
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 17 June 2021
Sec. Plant Metabolism and Chemodiversity
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.699326
This article is part of the Research Topic Exploring and Engineering Plant Specialized Metabolism: Latest Advances and New Horizons View all 10 articles
Transcription factors of the WRKY family play pivotal roles in plant defense responses, including the biosynthesis of specialized metabolites. Based on the previous findings of WRKY proteins regulating benzylisoquinoline alkaloid (BIA) biosynthesis, such as CjWRKY1—a regulator of berberine biosynthesis in Coptis japonica—and PsWRKY1—a regulator of morphine biosynthesis in Papaver somniferum—we performed genome-wide characterization of the WRKY gene family in Eschscholzia californica (California poppy), which produces various BIAs. Fifty WRKY genes were identified by homology search and classified into three groups based on phylogenetic, gene structure, and conserved motif analyses. RNA sequencing showed that several EcWRKY genes transiently responded to methyl jasmonate, a known alkaloid inducer, and the expression patterns of these EcWRKY genes were rather similar to those of BIA biosynthetic enzyme genes. Furthermore, tissue expression profiling suggested the involvement of a few subgroup IIc EcWRKYs in the regulation of BIA biosynthesis. Transactivation analysis using luciferase reporter genes harboring the promoters of biosynthetic enzyme genes indicated little activity of subgroup IIc EcWRKYs, suggesting that the transcriptional network of BIA biosynthesis constitutes multiple members. Finally, we investigated the coexpression patterns of EcWRKYs with some transporter genes and discussed the diversified functions of WRKY genes based on a previous finding that CjWRKY1 overexpression in California poppy cells enhanced BIA secretion into the medium.
Being sessile, plants have evolved a wide array of defense mechanisms to protect themselves from diverse environmental stresses. WRKY transcription factors (TFs), one of the most important transcriptional regulators, play pivotal roles in plant development, senescence, and defense responses (Eulgem and Somssich, 2007; Rushton et al., 2010). The WRKY family proteins harbor at least one highly conserved WRKY domain composed of 60 amino acid residues, which includes the conserved N-terminal WRKYGQK sequence followed by a C-terminal zinc finger motif (Eulgem et al., 2000). The WRKY family can be divided into three groups (I–III). Group I proteins generally harbor two WRKY domains and a C2H2-type zinc finger motif. Group II proteins harbor a single WRKY domain and a C2H2-type zinc finger, and these can be further classified into five subgroups (IIa–IIe). Group III proteins also harbor a single WRKY domain and a C2HC-type zinc finger-like motif. The WRKY proteins modulate the expression of target genes by binding to the W-box DNA motif (C/TTGACC/T) in their promoter regions (Ulker and Somssich, 2004). The structures of several WRKY proteins indicate that the conserved WRKYGQK motif with a β-sheet structure binds to the major groove of the DNA strand of W-box sequence (Yamasaki et al., 2013). The RKYGQK residues are directly involved in DNA binding through extensive hydrophobic contacts with the methyl groups of thymine (Yamasaki et al., 2012).
In several species, WRKY family proteins regulate plant-specific (secondary) metabolism related to defense response against biotic and abiotic stresses (Yamada and Sato, 2013). For instance, GaWRKY1 regulates sesquiterpene biosynthesis in Gossypium arboreum (Xu et al., 2004). AaWRKT1 and GLANDULAR TRICHOME-SPECIFIC WRKY1 (AaGSW1) positively regulate antimalarial artemisinin biosynthesis in Artemisia annua (Ma et al., 2009; Chen et al., 2017). CrWRKY1 acts as an activator of monoterpenoid indole alkaloid biosynthesis via binding to the tyrosine decarboxylase (TDC) gene promoter in Catharanthus roseus (Suttipanta et al., 2011). The expression of these WRKY genes could be clearly induced by methyl jasmonate (MeJA)—a crucial phytohormone involved in plant defense and plant-specific metabolism—indicating that the WRKY TFs involved in the regulation of specialized metabolism in plants play important roles in the jasmonic acid (JA) signaling cascade.
Furthermore, the biosynthesis of benzylisoquinoline alkaloids (BIAs), which are pharmaceutically important and structurally divergent specialized chemicals (e.g., analgesics morphine and codeine are found in Papaver somniferum, and antimicrobial berberine in Coptis japonica), is also regulated by the WRKY TFs CjWRKY1 and PsWRKY (Kato et al., 2007; Mishra et al., 2013). In C. japonica, belonging to the Ranunculaceae family, CjWRKY1 specifically regulates the expression of berberine biosynthetic enzyme genes by binding to several W-boxes in their promoters (Kato et al., 2007; Yamada et al., 2016). In P. somniferum, belonging to the Papaveraceae family, PsWRKY plays an important role in wound-induced regulation of morphine biosynthesis (Mishra et al., 2013). Although the functions of both WRKY genes were induced by MeJA, they were classified into different groups of the WRKY family: CjWRKY1 in subgroup IIc and PsWRKY in group I.
Eschscholzia californica (California poppy), belonging to the Papaveraceae family, produces various BIAs, such as sanguinarine, chelerythrine, and escholtzine, which are different types of BIAs from berberine and morphine, while a common biosynthetic pathway from L-tyrosine to (S)-reticuline is shared. The main BIA found in E. californica is sanguinarine, which also produced by P. somniferum cultured cells. The biosynthetic pathways of sanguinarine and related BIAs have been intensively investigated at the molecular level (Supplementary Figure 1). Furthermore, the basic helix–loop–helix TFs EcbHLH1-1 and EcbHLH1-2 have been identified as the positive regulators of sanguinarine biosynthesis (Yamada et al., 2015). Recently, the draft genome sequence of California poppy was compiled and various gene families related to BIA biosynthesis in the genome of this plant were explored (Hori et al., 2018; Yamada et al., 2021). In fact, novel cytochrome P450 enzymes involved in macarpine biosynthesis and possible AP2/ERF TFs involved in the regulation of sanguinarine biosynthesis have been identified (Hori et al., 2018; Yamada et al., 2020).
Heterologous CjWRKY1 expression in E. californica cells strongly enhanced BIA biosynthesis, suggesting the involvement of WRKY protein(s) in the regulation of the BIA biosynthetic pathway in California poppy (Yamada et al., 2017). Although CjWRKY1 has been identified as a comprehensive regulator of almost all genes encoding berberine biosynthetic enzymes in C. japonica, ectopic CjWRKY1 expression in California poppy cells upregulated only a few BIA biosynthetic enzyme genes. Interestingly, CjWRKY1 overexpression in cultured California poppy cells enhanced BIA accumulation in the culture medium. Together, these findings suggest that the potential WRKY TF(s) involved in the regulation of the BIA biosynthetic pathway might be functionally diversified in BIA-producing plant species and gained additional functions associated with BIA production and accumulation in E. californica.
In this study, we investigated the WRKY family genes in the California poppy genome using gene annotation data and compared their expression profiles with those of EcbHLH1 and some EcAP2/ERF genes involved in BIA biosynthesis. We classified the identified genes by phylogenetic analysis and performed gene structure and conserved motif analyses. The expression profiles of the EcWRKY genes in response to MeJA treatment were examined by RNA sequencing (RNA-Seq) and quantitative RT-PCR (qRT-PCR). Moreover, tissue-specific expression patterns of MeJA-responsive EcWRKY genes were investigated by qRT-PCR. We further searched for genes possibly involved in the efflux of BIAs and identified several transporter genes induced by MeJA, based on our previous finding of enhanced BIA secretion following CjWRKY1 overexpression in California poppy cells. The present characterization provides useful information on the physiological roles of EcWRKY genes and the transcriptional network of BIA biosynthesis in E. caifornica.
First, 76 putative WRKY genes were isolated from the E. californica draft genome based on annotated gene information in the Eschscholzia Genome Database.1 Next, 20 genes that did not contain complete WRKY domain-encoding sequences were removed based on domain search using the SMART database,2 and six genes were removed because they harbored partial open reading frames or abnormal sequences, probably due to assembly errors. After sequence validation using the PhytoMetaSyn transcriptomic database3 (Xiao et al., 2013) and the NCBI database4 using BLAST (Supplementary Table 1), 50 WRKY genes were identified in the California poppy genome.
The WRKY domain sequences of WRKY TFs from Arabidopsis thaliana and E. californica were obtained using the SMART database. Multiple sequence alignment was performed with ClustalW using BioEdit.5 An unrooted phylogenetic tree was created using MEGA 7.0.6 The neighbor-joining (NJ) method with the Jones–Thornton–Taylor (JTT) model and 1,000 bootstrap replications was used (Kumar et al., 2016).
The intron–exon organization of E. californica WRKY genes was visualized using the Gene Structure Display Server (GSDS)7 based on the predicted coding sequences and their corresponding genomic sequences. Conserved motifs of the EcWRKY proteins were predicted using MEME Suite (version 5.1.0)8 with the following parameters: maximum motif number of 15 and optimum motif width from ≥6 to ≤50 (Bailey et al., 2009). The topology of the phylogenetic tree was generated based on full-length WRKY protein sequences using MEGA 7.0.
California poppy seedlings (“Hitoezaki”; Takii Seed Co., Ltd.) were grown and treated with 0.1% dimethyl sulfoxide (DMSO) as a control or 100 μM MeJA, as previously described (Yamada et al., 2020). The California poppy plants for tissue expression and metabolite analyses were grown in flowerpots for 5–6 months.
Total RNA extraction and sequencing were performed as described previously (Yamada et al., 2020) (Hokkaido Biosystem Science Co., Ltd., Hokkaido, Japan). The fragments per kilobase of exon model per million fragments mapped (FPKM) values were calculated using Cufflinks to evaluate gene expression levels. Hierarchical clustering was performed and heat maps were constructed based on log2-transformed fold change (FC) values compared to the mock control (0 h) using R.9
Total RNA was extracted from six California poppy seedlings treated with 100 μM MeJA for 0, 0.5, 1, 2, 6, and 24 h, and tissues (leaf blade, petiole, root, flower bud, and flower) were obtained from nine plants using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). Single-stranded cDNA was synthesized from to 500–1,000 ng of total RNA with the ReverTra Ace qPCR RT Master Mix using the gDNA Remover Kit (TOYOBO, Osaka, Japan). Real-time PCR was performed with specific primer pairs (Supplementary Table 2) using the THUNDERBIRD Next SYBR qPCR Mix (TOYOBO, Osaka, Japan) on the LightCycler 96 system (Roche, Basel, Switzerland). The PCR conditions were 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. Gene expression levels were calculated using the 2–ΔΔCt method to analyze MeJA response or generate a standard curve for tissue expression analysis. The relative expression levels were standardized to those of actin as the internal control.
The promoter:LUC constructs of the Ec6OMT and EcCYP719A5 promoters have been constructed previously (Yamada et al., 2020). The full-length cDNAs of subgroup IIc EcWRKY genes were fused to the CaMV 35S promoter in the pBI221 vector, which was used as the effector construct. A dual-LUC reporter assay was then performed using C. japonica protoplasts, as previously described (Yamada and Sato, 2016).
California poppy tissues were ground in liquid nitrogen and extracted overnight with 4 μL mg–1 fresh weight methanol containing 0.01 N HCl at room temperature (20°C). After filtration, the filtrate was prepared for metabolite analysis. Ultra-performance liquid chromatography (UPLC) equipped with QDa mass spectrometry was performed using the ACQUITY UPLC BEH C18 column (2.1 mm × 100 mm, 1.7 μm; Waters Corp.) operated at 40°C. Mobile phase A comprised an aqueous solution of 0.01% acetic acid, whereas mobile phase B comprised acetonitrile containing 0.01% acetic acid. Gradient elution was performed as follows: 0–1 min, 5% B; 1–13 min, 5-30% B; 13–17 min, 30–80% B; 17–18 min, 80–5% B; and 18–20 min, 5% B. The flow rate and injection volume were set at 0.3 mL min–1 and 2 μL, respectively. The QDa conditions were set as follows: cone voltage, 15 V; capillary voltage, 0.8 kV; and source temperature, 600°C. The predicted pavine-type BIAs were detected using total ion chromatography and mass spectrometry in the single-ion recording mode, and the fragmentation spectra (50 V cone voltage) were compared with previous data (Fabre et al., 2000).
To identify the WRKY TF-encoding genes in California poppy, we searched the E. californica draft genome database with gene annotation information using the sequence of a typical WRKY domain. After the removal of incomplete and redundant sequences, a total of 50 putative WRKY genes were identified in the California poppy draft genome, which were designated as EcWRKY1 to EcWRKY50 (Table 1). Of the 50 putative EcWRKY proteins, eight proteins possessed two WRKY domains, while the remaining proteins possessed only a single WRKY domain.
To classify the 50 EcWRKY proteins, multiple sequence alignment using the WRKY domain of the 50 EcWRKY proteins and 72 AtWRKY proteins was performed, and an unrooted phylogenetic tree was constructed using the NJ method (Figure 1 and Supplementary Figure 2). Based on the classification of AtWRKY proteins and the phylogenetic tree, 11, 32, and 7 proteins were classified into groups I, II, and III, respectively (Table 1). Of the 32, respectively 1, 4, 13, 6, and 8 group II EcWRKY proteins were further divided into subgroups IIa, IIb, IIc, IId, and IIe. While majority of the group I WRKY proteins harbored two WRKY domains, three EcWRKY proteins in this group, namely EcWRKY4, EcWRKY15, and EcWRKY17, harbored only a single WRKY domain. Since the presence of group I WRKY proteins with a single WRKY domain has been reported in other plant species (Wei et al., 2012, 2016), these three EcWRKY proteins were classified as the group I WRKY proteins. California poppy has a similar number of WRKY groups to other plant species, with a similar number of genes in each group (Supplementary Table 3).
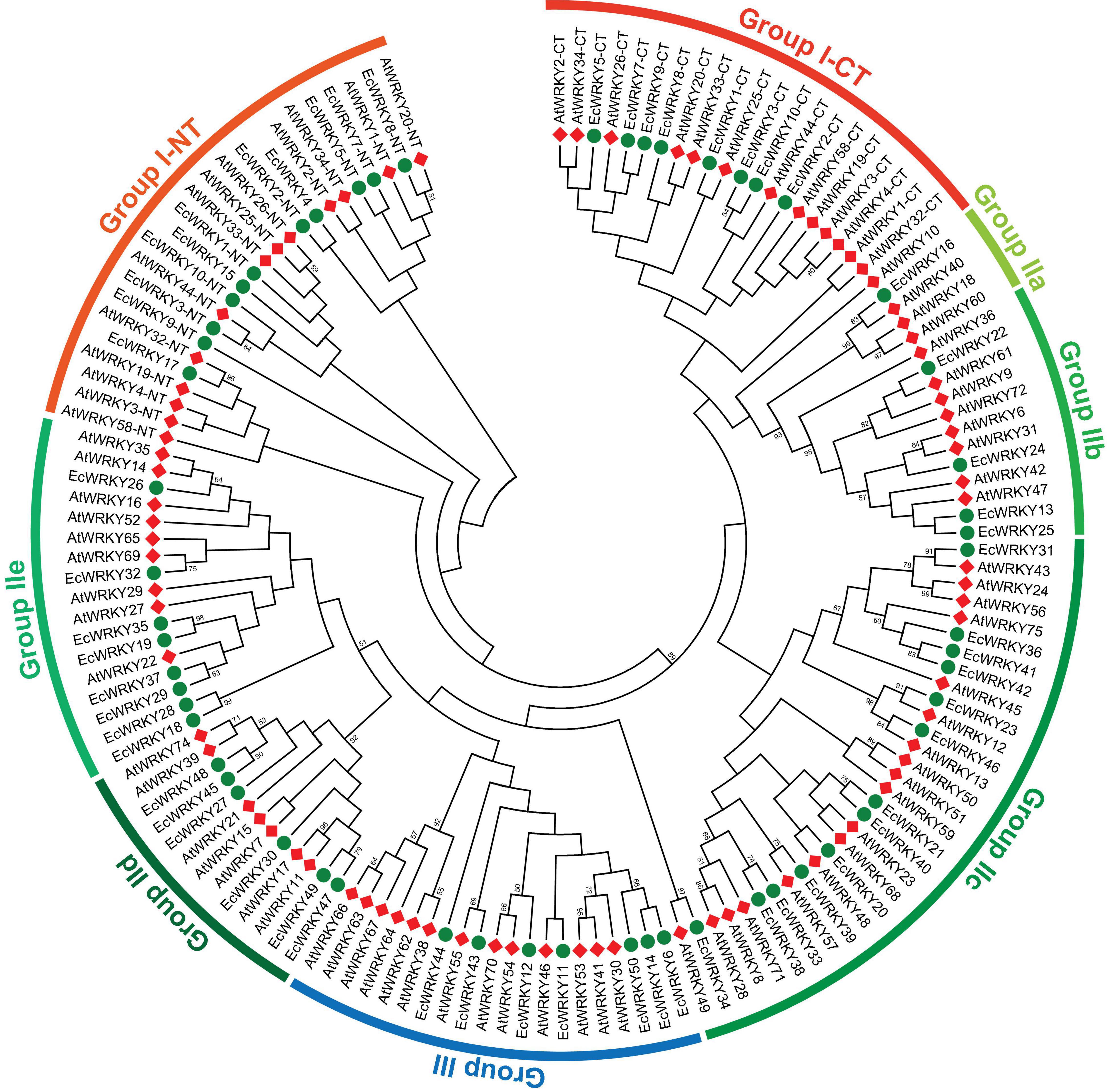
Figure 1. Phylogenetic tree of WRKY proteins in Eschscholzia californica and Arabidopsis thaliana. A neighbor-joining tree was constructed based on the amino acid sequences of the WRKY domains of 50 EcWRKY (green circle) and 72 AtWRKY (red diamond) proteins using MEGA 7.0. Bootstrap confidence values from 1,000 replicates are indicated at each branch.
Homology search using CjWRKY1 and PsWRKY amino acid sequences as queries in the Eschscholzia Genome Database revealed high similarity of CjWRKY1 with three subgroup IIc WRKY proteins, namely EcWRKY36, EcWRKY41, and EcWRKY42. PsWRKY showed the highest similarity to group I EcWRKY1. A phylogenetic tree constructed using the WRKY domain sequences of the 50 EcWRKY proteins, CjWRKY1, and PsWRKY also showed the same result as the homology search (Supplementary Figure 3).
To compare the genomic DNA sequences of 50 EcWRKY genes, we determined their intron–exon structures (Figure 2). All EcWRKY genes had at least two exons, with 43 of the 50 EcWRKY genes having more than three exons. The distributions of introns and exons in the genomic sequences were relatively similar in each group. Most of the group I genes had four to five exons, except EcWRKY4 and EcWRKY17, which had two exons. Furthermore, all subgroup IIb, IId, and III genes had six, three, and three exons, respectively. The phylogenetic tree indicated that subgroup IIc genes were divided into several clades. Four subgroup IIc genes (EcWRKY31, EcWRKY36, EcWRKY41, and EcWRKY42) in one clade had two exons, whereas the remaining nine genes in the other clades had three exons.
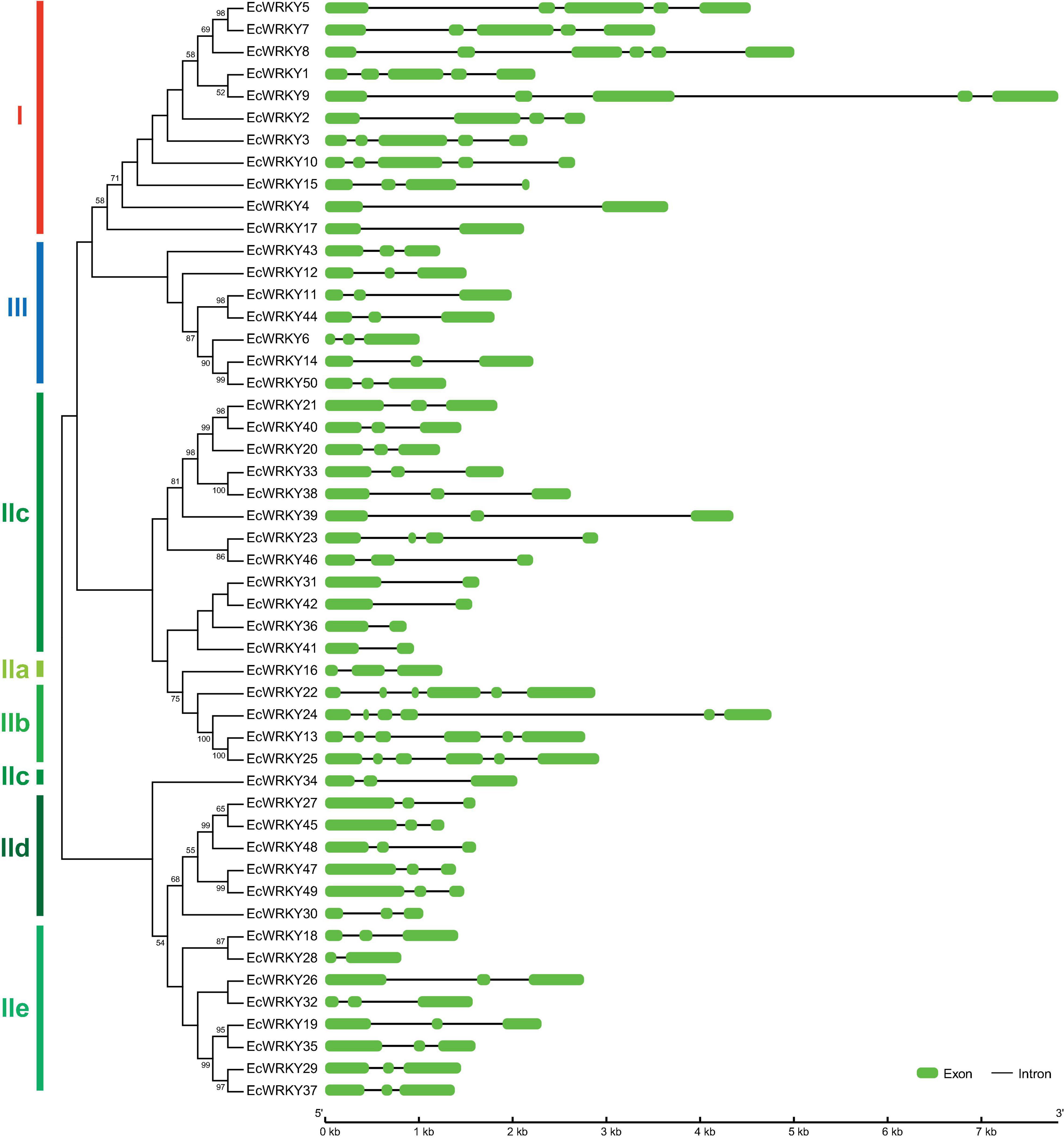
Figure 2. Structure of EcWRKY genes. A phylogenetic tree was constructed as shown in Figure 1, based on the amino acid sequences of the WRKY domains of 50 EcWRKY proteins. The intron–exon structure of the EcWRKY genes was visualized using GSDS. Green and black lines represent the exons and introns, respectively.
To examine the potential motifs of EcWRKY proteins in each family, we analyzed their conserved sequences using MEME Suite, a motif-based sequence analysis tool (Figure 3). Motifs 1, 2, and 3, which are components of the WRKY domain, were found in all EcWRKY proteins, while motif 4, which also contains the WRKYGQK core sequence, was only found in group I proteins, suggesting that motif 4 corresponds to the second WRKY domain. In addition to the WRKY domain, several conserved motifs were found in each EcWRKY family member. For example, motifs 7, 9, 11, and 15 were found only in (sub)group I, IIc, IIb, and IIe WRKY proteins, respectively. These conserved motifs might be important for the functional divergence of each protein group.

Figure 3. Conserved motifs of EcWRKY proteins. Fifteen motifs were identified using MEME and are indicated by colored rectangles. The height of the rectangles is proportional to the −log (p-value), truncated at the height of a motif with a p-value of 1e−10.
MeJA is an important phytohormone involved in defense response (Gundlach et al., 1992). Moreover, alkaloids play critical roles in protecting the plant body against pathogens and herbivores, and the expression of genes involved in the biosynthetic pathways of alkaloids, including BIAs, is strongly induced in response to MeJA (van der Fits and Memelink, 2000; Goossens et al., 2003; Ikezawa et al., 2007; Yamada et al., 2015). To investigate the MeJA responsiveness of EcWRKY genes, transcripts of California poppy seedlings treated with MeJA for 0, 0.5, 1, 3, 6, and 12 h were analyzed using RNA-Seq (Figure 4).
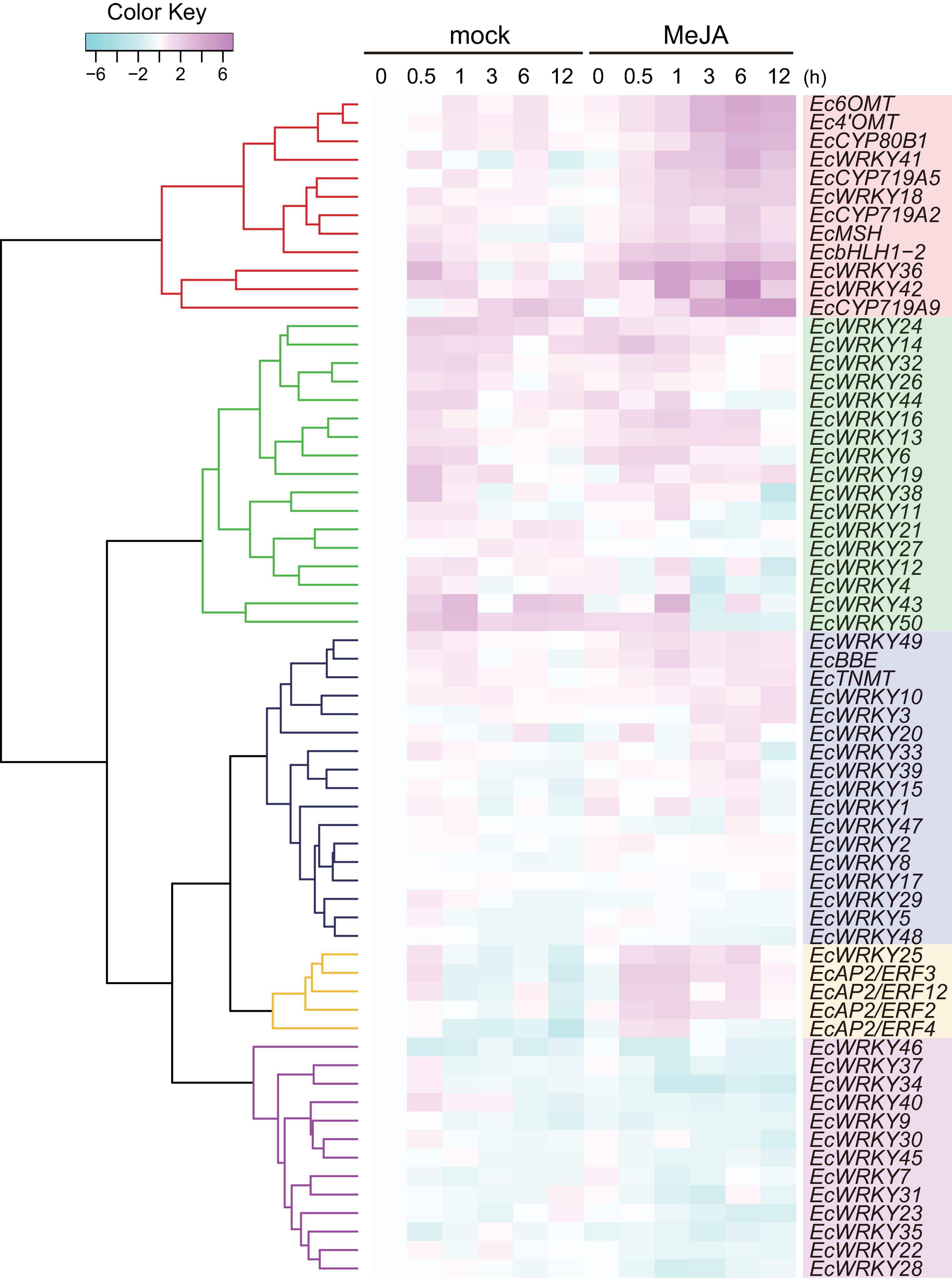
Figure 4. RNA sequencing-based expression profiles of EcWRKY genes following methyl jasmonate (MeJA) treatment. Heat maps showing the clustering of EcWRKY genes with EcbHLH1-2, Ec6OMT, EcCYP80B1, Ec4’OMT, EcBBE, EcCYP719A2, EcCYP719A5, EcCYP719A9, EcTNMT, EcMSH, EcAP2/ERF2, EcAP2/ERF3, EcAP2/ERF4, and EcAP2/ERF12 were created using log2-based FPKM values in R. Within each row, low and high values are indicated in light blue and pink, respectively. The scale represents the signal intensity of FPKM values.
The expression of BIA biosynthetic enzyme genes (Ec6OMT, Ec4’OMT, EcCYP80B1, EcCYP719A5, EcCYP719A2, EcMSH, and EcCYP719A9) and TF genes (EcbHLH1-2 and EcAP2/ERFs) were clearly induced in response to MeJA (Figure 4), as previously reported (Ikezawa et al., 2007, 2009; Yamada et al., 2015, 2020). The expression of EcWRKY18, EcWRKY36, EcWRKY41, and EcWRKY42 was clearly increased following MeJA treatment. Hierarchical clustering indicated that these EcWRKY genes belong to the same clade as the MeJA-responsive BIA biosynthetic enzyme genes and EcbHLH1-2, whereas EcAP2/ERF genes, which showed earlier induction following MeJA treatment (Yamada et al., 2020), were placed in different clades, as discussed later. The expression profile of four EcWRKY genes was rather similar to that of BIA biosynthetic enzyme genes, which were strongly upregulated at 0.5–6 h (log2 FC > 1) and showed the greatest increase in expression after 6 h. In particular, EcWRKY36 and EcWRKY42 showed a more than 5-fold increase in expression after 6 h. Among the four EcWRKY TFs, EcWRKY36, EcWRKY41, and EcWRKY42 showed high similarity to CjWRKY1, as mentioned above (Supplementary Figure 3).
EcBBE and EcTNMT involved in the later stages of BIA biosynthesis were also upregulated by MeJA, although they belonged to a different clade from other biosynthetic enzyme genes. The expression of EcWRKY3, EcWRKY10, and EcWRKY49 was weakly induced in response to MeJA, and these genes were placed in clade closely related to EcBBE and EcTNMT (Figure 4). EcWRKY49 showed the highest expression after 1 h, whereas EcWRKY3 and EcWRKY10 showed the highest expression after 12 h, which suggested that EcWRKY49 might act as an early regulator in the JA signaling cascade to control other MeJA-responsive genes including other EcWRKYs. Both EcWRKY3 and EcWRKY10, which encode group I proteins, showed relatively similar expression patterns in response to MeJA and their slower response to MeJA indicated that EcWRKY3 and EcWRKY10 might work further downstream of the JA signaling cascade.
Although EcWRKY13, EcWRKY16, and EcWRKY25 genes did not show similar expression patterns to BIA biosynthetic enzyme genes in response to MeJA, their expression was upregulated (log2 FC > 1). EcWRKY13 and EcWRKY25 encode subgroup IIb proteins, whereas EcWRKY16 encodes a subgroup IIa protein. Interestingly, the expression pattern of EcWRKY25 was similar to that of the MeJA-responsive group IX EcAP2/ERF genes, which are the possible early regulators of BIA biosynthesis (Yamada et al., 2020).
To verify the expression profiles of EcWRKY genes that showed a clear increase in response to MeJA in RNA-Seq analysis, qRT-PCR was performed using cDNA derived from California poppy seedlings treated with MeJA for 0, 0.5, 1, 2, 6, and 24 h, with three biological replicates (Figure 5). As previously described (Yamada et al., 2015), two EcbHLH1 and EcBBE genes were markedly upregulated in response to MeJA treatment, which is consistent with the results shown in Figure 4. The expression of EcWRKY18, EcWRKY36, EcWRKY41, and EcWRKY42 was highly upregulated in response to MeJA treatment. The expression patterns of subgroup IIc EcWRKY36, EcWRKY41, and EcWRKY42 were very similar. In contrast, MeJA did not strongly induce the expression of EcWRKY3, EcWRKY10, EcWRKY13, EcWRKY16, EcWRKY25, and EcWRKY49 because of variation in gene expression in each seedling sample. Overall, these results indicate that EcWRKY18, EcWRKY36, EcWRKY41, and EcWRKY42, which showed a clear response to MeJA, are candidate WRKY genes involved in BIA biosynthesis.
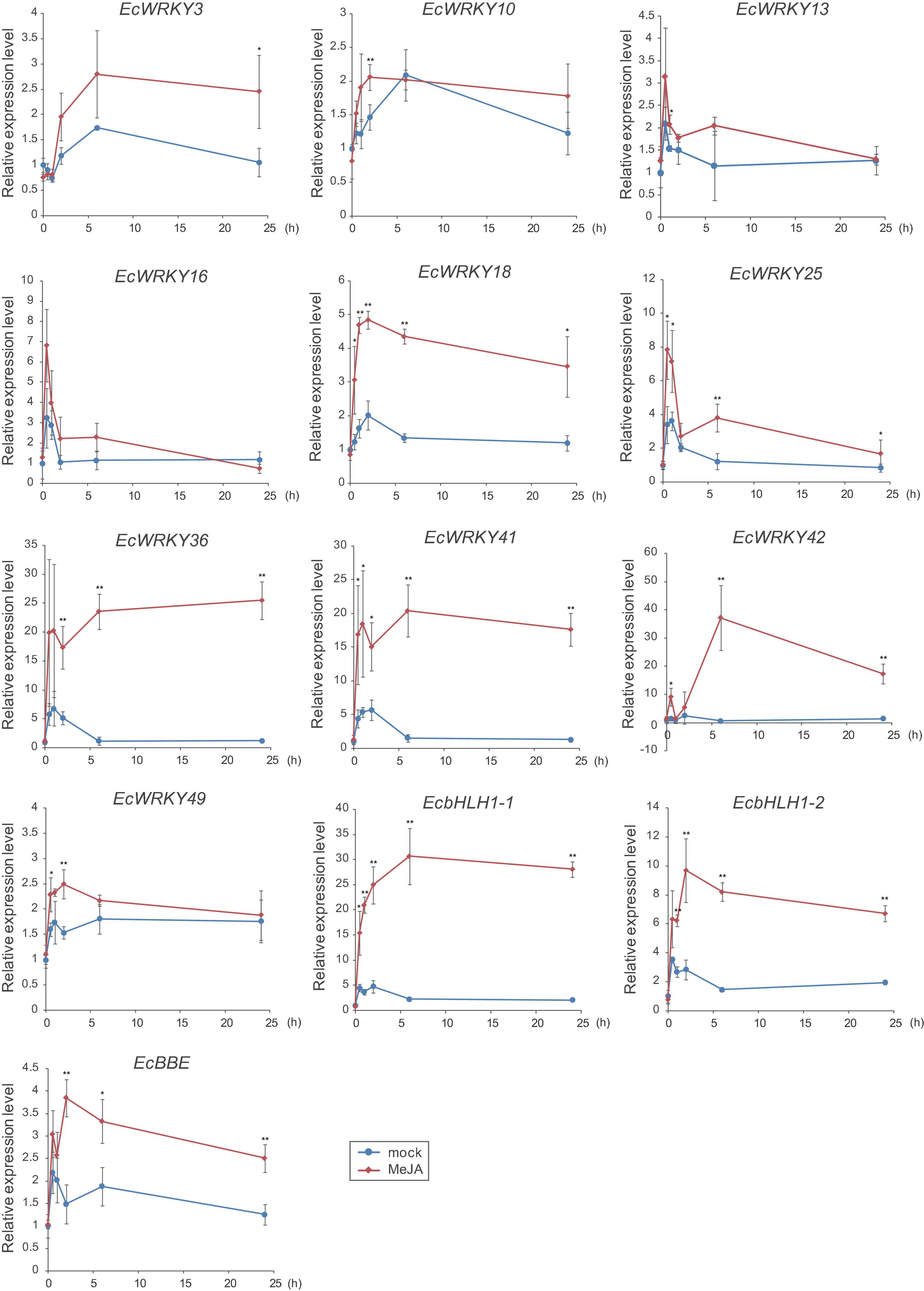
Figure 5. Expression levels of several EcWRKY genes in methyl jasmonate (MeJA)-treated seedlings. Expression levels of ten EcWRKY genes, EcbHLH1-1, EcbHLH1-2, and EcBBE were determined by qRT-PCR. The relative transcript levels represent the values standardized to those of the mock (0 h) samples set to 1. Error bars indicate the standard deviations calculated from three biological replicates. The asterisks denote significant differences according to Student’s t-test compared with the mocks: *P < 0.05; **P < 0.01.
California poppy produces several types of BIA, which are accumulated in specific tissues. For instance, sanguinarine and chelerythrine are commonly accumulated in the root, whereas pavine-type BIAs, such as caryachine, californidine, and escholtzine, are only accumulate in aerial parts (Supplementary Figure 4). A previous study also revealed that genes involved in sanguinarine biosynthesis were highly expressed in roots (Ikezawa et al., 2007; Yamada et al., 2015). To further investigate the involvement of MeJA-responsive EcWRKY genes in the regulation of BIA biosynthesis, we examined the expression profiles of EcWRKY18, EcWRKY36, EcWRKY41, and EcWRKY42 in different tissues, including leaf blades, petioles, roots, flower buds, and flowers (Figure 6) and compared them to the profiles of other TF genes involved in BIA biosynthesis, including EcbHLH and EcERFs. The expression profiles of EcWRKY18 and EcWRKY36 were highly similar to those of Ec6OMT and EcBBE, which encode sanguinarine biosynthetic enzymes; as such, these genes showed the highest expression in roots and relatively high expression in flowers. The expression profile of EcWRKY42 was also similar to that of EcWRKY18 and EcWRKY36, although it showed quite high expression in flowers as in roots. EcbHLH1-2, which is involved in sanguinarine biosynthesis, was highly and exclusively expressed in roots, as reported previously (Yamada et al., 2015). Meanwhile, group IX EcAP2/ERF genes were relatively highly expressed in leaves and roots, and these TF genes showed lower expression levels in flowers than EcWRKY genes. EcCYP719A9, encoding a possible enzyme involved in pavine-type BIA biosynthesis (Ikezawa et al., 2009), was highly expressed in aerial parts, particularly flower buds; however, the expression profile of any EcWRKY genes was not similar to that of EcCYP719A9. These results indicate that EcWRKY18 and subgroup IIc EcWRKYs, namely EcWRKY36, EcWRKY41, and EcWRKY42, are involved in the regulation of benzophenanthridine-type BIA biosynthesis.
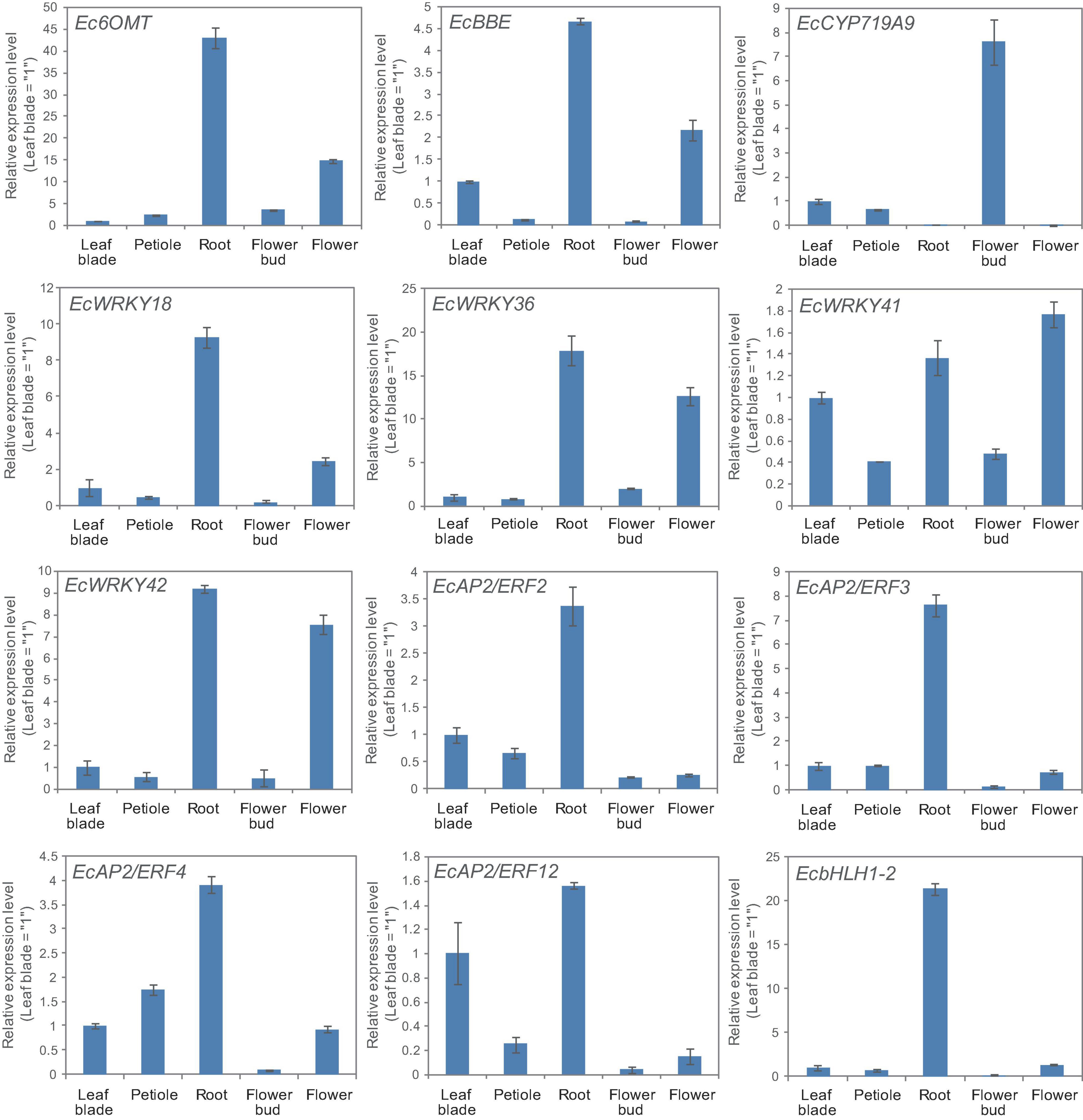
Figure 6. Tissue expression patterns of methyl jasmonate (MeJA)-responsive EcWRKY genes. The expression levels of four MeJA-responsive EcWRKY genes as well as those of Ec6OMT, EcBBE, EcCYP719A9, EcbHLH1-2, EcAP2/ERF2, EcAP2/ERF3, EcAP2/ERF4, and EcAP2/ERF12 were determined by qRT-PCR using cDNA derived from nine California poppy plants. The relative transcript levels represent the values standardized to those of the leaf blade or petiole samples set to 1. Error bars indicate the standard deviations calculated from three technical replicates.
Since EcWRKY36, EcWRKY41, and EcWRKY42 are putative CjWRKY1 homologs in the California poppy, we focused on these proteins and examined their transcriptional activity using a transient LUC reporter assay. While CjWRKY1 showed clear transcriptional activity in C. japonica cells (Kato et al., 2007; Yamada et al., 2016), EcWRKY36, EcWRKY41, and EcWRKY42 showed little induction of LUC activity derived from the Ec6OMT and EcCYP719A5 gene promoter:LUC constructs (Supplementary Figure 5). These results are consistent with our previous findings that the expression of many biosynthetic enzyme genes, including Ec6OMT and EcCYP719A5, was not significantly upregulated in CjWRKY1-overexpressing California poppy cells. These results also suggest that the regulatory role of WRKY TFs in BIA biosynthesis might be diversified between C. japonica and E. californica, and EcWRKY proteins serve different functions in the BIA biosynthetic pathway.
Our previous work revealed that heterologous expression of CjWRKY1 in California poppy cells increased BIA secretion into the culture medium (Yamada et al., 2017). This result suggests that the WRKY TFs modulate the expression of genes encoding transporter proteins involved in the efflux of alkaloids. To investigate the association between EcWRKY proteins involved in the regulation of genes encoding transporter proteins involved in the efflux of California poppy alkaloids, including BIAs, we explored transporter-encoding genes that showed similar expression patterns to EcWRKY18, EcWRKY36, and EcWRKY42 in response to MeJA. We screened 46 transporter-encoding genes that were upregulated (log2 FC > 1) following MeJA treatment for 1–12 h (Supplementary Table 4). These candidates included two genes encoding multidrug and toxic compound extrusion (MATE) transporters and three genes encoding B-type ATP-binding cassette (ABC) transporters (Table 2), have a possibility to be involved in the translocation of alkaloids, such as berberine in C. japonica and nicotine in Nicotiana tabacum (Shitan et al., 2003, 2013, 2014; Morita et al., 2009; Shoji et al., 2009; Takanashi et al., 2017). Hierarchical clustering analysis revealed that the expression patterns of Eca_sc001363.1_g1470.1 and Eca_sc100701.1_g2100.1, which are putative B-type ABC transporter genes in response to MeJA, were relatively similar to those of EcWRKY18 and EcWRKY36 and EcWRKY42, respectively (Figure 7). Therefore, these ABCB transporter genes might be involved in the transport of BIAs and regulated by MeJA-responsive EcWRKY transcription factors.
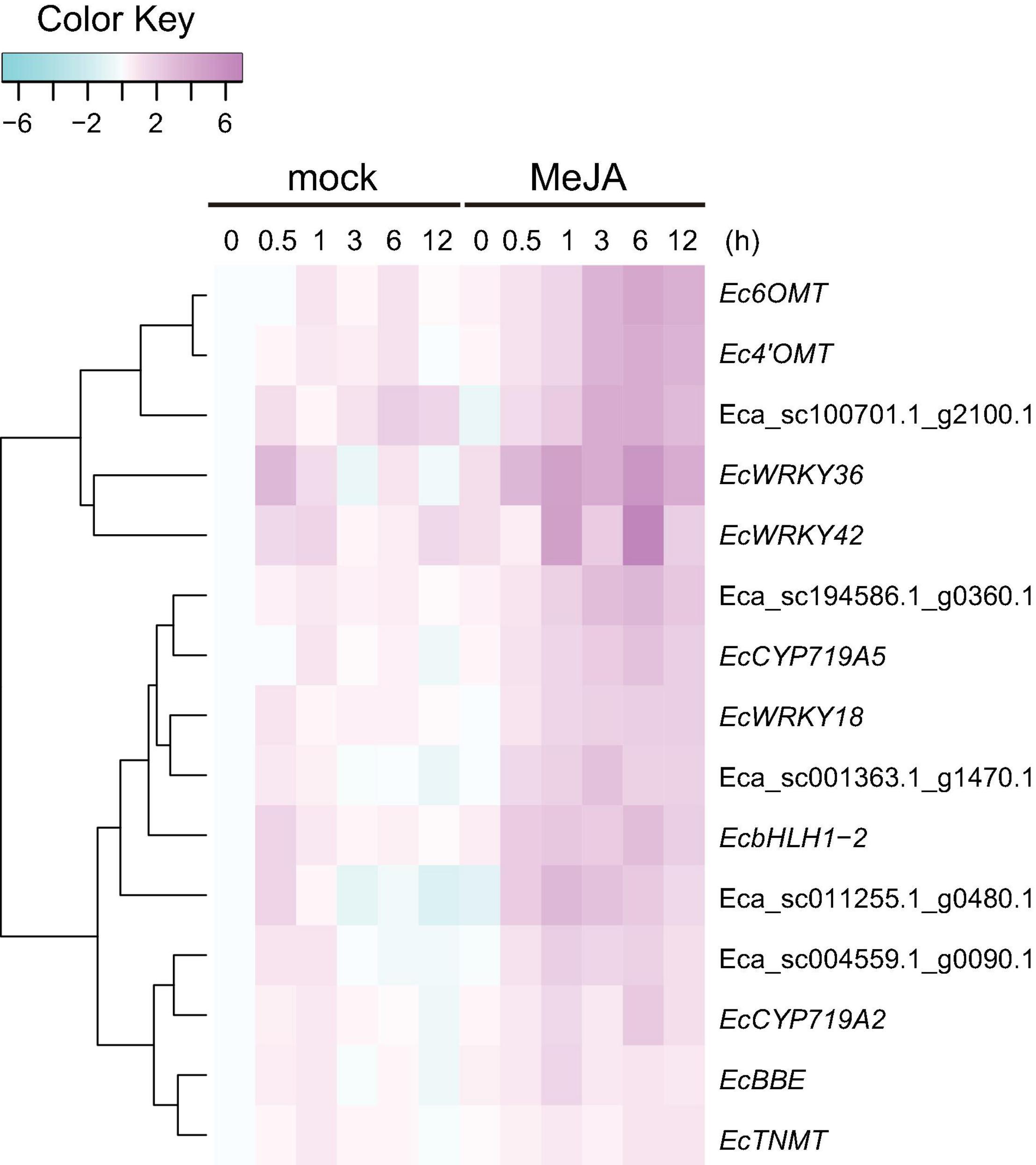
Figure 7. Expression profiles of putative transporter genes and EcWRKY genes following methyl jasmonate (MeJA) treatment. Heat maps showing the clustering of three ABCB transporter genes (Eca_sc001363.1_g1470.1, Eca_sc011255.1_g0480.1, and Eca_sc100701.1_g2100.1) and two MATE transporter genes (Eca_sc004559.1_g0090.1 and Eca_sc194586.1_g0360.1) with three EcWRKY genes (EcWRKY18, EcWRKY36, and EcWRKY42), EcbHLH1-2, Ec6OMT, Ec4’OMT, EcBBE, EcCYP719A2, EcCYP719A5, and EcTNMT were created using log2-based FPKM values. The scale represents the signal intensities of the FPKM values.
Genes of the WRKY superfamily, which is one of the largest groups of TFs involved in plant development and response to various stresses, have been identified in various plants (Rushton et al., 2010). Recent advances in whole-genome sequencing technologies have enabled us to perform genome-wide analysis of WRKY genes in many plant species. To date, 74 WRKY genes have been identified in A. thaliana (Ulker and Somssich, 2004), 52 in C. roseus (Schluttenhofer et al., 2014), 54 in Ananas comosus (Xie et al., 2018), 55 in Cucumis sativus (Ling et al., 2011), 85 in Manihot esculenta (Wei et al., 2016), 70 in Aquilaria sinensis (Xu et al., 2020), and 120 in Gossypium raimondii (Cai et al., 2014). However, our study is the first report on the genome-wide identification of WRKY TFs from E. californica, a BIA-producing plant of the Papaveraceae family. We identified 50 WRKY members in the California poppy draft genome (Table 1). The different number of WRKY genes among plant species may be implicated in differences in the size of the genome and functional diversification of WRKY family proteins during evolution. Based on phylogenetic analysis (Figure 1), the 50 EcWRKY proteins were classified into 11 group I proteins; 32 group II proteins, further divided into 1, 4, 13, 6, and 8 proteins in subgroup IIa, IIb, IIc, IId, and IIe, respectively; and 7 group III proteins. The distribution of each group of WRKY proteins in E. californica was quite similar to that in other plant species, although there were lower subgroup IIa and IIb proteins in California poppy than those in other species (Supplementary Table 3). Since E. californica is a basal eudicot of the Papaveraceae family, this difference likely reflects the evolutionary history of land plant subgroup IIa and IIb genes, which are considered to have evolved from group I genes due to deletion of the domain structure (Wang et al., 2014).
Gene structure and conserved motif analyses indicated that each protein group shared a similar number of introns and similar motifs (Figures 2, 3). All EcWRKY genes possessed more than one intron, which is consistent with reports in other plant species (Wei et al., 2016; Xie et al., 2018; Xu et al., 2020). These results suggest that gene duplication and structural diversification of WRKY genes may have occurred at the early stages of evolution. Furthermore, the similar motif compositions of each WRKY protein group indicate the potential functional similarity among them, as three subgroup IIc EcWRKY genes, namely EcWRKY36, EcWRKY41, and EcWRKY42, showed a marked response to MeJA.
Jasmonic acid signaling is a critical axis in defense response, including the biosynthesis of specialized metabolites, which act as chemical defense compounds against herbivores and pathogens. Alkaloid production is modulated by JA signaling, and many JA-responsive TFs, which play vital roles in the regulation of genes involved in the JA signaling cascade, have been identified and characterized (Yamada and Sato, 2013). Hence, JA-responsive WRKY genes in E. californica may regulate the expression of genes involved in the BIA biosynthetic pathway. The results of RNA-Seq and qRT-PCR revealed that four EcWRKY genes, namely EcWRKY18, EcWRKY36, EcWRKY41, and EcWRKY42, were upregulated following MeJA treatment (Figures 4, 5). The response pattern of EcWRKY genes was relatively similar to that of BIA biosynthetic enzyme genes, including group IX EcAP2/ERF and EcbHLH1-2, which showed a rapid MeJA response (Yamada et al., 2020). These results indicate that EcWRKY TFs may function downstream of group IX EcAP2/ERF and EcbHLH1-2 TFs in the JA signaling cascade (Figure 8). To investigate the detailed transcriptional network of BIA biosynthesis in California poppy, further functional characterization of bHLH, AP2/ERF, and WRKY TFs using stable transformants is warranted. Furthermore, EcWRKY36, EcWRKY41, and EcWRKY42, which are potential CjWRKY1 homologs, showed little transcriptional activity in transient LUC assay using the Ec6OMT and EcCYP719A5 gene promoters (Supplementary Figure 5), which is consistent with our previous results of CjWRKY1 overexpression in Californian poppy cells (Yamada et al., 2017). Therefore, the function of WRKY proteins involved in BIA biosynthesis may have diversified during evolution in E. californica and C. japonica, and EcWRKY proteins may serve additional functions contributing to BIA production. Additionally, post-transcriptional regulation might be involved in BIA biosynthesis such as protein phosphorylation and degradation (Yamada and Sato, 2016).
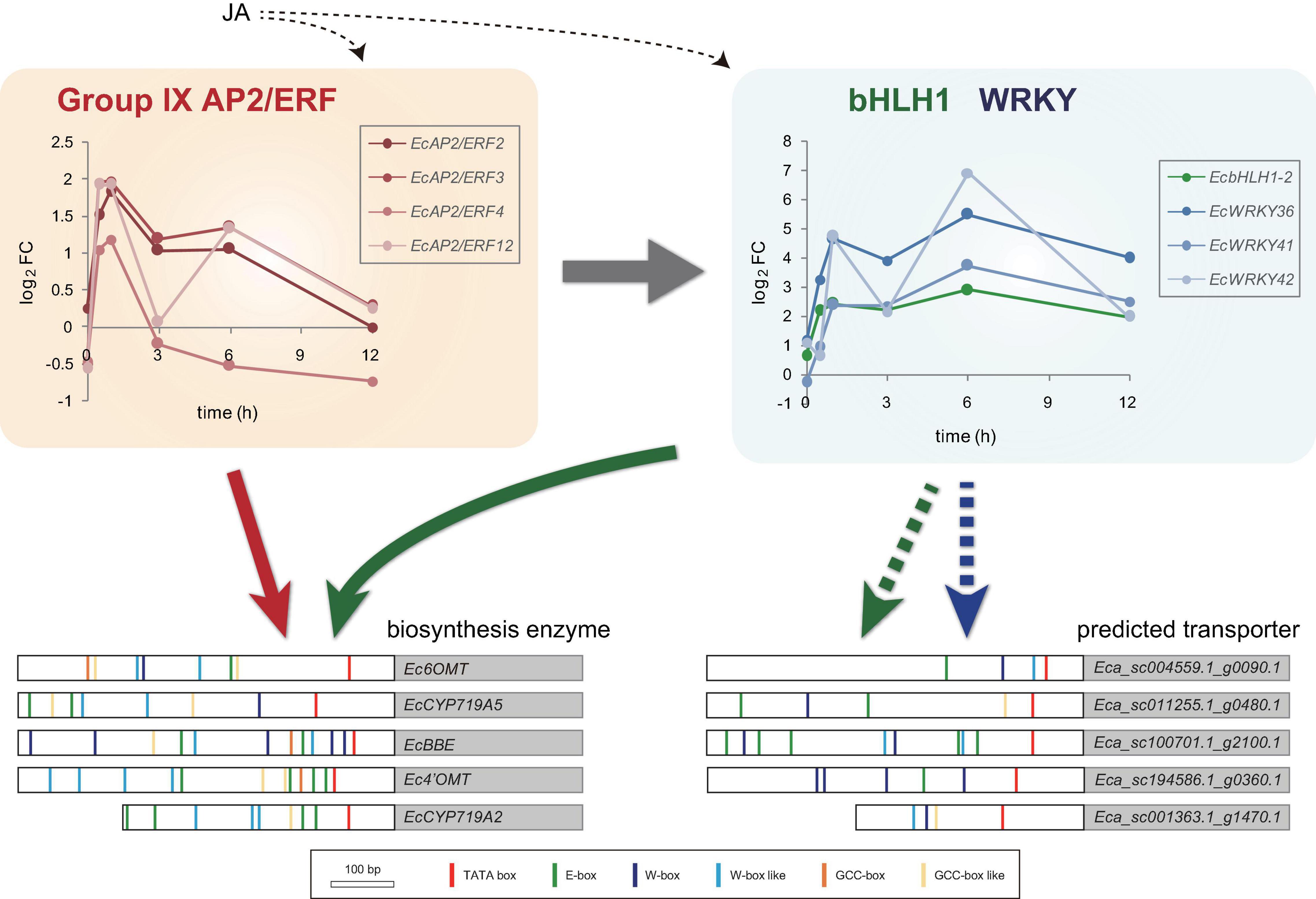
Figure 8. Predicted model of gene expression mechanism in benzylisoquinoline alkaloid biosynthesis via jasmonic acid (JA)-responsive transcription factors. Temporal expression patterns of group IX EcAP2/ERF, EcbHLH1-2, and EcWRKY genes are shown with log2 FC values based on the RNA-Seq data. Large white and gray boxes indicate the partial promoter and coding sequence, respectively. Small boxes with several colors represent the putative cis-acting elements. Based on the previous results, EcAP2/ERF and EcbHLH1 show transactivation of the promoters of biosynthetic enzyme genes (indicated by red and green arrows, respectively). Since the promoters of the predicted transporter genes harbor many putative E-box and W-box elements, EcbHLH and EcWRKY transcription factors may regulate them rather than EcAP2/ERF proteins (indicated by green and blue dotted arrows, respectively).
Group I WRKY proteins are involved in the regulation of BIA biosynthesis. For instance, PsWRKY has been identified as a potent transcriptional activator of BIA biosynthetic genes in P. somniferum (Mishra et al., 2013). Moreover, Apuya et al. (2008) reported that AtWRKY1 overexpression in P. somniferum and E. californica cultured cells enhanced BIA accumulation. In contrast, our RNA-Seq and qRT-PCR data revealed that the expression of group I EcWRKY genes, including EcWRKY1, the closest homologous gene of PsWRKY, was not or weakly induced in response to MeJA (Figures 4, 5). These results also suggest the functional diversification of group I WRKY proteins in the Papaveraceae family during evolution. However, whether PsWRKY modulates the expression of enzymes involved in sanguinarine or morphine biosynthetic pathways in P. somniferum remains unclear. Therefore, detailed functional characterization of group I WRKY proteins in BIA-producing plants is essential.
Our previous study revealed that CjWRKY1 overexpression in California poppy cultured cells enhanced BIA accumulation in culture medium (Yamada et al., 2017), suggesting that WRKY proteins regulate the expression of genes encoding potential transporters of BIAs in this plant. Several TFs regulating genes encoding transport proteins of specialized metabolites have been identified. For instance, A. thaliana MYB TFs regulate the expression of genes involved in the transport of proanthocyanidins (Sharma and Dixon, 2005), and grapevine MYB and WRKY TFs synergistically regulate the expression of genes involved in flavonoid accumulation (Amato et al., 2019). During alkaloid biosynthesis, the expression of genes encoding MATE transporters is regulated by bHLH and AP2/ERF TFs, which also control the expression of biosynthetic enzyme genes (Shoji et al., 2010; Takanashi et al., 2017). The compartmentalization of cytotoxic alkaloids in specific organs or organelles via transporters is important for protection against insects and herbivores, and the regulation of expression of such transporters is important; however, little is known regarding transporters involved in the compartmentalization of BIAs in E. californica cells. In this light, we investigated the coexpression patterns of WRKY and transporter genes in MeJA-treated E. californica seedlings. Three ABCB transporter and two MATE transporter genes that showed a clear MeJA response were coexpressed with EcWRKY18, EcWRKY36, and EcWRKY42. Interestingly, search for putative cis-elements using the New PLACE database10 in the promoter regions of MeJA-responsive transporter genes and biosynthetic enzyme genes (Higo et al., 1999) revealed that there were few GCC-box-like cis-elements, which are target sequences of group IX AP2/ERF TFs, in these transporter genes and at least one GCC-box or GCC-box-like nucleotide sequence was present in genes encoding biosynthetic enzymes (Figure 8). To reveal the direct interaction of these putative cis-elements with AP2/ERF, WRKY, and bHLH TFs, additional analyses are required in future studies. Furthermore, the predicted transporter-encoding genes that were highly upregulated by MeJA included many genes encoding nitrate transporter 1/peptide transporter family (NPF) proteins and purine permeases (Supplementary Table 4). CrNPF2.9 involved in the transport of strictosidine from vacuole to cytosol and BIA uptake purine permeases have recently been isolated from C. roseus and opium poppy, respectively (Payne et al., 2017; Dastmalchi et al., 2019). Therefore, MeJA-responsive NPF transporters and purine permeases might be involved in the translocation of BIAs in E. californica.
In conclusion, our genome-wide analysis and expression profiling of the WRKY family genes in E. californica would be useful for understanding the regulatory mechanisms underlying of BIA biosynthesis, accumulation, and translocation. Especially, different EcWRKY proteins might regulate the spatiotemporal expression patterns of genes related to BIA biosynthesis. Further characterization of EcWRKY TFs and transporters is required to elucidate regulatory mechanisms of BIA production and accumulation in California poppy. This information will contribute to the development of metabolic and transport engineering approaches for the efficient production of valuable alkaloids.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ddbj.nig.ac.jp/, BEHA01000001–BEHA01053253.
YY and FS conceived and designed the study and wrote the manuscript. YY and SN analyzed the genomic and transcriptomic data and performed the experiments. NS and FS supervised the project and discussed the results. All authors reviewed the manuscript.
This research was supported by the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT) [Grant-in-Aid for Scientific Research (S) 26221201 to FS].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Hideki Hirakawa (Kazusa DNA Research Institute, Japan) for depositing the draft genome sequence data of California poppy and establishing the Eschscholzia Genome DataBase. We thank Azusa Hirano and Asuka Tomita (Kobe Pharmaceutical University, Japan) for growing the California poppy plants in flowerpots.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.699326/full#supplementary-material
Amato, A., Cavallini, E., Walker, A. R., Pezzotti, M., Bliek, M., Quattrocchio, F., et al. (2019). The MYB5-driven MBW complex recruits a WRKY factor to enhance the expression of targets involved in vacuolar hyper-acidification and trafficking in grapevine. Plant J. 99, 1220–1241. doi: 10.1111/tpj.14419
Apuya, N. R., Park, J. H., Zhang, L., Ahyow, M., Davidow, P., Van Fleet, J., et al. (2008). Enhancement of alkaloid production in opium and California poppy by transactivation using heterologous regulatory factors. Plant Biotechnol. J. 6, 160–175. doi: 10.1111/j.1467-7652.2007.00302.x
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Cai, C., Niu, E., Du, H., Zhao, L., Feng, Y., and Guo, W. (2014). Genome-wide analysis of the WRKY transcription factor gene family in Gossypium raimondii and the expression of orthologs in cultivated tetraploid cotton. Crop J. 2, 87–101. doi: 10.1016/j.cj.2014.03.001
Chen, M., Yan, T., Shen, Q., Lu, X., Pan, Q., Huang, Y., et al. (2017). GLANDULAR TRICHOME-SPECIFIC WRKY 1 promotes artemisinin biosynthesis in Artemisia annua. New Phytol. 214, 304–316. doi: 10.1111/nph.14373
Dastmalchi, M., Chang, L., Chen, R., Yu, L., Chen, X., Hagel, J. M., et al. (2019). Purine permease-type benzylisoquinoline alkaloid transporters in opium poppy. Plant Physiol. 181, 916–933. doi: 10.1104/pp.19.00565
Eulgem, T., Rushton, P. J., Robatzek, S., and Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/s1360-1385(00)01600-9
Eulgem, T., and Somssich, I. E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10, 366–371. doi: 10.1016/j.pbi.2007.04.020
Fabre, N., Claparols, C., Richelme, S., Angelin, M. L., Fourasté, I., and Moulis, C. (2000). Direct characterization of isoquinoline alkaloids in a crude plant extract by ion-pair liquid chromatography-electrospray ionization tandem mass spectrometry: example of Eschscholtzia californica. J. Chromatogr. A. 904, 35–46. doi: 10.1016/s0021-9673(00)00919-5
Goossens, A., Häkkinen, S. T., Laakso, I., Seppänen-Laakso, T., Biondi, S., De Sutter, V., et al. (2003). A functional genomics approach toward the understanding of secondary metabolism in plant cells. Proc. Natl. Acad. Sci. U.S.A. 100, 8595–8600. doi: 10.1073/pnas.1032967100
Gundlach, H., Müller, M. J., Kutchan, T. M., and Zenk, M. H. (1992). Jasmonic acid is a signal transducer in elicitor-induced plant cell cultures. Proc. Natl. Acad. Sci. U.S.A. 89, 2389–2393. doi: 10.1073/pnas.89.6.2389
Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27, 297–300. doi: 10.1093/nar/27.1.297
Hori, K., Yamada, Y., Purwanto, R., Minakuchi, Y., Toyoda, A., Hirakawa, H., et al. (2018). Mining of the Uncharacterized Cytochrome P450 genes involved in alkaloid biosynthesis in california poppy using a draft genome sequence. Plant Cell Physiol. 59, 222–233. doi: 10.1093/pcp/pcx210
Ikezawa, N., Iwasa, K., and Sato, F. (2007). Molecular cloning and characterization of methylenedioxy bridge-forming enzymes involved in stylopine biosynthesis in Eschscholzia californica. FEBS J. 274, 1019–1035. doi: 10.1111/j.1742-4658.2007.05652.x
Ikezawa, N., Iwasa, K., and Sato, F. (2009). CYP719A subfamily of cytochrome P450 oxygenases and isoquinoline alkaloid biosynthesis in Eschscholzia californica. Plant Cell Rep. 28, 123–133. doi: 10.1007/s00299-008-0624-8
Kato, N., Dubouzet, E., Kokabu, Y., Yoshida, S., Taniguchi, Y., Dubouzet, J. G., et al. (2007). Identification of a WRKY protein as a transcriptional regulator of benzylisoquinoline alkaloid biosynthesis in Coptis japonica. Plant Cell Physiol. 48, 8–18. doi: 10.1093/pcp/pcl041
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Ling, J., Jiang, W., Zhang, Y., Yu, H., Mao, Z., Gu, X., et al. (2011). Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genom. 12:471. doi: 10.1186/1471-2164-12-471
Ma, D., Pu, G., Lei, C., Ma, L., Wang, H., Guo, Y., et al. (2009). Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha-4,11-diene synthase gene, a key gene of artemisinin biosynthesis. Plant Cell Physiol. 50, 2146–2161. doi: 10.1093/pcp/pcp149
Mishra, S., Triptahi, V., Singh, S., Phukan, U. J., Gupta, M. M., Shanker, K., et al. (2013). Wound induced tanscriptional regulation of benzylisoquinoline pathway and characterization of wound inducible PsWRKY transcription factor from Papaver somniferum. PLoS One 8:e52784. doi: 10.1371/journal.pone.0052784
Morita, M., Shitan, N., Sawada, K., Van Montagu, M. C., Inzé, D., Rischer, H., et al. (2009). Vacuolar transport of nicotine is mediated by a multidrug and toxic compound extrusion (MATE) transporter in Nicotiana tabacum. Proc. Natl. Acad. Sci. U.S.A. 106, 2447–2452. doi: 10.1073/pnas.0812512106
Payne, R. M., Xu, D., Foureau, E., Teto Carqueijeiro, M. I., Oudin, A., Bernonville, T. D., et al. (2017). An NPF transporter exports a central monoterpene indole alkaloid intermediate from the vacuole. Nat. Plants 3:16208. doi: 10.1038/nplants.2016.208
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Schluttenhofer, C., Pattanaik, S., Patra, B., and Yuan, L. (2014). Analyses of Catharanthus roseus and Arabidopsis thaliana WRKY transcription factors reveal involvement in jasmonate signaling. BMC Genom. 15:502. doi: 10.1186/1471-2164-15-502
Sharma, S. B., and Dixon, R. A. (2005). Metabolic engineering of proanthocyanidins by ectopic expression of transcription factors in Arabidopsis thaliana. Plant J. 44, 62–75. doi: 10.1111/j.1365-313X.2005.02510.x
Shitan, N., Bazin, I., Dan, K., Obata, K., Kigawa, K., Ueda, K., et al. (2003). Involvement of CjMDR1, a plant multidrug-resistance-type ATP-binding cassette protein, in alkaloid transport in Coptis japonica. Proc. Natl. Acad. Sci. U.S.A. 100, 751–756. doi: 10.1073/pnas.0134257100
Shitan, N., Dalmas, F., Dan, K., Kato, N., Ueda, K., Sato, F., et al. (2013). Characterization of Coptis japonica CjABCB2, an ATP-binding cassette protein involved in alkaloid transport. Phytochemistry 91, 109–116. doi: 10.1016/j.phytochem.2012.02.012
Shitan, N., Minami, S., Morita, M., Hayashida, M., Ito, S., Takanashi, K., et al. (2014). Involvement of the leaf-specific multidrug and toxic compound extrusion (MATE) transporter Nt-JAT2 in vacuolar sequestration of nicotine in Nicotiana tabacum. PLoS One 9:e108789. doi: 10.1371/journal.pone.0108789
Shoji, T., Inai, K., Yazaki, Y., Sato, Y., Takase, H., Shitan, N., et al. (2009). Multidrug and toxic compound extrusion-type transporters implicated in vacuolar sequestration of nicotine in tobacco roots. Plant Physiol. 149, 708–718. doi: 10.1104/pp.108.132811
Shoji, T., Kajikawa, M., and Hashimoto, T. (2010). Clustered transcription factor genes regulate nicotine biosynthesis in tobacco. Plant Cell 22, 3390–3409. doi: 10.1105/tpc.110.078543
Suttipanta, N., Pattanaik, S., Kulshrestha, M., Patra, B., Singh, S. K., and Yuan, L. (2011). The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 157, 2081–2093. doi: 10.1104/pp.111.181834
Takanashi, K., Yamada, Y., Sasaki, T., Yamamoto, Y., Sato, F., and Yazaki, K. (2017). A multidrug and toxic compound extrusion transporter mediates berberine accumulation into vacuoles in Coptis japonica. Phytochemistry 138, 76–82. doi: 10.1016/j.phytochem.2017.03.003
Ulker, B., and Somssich, I. E. (2004). WRKY transcription factors: from DNA binding towards biological function. Curr. Opin. Plant Biol. 7, 491–498. doi: 10.1016/j.pbi.2004.07.012
van der Fits, L., and Memelink, J. (2000). ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289, 295–297. doi: 10.1126/science.289.5477.295
Wang, L., Zhu, W., Fang, L., Sun, X., Su, L., Liang, Z., et al. (2014). Genome-wide identification of WRKY family genes and their response to cold stress in Vitis vinifera. BMC Plant Biol. 14:103. doi: 10.1186/1471-2229-14-103
Wei, K. F., Chen, J., Chen, Y. F., Wu, L. J., and Xie, D. X. (2012). Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res. 19, 153–164. doi: 10.1093/dnares/dsr048
Wei, Y., Shi, H., Xia, Z., Tie, W., Ding, Z., Yan, Y., et al. (2016). Genome-wide identification and expression analysis of the WRKY Gene family in cassava. Front. Plant Sci. 7:25. doi: 10.3389/fpls.2016.00025
Xiao, M., Zhang, Y., Chen, X., Lee, E. J., Barber, C. J., Chakrabarty, R., et al. (2013). Transcriptome analysis based on next-generation sequencing of non-model plants producing specialized metabolites of biotechnological interest. J. Biotechnol. 166, 122–134. doi: 10.1016/j.jbiotec.2013.04.004
Xie, T., Chen, C., Li, C., Liu, J., Liu, C., and He, Y. (2018). Genome-wide investigation of WRKY gene family in pineapple: evolution and expression profiles during development and stress. BMC Genom. 19:490. doi: 10.1186/s12864-018-4880-x
Xu, Y. H., Sun, P. W., Tang, X. L., Gao, Z. H., Zhang, Z., and Wei, J. H. (2020). Genome-wide analysis of WRKY transcription factors in Aquilaria sinensis (Lour.) Gilg. Sci. Rep. 10:3018. doi: 10.1038/s41598-020-59597-w
Xu, Y. H., Wang, J. W., Wang, S., Wang, J. Y., and Chen, X. Y. (2004). Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-delta-cadinene synthase-A. Plant Physiol. 135, 507–515. doi: 10.1104/pp.104.038612
Yamada, Y., Hirakawa, H., Hori, K., Minakuchi, Y., Toyoda, A., Shitan, N., et al. (2021). Comparative analysis using the draft genome sequence of California poppy (Eschscholzia californica) for exploring the candidate genes involved in benzylisoquinoline alkaloid biosynthesis. Biosci. Biotechnol. Biochem. 85, 851–859. doi: 10.1093/bbb/zbaa091
Yamada, Y., Motomura, Y., and Sato, F. (2015). CjbHLH1 homologs regulate sanguinarine biosynthesis in Eschscholzia californica cells. Plant Cell Physiol. 56, 1019–1030. doi: 10.1093/pcp/pcv027
Yamada, Y., Nishida, S., Shitan, N., and Sato, F. (2020). Genome-wide identification of AP2/ERF transcription factor-encoding genes in California poppy (Eschscholzia californica) and their expression profiles in response to methyl jasmonate. Sci. Rep. 10:18066. doi: 10.1038/s41598-020-75069-7
Yamada, Y., and Sato, F. (2013). Transcription factors in alkaloid biosynthesis. Int. Rev. Cell Mol. Biol. 305, 339–382. doi: 10.1016/B978-0-12-407695-2.00008-1
Yamada, Y., and Sato, F. (2016). Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis. Sci. Rep. 6:31988. doi: 10.1038/srep31988
Yamada, Y., Shimada, T., Motomura, Y., and Sato, F. (2017). Modulation of benzylisoquinoline alkaloid biosynthesis by heterologous expression of CjWRKY1 in Eschscholzia californica cells. PLoS One 12:e0186953. doi: 10.1371/journal.pone.0186953
Yamada, Y., Yoshimoto, T., Yoshida, S. T., and Sato, F. (2016). Characterization of the promoter region of biosynthetic enzyme genes involved in berberine biosynthesis in Coptis japonica. Front. Plant Sci. 7:1352. doi: 10.3389/fpls.2016.01352
Yamasaki, K., Kigawa, T., Seki, M., Shinozaki, K., and Yokoyama, S. (2013). DNA-binding domains of plant-specific transcription factors: structure, function, and evolution. Trends Plant Sci. 18, 267–276. doi: 10.1016/j.tplants.2012.09.001
Keywords: benzylisoquinoline alkaloid, Eschscholzia californica, California poppy, WRKY, methyl jasmonate, RNA sequencing, tissue expression
Citation: Yamada Y, Nishida S, Shitan N and Sato F (2021) Genome-Wide Profiling of WRKY Genes Involved in Benzylisoquinoline Alkaloid Biosynthesis in California Poppy (Eschscholzia californica). Front. Plant Sci. 12:699326. doi: 10.3389/fpls.2021.699326
Received: 23 April 2021; Accepted: 25 May 2021;
Published: 17 June 2021.
Edited by:
Yang Zhang, Sichuan University, ChinaReviewed by:
Thu Thuy Dang, University of British Columbia Okanagan, CanadaCopyright © 2021 Yamada, Nishida, Shitan and Sato. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yasuyuki Yamada, eXlhbWFkYUBrb2JlcGhhcm1hLXUuYWMuanA=; Fumihiko Sato, ZnNhdG9AbGlmLmt5b3RvLXUuYWMuanA=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.