
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 19 November 2021
Sec. Plant Pathogen Interactions
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.685533
This article is part of the Research Topic Elicitors, Secret Agents at the Service of the Plant Kingdom View all 10 articles
Agrobacterium tumefaciens can cause crown gall tumors by transferring both an oncogenic piece of DNA (T-DNA) and several effector proteins into a wide range of host plants. For the translocated effector VirE3 multiple functions have been reported. It acts as a transcription factor in the nucleus binding to the Arabidopsis thaliana pBrp TFIIB-like protein to activate the expression of VBF, an F-box protein involved in degradation of the VirE2 and VIP1 proteins, facilitating Agrobacterium-mediated transformation. Also VirE3 has been found at the plasma membrane, where it could interact with VirE2. Here, we identified AtJAZ8 in a yeast two-hybrid screening with VirE3 as a bait and confirmed the interaction by pull-down and bimolecular fluorescence complementation assays. We also found that the deletion of virE3 reduced Agrobacterium virulence in a root tumor assay. Overexpression of virE3 in Arabidopsis enhanced tumorigenesis, whereas overexpression of AtJAZ8 in Arabidopsis significantly decreased the numbers of tumors formed. Further experiments demonstrated that AtJAZ8 inhibited the activity of VirE3 as a plant transcriptional regulator, and overexpression of AtJAZ8 in Arabidopsis activated AtPR1 gene expression while it repressed the expression of AtPDF1.2. Conversely, overexpression of virE3 in Arabidopsis suppressed the expression of AtPR1 whereas activated the expression of AtPDF1.2. Our results proposed a novel mechanism of counter defense signaling pathways used by Agrobacterium, suggesting that VirE3 and JAZ8 may antagonistically modulate the salicylic acid/jasmonic acid (SA/JA)-mediated plant defense signaling response during Agrobacterium infection.
The plant pathogen Agrobacterium tumefaciens, a soil-borne gram negative bacterium, provokes crown gall disease in dicotyledonous plant species, which is characterized by neoplastic growth at infection sites (McCullen and Binns, 2006; Guo et al., 2019). During infection, Agrobacterium transfers a single-stranded DNA segment from its tumor-inducing (Ti) plasmid, the T-DNA, into the plant cell, where it integrates into the chromosomes (Beijersbergen and Hooykaas, 1993; Gelvin, 2000, 2003, 2021; Citovsky et al., 2007; Guo et al., 2019). Expression of the oncogenic genes located on T-DNA in transformed plant cells promotes uncontrolled cell proliferation and results in the formation of crown gall tumors (Nester et al., 1984). Under laboratory conditions, Agrobacterium is also able to transform non-plant organisms, such as the yeast Saccharomyces cerevisiae and fungi (Bundock and Hooykaas, 1996; Piers et al., 1996; de Groot et al., 1998). By taking advantage of this unique ability of interkingdom gene transfer, Agrobacterium has been widely used in creating transgenic plants and fungi for research and biotechnology through a method known as the Agrobacterium-mediated transformation (AMT).
Besides the T-region, the virulence (vir) region in the Ti plasmid embraces a series of genes that are essential for AMT, which facilitate T-DNA processing, delivery, and integration into host cell nucleus. Within Agrobacterium, the virulence protein VirD2, in conjunction with VirD1, recognizes and generates nicks in the border sequence surrounding the T-DNA region, releasing the T-strand from the Ti plasmid (Yanofsky et al., 1986; Herrera-Estrella et al., 1988). VirD2 remains covalently attached to the 5′ end of the T-strand in Agrobacterium, and then the VirD2–T-strand complex is delivered into host cells through a type IV secretion system (T4SS) composed of 11 VirB proteins and VirD4 (Young and Nester, 1988; Christie et al., 2005; Li and Christie, 2018). In host cells, the nuclear localization signal (NLS) of the VirD2 protein pilots the T-strand to the nucleus (Howard et al., 1992; Shurvinton et al., 1992; Tinland et al., 1992; Rossi et al., 1993). In addition, some of the virulence proteins, nowadays referred to as effector proteins, including VirE2, VirE3, VirD5, and VirF are directly translocated into host cells via the T4SS independent of the T-strand (Vergunst, 2000; Schrammeijer, 2003). VirE2 is a single-strand DNA binding protein which is supposed to coat the T-strand in the host cytoplasm to form a T-complex, protecting the T-strand from degradation by host nucleases (Citovsky et al., 1989; Gelvin, 1998; Abu-Arish et al., 2004). VirE2 can interact with VIP1, a host bZIP transcription factor, which relocalizes from the cytoplasm to the nucleus to activate defense-related genes upon Agrobacterium infection (Tzfira, 2001; Djamei et al., 2007). It has been reported that VIP1 assists in the nuclear entry of the T-complex and facilitates transformation (Tzfira, 2001; Li et al., 2005; Djamei et al., 2007). However, the importance of VIP in AMT is controversial, as no difference in transformation susceptibility between wild-type and vip1-1 mutant was observed, and a recent report pointed out that VIP1 and its homologs are not required for AMT (Shi et al., 2014; Lapham et al., 2018). The VirF protein is a host–range factor (Melchers et al., 1990; Jarchow et al., 1991) and contains an F-box motif by which it interacts with plant Arabidopsis Skp1–like (ASK1) to form an Skp1–Cullin–F-box (SCF) E3 ubiquitin ligase in the host cell (Jarchow et al., 1991; Schrammeijer et al., 2001). The SCFvirF is thought to use the host ubiquitin-26S proteasome system to promote proteolysis of VirE2, VIP1, and some other host proteins in nucleus (Tzfira et al., 2004; Zaltsman et al., 2012; García-Cano et al., 2018). Degradation of VirE2 and VIP1 uncoats the T-complex and thus, may enable integration of T-DNA into the host genome (Tzfira et al., 2004). In addition, proteasomal degradation of Arabidopsis transcription factor VIP1 and VFP4 may dampen the defense response of the host cells (Djamei et al., 2007; García-Cano et al., 2018). Some plant species like Arabidopsis, encode an endogenous F-box protein “VBF,” which can functionally replace VirF, and therefore, do not require VirF for transformation (Zaltsman et al., 2010).
VirE3 is another translocated effector that has been shown to interact with VirE2 and bind to plant importin-a, possibly acting as an “adapter” molecule between VirE2 and importin-a to assist the nuclear import of the T-complex, thereby mimicking the function of VIP1 (Lacroix et al., 2004). More recently, an interaction between VirE3 and VirE2 was found to take place at the plasma membrane, suggesting that VirE3 may be involved in the early stage of T-complex formation (Li et al., 2018). Besides, VirE3 can act as a transcriptional activator in the nucleus to modulate the plant gene expression profile through interaction with pBrp, a plant specific general transcription factor belonging to the TFIIB family (García-Rodríguez et al., 2006). One of the genes strongly induced by VirE3 was pointed out to be the gene encoding VBF, thus explaining why simultaneous deletion of virF and virE3 results in a much stronger deficiency of tumorigenicity than observed in the single mutant (Niu et al., 2015). Another gene highly induced by VirE3 was NIMIN1, which operates as a negative regulator by binding to NPR1 and reduces salicylic acid (SA)-mediated PR1 gene expression and attenuates the defense response (Weigel et al., 2005; Niu et al., 2015). Here, we found that Agrobacterium VirE3 can interact with Arabidopsis JAZ8 and that overexpression of virE3 promotes tumor formation, whereas JAZ8 overexpression makes roots recalcitrant to tumorigenesis by Agrobacterium. JAZ proteins are best known as repressors of the jasmonate response, which recruit general corepressors to prevent the transcriptional activity of MYC transcription factors (Pauwels and Goossens, 2011; Howe et al., 2018). We found that the activity of VirE3 as a plant transcription regulator was repressed by JAZ8. The jasmonic acid (JA)-regulated PDF1.2 gene expression was elevated in the virE3-overexpressor plant line, but the SA-mediated PR1 gene expression was reduced in the virE3-overexpressor plant line. Our results suggest the VirE3 and JAZ8 may antagonistically modulate each other’s activity, and that Agrobacterium uses VirE3 to shift the plant defense response during AMT from SA signaling to JA signaling, which is less deleterious for biotrophic pathogens.
Supplementary Tables 1–3 list the primer sequences, plasmids, and strains used in this study, respectively. The JAZ8 coding sequence was amplified by PCR using cDNA of Col-0 Arabidopsis thaliana with the primer set JAZ8-1302-F and JAZ8-1302-R. The PCR fragment was inserted into pCAMBIA1302 by homologous recombination, yielding JAZ8 overexpression construct JAZ8-1302. The JAZ8 sequence was amplified by PCR on plasmid JAZ8-1302 using primers listed in Supplementary Table 1 for JAZ8-pAS2.1, JAZ8-ZIM-pAS2.1, JAZ8NT-pAS2.1, JAZ8CT-pAS2.1, JAZ8-pMAL-c5X, JAZ8-cYFP, and pRT101-JAZ8 constructs. The PCR fragments were digested and inserted into pAS2.1, pMAL-c5X, pRTL2-HAYC, and pRT101, yielding corresponding constructs.
The virE3 coding sequence was amplified by PCR on plasmid pRT101-VirE3 (Niu et al., 2015) using primers listed in Supplementary Table 1 for virE3-pACT, virE3-pGEX6P-1, and virE3-nYFP constructs. The resulting PCR fragment was digested and inserted into the pACT2, pGEX-6P-1, and pRTL2-EEYN vector, yielding corresponding constructs. All constructs were verified by sequencing.
The genetic background of ecotype Col-0 was used as wild type Arabidopsis thaliana. JAZ8-1302 (pCAMBIA1302) construct and 1,302 empty vector were used for Arabidopsis transformation. All plasmids used in this work are listed in Supplementary Table 2. The construct JAZ8-1302 and the empty vector 1,302 (used as control) were introduced into GV3101 by electroporation. Arabidopsis were transformed using the floral dip method (Clough and Bent, 1998) and grown at 21°C in a growth chamber (16-h light/8-h dark, 2,500 lux). Seeds from the dipped plants were harvested and planted on MS medium containing 30 mg L–1 hygromycin. The transgenic plants were verified by PCR of Hph gene. The positive seedlings were transferred to soil and grown until new seeds could be harvested. The TAM-virE3 transgenic plant lines used in this study were from Niu et al. (2015).
The truncated VirE3 (pASE3ΔC) without autoactivation activity was used as a bait in yeast two-hybrid screening (García-Rodríguez et al., 2006). The Arabidopsis cDNA library representing 5.7 × 105 primary transformants was generated by Zhou (Zhou et al., 2017). The bait plasmid pASE3ΔC and Arabidopsis cDNA library was cotransformed into yeast strain PJ69-4A using the PEG-lithium acetate method (Gietz et al., 1992; James et al., 1996). Full-length JAZ8 (AT1G30135), JAZ8NT (aa 1–101), JAZ8CT (aa 101–127), and JAZ8-ZIM (aa 13–101) were cloned in pAS2.1 (acc. No. U30497). The corresponding vectors were cotransformed with empty pACT2 (acc. No. U29899) and pACT2-VirE3, respectively, to yeast strain PJ69-4A. Empty pAS2.1 and pACT2 were used as negative control. Yeast two-hybrid assays were conducted by the cotransformation of bait and prey plasmids into yeast strain PJ69-4A as described by Zhou (Zhou et al., 2017). Transformants were allowed to grow for 4 days at 28°C on synthetic defined (SD)-glucose plates lacking leucine, tryptophan, and histidine (-LWH). Then cells were incubated overnight in liquid SD–LWH medium and 4 μl of 100-fold dilutions were spotted on SD-LWH plate to grow for 4 days at 28°C. The experiments were repeated three times.
The JAZ8 coding sequence was cloned into pMAL-c5X through NdeI and BamHI sites for fusion with MBP tag. Full-length CDS of virE3 was cloned into pGEX6P-1 through BamHI and SalI sites for fusion with GST tag. Constructs JAZ8-pMAL-c5X and virE3-PGEX6P were transformed into E. coli strain BL21 (DE3) pLysS, separately and screened by 20 μg/ml chloramphenicol and 100 μg/ml carbenicillin. Protein expression of MBP-JAZ8 and GST-VirE3 were induced by 1 mM IPTG. The MBP-fusion proteins and GST-fusion proteins were extracted and immobilized onto MBP and GST agarose beads. For pull-down assays, proteins attached to the beads were subsequently examined by SDS-PAGE and detected with anti-MBP and anti-GST antibody, respectively. Detection was performed by incubating the blots in 8 ml western lighting reagent and then exposure to X-ray films.
virE3 was fused with the N-terminal part of the improved yellow fluorescent protein (YFP) in pRTL2-EEYN to produce VirE3-nYFP. JAZ8 was fused with the C-terminal part of YFP in pRTL2-HAYC to produce JAZ8-cYFP (Bracha-Drori et al., 2004). VirE3-nYFP was transiently cotransformed with JAZ8-cYFP into the Arabidopsis leaf protoplast. Images of transfected protoplasts were acquired with a Leica DM IRBE confocal laser scanning microscope equipped an excitation at 514 nm and a band mission was detected at 530–600 nm. Microscopic images were analyzed using Image J software.
The Agrobacterium tumorigenic strain LBA 1010 (WT) and LBA 2564 (-virE3) were used for tumorigenesis assays on Arabidopsis Col-0 wild-type, JAZ8-OE, and virE3-OE plant lines. Arabidopsis root tumor assay was carried out as described previously with minor modifications (Valvekens et al., 1988; Czako et al., 1993). In brief, Arabidopsis seeds of Col-0, JAZ8-OE, and virE3-OE plant lines, respectively, were sterilized and incubated in liquid B5 medium at 4°C for 2 days. Then, the seeds in liquid B5 medium were incubated on a shaker (120 rpm) in growth room (21°C, 16 h light/8hrs dark, 2,000 lux) for 10 days. The root segments pooled from 10-day-old plant of Col-0, JAZ8-OE, and virE3-OE plants, respectively, were incubated with Agrobacterium LBA1010 and LBA2564 (OD620 = 0.2) for 2 min. The infected roots were cut into 4 mm (named explants) and dried on sterile filter paper, transferred to B5 agar plants containing 100 μM acetosyringone for cocultivation for 2 days in the growth room (25°C, 2,000 lux). After cocultivation, root explants were washed with liquid B5 and then transferred to B5 plates supplemented with 100 μg/ml timentin for 3 weeks until calli/tumors were formed. The percentage of roots that form tumors was calculated by counting tumors on 100 to 120 explants. Three independent replicates of 100 to 120 explants were used for each line.
Plasmid VBF-promoter-GUS was obtained by inserting the promoter sequence of VBF (857bp) into plasmid GUSXX (Niu et al., 2015). Full length of virE3 and JAZ8 were inserted into plasmid pRT101. Plasmids carrying VBF-promoter-GUS and plasmids expressing VirE3 and/or JAZ8 were cotransformed into Arabidopsis Col-0 leaf protoplasts by polyethylene-glycol-mediated transformation as previously described (Schirawski et al., 2000). Cotransformation of pRT101 with GUSXX plasmids was used as control (Töpfer et al., 1987). Protoplasts were incubated at 25°C for 16 h and harvested by centrifugation. The GUS activity assays were performed as described (Niu et al., 2015). Relative expression levels of virE3, AtJAZ8, AtPR1, and AtPDF1.2 were measured.
Arabidopsis seeds harvested from three independent JAZ8-OE and 1,302 plant lines were grown vertically on MS plates. RNA isolated from 10-day-old seedlings were used for Illumina sequencing. Illumina mRNA-seq libraries were generated and sequenced by Majorbio Cloud Platform1. The resulting libraries were checked and quantified. The libraries were multiplexed, clustered, and sequenced on an Illumina HiSeq 2000. Sequence reads of low quality and reads containing adaptor sequences were removed. To validate the RNA-seq results, total RNA was extracted from three independent JAZ8-OE and 1,302 plant lines using the same protocol for RNA-seq. cDNA was generated from total RNA. qRT-PCR was performed by Universal SYBR Green Master mix according to the manufacturer’s instructions. The relative amount of gene expression level was calculated using 2–ΔΔCt method. EF1a (At5G09810) was used as the reference gene.
Arabidopsis seeds of Col-0, JAZ8-OE, and VirE3-OE were sterilized and incubated in liquid B5 medium at 4°C for 2 days. Then, the seeds in liquid B5 medium were incubated on a shaker (120 rpm) in growth room (21°C, 16-h light/8-h dark, 2,000 lux) for 10 days. Agrobacterium strains LBA1100 and LBA2564 were preadjusted to OD600 = 0.2. Seedlings of the Col-0, JAZ8-OE, and virE3-OE were infected by Agrobacterium strains LBA1100 and LBA2564 for 2 min. The infected seedlings were taken out and immediately frozen in liquid nitrogen at different time points. RNA was extracted for qRT-PCR analysis. Quantitative RT-PCR analysis was carried out using Bio-Rad iCycler iQ5. The first strand of cDNA was synthesized by HiScript® III RT SuperMix for qPCR (+ gDNA wiper) kit. Specific primers were designed for AtPR1, AtPDF1.2, and EF1a reference gene by primer 5. The primers are listed in Supplementary Table 1. The qRT-PCR reactions were as follows: 95°C 30 s; 95°C 10 s, 60°C 30 s, followed by 40 cycles. Relative expression levels of AtPR1 and AtPDF1.2 were calculated by 2–ΔΔCT methods. All dates are collected from three biological replicates.
The data were analyzed by Excel and SPSS software, and Duncan’s new complex range method was used to compare the differences among the treatments. P value < 0.05 was considered to be significant.
Early publications reported that VirE3 was bound with pBrp to facilitate AMT through modulating gene expression profiles in the host cell (García-Rodríguez et al., 2006). To gain further insight into the function of VirE3 within the host cells during Agrobacterium infection, we used the yeast two-hybrid system (Y2H) to screen for interactors of VirE3. Due to the transcriptional activation activity of the full-length VirE3 in yeast strain PJ69-4A, the truncated VirE3 (pASE3ΔC) without autoactivation activity (García-Rodríguez et al., 2006) was used as a bait and an Arabidopsis cDNA library (Zhou et al., 2017) was used as a prey. Five colonies were found to grow on minimal medium without histidine. Recovered prey plasmids were retransformed with pASE3ΔC into yeast strain PJ69-4A, and only two of these plasmids could grow on selective medium. From these candidate VirE3 interactors, only one cDNA sequence was in frame with the GAL4 activation domain and encoded the JAZ8 protein (AT1G30135). Our Y2H screening thus identified the Jasmonate ZIM-domain protein JAZ8 as a positive interactor of VirE3. To investigate which domain of JAZ8 was responsible for interaction with VirE3, we divided JAZ8 into N-terminal (JAZ8NT), ZIM domain containing (JAZ8-ZIM), and C-terminal fragments (JAZ8CT). JAZ8NT contained an EAR and a ZIM domain, whereas the JAZ8-ZIM and JAZ8CT only contained the ZIM domain and Jas domain, respectively (Figure 1A). We then cloned the full-length VirE3 into the activation domain (AD) and the different truncated versions of JAZ8 into the binding domain (BD) to verify the interaction in the Y2H system. We found that JAZ8, JAZ8NT, and JAZ8-ZIM interacted with VirE3 but not JAZ8CT. These results indicate that the ZIM domain of JAZ8 is responsible for the interaction between VirE3 and JAZ8 (Figure 1B). We further applied pull-down assays to verify the interaction between VirE3 and JAZ8 in vitro. GST-tagged VirE3 and maltose binding protein (MBP)-fused JAZ8 (MBP-JAZ8) were heterologously expressed in E. coli BL21 (DE3) cells and MBP-pull-down assays were performed using two fusion proteins. MBP-JAZ8 resin was incubated with GST-VirE3 and then separated on SDS-PAGE for immunoblotting with anti-GST antibody. As shown in Figure 1C, the negative control (MBP resin) was unable to pull-down GST-VirE3, whereas the GST-VirE3 was efficiently precipitated by MBP-JAZ8, indicating that VirE3 can physically interact with JAZ8 in vitro. We next used bimolecular fluorescence complementation (BiFC) assays to verify the interaction of VirE3 and JAZ8 proteins in planta. VirE3 was fused with the N-terminal part of the improved YFP in pRTL2-EEYN to produce VirE3-nYFP, and JAZ8 was fused with the C-terminal part of YFP in pRTL2-EEYN to produce JAZ8-cYFP. VirE3-nYFP was transiently coexpressed with JAZ8-cYFP in Arabidopsis protoplasts. We found that co-expression of VirE3-nYFP with JAZ8-cYFP resulted in strong YFP fluorescence in the nucleus of Arabidopsis mesophyll protoplasts (Figure 1D), whereas no YFP fluorescence was detected in negative controls (VirE3-nYFP coexpressed with cYFP or nYFP coexpressed with cYFP-JAZ8). Taken together, the Y2H assays, pull-down assays, and BiFC assays consistently demonstrate that VirE3 interacts with JAZ8 protein in vitro and in vivo.
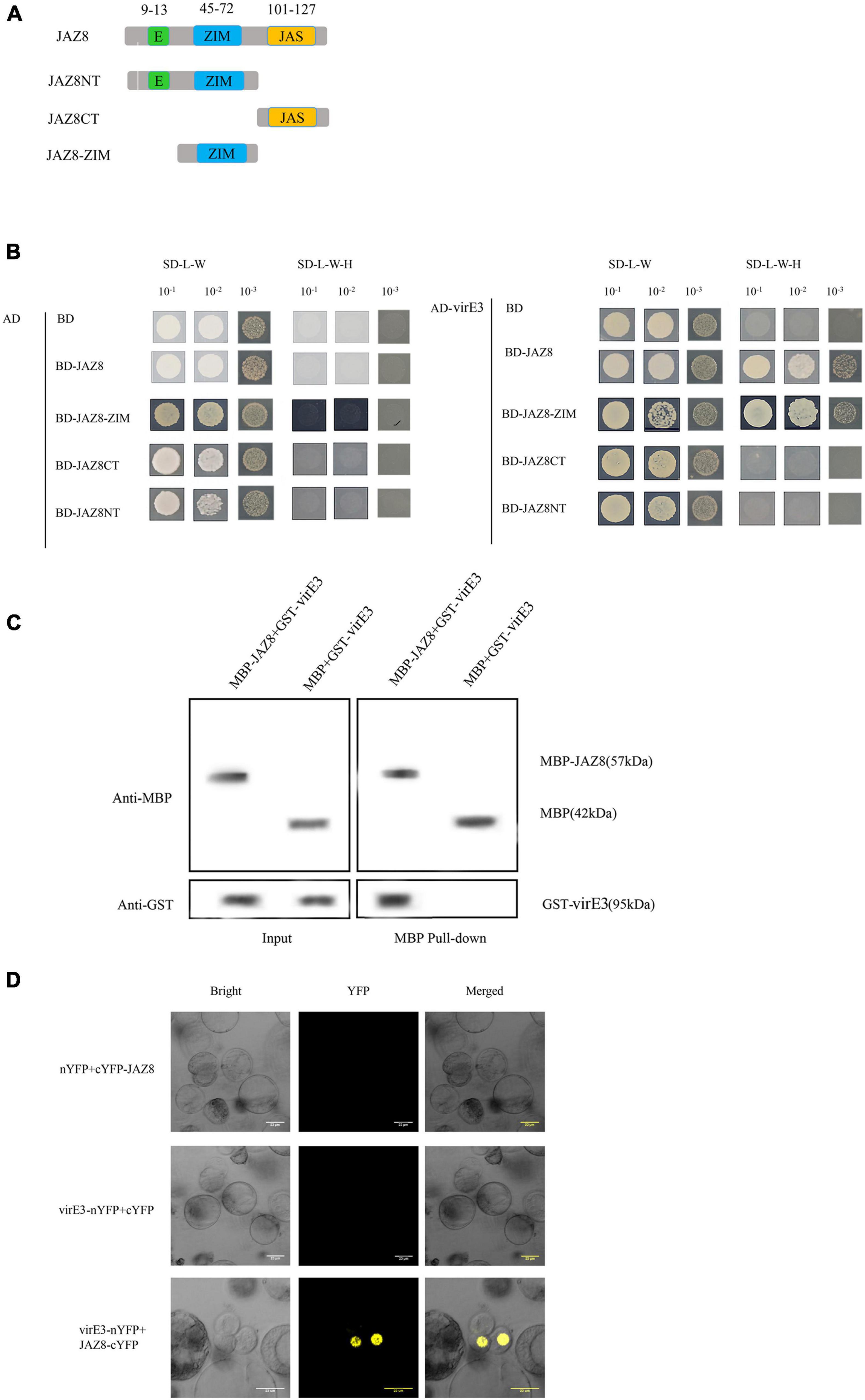
Figure 1. VirE3 interacts with AtJAZ8 in vivo and in vitro. (A) Schematic representation of JAZ8 constructs used for yeast two-hybrid assay. Numbers represent amino acid positions in full-length JAZ8. (B) Yeast two-hybrid assay. Full length of JAZ8 and the truncated fragments of JAZ8NT (aa 1–101), JAZ8CT (aa 101–127), JAZ8-ZIM (aa 13–101) were cloned into pAS2.1 (BD). The virE3 was cloned into pACT2 (AD). The indicated prey and bait constructs were cotransformed into yeast strain PJ69-4A. The transformants were first screened on synthetic defined (SD) medium minus tryptophan and leucine (SD/-Trp-Leu) and the positive yeast cells were subcultured on SD medium minus tryptophan, leucine and histidine at 30°C for 4 days. pAS2.1 and pACT2 were used as the negative control. (C) Maltose-binding protein (MBP) pulldown assay. The MBP pull-down proteins were detected by Western blotting (WB) using anti-GST or anti-MBP antibody. MBP alone was used as the negative control. Input, protein samples without pull-down were analyzed by Western blotting. (D) Bimolecular fluorescence complementation (BIFC) assay. The constructs 35S::VirE3-nYFP and 35S::AtJAZ8-cYFP were transiently co-transformed into Arabidopsis protoplast and the transformed cells were observed under a confocal microscope. 35S::nYFP and 35S:: cYFP were used as negative control.
It has been reported that VirE3 plays a role in tumorigenesis; however, mutation of virE3 led to hardly any attenuation in tumor formation on the stems of six plant species including Nicotiana glauca, Nicotiana tabacum, Helianthus annuus, Lycopersicon esculentum, Kalanchoe tubiflora, and Kalanchoe daigramontiana (García-Rodríguez et al., 2006). To better understand the role of VirE3 in tumorigenesis, the Agrobacterium wild-type strain (LBA1010) and a virE3 deletion strain (LBA2564) were compared in more quantitative root transformation assays on Arabidopsis Col-0. As shown in Figure 2A, the virE3 mutant developed a lower number of tumors compared with LBA1010 (average 32 versus 39% of inoculated roots showing tumors). Analysis by the Student’s t-test confirmed that tumorigenesis of LBA1010 and LBA2564 on Arabidopsis Col-0 was significantly different. To determine whether overexpression of virE3 in Arabidopsis could increase susceptibility to Agrobacterium infection, we used a transgenic line of Arabidopsis that expresses the virE3 coding sequence under the control of the tamoxifen-inducible promoter (Niu et al., 2015). After induction by tamoxifen (Figure 2C) the roots of virE3-OE line1 were transformed by LBA1010 and LBA2564 and the fractions of roots that formed tumors were calculated. As shown in Figure 2B, the virE3-OE line1 was much more susceptible to Agrobacterium LBA1010 and LBA2564 infection with tumors forming in 52 and 41% of the inoculated roots for wild type and virE3 mutant, respectively. Apparently, increasing the amount of VirE3 can enhance tumor formation even by the wild type Agrobacterium strain. The difference in tumorigenesis between LBA1010 and LBA2564 on virE3-OE line1 was statistically significant, suggesting that translocated VirE3 from LBA1010 also still contributes to tumor formation.
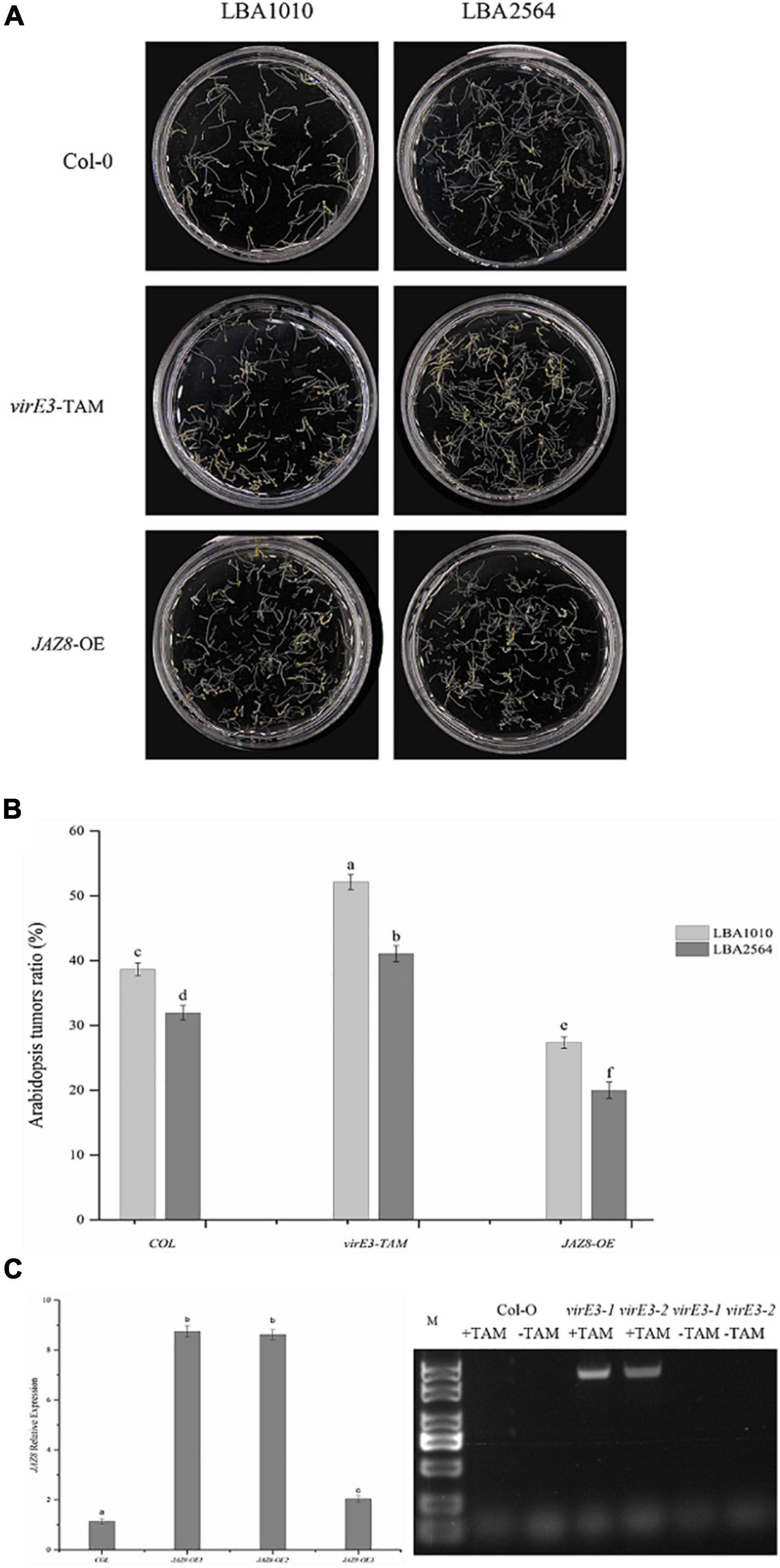
Figure 2. Efficiency of Agrobacterium-mediated root transformation in virE3 and JAZ8 overexpressor lines (virE3-TAM and JAZ8-OE lines) as compared with wild-type plants (Col-0) by Agrobacterium strain LBA1010 and virE3 deletion mutant LBA2564. (A) Tumors developed on root explants. Root segments of Columbia, virE3-TAM1 and JAZ8-OE1 lines were infected with tumorigenic Agrobacterium strain LBA1010 and its virE3 deletion mutant LBA2564. The root segments were photographed and scored 30 days after infection. (B) Quantification of the tumorigenicity showed in panel (A). Error bars in B indicate SEM. Differences in tumorigenicity indicated by different letters are statistically significant (P values < 0.05) and by the same letter are not statistically significant. (C) Gene expression analysis of JAZ8 and virE3 in transgenic over-expressor plant lines.
As we found that VirE3 can interact with JAZ8, we further investigated the role of JAZ8 in tumor formation by Agrobacterium. To this end, transgenic plant lines that constitutively expressed the JAZ8 coding sequence under the CaMV 35S promoter were generated. Two independent T1 transgenic lines, designated JAZ8-OE1 and JAZ8-OE2, both showed about ninefold higher levels of JAZ8 transcript accumulation in their roots compared with Col-0 wild type by qRT-PCR analysis. The roots of JAZ8-OE line1 were transformed by LBA1010 and LBA2564 and the fraction of roots on which tumors were formed was calculated. Interestingly, we found that on JAZ8-OE line1 much less tumors were formed by our wild type strain LBA1010 than on Arabidopsis Col-0 (27 versus 39% of inoculated roots showing tumors, statistically significant). When the virE3 mutant LBA2564 was used for inoculation of the JAZ8-OE roots, the fraction of roots forming tumors was even lower with tumorigenesis seen on only about 22% of the roots (Figure 2B). Our results indicate that JAZ8 counters Agrobacterium virulence, whereas VirE3 enhances Agrobacterium tumorigenicity on Arabidopsis roots.
VirE3 can act as a plant transcription factor which activates genes such as Arabidopsis VBF, which are needed for tumorigenesis (Niu et al., 2015). JAZ8 belongs to the plant-specific TIFY family of transcriptional regulators that repress JA-regulated defense responses by binding to MYC transcription factors (Shyu et al., 2012). To find out whether JAZ8 could reduce the transcriptional activity of VirE3, the effect of JAZ8 on the VirE3-dependent activation of VBF (At1G56250) promoter was investigated in a protoplast transactivation assay. The 857 bp of VBF promoter was fused to the GUS reporter, and the full length of virE3 or JAZ8 coding sequences wase cloned into the pRT101 plasmid, in which virE3 or JAZ8 was expressed from the CaMV 35S promoter. Arabidopsis protoplasts were cotransformed with the VBF-promoter-GUS reporter plasmid and at the same time with the effector plasmid pRT101-VirE3 and/or pRT101-JAZ8. First we measured whether the JAZ8 and virE3 construct were expressed in Arabidopsis protoplasts by RT-PCR. As shown in Figure 3B, the mRNA expression level of JAZ8 was around 30-fold higher in protoplasts transformed with plasmid pRT101-JAZ8 and also in protoplasts transformed by pRT101-JAZ8 together with pRT101-VirE3 compared with that in Col-0 protoplasts. The mRNA expression of virE3 was also confirmed by RT-PCR analysis in protoplasts transformed with plasmid pRT101-VirE3 and plasmids pRT101-VirE3 together with pRT101-JAZ8 (Figure 3C). These results indicate that the mRNAs of both genes are indeed expressed in the transformed protoplasts and that the expression of the JAZ8 gene does not negatively affect the expression of virE3 and vice versa. Subsequently, we tested the effect of JAZ8 and VirE3 on the activity of the VBF promoter. As shown in Figure 3A, expression of JAZ8 alone barely affected the activity of the VBF promoter, whereas VirE3, as expected, activated the VBF promoter by approximately threefold. In contrast, co-expression of VirE3 and JAZ8 together led to a twofold reduced activity of the VBF promoter as compared with that in the presence of VirE3 alone. As JAZ8 is known to affect the JA response and promote the SA defense response, we also tested whether VirE3 could affect these defense responses. To this end, we measured the mRNA levels of PR1 (SA response) and PDF1.2 (JA response) in the same transformed protoplasts. Interestingly, we found that the expression level of PR1 was reduced in protoplasts transformed with pRT101-VirE3 and elevated in protoplasts transformed with pRT101-JAZ8. Reversely, the expression level of PDF1.2 was elevated in protoplasts transformed with pRT101-VirE3 and decreased in protoplasts transformed with pRT101-JAZ8 (Figure 3B). In summary, our results indicate that JAZ8 can repress the transcriptional activity of VirE3 on activating the expression of VBF. Also overexpression of VirE3 reduced PR1 gene expression and elevated PDF1.2 expression, whereas overexpression of JAZ8 elevated PR1 gene expression and reduced PR1 expression.
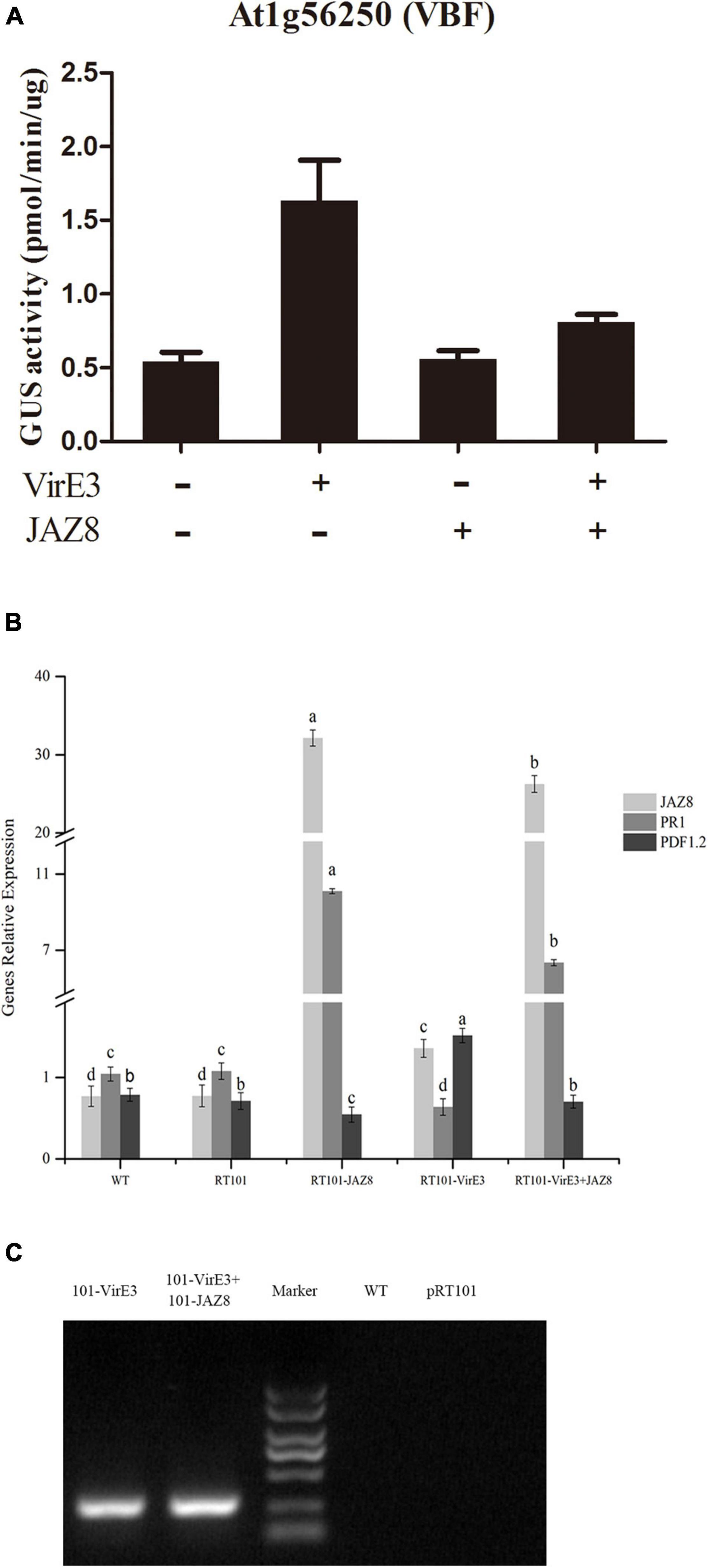
Figure 3. Protoplast expression analysis of the promoters of At1G56250 (VBF) and the gene expression analysis of PR1, PDF1.2, JAZ8, and virE3. (A) JAZ8 inhibits VirE3-dependent activation of the At1G56250 (VBF) (857 bp) promoters in Arabidopsis protoplasts. (B) qPCR analysis of the expression level of PR1, PDF1.2, JAZ8 in protoplasts transfected with corresponding plasmids. (C) RT-PCR analysis of the expression of virE3 in protoplast transformed with corresponding plasmids.
As a transcriptional repressor of the jasmonate downstream signaling pathway, JAZ8 most likely affects plant susceptibility to Agrobacterium by regulating the expression of plant defense responsive genes that interfere with the Agrobacterium infection. To investigate the role of JAZ8 on the transcription of defense responsive genes, we carried out transcriptome profiling by comparing AtJAZ8 overexpressor lines with control lines using RNA-seq analysis. Total RNA samples extracted from seedlings were analyzed for gene expression using RNA-seq. Using DESeq to calculate differentially expressed genes (DEGs), we identified a total of 696 genes that were either up- or down-regulated (FDR < 0.001 and log2FC > 2) in JAZ8-OE plant lines compared with the control lines (Supplementary Table 1). Raw data have been deposited in the NCBI SRA database (BioProject accession number: PRJNA719307). To validate the RNA-seq results, six DEGs from AtJAZ8 overexpressor plants were randomly selected for analysis by quantitative RT-PCR. As shown in Figure 4A, the results obtained from qRT-PCR analysis are in general agreement with the RNA-seq results. We then used GO annotation (TAIR10) to assign genes to functional categories and performed function enrichment analysis on the DEGs. The 428 genes upregulated in the JAZ8-OE line were enriched for genes implicated in regulation of SA metabolic process, regulation of defense response to bacterium, regulation of plant-type hypersensitive response, and also defense response (Figure 4B). By contrast, the 268 genes downregulated in the JAZ8-OE line were enriched for genes related to defense response, immune system process, response to JA, etc. (Figure 4C). Our results suggest that JAZ8 generally affects genes involved in plant defense responses. Interestingly, we found that the expression level of pathogenesis-related gene 1 (AtPR1) in JAZ8-OE was around five times (5.48) higher than in control lines, indicating JAZ8 may activate the SA-mediated signaling pathway against Agrobacterium infection.
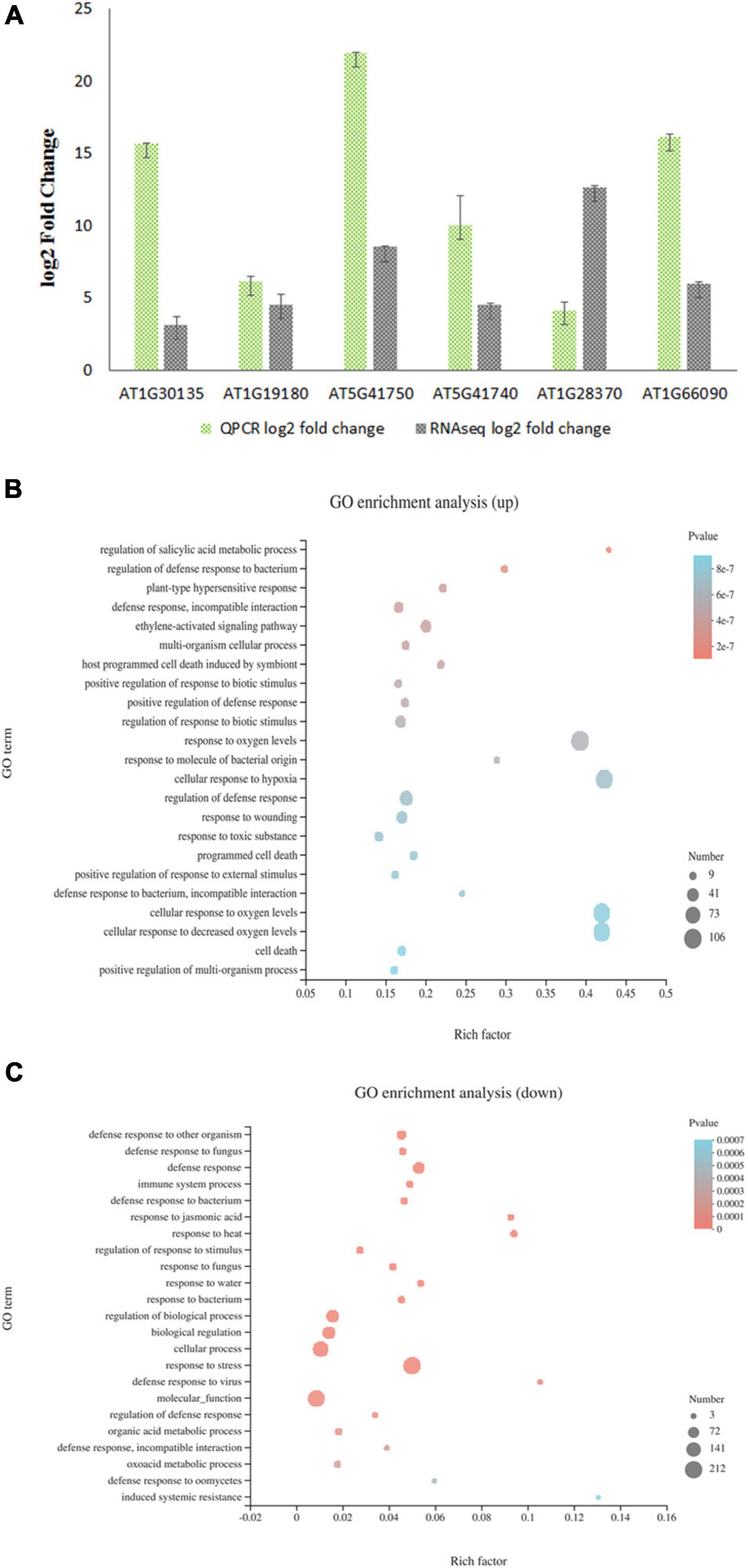
Figure 4. Transcript profiling of AtJAZ8 overexpressor plants. (A) Validation of the RNA-Seq data by quantitative RT-PCR. Six differential expression genes in AtJAZ8 overexpressor plants compared with control plant lines in the RNA-seq experiment were selected for validation by qRT-PCR. (B) GO term enrichment for 428 upregulated genes using Database for Annotation and Visualization. (C) GO term enrichment for 268 downregulated genes using Database for Annotation and Visualization.
We have shown that the percentage of root tumors was highly decreased in JAZ8-OE lines and increased in virE3-OE lines. To investigate the plant defense genes responsible for Agrobacterium mediated root tumorigenesis, the expression level of Arabidopsis pathogenesis-related GENE 1 (AtPR1) and Arabidopsis PLANT DEFENSIN 1.2(AtPDF1.2) were measured by qRT-PCR in wild type Col-0, JAZ8-OE, and virE3-OE plant lines infected by Agrobacterium strains LBA1100 (wild-type) and LBA2564 (virE3 deletion mutant) at different time points. To compare the gene expression level at different time points infected by two Agrobacterium strains LBA1100 and LBA2564, we normalized the expression level of AtPR1 and AtPDF1.2 to the Col-0 at 0 h before infection by qRT-PCR analysis. As shown in Figure 5A, 24 h after Agrobacterium infection, the expression level of AtPR1 was significantly reduced in virE3-OE plant line compared with that in Col-0 and JAZ8-OE plant line, whereas the expression level of AtPR1 gene was the highest in the JAZ8-OE plant line. Our results showed that VirE3 inhibits the expression of AtPR1 gene and JAZ8 promotes AtPR1 gene expression probably by repressing the JA-responsive defense pathway. The expression pattern of AtPR1 in plant lines was negatively well correlated with the root tumorigenicity by Agrobacterium. On the contrary, after Agrobacterium infection, the expression level of AtPDF1.2 was highest in virE3-OE plant line and lowest in JAZ8-OE plant line, indicating VirE3 might activate the JA-induced signaling pathway by repressing AtPR1 gene expression (Figure 5B). Our results showed that JAZ8 may antagonize VirE3 on activating the expression of AtPR1 gene, which in turn attenuates Agrobacterium tumorigenesis on Arabidopsis root.
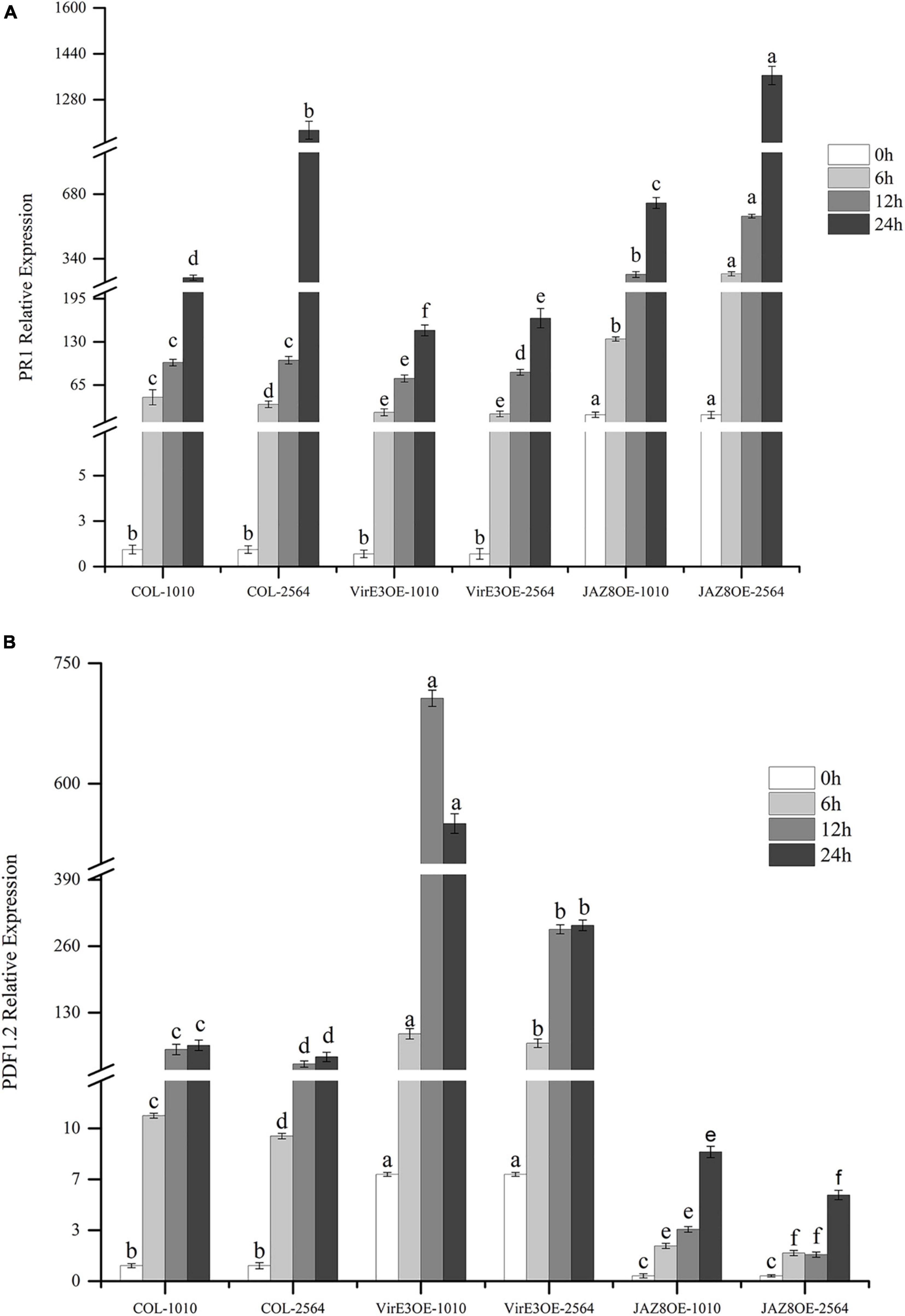
Figure 5. Quantification of AtPR1 and AtPDF1.2 gene expression level in Col-0, virE3-OE and JAZ8-OE1 lines after infection with Agrobacterium LBA1010 and LBA2564 at different time points. (A) qRT-PCR analysis of AtPR1 gene expression pattern. (B) qRT-PCR analysis of AtPDF1.2 gene expression pattern. EF1a was used as internal control for data normalization. The expression level of PR1 and PDF1.2 in the wild-type Col-0 at 0 h was set to 1.0, and error bars represent SEM of three independent biological replicates. The expression values of PR1 and PDF1.2 in virE3-OE and JAZ8-OE lines were compared to the control Col-0 at each corresponding condition. Differences indicated by different letters are statistically significant (P values < 0.05) and by the same letter are not statistically significant.
Agrobacterium delivers several virulence proteins, including VirD2, VirE2, VirE3, VirF, and VirD5 to plant cell for successful T-DNA integration and tumorigenesis (Lacroix and Citovsky, 2019). One of the translocated effector proteins, VirE3, is highly conserved among different Agrobacterium and rhizobia species, suggesting that VirE3 may play an important role during AMT. However, it was reported that VirE3 was not essential for AMT by octopine strains and a virE3 mutation did not significantly affect tumor formation on the stems of several plant species, and its contribution to tumorigenesis could only be seen in virE3 virF double mutants (García-Rodríguez et al., 2006). A recent report showed that VirE3 is required for full virulence of the agropine/succinamopine strain EHA105, which does not have a close homolog of octopine Ti virF. Deletion of virE3 resulted in a decreased transformation efficiency in the transient transformation assays of Nicotiana benthamiana leaves (Li et al., 2018, 2020). In contrast, another report showed constitutive overexpression of virE2 or virE3 under CaMV 35S promoter in Arabidopsis-induced plant defense responses and conferred resistance to Agrobacterium infection (Duan et al., 2018). Our results showed that VirE3 is required for tumorigenesis of oncogenic wild type Agrobacterium LBA1010. Deletion of virE3 attenuated Agrobacterium virulence in root tumor assay, whereas transient overexpression of virE3 enhanced Agrobacterium tumorigenesis (Figure 2A). The different results reported may be due to the different methods used for the transformation or to the different plant species or Agrobacterium strains used.
Upon attack by pathogens, plants possess highly sophisticated mechanisms to induce the expression of defense genes to restrict pathogen invasion. The phytohormones SA and JA, as key signaling molecules, play important roles in regulating the activation of the induced defense responses. Silencing of SA biosynthetic and signaling genes in N. benthamiana increases susceptibility to crown gall disease (Yuan et al., 2007; Anand et al., 2008). One of the crucial effects of SA in plant defense is to activate the expression of pathogenesis related genes such as PR1, resulting in both local and systemic acquired resistance to the pathogens. However, Agrobacterium has evolved strategies to dampen this host defense response. It has been reported that infection by Agrobacterium decreases the level of free SA in roots, leading to reduced PR1 gene expression and a diminished systemic-acquired resistance (SAR) (Gaspar et al., 2004). Otherwise it has been reported that Agrobacterium infection triggers the activation of mitogen-activated protein kinase (MAPK) MPK3, which phosphorylates the transcription factor VIP1, leading to its relocalization from the cytoplasm to the nucleus, and also in the of induction of stress-related genes. This can indirectly lead to increased PR1 gene expression (Djamei et al., 2007). However, the translocated effector VirE3 was shown to induce the expression of NIMIN1 (Niu et al., 2015). The NIMIN1 protein binds to NPR1/NIM1 and prevents the induction of PR1 and SAR (Niu et al., 2015). It has been shown that exogenous application of SA also affects Agrobacterium itself and inhibits induction of its virulence genes, and thus decreases its infectivity (Yuan et al., 2007). A recent publication revealed that SA is used by Agrobacterium to turn down the expression of its virulence genes late in infection. To this end Agrobacterium expresses the hydrolase SghA to liberate plant SA from the storage form, SA-glucoside (Wang et al., 2019).
Our new results showed that VirE3 enhances Agrobacterium tumorigenesis by reducing PR1 defense gene expression at the early stage (Figure 5A). Using yeast two-hybrid screening, we found that VirE3 can interact with Jasmonate ZIM domain (JAZ) repressor JAZ8 via its ZIM domain. JAZ proteins are suppressors of JA-induced transcriptional response, reducing the transcription of JA-responsive genes such as PDF1.2. Our results are in line with this and show that JAZ8 suppresses the JA-dependent defense response as seen by the downregulation of PDF1.2, and promotes the SA-dependent response as seen by enhanced PR1 gene expression with increased levels of JAZ8 expression (Figure 5B). JAZ8-OE in turn attenuates Agrobacterium tumorigenesis as shown in Figure 2B. We found that the effector proteinVirE3 binds to and thereby counteracts the activity of JAZ8 and thus promotes tumorigenesis by Agrobacterium. In the presence of VirE3, the expression of JA-defense genes is increased and the expression level of SA-defense genes decreased.
In conclusion, our data shown herein revealed that the effector protein VirE3 is required for the full virulence of Agrobacterium by counteracting the transcriptional repressor activity of JAZ8, thus promoting the JA defense response and diminishing the SA defense response. Such a strategy is not uncommon for plant pathogenic bacteria. Plant pathogenic Pseudomonas syringae bacteria secrete coronatine, a jasmonate mimic, to enhance the JA defense response. Other strains may deliver Hop effector proteins via a Type3 secretion system such as HopZ1 that bind to and modify JAZ proteins leading to their destruction (Jiang et al., 2013). Whether Agrobacterium VirE3 binding to JAZ8 similarly leads to its degradation requires further study.
The original contributions presented in the study are publicly available. This data can be found here: NCBI repository, accession number: PRJNA719307 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA719307).
BX, MZ, XN, and PH conceived and designed the experiment. SL performed the experiments. XN and SL wrote the manuscript. MZ, XL, and JC provided critical analysis of the design and data. All authors read and approved the final manuscript.
This work was financially supported by the Natural Science Foundation of Hainan Province (No. 2019RC013) and Hainan Provincial Department of Education (Hnjg2019ZD-2). The funding agencies had no role in the design, analysis, and interpretation of the data, or writing of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.685533/full#supplementary-material
Abu-Arish, A., Frenkiel-Krispin, D., Fricke, T., Tzfira, T., Citovsky, V., Wolf, S. G., et al. (2004). Three-dimensional reconstruction of Agrobacterium VirE2 protein with single-stranded DNA. J. Biol. Chem. 279, 25359–25363. doi: 10.1074/jbc.m401804200
Anand, A., Uppalapati, S. R., Ryu, C.-M., Allen, S. N., Kang, L., Tang, Y., et al. (2008). Salicylic acid and systemic acquired resistance play a role in attenuating crown gall disease caused by Agrobacterium tumefaciens. Plant Physiol. 146, 703–715. doi: 10.1104/pp.107.111302
Beijersbergen, A., and Hooykaas, P. J. J. (1993). The virulence system of Agrobacterium Tumefaciens. Methods Mol. Biol. 14, 37–49.
Bracha-Drori, K., Shichrur, K., Katz, A., Oliva, M., Angelovici, R., Yalovsky, S., et al. (2004). Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 40, 419–427.
Bundock, P., and Hooykaas, P. J. J. (1996). Integration of Agrobacterium tumefaciens T-DNA in the Saccharomyces cerevisiae genome by illegitimate recombination. Proc. Natl. Acad. Sci. U.S.A. 93, 15272–15275. doi: 10.1073/pnas.93.26.15272
Christie, P. J., Atmakuri, K., Krishnamoorthy, V., Jakubowski, S., and Cascales, E. (2005). Biogenesis, architecture, and function of bacterial type Iv secretion systems. Annu. Rev. Microbiol. 59, 451–485.
Citovsky, V., Kozlovsky, S. V., Lacroix, B., Zaltsman, A., Dafny-Yelin, M., Vyas, S., et al. (2007). Biological systems of the host cell involved in Agrobacterium infection. Cell Microbiol. 9, 9–20. doi: 10.1111/j.1462-5822.2006.00830.x
Citovsky, V., Wong, M. L., and Zambryski, P. (1989). Cooperative interaction of Agrobacterium VirE2 protein with single-stranded DNA: implications for the T-DNA transfer process. Proc. Natl. Acad. Sci. U.S.A. 86, 1193–1197. doi: 10.1073/pnas.86.4.1193
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Czako, M., Wilson, J., Yu, X., and Marton, L. (1993). Sustained root culture for generation and vegetative propagation of transgenic Arabidopsis thaliana. Plant Cell Rep. 12, 603–606. doi: 10.1007/BF00232807
de Groot, M. J. A., Bundock, P., Hooykaas, P. J. J., and Beijersbergen, A. G. M. (1998). Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat. Biotechnol. 16, 839–842. doi: 10.1038/nbt0998-839
Djamei, A., Pitzschke, A., Nakagami, H., Rajh, I., and Hirt, H. (2007). Trojan horse strategy in Agrobacterium transformation: abusing MAPK defense signaling. Science 318, 453–456. doi: 10.1126/science.1148110
Duan, K., Willig, C. J., De Tar, J. R., Spollen, W. G., and Zhang, Z. J. (2018). Transcriptomic analysis of arabidopsis seedlings in response to an agrobacterium-mediated transformation process. Mol. Plant Microbe Interact. 31, 445–459. doi: 10.1094/MPMI-10-17-0249-R
García-Cano, E., Hak, H., Magori, S., Lazarowitz, S. G., and Citovsky, V. (2018). The Agrobacterium F-Box protein effector VirF destabilizes the Arabidopsis GLABROUS1 enhancer/binding protein-like transcription factor VFP4, a transcriptional activator of defense response genes. Mol. Plant Microbe Interact. 31, 576–586. doi: 10.1094/MPMI-07-17-0188-FI
García-Rodríguez, F. M., Schrammeijer, B., and Hooykaas, P. J. J. (2006). The Agrobacterium VirE3 effector protein: a potential plant transcriptional activator. Nucleic Acids Res. 34, 6496–6504. doi: 10.1093/nar/gkl877
Gaspar, Y. M., Nam, J., Schultz, C. J., Lee, L.-Y., Gilson, P. R., Gelvin, S. B., et al. (2004). Characterization of the Arabidopsis lysine-rich Arabinogalactan-Protein AtAGP17 Mutant (rat1) that results in a decreased efficiency of agrobacterium transformation. Plant Physiol. 135, 2162–2171. doi: 10.1104/pp.104.045542
Gelvin, S. B. (1998). Agrobacterium VirE2 proteins can form a complex with T strands in the plant Cytoplasm. J. Bacteriol. 180, 4300–4302. doi: 10.1128/JB.180.16.4300-4302.1998
Gelvin, S. B. (2000). Agrobacteriumandplantgenesinvolved Int-DNA transfer andintegration. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 223–256.
Gelvin, S. B. (2003). Agrobacterium-mediated plant transformation: the biology behind the “Gene-Jockeying”. tool. Microbiol. Mol. Biol. Rev. 67, 16–37. doi: 10.1128/MMBR.67.1.16-37.2003
Gelvin, S. B. (2021). Plant DNA repair and Agrobacterium T-DNA integration. Int. J. Mol. Sci. 22:8458. doi: 10.3390/ijms22168458
Gietz, D., St Jean, A., Woods, R. A., and Schiestl, R. H. (1992). Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425.
Guo, M., Ye, J., Gao, D., Xu, N., and Yang, J. (2019). Agrobacterium-mediated horizontal gene transfer: mechanism, biotechnological application, potential risk and forestalling strategy. Biotechnol. Adv. 37, 259–270. doi: 10.1016/j.biotechadv.2018.12.008
Herrera-Estrella, A., Chen, Z. M., Van Montagu, M., and Wang, K. (1988). VirD proteins of Agrobacterium tumefaciens are required for the formation of a covalent DNA-protein complex at the 5’ terminus of T-strand molecules. EMBO J. 7, 4055–4062. doi: 10.1002/j.1460-2075.1988.tb03299.x
Howard, E. A., Zupan, J. R., Citovsky, V., and Zambryski, P. C. (1992). The VirD2 protein of a. tumefaciens contains a C-terminal bipartite nuclear localization signal: implications for nuclear uptake of DNA in plant cells. Cell 68, 109–118. doi: 10.1016/0092-8674(92)90210-4
Howe, G. A., Major, I. T., and Koo, A. J. (2018). Modularity in jasmonate signaling for multistress resilience. Annu. Rev. Plant Biol. 69, 387–415. doi: 10.1146/annurev-arplant-042817-040047
James, P., Halladay, J., and Craig, E. A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436. doi: 10.1093/genetics/144.4.1425
Jarchow, E., Grimsley, N. H., and Hohn, B. (1991). virF, the host-range-determining virulence gene of Agrobacterium tumefaciens, affects T-DNA transfer to Zea mays. Proc. Natl. Acad. Sci. U.S.A. 88, 10426–10430. doi: 10.1073/pnas.88.23.10426
Jiang, S., Yao, J., Ma, K. W., Zhou, H., Song, J., He, S. Y., et al. (2013). Bacterial effector activates jasmonate signaling by directly targeting JAZ transcriptional repressors. PLoS Pathog. 9:e1003715. doi: 10.1371/journal.ppat.1003715
Lacroix, B. T., Vaidya, M., Tzfira, T., and Citovsky, V. (2004). The VirE3 protein of Agrobacterium mimics a host cell function required for plant genetic transformation. EMBO J. 24, 428–437. doi: 10.1038/sj.emboj.7600524
Lacroix, B., and Citovsky, V. (2019). Pathways of DNA transfer to plants from Agrobacterium tumefaciens and related bacterial species. Annu. Rev. Phytopathol. 57, 231–251. doi: 10.1146/annurev-phyto-082718-100101
Lapham, R., Lee, L.-Y., Tsugama, D., Lee, S., Mengiste, T., and Gelvin, S. B. (2018). VIP1 and its homologs are not required for agrobacterium-mediated transformation, but play a role in botrytis and salt stress responses. Front. Plant Sci. 9:749.
Li, J., Krichevsky, A., Vaidya, M., Tzfira, T., and Citovsky, V. (2005). Uncoupling of the functions of the Arabidopsis VIP1 protein in transient and stable plant genetic transformation by Agrobacterium. Proc. Natl. Acad. Sci. U.S.A. 102, 5733–5738. doi: 10.1073/pnas.0404118102
Li, X., Tu, H., and Pan, S. Q. (2018). Agrobacterium delivers anchorage protein VirE3 for companion VirE2 to aggregate at host entry sites for T-DNA protection. Cell Rep. 25, 302–311.e306. doi: 10.1016/j.celrep.2018.09.023
Li, X., Zhu, T., Tu, H., and Pan, S. Q. (2020). Agrobacterium VirE3 uses its two tandem domains at the C-terminus to retain its companion VirE2 on the Cytoplasmic side of the host plasma membrane. Front. Plant Sci. 11:464. doi: 10.3389/fpls.2020.00464
Li, Y. G., and Christie, P. J. (2018). The Agrobacterium VirB/VirD4 T4SS: mechanism and architecture defined through in vivo mutagenesis and chimeric systems. Curr. Top. Microbiol. Immunol. 418, 233–260. doi: 10.1007/82_2018_94
McCullen, C. A., and Binns, A. N. (2006). Agrobacterium tumefaciens and plant cell interactions and activities required for interkingdom macromolecular transfer. Annu. Rev. Cell Dev. Biol. 22, 101–127. doi: 10.1146/annurev.cellbio.22.011105.102022
Melchers, L. S., Maroney, M. J., den Dulk-Ras, A., Thompson, D. V., van Vuuren, H. A., Schilperoort, R. A., et al. (1990). Octopine and nopaline strains of Agrobacterium tumefaciens differ in virulence; molecular characterization of the virF locus. Plant Mol. Biol. 14, 249–259. doi: 10.1007/BF00018565
Nester, E. W., Gordon, M. P., Amasino, R. M., and Yanofsky, M. F. (1984). Crown gall: a molecular and physiological analysis. Annu. Rev. Plant Physiol. 35, 387–413.
Niu, X., Zhou, M., Henkel, C. V., van Heusden, G. P. H., and Hooykaas, P. J. J. (2015). The Agrobacterium tumefaciens virulence protein VirE3 is a transcriptional activator of the F-box geneVBF. Plant J. 84, 914–924. doi: 10.1111/tpj.13048
Pauwels, L., and Goossens, A. (2011). The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 23, 3089–3100. doi: 10.1105/tpc.111.089300
Piers, K. L., Heath, J. D., Liang, X., Stephens, K. M., and Nester, E. W. (1996). Agrobacterium tumefaciens-mediated transformation of yeast. Proc. Natl. Acad. Sci. U.S.A. 93, 1613–1618.
Rossi, L., Hohn, B., and Tinland, B. (1993). The VirD2 protein of Agrobacterlum tumefaciens carries nuclear localization signals important for transfer of T-DNA to plants. Mol. Gen. Genet. MGG 239, 345–353. doi: 10.1007/BF00276932
Schirawski, J., Planchais, S., and Haenni, A.-L. (2000). An improved protocol for the preparation of protoplasts from an established Arabidopsis thaliana cell suspension culture and infection with RNA of turnip yellow mosaic tymovirus: a simple and reliable method. J. Virol. Methods 86, 85–94. doi: 10.1016/s0166-0934(99)00173-1
Schrammeijer, B. (2003). Analysis of Vir protein translocation from Agrobacterium tumefaciens using Saccharomyces cerevisiae as a model: evidence for transport of a novel effector protein VirE3. Nucleic Acids Res. 31, 860–868. doi: 10.1093/nar/gkg179
Schrammeijer, B., Risseeuw, E., Pansegrau, W., Regensburg-Tuïnk, T. J. G., Crosby, W. L., and Hooykaas, P. J. J. (2001). Interaction of the virulence protein VirF of Agrobacterium tumefaciens with plant homologs of the yeast Skp1 protein. Curr. Biol. 11, 258–262. doi: 10.1016/s0960-9822(01)00069-0
Shi, Y., Lee, L.-Y., and Gelvin, S. B. (2014). Is VIP1 important for Agrobacterium-mediated transformation? Plant J. 79, 848–860. doi: 10.1111/tpj.12596
Shurvinton, C. E., Hodges, L., and Ream, W. (1992). A nuclear localization signal and the C-terminal omega sequence in the Agrobacterium tumefaciens VirD2 endonuclease are important for tumor formation. Proc. Natl. Acad. Sci. U.S.A. 89, 11837–11841. doi: 10.1073/pnas.89.24.11837
Shyu, C., Figueroa, P., DePew, C. L., Cooke, T. F., Sheard, L. B., Moreno, J. E., et al. (2012). JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. Plant Cell 24, 536–550. doi: 10.1105/tpc.111.093005
Tinland, B., Koukolikova-Nicola, Z., Hall, M. N., and Hohn, B. (1992). The T-DNA-linked VirD2 protein contains two distinct functional nuclear localization signals. Proc. Natl. Acad. Sci. U.S.A. 89, 7442–7446. doi: 10.1073/pnas.89.16.7442
Töpfer, R., Matzeit, V., Gronenborn, B., Schell, J., and Steinbiss, H.-H. (1987). A set of plant expression vectors for transcriptional and translational fusions. Nucleic Acids Res. 15, 5890–5890. doi: 10.1093/nar/15.14.5890
Tzfira, T. (2001). VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J. 20, 3596–3607.
Tzfira, T., Vaidya, M., and Citovsky, V. (2004). Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature 431, 87–92. doi: 10.1038/nature02857
Valvekens, D., Van Montagu, M., and Van Lijsebettens, M. (1988). Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. U.S.A. 85, 5536–5540. doi: 10.1073/pnas.85.15.5536
Vergunst, A. C. (2000). VirB/D4-dependent protein translocation from agrobacterium into plant cells. Science 290, 979–982. doi: 10.1126/science.290.5493.979
Wang, C., Ye, F., Chang, C., Liu, X., Wang, J., Wang, J., et al. (2019). Agrobacteria reprogram virulence gene expression by controlled release of host-conjugated signals. Proc. Natl. Acad. Sci. U.S.A. 116, 22331–22340. doi: 10.1073/pnas.1903695116
Weigel, R. R., Pfitzner, U. M., and Gatz, C. (2005). Interaction of NIMIN1 with NPR1 modulates PR gene expression in arabidopsis. Plant Cell 17, 1279–1291. doi: 10.1105/tpc.104.027441
Yanofsky, M. F., Porter, S. G., Young, C., Albright, L. M., Gordon, M. P., and Nester, E. W. (1986). The virD operon of Agrobacterium tumefaciens encodes a site-specific endonuclease. Cell 47, 471–477. doi: 10.1016/0092-8674(86)90604-5
Young, C., and Nester, E. W. (1988). Association of the virD2 protein with the 5’ end of T strands in Agrobacterium tumefaciens. J. Bacteriol. 170, 3367–3374. doi: 10.1128/jb.170.8.3367-3374.1988
Yuan, Z. C., Edlind, M. P., Liu, P., Saenkham, P., Banta, L. M., Wise, A. A., et al. (2007). The plant signal salicylic acid shuts down expression of the vir regulon and activates quormone-quenching genes in Agrobacterium. Proc. Natl. Acad. Sci. U.S.A. 104, 11790–11795. doi: 10.1073/pnas.0704866104
Zaltsman, A., Krichevsky, A., Loyter, A., and Citovsky, V. (2010). Agrobacterium induces expression of a host F-Box protein required for tumorigenicity. Cell Host Microbe 7, 197–209. doi: 10.1016/j.chom.2010.02.009
Zaltsman, A., Lacroix, B., Gafni, Y., and Citovsky, V. (2012). Disassembly of synthetic Agrobacterium T-DNA-protein complexes via the host SCFVBF ubiquitin-ligase complex pathway. Proc. Natl. Acad. Sci. U.S.A. 110, 169–174. doi: 10.1073/pnas.1210921110
Keywords: Agrobacterium tumefaciens, Arabidopsis thaliana, VirE3, JAZ8, jasmonates
Citation: Li S, Xu B, Niu X, Lu X, Cheng J, Zhou M and Hooykaas PJJ (2021) JAZ8 Interacts With VirE3 Attenuating Agrobacterium Mediated Root Tumorigenesis. Front. Plant Sci. 12:685533. doi: 10.3389/fpls.2021.685533
Received: 25 March 2021; Accepted: 11 October 2021;
Published: 19 November 2021.
Edited by:
Brigitte Mauch-Mani, Université de Neuchâtel, SwitzerlandReviewed by:
Andrew N. Binns, University of Pennsylvania, United StatesCopyright © 2021 Li, Xu, Niu, Lu, Cheng, Zhou and Hooykaas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bingliang Xu, eHVibEBnc2F1LmVkdS5jbg==; Xiaolei Niu, bmludGVyeGxsQGhhaW5hbnUuZWR1LmNu; Meiliang Zhou, emhvdW1laWxpYW5nQGNhYXMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.