
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 09 July 2021
Sec. Plant Development and EvoDevo
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.697609
This article is part of the Research Topic Environmental and molecular control of bud dormancy and bud break in woody perennials: An integrative approach View all 23 articles
Plant-specific TEOSINTE BRANCHED 1, CYCLOIDEA, PROLIFERATING CELL FACTORS (TCP) transcription factors have versatile functions in plant growth, development and response to environmental stress. Despite blueberry’s value as an important fruit crop, the TCP gene family has not been systematically studied in this plant. The current study identified blueberry TCP genes (VcTCPs) using genomic data from the tetraploid blueberry variety ‘Draper’; a total of 62 genes were obtained. Using multiple sequence alignment, conserved motif, and gene structure analyses, family members were divided into two subfamilies, of which class II was further divided into two subclasses, CIN and TB1. Synteny analysis showed that genome-wide or segment-based replication played an important role in the expansion of the blueberry TCP gene family. The expression patterns of VcTCP genes during fruit development, flower bud dormancy release, hormone treatment, and tissue-specific expression were analyzed using RNA-seq and qRT-PCR. The results showed that the TB1 subclass members exhibited a certain level of expression in the shoot, leaf, and bud; these genes were not expressed during fruit development, but transcript levels decreased uniformly during the release of flower bud dormancy by low-temperature accumulation. The further transgenic experiments showed the overexpression of VcTCP18 in Arabidopsis significantly decreased the seed germination rate in contrast to the wild type. The bud dormancy phenomena as late-flowering, fewer rosettes and main branches were also observed in transgenic plants. Overall, this study provides the first insight into the evolution, expression, and function of VcTCP genes, including the discovery that VcTCP18 negatively regulated bud dormancy release in blueberry. The results will deepen our understanding of the function of TCPs in plant growth and development.
TCP genes are a plant-specific transcription factor family whose members play vital roles in plant growth and development by affecting cell proliferation and differentiation. The name of TCP transcription factor was based on the first four identified members: TEOSINTE BRANCHED1 (TB1) from Zea mays, CYCLOIDEA (CYC) from Antirrhinum majus, and PROLIFERATING CELL FACTORS 1 and 2 (PCF1 and PCF2) from Oryza sativa (Luo et al., 1996; Doebley et al., 1997). Predictions of the secondary structure show that the TCP domain is similar to the eukaryotic transcription factor bHLH. However, the TCP basic region is long, and one of the conserved regions contains a non-canonical basic-helix-loop-helix structure, which results in different DNA binding characteristics. Therefore, the TCP family was defined as a new family of transcription factors (Cubas et al., 1999; Kosugi and Ohashi, 2002; Navaud et al., 2007).
The 24 TCPs in Arabidopsis thaliana can be divided into two subfamilies, Class I (PCF or TCP-P) and Class II (TCP-C), based on their different domains. The most significant difference between these classes is four additional amino acids in the TCP domain of class II. Class II TCP proteins also have a specific sequence, namely, the R structure, which is rich in 18–20 arginine (Asp) (Cubas et al., 1999). Type II TCPs are further divided into two subcategories, CINCINNATA (CIN)-like TCP (CIN-TCP) and CYCLOIDEA/TEOSINTE BRANCHED1 (CYC/TB1) (Navaud et al., 2007). The former subcategory contains eight members (AtTCP2/3/4/5/10/13/17/24), five of which are regulated by miRNA319 (Palatnik et al., 2003).
Class I members in the TCP transcription factor family promote cell proliferation and growth, but class II members inhibit these processes (Herve et al., 2009; Li et al., 2019). Genes of the two classes play versatile functions in leaf type control, axillary bud meristem development, plant height development, asymmetry of floral organs, hormone signal transduction, and the response to biotic and abiotic stresses (Martin-Trillo and Cubas, 2010; Daviere et al., 2014; Kim et al., 2014; Nicolas and Cubas, 2016; Viola et al., 2016). TCP genes are also involved in the regulation of plant circadian rhythm. For example, TCP proteins regulate the circadian clock by binding to the TGGGC (C/T) element in Arabidopsis thaliana (Giraud et al., 2010). There are few studies on the functional information of Class I TCP members, which are involved in plant development, growth, and proliferation. For example, AtTCP14 and AtTCP15 regulate embryonic growth via the gibberellin signaling pathway during seed germination (Resentini et al., 2015), and AtTCP15 directly acts on GA20ox1, HBI1, and PRE6 to promote petiole growth and hypocotyl elongation (Ferrero et al., 2019). In Arabidopsis, a tcp23-knockout line has an early-flowering phenotype, while overexpression lines exhibit the reverse phenotype, which indicates that AtTCP23 participates in the regulation of flowering (Balsemao-Pires et al., 2013). AtTCP16 shows higher expression in developing microspores; 50% of aborted pollen was produced in TCP16 RNA interference (RNAi) plants, which indicates that AtTCP16 plays a crucial role in the early stages of pollen grain development (Takeda et al., 2006). Compared with class I members, the functions of most class II TCP genes have already been elucidated. Experimental evidence supports that the proteins may be involved in various plant life activities, such as axillary bud meristem development, leaf and flower development, branching, hormone signaling, and plant defense (Luo et al., 1996; Doebley et al., 1997; Aguilar-Martínez et al., 2007; Nicolas and Cubas, 2016). Previous research demonstrated that AtTCP18 and AtTCP12 were similar to tb1 in protein sequence, expression pattern, and mutant phenotype; these genes were named BRC1 and BRC2, respectively. Compared with wild-type plants, BRC1-mutant plants had more axillary bud germination, which demonstrated that BRC genes plays a negative regulatory role during germination and inhibits the elongation of axillary bud branches (Aguilar-Martínez et al., 2007). Environmental factors, such as light and plant density, also regulate BRC1 gene expression. The evidence demonstrated that the type II TCP transcription factor, together with the FT-FD complex, controls the initiation of inflorescence in Arabidopsis, and TF directly binds to the AP1 promoter to enhance its transcription and regulate flowering (Li et al., 2019). The BRC1 homologous gene in cucumber inhibits lateral bud outgrowth via the direct suppression of PIN3 functioning and auxin accumulation in axillary buds (Shen et al., 2019).
Many TCP family members have been characterized in other plants, such as grape (Jiu et al., 2019; Leng et al., 2019), tobacco (Chen et al., 2016), Moso Bamboo (Liu et al., 2018), wheat (Zhao et al., 2018), corn (Ding et al., 2019), upland cotton (Li et al., 2017), plum (Zhou et al., 2016), tea (Zhou et al., 2019), and strawberry (Wei et al., 2016). Blueberries (Vaccinium spp.) are known as the “king of berries,” with high nutritional and great commercial value. Despite the significant roles of TCP family genes in plant growth and development, few detailed analyses of these genes in blueberry are available. Dormancy is the stage that perennial woody plants must go through for the next seasonal cycles, and it is also the adaptive evolution of plants to survive the harsh winter. Previous studies identified the transcription factor SHORT VEGETATIVE PHASE-LIKE (SVL) in poplar, closely related to Arabidopsis SVP, and its downstream target was determined to be TCP18 by chromatin immunoprecipitation (ChIP) experiments from transgenic plants. SVL and TCP18 can be regulated by low temperature, which play a negative regulatory role in bud germination (Singh et al., 2018).
The present study analyzed blueberry TCP genes using the genomic data of the tetraploid blueberry variety ‘Draper’ and identified a total of 62 TCP genes. Comprehensive analyses were performed, including multisequence alignment, conserved motif, gene structure, cis-acting elements and syntenic analysis. The expression patterns of VcTCPs in diverse tissues, stages of fruit development, flower bud dormancy release, and in response to hormone treatment were analyzed using multiple transcriptome data. The expression patterns of TCP gene family members during the process of dormancy release in flower buds were verified using qRT-PCR. The further transgenic experiments showed the overexpression of VcTCP18 significantly decreased the seed germination rate in contrast to the wild type. The bud dormancy phenomena as late-flowering, fewer rosettes and main branches were also observed in transgenic plants. Overall, this study provides the first insight into the evolution, expression, and function of VcTCP genes, including the discovery that VcTCP18 negatively regulates bud dormancy release in blueberry. The results will broaden our understanding of the function of TCPs in plant growth and development.
Plant materials were taken from the blueberry orchard of the Zhejiang key laboratory of biotechnology onspecialty economic plants in 2019. The variety is South highbush ‘O'Neal’. Shoots (spring shoots), leaves (tender leaf), flower buds (endo-dormancy flower buds), flowers (flower blooms) and fruits (‘O'Neal’ S4 fruit) (Yang et al., 2018) were immediately placed in liquid nitrogen and stored at −80°C for future use.
Artificial cold accumulation: On November 19, 2018, 1-year-old shoots (the flower buds in endodormancy stage) of ‘O'Neal’ trees were collected, incubated at 15°C for 3 days, and transferred to 4°C. The following culture conditions were used: day/night temperature 4°C; 16/8 h day/night; light intensity of 320 μmol ⋅ m–2 ⋅ s–1, and relative air humidity of 75%. The water was changed every 2 days, and the base of the branches was cut off to reveal new stubble. Low-temperature treatment was applied for 28 days, and 30 flower buds were collected every 4 days and frozen in liquid nitrogen. Samples were stored at −80°C for later use. Other branches were exposed to forcing conditions for bud break measurements. The end of endodormancy is considered to have been reached when the percentage budbreak is 50% (Yooyongwech et al., 2009). Three biological replicates were performed for all of the above treatments.
Arabidopsis (Col-0) and the transgenic plants seeds were cultivated in 1/2 MS medium for 1 week then transferred to soil and grown in the incubators (16 h photoperiods, 22°C and 70% relative humidity).
Two different methods were used to identify TCP genes in the blueberry genome. First, the hidden Markov model (HMM) profile of the conserved TCP domain (PF03634) was downloaded from the Pfam database1, and HMMER software was used against a local blueberry protein sequence database. The E value was set to 0.01. Second, we used all of the Arabidopsis TCP protein sequences downloaded from the Arabidopsis Information Resource (TAIR) database2 as reference sequences, and a local BLAST comparison was used to screen the blueberry genome database. All VcTCP protein sequences were verified using the SMART database3. We used MEGA to remove repeated sequences, and the obtained non-redundant VcTCP protein sequences were further analyzed. The molecular weights (MWs), isoelectric points (pIs), and the grand average of hydrophilicity (GRAVY) of VcTCP proteins were analyzed using the ExPASy website4. The subcellular localizations of VcTCP proteins were predicted using WoLF PSORT5 (Leng et al., 2019).
Twenty-four Arabidopsis TCP protein sequences were retrieved from TAIR2, and poplar, rice and kiwifruit TCP protein sequences were downloaded from the Plant Transcription Factor Database6. Antirrhinum CYC and maize TB1 sequences were retrieved from the NCBI database7. MEGAX software was used to perform multisequence alignment of the full amino acid sequences of the TCP proteins of blueberry, Arabidopsis, rice, kiwifruit, Antirrhinum and maize. The program and operating parameters for the construction of an evolutionary tree were the neighbor-joining method with 1,000 bootstrap replicates using MEGAX. The replacement model was JTT + G + F.
The online tool MEME8 was used to identify and analyze the conserved motifs of TCP proteins (parameter setting: maximum number of motifs: 5; maximum motif width: 60) (Bailey and Elkan, 1994). The CDS and corresponding genomic DNA sequences of VcTCP genes were acquired from the blueberry genome. GSDS9 was used to display the exon-intron organization via comparison of the coding sequences with their corresponding genomic sequences (Hu et al., 2015).
The 1,500 bp upstream of the coding regions of the VcTCP genes were extracted from the blueberry genome data. PlantCARE10 was used to search for putative cis-acting elements (Lescot et al., 2002).
Based on the blueberry genome information, 62 VcTCP genes were located on the chromosome. For the collinearity analysis, first, BlastP was performed to search for potential homologous gene pairs in the blueberry genome and between blueberry and Arabidopsis (E < 10–5, first five matches). Second, the homologous pairs were used as an input file for MCScanX to identify syntenic chains and types of duplication mechanisms (Tang et al., 2008; Wang et al., 2012). Circos and Tbtools software11 were used for visualization (Krzywinski et al., 2009; Chen et al., 2020).
The expression values of VcTCPs in numerous tissues, root salt stress treatment and leaf methyl jasmonate (MeJA) treatment were obtained from the RNA sequencing data reported previously (Colle et al., 2019), the variety is “Draper.” The transcriptome data of ‘O'Neal’ flower bud endo-dormancy and eco-dormancy release from our laboratory were used to analyze the expression profiles of VcTCPs. The RNA into 21 samples (7 bud development stage × 3 replicates), which were sequenced on an Illumina HiSeqTM 4000 after the library was qualified by Agilent 2100 Bioanalyzer and ABI Step One Plus real-time PCR System. Transcriptomic sequencing was conducted on the hypanthia and young fruits of ‘O'Neal’ and ‘Bluerain’ during anthesis and the early developmental period, sequenced on an Illumina HiSeq-Xten platform, which were acquired from our laboratory were also used to analyze. Zero to one was used to calculate the expression values. The expression heatmap of VcTCP genes was generated using TBtools software (Chen et al., 2020).
For reverse transcription, HiScript III RT SuperMix for qPCR (+gDNA wiper) (Nanjing Nuoweizan Biotechnology Co., Ltd.) was used to construct the cDNA library according to the instructions. The reverse-transcribed cDNA was used as a template for qRT-PCR. Quantitative primers were designed using the software Premier 5.0. The specificity of primers was determined by BLAST sequence comparison. The primers were synthesized by Hangzhou Qingke Biotechnology Co., Ltd. The primer sequences are shown in Table 1. The efficiency of the primers was tested using the standard curve method, and primers with efficiencies between 80% and 120% were used for subsequent experiments. The quantitative kit was 2 × SYBR Green qPCR Mix (High ROX) (Beijing Adelaide Biotechnology Co., Ltd.), which uses a two-step method to perform qPCR amplification reactions. Blueberry GAPDH was used as an internal reference gene, and data analyses were performed using the relative quantitative method (2–ΔΔCt). SPSS Statistics 21 software was used for significant difference analyses, and Prism was used for drawing.
Full-length VcTCP18 cDNA was amplified using cDNA prepared from mRNA extracted from blueberry ‘O'Neal’ flower bud as templates. The primer sequences are shown in Table 2. VcTCP18 cDNA were cloned into the pMD 19-T vector and transferred into the E. coli DH5α, verified with PCR, finally transferred into the vector pCAMBIA2300-3 × flag, via XbaI, Hind III double digestion and recovered with T4 DNA ligase. The recombinant plasmid pCAMBIA2300-CaMV35S-VcTCP18-3 × flag-CaMV poly(A) signal, which was subsequently transferred into Agrobacterium strain GV3101. The Arabidopsis transformation was conducted by floral dipping method. T0 seeds were screened by kanamycin, and T1 plants were confirmed by RT-PCR analysis. T3 generation homozygous lines were used for all analyses.
The seeds of WT and transgenic plants were cultivated in 1/2 MS medium with and without stratification (4°C treatment 2 days). The germination rate was counted every 6 h. Flowering time was measured by counting the number of days from sowing until flower buds were visible by the naked eye, meanwhile the number of rosette leaves were counted, main branches were also recorded (Xue et al., 2018). At 30-day-old seedling time, statistical analysis of the number of main branches in Transgenic plants and WT.
All prospective VcTCP members were obtained from the blueberry genome using HMMER and BlastP. The domain integrity of the VcTCP candidate genes was identified using the SMART database (Supplementary Figure 1). A total of 62 TCP gene family members were identified on 30 chromosomes and named VcTCP1-VcTCP62 according to the order of the chromosomes on which they were located. The TCP domains of some Class II TCP members also included an R domain, which was found at the C-terminus of all VcTCP proteins from blueberry Class II CYC/TB1 and two members of VcTCPs from CIN. Using the online tool ExPASy website, the physical and chemical properties of the VcTCP proteins were analyzed (Table 3). The length of VcTCP proteins varies from 198 (VcTCP29/47) to 686 amino acids (VcTCP24). VcTCP47 has the lowest relative molecular mass (20.97 kDa) and the highest is VcTCP24 (76.58 kDa). Theoretical isoelectric points (pIs) range from 5.2 to 10.02. Aliphatic amino acid indexes are between 51.67 and 78.45, which indicates that the VcTCP proteins are rich in aliphatic amino acids. The average hydrophilicity coefficient of all VcTCP proteins is less than 0, which indicates that VcTCPs are hydrophilic proteins. According to the prediction of WoLF PSORT, most of the VcTCP proteins are located in the nucleus, with only VcTCP26 possibly located in chloroplasts.
To study the evolutionary and phylogenetic relationships between the blueberry TCP genes and other known TCPs, the full-length sequences of 150 TCP proteins from Arabidopsis, rice, kiwifruit, blueberry, snapdragon CYC and maize TB1 were used to construct a phylogenetic tree. According to previous studies of the classification of TCP genes in Arabidopsis and rice, phylogenetic analysis and TCP domain alignment all showed that VcTCP proteins can be divided into two categories: class I (PCF), which contains 30 genes; and class II, which contains 32 genes (Figure 1). The phylogenetic tree indicates that class II may be further divided into two subclades, CYC/TB1 (14) and CIN (18). The TCPs of Arabidopsis and rice had the same groups or branches, indicating the reliability of the phylogenetic tree. Class II proteins have four more amino acids in the basic domain than class I proteins, which is the most significant difference between these two classes (Figure 2A). The TCP domains of some Class II TCP members also included an R domain, which was found at the C-terminus of all VcTCP proteins from blueberry Class II CYC/TB1 and two members of VcTCPs from CIN (Figure 2B).
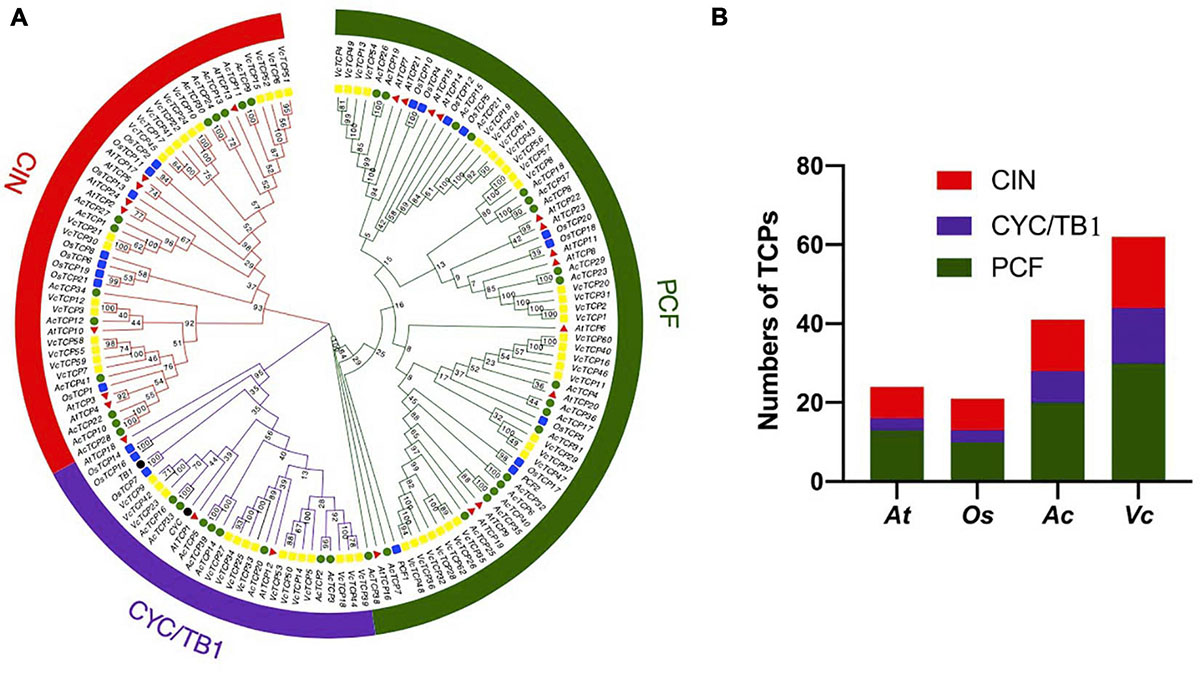
Figure 1. (A) Phylogenetic analysis of TCP members in blueberry (VcTCP rectangle, yellow), kiwifruit (AcTCP circle, green), Arabidopsis (AtTCP triangle, red), rice (OsTCP rectangle, blue), Antirrhinum CYC and maize TB1(circle, black). The phylogenetic tree was constructed using the neighbor-joining method with 1,000 bootstrap replicates in MEGAX. The branched lines of the subtrees are colored to indicate different TCP subgroups. The number on the branch denotes the corresponding bootstrap value. (B) Statistical analysis of TCP members from blueberry, rice, kiwifruit and Arabidopsis.
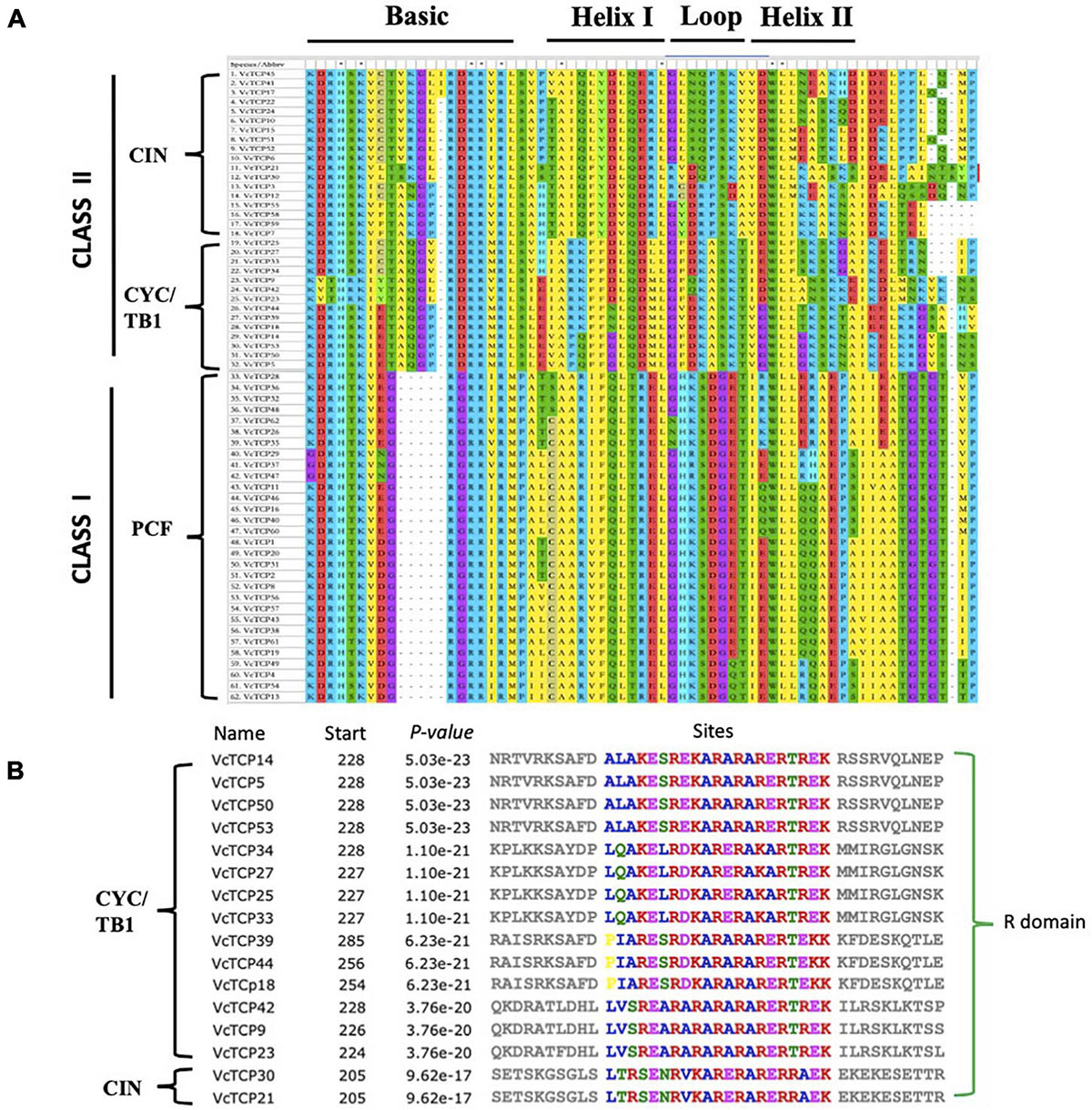
Figure 2. Conserved structural domain analysis of VcTCPs in blueberry. (A) Multiple alignment of VcTCPs protein sequences was performed using MEGA X software. (B) Multiple alignment of the R domain sequences.
To better understand the diversity of the motif composition of VcTCPs, the MEME online tool was used to predict the conserved motifs of the blueberry TCP gene family. A total of five conserved motifs were identified (Figure 3). The results showed that motif 1 existed in all VcTCP proteins. Motif 2 is located in the C-terminal TCP domain of Class I members with high specificity. Motif 4, 5 exist in members of the CIN subfamily, but it was not observed in the TB1 subfamily. By comparison, all class II TB1 proteins are characterized by motif 3 (R domain), two of the CIN subfamily members VcTCP21, VcTCP30, which are clustered with AtTCP2,24 also contain an R domain, which is consistent with previous research that the presence of certain motifs in a particular subgroup may play a specific role in these genes (Wei et al., 2016).
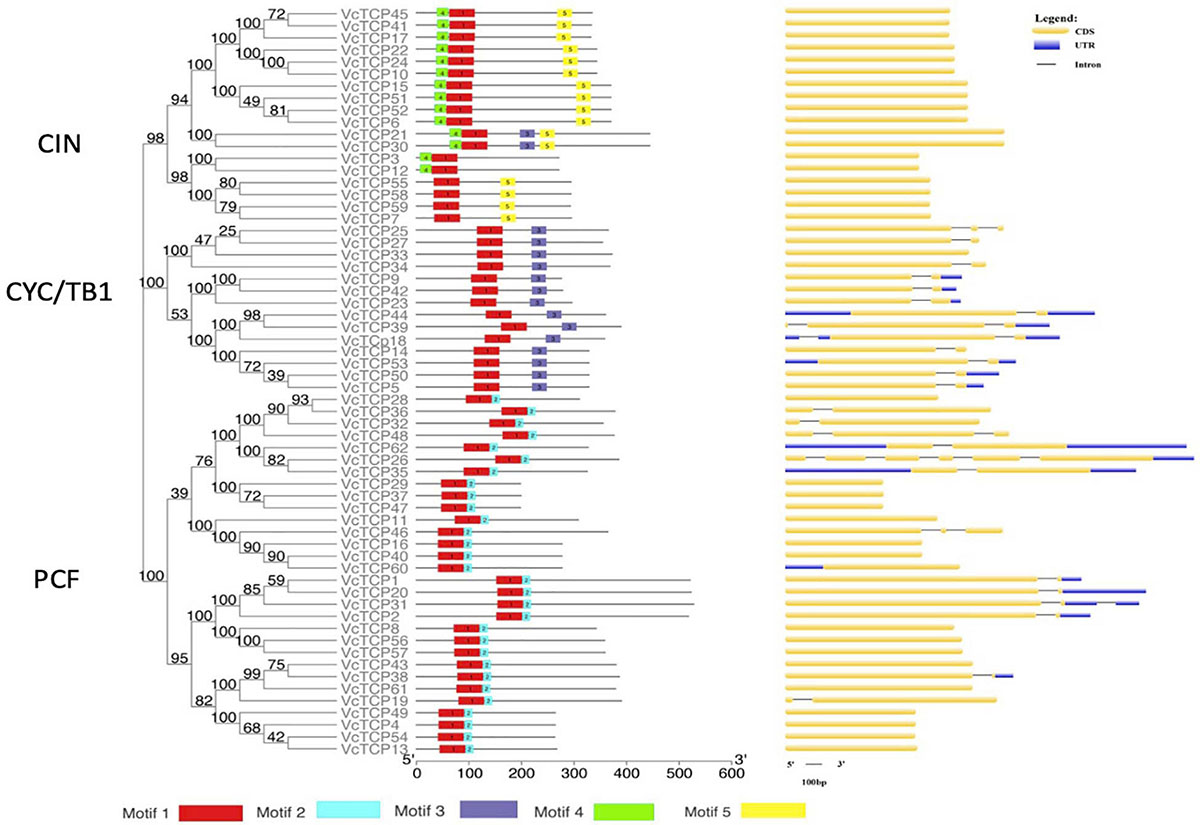
Figure 3. Phylogenetic analysis, conserved motifs and gene structure of the TCP family in blueberry. Conserved motifs of VcTCPs were identified using MEME. Different motifs are shown by different colors. CDS, introns and UTR are indicated by yellow rectangles, black lines and blue rectangles, respectively.
To investigate the evolutionary relationship and structural characteristics of VcTCP proteins, an evolutionary tree was constructed using the conserved domain sequences of VcTCPs, which were divided into three subfamilies (Figure 3). Almost all VcTCP genes exhibit a highly conserved exon-intron structure. Twenty-six of the 62 VcTCP genes have introns, 19 VcTCP genes have only one intron, and 7 VcTCP genes have two or more introns. Most of the VcTCP genes in the same subfamily show similar exon/intron distribution patterns, which supports the classification and evolutionary relationship of the subclade.
To better understand the response and regulation mechanisms of VcTCP genes, promoter cis-regulatory elements of the 62 VcTCP genes were analyzed. The sequences of 1,500 bp upstream of the genes were submitted to Plant CARE. In addition to the basic TATA and CAAT boxes, the promoters contain primarily light response, transcription factor binding (MYB and MYC, etc.), hormone response, injury response, low temperature response, zeatin metabolism, flavonoid metabolism regulation and other cis-acting elements. Among the 62 VcTCP gene promoters, light-responsive (TCT-motif, Sp1, GT1-motif), MYB-binding and MYC-binding elements predominate, and all VcTCP genes contain these three types of response elements. Hormone-response elements primarily include abscisic acid (ABRE), methyl jasmonate (CGTCA-motif, TGACG-motif), ethylene (ERE), auxin (TGA-element), salicylic acid (TCA-element, SARE), and gibberellin (P-box). Abscisic acid-responsive elements are the most common among the hormone-response elements, which suggests that abscisic acid regulates this family of transcription factors during growth and development. For the abiotic stress response elements, a large number of low temperature responsiveness (LTR), anaerobic induction (ARE), and drought induction (MBS) elements were detected. These results revealed that VcTCP genes play an important role in the response to biotic and abiotic stresses. Some cis-regulatory elements of growth and development, hormone response and biological abiotic stress in the present study are depicted in Figure 4.
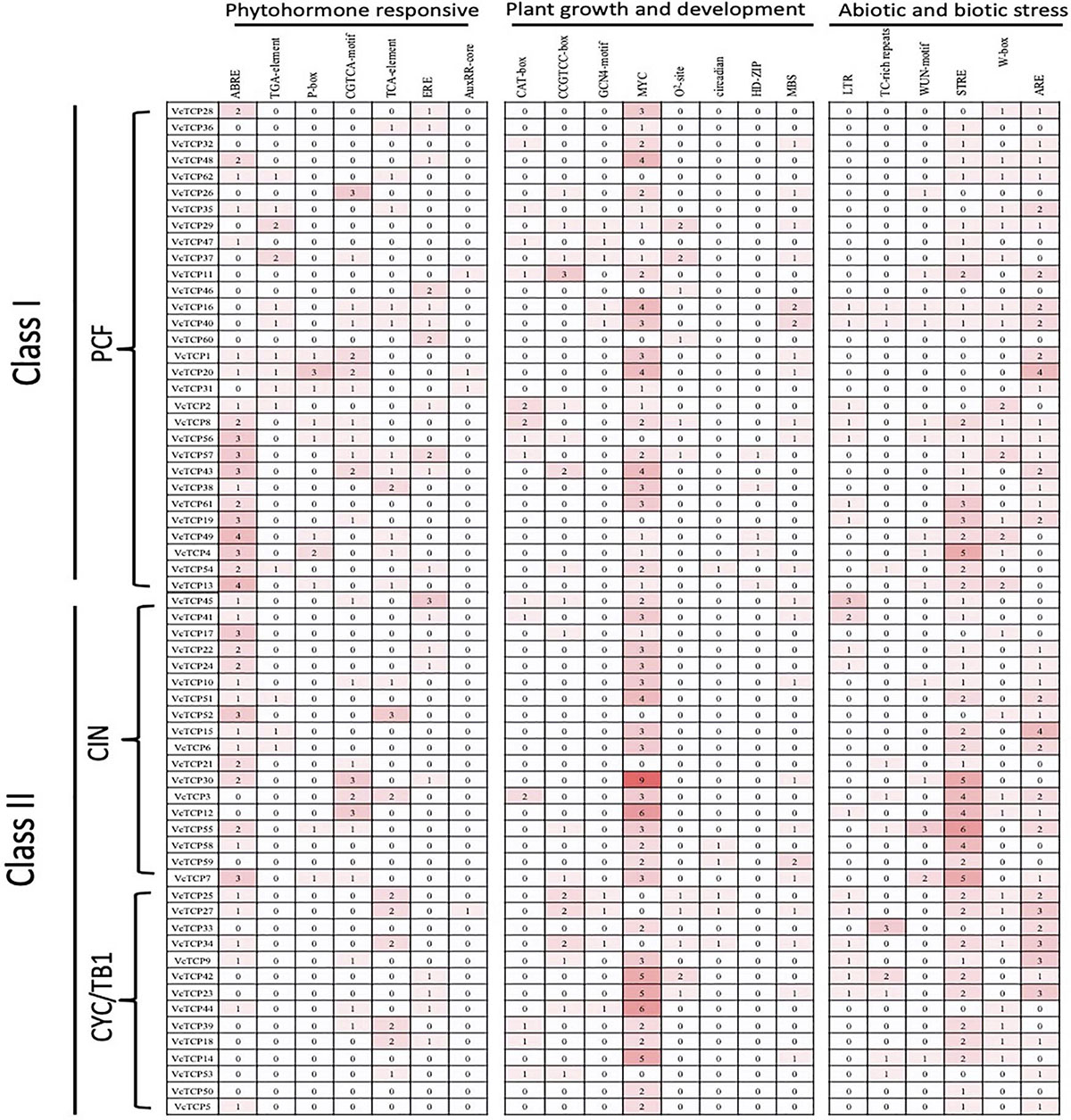
Figure 4. Cis-regulatory elements analysis of the promoter region of VcTCP genes. Numbers of each cis-regulatory element in the promoter region of VcTCP genes. Based on the functional annotation, the cis-acting elements were classified into three major classes: phytohormone responsive, plant growth and development, and abiotic and biotic stresses.
Gene replication and differentiation are important processes for gene family expansion and novel functional evolution in the plant genome. Some gene replication events, including tandem replication, WGD (Whole Genome Duplication)/segment duplication, and chromosomal and gene-level rearrangements, have driven the evolution of protein-coding gene families (Maher et al., 2006). To evaluate expansion of the VcTCP gene family, we used MCScanX software to analyze the origin of the VcTCP gene family repeat genes in the blueberry genome. In the gene type analysis, VcTCP58 exist in tandem, the other members derived from WGD/segment duplication (Supplementary Material Additional file 1) which demonstrated that the WGD/segment played an important role in the expansion of the VcTCP gene family.
To further examine the origin and possible evolutionary mechanism of the VcTCP gene family, the BlastP resulting files of blueberry with Arabidopsis and blueberry itself were entered into MCScanX, and the homology matching file was examined. The source pairings were very complicated, and no homologs were found between two genes VcTCP25 and VcTCP34. All other gene family members have collinear relationships, as shown by the red line (Figure 5). Homology analysis showed six homologous gene pairs between blueberry and Arabidopsis, as indicated by the blue line. The homology analysis of blueberry and Arabidopsis sequences demonstrated that these genes located in the corresponding homology region arose before the divergence of blueberry and Arabidopsis.
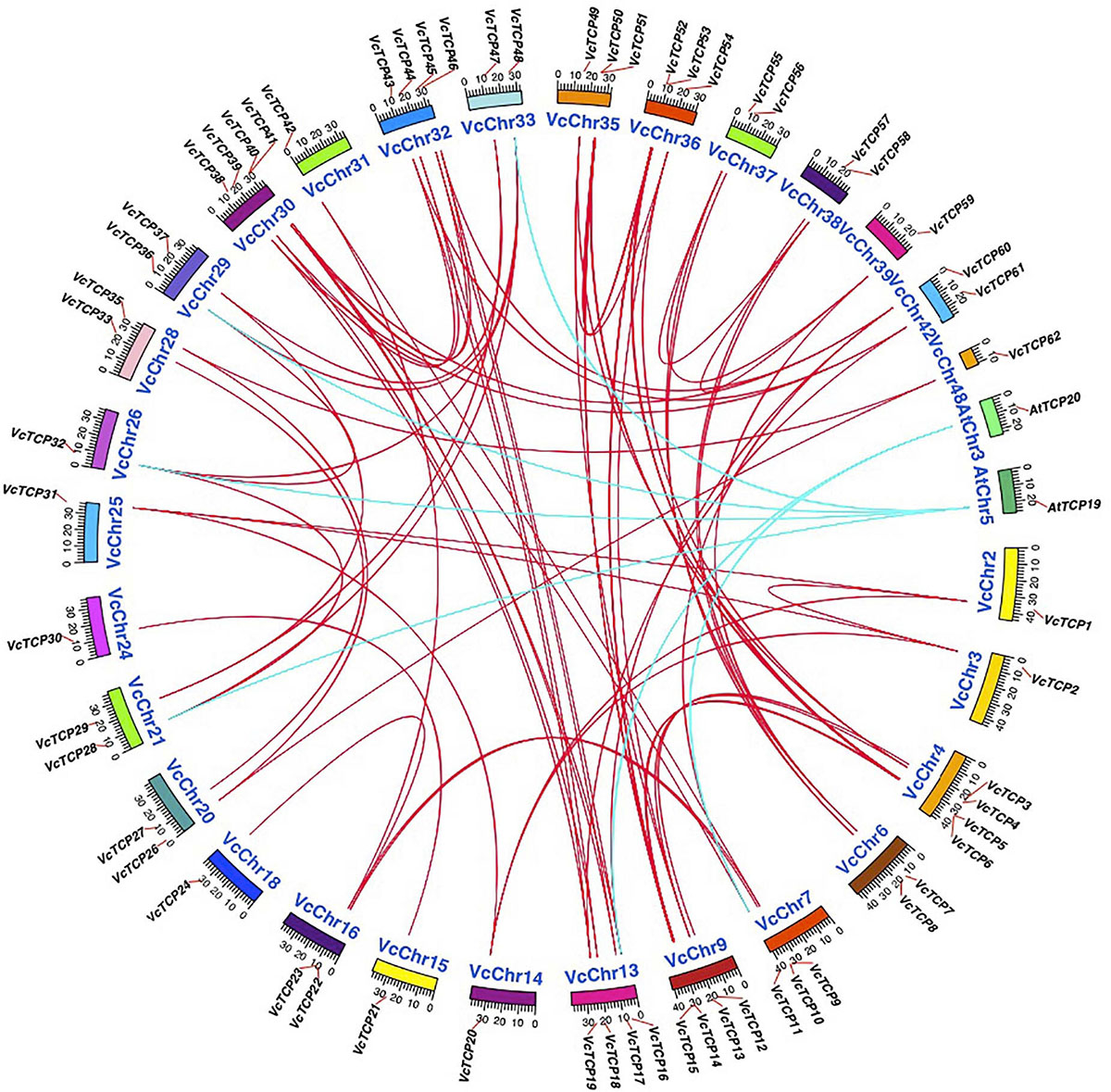
Figure 5. Chromosome distribution and synteny analysis of blueberry and A. thaliana TCP genes. Chromosomes of V. corymbosum and A. thaliana are shown in different colors and circular form. The approximate distribution of VcTCPs and AtTCPs are marked with a short red line on the circle. Red and blue curves denote the details of syntenic regions between blueberry and A. thaliana TCP genes.
The expression patterns of TCP genes were examined in different tissues, bud dormancy release, and fruit development in blueberry. As indicated in Figure 6, transcriptome profiling showed that the expression patterns of VcTCPs within the same subclade were not similar, which suggests that these genes play different roles in biological processes. The nine members of the class II subclades CYC/TB1 (VcTCP9/42/23/44/39/14/53/50/5) exhibited a certain level of expression in the shoot and leaf-night, and these genes were not expressed during fruit development. Before and after root salt treatment, members of this subclade were not expressed. After MeJA treatment, VcTCP23 was downregulated at 1 and 8 h and upregulated at 24 h. All members of the subclade decreased with cold, but the expression levels increased during the process of eco-dormancy release. During bud endo-dormancy release, the expression levels of the BRC1 homologous genes VcTCP5, VcTCP14, VcTCP18, VcTCP39, VcTCP44, VcTCP50, and VcTCP53 were obviously decreased; however, the change of expression levels of the BRC2 (AtTCP12) homologous genes VcTCP25, VcTCP27, VcTCP33, and VcTCP34 was not obvious, which is consistent with the results in the literature (Aguilar-Martínez et al., 2007).
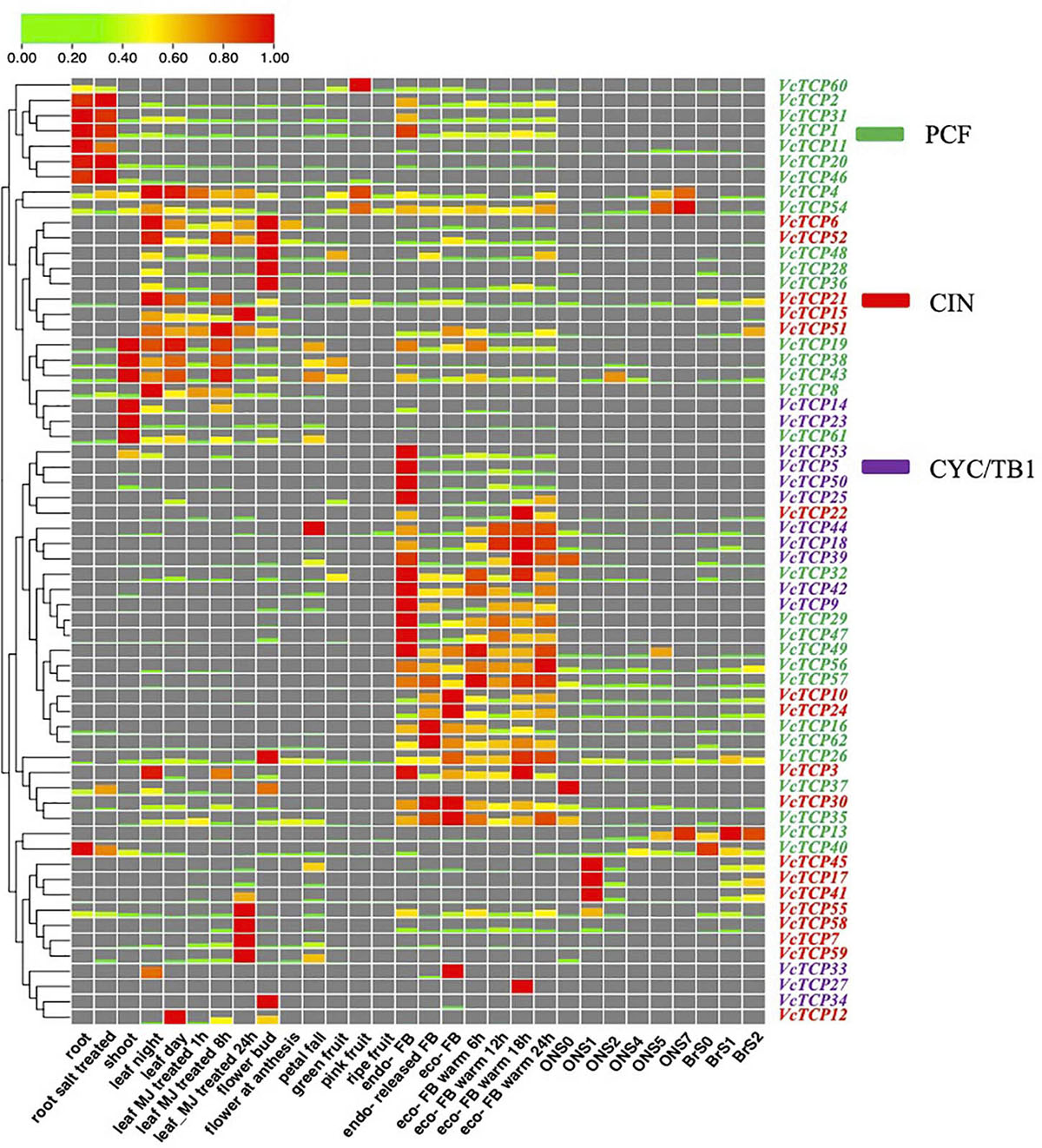
Figure 6. Expression profiles of VcTCP genes in diverse tissues, stages of fruit development, flower bud dormancy release, and in response to hormone treatment. The mean expression values were calculated using zero to one. Genes and expression patterns were hierarchically clustered based on the average Pearson’s metric. Green and red boxes show low and high expression levels, respectively, for every gene. Different colors of gene name belong to class I, CIN, and CYC/TB1, respectively. BrS0, S1, and S2 indicated the early fruit developmental stages of the blueberry variety “Blue rain”; ON S0–S7 indicated the whole fruit development of the blueberry variety ‘O'Neal.’ FB means flower bud. Leaf MJ-treatment: leaf methyl jasmonate (MeJA) treatment. Endo- means endo-dormancy; eco- means eco-dormancy.
Among the CIN members, the transcript levels of VcTCP21 and VcTCP30 were high in the root, leaf, flower, and fruit tissues, but the other members were not expressed in the root. The expression levels of VcTCP21 and VcTCP30 also exhibited significant differences during fruit development, which indicates that these genes may play a necessary role in the growth and development of root and fruit. The expression levels of VcTCP41, VcTCP17, VcTCP58, VcTCP59 and VcTCP7 were increased after MeJA treatment 24 h after treatment, and the transcript accumulation levels exhibited significant differences, which suggests that these genes play a significant role in the JA signal transduction pathway.
Most PCF members were expressed in all tissues. The expression levels were higher in root, shoot, leaf and bud. VcTCP31, VcTCP2, VcTCP8, VcTCP43, and VcTCP38 were downregulated during fruit development. VcTCP60, VcTCP4, and VcTCP54 were upregulated in pink fruit and downregulated in ripe fruit, which revealed that these genes participate in blueberry fruit development. The expression levels of VcTCP43, VcTCP38, VcTCP61, VcTCP19, and VcTCP49 decreased 1 h after MeJA treatment, increased after 8 h, and decreased after 24 h. This expression trend was consistent, whether this type of genes plays a specific role in JA signal transduction is not known.
To elucidate the roles of TCP genes in flower bud dormancy release, several VcTCP genes were selected for quantitative verification: VcTCP5, VcTCP18, VcTCP19, VcTCP21, VcTCP26, VcTCP54, VcTCP55, and VcTCP60. These genes belong to the CIN, TB1, and PCF subclasses and show different expression patterns during flower bud dormancy release, fruit development and hormone treatment in transcriptome profiling analysis.
To investigate the tissue-specific profiles of VcTCP genes in blueberry, we analyzed the expression levels of selected VcTCP genes using RT-qPCR to validate the results of transcriptomes of different tissues, including shoots, leaves, flower and fruits from the blueberry accession ‘O'Neal’. Eight VcTCP genes exhibited tissue-specific transcript accumulation patterns, indicating the functional divergence of VcTCP genes during blueberry growth and development (Figure 7A). For example, the expression levels of VcTCP5, VcTCP19, and VcTCP26 were very high in leaves, and there are significant differences compared with other tissues, indicating that these genes might play an important role in leaf development. VcTCP21 and VcTCP60 were highly expressed not only in the leaves but also in the buds, which indicated that these genes might play an important role in floral buds. All of these results were consistent with those in the transcriptome.
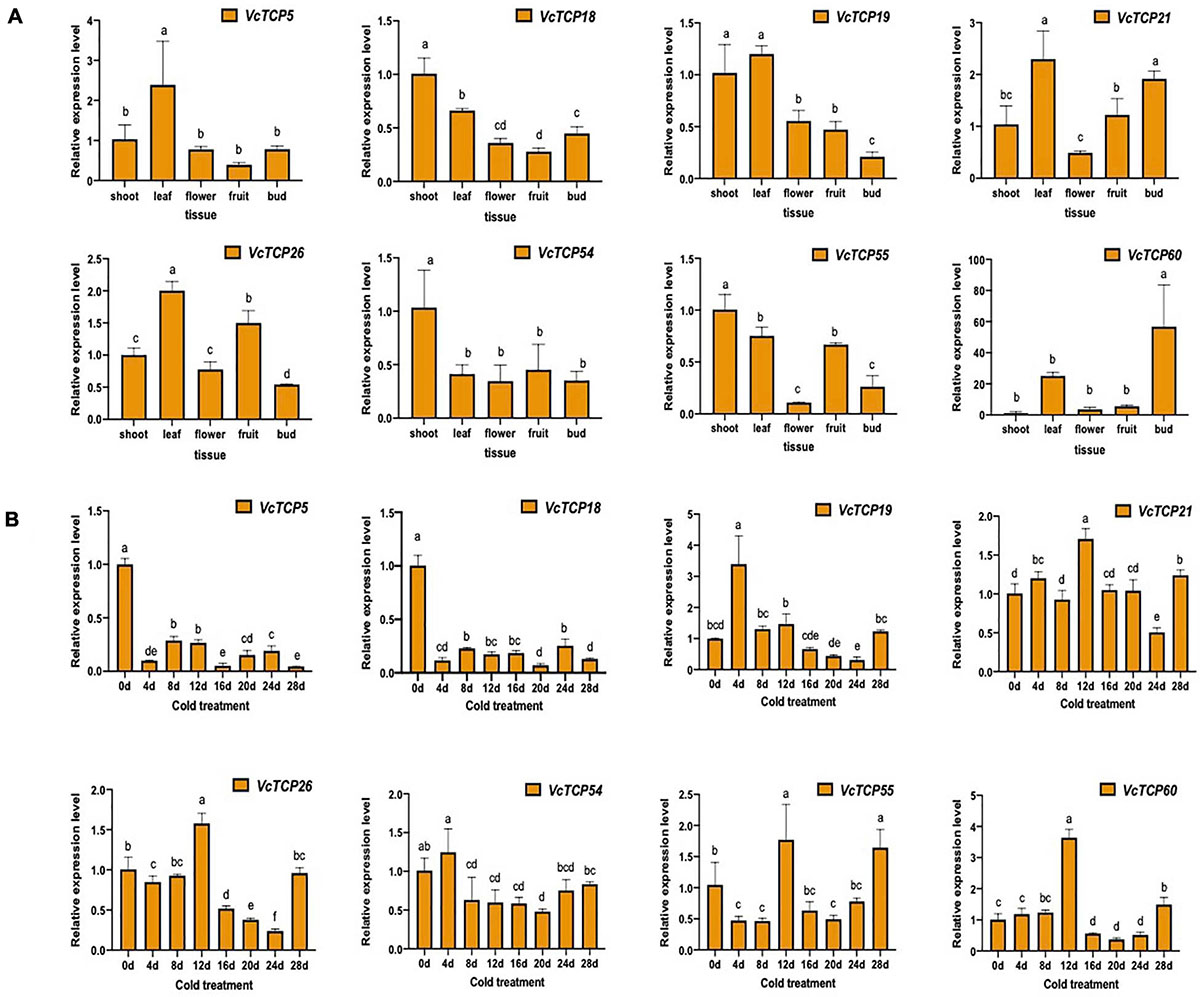
Figure 7. (A) Relative expression levels of VcTCPs in different tissues of ‘O'Neal’ blueberry. (B) Relative expression levels of VcTCPs during flower bud endodormancy release by artificial chilling treatment. Values were normalized against the expression data of VcGAPDH and given as the means ± standard error of three biological replicates. Different letters indicate significant differences between genes (p < 0.05) based on Duncan test. The expression levels were calculated based on the 2− ΔΔCt method.
As illustrated in Figure 8, average germination rates of ‘O'Neal’ flower bud increased with longer cold treatment times. The flower buds were in deep endo-dormancy stage before transfer to 4°C, and the germination rate was low. The sprouting rate of flower buds reached 65.89% after 12 days of cold treatment, which indicated the endodormancy was broken. After that point, flower buds have strong sprouting potential, then enter the eco-dormancy phase.
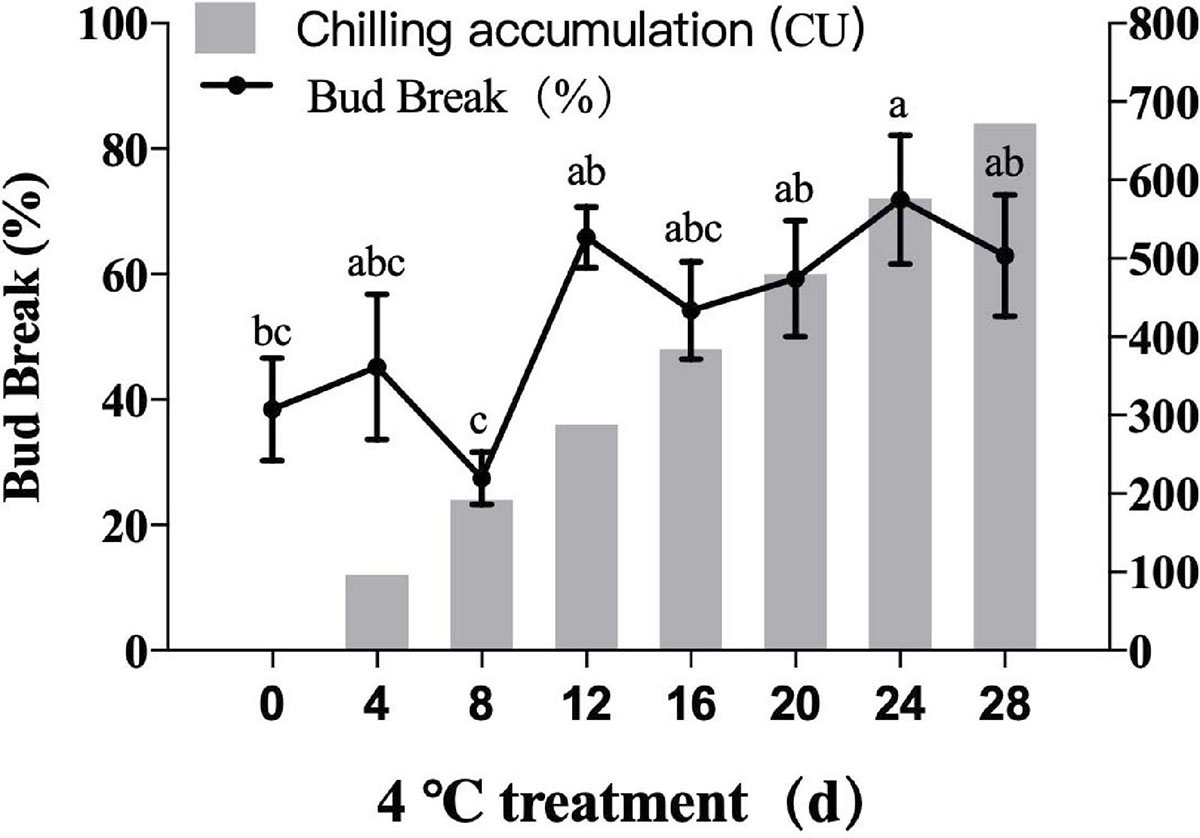
Figure 8. Germination rates of blueberry flower bud under different cold accumulation. All were the highest germination rates.
As shown in Figure 7B, VcTCP18 and VcTCP5 belong to the TB1 subclass and were highly expressed during the deep dormancy period (0 d); their expression levels decreased significantly when the dormant flower buds were subjected to 4 days of cold treatment. The expression of the two genes remained at a consistently low level with increasing duration of the cold treatment, which indicates that the two genes may inhibit bud germination. The expression levels were reduced after cold treatment, which restored the ability of the flower bud to restore germination.
VcTCP21 and VcTCP55 belong to the CIN subcategory, and their expression levels were increased significantly after 12 days of cold treatment. Notably, the germination of blueberry flower buds over 50% at this stage indicates that the genes may participate in flower bud endo-dormancy release. VcTCP26, VcTCP54, VcTCP19, and VcTCP60 belong to the PCF subclass, and the expression patterns in the cold treatment increased, decreased, then increased. The highest expression levels of VcTCP26 and VcTCP60 were observed after 12 days of cold treatment, but their expression levels increased a second time after cold treatment for 28 days. The flower buds were in the eco-dormancy stage at this time and germinate quickly if given the appropriate temperature. The expression patterns of VcTCP54 and VcTCP19 were the same as described above but occurred after 4 days of treatment, which indicates that these genes also responded to low temperature.
BRC1 has been shown to be the central integrator for multiple environmental and developmental factors that functions locally to inhibit bud break. Previous research has shown that DAM can be regulated by low temperature, which play a negative regulatory role in bud break, and TCP18/BRC1 is a downstream target gene of SVL/DAM (Singh et al., 2018). These observations prompted us to investigate BRC1s role in the blueberries bud dormancy release.
According to the analysis of gene expression in Figure 7B and the phylogenetic tree of CYC/TB1 subclade, shown in Supplementary Figure 2, although there are other putative BRC1-like genes in blueberry, VcTCP18 was selected in the present study, due to its profile that fit with an expected putative BRC1 gene for further analysis. A cDNA fragment with a length of 1,077 bp was obtained, as shown in Supplementary Figure 3A. The coding sequences were cloned into the vector pCAMBIA2300. Finally, the recombinant plasmid vector was transferred into Agrobacterium strain GV3101.
Through the Kana screening test, we obtained the T1 generation Arabidopsis transgenic seedlings. The further RT-PCR experiments and sequencing results showed that VcTCP18 was successfully expressed in Arabidopsis (Supplementary Figure 3B). Then the germination of T3 homozygous transgenic seeds were tested on 1/2 MS with and without stratification (Figure 9A). Without stratification treatment, the germination rate of transgenic seeds was much lower than that of WT. the germination rate of WT was first beyond 90% at 75 h after sowing, meanwhile, that of transgenic seeds was 2.5%, the rate of germination of WT was 59.48% until 147 h after sowing. Comparatively, when the seeds were treated by 4°C for 2 days before sowing, both of the seeds of WT and transgenic lines germinated faster than without stratification, however, transgenic seeds still germinated significantly slower than that of WT.
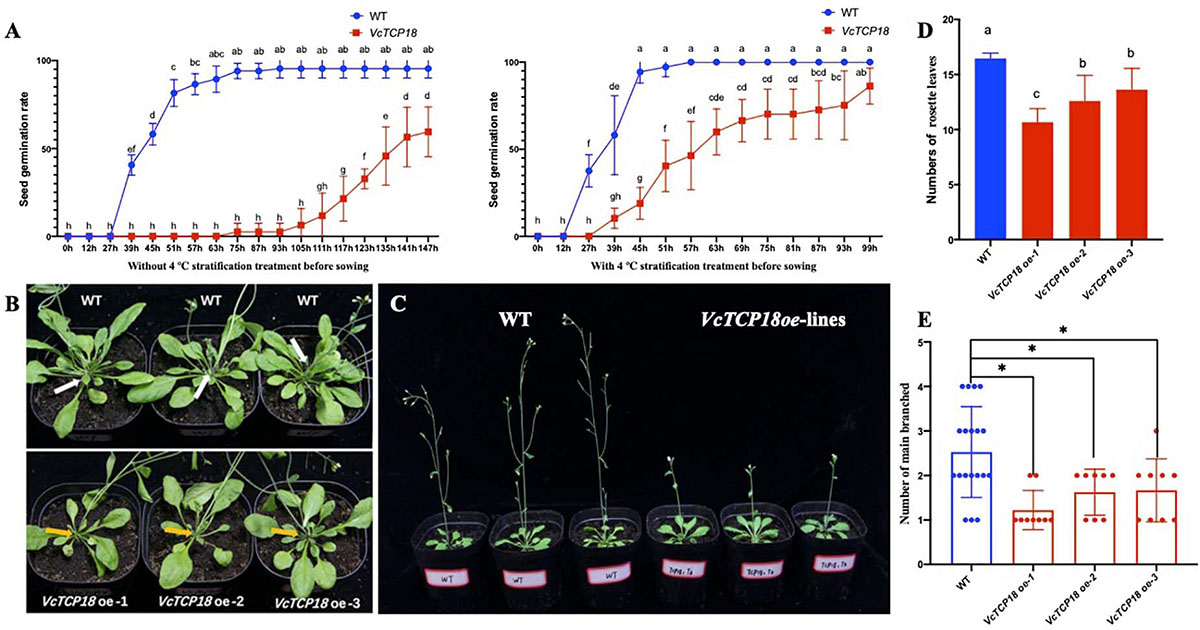
Figure 9. Phenotypes of transgenic Arabidopsis with overexpression of VcTCP18. (A) The seed germination rate of WT and transgenic lines with and without 4°C stratification treatment before sowing. (B) 30-day-old seedling observation of main branches, white and yellow arrows indicate main branches of WT and transgenic lines, respectively. (C) Flowering time observation, three lines on left are WT, the others on right are VcTCP18-overexpressed transgenic lines. (D) At first flowering time, the numbers of rosette leaves, different letters indicate significant differences between WT and transgenic lines (p < 0.05), statistical analysis was performed by one-way ANOVA followed by the Duncan test. (E) statistical analysis of the number of main branches in Transgenic plants and WT, each dot represents one plant. Asterisks (∗) indicate significant differences (P < 0.05) based on Duncan test.
When flower buds were visible by the naked eye, the numbers of rosette leaves were counted (Figures 9B,D). The rosette leaves of WT were 16.51, which significantly differed from VcTCP18 overexpression lines (P < 0.05). The T3 generation of VcTCP18 overexpressed plants displayed the late-flowering phenotype (Figure 9C). At 30-day-old seedlings time, the number of main branches in transgenic plants was lower than WT (Figures 9B,E). In the three transgenic lines, with the increase of expression efficiency of VcTCP18 (Supplementary Figure 4), the number of rosette leaves and main branches decreased, which significantly differed from that of WT. These VcTCP18−overexpressing lines exhibited bud break compared with the wild-type.
TCPs are a type of plant-specific transcription factor family. Previous studies have used the model plants Arabidopsis and rice as research objects to analyze the various functions of TCPs in plant growth and development. The present study identified 62 TCP gene family members in blueberry, compared with Arabidopsis (24 members), strawberry (19 members), tomato (20 members), apple (52 members), there were more numbers of TCP genes in blueberry. One possible reason is that whole-chromosome sequencing was performed, and blueberry has a large genome (1.63 GB). Another possible reason is that the blueberry variety ‘Draper’ is tetraploid, and thus gene replication events occurred during the evolution of the VcTCP genes. Gene replication and amplification events are widespread in the evolution of many plants, such as cotton (Ma et al., 2014), tomato (Parapunova et al., 2014), and apple (Xu et al., 2014).
Analysis of the structure of VcTCP genes showed that the genes in the same subclass had a similar gene structure, though the length of the VcTCP genes is quite different, which indicates that the VcTCP genes are evolutionarily conserved. There are also some VcTCP gene structures that are identical, but the other members are obviously different. This phenomenon has also been observed in the structural analysis of TCP gene family members in plants such as strawberry (Wei et al., 2016), cotton (Ma et al., 2014) and apple (Xu et al., 2014). These differences may be caused by the insertion or loss of introns during the evolution of the species, which indicates that the diverse functions and expression control methods of the TCP gene family have involved more replication fragments and gene doubling events during evolution. In our study, we found an arginine-rich R domain in the C-terminus of all VcTCP proteins from blueberry Class II CYC/TB1 and two members of VcTCPs (VcTCP21, VcTCP30) from CIN, which was predicted to promote protein interactions, as is the case for four genes in rice (OsTCP6, OsTCP7, OsTCP13, and OsTCP14), four genes (AtTCP2, AtTCP12, AtTCP18, and AtTCP24) in Arabidopsis, and maize TB1. Our result was concordant with previous studies in grape and tomato (Parapunova et al., 2014; Leng et al., 2019).
When plants suffer from environmental stresses, such as low temperature, drought, salinity and so on, these stresses trigger signaling cascades that activate transcriptional factors, by binding to specific cis-acting elements to regulate the expression of genes. The result showed that among 62 VcTCP promoters all contain light-responsive (TCT-motif, Sp1, and GT1-motif), MYB-binding and MYC-binding elements. MYB, MYC participates in drought, low temperature, salt, ABA, and GA stress responses (Griffiths et al., 2006), which indicate TCP genes appeared to be regulated by MYB and MYC. For the abiotic stress response elements, a large number of low temperature responsiveness (LTR), anaerobic induction (ARE), and drought induction (MBS) elements also were detected. These results revealed that VcTCP genes play an important role in the response to biotic and abiotic stresses.
Previous studies found that the expansion of gene families is mainly caused by gene duplication events, including WGD, segmental duplication, and tandem duplication, and so on (Maher et al., 2006). Our results showed that except for VcTCP58 from the tandem repeat event, the other members derived from WGD/segment duplication, which demonstrated that the WGD/segment duplication is a predominant type in the expansion of the VcTCP gene family in blueberry.
Analysis of the evolutionary tree of the homologs showed that VcTCP5, VcTCP14, VcTCP18, VcTCP39, VcTCP44, VcTCP50, and VcTCP53, were closely related to AtTCP18 (BRC1). These TCP genes encode proteins that may have the same biological functions in bud development. RT-qPCR experiments found that VcTCP5 and VcTCP18 had high expression during the stage of depth endodormancy. When the blueberries were subjected to low temperature, the expression levels decreased rapidly. During the process of endo-dormancy release, the expression levels were maintained at a relatively low level, indicating that low temperature affected these genes expression, which may participate in the release of endodormancy of flower buds.
The experiments with VcTCP18 transgene study in Arabidopsis suggested that the germination of transgenic seeds was markedly inhibited, however, the phenomenon can be alleviated to some extent by stratification. Earlier studies have indicated that there are similar mechanisms between seed and bud break (Fu et al., 2014; Wang et al., 2016). For example, the key ABA biosynthesis gene NCED3 controls normal ABA accumulation in flower buds, which seem to be key in regulating ABA biosynthesis to induce seed and bud dormancy. CYP707As control ABA inactivation, leading to dormancy release. A recent study found that BRC1 can bound to and positively regulates the transcription of three related genes, HB21, HB40, HB53, which belong to a transcription factor family that encodes a class of homeodomain leucine zipper protein (HD-ZIP). These genes are necessary and sufficient to enhance NCED3 expression (González-Grandío et al., 2017); This finding demonstrated the direct relationship between BRC1 and ABA signaling. Previous studies identified the SVL in poplar, which is similar to the DAM, and its downstream target gene is just TCP18/BRC1, indicating that SVL regulates TCP18/BRC1 and form a temperature-responsive transcription element to participate in dormancy release. low temperature induces the down expression of DAM genes, leading bud and seed dormancy release; however, overexpression of DAM genes can delay bud break. The results of seed germination experiments with transgenic Arabidopsis indicate that VcTCP18 is an important inhibitor in the release of seed dormancy, but the results of our study also proved that the VcTCP18 mediate the dormancy is not a long-term mechanism. If given chilling during the critical period, even the VcTCP18 gene still have a high expression, but could not alter the dormancy release process. Just as hypothesis of the previous (Singh et al., 2018), low temperature downregulates TCP18/BRC1 expression and prevents its binding to FT, leading to the normal functioning of FT. This positive feedback loop promotes bud germination; however, once the plant rhythm gene FT plays its role, it cannot prevent this transformation process. Studies in pears have shown that once endodormancy in the flower bud is lifted, it enters ecological dormancy, although exogenous ABA application cannot reverse this process (Yang et al., 2020).
Previous research has demonstrated that BRC1 physically interacts with FT2, and BRC1-FT interaction triggers growth cessation by antagonizing FT action (Maurya et al., 2020). Some studies also indicated that BRC1 interacted with FT and TSF (TWIN SISTER OF FT) to regulate the activity of florigen in axillary buds and prevented the premature flowering transformation of axillary meristems (Niwa et al., 2013; Li et al., 2019). BRC1 transcription is auxin responsive, and acts downstream of strigolactone in Arabidopsis, BRC1 determines bud activation potential but is dispensable for bud growth inhibition (Aguilar-Martínez et al., 2007). The mutant plants display an increasing amount of the rosette branches, on the contrary, little lateral branches were observed in over-expressed AtTCP18 Arabidopsis plants. The cucumber TCP family gene CsBRC1 directly inhibits the expression of CsPIN3, which promotes auxin accumulation in the axillary buds and inhibits the germination of the axillary buds (Shen et al., 2019). The main branches were observed at 30-day-old seedlings, the VcTCP18 transgenic plants have only one or two main branches, but there were three to four main branches in WT. The results indicated that VcTCP18 can inhibit the outgrowth of axillary buds.
By transgenic experiments, we have good reason to believe that VcTCP18 can negatively regulate the release of flower bud dormancy. In addition to the vegetative growth cycle, woody plants undergo flowering transitions. Flower buds go through dormancy, are released from endodormancy by low temperature accumulation, and eventually germinate. Therefore, the BRC1 transcription factor plays an important role in flowering transition, dormancy release in perennial plants, and its specific functions must be further studied.
The present study firstly identified 62 TCP genes in the blueberry genome and analyzed their phylogeny, multiple sequence alignments, gene structure, conserved motifs, and homologies. Transcriptome data were used to evaluate the tissue specificity of TCP gene family members, flower bud dormancy release, fruit development stage, and hormone treatment (Me-JA). qRT-PCR was employed to examine the expression patterns of TCP gene family members during the process of flower bud dormancy release at low temperature. the seed germination rate of VcTCP18 transgenic Arabidopsis was lower and later than that of wild type. The bud dormancy phenomena as later flowering, fewer rosettes and main branches were also observed in transgenic plants, which indicates that flower bud dormancy release can be negatively regulated by VcTCP18 in blueberry. Overall, our study will deepen our understanding of the function of TCPs in plant growth and development.
Publicly available datasets were analyzed in this study. This data can be found here: https://www.vaccinium.org/bio_data/968095 and https://www.vaccinium.org/node/854703.
YL: conceptualization, software, writing—original draft preparation, writing—review and editing, visualization, and project administration. YZ: methodology. SA: validation, formal analysis, and data curation. QC: investigation. WG: resources. WC: supervision. YL, LZ, and WG: funding acquisition. All authors have read and agreed to the published version of the manuscript.
This study was supported by Zhejiang Province Public Welfare Technology Application Research Project (LGN21C150011) and the Major Scientific and Technological Project of Zhejiang Province (2018C02007).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at Figshare via the doi: 10.6084/m9.figshare.14445723.
Supplementary Figure 1 | The domain integrity of the VcTCP candidate genes was identified using the SMART database.
Supplementary Figure 2 | Phylogenetic analysis of Class II CYC/TB1 TCPs in blueberry and AtTCP1, AtTCP12, and AtTCP18. The phylogenetic tree was constructed using the neighbor-joining method with 1,000 bootstrap replicates in MEGAX.
Supplementary Figure 3 | Analysis of electrophoretic profiles. (A) Bands of VcTCP18 full-length (1,077 bp) cDNA after gel electrophoresis. (B) Semi quantitative RT-PCR assay detected VcTCP18 products of 86 bp on partial T1 Arabidopsis transgenic seedlings.
Supplementary Figure 4 | Expression levels of VcTCP18 in different transgenic plants. (A) Semi quantitative RT-PCR assay, VcGADPH was used as internal reference. Lane 1, marker; lane 2–4, internal reference; lane 5–7, VcTCP18 oe-1, VcTCP18 oe-2, and VcTCP18 oe-3; bands of VcTCP18 full-length (1,077 bp). (B) Relative expression levels of VcTCP18 in three transgenic lines. Values were normalized against the expression data of VcGAPDH and are given as the means ± SD of three biological replicates. The expression levels were calculated based on the 2–ΔΔCt method.
Aguilar-Martínez, J. A., Poza-Carrión, C., and Cubas, P. (2007). Arabidopsis BRANCHED1 acts as an Integrator of branching signals within axillary buds. Plant Cell. 19, 458–472. doi: 10.1105/tpc.106.048934
Bailey, T. L., and Elkan, C. (1994). Fitting a Mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36.
Balsemao-Pires, E., Andrade, L. R., and Sachetto-Martins, G. (2013). Functional study of TCP23 in Arabidopsis thaliana during plant development. Plant Physiol. Biochem. 67, 120–125. doi: 10.1016/j.plaphy.2013.03.009
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, L., Chen, Y. Q., Ding, A. M., Chen, H., Xia, F., Wang, W. F., et al. (2016). Genome-wide analysis of TCP family in tobacco. Genet. Mol. Res. 15:15027728. doi: 10.4238/gmr.15027728
Colle, M., Leisner, C. P., Wai, C. M., Ou, S., Bird, K. A., Wang, J., et al. (2019). Haplotype-phased genome and evolution of phytonutrient pathways of tetraploid blueberry. Gigascience 8, 1–15. doi: 10.1093/gigascience/giz012
Cubas, P., Lauter, N., Doebley, J., and Coen, E. (1999). The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18, 215–222. doi: 10.1046/j.1365-313X.1999.00444.x
Daviere, J.-M., Wild, M., Regnault, T., Baumberger, N., Eisler, H., Genschik, P., et al. (2014). Class I TCP-DELLA Interactions in Inflorescence Shoot Apex Determine Plant Height. Curr. Biol. 24, 1923–1928. doi: 10.1016/j.cub.2014.07.012
Ding, S., Cai, Z., Du, H., and Wang, H. (2019). Genome-Wide Analysis of TCP Family Genes in Zea mays L. Identified a Role for ZmTCP42 in Drought Tolerance. Int. J. Mol. Sci. 20:2762. doi: 10.3390/ijms20112762
Doebley, J., Stec, A., and Hubbard, L. (1997). The evolution of apical dominance in maize. Nature 386, 485–488. doi: 10.1038/386485a0
Ferrero, L. V., Viola, I. L., Ariel, F. D., and Gonzalez, D. H. (2019). Class I TCP Transcription Factors Target the Gibberellin Biosynthesis Gene GA20ox1 and the Growth-Promoting Genes HBI1 and PRE6 during Thermomorphogenic Growth in Arabidopsis. Plant Cell. Physiol. 60, 1633–1645. doi: 10.1093/pcp/pcz137
Fu, X. L., Xiao, W., Wang, D. L., Chen, M., Tan, Q. P., Li, L., et al. (2014). Roles of Endoplasmic Reticulum Stress and Unfolded Protein Response Associated Genes in Seed Stratification and Bud Endodormancy during Chilling Accumulation in Prunus persica. PLoS One 9:e101808. doi: 10.1371/journal.pone.0101808
Giraud, E., Ng, S., Carrie, C., Duncan, O., Low, J., Lee, C. P., et al. (2010). TCP Transcription Factors Link the Regulation of Genes Encoding Mitochondrial Proteins with the Circadian Clock in Arabidopsis thaliana. Plant Cell. 22, 3921–3934. doi: 10.1105/tpc.110.074518
González-Grandío, E., Pajoro, A., Franco-Zorrilla, J. M., Tarancón, C., Immink, R. G. H., and Cubas, P. (2017). Abscisic acid signaling is controlled by a BRANCHED1/HD-ZIP I cascade in Arabidopsis axillary buds. Proc. Natl. Acad. Sci. U. S. A. 114, E245–E254. doi: 10.1073/pnas.1613199114
Griffiths, J., Murase, K., Rieu, I., Zentella, R., Zhang, Z.-L., Powers, S. J., et al. (2006). Genetic Characterization and Functional Analysis of the GID1 Gibberellin Receptors in Arabidopsis. Plant Cell. 18, 3399–3414. doi: 10.1105/tpc.106.047415
Herve, C., Dabos, P., Bardet, C., Jauneau, A., Auriac, M. C., Ramboer, A., et al. (2009). In Vivo Interference with AtTCP20 Function Induces Severe Plant Growth Alterations and Deregulates the Expression of Many Genes Important for Development. Plant Physiol. 149, 1462–1477. doi: 10.1104/pp.108.126136
Hu, B., Jin, J., Guo, A.-Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Jiu, S., Xu, Y., Wang, J., Wang, L., Wang, S., Ma, C., et al. (2019). Genome-Wide Identification, Characterization, and Transcript Analysis of the TCP Transcription Factors in Vitis vinifera. Front. Genet. 10:1276. doi: 10.3389/fgene.2019.01276
Kim, S. H., Son, G. H., Bhattacharjee, S., Kim, H. J., Nam, J. C., Nguyen, P. D. T., et al. (2014). The Arabidopsis immune adaptor SRFR1 interacts with TCP transcription factors that redundantly contribute to effector-triggered immunity. Plant J. 78, 978–989. doi: 10.1111/tpj.12527
Kosugi, S., and Ohashi, Y. (2002). DNA binding and dimerization specificity and potential targets for the TCP protein family. Cell. Mol. Biol. 30, 337–348. doi: 10.1046/j.1365-313X.2002.01294.x
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Leng, X., Wei, H., Xu, X., Ghuge, S. A., Jia, D., Liu, G., et al. (2019). Genome-wide identification and transcript analysis of TCP transcription factors in grapevine. BMC Genom. 20:786. doi: 10.1186/s12864-019-6159-2
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucl. Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, D., Zhang, H., Mou, M., Chen, Y., Xiang, S., Chen, L., et al. (2019). Arabidopsis Class II TCP Transcription Factors Integrate with the FT–FD Module to Control Flowering. Plant Physiol. 181, 97–111. doi: 10.1104/pp.19.00252
Li, W., Li, D.-D., Han, L.-H., Tao, M., Hu, Q.-Q., Wu, W.-Y., et al. (2017). Genome-wide identification and characterization of TCP transcription factor genes in upland cotton (Gossypium hirsutum). Sci. Rep. 7:10118. doi: 10.1038/s41598-017-10609-2
Liu, H.-L., Wu, M., Li, F., Gao, Y.-M., Chen, F., and Xiang, Y. (2018). TCP Transcription Factors in Moso Bamboo (Phyllostachys edulis): genome-Wide Identification and Expression Analysis. Front. Plant Sci. 9:1263. doi: 10.3389/fpls.2018.01263
Luo, D., Carpenter, R., Vincent, C., Copsey, L., and Coen, E. (1996). Origin of floral asymmetry in Antirrhinum. Nature 383, 794–799. doi: 10.1038/383794a0
Ma, J., Wang, Q., Sun, R., Xie, F., Jones, D. C., and Zhang, B. (2014). Genome-wide identification and expression analysis of TCP transcription factors in Gossypium raimondii. Sci. Rep. 4:6645. doi: 10.1038/srep06645
Maher, C., Stein, L., and Ware, D. (2006). Evolution of Arabidopsis microRNA families through duplication events. Genome Res. 16, 510–519. doi: 10.1101/gr.4680506
Martin-Trillo, M., and Cubas, P. (2010). TCP genes: a family snapshot ten years later. Trends Plant Sci. 15, 31–39. doi: 10.1016/j.tplants.2009.11.003
Maurya, J. P., Singh, R. K., Miskolczi, P. C., Prasad, A. N., Jonsson, K., Wu, F., et al. (2020). Branching Regulator BRC1 Mediates Photoperiodic Control of Seasonal Growth in Hybrid Aspen. Curr. Biol. 30, 122–126.e2. doi: 10.1016/j.cub.2019.11.001
Navaud, O., Dabos, P., Carnus, E., Tremousaygue, D., and Herve, C. (2007). TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 65, 23–33. doi: 10.1007/s00239-006-0174-z
Nicolas, M., and Cubas, P. (2016). TCP factors: new kids on the signaling block. Curr. Opinion Plant Biol. 33, 33–41. doi: 10.1016/j.pbi.2016.05.006
Niwa, M., Daimon, Y., Kurotani, K., Higo, A., Pruneda-Paz, J. L., Breton, G., et al. (2013). BRANCHED1 Interacts with FLOWERING LOCUS T to Repress the Floral Transition of the Axillary Meristems in Arabidopsis. Plant Cell. 25, 1228–1242. doi: 10.1105/tpc.112.109090
Palatnik, J. F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J. C., et al. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. doi: 10.1038/nature01958
Parapunova, V., Busscher, M., Busscher-Lange, J., Lammers, M., Karlova, R., Bovy, A. G., et al. (2014). Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol. 14:157. doi: 10.1186/1471-2229-14-157
Resentini, F., Felipo-Benavent, A., Colombo, L., Blázquez, M. A., Alabadí, D., and Masiero, S. (2015). TCP14 and TCP15 Mediate the Promotion of Seed Germination by Gibberellins in Arabidopsis thaliana. Mol. Plant 8, 482–485. doi: 10.1016/j.molp.2014.11.018
Shen, J., Zhang, Y., Ge, D., Wang, Z., Song, W., Gu, R., et al. (2019). CsBRC1 inhibits axillary bud outgrowth by directly repressing the auxin efflux carrier CsPIN3 in cucumber. Proc. Natl. Acad. Sci. U. S. A. 116, 17105–17114. doi: 10.1073/pnas.1907968116
Singh, R. K., Maurya, J. P., Azeez, A., Miskolczi, P., Tylewicz, S., Stojkoviè, K., et al. (2018). A genetic network mediating the control of bud break in hybrid aspen. Nat. Commun. 9:4173. doi: 10.1038/s41467-018-06696-y
Takeda, T., Amano, K., Ohto, M., Nakamura, K., Sato, S., Kato, T., et al. (2006). RNA Interference of the Arabidopsis Putative Transcription Factor TCP16 Gene Results in Abortion of Early Pollen Development. Plant Mol. Biol. 61, 165–177. doi: 10.1007/s11103-006-6265-9
Tang, H., Wang, X., Bowers, J. E., Ming, R., Alam, M., and Paterson, A. H. (2008). Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 18, 1944–1954. doi: 10.1101/gr.080978.108
Viola, I. L., Camoirano, A., and Gonzalez, D. H. (2016). Redox-Dependent Modulation of Anthocyanin Biosynthesis by the TCP Transcription Factor TCP15 during Exposure to High Light Intensity Conditions in Arabidopsis. Plant Physiol. 170, 74–85. doi: 10.1104/pp.15.01016
Wang, D., Gao, Z., Du, P., Xiao, W., Tan, Q., Chen, X., et al. (2016). Expression of ABA Metabolism-Related Genes Suggests Similarities and Differences Between Seed Dormancy and Bud Dormancy of Peach (Prunus persica). Front. Plant Sci. 6:1248. doi: 10.3389/fpls.2015.01248 eCollection 2015
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Wei, W., Hu, Y., Cui, M.-Y., Han, Y.-T., Gao, K., and Feng, J.-Y. (2016). Identification and Transcript Analysis of the TCP Transcription Factors in the Diploid Woodland Strawberry Fragaria vesca. Front. Plant Sci. 7:1937. doi: 10.3389/fpls.2016.01937
Xu, R., Sun, P., Jia, F., Lu, L., Li, Y., Zhang, S., et al. (2014). Genomewide analysis of TCP transcription factor gene family in Malus domestica. J. Genet. 93, 733–746. doi: 10.1007/s12041-014-0446-0
Xue, M., Long, Y., Zhao, Z., Huang, G., Huang, K., Zhang, T., et al. (2018). Isolation and Characterization of a Green-Tissue Promoter from Common Wild Rice (Oryza rufipogon Griff.). Int. J. Mol. Sci. 19:2009. doi: 10.3390/ijms19072009
Yang, L., Chen, M., Cai, K., Zhang, L., Zhu, Y., Ye, Q., et al. (2018). VcFAS, VcSUN and VcOVATE orchestrated the fruit morphogenesis in southern highbush blueberry during the pre-anthesis and fruit development. Sci. Horticul. 240, 109–115. doi: 10.1016/j.scienta.2018.05.023
Yang, Q., Yang, B., Li, J., Wang, Y., Tao, R., Yang, F., et al. (2020). ABA-responsive ABRE-BINDING FACTOR3 activates DAM3 expression to promote bud dormancy in Asian pear. Plant Cell. Environ. 43, 1360–1375. doi: 10.1111/pce.13744
Yooyongwech, S., Sugaya, S., Sekozawa, Y., and Gemma, H. (2009). Differential adaptation of high- and low-chill dormant peaches in winter through aquaporin gene expression and soluble sugar content. Plant Cell. Rep. 28, 1709–1715. doi: 10.1007/s00299-009-0770-7
Zhao, J., Zhai, Z., Li, Y., Geng, S., Song, G., Guan, J., et al. (2018). Genome-Wide Identification and Expression Profiling of the TCP Family Genes in Spike and Grain Development of Wheat (Triticum aestivum L.). Front. Plant Sci. 9:1282. doi: 10.3389/fpls.2018.01282
Zhou, Q. Y., Han, Y. H., Zhu, Y., Chen, S., Li, X. W., Peng, B., et al. (2019). Genome-Wide identification, classification and expression analysis of TCP gene family in tea plant. Acta Horti. Sin. 46, 2021–2036.
Keywords: blueberry, TCP transcription factors, expression profiles analysis, flower bud dormancy, transgenic plants
Citation: Li Y, An S, Cheng Q, Zong Y, Chen W, Guo W and Zhang L (2021) Analysis of Evolution, Expression and Genetic Transformation of TCP Transcription Factors in Blueberry Reveal That VcTCP18 Negatively Regulates the Release of Flower Bud Dormancy. Front. Plant Sci. 12:697609. doi: 10.3389/fpls.2021.697609
Received: 19 April 2021; Accepted: 15 June 2021;
Published: 09 July 2021.
Edited by:
Songling Bai, Zhejiang University, ChinaReviewed by:
Michael Nicolas, Centro Nacional de Biotecnología, Consejo Superior de Investigaciones Científicas (CSIC), SpainCopyright © 2021 Li, An, Cheng, Zong, Chen, Guo and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lu Zhang, emhhbmdsdTg1NkBtYWlsLmp4YXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.