
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 23 September 2021
Sec. Plant Proteomics and Protein Structural Biology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.709534
This article is part of the Research Topic Update on Plant Sexual Reproduction - a Proteomic Perspective View all 5 articles
 Yanhua Li1
Yanhua Li1 Liangjie Niu1
Liangjie Niu1 Xiaolin Wu1
Xiaolin Wu1 Claudia Faleri2
Claudia Faleri2 Fuju Tai1*
Fuju Tai1* Man Zhang1
Man Zhang1 Hui Liu1
Hui Liu1 Wei Wang1*
Wei Wang1* Giampiero Cai2*
Giampiero Cai2*Cysteine proteases, belonging to the C1-papain family, play a major role in plant growth and development, senescence, and immunity. There is evidence to suggest that pollen cysteine protease (CP) (ZmCP03) is involved in regulating the anther development and pollen formation in maize. However, there is no report on the genome-wide identification and comparison of CPs in the pollen coat and other tissues in maize. In this study, a total of 38 homologous genes of ZmCP03 in maize were identified. Subsequently, protein motifs, conserved domains, gene structures, and duplication patterns of 39 CPs are analyzed to explore their evolutionary relationship and potential functions. The cis-elements were identified in the upstream sequence of 39 CPs, especially those that are related to regulating growth and development and responding to environmental stresses and hormones. The expression patterns of these genes displayed remarked difference at a tissue or organ level in maize based on the available transcriptome data in the public database. Quantitative reverse transcription PCR (RT-qPCR) analysis showed that ZmCP03 was preferably expressed at a high level in maize pollen. Analyses by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot, immunofluorescence and immunogold electron microscopy all validated the cellular localization of ZmCP03 in both the pollen coat and pollen cytoplasm. In addition, 142 CP genes from Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa) and cotton (Gossypium hirsutum), together with 39 maize CPs, were retrieved to analyze their evolution by comparing with orthologous genes. The results suggested that ZmCP03 was relatively conservative and stable during evolution. This study may provide a referential evidence on the function of ZmCP03 in pollen development and germination in maize.
The pollen coat is the outermost surface of pollen grains that contain the haploid male gametes and actively participates in the pollination and fertilization in plants (Zhang et al., 2016). Its composition mainly includes lipids, pigments, proteins, and aromatic compounds, which provide the pollen coat with a hydrophobic feature. Thus, the pollen coat can protect pollen grains from dehydration during pollen release and germination in the stigmatic surface (Edlund et al., 2004; Murphy, 2006; Quilichini et al., 2015; Wu et al., 2015; Zhang et al., 2016).
Previous studies in Brassica, Arabidopsis, and Olea europaea have showed that the pollen coat serves in pollen-stigmatic adhesion, recognition, and hydration and pollen germination through pollen coat-derived proteins, such as S-locus cysteine-rich protein 11 (SCR/SP11), pollen coat protein B class, extracellular lipase 4, and caleosin (Pacini and Hesse, 2004; Updegraff et al., 2009; Marshall et al., 2011; Zienkiewicz et al., 2011; Rejón et al., 2016; Wang et al., 2017). In particular, SCR/SP11 plays a vital role in regulating self-incompatibility in Brassica (Schopfer et al., 1999; Takayama et al., 2000; Marshall et al., 2011). Pollen coat protein B class (PCP-Bs) are key regulators of compatible pollen hydration in Arabidopsis. After knocking out AtPCP-B genes, pollen hydration was impaired and pollen tube growth was obviously delayed in pcp-b mutants (Wang et al., 2017). In maize, two pollen coat rich-proteins, β-glucanase and xylanase, have been proved to regulate pollen germination and tube growth (Suen et al., 2003; Suen and Huang, 2007). Pollen coat β-expansin 1 (Zea m 1) has been identified as a cell wall-loosening agent (Yennawar et al., 2006), and the pollen tubes of expb1 mutant penetrated through the stigma and the style slower than the wild-type ones (Valdivia et al., 2007, 2009). Up to date, only a small number of pollen coat-derived proteins have been identified with definite functions in pollen-stigmatic interactions and pollen germination. Proteomic and bioinformatics analyses have implicated the important role of many functional-unknown pollen coat-derived proteins in maize (Gong et al., 2015; Wu et al., 2015) and rice (Dai et al., 2006). Recently, high-throughput sequencing and bioinformatic analyses have provided comprehensive perspectives in exploring the roles of pollen coat-derived proteins in cotton (Yang et al., 2020).
In the previous study, cysteine protease was found to be present at a high abundance in maize pollen coats (Wu et al., 2015). The enzyme, denoted as ZmCP03, belongs to the papain-like cysteine protease family (subfamily C1A) based on a domain structure analysis (Niño et al., 2020). Previously, ZmCP03 was found in the vacuoles of tapetum cells at a late stage of anther development (Li et al., 2012). Accumulating reports implicate that papain-like cysteine proteases play a role in tapetal programmed cell death (PCD) (Zhang et al., 2014; Shukla et al., 2019), biotic (Niño et al., 2020) and abiotic stresses (Pechan et al., 2000), and seed germination (Lu et al., 2015). For example, in Arabidopsis, a cysteine protease CEP1 has been found to participate in tapetal PCD and pollen development, while the cep1 mutant and cep1 overexpression both cause pollen infertility and abnormal tapetal PCD (Zhang et al., 2014).
At present, there is limited information on the role of ZmCP03 in maize pollen development and tube germination, and especially a lack of the genome-wide identification and comparison of cysteine protease (CPs) in pollen coats and other maize tissues. The evolutionary relationships of the CPs in maize and in three other plant species were performed to provide insights into the biological functions of ZmCP03, especially in pollen development and germination.
The sequences of ZmCP03 gene and its encoded protein were used as queries to identify the homologous genes and proteins in maize from the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/gene/?term=zea%20mays; February 15, 2021; species, zea mays; 68,633 sequences) using basic logical alignment search tool (BLAST) searches. After removing the redundant and incomplete sequences, 39 unique gene and protein sequences of maize CPs were collected. The sequences of CPs in Arabidopsis (Arabidopsis thaliana) were collected from the TAIR v.10 database (https://www.arabidopsis.org; February 15, 2021) (Richau et al., 2012), those for rice (Oryza sativa) from the NCBI database (https://www.ncbi.nlm.nih.gov/gene/?term=rice; February 15, 2021; species, Oryza sativa; 80,075 sequences) (Wang et al., 2018), and those for cotton (Gossypium hirsutum) from Cottongene (https://www.cottongen.org/organism/Gossypium/hirsutum; July 15, 2021; species, Gossypium hirsutum; 12,877,329 sequences) (Zhang et al., 2019). The definitions of the CP domains were based on a Pfam (PF00112) analysis (http://pfam.xfam.org/), and the domains of maize CPs were examined by Web CD-Search Tool (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) (Marchler-Bauer et al., 2017).
The structure of the ZmCP genes was created by the Gene Structure Display Server 2.0 (GSDS; http://gsds.gao-lab.org/) (Hu et al., 2015). The physiochemical properties of ZmCPs, especially theoretical isoelectric point (pI), molecular weights (MW), instability index, and grand average of hydropathy (GRAVY) values, were calculated by the ExPASy Protparam tool (https://web.expasy.org/protparam) (Artimo et al., 2012; Qanmber et al., 2019b).
The proteins sequences of 39 ZmCPs were aligned using “align by muscle” and the unrooted phylogenetic tree was created by MEGA X with neighbor-joining (NJ) algorithm. The parameters were set on 1,000 replicates for bootstrapping, p-distance, and 50% partial deletion site coverage cut-off (Kumar et al., 2018). The position information of ZmCPs on chromosomes was obtained from a gff3-file of maize genome annotation data (https://www.ncbi.nlm.nih.gov/genome/?term=maize) and presented using the TBtools toolkit (Chen et al., 2020). The collinear pairs of the CP genes were identified using MCScanX (Wang et al., 2012) and the collinearity map was built using the TBtools toolkit (Chen et al., 2020).
The protein motifs were analyzed using the MEME (Multiple EM for Motif Elicitation) tool (https://meme-suite.org/tools/meme) (Bailey et al., 2015). The parameters were set as follows: the motif width: 6–80 amino acids; the motif maximum number: 15 (Niu et al., 2020). The 2,000 bp upstream sequence regions of 39 ZmCP genes were collected to analyze the promoter cis-elements using the database PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002).
The number of synonymous (Ks) and non-synonymous (Ka) substitutions per site of duplicated cystatin genes were calculated by TBtools (Chen et al., 2020). Ka/Ks < 1 means negative selection, Ka/Ks = 1 means neutral selection, and Ka/Ks > 1 means positive selection.
Tissue-specific expression data were collected from MaizeGDB (https://www.maizegdb.org). The expression abundances were determined for replicated mRNA-sequencing datasets from 79 tissues and abiotic (salt stress, drought, and temperature stress) or biotic stress treatments (Colletotrichum graminicola and Cercosporazeina infection). The transcript abundances of ZmCPs were calculated as fragments per kilobase per million mapped fragments (FPKM) in an unstranded mode, a maximum intron length of 60 kb, and utilizing reference bias correction (Hoopes et al., 2019). The heatmap of the tissue-specific expression was created using expression values in different tissues by TBtools (Chen et al., 2020).
The root, stem, and leaves of maize inbred line B104 were sampled at the three-leaf seedling stage, and pollen grains were sampled at the pollen shed stage. Total RNA was extracted using RNA-Solv® reagent (Omega Bio-Tek, Norcross, GA, USA). Single-stranded cDNA was synthesized using a 5 × All-In-One MasterMix with an AccuRT Genomic DNA Removal Kit (Applied Biological Materials Inc., Richmond, Canada) according to the instructions of the manufacturer.
The gene-specific primers of ZmUBI and ZmCP03 were designed using the Primer Premier 5.0 software (http://www.premierbiosoft.com/) and commercially synthesized (Biomed Cooperation, Beijing, China) as follows: ZmUBI, 5′-TAAGCTGCCGATGTGCCTGCG-3′, and 5′-CTGAAAGACAGAACATAATGAGCACAG-3′; ZmCP03, 5′-AAGAAGCGGGCCAACGTATC-3′, and 5′-CCCTGTCGTGATCTTGGTGA-3′. The ZmUBI was used as an internal control to normalize the data.
A quantitative reverse transcription PCR (RT-qPCR) was carried out using the StepOnePlus™ Real-Time PCR Instrument Thermal Cycling Block (Applied Bio-systems). The PCR conditions contained an initial denaturation step at 95°C for 5 min, followed by 40 cycles at 95°C for 10 s and 60°C for 30 s. The quantification method 2−ΔΔCT (Livak and Schmittgen, 2001) was used to assess the relative expression level of ZmCP03 in different maize tissues. Data were presented as relative expression (mean ± SD) from three biological replicates after normalization based on ZmUBI expression. Significant differences in expression changes among tissues were analyzed by Student's t-test (*p < 0.05, **p < 0.01) in the GraphPad Prism 8.0 software (San Diego, CA, USA).
A 15 amino acid polypeptide, PVRRDAGKKRANVSS, from N115 to N130 of ZmCP03 was used to prepare anti-ZmCP03 antibodies. The synthesized peptide was sequenced by a matrix assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry analysis to verify its sequence. Mice were sequentially immunized on days 1, 15, 35, and 56 with the synthesized peptide. Splenocytes were harvested from mice on day 61 and fused with syngeneic myeloma cells to form hybridoma cells, which were selectively cultured. The positive hybridoma cells were then selected and clonally expanded before inoculation into the peritoneal cavity of mice. After 1–2 weeks, mouse ascites was removed and monoclonal antibodies were immunoaffinity purified.
As for immunoblot analysis, protein samples were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (12.5% gel), and electrophoretically transferred onto hydrophilic polyvinylidene fluoride (PVDF) membranes (Hybond-P, Amersham, Uppsala, Sweden). After blocking with bovine serum proteins, the blots were incubated with anti-ZmCP03 (1:5000 dilution) for 1 h. After rinsing, it was incubated with rabbit anti-mouse IgG (1:2000 dilution) labeled with horseradish peroxidase for 1 h and stained by AmershamTM ECLTM prime kit (Cytiva, Marlborough, MA, USA).
Pollens were fixed in paraformaldehyde, rinsed, dehydrated, and resin embedded. Sections (150-nm thick) were cut and placed on drops of water on glass slides and dried at 45°C. Sections were blocked with bovine serum albumin (BSA) and incubated with anti-ZmCP03 (1:100 dilution) for 1 h. After washing, sections were incubated with rabbit anti-mouse antibody labeled with fluorescein for 1 h, rinsed, and observed under a fluorescence microscope (Nikon C2-ER; Nikon Corp, Tokyo, Japan).
The immunogold electron microscopy localization of ZmCP03 was carried out as in our previous report (Wang et al., 2006). Briefly, pollen grains were fixed with paraformaldehyde, rinsed, dehydrated, and resin embedded. Ultrathin sections (80-nm thick) were blocked with bovine serum proteins and incubated with anti-ZmCP03 (1:100 dilution) for 1 h. After rinsing, it was incubated with gold particles (10 nm) conjugated with rabbit anti-mouse IgG for 1 h. Sections were sequentially negatively stained with uranyl acetate (10 min) and lead citrate (5 min), rinsed, and examined by transmission electron microscopy (Hitachi HT-7700, Tokyo, Japan).
The information about the protein length, MW, pI, and GRAVY values of 39 maize CP proteins is listed in Table 1, and the nucleotide and amino acid sequences are provided in Supplementary Dataset 1. Based on the instability index, 31 of 39 CPs were stable, whereas the others, including ZmCP03, were instable. Besides, their pIs ranged from 4.73 to 8.36, and eight CPs were alkaline proteins (pI > 7). All of the 39 CPs were hydrophilic with negative GRAVY values.
A total of 142 CP genes were identified in three plant species (Arabidopsis, cotton, and rice) as papain-like cysteine proteases. Among them, 31 were obtained from Arabidopsis (Richau et al., 2012), 78 from cotton (Zhang et al., 2019), and 33 from rice (Wang et al., 2018).
To examine the evolutionary events in the four plant species, a phylogenetic tree was created by MEGA X (Figure 1). The tree was divided into nine subfamilies, similar to the presentation of CPs in cotton (Zhang et al., 2019), namely, the subfamilies responsive to desiccation 21 (RD21), cysteine endopeptidase (CEP), xylem cysteine peptidase (XCP), xylem bark cysteine peptidase 3 (XBCP3), Th1 immune response-associated cysteine protease (THI), senescence-associated gene 12 (SAG12), responsive to desiccation19 (RD19), aleurain-like protease (ALP), and cathepsin B-like (CTB). Therefore, the present study used the same nomenclature as previous studies (Richau et al., 2012; Zhang et al., 2019).
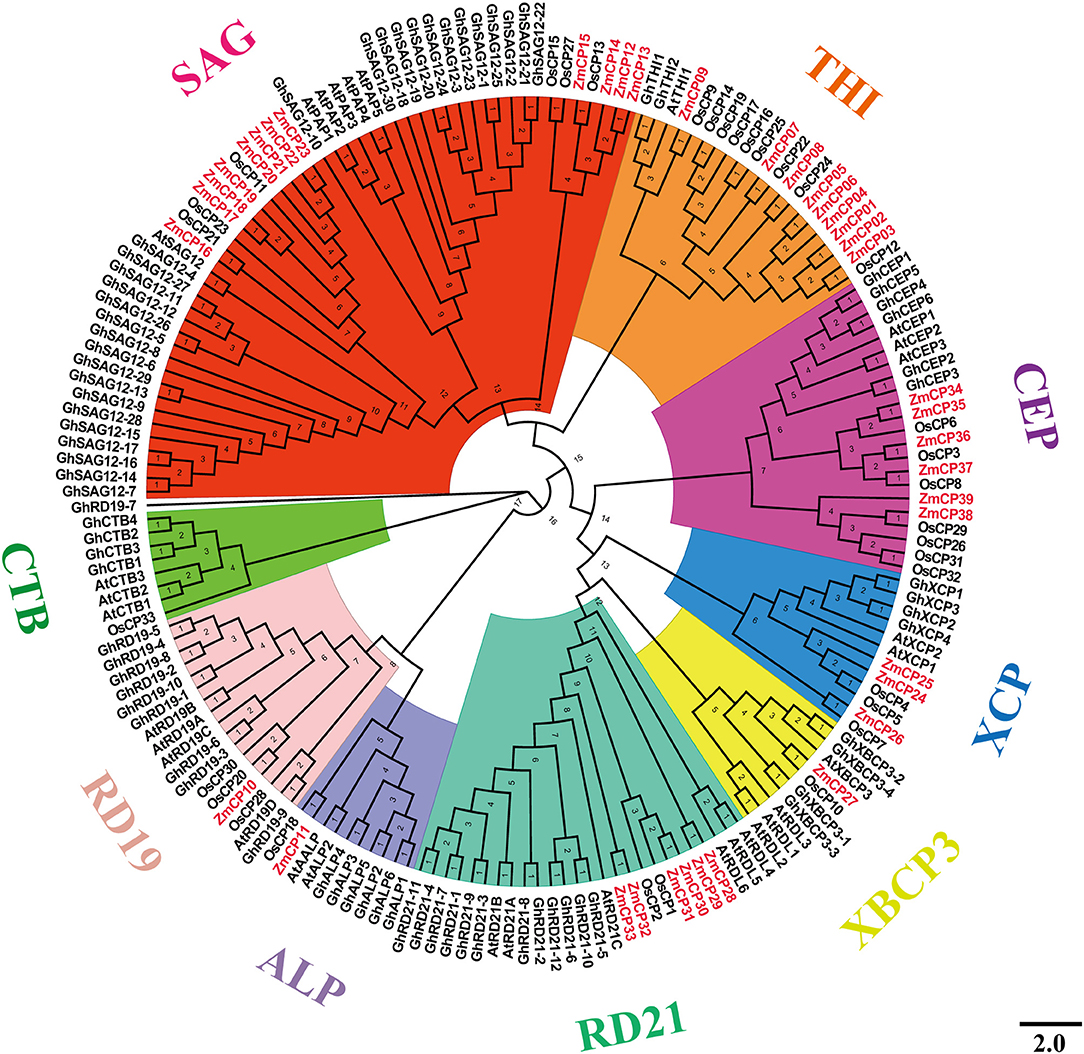
Figure 1. Evolutionary relations among 181 cysteine protease (CP) genes among the four plant species examined.
The SAG12 is the largest subfamily, whereas the subfamily XBCP3 had the fewest members (only eight). Previously, GhRD19-7 was in the same group with GhRD19-9 (Zhang et al., 2019), but it fell in the clade with GhRD19-9 in the results, possibly because the phylogenetic tree was built using four different plant species here, whereas the tree in the previous report (Zhang et al., 2019) was created using cotton and Arabidopsis.
To reveal the evolutionary relationship of the ZmCPs, a multiple sequence alignment of 39 maize CP proteins was carried out and used to construct an unrooted phylogenetic tree. As a result, the ZmCPs were divided into nine groups, namely, SAG12, THI, CEP, XCP, XBCP3, RD21, ALP, and RD19 (Figure 2A). Group SAG12 was composed of 12 ZmCPs, group THI had 9 members (including ZmCP03), group CEP had 6 members, while group XBCP3, ALP, and RD19 all had only one member. A similar phylogenetic tree was created using the maximum likelihood method (Supplementary Figure 1).
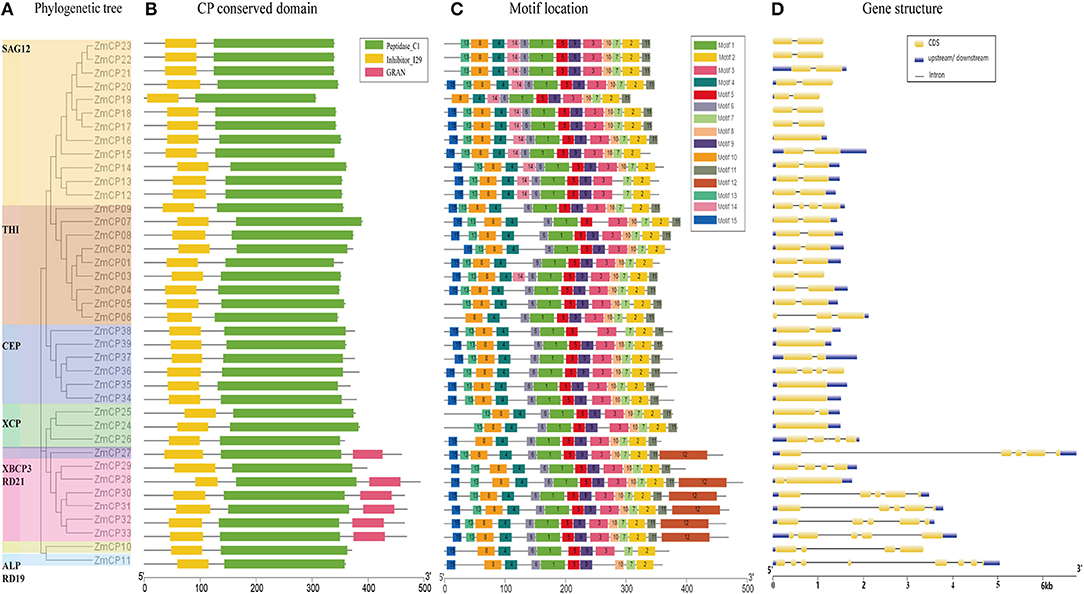
Figure 2. Evolutionary analysis, motif compositions, and gene structures of 39 ZmCPs. (A) The unrooted phylogenetic tree. (B) The conversed domain in ZmCPs. (C) The motif location in ZmCPs. (D) The gene structures of 39 ZmCPs.
Furthermore, the conserved domains of ZmCPs were visualized (Figure 2B). All ZmCPs contain the peptidase C1 domain, which is the known typical domain of peptidase family C1 (Rawlings et al., 2010). All ZmCPs also contained the inhibitor I29 domain (MEROPS peptidase inhibitor family I29). The I29 domain is also found at the N-terminus of a variety of peptidase precursors that belong to the MEROPS peptidase subfamily C1A (Groves et al., 1996). It forms an α-helical domain that runs through the substrate-binding site and prevents access. The removal of this region by proteolytic cleavage results in the activation of the enzyme (Rawlings et al., 2010). Besides, six ZmCP proteins (ZmCP27, 28, 30–33) from the subfamily XCPB3 and RD21 had a C-terminal extension domain named the GRAN domain (granulins). Granulins were first found in animals as growth hormones released after wounding events (Bateman and Bennett, 2009; Wang et al., 2018).
Fifteen different motifs with variable amino acid lengths and sequences were identified in 39 ZmCPs by the MEME search tool (https://meme-suite.org/tools/meme) (Figure 2C). Of them, motifs 1, 2, 6, and 7 existed in all examined ZmCPs, forming the catalytic triad Cys-His-Asn. The 38 ZmCPs, except for ZmCP37, possessed motifs 4 and 8, which were the conversed domain inhibitor I29 (Wang et al., 2018). The sequence of motif 8 is ExxxRxxxFxxNxxxI/VxxxN with one mismatch. Besides, motif 15 was special to the subfamily CEP except for ZmCP37. The sequence of motif 12 is Cx5Cx5CCCx7Cx4CCx6CCx5CCx6Cx6C (the GRAN domain), which existed in the C-terminal extensions of XBCP3 and five members from RD21.
The gene structure analysis showed that 5 genes contained no intron, 21 (most belonging to the subfamilies SAG12 and THI) contained only a single intron, and the remaining 13 contained 2–7 introns (Figure 2D). Obviously, the structures of the 39 ZmCP genes were similar to those reported in rice (Wang et al., 2018).
The upstream region of a gene, containing the promoter and the binding site of transcription factors, regulates the gene expression. Thus, a cis-element analysis helps to understand gene regulation and function (Higo et al., 1998, 1999; Qanmber et al., 2019a). To identify the putative cis-acting regulatory DNA elements, the 2,000-bp upstream sequences from the translation start codons of ZmCP genes were retrieved and analyzed in the PlantCARE database (Lescot et al., 2002). As a result, the promoters of ZmCP genes contained most elements or sites responsive to environmental factors (e.g., light, temperature, and moisture). The existence of defense and stress responsive elements and MYB binding sites suggested that ZmCPs genes were involved in drought and anaerobic responsive elements and may play an important role in counteracting abiotic stress (Figure 3). The hormone-responsive elements, ~40% of total elements, including abscisic acid (ABA), salicylic acid, gibberellin, auxin, and methyl jasmonate (MeJA) -responsive elements, were found within the ZmCP gene promoters.
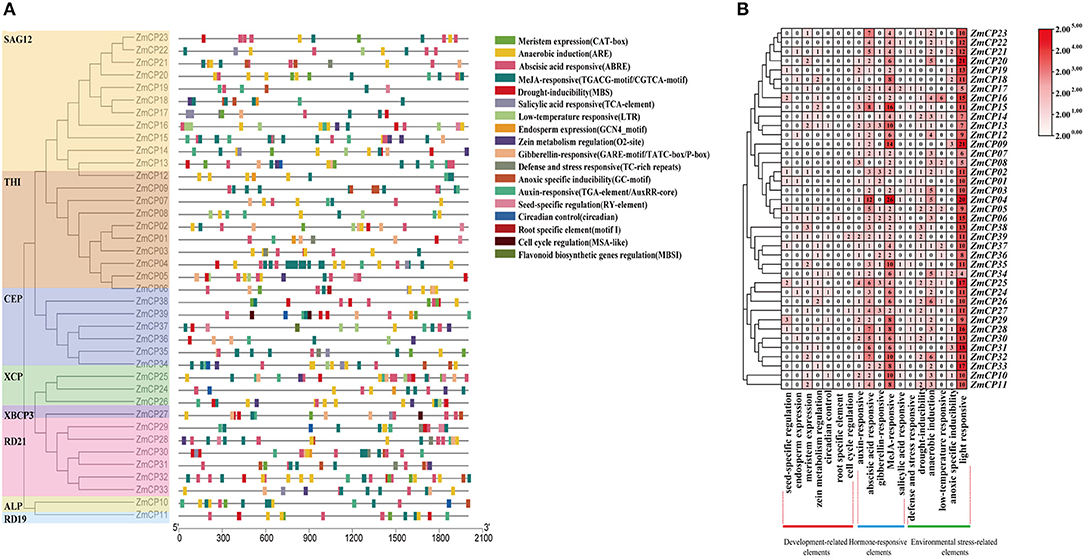
Figure 3. Promoter cis-elements analysis of ZmCP genes. (A) The position of elements in ZmCP promoters. (B) The number of occurrences cis-elements in the promoter region of ZmCP genes.
Moreover, 73 elements related to plant growth and development, namely, 23 meristem expression elements, 13 seed-specific regulation elements, 8 endosperm expression elements, 6 circadian control elements, 19 zein metabolism regulation elements, 3 cell cycle regulation elements, and 1 root specific element, were identified within the ZmCP gene promoters, indicating that some ZmCP genes may regulate plant growth and development.
The locations of ZmCP genes were mapped according to the data of gene locus, and they were unevenly distributed on chromosomes (Figure 4). Specifically, chromosome 2 contained more ZmCP genes (9), chromosomes 4, 7, and 8 each contained three, and chromosomes 1, 5, 6, 9, and 10 contained two, four, five, four, and five, respectively. Only ZmCP35 was localized on chromosome 3. The target gene ZmCP03 was located on chromosome 1. These indicated that genetic variations existed during the evolutionary process of maize. Similarly, in rice, 33 OsCPs were located on 10 chromosomes of 12 chromosomes in an unbalanced pattern (Wang et al., 2018). In cotton, GhCP genes were unequally distributed on 21 chromosomes (Zhang et al., 2019). Obviously, the number of CP genes differed, and their chromosomal distribution was unequal in the examined plant species.
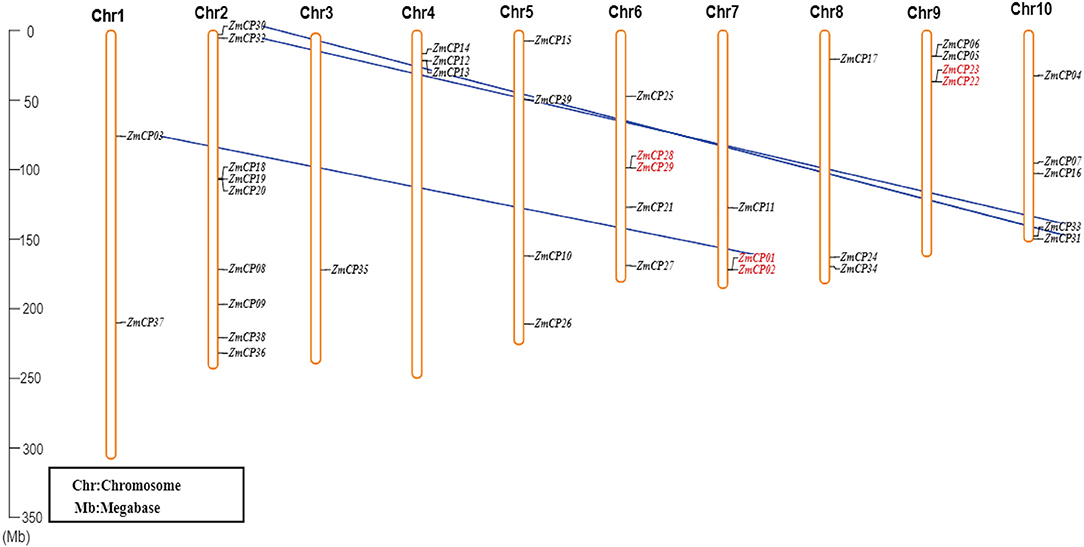
Figure 4. The chromosomal distribution of ZmCP genes. The left scale represents the length of the chromosomes. The chromosomal segmental duplications between two ZmCP genes are displayed by blue lines, and the two ZmCP genes that occurred as tandem duplication events are indicated in red.
Duplication is a major driving force for gene expansion during evolution. There are five types of gene duplication in evolution, which are singleton, dispersed, tandem, proximal, and segmental duplication (Zheng et al., 2020). The collinearity of ZmCP genes was examined in maize (Figure 4). As a result, three pairs (ZmCP01/ZmCP02, ZmCP22/ZmCP23, and ZmCP28/ZmCP29) from ZmCP genes were discovered in tandem repeats. Moreover, the three pairs (ZmCP01/ZmCP03, ZmCP30/ZmCP31, and ZmCP32/ZmCP33) probably resulted from the duplicated chromosomal segments, suggesting that ZmCPs were expanded by tandem and segmental duplication.
There are three types of the preservation of duplicated genes functional divergence during long-term evolution that are neo-functionalization (gaining new functions), sub-functionalization (partition of actual functions), or non-functionalization (loss of actual functions) (Prince and Pickett, 2002). The Ka and Ks were calculated for six gene pairs to explore whether Darwinian positive selection influenced the divergence of ZmCP members after the duplication. The Ka/Ks ratio indicates the gene divergence under selection pressure (Hurst, 2002). In maize, all duplicated gene pairs with Ka/Ks < 1 indicated negative selection (Table 2).
To analyze the expression patterns of the ZmCPs, the heatmap of their expression patterns in different maize tissues, including the shoot tip of the vegetative 5th leaf stage (V5), anther at silking stage (R1), primary root at vegetative emergence (VE) and vegetative first leaf stage (V1), pooled leaves from vegetative first leaf stage (V1), 11th leaf and 13th leaf at vegetative 9th leaf stage (V9), immature cob at vegetative 18th leaf stage (V18), silk at silking stage (R1), and seed from 4 DAP (days after pollination) (Figure 5), were built. The transcriptome data retrieved from MaizeGDB indicated the differential expression patterns of ZmCP genes during maize growth and development.
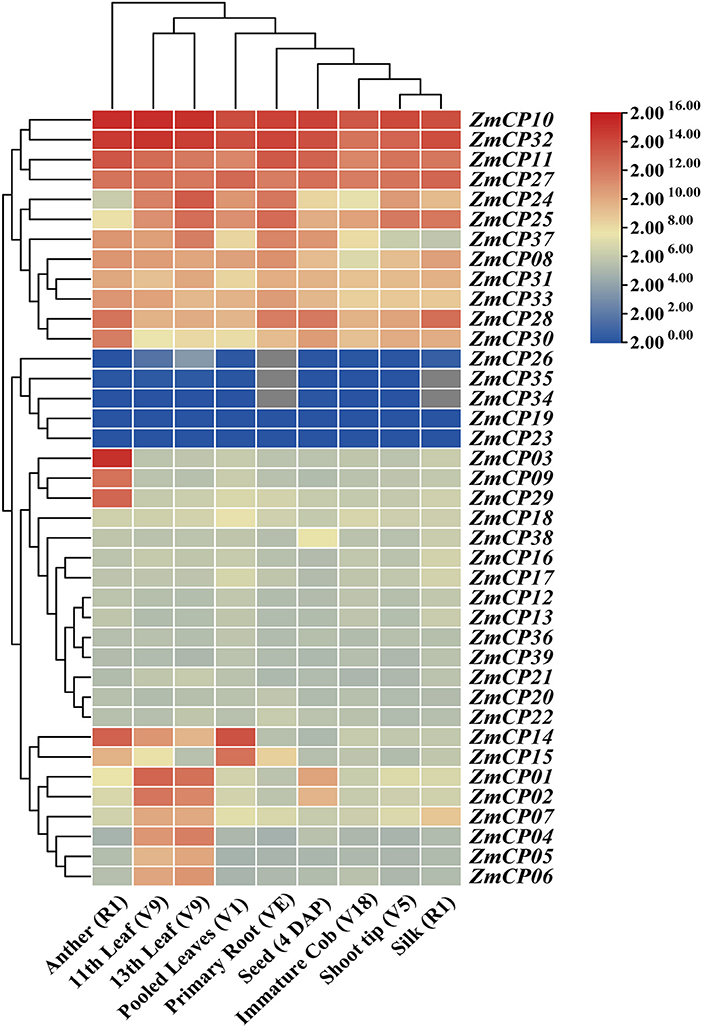
Figure 5. Heatmap of ZmCP genes expression levels in maize. R1: silking stage; VE: vegetative emergence; V1, vegetative 1st leaf stage; V5, vegetative 5th leaf stage; V9, vegetative 9th leaf stage; V18, vegetative 18th leaf stage; DAP, days after pollination.
Particularly, ZmCP10, ZmCP11, ZmCP27, and ZmCP32 displayed high expression levels in all examined tissues. ZmCP11 and ZmCP27 had relatively high expression levels in these tissues, especially ZmCP11 in anthers, primary roots (VE and V1), and ZmCP27 in leaves (V1). ZmCP14 and ZmCP15 represented relatively high expression levels in young leaves but low expression levels in elder leaves. However, ZmCP01, ZmCP02, and ZmCP04-07 were highly expressed in old leaves but lowly in young leaves. ZmCP34 and ZmCP35 had no expressional information in the primary root and silks and showed very low expression levels in other tissues. ZmCP26 had no expressional information in the primary root and other organs.
ZmCP03, the gene of interest, showed a tissue-specific expression pattern with unusually high expression levels in anthers but low expression levels in other tissues, suggesting it may be related with anther development. All members of the subfamily RD21 had high expression levels in anthers, as was the case in cotton (Zhang et al., 2019).
As a comparison with the transcriptome data retrieved from MaizeGDB, the expression levels of ZmCP03 in the root, stem, leaves, and pollen of maize were further analyzed using RT-qPCR. ZmCP03 had a high expression level in pollen grains but a low level in other tissues, consistent with the results of the transcriptome data (Figure 6).
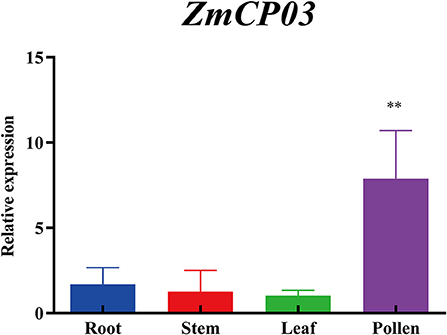
Figure 6. Quantitative reverse transcription PCR (RT-qPCR) analysis of ZmCP03 in different organs of maize. Data presented are relative expression (mean ± SD) from the three biological replicates after normalization based on ZmUBI expression levels. Student's t-test in the GraphPad Prism 8.0 software was used to compare significant differences (indicated by ** at p < 0.01) in expression levels among different tissues.
To examine the accumulation and localization of ZmCP03 in pollen grains, a monoclonal antibody (anti-ZmCP03) was prepared using a synthetic 15 peptide to immunize mice. Sodium dodecyl sulfate polyacrylamide gel electrophoresis and immunoblot analyses revealed the abundant accumulation of ZmCP03 in pollen grains, especially in the pollen coat component (Figure 7). In addition, fluorescence microscopy showed distinct bright spots detected on the pollen surface of intact pollen grains (Figures 8A–C). The presence of ZmCP03 was further examined by fluorescence microscopy using semi-thin sections of pollen grains. Obviously, ZmCP03 signals were most present on the pollen surface (coat) and in small quantities inside pollen grains (the cytoplasm) (Figures 8D,E).
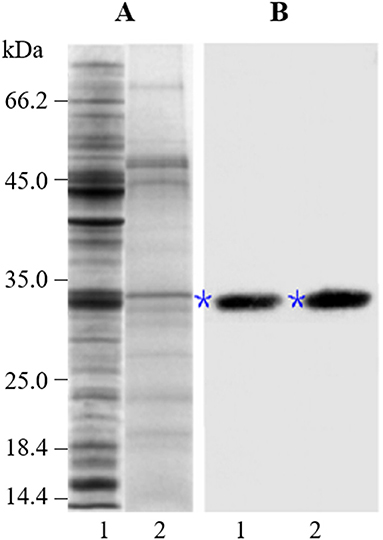
Figure 7. Detection of ZmCP03 in pollen grains of maize. Proteins were separated on an equal amount of pollen fresh weight. (A) Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), 12.5% resolving gel; (B) Immunoblotting, detection with anti-ZmCP03 (1: 5000 dilution). Lanes 1, pollen grains; 2, pollen coats. Asterisks indicate ZmCP03.
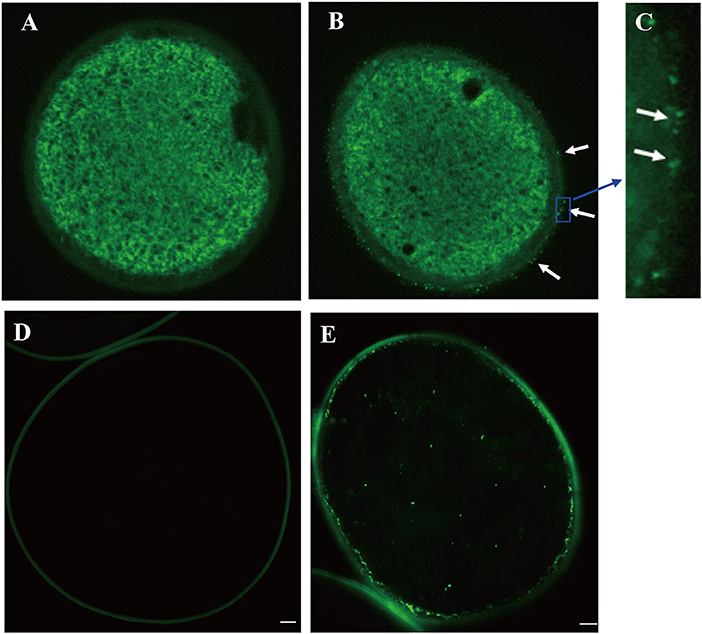
Figure 8. Localization of ZmCP03 in maize pollen by immunofluorescence microscopy. (A) Control without primary antibody, no signal is detected; (B) in the presence of primary antibody, several fluorescent dots can be seen on the surface of pollen grains (arrows); (C) a magnification picture of (B) highlighting some dots on the grain surface; (D) sections of maize pollen without a primary antibody; (E) sections of maize pollen with a primary antibody. Maize pollen sections were probed with (B,D) anti-CP (1:200 dilution, 4°C, overnight) or (A,D) negative control, and then incubated with fluorescence anti-rabbit lgG (1:100, 37°C, 45 min). Scale bar = 5 μm.
Finally, immunogold electron microscopy was carried out to examine the subcellular localization of maize pollen ZmCP03. Ultrathin sections of maize pollen grains were probed with the anti-ZmCP03. The results clearly demonstrated that ZmCP03 was located on the pollen surface (coat), but particularly in the inner layer of the wall (arrows). Furthermore, ZmCP03 signaling was also in the pollen cytoplasm (Figures 9B–D).
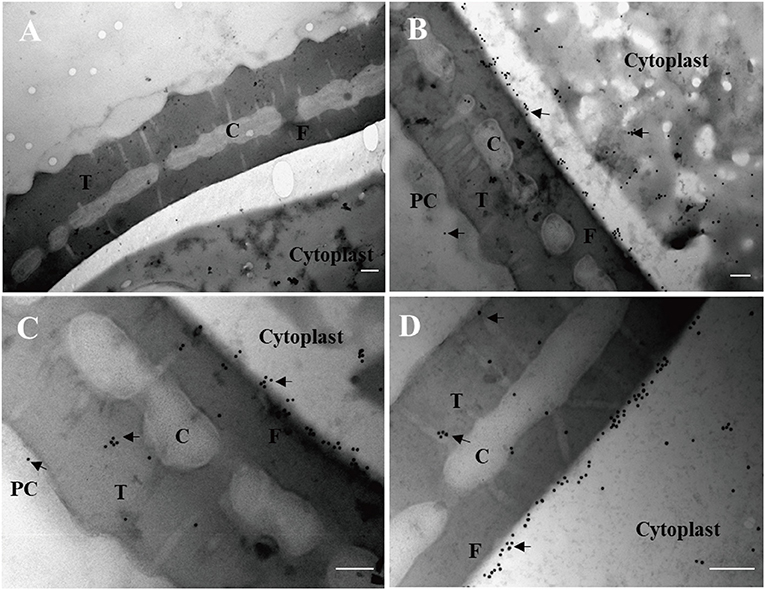
Figure 9. Subcellular localization of ZmCP03 in maize pollen by immunogold electron microscopy. (A) Negative control, preimmunized rabbit serum, and then incubated with 15-nm gold-conjugated anti-rabbit lgG (1:20, 45 min); (B–D) sections of maize pollen grains were probed with anti-CP (1:100 dilution, 4°C, overnight); arrows indicate the presence of ZmCP03; PC, pollen coat; T, tectum layer, C, columella; F, foot layer; The exine consists of the tectum layer, foot layer, and columella. Scale bar = 200 nm.
Several reports investigated the evolutionary relationship of papain-like cysteine proteases (PLCP) family in Arabidopsis (Richau et al., 2012), rubber (Zou et al., 2017), papaya (Liu et al., 2018), rice (Wang et al., 2018), castor bean and physic nut (Zou et al., 2018), and cotton (Zhang et al., 2019), and soybean (Yuan et al., 2020). In maize, a report analyzed the evolutionary relationship of CP genes only through building phylogenetic tree (Richau et al., 2012). In the present study, 39 CP genes in maize were analyzed using several bioinformatic tools, especially comparing CP in pollen and other tissues, to provide insights on the potential function of ZmCP03 in pollen development and germination.
The results here showed that the number of subfamilies of papain-like cysteine protease differed greatly in maize. SAG12 (12 members) and THI (9 members) were the two largest subfamilies, like in rice (Wang et al., 2018). However, the two subfamilies owned much more members in Ficus carica (Zhai et al., 2021) and Arabidopsis (Richau et al., 2012), implying that SAG12 may be generated earlier than the formation of individual species. Usually, SAG12 is significantly increased during leaf senescence, thus it is most widely used as a senescence-associated reference gene (James et al., 2018). The CTB subfamily has a negative role in regulating cryo-injury tolerance and cell viability via mediating the PCD event in plant cryopreservation in Arabidopsis and Agapanthus praecox (Chen et al., 2021). However, no CTB subfamily was identified in maize in the present study, perhaps being lost during evolution.
The results here further showed that the family of papain-like cysteine protease is relatively conservative during the evolution events based on their motif, conserved domain, and gene structure (Figure 2). All 39 ZmCP proteins are composed of the peptidase C1 domain and inhibitor I29 domain. In the same subfamily, most members have similar exon-intron structures, suggesting affinity and the conservation of evolutionary relationships. Meanwhile, segmental duplications and tandem duplications may have the same effect on expanding this family in maize. Ka/Ks < 1 indicates that duplicated gene pairs in ZmCPs were all purified selection, according to the theory that a species is more likely keeping protein as it is (purified selection) rather than a mutation occurring that changes the protein (Hurst, 2002). Moreover, ZmCP genes maintain conserved and stable evolutionary selection, suggesting that their functions were stabilized in maize. However, the CP genes in rice displayed different evolutionary selection ways, especially five out of seven pairs of tandem duplications were positive selection, and segmental duplications belong to purified selection in rice (Wang et al., 2018).
Previously, ZmCP03 was found to be synthesized in the rough endoplasmic reticulum (ER) and processed during or after its transfer to the vacuole at stage 4 of anther development (Li et al., 2012). In Arabidopsis, AtTHI1 (AtCP51) participates in pollen exine formation and anther development, and the pollen of transgenic CP51-RNAi plants was aborted due to defective exine and early degrading tapetum (Yang et al., 2014). According to the phylogenetic trees here, ZmCP03 was in the same clade with OsCP12 and in the same group with GhTHI1, GhTHI2, and AtTHI1. As OsCP12 displayed an organ-special expression pattern with a high expression level in anthers (Wang et al., 2018). ZmCP03 was also highly expressed in anthers but low in other tissues (Figure 6). Moreover, it was verified that ZmCP03 was mainly found in the pollen coat and, to a lesser extent, in the cytoplasm (Figures 8, 9). Therefore, both the previous studies (Li et al., 2012) and the present results supported the speculation that ZmCP03 may be related to the PCD of the tapetum and play a role in pollen development and germination.
In addition, the upstream region of the ZmCP03 contained the MYB binding site involved in drought-inducibility and the abscisic acid (ABA) -responsive element (ABRE), which is recognized as a necessary element controlling ABA-regulated gene expression (Fujita et al., 2005). A previous report indicated that a C1 cysteine protease was highly induced by drought in soybean (Cilliers et al., 2017), suggesting that ZmCP03 may be involved in the response to drought stress.
In conclusion, we characterized in this study the CP genes family in maize at a genome scale using comprehensive bioinformatic tools and further compared with Arabidopsis, cotton, and rice. Particularly, maize CPs in pollen coat and other tissues were emphatically compared. The evolutionary relationship, gene structure and duplication, chromosomal distribution, sequence characteristics, and expression profiles of ZmCP genes suggest that ZmCP03 is relatively conservative and stable in the process of evolution. This study may provide referential evidence on the function of ZmCP03 in maize pollen development and germination.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
WW and GC conceived the research. YL, LN, CF, GC, and MZ performed the experiments. XW, FT, WW, and GC analyzed the data. YL, HL, and WW drafted the manuscript. LN, GC, and WW edited the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the National Natural Science Foundation of China (Grant 31771700).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.709534/full#supplementary-material
Supplementary Figure 1. The unrooted phylogenetic tree was generated based on the amino acid sequences of 39 ZmCPs using a maximum likelihood method with 1,000 bootstrap values.
Supplementary Dataset 1. The nucleotide and amino acid sequences of 39 ZmCPs.
Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G., de Castro, E., et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40, W597–W603. doi: 10.1093/nar/gks400
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Bateman, A., and Bennett, H. P. (2009). The granulin gene family: from cancer to dementia. Bioessays 31, 1245–1254. doi: 10.1002/bies.200900086
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative Toolkit developed for interactive analyses of big biological data. Mol. Plant. 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, G., Zhang, D., Pan, J., Yue, J., and Shen, X. (2021). Cathepsin B-like cysteine protease ApCathB negatively regulates cryo-injury tolerance in transgenic Arabidopsis and Agapanthus praecox. Plant Sci. 308:110928. doi: 10.1016/j.plantsci.2021.110928
Cilliers, M., Wyk, S. V., Heerden, P. V., Kunert, K. J., and Vorster, B. J. (2017). Identification and changes of the drought-induced cysteine protease transcriptome in soybean (glycine max) root nodules. Environ. Exp. Bot. 148, 59–69. doi: 10.1016/j.envexpbot.2017.12.005
Dai, S., Li, L., Chen, T., Chong, K., Xue, Y., and Wang, T. (2006). Proteomic analyses of Oryza sativa mature pollen reveal novel proteins associated with pollen germination and tube growth. Proteomics 6, 2504–2529. doi: 10.1002/pmic.200401351
Edlund, A. F., Swanson, R., and Preuss, D. (2004). Pollen and stigma structure and function: the role of diversity in pollination. Plant Cell 16, S84–S97. doi: 10.1105/tpc.015800
Fujita, Y., Fujita, M., Satoh, R., Maruyama, K., Parvez, M. M., Seki, M., et al. (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17, 3470–3488. doi: 10.1105/tpc.105.035659
Gong, F., Wu, X., and Wang, W. (2015). Diversity and function of maize pollen coat proteins: from biochemistry to proteomics. Front. Plant Sci. 6:199. doi: 10.3389/fpls.2015.00199
Groves, M. R., Taylor, M. A., Scott, M., Cummings, N. J., Pickersgill, R. W., and Jenkins, J. A. (1996). The prosequence of procaricain forms an alpha-helical domain that prevents access to the substrate-binding cleft. Structure 4, 1193–1203. doi: 10.1016/S0969-2126(96)00127-X
Higo, K., Ugawa, Y., Iwamoto, M., and Higo, H. (1998). PLACE: a database of plant cis-acting regulatory DNA elements. Nucleic Acids Res. 26, 358–359. doi: 10.1093/nar/26.1.358
Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27, 297–300. doi: 10.1093/nar/27.1.297
Hoopes, G. M., Hamilton, J. P., Wood, J. C., Esteban, E., Pasha, A., Vaillancourt, B., et al. (2019). An updated gene atlas for maize reveals organ-specific and stress-induced genes. Plant J. 97, 1154–1167. doi: 10.1111/tpj.14184
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Hurst, L. D. (2002). The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 18:486. doi: 10.1016/S0168-9525(02)02722-1
James, M., Poret, M., Masclaux-Daubresse, C., Marmagne, A., Coquet, L., Jouenne, T., et al. (2018). SAG12, a major cysteine protease involved in nitrogen allocation during senescence for seed production in Arabidopsis thaliana. Plant Cell Physiol. 59, 2052–2063. doi: 10.1093/pcp/pcy125
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, Y., Suen, D. F., Huang, C. Y., Kung, S. Y., and Huang, A. H. (2012). The maize tapetum employs diverse mechanisms to synthesize and store proteins and flavonoids and transfer them to the pollen surface. Plant Physiol. 158, 1548–1561. doi: 10.1104/pp.111.189241
Liu, J., Sharma, A., Niewiara, M. J., Singh, R., Ming, R., and Yu, Q. (2018). Papain-like cysteine proteases in Carica papaya: lineage-specific gene duplication and expansion. BMC Genomics 19:26. doi: 10.1186/s12864-017-4394-y
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, H., Chandrasekar, B., Oeljeklaus, J., Misas-Villamil, J. C., Wang, Z., Shindo, T., et al. (2015). Subfamily-specific fluorescent probes for cysteine proteases display dynamic protease activities during seed germination. Plant Physiol. 168, 1462–1475. doi: 10.1104/pp.114.254466
Marchler-Bauer, A., Bo, Y., Han, L., He, J., Lanczycki, C. J., Lu, S., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203. doi: 10.1093/nar/gkw1129
Marshall, E., Costa, L. M., and Gutierrez-Marcos, J. (2011). Cysteine-rich peptides (CRPs) mediate diverse aspects of cell-cell communication in plant reproduction and development. J. Exp. Bot. 62, 1677–1686. doi: 10.1093/jxb/err002
Murphy, D. J. (2006). The extracellular pollen coat in members of the Brassicaceae: composition, biosynthesis, and functions in pollination. Protoplasma 228, 31–39. doi: 10.1007/s00709-006-0163-5
Niño, M. C., Kang, K. K., and Cho, Y. G. (2020). Genome-wide transcriptional response of papain-like cysteine protease-mediated resistance against Xanthomonas oryzaepv. oryzae in rice. Plant Cell Rep. 39, 457–472. doi: 10.1007/s00299-019-02502-1
Niu, L., Chu, H. D., Tran, C. D., Nguyen, K. H., Pham, H. X., Le, D. T., et al. (2020). The GATA gene family in chickpea: structure analysis and transcriptional responses to abscisic acid and dehydration treatments revealed potential genes involved in drought adaptation. J. Plant Growth Regul. 39, 1647–1660. doi: 10.1007/s00344-020-10201-5
Pacini, E., and Hesse, M. (2004). Cytophysiology of pollen presentation and dispersal. Flora Morphol. Distribut. Funct. Ecol. Plants 199, 273–285. doi: 10.1078/0367-2530-00156
Pechan, T., Ye, L., Chang, Y., Mitra, A., Lin, L., Davis, F. M., et al. (2000). A unique 33-kD cysteine proteinase accumulates in response to larval feeding in maize genotypes resistant to fall armyworm and other Lepidoptera. Plant Cell 12, 1031–1040. doi: 10.1105/tpc.12.7.1031
Prince, V. E., and Pickett, F. B. (2002). Splitting pairs: the diverging fates of duplicated genes. Nat. Rev. Genet. 3, 827–837. doi: 10.1038/nrg928
Qanmber, G., Ali, F., Lu, L., Mo, H., and Yang, Z. (2019a). Identification of histone h3 (hh3) genes in Gossypium hirsutum revealed diverse expression during ovule development and stress responses. Genes 10:355. doi: 10.3390/genes10050355
Qanmber, G., Liu, J., Yu, D., Liu, Z., Lu, L., Mo, H., et al. (2019b). Genome-wide identification and characterization of the PERK gene family in Gossypium hirsutum reveals gene duplication and functional divergence. Int. J. Mol. Sci. 20:1750. doi: 10.3390/ijms20071750
Quilichini, T. D., Grienenberger, E., and Douglas, C. J. (2015). The biosynthesis, composition and assembly of the outer pollen wall: a tough case to crack. Phytochemistry 113, 170–182. doi: 10.1016/j.phytochem.2014.05.002
Rawlings, N. D., Barrett, A. J., and Bateman, A. (2010). MEROPS: the peptidase database. Nucleic Acids Res. 38, D227–D233. doi: 10.1093/nar/gkp971
Rejón, J. D., Delalande, F., Schaeffer-Reiss, C., Alché, J. D., Rodríguez-García, M. I., Van Dorsselaer, A., et al. (2016). The pollen coat proteome: at the cutting edge of plant reproduction. Proteomes 4:5. doi: 10.3390/proteomes4010005
Richau, K. H., Kaschani, F., Verdoes, M., Pansuriya, T. C., Niessen, S., Stüber, K., et al. (2012). Subclassification and biochemical analysis of plant papain-like cysteine proteases displays subfamily-specific characteristics. Plant Physiol. 158, 1583–1599. doi: 10.1104/pp.112.194001
Schopfer, C. R., Nasrallah, M. E., and Nasrallah, J. B. (1999). The male determinant of self-incompatibility in Brassica. Science 286, 1697–1700. doi: 10.1126/science.286.5445.1697
Shukla, P., Gautam, R., Singh, N. K., Ahmed, I., and Kirti, P. B. (2019). A proteomic study of cysteine protease induced cell death in anthers of male sterile tobacco transgenic plants. Physiol. Mol. Biol. Plants 25, 1073–1082. doi: 10.1007/s12298-019-00642-y
Suen, D. F., and Huang, A. H. (2007). Maize pollen coat xylanase facilitates pollen tube penetration into silk during sexual reproduction. J. Biol. Chem. 282, 625–636. doi: 10.1074/jbc.M608567200
Suen, D. F., Wu, S. S., Chang, H. C., Dhugga, K. S., and Huang, A. H. (2003). Cell wall reactive proteins in the coat and wall of maize pollen: potential role in pollen tube growth on the stigma and through the style. J. Biol. Chem. 278, 43672–43681. doi: 10.1074/jbc.M307843200
Takayama, S., Shiba, H., Iwano, M., Shimosato, H., Che, F. S., Kai, N., et al. (2000). The pollen determinant of self-incompatibility in Brassica campestris. Proc. Natl. Acad. Sci. U.S.A. 97, 1920–1925. doi: 10.1073/pnas.040556397
Updegraff, E. P., Zhao, F., and Preuss, D. (2009). The extracellular lipase EXL4 is required for efficient hydration of Arabidopsis pollen. Sex. Plant Reprod. 22, 197–204. doi: 10.1007/s00497-009-0104-5
Valdivia, E. R., Stephenson, A. G., Durachko, D. M., and Cosgrove, D. (2009). Class B beta-expansins are needed for pollen separation and stigma penetration. Sex Plant Reprod. 22, 141–152. doi: 10.1007/s00497-009-0099-y
Valdivia, E. R., Wu, Y., Li, L. C., Cosgrove, D. J., and Stephenson, A. G. (2007). A group-1 grass pollen allergen influences the outcome of pollen competition in maize. PLoS ONE 2:e154. doi: 10.1371/journal.pone.0000154
Wang, L., Clarke, L. A., Eason, R. J., Parker, C. C., Qi, B., Scott, R. J., et al. (2017). PCP-B class pollen coat proteins are key regulators of the hydration checkpoint in Arabidopsis thaliana pollen-stigma interactions. New Phytol. 213, 764–777. doi: 10.1111/nph.14162
Wang, W., Milanesi, C., Faleri, C., and Cresti, M. (2006). Localization of group-1 allergen zea m 1 in the coat and wall of maize pollen. Acta Histochem. 108, 395–400. doi: 10.1016/j.acthis.2006.06.003
Wang, W., Zhou, X. M., Xiong, H. X., Mao, W. Y., Zhao, P., and Sun, M. X. (2018). Papain-like and legumain-like proteases in rice: genome-wide identification, comprehensive gene feature characterization and expression analysis. BMC Plant Biol. 18:87. doi: 10.1186/s12870-018-1298-1
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Wu, X., Cai, G., Gong, F., An, S., Cresti, M., and Wang, W. (2015). Proteome profiling of maize pollen coats reveals novel protein components. Plant Mol. Biol. Rep. 33, 975–986. doi: 10.1007/s11105-014-0812-3
Yang, Y., Dong, C., Yu, J., Shi, L., Tong, C., Li, Z., et al. (2014). Cysteine Protease 51 (CP51), an anther-specific cysteine protease gene, is essential for pollen exine formation in Arabidopsis. Plant Cell Tiss. Organ. Cult. 119, 383–397. doi: 10.1007/s11240-014-0542-0
Yang, Z., Qanmber, G., Wang, Z., Yang, Z., and Li, F. (2020). Gossypium genomics: trends, scope, and utilization for cotton improvement. Trends Plant Sci. 25, 488–500. doi: 10.1016/j.tplants.2019.12.011
Yennawar, N. H., Li, L. C., Dudzinski, D. M., Tabuchi, A., and Cosgrove, D. J. (2006). Crystal structure and activities of EXPB1 (Zea m 1), a beta-expansin and group-1 pollen allergen from maize. Proc. Natl. Acad. Sci. U.S.A. 103, 14664–14671. doi: 10.1073/pnas.0605979103
Yuan, S., Ke, D., Li, R., Li, X., Wang, L., Chen, H., et al. (2020). Genome-wide survey of soybean papain-like cysteine proteases and their expression analysis in root nodule symbiosis. BMC Plant Biol. 20:517. doi: 10.1186/s12870-020-02725-5
Zhai, Y., Cui, Y., Song, M., Vainstein, A., Chen, S., and Ma, H. (2021). Papain-like cysteine protease gene family in Fig (Ficus carica L.): genome-wide analysis and expression patterns. Front. Plant Sci. 12:681801. doi: 10.3389/fpls.2021.681801
Zhang, D., Liu, D., Lv, X., Wang, Y., Xun, Z., Liu, Z., et al. (2014). The cysteine protease CEP1, a key executor involved in tapetal programmed cell death, regulates pollen development in Arabidopsis. Plant Cell 26, 2939–2961. doi: 10.1105/tpc.114.127282
Zhang, D., Shi, J., and Yang, X. (2016). Role of lipid metabolism in plant pollen exine development. SubcellBiochem 86, 315–337. doi: 10.1007/978-3-319-25979-6_13
Zhang, S., Xu, Z., Sun, H., Sun, L., Shaban, M., Yang, X., et al. (2019). Genome-wide identification of papain-like cysteine proteases in gossypiumhirsutum and functional characterization in response to verticillium dahliae. Front. Plant Sci. 10:134. doi: 10.3389/fpls.2019.00134
Zheng, L., Wu, H., Qanmber, G., Ali, F., Wang, L., Liu, Z., et al. (2020). Genome-wide study of the GATL gene family in Gossypium hirsutum L. reveals that GhGATL genes act on pectin synthesis to regulate plant growth and fiber elongation. Genes 11:64. doi: 10.3390/genes11010064
Zienkiewicz, K., Zienkiewicz, A., Rodríguez-García, M. I., and Castro, A. J. (2011). Characterization of a caleosin expressed during olive (Olea europaea L.) pollen ontogeny. BMC Plant Biol. 11:122. doi: 10.1186/1471-2229-11-122
Zou, Z., Huang, Q., Xie, G., and Yang, L. (2018). Genome-wide comparative analysis of papain-like cysteine protease family genes in castor bean and physic nut. Sci. Rep. 8:331. doi: 10.1038/s41598-017-18760-6
Keywords: cysteine protease, expression analysis, immunofluorescence microscopy, immunogold electron microscopy, gene duplication, maize pollen, phylogenetic analysis
Citation: Li Y, Niu L, Wu X, Faleri C, Tai F, Zhang M, Liu H, Wang W and Cai G (2021) Genome-Wide Identification and Comparison of Cysteine Proteases in the Pollen Coat and Other Tissues in Maize. Front. Plant Sci. 12:709534. doi: 10.3389/fpls.2021.709534
Received: 14 May 2021; Accepted: 24 August 2021;
Published: 23 September 2021.
Edited by:
Klára Kosová, Crop Research Institute (CRI), CzechiaReviewed by:
Ghulam Qanmber, Cotton Research Institute (CAAS), ChinaCopyright © 2021 Li, Niu, Wu, Faleri, Tai, Zhang, Liu, Wang and Cai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fuju Tai, dGFpZnVqdUBoZW5hdS5lZHUuY24=; Wei Wang, d2FuZ3dlaUBoZW5hdS5lZHUuY24=; Giampiero Cai, Z2lhbXBpZXJvLmNhaUB1bmlzaS5pdA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.