- 1School of Applied Biosciences, Kyungpook National University, Daegu, South Korea
- 2Ministry of Agriculture State Company for Agricultural Supplies, Baghdad, Iraq
- 3Department of Entomology, Abdul Wali Khan University Mardan, Mardan, Pakistan
- 4Division of Applied Life Science (BK21 Program), Institute of Agriculture and Life Science (IALS), Plant Molecular Biology and Biotechnology Research Center (PMBBRC), Gyeongsang National University, Jinju, South Korea
Nitric oxide (NO) is a signaling molecule that regulates various processes, including plant growth and development, immunity, and environmental interactions. Using high throughput RNA-seq data, we explored the role of the NO-induced ATILL6 gene in plant growth and defense using functional genomics. The atill6 mutant and wild-types were challenged with either oxidative (H2O2, MV) or nitro-oxidative (CySNO, GSNO) stress conditions, and the phenotypic results showed that ATILL6 gene differentially regulates cotyledon development frequency (CDF) as well as the root and shoot lengths of the plants. To investigate whether ATILL6 plays a role in plant basal or resistance (R)-gene-mediated defense, the plants were challenged with either virulent or avirulent strains of Pseudomonas syringae pathovar tomato (Pst) DC3000. The atill6 line showed a susceptible phenotype, higher pathogen growth, and highly reduced transcript accumulation of PR1 and PR2 genes. These results suggested that ATILL6 positively regulates plant basal defense. Furthermore, after the inoculation of atill6 with avirulent Pst (DC3000), the expressions of the PR1 and PR2 genes decreased, suggesting a positive role in R-gene-mediated resistance in protecting the plant from further spread of disease. We also investigated the role of ATILL6 in systemic acquired resistance (SAR), and the results showed that ATILL6 positively regulates SAR, as the mutant line atill6 has significantly (p ≤ 0.05) lower transcript accumulation of PR, G3DPH, and AZI genes. Overall, these results indicate that the NO-induced ATILL6 gene differentially regulates plant growth and positively regulates plant basal defense, R-gene-mediated resistance, and SAR.
Introduction
Plants, due to their non-motile nature, are continuously subjected to biotic and abiotic stresses. This exposure induces the production of reactive oxygen species (ROS) and reactive nitrogen species (RNS) in plants, which are key signaling molecules (Domingos et al., 2015; Farnese et al., 2016; Khan et al., 2019). Due to the versatile role of Nitric oxide (NO) it was named as the “Molecule of the year” by the Science magazine in 1992 (Culotta and Koshland, 1992). Afterwards, in 1998 the Nobel Prize in Physiology or Medicine was awarded to Robert F. Furchgott, Louis J. Ignarro, and Ferid Murad jointly for their work on nitric oxide as a signaling molecule in the cardiovascular system. Although NO has been thoroughly studied in animals, its production was reported in plants in 1979 (Klepper, 1979). The organelles involved in NO production in plants are chloroplasts, peroxisomes, mitochondria, apoplasts (Roszer, 2012), endoplasmic reticula, and cell membranes (Fröhlich and Durner, 2011). NO is produced in plants by both enzymatic and non-enzymatic processes, which mainly include the oxidative and reductive release of NO (Khan et al., 2014). NO synthase has been identified in animals and other organisms, but it has not yet been identified in plants (Roszer, 2012). In plants, NO is produced by nitrate (NO3-) reductase (NR) activity on NO3- and nitrite (Rockel et al., 2002). The production of NO by NR is involved in different biological processes, such as plant immunity and environmental interactions (Mur et al., 2013). Hormones, chemicals, environmental interactions, and osmotic stress play stimulating roles in NO production in plants (Bright et al., 2006; Talwar et al., 2012; Wang et al., 2014). NO is also involved in plant growth and development (Sanz et al., 2014). Recently, the role of NO in plants has been extensively studied in both biotic (Hong et al., 2008; Yun et al., 2012; Khan et al., 2019; Rolly et al., 2020) and abiotic stress conditions (Cantrel et al., 2011; Camejo et al., 2013). To cope with pathogens/pests plants possess various defense mechanisms such as, pathogen associated molecular patterns (PAMPS)-triggered immunity (PTI, formerly called basal resistance), effector-triggered immunity [ETI, formerly called resistance (R)-gene-mediated], and SAR (Zipfel and Felix, 2005; Jones and Dangl, 2006). Furthermore, in response to pathogens, plants also rapidly produce ROS and RNS, which act as signaling molecules to activate several regulatory pathways (Burniston and Wilson, 2008). To explore the role of NO in plants, new technologies and techniques have been developed. NO can regulate protein function through a post-translational modification called nitrosation; a process through which NO reacts covalently with the cysteine (CyS-Fe-NO) residues of the target proteins (Wendehenne et al., 2001; Hess et al., 2005). The reservoirs of NO in the cells are glutathione and CyS-Fe-NO, which are degraded through cellular functions as and where required (Wendehenne et al., 2001; Graziano and Lamattina, 2005). In studies related to plants, the most commonly used NO donors are S-nitrosocysteine (CySNO), S-nitrosoglutathione (GSNO/SNOG) (Askew et al., 1995), and sodium nitroprusside (SNP) (Bivalacqua et al., 1999). Of these, CySNO and GSNO are preferred because they spontaneously release NO (Uehara et al., 2006; Cho et al., 2009). After infiltration into the plants, NO causes S-nitrosation, a post-translational modification of proteins (Zhu et al., 2008). For NO homeostasis, there are several scavengers, including CPTIO (Hogg et al., 1995), 2-4-carboxyphenyl-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (Amano and Noda, 1995), DTCS (Doi et al., 1996), and MGD (Komarov and Lai, 1995).
Plants possess an efficient antioxidant system for the homeostasis of ROS (Rao and Puppo, 2009; Mittler et al., 2011). In Arabidopsis ILR1-like (ILL) is a seven-gene family consisting of ILL1 (At5g56650), ILL2 (At5g56660), ILL3 (At5g54140), IAR3 (ILL4) (At1g51760), ILL5 (At1g51780), ILL6 (At1g44350), and ILR1 (ILL7) (At3g02875). In this family, the first gene to be described was IAR3 (ILL4), which encodes an indole acetic acid-alanine (IAA-Ala) hydrolase (Davies et al., 1999), a wound, and a jasmonate-induced gene named Jasmonate Responsive 3, and is used as a robust JA pathway marker (Titarenko et al., 1997). Auxins play a vital role in the growth and development of plants, but homeostasis of this hormone remains unknown. One important process of auxin homeostasis is the conjugation of the auxin IAA. The auxin conjugates that have been investigated in Arabidopsis thaliana seedlings are IAA-Leu, IAA-Ala, IAA-Asp, IAA-Glu, and IAA-Glc (Kowalczyk and Sandberg, 2001). These conjugates have several key functions in plants, including storage, transport, and the inactivation of IAA (Bartel et al., 2001). The auxin conjugates are converted into indole-3-acetate by the action of several enzymes, including IAR3, ILL5, and ILL6, and are eventually converted by several chemical reactions into camalexin (Truman et al., 2010). Camalexin is a secondary metabolite and is a vital phytoalexin that plays a crucial role during biotic stress in A. thaliana (Glawischnig, 2007). It is important to mention that in an earlier investigation involving transcriptomic analysis of Arabidopsis thaliana (Hussain et al., 2016) we found that the transcript accumulation of ILL6 increased by 97% following 1mM CySNO treatment. Therefore, in the current study, we investigated the role of the NO-induced ATILL6 (IAA-leucine resistant (ILR)-like gene) in plant growth and development under control, oxidative (H2O2 and MV), and nitro-oxidative (CySNO and GSNO) stress conditions. In addition, we investigated the role of NO-induced ATILL6 in plant basal defense, R-gene-mediated resistance, and systemic acquired resistance (SAR). For this purpose, the loss-of-function mutant atill6 and relative control genotypes were inoculated with the virulent and avirulent pathogenic bacteria Pseudomonas syringae (DC3000).
Materials and Methods
Plant Materials and Growth Conditions
Seeds of the A. thaliana wild-type (WT) ecotype columbia zero (Col-0) and the loss-of-function mutant lines atill6 (At1G44350), atgsnor1-3, atcat2, and atsid2 were obtained from Nottingham Arabidopsis Stock Center (http://arabidopsis.info/). The atill6 T-DNA insertion mutant line stock number SALK_22342.42.60.x in Col-0 background was ordered. The seeds were sown under long-day conditions (16 h light and 8 h dark) on either 1/2 Murashige and Skoog (MS) medium or soil at 23 ± 2°C. At the rosette stage (4-week-old plants), samples were collected for genotyping through PCR for the confirmation of T-DNA insertion. The atill6 mutant plants were genotyped to verify T-DNA insertion in the gene of interest (Supplementary Figure 1). The PCR products were sequenced to confirm T-DNA insertion (Supplementary Material). Furthermore, RTPCR was also performed to verify the abolishment of ATILL6 expression in the mutant line (Supplementary Figure 2). All genotypes used in this study were of the Col-0 background. The atgsnor1-3 line was used as a sensitive control. GSNOR1 has a well-established role in plant multiple developmental programs and plant immunity (Kwon et al., 2012). It is a representative line for studying Arabidopsis responses under variable nitro-oxidative environments. The atcat2 was used as a sensitive control for oxidative stress. CAT2 is a leaf, root and seed-expressed Class I catalase with significantly higher transcript abundance than the other catalases and shows circadian and photosynthetic-type rhythm in Arabidopsis (Zhong et al., 1994; Mhamdi et al., 2010). CAT2 is responsible for most of the catalase activity in Arabidopsis as knockout lines of cat1 and cat3 show much less decrease in leaf catalase activity than cat2 (Mhamdi et al., 2010. Journal of Experimental Botany, https://doi.org/10.1093/jxb/erq282). Therefore, cat2 line is regularly used as an oxidative stress-mimic model. For the salicylic acid (SA) pathway, atsid2 knockout mutant was used (Kim et al., 2013). The Salicylic Acid Induction Deficient 2 (SID2) encodes Icochorismate Synthase 1 (ICS1). The atisid2 mutant fails to accumulate SA and is deficient in SA-depenent defense responses.
Oxidative and Nitro-Oxidative Stress Conditions
To explore the role of ATILL6 under redox stress, plants were subjected to oxidative hydrogen peroxide (H2O2) and methyl viologen (MV) and nitro-oxidative stresses (CySNO and GSNO) based on the methods described previously (Khan et al., 2019). The seeds were surface sterilized in 50% commercial bleach with 0.1% Triton X-100 (Sigma Aldrich, USA) for 5 min. The seeds were then rinsed three times with sterilized distilled water and stratified at 4°C for 24 h. The seeds were sown on ½ MS medium supplemented with either 2 mM of H2O2 or 1 μM of methyl viologen for oxidative stress and 0.75 mM of GSNO and 0.75 mM of CySNO for nitro-oxidative stress. After 2 weeks, results were obtained for cotyledon development frequency (CDF) and root and shoot lengths with at least three replicates per treatment as previously described (Shahid et al., 2019). The CDF was used for green developed seedlings (Yun et al., 2011).
Pathogen Growth and Inoculation and Electrolyte Leakage Assay
The virulent and avirulent strains of P. syringae pv. tomato (Pst) DC3000 were grown and inoculated, as described previously (Yun et al., 2011). The bacterial strains were grown on Lauria–Bertani (LB) agar media supplemented with appropriate antibiotics for selection and incubated at 28°C overnight. The single colony was transferred to LB broth with appropriate antibiotics and incubated overnight at 28°C with continuous shaking. The strains were harvested by centrifugation at 8,000 rpm for 3 min and resuspended in 10 mM of MgCl2. To explore, the role of ATILL6 in the basal defense and R-gene-mediated resistance, the atill6 line along with the WT and other relevant controls; atgsnor1-3 and atsid2, were inoculated with virulent Pst DC3000 or avirulent Pst DC3000 with the avrB effector protein. The strains were infiltrated into the abaxial side of the leaves at a concentration of 5 × 105 colony-forming units (CFU). The control plants were infiltrated with only 10 mM of MgCl2. Leaf samples were collected at designated time points for the expressions of PR genes. To investigate further, we evaluated the pathogen growth in the WT and atill6 along with the relevant control mutants. The electrolyte leakage was measured as described previously (Dellagi et al., 1998) with slight modifications. The designated time points for the measurement of conductivity were 1, 2, 4, 6, 8, 12, and 24 h post-infiltration using a portable conductivity meter (HURIBA Twin Cond B-173, Japan).
Quantitative Real-Time PCR (qRT-PCR) Analysis
The total RNA extraction and qRT-PCR analyses were performed as described previously (Hussain et al., 2016). Briefly, total RNA was extracted from the inoculated leaves using TRIzol reagent (Invitrogen, USA) according to the manufacturer's instructions. Complementary DNA (cDNA) was synthesized using the DiaStar™ RT kit (SolGent, Korea) according to the manufacturer's instructions. For transcript accumulation analysis, cDNA was used as a template in the EcoTM real-time PCR machine (Illumina, USA) using the 2X Real-time PCR Master Mix (including SYBR® Green I BioFACT™, Korea) along with 100 ng of template DNA and 10 nM of each primer to a final volume of 20 mL. As a negative control, No Template Control was used, which contains only distilled water instead of template DNA. A two-step PCR reaction was established for 40 cycles under the following conditions: polymerase activation at 95°C for 15 min, denaturation at 95°C for 15 s, and annealing and extension at 60°C for 30 s. The melting curves were assessed at 60–95°C for the verification of amplicon specificity for each primer pair, and actin was used as an internal reference gene (Shahid et al., 2019). The primers used in this study are listed in Supplementary Table 1.
Statistical Analysis
For all assays, the experiments were performed more than twice and the representative results are presented. In media stress conditions, the data point is the mean of three replicates with five plants pooled in each replicate, while for the pathogenicity assay, the data point is the mean of three replicates. The significant difference between each treatment were analyzed by one-way ANOVA analysis of varience, followed by Duncan's multiple range test using statistical analysis system (SAS 9.1). The mean values, standard deviations, and standard errors were obtained in the Microsoft Excel program. The data were then visualized using GraphPad Prism software (version 6.0, San Diego, CA, USA).
Results
ATILL6 Differentially Regulates Root and Shoot Length Under Oxidative and Nitro-Oxidative Stress Conditions
To investigate the role of AtILL6 in plant growth and development, the assessed growth parameters were: CDF, shoot and root length. The atgsnor1-3 (deficient in S-nitrosoglutathione reductase enzyme -GSNOR1), and atcat2 (deficient in the CATALASE2 enzyme) were used as control plants due to their established role in plant growth and defense (Feechan et al., 2005a; Hu et al., 2010). There was no difference between the CDFs of atill6, atgsnor1-3, and atcat2 under control conditions compared to that of the WT (Supplementary Figure 3). In contrast, under H2O2 (oxidative stress) conditions, atill6 showed a significant increase (p ≤ 0.05) in CDF compared to that of the WT, but no significant difference was observed under MV (Supplementary Figure 3). On the other hand, under nitro-oxidative stress conditions (CySNO and GSNO), the CDF in atill6 significantly increased compared to that of the WT (Supplementary Figure 3). The shoot length of the atill6 was significantly longer (p ≤ 0.01) under control, oxidative (H2O2 only), and nitro-oxidative stress conditions induced by CySNO and GSNO as compared to that of the WT plants (Figure 1A and Supplementary Figure 4). The root length of atill6 was significantly shorter under control conditions, but a significant increase was recorded under both CySNO and GSNO stress conditions compared to that of the WT (Figure 1B and Supplementary Figure 4).
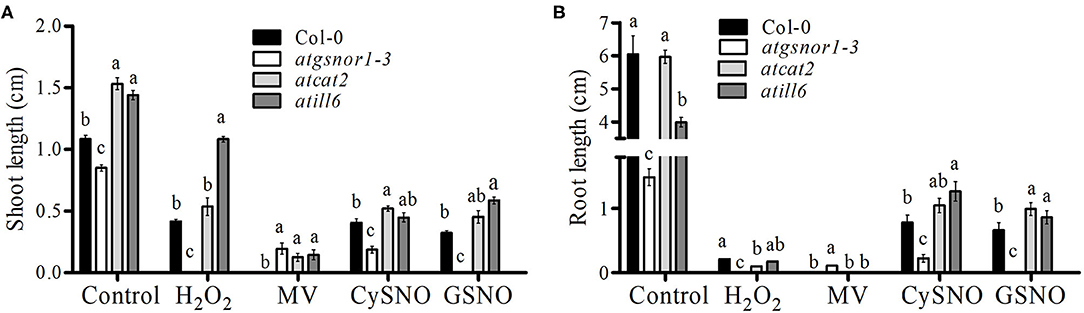
Figure 1. The response of atill6 and relevant control plants after exposure to oxidative (H2O2 and MV), and nitrosative (CySNO and GSNO) stress conditions. (A) Shoot and (B) Root lengths. All data points show the mean of at least three replicates, and the experiment was repeated twice with similar results. The significant difference between the treatments is represented by (a, b, c) one-way ANOVA analysis of varience, followed by Duncan's multiple range test using statistical analysis system (SAS 9.1).
ATILL6 Positively Regulates Plant Basal Defense
To explore the role of AtILL6 in the plant basal defense system, the atill6 line along with WT and genotypes, atgsnor1-3 and atsid2 were inoculated with virulent Pst DC3000. Previously, it has been reported (Feechan et al., 2005b), that atgsnor1-3 is susceptible to Pst DC3000. In the present study, we found that atill6 showed a susceptible phenotype compared to that of the WT (Supplementary Figure 5). To investigate further, we evaluated the pathogen growth in the WT and atill6 along with the relevant control mutants. No significant difference was observed in any genotype for bacterial growth at 0 days post-inoculation (dpi), but at 1, 2, 3, and 4 dpi, atill6 showed a significant increase (p ≤ 0.05) in pathogen growth compared to that of the WT (Figure 2A). These results indicate that ATILL6 plays a positive role in the basal defense system of the plants. As the response shown by plants to biotroph pathogens is induced by a significant plant hormone SA, we investigated the role of ATILL6 in the SA pathway and aimed to quantify the transcript accumulation of PR1 and PR2, the important marker genes in this pathway, in all genotypes used in this study. The results of qRT-PCR revealed significantly lower (p ≤ 0.05) expression of PR1 genes in atill6 after 12, and 24 h, as compared to that of the WT though no significant difference was observed after 48 h of inoculation (Figure 2B). Furthermore, a significant decrease (p ≤ 0.05) was observed in the expression of the PR2 gene at 24 and 48 h, but not at the 12 h time pint (Figure 2C). The other mutants atgsnor1-3 and atsid2 also showed reduced PR1 and PR2 transcript accumulation compared to that of the WT at all-time points (Figures 2B,C).
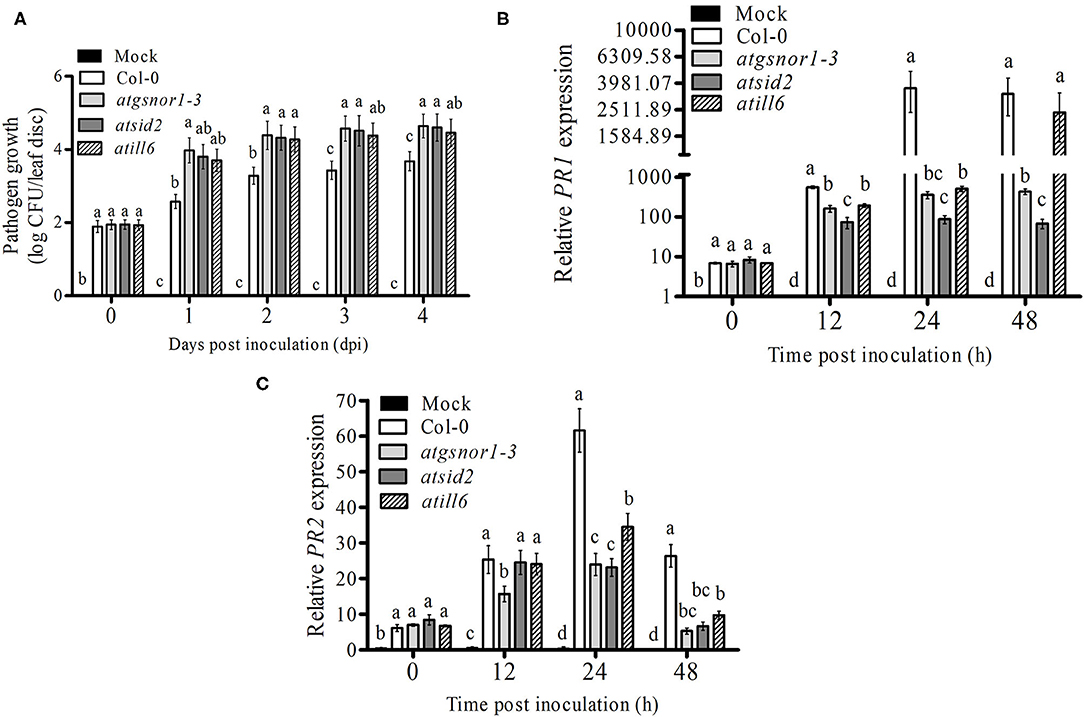
Figure 2. ATILL6 positively regulates the plant basal defense. (A) pathogen growth and (B,C) Relative gene expressions of PR1 and PR2 genes from infiltrated leaves in the indicated genotypes after inoculation with Pst DC3000 virulent bacteria. All data points are the means of three replicates, and error bars represent ± standard error. The significant difference between the treatments is represented by (a, b, c, d), one-way ANOVA analysis of varience, followed by Duncan's multiple range test using statistical analysis system (SAS 9.1).
ATILL6 Positively Regulates R-gene-Mediated Resistance
Plants recognize pathogen-released effector molecules by using R-genes encoded by the nucleotide-binding site leucine-rich repeats to induce R-gene-mediated resistance (Jones and Dangl, 2006). Following R-gene-mediated resistance, plant cells intentionally commit a type of cell suicide called the hypersensitive response (HR) to prevent further spread of disease (Jones and Dangl, 2006). Therefore, we further investigated the functional role of ATILL6 in R-gene-mediated resistance and the HR. For this purpose, all genotypes were challenged with an avirulent strain of Pst DC3000 with the avrB effector protein. After inoculation, the samples were collected from all genotypes at designated time points (0, 6, 12, and 24 h) to analyze the PR gene expression. The qRT-PCR results revealed a significant reduction (p ≤ 0.05) in the transcript accumulation of PR1 in atill6 after 6, 12, and 24 h. Similarly, a significant reduction was recorded in the expression of PR2 genes after 6 and 12 h post-inoculation compared to that of the WT (Figures 3A,B). Additionally, atill6 had higher levels of electrolyte leakage over time compared to that of the WT (Figure 3C). These results indicate that ATILL6 plays a positive role in R-gene-mediated resistance, a defense system of plants. Reduced PR1 and PR2 transcript accumulation was also observed for both atsid2 and atgsnor1-3 (Figures 3A,B).
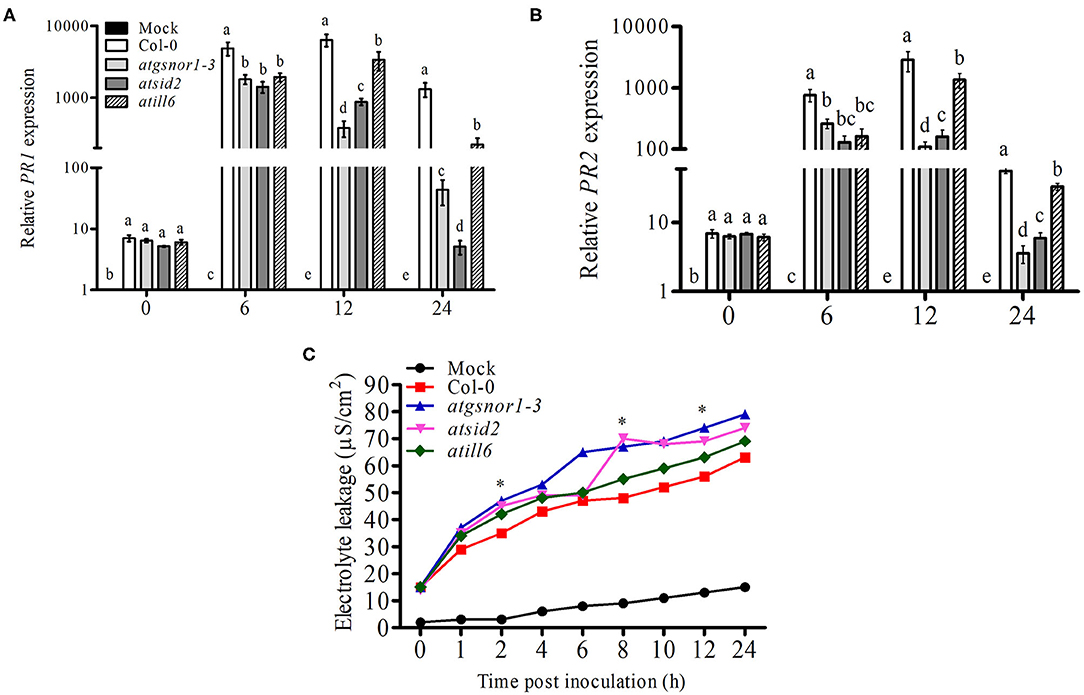
Figure 3. ATILL6 positively regulates plant R-gene-mediated resistance. (A,B) Relative gene expression of PR1 and PR2 genes and (C) electrolyte leakage in the indicated genotypes after inoculation with Pst DC3000 avirulent bacteria. All data points are the means of three replicates, and error bars represent ± standard error. The significant difference between the treatments is represented by (a, b, c, d, e), one-way ANOVA analysis of varience, followed by Duncan's multiple range test using statistical analysis system (SAS 9.1).
ATILL6 Positively Regulates SAR
SAR, a vital defense system of plants, is induced after local infection in the uninfected areas of plants. Both ROS and RNS are involved in the induction of SAR (Song et al., 2006). Therefore, we investigated the role of ATILL6 in SAR. For this purpose, plants were challenged with Pst DC3000 (avrB) at 5 × 106 CFU. After pathogen inoculation, samples were collected from non-inoculated leaves (systemic leaves) and the transcript accumulation of important SAR marker genes, such as PR1, PR2, glyceraldehyde 3-phosphate dehydrogenase, and azelaic acid inducer (AZI) were analyzed over time. The PR1 gene transcript accumulation was significantly (p ≤ 0.05) and highly significantly (p ≤ 0.01) lower after 6 and 12 h, and 24 h, respectively, in the systemic leaves of atill6, while the PR2 gene expression was significantly lower after 12 and 24 h compared to that of the WT (Figures 4A,B). Furthermore, atill6 showed a highly significant and significant decrease in the transcript accumulation of G3DPH after 6 and 12 h, and 24 h, respectively (Figure 4C). The expression of AZI significantly decreased after 6, 12, and 24 h, as shown in Figure 4D. The qRT-PCR results implied that ATILL6 plays a positive role in the activation of SAR when challenged with the avirulent pathogen Pst DC3000 (avrB) at 5 × 106 CFU.
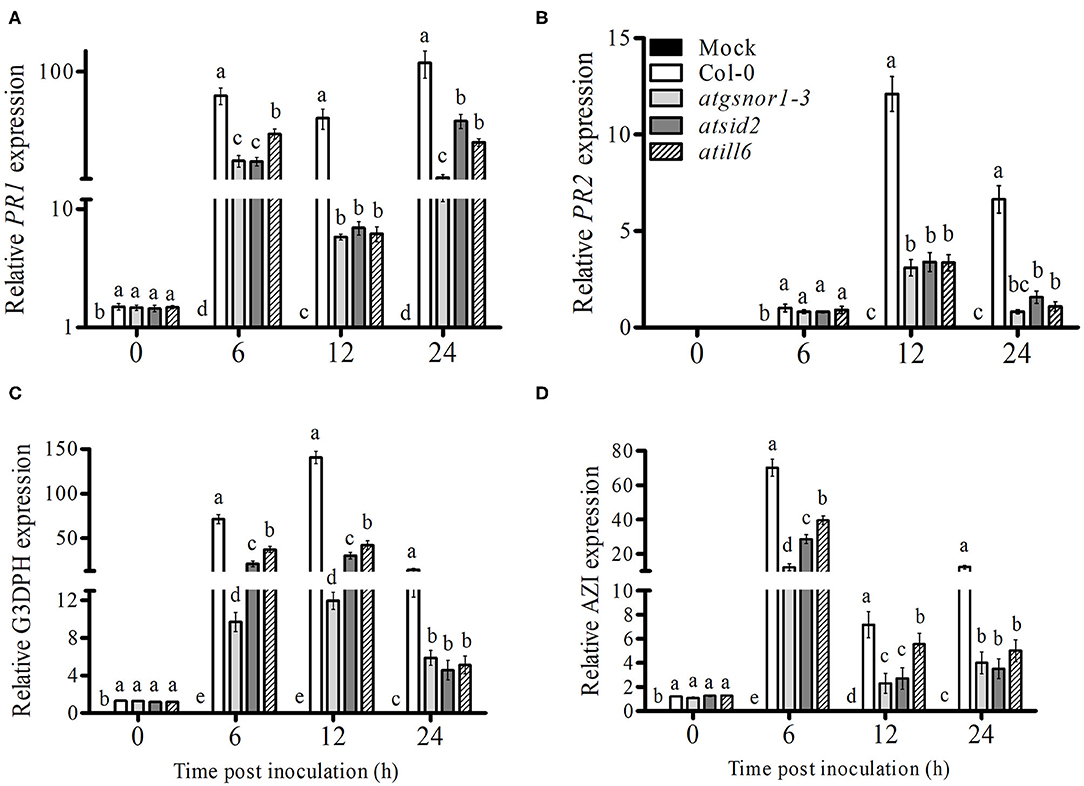
Figure 4. ATILL6 positively regulates plant systemic acquired resistance. (A–D) Relative gene expression of PR1, PR2, G3DPH, and AZI genes, respectively, in the systemic leaves of the indicated genotypes after inoculation with Pst DC3000 avirulent bacteria. All data points are the means of three replicates, and error bars represent ± standard error. The significant difference between the treatments is represented by (a, b, c, d, e), one-way ANOVA analysis of varience, followed by Duncan's multiple range test using statistical analysis system (SAS 9.1).
Discussion
The small redox-active molecule NO with high diffusivity plays a key role in multiple cellular processes, such as seed germination and the plant response to biotic and abiotic stress conditions (Garcia-Mata and Lamattina, 2002; Yun et al., 2011). NO can regulate the transcriptional machinery of certain genes to control different physiological processes. Global changes in gene expression in response to NO have been studied using microarrays (Parani et al., 2004), RNA-seq (Begara-Morales et al., 2014; Hussain et al., 2016), and qRT-PCR (Huang et al., 2002). The present study explored the functional role of NO-induced ATILL6 gene in plant growth and development under control, oxidative, and nitro-oxidative stress conditions. atgsnor1-3 deficient in AtGSNOR1, as well as atcat2 deficient in AtCATALAE2 were used as control plants due to their established role in plant growth and defense (Feechan et al., 2005a; Hu et al., 2010). The phenotypic results revealed that the loss-of-function mutant atill6 had significantly longer shoot length and shorter root length as comapred to control, which indicate that ATILL6 negatively regulates plant shoot length and positively regulates root length (Figures 1A,B and Supplementary Figure 3). Imposition of stress using H2O2 or CySNO and GSNO, the shoot length of atill6 significantly longer compared to WT (Figure 1A). The root length of the atill6 mutant line was also found significantly longer under CySNO and GSNO, as shown in Figure 1B. This phenotypic result implied that ATILL6 positively regulates root length under control conditions but negatively regulates it under CySNO and GSNO stress conditions, compared to that of the WT (Figure 1B). This may be due to the over production of ROS and RNS which leads to higher sensitivity of plants to oxidative stress (Garcia-Mata and Lamattina, 2002). Kopyra et al. (2004) also suggested that an increase in the shoot length under nitro-oxidative stress (GSNO) may be due to the possible role of NO in seed germination and seedling growth. Furthermore, Beligni and Lamattina (2000) suggested that NO may break seed dormancy to provide a good starting point for plants to grow which may be even better than GA3. To sum up, the current study revealed that ATILL6 differentially regulates the growth traits in A. thaliana under control and oxidative and nitro-oxidative stress conditions, depending upon the type of ROS and RNS donors.
We further evaluated the role of NO-induced ATILL6 in plant basal defense, R-gene-mediated resistance, and SAR. As indicated by Supplementary Figure 4, the loss-of-function mutant line atill6 plants were susceptible to infection when they were exposed to a virulent pathogen (Pst DC3000). This was further confirmed by the qRT-PCR results of the SA-dependent PR gene expressions, and the relative expressions of the PR1 and PR2 genes were significantly reduced in the loss-of-function mutant atill6 compared to those of the WT (Figure 2). The disease-susceptible phenotype, higher pathogen growth, and lower expression of PR genes indicated that NO-induced ATILL6 has a major role in plant basal defense. Therefore, we further evaluated the role of ATILL6 in ETI. Plants can identify the effector proteins of pathogens, such as avr, by the R-gene and cause R-gene-mediated resistance to reduce further spread of the disease (Glazebrook et al., 1996). The results indicate that, similar to plant basal defense, ATILL6 also positively regulated plant R-gene-mediated resistance. As shown by Figure 3, after the inoculation of an avirulent pathogen (Pst DC3000 expressing avrB effector), the expressions of the PR genes were highly reduced in the atill6 loss-of-function mutant compared to those of the WT. After the recognition of effector proteins by the R-gene of the host plant, the defense signals pass from local to systemic tissues of the plants due to the expression of PR, G3DPH, and AZI genes to activate plant SAR (Wang et al., 2014). SAR is a defense strategy of plants that assists with protecting the plants from secondary infections of virulent pathogens (El-Shetehy et al., 2015). Plants require SA for pathogen identification, the subsequent establishment of local resistance, and, eventually to the whole plant in order to protect them against biotic stress conditions (Tsuda et al., 2008). Thus, we also investigated the role of ATILL6 in SAR, and the results revealed that ATILL6 positively regulates SAR, as the mutant line atill6 had significantly lower transcript accumulation of PR, G3DPH, and AZI genes compared to that of the WT (Figures 4A–D).
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
B-WY and MK designed the experiments. MK performed the experiments and wrote the manuscript. AP and TNIAA drafted the manuscript. B-GM and D-SL conducted data analysis. B-HL and AH critically reviewed and edited the manuscript. B-WY provided supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (Grant number 2020R1I1A3073247).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.685156/full#supplementary-material
Supplementary Figure 1. Genotyping of atill6 mutant line.
Supplementary Figure 2. RTPCR confirmation for the abolishment of ILL6 expression in the ill6 T-DNA insertion mutant line.
Supplementary Figure 3. Cotyledon development frequency (CDF), after treatment with oxidative (H2O2 and MV), and nitrosative (CySNO and GSNO) stress conditions in atill6 mutant line and relevant control plants. All data points show the mean of at least three replicates, and the experiment was repeated twice with similar results. The significant difference between the treatments is represented by (a, b, c, d) one-way ANOVA analysis of varience, followed by Duncan's multiple range test using statistical analysis system (SAS 9.1).
Supplementary Figure 4. Phenotypic responses, after treatment with oxidative (H2O2 and MV), and nitrosative (CySNO and GSNO) stress conditions in atill6 mutant line and relevant control plants.
Supplementary Figure 5. Symptom development in atill6 and relevant control after inoculation with Pst DC3000 virulent bacteria.
References
Amano, F., and Noda, T. (1995). Improved detection of nitric oxide radical (NO•) production in an activated macrophage culture with a radical scavenger, car? y PTIO, and Griess reagent. FEBS Lett. 368, 425–428. doi: 10.1016/0014-5793(95)00700-J
Askew, S. C., Butler, A. R., Flitney, F. W., Kemp, G. D., and Megson, I. L. (1995). Chemical mechanisms underlying the vasodilator and platelet anti-aggregating properties of S-Nitroso-N-Acetyl-Dl-Penicillamine and S-Nitrosoglutathione. Bioorgan. Med. Chem. 3, 1–9. doi: 10.1016/0968-0896(94)00139-T
Bartel, B., Leclere, S., Magidin, M., and Zolman, B. K. (2001). Inputs to the active indole-3-acetic acid pool: de novo synthesis, conjugate hydrolysis, and indole-3-butyric acid b-oxidation. J. Plant Growth Regul. 20, 198–216. doi: 10.1007/s003440010025
Begara-Morales, J. C., Sanchez-Calvo, B., Luque, F., Leyva-Perez, M. O., Leterrier, M., Corpas, F. J., et al. (2014). Differential transcriptomic analysis by RNA-seq of GSNO-responsive genes between arabidopsis roots and leaves. Plant Cell Physiol. 55, 1080–1095. doi: 10.1093/pcp/pcu044
Beligni, M. V., and Lamattina, L. (2000). Nitric oxide stimulates seed germination and de-etiolation, and inhibits hypocotyl elongation, three light-inducible responses in plants. Planta 210, 215–221. doi: 10.1007/PL00008128
Bivalacqua, T. J., Champion, H. C., Wang, R., Kadowitz, P. J., Doherty, P. C., and Hellstrom, W. J. G. (1999). Feline penile erection induced by transurethral administration of sodium nitroprusside. Urol. Res. 27, 432–436. doi: 10.1007/s002400050132
Bright, J., Desikan, R., Hancock, J. T., Weir, I. S., and Neill, S. J. (2006). ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. Plant J. 45, 113–122. doi: 10.1111/j.1365-313X.2005.02615.x
Burniston, M. T., and Wilson, D. J. (2008). Radioiodine ablation outcomes after imaging with 123I or 131I: is no news good news? J. Nuclear Med. 49, 166–166. doi: 10.2967/jnumed.107.047076
Camejo, D., Romero-Puertas, M. D., Rodriguez-Serrano, M., Sandalio, L. M., Lazaro, J. J., Jimenez, A., et al. (2013). Salinity-induced changes in S-nitrosylation of pea mitochondrial proteins. J. Proteomics 79, 87–99. doi: 10.1016/j.jprot.2012.12.003
Cantrel, C., Vazquez, T., Puyaubert, J., Reze, N., Lesch, M., Kaiser, W. M., et al. (2011). Nitric oxide participates in cold-responsive phosphosphingolipid formation and gene expression in Arabidopsis thaliana. New Phytol. 189, 415–427. doi: 10.1111/j.1469-8137.2010.03500.x
Cho, D. H., Nakamura, T., Fang, J. G., Cieplak, P., Godzik, A., Gu, Z., et al. (2009). S-Nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science 324, 102–105. doi: 10.1126/science.1171091
Culotta, E., and Koshland, D. E Jr. (1992). NO news is good news. Science 258, 1862–1866. doi: 10.1126/science.1361684
Davies, R. T., Goetz, D. H., Lasswell, J., Anderson, M. N., and Bartel, B. (1999). IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell 11, 365–376. doi: 10.1105/tpc.11.3.365
Dellagi, A., Brisset, M. N., Paulin, J. P., and Expert, D. (1998). Dual role of desferrioxamine in Erwinia amylovora pathogenicity. Mol. Plant Microbe Interact. 11, 734–742. doi: 10.1094/MPMI.1998.11.8.734
Doi, K., Akaike, T., Horie, H., Noguchi, Y., Fujii, S., Beppu, T., et al. (1996). Excessive production of nitric oxide in rat solid tumor and its implication in rapid tumor growth. Cancer 77, 1598–1604. doi: 10.1002/(SICI)1097-0142(19960415)77:8+<1598::AID-CNCR3>3.0.CO
Domingos, P., Prado, A. M., Wong, A., Gehring, C., and Feijo, J. A. (2015). Nitric oxide: a multitasked signaling gas in plants. Mol. Plant 8, 506–520. doi: 10.1016/j.molp.2014.12.010
El-Shetehy, M., Wang, C. X., Shine, M. B., Yu, K. S., Kachroo, A., and Kachroo, P. (2015). Nitric oxide and reactive oxygen species are required for systemic acquired resistance in plants. Plant Signal. Behav. 10:998544. doi: 10.1080/15592324.2014.998544
Farnese, F. S., Menezes-Silva, P. E., Gusman, G. S., and Oliveira, J. A. (2016). When bad guys become good ones: the key role of reactive oxygen species and nitric oxide in the plant responses to abiotic stress. Front. Plant Sci. 7:471. doi: 10.3389/fpls.2016.00471
Feechan, A., Kwon, E., Yun, B.-W., Wang, Y., Pallas, J. A., and Loake, G. J. (2005a). A central role for S-nitrosothiols in plant disease resistance. Proc. Natl. Acad. Sci. U. S. A. 102, 8054–8059. doi: 10.1073/pnas.0501456102
Feechan, A., Kwon, E. J., Yuri, B. Y., Wang, Y., Pallas, J. A., and Loake, G. J. (2005b). A central role for S-nitrosothiols in plant disease resistance. Comp. Biochem. Physiol. 141:S241. Available online at: https://doi.org/10.1073/pnas.0501456102
Fröhlich, A., and Durner, J. (2011). The hunt for plant nitric oxide synthase (NOS): is one really needed? Plant Sci. 181, 401–404. doi: 10.1016/j.plantsci.2011.07.014
Garcia-Mata, C., and Lamattina, L. (2002). Nitric oxide and abscisic acid cross talk in guard cells. Plant Physiol. 128, 790–792. doi: 10.1104/pp.011020
Glazebrook, J., Rogers, E. E., and Ausubel, F. M. (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143, 973–982. doi: 10.1093/genetics/143.2.973
Graziano, M., and Lamattina, L. (2005). Nitric oxide and iron in plants: an emerging and converging story. Trends Plant Sci. 10, 4–8. doi: 10.1016/j.tplants.2004.12.004
Hess, D. T., Matsumoto, A., Kim, S.-O., Marshall, H. E., and Stamler, J. S. (2005). Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell Biol. 6, 150–166. doi: 10.1038/nrm1569
Hogg, N., Singh, R. J., Joseph, J., Neese, F., and Kalyanaraman, B. (1995). Reactions of nitric-oxide with nitronyl nitroxides and oxygen - prediction of nitrite and nitrate formation by kinetic simulation. Free Radic. Res. 22, 47–56. doi: 10.3109/10715769509147527
Hong, J. K., Yun, B. W., Kang, J. G., Raja, M. U., Kwon, E., Sorhagen, K., et al. (2008). Nitric oxide function and signalling in plant disease resistance. J. Exp. Bot. 59, 147–154. doi: 10.1093/jxb/erm244
Hu, Y. Q., Liu, S., Yuan, H. M., Li, J., Yan, D. W., Zhang, J., et al. (2010). Functional comparison of catalase genes in the elimination of photorespiratory H2O2 using promoter- and 3'-untranslated region exchange experiments in the Arabidopsis cat2 photorespiratory mutant. Plant Cell Environ. 33, 1656–1670. doi: 10.1111/j.1365-3040.2010.02171.x
Huang, X., Von Rad, U., and Durner, J. (2002). Nitric oxide induces transcriptional activation of the nitric oxide-tolerant alternative oxidase in Arabidopsis suspension cells. Planta 215, 914–923. doi: 10.1007/s00425-002-0828-z
Hussain, A., Mun, B.-G., Imran, Q. M., Lee, S.-U., Adamu, T. A., Shahid, M., et al. (2016). Nitric oxide mediated transcriptome profiling reveals activation of multiple regulatory pathways in Arabidopsis thaliana. Front. Plant Sci. 7:975. doi: 10.3389/fpls.2016.00975
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Khan, M., Imran, Q. M., Shahid, M., Mun, B.-G., Lee, S.-U., Khan, M. A., et al. (2019). Nitric oxide-induced AtAO3 differentially regulates plant defense and drought tolerance in Arabidopsis thaliana. BMC Plant Biol. 19, 1–19. doi: 10.1186/s12870-019-2210-3
Khan, M. N., Mobin, M., Mohammad, F., and Corpas, F. J. (2014). Nitric Oxide in Plants: Metabolism and Role in Stress Physiology. Cham: Springer. doi: 10.1007/978-3-319-06710-0
Kim, Y., Park, S., Gilmour, S. J., and Thomashow, M. F. (2013). Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of Arabidopsis. Plant J. 75, 364–376. doi: 10.1111/tpj.12205
Klepper, L. (1979). Nitric oxide (NO) and nitrogen dioxide (NO2) emissions from herbicide-treated soybean plants. Atmos. Environ. 13, 537–542.
Komarov, A. M., and Lai, C. S. (1995). Detection of nitric-oxide production in mice by spin-trapping electron-paramagnetic-resonance spectroscopy. Biochim. Biophys. Acta 1272, 29–36. doi: 10.1016/0925-4439(95)00061-8
Kopyra, M., Gwozdz, E., and Bialczak, E. (2004). The role of nitric oxide in plants treated with heavy metals. Acta Physiol. Plant. 26, 459–472. doi: 10.1007/s11738-004-0037-4
Kowalczyk, M., and Sandberg, G. (2001). Quantitative analysis of indole-3-acetic acid metabolites in Arabidopsis. Plant Physiol. 127, 1845–1853. doi: 10.1104/pp.010525
Kwon, E., Feechan, A., Yun, B.-W., Hwang, B.-H., Pallas, J. A., Kang, J.-G., et al. (2012). AtGSNOR1 function is required for multiple developmental programs in Arabidopsis. Planta 236, 887–900. doi: 10.1007/s00425-012-1697-8
Mhamdi, A., Queval, G., Chaouch, S., Vanderauwera, S., Van Breusegem, F., and Noctor, G. (2010). Catalase function in plants: a focus on Arabidopsis mutants as stress-mimic models. J. Exp. Bot. 61, 4197–4220. doi: 10.1093/jxb/erq282
Mittler, R., Vanderauwera, S., Suzuki, N., Miller, G., Tognetti, V. B., Vandepoele, K., et al. (2011). ROS signaling: the new wave? Trends Plant Sci. 16, 300–309. doi: 10.1016/j.tplants.2011.03.007
Mur, L. A. J., Mandon, J., Persijn, S., Cristescu, S. M., Moshkov, I. E., Novikova, G. V., et al. (2013). Nitric oxide in plants: an assessment of the current state of knowledge. Aob Plants 5:pls052. doi: 10.1093/aobpla/pls052
Parani, M., Rudrabhatla, S., Myers, R., Weirich, H., Smith, B., Leaman, D. W., et al. (2004). Microarray analysis of nitric oxide responsive transcripts in Arabidopsis (vol 2, pg 359, 2004). Plant Biotechnol. J. 2, 467–467. doi: 10.1111/j.1467-7652.2004.00085.x
Rao, L. A. D., and Puppo, A. (2009). Reactive Oxygen Species in Plant Signaling. Dordrecht; New York: Springer Verlag. doi: 10.1007/978-3-642-00390-5
Rockel, P., Strube, F., Rockel, A., Wildt, J., and Kaiser, W. M. (2002). Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J. Exp. Bot. 53, 103–110. doi: 10.1093/jexbot/53.366.103
Rolly, N. K., Imran, Q. M., Shahid, M., Imran, M., Khan, M., Lee, S.-U., et al. (2020). Drought-induced AtbZIP62 transcription factor regulates drought stress response in Arabidopsis. Plant Physiol. Biochem. 156, 384–395. doi: 10.1016/j.plaphy.2020.09.013
Roszer, T. (2012). “Nitric oxide synthesis in the chloroplast,” in The Biology of Subcellular Nitric Oxide, ed T. Roszer (Dordrecht: Springer), 49–66. doi: 10.1007/978-94-007-2819-6_3
Sanz, L., Fernandez-Marcos, M., Modrego, A., Lewis, D. R., Muday, G. K., Pollmann, S., et al. (2014). Nitric oxide plays a role in stem cell niche homeostasis through its interaction with auxin(1[W][OPEN]). Plant Physiol. 166, 1972–1984. doi: 10.1104/pp.114.247445
Shahid, M., Imran, Q. M., Hussain, A., Khan, M., Lee, S. U., Mun, B. G., et al. (2019). Comprehensive analyses of nitric oxide-induced plant stem cell-related genes in Arabidopsis thaliana. Genes 10:190. doi: 10.3390/genes10030190
Song, L. L., Ding, W., Zhao, M. G., Sun, B. T., and Zhang, L. X. (2006). Nitric oxide protects against oxidative stress under heat stress in the calluses from two ecotypes of reed. Plant Sci. 171, 449–458. doi: 10.1016/j.plantsci.2006.05.002
Talwar, P. S., Gupta, R., Maurya, A. K., and Deswal, R. (2012). Brassica juncea nitric oxide synthase like activity is stimulated by PKC activators and calcium suggesting modulation by PKC-like kinase. Plant Physiol. Biochem. 60, 157–164. doi: 10.1016/j.plaphy.2012.08.005
Titarenko, E., Rojo, E., Leon, J., and Sanchez-Serrano, J. J. (1997). Jasmonic acid-dependent and-independent signaling pathways control wound-induced gene activation in Arabidopsis thaliana. Plant Physiol. 115, 817–826. doi: 10.1104/pp.115.2.817
Truman, W. M., Bennett, M. H., Turnbull, C. G., and Grant, M. R. (2010). Arabidopsis auxin mutants are compromised in systemic acquired resistance and exhibit aberrant accumulation of various indolic compounds. Plant Physiol. 152, 1562–1573. doi: 10.1104/pp.109.152173
Tsuda, K., Sato, M., Glazebrook, J., Cohen, J. D., and Katagiri, F. (2008). Interplay between MAMP-triggered and SA-mediated defense responses. Plant J. 53, 763–775. doi: 10.1111/j.1365-313X.2007.03369.x
Uehara, T., Nakamura, T., Yao, D. D., Shi, Z. Q., Gu, Z. Z., Ma, Y. L., et al. (2006). S-Nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature 441, 513–517. doi: 10.1038/nature04782
Wang, C. X., El-Shetehy, M., Shine, M. B., Yu, K. S., Navarre, D., Wendehenne, D., et al. (2014). Free radicals mediate systemic acquired resistance. Cell Rep. 7, 348–355. doi: 10.1016/j.celrep.2014.03.032
Wendehenne, D., Pugin, A., Klessig, D. F., and Durner, J. (2001). Nitric oxide: comparative synthesis and signaling in animal and plant cells. Trends Plant Sci. 6, 177–183. doi: 10.1016/S1360-1385(01)01893-3
Yun, B. W., Feechan, A., Yin, M., Saidi, N. B., Le Bihan, T., Yu, M., et al. (2011). S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 478, 264–268. doi: 10.1038/nature10427
Yun, B. W., Spoel, S. H., and Loake, G. J. (2012). Synthesis of and signalling by small, redox active molecules in the plant immune response. Biochim. Biophys. Acta 1820, 770–776. doi: 10.1016/j.bbagen.2011.06.015
Zhong, H. H., Young, J. C., Pease, E. A., Hangarter, R. P., and Mcclung, C. R. (1994). Interactions between light and the circadian clock in the regulation of CAT2 expression in Arabidopsis. Plant Physiol. 104, 889–898. doi: 10.1104/pp.104.3.889
Zhu, J., Li, S., Marshall, Z. M., and Whorton, A. R. (2008). A cystine-cysteine shuttle mediated by xCT facilitates cellular responses to S-nitrosoalbumin. Am. J. Physiol. 294, C1012–C1020. doi: 10.1152/ajpcell.00411.2007
Keywords: Arabidopsis thaliana, growth under control, oxidative stress, nitro-oxidative stress, plant defense
Citation: Khan M, Al Azawi TNI, Pande A, Mun B-G, Lee D-S, Hussain A, Lee B-H and Yun B-W (2021) The Role of Nitric Oxide-Induced ATILL6 in Growth and Disease Resistance in Arabidopsis thaliana. Front. Plant Sci. 12:685156. doi: 10.3389/fpls.2021.685156
Received: 24 March 2021; Accepted: 07 June 2021;
Published: 02 July 2021.
Edited by:
Andrea Chini, Consejo Superior de Investigaciones Científicas (CSIC), SpainReviewed by:
Yiqin Wang, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, ChinaManda Yu, University of Wisconsin-Milwaukee, United States
Michael Skelly, University of Edinburgh, United Kingdom
Copyright © 2021 Khan, Al Azawi, Pande, Mun, Lee, Hussain, Lee and Yun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Byung-Wook Yun, Ynd5dW5Aa251LmFjLmty
 Murtaza Khan
Murtaza Khan Tiba Nazar Ibrahim Al Azawi2
Tiba Nazar Ibrahim Al Azawi2 Anjali Pande
Anjali Pande Bong-Gyu Mun
Bong-Gyu Mun Adil Hussain
Adil Hussain Byung-Wook Yun
Byung-Wook Yun