
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 06 July 2021
Sec. Plant Development and EvoDevo
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.709360
This article is part of the Research Topic Petal Development: From Cell Biology to EvoDevo View all 10 articles
Epigenetic modifications include histone modifications and DNA methylation; such modifications can induce heritable changes in gene expression by altering DNA accessibility and chromatin structure. A number of studies have demonstrated that epigenetic factors regulate plant developmental timing in response to environmental changes. However, we still have an incomplete picture of how epigenetic factors can regulate developmental events such as organogenesis. The small number of cell types and the relatively simple developmental progression required to form the Arabidopsis petal makes it a good model to investigate the molecular mechanisms driving plant organogenesis. In this minireview, we summarize recent studies demonstrating the epigenetic control of gene expression during various developmental transitions, and how such regulatory mechanisms can potentially act in petal growth and differentiation.
The formation of petals, like plant organogenesis in general, occurs via a period of cell division followed by post-mitotic cell expansion (Powell and Lenhard, 2012) and entails precise spatiotemporal control of gene expression. The Arabidopsis petal is a well- studied simple laminar organ with few cell types, making it an excellent model for understanding the mechanisms underlying plant organogenesis (Irish, 2008). A number of gene regulatory networks have been identified that act early in petal development to promote cell proliferation, and a largely distinct set of genes have been shown to participate in the later phases of cell expansion (Li et al., 2016). This transition is undoubtedly regulated by a wide variety of gene products, with a number of TEOSINTE BRANCHED1/CYCLOIDEA/PCF (TCP) transcription factors likely playing a prominent role in the temporal control of cell proliferation arrest (Czesnick and Lenhard, 2015). Despite the identification of genes and regulatory pathways regulating different phases of petal growth, the mechanisms that control this developmental shift are still largely unknown. We will discuss the possibility that epigenetic mechanisms underlie the regulation of this temporal shift, particularly in the maintenance of expression of key genes controlling early petal development.
“Epigenetic” refers to heritable changes in gene expression that are not due to changes in DNA sequence (Berger et al., 2009). In eukaryotes, DNA is packaged into chromatin, which is organized into nucleosomes that contain an octamer of histone proteins wrapped by 146 bp of DNA (Hollender and Liu, 2008). Alterations in the organization of chromatin, caused by post-translational modifications of histone proteins or DNA methylation, can affect the accessibility of chromatin to the transcriptional machinery, resulting in changes of gene expression (Sims and Reinberg, 2008). Such epigenetic changes can impact DNA replication, cell proliferation and gene transcription (Suganuma and Workman, 2008). In particular, histone modifications can alter chromatin structure directly by altering chromatin accessibility or by influencing the recruitment of effector proteins (Strahl and Allis, 2000; Karlic et al., 2010).
The amino termini of the core histones in nucleosomes are substrates of covalent modifications including acetylation, methylation, ubiquitylation, phosphorylation and SUMOylation (Roguev et al., 2001). These various modifications constitute a specific “histone code” to regulate gene expression by instructing the chromatin configuration to be either “open” or “closed” (Iñiguez-Lluhí, 2006; Lee et al., 2010). For example, histone acetylation allows the chromatin to relax and provides transcription factors and RNA polymerases access to the DNA, whereas SUMOylation appears to repress gene expression through compacting the chromatin (Iñiguez-Lluhí, 2006). By contrast, the effects of methylation and ubiquitylation depend on the residues being modified and their contexts. For example, trimethylation of lysine 27 of histone H3 (H3K27me3) catalyzed by PcG (Polycomb-group) proteins is a repressive histone modification mark, whereas methylation of lysine 4 of histone H3 (H3K4me) is an active histone modification in plants (Zhang et al., 2009).
Histone acetylation is widely studied and of particular importance to plant development, defense and adaptation (Hollender and Liu, 2008). Histone acetyltransferases (HATs) and histone deacetylases (HDACs) are enzymes required to catalyze histone acetylation and deacetylation, respectively. Many of the HATs and HDACs are components of large multisubunit complexes, which are recruited to gene promoters by DNA-bound proteins (Howe et al., 2001). For example, HDA19, a member of the RPD3/HDA1 family of HDACs, interacts with the TOPLESS (TPL) co-repressor complex and is recruited by transcription factors to specific sites on the DNA to repress gene transcription (Krogan et al., 2012; Wang et al., 2013; Oh et al., 2014). Characterization of HDAC mutants in Arabidopsis has indicated that the members of the RPD3/HDA1 family of HDACs play a vital role in regulating gene expression in various biological processes (Liu et al., 2014). For instance, HDA19 was shown to repress the transcription of AGAMOUS in the outer whorl floral organs (Krogan et al., 2012), and to repress the expression of CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) in controlling the circadian period (Wang et al., 2013); both processes relying on the interaction of HDA19 with the TPL co-repressor.
Plants can alter gene expression in response to environmental cues and such changes in gene expression can be maintained by epigenetic memory. Once the particular state of the cell is established, the epigenetic marks can act as codes to impart expression information from the mother cell to the daughter cell and even from generation to generation (D’Urso and Brickner, 2014). In Arabidopsis, cold exposure (vernalization) triggers epigenetic silencing of the floral repressor FLOWERING LOCUS C (FLC) and in turn makes the plants competent to flower (Angel et al., 2011). The silencing of FLC is achieved by the replacement of active histone modifications with repressive histone modifications across the FLC locus. Specifically, active histone modifications such as trimethylation of lysine 36 on histone H3 (H3K36me3) across the gene body are replaced by the repressive histone modification mark H3K27me3 (Berry and Dean, 2015; Tao et al., 2017). Upon returning to warmth, FLC repression is epigenetically maintained at the reproductive stage including in sperm and egg cells (Berry and Dean, 2015; Figure 1). Epigenetic marks that accumulate at the FLC locus during vernalization need to be reset to ensure proper development of the next generation (Feng et al., 2010; Moazed, 2011). The seed specific transcription factor LEAFY COTYLEDONI (LEC1) de novo activates gene expression of FLC in the pre-embryo stage by reversing the silenced chromatin inherited from gametes to an active state (Tao et al., 2017). Interestingly, the LEC1-induced active epigenetic memory on FLC can be transmitted to post-embryonic stages, even well after LEC1 expression dissipates (Tao et al., 2017; Figure 1). These observations indicate that the epigenetic memory of an initial transcriptional state can be maintained for some time during plant development.
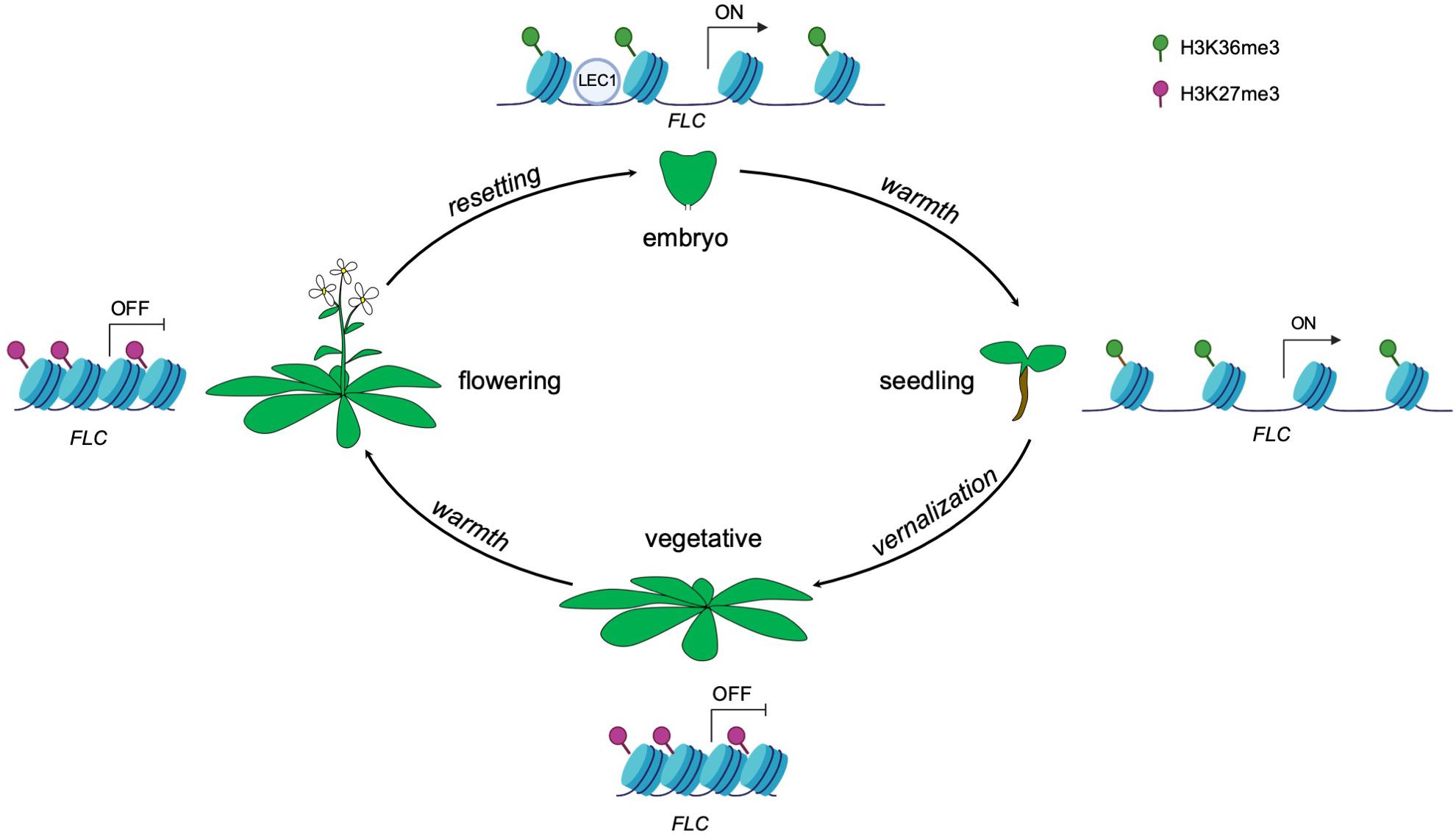
Figure 1. Epigenetic control of FLC expression by the seed-specific LEC1 protein throughout the Arabidopsis life cycle. Vernalization induces the deposition of the repressive histone modification mark H3K27me3, resulting in a silenced state of FLC. The repressive state is maintained even upon returning to warmth. The transcriptional status of FLC is reset in each generation during embryogenesis, resulting in the activation of FLC. The seed specific transcription factor LEC1 initially activates FLC expression by establishment of active histone modification H3K36me3 in early embryogenesis and this active memory is maintained and passed on to post-embryonic stages (seedlings).
Epigenetic modifiers also regulate the timing of floral meristem development. In floral meristems, the balance between the rates of stem cell proliferation and differentiation ensures a specific size and number of floral organs (sepals, petals, stamens and carpels) (Sun et al., 2009). In Arabidopsis, stem cell identity is maintained by WUSCHEL (WUS), whereas the termination of stem cell identity is achieved by zinc-finger protein KNUCKLES (KNU) induced repression of WUS (Sun et al., 2009). KNU is transcriptionally activated by the floral homeotic protein AGAMOUS (AG) (Figure 2A). In turn, the timing of the initiation of AG expression depends on the upregulation of the LEAFY transcription factor, which is itself upregulated in response to environmental signals that induce a feedforward loop to maintain the floral transition (Parcy et al., 1998; Busch et al., 1999; Jaeger et al., 2013). The timing of this activation is key in balancing stem cell proliferation and differentiation during floral organogenesis. The action of AG in inducing KNU expression requires about 2 days during which the repressive histone modification mark H3K27me3 across the KNU locus is removed (Sun et al., 2009). Furthermore, AG binding to the KNU promoter displaces PcG proteins from the locus and lead to cell division-dependent induction of KNU expression and further repression of WUS (Sun et al., 2014; Figure 2A). In sum, environmental signals induce a cascade of molecular events that culminate in the precisely orchestrated temporal transition from indeterminate to determinate growth.
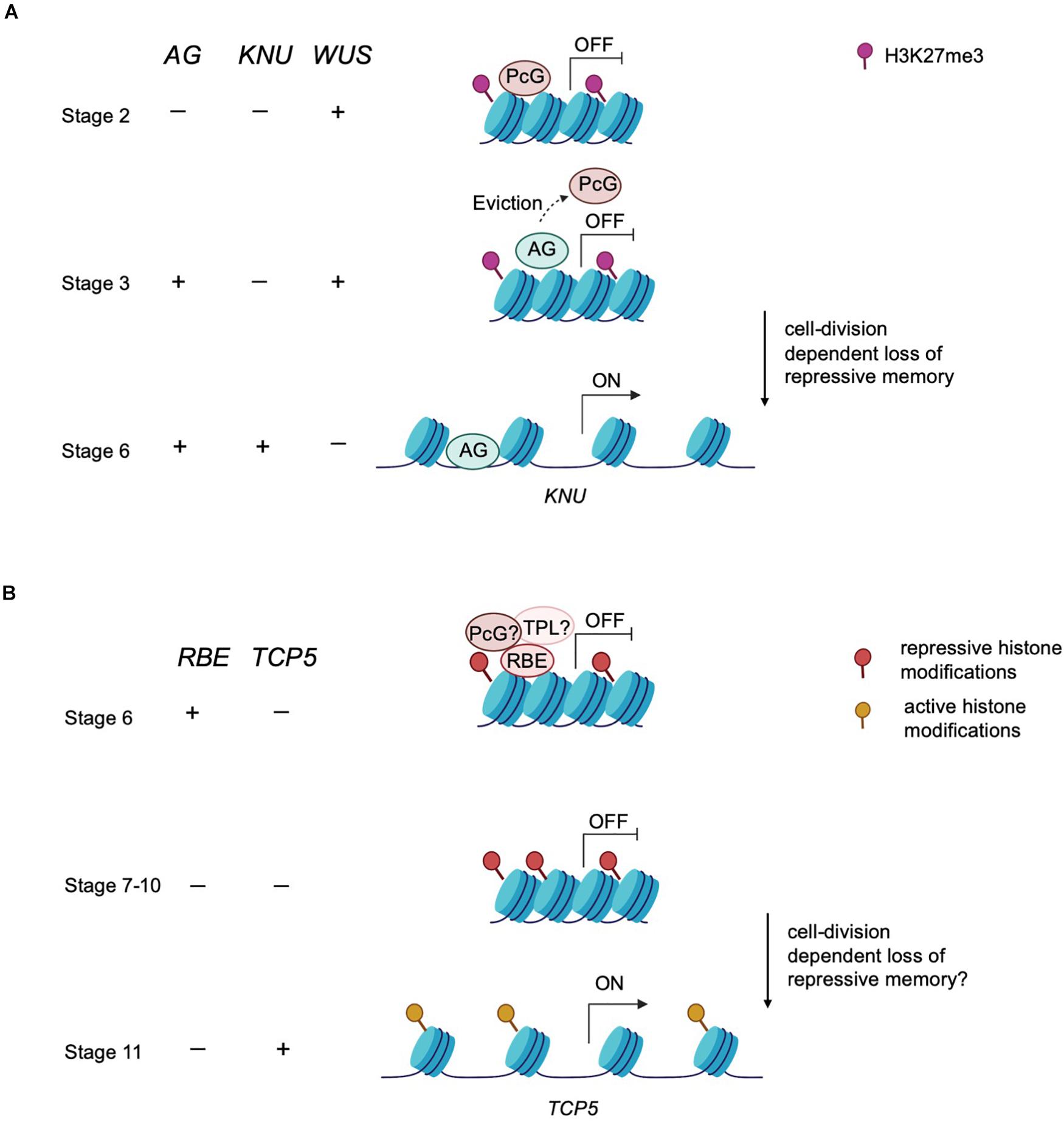
Figure 2. An epigenetic timer regulates floral development. (A) Epigenetic control of meristem determinacy. Prior to and during stage 2, repressive H3K27me3 modification catalyzed by the PcG protein is deposited across the KNU locus to silence KNU expression. At these early stages, WUS is expressed and maintains floral meristem stem cell proliferation. At stage 3, AG binds to the KNU promoter, evicting the PcG protein from KNU. The eviction of PcG protein induces cell division-dependent loss of H3K27me3 on KNU, which takes about 2 days. By stage 6, KNU is expressed and acts to repress WUS transcription and stem cell activity is then terminated. (B) Postulated model of epigenetic control of petal development. RBE confers repression on TCP5 to maintain cell proliferation at early stages of petal development. RBE could repress TCP5 transcription by recruiting epigenetic modifiers such as TPL or PcG proteins to establish repressive histone modifications across the TCP5 locus. This repressive memory is proposed to be maintained until about stage 10. By stage 11, TCP5 transcription is initiated, which is presumed to result from accrual of active histone modifications.
After petal primordia initiate, there is a period of cell division followed by a transition to cell expansion and cell differentiation to develop to mature petals (Krizek and Fletcher, 2005). The RABBIT EARS (RBE) gene encodes a C2H2 zinc finger transcription factor containing an EAR (ERF-associated amphiphilic repression) domain and regulates petal development in Arabidopsis. Loss of function rbe mutants exhibit underdeveloped petals and absence of petal initiation (Takeda et al., 2011). These strong petal defects make rbe an excellent genetic tool to disclose the molecular mechanisms underlying petal development. The TCP5 gene is a major downstream target of RBE as tcp5-1 largely rescues the petal defects of rbe-1 and RBE directly binds to the TCP5 regulatory region to repress its transcription (Huang and Irish, 2015). RBE transcripts are localized in the presumptive petal primordia from the stage 3 to stage 6 and act to promote cell division and establish enough cells with petal identity. This specific expression pattern of RBE implies that the alleviation of TCP5 repression by RBE commences at stage 6 (Huang and Irish, 2015). However, TCP5 transcription is only detectable from stage 11 on during petal development, which suggests a delay of approximately 6 days between alleviation of RBE-mediated repression and activation of TCP5 expression.
It is possible that RBE represses TCP5 by inducing chromatin-mediated silencing at that locus. The delay of transcriptional activation of TCP5 can be explained by the gradual elimination of this chromatin-mediated silencing, which may accompany the transition from cell division to cell expansion during petal development. It has been demonstrated that EAR motif containing proteins such as RBE recruit the TPL-HDA19 co-repressor complex to regulate gene transcription (Kagale and Rozwadowski, 2011). For instance, the EAR-motif containing APETALA2 (AP2) gene product recruits both TPL and HDA19 to repress the expression of multiple floral organ identity genes (Krogan et al., 2012). Additionally, EAR motif containing proteins can also interact with PcG proteins. SUPERMAN (SUP), encoding a similar C2H2 zinc finger protein to RBE, can interact with CURLY LEAF (CLF), a PRC2 component, to regulate floral whorl boundaries by controlling the expression of auxin biosynthetic genes (Xu et al., 2018). Furthermore, recent reports indicate that the EAR motif could act as a docking point for the TPL-HDAC and PRC2 complexes which in turn can result in chromatin remodeling at a target locus by increasing H3K27me3 and decreasing H3ac levels (Baile et al., 2020; Pelayo et al., 2021). Thus, EAR motif containing transcription factors might employ multiple layers of epigenetic modifications to maintain long-term repression.
Based on these observations, it is likely that RBE physically interacts with the TOPLESS-HDA19 complex and/or PcG proteins to temporally regulate the expression of TCP5 during petal development. Furthermore, TOPLESS and HDA19 have been shown to act in maintaining the balance of cell division and cell differentiation. For instance, the product of the WUSCHEL HOMEOBOX 5 (WOX5) gene maintains the root meristem niche and prevents cell type differentiation by recruiting TOPLESS and HDA19 to repress CYCLING DOF FACTOR 4 (CDF4) (Pi et al., 2015). Taken together, these results suggest RBE might orchestrate petal cell division and cell expansion by recruiting chromatin modifiers such as TPL or PcG proteins to temporally regulate TCP5 transcription (Figure 2B). Given the role of RBE in orchestrating many aspects of petal development, this presumptive epigenetic timing mechanism could potentially function to regulate the temporal control of expression of many downstream target genes involved in petal differentiation.
To our knowledge, epigenetic control mechanisms that regulate aspects of plant development are triggered in response to external cues such as daylength or temperature. We propose here that developmental timing can also occur through a gating mechanism that depends on protein half-life. In other words, the decay of RBE protein over time could function as a timer to alter the epigenetic status of target genes such as TCP5, and potentially other target genes as well. It is also possible that other petal regulators containing the EAR motif, such as JAGGED (JAG), might also function by recruiting epigenetic factors to confer temporal control of petal organogenesis (Ohno, 2004; Schiessl et al., 2014). In summary, the temporal control of petal development, and more broadly organ development, may rely in part on the stochastic process of alleviating repressive epigenetic marks over time to control the transition from cell division to post-mitotic cell expansion.
RH, TH, and VFI were contributed to the writing of this review. All authors contributed to the article and approved the submitted version.
Our work on petal organogenesis has been supported by NSF grant IOS-1354389 to VFI, and awards from the Guangdong Basic and Applied Basic Research Foundation (2021A1515011035) and Guangdong Special Support Program for Young Talents in Innovation Research of Science and Technology (2019TQ05N940) to TH.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank members of the Irish laboratory for discussions and comments during the course of writing this manuscript.
Angel, A., Song, J., Dean, C., and Howard, M. (2011). A Polycomb-based switch underlying quantitative epigenetic memory. Nature 476, 105–108. doi: 10.1038/nature10241
Baile, F., Merini, W., Hidalgo, I., and Calonje, M. (2020). Dissection of PRC1 and PRC2 recruitment in Arabidopsis connects EAR repressome to PRC2 anchoring. bioRxiv [Preprint]. doi: 10.1101/2020.08.28.271999
Berger, S. L., Kouzarides, T., Shiekhattar, R., and Shilatifard, A. (2009). An operational definition of epigenetics. Genes Dev. 23, 781–783. doi: 10.1101/gad.1787609
Berry, S., and Dean, C. (2015). Environmental perception and epigenetic memory: mechanistic insight through FLC. Plant J. 83, 133–148. doi: 10.1111/tpj.12869
Busch, M. A., Bomblies, K., and Weigel, D. (1999). Activation of a floral homeotic gene in Arabidopsis. Science 285, 585–587. doi: 10.1126/science.285.5427.585
Czesnick, H., and Lenhard, M. (2015). Size control in plants–lessons from leaves and flowers. Cold Spring Harb. Perspect. Biol. 7:a019190. doi: 10.1101/cshperspect.a019190
D’Urso, A., and Brickner, J. H. (2014). Mechanisms of epigenetic memory. Trends Genet. 30, 230–236. doi: 10.1016/j.tig.2014.04.004
Feng, S., Jacobsen, S. E., and Reik, W. (2010). Epigenetic reprogramming in plant and animal development. Science 330, 622–627. doi: 10.1126/science.1190614
Hollender, C., and Liu, Z. (2008). Histone deacetylase genes in Arabidopsis development. J. Integr. Plant Biol. 50, 875–885. doi: 10.1111/j.1744-7909.2008.00704.x
Howe, L., Auston, D., Grant, P., John, S., Cook, R. G., Workman, J. L., et al. (2001). Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes Dev. 15, 3144–3154. doi: 10.1101/gad.931401
Huang, T., and Irish, V. F. (2015). Temporal control of plant organ growth by TCP transcription factors. Curr. Biol. 25, 1765–1770. doi: 10.1016/j.cub.2015.05.024
Iñiguez-Lluhí, J. A. (2006). For a healthy histone code, a little SUMO in the tail keeps the acetyl away. ACS Chem. Biol. 1, 204–206. doi: 10.1021/cb600188m
Irish, V. F. (2008). The Arabidopsis petal: a model for plant organogenesis. Trends Plant Sci. 13, 430–436. doi: 10.1016/j.tplants.2008.05.006
Jaeger, K. E., Pullen, N., Lamzin, S., Morris, R. J., and Wigge, P. A. (2013). Interlocking feedback loops govern the dynamic behavior of the floral transition in Arabidopsis. Plant Cell 25, 820–833. doi: 10.1105/tpc.113.109355
Kagale, S., and Rozwadowski, K. (2011). EAR motif-mediated transcriptional repression in plants. Epigenetics 6, 141–146. doi: 10.4161/epi.6.2.13627
Karlic, R., Chung, H. R., Lasserre, J., Vlahovicek, K., and Vingron, M. (2010). Histone modification levels are predictive for gene expression. Proc. Natl. Acad. Sci. U.S.A. 107, 2926–2931. doi: 10.1073/pnas.0909344107
Krizek, B. A., and Fletcher, J. C. (2005). Molecular mechanisms of flower development: an armchair guide. Nat. Rev. Genet. 6, 688–698. doi: 10.1038/nrg1675
Krogan, N. T., Hogan, K., and Long, J. A. (2012). APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 139, 4180–4190. doi: 10.1242/dev.085407
Lee, J.-S., Smith, E., and Shilatifard, A. (2010). The language of histone crosstalk. Cell 142, 682–685. doi: 10.1016/j.cell.2010.08.011
Li, J., Wang, Y., Zhang, Y., Wang, W., Irish, V. F., and Huang, T. (2016). RABBIT EARS regulates the transcription of TCP4 during petal development in Arabidopsis. J. Exp. Bot. 67, 6473–6480. doi: 10.1093/jxb/erw419
Liu, X., Yang, S., Zhao, M., Luo, M., Yu, C.-W., Chen, C.-Y., et al. (2014). Transcriptional repression by histone deacetylases in plants. Mol. Plant 7, 764–772. doi: 10.1093/mp/ssu033
Moazed, D. (2011). Mechanisms for the inheritance of chromatin states. Cell 146, 510–518. doi: 10.1016/j.cell.2011.07.013
Oh, E., Zhu, J.-Y., Ryu, H., Hwang, I., and Wang, Z.-Y. (2014). TOPLESS mediates brassinosteroid-induced transcriptional repression through interaction with BZR1. Nat. Commun. 5:4140. doi: 10.1038/ncomms5140
Ohno, C. K. (2004). The Arabidopsis JAGGED gene encodes a zinc finger protein that promotes leaf tissue development. Development 131, 1111–1122. doi: 10.1242/dev.00991
Parcy, F., Nilsson, O., Busch, M. A., Lee, I., and Weigel, D. (1998). A genetic framework for floral patterning. Nature 395, 561–566. doi: 10.1038/26903
Pelayo, M. A., Yamaguchi, N., and Ito, T. (2021). One factor, many systems: the floral homeotic protein AGAMOUS and its epigenetic regulatory mechanisms. Curr. Opin. Plant Biol. 61:102009. doi: 10.1016/j.pbi.2021.102009
Pi, L., Aichinger, E., van der Graaff, E., Llavata-Peris, C. I., Weijers, D., Hennig, L., et al. (2015). Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Dev. Cell 33, 576–588. doi: 10.1016/j.devcel.2015.04.024
Powell, A. E., and Lenhard, M. (2012). Control of organ size in plants. Curr. Biol. 22, R360–R367. doi: 10.1016/j.cub.2012.02.010
Roguev, A., Schaft, D., Shevchenko, A., Pijnappel, W. W., Wilm, M., Aasland, R., et al. (2001). The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 20, 7137–7148. doi: 10.1093/emboj/20.24.7137
Schiessl, K., Muino, J. M., and Sablowski, R. (2014). Arabidopsis JAGGED links floral organ patterning to tissue growth by repressing Kip-related cell cycle inhibitors. Proc. Natl. Acad. Sci. U.S.A. 111, 2830–2835. doi: 10.1073/pnas.1320457111
Sims, R. J., and Reinberg, D. (2008). Is there a code embedded in proteins that is based on post-translational modifications? Nat. Rev. Mol. Cell Biol. 9, 815–820. doi: 10.1038/nrm2502
Strahl, B. D., and Allis, C. D. (2000). The language of covalent histone modifications. Nature 403, 41–45. doi: 10.1038/47412
Suganuma, T., and Workman, J. L. (2008). Crosstalk among histone modifications. Cell 135, 604–607. doi: 10.1016/j.cell.2008.10.036
Sun, B., Looi, L. S., Guo, S., He, Z., Gan, E. S., Huang, J., et al. (2014). Timing mechanism dependent on cell division is invoked by Polycomb eviction in plant stem cells. Science 343:1248559. doi: 10.1126/science.1248559
Sun, B., Xu, Y., Ng, K.-H., and Ito, T. (2009). A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes Dev. 23, 1791–1804. doi: 10.1101/gad.1800409
Takeda, S., Matsumoto, N., and Okada, K. (2011). RABBIT EARS, encoding a SUPERMAN-like zinc finger protein, regulates petal development in Arabidopsis thaliana. Development 138:3591. doi: 10.1242/dev.072058
Tao, Z., Shen, L., Gu, X., Wang, Y., Yu, H., and He, Y. (2017). Embryonic epigenetic reprogramming by a pioneer transcription factor in plants. Nature 551, 124–128. doi: 10.1038/nature24300
Wang, L., Kim, J., and Somers, D. E. (2013). Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. U.S.A. 110, 761–766. doi: 10.1073/pnas.1215010110
Xu, Y., Prunet, N., Gan, E. S., Wang, Y., Stewart, D., Wellmer, F., et al. (2018). SUPERMAN regulates floral whorl boundaries through control of auxin biosynthesis. EMBO J. 37:e97414. doi: 10.15252/embj.201797499
Keywords: petal, organogenesis, Arabidopsis, epigenetic regulation, histones
Citation: Huang R, Huang T and Irish VF (2021) Do Epigenetic Timers Control Petal Development? Front. Plant Sci. 12:709360. doi: 10.3389/fpls.2021.709360
Received: 13 May 2021; Accepted: 14 June 2021;
Published: 06 July 2021.
Edited by:
Patrick Laufs, Institut National de la Recherche Agronomique (INRA), FranceReviewed by:
Marcelo Carnier Dornelas, State University of Campinas, BrazilCopyright © 2021 Huang, Huang and Irish. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vivian F. Irish, dml2aWFuLmlyaXNoQHlhbGUuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.