
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 23 September 2021
Sec. Plant Development and EvoDevo
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.730007
Lily (Lilium spp.) is an important commercial flower crop, but its market popularity and applications are adversely affected by severe pollen pollution. Many studies have examined pollen development in model plants, but few studies have been conducted on flower crops such as lily. GAMYBs are a class of R2R3-MYB transcription factors and play important roles in plant development and biotic resistance; their functions vary in different pathways, and many of them are involved in anther development. However, their function and regulatory role in lily remain unclear. Here, the GAMYB homolog LoMYB33 was isolated and identified from lily. The open reading frame of LoMYB33 was 1620 bp and encoded a protein with 539 amino acids localized in the nucleus and cytoplasm. Protein sequence alignment showed that LoMYB33 contained a conserved R2R3 domain and three BOX motifs (BOX1, BOX2, and BOX3), which were unique to the GAMYB family. LoMYB33 had transcriptional activation activity, and its transactivation domain was located within 90 amino acids of the C-terminal. LoMYB33 was highly expressed during the late stages of anther development, especially in pollen. Analysis of the promoter activity of LoMYB33 in transgenic Arabidopsis revealed that the LoMYB33 promoter was highly activated in the pollen of stage 12 to 13 flowers. Overexpression of LoMYB33 in Arabidopsis significantly retarded growth; the excess accumulation of LoMYB33 also negatively affected normal anther development, which generated fewer pollen grains and resulted in partial male sterility in transgenic plants. Silencing of LoMYB33 in lily also greatly decreased the amount of pollen. Overall, our results suggested that LoMYB33 might play an important role in the anther development and pollen formation of lily.
Pollen is required for sexual reproduction in plants (Aizen and Harder, 2007). Normal pollen development is essential for ensuring the high yield and quality of crops and fruits, but deleterious effects are often inevitable. For example, many ornamental plants (e.g., Populus, lily, and chrysanthemum) contain high concentrations of pollen, which results in pollen contamination and anaphylactic reactions in susceptible populations and reduces the commercial value of these ornamental plants (Tong et al., 2013; Sui et al., 2015; Wang et al., 2016; Kim et al., 2018; Feng et al., 2019; Li et al., 2019; Teng et al., 2021). Thus, male-sterile or pollen-free ornamental plants are often highly useful. Although many studies have examined pollen development in model plants (Köhler et al., 2003; Millar and Gubler, 2005; Chu, 2006; Long et al., 2008), studies of flower crops are relatively scarce.
Gibberellins (GAs) are a major group of hormones in plants that play an essential role in plant vegetative growth and reproductive development (Hedden, 2001). GAMYBs are members of the R2R3-MYB family; in addition to the conserved R2R3 domain at the N-terminus (Woodger et al., 2003), the GAMYB transcription factors (TFs) contain three BOX motifs, an important feature of this family (Dubos et al., 2010). In 1995, GAMYBs were first reported in barley aleurone cells, and they were shown to be up-regulated by gibberellin (GA) and activated GA-regulated genes (Gubler et al., 1995). GAMYBs also play important roles in flower induction and flowering regulation. For example, AtMYB33 in Arabidopsis directly binds to the promoter of AtLFY to regulate flowering (Gocal et al., 2001); CsGAMYB affects the differentiation of male and female flowers in cucumber (Zhang et al., 2014). HvGAMYB also plays a role in the formation of male organs in barley (Murray et al., 2003). In addition, GAMYBs have been reported to regulate anther development in a variety of plants (Kaneko et al., 2004). Both AtMYB33 and AtMYB65 are GAMYB family members in Arabidopsis, which function redundantly to regulate anther development and pollen formation (Millar and Gubler, 2005). Rice gamyb mutants are sterile because the anthers are poorly developed and lack pollen (Aya et al., 2009).
In Arabidopsis, AtMYB33 and AtMYB65 are targeted by miRNA159; as the silencing or overexpression of miRNA159 can interfere with anther development, miRNA159 has been suggested to be a homeostatic modulator of GAMYB activity ensuring normal anther development (Achard et al., 2004). In rice, GA activates OsGAMYB in the tapetum through the GID1/DELLA sensing pathway, which leads to the up-regulation of OsCYP703A and OsKAR for sporopollen formation (Cheng et al., 2004; Kwon et al., 2015). Sporopollen is the key component of the pollen outer wall (Dickinson and Heslop-Harrison, 1968), and its absence causes pollen abortion. These results indicate that GAMYBs are required for normal pollen development in model plants, but their roles in ornamental plants remain unknown.
Lily (Lilium spp.) is an important commercial flower crop (Gong et al., 2014), but its market popularity and applications are adversely affected by severe pollen pollution. There is thus a need to explore the mechanism of pollen development in lilies with little or no pollen (Wang et al., 2019; Yuan et al., 2021). According to our previous transcriptome data of pollen abortion varieties (Wang et al., 2019), an MYB gene annotated as GAMYB (LoMYB33) is differentially expressed in two periods before and after pollen abortion, indicating that LoMYB33 may be involved in pollen development. Here, a GAMYB member in lily (LoMYB33) was isolated and identified. LoMYB33 showed transactivation ability, and it located in the cytoplasm and nucleus; its C-terminal contributed to its transactivation ability. LoMYB33 was highly expressed in the late stages of anther development, especially in pollen. LoMYB33 overexpression in Arabidopsis resulted in significant growth retardation and reduced the number of normal pollen grains, which caused male sterility in transgenic plants. The silencing of LoMYB33 in lily also reduced the number of pollen. These results indicate that LoMYB33 plays a role in pollen development; its function thus requires strict control under normal development conditions.
The Lilium Oriental hybrid “Siberia” and Lilium longiflorum “White Heaven” were used as the experimental materials. “Siberia” was planted in the Baguazhou Lily Germplasm Resource Base of Nanjing Agricultural University and planted in a greenhouse with sufficient sunlight at ~20°C. “White Heaven” was grown in a growth room at 22°C with a 16 h:8 h light:dark photoperiod. Both Arabidopsis (Col-0) and tobacco (Nicotiana benthamiana) were used as the testing platform. Arabidopsis and tobacco seeds were sterilized with 1% sodium hypochlorite for 15 min, washed with sterile water 5–6 times, and sowed on MS medium (Murashige and Skoog, 1962). The seeds were placed in the dark at 4°C for 3 days and then transferred to a light incubator at 22°C (16 h/8 h light period). After 10 days, seedlings with four true leaves were transferred into a pot filled with peat and vermiculite (1:1) and then cultured in a controlled environment as described above in subsequent experiments.
Anthers in the 12-cm length flower buds were collected, and total RNA was extracted with an RNAprep Pure Plant Kit (DP-103, Tiangen, China). cDNA was synthesized by M-MLV reverse transcriptase (R233-01, Vazyme, China) with oligo-dT primers. Based on transcriptome data (Wang et al., 2019), the specific primers for the LoMYB33 coding sequence (Supplementary Table 1) were designed, and the fragment was isolated using PrimeSTAR HS DNA polymerase (DR010A, Takara, Japan). The target fragment was constructed into the pMD18-T vector and sequenced.
The phylogenetic tree of different species, the phylogenetic tree of the MYB family of Arabidopsis, and the phylogenetic tree of LoMYB33 and some MYB proteins in rice were constructed using the neighbor-joining method in MEGA7 (Kumar et al., 2016). The sequences of Arabidopsis MYB members were downloaded from the TAIR database (https://www.arabidopsis.org/index.jsp). The sequences of rice MYB members were downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov/). The protein multiple alignment of LoMYB33 with other GAMYB members from different species was conducted using ClustalX 1.81 and BioEdit 7.0 software (Hall, 1999; Thompson et al., 2003).
The yeast system was used for transactivation analysis. The recombinant plasmids, the positive control GAL4, and the negative control pGBKT7 were transformed into yeast AH109 cells and spread on SD/-Trp medium. The transformed yeast cells were incubated at 30°C for 3 days. The transcriptional activity was evaluated by growing yeast colonies on SD/-Trp/-His and SD/-Trp/-His with 5 mM 3-AT (3-amino-1,2,4-triazole)-deficient solid media. The transformed yeast cells grown on SD/-Trp medium were also transferred to filter paper for transcriptional activity analysis, and X-α-Gal was added to observe the activity of β-galactosidase (Ding et al., 2021).
LoMYB33 or mLoMYB33 (with mutated target sites of miRNA159) was cloned into the pCAMBIA1300-green fluorescent protein (GFP) vector. The recombinant plasmid was then transformed into Agrobacterium tumefaciens strain GV3101. Five μL of Agrobacterium competent cells harboring plasmids was mixed gently, placed on ice for 5 min, and rapidly frozen for 1 min in liquid nitrogen, followed by treatment with 37°C for 5 min and cooled on ice for 2 min. The cells were then cultured with LB medium at 28°C and 200 rpm for 3–5 h. After centrifugation and resuspending the bacteria, they were evenly spread on solid LB medium (containing 50 μg·μL−1 Kan and 100 μg·μL−1 Rif) and cultured at 28°C for 2 days. The appropriate clones were then selected and cultured with liquid LB medium (containing 50 μg·μL−1 Kan and 100 μg·μL−1 Rif) for 12 h. The bacterial solution was resuspended with the injection buffer (10 mM MgCl2, 100 mM 2-morpholinoethanesulfonic acid, and 200 μM acetosyringone, pH = 5.8) and injected into tobacco leaves (Wu et al., 2019). After 48 h, the GFP signal was observed with a laser scanning confocal microscope (LSM800, Zeiss, Germany).
Total RNA was extracted from the bulbus, root, stem, leaf, petal, and anthers at different developmental stages, as well as the ovary of lily using the Trizol method (TaKaRa, Japan). Reverse transcription was performed using a HiScript II kit (R233-01, Vazyme, China). Real-time quantitative PCR (RT-qPCR) was used to determine the expression levels. The 18S rRNA of lily was used as the reference gene. The primers are shown in Supplementary Table 1. The SYBR® Green Realtime PCR reaction system was used for RT-qPCR. The cycling parameters were as follows: 95°C for 2 min; 95°C for 15 s, 55°C for 15 s, and 72°C for 20 s for 40 cycles. The CT value of each sample was obtained, and quantitative analysis of the relative level of expression was performed using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Lily “Siberia” plants at the same development stage were treated with 288 μM GA3 in a bottle; a water treatment group subjected to the same conditions was used as a control. Samples were taken at 0, 4, 8, 12, 24, and 48 h after treatment. Three anthers with bud lengths <5 cm were sampled for RT-qPCR analysis.
The LoMYB33 promoter was cloned using the hiTAIL-PCR method (Liu and Chen, 2007) with three special reverse primers at the 5′ end of the LoMYB33 and the five universal primers LAD1–5 and AC1 (Supplementary Table 1). The upstream fragment located 465 bp away from the ATG of LoMYB33 was isolated and identified. The promoter sequence was analyzed using New PLACE software (https://www.dna.affrc.go.jp/PLACE/?action=newplace).
The promoter fragment (465 bp) of LoMYB33 was inserted into the pCAMBIA1391 vector. The constructed vector was transformed into A. tumefaciens strain GV3101. The floral-dip method was used for Arabidopsis transformation (Clough and Bent, 1998). The proLoMYB33:GUS transgenic plants were screened on MS medium containing 35 mg L−1 hygromycin. For histochemical GUS analysis, plant tissues were cultured in GUS staining solution (Huayueyang, Beijing, China) overnight at 37°C. Chlorophyll was then removed with 70% ethanol for imaging.
The open reading frame (ORF) of LoMYB33 was cloned and inserted into the pCAMBIA1300 vector and transformed into GV3101. Five-week-old Arabidopsis plants were used for transformation by the floral-dip method. The transgenic lines were identified by RT-PCR; three T3-generation homozygous lines were selected for the functional analysis. The primers used for the transgene identification are listed in Supplementary Table 1.
The flower buds and anthers of wild-type and LoMYB33-overexpressing Arabidopsis plants at different developmental stages were vacuum-infiltrated with FAA solution. Fixed anthers were dehydrated in 50, 70, 85, 90, and 100% ethanol gradients (2 h each) and then embedded in paraffin. Paraffin sections (8 μm thick) were obtained with a microtome. The sections were stained with hematoxylin and observed using a light microscope (DM-6B, Leica, Germany).
Flowers were photographed under a stereomicroscope (M165FC, Leica, Germany). To determine pollen viability, anthers at stage 12 (Sanders et al., 1999) were collected and stained with Alexander solution (Alexander, 1969) and observed under an optical electron microscope (DM-6B, Leica, Germany).
The methods of Wang et al. (2018a) and Sun and Pan (2008) with slight modifications were used for pollen quantification. When Arabidopsis plants were in full bloom, six white or recently bloomed flowers on the main stem were placed in a 1.5-ml centrifuge tube and dried in an oven at 60°C. The anthers were completely cracked, and the pollen was released; 1 mL of 200 g L−1 (NaPO3)6 solution was added, and the solution was oscillated for 2 min on a micro vortex mixer. Next, two drops of 2.5 μL of suspension from each of the three samples were placed on a glass slide. The number of pollen grains in the suspension was counted under the objective lens (4×) of an optical microscope (DM-6B, Leica, Germany). The number of pollen grains in each droplet was counted, and the average value of six droplets was recorded. The same procedures were repeated for three lily anthers at the full bloom stage, except that 8 mL of 200 g L−1 (NaPO3)6 solution was added after the anthers were dried.
Amount of pollen in lily = anther number of single flower × number of pollen grains per slide × 3200.
To generate pTRV2-LoMYB33, a 300-bp gene-specific fragment was cloned into the pTRV2 vector using cDNA as a template. The primer pairs used to generate TRV vectors are shown in Supplementary Table 1. The recombinant plasmid was then transformed into A. tumefaciens GV3101. The bacterial solution was resuspended by the injection buffer (10 mM MgCl2, 100 mM 2-morpholinoethanesulfonic acid, 200 μM acetosyringone, pH = 5.8) (OD600 =1.0) and injected into “White heaven” leaves. Before injection, the mixture of 1:1 (v/v) A. tumefaciens culture containing pTRV1 and pTRV2, pTRV1, and pTRV2-LoMYB33 was stored in the dark at room temperature for 3 h. Lily “White heaven” plants that had not yet produced flower buds were injected; the anthers were photographed during flowering and then collected for identification (Chen et al., 2020).
Microsoft Excel 2010 (Microsoft Corp., USA) and Statistical Product and Service Solutions v 612 17.0 (SPSS, USA) were used to analyze the data. Student's t-test was used to test for significant differences, and the threshold for statistical significance was P < 0.05. Student–Newman–Keuls test (P < 0.05) was used to compare means after ANOVA.
The LoMYB33 ORF was 1,620 bp and encoded a protein with 539 amino acids. Phylogenetic analysis with 124 MYB family TFs from Arabidopsis indicated that LoMYB33 was closely related to AtMYB33 and AtMYB65 (Supplementary Figure 1), both of which are GAMYB family members (Millar and Gubler, 2005; Tsuji et al., 2006; Li et al., 2016). A BLAST search against the Arabidopsis TAIR database revealed that LoMYB33 was most closely related to AtMYB33 (Supplementary Figure 2); thus, this protein was named LoMYB33. A phylogenetic analysis of LoMYB33 with MYB members in rice revealed that it is more closely related to OsGAMYB (Supplementary Figure 3), suggesting that LoMYB33 is a member of the GAMYB family. The alignment of LoMYB33 with Phoenix dactylifera, Elaeis guineensis, Oryza sativa, Arabidopsis, Rosa chinensis and other GAMYB homologs revealed that LoMYB33 was clustered with the GAMYBs of P. dactylifera and E. guineensis, Musa acuminata, and Asparagus officinalis; P. dactylifera, E. guineensis, M. acuminata, A. officinalis, and lily are non-grass monocotyledonous plants (Figure 1A). Multiple protein alignment with the amino acid sequences of homologs from E. guineensis, O. sativa, Arabidopsis, R. chinensis, Zea mays, Glycine max, Hordeum vulgare, Triticum aestivum, Solanum lycopersicum, and Cucumis sativus showed that LoMYB33 contained the classical R2R3 domain of the R2R3-MYB family (Figure 1B), indicating that it belonged to the R2R3-MYB family. In addition, LoMYB33 contained three typical BOX1, BOX2, and BOX3 motifs of the GAMYB family. BOX1 is a QRaGLPxYPx (E/S) motif located near the C-terminal next to the R2R3 repeat DNA-binding domain (Kranz et al., 1998; Romero et al., 1998). BOX2 and BOX3 are also conserved regions of GAMYBs (Gocal et al., 2001). Protein alignment revealed that the BOX1, BOX2, and BOX3 motifs of LoMYB33 were not completely conserved, as some amino acid substitutions were detected; a similar pattern was observed for GmGAMYB1, GsGAMYB1, AtMYB65, and ZmGAMYB (Figure 1B).
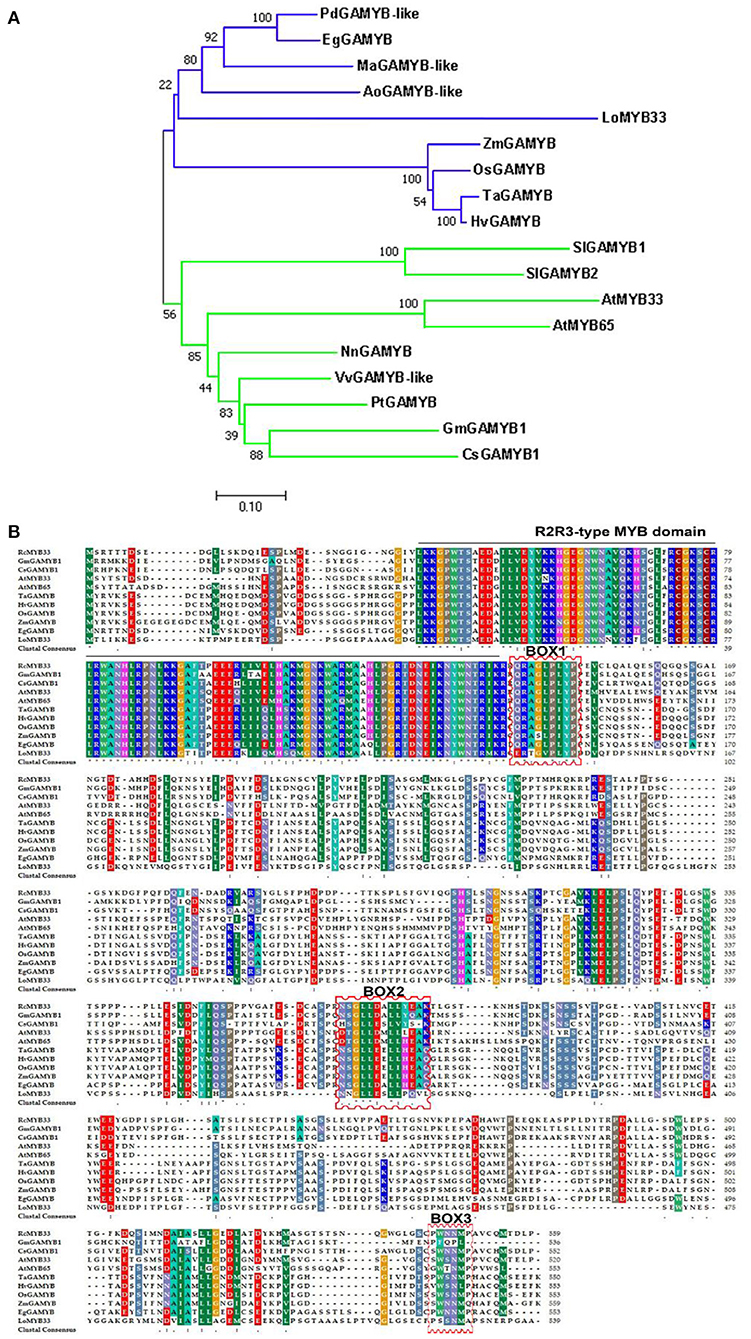
Figure 1. Amino acid sequence alignment, phylogenetic analysis, and bioinformatics analysis of LoMYB33. (A) Phylogenetic tree of LoMYB33 protein and GAMYB proteins from other plant species. Protein sequences of GAMYBs were taken from Phoenix dactylifera (PdGAMYB-like, XP_008792432.2), Elaeis guineensis (EgGAMYB, XP_010922970.1), Lilium spp. (LoMYB33), Oryza sativa (OsGAMYB, XP_015622335.1), Triticum aestivum (TaGAMYB, XP_037414515.1), Arabidopsis thaliana (AtMYB33, NP_001078537.1; AtMYB65, NP_001327042.1), Rosa chinensis (RcMYB33, XP_024166362.1), Zea mays (ZmGAMYB, NP_001241838.2), Glycine max (GmGAMYB1, NP_001304541.1), Hordeum vulgare (HvGAMYB, KAE8787635.1) and Cucumis sativus (CsGAMYB1, XP_004140923.1), Musa acuminata (MaGAMYB-like, XP_009398961.1), Nelumbo nucifera (NuGAMYB, XP_010251854.1), Populus tomentosa (PtGAMYB-like, AZQ25444.1), Vitis vinifera (VvGAMYB-like, XP_034705115.1), Asparagus officinalis (AoGAMYB-like, XP_020253654.1), and Solanum lycopersicum (SlGAMYB1, Solyc01g009070; SlGAMYB2, Solyc06g073640). (B) Protein sequences of GAMYBs were from Elaeis guineensis (EgGAMYB, XP_010922970.1), Lilium spp. (LoMYB33), Oryza sativa (OsGAMYB, XP_015622335.1), Arabidopsis thaliana (AtMYB33, NP_001078537.1; AtMYB65, NP_001327042.1), Rosa chinensis (RcMYB33, XP_024166362.1), Triticum aestivum (TaGAMYB, XP_037414515.1), Zea mays (ZmGAMYB, NP_001241838.2), Hordeum vulgare (HvGAMYB, KAE8787635.1), and Cucumis sativus (CsGAMYB1, XP_004140923.1); the R2R3 domain is indicated by black lines, and the BOX motifs are indicated by red frames.
We detected the expression of LoMYB33 in the bulbus, root, stem, leaf, petal, anther, and ovary of lily by RT-qPCR (Figure 2A). The expression of LoMYB33 was significantly higher in male organs such as anther and pollen than in female organs; LoMYB33 expression was also higher in male organs than in vegetative organs (Figure 2A). The expression of LoMYB33 was significantly higher in late developmental stages than in early developmental stages; LoMYB33 expression was highest in anthers collected from 11 cm flower buds, followed by buds of 9, 10, and 12 cm (Figure 2B). The expression of LoMYB33 was highest in mature pollen, especially during the early stage of pollen maturation (Figure 2C). In addition, there is a small expression peak in the tetrad stage of anther development (Figure 2C). These results indicated that LoMYB33 was mainly expressed in the late development stages of anthers; its transcripts were most highly accumulated in pollen, and it might also function in the tetrad stage of anther.
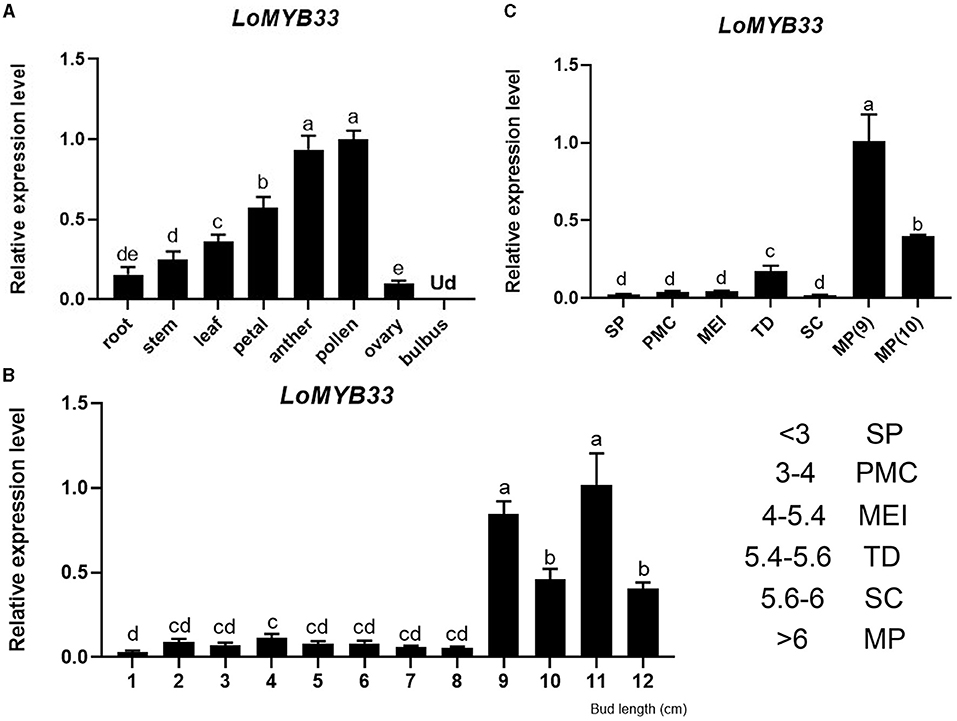
Figure 2. Expression analysis of LoMYB33. (A) Relative expression of LoMYB33 in different organs (9-cm flower bud) by RT-qPCR analysis. (B) Relative expression of LoMYB33 in anthers of 1 to 12-cm flower buds. (C) Relative expression of LoMYB33 in pollen from sporogenous cells to mature pollen. Data are mean ± SD of three independent experiments. Different letters indicate significant differences among these lines (Student–Newman–Keuls test, P < 0.05). SP, sporulation period; PMC, pollen mother cell stage; MEI, meiotic stage; TD, tetrad stage; SC, single-core stage; MP, mature pollen. Ud, Undetermined. The number indicates the length of the flower bud of “Siberia” lily, the unit is cm.
The 465-bp promoter of LoMYB33 was obtained and analyzed using PlantCARE online software (Supplementary Figure 4). The promoter contained 1 ARE, 5 CAAT-boxes, 2 GC-motifs, 1 TATA-box, and 2 TCCC-motifs. These elements were related to the photoresponse and anaerobic induction. The GUS gene driven by the LoMYB33 promoter was transformed into Arabidopsis; GUS histochemical staining analysis revealed that the LoMYB33 promoter was activated in the later stages of Arabidopsis anther development but was not activated in the early stages (Figures 3A–H), a pattern consistent with its expression in lily anthers (Figure 2). As expected, high GUS activity was observed in mature pollen (Figure 3I), which indicated that the promoter of LoMYB33 was highly activated in pollen.
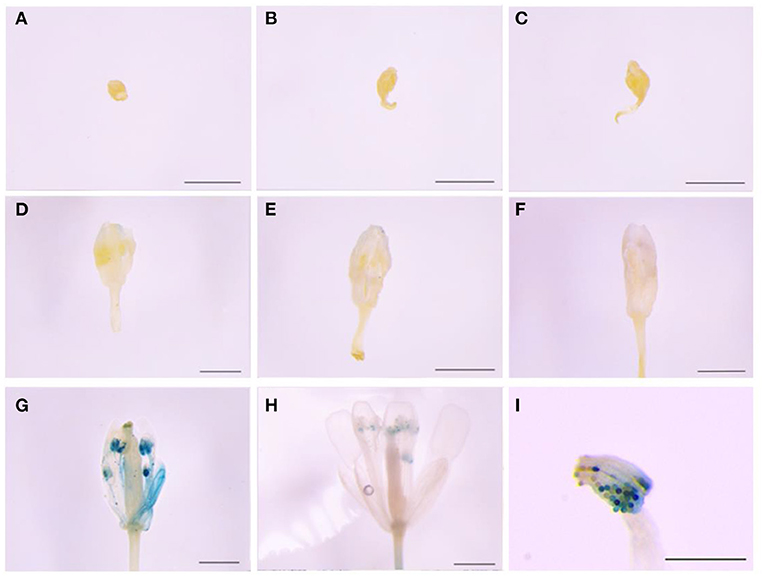
Figure 3. Analysis of LoMYB33 promoter activity. (A–H) show the results of the activity analysis of GUS for stages 1–13. (A) Sporulation stage (stage 2); (B) mother cell stage (stage 5); (C) start of the meiotic period (stage 6); (D) microspore stage (stage 8); (E) spore stage in which spores are small (stage 9); (F) microspore stage (stage 10); (G) mature pollen stage (stage 12); (H) mature pollen stage (stage 13); and (I) GUS staining of pollen in stage 12 flowers. Three independent experiments were performed, and one representative picture is shown. Bars are 1 mm.
The GFP signal of LoMYB33-GFP was not observed, but the GFP protein signal was observed following transient expression of the GFP fusion protein in tobacco leaves. Analysis of the gene sequence of LoMYB33 revealed that LoMYB33 might be a target of miRNA159; this was confirmed in a previous study of lily (Gao et al., 2020). In Arabidopsis, the GAMYB genes AtMYB33 and AtMYB65 are also miRNA159 targets (Rhoades et al., 2002; Allen et al., 2007). Analysis of the miRNA159 target sites of LoMYB33 revealed that the conserved TTGGAGCTCCCTTCATTCCAAAAT sequence of LoMYB33 might be the target sequence, which is located from 966 to 990 bp. Therefore, we speculated that wild-type LoMYB33 might be cut by miRNA159 in the tobacco cells. The miRNA159 target sites of LoMYB33 were then mutated to mLoMYB33, which was also transiently expressed in tobacco leaves. A large number of fluorescent signals appeared, and mLoMYB33-GFP was located in the nucleus and cytoplasm (Figure 4A), suggesting that LoMYB33 is a cytoplasm-nucleus localization protein.
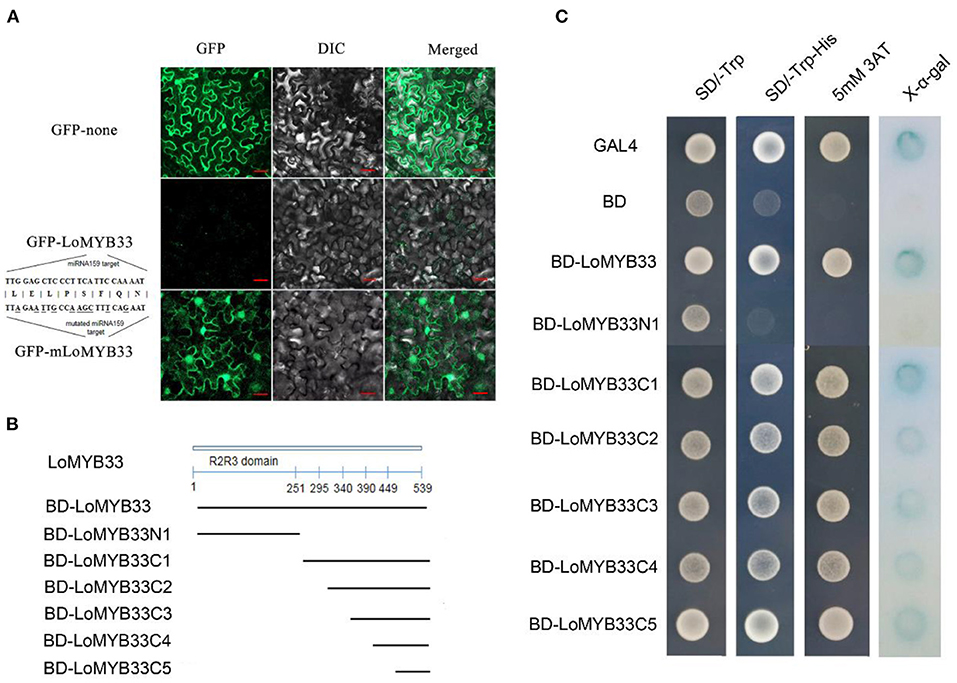
Figure 4. Analysis of LoMYB33 transcriptional activation activity and subcellular location. (A) The GFP (up) protein, LoMYB33-GFP (middle), and mLoMYB33-GFP (down) were transiently expressed in tobacco leaves. (B) The lines below the LoMYB33 diagram indicated the different regions that were used in the transactivation assay. All constructs fused with the GAL4 DNA-binding domain (BD) were inserted into the expression vector pGBKT7. (C) Transactivation activity of different constructs of LoMYB33 in yeast cells. The positive control was GAL4, and the negative control was BD vector. The SD-Trp medium (left panel) was used to detect transformation, the SD-Trp/His medium (middle panel) was used to examine transformant growth, and X-α-gal staining was used to detect β-galactosidase activity of transformed yeast cells (the right panel).
The full length and different portions of LoMYB33 were fused to the pGBKT7 vector and then transformed into the yeast strain AH109 (Figure 4B). Full-length LoMYB33 had transcriptional activation activity; all C1–C5 fragments showed transcriptional activation activity but not N1 (Figure 4C). Therefore, the transcriptional activation domain of LoMYB33 was within 90 amino acids of the C-terminal.
To study the effect of GA on the expression of LoMYB33 in anthers, lily “Siberia” plants showing normal growth were treated with GA3, and the expression of LoMYB33 was determined at several time points after treatment. The expression of LoMYB33 after GA3 treatment was up-regulated relative to the control (Figure 5). The expression of LoMYB33 in the control changed little following treatment, but the expression of LoMYB33 in the experimental group changed significantly after treatment; the expression of LoMYB33 was highest after 48 h of GA3 treatment, followed by 24 h of GA3 treatment (Figure 5). These results indicated that GA3 treatment could activate the expression of LoMYB33 in anthers.
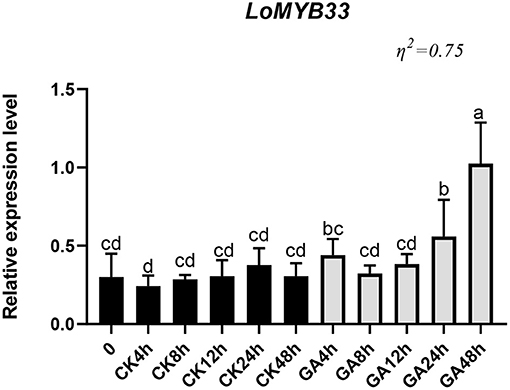
Figure 5. Expression of LoMYB33 in lily anthers after GA treatment. CK was the control group, and GA was the experimental group; the expression of LoMYB33 was determined at 0, 4, 8, 12, 24, and 48 h after GA treatment. Data are mean ± SD of three independent experiments. Different letters indicate significant differences among these lines (Student–Newman–Keuls test, P < 0.05).
To explore the function of LoMYB33, LoMYB33-overexpressing Arabidopsis plants were generated, and three independent overexpression (OE) lines were selected by RT-PCR (Supplementary Figure 5). RT-qPCR was performed on the inflorescence of wild-type plants and three LoMYB33-OE lines, and high expression levels of LoMYB33 were observed in the three overexpression lines. The expression levels of some pollen development-related genes, AtMYB33, AtMYB65, AtCYP703A2, AtCYP704B1, and AtACOS5, were significantly increased (Supplementary Figure 6). In addition, the normal growth of LoMYB33-OE plants was inhibited compared with wild-type plants, as these plants were shorter and later flowering (Figures 6A,B). The siliques of transgenic plants were significantly shorter compared with wild-type plants (Figures 6C,D), suggesting that the fertility of transgenic plants was reduced. Alexander staining revealed that the amount of pollen was lower in the three transgenic lines than in wild-type plants (Figure 6E). Transgenic plants contained less pollen in each anther compared with wild-type plants (Figure 6F), which might explain the reduction in fertility of transgenic plants.
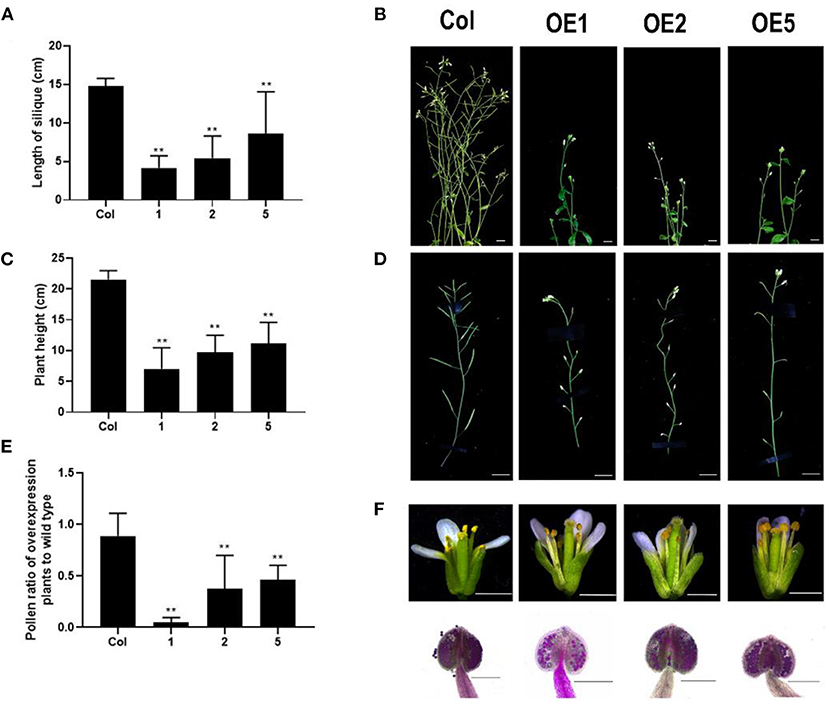
Figure 6. Morphological differences between LoMYB33-overexpressing and wild-type Arabidopsis plants. (A) Statistics of plant height of Col-0 and three overexpression lines (OE1, OE2, and OE5); (B) The main inflorescence of Col-0, OE1, OE2, and OE5; (C) Statistics of pod length of Col-0 and three overexpression lines. (D) Comparison of silique growth of Col-0, OE1, OE2, and OE5; (E) The relative ratio of the amount of pollen of three LoMYB33-OE lines and Col-0; (F) flowers at stage 13 of Col-0, OE1, OE2, and OE5. Bars are 1 mm. The anthers of Col-0, OE1, OE2, and OE5 stained by Alexander stain; the bars are 100 μM. At least three independent replicates were used for phenotypic identification; a representative result is presented. Data are mean ± SD of the tested plants. Data are mean ± SD of three independent experiments (t-test, **P < 0.01).
To further understand the effect of LoMYB33 overexpression on anther development, the anther development of wild-type and LoMYB33-OE Arabidopsis plants was observed in paraffin sections. The anther development of Arabidopsis was observed from stage 5 to 14 (Figure 7). When microspore mother cells underwent meiosis in four chambers and produced tetrads of haploid microspores in stage 7, the anther development of transgenic plants was abnormal; there was no tapetum in the anther compartment, and the middle layer cells did not degrade; and the development of the tapetum in OE-2 was delayed (Figures 7A–C,E–G). From stage 8 to 14, the pollen number of transgenic plants was lower than that of wild-type plants (Figures 7D,H,I–P). These results indicated that the overaccumulation of LoMYB33 might damage the normal development of pollen and anthers, suggesting that LoMYB33 might play a key role in pollen development; thus, an appropriate level of LoMYB33 expression is essential for normal development.
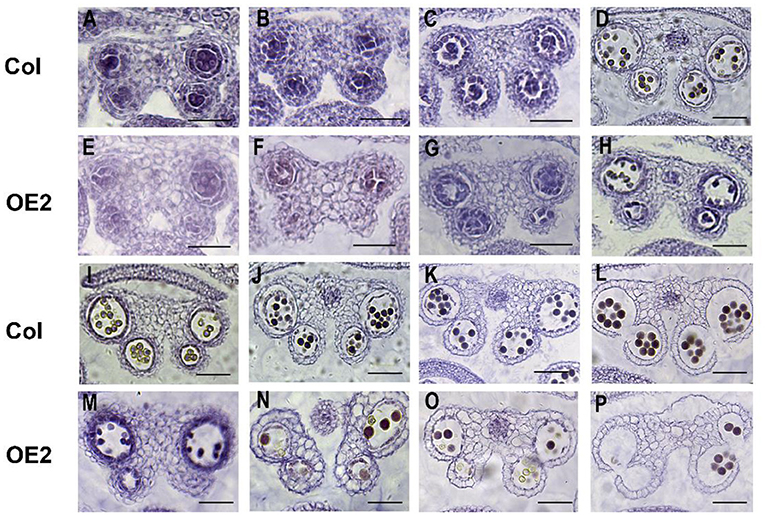
Figure 7. Cytological observations of different developmental stages of wild-type and LoMYB33-overexpressing Arabidopsis anthers. (A–D) Paraffin sections of anthers at stage 5–8 of wild-type plants. (E–H) Paraffin sections of anthers at stage 5–8 of OE2. (I–L) Paraffin sections of anthers at stages 9–14 of wild-type plants. (M–P) Paraffin sections of anthers at stage 9–14 of OE2. Three independent experiments were performed, and one representative picture is shown. Bars are 100 μm for stage 5–7. Bars are 50 μm for stage 8–14.
To determine whether LoMYB33 is involved in the development and formation of pollen in lily, VIGS of LoMYB33 was carried out in lily “White heaven” because its growth and development cycle is much shorter compared with other cultivars. Specific primers in the non-conservated region of LoMYB33 were designed to construct the TRV2-LoMYB33 silencing vector. RT-PCR showed that the TRV vectors had successfully spread to the anthers of TRV2-LoMYB33 lily lines (Supplementary Figure 7), and LoMYB33 expression in anthers was significantly decreased after silencing LoMYB33 (Figure 8A). The shape of lily flowers in TRV-control and LoMYB33-silenced lines did not differ, but the anthers became more withered in the LoMYB33-silenced lines. The amount of pollen was significantly reduced in these LoMYB33-silenced anthers compared with the TRV2-control (Figures 8B,C). These findings indicated that LoMYB33 might play an important role in anther development and pollen formation in lily.
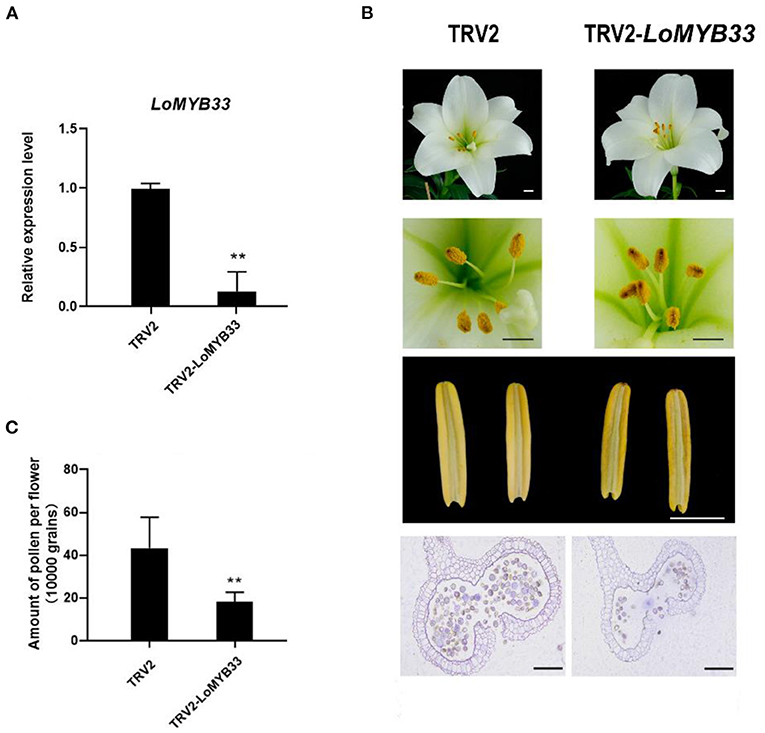
Figure 8. Silencing of LoMYB33 in lily anthers by VIGS. (A) LoMYB33 expression was determined in LoMYB33-silenced and TRV-control plants by RT-qPCR. Data are mean ± SD of three independent experiments (t-test, **P < 0.01). (B) Morphologies of flowers and anthers in TRV2 and TRV2-LoMYB33 lilies. Three independent experiments were performed, and one representative picture is shown. (C) Differences in the amount of pollen between LoMYB33-silenced and TRV-control plants were analyzed. Bars are 1 cm and 100 μm. Data are mean ± SD of three independent experiments (t-test, **P < 0.01).
Some R2R3-MYB members encoded by GAMYB genes have been shown to act as transcriptional activators of the GA signal pathway and play various roles in plants. In Arabidopsis, GAMYBs include three members: AtMYB33, AtMYB65, and AtMYB101. AtMYB33 and AtMYB65 are co-expressed in many tissues and have redundant functions. The myb33/myb65 double mutant shows defective anther development (Millar and Gubler, 2005). AtMYB101 expression is restricted to the subapical pith cells of both vegetative and flowering plants and to the hypocotyl hook and may be involved in GA-regulated stem elongation (Gocal et al., 2001). LtGAMYB plays an important signaling role in the flowering of ryegrass (Gocal et al., 1999). TaGAMYB expression is related to the length of wheat internodes (Chen et al., 2001). In germinated rice seeds, OsGAMYB induced the biological expression of α-amylase genes in the aleurone layer (Sutoh and Yamauchi, 2003). In tobacco, miR159 inhibition increases the expression of NtGAMYB, up-regulating disease resistance genes and enhancing resistance to Phytophthora (Zheng et al., 2020). In this study, we cloned and identified LoMYB33 from lily and showed that it may play a role in the development of anthers and pollen. Protein sequence analysis showed that LoMYB33 belonged to the R2R3-MYB family and that it contained three typical motifs of the GAMYB family (Figure 1) (Dubos et al., 2010). Phylogenetic analysis with Arabidopsis MYB members revealed that LoMYB33 is closely related to AtMYB33 and AtMYB65 (Supplementary Figure 1), both of which are GAMYB members. AtMYB33 and AtMYB65 have redundant functions in regulating anther and pollen development in Arabidopsis (Millar and Gubler, 2005). In rice, the homolog OsGAMYB also regulates the development of anthers (Aya et al., 2009). Based on the sequence alignment and phylogenetic analysis, we speculate that LoMYB33 is a GAMYB member in lily and that it may also play a role in anther development. According to previous studies, many species possess more than one member in the GAMYB family, such as Arabidopsis, tobacco, tomato, and rice, and these GAMYB members show redundant and special functions in different physiological processes (Gocal et al., 2001; Millar and Gubler, 2005; Tsuji et al., 2006; Gong and Bewley, 2008; da Silva et al., 2017; Zheng et al., 2020). In this study, LoMYB33 exhibited an expression pattern that was inconsistent with many reported GAMYBs of model plants (Figure 2), which suggested that the members of GAMYB family might be functionally differentiated in lily and that there might be other GAMYB members in lily.
Transcription activation activity analysis revealed that LoMYB33 had transactivation activity, the transactivation domain of R2R3-MYBs is generally distributed at the C-terminal (Zhao et al., 2017); as in previous studies, the C-terminal contributed to the transactivation ability of LoMYB33 (Figure 4).
Subcellular localization analysis of LoMYB33 was carried out, but no GFP fluorescence signal was observed when the wild-type ORF of LoMYB33 was used to construct the recombinant plasmid. Based on studies in model plants, GAMYBs are regulated by miRNA159 (Palatnik et al., 2007), miRNA159 is regulated by GA and regulates anther development by cutting GAMYB mRNA, and the target sites are conserved in different species (Achard et al., 2004; Csukasi et al., 2012; Zheng et al., 2020). Therefore, we mutated the predicted target sites of miRNA159 in LoMYB33 for subcellular localization analysis; as expected, GFP fluorescence was observed and located in the nucleus and cytoplasm (Figure 4), which suggests that LoMYB33 might also be regulated by the conserved miRNA159-GAMYB pathway in lily as in other plants.
After GA treatment of lily, the expression of LoMYB33 in anthers increased significantly (Figure 5), which indicated that GA could activate the expression of LoMYB33 in anthers. In barley, the expression of HvGAMYB was also activated by GA treatment (Gubler et al., 1995, 2002). Exogenous GA treatment resulted in the increased expression of AtMYB33 in Arabidopsis (Achard et al., 2004). Our results showed that LoMYB33 was activated by GA and thus that LoMYB33 might be a GA signal transduction factor similar to AtMYB33. However, additional research is needed to determine how LoMYB33 responds to GA signals.
According to the results of the RT-qPCR analysis in this study, LoMYB33 was the most highly expressed in pollen, followed by the anthers (Figure 2). LoMYB33 was highly expressed in the late anther development stages compared with the early anther development stages, which suggested that LoMYB33 might function in the late anther developmental stages. Contrary to expectation, LoMYB33 accumulated in mature pollen, especially in pollen collected from the anthers of 11-cm flower buds (Figure 2). LoMYB33 was also significantly higher in immature pollen at the tetrad stage than in other early stages of pollen development, although there was no difference in anthers from the same stage (Figure 2). High activity of the LoMYB33 promoter was detected in anthers at stages 12 to 13 of transgenic Arabidopsis plants, especially in mature pollen (Figure 3), which was consistent with the pattern of expression of LoMYB33 in lily anthers. In rice, the expression level of OsGAMYB was lowest in the mature pollen stage, and highest in the tetrad and mononuclear microspore stages (Aya et al., 2009). HvGAMYB in barley and both AtMYB33 and AtMYB65 in Arabidopsis are all weakly expressed in mature pollen grains (Murray et al., 2003; Kaneko et al., 2004; Millar and Gubler, 2005; Aya et al., 2009). The expression of LoMYB33 is inconsistent with the expression patterns of these genes. In cucumber, CsGAMYB1 is most highly expressed during the critical period when the stamen primordium and carpel primordium are initiated; it is also highly expressed in the mature pollen grains of male cucumber flowers (Zhang et al., 2014). These results indicate that the expression of LoMYB33 and its homologs may involve different conserved regulatory mechanisms. The function of LoMYB33 in the tetrad stage may be similar to that of OsGAMYB, which regulates the development of anthers by controlling the development of the tapetum (Figure 7) (Kaneko et al., 2004). In addition, the high expression of LoMYB33 in later stages may affect pollen maturation and germination. The presence of a certain amount of GA in pollen is required for pollen germination (Meeuse et al., 1976). GA also affects the biosynthesis of flavanols, which are important components in mature pollen (Koornneef and Veen, 1980; Cheng et al., 2009). LoMYB33 may participate in the GA signaling pathway in anthers and pollen and affect the germination and maturation of pollen.
In Arabidopsis, AtMYB33 is strongly expressed in the developing young anther chambers and weakly expressed in the pollen grains (Gocal et al., 2001; Millar and Gubler, 2005). The AtMYB33 promoter activity is strong in tapetum cells but weak in other anther wall layers and microspores (Aya et al., 2009). These results indicate that the activation pattern of LoMYB33 promoter in lily differed from that in Arabidopsis and rice, which suggests that LoMYB33 might be involved in pollen maturation and exine formation.
Overexpression of LoMYB33 in Arabidopsis significantly inhibits plant growth, results in late flowering, and decreases fertility (Figure 6). GAMYB homologs have been reported to regulate flowering time in Arabidopsis, ryegrass, barley, and rice (Blazquez et al., 1998; Blazquez and Weigel, 1999; Gocal et al., 1999; Murray et al., 2003; Kaneko et al., 2004). GAMYB is a promoter of plant flowering induction (Gocal et al., 1999), but SlMYB33 overexpression in tomato delays flowering (Zhang et al., 2020). Transgenic Arabidopsis plants with LoMYB33 are short and have short internodes; GmGAMYB overexpression accelerates flowering in soybean and increases plant height (Yang et al., 2021). In Arabidopsis, overexpression of resistant types of AtMYB33 and AtMYB65 with mutated miRNA159 target sites results in short plants and male sterility. However, no change in plant growth was noted in transgenic Arabidopsis plants overexpressing wild-type AtMYB33 or AtMYB65 because miRNA159 cleaves AtMYB33 and AtMYB65 in vegetative tissues (Palatnik et al., 2003; Li et al., 2016). Overexpression of wild-type LoMYB33 retarded plant growth (Figure 6). In poplar, no phenotypic changes were observed following overexpression of the miRNA159 target gene PtrMYB012; however, upward curling of the leaves, dwarfism, and male sterility were observed in PtrMYB012-transgenic Arabidopsis. Thus, PtrMYB012 may be completely degraded by miRNA159 in poplar but not in Arabidopsis (Kim et al., 2018). We speculated that LoMYB33 might not be completely degraded by Arabidopsis miRNA159 because of the species specificity of miRNA159.
In transgenic Arabidopsis plants, the expression of AtCYP703A2, AtCYP704B1, and AtACOS5 was significantly increased (Supplementary Figure 4), which suggested that these genes might be located downstream of LoMYB33 and that LoMYB33 activated their expression. In rice, OsGAMYB binds to the promoter of OsCYP703A3 through its MYB domain to activate its expression for the formation of the Ubisch body and the pollen outer wall (Aya et al., 2009). Acyl-CoA synthetase ACOS5 (a cytochrome P450 hydroxylase family member), CYP703A2, and CYP704B1 are all involved in pollen exine synthesis (Wang et al., 2018b), which suggests that LoMYB33 might play key roles in the maturation of pollen and the formation of pollen walls. In transgenic lines, the expression of AtMYB33 and AtMYB65 was also up-regulated, which might stem from their self-activating activity. The expression of AtMYB33, AtMYB65, and AtCYP704B1 is independent of LoMYB33 expression, and the expression pattern of AtCYP703A2 is opposite that of LoMYB33 (Wu et al., 2019; Ding et al., 2021). We speculate that this might be explained by a negative feedback regulation mechanism involving LoMYB33, wherein the high expression of LoMYB33 activates the feedback pathway to control the expression of these genes at a level appropriate for anther development. Our results indicate that LoMYB33 had a dose-regulating effect, as an appropriate expression level is required to ensure the normal development of the anthers (too high or too low expression leads to abnormal anther development).
When microspore mother cells undergo meiosis in four chambers and produce tetrads of haploid microspores in the 7th stage of anther development, anther development in LoMYB33-overexpressing plants was abnormal because the tapetum in the anther chamber and the cells in the middle layer were not degraded. In the tetrad stage of wild-type plants, the mesothelium cells had degraded, and the tapetum cells began to degenerate (Figure 7). From stage 8 to 14, the pollen number was lower in transgenic plants than in wild-type plants, and the morphology of some microspores or pollen was abnormal (Figure 7). The tapetum is the innermost layer of the anther wall, and it plays an important role in the development of microspores and pollen (Stevens and Murray, 1981; DeGuzman and Riggs, 2000; Taylor et al., 2010). In male sterile lines, pollen abortion is tied to tapetum abnormalities. The timely control of tapetal programmed cell death is essential for pollen maturation (Uzair et al., 2019). In Arabidopsis myb33/myb65 double mutant anthers, the tapetum experiences hypertrophy at the pollen mother cell stage, which leads to the termination of pollen development before meiosis (Millar and Gubler, 2005). In rice, the abnormal enlargement of tapetum cells and microspore collapse occur in gamyb-2 mutants at the mature pollen stage, which stemmed from the inability of tapetum cells to undergo programmed cell death (Aya et al., 2009). LoMYB33 overexpression may lead to male sterility by disturbing normal tapetal development. OsGAMYB is a major transcriptional regulator of meiosis and early tapetum and pollen development in rice (Ko et al., 2021). The tapetum of gamyb-4 and gamyb-5 mutants elongated abnormally during the late stage of anther development and occupied the entire anther chamber, which resulted in the male sterility of rice (Liu et al., 2010).
The flower shape of LoMYB33-silenced lines was not altered relative to wild-type plants, but the anthers became shriveled, and the amount of pollen significantly decreased (Figure 8), which is consistent with the findings of previous studies in Arabidopsis and rice. Mutations of AtMYB33 and AtMYB65 in Arabidopsis and GAMYB in rice cause male sterility because of a lack of pollen production (Millar and Gubler, 2005; Liu et al., 2010). LoMYB33 overexpression in Arabidopsis also leads to pollen reduction; over-accumulation of LoMYB33 resulted in more severe pollen reduction (Figure 6), indicating that both high and low levels of LoMYB33 lead to a serious decline in fertility. Transgenic barley plants overexpressing HvGAMYB may also cause male fertility, and the severity of the phenotype depends on the level of HvGAMYB accumulation (Murray et al., 2003). We speculate that the overexpression effects of LoMYB33 may also be dose-dependent. Genes related to pollen development, such as AtMYB24, show a similar pattern; overexpression of MYB24 cannot restore opr3 stamen development, and proper overexpression of MYB24 can restore stamen development and male fertility. The expression level of MYB24NT is related to male sterility (Huang et al., 2017). Overall, these findings suggest that LoMYB33 may be involved in both tapetum development and pollen formation. LoMYB33 may provide a useful candidate gene for the pollen-free breeding of lily.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: GenBank, MZ666106.
NT and ZW conceived and designed the experiments. XL, JF, GY, and LH performed the experiments under the supervision of NT. DZ provided technical help. XL and ZW analyzed the data and wrote the manuscript. All authors read and approved the final version of the manuscript.
This work was supported by a Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, the earmarked fund for germplasm resources of Nanjing Agricultural University (KYZZ2019020), the High Level Talent Project of the Top Six Talents in Jiangsu (Grant No. NY-077), and the Programs for New Century Excellent Talents in Universities, Ministry of Education of China (NCET-11-0669).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.730007/full#supplementary-material
Achard, P., Herr, A., Baulcombe, D. C., and Harberd, N. P. (2004). Modulation of floral development by a gibberellin-regulated microRNA. Development 131, 3357–3365. doi: 10.1242/dev.01206
Aizen, M., and Harder, L. (2007). Expanding the limits of the pollen-limitation concept: effects of pollen quantity and quality. Ecology 88, 271–281. doi: 10.1890/06-1017
Alexander, M. P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44, 117–122. doi: 10.3109/10520296909063335
Allen, R. S., Li, J., Stahle, M. I., Dubroue, A., Gubler, F., and Millar, A. A. (2007). Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proc. Natl. Acad. Sci. U.S.A. 104, 16371–16376. doi: 10.1073/pnas.0707653104
Aya, K., Ueguchi-Tanaka, M., Kondo, M., Hamada, K., Yano, K., Nishimura, M., et al. (2009). Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell 21, 1453–1472. doi: 10.1105/tpc.108.062935
Blazquez, M. A., Green, R., Nilsson, O., Sussman, M. R., and Weigel, D. (1998). Gibberellins promote flowering of Arabidopsis by activating the LEAFY promoter. Plant Cell 10, 791–800. doi: 10.1105/tpc.10.5.791
Blazquez, M. A., and Weigel, D. (1999). Independent regulation of flowering by phytochrome b and gibberellins in Arabidopsis. Plant Physiol. 120, 1025–1032. doi: 10.1104/pp.120.4.1025
Chen, C., Hussain, N., Wang, Y., Li, M., Liu, L., Qin, M., et al. (2020). An Ethylene-inhibited NF-YC transcription factor RhNF-YC9 regulates petal expansion in rose. Hortic. Plant J. 6, 419–427. doi: 10.1016/j.hpj.2020.11.007
Chen, L., Nishizawa, T., Higashitani, A., Suge, H., Wakui, Y., Takeda, K., et al. (2001). A variety of wheat tolerant to deep-seeding conditions: elongation of the first internode. Plant Cell Environ. 24, 469–476. doi: 10.1046/j.1365-3040.2001.00688.x
Cheng, H., Qin, L., Lee, S., Fu, X., Richards, D. E., Luo, D., et al. (2004). Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131, 1055–1064. doi: 10.1242/dev.00992
Cheng, H., Song, S., Xiao, L., Soo, H. M., Cheng, Z., Xie, D., et al. (2009). Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS. Genet. 5:e1000440. doi: 10.1371/journal.pgen.1000440
Chu, Z. (2006). Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 20, 1250–1255. doi: 10.1101/gad.1416306
Clough, S., and Bent, A. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Csukasi, F., Donaire, L., Casanal, A., Martinez-Priego, L., Botella, M. A., Medina-Escobar, N., et al. (2012). Two strawberry miR159 family members display developmental-specific expression patterns in the fruit receptacle and cooperatively regulate Fa-GAMYB. New Phytol. 195, 47–57. doi: 10.1111/j.1469-8137.2012.04134.x
da Silva, E. M., Silva, G. F. F. E., Bidoia, D. B., da Silva Azevedo, M., de Jesus, F. A., Pino, L. E., et al. (2017). microRNA159-targeted SlGAMYB transcription factors are required for fruit set in tomato. Plant J. 92, 95–109.doi: 10.1111/tpj.13637
DeGuzman, R., and Riggs, C. D. (2000). A survey of proteinases active during meiotic development. Planta 210, 921–924. doi: 10.1007/s004250050698
Dickinson, H. G., and Heslop-Harrison, J. (1968). Common mode of deposition for the sporopollenin of sexine and nexine. Nature 220:926. doi: 10.1038/220926a0
Ding, L., Wu, Z., Teng, R., Xu, S., Cao, X., Yuan, G., et al. (2021). LlWRKY39 is involved in thermotolerance by activating LlMBF1c and interacting with LlCaM3 in lily (Lilium longiflorum). Hortic. Res. 8:473. doi: 10.1038/s41438-021-00473-7
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Feng, J., Wu, Z., Wang, X., Zhang, Y., and Teng, N. (2019). Analysis of pollen allergens in lily by transcriptome and proteome data. Int. J. Mol. Sci. 20:5892. doi: 10.3390/ijms20235892
Gao, X., Zhang, Q., Zhao, Y. Q., Yang, J., He, H. B., and Jia, G. (2020). The lre-mir159a-LrGAMYB pathway mediates resistance to grey mould infection in Lilium regale. Mol. Plant Pathol. 21, 749–760. doi: 10.1111/mpp.12923
Gocal, G. F. W., Poole, A. T., Gubler, F., Watts, R. J., Blundell, C., King, R. W., et al. (1999). Long-day up-regulation of a GAMYB gene during Lolium temulentum inflorescence formation. Plant Physiol. 119, 1271–1278. doi: 10.1104/pp.119.4.1271
Gocal, G. F. W., Sheldon, C. C., Gubler, F., Moritz, T., Bagnall, D. J., MacMillan, C. P., et al. (2001). GAMYB-like Genes, Flowering, and Gibberellin Signaling in Arabidopsis. Plant Physiol. 127, 1682–1693. doi: 10.1104/pp.010442
Gong, B., Yi, J., Wu, J., Sui, J., Khan, M. A., Wu, Z., et al. (2014). LlHSFA1, a novel heat stress transcription factor in lily (Lilium longiflorum), can interact with LlHSFA2 and enhance the thermotolerance of transgenic Arabidopsis thaliana. Plant Cell Rep. 33, 1519–1533. doi: 10.1007/s00299-014-1635-2
Gong, X., and Bewley, J. D. (2008). A GAMYB-like gene in tomato and its expression during seed germination. Planta 228, 563–572. doi: 10.1007/s00425-008-0759-4
Gubler, F., Chandler, P. M., White, R. G., and Jacobsen, L. J. V. (2002). Gibberellin signaling in barley aleurone cells. control of sln1 and gamyb expression. Plant Physiol. 129, 191–200. doi: 10.2307/4280451
Gubler, F., Kalla, R., and Jacobsen, R. (1995). Gibberellin-regulated expression of a myb gene in barley aleurone cells: evidence for Myb transactivation of a high-pI alpha-amylase gene promoter. Plant Cell 7, 1879–1891. doi: 10.2307/3870195
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symposium Series 41, 95–98. doi: 10.1021/bk-1999-0734.ch008
Hedden, P. (2001). Gibberellin metabolism and its regulation. J. Plant Growth Regul. 20, 317–318. doi: 10.1007/s003440010039
Huang, H., Gao, H., Liu, B., Qi, T., Tong, J., Xiao, L., et al. (2017). Arabidopsis MYB24 regulates jasmonate-mediated stamen development. Front. Plant Sci. 8:1525. doi: 10.3389/fpls.2017.01525
Kaneko, M., Inukai, Y., Ueguchi-Tanaka, M., Itoh, H., Izawa, T., Kobayashi, Y., et al. (2004). Loss-of-function mutations of the rice GAMYB gene impair α-Amylase expression in aleurone and flower development. Plant Cell 16, 33–44. doi: 10.1105/tpc.017327
Kim, M., Cho, J., Lee, J., Bae, S., Choi, Y., Park, E., et al. (2018). Poplar MYB transcription factor PtrMYB012 and its Arabidopsis AtGAMYB orthologs are differentially repressed by the Arabidopsis miR159 family. Tree Physiol. 38, 801–812. doi: 10.1093/treephys/tpx164
Ko, S., Li, M., Ho, Y., Yu, C., Yang, T., Lin, Y., et al. (2021). Rice transcription factor GAMYB modulates bHLH142 and is homeostatically regulated by TDR during anther tapetal and pollen development. J. Exp. Bot. 72, 4888–4903. doi: 10.1093/jxb/erab190
Köhler, C., Hennig, L., Bouveret, R., Gheyselinck, J., Grossniklaus, U., and Gruissem, W. (2003). Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J. 22, 4804–4814. doi: 10.1093/emboj/cdg444
Koornneef, M., and Veen, J. (1980). Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) heynh. Theor. Appl. Genet. 58, 257–263. doi: 10.1007/BF00265176
Kranz, H., Denekamp, M., Greco, R., Jin, H., Leyva, A., Meissner, R., et al. (1998). Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 16, 263–276. doi: 10.1046/j.1365-313x.1998.00278.x
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kwon, C., Kim, S., Kim, D., and Paek, N. (2015). The rice floral repressor early flowering1 affects spikelet fertility by modulating gibberellin signaling. Rice 8:58. doi: 10.1186/s12284-015-0058-1
Li, Q., Wu, Z., Wu, H., Fang, W., Chen, F., and Teng, N. (2019). Transcriptome profiling unravels a vital role of pectin and pectinase in anther dehiscence in chrysanthemum. Int. J. Mol. Sci. 20:5865. doi: 10.3390/ijms20235865
Li, Y., Alonso-Peral, M., Wong, G., Wang, M., and Millar, A. A. (2016). Ubiquitous miR159 repression of MYB33/65 in Arabidopsis rosettes is robust and is not perturbed by a wide range of stresses. BMC Plant Biol. 16:867. doi: 10.1186/s12870-016-0867-4
Liu, Y. G., and Chen, Y. (2007). High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 43:649. doi: 10.2144/000112601
Liu, Z., Bao, W., Liang, W., Yin, J., and Zhang, D. (2010). Identification of gamyb-4 and analysis of the regulatory role of GAMYB in rice anther development. J. Integr. Plant Bio. 2, 670–678. doi: 10.1111/j.1744-7909.2010.00959.x
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using Real-Time quantitative PCR and the 2(-Delta Delta C(T)). Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Long, Y., Zhao, L., Niu, B., Su, J., Wu, H., Chen, Y., et al. (2008). Hybrid male sterility in rice controlled by interaction between divergent alleles of two adjacent genes. Proc. Natl. Acad. Sci. U.S.A. 105, 18871–18876. doi: 10.1073/pnas.0810108105
Meeuse, B., Stanley, R. G., and Linskens, H. F. (1976). Pollen: biology, biochemistry, management. Bioscience 25:7073. doi: 10.2307/1297073
Millar, A. A., and Gubler, F. (2005). The arabidopsis GAMYB-like genes, MYB33 and MYB65, are MicroRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17, 705–721. doi: 10.1105/tpc.104.027920
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol. Plants 15, 472–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Murray, F., Kalla, R., Jacobsen, J., and Gubler, F. (2003). A role for HvGAMYB in anther development. Plant J. 33, 481–491. doi: 10.1046/j.1365-313x.2003.01641.x
Palatnik, J. F., Allen, E., Wu, X., Schommer, C., Schwab, R., Weigel, D., et al. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. doi: 10.1038/nature01958
Palatnik, J. F., Wollmann, H., Schommer, C., and Weigel, D. (2007). Sequence and expression differences underlie functional specialization of arabidopsis MicroRNAs miR159 and miR319. Deve. Cell 13, 115–125, doi: 10.1016/j.devcel.2007.04.012
Rhoades, M. W., Reinhart, B. J., Lim, L. P., Burge, C. B., Bartel, B., and Bartel, D. (2002). Prediction of plant MicroRNA targets. Cell 110, 513–520. doi: 10.1016/S0092-8674(02)00863-2
Romero, I., Fuertes, A., Benito, M.J., Malpica, J.M., and Paz-Ares, J. (1998). More than 80R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J. 14, 273–284. doi: 10.1046/j.1365-313X.1998.00113.x
Sanders, P. M., Bui, A. Q., Weterings, K., McIntire, K. N., Hsu, Y., Lee, P., et al. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sexual Plant Reprod. 11, 297–322. doi: 10.1007/s004970050158
Stevens, V. A., and Murray, B. G. (1981). Studies on heteromorphic self-incompatibility systems: the cytochemistry and ultrastructure of the tapetum of Primula obconica. J. Cell Sci. 50, 419–431. doi: 10.1083/jcb.88.3.660
Sui, J., He, J., Wu, J., Gong, B., Cao, X., Seng, S., et al. (2015). Characterization and functional analysis of transcription factor LoMYB80 related to anther development in lily (Lilium Oriental Hybrids). J. Plant Growth Regul. 34, 545–557. doi: 10.1007/s00344-015-9489-6
Sun, C., and Pan, Y. (2008). Determination of pollen viability of Arabidopsis thaliana and its application in pollen development. Botanical Bull. 22, 268–275. doi: 10.3969/j.issn.1674-3466.2008.03.002
Sutoh, K., and Yamauchi, D. (2003). Two cis-acting elements necessary and sufficient for gibberellin-upregulated proteinase expression in rice seeds. Plant J. 34, 635–645. doi: 10.1046/j.1365-313x.2003.01753.x
Taylor, A. A., Horsch, A., Rzepczyk, A., Hasenkampf, C. A., and Riggs, C. D. (2010). Maturation and secretion of a serine proteinase is associated with events of late microsporogenesis. Plant J. 12, 1261–1271. doi: 10.1046/j.1365-313x.1997.12061261.x
Teng, R., Wu, Z., Xu, S., Hou, H., Chen, F., and Teng, N. (2021). A novel lateral organ boundary-domain factor CmLBD2 positively regulates pollen development via activating CmACOS5 in Chrysanthemum morifolium. Plant Cell Physiol. 124:pcab124. doi: 10.1093/pcp/pcab124
Thompson, J. D., Gibson, T. J., and Higgins, D. G. (2003). Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics. 2.3.1–2.3.22. doi: 10.1002/0471250953.bi0203s00
Tong, Z., Li, Q., Yang, Y., Dai, F., Gao, J., Hong, B., et al. (2013). Isolation and expression analysis of LoPIPP2, a lily (Lilium Oriental Hybrids) aquaporin gene involved in desiccation-induced anther dehiscence. Sci. Hortic. 164, 316–322. doi: 10.1016/j.scienta.2013.09.022
Tsuji, H., Aya, K., Ueguchi-Tanaka, M., Shimada, Y., Nakazono, M., Watanabe, R., et al. (2006). GAMYB controls different sets of genes and is differentially regulated by microRNA in aleurone cells and anthers. Plant J. 47, 427–444. doi: 10.1111/j.1365-313X.2006.02795.x
Uzair, M., Xu, D., Schreiber, L., Shi, J., Liang, W., Zhang, D., et al. (2019). PERSISTENT TAPETAL CELL2 is required for normal tapetal programmed cell death and pollen wall patterning. Plant Physiol. 182:688. doi: 10.1104/pp.19.00688
Wang, J. M., Ma, S. L. Y., Li, W. Q., Wang, Q., Cao, H. Y., Gu, J. H., et al. (2016). Genetic variability and diversity of the main resources of lily assessed via phenotypic characters, pollen morphology, and ISSR markers. Genet. Mol. Res. 15:7638. doi: 10.4238/gmr.15027638
Wang, K., Guo, Z., Zhou, W., Zhang, C., Zhang, Z., Yue, L., et al. (2018b). The regulation of sporopollenin biosynthesis genes for rapid pollen wall formation. Plant Physiol. 178, 283–294. doi: 10.1104/pp.18.00219
Wang, L., Wu, M., Wang, X., Fang, W., Chen, F., and Teng, N. (2018a). Determination of pollen content and analysis of loose pollen characteristics of 95 lily germplasm resources. J. Nanjing Agric. University 41, 1018–1028.
Wang, X., Wu, Z., Wang, L., Wu, M., Zhang, D., Fang, W., et al. (2019). Cytological and molecular characteristics of pollen abortion in lily with dysplastic tapetum. Hortic. Plant J. 5, 281–294. doi: 10.1016/j.hpj.2019.11.002
Woodger, F. J., Millar, A., Murray, F., Jacobsen, J. V., and Gubler, F. (2003). The Role of GAMYB transcription factors in GA-regulated gene expression. J. Plant Growth Regul. 22, 176–184. doi: 10.1007/s00344-003-0025-8
Wu, Z., Liang, J., Wang, C., Ding, L., Zhao, X., Cao, X., et al. (2019). Alternative splicing provides a mechanism to regulate LlHSFA3 function in response to heat stress in lily. Plant Physiol. 181, 1651–1667. doi: 10.1104/pp.19.00839
Yang, X., Li, X., Shan, J., Li, Y., and Zhang, Y. (2021). Overexpression of GmGAMYB accelerates the transition to flowering and increases plant height in soybean. Front. Plant Sci. 12:e667242. doi: 10.3389/fpls.2021.667242
Yuan, G., Wu, Z., Liu, X., Li, T., and Teng, N. (2021). Characterization and functional analysis of LoUDT1, a bHLH transcription factor related to anther development in the lily oriental hybrid Siberia (Lilium spp.). Plant Physiol. Bioch. 166, 1087–1095. doi: 10.1016/j.plaphy.2021.07.018
Zhang, Y., Zhang, B., Yang, T., Zhang, J., Liu, B., Zhan, X., et al. (2020). The GAMYB-like gene SlMYB33 mediates flowering and pollen development in tomato. Hortic. Res. 7:133. doi: 10.1038/s41438-020-00366-1
Zhang, Y., Zhang, X., Liu, B., Wang, W., Liu, X., Chen, C., et al. (2014). A GAMYB homologue CsGAMYB1 regulates sex expression of cucumber via an ethylene-independent pathway. J. Exp. Bot. 65, 3201–3213. doi: 10.1093/jxb/eru176
Zhao, Y., Tian, X., Wang, F., Zhang, L., Xin, M., Hu, Z., et al. (2017). Characterization of wheat MYB genes responsive to high temperatures. BMC Plant Biol. 17:1158. doi: 10.1186/s12870-017-1158-4
Keywords: lily, R2R3-MYB, LoMYB33, pollen, anther development
Citation: Liu X, Wu Z, Feng J, Yuan G, He L, Zhang D and Teng N (2021) A Novel R2R3-MYB Gene LoMYB33 From Lily Is Specifically Expressed in Anthers and Plays a Role in Pollen Development. Front. Plant Sci. 12:730007. doi: 10.3389/fpls.2021.730007
Received: 24 June 2021; Accepted: 27 August 2021;
Published: 23 September 2021.
Edited by:
Jian Wu, China Agricultural University, ChinaReviewed by:
Boas Pucker, University of Cambridge, United KingdomCopyright © 2021 Liu, Wu, Feng, Yuan, He, Zhang and Teng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nianjun Teng, bmp0ZW5nQG5qYXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.