- 1Department of Biochemistry, Molecular Biology, Entomology and Plant Pathology, Mississippi State University, Starkville, MS, United States
- 2State Key Laboratory of Hybrid Rice, Key Laboratory for Research and Utilization of Heterosis in Indica Rice, Ministry of Agriculture, College of Life Sciences, Wuhan University, Wuhan, China
- 3Key Laboratory of Crop Physiology, Ecology and Genetic Breeding, Ministry of Education of the People’s Republic of China, Jiangxi Agricultural University, Nanchang, China
- 4Key Laboratory of Agriculture Responding to Climate Change, Jiangxi Agricultural University, Nanchang, China
- 5Key Laboratory of Magnetic Resonance in Biological Systems, State Key Laboratory of Magnetic Resonance and Atomic and Molecular Physics, National Centre for Magnetic Resonance in Wuhan, Wuhan Institute of Physics and Mathematics, Innovation Academy of Precision Measurement Science and Technology, Chinese Academy of Sciences, Wuhan, China
- 6Youth League Committee, Jiangxi Agricultural University, Nanchang, China
UDP glucose pyrophosphorylase (UDPGP) family genes have been reported to play essential roles in cell death or individual survival. However, a systematic analysis on UDPGP gene family has not been performed yet. In this study, a total of 454 UDPGP proteins from 76 different species were analyzed. The analyses of the phylogenetic tree and orthogroups divided UDPGPs into three clades, including UDP-N-acetylglucosamine pyrophosphorylase (UAP), UDP-glucose pyrophosphorylase (UGP, containing UGP-A and UGP-B), and UDP-sugar pyrophosphorylase (USP). The evolutionary history of the UDPGPs indicated that the members of UAP, USP, and UGP-B were relatively conserved while varied in UGP-A. Homologous sequences of UGP-B and USP were found only in plants. The expression profile of UDPGP genes in Oryza sativa was mainly motivated under jasmonic acid (JA), abscisic acid (ABA), cadmium, and cold treatments, indicating that UDPGPs may play an important role in plant development and environment endurance. The key amino acids regulating the activity of UDPGPs were analyzed, and almost all of them were located in the NB-loop, SB-loop, or conserved motifs. Analysis of the natural variants of UDPGPs in rice revealed that only a few missense mutants existed in coding sequences (CDSs), and most of the resulting variations were located in the non-motif sites, indicating the conserved structure and function of UDPGPs in the evolution. Furthermore, alternative splicing may play a key role in regulating the activity of UDPGPs. The spatial structure prediction, enzymatic analysis, and transgenic verification of UAP isoforms illustrated that the loss of N- and C-terminal sequences did not affect the overall 3D structures, but the N- and C-terminal sequences are important for UAP genes to maintain their enzymatic activity. These results revealed a conserved UDPGP gene family and provided valuable information for further deep functional investigation of the UDPGP gene family in plants.
Introduction
UDP glucose pyrophosphorylase (UDPGP) is a big gene family with three groups, UDP-N-acetylglucosamine pyrophosphorylase (UAP), UDP-glucose pyrophosphorylase (UGP), and UDP-sugar pyrophosphorylase (USP). UAP prefers N-acetylglucosamine-1-P (GlcNAc-1-P) and N-acetylgalactosamine-1-P (GalNAc-1-P) as substrates (Yang et al., 2010; Decker and Kleczkowski, 2019). UGP is reported to be specific for uridine triphosphate (UTP) and glucose 1-phosphate (Glc-1-P) as substrates (Kleczkowski et al., 2010; Decker et al., 2012). For USP, previous studies reported that it had a broader substrate specificity, including galactose-1-phosphate (Gal-1-P), α-glucuronic acid 1-phosphate (GlcA-1-P), and glucose 1-phosphate (Glc-1-P) (Gronwald et al., 2008; Decker and Kleczkowski, 2017). The UDPGPs have been reported related to plant development such as programmed cell death in Arabidopsis thaliana (Chivasa et al., 2013), survival in insects (Arakane et al., 2011), and microorganisms (Yi and Huh, 2015), as well as cancers in humans (Itkonen et al., 2015; Wang et al., 2016).
Some studies on UAP were reported to illustrate its function in microorganisms, animals, and plants. UAP was reported in both prokaryotes and eukaryotes, while no homologous sequences were identified between them (Palaka et al., 2019). In fungi, a singular UDP-GlcNAc pyrophosphorylase gene was reported in yeast (Saccharomyces cerevisiae), and loss-of-function mutant (uap1Δ) exhibited aberrant morphology including swelled or lysed (Mio et al., 1998). In Moniliophthora perniciosa, the inhibition of this enzyme leads to cell death (Junior et al., 2013). In insects, the UAP enzyme plays a key role in chitin synthesis (Zhu K.Y. et al., 2016), protein glycosylation (Schimmelpfeng et al., 2006), growth, and development (Zhu K.Y. et al., 2016). Some insects have two members of UAP, and they usually account for different functions. For example, LdUAP1 regulated the chitin content, while LdUAP2 managed the development in Leptinotarsa decemlineata (Shi et al., 2016). Besides, in the Locusta migratoria, the LmUAP1 inhibited by RNAi resulted in mortality, while the LmUAP2 did not (Liu et al., 2013). In Drosophila, the gene mummy encodes a UDP-N-acetylglucosamine-dipohosphorylase, and the gene mutants exhibited central nervous system fasciculation, dorsal closure, and eye development defects (Schimmelpfeng et al., 2006). Besides, the mummy gene also acted as a BMP signaling antagonist (Humphreys et al., 2013). In human, the expression level of UAP1 is positively correlated with the androgen receptor, which is a main driver of prostate cancer. Inhibition of UAP1 can specifically sensitize prostate cancer cells to the inhibitors of N-linked glycosylation (Itkonen et al., 2015). Few UAP studies were reported in plants. In rice, functional inactivation of UAP1 was reported to be related to early leaf senescence, defense responses (Wang et al., 2015), and programmed cell death (Xiao et al., 2018).
UGP is a key enzyme in the metabolism of UDP-glucose, which plays an important role in cellulose, callose (Park et al., 2010), sucrose, and polysaccharide synthesis (Meng et al., 2009b). At first, only two homologous genes (ATUGP1 and ATUGP2) were identified belonging to UGP clades in A. thaliana with ATUGP1 predominantly expressed in most tissues (Meng et al., 2008; Meng et al., 2009b). Single mutants (atugp1 or atugp2) did not show any deficiency, while an atugp1/atugp2 double mutant exhibited extreme deficiency in plant growth and male sterility. Further experiments showed that the destruction of the callose wall around microspores at the tetrad stage gives rise to abnormal development of pollen, resulting in male sterility in the double mutant (Park et al., 2010). In another study, transfer-DNA gene-knockout plants proved that UDP-glucose pyrophosphorylase 1 (ATUGP1) regulates fumonisin B1-induced programmed cell death (Chivasa et al., 2013). Then, a novel UDP-glucose pyrophosphorylase 3 (AtUGP3) was reported in A. thaliana, which played a key role in sulfolipid biosynthesis (Okazaki et al., 2009). Similarly, two UDP-glucose pyrophosphorylase genes were identified in rice, UGP1 on chromosome 9 and UGP2 on chromosome 2 in early research (Chen et al., 2007). Both UGP1 and UGP2 were expressed ubiquitously in rice, and the expression level of UGP1 was much higher than that of UGP2. UGP1 is vital for callose deposition during the stage of pollen mother cells, and when UGP1 was silenced by RNA interference, the mutant plants exhibited both male sterility and chalky endosperm phenotypic characteristics (Chen et al., 2007). In other plants, such as potato and tobacco, the functions of UGP genes were also reported. Two UGPs (UGP3 and UGP5) were reported in potatoes (Katsube et al., 1990; Spychalla et al., 1994), and the studies demonstrated that the UGP was associated with cold-sweetening (Sowokinos et al., 2004; Gupta et al., 2008). In tobacco, plant height was significantly increased compared to control lines through overexpressing the UGP gene (Coleman et al., 2006). In Phaeodactylum tricornutum, UGP was reported to be associated with chrysolaminaran content, lipid biosynthesis, and carbon allocation (Zhu B.H. et al., 2016). In fungi, UGP was proven to be associated with oxidative stress response and long-term survival (Yi and Huh, 2015). In Dictyostelium, the UDP-glucose derivative plays a key role in autophagic cell death (Tresse et al., 2008). In humans, the loss of function of UGP2 caused a genetic disease (Roeben et al., 2006), and the UGP2 expression was correlated with clinicopathological and biological behaviors, which could be used as a biomarker for progression and poor prognosis of gallbladder cancer (Wang et al., 2016). There are a few studies focused on the function of USP. Only one USP gene was reported in Arabidopsis (AtUSP), and a knockout mutant of the USP gene exhibited an abnormal development in pollen, resulting in sterility (Schnurr et al., 2006; Geserick and Tenhaken, 2013).
Multiple UGP isoforms have been detected in soybean (Vella and Copeland, 1990), potato (Gupta and Sowokinos, 2003), and rice (Chen et al., 2007). Besides, isoforms of USP were also reported in Arabidopsis (Gronwald et al., 2008). However, isoforms of UAP have not been surveyed in plants yet. In the current study, we retrieved and classified UDPGP among plants, animals, and microorganisms for the sake of getting a better understanding of the evolution of the UDPGP gene family. Moreover, the expression profiles, motif and key amino acids, gene variation, 3D structures, and isoforms of UDPGPs in O. sativa were also surveyed for understanding the conserved function of the UDPGP genes.
Results and Discussion
Identification of UDP Glucose Pyrophosphorylase Genes in the Variety Species
To identify full-length UDPGP genes in different species, we searched the UDPGP genes using HMMER software (Potter et al., 2018). A total of 454 full-length primary protein sequences were identified in 76 organisms, including plants (58 species: 404 sequences), chlorophyte (seven species: 21 sequences), animals (six species: 16 sequences), fungi (three species: 11 sequences), kinetoplastid (one species: two sequences), and bacteria (one species: 0 sequence). The detailed information is in Supplementary Table 2.
Phylogenetic Classification of the UDP Glucose Pyrophosphorylase Gene Family Into Three Major Clades
To study the origin and evolutionary history of the UDPGP genes in different species, we constructed a phylogenetic tree with the Maximum Likelihood (ML) method. The phylogenetic tree with gene names and bootstrap values (UFBoot/SH-aLRT/aBayes) is shown in Supplementary Figure 1. The topology of the ML tree showed that UDPGP genes were clustered into three clades, which were defined as UAP, UGP, and USP. Besides, The UGP clade could be divided into two subgroups, UGP-A and UGP-B. These results were similar to the OrthoFinder analysis results (Supplementary Table 3). The orthogroup analysis divided the gene family into seven orthogroups, and the first four largest orthogroups, OG1, OG2, OG3, and OG4, were corresponding with the subgroup UGP-A, UAP, USP, and UGP-B, respectively, while three minor orthogroups (OG5, OG6, and OG7) contained UAP and UGP-A members. Notably, no homologous UGP-B and USP members were identified in animals, fungi, and kinetoplastid. Furthermore, Escherichia coli does not contain any homologous genes belonging to UDPGP in this study, which is corresponding to that of previous studies (Kleczkowski et al., 2004; Führing et al., 2013). Then, we analyzed the distribution of each subgroup in the 58 plants (Figure 1 and Supplementary Table 2). The result showed that the numbers of UDPGP genes significantly varied among the major lineages of plants, ranging from the two members detected in chlorophytes including Micromonas sp. RCC299 and Micromonas pusilla to the 19 sequences identified in Gossypium raimondii. In general, the numbers of UAP and USP genes were relatively stable among the plants, ranging from 0 to 5 (UAP) and 0 to 4 (USP), while the members in UGP (UGP-A and UGP-B) varied greatly, spanning from 0 to 14 (G. raimondii). Specifically, we did not find any full-length UAP genes in Kalanchoe fedtschenkoi or USP genes in Volvox carteri. In addition, Micromonas sp. RCC299, Micromonas pusilla, and Ostreococcus sp. Lucimarinus had no UGP (UGP-A or UGP-B) genes (Figure 1 and Supplementary Table 2).
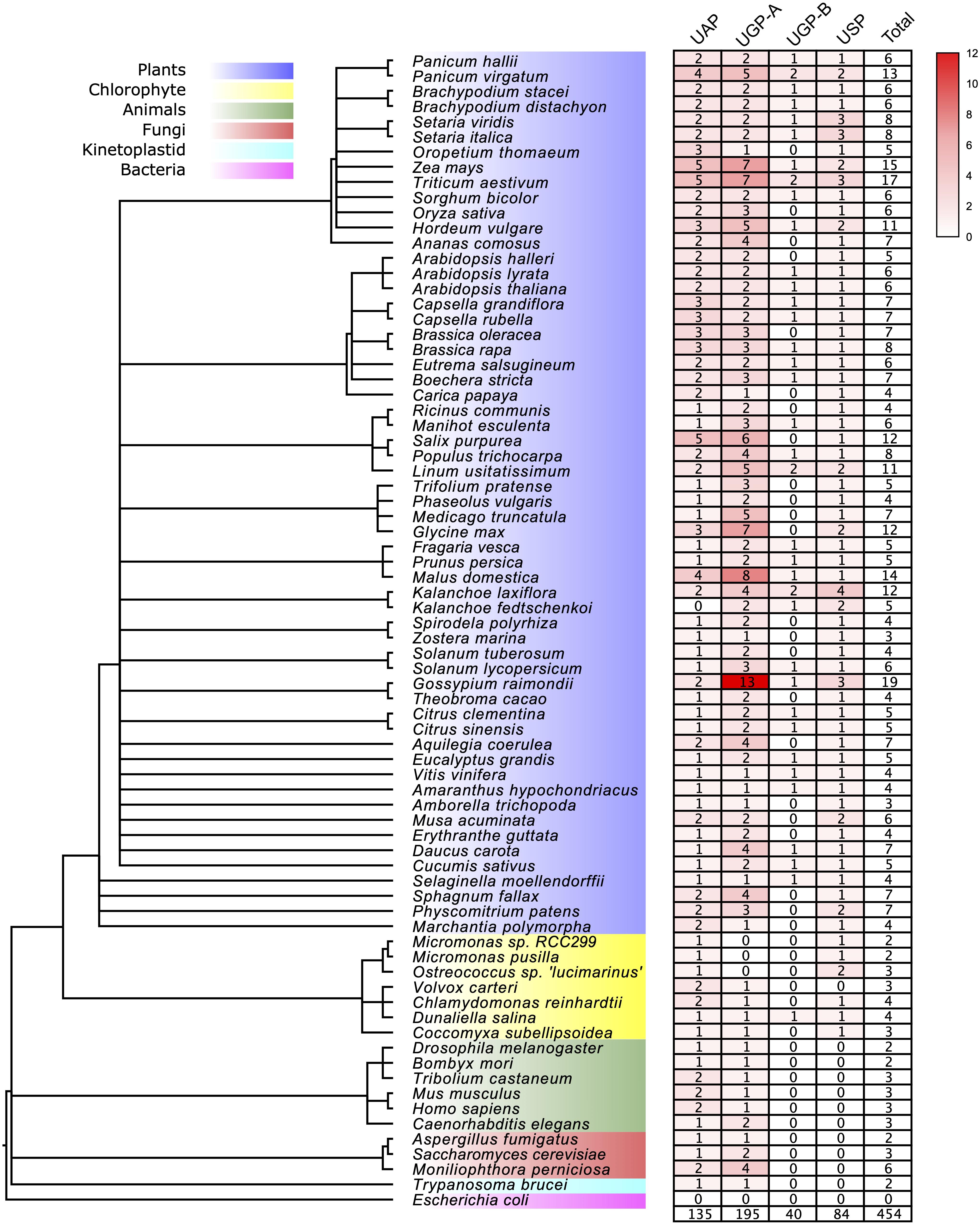
Figure 1. The UDP glucose pyrophosphorylase (UDPGP) genes were investigated in this study. The species tree was downloaded from the National Center for Biotechnology Information (NCBI) Taxonomy tree.
Physicochemical Features of UDP Glucose Pyrophosphorylase Gene Family
The calculated isoelectric point (pI) and molecular weight (MW) of each UDPGP sequence are shown in Supplementary Table 2. The average MWs of subgroups UAP, UGP-A, UGP-B, and USP were 56,234.91, 47,351.92, 93,678.22, and 66,044.22 Da, respectively. The results showed remarkable differences among the four subgroups, with subgroup UGP-A containing the smallest MW and subgroup UGP-B containing the heaviest MW. The pI for the UDPGP genes ranged from 4.33 to 9.99, implying a wide range of activity in microcellular environments. The subgroup UAP had more acidified pI with an average of 6.00 compared with the other subgroups, suggesting possible functional divergence of UAP (Supplementary Table 2). All these showed the conserved physicochemical features in UDPGP genes.
Gene Structure Difference in the Three Clades
The loss or gain of introns leads to different gene structures, makes genes more complex, and acts as the foundation of gene evolution (Fedorova and Fedorov, 2003). Previous studies have shown that introns play an important biological role in regulating gene expression (Castillo-Davis et al., 2002; Le Hir et al., 2003). In the current study, we constructed a phylogenetic tree using protein sequences from 16 representative plant species (Figure 2) and analyzed the exon–intron structures (Supplementary Figure 2). The exon–intron organization of UDPGP genes was significantly different among the four subgroups, while the structure within each subgroup was conserved, indicating the conservative characteristics in the UDPGP gene family (Supplementary Figure 2). Besides, the numbers of exons in each subgroup are similar, ranging from 15 in UAP to 21 in UGP-A typically.
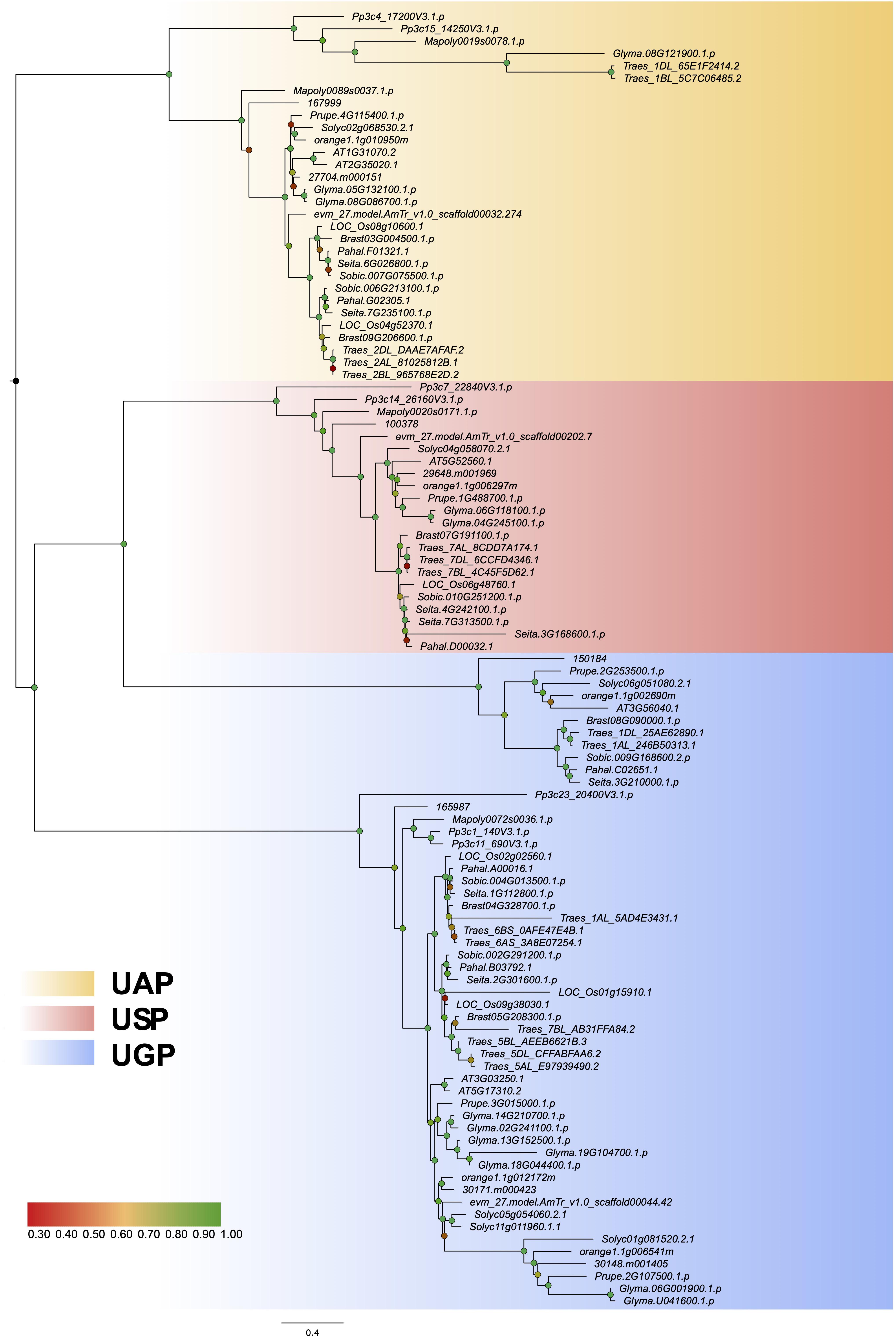
Figure 2. Maximum likelihood phylogenetic tree of UDP glucose pyrophosphorylases (UDPGPs). A maximum likelihood analysis phylogenetic tree illustrates the evolutionary relationships among UDPGP sequences from 16 species representing a wide variety of plant lineages and the ancestral homolog. The three phylogenetic clusters were designated as UDP-N-acetylglucosamine pyrophosphorylase (UAP; yellow), UDP-sugar pyrophosphorylase (USP; red), and UDP-glucose pyrophosphorylase (UGP; blue). Statistical support is shown in corresponding nodes at relevant clades according to the color of the label. Branch lengths in the tree are proportional to evolutionary distances between nodes, and the scale bar represents the number of inferred amino acid substitutions per site.
The Divergence and Segmental Duplication of UDP Glucose Pyrophosphorylase Family
In the evolutionary history of the UDPGP gene family, the number of members was relatively stable, resulting in an elementary gene family. As shown in Figure 3A, six of 12 chromosomes contain UDPGP genes in Oryza sativa. Three OsUGP genes (OsUGP1, OsUGP2, OsUGP3) posited on chromosome 9, chromosome 2, chromosome 3, respectively. Synteny analysis of the UDPGP family in rice showed that collinearity blocks between UDPGP members only existed in clade UAP (OsUAP1 and OsUAP2) and located on chromosome 8 and chromosome 4. Furthermore, the genome of O. sativa only contains a single OsUSP gene located on chromosome 6. The similarity of OsUAP1 and OsUAP2 indicated that these two genes originated from duplication. Among the five chromosomes in Arabidopsis, only chromosome 4 carried no UDPGP genes, and others had one or two UDPGP genes (Figure 3B). Chromosome 3 contained two ATUGP genes (ATUGP1 and ATUGP3), and chromosome 5 contained ATUSP and ATUGP2 genes at both arms. Besides, chromosome 1 and chromosome 2 separately hold GlcNA.UT1 (ATUAP1) and GlcNA.UT2 (ATUAP2) genes. In addition, we also detected duplication events, which were similar to O. sativa, in A. thaliana. The ATUAP1 and ATUAP2, ATUGP1 and ATUGP2 were two paired collinearity genes detected by McScanX, while no tandem gene pairs were identified in both O. sativa and A. thaliana. All these results showed that the members of UDPGP genes were stable and the expansions of the UDPGP gene family were only caused by segmental duplication.
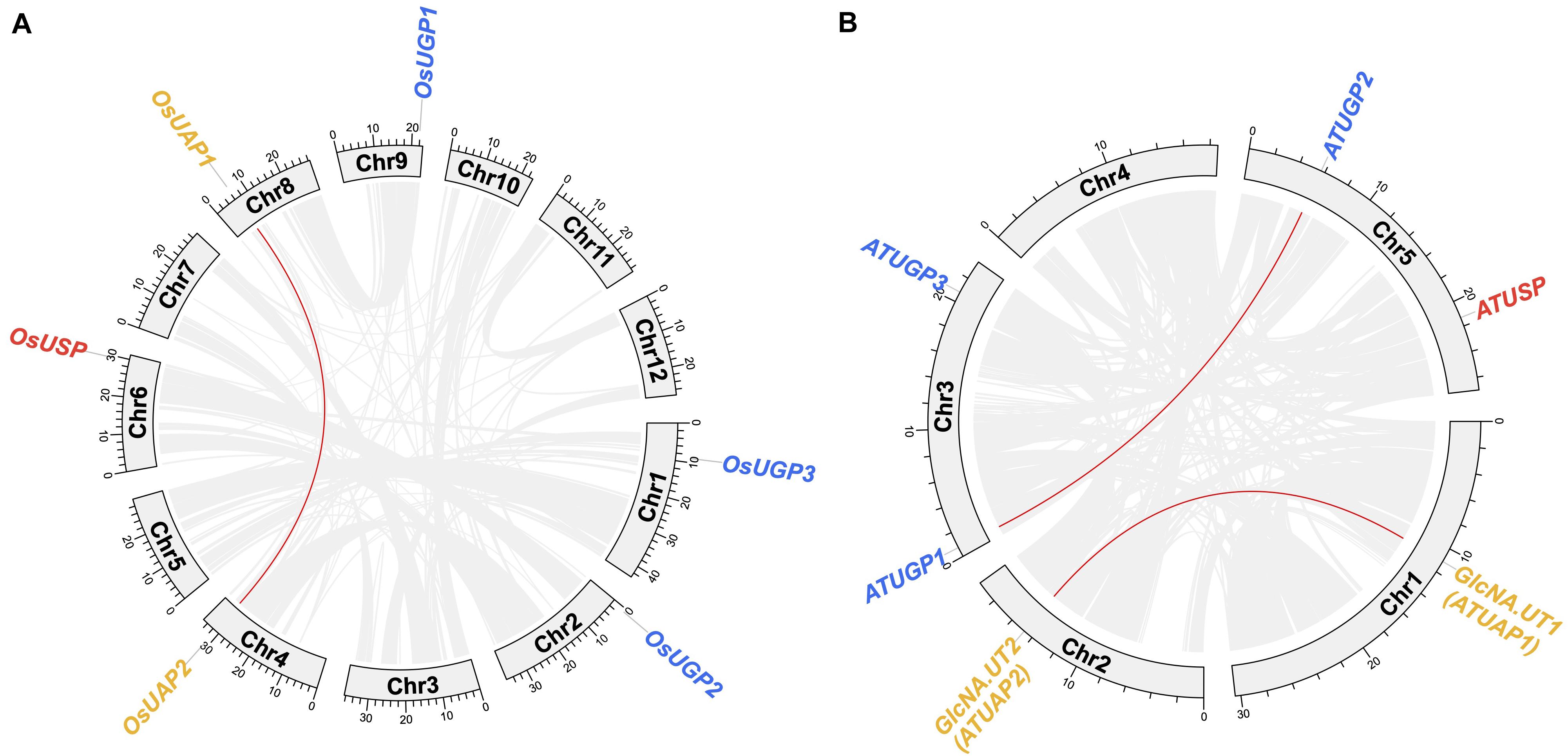
Figure 3. Chromosome distributions of UDP glucose pyrophosphorylases (UDPGPs). The chromosomal distributions of UDPGP genes in Oryza sativa (A) and Arabidopsis thaliana (B) are shown in the outer circle, where the numbers represent the chromosome length 10 Mb. The synteny and collinearity genes detected by MCScanX are connected by red arcs in the inner circle.
Expression Patterns of UDP Glucose Pyrophosphorylase Genes in Different Tissues
The transcriptomic profile reflects the tissue-specific function. RNA sequencing data from Rice Expression Database (RED) were downloaded to analyze the UDPGP genes in O. sativa expression pattern in eight (anther, callus, leaf, panicle, pistil, root, seed, shoot) different tissues (Figure 4). The results showed that the expression level of LOC_Os09g38030 (OsUGP1) was significantly higher than that of other gene members in the UDPGP family. Anther during flowering is the highest expressing tissue detected with OsUGP1. The LOC_Os02g02560 (OsUGP2) and LOC_Os01g15910 (OsUGP3) also belonged to the UGP clade in rice, whose expression levels were greatly lower than that in OsUGP1. For OsUGP2, anther (before and during flowering) and panicle (7 days before heading and 7 days after flowering tissues) in specific development stages exhibited higher expression levels than other tissues. Notably, OsUGP3 showed almost no expression in the seven tissues. Besides, for the UAP clade LOC_Os04g52370 (OsUAP2) and LOC_Os08g10600 (OsUAP1), the expression patterns were similar to each other and the expression level of OsUAP2 gene was slightly higher than OsUAP1 in some specific tissues, such as root and shoot from 7-day seedlings. In addition, for the OsUSP gene, a single member of the rice USP clade, the expression profile was ubiquitous in all tissues with a relatively low level. These results suggest that rice UDPGP genes can be expressed in various tissues to perform their roles with different expression levels as needed.
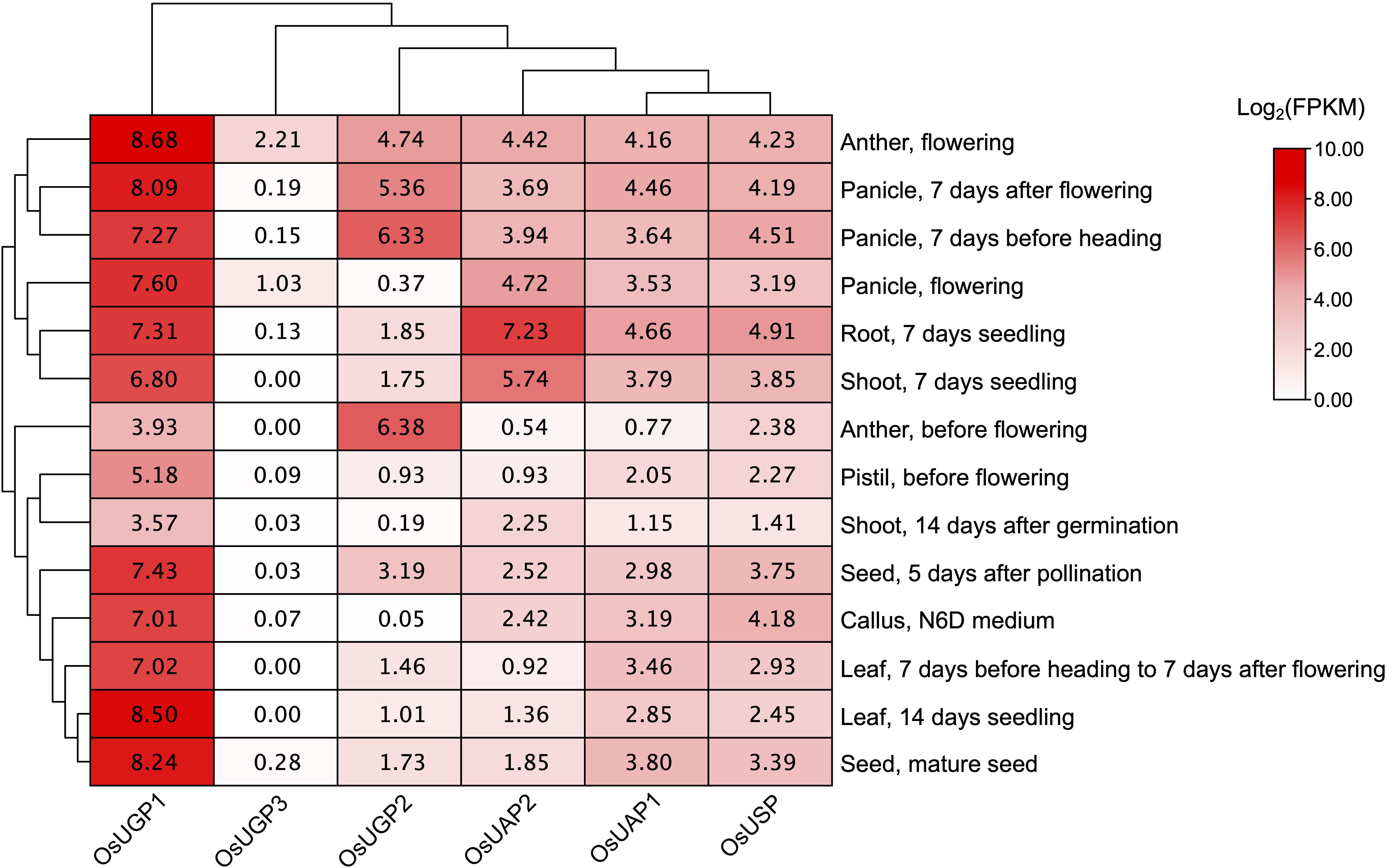
Figure 4. UDP glucose pyrophosphorylase (UDPGP) expression profiles in different tissues from Oryza sativa. The RNA sequencing expression data of UDPGPs from different tissues and developmental stages in O. sativa were downloaded from the Rice Expression Database and displayed as filled blocks from white to red.
Expression Profiles of UDP Glucose Pyrophosphorylases Under Hormones, Abiotic, Non-metal, and Heavy Metal Stress
Plant hormones, abscisic acid (ABA) and jasmonic acid (JA), are important regulatory factors involved in various biological processes, including cell death (Sreenivasulu et al., 2006; Reinbothe et al., 2009). The expression patterns of rice UDPGP genes under ABA and JA treatments are similar in both shoot (Figure 5A) and root (Figure 5B) tissues. Importantly, OsUAP1, OsUGP3, and OsUSP were generally upregulated in shoot tissue under the ABA and JA stress, and OsUSP was also upregulated in root tissue under the ABA and JA stress.
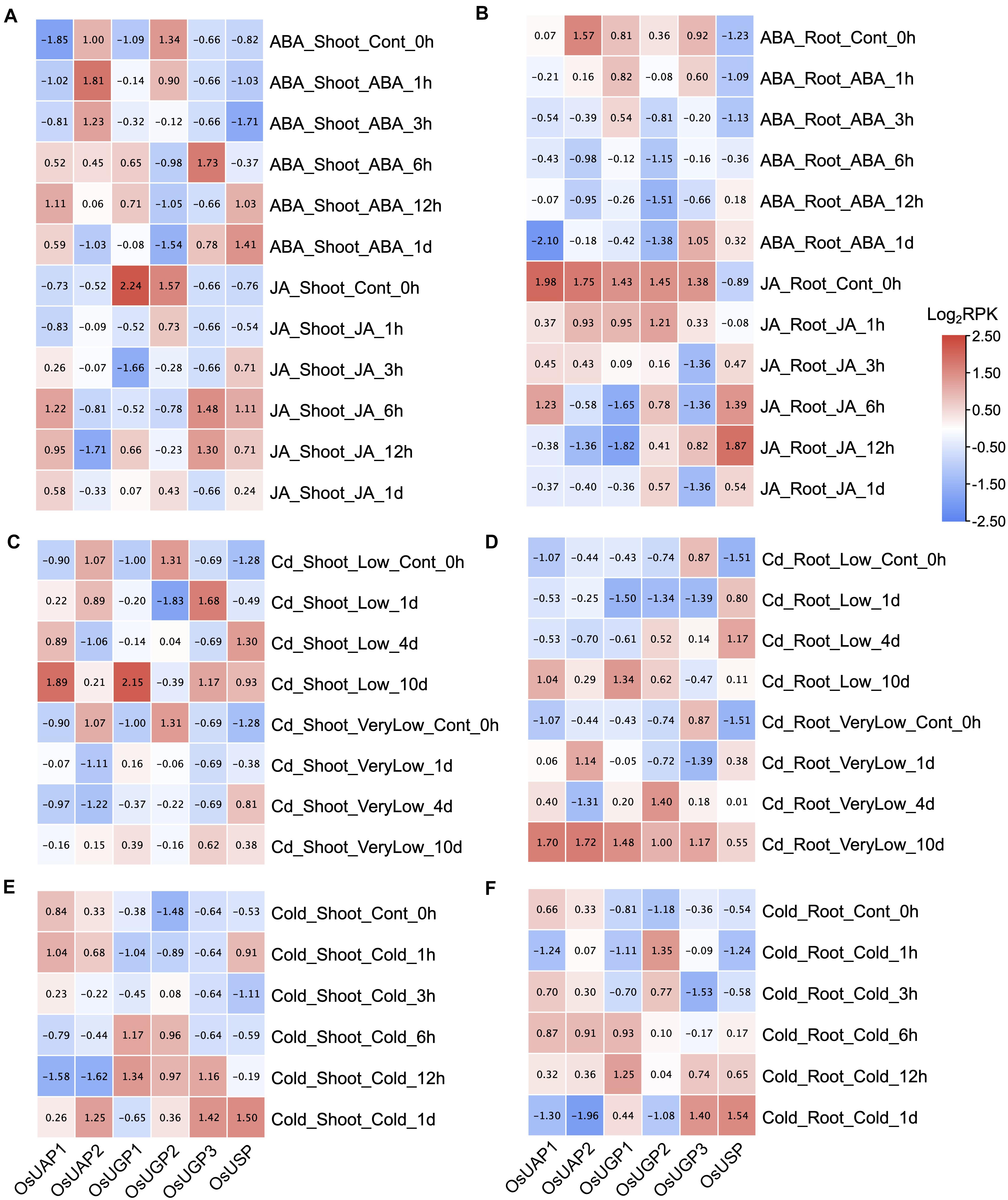
Figure 5. (A–F) UDP glucose pyrophosphorylase (UDPGP) expression profiles under abscisic acid (ABA), jasmonic acid (JA), cadmium, and cold treatment. Transcriptional expression changes of UDPGPs in Oryza sativa shoot under ABA (100 μM), JA (100 μM), low cadmium (1 μM CdSO4), very low cadmium (0.2 μM CdSO4), and cold (4°C) treatment were downloaded from the TENOR database and displayed as filled blocks from red (upregulated) to blue (downregulated). The scale on the right indicates the gene expression level transformed by log2(RPK). All the genes were normalized by columns.
Cadmium (Cd) contamination has become a big issue in food safety, especially in rice (Liu et al., 2020). Under the very low Cd and low Cd treatments, the expression level of all six rice UDPGP genes were uniformly upregulated in root tissue except for OsUGP3 (Figure 5D). The three genes (OsUAP1, OsUGP1, and OsUSP) also showed increased expression trend in shoot tissue (Figure 5C) under the same treatments. When stressed by cold, the four rice UDPGP genes (OsUGP1, OsUGP2, OsUGP3, and OsUSP) were raised both in shoot (Figure 5E) and root (Figure 5F).
Moreover, the expressions of UDPGP genes in O. sativa under abiotic (salinity, dry, flood, and osmotic) and non-metal [phosphate (P)] (Supplementary Figure 3) treatments were also investigated. In general, OsUAP1 and OsUSP were found to be upregulated under dry, flood, and osmotic stresses of shoot, and OsUSP was also upregulated under flood and osmotic stresses of root. Moreover, OsUAP2 and OsUGP2 separately showed upregulated expression under dry stress of shoot and osmotic stress of root. No significant expression changes were identified for UDPGP genes under salinity and P treatment in both shoot and root tissues.
The expression data above together implied that rice UDPGP genes may be involved in environmental endurance.
Conserved Motifs in UDP Glucose Pyrophosphorylase Family Members and the Key Amino Acids Affect Catalytic Activity
Phylogenetic analysis of the UDPGP homologs yielded three big different clades and four distinct subgroups (Figure 6A). In previous reports, UGP-A clade contained a nucleotide-binding loop (NB-loop) and substrate binding loop (SB-loop) at the active center and an insertion loop (I-loop) at the C-terminus site (Steiner et al., 2007; Führing et al., 2015; Chi et al., 2016; Figure 6B). For the UAP clade, only NB-loop and I-loop were reported (Peneff et al., 2001; Figure 6B). In the USP clade, NB-loop and SB-loop were described (Dickmanns et al., 2011; Figure 6B). Based on the NB-loop reported by Geisler et al. (2004), we identified the differences of the NB-loop in the three clades through multiple sequence alignment (MSA) (Figure 6C). The results showed that NB-loop was diverse in different subgroups. In general, all the NB-loops contained a start with “GGxG.”
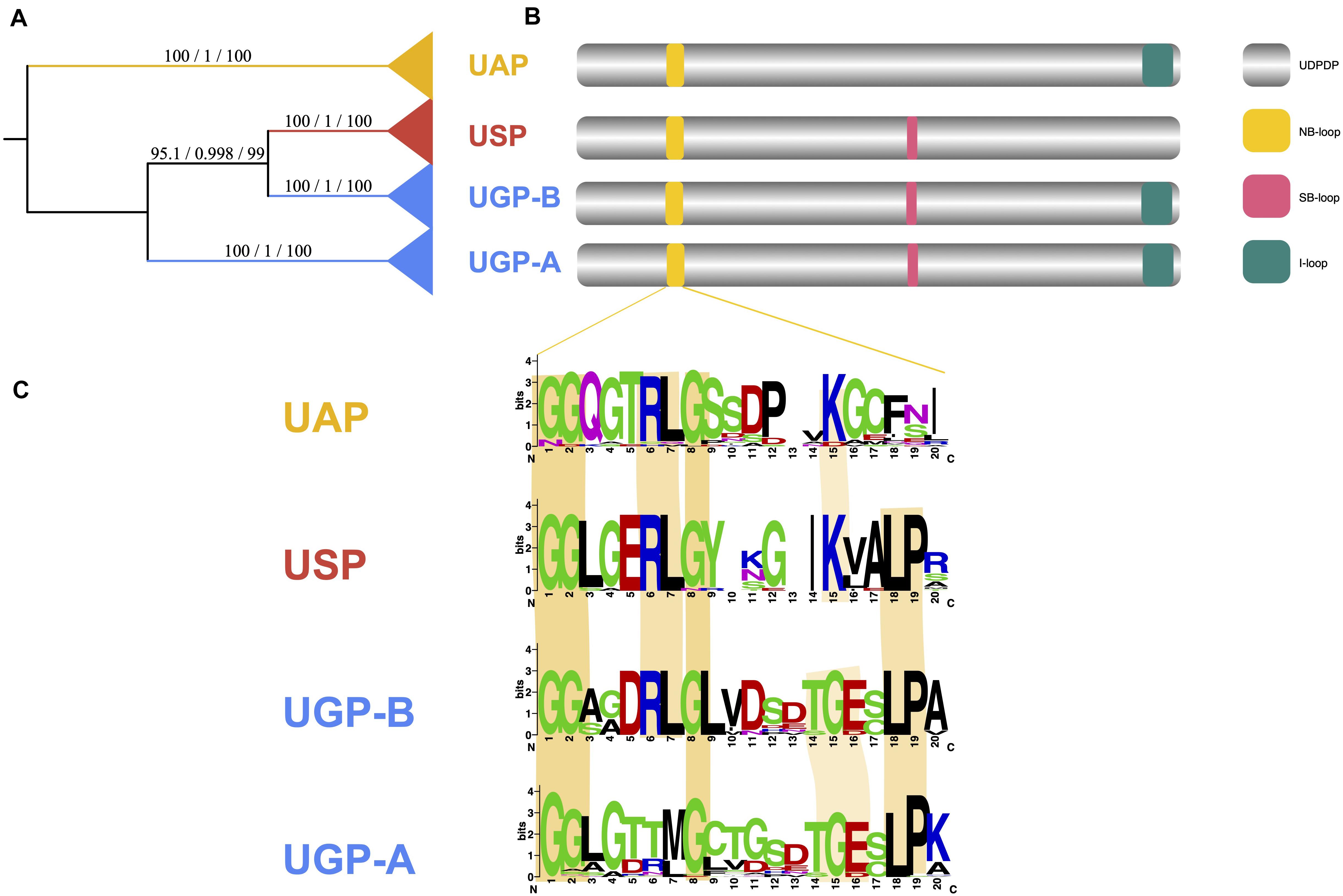
Figure 6. UDP glucose pyrophosphorylase (UDPGP) domain classification and conservation of functional motifs. (A) Phylogram showing the classification of the UDPGP gene family with the major clades labeled. (B) Typical UDPGP structure consists of an NB-loop, SB-loop, and I-loop with the approximate positions. (C) Conservation of the NB-loop functional motifs in different clades.
Then, the motifs from 16 represented species were analyzed based on the primary protein sequences using MEME software. As shown in Supplementary Figure 4, motifs 1, 4, 13, and 14 existed in all subgroups, while motifs 20, 18, and 15 were unique sequences at the N-terminus in UAP, USP, and UGP-A subgroups, respectively. Besides, motif 14 was a common motif at the C-terminus in all UDPGP clades except the UAP clade, which carried extra motifs 5, 9, and 17. Furthermore, motifs 6, 16, and 19 were only located in the USP clade, while the UGP-A subgroup contained distinctive motifs 7, 8, 10, 12, and 14. In our study, we defined a motif 8 with “NPSIELGPEFKKVGNFLSRFKSIPSIVELDSLKVSGDVWFG” sequence, which overlapped with a previously reported motif “RFKSIPSI,” and this motif was proven to be an essential element for the phosphorylation and binding with 14-3-3 protein (Toroser et al., 1998; Cotrim et al., 2018). Moreover, motif 2 “KLAVLLLAGGLGTRLGCTGPK” displayed in all clades except UGP-B; this motif was corresponding to that of a previous study that reported a high essential signature motif “LX2GXGTX6PK” (Mio et al., 1998) and three amino acids (G, R, and K) were proven vital for the activity of the enzyme. All the above contributed to the form of the UDPGP gene family and diverged it into three large clades (UAP, USP, and UGP). Thus, the conservation of these additional motifs in their respective clades may play a key role in their functional specificity.
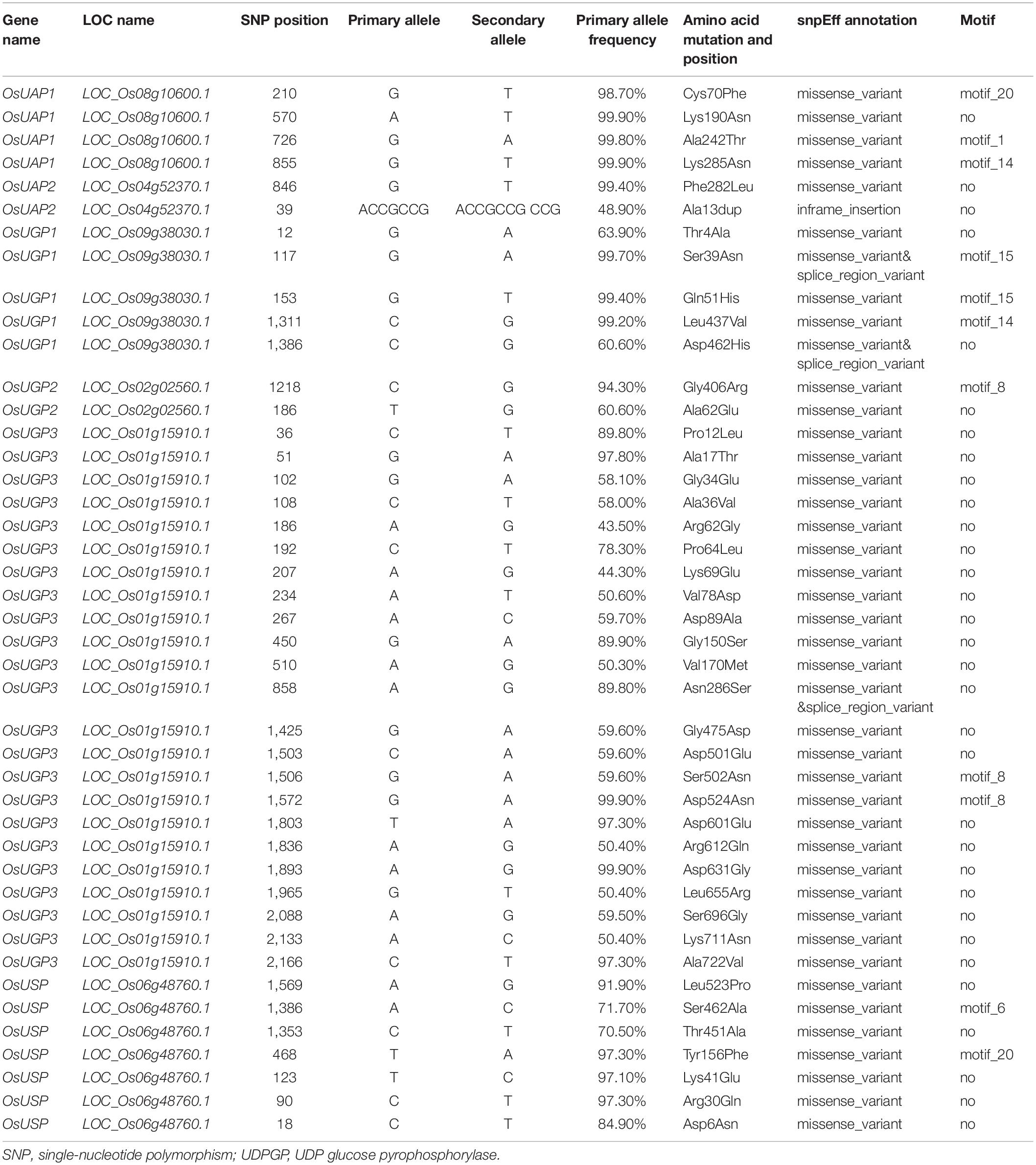
In OsUAP1 mutant osuap1, guanine (G) was replaced by thymine (T) at the position of 712 bp in the CDS, resulting in the 238th amino acid changed from glycine (Gly) to cysteine (Cys) and thus lost function of UAP enzymatic activities for the OsUAP1 protein (Wang et al., 2015; Supplementary Figure 5). This key amino acid site is located in motif 2, which exhibited in all clades except UGP-B, and this site might be involved in the uridine recognition region (Wang-Gillam et al., 2000). Some single amino acid and fragment deletion mutations were performed to study the key amino acid of UDPGPs. In the UAP clade, 14 site-directed mutants were used to study the key amino acid affecting the catalytic activity, including Giardia intestinalis (Mok and Edwards, 2005), Homo sapiens (Wang-Gillam et al., 2000; Peneff et al., 2001), Saccharomyces cerevisiae (Mio et al., 1998), and Aspergillus fumigatus (Raimi et al., 2020; Supplementary Figure 6 and Supplementary Table 4). In G. intestinalis (Mok and Edwards, 2005), when G108 (corresponding to G125 in OsUAP1) and G210 (corresponding to G236 in OsUAP1) were substituted by alanine, the enzymatic activity of UAP was significantly reduced. In H. sapiens (Wang-Gillam et al., 2000; Peneff et al., 2001), R115, P220, G222, G224, Y227, and G111 were replaced by alanine to study the catalytic property. The results showed that R115, G222, G224, and G111 were key amino acids to maintain the activity of the enzyme, corresponding to R129, G236, G238, and G125 in OsUAP1. Besides, the G111 and R115 were located in the NB-loop as well as motif 2, indicating the importance of the sequence. In Saccharomyces cerevisiae (Mio et al., 1998), three amino acids were studied, including G112, R116, and K123, which equated to G125, R129, and K136 in OsUAP1. The enzymatic activity was severely diminished when these three sites were replaced by alanine. In A. fumigatus, five site mutations were performed to study the key amino acids in AfUAP gene (Raimi et al., 2020). Three (K148, Y330, and K437) of them showed a key role in impacting Km value of UTP or GlcNAc-1P, corresponding respectively to K136, Y320, and K416 in OsUAP1 in rice. Several mutants in Hordeum vulgare (Martz et al., 2002; Meng et al., 2009a), Solanum tuberosum (Katsube et al., 1991), Cricetulus griseus (Flores-Díaz et al., 1997), and H. sapiens (Chang et al., 1996), including single amino acid and fragment deletion, were used to study the key amino acids of UGP (Supplementary Figure 7). In H. vulgare (Martz et al., 2002; Meng et al., 2009a), G91, C99, L117, I118, V119, K127, K128, L135, L136, L137, Y192, and K260 were key amino acids to provide the activity of proteins, which corresponded to G88, C96, L114, I115, V116, K124, K125, L132, L133, L134, Y189, and K257 in OsUGP1. And when K183, K332, K405 were replaced by alanine, the activity was not greatly impacted in HvUGP (HORVU5Hr1G087810.2). These three sites corresponded to the K180, K329, and K402 in OsUGP1. In addition, the authors trimmed different lengths of N and C terminals to study the key role in regulating the activity. Results showed that the majority (Ncut-21, Ncut-27, Ccut-8, Ccut-67, and Ccut-101) largely reduced the activity except for the cut 32 amino acids in C terminal, indicating the importance of N and C terminals for catalytic activity. In S. tuberosum (Katsube et al., 1991), five site mutants were studied through substituting with glutamine, including K263, K329, K367, K409, and K410. Among them, K263 and K367 were two important sites to keep the activity of UGP in S. tuberosum, which corresponded to K257 and K361 in OsUGP1. When G115 was replaced by aspartic acid (D) in Cricetulus griseus (Flores-Díaz et al., 1997), the activity of UGP was largely affected, which corresponded to G88 in OsUGP1. In H. sapiens (Chang et al., 1996), eight site mutants were developed including C123, W218, H266, W333, R389, R391, R422, and R445. Among them, C123, W333, R389, R391, and R422, corresponding to C96, W299, R354, R356, and R387 in OsUGP, were key amino acids for sustaining the activity. Among these, G115 in CgUAP was mapped to NB-loop, which corresponded to motif 2 assigned by MEME. And K236 in StUGP was a key amino acid located in SB-loop, which was in agreement with motif 3. The rest of the amino acids were located in motifs 4, 7, 8, and 11. Furthermore, no visible differences of activity were described when K183, K332, and K405 in HvUGP (Martz et al., 2002; Meng et al., 2009a) and W218, H266, and R445 in HsUGP (Chang et al., 1996) were substituted by other amino acids, indicating that these sites were not key amino acids in UGP protein. In addition, these four amino acids were not located in any motif assigned by MEME. The only one study reported on the key amino acid of USP was from Leishmania major (Prakash et al., 2019). V330, F383, and V199 were key sites for the activity of USP in LsUSP, corresponding to Q353, F405, and V239 in OsUSP (Supplementary Table 4 and Supplementary Figure 8). These sites were located in motifs 13, 16, and 19. Moreover, we compared the key amino acids, important loops (NB-loop, SB-loop, and I-loop), and motifs from MEME. The results showed that the amino acids in the loops or the motifs more likely contributed to the catalytic activity of UDPGPs.
Most UDP Glucose Pyrophosphorylase Genes Are Conserved in Natural Rice Variant
Previous studies showed that the mutants of UDPGP genes lead to aborting of enzymatic activity, resulting in cell death (Tresse et al., 2008). Besides, the family numbers of UAP and USP clades are conserved, and many species contained only two UAP genes or a single USP gene. To address the question if the amino acid sequences are conserved in UDPGP genes, we scanned the protein sequences of UDPGPs in 4,726 rice accessions in RiceVarMap1. A total of 4, 2, 5, 2, 23, and 7 gene variants for OsUAP1, OsUAP2, OsUGP1, OsUGP2, OsUGP3, and OsUSP were identified, respectively (Table 1). Furthermore, 32 of 43 variation sites located in the non-motif region, and only 11 posited on the motif sites. Among the 11 variants, only two of them showed a relatively higher proportion in nature. The 502nd amino acid (in Motif 8) changed from Ser to Asn in OsUGP3 with 40.40% and the 462nd amino acid changed (in Motif 6) from Ser to Ala in OsUSP with 28.30% (Table 1). In the previous study, one amino acid in Motif 8 was mutant in wheat, and the result showed it only slightly lowered the enzymatic activity (Meng et al., 2009a). In addition, the number of OsUGP3 variants was greater than other members (Table 1), but the expression levels of OsUGP3 were much lower than other members (Figure 4), which may indicate that this gene is under evolution. All above suggest that most UDPGP genes are conserved in the evolution.
Alternative Splicing Events Affect Protein Activities Among the UDP Glucose Pyrophosphorylase Gene Family
Alternative splicing events are important posttranscriptional regulatory mechanisms, which could reduce the enzyme activities or completely abolish the activity (Kelemen et al., 2013). Here we studied the alternative splicing events of the UDPGP gene family in O. sativa. Four (OsUAP1, OsUAP2, OsUSP, and OsUGP1) of six UDPGP genes carried more than one transcript sequence from the rice annotation file. The OsUAP1 gene had three different types of mRNA sequences, OsUAP1.1, OsUAP1.2, and OsUAP1.3. The OsUAP1.1 transcript was the longest form with 489 amino acids, while the OsUAP1.2 transcript lacked exon sequence near 5’ untranslated region (5’ UTR), and the OsUAP1.3 transcript lost exons at 3’ UTR (Figure 7A). The OsUAP2 gene had two similar transcripts with a difference of six nucleotides at the end of the fifth exon (Figure 7A and Supplementary Figure 9). The OsUSP gene and the OsUGP1 gene both had two transcripts with the exon differences closing to the 3’ UTR.
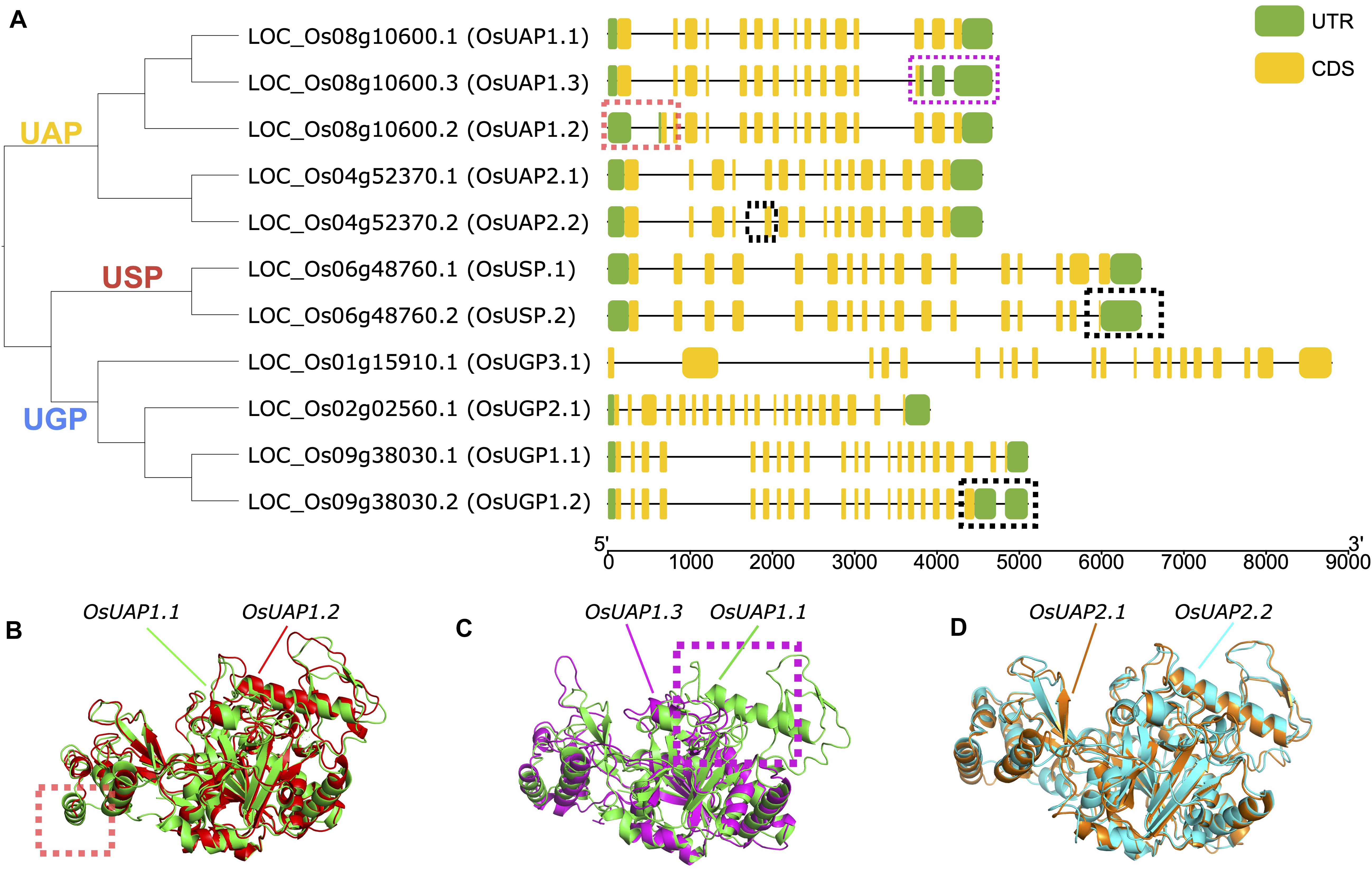
Figure 7. Alternative splicing of UDP glucose pyrophosphorylases (UDPGPs) in Oryza sativa. (A) Gene structure and phylogenetic tree of UDPGP genes in O. sativa, different alternative splicing events were marked with red (OsUAP1.2), purple (OsUAP1.3), or black dashed rectangles. (B) Comparison of the 3D protein structure of OsUAP1.1 (green) and OsUAP1.2 (red). (C) Comparison of the 3D protein structure of OsUAP1.1 (green) and OsUAP1.3 (purple). (D) Comparison of the 3D protein structure of OsUAP2.1 (orange) and OsUAP2.2 (blue).
To compare the structure differences for these gene isoforms, we predicted the 3D structures of proteins using I-TASSER. The results showed that the spatial structures among the three OsUAP1 isoforms were similar, while OsUAP1.2 (Figure 7B) and OsUAP1.3 (Figure 7C) lacked some folding due to the deficiency of exons at 5’ UTR and 3’ UTR. And for OsUAP2 isoforms, the predicted tertiary structures of OsUAP2.1 and OsUAP2.2 were almost the same (Figure 7D) due to the tiny difference between OsUAP2.1 and OsUAP2.2 variants, with OsUAP2.2 discarding two amino acids compared to OsUAP2.1 (Supplementary Figure 9). In addition, the structures of OsUGP1.1, OsUGP1.2, OsUSP.1, and OsUSP.2 were also predicted to compare differences in the spatial structures (Supplementary Figure 10). The results showed that the N and center domains were similar, while the C terminal was diverse because of the lack of sequences in OsUGP1.2 and OsUSP.2.
To understand if the alternative splicing events in UDPGP genes affect the catalytical characteristics, we performed the enzymatic activity analysis in the UAP clade. 1H-nuclear magnetic resonance (1H-NMR) spectroscopy was used to record the enzymatic reaction of OsUAP1.1, OsUAP1.2, and OsUAP1.3 in situ. In the time-gradient enzymatic progression at 60 min, forward conversion of GlcNAc-1-P (5.36 ppm) to UDP-GlcNAc (5.52 ppm) was observed with OsUAP1.1 (Figure 8A, line 2), but not with the glutathione S-transferase (GST) control (Figure 8A, line 1), OsUAP1.2 (Figure 8A, line 3), and OsUAP1.3 (Figure 8A, line 4). Besides, the reverse conversion of UDP-GlcNAc (5.52 ppm) to GlcNAc-1-P (5.36 ppm) was identified with OsUAP1.1 (Figure 8B, line 2), but not with the GST control (Figure 8B, line 1), OsUAP1.2 (Figure 8B, line 3), and OsUAP1.3 (Figure 8B, line 4). Furthermore, OsUAP1.1 could also catalyze the reverse conversion of UDP-GalNAc (5.55 ppm) to GalNAc-1-P (5.39 ppm) (Figure 8C, line 2), whereas GST (Figure 8C, line 1), OsUAP1.2 (Figure 8C, line 3), and OsUAP1.3 (Figure 8C, line 4) could not perform this reaction. Furthermore, we also conducted the same reaction in the OsUAP2 gene. The OsUAP2 gene was another member of the UAP clade in rice, which contained two isoforms (OsUAP2.1 and OsUAP2.2). The results showed that both OsUAP2.1 and OsUAP2.2 could catalyze the reaction from GlcNAc-1-P to UDP-GlcNAc, UDP-GlcNAc to GlcNAc-1-P, and UDP-GalNAc to GalNAc-1-P (Supplementary Figures 11A–C).
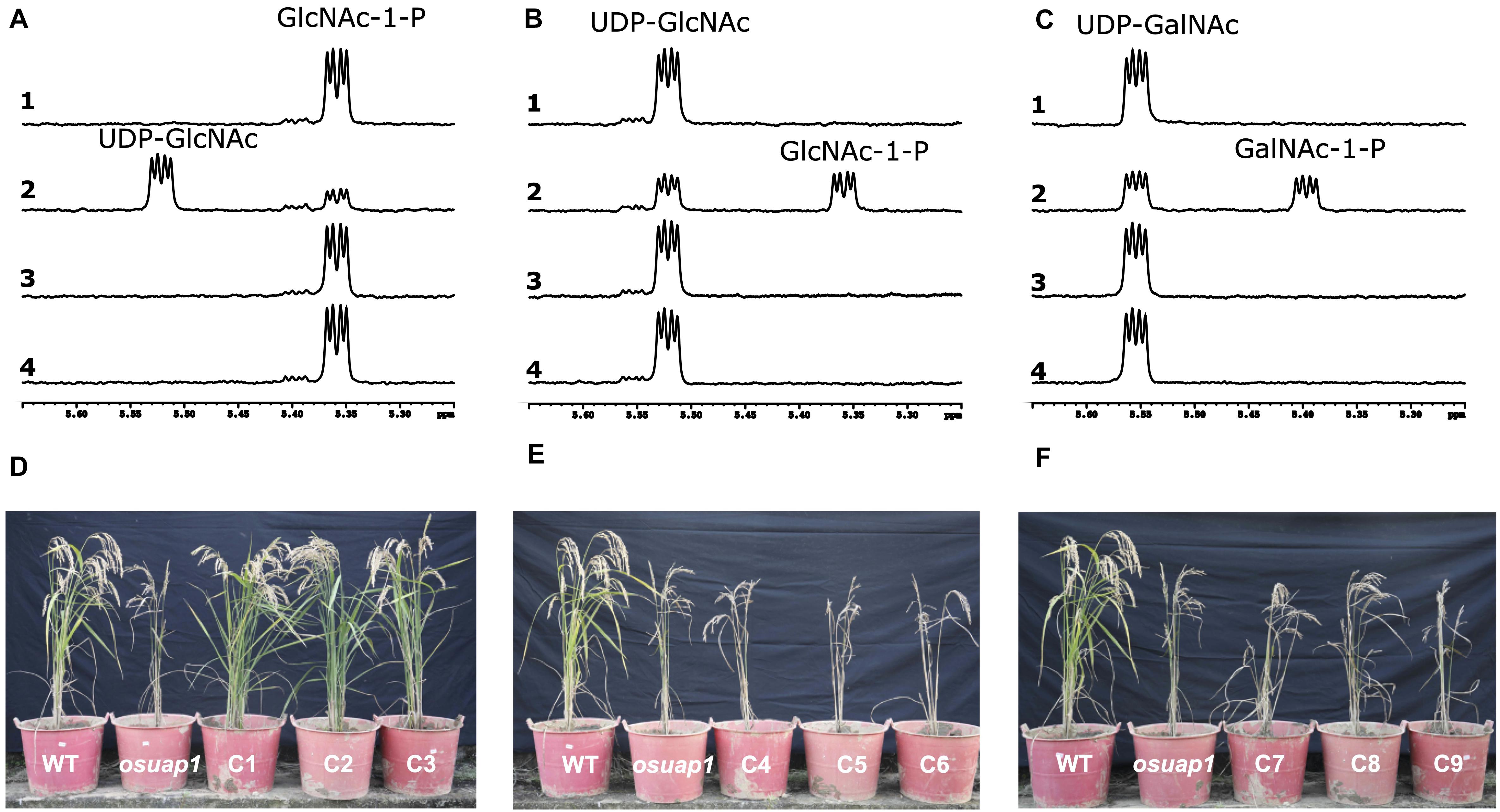
Figure 8. In vitro and in vivo activity of isoforms from OsUAP1. Enzymatic activities of three isoforms of OsUAP1 based on 1H-nuclear magnetic resonance (1H-NMR). (A) Forward activity: UTP + GlcNAc-1-P → UDP-GlcNAc + PPi. (B) Reverse activity: UDP-GlcNAc + PPi → GlcNAc-1-P + UTP. (C) Reverse activity: UDP-GalNAc + PPi → GalNAc-1-P + UTP. (A–C) Line 1, glutathione S-transferase (GST) control. Line 2, the protein of OsUAP1.1. Line 3, the protein of OsUAP1.2. Line 4, the protein of OsUAP1.3. (D) The phenotype of wild-type (WT), mutant (osuap1), and three independent complementary transgenic lines overexpressing OsUAP1.1. (E) The phenotype of WT, mutant (osuap1), and three independent complementary transgenic lines overexpressing OsUAP1.2. (F) The phenotype of WT, mutant (osuap1), and three independent complementary transgenic lines overexpressing OsUAP1.3.
Losing UAP enzymatic activity for OsUAP1 induces early leaf senescence (Wang et al., 2015). Actually, all three alternatives (OsUAP1.1, OsUAP1.2, and OsUAP1.3) were mutated, corresponding to the 238, 203, and 238 in amino acid sequences (Gly to Cys), respectively (Supplementary Figure 5). Accordingly, this mutant osuap1 was used to identify the function of OsUAP1.1, OsUAP1.2, OsUAP1.3, OsUAP2.1, and OsUAP2.2. Results showed that the osuap1 exhibited early leaf senescence, and overexpressing OsUAP1.1 in osuap1 could restore the healthy leaf (C1, C2, and C3 in Figure 8D) but not with OsUAP1.2 (C4, C5, C6 in Figure 8E) and OsUAP1.3 (C7, C8, C9 in Figure 8F). All the above indicated the key role of the N and C terminals in maintaining the activity of UAP. The UAP protein would lose its function without N or C terminal, which corresponded to a previous study on UGP protein (Supplementary Figure 7; Meng et al., 2009a). Then, we test whether the OsUAP2.1 and OsUAP2.2 could restore the osuap1 mutant. The results showed that both the OsUAP2.1 (Supplementary Figure 11D) and OsUAP2.2 (Supplementary Figure 11E) also complemented the function of OsUAP1.1, making the leaves of osuap1 grow normally without early leaf senescence.
The above results illustrated that the loss of N and C terminals in UDPGP gene isoforms did not affect the overall 3D structures, but these N- and C-terminal sequences may be important for the UDPGP gene isoforms to maintain or change their enzymatic activity. The molecular mechanisms for the existence of inactive UDPGP gene isoforms through alternative splicing are not clear. We proposed the following speculations: (1) the inactive UDPGP gene isoforms are mistake alternative splicing; (2) the inactive UDPGP gene isoforms are under evolution for new functions; (3) the inactive UDPGP gene isoforms have effective but unknown functions, for example, competing with active isoforms to bind the enzymatic substrates to regulate the enzymatic activities of active isoforms; (4) lack of the C terminal could reduce the spatial block of the UDPGP, resulting in forming an inactive dimer in the plant (Decker et al., 2012). All above represent potential regulating mechanisms of alternative splicing isoforms of UDPGP in the plant, and further experiments are needed to validate the hypothesis.
Conclusion
In the present study, the phylogenetic tree of the UDPGP gene family from 76 organism lineages divided this gene family into three clades, including UAP, UGP, and USP, and the UGP could be additionally separated into two subclades, UGP-A and UGP-B. This result was also supported by the diverse physicochemical features, gene structures, and motifs in different clades. Through scanning the UDPGP gene members in 76 species, we found that the number of the UGP-A is more variable among the clades, while UGP-B, UAP, and USP showed relatively conserved members, indicating the important role of these UDPGP genes. UDPGP genes significantly respond to cadmium, cold, ABA, and JA stresses in the shoot or root tissues in rice, while they do not exhibit obvious feedback to salinity, dry, flood, osmotic, and phosphate stimulation. Through MSA, we identified the key amino acids regulating the enzymatic activities of the UDPGP proteins, and many of them located in the NB-loop, SB-loop, and conserved motifs, demonstrating the key role of these structures. Alternative splicing may be a key mechanism to regulate the enzymatic activity of UDPGP genes. In the current study, in vitro enzymatic experiments showed that the OsUAP1 lost its catalytic activity without the N (OsUAP1.2) or C (OsUAP1.3) terminal, while OsUAP2.1 and OsUAP2.2 both maintained their catalytic activity with complete N and C terminals. The same results were also proven in in vivo transgenic experiments, where the early senescence phenotype of the mutant osuap1 could be rescued through overexpressing OsUAP1.1, OsUAP2.1, and OsUAP2.2, but not with OsUAP1.2 and OsUAP1.3. All above provide new insights into the evolution and function of the UDPGP gene family, which may lay a foundation to further investigate their molecular regulatory mechanisms in the plant.
Materials and Methods
Data Sources and Sequence Retrieval
Protein sequences, transcript sequences, genomic sequences, and GFF annotation files of 58 green plants, seven chlorophytes, six animals, three fungi, one kinetoplastid, and one bacterium were downloaded from Phytozome2 and Ensembl website3 (Supplementary Table 1). UDPGP homologs were identified by the following steps: (1) A hidden Markov model (HMM) of UDPGP (ACC: PF01704.19) was downloaded from the Pfam website. (2) The UDPGP HMM was used to direct HMMSEARCH with the parameters E-value < 0.1. (3) Pfam-A.hmm was a manually corrected database and was downloaded from the Pfam website (April 18, 2020). Hmmscan software was used to identify the UDPGP domain with default parameters, and a total of 454 non-redundant primary protein sequences were retrieved for further analysis.
Multiple Sequence Alignment and Phylogenetic Analysis
To explore the phylogenetic relationships of the UDPGP genes in the plant, animal, and microbial lineages, 454 full-length non-redundant primary protein sequences were used to perform MSA analysis using MAFFT V7.271 (Katoh and Standley, 2013) with default parameters. The MSA was submitted to IQ-TREE v2.1.1 (Minh et al., 2020) to be tested for the best substitution model, and the model with the lowest Bayesian Information Criterion (BIC) was selected as the best model (LG + I + G). Then, the phylogenetic tree was inferred by the ML method. We measured branch supports using the Ultrafast Bootstrap (UFBoot) algorithm with 1,000 replicates, the SH-aLRT, and approximate transformation Bayes test (aBayes). The tree was visualized using Interactive Tree of Life (iTOL)4 (Letunic and Bork, 2019) and FigTree v1.4.45.
Gene Structure, Sequence Motif Analysis, and Physicochemical Features
The coding sequence (CDS) information of the 105 full-length primary protein sequences from 16 representative plants was retrieved from the GFF annotation files and submitted to the TBtools (Chen et al., 2018) to visualize the exon–intron organization of UDPGP genes in representative species. Motif analysis was performed using MEME suite v5.3.0 (Bailey et al., 2009), which scans for motifs recurring in a set of sequences. Motif analysis was carried out using the MEME server6, keeping the width of the motif at 6–50 amino acids, the number of motifs was 20, and the other parameters set to default. The gene structure and conserved motif patterns were visualized by the TBtools (Chen et al., 2018). The pI/MW tool from ExPASy (Gasteiger et al., 2005) was used to compute the theoretical pI and MW of each sequence.
Transcriptomic Analyses
The UDPGP gene expression data of O. sativa were downloaded from the Rice Expression Database7 and TENOR database8. The expression abundance with different treatments was transformed by log2RPK and visualized using TBtools.
Orthogroup Generation
All full-length sequences from the 16 species (Amborella trichopoda, A. thaliana, Brachypodium stace, Citrus sinensis, Glycine max, Marchantia polymorpha, O. sativa, Panicum hallii, Physcomitrella patens, Prunus persica, Ricinus communis, Selaginella moellendorffii, Setaria italica, Solanum lycopersicum, Sorghum bicolor, and Triticum aestivum), including basal angiosperm, spikemoss, mosses, liverwort, monocots, and eudicots were selected to study the evolution of the UDPGP gene family (Supplementary Table 1). Orthologous genes were generated by OrthoFinder v2.2.7 (Emms and Kelly, 2015) with default parameters. In total, seven orthogroups were identified across the 16 species, and 105 (100%) of the input genes were assigned to orthogroups.
Collinearity and Analysis
The collinearity of O. sativa and A. thaliana was detected with MCScanX (Wang et al., 2012). The result was visualized using TBtools (Chen et al., 2020).
Evolutionary Expansion of UDP Glucose Pyrophosphorylase Gene Family
To understand and infer the evolutionary expansion history of the UDPGP family, we used a total of 105 full-length homologs from the 16 species.
3D Structure Modeling
The I-TASSER (Yang et al., 2014) was used to model the structures of all O. sativa UDPGP genes, including OsUAP1, OsUAP2, OsUGP1, OsUGP2, OsUGP3, and OsUSP. I-TASSER generates simulated protein structures depending on the pairwise structure similarity. Then, we selected the top models based on the C-score as representative structures. The 3D structures of proteins were visualized in PyMOL software.
Enzymatic Reaction Experiments for OsUAP Isoforms Examined by 1H-Nuclear Magnetic Resonance Analysis
The GST gene fusion constructs of UAP isoforms were generated. The full-length CDS of the OsUAP1.1, OsUAP1.2, and OsUAP1.3 isoforms were specifically amplified using primers GST-OsUAP1.1 (F: cgGGATCCatggcggagatcgtggtggc, R: cgGAATTCctaaaatgaaatctcactcggtgc), GST-OsUAP1.2 (F: cgGG ATCCatggatgtacacagcc, R: cgGAATTCctaaaatgaaatctcactcggtgc), and GST-OsUAP1.3 (F: cgGGATCCatggcggagatcgtggtggc, R: cgGAATTCtcaagctttaagcctgccgtg). The full-length CDS of the OsUAP2.1 and OsUAP2.2 isoforms were amplified using primers GST-OsUAP2 (F: cgGGATCCatgaaggagatagtggttgggtcg, R: cgGAATTCctagaaggaaatctcactcggcg). PCR products were inserted into pGEX-6P-1 using the restriction enzyme sites BamHI and EcoRI. Then, the recombinant vectors were transferred into E. coli DH5α and sequenced to check if the constructions were correct. Expression and purification of the fused GST-UAP isoforms were performed using the same method as described (Wang et al., 2015). The reaction of enzymatic activities of UAPs were performed as described previously (Wang et al., 2015), with 0.5 μg purified GST-UAP recombinant proteins used in the reaction mixture. Examination of GlcNAc-1-P/GalNAc-1-P and UDP-GlcNAc/UDP-GalNAc was performed by 1H-NMR as described (Zhang et al., 2011; Wang et al., 2015). Data acquisition started at 60 min respectively after the addition of an enzyme to the reaction mixture.
Transgenic Experiments
The overexpression vectors of UAP gene isoforms were constructed. The full-length CDSs of the OsUAP1.1, OsUAP1.2, and OsUAP1.3 isoforms were specifically amplified using primers OsUAP1.1-OE (F: cagtGGTCTCa gttgatggcggagatcgtggtggc, R: cagtGGTCTCaagagctaaaatgaaatctca ctcg), OsUAP1.2-OE (F: cagtGGTCTCagttgatggatgtacacagcccact, R: cagtGGTCTCaagagctaaaatgaaatctcactcg), and OsUAP1.3-OE (F: cagtGGTCTCagttgatggcggagatcgtggtggc, R: cagtGGTCTC aagagctaaaatgaaatctcactcg). The full-length CDSs of the OsUAP2.1 and OsUAP2.2 isoforms were amplified using primer OsUAP2-OE (F: cagtGGTCTCagttgatgaaggagatagtggttgg, R: cagtGGTCTCaagagctagaaggaaatctcactcg). PCR products were inserted into the binary vector pBWA(V)BU (reconstructed from pCAMBIA3300) using BsaI sites for the digesting-link one-step reaction. The recombinant vectors were transferred into E. coli DH5α and sequenced to check if the constructions were correct. Correct vectors were introduced into Agrobacterium tumefaciens EHA105 and then transformed into the osuap1 calli. Positive transgenic plants were confirmed using the phosphinothricin solution (20 mg/L) screening and then cultivated under natural conditions.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
ZW designed the research. SL, HuZ, QW, CL, and ZW performed the research and analyzed the data. SL, HuZ, and ZW wrote the manuscript. TL, ZP, YL, HoZ, JL, YH, and ZW revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (31760380, 32060071, 32071990), the Science and Technology Projects of Jiangxi Province (20181BAB214011), and the Open Research Fund of State Key Laboratory of Hybrid Rice (Wuhan University) (KF201909).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are thankful to Jessica Flowers and Craig Gentry from the Writing Center in Mississippi State University for helping us with proofing the grammar and language.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.681719/full#supplementary-material
Supplementary Figure 1 | Maximum likelihood tree of 75 species generated by IQ-Tree and branch supports using ultrafast bootstrap approximation/SH-aLRT/aBayes methods. UAP, UGP, and USP were marked in yellow, blue, and red, respectively.
Supplementary Figure 2 | Gene structures of 105 UDPGPs from 16 species. Phase 0 intron doesn’t interrupt a codon, phase 1 intron interrupts a codon between the 1st and 2nd bases, and phase 2 intron interrupts a codon between the 2nd and 3rd bases.
Supplementary Figure 3 | Expression profile of UDPGPs of the shoot and root tissues under salinity (150 mM NaCl), dry (grown without medium), flood (completely submerged in medium), osmotic (0.6 M Mannitol), and P (3 mM KH2PO4) treatment from Oryza sativa. The scale on the right indicates the gene expression level transformed by log2(RPK). All the genes were normalized by columns.
Supplementary Figure 4 | Motif architectures of 105 UDPGPs from 16 species.
Supplementary Figure 5 | CDS alignment of three isoforms (OsUAP1.1, OsUAP1.2, and OsUAP1.3) from OsUAP1.
Supplementary Figure 6 | Key amino acids affect UAP catalytic activities. The key amino acids were marked by black triangles. The black squares mean the deletion sites.
Supplementary Figure 7 | Key amino acids affect UGP catalytic activities. The key amino acids were marked by dark triangles.
Supplementary Figure 8 | Key amino acids affect USP catalytic activities. The key amino acids were marked by black triangles.
Supplementary Figure 9 | CDS alignment of three isoforms (OsUAP2.1 and OsUAP2.2) from OsUAP2.
Supplementary Figure 10 | Spatial structure comparison of isoforms from OsUGP1 and OsUSP. (A) The structure comparison of OsUGP1.1 (green) and OsUGP1.2 (red). (B) The structure comparison of OsUSP.1 (green) and OsUSP.2 (red).
Supplementary Figure 11 | In vitro and in vivo activities of OsUAP2. Enzymatic activities of two isoforms of OsUAP2 based on 1H-NMR. (A) Forward activity: UTP + GlcNAc-1-P → UDP-GlcNAc + PPi. (B) Reverse activity: UDP-GlcNAc + PPi → GlcNAc-1-P + UTP. (C) Reverse activity: UDP-GalNAc + PPi → GalNAc-1-P + UTP. (A–C) Line 1, GST control. Line 2, protein of OsUAP2.1. Line 3, protein of OsUAP2.2. (D) The phenotype of wild type (WT), mutant (osuap1), and three independent complementary transgenic lines overexpressing OsUAP2.1. (E) The phenotype of wild type (WT), mutant (osuap1), and three independent complementary transgenic lines overexpressing OsUAP2.2.
Supplementary Table 1 | Protein resource of UDPGP genes from 76 species.
Supplementary Table 2 | Characteristics of the 454 UDPGPs in 75 species.
Supplementary Table 3 | Orthogroups generated by McScanX.
Supplementary Table 4 | Key amino acid residues affect the activity of UDPGP based on reported mutants.
Footnotes
- ^ http://ricevarmap.ncpgr.cn/
- ^ https://phytozome-next.jgi.doe.gov/
- ^ http://uswest.ensembl.org/index.html
- ^ https://itol.embl.de/login.cgi
- ^ http://tree.bio.ed.ac.uk/software/figtree/
- ^ http://meme-suite.org/tools/meme
- ^ http://expression.ic4r.org/
- ^ https://tenor.dna.affrc.go.jp/
References
Arakane, Y., Baguinon, M. C., Jasrapuria, S., Chaudhari, S., Doyungan, A., Kramer, K. J., et al. (2011). Both UDP N-acetylglucosamine pyrophosphorylases of Tribolium castaneum are critical for molting, survival and fecundity. Insect Biochem. Mol. Biol. 41, 42–50. doi: 10.1016/j.ibmb.2010.09.011
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME suite: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Castillo-Davis, C. I., Mekhedov, S. L., Hartl, D. L., Koonin, E. V., and Kondrashov, F. A. (2002). Selection for short introns in highly expressed genes. Nat. Genet. 31, 415–418. doi: 10.1038/ng940
Chang, H. Y., Peng, H. L., Chao, Y. C., and Duggleby, R. G. (1996). The importance of conserved residues in human liver UDPglucose pyrophosphorylase. Eur. J. Biochem. 236, 723–728. doi: 10.1111/j.1432-1033.1996.t01-1-00723.x
Chen, C., Chen, H., He, Y., and Xia, R. (2018). TBtools, a toolkit for Biologists integrating various biological data handling tools with a user-friendly interface. bioRxiv [Preprint]. doi: 10.1101/289660
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, R., Zhao, X., Shao, Z., Zhu, L., and He, G. (2007). Multiple isoforms of UDP-glucose pyrophosphorylase in rice. Physiol. Plant 129, 725–736. doi: 10.1111/j.1399-3054.2007.00865.x
Chi, S., Feng, Y. J., and Liu, T. (2016). Molecular cloning, characterization, and comparison of UDP-glucose pyrophosphorylase from Gracilaria chouae and Saccharina japonica. J. Appl. Phycol. 28, 2051–2059. doi: 10.1007/s10811-015-0738-7
Chivasa, S., Tomé, D. F. A., and Slabas, A. R. (2013). UDP-glucose pyrophosphorylase is a novel plant cell death regulator. J. Proteome Res. 12, 1743–1753. doi: 10.1021/pr3010887
Coleman, H. D., Ellis, D. D., Gilbert, M., and Mansfield, S. D. (2006). Up-regulation of sucrose synthase and UDP-glucose pyrophosphorylase impacts plant growth and metabolism. Plant Biotechnol. J. 4, 87–101. doi: 10.1111/j.1467-7652.2005.00160.x
Cotrim, C. A., Soares, J. S. M., Kobe, B., and Menossi, M. (2018). Crystal structure and insights into the oligomeric state of UDP-glucose pyrophosphorylase from sugarcane. PLoS One 13:e0193667. doi: 10.1371/journal.pone.0193667
Decker, D., and Kleczkowski, L. A. (2017). Substrate specificity and inhibitor sensitivity of plant UDP-sugar producing pyrophosphorylases. Front. Plant Sci. 8:1610. doi: 10.3389/fpls.2017.01610
Decker, D., and Kleczkowski, L. A. (2019). UDP-sugar producing pyrophosphorylases: distinct and essential enzymes with overlapping substrate specificities, providing de novo precursors for glycosylation reactions. Front. Plant Sci. 9:1822. doi: 10.3389/fpls.2018.01822
Decker, D., Meng, M., Gornicka, A., Hofer, A., Wilczynska, M., and Kleczkowski, L. A. (2012). Substrate kinetics and substrate effects on the quaternary structure of barley UDP-glucose pyrophosphorylase. Phytochemistry 79, 39–45. doi: 10.1016/j.phytochem.2012.04.002
Dickmanns, A., Damerow, S., Neumann, P., Schulz, E. C., Lamerz, A. C., Routier, F. H., et al. (2011). Structural basis for the broad substrate range of the UDP-sugar pyrophosphorylase from Leishmania major. J. Mol. Biol. 405, 461–478. doi: 10.1016/j.jmb.2010.10.057
Emms, D. M., and Kelly, S. (2015). OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16:157. doi: 10.1186/s13059-015-0721-2
Fedorova, L., and Fedorov, A. (2003). Introns in gene evolution. Genetica 118, 123–131. doi: 10.1023/A:1024145407467
Flores-Díaz, M., Alape-Girón, A., Persson, B., Pollesello, P., Moos, M., Von Eichel-Streiber, C., et al. (1997). Cellular UDP-glucose deficiency caused by a single point mutation in the UDP-glucose pyrophosphorylase gene. J. Biol. Chem. 272, 23784–23791. doi: 10.1074/jbc.272.38.23784
Führing, J. I., Cramer, J. T., Schneider, J., Baruch, P., Gerardy-Schahn, R., and Fedorov, R. (2015). A quaternary mechanism enables the complex biological functions of octameric human UDP-glucose pyrophosphorylase, a key enzyme in cell metabolism. Sci. Rep. 5:9618. doi: 10.1038/srep09618
Führing, J., Damerow, S., Fedorov, R., Schneider, J., Münster-Kühnel, A. K., and Gerardy-Schahn, R. (2013). Octamerization is essential for enzymatic function of human UDP-glucose pyrophosphorylase. Glycobiology 23, 426–437. doi: 10.1093/glycob/cws217
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D., et al. (2005). “Protein identification and analysis tools on the ExPASy server,” in The Proteomics Protocols Handbook, ed. J. M. Walker (Totowa, NJ: Humana Press), 571–607.
Geisler, M., Wilczynska, M., Karpinski, S., and Kleczkowski, L. A. (2004). Toward a blueprint for UDP-glucose pyrophosphorylase structure/function properties: homology-modeling analyses. Plant Mol. Biol. 56, 783–794.
Geserick, C., and Tenhaken, R. (2013). UDP-sugar pyrophosphorylase is essential for arabinose and xylose recycling, and is required during vegetative and reproductive growth in Arabidopsis. Plant J. 74, 239–247. doi: 10.1111/tpj.12116
Gronwald, J. W., Miller, S. S., and Vance, C. P. (2008). Arabidopsis UDP-sugar pyrophosphorylase: evidence for two isoforms. Plant Physiol. Biochem. 46, 1101–1105. doi: 10.1016/j.plaphy.2008.07.001
Gupta, S. K., and Sowokinos, J. R. (2003). Physicochemical and kinetic properties of unique isozymes of UDP-Glc pyrophosphorylase that are associated with resistance to sweetening in cold-stored potato tubers. J. Plant Physiol. 160, 589–600. doi: 10.1078/0176-1617-01045
Gupta, S. K., Sowokinos, J. R., and Hahn, I. S. (2008). Regulation of UDP-glucose pyrophosphorylase isozyme UGP5 associated with cold-sweetening resistance in potatoes. J. Plant Physiol. 165, 679–690. doi: 10.1016/j.jplph.2007.09.001
Humphreys, G. B., Jud, M. C., Monroe, K. M., Kimball, S. S., Higley, M., Shipley, D., et al. (2013). Mummy, a UDP-N-acetylglucosamine pyrophosphorylase, modulates DPP signaling in the embryonic epidermis of Drosophila. Dev. Biol. 381, 434–445. doi: 10.1016/j.ydbio.2013.06.006
Itkonen, H. M., Engedal, N., Babaie, E., Luhr, M., Guldvik, I. J., Minner, S., et al. (2015). UAP1 is overexpressed in prostate cancer and is protective against inhibitors of N-linked glycosylation. Oncogene 34, 3744–3750. doi: 10.1038/onc.2014.307
Junior, M. C. S., de Assis, S. A., Góes-Neto, A., Duarte, ÂA., Alves, R. J., Junior, M. C., et al. (2013). Structure-based drug design studies of UDP-N-acetylglucosamine pyrophosphosrylase, a key enzyme for the control of witches’ broom disease. Chem. Cent. J. 7:48. doi: 10.1186/1752-153X-7-48
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Katsube, T., Kazuta, Y., Mori, H., Nakano, K., Tanizawa, K., and Fukui, T. (1990). UDP-glucose pyrophosphorylase from potato tuber: CDNA cloning and sequencing. J. Biochem. 108, 321–326. doi: 10.1093/oxfordjournals.jbchem.a123200
Katsube, T., Kazuta, Y., Tanizawa, K., and Fukui, T. (1991). Expression in Escherichia coli of UDP-glucose pyrophosphorylase cDNA from potato tuber and functional assessment of the five lysyl residues located at the substrate-binding site. Biochemistry 30, 8546–8551. doi: 10.1021/bi00099a008
Kelemen, O., Convertini, P., Zhang, Z., Wen, Y., Shen, M., Falaleeva, M., et al. (2013). Function of alternative splicing. Gene 514, 1–30. doi: 10.1016/j.gene.2012.07.083
Kleczkowski, L. A., Geisler, M., Ciereszko, I., and Johansson, H. (2004). UDP-glucose pyrophosphorylase. an old protein with new tricks. Plant Physiol. 134, 912–918. doi: 10.1104/pp.103.036053
Kleczkowski, L. A., Kunz, S., and Wilczynska, M. (2010). Mechanisms of UDP-glucose synthesis in plants. Crit. Rev. Plant Sci. 29, 191–203. doi: 10.1080/07352689.2010.483578
Le Hir, H., Nott, A., and Moore, M. J. (2003). How introns influence and enhance eukaryotic gene expression. Trends Biochem. Sci. 28, 215–220. doi: 10.1016/S0968-0004(03)00052-5
Letunic, I., and Bork, P. (2019). Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Liu, S., Zhong, H., Meng, X., Sun, T., Li, Y., Pinson, S. R. M., et al. (2020). Genome-wide association studies of ionomic and agronomic traits in USDA mini core collection of rice and comparative analyses of different mapping methods. BMC Plant Biol. 20:441. doi: 10.1186/s12870-020-02603-0
Liu, X., Li, F., Li, D., Ma, E., Zhang, W., Zhu, K. Y., et al. (2013). Molecular and functional analysis of UDP-N-acetylglucosamine pyrophosphorylases from the migratory locust, Locusta migratoria. PLoS One 8:e71970. doi: 10.1371/journal.pone.0071970
Martz, F., Wilczynska, M., and Kleczkowski, L. A. (2002). Oligomerization status, with the monomer as active species, defines catalytic efficiency of UDP-glucose pyrophosphorylase. Biochem. J. 367, 295–300. doi: 10.1042/BJ20020772
Meng, M., Fitzek, E., Gajowniczek, A., Wilczynska, M., and Kleczkowski, L. A. (2009a). Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley UDP-glucose pyrophosphorylase. Biochim. Biophys. Acta 1794, 1734–1742. doi: 10.1016/j.bbapap.2009.08.009
Meng, M., Geisler, M., Johansson, H., Harholt, J., Scheller, H. V., Mellerowicz, E. J., et al. (2009b). UDP-glucose pyrophosphorylase is not rate limiting, but is essential in Arabidopsis. Plant Cell Physiol. 50, 998–1011. doi: 10.1093/pcp/pcp052
Meng, M., Wilczynska, M., and Kleczkowski, L. A. (2008). Molecular and kinetic characterization of two UDP-glucose pyrophosphorylases, products of distinct genes, from Arabidopsis. Biochim. Biophys. Acta 1784, 967–972. doi: 10.1016/j.bbapap.2008.02.021
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., Von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Mio, T., Yabe, T., Arisawa, M., and Yamada-Okabe, H. (1998). The eukaryotic UDP-N-acetylglucosamine pyrophosphorylases: gene cloning, protein expression, and catalytic mechanism. J. Biol. Chem. 273, 14392–14397. doi: 10.1074/jbc.273.23.14392
Mok, M. T. S., and Edwards, M. R. (2005). Kinetic and physical characterization of the inducible UDP-N- acetylglucosamine pyrophosphorylase from Giardia intestinalis. J. Biol. Chem. 280, 39363–39372. doi: 10.1074/jbc.M509209200
Okazaki, Y., Shimojima, M., Sawada, Y., Toyooka, K., Narisawa, T., Mochida, K., et al. (2009). A chloroplastic UDP-Glucose pyrophosphorylase from Arabidopsis is the committed enzyme for the first step of sulfolipid biosynthesis. Plant Cell 21, 892–909. doi: 10.1105/tpc.108.063925
Palaka, B. K., Velmurugan Ilavarasi, A., Sapam, T. D., Kotapati, K. V., Nallala, V. S., Khan, M. B., et al. (2019). Molecular cloning, gene expression analysis, and in silico characterization of UDP-N-acetylglucosamine pyrophosphorylase from Bombyx mori. Biotechnol. Appl. Biochem. 66, 880–899. doi: 10.1002/bab.1802
Park, J. I., Ishimizu, T., Suwabe, K., Sudo, K., Masuko, H., Hakozaki, H., et al. (2010). UDP-glucose pyrophosphorylase is rate limiting in vegetative and reproductive phases in Arabidopsis thaliana. Plant Cell Physiol. 51, 981–996. doi: 10.1093/pcp/pcq057
Peneff, C., Ferrari, P., Charrier, V., Taburet, Y., Monnier, C., Zamboni, V., et al. (2001). Crystal structures of two human pyrophosphorylase isoforms in complexes with UDPGlc(Gal)NAc: role of the alternatively spliced insert in the enzyme oligomeric assembly and active site architecture. EMBO J. 20, 6191–6202. doi: 10.1093/emboj/20.22.6191
Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R., and Finn, R. D. (2018). HMMER web server: 2018 update. Nucleic Acids Res. 46, W200–W204. doi: 10.1093/nar/gky448
Prakash, O., Führing, J., Post, J., Shepherd, S. M., Eadsforth, T. C., Gray, D., et al. (2019). Identification of Leishmania major UDP-sugar pyrophosphorylase inhibitors using biosensor-based small molecule fragment library screening. Molecules 24:996. doi: 10.3390/molecules24050996
Raimi, O. G., Hurtado-Guerrero, R., Borodkin, V., Ferenbach, A., Urbaniak, M. D., Ferguson, M. A. J., et al. (2020). A mechanism-inspired UDP- N -acetylglucosamine pyrophosphorylase inhibitor. RSC Chem. Biol. 1, 13–25. doi: 10.1039/c9cb00017h
Reinbothe, C., Springer, A., Samol, I., and Reinbothe, S. (2009). Plant oxylipins: role of jasmonic acid during programmed cell death, defence and leaf senescence. FEBS J. 276, 4666–4681. doi: 10.1111/j.1742-4658.2009.07193.x
Roeben, A., Plitzko, J. M., Körner, R., Böttcher, U. M. K., Siegers, K., Hayer-Hartl, M., et al. (2006). Structural basis for subunit assembly in UDP-glucose pyrophosphorylase from Saccharomyces cerevisiae. J. Mol. Biol. 364, 551–560. doi: 10.1016/j.jmb.2006.08.079
Schimmelpfeng, K., Strunk, M., Stork, T., and Klämbt, C. (2006). mummy encodes an UDP-N-acetylglucosamine-dipohosphorylase and is required during Drosophila dorsal closure and nervous system development. Mech. Dev. 123, 487–499. doi: 10.1016/j.mod.2006.03.004
Schnurr, J. A., Storey, K. K., Jung, H. J. G., Somers, D. A., and Gronwald, J. W. (2006). UDP-sugar pyrophosphorylase is essential for pollen development in Arabidopsis. Planta 224, 520–532. doi: 10.1007/s00425-006-0240-1
Shi, J. F., Fu, J., Mu, L. L., Guo, W. C., and Li, G. Q. (2016). Two Leptinotarsa uridine diphosphate N-acetylglucosamine pyrophosphorylases are specialized for chitin synthesis in larval epidermal cuticle and midgut peritrophic matrix. Insect Biochem. Mol. Biol. 68, 1–12. doi: 10.1016/j.ibmb.2015.11.005
Sowokinos, J. R., Vigdorovich, V., and Abrahamsen, M. (2004). Molecular cloning and sequence variation of UDP-glucose pyrophosphorylase cDNAs from potatoes sensitive and resistant to cold sweetening. J. Plant Physiol. 161, 947–955. doi: 10.1016/j.jplph.2004.04.006
Spychalla, J. P., Scheffler, B. E., Sowokinos, J. R., and Bevan, M. W. (1994). Cloning, antisense RNA inhibition, and the coordinated expression of UDP-glucose pyrophosphorylase with starch biosynthetic genes in potato tubers. J. Plant Physiol. 144, 444–453. doi: 10.1016/S0176-1617(11)82121-8
Sreenivasulu, N., Radchuk, V., Strickert, M., Miersch, O., Weschke, W., and Wobus, U. (2006). Erratum: gene expression patterns reveal tissue-specific signalling networks controlling programmed cell death and ABA-regulated maturation in developing barley seeds (The Plant Journal (2006) 47, (310-327)). Plant J. 47:987. doi: 10.1111/j.1365-313X.2006.02899.x
Steiner, T., Lamerz, A. C., Hess, P., Breithaupt, C., Krapp, S., Bourenkov, G., et al. (2007). Open and closed structures of the UDP-glucose pyrophosphorylase from Leishmania major. J. Biol. Chem. 282, 13003–13010. doi: 10.1074/jbc.M609984200
Toroser, D., Athwal, G. S., and Huber, S. C. (1998). Site-specific regulatory interaction between spinach leaf sucrose-phosphate synthase and 14-3-3 proteins. FEBS Lett. 435, 110–114. doi: 10.1016/S0014-5793(98)01048-5
Tresse, E., Kosta, A., Giusti, C., Luciani, M. F., and Golstein, P. (2008). A UDP-glucose derivative is required for vacuolar autophagic cell death. Autophagy 4, 680–691. doi: 10.4161/auto.6084
Vella, J., and Copeland, L. (1990). UDP−glucose pyrophosphorylase from the plant fraction of nitrogen−fixing soybean nodules. Physiol. Plant. 78, 140–146. doi: 10.1111/j.1399-3054.1990.tb08728.x
Wang, Q., Yang, Z. L., Zou, Q., Yuan, Y., Li, J., Liang, L., et al. (2016). SHP2 and UGP2 are biomarkers for progression and poor prognosis of gallbladder cancer. Cancer Invest. 34, 255–264. doi: 10.1080/07357907.2016.1193745
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Wang, Z., Wang, Y., Hong, X., Hu, D., Liu, C., Yang, J., et al. (2015). Functional inactivation of UDP-N-acetylglucosamine pyrophosphorylase 1 (UAP1) induces early leaf senescence and defence responses in rice. J. Exp. Bot. 66, 973–987. doi: 10.1093/jxb/eru456
Wang-Gillam, A., Pastuszak, I., Stewart, M., Drake, R. R., and Elbein, A. D. (2000). Identification and modification of the uridine-binding site of the UDP- GalNAc (GlcNAc) pyrophosphorylase. J. Biol. Chem. 275, 1433–1438. doi: 10.1074/jbc.275.2.1433
Xiao, G., Zhou, J., Lu, X., Huang, R., and Zhang, H. (2018). Excessive UDPG resulting from the mutation of UAP1 causes programmed cell death by triggering reactive oxygen species accumulation and caspase-like activity in rice. New Phytol. 217, 332–343. doi: 10.1111/nph.14818
Yang, J., Yan, R., Roy, A., Xu, D., Poisson, J., and Zhang, Y. (2014). The I-TASSER suite: protein structure and function prediction. Nat. Methods 12, 7–8. doi: 10.1038/nmeth.3213
Yang, T., Echols, M., Martin, A., and Bar-Peled, M. (2010). Identification and characterization of a strict and a promiscuous N-acetylglucosamine-1-P uridylyltransferase in Arabidopsis. Biochem. J. 430, 275–284. doi: 10.1042/BJ20100315
Yi, D. G., and Huh, W. K. (2015). UDP-glucose pyrophosphorylase Ugp1 is involved in oxidative stress response and long-term survival during stationary phase in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 467, 657–663. doi: 10.1016/j.bbrc.2015.10.090
Zhang, J., Zhang, Y., Du, Y., Chen, S., and Tang, H. (2011). Dynamic metabonomic responses of tobacco (Nicotiana tabacum) plants to salt stress. J. Proteome Res. 10, 1904–1914. doi: 10.1021/pr101140n
Zhu, B. H., Shi, H. P., Yang, G. P., Lv, N. N., Yang, M., and Pan, K. H. (2016). Silencing UDP-glucose pyrophosphorylase gene in Phaeodactylum tricornutum affects carbon allocation. N. Biotechnol. 33, 237–244. doi: 10.1016/j.nbt.2015.06.003
Keywords: UDP glucose pyrophosphorylase family, UDP-N-acetylglucosamine pyrophosphorylase, UDP-glucose pyrophosphorylase, UDP-sugar pyrophosphorylase, conserved evolution, alternative splicing
Citation: Liu S, Zhong H, Wang Q, Liu C, Li T, Peng Z, Li Y, Zhang H, Liao J, Huang Y and Wang Z (2021) Global Analysis of UDP Glucose Pyrophosphorylase (UDPGP) Gene Family in Plants: Conserved Evolution Involved in Cell Death. Front. Plant Sci. 12:681719. doi: 10.3389/fpls.2021.681719
Received: 17 March 2021; Accepted: 26 April 2021;
Published: 10 June 2021.
Edited by:
Raju Datla, Global Institute for Food Security, CanadaReviewed by:
Leszek A. Kleczkowski, Umeå University, SwedenRaimund Tenhaken, University of Salzburg, Austria
Copyright © 2021 Liu, Zhong, Wang, Liu, Li, Peng, Li, Zhang, Liao, Huang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhaohai Wang, emhhb2hhaV93YW5nQDE2My5jb20=
†These authors have contributed equally to this work
 Shuai Liu
Shuai Liu Hua Zhong
Hua Zhong Qiang Wang
Qiang Wang Caixiang Liu
Caixiang Liu Ting Li6
Ting Li6 Zhaohua Peng
Zhaohua Peng Yangsheng Li
Yangsheng Li Hongyu Zhang
Hongyu Zhang Jianglin Liao
Jianglin Liao Yingjin Huang
Yingjin Huang Zhaohai Wang
Zhaohai Wang