
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 09 July 2021
Sec. Plant Abiotic Stress
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.704905
This article is part of the Research Topic Cellular Signaling Networks in Plant Heat Stress Responses View all 15 articles
A continuous increase in ambient temperature caused by global warming has been considered a worldwide threat. As sessile organisms, plants have evolved sophisticated heat shock response (HSR) to respond to elevated temperatures and other abiotic stresses, thereby minimizing damage and ensuring the protection of cellular homeostasis. In particular, for perennial trees, HSR is crucial for their long life cycle and development. HSR is a cell stress response that increases the number of chaperones including heat shock proteins (HSPs) to counter the negative effects on proteins caused by heat and other stresses. There are a large number of HSPs in plants, and their expression is directly regulated by a series of heat shock transcription factors (HSFs). Therefore, understanding the detailed molecular mechanisms of woody plants in response to extreme temperature is critical for exploring how woody species will be affected by climate changes. In this review article, we summarize the latest findings of the role of HSFs and HSPs in the HSR of woody species and discuss their regulatory networks and cross talk in HSR. In addition, strategies and programs for future research studies on the functions of HSFs and HSPs in the HSR of woody species are also proposed.
As one of the most harmful abiotic stresses, heat stress poses a threat to plant life by impacting plant metabolism directly (Rennenberg et al., 2006). In order to successfully survive the occurrence of a long heatwave, plants have evolved a series of adaptive mechanisms to cope with the rising ambient temperature (Bäurle, 2016). Among those mechanisms, heat shock response (HSR) is a rapid response mechanism that protects the proteome against elevated temperature and other stresses (Mittler et al., 2012). It is wildly accepted that a class of molecular chaperones, heat shock proteins (HSPs), are rapidly induced under the drive of heat shock transcription factors (HSFs) when plants are exposed to extremely high temperatures (Hu et al., 2009). In the HSR regulatory network, HSFs are the central components of the effective defense systems (Xue et al., 2014), whereas HSPs are directly responsible for protein folding, assembly, translocation, and degradation (Molinier et al., 2006).
Perennial woody plants are greatly significant components of the global ecosystem and play a key role in limiting the emissions of carbon dioxide (CO2) and other greenhouse gases. In addition, woody plants are the main biomass resources for biofuels. Due to the perennial and long life cycle, woody plants experience more severe and extreme abiotic stresses during their lives compared to herbaceous plants. Woody plants may have evolved more complex stress-responsive mechanisms (Anderegg et al., 2012). The ubiquitous and conserved HSR has been extensively studied in herbaceous plants. However, similar research studies on forest trees remain limited (Mittler et al., 2012). In this study, we review the recent progress on the roles of HSFs and HSPs and their regulatory network in the HSR of woody plants. Unraveling this underlying interconnected mechanism will help to understand the complex regulatory networks of the fitness and adaptive advantage of higher plants.
Heat shock proteins are found in all living organisms and are classified into at least six different types based on their molecular weight: sHSPs (small heat stress proteins), HSP40s, HSP60s, HSP70s, HSP90s, and HSP100s (Zhang et al., 2013, 2015a; Zandalinas et al., 2018). Massive production of HSPs in plants is a major characteristic response for the acquisition of thermotolerance.
Small heat stress protein is a class of alpha-crystallin domain (ACD) chaperons with a molecular mass of 15–30 kDa. According to the sequence homology and subcellular localization, sHSPs are classified into 11 different classes. Class I–VI sHSPs are localized in the nucleus or cytoplasm, and the rest five classes of sHSPs are localized in mitochondria, chloroplasts, peroxisomes, or ER (Waters et al., 2008). The diversity of plant sHSPs reflects the molecular adaptability to various biotic and abiotic stresses (Hilton et al., 2012).
Until now, most studies have focused on the class I cytoplasmic sHSP and have divulged that sHSPs are involved in regulating thermotolerance in woody plants (Table 1), such as RcHSP17.8 in Rosa chinensis (R. chinensis) (Jiang et al., 2009, 2020), ThHSP18.3 in Tamarix hispida (T. hispida) (Gao et al., 2012), PtHSP17.8 in Populus trichocarpa (P. trichocarpa) (Li et al., 2016), CsHSP17.2 in Camellia sinensis (C. sinensis) (Wang et al., 2017a), and MsHSP16.9 in Malus sieversii (M. sieversii) (Yang et al., 2017). Among these genes, RcHSP17.8 confers resistance to various stresses in Escherichia coli (E. coli), yeast, and Arabidopsis (Jiang et al., 2009). In addition, overexpression of RcHSP17.8 in transgenic tobacco seedlings exhibits significant resistance to high temperatures and osmotic stresses, manifested by low electrolyte leakage and higher proline content under stress conditions (Jiang et al., 2020). Heterologous expression of ThHSP18.3 protects yeast cells from salt, drought, heavy metals, and extreme temperatures (Gao et al., 2012). Overexpression of PtHSP17.8 in Arabidopsis increases survival rate and root length under heat and salt stresses (Li et al., 2016). The CsHSP17.2 in C. sinensis acts as a molecular chaperone to mediate heat tolerance by maintaining maximum photochemical efficiency and protein synthesis, enhancing the scavenging of reactive oxygen species (ROS), and inducing the expression of heat-responsive (HR) genes (Wang et al., 2017a). Overexpression of M. sieversii MsHSP16.9 in Arabidopsis improves the tolerance of a plant to heat by alleviating the damages of ROS and regulating the expression levels of stress-related genes (Yang et al., 2017). Moreover, cytoplasmic class II and III sHSPs have also been reported to be involved in the HSR in woody species. For example, SpHSP17.3, a cytoplasmic class II sHSP gene from Sorbus pohuashanensis (S. pohuashanensis), responds to high temperature, salt, and drought stresses and has a certain effect on the adaptability of introduction and domestication (Zhang et al., 2020). Overexpression of a Prunus mume (P. mume) cytoplasmic class III sHSP gene (PmHSP17.9) improves the thermotolerance of transgenic Arabidopsis by enhancing superoxide dismutase (SOD) activity (Wan et al., 2016). Transient overexpression of a Juglans regia (J. regia) sHSP gene JrsHSP17.3 in leaves enhances tolerance to cold, heat, and salt stresses by scavenging the accumulation of ROS and by accumulating osmotic adjustment substances (Zhai et al., 2016). In addition, the expressions of chloroplast small HSPs CsHSP17.7, CsHSP18.1, and CsHSP21.8 (cytoplasmic classes I, II, and IV, respectively) from C. sinensis could be highly induced by heat and cold stresses. Overexpression of these small CsHSPs confers heat and cold tolerances in transgenic Pichia pastoris (P. pastoris) and Arabidopsis (Wang et al., 2017b). These studies indicate that sHSPs exhibit considerable influence in the adaptation of woody plants under heat and other abiotic stresses, which may be related to the long-term adaptive evolution of woody species.
In contrast, the organelle-localized sHSPs have been more studied in herbaceous plants. For example, the mitochondria-localized GhHSP24.7 in cotton positively regulates seed germination through modulating the generation of ROS in a temperature-dependent manner (Ma et al., 2019). The chloroplast-localized AsHSP26.8a in creeping bentgrass plays a negative role in abiotic stresses through both abscisic acid (ABA)-dependent and ABA-independent signaling pathways and other stress signaling pathways (Sun et al., 2020). In Arabidopsis, an endoplasmic reticulum (ER)-localized sHSP, sHSP22, is involved in ABA and auxin signaling crosstalk (Li et al., 2018). However, the functional study of the organelle-localized sHSP in woody species has not been reported. In the future, more attention should be paid to these genes to reveal the response mechanisms of different organelle levels to high temperatures and other abiotic stresses in woody plants.
Compared with the sHSP genes, members in other HSP gene families, such as HSP40, HSP60, HSP70, HSP90, and HSP100, have not been well-studied in woody species. There is a lack of results about individual gene function validated through genetic modification, although the genome-wide identification of such genes has been performed in several woody species. For example, Zhang et al. (2013, 2015a) have provided a comprehensive analysis of the gene organization and expression of Populus HSF and other HSP genes, such as sHSP, HSP60, HSP70, HSP90, and HSP100, under different abiotic stresses. A complex transcriptional regulatory network between Populus HSFs and HSPs has been generated based on their transcription patterns in poplar.
Several studies indicated that members in these HSP families play pivotal roles in regulating the thermotolerance in herbaceous species. For instance, knockdown of CaHSP60-6 through virus-induced gene silencing in pepper [Capsicum annuum L. (C. annuum L.)] increases the heat sensitivity, which is manifested in the higher accumulation of ROS and lower membrane stability in silenced plants (Haq et al., 2019). Ectopic expression of a chrysanthemum CgHSP70 in Arabidopsis enhances tolerance to heat, drought, or salinity, thereby protecting the plants from total damage (Song et al., 2014). Cytosolic CaHSP70-1 from C. annuum is involved in the HSR through signal transduction pathways including calcium (Ca2+), hydrogen peroxide (H2O2), and putrescine (Guo et al., 2014). In addition, overexpression of a soybean GmHSP90A2 improves thermotolerance in Arabidopsis (Huang et al., 2019). However, the functional analysis of these high molecular weight HSPs in woody species is rarely reported. In order to explore the adaptability of woody species to elevated temperatures, more in-depth studies are needed.
The expression of the HSP genes is mainly regulated by HSFs. HSF/HSP transcriptional module-based HSR was recognized as an evolutionally conserved mechanism coping with heat stress (Scharf et al., 2012). Plants have evolved more complicated HSR compared to yeast and animals, for example, the HSF family of plants comprised of 18–52 members while yeast and Drosophila only have a single copy, and mammals have four HSFs (Andrási et al., 2020). Despite significant variability in size and sequence of HSFs, their structures and functions are highly conserved across plant species (Lin et al., 2011). HSFs have a modular structure comprising a DNA-binding domain, an oligomerization domain, and a C-terminal activation domain (Scharf et al., 2012). Based on the variations in these three domains, especially the oligomerization domain, HSFs of plants can be divided into three classes (A, B, and C). Because of the vital regulatory responses to different stresses and developmental processes, the HSF gene family has been extensively characterized in the model plant Arabidopsis, as well as in several woody plants such as apple (M. domestica) (Giorno et al., 2012), poplar (P. trichocarpa) (Zhang et al., 2015a; Liu et al., 2019), desert poplar (P. euphratica) (Zhang et al., 2016), willow [Salix suchowensis (S. suchowensis)] (Zhang et al., 2015b), pear [Pyrus bretschneideri (P. bretschneideri)] (Qiao et al., 2015), tea (C. sinensis) (Liu et al., 2016), and grape [Vitis vinifera (V. vinifera)] (Liu et al., 2018). In Arabidopsis, rice, and tomato, HSFA2 is strongly induced when plants are exposed to long-term heat stress or repeat-cycled heat stress and recovery (Scharf et al., 1998; Charng et al., 2007; Nishizawa-Yokoi et al., 2009), whereas HSFA1a seems to play a unique function as a master regulator of acquired thermotolerance and cannot be replaced by any other HSFs in tomato (Mishra et al., 2002). This implies that despite the existence of a certain conservative mechanism, HSF in different species may have functional specialization. Therefore, it is necessary to further study the gene function of HSF in various woody plants.
Transient overexpression of Betula platyphylla (B. platyphylla) BpHSFA4 gene in leaves improves the salt stress tolerance by increasing the ability to scavenge ROS, thereby reducing cell damage or cell death and enhancing birch resistance (Liu et al., 2020). In contrast to the activity of the transcriptional activation of class A HSFs, the class B HSF proteins lack the activation domain and have a repression domain in the C-terminus. Peng et al. (2013) identified a novel class B2 HSF gene from Chinese wild Vitis pseudoreticulata, VpHSF1, which plays a key role in biotic and abiotic stress responses. Overexpressing VpHSF1 in tobacco reduces the basal thermotolerance, improves the acquired thermotolerance, and enhances its susceptibility to osmotic stress and pathogen Phytophthora parasitica var. nicotianae Tucker (Table 1). Compared with herbaceous plants, the functional studies of HSFs in woody plants are still limited. CpHSFB1 from Carica papaya (C. papaya) (Tarora et al., 2010) and PsHSFB1 from Paeonia suffruticosa (P. suffruticosa) (Zhang et al., 2014) have been cloned from woody species, but their detailed biological functions have not been reported. Further unraveling the biological functions of each HSF can provide a better understanding of how woody plants can better cope with stresses during evolution.
Plants possess a complex HSR regulatory network consisting of multiple HSP and HSF genes. HSFs contain a conserved DNA-binding domain at the N-terminus, which can recognize the DNA motif, 5′-nGAAnnTTCn-3′. This heat shock element (HSE) motif is commonly found in the promoters of HSF target genes (Andrási et al., 2020). Hierarchical transcriptional network among HSFs, HSPs, and other heat stress-responsive genes has been well-built in the model plant Arabidopsis. HSFA1s are the master regulators that respond quickly to heat stress, and other transcription factors, such as HSFA2 and DREB2A, were directly activated by HSFA1s (Liu et al., 2011; Yoshida et al., 2011). HSPs and ROS-scavenging enzymes are the main heat stress-induced proteins regulated by HSFs. They are required for protein quality control and oxidative homeostasis under heat–stress conditions (Ohama et al., 2017). Compared to Arabidopsis, woody species have very limited studies on HSFs. Salt-inducible PeHSF from P. euphratica directly binds to the HSE motifs in the PeWRKY1 promoter and regulates its expression (Shen et al., 2015). HSFs regulate the expression of HSPs, and in turn, HSPs can physically interact with HSF proteins to affect the function of HSFs. In Arabidopsis, HSP90 and immunophilin ROF1 form a complex that co-imports HSFA2 into the nucleus and enhances its transcriptional activator activity under heat stress (Aviezer-Hagai et al., 2007; Meiri and Breiman, 2009). Another heat-inducible immunophilin ROF2, which is also a target of HSFA2, is recruited into the nucleus during accumulation and heterodimerizes with ROF1 in the complex, resulting in the inhibition of HSFA2. Therefore, the activity of HSFA2 in Arabidopsis is regulated by the two co-chaperones in both positive and negative manner by interacting with HSP90 (Meiri et al., 2010). In tomatoes, the constitutively expressed HSFA1a remains inactive in the complex formed with HSP70 and HSP90 under normal conditions. In contrast, the interaction of HSP90 with HSFB1 usually keeps the steady-state level of HSFB1 at a low level and targets HSFB1 to the degradation pathway of the 26S proteasome (Hahn et al., 2011). In addition, V. vinifera VvGOLS1 has been heterologously expressed in E. coli, showing that it encodes a functional galactinol synthase. Transient expression assays showed that the heat stress factor VvHSFA2 transactivates the promoter of VvGOLS1 in a heat stress-dependent manner (Pillet et al., 2012). However, the relationship between HSF and HSP, especially in woody species, remains unknown. With the rapid development of transcriptome-sequencing technology and the increase of big data in public databases, it is possible to use gene co-expression networks to explore the potential regulatory or interaction relationships between proteins. In this study, we use poplar as an example to construct an HSF-HSP co-expression network, due to the easy access to tremendous public expression data. As shown in Figure 1, we identified strong co-expressive relationships between HSP90 (PtHSP90-1a and PtHSP90-1b) and HSFA2 (PtHSF-A2), which is consistent with their protein–protein interactions in Arabidopsis and tomato (Meiri et al., 2010). This implies that HSR in woody species may share similar mechanisms across plant species. However, it is still an open question whether specific regulatory mechanisms or interaction relationships between HSFs and HSPs exist in woody plants.
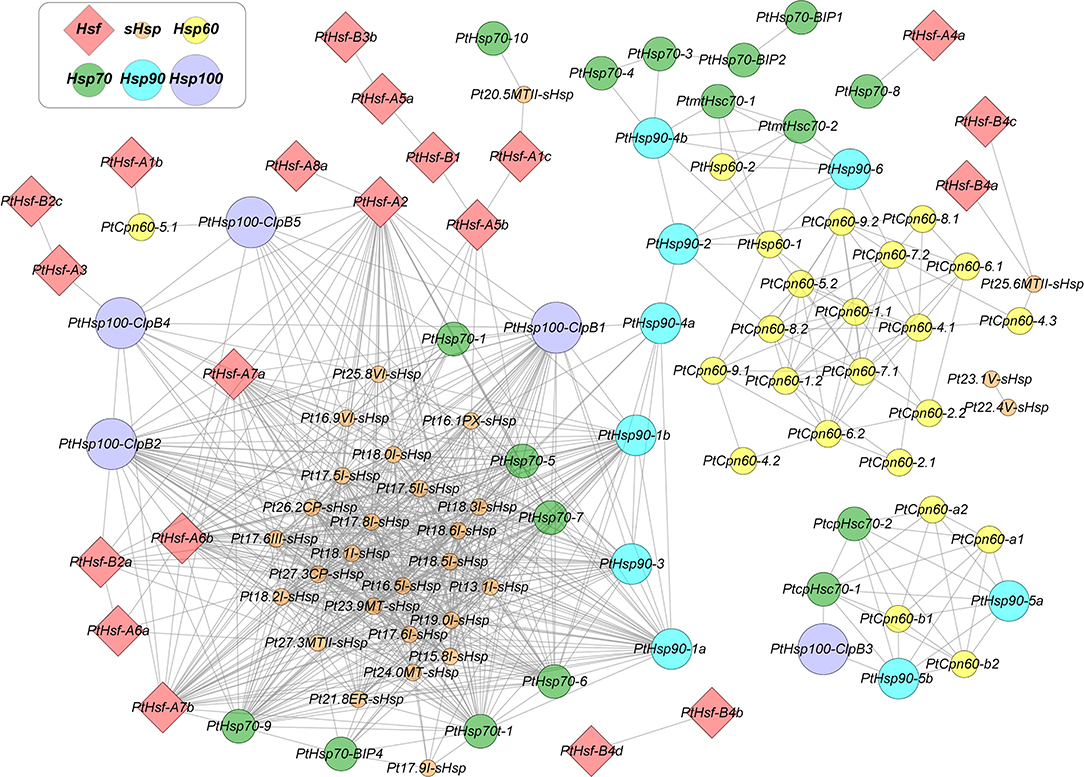
Figure 1. Co-expression network of the HSF and HSP genes in Populus. Nodes represent HSFs and HSPs genes in Populus, and edges indicate pairwise correlation constructed by weighted gene co-expression network analysis (WGCNA). Node color codes represent different gene families. Red diamonds indicate HSFs, and solid circles with orange, yellow, green, cyan, and purple indicate sHSPs, HSP60s, HSP70s, HSP90s, and HSP100s, respectively. Lines represent the co-expression interaction. The network was created using Cytoscape.
With ongoing greenhouse gas emissions, the ambient temperature is expected to rise over time. Woody plants have evolved more complex stress response mechanisms to cope with changing environments than herbaceous plants to guarantee a longer lifespan (Anderegg et al., 2012). However, the current knowledge of the functions of HSF and HSP genes during HSR in woody plants is still lacking. On the one hand, the transcriptional regulation or interaction between HSF and HSP in woody plants is unclear. Zhang et al. (2018) used the joint analysis of expression quantitative trait loci (eQTL) and co-expression network based on the P. trichocarpa natural variant populations to successfully identify the upstream regulatory transcription factor WRKY that controls the expression of the HCT2 gene. This strategy provides a new method to identify the transcriptional regulatory relationship between genes. Therefore, the public gene expression database of woody plants can be used to preliminarily examine whether there is a potential regulatory or interaction relationship between HSFs and HSPs in these species. In addition, the continuous maturity of ChIP-Seq, CUT&Tag-Seq, and other technologies have effectively improved the ability to analyze the interaction between protein and DNA. The recently developed reverse-ChIP technology (Wen et al., 2020) can reversely mine the upstream regulators of specific target genes. In the future, the combined use of these methods can provide technical supports for the clarification of gene regulatory relationships in the HSR of woody plants. On the other hand, the traditional genetic transformation system is unstable and time-consuming for many woody species, which leads to insufficient research studies on the function of individual genes. Most of the studies just rely on the ectopic expression of HSF or HSP genes obtained from woody species in the model plant Arabidopsis or yeast. Therefore, fast and efficient gene function validation systems are urgently needed. The protoplast transient expression system (Zhang et al., 2018) can be used to quickly verify the relationship between HSFs and HSPs. And the recently developed genome editing technology has been successfully applied in many woody species (Li et al., 2021). However, it is necessary to further improve the efficiency and accuracy of gene editing to overcome the limitation of its application in woody species. In addition, future research studies should also pay attention to the signals and cascade transduction pathways of woody plants after sensing high temperatures, how these signals further activate the HSR, and how the posttranscriptional and posttranslational regulatory pathways participate in the HSR of woody species. The combined use of these methods can provide a basis for further in-depth analysis of the function of HSFs and HSPs in HSR in woody plants.
JZ conceived the study. JZ, FT, and X-LH drafted the manuscript. TY, XY, J-GC, and M-ZL revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the Key Scientific Research Projects of Institutions of Higher Learning in Henan Province (No. 19A180004) to FT and the Zhejiang A&F University Research and Development Fund Talent Startup Project (2021LFR013) to JZ.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Anderegg, W.-R.-L., Berry, J.-A., and Field, C.-B. (2012). Linking definitions, mechanisms, and modeling of drought-induced tree death. Trends Plant Sci. 17, 693–700. doi: 10.1016/j.tplants.2012.09.006
Andrási, N., Pettkó-Szandtner, A., and Szabados, L. (2020). Diversity of plant heat shock factors: regulation, interactions, and functions. J. Exp. Bot. 72, 1558–1575. doi: 10.1093/jxb/eraa576
Aviezer-Hagai, K., Skovorodnikova, J., Galigniana, M., Farchi-Pisanty, O., Maayan, E., Bocovza, S., et al. (2007). Arabidopsis immunophilins ROF1 (AtFKBP62) and ROF2 (AtFKBP65) exhibit tissue specificity, are heat-stress induced, and bind HSP90. Plant Mol. Biol. 63, 237–255. doi: 10.1007/s11103-006-9085-z
Bäurle, I. (2016). Plant heat adaptation: priming in response to heat stress. fF1000res 5:694. doi: 10.12688/f1000research.7526.1
Charng, Y.-Y., Liu, H.-C., Liu, N.-Y., Chi, W.-T., Wang, C.-N., Chang, S.-H., et al. (2007). A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 143, 251–262. doi: 10.1104/pp.106.091322
Gao, C., Jiang, B., Wang, Y., Liu, G., and Yang, C. (2012). Overexpression of a heat shock protein (ThHSP18.3) from Tamarix hispida confers stress tolerance to yeast. Plant Mol. Biol. Rep. 39, 4889–4897. doi: 10.1007/s11033-011-1284-2
Giorno, F., Guerriero, G., Baric, S., and Mariani, C. (2012). Heat shock transcriptional factors in Malus domestica: identification, classification and expression analysis. BMC Genomics 13:639. doi: 10.1186/1471-2164-13-639
Guo, M., Zhai, Y.-F., Lu, J.-P., Chai, L., Chai, W.-G., Gong, Z.-H., et al. (2014). Characterization of CaHsp70-1, a pepper heat-shock protein gene in response to heat stress and some regulation exogenous substances in Capsicum annuum L. Int. J. Mol. Sci. 15, 19741–19759. doi: 10.3390/ijms151119741
Hahn, A., Bublak, D., Schleiff, E., and Scharf, K.-D. (2011). Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell 23, 741–755. doi: 10.1105/tpc.110.076018
Haq, S. U., Khan, A., Ali, M., Gai, W.-X., Zhang, H.-X., Yu, Q. H., et al. (2019). Knockdown of CaHSP60-6 confers enhanced sensitivity to heat stress in pepper (Capsicum annuum L.). Planta 250, 2127–2145. doi: 10.1007/s00425-019-03290-4
Hilton, G.-R., Lioe, H.-H., Stengel, F., Baldwin, A.-J., and Benesch, J.-L.-P. (2012). Small heat-shock proteins: paramedics of the cell. Topics Curr. Chem. 328, 69–98. doi: 10.1007/128-012-324
Hu, W., Hu, G., and Han, B. (2009). Genome-wide survey and expression profiling of heat shock proteins and heat shock factors revealed overlapped and stress specific response under abiotic stresses in rice. Plant Sci. 176, 583–590. doi: 10.1016/j.plantsci.2009.01.016
Huang, Y., Xuan, H., Yang, C., Guo, N., Wang, H., Zhao, J., et al. (2019). GmHsp90A2 is involved in soybean heat stress as a positive regulator. Plant Sci. 285, 26–33 doi: 10.1016/j.plantsci.2019.04.016
Jiang, C., Bi, Y., Li, M., Zhang, R., Feng, S., and Ming, F. (2020). A small heat shock protein gene (RcHSP17.8) from Chinese rose confers resistance to various abiotic stresses in transgenic tobacco. Plant Cell Tiss. Org. 141, 407–415. doi: 10.1007/s11240-020-01798-2
Jiang, C., Xu, J., Hao, Z., Zhang, X., and Ming, F. (2009). A cytosolic class I small heat shock protein, RcHSP17.8, of Rosa chinensis confers resistance to a variety of stresses to Escherichia coli, yeast and Arabidopsis thaliana. Plant Cell Environ. 32, 1046–1059. doi: 10.1111/j.1365-3040.2009.01987.x
Li, G., Sretenovic, S., Eisenstein, E., Coleman, G., and Qi, Y. (2021). Highly efficient C-to-T and A-to-G base editing in a Populus hybrid. Plant Biotech. J. 19, 1086–1088. doi: 10.1111/pbi.13581
Li, J., Zhang, J., Jia, H., Li, Y., Xu, X., Wang, L., et al. (2016). The Populus trichocarpa PtHSP17.8 involved in heat and salt stress tolerances. Plant Cell Rep. 35, 1587–1599. doi: 10.1007/s00299-016-1973-3
Li, Y., Li, Y., Liu, Y., Wu, Y., and Xie, Q. (2018). The sHSP22 heat shock protein requires the ABI1 protein phosphatase to modulate polar auxin transport and downstream responses. Plant Physiol. 176, 2406–2425. doi: 10.1104/pp.17.01206
Lin, Y.-X., Jiang, H.-Y., Chu, Z.-X., Tang, X.-L., Zhu, S.-W., and Cheng, B.-J. (2011). Genome-wide identification; classification and analysis of heat shock transcription factor family in maize. BMC Genomics 12:76. doi: 10.1186/1471-2164-12-76
Liu, B., Hu, J., and Zhang, J. (2019). Evolutionary divergence of duplicated Hsf genes in Populus. Cells 8:438. doi: 10.3390/cells8050438
Liu, G., Chai, F., Wang, Y., Jiang, J., Duan, W., Wang, Y., et al. (2018). Genome-wide identification and classification of HSF family in grape, and their transcriptional analysis under heat acclimation and heat stress. Horticult. Plant J. 4, 133–143. doi: 10.1016/j.hpj.2018.06.001
Liu, H. C., Liao, H. T., and Charng, Y. Y. (2011). The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 34, 738–751. doi: 10.1111/j.1365-3040.2011.02278.x
Liu, Z.-W., Wu, Z.-J., Li, X.-H., Huang, Y., Li, H., Wang, Y.-X., et al. (2016). Identification, classification, and expression profiles of heat shock transcription factors in tea plant (Camellia sinensis) under temperature stress. Gene 576, 52–59. doi: 10.1016/j.gene.2015.09.076
Liu, Z.-Y., Liu, Z., Xu, Y., Liu, S.-S., Tian, Z.-L., Xie, Q.-J., et al. (2020). Cloning and salt tolerance analysis of transcription factor HSFA4 from Betula platyphylla. Sci. Silvae Sinicae 56, 69–79. doi: 10.11707/j.1001-7488.20200508
Ma, W., Guan, X., Li, J., Pan, R., Wang, L., Liu, F., et al. (2019). Mitochondrial small heat shock protein mediates seed germination via thermal sensing. Proc. Natl. Acad. Sci. U.S.A. 116, 4716–4721. doi: 10.1073/pnas.1815790116
Meiri, D., and Breiman, A. (2009). Arabidopsis ROF1 (FKBP62) modulates thermotolerance by interacting with HSP90.1 and affecting the accumulation of HsfA2-regulated sHSPs. Plant J. 59, 387–399. doi: 10.1111/j.1365-313X.2009.03878.x
Meiri, D., Tazat, K., Cohen-Peer, R., Farchi-Pisanty, O., Aviezer-Hagai, K., Avni, A., et al. (2010). Involvement of Arabidopsis ROF2 (FKBP65) in thermotolerance. Plant Mol. Biol. 72, 191–203. doi: 10.1007/s11103-009-9561-3
Mishra, S.-K., Tripp, J., Winkelhaus, S., Tschiersch, B., Theres, K., Nover, L., et al. (2002). In the complex family of heat stress transcription factors, HSfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev. 16, 1555–1567. doi: 10.1101/gad.228802
Mittler, R., Finka, A., and Goloubinoff, P. (2012). How do plants feel the heat? Trends Biochem. Sci. 37, 118–125. doi: 10.1016/j.tibs.2011.11.007
Molinier, J., Ries, G., Zipfel, C., and Hohn, B. (2006). Trans generation memory of stress in plants. Nature 442, 1046–1049. doi: 10.1038/nature05022
Nishizawa-Yokoi, A., Yoshida, E., Yabuta, Y., and Shigeoka, S. (2009). Analysis of the regulation of target genes by an Arabidopsis heat shock transcription factor, HsfA2. Biosci. Biotechnol. Biochem. 73, 890–895. doi: 10.1271/bbb.80809
Ohama, N., Sato, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2017). Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 22, 53–65. doi: 10.1016/j.tplants.2016.08.015
Peng, S.-B., Zhu, Z.-G., Zhao, K.-S., Jiang, L., Yang, Y. Z., He, M.-Y., et al. (2013). A novel heat shock transcription factor, VpHsf1, from Chinese wild Vitis pseudoreticulata is involved in biotic and abiotic stresses. Plant Mol. Biol. Rep. 31, 240–247. doi: 10.1007/s11105-012-0463-1
Pillet, J., Egert, A., Pieri, P., Lecourieux, F., Kappel, C., Charon, J., et al. (2012). VvGOLS1 and VvHsfA2 are involved in the heat stress responses in Grapevine berries. Plant Cell Physiol. 53, 1776–1792. doi: 10.1093/pcp/pcs121
Qiao, X., Li, M., Li, L., Yin, H., Wu, J., and Zhang, S. (2015). Genome-wide identification and comparative analysis of the heat shock transcription factor family in Chinese white pear (Pyrus bretschneideri) and five other rosaceae species. BMC Plant Biol. 15:12. doi: 10.1186/s12870-014-0401-5
Rennenberg, H., Loreto, F., Polle, A., Brilli, F., Fares, S., Beniwal, R.-S., et al. (2006). Physiological responses of forest trees to heat and drought. Plant Biol. 8, 556–571. doi: 10.1055/s-2006-924084
Scharf, K.-D., Berberich, T., Ebersberger, I., and Nover, L. (2012). The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochim. Biophys. Acta. Gene Regul. Mech. 1819, 104–119. doi: 10.1016/j.bbagrm.2011.10.002
Scharf, K.-D., Heider, H., Hohfeld, I., Lyck, R., Schmidt, E., and Nover, L. (1998). The tomato HSF system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Mol. Cell Biol. 18, 2240–2251. doi: 10.1128/mcb.18.4.2240
Shen, Z., Yao, J., Sun, J., Chang, L., Wang, S., Ding, M., et al. (2015). Populus euphratica HSF binds the promoter of WRKY1 to enhance salt tolerance. Plant Science 235, 89–100. doi: 10.1016/j.plantsci.2015.03.006
Song, A., Zhu, X., Chen, F., Gao, H., Jiang, J., and Chen, S. (2014). A chrysanthemum heat shock protein confers tolerance to abiotic stress. Int. J. Mol. Sci. 15, 5063–5078. doi: 10.3390/ijms15035063
Sun, X., Zhu, J., Li, X., Li, Z., and Luo, H. (2020). AsHSP26.8a, a creeping bentgrass small heat shock protein integrates different signaling pathways to modulate plant abiotic stress response. BMC Plant Biol. 184, 2–19. doi: 10.1186/s12870-020-02369-5
Tarora, K., Tamaki, M., Shudo, A., Urasaki, N., Matsumura, H., and Adaniya, S. (2010). Cloning of a heat stress transcription factor, CphsfB1, that is constitutively expressed in radicles and is heat-inducible in the leaves of Carica papaya. Plant Cell Tiss. Org. 102, 69–77. doi: 10.1007/s11240-010-9707-7
Wan, X. L., Yang, J., Li, X. B., Zhou, Q., Guo, C., Bao, M.-Z., et al. (2016). Over-expression of PmHSP17.9 in transgenic Arabidopsis thaliana confers thermotolerance. Plant Mol. Biol. Rep. 34, 899–908. doi: 10.1007/s11105-016-0974-2
Wang, M., Zou, Z., Li, Q., Sun, K., Chen, X., and Li, X. (2017a). The CsHSP17.2 molecular chaperone is essential for thermotolerance in Camellia sinensis. Sci. Rep. 7:1237. doi: 10.1038/s41598-017-01407-x
Wang, M., Zou, Z., Li, Q., Xin, H., Zhu, X., Chen, X., et al. (2017b). Heterologous expression of three Camellia sinensis small heat shock protein genes confers temperature stress tolerance in yeast and Arabidopsis thaliana. Plant Cell Rep. 36, 1125–1135. doi: 10.1007/s00299-017-2143-y
Waters, E. R., Aevermann, B. D., and Sanders-Reed, Z. (2008). Comparative analysis of the small heat shock proteins in three angiosperm genomes identifies new subfamilies and reveals diverse evolutionary patterns. Cell Stress Chaperon. 13, 127–142. doi: 10.1007/s12192-008-0023-7
Wen, X., Wang, J., Zhang, D., Ding, Y., Ji, X., Tan, Z., et al. (2020). Reverse Chromatin Immunoprecipitation (R-ChIP) enables investigation of the upstream regulators of plant genes. Commun. Biol. 3:770. doi: 10.1038/s42003-020-01500-4
Xue, G.-P., Sadat, S., Drenth, J., and McIntyre, C.-L. (2014). The heat shock factor family from Triticum aestivum in response to heat and other major abiotic stresses and their role in regulation of heat shock protein genes. J. Exp. Bot. 65, 539–557. doi: 10.1093/jxb/ert399
Yang, M., Zhang, Y., Zhang, H., Wang, H., Wei, T., Che, S., et al. (2017). Identification of MsHsp20 gene family in Malus sieversii and functional characterization of MsHsp16.9 in heat tolerance. Front. Plant Sci. 8:1761. doi: 10.3389/fpls.2017.01761
Yoshida, T., Ohama, N., Nakajima, J., Kidokoro, S., Mizoi, J., Nakashima, K., et al. (2011). Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol. Genet. Genomics 286, 321–332. doi: 10.1007/s00438-011-0647-7
Zandalinas, S.-I., Mittlerb, R., Balfagóna, D., Arbona, V., and Gómez-Cadenas, A. (2018). Plant adaptations to the combination of drought and high temperatures. Physiol. Plantarum 162, 2–12. doi: 10.1111/ppl.12540
Zhai, M., Sun, Y., Jia, C., Peng, S., Liu, Z., and Yang, G. (2016). Over-expression of JrsHSP17.3 gene from Juglans regia confer the tolerance to abnormal temperature and NaCl stresses. J. Plant Biol. 59, 549–558. doi: 10.1007/s12374-015-0507-9
Zhang, J., Jia, H., Li, J., Li, Y., Lu, M., and Hu, J. (2013). Genome-wide analysis of the Populus Hsp90 gene family reveals differential expression patterns, localization, and heat stress responses. BMC Genomics 14:532. doi: 10.1186/1471-2164-14-532
Zhang, J., Jia, H., Li, J., Li, Y., Lu, M., and Hu, J. (2016). Molecular evolution and expression divergence of the Populus euphratica Hsf genes provide insight into the stress acclimation of desert poplar. Sci. Rep. 30050, 1–14. doi: 10.1038/srep30050
Zhang, J., Li, Y., Jia, H. X., Li, J. B., Huang, J., Lu, M. Z., et al. (2015b). The heat shock factor gene family in Salix suchowensis: a genome-wide survey and expression profiling during development and abiotic stresses. Front. Plant Sci. 6:748. doi: 10.3389/fpls.2015.00748
Zhang, J., Liu, B., Li, J., Zhang, L., Wang, Y., Zheng, H., et al. (2015a). Hsf and Hsp gene families in Populus: genome-wide identification, organization and correlated expression during development and in stress responses. BMC Genomics 16, 1–19. doi: 10.1186/s12864-015-1398-3
Zhang, J., Yang, Y., Zheng, K., Xie, M., Feng, K., Jawdy, S., et al. (2018). Genome-wide association studies and expression-based quantitative trait loci analyses reveal roles of HCT2 in caffeoylquinic acid biosynthesis and its regulation by defense-responsive transcription factors in Populus. New Phytol. 220, 502–516. doi: 10.1111/nph.15297
Zhang, Y.-Z., Cheng, Y.-W., Ya, H.-Y., Gao, K., and Han, J.-M. (2014). Identification and analysis of the heat shock transcriptor PsHsfB1 in tree peony. Appl. Mech. Mater. 675–677, 1133–1136. doi: 10.4028/www.scientific.net/AMM.675-677.1133
Keywords: heat stress, woody plants, signaling network, molecular response, heat shock transcription factor, heat shock protein
Citation: Tian F, Hu X-L, Yao T, Yang X, Chen J-G, Lu M-Z and Zhang J (2021) Recent Advances in the Roles of HSFs and HSPs in Heat Stress Response in Woody Plants. Front. Plant Sci. 12:704905. doi: 10.3389/fpls.2021.704905
Received: 04 May 2021; Accepted: 07 June 2021;
Published: 09 July 2021.
Edited by:
Rosa M. Rivero, Spanish National Research Council, SpainReviewed by:
Xin-Rong Ma, Chinese Academy of Sciences, ChinaCopyright © 2021 Tian, Hu, Yao, Yang, Chen, Lu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin Zhang, emhhbmdqQHphZnUuZWR1LmNu; orcid.org/0000-0002-8397-5078
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.