- School of Life Sciences, Jiangsu University, Zhenjiang, China
AT-hook motif nuclear localization (AHL) proteins belong to a family of transcription factors, and play important roles in plant growth and development and response to various stresses through protein-DNA and protein-protein interactions. To better understand the Brassica napus AHL gene family, AHL genes in B. napus and related species were analyzed. Using Arabidopsis as a reference, 122 AHL gene family members were first identified in B. napus. According to the phylogenetic tree and gene organization, the BnaAHLs were classified into two clades (Clade-A and Clade-B) and three types (Type-I, Type-II, and Type-III). Gene organization and motif distribution analysis suggested that the AHL gene family is relatively conserved during evolution. These BnaAHLs are unevenly distributed on 38 chromosomes and expanded by whole-genome duplication (WGD) or segmental duplication. And large-scale loss events have also occurred in evolution. All types of BnaAHLs are subject to purification or neutral selection, while some positive selection sites are also identified in Type-II and Type-III groups. At the same time, the purification effect of Type-I members are stronger than that of the others. In addition, RNA-seq data and cis-acting element analysis also suggested that the BnaAHLs play important roles in B. napus growth and development, as well as in response to some abiotic and biotic stresses. Protein-protein interaction analysis identified some important BnaAHL-binding proteins, which also play key roles in plant growth and development. This study is helpful to fully understand the origin and evolution of the AHL gene in B. napus, and lays the foundation for their functional studies.
Introduction
AT-hook motif nuclear localized (AHL) proteins contain two conserved structural units: the AT-hook motif and the plant and prokaryote conserved (PPC) domain. The latter is also annotated as the domain of un-known function #296 (DUF296) (Fujimoto et al., 2004). The AT-hook motif is a small DNA-binding motif that was first described as a high mobility group (HMG) non-histone chromosomal protein HMG-I/Y (Zhao et al., 1993). So far, it has been identified in various gene families of prokaryotes and eukaryotes, including the high mobility group A (HMGA) proteins of mammals (Aravind and Landsman, 1998). This AT-hook motif has an obvious feature: a conserved core sequence Arg-Gly-Arg amino acid (aa) with Arg-Lys or Pro residues on both sides. The AT-hook motif can use the conserved Arg-Gly-Arg residue to bind to the minor groove of stretches of AT-rich B-form DNA (Huth et al., 1997). Unlike most other transcription factors like leucine zipper and helix-loop-helix, AHLs mainly bind to the minor groove of duplex DNA (Aravind and Landsman, 1998; Reeves and Beckerbauer, 2001). When combined with DNA, the core sequence of Arg-Gly-Arg takes a concave conformation, and the side chains of two Arg residues should be firmly inserted into the minor groove, thereby modifying the chromatin structure (Huth et al., 1997). It is worth noting that the AT-hook motifs can recognize specific DNA structures rather than nucleotide sequences and activate or inhibit the transcription of many genes (Aravind and Landsman, 1998; Nagano et al., 2001). Proteins with PPC-like domains are also found in Bacteria and Archaea, but without AT-hook motifs (Fujimoto et al., 2004).
According to the conserved amino acid sequence, AT-hooks can be divided into two categories. AT-hooks containing Gly-Ser-Lys-Asn-Lys consensus sequences are Type-I motifs, and AT-hooks with Arg-Lys-Tyr consensus sequences at the end of the Arg-Gly-Arg core are Type-II motifs. The PPC/DUF296 conserved domain is located at the carboxy terminus of AHL proteins and contains about 120 aa (Fujimoto et al., 2004). According to sequence differences, PPC/DUF296 domain is divided into two types: PPC/DUF296 domain contains a core conserved sequence of Leu-Arg-Ser-His amino acids, which is defined as Type-A, while Type-B contains Phe-Thr-Pro-His in the upstream region (Zhao et al., 2014). All PPC/DUF296 domains also have a Gly-Arg-Phe-Glu-Ile-Leu conserved sequence, which is believed to mediate the interaction with transcription factors to promote the function of certain AHL proteins (Zhao et al., 2013).
Based on the evolution of AHL genes in land plants, they have been divided into two phylogenetic branches: Clades-A and Clades-B (Matsushita et al., 2007; Zhao et al., 2013, 2014). Clade-A has only one Type-I AT-hook motif and one intron-free Type-A PPC/DUF296 domain. Clade-B is further divided into two branches: one type of AHLs contains two AT-hooks (Type-I and Type-II) and a Type-B PPC/DUF296 domain; the other type of AHLs contain a Type-II AT-hook motif and a Type-B PPC/DUF296 domain (Zhao et al., 2014). In contrast to Clades-A, Clades-B contains introns (Bishop et al., 2020).
The AHL gene family is highly conserved in all land plants and play important roles in plant growth and development and stress response. Studies have shown that AHL gene family members can regulate a variety of developmental processes. AHL18 promotes elongation and cell division in root apical meristem. Overexpression of AHL18 makes the root system more developed, and promotes primary root growth (Sirl et al., 2020). An AT-hook protein, encoded by a palea defective mutant (dp1) of rice, can lead to a significant increase in the number of floral organs by affecting palea formation (Jin et al., 2011). Knockout of AHL16 gene will activate various transposons and delay the flowering time in Arabidopsis (Xu et al., 2013a,b). Overexpression of the AHL22 also delayed flowering by modifying FLOWERING LOCUS T (FT) chromatin in Arabidopsis (Xiao et al., 2009; Yun et al., 2012). TRANSPOSABLE ELEMENT SILENCING VIA AT-HOOK (TEK) gene can lead to male sterility by regulating the expression of Arabinogalactan (AGP) gene in anthers (Lou et al., 2014; Jia et al., 2015). AHL21 is a direct target of AGAMOUS (AG). AHL21 overexpression and loss of function can lead to reproductive defects (Ng et al., 2009). Barren stalk fastigiate1 (Baf1) encodes an AT-hook DNA binding protein, which is a transcriptional regulator of maize panicle development. In baf1 mutants, most plants failed to form panicles (Gallavotti et al., 2011). Overexpression of the AHL22, AHL27, and AHL29 will suppress hypocotyl growth of Arabidopsis (Street et al., 2008; Xiao et al., 2009; Zhao et al., 2013). Two interacting AT-hook factors, AHL3 and AHL4, facilitate intercellular trafficking and regulate vascular tissue boundaries in Arabidopsis roots (Zhou et al., 2013). ORE7/ESC is the member of the AHL gene organization, which ectopic overexpression may extend leaf longevity by altering chromatin architecture in plants (Lim et al., 2007). AHL15 gene is a suppressor of axillary meristems maturation in Arabidopsis, which can indirectly enhance the repressive effect on maturation of axillary meristems under short-day conditions, and prolong the lifespan of plant (Karami et al., 2020). In addition to regulating the growth and development of plants, some members of the AHL gene family also regulate the primary metabolism pathway including coordinating various responses to biotic and abiotic stresses in plants (Li and Kliebenstein, 2014). As an AT-hook protein, AGF1 can bind to the cis-elements of gibberellin and participate in its negative feedback regulation (Matsushita et al., 2007). Phosphorylation of AHL10 coordinates drought stress and regulates jasmonic acid and auxin-related gene expression (Rashotte et al., 2003; Vom Endt et al., 2007; Wong et al., 2019). AHL1 overexpression plants can better survive under drought, high salt and low temperature conditions (Zhou et al., 2016). The pathogen-associated molecular patterns (PAMPs)-triggered immunity (PTI) can resist the invasion of pathogenic microorganisms (Chisholm et al., 2006). This process will be inhibited by the over-expression of AHL20, thereby inhibiting the innate immune response (Lu et al., 2010). AHL18 also plays important roles in disease resistance by forming homodimer or heterodimer in plant nucleus (Kumar et al., 2018).
Recently, AHL gene family has been identified and analyzed in several species. 29 AHL genes (AtAHLs) were identified from the Arabidopsis genome (Matsushita et al., 2007). The sorghum genome contains 22 AHL genes (Zhao et al., 2014). Twenty AHL genes have been discovered in the rice genome (Kim et al., 2011). Thirty-seven AHLs were identified from the maize genome (Bishop et al., 2020). Forty-eight, fifty-one, and ninety-nine AHLs were identified from three cottons (Gossypium raimondii, G. arboretum, and G. hirsutum), respectively (Zhao et al., 2020). The soybean genome contains 63 AHL genes (Wang et al., 2021b). About 7,500 years ago, a natural hybridization occurred between Brassica rapa (AA, 2n = 20) and B. oleracea (CC, 2n = 18), forming a natural polyploid B. napus (AACC, 2n = 38) (Chalhoub et al., 2014). Brassica napus, an important oil crop, has emerged as a model plant for studying genetics, evolution, and other biological processes. Rape yield is continually affected by multiple biotic and abiotic stresses. While AHLs are expected to play important and diverse roles in these stress responses, a detailed genome-wide analysis of AHL gene family has not been performed. In this study, we identified 122 AHL genes (BnaAHLs) from the B. napus genome and then analyzed their molecular evolution characteristics.
Materials and Methods
Genome-Wide Identification of AHL Proteins in the B. napus Genome
We obtain the B. napus genome data from the B. napus database.1 Twenty-nine AHLs were acquired from the Arabidopsis database.2 According to the protein sequence of AtAHLs, we searched homologous proteins in B. napus with homology comparison in blast+ (Camacho et al., 2009) with above 10–5 e-value. Based on the conserved domain of AHLs, the pfam (Finn et al., 2014) and CD-Search (Marchler-Bauer and Bryant, 2004) were also used to identify the DUF296 domain in the predicted BnaAHLs. Some candidate proteins that do not contain DUF296 domain will be discarded. Next, ExPASy (Gasteiger et al., 2003)3 was also used to investigate the physical and chemical properties of these AHL proteins. CELLO (Yu et al., 2006)4 was used to predict the location of these AHL proteins.
Phylogenetic Analysis of AHL Proteins
The phylogenetic tree of the AHL proteins of B. napus and Arabidopsis was constructed using the neighbor-joining (NJ) method in MEGA 6.0 (Tamura et al., 2013)5 with the Poisson model and pairwise deletion. The reliability of phylogenetic tree was evaluated by 1,000 bootstrap duplications. At the same time, based on Jones Taylor Thornton (JTT) model and gamma distributed, we also constructed the phylogenetic tree using the maximum-likelihood (ML) method in MEGA 6.0. The phylogeny test is consistent with the NJ method, and other options are set to default values.
Genes Structure Analysis
The AT-hook motifs of the BnaAHL proteins were predicted using the MEME v5.3.06 (Bailey et al., 2015). The number of motifs should not exceed 7. The distribution of motifs occurs zero or one time in each sequence. In order to investigate the structural characteristics of the BnaAHLs, the gff3 file was downloaded from the B. napus database (see text footnote 1), which has the annotation information of the B. napus genome. The position information of introns and exons is obtained from the gff3 file. Meanwhile, the position information of the DUF296 conserved domain was also predicted with Batch CD-Search (Yang et al., 2020),7 and was submitted to TBtools8 (Chen et al., 2020) to graphically display gene structures and motif distributions.
BnaAHLs Localization and Duplications
The annotation information data of B. napus, Arabidopsis, B. rapa, and B. oleracea was download from public databases (see text footnotes 1, 2),9 respectively. In order to study the duplication events and collinearity relationship of B. napus, MCScanX of the TBtools (Chen et al., 2020) was used to draw the collinearity map of B. napus and B. napus, B. napus and Arabidopsis, B. napus and B. rapa, and B. napus and B. oleracea, respectively.
Two genes on a branch at the end of the phylogenetic tree were regarded as a gene pair. Based on the coding DNA (CDS) sequence of these genes, the Ks value of the gene pairs can be calculated with TBtools (Chen et al., 2020). Based on the T = Ks/2λ, duplication time of these gene pairs was calculated by the commonly adopted clock-like rates (λ) of 1.4 × 10–8 synonymous substitutions per site per year (Wang et al., 2011).
Cis-Acting Element Analysis
To identify the cis-acting element of BnaAHLs, TBtools (Chen et al., 2020) was used to obtain the 2,000 bp sequences in front of the genomic CDS. Then, the PlantCARE10 (Lescot et al., 2002) was used to predict the cis-acting elements on these promoters. Thus, the number and types of different cis-acting elements in BnaAHLs were classified and visualized with TBtools (Chen et al., 2020).
Selective Pressure Analysis
Likelihood-ratio test was used to compare the implemented models. We used four evolution models [M8 (ωs ≥ 1), M8a (ωs = 1), M7 (beta), and M5 (gamma)] to evaluate the selective pressure (Yang et al., 2000). BnaAHL29e, BnaAHL13j, and BnaAHL13g proteins are used as query sequences to display the analysis results for Type-I, Type-II, and Type-III groups, respectively. All CDSs on each branch are used to calculate their selective pressure with the Selecton (Doron-Faigenboim et al., 2005).11 In order to determine the positions of these identified positive selection sites, I-TASSER (Yang et al., 2015)12 was used to predict the tertiary structures of these proteins.
Gene Expression Analysis
The expression patterns of BnaAHLs in different tissues are obtained from the BnTIR (Brassica napus transcriptome information resource) database.13 RNA-seq data (accession number: GSE169299) of B. napus under the Sclerotinia sclerotiorum stress come from the GEO (Gene Expression Omnibus)14 (Clough and Barrett, 2016). Rape leaves were infected with S. sclerotiorum, and the epidermis (EPI), mesophyll (MES), and vascular (VAS) expression profiles were obtained. Next, RNA-seq data (accession number: GSE156029) of some abiotic stresses like drought and high temperature was also downloaded from the GEO (Clough and Barrett, 2016). All the RNA-seq data is standardized based on log2 scale, and clustered and visualized with TBtools (Chen et al., 2020).
Protein-Protein Interaction Network Analysis
Interaction network of AHL proteins was predicted by the STRING15 database (Szklarczyk et al., 2019). Since there is no relevant data of B. napus in the STRING, we used the B. rapa data to reflect the interaction of AHL proteins in B. napus. All BnaAHLs proteins are used as the query to search for the interaction network of their homologous proteins in the cabbage. The network type is set to full network, that is, the edges indicate both functional and physical protein associations. The minimum required interaction score selects medium confidence (0.400). The number of interactors in the first shell cannot exceed 20, and that of the second shell cannot exceed 5.
Results
Identification and Classification of AHL Gene Family
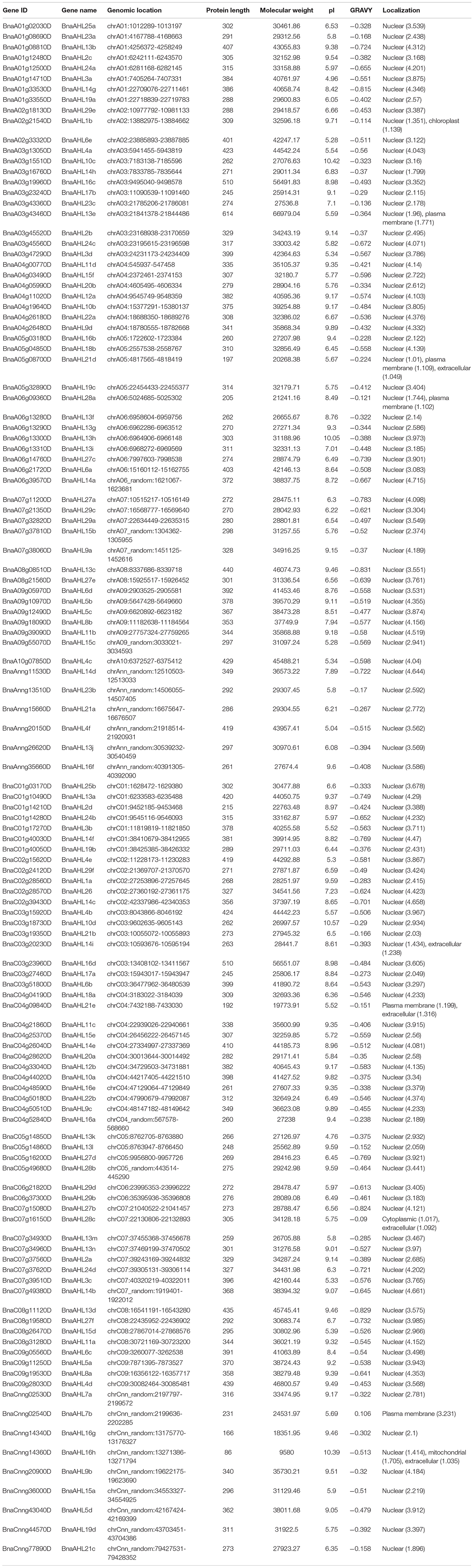
122 AHL protein sequences were first identified in the B. napus genome, which contain conserved PPC/DUF296 domains and AT-hook motifs. Next, their physical and chemical properties were also analyzed (Table 1). These genes encode the amino acids ranging in length from 86 to 614 with the molecular weight from 9,580 to 66,979.04 Dalton. Isoelectric point (pI) of these amino acids ranged from 4.76 to 10.57. The hydrophilic coefficient of BnaCnng02540D is the largest (0.106), while that of BnaAHL13c is the smallest (−0.831). Approximately 98% of AHL transcripts were predicted to be located in the nucleus. This finding is consistent with one previous report that AHLs are nuclear localization proteins (Zhao et al., 2014).
In order to understand the evolutionary relationship of AHLs in B. napus and Arabidopsis, their protein sequences were alignmented with Clustal X (Jeanmougin et al., 1998) and phylogenetic trees were constructed by ML and NJ methods. The evolutionary relationship between the ML tree and the NJ tree is very similar (Figure 1 and Supplementary Figure 1). It showed that AHLs can be clearly divided into two clades: Clade-A and Clade-B. Clade-A contains 56 AHL genes of B. napus and 15 AHL genes of Arabidopsis, and it is also classified into Type-I. There are 80 AHL genes in Clade-B, 66 of which are from the B. napus and other 14 are from the Arabidopsis. Clade-B is further divided into two types: Type-II and Type-III, of which Type-II contains 47 AHL genes, while Type-III includes 33 AHL genes (Figure 1).
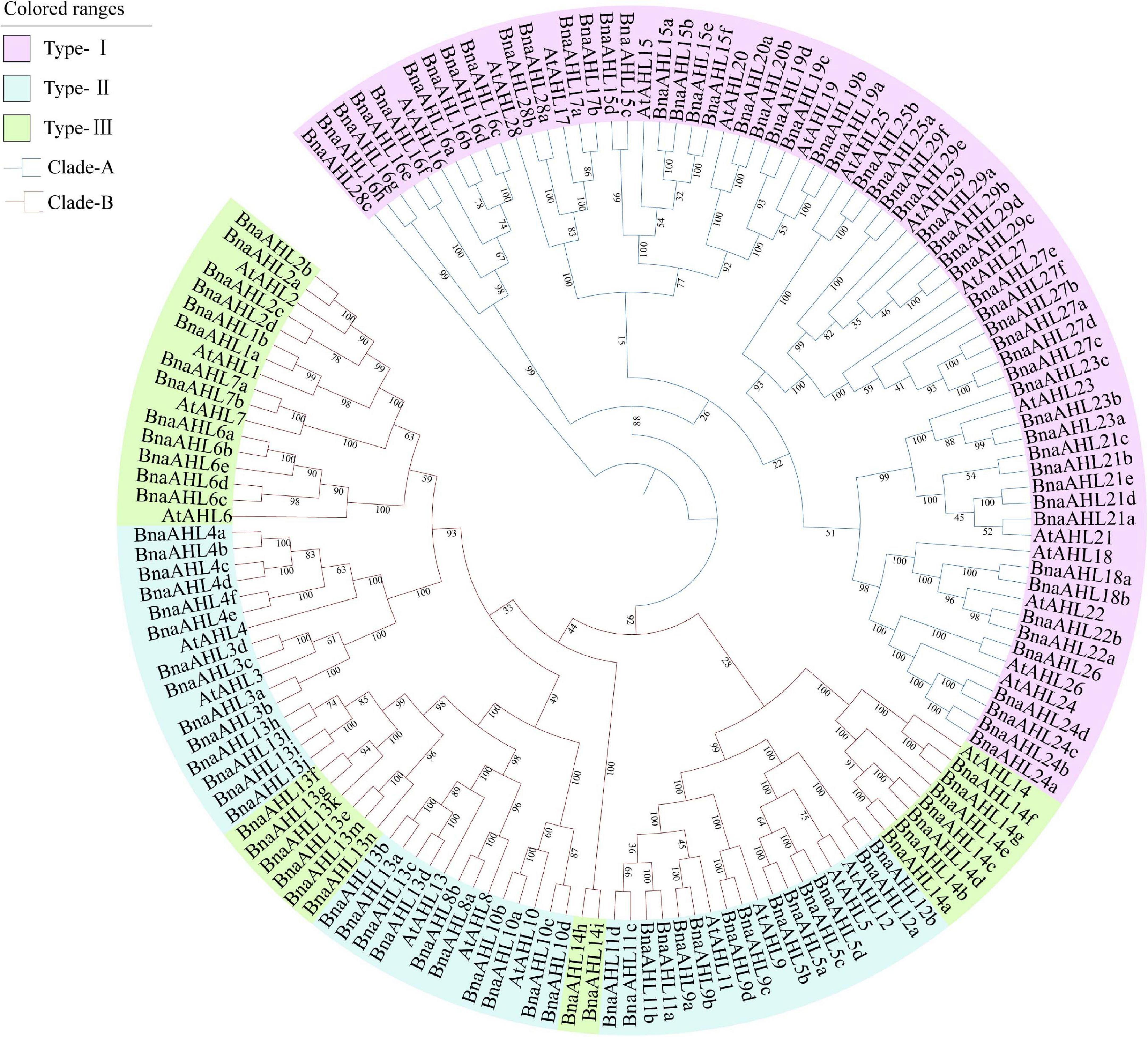
Figure 1. Phylogenetic relationship of AHL genes in B. napus and Arabidopsis. Branches of different colors indicate different evolutionary clades. Each branch has a number indicating the percentage of reliability of the evolutionary branch.
Gene Structures and Motif Analysis of AHLs in B. napus
In order to study the homology domain and conservation degree of the BnaAHLs, MEME (Bailey et al., 2015) and TBtools (Chen et al., 2020) were used to predict and visualize their conserved domain and gene organization, respectively. As a result, 7 conserved motifs were found in the 122 AHLs of B. napus (Figure 2). Motif 6 and motif 7 have highly conserved R-G-R-P and R-G-R-P-R-K-Y amino acid sequences, respectively. Among them, R-G-R-P is the core sequence shared by the two motifs (Figure 2). This finding is consistent with previous reports of the AHL structure (Zhao et al., 2014, 2020; Bishop et al., 2020; Wang et al., 2021a).
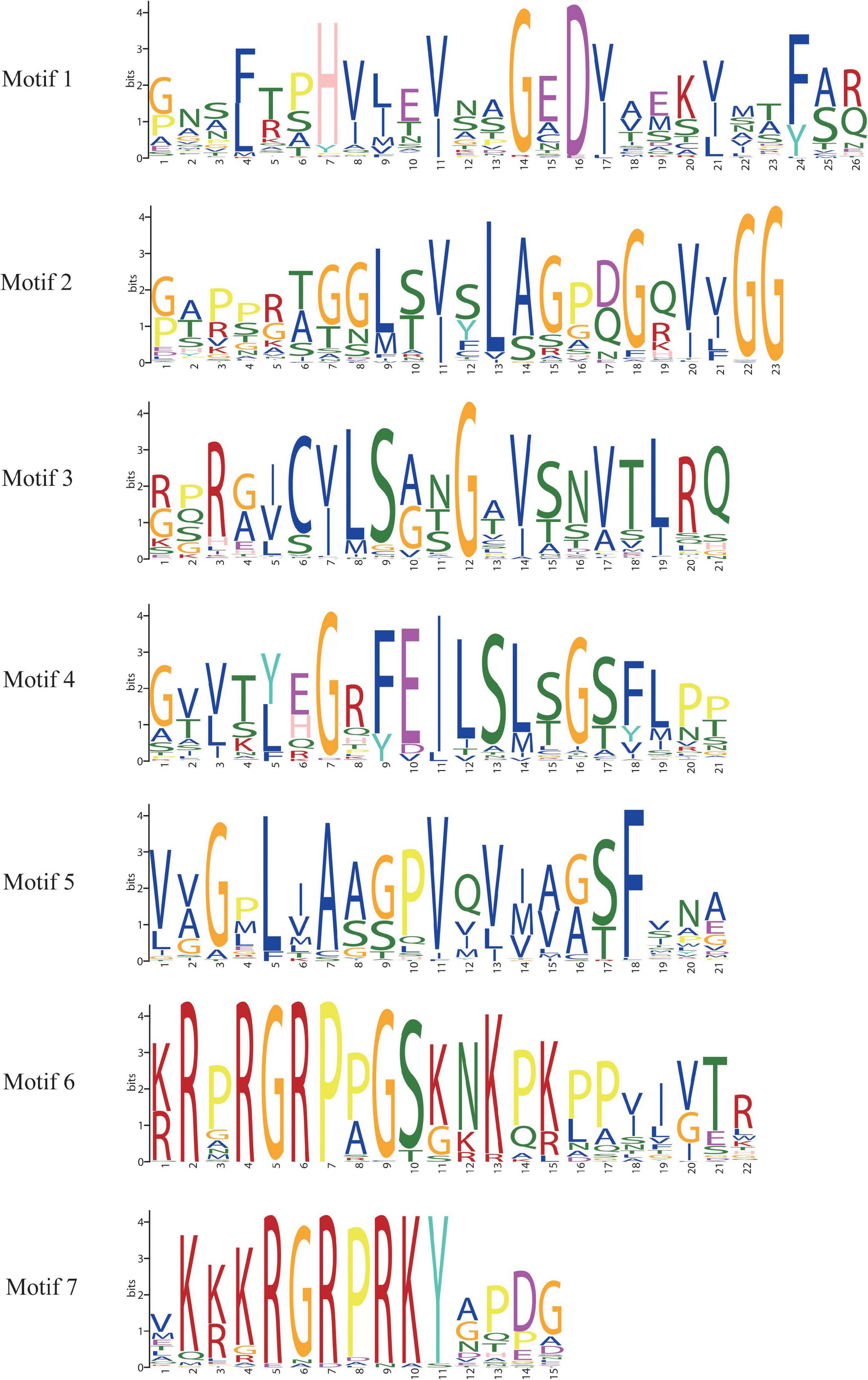
Figure 2. Sequence logos of conserved motifs in BnaAHLs. The logo indicates the sequence of seven motifs. A letter represents an amino acid, and the size represents its conservation at this position.
The appearance of introns can enhance the transcriptional activation of AHLs (Zhao et al., 2014). We also investigated the distribution of introns and exons to study the diversity of gene structure. The structure of AHLs can be clearly divided into two types: one contains multiple introns, and the other contains no or only one intron (Figure 3). The 66 genes in Clade-B have 3 introns, except for the 14 genes with 1 intron in Clade-A, the other 42 BnaAHLs without intron. While in other species, AHLs in Clade-A do not contain any intron (Zhao et al., 2014, 2020; Bishop et al., 2020).
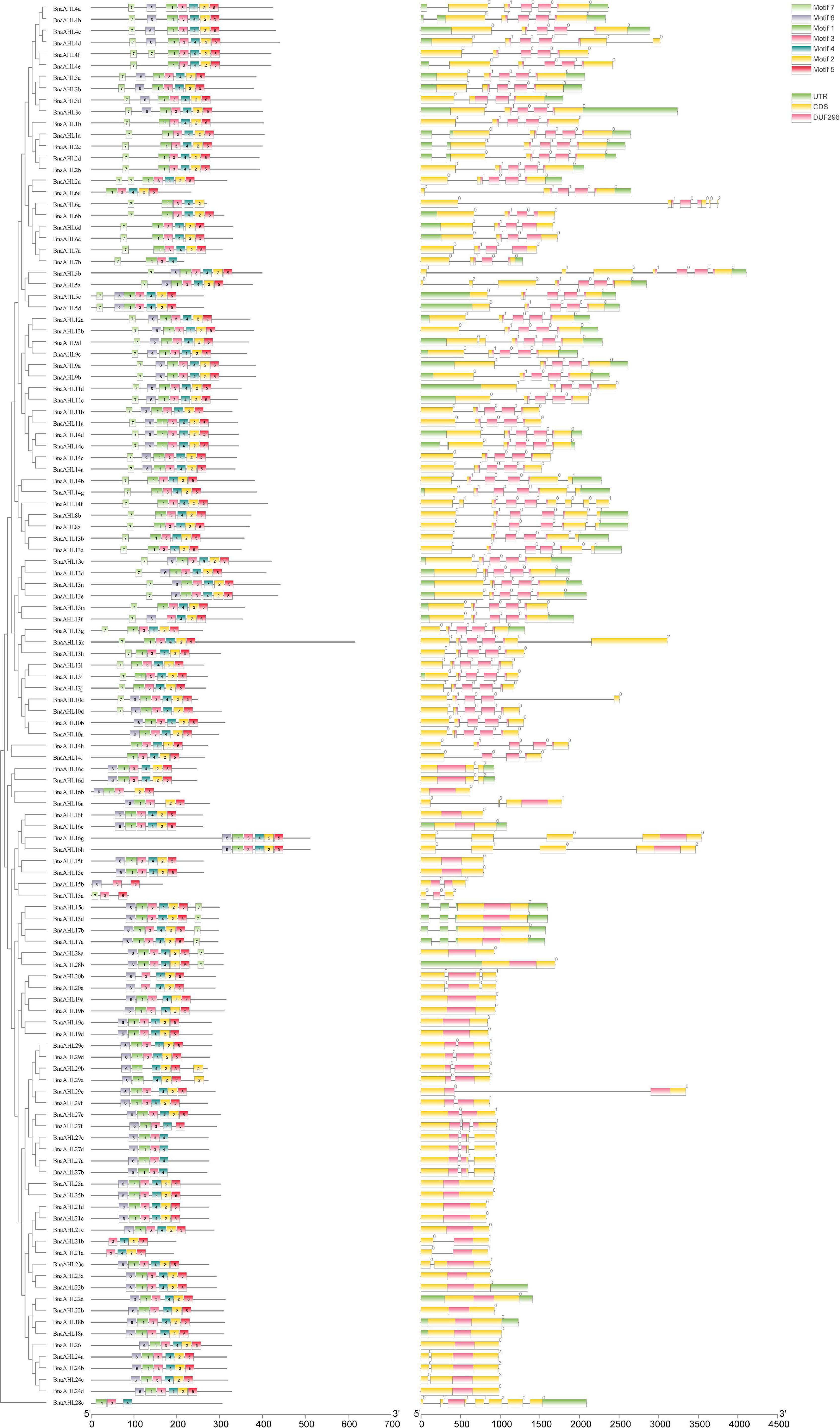
Figure 3. Motif distribution and exon-intron structure of AHLs. The neighbor-joining phylogenetic tree was constructed with MEGA 6.0 software, and the conserved motifs and gene structure were predicted with MEME and Tbtools, respectively. The scale at the bottom indicates the length of protein and DNA sequences, respectively. In the left picture, different colored boxes indicate different conserved motifs, and the black line indicates non-conserved amino acids. The black line on the right represents introns, the green box represents untranslated 5′ and 3′ regions; the yellow box represents exons, and the purple box represents the conserved domain of the AHLs.
In addition, we also found that the intron phases of BnaAHLs were highly conserved in the same clade, implicating the evolutionary similarity between these members. There are phase 0, 1, and 2 introns in all eukaryotic genomes, and the 0 phase intron accounts for the highest proportion and has the highest level of conservation (Long et al., 1995). The PPC/DUF296 domains of most AHL genes in Clade-A doesn’t have any intron, while 14 AHL genes have phase 0 introns. However, the PPC/DUF296 conserved domain of most AHL genes in Clade-B has the same intron phase distribution except BnaAHL1a, BnaAHL1b, BnaAHL2a, BnaAHL2b, BnaAHL2c, and BnaAHL2d. This result implies that the AHL gene family has a relatively conservative evolutionary relationship in each clade (Figure 3).
Chromosomal Distribution and Duplication of BnaAHLs
The A and C genomes of Brassica are not only highly conserved, but also characterized by a large number of gene loss, tandem duplication, polyploidy, and chromosome rearrangement (Struss et al., 1996; Howell et al., 2008; Mun et al., 2009; Liu et al., 2014). Genomic mapping of the BnaAHLs showed that 100 BnaAHLs were unevenly distributed on 38 chromosomes (Figure 4), while the other 22 genes were not accurately located. Among them, 50 BnaAHLs are distributed on the A and C subgenome, respectively. A large number of BnaAHL genes were distributed in chromosome ChrC04 and ChrA03, which contained 11 and 10 members, respectively. However, only one member was found in ChrA10.
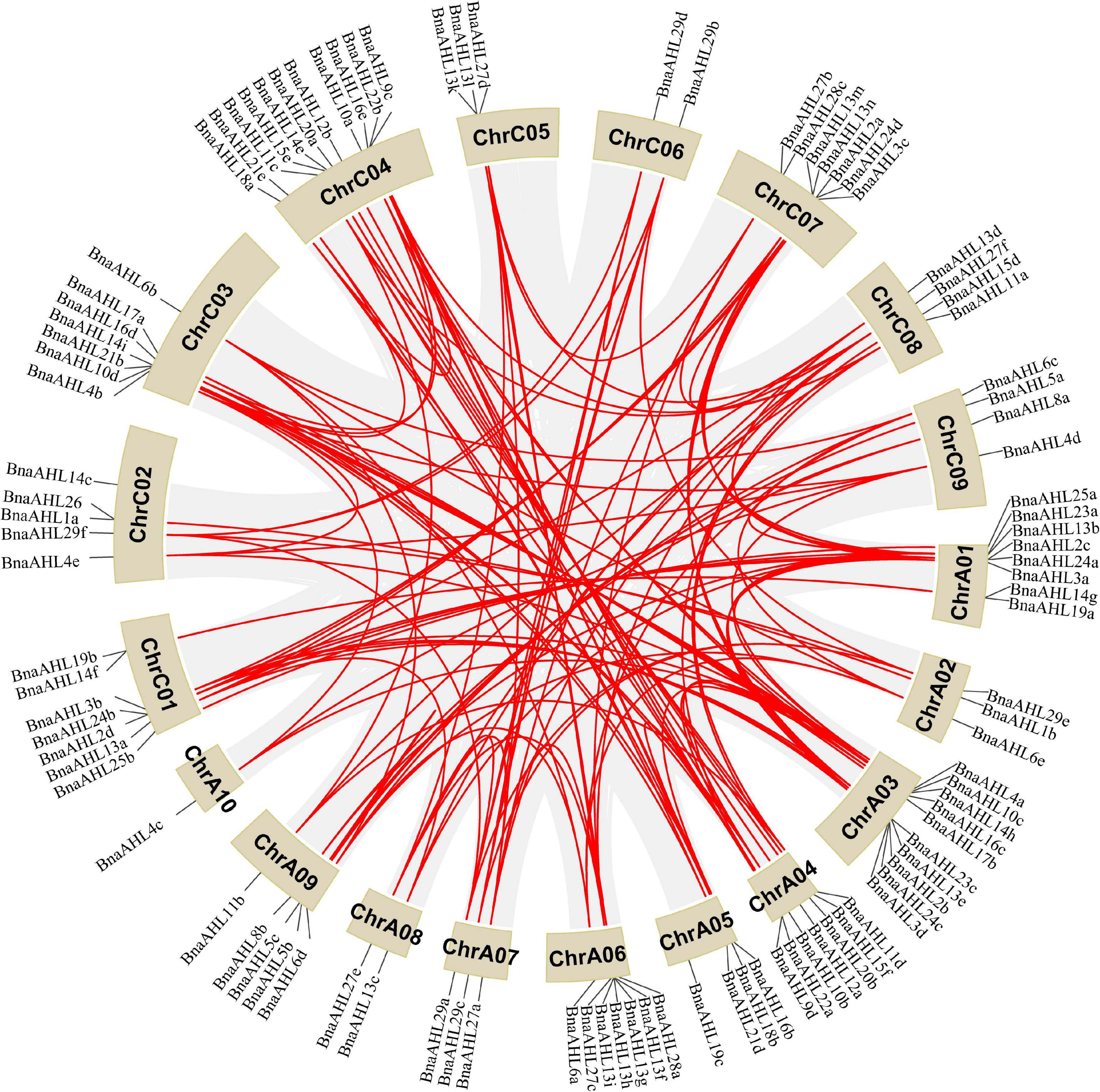
Figure 4. Distribution of BnaAHLs on chromosome. One hundred BnaAHLs are distributed on nineteen different chromosomes of B. napus. In addition, 22 BnaAHLs are not mapped to any chromosomes. The gray lines in the background indicate the collinear blocks in the genomes of B. napus, while the red lines emphasize the segmental duplication of the AHLs. The brown block indicates the chromosome and its length represents the size of the chromosome.
About 20 million years ago, Arabidopsis diverged from Brassica, a member of the cruciferous family (Yang et al., 1999). The whole Brassica genome triploidized about 13 million years ago (Yang et al., 1999; Lysak et al., 2005). The divergence time of A and C genome fragments in the diploid ancestors of B. napus spanned about 0.12–1.37 million years (Cheung et al., 2009). The common mutation rate of 1.4 × 10–8 synonym substitutions per site per year was used to estimate the time of BnaAHLs divergence (Wang et al., 2011). We found that these BnaAHLs had been duplicated about 0.63–6.55 million years ago (Supplementary Table 1). Among them, seven gene pairs (BnaAHL8b/BnaAHL8a, BnaAHL13i/BnaAHL13j, BnaAHL23a/BnaAHL23b, BnaAHL15b/BnaAHL15a, BnaAHL19a/BnaAHL 19b, BnaAHL16f/BnaAHL16e, BnaAHL29e/BnaAHL29f) are duplicated during the divergence of A and C genomes, while the other gene paires duplicated after triploidy of the whole genome, but before the divergence of A and C genomes. Therefore, whole genome duplication is the main reason of the BnaAHLs amplification. It can be seen from Supplementary Table 1 that the two BnaAHLs in a gene pair are from the A and C genomes, respectively. So it was further illustrated that the amplification of BnaAHLs is also caused by polyploidization. Among them, BnaAHL27e and BnaAHL27f were duplicated about 6.12 million years ago. The type of replication of some BnaAHLs is tandem duplication (Supplementary Table 2). In additional, BnaAHL13f, BnaAHL13g, BnaAHL13h, and BnaAHL13i are distributed in tandem on chromosomes (Figure 4 and Table 1), which is further confirmed that the BnaAHL genes have duplicated in tandem during the evolution.
Gene duplication events occur with very high frequency in the genomes of all organisms (Lynch and Conery, 2000). We analyzed the collinear relationship between B. napus and Arabidopsis, B. napus and B. rapa, and B. napus and B. oleracea, respectively (Figure 5). The result revealed that there are a large number of homologous AHLs between B. napus and Arabidopsis, B. napus and B. rapa, B. napus and B. oleracea, respectively. Among them, B. napus and B. rapa have the most collinear genes (Figure 5). More than 78 percent of BnaAHL genes are generated by the whole genome duplication (WGD) or segmental duplication (Supplementary Table 2). Therefore, WGD or segmental duplication is a main driver of BnaAHLs expansion in the B. napus genome.
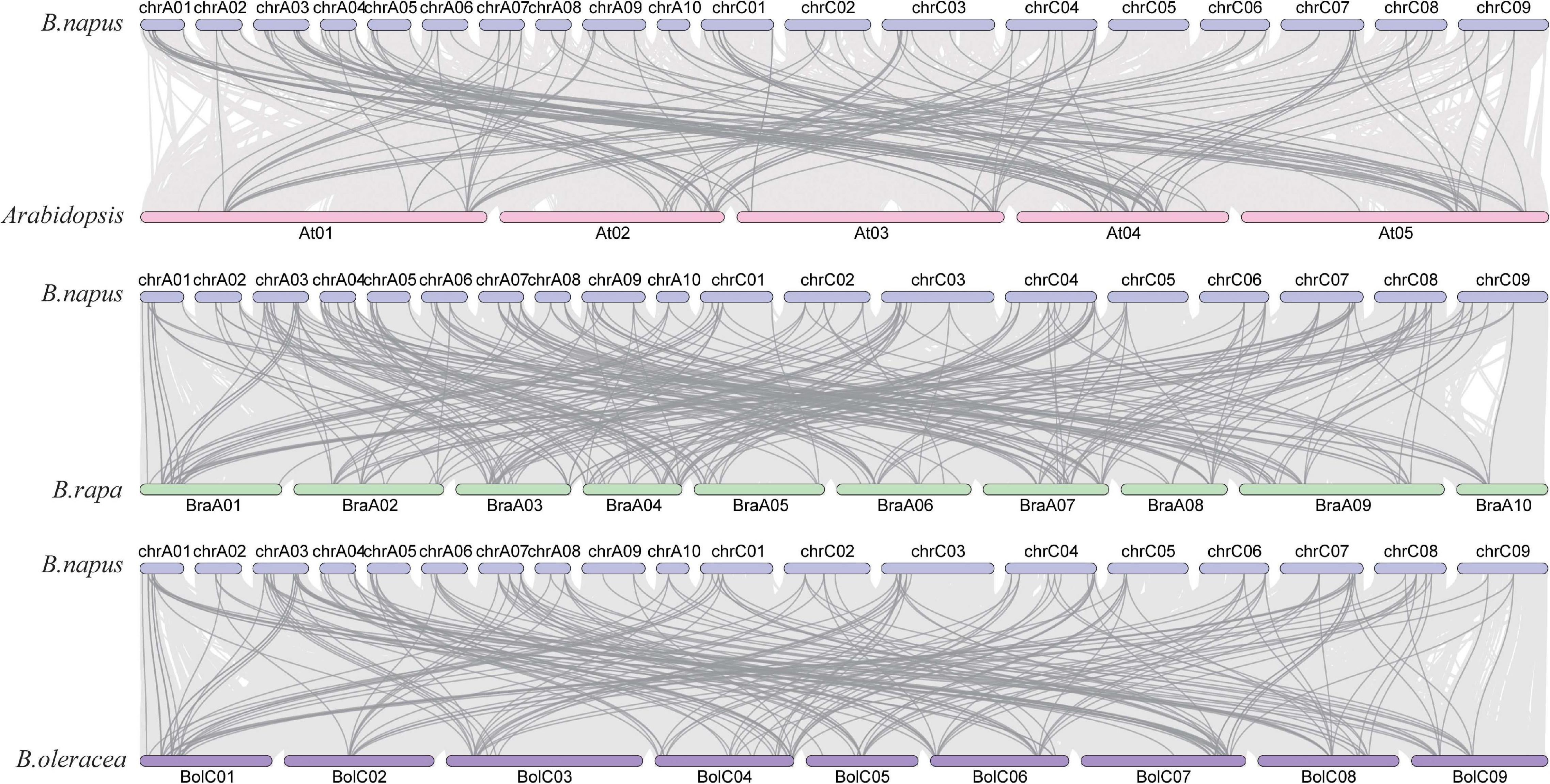
Figure 5. The collinear relationship of AHLs on chromosome. The blue, pink, green, and purple bars represent the chromosomes of B. napus, Arabidopsis, B. rapa, and B. oleracea, respectively. The gray lines in the background indicate the collinear blocks in the genomes of B. napus and Arabidopsis, B. rapa, and B. oleracea, while the darker colored lines emphasize the segmental duplication of the AHLs.
Cis-Acting Element Analysis of BnaAHLs
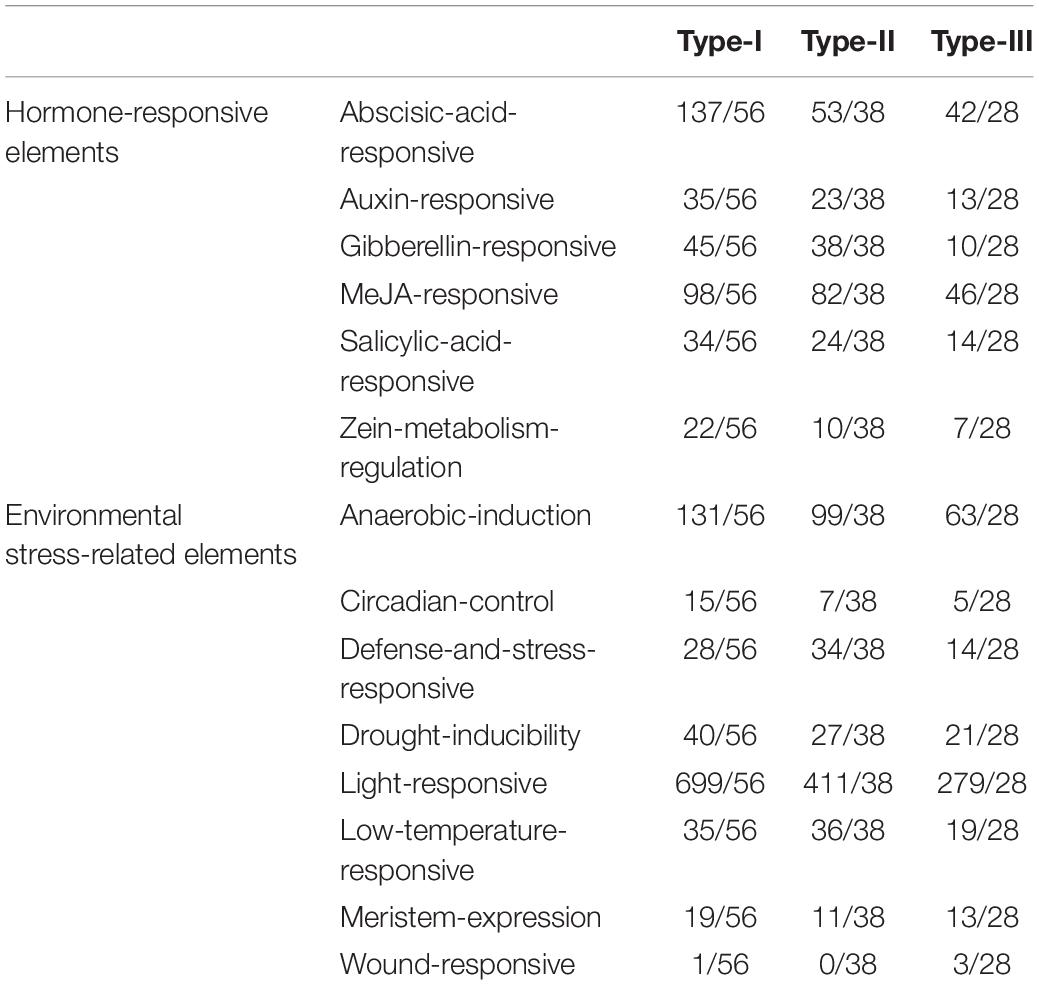
Cis-acting elements regulate the initiation and efficiency of gene transcription by binding to transcription factors (Maire et al., 1989). We analyzed the cis-acting elements of 122 AHLs promoters with PlantCARE (Lescot et al., 2002; Table 2 and Supplementary Figure 2). Most cis-acting elements of BnaAHLs promoters are associated with plant growth and development, hormone response and abiotic stress response. Some cis-acting elements, namely, anaerobic induction, circadian control, defense and stress responsive, drought induction, light response, low temperature response, meristem expression, and wound response, are the main members in regulating environmental stress. Among them, the most common elements are associated with light response, indicating that the growth and development of plants regulated by BnaAHLs is affected by light. At the same time, we also found that the elements related to hormone response, including abscisic acid, MeJA, gibberellin and so on, accounted for a large proportion. These results indicate that BnaAHLs not only regulate plant growth and development, but also play important roles in hormone response and abiotic stress. This is consistent with the report that AHLs can not only regulate growth, but also activate stress response (Favero et al., 2020). The promoters of some duplicated gene pairs like BnaAHL16b/BnaAHL16a, BnaAHL16g/BnaAHL16h, and BnaAHL13a/BnaAHL13b contained basically the same cis-acting elements. The cis-acting elements of other duplicated gene pairs are not exactly the same, such as BnaAHL22b/BnaAHL22a, BnaAHL23a/BnaAHL23b, BnaAHL13f/BnaAHL13g. In general, these differences are mainly caused by the gain and loss of cis-acting elements, including MeJA responsive element, abscisic acid responsive element, defense and stress responsive element, circadian control element, and low temperature responsive element (Table 2, Supplementary Table 1, and Supplementary Figure 2). The cis-acting elements in the upstream promoter region of genes are closely related to the function of downstream genes (Chow et al., 2018). The diversity of cis-acting elements in these duplicated genes indicates that the responses of genes to biotic and abiotic stresses have changed, which is the molecular basis of gene functional diversity.
Positive Selection in the AHL Gene Family
The ratio of non-synonymous substitution rate to synonymous substitution rate, namely Ka/Ks, is a method for diagnosing sequence evolution (Sueoka, 1992; Hurst, 2002). Ka/Ks = 1 means that the gene is subject to neutral selection. Ka/Ks > 1 is considered to have a positive selection effect, otherwise it means that the gene is subject to purify selection. We calculated the Ka/Ks ratio of the BnaAHLs to study their evolutionary forms (Table 3). According to the phylogenetic tree, we first align each evolutionary branch separately, and then use four evolution models [M8 (ωs ≥ 1), M8a (ωs = 1), M7 (beta), and M5 (gamma)] to calculate the Ka/Ks value of each codon. The results showed that the Ka/Ks value of the three AHL types were all less than 0.5, indicating that the AHL gene family had undergone strong purification selection during the evolutionary process. The Ka/Ks value of Type-I AHLs is the smallest, ranging from 0.28 to 0.33. While the Ka/Ks value of Type-III is the largest, ranging from 0.42 to 0.51. It suggested that the purification effect of Type-I AHLs is stronger than those of the other two groups. We also find that some sites are subject to positive selection in the Type-II and Type-III groups predicted by M5 model. The results indicate that these positive selection sites are not in the conserved domains or motifs, but mainly in the random coils of the protein (Figure 6).
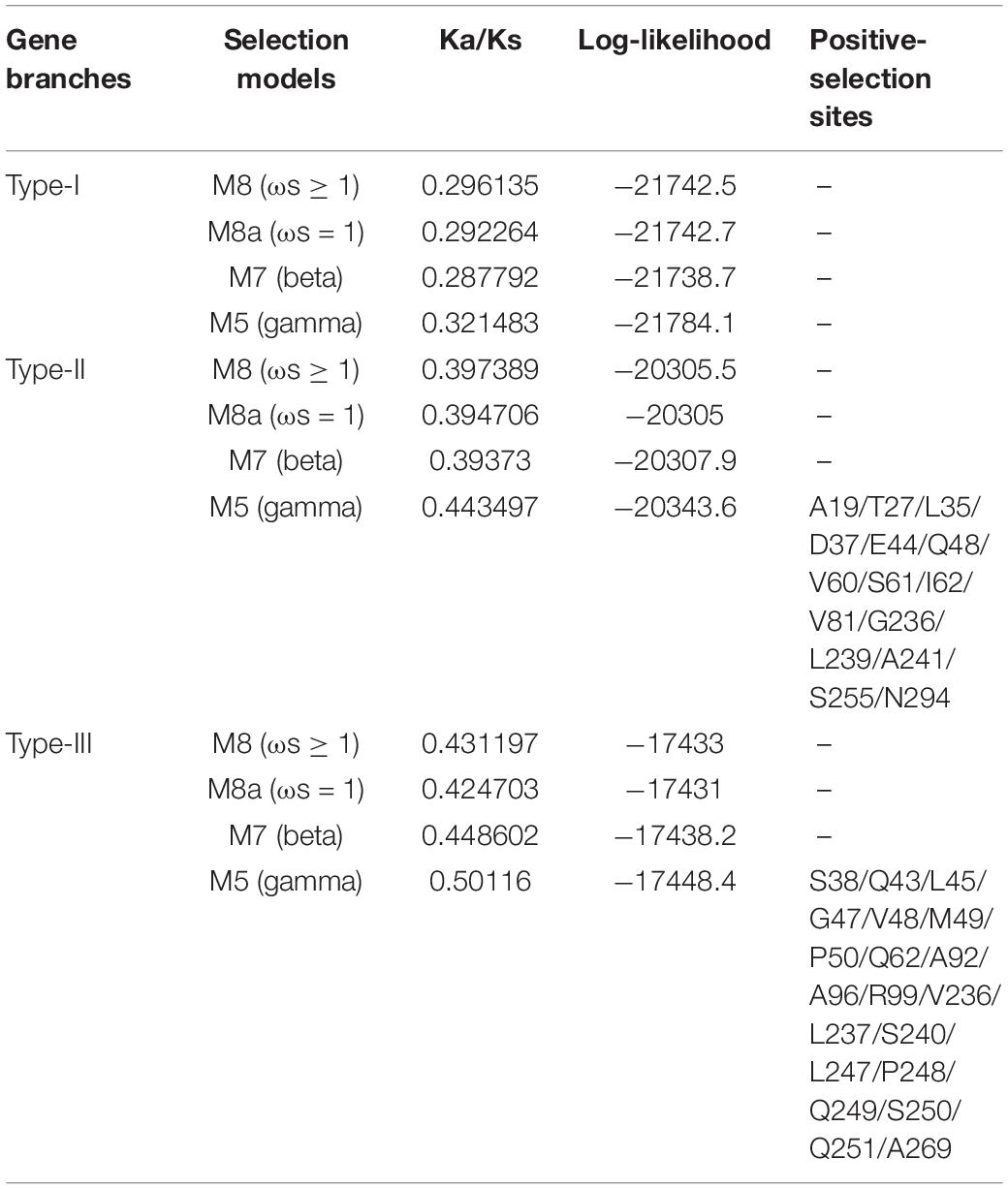
Table 3. Likelihood values and parameter estimation of positive selection among each codon of BnaAHLs.
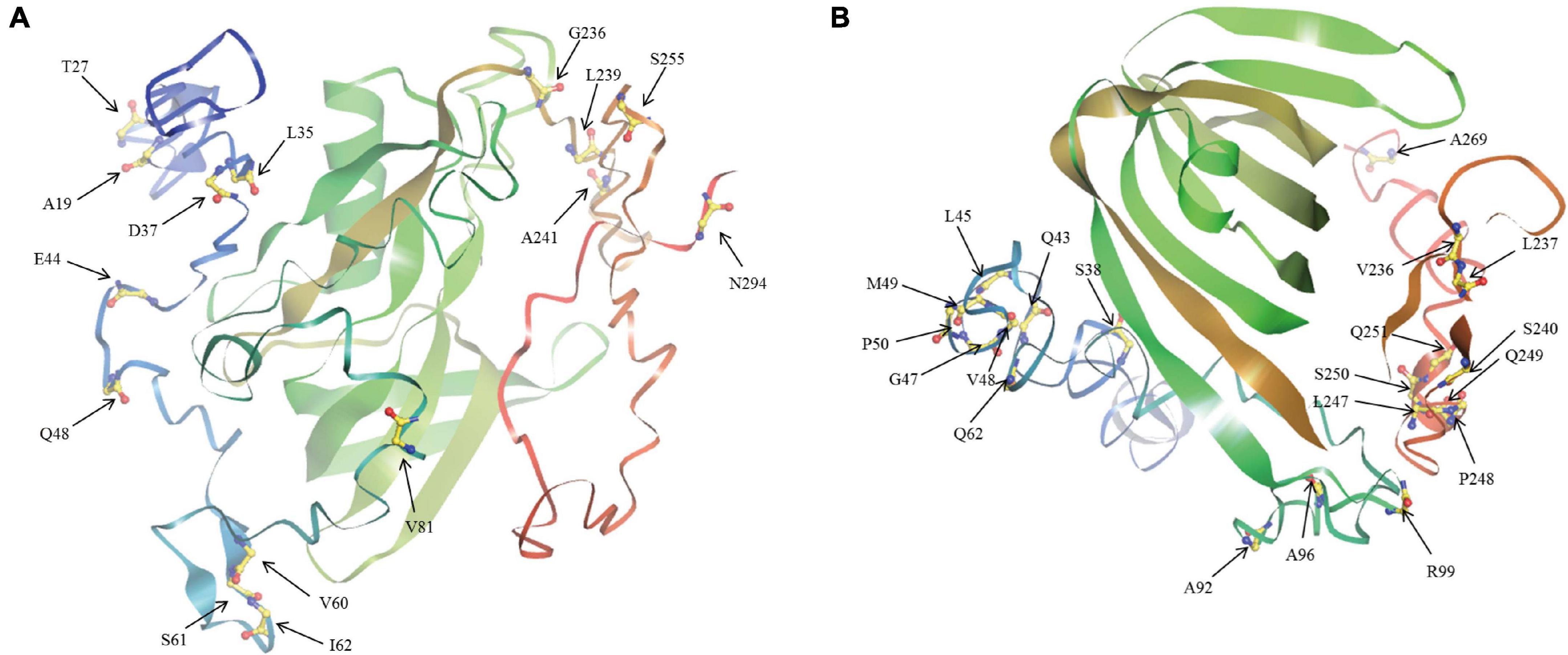
Figure 6. The tertiary structure of BnaAHL proteins. This is a schematic diagram of the tertiary spatial structure of the BnaAHL13j (A) and BnaAHL13g (B), which are the Type-II and Type-III protein query sequence, respectively. The arrow points to the amino acid of positive selection.
Analysis of Differential Expression of BnaAHLs
Studies have revealed that the AHL genes not only play significant roles in the growth and development of plants (Zhao et al., 2014), but also help plants to resist some biotic and abiotic stresses (Jin et al., 2011). We obtained RNA-seq data from B. napus database (see text footnote 13), and used TBtools (Chen et al., 2020) to analyze the expression patterns of the BnaAHLs in different tissues, including root, stem, cotyledon, vegetative rosette, bud, sepal, petal, filament and pollen (Figure 7). RNA-seq data of 20 BnaAHLs were not found in the database. On the whole, expression level of most BnaAHLs is up-regulated in the root and pollen, but down-regulated in the bud (Figure 7). Expression level of about 30% BnaAHLs is up-regulated in the stem. In sepal, petal and filament, about 12 and 14% of BnaAHLs are highly expressed and low expressed, respectively. The results suggested that BnaAHLs could regulate a variety of developmental processes, which is consistent with the reported literatures (Ng et al., 2009; Karami et al., 2020; Sirl et al., 2020).
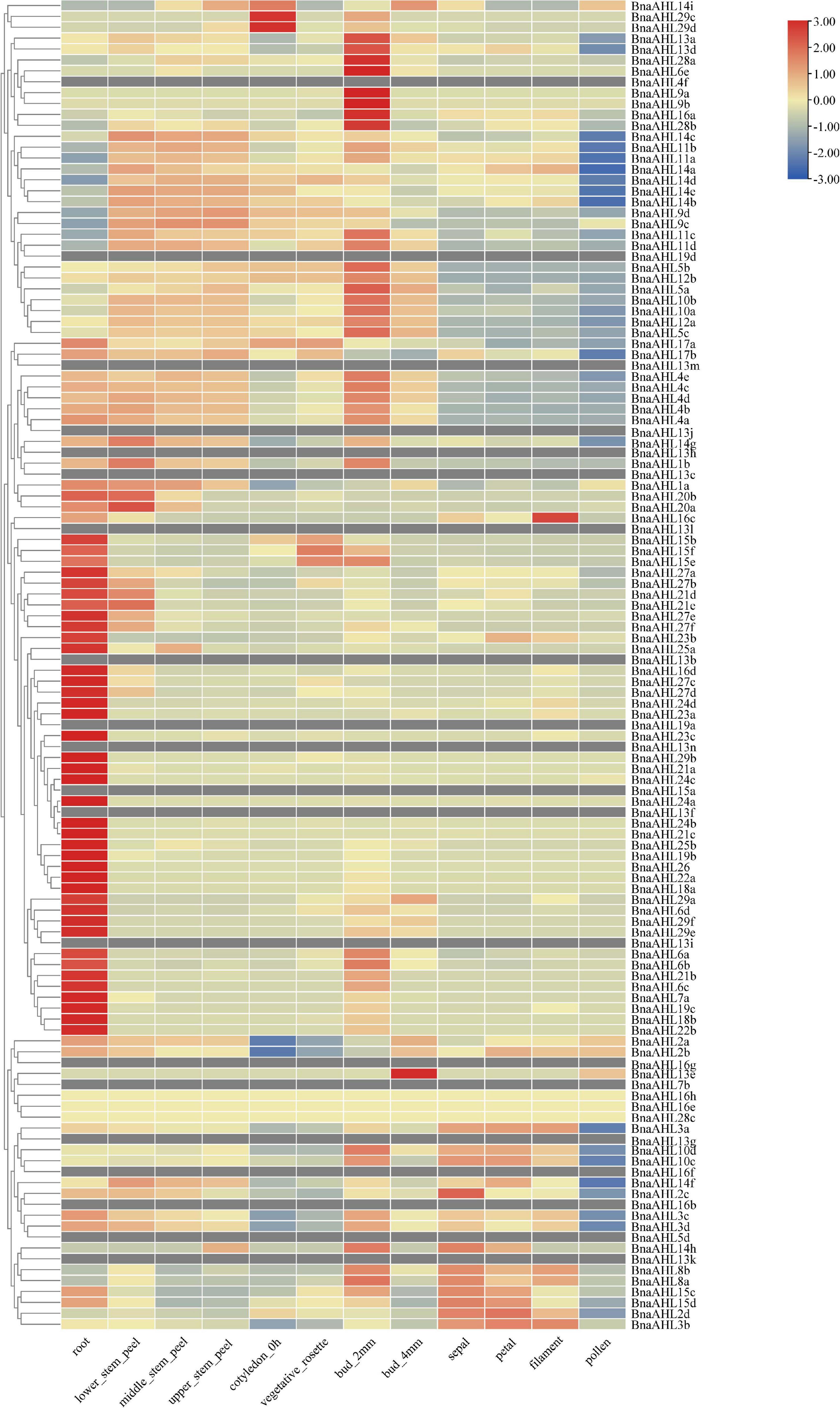
Figure 7. Expression patterns of BnaAHLs in different tissues. BnaAHLs are clustered by hierarchical clustering. The FPKM (the reads per kilobase per million mapped reads) method was used to normalize gene expression. The legend label is shown on the right side of the figure. Genes with high expression levels are shown in red, genes with ground expression levels are shown in blue. The gray box indicates that the expression level of the gene has not been found in the database.
Previous studies have shown that the AHLs have a response to some biological stresses (Lu et al., 2010; Kumar et al., 2018). In order to explore the biological functions of BnaAHLs under biological stress, we inspected the expression patterns of BnaAHLs based on the RNA-seq data infected by S. sclerotiorum (Figure 8A). Compared with the control group, the expression level of 10 BnaAHLs were up-regulated, while the expression pattern of 24 BnaAHLs were down-regulated in the epidermis, mesophyll and vascular of the leaf infected with S. sclerotiorum. 22 BnaAHLs are only highly expressed in mesophyll cells. These expression patterns indicate that BnaAHLs play important roles in regulating some biological stress.
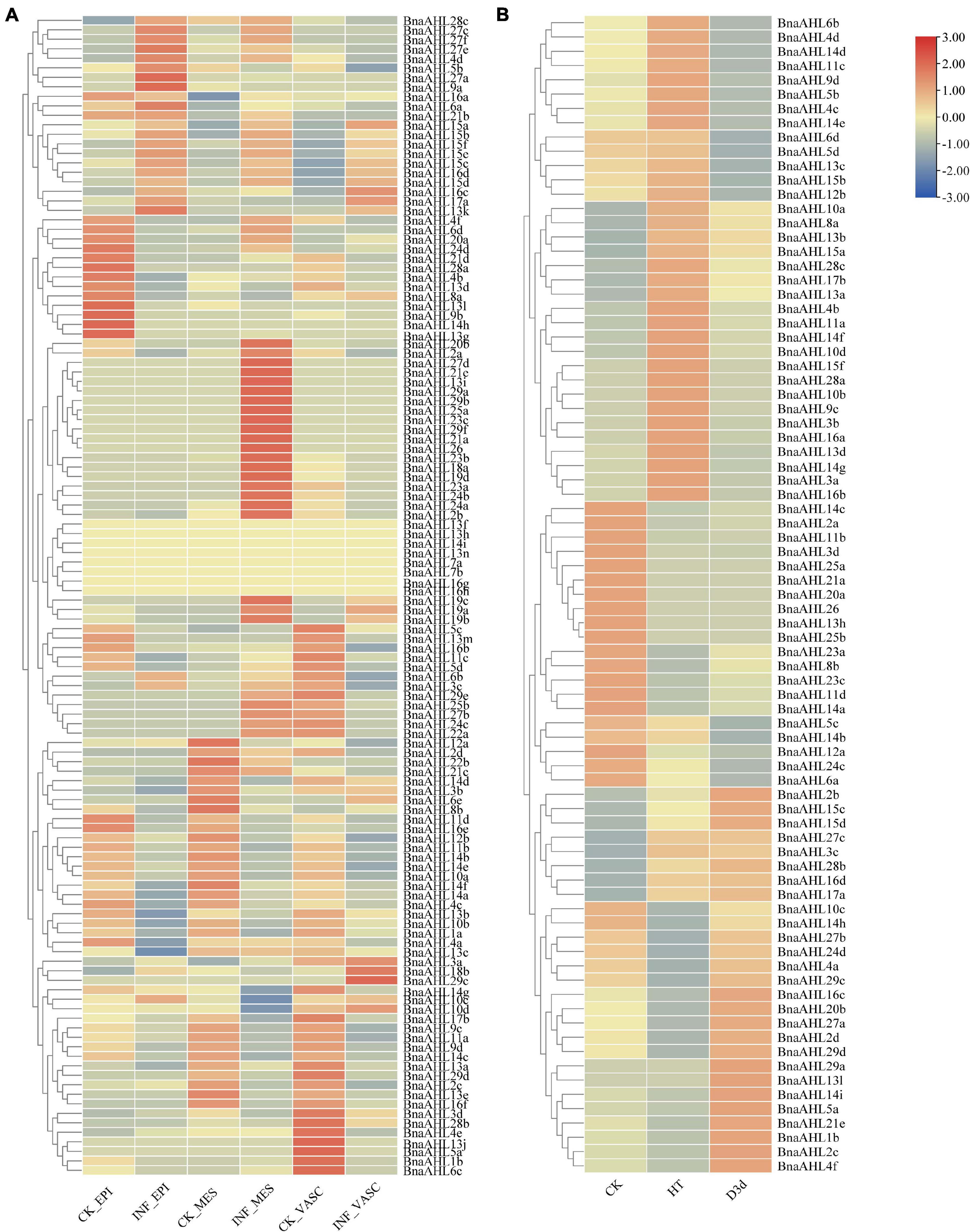
Figure 8. Expression patterns of BnaAHLs under infection by S. sclerotiorum (A) and under abiotic stresses (B). BnaAHLs are clustered by hierarchical clustering. The TPM (Transcripts Per Million) method was used to normalize gene expression. The legend label is shown on the right side of the figure. Genes with high expression levels are shown in red, and genes with low expression levels are shown in blue. CK is control check, INF is the sample infected with S. sclerotiorum, EPI, MES, and VASC is epidermis, mesophyll and vascular, respectively. HT and D3d represent the expression of B. napus under heat treatment and drought treatment after 3 days, respectively.
We also investigated the expression patterns of BnaAHLs under drought and heat stress (Figure 8B). Expression levels of 14 BnaAHLs are up-regulated under drought stress, while the expression of 20 BnaAHLs are down-regulated under drought stress. Similarly, the 14 BnaAHLs whose expression levels were up-regulated under drought stress were also highly expressed under heat stress, while the 20 BnaAHLs that were down-regulated under drought stress were equally low expressed under heat stress. 34 BnaAHLs were highly expressed under heat stress, and 13 BnaAHLs were lowly expressed under drought stress. At the same time, the expression levels of some BnaAHLs (BnaAHL10c, BnaAHL14h, BnaAHL27b, BnaAHL24d, BnaAHL4a, BnaAHL29c, BnaAHL16c, Bna AHL20b, BnaAHL27a, BnaAHL2d, BnaAHL29d, BnaAHL29a, BnaAHL13l, BnaAHL14i, BnaAHL5a, BnaAHL21e, BnaAHL1b, BnaAHL2c, and BnaAHL4f) decreased under heat stress, but increased under drought stress. It suggests that the AHLs play important roles in the adaptation of rapeseed to drought and heat stress, which can also help B. napus to survive in the harsh environments. Therefore, AHLs not only play important roles in plant growth and development, but also can resist various biological and abiotic stresses (Zhou et al., 2016).
Protein-Protein Interaction Analysis of BnaAHLs
Protein-protein interaction network can be used to analyze the complex life activities in cells according to the specific functions and processes of specific sub-networks (Hao et al., 2016). To better understand the functional roles of BnaAHLs, STRING (Szklarczyk et al., 2019) was used to investigate their interaction network. There are 7 members (BnaAHL28c, BnaAHL24d, BnaAHL18a, BnaAHL22a, BnaAHL16a, BnaAHL16h, and BnaAHL16g) of BnaAHLs involved in the construction of protein-protein interaction network. Some key proteins, like histone deacetylases (HDACs), arabinogalactan proteins (AGPs), α-tubulin (TUA), β-tubulin (TUB), ubiquitin thioesterase, arabidopsis pumilio (APUM), SIN3 ASSOCIATED POLYPEPTIDE P18 (SAP18), and HIGH-LEVEL EXPRESSION OF SUGAR-INDUCIBLE GENE2-LIKE1 (HSI2-LIKE1 or HSL1), were predicted to interact with these AHLs (Figure 9). HDACs can inhibit transcription by removing the acetyl groups in the tail core of histones and changing the chromatin structure (Hollender and Liu, 2008) and play key regulatory roles in plant development and stress response (Tian and Chen, 2001; Tian et al., 2003, 2005; Zhou et al., 2005; Chen et al., 2019). Research has demonstrated that AHL proteins can interact with HDACs to affect physiological processes such as flowering time (Yun et al., 2012). AGP6 and AGP11 are involved in a variety of signaling pathways (Costa et al., 2013) and developmental processes (Cheung et al., 1995; Willats and Knox, 1996; Suzuki et al., 2002; van Hengel and Roberts, 2002), especially pollen tube growth and pollen grain germination (Coimbra et al., 2010). AHL16 can directly regulate the expression of AGPs and activate the transcription of pollen development genes (Jia et al., 2015). Polymers of TUA and TUB guide the deposition of cellulose microfibrils during plant cell wall formation (Ledbetter and Porter, 1963). Studies have shown that TUA and TUB are specifically expressed in pollen (Carpenter et al., 1992), roots and leaves (Snustad et al., 1992; Cheng et al., 2001), and vegetative tissues (Kopczak et al., 1992; Abe et al., 2004). There is evidence that HSL1 can inhibit the ectopic expression of sugar-induced seed maturation genes during seed growth, and this protein play important roles in regulating seed germination (Tsukagoshi et al., 2007). As post-transcriptional regulators, APUM (Huh and Paek, 2014) and SAP18 (Song and Galbraith, 2006) were involved in some abiotic stress responses including salt stress. Here, some AHLs were predicted to interact with these proteins, indicating that AHL proteins, as a kind of transcription factor, are also involved in these plant growth, development and abiotic stress response.
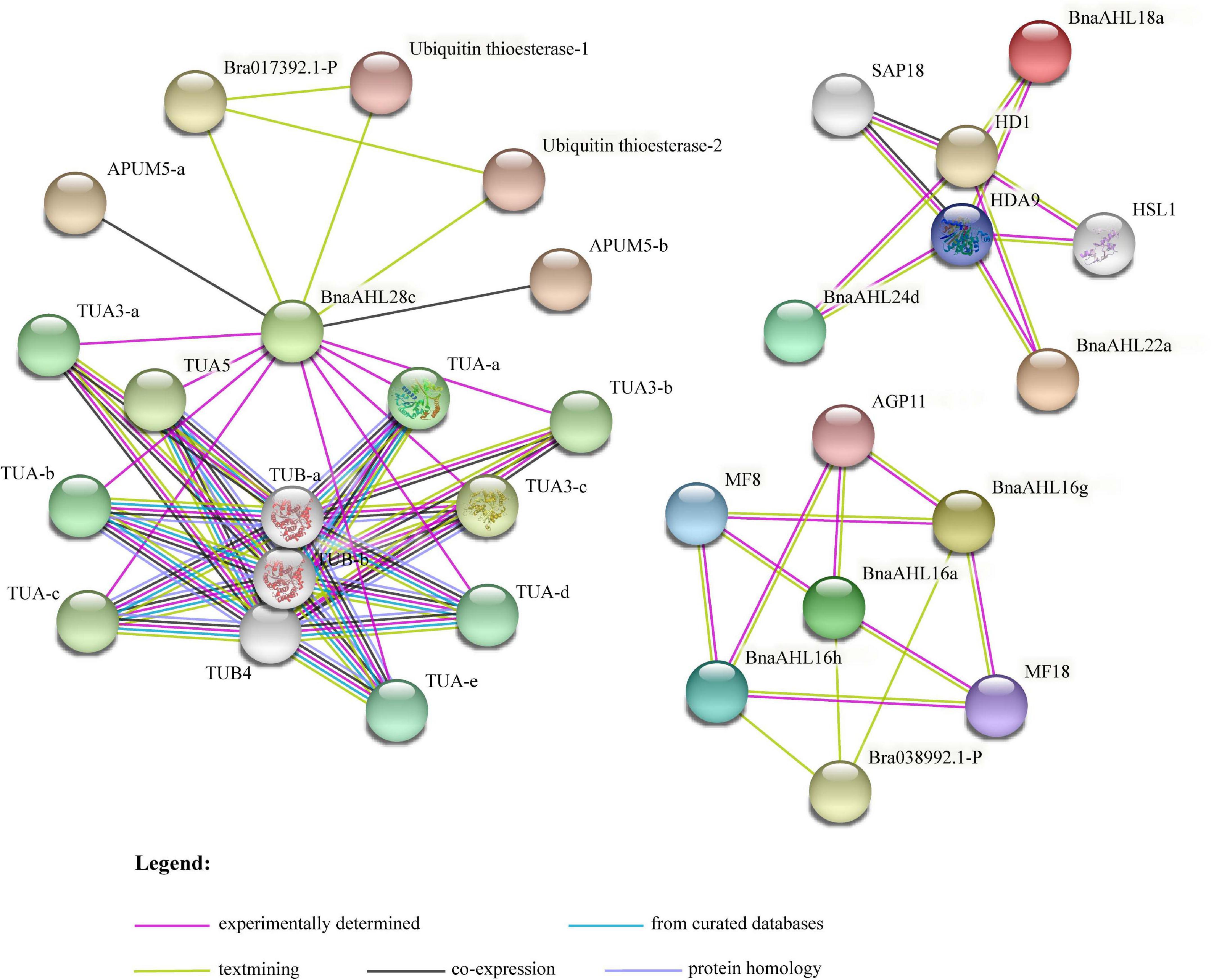
Figure 9. The interaction network of BnaAHL proteins. The line color indicates the type of interaction evidence. The legend is at the bottom of the figure.
Discussion
The R-G-R-P amino acid core sequence was present in the AT-hook motif of BnaAHLs. R-G-R-P can prevent the change of DNA conformation by combining with the minor groove of DNA, thereby promoting the binding of transcription factors with the major groove (Huth et al., 1997). AT-hook can form a C-shaped structure, and the concave back side of the C-shaped structure can be inserted into the small groove of the B-form DNA to combine with the AT-rich DNA sequence (Huth et al., 1997). Among them, proline residues are responsible for maintaining the rigid connection of domains, while the Arg-Gly-Arg conserved sequence is mainly involved in DNA-protein interactions (Aravind and Landsman, 1998), thereby changing the chromatin structure (Strick and Laemmli, 1995). Although BnaAHL7b, BnaAHL14h, BnaAHL14i, BnaAHL21d, BnaAHL21e, and BnaAHL28c proteins lack the R-G-R-P conserved motif, they have the PPC/DUF296 domain. There are a motif 6 and a motif 7 in Type-II members. Type-I and Type-III members contain a motif 6 and motif 7, respectively. In Clade-B, motif 6 was lost and regained more than once during the evolutionary process, therefore the Type-II and Type-III AHL genes have been exchanged several times (Zhao et al., 2014). According to the distribution of phylogenetic tree and conserved motifs, AHL genes of Type-II and Type-III were not distributed separately, but crossed with each other. This finding further illustrates that the AHLs have undergone multiple transitions from Type-II to Type-III and from Type-III to Type-II. The emergence of new motifs may be caused by the insertion and deletion of nucleotides, but this mutation is deleterious. When duplicated genes are present, redundant copies can compensate for the loss of function caused by frameshift mutations, resulting in functional differentiation of genes (Litt and Irish, 2003; Vandenbussche et al., 2003; Raes and Van de Peer, 2005; Kramer et al., 2006; Litt, 2007; Shan et al., 2007). Part of type-II AHLs are formed by individual members of type-III AHL acquiring motif 6. A portion of type-II AHLs lost motif 6 to form type-III AHL (Zhao et al., 2014). R-G-R-P conserved sequence contained in motif 6 is responsible for binding to DNA, and each amino acid is responsible for a different function. A research has shown that the binding of motif 6 to DNA will be affected when the second R of mutation occurs (Zhao et al., 2013).
Study has shown that the intron-containing AHLs are diverged from intron-less AHLs (Zhao et al., 2014). Clade-A consist of the intron-less AHLs, while Clade-B is the opposite. This differential distribution of introns among different members exists not only in the AHL gene family but also in other gene families. In the NADK (Li et al., 2014) and Sus (Wang et al., 2015) gene family, the number, arrangement, and phase of introns are significantly different between subfamilies. Introns are ubiquitous in eukaryotic genomes. More and more evidences show that introns affect gene function by affecting gene expression regulation (Castillo-Davis et al., 2002). At the same time, introns can increase the recombination within and between genes and regulate the rate of gene evolution (Roy and Gilbert, 2006; Rogozin et al., 2012). In addition, many introns contain specific functional elements for exon shuffling and selective splicing, generic functions of non-coding DNA, and signaling as mRNA output from the nucleus (Fedorova and Fedorov, 2003; Le Hir et al., 2003). It is suggested that the AHLs of Clade-B in the rape genome have diverged from Clade-A. In the long evolutionary process, the acquisition of introns may cause the structural and functional changes of BnaAHLs. In addition, the motif structure, exon-intron pattern and amino acid sequence of BnaAHLs may be generated by gene duplication events during evolution. So far, there is no exact explanation for the appearance of introns. The scientific community has two main views on the development of introns. One view is that introns may be the result of tandem duplication within exons and expand one or more times in a manner similar to transposable factors (Zhuo et al., 2007). Another view is that introns emerge through reverse splicing catalyzed by the splicing mechanism itself (Coghlan and Wolfe, 2004). The rate of nucleotide substitution between genes is usually higher than that between introns (Gaffney and Keightley, 2006). Studies have shown that selective pressure on introns play protective roles in genes (Majewski and Ott, 2002). The selective pressure was negatively correlated with the number of introns (Gaffney and Keightley, 2006). Introns are subject to less selection pressure than exons, so the gains and losses of intron sequences are higher than those of exons, and their sequences differentiate more quickly than exons. AHLs of Type-III are subject to less selective pressure due to the presence of introns, so it can be reasonably explained that the value of Ka/Ks is greater than the value of Type-I AHLs.
Although the lengths of ChrC04 and ChrA04 chromosomes are significantly different, the AHLs on these two chromosomes are collinear. Therefore, these two chromosomes are homologous. We speculate that the unequal length of these two chromosomes may be caused by different chromosome duplication. The number of BnaAHL on ChrA06 and ChrC07 is significantly higher than that of ChrC06 and ChrA07, respectively. There was a significant collinear relationship between ChrA07 and ChrC06 of B. napus genome, which may be caused by chromosomal rearrangement. However, the collinearity between ChrA06 and ChrC07 chromosomes is very low. AHL genes exist on these two chromosomes in the form of gene clusters, which may be caused by tandem duplication after transposition.
After the separation of Arabidopsis and Brassica, one ancestor species similar with Arabidopsis formed hexaploid by triploidization. This hexaploid ancestor diverged into B. rapa, B. oleracea, and B. nigra (Lysak et al., 2005; Cheng et al., 2013; Chalhoub et al., 2014). Brassica rapus is a heterotetraploid formed by crossing B. rapa and B. oleracea (Chalhoub et al., 2014). The Arabidopsis genome contains 29 AHLs. Theoretically, the B. napus genome should contain nearly about 174 AHLs, but only 122 AHLs have been identified. These results suggest that the some AHLs were lost on a large scale after the triploidization.
The tertiary structure prediction and selective pressure analysis showed that there were positive selective amino acid sites near the AT-hook motif. The extended AT-hook motif (eAT-hook) has a stronger binding affinity to RNA than with DNA, which length is about 3 times of the ordinary AT-hook motif, and the basic amino acids are located about 12–15 residue distance on both sides of the G-R-P core sequences (Filarsky et al., 2015). The positive selection sites V81 of Type-II group and Q62 and A92 of Type-III group are within this range. The change at these sites may cause the AT-hook loss of binding activity with DNA, but increase the ability to bind to RNA.
Drought and heat are two important factors that affecting plant growth and development. For a long time, plants have evolved a variety of survival mechanisms in response to adversity, such as developing specific morphological traits and regulating metabolic pathways (Vijayaraghavareddy et al., 2020). Abiotic stress changes plant physiological processes by affecting gene expression, RNA or protein stability, and ion transport, etc. (Kosova et al., 2015). The first plant organ to sense salt stress is the root, which immediately relays this signal to the leaves through the stem, causing stomata to close and reducing water to loss (Cosgrove and Hedrich, 1991; Christmann et al., 2013). The analysis of expression patterns and cis-acting elements showed that BnaAHLs were highly expressed in root, and the upstream of BnaAHLs cis-acting elements were also involved in the regulation of light, drought, temperature, oxygen and other stresses. In addition, plant hormones also play roles in regulating plant growth and development by sensing drought and other environmental stress and transmitting signals (Lymperopoulos et al., 2018). The upstream of BnaAHLs carries a large number of cis-acting elements, including abscisic acid and MeJA cis-acting elements.
The expression patterns of some duplicated genes, including BnaAHL10a/BnaAHL10b, BnaAHL9c/BnaAHL9d, and BnaAHL15c/BnaAHL15d, were consistent in various tissues and under stress of heat, drought, and S. sclerotiorum (Figures 7, 8 and Supplementary Table 1). For the duplicated gene pair BnaAHL1a/BnaAHL1b, the expression pattern of the former is down-regulated in bud_2mm, and up-regulated in bud_4mm and pollen, while the expression pattern of the latter is completely opposite (Figure 7). Others examples like BnaAHL4a/BnaAHL4b, BnaAHL10c/BnaAHL10d, BnaAHL11b/BnaAHL11a, BnaAHL11d/BnaAHL11c, and BnaAHL8b/BnaAHL8a, their expression patterns are also different under heat stress. In addition, the expression pattern of BnaAHL5a/BnaAHL5b and BnaAHL3c/BnaAHL3d gene pairs was significantly different. Immune receptors in the cell membrane and cytoplasm of plants are the main innate immune systems. The plasma membrane-embedded PRRs (Pattern Recognition Receptors) have evolved to detect microbial PAMPs and activate PTI (Akira et al., 2006; Chisholm et al., 2006; Boutrot and Zipfel, 2017; Tang et al., 2017; Albert et al., 2020). Sclerotinia sclerotiorum is a destructive pathogen that can infect many monocotyledonous and dicotyledonous plants. Our study showed that the expression of BnaAHLs was significantly altered in B. napus leaves after S. sclerotiorum infecting. Duplication genes with opposite expression patterns were also found in leafs under the stress of S. sclerotiorum (Figure 8). These different expression patterns may be caused by the changes of cis-acting elements upstream of the gene of allopolyploid formed by natural polyploidization and their adaptation to the environment. These changes are the result of many genetic variations occurring in allopolyploid genomes (Song et al., 1995).
Conclusion
In this study, we identified 122 AHLs in the B. napus genome and systematically analyzed them. Based on the phylogenetic analysis, the AHL gene family is divided into three branches, each of which has the same gene structure and motif. About 98% of BnaAHLs products were located in the nucleus. Collinearity analysis shows that the AHL gene family is relatively conserved in evolution of B. rapa, B. oleracea, Arabidopsis, and B. napus. BnaAHLs has high homology with AHLs of B. rapa, B. oleracea, and Arabidopsis. WGD duplication is the main method of BnaAHLs amplification. The AHL gene family has been subjected to purification or neutral selection during the evolution process, and the BnaAHLs of Type-I has the strongest purification effect. The cis-acting elements that regulate plant growth and development and assist plants to resist biotic and abiotic stresses are found in the BnaAHL promoters. This is also verified in tissue expression analysis. Some important BnaAHLs interacting proteins are found in the protein-protein interaction network, which indirectly indicates that AHL genes play important roles in plant growth and development. The identification and characterization of the AHL gene family in B. napus will contribute to further understand their role in these important oil crops.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
W-MZ was the executor of the study, mainly completing the basic analysis of data, and writing the first draft of the manuscript. DF and X-ZC participated in the writing and revision of the manuscript. JC and X-LT were the conceitor and principal of the project, directed the research design and final correction of the data analysis results, and was responsible for the writing and revision of the thesis. All authors have read and agreed to the final text.
Funding
This research was supported by grants from the National Natural Science Foundation of China (Nos. 31871655 and 32072019).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.714305/full#supplementary-material
Footnotes
- ^ https://www.genoscope.cns.fr/brassicanapus/
- ^ https://www.arabidopsis.org/
- ^ https://web.expasy.org/protparam/
- ^ http://cello.life.nctu.edu.tw/
- ^ https://www.megasoftware.net/
- ^ http://meme-suite.org/tools/meme
- ^ https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi
- ^ https://github.com/CJ-Chen/TBtools/releases
- ^ http://plants.ensembl.org/index.html
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
- ^ http://selecton.tau.ac.il/
- ^ https://zhanglab.ccmb.med.umich.edu/I-TASSER/
- ^ http://yanglab.hzau.edu.cn/BnTIR
- ^ https://www.ncbi.nlm.nih.gov/geo
- ^ https://string-db.org/cgi/input.pl
References
Abe, T., Thitamadee, S., and Hashimoto, T. (2004). Microtubule defects and cell morphogenesis in the lefty1lefty2 tubulin mutant of Arabidopsis thaliana. Plant Cell Physiol. 45, 211–220. doi: 10.1093/pcp/pch026
Akira, S., Uematsu, S., and Takeuchi, O. (2006). Pathogen recognition and innate immunity. Cell 124, 783–801. doi: 10.1016/j.cell.2006.02.015
Albert, I., Hua, C., Nurnberger, T., Pruitt, R. N., and Zhang, L. (2020). Surface sensor systems in plant immunity. Plant Physiol. 182, 1582–1596. doi: 10.1104/pp.19.01299
Aravind, L., and Landsman, D. (1998). AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 26, 4413–4421. doi: 10.1093/nar/26.19.4413
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Bishop, E. H., Kumar, R., Luo, F., Saski, C., and Sekhon, R. S. (2020). Genome-wide identification, expression profiling, and network analysis of AT-hook gene family in maize. Genomics 112, 1233–1244. doi: 10.1016/j.ygeno.2019.07.009
Boutrot, F., and Zipfel, C. (2017). Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu. Rev. Phytopathol. 55, 257–286. doi: 10.1146/annurev-phyto-080614-120106
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Carpenter, J. L., Ploense, S. E., Snustad, D. P., and Silflow, C. D. (1992). Preferential expression of an alpha-tubulin gene of Arabidopsis in pollen. Plant Cell 4, 557–571. doi: 10.1105/tpc.4.5.557
Castillo-Davis, C. I., Mekhedov, S. L., Hartl, D. L., Koonin, E. V., and Kondrashov, F. A. (2002). Selection for short introns in highly expressed genes. Nat. Genet. 31, 415–418. doi: 10.1038/ng940
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wang, X., et al. (2014). Plant genetics. early allopolyploid evolution in the post-neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, W. Q., Drapek, C., Li, D. X., Xu, Z. H., Benfey, P. N., and Bai, S. N. (2019). Histone deacetylase HDA19 affects root cortical cell fate by interacting with scarecrow. Plant Physiol. 180, 276–288. doi: 10.1104/pp.19.00056
Cheng, F., Mandakova, T., Wu, J., Xie, Q., Lysak, M. A., and Wang, X. (2013). Deciphering the diploid ancestral genome of the mesohexaploid Brassica rapa. Plant Cell 25, 1541–1554. doi: 10.1105/tpc.113.110486
Cheng, Z., Snustad, D. P., and Carter, J. V. (2001). Temporal and spatial expression patterns of TUB9, a beta-tubulin gene of Arabidopsis thaliana. Plant Mol. Biol. 47, 389–398. doi: 10.1023/a:1011628024798
Cheung, A. Y., Wang, H., and Wu, H. M. (1995). A floral transmitting tissue-specific glycoprotein attracts pollen tubes and stimulates their growth. Cell 82, 383–393. doi: 10.1016/0092-8674(95)90427-1
Cheung, F., Trick, M., Drou, N., Lim, Y. P., Park, J. Y., Kwon, S. J., et al. (2009). Comparative analysis between homoeologous genome segments of Brassica napus and its progenitor species reveals extensive sequence-level divergence. Plant Cell 21, 1912–1928. doi: 10.1105/tpc.108.060376
Chisholm, S. T., Coaker, G., Day, B., and Staskawicz, B. J. (2006). Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124, 803–814. doi: 10.1016/j.cell.2006.02.008
Chow, C. N., Chiang-Hsieh, Y. F., Chien, C. H., Zheng, H. Q., Lee, T. Y., Wu, N. Y., et al. (2018). Delineation of condition specific cis- and trans-acting elements in plant promoters under various endo- and exogenous stimuli. BMC Genomics 19(Suppl 2):85. doi: 10.1186/s12864-018-4469-4
Christmann, A., Grill, E., and Huang, J. (2013). Hydraulic signals in long-distance signaling. Curr. Opin. Plant Biol. 16, 293–300. doi: 10.1016/j.pbi.2013.02.011
Clough, E., and Barrett, T. (2016). The gene expression omnibus database. Methods Mol. Biol. 1418, 93–110. doi: 10.1007/978-1-4939-3578-9_5
Coghlan, A., and Wolfe, K. H. (2004). Origins of recently gained introns in caenorhabditis. Proc. Natl. Acad. Sci. U.S.A. 101, 11362–1367. doi: 10.1073/pnas.0308192101
Coimbra, S., Costa, M., Mendes, M. A., Pereira, A. M., Pinto, J., and Pereira, L. G. (2010). Early germination of Arabidopsis pollen in a double null mutant for the arabinogalactan protein genes AGP6 and AGP11. Sex. Plant Reprod. 23, 199–205. doi: 10.1007/s00497-010-0136-x
Cosgrove, D. J., and Hedrich, R. (1991). Stretch-activated chloride, potassium, and calcium channels coexisting in plasma membranes of guard cells of Vicia faba L. Planta 186, 143–153. doi: 10.1007/BF00201510
Costa, M., Nobre, M. S., Becker, J. D., Masiero, S., Amorim, M. I., Pereira, L. G., et al. (2013). Expression-based and co-localization detection of arabinogalactan protein 6 and arabinogalactan protein 11 interactors in Arabidopsis pollen and pollen tubes. BMC Plant Biol. 13:7. doi: 10.1186/1471-2229-13-7
Doron-Faigenboim, A., Stern, A., Mayrose, I., Bacharach, E., and Pupko, T. (2005). Selecton: a server for detecting evolutionary forces at a single amino-acid site. Bioinformatics 21, 2101–2103. doi: 10.1093/bioinformatics/bti259
Favero, D. S., Kawamura, A., Shibata, M., Takebayashi, A., Jung, J. H., Suzuki, T., et al. (2020). AT-hook transcription factors restrict petiole growth by antagonizing PIFs. Curr. Biol. 30, 1454–1466. doi: 10.1016/j.cub.2020.02.017
Fedorova, L., and Fedorov, A. (2003). Introns in gene evolution. Genetica 118, 123–131. doi: 10.1023/A:1024145407467
Filarsky, M., Zillner, K., Araya, I., Villar-Garea, A., Merkl, R., Langst, G., et al. (2015). The extended AT-hook is a novel RNA binding motif. RNA Biol. 12, 864–876. doi: 10.1080/15476286.2015.1060394
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230. doi: 10.1093/nar/gkt1223
Fujimoto, S., Matsunaga, S., Yonemura, M., Uchiyama, S., Azuma, T., and Fukui, K. (2004). Identification of a novel plant MAR DNA binding protein localized on chromosomal surfaces. Plant Mol. Biol. 56, 225–239. doi: 10.1007/s11103-004-3249-5
Gaffney, D. J., and Keightley, P. D. (2006). Genomic selective constraints in murid noncoding DNA. Plos Genet. 2:e204. doi: 10.1371/journal.pgen.0020204
Gallavotti, A., Malcomber, S., Gaines, C., Stanfield, S., Whipple, C., Kellogg, E., et al. (2011). Barren stalk fastigiate1 is an AT-hook protein required for the formation of maize ears. Plant Cell 23, 1756–1771. doi: 10.1105/tpc.111.084590
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Hao, T., Peng, W., Wang, Q., Wang, B., and Sun, J. (2016). Reconstruction and application of protein-protein interaction network. Int. J. Mol. Sci. 17:907. doi: 10.3390/ijms17060907
Hollender, C., and Liu, Z. (2008). Histone deacetylase genes in Arabidopsis development. J. Integr. Plant Biol. 50, 875–885. doi: 10.1111/j.1744-7909.2008.00704.x
Howell, E. C., Kearsey, M. J., Jones, G. H., King, G. J., and Armstrong, S. J. (2008). A and C genome distinction and chromosome identification in Brassica napus by sequential fluorescence in situ hybridization and genomic in situ hybridization. Genetics 180, 1849–1857. doi: 10.1534/genetics.108.095893
Huh, S. U., and Paek, K. H. (2014). APUM5, encoding a pumilio RNA binding protein, negatively regulates abiotic stress responsive gene expression. BMC Plant Biol. 14:75. doi: 10.1186/1471-2229-14-75
Hurst, L. D. (2002). The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 18:486. doi: 10.1016/s0168-9525(02)02722-1
Huth, J. R., Bewley, C. A., Nissen, M. S., Evans, J. N., Reeves, R., Gronenborn, A. M., et al. (1997). The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif. Nat. Struct. Biol. 4, 657–665. doi: 10.1038/nsb0897-657
Jeanmougin, F., Thompson, J. D., Gouy, M., Higgins, D. G., and Gibson, T. J. (1998). Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23, 403–405. doi: 10.1016/s0968-0004(98)01285-7
Jia, Q. S., Zhu, J., Xu, X. F., Lou, Y., Zhang, Z. L., Zhang, Z. P., et al. (2015). Arabidopsis AT-hook protein TEK positively regulates the expression of arabinogalactan proteins for nexine formation. Mol. Plant 8, 251–260. doi: 10.1016/j.molp.2014.10.001
Jin, Y., Luo, Q., Tong, H., Wang, A., Cheng, Z., Tang, J., et al. (2011). An AT-hook gene is required for palea formation and floral organ number control in rice. Dev. Biol. 359, 277–288. doi: 10.1016/j.ydbio.2011.08.023
Karami, O., Rahimi, A., Khan, M., Bemer, M., Hazarika, R. R., Mak, P., et al. (2020). A suppressor of axillary meristem maturation promotes longevity in flowering plants. Nat. Plants 6, 368–376. doi: 10.1038/s41477-020-0637-z
Kim, H. B., Oh, C. J., Park, Y. C., Lee, Y., Choe, S., An, C. S., et al. (2011). Comprehensive analysis of AHL homologous genes encoding AT-hook motif nuclear localized protein in rice. BMB Rep. 44, 680–685. doi: 10.5483/BMBRep.2011.44.10.680
Kopczak, S. D., Haas, N. A., Hussey, P. J., Silflow, C. D., and Snustad, D. P. (1992). The small genome of Arabidopsis contains at least six expressed alpha-tubulin genes. Plant Cell 4, 539–547. doi: 10.1105/tpc.4.5.539
Kosova, K., Vitamvas, P., Urban, M. O., Klima, M., Roy, A., and Prasil, I. T. (2015). Biological networks underlying abiotic stress tolerance in temperate crops–a proteomic perspective. Int. J. Mol. Sci. 16, 20913–20942. doi: 10.3390/ijms160920913
Kramer, E. M., Su, H. J., Wu, C. C., and Hu, J. M. (2006). A simplified explanation for the frameshift mutation that created a novel C-terminal motif in the APETALA3 gene lineage. BMC Evol. Biol. 6:30. doi: 10.1186/1471-2148-6-30
Kumar, K., Purayannur, S., Kaladhar, V. C., Parida, S. K., and Verma, P. K. (2018). Mqtl-seq and classical mapping implicates the role of an at-hook motif containing nuclear localized (AHL) family gene in Ascochyta blight resistance of chickpea. Plant Cell Environ 41, 2128–2140. doi: 10.1111/pce.13177
Le Hir, H., Nott, A., and Moore, M. J. (2003). How introns influence and enhance eukaryotic gene expression. Trends Biochem. Sci. 28, 215–220. doi: 10.1016/S0968-0004(03)00052-5
Ledbetter, M. C., and Porter, K. R. (1963). A “microtubule” in plant cell fine structure. J. Cell Biol. 19, 239–250. doi: 10.1083/jcb.19.1.239
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, B., and Kliebenstein, D. J. (2014). The AT-hook motif-encoding gene metabolic network modulator 1 underlies natural variation in Arabidopsis primary metabolism. Front. Plant Sci. 5:415. doi: 10.3389/fpls.2014.00415
Li, W. Y., Wang, X., Li, R., Li, W. Q., and Chen, K. M. (2014). Genome-wide analysis of the NADK gene family in plants. PLoS One 9:e101051. doi: 10.1371/journal.pone.0101051
Lim, P. O., Kim, Y., Breeze, E., Koo, J. C., Woo, H. R., Ryu, J. S., et al. (2007). Overexpression of a chromatin architecture-controlling AT-hook protein extends leaf longevity and increases the post-harvest storage life of plants. Plant J. 52, 1140–1153. doi: 10.1111/j.1365-313X.2007.03317.x
Litt, A. (2007). An evaluation of a-function: evidence from the APETALA1 and APETALA2 gene lineages. Int. J. Plant Sci. 168, 73–91. doi: 10.1086/509662
Litt, A., and Irish, V. F. (2003). Duplication and diversification in the Apetala1/Fruitfull floral homeotic gene lineage: implications for the evolution of floral development. Genetics 165, 821–833.
Liu, S., Liu, Y., Yang, X., Tong, C., Edwards, D., Parkin, I. A., et al. (2014). The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 5:3930. doi: 10.1038/ncomms4930
Long, M., Rosenberg, C., and Gilbert, W. (1995). Intron phase correlations and the evolution of the intron/exon structure of genes. Proc. Natl. Acad. Sci. U.S.A. 92, 12495–12499. doi: 10.1073/pnas.92.26.12495
Lou, Y., Xu, X. F., Zhu, J., Gu, J. N., Blackmore, S., and Yang, Z. N. (2014). The tapetal AHL family protein TEK determines nexine formation in the pollen wall. Nat. Commun. 5:3855. doi: 10.1038/ncomms4855
Lu, H., Zou, Y., and Feng, N. (2010). Overexpression of AHL20 negatively regulates defenses in Arabidopsis. J. Integr. Plant Biol. 52, 801–808. doi: 10.1111/j.1744-7909.2010.00969.x
Lymperopoulos, P., Msanne, J., and Rabara, R. (2018). Phytochrome and phytohormones: working in tandem for plant growth and development. Front. Plant Sci. 9:1037. doi: 10.3389/fpls.2018.01037
Lynch, M., and Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. doi: 10.1126/science.290.5494.1151
Lysak, M. A., Koch, M. A., Pecinka, A., and Schubert, I. (2005). Chromosome triplication found across the tribe Brassiceae. Genome Res. 15, 516–525. doi: 10.1101/gr.3531105
Maire, P., Wuarin, J., and Schibler, U. (1989). The role of cis-acting promoter elements in tissue-specific albumin gene expression. Science 244, 343–346. doi: 10.1126/science.2711183
Majewski, J., and Ott, J. (2002). Distribution and characterization of regulatory elements in the human genome. Genome Res. 12, 1827–1836. doi: 10.1101/gr.606402
Marchler-Bauer, A., and Bryant, S. H. (2004). CD-search: protein domain annotations on the fly. Nucleic Acids Res. 32, W327–W331. doi: 10.1093/nar/gkh454
Matsushita, A., Furumoto, T., Ishida, S., and Takahashi, Y. (2007). AGF1, an AT-hook protein, is necessary for the negative feedback of AtGA3ox1 encoding GA 3-oxidase. Plant Physiol. 143, 1152–1162. doi: 10.1104/pp.106.093542
Mun, J. H., Kwon, S. J., Yang, T. J., Seol, Y. J., Jin, M., Kim, J. A., et al. (2009). Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome Biol. 10:R111. doi: 10.1186/gb-2009-10-10-r111
Nagano, Y., Furuhashi, H., Inaba, T., and Sasaki, Y. (2001). A novel class of plant-specific zinc-dependent DNA-binding protein that binds to A/T-rich DNA sequences. Nucleic Acids Res. 29, 4097–4105. doi: 10.1093/nar/29.20.4097
Ng, K. H., Yu, H., and Ito, T. (2009). AGAMOUS controls GIANT KILLER, a multifunctional chromatin modifier in reproductive organ patterning and differentiation. PLoS Biol. 7:e1000251. doi: 10.1371/journal.pbio.1000251
Raes, J., and Van de Peer, Y. (2005). Functional divergence of proteins through frameshift mutations. Trends Genet. 21, 428–431. doi: 10.1016/j.tig.2005.05.013
Rashotte, A. M., Carson, S. D., To, J. P., and Kieber, J. J. (2003). Expression profiling of cytokinin action in Arabidopsis. Plant Physiol. 132, 1998–2011. doi: 10.1104/pp.103.021436
Reeves, R., and Beckerbauer, L. (2001). HMGI/Y proteins: flexible regulators of transcription and chromatin structure. Biochim. Biophys. Acta 1519, 13–29. doi: 10.1016/s0167-4781(01)00215-9
Rogozin, I. B., Carmel, L., Csuros, M., and Koonin, E. V. (2012). Origin and evolution of spliceosomal introns. Biol. Direct 7:11. doi: 10.1186/1745-6150-7-11
Roy, S. W., and Gilbert, W. (2006). The evolution of spliceosomal introns: patterns, puzzles and progress. Nat. Rev. Genet. 7, 211–221. doi: 10.1038/nrg1807
Shan, H. Y., Zhan, N., Liu, C. J., Xu, G. X., Zhang, J., Chen, Z. D., et al. (2007). Patterns of gene duplication and functional diversification during the evolution of the AP1/SQUA subfamily of plant MADS-box genes. Mol. Phylogenet. Evol. 44, 26–41. doi: 10.1016/j.ympev.2007.02.016
Sirl, M., Snajdrova, T., Gutierrez-Alanis, D., Dubrovsky, J. G., Vielle-Calzada, J. P., Kulich, I., et al. (2020). At-hook motif nuclear localised protein 18 as a novel modulator of root system architecture. Int. J. Mol. Sci. 21:1886. doi: 10.3390/ijms21051886
Snustad, D. P., Haas, N. A., Kopczak, S. D., and Silflow, C. D. (1992). The small genome of Arabidopsis contains at least nine expressed beta-tubulin genes. Plant Cell 4, 549–556. doi: 10.1105/tpc.4.5.549
Song, C. P., and Galbraith, D. W. (2006). AtSAP18, an orthologue of human SAP18, is involved in the regulation of salt stress and mediates transcriptional repression in Arabidopsis. Plant Mol. Biol. 60, 241–257. doi: 10.1007/s11103-005-3880-9
Song, K., Lu, P., Tang, K., and Osborn, T. C. (1995). Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl. Acad. Sci. U.S.A. 92, 7719–7723. doi: 10.1073/pnas.92.17.7719
Street, I. H., Shah, P. K., Smith, A. M., Avery, N., and Neff, M. M. (2008). The AT-hook-containing proteins SOB3/AHL29 and ESC/AHL27 are negative modulators of hypocotyl growth in Arabidopsis. Plant J. 54, 1–14. doi: 10.1111/j.1365-313X.2007.03393.x
Strick, R., and Laemmli, U. K. (1995). SARs are cis DNA elements of chromosome dynamics: synthesis of a SAR repressor protein. Cell 83, 1137–1148. doi: 10.1016/0092-8674(95)90140-x
Struss, D., Quiros, C. F., Plieske, J., and Robbelen, G. (1996). Construction of brassica B genome synteny groups based on chromosomes extracted from three different sources by phenotypic, isozyme and molecular markers. Theor. Appl. Genet. 93, 1026–1032. doi: 10.1007/BF00230120
Sueoka, N. (1992). Directional mutation pressure, selective constraints, and genetic equilibria. J. Mol. Evol. 34, 95–114. doi: 10.1007/BF00182387
Suzuki, Y., Kitagawa, M., Knox, J. P., and Yamaguchi, I. (2002). A role for arabinogalactan proteins in gibberellin-induced alpha-amylase production in barley aleurone cells. Plant J. 29, 733–741. doi: 10.1046/j.1365-313x.2002.01259.x
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613. doi: 10.1093/nar/gky1131
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tang, D., Wang, G., and Zhou, J. M. (2017). Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell 29, 618–637. doi: 10.1105/tpc.16.00891
Tian, L., and Chen, Z. J. (2001). Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc. Natl. Acad. Sci. U.S.A. 98, 200–205. doi: 10.1073/pnas.011347998
Tian, L., Fong, M. P., Wang, J. J., Wei, N. E., Jiang, H., Doerge, R. W., et al. (2005). Reversible histone acetylation and deacetylation mediate genome-wide, promoter-dependent and locus-specific changes in gene expression during plant development. Genetics 169, 337–345. doi: 10.1534/genetics.104.033142
Tian, L., Wang, J., Fong, M. P., Chen, M., Cao, H., Gelvin, S. B., et al. (2003). Genetic control of developmental changes induced by disruption of Arabidopsis histone deacetylase 1 (AtHD1) expression. Genetics 165, 399–409.
Tsukagoshi, H., Morikami, A., and Nakamura, K. (2007). Two B3 domain transcriptional repressors prevent sugar-inducible expression of seed maturation genes in Arabidopsis seedlings. Proc. Natl. Acad. Sci. U.S.A. 104, 2543–2547. doi: 10.1073/pnas.0607940104
van Hengel, A. J., and Roberts, K. (2002). Fucosylated arabinogalactan-proteins are required for full root cell elongation in Arabidopsis. Plant J. 32, 105–113. doi: 10.1046/j.1365-313x.2002.01406.x
Vandenbussche, M., Theissen, G., Van de Peer, Y., and Gerats, T. (2003). Structural diversification and neo-functionalization during floral MADS-box gene evolution by C-terminal frameshift mutations. Nucleic Acids Res. 31, 4401–4409. doi: 10.1093/nar/gkg642
Vijayaraghavareddy, P., Vemanna, R. S., Yin, X., Struik, P. C., Makarla, U., and Sreeman, S. (2020). Acquired traits contribute more to drought tolerance in wheat than in rice. Plant Phenomics 2020:5905371. doi: 10.34133/2020/5905371
Vom Endt, D., Soares e Silva, M., Kijne, J. W., Pasquali, G., and Memelink, J. (2007). Identification of a bipartite jasmonate-responsive promoter element in the Catharanthus roseus ORCA3 transcription factor gene that interacts specifically with AT-Hook DNA-binding proteins. Plant Physiol. 144, 1680–1689. doi: 10.1104/pp.107.096115
Wang, H., Leng, X., Yang, J., Zhang, M., Zeng, M., Xu, X., et al. (2021a). Comprehensive analysis of AHL gene family and their expression under drought stress and ABA treatment in Populus trichocarpa. PeerJ 9:e10932. doi: 10.7717/peerj.10932
Wang, M., Chen, B., Zhou, W., Xie, L., Wang, L., Zhang, Y., et al. (2021b). Genome-wide identification and expression analysis of the AT-hook motif nuclear localized gene family in soybean. BMC Genomics 22:361. doi: 10.1186/s12864-021-07687-y
Wang, X., Wang, H., Wang, J., Sun, R., Wu, J., Liu, S., et al. (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1039. doi: 10.1038/ng.919
Wang, Z., Wei, P., Wu, M., Xu, Y., Li, F., Luo, Z., et al. (2015). Analysis of the sucrose synthase gene family in tobacco: structure, phylogeny, and expression patterns. Planta 242, 153–166. doi: 10.1007/s00425-015-2297-1
Willats, W. G., and Knox, J. P. (1996). A role for arabinogalactan-proteins in plant cell expansion: evidence from studies on the interaction of beta-glucosyl Yariv reagent with seedlings of Arabidopsis thaliana. Plant J. 9, 919–925. doi: 10.1046/j.1365-313x.1996.9060919.x
Wong, M. M., Bhaskara, G. B., Wen, T. N., Lin, W. D., Nguyen, T. T., Chong, G. L., et al. (2019). Phosphoproteomics of Arabidopsis highly ABA-induced1 identifies AT-hook-like10 phosphorylation required for stress growth regulation. Proc. Natl. Acad. Sci. U.S.A. 116, 2354–2363. doi: 10.1073/pnas.1819971116
Xiao, C., Chen, F., Yu, X., Lin, C., and Fu, Y. F. (2009). Over-expression of an AT-hook gene, AHL22, delays flowering and inhibits the elongation of the hypocotyl in Arabidopsis thaliana. Plant Mol. Biol. 71, 39–50. doi: 10.1007/s11103-009-9507-9
Xu, Y., Gan, E. S., and Ito, T. (2013a). The AT-hook/PPC domain protein TEK negatively regulates floral repressors including MAF4 and MAF5. Plant Signal. Behav. 8:e25006. doi: 10.4161/psb.25006
Xu, Y., Wang, Y., Stroud, H., Gu, X., Sun, B., Gan, E. S., et al. (2013b). A matrix protein silences transposons and repeats through interaction with retinoblastoma-associated proteins. Curr. Biol. 23, 345–350. doi: 10.1016/j.cub.2013.01.030
Yang, J., Yan, R., Roy, A., Xu, D., Poisson, J., and Zhang, Y. (2015). The I-TASSER suite: protein structure and function prediction. Nat. Methods 12, 7–8. doi: 10.1038/nmeth.3213
Yang, M., Derbyshire, M. K., Yamashita, R. A., and Marchler-Bauer, A. (2020). NCBI’s conserved domain database and tools for protein domain analysis. Curr. Protoc. Bioinformatics 69:e90. doi: 10.1002/cpbi.90
Yang, Y. W., Lai, K. N., Tai, P. Y., Ma, D. P., and Li, W. H. (1999). Molecular phylogenetic studies of brassica, rorippa, Arabidopsis and allied genera based on the internal transcribed spacer region of 18S-25S rDNA. Mol. Phylogenet. Evol. 13, 455–462. doi: 10.1006/mpev.1999.0648
Yang, Z., Nielsen, R., Goldman, N., and Pedersen, A. M. (2000). Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155, 431–449.
Yu, C. S., Chen, Y. C., Lu, C. H., and Hwang, J. K. (2006). Prediction of protein subcellular localization. Proteins 64, 643–651. doi: 10.1002/prot.21018
Yun, J., Kim, Y. S., Jung, J. H., Seo, P. J., and Park, C. M. (2012). The AT-hook motif-containing protein AHL22 regulates flowering initiation by modifying Flowering locus T chromatin in Arabidopsis. J. Biol. Chem. 287, 15307–15316. doi: 10.1074/jbc.M111.318477
Zhao, J., Favero, D. S., Peng, H., and Neff, M. M. (2013). Arabidopsis thaliana AHL family modulates hypocotyl growth redundantly by interacting with each other via the PPC/DUF296 domain. Proc. Natl. Acad. Sci. U.S.A. 110, E4688–E4697. doi: 10.1073/pnas.1219277110
Zhao, J., Favero, D. S., Qiu, J., Roalson, E. H., and Neff, M. M. (2014). Insights into the evolution and diversification of the AT-hook motif nuclear localized gene family in land plants. BMC Plant Biol. 14:266. doi: 10.1186/s12870-014-0266-7
Zhao, K., Kas, E., Gonzalez, E., and Laemmli, U. K. (1993). SAR-dependent mobilization of histone H1 by HMG-I/Y in vitro: Hmg-I/Y is enriched in H1-depleted chromatin. EMBO J. 12, 3237–3247.
Zhao, L., Lu, Y., Chen, W., Yao, J., Li, Y., Li, Q., et al. (2020). Genome-wide identification and analyses of the AHL gene family in cotton (Gossypium). BMC Genomics 21:69. doi: 10.1186/s12864-019-6406-6
Zhou, C., Zhang, L., Duan, J., Miki, B., and Wu, K. (2005). Histone deacetylase19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell 17, 1196–1204. doi: 10.1105/tpc.104.028514
Zhou, J., Wang, X., Lee, J. Y., and Lee, J. Y. (2013). Cell-to-cell movement of two interacting AT-hook factors in Arabidopsis root vascular tissue patterning. Plant Cell 25, 187–201. doi: 10.1105/tpc.112.102210
Zhou, L., Liu, Z., Liu, Y., Kong, D., Li, T., Yu, S., et al. (2016). A novel gene OsAHL1 improves both drought avoidance and drought tolerance in rice. Sci. Rep. 6:30264. doi: 10.1038/srep30264
Keywords: Brassica napus, AT-hook motif, gene organization, gene duplication, cis-acting elements, selective pressure
Citation: Zhang W-M, Fang D, Cheng X-Z, Cao J and Tan X-L (2021) Insights Into the Molecular Evolution of AT-Hook Motif Nuclear Localization Genes in Brassica napus. Front. Plant Sci. 12:714305. doi: 10.3389/fpls.2021.714305
Received: 25 May 2021; Accepted: 19 July 2021;
Published: 09 September 2021.
Edited by:
Maoteng Li, Huazhong University of Science and Technology, ChinaReviewed by:
Liezhao Liu, Southwest University, ChinaQing-Yong Yang, Huazhong Agricultural University, China
Copyright © 2021 Zhang, Fang, Cheng, Cao and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Cao, Y2ppbmZvckAxNjMuY29t; Y2ppbmZvckB1anMuZWR1LmNu; Xiao-Li Tan, eGx0YW5AdWpzLmVkdS5jbg==
 Wei-Meng Zhang
Wei-Meng Zhang Jun Cao
Jun Cao Xiao-Li Tan
Xiao-Li Tan