
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 14 June 2021
Sec. Plant Breeding
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.684122
This article is part of the Research TopicRecent Advances in Hazelnut (Corylus spp.)View all 26 articles
Commercial production of hazelnut (Corylus avellana) in Oregon’s Willamette Valley is threatened by eastern filbert blight (EFB), a serious canker disease caused by the pyrenomycete Anisogramma anomala (Peck) E. Müller. The fungus also prevents the establishment of hazelnut orchards in eastern North America. Genetic resistance is considered the most effective way to control the disease. A high level of EFB resistance was first discovered in ’Gasaway’. This resistance is conferred by a dominant allele at a single locus on linkage group 6 (LG6). Resistance from several additional sources has been assigned to the same chromosomal region. In this study, new simple sequence repeat (SSR) markers were developed for the resistance region on LG6 and new sources of resistance were investigated. Forty-two new SSR markers were developed from four contigs in the genome sequence of ‘Jefferson’ hazelnut, characterized, and nine of them were placed on LG6 of the genetic map. Accessions representing 12 new sources of EFB resistance were crossed with susceptible selections resulting in 18 seedling populations. Segregation ratios in the seedling populations fit the expected 1:1 ratio for 10 sources, while one source showed an excess of resistant seedlings and another showed an excess of susceptible seedlings. Based on correlation of disease response and scores of SSR markers in the ‘Gasaway’ resistance region in the seedlings, eight resistance sources were assigned to LG6. Linkage maps were constructed for each progeny using SSR markers. The LG6 resistance sources include two selections (#23 and #26) from the Russian Research Institute of Forestry and Mechanization near Moscow, four selections from southern Russia, one selection (OSU 1185.126) from Crimea, one selection (OSU 533.129) from Michigan, Corylus heterophylla ‘Ogyoo’ from the South Korea, and the interspecific hybrid ’Estrella #1’. These new LG6 resistance sources and SSR markers should be useful in breeding new cultivars, including the pyramiding of resistance genes. For the other four resistance sources (Moscow #37, hybrid selection OSU 401.014, C. americana ‘Winkler’ and C. americana OSU 366.060), SSR marker scores on linkage groups 6, 7 and 2 were not correlated with disease response and merit further investigation.
Hazelnut is an important tree nut. Cultivars of European hazelnut (Corylus avellana), also known as filbert, are clonally propagated, highly heterozygous, monoecious, dichogamous, wind-pollinated and diploid with 11 pairs of chromosomes (2n = 2x = 22). Traditional propagation is by the rooted suckers that grow around the crown of the plant. Nearly all of the world’s hazelnuts are of the European species, Corylus avellana, which is a member of the family Betulaceae. The European hazelnut is widely distributed in Europe, Turkey and the Caucasus republics, but commercial production is limited to areas near large bodies of water at middle latitudes with moderate temperatures in winter and summer, and high humidity during mid-winter bloom (Mehlenbacher, 1991). These areas include the Black Sea coasts of Turkey and Georgia, areas in Italy and Spain near the Mediterranean Sea, southwestern France near the Bay of Biscay, and the Willamette Valley of Oregon, United States near the Pacific Ocean. Turkey produces 67% of the world crop, followed by Italy, Azerbaijan, and the United States1.
Simple sequence repeat (SSR) markers, also known as microsatellites, are short tandem repeats widely distributed throughout plant genomes. They are the marker type of choice for many studies due to their ease of amplification by the polymerase chain reaction (PCR), high level of polymorphism, experimental reproducibility, ease of sharing among labs, usefulness in many progenies, and transferability among related species (Vieira et al., 2016). The steps in developing SSR markers are identifying the desired type of repeat in nucleotide sequences, designing primers complementary to the regions flanking the SSR, amplification by PCR, separating the PCR products by electrophoresis, and detecting polymorphism among individuals (Mason, 2015). Next Generation Sequencing technology allows rapid and inexpensive SSR marker development from genome and transcriptome sequences (Tang et al., 2008; Hoffman and Nichols, 2011; Vukosavljev et al., 2015; Bhattarai and Mehlenbacher, 2017, 2018; Colburn et al., 2017; Khodaeiaminjan et al., 2018; Taheri et al., 2018). Post-PCR multiplexing of products reduces the cost of allele sizing by capillary electrophoresis. Akın et al. (2016) identified a set of 14 primer pairs for pre-PCR multiplexing, further simplifying and reducing the cost of the procedure. More than 900 polymorphic SSR markers have been developed in C. avellana (Bassil et al., 2005a,b, 2013; Boccacci et al., 2005, 2015; Gürcan and Mehlenbacher, 2010a,b; Gürcan et al., 2010; Bhattarai and Mehlenbacher, 2017, 2018; Colburn et al., 2017; Öztürk et al., 2017; Zhao et al., 2019; Kavas et al., 2020; Hill et al., 2021; Şekerli et al., 2021) of which ∼450 have been placed on the reference linkage map (Mehlenbacher et al., 2006; Mehlenbacher and Bhattarai, 2018).
Eastern filbert blight (EFB), caused by the pyrenomycete Anisogramma anomala, has prevented the establishment of commercial orchards in eastern North America (Thompson et al., 1996; Capik and Molnar, 2012) and is now present throughout the Willamette Valley where 99% of the United States hazelnut crop is produced2. The pathogen is native to eastern North America where it is found on the wild American hazelnut, C. americana, on which it causes only limited damage (Capik and Molnar, 2012). On most C. avellana cultivars, however, it causes large, perennial stem cankers, branch die-back, and eventual tree death after several years. The disease life cycle is now well-understood (Pinkerton et al., 1992, 1995, 1998, 2001; Stone et al., 1992; Johnson et al., 1994, 1996). The fungus is an obligate biotroph with a 2-year life cycle3. Ascospores are released in winter during periods of branch wetness and dispersed by rain and air currents. Hyphae from germinating spores penetrate young growing shoots in the spring, and then spread in the cambium and phloem. Cankers become visible about 15 months after infection. Controlling the disease with scouting, pruning infected branches 30 to 90 cm below the cankers, and fungicide applications is costly and labor-intensive. Alternative disease management strategies are desirable and host genetic resistance is considered the most cost-effective method (Mehlenbacher, 1995). The high level of EFB resistance first discovered in ‘Gasaway’ (Cameron, 1976) was shown to be controlled by a dominant allele at a single locus (Mehlenbacher et al., 1991). Random amplified polymorphic DNA (RAPD) markers linked to resistance were identified (Mehlenbacher et al., 2004) and the resistance locus was placed on the reference linkage map (Mehlenbacher et al., 2006). ‘Gasaway’ resistance has been extensively used in the hazelnut breeding program at Oregon State University (OSU), and several resistant cultivars and pollinizers have been released. Fungal isolates able to overcome ‘Gasaway’ resistance were recently reported in New Jersey (Molnar et al., 2010; Muehlbauer et al., 2018; Dunlevy et al., 2019). There is an urgent need to find new sources of resistance and use them in breeding. Previously studied resistance sources include OSU 408.040 from Minnesota, ‘Culpla’ from Spain, OSU 495.072 from southern Russia, and ‘Crvenje’ and ‘Uebov’ from Serbia. Resistance from all five of these sources was placed on linkage group 6 (LG6) in the ‘Gasaway’ resistance region (Sathuvalli et al., 2012; Colburn et al., 2015; Bhattarai et al., 2017b). On the other hand, resistance in ‘Ratoli’ from Spain, C. americana ‘Rush’ from Pennsylvania and interspecific hybrid ‘Yoder #5’ from Ohio was assigned to a region on LG7 (Sathuvalli et al., 2011a; Bhattarai et al., 2017a). Resistance in selection OSU 759.010 from the Republic of Georgia and Rutgers University selection H3R07P25 from southern Russia was assigned to a region on LG2 (Sathuvalli et al., 2011b; Honig et al., 2019). Additional sources of EFB resistance have been identified, including Moscow #26 (Sathuvalli et al., 2010), C. heterophylla ‘Ogyoo’ (Coyne et al., 1998), and interspecific hybrid ‘Estrella #1’ (Chen et al., 2007), as well as germplasm collected in Russia, Crimea and Poland (Molnar et al., 2007; Capik et al., 2013; Leadbetter et al., 2016). Muehlbauer et al. (2014) characterized a large collection of EFB-resistant selections at Rutgers University.
The goals of this study were to develop and characterize new SSR markers in the resistance region on LG6 and study EFB resistance from 12 new sources.
For characterization of the new SSR markers, a diversity panel of 50 hazelnut accessions (Table 1) including the parents of the reference mapping population (OSU 252.146 and OSU 414.062) was used. For the investigation of EFB resistance, 18 seedling populations segregating for resistance from 14 resistant parents representing 12 sources were created by crossing susceptible selections with them or an advanced selection carrying the same resistance (Table 2). The pedigrees are shown in Supplementary Material 1. The resistance sources include three selections (#23, #26, and #37) from the Russian Research Institute of Forestry and Mechanization near Moscow, Russia, four selections (H3R04P23, H3R04P28, H3R04P30, and H3R13P40) from Rutgers University that arose from seeds purchased in an outdoor market in the village of Holmskij, near Krasnodar, Russia, and one selection (OSU 1185.126) from seeds purchased near Simferopol, Crimea. The four Rutgers University selections from Holmskij and one OSU selection from Simferopol originated from seeds purchased on a collection trip in 2002 by Thomas Molnar, David Zaurov and Shawn Mehlenbacher. The seed lots were shared by the two institutions (Rutgers University and OSU) and the seed source ID numbers were listed by Molnar et al. (2007). The four Rutgers University selections had been inoculated with the EFB pathogen in greenhouse or field in New Jersey in 2005 and found to be resistant (Molnar et al., 2007; Capik et al., 2013). Additional sources of resistance include OSU 533.129, selected from a lot of open-pollinated seeds received from Cecil Farris in Lansing, Michigan, one cultivar (‘Ogyoo’) of Corylus heterophylla from the South Korea, and interspecific hybrid ’Estrella #1’. OSU 1181.002 carries resistance from ’Ogyoo’. ‘Estrella #1’ from private breeder Cecil Farris is a hybrid of a single accession of C. sutchuenensis (syn. C. heterophylla var. sutchuenensis) obtained from western China as the female parent and C. avellana ‘Holder’ as the pollen parent (Farris, 1974). The three remaining resistance sources investigated are two clones of C. americana (‘Winkler’ and OSU 366.060) and the interspecific hybrid OSU 401.010 that originated from open-pollinated seeds sent from New Carlisle, Ohio by Ken Bauman, a long-time member of the Northern Nut Growers Association. The phenotype of OSU 401.040 indicates that it is a C. americana × C. avellana hybrid. C. americana OSU 366.060, preserved as PI 433984 at the USDA National Clonal Germplasm Repository in Corvallis, OR, was selected from a seed lot received from Mississippi. Four resistant parents (Rutgers University selections H3R04P23 and H3R13P40, OSU 533.129 and ‘Estrella #1’) were each represented by two segregating progenies, and the remaining ten parents were each represented by a single progeny.
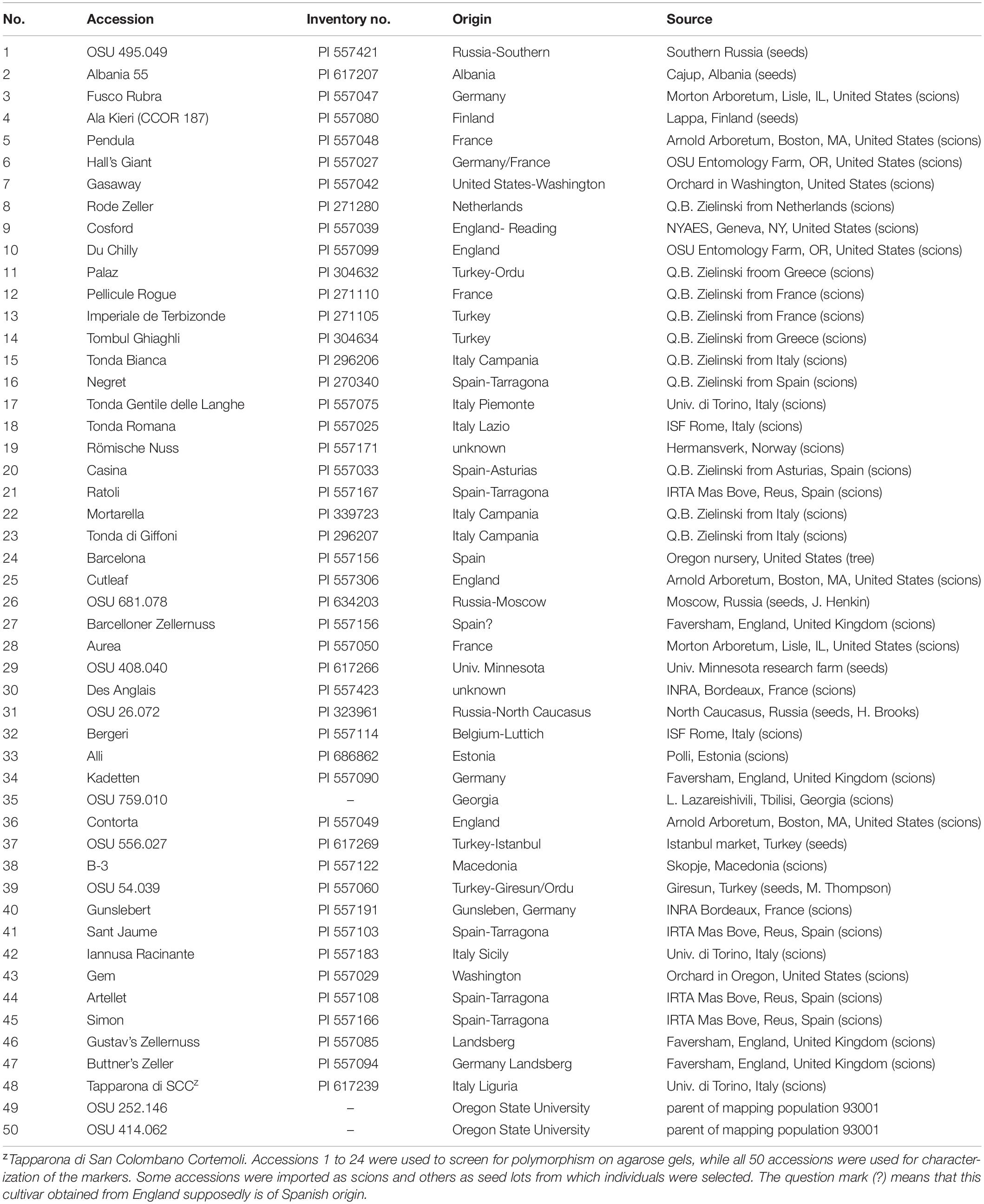
Table 1. Hazelnut accessions used for characterization of 42 new polymorphic simple sequence repeat (SSR) markers.
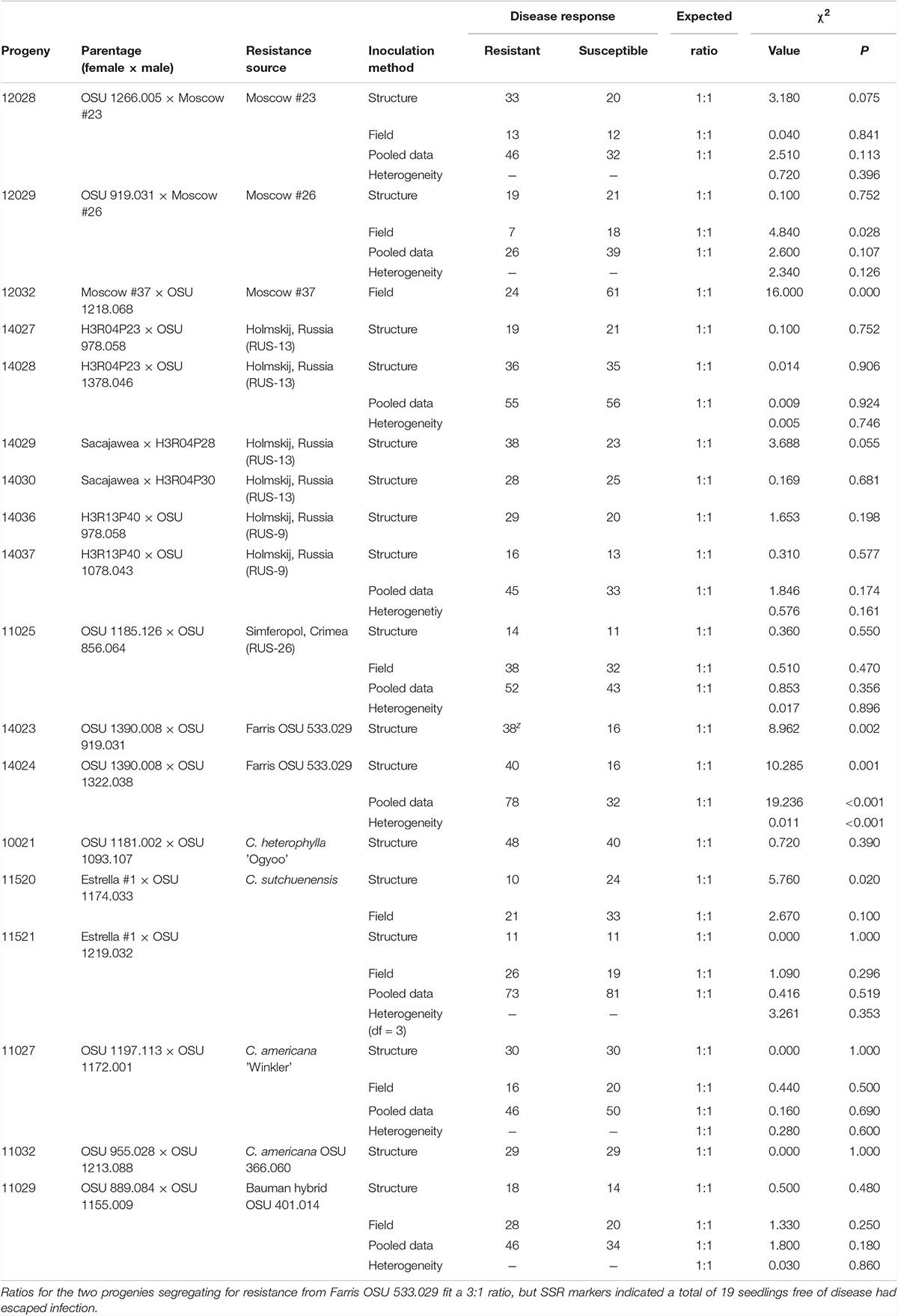
Table 2. Segregation for eastern filbert blight response in 18 hazelnut progenies from 14 resistant parents and 12 sources following structure or field inoculation.
The genome sequence of ‘Jefferson’ hazelnut (V2), assembled from Pacific Biosciences (PacBio, Menlo Park, CA, United States) sequences and error-corrected using Illumina (San Diego, CA, United States) reads, was used as the reference genome for marker development. Sequences of four types predicted to be near the ‘Gasaway’ EFB resistance locus on LG6 were aligned to the reference genome using the Basic Local Alignment Search Tool (BLAST)4. The sequences were of RAPD markers, SSR markers, Illumina sequences of bacterial artificial chromosomes (BACs), and BAC end sequences. The identified PacBio contigs (Table 3) were investigated for presence of SSRs using the Genome-wide Microsatellite Analyzing Tool (GMATo) (Wang et al., 2013) with minimum numbers of repeats for the di-, tri-, tetra-, penta- and hexa- repeats set at 8, 6, 4, 4 and 3, respectively. Repeat motifs containing only As and Ts were not pursued as experience has shown them to be difficult to score. Samtools (Li et al., 2009) were used to trim the SSR-containing fragments, retaining the repeat motif and 250 bp on either side. One at a time, the SSR-containing fragments from ‘Jefferson’ were used as the reference and aligned in silico with Illumina genome sequences of seven other cultivars (’Barcelona’, ’Ratoli’, ’Tonda Gentile delle Langhe’, ’Tonda di Giffoni’, ’Daviana’, ’Hall’s Giant’, and ’Tombul’) (Rowley et al., 2018). The aligned reads were visualized using Tablet software (Milne et al., 2010), inspected, and classified as “not polymorphic”, “slightly polymorphic”, or “clearly polymorphic”. Only those in the latter category were pursued. Forward and reverse primers (Supplementary Material 3) were designed from the conserved sequences that flanked each selected SSR using Websat (Martins et al., 2009) and Primer3 software (Untergasser et al., 2012) with parameters set at annealing temperature 60°C, a minimum GC content of 50%, and an amplicon size of 90-350 bp to facilitate post-PCR multiplexing of primer products for genotyping. A BLAST search against the NCBI database was used to verify that the sequences had not previously been used for SSR development. DNA of 24 accessions (Table 1) was amplified with each pair of primers in GeneAmp PCR system 9700 thermal cyclers (Applied Biosystems, Foster City, CA, United States) in 96-well plates as follows: denaturation at 95°C for 5 min followed by 40 cycles of 94°C for 40 s, 60°C for 40 s, 72°C for 40 s, extension at 72°C for 7 min, and a final infinite hold at 4°C. Polymorphic SSRs were identified by separating the PCR products for 2.5 h at 90V on 3% w/v agarose gels in TBE buffer. The gels were stained with ethidium bromide and images were recorded under ultraviolet light using a BioDoc-It® Imaging System (UVP, Upland, CA, United States).
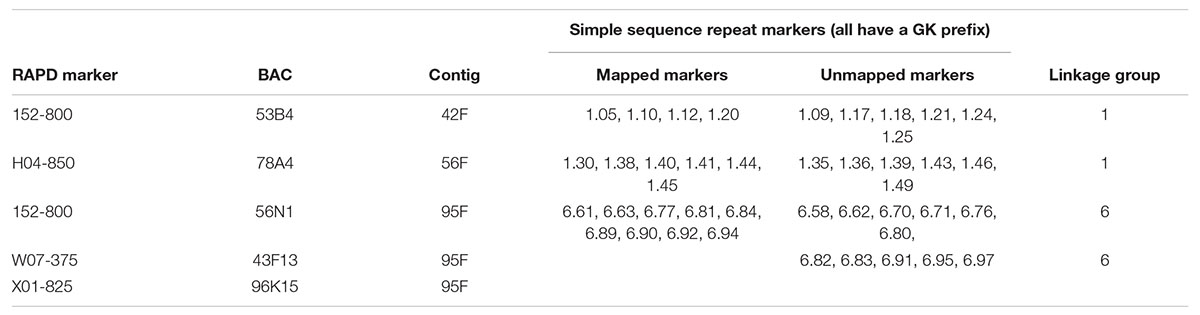
Table 3. New simple sequence repeat markers developed from three contigs in the ‘Jefferson’ hazelnut genome sequence (V2) and the random amplified polymorphic DNA (RAPD) marker and bacterial artificial chromosome (BAC) sequences used to identify the contigs.
Fluorescent forward primers with a label of FAM or HEX (Integrated DNA Technologies, Coralville, IA, United States) were ordered for the SSRs considered “clearly polymorphic” on agarose gels. DNA from 48 hazelnut accessions and the two parents of the reference mapping population (Table 1) was amplified with the fluorescent forward and non-fluorescent reverse primers. An aliquot of 2 μL of the PCR product of each primer pair was diluted with water to make a final volume of 150 μL. An average of six primer pairs was post-PCR multiplexed in a single well. An aliquot of 1.6 μL of the multiplex was submitted to the Core Labs of OSU’s Center for Genome Research and Biocomputing (CGRB) for fragment sizing by capillary electrophoresis on an ABI 3730 (Life Technologies, Carlsbad, CA, United States) with ROX-500 as the size standard. GeneMapper software (Life Technologies) was used for allele size determination followed by manual verification. Characterization of marker loci was carried out using PowerMarker (Liu and Muse, 2005) and Cervus software (Kalinowski et al., 2007). PowerMarker software was used to calculate the number of alleles (n), observed heterozygosity (Ho), expected heterozygosity (He), and polymorphism information content (PIC) for each locus. The frequency of null alleles was calculated with Cervus software (Table 4).
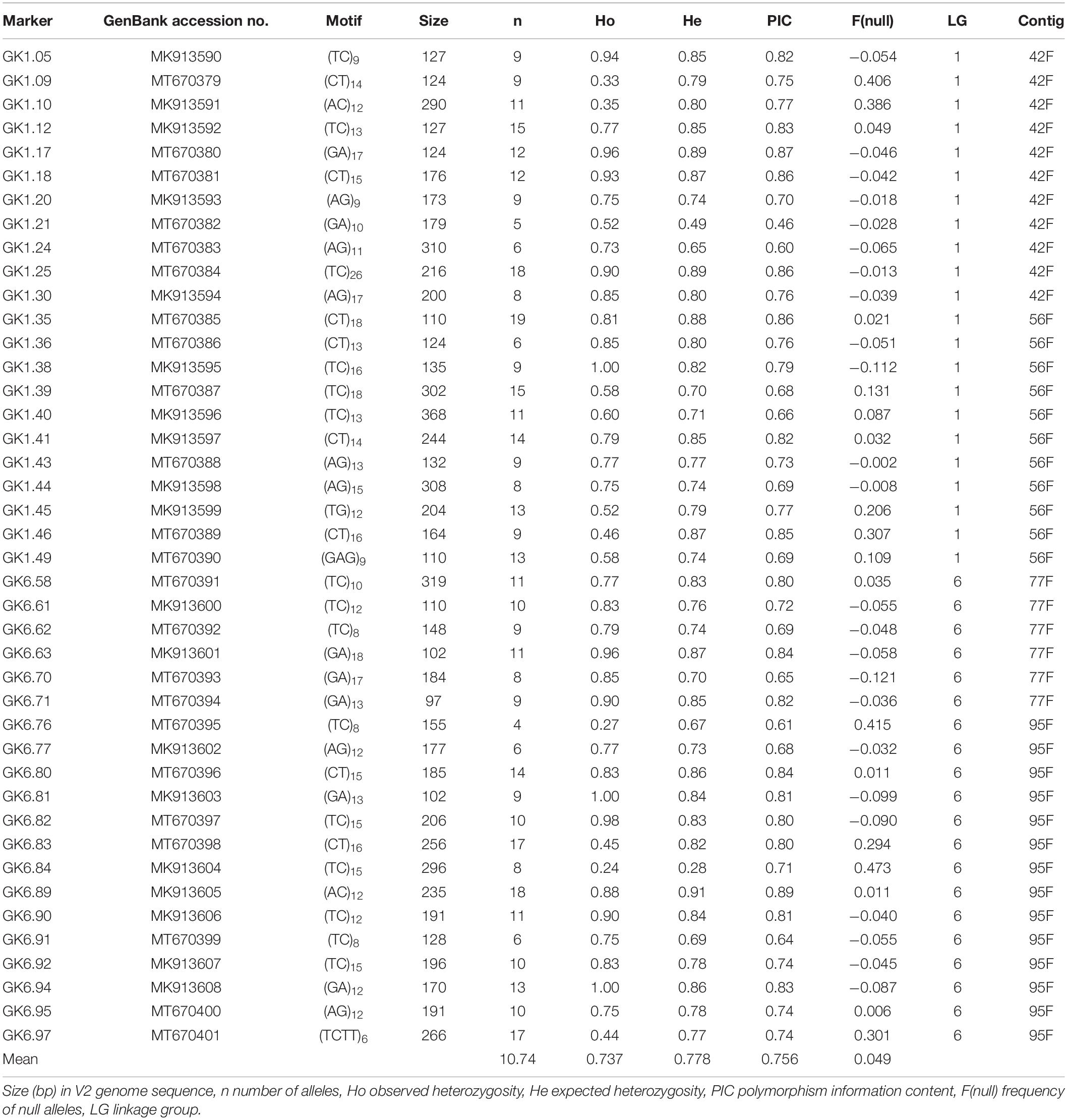
Table 4. Characteristics of 42 new simple sequence repeat markers developed from four contigs in the V2 genome sequence of ‘Jefferson’ hazelnut (Corylus avellana).
To place the new SSR markers on the reference linkage map, the two-way pseudotestcross approach (Grattapaglia and Sederoff, 1994) and all 138 seedlings in the reference mapping population (OSU 252.146 × OSU 414.062) were used. Each allele size was scored as present or absent in each seedling. For analysis with Join Map 4.0 (Van Ooijen, 2006) and the BC1 function, marker present was scored as “h”, marker absent was scored as “a”, and unknown status was scored as “u”. Scores for previously mapped markers and the new markers were combined and grouped at a LOD score of 12. Linkage maps were constructed using the maximum likelihood algorithm and distances shown in Haldane units (cM). “Dummy variables” were created to allow the merger of markers linked in coupling and repulsion.
Two approaches were used for disease inoculation: exposure of potted trees under an inoculation structure and planting seedlings in a field near a heavily diseased orchard. Potted seedlings grown in 5 L pots were inoculated with EFB by placing them under a structure topped with diseased branches in the spring, based on Pinkerton et al. (1993). The structure was located at the OSU Smith Horticulture Research Farm in Corvallis, OR, United States. Sprinklers on top of the structure kept the inoculum branches wet, allowing spores to drip down on the seedling trees shortly after leaf budbreak. The trees were lined out in a nursery row after exposure and scored for disease response 18 months after inoculation and again 12 months later. Rooted layers of check cultivars ‘Ennis’ (highly susceptible), ‘Lewis’ (moderately resistant), and ‘Tonda di Giffoni’ (high quantitative resistance) were grown in pots and included as controls in the inoculation. Structure-inoculated seedlings were observed for the presence of cankers and stromata, and disease severity was rated on a scale of 0 to 5, with a rating of 0 for absence of disease symptoms, 1 for presence of small sunken cankers without any stromata, 2 for presence of small cankers with few stromata, 3 for presence of cankers with mature stromata, 4 for cankers all over the tree but the tree was still alive, and 5 for cankers all over the tree and the top of the tree was dead. For the initial analysis of segregation for disease response following structure exposure, seedlings with an EFB score of 0, 1, and 2 were considered resistant and seedlings with a score of 3, 4, or 5 were considered susceptible. In the second method, seedlings were planted in the field adjacent to a highly diseased orchard. Each seedling was inspected annually in the winter for 4 years (2014-2017) and presence of EFB cankers was noted. EFB-susceptible seedlings in the plot adjacent to the infected orchard, susceptible seedlings in nearby plots and susceptible selections in a replicated trial 200 m to the northwest of the infected orchard were also inspected for EFB. The presence of dwarf seedlings was also noted in the field plot. Field-exposed seedlings were scored as resistant (no cankers with stromata) or susceptible (cankers with stromata). Chi-square tests were used to assess goodness-of-fit to the 1:1 segregation ratio expected for segregation at a single locus and a heterozygous resistant parent. Heterogeneity Chi-square tests were performed when the disease response of seedlings from the same progeny were investigated by structure and field exposure to determine if the data from the two methods could be pooled. Heterogeneity Chi-square tests were also performed when two progenies segregated for resistance from the same parent to determine if the data could be pooled.
Leaves were collected from trees growing in the field in two locations in Corvallis, OR [National Clonal Germplasm Repository of the United States Dept. of Agriculture-Agricultural Research Service and OSU’s Smith Horticultural Research Farm]. Young leaves were collected from the 50 accessions in the diversity panel (Table 1). For the progenies segregating for disease response following structure exposure, leaves were collected from seedlings in a nursery row 2 years after exposure and 4 years after the controlled cross had been made. For the progenies exposed in the field, leaves were collected in spring 2016 from progenies 10021, 11025, 11027, 11029, 11032, 11520, 11521, 12028, 12029, and 12032. Genomic DNA was extracted based on Lunde et al. (2000) with no RNAase treatment. A Synergy2 microplate reader and Gen5 software (Biotek Instruments, Winooski, VT, United States) were used to quantify the extracted DNA. The DNA was diluted with TE buffer to a concentration of 20 ng μL–1.
Polymerase chain reactions were performed in 10 μl final volumes with a mixture of 20 ng DNA, 1 × Biolase NH4 reaction buffer, 2.5 mM mix dNTP, 2 mM MgCl2, 0.3 μL of each forward and reverse primer (10 μM), and 0.25 units of Biolase DNA polymerase (Bioline USA Inc., Taunton, MA, United States). PCRs were in 96-well plates on GeneAmp PCR System 9700 thermal cyclers (Applied Biosystems, Foster City, CA, United States). The amplification program consisted of an initial denaturation for 5 min at 94°C followed by 40 cycles of 40 s at 94°C, 40 s at the annealing temperature (60 or 62°C), 40 s for elongation at 72°C, and a final extension step of 7 min at 72°C, then an infinite hold at 4°C. After PCR, products were multiplexed by mixing 2 μL from each product and diluted with water to make a final volume of 200 μL. A 1.8 μL aliquot was submitted to the Core Labs of OSU’s Center for Genome Research and Biocomputing (CGRB) for fragment sizing by capillary electrophoresis on an ABI 3730 instrument using ROX-500 as the size standard. Allele sizes were visualized and scored with ABI GeneMapper software (Life Technologies, Carlsbad, CA, United States). If amplification failed or the result was unclear, the PCR amplification and fragment sizing were repeated. Allele sizes at each SSR marker and disease scores for each seedling in each progeny were entered in a spreadsheet.
Three sets of SSR markers, one on LG6, a second set on LG2 and a third set on LG7 near previously mapped resistance loci were used to score the seedlings and their parents. When two progenies were available for the same resistance source, only one progeny was used for the correlation analysis. For 8 progenies (10021, 11025, 11027, 11029, 11032, 11520, 12028, and 12029), the markers were scored in 32 seedlings in the initial correlation analysis, and for the remaining five progenies (14023, 14028, 14029, 14030, and 14036), 46 seedlings were scored for the correlation analysis. In this analysis, the presence of an SSR allele or resistance was scored as 1 and absence or susceptibility was scored as 0. Pearson product-moment correlation coefficients were calculated. Coefficients ≥0.5 were interpreted as indicating linkage of disease response and the marker, while those <0.5 were interpreted as showing independence of disease response and marker scores.
Join Map 4.0 (Van Ooijen, 2006) and the BC1 function were used to construct maps for each source of resistance using the procedures described earlier for placing the new SSR markers on the reference linkage map. Resistance and presence of a marker allele were scored as “h”, susceptible and marker allele absent were scored as “a”, and unknown status was scored as “u”. Markers were grouped at a LOD score of 12 and linkage maps constructed using the maximum likelihood algorithm. Two progenies were available for each of three resistance sources (H3R04P23, H3R13P40, and OSU 533.129). Separate maps were made for each progeny in these pairs, and then the data merged and a new map constructed for that resistant parent.
Data were recorded in a spreadsheet, with presence of an SSR allele or disease resistance scored as “1” and absence or susceptibility scored as “0”. Scores for each marker and disease resistance were in separate columns, and the scores for a seedling were in a single row. The columns were then placed in order according to their positions on the reference linkage map. The SSR and disease resistance scores were inspected and individuals showing a conflict between disease score and adjacent SSR marker scores were identified. Disease scores for these seedlings were reentered as “u” for unknown, and new maps were created.
Alignment of the sequences of mapped SSR and RAPD markers, BAC end sequences, and Illumina sequences of BACs with the V2 reference ‘Jefferson’ genome identified four contigs (95F, 56F, 42F, and 77F) as being in the ‘Gasaway’ resistance region on LG6 (Table 3). A search of these four contigs for SSRs identified 896 di-, 187 tri-, 206 tetra-, 70 penta-, and 282 hexa-nucleotide repeats, with di-nucleotide repeats more abundant that the longer motifs. Removal of the repeats that contained only As and Ts reduced the number of unique fragments to 451. When the Illumina genome sequence reads of the 7 other cultivars were aligned with the trimmed ‘Jefferson’ fragments using Tablet software, 116 were identified as clearly polymorphic with variation in number of repeats but conserved flanking sequences. The alignments showed that the di-nucleotide repeats were more polymorphic than the tri-, tetra-, penta- and hexa-nucleotide repeats. After PCR amplification of 24 accessions and electrophoresis on 3% agarose gels, 60 of the 116 were scored as clearly polymorphic. A BLAST search against the NCBI database confirmed that all of the markers were different from previously developed SSR markers. Following amplification using fluorescent forward primers, allele sizes were scored in the 50 hazelnut accessions. Of the 60 markers scored as clearly polymorphic on agarose gels, 42 were easy to score while 18 were difficult to score and not pursued further. These 42 markers were characterized using 50 hazelnut accessions (Table 4). The number of alleles per locus ranged from 5 to 19 with an average of 10.7. The mean values for Ho, He and PIC were 0.74, 0.78, and 0.76, respectively. The frequency of null alleles at the 42 SSR markers averaged 0.049 and showed a range from −0.121 to 0.473. A null allele is defined as any allele that consistently fails to amplify due to primer template mismatch. The frequency of null alleles exceeded 0.20 at 8 markers (GK1.09, GK1.10, GK1.45, GK1.46, GK6.76, GK6.83, GK6.84, and GK6.97). Of the 42 markers, 20 segregated in the mapping population and were placed on the reference linkage map (Figures 1, 2). The map locations of the remaining markers were deduced based on the V2 contig from which they were developed and their coordinates in the V3 ‘Jefferson’ genome (Supplementary Material 3). Of the 20 mapped markers, nine were placed on LG1 and 11 were placed on LG6. Of the nine markers on LG1, six (GK1.05, GK1.30, GK1.40, GK1.41, GK1.44, and GK1.45) were placed on the maps of both the female and male parents and three markers (GK1.12, GK1.10, GK1.20) were placed only on the map of the female parent (Figure 1). Of the 11 markers on LG6, seven (GK6.61, GK6.63, GK6.81, GK6.89, GK6.90, GK6.92, and GK6.94) were placed on the maps of both the female and male parents, while one (LG6.84) was placed only on the map of the female parent and three (GK6.77, GK6.80, and GK6.91) were placed only on the map of the male parent (Figure 2). All of the markers assigned to LG1 were developed from contigs 56F and 42F, which had been identified using sequences of BACs 53B4 and 78A4 in the physical map of the eastern filbert blight resistance region (Sathuvalli et al., 2017). None of the markers developed from contigs 56F and 42F were placed on LG 6.
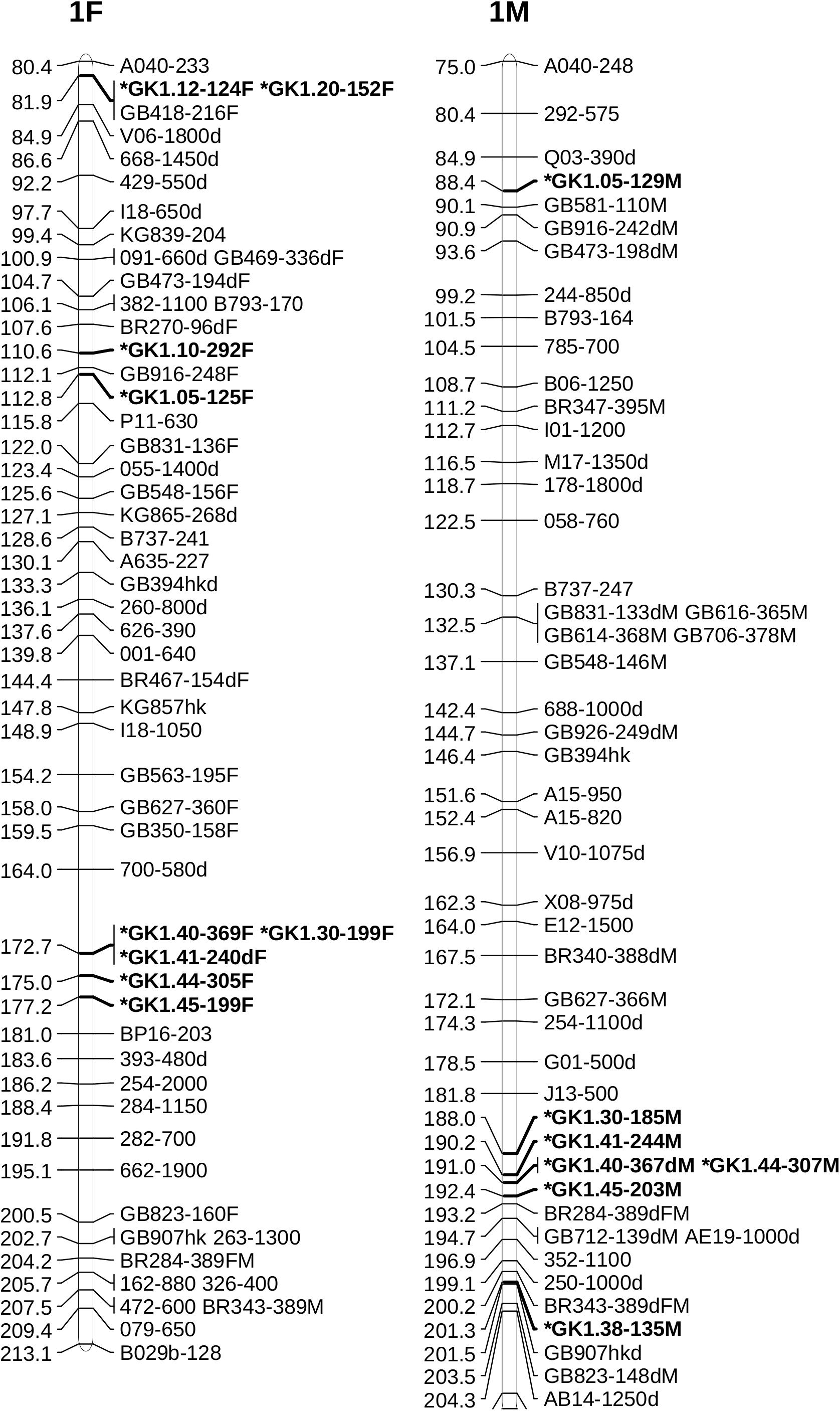
Figure 1. Maps of linkage group 1 (LG1) of female parent OSU 252.146 (left) and resistant parent OSU 414.062 (right) in the reference mapping population for hazelnut (Corylus avellana) with new simple sequence repeat markers indicated by *. Units are centimorgans (cM).
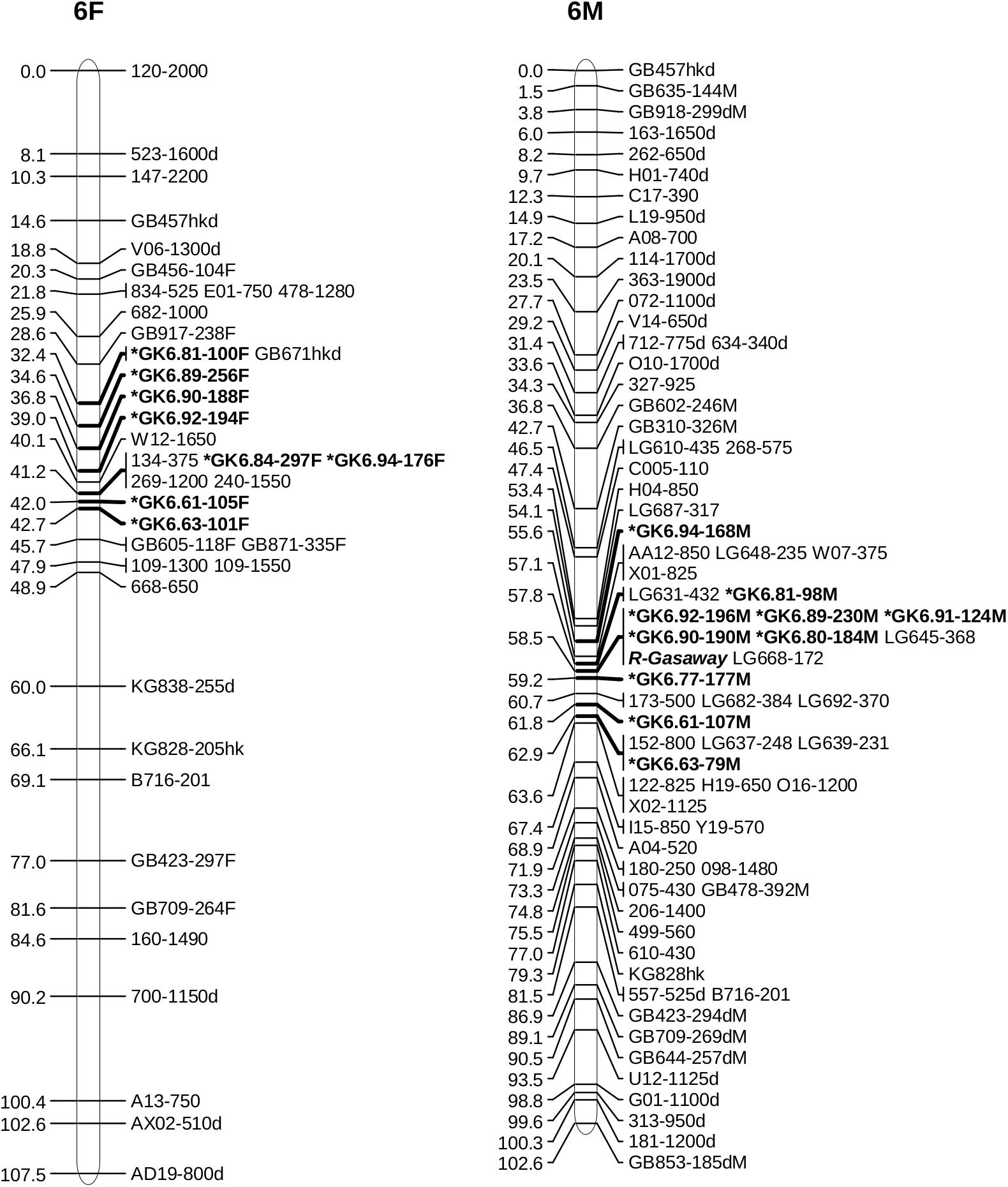
Figure 2. Maps of linkage group 6 (LG6) of female parent OSU 252.146 (left) and resistant parent OSU 414.062 (right) in the reference mapping population for hazelnut (Corylus avellana) with new simple sequence repeat markers indicated by *. Units are centimorgans (cM).
Segregation for disease response was studied in 18 progenies representing 14 resistant parents and 12 resistance sources (Table 2). Of the 14 parents, 12 fit the 1:1 ratio expected when resistance is dominant and under the control of a single locus. The two progenies segregating for resistance from Farris OSU 533.029 had an excess of resistant seedlings, and the heterogeneity Chi-square values indicated it was appropriate to pool the data. A single progeny segregating for resistance from ‘Moscow #37’ had an excess of susceptible seedlings. The field-grown progenies segregating for resistance from ‘Estrella #1’ included several very weak seedlings with no disease, 9 in progeny 11520 and 17 in progeny 11521, of which 7 and 4, respectively, died before DNA could be extracted. After the very week seedlings were removed, the Estrella #1 progenies in the EFB nursery and field fit the expected 1:1 ratio. In six progenies, some seedlings were structure-inoculated and others were field-inoculated; in all cases the heterogeneity chi-square indicated it was appropriate to pool the results of the two inoculation methods. For the RUS-13 (Holmskij) source, heterogeneity Chi-square values indicated that it was appropriate to pool the data of the two progenies of H3R04P23, and also to pool all four progenies with RUS-13 resistance.
Correlation coefficients for disease reaction and marker scores are presented with markers grouped by LG (Tables 5, 6). In 10 progenies, disease response was highly correlated (r > 0.70) with allele scores for markers on LG6, and thus EFB resistance was assigned to LG6 (Table 5). In progeny 12032, which segregated for resistance from Moscow #37, there was a surplus of susceptible seedlings. In progeny 11027 segregating for resistance from C. americana ’Winkler’, progeny 11032 segregating for resistance from C. americana OSU 366.060, and progeny 11029 segregating for resistance from Bauman hybrid OSU 401.014, the data fit the expected 1:1 ratio. In these four progenies, several markers on LG6, LG2, and LG7 were tested but disease response was not correlated with any marker allele scores, and EFB resistance could not be assigned to LG6, LG2, or LG7 (Table 6).
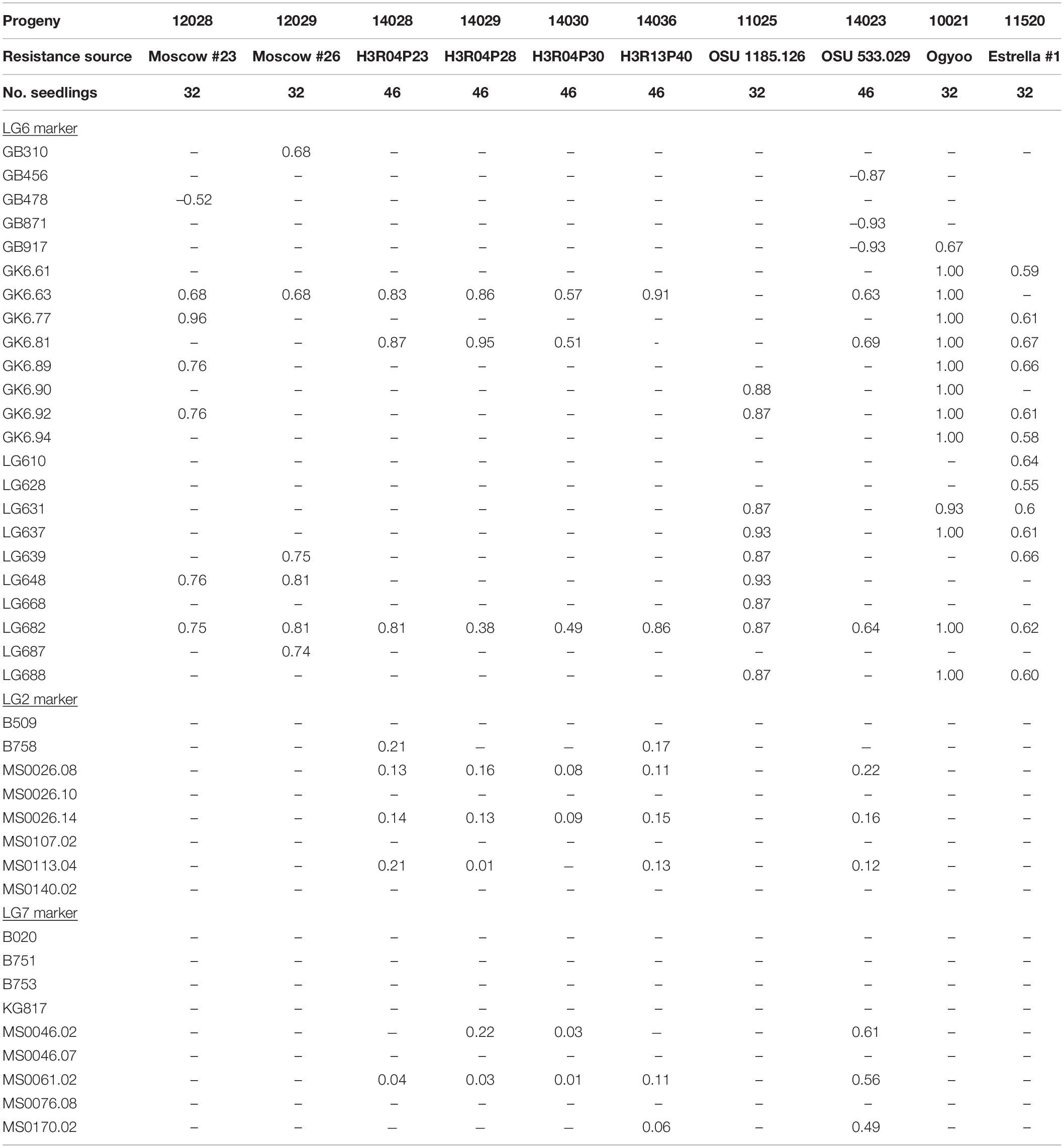
Table 5. Correlation coefficients for disease and simple sequence repeat marker scores in ten hazelnut progenies segregating for resistance to eastern filbert blight.
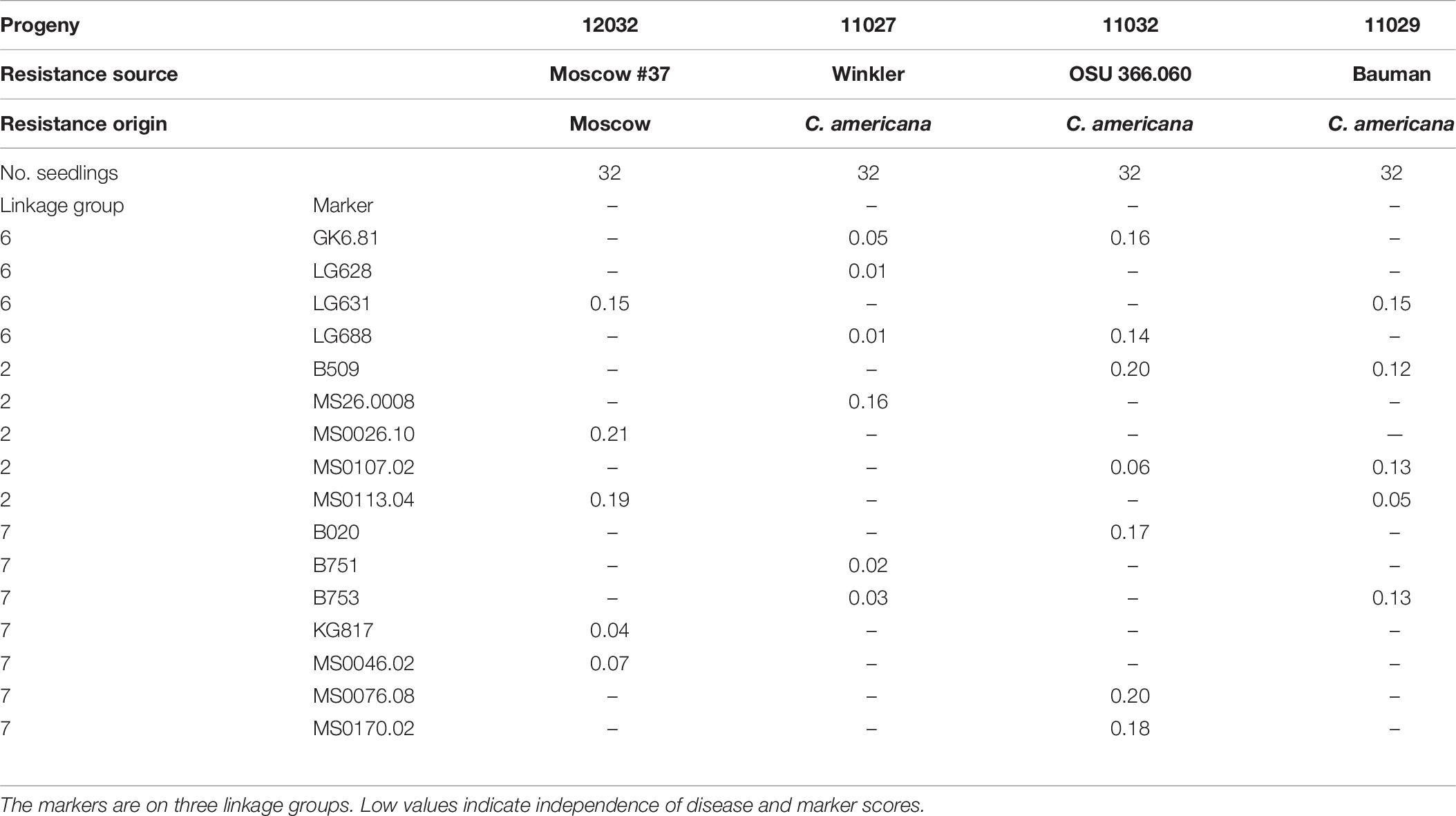
Table 6. Correlation coefficients for disease and simple sequence repeat marker scores in four hazelnut progenies segregating for resistance to eastern filbert blight.
Previously mapped and new SSR markers were used for construction of linkage maps for the eight EFB resistance sources on LG6. The initial maps were based on the marker and disease response scores recorded in a spreadsheet. Inspection identified very few conflicts between the score for disease response and for presence of adjacent SSR markers. A second version of the map was created after rescoring questionable disease scores as “unknown.”
The map for progeny 12028 (Figure 3A, n = 78), which segregates for resistance from Moscow #23, was drawn with six markers, and shows resistance co-segregating with LG648 and flanked by markers GK6.63 and LG682 at distances of 1.3 cM and 2.0 cM, respectively. The map for progeny 12029 (Figure 3B, n = 65), which segregates for resistance from Moscow #26, has 6 markers and spans 13.9 cM with the resistance locus at the end and markers LG687 and GB310 placed 1.9 and 3.6 cM away. Three selections (H3R04P23, H3R04P28 and H3R04P30) were selected from seed lot RUS-13 from Holmskij, Russia. H3R04P23 is a parent of progenies 14027 and 14028. Initially a separate map was drawn for each progeny, and then a single map (Figure 3C, n = 111) was drawn for the merged data. Resistant selections H3R04P28 and H3R04P30 are the parents of progenies 14029 and 14030, respectively, and the maps are presented (Figure 3D, n = 61; Figure 3E, n = 53). In the two progenies segregating for resistance from H3R13P40, selected from seed lot RUS-9, also from Holmskij, Russia, resistance co-segregated with seven markers and was flanked by markers A614 and KG821 at distances of 4.4 and 2.9 cM, respectively (Figure 3F, n = 78). The map for progeny 11025, which segregates for resistance from OSU 1185.126 (selected from a seed lot from Crimea), was drawn with nine markers. The map (Figure 3G, n = 95) spans 10.1 cM with LG648 co-segregating with resistance and flanking markers on both sides.
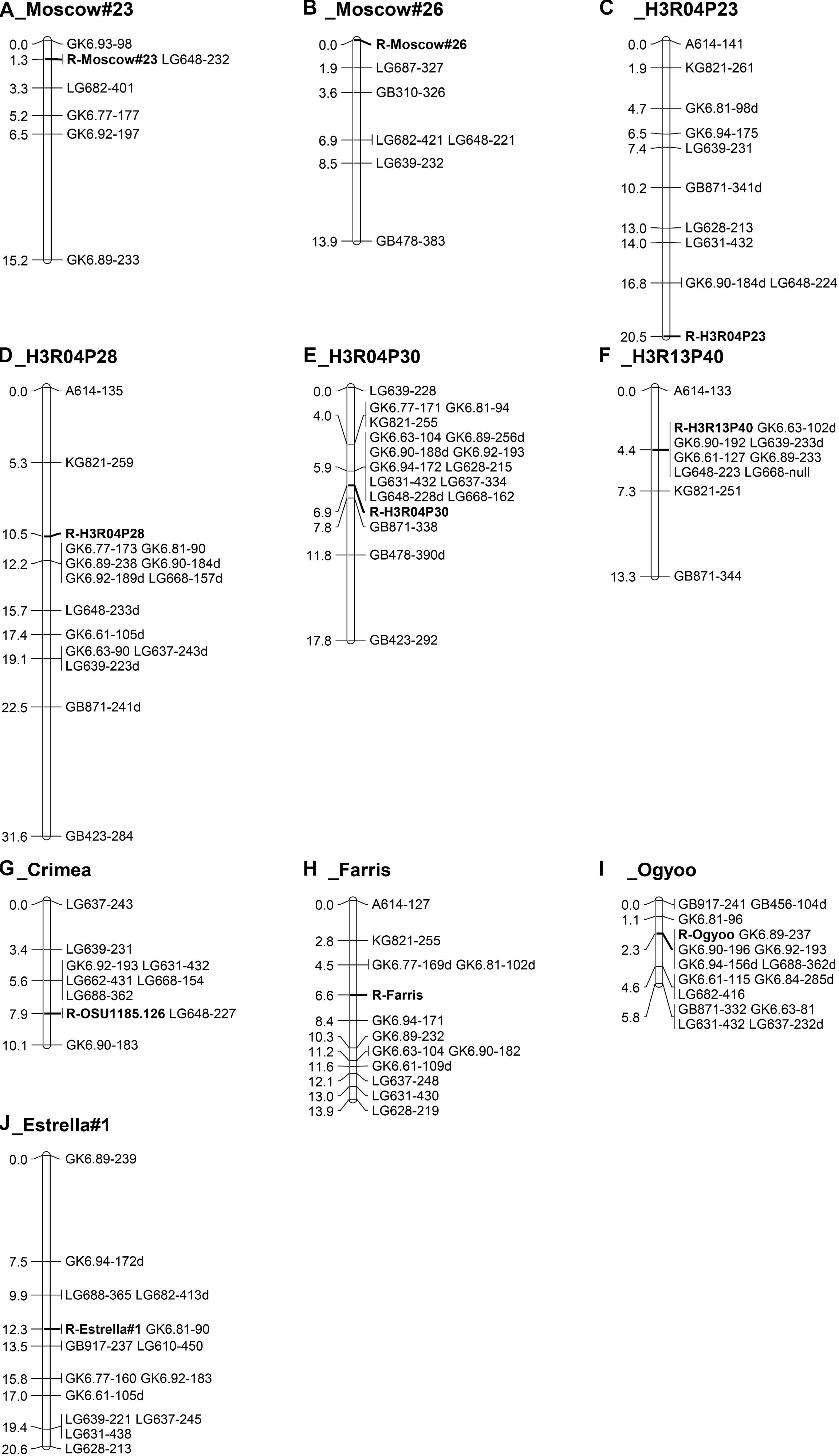
Figure 3. Maps of linkage group 6 (LG6) in hazelnut populations segregating for eastern filbert blight resistance from ten parents. (A) progeny 12028 for resistance from Moscow #23, (B) progeny 12029 for resistance from Moscow #26, (C) two progenies for resistance from H3R04P23 (RUS-13, Holmskij, Russia), (D) progeny 14029 for resistance from H3R04P28 (RUS-13), (E) progeny 14030 for resistance from H3R04P30 (RUS-13), (F) progeny 14036 for resistance from H3R04P30 (RUS-9, Holmskij, Russia), (G) progeny 11025 for resistance from OSU 1185.126 (RUS-26, Simferopol, Crimea), (H) two progenies for resistance from Farris OSU 533.029, (I) progeny 10021 for resistance from C. heterophylla ’Ogyoo’, and (J) progeny 11520 for resistance from ’Estrella #1’.
The two progenies (14023 and 14024) segregating for resistance from Farris OSU 533.029 showed a surplus of seedlings scored as resistant (Table 2), although in progeny 14023 the correlation coefficients with markers on LG6 were > 0.62. Alignment of disease and LG6 marker scores in progeny 14023 indicated that 10 of the 54 seedlings were likely “escapes”. In progeny 14024, 9 of the 56 seedlings appear to be “escapes” and two seedlings with intermediate disease ratings of 3 had marker alleles indicating resistance. The disease scores in these seedlings were recoded as “u” and the maps redrawn. After recoding the disease response in these 22 seedlings, a map was drawn from the merged data of the two progenies. This map (Figure 3H, n = 110) shows resistance from Farris OSU 533.029 in the middle, with four markers on one side and eight on the other side. After the disease responses of the seedlings were reassessed based on marker scores, a total of 19 escapes were reclassified as susceptible and three seedlings with small cankers but markers for resistance were reclassified as resistant. In contrast to the original segregation ratios (Table 2), the revised ratio of resistant to susceptible seedlings is 28:26 in progeny 14023, 33:23 in progeny 14024, and 61:49 overall, with all of these ratios fitting the expected 1:1 ratio. We conclude that resistance from Farris OSU 533.029 is conferred by a dominant allele at a single locus on LG6 but that several seedlings escaped infection in this structure inoculation.
The map for progeny 10021 (Figure 3I, n = 88), which segregates for resistance from C. heterophylla ’Ogyoo’, includes 15 SSR markers and spans 5.8 cM. Five markers co-segregate with resistance, and additional markers flank the resistance locus on both sides. Progenies 11520 and 11521 segregated for resistance from ‘Estrella #1’ and segregation ratios from structure exposure showed good fit to the 1:1 expected ratio. In the field, dwarf seedlings were noted in both progenies. In progeny 11520, seven weak seedlings died when they were very young, two others were severely stunted, and 53 were of normal size. In progeny 11521, four weak seedlings died when they were very young, 13 others were dwarfs, and 45 were of normal size. After removal of these weak seedlings, the segregation ratios in both progenies fit the1:1 expected ratio. Furthermore, the homogeneity chi-square values indicated that it was appropriate to pool the data for progenies 11520 and 11521 in both structure and field exposure, and all fit the 1:1 expected ratio. The map for progeny 11520 (Figure 3J, n = 88) includes 14 SSR markers and spans 20.6 cM, with marker GK6.81 co-segregating with resistance, LG688 and LG682 on one side and LG610 and GB917 on the other side. Inspection of marker scores revealed the same segregation seen for resistance, with a slight deficiency of SSR alleles linked to resistance in the seedlings.
In the progenies investigated by field exposure, a total of nine individuals were recorded as a resistant while the SSR data indicated susceptibility, and it is likely that these seedlings escaped infection from A. anomala by chance. An additional four seedlings were scored as susceptible but their SSR data indicated resistance. As noted earlier, disease scores for these seedlings were recoded as “unknown” before the final maps were drawn.
The ‘Jefferson’ genome sequence (V2) allowed efficient development of new SSR markers. The V2 sequence (Snelling et al., 2018) was from Pacific Biosciences reads, for which the V1 Illumina sequences (Rowley et al., 2018) were used for error correction. SSRs continue to be widely used in plant genetics. Polymorphic SSRs often segregate in many different progenies, map to a single point in the genome, and serve as anchor loci for the alignment of multiple linkage maps. In this study, 42 new polymorphic SSR markers were developed from four contigs in the ‘Jefferson’ genome sequence (V2), and 19 were placed on the reference linkage map. Markers developed from contigs 95F and 77F mapped to LG6 as expected, but markers developed from contigs 56F and 42F mapped to LG1. This was unexpected, as the physical map of the EFB resistance region in ‘Jefferson’ (Sathuvalli et al., 2017) include the BACs 53B4 and 78A4, which in this study identified contigs 42F and 56F, respectively. It is likely that these two BACs were false positives in the BAC library screening. Unexpected LG assignments were also reported by Sathuvalli and Mehlenbacher (2013), including SSR markers LG612 and LG613 developed from BAC 38N24, which mapped to LG1. Additional false positives in their study were LG605 developed from BAC 62A9, which mapped to LG4, and LG655 and LG657 from BAC 65G23, which mapped to LG5. SSR markers developed for hazelnut are transferable across Corylus species (Bassil et al., 2005a, 2013, Boccacci et al., 2005) and even the related genera Betula and Alnus in the family Betulaceae (Gürcan and Mehlenbacher, 2010b). Future research, including saturation of the LG6 map with single nucleotide polymorphism markers, and alignment of these markers and BAC end sequences with the V3 ‘Jefferson’ genome sequence will allow rapid identification of genes of interest. The V3 ‘Jefferson’ genome sequence is from PacBio sequencing plus Hi-C proximity ligation (Dovetail Genomics, Scotts Valley, CA, United States) and consists of 11 scaffolds that represent the haploid number of hazelnut.
Segregation for response to EFB inoculation was studied in 18 progenies from 14 resistant parents representing 12 resistance sources (Table 2) of diverse origins. Seven sources are of Russian origin (Moscow #23, Moscow #26, Moscow #37, and four selections from seeds purchased in Holmskij), one (OSU 1185.126) originated from seeds purchased near Simferopol, Crimea, one (OSU 533.029) is from a seed lot received from Michigan, United States, and two are hybrids with other Corylus species. ‘Ogyoo’ (HF13) was selected from wild C. heterophylla in the South Korea. ’Estrella #1’, an interspecific hybrid from C. sutchuenensis × C. avellana ‘Holder’ (Farris, 1974), showed complete resistance to EFB after greenhouse inoculation (Chen et al., 2007) and 6 years of exposure in a field planting with high disease pressure (Capik and Molnar, 2012). Farris (1974) grew out several dozen seedlings from the cross and they appeared to be true hybrids based on morphology, but some were dwarf and stunted while others were vigorous and healthy. He selected the five best plants and named them Estrella hybrids #1 to #5. The observation of weak seedlings in the offspring of ‘Estrella #1’ in this study is consistent with the breeder’s notes for the original cross. ‘Estrella #1’ yields well and produces medium-size nuts with a slightly long shape but is male-sterile. The phenotype of Bauman selection OSU 401.014 indicates that it is a hybrid of C. americana × C. avellana, which would not be surprising as the hazelnut collections of many members of the Northern Nut Growers Association include both parent species and interspecific hybrids. In progenies segregating for resistance from 12 of these 14 sources, the segregation ratio for disease response fit the 1:1 ratio expected for control by single loci at which the dominant alleles confer resistance, and the resistant parent is heterozygous. The two progenies segregating for resistance from Farris OSU 533.029 had an excess of seedlings lacking disease and SSR markers indicated that a high number of seedlings had escaped infection. When these escapes were reclassified, the segregation ratios fit the expected 1:1 ratio. The one progeny segregating for resistance from ‘Moscow #37’ had an excess of susceptible seedlings. An excess of susceptible seedlings was reported in previous EFB resistance studies in hazelnut (Colburn et al., 2015; Bhattarai et al., 2017b). Lunde et al. (2006) noted an excess of resistant seedlings in the offspring of ’Zimmerman’, and that even when SSR markers indicate that the resistance gene is present, small cankers occasionally develop. Correlation with scores for alleles at mapped SSR markers allowed resistance from ten sources to be assigned to LG6 in the same region as ‘Gasaway’ resistance, and linkage maps were constructed and compared using common SSR markers. Disease response in progenies segregating for the remaining four resistance sources (Moscow #37, C. americana ‘Winkler’ and OSU 366.060, and Bauman hybrid OSU 401.014) was not correlated with scores for markers on LG6 or LG7 or LG2. The resistance from these sources might be due to a major gene on one of the other 8 LGs, and will be investigated further. Resistance on different linkage groups will be especially useful for R-gene pyramiding.
Most resistance genes follow the gene-for-gene hypothesis (Flor, 1956), which states that for every resistance gene in the plant, there is a corresponding Avr gene in the pathogen that confers avirulence. The host’s resistance gene allows it to detect and defend against the invader. Of the categories of resistance genes, those with a nucleotide binding site and leucine rich repeat (NBS-LRR) are most common, are associated with resistance to several plant pathogens, and are the targets of many investigations (Hulbert et al., 2001; Morata and Puigdomènech, 2017). Most NBS-LRR genes are physically clustered in plant genomes (Meyers et al., 2003; Zhou et al., 2004), possibly the result of duplication or amplification of the gene families. ‘Jefferson’ carries resistance inherited from ‘Gasaway’ and is heterozygous at a resistance locus on LG6. Sathuvalli et al. (2017) performed fine mapping of the region and suggested candidates for the EFB resistance gene. Further research is needed to determine if the ten new sources of resistance on LG6 investigated in this study are the same gene as ‘Gasaway’ or different genes.
Several studies have described the pyramiding of major genes for disease resistance as an approach for more durable resistance (Joshi and Nayak, 2010; Zhu et al., 2012), a strategy that would be facilitated by molecular markers tightly linked to the different resistance alleles (Mundt, 2018). The new SSR markers developed in this study and placed on LG6 near the ‘Gasaway’ resistance locus will be useful in marker-assisted selection and cultivar fingerprinting. Of these, the most promising from contig 77F are KG6.63 and GK6.61 and from contig 95F are GK6.81 and GK6.92 as they have high PIC values, few null alleles, and are easy to score. As breeder-friendly markers, they are alternatives to the RAPD markers 152-800 and 268-580 currently used for MAS. All of the new sources of resistance to A. anomala investigated in this study are promising for use in breeding. Eight sources were assigned to LG6, for which the newly developed SSR markers will aid the pyramiding of resistance genes. Four additional sources are also useful for breeding but their resistance has not yet been mapped.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MK913590-MK913608 and MT670379-MT670401. Marker sequences are available from GenBank.
SM conceived, designed, and secured funding for the study and wrote the first draft of the manuscript. GK developed, characterized, and mapped the new markers. GK and MŞ studied segregation for disease response, correlation of disease response and markers, mapping of resistance regions, and statistical analysis. JS assisted in marker development and scoring. GK, MŞ, and JS wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
This research was supported by the Oregon Hazelnut Commission, the Oregon Agricultural Experiment Station, USDA-NIFA Specialty Crops Research Initiative Competitive Grant 2016-51181-25412, USDA-NIFA Agriculture and Food Research Initiative Competitive Grant 2014-67013-22421 and a USDA Specific Cooperative Agreement for Eastern Filbert Blight Research. This research partially fulfilled the requirement for GK Doctor of Philosophy degree and MŞ Master of Science degree.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank Senior Research Assistants David C. Smith and Rebecca L. McCluskey for managing the plant material and assistance with disease data collection. The authors also thank Kelly Vining for providing the ‘Jefferson’ hazelnut genome sequences (V2 and V3) used in this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.684122/full#supplementary-material
Supplementary Material 1 | Pedigrees of hazelnut (Corylus avellana) progenies segregating for resistance to eastern filbert blight. Resistant selections are underlined; all others are susceptible.
Supplementary Material 2 | Primer sequences, linkage group assignments, and coordinates in the ‘Jefferson’ hazelnut genome sequence of 42 new simple sequence repeat markers in hazelnut.
Supplementary Material 3 | Segregation and linkage group assignments of 19 simple sequence repeat markers in the reference hazelnut mapping population (OSU 252.146 × OSU 414.062).
EFB, eastern filbert blight; OSU, Oregon State University; SSR, simple sequence repeat.
Akın, M., Nyberg, A., Postman, J., Mehlenbacher, S., and Bassil, N. V. (2016). A multiplexed microsatellite fingerprinting set for hazelnut cultivar identification. Eur. J. Hortic. Sci. 81, 327–338. doi: 10.17660/eJHS.2016/81.6.6
Bassil, N. V., Botta, R., and Mehlenbacher, S. A. (2005a). Additional microsatellite markers of the European hazelnut. Acta Hortic. 686, 105–110. doi: 10.17660/ActaHortic.2005.686.13
Bassil, N. V., Botta, R., and Mehlenbacher, S. A. (2005b). Microsatellite markers in hazelnut: isolation, characterization and cross-species amplification. J. Am. Soc. Hortic. Sci. 130, 543–549. doi: 10.21273/JASHS.130.4.543
Bassil, N., Boccacci, P., Botta, R., Postman, J., and Mehlenbacher, S. (2013). Nuclear and chloroplast microsatellite markers to assess genetic diversity and evolution in hazelnut species, hybrids and cultivars. Genet. Resour. Crop Evol. 60, 543–568. doi: 10.1007/s10722-012-9857-z
Bhattarai, G., and Mehlenbacher, S. A. (2017). In silico development and characterization of tri-nucleotide simple sequence repeat markers in hazelnut (Corylus avellana L.). PLoS One 12:e0178061. doi: 10.1371/journal.pone.0178061
Bhattarai, G., and Mehlenbacher, S. A. (2018). Discovery, characterization, and linkage mapping of simple sequence repeat markers in hazelnut. J. Am. Soc. Hortic. Sci. 143, 347–362.
Bhattarai, G., Mehlenbacher, S. A., and Smith, D. C. (2017a). Eastern filbert blight disease resistance from Corylus americana ‘Rush’ and selection ‘Yoder #5’ maps to linkage group 7. Tree Genet. Genomes 13:45.
Bhattarai, G., Mehlenbacher, S. A., and Smith, D. C. (2017b). Inheritance and linkage mapping of eastern filbert blight disease resistance in ‘Uebov’ hazelnut. J. Am. Soc. Hortic. Sci. 142, 289–297. doi: 10.21273/jashs04145-17
Boccacci, P., Akkak, A., Bassil, N. V., Mehlenbacher, S. A., and Botta, R. (2005). Characterization and evaluation of microsatellite loci in European hazelnut (Corylus avellana L.) and their transferability to other Corylus species. Mol. Ecol. Notes 5, 934–937. doi: 10.1111/j.1471-8286.2005.01121.x
Boccacci, P., Beltramo, C., Sandoval Prando, M. A., Lembo, A., Sartor, C., Mehlenbacher, S. A., et al. (2015). In silico mining, characterization and cross-species transferability of EST-SSR markers for European hazelnut (Corylus avellana L.). Mol. Breed. 35:21. doi: 10.1007/s11032-015-0195-7
Cameron, H. R. (1976). Eastern filbert blight established in the Pacific Northwest. Plant Dis. Rep. 60, 737–740.
Capik, J. M., and Molnar, T. J. (2012). Assessment of host (Corylus sp.) resistance to eastern filbert blight in New Jersey. J. Am. Soc. Hortic. Sci. 137, 157–172. doi: 10.21273/JASHS.137.3.157
Capik, J. M., Muehlbauer, M., Novy, A., Honig, J. A., and Molnar, T. J. (2013). Eastern filbert blight-resistant hazelnuts from Russia. Ukraine, and Poland. Am. Soc. Hortic. Sci. 48, 466–473. doi: 10.21273/HORTSCI.48.4.466
Chen, H., Mehlenbacher, S. A., and Smith, D. C. (2007). Hazelnut accessions provide new sources of resistance to eastern filbert blight. HortScience 42, 466–469. doi: 10.21273/HORTSCI.42.3.466
Colburn, B. C., Mehlenbacher, S. A., and Sathuvalli, V. R. (2017). Development and mapping of microsatellite markers from transcriptome sequences of European hazelnut (Corylus avellana L.) and use for germplasm characterization. Mol. Breed. 37:16. doi: 10.1007/s11032-016-0616-2
Colburn, B. C., Mehlenbacher, S. A., Sathuvalli, V. R., and Smith, D. C. (2015). Eastern filbert blight resistance in hazelnut accessions ‘Culplà’, ‘Crvenje’, and OSU 495.072. J. Am. Soc. Hortic. Sci. 140, 191–200. doi: 10.21273/jashs.140.2.191
Coyne, C. J., Mehlenbacher, S. A., and Smith, D. C. (1998). Sources of resistance to eastern filbert blight in hazelnut. J. Am. Soc. Hortic. Sci. 123, 253–257. doi: 10.21273/JASHS.123.2.253
Dunlevy, A., Gandler, M. P., Hlubik, D., Capik, J. M., and Molnar, T. J. (2019). Examining Pathogenic Variation and Host Plant Response to Eastern Filbert Blight in Hazelnut Cultivars Protected by the ‘Gasaway’ Resistance Gene. Abstract retrieved from HortS54(9) Se 238. Available online at: https://ashs.confex.com/ashs/2019/webprogramarchives/Paper30640.html (accessed July 23, 2019).
Farris, C. W. (1974). An introduction to the stars–a new family of filbert hybrids. Annu. Rep. North Nut Grow. Assoc. 67, 80–82.
Grattapaglia, D., and Sederoff, R. (1994). Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics 137, 1121–1137. doi: 10.1093/genetics/137.4.1121
Gürcan, K., and Mehlenbacher, S. A. (2010a). Development of microsatellite marker loci for European hazelnut (Corylus avellana L.) from ISSR fragments. Mol. Breed. 26, 551–559. doi: 10.1007/s11032-010-9464-7
Gürcan, K., and Mehlenbacher, S. A. (2010b). Transferability of microsatellite markers in the Betulaceae. J. Am. Soc. Hortic. Sci. 135, 159–173. doi: 10.21273/JASHS.135.2.159
Gürcan, K., Mehlenbacher, S. A., Botta, R., and Boccacci, P. (2010). Development, characterization, segregation, and mapping of microsatellite markers for European hazelnut (Corylus avellana L.) from enriched genomic libraries and usefulness in genetic diversity studies. Tree Genet. Genomes 6, 513–531. doi: 10.1007/s11295-010-0269-y
Hill, R., Baldassi, C., Snelling, J. W., Vining, K. J., and Mehlenbacher, S. (2021). Fine mapping of the locus controlling self-incompatibility in European hazelnut. Tree Genet. Genomes 17:6. doi: 10.1007/s11295-020-01485-5
Hoffman, J. I., and Nichols, H. J. (2011). A novel approach for mining polymorphic microsatellite markers in silico. PLoS One 6:e23283. doi: 10.1371/journal.pone.0023283
Honig, J. A., Muehlbauer, M., Capik, J. M., Kubik, C., Vaiciunas, J. N., Mehlenbacher, S. A., et al. (2019). Identification and mapping of eastern filbert blight resistance quantitative trait loci in European hazelnut using double digestion restriction site associated DNA sequencing. J. Amer. Soc. Hort. Sci. 144, 1–10. doi: 10.21273/JASHS04694-19
Hulbert, S. H., Webb, C. A., Smith, S. M., and Sun, Q. (2001). Resistance gene complexes: evolution and utilization. Ann. Rev. Phytopathol. 39, 285–312. doi: 10.1146/annurev.phyto.39.1.285
Johnson, K. B., Mehlenbacher, S. A., Stone, J. K., Pscheidt, J. W., and Pinkerton, J. N. (1996). Eastern filbert blight of European hazelnut–it’s becoming a manageable disease. Plant Dis. 80, 1308–1316. doi: 10.1094/PD-80-1308
Johnson, K. B., Pinkerton, J. N., Gaudreault, S. M., and Stone, J. K. (1994). Infection of European hazelnut by Anisogramma anomala: site of infection and effect of host developmental stage. Phytopathology 84, 1465–1470. doi: 10.1094/Phyto-84-1465
Joshi, R. K., and Nayak, S. (2010). Gene pyramiding—a broad spectrum technique for developing durable stress resistance in crops. Biotechnol. Mol. Biol. Rev. 5, 51–60. doi: 10.5897/BMBR2010.0006
Kalinowski, S. T., Taper, M. L., and Marshall, T. C. (2007). Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 16, 1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x
Kavas, M., Yıldırım, K., Seçgin, Z., and Gökdemir, G. (2020). Discovery of simple sequence repeat (SSR) markers in hazelnut (Corylus avellana L.) by transcriptome sequencing and SSR-based characterization of hazelnut cultivars. Scand. J. For. Res. 35, 227–237. doi: 10.1080/02827581.2020.1797155
Khodaeiaminjan, M., Kafkas, S., Ziya Motalebipour, E., and Çoban, N. (2018). In silico polymorphic novel SSR marker development and the first SSR-based genetic linkage map in pistachio. Tree Genet. Genomes 14:45.
Leadbetter, C. W., Capik, J. M., Mehlenbacher, S. A., and Molnar, T. J. (2016). Hazelnut accessions from Russia and Crimea transmit resistance to eastern filbert blight. J. Am. Pomol. Soc. 70, 92–109.
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics (Oxf. Engl.) 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Liu, K., and Muse, S. V. (2005). PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21, 2128–2129. doi: 10.1093/bioinformatics/bti282
Lunde, C. F., Mehlenbacher, S. A., and Smith, D. C. (2000). Survey of hazelnut cultivars for response to eastern filbert blight inoculation. HortScience 35, 729–731. doi: 10.21273/HORTSCI.35.4.729
Lunde, C.F., Mehlenbacher, S.A., and Smith, D.C. (2006). Segregation for resistance to eastern filbert blight in progeny of ‘Zimmerman’ hazelnut. J. Amer. Soc. Hort. Sci. 131, 731–737. doi: 10.21273/JASHS.131.6.731
Martins, W. S., Lucas, D. C. S., Neves, D. C. S., de Souza Naves, K. F., and Bertioli, D. J. (2009). WebSat–A Web software for MicroSatellite marker development. Bioinformation 3, 282–283.
Mason, A. S. (2015). “SSR genotyping”, in Plant genotyping. Methods in molecular biology (Methods and Protocols), Vol. 1245, ed. J. Batley (New York, NY: Humana Press), 77–89. doi: 10.1007/978-1-4939-1966-6_6
Mehlenbacher, S. A. (1991). “Hazelnuts (Corylus)”, in Genetic Resources in Temperate fruit and Nut Crops, Vol. 290, eds J. N. Moore and J. R. Ballington (Leuven: ISHS Acta Horticulturae), 789–836. doi: 10.17660/ActaHortic.1991.290.18
Mehlenbacher, S. A. (1995). Classical and molecular approaches to breeding fruit and nut crops for disease resistance. HortScience 30, 466–477.
Mehlenbacher, S. A., and Bhattarai, G. (2018). An updated linkage map for hazelnut with new simple sequence repeat markers. Acta Hortic. 1226, 31–37. doi: 10.17660/ActaHortic.2018.1226.4
Mehlenbacher, S. A., Brown, R. N., Davis, J. W., Chen, H., Bassil, N. V., Smith, D. C., et al. (2004). RAPD markers linked to eastern filbert blight resistance in Corylus avellana. Theor. Appl. Genet. 108, 651–656. doi: 10.1007/s00122-003-1476-9
Mehlenbacher, S. A., Brown, R. N., Nouhra, E. R., Gökirmak, T., Bassil, N. V., and Kubisiak, T. L. (2006). A genetic linkage map for hazelnut (Corylus avellana L.) based on RAPD and SSR markers. Genome 49, 122–133.
Mehlenbacher, S. A., Thompson, M. M., and Cameron, H. R. (1991). Occurrence and inheritance of resistance to eastern filbert blight in ‘Gasaway’ hazelnut. HortScience 26, 410–411. doi: 10.21273/HORTSCI.26.4.410
Meyers, B. C., Kozik, A., Griego, A., Kuang, H., and Michelmore, R. W. (2003). Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15, 809–834. doi: 10.1105/tpc.009308
Milne, I., Bayer, M., Cardle, L., Shaw, P., Stephen, G., Wright, F., et al. (2010). Tablet–next generation sequence assembly visualization. Bioinformatics (Oxf. Engl.) 26, 401–402. doi: 10.1093/bioinformatics/btp666
Molnar, T. J., Capik, J., Zhao, S., and Zhang, N. (2010). First report of eastern filbert blight on Corylus avellana ‘Gasaway’ and ‘VR20-11’ caused by Anisogramma anomala in New Jersey. Plant Dis. 94, 1265–1265. doi: 10.1094/PDIS-06-10-0445
Molnar, T. J., Zaurov, D. E., Goffreda, J. C., and Mehlenbacher, S. A. (2007). Survey of hazelnut germplasm from Russia and Crimea for response to eastern filbert blight. HortScience 42, 51–56. doi: 10.21273/HORTSCI.42.1.51
Morata, J., and Puigdomènech, P. (2017). Variability among Cucurbitaceae species (melon, cucumber and watermelon) in a genomic region containing a cluster of NBS-LRR genes. BMC Genomics 18:138. doi: 10.1186/s12864-017-3529-5
Muehlbauer, M., Honig, J., Capik, J. M., Vaiciunas, J. N., and Molnar, T. (2014). Characterization of eastern filbert blight-resistant hazelnut germplasm using microsatellite markers. J. Am. Soc. Hortic. Sci. 139, 399–432. doi: 10.21273/JASHS.139.4.399
Muehlbauer, M., Capik, J. M., Molnar, T. J., and Mehlenbacher, S. A. (2018). Assessment of the ‘Gasaway’ source of resistance to eastern filbert blight in New Jersey. Sci. Hortic. 235, 367–372. doi: 10.1016/j.scienta.2018.03.019
Mundt, C. C. (2018). Pyramiding for resistance durability: theory and practice. Phytopathology 108, 792–802. doi: 10.1094/PHYTO-12-17-0426-RVW
Öztürk, S. C., Öztürk, S. E., Celik, I., Stampar, F., Veberic, R., Doganlar, S., et al. (2017). Molecular genetic diversity and association mapping of nut and kernel traits in Slovenian hazelnut (Corylus avellana) germplasm. Tree Genet. Genomes 13:16. doi: 10.1007/s11295-016-1098-4
Pinkerton, J. N., Johnson, K. B., Theiling, K. M., and Griesbach, J. A. (1992). Distribution and characteristics of the eastern filbert blight epidemic in western Oregon. Plant Dis. 76, 1179–1182. doi: 10.1094/Pd-76-1179
Pinkerton, J. N., Johnson, K. B., Mehlenbacher, S. A., and Pscheidt, J. W. (1993). Susceptibility of European hazelnut clones to eastern filbert blight. Plant Dis. 77, 261–266. doi: 10.1094/PD-77-0261
Pinkerton, J. N., Johnson, K. B., Aylor, D. E., and Stone, J. K. (2001). Spatial and temporal increase of eastern filbert blight in European hazelnut orchards in the Pacific Northwest. Phytopathology 91, 1214–1223. doi: 10.1094/phyto.2001.91.12.1214
Pinkerton, J. N., Johnson, K. B., Stone, J. K., and Ivors, K. L. (1998). Maturation and seasonal discharge pattern of ascospores of Anisogramma anomala. Phytopathology 88, 1165–1173. doi: 10.1094/phyto.1998.88.11.1165
Pinkerton, J. N., Stone, J. K., Nelson, S. J., and Johnson, K. B. (1995). Infection of European hazelnut by Anisogramma anomala–ascospore adhesion, mode of penetration of immature shoots, and host response. Phytopathology 85, 1260–1268. doi: 10.1094/Phyto-85-1260
Rowley, E. R., VanBuren, R., Bryant, D. W., Priest, H. D., Mehlenbacher, S. A., and Mockler, T. C. (2018). A draft genome and high-density genetic map of European hazelnut (Corylus avellana L.). bioRxiv [Preprint] doi: 10.1101/469015 bioRxiv:469015,
Sathuvalli, V. R., Mehlenbacher, S. A., and Smith, D. C. (2010). Response of hazelnut accessions to greenhouse inoculation with Anisogramma anomala. HortScience 45, 1116–1119. doi: 10.21273/HORTSCI.45.7.1116
Sathuvalli, V. R., Chen, H., Mehlenbacher, S. A., and Smith, D. C. (2011a). DNA markers linked to eastern filbert blight resistance in ‘Ratoli’ hazelnut (Corylus avellana L.). Tree Genet. Genomes 7, 337–345. doi: 10.1007/s11295-010-0335-5
Sathuvalli, V. R., Mehlenbacher, S. A., and Smith, D. C. (2011b). DNA markers linked to eastern filbert blight resistance from a hazelnut selection from the Republic of Georgia. J. Am. Soc. Hortic. Sci. 136, 350–357. doi: 10.21273/JASHS.136.5.350
Sathuvalli, V. R., Mehlenbacher, S. A., and Smith, D. C. (2012). Identification and mapping of DNA markers linked to eastern filbert blight resistance from OSU 408.040 hazelnut. HortScience 47, 570–573. doi: 10.21273/HORTSCI.47.5.570
Sathuvalli, V. R., and Mehlenbacher, S. A. (2013). De novo sequencing of hazelnut bacterial artificial chromosomes (BACs) using multiplex Illumina sequencing and targeted marker development for eastern filbert blight resistance. Tree Genet. Genomes 9, 1109–1118. doi: 10.1007/s11295-013-0626-8
Sathuvalli, V. R., Mehlenbacher, S. A., and Smith, D. C. (2017). High-resolution genetic and physical mapping of the eastern filbert blight resistance region in ‘Jefferson’ hazelnut (Corylus avellana L.). The Plant Genome 10, 1–10. doi: 10.3835/plantgenome2016.12.0123
Şekerli, M., Komaei Koma, G., Snelling, J. W., and Mehlenbacher, S. A. (2021). New simple sequence repeat markers on linkage groups 2 and 7 and investigation of new sources of eastern filbert blight resistance in hazelnut. J. Am. Soc. Hortic. Sci. 146. 1–18. doi: 10.21273/JASHS05040-21
Snelling, J. W., Sathuvalli, V. R., Colburn, B. C., Bhattarai, G., Rowley, E. R., Mockler, T. C., et al. (2018). Genomic resource development in hazelnut breeding. Acta Hortic. 1226, 39–45. doi: 10.17660/ActaHortic.2018.1226.5
Stone, J. K., Johnson, K. B., Pinkerton, J. N., and Pscheidt, J. W. (1992). Natural infection period and susceptibility of vegetative seedlings of European hazelnut to Anisogramma anomala. Plant Dis. 76, 348–352. doi: 10.1094/pd-76-0348
Taheri, S., Abdullah, T. L., Yusop, M., Hanafi, M., Sahebi, M., Azizi, P., et al. (2018). Mining and development of novel SSR markers using next generation sequencing (NGS) data in plants. Molecules 23:399. doi: 10.3390/molecules23020399
Tang, J., Baldwin, S. J., Jacobs, J. M. E., van der Linden, C. G., Voorrips, R. E., Leunissen, J. A., et al. (2008). Large-scale identification of polymorphic microsatellites using an in silico approach. BMC Bioinformatics 9:374. doi: 10.1186/1471-2105-9-374
Thompson, M. M., Lagerstedt, H. B., and Mehlenbacher, S. A. (1996). “Hazelnuts”, in Fruit Breeding, Vol. 3, eds J. Janick and J. N. Moore (New York, NY: Wiley), 125–184.
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3–new capabilities and interfaces. Nucleic Acids Res. 40:e115. doi: 10.1093/nar/gks596
Van Ooijen, J. W. (2006). JoinMap ® 4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations. 33(10.1371). Wageningen: Kyazma BV.
Vieira, M. L. C., Santini, L., Diniz, A. L., and Munhoz, C. D. F. (2016). Microsatellite markers: what they mean and why they are so useful. Genet. Mol. Biol. 39, 312–328. doi: 10.1590/1678-4685-GMB-2016-0027
Vukosavljev, M., Esselink, G. D., van’t Westende, W. P. C., Cox, P., Visser, R. G. F., Arens, P., et al. (2015). Efficient development of highly polymorphic microsatellite markers based on polymorphic repeats in transcriptome sequences of multiple individuals. Mol. Ecol. Resour. 15, 17–27. doi: 10.1111/1755-0998.12289
Wang, X., Lu, P., and Luo, Z. (2013). GMATo: a novel tool for the identification and analysis of microsatellites in large genomes. Bioinformation 9, 541–544. doi: 10.6026/97320630009541
Zhao, T., Li, J., Miao, D., Liang, L., Wang, G., and Ma, Q. (2019). Analysis of SSR information in the pistil transcriptome and the development and characterization of novel tri-nucleotide SSR markers in Corylus. Eur. J. Hortic. Sci. 84, 310–321. doi: 10.17660/eJHS.2019/84.5.7
Zhou, T., Wang, Y., Chen, J. Q., Araki, H., Jing, Z., Jiang, K., et al. (2004). Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol. Genet. Genomics 271, 402–415. doi: 10.1007/s00438-004-0990-z
Keywords: Anisogramma anomala, Corylus avellana, eastern filbert blight, hazelnut, simple sequence repeat, microsatellite, disease resistance, linkage map
Citation: Komaei Koma G, Şekerli M, Snelling JW and Mehlenbacher SA (2021) New Sources of Eastern Filbert Blight Resistance and Simple Sequence Repeat Markers on Linkage Group 6 in Hazelnut (Corylus avellana L.). Front. Plant Sci. 12:684122. doi: 10.3389/fpls.2021.684122
Received: 22 March 2021; Accepted: 17 May 2021;
Published: 14 June 2021.
Edited by:
Inaki Hormaza, Institute of Subtropical and Mediterranean Horticulture La Mayora, SpainReviewed by:
Ezio Portis, University of Turin, ItalyCopyright © 2021 Komaei Koma, Şekerli, Snelling and Mehlenbacher. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shawn A. Mehlenbacher, U2hhd24uTWVobGVuYmFjaGVyQG9yZWdvbnN0YXRlLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.