
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 02 December 2021
Sec. Plant Abiotic Stress
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.685054
This article is part of the Research TopicIdentification and Functional Dissection of Stress-responsive Genes in CottonView all 26 articles
 Kangtai Sun1†
Kangtai Sun1† Hui Fang1†
Hui Fang1† Yu Chen2†
Yu Chen2† Zhimin Zhuang1
Zhimin Zhuang1 Qi Chen1
Qi Chen1 Tingyu Shan1
Tingyu Shan1 Muhammad Kashif Riaz Khan3
Muhammad Kashif Riaz Khan3 Jun Zhang2*
Jun Zhang2* Baohua Wang1*
Baohua Wang1*Plant cytochrome P450 (P450) participates in a wide range of biosynthetic reactions and targets a variety of biological molecules. These reactions lead to various fatty acid conjugates, plant hormones, secondary metabolites, lignin, and various defensive compounds. In our previous research, transcriptome analysis was performed on the salt-tolerant upland cotton “Tongyan No. 1.” Many differentially expressed genes (DEGs) belong to the P450 family, and their domains occur widely in plants. In this current research, P450 genes were identified in Gossypium hirsutum with the aid of bioinformatics methods for investigating phylogenetic relations, gene structure, cis-elements, chromosomal localization, and collinearity within a genome. qRT-PCR was conducted to analyze P450 gene expression patterns under salt stress. The molecular weights of the 156 P450 genes were in the range of 5,949.6–245,576.3 Da, and the length of the encoded amino acids for all the identified P450 genes ranged from 51 to 2,144. P450 proteins are divided into four different subfamilies based on phylogenetic relationship, gene structure, and chromosomal localization of gene replication. The length of P450 genes in upland cotton differs greatly, ranging from 1,500 to 13,000 bp. The number of exons in the P450 family genes ranged from 1 to 9, while the number of introns ranged from 0 to 8, and there were similar trends within clusters. A total of 31 cis-acting elements were identified by analyzing 1,500 bp promoter sequences. Differences were found in cis-acting elements among genes. The consistency between qRT-PCR and previous transcriptome analysis of salt tolerance DEGs indicated that they were likely to be involved in the salt tolerance of cotton seedlings. Our results provide valuable information on the evolutionary relationships of genes and functional characteristics of the gene family, which is beneficial for further study of the cotton P450 gene family.
With the continuous expansion of activities related to human survival and the rapid development of industrial activities, earth’s environments are continuously deteriorating, which has led to great challenges in the development of agriculture. Land salinization is one of the important problems facing agricultural production. Salt stress reduces water uptake by the root system and leads to internal dehydration, and salt accumulation affects metabolic processes, especially in photosynthetic cells. Damage to plants can occur when transport tissues carry too much salt (Munns, 2020). It was reported that after cotton was exposed to salt stress, superoxide dismutase (SOD), glutathione reductase (GR), and peroxidase (POD) activity increased with the increase of salt concentration; meanwhile, net photosynthetic rate and stomatal conductance decreased with salt stress, and leaf chlorophyll content decreased significantly (Meloni et al., 2003).
Plant cytochrome P450 (P450) participates in a wide range of biosynthetic reactions and targets a variety of biological molecules. These reactions lead to various fatty acid conjugates, plant hormones, secondary metabolites, lignin, and various defensive compounds (Schuler and Werck-Reichhart, 2003; Tang et al., 2016), e.g., CYP706B1 and CYP82D113 are involved in the biosynthesis of gossypol in plants (Ma et al., 2016; Teh et al., 2017). CYP71AV1 is involved in the biosynthesis of artemisinin. Plant genome annotation shows that P450 genes account for up to 1% of plant genes. The number and diversity of P450 genes are partly responsible for the presence of multiple biologically active compounds (Mizutani and Sato, 2011).
P450 is the largest enzyme family that supports plant metabolism to date and is representative of metabolic structure and evolution. It is a very useful tool for discovering the basic functions of plant metabolism and guiding gene function research (Du et al., 2016; Ghosh, 2017; Li and Wei, 2020). With the development of biotechnology in recent years, a series of factors related to adversity have been identified (Gong et al., 2020; Wang et al., 2020), and the P450 family is widely studied.
In the P450 family, some genes are copied in large numbers (Nelson and Werck-Reichhart, 2011), e.g., genes involved in the phenylalanine pathway. Genes in the P450 family have been verified to be involved in the biosynthesis of jasmonic acid (JA) in plants such as soybeans and wheat, with locations in stems and/or roots (Yan et al., 2016). Studies have also shown that P45094B3 exerts a negative feedback control effect on JA-Ile (jasmonic acid and isoleucine) levels (Koo et al., 2011), and it plays a key role in the weakening of the jasmonate response. Biosynthetic P450 is used in the synthesis of lignin intermediates, sterols, terpenes, flavonoids, isoflavonoids, furanocoumarins and various other secondary plant products, in which it plays a vital role (Schuler and Werck-Reichhart, 2003; Carelli et al., 2011; Nelson, 2011). Some subfamily members are better retained. P450 genes also participate in the primary metabolism of plants involved in the biosynthesis of essential sterols and steroid hormones (Mills et al., 2018). They may even perform chemical defense against specific pathogens in specific species (Schuler, 2010).
Cotton contains gossypol and many terpenoids (Qian and Wang, 1984; Tian et al., 2016). Many terpenoids are used as signal molecules, participating in the interaction between organisms or as defensive compounds that protect plants and resist pathogen infection and animal ingestion (Pompon et al., 1996; Wang et al., 2004; Sunilkumar et al., 2006). Semiterpenoids have great complexity and diversity. They are closely related to the diverse enzyme genes, including P450, associated with secondary metabolic pathways (Sunilkumar et al., 2006). Studies have shown that RNA interference is used to inhibit the expression of CYP82D109 in upland cotton. The accumulation of related compounds can be detected in transgenic plants (Zhao et al., 2018). When virus-induced gene silencing (VIGS) inhibits the expression of CYP82D113 in cotton plants, the contents of gossypol and hemigossypolone decrease significantly (Ma et al., 2016). To date, the analysis of the gossypol biosynthetic pathway has not been completed. The functions of the four enzymes, including sesquiterpene synthase CDNC, short-chain alcohol dehydrogenase DH1 and two cytochrome P450 monooxygenases, have been identified (Bathe and Tissier, 2019). In biosynthesis, the hydrocarbon backbone produced by terpene synthase is usually oxidized by cytochrome P450 oxidase. In cotton, gossypol mainly accumulates in the epidermis of roots and aboveground tissues (leaves, stems, flowers, bolls, and seeds) (Guo et al., 2009). The discovery of key genes in the P450 family is helpful for research on stress tolerance in various species.
In our previous research, transcriptome analysis was performed on the salt-tolerant upland cotton line “Tongyan No. 1.” A batch of differentially expressed genes (DEGs) were obtained, some of which belong to the same P450 family. P450 belongs to a multigene family. To fully understand the gene structure and protein function of the P450 family in the upland cotton genome, bioinformatics methods were used to analyze the number of P450 genes in upland cotton and its gene structure, chromosome location, phylogeny, and expression of salt tolerance. The results of this research will help reveal the salt stress response mechanism in cotton.
A salt-tolerant upland cotton strain, “Tongyan No. 1,” was used for the following process. Plump and similarly sized seeds were selected and sterilized after dipping in dilute hydrochloric acid, and then they were planted in flowerpots filled with a mixture of nutrient soil and vermiculite at a ratio of 3:1 and cultivated in a greenhouse at a temperature of 23°C with a light:dark ratio of 16:8 h. When the second euphyll was fully expanded, the cotton plants were divided into the control and experimental groups. The control group was watered with 250 ml water solution, while the experimental group was watered with 250 mmol NaCl of the same volume every afternoon (Shaheen et al., 2012). Leaves were collected from the two cotton groups after 2 days in the treatments and stored in an ultralow temperature refrigerator at −80°C for RNA extraction and transcriptome sequencing.
The genome data of upland cotton were obtained from the Cotton Research Institute of Nanjing Agricultural University1. The Hidden Markov Model (HMM) database (protein families database of alignments) and HMM. The Pfam database2 was used to download the seed file (PF00067) of the P450 gene. HMMER 3.0 software and the local BLASTP program were applied to compare and search for sequences containing the P450 protein domain. After the screening, the protein sequences were uploaded to the online software Pfam, the smart database (Simple Modular Architecture Research Tool, SMART3) and the Conserved Domain Database (Conserved domain database, CDD4), and sequence alignment and analysis were carried out to remove unannotated genes and redundant sequences. Afterward, the gene IDs containing the P450 domain and its corresponding sequence were obtained. ExPASy Proteomics Server software5 was used to analyze the amino acid sequence of all members of the cotton P450 gene family and calculate the amino acid length and isoelectric point.
The MEGA program (MEGA7) was used to perform multiple sequence alignments of the obtained genes (Kumar et al., 2016). Based on the alignment results, an evolutionary tree was constructed with the adjacency joining algorithm. A bootstrap test (replications) with 1,000 iterations was performed. The obtained evolutionary tree was further modified using the Evolview6 website. Meme software7 was applied to carry out motif analysis based on the protein sequences of P450 family genes. The shortest motif length was six base pairs, and the greatest motif length was 50 base pairs.
The location information of all the P450 family genes, including the exons, introns, and UTR location information on the chromosome, were available in the reference genome of Gossypium hirsutum L. acc. TM-1. dataset. A gene structure diagram (Hu et al., 2015) was drawn using the online website GSDS8. To explore the regulation of gene expression, 1.5 kb sequences upstream of ATG in all the P450 family genes were extracted to analyze the cis-elements of the genes. Plant CARE (Cis-Acting Regulatory Element9) was used to further analyze the cis-elements, and the cis-element information obtained was mapped through the GSDS online website.
MapChart software was applied to analyze and draw the chromosome location map, and AI (Adobe Illustrator CS6) software was used to visualize the chromosome position map.
All the upland cotton protein sequences were BLAST compared by MCSCANX software, and collinear genes were found throughout the genome. The collinearity of genes in the gene family was discussed, and the gene family circle was drawn by MCSCANX software with special annotation.
P450 family genes were confirmed to be associated with salt tolerance in a previous study on the salt-tolerant transcriptome of the upland cotton ‘‘Tongyan No. 1.’’ The expression heatmap of all the P450 family genes in upland cotton was drawn with the heatmap website10, with units of FPKM (fragments per kilobase of exon model per million mapped fragments, that is, the fragments mapped per million transcribed per thousand base length).
Total RNA was extracted from cotton leaves in various stages after salt treatment of “Tongyan No. 1” using Aidlab’s EASYspin polysaccharide polyphenol plant total RNA extraction kit. Primer5 software was used to design primers for qRT-PCR experiment (Table 1). The M5 Super plus kit purchased from Beijing Polymer Company was used to synthesize first-strand cDNA. Quantitative real-time polymerase chain reaction (qRT-PCR) was carried out in triplicate for each sample using a 2*M5 HIPer Real-time PCR Supermix (SYBRGreen) kit with an ABI 7500 real-time PCR system. The upland cotton GhUBQ7 (DQ116441) gene was used as a control for normalization between samples. Relative transcript levels were calculated using the comparative threshold cycle method (Livak and Schmittgen, 2001).
Using domain information (PF00064) obtained from domain prediction to identify members of the whole cotton genome, more than 300 genes were obtained. After comparison of the CDD, SMART, and other databases, 156 pairs of genes were identified as belonging to the P450 gene family. Then, the physicochemical properties of the amino acid sequences of cotton P450 gene family members were analyzed (Supplementary Table 1). The results showed that the molecular weight of the 156 P450 genes was in the range of 5,949.6–245,576.3 Da. The length of the encoded amino acids for all the identified P450 genes ranged from 51 to 2,144, and the theoretical isoelectric point size of the coding protein ranged from 4.15 to 10.97, with an average of 10.23, which is strongly alkaline.
To analyze the evolutionary relationship between the various members of the P450 family, ClustalW in MEGA7 software was used to carry out alignment for the 156 obtained protein sequences, and then the sequences with genetic distances that were too long were removed. A rootless phylogeny was constructed using the neighbor-joining method in MEGA7 software (Figure 1). As shown in Figure 1, the P450 protein sequence is divided into four different subfamilies. The group marked in red is the largest subfamily, which contains 81 P450 genes. Genes from the same subgroup can be considered to have the same function. P450 proteins derived from homologous chromosomes A subgroups and D subgroups tend to aggregate in the same branch. The conserved motifs in the P450 proteins were analyzed by MEME and TBtools software (Figure 2). The P450 gene is a family that contains a total of eight conserved motifs. The motifs contained in different genes were the same, indicating the conservatism of the motif domain for almost all the identified P450 family genes. The overall expression pattern of the first subgroup of motifs was 3_5_2_6_1_8; the expression pattern of the second subgroup was 2_6_7_8; the expression pattern of the third subgroup was 3_2_6_1_8; and the expression pattern of the fourth subgroup was 3_2_6_1_8.
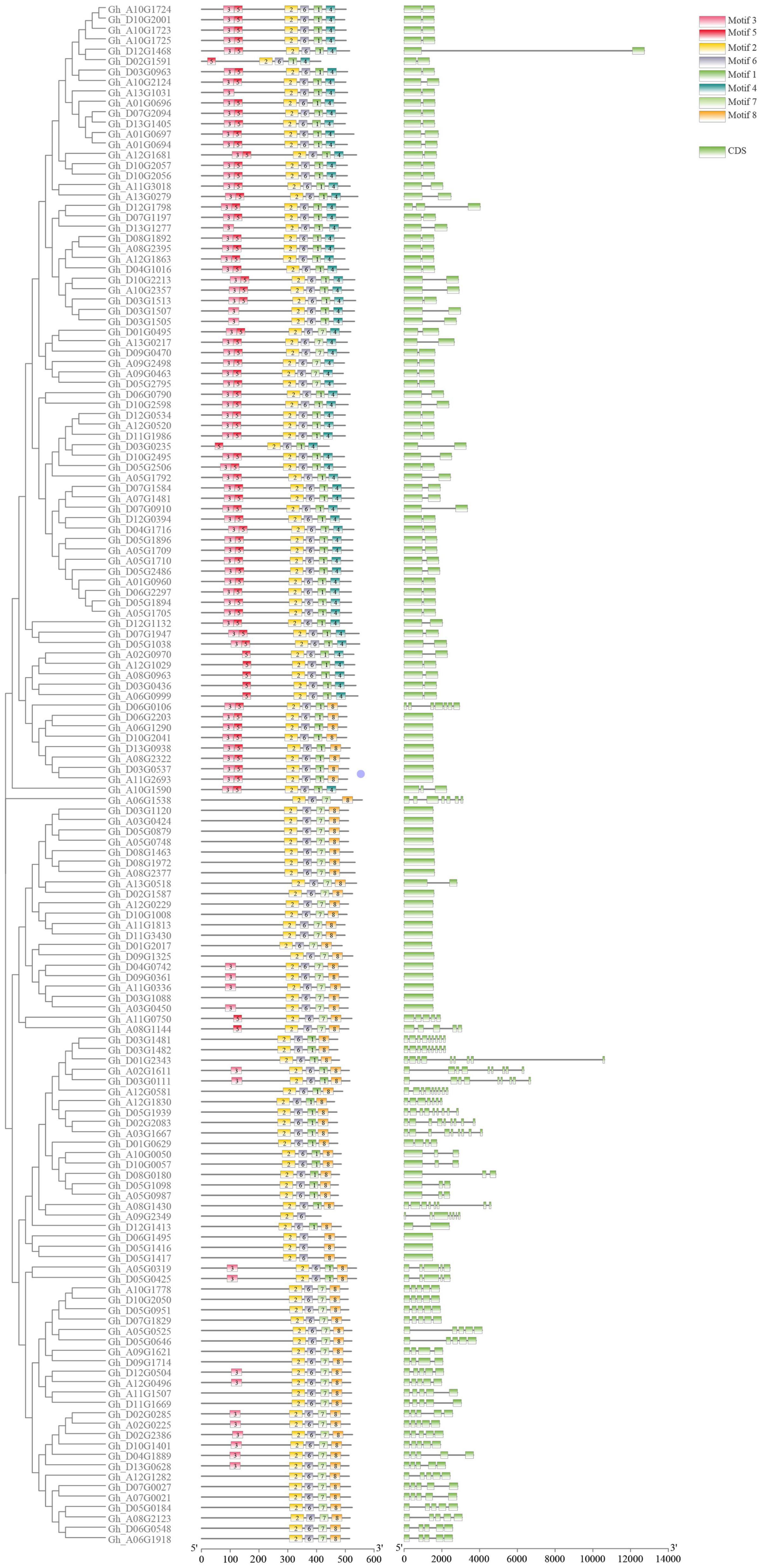
Figure 2. Gene structure diagram of the P450 family in upland cotton. The three columns in the diagram are phylogenetic tree, motif, and exon in order from left to right.
The length of P450 genes in upland cotton differs greatly, ranging from 1,500 to 13,000 bp. The number of exons in the P450 family genes ranged from 1 to 9, while the number of introns ranged from 0 to 8, and there were similar trends within clusters (Figure 2). These results further confirmed the reliability of the phylogenetic tree. A total of 31 cis-acting elements were identified by analyzing a 1,500 bp promoter sequence (Figure 3), and differences existed in the cis-acting elements of these genes (Table 2).
The 156 genes were randomly distributed on 25 chromosomes of the cotton genome (Figure 4), and the number of genes varied on each chromosome. Chromosome D05, which contained 17 genes, had the largest number of genes. There is no P450 gene on Chromosome A04.
A total of 40 tandem repeat genes were submitted to gene collinearity analysis in the upland cotton genome (Supplementary Table 2), and the results are shown in Figure 5. There were many double duplications of genes among the AD subgroups. Furthermore, four genes on chromosomes A05 and D05 were duplicated.
According to the salt-tolerant transcriptome data of upland cotton obtained in a previous study, 55 differentially expressed P450 family genes belonging to different subfamilies were identified (Figure 6). We selected six DEGs out of these 55, three genes with significantly upregulated expression and three genes with significantly downregulated expression of transcription, to perform qRT-PCR verification experiments. The expression of the three genes, Gh_D04G1016, Gh_D05G0425 and Gh_D12G0534, was significantly downregulated after 48 h in the salt treatment, whereas the expression of the remaining three genes, Gh_A09G0463, Gh_D04G1716 and Gh_A10G2124, was significantly upregulated after 48 h of treatment. The results of the qRT-PCR were consistent with the expression trends in the transcriptome analysis (Figure 7 and Supplementary Table 3).
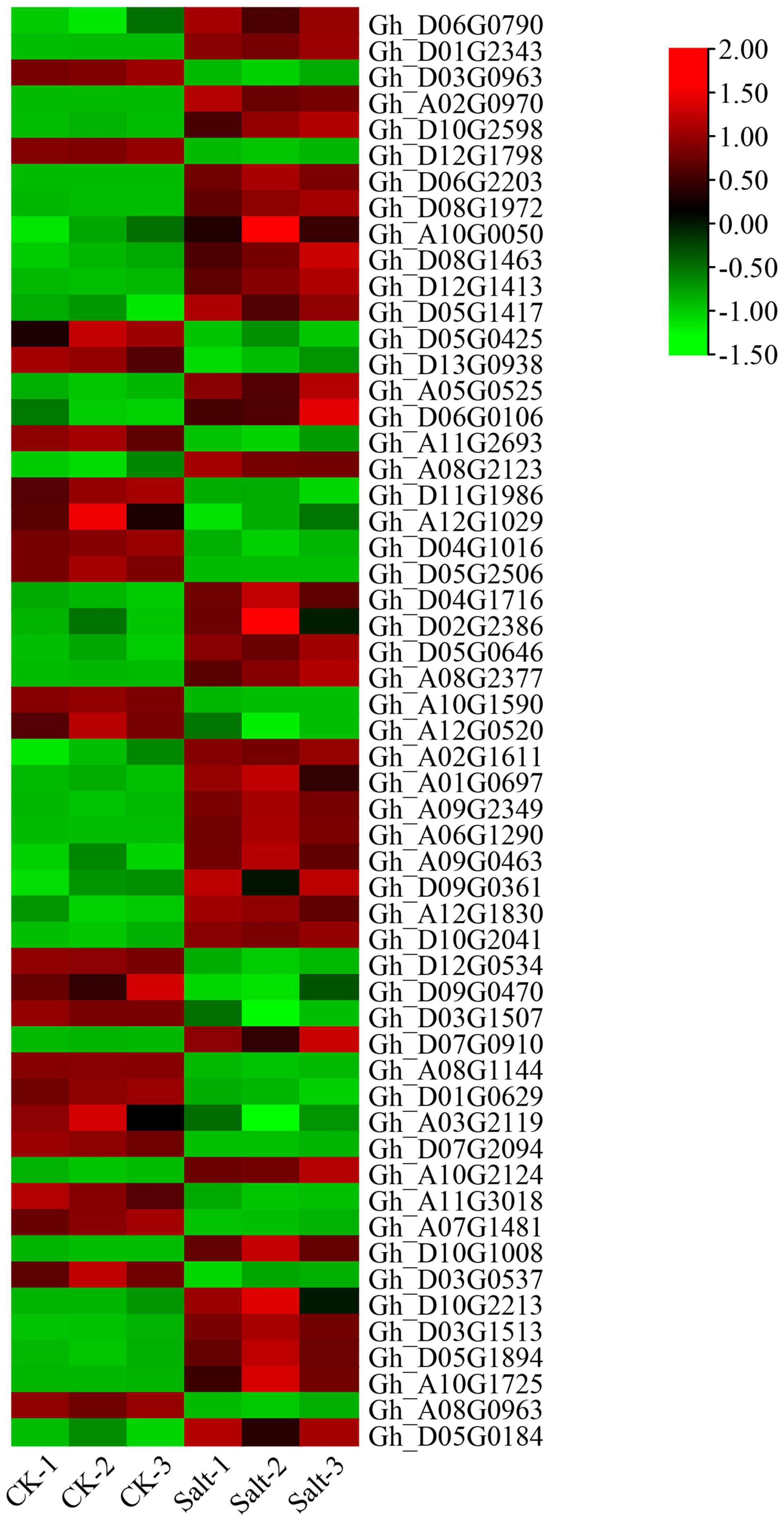
Figure 6. Expression patterns of the P450 family genes in the salt-tolerant transcriptome of upland cotton “Tongyan No. 1”.
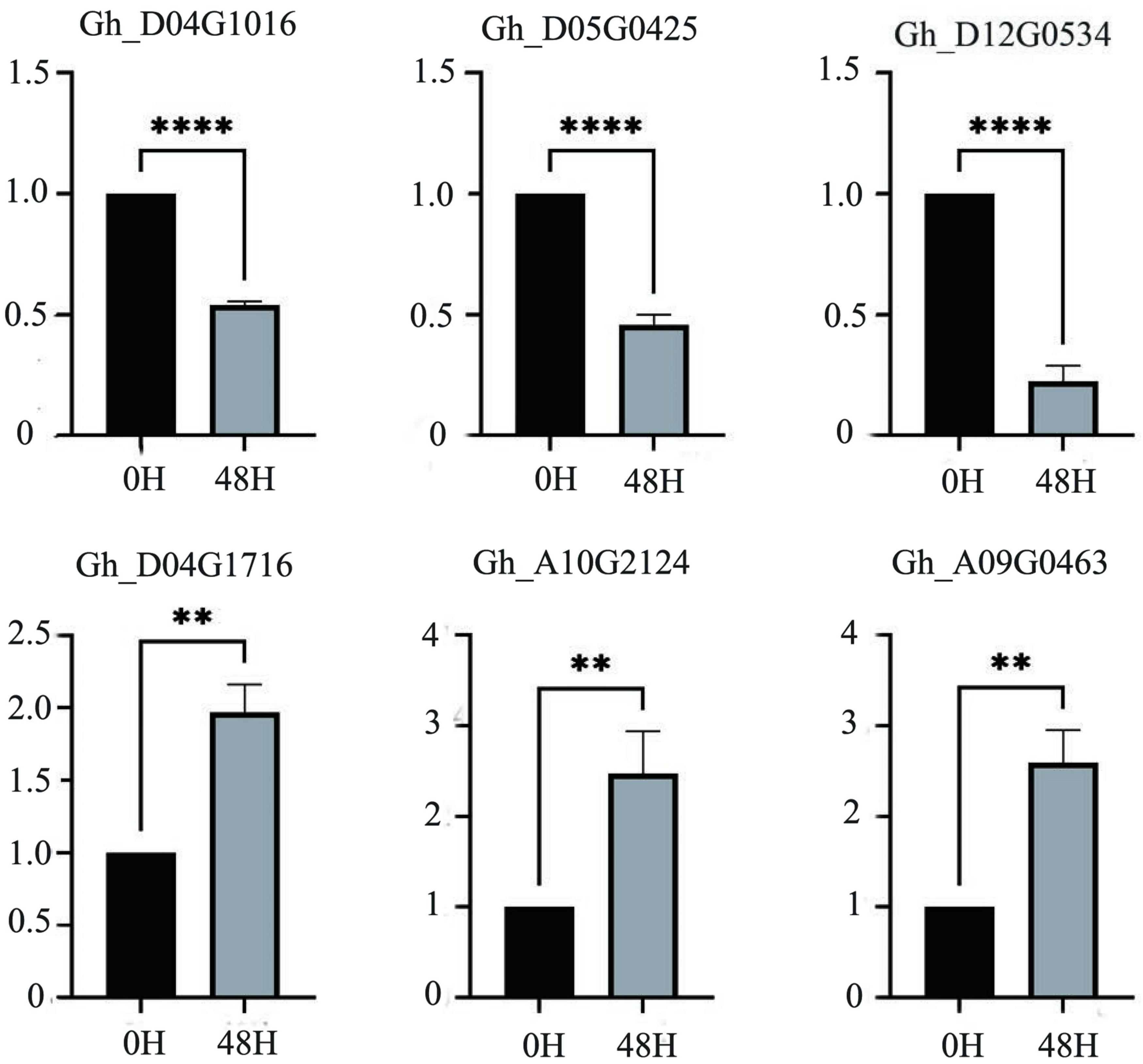
Figure 7. The relative expression levels of six candidate genes in upland cotton under salt stress. A asterisk on the bars indicate significant difference at 0.05 probability level by t test. **P ≤ 0.01, ****P ≤ 0.0001 in t-test.
Soil salinization seriously reduces crop yields. As cotton is one of the most important crops in the world, it is particularly important to develop salt-tolerant cotton varieties and rationally utilize saline-alkali land. With the release of the genome sequencing of different cotton species (Li et al., 2015; Zhang et al., 2015; Udall et al., 2019; Chen et al., 2020; Huang et al., 2020), it is possible to obtain the sequence and structure of gene families and compare the evolutionary relationship of different genes at the genome level.
According to the transcriptome data of the salt-tolerant upland cotton line “Tongyan No. 1” obtained in our previous study, P450 family genes have significant differential expression under salt stress, and according to the Kyoto Encyclopedia of Genes and Genomes (KEGG), a large number of genes are enriched in pathways related to sesquiterpenes and triterpenes. Cotton has a large amount of gossypol and related sesquiterpenoids and is a unique polyphenolic compound. The pigment glands of cotton are known to be support pest resistance and enable strong adaptability of cotton under adverse conditions. Sesquiterpenoids also serve as plant protection elements, resist pathogens, and prevent animals from consuming plants. Moreover, they can also be used as a signal molecule, participating in the interaction between organisms. Sesquiterpenoids are highly diverse and are closely related to the diversity of enzyme genes in secondary metabolic pathways, including P450. At present, there are relatively few studies on the function of the P450 gene family. According to the published genome information of the standard genetic line of upland cotton (TM-1), 156 protein sequences containing complete P450-specific domains were identified by BLASTP, and their molecular weights were 5,949.6–245,576.3 Da, while the corresponding number of encoded amino acids was 51–2,144. The P450 family genes encode proteins with different physical and chemical properties, with obvious differences in size and different functions and regulatory mechanisms, whereas they all possess a stable P450 domain.
Through analysis of the phylogenetic tree, the P450 family genes of the upland cotton species were divided into four subgroups. In the analysis of the structure of the P450 family genes and the conserved motifs, the four subgroups had similar gene structures and conserved sequence expression. Moreover, the existence of a large number of genes in the four subgroups also proves that the P450 family has an important and stable function in the genetic development of upland cotton. The analysis of collinearity within the genome also proved that there are a large number of tandem duplication effects between homologous chromosomes. Duplication is the most common means of amplification of gene families (Cannon et al., 2004). The presence of multiple gene copies can prevent the loss of function caused by genetic mutations, suggesting the importance of gene function. The large amount of duplication allows the P450 family to participate in a variety of stress resistance pathways and phytoalexin production. The degree of conservation in the P450 gene family is very high. The 156 genes are located and distributed on 25 chromosomes of cotton. Except for Chromosome A04, all the chromosomes have P450 family genes. Most of the genes are distributed in the two ends and the middle of the chromosome, with stable heritability, which also shows the importance and widespread nature of the P450 family.
By identifying the upstream cis-elements of each gene, it is possible to better study the regulatory mechanism of each gene. P450 family genes contain several cis-elements. These cis-elements do not encode proteins but can regulate gene expression. Differences exist in cis-acting elements in genes (Figure 3), which means that each gene has different regulatory mechanisms (Table 2). The existence of cis-elements increased the complexity of the gene-encoded protein, and the function of cis-acting elements needs further research. When comparing the P450 family genes we obtained with the salt-tolerant transcriptome data of upland cotton, 55 differentially expressed P450 family genes were obtained. These 55 genes have significant differential expression under salt stress, and they are used as the target genes of the P450 family participating in salt stress regulation.
We randomly selected and verified six of these genes by qRT-PCR according to the up- and downregulation of gene expression (Figure 7). The consistency between the results of the qRT-PCR and transcriptome analysis indicated that these genes were likely to be involved in the salt tolerance of the cotton seedlings. Among the downregulated genes, Gh_D04G1016 is the CYP76A2 gene. The CYP76 subfamily improves stress tolerance in Arabidopsis, can decompose toxic substances in plants, and participates in redox reactions in plants. After salt treatment, CYP76A2 was greatly downregulated (Siminszky, 2006; Matthes et al., 2011; Wang et al., 2012; Ilc et al., 2018). Gh_D05G0425 is the CYP711A1 gene, which is involved in the synthesis of strigolactone. Strigolactone is a new type of hormone. It was first discovered in cotton and has the effect of resisting salt stress and drought stress. After 48 h of salt stress, the expression of Gh_D05G0425 was significantly down-regulated (Benveniste et al., 2006; Wakabayashi et al., 2020). Gh_D12G0534 is the CYP81D1 gene. The CYP81D1 gene is considered to be a suitable marker gene for studying transcriptional activation mechanisms under chemical stress. It shares a common expression and synthesis pathway with jasmonic acid, which can induce plants to excrete harmful substances. We speculate that the synthesis of jasmonic acid is increased under salt stress to resist salt damage, so CYP81D11 gene expression is down-regulated (Didierjean et al., 2002; Siminszky, 2006; Matthes et al., 2011). Among the genes with upregulated expression levels, the function of Gh_A09G0463 is currently unknown, and the specific pathway needs further study. Gh_D04G1716 is the CYP82C4 gene, which is related to Fe ion transport. Because salt stress causes an imbalance of ions inside and outside the plant to increase iron ion transport, the expression of this gene is upregulated after salt treatment (Murgia et al., 2011; Ilc et al., 2018). Gh_A10G2124 is a CYP71A1 gene that is involved in the synthesis of indole acetic acid, and indole acetic acid can promote the growth of plants. Under salt stress, plants increase the expression of this gene to control the synthesis of indole acetic acid and promote growth (Wang et al., 2012). Regarding other genes in the P450 gene family, it was found that Gh_A08G0963 affected the size of the seed (Meng et al., 2015); Gh D13G0938 was identified for herbicide resistance; Gh A06G1290 and Gh_D10G2041 led to male sterility in cotton; Gh_D06G0790 regulated ROS activity and promoted auxin signaling pathway to enhance resilience by activating T31B5_170 expression; Gh_D09G0361 participated in two successive oxidation steps in the synthesis of the plant hormone jasmonoyl isoleucine, enabling it to promote catabolism. After all, P450 family has many genes, and the functions of unknown genes in the same family can be inferred through known genes. It is speculated that under salt stress, the P450 family genes enhanced the adaptability of cotton by regulating gossypol and sesquiterpenoids. However, the functions and regulatory mechanisms of individual genes need further research. The expression of the P450 family under other abiotic stresses also needs further study. The identification and analysis of the P450 gene family presented in this article is helpful for breeding salt-tolerant cotton varieties and provides a basis for the study of cotton salt stress response pathways.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
KS, HF, and YC performed most of the experiments and data analysis. ZZ, QC, and TS helped in sample preparation. MK and JZ helped design the experiments and revise the manuscript. BW designed the experiments and edited the manuscript. All authors read and approved the final manuscript.
This work was supported by the National Key R&D Program of China (2021YFE0101200), PSF/CRP/18thProtocol (07), and the National Natural Science Foundation of China (32072116).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.685054/full#supplementary-material
Bathe, U., and Tissier, A. (2019). Cytochrome P450 enzymes: a driving force of plant diterpene diversity. Phytochemistry 161, 149–162. doi: 10.1016/j.phytochem.2018.12.003
Benveniste, I., Saito, T., Wang, Y., Kandel, S., Huang, H., Pinot, F., et al. (2006). Evolutionary relationship and substrate specificity of Arabidopsis thaliana fatty acid omega-hydroxylase. Plant Sci. 170, 326–338. doi: 10.1016/j.plantsci.2005.08.028
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4:10. doi: 10.1186/1471-2229-4-10
Carelli, M., Biazzi, E., Panara, F., Tava, A., Scaramelli, L., Porceddu, A., et al. (2011). Medicago truncatula CYP716A12 is a multifunctional oxidase involved in the biosynthesis of hemolytic saponins. Plant Cell 23, 3070–3081. doi: 10.1105/tpc.111.087312
Chen, Z. J., Sreedasyam, A., Ando, A., Song, Q., De Santiago, L. M., Hulse-Kemp, A. M., et al. (2020). Genomic diversifications of five Gossypium allopolyploid species and their impact on cotton improvement. Nat. Genet. 52, 525–533. doi: 10.1038/s41588-020-0614-5
Didierjean, L., Gondet, L., Perkins, R., Lau, S. M., Schaller, H., O’Keefe, D. P., et al. (2002). Engineering herbicide metabolism in tobacco and Arabidopsis with CYP76B1, a cytochrome P450 enzyme from Jerusalem artichoke. Plant Physiol. 130, 179–189. doi: 10.1104/pp.005801
Du, H., Ran, F., Dong, H. L., Wen, J., Li, J. N., and Liang, Z. (2016). Genome-Wide analysis, classification, evolution, and expression analysis of the cytochrome P450 93 family in land plants. PLoS One 11:e0165020. doi: 10.1371/journal.pone.0165020
Ghosh, S. (2017). Triterpene structural diversification by plant cytochrome P450 enzymes. Front. Plant Sci. 8:1886. doi: 10.3389/fpls.2017.01886
Gong, Z., Xiong, L., Shi, H., Yang, S., Herrera-Estrella, L. R., Xu, G., et al. (2020). Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 63, 635–674. doi: 10.1007/s11427-020-1683-x
Guo, Y. H., Yu, Y. P., Wang, D., Wu, C. A., Yang, G. D., Huang, J. G., et al. (2009). GhZFP1, a novel CCCH-type zinc finger protein from cotton, enhances salt stress tolerance and fungal disease resistance in transgenic tobacco by interacting with GZIRD21A and GZIPR5. New Phytol. 183, 62–75. doi: 10.1111/j.1469-8137.2009.02838.x
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Huang, G., Wu, Z., Percy, R. G., Bai, M., Li, Y., Frelichowski, J. E., et al. (2020). Genome sequence of Gossypium herbaceum and genome updates of Gossypium arboreum and Gossypium hirsutum provide insights into cotton a-genome evolution. Nat. Genet. 52, 516–524. doi: 10.1038/s41588-020-0607-4
Ilc, T., Arista, G., Tavares, R., Navrot, N., Duchene, E., Velt, A., et al. (2018). Annotation, classification, genomic organization and expression of the Vitis vinifera CYPome. PLoS One 13:e0199902. doi: 10.1371/journal.pone.0199902
Koo, A. J., Cooke, T. F., and Howe, G. A. (2011). Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-L-isoleucine. Proc. Natl. Acad. Sci. U.S.A. 108, 9298–9303. doi: 10.1073/pnas.1103542108
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 7:33. doi: 10.1093/molbev/msw054
Li, F., Fan, G., Lu, C., Xiao, G., Zou, C., Kohel, R. J., et al. (2015). Genome sequence of cultivated upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 33, 524–530. doi: 10.1038/nbt.3208
Li, Y., and Wei, K. (2020). Comparative functional genomics analysis of cytochrome P450 gene superfamily in wheat and maize. BMC Plant Biol. 20:93. doi: 10.1186/s12870-020-2288-7
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, D., Hu, Y., Yang, C., Liu, B., Fang, L., Wan, Q., et al. (2016). Genetic basis for glandular trichome formation in cotton. Nat. Commun. 7:10456. doi: 10.1038/ncomms10456
Matthes, M., Bruce, T., Chamberlain, K., Pickett, J., and Napier, J. (2011). Emerging roles in plant defense for cis-jasmone-induced cytochrome P450 CYP81D11. Plant Signal. Behav. 6, 563–565. doi: 10.4161/psb.6.4.14915
Meloni, D. A., Oliva, M. A., Martinez, C. A., and Cambraia, J. (2003). Photosynthesis and activity of superoxide dismutase, peroxidase and glutathione reductase in cotton under salt stress. Environ. Exp. Bot. 49, 69–76. doi: 10.1016/S0098-8472(02)00058-8
Meng, M., Qian, W., Li, Z., Cheng, H., Li, Z., and Liu, X. (2015). Expression of tacyp78a3, a gene encoding cytochrome p450 cyp78a3 protein in wheat (triticum aestivum l.), affects seed size. Plant J. Cell Mol. Biol. 83, 312–325. doi: 10.1111/tpj.12896
Mills, E. L., Ryan, D. G., Prag, H. A., Dikovskaya, D., Menon, D., Zaslona, Z., et al. (2018). Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 556, 113–117. doi: 10.1038/nature25986
Mizutani, M., and Sato, F. (2011). Unusual P450 reactions in plant secondary metabolism. Arch. Biochem. Biophys. 507, 194–203. doi: 10.1016/j.abb.2010.09.026
Munns, R. (2020). Comparative physiology of salt and water stress. Plant Cell Environ. 25, 239–250. doi: 10.1046/j.0016-8025.2001.00808.x
Murgia, I., Tarantino, D., Soave, C., and Morandini, P. (2011). Arabidopsis CYP82C4 expression is dependent on Fe availability and circadian rhythm, and correlates with genes involved in the early Fe deficiency response. J. Plant Physiol. 168, 894–902. doi: 10.1016/j.jplph.2010.11.020
Nelson, D. R. (2011). Progress in tracing the evolutionary paths of cytochrome P450. Biochim. Biophys. Acta 1814, 14–18. doi: 10.1016/j.bbapap.2010.08.008
Nelson, D., and Werck-Reichhart, D. (2011). A P450-centric view of plant evolution. Plant J. 66, 194–211. doi: 10.1111/j.1365-313X.2011.04529.x
Pompon, D., Louerat, B., Bronine, A., and Urban, P. (1996). Yeast expression of animal and plant p450s in optimized redox environments. Methods Enzymol. 272351, 51–64. doi: 10.1016/S0076-6879(96)72008-6
Qian, S. Z., and Wang, Z. G. (1984). Gossypol: a potential antifertility agent for males. Annu. Rev. Pharmacol. Toxicol. 24, 329–360. doi: 10.1146/annurev.pa.24.040184.001553
Schuler, M. A. (2010). Plant Cytochrome P450 Monooxygenases. Crit. Plant Sci. 15, 235–284. doi: 10.1080/07352689609701942
Schuler, M. A., and Werck-Reichhart, D. (2003). Functional genomics of P450s. Annu. Rev. Plant Biol. 54, 629–667. doi: 10.1146/annurev.arplant.54.031902.134840
Shaheen, H. L., Shahbaz, M., Ullah, I., and Iqbal, M. Z. (2012). Morpho-physiological responses of cotton (Gossypium hirsutum) to salt stress. Int. J. Agtic. Biol. 14, 980–984. doi: 10.1021/jf3034804
Siminszky, B. (2006). Plant cytochrome P450-mediated herbicide metabolism. Phytochem. Rev. 5, 445–458. doi: 10.1007/s11101-006-9011-7
Sunilkumar, G., Campbell, L. M., Puckhaber, L., Stipanovic, R. D., and Rathore, K. S. (2006). Engineering cottonseed for use in human nutrition by tissue-specific reduction of toxic gossypol. Proc. Natl. Acad. Sci. U.S.A. 103:48. doi: 10.1073/pnas.0605389103
Tang, C., Yang, M., Fang, Y., Luo, Y., Gao, S., Xiao, X., et al. (2016). The rubber tree genome reveals new insights into rubber production and species adaptation. Nat. Plants 2:16073. doi: 10.1038/nplants.2016.73
Teh, B. T., Lim, K., Yong, C. H., Ng, C. C. Y., Rao, S. R., Rajasegaran, V., et al. (2017). The draft genome of tropical fruit durian (Durio zibethinus). Nat. Genet. 49, 1633–1641. doi: 10.1038/ng.3972
Tian, X., Ruan, J., Huang, J., Fang, X., Mao, Y., Wang, L., et al. (2016). Gossypol: phytoalexin of cotton. Sci. China Life Sci. 59, 122–129. doi: 10.1007/s11427-016-5003-z
Udall, J. A., Long, E., Hanson, C., Yuan, D., Ramaraj, T., Conover, J. L., et al. (2019). De novo genome sequence assemblies of Gossypium raimondii and Gossypium turneri. G3 (Bethesda) 9, 3079–3085. doi: 10.1534/g3.119.400392
Wakabayashi, T., Shida, K., Kitano, Y., Takikawa, H., Mizutani, M., and Sugimoto, Y. (2020). CYP722C from Gossypium arboreum catalyzes the conversion of carlactonoic acid to 5-deoxystrigol. Planta 251:97. doi: 10.1007/s00425-020-03390-6
Wang, J. Y., Cai, Y., Gou, J. Y., Mao, Y. B., Xu, Y. H., Jiang, W. H., et al. (2004). VdNEP, an elicitor from Verticillium dahliae, induces cotton plant wilting. Appl. Environ. Microbiol. 70, 4989–4995. doi: 10.1128/AEM.70.8.4989-4995.2004
Wang, Q., Hillwig, M. L., Okada, K., Yamazaki, K., Wu, Y., Swaminathan, S., et al. (2012). Characterization of CYP76M5-8 indicates metabolic plasticity within a plant biosynthetic gene cluster. J. Biol. Chem. 287, 6159–6168. doi: 10.1074/jbc.M111.305599
Wang, Q., Yu, F., and Xie, Q. (2020). Balancing growth and adaptation to stress: crosstalk between brassinosteroid and abscisic acid signaling. Plant Cell Environ. 43, 2325–2335. doi: 10.1111/pce.13846
Yan, Q., Cui, X., Lin, S., Gan, S., Xing, H., and Dou, D. (2016). GmCYP82A3, a soybean cytochrome P450 family gene involved in the jasmonic acid and ethylene signaling pathway, enhances plant resistance to biotic and abiotic stresses. PLoS One 11:e0162253. doi: 10.1371/journal.pone.0162253
Zhang, T., Hu, Y., Jiang, W., Fang, L., Guan, X., Chen, J., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–537. doi: 10.1038/nbt.3207
Keywords: P450, gene family, differentially expressed genes, salt tolerance, cotton
Citation: Sun K, Fang H, Chen Y, Zhuang Z, Chen Q, Shan T, Khan MKR, Zhang J and Wang B (2021) Genome-Wide Analysis of the Cytochrome P450 Gene Family Involved in Salt Tolerance in Gossypium hirsutum. Front. Plant Sci. 12:685054. doi: 10.3389/fpls.2021.685054
Received: 24 March 2021; Accepted: 09 November 2021;
Published: 02 December 2021.
Edited by:
Jinfa Zhang, New Mexico State University, United StatesReviewed by:
Jing Zhang, Nanjing Agricultural University, ChinaCopyright © 2021 Sun, Fang, Chen, Zhuang, Chen, Shan, Khan, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Zhang, emowOTI4QDEyNi5jb20=; Baohua Wang, Ymh3YW5nQG50dS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.