
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 14 January 2022
Sec. Plant Biotechnology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.715809
This article is part of the Research Topic Wood Development and Physiology in a Changing Climate View all 17 articles
 Yamei Zhuang1,2
Yamei Zhuang1,2 Sihui Chen2
Sihui Chen2 Wenjun Lian1
Wenjun Lian1 Li Xu1
Li Xu1 Dian Wang2,3
Dian Wang2,3 Congpeng Wang1,2
Congpeng Wang1,2 Jie Meng1,2
Jie Meng1,2 Xianfeng Tang2
Xianfeng Tang2 Hua Xu2
Hua Xu2 Shumin Wang2
Shumin Wang2 Lin Du2
Lin Du2 Yang Zhang2
Yang Zhang2 Gongke Zhou1,2,4*
Gongke Zhou1,2,4* Guohua Chai1,2,4*
Guohua Chai1,2,4*
Wood formation of trees is a complex and costly developmental process, whose regulatory network is involved in the protein-protein and protein-DNA interactions. To detect such interactions in wood development, we developed a high-throughput screening system with 517 Gal4-AD-wood-associated transcription factors (TFs) library from Populus alba × P. glandulosa cv “84K.” This system can be used for screening the upstream regulators and interacting proteins of targets by mating-based yeast-one hybrid (Y1H) and yeast-two-hybrid (Y2H) method, respectively. Multiple regulatory modules of lignin biosynthesis were identified based on this Populus system. Five TFs interacted with the 500-bp promoter fragment of PHENYLALANINE AMMONIA-LYASE 2 (PAL2), the first rate-limiting enzyme gene in the lignin biosynthesis pathway, and 10 TFs interacted with PaMYB4/LTF1, a key regulator of lignin biosynthesis. Some of these interactions were further validated by EMSA and BiFC assays. The TF-PaPAL2 promoter interaction and TF-PaMYB4 interaction revealed a complex mechanism governing the regulation of lignin synthesis in wood cells. Our high-throughput Y1H/Y2H screening system may be an efficient tool for studying regulatory network of wood formation in tree species.
The woody secondary cell walls of plants are the largest repository of renewable carbon biopolymers on the planet. They are widely used for timber, paper and pulp, and have potential as a source of bioenergy (Du and Groover, 2010). In tree species, wood (secondary xylem) is a complex biomass material constituted mainly of cellulose, hemicelluloses and lignin. Long cellulose microfibrils impart tensile strength, shorter hemicelluloses establish carbohydrate cross-linking, and lignin as a phenolic polymer fills in and cross-links the carbohydrate matrix (Albersheim et al., 2011). Understanding the molecular regulation of wood formation is required for the improvement of biomass and wood characteristics.
Wood formation is finely controlled by a hierarchical transcription factor network (HTFN), which is relatively conserved between Arabidopsis and trees (Wang and Dixon, 2012; Lin et al., 2013; Nakano et al., 2015; Chen et al., 2019). Functional characterization of a serial of Arabidopsis mutants shows a three-layered regulatory network for secondary cell wall formation (Zhong et al., 2010; Wang and Dixon, 2012; Nakano et al., 2015). In Arabidopsis stems, SND1 is the highest level regulator of HTFN, and it directly activates the second-layered master switches MYB46 and its paralog MYB83, inducing the expression of multiple cell wall biosynthetic genes. A wood-associated HTFN involving TF-DNA and TF-TF regulations has been recently constructed using quantitative transcriptomics, yeast one/two hybrid (Y1H/Y2H) and chromatin binding assays (Chen et al., 2019). Similar to Arabidopsis SND1-mediated HTFN, in Populus stems PtrSND1-B1 induces PtrMYB21, a homolog of Arabidopsis MYB46/83, to regulate the expression of a number of genes associated with cell wall component and wood biosynthesis. However, Populus undergoes multiple gene duplication events during evolutionary process, resulting in the ratio of 1.4∼1.6 Populus homologs to each Arabidopsis gene (Tuskan et al., 2006). Duplicated genes may have divergent fates such as subfunctionalization, neofunctionalization, or non-functionalization, causing more complex regulation of wood formation in Populus compared with Arabidopsis. For instance, the TF protein-complex regulators (dimers and trimers) are shown to cooperatively or combinatorially mediate the biosynthesis of specific types of lignin in Populus stems (Chen et al., 2019).
Lignin is the generic term for a large group of aromatic polymers, and is one of the most important limiting factors in the conversion of plant biomass to pulp or biofuels (Vanholme et al., 2010; Liu et al., 2014; Zhao, 2016). These lignin polymers are resulted from the oxidative combinatorial coupling of 4-hydroxyphenylpropanoids. In plants, the main biosynthetic route of lignin is generally conserved and involved in a battery of enzymes (Vanholme et al., 2010). PHENYLALANINE AMMONIA-LYASE (PAL) is the first rate-limiting enzyme in the lignin biosynthesis pathway, and catalyzes the deamination of L-phenylalanine to cinnamic acid (Bate et al., 1994). Five Populus PAL isoforms (PtrPAL1 to PtrPAL5) exhibit essentially identical catalytic activities in secondary differentiating xylem (SDX) based on the Michaelis-Menten kinetic parameters, indicating functional redundancy in the synthesis of monolignol (Wang et al., 2014). 4-COUMARATE:CoA LIGASE (4CL) is the last enzyme in the phenylpropionic acid pathway, activating the hydroxycinnamic acids to their corresponding esters with coenzyme A, a rate-limiting enzyme connecting the phenylpropionic acid pathway and the lignin biosynthesis pathway (Lu et al., 2004). In Populus, there are two 4CL isoforms (Ptr4CL3 and Ptr4CL5) with distinct reaction kinetic parameters (Chen et al., 2013). Ptr4CL3 has the highest conversion rate for 4-coumaric acid, while Ptr4CL5 most effectively metabolizes caffeic acid (Wang et al., 2014). Transcriptional regulation of PtrPAL2 and Ptr4CL3 expression were experimentally proved during lignin biosynthesis (Chen et al., 2019; Gui et al., 2019). In the Populus SDX protoplasts, the expression of PtrPAL2 is directly activated by PtrMYB21 in PtrSND1-B1-mediated network (Chen et al., 2019). LTF1/PdMYB4 was identified as an upstream regulator of Pd4CL3 by screening a Populus developing xylem library using Y1H (Gui et al., 2019). In response to environmental stimuli, phosphorylation of LTF1 by MPK3/6 functions as a sensory switch regulating lignin biosynthesis. Currently, it remains unclear how PtrPAL2 and LTF1/PdMYB4, two lignin-associated proteins, are precisely regulated in woody cells.
To understand the regulatory mechanism of wood formation at the transcriptional level, we here generated a wood-associated Gal4-AD–TF library that contains 517 TFs from poplar, a fast-growing tree species. High-throughput Y1H and Y2H screens were applied to verify the efficiency of this library using two lignin-associated genes (PaPAL2 and LTF1/PaMYB4) as the baits. The interactions between 5 TFs and the promoter fragment of PaPAL2 and between 10 TFs and LTF1/PaMYB4 offer a clue as to how lignin biosynthesis is precisely controlled. Our high-throughput Y1H/Y2H screening system is a powerful tool to help dissect transcriptional regulatory networks of wood formation in trees.
To generate the Gal4–AD-fused constructs, the coding regions of 517 TFs were amplified by PCR from the first-strand cDNA, that was prepared with the whole stem of 1.5-m-high Populus alba × P. glandulosa cv “84K” following the method described previously (Chai et al., 2014). After confirmation by sequencing, the PCR products were cloned into pGADT7 through the EcoRI site. The resulting constructs were transformed into the yeast strain Y187 using the PEG/LiAc method. The yeast cells containing the transformant was mixed with 30% sterilized glycerol in 2-ml 96-well plates and stored at −80°C for use.
The 500-bp promoter fragment of PaPAL2 was amplified from genomic DNA of “84K” and the coding region of LTF1/PaMYB4 was amplified from the xylem cDNA using gene-specific primers (Supplementary Table 1). Following the methods described by Ou et al. (2011), this PaPAL2 promoter fragment was inserted into the vector pHIS2.1 for mating-based Y1H screening, and the LTF1 coding region was inserted into pGBKT7 for mating-based Y2H screening. Yeast strain AH109 carrying the two baits was grown in selective medium and the Gal4–AD–TF strains were grown on SD-Leu medium in 96-well plates overnight. 30 μL/well of donor and host strains were transferred to a new 96-well plate with 100 μL YPAD medium in each well. Mating was conducted for 20–24 h by shaking at 30°C and 200 rpm. After 10-fold dilution with water, the mating products (5 μL well–1) were plated to different selective plates and incubated for 3 days at 30°C.
The coding regions of PaMYB21, PaMYBH, PaWRKY20 and PaDF1, four regulatory candidates of PaPAL2 identified by mating-based Y1H, were amplified from the xylem cDNA of “84K” (primers in Supplementary Table 1) and cloned into the pGADT7 vector. Each of these pGADT7-TFs (pGADT7 as control) and pHIS-proPaPAL2 were co-transformed into yeast stain Y187. The transformed cells were observed on the SD/-Leu-His plates and triple dropout plates (SD/-Trp-His-Leu) supplemented with 10 mM 3-amino-1,2,4-triazole (3-AT) for 3 days at 28°C, following the Y1H procedure described previously (Wang et al., 2020).
EMSA was performed following the method described previously (Chai et al., 2014). The coding regions of PaMYBH, PaWRKY20, and PaDF1 were fused in frame with MBP in pMAL-p4X and expressed in Escherichia coli. Recombinant protein was purified using amylose resin (New England Biolabs). The PaPAL2 promoter fragments covering the corresponding binding sites of PaMYBH, PaWRKY20, and PaDF1 were labeled with biotin at the 5’ end and used as the probes (Supplementary Figure 1, BGI). The same unlabeled oligos were annealed for competition. Binding reactions were performed with a LightShift® Chemiluminescent EMSA Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. The Pa MYBH-, PaWRKY20- or PaDF1-bound DNA fragments were separated from the unbound fragments by polyacrylamide gel electrophoresis. The DNA on a nitrocellulose membrane was detected by chemiluminescence. Three independent experiments were performed for each probe.
The coding regions of PaMYB21, PaGRAS2, PaWRKY20, PaANAC83, and PaDF1 identified by mating Y2H were amplified from the xylem cDNA of “84K” and cloned into the pGBKT7 vector (BD). Primers were shown in Supplementary Table 1. The pGADT7-LTF1/PaMYB4 and pGBKT7-TF were co-transformed into yeast strain AH109. The transformed cells were cultured on SD/-Leu/-Trp plates for 2 days, and then dropped on the SD/-Leu/-Trp/-Ade/-His medium with or without 5 mM 3-AT for 3 days at 30°C following the Y2H procedure described previously (Tang et al., 2020). The empty vector pGADT7 was used as blank control.
Bimolecular fluorescence complementation (BiFC) assays were conducted according to the method described previously (Tang et al., 2020). The coding region of LTF1/PaMYB4 was cloned into the pSAT1-nVenus (pE3228) vector, and that of PaMYB21, PaWRKY20 or PaANAC83 was cloned into pSAT1-cCFP (pE3242) (Walter et al., 2004). Arabidopsis leaf protoplasts were isolated and transfected following the method established by Sheen (2001). Confocal microscopy was performed with an Olympus FluoView FV1000 confocal microscope using an excitation wavelength of 488 nm and detection at 499–535 nm. The experiments were repeated three times.
To obtain wood-associated transcription factors (WTFs), we extracted 4287 Populus TFs covering 58 gene families from both PlantTFDB1 and Phytozome 122 (Figure 1). A total of 517 genes in 49 families were defined as WTFs based on the stringent filtering criteria as follows: genes show high (> 100) expression levels in xylem in Phytozome 12 and specific expression in wood tissues identified by the online software BAR3. Full-length ORFs of the 517 genes were amplified from the first-strand cDNA in the whole stems of Populus alba × P. glandulosa cv “84K.” After validation by sequencing, All ORFs were independently inserted into the pGADT7 vector through the EcoRI site at multiple clone sites (MCS) and then transformed into the yeast strain Y187.
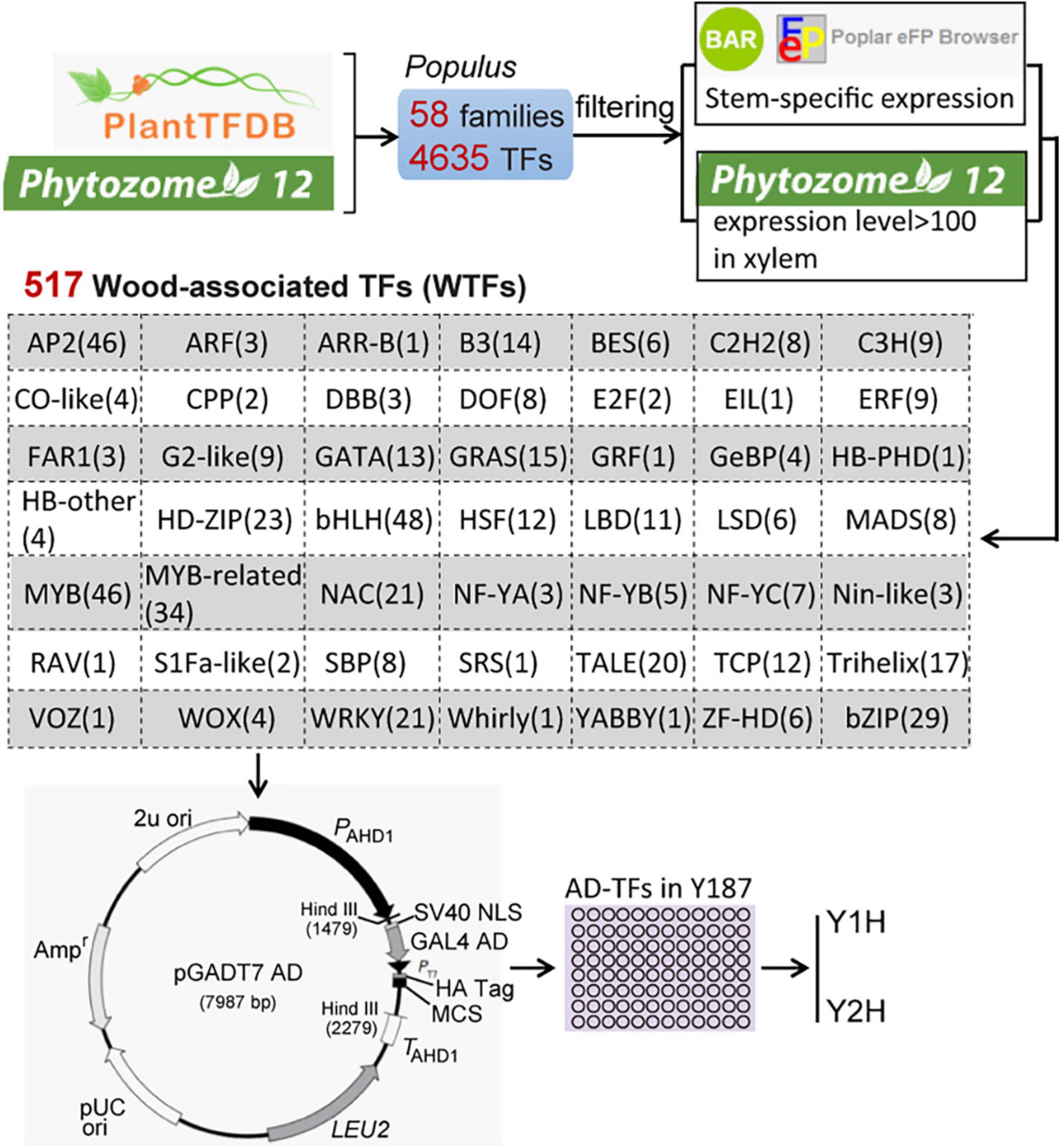
Figure 1. The scheme of the construction of a wood-associated transcription factors (TFs) library. A total of 517 Wood-associated TFs (WTFs) were identified from 4287 Populus TFs of 58 gene families in two database (PlantTDB and Phytozome 12) using the stringent filtering criteria. All pGADT7-TFs were independently transformed into the yeast strain Y187 for throughout mating-based Y1H or Y2H screening.
PaPAL2, the ortholog of Arabidopsis PAL1 in Populus, is a rate-limiting enzyme that controls the deamination of phenylpropanoid to cinnamic acid during monolignol biosynthesis in woody cells (Wang et al., 2014). Further, PaPAL2 is a direct target of PaMYB21 that is the second-layered master regulator of wood formation (Chen et al., 2019). Thus, PaPAL2 was selected to test the efficiency of this yeast library and to clarify the lignin biosynthesis pathway. Following the method described previously (Ou et al., 2011), we screened this library by mating the bait, a 500 bp promoter fragment upstream of ATG of PaPAL2, in Y1H assays. Yeast cells co-transformed with pHIS-proPaPAL2 and each of five pGADT7-TFs that were identified were able to grow normally on medium SD-Trp-Leu-His with 10 mM 3-AT (Figures 2A,B). These five WTFs belong to the MYB (PaMYB21), HD-ZIP (PaHB5), MYB-related (PaMYBH), Trihelix (PaDF1), and WRKY (PaWRKY20) family (Table 1). We further examined the cell-type expression patterns of PaPAL2 and its five regulatory candidates in stems, based on high-spatial-resolution transcriptome data that were sampled across secondary stem tissues in Populus (Sundell et al., 2017). As revealed in Supplementary Figure 2A, PaPAL2, PaMYB21, PaMYBH, PaHB5, PaDF1, or PaWRKY20 were expressed in wood-forming cells, including the cambial zone and secondary phloem/xylem cells.
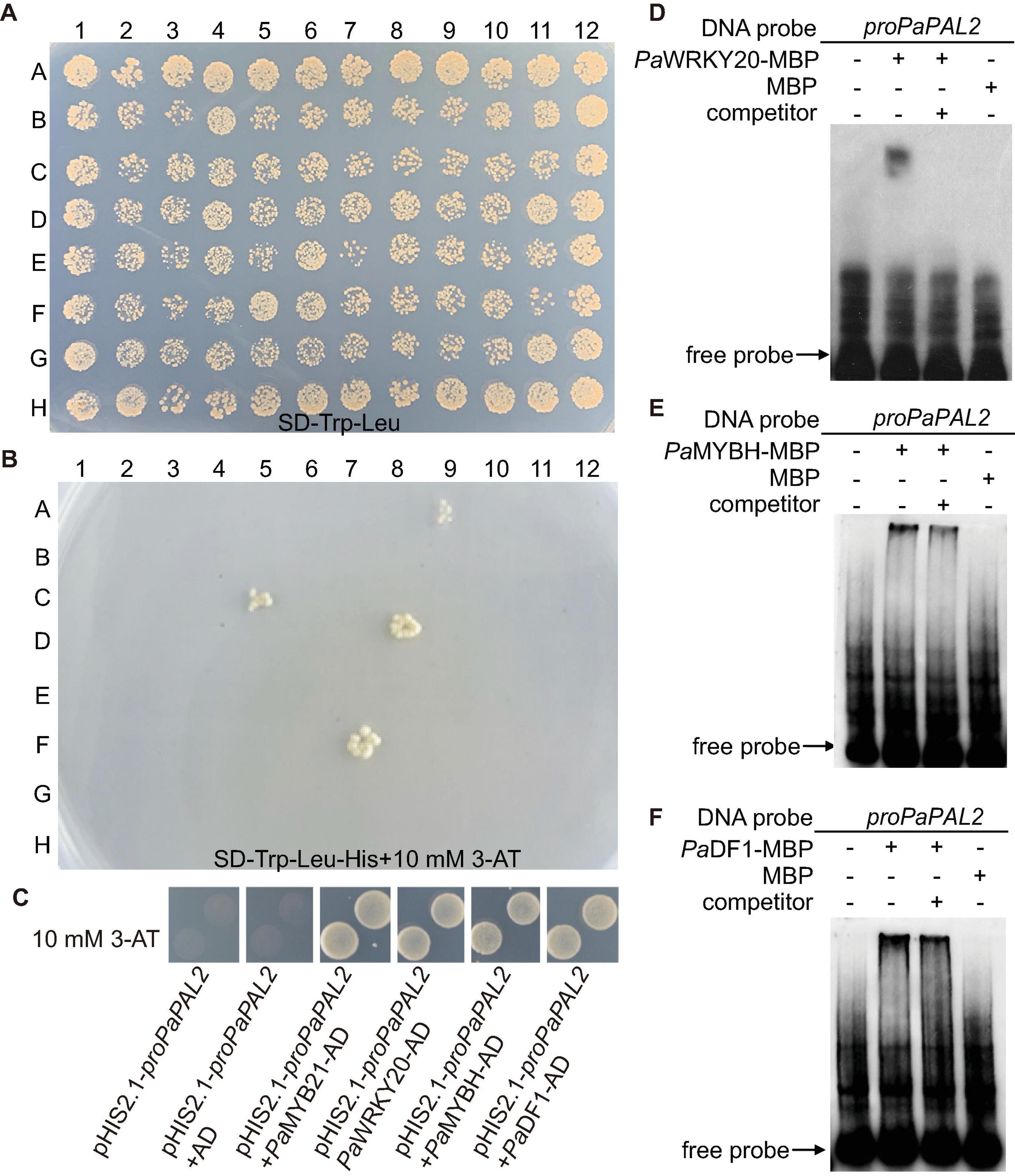
Figure 2. Identification of TFs that bind to the PaPAL2 promoter. (A,B) Y1H screening of the TFs library using 500-bp PaPAL2 promoter fragment as a bait. Yeast growth on the non-selective medium SD-Trp-Leu showed the mating efficiency. The candidate TFs can be detected on medium with 10 mM 3-AT. (C) Y1H assays verifying that PaMYB21, PaWRKY20, PaMYBH, or PaDF1 was able to bind to the PaPAL2 promoter fragment. Plasmids transformed into Y187 were screened on the SD/-Trp-Leu-His medium with 10 mM 3-AT. (D–F) EMSA results showing the specific bindings of PaWRKY20, PaMYBH, or PaDF1 protein to the PaPAL2 promoter in vitro. For competition assays, unlabeled probes (competitor) in 20-fold (+) molar excess relative to the labeled probe was include in the reaction.
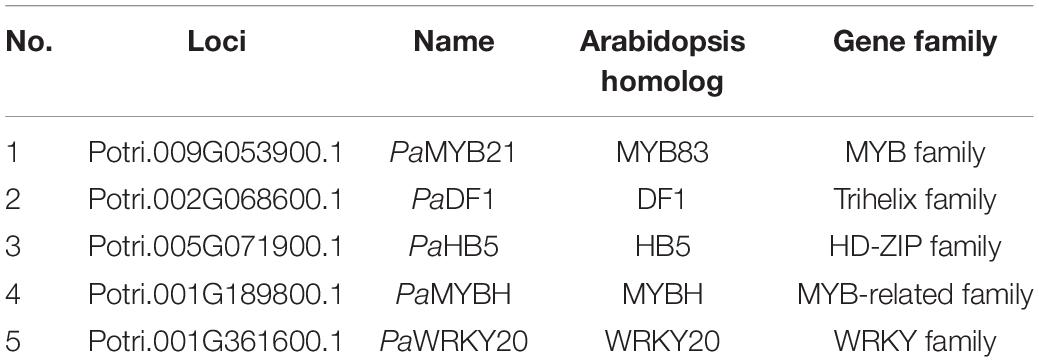
Table 1. Transcription factors that bind to the PaPAL2 promoter identified in the Populus wood-associated library using Y1H.
PaMYB21, PaDF1, PaMYBH, and PaWRKY20, like PaPAL2, exhibited high expression in lignified xylem, and they were selected for validation by Y1H method. Yeast cells co-transformed with pHIS-proPaPAL2 and pGADT7-TF grew normally on the SD-Trp-Leu-His medium supplemented with 10 mM 3-AT (Figure 2C), indicating the binding of PaMYB21, PaDF1, PaMYBH or PaWRKY20 to the PaPAL2 promoter in yeast. Analysis of the 500-bp PaPAL2 promoter fragment showed the DNA-binding motifs of PaDF1 (GT, Kaplan-Levy et al., 2012), PaMYBH (MRE, Du et al., 2013) and PaWRKY20 (W-box, Ulker and Somssich, 2004; Supplementary Figure 2). Not surprisingly, EMSA results revealed that PaDF1, PaMYBH, and PaWRKY20 proteins were able to bind to these cis-elements of the PaPAL2 promoter specifically in vitro (Figures 2D–F).
LTF1/PaMYB4 is a key negative regulator of lignin biosynthesis in wood formation (Gui et al., 2019; Holwerda et al., 2019). To elucidate the regulatory mechanism of lignin biosynthesis, we screened the LTF1/PaMYB4-interacting proteins in our TF library by mating the bait LTF1/PaMYB4. Ten TF clones were obtained (Figures 3A,B). Three of them were in the Trihelix family, three were in the GRAS family, and one in the MYB, NAC, WRKY, or SBP family (Table 2). Interestingly, these TFs included PaMYB21, PaDF1, and PaWRKY20, that were shown to directly regulate PaPAL2 expression (Figure 2). Analysis of high-spatial-resolution wood transcriptome profiles revealed the high expression of LTF1/PaMYB4 and its 10 interacting proteins in wood-forming tissues such as secondary phloem and lignified xylem (Supplementary Figure 1B).
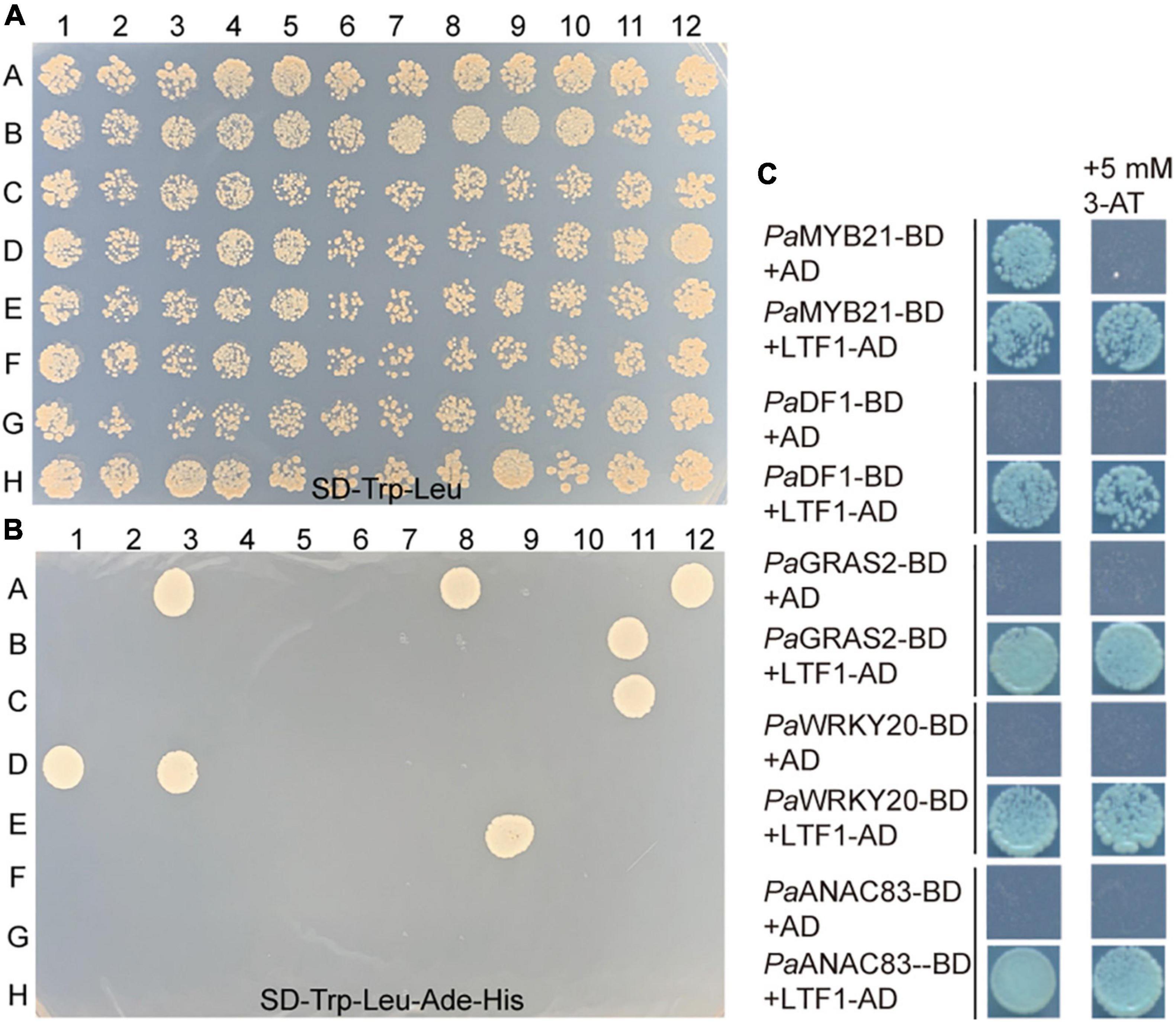
Figure 3. Identification of TFs that interact with PaMYB4/LTF1. (A,B) Y2H screening of this TFs library by mating the bait PaMYB4/LTF1. Strong interaction can be detected on the stringent medium SD-Leu-Trp-His-Ade. (C) Y2H assays in yeast confirming the interaction between PaMYB4/LTF1 and PaMYB21, PaDF1, PaGRAS2, PaWRKY20, or PaANAC83 in vivo. Plasmids transformed into AH109 were screened on the SD/-Trp-Leu-His-Ade medium with or without 5 mM 3-AT.
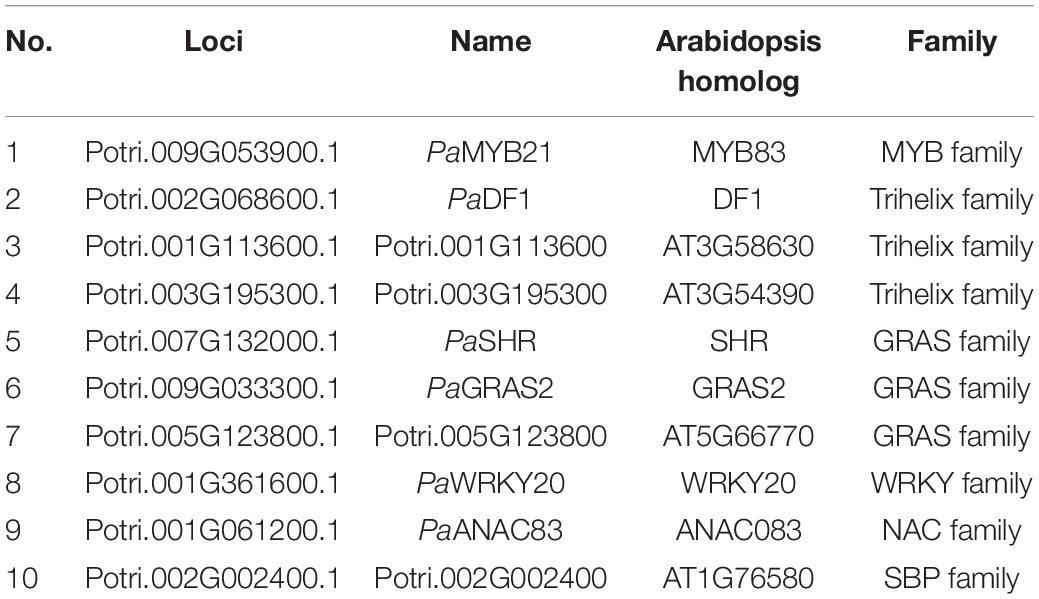
Table 2. Transcription factors that interact with PaMYB4/LTF1 identified in the Populus wood-associated library using Y2H.
Five TFs (PaMYB21, PaDF1, PaGRAS2, PaWRKY20, and PaANAC83) were selected to verify their interactions with PaMYB4 using Y2H (Figure 3C). Yeast cells transformed with TF-BD and PaMYB4-AD exhibited blue on SD/-Leu-Trp-His-Ade plate, confirming the interaction between the five TFs and LTF1/PaMYB4. Of the five interacting TFs, only PaMYB21 protein had self-activation in yeast, that can be effectively inhibited by addition of 5 mM 3-AT. BiFC assays were conducted in Arabidopsis leaf protoplasts to verify the in vivo interaction between PaMYB4/TFL1 and PaMYB21, PaDF1, or PaWRKY20 (Figure 4). Co-expression of PaMYB4/LTF1 fused to the amino-terminal half of YFP (YFPNE) and PaMYB21, PaDF1, or PaWRKY20 fused to the carboxy-terminal half (YFPCE) of yellow florescent protein led to visible fluorescence in the nucleus of co-transformed protoplasts. However, no YFP fluorescence was detected when PaMYB4-YFPNE was co-expressed with the carboxy-terminal half of YFP (cYFP) or PaMYB21-, PaDF1- or PaWRKY20-YFPCE was co-expressed with the amino-terminal half of YFP (nYFP).
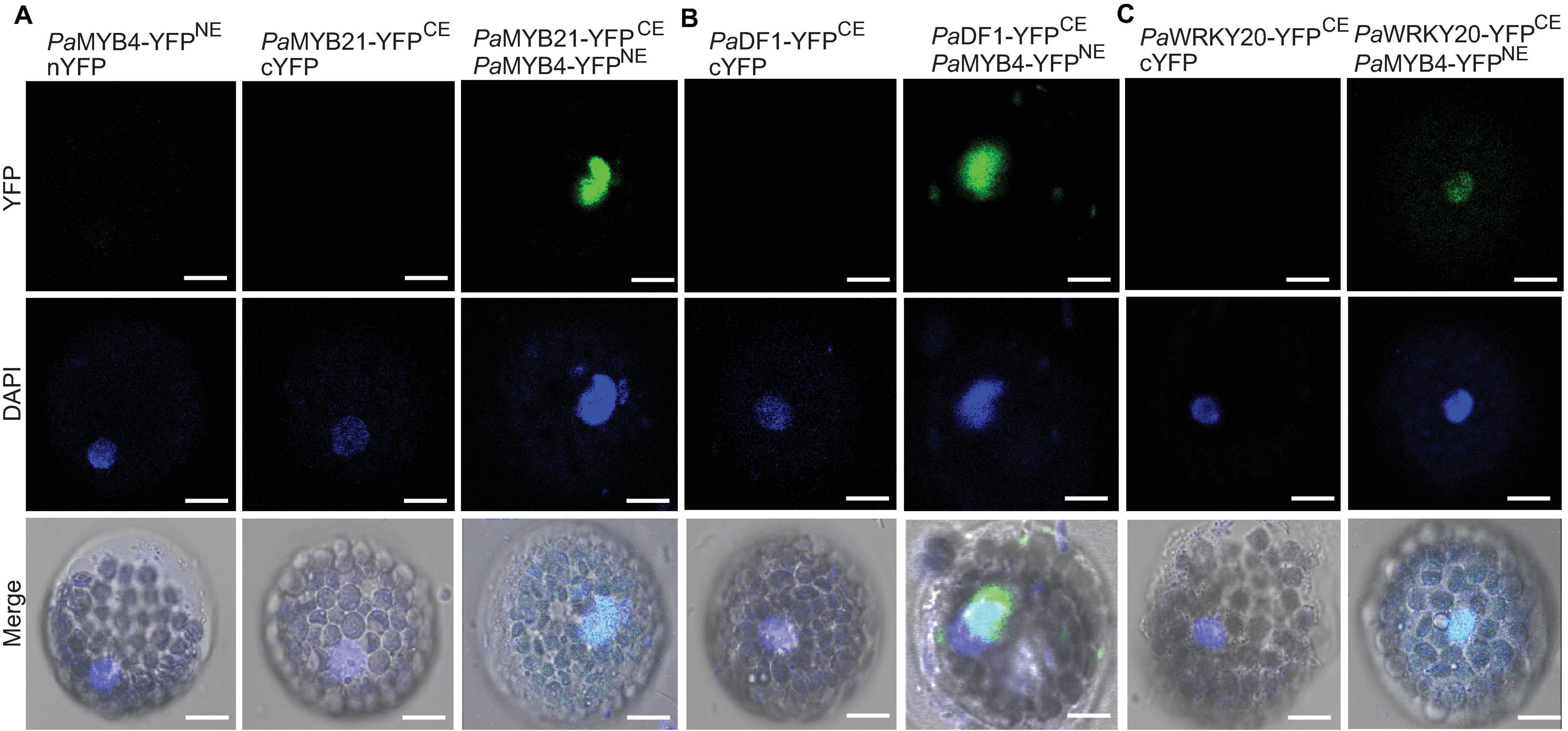
Figure 4. BiFC validation of the in vivo interaction between PaMYB21, PaDF1, or PaWRKY20 and PaMYB4/LTF1. (A–C) Pa MYB21-, Pa DF1-, or PaWRKY20-YFPCE and PaMYB4-YFPNE interact to form a functional YFP in the nucleus of Arabidopsis leaf protoplasts. The DNA fluorochrome 4′, 6-diamidino-2-phenylindole (DAPI) was used to stain the cell nucleus (blue). Bar = 10 μm.
Recent studies in the stem of Populus show that wood formation involves regulatory homeostasis determined by combinations of TF-DNA and TF-protein regulations (Petzold et al., 2017; Chen et al., 2019). Using Y1H and Y2H assays, 40 protein-DNA interactions involving 20 different TFs and 165 protein-protein interactions involving 162 different proteins are shown to be relevant to wood formation (Petzold et al., 2017). These interactions are incorporated into a network that includes 14 connected subnetworks, with the largest having 132 members. Integration of quantitative transcriptomics and chromatin binding data constructs a TRN, in which PtrSND1-B1 directs 57 TF-DNA interactions through 17 TFs transregulating 27 cell wall genes (Chen et al., 2019). Of the multiple methods for determining TF-DNA and TF-protein interactions, Y1H and Y2H screenings are the most widely used in trees. However, a major limitation for the two methods is low expression levels of TFs in wood-forming tissues. To overcome this disadvantage, we set up a high throughout mating-based screening system that includes a Gal4-AD-TF library of 517 wood-associated TFs (WTFs). The 517 WTFs were filtered by the stringent criteria and accounted for 11% of all Populus TFs (4287). By mating yeast strains (bait) with a series of strains (prey) expressing WTF-AD protein, we were able to identify protein-DNA or protein-protein interactions and identify connected networks during wood formation. Importantly, our system is still effective for Y1H screening using a single fragment of up to 500 bp as a bait, which has an advantage of minimizing the effort to find out the exact cis-element and increasing the screening specificity. Thus, our library is a resource for Y1H and Y2H assay and functional analyses of genes associated with poplar wood formation.
Lignin biosynthesis starts with the deamination of phenylalanine, followed by a series of hydroxylation, methylation and reduction resulting in the production of p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S), three basic units of the lignin complex in trees (Zhao, 2016). In the Populus lignin biosynthesis pathway, PtrMYB90, PtrMYB161, PtrNAC123, or PtrWBLH1 targets G-specific CCoAOMT gene family members, and PtrMYB90, PtrMYB161, or PtrWBLH1 targets CAld5H that is needed for the biosynthesis of the S subunits (Chen et al., 2019). In this study, we focus on PaPAL2 that is rate-limiting enzyme in the lignin synthesis pathway, and LTF1/PaMYB4 that targets Pa4CL3, a key enzyme of general phenylpropanoid metabolism that provides the precursors for both lignin biosynthesis (Lu et al., 2004; Chen et al., 2013, 2019; Wang et al., 2014; Gui et al., 2019).
With the help of a high-throughput Y1H/Y2H screening system, we identified five TFs that bound to the PaPAL2 promoter fragment and 10 TFs interacting with PaMYB4 in Populus, indicating a high efficiency of our library. Selected interactions were further verified by EMSA and BiFC independently. Of these TFs, PaMYB21, PaWRKY20, PaDF1 were found to interact with the PaPAL2 promoter or PaMYB4 protein. Consistently, they exhibited overlapping expression with PaPAL2 or PaMYB4 in wood-forming tissues predicted by high-spatial-resolution wood transcriptome data (Sundell et al., 2017). Of these interactors, PtrMYB21 is proven to directly activate PtrPAL2 expression, promoting lignin biosynthesis during wood formation (Chen et al., 2019). Other TFs are not functionally characterized in trees, but the homologs of PaDF1 and PaWRKY20 in Arabidopsis were identified as the components of the gene regulatory network for secondary cell wall synthesis in stems (Taylor-Teeples et al., 2015). These preliminary results revealed a coordinated role of the TF-proPaPAL2 and TF-PaMYB4 interactions in lignin biosynthesis in Populus stems. The TF-DNA interactions for Populus PAL4, a paralog of PaPAL2, were also identified in xylem cells by Y1H assays (Petzold et al., 2017). Six TFs, including PtrMYB2 that is a homolog of PtrMYB21, are shown to bind to the PtrPAL4 promoter. This suggests different TF-DNA interactions between homologs in woody cells. Our powerful system will facilitate to generate comprehensive interaction networks of TFs to understand the regulatory mechanism of wood formation in tree species.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
GC designed the experiments, performed data processing, and drafted the manuscript. YZhu, SC, WL, LX, DW, CW, JM, XT, HX, SW, LD, and YZha prepared the materials and performed the experiments. GZ conceived the study and revised the manuscript. All authors read and approved the final version of the manuscript.
Financial support for this work was obtained from the National Key Scientific Research Project of China (2021YFD2200205), the National Natural Science Foundation of China (31670606, 31972955, 31770315, 32101549, and 31972860), the Taishan Scholar Program of Shandong (to GC), and the “First Class Grassland Science Discipline” Program of Shandong Province.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Dr. Guoqing Dong (Wuhan Polytechnic University) for critical suggestions of this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.715809/full#supplementary-material
Albersheim, P., Darvill, A., Roberts, K., Sederoff, R., and Staehelin, A. (2011). “Cell walls and plant anatomy,” in Plant Cell Walls: From Chemistry to Biology, eds P. Albersheim, A. Darvill, K. Roberts, R. Sederoff, and A. Staehelin (New York: Garland Science), 1–42.
Bate, N. J., Orr, J., Ni, W., Meromi, A., Nadler-Hassar, T., Doerner, P. W., et al. (1994). Quantitative relationship between phenylalanine ammonia-lyase levels and phenylpropanoid accumulation in transgenic tobacco identifies a rate-determining step in natural product synthesis. Proc. Natl. Acad. Sci. U. S. A. 91, 7608–7612. doi: 10.1073/pnas.91.16.7608
Chai, G., Qi, G., Cao, Y., Wang, Z., Yu, L., Tang, X., et al. (2014). Poplar PdC3H17 and PdC3H18 are direct targets of PdMYB3 and PdMYB21, and positively regulate secondary wall formation in Arabidopsis and poplar. New Phytol. 203, 520–534. doi: 10.1111/nph.12825
Chen, H., Song, J., Williams, C. M., Shuford, C. M., Liu, J., Wang, J. P., et al. (2013). Monolignol pathway 4-coumaric acid:coenzyme A ligases in Populus trichocarpa: novel specificity, metabolic regulation, and simulation of coenzyme A ligation fluxes. Plant Physiol. 161, 1501–1516. doi: 10.1104/pp.112.210971
Chen, H., Wang, J. P., Liu, H., Li, H., Lin, Y. J., Shi, R., et al. (2019). Hierarchical transcription factor and chromatin binding network for wood formation in black cottonwood (Populus trichocarpa). Plant Cell 31, 602–626. doi: 10.1105/tpc.18.00620
Du, H., Wang, Y. B., Xie, Y., Liang, Z., Jiang, S. J., Zhang, S. S., et al. (2013). Genome-wide identification and evolutionary and expression analyses of MYB-related genes in land plants. DNA Res. 20, 437–448. doi: 10.1093/dnares/dst021
Du, J., and Groover, A. (2010). Transcriptional regulation of secondary growth and wood formation. J. Integr. Plant Biol. 52, 17–27. doi: 10.1111/j.1744-7909.2010.00901.x
Gui, J., Luo, L., Zhong, Y., Sun, J., Umezawa, T., and Li, L. (2019). Phosphorylation of LTF1, an MYB transcription factor in Populus, acts as a sensory switch regulating lignin biosynthesis in wood cells. Mol. Plant 12, 1325–1337. doi: 10.1016/j.molp.2019.05.008
Holwerda, E. K., Worthen, R. S., Kothari, N., Lasky, R. C., Davison, B. H., Fu, C., et al. (2019). Multiple levers for overcoming the recalcitrance of lignocellulosic biomass. Biotechnol. Biofuels 17:15. doi: 10.1186/s13068-019-1353-7
Kaplan-Levy, R. N., Brewer, P. B., Quon, T., and Smyth, D. R. (2012). The trihelix family of transcription factors-light, stress and development. Trends Plant Sci. 17, 163–171. doi: 10.1016/j.tplants.2011.12.002
Lin, Y. C., Li, W., Sun, Y. H., Kumari, S., Wei, H., Li, Q., et al. (2013). SND1 transcription factor-directed quantitative functional hierarchical genetic regulatory network in wood formation in Populus trichocarpa. Plant Cell 25, 4324–4341. doi: 10.1105/tpc.113.117697
Liu, C., Cai, Y., Zhang, X., Gou, M., and Yang, H. (2014). Tailoring lignin biosynthesis for efficient and sustainable biofuel production. Plant Biotechnol. J. 12, 1154–1162. doi: 10.1111/pbi.12250
Lu, H., Zhao, Y. L., and Jiang, X. N. (2004). Stable and specific expression of 4-coumarate: coenzyme A ligase gene (4CL1) driven by the xylem-specific Pto4CL1 promoter in the transgenic tobacco. Biotechnol. Lett. 26, 1147–1152. doi: 10.1023/B:BILE.0000035487.91628.9e
Nakano, Y., Yamaguchi, M., Endo, H., Rejab, N. A., and Ohtani, M. (2015). NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 6:288. doi: 10.3389/fpls.2015.00288
Ou, B., Yin, K., Liu, S., Yang, Y., Gu, T., Hui, J. M. W., et al. (2011). A high-throughput screening system for Arabidopsis transcription factors and its application to Med25-dependent transcriptional regulation. Mol. Plant 4, 546–555. doi: 10.1093/mp/ssr002
Petzold, E., Rigoulot, S. B., Zhao, C., Chanda, B., Sheng, X., Zhao, M., et al. (2017). Identification of new protein-protein and protein-DNA interactions linked with wood formation in Populus trichocarpa. Tree Physiol. 38, 362–377. doi: 10.1093/treephys/tpx121
Sheen, J. (2001). Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 127, 1466–1475.
Sundell, D., Street, N. R., Kumar, M., Mellerowicz, E. J., Kucukoglu, M., Johnsson, C., et al. (2017). AspWood: high-spatial-resolution transcriptome profiles reveal uncharacterized modularity of wood formation in Populus tremula. Plant Cell 29, 1585–1604. doi: 10.1105/tpc.17.00153
Tang, X., Wang, D., Liu, Y., Lu, M., Zhuang, Y., Xie, Z., et al. (2020). Dual regulation of xylem formation by an auxin-mediated PaC3H17 and PaMYB199 module in Populus. New Phytol. 225, 1545–1561. doi: 10.1111/nph.16244
Taylor-Teeples, M., Lin, L., de Lucas, M., Turco, G., Toal, T. W., Gaudinier, A., et al. (2015). An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517, 571–575. doi: 10.1038/nature14099
Tuskan, G. A., Difazio, S., Jansson, J., Grigoriev, I., Hellsten, U., Putnam, N., et al. (2006). The genome of black cottonwood, Populus trichocarpa. Science 313, 1596–1604. doi: 10.1126/science.1128691
Ulker, B., and Somssich, I. (2004). WRKY transcription factors: from DNA binding towards biological function. Curr. Opin. Plant Biol. 7, 491–498. doi: 10.1016/j.pbi.2004.07.012
Vanholme, R., Demedts, B., Morreel, K., Palph, J., and Boerjan, W. (2010). Lignin biosynthesis and structure. Plant Physiol. 153, 895–905. doi: 10.1104/pp.110.155119
Walter, M., Chaban, C., Schutze, K., Batistic, O., Weckermann, K., Nake, C., et al. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438. doi: 10.1111/j.1365-313X.2004.02219.x
Wang, D., Xu, H., Huang, J., Kong, Y., AbuQamar, S., Yu, D., et al. (2020). The Arabidopsis CCCH protein C3H14 contributes to basal defense against Botrytis cinerea mainly through the WRKY33-dependent pathway. Plant Cell Environ. 43, 1792–1806. doi: 10.1111/pce.13771
Wang, H. Z., and Dixon, R. A. (2012). On-off switches for secondary cell wall biosynthesis. Mol. Plant 5, 297–303. doi: 10.1093/mp/ssr098
Wang, J. P., Naik, P. P., Chen, H. C., Shi, R., Lin, C. Y., Liu, J., et al. (2014). Complete proteomic-based enzyme reaction and inhibition kinetics reveal how monolignol biosynthetic enzyme families affect metabolic flux and lignin in Populus trichocarpa. Plant Cell 26, 894–914. doi: 10.1105/tpc.113.120881
Zhao, Q. (2016). Lignification: flexibility, biosynthesis and regulation. Trends Plant Sci. 21, 713–721. doi: 10.1016/j.tplants.2016.04.006
Keywords: wood, transcription factors, Y1H, Y2H, lignin, Populus
Citation: Zhuang Y, Chen S, Lian W, Xu L, Wang D, Wang C, Meng J, Tang X, Xu H, Wang S, Du L, Zhang Y, Zhou G and Chai G (2022) A High-Throughput Screening System for Populus Wood-Associated Transcription Factors and Its Application to Lignin Regulation. Front. Plant Sci. 12:715809. doi: 10.3389/fpls.2021.715809
Received: 27 May 2021; Accepted: 09 December 2021;
Published: 14 January 2022.
Edited by:
Hong Luo, Clemson University, United StatesReviewed by:
Concepcion Avila, University of Malaga, SpainCopyright © 2022 Zhuang, Chen, Lian, Xu, Wang, Wang, Meng, Tang, Xu, Wang, Du, Zhang, Zhou and Chai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gongke Zhou, emhvdWdrQHFhdS5lZHUuY24=; Guohua Chai, Y2hhaWdoQHFhdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.