
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 17 September 2021
Sec. Plant Development and EvoDevo
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.726762
This article is part of the Research Topic Epigenetics in Plant Development View all 20 articles
Small RNAs (sRNAs) are a class of non-coding RNAs that consist of 21–24 nucleotides. They have been extensively investigated as critical regulators in a variety of biological processes in plants. sRNAs include two major classes: microRNAs (miRNAs) and small interfering RNAs (siRNAs), which differ in their biogenesis and functional pathways. Due to global warming, high-temperature stress has become one of the primary causes for crop loss worldwide. Recent studies have shown that sRNAs are involved in heat stress responses in plants and play essential roles in high-temperature acclimation. Genome-wide studies for heat-responsive sRNAs have been conducted in many plant species using high-throughput sequencing. The roles for these sRNAs in heat stress response were also unraveled subsequently in model plants and crops. Exploring how sRNAs regulate gene expression and their regulatory mechanisms will broaden our understanding of sRNAs in thermal stress responses of plant. Here, we highlight the roles of currently known miRNAs and siRNAs in heat stress responses and acclimation of plants. We also discuss the regulatory mechanisms of sRNAs and their targets that are responsive to heat stress, which will provide powerful molecular biological resources for engineering crops with improved thermotolerance.
As sessile organisms, plants are constantly exposed to a wide range of biotic and abiotic stresses that are unfavorable for their growth and development. Abiotic stresses, such as drought, salt, temperature, and heavy metals seriously impact the productivity of plants (Zhao et al., 2016; Zhu, 2016). Due to global warming, high-temperature stress has become one of the primary causes for crop loss (Liu et al., 2017a). Global yields of maize and wheat declined by 3.8 and 5.5%, respectively, due to temperature increases of approximately 0.13°C per decade since 1980 (Lobell et al., 2011). Consequently, the mechanisms for heat stress responses in plants have become a global concern and have received much attention.
Plants have evolved complex and diverse mechanisms to defend against ambient high-temperature stress and various factors are involved in plant thermotolerance, such as heat shock proteins (HSPs), reactive oxygen species (ROS)-scavenging enzymes, heat shock transcription factor (HSFs), and small RNAs (sRNAs; Ohama et al., 2017; Zhao et al., 2021). sRNAs are a class of non-coding RNAs that consist of 21–24 nucleotides (nt) and are critical regulators of gene expression by causing either transcriptional gene silencing (TGS) or post-transcriptional gene silencing (PTGS; Axtell, 2013; D’Ario et al., 2017; Yu et al., 2019). Recently, sRNAs have been reported to participate in heat stress responses and play important roles in plant thermotolerance (Ruiz-Ferrer and Voinnet, 2009; Khraiwesh et al., 2012; Shriram et al., 2016; Liu et al., 2017a; Pagano et al., 2021). In this review, we focus on the roles and the regulatory mechanisms of sRNAs, mainly microRNAs (miRNAs) and small interfering RNAs (siRNAs) underlying heat stress tolerance in plants.
Endogenous sRNAs in plants are classified into two major types based on the tertiary subdivision by Axtell (2013): hairpin RNAs (hpRNAs) and siRNAs. Both hpRNAs and siRNAs result from cleavage of a double-stranded duplex from the helical region of larger RNA precursors by Dicer-like (DCL) enzyme (Axtell, 2013). hpRNAs are derived from a single-stranded RNA (ssRNA) precursor with a stem-loop hairpin structure, whereas siRNAs are derived from a double-stranded RNA (dsRNA) precursors. miRNAs are a well-studied subset of hpRNAs. siRNAs can be divided into two major subgroups including heterochromatic siRNAs (hc-siRNAs) and phased siRNAs (phasiRNAs; Axtell, 2013; Yu et al., 2019). Furthermore, trans-acting siRNAs (tasiRNAs) are a particular class of phasiRNAs that silence targets in trans. All these sRNAs differ in their biogenesis and modes of action.
The biogenesis and processing of miRNAs occur in multiple steps in plants (Figure 1A). (i) Similar to protein-coding genes, miRNA-encoded MIR genes are transcribed by RNA polymerase II (Pol II) to generate a long single-stranded primary miRNA (pri-miRNA), which is capped and polyadenylated in its 5' and 3' terminal regions, respectively (Lee et al., 2004; Xie et al., 2005). The pri-miRNA is predicted to form a stem-loop or hairpin secondary structure and the imperfectly paired double-stranded stem region contains the miRNA and miRNA* (Meyers et al., 2010). (ii) The pri-miRNA is first cleaved into a stem-loop miRNA precursor miRNA (pre-miRNA) and then the pre-miRNA is processed into miRNA-miRNA* duplex (Schauer et al., 2002; Kurihara and Watanabe, 2004). In this complex process, the RNase III enzyme DCL1 forms a nuclear dicing bodies (D-bodies) in the nucleus with two other partner proteins, HYPONASTIC LEAVES1 (HYL1) and SERRATE (SE), which ensure accurate and efficient splicing of pre-miRNA, resulting in the base of the stem being sliced (Yu et al., 2020). Other cofactors are involved for proper processing of pri-miRNA, such as the Cap-binding complex (CBC; Laubinger et al., 2008) and the Forkhead-associated (FHA) domain-containing protein Dawdle (DDL; Yu et al., 2008). The pre-miRNA without the base is then processed into a miRNA-miRNA* duplex by DCL1, which removes the hairpin loop. (iii) The miRNA-miRNA* duplex is further methylated by methyltransferase HUA ENHANCER1 (HEN1) to protect the 3' ends from uridylation (Li et al., 2005; Yu et al., 2005). (iv) In most cases, the methylated guide strand (miRNA) is incorporated into Argonaute-1 (AGO1) with the aid of TRANSPORIN 1 (TRN1); while the passenger strand (miRNA*) of the duplex is degraded. The AGO1-miRNA complex is exported from the nucleus to the cytoplasm via HASTY (HST; Bologna et al., 2018; Yu et al., 2019). (v) The mature miRNA associated with the miRNA-induced silencing complex (miRISC) is guided to target mRNAs for mRNA cleavage by AGO1 (transcript cleavage) or via inhibition of protein synthesis (translational repression; Llave et al., 2002; Iki et al., 2010; Li et al., 2013). In addition, the exonucleases SMALL RNA DEGRADING NUCLEASE1 (SDN1), nucleotidyl transferase HEN1 SUPPRESSOR1 (HESO1), and UTP: RNA URIDYLYLTRANSFERASE 1 (URT1) play critical roles in the process of miRNA turnover to regulate its steady-state level (Ramachandran and Chen, 2008; Zhao et al., 2012; Tu et al., 2015).
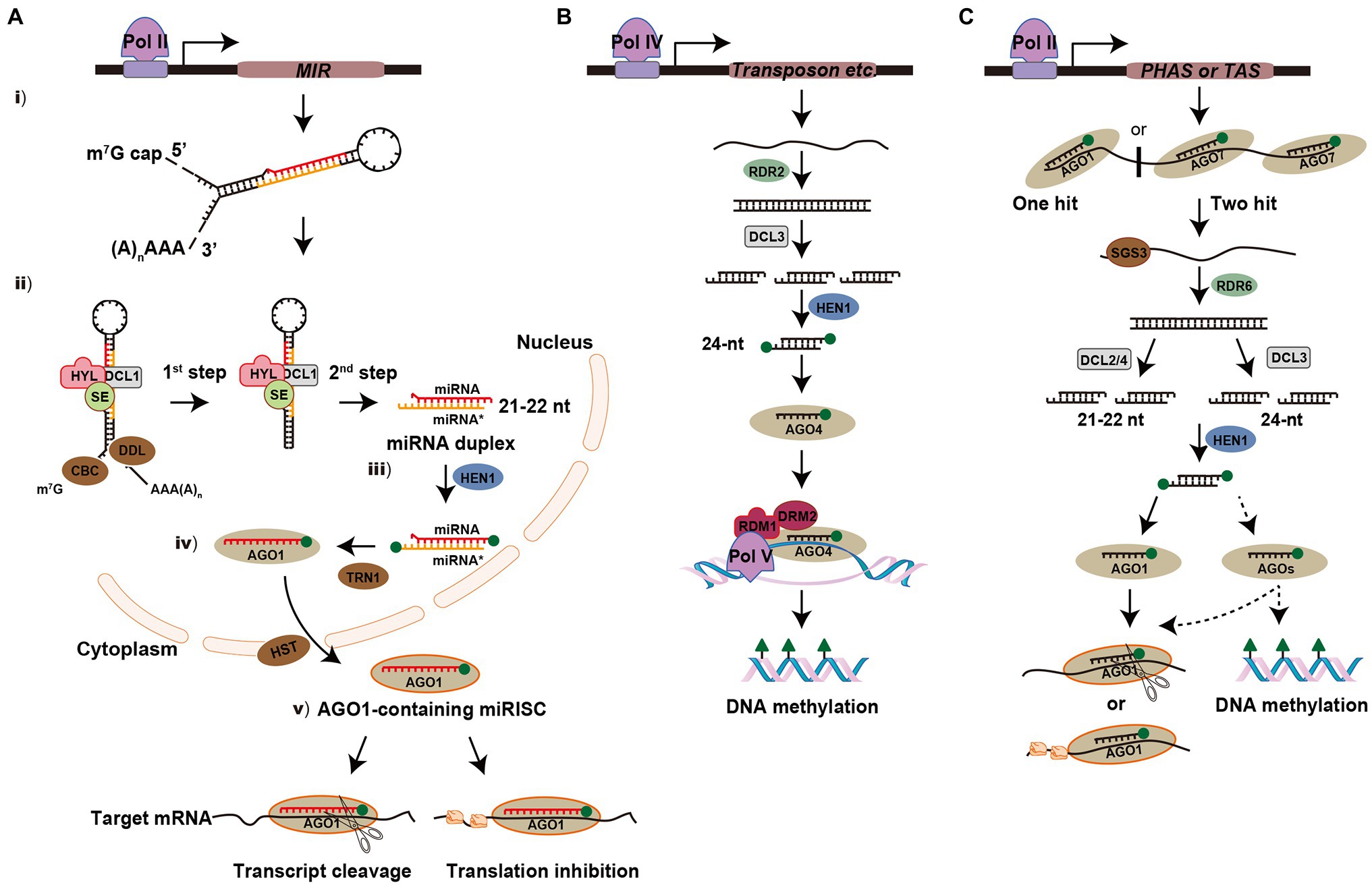
Figure 1. Biogenesis and modes of action of microRNAs (miRNA) and small interfering RNAs (siRNA) in plants. (A) Overview of miRNA pathway. MIR gene is transcribed by polymerase II (Pol II) to generate a long single-stranded primary miRNA (pri-miRNA) and form a stem-loop or hairpin secondary structure. The pri-miRNA is firstly cleaved into stem-loop precursor miRNA (pre-miRNA) and then processed into miRNA-miRNA* duplex. In this complex process, DCL1 forms a D-bodies with HYPONASTIC LEAVES1 (HYL1) and SE to ensure the accurate and efficient splicing of pre-mRNA. Other cofactors are involved for proper processing, such as cap-binding complex (CBC) and Dawdle (DDL). The miRNA-miRNA* duplex is then methylated by HUA ENHANCER1 (HEN1) to protect the 3' ends from uridylation. The guide strand (miRNA) is incorporated into Argonaute-1 (AGO1) with the aid of TRANSPORIN 1 (TRN1) and then exported from the nucleus to the cytoplasm via HASTY (HST). In the cytoplasm, mature miRNA directs post-transcriptional gene silencing (PTGS) via transcript cleavage or translational repression. The green balls at the ends of miRNA-miRNA* duplex represent 2'-O-methy groups. (B) Overview of the heterochromatic siRNA (hc-siRNA) pathway. hc-siRNAs are derived from transposable and repetitive elements. The biogenesis of hc-siRNA requires Polymerase IV (Pol IV) to transcribe a single-stranded RNA (ssRNA) and synthesis into a double-stranded RNA (dsRNA) by RDR2. The dsRNA is then processed into 24-nt siRNA by DCL3. After the siRNA is incorporated into AGO4, Pol V transcribes transcripts that base pairing with siRNA and the siRNA-AGO4 complex is recruited. AGO4, RNA-DIRECTED DNA METHYLATION 1 (RDM1) and DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) trigger DNA methylation at the transcriptional gene silencing (TGS) level. (C) Overview of the phased siRNA (phasiRNA) pathway. phasiRNAs originated from phasiRNA-generating (PHAS) or trans-acting siRNA (TAS) loci are transcribed by Pol II and their precursors are cleaved via the miRNA “one-hit” system (AGO1-miRNA complex) or the “two-hit” system (AGO7-miRNA complex). SUPPRESSOR OF GENE SILENCING3 (SGS3) stabilizes the 5'- or 3'-cleaved fragments to protect them from AGO-mediated slicing. Then the primary siRNAs are converted into dsRNAs by RDR6 and further processed into 21- or 22-nt siRNAs by DCL2 and DCL4 or 24-nt siRNAs by DCL3. For further processing, some of them will be recruited by the AGO1 complex and some of them may be incorporated into other uncertain AGOs to trigger the cleavage of target mRNA, translational repression or DNA methylation.
Heterochromatic siRNAs (hc-siRNAs) are derived from transposable and repetitive elements and are involved in DNA methylation or chromatin alteration by the canonical RNA-directed DNA methylation (RdDM) pathway (Matzke and Mosher, 2014; Borges and Martienssen, 2015). The biogenesis of hc-siRNAs requires RNA Polymerase IV (Pol IV) to transcribe the ssRNA, which is then synthesized into dsRNA by RDR2. The dsRNA is then processed into 24-nt siRNAs by DCL3 and methylated at their 3' ends by HEN1. After the siRNAs is incorporated into AGO4, RNA polymerase V (Pol V) transcribes transcripts that are complementary to the siRNA, and the siRNA-AGO4 complex is recruited. AGO4, RNA-DIRECTED DNA METHYLATION 1 (RDM1), and DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) trigger de novo DNA methylation at the TGS level at symmetric CG and CHG sites, or asymmetric CHH sites (H stands for C, T, or A; Figure 1B; Matzke and Mosher, 2014; Du et al., 2015).
Phased siRNAs (PhasiRNAs) originated from non-coding phasiRNA-generating (PHAS) loci in monocots or protein-coding genes in dicots, which are transcribed by Pol II (Deng et al., 2018; Yu et al., 2018). These target precursors are cleaved using a miRNA-mediated cleavage system: either “one-hit” (AGO1-miRNA complex) or “two-hit” (AGO7-miRNA complex) system. SUPPRESSOR OF GENE SILENCING3 (SGS3) stabilizes the primary siRNAs to protect them from AGO-mediated slicing (Yoshikawa et al., 2013). Then the primary siRNAs are converted into dsRNA by RDR6 and processed into 21- or 22-nt secondary siRNAs by DCL4 or DCL2, respectively. The 21-nt tasiRNAs, which originate from non-coding trans-acting siRNA (TAS) loci transcripts are recruited by the AGO1 complex to participate in the cleavage of target mRNAs (Adenot et al., 2006; Fukudome et al., 2011). The biogenesis of 21-nt phasiRNAs largely depends on DCL4 in rice; whereas a class of 24-nt phasiRNAs are processed by the DCL3 homolog DCL3b during the reproductive stage (Song et al., 2012; Komiya, 2017). However, little is known about the downstream processing of these phasiRNAs, such as which AGOs they interact with for silencing remains elusive (Figure 1C).
Global warming has diverse and profound effects on plant growth and development, and poses a serious threat to the global crop yields. Therefore, the plant response to high-temperature stress and the mechanism underlying plant thermotolerance have become focuses of research (Bita and Gerats, 2013). Recent studies have shown that plant miRNAs and siRNAs act as key regulators in response to high-temperature stress. Genome-wide studies for heat-responsive sRNAs have been conducted in many plant species using high-throughput sequencing and bioinformatics. A series of heat stress-responsive sRNAs have been identified from various plant species, suggesting that these sRNAs have persistent regulatory roles under extreme temperature (Supplementary Table S1).
Extensive studies have shown that miRNAs can target genes encoding a diverse range of regulatory proteins, including a large proportion of TFs, suggesting that miRNAs function at the core of gene regulatory networks. One miRNA family usually has multiple target genes and plays versatile roles in several aspects of plant development and stress resistance. Accumulating evidence has shown that miRNAs are involved in plant responses to heat stress, and act as critical factors in coordinating plant development and heat stress resistance.
miR156 and its targets, SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE (SPL) genes are highly conserved in plants, and regulate developmental phase transitions, including juvenile-to-adult and vegetative-to-reproductive transitions (Xu et al., 2016; He et al., 2019; Ma et al., 2020). miR156 is highly expressed in young seedlings and its expression declines when the shoot develops, displaying opposite changes to its targets (Xu et al., 2016). The miR156-SPL3 module regulates flowering locus T (FT) expression to control Arabidopsis flowering time in response to ambient temperature (16 and 23°C; Lee et al., 2010; Kim et al., 2012). Besides, miR156 was also found to response to high-temperature stress. Stief et al. (2014a) showed that miR156 isoforms were highly induced after recurring heat stress (37 and 44°C) and promoted sustained expression of heat stress responsive genes in Arabidopsis, suggesting miR156 was functionally important for heat stress memory. Heat stress memory refers to the maintenance of acquired thermotolerance that plants obtain after heat stress. Heat stress memory is one of the mechanisms for plants survival under recurring heat stress. Plants can withstand high temperature, which are lethal to them in a normal state. This acquired thermotolerance can be maintained several days after returning to non-stress temperatures (Lämke et al., 2016). miR156 mediated repression of SPL2 and other target genes enhanced and prolonged the heat stress memory, and this process was also regulated by the HSFA2 cascade, which required HEAT STRESS-ASSOCIATED 32 (HSA32) and ROF1 (Stief et al., 2014a,b). In addition, recent studies showed that miR156-SPL13 mediates heat stress response in alfalfa, and overexpression of soybean miR156b in Arabidopsis led to male sterility under heat stress (Matthews et al., 2019; Ding et al., 2021). These studies suggest that the role of the miR156-SPL module is conserved in plants and that plant development and heat stress responses are mediated by miRNAs.
In contrast to miR156, miR172 is a positive regulator in juvenile-to-adult developmental transition by targeting APETALA2 (AP2) family genes, such as AP2, TARGET OF EAT 1 (TOE1), TOE2, TOE3, SCHLAFMUTZE (SMZ), and SCHNARCHZAPFEN (SNZ; Gahlaut et al., 2018; Ma et al., 2020). However, miR156 is highly expressed in the juvenile phase; whereas miR172 is barely expressed in this stage (Wu et al., 2009). The targets of miR156, SPL9, and SPL10, directly promote the expression of miR172b, which indicates that miR172 acts downstream of miR156 to promote adult epidermal identity (Wu et al., 2009). miR172 has been reported to function in thermosensory pathway to regulate ambient temperature-responsive flowering under non-stress temperature conditions. The transgenic plants with overexpression of miR172 showed a temperature insensitive early flowering (Lee et al., 2010). Jung et al. (2012) discovered that RNA-binding protein FCA percept temperature fluctuation and promoted the processing of pri-miR172 via recognition of RNA motif in the stem-loop during the early stage of thermosensory flowering pathway. In addition, miR172 has also been revealed to response to high-temperature stress. In both rice post-meiosis panicle and safflower leaf tissues, miR172 was observed to be significantly downregulated, whereas its target AP2 genes were upregulated under heat stress, indicating an important role of miR172-AP2 module in plant heat stress response (Kouhi et al., 2020; Peng et al., 2020).
The miR159 and miR319 families are highly conserved in plants and have a high degree of sequence identity (Palatnik et al., 2019). miR159 targets several members of GIBBERELLIC ACID MYB (GAMYB) genes and plays important roles in flowering and male fertility. miR319 targets TEOSINTE BRANCHED/CYCLOIDEA/PCF (TCP) genes and several MYB genes to control leaf growth (Palatnik et al., 2019). miR159 was upregulated by heat stress in flowering Chinese cabbage (Ahmed et al., 2019). In contrast, miR159 was downregulated after heat stress in Triticum aestivum, and the transgenic rice overexpressing tae-miR159 showed sensitivity to heat stress (Wang et al., 2012). Heat stress caused a significant decrease of miR159 and an increase of its target genes, CsGAMYB1 and CsMYB29-like in cucumber (Li et al., 2016). In addition, ectopic expression of csa-miR159b in Arabidopsis decreased heat tolerance by targeting AtMYB33 (Li et al., 2016). For miR319, overexpression of sha-miR319d increased expression levels of heat stress-responsive genes and conferred heat stress tolerance in transgenic Solanum lycopersicum with increased activities of superoxide dismutase (SOD), catalase (CAT), and ascorbate peroxidase (APX; Shi et al., 2019).
The miR160 family targets the AUXIN RESPONSIVE FACTOR (ARF) gene family, which play vital roles in plant growth and development by regulating auxin signaling (Gahlaut et al., 2018). The miR160-ARF module is involved in phenotypic plasticity somatic embryo development, leaf development, root formation, and cell differentiation (Lin et al., 2015, 2018). In Gossypium hirsutum, overexpression of miR160 caused sensitivity to heat stress via suppressing the expression of ARF10 and ARF17 and caused anther indehiscence (Ding et al., 2017). Overexpression of a miR160 precursor presented increased thermotolerance, which shared similar phenotype with arf10, arf16, and arf17 mutants in Arabidopsis (Lin et al., 2018). Furthermore, miR160 also regulates seed germination, hypocotyl, and rachis growth under heat stress (Lin et al., 2018).
As described above, miR160 is related to auxin. miR393 is also involved in auxin-related development in plants by regulating the expression of the auxin receptors (TAARs) including TRANSPORT INHIBITOR RESPONSE1 (TIR1) and AUXIN SIGNALING F-BOX (AFBs). These TAARs can degrade the AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) genes and allow specific ARF TFs to active the auxin-responsive genes, which functions in the primary auxin-responsive pathway (Si-Ammour et al., 2011). Overexpression of osa-miR393a in transgenic creeping bentgrass increased heat tolerance by repressing its targets AsAFB2 and AsTIR1, and this enhanced heat stress tolerance was associated with induced expression of HSPs (Zhao et al., 2019).
Heat stress causes the accumulation of ROS. SODs including iron SOD (Fe-SOD), manganese SOD (Mn-SOD), and copper/zinc SOD (Cu/Zn-SOD) encoded by CSDs are important ROS-scavenging enzymes that catalyze the superoxide radicals in plants. miR398 family members were found to be rapidly induced by heat stress, leading to the downregulation of their target genes CSD1, CSD2, and Copper chaperone for SOD (CCS) in Arabidopsis (Guan et al., 2013). Furthermore, csd1, csd2, and ccs mutants showed heat stress tolerance with increased expression of HSFs and HSPs (Guan et al., 2013). Fang et al. (2019) showed that the induction of tocopherols and 3'-phosphoadenosine 5'-phosphate (PAP) are required for the increased accumulation of miR398 and acquisition of heat tolerance. In addition, HSFA1b and HSFA7b were revealed to be responsible for the heat induction of miR398 by binding directly to the promoter of MIR398 to activate its transcription (Guan et al., 2013). A recent study further revealed that the MIR398 genes possess their natural antisense transcripts (NATs) and uncovered a regulatory loop between them; the cis-NATs of MIR398 genes repress the processing of miR398 pri-miRNAs, which cause poorer thermotolerance due to the upregulation of miR398-targeted genes (Li et al., 2020). However, the underlying mechanism of how heat stress regulates the expression of MIR398 cis-NATs remains to be investigated.
Other development-related miRNAs are also associated with plant heat stress responses. miR169 family members can target Nuclear transcription factor Y subunit (NF-YA) genes and function at flowering stage in rice under heat stress, which was confirmed by overexpression of miR169r-5p (Liu et al., 2017b). The paradigmatic miR396-GROWTH-REGULATING FACTOR (GRF) model is well established and plays important roles in regulating the size of multiple plant tissues or organs (Liu et al., 2021). Interestingly, Giacomelli et al. (2012) reported that miR396 mediated the cleavage of HaWRKY6 in sunflower during early responses to high temperature. Heat stress reduced the accumulation of miR396, which showed opposite expression patterns to HaWRKY6, and expression of a miR396-resistant version of HaWRKY6 altered heat stress responses in Arabidopsis (Giacomelli et al., 2012). For MIR400 family, a heat stress-induced alternative splicing event was observed to occur in the intron of MIR400, which was co-transcribed with its host gene in Arabidopsis (Yan et al., 2012). Under heat stress, the alternative splicing of the MIR400 intron resulted in greater accumulation of miR400 primary transcripts and reduced level of mature miR400. In addition, overexpression of miR400 caused higher sensitivity to heat stress in transgenic plants compare to the wild type plants. These results demonstrated that miR400 acts as a negative regulator in plant heat stress resistance and revealed the essential role of alternative splicing in linking miRNA and high-temperature stress (Yan et al., 2012). A report of rice miR5144-3p showed that miR5144-3p plays a role in protein folding and abiotic stress during rice development by regulating the expression of OsPDIL1;1 (Xia et al., 2018). miR5144-3p was downregulated under heat stress, leading to the increased accumulation of OsPDIL1;1 mRNA in rice; STTM-miR5144-3p and OsPDIL1;1 overexpression transgenic rice exhibited improved heat tolerance (Xia et al., 2018).
A large number of miRNAs and putative siRNAs participate in plant responses to environmental stresses, such as dehydration, salinity, cold, and ABA (Sunkar and Zhu, 2004). Compared with the large number of miRNAs and siRNAs identified in responses to other environmental stresses in plants, hitherto siRNAs identified in response to heat stress is relatively few. Previous researches have reported that heat-induced copia-type retrotransposon ONSEN was accumulated in the siRNAs biogenesis impaired mutants, which revealed the potential roles of siRNAs in plant heat stress response (Ito et al., 2011). The accumulation of a particular class of phasiRNAs-tasiRNAs derived from Arabidopsis TAS loci were found to decrease significantly under heat stress, indicating their participation in plant heat stress responses, and their functions in thermotolerance were subsequently investigated (Zhong et al., 2013; Li et al., 2014). Overexpression of TAS1a-derived tasiRNAs in Arabidopsis downregulated target genes HEAT-INDUCED TAS1 TARGET1 (HTT1) and HTT2, and led to weaker thermotolerance in transgenic plants; whereas overexpression of HTT1 and HTT2 led to improved thermotolerance via upregulation of several HSFs (Li et al., 2014). Furthermore, heat-induced tasiRNA decrease was found to be involved in thermomemory of early flowering and attenuated immunity through targeting HTT5, which provides insights for understanding how heat exposure influence the fitness of plant progeny (Liu et al., 2019). Hu et al. (2020) have also identified a newly evolved phasiRNA locus that generated consecutive 21-nt phasiRNAs in response to heat stress in Camellia. Predictive bioinformatics and gene expression analysis showed that these secondary phasiRNAs could potentially target several genes including LIPOXYGENASE, RAN GTPase, XYLOGLUCAN ENDOTRANSGLUCOSYLASE, and ATPase to regulate their expression in a trans-acting manner. However, further genetic studies are required to verify these targets and elucidate the specific function of these phasiRNAs in thermal resistance. In a recent study, a large population of transposable element derived 24-nt siRNAs were significantly reduced in maize tassels and roots after exposure to high temperature, and genes nearby these transposable elements tended to be downregulated, indicating that the expression of heat-dependent gene is influenced by adjacent transposon sequences. However, the underlying mechanism controlling the relationship among transposable element, transposable element derived 24-nt siRNAs and nearby genes in response to heat stress remains elusive (He et al., 2019).
The findings above demonstrate that sRNAs can serve as vital regulators of plant responses to heat stress. The major miRNAs and siRNAs involved in heat stress resistance and their regulatory pathway have been summarized in Figure 2. Interestingly, some sRNAs tend to display species-dependent expression patterns under heat stress, indicating that they may have distinct regulatory mechanisms in different plant species. For example, miR397 was downregulated in rice and tomato, but upregulated in banana in response to high temperature stress (Liu et al., 2017b; Pan et al., 2017; Zhu et al., 2019). In addition, several studies have identified novel species-specific sRNAs by genome-wide deep sequencing. For example, Liu et al. (2015b) identified 25 novel heat stress-responsive miRNAs in Saccharina japonica, such as sja-novel-mir-5, sja-novel-mir-13, and sja-novel-mir-59. These species-specific sRNAs may have essential roles in plant heat stress responses and dissecting their functions will broaden our understanding of the underlying regulatory mechanisms governing thermotolerance in different plant species.
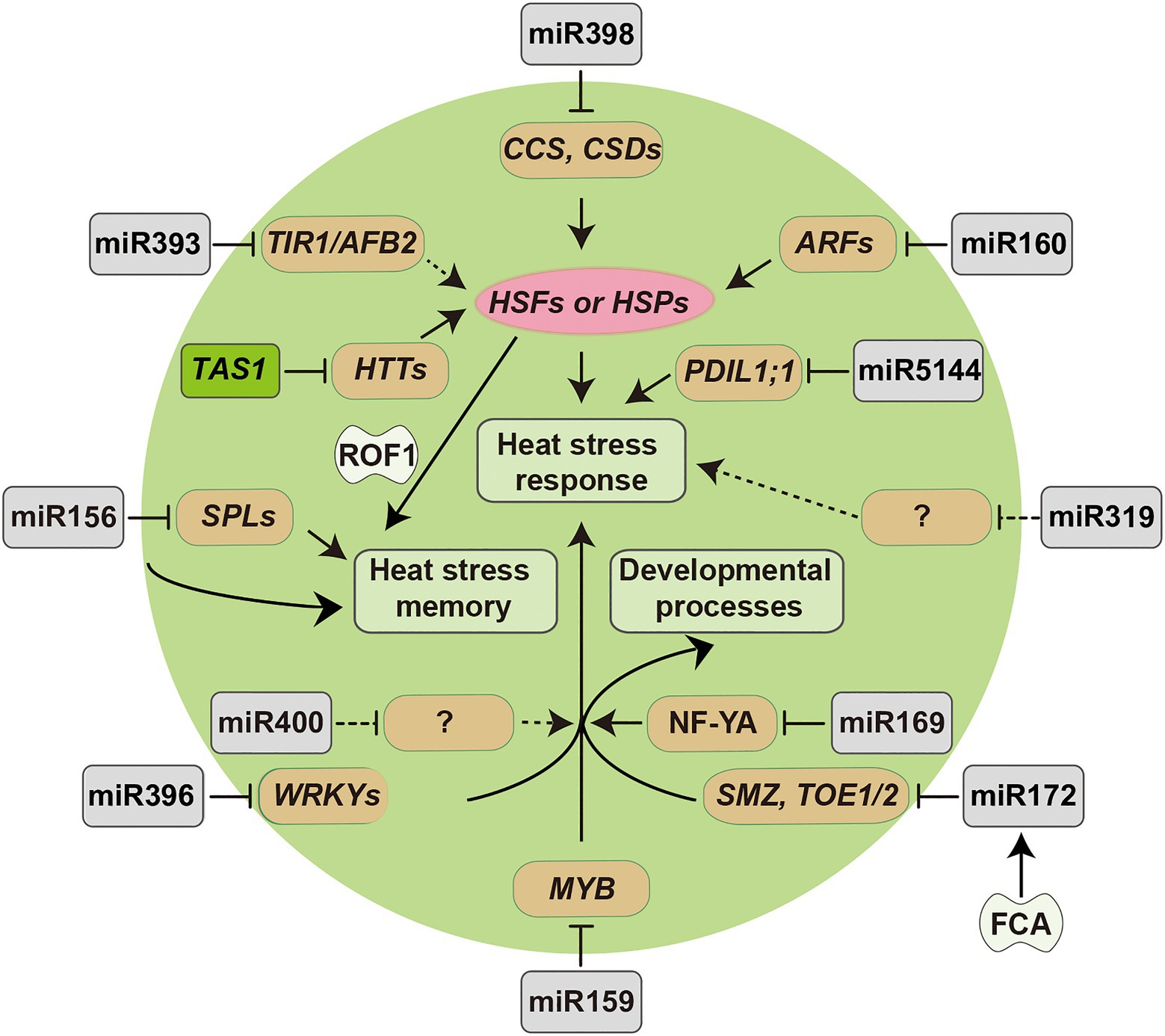
Figure 2. Diagram of miRNA-target modules involved in plant responses to heat stress. Arrowed lines represent promotional effects and flat lines represent repressive effects. Dashed lines indicate hypotheses that need to be confirmed.
Heat stress causes many adverse effects on plant growth, development, and physiological processes. Reduced water content, excess generation of ROS, and protein denaturation caused by heat stress greatly impede normal cellular functions (Hasanuzzaman et al., 2013; Jacob et al., 2017). Plants adapted to respond to and survive from heat stress by developing diverse mechanisms to cope with severe conditions, such as basal thermotolerance (Bokszczanin and Fragkostefanakis, 2013). Currently, heat stress response pathways have been extensively investigated in plants, and mainly include the HSP-based protective pathway, phospholipid pathway, ROS/redox signaling pathway, and phytohormone signaling pathway (Bokszczanin and Fragkostefanakis, 2013; Qu et al., 2013).
Heat-induced changes in plasma membrane fluidity cause a transient opening of Ca2+ channels, which induces Ca2+ influx to the cytoplasm. The increased levels of cytosolic Ca2+ activate multiple kinases, such as Calcium-dependent protein kinases (CDPKs), thereby evoking the expression of transcriptional regulators in response to heat stress. HSFs and HSPs play critical roles in this process (Qu et al., 2013; Guo et al., 2016). HSFs are evolutionarily grouped into A, B, and C classes. The HSFA subfamily genes, HSFA1-HSFA9 have been well studied. HSFA1s function as master regulators, which are indispensable in acquired thermotolerance of plants by activating downstream heat stress responsive transcription factors (TFs), such as MULTIPROTEIN BRIDGING FACTOR1C (MBF1C) and DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN2A (DREB2A; Liu et al., 2011; Yoshida et al., 2011). HSPs, including HSP70, HSP90, HSP100, and HSP101 are the main inducers of HSFs, which resolve large aggregates induced by unfolded proteins and reduce protein misfolding (Jacob et al., 2017). In addition, HSP70 and HSP90 repress the activity of HSFA1s under non-stress condition by protein–protein interactions (Ohama et al., 2017). Phosphorylation and dephosphorylation by CALMODULIN-BINDING PROTEIN KINASE3 (CBK3) and PROTEIN PHOSPHATASE7 (PP7), respectively, can activate HSFA1s via post-translational modification under heat stress.
Heat stress causes remodeling of lipids in the membrane, which may induce the phospholipid signaling and the key mediators in this pathway include phosphatidyl inositol 4,5-bisphosphosphate (PIP2) and phosphatidic acid (PA). Accumulation of lipid signaling molecules can in turn trigger Ca2+ influx through channels in the plasma membrane (Bokszczanin and Fragkostefanakis, 2013). The mechanisms behind phospholipid signaling, from heat signal initiation to transduction, and the relationship between lipid signaling and plasma membrane channels as yet are still much unknown in plants.
Heat stress induces unfolded or misfolded proteins in the cytosol and endoplasmic reticulum (ER) that may trigger the unfolded protein response (UPR). Cytosolic UPR is mainly associated with specific HSFs, such as HSFA2, which is induced by HSFA1s and regulated by one splice variant of its own, S-HSfA2 via post-transcriptional regulation (Sugio et al., 2009; Liu and Charng, 2013). Two signaling pathways are involved in UPR, proteolytic processing, mediated by the basic leucine zipper domain (bZIP) TFs, RNA splicing, mediated by Inositol-requiring enzyme 1 (IRE1; Deng et al., 2013). ROS that accumulates during heat stress responses, such as H2O2, can act as signaling molecules to trigger the ROS-scavenging pathway; Respiratory burst oxidase homologs (RBOHs) also play important roles for the initiation and signal propagation of this pathway (Baxter et al., 2014). Furthermore, ROS-scavenging antioxidant enzymes, Late embryogenesis abundant (LEA) proteins, osmolytes, and secondary metabolites are considered necessary for detoxification of ROS (Bokszczanin and Fragkostefanakis, 2013).
Phytohormones are key players in plant growth and development from seed germination to senescence, and are involved in plant adaptation to adverse ambient stresses. Strigolactone (SL), cytokinin (CK), abscisic acid (ABA), and ethylene (Et) regulate the leaf senescence under heat stress (Abdelrahman et al., 2017). In addition, salicylic acid (SA) and jasmonic acid (JA), which are responsive to abiotic stresses, are also involved in the regulation of plant HSF genes (Guo et al., 2016; Rai et al., 2020). One phytohormone signaling pathway can respond to multiple abiotic stresses via crosstalk because plants endure multiple stresses in nature. For example, heat stress is usually associated with high light.
Ambient temperature fluctuations affect plant functioning, geographical distribution, and agricultural production of crops (Proveniers and van Zanten, 2013). The phenotypic responses of plants to deal with high temperature include hypocotyl elongation, leaf hyponasty, and floral induction. The warm temperature induced basic helix–loop–helix (bHLH) TF, PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) plays a central role in warmth-mediated morphological acclimation (Franklin et al., 2011; Kumar et al., 2012; Kim et al., 2020). miRNAs are also involved in morphological adaptations of plant under heat stress. For example, overexpression of miR160 improved seed germination and increased the length of hypocotyl elongation and the rachis (Lin et al., 2018). The reproductive phase of flowering plants is highly sensitive to high temperature, which often contributes to the acceleration of flowering and results in poor seed set (Zinn et al., 2010). As indicated above, the heat-responsive miR156, miR159, miR172, and miR319 regulate flowering time or male and female fertility in different plant species, which illustrates that miRNAs play important roles in triggering the development of flower set (Supplementary Table S1; Wu et al., 2009; Lee et al., 2010; Stief et al., 2014a; Yin et al., 2018; Ahmed et al., 2019; Hu et al., 2019; Zhu et al., 2019; Kouhi et al., 2020). Furthermore, the heat-induced retrotransposon ONSEN was accumulated during flower development and before gametogenesis in mutants that are deficient in siRNA synthesis (Ito et al., 2011). Taken together, it is suggested that sRNAs play pivotal roles in regulating plant growth and reproductive tissue development under heat stress, which ensures transgenerational seed production (Figure 3).
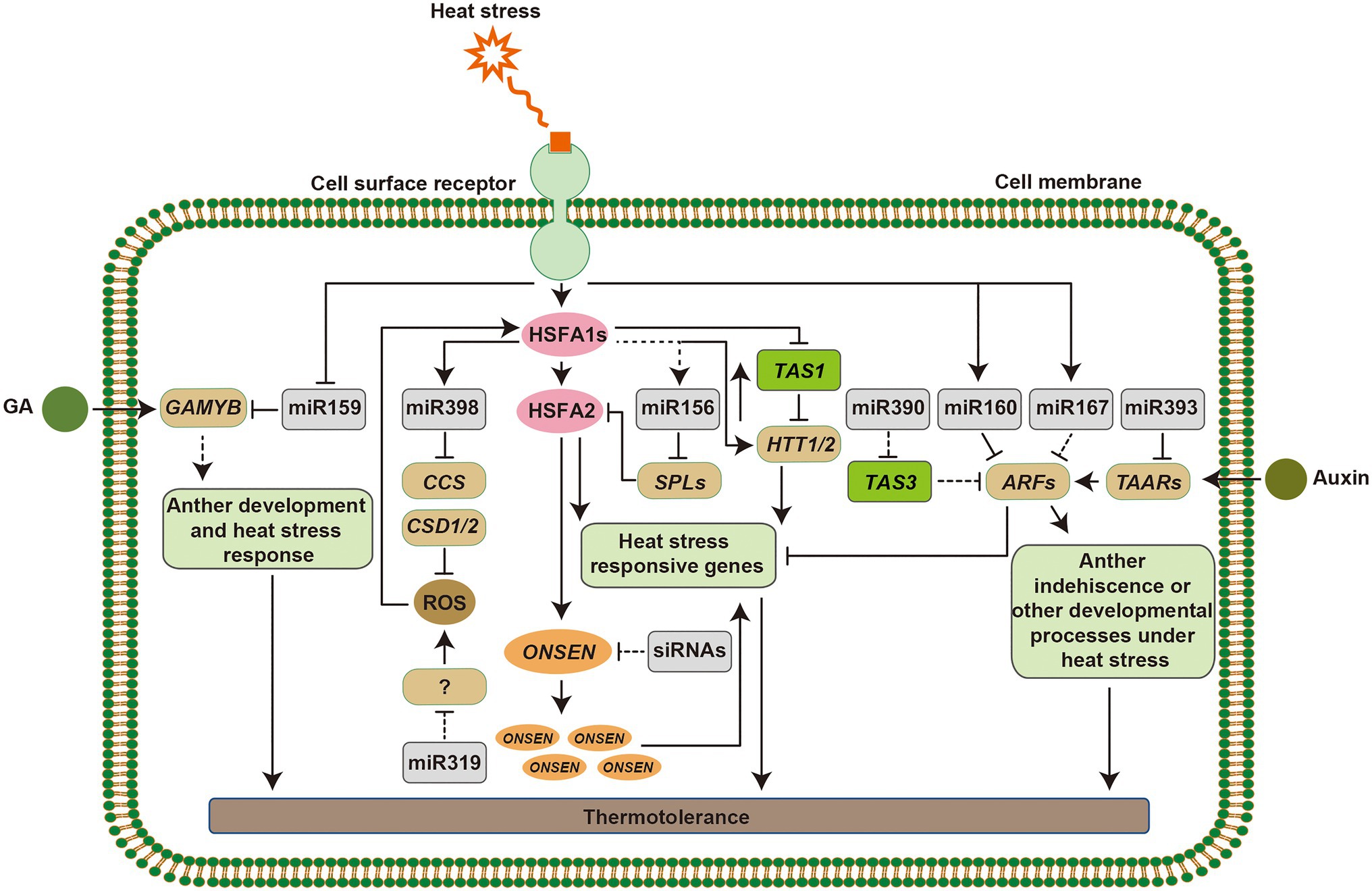
Figure 3. Mechanism of small RNA (sRNA) regulation in plant responses to heat stress. Heat stress signaling can be perceived by putative sensors on the cell membrane, which induce the expression of heat stress responsive genes. HSFA1s function as master regulators to active the downstream heat stress responsive transcription factors and genes. Heat-induced miR398 downregulates CSDs and CCS. Accumulation of reactive oxygen species (ROS) in the cells acts as signals to alter the expression of heat shock transcription factors (HSFs), and miR319 functions in this pathway via unknown targets. HSFA1s directly activate the expression of HTT genes, targets of TAS1, which upregulate HSF and heat shock protein (HSP) genes. The miR156-SPL module promotes sustained expression of heat stress-inducible genes in response to recurring heat stress and prolong the heat stress memory. In addition, the retrotransposition ONSEN is activated by HSFAs, although, siRNAs regulate its activity, which can be inherited to its progeny. miR159-GAMYB modules function in gibberellin (GA) signaling. miR160, miR167, miR390, and miR393 target AUXIN RESPONSIVE FACTOR (ARF) or TAARs that act in the auxin response pathway and show morphological adaptations under heat stress. Arrowed lines represent promotional effects and flat lines represent repressive effects. Dashed lines indicate hypotheses that need to be confirmed.
Heat shock proteins that act as molecular chaperones are major functional proteins in heat stress response via the activation of HSFs (Qu et al., 2013; Ohama et al., 2017). Various attempts have been made to increase thermotolerance in different host by overexpression of a single HSF or HSP gene, such as CaHSP25.9, ZmHSF05, TaHSP23.9, and OsHSP20, (Feng et al., 2019; Li et al., 2019; Guo et al., 2020; Wang et al., 2020). Nevertheless, AsHSP26.8a, a novel chloroplast-localized small HSP gene from creeping bentgrass negatively regulates heat stress resistance through modulating ABA and other stress signaling pathways (Sun et al., 2020). In addition, several miRNAs affect heat stress responses by targeting and activating HSF/HSP genes. The miR156-SPL module downregulates the expression of heat stress inducible genes, such as HEAT STRESS ASSOCIATED 32 (HSA32), HSP17.6A, and HSP22.0 during recovery from heat stress, which is functionally important for heat stress memory (Stief et al., 2014a). Overexpression of miR160 altered the expression of HSPs including HSP17.6A, HSP17.6II, HSP21, and HSP70B, which allow plants to survive under heat stress (Lin et al., 2018). In Solanum habrochaites, constitutive expression of sha-miR319d enhanced heat tolerance via upregulation of HSFA1a and HSFA1b, while overexpression of Osa-miR393a caused higher expression levels of AsHSP17.0 and AsHSP26.7a than in wild type plants (Shi et al., 2019; Zhao et al., 2019). Transgenic plants expressing miR398-resistant forms of CSD1, CSD2, or CCS showed reduced expression levels of HSF genes (HSFA1e, HSFA2, HSFA3, and HSFA7b) and HSP genes (HSP17.6, HSP70B, and HSP90.1), while csd1, csd2, and ccs loss-of-function mutants showed enhanced thermotolerance with increased expression of heat stress inducible genes (Guan et al., 2013). Furthermore, HSFA1b and HSFA7b bind to heat stress elements directly in the promoter region of miR398b, which constituted a positive regulatory feedback loop (Figure 3). Overexpression of HTT1 and HTT2, targets of TAS1, upregulated several HSF genes and enhanced thermotolerance in Arabidopsis (Li et al., 2014). Meanwhile, HSFA1a directly activates the expression of HTT genes, such as HTT1, which act as cofactors of HSP70-14 complexes in the thermotolerance pathway (Li et al., 2014). All of these discoveries reveal that sRNAs help plants achieve thermotolerance by regulating the expression of HSFs and HSPs (Figure 3).
Unfavorable environments trigger the production of ROS, which causes oxidative damages to proteins, lipids and stress-induced electrolyte leakage in plants. Plants have evolved an antioxidant defense system equipped with various enzymatic and non-enzymatic ROS-scavengers to maintain an equilibrium between the production of ROS and elimination of excessive ROS (Caverzan et al., 2019). Several antioxidant enzymes, such as SOD, APX, and CAT are involved in the heat stress responses in the ROS-scavenging pathway, where oxidative stress is produced as a secondary stress (Qu et al., 2013). As described above, downregulation of CSD1, CSD2, and CCS by heat-induced miR398 led to accumulation ROS in cells, which contributed to the expression of HSFs and other heat stress-responsive genes (Guan et al., 2013). This regulatory mechanism constitutes a regulatory loop for plant thermotolerance that involves miR398, HSF genes, and ROS-scavenging enzymes. A recent study showed that, tocopherols and PAP positively regulated the biogenesis of miR398 in chloroplasts and promoted plant thermotolerance (Fang et al., 2019). In addition, altered ROS under heat stress may act as signals in cells to induce the expression of HSF1Aa and HSF1Ab in miR319d transgenic plants, but the mechanisms of ROS, miR319d, and its putative targets in this signal transduction are much unknown (Shi et al., 2019; Figure 3). These findings suggest that sRNAs-mediated heat stress responses are partially dependent on the ROS signaling pathways in plants.
Phytohormones are produced via environmental signals, such as heat stress. For example, miR159-regulated GAMYB-like TFs function in gibberellin (GA) signaling and overexpression of tae-miR159 caused sensitivity to heat stress in transgenic plants, which suggested that tae-miR159 may participate in a heat stress related signaling pathway (Murray et al., 2003; Wang et al., 2012). Auxin orchestrates many morphogenetic processes, such as root formation and anther development, and endogenous auxin is also involved in heat stress responses. miR160 and miR393 target ARFs or TAARs, which are components in the auxin response pathway (Si-Ammour et al., 2011; Kruszka et al., 2014; Gahlaut et al., 2018). Overexpression of miR160 in cotton increased sensitivity to heat stress and caused anther indehiscence by activating the auxin response; while constitutive expression of miR157 suppressed the auxin signal, which also caused sensitivity to heat stress with microspore abortion and anther indehiscence (Ding et al., 2017). Overexpressing of an miR160 precursor in Arabidopsis significantly reduced the expression of its target genes ARF10, ARF16, and ARF17, and led to improved tolerance of transgenic plants. Furthermore, arf10, arf16, and arf17 mutants showed advanced thermotolerance by regulating the expression of HSPs (Lin et al., 2018). In addition, the miR167-ARFs and miR390-TAS3-ARFs models are involved in plants developmental processes; however, whether miR167 and miR390 function in heat stress response by targeting ARFs need to be tested (Figure 3). ABA induces the accumulation of miR168, and both plants overexpressing miR168a and loss-of-function mutant of its target AGO1, ago1-27, displayed ABA hypersensitivity and several abiotic stress tolerances (Li et al., 2012). miR168 also responds to heat stress in various species, such as Arabidopsis, rice, Brassica rapa, and flowering Chinese cabbage (Barciszewska-Pacak et al., 2015; Bilichak et al., 2015; Mangrauthia et al., 2017; Ahmed et al., 2019). Responses of plants to heat stress are complex, and may require physiological or metabolic changes from several phytohormone signaling pathways with a crosstalk, and there is no doubt that sRNAs are essential regulators in these processes.
As discussed above, a multi-layered regulatory signaling pathways are involved in plant response to heat stress. Heat stress memory as one of the mechanisms for plants survival under recurring heat stress included the regulation of chromatin modifications. For example, high levels of histone H3 lysine 4 (H3K4) methylation are related to hyper-induction of heat stress inducible genes after a recurring heat stress, which depends on HSFA2 (Yoshida et al., 2011; Liu and Charng, 2013; Lämke et al., 2016). miR156 promotes sustained expression of heat stress inducible genes in response to recurring heat stress, and increases of miR156 prolong heat stress memory (Stief et al., 2014a,b). miR156 targets, SPLs, are critical for heat stress memory, and may serve to integrate morphological acclimation with heat stress responses. Furthermore, plants display transgenerational memory mediated by transposons. Transgenerational memory refers to transmitting epigenetic states or environmental responses from one generation to the next that may offer the offspring an adaptive advantage for better fitness (Liu et al., 2015a). A surprisingly high frequency of new heat-induced retrotransposition, ONSEN insertions in progeny after heat stress revealed that the transgenerational memory of heat stress is maintained during differentiation of generative organs by priming ONSEN to transpose (Ito et al., 2011). In addition, the activation of ONSEN requires heat-induced TFs in the heat stress response pathway, such as HSFA2 (Cavrak et al., 2014). Although, the mechanisms of DNA methylation, sRNAs, and transposons in heat stress memory are obscure, epigenetic regulation is an important mechanism in response to heat stress (Figure 3).
Heat stress caused by the global warming affects the growth and development of plants, which increases the risk of yield reductions in agricultural crops. It is important to elucidate how plants respond to heat stress, but many questions remain to be answered, such as how do plants sense heat stress, and what kinds of signaling pathways the sensors use to transduce the signals into nucleus? Given that epigenetic regulation by sRNAs is crucial in gene regulatory networks for heat stress responses, we believe that an intensive understanding of their roles and functions will provide plentiful potential biological resources for plant engineering. Recently, various heat stress responsive sRNAs have been identified. However, it should be noted that many experimental factors affect the responses of sRNAs in plants, such as plant species, tissue, developmental stage, treatment time, and growth conditions, which means that a slight change can lead to different expression patterns of sRNAs. We summarized the literatures on sRNAs involved in heat stress responses of plants and listed the heat stress conditions used for treatments in Supplementary Table S1. Even though, a large number of diverse heat stress responsive sRNAs have been identified in plants, their roles and molecular mechanisms are still not fully elucidated, which may be due to the lack of genetic materials, especially for non-model plants and important crops. Future research should focus on the creation of genetically modified mutants and genetic manipulations of sRNAs to identify additional sRNA-target modules in the regulatory networks of heat stress responses. Notably, most miRNAs that have been functionally investigated so far are evolutionary conserved miRNAs, whereas the species- or tissue-specific miRNAs have been rarely studied. Thus, much more work is required to decipher the regulatory mechanisms of non-conserved miRNAs in response to heat stress in more crops. In addition, the investigation of upstream regulation of heat-responsive miRNA would also be an interesting research topic and worthy of more attention.
As discussed above, several sRNAs were shown to have potential in improving plant thermotalerance. One type of sRNAs is involved in the heat stress responses by targeting and activating HSF/HSP genes that act as molecular chaperones to prevent denaturation or aggregation of target proteins, such as miR160 and miR393 (Lin et al., 2018; Zhao et al., 2019). Thus, increasing the saturation level of HSFs and HSPs by overexpression of one specific miRNA can enhance heat tolerance to their host plants using transgenic approaches. The other type of sRNAs participate in heat stress response by triggering ROS-scavenging pathway in plants, such as miR319 and miR398 (Guan et al., 2013; Shi et al., 2019). High levels of ROS-scavenging enzymes accumulation by manipulating certain miRNAs can be another strategy to generate heat stress tolerant plants. Recently, new biotechnological tools have been successfully explored to investigate MIR genes or miRNA modulation, such as endogenous artificial target mimicry (Short tandem target mimicry, STTM), miRNA transient virus induced gene silencing (VIGS) and MIR genes editing using CRISPR/Cas9 system (Basso et al., 2019). Furthermore, next-generation sequencing, for example, sRNA sequencing (sRNA-seq), Parallel Analysis of RNA Ends (PARE) analysis (Zhai et al., 2014; Jiang et al., 2020), and recently developed single-cell sRNA-mRNA co-sequencing (Wang et al., 2019) have provided powerful methods for elucidating the functions of sRNAs and their target genes. These techniques will be essential in further research and will expand the range of sRNA applications for crop breeding in thermotolerance.
Ultimately, the gene-silencing mechanisms mediated by sRNAs explore a new vista in the application of genetic engineering, which can be used to revolutionize agriculture by controlling a wide array of crop traits, including thermotolerance. sRNAs can work efficiently and precisely to develop targeted gene-silencing approaches in plants for various requirements, which can be used not only for the study of the functional analysis of genes responsive to heat stress but also to improve crop plants by manipulating their target genes. All of these accumulated researches will enable the successful and extensive application of sRNA technology for the development of next generation crops.
Z-FZ and LL wrote the manuscript. WH, JL, and BM provided the critical comments and edited the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by Shenzhen Grant Plan for Science and Technology (JCYJ20190808112207542), Guangdong Innovation Research Team Fund (2014ZT05S078), Natural Science Foundation of Guangdong Province (2019A1515011222 and 2021A1515010482), and Guangdong Basic and Applied Basic Research Foundation (2019A1515110162).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are thankful to reviewers for their valuable suggestions to improve this article.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.726762/full#supplementary-material
Abdelrahman, M., El-Sayed, M., Jogaiah, S., Burritt, D. J., and Tran, L. P. (2017). The “STAY-GREEN” trait and phytohormone signaling networks in plants under heat stress. Plant Cell Rep. 36, 1009–1025. doi: 10.1007/s00299-017-2119-y
Adenot, X., Elmayan, T., Lauressergues, D., Boutet, S., Bouché, N., Gasciolli, V., et al. (2006). DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 16, 927–932. doi: 10.1016/j.cub.2006.03.035
Ahmed, W., Xia, Y., Zhang, H., Li, R., Bai, G., Siddique, K. H. M., et al. (2019). Identification of conserved and novel miRNAs responsive to heat stress in flowering Chinese cabbage using high-throughput sequencing. Sci. Rep. 9:14922. doi: 10.1038/s41598-019-51443-y
Axtell, M. J. (2013). Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 64, 137–159. doi: 10.1146/annurev-arplant-050312-120043
Ballén-Taborda, C., Plata, G., Ayling, S., Rodríguez-Zapata, F., Becerra Lopez-Lavalle, L. A., Duitama, J., et al. (2013). Identification of cassava microRNAs under abiotic stress. Int. J. Genomics 1–10. doi: 10.1155/2013/857986
Barciszewska-Pacak, M., Milanowska, K., Knop, K., Bielewicz, D., Nuc, P., Plewka, P., et al. (2015). Arabidopsis microRNA expression regulation in a wide range of abiotic stress responses. Front. Plant Sci. 6:410. doi: 10.3389/fpls.2015.00410
Basso, M. F., Ferreira, P. C. G., Kobayashi, A. K., Harmon, F. G., Nepomuceno, A. L., Molinari, H. B. C., et al. (2019). MicroRNAs and new biotechnological tools for its modulation and improving stress tolerance in plants. Plant Biotechnol. J. 17, 1482–1500. doi: 10.1111/pbi.13116
Baxter, A., Mittler, R., and Suzuki, N. (2014). ROS as key players in plant stress signalling. J. Exp. Bot. 65, 1229–1240. doi: 10.1093/jxb/ert375
Bilichak, A., Ilnytskyy, Y., Wóycicki, R., Kepeshchuk, N., Fogen, D., and Kovalchuk, I. (2015). The elucidation of stress memory inheritance in Brassica rapa plants. Front. Plant Sci. 6:5. doi: 10.3389/fpls.2015.00005
Bita, C. E., and Gerats, T. (2013). Plant tolerance to high temperature in a changing environment: scientific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 4:273. doi: 10.3389/fpls.2013.00273
Bokszczanin, K. L., and Fragkostefanakis, S. (2013). Perspectives on deciphering mechanisms underlying plant heat stress response and thermotolerance. Front. Plant Sci. 4:315. doi: 10.3389/fpls.2013.00315
Bologna, N. G., Iselin, R., Abriata, L. A., Sarazin, A., Pumplin, N., Jay, F., et al. (2018). Nucleo-cytosolic shuttling of ARGONAUTE1 prompts a revised model of the plant microRNA pathway. Mol. Cell 69, 709–719. doi: 10.1016/j.molcel.2018.01.007
Borges, F., and Martienssen, R. A. (2015). The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 16, 727–741. doi: 10.1038/nrm4085
Caverzan, A., Piasecki, C., Chavarria, G., Stewart, C. N., and Vargas, L. (2019). Defenses against ROS in crops and weeds: the effects of interference and herbicides. Int. J. Mol. Sci. 20:1086. doi: 10.3390/ijms20051086
Cavrak, V. V., Lettner, N., Jamge, S., Kosarewicz, A., Bayer, L. M., and Mittelsten Scheid, O. (2014). How a retrotransposon exploits the plant's heat stress response for its activation. PLoS Genet. 10:e1004115. doi: 10.1371/journal.pgen.1004115
Chen, L., Ren, Y., Zhang, Y., Xu, J., Sun, F., Zhang, Z., et al. (2012). Genome-wide identification and expression analysis of heat-responsive and novel microRNAs in populus tomentosa. Gene 504, 160–165. doi: 10.1016/j.gene.2012.05.034
D’Ario, M., Griffiths-Jones, S., and Kim, M. (2017). Small RNAs: big impact on plant development. Trends Plant Sci. 22, 1056–1068. doi: 10.1016/j.tplants.2017.09.009
Deng, P., Muhammad, S., Cao, M., and Wu, L. (2018). Biogenesis and regulatory hierarchy of phased small interfering RNAs in plants. Plant Biotechnol. J. 16, 965–975. doi: 10.1111/pbi.12882
Deng, Y., Srivastava, R., and Howell, S. H. (2013). Endoplasmic reticulum (ER) stress response and its physiological roles in plants. Int. J. Mol. Sci. 14, 8188–8212. doi: 10.3390/ijms14048188
Ding, X., Guo, J., Zhang, Q., Yu, L., Zhao, T., and Yang, S. (2021). Heat-responsive miRNAs participate in the regulation of male fertility stability in soybean CMS-based F1 under high temperature stress. Int. J. Mol. Sci. 22:2446. doi: 10.3390/ijms22052446
Ding, Y., Ma, Y., Liu, N., Xu, J., Hu, Q., Li, Y., et al. (2017). microRNAs involved in auxin signalling modulate male sterility under high-temperature stress in cotton (Gossypium hirsutum). Plant J. 91, 977–994. doi: 10.1111/tpj.13620
Du, J., Johnson, L. M., Jacobsen, S. E., and Patel, D. J. (2015). DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 16, 519–532. doi: 10.1038/nrm4043
Fang, X., Zhao, G., Zhang, S., Li, Y., Gu, H., Li, Y., et al. (2019). Chloroplast-to-nucleus signaling regulates microRNA biogenesis in Arabidopsis. Dev. Cell 48, 371.e4–382.e4. doi: 10.1016/j.devcel.2018.11.046
Feng, X. H., Zhang, H. X., Ali, M., Gai, W. X., Cheng, G. X., Yu, Q. H., et al. (2019). A small heat shock protein CaHsp25.9 positively regulates heat, salt, and drought stress tolerance in pepper (Capsicum annuum L.). Plant Physiol. Biochem. 142, 151–162. doi: 10.1016/j.plaphy.2019.07.001
Franklin, K. A., Lee, S. H., Patel, D., Kumar, S. V., Spartz, A. K., Gu, C., et al. (2011). Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. U. S. A. 108, 20231–20235. doi: 10.1073/pnas.1110682108
Fukudome, A., Kanaya, A., Egami, M., Nakazawa, Y., Hiraguri, A., Moriyama, H., et al. (2011). Specific requirement of DRB4, a dsRNA-binding protein, for the in vitro dsRNA-cleaving activity of Arabidopsis Dicer-like 4. RNA 17, 750–760. doi: 10.1261/rna.2455411
Gahlaut, V., Baranwal, V. K., and Khurana, P. (2018). miRNomes involved in imparting thermotolerance to crop plants. 3 Biotech 8:497. doi: 10.1007/s13205-018-1521-7
Giacomelli, J. I., Weigel, D., Chan, R. L., and Manavella, P. A. (2012). Role of recently evolved miRNA regulation of sunflower HaWRKY6 in response to temperature damage. New Phytol. 195, 766–773. doi: 10.1111/j.1469-8137.2012.04259.x
Goswami, S., Kumar, R. R., and Rai, R. D. (2014). Heat-responsive microRNAs regulate the transcription factors and heat shock proteins in modulating thermo stability of starch biosynthesis enzymes in wheat (Triticum aestivum L.) under the heat stress. Aust. J. Crop. Sci. 8:697.
Guan, Q., Lu, X., Zeng, H., Zhang, Y., and Zhu, J. (2013). Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J. 74, 840–851. doi: 10.1111/tpj.12169
Guo, L. M., Li, J., He, J., Liu, H., and Zhang, H. M. (2020). A class I cytosolic HSP20 of rice enhances heat and salt tolerance in different organisms. Sci. Rep. 10:1383. doi: 10.1038/s41598-020-58395-8
Guo, M., Liu, J. H., Ma, X., Luo, D. X., Gong, Z. H., and Lu, M. H. (2016). The plant heat stress transcription factors (HSFs): structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 7:114. doi: 10.3389/fpls.2016.00114
Hasanuzzaman, M., Nahar, K., Alam, M. M., Roychowdhury, R., and Fujita, M. (2013). Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 14, 9643–9684. doi: 10.3390/ijms14059643
He, J., Jiang, Z., Gao, L., You, C., Ma, X., Wang, X., et al. (2019). Genome-wide transcript and small RNA profiling reveals transcriptomic responses to heat stress. Plant Physiol. 181, 609–629. doi: 10.1104/pp.19.00403
Hivrale, V., Zheng, Y., Puli, C. O. R., Jagadeeswaran, G., Gowdu, K., Kakani, V. G., et al. (2016). Characterization of drought- and heat-responsive microRNAs in switchgrass. Plant Sci. 242, 214–223. doi: 10.1016/j.plantsci.2015.07.018
Hu, Z., Lyu, T., Yan, C., Wang, Y., Ye, N., Fan, Z., et al. (2020). Identification of alternatively spliced gene isoforms and novel noncoding RNAs by single-molecule long-read sequencing in camellia. RNA Biol. 17, 966–976. doi: 10.1080/15476286.2020.1738703
Hu, Z., Shen, X., Xiang, X., and Cao, J. (2019). Evolution of MIR159/319 genes in Brassica campestris and their function in pollen development. Plant Mol. Biol. 101, 537–550. doi: 10.1007/s11103-019-00920-z
Iki, T., Yoshikawa, M., Nishikiori, M., Jaudal, M. C., Matsumoto-Yokoyama, E., Mitsuhara, I., et al. (2010). In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol. Cell 39, 282–291. doi: 10.1016/j.molcel.2010.05.014
Ito, H., Gaubert, H., Bucher, E., Mirouze, M., Vaillant, I., and Paszkowski, J. (2011). An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472, 115–119. doi: 10.1038/nature09861
Jacob, P., Hirt, H., and Bendahmane, A. (2017). The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 15, 405–414. doi: 10.1111/pbi.12659
Jiang, P., Lian, B., Liu, C., Fu, Z., Shen, Y., Cheng, Z., et al. (2020). 21-nt phasiRNAs direct target mRNA cleavage in rice male germ cells. Nat. Commun. 11, 1–10. doi: 10.1038/s41467-020-19034-y
Jodder, J., Das, R., Sarkar, D., Bhattacharjee, P., and Kundu, P. (2018). Distinct transcriptional and processing regulations control miR167a level in tomato during stress. RNA Biol. 15, 130–143. doi: 10.1080/15476286.2017.1391438
Jung, J. H., Seo, P. J., Ahn, J. H., and Park, C. M. (2012). Arabidopsis RNA-binding protein FCA regulates microRNA172 processing in thermosensory flowering. J. Biol. Chem. 287, 16007–16016. doi: 10.1074/jbc.M111.337485
Khraiwesh, B., Zhu, J.-K., and Zhu, J. (2012). Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta 1819, 137–148. doi: 10.1016/j.bbagrm.2011.05.001
Kim, S., Hwang, G., Kim, S., Thi, T. N., Kim, H., Jeong, J., et al. (2020). The epidermis coordinates thermoresponsive growth through the phyB-PIF4-auxin pathway. Nat. Commun. 11:1053. doi: 10.1038/s41467-020-14905-w
Kim, J. J., Lee, J. H., Kim, W., Jung, H. S., Huijser, P., and Ahn, J. H. (2012). The microRNA156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 module regulates ambient temperature-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Plant Physiol. 159, 461–478. doi: 10.1104/pp.111.192369
Komiya, R. (2017). Biogenesis of diverse plant phasiRNAs involves an miRNA-trigger and Dicer-processing. J. Plant Res. 130, 17–23. doi: 10.1007/s10265-016-0878-0
Kouhi, F., Sorkheh, K., and Ercisli, S. (2020). MicroRNA expression patterns unveil differential expression of conserved miRNAs and target genes against abiotic stress in safflower. PLoS One 15:e0228850. doi: 10.1371/journal.pone.0228850
Kruszka, K., Pacak, A., Swida-Barteczka, A., Nuc, P., Alaba, S., Wroblewska, Z., et al. (2014). Transcriptionally and post-transcriptionally regulated microRNAs in heat stress response in barley. J. Exp. Bot. 65, 6123–6135. doi: 10.1093/jxb/eru353
Kumar, S. V., Lucyshyn, D., Jaeger, K. E., Alós, E., Alvey, E., Harberd, N. P., et al. (2012). Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484, 242–245. doi: 10.1038/nature10928
Kumar, R. R., Pathak, H., Sharma, S. K., Kala, Y. K., Nirjal, M. K., Singh, G. P., et al. (2015). Novel and conserved heat-responsive microRNAs in wheat (Triticum aestivum L.). Funct. Integr. Genomics 15, 323–348. doi: 10.1007/s10142-014-0421-0
Kurihara, Y., and Watanabe, Y. (2004). Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. U. S. A. 101, 12753–12758. doi: 10.1073/pnas.0403115101
Lämke, J., Brzezinka, K., Altmann, S., and Bäurle, I. (2016). A hit-and-run heat shock factor governs sustained histone methylation and transcriptional stress memory. EMBO J. 35, 162–175. doi: 10.15252/embj.201592593
Laubinger, S., Sachsenberg, T., Zeller, G., Busch, W., Lohmann, J. U., Rätsch, G., et al. (2008). Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 105, 8795–8800. doi: 10.1073/pnas.0802493105
Lee, Y., Kim, M., Han, J., Yeom, K.-H., Lee, S., Baek, S. H., et al. (2004). MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23, 4051–4060. doi: 10.1038/sj.emboj.7600385
Lee, H., Yoo, S. J., Lee, J. H., Kim, W., Yoo, S. K., Fitzgerald, H., et al. (2010). Genetic framework for flowering-time regulation by ambient temperature-responsive miRNAs in Arabidopsis. Nucleic Acids Res. 38, 3081–3093. doi: 10.1093/nar/gkp1240
Li, W., Cui, X., Meng, Z., Huang, X., Xie, Q., Wu, H., et al. (2012). Transcriptional regulation of Arabidopsis MIR168a and argonaute1 homeostasis in abscisic acid and abiotic stress responses. Plant Physiol. 158, 1279–1292. doi: 10.1104/pp.111.188789
Li, H., Hu, T., Amombo, E., and Fu, J. (2017). Genome-wide identification of heat stress-responsive small RNAs in tall fescue (Festuca arundinacea) by high-throughput sequencing. Plant Physiol. 213, 157–165. doi: 10.1016/j.jplph.2017.03.004
Li, Y., Li, X., Yang, J., and He, Y. (2020). Natural antisense transcripts of MIR398 genes suppress microR398 processing and attenuate plant thermotolerance. Nat. Commun. 11:5351. doi: 10.1038/s41467-020-19186-x
Li, S., Liu, J., Liu, Z., Li, X., Wu, F., and He, Y. (2014). Heat-induced TAS1 TARGET1 mediates thermotolerance via heat stress transcription factor A1a-directed pathways in Arabidopsis. Plant Cell 26, 1764–1780. doi: 10.1105/tpc.114.124883
Li, S., Liu, L., Zhuang, X., Yu, Y., Liu, X., Cui, X., et al. (2013). MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 153, 562–574. doi: 10.1016/j.cell.2013.04.005
Li, H., Wang, Y., Wang, Z., Guo, X., Wang, F., Xia, X. J., et al. (2016). Microarray and genetic analysis reveals that csa-miR159b plays a critical role in abscisic acid-mediated heat tolerance in grafted cucumber plants. Plant Cell Environ. 39, 1790–1804. doi: 10.1111/pce.12745
Li, J., Yang, Z., Yu, B., Liu, J., and Chen, X. (2005). Methylation protects miRNAs and siRNAs from a 3'-end uridylation activity in Arabidopsis. Curr. Biol. 15, 1501–1507. doi: 10.1016/j.cub.2005.07.029
Li, G. L., Zhang, H. N., Shao, H., Wang, G. Y., Zhang, Y. Y., Zhang, Y. J., et al. (2019). ZmHsf05, a new heat shock transcription factor from Zea mays L. improves thermotolerance in Arabidopsis thaliana and rescues thermotolerance defects of the athsfa2 mutant. Plant Sci. 283, 375–384. doi: 10.1016/j.plantsci.2019.03.002
Lin, J. S., Kuo, C. C., Yang, I. C., Tsai, W. A., Shen, Y. H., Lin, C. C., et al. (2018). MicroRNA160 modulates plant development and heat shock protein gene expression to mediate heat tolerance in Arabidopsis. Front. Plant Sci. 9:68. doi: 10.3389/fpls.2018.00068
Lin, Y., Lai, Z., Tian, Q., Lin, L., Lai, R., Yang, M., et al. (2015). Endogenous target mimics down-regulate miR160 mediation of ARF10, −16, and −17 cleavage during somatic embryogenesis in Dimocarpus longan lour. Front. Plant Sci. 6:956. doi: 10.3389/fpls.2015.00956
Liu, H. C., and Charng, Y. Y. (2013). Common and distinct functions of Arabidopsis class A1 and A2 heat shock factors in diverse abiotic stress responses and development. Plant Physiol. 163, 276–290. doi: 10.1104/pp.113.221168
Liu, J., Feng, L., Gu, X., Deng, X., Qiu, Q., Li, Q., et al. (2019). An H3K27me3 demethylase-HSFA2 regulatory loop orchestrates transgenerational thermomemory in Arabidopsis. Cell Res. 29, 379–390. doi: 10.1038/s41422-019-0145-8
Liu, J., Feng, L., Li, J., and He, Z. (2015a). Genetic and epigenetic control of plant heat responses. Front. Plant Sci. 6:267. doi: 10.3389/fpls.2015.00267
Liu, H. C., Liao, H. T., and Charng, Y. Y. (2011). The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 34, 738–751. doi: 10.1111/j.1365-3040.2011.02278.x
Liu, F., Wang, W., Sun, X., Liang, Z., and Wang, F. (2015b). Conserved and novel heat stress-responsive microRNAs were identified by deep sequencing in Saccharina japonica (Laminariales, Phaeophyta). Plant Cell Environ. 38, 1357–1367. doi: 10.1111/pce.12484
Liu, Y., Yan, J., Wang, K., Li, D., Yang, R., Luo, H., et al. (2021). MiR396-GRF module associates with switchgrass biomass yield and feedstock quality. Plant Biotechnol. J. doi: 10.1111/pbi.13567 [Epub ahead of print]
Liu, Q., Yan, S., Yang, T., Zhang, S., Chen, Y., and Liu, B. (2017a). Small RNAs in regulating temperature stress response in plants. J. Integr. Plant Biol. 59, 774–791. doi: 10.1111/jipb.12571
Liu, Q., Yang, T. F., Yu, T., Zhang, S., Mao, X., Zhao, J., et al. (2017b). Integrating small RNA sequencing with QTL mapping for identification of miRNAs and their target genes associated with heat tolerance at the flowering stage in rice. Front. Plant Sci. 8:43. doi: 10.3389/fpls.2017.00043
Llave, C., Xie, Z., Kasschau, K. D., and Carrington, J. C. (2002). Cleavage of scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297, 2053–2056. doi: 10.1126/science.1076311
Lobell, D. B., Schlenker, W., and Costa-Roberts, J. (2011). Climate trends and global crop production since 1980. Science 333, 616–620. doi: 10.1126/science.1076311
Ma, J., Zhao, P., Liu, S., Yang, Q., and Guo, H. (2020). The control of developmental phase transitions by microRNAs and their targets in seed plants. Int. J. Mol. Sci. 21:1971. doi: 10.3390/ijms21061971
Mangrauthia, S. K., Bhogireddy, S., Agarwal, S., Prasanth, V. V., Voleti, S. R., Neelamraju, S., et al. (2017). Genome-wide changes in microRNA expression during short and prolonged heat stress and recovery in contrasting rice cultivars. J. Exp. Bot. 68, 2399–2412. doi: 10.1093/jxb/erx111
Matthews, C., Arshad, M., and Hannoufa, A. (2019). Alfalfa response to heat stress is modulated by microRNA156. Physiol. Plant. 165, 830–842. doi: 10.1111/ppl.12787
Matzke, M. A., and Mosher, R. A. (2014). RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15, 394–408. doi: 10.1038/nrg3683
May, P., Liao, W., Wu, Y., Shuai, B., Mccombie, W. R., Zhang, M. Q., et al. (2013). The effects of carbon dioxide and temperature on microRNA expression in Arabidopsis development. Nat. Commun. 4:2145. doi: 10.1038/ncomms3145
Meyers, B. C., Simon, S. A., and Zhai, J. (2010). MicroRNA processing: battle of the bulge. Curr. Biol. 20, R68–R70. doi: 10.1016/j.cub.2009.12.008
Murray, F., Kalla, R., Jacobsen, J., and Gubler, F. (2003). A role for HvGAMYB in anther development. Plant J. 33, 481–491. doi: 10.1046/j.1365-313X.2003.01641.x
Ohama, N., Sato, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2017). Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 22, 53–65. doi: 10.1016/j.tplants.2016.08.015
Pagano, L., Rossi, R., Paesano, L., Marmiroli, N., and Marmiroli, M. (2021). miRNA regulation and stress adaptation in plants. Environ. Exp. Bot. 184:104369. doi: 10.1016/j.envexpbot.2020.104369
Palatnik, J. F., Wollmann, H., Schommer, C., Schwab, R., Boisbouvier, J., Rodriguez, R., et al. (2019). Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev. Cell 51:129. doi: 10.1016/j.devcel.2019.09.016
Pan, C., Ye, L., Zheng, Y., Wang, Y., Yang, D., Liu, X., et al. (2017). Identification and expression profiling of microRNAs involved in the stigma exsertion under high-temperature stress in tomato. BMC Genomics 18:843. doi: 10.1186/s12864-017-4238-9
Pandey, R., Joshi, G., Bhardwaj, A. R., Agarwal, M., and Katiyar-Agarwal, S. (2014). A comprehensive genome-wide study on tissue-specific and abiotic stress-specific mirnas in Triticum aestivum. PLoS One 9:e95800. doi: 10.1371/journal.pone.0095800
Peng, Y., Zhang, X., Liu, Y., and Chen, X. (2020). Exploring heat-response mechanisms of microRNAs based on microarray data of rice post-meiosis panicle. Int. J. Genomics 17:7582612. doi: 10.1155/2020/7582612
Proveniers, M. C., and Van Zanten, M. (2013). High temperature acclimation through PIF4 signaling. Trends Plant Sci. 18, 59–64. doi: 10.1016/j.tplants.2012.09.002
Qu, A. L., Ding, Y. F., Jiang, Q., and Zhu, C. (2013). Molecular mechanisms of the plant heat stress response. Biochem. Biophys. Res. Commun. 432, 203–207. doi: 10.1016/j.bbrc.2013.01.104
Ragupathy, R., Ravichandran, S., Mahdi, M. S., Huang, D., Reimer, E., Domaratzki, M., et al. (2016). Deep sequencing of wheat srna transcriptome reveals distinct temporal expression pattern of mirnas in response to heat, light and UV. Sci. Rep. 6:39373. doi: 10.1038/srep39373
Rai, K. K., Pandey, N., and Rai, S. P. (2020). Salicylic acid and nitric oxide signaling in plant heat stress. Physiol. Plant. 168, 241–255. doi: 10.1111/ppl.12958
Ramachandran, V., and Chen, X. (2008). Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science 321, 1490–1492. doi: 10.1126/science.1163728
Ravichandran, S., Ragupathy, R., Edwards, T., Domaratzki, M., and Cloutier, S. (2019). MicroRNA-guided regulation of heat stress response in wheat. BMC Genomics 20:488. doi: 10.1186/s12864-019-5799-6
Ruiz-Ferrer, V., and Voinnet, O. (2009). Roles of plant small RNAs in biotic stress responses. Annu. Rev. Plant Biol. 60, 485–510. doi: 10.1146/annurev.arplant.043008.092111
Sailaja, B., Voleti, S. R., Subrahmanyam, D., Sarla, N., Prasanth, V. V., Bhadana, V. P., et al. (2014). Prediction and expression analysis of miRNAs associated with heat stress in Oryza sativa. Rice Sci. 21, 3–12. doi: 10.1016/S1672-6308(13)60164-X
Schauer, S. E., Jacobsen, S. E., Meinke, D. W., and Ray, A. (2002). DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci. 7, 487–491. doi: 10.1016/S1360-1385(02)02355-5
Shi, X., Jiang, F., Wen, J., and Wu, Z. (2019). Overexpression of Solanum habrochaites microRNA319d (sha-miR319d) confers chilling and heat stress tolerance in tomato (S. lycopersicum). BMC Plant Biol. 19:214. doi: 10.1186/s12870-019-1823-x
Shriram, V., Kumar, V., Devarumath, R. M., Khare, T. S., and Wani, S. H. (2016). MicroRNAs as potential targets for abiotic stress tolerance in plants. Front. Plant Sci. 7:817. doi: 10.3389/fpls.2016.00817
Si-Ammour, A., Windels, D., Arn-Bouldoires, E., Kutter, C., Ailhas, J., Meins, F., et al. (2011). miR393 and secondary siRNAs regulate expression of the TIR1/AFB2 auxin receptor clade and auxin-related development of Arabidopsis leaves. Plant Physiol. 157, 683–691. doi: 10.1104/pp.111.180083
Song, X., Li, P., Zhai, J., Zhou, M., Ma, L., Liu, B., et al. (2012). Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. Plant J. 69, 462–474. doi: 10.1111/j.1365-313X.2011
Stief, A., Altmann, S., Hoffmann, K., Pant, B. D., Scheible, W. R., and Bäurle, I. (2014a). Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 26, 1792–1807. doi: 10.1105/tpc.114.123851
Stief, A., Brzezinka, K., Lämke, J., and Bäurle, I. (2014b). Epigenetic responses to heat stress at different time scales and the involvement of small RNAs. Plant Signal. Behav. 9:e970430. doi: 10.4161/15592316.2014.970430
Sugio, A., Dreos, R., Aparicio, F., and Maule, A. J. (2009). The cytosolic protein response as a subcomponent of the wider heat shock response in Arabidopsis. Plant Cell 21, 642–654. doi: 10.1105/tpc.108.062596
Sun, X., Zhu, J., Li, X., Li, Z., Han, L., and Luo, H. (2020). AsHSP26.8a, a creeping bentgrass small heat shock protein integrates different signaling pathways to modulate plant abiotic stress response. BMC Plant Biol. 20:184. doi: 10.1186/s12870-020-02369-5
Sunkar, R., and Zhu, J.-K. (2004). Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16, 2001–2019. doi: 10.1105/tpc.104.022830
Tu, B., Liu, L., Xu, C., Zhai, J., Li, S., Lopez, M. A., et al. (2015). Distinct and cooperative activities of HESO1 and URT1 nucleotidyl transferases in microRNA turnover in Arabidopsis. PLoS Genet. 11:e1005119. doi: 10.1371/journal.pgen.1005119
Wang, J., Gao, X., Dong, J., Tian, X., Wang, J., Palta, J. A., et al. (2020). Over-expression of the heat-responsive wheat gene TaHSP23.9 in transgenic Arabidopsis conferred tolerance to heat and salt stress. Front. Plant Sci. 11:243. doi: 10.3389/fpls.2020.00243
Wang, Q., Liu, N., Yang, X., Tu, L., and Zhang, X. (2016). Small RNA-mediated responses to low- and high-temperature stresses in cotton. Sci. Rep. 6:35558. doi: 10.1038/srep35558
Wang, Y., Sun, F., Cao, H., Peng, H., Ni, Z., Sun, Q., et al. (2012). TamiR159 directed wheat TaGAMYB cleavage and its involvement in anther development and heat response. PLoS One 7:e48445. doi: 10.1371/journal.pone.0048445
Wang, N., Zheng, J., Chen, Z., Liu, Y., Dura, B., Kwak, M., et al. (2019). Single-cell microRNA-mRNA co-sequencing reveals non-genetic heterogeneity and mechanisms of microRNA regulation. Nat. Commun. 10:95. doi: 10.1038/s41467-018-07981-6
Wong, J., Gao, L., Yang, Y., Zhai, J., Arikit, S., Yu, Y., et al. (2014). Roles of small RNAs in soybean defense against Phytophthora sojae infection. Plant J. 79, 928–940. doi: 10.1111/tpj.12590
Wu, G., Park, M. Y., Conway, S. R., Wang, J. W., Weigel, D., and Poethig, R. S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750–759. doi: 10.1016/j.cell.2009.06.031
Xia, K., Zeng, X., Jiao, Z., Li, M., Xu, W., Nong, Q., et al. (2018). Formation of protein disulfide bonds catalyzed by OsPDIL1; 1 is mediated by MicroRNA5144-3p in rice. Plant Cell Physiol. 59, 331–342. doi: 10.1093/pcp/pcx189
Xie, Z., Allen, E., Fahlgren, N., Calamar, A., Givan, S. A., and Carrington, J. C. (2005). Expression of Arabidopsis MIRNA genes. Plant Physiol. 138, 2145–2154. doi: 10.1104/pp.105.062943
Xin, M., Wang, Y., Yao, Y., Xie, C., Peng, H., Ni, Z., et al. (2010). Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC Plant Biol. 10:123. doi: 10.1186/1471-2229-10-123
Xu, M., Hu, T., Zhao, J., Park, M. Y., Earley, K. W., Wu, G., et al. (2016). Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genet. 12:e1006263. doi: 10.1371/journal.pgen.1006263
Yan, K., Liu, P., Wu, C. A., Yang, G. D., Xu, R., Guo, Q. H., et al. (2012). Stress-induced alternative splicing provides a mechanism for the regulation of microRNA processing in Arabidopsis thaliana. Mol. Cell 48, 521–531. doi: 10.1016/j.molcel.2012.08.032
Yin, Z., Li, Y., Zhu, W., Fu, X., Han, X., Wang, J., et al. (2018). Identification, characterization, and expression patterns of TCP genes and microRNA319 in cotton. Int. J. Mol. Sci. 19:3655. doi: 10.3390/ijms19113655
Yoshida, T., Ohama, N., Nakajima, J., Kidokoro, S., Mizoi, J., Nakashima, K., et al. (2011). Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol. Gen. Genomics. 286, 321–332. doi: 10.1007/s00438-011-0647-7
Yoshikawa, M., Iki, T., Tsutsui, Y., Miyashita, K., Poethig, R. S., Habu, Y., et al. (2013). 3' fragment of miR173-programmed RISC-cleaved RNA is protected from degradation in a complex with RISC and SGS3. Proc. Natl. Acad. Sci. U. S. A. 110, 4117–4122. doi: 10.1073/pnas.1217050110
Yu, B., Bi, L., Zheng, B., Ji, L., Chevalier, D., Agarwal, M., et al. (2008). The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc. Natl. Acad. Sci. U. S. A. 105, 10073–10078. doi: 10.1073/pnas.0804218105
Yu, Y., Mo, X., and Mo, B. (2020). “Introduction to plant small RNAs,” in Plant Small RNA. eds. P. Guleria and V. Kumar (London: Academic Press), 3–35.
Yu, X., Wang, H., Lu, Y., De Ruiter, M., Cariaso, M., Prins, M., et al. (2012). Identification of conserved and novel microRNAs that are responsive to heat stress in brassica rapa. J. Exp. Bot. 63, 1025–1038. doi: 10.1093/jxb/err337
Yu, B., Yang, Z., Li, J., Minakhina, S., Yang, M., Padgett, R. W., et al. (2005). Methylation as a crucial step in plant microRNA biogenesis. Science 307, 932–935. doi: 10.1126/science.1107130
Yu, Y., Zhang, Y., Chen, X., and Chen, Y. (2019). Plant noncoding RNAs: hidden players in development and stress responses. Annu. Rev. Cell Dev. Biol. 35, 407–431. doi: 10.1146/annurev-cellbio-100818-125218
Yu, Y., Zhou, Y., Zhang, Y., and Chen, Y. (2018). Grass phasiRNAs and male fertility. Sci. China Life Sci. 61, 148–154. doi: 10.1007/s11427-017-9166-3
Zhai, J., Arikit, S., Simon, S. A., Kingham, B. F., and Meyers, B. C. (2014). Rapid construction of parallel analysis of RNA end (PARE) libraries for Illumina sequencing. Methods 67, 84–90. doi: 10.1016/j.ymeth.2013.06.025
Zhao, J., He, Q., Chen, G., Wang, L., and Jin, B. (2016). Regulation of non-coding RNAs in heat stress responses of plants. Front. Plant Sci. 7:1213. doi: 10.3389/fpls.2016.01213
Zhao, J., Lu, Z., Wang, L., and Jin, B. (2021). Plant responses to heat stress: physiology, transcription, noncoding RNAs, and epigenetics. Int. J. Mol. Sci. 22:117. doi: 10.3390/ijms22010117
Zhao, Y., Yu, Y., Zhai, J., Ramachandran, V., Dinh, T. T., Meyers, B. C., et al. (2012). The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr. Biol. 22, 689–694. doi: 10.1016/j.cub.2012.02.051
Zhao, J., Yuan, S., Zhou, M., Yuan, N., Li, Z., Hu, Q., et al. (2019). Transgenic creeping bentgrass overexpressing Osa-miR393a exhibits altered plant development and improved multiple stress tolerance. Plant Biotechnol. J. 17, 233–251. doi: 10.1111/pbi.12960
Zhong, S. H., Liu, J. Z., Jin, H., Lin, L., Li, Q., Chen, Y., et al. (2013). Warm temperatures induce transgenerational epigenetic release of RNA silencing by inhibiting siRNA biogenesis in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 110, 9171–9176. doi: 10.1073/pnas.1219655110
Zhou, R., Wang, Q., Jiang, F., Cao, X., Sun, M., Liu, M., et al. (2016). Identification of miRNAs and their targets in wild tomato at moderately and acutely elevated temperatures by high-throughput sequencing and degradome analysis. Sci. Rep. 6:33777. doi: 10.1038/srep33777
Zhu, J.-K. (2016). Abiotic stress signaling and responses in plants. Cell 167, 313–324. doi: 10.1016/j.cell.2016.08.029
Zhu, H., Zhang, Y., Tang, R., Qu, H., Duan, X., and Jiang, Y. (2019). Banana sRNAome and degradome identify microRNAs functioning in differential responses to temperature stress. BMC Genomics 20:33. doi: 10.1186/s12864-018-5395-1
Keywords: microRNA, small interfering RNA, heat stress response, regulatory mechanism, thermotolerance
Citation: Zuo Z-F, He W, Li J, Mo B and Liu L (2021) Small RNAs: The Essential Regulators in Plant Thermotolerance. Front. Plant Sci. 12:726762. doi: 10.3389/fpls.2021.726762
Received: 17 June 2021; Accepted: 11 August 2021;
Published: 17 September 2021.
Edited by:
Mingli Xu, University of South Carolina, United StatesReviewed by:
Bin Yu, University of Nebraska-Lincoln, United StatesCopyright © 2021 Zuo, He, Li, Mo and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin Liu, bGlubGl1QHN6dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.