- Department of Entomology and Plant Pathology, Auburn University, Auburn, AL, United States
12-oxo-Phytodienoic acid (OPDA) is a primary precursor of (-)-jasmonic acid (JA), able to trigger autonomous signaling pathways that regulate a unique subset of jasmonate-responsive genes, activating and fine-tuning defense responses, as well as growth processes in plants. Recently, a number of studies have illuminated the physiol-molecular activities of OPDA signaling in plants, which interconnect the regulatory loop of photosynthesis, cellular redox homeostasis, and transcriptional regulatory networks, together shedding new light on (i) the underlying modes of cellular interfaces between growth and defense responses (e.g., fitness trade-offs or balances) and (ii) vital information in genetic engineering or molecular breeding approaches to upgrade own survival capacities of plants. However, our current knowledge regarding its mode of actions is still far from complete. This review will briefly revisit recent progresses on the roles and mechanisms of OPDA and information gaps within, which help in understanding the phenotypic and environmental plasticity of plants.
Phyto-Oxylipins: Jasmonate Signaling in Plants
Oxylipins, the oxygenated derivative of fatty acids (FAs), are critical signal molecules in diverse physiological processes in life, including plants and animals (Marnett, 2008). In plants, oxylipins are involved in a layer of defense and ontogenetic pathways, while mammalian oxylipins (eicosanoids) control intricate regulatory mechanisms in immunity, functioning as messengers in the central nervous system, and participating in the resolution process following tissue injury (Funk, 2001; Mosblech et al., 2010). Recent studies, moreover, have illuminated the medicinal values of phyto-oxylipins, presenting their anticancer, anti-inflammatory, and antioxidative activities (Flescher, 2007; Dang et al., 2008; Taki-Nakano et al., 2014). Noticeably, the molecular components and metabolic pathways, involved in oxylipin biogenesis and signaling, share common ancestry and evolutionary processes across Kingdoms (Marnett, 2008). Hence, uncovering the modes of actions associated with oxylipins will not only assist the development of agricultural strategies in advancing disease resistance and stress adaptation, as well as yield and biomass increases in plants, but also assist the improvement of drug development through facilitating the rational design of more potent and safe anticancer (and anti-inflammation) drugs. However, our current knowledge regarding oxylipin signaling is still incomplete, despite decades of investigations (Funk, 2001; Mosblech et al., 2010).
Lately, molecular underpinnings have been investigated for 12-oxo-phytodienoic acid (OPDA) signaling in plant defense responses. OPDA is a primary precursor of the jasmonate family of oxylipins, which includes jasmonic acid (JA) and its precursors and derivatives. Jasmonates are derived from trienoic-FA via the octadecanoid pathway in the chloroplasts. Lipase-mediated oxidation of trienoic-FA leads to the release of OPDA that travels to the peroxisomes through plastid envelope proteins (e.g., OPDAT1 and JASSY, Guan et al., 2019; Zhao et al., 2020) and/or peroxisomal ATP-binding transporters (e.g., COMATOSE, Theodoulou et al., 2005) and undergoes β-oxidations to form JA. JA can be further metabolized to several derivatives, including JA-isoleucine (JA-Ile), JA-tryptophan (JA-Trp), methyl-JA, and hydroxyl-JA. Signaling of these jasmonate molecules then controls a large number of gene expressions in the nucleus and mediates defense (adaptive) responses to various forms of biotic and abiotic stresses, including microbial pathogens and insect herbivores, tissue injury, and light damage. Jasmonate signaling also plays essential roles in reproduction and other developmental processes such as senescence, root growth and tuberization, fruit ripening, and tendril coiling (reviewed in Acosta and Farmer, 2010; Pieterse et al., 2012). These important, yet diverse activities of jasmonates must be tied to their versatility as major molecular and cellular modulators.
The most well-characterized jasmonate-associated signaling pathway revolves around JA-Ile. Once it is produced, JA-Ile binds a F-box protein, CORONATINE INSENSITIVE 1 (COI1, a part of SCF ubiquitin E3 ligase). This complex then binds and ubiquitinates jasmonate ZIM-domain (JAZ) proteins, which are negative transcription regulators of JA-responsive genes (JRGs). Thus, JAZ degradation by 26S proteasomes frees transcription factors (TFs, e.g., bHLH-containing MYCs) and allows subsequent gene expressions (Chini et al., 2007; Thines et al., 2007). Jasmonate signaling, however, must involve a much more complex network, given that a number of JRGs respond independently of COI1 (Devoto et al., 2005). For example, JA induction of GRX480 and AOC3 is mediated via a COI1-independent MYC2 regulatory pathway, whereas JA-activated MAP kinase cascades (e.g., MPK1, MPK2 and BIK1) and GST25 are regulated in a COI1- and/or MYC2-independent manner (Veronese et al., 2006; Ortiz-Masia et al., 2007; Stotz et al., 2013). In addition, OPDA is capable of triggering autonomous signaling pathways that regulate unique subsets of JRGs, coordinated with and without the canonical JA pathway (Taki et al., 2005). OPDA signaling is presumed to be independent of COI1, as it is unable to bind the COI1/JAZ complex (Thines et al., 2007). However, OPDA induction of PHO1;H10 needs COI1 activity (Ribot et al., 2008), suggesting additional layers of complexity in jasmonate signaling. In fact, ancestral plants such as the bryophyte Marchantia polymorpha are able to synthesize only a set of OPDAs (OPDA, dinor-cis-OPDA and dinor-iso-OPDA), but not JA/JA-Ile, though their genomes still express a functional COI1 (Monte et al., 2018). Hence, M. polymorpha deploys OPDAs, instead of JA-Ile, to activate COI1/JAZ signaling for defense activations and growth processes (Monte et al., 2018, 2019). The other study also established a distinct role of JA-Trp conjugate, linking jasmonate with auxin signaling (Staswick, 2009), further supporting the notion that distinct messages sent out by specific jasmonate coordinate essential molecular and cellular processes.
Biosynthesis of OPDA and its Derivatives
As alluded, jasmonates are synthesized in the chloroplasts from oxygenized FAs, linolenic acid (18:3) and hexadecatrienoic acid (16:3), that are stored mostly as the esterified monogalactosyldiacylglycerol (MGDG). The first step, hydroperoxidation, is began by 13-lipoxygenases adding molecular oxygen to 18:3 and 16:3 and forming 13(S)-hydroperoxy-octadecatrienoic acid and 11(S)-hydroperoxy-hexadecatrienoic acid, respectively. These compounds are then transformed via allene oxide synthase into (13S)12,13-epoxy-octadecatrienoic acid and (11S)10,11-epoxy-octadecatrienoic acid, which are subsequently cyclized through allene oxide cyclase to yield cis-(+)-OPDA and dinor-OPDA (collectively, OPDA); containing a reactive electrophilic α,β-unsaturated carbonyl group. These metabolic pathways are known to be activated in response to various herbivories and microbial pathogens, as well as abiotic stresses such as extreme temperatures and tissue injury (Stintzi et al., 2001; Kourtchenko et al., 2007; Vu et al., 2012; Bosch et al., 2014a,b; Monte et al., 2020). Some portion of OPDA is then further derivatized to a glutathione (GSH) conjugate, or galactolipids (later named “arabidopsides”) by its binding with MGDG and digalactosyl DG (Hisamatsu et al., 2003, 2005; Davoine et al., 2005, 2006; Andersson et al., 2006; Buseman et al., 2006). The biological roles of OPDA-GSH and arabidopsides are yet largely elusive, but have been hypothesized as the vacuolar delivery and storage forms, respectively, in maintaining the cellular-level homeostasis of OPDA to avoid their potential toxicity and/or negative effects on physiol-molecular processes in plants (Böttcher and Pollmann, 2009; Ohkama-Ohtsu et al., 2011). Alternatively, recent studies have been suggested that arabidopsides could interact with plant plasma membrane lipids such as glycosyl inositol phosphor ceramides, which thus lead them to locate and modify membrane organizations, and such changes could signal defense mechanism activations (Genva et al., 2019).
Signaling of OPDA in Plant Defense Responses
In plants, OPDA signaling plays intrinsic roles in activating and fine-tuning defense (adaptive) responses against an array of biotic and abiotic stresses, as well as growth processes (Böttcher and Pollmann, 2009; Dave and Graham, 2012; Maynard et al., 2018). Its distinctive activity in plant defense activations was first described by the pathoanalyses of a mutant Arabidopsis plant (opr3) arresting the conversion of OPDA to JA/JA-Ile (Stintzi et al., 2001). WT-like resistance of opr3, in contrast to enhanced susceptibility in other mutants disrupting trienoic-FA biosynthesis (fad3/7/8) and the octadecanoid pathway (dde2 and aos), against fungal pathogens (Alternaria brassicicola and Scerotinia sclerotiarum) and an insect herbivory (Bradysia impatiens), underlined a critical activity of OPDA signaling in plant disease resistance in the absence of JA/JA-Ile (Stintzi et al., 2001; Zhang and Turner, 2008; Stotz et al., 2011). Following studies with genetically modified (GM) plants reducing or impairing JA productions (OPR3-RNAi, SiOPR3s, and opr7opr8) or enhancing OPDA accumulations (OPR3ox) further substantiate that OPDA signaling is essential for the full activation of basal defense responses in tomato, maize, and rice against microbial and/or pest attacks such as Botrytis cinerea, tobacco hornworm (Manduca sexta larvae), beet armyworm (Spodoptera exigua larvae), brown plant hopper (Nilaparvata lugens), green peach aphid (Myxus persicae), and corn leaf aphid (Rhopalosiphum maidis) (Bosch et al., 2014a,b; Guo et al., 2014; Scalschi et al., 2015; Varsani et al., 2019; Grover et al., 2020b; Wang et al., 2020). Upon their infections, OPDA is induced rapidly in the chloroplasts and triggers the retrograde signaling toward the nucleus, which coordinates large-scale changes in defense gene expressions (Taki et al., 2005). These then lead to (i) the spatiotemporal induction of protease inhibitors (PIs) such as miraculin-like proteins, which likely serve as antinutrients against insect attackers by reducing their digestibility of dietary proteins (Felton, 2005; López-Galiano et al., 2017), (ii) the actuation of other hormone and metabolite biosynthesis (Figure 1) in maximizing defense capacity and survival of plants, and (iii) the stimulation of callose deposition (Scalschi et al., 2015; Varsani et al., 2019), a multifaceted cell wall barrier developed at the sites of infection, preventing the cell-to-cell spread of microbes and limiting the feeding capacity and colonization of insects (Luna et al., 2010; De Storme and Geelen, 2014). OPDA signaling appeared to trigger abscisic acid (ABA) accumulations (Dave et al., 2016) that activate a NADPH oxidase subunit of RBOHF (Respiratory Burst Oxidase Homolog Protein F) leading to transient reactive oxygen species (ROS) productions (Sirichandra et al., 2009; Figure 1) and in consequence stimulating callose synthesis (Luna et al., 2010). However, OPDA signal alone did not elevate the expression levels of callose synthase gene such as Tie-dyed2 in maize, suggesting rather the need of additional or alternative, yet unknown, defense and/or OPDA-inducible element(s), perhaps free thiols such as GSH and glucosinolates (Park et al., 2013; Zhou and Memelink, 2016; Varsani et al., 2019).
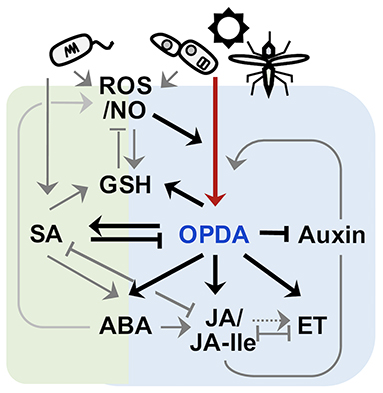
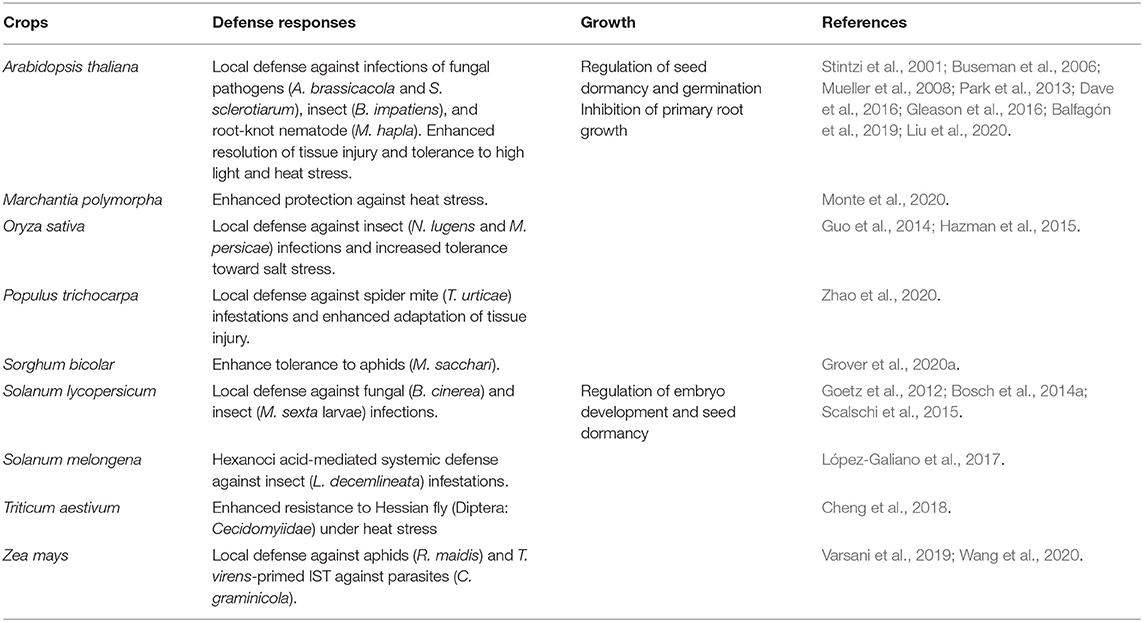
Figure 1. Schematic crosstalk of OPDA with the signaling pathways of other phytohormones. OPDA and JA/JA-Ile (collectively, jasmonate) signaling activates defense responses to various forms of biotic and abiotic stresses, including necrotroph infection, insect attack, wounding, and UV damage, whereas salicylic acid (SA) signaling confers disease resistance against mostly biotrophic pathogens. Once recognized “non-self,” plants rapidly produce ROS including nitric oxide (NO) (Huang et al., 2004; Wang et al., 2013), which in turn actuates jamonate biosynthesis (Palmieri et al., 2008). OPDA signaling then stimulates and coordinates with different defense signaling, e.g., JA/JA-Ile, SA, ABA, ethylene (ET), and GSH pathways (Taki et al., 2005; Park et al., 2013; Dave et al., 2016; Varsani et al., 2019; Scalschi et al., 2020), while suppressing growth hormone (e.g., auxin) signaling (Taki et al., 2005) to optimize defense capability of plants. On the other hand, JA/JA-Ile signaling can feedback induce own jasmonate biosynthesis (Taki et al., 2005), whereas SA signaling antagonizes both OPDA and JA/JA-Ile signaling (Leon-Reyes et al., 2010; Shim et al., 2013; Wei et al., 2014).
Indeed, it is still largely elusive how OPDA is perceived for signaling. Recently, in search of OPDA derivatives potentially binding SCFCOI, OPDA conjugated with Ile (OPDA-Ile) was identified in Arabidopsis (Floková et al., 2016) and described its ability to induce OPDA-responsive genes (ORGs) such as GRX480 and ZAT10 in JA/JA-Ile-deficient mutant (opr3 and jar1) plants (Arnold et al., 2016). The latter suggests that OPDA-Ile is a bioactive signal and conveys JA/JA-Ile-independent, OPDA-dependent signaling pathway. OPDA-Ile is though only active under specific conditions as it was found exclusively in wounded leaves of flowering plants (Floková et al., 2016). It would be interesting to delineate mechanisms underlying the perception of OPDA-Ile and its cross-networking with other OPDA signaling pathways (see “Summary: Mode of Action of OPDA Signaling”).
Besides its roles in local defenses, a new study has proposed that OPDA is a long-distance signal for “induced systemic resistance” (ISR) (Wang et al., 2020), a state of heightened defense that is activated throughout the plant following an initial encounter with plant growth-promoting rhizobacteria/fungi (PGPR/F) (Pieterse et al., 2014). Their oxylipin profiling of xylem saps collected from ISR-induced maize leaves detected uniquely OPDA and KODA (α-ketol-octadecadienoic acid). In addition, the transfusion of OPDA and KODA into naïve plants led to develop systemic resistance against an anamorphic fungus Collectotrichum graminicola in a dose-dependent manner (Wang et al., 2020), together proposing their role in conveying ISR signaling. A caveat is that OPDA appears to be stationary exhibiting little or no distal accumulation under pathogen attacks (Christensen et al., 2015). This speculates if an ISR receptor of OPDA is present in xylem. Alternatively, OPDA may be rapidly converted to and activate JA/JA-Ile signaling, upon arrival to systemic tissues, for priming systemic defense (Koo et al., 2009; Bosch et al., 2014a). In this context, an earlier grafting experiment using WT and JA/JA-deficient (OPR3-RNAi) plants showed that OPDA can substitute for JA/JA-Ile in the local induction of defense gene expressions, but the production of JA/JA-Ile is required for systemic responses (Bosch et al., 2014b). It will be intriguing to find out whether OPDA is “truly” moved from local to vascular to systemic tissues, and if OPDA can autonomously signal ISR priming or is converted to JA/JA-Ile for ISR development. In addition, we cannot still rule out a potential role of phloem in channeling a mobile signal of ISR development (Varsani et al., 2019). Perhaps, ISR may also involve multiple signals and transduction pathways as does in systemic acquired resistance (Klessig et al., 2018).
On the other hand, a recent report argued that only a biologically active jasmonate molecule is JA-Ile (Chini et al., 2018). Using a new mutant allele (opr3-3) completely lacking OPR3 reductase activity, the study demonstrated the increased accumulation levels (~fifteen-folds) of non-reduced cyclopente none, 4,5-didehydrojasmoante, in opr3 and its provisional reduction to JA by one of OPR3 isoforms, OPR2 reductase, together postulating that WT-like resistance of opr3 is actuated not by OPDA signaling, rather by COI1-dependent JA-Ile signaling. However, the OPR3-independent pathway of JA biosynthesis appeared to contribute to the accumulation of dismal amounts of JA-Ile (<2.0 % [less than its basal levels] of WT) under stress conditions, while conferring tenable strength defense responses against pathogen infections, prompting speculation that opr3-3 mutants may exert alternative, OPDA-associated defense pathways. In fact, coi1 mutants feedback suppress JA biosynthesis so that lack stress-induced OPDA and JA accumulations (Chung et al., 2008; Park et al., 2013). Thus, coi1-like increased susceptibility shown in coi1/opr3-3 against insect and fungal attacks (Chini et al., 2018) might be, not because WT-like resistance of opr3-3 requires COI1, due to auxiliary side effects led by double mutagenesis, perhaps lowering the level threshold of OPDA and JA-Ile signaling.
Signaling of OPDA in Plant Growth and Developmental Processes
An earlier study of COMATOSE, a peroxisomal ATP-binding cassette transporter, and its mutant plants (cts) disrupted the transport of OPDA into the peroxisome, where JA biosynthesis occurs, illuminated a critical activity of OPDA signaling in coordinating seed germination and dormancy (Russell et al., 2000). The mutant cts seeds exhibited increased accumulation level of OPDA and low germination rates (Russell et al., 2000; Dave et al., 2011), while exogenous OPDA applications stimulated the repression of the germination of WT seeds (Dave et al., 2011). Such an inhibitory effect of OPDA signaling is perhaps mediated through its activation of ABA biosynthesis by upregulating the expression of an ABA biosynthesis gene (ABA1 and ABA-deficient 1) and an inducer (RGL2, Repressor of Gibberellic Acid-like 2) of RING-H2 XERICO (ABA biosynthesis regulator) (Ko et al., 2006; Piskurewicz et al., 2008; Dave et al., 2016). OPDA and ABA both are then able to induce and/or stabilize the activity of GRL2 and ABI5 (ABA insensitive 5) bZIP TF, which in subsequence promotes the expression of MET (Mother-of FT and TFL1), an inhibitor of seed germination or early seedling growth (Skubacz et al., 2016; Vaistij et al., 2018), so that it suppresses seed germinations (Dave et al., 2016; Barros-Galvão et al., 2019). Two hormones, however, displayed different mechanistic outcomes that ABA signal ruptures seed coats and endosperm tissues, whereas OPDA-treated seeds keep intact endosperm and seed coats (Dave et al., 2011), indicating that OPDA signal, besides coordinating ABA biosynthesis/signaling, can execute its autonomous, unique regulatory metabolic pathways in plant organismal development.
OPDA-responsive ABA accumulations also convey the inhibition of root growth and morphogenesis in plants (Mueller et al., 2008; Park et al., 2013; Sun et al., 2018; Vissenberg et al., 2020). ABA could suppress primary root growth and lateral root branching, mediated via balancing the cellular homeostasis of several growth components (Arc et al., 2013; Sun et al., 2018) that enhance the production of ROS, Ca2+, and ethylene but reduce auxin levels (Wang et al., 2002; He et al., 2012; Jiao et al., 2013; Luo et al., 2014). These changes then stimulate the expression of PLETHORA TFs, rhizotatic regulators, and some cell cycle-related genes (e.g., Cyclin-dependent Protein Kinase and Cell Cycle B-type Cyclins), thus affecting DNA replication, cell division, and cell elongation in roots and inhibiting root growth (Wang et al., 2008, 2011; Yin et al., 2009; Xu et al., 2010; Hofhuis et al., 2013; Yao et al., 2013). However, the effects of OPDA signal on roots did also not entirely depend on ABA signaling. Our recent study indicated that OPDA signaling could act as a positive regulator in root growth and development (Figure 2). Disruption of OPDA signaling in Arabidopsis (cyp20-3, Park et al., 2013) engendered the impairment of root hair growth. It is though unclear if the opposite is correct; the increased accumulation level of OPDA under stress conditions could enhance root growth and branching; further studies are needed to reconstitute the complete, functional networks of OPDA signaling in plant growth and development.
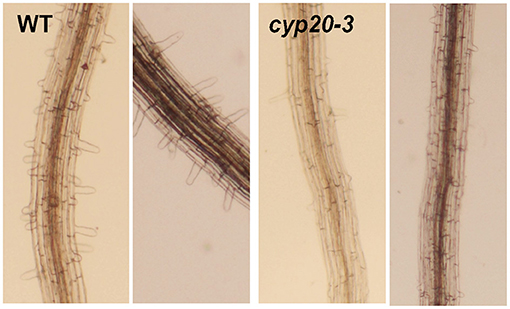
Figure 2. Important roles of OPDA signaling in root morphogenesis. OPDA signaling mutant Arabidopsis (cyp20-3) demonstrated the impairment of root hair growth.
A Mode of OPDA Signaling by its Binding Protein Cyclophilin 20-3
Previously, our search of jasmonate receptors uncovered that a small plastid protein, cyclophilin 20-3 (CYP20-3), can physically interact with OPDA, and its T-DNA insertion mutant Arabidopsis (cyp20-3) attenuates the expression of ORGs (Park et al., 2013). The Arabidopsis genome encodes 29 CYP and CYP-like proteins, belonging to the family of, namely, immunophilins that possess binding abilities toward immunosuppressive drugs, cyclosporin A, and functions in broad ranges of cellular processes, including transcriptional regulation, organogenesis, photosynthetic and hormone signaling pathways, stress adaptation, and defense responses (Dos Santos and Park, 2019). CYP20-3 is the only isoform localized in the chloroplast stroma and acts as a dual-enzyme able to chaperone protein folding (peptidyl-prolyl cis-trans isomerase; PPlase) and transfers electrons (e−) to peroxide substrates (reductase) in cysteine (Cys) biosynthesis (i.e., sulfur assimilation; Romano et al., 2004; Laxa et al., 2007; Dominguez-Solis et al., 2008; Park et al., 2013). In line with this scenario, OPDA, once accumulated under stress states, binds and stimulates CYP20-3 to form a complex with serine acetyltransferase1 (SAT1), which triggers the formation of a hetero-oligomeric Cys synthase complex (CSC) with O-acetylserine(thiol)lyase B (OASTL-B) (Figure 3, left side). CSC formation then leads to the production of Cys and subsequently thiol metabolites (e.g., GSH), which builds up cellular reduction potentials. The enhanced redox capacity in turn coordinates the expression of a subset of ORGs that activate and calibrate pathogen defense and stress adaptation processes. Thus, the KO of CYP20-3 (cyp20-3) displays enhanced susceptibility against necrotrophic fungal (e.g., A. brassicicola and B. cinerea) and oomycete (Pythium irregulare) infections, as well as nematode (Meloidogyne hapla) infestations, compared with WT (Park et al., 2013; Gleason et al., 2016; Dos Santos and Park, 2019), together concurring with the conclusion that OPDA is an autonomous metabolic messenger, connecting stress cues to the readjustment of cellular redox homeostasis in actuating retro-directional signaling from the chloroplasts to the nucleus for regulating defense gene expressions.
Figure 3. Proposed model of CYP20-3 as a regulatory hub in the interplay between light and OPDA signaling. In optimal conditions, CYP20-3 relays light signaling in buffering cellular redox homeostasis, whereas, in stressed states, CYP20-3 interfaces light and OPDA signaling, which fine-tunes plant fitness between growth (light-dependent detoxification and Calvin cycle) and defense response (redox-mediated retrograde signaling). Colored arrows indicate the enhanced interactions of CYP20-3 with TRXs (orange), SAT1 (red), and 2CPA (green) during stress (OPDA-signaling) defense responses. Hypothesized passage of electron (e−) transfers is noted in gray.
Cyp20-3 Relays OPDA Signaling Between Plant Defense and Growth Regulatory Pathways
Emerging evidence from a number of recent studies has underpinned that CYP20-3 is a versatile metabolite in plants, proposed to be a key regulator in controlling the interface between OPDA (defense) and light-dependent redox (growth) signaling (Dos Santos and Park, 2019, Figure 3). The latter, also known as the e− transport chain (ETC) photosystem I (PSI), is a primary metabolism converting solar energy into biologically useful chemical energies, necessary for the production of overall biomass of plants and living organisms (Chitnis, 2001; Jensen et al., 2007). When the PSI captures solar energy, it excites e− that reduce thioredoxins (TRXs) via a ferredoxin (Fd) and a Fd-TRX reductase (FTR). TRXs, small oxidoreductases, then deliver e− and activate target enzymes in the Calvin cycle that balances consumption in photosynthesis (Meyer et al., 2009; Serrato et al., 2013; Nikkanen and Rintamäki, 2014). Recent studies, however, have revealed that TRXs can also reduce other Calvin cycle-unrelated proteins, including CYP20-3, a regulator of OPDA defense signaling (see above), and photosynthetic ETC as an e− donor of 2-cysteine peroxiredoxin A (2CPA; Motohashi et al., 2001; Peltier et al., 2006; Laxa et al., 2007). Plastid 2CPA is a thiol-based peroxidase involved in protecting and optimizing photosynthesis. When arrived at the chloroplasts, 2CPA is activated by either different e− donors such as NADPH-dependent TRX reductase C (NTRC), TRXs, and CYP20-3, or—as recently proposed—oxidation folding with GSH (also called S-glutathionylation), which in turn reduces toxic by-products (e.g., H2O2) in photosynthesis or activates Calvin cycle enzymes such as a fructose 1,6-bisphosphatase (Konig et al., 2002; Peltier et al., 2006; Caporaletti et al., 2007; Laxa et al., 2007; Muthuramalingam et al., 2009; Liebthal et al., 2016; Pérez-Ruiz et al., 2017; Liu et al., 2020).
In this context, OPDA binding promotes the interaction of CYP20-3 with TRXs (e.g., type-f2 and x; Cheong et al., 2017), illuminating a mode of OPDA/CYP20-3 signaling in transferring e− from TRXs to 2CPA and/or SAT1 (Figure 3, orange arrow). The latter then stimulates plastid sulfur assimilations (e.g., GSH and thiol accumulations), which coordinate redox-resolved nucleus gene expressions in defense responses against biotic and abiotic stresses (Park et al., 2013), while accelerating the S-glutathionylation (activation) of 2CPA that promotes photosynthetic energy productions (Liu et al., 2020), postulating that OPDA/CYP20-3 signaling optimizes the growth, reproduction, and survival of plants under constant environmental stresses. Traditionally, the cost of resistance (often referred to as growth and defense tradeoff) has been typically described as a teeter-totter model where for defense to increase, growth must decrease and vice versa (Huot et al., 2014). This model well circumstantiates the response of plants to the persistent and excess surge of environmental stresses. However, in nature, plants are more often situated to encounter a consistent array of temporal and modest levels of environmental changes, while concurrently trying to ensure normal growth and developmental processes, in order to maximize their yields and production. For instance, a sorghum inbred line tolerant to sugarcane aphid (Melanaphis sacchari) accumulates significantly higher levels of OPDA, but little if any increase in JA/JA-Ile levels, compared to susceptible lines, as supporting insect growth (tolerance, defense) while concomitantly maintaining enhanced photosynthesis (growth, Grover et al., 2020a). Hence, recent studies have begun to elaborate an alternative model, “growth and defense coordination,” wherein a balancing act between growth and defense can synergistically optimize plant fitness (Kliebenstein, 2016). In agreement, plants' acclimations toward environmental changes and pressures causing oxidative stresses (e.g., insect and pathogen attack, tissue injury, excess light and temperature, and drought and salinity) accompany the accumulation of OPDA on a time sale of hours with attendant accumulation of reduced, non-protein thiols (Riemann et al., 2003; Kazan and Manners, 2011; Noctor et al., 2011; Savchenko and Dehesh, 2014; Hazman et al., 2015; Muench et al., 2016). This initial response subsequently reprograms cellular redox homeostasis and elevates net photosynthesis and nutrient acquisition, together providing plant tolerance to constant and multitudinous biological/ecological constraints (Koch et al., 2016).
Signaling of OPDA in the Plant Acclimation and Adaptation of Heat Shock (HS)
In parallel, our recent study has unveiled that OPDA/CYP20-3 signaling also conveys e− transfers toward peroxidatic (S-glutathionylated) 2CPA (2CPAGS) under HS stress (Figure 3, green arrow). The deglutathionylation then dimerized and inactivated 2CPA that removes HS- and PSI-induced H2O2 bursts and/or activates Calvin cycle enzymes. Hence, it enables the actuation of oxidative stress (defense) signaling and potentially counteracts plant growth, to some extent, elucidating the mode of OPDA in growth and defense tradeoff (Dave et al., 2011; Hazman et al., 2015). On the other hand, HS promoted the increased accumulation and S-glutathionylation of, in part through OPDA/CYP20-3 signaling, 2CPB (another 2CP isoform in the chloroplasts) that constitutes a stable, decameric conformation conferring a chaperone activity (Liu et al., 2020). The expression and activation of a battery of molecular chaperones (i.e., heat shock proteins, HSPs) in protecting native folding and/or assembly of cellular proteins are major defense machinery in plant acclimations of HS (Finka et al., 2011), supporting the critical role of OPDA signaling in thermotolerance (Dave et al., 2011; Muench et al., 2016; Monte et al., 2020). In Arabidopsis when HS occurred, OPDA is mainly, but not JA and JA-Ile, accumulated in the leaves and is only able to dramatically induce HSPs (Taki et al., 2005; Mueller et al., 2008; Balfagón et al., 2019). These HSPs are also induced in coi1 mutants further delineating that OPDA is involved in COI1-independent HS-responsive pathways (Monte et al., 2020). As known from the previous study (Mueller et al., 2008), ORG expressions are largely dependent on TGA TFs (a group of bZIP TFs containing the TGACG motif) TFs. However, the induction of HSPs does not employ TGA TFs but rather a key HS TFs (HSFA1s) or CYP20-3 (Muench et al., 2016). CYP20-3 relays HS-responsive OPDA signaling in the regulation of cellular redox homeostasis that induces and/or stabilizes HSPs (e.g., decameric 2CPBGS), while deglutathionylating 2CPAGS (suppressing peroxide detoxification) allowing to trigger the rapid ROS signaling, together enhancing heat tolerance in plants (Park et al., 2013; Hazman et al., 2015; Liu et al., 2020). This HS response subsequently leads to induce the short-term acquired tolerance and/or cross-defense responses against following abiotic and biotic stresses such as extreme temperatures, high light, salinity, drought, heavy metal, and insect (Mayetiola destuctor) herbivory (Cheng et al., 2018; Hossain et al., 2018; Balfagón et al., 2019), which highlights the vital activity of OPDA signaling in broad-spectrum, induced systemic tolerance/resistance against a wide range of environmental stresses, improving and optimizing growth and yield potential across economically important crops.
Summary: Mode of Action of OPDA Signaling
As sessile organisms, plants cope with constant encounters with a wide range of biotic competitors and consumers, and abiotic constraints, through mobilizing a number of primary and secondary metabolites, and intricate signaling networks that interconnect and orchestrate large-scale changes in transcriptome, proteome, and metabolome. As described in this review, the emerging evidence has espied that OPDA is a versatile signal molecule involved in a variety of metabolic pathways, coordinating plant growth and survival in optimal condition as well as under various forms of environmental stresses (Table 1). In the recent decade, a large number of efforts have been devoted and begun to delineate the mechanistical modus operandi of OPDA signaling; thus far, three working models have been proposed. Once it is produced in the chloroplasts, OPDA is i) conjugated with galactolipids, GSH, and/or amino acids (e.g., Ile) before/after being released to the cytosol, in turn targets yet unknown effector/receptor proteins, and conveys ORG expressions (Böttcher and Pollmann, 2009; Ohkama-Ohtsu et al., 2011; Floková et al., 2016). Alternatively, OPDA itself can ii) serve as a reactive electrophile that targets and modifies thiol residues of, e.g., cysteine, histidine, and lysine in proteins (Mueller and Berger, 2009; Monte et al., 2020) triggering downstream signal transductions and metabolic cascades, or iii) covalently bind a CYP20-3 receptor and builds up a reduction capacity that modulates the cellular activity of oxidoreductase cascades in controlling retrograde signaling, rapidly adjusting nuclear gene expressions (Tada et al., 2008; Park et al., 2013; Cheong et al., 2017). It is, however, still unclear how these signaling mechanisms ultimately stimulate global, spatiotemporal gene expression dynamics with both distinctive and redundant transcriptional outputs. Our study suggests though that OPDA can target and fine-tune an interface between photosynthesis-derived ETC and sulfur assimilation processes in the chloroplasts (Cheong et al., 2017; Liu et al., 2020). This interplay enables plants to make an adaptive decision in allocating resources (e−) between growth and defense responses (e.g., fitness trade-offs or balances) toward different ecological challenges such as pathogens, pests, tissue injury as well as light and oxidative stresses, in the end, ensuring optimal growth, reproduction, and survival of plants. Therefore, furthering our understanding of functional and biological activities of OPDA and associated molecular mechanisms (a) will not only provide new insights into a “broad-spectrum” defense responses and (b) can enrich plant breeding and engineering strategies for the selection of elite genetic traits that will maximize plant fitness, but also (c) will address fundamental gaps in the immune activation of a mammalian system, and (d) help in improving drug developments through facilitating the rational design of more potent and safe reagents.
Author Contributions
SWP and WL, designed and wrote the article. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This work was supported by the Alabama Agricultural Experiment Station (Auburn University), the Hatch Program of the National Institute of Food and Agriculture (United States Department of Agriculture), the Alabama Cotton Commission, and the Alabama Farmers Federation.
References
Acosta, I. F., and Farmer, E. E. (2010). Jasmonates, in Arabidopsis Book, eds Somerville, C.R., and Meyerowitz, E.M., (Rockville: American Society of Plant Biologists) 8, e0129.
Andersson, M. X., Hamberg, M., Kourtchenko, O., Brunnström, A., McPhail, K. L., Gerwick, W. H., et al. (2006). Oxylipin profiling of the hypersensitive response in Arabidopsis thaliana: formation of a novel oxo-phytodienoic acid-containing galactolipid, arabidopsides E. J. Biol. Chem. 281, 31528–31537. doi: 10.1074/jbc.M604820200
Arc, E., Sechet, J., Corbineau, F., Rajjou, L., and Marion-Poll, A. (2013). ABA crosstalk with ethylene and nitric oxide in seed dormancy and germination. Front. Plant Sci. 4:63. doi: 10.3389/fpls.2013.00063
Arnold, M. D., Gruber, C., Flokov,á, K., Miersch, O., Strnad, M., Novák, O., et al. (2016). The recently identified isoleucine conjugate of cis-12-oxo-phytodienoic acid is partially active in cis-12-oxo-phytodienoic acid-specific gene expression of Arabidopsis thaliana. PLoS ONE 11:e0162829. doi: 10.1371/journal.pone.0162829
Balfagón, D., Sengupta, S., Gómez-Cadenas, A., Fritschi, F. B., Azad, R. K., Mittler, R., et al. (2019). Jasmonic acid is required for plant acclimation to a combination of high light and heat stress. Plant Physiol. 181, 1668–1682. doi: 10.1104/pp.19.00956
Barros-Galvão, T., Dave, A., Cole, A., Harvey, D., Langer, S., Larson, T. R., et al. (2019). cis-12-Oxo phytodienoic acid represses Arabidopsis seed germination in shade conditions. J. Exp. Bot. 70, 5919–5927. doi: 10.1093/jxb/erz337
Bosch, M., Berger, S., Schaller, A., and Stintzi, A. (2014a). Jasmonate-dependent induction of polyphenol oxidase activity in tomato foliage is important for defense against Spodoptera exigua but not against Manduca sexta. BMC Plant Biol. 14:257. doi: 10.1186/s12870-014-0257-8
Bosch, M., Wright, L. P., Gershenzon, J., Wasternack, C., Hause, B., Schaller, A., et al. (2014b). Jasmonic acid and its precursor 12-oxophytodienoci acid control different aspects of constitutive and induced herbivore defenses in tomato. Plant Physiol. 116, 396–410. doi: 10.1104/pp.114.237388
Böttcher, C., and Pollmann, S. (2009). Plant oxylipins: Plant responses to 12-oxo-phytodienoic acid are governed by its specific structural and functional properties. FEBS J. 17, 4693–4703. doi: 10.1111/j.1742-4658.2009.07195.x
Buseman, C. M., Tamura, P., Sparks, A. A., Gaughman, E. J., Maatta, S., Zhao, J., et al. (2006). Wounding stimulates the accumulation of glycerolipids containing oxophytodienoic acid and dinor-oxophytodienoic acid in Arabidopsis leaves. Plant Physiol. 142, 28–39. doi: 10.1104/pp.106.082115
Caporaletti, D., D'Alessio, A. C., Rodriguez-Suarez, R. J., Senn, A. M., Duek, P. D., and Wolosiuk, R. A. (2007). Non-reductive modulation of chloroplast fructose-1,6-bisphos-phatase by 2-Cys peroxiredoxin. Biochem. Biophys. Res. Commun. 355, 722–727. doi: 10.1016/j.bbrc.2007.02.013
Cheng, G., Chen, M. S., and Zhu, L. (2018). 12-oxo-phytodienoic acid enhances wheat resistance to Hessian Fly (Diptera: Cecidomyiidae) under heat stress. J. Econ. Entomol. 111, 917–922. doi: 10.1093/jee/tox374
Cheong, H., Dos Santos, I. B., Liu, W., Gosse, H. N., and Park, S. W. (2017). Cyclophilin 20-3 is positioned as a regulatory hub between light-dependent redox and 12-oxo-phytodienoic acid signaling. Plant Signal. Behav. 12:e1362520. doi: 10.1080/15592324.2017.1362520
Chini, A., Fomseca, S., Fernández, G., Adie, B., Chico, J. M., Lorenzo, O., et al. (2007). The JAZ family of repressors is the missing link in jasmonate signaling. Nature 448, 666–671. doi: 10.1038/nature06006
Chini, A., Monte, I., Zamarreño, A. M., Hamberg, M., Lassueur, S., Reymond, P., et al. (2018). An OPR3-independent pathway uses 4,5-didehydrojasmonate for jasmonate synthesis. Nat. Chem. Biol. 14, 171–178. doi: 10.1038/nchembio.2540
Chitnis, P. R. (2001). Photosystme I: function and physiology. Annu. Rev. Plant Physiol. Mol. Biol. 52, 593–626. doi: 10.1146/annurev.arplant.52.1.593
Christensen, S. A., Huffaker, A., Kaplan, F., Slims, J., Ziemann, S., Doehlemann, G., et al. (2015). Maize death acids, 9-lipoxygenase-derived cyclopente(a)nones, display activity as cytotoxic phytoalexins and transcriptional mediators. Proc. Natl. Acad. Sci. U.S.A. 112, 11407–11412. doi: 10.1073/pnas.1511131112
Chung, H. S., Koo, A. J. K., Gao, X., Jayanty, S., Thines, B., Jones, A. D., et al. (2008). Regulation and function of Arabidopsis JAMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 146, 952–964. doi: 10.1104/pp.107.115691
Dang, H. T., Lee, H. J., Yoo, E. S., Hong, J., Bao, B., Choi, J. S., et al. (2008). New jasmonate analogues as potential anti-inflammatory agents. Bioorg. Med. Chem. 16, 10228–10235. doi: 10.1016/j.bmc.2008.10.050
Dave, A., and Graham, I. A. (2012). Oxylipin signaling: A distinct role for the jasmonic acid precursor cis-(+)-12-oxo-phytodienoic acid (cis-OPDA). Front. Plant Sci. 3, 42. doi: 10.3389/fpls.2012.00042
Dave, A., Hernández, M. L., He, Z., Andriotix, V. M. E., Vaistij, F. E., Larson, T. R., et al. (2011). 12-oxo-phytodienoic acid accumulation during seed development represses seed germination in Arabidopsis. Plant Cell 23, 583–599. doi: 10.1105/tpc.110.081489
Dave, A., Vaistij, F. E., Gilday, A. D., Penfield, S. D., and Graham, I. A. (2016). Regulation of Arabidopsis thaliana seed dormancy and germination by 12-oxo-phytodienoci acid. J. Exp. Bot. 67, 2277–2284. doi: 10.1093/jxb/erw028
Davoine, C., Douki, T., Iacazio, G., Montillet, J. L., and Triantaphylidès, C. (2005). Conjugation of keto fatty acids to glutathione in plant tissues. characterization and quantification by HPLC-tandem mass spectrometry. Anal. Chem. 77, 7366–7372. doi: 10.1021/ac051155y
Davoine, D., Falletti, O., Douki, T., Iacazio, G., Ennar, N., Montillet, J. L., et al. (2006). Adducts of oxylipin electrophiles to gluatathione reflect a 13 specificity of the downstream lipoxygenase pathway in the tobacco hypersensitive response. Plant Physiol. 140, 1484–01493. doi: 10.1104/pp.105.074690
De Storme, N., and Geelen, D. (2014). Callose homeostasis at plasmodesmata: molecular regulators and developmental relevance. Front. Plant Sci. 5, 138. doi: 10.3389/fpls.2014.00138
Devoto, A., Ellis, C., Magusin, A., Chang, H. S., Chilcott, C., Xhu, T., et al. (2005). Expression profiling reveals COI1 to be a key regulator of genes involved in wound- and methyl jasmonate-induced secondary metabolism, defence, and hormone interactions. Plant Mol. Biol. 58, 497–513. doi: 10.1007/s11103-005-7306-5
Dominguez-Solis, J. R., He, Z., Lima, A., Ting, J., Buchanan, B. B., and Luan, S. (2008). A cyclophilin links redox and light signals to cysteine biosynthesis and stress responses in chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 105, 16386–16391. doi: 10.1073/pnas.0808204105
Dos Santos, I. B., and Park, S. W. (2019). Versatility of cyclophilins in plant growth and survival: a case study in Arabidopsis. Biomolecules 9:20. doi: 10.3390/biom9010020
Felton, G. W. (2005). Indigestion is a plant's best defense. Proc. Natl. Acad. Sci. U.S.A. 92, 407–411. doi: 10.1073/pnas.0509895102
Finka, A., Mattoo, R. U., and Goloubinoff, P. (2011). Meta-analysis of heat- and chemically upregulated chaperone genes in plant and human cells. Cell Stress Chaperones 16, 15–31. doi: 10.1007/s12192-010-0216-8
Flescher, E. (2007). Jasmonates in cancer therapy. Cancer Lett. 245, 1–10. doi: 10.1016/j.canlet.2006.03.001
Floková, K., Feussner, K., Herrfurth, C., Miersch, O., Mik, V., Tarkowsk,á, D., et al. (2016). A previously undescribed jasmonate compound in flowering Arabidopsis thaliana—the identification of cis-(+)-OPDA-Ile. Phytochemistry 122, 230–237. doi: 10.1016/j.phytochem.2015.11.012
Funk, C. D. (2001). Prostaglandins and leukotrienes: advances in eicosanoid biology. Science. 294, 1871–1875. doi: 10.1126/science.294.5548.1871
Genva, M., Deleu, M., Andersson, M. X., Lins, L., and Fauconnier, M. L. (2019). “Evaluation of interactions between arabidopsides and plant plasma membrane lipids,” in Scientific Congresses and Symposiums. Available online at: https://orbi.uliege.be/bitstream/2268/222461/1/Bruxelles%20congress.pdf.
Gleason, C., Leelarasamee, N., Meldau, D., and Feussner, I. (2016). OPDA has key role in regulating plant susceptibility to the root-knot nematode Meloidogyne hapla in Arabidopsis. Front. Plant Sci. 7:1565. doi: 10.3389/fpls.2016.01565
Goetz, S., Hellwege, A., Stenzel, I., Kutter, C., Hauptmann, V., Forner, S., et al. (2012). Role of Cis-12-Oxo-Phytodienoic Acid in Tomato Embryo Development. Plant Physiol. 4, 1715–1727. doi: 10.1104/pp.111.192658
Grover, S., Agpawa, E., Sarath, G., Sattler, S. E., and Louis, J. (2020a). Interplay of phytohormones facilitate sorghum tolerance to aphids. Plant Mol. Biol. 1–12. doi: 10.1007/s11103-020-01083-y
Grover, S., Versani, S., Kolomietx, M. V., and Louis, J. (2020b). Maize defense elicitor, 12-oxo-phytodienoic acid, prolongs aphid salivation. Commun. Integr. Biol. 13, 63–66. doi: 10.1080/19420889.2020.1763562
Guan, L., Denkert, N., Eisa, A., Lehmann, M., Sjuts, I., Weiberg, A., et al. (2019). JASSY, a chloroplast outer membrane protein required for jasmonate biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 116, 10568–10575. doi: 10.1073/pnas.1900482116
Guo, H. M., Li, H. C., Zhou, S. H., Xue, H. W., and Miao, X. X. (2014). Cis-12-oxo- phytodienoic acid stimulates rice defense response to a piercing-sucking insect. Mol. Plant 7, 1683–1692. doi: 10.1093/mp/ssu098
Hazman, M., Hause, B., Eiche, E., Nick, P., and Riemann, M. (2015). Increased tolerance to salt stress in OPDA-deficient rice ALLENE OXDE CYCLASE mutants is linked to an increase ROS-scavenging activity. J. Exp. Bot. 66, 3339–3352. doi: 10.1093/jxb/erv142
He, J., Duan, Y., Hua, D., Fan, G., Wang, L., Liu, Y., et al. (2012). DEXH box RNA helicase-mediated mitochondrial reactive oxygen species production in Arabidopsis mediates crosstalk between abscisic acid and auxin signaling. Plant Cell 24, 1815–1833. doi: 10.1105/tpc.112.098707
Hisamatsu, Y., Goto, N., Hasegawa, K., and Shigemori, H. (2003). Arabidopsides A and B, two new oxylipins from Arabidopsis thaliana. Tetrahedron Lett. 44, 5553–5556. doi: 10.1016/S0040-4039(03)01148-1
Hisamatsu, Y., Goto, N., Sekiguchi, M., Hasegawa, K., and Shigemori, H. (2005). Oxylipins arabidopsides C and D from Arabidopsis thaliana. J. Nat. Prod. 68, 600–603. doi: 10.1021/np0495938
Hofhuis, H., Laskowski, M., Du, Y., Prasad, K., Grigg, S., Pinon, V., et al. (2013). Phyllotaxis and rhizotaxis in Arabidopsis are modified by three PLETHORA transcription factors. Curr. Biol. 23, 965–962. doi: 10.1016/j.cub.2013.04.048
Hossain, M. A., Li, A. G., Hoque, T. S., Burritt, D. J., Fujita, M., and Munnè-Bosch, S. (2018). Heat or cold priming-induced cross-tolerance to abiotic stresses in plants: key regulators and possible mechanisms. Protoplasma 255, 399–412. doi: 10.1007/s00709-017-1150-8
Huang, X., Stettmaier, K., Michel, C., Hutzler, P., Mueller, M. J., and Durner, J. (2004). Nitric oxide is induced by wounding and influences jasmonic acid signaling in Arabidopsis thaliana. Planta 218, 938–946. doi: 10.1007/s00425-003-1178-1
Huot, B., Yao, J., Montgomery, B. L., and He, S. Y. (2014). Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Mol. Plant 7, 1267–1287. doi: 10.1093/mp/ssu049
Jensen, P. E., Bassi, R., Boekema, E. J., Dekker, J. P., Jansson, S., Leister, D., et al. (2007). Structure, function and regulation of plant photosystem I. Biochim. Biophys. Acta. 1767, 335–352. doi: 10.1016/j.bbabio.2007.03.004
Jiao, Y., Sun, L., Song, Y., Wang, L., Liu, L., Zhang, L., et al. (2013). AtrbohD and atrbohF positively regulate abscisic acid-inhibited primary root growth by affecting Ca2+ signaling and auxin response of roots in Arabidopsis. J. Exp. Bot. 64, 4183–4192. doi: 10.1093/jxb/ert228
Kazan, K., and Manners, J. M. (2011). The interplay between light and jasmonate signaling during defence and development. J. Exp. Bot. 62, 4087–4100. doi: 10.1093/jxb/err142
Klessig, D. F., Choi, H. W., and Dempsey, D. A. (2018). Systemic acquired resistance and salicylic acid: past, present and future. Mol. Plant Microbe Interact. 31, 871–888. doi: 10.1094/MPMI-03-18-0067-CR
Kliebenstein, D. J. (2016). False idolatry of the mythical growth versus immunity tradeoff in molecular systems plant pathology. Physiol. Mol. Plant Pathol. 95, 55–59. doi: 10.1016/j.pmpp.2016.02.004
Ko, J. H., Yang, S. H., and Han, K. H. (2006). Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J. 47, 343–355. doi: 10.1111/j.1365-313X.2006.02782.x
Koch, K. G., Chapman, K., Louis, J., Heng-Moss, T., and Sarath, G. (2016). Plant tolerance: a unique approach to control hemipteran pests. Front. Plant Sci. 7:1363. doi: 10.3389/fpls.2016.01363
Konig, J., Baier, M., Horling, F., Kahmann, U., Harris, G., and Schurmann, P. (2002). The plant-specific function of 2-Cys peroxiredoxin-mediated detoxification of peroxides in the redox-hierarchy of photosynthetic electron flux. Proc. Natl. Acad. Sci. U.S.A. 99, 5738–5743. doi: 10.1073/pnas.072644999
Koo, A. J. K., Gao, X., Jones, A. D., and Howe, G. A. (2009). A rapid wound signal activates the systemic synthesis of bioactive jasmonates in Arabidopsis. Plant J. 59, 974–986. doi: 10.1111/j.1365-313X.2009.03924
Kourtchenko, O., Andersson, M. X., Hamberg, M., Brunnström, A., Göbel, C., McPhail, K. L., et al. (2007). Oxo-phytodienoic acid-containing galactolipids in Arabidopsis: jasmonate signaling dependence. Plant Physiol. 145, 1658–1669. doi: 10.1104/pp.107.104752
Laxa, M., Konig, J., Dietz, K. J., and Kandlbinder, A. (2007). Role of the cysteine residues in Arabidopsis thaliana cyclophilin CYP20-3 in peptidylprolyl cis-trans isomerase and redox-related function. Biochem. J. 401, 287–297. doi: 10.1042/BJ20061092
Leon-Reyes, A., Van der Does, D., De Lange, E. S., Delker, C., Wasternack, C., Van Wees, S. C., et al. (2010). Salicylate-mediated suppression of jasmonate-responsive gene expression in Arabidopsis is targeted downstream of the jasmonate biosynthesis pathway. Planta 232, 1423–1432. doi: 10.1007/s00425-010-1265-z
Liebthal, M., Strüve, M., Li, X., Hertle, Y., Maynard, D., Hellweg, T., et al. (2016). Redox-dependent conformational dynamics of decameric 2-cys-teine peroxiredoxin and its interaction with cyclophilin 20-3. Plant Cell Physiol. 57, 1415–1425. doi: 10.1093/pcp/pcw031
Liu, W., Barbosa dos Santos, I., Moye, A., and Park, S. W. (2020). CYP20-3 deglutathionylates 2-CysPRX A and suppresses peroxide detoxification during heat stress. Life Sci. Alliance 3:e202000775. doi: 10.26508/lsa.202000775
López-Galiano, M. J., Ruiz-Arroyo, V. M., Fernández-Crespo, E., Rausell, C., Real, M. D., García-Agustín, P., et al. (2017). Oxylipin mediated stress response of a miraculin-like protease inhibitor in hexanoic acid primed eggplant plants infested by Colorado potato beetle. J. Plant Physiol. 215, 59–64. doi: 10.1016/j.jplph.2017.04.013
Luna, E., Pastor, V., Robert, J., Flors, V., Mauch-Mani, B., and Ton, J. (2010). Callose deposition: a multifaceted plant defense response. Mol. Plant Microbe. Interact. 24, 183–193. doi: 10.1094/MPMI-07-10-0149
Luo, X., Chen, Z., Gao, J., and Gong, Z. (2014). Abscisic acid inhibits root growth in Arabidopsis through ethylene biosynthesis. Plant J. 79, 44–58. doi: 10.1111/tpj.12534
Maynard, D., Gröger, H., Dierks, T., and Dietz, K. J. (2018). The function of the oxylipin, 12-oxo-phytodienoic acid in cell signaling, stress acclimation, and development. J. Exp. Bot. 69, 5341–5354. doi: 10.1093/jxb/ery316
Meyer, Y., Buchanan, B. B., Vignols, F., and Reichheld, J. P. (2009). Thioredoxins and glutaredoxins: unifying elements in redox biology. Annu. Rev. Genet. 43, 335–367. doi: 10.1146/annurev-genet-102108-134201
Monte, I., Kneeshaw, S., Franco-Zorrilla, J. M., Chini, A., Zamarreño, A. M., García-Mina, J. M., et al. (2020). An ancient COI1-independent function for reactive electrophilic oxylipins in thermotolerance. Curr. Biol. 30, 962–971. doi: 10.1016/j.cub.2020.01.023
Monte, L., Franco-Zorrilla, J. M., García-Mina, J. M., Zamarreño, A. M., García-Mina, J. M., Nishihama, R., et al. (2019). A single JAZ repressor controls the jasmonate pathway in Marchantia polymorpha. Mol. Plant 12, 185–198. doi: 10.1016/j.molp.2018.12.017
Monte, L., Ishida, S., Zamarreño, A. M., Hamberg, M., Franco-Zorrilla, J. M., García-Mina, J. M., et al. (2018). Ligand-receptor co-evolution shaped the jasmonate pathway in land plants. Nat. Chem. Biol. 14, 480–488. doi: 10.1038/s41589-018-0033-4
Mosblech, A., Feussner, I., and Heilmann, I. (2010). Oxylipin signaling and plant growth, in Lipid Signaling in Plants, ed. Munnik, T., (Springer-Verlag, Berlin, Heidelberg) 277–291.
Motohashi, K., Kondoh, A., Stumpp, M. T., and Hisabori, T. (2001). Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc. Natl. Acad. Sci. U.S.A. 98, 11224–11229. doi: 10.1073/pnas.191282098
Mueller, S., and Berger, S. (2009). Reactive electrophilic oxylipins: Pattern recognition and signalling. Phytochemistry 70, 1511–1521. doi: 10.1016/j.phytochem.2009.05.018
Mueller, S., Hilbert, B., Dueckershoff, K., Roitsch, T., Krischke, M., Mueller, M. J., et al. (2008). General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell 20, 768–785. doi: 10.1105/tpc.107.054809
Muench, M., Hsin, C. H., Ferber, E., Berger, S., and Mueller, M. J. (2016). Reactive electrophilic oxylipins triggers a heat stress-like response through HSFA1 transcription factors. J. Exp. Bot. 67, 6139–6148. doi: 10.1093/jxb/erw376
Muthuramalingam, M., Seidel, T., Laxa, M., de Miranda, S. M. N., Gärtner, F., Ströher, E., et al. (2009). Multiple redox and non-redox interactions define 2-Cys peroxiredoxin as a regulatory hub in the chloroplast. Mol. Plant 2, 1273–1288. doi: 10.1093/mp/ssp089
Nikkanen, L., and Rintamäki, E. (2014). Thioredoxin-dependent regulatory networks in chloroplasts under fluctuating light conditions. Phil. Trans. R. Soc. B. 369, 20130224. doi: 10.1098/rstb.2013.0224
Noctor, G., Queval, G., Mhamdi, A., Chaouch, S., and Foyer, C. H. (2011). Glutathione, in Arabidopsis Book, eds. Somerville, C.R., and Meyerowitz, E.M., (Rockville: American Society of Plant Biologists) 9, e0142.
Ohkama-Ohtsu, N., Sasaki-Sekimoto, Y., Oikawa, A., Jikumaru, Y., Shinoda, S., Inoue, E., et al. (2011). 12-oxo-phytodienoic acid-glutathione conjugate is transported into the vacuole in Arabidopsis. Plant Cell Physiol. 52, 205–209. doi: 10.1093/pcp/pcq181
Ortiz-Masia, D., Perez-Amdor, M. A., Carbonell, J., and Marcote, M. J. (2007). Diverse stress signals activate the C1 subgroup MAP Kinases of Arabidopsis. FEBS Lett. 58, 1834–1840. doi: 10.1016/j.febslet.2007.03.075
Palmieri, M. C., Sell, S., Huang, X., Scherf, M., Werner, T., Durner, J., et al. (2008). Nitric oxide-resposive genes and promoters in Arabidopsis thaliana: a bioinformatics approach. J. Exp. Bot. 59, 177–186. doi: 10.1093/jxb/erm345
Park, S. W., Li, W., Voejjaiser, A., He, B., Kim, S., Nilsson, A. K., et al. (2013). Cyclophilin 20-3 relays a 12-oxo-phytodienoic acid signal during stress responsive regulation of cellular redox homeostasis. Proc. Natl. Acad. Sci. U.S.A. 110, 9559–9564. doi: 10.1073/pnas.1218872110
Peltier, J. B., Cai, Y., Sun, Q., Zabrouskov, V., Giacomelli, L., Rudella, A., et al. (2006). The oligomeric stromal proteome of Arabidopsis thaliana chloroplasts. Mol. Cell Proteomics 5, 114–133. doi: 10.1074/mcp.M500180-MCP200
Pérez-Ruiz, J. M., Naranjo, B., Ojeda, V., Guinea, M., and Cejudo, F. J. (2017). NTRC-dependent redox balance of 2-Cys peroxiredoxins is needed for optimal function of the photosynthetic apparatus. Proc. Natl. Acad. Sci. U.S.A. 114, 12069–12074. doi: 10.1073/pnas.1706003114
Pieterse, C. M. J., Van der Does, D., Zamioudis, C., Leon-Reyes, A., and Van Wees, S. C. M. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. doi: 10.1146/annurev-cellbio-092910-154055
Pieterse, C. M. J., Zamioudis, C., Berendsen, R. L., Weller, D. M., Van Wees, S. C. M., and Bakker, P. A. H. M. (2014). Induced systemic resistance by beneficial microbes. Annu. Rev. Phytophahtol. 52, 347–375. doi: 10.1146/annurev-phyto-082712-102340
Piskurewicz, U., Jikumaru, Y., Kinoshita, N., Nambara, E., Kamiya, Y., and Lopez-Molina, L. (2008). The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20, 2729–2745. doi: 10.1105/tpc.108.061515
Ribot, C., Zimmerili, C., Farmer, E. E., Reymond, P., and Poirier, Y. (2008). Induction of the Arabidopsis PHO1;H10 gene by 12-oxo-phytodienoic acid but not jasmonic acid via a CORONATINE INSENSITIVE1-dependent pathway. Plant Physiol. 147, 696–706. doi: 10.1104/pp.108.119321
Riemann, M., Müller, A., Korte, A., Furuya, M., Wiler, E. W., and Nick, P. (2003). Impaired induction of the jasmonate pathway in the rice mutant hebiba. Plant Physiol. 133, 1820–1830. doi: 10.1104/pp.103.027490
Romano, P. G. N., Horton, P., and Gray, J. E. (2004). The Arabidopsis cyclophilin gene family. Plant Physiol. 134, 1268–1282. doi: 10.1104/pp.103.022160
Russell, L., Larner, V., Kurup, S., Bougourd, S., and Holdsworth, M. (2000). The Arabidopsis COMATOSE locus regulates germination potential. Development 127, 3759–3767. doi: 10.1242/dev.127.17.3759
Savchenko, T., and Dehesh, K. (2014). Drought stress modulates oxylipin signature by eliciting 12-OPDA as a potent regulator of stomatal sperture. Plant Signal. Behav. 9:e28304. doi: 10.4161/psb.28304
Scalschi, L., Llorens, E., García-Agustín, P., and Vicedo, B. (2020). Role of jasmonic acid pathway in tomato plant-Pseudomonas syringae interaction. Plants 9, 136. doi: 10.3390/plants9020136
Scalschi, L., Sanmartin, M., Camañes, G., Troncho, P., Sánchez-Serrano, J. J., García-Agustín, P., et al. (2015). Silencing of OPR3 in tomato reveals the role of OPDA in callose deposition during the activation of defense responses against Botrytis cinerea. Plant J. 81, 304–315. doi: 10.1111/tpj.12728
Serrato, A. J., Fernández-Trijueque, J., Barajas-López, J., Chueca, A., and Sahrawy, M. (2013). Plastid thioredoxins: a “one-for-all” redox-signaling system in plants. Front. Plant Sci. 4, 463. doi: 10.3389/fpls.2013.00463
Shim, J. S., Jung, C., Lee, S., Min, K., Lee, Y. W., Choi, Y., et al. (2013). AtMYB44 regulates WRKY70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling. Plant J. 73, 483–495. doi: 10.1111/tpj.12051
Sirichandra, C., Gu, D., Hu, H. C., Davanture, M., Lee, S., Djaoui, M., et al. (2009). Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett. 583, 2982–2986. doi: 10.1016/j.febslet.2009.08.033
Skubacz, A., Daszkowska-Golec, A., and Szarejko, I. (2016). The role and regulation of ABI5 (ABA-insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front. Plant Sci. 7:1884. doi: 10.3389/fpls.2016.01884
Staswick, P. E. (2009). The tryptophan conjugates of jasmonic and idole-3-acetic acids are endoge-nous auxin inhibitors. Plant Physiol. 150, 1310–1321. doi: 10.1104/pp.109.138529
Stintzi, A., Weber, H., Reymond, P., Browse, J., and Farmer, E. E. (2001). Plant defense in the absence of jasmonic acid: The role of cyclopentenones. Proc. Natl. Acad. Sci. U.S.A. 97, 10625–10630. doi: 10.1073/pnas.190264497
Stotz, H. U., Jikumaru, Y., Shimada, Y., Sasaki, E., Stingl, N., Mueller, M. J., et al. (2011). Jasmonate-dependent and COI1-independent defense responses against Sclerotinia sclerotiorum in Arabidopsis thaliana: Auxin is part of COI1-independent defense signaling. Plant Cell Physiol. 52, 1941–1951. doi: 10.1093/pcp/pcr127
Stotz, H. U., Mueller, S., Zoeller, M., Mueller, M. J., and Berger, S. (2013). TGA transcription factors and jasmonate-independent COI1 signalling regulate specific plant responses to reactive oxylipins. J. Exp. Bot. 64, 963–975. doi: 10.1093/jxb/ers389
Sun, L. R., Wang, Y. B., He, S. B., and Hao, F. S. (2018). Mechanisms for abscisic acid inhibition of primary root growth. Plant Signal. Behav. 13, 9. doi: 10.1080/15592324.2018.1500069
Tada, Y., Spoel, S. H., Paferowska-Mukhtar, K., Mou, Z., Song, J., Wang, C., et al. (2008). Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science 321, 952–956. doi: 10.1126/science.1156970
Taki, N., Sasaki-Sekimoto, Y., Obayashi, T., Kikuta, A., Kobayashi, K., Ainai, T., et al. (2005). 12-oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiol. 139, 1268–1283. doi: 10.1104/pp.105.067058
Taki-Nakano, N., Ohzeki, H., Kotera, J., and Ohta, H. (2014). Cytoprotective effects of 12-oxo-phytodienoci acid, a plant-derived oxylipin jasmonate, on oxidative stress-induced toxicity in human neuroblastoma SH-SY5Y cells. Biochim. Biophys. Acta. 1840, 3413–3422. doi: 10.1016/j.bbagen.2014.09.003
Theodoulou, F. L., Job, K., Slocombe, S. P., Footitt, S., Holdsworth, M., Baker, A., et al. (2005). Jasmonic acid levels are reduced in COMATOSE ATP-binding cassette transporter mutants. Implications for transport of jasmonate precursors into peroxisomes. Plant Physiol. 137, 835–840. doi: 10.1104/pp.105.059352
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., et al. (2007). JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448, 661–665. doi: 10.1038/nature05960
Vaistij, F. E., Barros-Galvão, T., Cole, A. F., Gilday, A., He, Z., Li, Y., et al. (2018). MOTHER-OF-FT-AND-TFL1 represses seed germination under far-red light by modulating phytohormone responses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 115, 8442–8447. doi: 10.1073/pnas.1806460115
Varsani, S., Grover, S., Zhou, S., Koch, K. G., Huang, P. C., Kolomiets, M. V., et al. (2019). 12-oxo-phytodienoic acid acts as a regulator of maize defense against corn leaf aphid. Plant Physiol. 179, 1402–1415. doi: 10.1104/pp.18.01472
Veronese, P., Nakagami, H., Bluhm, B., Abuqamar, S., Chen, X., Salmeron, J., et al. (2006). The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18, 257–273. doi: 10.1105/tpc.105.035576
Vissenberg, K., Claeijs, N., Balcerowicz, D., and Schoenaers, S. (2020). Hormonal regulation of root hair growth and responses to the environment in Arabidopsis. J. Exp. Bot. 71, 2412–2427. doi: 10.1093/jxb/eraa048
Vu, H. S., Tamura, P., Galeva, N. A., Chaturvedi, R., Roth, M. R., Williams, T. D., et al. (2012). Direct infusion mass spectrometry of oxylipin-containing Arabidopsis membrane lipids reveals varied patterns in different stress responses. Plant Physiol. 158, 324–339. doi: 10.1104/pp.111.190280
Wang, H., Qi, Q., Schorr, P., Cutler, A. J., Crosby, W. L., and Fowke, L. C. (2008). ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J. 15, 501–510. doi: 10.1046/j.1365-313X.1998.00231.x
Wang, K. D., Borrego, E. J., Kenerley, C. M., and Kolemiets, M. V. (2020). Oxylipins other than jasmonic acid are xylem-resident signals regulating systemic resistance induced by Trichoderma virens in maize. Plant Cell 32, 166–185. doi: 10.1105/tpc.19.00487
Wang, K. L., Li, H., and Ecker, J. R. (2002). Ethylene biosynthesis and signaling networks. Plant Cell 14, S131–S151. doi: 10.1105/tpc.001768
Wang, L., Hua, D., He, J., Duan, Y., Chen, Z., Hong, X., et al. (2011). Auxin response factor2 (ARF2) and its regulated homeodomain gene HB33 mediate abscisic acid response in Arabidopsis. PLoS Genet. 7:e1002172. doi: 10.1371/journal.pgen.1002172
Wang, Y., Loake, G. J., and Chu, C. (2013). Cross-talk of nitric oxide and reactive oxygen species in plant programed cell death. Front Plant Sci. 4:314. doi: 10.3389/fpls.2013.00314
Wei, J., van Loon, J. J., Gols, R., Menzel, T. R., Li, N., Kang, L., et al. (2014). Reciprocal crosstalk between jasmonate and salicylate defence-signalling pathways modulates plant volatile emission and berbivore host-selection behaviour. J. Exp. Bot. 65, 3289–3298. doi: 10.1093/jxb/eru181
Xu, J., Gao, G. L., Du, J. J., Guo, Y., and Yang, C. W. (2010). Cell cycle modulation in response to the primary root of Arabidopsis. Pak. J. Bot. 42, 2703–2710.
Yao, T., Jin, D., Liu, Q., and Gong, Z. (2013). Abscisic acid suppresses the highly occurred somatic homologous recombination in Arabidopsis rfc1 mutant. J. Genet. Genomics 40, 465–471. doi: 10.1016/j.jgg.2013.05.006
Yin, H., Zhang, X., Liu, J., Wang, Y., He, J., Yang, T., et al. (2009). Epigenetic regulation, somatic homologous recombination, and abscisic acid signaling are influenced by DNA polymerase epsilon mutation in Arabidopsis. Plant Cell 21, 386–402. doi: 10.1105/tpc.108.061549
Zhang, Y., and Turner, J. G. (2008). Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS ONE 3:e3699. doi: 10.1371/journal.pone.0003699
Zhao, X., Li, N., Song, Q., Li, X., Meng, H., and Luo, K. (2020). OPDAT1, a plastid envelope protein involved in 12-oxo-phytodienoci acid export for jasmonic acid biosynthesis in Populus. Tree Physiol. 9:tpab037. doi: 10.1093/treephys/tpab037
Keywords: cyclophilin 20-3, fitness tradeoffs/balances, light-dependent redox reactions, redox signaling, retrograde signaling
Citation: Liu W and Park SW (2021) 12-oxo-Phytodienoic Acid: A Fuse and/or Switch of Plant Growth and Defense Responses? Front. Plant Sci. 12:724079. doi: 10.3389/fpls.2021.724079
Received: 11 June 2021; Accepted: 19 July 2021;
Published: 17 August 2021.
Edited by:
Koichi Sugimoto, University of Tsukuba, JapanReviewed by:
Tomonori Shinya, Okayama University, JapanSajjan Grover, University of Nebraska-Lincoln, United States
Copyright © 2021 Liu and Park. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sang-Wook Park, c3dwYXJrQGF1YnVybi5lZHU=
 Wenshan Liu
Wenshan Liu Sang-Wook Park
Sang-Wook Park