
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 22 June 2021
Sec. Plant Symbiotic Interactions
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.683601
This article is part of the Research Topic Nutrient Dependent Signaling Pathways Controlling the Symbiotic Nitrogen Fixation Process View all 7 articles
 Yanlin Ma1,2
Yanlin Ma1,2 Rujin Chen1,2*
Rujin Chen1,2*Nitrogen (N) and phosphorus (P) are the two predominant mineral elements, which are not only essential for plant growth and development in general but also play a key role in symbiotic N fixation in legumes. Legume plants have evolved complex signaling networks to respond to both external and internal levels of these macronutrients to optimize symbiotic N fixation in nodules. Inorganic phosphate (Pi) and nitrate (NO3−) are the two major forms of P and N elements utilized by plants, respectively. Pi starvation and NO3− application both reduce symbiotic N fixation via similar changes in the nodule gene expression and invoke local and long-distance, systemic responses, of which N-compound feedback regulation of rhizobial nitrogenase activity appears to operate under both conditions. Most of the N and P signaling and transport processes have been investigated in model organisms, such as Medicago truncatula, Lotus japonicus, Glycine max, Phaseolus vulgaris, Arabidopsis thaliana, Oryza sativa, etc. We attempted to discuss some of these processes wherever appropriate, to serve as references for a better understanding of the N and P signaling and transport during symbiosis.
Legume plants can establish symbiotic interactions with soil bacteria, collectively known as rhizobia, to form new organs, i.e., nodules, in which differentiated bacteroids enclosed in plant cell-derived membranes in nodule-infected cells reduce atmospheric dinitrogen (N2) to ammonium (NH3) as a nitrogen (N) source for the growth of the host. The establishment of symbiotic interactions involves extensive dialogs of the signaling processes between the plant host and bacterial partners (Oldroyd and Downie, 2008; Oldroyd et al., 2011; Oldroyd, 2013).
The nitrogenase complex of bacteroids in nodules catalyzes the reduction of N2 to NH3. This process is energy-intensive, which requires a significant input of photosynthates (approximately equivalent to 25% of the shoot dry matter at harvest) from the host (Schulze et al., 1999). To minimize the energy cost, legume plants preferentially use any alternative N sources for growth, and when alternative N sources are not available or environmental conditions are unfavorable for growth, they restrict the nodule N2 fixation to the minimal level necessary to support plant growth. For optimal growth, legume plants have evolved a long-distance (systemic) regulatory mechanism [termed as autoregulation of nodulation (AON)] to control the number of nodules formed on the roots of a plant, and therefore the total nitrogenase activity and mechanisms quickly and efficiently inhibit the N fixation activity of nodules when alternative N sources are available or under environmental stresses (Magori and Kawaguchi, 2009; Mortier et al., 2012; Cui et al., 2019; Kobayashi et al., 2020).
Phosphorus (P) is an essential macronutrient required for the biosynthesis of nucleic acids, phospholipids, and many other molecules and metabolites in plants (Richardson, 2009). Not surprisingly, a deficiency in inorganic phosphate (Pi) severely affects photosynthesis, photorespiration, and other essential processes, and thus reducing plant growth. A sufficient supply of Pi is also required for nodule development and symbiotic N fixation in legumes (Gunawardeba et al., 1992; Vance et al., 2003; Sulieman and Tran, 2015). Pi is largely immobile and has poor mobility in soil, and therefore it is often not easily accessible by plant roots (Holford, 1997). As a result of adaptation, plants have evolved various mechanisms to cope with growth under the condition of low Pi. These include morphological and growth changes, changes in the transcription of a large number of Pi starvation-responsive genes encoding transcription factors, Pi transporters, and scavengers. In legume plants, Pi starvation and nitrate (NO3−) application both inhibit symbiotic N fixation. In this review, we first provide a brief overview of a transcription factor NODULE INCEPTION (NIN) and its involvement in nodulation, followed by an overview of the role of NIN-like transcription factors in the NO3− inhibition of nodule development, NO3− and NH4+ transporters, nitric oxide (NO), and Pi signaling and transport in symbiotic N fixation. Finally, we provide a brief overview of the transcriptional regulation of Pi starvation and NO3− signaling.
Nodule development involves two separate processes, rhizobial infection, which takes place in root epidermal cells, and nodule organogenesis that is initiated in root cortical cells (Oldroyd and Downie, 2008). The nodule-specific transcription factor NIN plays an essential role in both processes (Schauser et al., 1999; Marsh et al., 2007). Under conditions of low soil N, NIN activation is downstream from the initial perception of rhizobial Nod factors [Nodulation factors (NFs)] by the plasma membrane-localized LysM domain receptor-like kinases NFR1 and NFR5 in Lotus japonicus and LYK3 and NFP in Medicago truncatula (Limpens et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003; Indrasumunar et al., 2010; Ried et al., 2014; Figure 1). The leucine-rich repeat receptor-like kinase SYMRK/DMI2 (L. japonicus/M. truncatula counterparts) also plays a key role in this process (Stracke et al., 2002; Limpens et al., 2005). The receptor-like cytoplasmic kinase NFR5-INTERACTING CYTOPLASMIC FORWARDING4 (NICK4) may link the NF signaling to downstream processes (Wong et al., 2019; Figure 1).
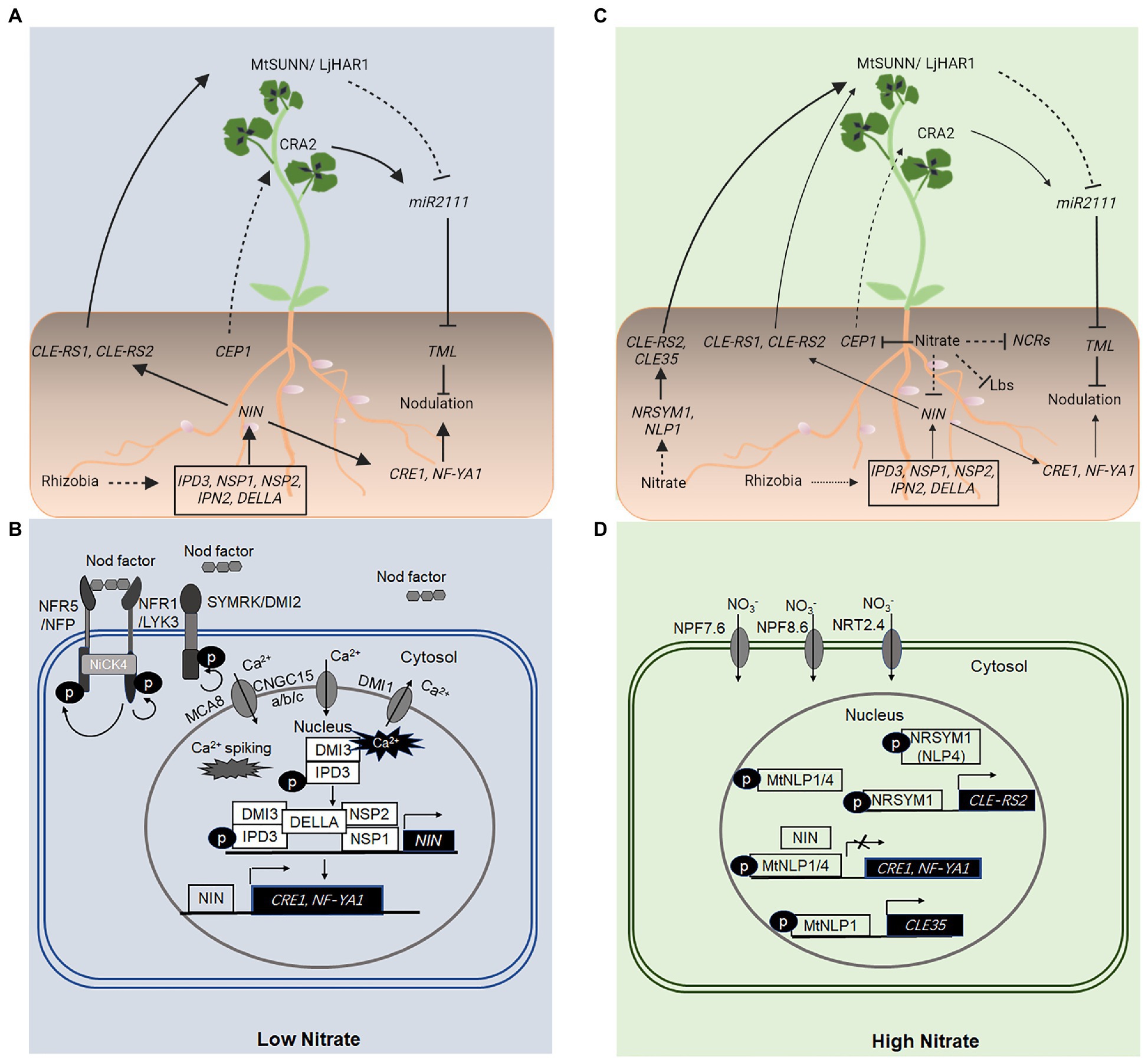
Figure 1. Local and long-distance, systemic regulation of nodulation using NODULE INCEPTION (NIN) and NIN-like proteins (NLPs). (A,B) Under conditions of low nitrate (NO3−), NIN activation is downstream from the initial perception of rhizobial nod factors by the LysM receptor-like kinases NFR1 and NFR5 in Lotus japonicus and LYK3 and NFP in Medicago truncatula, localized in the plasma membrane of root epidermal cells. Nodulation factor (NF) perception and signal transduction also require the LRR-RLK, SYMRK/DMI2, a cytoplasmic receptor-like kinase, NFR5-INTERACTING CYTOPLASMIC FORWARDING4 (NICK4), and a perinuclear Ca2+ oscillation (Ca2+ spiking) mediated by several proteins, including DOES NOT MAKE INFECTION1 (DMI1), CNGC15a/b/c, and MCA8 located in distinct nuclear membranes. The Ca2+ oscillation is perceived by the CALCIUM/CALMODULIN-DEPENDENT PROTEIN KINASE (CCaMK), DMI3, which interacts with and phosphorylates the transcription factor CYCLOP/IPD3. DELLA proteins promote DMI3–IPD3 interactions and phosphorylation of IPD3, and interactions between IPD3–NSP2–NSP1 complexes, which bind distinct cis-elements in the promoter sequence of NIN and activate NIN expression. The NF-induced NIN activation upregulates the expression of downstream genes including NF-YA1 and CRE1, which are essential for rhizobial infection and nodule organogenesis, respectively. NIN also binds directly to the promoter sequence and activates the expression of genes encoding CLE-like peptides such as CLE-RS1 and CLE-RS2 in L. japonicus, which are transported from the root to shoot, where they are perceived by the LRR-RLK, LjHAR1/MtSUNN. This leads to a reduced level of the mobile signal, miR2111 in the shoot and the root, and a corresponding increase in the transcript level of the miR2111 target, TML, which encodes a negative regulator of nodule initiation in the root. Conditions of low nitrogen (N) also activate a CEP1-CRA2 systemic pathway, which positively regulates the nodule number. (C,D) A high level of NO3− inhibits nodulation both locally and systemically. A high level of NO3− activates the expression of genes encoding the NLPs, NLP1, and NITRATE UNRESPONSIVE SYMBIOSIS1 (NRSYM1; NLP4), which directly activate the expression of genes encoding CLE35 and CLE-RS2 peptides in M. truncatula and L. japonicus, respectively. CLE-RS2 and CLE35 peptides function to inhibit the number of nodules formed on roots via the LjHAR1/MtSUNN-dependent autoregulation of nodulation (AON) pathway. A high level of NO3− also suppresses the CEP1 gene expression, diminishing the activity of a CEP1-CRA2 systemic pathway in the nodule number regulation. (D) Under conditions of high NO3−, NLPs (NLP1–NLP4 in M. truncatula) are activated and accumulate in the nucleus, where they interact with NIN and compete with NIN for binding sites in promoter sequences of target genes or both. As a result, the expression of NIN-activated genes including CRE1 and NF-YA1 is suppressed, inhibiting nodulation. A high level of NO3− also suppresses the expression of nodule cysteine-richs (NCRs) and LEGHEMOGLOBIN (Lbs), which results in the inhibition of nitrogenase activity. Solid and dashed lines indicate the processes that are supported by experimental evidence and putative, respectively. The size of the arrows indicates the relative strength of the signaling process.
Downstream from the NF signaling is a perinuclear Ca2+ oscillation (or calcium spiking) in root epidermal cells, which requires several proteins, including CYCLIC NUCLEOTIDE-GATED CHANNELs15a/b/c (CNGC15a/b/c), M. truncatula SERCA-Type Ca2+-ATPase8 (MtMCA8), and DOES NOT MAKE INFECTION1 (DMI1) located in distinct nuclear membranes (Capoen et al., 2011; Venkateshwaran et al., 2012; Charpentier et al., 2013, 2016; Leitao et al., 2019; Figure 1). The NF-induced perinuclear Ca2+ oscillation is perceived by the CALCIUM/CALMODULIN-DEPENDENT PROTEIN KINASE (CCaMK), DMI3 (Messinese et al., 2007), which specifically interacts with and phosphorylates the transcription factor CYCLOPS (also known as IPD3) in M. truncatula (Messinese et al., 2007; Yano et al., 2008). The phosphorylated IPD3 activates the transcription of NIN (Marsh et al., 2007; Messinese et al., 2007), which in turn upregulates the expression of downstream genes, including, among others, NF-YA1 and CRE1 required for rhizobial infection and nodule organogenesis, respectively (Vernie et al., 2015; Lin et al., 2018; Figure 1).
Notably, MtDELLA proteins (MtDELLA1, 2, 3), members of the plant-specific GRAS [GIBBERELLIN-INSENSITIVE (GAI), REPRESSOR OF ga1-3 (RGA) and SCARECROW (SCR)] protein family and key negative regulators of gibberellin signaling, promote the interaction between DMI3 (CCaMK) and CYCLOP/IPD3 and the phosphorylation of IPD3 (Jin et al., 2016). MtDELLA proteins also promote the interactions between IPD3 and NODULATION SIGNALING PATHWAY2 (NSP2), a different member of the GRAS family (Jin et al., 2016). Phosphorylated CYCLOP/IPD3 and NSP1-NSP2 complex can bind a CYC-box (TGCCATGTGGCA) and an NRE-box (AATTT) in the promoter sequence and activate the expression of NIN (Jin et al., 2016; Figure 1).
NODULE INCEPTION belongs to a NIN-like protein (NLP) subfamily of the plant-specific RWP-RK family of transcription factors (Wu et al., 2020). An NLP subfamily differs from the RWP-RK domain (RKD) subfamily in which, in addition to the RKD, they also contain in their C-terminal, the Phox and Bem1 (PB1) domain, which is involved in protein–protein interactions with other PB1 domain proteins. Although the functions of most RWP-RK proteins remain unknown, recent studies have shown that NLPs play an important role in NO3− signaling, and response in Arabidopsis and several other plant species (Mu and Luo, 2019). Lin et al. (2018) have identified five NLP genes in M. truncatula (Lin et al., 2018). MtNLP1–4 genes exhibit tissue-specific expression patterns, albeit most of them express in roots and nodules (Lin et al., 2018). Interestingly, M. truncatula transgenic hairy roots in which NLP1, NLP3, NLP4, or NLP5 gene is downregulated by using RNA interference (RNAi) developed mature nodules in the presence of a high concentration of NO3− (7.5 mM) that normally inhibits the nodule development in wild type plants. On the other hand, transgenic roots overexpressing the NLP1 gene exhibit hypersensitivity in nodule development in response to a high level of NO3−. The genetic analysis further shows that NLP1 and NLP4 functions are partially redundant in the NO3− inhibition of nodule development (Lin et al., 2018). These results indicate an important role of M. truncatula NIN-like proteins (MtNLPs) in the NO3− inhibition of nodule development.
In Arabidopsis thaliana, there are nine NLP proteins all of which have been shown to bind directly to a NO3−-responsive cis-element and regulate the expression of NO3−-responsive genes (Konishi and Yanagisawa, 2013). In L. japonicus, all five NLP proteins, including NIN, can bind to a NO3−-responsive cis-element and regulate downstream gene expression (Soyano et al., 2015). Interestingly, an amino-terminal of NLP proteins is responsible for the NO3− activation of NLP proteins. However, unlike other NLP proteins, an N-terminal domain of LjNIN is not responsive to NO3−. Furthermore, LjNIN antagonizes the NO3−-induced gene expression in L. japonicus (Soyano et al., 2015).
In A. thaliana, NLP7 is retained in the nucleus in response to NO3− to regulate the expression of early NO3−-responsive genes (Marchive et al., 2013). In M. truncatula, the mostly cytoplasmically localized NLP1 protein, whose gene expression is not induced by NO3−, accumulates in the nucleus after 10 min of NO3− treatments, and this NO3− response requires the N-terminal domain of MtNLP1. The NO3−-induced nuclear retention of NLP proteins appears to be conserved in A. thaliana (Marchive et al., 2013), L. japonicus (Nishida et al., 2018), and M. truncatula (Lin et al., 2018) and requires phosphorylation of the conserved Ser226 in the N-terminal domain of NLP proteins (Liu et al., 2017). MtNLP1ΔN (deletion of the N-terminal domain of MtNLP1) exhibits constitutive nuclear localization (Lin et al., 2018).
MtNIN interacts with all five MtNLPs via the PB1 domain (Lin et al., 2018). When co-expressed in Nicotiana benthamiana leaves, MtNLP1 suppresses the expression of MtNIN-activated genes, CRE1 and NF-YA1 only in the presence of NO3− (Lin et al., 2018). However, MtNLP1ΔN (containing the RWP-RK and PB1 domains) inhibits MtNIN-activated CRE1 and NF-YA1 expression in the absence of NO3−. Together with the genetic evidence, these results support the following model: when NO3− is below a threshold level, NLP1 and/or other NLP proteins remain largely in the cytosol. Nod factors induce the expression of NIN, which in turn activates the expression of downstream genes including NF-YA1 and CRE1 that are essential for rhizobial infection and nodule development, respectively. In the presence of a high level of NO3−, a Nod factor induction of NIN is lower, NLP1 and/or other NLP proteins accumulate in the nucleus, where NLP1 suppresses the NIN-activated gene expression by directly binding NIN, competing with NIN for binding sites in the promoters of target genes, or a combination of both, thus inhibiting nodule formation (Lin et al., 2018).
Nitrate greatly inhibits the NIN activation by rhizobia. In the nlp1 mutant, the NO3− inhibition of NIN expression in response to rhizobia is less severe, revealing a different mode of action of NLP1 in the NO3− regulation of nodulation (Lin et al., 2018). It is known that the NO3− inhibition of nodulation can occur locally or systemically via a long-range AON pathway (Nishida et al., 2018). Grafting experiments show that the role of MtNLP1 in the NO3− regulation of nodulation and lateral root development is determined by roots (Lin et al., 2018).
Recent studies have shown that NLP1 accumulates in the nucleus in response to NO3−, binds directly to the promoter sequence and activates the expression of CLE35, which plays a key role in the long-distance, systemic inhibition of nodule initiation in a SUNN-dependent manner (Luo et al., 2021; Figure 1).
In L. japonicus, NITRATE UNRESPONSIVE SYMBIOSIS1 (NRSYM1; LjNLP4) is involved in the NO3− inhibition of nodule development, nrsym1 mutant displays strong NO3− tolerance such that the number of nodules, infection threads, and nitrogenase activity are not reduced by NO3− (Nishida and Suzaki, 2018; Nishida et al., 2018). NRSYM1 accumulates in the nucleus in response to NO3− and binds directly to the promoter of CLE-RS2 and activates its expression in a NO3−-dependent manner (Nishida et al., 2018; Figure 1). LjNIN can also bind the same cis-element in a CLE-RS2 promoter to activate the CLE-RS2 expression. Ectopic expression of LjNIN interferes with the expression of NO3−-induced genes, and NO3− in turn, downregulates the expression of NIN target genes (Nishida et al., 2018; Figure 1). Genetic, biochemical, and grafting experiments have shown that NRSYM1 accumulated to the nucleus in response to NO3− activates CLE expression and reduces the number of nodules by an AON pathway (Nishida et al., 2018; Figure 1). Additionally, NRSYM1 is also involved in other nodulation processes, including the infection threads formation and nodule development and function (Nishida et al., 2018).
It is known that a low level of external NO3− promotes but a high level of NO3− inhibits symbiotic N fixation (Streeter and Wong, 1988; Nishida and Suzaki, 2018). The uptake and transport of NO3− in plants require two families of proteins, NITRATE TRANSPORTER1/PEPTIDE TRANSPORTER family (NPF; also called NRT1) and NITRATE TRANSPORTER2 (NRT2; Wang et al., 2020a). The larger NPF family consists of 53, 86, and 97 members in A. thaliana, L. japonicus, and M. truncatula, respectively, which are mostly low-affinity NO3− transporters with some exceptions such as Arabidopsis NRT1.1 and Medicago NRT1.3 exhibiting dual-affinity transporter activities (Morere-Le Paven et al., 2011; Wang et al., 2018, 2020b). The NRT2 family consists of only seven, four, and three members in A. thaliana, L. japonicus, and M. truncatula, respectively, and they are high-affinity NO3− transporters. Transcriptomic analyses have identified a large number of transporter-encoding genes that are induced in nodules (Colebatch et al., 2004; Takanashi et al., 2012; Laloum et al., 2014). This includes NPF and NRT2 genes (Criscuolo et al., 2012; Clarke et al., 2015) although the role of these proteins in nodule functioning has been reported only for a few of the family members.
Recently, M. truncatula NPF7.6 has been identified based on its nodule-specific expression pattern (Wang et al., 2020a). MtNPF7.6 is inducible by rhizobial infection or by a high level of NO3−, and in nodules, it is specifically expressed in nodule vascular tissues. MtNPF7.6 is localized to the plasma membrane of nodule transfer cells (NTCs) in the nodule vascular tissues. Experimental evidence support that it mediates the uptake of NO3− into nodules. Genetic analysis of MtNPF7.6 knockout mutants using the CRISPR-Cas9 gene-editing technique shows that npf7.6 mutants exhibit severely reduced responsiveness to NO3− at both low and high levels. Nodules developed on npf7.6 mutant roots exhibit the accumulation of a high level of NO, a severe reduction of LEGHEMOGLOBIN (Lb) gene expression, and impairment of nitrogenase activity, supporting a role for NPF7.6-mediated NO3− transport in regulating Lb gene expression, NO homeostasis, and nitrogenase activity. Because NPF7.6 is expressed in pericycle, xylem, and phloem tissues in nodules, with NPF7.6 proteins localized on the plasma membrane of NTCs of pericycle, xylem, and phloem, these results suggest that NPF7.6 mediates not only the uptake of NO3− from an external environment but also xylem-to-phloem transport of NO3−, and these processes appear to be critical for the NO3− regulation of nodule development and function (Wang et al., 2020a).
In L. japonicus, LjNRT2.4, a member of the NRT2 family of high-affinity NO3− transporters has been identified to play a role in the positive regulation of symbiotic N fixation by a low, permissible concentration of NO3− (Valkov et al., 2020). LjNRT2.4 is expressed in root and nodule vascular tissues. Comparing the symbiotic and nonsymbiotic growth in the presence of a low NO3− concentration (100 μM) of nrt2.4 mutants and wild type indicates that nrt2.4 mutants have a mild but significant reduction in the shoot biomass, nodule nitrogenase activity, and nodule NO3− content under the symbiotic condition. These phenotypes are even more pronounced when plants are grown in hydroponic culture (hypoxic condition). These phenotypic differences between wild type and nrt2.4 correlate well with a reduced level of NO in nodules of the nrt2.4 mutants in M. truncatula and L. japonicus (Horchani et al., 2011; Valkov et al., 2020). These studies support the role of NO3−-NO respiratory cycle in both mitochondria and bacteroids of nodule-infected cells as an alternative pathway for the energy production for symbiotic N fixation, which is particularly important under hypoxic conditions (Horchani et al., 2011; Valkov et al., 2020).
Ammonium levels in soils are usually 10–1,000 times less than that of NO3−, but some plants have preferentially taken up NH4+ when both NO3− and NH4+ are present (Gazzarrini et al., 1999). At a high level, NH4+ inhibits symbiotic N fixation although the regulatory mechanisms appear to be different from those of NO3− (Lin et al., 2018). There are two families of related transporter proteins, AMMONIUM TRANSPORTER1 and 2 (AMT1 and AMT2) that mediate NH4+ uptake from the soil and transport within the plants. In L. japonicus, AMT1;1, AMT1;2, and AMT1;3 can complement a yeast NH4+ transporter mutant and encoded proteins exhibit a high-affinity NH4+ transporter activity (D’Apuzzo et al., 2004). The expression of these three genes exhibits broad tissue specificity (D’Apuzzo et al., 2004). In nodules, the expression of LjAMT1;1 and LjAMT1;2 is in outer cortical cells and peripheral tissues, and the infected cells and vasculature tissues of the nodule, respectively (D’Apuzzo et al., 2004; Rogato et al., 2008). Furthermore, the expression of LjAMT1;1 and LjAMT1;2 is not responsive to NH4+ (Rogato et al., 2010). The expression of LjAMT1;3, however, is induced by NH4+ in a dose-dependent manner (Rogato et al., 2010). Knockdown of LjAMT1;1 expression using RNAi with a nodule-infected cell-specific Lbs promoter results in transgenic plants developing nodules with a compromised nitrogenase activity but a significantly increased number of nodules, supporting a role for LjAMT1;1-mediated NH4+ transport in nodule development and functioning (Rogato et al., 2008). In M. truncatula, three AMT2 family members, AMT2;3, AMT2;4, and AMT2;5, are induced in roots during symbiosis with arbuscular mycorrhizal fungi. Genetic analyses show that MtAMT2;3 but not MtAMT2;4 is required for the suppression of premature arbuscule degeneration (PAD) in phosphate transporter4 (pt4) mutants, supporting a role of the N status mediated by AMT2;3 in controlling arbuscule life span (Breuillin-Sessoms et al., 2015).
Nitric oxide, a reactive N species (RNS), is ubiquitous in plants and plays a signaling role in diverse developmental processes, including different stages during legume–rhizobial symbiosis. Both legumes and rhizobial partners contribute to the production and scavenging of NO during symbiotic N fixation. Enzymes, including NITRATE REDUCTASE (NR), PLASMA MEMBRANE-BOUND NITRITE: NO REDUCTASE, mitochondrial-electron transport chain (mETC)-dependent nitrite-reducing activity, NO synthase (NOS), POLYAMINE OXIDASE and HYDROXYLAMINE OXIDASE, and nonenzymatic reduction of nitrite have been suggested to contribute to NO production (Stohr et al., 2001; Bethke et al., 2004; Planchet et al., 2005; Tun et al., 2006; Corpas et al., 2009; Rumer et al., 2009; Horchani et al., 2011; Meilhoc et al., 2011; Blanquet et al., 2015; Astier et al., 2018; Leon and Costa-Broseta, 2020). NO production is enhanced by environmental stresses such as hypoxia and NO3−, and NO has been suggested to play a role in the NO3− inhibition of the nitrogenase activity (Kato et al., 2010).
Nitric oxide has been reported to both promote and inhibit symbiotic N fixation. These contradictory effects of NO may be associated with the level and pattern of NO production during legume–rhizobia interaction and nodule development (Signorelli et al., 2020). During symbiotic interactions between L. japonicus-Mesorhizobium loti and M. truncatula-Sinorhizobium meliloti, NO was induced at 4-h postinoculation (hpi; Nagata et al., 2008). Studies have shown that NO was induced in the infection pocket and specific scavenging of NO resulted in delayed development of nodules (del Giudice et al., 2011). Transcriptomic analyses revealed that NO can suppress defense responses, which would promote rhizobial infection and nodule development (Ferrarini et al., 2008; Boscari et al., 2013). Treatments with a low-level NO donor sodium nitroprusside (SNP, 0.1 mM) promoted the nitrogenase activity of nodules in L. japonicus although the mechanisms were not clear (Kato et al., 2010). These observations support a positive role of NO in the legume–rhizobia interaction and nodule development.
Treatments with a higher concentration of SNP (1 mM) inhibited the nitrogenase activity of nodules in L. japonicus (Kato et al., 2010). A similar situation was previously reported in treatments in vitro of nitrogenase isolated from the bacteroids of soybean (Trinchant and Rigaud, 1982). These observations support that NO can directly interact with and inhibit the activity of nitrogenase in soybean (Trinchant and Rigaud, 1982).
Nitric oxide can modulate the activity of proteins through S-nitrosylation, a reversible modification of a protein in which NO covalently binds to the thiol group of a cysteine residue of the protein to form an S-nitrosothiol. The modification results in structural changes and thereby may affect the activity of the protein. It is known that NO inhibits the activity of nitrogenase via the formation of a metal-nitrosyl complex (Michalski and Nicholas, 1987) or S-nitrosylation (Puppo et al., 2013). In addition to nitrogenase, many proteins related to the tricarboxylic acid (TCA) cycle and carbohydrate metabolism, amino acid metabolisms, such as ASPARAGINE SYNTHETASE, GLUTAMINE SYNTHETASE, S-ADENOSYLMETHIONINE SYNTHETASE, and redox regulation GLUTATHIONE PEROXIDASE1 are S-nitrosylated and inactivated by NO during symbiosis (Melo et al., 2011; Puppo et al., 2013; Castella et al., 2017).
LEGHEMOGLOBIN proteins transport and deliver a low but steady level of O2 to symbiosomes for bacteroid respiration, which is required for symbiotic N fixation but without inactivating the nitrogenase enzyme by O2. In addition to Lbs, plants also synthesize HEMOGLOBINs (Hbs), called Phytoglobins or Phytogbs, which can be divided into six groups based on the sequence and biochemical properties such as affinities to O2 (Hill et al., 2016). In L. japonicus nodules, all types of Phytogbs are highly expressed (Bustos-Sanmamed et al., 2011). At present, the function of Phytogbs remains to be investigated in detail. Under the microaerobic condition in nodules, NO produced by mETC is diffused to the cytosol and is oxidized to NO3− by OXYPHYTOGLOBIN [Phytogb(Fe2+)O2] and subsequently NO3− is reduced back to nitrite by NR and transported to mitochondria, and this cycle is repeated. This Phytogb-NO cycle is required for the re-oxidation of NAD(P)H to NAD(P)+, which is used in the glycolytic process (Igamberdiev and Hill, 2004).
In the N fixation zone of nodules, the NO level is high (Puppo et al., 2013). Both Lbs and Phytogbs contributed to the scavenging of NO and O2, and this is important for preventing the inactivation of the nitrogenase enzyme by NO (Herold and Puppo, 2005; Puppo et al., 2013; Berger et al., 2019). In S. meliloti, a FLAVOHEMOGLOBIN (HMP) has been shown to play a key role in detoxifying NO (Meilhoc et al., 2010; Cam et al., 2012).
Phosphate starvation response (PSR) includes both a long-distance, systemic response that involves sensing of cellular Pi level and a local, root tip inhibition response that involves sensing of extracellular Pi availability (Abel, 2017; Puga et al., 2017; Ried et al., 2021). Cellular Pi sensing relies on the MYB coiled-coil (MYB-CC) transcription factors, PHOSPHATE STARVATION RESPONSE1 (PHR1) and its related proteins PHLs (Rubio et al., 2001) and inositol pyrophosphates (PP-insPs)-binding SPX proteins that exhibit Pi-dependent inhibition of PHR activities (Puga et al., 2014; Wild et al., 2016; Ried et al., 2021). Under conditions of sufficient Pi, SPX binds to the coiled-coil domain of PHR and prevents the transcription factor from binding the cis-element in the promoter sequences of PSI genes and from activating their expression (Puga et al., 2014; Wang et al., 2014; Wild et al., 2016). Under the condition of Pi deficiency, PHR1 directly induces the transcription of PHT1 and PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 (PHF1), encoding an endoplasmic reticulum (ER) exit cofactor that interacts with and promotes PHT1 traffic to the plasma membrane (Gonzalez et al., 2005; Bayle et al., 2011; Chen et al., 2011).
Phosphate acquisition and transport are the key processes for efficient symbiotic N fixation in legumes (Nussaume et al., 2011; Gu et al., 2015; Sulieman and Tran, 2015). In a soybean, GmPT5, a high-affinity Pi transporter has been shown to transport Pi from roots to nodules, especially under conditions of limited P (Qin et al., 2012). Interestingly, another nodule-localized Pi transporter GmPT7 appears to play a role in delivering Pi from an external environment to the nodule fixation zone (Chen et al., 2019). PHR1, the constitutively expressed MYB-domain transcription factor, and its downstream target PHT1 constitute the key regulatory modules for symbiotic N fixation in legumes (Nussaume et al., 2011; Lu et al., 2020). Added to its complexity, it has been shown that a single soybean GmPHR has more than one GmPHT1 target, and one GmPHT1 is under the control of multiple GmPHRs (Lu et al., 2020). Therefore, different combinations of Pi transporters and Pi stress response factors form diverse regulatory networks in the N fixation and non-fixation zones to maintain Pi homeostasis in response to both systemic and local Pi availability for efficient symbiotic N fixation in soybean nodules (Lu et al., 2020). Eleven members of the PT1 family have been identified in M. truncatula, and most of them were highly expressed under conditions of low Pi (Cao et al., 2020). In addition to root epidermis, cortex, vascular tissues, and root tips, MtPT6 was also expressed in nodules, suggesting that it may play a role in Pi uptake from soil and transporting Pi to nodules from other tissues (Cao et al., 2020). Pi deficiency induces the expression of purple acid phosphatase (PAPs) in root nodules, which leads to an enhanced acquisition and utilization of P in root nodules (Li et al., 2017; Wang et al., 2020e). Furthermore, under conditions of Pi starvation, GmPHR1 can bind to the promoter sequence and activate the expression of GmPAP12 to maintain the P homeostasis and N fixation (Wang et al., 2020e).
Phosphate deficiency negatively impacts early molecular and physiological responses to rhizobial infection in common bean (Phaseolus vulgaris), including a significantly delayed activation of PvNSP2 and PvFLOT2 genes, compromised the root hair deformation and infection thread development, and the formation of a reduced number of nodules (Isidra-Arellano et al., 2018; Figure 2). These defects may be associated with an auxin-cytokinin imbalance, upregulation of ethylene, abscisic acid (ABA), and jasmonate-related signaling pathways under Pi deficiency in common bean (Isidra-Arellano et al., 2018). Recent studies have shown that the AON-related genes PvNIN, PvRIC1, PvRIC2, and PvTML are induced under conditions of Pi deficiency and their transcriptional activation is likely to be dependent on PHR1 (Isidra-Arellano et al., 2020; Figure 2). These studies suggest that Pi deficiency inhibits the root nodule symbiosis in common bean and soybean through constitutive activation of the AON pathway under Pi deficiency (Isidra-Arellano et al., 2020).
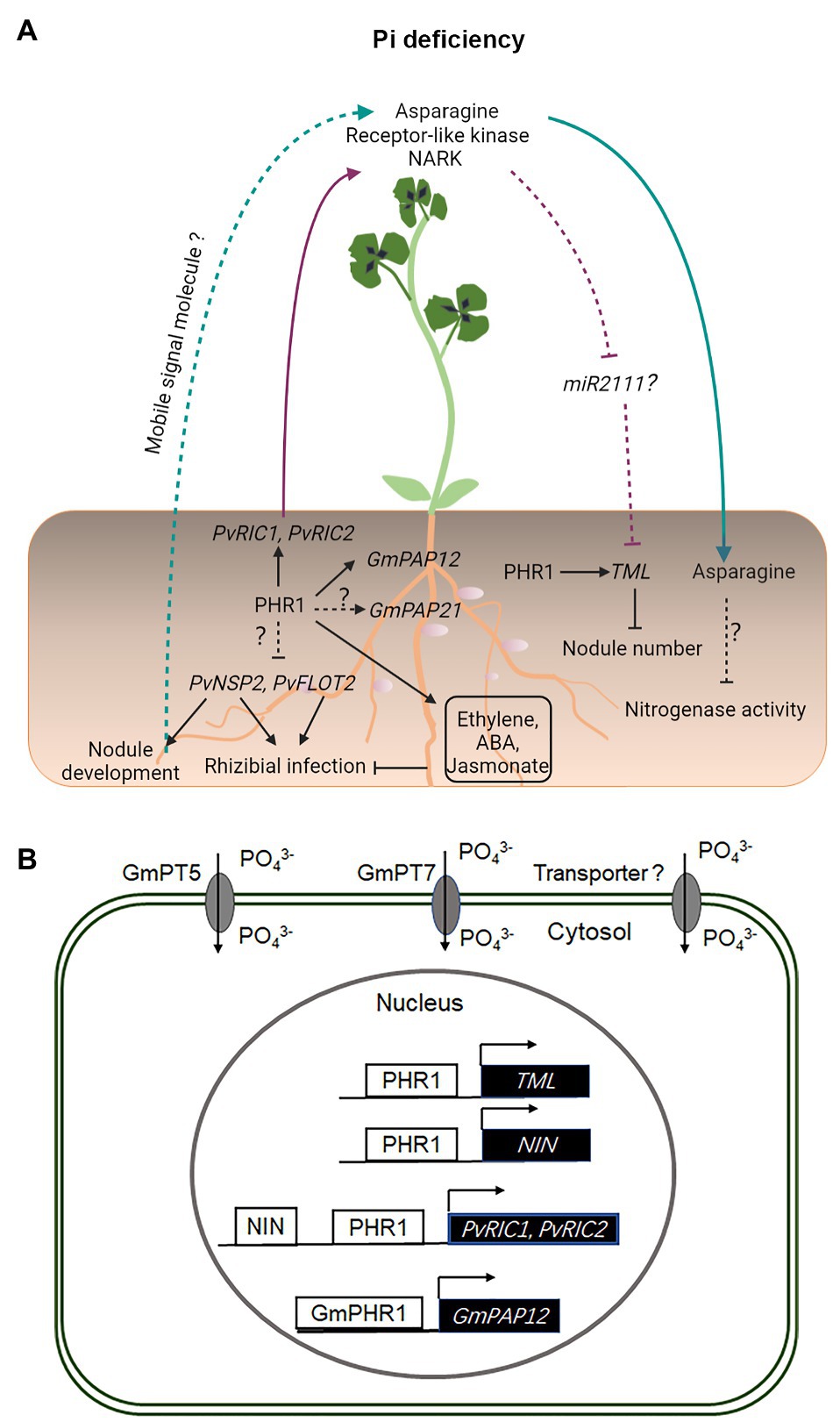
Figure 2. Legume–rhizobium symbiotic signaling under inorganic phosphate (Pi) deficiency. Under Pi deficiency, the expression of PvNSP2 and PvFLOT2 is inhibited, resulting in compromised rhizobial infection and nodule development (A). Pi deficiency induces the expression of GmPAP21 and GmPAP12, the latter of which is directly regulated by GmPHR1 (A,B). Pi deficiency also activates ethylene, abscisic acid (ABA), and jasmonate-related signaling processes, which inhibit rhizobial infection in Phaseolus vulgaris (A). The expression of NIN, RIC1, RIC2, and TML is directly activated by PHR1 in P. vulgaris under conditions of Pi deficiency (A,B). PvRIC1 and PvRIC2, two CLE-like peptides, negatively regulate the nodule number via the LRR-RLK, NARK-dependent AON pathway (A). It has not been shown whether miR2111 is involved in this process (A). Under Pi deficiency, the N-compound asparagine is significantly increased in the phloem sap and has been suggested to act as a systemic signal to inhibit the nitrogenase activity in nodules (A). Under Pi deficiency, GmPT5 and GmPT7 are activated and function to mobilize Pi from internal and external sources to nodules (B). Solid and dashed lines indicate the processes that are supported by experimental evidence and putative, respectively.
Phosphate deficiency leads to a decreased nodulation and nitrogenase activity in many legume species analyzed so far. Although the underlying molecular mechanisms remain largely unknown, studies have shown that Pi deficiency resulted in changes in the composition of free amino acids, particularly a significant increase of asparagine in the phloem sap, supporting the notion that the shoot-derived asparagine acting as a systemic signal to translocate to nodules and inhibit the nitrogenase activity in white clover (Trifolium repens) and M. truncatula (Almeida et al., 2000; Sulieman et al., 2013). Similarly, NO3− treatments also significantly increased the production of the N-compound asparagine in the shoot of nodulated soybean plants (Bacanamwo and Harper, 1997), supporting that the inhibition of nodulation by both Pi starvation and NO3− application may be due to an N-feedback regulation (Bacanamwo and Harper, 1997; Almeida et al., 2000; Sulieman et al., 2013).
Earlier studies have suggested that increased permeability of nodules to oxygen may explain the Pi deficiency-induced inhibition of the nodule function in common bean (Hernandez and Drevon, 1991; Lazali and Drevon, 2014). Physiological studies of different legumes have revealed that Pi is preferentially relocated from other organs to nodules under Pi deficiency (Sa and Israel, 1991; Tang et al., 2001). It has been shown that a high phytase activity in nodules constitutes a mechanism for faba bean (Vicia faba L.)-rhizobia symbiosis to adapt symbiotic N fixation to conditions of Pi deficiency (Makoudi et al., 2018). In white lupin (Lupinus albus), Pi deficiency results in enhanced nodulation in cluster root zones and an increased potential for organic acid production in root nodules, contributing to the resilience of white lupin to P-deficiency (Schulze et al., 2006).
A comprehensive transcriptome profiling analysis has shown that NO3− application and Pi starvation both reduce symbiotic N fixation via similar changes in the nodule gene expression in M. truncatula (Liese et al., 2017). Particularly, in both treatments, the activity of nitrogenase was targeted by a reduction in the expression of nodule cysteine-rich (NCR) genes (encoding nodule-specific cysteine-rich peptides), by a disturbance of iron allocation in nodule inner cells, and by a strong reduction in the expression of Lbs, which may restrict the supply of oxygen for respiration (Liese et al., 2017; Figures 2, 3).
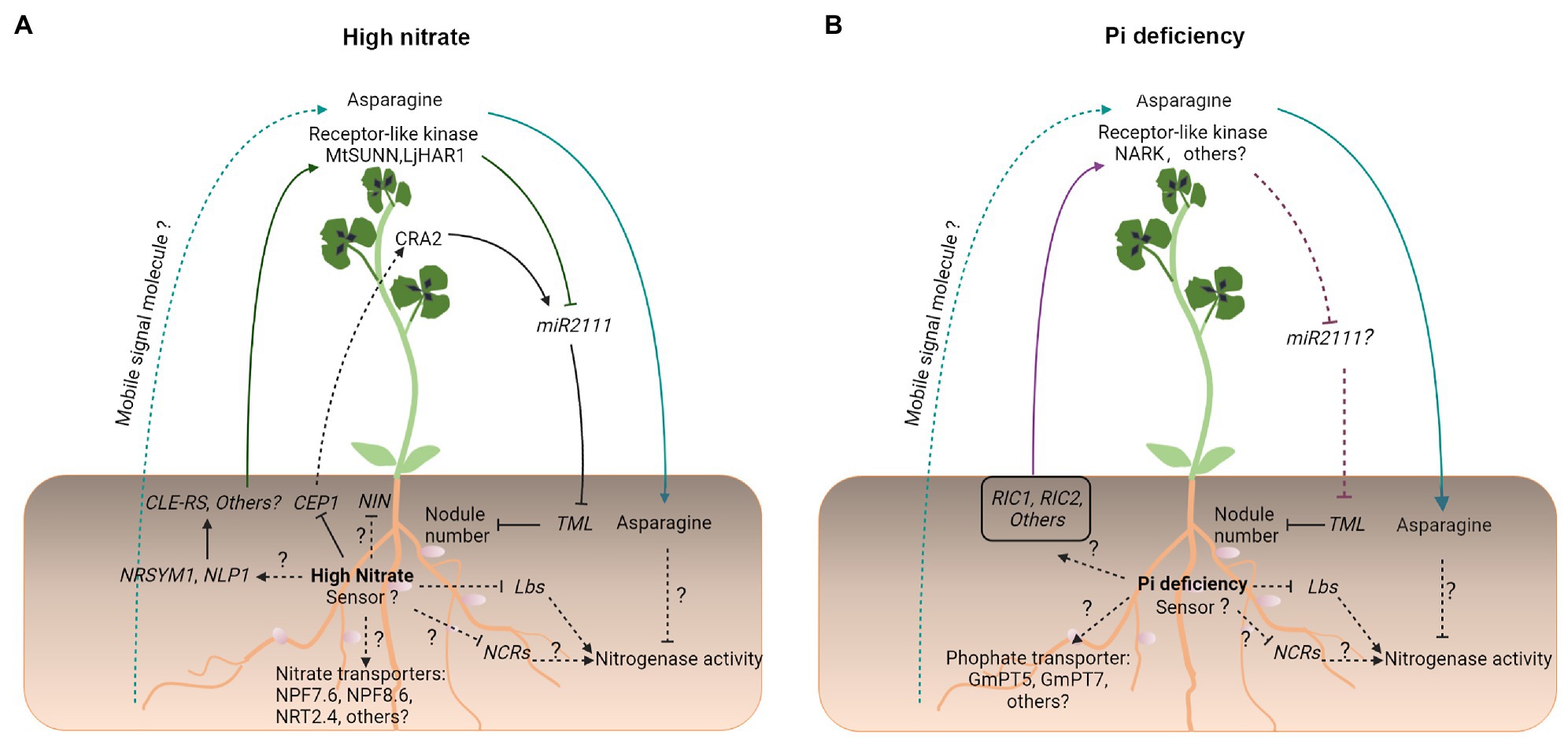
Figure 3. An overview of symbiotic signaling under high NO3− and Pi deficiency in legumes. Under conditions of (A) high NO3− and (B) Pi deficiency, compromised legume–rhizobial symbiotic interactions share both similar signaling processes and unique processes. Similar symbiotic signaling processes include enhanced CLE-like/LRR-RLK-dependent AON signaling, a significant increase in the production and transport of the N-compound asparagine from the shoot to root nodules, where it inhibits nitrogenase activity, and reduced the expression of genes encoding Lbs and NCR peptides, which are essential to maintain the nitrogenase activity. Unique symbiotic signaling processes include transport, initial signal perception, and transduction. Solid and dashed lines indicate the processes that are supported by experimental evidence and putative, respectively. Question marks indicate unknown pathways.
Under Pi starvation, plants upregulate Pi uptake but downregulate NO3− uptake (Wang et al., 2020c). An MYB-related GARP-type transcription factor NITRATE-INDUCIBLE, GARP-TYPE TRANSCRIPTIONAL REPRESSOR1.2 (NIGT1.2) has been identified to bind specific promoter sequences of PHT1;1 and PHT1;4, and also NRT1.1 in A. thaliana (Wang et al., 2020c). NIGT1.2 is induced by Pi-starvation and NO3− treatments (Wang et al., 2020b,c). Overexpression of NIGT1.2 results in an enhanced Pi uptake capacity and Pi levels in transgenic plants. On the contrary, nigt1.1 nigt1.2 double mutants exhibit a hypersensitive phenotype and significantly reduced Pi uptake capacity and levels under conditions of Pi deficiency although nigt1.2 single mutants were similar to wild type plants under the growth conditions of Pi sufficiency and Pi deficiency (Wang et al., 2020c). These results support a positive role of NIGT1.2 (also NIGT1.1) in Pi uptake under Pi-limitation.
Interestingly, Pi-limitation reduces the NO3− uptake and accumulation. The overexpression of NIGT1.2 results in a reduced NO3− influx and accumulation in transgenic plants. On the other hand, the nigt1.1 nigt1.2 double mutant exhibits a significantly increased NO3− influx only under Pi-limitation (Wang et al., 2020c). These results support a negative role of NIGT1.2 (also NIGT1.1) in the uptake of NO3− under Pi-starvation. NIGT1.2 binds AGANNNAAA and AAACNNAACC cis-elements in the promoter sequences of PHT1;1 and PHT1;4 both in vitro and in vivo, supporting a direct, transcriptional upregulation of PHT1;1 and PHT1;4 by NIGT1.2 under Pi-limitation (Medici et al., 2015; Wang et al., 2020c). Intriguingly, NIGT1.2 also binds AANNAGA, TGGGA, and GAGA cis-elements in the NRT1.1 promoter sequence, and downregulates the expression of NRT1.1 under Pi-starvation (Medici et al., 2015; Wang et al., 2020c).
Much progress has been made in our understanding of N and P signaling and transport during legume–rhizobium symbiosis (Figure 3). It is now known that the NLP family of transcription factors plays a key role in the inhibitory effect on legume–rhizobium symbiosis of a high level of NO3−. A high level of NO3− causes the retention and accumulation of NLP proteins in the nucleus, which inhibit the transcriptional activity of NIN in activating the expression of Nod factor-inducible genes such as CRE1 and NF-YA1 (Lin et al., 2018; Nishida et al., 2018; Figure 1). NLPs can interact directly with NIN, bind the same cis-elements as NIN, or both (Lin et al., 2018). Unlike those of NLPs, the N-terminal of NIN is not responsive to NO3− and the function of NIN also diverges from that of NLPs (Suzuki et al., 2013). These changes are thought to play an important role in the evolution of legume–rhizobium symbiosis. On the other hand, a high level of NO3− reduces the level of Nod factor-induced NIN expression (Barbulova et al., 2007). However, the mechanisms remain elusive.
A low level of NO3− promotes nodule development and symbiotic N fixation. However, the regulatory mechanisms remain largely unknown. In plants, there are two families of transporters for NO3− transport, a large NPF/NRT1 family of mostly low-affinity transporters (>80 members in L. japonicus and M. truncatula), and a small NRT2 family of high-affinity NO3− transporters (<5 in L. japonicus and M. truncatula; Criscuolo et al., 2012; Sol et al., 2019). The high-affinity LjNRT2.4 protein expressed in root and nodule vascular tissues has been identified to play a positive role in the regulation of nodule development and function by a low level of NO3−, supporting a NO3−–NO cycle being an alternative energy source for mitochondria and bacteroids and this is particularly important for symbiotic N fixation under a hypoxia condition (Valkov et al., 2020). The involvement in nodule development and function has been demonstrated only for a limited number of NO3− transporters. The presence of a large number of NO3− transporters supports the importance and complexities of these groups of proteins in regulating uptake and transport of NO3− during plant and/or nodule development and function and this needs to be demonstrated in future investigation.
Inorganic Pi signaling and transport have been investigated extensively in model organisms such as A. thaliana and rice. PSR appears to involve both a long-distance, systemic response that senses cellular Pi levels, and a local, root tip inhibition response that senses extracellular Pi availability (Wang et al., 2020d). Sensing of cellular Pi levels relies on the PHR1-PHT1 module (Puga et al., 2017; Lu et al., 2020), which is a multicomponent, transcriptional, and posttranscriptional regulatory network. The local, root tip inhibition response to Pi starvation includes both a rapid reduction of cell elongation at the root transition zone and a gradual inhibition of cell division at the root apical meristem (RAM) and is tightly linked to the accumulation of Fe3+ in the apoplast, production of ROS, and formation of callose at RAM and transition zone, which require LPR1 (multicopper oxidase), PDR2 (P5-type ATPase), ALMT1 (malate transporter), and STOP1 (zinc finger transcription factor; Ticconi et al., 2009; Muller et al., 2015; Balzergue et al., 2017; Naumann et al., 2019). The local response to Pi levels shares some components involved in aluminum toxicity response and DNA damage response, supporting a critical role of a sufficient level of Pi in maintaining the basic function of root tip cells.
Interestingly, there is an antagonistic interaction between Pi and NO3−. Under Pi- starvation, plants tend to upregulate Pi transport but downregulate NO3− transport. A GARP-type transcription factor, NIGT1.2 (also NIGT1.1) appears to directly upregulate the expression PHT1;1 and PHT1;4 genes and downregulate NRT1.1 expression through binding of different cis-elements in their promoter sequences (Wang et al., 2020c). Although much progress has been made in the field of N and P signaling and transport regulation in plant growth and development, only a limited number of transporter-encoding genes have been investigated during legume–rhizobium symbiosis (Figure 3). In addition to N and P, efficient symbiotic N fixation also requires sufficient supplies of other mineral elements, such as potassium, sulfur, calcium, magnesium, iron, and zinc (Garcia et al., 2020). With the development of some genetic resources in different legume species, including L. japonicus, M. truncatula, and Glycine max, and CRISPR/Cas9 gene-editing tools (Wolabu et al., 2020; Zhu et al., 2021), it is foreseeable that a better understanding of signaling, transport, and interactions of these mineral elements during nodule development will make it possible to improve the efficiency of symbiotic N fixation of legume crops to contribute to sustainable agriculture.
RC and YM conceived the project and wrote the manuscript. YM prepared the figures. All authors contributed to the article and approved the submitted version.
This work was supported by the Fundamental Research Funds for the Central Universities of China (grants lzujbky-2019-72 and lzujbky-2020-sp04).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abel, S. (2017). Phosphate scouting by root tips. Curr. Opin. Plant Biol. 39, 168–177. doi: 10.1016/j.pbi.2017.04.016
Almeida, J. P., Hartwig, U. A., Frehner, M., Nösberger, J., and Lüscher, A. (2000). Evidence that P deficiency induces N feedback regulation of symbiotic N2 fixation in white clover (Trifolium repens L.). J. Exp. Bot. 51, 1289–1297.
Astier, J., Gross, I., and Durner, J. (2018). Nitric oxide production in plants: an update. J. Exp. Bot. 69, 3401–3411. doi: 10.1093/jxb/erx420
Bacanamwo, M., and Harper, J. E. (1997). The feedback mechanism of nitrate inhibition of nitrogenase activity in soybean may involve asparagine and/or products of its metabolism. Physiol. Plant. 100, 371–377. doi: 10.1111/j.1399-3054.1997.tb04795.x
Balzergue, C., Dartevelle, T., Godon, C., Laugier, E., Meisrimler, C., Teulon, J. M., et al. (2017). Low phosphate activates STOP1-ALMT1 to rapidly inhibit root cell elongation. Nat. Commun. 8:15300. doi: 10.1038/ncomms15300
Barbulova, A., Rogato, A., D’Apuzzo, E., Omrane, S., and Chiurazzi, M. (2007). Differential effects of combined N sources on early steps of the nod factor-dependent transduction pathway in Lotus japonicus. Mol. Plant-Microbe Interact. 20, 994–1003. doi: 10.1094/MPMI-20-8-0994
Bayle, V., Arrighi, J. F., Creff, A., Nespoulous, C., Vialaret, J., Rossignol, M., et al. (2011). Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell 23, 1523–1535. doi: 10.1105/tpc.110.081067
Berger, A., Boscari, A., Frendo, P., and Brouquisse, R. (2019). Nitric oxide signaling, metabolism and toxicity in nitrogen-fixing symbiosis. J. Exp. Bot. 70, 4505–4520. doi: 10.1093/jxb/erz159
Bethke, P. C., Badger, M. R., and Jones, R. L. (2004). Apoplastic synthesis of nitric oxide by plant tissues. Plant Cell 16, 332–341. doi: 10.1105/tpc.017822
Blanquet, P., Silva, L., Catrice, O., Bruand, C., Carvalho, H., and Meilhoc, E. (2015). Sinorhizobium meliloti controls nitric oxide-mediated post-translational modification of a Medicago truncatula nodule protein. Mol. Plant-Microbe Interact. 28, 1353–1363. doi: 10.1094/MPMI-05-15-0118-R
Boscari, A., Del Giudice, J., Ferrarini, A., Venturini, L., Zaffini, A. L., Delledonne, M., et al. (2013). Expression dynamics of the Medicago truncatula transcriptome during the symbiotic interaction with Sinorhizobium meliloti: which role for nitric oxide? Plant Physiol. 161, 425–439. doi: 10.1104/pp.112.208538
Breuillin-Sessoms, F., Floss, D. S., Gomez, S. K., Pumplin, N., Ding, Y., Levesque-Tremblay, V., et al. (2015). Suppression of Arbuscule degeneration in Medicago truncatula phosphate transporter4 mutants is dependent on the ammonium transporter 2 family protein AMT2;3. Plant Cell 27, 1352–1366. doi: 10.1105/tpc.114.131144
Bustos-Sanmamed, P., Tovar-Mendez, A., Crespi, M., Sato, S., Tabata, S., and Becana, M. (2011). Regulation of nonsymbiotic and truncated hemoglobin genes of Lotus japonicus in plant organs and in response to nitric oxide and hormones. New Phytol. 189, 765–776. doi: 10.1111/j.1469-8137.2010.03527.x
Cam, Y., Pierre, O., Boncompagni, E., Herouart, D., Meilhoc, E., and Bruand, C. (2012). Nitric oxide (NO): a key player in the senescence of Medicago truncatula root nodules. New Phytol. 196, 548–560. doi: 10.1111/j.1469-8137.2012.04282.x
Cao, Y., Liu, J., Li, Y., Zhang, J., Li, S., An, Y., et al. (2020). Functional analysis of the phosphate transporter gene MtPT6 from Medicago truncatula. Front. Plant Sci. 11:620377. doi: 10.3389/fpls.2020.620377
Capoen, W., Sun, J., Wysham, D., Otegui, M. S., Venkateshwaran, M., Hirsch, S., et al. (2011). Nuclear membranes control symbiotic calcium signaling of legumes. Proc. Natl. Acad. Sci. U. S. A. 108, 14348–14353. doi: 10.1073/pnas.1107912108
Castella, C., Mirtziou, I., Seassau, A., Boscari, A., Montrichard, F., Papadopoulou, K., et al. (2017). Post-translational modifications of Medicago truncatula glutathione peroxidase 1 induced by nitric oxide. Nitric Oxide 68, 125–136. doi: 10.1016/j.niox.2017.02.004
Charpentier, M., Sun, J., Vaz Martins, T., Radhakrishnan, G. V., Findlay, K., Soumpourou, E., et al. (2016). Nuclear-localized cyclic nucleotide-gated channels mediate symbiotic calcium oscillations. Science 352, 1102–1105. doi: 10.1126/science.aae0109
Charpentier, M., Vaz Martins, T., Granqvist, E., Oldroyd, G. E., and Morris, R. J. (2013). The role of DMI1 in establishing Ca2+ oscillations in legume symbioses. Plant Signal. Behav. 8:e22894. doi: 10.4161/psb.22894
Chen, J., Liu, Y., Ni, J., Wang, Y., Bai, Y., Shi, J., et al. (2011). OsPHF1 regulates the plasma membrane localization of low- and high-affinity inorganic phosphate transporters and determines inorganic phosphate uptake and translocation in rice. Plant Physiol. 157, 269–278. doi: 10.1104/pp.111.181669
Chen, L., Qin, L., Zhou, L., Li, X., Chen, Z., Sun, L., et al. (2019). A nodule-localized phosphate transporter GmPT7 plays an important role in enhancing symbiotic N2 fixation and yield in soybean. New Phytol. 221, 2013–2025. doi: 10.1111/nph.15541
Clarke, V. C., Loughlin, P. C., Gavrin, A., Chen, C., Brear, E. M., Day, D. A., et al. (2015). Proteomic analysis of the soybean symbiosome identifies new symbiotic proteins. Mol. Cell. Proteomics 14, 1301–1322. doi: 10.1074/mcp.M114.043166
Colebatch, G., Desbrosses, G., Ott, T., Krusell, L., Montanari, O., Kloska, S., et al. (2004). Global changes in transcription orchestrate metabolic differentiation during symbiotic nitrogen fixation in Lotus japonicus. Plant J. 39, 487–512. doi: 10.1111/j.1365-313X.2004.02150.x
Corpas, F. J., Palma, J. M., Del Rio, L. A., and Barroso, J. B. (2009). Evidence supporting the existence of L-arginine-dependent nitric oxide synthase activity in plants. New Phytol. 184, 9–14. doi: 10.1111/j.1469-8137.2009.02989.x
Criscuolo, G., Valkov, V. T., Parlati, A., Alves, L. M., and Chiurazzi, M. (2012). Molecular characterization of the Lotus japonicus NRT1(PTR) and NRT2 families. Plant Cell Environ. 35, 1567–1581. doi: 10.1111/j.1365-3040.2012.02510.x
Cui, C., Wang, H., Hong, L., Xu, Y., Zhao, Y., and Zhou, C. (2019). MtBZR1 plays an important role in nodule development in Medicago truncatula. Int. J. Mol. Sci. 20:2941. doi: 10.3390/ijms20122941
D’Apuzzo, E., Rogato, A., Simon-Rosin, U., El Alaoui, H., Barbulova, A., Betti, M., et al. (2004). Characterization of three functional high-affinity ammonium transporters in Lotus japonicus with differential transcriptional regulation and spatial expression. Plant Physiol. 134, 1763–1774. doi: 10.1104/pp.103.034322
del Giudice, J., Cam, Y., Damiani, I., Fung-Chat, F., Meilhoc, E., Bruand, C., et al. (2011). Nitric oxide is required for an optimal establishment of the Medicago truncatula-Sinorhizobium meliloti symbiosis. New Phytol. 191, 405–417. doi: 10.1111/j.1469-8137.2011.03693.x
Ferrarini, A., De Stefano, M., Baudouin, E., Pucciariello, C., Polverari, A., Puppo, A., et al. (2008). Expression of Medicago truncatula genes responsive to nitric oxide in pathogenic and symbiotic conditions. Mol. Plant-Microbe Interact. 21, 781–790. doi: 10.1094/MPMI-21-6-0781
Garcia, K., Bucking, H., and Zimmermann, S. D. (2020). Editorial: importance of root symbiomes for plant nutrition: new insights, perspectives and future challenges. Front. Plant Sci. 11:594. doi: 10.3389/fpls.2020.00594
Gazzarrini, S., Lejay, L., Gojon, A., Ninnemann, O., Frommer, W. B., and von Wiren, N. (1999). Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into arabidopsis roots. Plant Cell 11, 937–947. doi: 10.1105/tpc.11.5.937
Gonzalez, E., Solano, R., Rubio, V., Leyva, A., and Paz-Ares, J. (2005). PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. Plant Cell 17, 3500–3512. doi: 10.1105/tpc.105.036640
Gu, M., Chen, A., Sun, S., and Xu, G. (2015). Complex regulation of plant phosphate transporters and the gap between molecular mechanisms and practical application: what is missing? Mol. Plant 9, 396–416. doi: 10.1016/j.molp.2015.12.012
Gunawardeba, S. F. B. N., Danso, S. K. A., and Zapata, F. (1992). Phosphorus requirements and nitrogen accumulation by three mungbean (Vigna radiata (L) Welzek) cultivars. Plant Soil 147, 267–274. doi: 10.1007/BF00029078
Hernandez, G., and Drevon, J.-J. (1991). Influence of oxygen and acetylene during in situ open-flow assays of nitrogenase activity (C2H2 reduction) in Phaseolus vulgaris root nodules. J. Plant Physiol. 138, 587–590. doi: 10.1016/S0176-1617(11)80246-4
Herold, S., and Puppo, A. (2005). Oxyleghemoglobin scavenges nitrogen monoxide and peroxynitrite: a possible role in functioning nodules? J. Biol. Inorg. Chem. 10, 935–945. doi: 10.1007/s00775-005-0046-9
Hill, R., Hargrove, M., and Arredondo-Peter, R. (2016). Phytoglobin: a novel nomenclature for plant globins accepted by the globin community at the 2014 XVIII conference on oxygen-binding and sensing proteins. F1000Res 5:212. doi: 10.12688/f1000research.8133.1
Holford, I. C. R. (1997). Soil phosphorus: its measurement, and its uptake by plants. Aust. J. Soil Res. 35:227. doi: 10.1071/S96047
Horchani, F., Prevot, M., Boscari, A., Evangelisti, E., Meilhoc, E., Bruand, C., et al. (2011). Both plant and bacterial nitrate reductases contribute to nitric oxide production in Medicago truncatula nitrogen-fixing nodules. Plant Physiol. 155, 1023–1036. doi: 10.1104/pp.110.166140
Igamberdiev, A. U., and Hill, R. D. (2004). Nitrate, NO and haemoglobin in plant adaptation to hypoxia: an alternative to classic fermentation pathways. J. Exp. Bot. 55, 2473–2482. doi: 10.1093/jxb/erh272
Indrasumunar, A., Kereszt, A., Searle, I., Miyagi, M., Li, D., Nguyen, C. D. T., et al. (2010). Inactivation of duplicated nod factor receptor 5 (NFR5) genes in recessive loss-of-function non-nodulation mutants of allotetraploid soybean (Glycine max L. Merr.). Plant Cell Physiol. 51, 201–214. doi: 10.1093/pcp/pcp178
Isidra-Arellano, M. C., Pozas-Rodriguez, E. A., Del Rocio Reyero-Saavedra, M., Arroyo-Canales, J., Ferrer-Orgaz, S., Del Socorro Sanchez-Correa, M., et al. (2020). Inhibition of legume nodulation by pi deficiency is dependent on the autoregulation of nodulation (AON) pathway. Plant J. 103, 1125–1139. doi: 10.1111/tpj.14789
Isidra-Arellano, M. C., Reyero-Saavedra, M. D. R., Sanchez-Correa, M. D. S., Pingault, L., Sen, S., Joshi, T., et al. (2018). Phosphate deficiency negatively affects early steps of the symbiosis between common bean and rhizobia. Gene 9:498. doi: 10.3390/genes9100498
Jin, Y., Liu, H., Luo, D., Yu, N., Dong, W., Wang, C., et al. (2016). DELLA proteins are common components of symbiotic rhizobial and mycorrhizal signalling pathways. Nat. Commun. 7:12433. doi: 10.1038/ncomms12433
Kato, K., Kanahama, K., and Kanayama, Y. (2010). Involvement of nitric oxide in the inhibition of nitrogenase activity by nitrate in Lotus root nodules. J. Plant Physiol. 167, 238–241. doi: 10.1016/j.jplph.2009.08.006
Kobayashi, R., Yamaguchi, S., and Iwasa, Y. (2020). Optimal control of root nodulation—prediction of life history theory of a mutualistic system. J. Theor. Biol. 510:110544. doi: 10.1016/j.jtbi.2020.110544
Konishi, M., and Yanagisawa, S. (2013). Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat. Commun. 4:1617. doi: 10.1038/ncomms2621
Laloum, T., Baudin, M., Frances, L., Lepage, A., Billault-Penneteau, B., Cerri, M. R., et al. (2014). Two CCAAT-box-binding transcription factors redundantly regulate early steps of the legume-rhizobia endosymbiosis. Plant J. 79, 757–768. doi: 10.1111/tpj.12587
Lazali, M., and Drevon, J. J. (2014). The nodule conductance to O2 diffusion increases with phytase activity in N2-fixing Phaseolus vulgaris L. Plant Physiol. Biochem. 80, 53–59. doi: 10.1016/j.plaphy.2014.03.023
Leitao, N., Dangeville, P., Carter, R., and Charpentier, M. (2019). Nuclear calcium signatures are associated with root development. Nat. Commun. 10:4865. doi: 10.1038/s41467-019-12845-8
Leon, J., and Costa-Broseta, A. (2020). Present knowledge and controversies, deficiencies, and misconceptions on nitric oxide synthesis, sensing, and signaling in plants. Plant Cell Environ. 43, 1–15. doi: 10.1111/pce.13617
Li, C., Li, C., Zhang, H., Liao, H., and Wang, X. (2017). The purple acid phosphatase GmPAP21 enhances internal phosphorus utilization and possibly plays a role in symbiosis with rhizobia in soybean. Physiol. Plant. 159, 215–227. doi: 10.1111/ppl.12524
Liese, R., Schulze, J., and Cabeza, R. A. (2017). Nitrate application or P deficiency induce a decline in Medicago truncatula N2-fixation by similar changes in the nodule transcriptome. Sci. Rep. 7:46264. doi: 10.1038/srep46264
Limpens, E., Franken, C., Smit, P., Willemse, J., Bisseling, T., and Geurts, R. (2003). LysM domain receptor kinases regulating rhizobial nod factor-induced infection. Science 302, 630–633. doi: 10.1126/science.1090074
Limpens, E., Mirabella, R., Fedorova, E., Franken, C., Franssen, H., Bisseling, T., et al. (2005). Formation of organelle-like N2-fixing symbiosomes in legume root nodules is controlled by DMI2. Proc. Natl. Acad. Sci. U. S. A. 102, 10375–10380. doi: 10.1073/pnas.0504284102
Lin, J. S., Li, X., Luo, Z., Mysore, K. S., Wen, J., and Xie, F. (2018). NIN interacts with NLPs to mediate nitrate inhibition of nodulation in Medicago truncatula. Nat. Plants 4, 942–952. doi: 10.1038/s41477-018-0261-3
Liu, K.-H., Niu, Y., Konishi, M., Wu, Y., Du, H., Sun Chung, H., et al. (2017). Discovery of nitrate–CPK–NLP signalling in central nutrient–growth networks. Nature 545, 311–316. doi: 10.1038/nature22077
Lu, M., Cheng, Z., Zhang, X. M., Huang, P., Fan, C., Yu, G., et al. (2020). Spatial divergence of PHR-PHT1 modules maintains phosphorus homeostasis in Soybean nodules. Plant Physiol. 184, 236–250. doi: 10.1104/pp.19.01209
Luo, Z., Lin, J.-S., Zhu, Y., Fu, M., Li, X., and Xie, F. (2021). NLP1 reciprocally regulates nitrate inhibition of nodulation through SUNN-CRA2 signaling in Medicago truncatula. Plant Commun. 2:100183. doi: 10.1016/j.xplc.2021.100183
Madsen, E. B., Madsen, L. H., Radutoiu, S., Olbryt, M., Rakwalska, M., Szczyglowski, K., et al. (2003). A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425, 637–640. doi: 10.1038/nature02045
Magori, S., and Kawaguchi, M. (2009). Long-distance control of nodulation: molecules and models. Mol. Cell 27, 129–134. doi: 10.1007/s10059-009-0016-0
Makoudi, B., Kabbadj, A., Mouradi, M., Amenc, L., Domergue, O., Blair, M., et al. (2018). Phosphorus deficiency increases nodule phytase activity of faba bean–rhizobia symbiosis. Acta Physiol. Plant. 40:63. doi: 10.1007/s11738-018-2619-6
Marchive, C., Roudier, F., Castaings, L., Brehaut, V., Blondet, E., Colot, V., et al. (2013). Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat. Commun. 4:1713. doi: 10.1038/ncomms2650
Marsh, J. F., Rakocevic, A., Mitra, R. M., Brocard, L., Sun, J., Eschstruth, A., et al. (2007). Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 144, 324–335. doi: 10.1104/pp.106.093021
Medici, A., Marshall-Colon, A., Ronzier, E., Szponarski, W., Wang, R., Gojon, A., et al. (2015). AtNIGT1/HRS1 integrates nitrate and phosphate signals at the Arabidopsis root tip. Nat. Commun. 6:6274. doi: 10.1038/ncomms7274
Meilhoc, E., Boscari, A., Bruand, C., Puppo, A., and Brouquisse, R. (2011). Nitric oxide in legume–rhizobium symbiosis. Plant Sci. 181, 573–581. doi: 10.1016/j.plantsci.2011.04.007
Meilhoc, E., Cam, Y., Skapski, A., and Bruand, C. (2010). The response to nitric oxide of the nitrogen-fixing symbiont Sinorhizobium meliloti. Mol. Plant-Microbe Interact. 23, 748–759. doi: 10.1094/MPMI-23-6-0748
Melo, P. M., Silva, L. S., Ribeiro, I., Seabra, A. R., and Carvalho, H. G. (2011). Glutamine synthetase is a molecular target of nitric oxide in root nodules of Medicago truncatula and is regulated by tyrosine nitration. Plant Physiol. 157, 1505–1517. doi: 10.1104/pp.111.186056
Messinese, E., Mun, J. H., Yeun, L. H., Jayaraman, D., Rouge, P., Barre, A., et al. (2007). A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula. Mol. Plant-Microbe Interact. 20, 912–921. doi: 10.1094/MPMI-20-8-0912
Michalski, W. P., and Nicholas, D. J. D. (1987). Inhibition of nitrogenase by nitrite and nitric oxide in Rhodopseudomonas sphaeroides f. sp. denitrificans. Arch. Microbiol. 147, 304–308. doi: 10.1007/BF00463493
Morere-Le Paven, M. C., Viau, L., Hamon, A., Vandecasteele, C., Pellizzaro, A., Bourdin, C., et al. (2011). Characterization of a dual-affinity nitrate transporter MtNRT1.3 in the model legume Medicago truncatula. J. Exp. Bot. 62, 5595–5605. doi: 10.1093/jxb/err243
Mortier, V., Holsters, M., and Goormachtig, S. (2012). Never too many? How legumes control nodule numbers. Plant Cell Environ. 35, 245–258. doi: 10.1111/j.1365-3040.2011.02406.x
Mu, X., and Luo, J. (2019). Evolutionary analyses of NIN-like proteins in plants and their roles in nitrate signaling. Cell. Mol. Life Sci. 76, 3753–3764. doi: 10.1007/s00018-019-03164-8
Muller, J., Toev, T., Heisters, M., Teller, J., Moore, K. L., Hause, G., et al. (2015). Iron-dependent callose deposition adjusts root meristem maintenance to phosphate availability. Dev. Cell 33, 216–230. doi: 10.1016/j.devcel.2015.02.007
Nagata, M., Murakami, E., Shimoda, Y., Shimoda-Sasakura, F., Kucho, K., Suzuki, A., et al. (2008). Expression of a class 1 hemoglobin gene and production of nitric oxide in response to symbiotic and pathogenic bacteria in Lotus japonicus. Mol. Plant-Microbe Interact. 21, 1175–1183. doi: 10.1094/MPMI-21-9-1175
Naumann, C., Muller, J., Sakhonwasee, S., Wieghaus, A., Hause, G., Heisters, M., et al. (2019). The local phosphate deficiency response activates endoplasmic reticulum stress-dependent autophagy. Plant Physiol. 179, 460–476. doi: 10.1104/pp.18.01379
Nishida, H., and Suzaki, T. (2018). Nitrate-mediated control of root nodule symbiosis. Curr. Opin. Plant Biol. 44, 129–136. doi: 10.1016/j.pbi.2018.04.006
Nishida, H., Tanaka, S., Handa, Y., Ito, M., Sakamoto, Y., Matsunaga, S., et al. (2018). A NIN-LIKE PROTEIN mediates nitrate-induced control of root nodule symbiosis in Lotus japonicus. Nat. Commun. 9:499. doi: 10.1038/s41467-018-02831-x
Nussaume, L., Kanno, S., Javot, H., Marin, E., Pochon, N., Ayadi, A., et al. (2011). Phosphate import in plants: focus on the PHT1 transporters. Front. Plant Sci. 2:83. doi: 10.3389/fpls.2011.00083
Oldroyd, G. E. (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11, 252–263. doi: 10.1038/nrmicro2990
Oldroyd, G. E., and Downie, J. A. (2008). Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59, 519–546. doi: 10.1146/annurev.arplant.59.032607.092839
Oldroyd, G. E., Murray, J. D., Poole, P. S., and Downie, J. A. (2011). The rules of engagement in the legume-rhizobial symbiosis. Annu. Rev. Genet. 45, 119–144. doi: 10.1146/annurev-genet-110410-132549
Planchet, E., Jagadis Gupta, K., Sonoda, M., and Kaiser, W. M. (2005). Nitric oxide emission from tobacco leaves and cell suspensions: rate limiting factors and evidence for the involvement of mitochondrial electron transport. Plant J. 41, 732–743. doi: 10.1111/j.1365-313X.2005.02335.x
Puga, M. I., Mateos, I., Charukesi, R., Wang, Z., Franco-Zorrilla, J. M., de Lorenzo, L., et al. (2014). SPX1 is a phosphate-dependent inhibitor of phosphate starvation response 1 in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 111, 14947–14952. doi: 10.1073/pnas.1404654111
Puga, M. I., Rojas-Triana, M., de Lorenzo, L., Leyva, A., Rubio, V., and Paz-Ares, J. (2017). Novel signals in the regulation of pi starvation responses in plants: facts and promises. Curr. Opin. Plant Biol. 39, 40–49. doi: 10.1016/j.pbi.2017.05.007
Puppo, A., Pauly, N., Boscari, A., Mandon, K., and Brouquisse, R. (2013). Hydrogen peroxide and nitric oxide: key regulators of the legume-rhizobium and mycorrhizal symbioses. Antioxid. Redox Signal. 18, 2202–2219. doi: 10.1089/ars.2012.5136
Qin, L., Zhao, J., Tian, J., Chen, L., Sun, Z., Guo, Y., et al. (2012). The high-affinity phosphate transporter GmPT5 regulates phosphate transport to nodules and nodulation in soybean. Plant Physiol. 159, 1634–1643. doi: 10.1104/pp.112.199786
Radutoiu, S., Madsen, L. H., Madsen, E. B., Felle, H. H., Umehara, Y., Gronlund, M., et al. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425, 585–592. doi: 10.1038/nature02039
Richardson, A. E. (2009). Regulating the phosphorus nutrition of plants: molecular biology meeting agronomic needs. Plant Soil 322, 17–24. doi: 10.1007/s11104-009-0071-5
Ried, M. K., Antolin-Llovera, M., and Parniske, M. (2014). Spontaneous symbiotic reprogramming of plant roots triggered by receptor-like kinases. eLife 3:e03891. doi: 10.7554/eLife.03891
Ried, M. K., Wild, R., Zhu, J., Pipercevic, J., Sturm, K., Broger, L., et al. (2021). Inositol pyrophosphates promote the interaction of SPX domains with the coiled-coil motif of PHR transcription factors to regulate plant phosphate homeostasis. Nat. Commun. 12:384. doi: 10.1038/s41467-020-20681-4
Rogato, A., D’Apuzzo, E., Barbulova, A., Omrane, S., Parlati, A., Carfagna, S., et al. (2010). Characterization of a developmental root response caused by external ammonium supply in Lotus japonicus. Plant Physiol. 154, 784–795. doi: 10.1104/pp.110.160309
Rogato, A., D’Apuzzo, E., Barbulova, A., Omrane, S., Stedel, C., Simon-Rosin, U., et al. (2008). Tissue-specific down-regulation of LjAMT1;1 compromises nodule function and enhances nodulation in Lotus japonicus. Plant Mol. Biol. 68, 585–595. doi: 10.1007/s11103-008-9394-5
Rubio, V., Linhares, F., Solano, R., Martin, A. C., Iglesias, J., Leyva, A., et al. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15, 2122–2133. doi: 10.1101/gad.204401
Rumer, S., Gupta, K. J., and Kaiser, W. M. (2009). Plant cells oxidize hydroxylamines to NO. J. Exp. Bot. 60, 2065–2072. doi: 10.1093/jxb/erp077
Sa, T. M., and Israel, D. W. (1991). Energy status and functioning of phosphorus-deficient soybean nodules. Plant Physiol. 97, 928–935. doi: 10.1104/pp.97.3.928
Schauser, L., Roussis, A., Stiller, J., and Stougaard, J. (1999). A plant regulator controlling development of symbiotic root nodules. Nature 402, 191–195. doi: 10.1038/46058
Schulze, J., Adgo, E., and Merbach, W. (1999). Carbon costs associated with N2 fixation in Vicia faba L and Pisum sativum 1. Over a 14-day period. Plant Biol. 1, 625–631. doi: 10.1111/j.1438-8677.1999.tb00273.x
Schulze, J., Temple, G., Temple, S. J., Beschow, H., and Vance, C. P. (2006). Nitrogen fixation by white lupin under phosphorus deficiency. Ann. Bot. 98, 731–740. doi: 10.1093/aob/mcl154
Signorelli, S., Sainz, M., Tabares-da Rosa, S., and Monza, J. (2020). The role of nitric oxide in nitrogen fixation by legumes. Front. Plant Sci. 11:521. doi: 10.3389/fpls.2020.00521
Sol, S., Valkov, V. T., Rogato, A., Noguero, M., Gargiulo, L., Mele, G., et al. (2019). Disruption of the Lotus japonicus transporter LjNPF2.9 increases shoot biomass and nitrate content without affecting symbiotic performances. BMC Plant Biol. 19:380. doi: 10.1186/s12870-019-1978-5
Soyano, T., Shimoda, Y., and Hayashi, M. (2015). NODULE INCEPTION antagonistically regulates gene expression with nitrate in Lotus japonicus. Plant Cell Physiol. 56, 368–376. doi: 10.1093/pcp/pcu168
Stohr, C., Strube, F., Marx, G., Ullrich, W. R., and Rockel, P. (2001). A plasma membrane-bound enzyme of tobacco roots catalyses the formation of nitric oxide from nitrite. Planta 212, 835–841. doi: 10.1007/s004250000447
Stracke, S., Kistner, C., Yoshida, S., Mulder, L., Sato, S., Kaneko, T., et al. (2002). A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417, 959–962. doi: 10.1038/nature00841
Streeter, J., and Wong, P. P. (1988). Inhibition of legume nodule formation and N2 fixation by nitrate. Crit. Rev. Plant Sci. 7, 1–23. doi: 10.1080/07352688809382257
Sulieman, S., Schulze, J., and Tran, L. S. (2013). Comparative analysis of the symbiotic efficiency of Medicago truncatula and Medicago sativa under phosphorus deficiency. Int. J. Mol. Sci. 14, 5198–5213. doi: 10.3390/ijms14035198
Sulieman, S., and Tran, L. S. (2015). Phosphorus homeostasis in legume nodules as an adaptive strategy to phosphorus deficiency. Plant Sci. 239, 36–43. doi: 10.1016/j.plantsci.2015.06.018
Suzuki, W., Konishi, M., and Yanagisawa, S. (2013). The evolutionary events necessary for the emergence of symbiotic nitrogen fixation in legumes may involve a loss of nitrate responsiveness of the NIN transcription factor. Plant Signal. Behav. 8:e25975. doi: 10.4161/jrn.25975
Takanashi, K., Takahashi, H., Sakurai, N., Sugiyama, A., Suzuki, H., Shibata, D., et al. (2012). Tissue-specific transcriptome analysis in nodules of Lotus japonicus. Mol. Plant-Microbe Interact. 25, 869–876. doi: 10.1094/MPMI-01-12-0011-R
Tang, C., Hinsinger, P., Drevon, J. J., and Jaillard, B. (2001). Phosphorus deficiency impairs early nodule functioning and enhances proton release in roots of Medicago truncatula L. Ann. Bot. 88, 131–138. doi: 10.1006/anbo.2001.1440
Ticconi, C. A., Lucero, R. D., Sakhonwasee, S., Adamson, A. W., Creff, A., Nussaume, L., et al. (2009). ER-resident proteins PDR2 and LPR1 mediate the developmental response of root meristems to phosphate availability. Proc. Natl. Acad. Sci. U. S. A. 106, 14174–14179. doi: 10.1073/pnas.0901778106
Trinchant, J. C., and Rigaud, J. (1982). Nitrite and nitric oxide as inhibitors of nitrogenase from soybean bacteroids. Appl. Environ. Microbiol. 44, 1385–1388. doi: 10.1128/AEM.44.6.1385-1388.1982
Tun, N. N., Santa-Catarina, C., Begum, T., Silveira, V., Handro, W., Floh, E. I., et al. (2006). Polyamines induce rapid biosynthesis of nitric oxide (NO) in Arabidopsis thaliana seedlings. Plant Cell Physiol. 47, 346–354. doi: 10.1093/pcp/pci252
Valkov, V. T., Sol, S., Rogato, A., and Chiurazzi, M. (2020). The functional characterization of LjNRT2.4 indicates a novel, positive role of nitrate for an efficient nodule N2-fixation activity. New Phytol. 228, 682–696. doi: 10.1111/nph.16728
Vance, C. P., Uhde-Stone, C., and Allan, D. L. (2003). Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol. 157, 423–447. doi: 10.1046/j.1469-8137.2003.00695.x
Venkateshwaran, M., Cosme, A., Han, L., Banba, M., Satyshur, K. A., Schleiff, E., et al. (2012). The recent evolution of a symbiotic ion channel in the legume family altered ion conductance and improved functionality in calcium signaling. Plant Cell 24, 2528–2545. doi: 10.1105/tpc.112.098475
Vernie, T., Kim, J., Frances, L., Ding, Y., Sun, J., Guan, D., et al. (2015). The NIN transcription factor coordinates diverse nodulation programs in different tissues of the Medicago truncatula root. Plant Cell 27, 3410–3424. doi: 10.1105/tpc.15.00461
Wang, Y., Chen, Y. F., and Wu, W. H. (2020d). Potassium and phosphorus transport and signaling in plants. J. Integr. Plant Biol. 63, 34–52. doi: 10.1111/jipb.13053
Wang, Y. Y., Cheng, Y. H., Chen, K. E., and Tsay, Y. F. (2018). Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 69, 85–122. doi: 10.1146/annurev-arplant-042817-040056
Wang, W., Hu, B., Li, A., and Chu, C. (2020b). NRT1.1s in plants: functions beyond nitrate transport. J. Exp. Bot. 71, 4373–4379. doi: 10.1093/jxb/erz554
Wang, Q., Huang, Y., Ren, Z., Zhang, X., Ren, J., Su, J., et al. (2020a). Transfer cells mediate nitrate uptake to control root nodule symbiosis. Nat. Plants 6, 800–808. doi: 10.1038/s41477-020-0683-6
Wang, Z., Ruan, W., Shi, J., Zhang, L., Xiang, D., Yang, C., et al. (2014). Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc. Natl. Acad. Sci. U. S. A. 111, 14953–14958. doi: 10.1073/pnas.1404680111
Wang, X., Wang, H. F., Chen, Y., Sun, M. M., Wang, Y., and Chen, Y. F. (2020c). The transcription factor NIGT1.2 modulates both phosphate uptake and nitrate influx during phosphate starvation in Arabidopsis and maize. Plant Cell 32, 3519–3534. doi: 10.1105/tpc.20.00361
Wang, Y., Yang, Z., Kong, Y., Li, X., Li, W., Du, H., et al. (2020e). GmPAP12 is required for nodule development and nitrogen fixation under phosphorus starvation in soybean. Front. Plant Sci. 11:450. doi: 10.3389/fpls.2020.00450
Wild, R., Gerasimaite, R., Jung, J. Y., Truffault, V., Pavlovic, I., Schmidt, A., et al. (2016). Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 352, 986–990. doi: 10.1126/science.aad9858
Wolabu, T. W., Park, J. J., Chen, M., Cong, L., Ge, Y., Jiang, Q., et al. (2020). Improving the genome editing efficiency of CRISPR/Cas9 in Arabidopsis and Medicago truncatula. Planta 252:15. doi: 10.1007/s00425-020-03415-0
Wong, J., Nadzieja, M., Madsen, L. H., Bucherl, C. A., Dam, S., Sandal, N. N., et al. (2019). A Lotus japonicus cytoplasmic kinase connects nod factor perception by the NFR5 LysM receptor to nodulation. Proc. Natl. Acad. Sci. U. S. A. 116, 14339–14348. doi: 10.1073/pnas.1815425116
Wu, Z., Liu, H., Huang, W., Yi, L., Qin, E., Yang, T., et al. (2020). Genome-wide identification, characterization, and regulation of RWP-RK gene family in the nitrogen-fixing clade. Plan. Theory 9:1178. doi: 10.3390/plants9091178
Yano, K., Yoshida, S., Muller, J., Singh, S., Banba, M., Vickers, K., et al. (2008). CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl. Acad. Sci. U. S. A. 105, 20540–20545. doi: 10.1073/pnas.0806858105
Keywords: legume–rhizobium symbiosis, nitrogen fixation, nitrogen and phosphorus signaling, transport, crosstalk
Citation: Ma Y and Chen R (2021) Nitrogen and Phosphorus Signaling and Transport During Legume–Rhizobium Symbiosis. Front. Plant Sci. 12:683601. doi: 10.3389/fpls.2021.683601
Received: 21 March 2021; Accepted: 25 May 2021;
Published: 22 June 2021.
Edited by:
Vladimir Totev Valkov, Italian National Research Council, ItalyReviewed by:
Ricardo A. Cabeza, University of Talca, ChileCopyright © 2021 Ma and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rujin Chen, cmpjaGVuQGx6dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.