- State Key Laboratory of Subtropical Silviculture, Zhejiang A&F University, Hangzhou, China
In natural systems, plant–symbiont–pathogen interactions play important roles in mitigating abiotic and biotic stresses in plants. Symbionts have their own special recognition ways, but they may share some similar characteristics with pathogens based on studies of model microbes and plants. Multi-omics technologies could be applied to study plant–microbe interactions, especially plant–endophyte interactions. Endophytes are naturally occurring microbes that inhabit plants, but do not cause apparent symptoms in them, and arise as an advantageous source of novel metabolites, agriculturally important promoters, and stress resisters in their host plants. Although biochemical, physiological, and molecular investigations have demonstrated that endophytes confer benefits to their hosts, especially in terms of promoting plant growth, increasing metabolic capabilities, and enhancing stress resistance, plant–endophyte interactions consist of complex mechanisms between the two symbionts. Further knowledge of these mechanisms may be gained by adopting a multi-omics approach. The involved interaction, which can range from colonization to protection against adverse conditions, has been investigated by transcriptomics and metabolomics. This review aims to provide effective means and ways of applying multi-omics studies to solve the current problems in the characterization of plant–microbe interactions, involving recognition and colonization. The obtained results should be useful for identifying the key determinants in such interactions and would also provide a timely theoretical and material basis for the study of interaction mechanisms and their applications.
Introduction
Plant–microbe interactions have occurred throughout the evolutionary history of the plants. The fossil record provides evidence that, over 450 million years since plants were first established on land, almost all plants in the natural ecosystem have been colonized by one or more microbial symbionts (Redecker et al., 2000; Krings et al., 2007; Bonfante and Genre, 2010; Kawaguchi and Minamisawa, 2010; Atsatt and Whiteside, 2014; Genre et al., 2020). Over this lengthy span of evolution, symbiotic microbes might have played a major role within the hosts to overcome the environmental changes. The symbiotic fungi or bacteria that live asymptomatically within a healthy plant tissue are called endophytes, and normally, these can provide benefits to their host in contrast to parasites (Khare et al., 2018; Sabra et al., 2018; Afzal et al., 2019; Mishra et al., 2021), by promoting the growth of host plants (Johnson et al., 2003; Rodriguez et al., 2008), eliciting metabolite production (Strobel et al., 2004; Kaul et al., 2012; Kusari et al., 2014; Wang et al., 2015), and mediating stress relief in plants (Yan et al., 2019). Thus, plant–endophyte interactions greatly contribute to the improved plant fitness. However, the interaction mechanism is complicated, and it remains difficult to clarify at the biochemical, physiological, and molecular levels why and how endophytes can invade via the defense systems of host plants and co-exist with them without causing any illness (Faeth and Fagan, 2002).
Early research on plant–endophyte interactions relied on host agronomic characteristics, composition determination, and strains’ isolation, cultivation, and characterization when speculating about the interaction mechanism involved. Nowadays, with the rapid development in high-throughput sequencing, the omics technologies (at the genome, transcriptome, and proteome levels) can provide new insights to elucidate the mechanism of interaction in a given system (Xu et al., 2021). Moreover, when coupled to metabolomic studies, omics-type investigations can gradually resolve aspects of the relationships between endophyte infection, metabolite accumulation, and stress mitigation in host plants. Transcriptomics uses next-generation sequencing (NGS) technology to reveal the presence and quantity of the individual RNA molecules in biological samples (Wang et al., 2009; Li and Li, 2018). Metabolomics is used to identify and quantify the changes in metabolites due to the deletion or overexpression of a given gene (Oliver et al., 1998; Wishart, 2007; Chen F. et al., 2019), including both endogenous metabolites that are naturally produced by an organism and by the exogenous chemicals that are not (Nordström et al., 2006). The combination of transcriptome and metabolome results can help to reveal the relationship between plants and their endophytes. This review aims to outline the use of omics techniques as an effective means to characterize the plant–endophyte interaction mechanism, and to identify the key factors that determine the effectiveness of this interaction. This work will provide a theoretical basis for elucidating the interaction mechanism, and the findings should prove useful for future applications, such as in exploring the relationships between other unknown plants and microbial species.
Roles of Plant–Endophyte Interactions
Since “endophyte” was first defined in 1809, endophytic microbes, including parasitic, mutualistic, and latent pathogenic fungi and bacteria have been continuously discovered. Mounting evidence points to the critical roles that endophytes played in the host plants. Thus, plant–endophyte interactions have become a research hotspot. Many studies have demonstrated that the relationships of endophytes living in plants ranges from mutualistic, where both plant and microbe benefit, to parasitic, where the microbe receives some benefit from the interaction at the expense of the host. Both mutualistic and parasitic interactions can be considered as symbiotic, according to the original definition of symbiosis. Latent pathogens might show pathogenicity only at certain stages of their life cycle or under specific circumstances, and the extent of damage caused by pathogens can greatly vary depending on the length of time they spend in their pathogenic phases (e.g., causing necrotic lesions) or in asymptomatic parasitic or mutualistic phases. Accordingly, microbes can transition between the trophic states of pathogenesis and mutualism and/or between mutualism and parasitism in response to internal host signals or environmental factors (Redman et al., 2001; Drew et al., 2021). Further, many convergent strategies are supported by symbiotic signaling and infection modules (Delaux and Schornack, 2021) between different endophytes and hosts. Here, we attempt to clarify the roles of symbiosis and pathogenesis as gleaned from the past and more recent studies.
Symbiosis
The term “symbiosis” simply means reciprocal interactions between plants and microbiota in narrow sense (Delaux and Schornack, 2021). In this context, the microbe enables the host plant to absorb nutrients (Oldroyd and Leyser, 2020) and mediates its stress tolerance (Carrión et al., 2019; Trivedi et al., 2020), thereby improving plant growth (Vandana et al., 2021). In return, the host plant actively recruits microbes to accelerate their colonization (Batstone et al., 2020) and protects them against autoimmunity (Huang et al., 2021) until the next generation. The symbiosis of plant and microbes entails several typical interaction models, including those of extracellular (ectomycorrhizas), intercellular (associations with cyanobacteria), and intracellular symbiosis [arbuscular mycorrhizas (AM), orchid mycorrhizas (OM), ericoid mycorrhizas (ERM), and nitrogen-fixing symbioses of rhizobia, and Frankia strains]. In AM and root nodule symbioses, an intracellular symbiotic interaction is established based on a common symbiosis signaling pathway (CSSP) governed by three core genes, namely SymRK, CCaMK, and CYCLOPS (Genre et al., 2020; Radhakrishnan et al., 2020). A recent study by Genre et al. (2020) tentatively links the CSSP to intracellular symbioses between plants and bacteria and fungi during the evolution of plants. Hence, intracellular symbiosis relationship is presumed to be most conserved at the evolutionary level, making it worthy of an in-depth study.
Two legumes forming nitrogen-fixing root nodules (namely, Lotus japonicus and Medicago truncatula) have served as the typical model plants to study the intracellular symbiotic interaction (Roy et al., 2020). The rhizobia enabled the nodulation to allow plants’ utilization of air dinitrogen under resource-poor conditions (Cook et al., 1997). The progress of nodulation involves several different incorporated lifestyles, including rhizosphere growth, root colonization, bacterial infection, N2-fixing bacteroids, and release from most types of legume nodules (Wheatley et al., 2020). Throughout the phase of growth in the rhizosphere, the rhizobia secret exopolysaccharide (EPS), amino acid, and cold shock protein compounds to respectively promote their entanglement with root hairs, transport, and survivability. For the uptake into plant roots and then into nodule primordium cells, followed by the differentiation into nitrogen fixing bacteroids, rhizobia depend on highly decorated lipochitooligosaccharide (LCO) Nod factors recognized by the extracellular lysine motif (LysM) domains of plant receptors and receptor kinases, homologs of the chitin elicitor receptor kinase 1 (CERK1) of Arabidopsis (Bozsoki et al., 2020). In response to the interaction with rhizobia, the roots of leguminous plants secrete a variety of amino acids and key compounds for the synthesis of purine, S-adenosyl-methionine, heme, vitamin B12, CoA, and riboflavin, all of which might be used by rhizobia. In particular, el Zahar Haichar et al. (2014) and Wheatley et al. (2020) identified a 5-aminoimidazole-4-carboxamide-1-β-D-ribofuranosyl 5′-monophosphate in the roots of legumes, a substance able to supplement the rhizobial purine biosynthesis mutants during growth in the rhizosphere. Further, nodulation is controlled by the SHR-SCR module for inducing cortical cell division that is regulated by the nodule inception (NIN) transcription factor, the transcription of which is activated by rhizobial Nod factors via the CSSP (Dong et al., 2021; Quilbé et al., 2021). After nodule establishment, the plant exploits nodule transfer cells to sense the environmental nitrate status, to regulate plasticity of nodule development, by specifically expressing transporters of the NFP (NRT1/PTR FAMILY) (Wang Q. et al., 2020).
Some non-legume plants can also establish a symbiotic relationship with rhizobia, namely members of the genus Parasponia, also using the CSSP (van Velzen et al., 2018). However, other mechanisms are involved in the extracellular endophytic symbiotic relationship between non-legume plants and rhizobia. A typical case is that of the non-expressor of pathogenesis-related (PR) genes 1 (NPR1), which is required to establish symbiotic relationship between barley and Rhizobium radiobacter (Kumar et al., 2021). In rice, bradyrhizobial strains use their quorum sensing system to sustain their symbiotic interaction with hosts (Pongdet et al., 2015; Cai et al., 2020). Non-rhizobial diazotrophic bacteria could also colonize rice, e.g., Azoarcus olearius which uses its flagella as a mediator of endophytic competence, or Herbaspirillum seropedicae (Pankievicz et al., 2021). Often, diazotrophic bacteria can fix air dinitrogen to produce ammonia that is usable by host plants (Peoples et al., 1995). The utilization of diazotrophs is a suitable and potential way for cereals to augment their nitrogen supply. An Azoarcus sp. strain was a typical diazotroph found in Karar grass plants (Leptochloa fusca L. Kunth), which can be co-cultured with rice (Hurek and Reinhold-Hurek, 2003). More recent work has demonstrated the utility of the diazotrophic bacterial associations of the grass Setaria viridis (e.g., with Azospirillum brasilense and H. seropedicae), as model systems for research aimed at enhancing biological nitrogen fixation for better nitrogen availability (Pankievicz et al., 2015, 2021).
Mycorrhizas are another typical model for the study of intracellular symbiotic interactions. The AM symbiosis is the most common mycorrhizal association, one that contributes to host-plant growth and development through nutrient absorption, in particular, of phosphate whose low solubility and mobility of phosphate in soil limits its accessibility to plants. In AM roots, plants make use of symbiotic interfaces provided by the fungal arbuscules or hyphal coils to take up the nutrients (Smith and Smith, 2011). The extraradical hyphal network explores the soil and takes up phosphates while inside the root, the plants employ mycorrhiza-specific phosphate transporters to transfer the phosphate released by the fungus into the root cortical cells; the fungal transporter that re-absorbs phosphate from the interfacial matrix acts as a sensor to control fungal growth and metabolism (Ferrol et al., 2019). The AM symbiosis can improve the growth of plants and enhance their tolerance against multiple stresses. In general, the symbiosis can be applied to phosphate-efficient farming systems aiming at sustainable agriculture (Ferrol et al., 2019).
Overall, the AM fungi colonize approximately 80% of land plants, while OM and ERM are specific for Orchidaceous and Ericoid hosts, respectively. To date, studies of ERM have sought basic knowledge of ERM symbiosis by using traditional methodologies, rendering details of its symbiotic mechanism that is mostly unknown (Vohník, 2020). By contrast, some symbiotic mechanisms for OM have been investigated. In their non-photosynthetic and photosynthetic stages, orchid plants respectively provide NH4+ and carbon to OM fungi to support fungal colonization (Dearnaley and Cameron, 2017). Orchid seeds harbor few reserves, so carbon sources, particularly lipids, provided by the fungus have to be harnessed for building a symbiotic structure, while the plant exports NH4+ (Bateman, 1995; Ghirardo et al., 2020). In the photosynthetic stage, the host accelerates fungal colonization by provision with sugars. As the fungus senesces and is digested in the plant cell, the host plant obtains nitrogen, phosphorus, and carbon from the hyphal coils (Dearnaley and Cameron, 2017). Research into the association between OM and orchid plants has focused mainly on fungal effects on plant growth, with few comprehensive metabolic and molecular mechanisms about their interaction actually studied yet. But based on the similar structure of nutrients exchange between AM and OM, some model of OM and their hosts could be proposed and investigated in depth with respect to interactive signaling pathways and the mechanisms of establishment of the symbiosis (Favre-Godal et al., 2020). Overall, these intracellular symbionts are crucial for the survival of several plant species, and will make a valuable contribution to broadening the biological and ecological strategies for use in agricultural applications in diverse and changing environments.
Some other plant–symbiotic fungi interactions were explored as well in recent years. It is generally known that Serendipita indica (formerly Piriformospora indica) can colonize a wide variety of terrestrial plants. The symbiotic relationship between S. indica and its host generally leads to a better crop productivity, strengthened tolerance against diverse stresses, and improved nutrient uptake and transport (Sharma and Varma, 2021). Therefore, the S. indica–host association can also serve as a reliable model system to explore the relevance of its interaction mechanism for crop improvements. Dark septate endophytes (DSEs) are a prominent part of the plant mutualistic symbiosis with plant roots, and are often capable of promoting tolerance of plants on certain abiotic stresses, such as low temperature (Chen et al., 2020), organic residues (He et al., 2020), and excess salinity (Yuan et al., 2021c). The DSEs show no host specificity and are capable of transferring nutrients from the soil into the plant cells. The benefits to hosts conferred by DSEs have emerged as an efficient strategy to augment the host growth and resistance given the low cost of culturing this type of microbe (Hidayat, 2019).
Pathogenesis
Parasitism sits at one end of the parasitism–mutualism continuum, taking place when after its colonization, the microbe obtains resources from its host and in the process, causes damage (Newton et al., 2010; Drew et al., 2021). Along the continuum, if the symbiont can obtain benefits from the host at no cost in the absence of any enemies, then this microbe is on course toward a parasitic lifestyle (Vorburger and Gouskov, 2011). Notably, the degree of damage incurred by the host and the extent of resource acquisition could be intensified depending on a nutritious environment for pathogens and driven by a great selection pressure to propagate given the limited life span of the microbe (Rafaluk-Mohr, 2019; Lekberg et al., 2021). Once the microbe overcomes all the defense barriers in its host plant, thus, putting the host into disease state, it is usually called a pathogen.
Pathogens will actively destroy hosts for their own nutritious benefits and their interaction with plants is another type of plant–endophyte interaction. For example, Pseudomonas syringae is one of the best-studied bacterial species for its pathogenicity and the molecular mechanisms underlying the plant–microbe interactions with model plants (i.e., Arabidopsis thaliana and tomato); adding to its relevance, P. syringae infects almost all important economic crops and threatens global crop production (Xin and He, 2013; Xin et al., 2018). Type III secretion system (T3SS) effectors (T3Es) are the main mediators for the infection and immune evasion of P. syringae. Therefore, many studies have concentrated on the anti-pathogenic and pathogenic mechanisms of T3Es. Besides the eight “core” T3Es that were already discovered (Cunnac et al., 2011), it was found recently that plasmodesmata are also controlled by T3Es for expanding the spread of the bacterium in the host plant (Aung et al., 2020). Additionally, understanding of the T3SS regulatory network based on the transcriptional network has led to the identification of new candidates of T3Es (Fan et al., 2020; Shao et al., 2021), as well as the mechanism by which pathogenic growth is inhibited (Nobori et al., 2020; Xing et al., 2021). Because infection by P. syringae is affected by external environmental conditions, complete knowledge of the multidimensional nature of plant–P. syringae–environment–microbiota interactions is urgently needed as it will help to prevent diseases of crop plants (Xin et al., 2018).
Rice is a staple food crop worldwide and a model plant for basic research, but its yields and quality are always influenced by various diseases, such as bacterial leaf blight and leaf streak caused by Xanthomonas oryzae pv. oryzae, or rice blast caused by Magnaporthe oryzae. To confer a broad-spectrum and a durable resistance in this cereal plant, currently, the best way forward is to select among the inherent resistance genes of rice to breed disease-resistant cultivars. In this respect, many discoveries have been made recently, such as the RNase P protein subunit Rpp30 with tRNA processing (Li W. et al., 2021), executor R proteins’ (Xa7 and Xa23 with EBEAvrxa23) response to T3Es of pathogens (Chen X. et al., 2021; Wei et al., 2021), and Bacterial Leaf Streak 1 (Ma Z. et al., 2021) which encodes a protein that acts against different pathogenic strains (Chen F. et al., 2021). As another economically important crop, wheat is susceptible to Fusarium head blight (Fhb) that is caused by several Fusarium strains. This disease severely limits wheat yields and causes the grains to accumulate deoxynivalenol, which poses problems for food safety and for the health of humans and animals (Bai and Shaner, 2004). To overcome the disease, broad-spectrum disease-resistant cultivars have been sought and bred by harnessing the Fhb7 gene found in endophytic Epichloë fungi and located in the genome of the grass Thinopyrum elongatum, via distant hybridization (Wang H. et al., 2020). The establishment of a metabolite-molecular regulatory network affected by Fhb in wheat could further enhance our understanding of the mechanism of this disease (Su et al., 2021).
Acquisition of Plant Stress Resistance From the Plant–Symbiont–Pathogen Relationship
In natural environments, certain microbiota assist plants in mitigating biotic and abiotic stress, mainly through two ways. The first is via horizontal gene transfer (HGT) (Li Y. et al., 2021). While transitioning from aquatic to terrestrial habitats, plants firstly underwent primary and secondary endosymbiosis involving HGT to withstand environmental challenges (Hiruma et al., 2016; Li et al., 2020). The HGT also took place outside the single evolutionary events of global significance: HGT The macro2 domain gene originated from mycorrhizal fungi and helped plants to adapt early on to life on land (Wang S. et al., 2020). The Fhb7 gene from endophytic Epichloë fungi, encoding a glutathione S-transferase for the detoxification of trichothecenes through de-epoxidation, has enhanced resistance of wheat to Fusarium head blight (Wang H. et al., 2020). On the other hand, the HGT of a syringopeptin synthetase homolog from Pseudomonas to Burkholderia glumae, the pathogen causing bacterial panicle blight, increased its virulence (Sai et al., 2021). The second is via a plant–symbiont–pathogen interaction that results in a continuous formation of novel defenses in hosts (Delaux and Schornack, 2021). In the course of evolution of host defense, plant–symbiont–pathogen interactions generated conserved gene modules, such as receptor-like kinases (Gong and Han, 2021), that are central to the plant innate immunity (Bentham et al., 2020). Notably, in the interaction with the fungal endophyte Colletotrichum tofieldiae, the fungus activates the plant’s PHT1 phosphate transporters which is beneficial under phosphorus-deficient conditions, while inducing plant defense by stimulating indole glucosinolate synthesis via the innate immune system (Hiruma et al., 2016). This would imply that symbiosis and pathogenesis can happen in the same interaction and also improved the host fitness.
The relationship between pathogens and hosts is not unlike an ‘arms race’ that reciprocally drives the immune system (mentioned above) and resistance to escalate because of the wide adaptability and heightened virulence of plant pathogens (Stringlis and Pieterse, 2021; Wang et al., 2021). However, plants could recruit some mutualistic microbes as partners to enhance their own immunity and to suppress pathogen infection. It seems then that microbes have adapted to be more cooperative with hosts for symbiosis and their co-existence confers better resistance to pathogens, which provides their host plants with superior growth and development over non-colonized ones (Batstone et al., 2020; McLaren and Callahan, 2020; Ma K.-W. et al., 2021). In other words, the plant, indeed, acts as a “microbial screening system” that distinguishes symbiotic microbes from pathogens.
Comparison of the Interaction Process Between Symbiotic Microbes and Pathogens
The microbial invasion of plants is divided into two stages: the “recognition” phase, where the microorganism actively grows toward the plant (indicative of host recognition) and grows externally on the surface, and the “colonization” phase, where the microbe invades the plant (Mukherjee et al., 2012; Vahabi et al., 2015b). The microbe must pass through four different host barriers: plant microbiota, physical cell barrier, and two plant immune systems, i.e., innate or microbe/pathogen-associated molecular patterns (M/PAMPs)-triggered immunity (PTI) and effector-triggered immunity (ETI), for a successful invasion to occur (Hacquard et al., 2017).
Recognition Phase
Plants possess a general stress response involving “core immunity responses” genes (Bjornson et al., 2021; Cole and Tringe, 2021), as well as the general “non-self” response (Maier et al., 2021). Plants possess various PRRs (pattern recognition receptors) that recognize M/PAMP ligands and initiate immune reactions. In response to microbial infection, plants can induce disease resistance genes that recognize pathogen-specific ligands. Both pathogens and symbionts can be recognized by PRRs because the M/PAMPs are not specific to pathogens; they include fragments of essential microbial protein and glycans.
To avoid recognition by the host plant followed by immune response, pathogens and symbionts have the evolved complex extracellular invasion strategies. Due to the similarity of pathogen and symbiont genomes (Reinhardt et al., 2021), common extracellular strategies exist between them, and they can be divided into three categories: avoiding accumulation of MAMP precursors, reducing hydrolytic MAMP release, and preventing MAMP perception (Buscaill and van der Hoorn, 2021), which can involve different microbial effectors. The effectors include the LysM effector (Sánchez-Vallet et al., 2020), salicylate hydroxylase (e.g., wheat and Fusarium graminearum) (Qi et al., 2019), cell wall degrading enzymes (CWDE) (A. thaliana and S. indica) (Hacquard et al., 2016), T3Es (e.g., A. thaliana and P. syringae), and EPS (e.g., the few basidiomycete-based lichen symbioses) (Wanke et al., 2021). For example, X. oryzae pv. oryzae secretes some effectors to upregulate the SWEET genes of rice so as to provide it with nutrition, especially AvrXa7 and AvrXa23 (Vera Cruz et al., 2000; Gupta, 2020), but they can be detected by Xa7 and Xa23 with EBEAvrxa23 in wild rice (Chen X. et al., 2021; Wei et al., 2021). Effector proteins are also secreted by oomycetes into the host to promote infection and suppress the immune system. This is exemplified by the “RxLR” motif family of effectors from Hyaloperonospora arabidopsidis, one of whose members, HaRxL21, interacts with the co-repressor Topless (Harvey et al., 2020), and an AM fungus whose secretion contains the LysM effector to subvert the chitin-triggered immunity response by the host plant (Zeng et al., 2020).
Furthermore, symbionts have developed various strategies to let their potential hosts better distinguish them from pathogens during the recognition phase. For example, lipochitooligosaccharide Nod Factors are perceived by legumes (Radutoiu et al., 2003; Bozsoki et al., 2020). In rice, short-chained chitotetraose triggers symbiotic signal transduction with the symbiotic complex receptor MYR1–CERK1, and this suppresses the formation of the CEBiP-CERK1 heteromer (Chiu and Paszkowski, 2021; Zhang et al., 2021). Besides, symbionts are also capable of colonizing hosts while overcoming the response to damage-associated molecular patterns (DAMPs) and MAMPs, while a response against pathogens is possible in the presence of non-pathogenic microbes (Zhou et al., 2020). Also, symbionts could induce jasmonic acid (JA) and suppress salicylic acid (SA) formation to induced systemic resistance (ISR), whereas pathogens typically enhance the SA biosynthesis to mediate systemic acquired resistance (SAR) in plants (Martínez-Medina et al., 2017).
Colonization Phase
After overcoming a plant’s microbiota and physical barriers, microbes will move across the different cell layers of the host and this movement elicits the response of cell-layer-specific programs of the plant (Fröschel et al., 2021). At this point in the colonization process, the growth of some pathogens will be suppressed by the endodermal barrier, while some will utilize molecular decoys to protect themselves (Ma Z. et al., 2017; Xia et al., 2020) and to penetrate the adjacent cell regions by regulating the pore size of plasmodesmata (Huang et al., 2019). Meanwhile, some pathogens inevitably trigger the NLR-mediated immunity involving a PTI-ETI crosstalk (Ngou et al., 2021; Yuan et al., 2021a,b). Finally, those that slip past and evade the immune response of the host will colonize it further to cause a disease.
For arbuscular mycorrhizal fungi (AMF), two receptor-like kinases called arbuscular receptor-like kinase 1 (ARK1) and ARK2 are employed to maintain the symbiotic interaction (Montero et al., 2021). In turn, plants employ a liquid-liquid phase separation to compartmentalize cellular activities in membrane-less organelles, protecting their microbial symbionts from the immune-response (Shin and Brangwynne, 2017; Huang et al., 2021). Plant autoimmunity and other physiological responses occur in the nucleus, leaving symbionts able to dwell safely in the intracellular matrix. Meanwhile, AMF are separated from the plant cytoplasm by a specialized host-derived membrane, which represents the main interface facilitating the bidirectional exchange of nutrients and information. The biosynthesis of this periarbuscular membrane is controlled by a gene called glucosamine inositol phosphorylceramide transferase 1 (Moore et al., 2021). The endophytic fungus S. indica secretes specific proteins to improve its access to micronutrients and to influence oxidative stress and reactive oxygen homeostasis to support the colonization of the host plant (Nostadt et al., 2020). A bacterial endophyte can utilize the hyphae of a fungal pathogen to gain access from the soil to plant roots, thereby protecting the host from infection (Palmieri et al., 2020), while others may use Type II secretion systems to modulate PTI to colonize the host (Teixeira et al., 2021). Symbiotic and pathogenic microbes do share some similar characteristics, but they also differ in certain ways; hence, plant–microbe interaction is a complicated phenomenon that requires multi-omics technologies for its in-depth research.
Background and Progress in the Development of Multi-Omics Technology
Omics can be used to perform a global analysis of biological samples in a high-throughput manner. In the early phase of molecular biology research, the invention of Sanger sequencing was a revolutionary breakthrough. However, it is only suitable and accurate for the study of short sequences. Since then, it has been improved to enable the sequencing of the first human genome, making it a great contribution to the treatment of medical diseases. However, the cost and time spent when using Sanger sequencing were enormous, which limits its accessibility and use by most studies. In stark contrast, next generation sequencing (NGS) allows for genetic sequencing in a high-throughput and less expensive way, thereby enabling the transcriptomes and genome of a species to be studied in detail. Nevertheless, the assembled sequence length is not sufficient to resolve the whole genome’s information. The development of third-generation sequencing, which is capable of long-read sequencing, is therefore a giant step forward in overcoming that problem (van Dijk et al., 2018). The continuous development of sequencing technology facilitates the discovery of new genes and makes the construction of regulatory networks of known genes possible. Meanwhile, other omics methodologies have been developed and are being used across multiple fields of research. Among these, metabolomics is a useful tool to identify and to quantify the full set of metabolites in an organism, which could be then related to its phenotypic features under dynamic circumstances.
Single omics provides a view of the biological processes limited to a single level and the analysis is, nonetheless, limited to correlations, which may generally conclude with consequential variation rather than causative ones. We now need to acquire global information integrated via several single omics technologies, which is called the multi-omics approach. For example, an interactome, generated by at least genomics and transcriptomics, is essential for studying mechanisms related to plant–microbe interactions. Genomic sequencing provides the pre-requisite genomic information, while the transcriptome findings link the gene function with specific conditions, and the metabolome information correlates the vast array of metabolites involved in multiple pathways, which in turn reflects the occurrence of corresponding gene expression events and their levels (Qin et al., 2016). When combined with other biological data, a genetic, protein, and metabolite multi-omics study is a complementary approach that can be utilized to decipher the individual signal molecules, proteins, genes, and gene regulation cascades, and their relationship in gene networks/pathways, thereby deepening our understanding of, e.g., the microbe-mediated mitigation of stresses in plants (Yin et al., 2014; Luan et al., 2015).
Transcriptomics Technology
The sequencing of mRNA is a useful approach to study the responses of different plants in relation to their endophytes, by generating the transcriptome-level information (Sheibani-Tezerji et al., 2015). The transcriptome analysis involves comparing endophyte-free (E−) and endophyte-inhabited (E+) plants to understand the basis of endophyte-mediated host responses. An adjustable transcriptomic workflow suitable for the study of plant–microbe interactions is summarized in Figure 1 (Conesa et al., 2016). Transcriptional profiling can be done using various methods, including microarray analysis, SOLiD-SAGE, and RNA sequencing (RNA-seq) (Table 1). In particular, RNA-seq has many recognized strengths: it allows differential expression analysis under different conditions, simultaneous detection of microbial and host gene expression, and the discovery of novel RNA candidates. Microarray analysis and SOLiD-SAGE are useful tools for conducting a differential gene expression analysis. With the emergence of third-generation sequencing, one can readily perform a global transcriptome analysis and obtain a host-specialized transcriptome, even when using a single-cell RNA sequencing and a spatial transcriptome to accurately study the transcriptional reaction of each cell. Through transcriptomic analysis, we can now uncover the dynamics and regulation of actively transcribed genes, thus, offering considerable advantages vis-à-vis a genomic analysis alone (Levy et al., 2018).
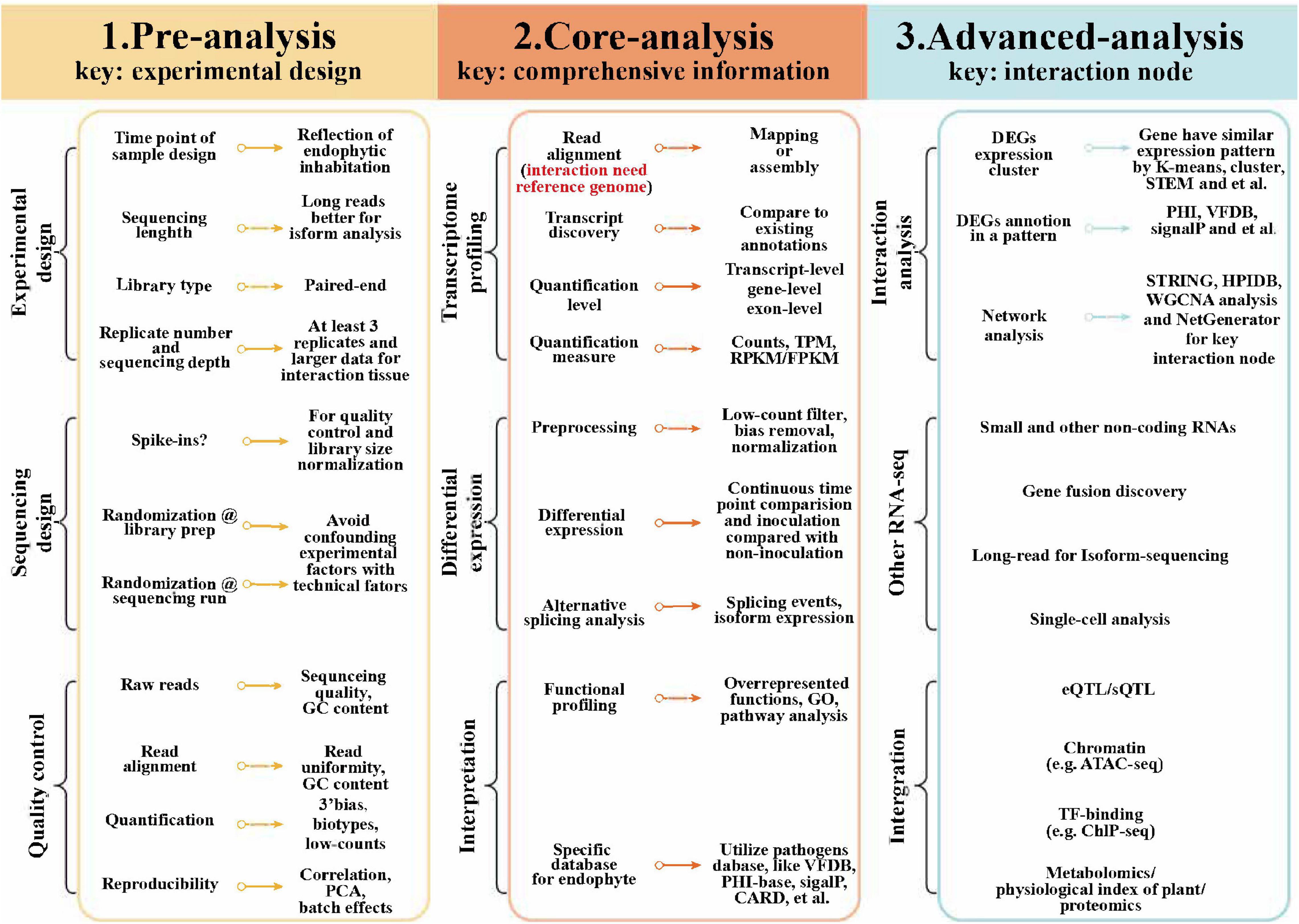
Figure 1. Transcriptomic workflow for plant–endophyte interaction studies. DEG, differential expressed gene; ChIP-seq, chromatin immunoprecipitation sequencing; eQTL, expression quantitative loci; FPKM, fragments per kilobase of exon model per million mapped reads; GSEA, gene set enrichment analysis; PCA, principal component analysis; PHI, pathogen host interactions; RPKM, reads per kilobase of exon model per million reads; sQTL, splicing quantitative trait loci; TF transcription factor; TPM, transcripts per million; HPIDB, host–pathogen interactions database; VFDB, virulence factors database; STRING, search tool for the retrieval of interacting genes; and WGCNA, weighted correlation network analysis.
Despite that, genome-based studies constitute a pivotal foundation for performing successful transcriptomic studies. Thus, pursuing genome and transcriptome analyses in tandem is more helpful for decoding the nature of plant–endophyte interactions (Kaul et al., 2016).
Metabolomics
Knowledge of plant–microbe interactions has increased substantially in recent years; however, the chemical communication leading to the priming is not yet well understood. Metabolomics, an array of advanced bioanalytical techniques in conjunction with chemometrics and bioinformatics tools, enables the characterization of perturbations to the metabolomes of interacting organisms (Mhlongo et al., 2018), thus providing a complement for other “-omics” techniques. Metabolomics can facilitate a greater understanding of the independent metabolisms of the plant and its endophyte, as well as the metabolic crosstalk that underpins the interactome (Jorge et al., 2016; Ofaim et al., 2017).
Several techniques are available to characterize metabolites extracted from fungi. An adjustable metabolomics flowchart includes sample preparation, data acquisition, data mining and analysis, statistical modeling, signatory biomarkers, and biochemical interpretation (Figure 2; Mhlongo et al., 2018). Different researchers have used different methods for metabolite profiling (Table 2). To separate and identify compounds, many detection techniques are widely used, such as column chromatography (CC), flash chromatography (FC), Fourier transform infrared spectroscopy (FTIR), thin layer chromatography (TLC), gas chromatography–mass spectrometry (GC-MS), liquid chromatography–mass spectrometry (LC-MS), LC-MS/MS, liquid chromatography-ultraviolet, and visible spectrum or diode array detection [LC-UV (DAD)], different types of high-performance liquid chromatography (HPLC), gas–liquid chromatography, nuclear magnetic resonance (NMR) imaging, and liquid chromatography/time of flight-mass spectrometry (LC-TOF/MS) (Khoomrung et al., 2017; Maheshwari, 2017; Xie et al., 2017). When traditional MS is not able to accurately detect spatial-temporal occurrences of metabolites, the spatial metabolome may be exploited based on desorption electrospray ionization-imaging mass spectrometry (DESI-IMS) and matrix-assisted laser desorption/ionization-IMS (MALDI-IMS), as well as using an air flow-assisted desorption electrospray ionization mass spectrometry imaging (AFADESI-MSI) (He et al., 2018).
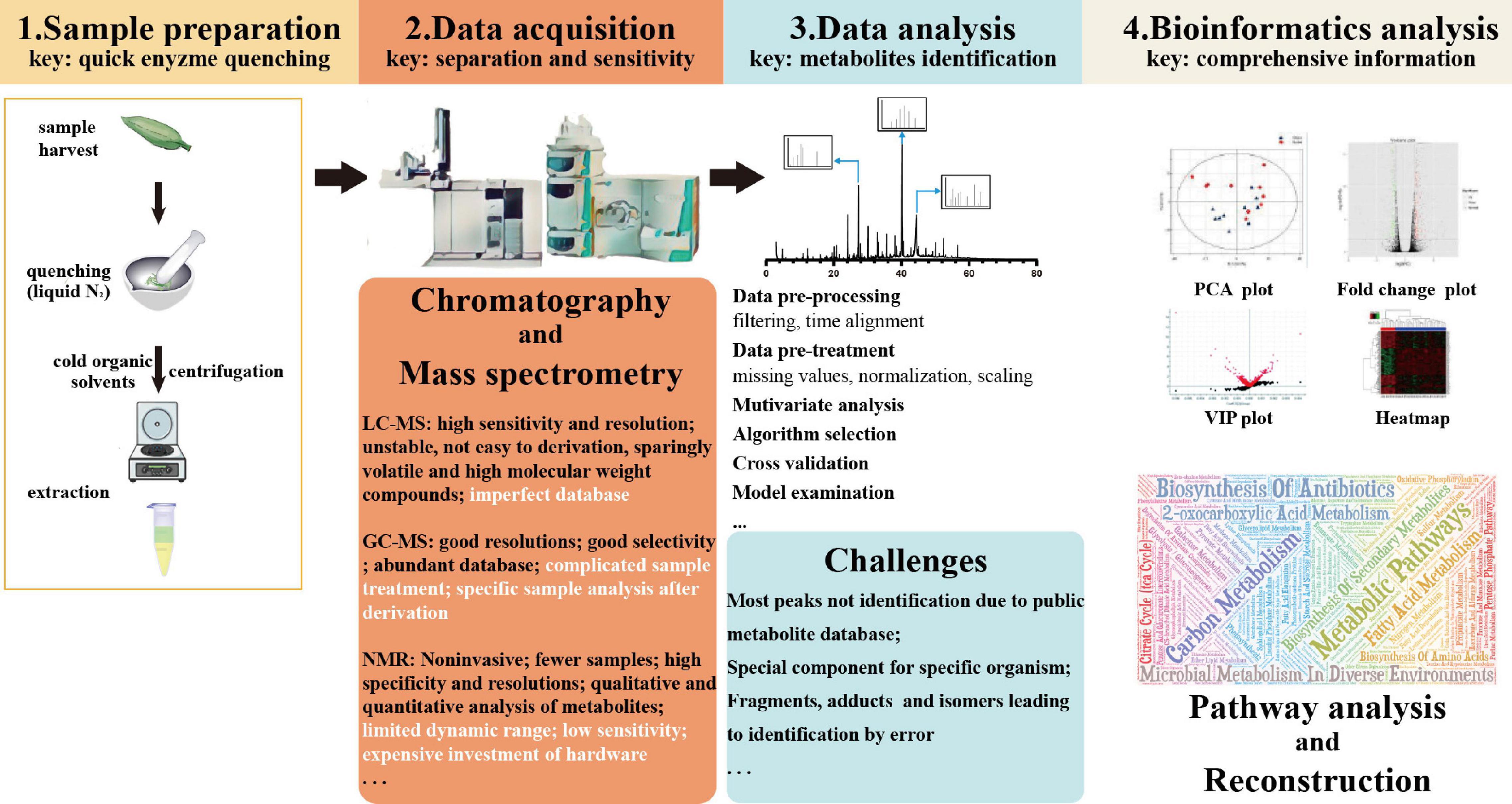
Figure 2. Flowchart for plant metabolomic studies. N2, nitrogen; LC-MS, liquid chromatography-mass spectrometry; GC-MS, chromatography-mass spectrometry; NMR, nuclear magnetic resonance; PCA, principal component analysis; and VIP, variable importance for the projection.
Transcriptomics and Metabolomics Applied to Plant–Endophyte Interactions
Numerous reports have elucidated how endophytes can promote plant growth, accelerate metabolism, and strengthen stress resistance in hosts. However, plant–endophyte interactions require complex and involve various response mechanisms. Nevertheless, it is now feasible to glean in-depth knowledge of these mechanisms by implementing transcriptomic and metabolomic techniques.
The Plant–Endophyte Interaction: From Inhabitation to Stress Resistance
Transcriptomic studies have addressed different aspects of the plant–endophyte interaction, spanning endophyte inhabitation (recognition and colonization) to stress resistance (Figure 3).
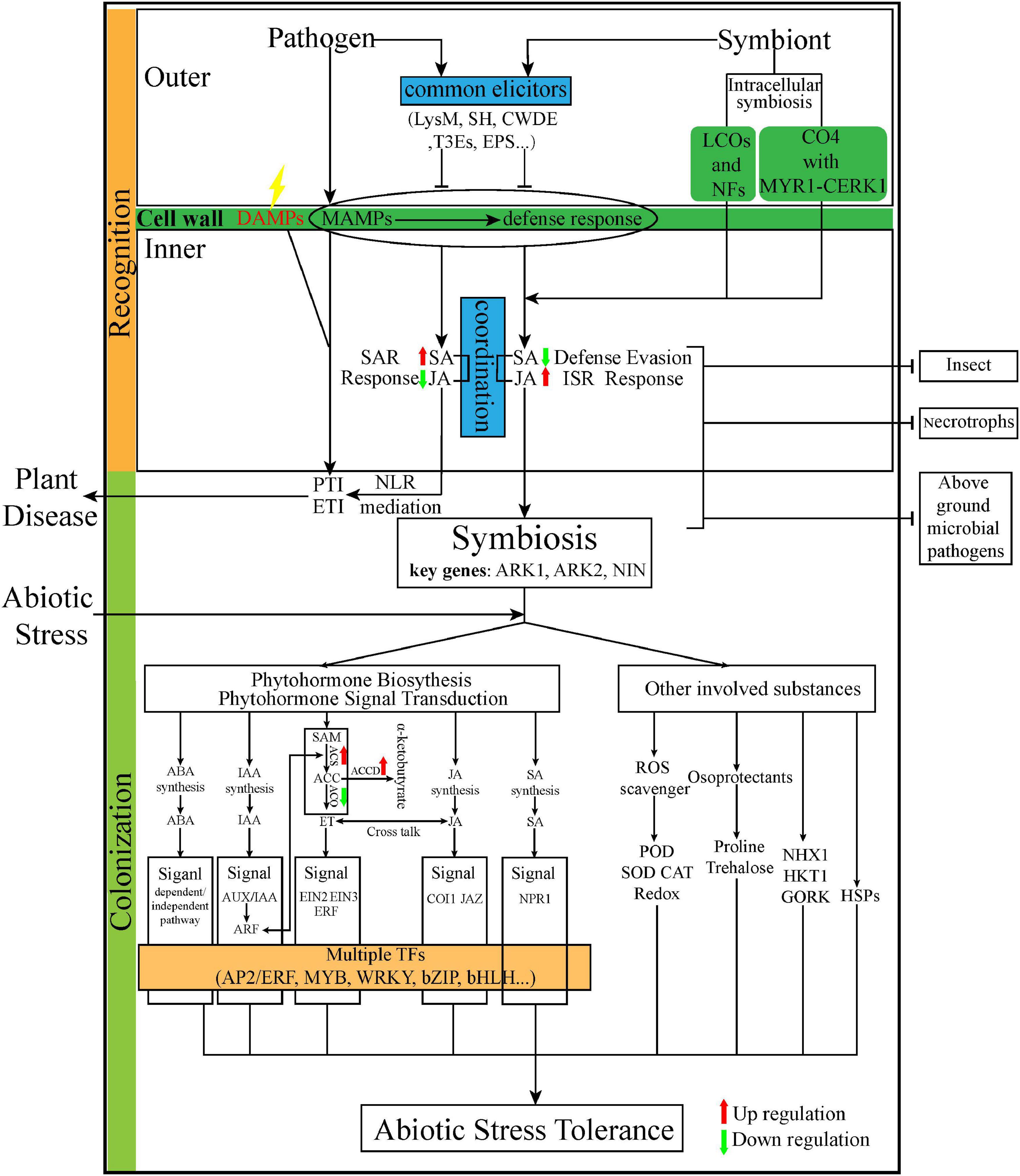
Figure 3. The summarization of the interaction from infection to tolerance of diverse abiotic stress. Red dotted arrow represents that elicitors/effector are one step ahead of microbes. Red and green arrows represent up and downregulation of genes or compounds, respectively. LysM, lysine motifs; SH, salicylate hydroxylase; CWDE, cell wall degrading enzymes; T3Es, Type III secretion system effectors; NFs, Nod factors; LCOs, lipochitooligosaccharides; CO4, short-chain chitotetraose; CERK1, chitin elicitor receptor kinase 1; SAR, systemic acquired resistance; ISR, induced systemic resistance; NLR, nucleotide-binding leucine-rich repeat receptor; PTI, pathogen-associated molecular patterns (PAMPs)-triggered immunity; ETI, effector-triggered immunity; ARK, arbuscular receptor-like kinase; NIN, nodule inception transcript factor; SA, salicylic acid; JA, jasmonic acid; ABA, abscisic acid; IAA, indole-3-acetic acid; ET, ethylene; SAM, S-adenosyl methionine; ACC, 1-aminocyclopropane-1-carboxylate; ACS, ACC synthase; ACO, ACC oxidase; ACCD, ACC deaminase;NPR1, non-expressor of pathogenesis-related genes 1 ROS, reactive oxygen species; POD, peroxidase; SOD, superoxide dismutase; CAT, catalase; NHX1, Na+/H+ antiporter; HKT1: high-affinity K+ transporter 1; GORK, gated outwardly rectifying K+ channel; HSP, heat shocked protein; and TFs, Transcriptional Factors.
Recognition Phase
During the recognition phase, endophytic microbes exhibit different mechanisms of interaction with hosts to evade host defenses before colonization. For example, a number of fungal endophytes mask chitin to forestall further defensive reactions (Sánchez-Vallet et al., 2013, 2015), to repress plant SA production (Navarro-Meléndez and Heil, 2014), and to promote JA biosynthesis (Straub et al., 2013; Pinski et al., 2019). Some endophytes employ miRNA-targeted regulating pathways to turn off the defense system in the host plant (Plett and Martin, 2018). In the recognition phase, compounds secreted by endophytes enter the host as messengers before microbial entry, to promote plant growth (Jiang C. H. et al., 2019; Lu et al., 2019), and they activate both stress and defense responses against pathogens (Vahabi et al., 2015b; Lu et al., 2019). In a study of Arabidopsis inoculated with S. indica, after this physical contact, the fold-change in the JA content was much lower than that of SA, reversing the situation that existed before the physical contact (Vahabi et al., 2015a,b). A recent study of the Trichoderma virens–maize interaction (Malinich et al., 2019) showed that T. virens reduced a plant SA formation. Meanwhile, the reduced SA formation prevented the production of catechol from SA, which increased the plant susceptibility to pathogen attack (Ambrose et al., 2015), while JA biosynthesis genes were upregulated to stimulate the establishment of ISR against necrotrophs. The SA levels in colonized plants, however, were not significantly different from those in non-colonized plants (Ambrose et al., 2015). That is, the physical contact between the two symbionts might not result in the increase of SA levels, but it may nonetheless suffice to establish a systemic defense, depending on the coordination between SA- and ethylene/JA-induced plant defenses (Tsuda et al., 2009; Hillmer et al., 2017; Constantin et al., 2019).
Colonization Phase
After overcoming a plant’s competing microbiota and physical barriers, endophytes may adopt different strategies to prevent themselves from being targeted by host defense responses. Endophytic bacteria format their own MAMPs (Vandenkoornhuyse et al., 2015) or produce enzymes in response to the immune system of their host (Trdá et al., 2015), while fungal endophytes rely on secreting chitin deacetylase or fungal lectins (Lahrmann and Zuccaro, 2012). In contrast to pathogenic microbes which elicit SAR through SA signaling, endophytes regulate JA/ET signaling to induce ISR which enhances the tolerance of above-ground plant parts against biotic stress (Bastias et al., 2017; Constantin et al., 2019; De Palma et al., 2019). Further, they limit their own colonization space by not “overstepping” and “overpowering” the plant in the critical colonization phase.
As the colonization phase continues, the endosymbiotic relationship that confers benefits to both organisms is developed. Host plants maintain endophytic communities in their specific tissues by supplying them with nutrients (Hao et al., 2017). Early in colonization, hosts will assist endophytes in colonization by promoting the expression of genes related to motility, chemotaxis, and adhesion by secreting more attachment proteins, cell-wall re-modeling proteins, and ABC transporters (Coutinho et al., 2015; Pankievicz et al., 2016; Sarkar et al., 2017). For instance, Azoarcus sp. BH72 released the type V autotransporters Azo1684 and Azo1653 for its establishment in rice hosts (Sarkar et al., 2017). Then, in turn, the hosts provided tissue sites suitable for endophytic growth (Utturkar et al., 2016; Schmid et al., 2017). This phenomenon indicates that a co-cultured endophyte displays more upregulated genes capable of promoting growth fitness of both organisms than non-co-cultured endophytes (Ambrose and Belanger, 2012; Yi et al., 2017; Zhou et al., 2017; Trovero et al., 2018).
Colonization With Promotion of Plant Growth and Metabolite Production
Endophytic microorganisms can promote growth of a plant in addition to accelerating its production of metabolites and enhancing its tolerance to stress. Numerous endophytes promote plant growth, even under stress conditions for hosts, because of this complicated interaction. Vahabi et al. (2015b) reported that the plant gene encoding NRT2.5, which belongs to the nitrate transporter family, was upregulated by approximately fourfold in the root-to-shoot signal transduction process after co-cultivation of Arabidopsis with S. indica, and this stimulated the growth of Arabidopsis. A DSE strain, S16, promoted the growth of sweet cherry by regulating nitrate uptake and plant hormone biosynthesis pathways (Wu et al., 2021). Another DSE strain, T010, also positively influenced plant genes involved in phytohormone and flavonoid metabolism to enhance host plant growth (Wu et al., 2020). The presence of S. indica also led to better root development by increasing the expression of indole-3-acetic acid (IAA) biosynthesis genes AXR1 and AUX1 in barley (Ghaffari et al., 2016), and the upregulation of genes encoding enzymes related to fatty acid biosynthesis, which resulted in better reproductive growth and improved oil quality of Brassica napus (Su et al., 2017). Inoculation of banana (Musa acuminata) with the plant growth promoting (PGP) rhizobacterium Bacillus amyloliquefaciens Bs006 led to the induction of many genes involved in nutrient absorption and upregulation of the plant cytochrome C (CYTC) gene in common with Pseudomonas fluorescens Ps006 which improves for root development (Gamez et al., 2019). Endophytic fungi isolated from Noccaea species enhanced the expression of ion transporters and fatty acid biosynthesis related genes to promote plant growth (Ważny et al., 2021).
Apart from the upregulated genes related to plant growth, an endophyte may also influence plants’ primary and secondary metabolite accumulation to increase plant growth. It is known that medicinal plants contain abundant metabolites and endophytes. Some medicinal plants, such as Salvia miltiorrhiza, ginseng, and Echinacea purpurea, have been studied as model plants for their interactions with endophytes (Maggini et al., 2020; Chu and Bae, 2021; Lu, 2021). In research on S. miltiorrhiza Bunge, the endophytic fungi U104 was able to induce the accumulation of tanshinone in the host after its infection (Jiang Y. et al., 2019). Although the endophytic interaction with medicinal plants is rarely studied at the transcriptomic level, some studies have linked these two phenomena. For example, Li et al. (2017a,b) reported that the increment of two metabolite types was associated with thickening of the stem, in comparison with non-inoculated plants. The polysaccharide level in Dendrobium nobile rose when D. nobile was co-cultivated with MF23 (Mycena sp.), as genes involved in photosynthesis, sucrose, fructose, and mannose metabolism were induced, whereas those involved in glycolysis and citrate cycle (TCA) were repressed (Li et al., 2017b). Also, MF23 might stimulate dendrobine biosynthesis by regulating the expression of key genes involved in the mevalonate (MVA) pathway, such as AACT, MVD, and PMK. Moreover, it was found that Ceratobasidium sp. AR2 was able to hasten the accumulation of flavonoids in Anoectochilus roxburghii by upregulating flavonoid biosynthesis genes (Zhang et al., 2020). In Atractylodes lancea, inoculation with Gilmaniella sp. AL12 upregulated sesquiterpenoids biosynthesis to improve that plant’s growth (Yuan et al., 2019).
A comparison of microbial effects confirms the growth benefits to hosts from endophytes. Manganiello et al. (2018) used the endophytic fungus Trichoderma harzianum and pathogenic fungus Rhizoctonia solani to invade tomato, finding that levels of steroidal glycoalkaloids were increased in the T. harzianum-colonized tomato, relative to the R. solani-colonized host after ingress into the host, which favored plant growth and defense. Besides growth stimulation, the accumulation of bioactive compounds in response to colonization by endophytes also assisted their host in responding to environmental stresses (Herburger et al., 2019; Yan et al., 2019).
Colonization With Enhancement of Abiotic Stress
Stress tolerance can also be enhanced by the modulation of phytohormones and reactive oxygen species (ROS) as induced by co-cultivation of plants with endophytes. Under non-stressed conditions, the inoculated endophyte positively regulates stress response genes of the host. The Trichoderma asperellum-treated rice plants showed increased expression levels of genes involved in photosynthesis, cell wall, and ROS modulation systems (Doni et al., 2019). Another study showed that, upon infection by H. seropedicae, rice activated iron uptake for the promotion of iron homeostasis, and also stimulated the expression of bacterial genes involved in nitrogen fixation, cell motility, and cell wall synthesis; conversely, certain plant defense genes were repressed, which allowed the endophyte to enter and rapidly colonize the intercellular space and xylem (Brusamarello-Santos et al., 2019). Moreover, in barley, endophytic Serendipita vermifera led to an extension of the plant protection barrier via the specific induction of genes involved in detoxification and redox homeostasis (Sarkar et al., 2019). Similarly, Epichloë coenophiala affected the expression of various WRKY transcription factors associated with the enhanced resistance in its host Lolium arundinaceum (Dinkins et al., 2017). Taken together, these studies suggest that the symbiosis continues to foster stress resistance in the host.
Stress phytohormones, including their biosynthesis pathways and signal transduction, are major endogenous effectors that modulate plant physiological responses, leading to environmental adaptation. The ROS, which are generated under a variety of abiotic stresses, accumulate as defense activators and act as signaling molecules in plant defense responses at low levels. Under salinity stress, endophytic microorganisms mainly induce genes involved in biosynthesis and signaling of phytohormones (auxin, JA and ET), iron transport, and ROS scavenging (Brotman et al., 2013; Ghaffari et al., 2016; Liu et al., 2017; Qin S. et al., 2018; Eida et al., 2019). Slight differences in various endophyte–plant interactions have also been identified. Colonization by Glutamicibacter halophytocola increased the biosynthesis of lignin and flavonoid in leaves (Qin S. et al., 2018). Furthermore, colonization by B. amyloliquefaciens was found to upregulate the 1-aminocyclopropane-1-carboxylate (ACC) synthase and ACC oxidase genes in conjunction with the abscisic acid (ABA)-independent pathway of salt tolerance in A. thaliana (Liu et al., 2017). Under drought stress, endophytic fungi function differently from endophytic bacteria. For example, S. indica enhanced auxin, ABA, SA, and cytokinin levels to enhance the expression of genes involved in the drought stress response of maize hosts (Zhang et al., 2018), while T. harzianum improved drought tolerance in rice by modulating the activity of genes for aquaporin and dehydrin, dehydration responsive element binding protein, and superoxide dismutase (SOD) (Pandey et al., 2016). However, in sugarcane plants treated with endophytic bacteria, Gluconacetobacter diazotrophicus has suppressed auxin, ABA, and ethylene (ET) hormone pathways in the roots, and activated only the genes involved in ABA-dependent signaling in the shoots (Vargas et al., 2014). In A. thaliana colonized by Pseudomonas chlororaphis, plant defense-related genes were activated and genes involved in drought response signaling (ABA and ET) were downregulated (Cho et al., 2013). In response to inoculation with Burkholderia phytofirmans, potato plants increased their expression of genes involved in extracytoplasmatic function group IV sigma factors, oxidative phosphorylation, transcription regulation, cellular homeostasis, and cell redox homeostasis (Sheibani-Tezerji et al., 2015). Moreover, in a study of the interaction between E. coenophiala and tall fescue, the endophyte had a small but potentially important effect on its host transcription that may enhance the plant resistance to stress, which entailed the repression of some genes involved in defense against fungi coupled with the priming of genes that may strengthen drought tolerance in various fungal strain/plant genotype combinations under stress conditions (Dinkins et al., 2019). Yet, surprisingly, little is known about the effects on other abiotic stresses, including cold, heavy metal, and heat stress. Epichloe gansuensis increased the biosynthesis of alkaloids and unsaturated fatty acids during the seed germination of Achnatherum inebrians, thereby increasing tolerance to cold stress (Chen et al., 2016), while B. phytofirmans induced the upregulation of some cold stress-related genes by Vitis vinifera (Theocharis et al., 2012). The endophytic smut fungus Thecaphora thlaspeos, a biotrophic pathogen with balanced virulence, has the ability to overwinter with its perennial hosts while seemingly suppressing cold acclimation (Courville et al., 2019). In plants colonized by Curvularia protuberata that is infected with Curvularia thermal tolerance virus, imposition of heat stress increased the expression levels of genes encoding trehalose phosphatase, betaine aldehyde dehydrogenase, taurine catabolism dioxygenase, and scytalone dehydratase (Morsy et al., 2010), while an endophytic Thermomyces strain induced the expression of phenylalanine ammonia-lyase in cucumber under heat stress (Ali et al., 2018). Under heavy metal stress, endophyte–plant interactions can affect several pathways, including metal binding and transporting processes; organic acid metabolic processes; and transporting, transcription factor, sulfate assimilation, DNA repair, and cell-wall metabolic processes (Zhao et al., 2015). The ROS scavengers, including glutathione S-transferases, peroxidase, SOD, and catalase, work to detoxify ROS and to balance the redox homeostasis of the hosts under stress conditions. In addition, levels of heat shock proteins, pyrroline-5-carboxylate synthase and its product, proline, as well as trehalose-6-phosphatase phosphatase and its product, trehalose, were enhanced in symbiosis when compared with non-inoculated plants under the above-mentioned stress conditions (Morsy et al., 2010; Dinkins et al., 2019).
These interconnected modules, pathways, compounds, and system-level patterns of molecular plant–endophyte interactions, from recognition to colonization, have been dissected partially by transcriptomics. However, the sophisticated underlying mechanisms involved in these symbioses make it challenging to distinguish the direct contribution of genetics from indirect factors, including determinants that enable plants to differentiate between endophytes and pathogens and to process information, to stimulate interactome networks, and to engage in signal transduction (Rodriguez et al., 2019). Meanwhile, transcriptomic studies are still constrained by experimental design, transcriptomic technology, costly analysis, and data storage. Furthermore, in some cases, gene annotation is a formidable endeavor due to the lack of high-quality reference genomes, especially those of co-expressed genes from hosts and endophytes, which may be difficult to identify in planta due to, e.g., the low abundance of bacterial mRNA (Levy et al., 2018). The study of molecular mechanisms of plant–endophyte interactions is nascent and further investigation is needed to deepen our understanding.
Metabolomics Applied to the Plant–Endophyte Interaction
Modulation of elemental stoichiometry and the metabolome are promising mechanisms to study plant–endophyte interactions (Rivas-Ubach et al., 2012). Quantitative metabolomics studies permit the analysis of root exudates and microbial signal molecules with high precision (Oldroyd and Downie, 2008; Inceoğlu et al., 2011; Peiffer et al., 2013), thus, clarifying how diverse biomolecules participate in a given cascade and the ensuing response of different microbes. Metabolomic research on plant–endophyte interactions indicates that endophytes, directly or indirectly, can synthesize plant growth regulators, plant secondary metabolites, and defense compounds, all of which are closely connected.
Growth-Promoting Metabolites
Endophytic microbes can secrete their own components to facilitate various plant growth regulators, such as IAA, cytokinins, and gibberellins, and also siderophores (Swarnalakshmi et al., 2019). Generally, endophytes may produce phytohormones or phytostimulants and signaling compounds to enhance the growth of their host plants (de Santi Ferrara et al., 2012; Khan et al., 2012; Mehmood et al., 2019; Table 2). Trichoderma can produce auxins (Angel Contreras-Cornejo et al., 2009) and secrete auxin-like substances, the plant growth regulator harzianolide, and compounds of unknown function like 6-pentyl-a-pyrone (Vinale et al., 2008). The B. amyloliquefaciens RWL-1, a PGP endophyte, can produces high levels of IAA by utilizing methanol (Shahzad et al., 2019). In a recent study of Bacillus sp. JC03 secreta, GC-MS identified volatile organic compounds, some of which, 2-ethyl-1-hexanol, tetrahydrofuran-3-ol, and 2-heptanone, affected phytohormone biosynthesis and metabolism to promote plant growth (Jiang C. H. et al., 2019); extracts of Paecilomyces variotii had the same effect but also promoted plant defense (Lu et al., 2019). In marine algae, co-cultivation with S. indica led to a significant modulation of the amounts of metabolites belonging to the GABA shunt pathway, to ameliorate biomass and lipid production (Bhatnagar et al., 2019). When tall fescue was co-cultivated with different E. coenophiala strains, root exudate composition was affected regarding lipids, carbohydrates, and carboxylic acids, and plant growth was increased (Guo et al., 2015). Numerous studies have shown that endophytes can produce siderophores which are then exploited by many plants, such as cowpea, silver birch, and black alder (Maksimova et al., 2018; Albelda-Berenguer et al., 2019). For example, P. fluorescens C7 improved iron supply to A. thaliana by producing siderophores, thereby promoting plant growth (Vansuyt et al., 2007).
Host-Related Metabolites
The chemical diversity of plants is enormous. Plants’ evolution has enabled the biosynthesis of a cornucopia of novel chemicals to enable them to survive and communicate in a complex ecological environment (Wurtzel and Kutchan, 2016). However, metabolome studies tend to focus mainly on small molecules found in a biological sample. Secondary metabolites are small organic compounds that are not only directly involved in normal growth but are also implicated in important plant functions (Table 2). Various endophytes produce different secondary metabolites and mediate an elicitor-induced increase in secondary metabolite biosynthesis in specific species and organs, during different stages of plant development (Zhai et al., 2017). Early work by Stierle et al. (1993) reported that Taxomyces andreanae isolated from the bark of Taxus brevifolia produced taxol during axenic conditions in vitro, thus, drawing attention to the compounds produced by endophytes (Jia et al., 2016). Fungal inoculations were shown to promote the content of reducing sugars, total flavonoids, total phenols, trans-resveratrol, and activities of phenylalanine ammonia-lyase in both leaves and berries of grapevine, such that inoculation with the endophytic fungal strains CXB-11 (Nigrospora sp.) and CXC-13 (Fusarium sp.) affected the metabolome of grapes much stronger than other fungal strains (Yang et al., 2016; Huang et al., 2018). Two endophytes originally isolated from grapevine roots, B. amyloliquefaciens SB-9 and P. fluorescens RG11, increased the melatonin synthesis as well as abundance of the intermediates 5-hydroxytryptophan, serotonin, and N-acetylserotonin (Jiao et al., 2016). This co-metabolism between plant and endophyte has significant implications in studies of biochemical pathways, particularly those involved in the metabolism of polycyclic and polyaromatic compounds (Horvath, 1972; Chauhan et al., 2008). In a study of the full metabolic potential the endophytic fungus Botryosphaeria mamane by addition of histone deacetylase inhibitors, a broad spectrum of metabolites could be identified (Triastuti et al., 2019). An investigation of the response of different members of the plant genus Hypericum to 18 biotic elicitors revealed that elicitors of Fusarium oxysporum and Trichoderma crassum predominantly stimulated the synthesis of naphthodianthrones and emodin, while those from S. indica promoted phloroglucinol production (Bálintová et al., 2019). Furthermore, compared to co-cultivation with S. indica, its cell wall extract acted as a more potent elicitor for asiaticoside production in Centella asiatica (Jisha et al., 2018). Taken together, these studies suggest that microbial endophytes modulate the content and type of secondary metabolite production in their host plants (Lucini et al., 2019).
Plant Stress Resistance-Related Metabolites
The scope of metabolomics involves the characterization of all metabolites of an organism under the influence of local environmental conditions. The metabolome varies greatly with alterations in the surrounding environment, which induce direct physiological changes correlated with diverse pathways in an organism (Bundy et al., 2005; Table 2). A study of time-series experiments with A. thaliana suggested that metabolic activities respond more quickly than that of transcriptional activities to abiotic alterations in real time (Caldana et al., 2011), demonstrating that metabolome studies are of vital importance. Under saline conditions, IAA concentrations of 0.22–25.58 μg mL–1, which can be produced by Pseudomonas sp., Rhizobium sp., Enterobacter sp., Pantoea sp., Marinobacterium sp., Acinetobacter sp., and Sinorhizobium sp., were shown to influence germination and seedling growth in wheat (Sorty et al., 2016). In another study of wheat under water deficient conditions, two endophytes, Acremonium sclerotigenum and Sarocladium implicatum, reduced the levels of stress damage markers and lessened the accumulation of metabolites involved in stress adaptation, particularly via changes to 2-oxocarboxylic acid metabolism and ABA levels (Llorens et al., 2019). The S. indica improved the adaptation of barley to drought stress by regulating amino acid and soluble sugar metabolism (Ghaffari et al., 2019). Some endophytic bacteria can solubilize phosphorus and/or potassium; for example, strains of Bacillus sp., which have phosphate-solubilizing potential, successfully improved the yield and quality of fennel in semiarid saline soil (Mishra et al., 2016). In other studies, Nagabhyru et al. (2013) used ion-exchange chromatography and LC-MS to analyze three colonized and non-colonized clones of tall fescue; colonization affected metabolite levels in that higher levels of free glucose, fructose, trehalose, sugar alcohols, proline, and glutamic acid were found in both shoots and roots after 2–3 days of water deprivation. In addition, asexual Epichloë endophytes with the perA gene for biosynthesis of the insect feeding deterrent peramine, conferred plant resistance to herbivory (Hettiarachchige et al., 2019).
Endophytes provide a rich source of novel bioactive natural products and their interaction with plants under specific conditions can induce the accumulation of numerous metabolites, which can facilitate the growth of plants and stress resistance. To identify those alterations of metabolite profiles induced either within a system or by a specific gene, metabolomic technology offers a powerful tool to quantitatively and qualitatively detect small molecules and their changes at the plant–endophyte interface, thereby enabling identification of novel metabolites for further endophyte applications.
Nevertheless, there are several challenges associated with conducting a metabolome analysis in plant–microbe systems. The equipment for metabolomic research is less accessible than those of DNA sequencing services. Furthermore, the availability of public metabolite reference databases is still limited, which further complicates the elucidaton and interpretation of metabolite networks and the assignment of detected metabolites to specific organisms (Levy et al., 2018). Furthermore, in the absence of a model compound, the identification of some isomeric or unknown compounds in the metabolite profile remains a challenging task.
Conclusion
Plant–microbe interactions involving pathogens and symbionts have always been complicated and difficult to understand, because these two categories of microbes share some similarities despite their other differences, for which only multi-omics studies can provide comprehensive global information. Research has addressed the plant–endophyte interactions from the perspective of recognition to signal exchange to stress the tolerance at both their transcriptomic and metabolomic levels. Such studies, based on model plants, have strengthened our understanding of the basic mechanisms underlying the relationship between plants and endophytes, including gene cascades and metabolic pathways, and the accumulation and enhancement of various metabolites, proteins, enzymes, and differentially expressed genes (DEGs). In the recognition phase especially, endophytes will secrete some elicitors/effectors, which are perceived as symbiotic signal molecules by host receptors, initiating the signaling cascades that lead to evading a plant immunity and inducing ISR to enhance coordination via phytohormone homeostasis. These secretions of endophytes and receptors of plants could be utilized to enhance plant resistance and moderate fertilizer use towards improving the nutrient captured by plants for enhancing sustainable crop production, which contributes to future applications of plant–microbe interactions for accelerating crops’ improvement in agriculture.
In fact, however, we still lack accurate and complete information on many gene functions and metabolites which could lead to overlooking the potential key elements involved in endophytic recognition and colonization. Little is known about core recognition sites in diverse mechanisms that have arisen in non-model plant–endophyte interactions. Nonetheless, combing multi-omics technologies to study endophyte-treated plants, especially those under various abiotic stress factors, will bridge gaps in our current understanding of the potential physiological development of endophytes. Persistent cultivation-independent problems need to be solved to make better use of endophyte resources (Meena et al., 2017) and cultivation-dependent endophytes. It is expected that a more high-throughput biological data will help to focus efforts upon the study of core recognition sites that will generate huge databases and methodologies for the discovery of myriad unknown genes, microbial species, and metabolites, which will collectively enable us to manipulate endophytes to harness their beneficial effects. High-resolution technology, such as single cell RNA sequencing, spatial transcriptome, and metabolome, will increase in usefulness in the near future. Forward and reverse genetic approaches should be applied to characterize detailed gene functions (Song et al., 2021). Meanwhile, the integration of multi-omics, cultivation, and ecological characterization (metabolic modeling) (Vilanova and Porcar, 2016) will create a new nexus capable of generating “holo-mics.” This is essential for building robust models that reveal both genetic and metabolic potential, as well as the ecology and evolution, of endophytes, the network of the complex interactions of endophytes with their host plants, and other associated microbes, and to let us better understand their respective roles in nature. We anticipate that full applications of these models would contribute to maintaining plant health in a stable living environment in the years to come.
Author Contributions
JS, LW, and SC reviewed and finalized the manuscript. XC completed the manuscript writing. XC and MS integrated the information of tables, analyzed the data, and made the pictures. All authors reviewed and approved the manuscript.
Funding
This work was supported by the Major Science and Technology Projects of Breeding New Varieties of Agriculture in Zhejiang Province (No. 2021C02074) and the National Natural Science Foundation of China (No. 31600259).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adhikari, K. B., Boelt, B., and Fomsgaard, I. S. (2016). Identification and quantification of loline-type alkaloids in endophyte-infected grasses by LC-MS/MS. J. Agric. Food Chem. 64, 6212–6218. doi: 10.1021/acs.jafc.6b02616
Afzal, I., Shinwari, Z. K., Sikandar, S., and Shahzad, S. (2019). Plant beneficial endophytic bacteria: mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 221, 36–49. doi: 10.1016/j.micres.2019.02.001
Aguiar, N. O., Olivares, F. L., Novotny, E. H., and Canellas, L. P. (2018). Changes in metabolic profiling of sugarcane leaves induced by endophytic diazotrophic bacteria and humic acids. PeerJ 6:e5445. doi: 10.7717/peerj.5445
Albelda-Berenguer, M., Monachon, M., and Joseph, E. (2019). Siderophores: From Natural Roles to Potential Applications, 1st Edn. Amsterdam: Elsevier Inc, doi: 10.1016/bs.aambs.2018.12.001
Ali, A. H., Abdelrahman, M., Radwan, U., El-Zayat, S., and El-Sayed, M. A. (2018). Effect of Thermomyces fungal endophyte isolated from extreme hot desert-adapted plant on heat stress tolerance of cucumber. Appl. Soil Ecol. 124, 155–162. doi: 10.1016/j.apsoil.2017.11.004
Ambrose, K. V., and Belanger, F. C. (2012). SOLiD-SAGE of endophyte-infected red fescue reveals numerous effects on host transcriptome and an abundance of highly expressed fungal secreted proteins. PLoS One 7:e53214. doi: 10.1371/journal.pone.0053214
Ambrose, K. V., Tian, Z., Wang, Y., Smith, J., Zylstra, G., Huang, B., et al. (2015). Functional characterization of salicylate hydroxylase from the fungal endophyte Epichloë festucae. Sci. Rep. 5:10939. doi: 10.1038/srep10939
Angel Contreras-Cornejo, H., Macías-Rodríguez, L., Cortés-Penagos, C., and López-Bucio, J. (2009). Trichoderma virens, a plant beneficial fungus, enhances biomass production and promotes lateral root growth through an auxin-dependent mechanism in Arabidopsis. Plant Physiol. 149, 1579–1592. doi: 10.1104/pp.108.130369
Anne-Emmanuelle, H., Hasna, B., Antoine, B., Marjolaine, R., Guillaume, M., Laetitia, C.-G., et al. (2017). Control of endophytic frankia sporulation by alnus nodule metabolites. Mol. Plant Microbe Interact. 30, 205–214. doi: 10.1094/MPMI-11-16-0235-R
Atsatt, P. R., and Whiteside, M. D. (2014). Novel symbiotic protoplasts formed by endophytic fungi explain their hidden existence, lifestyle switching, and diversity within the plant kingdom. PLoS One 9:e95266. doi: 10.1371/journal.pone.0095266
Aung, K., Kim, P., Li, Z., Joe, A., Kvitko, B., Alfano, J. R., et al. (2020). Pathogenic bacteria target plant plasmodesmata to colonize and invade surrounding tissues[CC-BY]. Plant Cell 32, 595–611. doi: 10.1105/tpc.19.00707
Bai, G., and Shaner, G. (2004). Management and resistance in wheat and barley to fusarium head blight. Annu. Rev. Phytopathol. 42, 135–161. doi: 10.1146/annurev.phyto.42.040803.140340
Bálintová, M., Bruòáková, K., Petijová, L., and Èellárová, E. (2019). Targeted metabolomic profiling reveals interspecific variation in the genus Hypericum in response to biotic elicitors. Plant Physiol. Biochem. 135, 348–358. doi: 10.1016/j.plaphy.2018.12.024
Bastias, D. A., Martínez-Ghersa, M. A., Ballaré, C. L., and Gundel, P. E. (2017). Epichloë fungal endophytes and plant defenses: not just alkaloids. Trends Plant Sci. 22, 939–948. doi: 10.1016/j.tplants.2017.08.005
Bateman, R. M. (1995). Terrestrial orchids: from seed to mycotrophic plant. Edinburgh J. Bot. 54, 357–359. doi: 10.1017/S0960428600004194
Batstone, R. T., O’Brien, A. M., Harrison, T. L., and Frederickson, M. E. (2020). Experimental evolution makes microbes more cooperative with their local host genotype. Science 370, 23–26. doi: 10.1126/science.abb7222
Bentham, A. R., de la Concepcion, J. C., Mukhi, N., Zdrzałek, R., Draeger, M., Gorenkin, D., et al. (2020). A molecular roadmap to the plant immune system. J. Biol. Chem. 295, 14916–14935. doi: 10.1074/jbc.REV120.010852
Bhatnagar, V. S., Bandyopadhyay, P., Rajacharya, G. H., Sarkar, S., Poluri, K. M., and Kumar, S. (2019). Amelioration of biomass and lipid in marine alga by an endophytic fungus Piriformospora indica. Biotechnol. Biofuels 12, 176. doi: 10.1186/s13068-019-1516-6
Bibi, F., Strobel, G. A., Naseer, M. I., Yasir, M., Al-Ghamdi, A. A. K., and Azhar, E. I. (2018). Microbial flora associated with the Halophyte-Salsola imbricate and its biotechnical potential. Front. Microbiol. 9:65. doi: 10.3389/fmicb.2018.00065
Bjornson, M., Pimprikar, P., Nürnberger, T., and Zipfel, C. (2021). The transcriptional landscape of Arabidopsis thaliana pattern-triggered immunity. Nat. Plants 7, 579–586. doi: 10.1038/s41477-021-00874-5
Bonfante, P., and Genre, A. (2010). Mechanisms underlying beneficial plant-fungus interactions in mycorrhizal symbiosis. Nat. Commun. 1:48. doi: 10.1038/ncomms1046
Bozsoki, Z., Gysel, K., Hansen, S. B., Lironi, D., Krönauer, C., Feng, F., et al. (2020). Ligand-recognizing motifs in plant LysM receptors are major determinants of specificity. Science 369, 663–670. doi: 10.1126/science.abb3377
Brotman, Y., Landau, U., Cuadros-Inostroza, Á, Takayuki, T., Fernie, A. R., Chet, I., et al. (2013). Responses and activation of the antioxidant machinery for saline stress tolerance. PLoS Pathog. 9:e1003221. doi: 10.1371/journal.ppat.1003221
Brusamarello-Santos, L. C. C., Alberton, D., Valdameri, G., Camilios-Neto, D., Covre, R., Lopes, K., et al. (2019). Modulation of defence and iron homeostasis genes in rice roots by the diazotrophic endophyte Herbaspirillum seropedicae. Sci. Rep. 9:10573. doi: 10.1038/s41598-019-45866-w
Bundy, J. G., Willey, T. L., Castell, R. S., Ellar, D. J., and Brindle, K. M. (2005). Discrimination of pathogenic clinical isolates and laboratory strains of Bacillus cereus by NMR-based metabolomic profiling. FEMS Microbiol. Lett. 242, 127–136. doi: 10.1016/j.femsle.2004.10.048
Buscaill, P., and van der Hoorn, R. A. L. (2021). Defeated by the nines: nine extracellular strategies to avoid MAMP recognition in plants. Plant Cell 33, 2116–2130. doi: 10.1093/plcell/koab109
Cai, W., Ou, F., Staehelin, C., and Dai, W. (2020). Bradyrhizobium sp. strain ORS278 promotes rice growth and its quorum sensing system is required for optimal root colonization. Environ. Microbiol. Rep. 12, 656–666. doi: 10.1111/1758-2229.12885
Caldana, C., Degenkolbe, T., Cuadros-Inostroza, A., Klie, S., Sulpice, R., Leisse, A., et al. (2011). High-density kinetic analysis of the metabolomic and transcriptomic response of Arabidopsis to eight environmental conditions. Plant J. 67, 869–884. doi: 10.1111/j.1365-313X.2011.04640.x
Carrión, V. J., Perez-Jaramillo, J., Cordovez, V., Tracanna, V., de Hollander, M., Ruiz-Buck, D., et al. (2019). Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science 366, 606–612. doi: 10.1126/science.aaw9285
Chauhan, A., Fazlurrahman, Oakeshott, J. G., and Jain, R. K. (2008). Bacterial metabolism of polycyclic aromatic hydrocarbons: strategies for bioremediation. Indian J. Microbiol. 48, 95–113. doi: 10.1007/s12088-008-0010-9
Chen, E., Blaze, J. A., Smith, R. S., Peng, S., and Byers, J. E. (2020). Freeze tolerance of poleward-spreading mangrove species weakened by soil properties of resident salt marsh competitor. J. Ecol. 108, 1725–1737. doi: 10.1111/1365-2745.13350
Chen, F., Ma, R., and Chen, X. L. (2019). Advances of metabolomics in fungal pathogen–plant interactions. Metabolites 9:169. doi: 10.3390/metabo9080169
Chen, F., Yan, B., Gong, X., Li, H., and He, Z. (2021). Genome sequencing of the bacterial blight pathogen DY89031 reveals its diverse virulence and origins of Xanthomonas oryzae pv. oryzae strains. Sci. China Life Sci. 64, 2175–2185. doi: 10.1007/s11427-020-1917-x
Chen, N., He, R., Chai, Q., Li, C., and Nan, Z. (2016). Transcriptomic analyses giving insights into molecular regulation mechanisms involved in cold tolerance by Epichloë endophyte in seed germination of Achnatherum inebrians. Plant Growth Regul. 80, 367–375. doi: 10.1007/s10725-016-0177-8
Chen, X., Liu, P., Mei, L., He, X., Chen, L., Liu, H., et al. (2021). Xa7, a new executor R gene that confers durable and broad-spectrum resistance to bacterial blight disease in rice. Plant Commun. 2:100143. doi: 10.1016/j.xplc.2021.100143
Chiu, C. H., and Paszkowski, U. (2021). How membrane receptors tread the fine balance between symbiosis and immunity signaling. Proc. Natl. Acad. Sci. U.S.A. 118:e2106567118. doi: 10.1073/pnas.2106567118
Cho, S., Kang, B. R., and Kim, Y. C. (2013). Transcriptome analysis of induced systemic drought tolerance elicited by Pseudomonas chlororaphis O6 in Arabidopsis thaliana. Plant Pathol. J. 29, 209–220. doi: 10.5423/PPJ.SI.07.2012.0103
Chu, L. L., and Bae, H. (2021). Bacterial endophytes from ginseng and their biotechnological application. J. Ginseng Res. (in press). doi: 10.1016/j.jgr.2021.04.004
Cole, B. J., and Tringe, S. G. (2021). Different threats, same response. Nat. Plants 7, 544–545. doi: 10.1038/s41477-021-00915-z
Conesa, A., Madrigal, P., Tarazona, S., Gomez-Cabrero, D., Cervera, A., McPherson, A., et al. (2016). A survey of best practices for RNA-seq data analysis. Genome Biol. 17, 13. doi: 10.1186/s13059-016-0881-8
Constantin, M. E., de Lamo, F. J., Vlieger, B. V., Rep, M., and Takken, F. L. W. (2019). Endophyte-mediated resistance in tomato to Fusarium oxysporum Is Independent of ET. JA, and SA. Front. Plant Sci. 10:979. doi: 10.3389/fpls.2019.00979
Cook, D. R., VandenBosch, K., de Bruijn, F. J., and Huguet, T. (1997). Model Legumes Get the Nod. Plant Cell 9, 275–280. doi: 10.1105/tpc.9.3.275
Courville, K. J., Frantzeskakis, L., Gul, S., Haeger, N., Kellner, R., Heßler, N., et al. (2019). Smut infection of perennial hosts: the genome and the transcriptome of the Brassicaceae smut fungus Thecaphora thlaspeos reveal functionally conserved and novel effectors. New Phytol. 222, 1474–1492. doi: 10.1111/nph.15692
Coutinho, B. G., Licastro, D., Mendonça-Previato, L., Cámara, M., and Venturi, V. (2015). Plant-influenced gene expression in the rice endophyte burkholderia kururiensis M130. Mol. Plant Microbe Interact. 28, 10–21. doi: 10.1094/MPMI-07-14-0225-R
Cunnac, S., Chakravarthy, S., Kvitko, B. H., Russell, A. B., Martin, G. B., and Collmer, A. (2011). Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae. Proc. Natl. Acad. Sci. U.S.A. 108, 2975–2980. doi: 10.1073/pnas.1013031108
De Palma, M., Salzano, M., Villano, C., Aversano, R., Lorito, M., Ruocco, M., et al. (2019). Transcriptome reprogramming, epigenetic modifications and alternative splicing orchestrate the tomato root response to the beneficial fungus Trichoderma harzianum. Hortic. Res. 6:5. doi: 10.1038/s41438-018-0079-1
de Santi Ferrara, F. I., Oliveira, Z. M., Gonzales, H. H. S., Floh, E. I. S., and Barbosa, H. R. (2012). Endophytic and rhizospheric enterobacteria isolated from sugar cane have different potentials for producing plant growth-promoting substances. Plant Soil 353, 409–417. doi: 10.1007/s11104-011-1042-1
Dearnaley, J. D. W., and Cameron, D. D. (2017). Nitrogen transport in the orchid mycorrhizal symbiosis – further evidence for a mutualistic association. New Phytol. 213, 10–12. doi: 10.1111/nph.14357
Delaux, P. M., and Schornack, S. (2021). Plant evolution driven by interactions with symbiotic and pathogenic microbes. Science 37:eaba6605. doi: 10.1126/science.aba6605
Dinkins, R. D., Nagabhyru, P., Graham, M. A., Boykin, D., and Schardl, C. L. (2017). Transcriptome response of Lolium arundinaceum to its fungal endophyte Epichloë coenophiala. New Phytol. 213, 324–337. doi: 10.1111/nph.14103
Dinkins, R. D., Nagabhyru, P., Young, C. A., West, C. P., and Schardl, C. L. (2019). Transcriptome analysis and differential expression in tall fescue harboring different endophyte strains in response to water deficit. Plant Genome 12:180071. doi: 10.3835/plantgenome2018.09.0071
Dong, W., Zhu, Y., Chang, H., Wang, C., Yang, J., Shi, J., et al. (2021). An SHR–SCR module specifies legume cortical cell fate to enable nodulation. Nature 589, 586–590. doi: 10.1038/s41586-020-3016-z
Doni, F., Fathurrahman, F., Mispan, M. S., Suhaimi, N. S. M., Yusoff, W. M. W., and Uphoff, N. (2019). Transcriptomic profiling of rice seedlings inoculated with the symbiotic fungus Trichoderma asperellum SL2. J. Plant Growth Regul. 38, 1507–1515. doi: 10.1007/s00344-019-09952-7
Drew, G. C., Stevens, E. J., and King, K. C. (2021). Microbial evolution and transitions along the parasite–mutualist continuum. Nat. Rev. Microbiol. 19, 623–638. doi: 10.1038/s41579-021-00550-7
Eida, A. A., Alzubaidy, H. S., de Zélicourt, A., Synek, L., Alsharif, W., Lafi, F. F., et al. (2019). Phylogenetically diverse endophytic bacteria from desert plants induce transcriptional changes of tissue-specific ion transporters and salinity stress in Arabidopsis thaliana. Plant Sci. 280, 228–240. doi: 10.1016/j.plantsci.2018.12.002
el Zahar Haichar, F., Santaella, C., Heulin, T., and Achouak, W. (2014). Root exudates mediated interactions belowground. Soil Biol. Biochem. 77, 69–80. doi: 10.1016/j.soilbio.2014.06.017
Faeth, S. H., and Fagan, W. F. (2002). Fungal endophytes: common host plant symbionts but uncommon mutualists. Integr. Comp. Biol. 42, 360–368. doi: 10.1093/icb/42.2.360
Fan, L., Wang, T., Hua, C., Sun, W., Li, X., Grunwald, L., et al. (2020). A compendium of DNA-binding specificities of transcription factors in Pseudomonas syringae. Nat. Commun. 11:4947. doi: 10.1038/s41467-020-18744-7
Favre-Godal, Q., Gourguillon, L., Lordel-Madeleine, S., Gindro, K., and Choisy, P. (2020). Orchids and their mycorrhizal fungi: an insufficiently explored relationship. Mycorrhiza 30, 5–22. doi: 10.1007/s00572-020-00934-2
Ferrol, N., Azcón-Aguilar, C., and Pérez-Tienda, J. (2019). Review: Arbuscular mycorrhizas as key players in sustainable plant phosphorus acquisition: an overview on the mechanisms involved. Plant Sci. 280, 441–447. doi: 10.1016/j.plantsci.2018.11.011
Fröschel, C., Komorek, J., Attard, A., Marsell, A., Lopez-Arboleda, W. A., Le Berre, J., et al. (2021). Plant roots employ cell-layer-specific programs to respond to pathogenic and beneficial microbes. Cell Host Microbe 29, 299–310.e7. doi: 10.1016/j.chom.2020.11.014
Gamez, R. M., Rodríguez, F., Vidal, N. M., Ramirez, S., Vera Alvarez, R., Landsman, D., et al. (2019). Banana (Musa acuminata) transcriptome profiling in response to rhizobacteria: Bacillus amyloliquefaciens Bs006 and Pseudomonas fluorescens Ps006. BMC Genomics 20:378. doi: 10.1186/s12864-019-5763-5
Gargallo-Garriga, A., Sardans, J., Pérez-Trujillo, M., Guenther, A., Llusià, J., Rico, L., et al. (2016). Shifts in plant foliar and floral metabolomes in response to the suppression of the associated microbiota. BMC Plant Biol. 16:78. doi: 10.1186/s12870-016-0767-7
Genre, A., Lanfranco, L., Perotto, S., and Bonfante, P. (2020). Unique and common traits in mycorrhizal symbioses. Nat. Rev. Microbiol. 18, 649–660. doi: 10.1038/s41579-020-0402-3
Ghaffari, M. R., Ghabooli, M., Khatabi, B., Hajirezaei, M. R., Schweizer, P., and Salekdeh, G. H. (2016). Metabolic and transcriptional response of central metabolism affected by root endophytic fungus Piriformospora indica under salinity in barley. Plant Mol. Biol. 90, 699–717. doi: 10.1007/s11103-016-0461-z
Ghaffari, M. R., Mirzaei, M., Ghabooli, M., Khatabi, B., Wu, Y., Zabet-Moghaddam, M., et al. (2019). Root endophytic fungus Piriformospora indica improves drought stress adaptation in barley by metabolic and proteomic reprogramming. Environ. Exp. Bot. 157, 197–210. doi: 10.1016/j.envexpbot.2018.10.002
Ghirardo, A., Fochi, V., Lange, B., Witting, M., Schnitzler, J.-P., Perotto, S., et al. (2020). Metabolomic adjustments in the orchid mycorrhizal fungus Tulasnella calospora during symbiosis with Serapias vomeracea. New Phytol. 228, 1939–1952. doi: 10.1111/nph.16812
Gong, Z., and Han, G. Z. (2021). Flourishing in water: the early evolution and diversification of plant receptor-like kinases. Plant J. 106, 174–184. doi: 10.1111/tpj.15157
Guo, J., McCulley, R. L., and McNear, D. H. (2015). Tall fescue cultivar and fungal endophyte combinations influence plant growth and root exudate composition. Front. Plant Sci. 6:183. doi: 10.3389/fpls.2015.00183
Gupta, P. K. (2020). SWEET genes for disease resistance in plants. Trends Genet. 36, 901–904. doi: 10.1016/j.tig.2020.08.007
Hacquard, S., Kracher, B., Hiruma, K., Münch, P. C., Garrido-Oter, R., Thon, M. R., et al. (2016). Survival trade-offs in plant roots during colonization by closely related beneficial and pathogenic fungi. Nat. Commun. 7:11362. doi: 10.1016/j.disc.2013.08.028
Hacquard, S., Spaepen, S., Garrido-Oter, R., and Schulze-Lefert, P. (2017). Interplay between innate immunity and the plant microbiota. Annu. Rev. Phytopathol. 55, 565–589. doi: 10.1146/annurev-phyto-080516-035623
Hao, K., Wang, F., Nong, X., McNeill, M. R., Liu, S., Wang, G., et al. (2017). Response of peanut Arachis hypogaea roots to the presence of beneficial and pathogenic fungi by transcriptome analysis. Sci. Rep. 7:964. doi: 10.1038/s41598-017-01029-3
Hartley, S. E., Eschen, R., Horwood, J. M., Gange, A. C., and Hill, E. M. (2015). Infection by a foliar endophyte elicits novel arabidopside-based plant defence reactions in its host, Cirsium arvense. New Phytol. 205, 816–827. doi: 10.1111/nph.13067
Harvey, S., Kumari, P., Lapin, D., Griebel, T., Hickman, R., Guo, W., et al. (2020). Downy Mildew effector HaRxL21 interacts with the transcriptional repressor TOPLESS to promote pathogen susceptibility. PLoS Pathog. 16:e1008835. doi: 10.1371/JOURNAL.PPAT.1008835
He, C., Cui, J., Chen, X., Wang, W., and Hou, J. (2020). Effects of enhancement of liquorice plants with dark septate endophytes on the root growth, glycyrrhizic acid and glycyrrhizin accumulation amended with organic residues. Curr. Plant Biol. 23, 100154. doi: 10.1016/j.cpb.2020.100154
He, J., Sun, C., Li, T., Luo, Z., Huang, L., Song, X., et al. (2018). A sensitive and wide coverage ambient mass spectrometry imaging method for functional metabolites based molecular histology. Adv. Sci. 5:1800250. doi: 10.1002/advs.201800250
Herburger, K., Xin, A., and Holzinger, A. (2019). Homogalacturonan accumulation in cell walls of the green alga Zygnema sp. (charophyta) increases desiccation resistance. Front. Plant Sci. 10:540. doi: 10.3389/fpls.2019.00540
Hettiarachchige, I. K., Elkins, A. C., Reddy, P., Mann, R. C., and Guthridge, K. M. (2019). Genetic modification of asexual Epichloë endophytes with the perA gene for peramine biosynthesis. Mol. Genet. Genomics 294, 315–328. doi: 10.1007/s00438-018-1510-x
Hidayat, I. (2019). “Dark septate endophytes and their role in enhancing plant resistance to abiotic and biotic stresses,” in Plant Growth Promoting Rhizobacteria for Sustainable Stress Management, eds R. Z. Sayyed, N. K. Arora, and M. S. Reddy (Berlin: Springer), 35–63. doi: 10.1007/978-981-13-6536-2_3
Hillmer, R. A., Tsuda, K., Rallapalli, G., Asai, S., Truman, W., Papke, M. D., et al. (2017). The highly buffered Arabidopsis immune signaling network conceals the functions of its components. PLoS Genet. 13:e1006639. doi: 10.1371/journal.pgen.1006639
Hiruma, K., Gerlach, N., Sacristán, S., Nakano, R. T., Hacquard, S., Kracher, B., et al. (2016). Root Endophyte Colletotrichum tofieldiae confers plant fitness benefits that are phosphate status dependent. Cell 165, 464–474. doi: 10.1016/j.cell.2016.02.028
Horvath, R. S. (1972). Microbial co-metabolism and the degradation of organic compounds in nature. Bacteriol. Rev. 36, 146–155. doi: 10.1128/br.36.2.146-155.1972
Huang, D., Sun, Y., Ma, Z., Ke, M., Cui, Y., Chen, Z., et al. (2019). Salicylic acid-mediated plasmodesmal closure via Remorin-dependent lipid organization. Proc. Natl. Acad. Sci. U.S.A. 116, 21274–21284. doi: 10.1073/pnas.1911892116
Huang, L. H., Yuan, M. Q., Ao, X. J., Ren, A. Y., Zhang, H. B., and Yang, M. Z. (2018). Endophytic fungi specifically introduce novel metabolites into grape flesh cells in vitro. PLoS One 13:e0196996. doi: 10.1371/journal.pone.0196996
Huang, S., Zhu, S., Kumar, P., and MacMicking, J. D. (2021). A phase-separated nuclear GBPL circuit controls immunity in plants. Nature 594, 424–429. doi: 10.1038/s41586-021-03572-6
Hurek, T., and Reinhold-Hurek, B. (2003). Azoarcus sp. strain BH72 as a model for nitrogen-fixing grass endophytes. J. Biotechnol. 106, 169–178. doi: 10.1016/j.jbiotec.2003.07.010
Inceoğlu, Ö, Al-Soud, W. A., Salles, J. F., Semenov, A. V., and van Elsas, J. D. (2011). Comparative analysis of bacterial communities in a potato field as determined by pyrosequencing. PLoS One 6:e23321. doi: 10.1371/journal.pone.0023321
Jia, M., Chen, L., Xin, H. L., Zheng, C. J., Rahman, K., Han, T., et al. (2016). A friendly relationship between endophytic fungi and medicinal plants: a systematic review. Front. Microbiol. 7:906. doi: 10.3389/fmicb.2016.00906
Jiang, C. H., Sheng, Y., Kai, X., Ning, Z., Zi, W., Li, J., et al. (2019). Volatile organic compounds emitted by Bacillus sp. JC03 promote plant growth through the action of auxin and strigolactone. Plant Growth Regul. 87, 317–328. doi: 10.1007/s10725-018-00473-z
Jiang, Y., Wang, L., Lu, S., Xue, Y., Wei, X., Lu, J., et al. (2019). Transcriptome sequencing of Salvia miltiorrhiza after infection by its endophytic fungi and identification of genes related to tanshinone biosynthesis. Pharm. Biol. 57, 760–769. doi: 10.1080/13880209.2019.1680706
Jiao, J., Ma, Y., Chen, S., Liu, C., Song, Y., Qin, Y., et al. (2016). Melatonin-producing endophytic bacteria from grapevine roots promote the abiotic stress-induced production of endogenous melatonin in their hosts. Front. Plant Sci. 7:1387. doi: 10.3389/fpls.2016.01387
Jisha, S., Gouri, P. R., Anith, K. N., and Sabu, K. K. (2018). Piriformospora indica cell wall extract as the best elicitor for asiaticoside production in Centella asiatica (L.) Urban, evidenced by morphological, physiological and molecular analyses. Plant Physiol. Biochem. 125, 106–115. doi: 10.1016/j.plaphy.2018.01.021
Johnson, L. J., Johnson, R. D., Schardl, C. L., and Panaccione, D. G. (2003). Identification of differentially expressed genes in the mutualistic association of tall fescue with Neotyphodium coenophialum. Physiol. Mol. Plant Pathol. 63, 305–317. doi: 10.1016/j.pmpp.2004.04.001
Jorge, T. F., Rodrigues, J. A., Caldana, C., Schmidt, R., van Dongen, J. T., Thomas-Oates, J., et al. (2016). Mass spectrometry-based plant metabolomics: metabolite responses to abiotic stress. Mass Spectrom. Rev. 35, 620–649. doi: 10.1002/mas.21449
Kaul, S., Gupta, S., Ahmed, M., and Dhar, M. K. (2012). Endophytic fungi from medicinal plants: a treasure hunt for bioactive metabolites. Phytochem. Rev. 11, 487–505. doi: 10.1007/s11101-012-9260-6
Kaul, S., Sharma, T., and Dhar, K. M. (2016). “Omics” Tools for Better Understanding the Plant–Endophyte Interactions. Front. Plant Sci. 7:955. doi: 10.3389/fpls.2016.00955
Kawaguchi, M., and Minamisawa, K. (2010). Plant-microbe communications for symbiosis. Plant Cell Physiol. 51, 1377–1380. doi: 10.1093/pcp/pcq125
Khan, A. L., Hamayun, M., Kang, S.-M., Kim, Y.-H., Jung, H.-Y., Lee, J.-H., et al. (2012). Endophytic fungal association via gibberellins and indole acetic acid can improve plant growth under abiotic stress: an example of Paecilomyces formosus LHL10. BMC Microbiol. 12:3. doi: 10.1186/1471-2180-12-3
Khare, E., Mishra, J., and Arora, N. K. (2018). Multifaceted interactions between endophytes and plant: developments and prospects. Front. Microbiol. 9:2732. doi: 10.3389/fmicb.2018.02732
Khoomrung, S., Wanichthanarak, K., Nookaew, I., Thamsermsang, O., Seubnooch, P., Laohapand, T., et al. (2017). Metabolomics and integrative omics for the development of Thai traditional medicine. Front. Pharmacol. 8:474. doi: 10.3389/fphar.2017.00474
Krings, M., Taylor, N. T., Hass, H., Kerp, H., Dotzler, N., and Hermsen, E. (2007). Fungal endophytes in a 400-million-yr-old land plant: infection pathways, spatial distribution, and host responses. New Phytol. 174, 648–657. doi: 10.1111/j.1469-8137.2007.02008.x
Kumar, N., Galli, M., Dempsey, D., Imani, J., Moebus, A., and Kogel, K.-H. (2021). NPR1 is required for root colonization and the establishment of a mutualistic symbiosis between the beneficial bacterium Rhizobium radiobacter and barley. Environ. Microbiol. 23, 2102–2115. doi: 10.1111/1462-2920.15356
Kusari, S., Singh, S., and Jayabaskaran, C. (2014). Biotechnological potential of plant-associated endophytic fungi: hope versus hype. Trends Biotechnol. 32, 297–303. doi: 10.1016/j.tibtech.2014.03.009
Lahrmann, U., and Zuccaro, A. (2012). Opprimo ergo sum-evasion and suppression in the root endophytic fungus Piriformospora indica. Mol. Plant Microbe Interact. 25, 727–737. doi: 10.1094/MPMI-11-11-0291
Lekberg, Y., Arnillas, C. A., Borer, E. T., Bullington, L. S., Fierer, N., Kennedy, P. G., et al. (2021). Nitrogen and phosphorus fertilization consistently favor pathogenic over mutualistic fungi in grassland soils. Nat. Commun. 12:3484. doi: 10.1038/s41467-021-23605-y
Levy, A., Conway, J. M., Dangl, J. L., and Woyke, T. (2018). Elucidating bacterial gene functions in the plant microbiome. Cell Host Microbe 24, 475–485. doi: 10.1016/j.chom.2018.09.005
Li, L., Wang, S., Wang, H., Sahu, S. K., Marin, B., Li, H., et al. (2020). The genome of Prasinoderma coloniale unveils the existence of a third phylum within green plants. Nat. Ecol. Evol. 4, 1220–1231. doi: 10.1038/s41559-020-1221-7
Li, Q., Ding, G., Li, B., and Guo, S. X. (2017a). Transcriptome analysis of genes involved in dendrobine biosynthesis in Dendrobium nobile lindl. infected with mycorrhizal fungus MF23 (Mycena sp.). Sci. Rep. 7, 1–16. doi: 10.1038/s41598-017-00445-9
Li, Q., Li, B., Zhou, L. S., Ding, G., Li, B., and Guo, S. X. (2017b). Molecular analysis of polysaccharide accumulation in: Dendrobium nobile infected with the mycorrhizal fungus Mycena sp. RSC Adv. 7, 25872–25884. doi: 10.1039/c7ra02010d
Li, W., Xiong, Y., Lai, L. B., Zhang, K., Li, Z., Kang, H., et al. (2021). The rice RNase P protein subunit Rpp30 confers broad-spectrum resistance to fungal and bacterial pathogens. Plant Biotechnol. J. 19, 1988–1999. doi: 10.1111/pbi.13612
Li, W. V., and Li, J. J. (2018). Modeling and analysis of RNA-seq data: a review from a statistical perspective. Quant. Biol. 6, 195–209. doi: 10.1007/s40484-018-0144-7
Li, Y., Steenwyk, J. L., Chang, Y., Wang, Y., James, T. Y., Stajich, J. E., et al. (2021). A genome-scale phylogeny of the kingdom Fungi. Curr. Biol. 31, 1653–1665.e5. doi: 10.1016/j.cub.2021.01.074
Liu, S., Hao, H., Lu, X., Zhao, X., Wang, Y., Zhang, Y., et al. (2017). Transcriptome profiling of genes involved in induced systemic salt tolerance conferred by Bacillus amyloliquefaciens FZB42 in Arabidopsis thaliana. Sci. Rep. 7:10795. doi: 10.1038/s41598-017-11308-8
Llorens, E., Sharon, O., Camañes, G., García-Agustín, P., and Sharon, A. (2019). Endophytes from wild cereals protect wheat plants from drought by alteration of physiological responses of the plants to water stress. Environ. Microbiol. 21, 3299–3312. doi: 10.1111/1462-2920.14530
Lu, C., Liu, H., Jiang, D., Wang, L., Jiang, Y., Tang, S., et al. (2019). Paecilomyces variotii extracts (ZNC) enhance plant immunity and promote plant growth. Plant Soil 441, 383–397. doi: 10.1007/s11104-019-04130-w
Lu, S. (2021). Biosynthesis and Regulatory Mechanisms of Bioactive Compounds in Salvia miltiorrhiza, a Model System for Medicinal Plant Biology. CRC. Crit. Rev. Plant Sci. 40, 243–283. doi: 10.1080/07352689.2021.1935719
Luan, Y., Cui, J., Zhai, J., Li, J., Han, L., and Meng, J. (2015). High-throughput sequencing reveals differential expression of miRNAs in tomato inoculated with Phytophthora infestans. Planta 241, 1405–1416. doi: 10.1007/s00425-015-2267-7
Lucini, L., Colla, G., Miras Moreno, M. B., Bernardo, L., Cardarelli, M., Terzi, V., et al. (2019). Inoculation of Rhizoglomus irregulare or Trichoderma atroviride differentially modulates metabolite profiling of wheat root exudates. Phytochemistry 157, 158–167. doi: 10.1016/j.phytochem.2018.10.033
Ma, K.-W., Niu, Y., Jia, Y., Ordon, J., Copeland, C., Emonet, A., et al. (2021). Coordination of microbe–host homeostasis by crosstalk with plant innate immunity. Nat. Plants 7, 814–825. doi: 10.1038/s41477-021-00920-2
Ma, Y., Jiao, J., Fan, X., Sun, H., Zhang, Y., Jiang, J., et al. (2017). Endophytic bacterium Pseudomonas fluorescens RG11 may transform tryptophan to melatonin and promote endogenous melatonin levels in the roots of four grape cultivars. Front. Plant Sci. 7:2068. doi: 10.3389/fpls.2016.02068
Ma, Z., Qin, G., Zhang, Y., Liu, C., Wei, M., Cen, Z., et al. (2021). Bacterial Leaf Streak 1 encoding a mitogen-activated protein kinase confers the rice resistance to bacterial leaf streak. Plant J. 107, 1084–1101. doi: 10.1111/tpj.15368
Ma, Z., Zhu, L., Song, T., Wang, Y., Zhang, Q., Xia, Y., et al. (2017). A paralogous decoy protects Phytophthora sojae apoplastic effector PsXEG1 from a host inhibitor. Science 355, 710–714. doi: 10.1126/science.aai7919
Maggini, V., Mengoni, A., Bogani, P., Firenzuoli, F., and Fani, R. (2020). Promoting model systems of microbiota–medicinal plant interactions. Trends Plant Sci. 25, 223–225. doi: 10.1016/j.tplants.2019.12.013
Maheshwari, D. K. (2017). Endophytes: Biology and Biotechnology. New York, NY: Springer International Publishing, doi: 10.1007/978-3-319-66541-2
Maier, B. A., Kiefer, P., Field, C. M., Hemmerle, L., Bortfeld-Miller, M., Emmenegger, B., et al. (2021). A general non-self response as part of plant immunity. Nat. Plants 7, 696–705. doi: 10.1038/s41477-021-00913-1
Maksimova, T. I., Popov, V. O., Sarvarova, E. R., Blagova, D. K., and Maksimov, I. V. (2018). Endophytic bacteria as effective agents of new-generation biopesticides (Review). Appl. Biochem. Microbiol. 54, 128–140. doi: 10.1134/s0003683818020072
Malinich, E. A., Wang, K., Mukherjee, P. K., Kolomiets, M., and Kenerley, C. M. (2019). Differential expression analysis of Trichoderma virens RNA reveals a dynamic transcriptome during colonization of Zea mays roots. BMC Genomics 20:289. doi: 10.1186/s12864-019-5651-z
Manganiello, G., Sacco, A., Ercolano, M. R., Vinale, F., Lanzuise, S., Pascale, A., et al. (2018). Modulation of tomato response to Rhizoctonia solani by Trichoderma harzianum and its secondary metabolite harzianic acid. Front. Microbiol. 9:1966. doi: 10.3389/fmicb.2018.01966
Martínez-Medina, A., Fernandez, I., Lok, G. B., Pozo, M. J., Pieterse, C. M. J., and Van Wees, S. C. M. (2017). Shifting from priming of salicylic acid-to jasmonic acid-regulated defences by Trichoderma protects tomato against the root knot nematode Meloidogyne incognita. New Phytol. 213, 1363–1377. doi: 10.1111/nph.14251
McLaren, M. R., and Callahan, B. J. (2020). Pathogen resistance may be the principal evolutionary advantage provided by the microbiome. Philos. Trans. R. Soc. B Biol. Sci. 375:20190592. doi: 10.1098/rstb.2019.0592rstb20190592
Meena, K. K., Sorty, A. M., Bitla, U. M., Choudhary, K., Gupta, P., Pareek, A., et al. (2017). Abiotic stress responses and microbe-mediated mitigation in plants: the omics strategies. Front. Plant Sci. 8:172. doi: 10.3389/fpls.2017.00172
Mehmood, A., Hussain, A., Irshad, M., Hamayun, M., Iqbal, A., Rahman, H., et al. (2019). Cinnamic acid as an inhibitor of growth, flavonoids exudation and endophytic fungus colonization in maize root. Plant Physiol. Biochem. 135, 61–68. doi: 10.1016/j.plaphy.2018.11.029
Mhlongo, M. I., Piater, L. A., Madala, N. E., Labuschagne, N., and Dubery, I. A. (2018). The chemistry of plant–microbe interactions in the rhizosphere and the potential for metabolomics to reveal signaling related to defense priming and induced systemic resistance. Front. Plant Sci. 9:112. doi: 10.3389/fpls.2018.00112
Mishra, B. K., Meena, K. K., Dubey, P. N., Aishwath, O. P., Kant, K., Sorty, A. M., et al. (2016). Influence on yield and quality of fennel (Foeniculum vulgare Mill.) grown under semi-arid saline soil, due to application of native phosphate solubilizing rhizobacterial isolates. Ecol. Eng. 97, 327–333. doi: 10.1016/j.ecoleng.2016.10.034
Mishra, S., Bhattacharjee, A., and Sharma, S. (2021). An ecological insight into the multifaceted world of plant-endophyte association. CRC. Crit. Rev. Plant Sci. 40, 127–146. doi: 10.1080/07352689.2021.1901044
Montero, H., Lee, T., Pucker, B., Ferreras-Garrucho, G., Oldroyd, G., Brockington, S. F., et al. (2021). A mycorrhiza-associated receptor-like kinase with an ancient origin in the green lineage. Proc. Natl. Acad. Sci. U.S.A. 118:e2105281118. doi: 10.1073/pnas.2105281118
Moore, W. M., Chan, C., Ishikawa, T., Rennie, E. A., Wipf, H. M. L., Benites, V., et al. (2021). Reprogramming sphingolipid glycosylation is required for endosymbiont persistence in Medicago truncatula. Curr. Biol. 31, 2374–2385.e4. doi: 10.1016/j.cub.2021.03.067
Morsy, M. R., Oswald, J., He, J., Tang, Y., and Roossinck, M. J. (2010). Teasing apart a three-way symbiosis: transcriptome analyses of Curvularia protuberata in response to viral infection and heat stress. Biochem. Biophys. Res. Commun. 401, 225–230. doi: 10.1016/j.bbrc.2010.09.034
Mukherjee, M., Mukherjee, P. K., Horwitz, B. A., Zachow, C., Berg, G., and Zeilinger, S. (2012). Trichoderma-plant-pathogen interactions: advances in genetics of biological control. Indian J. Microbiol. 52, 522–529. doi: 10.1007/s12088-012-0308-5
Nagabhyru, P., Dinkins, R. D., Wood, C. L., Bacon, C. W., and Schardl, C. L. (2013). Tall fescue endophyte effects on tolerance to water-deficit stress. BMC Plant Biol. 13:127. doi: 10.1186/1471-2229-13-127
Navarro-Meléndez, A. L., and Heil, M. (2014). Symptomless endophytic fungi suppress endogenous levels of salicylic acid and interact with the jasmonate-dependent indirect defense traits of their host, lima bean (Phaseolus lunatus). J. Chem. Ecol. 40, 816–825. doi: 10.1007/s10886-014-0477-2
Newton, A. C., Fitt, B. D. L., Atkins, S. D., Walters, D. R., and Daniell, T. J. (2010). Pathogenesis, parasitism and mutualism in the trophic space of microbe-plant interactions. Trends Microbiol. 18, 365–373. doi: 10.1016/j.tim.2010.06.002
Ngou, B. P. M., Ahn, H.-K., Ding, P., and Jones, J. D. G. (2021). Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592, 110–115. doi: 10.1038/s41586-021-03315-7
Nobori, T., Wang, Y., Wu, J., Stolze, S. C., Tsuda, Y., Finkemeier, I., et al. (2020). Multidimensional gene regulatory landscape of a bacterial pathogen in plants. Nat. Plants 6, 883–896. doi: 10.1038/s41477-020-0690-7
Nordström, A., O’Maille, G., Qin, C., and Siuzdak, G. (2006). Nonlinear data alignment for UPLC-MS and HPLC-MS based metabolomics: quantitative analysis of endogenous and exogenous metabolites in human serum. Anal. Chem. 78, 3289–3295. doi: 10.1021/ac060245f
Nostadt, R., Hilbert, M., Nizam, S., Rovenich, H., Wawra, S., Martin, J., et al. (2020). A secreted fungal histidine- and alanine-rich protein regulates metal ion homeostasis and oxidative stress. New Phytol. 227, 1174–1188. doi: 10.1111/nph.16606
Ofaim, S., Ofek-Lalzar, M., Sela, N., Jinag, J., Kashi, Y., Minz, D., et al. (2017). Analysis of microbial functions in the rhizosphere using a metabolic-network based framework for metagenomics interpretation. Front. Microbiol. 8:1606. doi: 10.3389/fmicb.2017.01606
Oldroyd, G. E. D., and Downie, J. A. (2008). Coordinating nodule morphogenesis with rhizobial. Infection in legumes. Annu. Rev. Plant Biol. 59:519. doi: 10.1146/annurev.arplant.59.032607.092839
Oldroyd, G. E. D., and Leyser, O. (2020). A plant’s diet, surviving in a variable nutrient environment. Science 368:eaba0196. doi: 10.1126/science.aba0196
Oliver, S. G., Winson, M. K., Kell, D. B., and Baganz, F. (1998). Systematic functional analysis of the yeast genome. Trends Biotechnol. 16, 373–378. doi: 10.1016/S0167-7799(98)01214-1
Palmieri, D., Vitale, S., Lima, G., Di Pietro, A., and Turrà, D. (2020). A bacterial endophyte exploits chemotropism of a fungal pathogen for plant colonization. Nat. Commun. 11:5264. doi: 10.1038/s41467-020-18994-5
Pandey, V., Ansari, M. W., Tula, S., and Yadav, S. (2016). Dose-dependent response of Trichoderma harzianum in improving drought tolerance in rice genotypes. Planta 243, 1251–1264. doi: 10.1007/s00425-016-2482-x
Pankievicz, V. C. S., Camilios-Neto, D., Bonato, P., Balsanelli, E., Tadra-Sfeir, M. Z., Faoro, H., et al. (2016). RNA-seq transcriptional profiling of Herbaspirillum seropedicae colonizing wheat (Triticum aestivum) roots. Plant Mol. Biol. 90, 589–603. doi: 10.1007/s11103-016-0430-6
Pankievicz, V. C. S., do Amaral, F. P., Ané, J.-M., and Stacey, G. (2021). Diazotrophic bacteria and their mechanisms to interact and benefit cereals. Mol. Plant Microbe Interact. 34, 491–498. doi: 10.1094/MPMI-11-20-0316-FI
Pankievicz, V. C. S., do Amaral, F. P., Santos, K. F. D. N., Agtuca, B., Xu, Y., Schueller, M. J., et al. (2015). Robust biological nitrogen fixation in a model grass–bacterial association. Plant J. 81, 907–919. doi: 10.1111/tpj.12777
Peiffer, J. A., Spor, A., Koren, O., Jin, Z., Tringe, S. G., Dangl, J. L., et al. (2013). Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. U.S.A. 110, 6548–6553. doi: 10.1073/pnas.1302837110
Peoples, M. B., Herridge, D. F., and Ladha, J. K. (1995). Biological nitrogen fixation: an efficient source of nitrogen for sustainable agricultural production? Plant Soil 174, 3–28. doi: 10.1007/BF00032239
Pinski, A., Betekhtin, A., Hupert-Kocurek, K., Mur, L. A. J., and Hasterok, R. (2019). Defining the genetic basis of plant-endophytic bacteria interactions. Int. J. Mol. Sci. 20:1947. doi: 10.3390/ijms20081947
Plett, J. M., and Martin, F. M. (2018). Know your enemy, embrace your friend: using omics to understand how plants respond differently to pathogenic and mutualistic microorganisms. Plant J. 93, 729–746. doi: 10.1111/tpj.13802
Pongdet, P., Teerana, G., Kamonluck, T., Takashi, O., Ryo, S., Achara, N., et al. (2015). Preferential association of endophytic bradyrhizobia with different rice cultivars and its implications for rice endophyte evolution. Appl. Environ. Microbiol. 81, 3049–3061. doi: 10.1128/AEM.04253-14
Qi, P. F., Zhang, Y. Z., Liu, C. H., Chen, Q., Guo, Z. R., Wang, Y., et al. (2019). Functional analysis of FgNahG clarifies the contribution of salicylic acid to wheat (Triticum aestivum) resistance against fusarium head blight. Toxins (Basel) 11:59. doi: 10.3390/toxins11020059
Qin, D., Wang, L., Han, M., Wang, J., Song, H., Yan, X., et al. (2018). Effects of an endophytic fungus Umbelopsis dimorpha on the secondary metabolites of host–plant Kadsura angustifolia. Front. Microbiol. 9:2845. doi: 10.3389/fmicb.2018.02845
Qin, S., Feng, W. W., Zhang, Y. J., Wang, T. T., Xiong, Y. W., and Xing, K. (2018). Diversity of Bacterial microbiota of coastal halophyte limonium sinense and amelioration of salinity stress damage by symbiotic plant growth-promoting actinobacterium Glutamicibacter halophytocola KLBMP 5180. Appl. Environ. Microbiol. 84:e01533-18. doi: 10.1128/AEM.01533-18
Qin, Y., Druzhinina, I. S., Pan, X., and Yuan, Z. (2016). Microbially mediated plant salt tolerance and microbiome-based solutions for saline agriculture. Biotechnol. Adv. 34, 1245–1259. doi: 10.1016/j.biotechadv.2016.08.005
Quilbé, J., Lamy, L., Brottier, L., Leleux, P., Fardoux, J., Rivallan, R., et al. (2021). Genetics of nodulation in Aeschynomene evenia uncovers mechanisms of the rhizobium–legume symbiosis. Nat. Commun. 12:829. doi: 10.1038/s41467-021-21094-7
Radhakrishnan, G. V., Keller, J., Rich, M. K., Vernié, T., Mbadinga Mbadinga, D. L., Vigneron, N., et al. (2020). An ancestral signalling pathway is conserved in intracellular symbioses-forming plant lineages. Nat. Plants 6, 280–289. doi: 10.1038/s41477-020-0613-7
Radutoiu, S., Madsen, L. H., Madsen, E. B., Felle, H. H., Umehara, Y., Grønlund, M., et al. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425, 585–592. doi: 10.1038/nature02039
Rafaluk-Mohr, C. (2019). The relationship between parasite virulence and environmental persistence: a meta-analysis. Parasitology 146, 897–902. doi: 10.1017/S0031182019000015
Redecker, D., Kodner, R., and Graham, L. E. (2000). Glomalean fungi from the Ordovician. Science 289, 1920–1921. doi: 10.1126/science.289.5486.1920
Redman, R. S., Dunigan, D. D., Rodriguez, R. J., and Rodriguez, R. (2001). Fungal symbiosis from mutualism to parasitism: who controls the outcome, host or invader? New Phytol. 151, 705–716. doi: 10.1046/j.0028-646x.2001.00210.x
Reinhardt, D., Roux, C., Corradi, N., and Di Pietro, A. (2021). Lineage-specific genes and cryptic sex: parallels and differences between arbuscular mycorrhizal fungi and fungal pathogens. Trends Plant Sci. 26, 111–123. doi: 10.1016/j.tplants.2020.09.006
Rivas-Ubach, A., Sardans, J., Pérez-Trujillo, M., Estiarte, M., and Peñuelas, J. (2012). Strong relationship between elemental stoichiometry and metabolome in plants. Proc. Natl. Acad. Sci. U.S.A. 109, 4181–4186. doi: 10.1073/pnas.1116092109
Rodriguez, P. A., Rothballer, M., Chowdhury, S. P., Nussbaumer, T., Gutjahr, C., and Falter-Braun, P. (2019). Systems biology of plant-microbiome interactions. Mol. Plant 12, 804–821. doi: 10.1016/j.molp.2019.05.006
Rodriguez, R. J., Henson, J., Van Volkenburgh, E., Hoy, M., Wright, L., Beckwith, F., et al. (2008). Stress tolerance in plants via habitat-adapted symbiosis. ISME J. 2, 404–416. doi: 10.1038/ismej.2007.106
Roy, S., Liu, W., Nandety, R. S., Crook, A., Mysore, K. S., Pislariu, C. I., et al. (2020). Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation[OPEN]. Plant Cell 32, 15–41. doi: 10.1105/tpc.19.00279
Sabra, M., Aboulnasr, A., Franken, P., Perreca, E., Wright, L. P., and Camehl, I. (2018). Beneficial root endophytic fungi increase growth and quality parameters of sweet basil in heavy metal contaminated soil. Front. Plant Sci. 9:1726. doi: 10.3389/fpls.2018.01726
Sai, W., Pei-hong, W., Wen-han, N. I. E., Zhou-qi, C. U. I., Hong-yu, L. I., and Yan, W. U. (2021). Horizontal gene transfer of a syp homolog contributes to the virulence of Burkholderia glumae. J. Integr. Agric. 20, 3222–3229. doi: 10.1016/S2095-3119(20)63553-5
Sánchez-Vallet, A., Mesters, J. R., and Thomma, B. P. H. J. (2015). The battle for chitin recognition in plant-microbe interactions. FEMS Microbiol. Rev. 39, 171–183. doi: 10.1093/femsre/fuu003
Sánchez-Vallet, A., Saleem-Batcha, R., Kombrink, A., Hansen, G., Valkenburg, D.-J., Thomma, B. P. H. J., et al. (2013). Fungal effector Ecp6 outcompetes host immune receptor for chitin binding through intrachain LysM dimerization. eLife 2, e00790. doi: 10.7554/eLife.00790
Sánchez-Vallet, A., Tian, H., Rodriguez-Moreno, L., Valkenburg, D.-J., Saleem-Batcha, R., Wawra, S., et al. (2020). A secreted LysM effector protects fungal hyphae through chitin-dependent homodimer polymerization. PLoS Pathog. 16:e1008652. doi: 10.1371/journal.ppat.1008652
Sarkar, A., Marszalkowska, M., Schäfer, M., Pees, T., Klingenberg, H., Macht, F., et al. (2017). Global expression analysis of the response to microaerobiosis reveals an important cue for endophytic establishment of Azoarcus sp. BH72. Environ. Microbiol. 19, 198–217. doi: 10.1111/1462-2920.13569
Sarkar, D., Rovenich, H., Jeena, G., Nizam, S., Tissier, A., Balcke, G. U., et al. (2019). The inconspicuous gatekeeper: endophytic Serendipita vermifera acts as extended plant protection barrier in the rhizosphere. New Phytol. 224, 886–901. doi: 10.1111/nph.15904
Schmid, J., Day, R., Zhang, N., Dupont, P.-Y., Cox, M. P., Schardl, C. L., et al. (2017). Host tissue environment directs activities of an Epichloë endophyte, while it induces systemic hormone and defense responses in its native perennial ryegrass host. Mol. Plant-Microbe Interact. 30, 138–149. doi: 10.1094/MPMI-10-16-0215-R
Shahzad, R., Latif, A., Muhammad, K., Ihsan, W., Saqib, U., Yoon, B., et al. (2019). Metabolic and proteomic alteration in phytohormone-producing endophytic Bacillus amyloliquefaciens RWL-1 during methanol utilization. Metabolomics 15:16. doi: 10.1007/s11306-018-1467-0
Shao, X., Tan, M., Xie, Y., Yao, C., Wang, T., Huang, H., et al. (2021). Integrated regulatory network in Pseudomonas syringae reveals dynamics of virulence. Cell Rep. 34:108920. doi: 10.1016/j.celrep.2021.108920
Sharma, N., and Varma, A. (2021). “Role of endophytic fungus Piriformospora indica in nutrient acquisition and plant health,” in Symbiotic Soil Microorganisms: Biology and Applications, eds N. Shrivastava, S. Mahajan, and A. Varma (Cham: Springer International Publishing), 161–169. doi: 10.1007/978-3-030-51916-2_10
Sheibani-Tezerji, R., Rattei, T., Sessitsch, A., Trognitz, F., and Mitter, B. (2015). Transcriptome profiling of the endophyte Burkholderia phytofirmans PsJN indicates sensing of the plant environment and drought stress. MBio 6, e00621-15. doi: 10.1128/mBio.00621-15
Shin, Y., and Brangwynne, C. P. (2017). Liquid phase condensation in cell physiology and disease. Science 357:eaaf4382. doi: 10.1126/science.aaf4382
Smith, S. E., and Smith, F. A. (2011). Roles of arbuscular mycorrhizas in plant nutrition and growth: new paradigms from cellular to ecosystem scales. Annu. Rev. Plant Biol. 62, 227–250. doi: 10.1146/annurev-arplant-042110-103846
Song, S., Liu, Y., Wang, N. R., and Haney, C. H. (2021). Mechanisms in plant–microbiome interactions: lessons from model systems. Curr. Opin. Plant Biol. 62:102003. doi: 10.1016/j.pbi.2021.102003
Sorty, A. M., Meena, K. K., Choudhary, K., Bitla, U. M., Minhas, P. S., and Krishnani, K. K. (2016). Effect of plant growth promoting bacteria associated with halophytic weed (Psoralea corylifolia L) on germination and seedling growth of wheat under saline conditions. Appl. Biochem. Biotechnol. 180, 872–882. doi: 10.1007/s12010-016-2139-z
Stierle, A., Strobel, G., and Stierle, D. (1993). Taxol and taxane production by Taxomyces andreanae, an endophytic fungus of Pacific yew. Science 260:214. doi: 10.1126/science.8097061
Straub, D., Yang, H., Liu, Y., Tsap, T., and Ludewig, U. (2013). Root ethylene signalling is involved in Miscanthus sinensis growth promotion by the bacterial endophyte Herbaspirillum frisingense GSF30T. J. Exp. Bot. 64, 4603–4615. doi: 10.1093/jxb/ert276
Stringlis, I. A., and Pieterse, C. M. J. (2021). Evolutionary “hide and seek” between bacterial flagellin and the plant immune system. Cell Host Microbe 29, 548–550. doi: 10.1016/j.chom.2021.03.010
Strobel, G., Daisy, B., Castillo, U., and Harper, J. (2004). Natural Products from Endophytic Microorganisms. J. Nat. Prod. 67, 257–268. doi: 10.1021/np030397v
Su, P., Zhao, L., Li, W., Zhao, J., Yan, J., Ma, X., et al. (2021). Integrated metabolo-transcriptomics and functional characterization reveals that the wheat auxin receptor TIR1 negatively regulates defense against Fusarium graminearum. J. Integr. Plant Biol. 63, 340–352. doi: 10.1111/jipb.12992
Su, Z. Zhu, Wang, T., Shrivastava, N., Chen, Y. Y., Liu, X., Sun, C., et al. (2017). Piriformospora indica promotes growth, seed yield and quality of Brassica napus L. Microbiol. Res. 199, 29–39. doi: 10.1016/j.micres.2017.02.006
Swarnalakshmi, K., Rajkhowa, S., Senthilkumar, M., and Dhar, D. W. (2019). “Influence of endophytic bacteria on growth promotion and protection against diseases in associated plants,” in Microbial Interventions in Agriculture and Environment: Volume 3: Soil and Crop Health Management, eds D. P. Singh and R. Prabha (Berlin: Springer), 263–287. doi: 10.1007/978-981-32-9084-6_12
Teixeira, P. J. P. L., Colaianni, N. R., Law, T. F., Conway, J. M., Gilbert, S., Li, H., et al. (2021). Specific modulation of the root immune system by a community of commensal bacteria. Proc. Natl. Acad. Sci. U.S.A. 118:e2100678118. doi: 10.1073/pnas.2100678118
Theocharis, A., Bordiec, S., Fernandez, O., Paquis, S., Dhondt-Cordelier, S., Baillieul, F., et al. (2012). Burkholderia phytofirmans PsJN primes Vitis vinifera L. and confers a better tolerance to low nonfreezing temperatures. Mol. Plant-Microbe Interact. 25, 241–249. doi: 10.1094/MPMI-05-11-0124
Tian, Y., Amand, S., Buisson, D., Kunz, C., Hachette, F., Dupont, J., et al. (2014). The fungal leaf endophyte Paraconiothyrium variabile specifically metabolizes the host-plant metabolome for its own benefit. Phytochemistry 108, 95–101. doi: 10.1016/j.phytochem.2014.09.021
Trdá, L., Boutrot, F., Ciaverie, J., Brulé, D., Dorey, S., and Poinssot, B. (2015). Perception of pathogenic or beneficial bacteria and their evasion of host immunity: pattern recognition receptors in the frontline. Front. Plant Sci. 6:219. doi: 10.3389/fpls.2015.00219
Triastuti, A., Vansteelandt, M., Barakat, F., Trinel, M., Jargeat, P., Fabre, N., et al. (2019). How histone deacetylase inhibitors alter the secondary metabolites of Botryosphaeria mamane, an endophytic fungus isolated from Bixa orellana. Chem. Biodivers. 16:e1800485. doi: 10.1002/cbdv.201800485
Trivedi, P., Leach, J. E., Tringe, S. G., Sa, T., and Singh, B. K. (2020). Plant–microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol. 18, 607–621. doi: 10.1038/s41579-020-0412-1
Trovero, M. F., Scavone, P., Platero, R., de Souza, E. M., Fabiano, E., and Rosconi, F. (2018). Herbaspirillum seropedicae differentially expressed genes in response to iron availability. Front. Microbiol. 9:1430. doi: 10.3389/fmicb.2018.01430
Tsuda, K., Sato, M., Stoddard, T., Glazebrook, J., and Katagiri, F. (2009). Network properties of robust immunity in plants. PLoS Genet. 5:e1000772. doi: 10.1371/journal.pgen.1000772
Utturkar, S. M., Cude, W. N., Robeson, M. S., Yang, Z. K., Klingeman, D. M., Land, M. L., et al. (2016). Enrichment of root endophytic bacteria from populus deltoides and single-cell-genomics analysis. Appl. Environ. Microbiol. 82, 5698–5708. doi: 10.1128/AEM.01285-16
Vahabi, K., Sherameti, I., Bakshi, M., Mrozinska, A., Ludwig, A., Reichelt, M., et al. (2015b). The interaction of Arabidopsis with Piriformospora indica shifts from initial transient stress induced by fungus-released chemical mediators to a mutualistic interaction after physical contact of the two symbionts. BMC Plant Biol. 15:58. doi: 10.1186/s12870-015-0419-3
Vahabi, K., Sherameti, I., Bakshi, M., Mrozinska, A., Ludwig, A., and Oelmüller, R. (2015a). Microarray analyses during early and later stages of the Arabidopsis/Piriformospora indica interaction. Genomics Data 6, 16–18. doi: 10.1016/j.gdata.2015.07.019
van Dijk, E. L., Jaszczyszyn, Y., Naquin, D., and Thermes, C. (2018). The third revolution in sequencing technology. Trends Genet. 34, 666–681. doi: 10.1016/j.tig.2018.05.008
van Velzen, R., Holmer, R., Bu, F., Rutten, L., van Zeijl, A., Liu, W., et al. (2018). Comparative genomics of the nonlegume Parasponia reveals insights into evolution of nitrogen-fixing rhizobium symbioses. Proc. Natl. Acad. Sci. U.S.A. 115, E4700–E4709. doi: 10.1073/pnas.1721395115
Vandana, U. K., Rajkumari, J., Singha, L. P., Satish, L., Alavilli, H., Sudheer, P. D. V. N., et al. (2021). The endophytic microbiome as a hotspot of synergistic interactions, with prospects of plant growth promotion. Biology (Basel) 10:101. doi: 10.3390/biology10020101
Vandenkoornhuyse, P., Quaiser, A., Duhamel, M., Le Van, A., and Dufresne, A. (2015). The importance of the microbiome of the plant holobiont. New Phytol. 206, 1196–1206. doi: 10.1111/nph.13312
Vansuyt, G., Robin, A., Briat, J.-F., Curie, C., and Lemanceau, P. (2007). Iron Acquisition from Fe-Pyoverdine by Arabidopsis thaliana. Mol. Plant Microbe Interact. 20, 441–447. doi: 10.1094/MPMI-20-4-0441
Vargas, L., Brígida, A. B. S., Mota Filho, J. P., De Carvalho, T. G., Rojas, C. A., Vaneechoutte, D., et al. (2014). Drought Tolerance Conferred to Sugarcane by Association with Gluconacetobacter diazotrophicus: a Transcriptomic View of Hormone Pathways. PLoS One 9:e114744. doi: 10.1371/journal.pone.0114744
Vera Cruz, C. M., Bai, J., Oña, I., Leung, H., Nelson, R. J., Mew, T.-W., et al. (2000). Predicting durability of a disease resistance gene based on an assessment of the fitness loss and epidemiological consequences of avirulence gene mutation. Proc. Natl. Acad. Sci. U.S.A. 97, 13500–13505. doi: 10.1073/pnas.250271997
Vilanova, C., and Porcar, M. (2016). Are multi-omics enough? Nat. Microbiol. 1:16101. doi: 10.1038/nmicrobiol.2016.101
Vinale, F., Sivasithamparam, K., Ghisalberti, E. L., Marra, R., Barbetti, M. J., Li, H., et al. (2008). A novel role for Trichoderma secondary metabolites in the interactions with plants. Physiol. Mol. Plant Pathol. 72, 80–86. doi: 10.1016/j.pmpp.2008.05.005
Vohník, M. (2020). Ericoid mycorrhizal symbiosis: theoretical background and methods for its comprehensive investigation. Mycorrhiza 30, 671–695. doi: 10.1007/s00572-020-00989-1
Vorburger, C., and Gouskov, A. (2011). Only helpful when required: a longevity cost of harbouring defensive symbionts. J. Evol. Biol. 24, 1611–1617. doi: 10.1111/j.1420-9101.2011.02292.x
Wang, H., Sun, S., Ge, W., Zhao, L., Hou, B., Wang, K., et al. (2020). Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 368:eaba5435. doi: 10.1126/science.aba5435
Wang, Q., Huang, Y., Ren, Z., Zhang, X., Ren, J., Su, J., et al. (2020). Transfer cells mediate nitrate uptake to control root nodule symbiosis. Nat. Plants 6, 800–808. doi: 10.1038/s41477-020-0683-6
Wang, Q., Shakoor, N., Boyher, A., Veley, K. M., Berry, J. C., Mockler, T. C., et al. (2021). Escalation in the host-pathogen arms race: a host resistance response corresponds to a heightened bacterial virulence response. PLoS Pathog. 17:e1009175. doi: 10.1371/journal.ppat.1009175
Wang, S., Guan, Y., Wang, Q., Zhao, J., Sun, G., Hu, X., et al. (2020). A mycorrhizae-like gene regulates stem cell and gametophore development in mosses. Nat. Commun. 11:2896. doi: 10.1038/s41467-020-16421-3
Wang, X., Zhang, X., Liu, L., Xiang, M., Wang, W., Sun, X., et al. (2015). Genomic and transcriptomic analysis of the endophytic fungus Pestalotiopsis fici reveals its lifestyle and high potential for synthesis of natural products. BMC Genomics 16:28. doi: 10.1186/s12864-014-1190-9
Wang, Z., Gerstein, M., and Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63. doi: 10.1038/nrg2484
Wanke, A., Malisic, M., Wawra, S., and Zuccaro, A. (2021). Unraveling the sugar code: the role of microbial extracellular glycans in plant–microbe interactions. J. Exp. Bot. 72, 15–35. doi: 10.1093/jxb/eraa414
Ważny, R., Rozpa̧dek, P., Domka, A., Jȩdrzejczyk, R. J., Nosek, M., Hubalewska-Mazgaj, M., et al. (2021). The effect of endophytic fungi on growth and nickel accumulation in Noccaea hyperaccumulators. Sci. Total Environ. 768:144666. doi: 10.1016/j.scitotenv.2020.144666
Wei, Z., Abdelrahman, M., Gao, Y., Ji, Z., Mishra, R., Sun, H., et al. (2021). Engineering broad-spectrum resistance to bacterial blight by CRISPR/Cas9-mediated precise homology directed repair in rice. Mol. Plant 14, 1215–1218. doi: 10.1016/j.molp.2021.05.012
Wheatley, R. M., Ford, B. L., Li, L., Aroney, S. T. N., Knights, H. E., Ledermann, R., et al. (2020). Lifestyle adaptations of Rhizobium from rhizosphere to symbiosis. Proc. Natl. Acad. Sci. U.S.A. 117, 23823–23834. doi: 10.1073/pnas.2009094117
Wishart, D. S. (2007). Current progress in computational metabolomics. Brief. Bioinform. 8, 279–293. doi: 10.1093/bib/bbm030
Wu, F., Qu, D., Tian, W., Wang, M., Chen, F., Li, K., et al. (2021). Transcriptome analysis for understanding the mechanism of dark septate endophyte S16 in promoting the growth and nitrate uptake of sweet cherry. J. Integr. Agric. 20, 1819–1831. doi: 10.1016/S2095-3119(20)63355-X
Wu, F.-L., Li, Y., Tian, W., Sun, Y., Chen, F., Zhang, Y., et al. (2020). A novel dark septate fungal endophyte positively affected blueberry growth and changed the expression of plant genes involved in phytohormone and flavonoid biosynthesis. Tree Physiol. 40, 1080–1094. doi: 10.1093/treephys/tpaa047
Wurtzel, E. T., and Kutchan, T. M. (2016). Plant metabolism, the diverse chemistry set of the future. Science 353, 1232–1236. doi: 10.1126/science.aad2062
Xia, Y., Ma, Z., Qiu, M., Guo, B., Zhang, Q., Jiang, H., et al. (2020). N-glycosylation shields Phytophthora sojae apoplastic effector PsXEG1 from a specific host aspartic protease. Proc. Natl. Acad. Sci. U.S.A. 117, 27685–27693. doi: 10.1073/pnas.2012149117
Xie, J., Zhang, A., and Wang, X. (2017). Metabolomic applications in hepatocellular carcinoma: toward the exploration of therapeutics and diagnosis through small molecules. RSC Adv. 7, 17217–17226. doi: 10.1039/c7ra00698e
Xin, X. F., and He, S. Y. (2013). Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu. Rev. Phytopathol. 51, 473–498. doi: 10.1146/annurev-phyto-082712-102321
Xin, X. F., Kvitko, B., and He, S. Y. (2018). Pseudomonas syringae: what it takes to be a pathogen. Nat. Rev. Microbiol. 16, 316–328. doi: 10.1038/nrmicro.2018.17
Xing, Y., Xu, N., Bhandari, D. D., Lapin, D., Sun, X., Luo, X., et al. (2021). Bacterial effector targeting of a plant iron sensor facilitates iron acquisition and pathogen colonization. Plant Cell 33, 2015–2031. doi: 10.1093/plcell/koab075
Xu, L., Pierroz, G., Wipf, H. M.-L., Gao, C., Taylor, J. W., Lemaux, P. G., et al. (2021). Holo-omics for deciphering plant-microbiome interactions. Microbiome 9:69. doi: 10.1186/s40168-021-01014-z
Xu, X. H., Wang, C., Li, S. X., Su, Z. Z., Zhou, H. N., Mao, L. J., et al. (2015). Friend or foe: differential responses of rice to invasion by mutualistic or pathogenic fungi revealed by RNAseq and metabolite profiling. Sci. Rep. 5:13624. doi: 10.1038/srep13624
Yan, L., Zhu, J., Zhao, X., Shi, J., Jiang, C., and Shao, D. (2019). Beneficial effects of endophytic fungi colonization on plants. Appl. Microbiol. Biotechnol. 103, 3327–3340. doi: 10.1007/s00253-019-09713-2
Yang, M. Z., Di Ma, M., Yuan, M. Q., Huang, Z. Y., Yang, W. X., Zhang, H. B., et al. (2016). Fungal endophytes as a metabolic fine-tuning regulator for wine grape. PLoS One 11:e0163186. doi: 10.1371/journal.pone.0163186
Yi, Y., de Jong, A., Frenzel, E., and Kuipers, O. P. (2017). Comparative Transcriptomics of Bacillus mycoides strains in response to potato-root exudates reveals different genetic adaptation of endophytic and soil isolates. Front. Microbiol. 8:1487. doi: 10.3389/fmicb.2017.01487
Yin, R., Han, K., Heller, W., Albert, A., Dobrev, P. I., Zažímalová, E., et al. (2014). Kaempferol 3-O-rhamnoside-7-O-rhamnoside is an endogenous flavonol inhibitor of polar auxin transport in Arabidopsis shoots. New Phytol. 201, 466–475. doi: 10.1111/nph.12558
Yuan, J., Zhang, W., Sun, K., Tang, M.-J., Chen, P.-X., Li, X., et al. (2019). Comparative Transcriptomics and Proteomics of Atractylodes lancea in response to endophytic fungus Gilmaniella sp. AL12 reveals regulation in plant metabolism. Front. Microbiol. 10:1208. doi: 10.3389/fmicb.2019.01208
Yuan, Z., Druzhinina, I. S., Gibbons, J. G., Zhong, Z., Van de Peer, Y., Rodriguez, R. J., et al. (2021c). Divergence of a genomic island leads to the evolution of melanization in a halophyte root fungus. ISME J. 15, 3468–3479. doi: 10.1038/s41396-021-01023-8
Yuan, M., Jiang, Z., Bi, G., Nomura, K., Liu, M., Wang, Y., et al. (2021a). Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592, 105–109. doi: 10.1038/s41586-021-03316-6
Yuan, M., Ngou, B. P. M., Ding, P., and Xin, X.-F. (2021b). PTI-ETI crosstalk: an integrative view of plant immunity. Curr. Opin. Plant Biol. 62:102030. doi: 10.1016/j.pbi.2021.102030
Zeng, T., Rodriguez-Moreno, L., Mansurkhodzaev, A., Wang, P., van den Berg, W., Gasciolli, V., et al. (2020). A lysin motif effector subverts chitin-triggered immunity to facilitate arbuscular mycorrhizal symbiosis. New Phytol. 225, 448–460. doi: 10.1111/nph.16245
Zhai, X., Jia, M., Chen, L., Zheng, C. J., Rahman, K., Han, T., et al. (2017). The regulatory mechanism of fungal elicitor-induced secondary metabolite biosynthesis in medical plants. Crit. Rev. Microbiol. 43, 238–261. doi: 10.1080/1040841X.2016.1201041
Zhang, C., He, J., Dai, H., Wang, G., Zhang, X., Wang, C., et al. (2021). Discriminating symbiosis and immunity signals by receptor competition in rice. Proc. Natl. Acad. Sci. U.S.A. 11:, e2023738118. doi: 10.1073/pnas.2023738118
Zhang, W., Wang, J., Xu, L., Wang, A., Huang, L., Du, H., et al. (2018). Drought stress responses in maize are diminished by Piriformospora indica. Plant Signal. Behav. 13:e1414121. doi: 10.1080/15592324.2017.1414121
Zhang, Y., Li, Y., Chen, X., Meng, Z., and Guo, S. (2020). Combined metabolome and transcriptome analyses reveal the effects of mycorrhizal fungus Ceratobasidium sp. AR2 on the flavonoid accumulation in Anoectochilus roxburghii during different growth stages. Int. J. Mol. Sci. 21:564. doi: 10.3390/ijms21020564
Zhao, D., Li, T., Shen, M., Wang, J., and Zhao, Z. (2015). Diverse strategies conferring extreme cadmium (Cd) tolerance in the dark septate endophyte (DSE), Exophiala pisciphila: evidence from RNA-seq data. Microbiol. Res. 170, 27–35. doi: 10.1016/j.micres.2014.09.005
Zhou, F., Emonet, A., Dénervaud Tendon, V., Marhavy, P., Wu, D., Lahaye, T., et al. (2020). Co-incidence of damage and microbial patterns controls localized immune responses in roots. Cell 180, 440–453.e18. doi: 10.1016/j.cell.2020.01.013
Keywords: metabolome, plant–endophyte interaction, plant growth promotion (PGP), stress resistance, transcriptome
Citation: Chen X-l, Sun M-c, Chong S-l, Si J-p and Wu L-s (2022) Transcriptomic and Metabolomic Approaches Deepen Our Knowledge of Plant–Endophyte Interactions. Front. Plant Sci. 12:700200. doi: 10.3389/fpls.2021.700200
Received: 25 April 2021; Accepted: 22 December 2021;
Published: 27 January 2022.
Edited by:
Katharina Pawlowski, Stockholm University, SwedenReviewed by:
Neung Teaumroong, Suranaree University of Technology, ThailandAjar Nath Yadav, Eternal University, India
Copyright © 2022 Chen, Sun, Chong, Si and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ling-shang Wu, c2hhbmcyMDAyMDEyQDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
 Xue-liang Chen
Xue-liang Chen Mei-chen Sun
Mei-chen Sun Sun-li Chong
Sun-li Chong Jin-ping Si
Jin-ping Si Ling-shang Wu
Ling-shang Wu