
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 14 June 2021
Sec. Plant Pathogen Interactions
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.688003
This article is part of the Research TopicThe Role of Epigenetics in Biotic Stress in PlantsView all 7 articles
Epigenetic marks such as covalent histone modification and DNA methylation are crucial for mitotically and meiotically inherited cellular memory-based plant immunity. However, the roles of individual players in the epigenetic regulation of plant immunity are not fully understood. Here we reveal the functions of two Arabidopsis thaliana homologs of human lysine-specific demethylase1-like1, LDL1 and LDL2, in the maintenance of methyl groups at lysine 4 of histone H3 and in plant immunity to Pseudomonas syringae infection. The growth of virulent P. syringae strains was reduced in ldl1 and ldl2 single mutants compared to wild-type plants. Local and systemic disease resistance responses, which coincided with the rapid, robust transcription of defense-related genes, were more stably expressed in ldl1 ldl2 double mutants than in the single mutants. At the nucleosome level, mono-methylated histone H3K4 accumulated in ldl1 ldl2 plants genome-wide and in the mainly promoter regions of the defense-related genes examined in this study. Furthermore, in silico comparative analysis of RNA-sequencing and chromatin immunoprecipitation data suggested that several WRKY transcription factors, e.g., WRKY22/40/70, might be partly responsible for the enhanced immunity of ldl1 ldl2. These findings suggest that LDL1 and LDL2 control the transcriptional sensitivity of a group of defense-related genes to establish a primed defense response in Arabidopsis.
Plants have evolved various disease resistance responses and the plasticity of these responses helps ensure plant survival in the face of biotic stress. Plant immunity is divided into two categories based on the types of immune receptors employed (Jones and Dangl, 2006; Han and Jung, 2013). Microbe/pathogen-associated molecular pattern (MAMP/PAMP, hereafter MAMP)-triggered immunity (PTI) is initiated by pattern recognition receptors (PRRs) located on the plasma membrane that recognize MAMPs. Effector-triggered immunity (ETI), which is controlled by intracellular Nucleotide-binding Oligomerization Domain (NOD)-like receptors (NLRs), also known as plant disease resistance (R) proteins, is accompanied by a hypersensitive response (HR) (Chisholm et al., 2006; Jones and Dangl, 2006). Additionally, since PTI and ETI are associated with each other, the former can intensify the latter and vice versa (Qi et al., 2011; Jung et al., 2020; Ngou et al., 2021; Yuan et al., 2021). Local disease resistance governed by PRRs and NLRs also induces systemic acquired resistance (SAR), which effectively restrains the colonization of pathogens throughout most of the plant in the face of subsequent pathogen infection (Durrant and Dong, 2004; Fu and Dong, 2013). These different immune responses require specialized signaling networks and share conserved defense-related signaling and responses, including rapid, robust transcriptional changes in local and/or systemic tissues during infection (Tsuda et al., 2009; Tsuda and Katagiri, 2010; Gruner et al., 2013; Tsuda and Somssich, 2015).
Immune signaling can also induce cellular memory, which helps prime the plant to respond to future infections. Cellular memory is inherited both mitotically and meiotically from parental cells to daughter cells and leads to phenotypic variation in plants by regulating gene expression (Saze, 2008; Holeski et al., 2012). Differential gene expression, which leads to epigenetic variation in eukaryotes, is controlled by DNA methylation and several covalent modifications on the N-terminal tails of histone proteins such as acetylation, methylation, ubiquitination, phosphorylation, ADP-ribosylation, and sumoylation (Pikaard and Mittelsten Scheid, 2014; Ramirez-Prado et al., 2018). These chromatin modifications affect the transcriptional state of genes associated with specific changes.
‘Defense priming’ (such as SAR) is an adaptive strategy that fosters a faster and stronger defense response against subsequent challenge (Conrath et al., 2006, 2015; Beckers et al., 2009; Jung et al., 2009; Conrath, 2011). The immunization of the local leaves of plants with SAR-inducing stimuli encourages chromatin remodeling in distal systemic leaves (Jaskiewicz et al., 2011; Luna et al., 2012; Ramirez-Prado et al., 2018). For example, histone H3 and H4 lysine methylation (especially di- and tri-methylation) and acetylation on the promoter regions of WRKY6/29/53 were induced in distal leaves after local infection with Pseudomonas syringae pv. maculicola. These modifications act as histone memory to help plants adapt to subsequent stresses. Hence, these genes are rapidly transcribed in the distal leaves of plants exposed to a second challenge (Jaskiewicz et al., 2011).
Induced resistance is epigenetically heritable in plants: disease-exposed plants can produce progeny that are primed for pathogen infection. In fact, the increased expression of salicylic acid (SA)-induced defense genes, such as PATHOGENESIS-RELATED PROTEIN1 (PR1) and WRKY6/53/70, is correlated with specific histone modifications (such as H3K6ac and H3K27me) at the promoter regions of these genes in progeny from pathogen-infected plants, suggesting that plants memorize changes in histone marks in a transgenerational manner (Luna et al., 2012). Furthermore, open chromatin regions were identified in systemic leaves of Arabidopsis following challenge infection and were used to isolate SAR regulators (Baum et al., 2019). Repetitive exposure to environmental stresses also induces changes to histone modifications, which confer resistance/tolerance to biotic and abiotic stress in plants (Singh P. et al., 2014; Brzezinka et al., 2016; Raxwal et al., 2020). Moreover, the failure to add specific histone modifications perturbs resistance to (hemi)biotrophs and necrotrophs (reviewed in Ding and Wang, 2015), indicating that the covalent modification of histone proteins is crucial for plant immunity.
Histone methylation and demethylation primarily occur at specific lysine (Lys, K) residues on histone H3 and H4 proteins (H3K4, H3K9, H3K27, H3K36, H3K79, and H4K20) and affect the transcription of target genes (Hyun et al., 2017). In animal cells, human lysine-specific histone demethylase1 (LSD1), also known as lysine-specific demethylase 1A (KDM1A), removes mono- and di-methyl groups from the lysine residues of histone H3, specifically H3K4 and H3K9, and participates in various biological processes, e.g., cell proliferation and tumor development (Shi et al., 2004; Chen et al., 2012). Therefore, human LSD1 is a proposed target for therapeutic purposes (Ellis and Loda, 2018; Tu et al., 2020). In plants, methyl groups of H3K4 are erased by Jumonji C domain-containing demethylases and plant LSD1-like (LDL) proteins (Spedaletti et al., 2008; Martignago et al., 2019).
The Arabidopsis thaliana genome contains four LDL genes [At1g62830 (LDL1), At3g13682 (LDL2), At4g16310 (LDL3), and At3g10390 (LDL4)], which are engaged in flowering time control, circadian clock regulation, homologous recombination repair, hormone responses, and systemic resistance (Jiang et al., 2007; Krichevsky et al., 2009; Singh et al., 2012, 2013; Shafiq et al., 2014; Zhao et al., 2015; Hung et al., 2018, 2019; Hirakawa et al., 2019; Martignago et al., 2019). For example, Arabidopsis LDL1/SWIRM domain PAO PROTEIN1 (LDL1/SWP1) represses the expression of FLOWERING LOCUS C (FLC), encoding a floral repressor, and LATERAL ROOT PRIMODIUM1, encoding a transcriptional activator, to promote auxin homeostasis-regulated gene expression in root primordia during early root development. Accordingly, the recessive ldl1 mutants exhibit late flowering and enhanced root elongation and lateral root formation (Jiang et al., 2007; Singh et al., 2012, 2020; Shafiq et al., 2014). LDL1 also regulates seed dormancy by controlling the expression of DELAY OF GERMINATION1, encoding a regulator of primary dormancy and the abscisic acid (ABA) signaling pathway (Zhao et al., 2015). LDL1 also controls the dissociation of RAD54 from damaged DNA sites by recognizing H3K4me2 during homologous recombination repair and maintains gene stability and integrity (Hirakawa et al., 2019). LDL2 regulates primary seed dormancy and the circadian clock in cooperation with LDL1 (Zhao et al., 2015; Hung et al., 2018, 2019). Interestingly, LDL4/FLOWERING LOCUS D/REDUCED SYSTEMIC IMMUNITY1 (LDL4/FLD/RSI1) positively regulates systemic resistance against Pseudomonas infection, and ldl4/fld/rsi1 mutants show hyper-susceptibility to necrotrophic fungal infection in local tissues (Singh et al., 2013, 2019; Singh P. et al., 2014). These findings suggest that other Arabidopsis LDL proteins also participate in plant immunity.
Here we uncovered the roles of Arabidopsis LDL1 and LDL2 in plant immune responses against Pseudomonas infection. Loss-of-function mutations in LDL1 and LDL2, which act in a partially redundant manner, led to local and systemic disease resistance to phytopathogenic bacterial infection, along with increased expression of defense-related genes involved in SA- and MAMP-dependent signaling following Pseudomonas infection. In silico comparative analysis of RNA-sequencing (RNA-seq) and previously published chromatin immunoprecipitation (ChIP)-seq data identified 39 differentially expressed genes (DEGs) whose nucleosome modifications might be controlled by LDL1 and LDL2. Monomethylated H3K4 (H3K4me1) strongly accumulated in several defense-related genes in ldl1 ldl2 double mutants compared with wild-type (WT) plants. Our findings suggest that LDL1 and LDL2 are internal targets that establish primed defense responses in Arabidopsis.
The Arabidopsis thaliana mutants used in this study are as follows: ldl1-1 (ldl1, Salk_048276), ldl1-2 (Salk_034869), ldl1-3 (Salk_108984), ldl2-1 (ldl2, Salk_135831), ldl2-2 (Salk_138820), ldl3 (SALK_146733), and ldl4/fld (Salk_015053). All mutants are in the Columbia-0 (Col-0) background. The ldl1 ldl2 double mutant was generated by crossing ldl1-1 to ldl2-1. Arabidopsis plants were grown in soil (Nongwoo Bio) under neutral day conditions (12 h light/12 h dark cycles, relative humidity 60–70%, 120 μmol m–2s–1, 22 ± 1°C) in a walk-in growth chamber (Jung et al., 2020).
For the root growth assay, 5-day-old Arabidopsis seedlings grown on half-strength Murashige and Skoog (MS) medium were transferred to MS medium supplemented with 25 and 50 μM salicylic acid (SA, Sigma-Aldrich). The seedlings were grown in medium under neutral day conditions for 2 weeks, and root lengths were measured using the Fiji program (Schneider et al., 2012).
Pseudomonas syringae pv. maculicola ES4326 (PsmES4326) [newly classified as P. cannabina pv. alisalensis (Bull et al., 2010)] and P. syringae pv. tomato DC3000 (PstDC3000) were used as virulent Pseudomonas strains. The avirulent derivatives of P. syringae were PsmES4326 carrying AvrRpt2 (PsmES4326/AvrRpt2, DG6) and PsmES4326 carrying AvrRpm1 (PsmEs4326/AvrRpm1, DG34) (Guttman and Greenberg, 2001). As virulence-deficient and attenuated strains of P. syringae, we employed PstDC3000 hrcC– and PstDC3000△AvrPto△AvrPtoB, respectively (Roine et al., 1997; Guttman et al., 2002; Lin and Martin, 2005) (Supplementary Table 1).
Bacterial strains were freshly prepared in King’s B medium supplemented with the appropriate antibiotics (Supplementary Table 1) and diluted to various concentrations in 10 mM MgSO4 as follows: OD600 = 0.01 for immunization with an avirulent pathogen (PsmES4326/AvrRpt2, DG6) and pathophysiological studies (PsmES4326 and PstDC3000); OD600 = 0.0001 to test disease responses (all strains). The strains were inoculated on three fully expanded leaves of 4-week-old Arabidopsis plants to evaluate pathophysiological responses and to count bacterial growth in leaf discs. The number of bacteria in infected leaves was determined 3 d after inoculation using a typical serial dilution method, and the infected leaves were photographed on the same day.
Leaves of 4-week-old Arabidopsis (∼0.1 g) plants were harvested at the indicated time points after Pseudomonas inoculation. Total RNA was extracted from the samples using TRIzol reagent (Thermo Fisher Scientific), and contaminating genomic DNA was removed using TURBO DNase (Ambion). First-strand cDNA was synthesized from 5 μg of total RNA using SuperScript-II Reverse Transcriptase (Thermo Fisher Scientific) according to the manufacturer’s instructions. Quantitative PCR (qPCR) was performed using SYBR Premix Ex Taq (TaKaRa Bio) and a CFX384 Real-time PCR Detection System (Bio-Rad). The cycling conditions were 95°C for 10 min, and 50 cycles of 95°C for 5 s, 60°C for 10 s, and 72°C for 35 s. ACTIN2 (At3g18780) was used as the reference gene to normalize transcript levels. Relative expression levels were analyzed using the comparative cycle threshold (ΔΔCt) method (Livak and Schmittgen, 2001) and are shown as mean ± SD (standard deviation). All experiments were performed with at least three biological replicates with two or three technical repeats unless otherwise noted. Asterisks indicate statistically significant differences from WT plants (∗p < 0.05, ∗∗p < 0.01, two-tailed Student’s t-test). The oligonucleotide sequences of the primers are shown in Supplementary Table 2.
Total RNA was isolated from the leaves of 3-week-old WT (Col-0) and ldl1 ldl2 plants grown under neutral day conditions, using TRIzol reagent. Precipitated and dissolved total RNA was cleaned using an RNeasy spin column (Qiagen). After confirming the purity of the RNA (Bioanalyzer, Agilent), the total RNA was used to construct an mRNA sequencing library using a TruSeq Stranded mRNA Sample Preparation Kit according to the manufacturer’s instructions (Illumina). Each library (from two biological replicates per genotype) was subjected to 100 bp paired-end sequencing on the HiSeq 2000 platform (Illumina), as described previously (Jung et al., 2020). The RNA-seq data were processed with TopHat2 and Bowtie2 (Trapnell et al., 2009; Langmead and Salzberg, 2012). To quantify the total transcript mass in fragments per kilobase of transcript per million mapped reads (FPKM), data from biological replicates of WT and ldl1 ldl2 plants were separately aligned with the A. thaliana TAIR10 gene model using the Cufflinks package (Trapnell et al., 2010). RNA-seq data have been deposited in the National Agricultural Biotechnology Information Center (NABIC)1 under accession numbers NN-1560, NN-1561, NN-1578, and NN-1580 and at the GEO datasets in National Center for Biotechnology Information (NCBI) (GSE171433).
Total proteins were extracted from 4-week-old Arabidopsis leaves using protein extraction buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.1% [v/v] Triton X-100, 0.1% [w/v] SDS, 5 mM DTT and proteinase inhibitors [Pierce Protease inhibitor, Thermo Fisher Scientific]) (Jung et al., 2020). SDS-polyacrylamide gel electrophoresis using Tris-glycine electrophoresis buffer and immunoblot analysis were carried out as described (Green and Sambrook, 2012). The antibodies used in this study are as follows: α-BAK1 (AS12 1858, Agrisera), α-H3K4me1 (PA5-17418, Invitrogen), α-H3K4me2 (701764, Invitrogen), α-H3K4me3 (PA5-17420, Invitrogen), and α-H3 (AS10 710, Agrisera) antibodies. The signal was visualized with SuperSignal Chemiluminescent Substrate (Thermo Fisher Scientific).
Free SA levels in infected leaves were measured using a high-performance liquid chromatograph coupled with a fluorescence detector (Agilent 1100) as described previously (Seskar et al., 1998; Jung et al., 2009).
Four-week-old Arabidopsis plants grown under neutral day conditions were inoculated with 10 mM MgSO4 and PstDC3000 (OD600 = 0.01), and 10 infected leaves were detached at 24 h post-inoculation (hpi). The leaf tissues were submerged in destaining solution (acetic acid:ethanol = 1:3) overnight. After rinsing the cleared leaves with 150 mM K2HPO4 for 30 min, the leaves were incubated in 0.01% aniline blue solution in 150 mM K2HPO4 for 2 h (Schenk et al., 2014). Deposited callose in infected leaves was detected by confocal microscopy (LSM 700 laser scanning confocal microscope, Carl-Zeiss), and the callose deposits were quantified using the Fiji program (Schneider et al., 2012).
Leaf tissues were collected from 3-week-old WT and ldl1 ldl2 plants grown in a walk-in growth chamber and used to extract nuclei, as previously described (Jaskiewicz et al., 2011; Jung et al., 2020). Extracted nuclei were homogenized in nuclei lysis buffer [50 mM Tris-HCl (pH 8.0), 10 mM EDTA, 1% (w/v) SDS, and proteinase inhibitors (Roche)], and the resulting chromatin was sheared by sonication to obtain fragment sizes of 200–800 bp (Bioruptor, Diagenode). ChIP was performed using α-H3K4me1, α-H3K4me2, and α-H3K4me3 antibodies (Invitrogen) according to the manufacturer’s instructions (Pierce Agarose ChIP Kit, Thermo Fisher Scientific). The enrichment of modified histone proteins in the genes of interest was determined by qPCR (Jung et al., 2020) and calculated by the percent input method (Lin et al., 2012). The primers used in this study are listed in Supplementary Table 2.
To explore whether Arabidopsis LDL genes are involved in plant immune responses, we examined the mRNA expression pattern of LDLs upon bacterial pathogen infection using the visualized meta-analysis database eFP2 (Winter et al., 2007). Leaves of 4-week-old Arabidopsis WT plants were inoculated with 10 mM MgSO4 (mock, M), an avirulent pathogen (PsmES4326/AvrRpt2, A), or a virulent pathogen (PsmES4326, V). The transcript levels of the LDLs were examined using ATH1 GeneChip at the indicated hours after inoculation and normalized by ACTIN2 (Supplementary Figure 1A). Mock (10 mM MgSO4) treatment did not affect the expression of any of the four LDL homologs: LDL1, LDL2, LDL3, and LDL4/FLD. Both avirulent and virulent bacterial infection had little effect on the expression of LDL1 and LDL4/FLD. Infection with the avirulent strain PsmES4326/AvrRpt2 induced LDL2 expression and reduced LDL3 expression. However, since these differences in expression were within 1.5-fold and 0.5-fold, these findings suggest that the expression of LDLs in local leaves is not altered by bacterial infection.
We also examined the expression of the LDLs in the distal systemic leaves of plants immunized with the SAR-inducing Pseudomonas strain PsmES4326/AvrRpt2 (DG6) (Supplementary Figure 1B). At 2 days after local infection (F) with the avirulent strain PsmES4326/AvrRpt2, we inoculated distal leaves (S) with the virulent PsmES4326 strain and examined the expression of the LDLs by qRT-PCR (Supplementary Figure 1C). The LDL genes in systemic leaves were not significantly up- or down-regulated compared to non-infected leaves, as their changes in expression were within 0.5-fold. However, at 10 h after challenge-inoculation, LDL1 and LDL3 expression significantly decreased in systemic leaves compared to the control.
Next, we tested the disease resistance responses of ldl mutants against PsmES4326 infection. The ldl4/fld/rsi1 mutants are defective in systemic resistance, but they still show WT-like local susceptibility to Pseudomonas infection (Singh et al., 2013). Under the experimental conditions we used to identify mutants exhibiting enhanced disease resistance or susceptibility against Pseudomonas infection, the titers of the virulent PsmES4326 strain decreased in ldl1 and ldl2, but not in ldl3 or ldl4/fld, compared to WT plants (Supplementary Figure 1D). To confirm the enhanced disease resistance of the ldl1 and ldl2 mutants, we inoculated different mutant alleles with PsmES4326 and PstDC3000. The mutants were resistant to both of these virulent Pseudomonas strains (Supplementary Figure 1E). These results indicate that both LDL1 and LDL2 suppress the immune response against Pseudomonas infection.
To exclude the possibility of functional redundancy between LDL1 and LDL2, as these proteins share 53% identity and 68% similarity, we crossed ldl1-1 with ldl2-1 and tested the disease resistance of the ldl1 ldl2 double mutants against infection with PsmES4326 and PstDC3000 (Figures 1A,B). The ldl1 and ldl2 single mutants exhibited disease resistance 6 and 4 times out of 7 individual infections with PsmES4326, respectively. The disease resistance response was more stable and firmly established in the ldl1 ldl2 mutants (7 out of 7 replicates) than the single mutants, even though the extent of resistance observed in ldl1 ldl2 was not significantly different from that of the single mutants (Figure 1A). We repeated the bacterial growth test in the ldl1 ldl2 mutants after PstDC3000 infection eight times. Like the stable resistance response against PsmES4326, the double mutants were resistant to PstDC3000 infection in all eight independent experiments compared to the WT (Figure 1B).
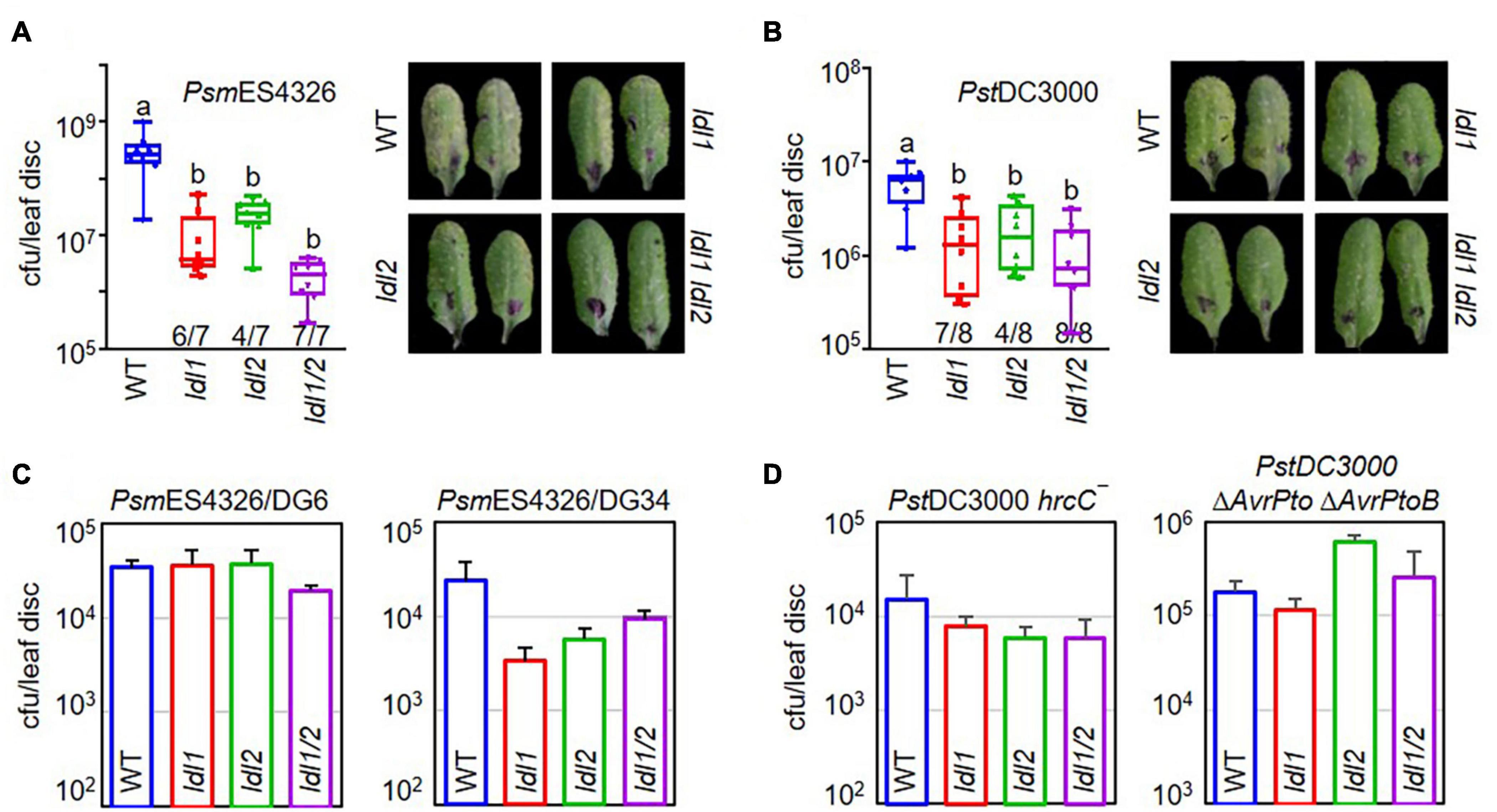
Figure 1. The hyper-resistance response of ldl1 ldl2 double mutants to virulent Pseudomonas infection is more stable than that of the single mutants. (A,B) The growth of P. syringae pv. maculicola ES4326 (PsmES4326, OD600 = 0.0001) (A) and P. syringae pv. tomato DC3000 (PstDC3000, OD600 = 0.0001) (B) in WT, ldl1, ldl2, and ldl1 ldl2 plants at 3 days post inoculation (dpi). The denominators and numerators under the box plots indicate the number of total repeats and the number of trials showing differences from WT, respectively. Box plots show the minimum, first quartile, median, third quartile, and maximum values. The right panels show disease symptoms of each genotype caused by P. syringae infection. (C,D) WT-like disease responses of single and double mutants against infection with different avirulent derivatives of PsmES4326 carrying AvrRpt2 (PsmES4326/DG6, OD600 = 0.0001) and AvrRpm1 (PsmES4326/DG34, OD600 = 0.0001) (C) and attenuated mutants of PstDC3000 hrcC– (OD600 = 0.001) and PstDC3000 ΔAvrPto ΔAvrPtoB (OD600 = 0.001) (D). The area of a leaf disc is 0.78 cm2. Bar indicates the average ± standard error (SEM) (p < 0.05, ANOVA-Tukey, n = 8). The experiments were repeated 3, 6, and 3 times for the avirulent derivatives (PsmES4326/DG6 and PsmES4326/DG34), PstDC3000 hrcC–, and PstDC3000 ΔAvrPto ΔAvrPtoB, respectively, with similar results.
The multiplication of avirulent pathogens can be restrained via an ETI-mediated pathway involving R proteins and various essential components, such as NDR1 (NON-RACE SPECIFIC DISEASE RESISTANCE1) and EDS1 (ENHANCED DISEASE SUSCEPTIBILITY1) (Parker et al., 1996; Century et al., 1997; Aarts et al., 1998; Falk et al., 1999). Therefore, we evaluated the resistance response of the ldl1 ldl2 mutants to avirulent pathogen infection to examine whether LDL1 and LDL2 are engaged in ETI (Figures 1C,D). PsmES4326/AvrRpt2 (DG6) and PsmES4326/AvrRpm1 (DG34) were inoculated into the leaves of WT, ldl1, ldl2, and ldl1 ldl2 plants. All plants with mutations in LDL1 or LDL2 showed comparable (similar) disease susceptibility to WT plants, suggesting that LDL1 and LDL2 are not involved in RPS2- and RPM1-mediated ETI in Arabidopsis (Figure 1C).
Phytopathogenic P. syringae uses the Type III secretion system (T3SS) to deliver effector proteins into plant cells to manipulate and/or inhibit host proteins (Collmer et al., 2002; Lindeberg et al., 2009, 2012). Thus, attenuated derivatives of P. syringae with defects in the T3SS machinery or effectors fail to colonize plants, since these strains cannot overcome the basal immune response initiated by the PRR extracellular immune receptors (Deng et al., 2017). Both the ldl single and double mutants showed WT-like resistance against infection with two attenuated P. syringae strains, PstDC3000 hrcC– and PstDC3000 ΔAvrPto ΔAvrPtoB (Figure 1D). These results demonstrate that individual mutations of LDL1 and LDL2 confer resistance to infection with virulent Pseudomonas and that each gene can compensate for the other.
SA accumulation upon bacterial infection occurs as part of the plant immune response to (hemi)biotrophic microbes. To investigate whether the resistance responses of the ldl mutants function via SA-dependent immunity, we measured free SA levels in the leaves of WT and mutant plants infected with PsmES4326. Following pathogen infection, free SA accumulated in infected leaves regardless of plant genotype, and these levels did not significantly differ in the different genotypes (Figure 2A).
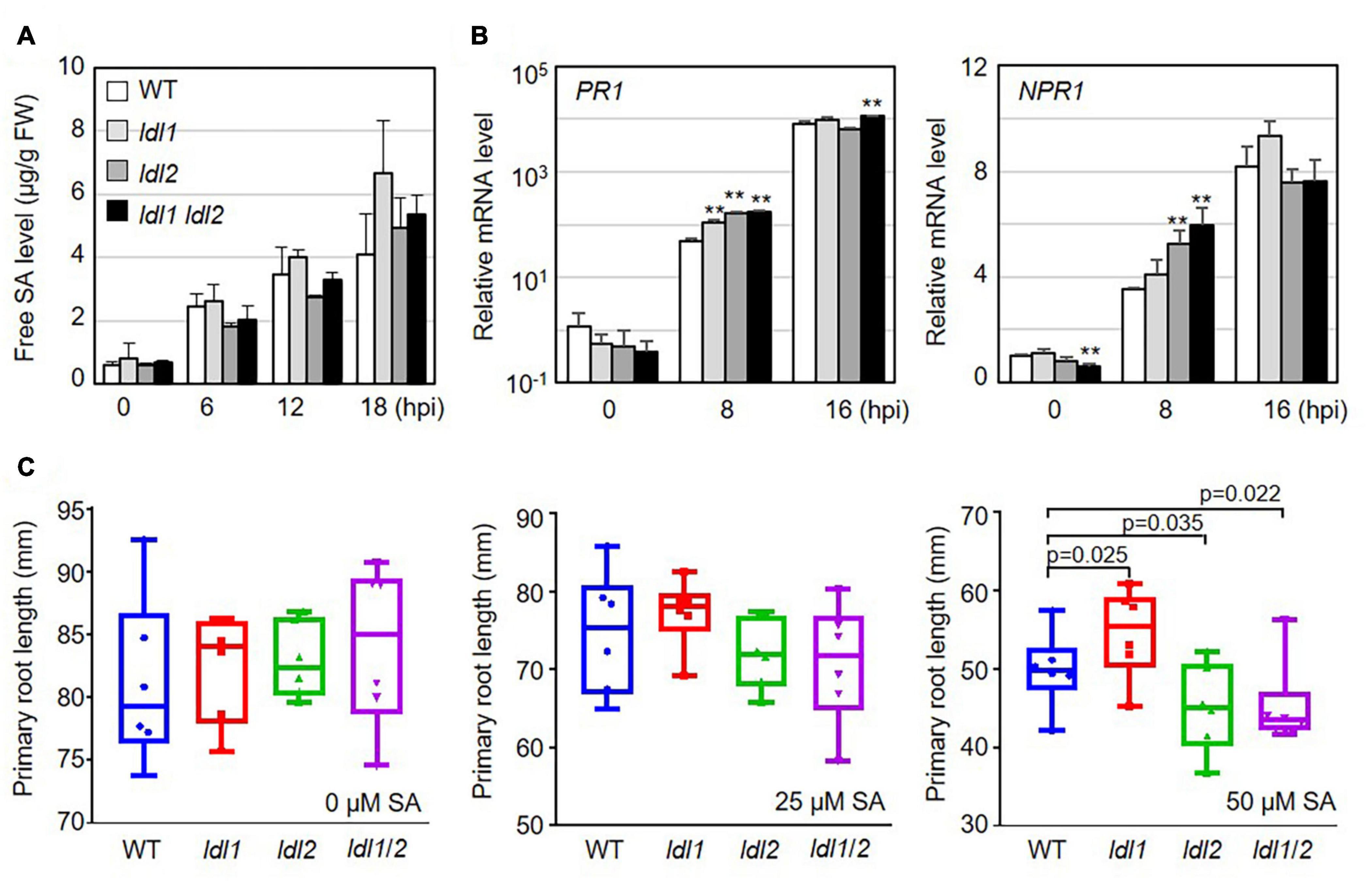
Figure 2. Transcription of salicylic acid (SA)-dependent signaling genes is more rapidly induced in ldl mutants than in WT plants after Pseudomonas infection. (A) Free SA levels in the leaves of Arabidopsis plants infected with the virulent PsmES4326 strain (OD600 = 0.01). The experiments were repeated three times with similar results. (B) Rapid expression of SA-responsive genes in PsmES4326 (OD600 = 0.01)-infected leaves of ldl1, ldl2, and ldl1 ldl2 plants. The averages ± SD (standard deviation) were plotted (**p < 0.01, two-tailed Student’s t-test, n = 3). Five biologically independent experiments with 3 technical repeats were performed, with similar trends, as shown in Supplementary Figure 2. (C) Primary root growth of WT, ldl1, ldl2, and ldl1 ldl2 plants grown in half-strength MS medium supplemented with 25 and 50 μM SA for 14 days. Box plots show the minimum, first quartile, median, third quartile, and maximum values. Each dot represents the average of independent biological replicates (n = 6), with fives samples per individual replicate (p-Values from two-tailed Student’s t-test, n = 6).
We then measured the transcript levels of genes essential for SA-dependent immunity, including the following: PR1 (SA responsiveness), NON-EXPRESSOR OF PR1 (NPR1, SA perception/responsiveness), AGD2-LIKE DEFENSE1 (ALD1, SA regulation), and WRKY70 (encoding an activator of SA-dependent defense genes and a modulator of antagonistic interactions between SA and JA signaling). All genes tested in this study were actively transcribed in the ldl single and double mutants, compared with WT plants, at the early phase (8 hpi) after local infection of PsmES4326 (Figure 2B and Supplementary Figure 2). To avoid overestimating the expression levels, we independently repeated the experiments five times, as summarized in Supplementary Figure 2. The transcript levels of the genes before infection (0 hpi) and at 16 hpi were not consistent among the experiments. However, despite these experimental variables, mRNA levels were higher in the ldl mutants, especially the ldl1 ldl2 double mutants, than in WT plants at 8 hpi (Figure 2B and Supplementary Figure 2).
The application of high concentrations of exogenous SA to Arabidopsis seedlings retards primary root elongation (Pasternak et al., 2019). Thus, we expected that the primary roots of the ldl mutants would be shorter than those of WT plants. To exclude a false positive effect due to the increased seed dormancy of ldl1 ldl2 mutants (Zhao et al., 2015), we transferred 5-day-old seedlings grown on 1/2 MS medium to 1/2 MS medium containing different concentrations of SA (0, 25, and 50 μM) and grew the seedlings under neutral day conditions for 14 days (12-h light/12 h dark). Primary root growth on 0 and 25 μM SA was not altered by the mutation of LDL1 and/or LDL2 (left and middle panels in Figure 2C). However, at 50 μM SA, the primary roots were significantly longer in ldl1 seedlings but shorter in ldl2 and ldl1 ldl2 seedlings compared to the WT (right panel in Figure 2C). These findings demonstrate that the ldl1 ldl2 mutants are hypersensitive to SA-dependent signaling, as confirmed by the finding that the expression of SA-responsive genes occurred rapidly in these plants during the early infection phase (Figure 2B and Supplementary Figure 2).
PTI represents a frontline defense barrier that protects plants from pathogen infection along with SA-related events. Since the growth rates of two different attenuated P. syringae strains in ldl mutants were similar to those in WT plants, we reasoned that the PTI response might not be dramatically altered in the ldl mutants. As expected, the protein abundance of BRASSINOSTEROID INSENSITIVE1-ASSOCIATED RECEPTOR KINASE1 (BAK1) and the amount of deposited callose in infected leaves of the mutants were comparable to those in WT plants (Figures 3A,B). Next, we examined the transcriptional regulation of MAMP-responsive genes in WT and mutant plants after PstDC3000 infection. The transcript levels of NDR/HIN1-LIKE 10 (NHL10) and At1g51890 (encoding a leucine-rich repeat protein kinase) increased at 8 hpi in the ldl1 and ldl1 ldl2 mutants, with similar results in three out of four independent experiments. At 8 hpi, FLG22-INDUCED RECEPTOR-LIKE KINASE1 (FRK1) transcript levels in the ldl1 ldl2 mutants were similar to or higher than those of WT plants (Figure 3C and Supplementary Figure 3). By contrast, at 16 hpi, the expression patterns of FRK1, NHL10, and At1g51890 varied depending on the gene, plant genotype, or biological repeat, and therefore, it appears that LDL1 and LDL2 have little or no effect on regulating the transcription of these genes (Figure 3C and Supplementary Figure 3). These results indicate that the transcription of some MAMP-responsive genes, as well as SA-responsive genes, is more sensitive in the ldl1 ldl2 mutants than the WT during the early phase of infection.
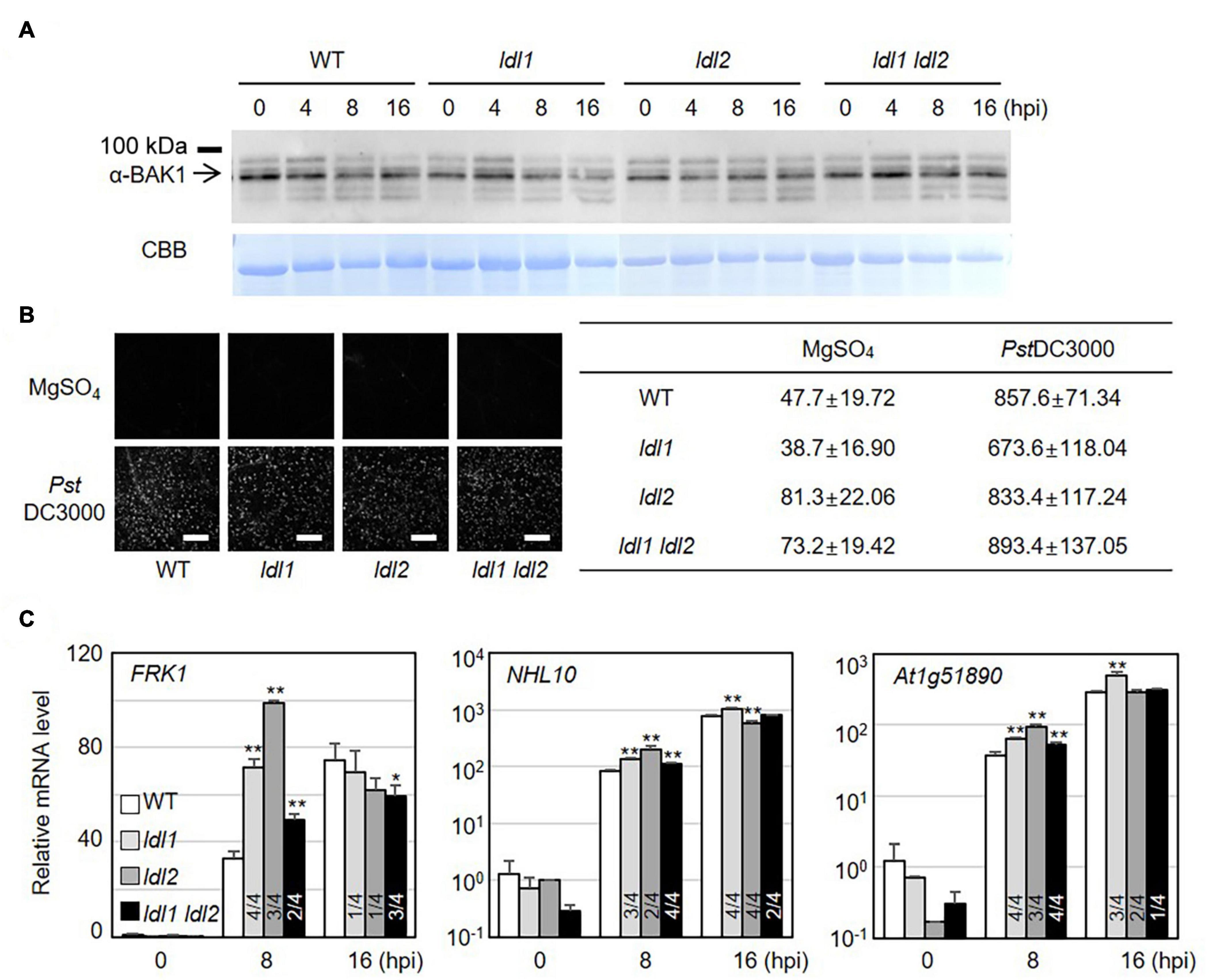
Figure 3. Transcript levels of MAMP-responsive genes increase in ldl mutants after Pseudomonas infection. (A) BAK1 protein abundance in WT, ldl1, ldl2, and ldl1 ldl2 during PstDC3000 infection (OD600 = 0.01). (B) Callose deposition in Arabidopsis leaves infiltrated with PstDC3000 at 24 h post-inoculation (hpi). Equivalent volumes of 10 m MgSO4 were used for mock conditions. Left panel: representative portions of leaves from WT, ldl1, ldl2, and ldl1 ldl2 plants stained with aniline blue to visualize deposited callose. Right panel: average ± SD of the number of callose deposits per field (n = 10) (Scale bars are 300 μm). No significant differences from WT plants were observed when analyzed with a one-way ANOVA-Tukey test (p < 0.05). (C) Transcript levels of FRK1, NHL10, and At1g51890 in local leaves of WT, ldl1, ldl2, and ldl1 ldl2 plants at 0, 8, and 16 hpi with PstDC3000 (OD600 = 0.01). Bar indicates the SD (*p < 0.05, **p < 0.01, two-tailed Student’s t-test, n = 3). The experiments were repeated 4 times; the numerators of the fractions below each plot indicate the number of trials showing differences from WT plants.
Specific covalent modifications of histone N-terminal tails, such as H3 and H4 acetylation and H3K4 methylation, reflect the establishment of systemic resistance (Jaskiewicz et al., 2011; Ding and Wang, 2015). As mentioned above, the ldl1 ldl2 double mutants showed an enhanced disease resistance response against local infection by virulent Pseudomonas strains. To investigate whether the enhanced local resistance in the mutants leads to accelerated systemic resistance, we performed SAR assays in which plants (WT, ldl1, ldl2, and ldl1 ldl2 plants) were exposed to a priming infection with avirulent strain PsmES4326/DG6 in local leaves, followed 2 days later by challenge inoculation with PsmES4326 in distal leaves (Figure 4A). The priming effect with successfully triggered systemic resistance was observed in WT and ldl2 plants (Figure 4B). However, we noted that the local resistance of ldl2 was not stable (Figures 1A,B). Bacterial growth and symptom development in distal leaves of ldl1 and ldl1 ldl2 mutants were also effectively restricted after challenge inoculation regardless of immunization (Figure 4B).
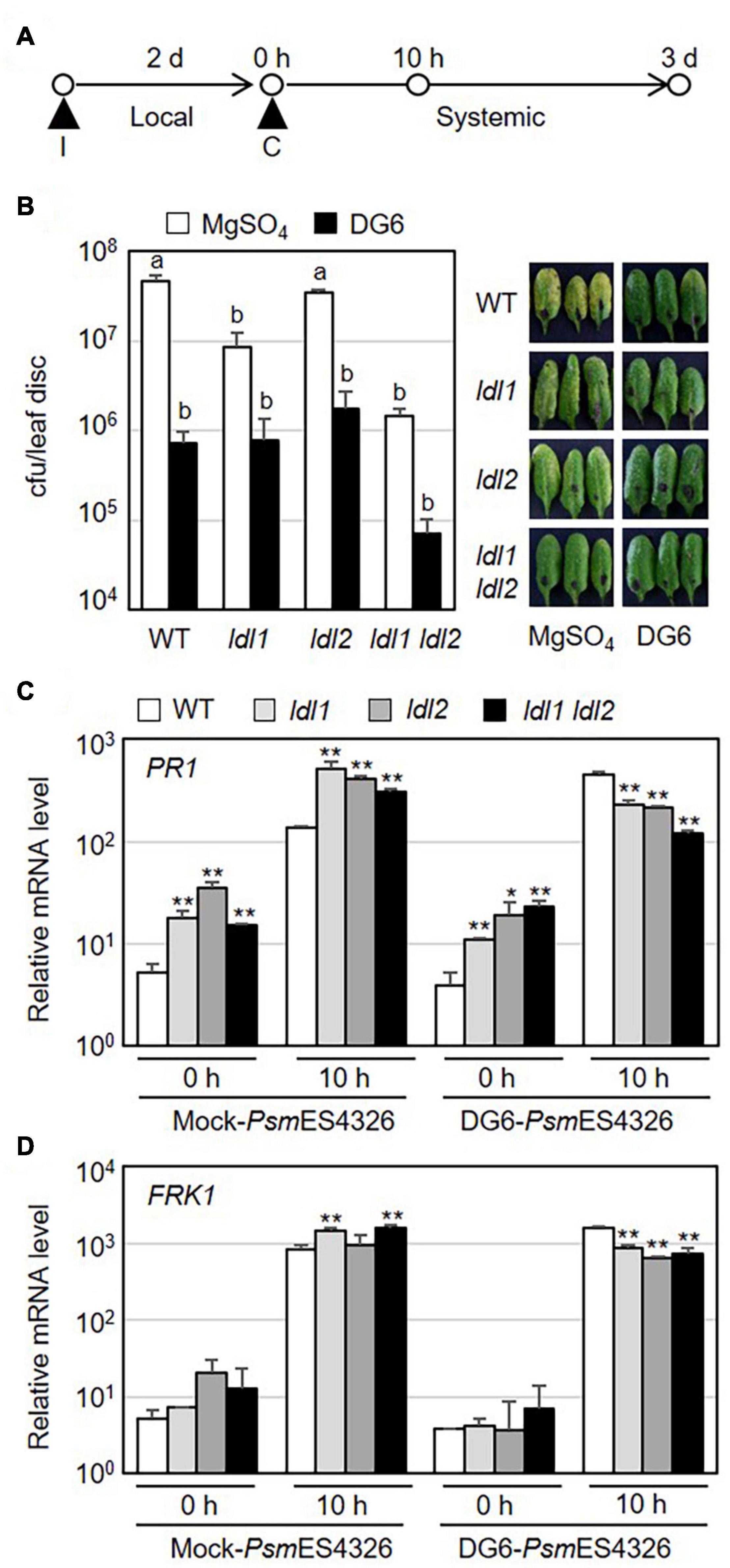
Figure 4. Systemic resistance is not further enhanced in ldl1 ldl2 double mutants. (A) Schematic diagram of the timepoints for pre-immunization (I) with avirulent strain PsmES4326/AvrRpt2 (DG6) (OD600 = 0.01) or 10 mM MgSO4 (mock), the second challenge inoculation (C) with virulent PsmES4326 (OD600 = 0.0001 or 0.01), and tissue samplings to evaluate the expression of defense-related genes and bacterial growth. (B) The growth of virulent PsmES4326 in distal leaves of plants treated with 10 mM MgSO4 or avirulent PsmES4326/AvrRpt2 (DG6). Bars indicate SEM (n = 8). Different letters indicate significant differences (p < 0.05, one-way ANOVA-Tukey test). Photographs of infected leaves were taken at 3 days post-challenge inoculation. The experiments were repeated at least three times with similar results. (C,D) Transcript levels of PR1 and FRK1 in distal leaves of immunized plants before (0 h) and after challenge inoculation (10 h) with PsmES4326 (OD600 = 0.01). Bar indicates the SD (*p < 0.05, **p < 0.01, two-way Student’s t-test, n = 3). The experiments were repeated twice with similar results (Supplementary Figure 4).
Systemic resistance usually correlates with stronger expression of defense-related genes in immunized plants than non-immunized plants (Conrath et al., 2015; Mauch-Mani et al., 2017; Jung et al., 2020). To test if the mutations of LDL1 and LDL2 lead to increased transcription of defense-related genes during SAR, we measured the mRNA levels of defense-related genes in distal leaves of immunized plants before and after challenge inoculation with PsmES4326. The expression levels of PR1, NPR1, WRKY70, and FRK1 did not consistently differ among WT and mutant plants before subsequent pathogen infection, although they were sometimes higher in mutant vs. WT plants (0 h in Figures 4C,D and Supplementary Figure 4). In the distal leaves of PsmES4326/DG6-immunized plants, the expression patterns of these genes were also comparable (and sometimes even lower) in mutant vs. WT plants (10 h in Figures 4C,D and Supplementary Figure 4). However, mock-immunization in local leaves triggered strong transcription of these genes in mutant plants, especially ldl1 and ldl1 ldl2, compared to WT plants (10 h in Figures 4C,D and Supplementary Figure 4). These results, together with the increased transcript levels in local infected leaves (Figure 2 and Supplementary Figure 2), indicate that the upregulation of defense-related genes reflects the enhanced disease resistance of the ldl mutants after pathogen infection.
To identify genes whose expression was affected by the simultaneous mutation of LDL1 and LDL2, we compared the whole transcriptomes of ldl1 ldl2 vs. WT plants. By analyzing mRNA-seq data from two biological replicates, we identified 273 DEGs in the ldl1 ldl2 mutants (Supplementary Table 2). We performed Gene Ontology (GO) analysis of these DEGs to identify GO terms that are enriched among up- or downregulated DEGs in the ldl1 ldl2 mutants (Mi et al., 2013). Of the 273 DEGs, 188 were successfully were mapped to GO IDs. A considerable portion of these DEGs (129 genes) appear to play roles in ‘response to stimulus’ (p < 1.08E-42, Fisher’s exact test). The top 10 biological processes included ‘cellular response to hypoxia,’ ‘responses to biotic and abiotic stress,’ ‘defense response,’ and ‘regulation of transcription’ (p < 1.E-04, Fisher’s exact test) (Supplementary Figure 5A). Two major functions of over-represented genes in ldl1 ldl2 were ‘regulation of transcription’ and ‘protein binding’ (p < 1.E-04, Fisher’s exact test) (Supplementary Figure 5B). Similarly, 30 DEGs (p < 1.78E-07, Fisher’s exact test) and 6 DEGs (p < 2.51E-03, Fisher’s exact test) encode ‘transcription factors’ and ‘calmodulin-related calcium-binding proteins,’ respectively (Supplementary Figure 5C).
Previous microarray analysis identified 449 misregulated genes in the ldl1 ldl2 double mutant (Berr et al., 2015). In addition, a more recent ChIP-seq analysis obtained from plants expressing LDL1-GFP (under its own promoter) revealed that LDL1 regulates the accumulation of methylated histone on the chromatin regions of 3,962 genes in the Arabidopsis genome (Hung et al., 2018). To narrow down LDL1 and LDL2 target genes in Arabidopsis, we compared these putative targets of LDL1 and LDL2 (Berr et al., 2015; Hung et al., 2018) with the 273 DEGs identified in the current study. Since the subset in common between the microarray data and RNA-seq data was too small to analyze, we focused on 39 DEGs that overlapped with LDL1 target genes identified by ChIP-seq (Table 1 and Supplementary Figure 5D). Since the well-known LDL1/LDL2-dependent genes MADS AFFECTING FLOWERING 4 (MAF4) and MAF5/AGAMOUS-LIKE68 (AGL68) (Berr et al., 2015; Hung et al., 2018) were identified in the in silico comparative analysis, we further analyzed these 39 DEGs to obtain clues to help explain the phenotypes of the ldl1 ldl2 mutants.
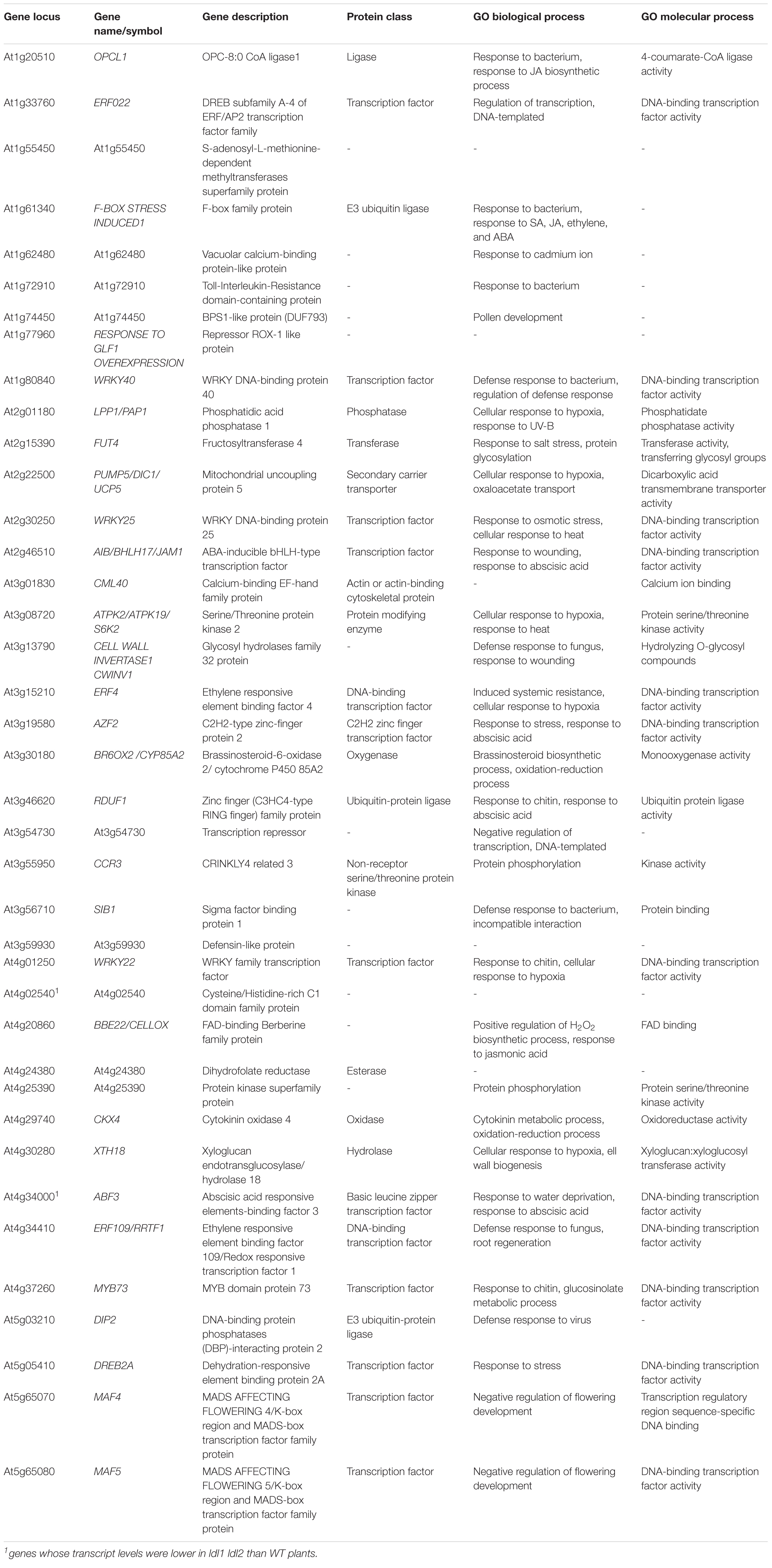
Table 1. List of DEGs in ldl1 ldl2 mutants overlapping with putative LDL1 target genes described by Hung et al. (2018).
To analyze the functional relationships among these 39 DEGs, we carried out STRING analysis3 (Szklarczyk et al., 2019). The primary biological process of 23 of the 39 genes was ‘response to stimulus’ [p = 6.07e-07, false discovery rate (FDR)], and among these genes, 8 were involved in ‘response to chitin and bacterium’ (p = 4.55e-09, FDR) (Supplementary Figure 5E). Eleven genes, including 4 AP2 domain-containing transcription factor genes (ERF4, ERF22, ERF109/RRTF1, and DREB2A) and 3 WRKY transcription factor genes (WRKY22, WRKY25, and WRKY40), were misregulated in the ldl1 ldl2 mutants at the transcriptional level (p = 4.55e-09, FDR) (Supplementary Figure 5E). In the small network composed of the 39 DEGs generated by STRING analysis, WRKY40 might act as a core protein, as it is co-expressed with 13 genes (Supplementary Figure 6A). Additionally, even outliers in this network such as At1g72910, At1g77960, CCR3, and CKX4 were also differentially transcribed in plants with enhanced disease resistance or whose corresponding mutants were susceptible to Pseudomonas infection [Table 1 and Supplementary Figure 6A (gray dot)] (Adams-Phillips et al., 2008; Thatcher et al., 2015; Mendy et al., 2017; Nasim et al., 2020). We also performed STRING network analysis to examine if these WRKYs and ERFs were co-expressed with some of the defense-related genes tested in this study. Most genes were co-expressed with WRKY40 and WRKY70 in the small network (Supplementary Figure 6B). Thus, we propose that the rapid and robust expression of defense-related genes in ldl1 ldl2 is due to the upregulation of these genes, e.g., WRKY22, WRKY40, and WRKY70, and that these 39 DEGs play major roles in the enhanced immunity of the ldl1 ldl2 mutants.
Plant LDL proteins remove methyl groups from histone H3K4 (Hung et al., 2018, 2019). To examine which specific methylation modifications were significantly altered by the simultaneous mutation of LDL1 and LDL2 in the absence of pathogen infection, we isolated nuclei from WT and ldl1 ldl2 plants and performed immunoblot analysis with α-H3K4me1, α-H3K4me2, and α-H3K4me3 antibodies to analyze the protein abundance of modified histone H3 proteins. Lys-4 monomethylated histone H3 (H3K4me1) proteins were present at significantly higher levels in the ldl1 ldl2 mutants compared to WT plants (Figure 5A and Supplementary Figure 7). The levels of histone H3K4me2 proteins also appeared to be higher in ldl1 ldl2 than WT plants in two out of three independent experiments, although these differences were not significant (Figure 5A and Supplementary Figure 7B). By contrast, no differences in the levels of histone H3K4me3 proteins were detected between WT and ldl1 ldl2 (Figure 5A and Supplementary Figure 7C). These observations support the notion that Arabidopsis LDL1 and LDL2 preferentially regulate H3K4 monomethylation, as previously described (Hung et al., 2018, 2019).
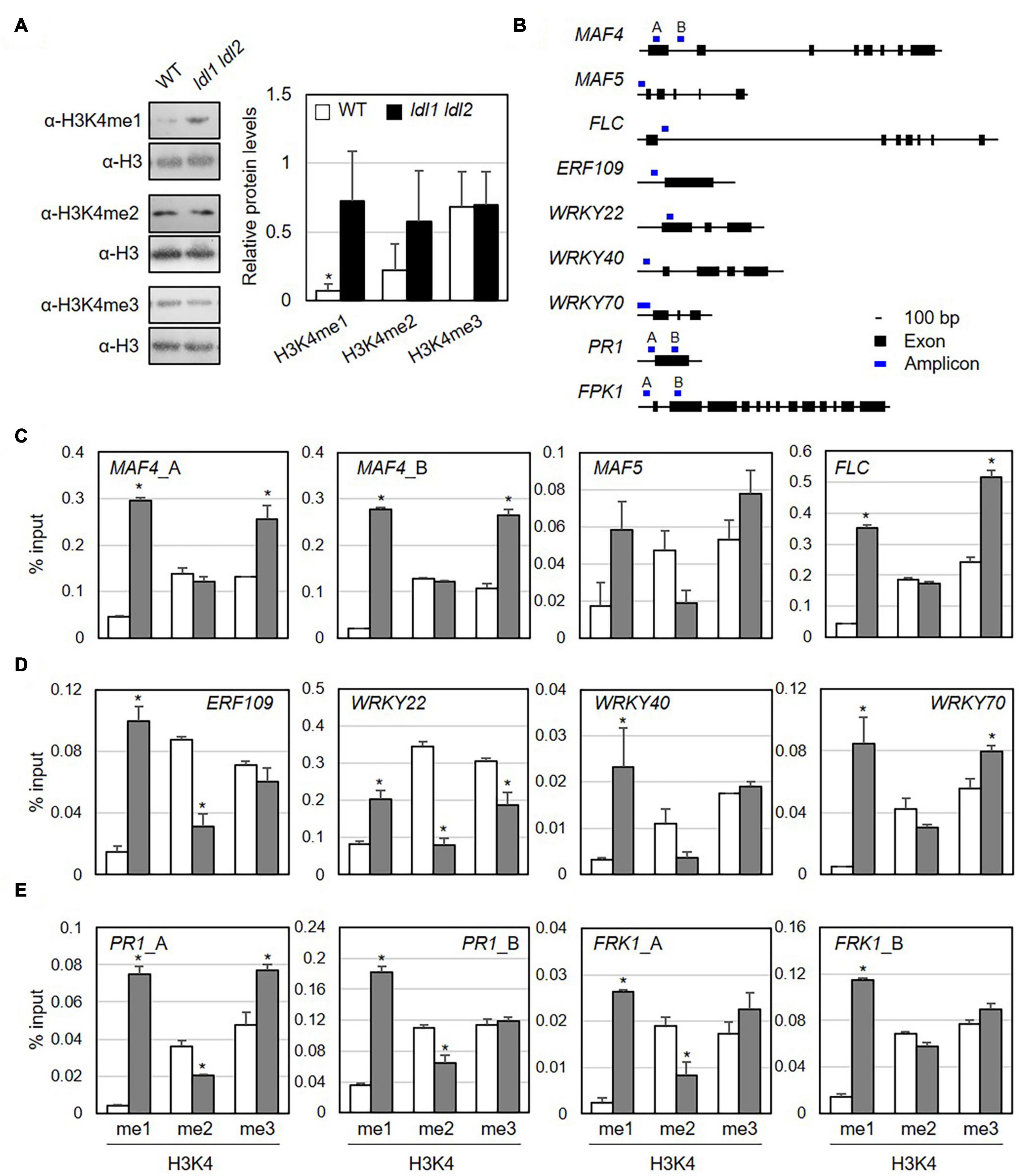
Figure 5. Monomethylation of histone H3K4 is associated with the transcription of genes encoding transcription factors in Arabidopsis. (A) Abundance of H3K4me1, H3K4me2, and H3K4me3 marked proteins in WT and ldl1 ldl2 plants. The right panel shows modified histone H3 protein levels in Arabidopsis (average ± SD, *p < 0.05, Student’s t-test, n = 3). The results of all independent experiments are shown in Supplementary Figure 6. (B) Genomic DNA regions of gene-specific primers used in panels (C-E). (C-E) ChIP-qPCR to examine the enrichment of modified histone H3 proteins in the identified LDL1-target genes (C), transcription factors overrepresented in the ldl1 ldl2 mutants (D), and PR1 and FRK1 genes (E). Enrichment of Lys 4- mono-, Lys 4- di-, and Lys 4-trimethylated histone H3 protein at the chromatin regions of genes was measured in wild type (white bars) and ldl1 ldl2 (gray bars). The amount of DNA after ChIP was quantified by qPCR, and the means represent the average immunoprecipitation efficiencies (%) against total input DNA used. Each plot shows a representative of two independent biological replications (with similar results), and the data points are the average values of technical triplicates (average ± SEM, *p < 0.05, Student’s t-test, n = 3). The blue bars are the amplicon regions used for ChiP-qPCR, and the primer sequences used in this study are shown in Supplementary Table 2.
To determine if specific modifications occur in loci harboring putative target genes, we examined the enrichment of modified histone H3 proteins in their chromatin regions by performing ChIP-qPCR using α-H3K4me1, α-H3K4me2, and α-H3K4me3 antibodies (Figure 5B and Table 1). The promoter and first exon and intron regions of the MADS-box genes MAF4 and MAF5/AGL68 and FLC are subject to LDL4/FLD-mediated modification, as the trimethylated histone (H3K4me3) levels in these DNA regions were higher in the fld mutant vs. the WT (Yu et al., 2011). Furthermore, LDL1 and LDL2 redundantly repress FLC expression via H3K4 demethylation (Jiang et al., 2007).
Next, we measured the accumulation of Lys-4 methylated histone H3 proteins on the MAF4, MAF5, and FLC DNA regions as positive controls to verify our experimental procedure. The levels of H3K4me1 and H3K4me3 proteins were higher at MAF4, MAF5, and FLC chromatin in ldl1 ldl2 vs. WT plants, supporting the previous observation that LDL1 and LDL2 function as histone demethylases (Figure 5C and Supplementary Figure 8) (Jiang et al., 2007; Hung et al., 2018). Hence, we analyzed the level of methylated H3K4 on a few putative targets of LDL1 and LDL2, including ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR109/REDOX RESPONSIVE TRANSCRIPTION FACTOR1 (ERF109/RRTF1) and the WRKY transcription factors genes WRKY22, WRKY40, and WRKY70 (Figure 5D and Supplementary Figure 8). Indeed, more Lys-4 methylated histone H3, especially H3K4me1, accumulated on the promoter regions of these genes in ldl1 ldl2 vs. WT plants (Figure 5D and Supplementary Table 3).
Finally, because the ldl mutants exhibited higher disease resistance due to rapid and higher expression of PR1 (Figure 2B) and FRK1 (Figure 3) vs. the WT, we examined whether LDL1 and LDL2 are involved in histone methylation of the promoter and coding regions of these genes. The level of H3K4me1 increased in ldl1 ldl2 on all amplicons of PR1 and FRK1. Interestingly, the expression of PR1 and FRK1 before bacterial infection (0 hpi) in the ldl single and double mutants was not significantly different from that of WT plants, likely because LDL1 and LDL2 act synergistically with histone deacetylase complex (HDAC) to repress gene expression (Hung et al., 2018, 2019, 2020). These results indicate that the H3K4me1 state in several defense-related genes primes plants to respond to subsequent Pseudomonas infection in a sensitive manner.
Our findings suggest that the Arabidopsis histone demethylase proteins LDL1 and LDL2 are required to maintain the switched-off state of the immune response under uninfected conditions. The individual and simultaneous mutations of both genes render Arabidopsis plants resistant to virulent Pseudomonas infection via the increased expression of SA- and MAMP-responsive genes after infection. These LDL1 and LDL2 proteins are key players that detach the monomethyl group from histone H3K4 proteins that occupy defense-related genes, such as WRKYs, ERFs, PR1, and FRK1. Thus, we hypothesized that an epigenetic eraser(s) associated with LDL1 and LDL2, which removes the methyl groups at histone H3K4 residues in the chromatin regions of defense-related genes, is vital for the primed defense response in Arabidopsis.
Unlike other histone methylation marks (H3K9, H3K27, and H4K20), histone H3K4, H3K36, and H3K79 methylation are euchromatic histone modifications (Black et al., 2012). Changes in H3K4 methylation patterns influence the immune responses of Arabidopsis and rice plants (Lee et al., 2016; Ramirez-Prado et al., 2018). The mutation of Arabidopsis TRITHORAX1/SET DOMAIN GROUP27 (ATX1/SDG27), encoding a H3K4 methyltransferase, led to reduced H3K4me2 and H3K4me3 levels. The atx1 mutants, which exhibit low PR1 and WRKY70 transcript levels, are susceptible to PstDC3000 infection (Alvarez-Venegas et al., 2003, 2007; Alvarez-Venegas, 2005). ATX-RELATED7/SET DOMAIN GROUP 25 (ATX7/SDG25) also plays a role in H3K4 methylation. The atx7/sdg25 mutants, with impaired monomethylation of H3K4, show increased susceptibility to infection by PstDC3000 and Botrytis cinerea (Lee et al., 2016). The demethylase JMJ704 positively regulates the immunity response of rice by suppressing the expression of genes encoding negative regulators of defense responses (Hou et al., 2015). By contrast, we demonstrated that ldl1 ldl2 double mutants showed increased H3K4me1 accumulation at the whole chromatin level and were enriched for H3K4me1 at the defense-related genes examined in this study. Together, these findings demonstrate that methylation of histone H3K4 is vital for the expression of defense-related genes and disease resistance responses during infection.
Since LDLs erase histone H3K4 methylation marks, especially mono-methylation, we cautiously suggest that LDL1 and LDL2 function as counterparts of ATX1/SDG27 and ATX7/SDG25 to fine-tune the methylation levels of histone H3K4 at the chromatin of defense-related genes in Arabidopsis. SAR-defective ldl4/fld/rsi1 mutants displayed reduced H3K4me2 levels at the WRKY6 and WRKY29 loci, which encode positive regulators of the immune response (Singh et al., 2013; Singh V. et al., 2014). Therefore, Arabidopsis LDLs may fine-tune plant immune responses in a sophisticated manner by targeting different genes. Indeed, ldl1, ldl2, and ldl4/fld exhibit a late-flowering phenotype, although the tissue- and organ-specific expression patterns of these genes and the targets of the encoded proteins are different (Greenberg et al., 2013; Yu et al., 2016; Hung et al., 2018; Martignago et al., 2019). For example, 2.3% of DEGs (60 genes out of 2539 DEGs) and 5.8% of DEGs (17 genes out of 273) appear to be involved in biotic stress responses in ldl4/fld and ldl1 ldl2, respectively (Yu et al., 2016; Supplementary Figure 5A). In addition, among WRKY genes (WRKY22, 25, 33, 40, 26, 48, and 53) whose expression increased in the ldl1 ldl2 double mutants, the expression of four WRKYs (WRKY22, 33, 40, and 53) was reduced in the ldl4/fld mutants (Yu et al., 2016; Supplementary Table 3). These findings suggest that all LDL histone demethylases function as epigenetic erasers but target different genes in the immune response.
Among the four homologous LDL group members, LDL1 and LDL2 are highly similar to LDL4/FLD structurally, with an N-terminal SWIRM domain and an amine oxidase domain, while the structure of LDL3 is different from the others (Martignago et al., 2019). Like the ldl4/fld mutant, mutations of LDL1 and LDL2 result in late flowering due to high expression of FLC and FWA, but the ldl3 mutants flower earlier and express FLC at lower levels than WT plants (Martignago et al., 2019). LDL4/FLD is involved in regulating flowering time in cooperation with the histone deacetylase HDA6 by controlling the occupation of acetylated and methylated histone proteins (H3K9K14Ac and H3K4Me3) on the DNA regions of FLC, MAF4, and MAF5, encoding floral repressors (Yu et al., 2011). LDL1/LDL2 also form a complex with HDA6, which functions as a negative transcriptional regulator of its target genes by switching its interacting partners. For example, the HDA6-LDL1/LDL2 complex associates with transcription factors such as CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), LATE ELONGATED HYPOCOTYL (LHY), and TIMING OF CAB EXPRESSION 1 (TOC1), which function as key circadian clock oscillators in a nested feedback loop to maintain circadian rhythms (Hung et al., 2018, 2019). The occupation of acetylated and/or methylated histone 3 proteins is significantly higher in hda6 and ldl mutants compared to WT. H3Ac and H3K4me level are likely higher in ldl1 ldl2 than the WT. Furthermore, the levels of H3Ac and H3K4me2 are higher in hda6 ldl1 ldl2 than in hda6 and ldl1 ldl2. In addition, TOC1 transcript levels are much higher in hda6 ldl1 ldl2 than in the single mutants (Hung et al., 2018). These findings suggest that histone deacetylase and histone demethylase stabilize the histone modification complex and/or regulate the transcription of their target genes in an additive manner. When we compared the mRNA-seq data generated in the current study to previous RNA-seq data (Yu et al., 2016), even though the plant growth conditions were different, 32 DEGs identified in ldl1 ldl2 (Supplementary Table 3) were also highly expressed in the hda6 mutant. Thus, we propose that the transcription and histone modification required for plant immunity are controlled by the HDA6-LDL1/LDL2 complex. The finding that hda6 and ldl mutants are resistant to pathogen infection (Wang et al., 2017 and this study) supports this idea.
The enhanced disease resistance of the ldl1 ldl2 mutants appears to be due to the upregulation of their putative target genes (Supplementary Figure 6B), including WRKYs and ERFs. WRKYs are important transcriptional regulators that function in plant defense. WRKY22 expression is induced during the early stage of bacterial pathogen infection (Dong et al., 2003). Pre-submerged wrky22 mutants were susceptible to bacterial infection with PstDC3000 compared to WT plants due to the downregulation of its target genes such as WRKY53 and FRK1, which confer innate immunity (Hsu et al., 2013). WRKY25 is involved in various stress responses (heat, salt, oxidative stress, and malnutrition) in addition to the response to bacterial pathogen infection (Zheng et al., 2007; Jiang and Deyholos, 2009; Li et al., 2009; Doll et al., 2020; Wu et al., 2020). WRKY40 negatively regulates PTI and attenuates early defense-induced genes during PTI (Lozano-Durán et al., 2013), but it positively regulates ETI via the Toll/Interleukin 1 Receptor (TIR)-type NLR RPS4 (Schön et al., 2013), whereas wrky70 mutants exhibited upregulated expression of disease-responsive genes such as PR1 and PDF1.2 (Ülker et al., 2007). Another putative target of LDL1 and LDL2, ERF109/RRTF1 (encoding an ERF/AP transcription factor involved in redox homeostasis), is under the control of WRKY40 (Pandey et al., 2010), and its promoter region appears to be subject to epigenetic changes (Soliman and Meyer, 2019). In addition, ERF109 promotes the expression of S-adenosyl-L-Met-dependent methyltransferase1 (BSMT1), which methylates SA and benzoic acid (Lin et al., 2020) and is thought to mediate crosstalk between jasmonic acid and auxin signaling to regulate lateral root formation (Cai et al., 2014) and various biotic and abiotic stress responses (Bahieldin et al., 2018). Thus, the altered expression of several WRKYs and ERFs, which are targets of LDL1 and LDL2, can explain the disease phenotype of the ldl mutants.
Notably, unlike the ldl4/fld/rsi1 mutants, which are defective in SAR but show WT-like local resistance against both virulent and avirulent Pseudomonas strains (Singh et al., 2013), loss-of-function mutations in LDL1 and LDL2 resulted in disease resistance against virulent bacterial infection along with the upregulated expression of defense-related genes without any increase in SA levels or callose deposition after pathogen infection (Figures 1–3 and Supplementary Figures 2, 3). Unlike well-characterized mutants with enhanced disease resistance, in which SA signaling and defense gene expression are constitutively active (Lorrain et al., 2003), analysis of ldl1 and ldl2 mutants indicated that LDL1 and LDL2 do not appear to directly control SA- and MAMP-dependent defense responses. The current observations point to the possibility that the ldl1 and ldl2 mutants might be in a defense priming state in the absence of any stimuli. Systemic resistance also results from defense priming, rendering plants sensitive to subsequent external stimuli, a process involving chromatin modification (Bruce et al., 2007; Conrath et al., 2015; Ding and Wang, 2015). Local immunization with SAR-inducing P. syringae pv. maculicola induced several covalent modifications of histone proteins, such as H3K4me2, H3K4me3, H4K5ac, H4K8ac, and H4K12ac, at the promoter regions of WRKY6, WRKY29, and WRKY53 in systemic leaves (Jaskiewicz et al., 2011). WRKY29 transcript levels, however, were not altered in the systemic leaves of immunized plants prior to secondary challenge-inoculation. Similar histone modifications and gene expression patterns were observed in benzothiadiazole-treated leaves before and after exposure to a secondary stimulus (Jaskiewicz et al., 2011). In line with these results, the transcription of defense-related genes increased significantly in local leaves of ldl1, ldl2, and ldl1 ldl2 compared to WT plants after pathogen infection (Figures 2–4 and Supplementary Figures 2–6). Furthermore, monomethylated histone H3K4 proteins predominantly occupied the promoter regions of defense-related genes (Figure 5 and Supplementary Figure 8). Our findings support the idea that histone modification, especially methylation of histone H3K4, reconstructs a docking region on chromatin for specific transcriptional activators, which would be rapidly activated upon subsequent stimuli (de la Cruz et al., 2005; Vermeulen et al., 2007; Ding et al., 2012). Considering the intensified transcription that occurs after local pathogen infection, it seems reasonable that transcriptional activation could occur in the systemic leaves of mock-immunized mutant plants. Taken together, these findings suggest that histone eraser complexes harboring LDL1 and LDL2 are involved in the proper growth/development and immunity responses of plants, likely in conjunction with various transcription factors (Supplementary Figure 9).
The original contributions presented in the study are publicly available. This data can be found here: NCBI repository, Accession Number: GSE171433.
SWN, R-RS, and HJP performed the experiments. HJP and HWJ analyzed the data and wrote the manuscript. HWJ designed the study. All authors contributed to the article and approved the submitted version.
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2020R1A6A1A03047729) to HJP and HWJ.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.688003/full#supplementary-material
Supplementary Figure 1 | Mutations of two different LDL genes render Arabidopsis plants resistant against virulent Pseudomonas infection. (A) Transcriptional changes in Arabidopsis LDL genes in local leaves of WT plants during pathogenesis. The expression levels of four Arabidopsis LDL homologs in local leaves upon bacterial infection were examined using eFP visualized meta-analysis (http://www.bar.utoronto.ca/) (Winter et al., 2007). Input data were obtained from microarray data set using ATH1 GeneChip showing mRNA levels in leaves of 4-week-old WT plants inoculated with 10 mM MgSO4 as the mock treatment (M), P. syringae pv. maculicola ES4326 carrying AvrRpt2 (PsmES4326/AvrRpt2) as the avirulent pathogen (A), and PsmES4326 as the virulent pathogen (V). (B) Scheme for pathogen inoculation and sampling times of plant tissue in panel (C). Leaves of 4-week-old WT plants were pre-immunized with PsmES4326 carrying AvrRpt2 (PsmES4326/AvrRpt2, DG6) (OD600 = 0.01, F), and 2 days later, distal leaves were infiltrated with PsmES4326 (OD600 = 0.01, S) using a needleless syringe. (C) RT-qPCR to measure the transcript levels of LDL homologous genes in non-infected local leaves (F) and distal leaves of immunized plants (S) at 0, 10, and 20 h post-inoculation (hpi). ACTIN2 was used to normalize the expression of the LDLs. Relative expression is shown as the average ± SD. Gray arrows indicate significant differences from non-infected local leaves (p > 0.05, two-tailed Student’s t-test, n = 11, each biological replicate contained three technical repeats). (D) Pseudomonas growth in the leaves of WT, ldl1, ldl2, ldl3, and ldl4/fld plants after PsmES4326 infection. (E) Enhanced resistance of different alleles of ldl1 and ldl2 mutants to virulent Pseudomonas infection. Virulent PsmES4326 (D,E) and P. syringae pv. tomato DC3000 (PstDC3000) (E) strains (OD600 = 0.0001) were used in these studies. Bacterial titers were evaluated at 3 dpi. The area of a leaf disc is 0.78 cm2. Averages ± SEM are plotted, and asterisks indicate significant differences from WT plants, as determined by a two-tailed Student’s t-test (∗p < 0.05, ∗∗p < 0.01, n = 8). The experiments were repeated over 3 times with similar results. (F) List of tested T-DNA insertion mutants corresponding to each LDL gene in Arabidopsis.
Supplementary Figure 2 | Transcript levels of SA-responsive genes in local leaves of WT and mutant plants upon infection with virulent strain PsmES4326 (OD600 = 0.01). (A,B) Each plot is representative of the five biological replicates of PR1 (A), and NPR1 (B) shown in Figure 2B. (C,D) Transcript levels of ALD1 (C) and WRKY70 (D) in Arabidopsis leaves during infection. Tables below each plot indicate the number of independent trials with significant differences between WT and a given plant genotype. The symbols represent the following: ↑, mRNA levels were upregulated in the mutants; =, mRNA levels in the mutants were comparable to those of WT plants; ↓, mRNA levels were downregulated in the mutants. The averages ± SD (standard deviation) were plotted (∗p < 0.05, ∗∗p < 0.01, two-tailed Student’s t-test, n = 3).
Supplementary Figure 3 | Transcript levels of MAMP-responsive genes in local leaves of WT and mutant plants upon infection with virulent strain PsmES4326 (OD600 = 0.01). (A-C) Each plot is representative of one of the four independent biological replicates of FRK1 (A), NHL10 (B), and At1g51890 (C) shown in Figure 3C. Bar indicates the SD (∗p < 0.05, ∗∗p < 0.01, two-tailed Student’s t-test, n = 3).
Supplementary Figure 4 | Transcript levels of defense-related genes in distal systemic leaves of WT and mutant plants upon challenge-inoculation with virulent Pseudomonas. (A,B) Each plot represents the other biological repeat of PR1 (A), and FRK1 (B) in Figures 4C,D. (C,D) Transcript levels of NPR1 (C) and WRKY70 (D) in distal leaves during SAR. Bar indicates the SD (∗p < 0.05, ∗∗p < 0.01, two-tailed Student’s t-test, n = 3).
Supplementary Figure 5 | GO analysis of DEGs in ldl1 ldl2 mutants and identification of LDL1- and LDL2-target genes that function in plant immunity. (A-C) Gene Ontology (GO) enrichment by the Panther classification system (Szklarczyk et al., 2019) of 273 DEGs in the biological process (A), molecular function (B), and protein class (C) categories in ldl1 ldl2 compared with WT. Annotated terms with p < 1.E-04 (A,B) and p < 2.51E-03 (C) are listed. (D) Venn diagram of DEGs in ldl1 ldl2 and LDL1-target genes. DEGs identified by microarray analysis of ldl1 ldl2 mutants (Berr et al., 2015) and using ChIP-seq data obtained from LDL1-GFP expressing plants (Hung et al., 2018) were compared with DEGs in the ldl1 ldl2 mutant identified by RNA-seq analysis. Numbers in parentheses are the total numbers of genes identified in each study. (E) 23, 11, and 8 genes involved in ‘response to stimulus’ (red dot), ‘DNA-binding transcription factor activity’ (blue dot), and ‘response to chitin and to bacterium’ (green dot), respectively, and their False Discovery Rates (FDR) are shown in the table.
Supplementary Figure 6 | STRING analysis of 39 DEGs and representative defense-related genes. (A) Thirty-nine proteins that are putative targets of LDLs (Table 1) were subjected to protein-protein interaction network functional enrichment analysis via STRING (Mi et al., 2013). Dots represent the putative functions of these proteins, including ‘response to stimulus’ (red dots), ‘DNA-binding transcription factor activity’ (blue dots), and ‘response to chitin and to bacterium’ (green dots) (Supplementary Figure 5E) (B) Proteins involved in SA signaling pathways (PR1, NPR1, and ALD1), the PTI pathway (WRKY70, WRKY25, RFK1, YLS9, and AT1G51890), and a few putative targets of LDLS (WRKY22, WRKY40, ERF1, and RRTF1) were subjected to STRING analysis.
Supplementary Figure 7 | Levels of modified histone H3 protein in WT and ldl1 ldl2. (A-C) Nuclear proteins extracted from three independent biological replications (1o, 2o, and 3o) were subjected to SDS-PAGE, and immunoblot analysis was performed using α-H3K4me1 (A), α-H3K4me2 (B), and α-H3K4me3 (C) antibodies. Numbers indicate independent experimental trials. w, WT plants; m, ldl1 ldl2. The normalized and processed data were plotted in Figure 5A.
Supplementary Figure 8 | Monomethylation of histone H3K4 is associated with the transcription of genes encoding transcription factors in Arabidopsis. The plots represent the second of two independent biological replications of ChIP-PCR to examine the enrichment of modified histone H3 proteins in the identified LDL1-target genes (A), transcription factors overrepresented in ldl1 ldl2 mutants (B), and FRK1 genes (C).
Supplementary Figure 9 | A proposed model about the role of LDL1 and LDL2 in the transcription of defense-related genes in Arabidopsis. A previous study reveals that Arabidopsis requires ATX1/SDG25 for monomethylation of histone H3K4, and its KO mutants were susceptible to PstDC3000 infection. On another side, both LDL1 and LDL2 are responsible for suppressing the immoderate expression of defense-related genes in the absence of pathogen infection. Thus, it is likely that the prevention of detaching methyl group of H3K4 in nucleosome associated with defense-related genes is enough to prime plants against subsequent pathogen infection.
Supplementary Table 1 | Bacterial strains used in this study.
Supplementary Table 2 | Oligonucleotide sequences used in this study.
Supplementary Table 3 | DEGs in the ldl1 ldl2 mutants, including DEGs that overlap with LDL1-target genes.
Aarts, N., Metz, M., Holub, E., Staskawicz, B. J., Daniels, M. J., and Parker, J. E. (1998). Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 95, 10306–10311. doi: 10.1073/pnas.95.17.10306
Adams-Phillips, L., Wan, J., Tan, X., Dunning, F. M., Meyers, B. C., Michelmore, R. W., et al. (2008). Discovery of ADP-Ribosylation and other plant defense pathway elements through expression profiling of four different Arabidopsis–Pseudomonas R-avr Interactions. Mol. Plant Microbe Int. 21, 646–657. doi: 10.1094/MPMI-21-5-0646
Alvarez-Venegas, R. (2005). Methylation patterns of histone H3 Lys 4, Lys 9 and Lys 27 in transcriptionally active and inactive Arabidopsis genes and in atx1 mutants. Nucleic Acids Res. 33, 5199–5207. doi: 10.1093/nar/gki830
Alvarez-Venegas, R., Abdallat, A. A., Guo, M., Alfano, J. R., and Avramova, Z. (2007). Epigenetic control of a transcription factor at the cross section of two antagonistic pathways. Epigenetics 2, 106–113. doi: 10.4161/epi.2.2.4404
Alvarez-Venegas, R., Pien, S., Sadder, M., Witmer, X., Grossniklaus, U., and Avramova, Z. (2003). ATX-1, an Arabidopsis homolog of Trithorax, activates flower homeotic genes. Curr. Biol. 13, 627–637. doi: 10.1016/S0960-9822(03)00243-4
Bahieldin, A., Atef, A., Edris, S., Gadalla, N. O., Ramadan, A. M., Hassan, S. M., et al. (2018). Multifunctional activities of ERF109 as affected by salt stress in Arabidopsis. Sci. Rep. 8:6403. doi: 10.1038/s41598-018-24452-6
Baum, S., Reimer-Michalski, E.-M., Bolger, A., Mantai, A. J., Benes, V., Usadel, B., et al. (2019). Isolation of open chromatin identifies regulators of systemic acquired resistance. Plant Physiol. 181, 817–833. doi: 10.1104/pp.19.00673
Beckers, G. J. M., Jaskiewicz, M., Liu, Y., Underwood, W. R., He, S. Y., Zhang, S., et al. (2009). Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell 21, 944–953. doi: 10.1105/tpc.108.062158
Berr, A., Shafiq, S., Pinon, V., Dong, A., and Shen, W.-H. (2015). The trxG family histone methyltransferase SET DOMAIN GROUP 26 promotes flowering via a distinctive genetic pathway. Plant J. 81, 316–328. doi: 10.1111/tpj.12729
Black, J. C., Van Rechem, C., and Whetstine, J. R. (2012). Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol. Cell 48, 491–507. doi: 10.1016/j.molcel.2012.11.006
Bruce, T. J. A., Matthes, M. C., Napier, J. A., and Pickett, J. A. (2007). Stressful “memories” of plants: Evidence and possible mechanisms. Plant Sci. 173, 603–608. doi: 10.1016/j.plantsci.2007.09.002
Brzezinka, K., Altmann, S., Czesnick, H., Nicolas, P., Gorka, M., Benke, E., et al. (2016). Arabidopsis FORGETTER1 mediates stress-induced chromatin memory through nucleosome remodeling. eLife 5:e17061. doi: 10.7554/eLife.17061
Bull, C. T., Manceau, C., Lydon, J., Kong, H., Vinatzer, B. A., and Fischer-Le Saux, M. (2010). Pseudomonas cannabina pv. cannabina pv. nov., and Pseudomonas cannabina pv. alisalensis (Cintas Koike and Bull, 2000) comb. nov., are members of the emended species Pseudomonas cannabina (ex Šutič & Dowson 1959) Gardan, Shafik, Belouin, Brosch, Grimont & Grimont 1999. Syst. Appl. Microbiol. 33, 105–115. doi: 10.1016/j.syapm.2010.02.001
Cai, X.-T., Xu, P., Zhao, P.-X., Liu, R., Yu, L.-H., and Xiang, C.-B. (2014). Arabidopsis ERF109 mediates cross-talk between jasmonic acid and auxin biosynthesis during lateral root formation. Nat. Commun. 5:5833. doi: 10.1038/ncomms6833
Century, K. S., Shapiro, A. D., Repetti, P. P., Dahlbeck, D., Holub, E., and Staskawicz, B. J. (1997). NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science 278:1963. doi: 10.1126/science.278.5345.1963
Chen, Y., Jie, W., Yan, W., Zhou, K., and Xiao, Y. (2012). Lysine-specific histone demethylase 1 (LSD1): A potential molecular target for tumor therapy. Crit. Rev. Eukaryot Gene Expr. 22, 53–59. doi: 10.1615/critreveukargeneexpr.v22.i1.40
Chisholm, S. T., Coaker, G., Day, B., and Staskawicz, B. J. (2006). Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124, 803–814. doi: 10.1016/j.cell.2006.02.008
Collmer, A., Lindeberg, M., Petnicki-Ocwieja, T., Schneider, D. J., and Alfano, J. R. (2002). Genomic mining type III secretion system effectors in Pseudomonas syringae yields new picks for all TTSS prospectors. Trends Microbiol. 10, 462–469. doi: 10.1016/S0966-842X(02)02451-4
Conrath, U. (2011). Molecular aspects of defence priming. Trends Plant Sci. 16, 524–531. doi: 10.1016/j.tplants.2011.06.004
Conrath, U., Beckers, G. J. M., Flors, V., García-Agustín, P., Jakab, G., Mauch, F., et al. (2006). Priming: getting ready for battle. Mol. Plant Microbe Int. 19, 1062–1071. doi: 10.1094/MPMI-19-1062
Conrath, U., Beckers, G. J. M., Langenbach, C. J. G., and Jaskiewicz, M. R. (2015). Priming for enhanced defense. Annu. Rev. Phytopathol. 53, 97–119. doi: 10.1146/annurev-phyto-080614-120132
de la Cruz, X., Lois, S., Sánchez-Molina, S., and Martínez-Balbás, M. A. (2005). Do protein motifs read the histone code?: Review articles. BioEssays 27, 164–175. doi: 10.1002/bies.20176
Deng, W., Marshall, N. C., Rowland, J. L., McCoy, J. M., Worrall, L. J., Santos, A. S., et al. (2017). Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 15, 323–337. doi: 10.1038/nrmicro.2017.20
Ding, B., and Wang, G.-L. (2015). Chromatin versus pathogens: the function of epigenetics in plant immunity. Front. Plant Sci. 6:675. doi: 10.3389/fpls.2015.00675
Ding, Y., Fromm, M., and Avramova, Z. (2012). Multiple exposures to drought “train” transcriptional responses in Arabidopsis. Nat. Commun. 3:740. doi: 10.1038/ncomms1732
Doll, J., Muth, M., Riester, L., Nebel, S., Bresson, J., Lee, H.-C., et al. (2020). Arabidopsis thaliana WRKY25 transcription factor mediates oxidative stress tolerance and regulates senescence in a redox-dependent manner. Front. Plant Sci. 10:1734. doi: 10.3389/fpls.2019.01734
Dong, J., Chen, C., and Chen, Z. (2003). Expression pro?les of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 51, 21–37. doi: 10.1023/a:1020780022549
Durrant, W. E., and Dong, X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209. doi: 10.1146/annurev.phyto.42.040803.140421
Ellis, L., and Loda, M. (2018). LSD1: A single target to combat lineage plasticity in lethal prostate cancer. Proc. Natl. Acad. Sci. U. S. A. 115, 4530–4531. doi: 10.1073/pnas.1804205115
Falk, A., Feys, B. J., Frost, L. N., Jones, J. D. G., Daniels, M. J., and Parker, J. E. (1999). EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. U. S. A. 96, 3292–3297. doi: 10.1073/pnas.96.6.3292
Fu, Z. Q., and Dong, X. (2013). Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. doi: 10.1146/annurev-arplant-042811-105606
Green, M. R., and Sambrook, J. (2012). Molecular Cloning: A Laboratory Manual. 4th ed. Harbor, N.Y: Cold Spring Harbor Laboratory Press.
Greenberg, M. V. C., Deleris, A., Hale, C. J., Liu, A., Feng, S., and Jacobsen, S. E. (2013). Interplay between active chromatin marks and RNA-directed DNA methylation in Arabidopsis thaliana. PLoS Genet. 9:e1003946. doi: 10.1371/journal.pgen.1003946
Gruner, K., Griebel, T., Návarová, H., Attaran, E., and Zeier, J. (2013). Reprogramming of plants during systemic acquired resistance. Front. Plant Sci. 4:252. doi: 10.3389/fpls.2013.00252
Guttman, D. S., and Greenberg, J. T. (2001). Functional analysis of the type III effectors AvrRpt2 and AvrRpm1 of Pseudomonas syringae with the use of a single-copy genomic integration system. Mol. Plant Microbe Int. 14, 145–155. doi: 10.1094/MPMI.2001.14.2.145
Guttman, D. S., Vinatzer, B. A., Sarkar, S. F., Ranall, M. V., Kettler, G., and Greenberg, J. T. (2002). A Functional screen for the type III (Hrp) secretome of the plant pathogen. Science 295, 1722–1726. doi: 10.1126/science.295.5560.1722
Han, S.-W., and Jung, H. W. (2013). Molecular sensors for plant immunity; pattern recognition receptors and race-specific resistance proteins. J. Plant Biol. 56, 357–366. doi: 10.1007/s12374-013-0323-z
Hirakawa, T., Kuwata, K., Gallego, M. E., White, C. I., Nomoto, M., Tada, Y., et al. (2019). LSD1-LIKE1-mediated H3K4me2 demethylation is required for homologous recombination repair. Plant Physiol. 181, 499–509. doi: 10.1104/pp.19.00530
Holeski, L. M., Jander, G., and Agrawal, A. A. (2012). Transgenerational defense induction and epigenetic inheritance in plants. Trends Ecol. Evol. 27, 618–626. doi: 10.1016/j.tree.2012.07.011
Hou, Y., Wang, L., Wang, L., Liu, L., Li, L., Sun, L., et al. (2015). JMJ704 positively regulates rice defense response against Xanthomonas oryzae pv. oryzae infection via reducing H3K4me2/3 associated with negative disease resistance regulators. BMC Plant Biol. 15:286. doi: 10.1186/s12870-015-0674-3
Hsu, F.-C., Chou, M.-Y., Chou, S.-J., Li, Y.-R., Peng, H.-P., and Shih, M.-C. (2013). Submergence confers immunity mediated by the WRKY22 transcription factor in Arabidopsis. Plant Cell 25, 2699–2713. doi: 10.1105/tpc.113.114447
Hung, F.-Y., Chen, C., Yen, M.-R., Hsieh, J.-W. A., Li, C., Shih, Y.-H., et al. (2020). The expression of long non-coding RNAs is associated with H3Ac and H3K4me2 changes regulated by the HDA6-LDL1/2 histone modification complex in Arabidopsis. NAR Genom. Bioinform. 2:lqaa066. doi: 10.1093/nargab/lqaa066
Hung, F.-Y., Chen, F.-F., Li, C., Chen, C., Chen, J.-H., Cui, Y., et al. (2019). The LDL1/2-HDA6 histone modification complex interacts with TOC1 and regulates the core circadian clock components in Arabidopsis. Front. Plant Sci. 10:233. doi: 10.3389/fpls.2019.00233
Hung, F.-Y., Chen, F.-F., Li, C., Chen, C., Lai, Y.-C., Chen, J.-H., et al. (2018). The Arabidopsis LDL1/2-HDA6 histone modification complex is functionally associated with CCA1/LHY in regulation of circadian clock genes. Nucleic Acids Res. 46, 10669–10681. doi: 10.1093/nar/gky749
Hyun, K., Jeon, J., Park, K., and Kim, J. (2017). Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 49:e324. doi: 10.1038/emm.2017.11
Jaskiewicz, M., Conrath, U., and Peterhänsel, C. (2011). Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep. 12, 50–55. doi: 10.1038/embor.2010.186
Jiang, D., Yang, W., He, Y., and Amasino, R. M. (2007). Arabidopsis relatives of the human Lysine-Specific Demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell 19, 2975–2987. doi: 10.1105/tpc.107.052373
Jiang, Y., and Deyholos, M. K. (2009). Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 69, 91–105. doi: 10.1007/s11103-008-9408-3
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Jung, H. W., Panigrahi, G. K., Jung, G. Y., Lee, Y. J., Shin, K. H., Sahoo, A., et al. (2020). Pathogen-associated molecular pattern-triggered immunity involves proteolytic degradation of core nonsense-mediated mRNA decay factors during the early defense response. Plant Cell 32, 1081–1101. doi: 10.1105/tpc.19.00631
Jung, H. W., Tschaplinski, T. J., Wang, L., Glazebrook, J., and Greenberg, J. T. (2009). Priming in systemic plant immunity. Science 324, 89–91. doi: 10.1126/science.1170025
Krichevsky, A., Zaltsman, A., Kozlovsky, S. V., Tian, G.-W., and Citovsky, V. (2009). Regulation of root elongation by histone acetylation in Arabidopsis. J. Mol. Biol. 385, 45–50. doi: 10.1016/j.jmb.2008.09.040
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lee, S., Fu, F., Xu, S., Lee, S. Y., Yun, D.-J., and Mengiste, T. (2016). Global regulation of plant immunity by histone lysine methyl transferases. Plant Cell 28, 1640–1661. doi: 10.1105/tpc.16.00012
Li, S., Fu, Q., Huang, W., and Yu, D. (2009). Functional analysis of an Arabidopsis transcription factor WRKY25 in heat stress. Plant Cell Rep. 28, 683–693. doi: 10.1007/s00299-008-0666-y
Lin, N.-C., and Martin, G. B. (2005). An avrPto/avrPtoB mutant of Pseudomonas syringae pv. tomato DC3000 does not elicit Pto-mediated resistance and is less virulent on tomato. Mol. Plant Microbe Int. 18, 43–51. doi: 10.1094/MPMI-18-0043
Lin, W., Zhang, H., Huang, D., Schenke, D., Cai, D., Wu, B., et al. (2020). Dual-localized WHIRLY1 affects salicylic acid biosynthesis via coordination of Isochorismate synthase1, Phenylalanine ammonia lyase1, and S -Adenosyl-L-Methionine-dependent methyltransferase1. Plant Physiol. 184, 1884–1899. doi: 10.1104/pp.20.00964
Lin, X., Tirichine, L., and Bowler, C. (2012). Protocol: Chromatin immunoprecipitation (ChIP) methodology to investigate histone modifications in two model diatom species. Plant Methods 8:48. doi: 10.1186/1746-4811-8-48
Lindeberg, M., Cunnac, S., and Collmer, A. (2009). The evolution of Pseudomonas syringae host specificity and type III effector repertoires. Mol. Plant Pathol. 10, 767–775. doi: 10.1111/j.1364-3703.2009.00587.x
Lindeberg, M., Cunnac, S., and Collmer, A. (2012). Pseudomonas syringae type III effector repertoires: last words in endless arguments. Trends Microbiol. 20, 199–208. doi: 10.1016/j.tim.2012.01.003
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lorrain, S., vailleau, F., Balague, C., and Roby. (2003). Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 8, 263–271. doi: 10.1016/S1360-1385(03)00108-0
Lozano-Durán, R., Macho, A. P., Boutrot, F., Segonzac, C., Somssich, I. E., and Zipfel, C. (2013). The transcriptional regulator BZR1 mediates trade-off between plant innate immunity and growth. eLife 2:e00983. doi: 10.7554/eLife.00983
Luna, E., Bruce, T. J. A., Roberts, M. R., Flors, V., and Ton, J. (2012). Next-generation systemic acquired resistance. Plant Physiol. 158, 844–853. doi: 10.1104/pp.111.18746
Martignago, D., Bernardini, B., Polticelli, F., Salvi, D., Cona, A., Angelini, R., et al. (2019). The four FAD-dependent histone demethylases of Arabidopsis are differently involved in the control of flowering time. Front. Plant Sci. 10:669. doi: 10.3389/fpls.2019.00669
Mauch-Mani, B., Baccelli, I., Luna, E., and Flors, V. (2017). Defense priming: An adaptive part of induced resistance. Annu. Rev. Plant Biol. 68, 485–512. doi: 10.1146/annurev-arplant-042916-041132
Mendy, B., Wang’ombe, M. W., Radakovic, Z. S., Holbein, J., Ilyas, M., Chopra, D., et al. (2017). Arabidopsis leucine-rich repeat receptor–like kinase NILR1 is required for induction of innate immunity to parasitic nematodes. PLoS Pathog. 13:e1006284. doi: 10.1371/journal.ppat.1006284
Mi, H., Muruganujan, A., Casagrande, J. T., and Thomas, P. D. (2013). Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8, 1551–1566. doi: 10.1038/nprot.2013.092
Nasim, Z., Fahim, M., Gawarecka, K., Susila, H., Jin, S., Youn, G., et al. (2020). Role of AT1G72910, AT1G72940, and ADR1-LIKE 2 in plant immunity under nonsense-mediated mRNA decay-compromised conditions at low temperatures. Int. J. Mol. Sci. 21:7986. doi: 10.3390/ijms21217986
Ngou, B. P. M., Ahn, H.-K., Ding, P., and Jones, J. D. G. (2021). Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592, 110–115. doi: 10.1038/s41586-021-03315-7
Pandey, S. P., Roccaro, M., Schön, M., Logemann, E., and Somssich, I. E. (2010). Transcriptional reprogramming regulated by WRKY18 and WRKY40 facilitates powdery mildew infection of Arabidopsis: WRKY18/40 in powdery mildew susceptibility. Plant J. 64, 912–923. doi: 10.1111/j.1365-313X.2010.04387.x
Parker, J. E., Holub, E., Frost, L. N., Falk, A., Gunn, N. D., and Daniels, M. J. (1996). Characterization of edsl, a mutation in Arabidopsis suppressing resistance to Peronospora parasíiíca specified by severa1 different RPP genes. Plant Cell 9, 2033–2046. doi: 10.1105/tpc.8.11.2033
Pasternak, T., Groot, E. P., Kazantsev, F. V., Teale, W., Omelyanchuk, N., Kovrizhnykh, V., et al. (2019). Salicylic acid affects root meristem patterning via auxin distribution in a concentration-dependent manner. Plant Physiol. 180, 1725–1739. doi: 10.1104/pp.19.00130
Pikaard, C. S., and Mittelsten Scheid, O. (2014). Epigenetic regulation in plants. Cold Spring Harb. Perspect. Biol. 6:a019315. doi: 10.1101/cshperspect.a019315
Qi, Y., Tsuda, K., Glazebrook, J., and Katagiri, F. (2011). Physical association of pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) immune receptors in Arabidopsis: Physical association of PTI and ETI receptors. Mol. Plant Pathol. 12, 702–708. doi: 10.1111/j.1364-3703.2010.00704.x
Ramirez-Prado, J. S., Piquerez, S. J. M., Bendahmane, A., Hirt, H., Raynaud, C., and Benhamed, M. (2018). Modify the histone to win the battle: Chromatin dynamics in plant-pathogen interactions. Front. Plant Sci. 9, 355. doi: 10.3389/fpls.2018.00355
Raxwal, V. K., Ghosh, S., Singh, S., Katiyar-Agarwal, S., Goel, S., Jagannath, A., et al. (2020). Abiotic stress-mediated modulation of the chromatin landscape in Arabidopsis thaliana. J. Exp. Bot. 71, 5280–5293. doi: 10.1093/jxb/eraa286
Roine, E., Wei, W., Yuan, J., Nurmiaho-Lassila, E. L., Kakkinen, N., Romantschuk, M., et al. (1997). Hrp pilus: an hrp-dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. U. S. A. 94, 3459–3464.
Saze, H. (2008). Epigenetic memory transmission through mitosis and meiosis in plants. Semin. Cell Dev. Biol. 19, 527–536. doi: 10.1016/j.semcdb.2008.07.017
Schenk, S. T., Hernández-Reyes, C., Samans, B., Stein, E., Neumann, C., Schikora, M., et al. (2014). N -Acyl-homoserine lactone primes plants for cell wall reinforcement and induces resistance to bacterial pathogens via the salicylic acid/oxylipin pathway. Plant Cell 26, 2708–2723. doi: 10.1105/tpc.114.126763
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Schön, M., Töller, A., Diezel, C., Roth, C., Westphal, L., Wiermer, M., et al. (2013). Analyses of wrky18 wrky40 plants reveal critical roles of SA/EDS1 signaling and Indole-Glucosinolate biosynthesis for Golovinomyces orontii resistance and a loss-of resistance towards Pseudomonas syringae pv. tomato AvrRPS4. Mol. Plant Microbe Int. 26, 758–767. doi: 10.1094/MPMI-11-12-0265-R
Seskar, M., Shulaev, V., and Raskin, I. (1998). Endogenous methyl salicylate in pathogen-inoculated tobacco plants. Plant Physiol. 116, 387–392. doi: 10.1104/pp.116.1.387
Shafiq, S., Berr, A., and Shen, W.-H. (2014). Combinatorial functions of diverse histone methylations in Arabidopsis thaliana flowering time regulation. N. Phytol. 201, 312–322. doi: 10.1111/nph.12493
Shi, Y., Lan, F., Matson, C., Mulligan, P., Whetstine, J. R., Cole, P. A., et al. (2004). Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953. doi: 10.1016/j.cell.2004.12.012
Singh, P., Yekondi, S., Chen, P.-W., Tsai, C.-H., Yu, C.-W., Wu, K., et al. (2014). Environmental history modulates Arabidopsis pattern-triggered immunity in a Histone acetyltransferase1–dependent manner. Plant Cell 26, 2676–2688. doi: 10.1105/tpc.114.123356
Singh, V., Roy, S., Singh, D., and Nandi, A. K. (2014). Arabidopsis FLOWERING LOCUS D influences systemic-acquired-resistance-induced expression and histone modifications of WRKY genes. J. Biosci. 39, 119–126. doi: 10.1007/s12038-013-9407-7
Singh, S., Singh, A., Roy, S., and Sarkar, A. K. (2012). SWP1 negatively regulates lateral root initiation and elongation in Arabidopsis. Plant Signal. Behav. 7, 1522–1525. doi: 10.4161/psb.22099
Singh, S., Yadav, S., Singh, A., Mahima, M., Singh, A., Gautam, V., et al. (2020). Auxin signaling modulates LATERAL ROOT PRIMORDIUM 1 (LRP 1) expression during lateral root development in Arabidopsis. Plant J. 101, 87–100. doi: 10.1111/tpj.14520
Singh, V., Roy, S., Giri, M. K., Chaturvedi, R., Chowdhury, Z., Shah, J., et al. (2013). Arabidopsis thaliana FLOWERING LOCUS D is required for systemic acquired resistance. Mol. Plant Microbe Int. 26, 1079–1088. doi: 10.1094/MPMI-04-13-0096-R
Singh, V., Singh, D., Gautam, J. K., and Nandi, A. K. (2019). RSI1/FLD is a positive regulator for defense against necrotrophic pathogens. Physiol. Mol. Plant Pathol. 107, 40–45. doi: 10.1016/j.pmpp.2019.04.005
Soliman, E. R. S., and Meyer, P. (2019). Responsiveness and adaptation to salt stress of the REDOX-RESPONSIVE TRANSCRIPTION FACTOR 1 (RRTF1) gene are controlled by its promoter. Mol. Biotechnol. 61, 254–260. doi: 10.1007/s12033-019-00155-9
Spedaletti, V., Polticelli, F., Capodaglio, V., Schininà, M. E., Stano, P., Federico, R., et al. (2008). Characterization of a lysine-specific histone demethylase from Arabidopsis thaliana. Biochemistry 47, 4936–4947. doi: 10.1021/bi701969k
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613. doi: 10.1093/nar/gky1131
Thatcher, L. F., Kamphuis, L. G., Hane, J. K., Oñate-Sánchez, L., and Singh, K. B. (2015). The Arabidopsis KH-domain RNA-binding protein ESR1 functions in components of jasmonate signalling, unlinking growth restraint and resistance to stress. PLoS One 10:e0126978. doi: 10.1371/journal.pone.0126978
Trapnell, C., Pachter, L., and Salzberg, S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. doi: 10.1093/bioinformatics/btp120
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Tsuda, K., and Katagiri, F. (2010). Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 13, 459–465. doi: 10.1016/j.pbi.2010.04.006
Tsuda, K., Sato, M., Stoddard, T., Glazebrook, J., and Katagiri, F. (2009). Network properties of robust immunity in Plants. PLoS Genet. 5:16.
Tsuda, K., and Somssich, I. E. (2015). Transcriptional networks in plant immunity. New Phytol. 206, 932–947. doi: 10.1111/nph.13286
Tu, W. J., McCuaig, R. D., Tan, A. H. Y., Hardy, K., Seddiki, N., Ali, S., et al. (2020). Targeting nuclear LSD1 to reprogram cancer cells and reinvigorate exhausted T cells via a novel LSD1-EOMES switch. Front. Immunol. 11:1228. doi: 10.3389/fimmu.2020.01228
Ülker, B., Shahid Mukhtar, M., and Somssich, I. E. (2007). The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta 226, 125–137. doi: 10.1007/s00425-006-0474-y
Vermeulen, M., Mulder, K. W., Denissov, S., Pijnappel, W. W. M. P., van Schaik, F. M. A., Varier, R. A., et al. (2007). Selective anchoring of TFIID to nucleosomes by trimethylation of Histone H3 Lysine 4. Cell 131, 58–69. doi: 10.1016/j.cell.2007.08.016
Wang, Y., Hu, Q., Wu, Z., Wang, H., Han, S., Jin, Y., et al. (2017). HISTONE DEACETYLASE 6 represses pathogen defence responses in Arabidopsis thaliana. Plant Cell Environ. 40, 2972–2986. doi: 10.1111/pce.13047
Winter, D., Vinegar, B., Nahal, H., Ammar, R., Wilson, G. V., and Provart, N. J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2:e718. doi: 10.1371/journal.pone.0000718
Wu, T.-Y., Krishnamoorthi, S., Goh, H., Leong, R., Sanson, A. C., and Urano, D. (2020). Crosstalk between heterotrimeric G protein-coupled signaling pathways and WRKY transcription factors modulating plant responses to suboptimal micronutrient conditions. J. Exp. Bot. 71, 3227–3239. doi: 10.1093/jxb/eraa108
Yu, C.-W., Chang, K.-Y., and Wu, K. (2016). Genome-wide analysis of gene regulatory networks of the FVE-HDA6-FLD complex in Arabidopsis. Front. Plant Sci. 7:555. doi: 10.3389/fpls.2016.00555
Yu, C.-W., Liu, X., Luo, M., Chen, C., Lin, X., Tian, G., et al. (2011). Histone deacetylase6 interacts with FLOWERING locus D and regulates flowering in Arabidopsis. Plant Physiol. 156, 173–184. doi: 10.1104/pp.111.174417
Yuan, M., Jiang, Z., Bi, G., Nomura, K., Liu, M., Wang, Y., et al. (2021). Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592, 105–109. doi: 10.1038/s41586-021-03316-6
Zhao, M., Yang, S., Liu, X., and Wu, K. (2015). Arabidopsis histone demethylases LDL1 and LDL2 control primary seed dormancy by regulating DELAY OF GERMINATION 1 and ABA signaling-related genes. Front. Plant Sci. 6:159. doi: 10.3389/fpls.2015.00159
Keywords: Arabidopsis, defense priming, epigenetic regulation, histone methylation, immunity, WRKY transcription factors
Citation: Noh SW, Seo R-R, Park HJ and Jung HW (2021) Two Arabidopsis Homologs of Human Lysine-Specific Demethylase Function in Epigenetic Regulation of Plant Defense Responses. Front. Plant Sci. 12:688003. doi: 10.3389/fpls.2021.688003
Received: 30 March 2021; Accepted: 18 May 2021;
Published: 14 June 2021.
Edited by:
Ana López Sánchez, Department of Molecular Genetics of Plants, National Center of Biotechnology, Spanish National Research Council (CSIC), SpainReviewed by:
Sang-Wook Han, Chung-Ang University, South KoreaCopyright © 2021 Noh, Seo, Park and Jung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hee Jin Park, Y2thdG93b3JAZGF1LmFjLmty; Ho Won Jung, aHdqdW5nQGRhdS5hYy5rcg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.