- 1College of Horticulture, Shenyang Agricultural University, Shenyang, China
- 2Key Laboratory of Protected Horticulture, Ministry of Education, Shenyang, China
- 3National & Local Joint Engineering Research Center of Northern Horticultural Facilities Design & Application Technology, Shenyang, China
- 4College of Land and Environment, Shenyang Agricultural University, Shenyang, China
- 5College of Agronomy, Jiangxi Agricultural University, Nanchang, China
- 6College of Forestry, Henan University of Science and Technology, Luoyang, China
Perceiving incoming environmental information is critical for optimizing plant growth and development. Multiple B-box proteins (BBXs) play essential roles in light-dependent developmental processes in plants. However, whether BBXs function as a signal integrator between light and temperature in tomato plants remains elusive. In this study, 31 SlBBX genes were identified from the newly released tomato (Solanum lycopersicum) genome sequences and were clustered into five subgroups. Gene structure and protein motif analyses showed relatively high conservation of closely clustered SlBBX genes within each subgroup; however, genome mapping analysis indicated the uneven distribution of the SlBBX genes on tomato chromosomes. Promoter cis-regulatory elements prediction and gene expression indicated that SlBBX genes were highly responsive to light, hormones, and stress conditions. Reverse genetic approaches revealed that disruption of SlBBX7, SlBBX9, and SlBBX20 largely suppressed the cold tolerance of tomato plants. Furthermore, the impairment of SlBBX7, SlBBX9, and SlBBX20 suppressed the photosynthetic response immediately after cold stress. Due to the impairment of non-photochemical quenching (NPQ), the excess photon energy and electron flow excited by low temperature were not consumed in SlBBX7-, SlBBX9-, and SlBBX20- silenced plants, leading to the over reduction of electron carriers and damage of the photosystem. Our study emphasized the positive roles of light signaling transcription factors SlBBXs in cold tolerance in tomato plants, which may improve the current understanding of how plants integrate light and temperature signals to adapt to adverse environments.
Highlight
- SlBBXs function as a signal integrator between light and temperature in tomato.
Introduction
The B-box (BBX) proteins represent a unique class of zinc-finger transcription factors (TFs) that possess single or double B-box domains in their N termini and a CCT (CO, CO-like, and TOC1) domain in their C termini in some cases (Gangappa and Botto, 2014). The B-box domains are of two classes, and each of them coordinates two zinc atoms (Khanna et al., 2009). The dissimilarities in the consensus sequences of the two B-box domains are the results of evolution through the segmental duplication and deletion events (Crocco and Botto, 2013). Studies suggest that the highly conserved CCT domain is important for transcriptional regulation and nuclear transport (Gendron et al., 2012). Furthermore, the valine-proline (VP) motif of six amino acids (G-I/V-V-P-S/T-F) contained by some BBX proteins, plays a crucial role in the interaction with CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1) (Holm et al., 2001; Datta et al., 2006). Based on the domain structures, 32 BBX proteins are divided into five subfamilies in Arabidopsis (Crocco and Botto, 2013; Gangappa and Botto, 2014).
A variety of wavelength-specific photoreceptors are involved in perceiving the light signals in plants, including phytochromes (phys), cryptochromes (CRYs), phototropins (PHOTs), ZEITLUPE family members, and UV-B resistance locus 8 (UVR8) (Galvao and Fankhauser, 2015; Paik and Huq, 2019). Light-activated photoreceptors inhibit the COP1-SUPPRESSOR OF PHYA-105 (SPA) E3 ubiquitin ligase complex, which functions for the degradation of the positive regulators of photomorphogenesis (Galvao and Fankhauser, 2015; Paik and Huq, 2019). Notably, HY5, a target of COP1-mediated protein degradation, plays a vital role in light-regulated plant growth and development (Osterlund et al., 2000; Ahammed et al., 2020). Thus, the light-dependent regulation of COP1–HY5 mediates the plant developmental transition from dark to light.
Upon light irradiation, BBX21 directly binds to BBX22, HY5 and its own promoter regions and activates its transcription (Xu et al., 2016, 2018; Xu, 2020). Moreover, both BBX21 and HY5 can associate with the BBX11 promoter to promote its transcription, while BBX11 binds to the HY5 promoter to activate its transcription (Zhao et al., 2020). Thus, these three TFs (BBX21, HY5, and BBX11) form a positive feedback loop that precisely regulates plant photomorphogenesis. OsBBX14 induces OsHY5L1 gene expression to stimulate photomorphogenesis in rice (Bai et al., 2019). MdBBX37 associates with MdHY5 promoter to inhibit its expression in apple (An et al., 2019a). Additionally, MdBBX22 and MdBBX25/MdCOL4 bind to the MdHY5 promoter to increase and decrease the transcriptional activation of MdHY5, respectively (An et al., 2019b). Both PpBBX16 and PpBBX18 interact with PpHY5 to increase the biochemical activity of PpHY5, while PpHY5 binds to the promoter region of PpBBX18 to promote the transcription of PpBBX18 in pear (Bai et al., 2019a,b). Furthermore, the interaction of PpBBX21 with PpHY5 and PpBBX18 affects the bioactive heterodimer formation of PpHY5-PpBBX18 (Bai et al., 2019b). Tomato RIPENING INHIBITOR (SlRIN) binds to the ripening-induced SlBBXs (i.e., SlBBX19, SlBBX20, and SlBBX26) promoter (Lira et al., 2020). SlBBX20 modulates carotenoid biosynthesis by directly activating PHYTOENE SYNTHASE 1, and is targeted for 26S proteasome-mediated degradation in tomato (Xiong et al., 2019). Therefore, specific BBXs and HY5 constitute an important regulatory network to precisely control normal plant growth and development.
In the darkness, CO/BBX1, BBX4, BBX10, BBX19, BBX20, BBX21, BBX22, BBX23, BBX24, BBX25, BBX28, and BBX29 are ubiquitinated by COP1 and subsequently degraded by the 26S proteasome system (Fan et al., 2012; Gangappa et al., 2013; Wang et al., 2015; Xu et al., 2016; Zhang et al., 2017; Lin et al., 2018; Ordoñez-Herrera et al., 2018; Heng et al., 2019a; Song et al., 2020). Moreover, BBX2 to 9 and BBX13 to 16 interacts with COP1 in vitro, indicating a role for COP1 in controlling the stability of these proteins in darkness (Ordoñez-Herrera et al., 2018). Nevertheless, COP1 preferentially stabilizes BBX11 instead of promoting its degradation (Zhao et al., 2020), which suggests that COP1 likely regulates a yet unidentified protein degrading BBX11. These studies suggest that numerous BBX proteins, along with COP1 and HY5, play critical roles in light-dependent development in plants.
BBX proteins also play vital roles in regulatory networks that control plant adaption to abiotic stress. Previous studies show that both BBX5 and BBX21 positively regulate plant tolerance to drought and salt stress in Arabidopsis (Nagaoka and Takano, 2003; Min et al., 2015). BBX24/STO directly interacts with H-protein promoter binding factor 1 (HPPBF-1), which is a salt-responsive MYB transcription factor, to enhance the root growth and salt tolerance in Arabidopsis (Nagaoka and Takano, 2003). CmBBX22 also positively regulates the plant drought tolerance (Liu Y. N. et al., 2019). In addition, MdBBX10 enhances tolerance to salt and drought by modulating ABA signaling and ROS accumulation (Liu X. et al., 2019). In Arabidopsis, BBX18 and BBX23 control thermomorphogenesis (Ding et al., 2018). Both MdBBX20 and MaCOL1 are responsive to low temperature in apple and banana, respectively (Chen et al., 2012; Fang et al., 2019). Furthermore, ZFPL, a homologous gene of AtBBX32, enhances cold tolerance in the grapevine (Takuhara et al., 2011). CmBBX24 also increases plant cold tolerance in Chrysanthemum (Yang et al., 2014). However, whether SlBBXs are involved in light and cold response in tomato remains to be explored.
In the present study, 31 SlBBX genes were identified and characterized in tomato. Gene distribution, synteny analyses, the architecture of exon-intron and motifs differences were investigated. Promoter and gene expression analysis showed that SlBBXs played important roles in plant response to light and low temperature signaling. Meanwhile, we found that the impairment of SlBBX7, SlBBX9, and SlBBX20 suppressed the photosynthetic response and non-photochemical quenching (NPQ) immediately after cold stress, which resulted in excess photon energy and electron flow in SlBBX7-, SlBBX9-, and SlBBX20- silenced plants, leading to the overreduction of electron carriers and damage of photosystem. Our study indicated that light signaling transcription factors SlBBX7-, SlBBX9-, and SlBBX20- play positive roles in cold tolerance in tomato plants, which may improve the current understanding of plant integrated light and temperature signals to adapt the adverse environments.
Materials and Methods
Genome-Wide Identification of SlBBX Genes in Tomato
The BBXs proteins in tomato were identified based on protein homology searches from Arabidopsis using the Hidden Markov Model (HMM) as previously described (Upadhyay and Mattoo, 2018). The protein sequence of Arabidopsis BBXs were downloaded from the TAIR database (https://www.arabidopsis.org/). Tomato BBX proteins were searched and downloaded from three public databases, including the NCBI database (http://www.ncbi.nlm.nih.gov/), the Phytozome 13.0 database (https://phytozome.jgi.doe.gov/pz/portal.html), and the Sol Genomics Network tomato database (version ITAG 4.0, https://solgenomics.net/). Tomato BBX proteins resulting from both searches (E-value, 10−5) were pooled and redundant sequences were removed. InterProScan database (http://www.ebi.ac.uk/interpro/), SMART (http://smart.embl-heidelberg.de/), and Conserved Domains Database (http://www.ncbi.nlm.nih.gov/cdd/) were used to further confirm the existence of B-box domain in retrieved BBX sequences.
Protein Properties, Multiple Sequence Alignment, Phylogenetic, and Conserved Motifs Analysis
The various physiochemical properties of tomato BBX proteins, such as MW, polypeptide length, pI, instability index, aliphatic index, and GRAVY were investigated using the ExPASy online tool (http://web.expasy.org/protparam/). To estimate the subcellular localization of tomato BBX proteins, we used CELLO v.2.5: sub-cellular localization predictor (http://cello.life.nctu.edu.tw/) (Yu et al., 2006) and pSORT prediction software (http://www.genscript.com/wolf-psort.html) (Horton et al., 2007). Open Reading Frame (ORF) numbers were calculated using the NCBI website (https://www.ncbi.nlm.nih.gov/orffinder/).
A multiple sequence alignment of the identified tomato BBX proteins and the known BBX families from Arabidopsis, rice, and potato, was performed with the MUSCLE (https://www.ebi.ac.uk/Tools/msa/muscle/) (Edgar, 2004) and DNAMAN software (Version 5.2.2). We constructed phylogenetic tree using MEGA 7.0 with 1,000 bootstrap value and the maximum likelihood method (Kumar et al., 2016), and the phylogenetic tree was displayed with an online website Evolview (http://www.evolgenius.info/evolview/#mytrees/) (Zhang et al., 2012).
The presence of conserved BBX_N and CCT_C—domains were identified by NCBI (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), and drawn with IBS software (Illustrator for Biological Sequences, http://ibs.biocuckoo.org/online.php) (Ren et al., 2009). We performed the protein structural motif annotation using the Meme program (http://meme-suite.org/tools/meme) (Bailey et al., 2009; Upadhyay and Mattoo, 2018) and limited our search to a maximum of 20 motifs.
Chromosomal Location, Gene Structure, and Synteny Analysis
SlBBX genes were mapped to tomato chromosomes according to the Phytozome 13.0 database with the MapChart software. The chromosome distribution diagram was drawn by the online website MG2C (http://mg2c.iask.in/mg2c_v2.1/) with the information from Sol Genomics Network (http://www.solgenomics.net).
Genomic DNA sequence and CDS corresponding to each identified SlBBX gene were retrieved from the tomato genome database. Intron-exon were displayed by comparing CDS to genomic sequences with the gene structure display server (GSDS, http://gsds.cbi.pku.edu.cn/) (Hu et al., 2015).
The syntenic blocks were designed from the Plant Genome Duplication Database (http://chibba.agtec.uga.edu/duplication/). The synteny figures were drawn by Circos-0.69 (http://circos.ca/) with E-value setting to 1e−10 and output format as tabular (-m 8).
Cis-Elements of Promoters Analysis
To identify potential light-, stress-, hormone-, and development-related cis-elements, the 2,000-bp genomic DNA sequence upstream of the start codon (ATG) of SlBBX genes were obtained from the tomato genome database. The cis-elements in these SlBBX genes promoter were identified by using the Plant Cis-Acting Regulatory Element (PlantCARE; http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002).
Plant Material and Growth Conditions
Seeds of tomato (Solanum lycopersicum) in the cv “Ailsa Craig” (Accession: LA2838A) background were obtained from the Tomato Genetics Resource Center (http://tgrc.ucdavis.edu) as previously reported (Wang et al., 2016). Seedlings, which grown in pots with a mixture of one part vermiculite to three parts peat, receive Hoagland nutrient solution. The growth conditions for tomato seedlings were 25/20°C (day/night) temperature with a 12 h photoperiod, light intensity of 600 μmol m−2 s−1, and 65% relative humidity. Tobacco rattle virus-based vectors (pTRV1/2) were used for the VIGS of SlBBX genes in tomato with the specific PCR-amplified primers listed in Supplementary Table 3. The PCR-amplified fragment was cloned into the pTRV2 vector. The empty vector of pTRV2 was used as the control. All constructs were confirmed by sequencing and subsequently transformed into Agrobacterium tumefaciens strain GV3101. VIGS was performed as described previously (Wang et al., 2016, 2019). The inoculated plants were grown under a 12 h photoperiod at 22/20°C (day/night).
Light and Cold Treatments
The six-leaf stage plants were used for all experiments. Plants grown under white light were exposed to a temperature of 25 or 4°C for the control or cold treatment, respectively, in environment-controlled growth chambers (Ningbo Jiangnan instrument factory, Ningbo, China). For different light quality treatments, plants were exposed to dark (D), white light (W), or different wavelength [purple (P), 394 nm; blue (B), 450 nm; green (G), 522 nm; yellow (Y), 594 nm; red (R), 660 nm and far-red (FR), 735 nm, Philips] light from 6:00 a.m. to 6:00 p.m. The light intensity was 100 μmol m−2 s−1. The Lighting Passport (Asensetek, Model No. ALP-01, Taiwan) was used to measure light intensity and light quality as a previous study (Wang et al., 2020b).
Gene Expression Analysis
Total RNA was extracted from tomato leaves as described previously (Wang et al., 2020a). The cDNA template for real-time qRT-PCR was synthesized using a Rever-Tra Ace qPCR RT Kit with a genomic DNA-removing enzyme (Toyobo). qRT-PCR experiments were carried out with an SYBR Green PCR Master Mix Kit (TaKaRa) using an Applied Biosystems 7500 Real-Time PCR System (qTOWER3G, Germany). The primer sequences are listed in Supplementary Table 2. The PCR was run at 95°C for 3 min, followed by 40 cycles of 30 s at 95oC, 30 s at 58°C, and 1 min at 72°C. The tomato ACTIN gene was used as an internal control. The relative gene expression was calculated as described previously (Livak and Schmittgen, 2001).
Cold Tolerance Assays
The relative electrolyte leakage (REL), an indicator for cellular membrane permeability, was measured as described previously (Cao et al., 2007). Chlorophyll fluorescence and P700 absorption changes were determined simultaneously using a DUAL-PAM-100 (Heinz Walz, Effeltrich, Germany) as previously described (Wang et al., 2020b). Before measurements, plants were dark-acclimated for 30 min. PSII and PSI parameters were calculated as follows: the maximum quantum yield of PSII (Fv/Fm) as (Fm - Fo)/Fm, the effective quantum yield of PSII [Y(II)] as (Fm' - Fs)/Fm', the quantum yield of non-regulated energy dissipation of PSII [Y(NO)] as Fs/Fm, the quantum yield of regulated energy dissipation of PSII [Y(NPQ)] as 1- Y(II) - Y(NO), NPQ as (Fm – Fm')/Fm', photochemical quenching coefficient (qP) as (Fm' - Fs)/(Fm' – Fo'), the donor limitation of PSI [Y(ND)] as 1 – P700red, or P/Pm, the acceptor side limitation of PSI [Y(NA)] as (Pm – Pm')/Pm, the quantum yield of PSI [Y(I)] as 1 – Y(ND) – Y(NA), where Fm and Fo represent the maximum and minimum fluoresce yields, whereas Pm and Pm′ represent the P700 signals recorded just before (P) then briefly after the onset of a saturating pulse (Pm′). P700red, which represents the fraction of overall P700 that is reduced in a given state, was determined with the help of a saturation pulse. The electron transport rate (ETRI or ETRII) was calculated as 0.5 × abs I × Y(I) or 0.5 × abs I × Y(II), where 0.5 is the proportion of absorbed light reaching PSI or PSII, and abs I is absorbed irradiance taken as 0.84 of incident irradiance (Wang et al., 2020b).
Statistical Analysis
All statistics were calculated using SPSS software. To determine statistical significance, we employed Tukey's test. A value of P < 0.05 was considered to indicate statistical significance.
Results
Identification and Characterization of SlBBX Genes in Tomato
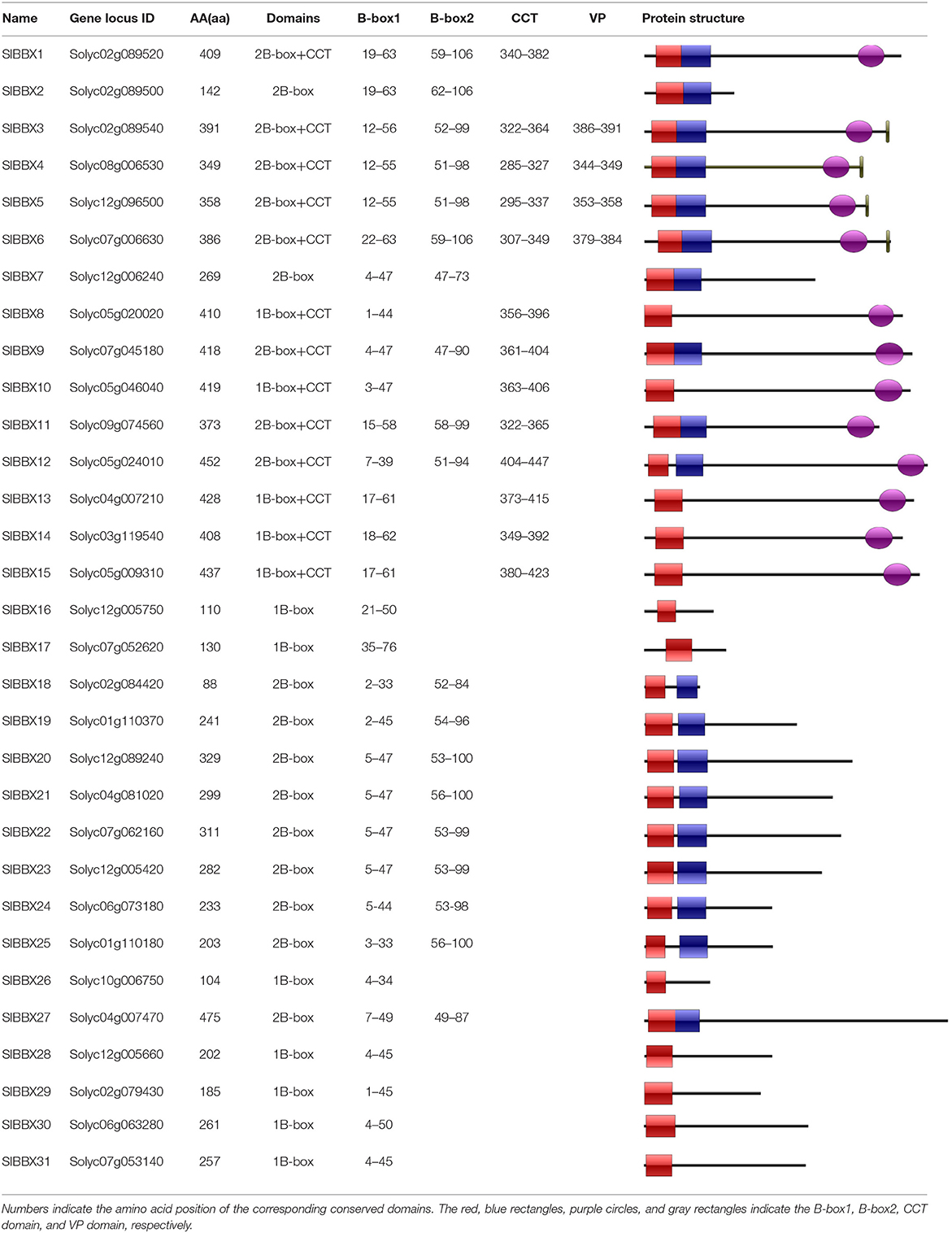
Based on the gene annotation as well as the conserved B-box motif characteristic of the BBX members, a total of 31 SlBBX genes were identified. The detailed information of each SlBBX is presented in Table 1. The lengths of amino acids (AA) of 31 SlBBXs range from 88 aa (SlBBX18) to 475 aa (SlBBX27). Thus, varied molecular weight (MW) and theoretical isoelectric point (pI) were observed among SlBBX proteins. The MW of SlBBXs varies from 9.57 (SlBBX18) to 53.14 kDa (SlBBX27). The pI ranged from 4.25 (SlBBX5 and SlBBX7) to 9.28 (SlBBX26), with 74.2% SlBBXs with a pI lower than seven, which indicated that most of the SlBBX proteins were acidic in nature. The pI ranged from 4 to 9 in SlBBX proteins contained one (single) or two (double) B-box domains; however, the pI ranged from 4 to 7 when SlBBX proteins contained a CCT domain (Supplementary Figure 1), suggesting that the CCT domain in SlBBX proteins may decrease its pI. The majority of SlBBXs were grouped into unstable proteins because their instability index was greater than 40, except for SlBBX6 in this family (Table 1). The predicted aliphatic index ranged from 50.05 to 97.43 in SlBBX proteins. All SlBBX proteins, with the exception of SlBBX18, were predicted to be hydrophilic due to the GRAVY value (<0). Subcellular localization predicted that 23 SlBBX proteins are located in nuclei. Among the rest 8 tomato BBXs, five (SlBBX5, SlBBX6, SlBBX17, SlBBX25, and SlBBX31), two (SlBBX16 and SlBBX18), and one (SlBBX19) SlBBX proteins are located in chloroplast, cytoplasm, and peroxisome, respectively (Table 1).
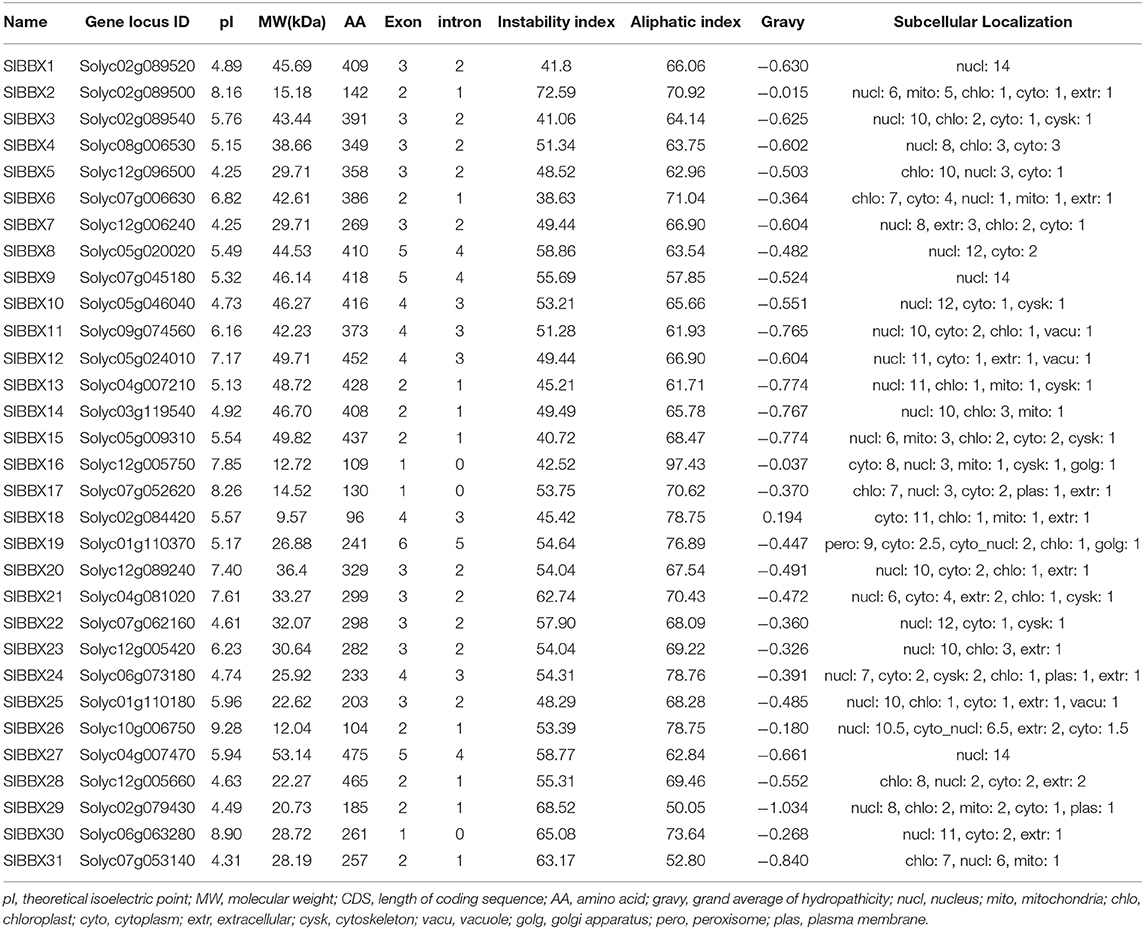
Table 1. Nomenclature, identification, chromosomal location, theoretical isoelectric point (pI), molecular weight (MW), CDS, peptide length, number of exon and intron, instability and aliphatic index, gravy and subcellular localization of BBX gene family in tomato.
Protein Sequences and Phylogenetic Analysis of SlBBXs
The domains logos and the sequences of the B-box1, B-box2, and CCT domain of the SlBBX proteins are shown in Figure 1. Eight members out of the 31 SlBBXs, were characterized by the occurrence of two B-box domains and also a conserved CCT domain, whereas four members of them had a valine-proline (VP) motif (Table 2). Only two B-box domains were found in 10 SlBBXs, whereas five members had one B-box domain and also a CCT domain, and eight members had only one B-box domain (Table 2). Among the three domains, we found that each tomato B-box motif contained ~40 residues with the consensus sequence C-X2-C-X8-C-X2-D-X4-C-X2-C-D-X3-H-X8-H (Figure 1). The conserved C, C, D, and H residues ligated two zinc ions (Khanna et al., 2009). Additionally, the consensus sequence of the conserved CCT domain was R-X5-R-Y-X-E-K-X3-R-X3-K-X2-R-Y-X2-R-K-X2-A-X2-R-X-R-X-K-G-R-F-X-K (Figure 1).
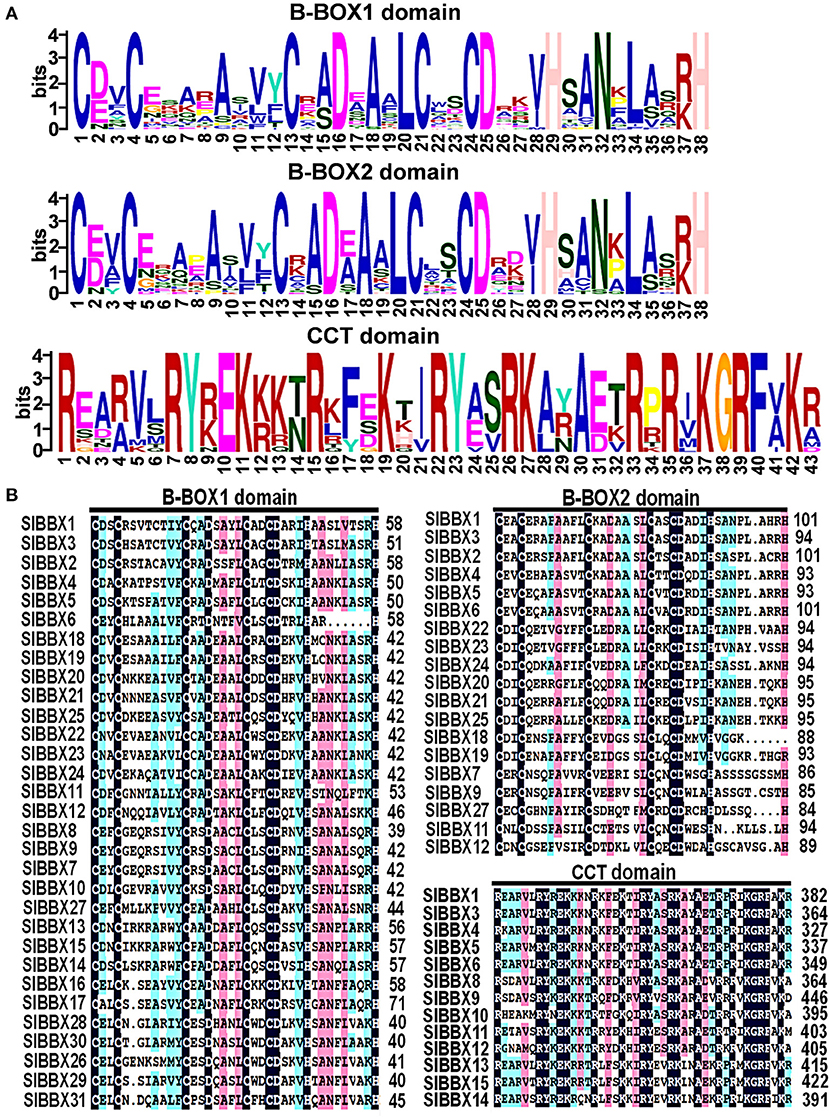
Figure 1. Domain composition of SlBBX proteins. (A) The amino acid sequence alignment of the B-box1, B-box2, and CCT domain. The y-axis and x-axis indicated the conservation rate of each amino acid and the conserved sequences of the domain, respectively. (B) Multiple sequence alignments of the domains of the SlBBXs. Multiple sequence alignments of the B-box1, B-box2, and CCT domains are shown. The identical conserved amino acids were represented by black and pink shaded.
To better reveal the evolutionary relationships, we generated a phylogenetic tree with the known BBX families from Arabidopsis, rice and potato, and the identified tomato BBX protein sequences (Figure 2, Supplementary Figure 2). All sequences of tomato BBX proteins were clustered into five subfamilies (Figure 2). The BBX genes in group 1 had two concatenated B-box domains, a CCT domain and a VP motif except for SlBBX1 and SlBBX2, which did not have a VP motif and a CCT domain. The members of group 2 were characterized by two B-box domains and also a CCT domain with the exception for SlBBX7 and SlBBX27, which contained two B-box domains only, and SlBBX8 and SlBBX10, which only had one B-box domain and a CCT domain. In group 3, all the members contained one B-box domain as well as a CCT domain. Group 4 and 5 possessed two and one B-box domain, respectively. Additionally, BBX proteins from two species showed scattered distribution across the branches of the evolutionary tree, which implies that the duplication events occurred after the lineages diverged.
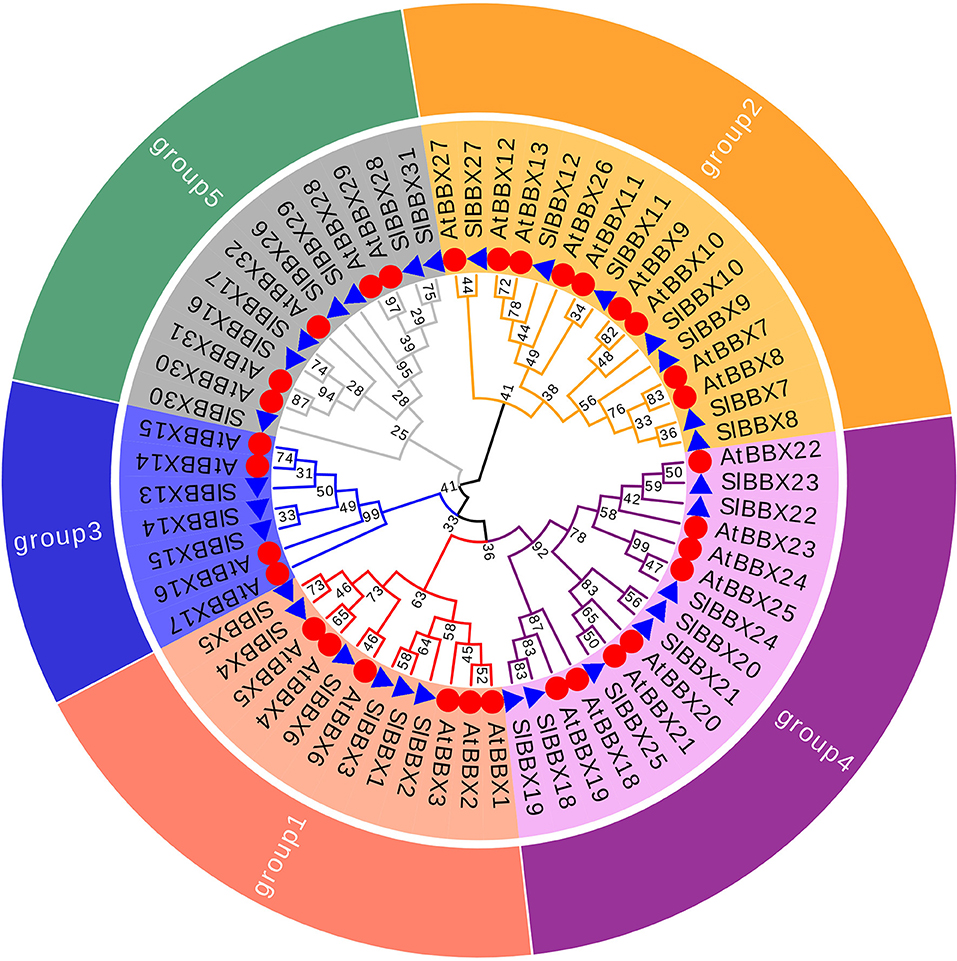
Figure 2. Molecular phylogenetic analysis of SlBBX genes in tomato. All SlBBX proteins were divided into five subclasses represented by different colored clusters. Red, orange, bule, purple, and green clusters represent subclasses I, II, III, IV, and V, respectively. The evolutionary history was inferred by using the Maximum Likelihood method using MEGA7 software with 1,000 bootstrap replicates. The red circles and blue triangles represent Arabidopsis and tomato, respectively.
Gene Structure, Conserved Motifs, Chromosomal Localization, and Synteny Analysis of SlBBXs
The evolution of multigene families can be driven by gene structural diversity. Examination of the genomic DNA sequences revealed that most SlBBXs contained one to five introns, while SlBBX16, SlBBX17, and SlBBX30 had no introns (Figure 3B, Table 1, Supplementary Figure 3). Among them, nine SlBBXs had one intron, followed by 10 SlBBXs with two introns, five SlBBXs with three introns, three SlBBXs with four introns, and one SlBBXs with five introns. Generally, members of each subclass, which are most closely related, exhibited analogous exon-intron structures. For instance, the members in groups 1 and 4 had one to two, and zero to one intron, respectively (Figures 3A,B, Supplementary Figure 3). However, a few SlBBX genes showed dissimilar exon-intron arrangements. For instance, SlBBX18 and SlBBX19 had high sequence similarity, but SlBBX18 and SlBBX19 contained two and five introns, respectively (Figures 3A,B, Supplementary Figure 3). These divergences indicated that both the gain and loss of introns during evolution, may better explain the functional diversity of SlBBX homologous genes.
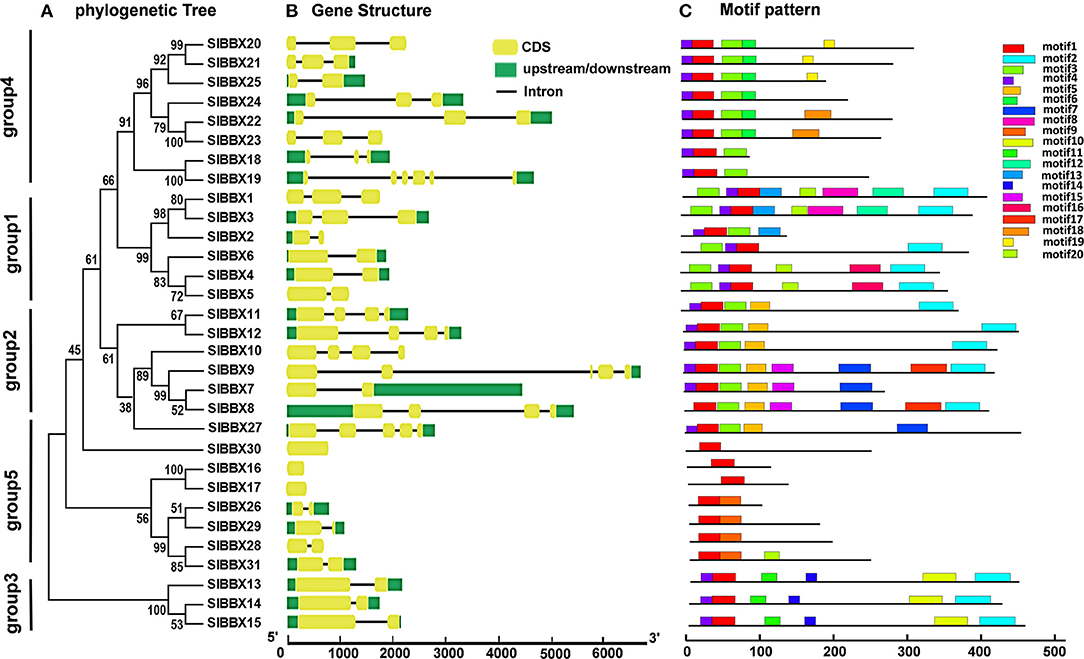
Figure 3. Phylogenetic relationship, gene structure and architecture of the conserved protein motifs in SlBBXs. (A) The phylogenetic tree was constructed based on the full-length sequences of SlBBX proteins. (B) Exon-intron structure of SlBBXs. Green boxes indicate untranslated 5′- and 3′-regions, yellow boxes indicate exons; and black lines indicate introns. (C) The motifs composition. The motifs, numbered 1–20, were displayed in different colored boxes. The sequence information for each motif is provided in Supplementary Table 1.
To further examine the structural features of SlBBXs, the conserved motif distributions were analyzed. Twenty conserved motifs were predicted (Figure 3C), while multilevel consensus sequences and the E-value of them were shown in Supplementary Table 1. The results showed that motif 17 was the largest motif depending on the width, followed by motif 8 and motif 2 (Supplementary Table 1). Motif 1 was found in all the SlBBXs (Figure 3C). Notably, 74.2% and 70.1% SlBBXs contained motif 4 and motif 3, respectively. Motif 2 was unique to the group 1, 2, and 3 of SlBBXs, while motif 5 was unique to group 2 except for the SlBBX27. Motif 10 was found only in group 3 of SlBBXs. Our results showed that members that are most closely related in the phylogenetic tree contained common motifs on the basis of alignment and position, which indicated that they may have a similar biological function.
Chromosomal locations showed that 31 SlBBX genes were unevenly distributed on the 12 chromosomes (Figure 4A). A maximum number of SlBBX genes were found on chromosome 12 (Chr12), comprising six genes. Five genes were located on Chr2 and Chr7. Four and three SlBBX genes were located on Chr5 and Chr4, respectively. Both Chr1 and Chr6 contained two members of SlBBX genes, whereas only one gene was detected on Chr3, 8, 9, and 10. Additionally, no SlBBX genes were found on Chr11.
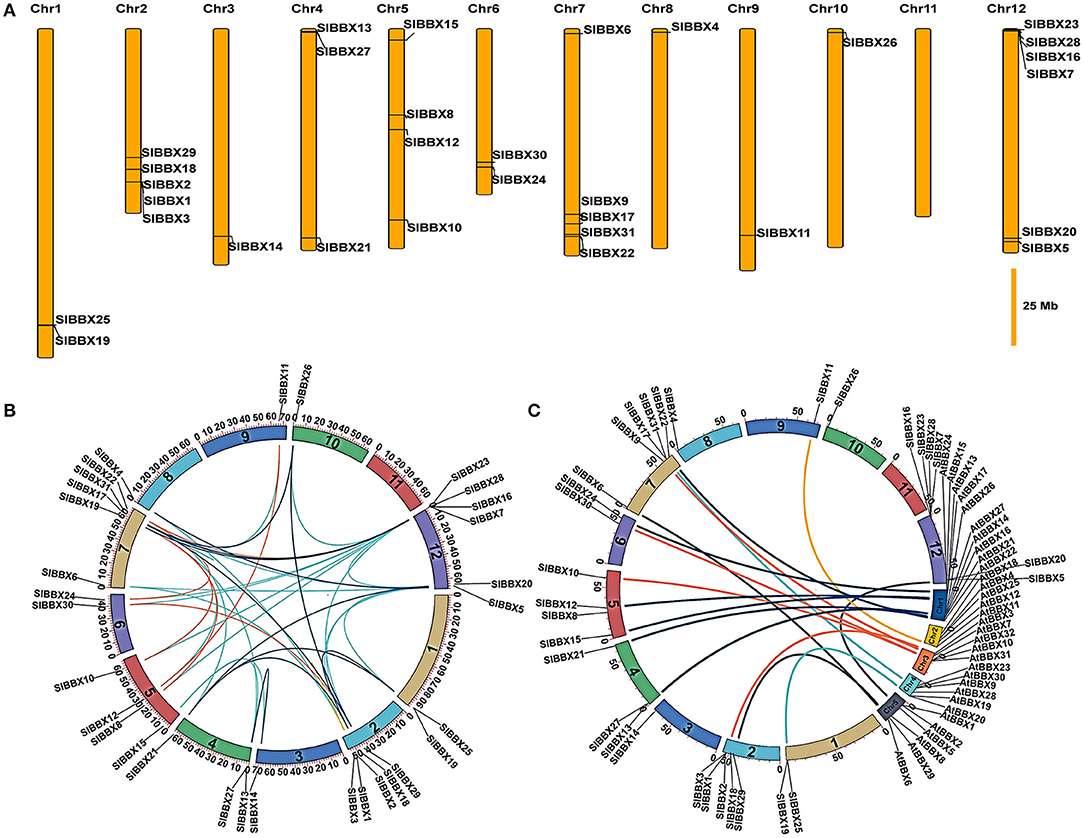
Figure 4. Chromosome distribution and syntenic analysis of SlBBX genes in tomato. (A) Positions of SlBBX genes family members on tomato chromosomes. (B) Segmental duplication of tomato BBX genes. Gene pairs located in the segmental duplicated chromosomal regions were linked using different lines. (C) Syntenic analysis of tomato and Arabidopsis BBX genes. Colored curves denote the syntenic relationships between tomato and Arabidopsis BBX genes.
Thirty-six pairs of SlBBXs were identified as segmental duplication in the tomato genome (Figure 4B). Chr2, 7, and 12 had more duplication regions, which partially explain the greater numbers of SlBBX genes that were located on these three chromosomes. Although SlBBX1 and SlBBX3 were located on the same chromosome (Figure 4A), and their sequence identity was 83% (Supplementary Figure 4), they were not tandem duplication. To further examine the evolutionary relationships between SlBBXs and AtBBXs, a synteny analysis was performed. A total of 16 of SlBBX-AtBBX orthologous pairs were identified (Figure 4C), which indicated the existence of numerous SlBBX genes prior to the divergence of Arabidopsis and tomato. Some members of SlBBXs were not localized in the syntenic block, suggesting that these genes might have certain specificity due to their evolution time.
Analysis of cis-Elements in the Promoter Region of SlBBXs
A total of 61 major cis-elements were predicted in promters of SlBBX genes (Figure 5A), including 22 light responsive, 12 hormone responsive, 11 stress responsive, and 16 development. The number of light responsive cis-elements was the largest in the promoters of 31 SlBBX genes (Figure 5B). The number of cis-elements in the promoters of SlBBX17 and SlBBX2 was the largest and least, respectively. The major light responsive elements contained box4 (21%), G-box (17.9%), and CMA1a/2a/2b (14.3%), which were located on 87.1% (27/31), 83.9% (26/31), and 96.8% (30/31) of SlBBXs promoters, respectively (Figure 5C). The most common motifs were the JA-responsive elements (MYC), abscisic acid (ABA)-responsive element (ABRE), and ethylene-responsive element (ERE), accounting for 24.8%, 21.5%, and 17.2% of the scanned hormone responsive motifs, respectively. The stress responsive elements MYB, STRE (stress-related elements) and WUN were located on 96.8% (30/31), 90.3% (28/31), and 77.4% (24/31) of 31 SlBBX genes promoters, respectively. In the development category, various growth and development related elements, such as AT-rich element (19.2%), O2-site for zein metabolism regulation (13.7%), CAT-box for meristem expression (12.3%), GCN4_motif required for endosperm expression (9.6%), were found. Our findings suggest that the promoter regions of SlBBX genes that contained the cis-elements played a critical role in the light and stress responses.
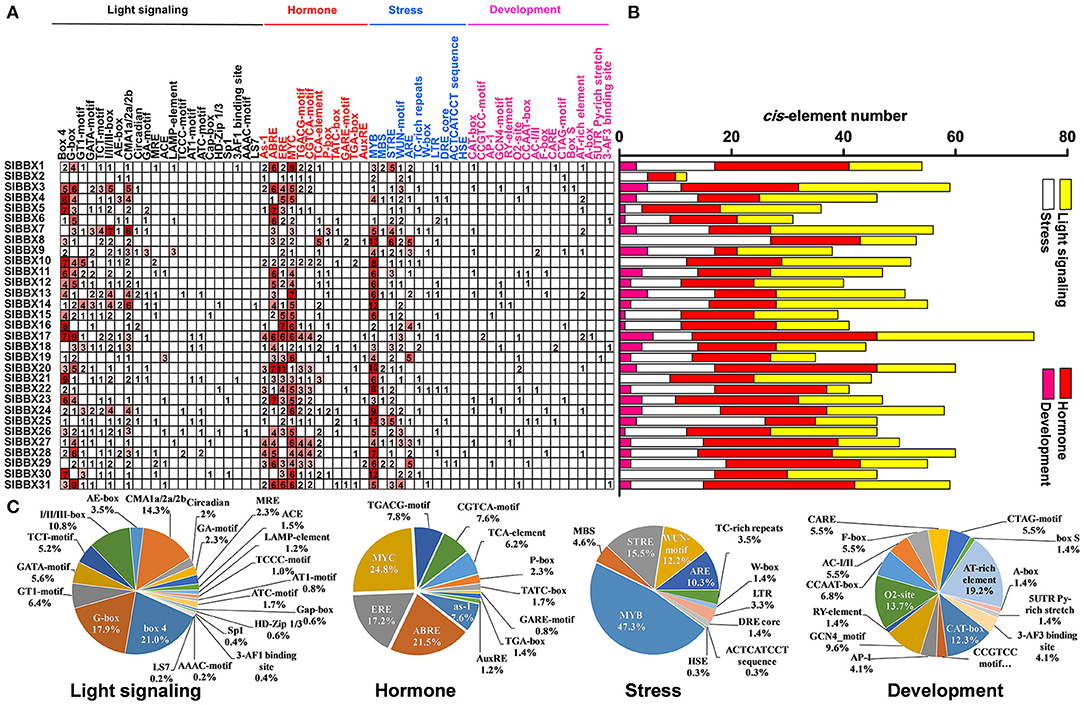
Figure 5. Inspection of cis-acting elements in tomato BBX genes. (A) The numbers of different promoter elements in these SlBBX genes were indicated by different colors and numbers of the grid. (B) The sum of the cis-acting elements in each category was represented by different colored histograms. (C) Pie charts with different sizes represented the ratio of each promoter element in each category.
SlBBX Genes Expression in Response to Different Light Quality
To assess whether light signaling regulates SlBBXs, we investigated the gene expression of SlBBXs in tomato plants grown at dark (D), white (W), and different light quality [purple (P), blue (B), green (G), yellow (Y), red (R), and far-red (FR)] conditions. In comparison with D, light decreased the transcripts of SlBBX1, SlBBX8, SlBBX10, and SlBBX12, while it increased the transcripts of SlBBX7, SlBBX13, and SlBBX15 (Figure 6). Plants grown at R light conditions showed higher expression of SlBBX4, SlBBX14, SlBBX23, SlBBX24, and SlBBX29 than those grown at other light qualities. Whereas, FR light significantly up-regulated the transcripts of SlBBX7, SlBBX13, SlBBX15, SlBBX21, SlBBX25, SlBBX26, and SlBBX27, it obviously down-regulated the transcripts of SlBBX14, SlBBX16, SlBBX18, SlBBX24, SlBBX28, SlBBX30, and SlBBX31 (Figure 6). Transcripts of SlBBX16, SlBBX17, SlBBX18, SlBBX30, and SlBBX31 were induced, while transcripts of SlBBX5, SlBBX6, SlBBX19, and SlBBX20 were inhibited in plants when grown at B light conditions. SlBBX15 was induced by G light irradiation at 6 h, whereas SlBBX9 and SlBBX28 were repressed (Figure 6). Y light led to an obvious reduction in expression of SlBBX9 and SlBBX31. Obviously, the P light increased the expression of SlBBX3, SlBBX5, SlBBX6, SlBBX15, SlBBX19, SlBBX20, SlBBX21, SlBBX26, and SlBBX27, but decreased the expression of SlBBX10 and SlBBX16. Interestingly, SlBBX4, SlBBX23, and SlBBX29 were only responsive to R light, while SlBBX7, SlBBX13, and SlBBX25 were induced just in response to FR light. Meanwhile, R light induced the expression of SlBBX14 and SlBBX24, but FR light inhibited their expression (Figure 6). In general, the results showed that SlBBX genes might act a critical role in response to light quality signaling.
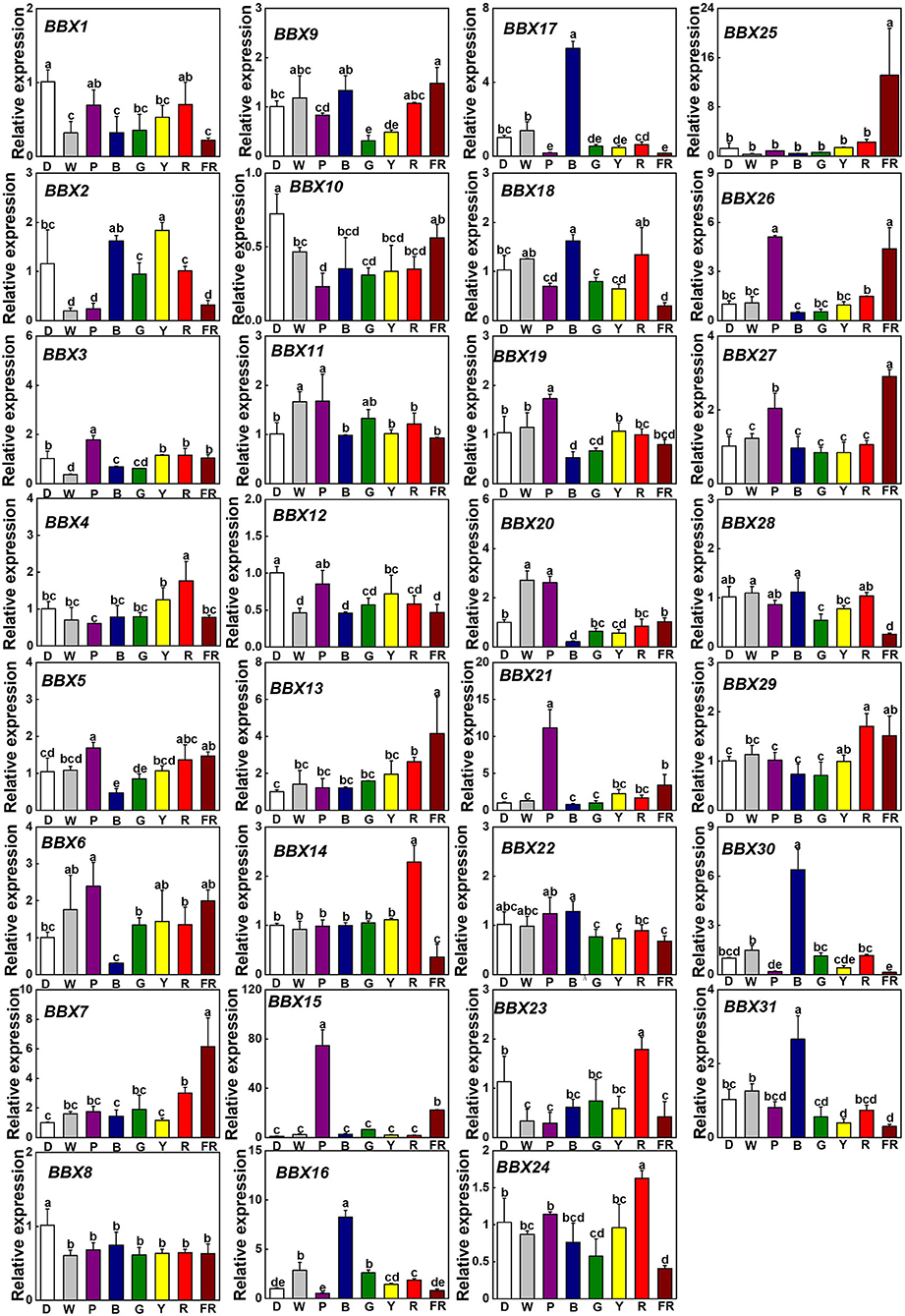
Figure 6. Gene expression of SlBBXs in tomato leaves after the exposure of plants to different light quality for 6 h from the dark. Light quality treatments include dark (D), white light (W) or purple (P), blue (B), green (G), yellow (Y), red (R), and far-red (FR) light. The light intensity was 100 μmol m−2 s−1. Data are presented as the means of three biological replicates (±SD). Different letters indicate significant differences (P < 0.05) according to Tukey's test.
SlBBXs Act Critical Roles in Regulation of Cold Tolerance in Tomato
To investigate whether SlBBX genes participated in cold stress, we analyzed the relative expression data of SlBBX genes in tomato plants (Chu et al., 2016), chose five genes, including SlBBX4, SlBBX7, SlBBX9, SlBBX18, and SlBBX20, and performed virus-induced gene silencing (VIGS) experiments to study their function under cold stress. After cold stress, the levels of relative electrolyte leakage (REL) in SlBBX7-silenced plants (pTRV-BBX7), SlBBX9-silenced plants (pTRV-BBX9) and SlBBX20-silenced plants (pTRV-BBX20) were higher than wild-type (pTRV) (Figure 7A), meanwhile, these silenced plants showed an increased sensitivity to cold stress compared with pTRV, as evidenced by a decrease in the maximum photosystem II efficiency (Fv/Fm) (Figure 7B, Supplementary Figure 5). In addition, the transcription of COLD RESPONSIVE (COR) genes, including COR47-like COR413-like was markedly lower in SlBBXs-silenced plants than those in control plants (pTRV) under cold stress (Figure 7C), which indicated that SlBBX7, SlBBX9, and SlBBX20 induced cold responsive genes under cold stress. Together, these results indicated that SlBBX7, SlBBX9, and SlBBX20 play positive roles in cold tolerance in tomato plants.
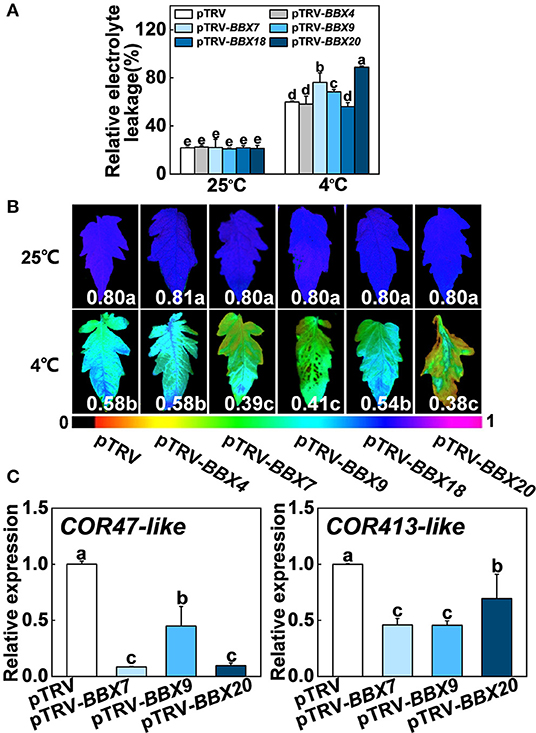
Figure 7. Roles of SlBBXs in cold tolerance in tomato plants. (A,B) The relative electrolyte leakage (A) and the maximum quantum yield of PSII (Fv/Fm) (B) in tomato wild-type (pTRV) and BBXs-silenced plants (pTRV-BBXs) after exposure to cold stress for 7 d. The false color code depicted at the bottom of the image ranges from 0 (black) to 1.0 (purple), representing the level of damage in leaves. (C) Cold responsive gene expression in tomato plants exposed 4°C for 6 h. Data are presented as the means of three biological replicates (±SD). Different letters indicate significant differences (P < 0.05) according to Tukey's test.
Roles of SlBBXs in Alleviation of Photoinhibition Under Cold Stress in Tomato
We analyzed the roles of SlBBXs in cold-induced photoinhibition. The maximum quantum yield of PSII (Fv/Fm) and the maximum level of the P700 signal (Pm, full oxidation of P700) in the dark were measured before and after cold treatment. Before treatment, Fv/Fm and Pm were similar among SlBBX7-, SlBBX9-, SlBBX20- silenced plants, and WT (pTRV) plants (Figures 8A,F). Fv/Fm was decreased by 27% in the pTRV plants after cold stress, while it was decreased by 51%, 48%, and 52% in SlBBX7-, SlBBX9-, and SlBBX20- silenced plants, respectively, after cold stress, which indicated that disruption of these three SlBBXs caused photoinhibition of PSII during cold stress. Furthermore, the Pm was decreased by 25% after cold stress, whereas it was decreased by 51%, 48%, and 33%, respectively, in SlBBX7-, SlBBX9-, and SlBBX20- silenced plants after cold stress, which suggested that the disruption of SlBBX7 and SlBBX9 caused obvious photoinhibition of PSI after cold stress, but the photoinhibition of PSI was slight in SlBBX20- silenced plants after cold stress. Together, these results indicated that these three SlBBXs play important roles in alleviating the photoinhibition of both photosystems during cold stress.
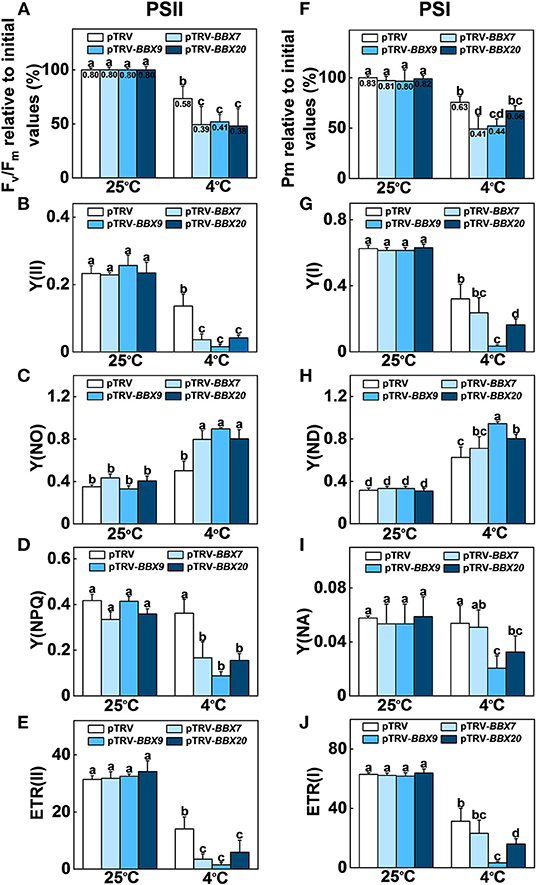
Figure 8. SlBBXs play important roles in alleviating cold-induced photoinhibition in tomato plants. (A–E) Changes in PSII parameters, including Fv/Fm (A), Y(II) (B), Y(NO) (C), Y(NPQ) (D), and ETR(II) (E) in tomato wild-type (pTRV) and BBXs-silenced plants (pTRV-BBXs) after exposure to cold stress for 5 d. (F–J) Changes in PSI parameters, including Pm (F), Y(I) (G), Y(ND) (H), Y(NA) (I), and ETR(I) (J) in tomato wild-type and BBXs-silenced plants after exposure to cold stress for 5 d. Data are presented as the means of three biological replicates (±SD). Different letters indicate significant differences (P < 0.05) according to Tukey's test.
In order to obtain a more detailed insight into the processes affecting photoinhibition in the SlBBX-silenced plants after cold stress, some electron transport parameters of photosystems I and II were measured. Both ETR II (Figures 8E, 9D) and ETR I (Figures 8J, 9H) were significantly reduced after cold stress in tomato plants, showing a decrease of 50%–55% over those in plants grown at normal temperature. The ETR II and ETR I in SlBBXs-silenced plants were lower than those in control plants (pTRV) after cold stress, except SlBBX7-silenced plants, whose ETR I is close to the values of pTRV. To further explore the decrease in the ETRs, we measured Y(II), Y(NPQ), and Y(NO) for PSII and Y(I), Y(ND), and Y(NA) for PSI. We found that Y(II) values were significantly lower in pTRV-BBX7, pTRV-BBX9, and pTRV-BBX20 plants than those in pTRV plants under cold stress (Figures 8B, 9A). This decrease in Y(II) appeared to be primarily due to a decrease in quantum yield of regulated energy dissipation of PSII [Y(NPQ)] and photochemical quenching coefficient (qP), and an increase in quantum yield of non-regulated energy dissipation of PSII [Y(NO)] in SlBBXs-silenced plants, especially in pTRV-BBX7 and pTRV-BBX9 plants (Figures 8B–D, 9A–C). Like Y(II), Y(I) was also decreased in SlBBXs-silenced plants, except for in pTRV-BBX7 plants (Figures 8G, 9E). These decrease seemed to be due to obvious donor side limitation of PSI (due to lower ETR II in PSII) as reflected by the elevated Y(ND), which were 29% and 47% higher in pTRV-BBX9 and pTRV-BBX20 plants, respectively, than in pTRV plants (Figures 8H, 9F). Although Y(ND) was 14% higher in pTRV-BBX7 plants than in pTRV plants, its Y(I) was similar to the control plants (pTRV), which indicated that disruption of SlBBX7 damaged the PSII rather than PSI during cold stress. Therefore, cold stress seriously reduced the capacity for photochemical energy conversion, electron transport rate and photoprotection in SlBBX7-, SlBBX9-, SlBBX20- silenced plants, suggesting that these three SlBBXs play critical roles in alleviating photoinhibition under cold stress.
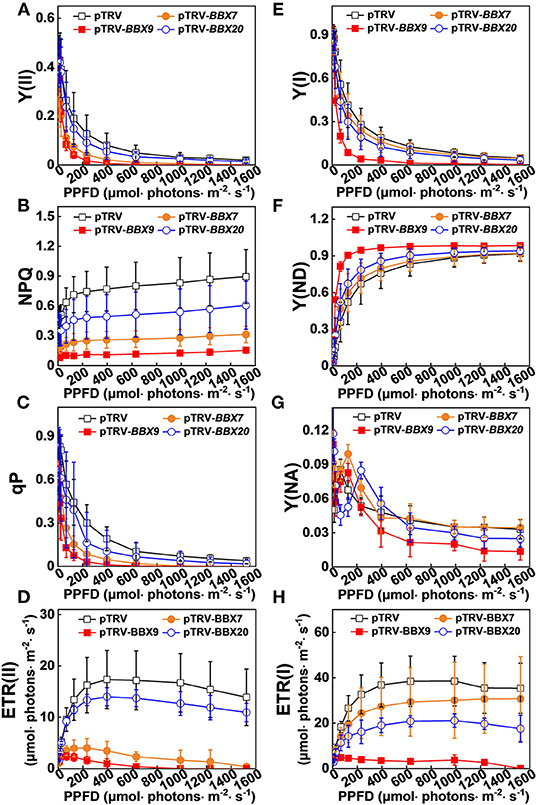
Figure 9. Impact of disruption of SlBBXs on the electron transport during steady-state photosynthesis in tomato plants grown under cold stress. The light intensity dependence of PSII and PSI photosynthetic parameters was monitored in the tomato wild type (WT) and BBXs-silenced plants (pTRV-BBXs). (A–D) PSII parameters Y(II) (A), NPQ (B), qP (C), and ETR(II) (D) in tomato wild-type (pTRV) and BBXs-silenced plants (pTRV-BBXs) after exposure to cold stress for 5 d, respectively. (E–H) PSI parameters Y(I) (E), Y(ND) (F), Y(NA) (G), and ETR(I) (H) in tomato wild-type (pTRV) and BBXs-silenced plants (pTRV-BBXs) after exposure to cold stress for 5 d, respectively. Data are presented as the means of three biological replicates (±SD). Different letters indicate significant differences (P < 0.05) according to Tukey's test.
Discussion
In this study, we identified and characterized 31 SlBBX genes in tomato (Figure 1, Tables 1, 2), which contained two additional loci encoding BBX proteins in the tomato genome, that were named SlBBX30 and SlBBX31, in comparison with the previous studies (Chu et al., 2016). BBX proteins are characterized by one or two B-box domains at the N-terminal and, in some cases, a CCT domain at the C-terminal (Gangappa and Botto, 2014). Here, we found both the newly retrieved SlBBX proteins (SlBBX30 and SlBBX31) contain a B-box domain at the N-terminal (Figure 1, Table 2), and they were also clustered in group 5 (Figure 2, Supplementary Figure 2), which further indicated these two proteins were new SlBBX proteins. The release of new tomato genomes and database updates may be the primary causes of this phenomenon. There were five subfamilies in the 32 members of Arabidopsis BBXs according to the combination of different conserved domains (Khanna et al., 2009). However, the conserved domain-based classification of BBX proteins in tomato was rather complex. As shown in Figure 2, SlBBX1 to SlBBX6 were classified into group 1, which had two B-boxes and a CCT plus a VP domains, whereas SlBBX1 lacked a VP domain, and SlBBX2 only contained two B-boxes (Table 2). Meanwhile, SlBBX7 to SlBBX12 and SlBBX27 were clustered into group 2, which possessed two B-boxes and a CCT domains; however, SlBBX8 and SlBBX10 had one B-box and a CCT domains, while SlBBX7 and SlBBX27 contained two B-boxes. Group 4 contained only one B-box. We investigated the detail of sequence alignment in SlBBXs (Figure 1), and found a high degree of conservation of the B-box1 domain among SlBBX7 to SlBBX12, thus the clustering results of these proteins were similar to that based on B-box1. These results indicated that during the process of evolution, some SlBBX proteins lost the B-box2 domain.
Accumulating evidence showed that some BBX proteins act as central players in a variety of light-regulated physiological processes in plants. Here, we found that the number of light responsive cis-elements was the largest in the promoters of 31 SlBBX genes (Figure 5), which indicated that SlBBX genes were regulated by light signaling. Thus, we examined the gene expression of all the SlBBXs in response to different light quality. Results showed that light decreased the transcripts of SlBBX1, SlBBX8, SlBBX10, and SlBBX12, while increased the transcripts of SlBBX7, SlBBX13, and SlBBX15 compared with dark (Figure 6). Previous studies had demonstrated that COP1, which is degraded after illumination, works as an E3 ubiquitin ligase that targets a variety of light signaling factors for ubiquitination and degradation in darkness (Osterlund et al., 2000; Han et al., 2020). For example, COP1 interacts with multiple BBXs, such as CO/BBX1 and BBX10, and subsequently degrades them by the 26S proteasome system (Liu et al., 2008; Ordoñez-Herrera et al., 2018). Nevertheless, COP1 stabilizes BBX11 rather than degrading it (Zhao et al., 2020), which suggests that COP1 likely degrades a yet unidentified component(s) targeting BBX11. Thus, COP1 may also control the stability of SlBBX proteins, including SlBBX1, SlBBX7, SlBBX8, SlBBX10, SlBBX12, SlBBX13, and SlBBX15, in the transition from dark to light. Interestingly, we found that SlBBX4, SlBBX23, and SlBBX29 were expressed only in response to R light, while SlBBX7, SlBBX13, and SlBBX25 were expressed just in response to FR light (Figure 6). These results indicate that these SlBBX proteins might directly interact with the photoreceptors, which sense R and FR light signals. Similarly, recent work has revealed that phyB directly interacts with BBX4 and positively regulates its accumulation in red light in Arabidopsis (Heng et al., 2019a), which demonstrates that photoreceptors may directly control some BBX proteins. In addition, the results showed that R light induced the expression of SlBBX14 and SlBBX24, whereas FR light inhibited their expression (Figure 6), which implied that these two SlBBX proteins might function antagonistically to regulate some plant physiological processes, such as shade avoidance and the elongation of hypocotyls. Here, we observed that B light induced the gene expression of SlBBX16, SlBBX17, SlBBX18, SlBBX30, and SlBBX31, whereas inhibited the transcripts of SlBBX5, SlBBX6, SlBBX19, and SlBBX20 (Figure 6). Previous work demonstrated that BBX28/BBX29 and BBX30/BBX31 could precisely control each other by forming a feedback loop in Arabidopsis (Lin et al., 2018; Heng et al., 2019b; Yadav et al., 2019; Song et al., 2020). Thus, these SlBBX proteins may work in concert with each other and some unidentified factors to regulate the plant growth in response to light signaling.
Light and temperature are more or less inter-related during plant growth and stress response (Wang et al., 2016). Here, we observed that the disruption of SlBBX7, SlBBX9, and SlBBX20 largely reduced the cold tolerance in tomato plants as evidenced by phenotypes, REL, Fv/Fm, and cold responsive genes (Figure 7, Supplementary Figure 5), which indicated that SlBBX7, SlBBX9, and SlBBX20 positively regulate cold tolerance in tomato plants. In addition, far-red light (FR) induced the transcription of SlBBX7 and SlBBX9 (Figure 6). and enhanced the cold tolerance in tomato plants (Wang et al., 2016, 2018, 2019, 2020a,b), which indicate that SlBBXs may play critical roles in the links between cold response and light signaling. Recent studies also showed that BBX18 and BBX23 are involved in the thermomorphogenesis in Arabidopsis (Ding et al., 2018). Both MdBBX20 and MaCOL1 are responsive to low temperature in apple and banana, respectively (Chen et al., 2012; Fang et al., 2019). ZFPL, a homologous gene of AtBBX32, enhances cold tolerance in the grapevine (Takuhara et al., 2011). CmBBX24 also increases plant cold tolerance in Chrysanthemum (Yang et al., 2014). Furthermore, MdBBX37 positively regulates JA-mediated cold-stress resistance in apples (An et al., 2021). However, whether BBXs are involved in cold stress–induced photoinhibition and the regulation of photoprotection during cold stress remains elusive.
Interestingly, the impairment of SlBBX7, SlBBX9, and SlBBX20 significantly suppressed the photochemical efficiencies and energy conversion in tomato plants under cold stress, leading to the overreduction of electron carriers and damage of photosystem in tomato plants (Figures 8, 9). More recently, it has been demonstrated that heterologous expression of Arabidopsis BBX21 in potato plants increases photosynthetic efficiency and reduces photoinhibition (Crocco et al., 2018). Therefore, BBXs play critical roles in photosynthesis and photoinhibition, however, the regulation mechanism is poorly understood. Previous works have revealed that BBX20, BBX21, BBX22, and BBX23 interact with HY5 to increase its transcriptional activity toward the target genes (Datta et al., 2008; Zhang et al., 2017; Job et al., 2018), whereas BBX24, BBX25, BBX28, and BBX29 suppress HY5 activity in Arabidopsis (Gangappa et al., 2013; Job et al., 2018; Lin et al., 2018; Song et al., 2020). HY5 also positively controls BBX22 at the transcriptional level (Chang et al., 2008), while repressing BBX30 and BBX31 gene expression by binding to the promoters of these two genes (Heng et al., 2019b; Yadav et al., 2019). In addition, direct interactions between BBX32 and BBX21 lead to inhibition in the BBX21-HY5 (Holtan et al., 2011). Therefore, SlBBXs might alleviate the photoinhibition in tomato plants during cold stress through an HY5-dependent photoprotection pathway. Our previous results demonstrated that SlHY5 alleviated photoinhibition in tomato plants under cold stress by induction of photoprotection, including increased NPQ, cyclic electron flux (CEF) around PSI and the activities of Foryer-Halliwell-Asada cycle enzymes (Wang et al., 2018). Here, we showed that the over-reduction in the flow of electrons from PSII to PSI and considerably low levels of NPQ caused a high excitation pressure in SlBBXs-silenced plants against cold stress, leading to a severe photoinhibition (Figures 8, 9). Thus, NPQ might function as a protective measure to prevent damage from high excitation pressure against photosynthesis and plant development (Upadhyay et al., 2013).
Conclusions
In this study, SlBBX family genes were identified and characterized in tomato by systematic analysis of conserved domains, phylogenetic relationship, gene structure, chromosome location. Two new members, SlBBX30 and SlBBX31, were identified from the newly released tomato genome sequences. The promoter responsive cis-acting regulatory elements and gene expression analysis indicated that multiple SlBBX genes were highly responsive to light quality and low temperature. Furthermore, we found that SlBBX7, SlBBX9, and SlBBX20 positively regulate cold tolerance in tomato plants via the prevention of photoinhibition and enhancing photoprotection. Our study emphasized the positive roles of light signaling transcription factors SlBBXs in cold tolerance in tomato plants, which may improve the current understanding of the integration of light and temperature signals by plants to adapt to adverse environments.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
FW and TL designed the research. FW, XB, XW, JY, YZ, SZ, and XS performed the experiments. FW, YL, MQ, and GA analyzed the data. FW, GA, and TL wrote and revised the paper. All authors have read and approved the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (31801904), the Liao Ning Revitalization Talents Program (XLYC1807020), the Shenyang Young and Middle-aged Science and Technology Innovation Talent Support Program (RC200449), National Key Research and Development Program of China (2018YFD1000800; 2019YFD1000300). Liaoning BaiQianWan Talents Program and China Agriculture Research System of MOF and MARA.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to the Tomato Genetics Resource Center at the California University for tomato seeds, and Yaofang Niu (Guhe Info technology Co. Ltd, Hangzhou, China) for assistance with syntenic analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.698525/full#supplementary-material
References
Ahammed, G. J., Gantait, S., Mitra, M., Yang, Y., and Li, X. (2020). Role of ethylene crosstalk in seed germination and early seedling development: a review. Plant Physiol. Biochem. 151, 124–131. doi: 10.1016/j.plaphy.2020.03.016
An, J. P., Wang, X. F., Espley, R. V., Wang, K. L., Bi, S. Q., You, C. X., et al. (2019a). An apple b-box protein MdBBX37 modulates anthocyanin biosynthesis and hypocotyl elongation synergistically with MdMYBs and MdHY5. Plant Cell Physiol. 61, 130–143. doi: 10.1093/pcp/pcz185
An, J. P., Wang, X. F., Zhang, X. W., Bi, S. Q., You, C. X., and Hao, Y. J. (2019b). MdBBX22 regulates UV-B-induced anthocyanin biosynthesis through regulating the function of MdHY5 and is targeted by MdBT2 for 26S proteasome-mediated degradation. Plant Biotechnol. J. 17, 2231–2233. doi: 10.1111/pbi.13196
An, J. P., Wang, X. F., Zhang, X. W., You, C. X., and Hao, Y. J. (2021). Apple B-box protein BBX37 regulates jasmonic acid mediated cold tolerance through the JAZ-BBX37-ICE1-CBF pathway and undergoes MIEL1-mediated ubiquitination and degradation. New Phytol. 229, 2707–2729. doi: 10.1111/nph.17050
Bai, B., Lu, N. N., Li, Y. P., Guo, S. L., Yin, H. B., He, Y. N., et al. (2019). OsBBX14 promotes photomorphogenesis in rice by activating OsHY5L1 expression under blue light conditions. Plant Sci. 284, 192–202. doi: 10.1016/j.plantsci.2019.04.017
Bai, S. L., Tao, R. Y., Tang, Y. X., Yin, L., Ma, Y. J., Ni, J. B., et al. (2019a). BBX16, a B-box protein, positively regulates light-induced anthocyanin accumulation by activating MYB10 in red pear. Plant Biotechnol. J. 17, 1985–1997. doi: 10.1111/pbi.13114
Bai, S. L., Tao, R. Y., Yin, L., Ni, J. B., Yang, Q. S., Yan, X. H., et al. (2019b). Two B-box proteins, PpBBX18 and PpBBX21, antagonistically regulate anthocyanin biosynthesis via competitive association with Pyrus pyrifolia ELONGATED HYPOCOTYL 5 in the peel of pear fruit. Plant J. 100, 1208–1223. doi: 10.1111/tpj.14510
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Cao, W. H., Liu, J., He, X. J., Mu, R. L., Zhou, H. L., Chen, S. Y., et al. (2007). Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol. 143, 707–719. doi: 10.1104/pp.106.094292
Chang, C. S. J., Li, Y. H., Chen, L. T., Chen, W. C., Hsieh, W. P., Shin, J., et al. (2008). LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. Plant J. 54, 205–219. doi: 10.1111/j.1365-313X.2008.03401.x
Chen, J., Chen, J. Y., Wang, J. N., Kuang, J. F., Shan, W., and Lu, W. J. (2012). Molecular characterization and expression profiles of MaCOL1, a CONSTANS-like gene in banana fruit. Gene 496, 110–117. doi: 10.1016/j.gene.2012.01.008
Chu, Z. N., Wang, X., Li, Y., Yu, H. Y., Li, J. H., Lu, Y. E., et al. (2016). Genomic organization, phylogenetic and expression analysis of the B-BOX gene family in tomato. Front. Plant Sci. 7:1552. doi: 10.3389/fpls.2016.01552
Crocco, C. D., and Botto, J. F. (2013). BBX proteins in green plants: insights into their evolution, structure, feature and functional diversification. Gene 531, 44–52. doi: 10.1016/j.gene.2013.08.037
Crocco, C. D., Ocampo, G. G., Ploschuk, E. L., Mantese, A., and Botto, J. F. (2018). Heterologous expression of AtBBX21 enhances the rate of photosynthesis and alleviates photoinhibition in Solanumtuberosum. Plant Physiol. 177, 369–380. doi: 10.1104/pp.17.01417
Datta, S., Hettiarachchi, G. H. C. M., Deng, X. W., and Holm, M. (2006). Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 18, 70–84. doi: 10.1105/tpc.105.038182
Datta, S., Johansson, H., Hettiarachchi, C., Irigoyen, M. L., Desai, M., Rubio, V., et al. (2008). LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell 20, 2324–2338. doi: 10.1105/tpc.108.061747
Ding, L., Wang, S., Song, Z. T., Jiang, Y. P., Han, J. J., Lu, S. J., et al. (2018). Two B-Box domain proteins, BBX18 and BBX23, interact with ELF3 and regulate thermomorphogenesis in Arabidopsis. Cell Rep. 25, 1718–1728. doi: 10.1016/j.celrep.2018.10.060
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Fan, X. Y., Dong, Y. S., Cao, D. M., Bai, M. Y., Luo, X. M., Yang, H. J., et al. (2012). BZS1, a B-box protein, promotes photomorphogenesis downstream of both brassinosteroid and light signaling pathways. Mol. Plant. 5, 591–600. doi: 10.1093/mp/sss041
Fang, H. C., Dong, Y. H., Yue, X. X., Hu, J. F., Jiang, S. H., Xu, H. F., et al. (2019). The B-Box zinc finger protein MdBBX20 integrates anthocyanin accumulation in response to ultraviolet radiation and low temperature. Plant Cell Environ. 42, 2090–2104. doi: 10.1111/pce.13552
Galvao, V. C., and Fankhauser, C. (2015). Sensing the light environment in plants: photoreceptors and early signaling steps. Curr. Opin. Neurobiol. 34C, 46–53. doi: 10.1016/j.conb.2015.01.013
Gangappa, S. N., and Botto, J. F. (2014). The BBX family of plant transcription factors. Trends Plant Sci. 19, 460–470. doi: 10.1016/j.tplants.2014.01.010
Gangappa, S. N., Crocco, C. D., Johansson, H., Datta, S., Hettiarachchi, C., Holm, M., et al. (2013). The Arabidopsis B-box protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell 25, 1243–1257. doi: 10.1105/tpc.113.109751
Gendron, J. M., Pruneda-Paz, J. L., Doherty, C. J., Gross, A. M., Kang, S. E., and Kay, S. A. (2012). Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. U.S.A. 109, 3167–3172. doi: 10.1073/pnas.1200355109
Han, X., Huang, X., and Deng, X. W. (2020). The photomorphogenic central repressor COP1: conservation and functional diversification during evolution. Plant Commun. 1, 2590–3462. doi: 10.1016/j.xplc.2020.100044
Heng, Y. Q., Jiang, Y., Zhao, X. H., Zhou, H., Wang, X. C., Deng, X. W., et al. (2019a). BBX4, a phyB-interacting and modulated regulator, directly interacts with PIF3 to fine tune red light-mediated photomorphogenesis. Proc. Natl. Acad. Sci. U.S.A. 116, 26049–26056. doi: 10.1073/pnas.1915149116
Heng, Y. Q., Lin, F., Jiang, Y., Ding, M. Q., Yan, T. T., Lan, H. X., et al. (2019b). B-Box containing proteins BBX30 and BBX31, acting downstream of HY5, negatively regulate photomorphogenesis in Arabidopsis. Plant Physiol. 180, 497–508. doi: 10.1104/pp.18.01244
Holm, M., Hardtke, C. S., Gaudet, R., and Deng, X. W. (2001). Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J. 20, 118–127. doi: 10.1093/emboj/20.1.118
Holtan, H. E., Bandong, S., Marion, C. M., Adam, L., Tiwari, S., Shen, Y., et al. (2011). BBX32, an Arabidopsis B-Box protein, functions in light signaling by suppressing HY5-regulated gene expression and interacting with STH2/BBX21. Plant Physiol. 156, 2109–2123. doi: 10.1104/pp.111.177139
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587. doi: 10.1093/nar/gkm259
Hu, B., Jin, J. P., Guo, A. Y., Zhang, H., Luo, J. C., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Job, N., Yadukrishnan, P., Bursch, K., Datta, S., and Johansson, H. (2018). Two B-Box proteins regulate photomorphogenesis by oppositely modulating HY5 through their diverse C-terminal domains. Plant Physiol. 176, 2963–2976. doi: 10.1104/pp.17.00856
Khanna, R., Kronmiller, B., Maszle, D. R., Coupland, G., Holm, M., Mizuno, T., et al. (2009). The Arabidopsis B-box zinc finger family. Plant Cell 21, 3416–3420. doi: 10.1105/tpc.109.069088
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de. Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Lin, F., Jiang, Y., Li, J., Yan, T. T., Fan, L. M., Liang, J. S., et al. (2018). B-BOX DOMAIN PROTEIN28 negatively regulates photomorphogenesis by repressing the activity of transcription factor HY5 and undergoes COP1-mediated degradation. Plant Cell 30, 2006–2019. doi: 10.1105/tpc.18.00226
Lira, B. S., Oliveira, M. J., Shiose, L., Wu, R. T. A., Rosado, D., Lupi, A. C. D., et al. (2020). Light and ripening-regulated BBX protein-encoding genes in Solanum lycopersicum. Sci. Rep. 10:19235. doi: 10.1038/s41598-020-76131-0
Liu, L. J., Zhang, Y. C., Li, Q. H., Sang, Y., Mao, J., Lian, H. L., et al. (2008). COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20, 292–306. doi: 10.1105/tpc.107.057281
Liu, X., Li, R., Dai, Y., Yuan, L., Sun, Q. H., Zhang, S. Z., et al. (2019). A B-box zinc finger protein, MdBBX10, enhanced salt and drought stresses tolerance in Arabidopsis. Plant Mol. Biol. 99, 437–447. doi: 10.1007/s11103-019-00828-8
Liu, Y. N., Chen, H., Ping, Q., Zhang, Z. X., Guan, Z. Y., Fang, W. M., et al. (2019). The heterologous expression of CmBBX22 delays leaf senescence and improves drought tolerance in Arabidopsis. Plant Cell Rep. 38, 15–24. doi: 10.1007/s00299-018-2345-y
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001
Min, J. H., Chung, J. S., Lee, K. H., and Kim, C. S. (2015). The CONSTANS-like 4 transcription factor, AtCOL4, positively regulates abiotic stress tolerance through an abscisic acid-dependent manner in Arabidopsis. J. Integr. Plant Biol. 57, 313–324. doi: 10.1111/jipb.12246
Nagaoka, S., and Takano, T. (2003). Salt tolerance-related protein STO binds to a Myb transcription factor homologue and confers salt tolerance in Arabidopsis. J. Exp. Bot. 54, 2231–2237. doi: 10.1093/jxb/erg241
Ordoñez-Herrera, N., Trimborn, L., Menje, M., Henschel, M., Robers, L., Kaufholdt, D., et al. (2018). The transcription factor COL12 is a substrate of the COP1/SPA E3ligase and regulates flowering time and plant architecture. Plant Physiol. 176, 1327–1340. doi: 10.1104/pp.17.01207
Osterlund, M. T., Hardtke, C. S., Wei, N., and Deng, X. W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466. doi: 10.1038/35013076
Paik, I., and Huq, E. (2019). Plant photoreceptors: multi-functional sensory proteins and their signaling networks. Semin. Cell. Dev. Biol. 92, 114–121. doi: 10.1016/j.semcdb.2019.03.007
Ren, J., Wen, L. P., Gao, X. J., Jin, C. J., Xue, Y., and Yao, X. B. (2009). DOG 1.0: illustrator of protein domain structures. Cell Res. 19, 271–2733. doi: 10.1038/cr.2009.6
Song, Z. Q., Yan, T. T., Liu, J. J., Bian, Y. T., Heng, Y. Q., Lin, F., et al. (2020). BBX28/BBX29-HY5-BBX30/31 form a feedback loop to fine-tune photomorphogenic development. Plant J. 104, 377–390. doi: 10.1111/tpj.14929
Takuhara, Y., Kobayashi, M., and Suzuki, S. (2011). Low-temperature induced transcription factors in grapevine enhance cold tolerance in transgenic Arabidopsis plants. J. Plant Physiol. 168, 967–975. doi: 10.1016/j.jplph.2010.11.008
Upadhyay, R. K., and Mattoo, A. K. (2018). Genome-wide identification of tomato (Solanum lycopersicum L.) lipoxygenases coupled with expression profiles during plant development and in response to methyl-jasmonate and wounding. J. Plant Physiol. 231, 318–328. doi: 10.1016/j.jplph.2018.10.001
Upadhyay, R. K., Soni, D. K., Singh, R., Dwivedi, U. N., Pathre, U. V., Nath, P., et al. (2013). SlERF36, an EAR-motif-containing ERF gene from tomato, alters stomatal density and modulates photosynthesis and growth. J. Exp. Bot. 64, 3237–3247. doi: 10.1093/jxb/ert162
Wang, C. Q., Sarmast, M. K., Jiang, J., and Dehesh, K. (2015). The transcriptional regulator BBX19 promotes hypocotyl growth by facilitating COP1-Mediated EARLY FLOWERING3 degradation in Arabidopsis. Plant Cell 27, 1128–1139. doi: 10.1105/tpc.15.00044
Wang, F., Chen, X. X., Dong, S. J., Jiang, X. C., Wang, L. Y., Yu, J. Q., et al. (2020a). Crosstalk of PIF4 and DELLA modulates CBF transcript and hormone homeostasis in cold response in tomato. Plant Biotechnol. J. 18, 1041–1055. doi: 10.1111/pbi.13272
Wang, F., Guo, Z. X., Li, H. Z., Wang, M. M., Onac, E., Zhou, J., et al. (2016). Phytochrome A and B function antagonistically to regulate cold tolerance via abscisic acid-dependent jasmonate signaling. Plant Physiol. 170, 459–471. doi: 10.1104/pp.15.01171
Wang, F., Wu, N., Zhang, L. Y., Ahammed, G. L., Chen, X. X., Xiang, X., et al. (2018). Light signaling-dependent regulation of photoinhibition and photoprotection in tomato. Plant Physiol. 176, 1311–1326. doi: 10.1104/pp.17.01143
Wang, F., Yan, J. R., Ahammed, G. J., Wang, X. J., Bu, X., Xiang, H. Z., et al. (2020b). PGR5/PGRL1 and NDH mediate far-red light-induced photoprotection in response to chilling stress in tomato. Front. Plant Sci. 11:669. doi: 10.3389/fpls.2020.00669
Wang, F., Zhang, L. Y., Chen, X. X., Wu, X. D., Xiang, X., Zhou, J., et al. (2019). SlHY5 integrates temperature, light and hormone signaling to balance plant growth and cold tolerance. Plant Physiol. 179, 749–760. doi: 10.1104/pp.18.01140
Xiong, C., Luo, D., Lin, A. H., Zhang, C. L., Shan, L. B., He, P., et al. (2019). A tomato B-box protein SlBBX20 modulates carotenoid biosynthesis by directly activating PHYTOENE SYNTHASE 1, and is targeted for 26S proteasome-mediated degradation. New Phytol. 221, 279–294. doi: 10.1111/nph.15373
Xu, D. Q. (2020). COP1 and BBXs-HY5-mediated light signal transduction in plants. New Phytol. 228, 1748–1753. doi: 10.1111/nph.16296
Xu, D. Q., Jiang, Y., Li, J., Holm, M., and Deng, X. W. (2018). The B-Box domain protein BBX21 promotes photomorphogenesis. Plant Physiol. 176, 2365–2375. doi: 10.1104/pp.17.01305
Xu, D. Q., Jiang, Y., Li, J. G., Lin, F., Holm, M., and Deng, X. W. (2016). BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome mediated degradation. Proc. Natl. Acad. Sci. U.S.A. 113, 7655–7660. doi: 10.1073/pnas.1607687113
Yadav, A., Bakshi, S., Yadukrishnan, P., Lingwan, M., Dolde, U., Wenkel, S., et al. (2019). The B-Box-containing microprotein miP1a/BBX31 regulates photomorphogenesis and UV-B protection. Plant Physiol. 179, 1876–1892. doi: 10.1104/pp.18.01258
Yang, Y. J., Ma, C., Xu, Y. J., Wei, Q., Imtiaz, M., Lan, H. B., et al. (2014). A zinc finger protein regulates flowering time and abiotic stress tolerance in Chrysanthemum by modulating gibberellin biosynthesis. Plant Cell 26, 2038–2054. doi: 10.1105/tpc.114.124867
Yu, C. S., Chen, Y. C., Lu, C. H., and Hwang, J. K. (2006). Prediction of protein subcellular localization. Proteins 64, 643–651. doi: 10.1002/prot.21018
Zhang, H. K., Gao, S. H., Lercher, M. J., Hu, S. N., and Chen, W. H. (2012). EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 40, W569–W572. doi: 10.1093/nar/gks576
Zhang, X. Y., Huai, J. L., Shang, F. F., Xu, G., Tang, W. J., Jing, Y. J., et al. (2017). A PIF1/PIF3-HY5-BBX23 transcription factor cascade affects photomorphogenesis. Plant Physiol. 174, 2487–2500. doi: 10.1104/pp.17.00418
Keywords: BBX, light, cold stress, Solanum lycopersicum, photoinhibition
Citation: Bu X, Wang X, Yan J, Zhang Y, Zhou S, Sun X, Yang Y, Ahammed GJ, Liu Y, Qi M, Wang F and Li T (2021) Genome-Wide Characterization of B-Box Gene Family and Its Roles in Responses to Light Quality and Cold Stress in Tomato. Front. Plant Sci. 12:698525. doi: 10.3389/fpls.2021.698525
Received: 21 April 2021; Accepted: 28 May 2021;
Published: 05 July 2021.
Edited by:
Rosalyn B. Angeles-Shim, Texas Tech University, United StatesReviewed by:
Ertugrul Filiz, Duzce University, TurkeyRakesh K. Upadhyay, United States Department of Agriculture (USDA), United States
Yushi Luan, Dalian University of Technology, China
Suhas Karkute, Indian Council of Agricultural Research (ICAR), India
Copyright © 2021 Bu, Wang, Yan, Zhang, Zhou, Sun, Yang, Ahammed, Liu, Qi, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng Wang, ZmVuZ3dhbmdAc3lhdS5lZHUuY24=; orcid.org/0000-0001-5351-1531; Tianlai Li, dGlhbmxhaWxpQDEyNi5jb20=
†These authors have contributed equally to this work
 Xin Bu1,2,3†
Xin Bu1,2,3† Golam Jalal Ahammed
Golam Jalal Ahammed Yufeng Liu
Yufeng Liu Mingfang Qi
Mingfang Qi Feng Wang
Feng Wang