
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 26 July 2021
Sec. Plant Breeding
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.715633
The fatty acid desaturase FAD2 genes are the main contributors to oleic acid content, and different FAD2 alleles can result in different oleic acid contents in rapeseed oil. Hence, identification of allelic variation in FAD2 is an extremely desirable breeding goal. By performing QTL mapping using 190 F2:3 lines genotyped by genome-wide single nucleotide polymorphism (SNP) markers assayed by the Brassica 60 K Infinium BeadChip Array, four quantitative trait loci (QTL) for C18:1 content were mapped on chromosomes A01, A05, A09 and C05 over 3 years in a population segregating for oleic acid content. Two BnFAD2 genes on A05 and C05 were anchored within the QTL intervals, explaining 45–52 and 15–44% of the observed variation for C18:1 content. Sequence polymorphisms between the corresponding coding regions of the parental lines found two single-nucleotide polymorphisms (SNPs) in BnFAD2.A05 and BnFAD2.C05, respectively, which led to the amino acid changes (C421T and G1073E) in the corresponding proteins. The mutation sites of Bnfad2.A05 and Bnfad2.C05 alleles were located within the second H-box and near the third H-box motif of the protein, respectively, and were found to be novel mutant alleles. Lines resulting from the combination of these two alleles contained up to 88% oleic acid in their seed oil, compared with 63% in wild-type controls. Two competitive allele-specific PCR (KASP) markers based on these two mutation sites were successfully developed and validated in segregating F2 populations. These markers will facilitate breeding for ultra-high seed oleic acid content in oilseed rape.
Oilseed rape (Brassica napus L.) is one of the most important oil crops worldwide. The quality of rapeseed oil is mainly determined by the fatty acid composition of the seeds (Napier et al., 2014), in particular by three unsaturated fatty acids: oleic acid (C18:1), linoleic acid (C18:2) and linolenic acid (C18:3) (Micha and Mozaffarian, 2009; Gillingham et al., 2011). High-oleic acid (>75%) oils have several benefits compared to non-high oleic acid oils, including decreasing low-density lipoprotein levels and putatively the risk of cardiovascular disease in humans (Chang and Huang, 1998) as well as superior anti-oxidative ability leading to a longer shelf life of the oil product (Browse et al., 1998; Lauridsen et al., 1999; Przybylski et al., 2013). Currently, the majority of oilseed rape cultivars worldwide are of canola quality (low erucic acid and glucosinolate content) and contain ~55–65% oleic acid content (Long et al., 2018). Identifying novel high oleic acid germplasm to further increase oleic acid content is a major goal for quality breeding of oilseed rape.
To date, several quantitative trait loci (QTL) for oleic acid content have been identified in Brassica (Burns et al., 2003; Hu et al., 2006; Zhao et al., 2008; Smooker et al., 2011; Yan et al., 2011; Yang et al., 2012; Wen et al., 2015; Bao et al., 2018; Chen et al., 2018) and association mapping studies (Niklas et al., 2016; Qu et al., 2017; Bao et al., 2018; Zhao et al., 2019). Of these, one QTL on chromosome C01 (Hu et al., 2006) and one major QTL located on A05 (Hu et al., 2006; Yang et al., 2012) were found to contain homologs to Arabidopsis AtFAD2, which catalyzes oleic acid (C18:1) into linoleic acid (C18:2), and hence plays essential role in regulating oleic acid content in seeds. In B. napus, four AtFAD2 orthologs have been identified in total: the major-effect C01 and A05 QTL plus their homoeologous copies on chromosomes A01 and C05 (Yang et al., 2012). Although BnFAD2.A01 appears to be a pseudogene, the three other copies are functional.
Variants in the B. napus FAD2 copies have previously been found to affect oleic acid content. A single nucleotide polymorphism in the BnFAD2.C01 coding region resulted in an increase in oleic acid content up to 77%, while for BnFAD2.A05, a single-nucleotide substitution (Hu et al., 2006) or a 4-bp insertion (Yang et al., 2012) in the coding region resulted in an increase in oleic acid content up to 75%. A new high oleic acid mutant reported by Long et al. (2018) showed two SNPs in BnFAD2.A05 and BnFAD2.C05, again confirming the importance of BnFAD2 for oleic acid content. Moreover, loss of function of BnFAD2 in B. napus via gene knockout (Wells et al., 2014), RNA interference (Peng et al., 2010), and CRISPR/Cas9-mediated genome editing (Okuzaki et al., 2018), all result in an increase in oleic acid content of up to 84–85%. Although great success has been achieved by manipulating BnFAD2 to obtain high oleic acid oilseed rape, most high oleic germplasm to date relies on mutation of BnFAD2.A5, and seems to be linked to poor agronomic performance of the plant, particularly at lower temperatures (Kinney, 1994; Bai et al., 2018). Development of novel genetic resources of oilseed rape with both high oleic acid content and superior field performance is thus highly desirable.
In the present study, QTL mapping was performed in B. napus using super-high (~85%) and normal-oleic acid (~65%) lines as parental lines. Two major QTLs were identified on A05 and C05, corresponded to the genes BnFAD2.A05 and BnFAD2.C05. Two previously unreported mutant alleles were identified for each of these genes. Subsequently, we developed and validated functional KASP markers for these genetic variants. Our results enhance our knowledge of the role of allelic variation in determining high oleic acid content and will facilitate breeding for high oleic acid varieties in oilseed rape.
A biparental population of 190 segregating F2:3 lines was derived from F2 offspring of a cross between a super-high (“FC81”, a homozygous M6 mutated line obtained from the 60Co-γ radiation with 85% seed oleic acid content) and normal-oleic acid (E183, 65%) lines, and was used for QTL mapping of oleic acid (Figure 1). The validation population was an F2 population also derived from crosses between super-high (FM5, a M6 mutated line which origin from the same high oleic acid mutant with “FC81,” 83%) and normal-oleic acid (FM6, 65%) lines, which was used for functional marker validation (Figure 1). Seed oleic acid contents of the F2:3 lines were measured on self-pollinated seed samples collected from field evaluation of the F2:3 population over 3 years. The inflorescences of 10 randomly-chosen plants in each plot were covered in pollen-proof bags at the onset of flowering to prevent cross-pollination. Self-pollinated seeds were collected in the bags at maturity for quality analysis. Seed oleic acid contents of the F2 validation populations were measured from the self-pollinated seeds from the F2 lines. All trials were grown at the agricultural field station of Zhejiang Academy of Agricultural Sciences (Hangzhou, China), with a plot size of 4.5 m2 (1.5 m plot size of rows per plot).
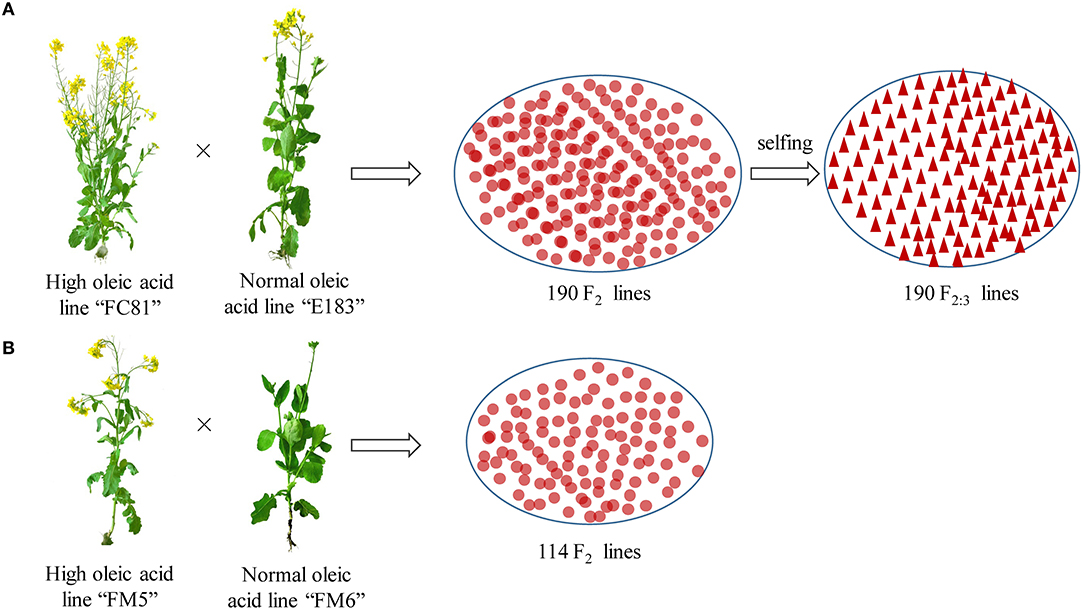
Figure 1. Experimental material and crossing design. (A, B) Represent the QTL mapping population and validation population, respectively.
Measurements for seed oleic acid content on the self-pollinated seeds were obtained by gas–liquid chromatography (GC) analysis with the Model 6890 GC analyser (Agilent Technologies, Inc., Wilmington, DE), following protocol described by Thies (1971), the phenotypic data of the 190 F2:3 lines was shown in Supplementary Table 1.
The Illumina Infinium Brassica 60K SNP Array (Clarke et al., 2016) was used for genotyping 190 F2 lines and two parental lines (Supplementary Table 2). Total genomic DNA was extracted using DP321-03 DNA extraction kits (Tiangen, Beijing, China). SNP genotyping was performed in the National Key Laboratory of Crop Genetic Improvement, National Subcenter of Rapeseed Improvement in Wuhan, Huazhong Agricultural University, Wuhan, China, according to the Infinium HD Assay Ultra manual protocols. Illumina HiSCAN scanner was used for imaging the hybridized chips. GenomeStudio v2011 (Illumina, Inc.) genotyping software was used for allele calling. SNP markers were named using the SNP plus index numbers assigned by GenomeStudio from the chip information (Clarke et al., 2016), followed by chromosome number. SNP positions were obtained by BLAST search (e ≤ 1e−50) against the B. napus genome reference Darmor v4.1 (Chalhoub et al., 2014).
Genetic linkage groups were constructed using the packages MSTmap (Wu et al., 2008) and JoinMap 4.0 softwares (Van Ooijen and Voorrips, 2006). Polymorphic SNP markers were grouped firstly by LOD 5.0, and marker orders were determined based on pairwise recombination frequencies, using MSTmap. Then, by applying the mapping function of Kosambi (1943) and a minimum LOD score of 3.0 using Joinmap 4.0, the marker order and distance were recalculated and confirmed. Markers with zero recombination were assigned to the same bin. QTL Detection for seed oleic content was performed in the F2:3 population using the composite interval mapping (CIM) procedure of the WinQTL Cartographer 2.5 software (Wang et al., 2005). A 1,000-permutation test was performed to estimate the significance threshold of the test statistic for each QTL based upon a 5% experiment-wise error rate (Churchill and Doerge, 1994).
The standard molecular cloning procedure described by Sambrook and Russell (2001) was followed to isolate the genomic sequence of the BnFAD2 genes in the two parents. The sequences were aligned among clones using the software VectorNTI (www.invitrogen.com/VectorNTI).
For each functional SNP of the BnFAD2 genes, two allele-specific forward primers and one common reverse primer were designed using the Primer Premier 5.0 program according to the standard KASP guidelines (Singh et al., 1998), and were named KASP-421 for the SNP BnaA.FAD2.a and KASP-1073 for the SNP BnaC.FAD2.a. For KASP-421, the two allele-specific primers were added with the standard FAM (5′-gttggaatggtggcgtcgatg-3′) and HEX (5′-gtgttggaatggtggcgtcgata-3′) tails respectively at the 5′ end, with a common reverse primer as follows: 5′-ggacgacaccgtcggcctca-3′. For KASP-1073, the two allele-specific primers were added with the standard FAM (5′-ggtggttaaggcgatgtggag-3′) and HEX (5′-cggtggttaaggcgatgtggaa-3′) tails respectively at the 5′ end, with the common reverse primer as follows: 5′-ccggttccacatagatacactcctt-3′.
The polymorphism of the developed KASP markers were firstly validated in four sequenced lines. The primer assay mixture comprised 46 μL ddH2O, 30 μL common reverse primer (μM), and 12 μL of each allele-specific forward primer (μM). KASP assays were carried out in 96-well plate formats using a Roche LightCycler 480-II instrument (Roche Applied Sciences, Beijing, China), and set up using 8 μl reaction volumes: 2.5 μL of 60 ng/μL DNA template, 2.5 μL of 2 × KASP master mixture, 0.07 μL of primer assay mixture, and 2.93 μL of ddH2O. PCR cycling was carried out as 94°C for 15 mins, followed by 10 touchdown cycles (94°C for 20 s; touchdown at 61°C initially and decreasing by 0.6°C per cycle for 60 s), followed by 26 additional cycles of annealing (94°C for 20 s; 55°C for 60 s). Fluorescence was read by the Pherastar and analyzed using the BMGPHERAstar Software.
Over 3 years, the high oleic acid content parental line “FC81” exhibited 84.4–86.9% oleic acid in self-pollinated seeds from the field trials, while the normal-oleic acid parental line “E183” exhibited 62.7–66.2% oleic acid. Transgressive segregation of seed oleic acid content was detected in the F2:3 population, with normal distribution of values ranging from 59.4 to 87.6% (Figure 2). Significant differences between genotypes, but no environment-specific or genotype-by-environment interactions were found for oleic acid content (ANOVA, P < 0.01) (Supplementary Table 3). Similarly, relatively high heritability of oleic acid content was observed across environments (H2 = 72.9%). For the other fatty acid production, significant difference was observed for C18:2 content between low oleic acid content (<75%) and high oleic acid content (>75%) (P < 0.05), with 14.62 and 9.10% of C18:2 content between two groups, while no significant difference was observed for C18:0 and C18:3 between two groups, with 1.45 and 1.50% of C18:0 content, 6.08 and 5.07% of C18:3 content between two groups, respectively. These results suggested that no other fatty acid production changes except the C18:1 and C18:2 content.
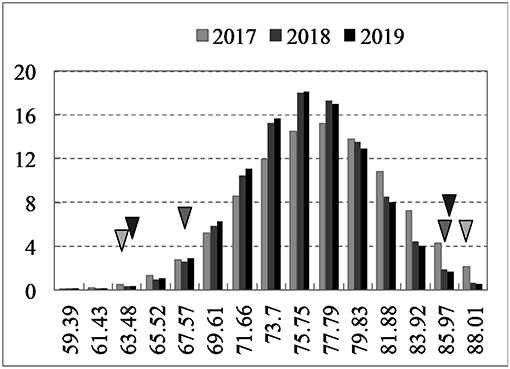
Figure 2. Frequency distributions of seed oleic acid content from 2017 to 2019. Triangles on the left and right of the figure represent the low oleic acid content line “E183” and high oleic acid content line “FC81”, respectively.
A total of 9728 SNP markers from the Brassica 60 K array showed expected segregation 1:1 ratio in the F2:3 population, which were used for genetic linkage construction. A set of 8128 SNP markers were successfully assigned to the 19 chromosomes of the A genome (A01–A10) and C genome (C01–C09) of B. napus, respectively. The genetic map spanned a genetic distance of 2311.16 cM, with an average distance of 0.28 cM between adjacent markers. As expected from previous genetic mapps of B. napus and the different size of chromosomes (Snowdon et al., 2002), the marker density and distribution considerably varied across chromosomes (Supplementary Figure 1). The highest marker density was found on chromosome A07, with 889 markers distributed over a genetic map distance of 75.53 cM. In the A genome four chromosomes showed gaps more than 20 cM, while only 1 chromosome in the C genome showed a gap over 20 cM.
The QTL analysis via CIM procedure in the software WinQTL Cartographer 2.5 revealed a total of four individual QTL for seed oleic acid content in individual environments, located across four chromosomes (A01, A05, A09 and C05) and each explaining between 3.00 and 52.09% of the phenotypic variation (Table 1). Of these, the QTL on A05 and C05 were major QTL, explained 45.02–52.09% and 14.74–44.29% of phenotype variations, respectively. The QTL on A05, A09 and C05 overlapped across environments, suggesting the reliability of these QTL across year environments. Negative additive effects were detected for all of these four QTL, indicating that the parent FC81 contributed to a strong increase in seed oleic acid content.
By comparing of the QTL region with the reference genome of B. napus, we found that the two BnFAD2 genes on A05 and C05, with key roles in controlling the oleic acid content, were physically anchored within the QTL intervals on A05 and C05 of the B. napus reference genome. Gene sequence comparison showed that all four cloned BnFAD2 gene sequences from the two parental lines exhibited more than 95% sequence similarity to the corresponding gene sequences from the reference genome. Two SNPs across the entire coding region, C421T in the BnFAD2.A05 and G1073A in the BnFAD2.C05, were detected by comparing the two parental lines (Figure 3), and resulted in the amino acid changes H141Y and R358K, respectively.

Figure 3. Nucleotide and protein sequence of BnFAD2.A05 and BnFAD2.C05 containing single nucleotide polymorphisms (SNPs) resulting in high oleic acid content in Brassica napus. Red bold font represented the positions of the SNP sites, marked as “wild type/mutant”.
Allelic KASP primers were designed to detect the two BnFAD2 SNPs (C421T and G1073A). Both the KASP421 primer pair (specific to C421T in the BnFAD2.A05) and the KASP1073 primer pair (specific to G1073A in the BnFAD2.C05) showed perfect segregation between the two parental lines as functional markers. In order to test the phenotypic prediction effect of the two KASP markers on seed oleic acid content, we genotyped a new segregation F2 population containing 114 lines using both markers (Figure 4). For the marker KASP421, the seed oleic acid content in the F2 group with the allele from “FC81” averaged 83.6%, significantly higher than that of “E183” (P < 0.01), with average oleic acid content of 68.5%. For the marker KASP1073, the seed oleic acid content in the F2 group with the allele from “FC81” showed average oleic acid content of 82.0%, significantly higher than that of “E183” (P < 0.01) with average oleic acid content of 70.7%. The lines containing both alleles from “FC81” showed significantly higher oleic acid content than lines containing both alleles from “E183” or lines containing one or the other allele (P < 0.01) (Figure 5).
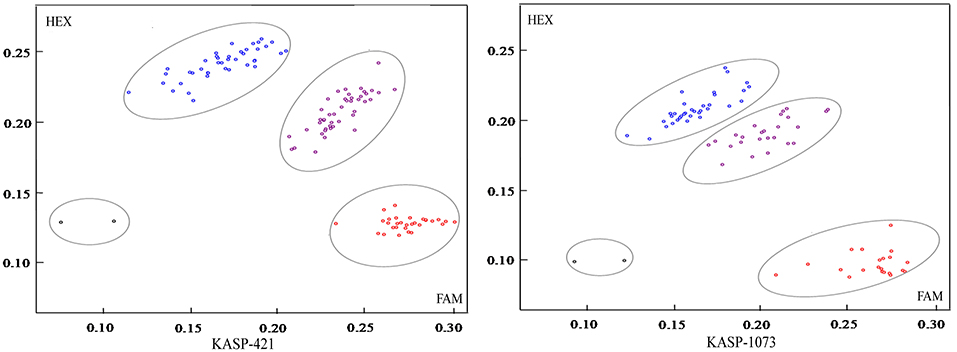
Figure 4. KASP marker genotyping in a segregating Brassica napus F2 population using functional markers KASP421 and KASP 1073 for high oleic acid content. The red circle represents homozygous alleles derived from parent line “E183” with normal oleic acid content, the blue circle represents homozygous alleles derived from parent line “FC81” with high oleic acid content, and the purple circle represents heterozygous loci. The black circle represents negative control.
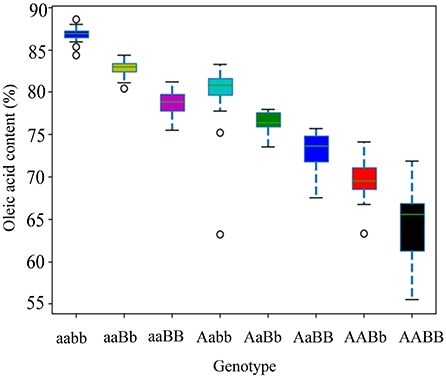
Figure 5. Seed oleic acid content of different BnFAD2 genotypes distinguished by allelic KASP primers KASP421 and KASP 1073 in an F2 population. The “A” and “B” represents wild type alleles by allelic KASP primers KASP421 and KASP 1073, respectively, while “a” and “b” represents mutated alleles by allelic KASP primers KASP421 and KASP 1073, respectively.
In the previous studies, most high oleic germplasm (>80%) seems to be linked to poor agronomic performance of the plant (Kinney, 1994; Bai et al., 2018). However, the high oleic lines in our populations did not show obvious negative effect on agronomic traits such as yield, flowering time, and oil content, which provided excellent germplasm for high oleic acid breeding in rapeseed. Based on this material, we identified four QTL explaining oleic acid content, two of which (A05 and C5) turned out to be due to novel variants of the well-characterized Brassica FAD2 genes, which convert oleic into linoleic acid, and two of which (A01 and A09) had unknown causal genes. Of these, the QTL on A05 and C05 were major QTL, explained 45.02–52.09% and 14.74–44.29% of phenotype variations, respectively. The QTL on A01 and A09 were minor QTL, explained phenotype variations <10%. The total phenotype contributions of QTL on A05 and C05 in 3 years were 89.31, 67.58 and 61.27%. Thus, the QTL on A05 and C05 will be the key loci for genetic improvement of high oleic acid breeding in rapeseed.
A number of studies have previously undertaken to map the genetic factors responsible for oleic acid content in rapeseed via either linkage mapping studies or by association approaches (Burns et al., 2003; Hu et al., 2006; Zhao et al., 2008, 2019; Smooker et al., 2011; Yan et al., 2011; Yang et al., 2012; Wen et al., 2015; Niklas et al., 2016; Qu et al., 2017; Bao et al., 2018; Chen et al., 2018). Of the QTL we identified, the major QTL on A05 (BnFAD2) has been repeatedly identified across previous studies (Hu et al., 2006; Yang et al., 2012). A minor QTL on A01/C1 has also been detected previously for seed oleic acid content (Hu et al., 2006), but we are unable to confirm if this is the same one that we found due to the lack of common flanking markers between studies and the unknown physical location of the markers from the earlier study. A minor QTL on A09 controlling oleic acid content was reported and fine-mapped by Zhao et al. (2019). However, the physical location of this QTL was at 28.00–28.07 Mb on chromosome A09, while our A09 QTL was located at 30.47–32.86 Mb. As both studies used the same SNP array and reference genome, it seems this could possibly indicate two different QTL. Although QTL for oleic acid content on chromosome C05 have not previously been identified, the direct mutation of the underlying BnFAD2-2 gene has been positively associated with elevated oleic acid levels (Long et al., 2018). Comparative to previous work, most of the QTL we identified appear to play a major role in determining oleic acid content across other populations and environments, indicative of a common genetic control. The QTL we identified on chromosome A09 (30.47–32.86 Mb) appears to be a novel locus controlling seed oleic acid in oilseed rape, and could be more genotype- or environment-specific.
The Bnfad2.A05 and Bnfad2.C05 alleles in this study were novel relative to previously identified variants (Table 2). However, beneficial fatty acids facilitate the utilization of rapeseed oil (Napier et al., 2014), and the relationship between BnFAD2 mutations and oleic acid content in rapeseed seeds is well known (Hu et al., 2006; Yang et al., 2012; Long et al., 2018); hence, several high oleic acid rapeseed lines have previously been created by mutagenesis, almost all via mutation of FAD2 genes (Auld et al., 1992; Schierholt et al., 2001; Hu et al., 2006; Spasibionek, 2006; Yang et al., 2012). BnFAD2 is a trans-membrane protein with three conserved histidine-rich motifs (also called histidine boxes, H-boxes), which form the active center of the enzyme (Shanklin et al., 1994). These three H-boxes are 105-HECGHHAF-111, 137-WKYSHRRHH-145, and 315-HVAHHLFS-323 (Tanhuanpää et al., 1998). Mutations in or near the H-boxes have a higher probability of inhibiting BnFAD2 enzyme activity. Although confirmation of the effects of different mutated positions requires additional functional analyses, for breeding purposes artificial mutation in the active H-box centers should be the most efficient for elevating seed oleic acid levels. In our study, the mutation Bnfad2.A05 was H141Y, which was located within the second H-box. The mutation site of Bnfad2-C05 was R358K, near the third H-box motif. Therefore, it is not surprising that these two alleles resulted in super high seed oleic acid content. Additionally, in soybean, Ser-185 just outside the BnFAD2 H-boxes was validated to play a key role in regulating post-translational modifications that directly affected FAD enzyme activity (Tang et al., 2005). Future work should further explore the relationship between mutation sites and oleic acid content in B. napus.
One of the goals of this research was to develop genetic markers for high oleic acid breeding. Clearly distinguishing the homologous from homoeologous genotypes was a key factor. As the Brassica A and C genome FAD2 copies have very high sequence similarity (Lee et al., 2013), it is difficult to design specific primers to amplify the mutant SNP sites in BnFAD2 genes. In this study, we successfully developed SNP-based KASP markers corresponding to each SNP site and clearly genotyped lines in a segregating population. The developed KASP markers were stable and can unambiguously differentiate the parent and hybrid genotypes, facilitating marker-assisted selection for the high oleic acid content trait. Given that the Bnfad2.A05 and Bnfad2.C05 alleles are located on different chromosomes, no linkage effects exist between these genes. Moreover, additive genetic effects contributed much more to oleic acid content than dominance and epistasis effects: as additive effects are the most highly heritable, high oleic acid content can therefore be easily transferred in breeding. The developed KASP markers are predicted to facilitate breeding for high oleic acid rapeseed varieties.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
YF conducted the whole experiment and wrote the manuscript, AM assisted with interpretation of results, manuscript writing and revision, YZ designed primers and participated in the field experiment and seed quality analysis, HY directed the project and contributed to the writing. All authors contributed to the article and approved the submitted version.
This study was supported by the Youth Fund of the Natural Science Foundation of Zhejiang Province (LQ19C130002); Project of Mechanized Oilseed rape Breeding of the Downstream of Yangtze River (2018YFD0100602); The Key Project of Novel Variety Breeding of Zhejiang Province (2016C02050-8).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.715633/full#supplementary-material
Auld, D. L., Heikkinen, M. K., Erickson, D. A., Sernyk, J. L., and Romero, J. E. (1992). Rapeseed mutants with reduced levels of polyunsaturated fatty acids and increased levels of oleic acid. Crop Sci. 32, 657–662. doi: 10.2135/cropsci1992.0011183X003200030016x
Bai, S., Engelen, S., Denolf, P., Wallis, J. G., Lynch, K., Bengtsson, J. D., et al. (2018). Identification, characterization and field testing of Brassica napus mutants producing high-oleic oils. Plant J. 98, 33–41. doi: 10.1111/tpj.14195
Bao, B., Chao, H., Wang, H., Zhao, W., Zhang, L., Raboanatahiry, N., et al. (2018). Stable, environmental specific and novel QTL identification as well as genetic dissection of fatty acid metabolism in Brassica napus. Front. Plant Sci. 9:1018. doi: 10.3389/fpls.2018.01018
Browse, J., Spychalla, J., Okuley, J., and Lightner, J. (1998). Altering the Fatty Acid Composition of Vegetable Oils. New York, NY: Cambridge University Press.
Burns, M. J., Barnes, S. R., Bowman, J. G., Clarke, M., Werner, C. P., and Kearsey, M. J. (2003). QTL analysis of an intervarietal set of substitution lines in Brassica napus: (i) Seed oil content and fatty acid composition. Heredity 90, 39–48. doi: 10.1038/sj.hdy.6800176
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Chang, N. W., and Huang, P. C. (1998). Effects of the ratio of polyunsaturated and monounsaturated fatty acid to saturated fatty acid on rat plasma and liver lipid concentrations. Lipids 33, 48–487. doi: 10.1007/s11745-998-0231-9
Chen, F., Zhang, W., Yu, K., Sun, L., Gao, J., and Zhou, X. (2018). Unconditional and conditional QTL analyses of seed fatty acid composition in Brassica napus L. BMC Plant Biol. 18:49. doi: 10.1186/s12870-018-1268-7
Churchill, G. A., and Doerge, R. W. (1994). Empirical threshold values for quantitative trait mapping. Genetics 138, 963–971. doi: 10.1093/genetics/138.3.963
Clarke, W. E., Higgins, E. E., Plieske, J., Wieseke, R., Sidebottom, C., and Khedikar, Y. (2016). A high-density SNP genotyping array for Brassica napus and its ancestral diploid species based on optimised selection of single-locus markers in the allotetraploid genome. Theor. Appl. Genet. 129, 1887–1899. doi: 10.1007/s00122-016-2746-7
Gillingham, L. G., Harris-Janz, S., and Jones, P. (2011). Dietary monounsaturated fatty acids are protective against metabolic syndrome and cardiovascular disease risk factors. Lipids 46, 209–228. doi: 10.1007/s11745-010-3524-y
Guan, C., Liu, C., Chen, S., Peng, Q., and Guan, M. (2006). High oleic acid content materials of rapeseed (Brassica napus) produced by radiation breeding. Acta Agronom. Sin. 32, 1625–1629.
Hu, X., Sullivan-Gilbert, M., Gupta, M., and Thompson, S. A. (2006). Mapping of the loci controlling oleic and linolenic acid contents and development of fad2 and fad3 allele-specific markers in canola (Brassica napus L.). Theor. Appl. Genet. 113, 497–507. doi: 10.1007/s00122-006-0315-1
Kinney, A. J. (1994). Genetic modification of the storage lipids of plants. Curr. Opin. Biotechnol. 5, 144–151. doi: 10.1016/S0958-1669(05)80027-8
Kosambi, D. D. (1943). The estimation of map distances from recombination values. Ann. Eugen. 12, 172–175. doi: 10.1111/j.1469-1809.1943.tb02321.x
Lauridsen, C., Nielsen, J. H., Henckel, P., and SoRensen, M. T. (1999). Antioxidative and oxidative status in muscles of pigs fed rapeseed oil, vitamin E, and copper. J. Anim. Sci. 77, 105–115. doi: 10.2527/1999.771105x
Lee, K. R., In Sohn, S., Jung, J. H., Kim, S. H., Roh, K. H., Kim, J. B., et al. (2013). Functional analysis and tissue-differential expression of four FAD2 genes in amphidiploid Brassica napus derived from Brassica rapa and Brassica oleracea. Gene 531, 253–262. doi: 10.1016/j.gene.2013.08.095
Long, W., Hu, M., Gao, J., Chen, S., Zhang, J., Cheng, L., et al. (2018). Identification and functional analysis of two new mutant BnFAD2 alleles that confer elevated oleic acid content in rapeseed. Front. Genet. 9:00399. doi: 10.3389/fgene.2018.00399
Micha, R., and Mozaffarian, D. (2009). Trans fatty acids: effects on metabolic syndrome, heart disease and diabetes. Nat. Rev. Endocrinol. 5, 335–344. doi: 10.1038/nrendo.2009.79
Napier, J. A., Haslam, R. P., Beaudoin, F., and Cahoon, E. B. (2014). Understanding and manipulating plant lipid composition: metabolic engineering leads the way. Curr. Opin. Plant Biol. 19, 68–75. doi: 10.1016/j.pbi.2014.04.001
Niklas, K., Anja, B., Li, J., Parkin, I., Benjamin, W., Snowdon, R. J., et al. (2016). Agronomic and seed quality traits dissected by genome-wide association mapping in Brassica napus. Front. Plant Sci. 7:386. doi: 10.3389/fpls.2016.00386
Okuzaki, A., Ogawa, T., Koizuka, C., Kaneko, K., Inaba, M., Imamura, J., et al. (2018). CRISPR/Cas9-mediated genome editing of the fatty acid desaturase 2 gene in Brassica napus. Plant Physiol. Biochem. 131, 63–69 doi: 10.1016/j.plaphy.2018.04.025
Peng, Q., Hu, Y., Wei, R., Zhang, Y., Guan, C., Ruan, Y., et al. (2010). Simultaneous silencing of FAD2 and FAE1 genes affects both oleic acid and erucic acid contents in Brassica napus seeds. Plant Cell Rep. 29, 317–325. doi: 10.1007/s00299-010-0823-y
Przybylski, R., Gruczynska, E., and Aladedunye, F. (2013). Performance of regular and modified canola and soybean oils in rotational frying. J. Am. Oil Chem. Soc. 90, 1271–1280. doi: 10.1007/s11746-013-2278-0
Qu, C., Jia, L., Fu, F., Zhao, H., Lu, K., Wei, L., et al. (2017). Genome-wide association mapping and Identification of candidate genes for fatty acid composition in Brassica napus L. using SNP markers. BMC Genom. 18:232. doi: 10.1186/s12864-017-3607-8
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning: A Laboratory Manual (3-Volume Set). Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press
Schierholt, A., Rücker, B., and Becker, H. C. (2001). Inheritance of high oleic acid mutations in winter oilseed rape (Brassica napus L.). Crop Sci. 41, 1444–1449. doi: 10.2135/cropsci2001.4151444x
Shanklin, J., Whittle, E., and Fox, B. G. (1994). Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry 33, 12787–12794. doi: 10.1021/bi00209a009
Singh, V. K., Mangalam, A. K., Dwivedi, S., and Naik, S. (1998). Primer premier: program for design of degenerate primers from a protein sequence. BioTechniques 24, 318–319. doi: 10.2144/98242pf02
Smooker, A. M., Wells, R., Morgan, C., Beaudoin, F., Cho, K., Fraser, F., et al. (2011). The identification and mapping of candidate genes and QTL involved in the fatty acid desaturation pathway in Brassica napus. Theor. Appl. Genet. 122, 1075–1090. doi: 10.1007/s00122-010-1512-5
Snowdon, R. J., Friedrich, T., Friedt, W., and Köhler, W. (2002). Identifying the chromosomes of the A and C genome diploid Brassica species B. rapa and B. oleracea in their amphidiploid B. napus. Theor. Appl. Genet. 104, 533–538. doi: 10.1007/s00122-001-0787-y
Spasibionek, S. (2006). New mutants of winter rapeseed (Brassica napus L.) with changed fatty acid composition. Plant Breed. 125, 259–267. doi: 10.1111/j.1439-0523.2006.01213.x
Tang, G. Q., Novitzky, W. P., Griffin, H. C., Huber, S. C., and Dewey, R. E. (2005). Oleate desaturase enzymes of soybean: evidence of regulation through differential stability and phosphorylation. Plant J 44, 433–446. doi: 10.1111/j.1365-313X.2005.02535.x
Tanhuanpää, P, Vilkki, J., and Vihinen, M. (1998). Mapping and cloning of FAD2 gene to develop allele-specific PCR for oleic acid in spring turnip rape (Brassica rapa ssp. oleifera). Mol. Breed. 4, 54–550. doi: 10.1023/A:1009642317634
Thies, W. (1971). Schnelle und einfache Analysen der Fettsaurezusammensetzung in einzelnen Raps-Kotyledonen. I. Gaschromatographische und papierchromatographische Methoden. Z. Pflanzenzucht 65, 181–202.
Van Ooijen, J., and Voorrips, R. (2006). JoinMap 4.0. Software for the Calculation of Genetic Linkage Maps in Experimental Populations. Wageningen: Kyazma BV.
Wang, S., Basten, C. J., and Zeng, Z. B. (2005). Windows QTL Cartographer Version 2.5. Statistical genetics. Raleigh: North Carolina State University.
Wells, R., Trick, M., Soumpourou, E., Clissold, L., and Bancroft, I. (2014). The control of seed oil polyunsaturate content in the polyploid crop species Brassica napus. Mol. Breed. 33, 349–362. doi: 10.1007/s11032-013-9954-5
Wen, J., Xu, J., Long, Y., Xu, H., Wu, J., Meng, J., et al. (2015). Mapping QTLs controlling beneficial fatty acids based on the embryo and maternal plant genomes in Brassica napus L. J. Am. Oil Chem. Soc. 92, 541–552. doi: 10.1007/s11746-015-2618-3
Wu, Y., Bhat, P. R., Close, T. J., Lonardi, S., and Kruglyak, L. (2008). Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph. PLoS Genet. 4:e1000212. doi: 10.1371/journal.pgen.1000212
Yan, X. Y., Li, J. N., Wang, R., Jin, M. Y., Chen, L., Qian, W., et al. (2011). Mapping of QTLs controlling content of fatty acid composition in rapeseed (Brassica napus). Genes Genom. 33, 365–371. doi: 10.1007/s13258-010-0149-8
Yang, Q., Fan, C., Guo, Z., Jie, Q., and Zhou, Y. (2012). Identification of FAD2 and FAD3 genes in Brassica napus genome and development of allele-specific markers for high oleic and low linolenic acid contents. Theor. Appl. Genet. 125, 715–729. doi: 10.1007/s00122-012-1863-1
Zhang, H. J., Xiao, G., Tan, T. L., Xun, L. I., and Guan, C. Y. (2008). High oleate material of rapeseed (Brassica napus) produced by EMS treatment. Sci. Agric. Sin. 41, 4016–4022. doi: 10.3864/j.issn.0578-1752.2008.12.010
Zhao, J., Dimov, Z., Becker, H. C., Ecke, W., and Möllers, C. (2008). Mapping QTL controlling fatty acid composition in a doubled haploid rapeseed population segregating for oil content. Mol. Breed. 21, 115–125. doi: 10.1007/s11032-007-9113-y
Keywords: Brassica napus, seed oleic acid, quantitative trait loci, BnFAD2, kompetitive allele specific PCR
Citation: Fu Y, Mason AS, Zhang Y and Yu H (2021) Identification and Development of KASP Markers for Novel Mutant BnFAD2 Alleles Associated With Elevated Oleic Acid in Brassica napus. Front. Plant Sci. 12:715633. doi: 10.3389/fpls.2021.715633
Received: 27 May 2021; Accepted: 21 June 2021;
Published: 26 July 2021.
Edited by:
Kun Lu, Southwest University, ChinaReviewed by:
Fu Donghui, Jiangxi Agricultural University, ChinaCopyright © 2021 Fu, Mason, Zhang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huasheng Yu, eXVodWFzaGVuZy0wQDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.