- 1Institute of Integrative Plant Biology, School of Life Sciences, Jiangsu Normal University, Xuzhou, China
- 2Jiangsu Key Laboratory of Phylogenomics & Comparative Genomics, School of Life Sciences, Jiangsu Normal University, Xuzhou, China
- 3Xuzhou Institute of Agricultural Sciences in Jiangsu Xuhuai District, Key Laboratory for Biology and Genetic Breeding of Sweetpotato (Xuzhou), Ministry of Agriculture/Jiangsu Xuzhou Sweetpotato Research Center, Xuzhou, China
Plant-specific TCP transcription factors play vital roles in the controlling of growth, development, and the stress response processes. Extensive researches have been carried out in numerous species, however, there hasn’t been any information available about TCP genes in sweet potato (Ipomoea batatas L.). In this study, a genome-wide analysis of TCP genes was carried out to explore the evolution and function in sweet potato. Altogether, 18 IbTCPs were identified and cloned. The expression profiles of the IbTCPs differed dramatically in different organs or different stages of leaf development. Furthermore, four CIN-clade IbTCP genes contained miR319-binding sites. Blocking IbmiR319 significantly increased the expression level of IbTCP11/17 and resulted in a decreased photosynthetic rate due to the change in leaf submicroscopic structure, indicating the significance of IbmiR319-targeted IbTCPs in leaf anatomical morphology. A systematic analyzation on the characterization of the IbTCPs together with the primary functions in leaf anatomical morphology were conducted to afford a basis for further study of the IbmiR319/IbTCP module in association with leaf anatomical morphology in sweet potato.
Introduction
TCP transcription factors (TFs) belong to a small family of plant-specific TFs (Yin et al., 2018) named after its initial members TEOSINTE BRANCHED1 (TB1) in Zea mays L. (Doebley et al., 1997), CYCLOIDEA (CYC) in Antirrhinum majus L. (Luo et al., 1996), and PROLIFERATING CELL FACTORS 1 and 2 (PCF1 and PCF2) in Oryza sativa L. (Kosugi and Ohashi, 1997). TCPs have been ascertained in numerous plant species, such as Medicago truncatula Gaertn. (Wang et al., 2018), Gossypium barbadense L. (Zheng et al., 2018), Citrullus lanatus (Thunb.) Matsum. and Nakai (Shi et al., 2016), Malus domestica Borkh. (Xu et al., 2014), Cucumis sativus L. (Wei et al., 2014), Solanum tuberosum L. (Bao et al., 2019; Wang et al., 2019b), Solanum lycopersicum L. (Parapunova et al., 2014), Populus euphratica Oliv. (Ma et al., 2016), Z. mays (Chai et al., 2017), and Panicum virgatum L. (Zheng et al., 2019). Despite the wide-ranging identification of TCPs, just a few of them have been intensively studied. A non-canonical basic-helix-loop-helix (bHLH) motif was at the N-terminal (about 59 amino acids in length) of all the TCP TFs, commonly referred to as the TCP domain. The TCP domain probably associated with the modulation of DNA binding (Kosugi and Ohashi, 2002) as well as protein–protein interactions (Dhaka et al., 2017). Based on the amino acid residue sequences of the TCP domain, TCPs can be categorized into two classes. For class I, it also referred to as the PCF or TCP-P class, while class II referred to the TCP-C class. The consensus DNA-binding site of class I TCPs is GGNCCCAC, however, which of class II is GTGGNCCC (Kosugi and Ohashi, 2002). Furthermore, class II was categorized into two different subclasses: CIN and CYC/TB1. The R domain rich in arginine with unknown function is present in all members of CYC/TB1 and probably mediating protein interactions through a coiled coil (Lupas et al., 1991; Cubas et al., 1999).
Increasing experimental evidence has showed that TCPs play a versatile regulatory role at the different stage of plant growth and development, for instance in leaf morphogenesis (Palatnik et al., 2003), trichome formation (Vadde et al., 2018), flower and fruit development (Nag et al., 2009; Koyama et al., 2011), and hormone biosynthesis, as well as in the response to varies stresses (Danisman, 2016; Zhang et al., 2018; Bao et al., 2019; Liu et al., 2019). Twenty-four AtTCPs have been identified in A. thaliana. AtTCP18, a member of the CYC/TB1 subgroup, regulated two florigen proteins (FT and TSF) to repress the switching from axillary meristems to premature floral (Niwa et al., 2013). AtTCP21 regulated the transcription of TOC1 and suppresses the expression of CCA1 to control the circadian clock (Pruneda-Paz et al., 2009; Giraud et al., 2010). Intriguingly, TCP regulated plant growth and development mainly by regulating the biosynthesis of bioactive substances, especially auxin (Challa et al., 2019), ethylene (Liu et al., 2019), gibberellins (Ferrero et al., 2019), brassinosteroids (Guo et al., 2010), jasmonic acid (Danisman et al., 2012), and flavonoids (Viola et al., 2016; Chahel et al., 2019). AtTCP14/15 directly interacted with GA20ox1, a key gene in gibberellin biosynthesis, to control the length of petiole and hypocotyl (Ferrero et al., 2019). In addition, AtTCP15 also directly activates the SAUR63 gene subfamily to participate in gibberellin-dependent stamen filament elongation (Gastaldi et al., 2020). AtTCP15 also serves as a repressor of anthocyanin accumulation by influencing the expression of anthocyanin biosynthesis genes and upstream transcriptional regulators (Viola et al., 2016).
MicroRNAs (miRNAs) are small single-stranded, non-coding RNAs that are typically 20–24 nucleotides (nt) in length. Usually, miRNAs regulate target gene expression through mRNA cleavage at the post-transcriptional level or translational repression (Bartel, 2009; Taylor et al., 2014; Cui et al., 2017). Since most of the target genes of miRNAs are TFs, which can realize the cascade amplification of signals through transcriptional regulation of their target genes, miRNAs have gradually become a potential target gene for crop improvement due to their function in plant growth, development (Axtell et al., 2007), and stress response (Silvestri et al., 2019; Wang et al., 2019a; Kang et al., 2020). In Arabidopsis, eight CIN-like TCP genes out of 24 AtTCPs, including miRJAW–targeted AtTCP2, AtTCP3, AtTCP4, AtTCP10, and AtTCP24, as well as miRJAW–resistant AtTCP5, AtTCP13, and AtTCP17, were found to chord with different pathways to regulate leaf development (Palatnik et al., 2003; Koyama et al., 2007). The down-regulated expression of AtTCP2/4 caused by miR319 target-cleavage resulted in serrated leaves (Palatnik et al., 2003). In switchgrass, miR319 negatively regulated PvTCPs to promote ethylene accumulation and enhance salt tolerance (Liu et al., 2019). In cotton, high levels of GhTCP4, the target of miR319, repressed a homeobox-containing factor, GhHOX3 transcriptional activity, to promote the secondary cell wall biosynthetic pathway in fiber cells, resulting in shorter fibers and thicker walls (Cao et al., 2020).
Allohexaploid sweet potato, a major global root and tuber crop, is a significant component as to subsistence agriculture because of its capability to guarantee food security and improving nutrition status regionally (Liu, 2014). However, due to the largeness and complexity of the genome, research on the molecular genetics of sweet potato has lagged behind that of other crops, such as rice and potato. Recently, its whole genome was sequenced (Yang et al., 2017). Additionally, its diploid relative wild species Ipomoea trifida (Kunth) G. Don and Ipomoea triloba L. have also been sequenced (Wu et al., 2018). These genomic sequences can serve as a reference for the hexaploid sweet potato genome. As we still know little about the TCPs family in sweet potato, an analysis concerning TCP gene family in sweet potato was performed globally in the present study. Eighteen IbTCP genes were ascertained and cloned, and phylogenetic relationships, chromosomal locations, and tissue-specific expression were carried out. Furthermore, we found that the transcription patterns of IbTCPs discrepant during leaf development. IbTCP11 and IbTCP17 were significantly up-regulated in the mature leaves. Blocking IbmiR319 led to an elongated chloroplast and decreased net photosynthetic rate, which further confirmed the critical roles of IbmiR319-targeted IbTCPs in leaf anatomical morphology. As a result, our data offers detailed information of IbTCPs category and helps elucidate the function of IbTCPs in leaf anatomical morphology in sweet potato.
Materials and Methods
Plant Materials
The wild-type (WT) sweet potato (Ipomoea batatas L.) cultivar “Xushu 22” (Xu22), developed by the Sweet Potato Research Institute of the China Agriculture Academy of Science, was used for IbTCP gene cloning. MIM319, overexpressing an artificial IbmiR319 target mimicry to sequester the normal expression of native IbmiR319, was generated using an Agrobacterium-mediated embryogenic calli transformation. Untransformed (WT) and transgenic plants transplanted and planted as described before (Yang et al., 2011). The cuttings of 3–4 cm height plants were transplanted grown into the field at the experimental station of Jiangsu Normal University (E 117°17.48′, N 34°16.95′, Jiangsu, China) were used for the assessment of the phenotype and agronomic traits. The plants grown in greenhouses were used for RNA extraction, and those grown in the field were used for morphological observations and photosynthesis parameter measurements.
Identification and Evolutionary Analysis
The TCP protein sequences of Arabidopsis and rice retrieved from the database PlantTFDB1 or TAIR2 were used to perform protein to protein BLASTP searching with the e-value of 10–5 in the I. batatas genome database3 and its two wild ancestors (I. trifida and I. triloba) genomics database.4 All sequences were further validated by the conserved domains database (CDD).5 We named IbTCP1 to IbTCP18 in the light of their allocations in the genome and relative orders on each chromosome. To study the evolutionary relationships of IbTCPs, Clustal X 2.0 was used to align the entire protein sequences of TCPs from sweet potato, Arabidopsis, and rice, which were then used to construct an evolutionary tree in MEGA 6.0 as described before (Tamura et al., 2013).
Sequence Analysis of IbTCP Genes
Using the ExPASy proteomics server,6 the molecular weight (MW) and isoelectric points (pI) of the IbTCPs were predicted.7 The conserved motifs of the IbTCPs were ascertained with CDD and the ExPASy proteomics server.
Subcellular Localization of IbTCPs
The coding sequence (CDS) without the termination codon of the IbTCP11 and IbTCP17 were amplified through PCR, and then purified and cloned into pCAMBIA2300-35S-eGFP to obtain subcellular localization vectors 35S: IbTCP11-GFP and 35S:GFP-IbTCP17. The Agrobacterium tumefaciens EHA105 strain harboring 35S: IbTCP11-GFP or 35S:GFP-IbTCP17 was infected into leaf of N. benthamiana. The IbTCP-GFP fusion proteins were detected as described before (Wang et al., 2018). The sequences of primers were also listed in Supplementary Table 1.
Identification of IbmiR319 and Prediction of IbmiR319 Target Genes
The sequence of IbmiR319 was acquired from our microRNA library of sweet potato based on high-throughput sequencing (Xie et al., 2017; Tang et al., 2020). To forecast the target sites of IbmiR319, the CDS of the IbTCPs was analyzed using the psRNATarget online tool.8
Plasmid and Agrobacterium-Mediated Sweet Potato Transformation
The miR319 target mimicry vector p35S-MIM319 was constructed as previously described (Franco-Zorrilla et al., 2007). Genetic transformation of sweet potato was implement as previously described by Yang et al. (2011) to obtained the transgenic plant MIM319.
Gene Expression Analysis
qRT-PCR was carried out to check the expression profiles of genes in different tissues, including the shoot buds (Sb), young leaf (YL), mature leaf (ML), stem (S), fibrous roots (FR), pencil roots (PR), and developing storage roots (DR), or leaves at the different developmental stages (L1–L10: the 1st through 10th leaves counted from the stem tip). Total RNAs was extracted from the above samples using TRIzol (Invitrogen) as described before (Meng et al., 2018). The inverse transcription and the qRT-PCR were performed as described before (Meng et al., 2018). The IbActin gene was used as an internal control. Data from three biological samples were collected, and the mean values were normalized to IbActin. NRT (no reverse transcription control) and NTC (no template control) were also implemented for each gene analysis. The 2–ΔΔCt method was used to judge the relative expression level of IbTCPs (Livak and Schmittgen, 2001). The sequences for the primers are listed in Supplementary Table 1.
Leaf Anatomical Morphology
After 2 months of growth, the third fully-expanded leaf counted from the terminal bud was removed and cross sectioned by hand for optical microscopic examinations and measurement of leaf thickness.
For transmission electron microscopy (TEM) observation, the first fully-expanded fresh leaves were resected and cut into small pieces immediately, fixed with electron microscope fixator (Servicebio, G1102, Wuhan Sevier Biotechnology Co. Ltd.). All the procedure were performed according the described online.9
Chlorophyll Content and Photosynthesis Parameter Measurements
The cuttings of WT and MIM319 with four to six expanded leaves transplanted to the field for 4 weeks were used for analysis. The chlorophyll content assay was performed using the third fully-expanded leaves of WT and MIM319 according to the described before (Kuo et al., 2019). The total chlorophyll content was figured out according to the formula in CT = 20.29A645−8.05A663. CT: total chlorophyll content. A645, A663: the absorbance of the chlorophyll solution at 645 nm and 663 nm, respectively.
Leaf photosynthesis parameters on the third fully-expanded leaves were scaled using a Li-6400 portable photosynthetic system (Ll-6400, Li-COR) on a sunny day. Ten seedlings per line were scaled.
Chlorophyll fluorescence was measured as described (Kuo et al., 2019). Briefly, after adapting to the dark for 30 min, the Fv/Fm values of the third fully-expanded leaves were measured using a Li-6400 system. Ten seedlings per line were measured in total.
Results
Identification and Cloning of the IbTCP Gene Family in Sweet Potato
To obtain TCP genes in sweet potato, the TCP amino acid sequences of Arabidopsis and O. sativa were utilized as a reference to BLAST against the hexaploid sweet potato Genome Database, its diploid wild ancestor Sweetpotato Genomics Resource database, and PlantTFDB. After removing the redundant sequences, a total of 18 predicted non-redundant IbTCP sequences were identified and further cloned, all of which comprised the conserved TCP domain (Table 1 and Figures 1A,B). Their sequences were listed in Supplementary Table 2. The CDS lengths of the IbTCPs varied from 396 bp (IbTCP8) to 1527 bp (IbTCP5). IbTCP8, with 131 amino acid residues (aa), was regarded as the shortest IbTCP, whereas the largest was IbTCP5 with 508 amino acid residues. The MW was in the range of 14.703–52.774 kDa, and the variation of pI is from 5.53 to 9.83 (Table 1).
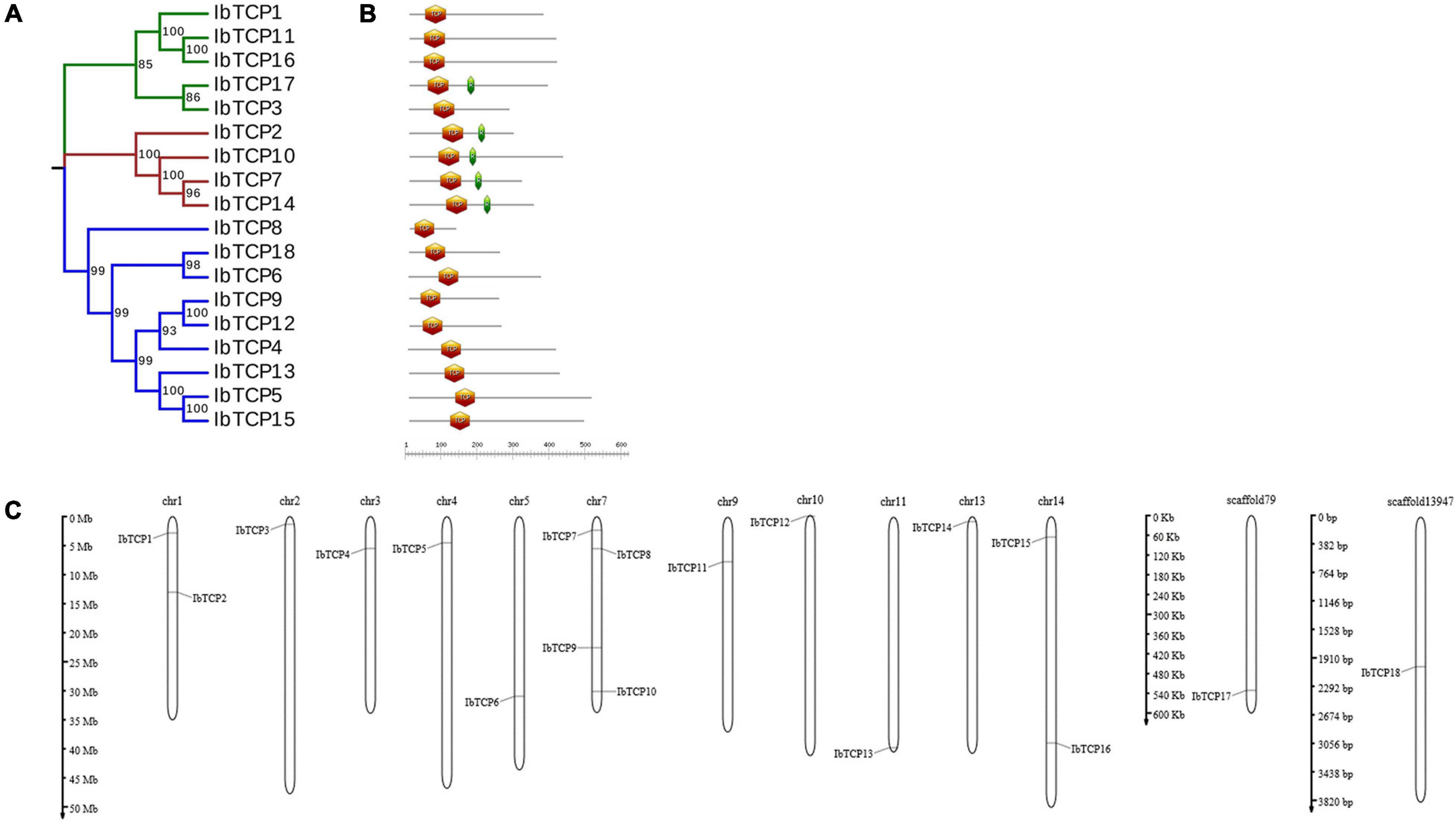
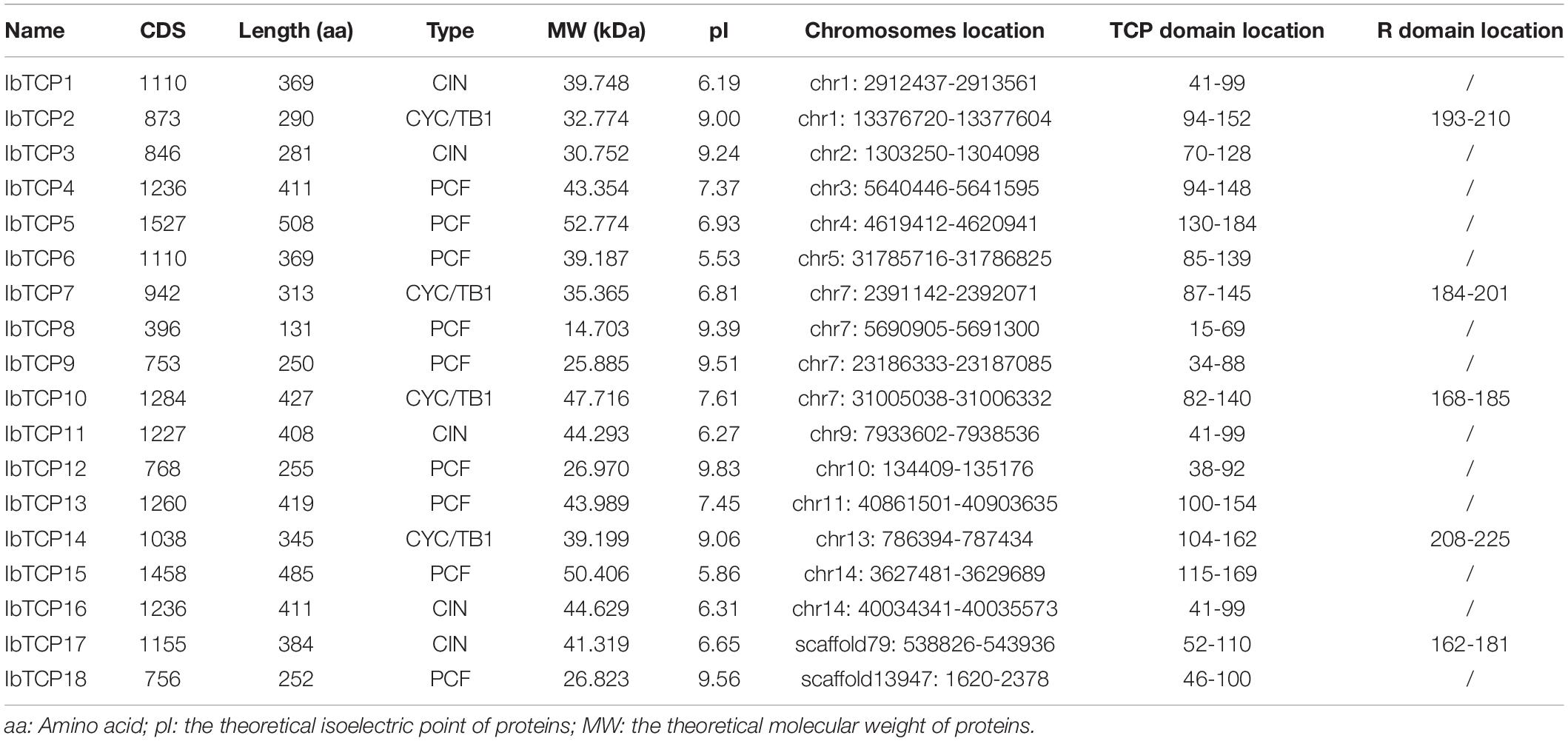
Figure 1. Distribution, phylogenetic tree, and conserved motifs analysis of IbTCPs in sweet potato. (A) Phylogenetic tree of 18 IbTCP genes. The phylogenetic tree was generated using the neighbor-joining method with 1000 bootstrap iterations. Numbers at each node represent the bootstrap values. The blue, green, and red colors indicate the PCF, CIN, and CYC/TB1 clades. (B) Conserved motifs analysis. Orange hexagon indicated the TCP domain, and green prism indicated the R domain. The scale refers to the amino acid residues lengths of the TCP proteins. (C) Distribution of IbTCPs on sweet potato chromosomes or scaffolds. The scale refers to the lengths of the chromosomes or scaffolds in hexaploid sweet potato.
To determine gene allocation pattern on chromosome, investigation of the exact genomic positions of them were carried out. These 18 IbTCPs were scattered across 11 chromosomes and two scaffolds of sweet potato (Table 1 and Figure 1C). Among them, the IbTCPs on chromosomes 2, 10, and 13 were tightly located on the upper end of the arm, while on chromosome 11, they were located tightly on the lower end of the arm. Chromosome 7 contained the highest number of IbTCPs, including IbTCP7, IbTCP8, IbTCP9, and IbTCP10. Chromosomes 1 and 14 contained two loci each.
Phylogenetic Analysis and Category of the IbTCP Family
A phylogenetic tree was structured aimed to investigating about the categorization and evolutionary relationships of sweet potato TCP proteins. A total of 88 TCP full-length amino acid sequences, including 18 IbTCPs, 31 AtTCPs, 23 ZmTCP, seven OsTCPs, and nine SlTCPs, were assembled to structure a phylogenetic tree using the NJ method with 1000 bootstrap replicates (Figure 2 and Supplementary Table 3). In accordance to the categorization in Arabidopsis, the IbTCPs may be divided into two TCP classes as well. Nine IbTCPs comprised Class I, while the other nine pertain to Class II, which could be classified into two subclasses: four members of the CYC/TB1 clade and the five members of the CIN clade (Figures 1A, 2). The results suggested that all the TCPs were evolutionarily conserved.
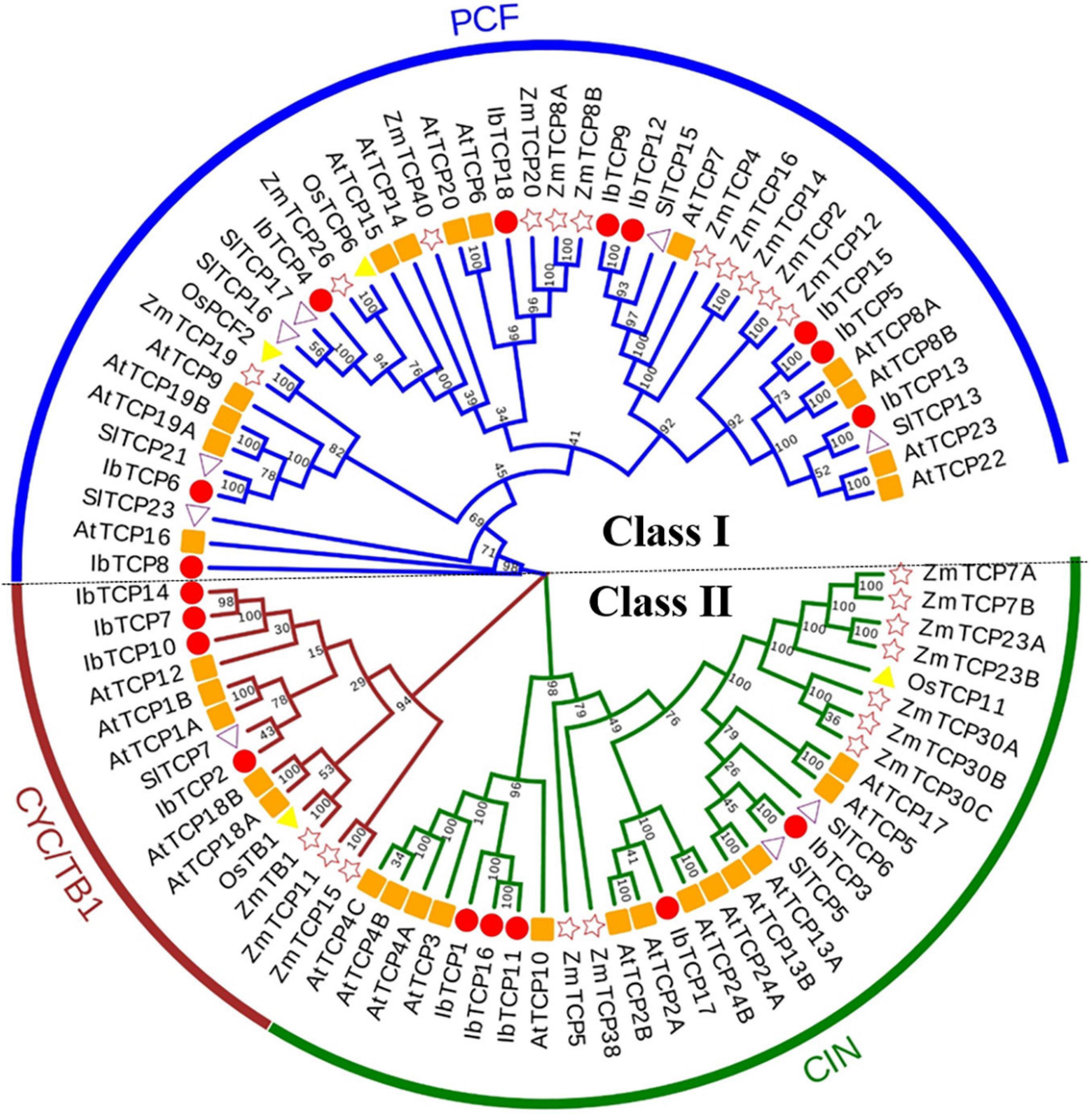
Figure 2. Phylogenetic analysis of TCP TFs from I. batata (IbTCP), Arabidopsis (AtTCP), rice (OsTCP), maize (ZmTCP), and S. lycopersicum (SlTCP). The phylogenetic tree was constructed from 88 full-length protein sequences from I. batata (18), Arabidopsis (31), rice (7), maize (23), and S. lycopersicum (9) using the neighbor-joining method in MEGA 6.0 with 1000 bootstrap replicates.
Conserved Domain Analysis and Recognition Sequence of miR319
To further comprehend the evolutionary relationships of IbTCPs in sweet potato, the domains of the IbTCPs were confirmed using the ScanProsite tool.10 As expected, all 18 IbTCPs displayed a highly conserved TCP domain that incorporated a bHLH-type motif located near the N-terminal (Figures 1B, 3A). There’s a large difference in the components of the loop and helixes I and II between the class I and II. By analyzing the phylogenetic tree and aligning the TCP domains, the IbTCP proteins are anticipated to be categorized into two classes (Figure 1A), as has been indicated for all species analyzed up to now. The conserved R domain was only found in IbTCP2, IbTCP7, IbTCP10, IbTCP14, and IbTCP17, all of which are members of Class II (Figures 1B, 3B).
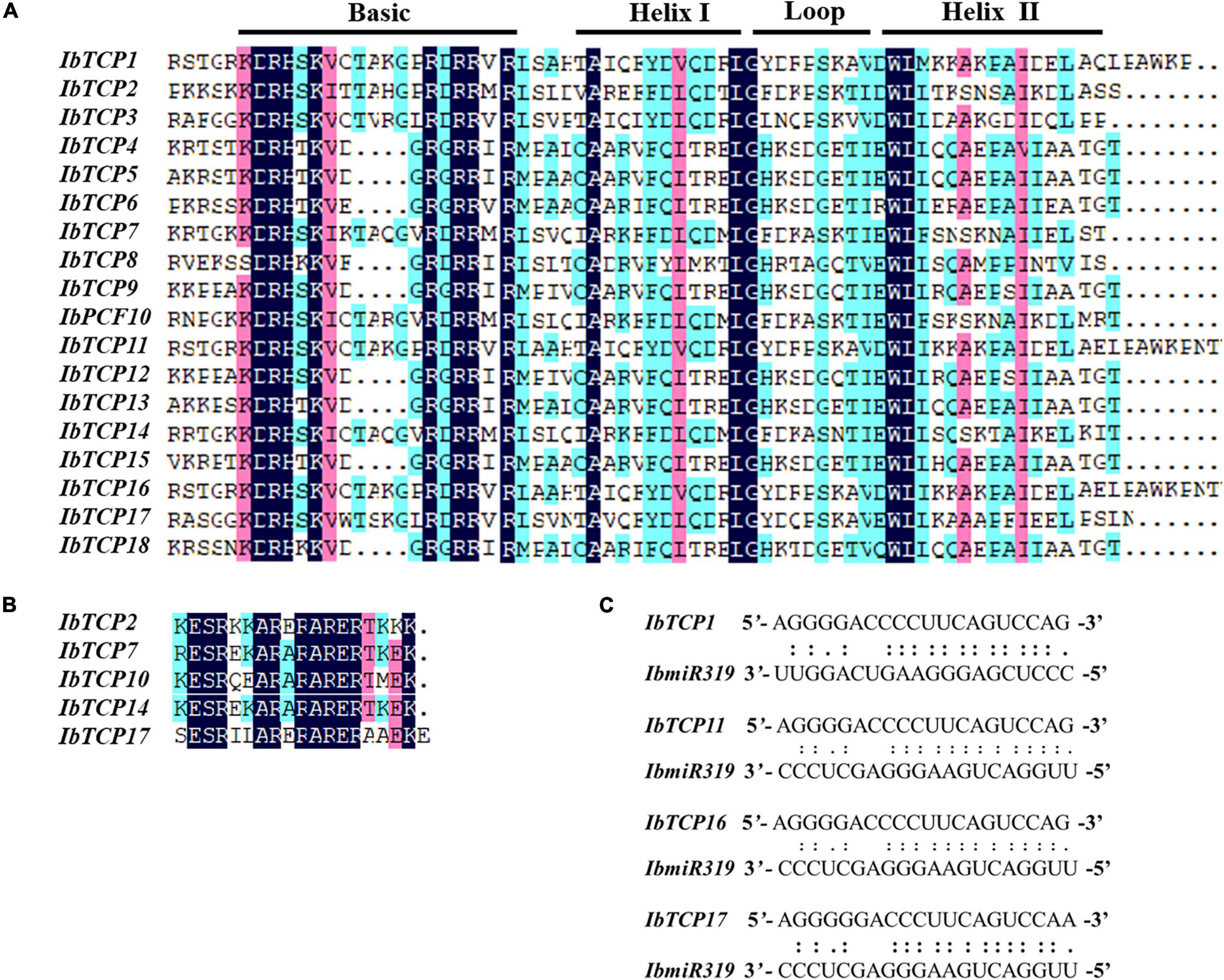
Figure 3. Multiple sequence alignment of IbTCP proteins and the IbmiR319-targeted IbTCPs. (A) Alignment of the TCP domain of the 18 TCP proteins in sweet potato. Shaded in different colors indict the conserved amino acids. Conserved domains, including Basic, Helix I, Loop, and Helix II, are shown at the top. (B) Alignment of the R-domain of the class II members. Shaded in different colors indict the conserved amino acids. (C) Complementarities of IbmiR319 and its target IbTCP1, IbTCP11, IbTCP16, and IbTCP17 mRNAs.
MicroRNA319 (miR319) pertains to one of the most ancient and conserved miRNA families. Previous studies have verified that miR319, targeting TF TCP genes, plays important roles in plant growth, morphogenesis, and reproduction (Palatnik et al., 2003; Sun et al., 2017; Cao et al., 2020). The IbmiR319 target sites among the 18 IbTCPs were analyzed using psRNATarget online. The putative recognition sites of IbmiR319 were found in IbTCP1, IbTCP11, IbTCP16, and IbTCP17 (Figure 3C). These four IbTCP genes all belonged to class II, which corroborates a previous study in Arabidopsis. Although mismatches existed at the 3′ end of the IbmiR319 and 5′ end of the targeted IbTCP, kernel sequences (3′-GGGAAGUCAGGU-5′) were conserved (Figure 3C). These data illustrate that miR319 maintained homologous target interactions in time of the evolution and diversification of plants.
Expression Profiles of IbTCP Genes in Different Tissues
To obtain credible information of the growth and developmental functions of IbTCP in sweet potato, their organic-specific expression patterns, containing the shoot buds, young leaf, mature leaf, stem, fibrous roots, pencil roots, and developing storage roots, were analyzed by qRT-PCR. As indicated in Figure 4, even though all of IbTCPs were expressed in all seven tissues tested, there’s considerable variation in transcription levels of different genes among different tissues. Generally speaking, the expression levels of IbTCPs were relatively low in the belowground organs, whereas constitutively high expression in the aboveground organs examined, especially ten IbTCPs (IbTCP2, IbTCP3, IbTCP5, IbTCP7, IbTCP8, IbTCP9, IbTCP10, IbTCP12, IbTCP17, and IbTCP19) showed highly expression levels in the mature leaves. This finding implies that these IbTCP genes probably perform different functions during growth and development. Interestingly, the relatively higher expression of all the IbmiR319-targeted CIN subclass IbTCPs (IbTCP1, IbTCP11, IbTCP16, and IbTCP17) were detected in the shoot bud, young leaf, and mature leaf, while expression levels of the other IbTCPs were high mainly in the mature leaf, suggesting that they may play similar or different roles in leaf development.
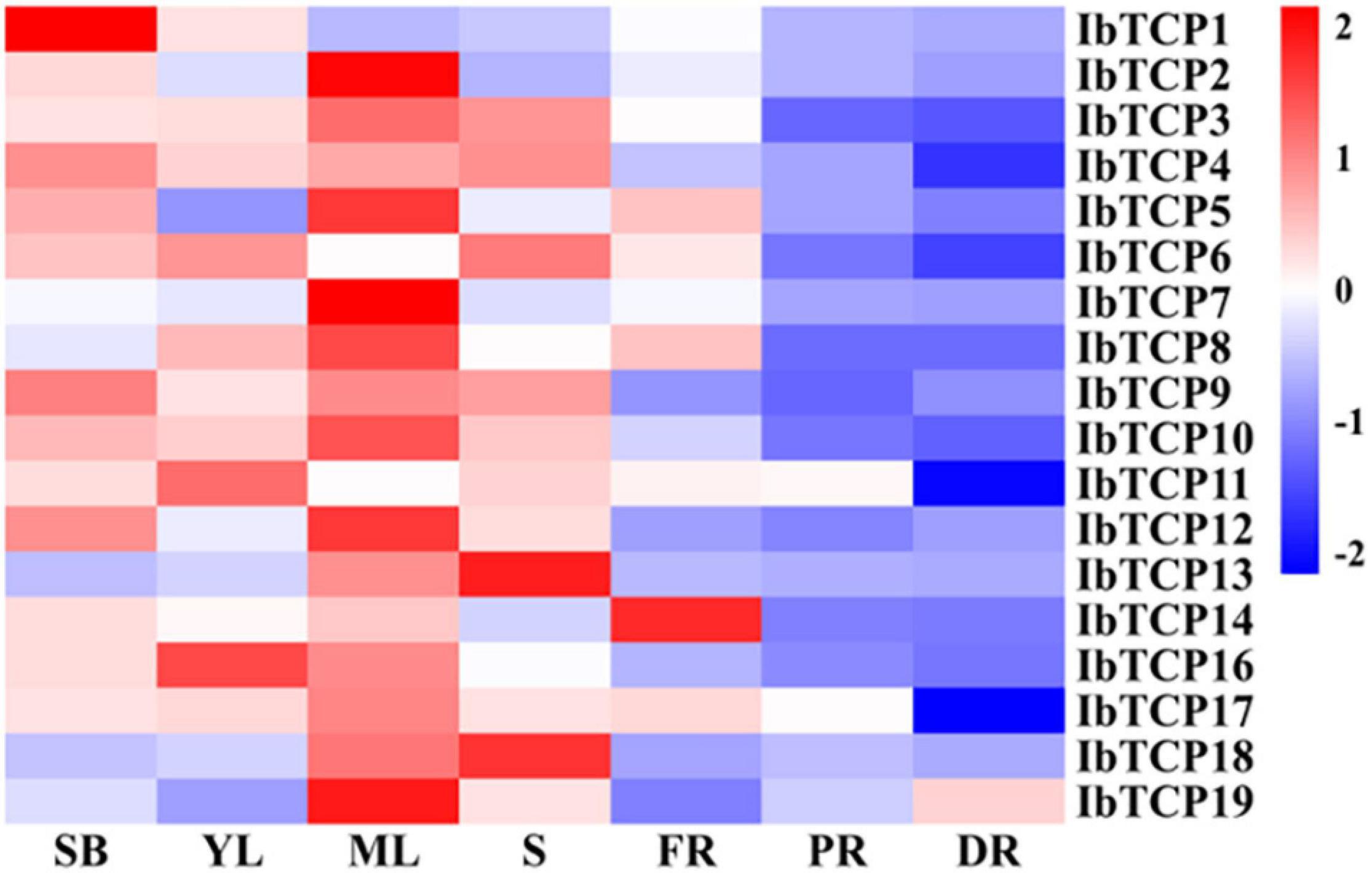
Figure 4. Heatmap representation of the gene expression of 18 IbTCPs in various tissues. The tissues used for transcription pattern analysis are listed at the bottom, shoot buds (SB), young leaves (YL), mature leaves (ML), stems (S), fibrous roots (FR), pencil roots (PR), and developing roots (DR). The genes are listed on the right of the expression bars.
IbmiR319-Targeted IbTCPs Play Vital Roles in Leaf Anatomical Morphology
Leaves, as the major photosynthetic organs of plants, take up a crucial position in plant growth and development. Leaves are also important for tuber crops, since the photosynthetic rate directly affects dry matter accumulation and storage root swelling (Sawada et al., 1999). To understand the function of IbTCPs in leaf development, their transcription levels of 18 IbTCPs at the different developmental stages of the leaves were investigated (Figure 5). The expression levels of most IbTCPs exhibited no signification changes at different developmental stages (Figures 5A,B). Yet, the transcription levels of IbmiR319-targeted IbTCPs, especially IbTCP11 and IbTCP17, presented a significant increase (Figure 5C). These data indicate that the IbmiR319-IbTCPs module is likely to associate with leaf development.
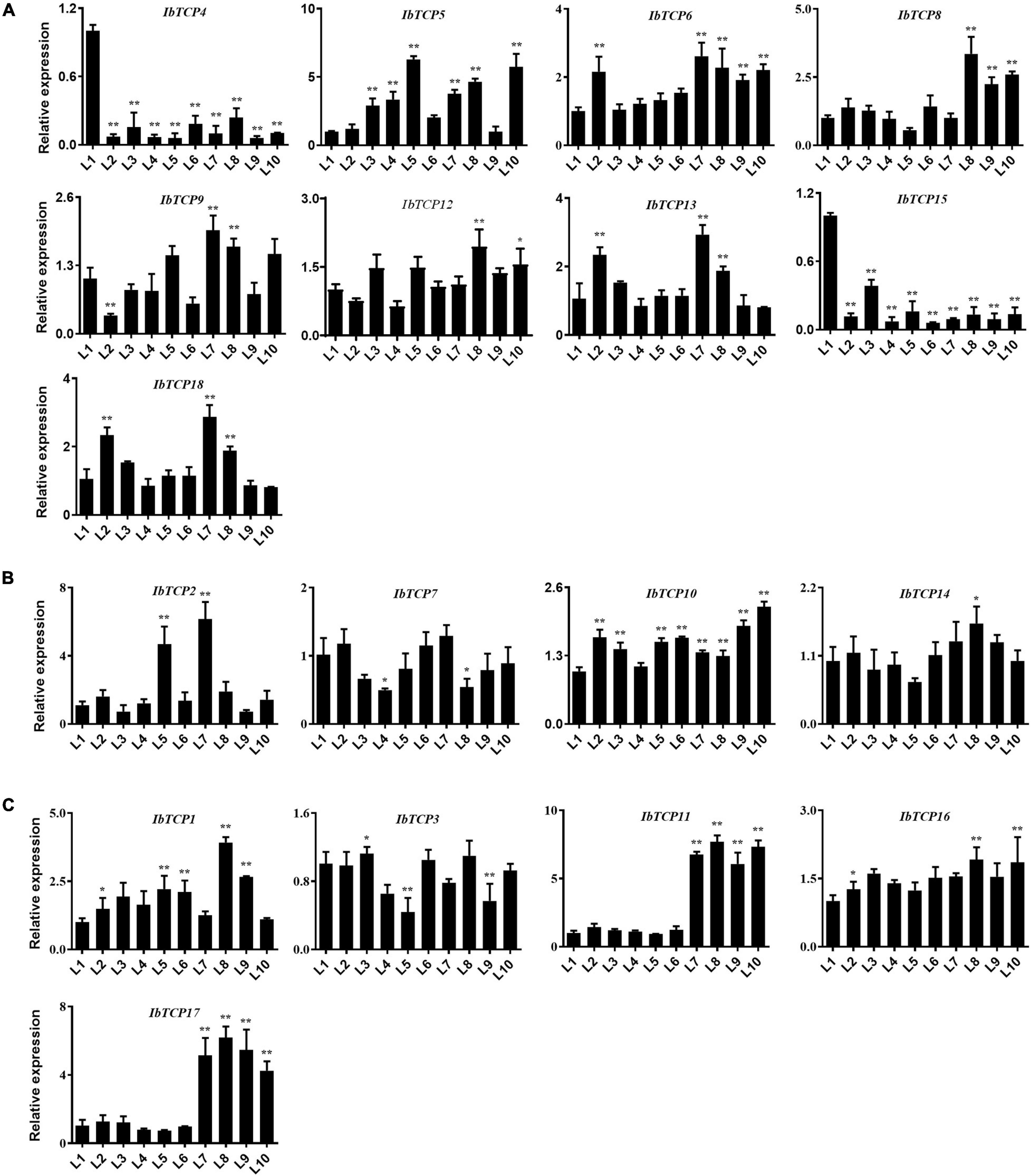
Figure 5. Expression pattern analysis of IbTCPs in varying leaf developmental periods. The qRT-PCR transcript analysis of IbTCPs of Class I (A), Class II CIN clade (B), and CYC/TB1 clade (C) in the 1st, 2nd, 3rd, to 10th leaf (L1–L10). The expression profiles were normalized using IbActin. Values are the mean and SD of three replicates. The asterisks indicate statistical significance to L1 (Student’s t-test; *P < 0.05, **P < 0.01).
For a more thorough evaluation of the duty of the IbmiR319-IbTCPs module, the subcellular localization of the IbmiR319-targeted IbTCPs was performed. IbTCP11 and IbTCP17 were fused with green fluorescence protein and infiltrated into N. tabacum epidermal cells. According to the distribution of green fluorescence signal, these two IbmiR319-target IbTCPs were specifically lay within the nuclei, implying that they are functional TFs (Figure 6). The IbmiR319 target mimicry vector p35S-MIM319 was then constructed and introduced into WT, and the stable transgenic line MIM319, which blocked IbmiR319, was generated (Supplementary Figure 1A). The expression levels of IbTCP11 and IbTCP17 were dramatically up-regulated in MIM319 compared with WT (Supplementary Figure 1B), indicating that these two IbTCPs were the targets of IbmiR319.
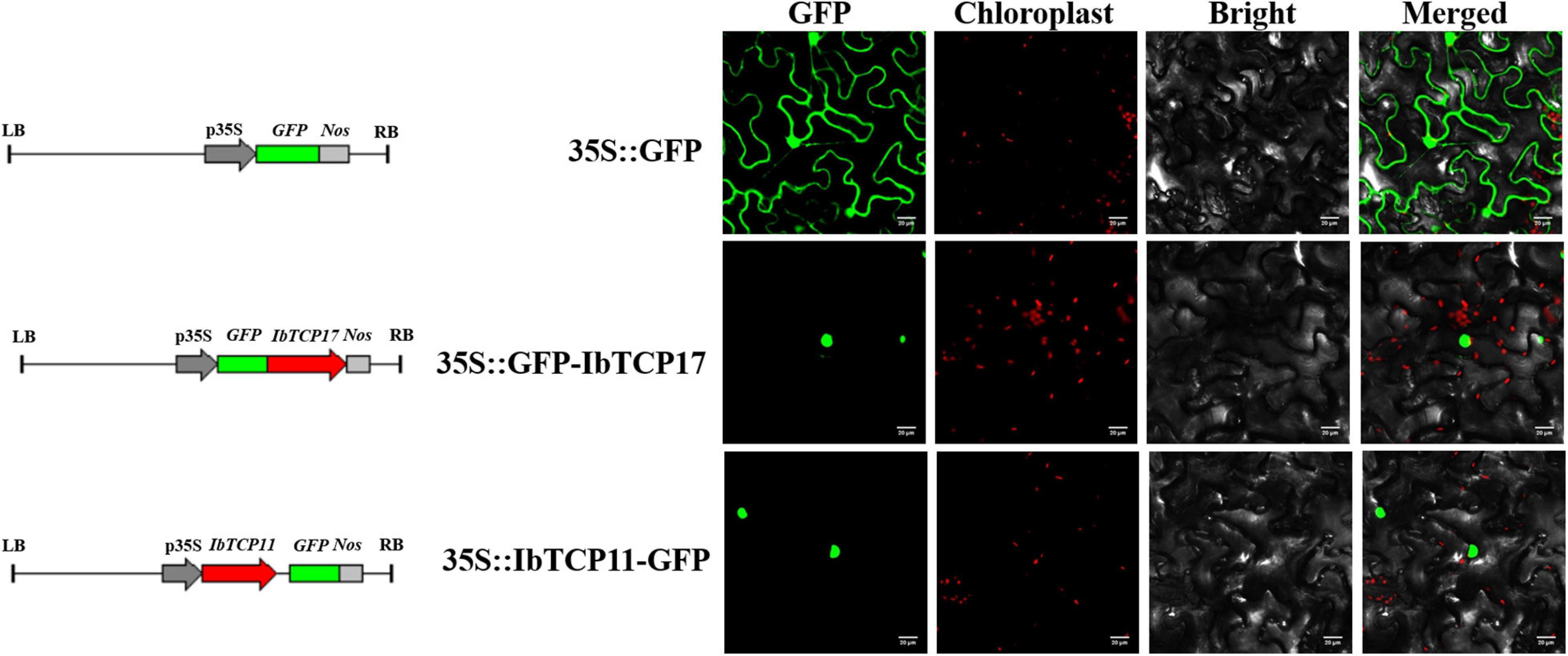
Figure 6. Subcellular localization of two GFP-fused sweet potato TCP proteins. The two IbTCP-GFP fusion proteins (IbTCP11-GFP and IbTCP17-GFP) were transiently expressed in tobacco leaves and observed by confocal microscopy 72 h later. GFP driven by the 35S promoter was used as a control. The left, middle-left, middle-right, and right panels stand for photoes taken in dark, chloroplast, bright, and merged views, respectively. Bars are 20 μm.
The up-regulation of IbTCP11 and IbTCP17 caused by blocking IbmiR319 in sweet potato led to dramatic changes in leaf size and leaf shape (Figures 7A,B). Quantitative analysis revealed that the width of the third fully-expanded leaves of MIM319 transgenic sweet potato ranged from 14.7 to 18.0 cm, which was dramatically less than that of WT (Figure 7C), and the leaf area ranged from 55.50 to 63.25 cm2, which was also significantly less than that of WT (Figure 7E). Conversely, there was no obvious difference in the length of the fully-expanded leaves between MIM319 and WT (Supplementary Figure 2). As a result, the ratio of the length to width of MIM319 was greater than that of WT (Figure 7D). Taken together, the data indicated that IbmiR319-targeted IbTCPs may have conserved functions during leaf development, similar to their functions in Arabidopsis and rice (Palatnik et al., 2003; Nag et al., 2009; Yang et al., 2013).
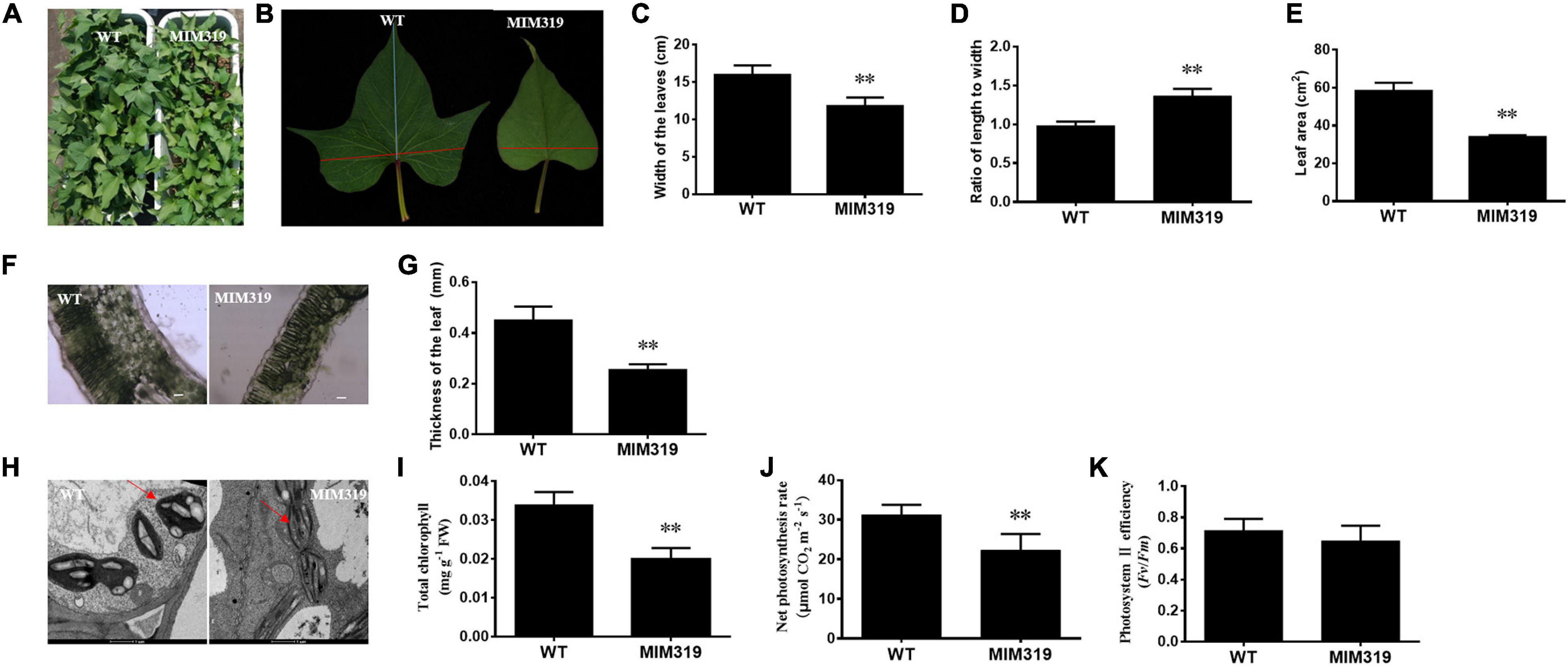
Figure 7. The phenotypes and anatomical morphology of MIM319 and WT sweet potato leaf. (A) Phenotype of the whole plant of WT and MIM319. (B) Phenotype of the third fully-expanded leaf of WT and MIM319. (C–E) The width, ratio of length to width, and area of the third fully-expanded leaf of WT and MIM319. (F) The microstructure of the hand-sliced third-fully expanded leaves of WT and MIM319 observed using an optical microscope. Bars are 50 μm. (G) The thickness of the third fully-expanded leaf of WT and MIM319. (H) The TEM image of the third fully-expanded leaf of WT and MIM319. The red arrows indicate the chloroplast. Bars indicate 1 μm in length. (I–K) The total chlorophyll, net photosynthesis rate, and photosystem II efficiency of WT and MIM319. The error bars and asterisks indicate the SD and statistical significance of 10 biological replicates (Student’s t-test; ∗∗P < 0.01), respectively.
To elucidate the cellular basis of the morphological changes, microstructure and ultrastructure observations of the leaves were carried out by electron microscopy using hand-sliced paraffin sections. Microscopic analysis of the leaf blade cross-sections revealed fewer mesophyll cells (including the palisade tissue and spongy tissue) and thinner layers of mesophyll cells in MIM319 than in WT (Figure 7F). The thickness of MIM319 leaves (0.264 mm on average) was significantly thinner than that of WT (0.464 mm on average) (Figure 7G). The chloroplasts were flatter, more elongated, and contained less starch grains compared with WT (Figure 7H). In combination, the data indicated that IbmiR319-target IbTCPs also play vital roles in leaf anatomical morphology.
Leaf size and structure are important since they impact photosynthetic efficiency (Marcotrigiano, 2010). Thinner leaves with less chlorophyll content are considered to have a detriment for the efficient usage of light energy, and lead to less photosynthetic efficiency (Higuchi et al., 1999). To address whether the changes in leaf anatomical morphology of MIM319 affect photosynthesis, some photosynthetic parameters were quantitatively measured. The total chlorophyll content of the MIM319 transgenic plants was 0.0199 mg g–1 FW on average, while that of WT was 0.0331 mg g–1 FW. The total chlorophyll contents of MIM319 were dramatically lower than that in WT (Figure 7I). Furthermore, the photosynthetic rate of the MIM319 transgenic sweet potato was 22.33 μmol CO2 m–2 s–1 on average, which was dramatically lower than 31.24 μmol CO2 m–2 s–1 in the WT (Figure 7J). Meanwhile, the efficiency of PSII in MIM319 transgenic sweet potato leaves was also examined based on the Fv/Fm values. However, there were no significant differences between MIM319 and WT (Figure 7K). Taken together, these data suggested that IbmiR319-targeted IbTCPs may play key roles in the anatomical morphology of the leaves to affect photosynthesis.
Discussion
As plant-specific TFs, TCPs have major functions during growth and development, such as leaf (including single leaf and compound leaf) development, shoot development, flower development (Ori et al., 2007; Broholm et al., 2008; Koyama et al., 2011; Bai et al., 2012), and root development (Hao et al., 2012), and are also involved in phytohormone biosynthesis (Danisman et al., 2012), lignin biosynthesis (Sun et al., 2017; Chahel et al., 2019) and the circadian clock (Giraud et al., 2010; Zhang et al., 2018). Nevertheless, there’s scarcely any precise exhaustive information of the TCP TF family in sweet potato which is the seventh largest food crop in terms of production globally and a vital tuber crop. In this study, 18 IbTCPs were identified and cloned in sweet potato and were discovered to be distributed on 11 chromosomes and two scaffolds with different densities (Figure 1C). Phylogenetic analysis revealed that 18 IbTCPs clustered into two classes according to their evolutionary relationships (Figures 1A, 2). These findings are consistent with previous classifications of TCPs from Arabidopsis, Z. mays, and S. lycopersicum (Kim et al., 2014; Parapunova et al., 2014; Chai et al., 2017). Conserved motif analysis showed that TCP domain with a bHLH-type motif existed in both class I and class II of IbTCPs, while the R domain only existed in class II (Table 1 and Figures 1B, 3A,B). Collectively, this evidence supports the classification and conservation of the sweet potato TCP TF family.
It is well-known that class II TCPs precisely regulate the transition from division to expansion (Tsukaya, 2010; Qi et al., 2017). For the leaf lamina, the timing of the transition is a significant determinant of the final size, shape, flatness, and complexity. Class II CYC/TB1 clade TCPs basically participate in the developmental regulation of axillary meristems, which results in the growth of either lateral branches or flowers. This clade included AtTCP2/3/4/5/10 and IbTCP11/17 (Figure 2). We therefore examined the expression patterns of IbTCPs to investigate whether they have essential functions in the growth and development of these organs or tissues. The expression in different tissues and different organs varied widely among IbTCP genes and different development stages of the leaf for individual TCP genes (Figures 4, 5). This states the functional divergence of IbTCPs at different stages of developmental processes, especially leaf development in sweet potato. In our study, all 18 IbTCPs showed relatively weak expression in the belowground organs, but high expression in the aboveground organs, with even IbTCP11 and IbTCP17 showing higher expression in the mature leaves (Figure 5C). AtTCP3 and AtTCP4, orthologs of IbTCP11, act as decisive modulators of de novo shoot organogenesis (Woorim et al., 2020). A previous study demonstrated that the overexpression of AtTCP4 led to in miniature cotyledons and shoot apical-meristem termination (Efroni et al., 2008), and the ectopic expression of AtTCP3 caused the failure of shoot-meristem formation (Koyama et al., 2007). Double, triple, or even quadruple knockouts for the miR319-regulated TCP genes, AtTCP2, AtTCP3, AtTCP4, and AtTCP10, demonstrate leaf crinkling to varying degrees, yet single knockouts present only slight effects on leaf morphology (Schommer et al., 2008; Bresso et al., 2018). AtTCP5 represses the initiation and outgrowth of leaf serrations by directly promoting the expression of KNAT3 and indirectly activating the transcription of SAW1 (Yu et al., 2020). Together, these data reveal the tissue expression diversity of TCPs in various plants. Compared with other plants, the great number of highly-expressed IbTCPs in the aboveground organs indicates their important function in sweet potato development, especially leaf development.
Previous study recorded that TCPs are fundamental regulators of plant growth and development, especially leaf development and senescence (Schippers, 2015). The leaf is the pivotal organ of photosynthesis, and its size and anatomical morphology directly affect photosynthetic efficiency (Marcotrigiano, 2010; Pan et al., 2018). Many genes and pathways can regulate leaf morphogenesis so as to generate organs of a wide variety of sizes and shapes (Rodriguez et al., 2014). Meanwhile, more and more experimental data show that miRNAs regulate the TFs which involved in plant development. The MiR164 family includes post-transcriptionally regulated CUC1, CUC2, and other NAC TFs. The overexpression of the miR164 family in Arabidopsis compromised organ separation, causing fusion between cotyledons and leaves, and also between the inflorescence stem and cauline leaves (Laufs et al., 2004; Mallory et al., 2004; Nikovics et al., 2006). MiR165/166 precisely regulated the expression pattern of polarity genes during leaf development in maize (Juarez et al., 2004). Increased miR408 expression leads to enhanced photosynthesis by elevating the abundance of plastocyanin to improve irradiation utilization efficiency in Arabidopsis, tobacco, and rice (Pan et al., 2018). This study indicates that blocking miR319 in sweet potato increases TCP gene expression to limit photosynthetic performance through thinner and flatter chloroplasts, a lower chlorophyll content, and the development of thinner and smaller leaves.
Our findings provide evidence for the connection between structure and function in the IbTCPs as well as lay a basis for further illustrating the functions of IbTCPs and their relationship with miR319 in sweet potato.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding authors.
Author Contributions
LR, LZ, DM, and AW designed the research. LR, HW, and TZ implemented most of the research and analysis. XG, WZ, and TW implemented IbTCPs identification, cloning, and bioinformatics analysis. LZ contributed analytical tools. LR, LZ, and AW wrote the manuscript. LZ and DM modified the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key R&D Program of China (2018YFD1000705 and 2018YFD1000700), the China Agriculture Research System of MOF and MARA (Grant No. CARS-10-B03), the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (Grant No. 18KJB180005), the Project of Xuzhou Science and Technology Key R&D Program (Grant No. KC18143), the Research Support Program for Teachers’ Doctoral Degree of Jiangsu Normal University (Grant No. 18XLRS014), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), the Undergraduate Innovation and Entrepreneurship Training Program of Jiangsu (Grant No. 201910320114Y), and the Research Innovation Program for College Graduates of Jiangsu Normal University (Grant No. 2020XKT497).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.686698/full#supplementary-material
Supplementary Figure 1 | The expression of IbmiR319 (A) and its target genes IbTCP11, 17 (B) in MIM319.
Supplementary Figure 2 | The length of leaf in MIM319.
Footnotes
- ^ http://planttfdb.gao-lab.org/
- ^ https://www.arabidopsis.org/
- ^ http://public-genomes-ngs.molgen.mpg.de/SweetPotato/
- ^ http://sweetpotato.plantbiology.msu.edu/
- ^ https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
- ^ https://prosite.expasy.org/scanprosite/
- ^ https://web.expasy.org/compute_pi/
- ^ http://plantgrn.noble.org/psRNATarget/home
- ^ https://www.servicebio.cn/data-detail?id=4305&code=DJSYBG
- ^ https://prosite.expasy.org/prosite.html
References
Axtell, M. J., Snyder, J. A., and Bartell, D. P. (2007). Common functions for diverse small RNAs of land plants. Plant Cell. 19, 1750–1769. doi: 10.1105/tpc.107.051706
Bai, F., Reinheimer, R., Durantini, D., Kellogg, E. A., and Schmidt, R. J. (2012). TCP transcription factor, BRANCH ANGLE DEFECTIVE 1 (BAD1), is required for normal tassel branch angle formation in maize. Proc. Natl. Acad. Sci. U. S. A. 109, 12225–12230. doi: 10.1073/pnas.1202439109
Bao, S., Zhang, Z., Lian, Q., Sun, Q., and Zhang, R. (2019). Evolution and expression of genes encoding TCP transcription factors in Solanum tuberosum reveal the involvement of StTCP23 in plant defence. BMC Genet. 20:91. doi: 10.1186/s12863-019-0793-1
Bartel, D. P. (2009). MicroRNAs: target Recognition and Regulatory Functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Bresso, E. G., Chorostecki, U., Rodriguez, R. E., Palatnik, J. F., and Schommer, C. (2018). Spatial Control of Gene Expression by miR319-Regulated TCP Transcription Factors in Leaf Development. Plant Physiol. 176, 1694–1708. doi: 10.1104/pp.17.00823
Broholm, S. K., Tahtiharju, S., Laitinen, R. A. E., Albert, V. A., Teeri, T. H., and Elomaa, P. (2008). A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc. Natl. Acad. Sci. U. S. A. 105, 9117–9122. doi: 10.1073/pnas.0801359105
Cao, J.-F., Zhao, B., Huang, C.-C., Chen, Z.-W., Zhao, T., Liu, H.-R., et al. (2020). The miR319-Targeted GhTCP4 Promotes the Transition from Cell Elongation to Wall Thickening in Cotton Fiber. Mol. Plant 13, 1063–1077. doi: 10.1016/j.molp.2020.05.006
Chahel, A. A., Zeng, S., Yousaf, Z., Liao, Y., Yang, Z., Wei., X., et al. (2019). Plant-specific transcription factor LrTCP4 enhances secondary metabolite biosynthesis in Lycium ruthenicum hairy roots. Plant Cell Tissue Org. Cult. 136, 323–337. doi: 10.1007/s11240-018-1518-2
Chai, W., Jiang, P., Huang, G., Jiang, H., and Li, X. (2017). Identification and expression profiling analysis of TCP family genes involved in growth and development in maize. Physiol. Mol. Biol. Plants 23, 779–791. doi: 10.1007/s12298-017-0476-1
Challa, K. R., Rath, M., and Nath, U. (2019). The CIN-TCP transcription factors promote commitment to differentiation in Arabidopsis leaf pavement cells via both auxin-dependent and independent pathways. PLoS Genet. 15:e1007988. doi: 10.1371/journal.pgen.1007988
Cubas, P., Lauter, N., Doebley, J., and Coen, E. (1999). The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18, 215–222. doi: 10.1046/j.1365-313x.1999.00444.x
Cui, J., You., C., and Chen, X. (2017). The evolution of microRNAs in plants. Curr. Opin. Plant Biol. 35, 61–67. doi: 10.1016/j.pbi.2016.11.006
Danisman, S. (2016). TCP Transcription Factors at the Interface between Environmental Challenges and the Plant’s Growth Responses. Front. Plant Sci. 7:1930. doi: 10.3389/fpls.2016.01930
Danisman, S., van der Wal, F., Dhondt, S., Waites, R., de Folter, S., Bimbo, A., et al. (2012). Arabidopsis Class I and Class II TCP Transcription Factors Regulate Jasmonic Acid Metabolism and Leaf Development Antagonistically. Plant Physiol. 159, 1511–1523. doi: 10.1104/pp.112.200303
Dhaka, N., Bhardwaj, V., Sharma, M. K., and Sharma, R. (2017). Evolving Tale of TCPs: new Paradigms and Old Lacunae. Front. Plant Sci. 8:479. doi: 10.3389/fpls.2017.00479
Doebley, J., Stec, A., and Hubbard, L. (1997). The evolution of apical dominance in maize. Nature 386, 485–488. doi: 10.1038/386485a0
Efroni, I., Blum, E., Goldshmidt, A., and Eshed, Y. (2008). A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20, 2293–2306. doi: 10.1105/tpc.107.057521
Ferrero, L. V., Viola, I. L., Ariel, F. D., and Gonzalez, D. H. (2019). Class I TCP Transcription Factors Target the Gibberellin Biosynthesis Gene GA20ox1 and the Growth-Promoting Genes HBI1 and PRE6 during Thermomorphogenic Growth in Arabidopsis. Plant Cell Physiol. 60, 1633–1645. doi: 10.1093/pcp/pcz137
Franco-Zorrilla, J. M., Valli, A., Todesco, M., Mateos, I., Puga, M. I., Rubio-Somoza, I., et al. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39, 1033–1037. doi: 10.1038/ng2079
Gastaldi, V., Lucero, L. E., Ferrero, L. V., Ariel, F. D., and Gonzalez, D. H. (2020). Class-I TCP Transcription Factors Activate the SAUR63 Gene Subfamily in Gibberellin-Dependent Stamen Filament Elongation(1)(OPEN). Plant Physiol. 182, 2096–2110. doi: 10.1104/pp.19.01501
Giraud, E., Ng, S., Carrie, C., Duncan, O., Low, J., Lee, C. P., et al. (2010). TCP Transcription Factors Link the Regulation of Genes Encoding Mitochondrial Proteins with the Circadian Clock in Arabidopsis thaliana. Plant Cell 22, 3921–3934. doi: 10.1105/tpc.110.074518
Guo, Z., Fujioka, S., Blancaflor, E. B., Miao, S., Gou, X., and Li, J. (2010). TCP1 Modulates Brassinosteroid Biosynthesis by Regulating the Expression of the Key Biosynthetic Gene DWARF4 in Arabidopsis thaliana. Plant Cell 22, 1161–1173. doi: 10.1105/tpc.109.069203
Hao, J., Tu, L., Hu, H., Tan, J., Deng, F., Tang, W., et al. (2012). GbTCP, a cotton TCP transcription factor, confers fibre elongation and root hair development by a complex regulating system. J. Exp. Bot. 63, 6267–6281. doi: 10.1093/jxb/ers278
Higuchi, H., Sakuratani, T., and Utsunomiya, N. (1999). Photosynthesis, leaf morphology, and shoot growth as affected by temperatures in cherimoya (Annona cherimola Mill.) trees. Sci. Horticult. 80, 91–104. doi: 10.1016/s0304-4238(98)00221-0
Juarez, M. T., Kui, J. S., Thomas, J., Heller, B. A., and Timmermans, M. C. P. (2004). microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428, 84–88. doi: 10.1038/nature02363
Kang, T., Yu, C.-Y., Liu, Y., Song, W.-M., Bao, Y., Guo, X.-T., et al. (2020). Subtly Manipulated Expression of ZmmiR156 in Tobacco Improves Drought and Salt Tolerance Without Changing the Architecture of Transgenic Plants. Front. Plant Sci. 10:1664. doi: 10.3389/fpls.2019.01664
Kim, H. E., Choi, Y. G., Lee, A. R., Seo, Y. J., Kwon, M. Y., and Lee, J. H. (2014). Temperature dependent hydrogen exchange study of DNA duplexes containing binding sites for Arabidopsis TCP transcription factors. J. Korean Magn. Reson. Soc. 18, 52–57. doi: 10.6564/jkmrs.2014.18.2.052
Kosugi, S., and Ohashi, Y. (1997). PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9, 1607–1619. doi: 10.2307/3870447
Kosugi, S., and Ohashi, Y. (2002). , DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30, 337–348. doi: 10.1046/j.1365-313x.2002.01294.x
Koyama, T., Furutani, M., Tasaka., M., and Ohme-Takagi, M. (2007). TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19, 473–484. doi: 10.1105/tpc.106.044792
Koyama, T., Ohme-Takagi, M., and Sato, F. (2011). Generation of serrated and wavy petals by inhibition of the activity of TCP transcription factors in Arabidopsis thaliana. Plant Signal. Behav. 6, 697–699. doi: 10.4161/psb.6.5.14979
Kuo, Y. W., Lin, J. S., Li, Y. C., Jhu, M. Y., King, Y. C., and Jeng, S. T. (2019). MicroR408 regulates defense response upon wounding in sweet potato. J. Exp. Bot. 70, 469–483. doi: 10.1093/jxb/ery381
Laufs, P., Peaucelle, A., Morin, H., and Traas, J. (2004). MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131, 4311–4322. doi: 10.1242/dev.01320
Liu, Y., Li, D., Yan, J., Wang, K., Luo, H., and Zhang, W. (2019). MiR319-mediated ethylene biosynthesis, signalling and salt stress response in switchgrass. Plant Biotechnol. J. 17, 2370–2383. doi: 10.1111/pbi.13154
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luo, D., Carpenter, R., Vincent, C., Copsey, L., and Coen, E. (1996). Origin of floral asymmetry in Antirrhinum. Nature 383, 794–799. doi: 10.1038/383794a0
Lupas, A., and Dyke., M. Van, and Stock, J. (1991). Predicting coiled coils from protein sequences. Science 252, 1162–1164. doi: 10.1126/science.252.5009.1162
Ma, X., Ma, J., Fan, D., Li, C., Jiang, Y., and Luo, K. (2016). Genome-wide Identification of TCP Family Transcription Factors from Populus euphratica and Their Involvement in Leaf Shape Regulation. Sci. Rep. 6:32795.
Mallory, A. C., Dugas, D. V., Barte, D. P., and Bartel, lB (2004). MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. 14, 1035–1046. doi: 10.1016/j.cub.2004.06.022
Marcotrigiano, M. (2010). A role for leaf epidermis in the control of leaf size and the rate and extent of mesophyll cell division. Am. J. Bot. 97, 224–233. doi: 10.3732/ajb.0900102
Meng, X., Li, G., Yu, J., Cai, J., Dong, T., Sun, J., et al. (2018). Isolation, Expression Analysis, and Function Evaluation of 12 Novel Stress-Responsive Genes of NAC Transcription Factors in Sweetpotato. Crop Sci. 58, 1328–1341. doi: 10.2135/cropsci2017.12.0738
Nag, A., King, S., and Jack, T. (2009). miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 106, 22534–22539. doi: 10.1073/pnas.0908718106
Nikovics, K., Blein, T., Peaucelle, A., Ishida, T., Morin, H., Aida, M., et al. (2006). The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18, 2929–2945. doi: 10.1105/tpc.106.045617
Niwa, M., Daimon, Y., Kurotani, K.-I., Higo, A., Pruneda-Paz, J. L., Breton, G., et al. (2013). BRANCHED1 Interacts with FLOWERING LOCUS T to Repress the Floral Transition of the Axillary Meristems in Arabidopsis. Plant Cell 25, 1228–1242. doi: 10.1105/tpc.112.109090
Ori, N., Cohen, A. R., Etzioni, A., Brand, A., Yanai, O., Shleizer, S., et al. (2007). Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nature Genet. 39, 787–791. doi: 10.1038/ng2036
Palatnik, J. F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J. C., et al. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. doi: 10.1038/nature01958
Pan, J. W., Huang, D. H., Guo, Z. L., Kuang, Z., Zhang, H., Xie, X. Y., et al. (2018). Overexpression of microRNA408 enhances photosynthesis, growth, and seed yield in diverse plants. J. Integr. Plant Biol. 60, 323–340. doi: 10.1111/jipb.12634
Parapunova, V., Busscher, M., Busscher-Lange, J., Lammers, M., Karlova, R., Bovy, A. G., et al. (2014). Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol. 14:157. doi: 10.1186/1471-2229-14-157
Pruneda-Paz, J. L., Breton, G., Para, A., and Kay, S. A. (2009). A Functional Genomics Approach Reveals CHE as a Component of the Arabidopsis Circadian Clock. Science 323, 1481–1485. doi: 10.1126/science.1167206
Qi, J., Wu, B., Feng, S., Lü, S., Guan, C., Zhang, X., et al. (2017). Mechanical regulation of organ asymmetry in leaves. Nat. Plants 3, 724–733. doi: 10.1038/s41477-017-0008-6
Rodriguez, R. E., Debernardi, J. M., and Palatnik, J. F. (2014). Morphogenesis of simple leaves: regulation of leaf size and shape. Wiley Interdiscip. Rev. Dev. Biol. 3, 41–57. doi: 10.1002/wdev.115
Sawada, S., Sakai, N., and Sano, T. (1999). Feed-Forward Effects on the Photosynthetic Source-Sink Balance in Single-Rooted Leaves of Sweet Potato. Plant Prod. Sci. 2, 87–91. doi: 10.1626/pps.2.87
Schippers, J. H. M. (2015). Transcriptional networks in leaf senescence. Curr. Opin. Plant Biol. 27, 77–83. doi: 10.1016/j.pbi.2015.06.018
Schommer, C., Palatnik, J. F., Aggarwal, P., Chételat, A., Cubas, P., Farmer, E. E., et al. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6:e230. doi: 10.1371/journal.pbio.0060230
Shi, P., Guy, K. M., Wu, W., Fang, B., Yang, J., Zhang, M., et al. (2016). Genome-wide identification and expression analysis of the ClTCP transcription factors in Citrullus lanatus. BMC Plant Biol. 16:85. doi: 10.1186/s12870-016-0765-9
Silvestri, A., Fiorilli, V., Miozzi, L., Accotto, G. P., Turina, M., and Lanfranco, L. (2019). In silico analysis of fungal small RNA accumulation reveals putative plant mRNA targets in the symbiosis between an arbuscular mycorrhizal fungus and its host plant. BMC Genom. 20:169. doi: 10.1186/s12864-019-5561-0
Sun, X., Wang, C., Xiang, N., Li, X., Yang, S., Du, J., et al. (2017). Activation of secondary cell wall biosynthesis by miR319-targeted TCP4 transcription factor. Plant Biotechnol. J. 15, 1284–1294. doi: 10.1111/pbi.12715
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tang, C., Han, R., Zhou, Z., Yang, Y., Zhu, M., Xu, T., et al. (2020). Identification of candidate miRNAs related in storage root development of sweet potato by high throughput sequencing. J. Plant Physiol. 251, 153224–153224. doi: 10.1016/j.jplph.2020.153224
Taylor, R. S., Tarver, J. E., Hiscock, S. J., and Donoghue, P. C. J. (2014). Evolutionary history of plant microRNAs. Trends Plant Sci. 19, 175–182. doi: 10.1016/j.tplants.2013.11.008
Tsukaya, H. (2010). The Mechanism of Cell Cycle Arrest Front Progression Explained by a KLUH/CYP78A5-dependent Mobile Growth Factor in Developing Leaves of Arabidopsis thaliana. Plant Cell Physiol. 51, 1046–1054. doi: 10.1093/pcp/pcq051
Vadde, B. V. L., Challa, K. R., and Nath, U. (2018). The TCP4 transcription factor regulates trichome cell differentiation by directly activating GLABROUS INFLORESCENCE STEMS in Arabidopsis thaliana. Plant J. 93, 259–269. doi: 10.1111/tpj.13772
Viola, I. L., Camoirano, A., and Gonzalez, D. H. (2016). Redox-Dependent Modulation of Anthocyanin Biosynthesis by the TCP Transcription Factor TCP15 during Exposure to High Light Intensity Conditions in Arabidopsis. Plant Physiol. 170, 74–85. doi: 10.1104/pp.15.01016
Wang, H., Wang, H., Liu, R., Xu, Y., Lu, Z., and Zhou, C. (2018). Genome-Wide Identification of TCP Family Transcription Factors in Medicago truncatula Reveals Significant Roles of miR319-Targeted TCPs in Nodule Development. Front. Plant Sci. 9:774. doi: 10.3389/fpls.2018.00774
Wang, W., Liu, D., Chen, D., Cheng, Y., Zhang, X., Song, L., et al. (2019a). MicroRNA414c affects salt tolerance of cotton by regulating reactive oxygen species metabolism under salinity stress. RNA Biol. 16, 362–375. doi: 10.1080/15476286.2019.1574163
Wang, Y., Zhang, N., Li, T., Yang, J., Zhu, X., Fang, C., et al. (2019b). Genome-wide identification and expression analysis of StTCP transcription factors of potato (Solarium tuberosum L.). Comput. Biol. Chem. 78, 53–63. doi: 10.1016/j.compbiolchem.2018.11.009
Wei, Y., Gu, Z., Chu, W., Ye, L., and Yang, G. (2014). Identification and Structure Analysis of TCP Transcription Factors in Cucumber. Mol. Plant Breed. 12, 287–295.
Woorim, Y., Myung-Hwan, C., Bos, N., and Yoo-Sun, lN (2020). De Novo Shoot Regeneration Controlled by HEN1 and TCP3/4 in Arabidopsis. Plant Cell Physiol. 9:9.
Wu, S., Lau, K. H., Cao, Q. H., Hamilton, J. P., Sun, H. H., Zhou, C. X., et al. (2018). Genome sequences of two diploid wild relatives of cultivated sweetpotato reveal targets for genetic improvement. Nat. Commun. 9:12.
Xie, Z., Wang, A., Li, H., Yu, J., Jiang, J., Tang, Z., et al. (2017). High throughput deep sequencing reveals the important roles of microRNAs during sweetpotato storage at chilling temperature. Sci. Rep. 7:16578.
Xu, R., Sun, P., Jia, F., Lu, L., Li, Y., Zhang, S., et al. (2014). Genomewide analysis of TCP transcription factor gene family in Malus domestica. J. Genet. 93, 733–746. doi: 10.1007/s12041-014-0446-0
Yang, C., Li, D., Mao, D., Liu, X., Ji, C., Li, X., et al. (2013). Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.). Plant Cell Environ. 36, 2207–2218. doi: 10.1111/pce.12130
Yang, J., Bi, H.-P., Fan, W.-J., Zhang, M., Wang, H.-X., and Zhang, P. (2011). Efficient embryogenic suspension culturing and rapid transformation of a range of elite genotypes of sweet potato (Ipomoea batatas L. Lam.). Plant Sci. 181, 701–711. doi: 10.1016/j.plantsci.2011.01.005
Yang, J., Moeinzadeh, M. H., Kuhl, H., Helmuth, J., Xiao, P., Haas, S., et al. (2017). Haplotype-resolved sweet potato genome traces back its hexaploidization history. Nat. Plants 3, 696–703. doi: 10.1038/s41477-017-0002-z
Yin, Z., Li, Y., Zhu, W., Fu, X., Han, X., Wang, J., et al. (2018). Identification, Characterization, and Expression Patterns of TCP Genes and microRNA319 in Cotton. Int. J. Mol. Sci. 19:3655. doi: 10.3390/ijms19113655
Yu, H., Zhang, L., Wang, W., Tian, P., Wang, W., Wang, K., et al. (2020). TCP5 controls leaf margin development by regulating the KNOX and BEL-like transcription factors in Arabidopsis. J. Exp. Bot. 72, 1809–1821. doi: 10.1093/jxb/eraa569
Zhang, N., Wang, Z., Bao, Z., Yang, L., Wu, D., Shu, X., et al. (2018). MOS1 functions closely with TCP transcription factors to modulate immunity and cell cycle in Arabidopsis. Plant J. 93, 66–78. doi: 10.1111/tpj.13757
Zheng, A., Sun, F., Cheng, T., Wang, Y., Xie, K., Zhang, C., et al. (2019). Genome-wide identification of members of the TCP gene family in switchgrass (Panicum virgatum L.) and analysis of their expression. Gene 702, 89–98. doi: 10.1016/j.gene.2019.03.059
Keywords: Ipomoea batatas L., TCP transcription factors, miR319, chloroplast development, expression analysis
Citation: Ren L, Wu H, Zhang T, Ge X, Wang T, Zhou W, Zhang L, Ma D and Wang A (2021) Genome-Wide Identification of TCP Transcription Factors Family in Sweet Potato Reveals Significant Roles of miR319-Targeted TCPs in Leaf Anatomical Morphology. Front. Plant Sci. 12:686698. doi: 10.3389/fpls.2021.686698
Received: 27 March 2021; Accepted: 21 June 2021;
Published: 06 August 2021.
Edited by:
Turgay Unver, FicusBio, TurkeyReviewed by:
Anja Schneider, Ludwig Maximilian University of Munich, GermanyHuseyin Tombuloglu, Imam Abdulrahman Bin Faisal University, Saudi Arabia
Copyright © 2021 Ren, Wu, Zhang, Ge, Wang, Zhou, Zhang, Ma and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aimin Wang, YWltaW53YW5nQGpzbnUuZWR1LmNu; Daifu Ma, ZGFpZnVtYUAxNjMuY29t; Lei Zhang, bGVpemhhbmdAanNudS5lZHUuY24=
†These authors have contributed equally to this work
 Lei Ren1,2†
Lei Ren1,2† Lei Zhang
Lei Zhang Aimin Wang
Aimin Wang