicae inoculation, the relative abundance of 21-nt sRNAs increased in both 409R and 409S (Figure 1A). The miRNAs identified in this study included members of known miRNA families of rapeseed, as well as 17 novel members of conserved miRNA families and 55 completely new miRNAs (Supplementary Table 1). The identification of these new miRNAs expanded the current knowledge of the miRNA pool in B. napus. Interestingly, there were 18 differentially expressed miRNAs (DE miRNAs) in 409R after P. brassicae infection, while there was only one DE miRNA in 409S (Table 3). Many of the DE miRNAs are novel candidates that have not been reported before (Supplementary Table 1). The fact that the resistant line (409R) had more DE miRNAs than its isogenic counterpart (409S) upon infection highlights a possible regulatory role of R genes in clubroot disease resistance.
The GO and KEGG enrichment analyses identified pathways related to fatty acid metabolism in 409S (Figure 3C, Table 5). Two miRNA-target pairs associated with lipid biosynthesis pathways, novel_51-CAC3 and novel_149-LACS6, were identified only in 409S upon infection (Figure 3C, Table 5). It has been reported that resting spores of P. brassicae can accumulate lipid droplets as an energy source for future sporulation and that many genes related to the fatty acid metabolism of P. brassicae have been identified (Bi et al., 2016). It has also been shown that fatty acids, as carbon source nutrients, play an important role in plant-powdery mildew interaction (Jiang et al., 2017). We speculate that fatty acids also serve as a potential determinant of the interaction between P. brassicae and B. napus. P. brassicae might take advantage of plant fatty acid biosynthesis for successful and systematic invasion. More studies are needed to unravel the function of fatty acid biosynthesis in clubroot disease progression.
We found that the abundance of miRNAs changed more dramatically in 409R than in 409S upon infection (Table 3). Combining transcriptome and degradome data, we constructed a miRNA-target regulatory network in clubroot-resistant B. napus in response to P. brassicae infection (Figure 5). miRNA biogenesis is known to be important for PTI response (Agorio and Vera, 2007; Navarro et al., 2008), but relevant miRNAs have not been identified in B. napus. In the novel_246-HSP90-2 pair (Figure 5A), novel_246 was upregulated in 409R (Table 3), and HSP90-2 encoded a molecular chaperone that regulates RPM1/RPP4-mediated defense response (Bao et al., 2014; Huang et al., 2014). Similarly, bna-miR168b was upregulated in 409R (Figure 5B), which targets the AGO1 and Zinc finger transcription factor CCCH4. In Malus hupehensis, miR168 targets AGO1 and contributes to resistance against Botryosphaeria dothidea infection (Yu et al., 2017). Therefore, some of these identified DE miRNAs may regulate the target key transcripts/proteins through miRNA interference silencing complex (miRISC) to mediate host response to clubroot disease.
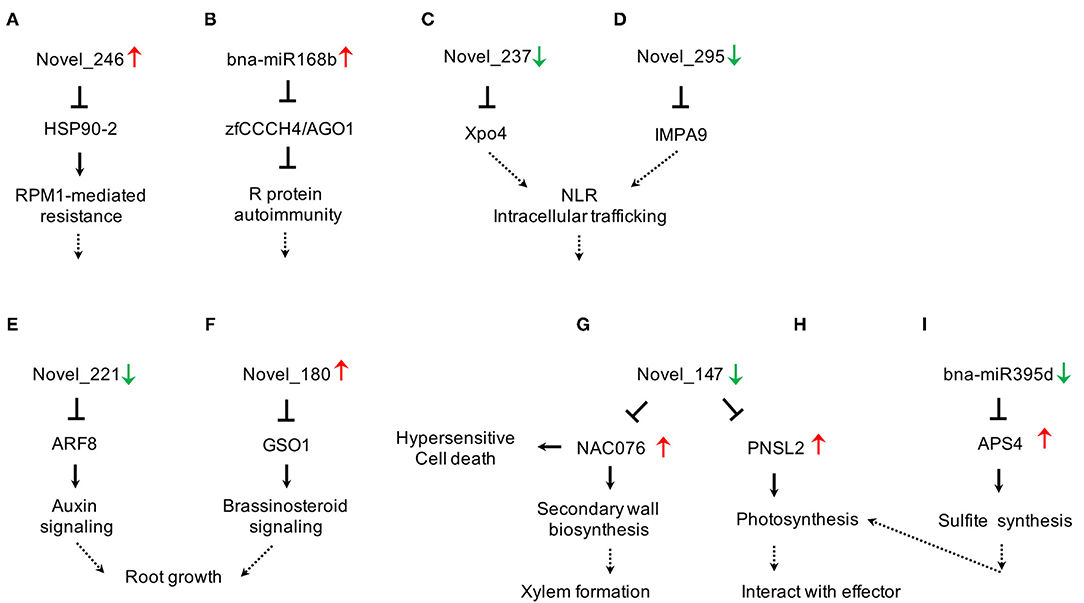
Figure 5. Diverse cellular pathways regulated by miRNA-target modules in clubroot-resistant B. napus in response to P. brassicae. (A,B) Novel_246-HSP90-2 and bna-miRA168b-zfCCCH4/AGO1 for miRNA biogenesis; (C,D) Novel_237-Xpo4 and novel_295-IMPA9 for NLR signalling; (E,F) Novel_221-ARF8 and Novel_180-GSO1 for root growth; (G,H) Novel_147-NAC076/PNSL2 for secondary wall biosynthesis and photosynthesis; (I) bna-miR395d-APS4 for sulphite synthesis. Up- or Down- regulation of miRNAs or target transcripts were indicated by red or green arrows, respectively.
nucleotide binding and leucine rich repeat (NLR) proteins are nucleic acid-binding proteins involved in pathogen-induced signaling (Jones and Dangl, 2006). Two NLR-type R genes for P. brassicae have been identified (Hatakeyama et al., 2013, 2017). Many studies have shown that NLRs are localized to both the cytoplasm and nucleus and that their nuclear accumulation is necessary for pathogen resistance (Shen et al., 2007; Bai et al., 2012; Inoue et al., 2013). Importins and exportins, which act as transport receptors, play important roles in the nuclear pore complex (NPC)-directed partitioning of nucleocytoplasmic NLRs (Garcia and Parker, 2009; Meier and Somers, 2011). In this study, two novel miRNAs (novel_237 and novel_295) that target exportins4 (Xpo4) and importin subunit alpha-9-like (IMPA9) were found to be downregulated in 409R upon infection (Table 3, Figure 4E). It can be inferred that the changes in these miRNAs and further in importins and/or exportins can affect the nucleocytoplasmic partitioning of NLRs. NLRs function together with other cellular metabolic or signaling pathways to contribute to the disease resistance of 409R after P. brassicae inoculation (Figures 5C,D).
The root is the source organ for P. brassicae infection, and its morphology and development, especially those of root hair and root cortex, are critical for clubroot disease progression (Kageyama and Asano, 2009). ARF8 is known as an auxin response factor that inhibits root elongation and promotes lateral root initiation (Wang et al., 2015). We found that novel_221, which regulates ARF8, was downregulated in 409R after P. brassicae infection (Table 3, Figure 5E). Similarly, the novel_180-GSO1 pair is associated with root growth (Figure 5F). GSO1 works in coordination with GSO2 to regulate root growth through cell division and specification (Racolta et al., 2014; Nakayama et al., 2017). Our data suggest that the abundance of novel_180 increased in 409R after P. brassicae infection (Table 3). Therefore, novel_221-ARF8 and novel_180-GSO1 might regulate root growth and suppress gall formation in 409R via hormone signaling (Figures 5E,F).
Plant secondary cell wall thickening is a powerful way to prevent the systematic spreading of pathogens after being attacked. NAC-containing protein is a master transcription activator for xylem formation and SCW thickening (Zhou et al., 2014). Another study has reported that Arabidopsis miR164a and its target NAC4 play important roles in regulating hypersensitive (HR) cell death in response to avirulent bacterial pathogens (Lee et al., 2017). In our research, NAC076 expression was also under the control of novel_147 and miR164 (Figure 4D). We found that novel_147 was downregulated in 409R after P. brassicae infection, and NAC076 transcript level was also upregulated accordingly (Table 6). The activation of NAC076 could lead to the induction of SCW genes and subsequent cell wall thickening or HR cell death, which will then block the invasion of P. brassicae and confer disease resistance (Figure 5G).
Another target of novel_147 was PNSL2, which acts as a chloroplast NAD(P)H dehydrogenase (NDH) complex (Marjaana et al., 2010; Shinya et al., 2010; Figure 5H). Some studies have suggested interplay between photosynthesis and plant defense (Xu et al., 2011; Rodríguez-Herva et al., 2012). In Arabidopsis, the PSII subunit PsbP interacts with the coat protein of the Alfalfa mosaic virus to inhibit viral replication (Balasubramaniam et al., 2014). In another case, P. syringae effectors, HopI1 and HopN1, can remodel host chloroplasts by interacting with PsbQ of PSII to suppress immunity response (Jelenska et al., 2007). In this study, with the downregulation of novel_147 in 409R upon infection, the transcripts of PNSL2 were upregulated (Table 6). We hypothesize that miRNAs competitively target and modulate photosynthesis-related genes in chloroplasts, thereby indirectly inhibiting proteins interacting with P. brassicae effectors and then mediating resistance to P. brassicae in 409R. Lastly, the miR395d-APS4 pair may also mediate clubroot disease progression via photosynthesis (Figure 5I). Inorganic sulfate from the soil is absorbed by root hairs and then transported to leaves, where it is activated into adenosine 5'-phosphosulfate by APSs (ATP sulfurylases) in chloroplasts (Liang et al., 2010; Jagadeeswaran et al., 2014). We found that after P. brassicae infection, 409R showed a decrease in the abundance of bna-miR395d and an increase in the transcript level of APS4 (Table 6). The effect of sulfite synthesis on chloroplast physiology and disease progression in roots remains to be further explored (Figure 5I).
The data presented in the study are deposited in the BioSample repository (https://www.ncbi.nlm.nih.gov/biosample/), accession number PRJNA742780. Accessions from SAMN19977475-SAMN19977486 are data from sRNAseq; accessions SAMN19977487-SAMN19977490 are data from degradome sequencing.
QL analyzed the data, performed the experiments, prepared the figures, and drafted the manuscript. XZ helped analyzed the data. CZ and PC conceived the study and participated in its coordination. PC helped to draft the manuscript. All authors have read and approved the final version of the manuscript.
This study was supported by the National Natural Science Foundation of China (Grant No: 31871659) and CARS-12 to CZ.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.734419/full#supplementary-material
Agorio, A., and Vera, P. (2007). ARGONAUTE4 is required for resistance to Pseudomonas syringae in Arabidopsis. Plant Cell 19, 3778–3790. doi: 10.1105/tpc.107.054494
Bai, S., Liu, J., Chang, C., Zhang, L., Maekawa, T., Wang, Q., et al. (2012). Structure- function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathog. 8:e1002752. doi: 10.1371/journal.ppat.1002752
Balasubramaniam, M., Kim, B. S., Hutchens-Williams, H. M., and Loesch-Fries, L. S. (2014). The photosystem II oxygen-evolving complex protein PsbP interacts with the coat protein of Alfalfa mosaic virus and inhibits virus replication. Mol. Plant Microbe Interact. 27, 1107–1118. doi: 10.1094/MPMI-02-14-0035-R
Bao, F., Huang, X., Zhu, C., Zhang, X., Li, X., and Yang, S. (2014). Arabidopsis HSP90 protein modulates RPP4-mediated temperature-dependent cell death and defense responses. New Phytol. 202, 1320–1334. doi: 10.1111/nph.12760
Bao, N., Lye, K. W., and Barton, M. K. (2004). MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev. Cell 7, 653–662. doi: 10.1016/j.devcel.2004.10.003
Baumberger, N., and Baulcombe, D. C. (2005). Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. U.S.A. 102, 11928–11933. doi: 10.1073/pnas.0505461102
Bhattacharya, I., Dutta, S., Mondal, S., and Mondal, B. (2014). Clubroot disease on brassica crops in India. Can J. Plant Pathol. 36, 154–160. doi: 10.1080/07060661.2013.875064
Bi, K., He, Z., Gao, Z., Zhao, Y., Fu, Y., Cheng, J., et al. (2016). Integrated omics study of lipid droplets from Plasmodiophora brassicae. Sci. Rep. 6:36965. doi: 10.1038/srep36965
Boller, T., and Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern- recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi: 10.1146/annurev.arplant.57.032905.105346
Brodersen, P., Sakvarelidze-Achard, L., Bruun-Rasmussen, M., Dunoyer, P., Yamamoto, Y. Y., Sieburth, L., et al. (2008). Widespread translational inhibition by plant miRNAs and siRNAs. Science 320, 1185–1190. doi: 10.1126/science.1159151
Carbonell, A., Fahlgren, N., Garcia-Ruiz, H., Gilbert, K. B., Montgomery, T. A., Nguyen, T., et al. (2012). Functional analysis of three Arabidopsis ARGONAUTES using slicer-defective mutants. Plant Cell 24, 3613–3629. doi: 10.1105/tpc.112.099945
Chai, A., Xie, X., Shi, Y., and Li, B. (2014). Research status of clubroot (Plasmodiophora brassicae) on cruciferous crops in China. Can. J. Plant Pathol. 36, 142–153. doi: 10.1080/07060661.2013.868829
Chen, J., Jing, J., Zhan, Z., Zhang, T., Zhang, C., and Piao, Z. (2013). Identification of novel QTLs for isolate-specific partial resistance to Plasmodiophora brassicae in Brassica rapa. PLoS ONE 8:e85307. doi: 10.1371/journal.pone.0085307
Chu, M., Song, T., Falk, K. C., Zhang, X., Liu, X., Chang, A., et al. (2014). Fine mapping of Rcr1 and analyses of its effect on transcriptome patterns during infection by Plasmodiophora brassicae. BMC Genomics 15:1166. doi: 10.1186/1471-2164-15-1166
Cui, C., Wang, J., Zhao, J., Fang, Y., He, X., Guo, H., et al. (2020). A Brassica miRNA regulates plant growth and immunity through distinct modes of action. Mol. Plant 13, 231–245. doi: 10.1016/j.molp.2019.11.010
Dangl, J. L. (2013). Pivoting the plant immune system from dissection to deployment. Science 341, 746–751. doi: 10.1126/science.1236011
de Vries, S., Kloesges, T., and Rose, L. E. (2015). Evolutionarily dynamic, but robust, targeting of resistance genes by the miR482/2118 gene family in the Solanaceae. Genome Biol. Evol. 7, 3307–3321. doi: 10.1093/gbe/evv225
Deng, Y., Liu, M., Li, X., and Li, F. (2018). microRNA-mediated R gene regulation: molecular scabbards for double-edged swords. Sci. China Life Sci. 61, 138–147. doi: 10.1007/s11427-017-9237-4
Deng, Y., Zhai, K., Xie, Z., Yang, D., Zhu, X., Liu, J., et al. (2017). Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 355, 962–965. doi: 10.1126/science.aai8898
Diederichsen, E., Frauen, M., Linders, E. G. A., Hatakeyama, K., and Hirai, M. (2009). Status and perspectives of clubroot resistance breeding in crucifer crops. J. Plant Growth Regul. 28, 265–281. doi: 10.1007/s00344-009-9100-0
Dixon, G. R. (2009). The occurrence and economic impact of Plasmodiophora brassicae and clubroot disease. J. Plant Growth Regul. 28, 194–202. doi: 10.1007/s00344-009-9090-y
Dixon, G. R. (2014). Clubroot (Plasmodiophora brassicae woronin)–an agricultural and biological challenge worldwide. Can. J. Plant Pathol. 36, 5–18. doi: 10.1080/07060661.2013.875487
Fei, Q., Xia, R., and Meyers, B. C. (2013). Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25, 2400–2415. doi: 10.1105/tpc.113.114652
Garcia, A. V., and Parker, J. E. (2009). Heaven's gate: nuclear accessibility and activities of plant immune regulators. Trends Plant Sci. 14, 479–487. doi: 10.1016/j.tplants.2009.07.004
German, M. A., Pillay, M., Jeong, D. H., Hetawal, A., Luo, S., Janardhanan, P., et al. (2008). Global identification of microRNA–target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 26, 941–946. doi: 10.1038/nbt1417
Gonzalez, V. M., Muller, S., Baulcombe, D., and Puigdomenech, P. (2015). Evolution of NBS-LRR gene copies among dicot plants and its regulation by members of the miR482/2118 superfamily of miRNAs. Mol. Plant 8, 329–331. doi: 10.1016/j.molp.2014.11.013
Greene, G. H., and Dong, X. (2018). To grow and to defend. Science 361, 976–977. doi: 10.1126/science.aau9065
Hatakeyama, K., Niwa, T., Kato, T., Ohara, T., Kakizaki, T., and Matsumoto, S. (2017). The tandem repeated organization of NB-LRR genes in the clubroot-resistant CRb locus in Brassica rapa L. Mol. Genet. Genomics 292, 397–405. doi: 10.1007/s00438-016-1281-1
Hatakeyama, K., Suwabe, K., Tomita, R. N., Kato, T., Nunome, T., Fukuoka, H., et al. (2013). Identification and characterization of Crr1a, a gene for resistance to clubroot disease (Plasmodiophora brassicae Woronin) in Brassica rapa L. PLoS ONE 8:e54745. doi: 10.1371/journal.pone.0054745
Hirai, M., Harada, T., Kubo, N., Tsukada, M., Suwabe, K., and Matsumoto, S. (2004). A nobel locus for clubroot resistance in Brassica rapa and its linkage markers. Theor. Appl. Genet. 108, 639–643. doi: 10.1007/s00122-003-1475-x
Hirani, A. H., Gao, F., Liu, J., Fu, G., Wu, C., Yuan, Y., et al. (2016). Transferring clubroot resistance from Chinese cabbage (Brassica rapa) to canola (B. napus). Can. J. Plant Patho. 38, 82–90. doi: 10.1080/07060661.2016.1141799
Hu, Q., Hua, W., Yin, Y., Zhang, X., Liu, L., Shi, J., et al. (2017). Rapeseed research and production in China. Crop J. 5, 127–135. doi: 10.1016/j.cj.2016.06.005
Huang, S., Monaghan, J., Zhong, X., Lin, L., Sun, T., Dong, O. X., et al. (2014). HSP90s are required for NLR immune receptor accumulation in Arabidopsis. Plant J. 79, 427–439. doi: 10.1111/tpj.12573
Hwang, S. F., Strelkov, S. E., Gossen, B. D., Turnbull, G. D., Ahmed, H. U., and Manolii, V. P. (2011). Soil treatments and amendments for amelioration of clubroot of canola. Can. J. Plant Sci. 91, 999–1010. doi: 10.4141/cjps2011-028
Inoue, H., Hayashi, N., Matsushita, A., Liu, X., Nakayama, A., Sugano, S., et al. (2013). Blast resistance of CC-NB-LRR protein Pb1 is mediated by WRKY45 through protein-protein interaction. Proc. Natl. Acad. Sci. U.S.A. 110, 9577–9582. doi: 10.1073/pnas.1222155110
Iwakawa, H. O., and Tomari, Y. (2013). Molecular insights into microRNA-mediated translational repression in plants. Mol. Cell 52, 591–601. doi: 10.1016/j.molcel.2013.10.033
Jagadeeswaran, G., Li, Y., and Sunkar, R. (2014). Redox signaling mediates the expression of a sulfate deprivation-inducible microRNA395 in Arabidopsis. Plant J. 77, 85–96. doi: 10.1111/tpj.12364
Jelenska, J., Yao, N., Vinatzer, B. A., Wright, C. M., Brodsky, J. L., and Greenberg, J. T. (2007). A J domain virulence effector of Pseudomonas syringae remodels host chloroplasts and suppresses defenses. Curr. Biol. 17, 499–508. doi: 10.1016/j.cub.2007.02.028
Jiang, Y., Wang, W., Xie, Q., Liu, N., Wang, D., Zhang, X., et al. (2017). Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 356, 1172–1175. doi: 10.1126/science.aam9970
Jin, D., Wang, Y., Zhao, Y., and Chen, M. (2013). MicroRNAs and their cross-talks in plant development. J. Genet. Genomics 40, 161–170. doi: 10.1016/j.jgg.2013.02.003
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kageyama, K., and Asano, T. (2009). Life cycle of Plasmodiophora brassicae. J. Plant Growth Regul. 28, 203–211. doi: 10.1007/s00344-009-9101-z
Kulshrestha, C., Pathak, H., Kumar, D., Dave, S., and Sudan, J. (2020). Elucidating micro RNAs role in different plant-pathogen interactions. Mol. Biol. Rep. 47, 8219–8227. doi: 10.1007/s11033-020-05810-y
Lee, M. H., Jeon, H. S., Kim, H. G., and Park, O. K. (2017). An Arabidopsis NAC transcription factor NAC4 promotes pathogen-induced cell death under negative regulation by microRNA164. New Phytol. 214, 343–360. doi: 10.1111/nph.14371
Li, F., Pignatta, D., Bendix, C., Brunkard, J. O., Cohn, M. M., Tung, J., et al. (2012). MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. U.S.A. 109, 1790–1795. doi: 10.1073/pnas.1118282109
Li, Q., Nadil, S., Zhou, Y., Hou, Z., Gong, J., Liu, Y., et al. (2021). Breeding of a novel clubroot disease-resistant Brassica napus variety Huayouza 62R. Acta Agron. Sin. 47, 210–223. doi: 10.3724/SP.J.1006.2021.04086
Li, S., Liu, L., Zhuang, X., Yu, Y., Liu, X., Cui, X., et al. (2013). MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 153, 562–574. doi: 10.1016/j.cell.2013.04.005
Liang, G., Yang, F., and Yu, D. (2010). MicroRNA395 mediates regulation of sulfate accumulation and allocation in Arabidopsis thaliana. Plant J. 62, 1046–1057. doi: 10.1111/j.1365-313X.2010.04216.x
Liu, J., Cheng, X., Liu, D., Xu, W., Wise, R., and Shen, Q. (2014). The miR9863 family regulates distinct Mla alleles in barley to attenuate NLR receptor-triggered disease resistance and cell-death signaling. PLoS Genet. 10:e1004755. doi: 10.1371/journal.pgen.1004755
Liu, M., Shi, Z., Zhang, X., Wang, M., Zhang, L., Zheng, K., et al. (2019). Inducible overexpression of ideal plant architecture1 improves both yield and disease resistance in rice. Nat. Plants 5, 389–400. doi: 10.1038/s41477-019-0383-2
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta schnittger C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, C., Meyers, B. C., and Green, P. J. (2007). Construction of small RNA cDNA libraries for deep sequencing. Methods 43, 110–117. doi: 10.1016/j.ymeth.2007.05.002
Ma, Z., Coruh, C., and Axtell, M. J. (2010). Arabidopsis lyrata small RNAs: transient MIRNA and small interfering RNA loci within the Arabidopsis genus. Plant Cell 22, 1090–1103. doi: 10.1105/tpc.110.073882
Marjaana, S., Sari, S., Virpi, P., Nilima, K., Maija, H., and Eva-Mari, A. (2010). Two Proteins homologous to PsbQ are novel subunits of the chloroplast NAD(P)H dehydrogenase. Plant Cell Physiol. 51, 877–883. doi: 10.1093/pcp/pcq070
Matsumoto, E., Yasui, C., Ohi, M., and Tsukada, M. (1998). Linkage analysis of RFLP markers for clubroot resistance and pigmentation in Chinese cabbage (Brassica rapa ssp. pekinensis). Euphytica 104, 79–86. doi: 10.1023/A:1018370418201
Meier, I., and Somers, D. E. (2011). Regulation of nucleocytoplasmic trafficking in plants. Curr. Opin. Plant Biol. 14, 538–546. doi: 10.1016/j.pbi.2011.06.005
Nakayama, T., Shinohara, H., Tanaka, M., Baba, K., Ogawa-Ohnishi, M., and Matsubayashi, Y. (2017). A peptide hormone required for casparian strip diffusion barrier formation in Arabidopsis roots. Science 355, 284–286. doi: 10.1126/science.aai9057
Navarro, L., Dunoyer, P., Jay, F., Arnold, B., Dharmasiri, N., Estelle, M., et al. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, :436–439. doi: 10.1126/science.1126088
Navarro, L., Jay, F., Nomura, K., He, S. Y., and Voinnet, O. (2008). Suppression of the microRNA pathway by bacterial effector proteins. Science 321, 964–967. doi: 10.1126/science.1159505
Pang, W., Fu, P., Li, X., Zhan, Z., Yu, S., and Piao, Z. (2018). Identification and mapping of the clubroot resistance gene CRd in Chinese cabbage (Brassica rapa ssp. pekinensis). Front. Plant Sci. 9:653. doi: 10.3389/fpls.2018.00653
Pang, W., Liang, Y., Zhan, Z., Li, X., and Piao, Z. (2020). Development of a sinitic clubroot differential set for the pathotype classification of Plasmodiophora brassicae. Front. Plant Sci. 11:568771. doi: 10.3389/fpls.2020.568771
Peng, Y., van Wersch, R., and Zhang, Y. (2018). Convergent and divergent signaling in PAMP-triggered immunity and effector-triggered immunity. Mol. Plant Microbe Interact. 31, 403–409. doi: 10.1094/MPMI-06-17-0145-CR
Piao, Z., Deng, Y., Choi, S., Park, Y. J., and Lim, Y. P. (2004). SCAR and CAPS mapping of CRb, a gene conferring resistance to Plasmodiophora brassicae in Chinese cabbage (Brassica rapa ssp. pekinensis). Theor. Appl. Genet. 108, 1458–1465. doi: 10.1007/s00122-003-1577-5
Racolta, A., Bryan, A. C., and Tax, F. E. (2014). The receptor-like kinases GSO1 and GSO2 together regulate root growth in Arabidopsis through control of cell division and cell fate specification. Dev. Dyn. 243, 257–278. doi: 10.1002/dvdy.24066
Rahman, H., Peng, G., Yu, F., Falk, K. C., Kulkarni, M., and Selvaraj, G. (2014). Genetics and breeding for clubroot resistance in Canadian spring canola (Brassica napus L.). Can. J. Plant Pathol. 36, 122–134. doi: 10.1080/07060661.2013.862571
Reis, R. S., Hart-Smith, G., Eamens, A. L., Wilkins, M. R., and Waterhouse, P. M. (2015). Gene regulation by translational inhibition is determined by dicer partnering proteins. Nat. Plants 1, 14027. doi: 10.1038/nplants.2014.27
Rodríguez-Herva, J. J., González-Melendi, P., Cuartas-Lanza, R., Antúnez-Lamas, M., Río-Alvarez, I., Li, Z., et al. (2012). Bacterial cysteine protease effector protein interferes with photosynthesis to suppress plant innate immune responses. Cell Microbiol. 14, 669–681. doi: 10.1111/j.1462-5822.2012.01749.x
Sakamoto, K., Saito, A., Hayashida, N., Taguchi, G., and Matsumoto, E. (2008). Mapping of isolate-specific QTL for clubroot resistance in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Theo. Appl. Genet. 117, 759–767. doi: 10.1007/s00122-008-0817-0
Shen, Q., Saijo, Y., Mauch, S., Biskup, C., Bieri, S., Keller, B., et al. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315, 1098–1103. doi: 10.1126/science.1136372
Shinya, Y., Kentaro, I., Atsushi, T., Seiko, I., Kunio, I., Noriko, I., et al. (2010). Three PsbQ-Like proteins are required for the function of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant Cell Physiol. 51, 866–876. doi: 10.1093/pcp/pcq060
Shivaprasad, P. V., Chen, H. M., Patel, K., Bond, D. M., Santos, B. A., and Baulcombe, D. C. (2012). A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24, 859–874. doi: 10.1105/tpc.111.095380
Song, J. J., Smith, S. K., Hannon, G. J., and Joshua-Tor, L. (2004). Crystal structure of argonaute and its implications for RISC slicer activity. Science 305, 1434–1437. doi: 10.1126/science.1102514
Song, X., Li, Y., Cao, X., and Qi, Y. (2019). MicroRNAs and their regulatory roles in plant–environment interactions. Annu. Rev. Plant Biol. 70, 489–525. doi: 10.1146/annurev-arplant-050718-100334
Sunkar, R., Li, Y. F., and Jagadeeswaran, G. (2012). Functions of microRNAs in plant stress responses. Trends Plant Sci. 17, 196–203. doi: 10.1016/j.tplants.2012.01.010
Suwabe, K., Tsukazaki, H., Iketani, H., Hatakeyama, K., Fujimura, M., Nunome, T., et al. (2003). Identification of two loci for resistance to clubroot (Plasmodiophora brassicae Woronin) in Brassica rapa L. Theo. Appl. Genet. 107, 997–1002. doi: 10.1007/s00122-003-1309-x
Suwabe, K., Tsukazaki, H., Iketani, H., Hatakeyama, K., Kondo, M., Fujimura, M., et al. (2006). Simple sequence repeat-based comparative genomics between Brassica rapa and Arabidopsis thaliana: the genetic origin of clubroot resistance. Genetics 173, 309–319. doi: 10.1534/genetics.104.038968
Tang, C., Yang, M., Wu, F., Zhao, H., Pang, Y., Yang, R., et al. (2015). Identification of miRNAs and their targets in transgenic Brassica napus and its acceptor (Westar) by high-throughput sequencing and degradome analysis. RSC Adv. 5, 85383–85394. doi: 10.1039/C5RA14672K
Tang, J., and Chu, C. (2017). MicroRNAs in crop improvement: fine-tuners for complex traits. Nat. Plants 3:17077. doi: 10.1038/nplants.2017.77
Tian, D., Traw, M. B., Chen, J., Kreitman, M., and Bergelson, J. (2003). Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423, 74–77. doi: 10.1038/nature01588
Verma, S. S., Rahman, M. H., Deyholos, M. K., Basu, U., and Kav, N. N. V. (2014). Differential expression of miRNAs in Brassica napus root following infection with Plasmodiophora brassicae. PLoS ONE 9:e86648. doi: 10.1371/journal.pone.0086648
Wallenhammar, A. C., Almquist, C., Schwelm, A., Roos, J., Marzec-Schmidt, K., Jonsson, A., et al. (2014). Clubroot, a persistent threat to Swedish oilseed rape production. Can. J. Plant Pathol. 36, 135–141. doi: 10.1080/07060661.2013.870606
Wang, J., Zhou, L., Shi, H., Chern, M., Yu, H., Yi, H., et al. (2018). A single transcription factor promotes both yield and immunity in rice. Science 361, 1026–1028. doi: 10.1126/science.aat7675
Wang, Y., Li, K., Chen, L., Zou, Y., Liu, H., Tian, Y., et al. (2015). MicroRNA167-directed regulation of the auxin response factors GmARF8a and GmARF8b is required for soybean nodulation and lateral root development. Plant Physiol. 168, 984–999. doi: 10.1104/pp.15.00265
Wei, X., Xu, W., Yuan, Y., Yao, Q., Zhao, Y., Wang, Z., et al. (2016). Genome-wide investigation of microRNAs and their targets in Brassica rapa ssp. pekinensis root with Plasmodiophora brassicae infection. Hortic. Plant J. 2, 209–216. doi: 10.1016/j.hpj.2016.11.004
Williams, P. H. (1966). A system for the determination of races of Plasmodiophora brassicae that infect cabbage and rutabaga. Phytopathology 56,624–626
Wong, J., Gao, L., Yang, Y., Zhai, J., Arikit, S., Yu, Y., et al. (2014). Roles of small RNAs in soybean defense against Phytophthora sojae infection. Plant J. 79, 928–940. doi: 10.1111/tpj.12590
Wu, L., Zhou, H., Zhang, Q., Zhang, J, Ni, F., Liu, C., et al. (2010). DNA methylation mediated by a microRNA pathway. Mol. Cell 38, 465–475. doi: 10.1016/j.molcel.2010.03.008
Xu, W., Meng, Y., and Wise, R. P. (2014). Mla- and Rom1-mediated control of microRNA398 and chloroplast copper/zinc superoxide dismutase regulates cell death in response to the barley powdery mildew fungus. New Phytol. 201, 1396–1412. doi: 10.1111/nph.12598
Xu, Y., Liu, R., Yan, L., Liu, Z., Jiang, S., Shen, Y., et al. (2011). Light-harvesting chlorophyll a/b-binding proteins are required for stomatal response to abscisic acid in Arabidopsis. J. Exp. Bot. 63, 1095–1106. doi: 10.1093/jxb/err315
Yu, X., Hou, Y., Chen, W., Wang, S., Wang, P., and Qu, S. (2017). Malus hupehensis miR168 targets to ARGONAUTE1 and contributes to the resistance against Botryosphaeria dothidea infection by altering defense responses. Plant Cell Physiol. 58, 1541–1557. doi: 10.1093/pcp/pcx080
Zajac, T., Klimek-Kopyra, A., Oleksy, A., Lorenc-Kozik, A., and Ratajczak, K. (2016). Analysis of yield and plant traits of oilseed rape (Brassica napus L.) cultivated in temperate region in light of the possibilities of sowing in arid areas. Acta Agrobot 69:1696. doi: 10.5586/aa.1696
Zhai, J., Arikit, S., Simon, S. A., Kingham, B. F., and Meyers, B. C. (2014). Rapid construction of parallel analysis of RNA end (PARE) libraries for Illumina sequencing. Methods 67, 84–90. doi: 10.1016/j.ymeth.2013.06.025
Zhai, J., Jeong, D. H., Paoli, D. E., Park, S., Rosen, B. D., Li, Y., et al. (2011). MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25, 2540–2553. doi: 10.1101/gad.177527.111
Zhang, X., Zhao, H., Gao, S., Wang, W., Katiyar-Agarwal, S., Huang, H. D., et al. (2011). Arabidopsis Argonaute 2 regulates innate immunity via miRNA393*-mediated silencing of a Golgi-localized SNARE gene, MEMB12. Mol. Cell 42, 356–366. doi: 10.1016/j.molcel.2011.04.010
Zhang, Y., Xia, R., Kuang, H., and Meyers, B. C. (2016). The diversification of plant NBS-LRR defense genes directs the evolution of microRNAs that target them. Mol. Biol. Evol. 33, 2692–2705. doi: 10.1093/molbev/msw154
Keywords: Plasmodiophora brassicae, clubroot resistance, miRNA, degradome, Brassica napus
Citation: Li Q, Shah N, Zhou X, Wang H, Yu W, Luo J, Liu Y, Li G, Liu C, Zhang C and Chen P (2021) Identification of Micro Ribonucleic Acids and Their Targets in Response to Plasmodiophora brassicae Infection in Brassica napus. Front. Plant Sci. 12:734419. doi: 10.3389/fpls.2021.734419
Received: 01 July 2021; Accepted: 21 September 2021;
Published: 28 October 2021.
Edited by:
Ryo Fujimoto, Kobe University, JapanReviewed by:
Jian Wu, Yangzhou University, ChinaCopyright © 2021 Li, Shah, Zhou, Wang, Yu, Luo, Liu, Li, Liu, Zhang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peng Chen, Y2hlbnBlbmdAbWFpbC5oemF1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.